- 1Department of Orthodontics, Hospital of Stomatology, Jilin University, Changchun, China
- 2Jilin Provincial Key Laboratory of Tooth Development and Bone Remodeling, Changchun, China
Bone metabolism consists of bone formation and resorption and maintains a dynamic balance in vivo. When bone homeostasis is broken, it can manifest as osteoarthritis (OA), rheumatoid arthritis (RA), osteosarcoma (OS), etc. MiR-671, an important class of non-coding nucleotide sequences in vivo, is regulated by lncRNA and regulates bone metabolism balance by regulating downstream target proteins and activating various signaling pathways. Based on the structure and primary function of miR-671, this paper summarizes the effect and mechanism of miR-671 in bone-related inflammation and cancer diseases, and prospects the application possibility of miR-671, providing reference information for targeted therapy of bone-related disorders.
Introduction
Bone metabolism includes bone formation and bone resorption. Osteoblasts and osteoclasts are essential cells in maintaining bone homeostasis. Bone components maintain dynamic balance by regulating cytokines and metal ions in the body. When there is exogenous or endogenous stimulation, abnormal metabolism of osteoblasts and osteoclasts can be manifested as osteoarthritis (OA), rheumatoid arthritis (RA), osteosarcoma (OS), etc. MicroRNAs (miRNAs) are non-coding RNAs ranging from 18 to 25 nucleotides in length, which play an essential role in biological processes such as cell proliferation, differentiation, migration, apoptosis, and tumorigenesis. MiRNAs can also regulate signaling pathways in the cytoskeleton, inflammation, and metabolism. MiRNAs have gradually become biomarkers for diagnosing and prognostic evaluating many diseases (1). With continuous research, microRNA-671 (miR-671) has been found to reflect the degree of inflammation and tumorigenesis in bone metabolism and related disorders. Its influence on bone metabolism and bone-related diseases has been gradually clarified. This paper mainly reviews the impact of miR-671 on bone metabolism and the occurrence and development of bone-related diseases to make early diagnosis and targeted therapy based on the changes of miR-671 content in vivo in clinical practice.
Structure and basic functions of miR-671
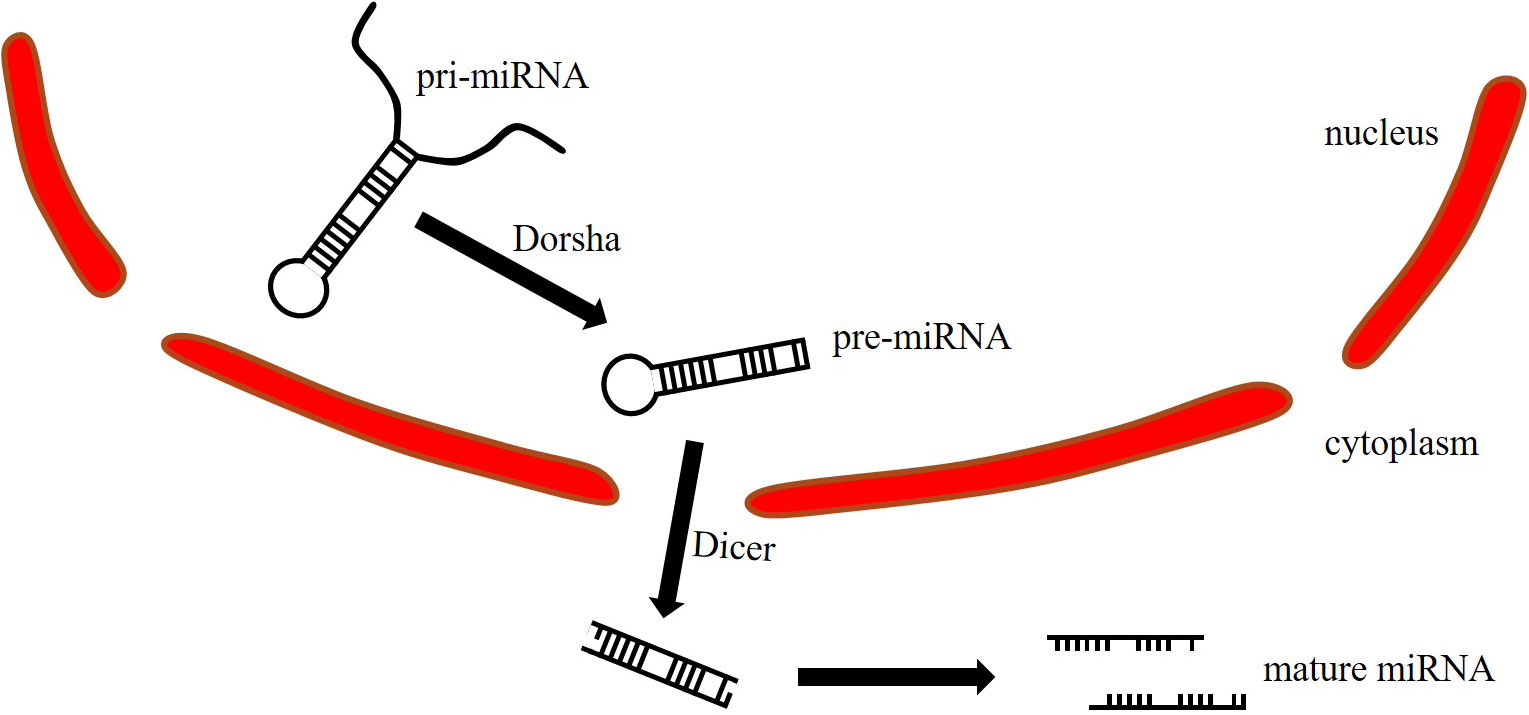
MiRNA maturation begins with the primary miRNA (pri-miRNA) in the nucleus, and the length is between 300 and 10,000 bp (Figure 1). Pri-miRNA is cleaved by Dorsha to generate precursor miRNA (pre-miRNA). The pre-miRNA is 70-90bp in length and transferred from the nucleus to the cytoplasm. Then, the Dicer opens the hairpin structure of the pre-miRNA to generate double-stranded miRNA, which is further cleaved to generate two single-stranded miRNAs (2). The passenger strand is removed and degraded, while the guide strand is retained to form the RNA-induced silencing complex (RISC) (3). The guide strand is stable under Argonaute (AGO) protein protection from cellular exoribonucleases (4, 5). MiRNA regulates gene expression by fully or incompletely binding to the 3’UTR seed sequence of the target gene mRNA, resulting in degradation or translation (6).
