
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 12 July 2011
Sec. Cancer Epidemiology and Prevention
Volume 1 - 2011 | https://doi.org/10.3389/fonc.2011.00013
 Shu-Chun Chuang1,2
Shu-Chun Chuang1,2 Antonio Agudo3
Antonio Agudo3 Wolfgang Ahrens4
Wolfgang Ahrens4 Devasena Anantharaman5
Devasena Anantharaman5 Simone Benhamou6
Simone Benhamou6 Stefania Boccia7,8
Stefania Boccia7,8 Chu Chen9
Chu Chen9 David I. Conway10
David I. Conway10 Eleonora Fabianova11
Eleonora Fabianova11 Richard B. Hayes12
Richard B. Hayes12 Claire M. Healy13
Claire M. Healy13 Ivana Holcatova14
Ivana Holcatova14 Kristina Kjaerheim15
Kristina Kjaerheim15 Pagona Lagiou16
Pagona Lagiou16 Philip Lazarus17
Philip Lazarus17 Tatiana V. Macfarlane18
Tatiana V. Macfarlane18 Manoj B. Mahimkar5
Manoj B. Mahimkar5 Dana Mates19
Dana Mates19 Keitaro Matsuo20
Keitaro Matsuo20 Franco Merletti21
Franco Merletti21 Andres Metspalu22
Andres Metspalu22 Hal Morgenstern23,24
Hal Morgenstern23,24 Joshua Muscat17
Joshua Muscat17 Gabriella Cadoni25
Gabriella Cadoni25 Andrew F. Olshan26
Andrew F. Olshan26 Mark Purdue27
Mark Purdue27 Heribert Ramroth28
Heribert Ramroth28 Peter Rudnai29
Peter Rudnai29 Stephen M. Schwartz9
Stephen M. Schwartz9 Lorenzo Simonato30
Lorenzo Simonato30 Elaine M. Smith31
Elaine M. Smith31 Erich M. Sturgis32
Erich M. Sturgis32 Neonilia Szeszenia-Dabrowska33
Neonilia Szeszenia-Dabrowska33 Renato Talamini34
Renato Talamini34 Peter Thomson35
Peter Thomson35 Qingyi Wei32
Qingyi Wei32 David Zaridze36
David Zaridze36 Zuo-Feng Zhang37
Zuo-Feng Zhang37 Ariana Znaor38
Ariana Znaor38 Paul Brennan1
Paul Brennan1 Paolo Boffetta1,39,40
Paolo Boffetta1,39,40 Mia Hashibe1,41*
Mia Hashibe1,41*Previous molecular epidemiological studies on head and neck cancer have examined various single nucleotide polymorphisms (SNPs), but there are very few documented associations. In the International head and neck cancer epidemiology (INHANCE) consortium, we evaluated associations between SNPs in the metabolism, cell cycle, and DNA repair pathways and the risk of head and neck cancer. We analyzed individual-level pooled data from 14 European, North American, Central American, and Asia case–control studies (5,915 head and neck cancer cases and 10,644 controls) participating in the INHANCE consortium. Unconditional logistic regression was used to estimate odds ratios (OR) and 95% confidence intervals (CI) for SNP effects, adjusting for age, sex, race, and country. We observed an association between head and neck cancer risk and MGMT Leu84Phe heterozygotes (OR = 0.79, 95% CI = 0.68–0.93), XRCC1 Arg194Trp homozygotes Arg/Arg (OR = 2.3, 95% CI = 1.1–4.7), ADH1B Arg48His homozygotes Arg/Arg (OR = 2.7, 95% CI = 1.9–4.0), ADH1C Ile350Val homozygotes Ile/Ile (OR = 1.2, 95% CI = 1.1–1.4), and the GSTM1 null genotype (OR = 1.1, 95% CI = 1.0–1.2). Among these results, MGMT Leu84Phe, ADH1B Arg48His, ADH1C Ile350Arg, and the GSTM1 null genotype had fairly low false positive report probabilities (<20%). We observed associations between ADH1B Arg48His, ADH1C Ile350Arg, and GSTM1 null genotype and head and neck cancer risk. No functional study currently supports the observed association for MGMT Leu84Phe, and the association with XRCC1 Arg194Trp may be a chance finding.
Head and neck cancer, including cancers in oral cavity, pharynx (other than nasopharynx), and larynx, is the sixth most common cancer in the world (Parkin et al., 2005). It accounted for about 900,000 of cases and 300,000 deaths in 2008 (Ferlay et al., 2010). The 5-year survival rate was about 61% in the US for all sites (Altekruse et al., 2010) and 26 to 63% in Europe depending on the subsite (Berrino et al., 2007). The major risk factors for head and neck cancer are tobacco smoking and alcohol drinking (Hashibe et al., 2007). The interaction between tobacco smoking and alcohol drinking is greater than the expected multiplicative null with an overall attributable risk of 72% (Hashibe et al., 2009). Other risk factors include human papillomavirus (HPV) infection (IARC Working Group/Human Papillomaviruses, 2007), passive smoking (Lee et al., 2008), low body mass index (BMI; Gaudet et al., 2010), poor diet (World Cancer Research Fund/American Institute for Cancer Research, 2007), and family history of cancer (Negri et al., 2009).
Previous molecular epidemiological studies on head and neck cancer have examined single nucleotide polymorphisms (SNPs), focusing on metabolic and DNA repair genes (Sturgis and Wei, 2002; Hashibe et al., 2003, 2008; Canova et al., 2009); however, very few SNP associations have been consistent for head and neck cancer risk. The heterogeneity may result from study design, e.g., population- or hospital-based controls, study sample size, and study populations, e.g., race/ethnicity (Hashibe et al., 2003; Lohmueller et al., 2003). The International head and neck cancer epidemiology (INHANCE) consortium is a collaboration of research groups leading large molecular epidemiology studies of head and neck cancer. Since larger sample sizes can increase the precision of the effect measure estimates and the ability to detect statistically significant moderate associations, we pooled data on SNPs that were genotyped in common across the INHANCE studies, to evaluate the associations between the SNPs in several pathways and the risk of head and neck cancer.
The INHANCE consortium (http://inhance.iarc.fr/) was established in 2004. Fourteen studies participating in the consortium contributed SNP data: France (Benhamou et al., 2004), Central Europe (Hashibe et al., 2006), Seattle (Schwartz et al., 2001; Huang et al., 2005), Iowa (Wang et al., 2005), North Carolina (Olshan et al., 2000), Los Angeles (Cui et al., 2006), Houston (Zhang et al., 2005), Puerto Rico (Hayes et al., 1999), Rome (Boccia et al., 2008), Western Europe (Canova et al., 2009), Heidelberg (Risch et al., 2003), Japan (Suzuki et al., 2007), Northeast US (Park et al., 2003), and India (Anantharaman et al., 2007). Descriptions of the individual studies are presented in the Table A1 in Appendix. To increase power, we included all controls selected for lung and kidney cancers in the Central Europe multicenter case–control study, in addition to the head and neck cancer controls. There were 6,694 head and neck cancer cases and 12,601 controls.
Cases and controls with missing data on age, sex, race/ethnicity, or SNP information, and cases with missing information on the site of origin of their cancer were excluded (779 cases and 1,957 controls). In total, 5,915 cases and 10,644 controls were included in the analysis. Among the cases, 1,901 were oral cancer, 1,751 were pharyngeal cancer, 440 were cancers of the oral cavity or pharynx not otherwise specified, 1,632 were laryngeal cancer and 191 overlapping or subsite missing.
Written informed consent was obtained from all study subjects and the investigations were approved by institutional review boards at each of the institutes involved. Questionnaires were collected from all the individual studies, to assess the comparability of the collected data and of the wording of interview questions among the studies. Each data item was checked for illogical or missing values and inconsistencies were resolved as necessary.
