
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 17 November 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.783211
This article is part of the Research Topic Molecular Mechanisms in Breast Cancer Progression and Metastasis View all 9 articles
The long-non-coding HOX transcript antisense intergenic RNA (HOTAIR) was identified as significantly upregulated in breast ductal carcinoma in situ (DCIS). The aim of this study was to characterize the phenotypic effects and signaling pathways modulated by HOTAIR in early-stage breast cancer progression. We determined that HOTAIR induces premalignant phenotypic changes by increasing cell proliferation, migration, invasion and in vivo growth in normal and DCIS breast cell lines. Transcriptomic studies (RNA-seq) identified the main signaling pathways modulated by HOTAIR which include bioprocesses related to epithelial to mesenchymal transition, cell migration, extracellular matrix remodeling and activation of several signaling pathways (HIF1A, AP1 and FGFR). Similar pathways were identified as activated in primary invasive breast carcinomas with HOTAIR over-expression. We conclude that HOTAIR over-expression behaves as a positive regulator of cell growth and migration both in normal and DCIS breast cells involved with early-stage breast cancer progression.
Ductal carcinoma in situ (DCIS) is a premalignant lesion and non-obligate precursor to most invasive breast carcinomas (IBC). It has been estimated that more than one third of DCIS lesions have the potential to progress to invasive ductal carcinoma if left untreated (1). The reasons on why only some DCIS lesions progress to the invasive stage remain unclear. In a previous study, we performed the first comprehensive molecular profiling of pure high-grade (HG) DCIS lesions, thus identifying the main genomic, transcriptomic, methylation and gene pathway changes occurring at this pre-invasive breast cancer stage (2). RNA-seq profiling allowed us to identify HG-DCIS lesions with the most aggressive phenotypes, based on tumor intrinsic subtypes, proliferative, immune scores and in the activity of specific signaling pathways. Among the transcriptomic signatures of the most aggressive DCIS lesions, we identified the deregulated expression of almost 200 long-non-coding RNAs (lncRNAs), many of which might be associated with breast cancer progression. HOX transcript antisense intergenic RNA (HOTAIR) was one of the most significantly upregulated lncRNAs in aggressive DCIS lesions (2). LncRNAs are defined as non-coding RNAs exceeding 200 nucleotides in length and without evident protein coding functions (3). Over ten thousand lncRNAs have been annotated in the human genome, and although they have been increasingly implicated in neoplastic diseases, only a few have been functionally characterized (4).
HOTAIR belongs to the first lncRNAs described as aberrantly expressed in invasive breast carcinomas. Since its identification in breast cancer, HOTAIR overexpression has been reported in almost all solid tumor sites (5). HOTAIR is transcribed from the anti-sense strand of the HOXC gene cluster located on chromosome 12q13.13 and it serves as scaffold to epigenetically repress expression of the more distal HOXD gene cluster and genes in other chromosomes (6). HOTAIR is able to bind two different chromatin modifiers: the Polycomb Repressive Complex (PRC2) at the 5’ end, and the Lysine-Specific histone Demethylase 1 complex (LSD1) at the 3’ end (6, 7). Hence, HOTAIR has bifunctional modulation on chromatin status epigenetically repressing the transcription of their target genes. In addition, HOTAIR is also implicated in post-transcriptional and post-translational modulation by interaction with multiple miRNAs (e.g. miR-7, miR-148a, miR-204) or binding to E3 ubiquitin ligases, such as Mex3b and Dzip3, and promoting target degradation (8, 9). HOTAIR overexpression has been extensively described in primary and metastatic breast cancer. Early studies associated overexpression of this lncRNA in primary breast carcinomas with high metastatic potential and poor overall patient survival (10). Further studies postulated HOTAIR upregulation as a prognostic marker of lymph node metastases in ER-negative breast cancer patients (11). HOTAIR has been shown to modulate critical molecular pathways related to breast cancer development and progression such as autophagy, epithelial mesenchymal transition (EMT), and drug resistance (12). HOTAIR overexpression increases the invasive ability of breast cancer cells in vitro and in vivo (10). Notably, in murine xenograft models, HOTAIR knockout can reduce tumor growth in vivo. Thus, HOTAIR has been postulated as putative breast cancer oncogene (5).
Here we characterized for the first time the phenotypic and molecular effects of HOTAIR overexpression in non-invasive breast cancer models. Overall, we also demonstrated the relevance of its pro-oncogenic behavior at early-stages of breast cancer progression.
MCF10A cell line was obtained from the American Type Culture Collection (#CRL-10318; ATCC, VA, USA) and validated by DNA fingerprinting. MCF10A cells were cultured in Dulbecco’s Modified Eagle Medium F-12 (DMEM/F-12, Sigma-Aldrich) supplemented with 5% horse serum, 20 ng/mL epidermal growth factor (Sigma-Aldrich), 100 µg/mL hydrocortisone (Sigma-Aldrich), 10 µg/mL insulin (Sigma-Aldrich), 100 ng/mL cholera toxin (Sigma-Aldrich) and 100 U/ml penicillin - 100 μg/ml streptomycin (Sigma-Aldrich). MCF10 DCIS.COM (hereafter DCIS.COM) cells were a kind gift from Dr. Daniel Medina (13) and were maintained in DMEM/F-12 supplemented with 5% horse serum. Cell lines were maintained to 37°C with 5% CO2.
The full-length sequence of HOTAIR (2146 bp spanning six exons) was obtained from Addgene (Plasmid #26110, Watertown, MA USA), sequenced verified and subsequently cloned into the pCDH lentiviral expression vector. Virus particles were produced using packaging line Lenti-X 293T (Takara Bio, CA USA). Normal breast epithelial cell lines MCF10A and DCIS cell line DCIS.COM were stably transduced and selected with 10µg/ml puromycin.
MCF10A stably transduced to overexpress HOTAIR or an empty vector control were plated (1,000 cells per well) on 96 well plates in triplicate and cell proliferation was determined by means of the colorimetric MTT assay kit (Cell Proliferation Kit, Roche) and measuring optical density (OD). For clonal growth assays, MCF10A stably transduced to overexpress HOTAIR or vector control were plated at clonal density (500 cells/dish) in individual wells of 6-well plates and maintained in adequate media as described above. After 9 days of growth, cells were fixed and colonies stained with crystal violet. Digital images of individual wells were obtained and used to determine the number and area of growing colonies using ImageJ software. Transwell migration assays were performed using standard Boyden chambers containing 12 μm pore divider membranes, 5% FBS was used in the lower chamber as chemoattractant. Statistical significance was determined using Mann-Whitney-Wilcoxon test.