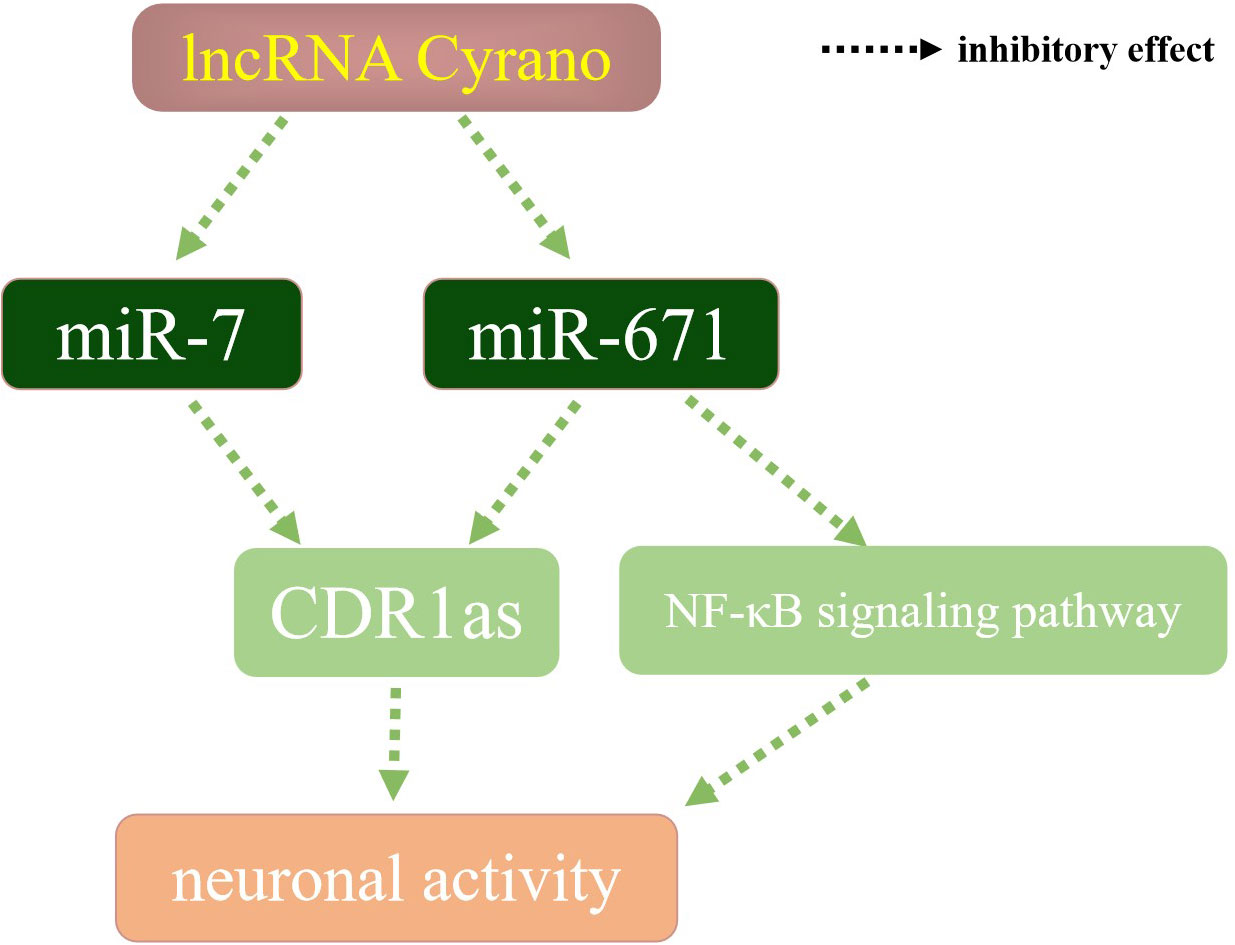
As miRNA can be derived from different stem arms, pre-miR-671 generates two miRNAs, miR-671-5p and miR-671-3p (Figure 2), respectively, both of which play an essential role in the stable regulation of the human body environment by mainly binding to the target mRNA (8). Content change of components in the lncRNA-miRNA-mRNA axis could affect the downstream pathways, such as TGF-β signaling (9) and Wnt/β-catenin signaling (10). In terms of nervous system regulation (Figure 3), miRNA-671 can affect the content of mRNA and circRNA in mammalian brain signal networks (11) and regulates neuronal apoptosis with lncRNA and circRNA (12). MiR-7 is one of the first known and also most investigated miRNAs. It is considered a network stabilizer and connects different signaling pathways, especially in the central nervous system (13, 14). LncRNA Cyrano can degrade miR-7 through a wide range of pairing sites, thereby increasing the content of CDR1as and negatively regulating neuronal activity. When the content of lncRNA Cyrano is decreased, miR-7 and miR-671 will increase. MiR-7 and miR-671 inhibit downstream CDR1as together (15, 16), thereby increasing neuronal activity. Reduced expression of miRNA-671-5p has been detected in Parkinson’s disease, ischemic stroke, and other diseases (17, 18), which weakens the inhibitory effect of NF-κB, and the macroscopic manifestations are increasing neuroinflammatory responses. MiR-671 also plays a vital role in maintaining the dynamic balance of bone metabolism. Studies on the osteogenic differentiation of dental pulp stem cells and bone marrow stem cells showed that miR-671-3p and miR-671-5p expressions were up-regulated (19). When bone metabolism is abnormal and the bone microenvironment is destroyed, the content of miR-671 changes in varying degrees.
