- 1Department of Genomic Medicine in Hepatology, Civil Hospital of Guadalajara, Fray Antonio Alcalde, Health Sciences Center, University of Guadalajara, Guadalajara, Jalisco, Mexico
- 2Department of Medical and Surgical Sciences and Advanced Technologies “GF Ingrassia”, University of Catania, Catania, Italy
- 3Medicine and Psychology School, Autonomous University of Baja California, Tijuana, Baja California, Mexico
Chronic liver disease is a global health issue. Patients with chronic liver disease require a fresh approach that focuses on the genetic and environmental factors that contribute to disease initiation and progression. Emerging knowledge in the fields of Genomic Medicine and Genomic Nutrition demonstrates differences between countries in terms of genetics and lifestyle risk factors such as diet, physical activity, and mental health in chronic liver disease, which serves as the foundation for the implementation of Personalized Medicine and Nutrition (PerMed-Nut) strategies. Most of the world’s populations have descended from various ethnic groupings. Mexico’s population has a tripartite ancestral background, consisting of Amerindian, European, and African lineages, which is common across Latin America’s regional countries. The purpose of this review is to discuss the genetic and environmental components that could be incorporated into a PerMed-Nut model for metabolic-associated liver disease, viral hepatitis B and C, and hepatocellular carcinoma in Mexico. Additionally, the implementation of the PerMed-Nut approach will require updated medicine and nutrition education curricula. Training and equipping future health professionals and researchers with new clinical and investigative abilities focused on preventing liver illnesses in the field of genomic hepatology globally is a vision that clinicians and nutritionists should be concerned about.
1 Introduction
Modern humans, taxonomically known as Homo sapiens sapiens, emerged in Africa 200,000 years ago and began the migration out-of-Africa to colonize the globe between 50,000 and 70,000 years ago (1, 2). Studies on human evolutionary history relate that most of the world’s populations have descended from 19 ancestral human ethnic groups (Figures 1A,B) (2, 4). However, the human genome, which is constantly evolving, is a melting pot of past, present, and even future genetic signs of adaptations to diverse environmental niches, and it appears that they are not always meant to create life-threatening disorders (5, 6). These differences are explained by genetic variants including single nucleotide polymorphisms (SNPs), copy number variations (CNVs), and deletion or insertion (Del/Ins) polymorphisms (7). Other non-genetic mechanisms, such as transgenerational plasticity and cultural evolution, were undoubtedly additional powerful forces that contributed to human adaptation (8). However, discrete changes in the nature and frequency of specific genetic variants might influence health triggered by environmental factors (9). Such variants have been revealed with the completion of the Human Genome Project’s mapping of the human haploid reference genome (10) and the incorporation of other worldwide genomic databases (11) so that the understanding of the genetic basis of human disease and, ultimately, the prevention of many diseases may be achieved (12).
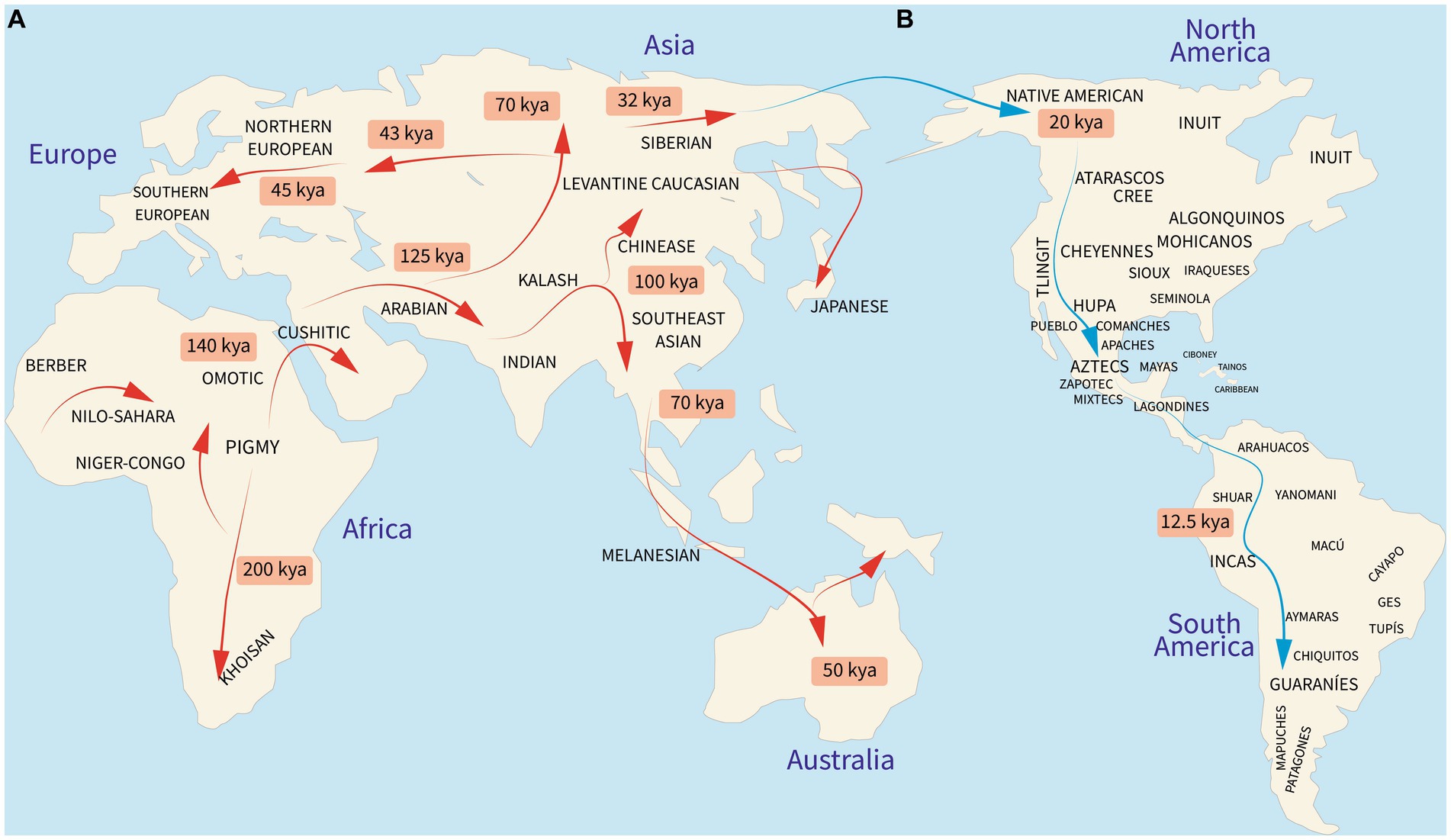
Figure 1. Peopling of the world. (A) Migration of the 19 ancestral human populations from which Native Americans made their way to the Americas. (B) Peopling and main settlements of the Americas in pre-Colombian times by the ancestral Native American component. kya, kilo years ago. Reproduced from references (1–3).
Currently, the paradigm shift in medicine from reactive to predictive is focused in providing models of prevention strategies based on the population’s genetic makeup and the risk factors that contribute to the emergence of chronic diseases, whether infectious or non-communicable (13). Clinicians need to reconsider their traditional approach to managing major diseases (such as hypertension, heart disease, type 2 diabetes, cancer, renal, and liver disease) from a bench to bedside/translational perspective, combining novel omics data with pathological clinical phenotypes in search of an individualized therapy (14). This has resulted in a multitude of terms, including Predictive Medicine, Individualized Medicine, Personalized Medicine, and Precision Medicine (15, 16), all of which acknowledge that each person is unique but still shares ancestral genetic variants based on ethnicity and environmental circumstances. These characteristics may explain why certain people are susceptible or prone to diseases when exposed to risky conditions or behaviors, but others in similar circumstances (i.e., family members) are unaffected (17). Personalized Medicine “omics” techniques have been proposed for nearly all medical fields including Nutrition, where Genomic Nutrition, Personalized Nutrition, and Precision Nutrition are becoming increasingly important (18).
The Personalized Medicine and Nutrition (PerMed-Nut) approach in Hepatology will inevitably include managing the main etiologies that cause chronic liver disease worldwide (19), such as alcohol persistence, chronic viral hepatitis B and C infections, metabolic-associated steatotic liver disease, hepatotoxicity (drugs or toxins), and auto-immune conditions. However, the prevalence of these etiologies varies greatly between countries due to genetic, environmental, and cultural differences in the host populations (20). As a result, the extent of liver damage might be immediate and self-limiting, or it can progress to chronic phases in which the inflammatory process of injury causes fibrosis, cirrhosis, and, in rare cases, hepatocellular carcinoma (HCC).
How can we begin using the PerMed-Nut approach as a strategy to prevent chronic liver diseases? Firstly, it is essential to understand the impact of genes and the environment at a regional level to develop measures to prevent liver damage. Hence, this work aims to define these factors for the prevention and management of liver illnesses using a PerMed-Nut model and the need for training in genomic education. We will address some polymorphic genes associated with the clinical outcomes of liver diseases as well as sociocultural features of the Mexican population in the sections that follow.
2 Mexico’s genetic ancestry and sociodemographic background
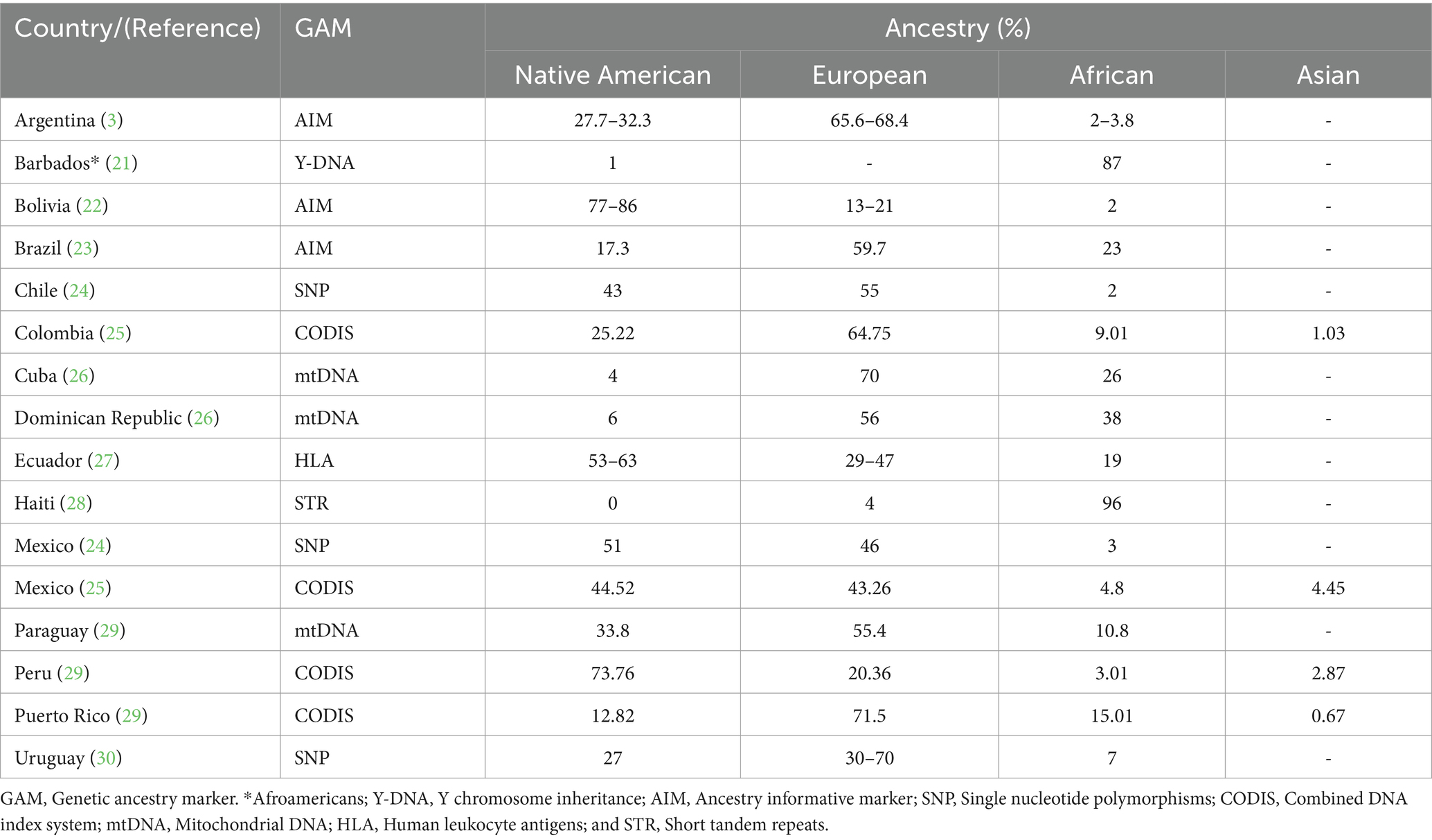
Studies examining the genetic ancestry of the Latin American (LATAM) populations including Mexico have revealed heterogeneous proportions of three main ancestral lineages: Amerindian, European, and African. As shown in Table 1, most LATAM countries contain all three with some having a higher degree of African (21, 28) and Native American (22, 27, 29, 30) ancestry than European (3, 23–26, 29). The indigenous people of Mexico are part of the third Native American migration wave that crossed the Bering Strait to the American continent around 20,000 years ago (2) (Figure 1B, blue arrow). These Native Americans settled primarily in Mexico (Tenochtitlan) and Peru (Cuzco) followed by Brazil, the United States, Bolivia, Columbia, and in lesser proportions Argentina, Venezuela, Ecuador, Canada, and Alaska (31). The current 68 ethnic groups of Mexican Amerindians are descendants of the original First Nations who followed either a nomadic or agricultural lifestyle, primarily in Aridoamerica and Mesoamerica, respectively (3, 32). During the Spanish colony (from 1521 to 1826), a genetic and cultural admixture occurred among Mexican Amerindians, Spaniards, and enslaved Africans (21). The Spanish Catholic Church recognized new births using a caste system, with Amerindian and Black heritage overlooked that has marked the social distribution of the ancestral lineages since colonial up until modern times.
