- 1School of Nursing, Hunan University of Chinese Medicine, Changsha, Hunan, China
- 2Key Laboratory of Hunan Province for Prevention and Treatment of Integrated Traditional Chinese and Western Medicine on Cardiocerebral Diseases, Hunan University of Chinese Medicine, Changsha, Hunan, China
Objective: Cerebral ischemia can cause mild damage to local brain nerves due to hypoxia and even lead to irreversible damage due to neuronal cell death. However, the underlying pathogenesis of this phenomenon remains unclear. This study utilized bioinformatics to explore the role of cuproptosis in cerebral ischemic disease and its associated biomarkers.
Method: R software identified the overlap of cerebral ischemia and cuproptosis genes, analyzed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG), and explored hub genes. Expressions and localizations of hub genes in brain tissue, cells, and immune cells were analyzed, along with predictions of protein structures, miRNAs, and transcription factors. A network was constructed depicting hub gene co-expression with miRNAs and interactions with transcription factors. Ferredoxin 1 (FDX1) expression was determined using qRT-PCR.
Results: Ten cuproptosis-related genes in cerebral ischemia were identified, with GO analysis revealing involvement in acetyl-CoA synthesis, metabolism, mitochondrial function, and iron–sulfur cluster binding. KEGG highlighted processes like the tricarboxylic acid cycle, pyruvate metabolism, and glycolysis/gluconeogenesis. Using the Human Protein Atlas, eight hub genes associated with cuproptosis were verified in brain tissues, hippocampus, and AF22 cells. Lipoyl(octanoyl) transferase 1 (LIPT1), was undetected, while others were found in mitochondria or both nucleus and mitochondria. These genes were differentially expressed in immune cells. FDX1, lipoic acid synthetase (LIAS), dihydrolipoamide S-acetyltransferase (DLAT), pyruvate dehydrogenase E1 component subunit alpha 1 (PDHA1), PDHB, and glutaminase (GLS) were predicted to target 111 miRNAs. PDHA1, FDX1, LIPT1, PDHB, LIAS, DLAT, GLS, and dihydrolipoamide dehydrogenase (DLD) were predicted to interact with 11, 10, 10, 9, 8, 7, 5, and 4 transcription factors, respectively. Finally, FDX1 expression was significantly upregulated in the hippocampus of ovariectomized rats with ischemia.
Conclusion: This study revealed an association between cerebral ischemic disease and cuproptosis, identifying eight potential target genes. These findings offer new insights into potential biomarkers for the diagnosis, treatment, and prognosis of cerebral ischemia, and provide avenues for the exploration of new medical intervention targets.
Background
Cerebral ischemia involves complex pathophysiological processes in cells, such as oxidative stress, calcium overload, mitochondrial damage, and excitatory amino acid toxicity. These processes can activate cell death pathways, including apoptosis, programmed necrosis, autophagy, ferroptosis, and pyroptosis (1, 2). Recently, copper-induced cell death, termed cuproptosis, has been established as a novel form of cell death, differing from other programmed cell death mechanisms. Cuproptosis is induced by the binding of copper ions to thioacylated proteins in the tricarboxylic acid (TCA) cycle. This binding leads to abnormal oligomerization of thioacylated proteins, downregulation of Fe-S cluster protein levels, protein toxicity stress, and ultimately cell death (3).
Previous studies have demonstrated that copper ions are involved in the onset and progression of ischemic stroke (IS) (4, 5). Furthermore, plasma copper levels are positively correlated with the risk of initial IS (6, 7), and elevated plasma copper levels are significantly associated with an increased risk of IS (8). These findings underscore the importance of better understanding the relationship between cerebral ischemia and cuproptosis. However, cuproptosis biomarkers in cerebral ischemia have not been fully characterized. Therefore, this study aimed to investigate the hub genes and biomarkers related to cuproptosis in cerebral ischemia using bioinformatics. Furthermore, the research has not only preliminarily validated through Human Protein Atlas (HPA), but also further experimentally validated in model animals to ensure the accuracy and reliability of the research conclusions.
Methods
Intersection of cerebral ischemia and cuproptosis datasets
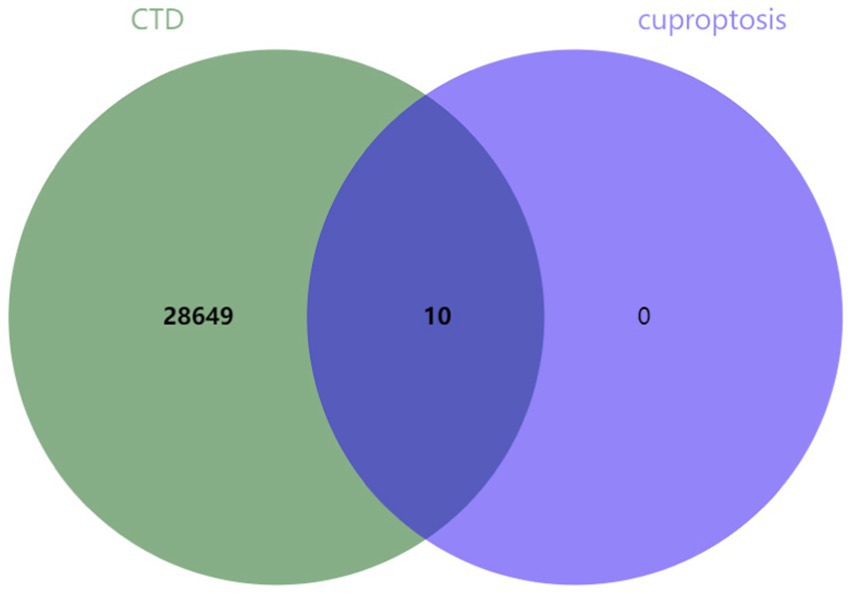
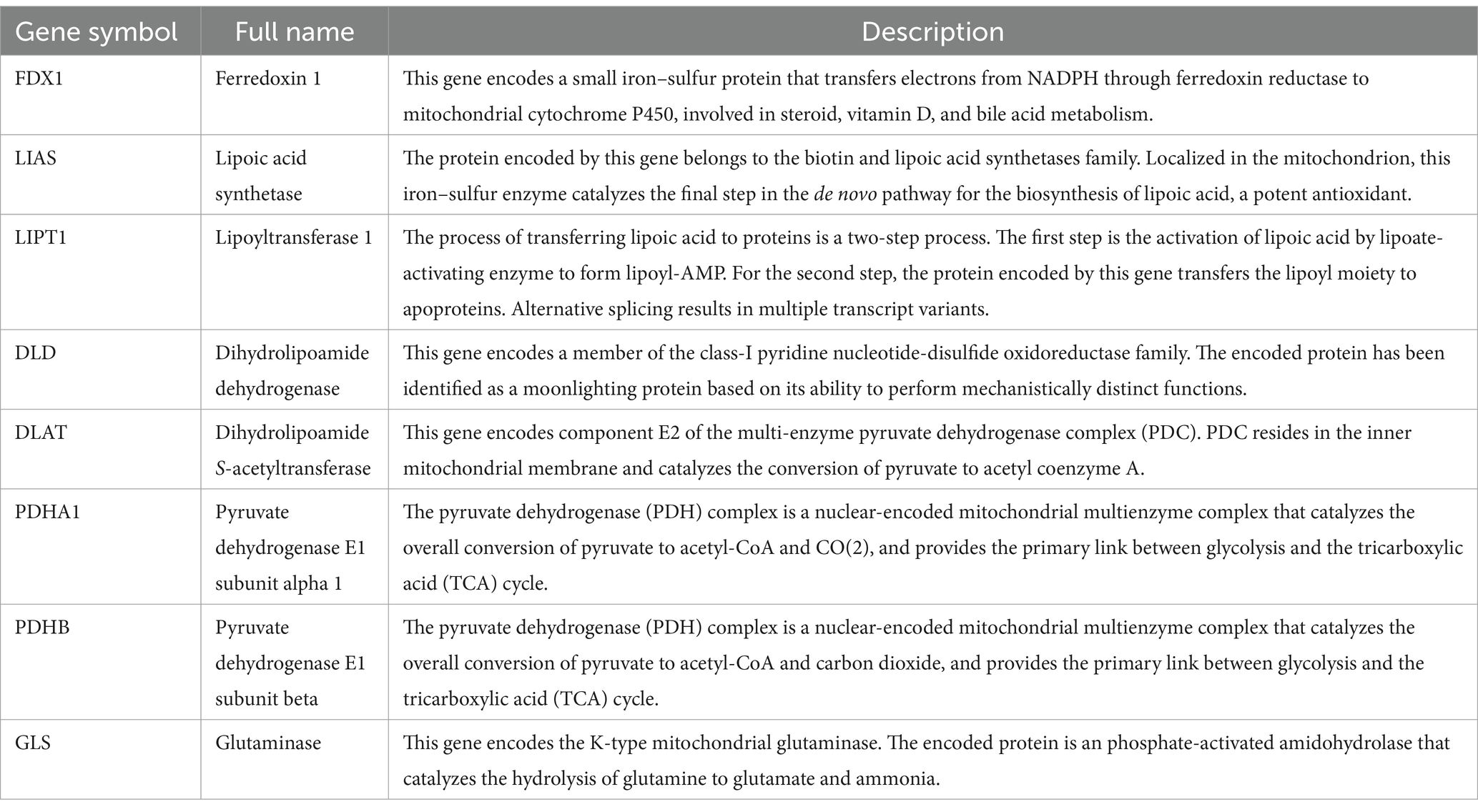
Select “Diseases” through keyword search, enter “cerebral ischemia,” and click search, cerebral ischemia-related genes were downloaded from the Comparative Toxicogenomics Database (CTD).1 Ten genes associated with cuproptosis, namely ferredoxin 1 (FDX1), lipoyl(octanoyl) transferase 1 (LIPT1), lipoic acid synthetase (LIAS), dihydrolipoamide dehydrogenase (DLD), metal regulatory transcription factor 1 (MTF1), dihydrolipoamide S-acetyltransferase (DLAT), pyruvate dehydrogenase E1 component subunit alpha 1 (PDHA1), PDHB, glutaminase (GLS), and cyclin-dependent kinase inhibitor 2A (CDKN2A), were identified in a previous study by Tsvetkov et al. (3). The genes obtained from both datasets were intersected and visualized using jvenn online tool.2
Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses
The GO/KEGG analyses were performed using clusterProfiler (4.4.4) packages in R (4.2.1) software. The ID conversion of the input molecular lists was converted via the org.Hs.eg.db package in the ID conversion library (9). Enrichment analysis and visualization were performed using the ggplot2 (3.3.6), igraph (1.4.1), and ggraph packages (2.1.0) (10).
