- 1Department of Urology, Huashan Hospital, Fudan University, Shanghai, China
- 2Fudan Institute of Urology, Huashan Hospital, Fudan University, Shanghai, China
- 3National Clinical Research Center for Aging and Medicine, Huashan Hospital, Fudan University, Shanghai, China
Background: The influences of blood lipids and lipid-regulatory medications on the risk of bladder cancer have long been suspected, and previous findings remain controversial. We aimed to assess the causality between blood lipids or lipid-regulatory medications and bladder cancer susceptibility by means of a comprehensive Mendelian Randomization (MR) study.
Methods: Genetic proxies from genome-wide association studies (GWAS) of four blood lipid traits and lipid-lowering variants in genes encoding the targets of lipid-regulatory medications were employed. The largest ever GWAS data of blood lipids and bladder cancer involving up to 440,546 and 205,771 individuals of European ancestry were extracted from UK Biobank and FinnGen Project Round 6, respectively. A two-sample bidirectional MR study was performed using the inverse variance weighted as the main method. The heterogeneity, horizontal pleiotropy, MR Steiger, and leave-one-out analyses were also conducted as sensitivity tests.
Results: There was indicative evidence that genetically predicted low-density lipoprotein cholesterol (LDL-C) affected bladder cancer susceptibility based on 146 single nucleotide polymorphisms (SNPs) with an odds ratio (OR) of 0.776 (95% confidence interval [CI] = 0.625–0.965, p = 0.022). However, this result became non-significant after two SNPs that possibly drove the effect were removed as demonstrated by leave-one-out analysis. The reversed MR analysis suggested that bladder cancer could not affect serum lipid levels. No causal relationship was found between the lipid-lowering effect of lipid-regulatory medications (fibrates, probucol, statins, ezetimibe, proprotein convertase subtilisin/kexin type 9 [PCSK9] inhibitors, and evinacumab) and the risk of bladder cancer. No heterogeneity or pleiotropy was found (all p > 0.05).
Conclusion: This MR study revealed for the first time, using the most recent and comprehensive GWAS data to date, that genetically predicted total cholesterol (TC) and the lipid-lowering effect of lipid-regulatory medications had no causal association with bladder cancer susceptibility. We also verified claims from early studies that low-density lipoprotein cholesterol (HDL-C), LDL-C, and triglyceride (TG) are not related to bladder cancer susceptibility either. The current study indicated that lipid metabolism may not be as important in the tumorigenesis of bladder cancer as previously believed.
Introduction
According to the estimation of the International Agency for Research on Cancer and the American Cancer Society, there were approximately 573,000 newly diagnosed cases and 212,000 disease-related deaths of bladder cancer around the globe in 2020 alone, making it the tenth most common cancer worldwide and the most likely encountered malignancy for urologists (1). Although the incidence of bladder cancer is far higher in men, for whom it is the sixth most prevalent cancer, the past 2 decades have witnessed a rise in women’s tobacco consumption, which has in turn contributed to the increased trend in incidence among women, while the trend for men has remained stable (2, 3). Non-muscle invasive bladder cancer is characterized by a high risk of recurrence after the initial transurethral resection and requires frequent imaging examinations or even, when necessary, a second transurethral cystoscopy, albeit being confined to the mucosa or submucosa at the early stage (4). Eventually, approximately 30% of non-muscle invasive bladder cancer will progress into muscle-invasive one during the course of the disease (5). While radical cystectomy with postoperative platinum-based chemotherapy has been the first-line treatment for advanced-stage patients for decades, the illness remains refractory and often metastasizes to adjacent organs, lungs, or the liver (2). The emergence of immunotherapy with checkpoint inhibitors has somewhat improved the prognosis of terminal bladder cancer (6, 7). However, its response rate is still modest (8), not to mention the local and systemic side effects it brings about, resulting in treatment discontinuation (7). Therefore, primary prevention of bladder cancer is of crucial importance not only for patients themselves but also for the reduction of socio-economic burdens for families and society alike.
It is widely recognized that smoking is a key risk factor for bladder cancer (3) though other factors such as occupational exposure to aromatic amines and infection with Schistosoma haematobium may also be major contributors in certain populations (2, 9). The roles of dyslipidemia and lipid-regulating therapy in the etiology of bladder cancer have long been suspected. A Chinese case–control study recruiting 972 patients with pathologically diagnosed urothelial carcinoma of the bladder and 1,098 cancer-free controls discovered that hypertriglyceridemia was positively related to bladder cancer (10), while a previous MR study using limited SNPs showed no correlations between TG, LDL-C or HDL-C, and bladder cancer susceptibility (11). The different and even controversial outcomes among studies may possibly attribute to the size of study populations, statistical power, and biases. Thus, higher-quality, well-designed research with a broader subject scale and fewer biases of any kind is urgently required to address this issue.
Mendelian randomization, a novel statistical strategy that makes use of genetic variants associated with the exposure of interest as instruments, has been increasingly applied to estimate the causal effects of risk factors on the outcome (12). Due to the random allocation of genes during conception and gamete formation, MR is less susceptible to confounding influences that emerge over one’s lifespan. As a result, it overcomes the drawbacks of conventional observational studies (13). Moreover, genetic variants adjacent to or within the genes that code for drug targets can regulate their expression and, therefore, affect the activities of pharmacological targets. These genetic variants can anticipate and reflect the therapeutic effects of drugs on individuals as previously described in antihypertensive medicine (14). To the best of our knowledge, there has only been one MR study that employed merely one SNP named rs12916 as a surrogate for 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA reductase, HMGCR), the target of statins, to investigate the association between statins use and the risk of bladder cancer (11).