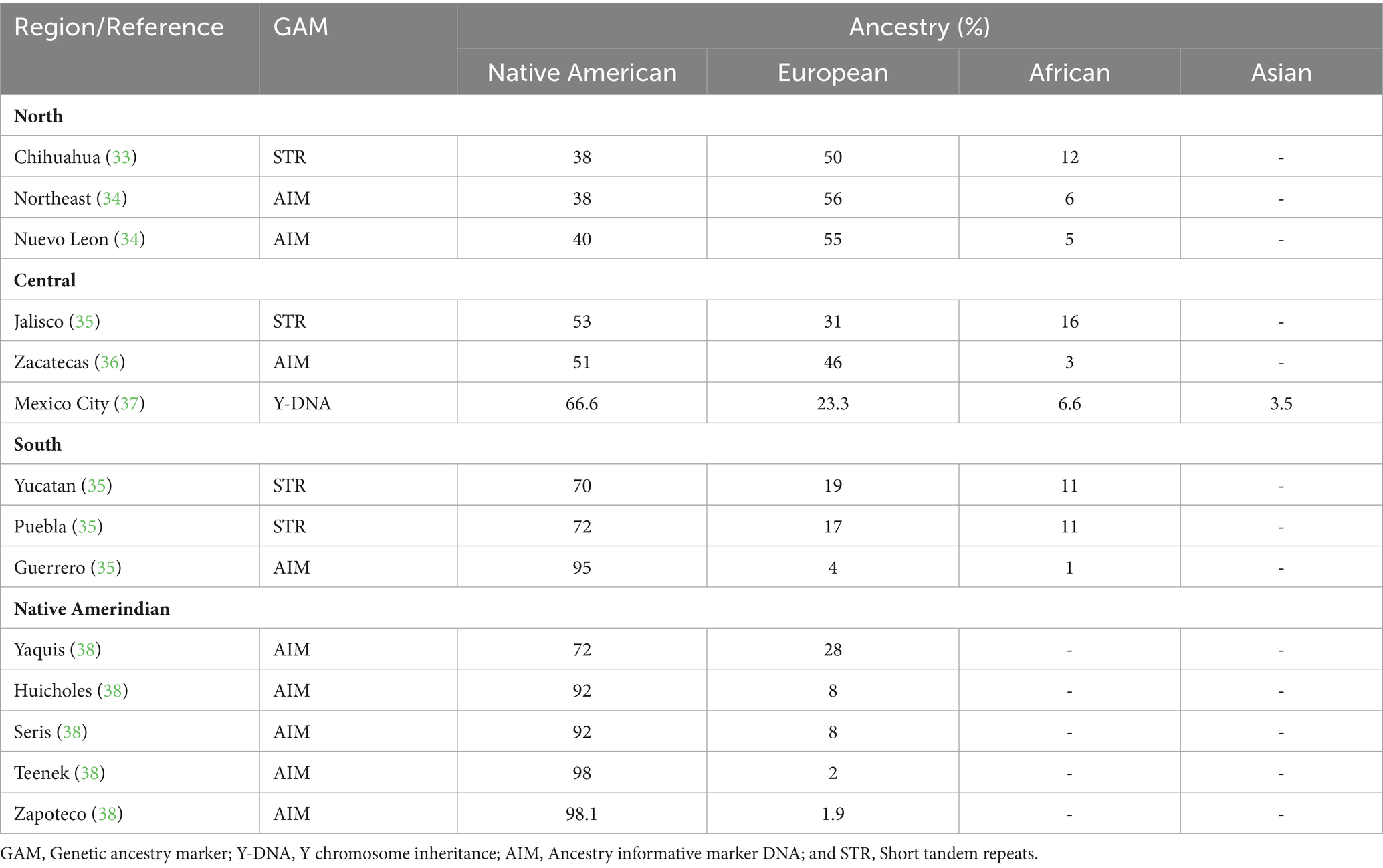
Most Mexicans are admixed containing practically half of the main lineages (24, 25). However, as illustrated in Table 2, the degree of admixture varies across the country. Northern Mexico has a higher rate of European trait carriers (33, 34) and intermediate rates in the Central Region (35–37), contrasting with Southern Mexico, who are more likely to be carriers of Amerindian traits (35, 39) including the true Native Americans per se (38). African ancestry is present in varying quantities among the admixed general population and Afro-Mexicans who reside along the South Pacific and the Gulf of Mexico coastal communities (40). Further genetic studies show indigenous mitochondrial DNA as the maternal source of inheritance, while Y-STR analysis tracks European paternal lineage (37, 41).
Among the Genome-Wide Association Study (GWAS) database catalog, a total of 284 studies related to liver diseases have been conducted, identifying 1,546 associations in over 606 genes (42, 43). They are mainly reported in European, Asian, and African populations and scarcely in Native American ancestry (Figure 2A). However, one GWAS using the Mexican Biobank have shown 25 associations in 15 genes involved in lipid metabolism and transportation, vitamin A metabolism, protein folding, regulation of gene expression, cellular signaling, embryonic development, and heart development (Figure 2B). Among the most important clinical associations are hypercholesterolemia, hyper triglyceridemia, body weight, creatinine levels, hypertension, and arthritis. Furthermore, this study confirmed earlier findings of the north–south gradient of European-Amerindian ancestry (44).
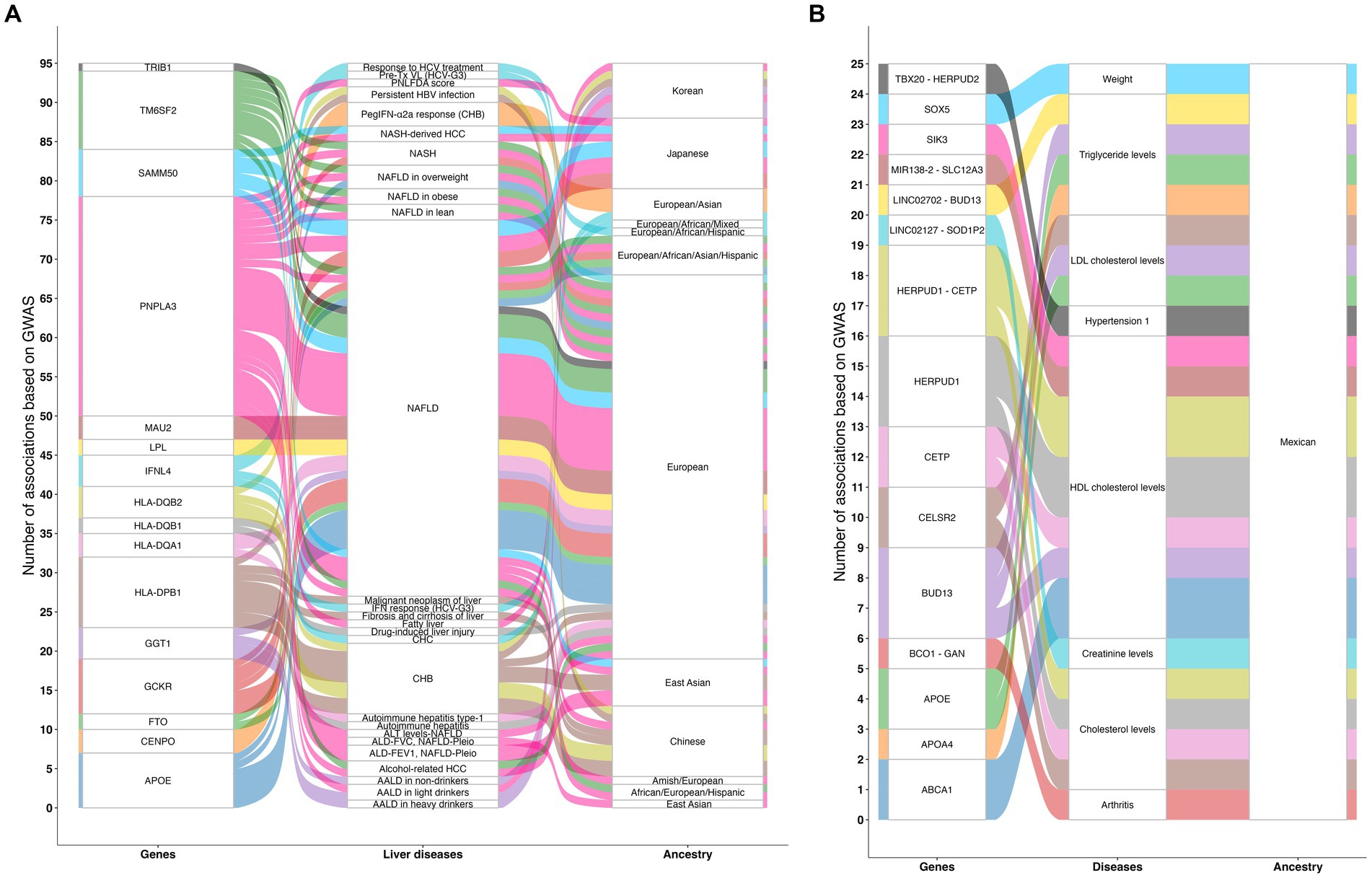
Figure 2. Genomic studies related to liver diseases. (A) Illustration of 284 GWAS related to liver diseases that have been conducted, identifying 1,546 associations in over 606 genes that involve the regulation of body weight and fat mass, glucose metabolism, peptide metabolism, lipid metabolism and transport, centromere structure and function, immune system, response to viral infections, meiotic cell division, mitochondrial membrane protein assembly, and cellular proliferation. (B) Illustration of the results of the Mexican Biobank analysis showing 25 associations in 15 genes involved in lipid metabolism and transport, vitamin A metabolism, protein folding, regulation of gene expression, cellular signaling, embryonic development, and heart development. Among the most important clinical associations are hypercholesterolemia, hyper triglyceridemia, body weight, creatinine levels, hypertension, and arthritis. The lines on the chart indicate the number of associations found between each gene, disease, and ancestral group. GWAS, Genome-wide association study; ALD-FEV1, NAFLD-Pleio, Accelerated lung function decline (FEV1) and worsened/persistent change in fatty liver (pleiotropy); ALD-FVC, NAFLD-Pleio, Accelerated lung function decline (FVC) and worsened/persistent change in fatty liver (pleiotropy); ALT levels-NAFLD, Alanine aminotransferase levels in non-alcoholic fatty liver disease; AALD in heavy drinkers, Alcohol-associated liver disease in heavy drinkers; AALD in light drinkers, Alcohol-associated liver disease in light drinkers; AALD in non-drinkers, Alcohol-associated liver disease in non-drinkers; Alcohol-related HCC, Alcohol-related hepatocellular carcinoma; Autoimmune hepatitis, Autoimmune hepatitis in primary sclerosing cholangitis; Autoimmune hepatitis type-1, Autoimmune hepatitis type-1; CHB, Chronic hepatitis B infection; CHC, Chronic hepatitis C infection; Drug-induced liver injury, Drug-induced liver injury (amoxicillin-clavulanate); Fatty liver, Fatty liver; Malignant neoplasm of liver: ICD10 C22, Malignant neoplasm of liver and intrahepatic bile ducts; Fibrosis and cirrhosis of liver: ICD10 K74, Fibrosis and cirrhosis of liver; NAFLD, Nonalcoholic fatty liver disease; NASH, Nonalcoholic steatohepatitis; NASH-derived HCC, Nonalcoholic steatohepatitis-derived hepatocellular carcinoma; PNLFDA score, Pediatric non-alcoholic fatty liver disease activity score; PegIFN-α2a response (CHB), Peginterferon alfa-2a treatment response in chronic hepatitis B infection; Persistent HBV infection, Persistent hepatitis B virus infection; Pre-Tx VL (HCV-G3), Pre-treatment viral load in hepatitis C virus genotype 3; Response to HCV treatment, Response to hepatitis C treatment; IFN response (HCV-G3), Response to interferon treatment in hepatitis C virus genotype 3. Source: GWAS catalog liver diseases, licensed under CC0. Summary statistics were downloaded on March 11, 2024 from the NHGRI-EBI GWAS Catalog. Accession ID: EFO_0001421.
Overall, the underlying genetic heterogeneity of most LATAM populations including Mexico constitutes a key aspect that makes sub-structure analysis and GWAS combined important tools to reveal which lineage is carrying the disease-related genetic polymorphisms and, consequently, the genetic foundation of disease vulnerability (17, 45). As shown in Figure 3A, regional variations in the ancestral component carrying the genetic variants as well the interactions with environmental and lifestyle factors support the need of regional PerMed-Nut strategies to address these differences (46).