Hub gene analysis
Protein interactions were analyzed using the STRING Database (Search Tool for the Retrieval of Interacting Genes/Proteins; version 11.0, http://string-db.org) (11). The gene cluster was analyzed using the maximum clique centrality and degree methods (MCODE) plug-in of the Cytoscape software (3.7.2). The full names and descriptions of the hub genes were extracted from Gene Cards.3
Expression, localization, and protein structure of hub genes in brain tissues, brain cells, and immune cells
The expression of hub genes was verified using the HPA online database in brain tissues, human neuroepithelial stem cells (AF22 cells), and immune cells.4 Subcellular localization of hub genes and prediction of their protein structure are shown. Regarding the confidence of the predicted protein structure, deep blue is used to indicate very high confidence (pLDDT >90), light blue is used to indicate high confidence (90 > pLDDT >70), yellow is used to indicate low confidence (70 > pLDDT >50), and orange is used to indicate very low confidence (pLDDT <50) (see Footnote 4).
Co-expression network of hub genes and micro RNAs (miRNAs)
The target miRNAs associated with hub genes related to cuproptosis were predicted using the miRTarBas9.0 database,5 and two validation methods were selected to verify the predicted target miRNAs. Subsequently, a co-expression network was constructed using Cytoscape software.
Transcription factor (TF)–gene interactions
The TFs of hub genes related to cuproptosis were searched using the NetworkAnalyst database.6 The JASPAR database7 was selected as the TF and gene interaction database, and the interaction relationships were visualized using the JASPAR database (see Footnote 7).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Previous studies on cerebral ischemia often used male rats to minimize estrogen influence. However, the soaring incidence of IS among postmenopausal women (12), with enlarged infarction and aggravated brain damage in ovariectomized rats (13), poses a critical threat. Our study employed female rats with ovariectomy followed by cerebral ischemia induction for animal experimentation.
According to body weight, 8-week-old female SD rats were randomly divided into sham and model groups, each group with three rats. Rats were adapted to an SPF environment at the Experimental Animal Center of Hunan University of Chinese Medicine for 1 week. Referring to previous literature (14), the ovariectomy was performed and the cerebral ischemia model was prepared, the specific steps are as follows: the rats were fasted for 12 h before surgery and anesthetized with 2% pentobarbital sodium via intraperitoneal injection. The model group rats quickly made approximately 3 cm incisions on both sides of their backs under sterile conditions, and after tightening the fallopian tubes, the ovaries were removed and the incisions were sutured. The back skin of rats in the sham group was cut open, bilateral ovaries were separated, and then directly sutured. After surgery, rats were administered with 160,000 units of penicillin via intraperitoneal injection daily for a total of 3 days. Starting from the 5th day post-surgery, vaginal smears were collected from the rats once daily for a total of 5 days. The success of modeling was determined by the absence of an estrous cycle response during this period. Starting from the 5th day after surgery, vaginal secretion smears from rats were taken for smear testing once a day for a total of 5 days, and the successful modeling was considered as no occurrence of estrous cycle reactions. After the successful ovariectomy model in rats, fasting for 12 h was performed. On the 12th day post-surgery, they were anesthetized by intraperitoneal injection of 2% pentobarbital sodium. A midline incision was made on the neck, blunt dissection was performed, and the right common carotid artery was exposed. The internal and external carotid arteries were separated, and the proximal and distal ends of the external carotid artery were ligated. A monofilament nylon suture was inserted into the internal carotid artery to a depth of (18.5 ± 0.5) mm until slight resistance was felt, and then the suture was fixed. The incision was sutured layer by layer. Neurological function was assessed using the Longa score 24 h after cerebral ischemia, with scores of 1 to 3 indicating a successful model. The skin of the rats in the sham surgery group was cut open, and the right common carotid artery was separated, and sutured.
After successful modeling, the rats were anesthetized via intraperitoneal injection of 2% pentobarbital sodium and sacrificed, and their hippocampal tissue was collected. Total RNA was extracted and synthesized into cDNA using the total RNA Extraction Kit (Jiangsu Cowin Biotech Co., Ltd., CW0581S) and qRT-PCR. The expression of FDX1 was assessed using the 2-ΔΔCt method (forward: AAGAACCGAGATGGTGAAAC, reverse: AGAGCAAGCCAAAGTCCC, 126 bp).
Statistical analysis
The data calculations and statistical analysis were performed using IBM SPSS Statistics 25.0. Independent sample t-test was used for intergroup comparisons (p < 0.05).
Results
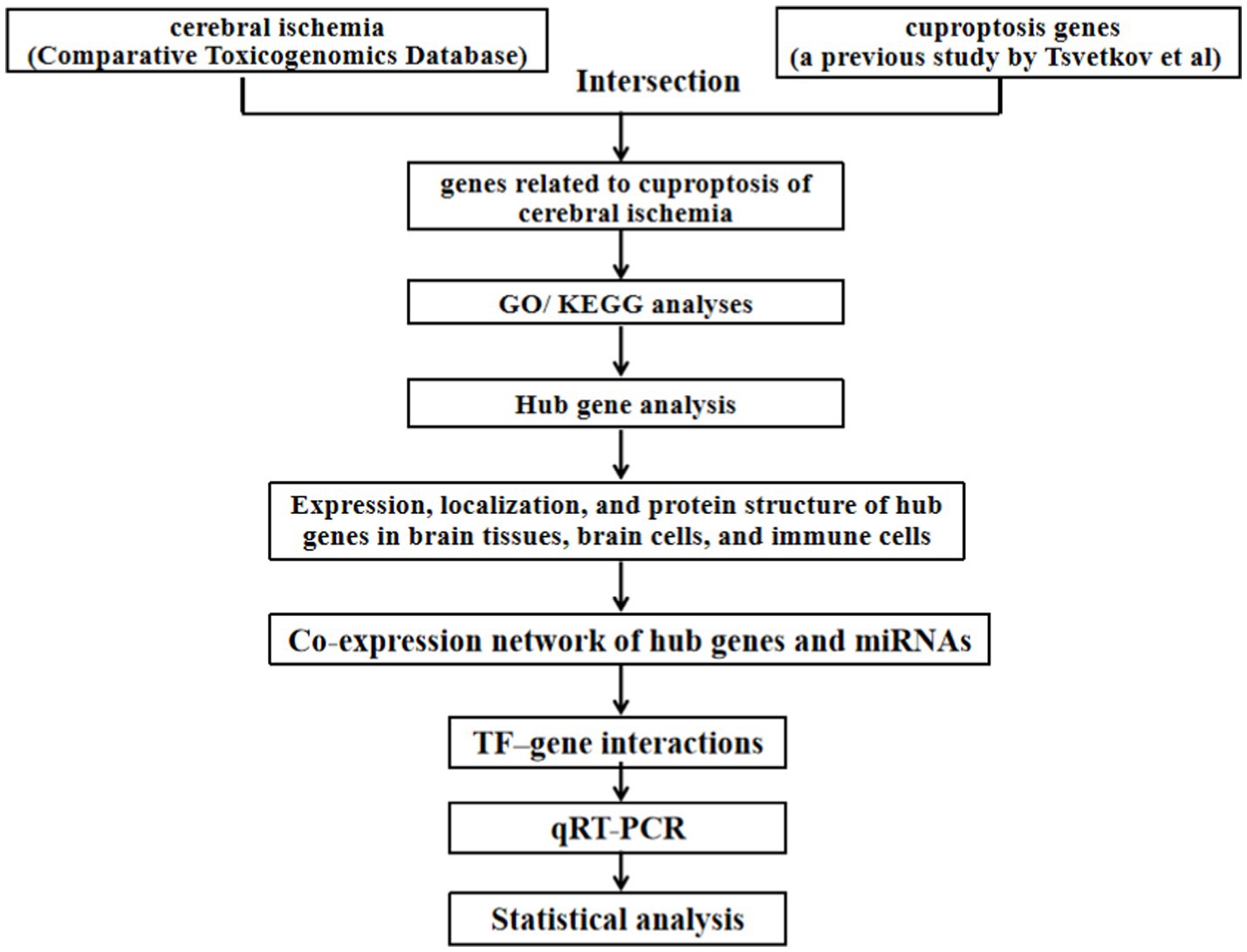
The flowchart of this study was shown in Figure 1.
GO and KEGG enrichment analyses for cuproptosis genes in cerebral ischemia
Ten genes (FDX1, LIPT1, LIAS, DLD, MTF1, DLAT, PDHA1, PDHB, GLS, and CDKN2A) associated with cuproptosis were intersected with genes related to brain ischemia, revealing that all 10 genes were brain ischemia-related genes (refer to Figure 2).
The significance cut-off value for the GO/KEGG analyses was set at a corrected p-value (p.adj) of <0.05. The results of the identified biological processes (BPs), cellular components (CCs), and molecular functions (MFs) were 81, 6, and 15, respectively. KEGG analysis revealed enrichment in 8 pathways. The top 5 GO and KEGG pathways are presented in Tables 1, 2, as well as Figure 3.
