- 1Division of Applied Nutrition, Faculty of Human Ecology, Department of Community Nutrition, IPB University, Bogor, Indonesia
- 2Department of Biological Sciences, Faculty of Sciences and Technology, State Islamic University of Sunan Kalijaga (UIN Sunan Kalijaga Yogyakarta), Yogyakarta, Indonesia
- 3Alumnus of Department of Internal Medicine, Faculty of Medicine, University of Indonesia–Cipto Mangunkusumo Hospital, Jakarta, Indonesia
- 4Alumnus of Department of Nutrition Science, Faculty of Medicine, Diponegoro University, Semarang, Indonesia
- 5Medical Study Programme, Faculty of Medicine, Brawijaya University, Malang, Indonesia
- 6Medical Student of Faculty of Medicine, University of Jember–Soebandi Regional Hospital, Jember, Indonesia
- 7Medical Student of Faculty of Medicine, Universitas Airlangga, Surabaya, Indonesia
1. Introduction
Microbiome has become a topic that is developing rapidly in the health sector for the past 2 decades (1). Previous research has found that the microbiome was associated with a variety of diseases, such as metabolic, gastrointestinal, and immunity disorders (2). The microbiome is a collection of genomes or genetic materials from all microorganisms, symbiotics, and pathogens that live in vertebrates such as bacteria, viruses, archaea, fungi, and small protists (1, 3). About 100 trillion microbiomes called the gut microbiome to reside in the digestive tract (4). The gut microbiome plays a role in nutrient and drug metabolism, immune modulation, and maintenance of gut integrity (3).
In addition to the gut, the second highest number of microbiomes is located in the mouth and called oral microbiome or salivary bacteriome, considering that there is a link between the two both physically and chemically (5, 6). An oral cavity is an ideal place for the survival of the oral microbiome since it has an average temperature of 37°C and saliva with a stable pH of 6.5–7, causing the bacteria to hydrate transporting the nutrients to microorganisms (7). The oral cavity is the first part of the gastrointestinal tract and the place where it meets with food, exogenous microbes, and allergens before it gets into the gastrointestinal tract. The presence of direct exposure in the absence of fibrous epithelium makes the oral cavity susceptible to infection (8). Those conditions highlight the important role of the microbiome in maintaining ecological balance and preventing oral cavities (9). One of the mechanisms is resistance to the colonization of pathogens by defeating pathogenic species and lowering the chance of integration by exogenous pathogens. Some microbiomes are also able to fight acids produced by caries-causing bacteria by increasing the pH of saliva through the production of alkaline metabolic byproducts (8, 10). Disorder in this system may cause dysbiosis triggered by various factors such as diet, inflammatory response, systemic disorders, and alcohol which will induce oral diseases (5, 8).
Recently, it has been found that the human microbiome has an important role in the occurrence and development of diabetes mellitus (11). Several studies have shown that type 2 diabetes mellitus (T2DM) is associated with changes in the diversity and number of bacteria in supragingival plaques, and oral microbiota changes have also been found in various glycemic states (12, 13). The phylum Actinobacteria was found to have decreased in the T2DM group compared to the control group, and the increase was associated with a reduced risk of T2DM (11, 14). The phylum Actinobacteria is also associated with the prevalence of obesity, suggesting that the oral microbiome may have an important role in the etiology of diabetes (14). The number of microbiotas of the genus Prevotella was decreased in the T2DM group, and Prevotella spp. was reduced in high-glucose salivary conditions (11, 12). Bacteria of the genus Rothia experienced a decrease in T2DM and potential pre-diabetic conditions (11, 15). Bacteria in the phylum Firmicutes, one of which is the Streptococcus, was significantly increased in the T2DM and pre-diabetic groups compared to the non-diabetic group (11, 13, 16).
To the best of our knowledge, there has been no review study or opinion article related to the use of oral or salivary microbiome as a biodetector of T2DM disease. Therefore, the main purpose of this critical opinion is to summarize the findings regarding the oral microbiome as a biodetector of T2DM and explain its opportunities, implications, and strategies for future use.
