
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 11 May 2023
Sec. Nutrition and Microbes
Volume 10 - 2023 | https://doi.org/10.3389/fnut.2023.1181492
 Weihua Tan1,2,3†
Weihua Tan1,2,3† Yuanyuan Tian1,2†
Yuanyuan Tian1,2† Qingya Zhang2,3
Qingya Zhang2,3 Siwei Miao4
Siwei Miao4 Wenrong Wu2,3
Wenrong Wu2,3 Xiaoqing Miao2,4
Xiaoqing Miao2,4 Haiou Kuang4,5
Haiou Kuang4,5 Wenchao Yang1,2,3,4*
Wenchao Yang1,2,3,4*Salmonella enterica serovar Typhimurium (S. Typhimurium) is a common food-borne pathogen that commonly causes gastroenteritis in humans and animals. Apis laboriosa honey (ALH) harvested in China has significant antibacterial activity against Staphylococcus aureus, Escherichia coli, and Bacillus subtilis. We hypothesize that ALH has antibacterial activity against S. Typhimurium. The physicochemical parameters, minimum inhibitory and bactericidal concentrations (MIC and MBC) and the possible mechanism were determined. The results showed that there were significantly different physicochemical parameters, including 73 phenolic compounds, among ALH samples harvested at different times and from different regions. Their antioxidant activity was affected by their components, especially total phenol and flavonoid contents (TPC, TFC), which had a high correlation with antioxidant activities except for the O2- assay. The MIC and MBC of ALH against S. Typhimurium were 20–30% and 25–40%, respectively, which were close to those of UMF5+ manuka honey. The proteomic experiment revealed the possible antibacterial mechanism of ALH1 at IC50 (2.97%, w/v), whose antioxidant activity reduced the bacterial reduction reaction and energy supply, mainly by inhibiting the citrate cycle (TCA cycle), amino acid metabolism pathways and enhancing the glycolysis pathway. The results provide a theoretical basis for the development of bacteriostatic agents and application of ALH.
Honey is a naturally sweet substance made from the nectar of blossoms or the excretions of plant-sucking insects living on parts of plants by honey bees. Honey is composed of carbohydrates (90–95% of dry mass), approximately 17% water, and minor components (approximately 3% in total), for instance, proteins, amino acids, organic acids, vitamins, minerals, polyphenols, and volatile compounds, which content vary depending on the floral source, type of bee, environmental conditions, and/or extraction, processing, and storage conditions of packaged honey (1). Honey is used not only as a natural food but also as a treatment for diseases and wounds (1–3). Furthermore, honey has antioxidant (4), antibacterial (5), anticancer (6), anti-inflammatory (7), antivirus (2), and prebiotic activities (1).
The antioxidant activity of honey is a popular topic. Different antioxidant activities of honey samples are variable depending on their botanical sources (8), geographic regions (9), processing and storage (10) and determination assays (11), which include diphenyl-1-picrylhydrazyl (DPPH), ferric reducing antioxidant power (FRAP), oxygen radical absorbance capacity (ORAC), 2,2-azinobis (3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS), 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox)-equivalent antioxidant capacity (TEAC), ascorbic acid content and different enzyme assays, such as catalase (CAT), glutathione peroxidase (GPO), and superoxide dismutase (SOD) assays. Phenolic compounds are mainly responsible for the antioxidant activity of honey (8, 12–15). Total flavonoid content, total water-soluble vitamins, mineral content and proteins also contribute to the antioxidant activity of honey (8, 9, 12). There are only a few reports about the antioxidant activities of Apis laboriosa honey (ALH), a wild honey made from the nectar of blossoms or the excretions of plant-sucking insects living on parts of plants by Apis laboriosa workers, which also vary by harvesting season, region and botanical source (16).
On the other hand, the antibacterial activity of honey has been widely studied. Honey has potential antibacterial effects against a wide range of bacteria and even against several antibiotic-resistant bacteria (17). The osmotic pressure, hydrogen peroxide content, phytochemical factors, low pH, phenolic acid level and flavonoid compounds of honey contribute to its antibacterial effect (18, 19). Phenolic compounds in honey play an important role in its biological and antibacterial abilities (20). At the same time, honey may contain some unknown components that inhibit the growth of bacteria (21–23). Our previous research demonstrated that ALH harvested in China has significant antibacterial activity against Staphylococcus aureus, Escherichia coli, and Bacillus subtilis when diluted to 90% moisture content (24). It was reported that ALH from Nepal has an antibacterial effect against S. aureus and E. coli (25). The differences in the antibacterial activity of ALH were attributed to differences in its botanical sources and entomological proteins (24). The antibacterial activity of ALH against Salmonella enterica serovar Typhimurium (S. Typhimurium) is unknown.
Salmonella Typhimurium is a common food-borne pathogen that commonly causes gastroenteritis in humans and animals. The replication of S. Typhimurium in the intestinal tract is sustained by the secretion of effector proteins by type III secretion systems to trigger an inflammatory response without the engagement of innate immune receptors (26). S. Typhimurium was listed as “the most threatening to public health” by the Centers for Disease Control and Prevention due to its frequent adulteration of beef and poultry food products and its association with multidrug resistance (27, 28). Therefore, developing new strategies to inhibit this bacterium is urgent. Previous reports showed that silver (Ag) and citric acid coated iron oxide (Fe3O4) nanoparticles (29), olive oil polyphenol extract (30), cocktails of phages targeting multiple host receptors (31), novel deoxy-tetradeuterio-curcumin derivative (32), linalool nanoemulsions (33), zinc oxide nanoparticles (34), etc., had antibacterial activities against S. Typhimurium. At present, research data on the antibacterial ability and antibacterial mechanisms of ALH against S. Typhimurium are limited.
In this study, the antioxidant and antibacterial activities and possible antibacterial mechanism of ALH was investigated, which will provide an alternative bacteriostatic strategy against S. Typhimurium.
The study was performed between March 2021 and October 2022 at the Bee Product Processing and Application Research Center of the Ministry of Education (Fuzhou) at Fujian Agricultural and Forestry University.
ALH samples were harvested from Yunnan Province, China. The harvesting location and date information are listed in Table 1. Manuka honey (MH: UMF5+, 10+, and 15+: MH1, MH2, and MH3, respectively), which UMF data means the antibacterial activity of honey is the same as that of phenol 5, 10, and 15% aqueous solution respectively, was purchased from Comvita ® News Land in August 2019 and mailed to our lab. These honey samples were stored in a refrigerator at 4°C until testing.
The bacterial strain Salmonella enterica serovar Typhimurium (ATCC14028) (S. Typhimurium) used in this experiment was purchased from Guangdong Huankai Microbial Sci. & Tech. Co., Ltd., Guangzhou, China.
Chemicals of 1,1-diphenyl-2-trinitrophenylhydrazine, 6-hydroxy-2,5,7,8-tetramethyltryptophan-2-carboxylic acid, 2,2-diazodi (3-ethyl-benzothiazole-6-sulfonic acid) diammonium salt, phenazine methyl sulfate β-Nicotinamide adenine dinucleotide disodium salt, nitrotetrazolium chloride blue, 2,4,6-tripyridyl triazine, and analytical methanol were purchased from Shanghai McLean Biochemical Technology Co., Ltd. Shanghai, China. Chemicals of ferric chloride, hydrochloric acid, acetic acid, sodium acetate, sodium dihydrogen phosphate and disodium hydrogen phosphate were purchased from Sinopharm Chemical Reagent Co., Ltd. Shanghai, China. LB broth medium was purchased from Bioengineering (Shanghai) Co., Ltd. Shanghai, China. TTC nutritional agar, folinol and rutin were purchased from Beijing Solebar Technology Co., Ltd. Beijing, China. Gallic acid was purchased from Shanghai Yuanye Co., Ltd. Shanghai, China. Hieff UNICON Universal Blue qPCR SYBR Green Master Mix was purchased from Yeasen Biotechnology Co., Ltd. Shanghai, China.
Determinations of the moisture, ash, pH, glucose, fructose, sucrose and 5-HMF in ALH were performed according to the method of our earlier report (24). The total phenol and flavonoid contents were determined according to a previous method (35). The data were calculated by the dry weight of ALH.
The phenolic components were determined by untargeted metabolomics at Novogene Co., Ltd. (Beijing, China). The honey samples (100 μl) were placed in EP tubes and resuspended in prechilled 80% methanol by vortexing well. Then, the samples were incubated on ice for 5 min and centrifuged at 15,000 g and 4°C for 20 min. The supernatant was diluted to a final concentration containing 53% methanol by LC–MS grade water and then centrifuged at 15,000 g and 4°C for 20 min. The supernatant was injected into the LC–MS/MS system for analysis. UHPLC–MS/MS analyses were performed using a Vanquish UHPLC system (Thermo Fisher, Germany) coupled with an Orbitrap Q Exactive TM HF-X mass spectrometer (Thermo Fisher, Germany). Samples were injected onto a Hypersil Gold column (100 mm × 2.1 mm, 1.9 μm) using a 17-min linear gradient at a flow rate of 0.2 ml/min. The eluents for the positive polarity mode were eluent A (0.1% formic acid in water) and eluent B (methanol). The eluents for the negative polarity mode were eluent A (5 m ammonium acetate, pH 9.0) and eluent B (methanol). The solvent gradient was set as follows: 2% B for 1.5 min, 2–85% B for 3 min, 85–100% B for 10 min, 100–2% B for 10.1 min, and 2% B for 12 min. A Q Exactive TM HF-X mass spectrometer was operated in positive/negative polarity mode with a spray voltage of 3.5 kV, capillary temperature of 320°C, sheath gas flow rate of 35 psi, aux gas flow rate of 10 l/min, S-lens RF level of 60, and aux gas heater temperature of 350°C. The raw data files were obtained for further analysis.
