- 1Department of Oncology, Institute of Tumor, Guangzhou University of Chinese Medicine, Guangzhou, China
- 2Department of Oncology, Lingnan Medical Research Center of Guangzhou University of Chinese Medicine, Guangzhou, China
- 3Department of Oncology, Guangzhou University of Chinese Medicine, Guangzhou, China
- 4Department of Oncology, The First Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou University of Chinese Medicine, Guangzhou, China
- 5Department of Hepatopancreatobiliary, Cancer Center, Southern Medical University, Guangzhou, China
- 6Department of Hepatology, TCM-Integrated Hospital of Southern Medical University, Guangzhou, China
Circular RNAs (circRNAs) are stable covalently closed non-coding RNAs (ncRNAs). Many studies indicate that circRNAs are involved in the pathological and physiological processes of liver cancer. However, the functions of circRNAs in liver cancer immunity are less known. In this review, we summarized the functions of circRNAs in liver cancer, including proliferative, metastasis and apoptosis, liver cancer stemness, cell cycle, immune evasion, glycolysis, angiogenesis, drug resistance/sensitizer, and senescence. Immune escape is considered to be one of the hallmarks of cancer development, and circRNA participates in the immune escape of liver cancer cells by regulating natural killer (NK) cell function. CircRNAs may provide new ideas for immunotherapy in liver cancer.
Introduction
Liver cancer, a disease with high mortality and poor prognosis, is one of the most common malignant tumors in the world (1). Statistics show that liver cancer ranks the fifth in cancer incidence, the second in all cancer deaths, and the third in cancer mortality (2). Liver cancer includes three major pathological types: hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC), and HCC-ICC mixed type (3). The occurrence of liver cancer is closely related to hepatitis B, hepatitis C, and non-alcoholic fatty liver disease (4–6). According to the patient’s overall conditions, a range of therapies have been utilized in the liver cancer treatment, such as surgical resection, liver transplantation, immunotherapy, local ablative therapies, and systemic chemotherapy. However, liver cancer is generally detected at the late stage because the patients might not perform the clinical symptoms at the beginning. Its recurrence is approximately 50–80% after treatment within 5 years (7). A better understanding of the molecular mechanisms of liver cancer is essential to largely improve the overall prognosis and discover novel effective therapies of liver cancer.
Immune escape refers to the growth and metastasis of tumor cells through various mechanisms to avoid recognition and attack by the immune system (8). The mechanisms of immune escape are mainly related to modifications, changes in tumor cells and alterations in the tumor immune microenvironment. Through the mechanisms of modification and change, tumor cells themselves can enhance their ability to evade immune surveillance and attack. Tumor has a highly heterogeneous structure, and tumor cells interact with many cells and factors including immune cells and immune factors to form a complex tumor immune microenvironment. The tumor microenvironment is the place where the immune system interacts with tumor cells. Natural killer (NK) cells are involved in tumor immune escape through multiple mechanisms (9–11). Various soluble factors and cytokines released by tumor cells or the tumor microenvironment reduce the activity of NK cells and their cytotoxic activity (12–14). Therefore, restoration of NK cell function is an important area of research in antitumor immunotherapy. Various strategies have been developed to restore NK cell function, including cytokine therapies, monoclonal antibodies, and adoptive cell transfer (15–18). NK cells can be divided into CD56bright and CD56dim based on the expression of CD56. Two subpopulations of CD56 are present, of which the CD56bright subpopulation can be amplified by IL-2 stimulation. The CD56bright subpopulation can be amplified by IL-2 stimulation, and about 10% of them express killer cells. Immunoglobulin-like receptor secretes synthetic TNF-associated apoptosis-inducing ligand (TRAIL). CD56dim subpopulation is insensitive to IL-2 stimulation, and 85% of CD56dim are KIR+ (19–22). In HCC, Rae1 is expressed on the surface of HCC cells, and this factor, as a ligand of NKG2D, the NK cell activation receptor, can activate NK cells and promote their anti-tumor immunity. On the other hand, the immune function of NK cells is limited, and the subsets of CD56dimNK cells in the peripheral blood of HCC patients were significantly lower than those in the healthy control group (23). CD56dimNK cells in the tumor area of HCC patients expressed fewer IFN-γ than non-CD56dimNK cells, which was associated with CD4+CD25+Tregs in vitro. During hepatocarcinogenesis, changes in the microenvironment of the extracellular matrix and the secretion of TGF-β by hepatic stellate cells can inhibit the activity and function of NK cells, thus weakening their monitoring function of hepatocytes (24). TGF-β secreted by Treg can inhibit NK cell activation by down-regulating NKG2D, affecting its immune killing function against liver cancer cells (25). Studies have reported that in liver cancer, circular RNAs (circRNAs) are involved in NK cell-associated immune evasion. Targeting circRNAs to restore NK cell function may provide new directions for the treatment of liver cancer.
Non-coding RNAs (ncRNAs), without the ability to translate into protein, were seen as ‘junk DNA’ by scientists for years. However, an array of ncRNAs nowadays has been discovered based on advances in sequencing technologies. In addition, accumulating lines of evidence indicate that ncRNAs play major roles in the processes of carcinoma initiation, progression, and metastasis by regulating proliferation, apoptosis, and cell cycle (26, 27). Based on the length and shape of RNA molecules, the ncRNAs are divided into three types including short ncRNAs (<200 nucleotides) and long ncRNAs (lncRNAs, >200 nucleotides) and circRNAs. CircRNAs, a novel category of endogenous ncRNAs, come from non-canonical back-splicing events of precursor mRNAs (pre-mRNAs) (28). CircRNAs were originally discovered in an RNA virus in 1976 and observed in eukaryotic cells in 1979 (29–31). CircRNAs have been recognized as ‘splicing noise’ or aberrant byproducts for a long time because they present a covalently joined continuous loop structure without 5′caps and 3′tails (32, 33). However, high-throughput sequencing and bioinformatics algorithms have clearly shown that circRNAs are not the accidental byproducts (34–37). Besides, circRNAs have been proved to be abundant and evolutionarily conserved, and are expressed in different types of tumors (38, 39). CircRNAs can not only regulate the expression of host genes by acting as transcriptional regulators, but also serve as microRNA (miRNA) sponge to fine-tune the regulatory axis of miRNA-mRNA (40–45). It has been confirmed that circRNAs can be used as prognostic biomarkers because they have remarkably stable characteristics (46). Furthermore, studies demonstrated that circRNAs can encode hidden peptides, and serve as a new drug targets resource bank (47–50). We herein illustrated the circRNAs molecular mechanisms connected to liver cancer, offered a novel perspective and a new horizon for cancer treatment and diagnosis. CircRNAs provide new ideas for the study of immune escape in liver cancer.
Biogenesis of CircRNAs
CircRNAs are stable RNAs that are resistant to RNase R, circRNAs are mainly produced by the pre-mRNA through backsplicing. Although backsplicing is considered as an alternative splicing, it has different molecular mechanisms from linear alternative splicing. The hypothesis of backsplicing is that the downstream splicing site is reversed, and the upstream splicing site is connected to form a closed circRNA molecule. According to the region of origin, circRNAs can be divided into three types: (a) exon–intron circRNAs (EIcircRNAs), (b) exon circRNAs (ecircRNAs), (c) Intronic RNAs (ciRNAs) (Figure 1) (51).
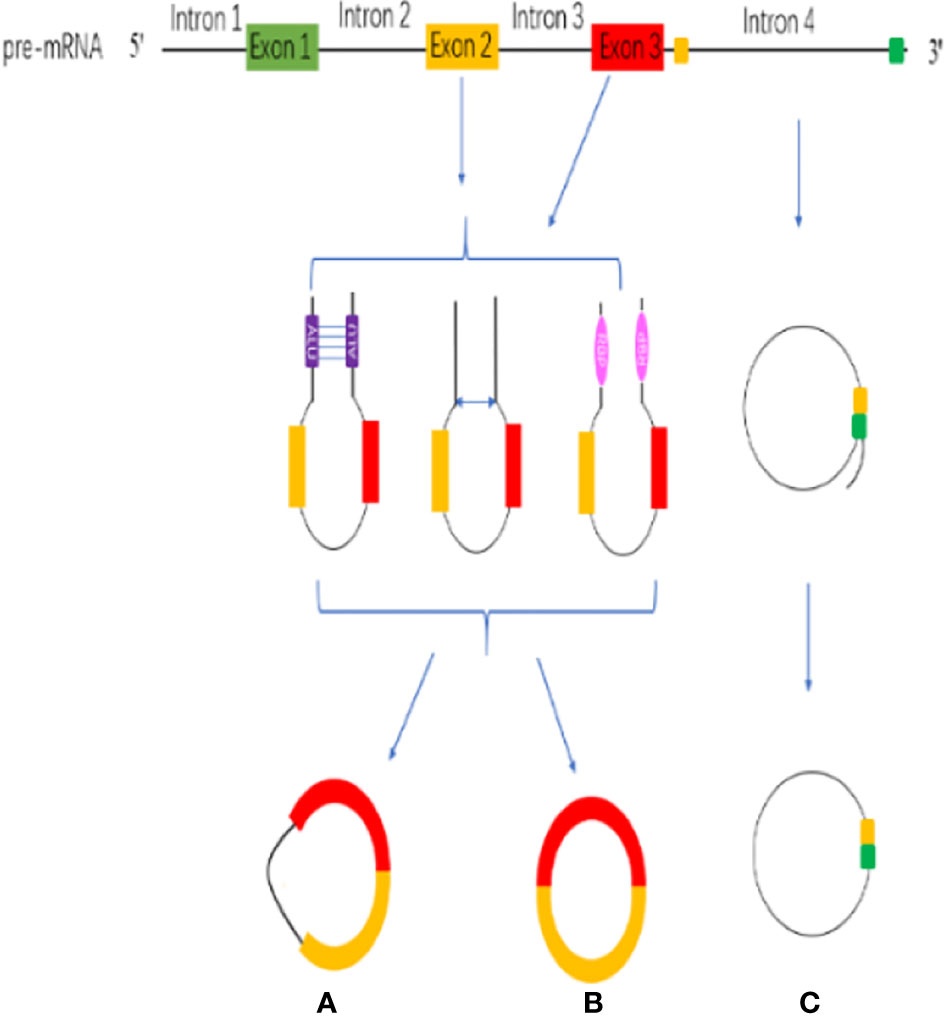
Figure 1 CircRNA biogenesis. (A) exon–intron circRNAs (EIcircRNAs). (B) exon circRNAs (ecircRNAs). (C) Intronic RNAs (ciRNAs).