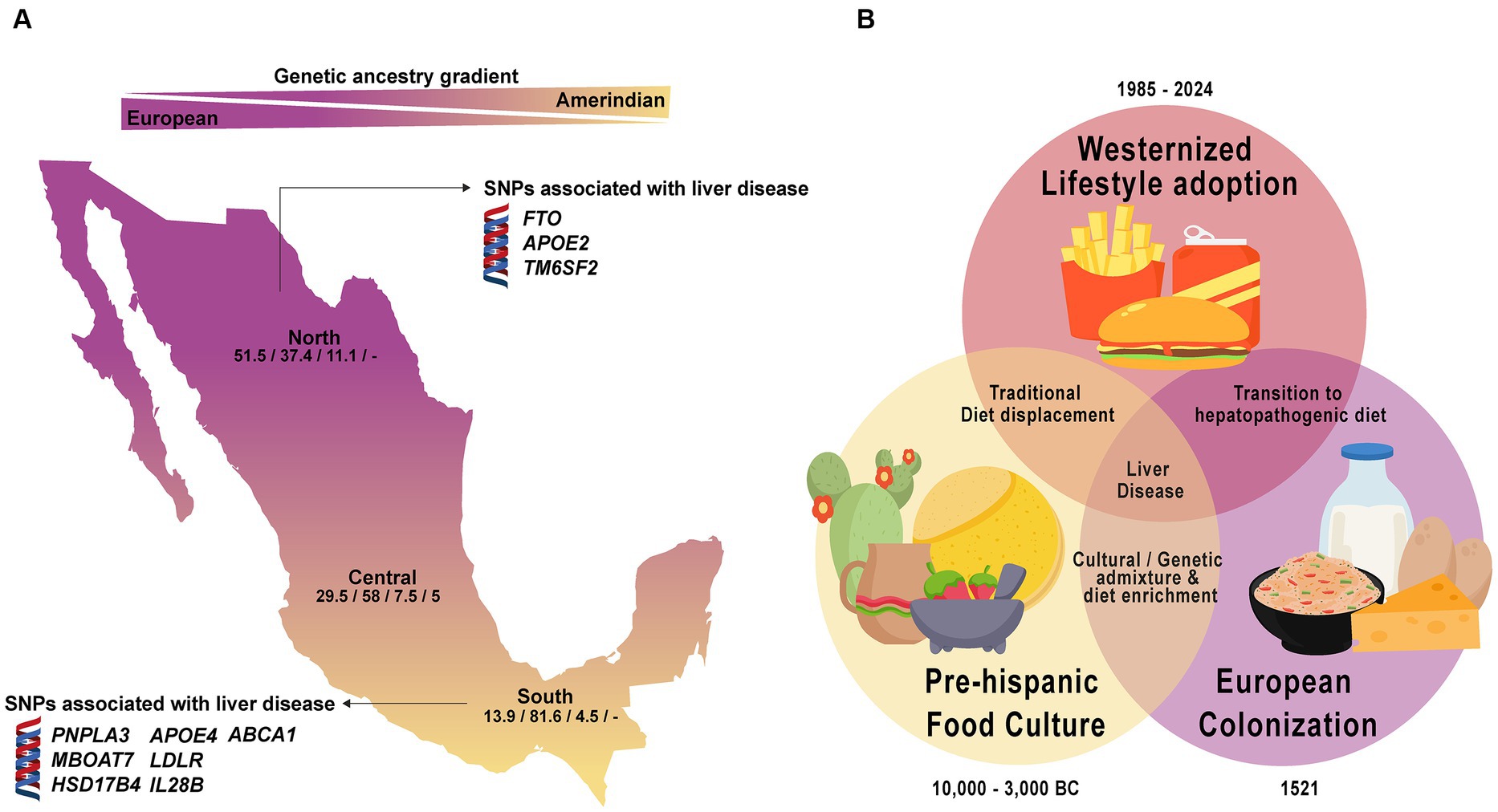
Figure 3. Ancestral genetic gradient and nutrition transition in Mexico. (A) Contemporary Mexicans show a tripartite ancestry admixture with predominant European ancestry in the North and increasing Amerindian ancestry towards the southern region. Genetic variants linked to liver disease vary based on population ancestry. Numbers in the map represent the average percentage of the European/Amerindian/African/Asian component. (B) Dietary transition in Mexico reflects a shift from traditional to a westernized hepatopathogenic diet, contributing to a higher liver disease prevalence in the population.
3 Environmental changes and evolutionary genetic mismatch
One important factor in the gene–environment interaction equation is diet. The domestication of endemic wild plants and animals by the prehispanic groups of ancient Mexico, approximately 6,000 years ago, is a historic milestone for the peopling and development of the Aztec civilization and other ethnic groups because it provided the primary source of nutrients for the indigenous communities (47). Mexico is noted for three staple plants: maize, beans, and chili, along with squash, tomato, amaranth, and chia growing in an eco-agricultural style known as “milpa” using the “chinampas” system (48). The inverted Mesoamerican food pyramid contains edible leafy green vegetables and wild fruits at the top, cereals/grains at the middle, and animal meat at the bottom, implying that it was consumed in smaller quantities because animal rearing was uncommon. The exposure to this diet over millennia may have exerted positive selective pressure on the Mexican genome, favoring the dominance of diet-related adaptive gene (DRAG) polymorphisms that regulate essential energy, immunological, and nutritional pathways (47). Based on the earlier environmental conditions, the adaptive allele, which was selected over the wild allele, is a risk allele for disease in unfavorable conditions such as the occurring globalized nutrition transition (48–50). Different allelic frequencies of these DRAGs may influence the risk of complex diseases among populations based on interactions with dietary contexts, such as the current obesity endemic. For example, the cholesterol transporter ATP-binding cassette transporter A1 (ABCA1) gene variant (R230C, rs9282541) is unique to Native American individuals and has been associated with low high-density lipoprotein cholesterol (HDL-C) levels, obesity and type 2 diabetes in admixed Mexicans (51, 52). The positive selection of the C allele among the Native American population may be related to energy-saving processes among the ancient Mesoamericans. Overall, the current frequency of the ABCA1 230RC + CC genotypes ranges from 6 to 20% (46) and studies performed in central west Mexico have shown that up to 41% of Native American individuals who are carriers of the ABCA1 C allele have low HDL-c levels compared to admixed Mexicans in 7% (53), suggesting the need of targeted strategies among the population.
In this sense, populations that maintain their historical staple foods are less likely to develop nutrition-related disorders (54). However, when combined with imbalanced modern diets, these adaptive genes may generate pathological processes. In Mexico, globalization and urbanization have driven the nutrition transition, leading to a shift toward a hepatopathogenic diet containing ultra-processed foods, unhealthy dietary habits, and an imbalance in essential fatty acids, vitamins, and minerals (48, 55) (Figure 3B). Notably, a recent study analyzing data from the National Health and Nutrition Survey (2018–2019) revealed that 68% of the Mexican population consumes a western dietary pattern high in processed foods compared to only 7% of those following a traditional Mexican diet (56). Currently, 72.1% of the Mexican population is overweight or obese; this risk factor, in combination with genetic vulnerability, has increased the incidence of cardiovascular disease, type 2 diabetes, cerebrovascular events, hepatopathologies, and cancer (breast and prostate) (57). Thus, understanding the ancestral and historical transitions is the foundation for developing a regionalized PerMed-Nut approach to address the dynamic interplay between genetic and environmental features that could affect the outcome of liver diseases in this region.
4 Managing chronic liver disease using a PERMED-NUT approach
4.1 Metabolic-associated liver disease
Genetic susceptibility (Figure 2) and the consumption of an unhealthy diet are the two most important risk factors related to metabolic-associated steatotic liver disease (MASLD) (formerly known as non-alcoholic fatty liver disease, NAFLD), which encompasses fatty liver or steatosis and metabolic-associated steatohepatitis (MASH) (formerly non-alcoholic steatohepatitis, NASH). Hence, a PerMed-Nut approach for the management of MAFLD would require knowledge of the allele/genotype frequency of several genes involved in food preferences and lipid/carbohydrate metabolism.
Food preference genes may partially explain the risk of abnormal lipid parameters and liver damage among the Mexican population. In this regard, some candidate-gene studies have been performed one example is the A allele (rs1761667) of the class B scavenger (CD36) receptor gene, which was found to be associated with higher fat intake and hypercholesterolemia in overweight individuals and more instances of liver damage in chronic hepatitis C patients (58, 59). Moreover, Mexicans carrying the Val allele (rs35874116) of the sweet taste receptor (TAS1R2) gene consumed more carbohydrates and had increased serum triglyceride levels (60). Furthermore, the novel AVV haplotype of the bitter taste receptor (TAS2R38) gene was first identified in the Mexican population and was associated with high alcohol intake (61). However, because genome structure corrections were not performed in these studies, further research is required to confirm these results.
Apolipoprotein E (APO E) is a plasma protein that transfers lipids from circulating lipoproteins to tissues through binding membrane receptors (62). The APOE gene encodes three major protein isoforms, E2, E3, and E4 with differential binding receptor affinities (63). The frequency of the APOE alleles differs worldwide (64). Recently, we identified a differential distribution of lipid genes among several ethnic groups showing that the E4 allele associated with hypercholesterolemia was more frequent in Mexican Amerindians, whereas those associated with hypertriglyceridemia such as E2 are common among European populations ancestry (53).
Another functional gene is the Patatin-like phospholipase domain-containing protein 3 (PNPLA3) I148M (rs738409 C > G) variant, one of the most studied genetic determinants implicated in liver damage associated with the generation of a pro-inflammatory environment and oxidative stress (65, 66). This variant has shown a geographic relationship with the prevalence of MASH and it has been documented that the PNPLA3 risk G allele (148 M) highly prevails in Mexican Amerindians compared to admixed populations (67). This variant has been strongly associated with elevated alanine transaminase (ALT) levels in normal weight and overweight/obese Mexican children suggesting that it may be a risk factor for liver damage (68). Furthermore, the Transmembrane 6 superfamily 2 (TM6SF2) gene is involved in plasma lipid regulation by influencing triglyceride secretion and hepatic lipid droplet content. The TM6SF2 E167K (rs58542926) polymorphism has been associated with impaired hepatic lipid synthesis in NAFLD patients (69). Additionally, the fat mass and obesity (FTO) associated gene encoding the 2-oxoglutarate dependent DNA/RNA methylase interacts with lifestyle factors. The A allele of the FTO A > T (rs9939609) polymorphism in interaction with modifiable lifestyle factors has been associated with energy homeostasis, eating behavior, and appetite with an impact on body weight. Notably, some populations with European lineage have shown a higher risk allele frequency, thus predisposing them to extreme obesity (70).
Therefore, screening these genetic variants could be useful to identify genetically susceptible groups and prescribe tailored genome-based nutritional advice under a regionalized and personalized scope. On the other hand, the nutritional composition of the hepatopathogenic diet may lead to dyslipidemia (including hypertriglyceridemia), insulin resistance, oxidative stress, and the induction of a pro-inflammatory state, which jointly may contribute to the development of liver damage. In this context, a high prevalence of MASH and abnormal liver stiffness was reported in young Mexicans with obesity, even in normal-weight individuals consuming a high-fat/high-sugar diet (71). Altogether, these data reveal that among the admixed population of Mexico, the interaction between genes and lifestyle factors such as diet significantly impact liver health. Preventing the onset and progression of liver injury should be a priority by healthcare providers to avoid further disease. In this context, intervention studies have been conducted to avoid plausible diet-related chronic diseases. By using a genome-based nutrigenetic strategy, a Genomex diet was implemented to provide a resource for managing patients with a risk of chronic disease (72). This diet provided the appropriate recommended daily allowances of macro-, and micronutrients considering the staple foods and culture of the Mexican population and was based on the prevalence of some adaptive alleles that are predominant among the population. In this manner, patients who had altered metabolic parameters were normalized in 24 weeks with a significant reduction in body weight, BMI, insulin resistance, hypertriglyceridemia, and VLDL levels. Although this was a quasi-experimental study evaluating the effect on metabolic and clinical parameters before and after the Genomex intervention, a randomized trial evaluating the effect of a dietary intervention integrating some Mexican staples vs. a habitual diet on metabolic syndrome parameters yielded similar results. Only patients on the Mexican food-enriched diet decreased their triglyceride levels, glucose intolerance, and area under the curve for insulin, in addition, carriers of the ABCA1 230C variant showed greater weight loss and increased serum adiponectin (73).
4.2 Alcoholic liver disease
Alcoholic liver disease (ALD) is a chronic condition that requires a multidisciplinary approach involving medical, nutritional, and psychosocial interventions, which are key to avoiding disease recurrence and managing complications. Early intervention and adherence to treatment plans are crucial for improving outcomes in patients with ALD (74). The development of the disease is multifactorial, involving interactions between genetic susceptibility and lifestyle factors, such as alcohol consumption. Thus, PerMed-Nut strategies could be beneficial to tailor patient’s needs based on genetic and environmental factors.
Genes involved in alcoholism and the risk for chronic liver disease include taste receptors, neurotransmitter receptors, alcohol-metabolizing enzymes, and steatotic genes that are highly polymorphic worldwide (75, 76). Notably, some of these genes overlap with the risk for MAFLD and HCC. In a Mexican cohort, a 36% TAS2R38 AVV haplotype homozygosity (non-taster phenotype) was found among drinkers compared to non-drinkers (61). Additionally, the DRD2/ANKK1 Taq1A1 (rs1800497) polymorphism involved in addictive behaviors ranges up to 67% among the Amerindian subpopulations compared to 47% in the admixed groups (77). Likewise, a cohort of subjects displaying unhealthy food choices and altered biochemical parameters showed a high prevalence of this genetic variant (78). Thus, the riskier TAS2R38/DRD2/ANKK1 genetic profile (affecting bitter taste perception and food reward, respectively) predisposes one to consume significantly more calories (alcohol or high-dense food), which may enhance hepatic damage. However, these associations need to be explored in other regions across Mexico and even in other populations worldwide.
Additionally, the genetic profile of the alcohol-oxidizing liver enzymes among the Mexicans reveals a high rate of the A1 allele (rs1229984) of the alcohol dehydrogenase 1B (ALD1B) gene, contrasting with a low rate of the protective A2 allele (rs671) of the aldehyde dehydrogenase (ALDH2) gene whereas the cytochrome P450 2E1 (CYP2E1) C2 allele (rs2031920) is highly present in the Amerindian populations (79). It is noteworthy to mention that despite a high-risk genetic profile tending toward liver damage, the peril will depend on both the specific combination of the risk or protective alleles (slow, intermediate, or fast metabolizers) and the pattern of alcohol consumption influenced by socio-cultural factors (75).
In this sense, the history of alcohol consumption among the Mexicans has evolved over the centuries in which distilled liquor was not regularly consumed by the Mesoamericans until the arrival of the Europeans. Most alcoholic beverages were fermented and obtained mainly from maize (tesgüino) or fruits (pulque from the agave plant), and mostly nobles (emperors or priests) had access to them. Only in circumstances such as illness o festivities were the laypeople allowed to consume alcoholic beverages. In contrast, the national alcohol consumption is currently an average of 4.4 L/per capita in young adults who begin to drink at early ages during the weekends despite public health warning measures, and eventually, alcohol intake increases (79). Thus, risk scores containing both the genetic and social factors involved in alcohol consumption should be taken into account to devise preventive strategies.