2. Oral microbiome in general
The human mouth is inhabited by various microorganisms such as bacteria, viruses, fungi, and protozoa called oral microbiota. The diversity of the human oral microbiota is one of the effects of accelerated regeneration and non-keratinized epithelium types found in the oral cavity (17, 18) since these factors increase the process of molecular absorption, implying that the microbiota is more likely to reach other organs and has a broad metabolic effect. In addition, the diversity of the oral microbiota can also be caused by the high intensity of contact with the external world via air and food (18, 19). Assuming the health-related consequences of the composition of the oral microbiota, the oral cavity would be an ideal place to analyze biomarkers since the samples will be easier to obtain than other organs (20). To date, 445 types of oral microbiota have been recognized in literature, which is ranked second after the intestine and 57% of them have been named and perfectly cultured (21, 22).
Due to its diverse and easily detectable natures, oral microbiota can be used as a biomarker of some diseases in humans (18). In diseases related to children’s mouths, some fungal species such as Candida dubliniensis and Candida tropicalis in saliva are indicators that children are at risk of dental caries (23). Changes in the oral microbiota to acidogenic and acidic cariogenic bacteria can indicate tooth decay and enamel demineralization in children (24). In adults, the oral microbiota can be an indicator of the incidence of oral cancer since the bacterial composition changes due to a cluster of factors that cause oral cancer, including alcohol and cigarettes. The byproducts of oral bacteria can induce genetic changes in mucosal epithelial cells that are predictors of Squamous cell carcinoma in the mouth (25). Further findings found that the use of oral microbiota was able to detect the presence of oropharyngeal cancer and malignancy in the human gastrointestinal tract (7, 26).
In addition, oral microbiota can also be an important biomarker of systemic diseases in other organs (27). Oral microbiota has also been detected in the lungs in cystic fibrosis patients and can cause various soft tissue infections in the event of wound bites (28, 29). Selenomonas (S. artemidis and S. infelix) can be used as important biomarkers of lung infections associated with acute respiratory distress syndrome (ARDS) (30). The oral microbiota has also long been known as a reservoir of infections in various parts of the body. The appearance of abscesses in the brain can also be detected with the discovery of an increase in Porphyromonas gingivalis which is an etiological agent in periodontitis (31). The study with 228 subjects diagnosed with metabolic syndrome disease in Korea also showed significant differences where the group with metabolic syndrome had slightly more Firmicutes (37.9%) and slightly fewer Proteobacteria (29.2%) than the healthy group (21). In other studies, several dominant oral microbiotas were found such as Fusobacterium nucleatum, Veillonella, Streptococcus anginosus, Streptococcus oralis, and Actinomyces meyeri in pyogenic infectious diseases of the brain and spinal cord (30, 32). Looking at these findings drew attention to the question “Can the oral profile of the microbiota be used as a diagnostic biomarker or biodetector of T2DM?” Following the purpose of this opinion article, the author tries to summarize the findings regarding the oral microbiome as a biodetector of T2DM.
3. Is the oral microbiome effective in detecting T2DM?
3.1. In vivo or preclinical trial studies
The decline of the body’s immune system in people with diabetes mellitus occurs due to polymorphonuclear cell dysfunction (PMN) and various other inflammatory cytokines, which causes a shift in the normal flora of the oral mucosa and triggers the growth and development of various pathogenic germs (33). Xiao et al. conducted in vivo experiments on rats treated to be hyperglycemic. This study found elevated levels of Enterobacteriaceae, Aerococcus, Enterococcus, and Staphylococcus which are often associated with T2DM (34). The human and mouse microbiomes have identical compositions at the phylum level, including Firmicutes and Bacteroidetes, followed by Actinobacteria and Proteobacteria In addition, at the genus level, Lactobacillus is the dominant genus at 8 weeks of age and remains dominant during the development of T2DM (35, 36).