Herein, we aimed to perform a two-sample, bidirectional MR analysis to assess the causal effects of genetically predicted blood lipids and genetic proxies for lipid-regulatory medications on the risk of bladder cancer in a comprehensive and reliable manner using the largest GWAS data for blood lipids and bladder cancer to date.
Methods
Data sources
The biggest publicly available genome-wide association study for blood lipids to date was acquired from the UK Biobank1 and IEU-GWAS summary data2 in order to enlarge the sample size and avoid biases to the greatest extent. The details of the study protocol have been published in their previous study (15). For four representatives of circulating lipids, namely, HDL-C, LDL-C, TC, and TG, 403,943, 440,546, 115,078, and 115,078 European participants were enlisted, respectively (16). Since all the participants from UK Biobank went through the same standard blood lipid testing procedures with laboratory results being continuous variables, the effects of any change in blood lipids on the outcome were represented as one standard deviation.
GWAS summary association statistics for bladder cancer were downloaded from the FinnGen Project.3 It is a gigantic initiative first launched in 2017, encompassing everything from genetic information to digital health care data, much to the UK Biobank database. In-depth quality control procedures were clarified by its earlier studies (17). The most recent data from Round 6 was released on January 24, 2022, which contained 1,701 cases of malignant neoplasm of the bladder and 204,070 controls (all types of cancers were excluded from controls) of European ancestry. While the UK Biobank datasets of blood lipids were large enough to support sufficient statistical power, a modest degree of statistical power was expected for the outcome due to the relatively small number of cases of bladder cancer in the FinnGen Project compared to that of controls (18).
In general, all the data utilized in this study is open to the public. Informed consent was obtained from all participants in the original genome-wide association studies which were authorized by their corresponding ethics committee. Information on data sources is listed in Supplementary File 1.
Instrumental variables identification
SNPs with p-values less than 5 × 10−8 were selected from the UK Biobank GWAS for the four blood lipid traits. Those who were palindromic or in linkage disequilibrium (LD, threshold: r2 < 0.001, kb = 10,000) based on the 1,000 Genomes Project reference panel were discarded. By identifying confounder-related SNPs in an online phenome-wide association study database4 and the Phenotype Scanner5, SNPs closely related to the potential confounders, including smoking, body mass index, waist-to-hip ratio, and type 2 diabetes mellitus, were also excluded. The remaining SNPs were correspondingly chosen as instrumental variables (IVs) for their genetic associations with HDL-C, LDL-C, TC, and TG.
As for the causal effect of pharmacological interventions on bladder cancer, we adopted genetic IVs as proxies for the LDL-C lowering effect of lipid-regulatory medications commonly used in clinical practice. The following drugs with indications concerning lipid metabolism were not recruited for either exerting multisystemic effects (e.g., niacin) or are rarely prescribed by physicians (e.g., Mipomersen, a binder to the mRNA encoding apolipoprotein B, is only available to patients with homozygous familial hypercholesterolemia under a restricted clinical program due to its liver toxicity). Then, we searched for the validated targets implicated in the lipid metabolic process of these drugs at the DrugBank6 and collected their SNPs recorded by GeneCards.7 Similarly, SNPs that were genetically correlated with LDL-C (p < 5 × 10−8) and underwent LD-clumping (threshold r2 < 0.4, kb =10,000) were identified as final IVs for each class of lipid-regulatory medication upon the exclusion of confounder-related SNPs mentioned above. Such a relatively loose threshold allowed for an increase in the number of IVs, the proportion of variance explained, and hence the statistical power. In addition, we also set a stricter LD-clumping threshold of r2 < 0.001.
Statistical analysis
After harmonizing the alleles for consistency, MR analyses were conducted for estimations of causal effects by five complementary methods: the inverse variance weighted (IVW), MR-Egger, weighted median, weighted mode, and simple mode, with the IVW being the main approach established on the assumption that instrumental variables affected the outcome solely through the exposure of interest (19). When only one SNP was available, the Wald ratio was calculated instead. The estimates of median-based approaches remain robust even when half of the SNPs are weak instruments (20). However, as these methods were still susceptible to pleiotropy, we examined the intercept of MR-Egger regression and employed MR-Pleiotropy Residual Sum and Outlier methods (MR-PRESSO) for detection of horizontal pleiotropy despite having minimized confounding biases (21, 22). To assess for the underlying heterogeneity, Cochran’s Q-value was estimated by the IVW and MR-Egger (23). Leave-one-out analysis, where an instrumental SNP was removed one at a time to analyze the causal effects of the remaining SNPs on the outcome, was also performed as a sensitivity analysis to determine if the MR results were driven by any particular SNP. Furthermore, we performed the MR Steiger test of directionality to judge whether the results yielded were in the same direction as our hypothesis. The causal direction was evaluated as true if all of the IVs together explained more variance in the exposure than the outcome (24). The F statistics were generated for all the exposure in case of weak instrument biases caused by SNPs with F statistics less than 10 (25).
The current MR study consisted of two stages. First of all, the causal effects of the four blood lipid traits on the risk of bladder cancer were investigated, followed by a reversed MR analysis where bladder cancer was treated as the exposure, while the blood lipids as the outcomes in order to rule out reverse causality. We loosened the standard of significance for IVs to p < 1 × 10−6 in the reversed procedure so as to enhance statistical power and lower the false-negative rate. Subsequently, a second MR analysis involving lipid-lowering therapy and bladder cancer was performed. The correlation was considered statistically significant after the Bonferroni correction for multiple hypothesis testing for four blood lipid traits (p < 0.0125) and six types of medications (p < 0.0083). A p-value greater than the adjusted cutoff while less than 0.05 was considered indicative of evidence for probable causality. The statistical significance of the analyses above was indicated as a two-tailed p-value less than 0.05. All of the analyses were carried out in R (version 4.0.3) with “TwoSampleMR” and “MR-PRESSO” being the primary packages used (26). Figure 1 depicts the conceptual framework for the MR analysis of lipid-regulatory medications and the risk of bladder cancer as well as the entire design of the current study.