4.3 Hepatitis B infection
The first step in using PerMed-Nut strategies in the management of hepatitis B patients is to make an appropriate diagnosis. Personalized therapy is highly dependent on the patient’s genetic features and HBV genotype (80). HBV is divided into 10 HBV genotypes (A-J) based on the whole genome, each with a unique global distribution and clinical outcomes based on the interactions between the host and the virus (81). To date, five HBV genotypes may be significant for PerMed-Nut in Mexicans with different clinical implications. Among these, HBV genotypes H is predominant followed by G, F, A, and D which circulate in patients according to genetic and sociodemographic backgrounds (82, 83). Based on molecular epidemiology data, HBV genotypes A and D discovered in Mexico are deemed exotic when compared to the Americas’ prevalent H and F. Indeed, phylogenetic examination of those genotypes suggested that A2 originated in Europe and D4 in Africa. These findings are consistent with the demographic history of the Mexican population, as previously stated.
Regarding the clinical result, HBV genotypes A and D have been identified in individuals with chronic infection, but genotype H is common in occult hepatitis B cases and exhibits a high degree of adaptation among the local people (82). Interestingly, sub-genotype F1b native to South America and linked to HCC in Native Alaskans (84) and Native Peruvians (85) has been found in Mexican patients (86). However, the potential association of liver cancer in patients acutely infected with F1b has not been tested due to its low incidence (87). Nonetheless, among HIV or men who have sex with men (MSM) individuals, the presence of three HBV genotypes (in various combinations) increased the likelihood of severe liver fibrosis by 15-fold compared to dual-mixtures or single HBV genotype infections (88). As a result, in these populations, early diagnosis of the HBV genotype may help determine the risk of liver injury. Likewise, chronic hepatitis B patients often remain asymptomatic until decompensation, making late diagnosis of cirrhosis a challenge (89). Genetic markers could improve early diagnosis and prevent end-stage liver disease. Genome-wide association (Figure 2) and SNP studies reveal associations with clinical outcomes (90).
Pharmacological treatment is the primary target for managing chronic liver disease, considering mutations in the reverse transcriptase domain of the polymerase. The use of lamivudine should be avoided in samples with M204V/I or L180M + M204V mutations, and tenofovir should be monitored when quadruple mutation CYEI is detected (91). The main mutations associated with resistance to adefovir are A181T/V/S and N236T (92). Also, response to long-term lamivudine may be decreased in patients with genotype A since it is more susceptible to YMDD motif mutations (93).
Together with pharmacological treatment, adjuvant nutritional support, including vitamins and bioactive components like resveratrol, vitamin E, lactoferrin, selenium, curcumin, luteolin-7-O-glucoside, moringa extracts, chlorogenic acid, and epigallocatechin-3-gallate showing in vitro and in vivo anti-HBV activity may be beneficial for managing chronic hepatitis B patients. These nutrients are accessible and found in native American foods (94). The effectiveness of nutritional therapy will depend on the patient’s capacity to absorb anti-HBV nutrients, HBV genotype, and population ancestry.
4.4 Hepatitis C infection
4.4.1 Immune response
PerMed-Nut strategies should include early detection of patients at risk of chronic HCV infection. Next-generation sequencing-based studies have identified human variants that predispose to susceptibility or are associated with self-resolution (95, 96). Some genetic markers are associated with HCV infection outcome, as they influence the immune response and lipid metabolic pathways. The interleukin 28-B (IFNL3) gene belonging to the type III IFN-β family is one of the best predictors of HCV infection outcome and induces antiviral and antitumor states through innate and adaptive immune responses (97, 98). The polymorphism, rs12979860 C/T, is located 3 kb upstream of the IFNL3 gene. Patients homozygous for the CC genotype are more likely to achieve a sustained virological response (SVR) following a PEG-IFN-β plus ribavirin than CT and TT genotypes (99). The rs12979860-CC genotype, associated with spontaneous clearance in white, African, and Hispanic HCV patients, was found to be proportional to the SVR rate across ethnic groups (100, 101). In Mexico, the C allele has a frequency of 56.5% in admixed patients and was associated with self-clearance and less liver damage (101). Similarly, a polymorphism in the IFNL4 rs368234815 (TT) gene was also associated with HCV clearance in Mexican mestizo patients. When these polymorphisms were analyzed in haplotype with the IFNL3 rs8099917 SNP, the beneficial haplotype C/T/TT (rs12979860, rs8099917, and rs368234815) was associated with spontaneous clearance and less liver damage in Mexican patients (101).
4.4.2 Lipid metabolism
APOE has several ligands in the hepatocyte, including low-density lipoprotein receptor (LDLR), syndecan-1 and syndecan-2, and heparan sulfate proteoglycans (102). APOE is also a structural component of the HCV envelope and mediates virion entry, assembly, and production (62). The E4 allele is protective against HCV, as carriers have a higher chance of spontaneous recovery after infection and treatment with interferon plus ribavirin (103, 104). The E2 allele protects against chronic infection, while the E3 is associated with chronic infection (105). In Mexico, the E4 allele has been associated with spontaneous clearance and less liver damage, while E3 was associated with advanced fibrosis in chronic patients, which is present in 85% of the mestizo population (72), however, the E2 allele exerted no effect (106). Therefore, there is a higher risk of developing advanced liver damage induced by HCV.
Clustering differentiation 36 (CD36) is a candidate receptor involved in the gustatory detection of lipids. Evidence suggests that genetic variation in the CD36 gene can modulate the uptake of fatty acids. A promoter polymorphism -31118G > A, rs1761667 was studied among admixed Mexican HCV patients. It was demonstrated that the AA genotype had higher values of total fat and saturated fatty acids than non-GG genotypes. Additionally, AA genotype increased AST and liver fibrosis among chronic HCV-infected patients (59). It is known that the AA genotype decreases fat taste perception, thus, AA carriers could need an increased amount of fat to reach satiety. Thus, triggering the onset of fat liver accumulation that in conjunction with HCV infection potentializes liver damage among HCV patients.
The low-density lipoprotein receptor (LDLR) is a protein involved in the trafficking of lipoproteins containing APOE, such as VLDL and chylomicrons (107, 108). The rs688 C/T polymorphism protects against infection development, affects cholesterol levels and mRNA splicing, and decreases receptor surface expression levels. This makes LDLR less capable of lipid uptake, making it less susceptible to HCV.
As mentioned above, nutrition also plays a crucial role in HCV infection because it can modify serum lipid components and provide anti-HCV micronutrients. In vitro studies have shown the effect of certain nutrients with anti-HCV properties on the outcome of HCV infection, but no potential diet intervention has been performed in patients (55). A recent study in Mexican patients revealed that adherence to a fish-rich diet, mainly consisting of fish, seafood, and vegetable oils, had low viral loads and significant consumption of PUFAs ≥4.9% (109). Therefore, diets rich in anti-HCV macro- and micronutrients may affect HCV infection outcomes. Further dietary interventions are needed to clarify the role of diet in the management of HCV infection.
4.5 Hepatocellular carcinoma
Hepatocellular carcinoma seen as the end-stage outcome of chronic liver disease is influenced by various gene–environment interactions that vary depending on the population (81). In Mexico, HCC is low in prevalence which may be due to the population’s immune response to the HBV/H genotype, despite the high rate of occult HBV infection (82). Nonetheless, the influence of the emergent F1b, A, and D genotypes on the natural history of HCC has not been thoroughly studied (86).
Genetic variations involved in the development of HCC are very diverse, and each example correlates with specific susceptibility. The aforementioned PNPLA3 I148M allele associated with MAFLD/MASH has been linked to the development of HCC in patients with obesity (65, 110). In Europe, individuals with the PNPLA3 GG genotype, particularly those with severe obesity, have a higher risk of developing HCC (111, 112). In Japan, patients with obesity/MAFLD or chronic hepatitis C infection have a higher frequency of the GG genotype, making them more susceptible to HCC (113, 114). Furthermore, association of rs738409 PNPLA3 and rs58542926 TM6SF2 as a risk factor for HCC was reported in patients with different liver disease etiologies (115), suggesting that underlying altered lipid metabolism influences the outcome of chronic liver disease.
The Membrane-bound O-acyltransferase domain containing 7 (MBOAT7), also known as lysophosphatidylinositol acyltransferase 1 is involved in the remodeling phospholipid chains and controlling cell membrane desaturation (116). The reduced expression of the MBOAT7 rs641738 T allele is associated with altered cell membranes and plasma composition, including cell fat accumulation, pro-inflammatory environment, MAFLD, MASH, severe fibrosis, and HCC (117–121). An Italian cohort showed an 80% increased risk of HCC due to the presence of the MBOAT7 rs641738 T allele, contributing to the evolution of liver disease (118). However, the rs641738 T allele was not associated neither with HCC risk nor HBV infection in Chinese patients. (122).
Likewise, the hydroxysteroid 17-Beta dehydrogenase 13 protein encoded by the HSD17B13 gene is involved in the metabolism of steroid hormones, prostaglandins, lipids, xenobiotics, and retinoids (123). This liver lipid droplet-associated enzyme is markedly upregulated in patients with MAFLD (124). HSD17B13 expression has been linked to steatosis, MASH, type 2 diabetes, and liver cancer (125) and was shown to be downregulated in an HCC model (126). Some HSD17B13 variants such as the rs72613567 T > A have been associated with a protective effect in cirrhotic and HCC patients (127) as well as in European patients with ALD (128, 129) and HCV patients (130). In a recent study, this polymorphism was detected in patients with extreme obesity and MASH among other gen variants related to HCC (131). However, further genetic studies are also needed for LATAM populations, including Mexico, to confirm these associations.
Finally, a healthy diet can be potentially protective against HCC. Recent studies have identified nutrients, dietary patterns, and food groups with reduced, neutral, and high risk of developing HCC (132, 133). The nutrients reducing the risk of HCC are monounsaturated fatty acids, vitamin E, vitamin B9, beta-carotene, manganese, and potassium. Conversely, sodium, processed red meat, and sugar-sweetened beverages increase the risk. Currently, Mexicans consume a hepatopathogenic diet (48) with a potentially high risk of developing MAFLD/MASH setting the scenario that HCC may be underdiagnosed or increase in the future years (71, 134). The Genomex diet, containing a high content of protective nutrients has been implemented to prevent developing chronic liver disease (72). Furthermore, it has been reported that the treatment of alkaline-brined corn dough used to make the widely consumed Mexican tortillas can potentially protect against aflatoxin-induced HCC (135). However, further studies are needed to validate the role of diet in patients at risk of HCC, given the current prevalence of overweight and obesity.
5 Genomic education
Medical specialties and subspecialties, in their first stage of scientific medicine focused more on the complications of chronic disease, prolonging the patients’ life and improving their quality of life, but do not prevent in all cases disease remission (136). Faced with this situation, PerMed-Nut approaches based on genomic medicine/nutrition need to integrate medical, nutritional, sociocultural, and emotional/spiritual aspects of health (137). However, new medical education and training for medical professionals are required (138, 139). In LATAM and Mexico, updating the medical and other health sciences education curriculum both at the undergraduate and graduate levels must be considered a priority. This curriculum needs to integrate the so-called basic subjects: Biochemistry, Cell Biology, Genetics/Genomics, and Molecular Biology at different levels with bioinformatics, technological/digital strategies (Telemedicine) and novel clinical approaches focused on preventing chronic diseases and long-term complications (140). These basic subjects can be integrated sequentially during the first years of schooling, not independently, as in the past. In the last century, most medical specialties and careers in Nutrition were created separately. However, with this new understanding, physicians/clinicians need to integrate nutrition, while the nutritionist should handle the knowledge of disease the same way as the doctors work (141).
Likewise, hepatology was not born when most medical specialties were created (142). Although some hepatology postgraduate courses have been sponsored by the associations for the study of the liver to update medical specialists, pre-graduate doctors and nutritionists need to understand the molecular physiology of the liver, while more postgraduate courses in MedPer-Nut are required to advance in Genomic Hepatology (143). Furthermore, the capability to analyze hundreds or thousands of these genes simultaneously leads us to large-scale data management (Big Data) and specialized bioinformatics (machine learning and other artificial intelligence methods), together with advanced omics biotechnologies (epigenetics, metagenomics, proteomics, lipidomics, and metabolomics), digital media, and electronic medical records (144). These applications are not the Medicine of the future, they are the present Medicine that will require training and re-constructing health career curriculums.
Lastly, the major benefit of re-engineering the educational curriculum is to promote the formation of researchers in LATAM and Mexico, thus creating novel knowledge in the field of MedPer-Nut in this region (145). In this sense, transforming the present clinical practice guidelines is an ongoing challenge because most recommendations are based on studies carried out in foreign populations (146). As illustrated in Figure 4, the genetic profile interacting with the environmental factors that concur in Mexico, may not replicate in other regions and vice versa. Thus, the vision is to prevent the onset and progression of chronic liver disease using MedPer-Nut approaches concordant with the genetic and environmental characteristics of the population (147).