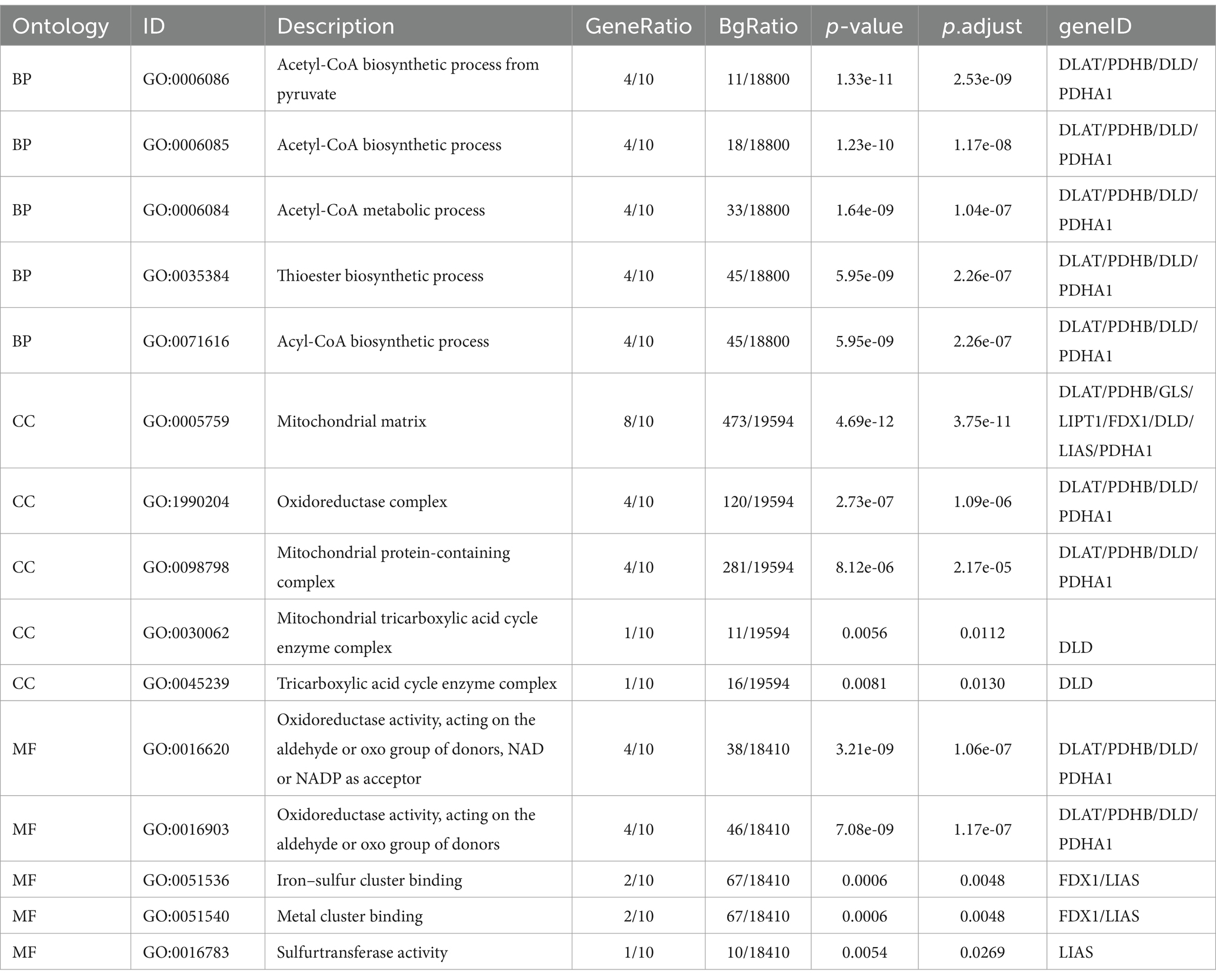
Table 1. Biological process, cellular composition, and molecular function analysis results for genes related to cuproptosis in cerebral ischemia.
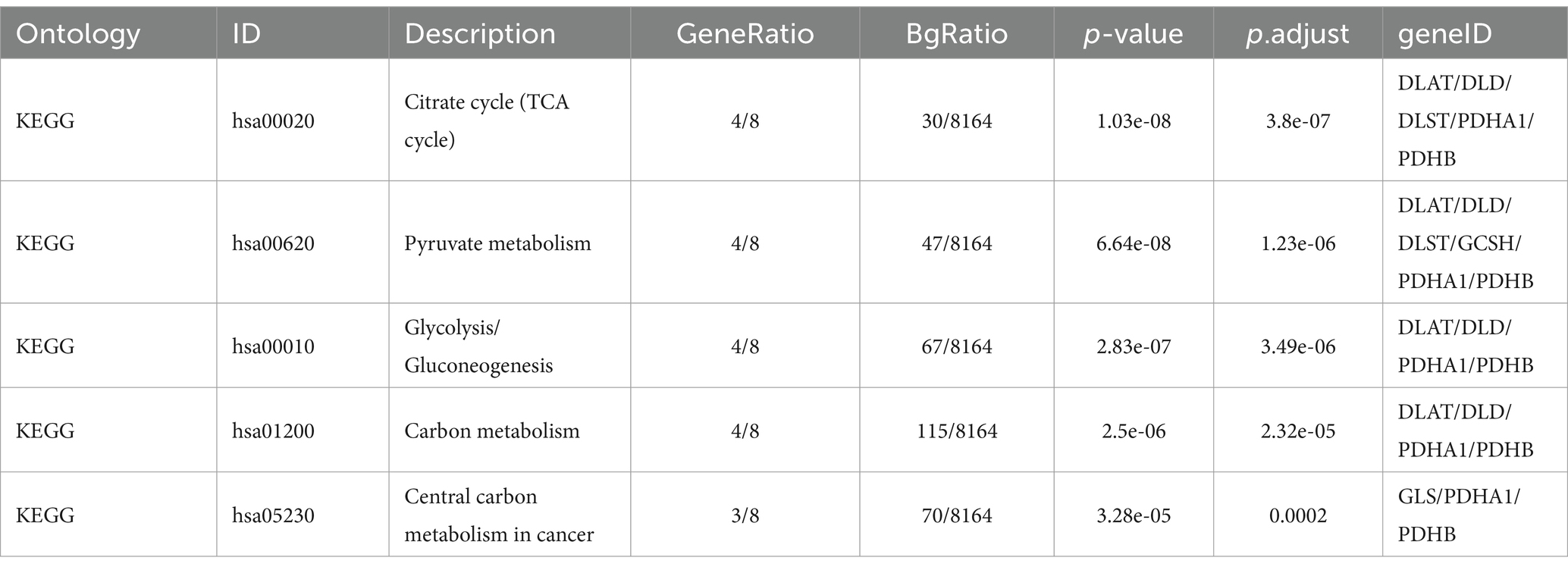
Table 2. Kyoto Encyclopedia of Genes and Genomes analysis results for genes related to cuproptosis in cerebral ischemia.
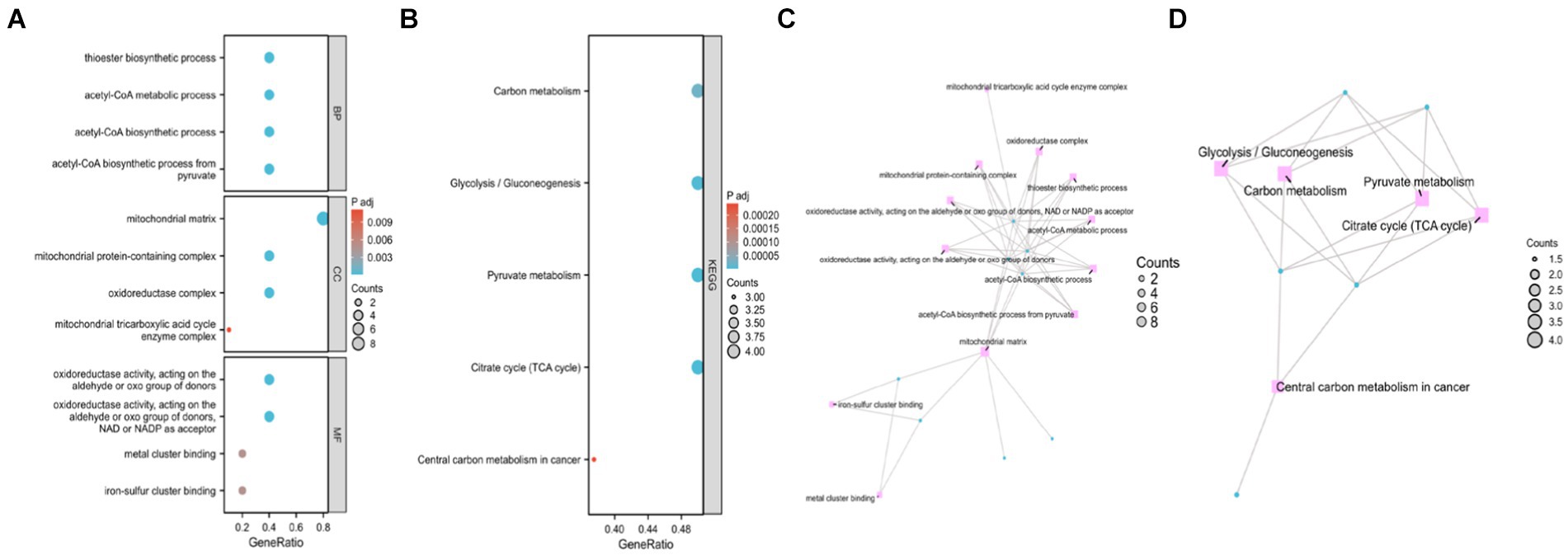
Figure 3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of genes related to cuproptosis in cerebral ischemia. (A) Bubble Chart of top four enriched biological process (BP) terms, cellular components (CCs), and molecular function (MF) terms for genes related to cuproptosis on cerebral ischemia. (B) Bubble Chart of top five KEGG pathways for genes related to cuproptosis in cerebral ischemia. (C) Network diagram of top four enriched BP, (CC), and MF terms for genes related to cuproptosis in cerebral ischemia. (D) Network diagram of five KEGG pathways of genes related to cuproptosis in cerebral ischemia.
Enriched BPs included acetyl-CoA biosynthetic process from pyruvate, acetyl-CoA metabolic process, acetyl-CoA metabolic process, acetyl-CoA biosynthetic process, acetyl-CoA biosynthetic process, and thioester biosynthetic process. CCs encompassed various complexes, such as mitochondrial matrix, oxidoreductase complex, mitochondrial protein-containing complex, mitochondrial tricarboxylic acid cycle enzyme complex, and tricarboxylic acid cycle enzyme complex. MFs included activities such as oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor, oxidoreductase activity, acting on the aldehyde or oxo group of donors, iron–sulfur cluster binding, metal cluster binding, and sulfurtransferase activity. Moreover, KEGG analysis revealed involvement in processes such as pyruvate metabolism, citrate cycle (TCA cycle), carbon metabolism, glycolysis/gluconeogenesis, and central carbon metabolism in cancer.