The DPPH radical scavenging activity and FRAP total antioxidant capacity were determined according to a previous method (36) at 517 and 593 nm, respectively. Trolox was used as the standard, and the result is expressed in Trolox equivalents, mg TE/100 g.
According to a previous method (37), ABTS cation radical scavenging activity was determined at 734 nm. Trolox was used as the standard and expressed as the result using its equivalent, mg TE/100 g.
The reduction ability of the ALH solution in the system was determined by detecting the absorbance value with an ultraviolet spectrophotometer (T6, Beijing Puxi General Instrument Co., Ltd. Beijing, China) at 560 nm, with NADH-PMS-NBT used as the superoxide anion (O2·-) generation system (38).
All antioxidant activities were expressed by the dry weight of ALH.
The bacterial suspension, which was adjusted to 1 × 108 CFU/ml with physiological saline, was added to the 96-well plate (10 μl). Then, 190 μl of culture medium (ALH and broth culture) was added and mixed well. It was placed in a shaker incubator at 80 r/min at 37°C for 24 h.
Minimum inhibitory concentration (MIC) determination was performed according to a previous method (39). The ALH and broth culture medium were mixed evenly and prepared at different concentrations of 0, 5, 10, 15, 20, 25, 30, 35, and 40% (dry weight of honey/volume of mixture); other ALH concentrations for antibacterial activity determination were the same. The mixed liquid after shaking was filtered with a 0.22 μm filter membrane for further determination.
The liquid (200 μl) in the 96-well plate was transferred to LB solid medium, incubated at 37°C for 24 h, and the growth of the colony in the solid culture was observed. MBC is the lowest honey concentration without colony growth (39).
The honey sample ALH1 has medium IC50 against S. Typhimurium (Table 2). ALH1 was selected for further antibacterial mechanism experiment.
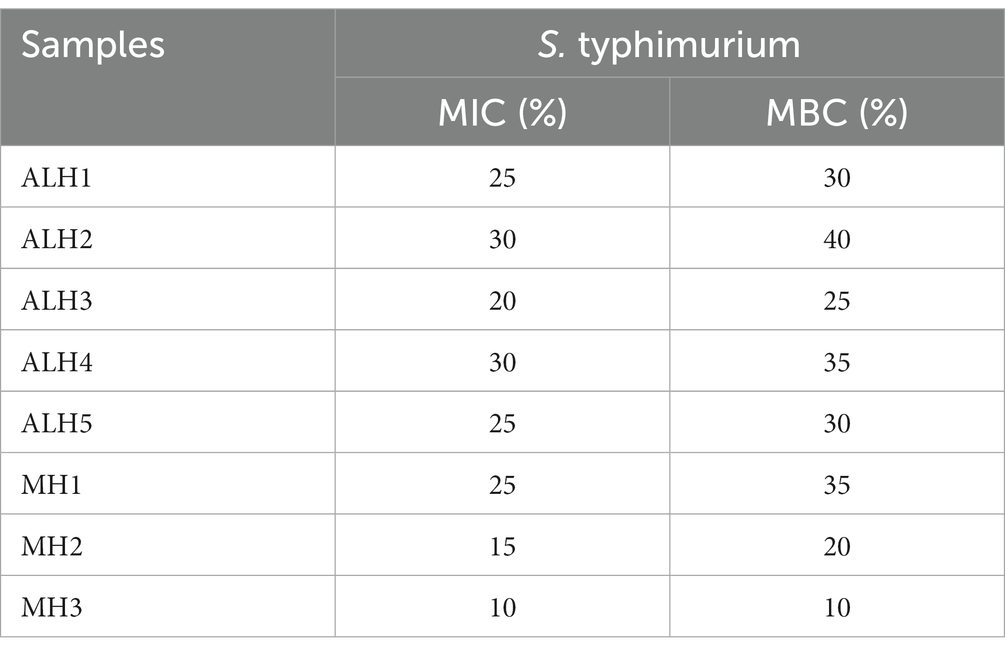
Table 2. Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of honey samples against S. typhimurium.
Broth medium with 0, 2, 4, 6, and 8% (w/v) ALH1 was prepared and filtered through a 0.22 μm filter. Their inhibition rates were calculated according to their optical densities. The IC50 was calculated using GraphPad Prism 8.4.3 for Windows (GraphPad Software, Inc.) according to the inhibition rates of different concentrations of ALH1 against S. Typhimurium.
Salmonella Typhimurium was cultured as the treatment group (IC50 of ALH1, 2.97% w/v) and the control group (no ALH1). After culturing, S. Typhimurium was collected and snap-frozen in liquid nitrogen and stored at −80°C for subsequent experiments. The proteins were determined using LC–MS–MS with a Q Exactive HF-X mass spectrometer (Thermo Fisher, Germany) with a Nanospray Flex™ electrospray ionization (ESI) source by Novgene Biotech Co., Ltd., China.
Salmonella Typhimurium was cultured with IC50 ALH1. RNA was extracted using the TransZol UP kit, and its concentration and purity were determined by an ultramicro UV spectrophotometer (NanoDrop One; Thermo Fisher Scientific, United States). The relative expression levels of gene-coded proteins in protein interactions in the citrate cycle (TCA cycle) and bacterial chemotaxis (icdA, gltA, fumC, nuoC, sucB, lpdA, motA, FliG, motB, pgi) were determined using RT-PCR assays. cDNA synthesis was performed using the HiScript II Q RT SuperMix for qPCR (+gDNA wiper) kit. The internal reference gene was 5S (40). Their primer sequences were designed using Primer Premier 5.0 (Primer) and were listed in Supplementary Table S1, which synthesis were performed by Sangon Biotech (Shanghai, China) Co., Ltd. These primers were identified by RT-PCR procedure was performed as previously described (41). Finally, a SYBR mixture was used for gene expression quantification.
All experiments were performed in triplicate. The experimental results were expressed as the mean ± standard error. Percentages (p) were then transformed to arc sin (degree) values (according to the formula arc sin√p) before ANOVA. One-way ANOVA was used to analyze the significance differences using GraphPad Prism 8.0.2 for Windows (GraphPad Software, Inc.). The correlations between TPC, TFC and antioxidant activity were analyzed using IBM SPSS Statistics 26.0 (IBM). The relative gene expression was expressed by the ratio of the target gene to the internal reference gene.
The raw data files of phenolic compounds were processed using Compound Discoverer 3.1 (CD3.1, Thermo Fisher) to perform peak alignment, peak picking, and quantitation for each metabolite. The main parameters were set as follows: retention time tolerance of 0.2 min, actual mass tolerance of 5 ppm, signal intensity tolerance of 30%, signal/noise ratio of 3, and minimum intensity. The peak intensities were normalized to the total spectral intensity. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks and fragment ions. These peaks were matched with the mzCloud1, mzVault and MassList databases to obtain accurate qualitative and relative quantitative results (r2 ≥ 0.99). Statistical analyses were performed using the statistical software R (R-3.4.3), Python (Python V2.7.6) and CentOS (CentOS release 6.6). Compounds whose CVs of relative peak areas in QC samples were greater than 30% were removed, and finally, the metabolites’ identification and relative quantification results were obtained. All the phenolic components were screened according to a previous method (42).
The spectra of proteins obtained from LC–MS/MS were searched against the UniProt database blasted with the NCBI database by Proteome Discoverer 2.2 (Thermo) with a credibility of more than 99% Peptide Spectrum Matches. Protein identification, differential protein definition (fold change ≥2 or ≤0.5; p < 0.05), GO term enrichment, KEGG pathway enrichment and protein–protein interactions (PPI, interaction score 0.900) were explored as described in our previous report (43).
The physicochemical parameters of the ALH samples were shown in Table 3. The moisture, ash, pH, glucose, fructose, sucrose, 5-HMF contents, total phenols and total flavonoids in these samples were significantly different and are labeled with different lowercase letters. The physicochemical parameters of MH1, MH2, and MH3 were the same as those in our previous report (24). There were 73 phenolic compounds (Table 4), which spectrogram are shown in Supplementary Figure S1.
The antioxidant activities of the ALH samples are shown in Table 5. The DPPḤ, ABTS·+, FRAP and superoxide anion scavenging activities of these samples were significantly different and are labeled with different lowercase letters.
The correlation ships among TPC, TFC and antioxidant activities are shown in Table 6. They were strongly correlated with DPPH free radical scavenging activity, ABTS free radical scavenging activity, and FRAP total antioxidant capacity (p < 0.05).