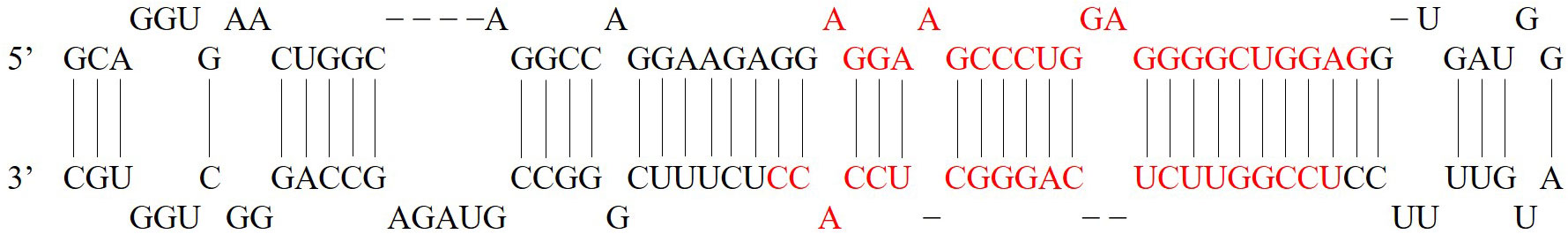
Figure 2 pre-miR-671 stem-loop structure (7). The red sequence is the mature miRNA. miR-671-5p: 29-AGGAAGCCCUGGAGGGGCUGGAG-51. miR-671-3p: 68-UCCGGUUCUCAGGGCUCCACC-88.
MiR-671 in bone-related inflammatory diseases
When inflammation occurs in vivo, miR-671-5p, as a critical miRNA, can affect the expression of inflammatory genes. It can be used as a biomarker to diagnose inflammation, such as Kawasaki disease and hepatitis (20–23), reflecting the degree of inflammation and affecting the healing time of injury (24). Inflammation is often related to the development and progression of cancer. Inflammation-related cells are genetically stable and do not rapidly develop drug resistance and invasiveness (25). As inflammation evolves, repeated stimulation affects normal cells, leading to genetic changes and increasing cancer risk, similar to bone inflammatory diseases. Therefore, to diagnose and treat bone inflammatory diseases early and reduce the risk of canceration, the content of miR-671 can be detected to clarify the process of relevant inflammatory reactions.
Mechanisms of miR-671 regulating the inflammatory response
Inflammation will increase the permeability of local blood vessels, and liquid and cellular components exudate. It releases many cytokines and increases the number of white blood cells. Also, inflammation-related factors can affect cell functions through miRNA (26). The effective elimination of pathogens depends on several signaling pathways, such as Toll-like-receptor (TLR) signaling; many of these are regulated by miRNA (27). MiR-671, as a biomarker, is involved in macrophage immune signal transduction in the early stage of the inflammatory response (28–30). Activated macrophages release various immune regulatory factors and reduce the expression of miR-671 (31).
Significant up-regulation of miR-671-5p can be detected in both periodontitis and late stage of hepatitis B, thus inhibiting inflammatory response (22, 23). The relative fold change significantly increased in advanced fibrosis versus early fibrosis. MiR-671-5p can also act as a target to mediate immune regulation, affect cytokine production, and changes in antigen-related marker levels (32). Meanwhile, miR-671-3p exists in the miRNA-mRNA-immune cell regulatory network (33). It can be a biomarker for predicting and diagnosing grades II-IV acute graft-versus-host disease (34). MiR-671-3p RQ level<0.5 (down expressed) could also be observed when inflammation is reducing (35). Therefore, miR-671 level is usually up-regulated during the middle and late stages of inflammatory responses. Thus, there is a positive correlation between the miR-671 level and inflammation.
Mechanisms of miR-671 regulating the bone-related inflammatory diseases
OA is a degenerative joint disease characterized by articular cartilage degeneration and inflammation. Many studies have confirmed that miR-671 is closely related to OA. Through miRNA sequencing analysis of tissues, plasma, and chondrocytes of OA patients, the results showed that miR-671-3p level was down-regulated during the rapid progression of the disease (36), and the expression of the downstream target gene, TNF receptor-associated factor, was enhanced (37). In the late stage of the disease, the increase of miR-671 can reduce the inhibition of cell proliferation and apoptotic stimulation induced by IL-1β and reduce the expression of type II collagen in chondrocytes, which can reduce the progression of OA (38). Therefore, miR-671-3p reflects the progressive state of bone metabolism and can be used as a potential biomarker to assess the risk and progression of OA (39). The circRNA-miRNA-mRNA regulatory network plays a crucial role in the regulation of OA (40). Upstream circRNA usually acts as a miRNA sponge in cells and weakens the inhibitory effect of miR-671 on inflammation through the competitive binding of miR-671. Compared with healthy people, the expression of circ-IQGAP1 and circ_0114876 increases in cartilage tissues, while the expression of miR-671-5p decreases (41). Reducing circRNA content can reduce chondrocyte apoptosis, inflammatory cytokine secretion, and extracellular matrix degradation induced by IL-1β.
In terms of regulating RA, circRNA pituitary tumor-transforming 1 interacting protein (circ-PTTG1IP) regulates toll-like receptor 4 (TLR4), an upstream regulator of the nuclear factor-kappa B (NF-κB), by directly binding to miR-671-5p. CircRNAs could act as a miRNA sponge to absorb the functional miRNAs, such as miR-671-5p, which could suppress the function of TLR4. If the expression of circRNA is increased, the proliferation, migration, and invasion of RA-flss can be promoted by activating the NF-κB pathway, and local inflammatory response can be affected (42). Similarly, the upstream target of miR-671-5p, circ_0001947, can also up-regulate the expression of downstream target STAT3 by absorbing miR-671-5p, thus promoting the progression of RA (43) (Figure 4). Therefore, drugs for RA can reduce the expression of circRNA by influencing the circRNA-miRNA-mRNA regulatory network, thus enhancing the expression of miR-671-5p, inhibiting the inflammatory response, and promoting RA healing (44).