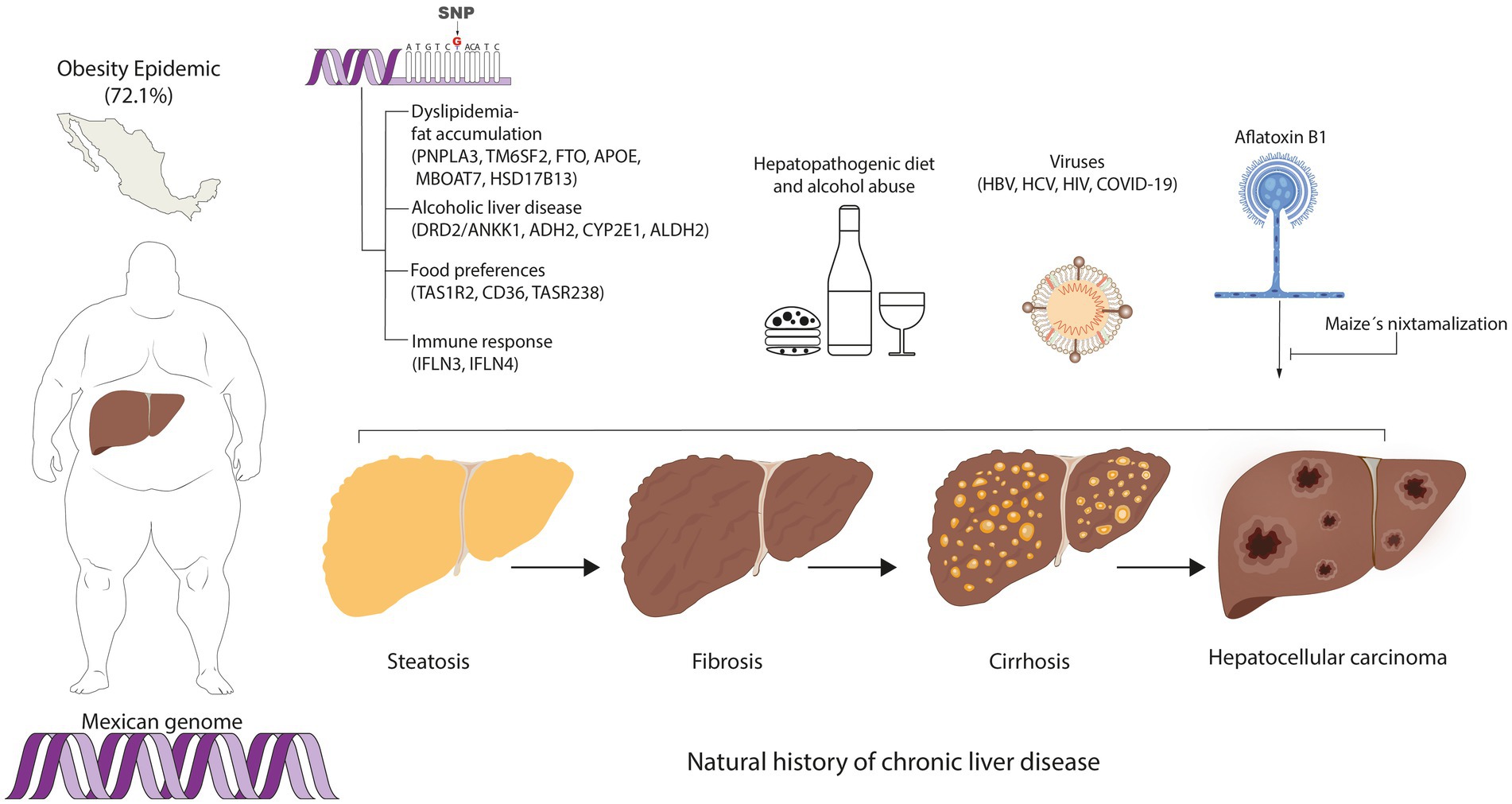
Figure 4. Personalized medicine and nutrition in hepatology among the Mexican population. The concurring genetic and environmental factors involved in chronic liver disease among the Mexican population are components to consider for the implementation of regionalized prevention strategies for chronic liver disease.
6 Discussion
In the field of Medicine, we are now moving toward a new landscape of what exactly health means and how the pathological process begins. A primary goal is to avoid the onset of chronic diseases such as obesity, type 2 diabetes, and cardiovascular and liver diseases with their associated complications in the advanced stages. Reaching this goal will require detecting the genetic susceptibility to such diseases at young ages or at early stages, avoiding the pathology’s development as is the case of liver cirrhosis due to alcohol, viral hepatitis, or MASLD. Such studies are the beginning of the pathway toward the new age of PerMed-Nut in liver diseases.
In perspective, 20 years ago, it was commonly said that genes and environment interact with each other, but it was not clear how such interactions took place, nor which were the main environmental factors. Currently, it is recognized that at least three environmental factors constantly interact with our genes: diet (micro and macronutrients), physical activity (exercise), and emotions (stressors) (137). Human adaptability to these elements lies in the genetic variations gained through evolution that mark the differences between individuals or populations at the genomic and cultural levels (47, 148, 149). As shown in this review, within LATAM, despite having a common social history of the peopling of the continent, a wide spectrum of ancestral inter-variability is notable as well a distinct environmental conditions (24, 25, 28, 33). In the case of Mexico, it has been shown that the North–South gradient of the European-Amerindian ancestry impacts importantly in the distribution of the several genes with biomedical implications for the risk of chronic liver diseases (Tables 2, 3; Figure 3) (32, 44, 45, 153). More so, this pattern of distribution could also be replicated intra-regionally in light of the social-demographic movements that occurred over time as in the case of the ABCA1 polymorphism (46). This feature is the reason for further studies regarding the plausible association of certain genetic alleles with the clinical outcomes to tailor preventive strategies.
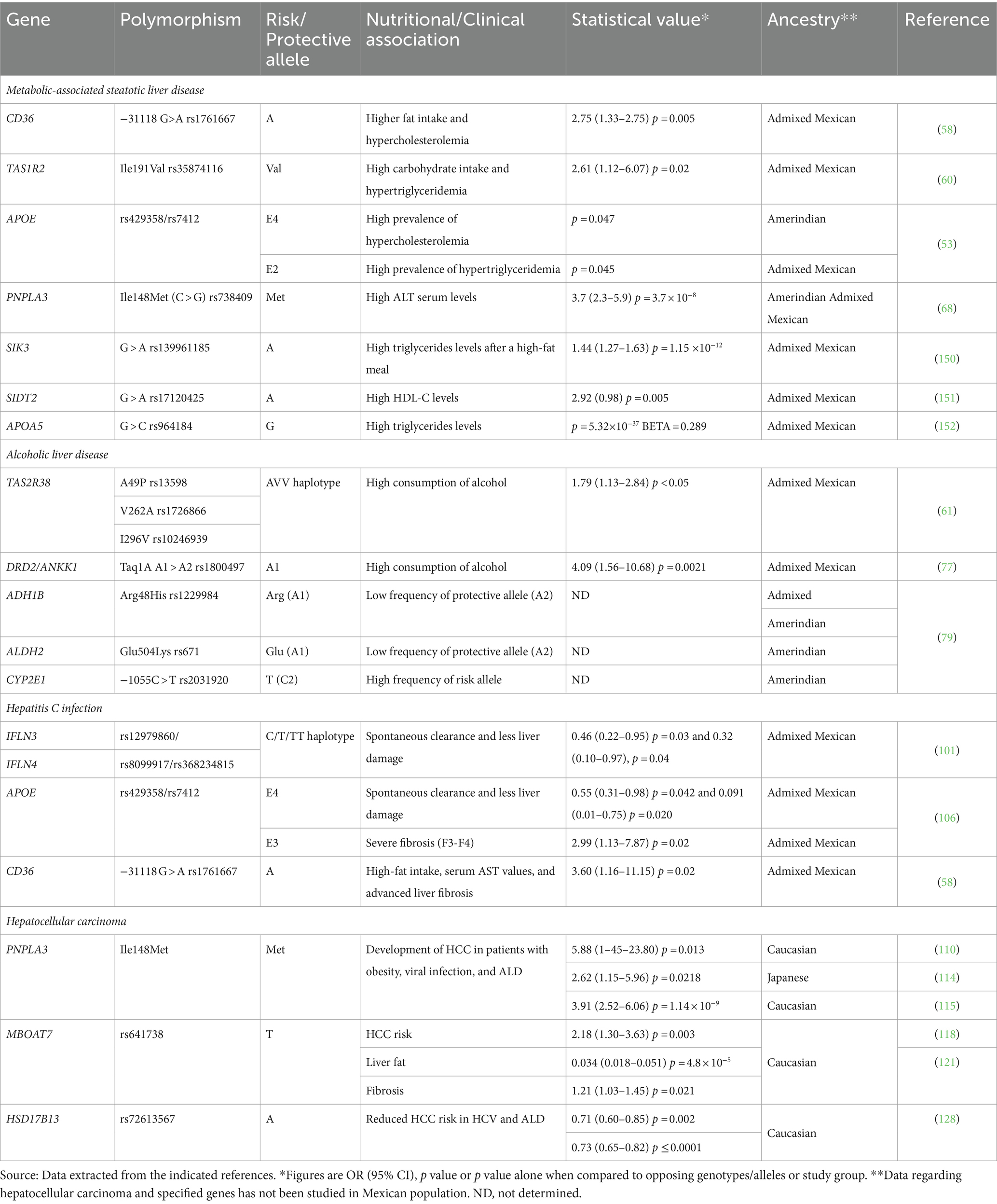
Table 3. Summary of genetic variants related to etiologies of liver conditions or disease based on ancestry.
The diet-related alleles that have been studied to date among the Mexican population are key elements in the development of chronic illnesses including liver disease. As shown in Table 3, some polymorphisms are related to high food preferences for carbohydrates or lipids, altered food behaviors (excessive alcohol consumption), and dyslipidemias that can partly explain the increasing onset of MAFLD among the Mexican population. Interestingly, several genomic studies in Mexican Amerindian and admixed cohorts have identified the signature of positive selection of some serum lipid-modulating gene traits involved in cardiovascular diseases (150–152) that plausibly could overlap with the natural history of MAFLD. Overall, these genes can be useful for the development and validation of polygenic risk scores testing for the susceptibility in liver diseases among the Mexican population in the context of the tripartite ancestry (154–156). On the other hand, the impact of nutrition in patients with chronic viral hepatitis, specifically hepatitis B and C is an opportunity of exploration since patients with these pathologies have unmet needs in terms of their nutrition care to prevent further liver damage due to genetic susceptibility and harmful eating habits. Ultimately, understanding the molecular basis of these pathologies and the interactions that take place with the environment will derive in the making of a PerMed-Nut strategy appropriate for the Mexican population focused on the implementation of preventive strategies based on nutrition, exercise, and mental health instead of treating advanced diseases (157).
7 Conclusion
LATAM including Mexico shares a common ancestry in terms of genetics and culture that have an impact on health. However, further research regarding the distribution of genetic risk alleles and the interaction with environmental factors is required in this region. PerMed-Nut strategies based on these factors are the forthcoming trend in the field of Genomic Hepatology and other fields of medical specialties for preventing and managing chronic liver diseases. Knowledge of the genetic and lifestyle factors involved in the onset and progression of the major etiologies of chronic liver diseases needs to be generated by local researchers to provide regional clinical practice guidelines concordant with the features of the population. Achieving this PerMed-Nut approach is not without challenges since updated Medicine and Nutrition education curriculums are required. Training and preparing future health professionals and researchers with new clinical and investigative skills focused on preventing liver diseases in the field of Genomic Hepatology globally is a vision that clinicians and nutritionists should be concerned about.
Author contributions
AP: Conceptualization, Investigation, Visualization, Writing – original draft, Writing – review & editing. SR: Conceptualization, Investigation, Visualization, Writing – original draft, Writing – review & editing. IM-M: Investigation, Visualization, Writing – review & editing. AJ-A: Investigation, Visualization, Writing – review & editing. KG-A: Investigation, Visualization, Writing – review & editing. CO-G: Investigation, Visualization, Writing – review & editing. OR-L: Investigation, Visualization, Writing – review & editing. LT-R: Investigation, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Shriner, D, Tekola-Ayele, F, Adeyemo, A, and Rotimi, CN. Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Sci Rep. (2014) 4:6055. doi: 10.1038/srep06055
2. Shriner, D, Tekola-Ayele, F, Adeyemo, A, and Rotimi, CN. Ancient human migration after out-of-Africa. Sci Rep. (2016) 6:26565. doi: 10.1038/srep26565
3. Adhikari, K, Chacón-Duque, JC, Mendoza-Revilla, J, Fuentes-Guajardo, M, and Ruiz-Linares, A. The genetic diversity of the Americas. Annu Rev Genomics Hum Genet. (2017) 18:277–96. doi: 10.1146/annurev-genom-083115-022331
4. Harcourt, AH . Human phylogeography and diversity. Proc Natl Acad Sci USA. (2016) 113:8072–8. doi: 10.1073/pnas.1601068113
5. Fan, S, Hansen, MEB, Lo, Y, and Tishkoff, SA. Going global by adapting local: a review of recent human adaptation. Science. (2016) 354:54–9. doi: 10.1126/science.aaf5098
6. Fu, W, and Akey, JM. Selection and adaptation in the human genome. Annu Rev Genomics Hum Genet. (2013) 14:467–89. doi: 10.1146/annurev-genom-091212-153509
7. National Institute of Health. National Human Genome Research Institute Human Genomic Variation (2024). Available at: https://www.genome.gov/about-genomics/educational-resources/fact-sheets/human-genomic-variation (Accessed March 10, 2024)
8. O’Dea, RE, Noble, DWA, Johnson, SL, Hesselson, D, and Nakagawa, S. The role of non-genetic inheritance in evolutionary rescue: epigenetic buffering, heritable bet hedging and epigenetic traps. Environ. Epigenetics. (2016) 2:dvv014. doi: 10.1093/eep/dvv014
9. Ottman, R . Gene-environment interaction: definitions and study designs. Prev Med. (1996) 25:764–70. doi: 10.1006/pmed.1996.0117
10. Chial, H. (2024). Human Genome Project: Sequencing the Human Genome | Learn Science at Scitable. Available at: http://www.nature.com/scitable/topicpage/dna-sequencing-technologies-key-to-the-human-828 (Accessed January 24, 2024).
11. Lathe, W., Williams, J., Mangan, M., and Karolchik, D. (2024). Genomic Data Resources: Curation, Databasing, and Browsers | Learn Science at Scitable. Available at: https://www.nature.com/scitable/topicpage/genomic-data-resources-challenges-and-promises-743721/# (Accessed March 8, 2024).
12. Gonzaga-Jauregui, C, Lupski, JR, and Gibbs, RA. Human genome sequencing in health and disease. Annu Rev Med. (2012) 63:35–61. doi: 10.1146/annurev-med-051010-162644
13. Golubnitschaja, O, Baban, B, Boniolo, G, Wang, W, Bubnov, R, Kapalla, M, et al. Medicine in the early twenty-first century: paradigm and anticipation—EPMA position paper 2016. EPMA J. (2016) 7:23. doi: 10.1186/s13167-016-0072-4
14. Goldblatt, EM, and Lee, W-H. From bench to bedside: the growing use of translational research in cancer medicine. Am J Transl Res. (2010) 2:1–18.