As shown in Table 2, the MIC and MBC of ALH against S. Typhimurium were 20–30% and 25–40%, respectively. They were higher than those of MH2 and MH3.
The inhibition rates of different doses of honey samples against S. Typhimurium are shown in Figure 1. The IC50 of ALH1 was 2.97% (w/v), 1.99% for MH1, 1.43% for MH2 and 1.50% for MH3.
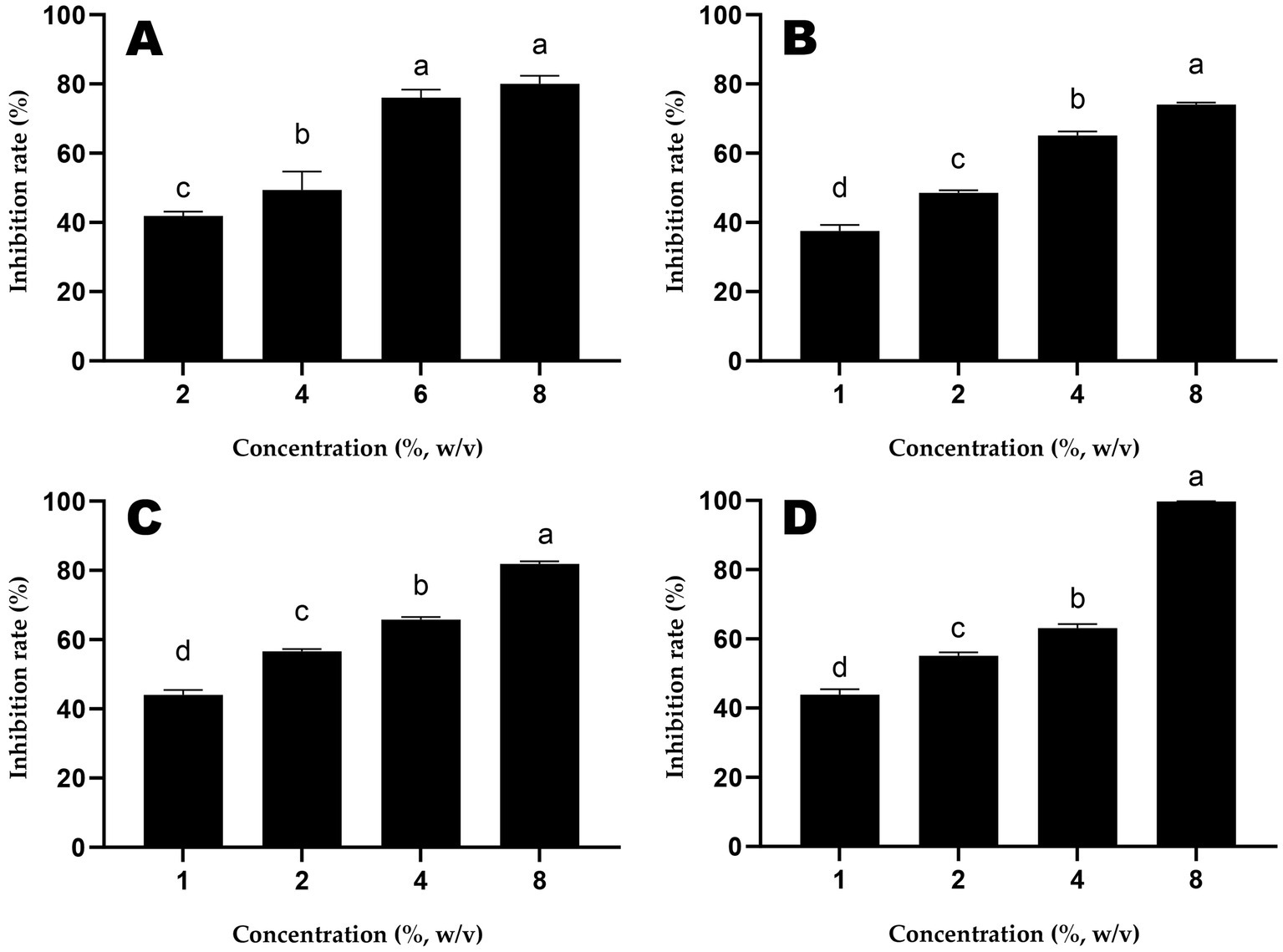
Figure 1. The inhibition rates of different doses of honey samples (A: ALH1; B: MH1; C: MH2; D: MH3) against S. Typhimurium. Different lowercases mean significant differences among inhibition rates of different doses of honey.
There were 2,606 proteins and 708 differentially expressed proteins, which included 130 upregulated proteins (the difference in protein abundance reached 2 times or more, p < 0.05) and 578 downregulated proteins (the difference in protein abundance was 0.5 times or less, p < 0.05) (Figure 2).
There were 44 enriched Gene Ontology terms of differential proteins between the treatment and control groups (p < 0.05, Figure 3). If the adjusted value of p was considered, 5 terms enriched more differential proteins (adjusted p < 0.05): single-organism process (171 differential proteins; p = 0.000149), single-organism metabolic process (117 differential proteins; p = 0.00574), membrane (77 differential proteins; p = 0.0352), oxidation–reduction process (70 differential proteins; p = 0.000196) and oxidoreductase activity (59 differential proteins; p = 0.000550).
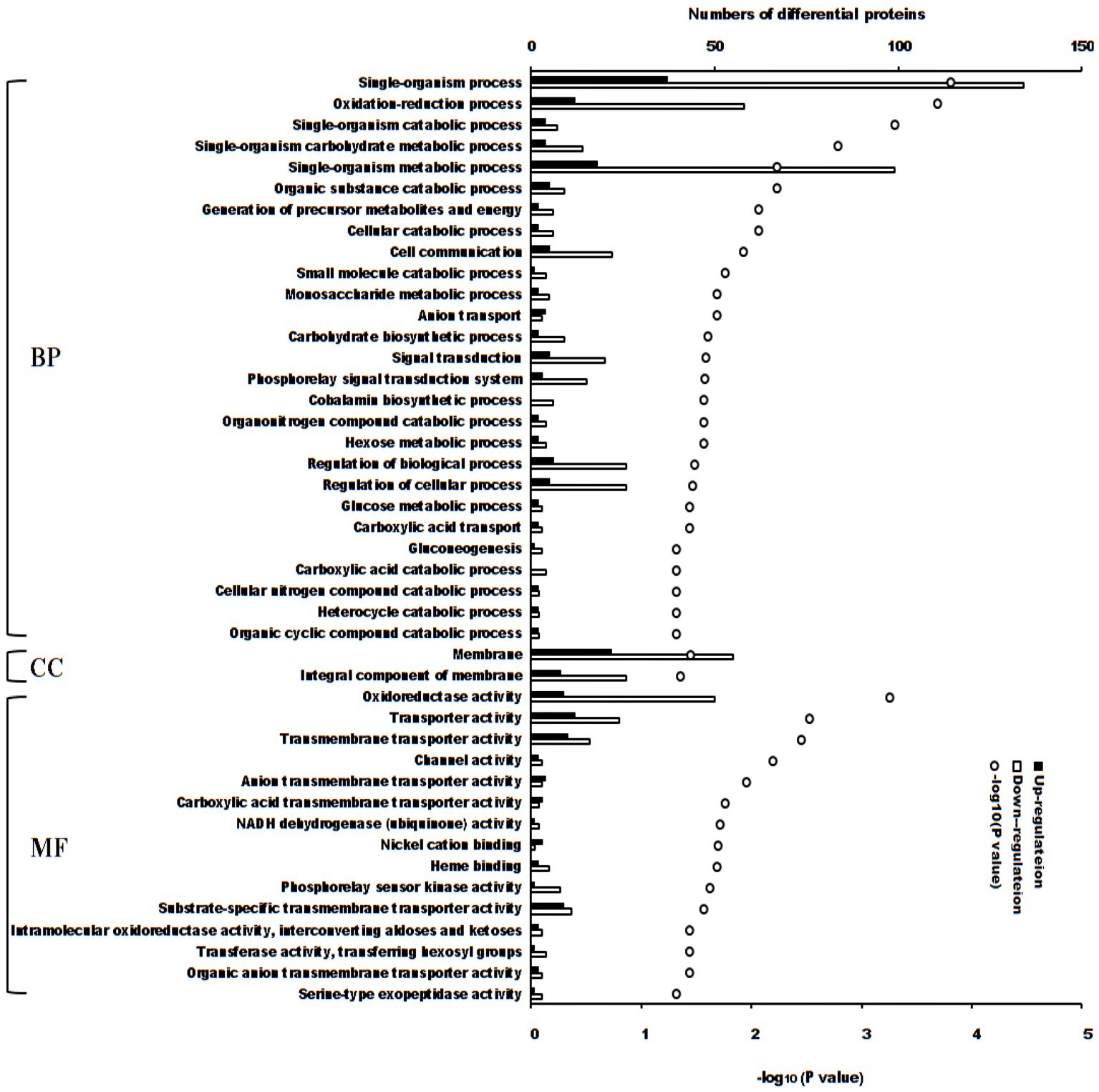
Figure 3. Enriched terms of Gene Ontology of differential proteins between S. Typhimurium treated with ALH1 and control groups. The blank and black bars mean down- or up-regulated differential proteins, repetitively. The open circle means –log10 (p-value). BP, biological process; CC, cellular component; MF, molecular function.