The circularization model of circRNA is divided into intron circularization and exon circularization. There are three models for the circularization of EIcircRNAs and ecircRNAs: Intron pairing, Lariat and RNA-binding protein (RBP) (Figure 1) (52). Intron pairing-driven circularization, which known as direct backsplicing, is achieved by direct base pairs of intron flanking complementary sequences or reverse repeats (53, 54). The main component of intron pairing-driven circularization is the cis-acting elements, which enable direct base pairing between flank introns, either as short interspersed nuclear elements or as non-repeating complementary sequences (55, 56). Lariat-driven circularization, which is known as exon-skipping, is formed during linear splicing. During the transcription, the pre-mRNA can be partially folded, which formed an RNA lariat containing a 7 nt GU-rich element adjacent to the 5′ splice site and an 11 nt C-rich element closed to the branch point site consensus motif (28, 57). In addition, the third pattern is RBP-driven circularization. Through protein-protein interactions or the dimerization of the RBPs, the splicing sites are pulled closer, and the spliceosomes participate in the backsplicing reaction (40, 58). RBP-driven circularization is guided by two flanking intron pairs that are close to the flanking intron reverse complementary sequences (59). Above all, biogenesis of circRNA is a complicated process, and there are many regulatory details need to dig into.
Functions of CircRNAs
CircRNAs have become a hot topic in the field of ncRNA. The function of circRNAs has been extensively studied. Different types of circRNAs have different characteristics. EIcircRNAs and ciRNAs are usually located in the nucleus, and ecircRNAs are usually located in the cytoplasm. Different locations make them play different functions. The main mechanisms and biological functions of circRNAs are shown in Figure 2 and discussed below.
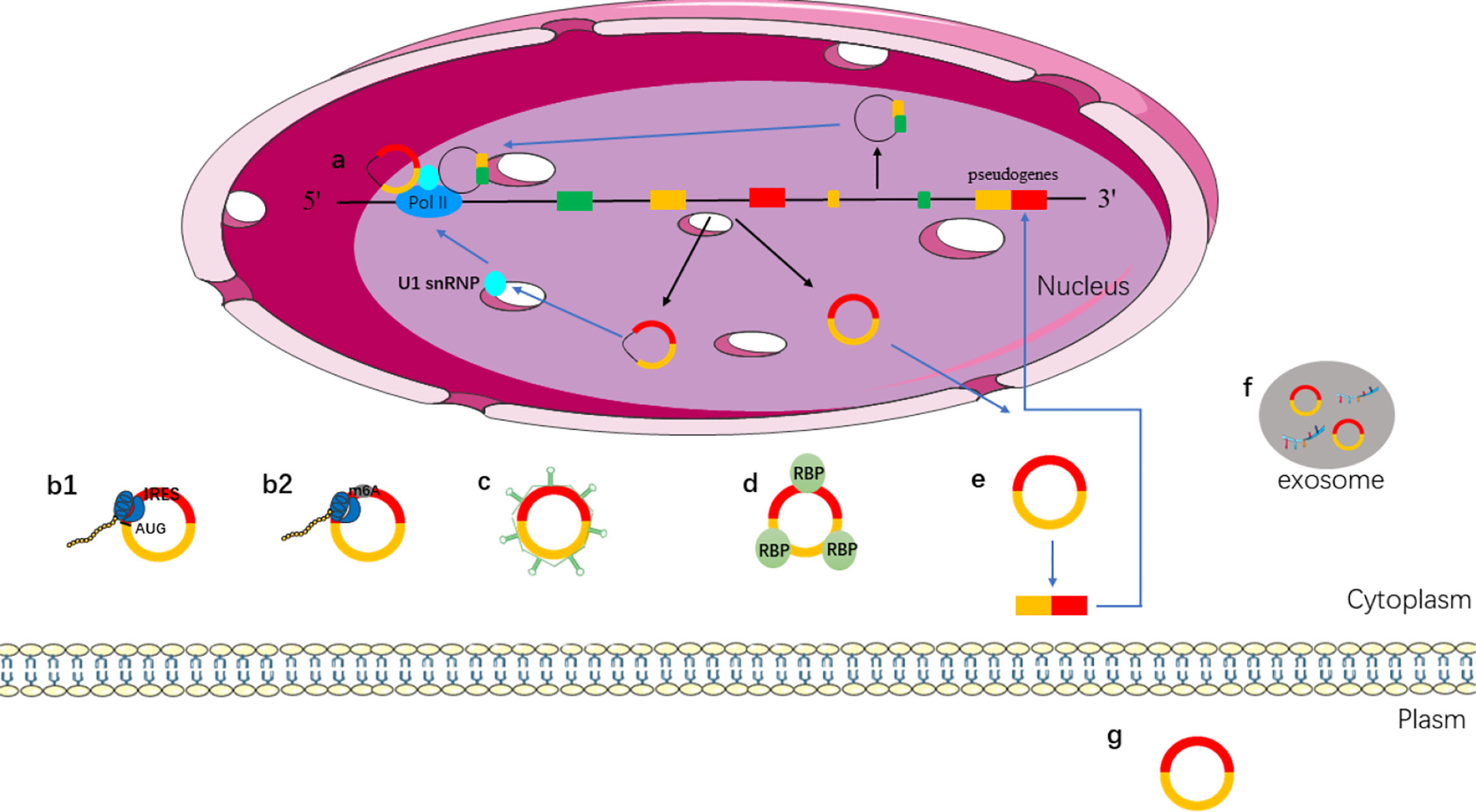
Figure 2 Functions of circRNAs. (a) EIcircRNAs and ciRNAs can regulate gene transcription via binding to U1 snRNP and RNA pol II in the nucleus; (b) ecircRNAs exported into cytoplasm; (b1) IRES-mediated cap-independent translation of ecircRNAs; (b2) m6A-mediated cap-independent translation of ecircRNAs; (c) ecircRNAs act as miRNA sponges; (d) ecircRNAs interact with RBP motifs; (e) ecircRNAs can form pseudogenes; (f) ecircRNAs can be secreted from the cell to outside via exosomes; (g) ecircRNAs act as biomarkers.
Transcription Regulation
A growing number of studies have shown that circRNAs play a role in regulating gene expression. CircRNAs, ciRNA, and EIciRNAs are located in the nucleus that can regulate protein expression by regulating transcription or post-transcription (59, 60). EIciRNAs can regulate transcription because they retain intronic sequences of host genes (61). For instance, circEIF3J and circPAIP2, which are located in the nucleus, can interact with U1 small nuclear ribosomal nucleoprotein (snRNP) to promote the transcription of host genes by binding to RNA polymerase II (RNA pol II) (41). Although EIciRNA and ciRNA do not have the function of miRNA sponge, they can regulate gene transcription and expression in transcription or post-transcription (Figure 2a).
Translation
Traditionally, the 5′ and 3′ untranslated regions (UTRs) have been regarded as the basic elements of translation in eukaryotic cells. Although circRNAs contain exons, the absence of a 5′ cap structure and a poly A tail is considered to be ncRNA that does not encode proteins. However, an increasing number of researches have shown that some circRNAs can be translated into proteins. The researchers constructed artificial circRNAs containing an infinite reading frame to recruit 40s ribosomal subunits and translation into peptides in vitro (62). In 2017, Legnini et al. found that circ-ZNF609, a backsplicing product of ZNF609 exon 2, can be translated into proteins based on high-throughput phenotype screening. And, it can be translated into proteins in a splice-dependent and cap-independent manner (63).
More and more circRNAs are found to be able to translate into proteins, so how do the circRNAs initiate the translation mechanism? In some conditions, such as viral infection, mRNA translation can be initiated via internal ribosome entry site (IRES), which is an alternative mechanism for cap-independent translation (64, 65). IRESs can recruit ribosomes directly to initiate translation. IRES mediated translation is a widely accepted mechanism for initiating translation of circRNAs (66–68). Studies have shown that circRNA can be translated when an IRES is introduced into it (62). Both IRES and N6-methyladenosines (m6A) can drive circRNA translation (Figure 2b). The presence of methylated adenosine residues in m6A form is another cap-independent translation mechanism (69). Studies have shown that m6A can directly bind to eukaryotic initiation factor 3 and initiate the translation of circRNAs into proteins in human cells (69, 70).
