
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 27 October 2020
Sec. Molecular and Cellular Oncology
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.585288
One common and reversible type of post-translational modification (PTM) is the addition of O-linked β-N-acetylglucosamine (O-GlcNAc) modification (O-GlcNAcylation), and its dynamic balance is controlled by O-GlcNAc transferase (OGT) and glycoside hydrolase O-GlcNAcase (OGA) through the addition or removal of O-GlcNAc groups. A large amount of research data confirms that proteins regulated by O-GlcNAcylation play a pivotal role in cells. In particularly, imbalanced levels of OGT and O-GlcNAcylation have been found in various types of cancers. Recently, increasing evidence shows that imbalanced O-GlcNAcylation directly or indirectly impacts the process of cancer metastasis. This review summarizes the current understanding of the influence of O-GlcNAc-proteins on the regulation of cancer metastasis. It will provide a theoretical basis to further elucidate of the molecular mechanisms underlying cancer emergence and progression.
As one of the post-translational modifications (PTMs), O-GlcNAcylation often occurs on serine (Ser) and threonine (Thr) residues of specific substrate cellular proteins including transcription factors, signaling pathway members and metabolic enzymes (1). O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) are responsible for adding or removing O-GlcNAc groups at the serine/threonine (Ser/Thr) residues of the target proteins to maintain the dynamic balance of intracellular O-GlcNAcylation (2). The sugar nucleotide uridine diphospho-N-acetylglucosamine (UDP-GlcNAc) which is generated by the nutrient-dependent hexosamine biosynthetic pathway (HBP), serves as a donor for O-GlcNAc addition to specific substrate proteins, demonstrating the link between glucose metabolism and O-GlcNAcylation (3). Therefore, O-GlcNAcylation is often referred to as a nutrient sensor. It has been found that O-GlcNAcylation is involved in diverse fundamental cellular processes, including cell signaling as well as tumorigenesis and tumor progression (4). A decade of research regarding the role of O-GlcNAcylation in cancer progression has resulted in accumulating studies on its potential roles in metastasis. Here, the role of O-GlcNAcylation in cancer metastasis will be summarized. In addition, the potential roles of the O-GlcNAcylation-PTMs axis in metastasis and small molecules that target O-GlcNAcylation are discussed.
As mentioned previously, intracellular O-GlcNAcylation is dynamically regulated by OGT and OGA. Notably, OGT and OGA are the only enzymes found to be involved in the addition and removal of O-GlcNAc groups to or from Ser/Thr residues of the substrate proteins (5). Human cells express three isoforms of OGT—nucleocytoplasmic (ncOGT, 116kDa), mitochondrial (mOGT, 103kDa), and short (sOGT, 75kDa)—which differ only in their subcellular location and number of N-terminal tetratricopeptide-repeats (TPRs), three different transcripts contain 13.5 (ncOGT), 9 (mOGT), and 3 (sOGT) TPRs, respectively. It is already clear that OGT is divided into two highly conserved functional domains (Table 1) (6, 7, 14). The N-terminal TPR domain binds the substrate protein, while the C-terminal catalytic domain binds UDP-GlcNAc and catalyzes O-GlcNAcylation of the substrate (8, 15–17). And OGA was initially isolated from crude cellular extract, and it catalyzes hydrolytic cleavage of O-GlcNAc from proteins (18). There are two alternative OGA splicing isoforms as follows: OGA-L (916 amino acids) predominantly localizes in the cytoplasm, and OGA-S (677 amino acids) localizes to the nucleus and lipid-droplets (8, 15–17, 19). OGA is also divided into two functional domains, N-terminus N-acetyl-β-D-glucosaminidase domain and C-terminal pseudo-histone acetyltransferase (HAT) domain (20). In cells, OGA can interact with OGT to form an “O-GlcNAczyme” complex under high glucose conditions (21), however disrupting this balance will lead to abnormal cell function and possibly even cancer.
O-GlcNAcylation harboring many substrates is involved in various cellular processes, including gene transcription regulation, stem cell differentiation, enzyme activity, and protein stability, among others (21–26). In view of the important roles of O-GlcNAcylation in multiple fundamental cellular processes, it is unsurprising that imbalanced profiles of OGT/O-GlcNAcylation frequently lead to the occurrence of many diseases such as diabetes, neurological disorders, cardiovascular disease, and even cancer (27, 28). In many types of cancer such as breast, prostate, lung, colorectal, and esophageal cancers, higher levels of OGT/O-GlcNAcylation are observed (29), suggesting that alterations of the intracellular level of OGT and O-GlcNAcylation are tightly associated with tumorigenesis, which might further participate directly or indirectly in the regulation of the biological processes associated with cancer metastasis. For example, the increased levels of OGT/O-GlcNAcylation in patients with lung cancer or colon cancer are closely correlated with poor overall survival, as well as the anchorage-independent growth, migration, and invasion ability of lung and colon cancer cell lines (22, 23, 30, 31). Elevated OGT proteins, as well as O-GlcNAcylation level, are also found in both breast cancer cells and tumor tissues (24, 25). Further research has revealed that O-GlcNAcylation of progesterone receptor (PR) by OGT transcriptionally activates its target genes, and PR-positive breast cancers express higher levels of OGT (26). In addition, 22 of 56 prostate cancer biopsy specimens were found to show increased O-GlcNAcylation, which correlated with poor prognosis (28). Furthermore, in prostate carcinoma and bladder cancer cells, while the level of OGT/O-GlcNAcylation increased, the level of deglycosylase OGA decreased (32, 33), prompting a dynamic imbalance between OGT and OGA. More in-depth research results confirmed the correlation between the OGT protein level and tumor metastatic progression in prostate cancer cells (32). In addition, downregulation of O-GlcNAcylation induced by OGT silencing results in cell cycle arrest, as well as the induction of autophagy and apoptosis, in bladder cancer cells (34, 35). However, in rare cases, O-GlcNAcylation is decreased in cancer tissues such as ovarian cancer tissues which harbor high rates of p53 mutations (36). In ovarian cell lines expressing wild-type p53, the high level of OGT/O-GlcNAcylation can stabilize the tumor suppressor p53, and stabilized p53 further promotes the acquisition of new pro-oncogenic activities including cell proliferation and metabolic changes, whereas the stabilization of p53 was not detected in cell lines with mutated p53 (36, 37), indicating a role of O-GlcNAcylation in regulating ovarian cancer proliferation and progression. Moreover, data of aberrant OGT level in various cancer tissues is also collected and analyzed by UALCAN based on TCGA datasets (Figure 1) (38–40). Taken together, the changes in OGT/O-GlcNAcylation level directly affect tumor occurrence and progression.