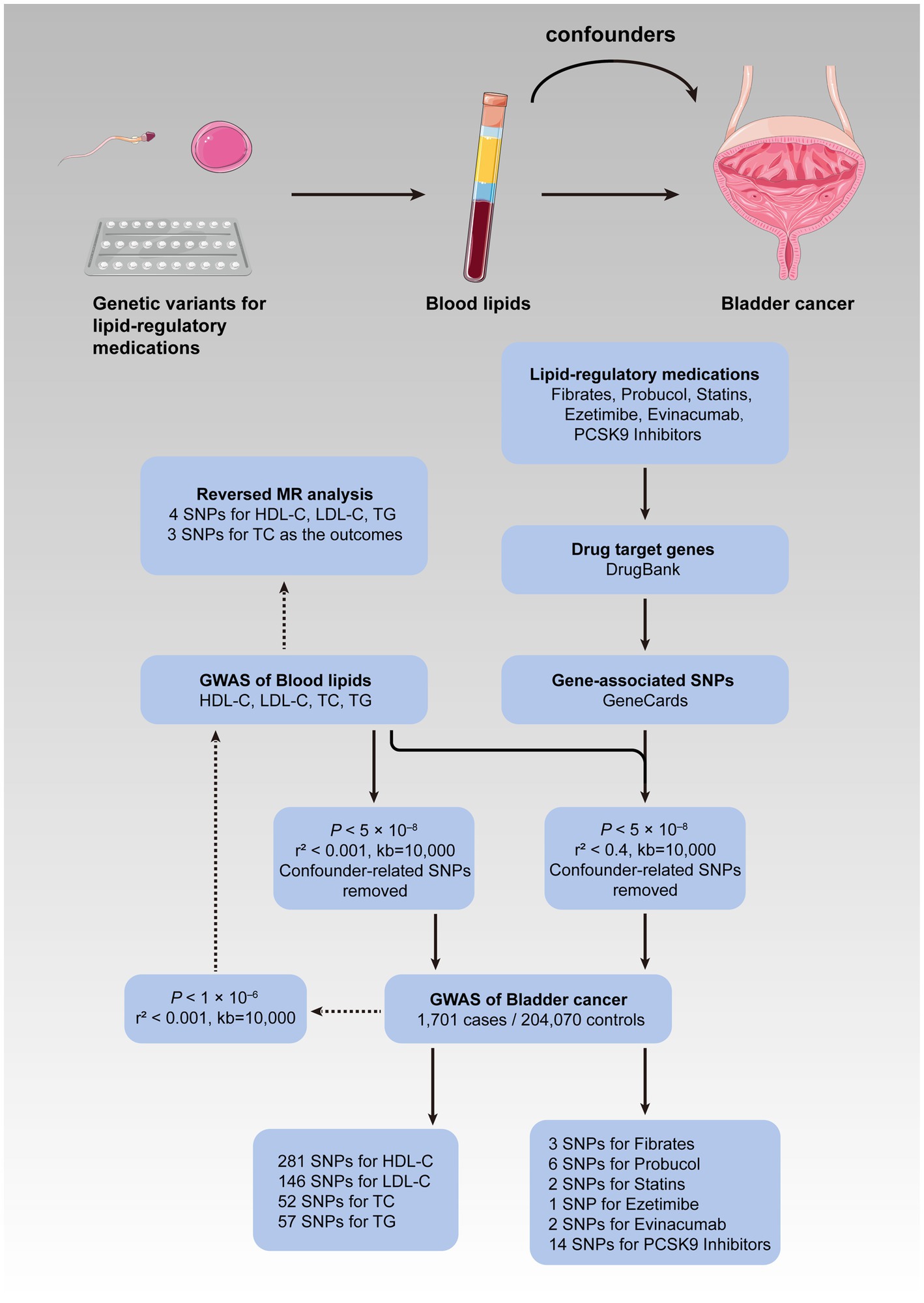
Figure 1. Flow diagram of the current MR study. The upper panel illustrated the conceptual framework for the MR analysis of lipid-regulatory medications and the risk of bladder cancer, while the lower panel showed the entire design of the current study. MR, Mendelian randomization; GWAS, genome-wide association study; SNPs, single nucleotide polymorphisms; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; PCSK9, proprotein convertase subtilisin/kexin type 9.
Results
Genetically determined blood lipids and the risk of bladder cancer
To start with, we examined the causal relations between genetically determined blood lipids and the risk of bladder cancer. After the initial filtering and complete exclusion of confounder-related SNPs, a total of 281, 146, 52, and 57 instrumental variants in close connection with HDL-C, LDL-C, TC, and TG, respectively, were identified. All the 536 IVs are displayed in Supplementary File 2. There was no causal effect of genetically predicted HDL-C, TC, or TG on the risk of bladder cancer with all p-values >0.05. Nevertheless, the result indicated a causal link of suggestive evidence between LDL-C and bladder cancer via the IVW approach with an OR of 0.776 (95% CI = 0.625–0.965, p = 0.022). This was further corroborated by the weighted mode (OR = 0.723, 95% CI = 0.554–0.945, p = 0.019; Figure 2). There were two single SNPs closely linked to LDL-C that plausibly drove the connection as demonstrated by the leave-one-out analysis. With these two instrumental variants removed, the result became insufficient to draw a conclusion of causality between LDL-C and bladder cancer susceptibility with an OR of 0.832 (95% CI = 0.659–1.050, p = 0.121). There was neither evidence of heterogeneity (IVW and MR-Egger) nor horizontal pleiotropy (MR-Egger intercept and MR-PRESSO) in all of the analyses (Table 1). Scatter plots and leave-one-out plots are presented in Supplementary File 3.