3.2. Is there sufficient clinical evidence or clinical trials?
Table 1 showed that some of these studies showed contrasting findings. Almeida-Santos et al. and Lee et al. found no difference in oral microbiome profiles between diabetic and non-diabetic groups (37, 38). However, it can be concluded that several oral microbiomes are quite often found in T2DM, including the Firmicutes and Bacteroidetes phylum, followed by Actinobacteria and Proteobacteria. Based on in vivo and clinical studies, the mechanism of identification and the correlation between the oral microbiome and the occurrence of T2DM still need to be further researched, especially considering the possible differences between ethnicities or populations (43). It is realized, the insignificant of studies in Table 1 were likely underpowered because perhaps the number of samples was relatively small (<50 patients). Future clinical trial studies or RCTs should be conducted with more power based on sample size, may more than fifty patients or subject.
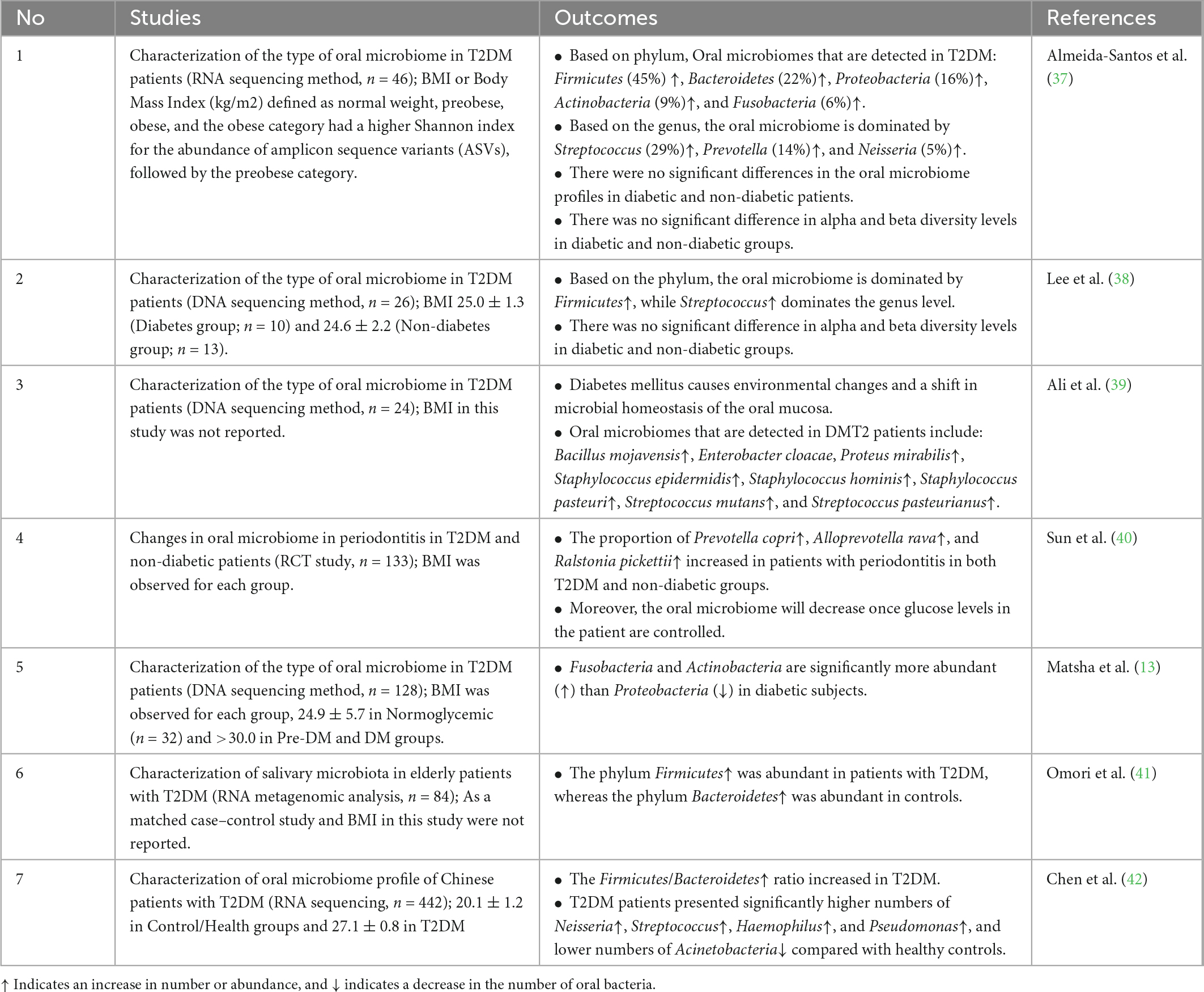
Table 1. List of clinical evidence regarding the effectivity of oral microbiome in detecting type 2 diabetes mellitus (T2DM).