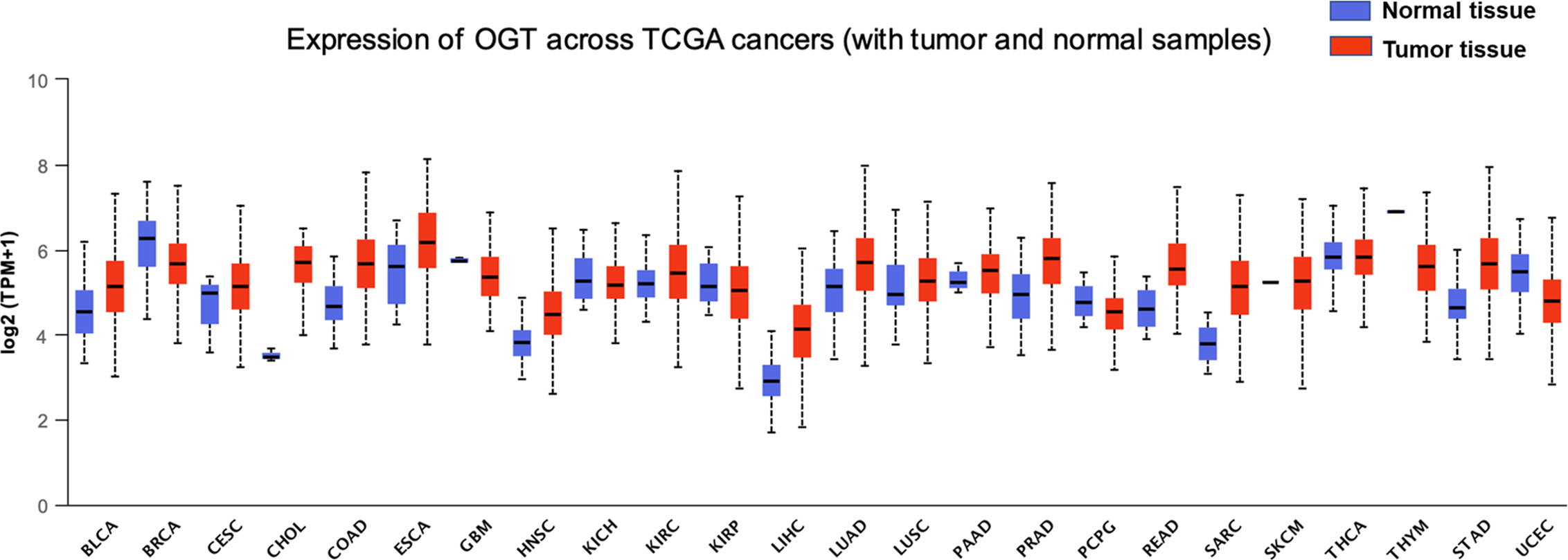
Figure 1 Expression of OGT across multiple cancer types. Pharmacological network analysis was performed by UALCAN (http://ualcan.path.uab.edu) on data extracted from The Cancer Genome Atlas (TCGA). BLCA, bladder urothelial carcinoma, normal tissue = 19, tumor tissue = 408, statistical significance = 2.803800E-03; BRCA, breast invasive carcinoma, normal tissue = 114, tumor tissue = 1,097, statistical significance = 3.12030001836661E-09; CESC, cervical cancer, normal tissue = 3, tumor tissue = 305, statistical significance = 4.214600E-01; CHOL, cholangiocarcinoma, normal tissue = 9, tumor tissue = 36, statistical significance = 2.6247892748188E-12; COAD, chronic obstructive pulmonary disease, normal tissue = 41, tumor tissue = 286, statistical significance = 1.62436730732907E-12; ESCA, esophageal carcinoma, normal tissue = 11, tumor tissue = 184, statistical significance = 6.667600E-02; GBM, glioblastoma, normal tissue = 5, tumor tissue = 156, statistical significance = 2.163600E-01; HNSC, head and neck squamous cell carcinoma, normal tissue = 44, tumor tissue = 520, statistical significance = 2.14550044397299E-10; KICH, kidney chromophobe, normal tissue = 25, tumor tissue = 67, statistical significance = 4.953000E-01; KIRC, kidney renal clear cell carcinoma, normal tissue = 72, tumor tissue = 533, statistical significance = 1.84977999999614E-05; KIRP, kidney renal papillary cell carcinoma, normal tissue = 32, tumor tissue = 290, statistical significance = 7.601200E-01; LIHC, liver hepatocellular carcinoma, normal tissue = 50, tumor tissue = 371, statistical significance = <1E-12; LUAD, lung adenocarcinoma, normal tissue = 59, tumor tissue = 515, statistical significance = 6.66022792472631E-12; LUSC, lung squamous cell carcinoma, normal tissue = 52, tumor tissue = 503, statistical significance = 2.798400E-02; PAAD, pancreatic adenocarcinoma, normal tissue = 4, tumor tissue = 178, statistical significance = 4.355800E-01; PCPG, pheochromocytoma and paraganglioma, normal tissue = 3, tumor tissue = 179, statistical significance = 1.192150E-01; PRAD, prostate adenocarcinoma, normal tissue = 52, tumor tissue = 497, statistical significance = 1.62458935193399E-12; READ, rectum adenocarcinoma, normal tissue = 10, tumor tissue = 166, statistical significance = 8.70579999978638E-07; SARC, sarcoma, normal tissue = 2, tumor tissue = 260, statistical significance = 1.889990E-01; SKCM, skin cutaneous melanoma, normal tissue = 1, tumor tissue = 104, statistical significance = 3.755700E-03; STAD, stomach adenocarcinoma, normal tissue = 3, tumor tissue = 415, statistical significance = <1E-12; THCA, thyroid carcinoma, normal tissue = 59, tumor tissue = 505, statistical significance = 4.631800E-01; THYM, thymoma, normal tissue = 2, tumor tissue = 120, statistical significance = 1.718990E-03; UCEC, uterine corpus endometrial carcinoma, normal tissue = 35, tumor tissue = 546, statistical significance = 1.66160000003579E-06.
Tumor cells are characterized by high metabolic rates, rapid growth, and high proliferative capacity. They, therefore, exhibit a high energy demand, necessitating anaerobic metabolism within the hypoxic tumor microenvironment (TME). Accumulating evidence indicates that OGT-mediated O-GlcNAcylation on a variety of substrates including transcription factors, oncoproteins, and proteins associated with epithelial mesenchymal transition (EMT) promotes tumor metastatic capacity in numerous cancer cells, including those derived from colorectal cancer (CRC), breast cancer, gastric cancer, pancreatic cancer, and cholangiocarcinoma (CCA) (31, 41–45). Of the proteins associated with CCA progression, 21 display O-GlcNAcylation sites (46). There are already research data confirming that CRC patients with high O-GlcNAcylation are typically diagnosed with greater lymph node metastasis potential (41, 47). Abolishing such modification of actin-binding protein cofilin at Ser108 suppresses the invasive capability of breast cancer cells (48). Moreover, decreasing O-GlcNAcylation levels via OGT knockdown or microRNA (miRNA; e.g., miR-483 and miR-24-1)-mediated depletion suppresses the growth, migration, and invasive capability of cancer cells (31, 39, 40).
Many genes are involved in the process of cancer metastasis. Therefore, altered global cellular O-GlcNAcylation profiles can directly or indirectly impact the expression and activation of transcription factors, and this further change the biological behavior of those regulatory factors (Figure 2).
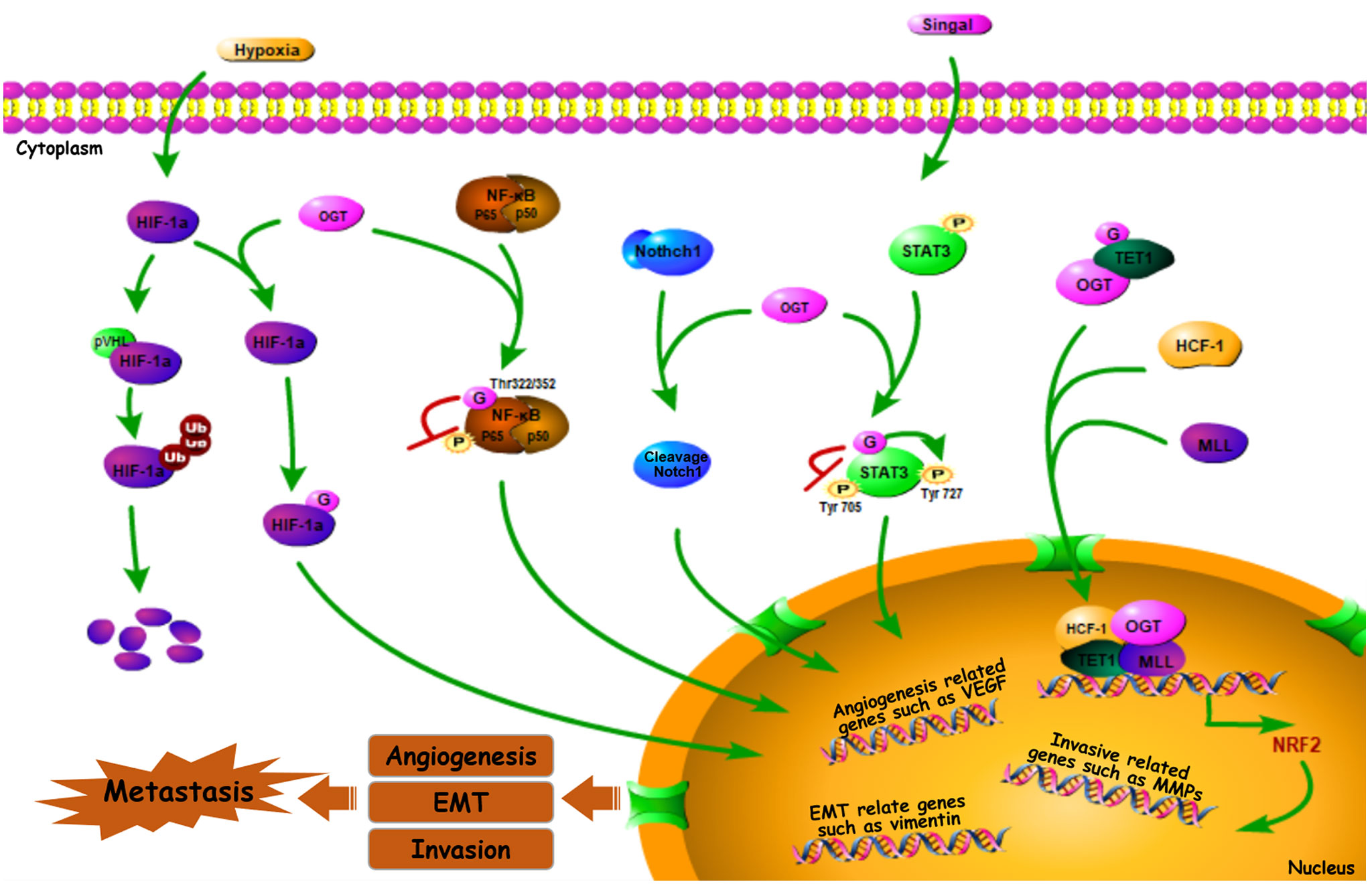
Figure 2 O-GlcNAc-modified transcription factors in cancer metastasis. Some O-GlcNAc-modified metastasis related transcription factors such as HIF-1α, Notch1, NF-κB, STAT3, Nrf2 can impact cancer metastasis by affecting downstream genes (or proteins) (49–61).