Cuproptosis-related hub gene identification and their expression in the brain
Eight hub genes (FDX1, LIPT1, LIAS, DLD, PDHA1, DLAT, PDHB, and GLS) were identified as potential core targets of cerebral ischemia (Figure 4A and Table 3). Using the HPA database (see footnote 4), the expression of the hub genes was analyzed in brain tissues, revealing that these genes associated with cuproptosis were expressed in various regions of the human brain, such as the cortex, hippocampus, amygdala, basal ganglia, thalamus, hypothalamus, midbrain, white matter of the spinal cord, and other areas (Figure 4B). Furthermore, these genes were expressed in specific regions within the internal structure of the hippocampus (Figure 4C).
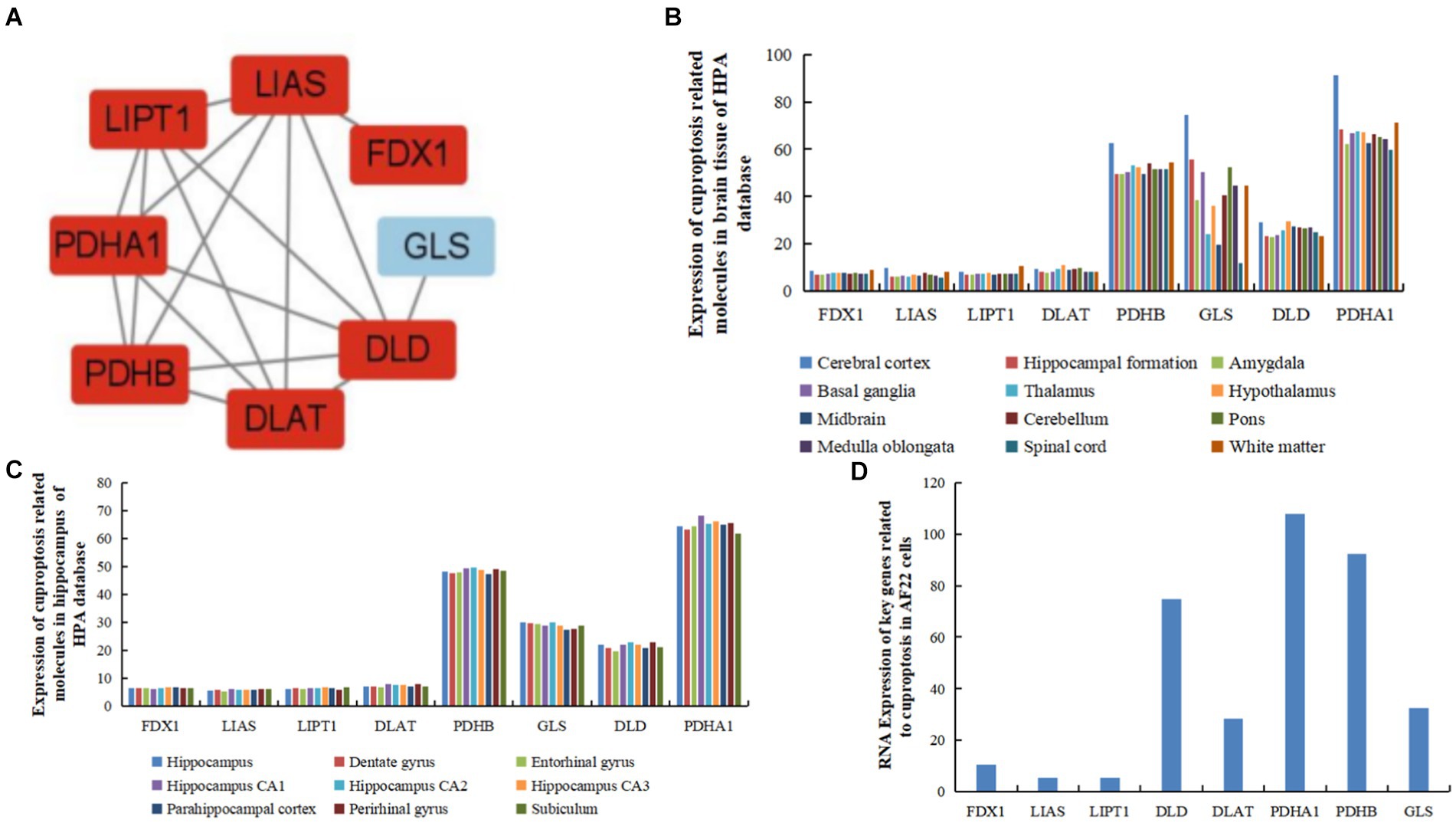
Figure 4. Expression of hub genes related to cuproptosis and cerebral ischemia in brain tissues, and human and AF22 cells. (A) Hub genes of genes related to cuproptosis in cerebral ischemia identified using MCODE. (B) Expression of hub genes related to cuproptosis in brain tissues in the HPA database. (C) Expression of hub genes related to cuproptosis in the hippocampus in the HPA database. (D) RNA Expression of hub genes related to cuproptosis in human neuroepithelial stem cells (AF22 cells).
In human neuroepithelial stem cells (AF22 cells) from the HPA database, PDHA1 exhibited the highest expression, followed by PDHB, DLD, GLS, DLAT, and FDX1. In contrast, LIAS and LIPT1 exhibited the lowest expression levels (Figure 4D).
Subcellular localization and structural prediction of cuproptosis-related hub gene
The subcellular localization results indicated that FDX1, DLAT, PDHA1, and GLS were detected only in the mitochondria, whereas LIAS, DLD, and PDHB were detected in both the nucleus and mitochondria (Figure 5). The predicted structure diagrams of the hub genes and predicted structures of variant populations are shown in Figures 6A,B, respectively.
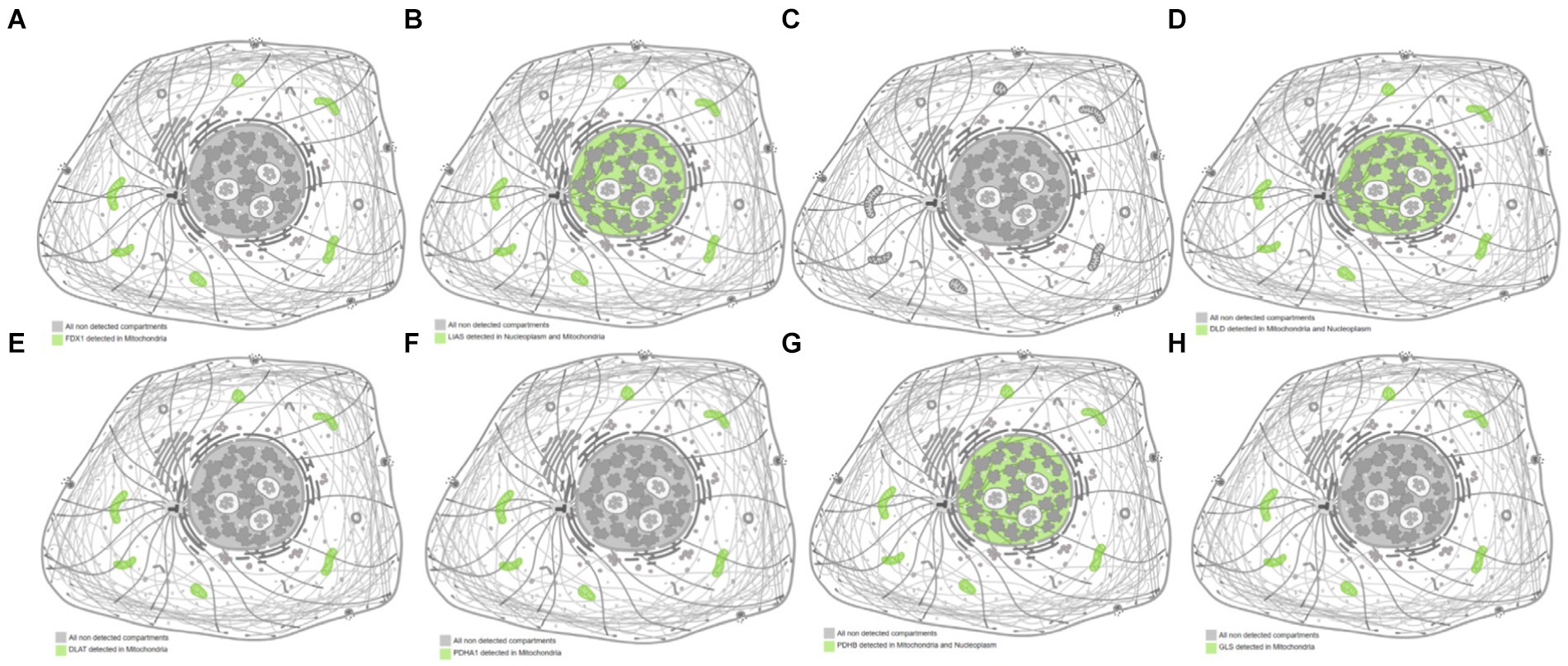
Figure 5. Subcellular localization of hub genes related to cuproptosis and cerebral ischemia. (A) FDX1; (B) LIAS; (C) LIPT1; (D) DLD; (E) DLAT; (F) PDHA1; (G) PDHB; (H) GLS.