4. Discussion with future implications and strategies
Previous studies has highlighted the interesting bidirectional relationship between the salivary microbiome and T2DM (44). The abundance of Firmicutes and increased ratio of Firmicutes/Bacteroidetes were found in most studies. It has been suggested that the Firmicutes were more efficient than the Bacteroidetes at obtaining energy from food, resulting in more effective absorption of calories and the ensuing weight gain (45). On the other hand, dietary intake and obesity were correlated with the incidence of T2DM (46). A lower level of Bacteroidetes has also been linked with the incidence of inflammatory disease (47), while inflammation also initiates the pathophysiology of T2DM (48). However, even this fundamental theory was found to be contradicted as Firmicutes was argued to be health-promoting bacteria through the synthesis of butyrate (49). Lee et al., Chen et al. and Ali et al. also mentioned that Streptococcus was one of the abundant bacteria in the oral microbiome of T2DM subjects (Figure 1) (38, 39, 42). Lower Prevotella and higher Streptococcus may be explained by low salivary pH that is caused by prolonged elevated blood glucose (12). However, a clinical research report stated that salivary microbiome profiles were prone to change, as they might be influenced by different stages of oral diseases (13). This was confirmed by the study of Sun et al. (40) as the changes of bacteria between diabetic and non-diabetic groups were similar in the presence of periodontitis.