Hypoxia-inducible factor-1α (HIF-1α) is a well-known transcription factor that was originally identified as mediating adaptation to the hypoxic TME (62). It is clear that HIF-1α, the expression of which is induced by hypoxia, further activates the expression of its numerous targets—including matrix metalloproteinases (MMPs), E-cadherin, and transcription factor 3 (TCF3), among others—that enhance cancer metastasis via multiple mechanisms, favoring invasion, extravasation, and metastatic niche formation (63–65). OGT stabilizes HIF-1α by suppressing its interaction with von Hippel-Lindau tumor suppressor protein (pVHL), a E3-ubiquitin ligase mediates HIF-1α degradation. Thereby O-GlcNAcylation stabilizes HIF-1α and activates its transcriptional activity (49). Meanwhile, decreased OGT expression and O-GlcNAcylation level were observed when the protein levels and transactivation of HIF-1α were inhibited (66). This sets up a positive feedback loop, facilitating hypoxic adaptation, which further regulates processes such as immortalization, angiogenesis, invasive capability, and metastasis of breast cancer (49, 50). Indeed, reduced OGT results in a lower angiogenic potential and decreased vascular endothelial growth factor (VEGF) mRNA level in prostate cancer cell line (32). However, VEGF-mediated angiogenesis within tumors can be driven by HIF-1α activation (47). Furthermore, HIF-1α-induced VEGF upregulation promotes retinal angiogenesis in rats (67). Suggesting that by mediating the stabilization and activation of HIF-1α, OGT regulates HIF-1α target genes and functions in angiogenesis, as well as cancer metastasis (56).
Notch receptor 1 (Notch1), a type 1 trans-membrane receptor, is a key regulator of tumor angiogenesis and metastasis. It exhibits sustained activation in pre-metastatic lesions, which promotes migration in various types of tumor cells, including those derived from CRC, lung cancer, and melanoma (68, 69). In addition, Notch1 signaling plays a critical role in metastasis, including metastatic initiation in medulloblastoma and the promotion of highly-penetrant metastases in CRC (70, 71). Recently, it was demonstrated that OGT can O-GlcNAcylate Notch1, a process enhanced by glucosamine, resulting in the cleavage and nuclear translocation of Notch1 (51), thereby regulating the transcription of target genes, suggesting the importance of Notch1 transcriptional activity in cancer metastasis by modulating its O-GlcNAcylation.
Based on one report, nuclear factor κB (NF-κB), a transcription factor, is not only critically involved in the inflammatory response (including regulating IL-1β and IL-6 expression), but also contributes to tumor hematologic and lymphatic metastases, suggesting the correlation between NF-κB signal pathway and cancer metastasis (65). In breast cancer cells, one of the most common NF-κB dimeric forms, RELA (p65)/p50, can be O-GlcNAcylated at Thr322 and Thr352 residues of p65, which competitively inhibits p65 Ser536 phosphorylation, thus facilitating activated NF-κB-mediated gene transcription (52–54). Further, inflammation has timing- and context- specific roles during tumorigenesis and progression to cancer. For example, while NF-κB O-GlcNAcylation promotes MMP-mediated migration and invasive capability of CCA cells (55), NF-κB p65 O-GlcNAcylation downregulates C-X-C chemokine receptor 4 (CXCR4) to inhibit cervical cancer (CESC) cell metastasis to the lungs (56), and NF-κB activation-mediated upregulation of inducible nitric oxide synthase (iNOS) modulates immune suppression and tumor progression (57, 58). Activation of NF-κB via O-GlcNAcylation, therefore, modulates the expression of a variety of downstream genes involved in both tumor suppression and progression (72), indicating a role of OGT in regulating cancer metastasis by changing the NF-κB activation through its O-GlcNAc modification.
The transcription factor signal transducer and activator of transcription 3 (STAT3) is constitutively activated in tumors of different origins. Phosphorylation can activate STAT3, resulting in its translocation to the nucleus to regulate gene expression, further enhancing tumor angiogenic and invasive capability (73). For instance, phosphorylated STAT3 promotes proliferation and metastasis in epithelial ovarian cancer (74). Cross-talk between STAT3 O-GlcNAcylation and phosphorylation also occurs, with the former inhibiting the latter (75–77). Phosphorylation targets STAT3 residues, Tyr705 and Ser727, and these two modifications demonstrate a negative relationship to maintain its activity (78). Whereas STAT3 O-GlcNAcylation promotes Tyr705 phosphorylation, it inhibits Ser727 phosphorylation (59, 60), thereby enhancing metastasis by regulating STAT3 signaling and target gene transcription.Nuclear factor erythroid 2-related factor 2 (Nrf2) is another transcription factor, the activation of which plays a critical role in sustained angiogenesis, tumor invasion and metastasis (79). For example, activated Nrf2 stabilizes BTB domain and CNC homolog 1 (BACH1), accelerating lung cancer metastasis (80). Similarly, Nrf-2 activation promotes CRC and hepatic carcinoma metastasis (81). Nrf-2 activation is likely modulated by OGT, cause in Caenorhabditis elegans, the ortholog of human Nrf-2, is O-GlcNAcylated at Ser470 and Thr493 (82). Moreover, Nrf-2 transcriptional level is initiated when OGT is recruited at the promoter region by ten-eleven translocation 1 (TET1), and form a complex with host cell factor 1(HCF1) and mixed-lineage leukemia (MLL) (61).
Cancer cells undergoing EMT acquire the characteristics of aggressive, more invasive, stem-like features, with increased ability for cell migration, invasion and metastasis (83). In cancer cells and embryonic stem cells, O-GlcNAc-modification frequently facilitates the occurrence of EMT. For example, OGT is required for the induction and maintenance of EMT in NSCLC (84). In addition, hyper-O-GlcNAcylation contributes to the EMT of EC (85). The cell surface protein E-cadherin mediates cell-cell interactions, which is directly correlated with cancer cell adhesiveness, and this therefore mediates the invasive and metastatic capabilities of cells (86). While high levels of soluble E-cadherin in ovarian cancer-associated ascitic fluid promote tumor angiogenesis (87), decreased surface E-cadherin levels promote metastasis of breast cancer cells and lung adenocarcinoma cells (88, 89). Increased OGT expression and higher global O-GlcNAcylation levels suppress E-cadherin expression, thereby promoting breast cancer metastasis to the lungs (25). This inverse relationship between E-cadherin expression and metastatic potential also exists in ovarian cancer and CRC cells (90, 91). Notably, transcriptional expression of E-cadherin can be regulated by upstream proteins. For instance, Snail as an E-cadherin repressor can stabilize E-cadherin via Ser112 O-GlcNAcylation and enhance the migration and invasive capability of cancer cells (92). In addition, the cytoskeletal protein vimentin is a substrate of OGT, and the stabilization of E-cadherin is regulated by the O-GlcNAcylation status of vimentin (93, 94). Moreover, E-cadherin can be directly O-GlcNAcylated in breast cancer cells during drug-induced apoptosis, and this modification inhibits its transport to the cell surface, thereby decreasing cell-cell interactions and promoting EMT. Decreased surface E-cadherin levels increase infiltrative capacity, and cancer cell proliferation and survival are simultaneously decreased (95). O-GlcNAcylation of EMT-Related Proteins in Cancer Metastasis
During EMT, reduced E-cadherin expression and elevated snail, vimentin, fibronectin, and N-cadherin expression levels can be observed, thus these proteins are considered EMT markers (96). Beyond these markers, many EMT-related proteins including transcriptional factors are involved in the process of EMT. Receptor for activated protein kinase C (RACK1), encoded by GNB2L1, is a scaffold protein. (97). RACK1 induces EMT, further promotes the progression of esophageal squamous cell carcinoma (ESCC) and glioma (98, 99). Moreover, O-GlcNAcylation of RACK1 by OGT stabilizes RACK1, and results in a reduction of N-cadherin and upregulation of E-cadherin, indicating the induction of EMT and suppression of metastasis in chemoresistant gastric cancer (79, 100, 101). Numerous transcriptional factors including HIF-1α, Notch1, NF-κB have a critical role in EMT procession (102). For example, STAT3 regulates the expression of mesenchymal-related molecules including vimentin, the inhibition of which suppresses EMT-mediated lung cancer cell invasion (103). By regulating these transcriptional factors, the role of O-GlcNAcylation in EMT could be understood.