MiRNA Sponge
MiRNA is a type of ncRNA with a length of about 19–25 nt, which regulates the transcription of the target gene by binding to the 3′ UTR of the target gene through its seed sequence (71). Studies have shown that circRNAs contain miRNA response elements (MREs), which can competitively bind to miRNA (72). That is, circRNAs can bind to miRNA as miRNA sponges and then regulate the expression of target genes (71), such as, overexpression of circITCH can bind miR-17 and miR-224 to regulate p21 and PTEN genes to inhibit the development of breast cancer (73). CircHIPK3 inhibits the growth of cancer cells by binding to various miRNAs such as the tumor suppressor miR-124 (74, 75) (Figure 2c).
Protein Regulation
Some circRNAs have been shown to bind to RBP, and can isolate RBP and transfer proteins to specific subcellular sites (76). The combination of circPABPN1 and RBP (HuR) prevented the interaction between HuR and PABPN1 (mRNA) and inhibited the translation of PABPN1 (77). High expression of circANRIL can be combined with peccadillo ribosomal biogenesis factor 1 (PES1) to control ribosomal RNA maturation (78). CircAmotl1 can promote the nuclear translocation of PDK1, AKT1, STAT3, c-myc, and other proteins by interacting with RBP and regulate the expression of corresponding target genes (79–81). The above lines of evidence suggest that circRNAs can regulate the function of proteins by binding to PBP instead of a single protein (Figure 2d).
Form Pseudogenes
Pseudogenes are typically derived from reverse-transcriptional of linear mRNA, which integration into the host genome. In the human genome, thousands of pseudogenes are found at about 10% of the gene sites (82, 83). In 2016, the research revealed for the first time that mammalian genomes contain pseudogenes derived from circRNA by establishing a new type of computing analysis process (CIRCpseudo). It revealed that mice circSATB1 source of pseudogenes can be combined with CTCF, which prompts the pseudogenes derived from circRNAs to have the potential to control gene expression. This study showed a fresh perspective on the fact that circRNAs can be inserted into the genome via reverse transcription to alter genomic genetic information and regulate gene expression. Furthermore, many pseudogenes derived from circRNAs have been identified by searching for non-collinear backsplicing in both mouse and human genomes (84). In mice, the reverse transcription of circRFWD2 produced pseudogenes associated with long terminal repeats. The molecular mechanism of circRNA reverse transcription remains to be further studied (Figure 2e).
Other Functions
Exosomes are a type of vesicles with a diameter of 40–150 nm; it is released by the majority of cell types (85). Exosome contains miRNA, lncRNA, circRNA, mRNA, transcription factors, lipids, and proteins (86). Exosomes can be used for liquid biopsy to monitor the development and metastasis of tumors. CircRNAs can be transported to the extracellular via exosomes (Figure 2f) (87). It has been found that exosomes contain abundant circRNAs, and the role of exosomal circRNAs remains to be further explored. CircRNA can be secreted into the blood, saliva, and other body fluids as a biomarker for disease prediction (Figure 2g). CircRNA is stable in body fluids because of its properties, and it is a promising biomarker for the diagnosis of cancer (88).
The Functions of CircRNAs in Liver Cancer
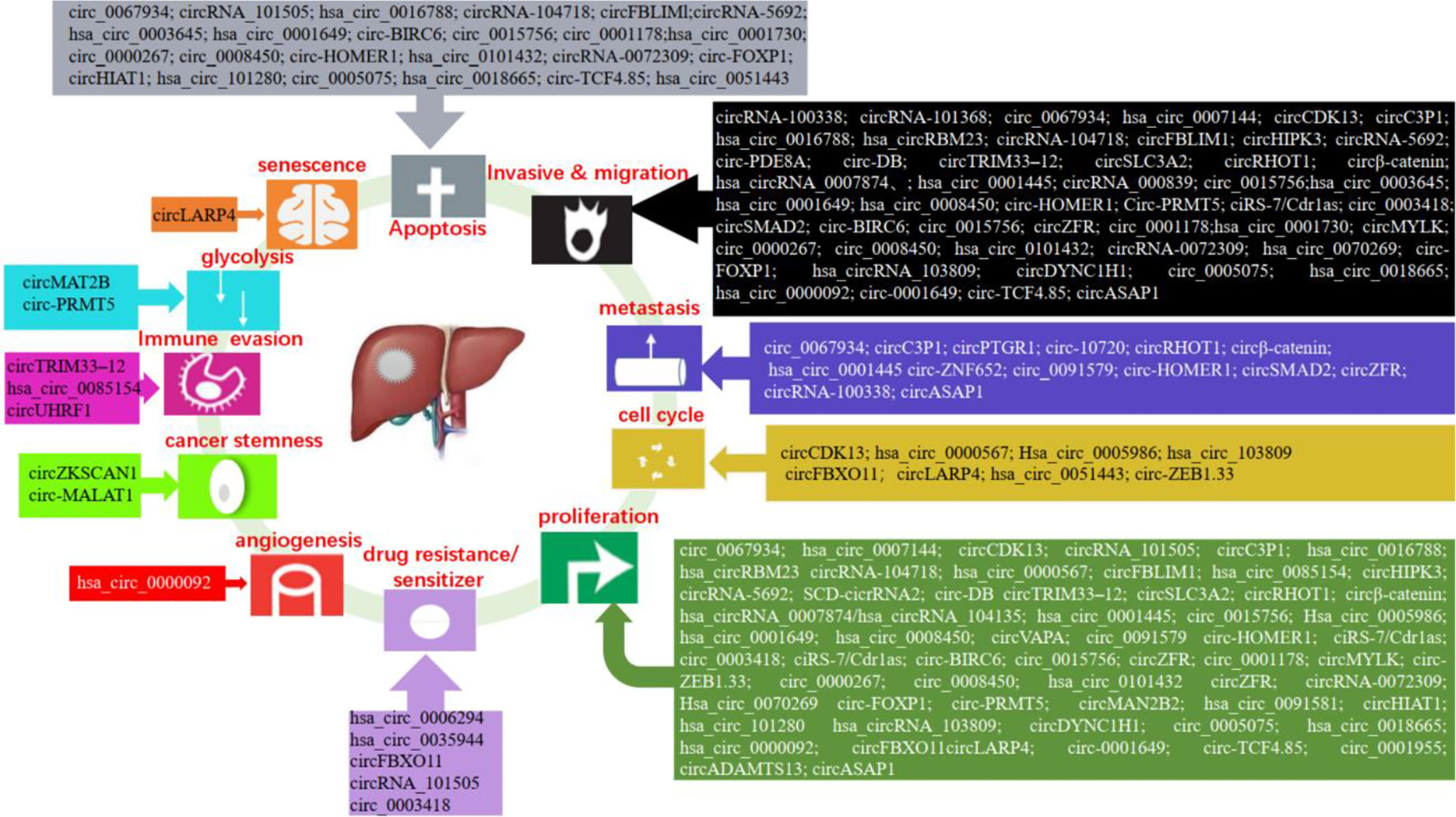
The high mortality, poor prognosis, and lack of effective treatment methods of liver cancer force us to search for effective therapeutic targets and better tumor biomarkers. The studies have shown that a large number of circRNAs are abnormally expressed in liver cancer, which play a regulatory role in the development of liver cancer. The expression and function of circRNA in liver cancer are shown in Figure 3.
Proliferative, Metastasis, and Apoptosis
Studies have shown that circRNAs can regulate the proliferation, migration, invasion, apoptosis, and metastasis of liver cancer cells. In liver cancer, hsa_circ_0000567, hsa_circ_0085154, hsa_circRNA_0007874, hsa_circ_0005986, hsa_circ_0001730, circRNA-0072309, hsa_circ_0070269, circHIAT1, circADAMTS13, ciRS-7/Cdr1as, and hsa_circ_0018665 suppressed cell proliferation (44, 89–98). While, hsa_circ_0101432, SCD-circRNA 2, circVAPA, circ_0015756, circ_0001178, circMYLK, circ-ZEB1.33, circZFR, circ-FOXP1, circMAN2B2, hsa_circ_0091581, hsa_circ_0005075, hsa_circ_101280, hsa_circ_103809, circDYNC1H1, hsa_circ_0000092, circFBXO11, circLARP4, hsa_circ_0001649, circ_0001955, and circ-TCF4.85 promoted tumor growth (99–120). Circ_0067934, hsa_circ_0007144, hsa_circRBM23, circHIPK3, circSLC3A2, circRHOT1, circβ-Catenin, circRNA-104718, circ-PRMT5, ciRS-7/Cdr1as, exosomal circ-DB, circ_0015756, circ_0091579, and circZFR enhanced cell proliferation, migration, and invasion (121–134). However, circCDK13, circC3P1, circ_0003418, circTRIM33-12, hsa_circ_0001445, and hsa_circ_0008450 inhibited cell proliferation, migration, and invasion (135–140). The study showed that circRNA_100338 increased cell invasive (141). CircRNA_000839 enhanced cell invasion and migration (142). Exosomal circPTGR1, circASAP1, and exosomal circRNA-100338 increased cell metastasis (143–145). Circ-10720 and circ-ZNF652 induced epithelial–mesenchymal transition (EMT) (146, 147). CircSMAD2 suppressed the EMT (148). And, circRNA_101368 suppressed the migration (149). The research showed that circRNA_101505 decreased cell proliferation and induced apoptosis (150). Conversely, hsa_circ_0016788, circFBLIM1, circ-BIRC6, circ_0000267, and circ_0008450 promoted cell proliferation, invasion, and suppressed the apoptosis (151–155). The research validated that circRNA_5692 suppressed the progression and invasion, induced apoptosis (156). On the contrary, hsa_circ_0003645 promoted cell migration, invasion and suppressed cell apoptosis (157). Circ-HOMER1 enhanced the proliferation, migration, invasion, and suppressed apoptosis (158). In addition, exosomal hsa_circ_0051443 enhanced cell apoptosis (159).