15. Roth, SC . What is genomic medicine? J Med Libr Assoc. (2019) 107:442–8. doi: 10.5195/jmla.2019.604
16. Pokorska-Bocci, A, Stewart, A, Sagoo, GS, Hall, A, Kroese, M, and Burton, H. “Personalized medicine”: what’s in a name? Pers Med. (2014) 11:197–210. doi: 10.2217/pme.13.107
17. Romero-Hidalgo, S, Flores-Rivera, J, Rivas-Alonso, V, Barquera, R, Villarreal-Molina, MT, Antuna-Puente, B, et al. Native American ancestry significantly contributes to neuromyelitis optica susceptibility in the admixed Mexican population. Sci Rep. (2020) 10:13706. doi: 10.1038/s41598-020-69224-3
18. Ramos-Lopez, O, Milagro, FI, Allayee, H, Chmurzynska, A, Choi, MS, Curi, R, et al. Guide for current nutrigenetic, nutrigenomic, and nutriepigenetic approaches for precision nutrition involving the prevention and management of chronic diseases associated with obesity. J Nutr. (2017) 10:43–62. doi: 10.1159/000477729
19. Cheemerla, S, and Balakrishnan, M. Global epidemiology of chronic liver disease. Clin Liver Dis. (2021) 17:365–70. doi: 10.1002/cld.1061
20. Teng, ML, Ng, CH, Huang, DQ, Chan, KE, Tan, DJ, Lim, WH, et al. Global incidence and prevalence of nonalcoholic fatty liver disease. Clin Mol Hepatol. (2023) 29:S32–42. doi: 10.3350/cmh.2022.0365
21. Rotimi, CN, Tekola-Ayele, F, Baker, JL, and Shriner, D. The African diaspora: history, adaptation and health. Curr Opin Genet Dev. (2016) 41:77–84. doi: 10.1016/j.gde.2016.08.005
22. Heinz, T, Álvarez-Iglesias, V, Pardo-Seco, J, Taboada-Echalar, P, Gómez-Carballa, A, Torres-Balanza, A, et al. Ancestry analysis reveals a predominant native American component with moderate European admixture in Bolivians. Forensic Sci Int Genet. (2013) 7:537–42. doi: 10.1016/j.fsigen.2013.05.012
23. Coelho, AVC, Moura, RR, Cavalcanti, C, Guimarães, RL, Sandrin-Garcia, P, Crovella, S, et al. A rapid screening of ancestry for genetic association studies in an admixed population from Pernambuco, Brazil. Genet Mol Res. (2015) 14:2876–84. doi: 10.4238/2015.March.31.18
24. Norris, ET, Wang, L, Conley, AB, Rishishwar, L, Mariño-Ramírez, L, Valderrama-Aguirre, A, et al. Genetic ancestry, admixture and health determinants in Latin America. BMC Genomics. (2018) 19:861. doi: 10.1186/s12864-018-5195-7
25. Hughes, CE, Algee-Hewitt, BFB, and Konigsberg, LW. Population identifiability from forensic genetic markers: ancestry variation in Latin America. Hum Biol. (2018) 90:161–75. doi: 10.13110/humanbiology.90.3.03
26. Fernandes, DM, Sirak, KA, Ringbauer, H, Sedig, J, Rohland, N, Cheronet, O, et al. A genetic history of the pre-contact Caribbean. Nature. (2021) 590:103–10. doi: 10.1038/s41586-020-03053-2
27. Galarza, JM, Barquera, R, Álvarez, AMT, Hernández Zaragoza, DI, Sevilla, GP, Tamayo, A, et al. Genetic diversity of the HLA system in human populations from the Sierra (Andean), Oriente (Amazonian) and costa (coastal) regions of Ecuador. Hum Immunol. (2018) 79:639–50. doi: 10.1016/j.humimm.2018.06.004
28. Salzano, FM, and Sans, M. Interethnic admixture and the evolution of Latin American populations. Genet Mol Biol. (2014) 37:151–70. doi: 10.1590/S1415-47572014000200003
29. Simão, F, Ribeiro, J, Vullo, C, Catelli, L, Gomes, V, Xavier, C, et al. The ancestry of eastern Paraguay: a typical south American profile with a unique pattern of admixture. Gene. (2021) 12:1788. doi: 10.3390/genes12111788
30. Spangenberg, L, Fariello, MI, Arce, D, Illanes, G, Greif, G, Shin, J-Y, et al. Indigenous ancestry and admixture in the Uruguayan population. Front Genet. (2021) 12:733195. doi: 10.3389/fgene.2021.733195
31. Andrey, E. (2024). Civilizaciones Prehispanicas de America. Available at: https://www.academia.edu/19510611/Osvaldo_Silva_Galdames_Civilizaciones_Prehispanicas_de_America (Accessed January 24, 2024).
32. García-Ortiz, H, Barajas-Olmos, F, Contreras-Cubas, C, Reynolds, AW, Flores-Huacuja, M, Snow, M, et al. Unraveling signatures of local adaptation among indigenous groups from Mexico. Gene. (2022) 13:2251. doi: 10.3390/genes13122251
33. Chacón-Duque, J-C, Adhikari, K, Fuentes-Guajardo, M, Mendoza-Revilla, J, Acuña-Alonzo, V, Barquera, R, et al. Latin Americans show wide-spread converso ancestry and imprint of local native ancestry on physical appearance. Nat Commun. (2018) 9:5388. doi: 10.1038/s41467-018-07748-z
34. Martinez-Fierro, ML, Beuten, J, Leach, RJ, Parra, EJ, Cruz-Lopez, M, Rangel-Villalobos, H, et al. Ancestry informative markers and admixture proportions in northeastern Mexico. J Hum Genet. (2009) 54:504–9. doi: 10.1038/jhg.2009.65
35. Rubi-Castellanos, R, Anaya-Palafox, M, Mena-Rojas, E, Bautista-España, D, Muñoz-Valle, JF, and Rangel-Villalobos, H. Genetic data of 15 autosomal STRs (Identifiler kit) of three Mexican mestizo population samples from the states of Jalisco (west), Puebla (Center), and Yucatan (southeast). Forensic Sci Int Genet. (2009) 3:e71–6. doi: 10.1016/j.fsigen.2008.07.006
36. Silva-Zolezzi, I, Hidalgo-Miranda, A, Estrada-Gil, J, Fernandez-Lopez, JC, Uribe-Figueroa, L, Contreras, A, et al. Analysis of genomic diversity in Mexican mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci USA. (2009) 106:8611–6. doi: 10.1073/pnas.0903045106
37. González-Sobrino, BZ, Pintado-Cortina, AP, Sebastián-Medina, L, Morales-Mandujano, F, Contreras, AV, Aguilar, YE, et al. Genetic diversity and differentiation in urban and indigenous populations of Mexico: patterns of mitochondrial DNA and Y-chromosome lineages. Biodemography Soc Biol. (2016) 62:53–72. doi: 10.1080/19485565.2015.1117938
38. Granados-Silvestre, MA, Ortiz-López, MG, Granados, J, Canizales-Quinteros, S, Peñaloza-Espinosa, RI, Lechuga, C, et al. Susceptibility background for type 2 diabetes in eleven Mexican indigenous populations: HNF4A gene analysis. Mol Genet Genom. (2017) 292:1209–19. doi: 10.1007/s00438-017-1340-2
39. Bonilla, C, Gutiérrez, G, Parra, EJ, Kline, C, and Shriver, MD. Admixture analysis of a rural population of the state of Guerrero, Mexico. Am J Phys Anthropol. (2005) 128:861–9. doi: 10.1002/ajpa.20227
40. Torre-Cantalapiedra, E, Sánchez-Soto, G, Torre-Cantalapiedra, E, and Sánchez-Soto, G. Afro-descendants and social stratification in Mexico. New evidence from the 2015 intercensal survey. Papeles Poblac. (2019) 25:273–302. doi: 10.22185/24487147.2019.100.20
41. López-Ramírez, YL, Aguilar-Velázquez, JA, López-Armenta, M, Ruiz-Hernández, M, and Rangel-Villalobos, H. Paternal lineages and forensic parameters based on 23 Y-STRs (Powerplex® Y23) in mestizo males from Mexico City. Int J Legal Med. (2020) 134:199–202. doi: 10.1007/s00414-019-02183-1
42. GWAS (2024). Catalog. Available at: https://www.ebi.ac.uk/gwas/efotraits/EFO_0001421 (Accessed March 9, 2024).
43. Buniello, A, MacArthur, JAL, Cerezo, M, Harris, LW, Hayhurst, J, Malangone, C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. (2019) 47:D1005–12. doi: 10.1093/nar/gky1120
44. Sohail, M, Palma-Martínez, MJ, Chong, AY, Quinto-Cortés, CD, Barberena-Jonas, C, Medina-Muñoz, SG, et al. Mexican biobank advances population and medical genomics of diverse ancestries. Nature. (2023) 622:775–83. doi: 10.1038/s41586-023-06560-0
45. Moreno-Estrada, A, Gignoux, CR, Fernández-López, JC, Zakharia, F, Sikora, M, Contreras, AV, et al. Human genetics. The genetics of Mexico recapitulates native American substructure and affects biomedical traits. Science. (2014) 344:1280–5. doi: 10.1126/science.1251688
46. Ojeda-Granados, C, Panduro, A, Gonzalez-Aldaco, K, Sepulveda-Villegas, M, Rivera-Iñiguez, I, and Roman, S. Tailoring nutritional advice for Mexicans based on prevalence profiles of diet-related adaptive gene polymorphisms. J Pers Med. (2017) 7:16. doi: 10.3390/jpm7040016
47. Ojeda-Granados, C, Abondio, P, Setti, A, Sarno, S, Gnecchi-Ruscone, GA, González-Orozco, E, et al. Dietary, cultural, and pathogens-related selective pressures shaped differential adaptive evolution among native Mexican populations. Mol Biol Evol. (2021) 39:msab290. doi: 10.1093/molbev/msab290
48. Roman, S, Ojeda-Granados, C, Ramos-Lopez, O, and Panduro, A. Genome-based nutrition: an intervention strategy for the prevention and treatment of obesity and nonalcoholic steatohepatitis. World J Gastroenterol. (2015) 21:3449–61. doi: 10.3748/wjg.v21.i12.3449
49. Shetty, P . Nutrition transition and its health outcomes. Indian J Pediatr. (2013) 80:S21–7. doi: 10.1007/s12098-013-0971-5
50. Popkin, BM, and Gordon-Larsen, P. The nutrition transition: worldwide obesity dynamics and their determinants. Int J Obes Relat Metab Disord J Int Assoc Study Obes. (2004) 28:S2–9. doi: 10.1038/sj.ijo.0802804
51. Acuña-Alonzo, V, Flores-Dorantes, T, Kruit, JK, Villarreal-Molina, T, Arellano-Campos, O, Hünemeier, T, et al. A functional ABCA1 gene variant is associated with low HDL-cholesterol levels and shows evidence of positive selection in native Americans. Hum Mol Genet. (2010) 19:2877–85. doi: 10.1093/hmg/ddq173
52. Villarreal-Molina, T, Aguilar-Salinas, C, Rodríguez-Cruz, M, Barros, D, Villalobos-Comparan, M, Coral, R, et al. The ATP-binding cassette transporter A1 R230C variant affects HDL cholesterol levels and BMI in the Mexican population association with obesity and obesity-related comorbidities. Diabetes. (2007) 56:1881–7. doi: 10.2337/db06-0905
53. Torres-Valadez, R, Roman, S, Ojeda-Granados, C, Gonzalez-Aldaco, K, and Panduro, A. Differential distribution of gene polymorphisms associated with hypercholesterolemia, hypertriglyceridemia, and hypoalphalipoproteinemia among native American and mestizo Mexicans. World J Hepatol. (2022) 14:1408–20. doi: 10.4254/wjh.v14.i7.1408
54. Aguilar, CA, Talavera, G, Ordovas, JM, Barriguete, JA, Guillén, LE, Leco, ME, et al. The apolipoprotein E4 allele is not associated with an abnormal lipid profile in a native American population following its traditional lifestyle. Atherosclerosis. (1999) 142:409–14. doi: 10.1016/S0021-9150(98)00251-2
55. Roman, S, Rivera-Iñiguez, I, Ojeda-Granados, C, Sepulveda-Villegas, M, and Panduro, A. Genome-based nutrition in chronic liver disease In: RR Watson and VR Preedy, editors. Dietary Interventions in Liver Disease : Academic Press, Elsevier, USA (2019). 3–14.