DCIS.COM stably transduced cells with HOTAIR (n=3) or empty vector as control (n=3 mice) were inoculated via the nipple using a 30-gauge Hamilton syringe into the intact main mammary duct of both inguinal mammary glands of female SCID mice 6-8 wks of age. Tumor growth was monitored, and after an observation period of 10 wks. post injection, mice were euthanized and both inguinal mammary glands were dissected. Xenografts of wild-type DCIS.COM cell line result in the formation of DCIS-like tumors but do not invade (13).
MCF10A and DCIS.COM stably transduced cells were used for RNA isolation from subconfluent plates using the RNeasy kit (Qiagen, CA, USA). RNA concentration and integrity were measured on an Agilent 2100 Bioanalyzer (Agilent Technologies). Only RNA samples with RNA integrity values (RIN) over 8.0 were considered for subsequent analysis. RNA-seq library construction was performed using the ScriptSeq v2 RNA-seq Library Preparation Kit (Epicentre) according to the manufacturer’s protocol. We performed 76 nt paired-end sequencing using an Illumina HiSeq2000 platform and ~20 million reads per sample were obtained. The short-sequenced reads were mapped to the human reference genome (hg19) by the splice junction aligner Rsubread package. We employed several R/Bioconductor packages to accurately calculate the gene expression abundance at the whole-genome level using the aligned records (BAM files) and to identify differentially expressed genes between cells stably transduced with HOTAIR and empty vector. Briefly, the number of reads mapped to each gene based on the UCSC.hg19.KnownGene database were counted, reported and annotated using the featureCounts and org.Hs.eg.db packages. Data are available at GEO under accession number GSE183058. To identify differentially expressed genes (log2 fold change [FC] > ± 1.5, False Discovery Rate [FDR] <0.05) between the empty vector and HOTAIR overexpressing counterparts, we utilized the edgeR Bioconductor package based on the normalized log2 based count per million values. For functional enrichment analyses, we used R/Bioconductor clusterProfiler package and the InnateDB resource (http://www.innatedb.com/) based on the list of dysregulated transcripts. Data integration and visualization of differentially expressed transcripts were done with R/Bioconductor and the MultiExperiment Viewer software (MeV v4.9).
Pre-processed HOTAIR expression profiles among five early-stage breast cancer datasets: GSE69994 (2), GSE59246 (14), GSE41228 (15), GSE66301 (16) and GSE47462 (17) were obtained from GEO and analyzed using R software. In addition, pre-processed HOTAIR RNA-seq expression levels among primary breast carcinomas with intrinsic subtype data and their integrated pathway activities (pathway activity – z score of 1387 constituent PARADIGM pathways) were obtained from the TCGA Breast Cancer (BRCA) dataset through the UCSC Xena browser (http://xena.ucsc.edu/). The PARADIGM algorithm integrates pathway, expression, and copy number data to infer activation of pathway features within a superimposed pathway network structure extracted from NCI-PID, BioCarta, and Reactome (18). Briefly, primary breast carcinomas (n = 1097) were divided into low (n=191) or high (n=392) HOTAIR expression levels according to the StepMiner one-step algorithm (19). These two groups were then compared at their integrated pathway activities to identify the most relevant signaling pathways associated with HOTAIR expression using the T-test (p-adj. < 0.01) with MultiExperiment Viewer Software (MeV 4.9). Statistical analysis was performed using the computing environment R.
In a previous study, we performed a comprehensive molecular profiling of ‘pure’ high-grade DCIS lesions, providing the first catalogue of genomic, transcriptomic, methylation and gene pathway changes occurring at this pre-invasive breast cancer stage (2). Among the most significantly upregulated lncRNAs we found HOTAIR (fold change (FC) = 32.7; false discovery rate (FDR) < 0.0001) when DCIS were compared with normal breast tissue (Figure 1A). Therefore, we hypothesize that HOTAIR might have a relevant role also in early-stage breast development and not just in later stages of tumor progression as previously described. In this study we characterized the molecular and phenotypic effects of HOTAIR expression in normal and non-invasive breast cancer models. In silico analysis of HOTAIR expression among five early-stage breast cancer datasets obtained from Gene Expression Omnibus (GEO) showed significant upregulation of this transcript in DCIS and IBC when compared to normal samples (p < 0.01; Figure 1B). However, non-significant differences were observed in HOTAIR expression levels when DCIS was compared with IBC samples as seen in analyses of three independent DCIS-IBC datasets (p > 0.05; Figure 1B) (14–16). HOTAIR expression levels were also evaluated in normal and early stage neoplasias (including columnar cell lesions and atypical ductal hyperplasia) obtained from the GSE47462 dataset (17). Interestingly, the DCIS precursor lesions (described as early-neoplasia in Figure 1C), showed significant HOTAIR overexpression when compared with normal samples (p<0.001; Figure 1C). HOTAIR expression was also compared across DCIS intrinsic subtypes in two independent datasets (2, 14). HER2 and luminal A DCIS intrinsic subtypes showed significantly higher HOTAIR expression levels compared with the luminal B and basal-like subtypes (p<0.01; Figure 1D).