Details on harmonizing questionnaire data have been published previously (Hashibe et al., 2007). Briefly, the definitions for ever smoking and drinking are different across studies. We reclassified ever tobacco smokers as those who have smoked at least 100 cigarettes or 100 cigars or 100 pipes in their lifetime. In our previous analyses, drinking (≥3 alcoholic drinks/day) was associated increased HNC risks (Hashibe et al., 2007), we thus classified heavy drinkers as those who have consumed three or more alcoholic drinks per day.
Single nucleotide polymorphisms reported in more than two studies were included in the current pooled analyses. In total, 28 SNPs in cell cycle (p21 Ser31Arg rs1801270 and p53 Pro72Arg rs1042522), DNA repair (ERCC2 Lys751Gln rs28365048, MGMT Leu84Phe rs12917, Ile143Val rs2308321, 171C > T rs1803965, OGG1 Ser326Cys rs1052133, XRCC1 Arg194Trp rs1799782, Arg280His rs25489, Arg399Gln rs25487, and XRCC3 Thr241Met rs861539), folate metabolism (MTHFR Ala222Val rs1801133 and Glu429Ala rs1801131), and carcinogen metabolism (ADH1B Arg48His rs1229984, ADH1C Ile350Val rs698, CYP1A1 Ile462Val rs1048943, 3801T > C E0322, CYP2E1 1054C > T rs2031920, 1143A > T rs6413432, 1293G > C rs3813867, EPHX1 Try113His rs1051740, His139Arg rs2234922, GSTM1 null, GSTM3 Mnl AGG deletion rs1799735, GSTP1 Ile105Val rs947894, Ala114Val rs1799811, GSTT1 null, NQO1 Pro187Ser rs1800566) pathways were included. Hardy–Weinberg equilibrium was tested in the controls by study. SNPs which did not pass the test in a specific study were excluded from the analyses.
The associations between SNPs and head and neck cancer risk were assessed by estimating odds ratios (OR) and 95% confidence intervals (CI) with unconditional logistic regression models for each study. The models included age (categories), sex, race/ethnicity (categories), and country (categories). In the Central Europe study, information on ethnicity was not collected and all subjects were classified as non-Hispanic white, since the majority of these populations were expected to be white. Pooled ORs were estimated with a fixed effects and random effects (DerSimonian and Laird, 1986) logistic regression model. We assessed heterogeneity across studies by the likelihood ratio test. Stratified analyses were conducted by cancer site (oral, pharynx, oral/pharynx not specified, and larynx), age (<45 and ≥45 years), sex, race (White, Black, Hispanic, and Asian) geographic region (Europe, North America, and Latin America), study type (hospital-based and population-based), study size (<300 cases and ≥300 cases), smoking (never and ever), drinking [≤3 and >3 drinks/day (Hashibe et al., 2007)] and fruit and vegetable intake (lower and higher than center-specific median among controls). The analyses were performed using SAS 9 and significant associations were defined as a two-sided p-value less than 0.05.
False positive report probability (FPRP; Wacholder et al., 2004) was assessed for all associations in which the two-sided null p-value was less than 0.05. FPRP was assessed for OR = 1.5 for probability of true association were 0.25, 0.1, 0.01, and 0.001. For the haplotype analysis, we selected the studies that had information on the multiple SNPs for the gene. We used PHASE v2. (Stephens et al., 2001; Stephens and Donnelly, 2003) to reconstruct the haplotype for genes with multiple SNPs available. Overall, haplotypes for four genes, CYP2E1, EPHX1, GSTP1, and MTHFR, could be reconstructed. The most frequent haplotype was treated as the referent group.
A total of 5,915 cases and 10,644 controls were pooled from 14 studies in Europe (2,759 cases and 4,629 controls), North America (2,234 cases and 3,290 controls), Central America (147 cases and 149 controls), and Asia (775 cases and 2,576 controls). Demographic characteristics of the cases and controls are presented in Table 1. Among the cell cycle and DNA repair SNPs, we observed associations between head and neck cancer risk and MGMT Leu84Phe heterozygotes (OR = 0.79, 95% CI = 0.68–0.93) and XRCC1 Arg194Trp rare homozygotes (OR = 2.3, 95% CI = 1.1–4.7; Table 2).
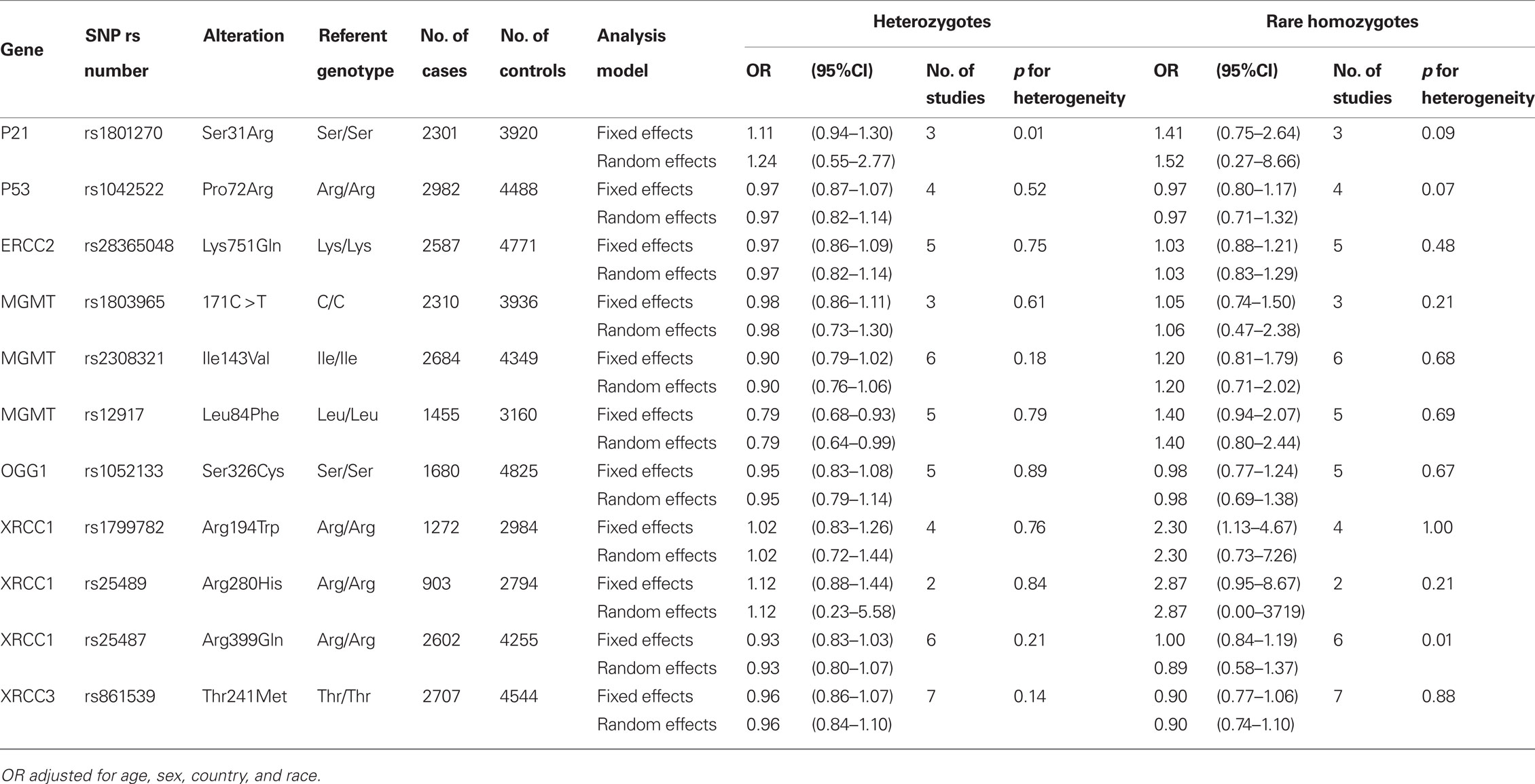
Table 2. Cell cycle and DNA repair SNPs and the risk of head and neck cancer in the INHANCE consortium.