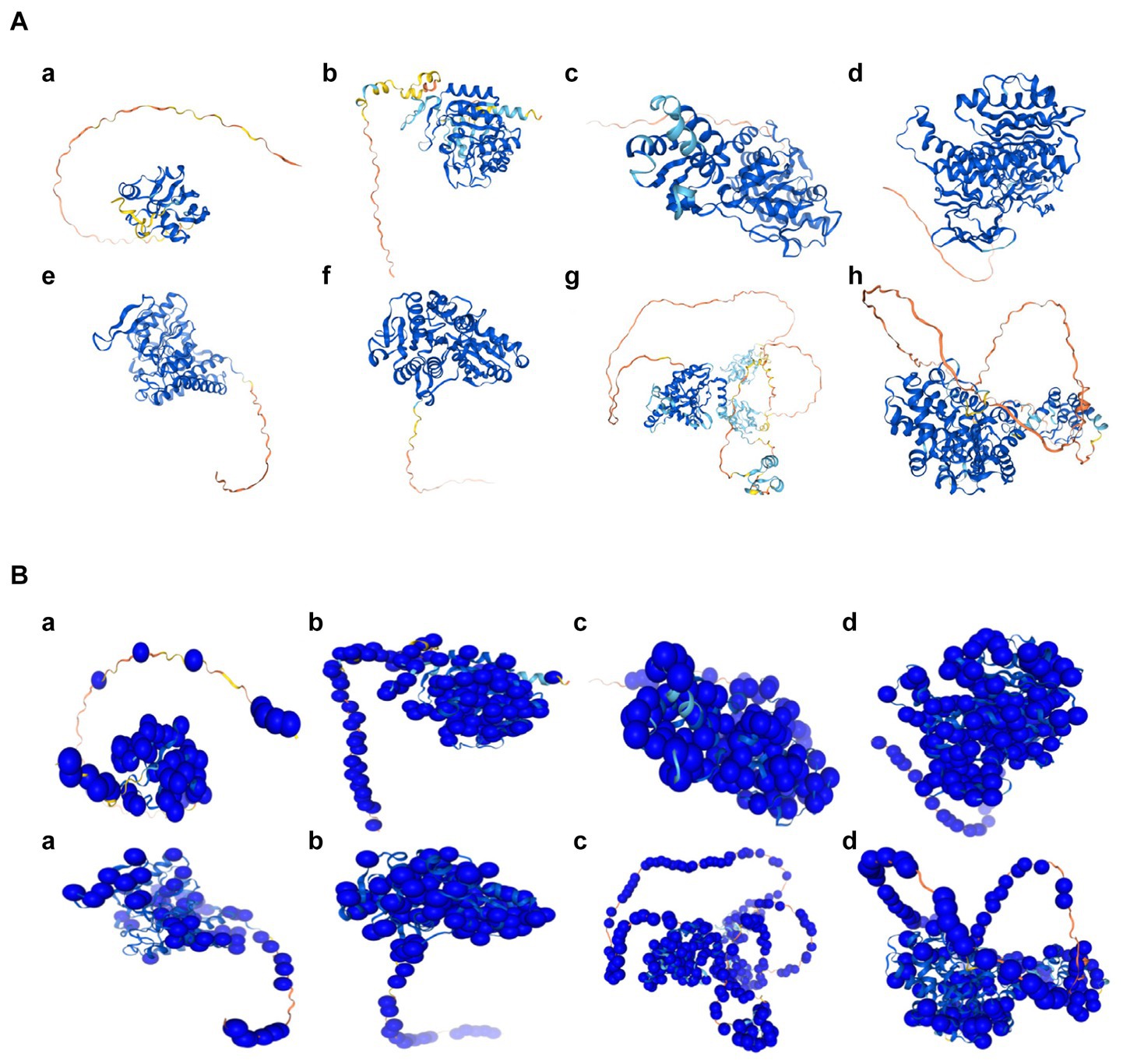
Figure 6. The predicted structures of hub genes related to cuproptosis of cerebral ischemia. (A) The predicted structure diagrams, colored based on the confidence score per residue (pLDDT); dark blue represents the highest confidence level, and orange represents the lowest confidence level. a, FDX1; b, LIAS; c, LIPT1; d, DLD; e, PDHA1; f, PDHB; g, DLAT; h, GLS. (B) The predicted structures of the variable populations. a, FDX1; b, LIAS; c, LIPT1; d, DLD; e, PDHA1; f, PDHB; g, DLAT; h, GLS.
Expression of cuproptosis-related hub genes in immune cells
The RNA expression data from the immune cells of the HPA database9 revealed that FDX1 exhibited the highest expression in B cells. In contrast, LIAS, LIPT1, and GLS exhibited the highest expression in T cells, DLAT and DLD in granulocytes, PDHA1 in monocytes, and PDHB in dendritic cells. FDX1 and GLS exhibited the lowest expression levels in peripheral blood mononuclear cells (PBMCs), LIAS and PDHB in granulocytes, LIPT1 in dendritic cells, PDHA1 in T cells, DLD in B cells, and DLAT in natural killer (NK) cells. Moreover, FDX1 and DLAT exhibited the lowest expression levels in granulocytes, and GLS in monocytes, B cells, NK cells, total PBMC, and dendritic cells (Figure 7A).
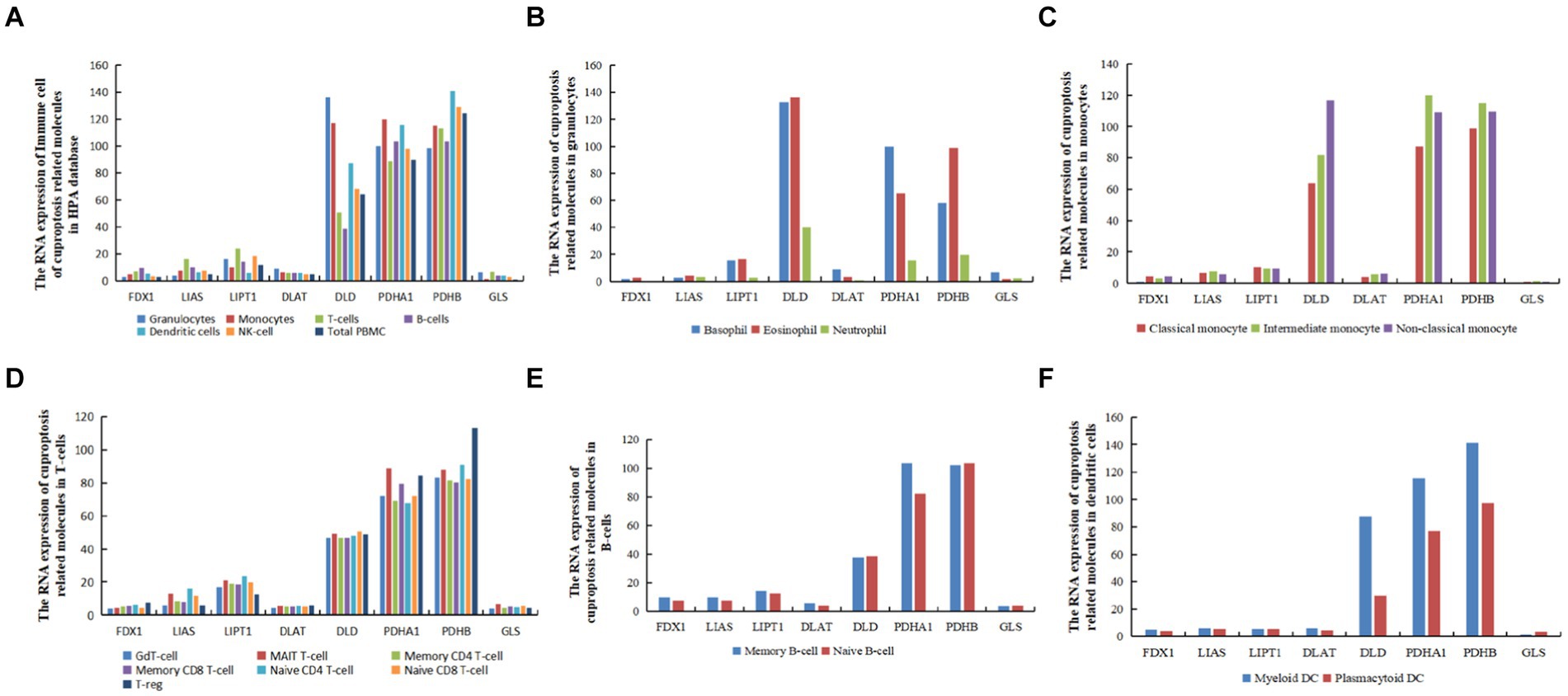
Figure 7. The RNA levels of hub genes related to cuproptosis and cerebral ischemia in immune cells in the HPA database (nTPM, transcripts per million). (A) The RNA expression of immune cell of cuproptosis related molecules in HPA database. (B) The RNA expression of cuproptosis related molecules in granulocytes. (C) The RNA expression of cuproptosis related molecules in monocytes. (D) The RNA expression of cuproptosis related molecules in T-cells. (E) The RNA expression of cuproptosis related molecules in dendritic B-cells. (F) The RNA expression of cuproptosis related molecules in dendritic cells.
In granulocytes, FDX1, LIAS, LIPT1, DLD, and PDHB exhibited the highest expression in eosinophils, whereas PDHA1, DLAT, and GLS genes were most highly expressed in basophils (Figure 7B). In monocytes, FDX1, DLAT, and DLD were most highly expressed in non-classical monocytes, LIPT1 was most highly expressed in classical monocytes, and LIAS,GLS, PDHA1, LIAS, and PDHB were most highly expressed in intermediate monocytes (Figure 7C). In T cells, FDX1, DLAT, and PDHB were most highly expressed in T regulatory cells (T-regs), LIAS and LIPT1 in naive CD4 T cells, DLD in memory CD8 T cells, and GLS and PDHA1 in mucosal-associated invariant (MAIT) T cells (Figure 7D). In B cells, except for DLD and GLS, which exhibited the highest expression in naive B cells, the remaining six genes were most highly expressed in memory B cells (Figure 7E). In dendritic cells (DCs), except for GLS, which was most highly expressed in plasmacytoid DCs, and LIPT1, which was equally expressed in myeloid DCs and plasmacytoid DCs, all other genes were most highly expressed in myeloid DCs (Figure 7F).
Co-expression network of hub genes and miRNAs
The miRNA targets of the eight cuproptosis-related hub genes were predicted using the miRTarBas9.0 database. Among these genes, six, including FDX1, LIAS, DLAT, PDHA1, PDHB, and GLS, were predicted to target 111 miRNAs. A co-expression network was then constructed using Cytoscape software, as shown in Figure 8A.