These differentially expressed proteins were significantly enriched in 12 KEGG pathways (Figures 4, p < 0.05). There were 3 KEGG pathways if the adjusted p-value was considered: carbon metabolism (52 differential proteins, adjusted p = 5.47 × 10−5), citrate cycle (TCA cycle) (17 differential proteins, adjusted p = 1.47 × 10−3) and microbial metabolism in diverse environments (79 differential proteins, adjusted p = 0.0221).
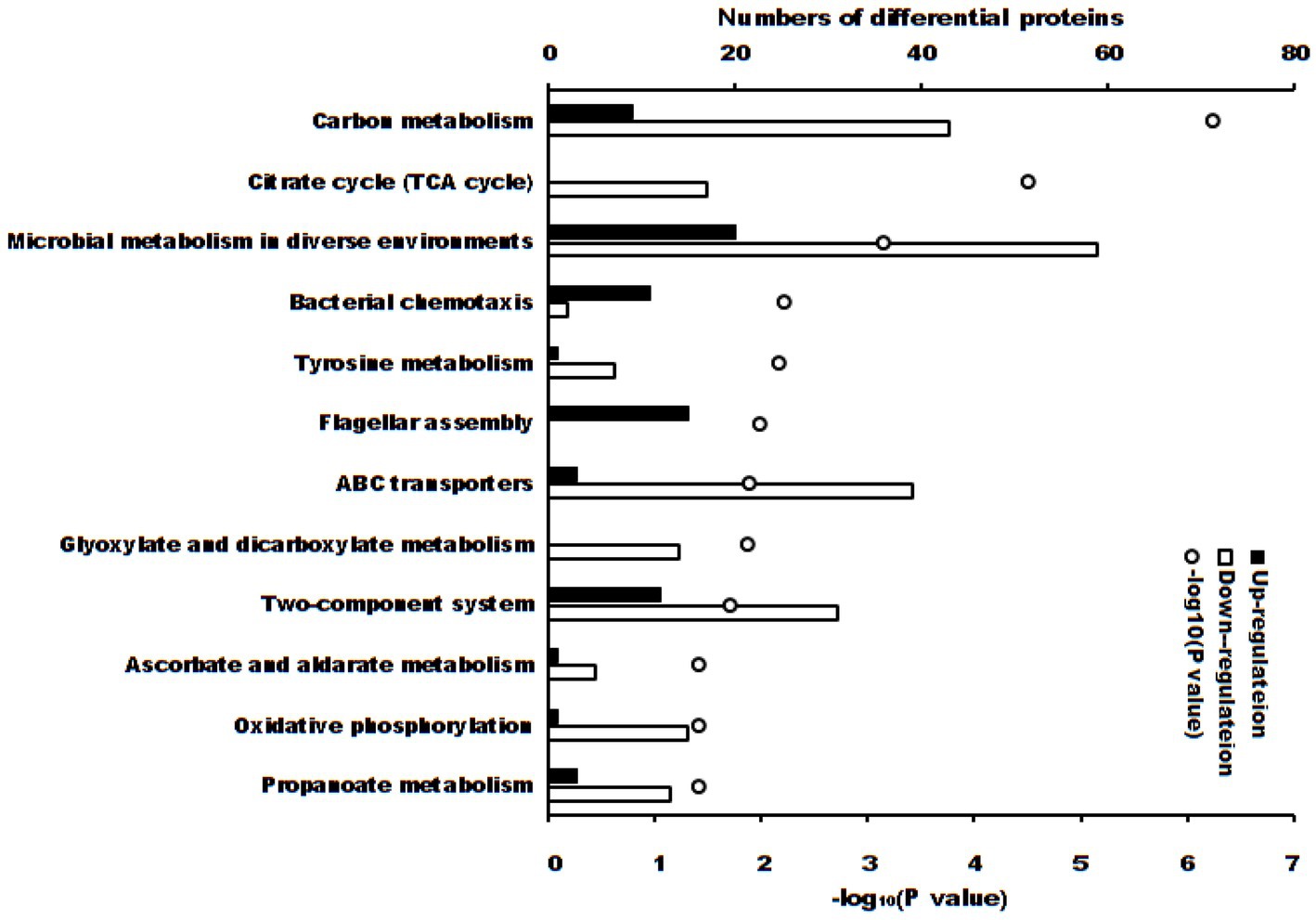
Figure 4. KEGG pathways (p < 0.05) of differential proteins between S. Typhimurium in ALH1 and control groups. The blank and solid bars mean down- or up-regulated differential proteins, repetitively. The open circle means –log10 (p-value).
The protein–protein interaction network analysis is shown in Figure 5. The top 4 interacting proteins were aldehyde-alcohol dehydrogenase (A0A0F6B241, upregulated, p = 0.000158), putative pyruvate-flavodoxin oxidoreductase (A0A0F6B1S9, downregulated, p = 0.012980), N-acetyl glucosamine-specific PTS system components IIABC (A0A0F6AYI0, downregulated, p = 0.014904) and malic enzyme (A0A0F6B4M2, downregulated, p = 0.006444).
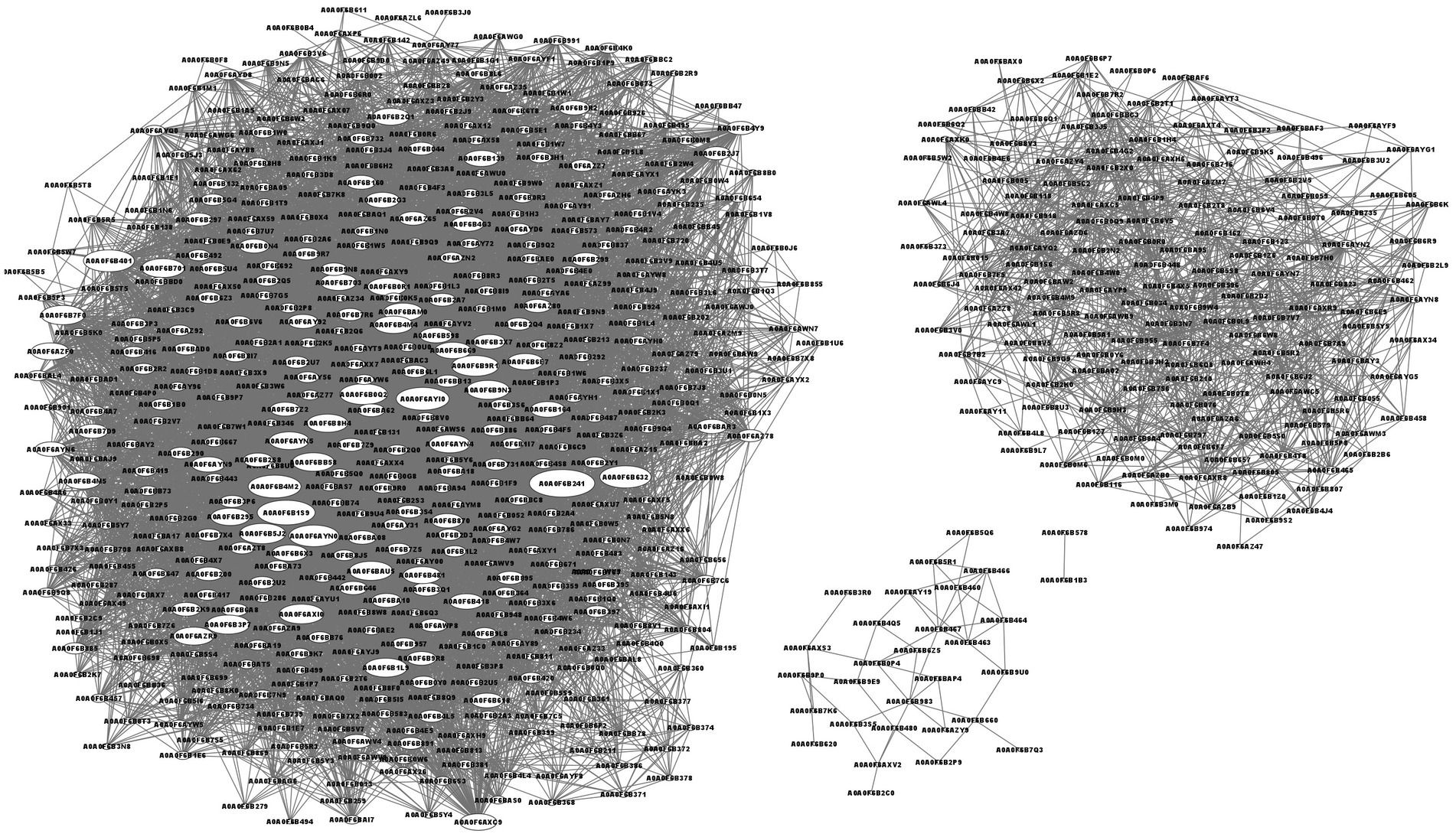
Figure 5. Network of protein–protein interactions. The characters in oval means protein ID. The line means the interaction of two proteins. The bigger size of the oval means more proteins interacted.