The MMP family plays a critical role in cancer cell migration. For example, MMP-9 overexpression is often observed across numerous malignant tumor types, and MMP-9 has been investigated for its potential as a cancer biomarker (104). Decreased global cellular O-GlcNAcylation levels result in decreased MMP-9 mRNA and protein levels, concurrently decreasing migration, invasive, and metastatic capability of gastric and EC cells (105, 106). Sirtuin1 (SIRT1) is a histone de-acetylase and O-GlcNAcylation of SIRT1/Ser549 promotes its enzymatic activity (107). Decreasing O-GlcNAcylation of this protein via OGT inhibition or knockdown in breast cancer cells increases both SIRT1 level and activity, thereby regulating forkhead box M1 (FOXM1), MMP-2, and MMP-9 protein level, and modulating breast cancer cell invasive and metastatic capability in vitro and in vivo (42). Via MMP targeting, O-GlcNAcylation plays an important role in cancer metastasis (47, 95, 100, 101, 106–121).
In summary, based on substrates of OGT, as well as their downstream effectors, which have key roles in regulating hypoxia, gene transcription, EMT, and metastasis, O-GlcNAcylation significantly modulates cancer progression.
Various PTMs of intracellular proteins rely on epigenetic regulatory enzymes with different catalytic functions. Generally, different PTMs often coordinate with each other to adapt to the process of complex biological functions in cells. O-GlcNAcylation is no exception. There has been much evidence confirming the interactions between O-GlcNAcylation and other PTMs. As a typical example (17, 122–127), both O-GlcNAcylation and phosphorylation occur on Ser/Thr residues of substrate proteins, and extensive crosstalk between two PTMs through mutual inhibition of the same or nearby residues has been identified (108). 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3), a glycolytic regulator, can be O-GlcNAcylated and phosphorylated at Ser172, and the competition between these two PTMs regulates the function of PFKFB3 in promoting nasopharyngeal carcinoma and gastric cancer proliferation, as well as migration (109–111). The enhancer of zeste homolog (EZH2) is responsible for H3K27me3, which promotes the metastasis of cancers such as melanoma and breast cancer (112, 113). O-GlcNAcylation at Ser729 of EZH2 plays a key role in maintaining the stabilization and methylation activity of its target protein (114, 118). Further, ubiquitination-mediated degradation of EZH2 suppresses breast cancer invasion and metastasis (119), and O-GlcNAc-modified EZH2 could reverse this degradation. EZH2 is stabilized by OGT via O-GlcNAcylation and promotes EMT and metastasis of CRC (41). The critical roles of histone deacetylases (HDACs) in tumorigenesis and tumor progression have been widely studied. Among them, HDAC1 and SIRT1 were identified as being O-GlcNAcylated at certain residues, and O-GlcNAcylation on specific residues further promotes the histone deacetylase activity of HDAC1 and SIRT1 (107, 116). In breast cancer cells, Nrf1 can be stabilized by OGT through O-GlcNAcylation at Ser448 and Ser451, a modification that suppresses the ubiquitin-proteasome mediated degradation of Nrf1. In contrast, reduced expression of Nrf1 suppresses its invasion and migration ability (115, 128). In summary, crosstalk between O-GlcNAcylation and other PTMs plays critical roles in regulating cancer metastasis.
Tumorigenesis and tumor progression are often accompanied by higher O-GlcNAcylation, which likely drives a range of oncogenic adaptations made by cancer cells, including rapid proliferation. Therefore, inhibiting global O-GlcNAcylation levels may also be an effective anti-cancer approach. In line with this, reducing intracellular OGT levels has been shown to inhibit the growth of lung cancer cells (23). A similar phenomenon was revealed in bladder cancer cells and renal cell carcinoma (RCC). Knocking down OGT results in cell cycle arrest as well as induction of autophagy and apoptosis (34, 35, 117). Considering the critical function of aberrant O-GlcNAcylation in cancer progression and metastasis which has been summarized previously herein, it is likely that downregulation of hyper O-GlcNAcylation via OGT inhibition might not only slow cancer proliferation, but also cancer metastasis.
In light of the findings that high levels of O-GlcNAcylation and OGT can affect multiple targets and signaling pathways during tumorigenesis, efforts are being made to find small molecules that can inhibit the activity of OGT. By rebalancing global O-GlcNAcylation profiles or targeting specific O-GlcNAcylated proteins, small molecules targeting OGT have been identified as exhibiting anti-cancer therapeutic potential. For example, miRNA-24, miRNA-101, and miRNA-483, all of which decrease OGT transcription, have been shown to inhibit the invasive ability of breast cancer, CRC, and gastric cancer, respectively (40, 41, 100). Similarly, ST045849, an OGT inhibitor, suppresses prostate cancer cell proliferation via metabolic reprogramming, and has been shown to inhibit hepatocellular carcinoma (HCC) cell proliferation (101, 120). Another OGT inhibitor, OSMI-1, developed via high-throughput screening, inhibits protein O-GlcNAcylation (121) and decreases tumor volume (129). Furthermore, the OGT inhibitor OSMI-2 decreases global chromatin O-GlcNAcylation and inhibits the proliferation of prostate cancer cells as a single drug. This suppression is also observed in organoids derived from patients with metastatic prostate cancer but not normal prostate cells, when OSMI-2 was combined with a CDK9 inhibitor (130, 131). In addition, Ac-5SGlcNAc, an OGT inhibitor that decreased global O-GlcNAcylation, but not N-glycosylation or N-glycosylation, suppresses the proliferation of pancreatic and breast cancer cells (54, 122, 132). Ac-5SGlcNAc treatment also blocks serum-stimulated cyclin D1 synthesis during the G0/G1 transition of breast cancer cells, suggesting that the role of OGT inhibitors in regulating the cell cycle further affects cell proliferation (123). Novel OGT-targeting small molecules are regularly identified. For instance, BZX2, OSMI-3, OSMI-4, L01, and ES1 have been identified as OGT inhibitors, but their broader biological impact is yet to be explored (124–127). Given the critical roles of OGT, such small molecule inhibitors may contribute substantially towards clarifying the function of OGT in cancer metastasis, and may be developed as clinically applicable anti-cancer therapeutic agents that can be used alone or in combination with other drugs (Table 2). However, considering the key roles of OGT in normal cell processes (e.g., energy metabolism), small molecule inhibitors of OGT might also impact normal physiology. Thus, studies focused on correcting aberrant O-GlcNAcylation to normal levels will need, to prevent or mitigate such off-target and potentially adverse effects.
O-GlcNAcylation is implicated in various fundamental cellular processes via the regulation of gene transcription, metabolism, and various signaling pathways. Several potential mechanisms by which OGT-mediated O-GlcNAcylation of substrate proteins modulates cancer progression include the following cellular processes: (1) creating recognition sites for recruitment to initiate cascades leading to the activation of downstream effectors, (2) cross-talk with PTMs to modulate substrate stabilization and activation, (3) integration of EMT/transcription factors/metastasis-associated protein activities, and (4) directing cancers towards metastasis via high levels of protein O-GlcNAcylation (Figure 3).
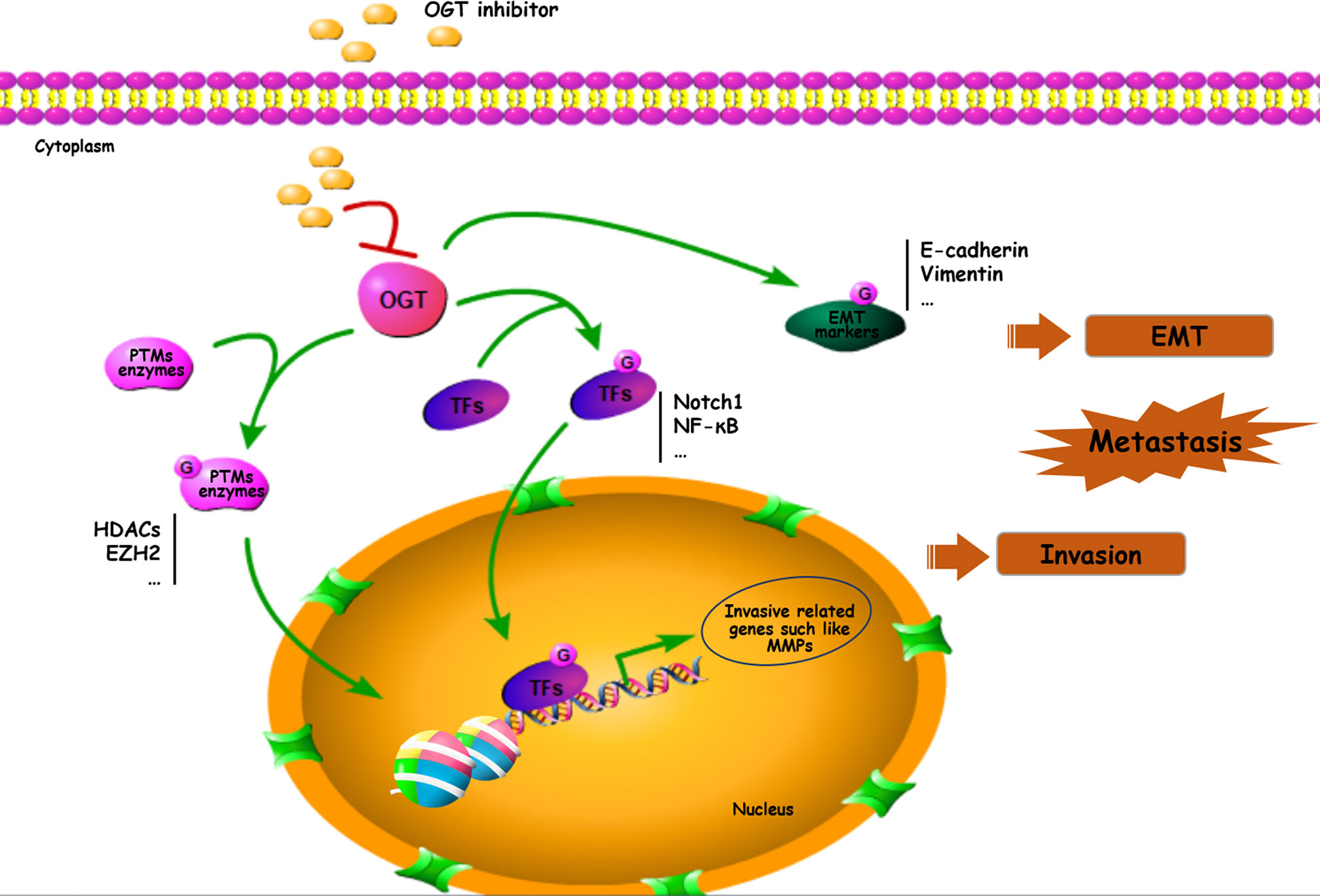
Figure 3 Important roles of O-GlcNAcylation on EMT/PTMs-related proteins in cancer metastasis. Many proteins including transcriptional factors, EMT related proteins, MMPs and PTMs enzymes can be O-GlcNAc-modified and participated in cancer metastasis. TFs, transcriptional factors.