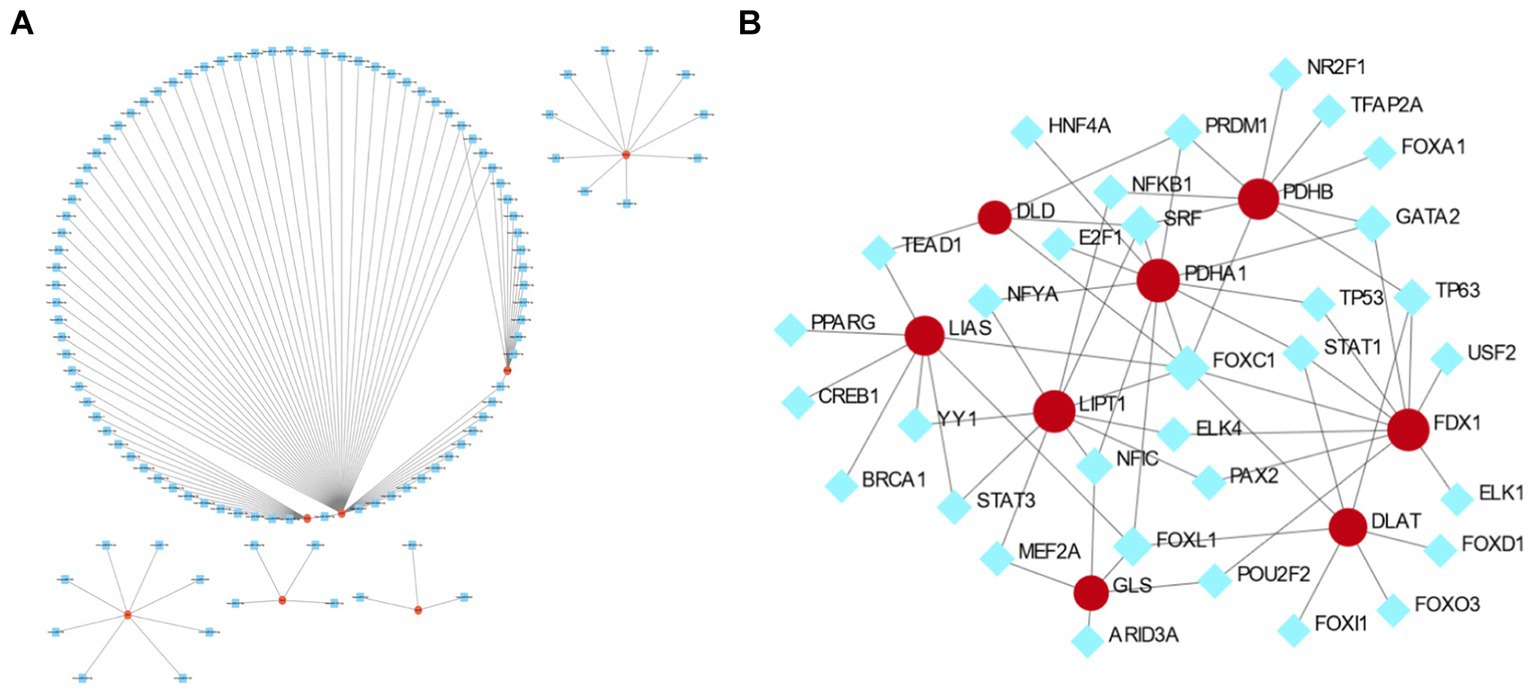
Figure 8. The co-expression network of mRNA–microRNA (miRNA) and transcription factor (TF)–mRNA interactions of hub genes related to cuproptosis in cerebral ischemia. (A) The co-expression network of mRNA–miRNA of hub genes related to cuproptosis in cerebral ischemia. (B) The TF–mRNA interaction of hub genes related to cuproptosis in cerebral ischemia.
TF–gene interactions
Analysis of TF and gene interactions using the NetworkAnalyst database revealed that PDHA1, FDX1, LIPT1, PDHB, LIAS, DLAT, GLS, and DLD interact with 11, 10, 10, 9, 8, 7, 5, and 4 TFs, respectively. These interactions are visualized in Figure 8B.
Expression level of FDX1
To further validate the aforementioned results, the expression of FDX1 was determined through qRT-PCR. Compared to that in the sham group (1.68 ± 0.53), FDX expression in the hippocampus of ovariectomized rats with ischemia (5.05 ± 0.03) was significantly upregulated (t = −15.58, p = 0.00), with 95% confidence interval for the difference (−3.84, −2.88).
Discussion
Copper is essential for metal signaling regulation, metal allosteric regulation, mitochondrial respiration, antioxidant defense, and neurotransmitter function, also influencing cell fate through metabolic reprogramming (15, 16). Recent studies have revealed that abnormal accumulation of copper ions in human cells induces a distinct form of cell death, distinct from known regulated cell death mechanisms. Copper ions could still trigger cell death even when known cell death modes such as apoptosis, pyroptosis, ferroptosis, and necrotic apoptosis are blocked, relying on mitochondrial respiration (3).
This study identified FDX1, LIAS, LIPT1, DLD, DLAT, PDHA1, PDHB, and GLS as eight hub genes associated with cuproptosis in cerebral ischemia. Expression analysis in the HPA database revealed that these hub genes were expressed in the hippocampus, amygdala, basal ganglia, thalamus, hypothalamus, midbrain, spinal cord white matter, as well as in different regions within the internal structure of the hippocampus and immune cells.
FDX1 encodes a small iron–sulfur protein involved in mitochondrial cytochrome reduction, Fe-S cluster biosynthesis, and synthesis of various steroid hormones (17, 18). It serves as a key regulator of cuproptosis (3), primarily localized within the cytosol, endoplasmic reticulum, and nucleus, but its highest activity is observed in the mitochondria (19). The results of this study indicated that its subcellular localization was exclusively in the mitochondria, further elucidating the involvement of specific mitochondrial enzymes in the process of cuproptosis and their relationship with mitochondrial respiration. Moreover, the GO analysis revealed the mitochondrial matrix as the CC, aligning with the subcellular localization predictions and existing literature (19). Previous research demonstrated that FDX1 knockdown resulted in metabolic alterations, particularly in glucose metabolism, fatty acid oxidation, and amino acid metabolism (7), consistent with the Reactome analysis prediction of the signaling pathway involving the human FDX1 gene (20). Given that prior literature has conclusively demonstrated and reported FDX1 as a crucial gene in the process of ferroptosis (3), We had prioritized validating the expression of FDX1 through animal experiments in the present study, our research findings indicated an elevated expression level of FDX in the hippocampus of castrated ovariectomized rats with ischemia, which aligns with the previously reported increase of FDX expression in the hippocampus of rats with ischemic injury (21). Overexpression of FDX1 in vitro partially reversed the protective effect of dexamethasone (DEX) on rat cerebral infarction, including the DEX-induced significant alleviation of rat cerebral infarction, reduced copper levels, mitochondrial function maintenance, increased GSH levels, and decreased levels of key proteins associated with copper toxicity (21).
The LIAS is primarily expressed in the mitochondria (20), and the present study revealed its localization both in the mitochondria and nucleus. This finding aligns with the CC localization results of the mitochondrial matrix, as shown in the GO analysis. Research indicates that LIAS mutations could lead to mitochondrial energy metabolism defects (22). Moreover, LIAS is involved in the synthesis of mitochondria-related metabolic enzymes, the biosynthesis of endogenous fatty acids, energy metabolism, and antioxidant reactions (23, 24). Notably, in the present study, the GO-BP analysis revealed that LIAS participates in complex biosynthetic processes. GO-MF analysis demonstrated the ability of LIAS to bind to iron, sulfur, and metal clusters. FDX1 is a critical upstream regulatory factor for protein thioacylation, which plays a vital role in cells relying on mitochondrial metabolism. FDX1 also acts as a key regulator of steatosis by directly binding to LIAS, thereby exerting a lethal metabolic effect on cancer cells (25). The GO-MF analysis in this study indicated that LIAS can bind to iron–sulfur clusters and metal clusters. Moreover, FDX1 serves as a key regulatory factor for protein lipoylation through direct binding to LIAS and plays a role in metabolic conditioning-induced cell death in cancer cells (25).
LIPT1, an enzyme specific to lipid esters, plays a significant role in copper homeostasis. It is an essential enzyme for activating mitochondrial 2-ketoacid dehydrogenases and is involved in maintaining the oxidative and reductive metabolism of glutamine (26). LIPT1 participates in the biosynthesis and function of lipoic acid and fatty acylation (27, 28). These previous findings align with the results of CC localization in the GO analysis of the present study, indicating mitochondrial matrix as the subcellular localization of LIPT1.
The DLD is an important component of various mitochondrial multienzyme complexes, participating in the composition of complexes such as α-ketoglutarate dehydrogenase, α-ketohexanoate dehydrogenase, and glycine decarboxylase (29). It is also involved in the decarboxylation of pyruvate, converting the product into acetyl-CoA in the TCA cycle (30). The results of this study indicated that DLD was localized in both mitochondria and nuclei. GO-CC analysis results suggested associations with mitochondrial matrix, redox enzyme complex mitochondria, and mitochondrial tricarboxylate cyclase complex. Moreover, KEGG analysis indicated its involvement in pathways such as the TCA cycle and pyruvate metabolism. Research has shown that DLD downregulation could affect mitochondrial metabolism, leading to reduced levels of downstream metabolites in the TCA cycle and inducing melanoma cell death (31). Moreover, DLD is known to promote cell death, such as apoptosis (32), copper poisoning-induced cell death (3), and cuproptosis. DLD serves as a key gene in cuproptosis and is a positive regulator, enhancing copper-dependent cell death (30). The present study also identified DLD as one of the hub genes responsible for cuproptosis following cerebral ischemia.
DLAT, a mitochondrial protein involved in glucose metabolism, is located in the inner mitochondrial membrane and plays a role in the conversion of pyruvate to acetyl acetyl-CoA (33). Furthermore, DLAT overexpression inhibits the production of acetyl-CoA (34). These findings are consistent with the GO/KEGG analyses results of this study. Notably, DLAT is associated with cuproptosis, as copper promotes the oligomerization of DLAT, thereby increasing the insoluble DLAT, leading to protein toxicity stress and cell death (3). This oligomerization is due to the integration of copper with lipoylated proteins in the TCA cycle (3). Cu-induced neuronal degeneration and oxidative damage have been shown to promote the expression of FDX1, DLAT, and HSP70 while reducing that of Fe-S cluster proteins (35). Noteworthily, proteins such as ATP6V1A, DLAT, and HSP70 are expressed in the hippocampus of patients with temporal lobe epilepsy (36). In contrast, patients with acute myocardial infarction exhibit decreased levels of LIAS, PDHB, LIPT1, DLAT, and GLS, along with increased MTF1 levels. A previous Kaplan–Meier analysis indicated the prognostic value of DLAT in ischemic events (37). The present study also identified DLAT as one of the hub genes responsible for cuproptosis after cerebral ischemia.