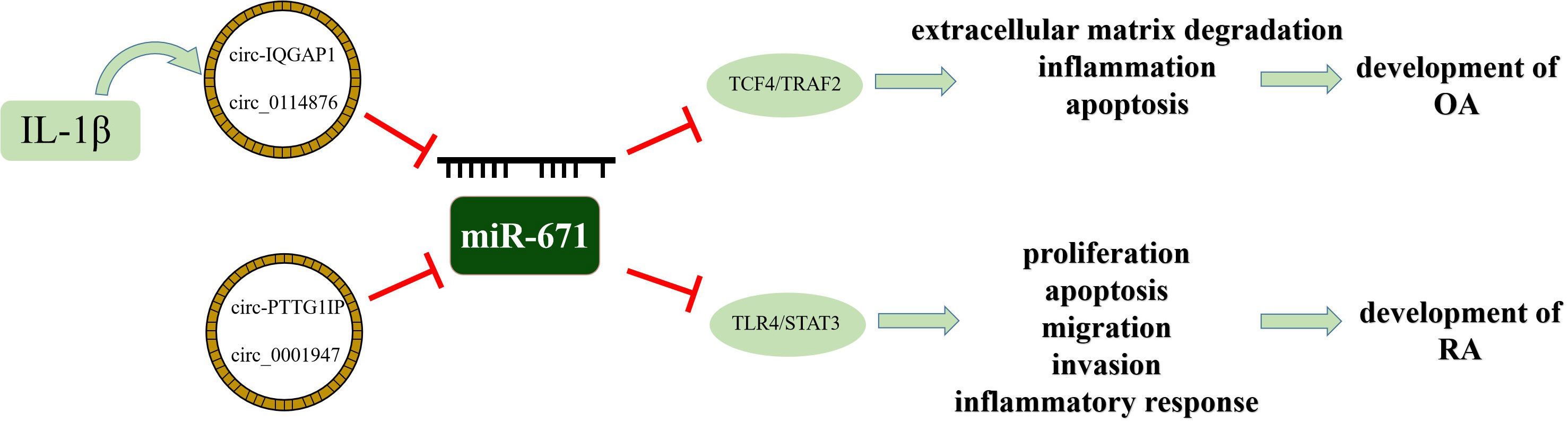
Figure 4 The schematic diagram of the involvement of the circRNA-miRNA-mRNA axis in OA and RA development.
MiR-671 and OS
Persistent inflammation can lead to cell damage and fibrosis, increasing the risk of canceration (45). Cytogenetic changes often accompany the generation of cancer. MiRNAs participate in post-transcriptional regulation and interfere with oncogenes or tumor suppressor genes by binding to the 3′UTR region of mRNA, thus affecting cancer development (46).
OS is a highly invasive bone tumor that occurs mainly in young patients. Like most cancers, miR-671, as a tumor suppressor gene, plays a significant role in OS. The expression of miR-671-5p is significantly decreased in tissues and cells of patients with OS. Overexpression of miR-671-5p inhibits the proliferation, migration, and invasion of OS cells. MiR-671-5p directly binds cyclin D1 and CDC34 to inhibit cell cycle progression, thus inhibiting the progression of cancer (47). TUFT1 and SMAD3, the targets of miR-671-5p, can enhance the migration and invasion of OS cells. Low expression of miR-671-5p and high expression of TUFT1 and SMAD3 will lead to poor prognosis (48, 49). LncRNA DLEU1, an upstream regulator of miR-671, is highly expressed in the tissues and cells of patients with OS. Through directly absorbing miR-671-5p, lncRNA DLEU1 regulates the expression of DEAD-box helicase 5 and plays an oncogenic role in OS (50). Therefore, by targeting the upstream and downstream genes of miR-671-5p, the regulation of the lncRNA-miRNA-mRNA network can be a targeted therapy for OS.
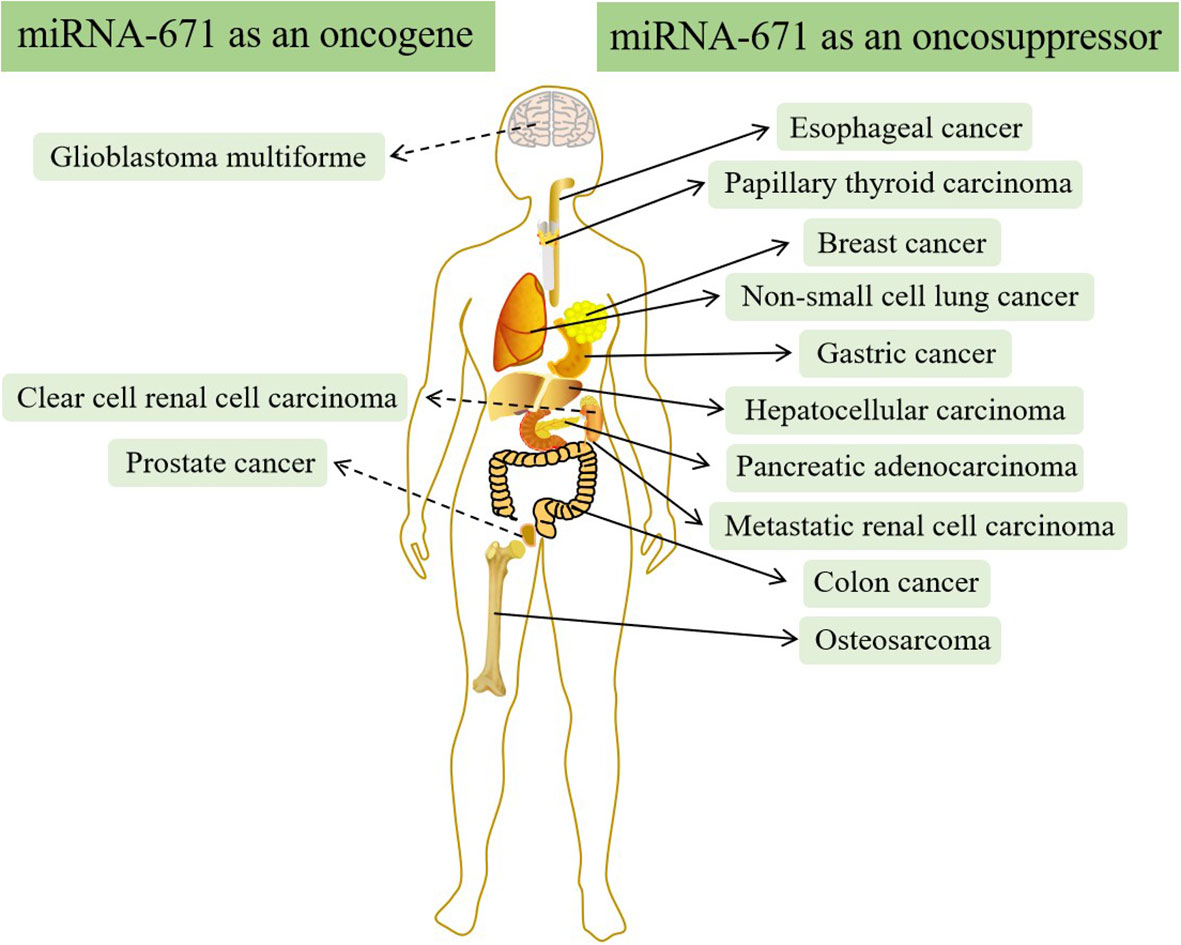
In addition to OS, there is increasing evidence that miR-671 is often abnormally expressed in other types of cancer and can regulate the structure and expression of mRNA and protein (51–53). LncRNA-miRNA-mRNA regulatory network plays a significant role in cancer’s occurrence, development, and prognosis by regulating downstream target genes and proteins (54–57). Therefore, targeted therapy drugs can inhibit cancer growth by influencing the regulatory network (58, 59). In most types of cancer, such as breast cancer, esophageal cancer, non-small cell lung cancer, hepatocellular carcinoma, etc., miR-671 acts as a tumor suppressor and is down-regulated in cancer (60–69). However, in a few kinds of cancer, miR-671 acts as an oncogene and is up-regulated in vivo (70–77) (Figure 5).
Conclusion
Abnormal bone metabolism affects bone quality and leads to an increased incidence of fractures. The continuous exploration of miR-671 in clinical and basic experiments will promote the application of miR-671 in the screening and treatment of abnormal bone metabolism more and more widely. In this review, we collect the studies on miR-671 in recent years and summarize the role of miR-671 in bone metabolism and bone-related diseases. However, there are few studies on the mechanism of miR-671 regulating bone metabolism through various signaling pathways. In the future, searching for critical genes and using miRNA-targeted therapy can block the development of the diseases by studying the pathogenesis. Thus, doctors can reduce the cost of treatment and the damage of inflammation and cancer.
Author contributions
SW, DS and LT contributed to the research retrieval and drafting. MH and HJ contributed to the outline drafting and critically revised the manuscript. Every author gave final approval and agreed to be accountable for all aspects of the work. All authors have read and agreed to the published version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study is partly supported by the National Natural Science Foundation of China (No.81870795), the Finance department Project of Jilin Province (JCSZ2019378-19), and the National Science Foundation of Jilin Province (20200201348JC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Terrinoni A, Calabrese C, Basso D, Aita A, Caporali S, Plebani M, et al. The circulating miRNAs as diagnostic and prognostic markers. Clin Chem Lab Med (2019) 57(7):932–53. doi: 10.1515/cclm-2018-0838
2. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (2018) 9:402. doi: 10.3389/fendo.2018.00402
3. Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell (2003) 115(2):199–208. doi: 10.1016/S0092-8674(03)00759-1
4. Sheu-Gruttadauria J, Pawlica P, Klum SM, Wang S, Yario TA, Schirle Oakdale NT, et al. Structural basis for target-directed MicroRNA degradation. Mol Cell (2019) 75(6):1243–1255.e1247. doi: 10.1016/j.molcel.2019.06.019
5. Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature (2008) 456(7219):209–13. doi: 10.1038/nature07315
6. Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev biophysics (2013) 42:217–39. doi: 10.1146/annurev-biophys-083012-130404
7. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res (2019) 47(D1):D155–d162. doi: 10.1093/nar/gky1141
8. Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med (2016) 14(1):143. doi: 10.1186/s12967-016-0893-x
9. Wang X, Zhu Y, Wu C, Liu W, He Y, Yang Q. Adipose-derived mesenchymal stem cells-derived exosomes carry MicroRNA-671 to alleviate myocardial infarction through inactivating the TGFBR2/Smad2 axis. Inflammation (2021) 44(5):1815–30. doi: 10.1007/s10753-021-01460-9
10. Wang C, Liu J, Zhang X, Chen Q, Bai X, Hong X, et al. Role of miRNA-671-5p in mediating wnt/β-Catenin-Triggered podocyte injury. Front Pharmacol (2021) 12:784489. doi: 10.3389/fphar.2021.784489
11. Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J (2011) 30(21):4414–22. doi: 10.1038/emboj.2011.359
12. Nan A, Chen L, Zhang N, Liu Z, Yang T, Wang Z, et al. A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch Toxicol (2017) 91(4):1671–84. doi: 10.1007/s00204-016-1837-1
13. Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell (2009) 137(2):273–82. doi: 10.1016/j.cell.2009.01.058
14. Caygill EE, Brand AH. miR-7 buffers differentiation in the developing drosophila visual system. Cell Rep (2017) 20(6):1255–61. doi: 10.1016/j.celrep.2017.07.047
15. Kleaveland B, Shi CY, Stefano J, Bartel DP. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell (2018) 174(2):350–362.e317. doi: 10.1016/j.cell.2018.05.022
16. Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science (2017) 357(6357):eaam8526. doi: 10.1126/science.aam8526
17. Uwatoko H, Hama Y, Iwata IT, Shirai S, Matsushima M, Yabe I, et al. Identification of plasma microRNA expression changes in multiple system atrophy and parkinson's disease. Mol Brain (2019) 12(1):49. doi: 10.1186/s13041-019-0471-2
18. Deng L, Guo Y, Liu J, Wang X, Chen S, Wang Q, et al. miR-671-5p attenuates neuroinflammation via suppressing NF-κB expression in an acute ischemic stroke model. Neurochemical Res (2021) 46(7):1801–13. doi: 10.1007/s11064-021-03321-1
19. Gaus S, Li H, Li S, Wang Q, Kottek T, Hahnel S, et al. Shared genetic and epigenetic mechanisms between the osteogenic differentiation of dental pulp stem cells and bone marrow stem cells. BioMed Res Int (2021) 2021:6697810. doi: 10.1155/2021/6697810
20. Zhang X, Xin G, Sun D. Serum exosomal miR-328, miR-575, miR-134 and miR-671-5p as potential biomarkers for the diagnosis of Kawasaki disease and the prediction of therapeutic outcomes of intravenous immunoglobulin therapy. Exp Ther Med (2018) 16(3):2420–32. doi: 10.3892/etm.2018.6458
21. Jia HL, Liu CW, Zhang L, Xu WJ, Gao XJ, Bai J, et al. Sets of serum exosomal microRNAs as candidate diagnostic biomarkers for Kawasaki disease. Sci Rep (2017) 7:44706. doi: 10.1038/srep44706
22. Wang BH, Zhao YF, Shen LR, Zhuang Q. [Differential screening and functional prediction analysis of miRNA expression profiles in periodontitis]. Shanghai kou qiang yi xue = Shanghai J stomatol (2019) 28(4):408–11.
23. Singh AK, Rooge SB, Varshney A, Vasudevan M, Bhardwaj A, Venugopal SK, et al. Global microRNA expression profiling in the liver biopsies of hepatitis b virus-infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatol (Baltimore Md) (2018) 67(5):1695–709. doi: 10.1002/hep.29690