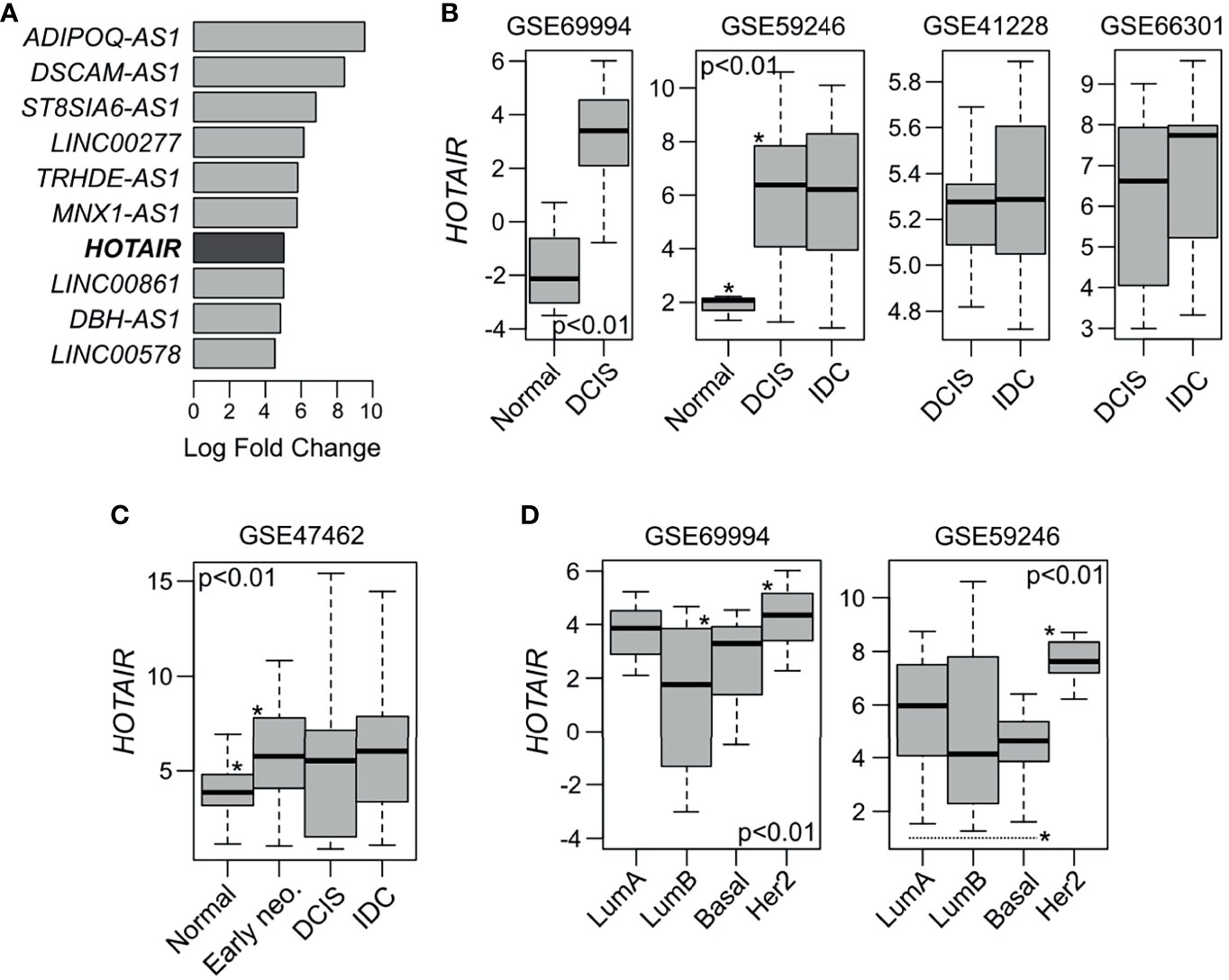
Figure 1 HOTAIR expression in normal, pre-invasive and invasive breast samples. (A) Top ten most upmodulated lncRNAs in DCIS lesions compared with normal breast samples according to GSE69994 study (2). (B) In silico HOTAIR expression analysis among normal, DCIS and IBC samples obtained from four independent GEO dataset (2, 14–16). HOTAIR expression was significant upregulated in DCIS and IBC samples compared with normal samples (p < 0.01), while non-significant differences were observed between DCIS and IBC cases (p > 0.05). (C) HOTAIR expression analysis among normal and DCIS precursor lesions (early neo.) such as columnar cell lesions and atypical ductal hyperplasia, obtained from GSE47462 dataset (17). (D) HOTAIR expression analysis across DCIS intrinsic subtype obtained from two independent GEO datasets (2, 14). ANOVA or T-test were used to compare the HOTAIR expression among groups. *Statistical significance differences.
HOTAIR expression is modulated by multiple signaling pathways. Its promoter sequence contains binding sites for diverse transcription factors, such as estrogen response elements (EREs), hypoxia response elements (HREs), AP1 response elements (TREs) among others (20). HOTAIR expression can be induced by estradiol (E2) in an estrogen receptor dependent manner through EREs or independent via interaction with G-protein-coupled estrogen receptor-1 (GPER) (21, 22). HER2 has also been recently described as an activator of HOTAIR expression by acting on the effector mitogen-activated protein kinase (MAPK) in primary invasive breast carcinomas and invasive breast cancer cells (23). In agreement with these observations, the HER2 DCIS intrinsic subtype appears as the group with highest HOTAIR expression levels followed by the E2/ER responsive luminal A subtype (Figure 1D).
Overall, these data suggest that HOTAIR over-expression might be a critical molecular event promoting breast cancer development at early pre-invasive stages, remaining up-modulated in invasive and metastatic carcinomas in specific molecular subtypes.
To better understand the mechanism of action of HOTAIR and their phenotypic impact in normal and DCIS cells, MCF10A and DCIS.COM cells were stably transduced for HOTAIR overexpression for further transcriptomic, in vitro and in vivo characterization. Whole-transcriptome unsupervised analysis from RNA-Seq data demonstrates a clear segregation of transduced cells in MCF10A and DCIS.COM groups (Figure 2A).