Liver Cancer Stemness
Both cancer stem cells (CSCs) and circRNAs could affect the carcinogenesis and development of liver cancer, but there are few studies on the relationship between CSCs and circRNAs. Recent studies have found that circ-MALAT1, generated by the backsplicing of lncRNA, promoted the self-renewal of liver cancer CSCs (160). In addition, the researchers found that circZKSCAN1 can regulate the CSCs of HCC via Qki5/circZKSCAN1/FMRP/CCAR1/Wnt signaling axis (161). These findings revealed the role of circRNA in regulating stem cells and enrich the function of circRNA.
Cell Cycle
An increasing number of studies have shown that circRNAs can be involved in the regulation of the cell cycle in liver cancer. For instance, hsa_circ_0000567 induced G1/S arrest in HCC cells by sponging miRNA-421 (89). Hsa_circ_0005986 suppressed the cell proliferation of HCC through promoting the G0/G1 to S phase transition (91). Circ-ZEB1.33 increased the percentage of S phase by regulating CDK6/Rb (105). Inhibition of hsa_circRNA_103809 significantly induced G1/S arrest (113). Down-regulation of circFBXO11 induced G1/G0 arrest (116). Furthermore, exosome-derived circRNA could be involved in the regulation of the cell cycle. Such as, exosome-transmitted hsa_circ_0051443 arrested the cell cycle in HCC (159).
Glycolysis
Hepatoma cells required glycolysis to meet their proliferation needs under hypoxia conditions, and glucose reprogramming is a feature of cancers. Under the hypoxia environment, circMAT2B enhanced glycolysis of HCC via the miR-338-3p/PKM2 axis (162). Furthermore, circ-PRMT5 increased glycolysis of HCC by the miR-188-5p/HK2 axis (129).
Angiogenesis
Cancer cells secrete the angiogenic factors that lead to the formation of abnormal vascular networks. Tumor blood vessel is the key target of tumor treatment. A recent study found that hsa_circ_0000092 promoted angiogenesis in HCC (115).
Drug Resistance/Sensitizer
Resistance to chemotherapy is one of the causes of failure in the treatment of hepatocellular carcinoma. The research showed that circRNA_101505 inhibited cisplatin chemoresistance through miR103/Oxidored-Nitro Domain-Containing Protein 1 pathway (150). In addition, circ_0003418 sensitized HCC to cisplatin by Wnt/β-Catenin pathway (137). CircFBXO11 regulated oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis in HCC (116). The expression of hsa_circ_0006294 and hsa_circ_0035944 was decreased in resistant HCC cells, and they may play a key role in sorafenib-resistant HCC cells (163). Therefore, circRNAs may provide us with a new strategy for the treatment of HCC.
Senescence
Cell senescence is a defense mechanism to prevent and control cell damage and a barrier to prevent tumorigenesis. p53 and p21 are regulatory molecules in the senescence process. Research has found that circLARP4 promoted cellular senescence by regulating miR-761/RUNX3/p53/p21 signaling in HCC (117).
Immune Evasion
Dysfunction of the immune system can lead to abnormal immune surveillance of liver cancer, and liver cancer cells can also act on the immune system to lead the immune escape. NK cells account for 50% of the total number of hepatic lymphocytes and are cytotoxic cells with antitumor functions mediated by the release of cytotoxic granules, FasL and TRAIL (164). NK cells do not rely on antigen presentation; this allows NK cells to target stress and damaged self-cells (165).
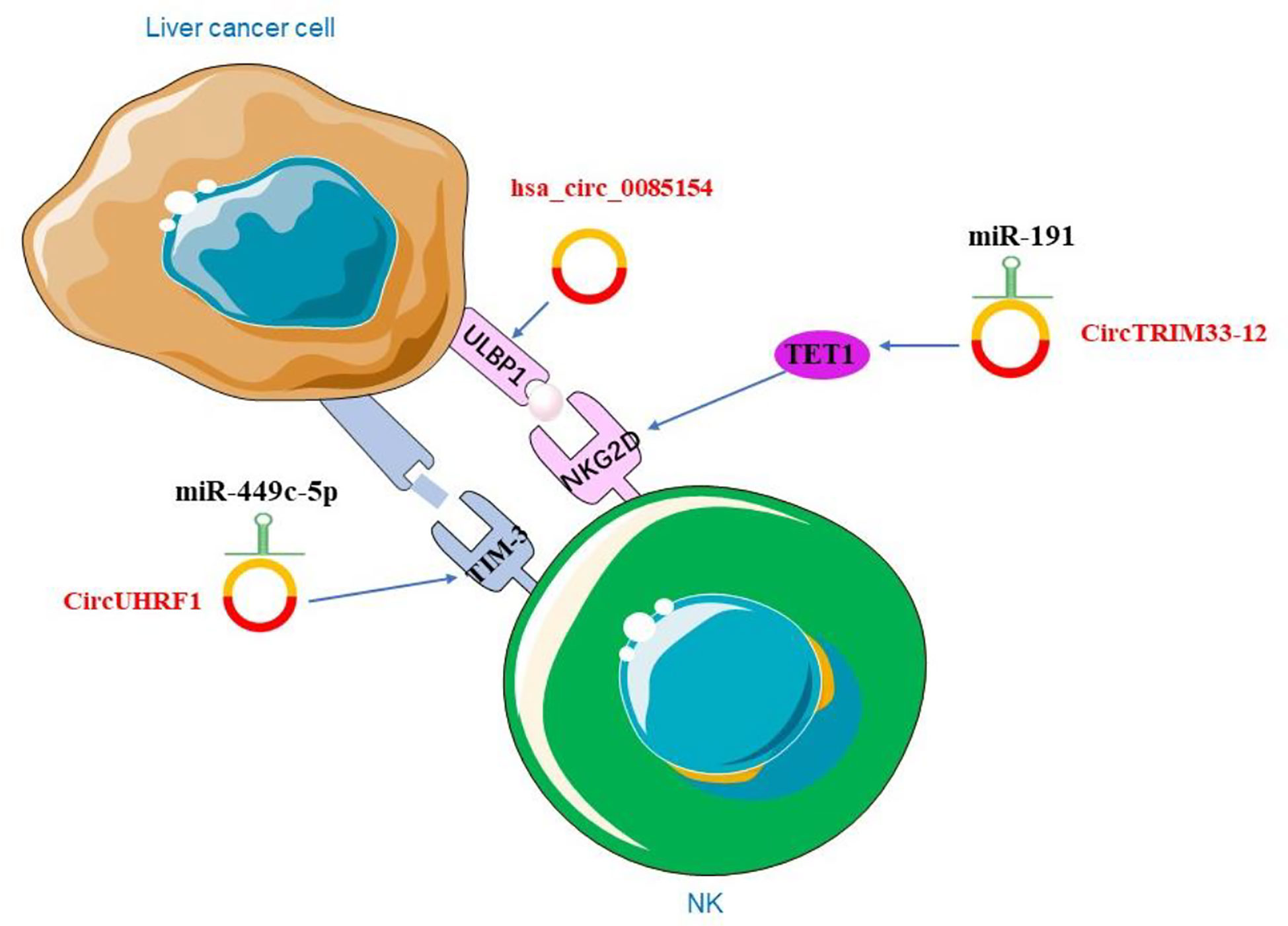
Liver cancer cells avoid being destroyed by immune escape. Studies showed that circRNA could be involved in immune escape. Activation receptor natural killer group 2 member D (NKG2D) and its ligands in NK cells play a crucial role in cell-mediated immune responses to cancer (166). The researchers examined the expression of NKG2D in 200 patients with HCC and showed that the number of NKG2D-positive cells in HCC tissues was significantly reduced compared to adjacent non-tumor tissues. The expression of circTRIM33-12 was positively correlated with the number of NKG2D-positive cells in HCC. The result showed that circTRIM33-12 may enhance immune function by protecting Ten eleven translocation 1 (TET1) via sponging miR-191 (138). TET1, one of the 2-OG-dependent dioxygenases, is involved in regulating the formation of 5-hydroxymethylcytosine (5hmC) and has been proposed to be involved in DNA demethylation process (167). The studies have indirectly linked TET1 as a tumor suppressor in HCC (168). Hsa_circ_0085154 could enhance the innate immune monitoring effect of NK cells by up-regulating UL16 binding protein 1 (ULBP1), which suggests that circRNA may play a role in tumor immunity (169). In HCC, hsa_circ_0085154 promoted ULBP1 expression and assisted NK cells to recognize target tumor cells (169). ULBP1 is an NKG2D ligand that activates receptors expressed by NK cells (170) (Figure 4). NKG2D is a basic activation receptor belonging to the C-type lectin-like family that is constitutively expressed on NK cells (171). The apparently invariant activation receptor NKG2D binds promiscuously to a variety of ligands, such as major histocompatibility complex class I-associated chains A and B (MICA/B) and a unique family of long 16 binding proteins (ULBPs), which are poorly expressed on healthy cells, but they are up-regulated under DNA damage (172). The up-regulation of these ligands may lead to a shift in NK cell homeostasis from inhibition to activation. The research revealed that ULBP1, one of the NKG2D ligands, was not expressed in poorly differentiated human hepatoma tissues and cell lines, but was abundantly expressed in hyperplastic abnormal nodules and well to moderately differentiated HCC cells (172). These findings provided conclusive evidences for the role of NK cells and the NKG2D receptor pathway in immune surveillance of HCC. In addition, HCC-derived exosomal circUHRF1 induced impairment of IFN-γ and TNF-α secretion in NK cells. In HCC, high level of circUHRF1 suggested poor clinical prognosis and dysfunction of NK cells. CircUHRF1 inhibited the secretion of NK cell-derived IFN-γ and TNF-α. High level of plasma exosomal circUHRF1 was associated with a decreased proportion of NK cells and decreased NK cells tumor infiltration. In addition, circUHRF1 up-regulated the expression of T cell immunoglobulin and mucin domain 3 (TIM-3) by degrading miR-449c-5p, thereby inhibiting the function of NK cells (173) (Figure 4). TIM-3 plays an important role in cell immunity, it was expressed in NK cells and affects cellular immune responses (174). In recent years, many studies have focused on the expression of TIM-3 in HCC and its mechanism (175). TIM-3 polymorphisms have been found to play an important role in the susceptibility and characteristics of HCC. The TIM-3 promoter region is associated with certain features of HCC, including lymph node metastasis and tumor stage (176). Modulation of the role of TIM-3 in innate immunity offers new directions for HCC treatment.