24. Nunes S, Silva IB, Ampuero MR, de Noronha ALL, de Souza LCL, Correia TC, et al. Integrated analysis reveals that miR-193b, miR-671, and TREM-1 correlate with a good response to treatment of human localized cutaneous leishmaniasis caused by leishmania braziliensis. Front Immunol (2018) 9:640. doi: 10.3389/fimmu.2018.00640
25. Singh N, Baby D, Rajguru JP, Patil PB, Thakkannavar SS, Pujari VB. Inflammation and cancer. Ann Afr Med (2019) 18(3):121–6. doi: 10.4103/aam.aam_56_18
26. Sánchez-Jiménez C, Carrascoso I, Barrero J, Izquierdo JM. Identification of a set of miRNAs differentially expressed in transiently TIA-depleted HeLa cells by genome-wide profiling. BMC Mol Biol (2013) 14:4. doi: 10.1186/1471-2199-14-4
27. Alam MM, O'Neill LA. MicroRNAs and the resolution phase of inflammation in macrophages. Eur J Immunol (2011) 41(9):2482–5. doi: 10.1002/eji.201141740
28. Zur Bruegge J, Backes C, Gölz G, Hemmrich-Stanisak G, Scharek-Tedin L, Franke A, et al. MicroRNA response of primary human macrophages to arcobacter butzleri infection. Eur J Microbiol Immunol (2016) 6(2):99–108. doi: 10.1556/1886.2016.00015
29. Zhu H, Jiao H, Nie X, Li B, Xu K, Pang F, et al. Alterations of microRNAs and their predicted targeting mRNAs expression in RAW264.7 macrophages infected with Omp25 mutant brucella melitensis. Innate Immun (2018) 24(6):382–9. doi: 10.1177/1753425918792298
30. Singh N, Chauhan IS. MicroRNA expression profiling of dibenzalacetone (DBA) treated intracellular amastigotes of leishmania donovani. Exp Parasitol (2018) 193:5–19. doi: 10.1016/j.exppara.2018.07.018
31. Lien GS, Liu JF, Chien MH, Hsu WT, Chang TH, Ku CC, et al. The ability to suppress macrophage-mediated inflammation in orbital fat stem cells is controlled by miR-671-5p. Stem Cell Res Ther (2014) 5(4):97. doi: 10.1186/scrt486
32. Bravo-Parra M, Arenas-Padilla M, Brcenas-Preciado V, Hernández J, Mata-Haro V. The probiotic BB12 induces MicroRNAs involved in antigen processing and presentation in porcine monocyte-derived dendritic cells. Int J Mol Sci (2020) 21(3):687. doi: 10.3390/ijms21030687
33. Qi X, Lin H, Hou Y, Su X, Gao Y. Comprehensive analysis of potential miRNA-target mRNA-immunocyte subtype network in cerebral infarction. Eur Neurol (2022) 85(2):148–61. doi: 10.1159/000518893
34. Zhang C, Bai N, Huang W, Zhang P, Luo Y, Men S, et al. The predictive value of selected serum microRNAs for acute GVHD by TaqMan MicroRNA arrays. Ann Hematol (2016) 95(11):1833–43. doi: 10.1007/s00277-016-2781-0
35. Nardelli C, Iaffaldano L, Pilone V, Labruna G, Ferrigno M, Carlomagno N, et al. Changes in the MicroRNA profile observed in the subcutaneous adipose tissue of obese patients after laparoscopic adjustable gastric banding. J Obes (2017) 2017:6754734. doi: 10.1155/2017/6754734
36. Ali SA, Gandhi R, Potla P, Keshavarzi S, Espin-Garcia O, Shestopaloff K, et al. Sequencing identifies a distinct signature of circulating microRNAs in early radiographic knee osteoarthritis. Osteoarthritis cartilage (2020) 28(11):1471–81. doi: 10.1016/j.joca.2020.07.003
37. Liu Z, Chen S, Yang Y, Lu S, Zhao X, Hu B, et al. MicroRNA−671−3p regulates the development of knee osteoarthritis by targeting TRAF3 in chondrocytes. Mol Med Rep (2019) 20(3):2843–50. doi: 10.3892/mmr.2019.10488
38. Zhang B, Sun M, Wang J, Ma C, Hao T, Liu G, et al. MiR-671 ameliorates the progression of osteoarthritis in vitro and in vivo. Pathology Res Pract (2019) 215(7):152423. doi: 10.1016/j.prp.2019.04.015
39. Ntoumou E, Tzetis M, Braoudaki M, Lambrou G, Poulou M, Malizos K, et al. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenet (2017) 9:127. doi: 10.1186/s13148-017-0428-1
40. Wu Y, Lu X, Shen B, Zeng Y. The therapeutic potential and role of miRNA, lncRNA, and circRNA in osteoarthritis. Curr Gene Ther (2019) 19(4):255–63. doi: 10.2174/1566523219666190716092203
41. Xi P, Zhang CL, Wu SY, Liu L, Li WJ, Li YM. CircRNA circ-IQGAP1 knockdown alleviates interleukin-1β-Induced osteoarthritis progression via targeting miR-671-5p/TCF4. Orthopaedic Surg (2021) 13(3):1036–46. doi: 10.1111/os.12923
42. Chen L, Huang H, Chen L, Xu L, Chen J, Lu Q. Circ-PTTG1IP/miR-671-5p/TLR4 axis regulates proliferation, migration, invasion and inflammatory response of fibroblast-like synoviocytes in rheumatoid arthritis. Gen Physiol biophysics (2021) 40(3):207–19. doi: 10.4149/gpb_2021014