56. Guibrunet, L, Ortega-Avila, AG, Arnés, E, and Mora, AF. Socioeconomic, demographic and geographic determinants of food consumption in Mexico. PLoS One. (2023) 18:e0288235. doi: 10.1371/journal.pone.0288235
57. Barquera, S, and Rivera, JA. Obesity in Mexico: rapid epidemiological transition and food industry interference in health policies. Lancet Diabetes Endocrinol. (2020) 8:746–7. doi: 10.1016/S2213-8587(20)30269-2
58. Ramos-Lopez, O, Panduro, A, Martinez Lopez, E, Fierro Nora, A, and Ojeda-Granados, C. Genetic variant in the CD36 gene (rs1761667) is associated with higher fat intake and high serum cholesterol among the population of West Mexico. J Nutr Food Sci. (2014) 5:1000353. doi: 10.4172/2155-9600.1000353
59. Ramos-Lopez, O, Roman, S, Martinez-Lopez, E, Fierro, NA, Gonzalez-Aldaco, K, Jose-Abrego, A, et al. CD36 genetic variation, fat intake and liver fibrosis in chronic hepatitis C virus infection. World J Hepatol. (2016) 8:1067–74. doi: 10.4254/wjh.v8.i25.1067
60. Ramos-Lopez, O, Panduro, A, Martinez-Lopez, E, and Roman, S. Sweet taste receptor TAS1R2 polymorphism (Val191Val) is associated with a higher carbohydrate intake and hypertriglyceridemia among the population of West Mexico. Nutrients. (2016) 8:101. doi: 10.3390/nu8020101
61. Ramos-Lopez, O, Roman, S, Martinez-Lopez, E, Gonzalez-Aldaco, K, Ojeda-Granados, C, Sepulveda-Villegas, M, et al. Association of a novel TAS2R38 haplotype with alcohol intake among Mexican-mestizo population. Ann Hepatol. (2015) 14:729–34. doi: 10.1016/S1665-2681(19)30768-9
62. Qiao, L, and Luo, GG. Functional characterization of apolipoproteins in the HCV life cycle. Methods Mol Biol. (2019) 1911:235–46. doi: 10.1007/978-1-4939-8976-8_16
63. Seripa, D, D’Onofrio, G, Panza, F, Cascavilla, L, Masullo, C, and Pilotto, A. The genetics of the human APOE polymorphism. Rejuvenation Res. (2011) 14:491–500. doi: 10.1089/rej.2011.1169
64. Eisenberg, DTA, Kuzawa, CW, and Hayes, MG. Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. Am J Phys Anthropol. (2010) 143:100–11. doi: 10.1002/ajpa.21298
65. Romeo, S, Kozlitina, J, Xing, C, Pertsemlidis, A, Cox, D, Pennacchio, LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. (2008) 40:1461–5. doi: 10.1038/ng.257
66. Pingitore, P, Pirazzi, C, Mancina, RM, Motta, BM, Indiveri, C, Pujia, A, et al. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim Biophys Acta. (2014) 1841:574–80. doi: 10.1016/j.bbalip.2013.12.006
67. Panduro, A, Sepulveda-Villegas, M, Andrea, P, Guevara-Gonzalez, AP, Milan, RG, Gonzalez-Aldaco, K, et al. Influence of ancestry on the FTO, PNPLA3, and TM6SF2 risk alleles and their association with metabolic abnormalities in admixed Mexicans with NASH and extreme obesity. Hepatology. (2022) 76:S1–S1564. doi: 10.1002/hep.32697
68. Larrieta-Carrasco, E, León-Mimila, P, Villarreal-Molina, T, Villamil-Ramírez, H, Romero-Hidalgo, S, Jacobo-Albavera, L, et al. Association of the I148M/PNPLA3 variant with elevated alanine transaminase levels in normal-weight and overweight/obese Mexican children. Gene. (2013) 520:185–8. doi: 10.1016/j.gene.2013.03.038
69. Xue, W-Y, Zhang, L, Liu, C-M, Gao, Y, Li, S-J, Huai, Z-Y, et al. Research progress on the relationship between TM6SF2 rs58542926 polymorphism and non-alcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. (2022) 16:97–107. doi: 10.1080/17474124.2022.2032661
70. Chermon, D, and Birk, R. FTO common obesity SNPs interact with actionable environmental factors: physical activity, sugar-sweetened beverages and wine consumption. Nutrients. (2022) 14:4202. doi: 10.3390/nu14194202
71. Sepulveda-Villegas, M, Roman, S, Rivera-Iñiguez, I, Ojeda-Granados, C, Gonzalez-Aldaco, K, Torres-Reyes, LA, et al. High prevalence of nonalcoholic steatohepatitis and abnormal liver stiffness in a young and obese Mexican population. PLoS One. (2019) 14:e0208926. doi: 10.1371/journal.pone.0208926
72. Ojeda-Granados, C, Panduro, A, Rivera-Iñiguez, I, Sepúlveda-Villegas, M, and Roman, S. A regionalized genome-based Mexican diet improves anthropometric and metabolic parameters in subjects at risk for obesity-related chronic diseases. Nutrients. (2020) 12:645. doi: 10.3390/nu12030645
73. Guevara-Cruz, M, Tovar, AR, Aguilar-Salinas, CA, Medina-Vera, I, Gil-Zenteno, L, Hernández-Viveros, I, et al. A dietary pattern including nopal, chia seed, soy protein, and oat reduces serum triglycerides and glucose intolerance in patients with metabolic syndrome. J Nutr. (2012) 142:64–9. doi: 10.3945/jn.111.147447
74. Dugum, M, and McCullough, A. Diagnosis and Management of Alcoholic Liver Disease. J Clin Transl Hepatol. (2015) 3:109–16. doi: 10.14218/JCTH.2015.00008
75. Panduro, A, Rivera-Iñiguez, I, Ramos-Lopez, O, and Roman, S. Chapter 50—genes and alcoholism: taste, addiction, and metabolism In: VR Preedy , editor. Neuroscience of Alcohol : Academic Press, Elsevier, USA (2019). 483–91.
76. Trépo, E, and Valenti, L. Update on NAFLD genetics: from new variants to the clinic. J Hepatol. (2020) 72:1196–209. doi: 10.1016/j.jhep.2020.02.020
77. Panduro, A, Ramos-Lopez, O, Campollo, O, Zepeda-Carrillo, EA, Gonzalez-Aldaco, K, Torres-Valadez, R, et al. High frequency of the DRD2/ANKK1 A1 allele in Mexican native Amerindians and mestizos and its association with alcohol consumption. Drug Alcohol Depend. (2017) 172:66–72. doi: 10.1016/j.drugalcdep.2016.12.006
78. Rivera-Iñiguez, I, Panduro, A, Ramos-Lopez, O, Villaseñor-Bayardo, SJ, and Roman, S. DRD2/ANKK1 TaqI A1 polymorphism associates with overconsumption of unhealthy foods and biochemical abnormalities in a Mexican population. Eat Weight Disord. (2019) 24:835–44. doi: 10.1007/s40519-018-0596-9
79. Roman, S, Zepeda-Carrillo, EA, Moreno-Luna, LE, and Panduro, A. Alcoholism and liver disease in Mexico: genetic and environmental factors. World J Gastroenterol. (2013) 19:7972–82. doi: 10.3748/wjg.v19.i44.7972
80. Lok, AS . Personalized treatment of hepatitis B. Clin Mol Hepatol. (2015) 21:1–6. doi: 10.3350/cmh.2015.21.1.1
81. Jose-Abrego, A, Roman, S, Laguna-Meraz, S, and Panduro, A. Host and HBV interactions and their potential impact on clinical outcomes. Pathogens. (2023) 12:1146. doi: 10.3390/pathogens12091146
82. Panduro, A, Roman, S, Laguna-Meraz, S, and Jose-Abrego, A. Hepatitis B virus genotype H: epidemiological, molecular, and clinical characteristics in Mexico. Viruses. (2023) 15:2186. doi: 10.3390/v15112186
83. Roman, S, Jose-Abrego, A, Fierro, NA, Escobedo-Melendez, G, Ojeda-Granados, C, Martinez-Lopez, E, et al. Hepatitis B virus infection in Latin America: a genomic medicine approach. World J Gastroenterol. (2014) 20:7181–96. doi: 10.3748/wjg.v20.i23.7181
84. Hayashi, S, Khan, A, Simons, BC, Homan, C, Matsui, T, Ogawa, K, et al. An association between Core mutations in hepatitis B virus genotype F1b and hepatocellular carcinoma in Alaskan native people. Hepatol Baltim. (2019) 69:19–33. doi: 10.1002/hep.30111
85. Cerapio, JP, Marchio, A, Cano, L, López, I, Fournié, J-J, Régnault, B, et al. Global DNA hypermethylation pattern and unique gene expression signature in liver cancer from patients with indigenous American ancestry. Oncotarget. (2021) 12:475–92. doi: 10.18632/oncotarget.27890
86. Jose-Abrego, A, Panduro, A, Fierro, NA, and Roman, S. High prevalence of HBV infection, detection of subgenotypes F1b, A2, and D4, and differential risk factors among Mexican risk populations with low socioeconomic status. J Med Virol. (2017) 89:2149–57. doi: 10.1002/jmv.24913
87. Panduro, A, Roman, S, Fierro, NA, and Rebello-Pinho, JR. Viral kinetics of an acute hepatitis B virus subgenotype F1b infection in a Mexican subject. Clin Liver Dis. (2022) 19:41–8. doi: 10.1002/cld.1178
88. Jose-Abrego, A, Roman, S, Rebello Pinho, JR, de Castro, VFD, and Panduro, A. Hepatitis B virus (HBV) genotype mixtures, viral load, and liver damage in HBV patients co-infected with human immunodeficiency virus. Front Microbiol. (2021) 12:640889. doi: 10.3389/fmicb.2021.640889
89. Heidelbaugh, JJ, and Bruderly, M. Cirrhosis and chronic liver failure: part I. Diagnosis and evaluation. Am Fam Physician. (2006) 74:756–62.
90. Zhang, Z, Wang, C, Liu, Z, Zou, G, Li, J, and Lu, M. Host genetic determinants of hepatitis B virus infection. Front Genet. (2019) 10:696. doi: 10.3389/fgene.2019.00696
91. Terrault, NA, Lok, ASF, McMahon, BJ, Chang, K-M, Hwang, JP, Jonas, MM, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatol Baltim. (2018) 67:1560–99. doi: 10.1002/hep.29800
92. Park, E-S, Lee, AR, Kim, DH, Lee, J-H, Yoo, J-J, Ahn, SH, et al. Identification of a quadruple mutation that confers tenofovir resistance in chronic hepatitis B patients. J Hepatol. (2019) 70:1093–102. doi: 10.1016/j.jhep.2019.02.006
93. Kobayashi, M, Suzuki, F, Akuta, N, Suzuki, Y, Arase, Y, Ikeda, K, et al. Response to long-term lamivudine treatment in patients infected with hepatitis B virus genotypes a, B, and C. J Med Virol. (2006) 78:1276–83. doi: 10.1002/jmv.20701
94. Jose-Abrego, A, Rivera-Iñiguez, I, Torres-Reyes, LA, and Roman, S. Anti-hepatitis B virus activity of food nutrients and potential mechanisms of action. Ann Hepatol. (2023) 28:100766. doi: 10.1016/j.aohep.2022.100766
95. Vergara, C, Duggal, P, Thio, CL, Valencia, A, O’Brien, TR, Latanich, R, et al. Multi-ancestry fine mapping of interferon lambda and the outcome of acute hepatitis C virus infection. Genes Immun. (2020) 21:348–59. doi: 10.1038/s41435-020-00115-3
96. Rauch, A, Kutalik, Z, Descombes, P, Cai, T, Di Iulio, J, Mueller, T, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. (2010) 138:1345.e1–7. doi: 10.1053/j.gastro.2009.12.056
97. Prokunina-Olsson, L . Genetics of the human interferon lambda region. J Int Soc Interferon Cytokine Res. (2019) 39:599–608. doi: 10.1089/jir.2019.0043
98. Bruening, J, Weigel, B, and Gerold, G. The role of type III interferons in hepatitis C virus infection and therapy. J Immunol Res. (2017) 2017:7232361. doi: 10.1155/2017/7232361
99. Hayes, CN, Kobayashi, M, Akuta, N, Suzuki, F, Kumada, H, Abe, H, et al. HCV substitutions and IL28B polymorphisms on outcome of peg-interferon plus ribavirin combination therapy. Gut. (2011) 60:261–7. doi: 10.1136/gut.2010.223495
100. Thomas, DL, Thio, CL, Martin, MP, Qi, Y, Ge, D, O’Huigin, C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. (2009) 461:798–801. doi: 10.1038/nature08463
101. Gonzalez-Aldaco, K, Rebello Pinho, JR, Roman, S, Gleyzer, K, Fierro, NA, Oyakawa, L, et al. Association with spontaneous hepatitis C viral clearance and genetic differentiation of IL28B/IFNL4 haplotypes in populations from Mexico. PLoS One. (2016) 11:e0146258. doi: 10.1371/journal.pone.0146258
102. Wilsie, LC, Gonzales, AM, and Orlando, RA. Syndecan-1 mediates internalization of apoE-VLDL through a low density lipoprotein receptor-related protein (LRP)-independent, non-clathrin-mediated pathway. Lipids Health Dis. (2006) 5:23. doi: 10.1186/1476-511X-5-23
103. Gomaa, HE, Mahmoud, M, Saad, NE, Saad-Hussein, A, Ismail, S, Thabet, EH, et al. Impact of apo E gene polymorphism on HCV therapy related outcome in a cohort of HCV Egyptian patients. J Genet Eng Biotechnol. (2018) 16:47–51. doi: 10.1016/j.jgeb.2017.10.008
104. Mueller, T, Fischer, J, Gessner, R, Rosendahl, J, Böhm, S, van Bömmel, F, et al. Apolipoprotein E allele frequencies in chronic and self-limited hepatitis C suggest a protective effect of APOE4 in the course of hepatitis C virus infection. Liver Int Off J Int Assoc Study Liver. (2016) 36:1267–74. doi: 10.1111/liv.13094
105. Price, DA, Bassendine, MF, Norris, SM, Golding, C, Toms, GL, Schmid, ML, et al. Apolipoprotein epsilon3 allele is associated with persistent hepatitis C virus infection. Gut. (2006) 55:715–8. doi: 10.1136/gut.2005.079905
106. Gonzalez-Aldaco, K, Roman, S, Torres-Valadez, R, Ojeda-Granados, C, Torres-Reyes, LA, and Panduro, A. Hepatitis C virus clearance and less liver damage in patients with high cholesterol, low-density lipoprotein cholesterol and APOE ε4 allele. World J Gastroenterol. (2019) 25:5826–37. doi: 10.3748/wjg.v25.i38.5826
107. Syed, GH, Tang, H, Khan, M, Hassanein, T, Liu, J, and Siddiqui, A. Hepatitis C virus stimulates low-density lipoprotein receptor expression to facilitate viral propagation. J Virol. (2014) 88:2519–29. doi: 10.1128/JVI.02727-13