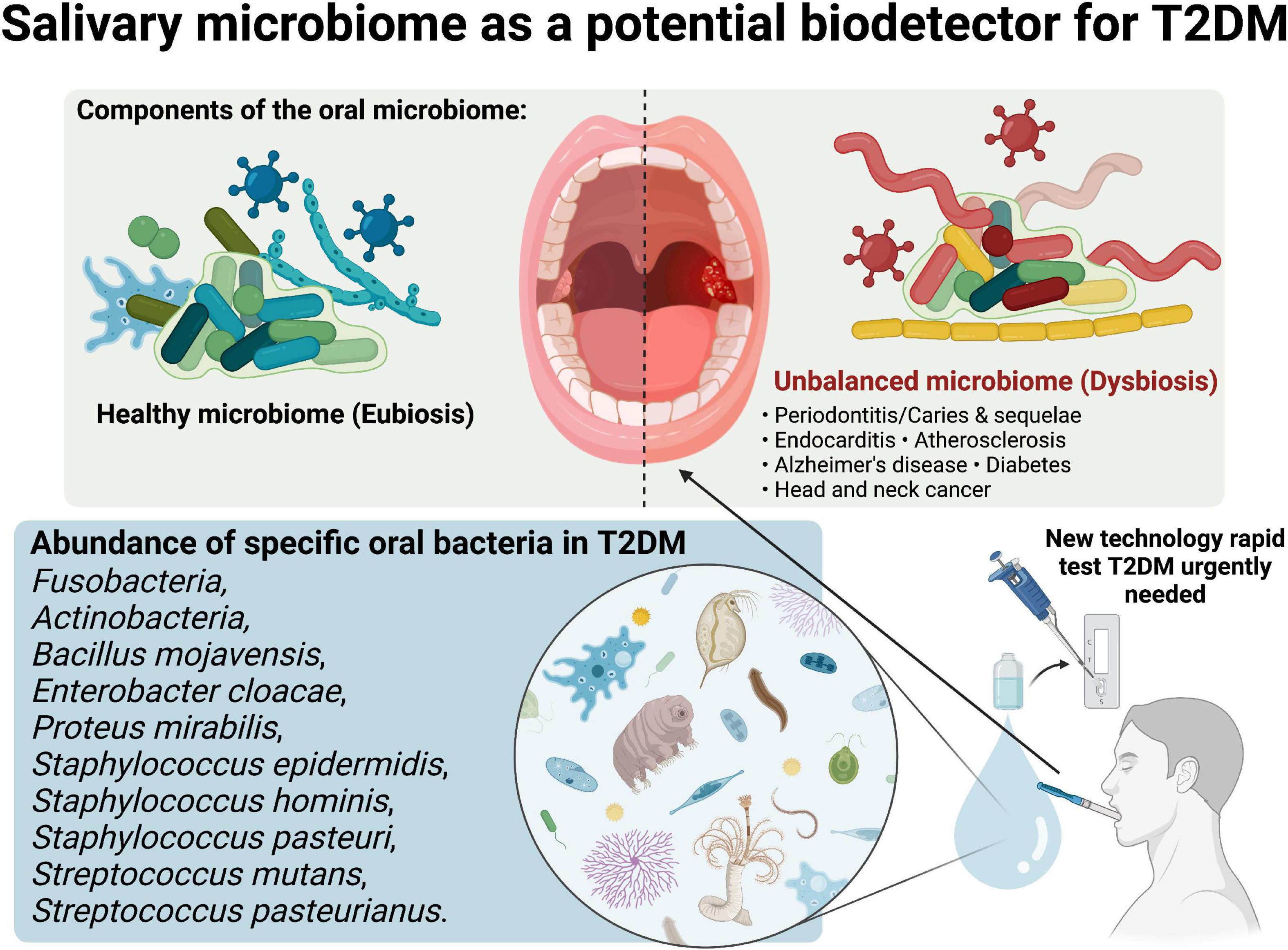
Figure 1. Salivary microbiome as a potential biodetector for type 2 diabetes mellitus (T2DM). Created with BioRender.com premium license by Fahrul Nurkolis.
Even though oral microbial profiles exhibited wide metabolic implications that were similar but at a lower level compared to gut microbiome (50), they may contribute an important role as biodetector of diseases, especially T2DM. Learning from the studies reviewed in this opinion, it can be concluded that randomized controlled trials (RCTs) are needed to consolidate the evidence regarding the link between oral microbiome and T2DM. These studies presented us with insights regarding related bacteria that can be targeted during upcoming RCTs, as these bacteria have been studied for their correlation with impaired metabolical conditions such as T2DM. Profiling the salivary microbiome may become a feasible rapid test in detecting T2DM (Figure 1), considering the fact that bio samples of the oral cavity will be easier to procure compared to gut (fecal) microbiota for future use, compared to detection using blood which some people are afraid of needles.