Conclusion
Structurally and mechanistically, the liver is an immune organ. It is rich in immune cells. Immune cells including dendritic cell (DC), NK cell, myeloid-derived suppressor cell (MDSC), CD8+ T cell, CD4 +T cell, regulatory T cell (Treg), T helper cell 1 (Th1), T helper cell 2 (Th2), T helper cell 17 (Th17), and tumor-associated macrophages (TAMs) (177–179). Liver cancer is one of the most common malignancies in the world. It is seriously threatening the health of Chinese people. In recent years, tumor immunotherapy is a major advance in cancer treatment, and targeted blocking of PD-1/PD-L1 immune checkpoints antibody-therapy is a milestone in the development of cancer immunotherapy. Currently, the FDA approved PD-1 antibody drug nivolumab (Opdivo) is being used in the treatment of cancer, and pembrolizumab (Keytruda) with the popular anti-cancer drug docetaxel (Sorafenib) combination for the treatment of HCC has been effective (180, 181). However, due to the primary/secondary drug resistance, immune escape, and antibody-drug effectiveness, the survival rate did not increase significantly in liver cancer. Therefore, it is of great significance to find a new way to improve the immunotherapy of liver cancer. NK cells are a new target for immunotherapy. There is a growing body of research using NK cell-related therapies to fight cancer (182, 183). NK cell-mediated immune surveillance is an important mechanism for tumor suppression. NK cells kill tumor cells through the release of perforin and granzyme and the secretion of pro-inflammatory cytokines and chemokines (184).
More and more studies have shown that circRNAs are involved in the carcinogenesis and progression of liver cancer. In this review, we summarized the functions of circRNAs in liver cancer. We found that circRNAs affect the cytotoxicity of NK cells. CircUHRF1 up-regulated TIM-3, the immune checkpoint, to inhibit the function of NK cells. Binding of TIM-3 to its ligand induces immune tolerance by depletes NK cells (185). It has been found that in tumor cells, immune checkpoints can lead to NK cell dysfunction, blocking these immune checkpoints (e.g. TIM-3, NKG2A, CTLA-4, PD-1, KIR2DL-1/2/3, CD96, TIGIT) can restore the function of NK cells (186). We can inhibit circUHRF1 to enhance NK cell function by down-regulating the expression of TIM-3. CircUHRF1 may provide a potential therapeutic strategy for immune checkpoints in liver cancer. More circRNAs regulating immune checkpoints are yet to be discovered, and targeting circRNAs provided a new direction for immune checkpoint therapy. Furthermore, circRNAs can affect the function of NK cells by regulating the receptor and ligand of NK cells. However, the relationship between circRNAs and other immune cells still needs further study. Understanding the mechanism of circRNAs in HCC patients is important in the design of effective immunotherapeutic protocols. Although circRNAs have shown an important role in liver cancer, many fields remain to be studied. For instance, the mechanism that ecircRNAs transported from the nucleus to the cytoplasm? the degradation of circRNAs?
Author Contributions
YT and H-MJ wrote the draft of the manuscript. ZY and Y-SH collected the data. R-SZ contributed to the discussion. D-YZ and H-FP organized the structure of the manuscript. YT and D-YZ contributed to the conception of the work. X-SL and B-YQ contributed to the revision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Project of Administration of Traditional Chinese Medicine of Guangdong Province of China (Project No. 20201109) and National Natural Science Foundation of China (Project No. 81973816).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Singal AG, El-Serag HB. Hepatocellular Carcinoma From Epidemiology to Prevention: Translating Knowledge into Practice. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterological Association. (2015) 13(12):2140–51. doi: 10.1016/j.cgh.2015.08.014
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Mol Cell (2018) 68: (1):7–30. doi: 10.3322/caac.21442
3. Orcutt ST, Anaya DA. Liver Resection and Surgical Strategies for Management of Primary Liver Cancer. Cancer Control (2018) 25(1):1073274817744621. doi: 10.1177/1073274817744621
4. Wong MC, Jiang JY, Goggins WB, Liang M, Fang Y, Fung FD, et al. International incidence and mortality trends of liver cancer: a global profile. Sci Rep (2017) 7:45846. doi: 10.1038/srep45846
5. Petrick JL, Kelly SP, Altekruse SF, McGlynn KA, Rosenberg PS. Future of Hepatocellular Carcinoma Incidence in the United States Forecast Through 2030. J Clin Oncol (2016) 34(15):1787–94. doi: 10.1200/jco.2015.64.7412
6. McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clinics Liver Dis (2015) 19(2):223–38. doi: 10.1016/j.cld.2015.01.001
7. Villanueva A, Hernandez-Gea V, Llovet JM. Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol (2013) 10(1):34–42. doi: 10.1038/nrgastro.2012.199
8. Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer (2019) 18(1):10. doi: 10.1186/s12943-018-0928-4
9. Vitale M, Cantoni C, Pietra G, Mingari MC, Moretta L. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur J Immunol (2014) 44(6):1582–92. doi: 10.1002/eji.201344272
10. Stojanovic A, Correia MP, Cerwenka A. Shaping of NK cell responses by the tumor microenvironment. Cancer Microenviron (2013) 6(2):135–46. doi: 10.1007/s12307-012-0125-8
11. Terrén I, Orrantia A, Vitallé J, Zenarruzabeitia O, Borrego F. NK Cell Metabolism and Tumor Microenvironment. Front Immunol (2019) 10:2278. doi: 10.3389/fimmu.2019.02278
12. Park A, Lee Y, Kim MS, Kang YJ, Park YJ, Jung H, et al. Prostaglandin E2 Secreted by Thyroid Cancer Cells Contributes to Immune Escape Through the Suppression of Natural Killer (NK) Cell Cytotoxicity and NK Cell Differentiation. Front Immunol (2018) 9:1859. doi: 10.3389/fimmu.2018.01859
13. Zingoni A, Vulpis E, Nardone I, Soriani A, Fionda C, Cippitelli M, et al. Targeting NKG2D and NKp30 Ligands Shedding to Improve NK Cell-Based Immunotherapy. Crit Rev Immunol (2016) 36(6):445–60. doi: 10.1615/CritRevImmunol.2017020166
14. Krzywinska E, Kantari-Mimoun C, Kerdiles Y, Sobecki M, Isagawa T, Gotthardt D, et al. Loss of HIF-1α in natural killer cells inhibits tumour growth by stimulating non-productive angiogenesis. Immunol Cell Biol (2017) 8(1):1597. doi: 10.1038/s41467-017-01599-w
15. Chiossone L, Dumas PY. Natural killer cells and other innate lymphoid cells in cancer. Cell (2018) 18(11):671–88. doi: 10.1038/s41577-018-0061-z
16. Chiossone L, Vienne M, Kerdiles YM, Vivier E. Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more. Semin Immunol (2017) 31:55–63. doi: 10.1016/j.smim.2017.08.003
17. Guillerey C, Huntington ND. Targeting natural killer cells in cancer immunotherapy. Immunol Rev (2016) 17(9):1025–36. doi: 10.1038/ni.3518
18. Rezvani K, Rouce R, Liu E, Shpall E. Engineering Natural Killer Cells for Cancer Immunotherapy. Mol Ther J Am Soc Gene Ther (2017) 25(8):1769–81. doi: 10.1016/j.ymthe.2017.06.012
19. Michel T, Poli A, Cuapio A, Briquemont B, Iserentant G, Ollert M, et al. Human CD56bright NK Cells: An Update. J Immunol (Baltimore Md 1950) (2016) 196(7):2923–31. doi: 10.4049/jimmunol.1502570
20. Batoni G, Esin S, Favilli F, Pardini M, Bottai D, Maisetta G, et al. Human CD56bright and CD56dim natural killer cell subsets respond differentially to direct stimulation with Mycobacterium bovis bacillus Calmette-Guérin. Scand J Immunol (2005) 62(6):498–506. doi: 10.1111/j.1365-3083.2005.01692.x
21. Pesce S, Moretta L, Moretta A, Marcenaro E. Human NK Cell Subsets Redistribution in Pathological Conditions: A Role for CCR7 Receptor. Front Immunol (2016) 7:414. doi: 10.3389/fimmu.2016.00414
22. Cichocki F, Grzywacz B, Miller JS. Human NK Cell Development: One Road or Many? Front Immunol (2019) 10:2078. doi: 10.3389/fimmu.2019.02078
23. Gao B, Radaeva S, Park O. Liver natural killer and natural killer T cells: immunobiology and emerging roles in liver diseases. J Leukoc Biol (2009) 86(3):513–28. doi: 10.1189/jlb.0309135
24. Muhanna N, Doron S, Wald O, Horani A, Eid A, Pappo O, et al. Activation of hepatic stellate cells after phagocytosis of lymphocytes: A novel pathway of fibrogenesis. Hepatology (Baltimore Md) (2008) 48(3):963–77. doi: 10.1002/hep.22413