PDHA1 serves as a crucial component of the pyruvate dehydrogenase (PDH) complex, an enzyme complex that regulates the TCA cycle (38). PDHA1 plays a crucial role in glucose metabolism, oxidative phosphorylation, and the TCA cycle in the mitochondria (39). The results of this study indicated the involvement of PDHA1 in the biosynthesis of acetyl-CoA from pyruvate, the biosynthesis process of acetyl-CoA, and acetyl-CoA metabolism processes. KEGG analysis results indicated that these processes were related to pathways such as the TCA cycle, pyruvate metabolism, and glycolysis/gluconeogenesis. These results are consistent with previous reports (39). In mice, PDHA1 knockout resulted in ultrastructural disruptions of hippocampal neurons and lactate accumulation (40), which subsequently caused impaired neuronal function (41), ultimately resulting in hippocampal dysfunction. Moreover, prostate cancer cells and human esophageal squamous cancer cells lacking PDHA1 exhibit impaired normal mitochondrial oxidative phosphorylation and a reliance on glycolysis (42, 43). These previous data are consistent with the results of the present study in terms of the subcellular localization to the mitochondria, and the cellular component annotations in GO/KEGG analyses corroborate the preservation of the CC.
The PDHB catalyzes the conversion of pyruvate to acetyl-CoA, bridging the TCA cycle and glycolytic pathway (44). Its expression is predominantly distributed within the mitochondria (45). Consistently, in the present study, the GO-BP results indicated the involvement of PDHB d in the biosynthesis process of acetyl-CoA from pyruvate, the biosynthesis process of acetyl-CoA, and the metabolism of acetyl-CoA. Moreover, KEGG analysis revealed that PDHB participates in BPs related to the TCA cycle, pyruvate metabolism, and glycolysis pathways. In a previous study, PDHB knockdown in primary human muscle inhibited pyruvate metabolism and upregulated Ariadne RBR E3 ubiquitin protein ligase 2 (Arih2) in the cellular catabolic pathway (46). Moreover, dPDHB knockout shortened the lifespan of adult flies, leading to rough eye phenotypes and abnormal photoreceptor axon targeting (47). Mutations in PDHA1 and PDHB are associated with coenzyme Q10 levels and mitochondrial homeostasis imbalance, severely affecting the brain (48). Currently, research is primarily focusing on the role of PDHB in cancer, particularly in the cuproptosis pathway associated with non-alcoholic fatty liver disease (NAFLD), where both DLD and PDHB are potential candidate genes for NAFLD diagnosis and treatment options (49). Additionally, five genes related to cuproptosis (FDX1, LIPT1, PDHA1, PDHB, and CDKN2A) are considered candidate biomarkers or therapeutic targets for osteoarthritis synovitis (50). The present study also reported the involvement of PDHB in the cuproptosis pathway in cerebral ischemia.
The GLS catalyzes the hydrolysis of glutamine to produce glutamic acid (50). It exists in mammals in two isoforms: GLS1 and GLS2. GLS1 is primarily expressed in organs such as the brain, heart, pancreas, and kidneys, while GLS2 is mainly expressed in the liver (51). The subcellular localization of GLS1 is in the mitochondria, while GLS2 is located in the nucleus (51). However, the results of this study indicated that GLS was only localized to the mitochondria. Dysregulated expression or dysfunction of GLS results in the overproduction of glutamate, alterations in the expression of inflammatory factors, and the disruption of metabolic homeostasis, resulting in the activation of microglia (51). Upregulation of GLS1 expression leads to excessive production of glutamate in microglial cells, increasing extracellular glutamate levels, which in turn cause excitotoxicity and neuronal degeneration (51). Notably, cuproptosis is associated with the TCA cycle and the aggregation of lipoylated proteins in the TCA cycle (3, 51). In rats with cerebral artery occlusion, the expression of GLS1 was substantially upregulated, and the GLS inhibitor CB-839 markedly reduced the expression of pro-inflammatory factors, thereby alleviating neuroinflammation and brain damage (52). The present study also identified GLS as a hub gene associated with copper-induced cell death during cerebral ischemia.
This study presents novel insights into hub genes involved in cuproptosis during cerebral ischemia, validating them through multiple approaches including brain tissue, immune cells, localization, structure, and prediction of miRNAs and TFs. However, some limitations should be noted; for example, currently, only one of the hub genes, FDX1, has been validated in animal experiments and the hippocamp of rats, while other hub genes and the expression of brain regions need to be validated in experiments. In addition, there is a lack of clinical research on the hub genes associated with cuproptosis. In the future, we will strive to incorporate a wider range of brain regions and multiple gene targets into our research endeavors, further deepening our understanding of the intricate mechanisms underlying ischemic injury. However, in subsequent studies, we are predicting and understanding the binding interactions between small molecules (ligands) and target proteins related to copper death in cerebral ischemia; Using computer simulation technology, small molecule drug molecules are docked to the surface of proteins to search for possible binding sites for drug molecule docking. This part of the research work is currently underway, and once progress is made in the next stage, we will share our findings with everyone.
Conclusion
In summary, this study utilized genes related to cerebral ischemic and cuproptosis to identify and validate genes implicated in both cerebral ischemic and cuproptosis. Eight hub genes with the potential to serve as novel markers were identified, holding promise as specific biomarkers for the diagnosis, treatment, and prognosis of cerebral ischemia in clinical applications.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal studies were approved by Experimental Animal Ethics Committee of Hunan University of Traditional Chinese Medicine. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
LQ: Conceptualization, Formal analysis, Funding acquisition, Writing – original draft. XC: Visualization, Writing – original draft. TH: Data curation, Methodology, Software, Writing – original draft. YL: Formal analysis, Writing – original draft. SL: Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China [Nos. 82374437 and 81904180], Hunan Provincial Natural Science Foundation of China [Nos. 2023JJ50035 and 2024JJ8124], Scientific Research Project of Hunan Provincial Health Commission [No. B202319018677], “Disciplinary Reveal System” project of Hunan University of Chinese Medicine [No. 22JBZ041], Hunan Province Traditional Chinese Medicine Research Plan Project [No. A2024001]. Discipline construction at Hunan University of Chinese Medicine.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
2. ^https://jvenn.toulouse.inrae.fr/app/index.html
4. ^https://www.proteinatlas.org/
5. ^https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php
6. ^https://www.networkanalyst.ca/
References
1. Shi, XH, Mang, J, and Xu, ZX. Research progress in cell death modes of cerebral ischemia-reperfusion injury. J Jilin Univ Med Edit. (2022) 48:1635–43. doi: 10.13481/j.1671-587X.20220633
2. Hu, WX, and Xie,. Ferroptosis and its roles in cerebral ischemia-reperfusion injury. Prog Physiol Sci. (2021) 52:169–75.
3. Tsvetkov, P, Coy, S, Petrova, B, Dreishpoon, M, Verma, A, Abdusamad, M, et al. Copper induces cell death by targeting lipoylated TCA cycle proteinsJ. Science. (2022) 375:1254–61. doi: 10.1126/science.abf0529
4. Wu, YJ, Sun, ZR, and Zhang, Y. Effect of acupuncture on serum copper and chromium content in rats with acute focal cerebral ischemia. Inform Tradit Chin Med. (2004) 6:35–7.
5. Wang, TT, Hu, LC, and Fan, H. Cuproptosis and its possible role in neuronal cell death in ischemic stroke. Chin J of Emerg Med. (2022) 31:1724–9.
6. Valentine, RC . Bacterial ferredoxin. Bacteriol Rev. (1964) 28:497–517. doi: 10.1128/br.28.4.497-517.1964
7. Zhang, Z, Ma, Y, Guo, X, du, Y, Zhu, Q, Wang, X, et al. FDX1 can impact the prognosis and mediate the metabolism of lung adenocarcinoma. Front Pharmacol. (2021) 12:749134. doi: 10.3389/fphar.2021.749134
8. Xiao, Y, Yuan, Y, Liu, Y, Yu, Y, Jia, N, Zhou, L, et al. Circulating multiple metals and incident stroke in Chinese adults. Stroke. (2019) 50:1661–8. doi: 10.1161/STROKEAHA.119.025060
9. Yu, G, Wang, LG, Han, Y, and He, QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
10. Csardi, G, and Tamas, N. The igraph software package for complex network research. Inter J Complex Syst. (2006) 1695:1–9.
11. Szklarczyk, D, Gable, AL, Nastou, KC, Lyon, D, Kirsch, R, Pyysalo, S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021) 49:D605–12. doi: 10.1093/nar/gkaa1074
12. Spychala, MS, Honarpisheh, P, McCullough, LD, and She, J. Sex differences in neuroinflammation and neuroprotection in ischemic stroke. J Neurosci Res. (2017) 95:462–71. doi: 10.1002/jnr.23962
13. Qin, LH, Liu, Y, Huang, J, Cheng, SW, Liu, l, Li, S, et al. Effect of Jiawei Naotaifang on cerebral infarction area and level of estrogen of ovariectomized rats with cerebral ischemia and its correlation. Chin Pharm Bull. (2018) 34:428–31.
14. Qin, LH, Wang, GZ, Liu, L, Huang, J, Liu, Y, and Yi, YQ. Effects of estrogen inhibitor on ATF4/CHOP/Puma pathway in ovariectomized rats with cerebral ischemia and the intervention effect of Jiawei Naotai formula. Chin J Tradit Chin Med Pharm. (2020) 35:3594–7.