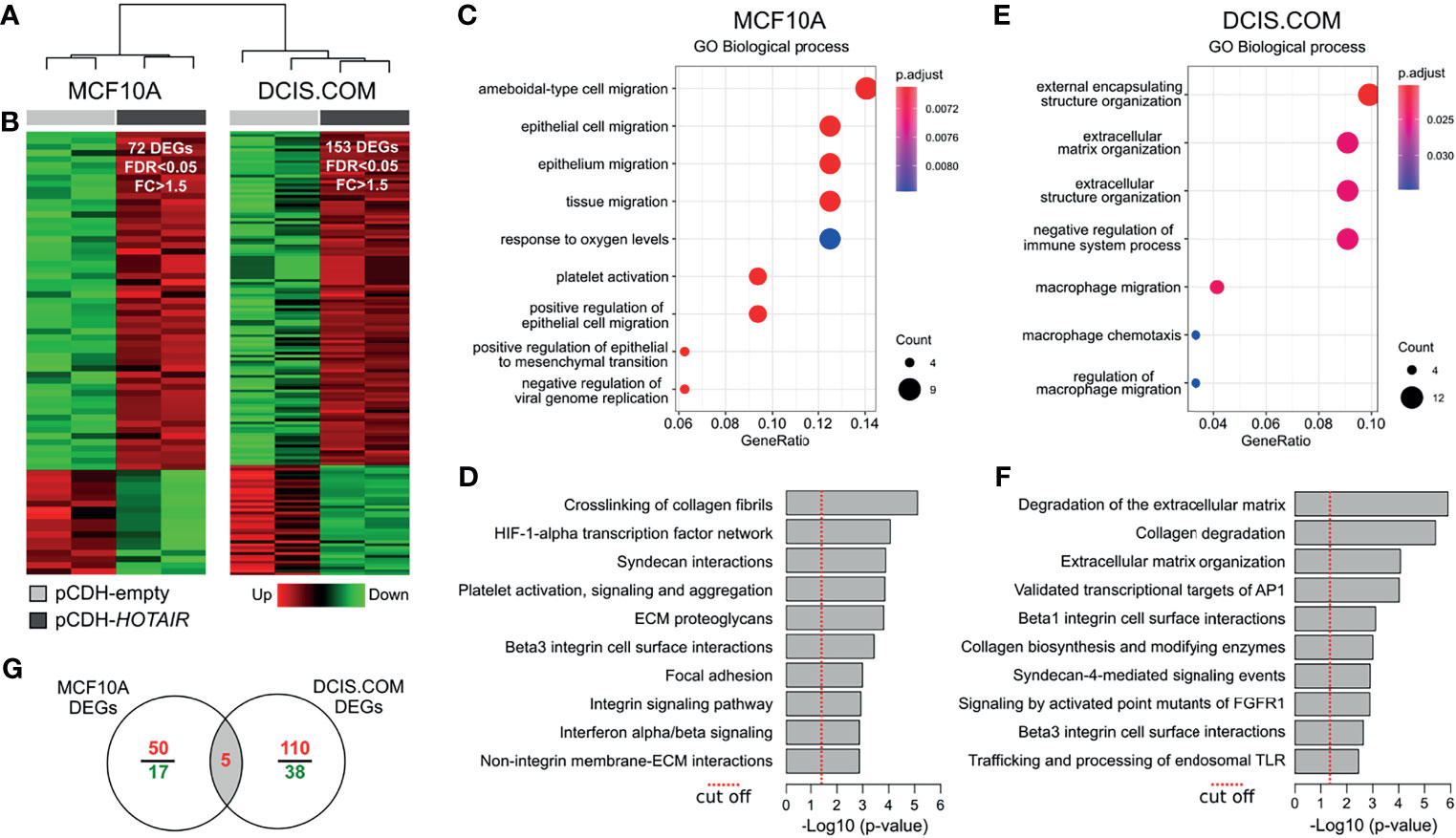
Figure 2 Transcriptomic analysis of HOTAIR overexpressing cells. (A) Hierarchical clustering of MCF10A and DCIS.COM stably transduced cells with either vector control (pCDH-empty) or lentivirus expressing HOTAIR (pCDH-HOTAIR) based on RNA-seq profiles. (B) Heat map representation of the differentially expressed genes (DEG) obtained by RNA-seq analysis (FDR<0.05; FC>1.5). Red represents upregulated genes and green downregulated genes. (C, D) Functional enrichment analysis of GO biological processes and pathways identified as affected by expression of HOTAIR in MCF10A cells. (E, F) Functional enrichment analysis of GO biological processes and pathways identified as affected by expression of HOTAIR in DCIS.COM cells. (G) Venn diagram of transcripts commonly modulated among MCF10 and DCIS.COM cells stably transduced with HOTAIR.
RNA-seq analysis of MCF10A cells identified 72 differentially expressed genes (DEG) of which 55 were upregulated and 17 were downregulated comparing HOTAIR expressing cells with the empty-vector cells (FC ≥ 1.5; FDR ≤ 0.05; Figure 2B and Supplementary Data 1). Functional enrichment analysis of DEG in MCF10A cells indicated a robust association with positive regulation of cell migration and EMT gene ontology (GO) biological processes (p < 0.001; Figure 2C). Remarkably, extracellular matrix (ECM) interaction/remodeling and HIF1A transcription factor signaling were among the most significantly modulated pathways in MCF10A cells stably transduced with HOTAIR (Figure 2D). Among the upregulated genes in MCF10A, we found AXL (AXL receptor tyrosine kinase), ANGPTL4 (Angiopoietin like 4), MALAT1 (Metastasis associated lung adenocarcinoma transcript 1), VIM (Vimentin) and CDH2 (Cadherin 2). These genes are involved in pro-tumorigenic processes including EMT, cell migration, invasion and stemness (24–26). EMT is a dynamic and reversible process modulated by epigenetic regulators (PRC2, NuRD, LSD1 and PHF2) and gene expression changes (27, 28). HOTAIR interacts with PRC2 to trigger H3K27 methylation of EMT gene promoters (10, 29).
Furthermore, a recent study has shown that HOTAIR negatively regulates the function of LSD1 in maintaining epithelial identity demonstrating that most of the transcriptome changes induced by HOTAIR require both PRC2- and LSD1-interacting domains (30). In this sense, the upmodulation of mesenchymal markers such as VIM and CDH2 in MCF10A HOTAIR transduced cells, clearly suggests HOTAIR involvement in EMT regulation at early stages of breast cancer progression. As mentioned, the lncRNA MALAT1 was also detected as upregulated in association with HOTAIR overexpresion in MCF10A cells. This lncRNA was initially identified as upregulated in primary non-small cell lung cancer cells with higher metastasis ability and subsequently associated with other tumor types (24, 31). Recently, MALAT1 was identified as a hypoxia-induced transcript that could promote cellular migration and proliferation of breast cancer cells (32). Interestingly, the HIF-1 alpha transcription factor network was among the most significantly enriched pathways in MCF10A HOTAIR transduced cells suggesting a cooperative role between both oncogenic lncRNAs.