108. Go, G-W, and Mani, A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J Biol Med. (2012) 85:19–28.
109. Ojeda-Granados, C, Panduro, A, Gonzalez-Aldaco, K, Rivera-Iñiguez, I, Campos-Medina, L, and Roman, S. Adherence to a fish-rich dietary pattern is associated with chronic hepatitis C patients showing low viral load: implications for nutritional management. Nutrients. (2021) 13:3337. doi: 10.3390/nu13103337
110. Li, J-F, Zheng, E-Q, and Xie, M. Association between rs738409 polymorphism in patatin-like phospholipase domain-containing protein 3 (PNPLA3) gene and hepatocellular carcinoma susceptibility: evidence from case-control studies. Gene. (2019) 685:143–8. doi: 10.1016/j.gene.2018.11.012
111. Liu, Y-L, Patman, GL, Leathart, JBS, Piguet, A-C, Burt, AD, Dufour, J-F, et al. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. (2014) 61:75–81. doi: 10.1016/j.jhep.2014.02.030
112. Burza, MA, Pirazzi, C, Maglio, C, Sjöholm, K, Mancina, RM, Svensson, P-A, et al. PNPLA3 I148M (rs738409) genetic variant is associated with hepatocellular carcinoma in obese individuals. Dig Liver Dis Off J Ital Soc Gastroenterol Ital Assoc Study Liver. (2012) 44:1037–41. doi: 10.1016/j.dld.2012.05.006
113. Takeuchi, Y, Ikeda, F, Moritou, Y, Hagihara, H, Yasunaka, T, Kuwaki, K, et al. The impact of patatin-like phospholipase domain-containing protein 3 polymorphism on hepatocellular carcinoma prognosis. J Gastroenterol. (2013) 48:405–12. doi: 10.1007/s00535-012-0647-3
114. Nakaoka, K, Hashimoto, S, Kawabe, N, Nitta, Y, Murao, M, Nakano, T, et al. PNPLA3 I148M associations with liver carcinogenesis in Japanese chronic hepatitis C patients. Springerplus. (2015) 4:83. doi: 10.1186/s40064-015-0870-5
115. Yang, J, Trépo, E, Nahon, P, Cao, Q, Moreno, C, Letouzé, E, et al. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int J Cancer. (2019) 144:533–44. doi: 10.1002/ijc.31910
116. Gijón, MA, Riekhof, WR, Zarini, S, Murphy, RC, and Voelker, DR. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. (2008) 283:30235–45. doi: 10.1074/jbc.M806194200
117. Mancina, RM, Dongiovanni, P, Petta, S, Pingitore, P, Meroni, M, Rametta, R, et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. (2016) 150:1219–1230.e6. doi: 10.1053/j.gastro.2016.01.032
118. Donati, B, Dongiovanni, P, Romeo, S, Meroni, M, McCain, M, Miele, L, et al. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep. (2017) 7:4492. doi: 10.1038/s41598-017-04991-0
119. Meroni, M, Longo, M, Fracanzani, AL, and Dongiovanni, P. MBOAT7 down-regulation by genetic and environmental factors predisposes to MAFLD. EBioMedicine. (2020) 57:102866. doi: 10.1016/j.ebiom.2020.102866
120. Pelusi, S, Baselli, G, Pietrelli, A, Dongiovanni, P, Donati, B, McCain, MV, et al. Rare pathogenic variants predispose to hepatocellular carcinoma in nonalcoholic fatty liver disease. Sci Rep. (2019) 9:3682. doi: 10.1038/s41598-019-39998-2
121. Teo, K, Abeysekera, KWM, Adams, L, Aigner, E, Anstee, QM, Banales, JM, et al. rs641738C>T near MBOAT7 is associated with liver fat, ALT and fibrosis in NAFLD: a meta-analysis. J Hepatol. (2021) 74:20–30. doi: 10.1016/j.jhep.2020.08.027
122. Wang, P, Li, Y, Li, L, Zhong, R, and Shen, N. MBOAT7-TMC4 rs641738 is not associated with the risk of hepatocellular carcinoma or persistent hepatitis B infection. Front Oncol. (2021) 11:639438. doi: 10.3389/fonc.2021.639438
123. Marchais-Oberwinkler, S, Henn, C, Möller, G, Klein, T, Negri, M, Oster, A, et al. 17β-hydroxysteroid dehydrogenases (17β-HSDs) as therapeutic targets: protein structures, functions, and recent progress in inhibitor development. J Steroid Biochem Mol Biol. (2011) 125:66–82. doi: 10.1016/j.jsbmb.2010.12.013
124. Su, W, Wang, Y, Jia, X, Wu, W, Li, L, Tian, X, et al. Comparative proteomic study reveals 17β-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA. (2014) 111:11437–42. doi: 10.1073/pnas.1410741111
125. Su, W, Mao, Z, Liu, Y, Zhang, X, Zhang, W, Gustafsson, J-A, et al. Role of HSD17B13 in the liver physiology and pathophysiology. Mol Cell Endocrinol. (2019) 489:119–25. doi: 10.1016/j.mce.2018.10.014
126. Chen, J, Zhuo, J-Y, Yang, F, Liu, Z-K, Zhou, L, Xie, H-Y, et al. 17-beta-hydroxysteroid dehydrogenase 13 inhibits the progression and recurrence of hepatocellular carcinoma. Hepatobil Pancreat Dis Int. (2018) 17:220–6. doi: 10.1016/j.hbpd.2018.04.006
127. Gellert-Kristensen, H, Nordestgaard, BG, Tybjaerg-Hansen, A, and Stender, S. High risk of fatty liver disease amplifies the alanine transaminase-lowering effect of a HSD17B13 variant. Hepatol Baltim. (2020) 71:56–66. doi: 10.1002/hep.30799
128. Yang, J, Trépo, E, Nahon, P, Cao, Q, Moreno, C, Letouzé, E, et al. A 17-Beta-hydroxysteroid dehydrogenase 13 variant protects from hepatocellular carcinoma development in alcoholic liver disease. Hepatol Baltim. (2019) 70:231–40. doi: 10.1002/hep.30623
129. Stickel, F, Lutz, P, Buch, S, Nischalke, HD, Silva, I, Rausch, V, et al. Genetic variation in HSD17B13 reduces the risk of developing cirrhosis and hepatocellular carcinoma in alcohol misusers. Hepatol Baltim. (2020) 72:88–102. doi: 10.1002/hep.30996
130. De Benedittis, C, Bellan, M, Crevola, M, Boin, E, Barbaglia, MN, Mallela, VR, et al. Interplay of PNPLA3 and HSD17B13 variants in modulating the risk of hepatocellular carcinoma among hepatitis C patients. Gastroenterol Res Pract. (2020) 2020:4216451. doi: 10.1155/2020/4216451
131. Torres-Reyes, LA, Gonzalez-Aldaco, K, Panduro, A, Jose-Abrego, A, and Roman, S. Whole-exome sequencing identified olfactory receptor genes as a key contributor to extreme obesity with progression to nonalcoholic steatohepatitis in Mexican patients: olfactory receptor genes in obese NASH patients. Ann Hepatol. (2022) 27:100767. doi: 10.1016/j.aohep.2022.100767
132. Wiseman, MJ . Nutrition and cancer: prevention and survival. Br J Nutr. (2019) 122:481–7. doi: 10.1017/S0007114518002222
133. George, ES, Sood, S, Broughton, A, Cogan, G, Hickey, M, Chan, WS, et al. The association between diet and hepatocellular carcinoma: a systematic review. Nutrients. (2021) 13:172. doi: 10.3390/nu13010172
134. Ramos-Lopez, O, Martinez-Lopez, E, Roman, S, Fierro, NA, and Panduro, A. Genetic, metabolic and environmental factors involved in the development of liver cirrhosis in Mexico. World J Gastroenterol. (2015) 21:11552–66. doi: 10.3748/wjg.v21.i41.11552
135. Gomez-Quiroz, LE, and Roman, S. Influence of genetic and environmental risk factors in the development of hepatocellular carcinoma in Mexico. Ann Hepatol. (2022) 27:100649. doi: 10.1016/j.aohep.2021.100649
136. Cassel, CK, and Reuben, DB. Specialization, subspecialization, and Subsubspecialization in internal medicine. N Engl J Med. (2011) 364:1169–73. doi: 10.1056/NEJMsb1012647
137. Panduro, A, Rivera-Iñiguez, I, Sepulveda-Villegas, M, and Roman, S. Genes, emotions and gut microbiota: the next frontier for the gastroenterologist. World J Gastroenterol. (2017) 23:3030–42. doi: 10.3748/wjg.v23.i17.3030
138. Slade, I, Subramanian, DN, and Burton, H. Genomics education for medical professionals—the current UK landscape. Clin Med Lond Engl. (2016) 16:347–52. doi: 10.7861/clinmedicine.16-4-347
139. Simpson, S, Seller, A, and Bishop, M. Using the findings of a National Survey to inform the work of England’s genomics education programme. Front Genet. (2019) 10:1265. doi: 10.3389/fgene.2019.01265
140. Roman, S, and Panduro, A. Genomic medicine in gastroenterology: a new approach or a new specialty? World J Gastroenterol. (2015) 21:8227–37. doi: 10.3748/wjg.v21.i27.8227
141. Donini, LM, Leonardi, F, Rondanelli, M, Banderali, G, Battino, M, Bertoli, E, et al. The domains of human nutrition: the importance of nutrition education in academia and medical schools. Front Nutr. (2017) 4:2. doi: 10.3389/fnut.2017.00002
142. Bacon, BR, and Adams, PC. Hepatology as a distinct specialty—born in the USA? Can J Gastroenterol. (2007) 21:421. doi: 10.1155/2007/320986
143. Panduro, A . Training in hepatology: from medical school to a Ph.D. and clinical specialty program. Ann Hepatol. (2022) 27:100682. doi: 10.1016/j.aohep.2022.100682
144. Panduro, A . Viruses and the liver 2020: before COVID-19 and the beginning of a new age in medicine. Ann Hepatol. (2021) 20:100293. doi: 10.1016/j.aohep.2020.100293
145. Panduro, A, and Roman, S. Personalized medicine in Latin America. Pers Med. (2020) 17:339–43. doi: 10.2217/pme-2020-0049
146. Panduro, A, and Roman, S. Advancements in genomic medicine and the need for updated regional clinical practice guidelines in the field of hepatology. Ann Hepatol. (2020) 19:1–2. doi: 10.1016/j.aohep.2019.12.002
147. Ojeda-Granados, C, and Roman, S. Mediterranean diet or genome-based nutrition diets in Latin America’s clinical practice guidelines for managing chronic liver diseases? Ann Hepatol. (2021) 20:100291. doi: 10.1016/j.aohep.2020.100291
148. Alt, KW, Al-Ahmad, A, and Woelber, JP. Nutrition and health in human evolution–past to present. Nutrients. (2022) 14:3594. doi: 10.3390/nu14173594
149. Benton, ML, Abraham, A, LaBella, AL, Abbot, P, Rokas, A, and Capra, JA. The influence of evolutionary history on human health and disease. Nat Rev Genet. (2021) 22:269–83. doi: 10.1038/s41576-020-00305-9
150. Ko, A, Cantor, RM, Weissglas-Volkov, D, Nikkola, E, Reddy, PMVL, Sinsheimer, JS, et al. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat Commun. (2014) 5:3983. doi: 10.1038/ncomms4983
151. León-Mimila, P, Villamil-Ramírez, H, Macías-Kauffer, LR, Jacobo-Albavera, L, López-Contreras, BE, Posadas-Sánchez, R, et al. Genome-wide association study identifies a functional SIDT2 variant associated with HDL-C (high-density lipoprotein cholesterol) levels and premature coronary artery disease. Arterioscler Thromb Vasc Biol. (2021) 41:2494–508. doi: 10.1161/ATVBAHA.120.315391
152. Below, JE, Parra, EJ, Gamazon, ER, Torres, J, Krithika, S, Candille, S, et al. Meta-analysis of lipid-traits in Hispanics identifies novel loci, population-specific effects, and tissue-specific enrichment of eQTLs. Sci Rep. (2016) 6:19429. doi: 10.1038/srep19429
153. García-Ortiz, H, Barajas-Olmos, F, Contreras-Cubas, C, Cid-Soto, MÁ, Córdova, EJ, Centeno-Cruz, F, et al. The genomic landscape of Mexican indigenous populations brings insights into the peopling of the Americas. Nat Commun. (2021) 12:5942. doi: 10.1038/s41467-021-26188-w
154. Serra-Burriel, M, Juanola, A, Serra-Burriel, F, Thiele, M, Graupera, I, Pose, E, et al. Development, validation, and prognostic evaluation of a risk score for long-term liver-related outcomes in the general population: a multicohort study. Lancet. (2023) 402:988–96. doi: 10.1016/S0140-6736(23)01174-1
155. Nature (2024). Polygenic risk: What’s the score? Available at: https://www.nature.com/articles/d42473-019-00270-w (Accessed March 12, 2024).
156. Busby, GB, Kulm, S, Bolli, A, Kintzle, J, Domenico, PD, and Bottà, G. Ancestry-specific polygenic risk scores are risk enhancers for clinical cardiovascular disease assessments. Nat Commun. (2023) 14:7105. doi: 10.1038/s41467-023-42897-w
Keywords: Mexico, genetics, diet-related adaptive genes, hepatopathogenic diet, Genomex diet, nutrients, training, genomic medicine in hepatology
Citation: Panduro A, Roman S, Mariscal-Martinez IM, Jose-Abrego A, Gonzalez-Aldaco K, Ojeda-Granados C, Ramos-Lopez O and Torres-Reyes LA (2024) Personalized medicine and nutrition in hepatology for preventing chronic liver disease in Mexico. Front. Nutr. 11:1379364. doi: 10.3389/fnut.2024.1379364
Edited by:
Katia Petroni, University of Milan, ItalyReviewed by:
Félix Gómez-Gallego, Rey Juan Carlos University, SpainLuis Rodrigo Macias Kauffer, Universität zu Lübeck, Germany
Copyright © 2024 Panduro, Roman, Mariscal-Martinez, Jose-Abrego, Gonzalez-Aldaco, Ojeda-Granados, Ramos-Lopez and Torres-Reyes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arturo Panduro, YXBhbmR1cm9AcHJvZGlneS5uZXQubXg=;; YXBhbmR1cm81M0BnbWFpbC5jb20=
 Arturo Panduro
Arturo Panduro Sonia Roman
Sonia Roman Irene M. Mariscal-Martinez
Irene M. Mariscal-Martinez Alexis Jose-Abrego
Alexis Jose-Abrego Karina Gonzalez-Aldaco
Karina Gonzalez-Aldaco Claudia Ojeda-Granados
Claudia Ojeda-Granados Omar Ramos-Lopez
Omar Ramos-Lopez Luis A. Torres-Reyes
Luis A. Torres-Reyes