15. Ruiz, LM, Libedinsky, A, and Elorza, AA. Role of copper on mitochondrial function and metabolism. Front Mol Biosci. (2021) 8:711227. doi: 10.3389/fmolb.2021.711227
16. Ge, EJ, Bush, AI, Casini, A, Cobine, PA, Cross, JR, DeNicola, GM, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer. (2022) 22:102–13. doi: 10.1038/s41568-021-00417-2
17. Sheftel, AD, Stehling, O, Pierik, AJ, Elsässer, HP, Mühlenhoff, U, Webert, H, et al. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci USA. (2010) 107:11775–80. doi: 10.1073/pnas.1004250107
18. Strushkevich, N, MacKenzie, F, Cherkesova, T, Grabovec, I, Usanov, S, and Park, HW. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc Natl Acad Sci USA. (2011) 108:10139–43. doi: 10.1073/pnas.1019441108
19. Tang, D, Chen, X, and Kroemer, G. Cuproptosis: a copper-triggered modality of mitochondrial cell death. Cell Res. (2022) 32:417–8. doi: 10.1038/s41422-022-00653-7
20. Guan, YL . Bioinformatics analysis of human ferredoxin 1, the key regulatory gene of Cuproptosis. J Jiangsu Univ :1–11. doi: 10.13312/j.issn.1671-7783.y220159
21. Guo, Q, Ma, M, Yu, H, Han, Y, and Zhang, D. Dexmedetomidine enables copper homeostasis in cerebral ischemia/reperfusion via ferredoxin 1. Ann Med. (2023) 55:2209735. doi: 10.1080/07853890.2023.2209735
22. Habarou, F, Hamel, Y, Haack, TB, Feichtinger, RG, Lebigot, E, Marquardt, I, et al. Biallelic mutations in LIPT2 cause a mitochondrial lipoylation defect associated with severe neonatal encephalopathy. Am J Hum Genet. (2017) 101:283–90. doi: 10.1016/j.ajhg.2017.07.001
23. Lu, WF, Cao, JJ, Guo, YJ, Zhong, K, Zha, GM, Wang, LF, et al. Expression of the porcine lipoic acid synthase (LIAS) gene in Escherichia coli. Genet Mol Res. (2014) 13:5369–77. doi: 10.4238/2014.July.24.16
24. Yi, X, Kim, K, Yuan, W, Xu, L, Kim, HS, Homeister, JW, et al. Mice with heterozygous deficiency of lipoic acid synthase have an increased sensitivity to lipopolysaccharide-induced tissue injury. J Leukoc Biol. (2009) 85:146–53. doi: 10.1189/jlb.0308161
25. Dreishpoon, MB, Bick, NR, Petrova, B, Warui, DM, Cameron, A, Booker, SJ, et al. FDX1 regulates cellular protein lipoylation through direct binding to LIAS. [Epubh ahead of preprint]. (2023). doi: 10.1016/j.jbc.2023.105046
26. Ni, M, Solmonson, A, Pan, C, Yang, C, Li, D, Notzon, A, et al. Functional assessment of Lipoyltransferase-1 deficiency in cells, mice, and humans. Cell Rep. (2019) 27:1376–1386.e6. doi: 10.1016/j.celrep.2019.04.005
27. Liu, Y, Luo, G, Yan, Y, and Peng, J. A pan-cancer analysis of copper homeostasis-related gene lipoyltransferase 1: its potential biological functions and prognosis values. Front Genet. (2022) 13:1038174. doi: 10.3389/fgene.2022.1038174
28. Taché, V, Bivina, L, White, S, Gregg, J, Deignan, J, Boyadjievd, SA, et al. Lipoyltransferase 1 gene defect resulting in fatal lactic acidosis in two siblings. Case Rep Obstet Gynecol. (2016) 2016:1–4. doi: 10.1155/2016/6520148
29. Duarte, IF, Caio, J, Moedas, MF, Rodrigues, LA, Leandro, AP, Rivera, IA, et al. Dihydrolipoamide dehydrogenase, pyruvate oxidation, and acetylation-dependent mechanisms intersecting drug iatrogenesis. Cell Mol Life Sci. (2021) 78:7451–68. doi: 10.1007/s00018-021-03996-3
30. Yang, W, Guo, Q, Wu, H, Tong, L, Xiao, J, Wang, Y, et al. Comprehensive analysis of the cuproptosis-related gene DLD across cancers: a potential prognostic and immunotherapeutic target. Front Pharmacol. (2023) 14:1111462. doi: 10.3389/fphar.2023.1111462
31. Yumnam, S, Kang, MC, Oh, SH, Kwon, HC, Kim, JC, Jung, ES, et al. Downregulation of dihydrolipoyl dehydrogenase by UVA suppresses melanoma progression via triggering oxidative stress and altering energy metabolism. Free Radic Biol Med. (2021) 162:77–87. doi: 10.1016/j.freeradbiomed.2020.11.037
32. Dayan, A, Fleminger, G, and Ashur-Fabian, O. Targeting the Achilles' heel of cancer cells via integrin-mediated delivery of ROS-generating dihydrolipoamide dehydrogenase. Oncogene. (2019) 38:5050–61. doi: 10.1038/s41388-019-0775-9
33. Goh, WQ, Ow, GS, Kuznetsov, VA, Chong, S, and Lim, YP. DLAT subunit of the pyruvate dehydrogenase complex is upregulated in gastric cancer-implications in cancer therapy. Am J Transl Res. (2015) 7:1140–51.
34. Chen, Q, Wang, Y, Yang, L, Sun, L, Wen, Y, Huang, Y, et al. PM2.5 promotes NSCLC carcinogenesis through translationally and transcriptionally activating DLAT-mediated glycolysis reprograming. J Exp Clin Cancer Res. (2022) 41:229. doi: 10.1186/s13046-022-02437-8
35. Zhang, Y, Zhou, Q, Lu, L, Su, Y, Shi, W, Zhang, H, et al. Copper induces cognitive impairment in mice via modulation of Cuproptosis and CREB signaling. Nutrients. (2023) 15:972. doi: 10.3390/nu15040972
36. Persike, DS, Marques-Carneiro, JE, Stein, MLL, Yacubian, EMT, Centeno, R, Canzian, M, et al. Altered proteins in the Hippocampus of patients with mesial temporal lobe epilepsy. Pharmaceuticals. (2018) 11:95. doi: 10.3390/ph11040095
37. Liu, Z, Wang, L, Xing, Q, Liu, X, Hu, Y, Li, W, et al. Identification of GLS as a cuproptosis-related diagnosis gene in acute myocardial infarction. Front Cardiovasc Med. (2022) 9:1016081. doi: 10.3389/fcvm.2022.1016081
38. Deng, L, Jiang, A, Zeng, H, Peng, X, and Song, L. Comprehensive analyses of PDHA1 that serves as a predictive biomarker for immunotherapy response in cancer. Front Pharmacol. (2022) 13:947372. doi: 10.3389/fphar.2022.947372
39. Patel, MS, Nemeria, NS, Furey, W, and Jordan, F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem. (2014) 289:16615–23. doi: 10.1074/jbc.R114.563148
40. Chen, W, Sun, X, Zhan, L, Zhou, W, and Bi, T. Conditional knockout of Pdha1 in mouse Hippocampus impairs cognitive function: the possible involvement of lactate. Front Neurosci. (2021) 15:767560. doi: 10.3389/fnins.2021.767560
41. Scandella, V, and Knobloch, M. Sensing the environment: extracellular lactate levels control adult neurogenesis. Cell Stem Cell. (2019) 25:729–31. doi: 10.1016/j.stem.2019.11.008
42. Zhong, Y, Li, X, Ji, Y, Li, X, Li, Y, Yu, D, et al. Pyruvate dehydrogenase expression is negatively associated with cell stemness and worse clinical outcome in prostate cancers. Oncotarget. (2017) 8:13344–56. doi: 10.18632/oncotarget.14527
43. Liu, L, Cao, J, Zhao, J, Li, X, Suo, Z, and Li, H. PDHA1 gene knockout in human esophageal squamous Cancer cells resulted in greater Warburg effect and aggressive features in vitro and in vivo. Onco Targets Ther. (2019) 12:9899–913. doi: 10.2147/OTT.S226851
44. Li, A, Zhang, Y, Zhao, Z, Wang, M, and Zan, L. Molecular characterization and transcriptional regulation analysis of the bovine PDHB gene. PLoS One. (2016) 11:e0157445. doi: 10.1371/journal.pone.0157445
45. Taylor, SI, Mukherjee, C, and Jungas, RL. Regulation of pyruvate dehydrogenase in isolated rat liver mitochondria. Effects of octanoate, oxidation-reduction state, and adenosine triphosphate to adenosine diphosphate ratio. J Biol Chem. (1975) 250:2028–35. doi: 10.1016/S0021-9258(19)41679-7
46. Jiang, X, Ji, S, Yuan, F, Li, T, Cui, S, Wang, W, et al. Pyruvate dehydrogenase B regulates myogenic differentiation via the FoxP1-Arih2 axis. J Cachexia Sarcopenia Muscle. (2023) 14:606–21. doi: 10.1002/jcsm.13166
47. Dung, VM, Suong, DNA, Okamaoto, Y, Hiramatsu, Y, Thao, DTP, Yoshida, H, et al. Neuron-specific knockdown of Drosophila PDHB induces reduction of lifespan, deficient locomotive ability, abnormal morphology of motor neuron terminals and photoreceptor axon targeting. Exp Cell Res. (2018) 366:92–102. doi: 10.1016/j.yexcr.2018.02.035
48. Asencio, C, Rodríguez-Hernandez, MA, Briones, P, Montoya, J, Cortés, A, Emperador, S, et al. Severe encephalopathy associated to pyruvate dehydrogenase mutations and unbalanced coenzyme Q10 content. Eur J Hum Genet. (2016) 24:367–72. doi: 10.1038/ejhg.2015.112
49. Wu, C, Liu, X, Zhong, L, Zhou, Y, Long, L, Yi, T, et al. Identification of Cuproptosis-related genes in non-alcoholic fatty liver disease. Oxidative Med Cell Longev. (2023) 2023:1–18. doi: 10.1155/2023/9245667
50. Chang, B, Hu, Z, Chen, L, Jin, Z, and Yang, Y. Development and validation of cuproptosis-related genes in synovitis during osteoarthritis progress. Front Immunol. (2023) 14:1090596. doi: 10.3389/fimmu.2023.1090596
51. Ding, L, Xu, X, Li, C, Wang, Y, Xia, X, and Zheng, JC. Glutaminase in microglia: A novel regulator of neuroinflammation. Brain Behav Immun. (2021) 92:139–56. doi: 10.1016/j.bbi.2020.11.038
Keywords: cerebral ischemia, cuproptosis, biomarkers, gene expression, bioinformatics
Citation: Qin L, Cao X, Huang T, Liu Y and Li S (2024) Identification of potential biomarkers of cuproptosis in cerebral ischemia. Front. Nutr. 11:1410431. doi: 10.3389/fnut.2024.1410431
Edited by:
Adriana Ximenes-da-Silva, Federal University of Alagoas, BrazilReviewed by:
Yanyao Liu, The First Affiliated Hospital of Chongqing Medical University, ChinaQin Hu, Shanghai Jiao Tong University, China
Copyright © 2024 Qin, Cao, Huang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Li, NDIwNjQ4MTYxQHFxLmNvbQ==
 Lihua Qin
Lihua Qin Xi Cao1
Xi Cao1