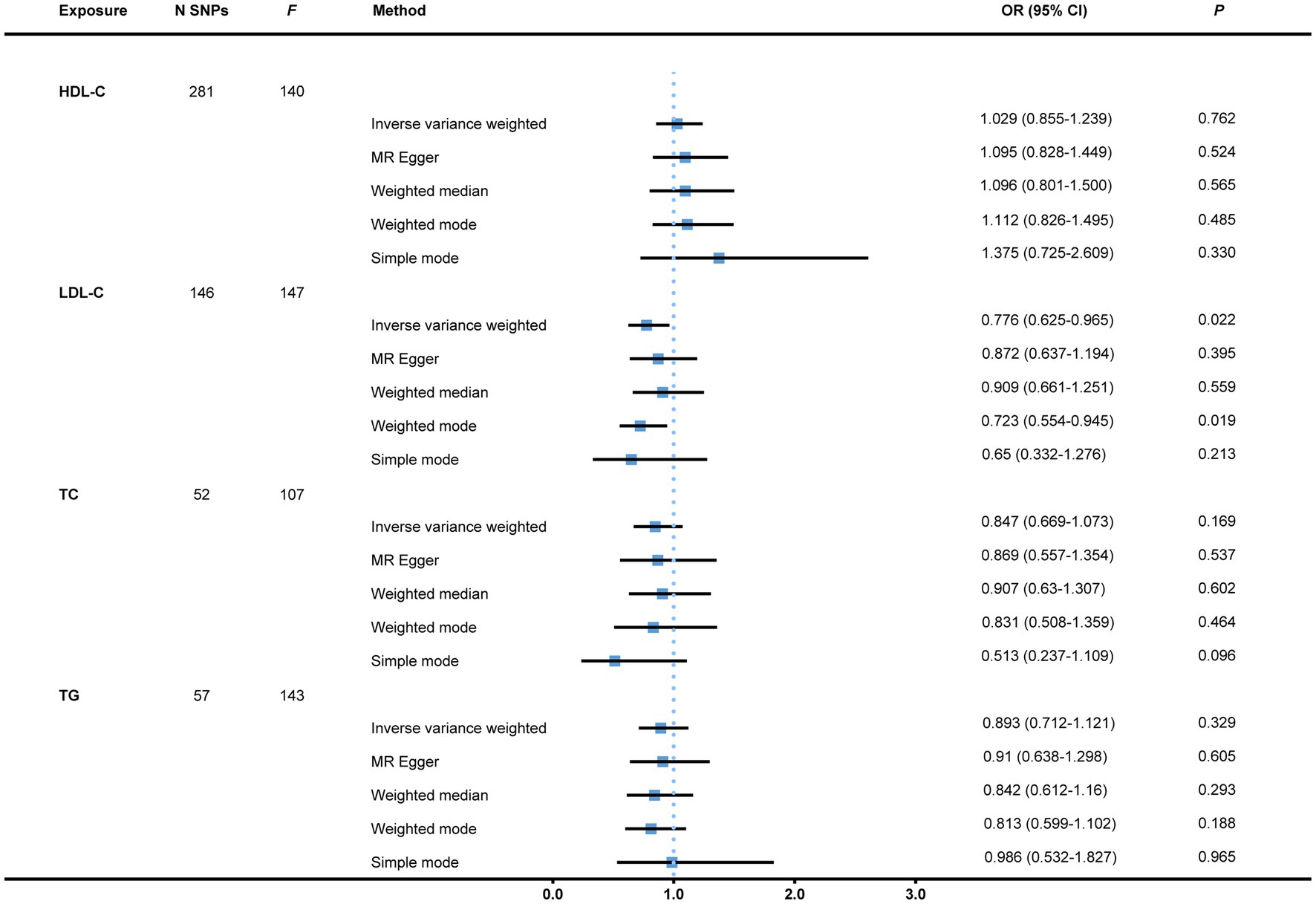
Figure 2. MR analysis of causal effects of genetically predicted blood lipids on the risk of bladder cancer. Genome-wide significantly associated (p < 5 × 10−8) independent (LD-clumping r2 < 0.001, kb = 10,000) SNPs were selected as instrumental variables. However, the causal effect of LDL-C on bladder cancer susceptibility became non-significant with an OR of 0.832 (95% CI = 0.659–1.050, p = 0.121, as not shown in the figure) after two SNPs possibly driving the effect as demonstrated by the leave-one-out analysis were removed. MR, Mendelian randomization; N SNPs, number of single nucleotide polymorphisms; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; OR, Odds ratio; CI, confidence interval; LD, linkage disequilibrium.
In the reversed MR stage, we gained 141 SNPs whose p-values were less than 1 × 10−6 out of a total of 16,355,128 SNPs from Round 6 data of the FinnGen Project, among which we finally confirmed four independent SNPs as genetic instruments for bladder cancer as the exposure. No significant results were detected albeit the heterogeneity arose when total serum cholesterol was treated as the endpoint. The findings remained the same after a single SNP analysis and the removal of an outlier (Figure 3 and Supplementary File 4). All of the heterogeneity and pleiotropy tests came out negative (Supplementary File 5).