In DCIS.COM cells HOTAIR overexpression caused the deregulation of 153 genes, of which 115 were upregulated and 38 were downregulated (FC > 1.5; FDR < 0.05; Figure 2B and Supplementary Data 1). Functional enrichment analysis of DEG in DCIS.COM showed a significant enrichment of ECM organization and immune related GO biofunctions (Figure 2E). Consistently, ECM/Collagen degradation and AP1 and FGFR1 signaling pathways were significantly dysregulated in DCIS.COM cells stably transduced with HOTAIR (Figure 2F). Among the upregulated genes in DCIS.COM, we found several matrix metallopeptidases (e.g.: MMP2, MMP14, MMP28), and fibrogenic ECM (e.g.: COL7A1, COL9A3, COL16A1, COL17A1) and Beta1/3 integrin related genes (e.g.: COL7A1, MDK, PLAU). The ECM is composed of a complex meshwork of highly cross-linked components, including fibrous proteins, glycoproteins, proteoglycans, and polysaccharides. Matrix metalloproteinases are zinc-dependent endopeptidases involved in ECM degradation and tissue remodeling. These endopeptidases are capable of degrading both the ECM and basement membrane, physical barriers that prevent expanding growth and migration of cancer cells (33). In addition, several studies have involved the high collagen and integrins expression levels with the tumor stroma-associated fibrosis (also called desmoplasia), a process that promotes tumor cells migration and metastasis (34). In this sense, increased HOTAIR expression in DCIS could facilitate acquiring the invasiveness capability to progress to the malignant stages. Despite the small number of genes commonly modulated between MCF10A and DCIS.COM cells (HOTAIR, GNG2, ENPP2, PPFIA4 and NDRG1) (Figure 2G), several pathways related with ECM organization, collagen degradation, and Beta integrin cell surface interactions were commonly modulated between normal and DCIS HOTAIR transduced cells (Supplementary Data 1).
To investigate the phenotypic impact of HOTAIR overexpression in normal breast epithelial cells, we conducted cell proliferation, colony formation, and transwell migration assays on stably transduced MCF10A cells (Figure 3A). We first determined the effects of stable HOTAIR expression on cell proliferation by means of the MTT assay. As can be observed in Figure 3B, stable HOTAIR expression behaved as a pro-oncogenic stimulus inducing increased cell proliferation in normal breast cells after a week of cell culture (p < 0.01). The positive effect of HOTAIR on cell proliferation was further confirmed by means of colony formation assays. MCF10A cells stably transduced to overexpress HOTAIR displayed dramatic increase in colony growth when seeded at clonal density (Figure 3C). MCF10A cell line showed increased percentage area covered by colonies (p < 0.01) indicating increased cell growth (cell proliferation) as consequence of HOTAIR overexpression. Furthermore, MCF10A cells stably transduced with HOTAIR encoding lentivirus were also characterized by effects in the transwell migration assay (p < 0.01) (Figure 3D). The described results demonstrate that HOTAIR indeed behaves as a positive regulator of cell growth and migration in normal breast cells.
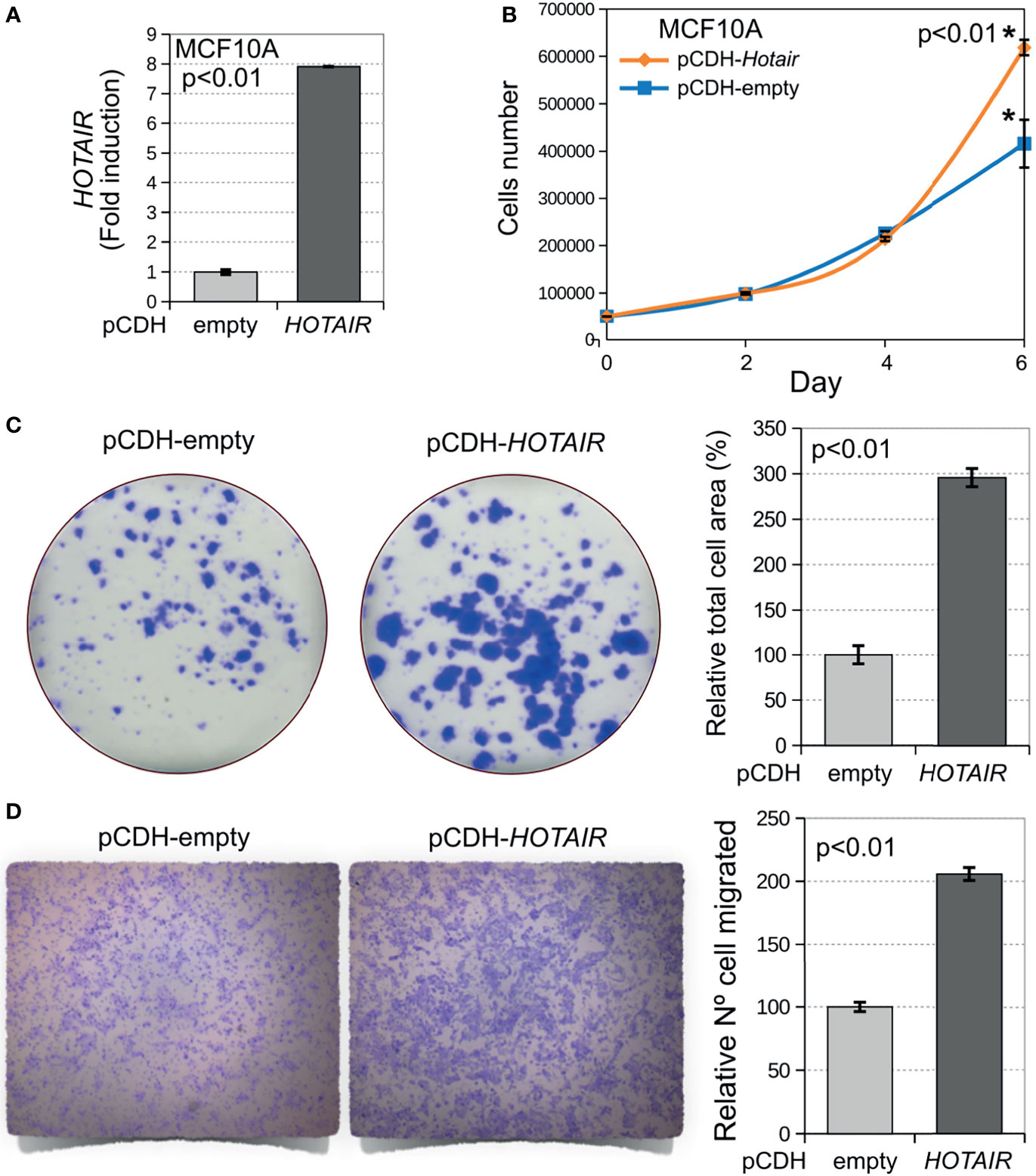
Figure 3 Stable overexpression of HOTAIR induces increased cell proliferation, colony growth and invasion in normal breast cells. (A) Levels of HOTAIR expression in stably transduced MCF10A cells based on their RNA-seq profiles (log2 CPM). (B) Stable overexpression of HOTAIR increases cell proliferation in normal breast cells (p < 0.01). Cells were plated 1,000 cells per well on 96 well plates in triplicate and cell proliferation was determined by means of the MTT colorimetric assay and measuring optical density (OD). (C) Cells stably transduced with lentivirus expressing HOTAIR or vector control were plated at clonal density in 6-well plates. Cells were allowed to grow for 9 days, fixed and stained with crystal violet. Bar chart displays increased area occupied by colonies for HOTAIR stably transduced cells compared with vector control. (D) Transwell migration assay of DCIS.COM cells stably transduced with HOTAIR. On the left comparative pictures of cells that migrated through the membrane, on the right bar chart of the relative numbers of cells per membrane for HOTAIR stably transduced cells compared with vector control (p < 0.01). Statistical significance was determined using Mann-Whitney-Wilcoxon test. *Statistical significance differences.