For the carcinogen metabolism SNPs, associations were observed between head and neck cancer risk and ADH1B Arg48His His/His homozygotes (OR = 2.7, 95% CI = 1.9–4.0), ADH1C Ile350Val rare homozygotes (OR = 1.2, 95% CI = 1.1–1.4), and the GSTM1 null type (OR = 1.1, 95% CI = 1.0–1.2; Table 3). Further adjusting for cigarette smoking and alcohol consumption (Table A2 in Appendix) did not change the results greatly. In addition, XRCC1 Arg280His rare homozygotes showed an association with head and neck cancer risk after adjustment of cigarette smoking and alcohol consumption (OR = 3.3, 95% CI = 1.1–10).
Table 4 shows the FPRP for the observed associations for the selected SNPS under a given range of probability of true associations. If the prior probability is greater than 10% and we assume the expected OR = 1.5, the FPRPs for MGMT Leu84Phe, ADH1B Arg48His, ADH1C Ile350Val, and GSTM1 were lower than 20%.
Odds ratios for selected SNPs by head and neck cancer subsite are shown in Table 5. ADH1B His48Arg and ADH1C Ile350Val were consistently associated with oral, pharyngeal, and oral/pharyngeal NOS cancer, but ADH1C Ile350Val was not associated with laryngeal cancer. The association of GSTM1 null genotype was observed only with oral cancer (OR = 1.2 95% CI = 1.0–1.3).
Figure 1 shows the stratified results for the selected SNPs, comparing the rare homozygotes to the common homozygotes. The effects from ADH1B Arg48His tended to be stronger among ever smokers and heavy drinkers than non-smokers and light drinkers. The associations from ADH1C Ile350Val were much stronger among Hispanic and Asian; however, these were based on relatively small numbers (case/control in the rare homozygotes: 16/10 for Hispanic and 2/3 for Asian).
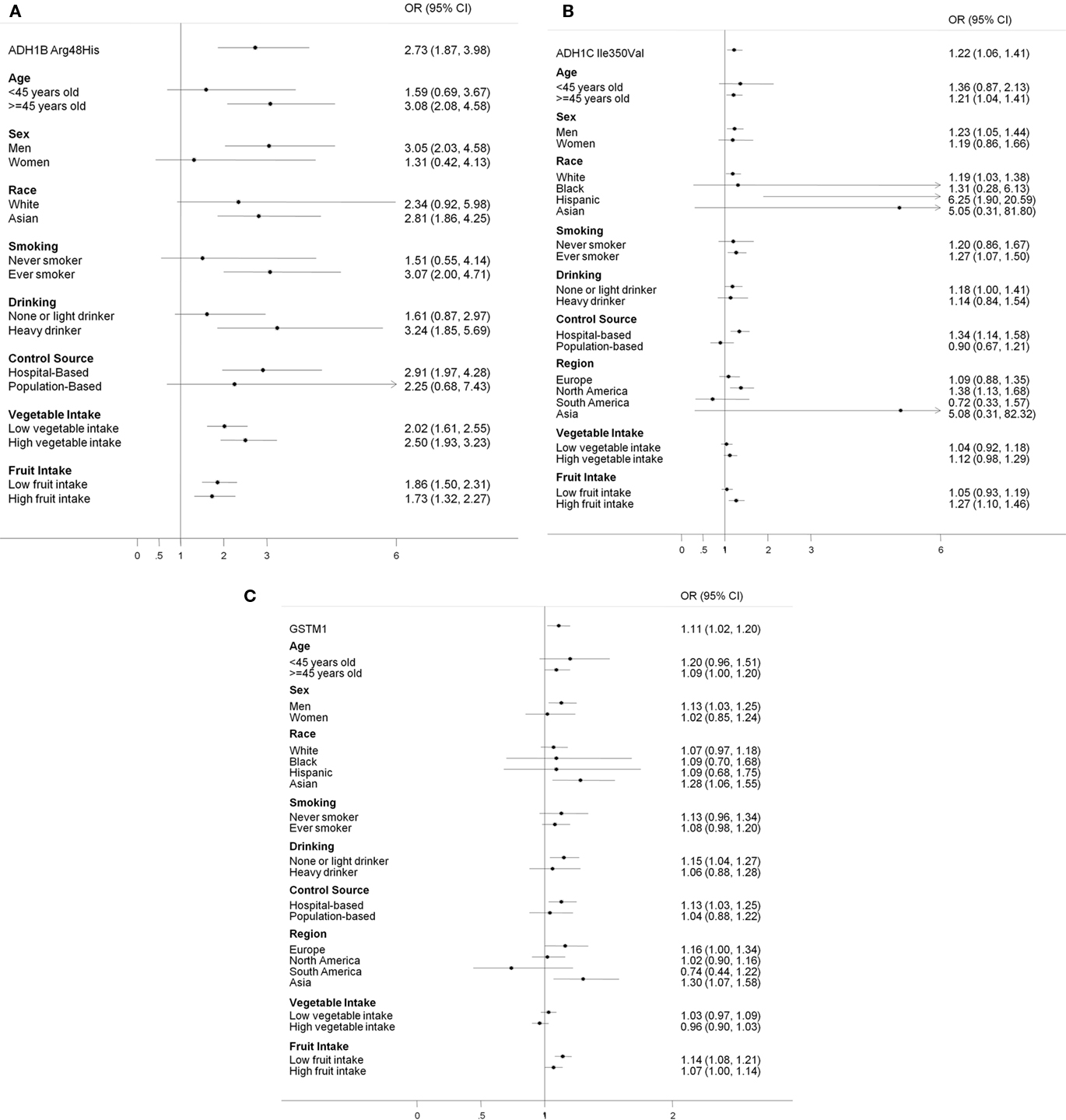
Figure 1. Stratified analysis for selected SNPs: (A) ADH1B Arg48His; His/His vs. Arg/Arg (B) ADH1C Ile350Val: Val/Val vs. Ile/Ile; (C) GSTM1 null genotype.
Table 6 further evaluated the joint effects among smoking, drinking, and the selected SNPs. There was no evidence for the interaction between drinking and ADH1B Arg48His and ADH1C Ile350Val, and smoking and GSTM1 on the risk of head and neck cancer.
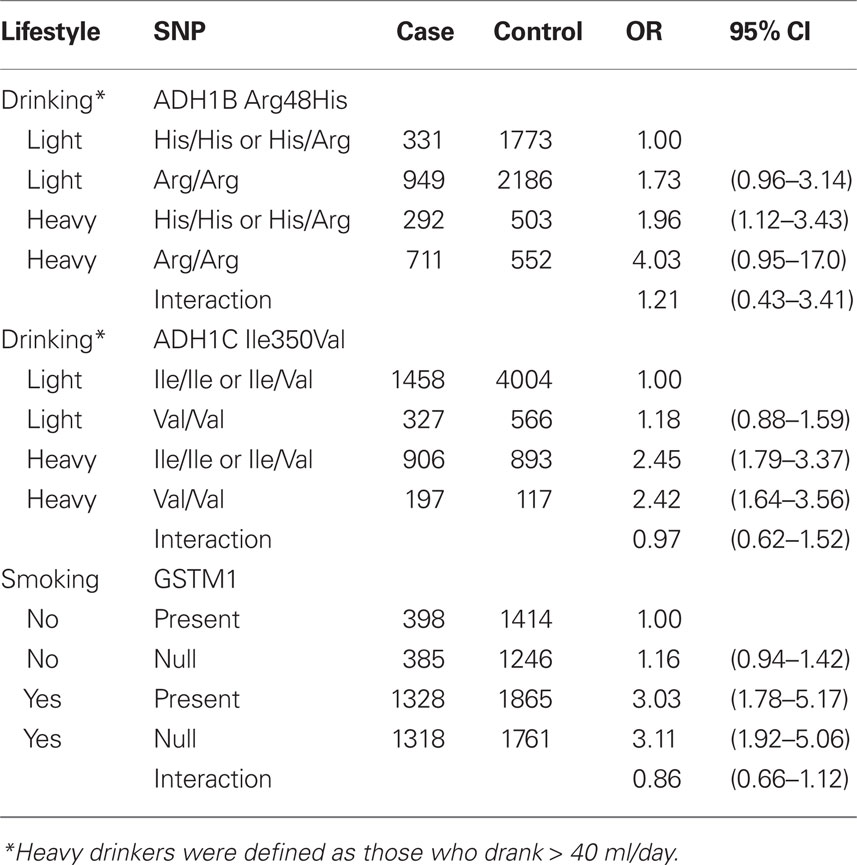
Table 6. Joint effects of drinking and ADH1B Arg48His and ADH1C Ile350Val and smoking and GSTM1 on the risk of head and neck cancer.