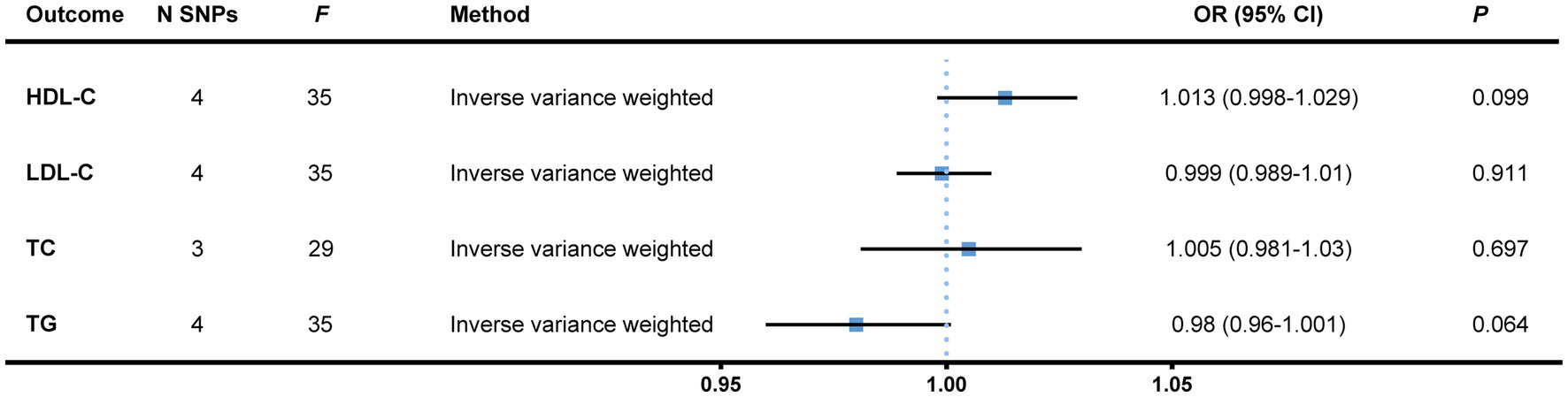
Figure 3. Reversed MR analysis with bladder cancer treated as the exposure, and blood lipids, the outcomes. Independent (LD-clumping r2 < 0.001, kb = 10,000) SNPs of bladder cancer from the FinnGen Project with a relatively loose standard of genetical significance (p < 1 × 10−6) were selected as instrumental variables. The IVW method was employed to calculate causal effects. MR, Mendelian randomization; N SNPs, number of single nucleotide polymorphisms; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; OR, odds ratio; CI, confidence interval; LD, linkage disequilibrium; IVW, inverse variance weighted.
Genetic proxies for lipid-regulatory medications and the risk of bladder cancer
Second, we chose SNPs that genetically predicted the lipid-lowering effect of genes that encode targets aimed by lipid-regulatory medications as genetic proxies. After eliminating confounder-related SNPs, a total of 28 IVs were identified, including three for fibrates, six for probucol, two for statins, one for ezetimibe, 14 for PCSK9 inhibitors (alirocumab, evolocumab, and inclisiran), as well as two for evinacumab (Supplementary File 6). Nonetheless, we failed to acquire the proxies for adenosine triphosphate-citrate lyase (ACLY), the target of bempedoic acid. All IVs had F statistics greater than 10. There was no causation found between lipid-lowering therapy and the risk of bladder cancer (Figure 4 and Supplementary File 7). The Cochran’s Q statistics of both the IVW and MR-Egger methods showed little heterogeneity among genetic variants. Neither did the pleiotropy tests exhibit notable horizontal pleiotropy (Table 1).
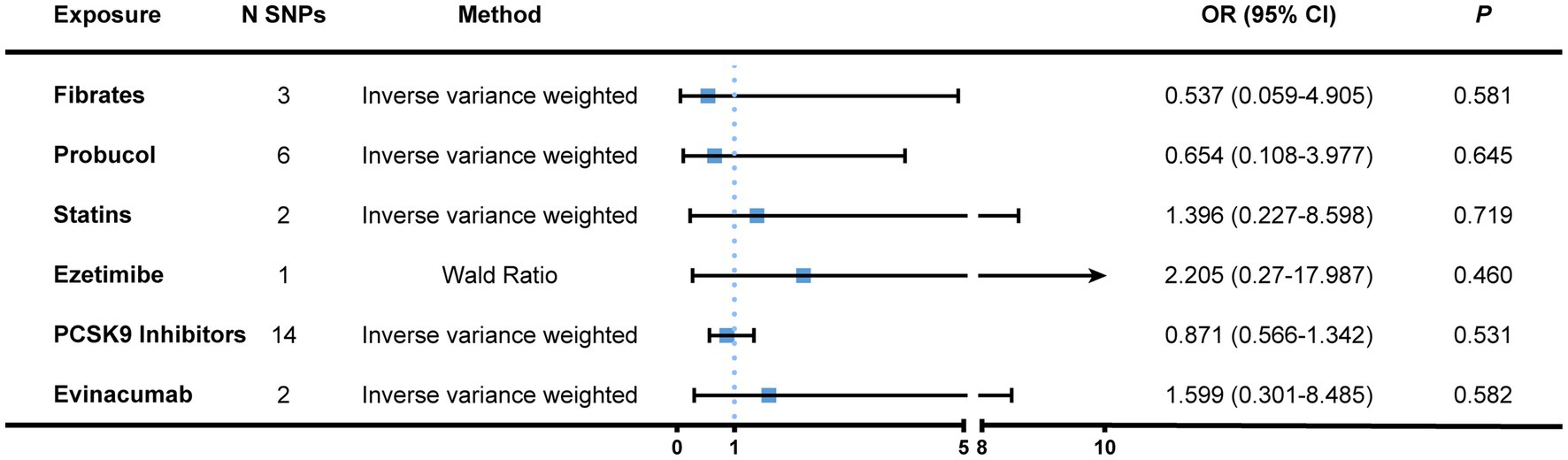
Figure 4. MR association between genetically proxied lipid-regulatory medications and the risk of bladder cancer. Genome-wide significantly associated (p < 5 × 10−8) SNPs of LDL-C with a relatively loose standard of LD-clumping (r2 < 0.4, kb = 10,000) were selected as instrumental variables. The IVW and Wald ratio (when only one SNP was available) was employed as the main approach to calculate causal effects. Complete results were displayed in Supplementary File 5. MR, Mendelian randomization; N SNPs, number of single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval; PCSK9, proprotein convertase subtilisin/kexin type 9; LD, linkage disequilibrium; IVW, inverse variance weighted.