To investigate the effects of HOTAIR on the in vivo progression of the stably transduced DCIS.COM cell line (Figure 4A), the mammary intraductal DCIS model (MIND) was employed (13). Briefly, the MIND assay consists in the inoculation with DCIS.COM cells (HOTAIR transduced or empty vector) via the nipple into the intact main mammary duct of both inguinal mammary glands of female NSG mice. Behbod et al. have described that DCIS.COM cells when injected via intra-nipple in NSG mice, grow intraductally and do not invade, thus mimicking the non-invasive features of DCIS growth and lesions look histologically almost identical to clinical DCIS (13). DCIS.COM cells stably transduced with HOTAIR produced in vivo growth and development of invasive lesions in 2 out of 3 injected mice (Figures 4B, C), while DCIS.COM invasive growth was not observed in mice injected with DCIS.COM cells transduced with empty vector (n= 3). These data suggest that HOTAIR overexpression may behave as a driver of growth in vitro and in vivo at premalignant stages of breast cancer progression.

Figure 4 DCIS.COM stably transduced cells with either vector control or lentivirus expressing HOTAIR were compared using the in vivo MIND model. (A) HOTAIR expression levels in stably transduced DCIS.COM cells based on their RNA-seq profiles (log2 CPM). (B) DCIS growth was detected among injected mice. (C) Representative invasive ductal carcinoma (10X H&E staining) induced by DCIS.COM cells stably transduced with HOTAIR. Scale bar: 200 μm.
To further evaluate the relevance of the HOTAIR expression and their modulated pathways identified in normal and pre-invasive models with the invasive stage, we performed an in silico analysis on invasive breast carcinomas obtained from TCGA (n=1097). The Step-miner algorithm (19) allowed us to identify primary tumors with high (n=392) or low (n=191) HOTAIR expression (Figure 5A). Interestingly, a significantly larger number of tumors with high HOTAIR expression were detected in HER2+ (98%) and basal-like (82%) subtypes compared with luminal A (60%) and luminal B (53%) breast cancer subtypes (p < 0.0001; Figures 5B, C). These results are in agreement with higher HOTAIR expression levels detected in HER2+ DCIS than any of the other subtypes (Figure 1B). Analysis of pathway-based representation analysis (PARADIGM) identified 68 activated signaling pathways in invasive carcinomas with high HOTAIR expression compared with low expression counterparts (p-adj. < 0.01; Figure 5C). Interestingly, several of the activated signaling pathways identified in invasive carcinomas with high HOTAIR expression (Figure 5D) were detected in normal and DCIS HOTAIR stably transduced cells such as: Syndecan signaling, HIF1A transcription factor network, FGFR signaling, degradation of collagen, AP1 transcriptional targets, among others (Supplementary Data 2). However, other activities such as p53/p63, Wnt and nuclear B-catenin signaling were only detected in the invasive stage associated with HOTAIR overexpression. Nevertheless, our results revealed that multiple signaling pathways associated with HOTAIR overexpression in invasive breast carcinomas were also modulated in normal and DCIS HOTAIR transduced cells. Overall, the comparative transcriptomic analysis suggests that HOTAIR is probably a critical mediator of the EMT, cell migration, and ECM remodeling programs to drive breast cancer progression at premalignant stages.