43. Yang Y, Lin S, Yang Z, Huang Y, Zhan F. Circ_0001947 promotes cell proliferation, invasion, migration and inflammation and inhibits apoptosis in human rheumatoid arthritis fibroblast-like synoviocytes through miR-671-5p/STAT3 axis. J orthopaedic Surg Res (2022) 17(1):54. doi: 10.1186/s13018-022-02939-3
44. Ma J, Meng Q, Zhan J, Wang H, Fan W, Wang Y, et al. Paeoniflorin suppresses rheumatoid arthritis development via modulating the circ-FAM120A/miR-671-5p/MDM4 axis. Inflammation (2021) 44(6):2309–22. doi: 10.1007/s10753-021-01504-0
45. Germolec DR, Shipkowski KA, Frawley RP, Evans E. Markers of inflammation. Methods Mol Biol (Clifton NJ) (2018) 1803:57–79. doi: 10.1007/978-1-4939-8549-4_5
46. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci (2020) 21(5):1723. doi: 10.3390/ijms21051723
47. Xin C, Lu S, Li Y, Zhang Y, Tian J, Zhang S, et al. miR-671-5p inhibits tumor proliferation by blocking cell cycle in osteosarcoma. DNA Cell Biol (2019) 38(9):996–1004. doi: 10.1089/dna.2019.4870
48. Ma C, Nie ZK, Guo HM, Kong Y. MiR-671-5p plays a promising role in restraining osteosarcoma cell characteristics through targeting TUFT1. J Biochem Mol Toxicol (2020) 34(7):e22490. doi: 10.1002/jbt.22490
49. Hu Y, Liang D, Chen X, Chen L, Bai J, Li H, et al. [MiR-671-5p negatively regulates SMAD3 to inhibit migration and invasion of osteosarcoma cells]. Nan fang yi ke da xue xue bao = J South Med Univ (2021) 41(10):1562–8. doi: 10.12122/j.issn.1673-4254.2021.10.16
50. Chen X, Zhang C, Wang X. Long noncoding RNA DLEU1 aggravates osteosarcoma carcinogenesis via regulating the miR-671-5p/DDX5 axis. Artif cells nanomed Biotechnol (2019) 47(1):3322–8. doi: 10.1080/21691401.2019.1648285
51. Gartner JJ, Parker SC, Prickett TD, Dutton-Regester K, Stitzel ML, Lin JC, et al. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci United States America (2013) 110(33):13481–6. doi: 10.1073/pnas.1304227110
52. Papp G, Krausz T, Stricker TP, Szendrői M, Sápi Z. SMARCB1 expression in epithelioid sarcoma is regulated by miR-206, miR-381, and miR-671-5p on both mRNA and protein levels. Genes Chromosomes Cancer (2014) 53(2):168–76. doi: 10.1002/gcc.22128
53. Malgulwar PB, Pathak P, Singh M, Kale SS, Suri V, Sarkar C, et al. Downregulation of SMARCB1/INI1 expression in pediatric chordomas correlates with upregulation of miR-671-5p and miR-193a-5p expressions. Brain tumor Pathol (2017) 34(4):155–9. doi: 10.1007/s10014-017-0295-7
54. Nagini S. Breast cancer: Current molecular therapeutic targets and new players. Anti-cancer Agents medicinal Chem (2017) 17(2):152–63. doi: 10.2174/1871520616666160502122724
55. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci (2019) 20(22):5758. doi: 10.3390/ijms20225758
56. Han TS, Hur K, Cho HS, Ban HS. Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular carcinoma. Cancers (2020) 12(9):2622. doi: 10.3390/cancers12092622
57. Yin TF, Zhao DY, Zhou YC, Wang QQ, Yao SK. Identification of the circRNA-miRNA-mRNA regulatory network and its prognostic effect in colorectal cancer. World J Clin cases (2021) 9(18):4520–41. doi: 10.12998/wjcc.v9.i18.4520
58. Zhang B. Guizhi fuling pills inhibit the proliferation, migration and invasion of human cutaneous malignant melanoma cells by regulating the molecular axis of LncRNA TPT1-AS1 / miR-671-5p. Cell Mol Biol (Noisy-le-Grand France) (2020) 66(5):148–54. doi: 10.14715/cmb/2020.66.5.26
59. Wang C, Su Z, Sanai N, Xue X, Lu L, Chen Y, et al. microRNA expression profile and differentially-expressed genes in prolactinomas following bromocriptine treatment. Oncol Rep (2012) 27(5):1312–20. doi: 10.3892/or.2012.1690
60. Borze I, Scheinin I, Siitonen S, Elonen E, Juvonen E, Knuutila S. miRNA expression profiles in myelodysplastic syndromes reveal Epstein-Barr virus miR-BART13 dysregulation. Leukemia lymphoma (2011) 52(8):1567–73. doi: 10.3109/10428194.2011.568652