The results showed that the relative expression levels of motA and FliG were upregulated, and icdA, gltA, fumC, nuoC, sucB, lpdA, motB, and pgi were downregulated (Figure 6).
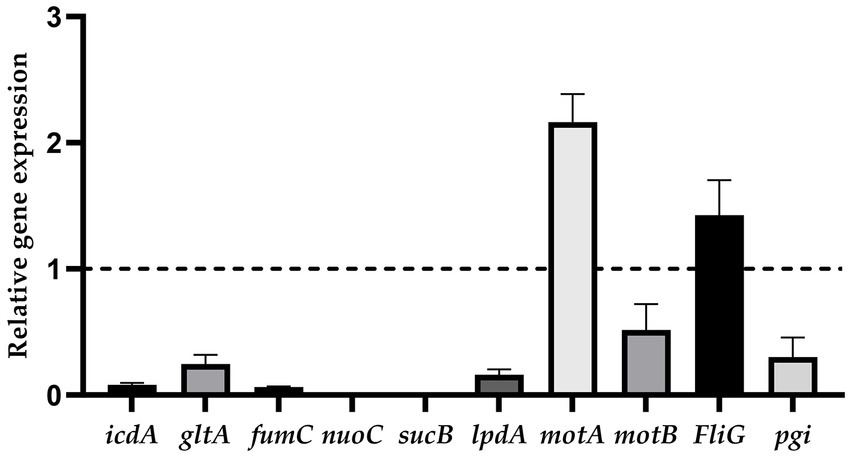
Figure 6. The relative expression of genes (ratio of the target gene to the internal reference gene).
The physicochemical parameters of the ALH samples varied over a large range. These compounds were within the range in previous reports (17, 24, 25). The moisture content varied from 16.96 to 26.41%, which was influenced by the geographic locations, which have different relative humidity of air (1, 44) and maturity degrees (45). The relative humidity of air and maturity degrees of ALH were not controlled by us because the migratory habit of Apis laboriosa. Other parameters of samples with higher moisture, showed in dry weight, were in reasonable scopes (Table 3). Other parameters also had significant differences, which is in line with our previous report (24). They are affected by botanical sources, geographic locations and storage or transport conditions (24, 45–47). The different physicochemical parameters caused different antioxidant activities when they were determined using different assays (Table 5). However, except for O2·-, they have a high correlation (Table 6). Among these parameters, TPC and TFC were highly correlated with antioxidant activity (Table 6) determined using DPPḤ, ABTS· + and FRAP assays, agreed with other references (8, 12–15). The result of the superoxide anion scavenging activity assay was not in line with those of other antioxidant assays, which was also found in a previous report (48). These differences may be caused by different chemical components in honey and their detection principle (11). It was reported that TFC accounted for approximately 25–70% of TPC depending on the honey botanical source (49), which in ALH samples were 23–44%, except for that in ALH5. The high moisture content in ALH5 indicates a low ripening degree (45), which resulted in a low TFC.
The TPC and TFC in honey were also responsible for its antibacterial activity. Polyphenols with pyrogallol groups (50), methyl gallate and gallic acid (51, 52) showed strong antibacterial activity against various kinds of bacteria. Polyphenols with pyrogallol groups had strong antibacterial activity against 26 species of bacteria, but those with catechol and resorcinol rings showed lower activity (50). The MBC of methyl gallate and gallic acid against different bacteria varied from 0.5 to less than 8 mg/ml (51, 52). The MIC against S. Typhimurium varied from 20 to 30%, and the MBC varied from 25 to 40% ALH, with TPCs of 49–123 mg GAE/100 g (Table 3). The calculated TPCs in MIC or MBC were close to these reports (51, 52), even though the data were determined using differential species of bacteria. The antibacterial activities of honey depend on the type of flowers foraged by the bees and its geographical origin rather than its monofloral or polyfloral nature (53). However, there was no correlation between TPC and the well diffusion assay for S. aureus (54). The antibacterial activity of honey is attributed to many factors, including sugar, polyphenol compounds, hydrogen peroxide, 1, 2-dicarbonyl compounds, and bee defensin-1 contents (18). The phenolic compositions varied depending on the botanical sources of the ALH samples (Table 4), causing different antibacterial activities against S. Typhimurium.
The antibacterial activity can be determined using MIC, MBC or IC50, etc. For further proteomic experiments, the IC50 of ALH1 against S. Typhimurium was determined to be 2.97% (w/v) after 24 h of treatment (Figure 1). Then, this concentration was used for proteomic and RT–PCR experiments to determine the possible mechanism of the antibacterial activity. The differential proteins in S. Typhimurium between the ALH1-treated and control groups played an important role in its antibacterial activity. There were 160 differential proteins with p = 0 (Supplementary Table S2), of which there were 14 upregulated proteins. All the differential proteins were significantly enriched in 3 KEGG pathways (adjusted p < 0.05): carbon metabolism, TCA cycle and microbial metabolism in diverse environments.
First, carbon metabolism (adjusted p = 5.47 × 10−5) included 52 differential proteins (9 up- and 43 downregulations), whose proportion in total proteins of the pathway was 50.49% (Figure 4). There are six key oxidoreductases of central carbon metabolism (CCM): glucose-6-phosphate dehydrogenase, pyruvate dehydrogenase, 2-ketoglutarate dehydrogenase, malate dehydrogenase, malic enzyme, and isocitrate dehydrogenase (55). In this experiment, malate dehydrogenase (p = 0.00763), malic enzyme (p = 0.00644) and isocitrate dehydrogenase (p = 0.0113) were downregulated, which indicated that ALH1 inhibited central carbon metabolism via a decrease in oxidoreductase activity. Malic enzyme and isocitrate dehydrogenase are necessary for the normal growth and/or persistence of bacteria (56). ALH1 repressed central carbon metabolism and inhibited proliferation and survival via its antioxidant substances. The irreversible reaction glycolysis (one of CCM pathways) is glucokinase, which was downregulated (p = 0.00384). One of the key enzymes in the pentose phosphate pathway (PPP) of CCM, the downregulated transketolase (p = 0.045), decreased the process of glucose turnover that produces NADPH as reducing equivalents and pentoses as essential parts of nucleotides. Other key enzymes, such as glucose 6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase and NOX2 (57), were not differentially expressed proteins. Triosephosphate isomerase and phosphogluconate dehydratase were the only two proteins that were upregulated in the Entner-Doudoroff pathway to speed up the metabolism of glucose (58). There were no differential proteins in the semi or non-phosphorylative Entner-Doudoroff pathway. Another important part of CCM was the TCA cycle, which was also an enrichment pathway parallel to carbon metabolism.
The TCA cycle (adjusted p = 1.47 × 10−3) included 17 differentially expressed proteins (total downregulation, 13 differentially expressed proteins with p = 0), and the proportion of total proteins in the pathway was 68% (Figure 4). The TCA cycle, which is also one part of CCM, is a core process in aerobic respiration for energy production and the production of carbon-based precursor molecules for many biosynthetic pathways in aerobic organisms. It is also of vital importance for the survival of lifeforms (59). There are three key regulators of the catalytic reaction in the TAC cycle, citrate synthase, isocitrate dehydrogenase and α-ketoglutarate dehydrogenase (α-KGDH), because their reactions are irreversible (60). Citrate synthase, which catalyzes the synthesis of citrate from acetyl-CoA and oxaloacetate, is the first step of the TCA cycle (54). In this experiment, citrate synthase was downregulated in ALH1-treated S. Typhimurium (p = 0.0398), which decreased the speed of citrate synthesis. Another key regulator in the TAC cycle, isocitrate dehydrogenase, was also downregulated (p = 0.0113). Therefore, the amount of alpha-ketoglutarate converted to succinyl-CoA was decreased. These two downregulated key regulators decreased respiration for energy production and production of carbon-based precursor molecules in the TCA cycle, which indicates that ALH1 can effectively inhibit the oxidative respiratory metabolism of S. Typhimurium through the TCA pathway inhibiting bacterial survival. This result agreed with that of a previous report, such as that oregano essential oil against methicillin-resistant S. aureus decreased citrate synthase, isocitrate dehydrogenase and α-KGDH (61), Ag+ against E. coli decreased isocitrate dehydrogenase (62), kendomycin against methicillin-resistant S. aureus decreased α-ketoglutarate dehydrogenase (63), and clove essential oil against S. aureus decreased citrate synthase, isocitrate dehydrogenase and α-KGDH (64). Other carbohydrate metabolism pathways, including the glyoxylate cycle, ethylmalonyl pathway, methylaspartate cycle, photorespiration, malonate semialdehyde pathway, and propanoyl-CoA metabolism, were enriched in multiple downregulated and not upregulated proteins.