No obvious associations were observed for the haplotypes of CYP2E1, EPHX1, GSTP1, and MTHFR (data not shown).
Our results showed associations between head and neck cancer risk and genetic variants in MGMT Leu84Phe, XRCC1 Arg194Trp, ADH1B Arg48His, ADH1C Ile350Val, and the GSTM1. Among them, the results for XRCC1 Arg194Trp might be a false finding because the FPRP was high even if the prior probability was set to be high (0.25).
Among the four studies reported XRCC1 Arg194Trp, none of them were statistically significant with a total of 16 cases and 21 controls in the Trp/Trp group (Table 4). In the International lung cancer consortium, which pooled five studies with 26 cases and 28 controls in the rare homozygote group, reported a non-statistically significant association with increased lung cancer risk (Hung et al., 2008). In a meta-analysis on DNA damage response genes and head and neck cancer reported an increased oral cancer risk for XRCC1 194Trp in the Asian population; however, no association was observed in the Caucasian population (Flores-Obando et al., 2010). A meta-analysis investigated the associations between DNA repair gene polymorphisms and smoking and found that XRCC1 194Trp, 280His, and 399Gln were associated with smoking behaviors (Hodgson et al., 2010). Nevertheless, further adjusted for smoking and drinking behaviors did not change the results materially, even though the association with 280His becomes statistically significant (Table A2 in Appendix). Further investigation into the XRCC1 function and its association in tobacco induced gene repair might be helpful to elucidate the associations with head and neck cancer risks.
Association between MGMT Leu84Phe and head and neck cancer risk was observed in the present report. The effects were similar regarding the cigarette smoking and alcohol drinking status, age, sex, and race (data not shown). The FPRP was 0.787 if the priority probability was very low (0.001). However, the associations between heterozygotes and rare homozygotes and head and neck cancer risk were in different directions. The results for the rare homozygotes were imprecise across the five studies provided the SNP (OR = 1.60, 0.90, 0.80, 1.55, and 0.66). Excluding the largest study which provided 608 cases and 1925 controls (OR = 1.60) in the analyses yield an OR 0f 0.99 (0.33–3.02). Nevertheless, the function of the MGMT Leu84Phe rare allele is not thought to differ from that of common homozygotes (based on the purified protein in the repair of O6-methylguanine in vitro; Pegg et al., 2007). In addition, more than 500 SNPs have been identified in MGMT in Caucasians and ∼60% of the identified SNPs have minor allele frequency greater than 0.05 (Bugni et al., 2007). The MGMT polymorphism might be associated with MGMT hypermethylation (Ogino et al., 2007), which could cause mutations in other critical genes (Mukai and Sekiguchi, 2002). The observed association might be a result of the mutated gene or an unidentified causal SNP. More studies would be needed to explore the associations and the mechanisms between MGMT polymorphisms and head and neck cancer.
Both ADH1B and ADH1C gene belongs to the ADH class I family, which plays a major role in ethanol metabolism (Edenberg, 2000). ADH1C 350Val and 272Arg result in a faster metabolism of ethanol (Hoog et al., 1986; Carr et al., 1989). Earlier associations on ADH1B and head and neck cancer were based on Asian studies (Hori et al., 1997; Yokoyama et al., 1999, 2001; Asakage et al., 2007; Hiraki et al., 2007). Recently, two large case–control studies further confirmed the association in European populations (Hashibe et al., 2006, 2008). The current analysis included the above studies; however, none of the study is influential on the overall pooled estimates. The differences in the published results could be because the frequency of the His allele is extremely low in the European population (International HapMap Project, 2008). In our study, only 1.2% European subjects carried the His/His genotype while 61.3% Asian participants had the polymorphism. Despite the differences in distribution, the effects from the SNP did not differ by populations (Figure 1). The effects from ADH1B Arg/Arg were stronger among ever smokers or heavy drinkers. People who carry the ADH1B Arg/Arg genotype also tended to consume more alcohol than those who carried the His/His or His/Arg genotype (Table A3 in Appendix).
Earlier studies found no association between ADH1C Ile350Val and head and neck cancers (Coutelle et al., 1997; Bouchardy et al., 2000; Olshan et al., 2001; Sturgis et al., 2001; Wang et al., 2005) while some recent studies reported associations as main effects or combined effects with alcohol drinking (Harty et al., 1997; Peters et al., 2005; Hashibe et al., 2006). A large study (Hashibe et al., 2008) combining data from Europe and Latin America further suggested associations of ADH1B, ADH1C, and ADH7 genes with head and neck cancer risk. In the present pooled analysis, which combined data from Europe, North America, Latin America, and Asia, we observed an overall 25% increased risk of head and neck cancer on ADH1C Ile350Val, especially among oral and pharyngeal cancer patients. The result from Asia was very unstable possibly due to the low frequency of the homozygotes in the population. Excluding the Asian study from the analysis did not change the results materially.
ADH1B Arg48His and ADH1C Ile350Val genes are only 100 kb away and have strong linkage disequilibrium (LD > 0.65; International HapMap Project, 2008). However, after restriction to the ADH1B His/His or His/Arg population, ADH1C Val/Val still showed strong association on head and neck cancer risk. The frequency was very low in the European population and thus the statistical power was low (data not shown).
GSTM1 belongs to the glutathione S-transferases (GST) family producing phase II xenobiotic metabolic enzymes. The GSTs play a role in the metabolism of chemical carcinogens, especially with regard to those present in tobacco smoke (Peters et al., 2006). However, epidemiological studies on the associations between GSTs and head and neck cancer have been inconsistent. The inconsistency could be attributed to study design, for example, population-based controls vs. hospital-based controls, matching criteria, and incident cases vs. prevalent cases (Geisler and Olshan, 2001). The GST enzymes are also known to be expressed differently by site (Pacifici et al., 1988; Moscow et al., 1989; Howie et al., 1990). In the present study, GSTM1 null genotype was associated with oral cavity cancer but not the other head and neck cancer sites.
Interestingly, race seems to be a potential effect modifier for the association between GSTM1 and head and neck cancer, with ORs of 1.1 for Whites, Blacks, and Hispanic and 1.3 for Asians, even though the CI overlapped. The OR for Asians in North America was 1.3 (case/control: 28/64, 95% CI = 0.11–16).
In addition, significant results were only observed in the hospital-based studies for the association between GSTM1 and head and neck cancer. Hospital-based studies in our study exclude individuals with previous or malignant disease, including respiratory disease (France, Rome, North Carolina, Northeast US, India), tobacco or alcohol related diseases (Central Europe and Western Europe) or from hospital visitors (Houston). The GSTM1 genotype distribution among the hospital-based controls may not reflect those of the base population in our study. Among controls, the hospital-based studies had lower frequency of the null type GSTM1 than population-based studies (45 vs. 52%, p < 0.0001).
A limitation of our study is that the pooled analysis is based on a heterogeneous population from different geographic regions and ethnicities. However, heterogeneity across studies was not significant for most of our results. In addition, stratified analysis was conducted to assess whether our observations were due to a specific geography and ethnicity. In addition, individual data was available and were harmonized according to standard protocol; thus, we were able to control the potential confounders consistently.