25. Ghiringhelli F, Ménard C, Martin F, Zitvogel L. The role of regulatory T cells in the control of natural killer cells: relevance during tumor progression. Immunol Rev (2006) 214:229–38. doi: 10.1111/j.1600-065X.2006.00445.x
26. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell (2014) 157(1):77–94. doi: 10.1016/j.cell.2014.03.008
27. Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer (2016) 139(2):269–80. doi: 10.1002/ijc.30039
28. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife (2015) 4:e07540. doi: 10.7554/eLife.07540
29. Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell (1976) 8(4):547–55. doi: 10.1016/0092-8674(76)90223-3
30. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
31. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature (1979) 280(5720):339–40. doi: 10.1038/280339a0
32. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell (1993) 73(5):1019–30. doi: 10.1016/0092-8674(93)90279-y
33. Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA (New York NY) (1996) 2(6):603–10.
34. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PloS One (2014) 9(6):e90859. doi: 10.1371/journal.pone.0090859
35. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PloS Genet (2013) 9(9):e1003777. doi: 10.1371/journal.pgen.1003777
36. Maass PG, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berlin Germany) (2017) 95(11):1179–89. doi: 10.1007/s00109-017-1582-9
37. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings Bioinform (2017) 18(6):984–92. doi: 10.1093/bib/bbw081
38. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development (Cambridge England) (2016) 143(11):1838–47. doi: 10.1242/dev.128074
39. Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics (2017) 18(Suppl 6):680. doi: 10.1186/s12864-017-4029-3
40. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
41. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
42. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
43. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
44. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (Baltimore Md) (2017) 66(4):1151–64. doi: 10.1002/hep.29270
45. Kleaveland B, Shi CY, Stefano J, Bartel DP. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell (2018) 174(2):350–62.e17. doi: 10.1016/j.cell.2018.05.022
46. Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci (2019) 16(2):292–301. doi: 10.7150/ijms.28047
47. Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta (2016) 1859(10):1245–51. doi: 10.1016/j.bbagrm.2016.07.009
48. Dhamija S, Menon MB. Non-coding transcript variants of protein-coding genes - what are they good for? Nat Immunol (2018) 15(8):1025–31. doi: 10.1080/15476286.2018.1511675
49. Zhu S, Wang J, He Y, Meng N, Yan GR. Peptides/Proteins Encoded by Non-coding RNA: A Novel Resource Bank for Drug Targets and Biomarkers. Front Pharmacol (2018) 9:1295. doi: 10.3389/fphar.2018.01295
50. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer (2019) 18(1):6. doi: 10.1186/s12943-018-0934-6
51. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, et al. Exon circularization requires canonical splice signals. Cell Rep (2015) 10(1):103–11. doi: 10.1016/j.celrep.2014.12.002
52. Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nature (2019) 16(8):503–14. doi: 10.1038/s41569-019-0185-2
53. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell (2014) 159(1):134–47. doi: 10.1016/j.cell.2014.09.001
54. Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem (2015) 84:291–323. doi: 10.1146/annurev-biochem-060614-034316
55. Zeng X, Lin W. A comprehensive overview and evaluation of circular RNA detection tools. Hepatology (2017) 13: (6):e1005420. doi: 10.1371/journal.pcbi.1005420
56. Kamens J. The Addgene repository: an international nonprofit plasmid and data resource. Nucleic Acids Res (2015) 43(Database issue):D1152–7. doi: 10.1093/nar/gku893
57. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
58. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell (2015) 160(6):1125–34. doi: 10.1016/j.cell.2015.02.014
59. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York NY) (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
60. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One (2012) 7(2):e30733. doi: 10.1371/journal.pone.0030733
61. Wang M, Yu F, Li P. Circular RNAs: Characteristics, Function and Clinical Significance in Hepatocellular Carcinoma. Cancers (2018) 10(8):258–80. doi: 10.3390/cancers10080258
62. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science (New York NY) (1995) 268(5209):415–7. doi: 10.1126/science.7536344
63. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell (2017) 66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017
64. Godet AC, David F, Hantelys F, Tatin F, Lacazette E, Garmy-Susini B. IRES Trans-Acting Factors, Key Actors of the Stress Response. Int J Mol Sci (2019) 20(4):924–53. doi: 10.3390/ijms20040924
65. Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim Biophys Acta (2009) 1789(9-10):518–28. doi: 10.1016/j.bbagrm.2009.07.004
66. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
67. Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res (2015) 43(4):2454–65. doi: 10.1093/nar/gkv045
68. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA (New York NY) (2015) 21(2):172–9. doi: 10.1261/rna.048272.114
69. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
70. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
71. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5
72. Bezzi M, Guarnerio J, Pandolfi PP. A circular twist on microRNA regulation. Cell Res (2017) 27(12):1401–2. doi: 10.1038/cr.2017.136
73. Yang C, Yuan W, Yang X, Li P, Wang J, Han J, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer (2018) 17(1):19. doi: 10.1186/s12943-018-0771-7
74. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun (2016) 7:11215. doi: 10.1038/ncomms11215
75. Wang Y, Chen L, Wu Z, Wang M, Jin F, Wang N, et al. miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer (2016) 16(1):826. doi: 10.1186/s12885-016-2862-4
76. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and Characterizing circRNA-Protein Interaction. Theranostics (2017) 7(17):4183–91. doi: 10.7150/thno.21299
77. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol (2017) 14(3):361–9. doi: 10.1080/15476286.2017.1279788
78. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Cancer Cell Int (2016) 7:12429. doi: 10.1038/ncomms12429
79. Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ (2017) 24(9):1609–20. doi: 10.1038/cdd.2017.86
80. Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu D, et al. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther (2017) 25(9):2062–74. doi: 10.1016/j.ymthe.2017.05.022
81. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics (2017) 7(16):3842–55. doi: 10.7150/thno.19764
82. Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res (2003) 13(12):2541–58. doi: 10.1101/gr.1429003
83. Zhang Z, Carriero N, Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet TIG (2004) 20(2):62–7. doi: 10.1016/j.tig.2003.12.005
84. Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL, Yang L. CircRNA-derived pseudogenes. Cell Res (2016) 26(6):747–50. doi: 10.1038/cr.2016.42
85. Barile L, Moccetti T, Marban E, Vassalli G. Roles of exosomes in cardioprotection. Eur Heart J (2017) 38(18):1372–9. doi: 10.1093/eurheartj/ehw304
86. Zhang W, Jiang X, Bao J, Wang Y, Liu H, Tang L. Exosomes in Pathogen Infections: A Bridge to Deliver Molecules and Link Functions. Front Immunol (2018) 9:90. doi: 10.3389/fimmu.2018.00090
87. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
88. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem (2015) 61(1):221–30. doi: 10.1373/clinchem.2014.230433
89. Xu L, Feng X, Hao X, Wang P, Zhang Y, Zheng X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res (2019) 38(1):98. doi: 10.1186/s13046-019-1041-2
90. Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou S, et al. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis (2017) 8(11):e3171. doi: 10.1038/cddis.2017.556
91. Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget (2017) 8(27):43878–88. doi: 10.18632/oncotarget.16709