Additionally, carbon fixation pathways have multiple submodules. One of them was the phosphate acetyltransferase-acetate kinase pathway. Acetyl-CoA was catalyzed by the upregulated acetate kinase (p = 0.00119) and phosphate acetyltransferase (p = 7.23 × 10−5) to provide more ATP (65), and their reversibility enhances the use of acetate as the carbon source to be crucial for growth and substrate assimilation (66). These two enzymes were also involved in the methanogenesis pathway in methane metabolism. These results indicated that energy for bacteria treated with ALH1 was supplied via the phosphate acetyltransferase-acetate kinase pathway when the TCA cycle was inhibited. The upregulated methylenetetrahydrofolate reductase in the reductive acetyl-CoA pathway (wood-Ljungdahl pathway), which is the rate-limiting enzyme in the methyl cycle, increasingly committed tetrahydrofolate-bound one-carbon units to use in the methylation of homocysteine to form methionine (67). In methane metabolism, another upregulated protein was ATP-dependent 6-phosphofructokinase in formaldehyde assimilation, the ribulose monophosphate pathway, which catalyzes the phosphorylation of D-fructose 6-phosphate to fructose 1, 6-bisphosphate by ATP as the first committing step, the key regulatory step of glycolysis (68). Other differentially expressed proteins in carbon fixation pathways and methane metabolism were downregulated. There were 4 differentially expressed proteins in amino acid metabolism, which were downregulated to inhibit transporters for uptake, biosynthesis and degradation and extraction of amino acids.
The third significantly different pathway (adjusted p = 0.0221) was microbial metabolism in diverse environments, which had 20 up- and 59 downregulated proteins and 38.73% of total proteins in the pathway. Metabolism diversity reflects different metabolic strategies of bacteria to adapt to different environments and obtain the energy and carbon needed for the production of cellular constituents necessary for growth, survival, and reproduction. All of the differential proteins in this pathway were also enriched in other pathways because many of the compounds played an important role in both catabolic and anabolic reactions in cells (69). Therefore, differentially expressed proteins interact with each other.
The top 4 interacting proteins were aldehyde-alcohol, putative pyruvate-flavodoxin oxidoreductase, N-acetyl glucosamine specific PTS system components IIABC and malic enzyme (Figure 5). Aldehyde-alcohol dehydrogenase is a NAD(H)-dependent bifunctional and highly conserved enzyme in bacteria. This is essential for the fermentation of glucose to sustain the glycolytic pathway of acetyl-CoA to ethanol (70–72), utilization of ethanol as a substrate to generate NADH and other carbon intermediates (71, 72), and a functional enzyme such as pyruvate formate-lyase to catalyze the conversion of pyruvate and coenzyme A to formate and acetyl-CoA (70). Due to its multifunctional activity, aldehyde-alcohol dehydrogenase has the most interacting proteins. Putative pyruvate-flavodoxin oxidoreductase was enriched in glycolysis/gluconeogenesis, the citrate cycle (TCA cycle), pyruvate metabolism, butanoate metabolism, metabolic pathways, the biosynthesis of secondary metabolites, microbial metabolism in diverse environments and carbon metabolism pathways to produce acetyl-CoA from pyruvate (73, 74). N-acetyl glucosamine-specific PTS system component IIABC was enriched in amino sugar and nucleotide sugar metabolism and the phosphotransferase system (PTS) (75). Malic enzyme, a major contributor to cellular reducing power and carbon flux, was engaged in NADP - malic enzyme type C4-dicarboxylic acid cycle, pyruvate metabolism, metabolic pathways and microbial metabolism in diverse environments to fit the new component in culture medium (76). The number of interacting proteins were determined by the complexity of the metabolic system and the pathway it engaged.
The different relative expressions of proteins were not the same as the relative expression trend of genes selected to undergo RT-PCR (Figure 6). The relative expression of genes is influenced by posttranscriptional regulation and stability of mRNA (77).
There are some defects in this research on the possible mechanism of ALH1 antibacterial activity against S. Typhimurium but other ALH samples were not employed for this purpose because of the high cost of the proteomic experiment. It is difficult to refer to the key or an absolute advantage pathway to explain the possible mechanism because there were too many differential proteins obtained and too much diversity in the metabolism of bacteria. In particular, carbon metabolism has 47 submodules, and proteins participate in more than 1 or 2 pathways. Therefore, the C13-carboxyl-labeled method can be used to confirm the mechanism precisely. Metabolomics, single components separated from phenols or flavonoids as treatments, single-cell transcriptomics, deletion genes of key proteins, or other methods can be employed to more accurately explore the regulation of ALH against S. Typhimurium.
The different antioxidant activities of ALH samples harvested at different times and from different regions were affected by their physicochemical parameters, especially TPC and TFC, which have a high correlation between TPC or TFC and antioxidant activity except for the O2·- assay. The MIC and MBC of ALH against S. Typhimurium were 20–30% and 25–40%, respectively. Proteomic analysis revealed the possible antibacterial mechanism of ALH1 at IC50 (2.97%, w/v), whose antioxidant activity can reduce the bacterial reduction reaction and energy supply, inhibit the TCA cycle and amino acid metabolism and enhance glycolysis. Overall, the results provide a theoretical basis for the treatment of S. Typhimurium using ALH.
The original contributions presented in the study are publicly available. This data can be found here: https://www.iprox.cn//page/project.html?id=IPX0006322000.
HK and WY: conceptualization. WT, YT, and QZ: methodology. WT and YT: formal analysis. HK and SM: investigation and resources. WT and WW: writing – original draft preparation. WY: writing – review and editing, project administration, and funding acquisition. WY and XM: supervision. All authors have read and agreed to the published version of the manuscript.
This research was funded by Fujian Provincial Natural Science Funding for WY, grant number 2020 J01540.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2023.1181492/full#supplementary-material
1. Ali, A, Paramanya, A, Poojari, P, Arslan-Acaroz, D, Acaroz, U, and Kostić, AŽ. The utilization of bee products as a holistic approach to managing polycystic ovarian syndrome-related infertility. Nutrients. (2023) 15:1165. doi: 10.3390/nu15051165
2. Asma, ST, Bobiş, O, Bonta, V, Acaroz, U, Shah, SRA, Istanbullugil, FR, et al. General nutritional profile of bee products and their potential antiviral properties against mammalian viruses. Nutrients. (2022) 14:3579. doi: 10.3390/nu14173579
3. Vandamme, L, Heyneman, A, Hoeksema, H, Verbelen, J, and Monstrey, S. Honey in modern wound care: a systematic review. Burns. (2013) 39:1514–25. doi: 10.1016/j.burns.2013.06.014
4. Ahmed, S, Sulaiman, SA, Baig, AA, Ibrahim, M, Liaqat, S, Fatima, S, et al. Honey as a potential natural antioxidant medicine: an insight into its molecular mechanisms of action. Oxidative Med Cell Longev. (2018) 2018:1–19. doi: 10.1155/2018/8367846
5. Majtan, J, Bucekova, M, Kafantaris, I, Szweda, P, Hammer, K, and Mossialos, D. Honey antibacterial activity: a neglected aspect of honey quality assurance as functional food. Trends Food Sci Technol. (2021) 118:870–86. doi: 10.1016/j.tifs.2021.11.012
6. Waheed, M, Hussain, MB, Javed, A, Mushtaq, Z, Hassan, S, Shariati, MA, et al. Honey and cancer: a mechanistic review. Clin Nutr. (2019) 38:2499–503. doi: 10.1016/j.clnu.2018.12.019
7. Tomblin, V, Ferguson, LR, Han, DY, Murray, P, and Schlothauer, R. Potential pathway of anti-inflammatory effect by New Zealand honeys. Int J Gen Med. (2014) 7:149–58. doi: 10.2147/ijgm.s45839
8. Escuredo, O, Míguez, M, Fernández-González, M, and Seijo, MC. Nutritional value and antioxidant activity of honeys produced in a European Atlantic area. Food Chem. (2013) 138:851–6. doi: 10.1016/j.foodchem.2012.11.015
9. Jara-Palacios, MJ, Ávila, FJ, Escudero-Gilete, ML, Gómez Pajuelo, A, Heredia, FJ, Hernanz, D, et al. Physicochemical properties, colour, chemical composition, and antioxidant activity of Spanish Quercus honeydew honeys. Eur Food Res Technol. (2019) 245:2017–26. doi: 10.1007/s00217-019-03316-x
10. Wang, XH, Gheldof, N, and Engeseth, NJ. Effect of processing and storage on antioxidant capacity of honey. J Food Sci. (2004) 69:fct96–fct101. doi: 10.1111/j.1365-2621.2004.tb15509.x
11. Moniruzzaman, M, Khalil, MI, Sulaiman, SA, and Gan, SH. Advances in the analytical methods for determining the antioxidant properties of honey: a review. Afr J Tradit Complementary Altern Med. (2012) 9:36–42. doi: 10.4314/ajtcam.v9i1.5
12. Chua, LS, Rahaman, NLA, Adnan, NA, and Eddie Tan, TT. Antioxidant activity of three honey samples in relation with their biochemical components. J Anal Methods Chem. (2013) 2013:1–8. doi: 10.1155/2013/313798
13. Becerril-Sánchez, AL, Quintero-Salazar, B, Dublán-García, O, and Escalona-Buendía, HB. Phenolic compounds in honey and their relationship with antioxidant activity, botanical origin, and color. Antioxidants. (2021) 10:1700. doi: 10.3390/antiox10111700
14. Al-Mamary, M, Al-Meeri, A, and Al-Habori, M. Antioxidant activities and total phenolics of different types of honey. Nutr Res. (2002) 22:1041–7. doi: 10.1016/s0271-5317(02)00406-2
15. Bertoncelj, J, Doberšek, U, Jamnik, M, and Golob, T. Evaluation of the phenolic content, antioxidant activity and colour of Slovenian honey. Food Chem. (2007) 105:822–8. doi: 10.1016/j.foodchem.2007.01.060
16. Bajgain, A, Neupane, BD, Sarraf, D, Karmacharya, J, Ranjitkar, S, Shrestha, R, et al. Quality evaluation of Apis laboriosa and Apis mellifera honey collected from Bagmati Province. Nepal J Biotechnol. (2022) 10:32–9. doi: 10.54796/njb.v10i1.228
17. Asma, ST, Imre, K, Morar, A, Imre, M, Acaroz, U, Shah, SRA, et al. Natural strategies as potential weapons against bacterial biofilms. Life. (2022) 12:1618. doi: 10.3390/life12101618
18. Almasaudi, S. The antibacterial activities of honey. Saudi J. Biol. Sci. (2021) 28:2188–96. doi: 10.1016/j.sjbs.2020.10.017
19. Leyva-Jimenez, FJ, Lozano-Sanchez, J, Borras-Linares, I, de la Luz, C-GM, and Mahmoodi-Khaledi, E. Potential antimicrobial activity of honey phenolic compounds against gram positive and gram negative bacteria. LWT--Food Sci Technol. (2019) 101:236–45. doi: 10.1016/j.lwt.2018.11.015
20. Ciucure, CT, and Geană, EI. Phenolic compounds profile and biochemical properties of honeys in relationship to the honey floral sources. Phytochem Anal. (2019) 30:481–92. doi: 10.1002/pca.2831
21. Alvarez-Suarez, J, Gasparrini, M, Forbes-Hernández, T, Mazzoni, L, and Giampieri, F. The composition and biological activity of honey: a focus on manuka honey. Foods. (2014) 3:420–32. doi: 10.3390/foods3030420
22. Aljadi, AM, and Yusoff, KM. Isolation and identification of phenolic acids in Malaysian honey with antibacterial properties. Turk J Med Sci. (2003) 33:229–36.