A genome-wide association study (GWAS) was performed independently within the INHANCE consortium using HumanHap300 platform (McKay et al., 2011). The GWAS used the Central Europe and Western Europe studies for the discovery phase and replicated the findings in another subset of the INHANCE consortium: Los Angeles, Houston, Latin America, IARC Multicenter, Boston, and Rome studies, in combination with additional studies not in the current pooled analyses. The ADH1C Ile350Val was strongly associated with the head and neck cancer in the discovery phase (p < 10−5) and was replicated in the replication phase. The ADH1B His48Arg was not tagged on HumanHap300 but was selected as a candidate gene for the replication and showed the strongest association in the replication phase (p = 3 × 10−12). Inconsistency has been observed between results from candidate gene approach and from GWAS in previous studies (Siontis et al., 2010). The consistent findings in the two ADH SNPs in both GWAS (McKay et al., 2011) and a previous analyses (Hashibe et al., 2006) as well as the current pooled analyses based on candidate gene approach further support the role of alcohol metabolism genes on the head and neck cancer etiology. Our observations suggest that, while GWAS may lead to novel hypotheses by elucidating new disease associated loci, traditional hypotheses-driven associations should not be ignored.
In summary, we observed associations between ADH1B Arg48His, ADH1C Ile350Arg, GSTM1 null and head and neck cancer risk. An association for XRCC1 Arg194Trp was not well supported based on the FPRPs. Further investigation is needed for MGMT Leu84Phe for the mechanism. ADH1B Arg48His and ADH1C Ile350Arg could be risk factors or markers of other risk genes for head and neck cancer. Effect of GSTM1 is slightly associated with increased risk of head and neck cancer but inconsistent across subsites.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This pooled analysis of SNPs within INHANCE consortium was supported by a grant from the US National Institutes of Health (NIH), National Institute of Dental, and Craniofacial Research (NIDCR; R03DE016611). Shu-Chun Chuang worked on this project during the tenure of a Special Training Award from the International Agency for Research on Cancer. The individual studies were funded by the following grants: France study: Swiss League against Cancer (KFS1069-09-2000), Fribourg League against Cancer (FOR381.88), Swiss Cancer Research (AKT 617), and Gustave-Roussy Institute (88D28). Central Europe study: World Cancer Research Fund and the European Commission’s INCO-COPERNICUS Program (Contract No. IC15-CT98-0332). Seattle study: National Institutes of Health (NIH) US (R01CA048996, R01DE012609). Iowa: National Institutes of Health (NIH) US (NIDCR R01DE11979, NIDCR R01DE13110, NIH FIRCA TW01500) and Veterans Affairs Merit Review Funds. North Carolina study: National Institutes of Health (NIH) US (R01CA61188), and in part by a grant from the National Institute of Environmental Health Sciences (P30ES010126). Los Angeles study: National Institute of Health (NIH) US (P50CA90388, R01DA11386, R03CA77954, T32CA09142, U01CA96134, R21ES011667) and the Alper Research Program for Environmental Genomics of the UCLA Jonsson Comprehensive Cancer Center. Houston: National Institutes of Health (NIH) US (R01ES11740, R01CA100264). Puerto Rico study: jointly funded by National Institutes of Health (NCI) US and NIDCR intramural programs. Rome study: AIRC (Italian Agency for Research on Cancer). Western Europe Study: European Commission’s 5th Framework Program (Contract No. QLK1-2001-00182), Italian Association for Cancer Research, Compagnia di San Paolo/FIRMS, Region Piemonte, and Padova University (Contract No. CPDA057222). Germany-Heidelberg study: grant No. 01GB9702/3 from the German Ministry of Education and Research. Japan study: Scientific Research grant from the Ministry of Education, Science, Sports, Culture, and Technology of Japan (17015052) and grant for the Third-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labor, and Welfare of Japan (H20-002). Northeast US: National Institutes of Health (NIH) US (R01DE13158). India: Department of Biotechnology Government of India (DBT Grant no: BT/PR2277/MED/09/333/2000 and BT/PR6958/MED/14/912/2005).
International HapMap Project (2008). International HapMap Project. Available at http://hapmap.ncbi.nlm.nih.gov/
Altekruse, S. F., Kosary, C. L., Krapcho, M., Neyman, N., Aminou, R., Waldron, W., Ruhl, J., Howlader, N., Tatalovich, Z., Cho, H., Mariotto, A., Eisner, M. P., Lewis, D. R., Cronin, K., Chen, H. S., Feuer, E. J., Stinchcomb, D. G., and Edwards, B. K. (eds). (2010). SEER Cancer Statistics Review, 1975-2007. Bethesda, MD: National Cancer Institute. Available at: http://seer.cancer.gov/csr/1975_2007/, based on November 2009 SEER data submission, posted to the SEER web site, 2010
Anantharaman, D., Chaubal, P. M., Kannan, S., Bhisey, R. A., and Mahimkar, M. B. (2007). Susceptibility to oral cancer by genetic polymorphisms at CYP1A1, GSTM1 and GSTT1 loci among Indians: tobacco exposure as a risk modulator. Carcinogenesis 7, 1455–1462.
Asakage, T., Yokoyama, A., Haneda, T., Yamazaki, M., Muto, M., Yokoyama, T., Kato, H., Igaki, H., Tsujinaka, T., Kumagai, Y., Yokoyama, M., Omori, T., and Watanabe, H. (2007). Genetic polymorphisms of alcohol and aldehyde dehydrogenases, and drinking, smoking and diet in Japanese men with oral and pharyngeal squamous cell carcinoma. Carcinogenesis 4, 865–874.
Benhamou, S., Tuimala, J., Bouchardy, C., Dayer, P., Sarasin, A., and Hirvonen, A. (2004). DNA repair gene XRCC2 and XRCC3 polymorphisms and susceptibility to cancers of the upper aerodigestive tract. Int. J. Cancer 5, 901–904.
Berrino, F., De Angelis, R., Sant, M., Rosso, S., Bielska-Lasota, M., Coebergh, J. W., and Santaquilani, M. (2007). Survival for eight major cancers and all cancers combined for European adults diagnosed in 1995-99: results of the EUROCARE-4 study. Lancet Oncol. 9, 773–783.
Boccia, S., Cadoni, G., Sayed-Tabatabaei, F. A., Volante, M., Arzani, D., De Lauretis, A., Cattel, C., Almadori, G., van Duijn, C. M., Paludetti, G., and Ricciardi, G. (2008). CYP1A1, CYP2E1, GSTM1, GSTT1, EPHX1 exons 3 and 4, and NAT2 polymorphisms, smoking, consumption of alcohol and fruit and vegetables and risk of head and neck cancer. J. Cancer Res. Clin. Oncol. 1, 93–100.
Bouchardy, C., Hirvonen, A., Coutelle, C., Ward, P. J., Dayer, P., and Benhamou, S. (2000). Role of alcohol dehydrogenase 3 and cytochrome P-4502E1 genotypes in susceptibility to cancers of the upper aerodigestive tract. Int. J. Cancer 5, 734–740.
Bugni, J. M., Han, J., Tsai, M. S., Hunter, D. J., and Samson, L. D. (2007). Genetic association and functional studies of major polymorphic variants of MGMT. DNA Repair (Amst.) 8, 1116–1126.
Canova, C., Hashibe, M., Simonato, L., Nelis, M., Metspalu, A., Lagiou, P., Trichopoulos, D., Ahrens, W., Pigeot, I., Merletti, F., Richiardi, L., Talamini, R., Barzan, L., Macfarlane, G. J., Macfarlane, T. V., Holcatova, I., Bencko, V., Benhamou, S., Bouchardy, C., Kjaerheim, K., Lowry, R., Agudo, A., Castellsague, X., Conway, D. I., McKinney, P. A., Znaor, A., McCartan, B. E., Healy, C. M., Marron, M., and Brennan, P. (2009). Genetic associations of 115 polymorphisms with cancers of the upper aerodigestive tract across 10 European countries: the ARCAGE project. Cancer Res. 7, 2956–2965.