92. Tan Y, Du B, Zhan Y, Wang K, Wang X, Chen B, et al. Antitumor effects of circ-EPHB4 in hepatocellular carcinoma via inhibition of HIF-1alpha. Cancer Res (2019) 58(6):875–86. doi: 10.1002/mc.22976
93. Yu Q, Dai J, Shu M. Circular RNA-0072309 has antitumor influences in Hep3B cell line by targeting microRNA-665. Biofactors (2020) 46:1618–29. doi: 10.1002/biof.1618
94. Su X, Su J, He H, Zhan Y, Liu H. Hsa_circ_0070269 inhibits hepatocellular carcinoma progression through modulating miR-182/NPTX1 axis. Biomed Pharmacother (2019) 120:109497. doi: 10.1016/j.biopha.2019.109497
95. Wang Z, Zhao Y, Wang Y, Jin C. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed Pharmacother (2019) 116:108932. doi: 10.1016/j.biopha.2019.108932
96. Qiu L, Huang Y, Li Z, Dong X, Chen G, Xu H, et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol (2019) 13(2):441–55. doi: 10.1002/1878-0261.12424
97. Yang X, Xiong Q, Wu Y, Li S, Ge F. Quantitative Proteomics Reveals the Regulatory Networks of Circular RNA CDR1as in Hepatocellular Carcinoma Cells. Eur Rev Med Pharmacol Sci (2017) 16(10):3891–902. doi: 10.1021/acs.jproteome.7b00519
98. Song C, Li D, Liu H, Sun H, Liu Z, Zhang L. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. JMCC (2019) 234(3):2460–70. doi: 10.1002/jcp.26764
99. Zou H, Xu X, Luo L, Zhang Y, Luo L, Yao Y, et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Mol Cell (2019) 18(19):2398–413. doi: 10.1080/15384101.2019.1618120
100. Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine (2019) 45:155–67. doi: 10.1016/j.ebiom.2019.06.030
101. Liu C, Zhong X, Li J, Xu F. Circular RNA circVAPA Promotes Cell Proliferation in Hepatocellular Carcinoma. Hum Gene Ther Clin Dev (2019) 30(4):152–9. doi: 10.1089/humc.2019.079
102. Liu L, Yang X, Li NF, Lin L, Luo H. Circ_0015756 promotes proliferation, invasion and migration by microRNA-7-dependent inhibition of FAK in hepatocellular carcinoma. Cell Cycle (Georgetown Tex) (2019) 18(21):2939–53. doi: 10.1080/15384101.2019.1664223
103. Gao S, Hu W, Huang X, Huang X, Chen W, Hao L, et al. Circ_0001178 regulates miR-382/VEGFA axis to facilitate hepatocellular carcinoma progression. Cell Signal (2020) 72:109621. doi: 10.1016/j.cellsig.2020.109621
104. Li Z, Hu Y, Zeng Q, Wang H, Yan J, Li H, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int (2019) 19:211. doi: 10.1186/s12935-019-0926-7
105. Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int (2018) 18:116. doi: 10.1186/s12935-018-0602-3
106. Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/beta-catenin pathway. Arch Biochem Biophys (2019) 661:196–202. doi: 10.1016/j.abb.2018.11.020
107. Wang W, Li Y, Li X, Liu B, Han S, Li X, et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed Pharmacother (2020) 121:109517. doi: 10.1016/j.biopha.2019.109517
108. Fu X, Zhang J, He X, Yan X, Wei J, Huang M, et al. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J Cancer (2020) 11(11):3318–26. doi: 10.7150/jca.36500
109. Wei X, Zheng W, Tian P, He Y, Liu H, Peng M, et al. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle (Georgetown Tex) (2020) 19(7):817–24. doi: 10.1080/15384101.2020.1731945
110. Li MF, Li YH, He YH, Wang Q, Zhang Y, Li XF, et al. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed Pharmacother (2018) 99:848–58. doi: 10.1016/j.biopha.2018.01.150
111. Yang X, Song H, Zi Z, Kou J, Chen S, Dai Y, et al. Circ_0005075 promotes hepatocellular carcinoma progression by suppression of microRNA-335. Cell Mol Immunol (2019) 234: (12):21937–46. doi: 10.1002/jcp.28757
112. Cao S, Wang G, Wang J, Li C, Zhang L. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Trend Immunol (2019) 97(2):218–28. doi: 10.1111/imcb.12213
113. Zhan W, Liao X, Chen Z, Li L, Tian T, Yu L, et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. PLoS Genet (2020) 235(2):1733–45. doi: 10.1002/jcp.29092
114. Wang ZY, Zhu Z, Wang HF, Qin B, Liu J, Yao XH, et al. Downregulation of circDYNC1H1 exhibits inhibitor effect on cell proliferation and migration in hepatocellular carcinoma through miR-140-5p. PloS One (2019) 234(10):17775–85. doi: 10.1002/jcp.28403
115. Pu J, Wang J, Li W, Lu Y, Wu X, Long X, et al. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J Cell Mol Med (2020) 22:1–13. doi: 10.1111/jcmm.15010
116. Li J, Qin X, Wu R, Wan L, Zhang L, Liu R. Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis. J Cell Mol Med (2020) 24:5152–61. doi: 10.1111/jcmm.15162
117. Chen Z, Zuo X, Pu L, Zhang Y, Han G, Zhang L, et al. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cell Death Dis (2019) 110(2):568–81. doi: 10.1111/cas.13901
118. Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging (2019) 11(10):3362–75. doi: 10.18632/aging.101988
119. Gao J, Dai C, Yu X, Yin XB, Zhou F. Circ-TCF4.85 silencing inhibits cancer progression through microRNA-486-5p-targeted inhibition of ABCF2 in hepatocellular carcinoma. Clin Gastroenterol Hepatol (2020) 14(2):447–61. doi: 10.1002/1878-0261.12603
120. Yao Z, Xu R, Yuan L, Xu M, Zhuang H, Li Y, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis (2019) 10(12):945. doi: 10.1038/s41419-019-2176-y
121. Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun (2018) 497(2):626–32. doi: 10.1016/j.bbrc.2018.02.119
122. Luo Z, Mao X, Cui W. Circular RNA expression and circPTPRM promotes proliferation and migration in hepatocellular carcinoma. Med Oncol (2019) 36(10):86. doi: 10.1007/s12032-019-1311-z
123. Wang B, Chen H, Zhang C, Yang T, Zhao Q, Yan Y, et al. Effects of hsa_circRBM23 on Hepatocellular Carcinoma Cell Viability and Migration as Produced by Regulating miR-138 Expression. Cancer Biother Radiopharm (2018) 33(5):194–202. doi: 10.1089/cbr.2017.2424
124. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis (2018) 9(2):175. doi: 10.1038/s41419-017-0204-3
125. Wang H, Chen W, Jin M, Hou L, Chen X, Zhang R, et al. CircSLC3A2 functions as an oncogenic factor in hepatocellular carcinoma by sponging miR-490-3p and regulating PPM1F expression. Arch Biochem Biophys (2018) 17(1):165. doi: 10.1186/s12943-018-0909-7
126. Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Carcinog (2019) 18(1):119. doi: 10.1186/s12943-019-1046-7
127. Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol (2019) 20(1):84. doi: 10.1186/s13059-019-1685-4
128. Yu J, Yang M, Zhou B, Luo J, Zhang Z, Zhang W, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Nature Rev (2019) 133(13):1487–503. doi: 10.1042/cs20190394
129. Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol (2020) 19:269–79. doi: 10.1016/j.aohep.2020.01.002
130. Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PloS One (2016) 11(7):e0158347. doi: 10.1371/journal.pone.0158347
131. Yang X, Liu L, Zou H, Zheng YW, Wang KP. circZFR promotes cell proliferation and migration by regulating miR-511/AKT1 axis in hepatocellular carcinoma. Dig Liver Dis (2019) 51(10):1446–55. doi: 10.1016/j.dld.2019.04.012
132. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene (2019) 38(15):2844–59. doi: 10.1038/s41388-018-0619-z
133. Liu BH, Zhang BB, Liu XQ, Zheng S, Dong KR, Dong R. Expression Profiling Identifies Circular RNA Signature in Hepatoblastoma. Cell Physiol Biochem (2018) 45(2):706–19. doi: 10.1159/000487163
134. Niu WY, Chen L, Zhang P, Zang H, Zhu B, Shao WB. Circ_0091579 promotes proliferative ability and metastasis of liver cancer cells by regulating microRNA-490-3p. Eur Rev Med Pharmacol Sci (2019) 23(23):10264–73. doi: 10.26355/eurrev_201912_19664
135. Lin Q, Ling YB, Chen JW, Zhou CR, Chen J, Li X, et al. Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int J Oncol (2018) 53(1):246–56. doi: 10.3892/ijo.2018.4371
136. Zhong L, Wang Y, Cheng Y, Wang W, Lu B, Zhu L, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun (2018) 499(4):1044–9. doi: 10.1016/j.bbrc.2018.03.221
137. Chen H, Liu S, Li M, Huang P, Li X. circ_0003418 Inhibits Tumorigenesis And Cisplatin Chemoresistance Through Wnt/beta-Catenin Pathway In Hepatocellular Carcinoma. Onco Targets Ther (2019) 12:9539–49. doi: 10.2147/ott.s229507
138. Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. BMC Cancer (2019) 18:(1):105. doi: 10.1186/s12943-019-1031-1
139. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol (2018) 68(6):1214–27. doi: 10.1016/j.jhep.2018.01.012