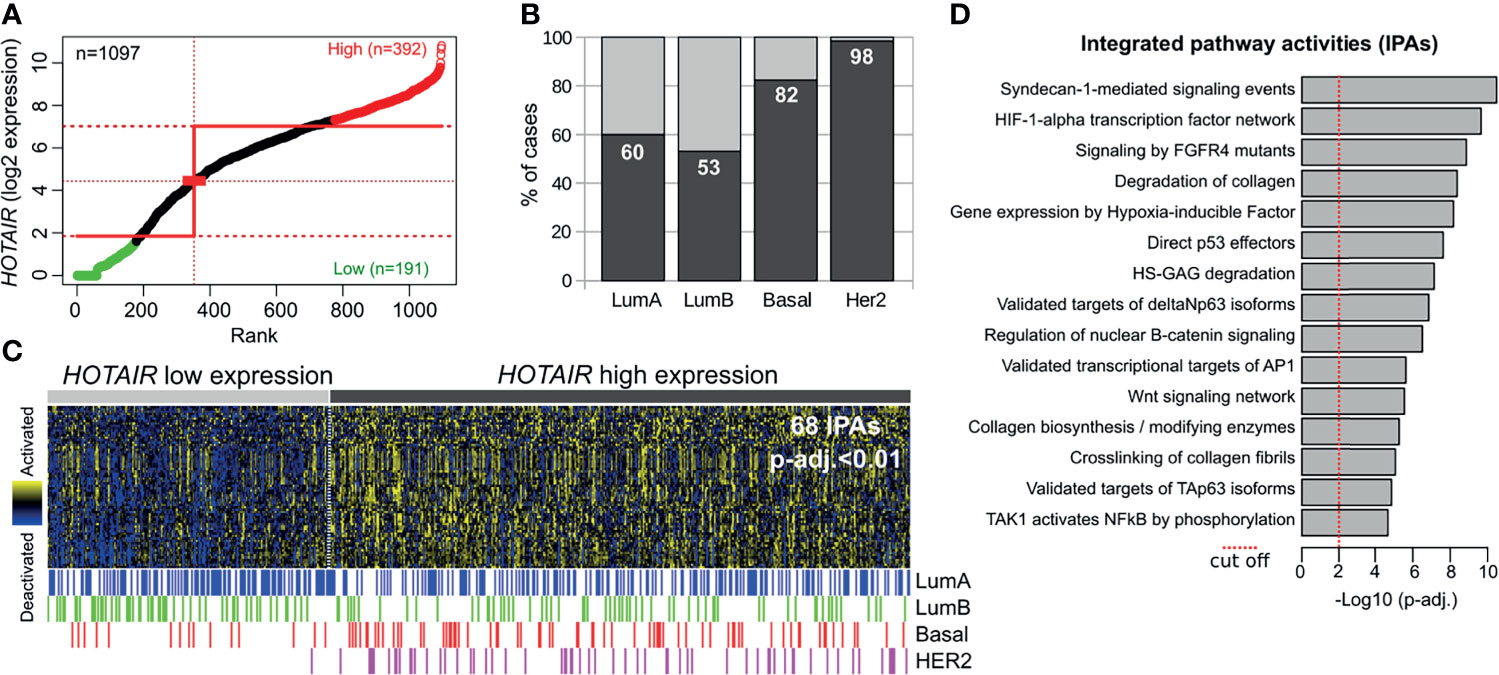
Figure 5 HOTAIR expression and pathway activity analysis in primary invasive breast carcinomas. (A) Primary breast carcinomas were divided into HOTAIR low or high expression levels based on the StepMiner algorithm using TCGA RNA-seq datasets obtained from the UCSC Xena resource (https://xenabrowser.net/). (B) Percentage of cases with high or low HOTAIR expression among intrinsic subtypes showing a consistent up-regulation in basal-like and HER2 subtypes compared with luminal-like tumors. (C) Heat map with the 68 significantly activated PARADIGM Integrated Pathways Activities (IPAs) among HOTAIR high expression tumors (p-adj.<0.01). (D) Bar plot of the top fifteen activated pathways determined by PARADIGM algorithm in primary invasive breast carcinomas with HOTAIR overexpression.
In conclusion, the described results indicate that HOTAIR overexpression induces premalignant phenotypic changes in normal breast epithelial and DCIS cells compatible with the necessary steps towards malignancy, such as increase in cell proliferation, migration and invasion. In agreement with the in vitro and in vivo observations, we identified that HOTAIR upmodulates the expression of transcripts associated with the epithelial to mesenchymal transition, cell migration, and extracellular matrix degradation among other bioprocesses. Finally, HOTAIR overexpression was significantly associated with HER2+ DCIS and IBC subtypes. Further mechanistic characterization of HOTAIR in preinvasive in vitro and in vivo models may provide insights into how this oncogenic lncRNA could contribute to the early stages of breast cancer development and progression.
The transcriptomic data are available at GEO under accession number GSE183058. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE183058.
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of UT-MDACC.
MCA and CMA contributed the conception of the project and the design of all experiments. Experiments were conducted by JL, PT, and HK. MCA and MF carried out all bioinformatic analyses. MCA and CMA wrote the main body of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Breast Cancer Research Program under Award No. W81XWH-16-1-0027, BC150021, the MD Anderson Cancer Center Support Grant P30 NIH CA16672, and the CPRIT Core Facility Support Grant RP170002 to CMA, and the Argentine National Agency of Scientific and Technological Promotion, grant PICT-2018-01403 to MCA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.783211/full#supplementary-material
Supplementary Data Sheet 1 | Differentially expressed genes and deregulated pathways in MCF10A and DCIS.COM stably transduced cells.
Supplementary Data Sheet 2 | Activated pathways in HOTAIR high vs. low expression invasive breast carcinomas.
1. Allred DC. Ductal Carcinoma in Situ: Terminology, Classification, and Natural History. J Natl Cancer Inst Monogr (2010) 2010(41):134–8. doi: 10.1093/jncimonographs/lgq035
2. Abba MC, Gong T, Lu Y, Lee J, Zhong Y, Lacunza E, et al. A Molecular Portrait of High-Grade Ductal Carcinoma In Situ. Cancer Res (2015) 75(18):3980–90. doi: 10.1158/0008-5472.CAN-15-0506
3. Amelio I, Bernassola F, Candi E. Emerging Roles of Long Non-Coding RNAs in Breast Cancer Biology and Management. In: Sem Cancer Biol (2021) 72:36–45. doi: 10.1016/j.semcancer.2020.06.019
4. Ransohoff JD, Wei Y, Khavari PA. The Functions and Unique Features of Long Intergenic non-Coding RNA. Nat Rev Mol Cell Biol (2018) 19:143. doi: 10.1038/nrm.2017.104
5. Yu X, Li Z. Long non-Coding RNA HOTAIR: A Novel Oncogene. Mol Med Rep (2015) 12(4):5611–8. doi: 10.3892/mmr.2015.4161
6. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
7. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science (2010) 329:689–93. doi: 10.1126/science.1192002
8. Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold Function of Long Non-Coding RNA HOTAIR in Protein Ubiquitination. Nat Commun (2013) 4:2939. doi: 10.1038/ncomms3939
9. Cantile M, Di Bonito M, Tracey De Bellis M, Botti G. Functional Interaction Among lncRNA HOTAIR and MicroRNAs in Cancer and Other Human Diseases. Cancers (2021) 13:570. doi: 10.3390/cancers13030570
10. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464:1071–6. doi: 10.1038/nature08975
11. Gökmen-Polar Y, Vladislav IT, Neelamraju Y, Janga SC, Badve S. Prognostic Impact of HOTAIR Expression Is Restricted to ER-Negative Breast Cancers. Sci Rep (2015) 5:8765. doi: 10.1038/srep08765
12. Cantile M, Di Bonito M, Cerrone M, Collina F, De Laurentiis M, Botti G. Long non-Coding RNA HOTAIR in Breast Cancer Therapy. Cancers (2020) 12(5):1197. doi: 10.3390/cancers12051197
13. Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, et al. An Intraductal Human-in-Mouse Transplantation Model Mimics the Subtypes of Ductal Carcinoma in Situ. Breast Cancer Res (2009) 11(5):R66. doi: 10.1186/bcr2358