Elucidating the functional mechanisms through which O-GlcNAcylation promotes cancer metastasis will provide a theoretical basis for future rational research. Considering the close relationship between O-GlcNAcylation and cancer progression-associated pathways, small molecules targeting OGT may have potential as anti-cancer therapies, especially in the inhibition of metastasis. In particular, the anti-cancer activities of more specific OGT inhibitors, alone or in combination with other drugs, as well as the side effects should be further investigated.
DW, JJ, and DL participated in writing, editing, and making figures. ZQ and HL read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
National Natural Science Foundation of China, (grant NO. 81903876, 81803680, 81973712, 81973468, 81803649). Jilin Province Traditional Chinese Medicine Technology Project (grant No. 2019051, 2020041).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Editage (www.editage.cn) for English language editing.
CCA, cholangiocarcinoma; CESC, cervical cancer; CRC, colorectal cancer; CXCR4, C-X-C chemokine receptor 4; EC, esophageal cancer; EMT, epithelial-mesenchymal transition; EZH2, zeste homolog; HBP, hexosamine biosynthetic pathway; HCC, hepatocellular carcinoma; HDAC, histone deacetylase; HIF-1α, hypoxia-inducible factor 1 alpha; H3K27, histone 3 lysine 27; iNOS, Inducible Nitric Oxide Synthase; MMPs, Matrix metalloproteinases; NF-κB, Nuclear factor κB; Notch1, notch receptor 1; Nrf, Nuclear factor erythroid 2-related factor; O-GlcNAc, O-linked β-N-Acetylglucosamine; O-GlcNAcylation, O-linked β-N-Acetylglucosamine modification, O-GlcNAc modification; OGT, O-GlcNAc transferase; OGA, O-GlcNAcase; PFKFB3, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3; PTM, post-translational modification; RACK1, Receptor for activated protein kinase C; RCC, renal cell carcinoma; Ser, Serine; SIRT1, sirtuin1; STAT3, Signal transducer and activator of transcription 3; Thr, Threonine; THYM, thymoma; TME, tumor microenvironment; UDP-GlcNAc, Uridine diphospho-N-acetylglucosamine; VEGF, vascular endothelial growth factor.
1. Liu J, Qian C, Cao X. Post-Translational Modification Control of Innate Immunity. Immunity (2016) 45:15–30. doi: 10.1016/j.immuni.2016.06.020
2. Copeland RJ, Bullen JW, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab (2008) 295:E17–28. doi: 10.1152/ajpendo.90281.2008
3. Hart GW. Nutrient regulation of signaling and transcription. J Biol Chem (2019) 294:2211–31. doi: 10.1074/jbc.AW119.003226
4. Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol (2015) 208:869–80. doi: 10.1083/jcb.201501101
5. Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem (1984) 259:3308–17.
6. Seo HG, Kim HB, Kang MJ, Ryum JH, Yi EC, Cho JW. Identification of the nuclear localisation signal of O-GlcNAc transferase and its nuclear import regulation. Sci Rep (2016) 6:34614–. doi: 10.1038/srep34614
7. Griffin ME, Jensen EH, Mason DE, Jenkins CL, Stone SE, Peters EC, et al. Comprehensive mapping of O-GlcNAc modification sites using a chemically cleavable tag. Mol BioSyst (2016) 12:1756–9. doi: 10.1039/C6MB00138F
8. Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature (2011) 469:564–7. doi: 10.1038/nature09638
9. Love DC, Kochan J, Cathey RL, Shin SH, Hanover JA. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci (2003) 116:647–54. doi: 10.1242/jcs.00246
10. Trapannone R, Mariappa D, Ferenbach AT, van Aalten DM. Nucleocytoplasmic human O-GlcNAc transferase is sufficient for O-GlcNAcylation of mitochondrial proteins. Biochem J (2016) 473:1693–702. doi: 10.1042/BCJ20160092
11. Sacoman JL, Dagda RY, Burnham-Marusich AR, Dagda RK, Berninsone PM. Mitochondrial O-GlcNAc Transferase (mOGT) Regulates Mitochondrial Structure, Function, and Survival in HeLa Cells. J Biol Chem (2017) 292:4499–518. doi: 10.1074/jbc.M116.726752
12. Shin SH, Love DC, Hanover JA. Elevated O-GlcNAc-dependent signaling through inducible mOGT expression selectively triggers apoptosis. Amino Acids (2011) 40:885–93. doi: 10.1007/s00726-010-0719-8
13. Riu IH, Shin IS, Do SI. Sp1 modulates ncOGT activity to alter target recognition and enhanced thermotolerance in E. coli. Biochem Biophys Res Commun (2008) 372:203–9. doi: 10.1016/j.bbrc.2008.05.034
14. Wu D, Cai Y, Jin J. Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell (2017) 8:713–23. doi: 10.1007/s13238-017-0416-4
15. Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol (2004) 11:1001–7. doi: 10.1038/nsmb833
16. Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem (1999) 274:32015–22. doi: 10.1074/jbc.274.45.32015
17. Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem (2000) 275:10983–8. doi: 10.1074/jbc.275.15.10983
18. Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem (2001) 276:9838–45. doi: 10.1074/jbc.M010420200
19. Keembiyehetty CN, Krzeslak A, Love DC, Hanover JA. A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome. J Cell Sci (2011) 124:2851–60. doi: 10.1242/jcs.083287
20. Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem (2004) 279:53665–73. doi: 10.1074/jbc.M410406200
21. Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol (2017) 18:452–65. doi: 10.1038/nrm.2017.22
22. Yu M, Chu S, Fei B, Fang X, Liu Z. O-GlcNAcylation of ITGA5 facilitates the occurrence and development of colorectal cancer. Exp Cell Res (2019) 382:111464. doi: 10.1016/j.yexcr.2019.06.009
23. Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X, et al. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta (2011) 1812:514–9. doi: 10.1016/j.bbadis.2011.01.009
24. Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene (2010) 29:2831–42. doi: 10.1038/onc.2010.41
25. Gu Y, Mi W, Ge Y, Liu H, Fan Q, Han C, et al. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res (2010) 70:6344–51. doi: 10.1158/0008-5472.CAN-09-1887
26. Trinca GM, Goodman ML, Papachristou EK, D’Santos CS, Chalise P, Madan R, et al. O-GlcNAc-Dependent Regulation of Progesterone Receptor Function in Breast Cancer. Horm Cancer (2018) 9:12–21. doi: 10.1007/s12672-017-0310-9
27. Zhao L, Shah JA, Cai Y, Jin J. ‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins. Molecules (2018) 23(8):1967. doi: 10.3390/molecules23081967
28. Kamigaito T, Okaneya T, Kawakubo M, Shimojo H, Nishizawa O, Nakayama J. Overexpression of O-GlcNAc by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis (2014) 17:18–22. doi: 10.1038/pcan.2013.56
29. Ferrer CM, Sodi VL, Reginato MJ. O-GlcNAcylation in Cancer Biology: Linking Metabolism and Signaling. J Mol Biol (2016) 428:3282–94. doi: 10.1016/j.jmb.2016.05.028
30. Lin YC, Lin CH, Yeh YC, Ho HL, Wu YC, Chen MY, et al. High O-linked N-acetylglucosamine transferase expression predicts poor survival in patients with early stage lung adenocarcinoma. Oncotarget (2018) 9:31032–44. doi: 10.18632/oncotarget.25772
31. Xu D, Wang W, Bian T, Yang W, Shao M, Yang H. Increased expression of O-GlcNAc transferase (OGT) is a biomarker for poor prognosis and allows tumorigenesis and invasion in colon cancer. Int J Clin Exp Pathol (2019) 12:1305–14.
32. Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-Linked β-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem (2012) 287:11070–81. doi: 10.1074/jbc.M111.302547
33. Jin L, Lu MH, Dai GC, Yao Q, Xiang H, Wang LX, et al. O-GlcNAcylation promotes malignant phenotypes of bladder cancer cells. Neoplasma (2020) 67:880–8. doi: 10.4149/neo_2020_191006N1009
34. Wang L, Chen S, Zhang Z, Zhang J, Mao S, Zheng J, et al. Suppressed OGT expression inhibits cell proliferation while inducing cell apoptosis in bladder cancer. BMC Cancer (2018) 18:1141. doi: 10.1186/s12885-018-5033-y
35. Jin L, Yuan F, Dai G, Yao Q, Xiang H, Wang L, et al. Blockage of O-linked GlcNAcylation induces AMPK-dependent autophagy in bladder cancer cells. Cell Mol Biol Lett (2020) 25:17. doi: 10.1186/s11658-020-00208-x
36. de Queiroz RM, Madan R, Chien J, Dias WB, Slawson C. Changes in O-Linked N-Acetylglucosamine (O-GlcNAc) Homeostasis Activate the p53 Pathway in Ovarian Cancer Cells. J Biol Chem (2016) 291:18897–914. doi: 10.1074/jbc.M116.734533
37. Parrales A, Iwakuma T. Targeting Oncogenic Mutant p53 for Cancer Therapy. Front Oncol (2015) 5:288. doi: 10.3389/fonc.2015.00288
38. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (N Y NY) (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002
39. Liu Y, Huang H, Liu M, Wu Q, Li W, Zhang J. MicroRNA-24-1 suppresses mouse hepatoma cell invasion and metastasis via directly targeting O-GlcNAc transferase. Biomed Pharmacother Biomed Pharmacother (2017) 91:731–8. doi: 10.1016/j.biopha.2017.05.007
40. Yu FY, Zhou CY, Liu YB, Wang B, Mao L, Li Y. miR-483 is down-regulated in gastric cancer and suppresses cell proliferation, invasion and protein O-GlcNAcylation by targeting OGT. Neoplasma (2018) 65:406–14. doi: 10.4149/neo_2018_170608N411
41. Jiang M, Xu B, Li X, Shang Y, Chu Y, Wang W, et al. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene (2019) 38:301–16. doi: 10.1038/s41388-018-0435-5
42. Ferrer CM, Lu TY, Bacigalupa ZA, Katsetos CD, Sinclair DA, Reginato MJ. O-GlcNAcylation regulates breast cancer metastasis via SIRT1 modulation of FOXM1 pathway. Oncogene (2017) 36:559–69. doi: 10.1038/onc.2016.228
43. Phoomak C, Silsirivanit A, Park D, Sawanyawisuth K, Vaeteewoottacharn K, Wongkham C, et al. O-GlcNAcylation mediates metastasis of cholangiocarcinoma through FOXO3 and MAN1A1. Oncogene (2018) 37:5648–65.doi: 10.1038/s41388-018-0366-1
44. Jang TJ, Kim UJ. O-GlcNAcylation is associated with the development and progression of gastric carcinoma. Pathol Res Pract (2016) 212:622–30.doi: 10.1016/j.prp.2016.04.002
45. Munkley J. The glycosylation landscape of pancreatic cancer. Oncol Lett (2019) 17:2569–75. doi: 10.3892/ol.2019.9885
46. Phoomak C, Park D, Silsirivanit A, Sawanyawisuth K, Vaeteewoottacharn K, Detarya M, et al. O-GlcNAc-induced nuclear translocation of hnRNP-K is associated with progression and metastasis of cholangiocarcinoma. Mol Oncol (2019) 13:338–57. doi: 10.1002/1878-0261.12406
47. Morfoisse F, Kuchnio A, Frainay C, Gomez-Brouchet A, Delisle MB, Marzi S, et al. Hypoxia induces VEGF-C expression in metastatic tumor cells via a HIF-1α-independent translation-mediated mechanism. Cell Rep (2014) 6:155–67. doi: 10.1016/j.celrep.2013.12.011
48. Huang X, Pan Q, Sun D, Chen W, Shen A, Huang M, et al. O-GlcNAcylation of cofilin promotes breast cancer cell invasion. J Biol Chem (2013) 288:36418–25. doi: 10.1074/jbc.M113.495713
49. Ferrer CM, Lynch TP, Sodi VL, Falcone JN, Schwab LP, Peacock DL, et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell (2014) 54:820–31. doi: 10.1016/j.molcel.2014.04.026
50. Yang YR, Jang HJ, Lee YH, Kim IS, Lee H, Ryu SH, et al. O-GlcNAc cycling enzymes control vascular development of the placenta by modulating the levels of HIF-1α. Placenta (2015) 36:1063–8. doi: 10.1016/j.placenta.2015.08.001
51. Jeon JH, Suh HN, Kim MO, Ryu JM, Han HJ. Glucosamine-induced OGT activation mediates glucose production through cleaved Notch1 and FoxO1, which coordinately contributed to the regulation of maintenance of self-renewal in mouse embryonic stem cells. Stem Cells Dev (2014) 23:2067–79. doi: 10.1089/scd.2013.0583
52. Yang WH, Park SY, Nam HW, Kim DH, Kang JG, Kang ES, et al. NFkappaB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc Natl Acad Sci U S A (2008) 105:17345–50. doi: 10.1073/pnas.0806198105
53. Yang YR, Kim DH, Seo YK, Park D, Jang HJ, Choi SY, et al. Elevated O-GlcNAcylation promotes colonic inflammation and tumorigenesis by modulating NF-κB signaling. Oncotarget (2015) 6:12529–42. doi: 10.18632/oncotarget.3725
54. Ma Z, Vocadlo DJ, Vosseller K. Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J Biol Chem (2013) 288:15121–30. doi: 10.1074/jbc.M113.470047
55. Phoomak C, Vaeteewoottacharn K, Sawanyawisuth K, Seubwai W, Wongkham C, Silsirivanit A, et al. Mechanistic insights of O-GlcNAcylation that promote progression of cholangiocarcinoma cells via nuclear translocation of NF-κB. Sci Rep (2016) 6:27853. doi: 10.1038/srep27853
56. Ali A, Kim SH, Kim MJ, Choi MY, Kang SS, Cho GJ, et al. O-GlcNAcylation of NF-kappaB Promotes Lung Metastasis of Cervical Cancer Cells via Upregulation of CXCR4 Expression. Mol Cells (2017) 40:476–84. doi: 10.14348/molcells.2017.2309
57. Kielbik M, Szulc-Kielbik I, Klink M. The Potential Role of iNOS in Ovarian Cancer Progression and Chemoresistance. Int J Mol Sci (2019) 20(7):1751. doi: 10.3390/ijms20071751
58. Simon PS, Sharman SK, Lu C, Yang D, Paschall AV, Tulachan SS, et al. The NF-κB p65 and p50 homodimer cooperate with IRF8 to activate iNOS transcription. BMC Cancer (2015) 15:770. doi: 10.1186/s12885-015-1808-6
59. Xu C, Liu GD, Feng L, Zhang CH, Wang F. Identification of O-GlcNAcylation Modification in Diabetic Retinopathy and Crosstalk with Phosphorylation of STAT3 in Retina Vascular Endothelium Cells. Cell Physiol Biochem (2018) 49:1389–402. doi: 10.1159/000493444
60. Zimmerman AD, Harris RB. In vivo and in vitro evidence that chronic activation of the hexosamine biosynthetic pathway interferes with leptin-dependent STAT3 phosphorylation. Am J Physiol Regul Integr Comp Physiol (2015) 308:R543–55. doi: 10.1152/ajpregu.00347.2014
61. Kang KA, Piao MJ, Ryu YS, Kang HK, Chang WY, Keum YS, et al. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget (2016) 7:40594–620. doi: 10.18632/oncotarget.9745
62. Balamurugan K. HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int J Cancer (2016) 138:1058–66. doi: 10.1002/ijc.29519
63. Liu ZJ, Semenza GL, Zhang HF. Hypoxia-inducible factor 1 and breast cancer metastasis. J Zhejiang Univ Sci B (2015) 16:32–43. doi: 10.1631/jzus.B1400221
64. Soni S, Padwad YS. HIF-1 in cancer therapy: two decade long story of a transcription factor. Acta Oncol (2017) 56:503–15. doi: 10.1080/0284186X.2017.1301680
65. Yan M, Xu Q, Zhang P, Zhou XJ, Zhang ZY, Chen WT. Correlation of NF-kappaB signal pathway with tumor metastasis of human head and neck squamous cell carcinoma. BMC Cancer (2010) 10:437. doi: 10.1186/1471-2407-10-437
66. Li YN, Hu JA, Wang HM. Inhibition of HIF-1alpha Affects Autophagy Mediated Glycosylation in Oral Squamous Cell Carcinoma Cells. Dis Markers (2015) 2015:239479. doi: 10.1155/2015/239479