An additional investigation into the causal effect of lipid-lowering intervention on the risk of bladder cancer using a more stringent LD criterion of r2 < 0.001 displayed comparable results (Supplementary File 8) though there was no suitable genetic instrument for statins under this LD threshold. The reason why we did not mix multiple types of lipid-regulatory medications to conduct a multivariant MR was that these drugs are usually administered alone or in combination with therapies treated for other chronic conditions. All of the aforementioned analyses passed the MR Steiger test, indicating that the directionality was correct (Supplementary File 9).
Discussion
In general, an MR study was based on three core assumptions. First, the SNPs chosen as IVs are closely related to the exposure of interest. Second, the IVs have no relationship with any confounding factors. Third, the risk of the outcome is not affected except by the exposure (12, 13, 27). Realizing only the first assumption could be fully satisfied, we managed to circumvent biases to the largest extent by excluding SNPs closely associated with confounders, i.e., smoking, body mass index, waist-to-hip ratio, and type 2 diabetes mellitus.
In the current two-sample MR study, we used genetic variants as proxies for four circulating lipids and the lipid-lowering effect of six lipid-modulatory medications to probe into their causal effects on the risk of bladder cancer. The result of LDL-C initially showed suggestive evidence of the risk of bladder cancer; however, it became non-significant after omitting two SNPs identified as driving SNPs by the leave-one-out analysis. Although the analysis yielded little evidence of causal impacts for HDL-C, LDL-C, and TG on bladder cancer, which was consistent with a previous MR study (11), it was, to the best of our knowledge, the first MR study to shed light on the causal effects of TC as well as various classes of lipid-regulatory medications on bladder cancer susceptibility.
A case–control study in 2015 enrolling 2,070 Chinese people, including 972 newly diagnosed urothelial carcinoma of the bladder and 1,098 cancer-free controls, revealed a positive association between hypertriglyceridemia and bladder cancer (adjusted OR 1.254, 95% CI 1.020–1.542, p = 0.032) (10). In addition, triglycerides were found to be positively linked with the overall bladder cancer risk in men in a sex-stratified prospective study involving six European cohorts, regardless of whether the tumor was muscle invasive or not (hazard ratio [HR] = 1.17, 95% CI = 1.06–1.27). Teleka et al. also discovered in 2018 that an elevated level of triglycerides (HR = 1.30, 95% CI = 1.12–1.48) and cholesterol (HR = 1.14, 95% CI = 1.02–1.25) were associated with a higher incidence of non-muscle invasive bladder cancer among men (28). In 2018, however, Orho-Melander et al. observed no correlations between TG (OR = 0.96, 95% CI = 0.68–1.37), LDL-C (OR = 0.89, 95% CI = 0.65–1.22) or HDL-C (OR = 0.86, 95% CI = 0.64–1.16) and bladder cancer risk in a prior MR study, which used 26, 32, and 41 SNPs as proxies for three serum lipid characteristics, respectively (11). What we unraveled in our study verified the results of Orho-Melander’s with the most recent and comprehensive GWAS statistics available to date. The statistical power of our study was further enhanced by the enrollment of 1,701 patients with bladder malignant neoplasms as opposed to only 400 bladder cancer cases in Orho-Melander M’s MR investigation.
Statins, which inhibit the activity of HMGCR, are the most widely prescribed lipid-regulatory medications for the treatment of dyslipidemia, cardiovascular diseases, chronic kidney disease, and so on. A retrospective study found no link between statin use and the risk of bladder cancer despite the small number of participants enrolled (29). On behalf of statin use, only one SNP, rs12916 in the HMGCR gene, was employed in Orho–Menlander’s MR study and suggested no relation with bladder cancer risk (11). A more recent investigation found that 17,708 post-coronary syndrome patients, either randomized to the ezetimibe group, a cholesterol absorption inhibitor that blocks the sterol transporter Niemann-Pick C1-Like 1 (NPC1L1) on the intestinal villi of the enterocytes, or the matching placebo group, had similar incidences of malignancy and malignancy-related death, including bladder cancer (30).
There was very little research that focused on the influence of other lipid-regulatory medications or their targets on the risk of bladder cancer, let alone those using Mendelian Randomization. Hence, what we presented here was a novel study that not only probed into the causal effects of representative traits for blood lipids on the risk of bladder cancer through the largest cohorts to date but also for the first time elucidated the causal relationship between the lipid-lowering effect and bladder cancer susceptibility using genetically proxied lipid-modulatory medications. We thoroughly explored the targets of commonly prescribed lipid-regulatory drugs in clinical practice and strictly adhered to MR procedures by discarding confounding-related SNPs, applying sensitivity tests of heterogeneity, horizontal pleiotropy, leave-one-out analysis, and so forth.