14. Lesurf R, Aure MR, Mørk HH, Vitelli V, Sauer T, Geisler J, et al. Molecular Features of Subtype-Specific Progression From Ductal Carcinoma in Situ to Invasive Breast Cancer. Cell Rep (2016) 16(4):1166–79. doi: 10.1016/j.celrep.2016.06.051
15. Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, et al. Differentially Expressed Genes Regulating the Progression of Ductal Carcinoma in Situ to Invasive Breast Cancer. Cancer Res (2012) 72(17):4574–86. doi: 10.1158/0008-5472.CAN-12-0636
16. DeVaux RS, Ropri AS, Grimm SL, Hall PA, Herrera EO, Chittur SV, et al. Long Noncoding RNA BHLHE40-AS1 Promotes Early Breast Cancer Progression Through Modulating IL-6/STAT3 Signaling. J Cell Biochem (2020) 121(7):3465–78. doi: 10.1002/jcb.29621
17. Brunner AL, Li J, Guo X, Sweeney RT, Varma S, Zhu SX, et al. A Shared Transcriptional Program in Early Breast Neoplasias Despite Genetic and Clinical Distinctions. Genome Biol (2014) 15(5):1–6. doi: 10.1186/gb-2014-15-5-r71
18. Vaske CJ, Benz SC, Sanborn CJ, Earl D, Szeto C, Zhu J, et al. Inference of Patient-Specific Pathway Activities From Multidimensional Cancer Genomics Data Using PARADIGM. Bioinformatics (2010) 26:i237–45. doi: 10.1093/bioinformatics/btq182
19. Sahoo D, Dill DL, Tibshirani R, Plevritis SK. Extracting Binary Signals From Microarray Time-Course Data. Nucleic Acids Res (2007) 35(11):3705–12. doi: 10.1093/nar/gkm284
20. Bhan A, Mandal SS. LncRNA HOTAIR: A Master Regulator of Chromatin Dynamics and Cancer. Biochim Biophys Acta (2015) 1856:151–64. doi: 10.1016/j.bbcan.2015.07.001
21. Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR Is Transcriptionally Induced by Estradiol. J Mol Biol (2013) 425:3707–22. doi: 10.1016/j.jmb.2013.01.022
22. Tao S, He H, Chen Q. Estradiol Induces HOTAIR Levels via GPER-Mediated miR-148a Inhibition in Breast Cancer. J Transl Med (2015) 13:131. doi: 10.1186/s12967-015-0489-x
23. Wang YL, Liu LC, Hung Y, Chen CJ, Lin YZ, Wu WR, et al. Long non-Coding RNA HOTAIR in Circulatory Exosomes Is Correlated With ErbB2/HER2 Positivity in Breast Cancer. Breast (2019) 46:64–9. doi: 10.1016/j.breast.2019.05.003
24. Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 Contributes to Bladder Cancer Cell Migration by Inducing Epithelial-to-Mesenchymal Transition. Mol Biosyst (2012) 8(9):2289–94. doi: 10.1039/c2mb25070e
25. Carbone C, Piro G, Merz V, Simionato F, Santoro R, Zecchetto C, et al. Angiopoietin-Like Proteins in Angiogenesis, Inflammation and Cancer. Int J Mol Sci (2018) 19(2):431. doi: 10.3390/ijms19020431
26. Khera L, Lev S. Accelerating AXL Targeting for TNBC Therapy. Int J Biochem Cell Biol (2021) 14:106057. doi: 10.1016/j.biocel.2021.106057
27. Tam WL, Weinberg RA. The Epigenetics of Epithelial-Mesenchymal Plasticity in Cancer. Nat Med (2013) 19(11):1438–49. doi: 10.1038/nm.3336
28. Lamouille S, Xu J, Derynck R. Molecular Mechanisms of Epithelial–Mesenchymal Transition. Nat Rev Mol Cell Biol (2014) 15(3):178–96. doi: 10.1038/nrm3758
29. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long Noncoding RNA HOTAIR Regulates Polycomb-Dependent Chromatin Modification and is Associated With Poor Prognosis in Colorectal Cancers. Cancer Res (2011) 71(20):6320–6. doi: 10.1158/0008-5472.CAN-11-1021
30. Jarroux J, Foretek D, Bertrand C, Gabriel M, Szachnowski U, Saci Z, et al. HOTAIR lncRNA Promotes Epithelial–Mesenchymal Transition by Redistributing LSD1 at Regulatory Chromatin Regions. EMBO Rep (2021) 6:e50193. doi: 10.15252/embr.202050193
31. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM. MALAT-1, a Novel Noncoding RNA, and Thymosin Beta4 Predict Metastasis and Survival in Early-Stage non-Small Cell Lung Cancer. Oncogene (2003) 22(39):8031–41. doi: 10.1038/sj.onc.1206928
32. Shih CH, Chuang LL, Tsai MH, Chen LH, Chuang EY, Lu TP, et al. Hypoxia-Induced MALAT1 Promotes the Proliferation and Migration of Breast Cancer Cells by Sponging MiR-3064-5p. Front Oncol (2021) 11. doi: 10.3389/fonc.2021.658151
33. Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell (2010) 141(1):52–67. doi: 10.1016/j.cell.2010.03.015
Keywords: HOTAIR, lncRNA, breast cancer, DCIS, proliferation, invasion
Citation: Abba MC, Fabre ML, Lee J, Tatineni P, Kil H and Aldaz CM (2021) HOTAIR Modulated Pathways in Early-Stage Breast Cancer Progression. Front. Oncol. 11:783211. doi: 10.3389/fonc.2021.783211
Received: 27 September 2021; Accepted: 29 October 2021;
Published: 17 November 2021.
Edited by:
César López-Camarillo, Universidad Autónoma de la Ciudad de México, MexicoReviewed by:
Hong Zheng, Stanford University, United StatesCopyright © 2021 Abba, Fabre, Lee, Tatineni, Kil and Aldaz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin C. Abba, bWNhYmJhQGdtYWlsLmNvbQ==; C. Marcelo Aldaz, bWFhbGRhekBtZGFuZGVyc29uLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.