23. Mundo, MA, Padilla-Zakou, OI, and Worobo, RW. Growth inhibition of foodborne pathogens and food spoilage organisms by select raw honeys. Int J Food Microbiol. (2004) 97:1–8. doi: 10.1016/j.ijfoodmicro.2004.03.025
24. Yang, W, Shen, M, Kuang, H, Liu, X, Zhang, C, Tian, Y, et al. The botanical sources, entomological proteome and antibiotic properties of wild honey. Innov Food Sci Emerg Technol. (2021) 67:102589. doi: 10.1016/j.ifset.2020.102589
25. Neupane, BP, Chaudhary, D, Paudel, S, Timsina, S, Chapagain, B, Jamarkattel, N, et al. Himalayan honey loaded iron oxide nanoparticles: synthesis, characterization and study of antioxidant and antimicrobial activities. Int J Nanomedicine. (2019) 14:3533–41. doi: 10.2147/IJN.S196671
26. Galán, JE. Salmonella Typhimurium and inflammation: a pathogen-centric affair. Nat Rev Microbiol. (2021) 19:716–25. doi: 10.1038/s41579-021-00561-4
27. CDC. National Enteric Disease Surveillance: Salmonella annual summary; US Department of Health and Human Services, vol. 2016. Atlanta, GA: CDC (2016).
28. Xiang, Y, Li, F, Dong, N, Tian, S, Zhang, H, Du, X, et al. Investigation of a salmonellosis outbreak caused by multidrug resistant Salmonella Typhimurium in China. Front Microbiol. (2020) 11:1–12. doi: 10.3389/fmicb.2020.00801
29. Gabrielyan, L, Badalyan, H, Gevorgyan, V, and Trchounian, A. Comparable antibacterial effects and action mechanisms of silver and iron oxide nanoparticles on Escherichia coli and Salmonella typhimurium. Sci Rep. (2020) 10:13145–12. doi: 10.1038/s41598-020-70211-x
30. Guo, L, Gong, S, Wang, Y, Sun, Q, Duo, K, and Fei, P. Antibacterial activity of olive oil polyphenol extract against Salmonella Typhimurium and Staphylococcus aureus: possible mechanisms. Foodborne Pathog Dis. (2020) 17:396–403. doi: 10.1089/fpd.2019.2713
31. Bai, J, Jeon, B, and Ryu, S. Effective inhibition of Salmonella Typhimurium in fresh produce by a phage cocktail targeting multiple host receptors. Food Microbiol. (2019) 77:52–60. doi: 10.1016/j.fm.2018.08.011
32. Debroy, R, and Ramaiah, S. MurC ligase of multi-drug resistant Salmonella Typhi can be inhibited by novel curcumin derivative: evidence from molecular docking and dynamics simulations. Int J Biochem Cell Biol. (2022) 151:106279. doi: 10.1016/j.biocel.2022.106279
33. Prakash, A, Vadivel, V, Rubini, D, and Nithyanand, P. Antibacterial and antibiofilm activities of linalool nanoemulsions against Salmonella Typhimurium. Food Biosci. (2019) 28:57–65. doi: 10.1016/j.fbio.2019.01.018
34. Akbar, A, Sadiq, MB, Ali, I, Muhammad, N, Rehman, Z, Khan, MN, et al. Synthesis and antimicrobial activity of zinc oxide nanoparticles against foodborne pathogens Salmonella typhimurium and Staphylococcus aureus. Biocatal Agric Biotechnol. (2018) 17:36–42. doi: 10.1016/j.bcab.2018.11.005
35. Wang, L, Ning, F, Liu, T, Huang, X, Zhang, J, Liu, Y, et al. Physicochemical properties, chemical composition, and antioxidant activity of Dendropanax dentiger honey. LWT Food Sci Technol. (2021) 147:111693. doi: 10.1016/j.lwt.2021.111693
36. Anand, S, Pang, E, Livanos, G, and Mantri, N. Characterization of physico-chemical properties and antioxidant capacities of bioactive honey produced from Australian grown Agastache rugosa and its correlation with colour and poly-phenol content. Molecules. (2018) 23:108. doi: 10.3390/molecules23010108
37. Bellik, Y, and Selles, SMA. In vitro synergistic antioxidant activity of honey-Mentha spicata combination. J Food Meas Charact. (2017) 11:111–8. doi: 10.1007/s11694-016-9377-1
38. El-Guendouz, S, Aazza, S, Lyoussi, B, Bankova, V, Popova, M, Neto, L, et al. Moroccan propolis: a natural antioxidant, antibacterial, and antibiofilm against Staphylococcus aureus with no induction of resistance after continuous exposure. J Evid Based Complement Altern Med. (2018) 2018:1–19. doi: 10.1155/2018/9759240
39. Girma, A, Seo, W, and She, RC. Antibacterial activity of varying UMF-graded Manuka honeys. PLoS One. (2019) 14:e0224495. doi: 10.1371/journal.pone.0224495
40. Chen, L, Gu, L, Geng, X, Xu, G, Huang, X, and Zhu, X. A novel cis antisense RNA AsfD promotes Salmonella enterica serovar Typhi motility and biofilm formation. Microb Pathog. (2020) 142:104044. doi: 10.1016/j.micpath.2020.104044
41. Xu, X, Pu, R, Li, Y, Wu, Z, Li, C, Miao, X, et al. Chemical compositions of proplis from China and the United States and their antimicrobial activities against Penicillium notatum. Molecules. (2019) 24:3576. doi: 10.3390/molecules24193576
42. Al Mamari, HH. Phenolic compounds: classification, chemistry, and updated techniques of analysis and synthesis In: Phenolic compounds-chemistry, synthesis, diversity, non-conventional industrial, pharmaceutical and therapeutic applications, vol. 26 (2021)
43. Liu, X, Tian, Y, Yang, A, Zhang, C, Miao, X, and Yang, W. Antitumor effects of poplar propolis on DLBCL SU-DHL-2 cells. Foods. (2023) 12:283. doi: 10.3390/foods12020283
44. Martin, EC. Some aspects of hygroscopic properties and fermentation of honey. Bee World. (1958) 39:165–78. doi: 10.1080/0005772X.1958.11095058
45. Zhang, G, Tian, J, Zhang, Y, Li, S, Zheng, H, and Hu, F. Investigation of the maturity evaluation indicator of honey in natural ripening process: the case of rape honey. Foods. (2021) 10:2882. doi: 10.3390/foods10112882
46. Sajid, M, Yamin, M, Asad, F, Yaqub, S, Ahmad, S, Mubarik, M, et al. Comparative study of physio-chemical analysis of fresh and branded honeys from Pakistan. Saudi J Biol Sci. (2019) 27:173–6. doi: 10.1016/j.sjbs.2019.06.014
47. Kahraman, T, Buyukunal, SK, Vural, A, and Altunatmaz, SS. Physico-chemical properties in honey from different regions of Turkey. Food Chem. (2010) 123:41–4. doi: 10.1016/j.foodchem.2010.03.123
48. Kishore, RK, Halim, AS, Syazana, MN, and Sirajudeen, KNS. Tualang honey has higher phenolic content and greater radical scavenging activity compared with other honey sources. Nutr Res. (2011) 31:322–5. doi: 10.1016/j.nutres.2011.03.001
49. Gośliński, M, Nowak, D, and Kłębukowska, L. Antioxidant properties and antimicrobial activity of manuka honey versus polish honeys. J Food Sci Technol. (2020) 57:1269–77. doi: 10.1007/s13197-019-04159-w
50. Taguri, T, Tanaka, T, and Kouno, I. Antibacterial spectrum of plant polyphenols and extracts depending upon hydroxyphenyl structure. Biol Pharm Bull. (2006) 29:2226–35. doi: 10.1248/bpb.29.2226
51. Borges, A, Saavedra, MJ, and Simões, M. The activity of ferulic and gallic acids in biofilm prevention and control of pathogenic bacteria. Biofouling. (2012) 28:755–67. doi: 10.1080/08927014.2012.706751