The findings of this study shall be interpreted in light of its limitations. First, despite having access to the largest and latest GWAS data, the study could still be prone to weak instrument biases, particularly in the case of lipid-regulatory medications, which had a restricted number of genetic proxies available. Larger GWAS for both bladder cancer and diverse sorts of blood lipid characteristics with more reliable genetic tools may enable us to evaluate the causal effects more precisely. Second, due to the original GWAS statistics, we were unable to divide the cohorts by gender and make use of sex-specific genetic IVs for blood lipid traits and lipid-regulatory medications, which might not be a pivotal influencing factor in the results because a recently published gender-stratified GWAS covering 33 biomarkers from the UK Biobank discovered that the gender actually exerted a limited impact on the genetics of most traits (31). Similarly, as the outcome of this study, bladder cancer owns distinct types of histopathology, such as the most prevalent urothelial carcinoma, and the squamous carcinoma, which mainly occurred in sub-Saharan African populations, etc. Future GWAS with specific pathological types of bladder cancer are needed. Third, we selected genetic proxies for lipid-regulatory medications based on key MR assumptions. In other words, it reflected the life-long modulation of the lipid-lowering effect, making it impossible to estimate the effect on the risk of bladder cancer in the short run. Fourth, it has been reported that the gut microbiota might contribute to blood lipid fluctuation in the host as well (32), but the MR study focuses solely on the human genome to explore the causality. Fifth, the GWAS data analyzed in the present study came entirely from the European population, which could lead to biases based on race, geographic environment, and diet. We are looking forward to more GWAS with populations of diverse races in future as well as growing international cooperation to benefit the health of mankind through Mendelian randomization.
Conclusion
In summary, using the latest and largest GWAS data to date, the current MR study discovered for the first time that genetically predicted TC and lipid-lowering effect of lipid-regulatory medications had no causal relationship with the risk of bladder cancer. We confirmed that HDL-C, LDL-C, and TG are not associated with bladder cancer susceptibility either. The results indicated that blood lipids may not play a crucial role in the tumorigenesis of bladder cancer and should be interpreted with caution. Further large-scale randomized controlled trials with various ethnic groups are warranted to validate our MR results.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
The study was conceived, designed, and analyzed by ZC. The manuscript was drafted by ZC and FY. HJ revised the manuscript and supervised the whole research. Others contributed to the summary of existing literature. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of P.R. of China (82373222), Leading Talent Program of Shanghai Municipal Health Commission (2022LJ008), and Medical Innovation Research Special Project of Science and Technology Commission of Shanghai Municipality (22Y21900200).
Acknowledgments
The authors are extremely grateful to the staff and investigators of UK Biobank and the FinnGen Project, without whose unremitting efforts and selflessness the researchers around the world would never have access to such large, detailed, and accurate datasets. Zhang Cheng, the first author, would like to express sincere gratitude to Shidong Chen from the Department of Neurology, Huashan Hospital, who is Zhang Cheng’s dearest roommate and guided Zhang Cheng into the terrific field of Mendelian Randomization.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2023.992608/full#supplementary-material
Abbreviations
MR, Mendelian randomization; GWAS, genome-wide association study; SNP, single nucleotide polymorphism; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; LD, linkage disequilibrium; OR, odds ratio; HR, hazard ratio; CI, confidence interval; ACLY, adenosine triphosphate-citrate lyase; HMGCR, hydroxy-methylglutaryl-coenzyme A reductase; NPC1L1, Niemann-Pick C1-Like 1; PCSK9, proprotein convertase subtilisin/kexin type 9.
Footnotes
1. ^https://www.ukbiobank.ac.uk/
2. ^https://gwas.mrcieu.ac.uk/
4. ^https://gwas.mrcieu.ac.uk/phewas/
References
1. Sung, H, Ferlay, J, Siegel, RL, Laversanne, M, Soerjomataram, I, Jemal, A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Antoni, S, Ferlay, J, Soerjomataram, I, Znaor, A, Jemal, A, and Bray, F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. (2017) 71:96–108. doi: 10.1016/j.eururo.2016.06.010
3. Teoh, JYC, Huang, J, Ko, WYK, Lok, V, Choi, P, Ng, CF, et al. Global trends of bladder cancer incidence and mortality, and their associations with tobacco use and gross domestic product per capita. Eur Urol. (2020) 78:893–906. doi: 10.1016/j.eururo.2020.09.006
4. Chamie, K, Litwin, MS, Bassett, JC, Daskivich, TJ, Lai, J, Hanley, JM, et al. Recurrence of high-risk bladder cancer: a population-based analysis. Cancer. (2013) 119:3219–27. doi: 10.1002/cncr.28147
5. van der Heijden, AG, and Witjes, JA. Recurrence, progression, and follow-up in non-muscle-invasive bladder cancer. Eur Urol Suppl. (2009) 8:556–62. doi: 10.1016/j.eursup.2009.06.010
6. Vale, CL. Adjuvant chemotherapy in invasive bladder cancer: a systematic review and meta-analysis of individual patient data. Eur Urol. (2005) 48:189–99. doi: 10.1016/j.eururo.2005.04.005
7. Singh, P, and Black, P. Emerging role of checkpoint inhibition in localized bladder cancer. Urol Oncol. (2016) 34:548–55. doi: 10.1016/j.urolonc.2016.09.004