Carr, L. G., Xu, Y., Ho, W. H., and Edenberg, H. J. (1989). Nucleotide sequence of the ADH2(3) gene encoding the human alcohol dehydrogenase beta 3 subunit. Alcohol. Clin. Exp. Res. 4, 594–596.
Coutelle, C., Ward, P. J., Fleury, B., Quattrocchi, P., Chambrin, H., Iron, A., Couzigou, P., and Cassaigne, A. (1997). Laryngeal and oropharyngeal cancer, and alcohol dehydrogenase 3 and glutathione S-transferase M1 polymorphisms. Hum. Genet. 3, 319–325.
Crump, C., Chen, C., Appelbaum, F. R., Kopecky, K. J., Schwartz, S. M., Willman, C. L., Slovak, M. L., and Weiss, N. S. (2000). Glutathione S-transferase theta 1 gene deletion and risk of acute myeloid leukemia. Cancer Epidemiol. Biomarkers Prev. 5, 457–460.
Cui, Y., Morgenstern, H., Greenland, S., Tashkin, D. P., Mao, J., Cao, W., Cozen, W., Mack, T. M., and Zhang, Z. F. (2006). Polymorphism of Xeroderma pigmentosum group G and the risk of lung cancer and squamous cell carcinomas of the oropharynx, larynx and esophagus. Int. J. Cancer 3, 714–720.
DerSimonian, R., and Laird, N. (1986). Meta-analysis in clinical trials. Control. Clin. Trials 3, 177–188.
Edenberg, H. J. (2000). Regulation of the mammalian alcohol dehydrogenase genes. Prog. Nucleic Acid Res. Mol. Biol. 295–341.
Ferlay, J., Shin, H. R., Bray, F., Forman, D., Mathers, C., and Parkin, D. M. (2010). GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC Cancer Base No. 10, International Agency for Research on Cancer, Lyon. Available at: http://globocan.iarc.fr
Flores-Obando, R. E., Gollin, S. M., and Ragin, C. C. (2010). Polymorphisms in DNA damage response genes and head and neck cancer risk. Biomarkers 5, 379–399.
Gaudet, M. M., Olshan, A. F., Chuang, S. C., Berthiller, J., Zhang, Z. F., Lissowska, J., Zaridze, D., Winn, D. M., Wei, Q., Talamini, R., Szeszenia-Dabrowska, N., Sturgis, E. M., Schwartz, S. M., Rudnai, P., Eluf-Neto, J., Muscat, J., Morgenstern, H., Menezes, A., Matos, E., Bucur, A., Levi, F., Lazarus, P., La, V. C., Koifman, S., Kelsey, K., Herrero, R., Hayes, R. B., Franceschi, S., Wunsch-Filho, V., Fernandez, L., Fabianova, E., Daudt, A. W., Dal, M. L., Curado, M. P., Chen, C., Castellsague, X., Benhamou, S., Boffetta, P., Brennan, P., and Hashibe, M. (2010). Body mass index and risk of head and neck cancer in a pooled analysis of case-control studies in the International head and neck cancer epidemiology (INHANCE) consortium. Int. J. Epidemiol. 4, 1091–1102.
Geisler, S. A., and Olshan, A. F. (2001). GSTM1, GSTT1, and the risk of squamous cell carcinoma of the head and neck: a mini-HuGE review. Am. J. Epidemiol. 2, 95–105.
Harty, L. C., Caporaso, N. E., Hayes, R. B., Winn, D. M., Bravo-Otero, E., Blot, W. J., Kleinman, D. V., Brown, L. M., Armenian, H. K., Fraumeni, J. F. Jr., and Shields, P. G. (1997). Alcohol dehydrogenase 3 genotype and risk of oral cavity and pharyngeal cancers. J. Natl. Cancer Inst. 22, 1698–1705.
Hashibe, M., Boffetta, P., Zaridze, D., Shangina, O., Szeszenia-Dabrowska, N., Mates, D., Janout, V., Fabianova, E., Bencko, V., Moullan, N., Chabrier, A., Hung, R., Hall, J., Canzian, F., and Brennan, P. (2006). Evidence for an important role of alcohol- and aldehyde-metabolizing genes in cancers of the upper aerodigestive tract. Cancer Epidemiol. Biomarkers Prev. 4, 696–703.
Hashibe, M., Brennan, P., Benhamou, S., Castellsague, X., Chen, C., Curado, M. P., Dal Maso, L., Daudt, A. W., Fabianova, E., Fernandez, L., Wunsch-Filho, V., Franceschi, S., Hayes, R. B., Herrero, R., Koifman, S., La Vecchia, C., Lazarus, P., Levi, F., Mates, D., Matos, E., Menezes, A., Muscat, J., Eluf-Neto, J., Olshan, A. F., Rudnai, P., Schwartz, S. M., Smith, E., Sturgis, E. M., Szeszenia-Dabrowska, N., Talamini, R., Wei, Q., Winn, D. M., Zaridze, D., Zatonski, W., Zhang, Z. F., Berthiller, J., and Boffetta, P. (2007). Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International head and neck cancer epidemiology consortium. J. Natl. Cancer Inst. 10, 777–789.
Hashibe, M., Brennan, P., Chuang, S. C., Boccia, S., Castellsague, X., Chen, C., Curado, M. P., Dal, M. L., Daudt, A. W., Fabianova, E., Fernandez, L., Wunsch-Filho, V., Franceschi, S., Hayes, R. B., Herrero, R., Kelsey, K., Koifman, S., La, V. C., Lazarus, P., Levi, F., Lence, J. J., Mates, D., Matos, E., Menezes, A., McClean, M. D., Muscat, J., Eluf-Neto, J., Olshan, A. F., Purdue, M., Rudnai, P., Schwartz, S. M., Smith, E., Sturgis, E. M., Szeszenia-Dabrowska, N., Talamini, R., Wei, Q., Winn, D. M., Shangina, O., Pilarska, A., Zhang, Z. F., Ferro, G., Berthiller, J., and Boffetta, P. (2009). Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International head and neck cancer epidemiology consortium. Cancer Epidemiol. Biomarkers Prev. 2, 541–550.
Hashibe, M., Brennan, P., Strange, R. C., Bhisey, R., Cascorbi, I., Lazarus, P., Oude Ophuis, M. B., Benhamou, S., Foulkes, W. D., Katoh, T., Coutelle, C., Romkes, M., Gaspari, L., Taioli, E., and Boffetta, P. (2003). Meta- and pooled analyses of GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes and risk of head and neck cancer. Cancer Epidemiol. Biomarkers Prev. 12, 1509–1517.
Hashibe, M., McKay, J. D., Curado, M. P., Oliveira, J. C., Koifman, S., Koifman, R., Zaridze, D., Shangina, O., Wunsch-Filho, V., Eluf-Neto, J., Levi, J. E., Matos, E., Lagiou, P., Lagiou, A., Benhamou, S., Bouchardy, C., Szeszenia-Dabrowska, N., Menezes, A., Dall’Agnol, M. M., Merletti, F., Richiardi, L., Fernandez, L., Lence, J., Talamini, R., Barzan, L., Mates, D., Mates, I. N., Kjaerheim, K., Macfarlane, G. J., Macfarlane, T. V., Simonato, L., Canova, C., Holcatova, I., Agudo, A., Castellsague, X., Lowry, R., Janout, V., Kollarova, H., Conway, D. I., McKinney, P. A., Znaor, A., Fabianova, E., Bencko, V., Lissowska, J., Chabrier, A., Hung, R. J., Gaborieau, V., Boffetta, P., and Brennan, P. (2008). Multiple ADH genes are associated with upper aerodigestive cancers. Nat. Genet. 6, 707–709.
Hayes, R. B., Bravo-Otero, E., Kleinman, D. V., Brown, L. M., Fraumeni, J. F. Jr., Harty, L. C., and Winn, D. M. (1999). Tobacco and alcohol use and oral cancer in Puerto Rico. Cancer Causes Control 1, 27–33.