52. Kang, MS, Oh, JS, Kang, IC, Hong, SJ, and Choi, CH. Inhibitory effect of methyl gallate and gallic acid on oral bacteria. JMicrobiol. (2008) 46:744–50. doi: 10.1007/s12275-008-0235-7
53. Otmani, A, Amessis-Ouchemoukh, N, Birinci, C, Yahiaoui, S, Kolayli, S, Rodríguez-Flores, MS, et al. Phenolic compounds and antioxidant and antibacterial activities of Algerian honeys. Food Biosci. (2021) 42:101070. doi: 10.1016/j.fbio.2021.101070
54. Stagos, D, Soulitsiotis, N, Tsadila, C, Papaeconomou, S, Arvanitis, C, Ntontos, A, et al. Antibacterial and antioxidant activity of different types of honey derived from Mount Olympus in Greece. Int J Mol Med. (2018) 42:726–34. doi: 10.3892/ijmm.2018.3656
55. Wen, W, Wang, S, Zhou, X, and Fang, B. Central carbon metabolism in marine bacteria examined with a simplified assay for dehydrogenases. Appl Biochem Biotechnol. (2013) 170:473–82. doi: 10.1007/s12010-013-0200-8
56. Muñoz-Elías, EJ, and McKinney, JD. Carbon metabolism of intracellular bacteria. Cell Microbiol. (2006) 8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x
57. Zhu, X, Guo, Y, Liu, Z, Yang, J, Tang, H, and Wang, Y. Itaconic acid exerts anti-inflammatory and antibacterial effects via promoting pentose phosphate pathway to produce ROS. Sci Rep. (2021) 11:18173–9. doi: 10.1038/s41598-021-97352-x
58. Wilkes, RA, Mendonca, CM, and Aristilde, L. A cyclic metabolic network in Pseudomonas protegens Pf-5 prioritizes the Entner-Doudoroff pathway and exhibits substrate hierarchy during carbohydrate co-utilization. Appl Environ Microbiol. (2019) 85:e02084–18. doi: 10.1128/AEM.02084-18
59. Akram, M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem Biophys. (2014) 68:475–8. doi: 10.1007/s12013-013-9750-1
60. Wan, B, LaNoue, KF, Cheung, JY, and Scaduto, RC. Regulation of citric acid cycle by calcium. J Biol Chem. (1989) 264:13430–9. doi: 10.1016/S0021-9258(18)80015-1
61. Cui, H, Zhang, C, Li, C, and Li, L. Antibacterial mechanism of oregano essential oil. Ind Crop Prod. (2019) 139:111498. doi: 10.1016/j.indcrop.2019.111498
62. Wang, H, Yan, A, Liu, Z, Yang, X, Xu, Z, Wang, Y, et al. Deciphering molecular mechanism of silver by integrated omic approaches enables enhancing its antimicrobial efficacy in E. coli. PLoS Biol. (2019) 17:e3000292. doi: 10.1371/journal.pbio.3000292
63. Elnakady, Y, Chatterjee, I, Bischoff, M, Rohde, M, Josten, M, Sahl, HG, et al. Investigations to the antibacterial mechanism of action of kendomycin. PLoS One. (2016) 11:e0146165. doi: 10.1371/journal.pone.0146165
64. Li, J, Li, C, Shi, C, Aliakbarlu, J, Cui, H, and Lin, L. Antibacterial mechanisms of clove Essen-tial oil against Staphylococcus aureus and its application in pork. Int J Food Microbiol. (2022) 380:109864. doi: 10.1016/j.ijfoodmicro.2022.109864
65. Kofoid, E, Rappleye, C, Stojiljkovic, I, and Roth, J. The 17-gene ethanolamine (eut) operon of Salmonella Typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. (1999) 181:5317–29. doi: 10.1128/jb.181.17.5317-5329.1999
66. Castaño-Cerezo, S, Pastor, JM, Renilla, S, Bernal, V, Iborra, JL, and Cánovas, M. An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli. Microb Cell Factor. (2009) 8:1–19. doi: 10.1186/1475-2859-8-54
67. Sheppard, CA, Trimmer, EE, and Matthews, RG. Purification and properties of NADH-dependent 5, 10-methylenetetrahydrofolate reductase (MetF) from Escherichia coli. J Bacteriol. (1999) 181:718–25. doi: 10.1128/jb.181.3.718-725.1999
68. Siebers, B, Klenk, HP, and Hensel, R. PPi-dependent phosphofructokinase from Thermoproteus tenax, an archaeal descendant of an ancient line in phosphofructokinase evolution. J Bacteriol. (1998) 180:2137–43. doi: 10.1128/jb.180.8.2137-2143.1998
69. Fuhrmann, JJ. “Microbial metabolism,” in Principles and Applications of Soil Microbiology 3e. eds. T. J. Gentry, J. J. Fuhrmann, and D. A. Zuberer (Elsevier Inc.), (2021):57–87.
70. Kessler, D, Leibrecht, I, and Knappe, J. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. (1991) 281:59–63. doi: 10.1016/0014-5793(91)80358-A
71. Yang, X, Teng, K, Su, R, Li, L, Zhang, T, Fan, K, et al. AcrR and Rex control mannitol and sorbitol utilization through their cross-regulation of aldehyde-alcohol dehydrogenase (AdhE) in Lactobacillus plantarum. Appl Environ Microbiol. (2019) 85:e02035–18. doi: 10.1128/aem.02035-18
72. Kim, G, Azmi, L, Jang, S, Jung, T, Hebert, H, Roe, AJ, et al. Aldehyde-alcohol dehydrogenase forms a high-order spirosome architecture critical for its activity. Nat Commun. (2019) 10:4527–11. doi: 10.1038/s41467-019-12427-8
73. Bertsch, J, Siemund, AL, Kremp, F, and Muller, V. A novel route for ethanol oxidation in the acetogenic bacterium Acetobacterium woodii: the acetaldehyde/ethanol dehydrogenase pathway. Environ Microbiol. (2016) 18:2913–22. doi: 10.1111/1462-2920.13082
74. Encheva, V, Shah, HN, and Gharbia, SE. Proteomic analysis of the adaptive response of Salmonella enterica serovar Typhimurium to growth under anaerobic conditions. Microbiology. (2009) 155:2429–41. doi: 10.1099/mic.0.026138-0
75. Crigler, J, Bannerman-Akwei, L, Cole, AE, Eiteman, MA, and Altman, E. Glucose can be transported and utilized in Escherichia coli by an altered or overproduced N-acetylglucosamine phosphotransferase system (PTS). Microbiology. (2018) 164:163–72. doi: 10.1099/mic.0.000596
76. Diacovich, L, Lorenzi, L, Tomassetti, M, Méresse, S, and Gramajo, H. The infectious intracellular lifestyle of Salmonella enterica relies on the adaptation to nutritional conditions within the Salmonella-containing vacuole. Virulence. (2017) 8:975–92. doi: 10.1080/21505594.2016.1270493
Keywords: Apis laboriosa honey, antioxidant activity, antibacterial activity, Salmonella Typhimurium, proteomics
Citation: Tan W, Tian Y, Zhang Q, Miao S, Wu W, Miao X, Kuang H and Yang W (2023) Antioxidant and antibacterial activity of Apis laboriosa honey against Salmonella enterica serovar Typhimurium. Front. Nutr. 10:1181492. doi: 10.3389/fnut.2023.1181492
Received: 07 March 2023; Accepted: 18 April 2023;
Published: 11 May 2023.
Edited by:
Santos Garcia, Universidad Autónoma de Nuevo León, MexicoReviewed by:
Juraj Majtan, Institute of Molecular Biology (SAS), SlovakiaCopyright © 2023 Tan, Tian, Zhang, Miao, Wu, Miao, Kuang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenchao Yang, MDAwcTA2MTAwNUBmYWZ1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.