8. Gakis, G. Management of muscle-invasive bladder cancer in the 2020s: challenges and perspectives. Eur Urol Focus. (2020) 6:632–8. doi: 10.1016/j.euf.2020.01.007
9. Cumberbatch, MGK, Cox, A, Teare, D, and Catto, JWF. Contemporary occupational carcinogen exposure and bladder cancer. JAMA Oncol. (2015) 1:1282–90. doi: 10.1001/jamaoncol.2015.3209
10. Xu, S, Zhang, GM, Guan, FJ, Dong, DH, Luo, L, Li, B, et al. The association between metabolic syndrome and the risk of urothelial carcinoma of the bladder: a case-control study in China. World J Surg Oncol. (2015) 13:236. doi: 10.1186/s12957-015-0631-5
11. Orho-Melander, M, Hindy, G, Borgquist, S, Schulz, CA, Manjer, J, Melander, O, et al. Blood lipid genetic scores, the HMGCR gene and cancer risk: a Mendelian randomization study. Int J Epidemiol. (2018) 47:495–505. doi: 10.1093/ije/dyx237
12. Smith, GD, and Hemani, G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. (2014) 23:R89–98. doi: 10.1093/hmg/ddu328
13. Davies, NM, Holmes, MV, and Davey Smith, G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ. (2018) 362:k601. doi: 10.1136/bmj.k601
14. Ou, YN, Yang, YX, Shen, XN, Ma, YH, Chen, SD, Dong, Q, et al. Genetically determined blood pressure, antihypertensive medications, and risk of Alzheimer’s disease: a Mendelian randomization study. Alzheimers Res Ther. (2021) 13:41. doi: 10.1186/s13195-021-00782-y
15. Sudlow, C, Gallacher, J, Allen, N, Beral, V, Burton, P, Danesh, J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. (2015) 12:e1001779. doi: 10.1371/journal.pmed.1001779
16. Richardson, TG, Sanderson, E, Palmerid, TM, Korpelaid, MA, Ference, BA, Smith, GD, et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med. (2020) 17:e1003062. doi: 10.1371/JOURNAL.PMED.1003062
17. Kurki, MI, Karjalainen, J, Palta, P, Sipilä, TP, Kristiansson, K, Donner, K, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. MedRxiv. (2022)
18. Burgess, S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. (2014) 43:922–9. doi: 10.1093/ije/dyu005
19. Bowden, J, Smith, GD, and Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
20. Bowden, J, Davey Smith, G, Haycock, PC, and Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
21. Bowden, J, Fabiola Del Greco, M, Minelli, C, Smith, GD, Sheehan, NA, and Thompson, JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-egger regression: the role of the I 2 statistic. Int J Epidemiol. (2016) 45:1961–74. doi: 10.1093/ije/dyw220
22. Verbanck, M, Chen, CY, Neale, B, and Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
23. Burgess, S, and Thompson, SG. Interpreting findings from Mendelian randomization using the MR-egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
24. Hemani, G, Tilling, K, and Davey, SG. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. (2017) 13:e1007081. doi: 10.1371/journal.pgen.1007081
25. Burgess, S, and Thompson, SG. Avoiding bias from weak instruments in mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036
26. Hemani, G, Zheng, J, Elsworth, B, Wade, KH, Haberland, V, Baird, D, et al. The MR-base platform supports systematic causal inference across the human phenome. elife. (2018) 7:e34408. doi: 10.7554/eLife.34408
27. Tin, A, and Kottgen, A. Mendelian randomization analysis as a tool to gain insights into causes of diseases: a primer. J Am Soc Nephrol. (2021) 32:2400–7. doi: 10.1681/ASN.2020121760
28. Teleka, S, Häggström, C, Nagel, G, Bjørge, T, Manjer, J, Ulmer, H, et al. Risk of bladder cancer by disease severity in relation to metabolic factors and smoking: a prospective pooled cohort study of 800,000 men and women. Int J Cancer. (2018) 143:3071–82. doi: 10.1002/ijc.31597
29. Kuo, CC, Chiu, HF, Lee, IM, Kuo, HW, Lee, CT, and Yang, CY. Statin use and the risk of bladder cancer: a population-based casecontrol study. Expert Opin Drug Saf. (2012) 11:733–8. doi: 10.1517/14740338.2012.712960
30. Giugliano, RP, Gencer, B, Wiviott, SD, Park, JG, Fuchs, CS, Goessling, W, et al. Prospective evaluation of malignancy in 17,708 patients randomized to Ezetimibe versus placebo: analysis from IMPROVE-IT. JACC CardioOncol. (2020) 2:385–96. doi: 10.1016/j.jaccao.2020.07.008
31. Flynn, E, Tanigawa, Y, Rodriguez, F, Altman, RB, Sinnott-Armstrong, N, and Rivas, MA. Sex-specific genetic effects across biomarkers. Eur J Hum Genet. (2021) 29:154–63. doi: 10.1038/s41431-020-00712-w
Keywords: blood lipids, lipid-regulatory medications, bladder cancer, Mendelian randomization, UK Biobank, FinnGen
Citation: Cheng Z, Ye F, Liang Y, Xu C, Zhang Z, Ou Y, Chen X, Dai X, Mou Z, Li W, Chen Y, Zhou Q, Zou L, Mao S and Jiang H (2023) Blood lipids, lipid-regulatory medications, and risk of bladder cancer: a Mendelian randomization study. Front. Nutr. 10:992608. doi: 10.3389/fnut.2023.992608
Edited by:
Manja Zec, University of Colorado Anschutz Medical Campus, United StatesReviewed by:
Jian Yin, Chinese Academy of Medical Science and Peking Union Medical College, ChinaJeff M. P. Holly, University of Bristol, United Kingdom
Copyright © 2023 Cheng, Ye, Liang, Xu, Zhang, Ou, Chen, Dai, Mou, Li, Chen, Zhou, Zou, Mao and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haowen Jiang, dXJvbG9neV9oc0AxNjMuY29t; Shanhua Mao, bWFvc2hhbmh1YUAxNjMuY29t
†These authors have contributed equally to this work
 Zhang Cheng
Zhang Cheng Fangdie Ye
Fangdie Ye Yingchun Liang
Yingchun Liang Chenyang Xu
Chenyang Xu Zheyu Zhang
Zheyu Zhang Yuxi Ou
Yuxi Ou Xinan Chen
Xinan Chen Xiyu Dai
Xiyu Dai Zezhong Mou
Zezhong Mou Weijian Li
Weijian Li Yiling Chen
Yiling Chen Quan Zhou
Quan Zhou Lujia Zou
Lujia Zou Shanhua Mao
Shanhua Mao Haowen Jiang
Haowen Jiang