Hiraki, A., Matsuo, K., Wakai, K., Suzuki, T., Hasegawa, Y., and Tajima, K. (2007). Gene-gene and gene-environment interactions between alcohol drinking habit and polymorphisms in alcohol-metabolizing enzyme genes and the risk of head and neck cancer in Japan. Cancer Sci. 7, 1087–1091.
Hodgson, M. E., Poole, C., Olshan, A. F., North, K. E., Zeng, D., and Millikan, R. C. (2010). Smoking and selected DNA repair gene polymorphisms in controls: systematic review and meta-analysis. Cancer Epidemiol. Biomarkers Prev. 12, 3055–3086.
Hoog, J. O., Heden, L. O., Larsson, K., Jornvall, H., and Bahr-Lindstrom, H. (1986). The gamma 1 and gamma 2 subunits of human liver alcohol dehydrogenase. cDNA structures, two amino acid replacements, and compatibility with changes in the enzymatic properties. Eur. J. Biochem. 2, 215–218.
Hori, H., Kawano, T., Endo, M., and Yuasa, Y. (1997). Genetic polymorphisms of tobacco- and alcohol-related metabolizing enzymes and human esophageal squamous cell carcinoma susceptibility. J. Clin. Gastroenterol. 4, 568–575.
Howie, A. F., Forrester, L. M., Glancey, M. J., Schlager, J. J., Powis, G., Beckett, G. J., Hayes, J. D., and Wolf, C. R. (1990). Glutathione S-transferase and glutathione peroxidase expression in normal and tumour human tissues. Carcinogenesis 3, 451–458.
Huang, W. Y., Olshan, A. F., Schwartz, S. M., Berndt, S. I., Chen, C., Llaca, V., Chanock, S. J., Fraumeni, J. F. Jr., and Hayes, R. B. (2005). Selected genetic polymorphisms in MGMT, XRCC1, XPD, and XRCC3 and risk of head and neck cancer: a pooled analysis. Cancer Epidemiol. Biomarkers Prev. 7, 1747–1753.
Hung, R. J., Christiani, D. C., Risch, A., Popanda, O., Haugen, A., Zienolddiny, S., Benhamou, S., Bouchardy, C., Lan, Q., Spitz, M. R., Wichmann, H. E., LeMarchand, L., Vineis, P., Matullo, G., Kiyohara, C., Zhang, Z. F., Pezeshki, B., Harris, C., Mechanic, L., Seow, A., Ng, D. P., Szeszenia-Dabrowska, N., Zaridze, D., Lissowska, J., Rudnai, P., Fabianova, E., Mates, D., Foretova, L., Janout, V., Bencko, V., Caporaso, N., Chen, C., Duell, E. J., Goodman, G., Field, J. K., Houlston, R. S., Hong, Y. C., Landi, M. T., Lazarus, P., Muscat, J., McLaughlin, J., Schwartz, A. G., Shen, H., Stucker, I., Tajima, K., Matsuo, K., Thun, M., Yang, P., Wiencke, J., Andrew, A. S., Monnier, S., Boffetta, P., and Brennan, P. (2008). International lung cancer consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol. Biomarkers Prev. 11, 3081–3089.
IARC Working Group/Human Papillomaviruses. (2007). IARC Monograph on the Evaluation of Carcinogenic Risks to Humans, Vol. 90. Lyon: IRAC Press, 1–1438.
Lee, Y. C., Boffetta, P., Sturgis, E. M., Wei, Q., Zhang, Z. F., Muscat, J., Lazarus, P., Matos, E., Hayes, R. B., Winn, D. M., Zaridze, D., Wunsch-Filho, V., Eluf-Neto, J., Koifman, S., Mates, D., Curado, M. P., Menezes, A., Fernandez, L., Daudt, A. W., Szeszenia-Dabrowska, N., Fabianova, E., Rudnai, P., Ferro, G., Berthiller, J., Brennan, P., and Hashibe, M. (2008). Involuntary smoking and head and neck cancer risk: pooled analysis in the International head and neck cancer epidemiology consortium. Cancer Epidemiol. Biomarkers Prev. 8, 1974–1981.
Lohmueller, K. E., Pearce, C. L., Pike, M., Lander, E. S., and Hirschhorn, J. N. (2003). Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2, 177–182.
McKay, J. D., Truong, T., Gaborieau, V., Chabrier, A., Chuang, S. C., Byrnes, G., Zaridze, D., Shangina, O., Szeszenia-Dabrowska, N., Lissowska, J., Rudnai, P., Fabianova, E., Bucur, A., Bencko, V., Holcatova, I., Janout, V., Foretova, L., Lagiou, P., Trichopoulos, D., Benhamou, S., Bouchardy, C., Ahrens, W., Merletti, F., Richiardi, L., Talamini, R., Barzan, L., Kjaerheim, K., Macfarlane, G. J., Macfarlane, T. V., Simonato, L., Canova, C., Agudo, A., Castellsague, X., Lowry, R., Conway, D. I., McKinney, P. A., Healy, C. M., Toner, M. E., Znaor, A., Curado, M. P., Koifman, S., Menezes, A., Wunsch-Filho, V., Neto, J. E., Garrote, L. F., Boccia, S., Cadoni, G., Arzani, D., Olshan, A. F., Weissler, M. C., Funkhouser, W. K., Luo, J., Lubinski, J., Trubicka, J., Lener, M., Oszutowska, D., Schwartz, S. M., Chen, C., Fish, S., Doody, D. R., Muscat, J. E., Lazarus, P., Gallagher, C. J., Chang, S. C., Zhang, Z. F., Wei, Q., Sturgis, E. M., Wang, L. E., Franceschi, S., Herrero, R., Kelsey, K. T., McClean, M. D., Marsit, C. J., Nelson, H. H., Romkes, M., Buch, S., Nukui, T., Zhong, S., Lacko, M., Manni, J. J., Peters, W. H., Hung, R. J., McLaughlin, J., Vatten, L., Njolstad, I., Goodman, G. E., Field, J. K., Liloglou, T., Vineis, P., Clavel-Chapelon, F., Palli, D., Tumino, R., Krogh, V., Panico, S., Gonzalez, C. A., Quiros, J. R., Martinez, C., Navarro, C., Ardanaz, E., Larranaga, N., Khaw, K. T., Key, T., Bueno-de-Mesquita, H. B., Peeters, P. H., Trichopoulou, A., Linseisen, J., Boeing, H., Hallmans, G., Overvad, K., Tjonneland, A., Kumle, M., Riboli, E., Valk, K., Voodern, T., Metspalu, A., Zelenika, D., Boland, A., Delepine, M., Foglio, M., Lechner, D., Blanche, H., Gut, I. G., Galan, P., Heath, S., Hashibe, M., Hayes, R. B., Boffetta, P., Lathrop, M., and Brennan, P. (2011). A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 3, e1001333.
Moscow, J. A., Fairchild, C. R., Madden, M. J., Ransom, D. T., Wieand, H. S., O’Brien, E. E., Poplack, D. G., Cossman, J., Myers, C. E., and Cowan, K. H. (1989). Expression of anionic glutathione-S-transferase and P-glycoprotein genes in human tissues and tumors. Cancer Res. 6, 1422–1428.
Mukai, T., and Sekiguchi, M. (2002). Gene silencing in phenomena related to DNA repair. Oncogene 58, 9033–9042.
Negri, E., Boffetta, P., Berthiller, J., Castellsague, X., Curado, M. P., Dal Maso, L., Daudt, A. W., Fabianova, E., Fernandez, L., Wunsch-Filho, V., Franceschi, S., Hayes, R. B., Herrero, R., Koifman, S., Lazarus, P., Lence, J. J., Levi, F., Mates, D., Matos, E., Menezes, A., Muscat, J., Eluf-Neto, J., Olshan, A. F., Rudnai, P., Shangina, O., Sturgis, E. M., Szeszenia-Dabrowska, N., Talamini, R., Wei, Q., Winn, D. M., Zaridze, D., Lissowska, J., Zhang, Z. F., Ferro, G., Brennan, P., La Vecchia, C., and Hashibe, M. (2009). Family history of cancer: pooled analysis in the International head and neck cancer epidemiology consortium. Int. J. Cancer 2, 394–401.