61. Tan X, Fu Y, Chen L, Lee W, Lai Y, Rezaei K, et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget (2016) 7(1):293–307. doi: 10.18632/oncotarget.6344
62. Tan X, Li Z, Ren S, Rezaei K, Pan Q, Goldstein AT, et al. Dynamically decreased miR-671-5p expression is associated with oncogenic transformation and radiochemoresistance in breast cancer. Breast Cancer Res BCR (2019) 21(1):89. doi: 10.1186/s13058-019-1173-5
63. Li X, Nie C, Tian B, Tan X, Han W, Wang J, et al. miR-671-5p blocks the progression of human esophageal squamous cell carcinoma by suppressing FGFR2. Int J Biol Sci (2019) 15(9):1892–904. doi: 10.7150/ijbs.32429
64. Chen B, Gao T, Yuan W, Zhao W, Wang TH, Wu J. Prognostic value of survival of MicroRNAs signatures in non-small cell lung cancer. J Cancer (2019) 10(23):5793–804. doi: 10.7150/jca.30336
65. Harrison EB, Porrello A, Bowman BM, Belanger AR, Yacovone G, Azam SH, et al. A circle RNA regulatory axis promotes lung squamous metastasis via CDR1-mediated regulation of golgi trafficking. Cancer Res (2020) 80(22):4972–85. doi: 10.1158/0008-5472.CAN-20-1162
66. Liu Y, Feng X, Kang S, Lv F, Ni Y, Wu H. CircRIP2 promotes NSCLC progression by sponging for miR-671-5p to regulate FOXM1 expression. Histol histopathol (2022) 37(2):117–24. doi: 10.14670/HH-18-360
67. Ye J, Luo W, Luo L, Zhai L, Huang P. MicroRNA−671−5p inhibits cell proliferation, migration and invasion in non−small cell lung cancer by targeting MFAP3L. Mol Med Rep (2022) 25(1):30. doi: 10.3892/mmr.2021.12546
68. Li D, Zhang J, Li J. Role of miRNA sponges in hepatocellular carcinoma. Clinica chimica acta; Int J Clin Chem (2020) 500:10–9. doi: 10.1016/j.cca.2019.09.013
69. Dou C, Zhou Z, Xu Q, Liu Z, Zeng Y, Wang Y, et al. Hypoxia-induced TUFT1 promotes the growth and metastasis of hepatocellular carcinoma by activating the Ca(2+)/PI3K/AKT pathway. Oncogene (2019) 38(8):1239–55. doi: 10.1038/s41388-018-0505-8
70. Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol (2019) 234(8):13807–19. doi: 10.1002/jcp.28061
71. Barbagallo D, Condorelli A, Ragusa M, Salito L, Sammito M, Banelli B, et al. Dysregulated miR-671-5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme. Oncotarget (2016) 7(4):4746–59. doi: 10.18632/oncotarget.6621
72. Lu GF, You CY, Chen YS, Jiang H, Zheng X, Tang WW, et al. MicroRNA-671-3p promotes proliferation and migration of glioma cells via targeting CKAP4. OncoTargets Ther (2018) 11:6217–26. doi: 10.2147/OTT.S177325
73. Chi XG, Meng XX, Ding DL, Xuan XH, Chen YZ, Cai Q, et al. HMGA1-mediated miR-671-5p targets APC to promote metastasis of clear cell renal cell carcinoma through wnt signaling. Neoplasma (2020) 67(1):46–53. doi: 10.4149/neo_2019_190217N135
74. Ge YZ, Xu LW, Zhou CC, Lu TZ, Yao WT, Wu R, et al. A BAP1 mutation-specific MicroRNA signature predicts clinical outcomes in clear cell renal cell carcinoma patients with wild-type BAP1. J Cancer (2017) 8(13):2643–52. doi: 10.7150/jca.20234
75. Zhu Z, Luo L, Xiang Q, Wang J, Liu Y, Deng Y, et al. MiRNA-671-5p promotes prostate cancer development and metastasis by targeting NFIA/CRYAB axis. Cell Death Dis (2020) 11(11):949. doi: 10.1038/s41419-020-03138-w
76. Yu Y, Wang Z, Sun D, Zhou X, Wei X, Hou W, et al. miR-671 promotes prostate cancer cell proliferation by targeting tumor suppressor SOX6. Eur J Pharmacol (2018) 823:65–71. doi: 10.1016/j.ejphar.2018.01.016
Keywords: bone metabolism, miR-671, lncRNA, inflammation, cancer
Citation: Wang S, Hu M, Song D, Tang L and Jiang H (2023) Research progress on the role and mechanism of miR-671 in bone metabolism and bone-related diseases. Front. Oncol. 12:1018308. doi: 10.3389/fonc.2022.1018308
Received: 13 August 2022; Accepted: 20 December 2022;
Published: 11 January 2023.
Edited by:
Shiwei Duan, Zhejiang University City College, ChinaReviewed by:
Simmi Kharb, Pandit Bhagwat Dayal Sharma University of Health Sciences, IndiaCopyright © 2023 Wang, Hu, Song, Tang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Hu, aHVtaW5Aamx1LmVkdS5jbg==; Huan Jiang, amlhbmdodWFuQGpsdS5lZHUuY24=
 Shaotai Wang
Shaotai Wang Min Hu
Min Hu Dongsheng Song1,2
Dongsheng Song1,2 Huan Jiang
Huan Jiang