67. Zhang D, Lv FL, Wang GH. Effects of HIF-1α on diabetic retinopathy angiogenesis and VEGF expression. Eur Rev Med Pharmacol Sci (2018) 22:5071–6. doi: 10.26355/eurrev_201808_15699
68. Wieland E, Rodriguez-Vita J, Liebler SS, Mogler C, Moll I, Herberich SE, et al. Endothelial Notch1 Activity Facilitates Metastasis. Cancer Cell (2017) 31:355–67. doi: 10.1016/j.ccell.2017.01.007
69. Ruland J. Colon Cancer: Epithelial Notch Signaling Recruits Neutrophils to Drive Metastasis. Cancer Cell (2019) 36:213–4. doi: 10.1016/j.ccell.2019.08.010
70. Kahn SA, Wang X, Nitta RT, Gholamin S, Theruvath J, Hutter G, et al. Notch1 regulates the initiation of metastasis and self-renewal of Group 3 medulloblastoma. Nat Commun (2018) 9:4121. doi: 10.1038/s41467-018-07182-1
71. Jackstadt R, van Hooff SR, Leach JD, Cortes-Lavaud X, Lohuis JO, Ridgway RA, et al. Epithelial NOTCH Signaling Rewires the Tumor Microenvironment of Colorectal Cancer to Drive Poor-Prognosis Subtypes and Metastasis. Cancer Cell (2019) 36:319–36.e7. doi: 10.1016/j.ccell.2019.08.003
72. Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol (2018) 18:309–24. doi: 10.1038/nri.2017.142
73. Dubový P, Hradilová-Svíženská I, Klusáková I, Kokošová V, Brázda V, Joukal M. Bilateral activation of STAT3 by phosphorylation at the tyrosine-705 (Y705) and serine-727 (S727) positions and its nuclear translocation in primary sensory neurons following unilateral sciatic nerve injury. Histochem Cell Biol (2018) 150:37–47. doi: 10.1007/s00418-018-1656-y
74. Wu CJ, Sundararajan V, Sheu BC, Huang RY, Wei LH. Activation of STAT3 and STAT5 Signaling in Epithelial Ovarian Cancer Progression: Mechanism and Therapeutic Opportunity. Cancers (Basel) (2019) 12(1):24. doi: 10.3390/cancers12010024
75. Li X, Zhang Z, Li L, Gong W, Lazenby AJ, Swanson BJ, et al. Myeloid-derived cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. J Exp Med (2017) 214:1093–109. doi: 10.1084/jem.20161105
76. Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem (2004) 279:3563–72. doi: 10.1074/jbc.M306449200
77. Miguez JSG, Dela Justina V, Bressan AFM, Marchi PGF, Honorio-França AC, Carneiro FS, et al. O-Glycosylation with O-linked β-N-acetylglucosamine increases vascular contraction: Possible modulatory role on Interleukin-10 signaling pathway. Life Sci (2018) 209:78–84. doi: 10.1016/j.lfs.2018.07.058
78. Wakahara R, Kunimoto H, Tanino K, Kojima H, Inoue A, Shintaku H, et al. Phospho-Ser727 of STAT3 regulates STAT3 activity by enhancing dephosphorylation of phospho-Tyr705 largely through TC45. Genes Cells (2012) 17:132–45. doi: 10.1111/j.1365-2443.2011.01575.x
79. Cheng S, Mao Q, Dong Y, Ren J, Su L, Liu J, et al. GNB2L1 and its O-GlcNAcylation regulates metastasis via modulating epithelial-mesenchymal transition in the chemoresistance of gastric cancer. PloS One (2017) 12:e0182696. doi: 10.1371/journal.pone.0182696
80. Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, et al. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell (2019) 178:316–29.e18. doi: 10.1016/j.cell.2019.06.003
81. Wang H, Liu X, Long M, Huang Y, Zhang L, Zhang R, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med (2016) 8:334ra51. doi: 10.1126/scitranslmed.aad6095
82. Li H, Liu X, Wang D, Su L, Zhao T, Li Z, et al. O-GlcNAcylation of SKN-1 modulates the lifespan and oxidative stress resistance in Caenorhabditis elegans. Sci Rep (2017) 7:43601. doi: 10.1038/srep43601
83. Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol (2016) 43:7–13. doi: 10.1016/j.ceb.2016.06.002
84. Szymura SJ, Zaemes JP, Allison DF, Clift SH, D’Innocenzi JM, Gray LG, et al. NF-κB upregulates glutamine-fructose-6-phosphate transaminase 2 to promote migration in non-small cell lung cancer. Cell Commun Signaling CCS (2019) 17:24. doi: 10.1186/s12964-019-0335-5
85. Jaskiewicz NM, Townson DH. Hyper-O-GlcNAcylation promotes epithelial-mesenchymal transition in endometrial cancer cells. Oncotarget (2019) 10:2899–910. doi: 10.18632/oncotarget.26884
86. Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci (2013) 126:393–401. doi: 10.1242/jcs.100115
87. Tang MKS, Yue PYK, Ip PP, Huang RL, Lai HC, Cheung ANY, et al. Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat Commun (2018) 9:2270. doi: 10.1038/s41467-018-04695-7
88. Sinkevicius KW, Bellaria KJ, Barrios J, Pessina P, Gupta M, Brainson CF, et al. E-Cadherin Loss Accelerates Tumor Progression and Metastasis in a Mouse Model of Lung Adenocarcinoma. Am J Respir Cell Mol Biol (2018) 59:237–45. doi: 10.1165/rcmb.2017-0210OC
89. Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res (2008) 68:3645–54. doi: 10.1158/0008-5472.CAN-07-2938
90. Biwi J, Clarisse C, Biot C, Kozak RP, Madunic K, Mortuaire M, et al. OGT Controls the Expression and the Glycosylation of E-cadherin, and Affects Glycosphingolipid Structures in Human Colon Cell Lines. Proteomics (2019) 19:e1800452. doi: 10.1002/pmic.201800452
91. Jin FZ, Yu C, Zhao DZ, Wu MJ, Yang Z. A correlation between altered O-GlcNAcylation, migration and with changes in E-cadherin levels in ovarian cancer cells. Exp Cell Res (2013) 319:1482–90. doi: 10.1016/j.yexcr.2013.03.013
92. Park SY, Kim HS, Kim NH, Ji S, Cha SY, Kang JG, et al. Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J (2010) 29:3787–96. doi: 10.1038/emboj.2010.254
93. Slawson C, Lakshmanan T, Knapp S, Hart GW. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol Biol Cell (2008) 19:4130–40. doi: 10.1091/mbc.e07-11-1146
94. Phoomak C, Vaeteewoottacharn K, Silsirivanit A, Saengboonmee C, Seubwai W, Sawanyawisuth K, et al. High glucose levels boost the aggressiveness of highly metastatic cholangiocarcinoma cells via O-GlcNAcylation. Sci Rep (2017) 7:43842. doi: 10.1038/srep43842
95. Padmanaban V, Krol I, Suhail Y, Szczerba BM, Aceto N, Bader JS, et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature (2019) 573:439–44. doi: 10.1038/s41586-019-1526-3
96. Carvalho-Cruz P, Alisson-Silva F, Todeschini AR, Dias WB. Cellular glycosylation senses metabolic changes and modulates cell plasticity during epithelial to mesenchymal transition. Dev Dyn (2018) 247:481–91. doi: 10.1002/dvdy.24553
97. McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol (2002) 62:1261–73. doi: 10.1124/mol.62.6.1261
98. Lv QL, Huang YT, Wang GH, Liu YL, Huang J, Qu Q, et al. Overexpression of RACK1 Promotes Metastasis by Enhancing Epithelial-Mesenchymal Transition and Predicts Poor Prognosis in Human Glioma. Int J Environ Res Public Health (2016) 13(10):1021. doi: 10.3390/ijerph13101021
99. Wang N, Liu F, Cao F, Jia Y, Wang J, Ma W, et al. RACK1 predicts poor prognosis and regulates progression of esophageal squamous cell carcinoma through its epithelial-mesenchymal transition. Cancer Biol Ther (2015) 16:528–40. doi: 10.1080/15384047.2015.1016687
100. Liu Y, Huang H, Cao Y, Wu Q, Li W, Zhang J. Suppression of OGT by microRNA24 reduces FOXA1 stability and prevents breast cancer cells invasion. Biochem Biophys Res Commun (2017) 487:755–62. doi: 10.1016/j.bbrc.2017.04.135
101. Itkonen HM, Gorad SS, Duveau DY, Martin SES, Barkovskaya A, Bathen TF, et al. Inhibition of O-GlcNAc transferase activity reprograms prostate cancer cell metabolism. Oncotarget (2016) 7:12464–76. doi: 10.18632/oncotarget.7039