Ogino, S., Hazra, A., Tranah, G. J., Kirkner, G. J., Kawasaki, T., Nosho, K., Ohnishi, M., Suemoto, Y., Meyerhardt, J. A., Hunter, D. J., and Fuchs, C. S. (2007). MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis 9, 1985–1990.
Olshan, A. F., Weissler, M. C., Watson, M. A., and Bell, D. A. (2000). GSTM1, GSTT1, GSTP1, CYP1A1, and NAT1 polymorphisms, tobacco use, and the risk of head and neck cancer. Cancer Epidemiol. Biomarkers Prev. 2, 185–191.
Olshan, A. F., Weissler, M. C., Watson, M. A., and Bell, D. A. (2001). Risk of head and neck cancer and the alcohol dehydrogenase 3 genotype. Carcinogenesis 1, 57–61.
Pacifici, G. M., Franchi, M., Bencini, C., Repetti, F., Di Lascio, N., and Muraro, G. B. (1988). Tissue distribution of drug-metabolizing enzymes in humans. Xenobiotica 7, 849–856.
Park, J. Y., Schantz, S. P., and Lazarus, P. (2003). Epoxide hydrolase genotype and orolaryngeal cancer risk: interaction with GSTM1 genotype. Oral Oncol. 5, 483–490.
Parkin, D. M., Bray, F., Ferlay, J., and Pisani, P. (2005). Global cancer statistics, 2002. CA Cancer J. Clin. 2, 74–108.
Pegg, A. E., Fang, Q., and Loktionova, N. A. (2007). Human variants of O6-alkylguanine-DNA alkyltransferase. DNA Repair (Amst.) 8, 1071–1078.
Peters, E. S., McClean, M. D., Liu, M., Eisen, E. A., Mueller, N., and Kelsey, K. T. (2005). The ADH1C polymorphism modifies the risk of squamous cell carcinoma of the head and neck associated with alcohol and tobacco use. Cancer Epidemiol. Biomarkers Prev. 2, 476–482.
Peters, E. S., McClean, M. D., Marsit, C. J., Luckett, B., and Kelsey, K. T. (2006). Glutathione S-transferase polymorphisms and the synergy of alcohol and tobacco in oral, pharyngeal, and laryngeal carcinoma. Cancer Epidemiol. Biomarkers Prev. 11, 2196–2202.
Risch, A., Ramroth, H., Raedts, V., Rajaee-Behbahani, N., Schmezer, P., Bartsch, H., Becher, H., and Dietz, A. (2003). Laryngeal cancer risk in Caucasians is associated with alcohol and tobacco consumption but not modified by genetic polymorphisms in class I alcohol dehydrogenases ADH1B and ADH1C, and glutathione-S-transferases GSTM1 and GSTT1. Pharmacogenetics 4, 225–230.
Schwartz, S. M., Doody, D. R., Fitzgibbons, E. D., Ricks, S., Porter, P. L., and Chen, C. (2001). Oral squamous cell cancer risk in relation to alcohol consumption and alcohol dehydrogenase-3 genotypes. Cancer Epidemiol. Biomarkers Prev. 11, 1137–1144.
Siontis, K. C., Patsopoulos, N. A., and Ioannidis, J. P. (2010). Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur. J. Hum. Genet. 7, 832–837.
Stephens, M., and Donnelly, P. (2003). A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 5, 1162–1169.
Stephens, M., Smith, N. J., and Donnelly, P. (2001). A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 4, 978–989.
Sturgis, E. M., Dahlstrom, K. R., Guan, Y., Eicher, S. A., Strom, S. S., Spitz, M. R., and Wei, Q. (2001). Alcohol dehydrogenase 3 genotype is not associated with risk of squamous cell carcinoma of the oral cavity and pharynx. Cancer Epidemiol. Biomarkers Prev. 3, 273–275.
Sturgis, E. M., and Wei, Q. (2002). Genetic susceptibility–molecular epidemiology of head and neck cancer. Curr. Opin. Oncol. 3, 310–317.
Suzuki, T., Matsuo, K., Hasegawa, Y., Hiraki, A., Wakai, K., Hirose, K., Saito, T., Sato, S., Ueda, R., and Tajima, K. (2007). One-carbon metabolism-related gene polymorphisms and risk of head and neck squamous cell carcinoma: case-control study. Cancer Sci. 9, 1439–1446.
Wacholder, S., Chanock, S., Garcia-Closas, M., El Ghormli, L., and Rothman, N. (2004). Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 6, 434–442.
Wang, D., Ritchie, J. M., Smith, E. M., Zhang, Z., Turek, L. P., and Haugen, T. H. (2005). Alcohol dehydrogenase 3 and risk of squamous cell carcinomas of the head and neck. Cancer Epidemiol. Biomarkers Prev. 3, 626–632.
World Cancer Research Fund/American Institute for Cancer Research. (2007). Food, Nutrition, Physical Activity, and the Prevention of Cancer: a Global Perspective, AICR, Washington, DC.
Yokoyama, A., Muramatsu, T., Omori, T., Matsushita, S., Yoshimizu, H., Higuchi, S., Yokoyama, T., Maruyama, K., and Ishii, H. (1999). Alcohol and aldehyde dehydrogenase gene polymorphisms influence susceptibility to esophageal cancer in Japanese alcoholics. Alcohol. Clin. Exp. Res. 11, 1705–1710.
Yokoyama, A., Muramatsu, T., Omori, T., Yokoyama, T., Matsushita, S., Higuchi, S., Maruyama, K., and Ishii, H. (2001). Alcohol and aldehyde dehydrogenase gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis 3, 433–439.
Keywords: SNP, head and neck cancer, INHANCE
Citation: Chuang S-C, Agudo A, Ahrens W, Anantharaman D, Benhamou S, Boccia S, Chen C, Conway DI, Fabianova E, Hayes RB, Healy CM, Holcatova I, Kjaerheim K, Lagiou P, Lazarus P, Macfarlane TV, Mahimkar MB, Mates D, Matsuo K, Merletti F, Metspalu A, Morgenstern H, Muscat J, Cadoni G, Olshan AF, Purdue M, Ramroth H, Rudnai P. Schwartz SM, Simonato L, Smith EM, Sturgis EM, Szeszenia-Dabrowska N. Talamini R, Thomson P, Wei Q, Zaridze D, Zhang Z-F, Znaor A, Brennan P, Boffetta P and Hashibe M (2011) Sequence variants and the risk of head and neck cancer: pooled analysis in the INHANCE consortium. Front. Oncol. 1:13. doi: 10.3389/fonc.2011.00013
Received: 20 March 2011; Paper pending published: 11 April 2011;
Accepted: 13 June 2011; Published online: 12 July 2011.
Edited by:
Min Dai, Cancer Institute and Hospital Chinese Academy of Medical Sciences, ChinaReviewed by:
Lifang Hou, Northwestern University, USACopyright: © 2011 Chuang, Agudo, Ahrens, Anantharaman, Benhamou, Boccia, Chen, Conway, Fabianova, Hayes, Healy, Holcatova, Kjaerheim, Lagiou, Lazarus, Macfarlane, Mahimkar, Mates, Matsuo, Merletti, Metspalu, Morgenstern, Muscat, Cadoni, Olshan, Purdue, Ramroth, Rudnai, Schwartz, Simonato, Smith, Sturgis, Szeszenia-Dabrowska, Talamini, Thomson, Wei, Zaridze, Zhang, Znaor, Brennan, Boffetta and Hashibe. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Mia Hashibe, Division of Public Health, Department of Family and Preventive Medicine, University of Utah School of Medicine, 375 Chipeta Way, Suite A, Salt Lake City, UT 84108, USA. e-mail:bWlhLmhhc2hpYmVAdXRhaC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.