140. Lin T, Dai Y, Guo X, Chen W, Zhao J, Cao L, et al. Silencing Of hsa_circ_0008450 Represses Hepatocellular Carcinoma Progression Through Regulation Of microRNA-214-3p/EZH2 Axis. Cancer Manage Res (2019) 11:9133–43. doi: 10.2147/cmar.s222716
141. Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep (2017) 7(1):5428. doi: 10.1038/s41598-017-05432-8
142. Wang BG, Li JS, Liu YF, Xu Q. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumour Biol (2017) 39(7):1010428317719577. doi: 10.1177/1010428317719577
143. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062
144. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res (2020) 39(1):20. doi: 10.1186/s13046-020-1529-9
145. Hu ZQ, Zhou SL. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology (2019) 72:906–922. doi: 10.1002/hep.31068
146. Meng J, Chen S, Han JX, Qian B, Wang XR, Zhong WL, et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res (2018) 78(15):4150–62. doi: 10.1158/0008-5472.can-17-3009
147. Guo J, Duan H, Li Y, Yang L, Yuan L. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem Biophys Res Commun (2019) 513(4):812–9. doi: 10.1016/j.bbrc.2019.03.214
148. Zhang X, Luo P, Jing W, Zhou H, Liang C, Tu J. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther (2018) 11:2853–63. doi: 10.2147/ott.s158008
149. Li S, Gu H, Huang Y, Peng Q, Zhou R, Yi P, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle (Georgetown Tex) (2018) 17(19-20):2349–59. doi: 10.1080/15384101.2018.1526599
150. Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R, et al. CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein 1 expression. Cell Death Discov (2019) 5:121. doi: 10.1038/s41420-019-0202-6
151. Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao Y, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res (2018) 37(1):172. doi: 10.1186/s13046-018-0838-8
152. Guan Z, Tan J, Gao W, Li X, Yang Y, Li X, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. Cell Death Differ (2018) 234: (1):500–8. doi: 10.1002/jcp.26612
153. Yang G, Wang X, Liu B, Lu Z, Xu Z, Xiu P, et al. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle (Georgetown Tex) (2019) 18(9):976–89. doi: 10.1080/15384101.2019.1601477
154. Pan H, Tang L, Jiang H, Li X, Wang R, Gao J, et al. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J Cell Biochem (2019) 120:11350–7. doi: 10.1002/jcb.28411
155. Zhang J, Chang Y, Xu L, Qin L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J Proteome Res (2019) 120(6):9487–94. doi: 10.1002/jcb.28224
156. Liu Z, Yu Y, Huang Z, Kong Y, Hu X, Xiao W, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis (2019) 10(12):900. doi: 10.1038/s41419-019-2089-9
157. Yu Q, Dai J, Shu M. Hsa_circ_0003645 shows an oncogenic role by sponging microRNA-1299 in hepatocellular carcinoma cells. J Clin Lab Anal (2020) 34:e23249. doi: 10.1002/jcla.23249
158. Zhao M, Dong G, Meng Q, Lin S, Li X. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J Cell Biochem (2020) 121:4440–9. doi: 10.1002/jcb.29672
159. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
160. Chen L, Kong R, Wu C, Wang S, Liu Z, Liu S, et al. Circ-MALAT1 Functions as Both an mRNA Translation Brake and a microRNA Sponge to Promote Self-Renewal of Hepatocellular Cancer Stem Cells. Clin Sci (2020) 7(4):1900949. doi: 10.1002/advs.201900949
161. Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics (2019) 9(12):3526–40. doi: 10.7150/thno.32796
162. Li Q, Pan X, Zhu D, Deng Z, Jiang R. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. BioFactors (2019) 70(4):1298–316. doi: 10.1002/hep.30671
163. Wu MY, Tang YP, Liu JJ, Liang R, Luo XL. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J Cancer (2020) 11(10):2993–3001. doi: 10.7150/jca.39854
164. Robinson MW, Harmon C, O’Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol (2016) 13(3):267–76. doi: 10.1038/cmi.2016.3
165. Jenne CN, Kubes P. Immune surveillance by the liver. Nat Immunol (2013) 14(10):996–1006. doi: 10.1038/ni.2691
166. Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science (New York NY) (1999) 285(5428):727–9. doi: 10.1126/science.285.5428.727
167. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature (2013) 502(7472):472–9. doi: 10.1038/nature12750
168. Chuang KH, Whitney-Miller CL, Chu CY, Zhou Z, Dokus MK, Schmit S, et al. MicroRNA-494 is a master epigenetic regulator of multiple invasion-suppressor microRNAs by targeting ten eleven translocation 1 in invasive human hepatocellular carcinoma tumors. Hepatology (Baltimore Md) (2015) 62(2):466–80. doi: 10.1002/hep.27816
169. Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett (2019) 17(1):388–97. doi: 10.3892/ol.2018.9606
170. Huergo-Zapico L, Acebes-Huerta A, López-Soto A, Villa-Álvarez M, Gonzalez-Rodriguez AP, Gonzalez S. Molecular Bases for the Regulation of NKG2D Ligands in Cancer. Front Immunol (2014) 5:106. doi: 10.3389/fimmu.2014.00106
171. Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene (2008) 27(45):5944–58. doi: 10.1038/onc.2008.272
172. Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev (2010) 235(1):267–85. doi: 10.1111/j.0105-2896.2010.00893.x
173. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Cell (2020) 19: (1):110. doi: 10.1186/s12943-020-01222-5
174. Sakuishi K, Jayaraman P, Behar SM, Anderson AC, Kuchroo VK. Emerging Tim-3 functions in antimicrobial and tumor immunity. Trends Immunol (2011) 32(8):345–9. doi: 10.1016/j.it.2011.05.003
175. Li H, Wu K, Tao K, Chen L, Zheng Q, Lu X, et al. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology (Baltimore Md) (2012) 56(4):1342–51. doi: 10.1002/hep.25777
176. Li Z, Liu Z, Zhang G, Han Q, Li N, Zhu Q, et al. TIM3 gene polymorphisms in patients with chronic hepatitis B virus infection: impact on disease susceptibility and hepatocellular carcinoma traits. Tissue Antigens (2012) 80(2):151–7. doi: 10.1111/j.1399-0039.2012.01898.x
177. Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol (2018) 19(3):222–32. doi: 10.1038/s41590-018-0044-z
178. Piñeiro Fernández J, Luddy KA, Harmon C, O’Farrelly C. Hepatic Tumor Microenvironments and Effects on NK Cell Phenotype and Function. Int J Mol Sci (2019) 20(17):4131–48. doi: 10.3390/ijms20174131
179. Han Q, Zhao H, Jiang Y, Yin C, Zhang J. HCC-Derived Exosomes: Critical Player and Target for Cancer Immune Escape. Cells (2019) 8(6):558–69. doi: 10.3390/cells8060558
180. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet (London England) (2017) 389(10088):2492–502. doi: 10.1016/s0140-6736(17)31046-2
181. Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol (2018) 19(7):940–52. doi: 10.1016/s1470-2045(18)30351-6
182. Xu T, Cui T, Peng L, Kong S, Zou J, Tian X. The anti-hepatocellular carcinoma activity of Mel-P15 is mediated by natural killer cells. Oncol Lett (2017) 14(6):6901–6. doi: 10.3892/ol.2017.7018
183. Zhu L, Li XJ, Kalimuthu S, Gangadaran P, Lee HW, Oh JM, et al. Natural Killer Cell (NK-92MI)-Based Therapy for Pulmonary Metastasis of Anaplastic Thyroid Cancer in a Nude Mouse Model. Front Immunol (2017) 8:816. doi: 10.3389/fimmu.2017.00816
184. Grossenbacher SK, Aguilar EG, Murphy WJ. Leveraging natural killer cells for cancer immunotherapy. Immunotherapy (2017) 9(6):487–97. doi: 10.2217/imt-2017-0013
185. Xu L, Huang Y, Tan L, Yu W, Chen D, Lu C, et al. Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma. Int Immunopharmacol (2015) 29(2):635–41. doi: 10.1016/j.intimp.2015.09.017
Keywords: liver cancer, circRNA, immune evasion, natural killer (NK) cells, innate immunity
Citation: Tang Y, Jiang M, Jiang H-M, Ye ZJ, Huang Y-S, Li X-S, Qin B-Y, Zhou R-S, Pan H-F and Zheng D-Y (2021) The Roles of circRNAs in Liver Cancer Immunity. Front. Oncol. 10:598464. doi: 10.3389/fonc.2020.598464
Received: 24 August 2020; Accepted: 03 December 2020;
Published: 04 February 2021.
Edited by:
Weijia Fang, Zhejiang University, ChinaReviewed by:
Zhigang Ren, First Affiliated Hospital of Zhengzhou University, ChinaZheng Wang, University of Texas Health Science Center at Houston, United States
Copyright © 2021 Tang, Jiang, Jiang, Ye, Huang, Li, Qin, Zhou, Pan and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua-Feng Pan, gzphf@gzucm.edu.cn; Da-Yong Zheng, zhengdy@smu.edu.cn
†These authors have contributed equally to this work
 Ying Tang
Ying Tang