102. De Francesco EM, Maggiolini M, Musti AM. Crosstalk between Notch, HIF-1α and GPER in Breast Cancer EMT. Int J Mol Sci (2018) 19(7):2011. doi: 10.3390/ijms19072011
103. Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer (2019) 18:40. doi: 10.1186/s12943-019-0959-5
104. Huang H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances. Sens (Basel Switzerland) (2018) 18(10):3249. doi: 10.3390/s18103249
105. Akter H, Park M, Kwon OS, Song EJ, Park WS, Kang MJ. Activation of matrix metalloproteinase-9 (MMP-9) by neurotensin promotes cell invasion and migration through ERK pathway in gastric cancer. Tumour Biol (2015) 36:6053–62. doi: 10.1007/s13277-015-3282-9
106. Qiao Z, Dang C, Zhou B, Li S, Zhang W, Jiang J, et al. Downregulation of O-linked N-acetylglucosamine transferase by RNA interference decreases MMP9 expression in human esophageal cancer cells. Oncol Lett (2016) 11:3317–23. doi: 10.3892/ol.2016.4428
107. Han C, Gu Y, Shan H, Mi W, Sun J, Shi M, et al. O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat Commun (2017) 8:1491–. doi: 10.1038/s41467-017-01654-6
108. Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem (2011) 80:825–58. doi: 10.1146/annurev-biochem-060608-102511
109. Lei Y, Chen T, Li Y, Shang M, Zhang Y, Jin Y, et al. O-GlcNAcylation of PFKFB3 is required for tumor cell proliferation under hypoxia. Oncogenesis (2020) 9:21–. doi: 10.1038/s41389-020-0208-1
110. Gu M, Li L, Zhang Z, Chen J, Zhang W, Zhang J, et al. PFKFB3 promotes proliferation, migration and angiogenesis in nasopharyngeal carcinoma. J Cancer (2017) 8:3887–96. doi: 10.7150/jca.19112
111. Han J, Meng Q, Xi Q, Wang H, Wu G. PFKFB3 was overexpressed in gastric cancer patients and promoted the proliferation and migration of gastric cancer cells. Cancer Biomark (2017) 18:249–56. doi: 10.3233/CBM-160143
112. Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun (2015) 6:6051. doi: 10.1038/ncomms7051
113. Anwar T, Arellano-Garcia C, Ropa J, Chen YC, Kim HS, Yoon E, et al. p38-mediated phosphorylation at T367 induces EZH2 cytoplasmic localization to promote breast cancer metastasis. Nat Commun (2018) 9:2801. doi: 10.1038/s41467-018-05078-8
114. Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A (2014) 111:1355–60. doi: 10.1073/pnas.1323226111
115. Zhou Y, Xu Z, Quan D, Zhang F, Zhang H, Xiao T, et al. Nuclear respiratory factor 1 promotes spheroid survival and mesenchymal transition in mammary epithelial cells. Oncogene (2018) 37:6152–65. doi: 10.1038/s41388-018-0349-2
116. Zhu G, Tao T, Zhang D, Liu X, Qiu H, Han L, et al. O-GlcNAcylation of histone deacetylases 1 in hepatocellular carcinoma promotes cancer progression. Glycobiology (2016) 26:820–33. doi: 10.1093/glycob/cww025
117. Wang L, Chen S, Zhang J, Mao S, Mao W, Zhang W, et al. Suppressed OGT expression inhibits cell proliferation and modulates EGFR expression in renal cell carcinoma. Cancer Manag Res (2019) 11:2215–23. doi: 10.2147/CMAR.S190642
118. Lo PW, Shie JJ, Chen CH, Wu CY, Hsu TL, Wong CH. O-GlcNAcylation regulates the stability and enzymatic activity of the histone methyltransferase EZH2. Proc Natl Acad Sci U S A (2018) 115:7302–7. doi: 10.1073/pnas.1801850115
119. Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ (2017) 24:59–71. doi: 10.1038/cdd.2016.95
120. Xu W, Zhang X, Wu JL, Fu L, Liu K, Liu D, et al. O-GlcNAc transferase promotes fatty liver-associated liver cancer through inducing palmitic acid and activating endoplasmic reticulum stress. J Hepatol (2017) 67:310–20. doi: 10.1016/j.jhep.2017.03.017
121. Ortiz-Meoz RF, Jiang J, Lazarus MB, Orman M, Janetzko J, Fan C, et al. A small molecule that inhibits OGT activity in cells. ACS Chem Biol (2015) 10:1392–7. doi: 10.1021/acschembio.5b00004
122. Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol (2011) 7:174–81. doi: 10.1038/nchembio.520
123. Olivier-Van Stichelen S, Drougat L, Dehennaut V, El Yazidi-Belkoura I, Guinez C, Mir AM, et al. Serum-stimulated cell cycle entry promotes ncOGT synthesis required for cyclin D expression. Oncogenesis (2012) 1:e36. doi: 10.1038/oncsis.2012.36
124. Liu Y, Ren Y, Cao Y, Huang H, Wu Q, Li W, et al. Discovery of a Low Toxicity O-GlcNAc Transferase (OGT) Inhibitor by Structure-based Virtual Screening of Natural Products. Sci Rep (2017) 7:12334–. doi: 10.1038/s41598-017-12522-0
125. Worth M, Hu C-W, Li H, Fan D, Estevez A, Zhu D, et al. Targeted covalent inhibition of O-GlcNAc transferase in cells. Chem Commun (Camb) (2019) 55:13291–4. doi: 10.1039/c9cc04560k
126. Lim S, Haque MM, Nam G, Ryoo N, Rhim H, Kim YK. Monitoring of Intracellular Tau Aggregation Regulated by OGA/OGT Inhibitors. Int J Mol Sci (2015) 16:20212–24. doi: 10.3390/ijms160920212
127. Martin SES, Tan ZW, Itkonen HM, Duveau DY, Paulo JA, Janetzko J, et al. Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors. J Am Chem Soc (2018) 140:13542–5. doi: 10.1021/jacs.8b07328
128. Sekine H, Okazaki K, Kato K, Alam MM, Shima H, Katsuoka F, et al. O-GlcNAcylation Signal Mediates Proteasome Inhibitor Resistance in Cancer Cells by Stabilizing NRF1. Mol Cell Biol (2018) 38(17):e00252–18. doi: 10.1128/MCB.00252-18
129. Sharma NS, Gupta VK, Dauer P, Kesh K, Hadad R, Giri B, et al. O-GlcNAc modification of Sox2 regulates self-renewal in pancreatic cancer by promoting its stability. Theranostics (2019) 9:3410–24. doi: 10.7150/thno.32615
130. Itkonen HM, Urbanucci A, Martin SE, Khan A, Mathelier A, Thiede B, et al. High OGT activity is essential for MYC-driven proliferation of prostate cancer cells. Theranostics (2019) 9:2183–97. doi: 10.7150/thno.30834
131. Itkonen HM, Poulose N, Steele RE, Martin SES, Levine ZG, Duveau DY, et al. Inhibition of O-GlcNAc Transferase Renders Prostate Cancer Cells Dependent on CDK9. Mol Cancer Res (2020) 18(10):1512–21. doi: 10.1158/1541-7786.MCR-20-0339
132. Sodi VL, Khaku S, Krutilina R, Schwab LP, Vocadlo DJ, Seagroves TN, et al. mTOR/MYC Axis Regulates O-GlcNAc Transferase Expression and O-GlcNAcylation in Breast Cancer. Mol Cancer Res (2015) 13:923–33. doi: 10.1158/1541-7786.MCR-14-0536
133. Steenackers A, Olivier-Van Stichelen S, Baldini SF, Dehennaut V, Toillon RA, Le Bourhis X, et al. Silencing the Nucleocytoplasmic O-GlcNAc Transferase Reduces Proliferation, Adhesion, and Migration of Cancer and Fetal Human Colon Cell Lines. Front Endocrinol (2016) 7:46. doi: 10.3389/fendo.2016.00046
Keywords: O-GlcNAcylation, O-GlcNAc transferase (OGT), cancer metastasis, transcriptional factors, post-translational modifications (PTMs)
Citation: Wu D, Jin J, Qiu Z, Liu D and Luo H (2020) Functional Analysis of O-GlcNAcylation in Cancer Metastasis. Front. Oncol. 10:585288. doi: 10.3389/fonc.2020.585288
Received: 30 July 2020; Accepted: 06 October 2020;
Published: 27 October 2020.
Edited by:
Daniel Christian Hoessli, University of Karachi, PakistanReviewed by:
Mauricio Reginato, Drexel University, United StatesCopyright © 2020 Wu, Jin, Qiu, Liu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haoming Luo, bHVvLmhhb21pbmdAMTYzLmNvbQ==; Da Liu, bGl1ZGFfMTk4NkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.