Author contributions
HH and FN: conception and design of opinion studies. FN: figure visualization and BioRender license holder. All authors wrote, edited, and revised the manuscript, and approved the final version of the submitted manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Berg G, Rybakova D, Fischer D, Cernava T, Vergès M, Charles T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. (2020) 8:103. doi: 10.1186/s40168-020-00875-0
2. Rajagopala SV, Vashee S, Oldfield L, Suzuki Y, Venter J, Telenti A, et al. The human microbiome and cancer. Cancer Prev Res (Phila). (2017) 10:226–34. doi: 10.1158/1940-6207.CAPR-16-0249
3. Coker O. Non-bacteria microbiome (virus, fungi, and archaea) in gastrointestinal cancer. J Gastroenterol Hepatol. (2022) 37:256–62. doi: 10.1111/jgh.15738
4. Valdes A, Walter J, Segal E, Spector T. Role of the gut microbiota in nutrition and health. BMJ. (2018) 361:k2179. doi: 10.1136/bmj.k2179
5. Deo P, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol JOMFP. (2019) 23:122–8. doi: 10.4103/jomfp.JOMFP_152_19
6. Park S, Hwang B, Lim M, Ok S, Lee S, Chun K, et al. Oral-gut microbiome axis in gastrointestinal disease and cancer. Cancers. (2021) 13:2124. doi: 10.3390/cancers13092124
7. Lim Y, Totsika M, Morrison M, Punyadeera C. Oral microbiome: a new biomarker reservoir for oral and oropharyngeal cancers. Theranostics. (2017) 7:4313–21. doi: 10.7150/thno.21804
8. Sedghi L, DiMassa V, Harrington A, Lynch SV, Kapila Y. The oral microbiome: role of key organisms and complex networks in oral health and disease. Periodontol 2000. (2021) 87:107–31. doi: 10.1111/prd.12393
9. Jia G, Zhi A, Lai P, Wang G, Xia Y, Xiong Z, et al. The oral microbiota - a mechanistic role for systemic diseases. Br Dent J. (2018) 224:447–55. doi: 10.1038/sj.bdj.2018.217
10. Senneby A, Davies J, Svensäter G, Neilands J. Acid tolerance properties of dental biofilms in vivo. BMC Microbiol. (2017) 17:165. doi: 10.1186/s12866-017-1074-7
11. Liu YK, Chen V, He JZ, Zheng X, Xu X, Zhou XD. A salivary microbiome-based auxiliary diagnostic model for type 2 diabetes mellitus. Arch Oral Biol. (2021) 126:105118.
12. Goodson J, Hartman M, Shi P, Hasturk H, Yaskell T, Vargas J, et al. The salivary microbiome is altered in the presence of a high salivary glucose concentration. PLoS One. (2017) 12:e0170437. doi: 10.1371/journal.pone.0170437
13. Matsha T, Prince Y, Davids S, Chikte U, Erasmus R, Kengne A, et al. Oral microbiome signatures in diabetes mellitus and periodontal disease. J Dent Res. (2020) 99:658–65. doi: 10.1177/0022034520913818
14. Long J, Cai Q, Steinwandel M, Hargreaves M, Bordenstein S, Blot W, et al. Association of oral microbiome with type 2 diabetes risk. J Periodontal Res. (2017) 52:636–43. doi: 10.1111/jre.12432
15. Rungrueang K, Yuma S, Tantipoj C, Khovidhunkit S, Fuangtharnthip P, Thuramonwong T, et al. Oral bacterial microbiomes in association with potential prediabetes using different criteria of diagnosis. Int J Environ Res Public Health. (2021) 18:7436. doi: 10.3390/ijerph18147436
16. Anbalagan R, Srikanth P, Mani M, Barani R, Seshadri K, Janarthanan R. Next generation sequencing of oral microbiota in type 2 diabetes mellitus prior to and after neem stick usage and correlation with serum monocyte chemoattractant-1. Diabetes Res Clin Pract. (2017) 130:204–10. doi: 10.1016/j.diabres.2017.06.009
17. Mark Welch J, Ramírez-Puebla S, Borisy G. Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe. (2020) 28:160–8. doi: 10.1016/j.chom.2020.07.009
18. Sharma N, Bhatia S, Sodhi A, Batra N. Oral microbiome and health. AIMS Microbiol. (2018) 4:42–66. doi: 10.3934/microbiol.2018.1.42
19. Dong J, Li W, Wang Q, Chen J, Zu Y, Zhou X, et al. Relationships between oral microecosystem and respiratory diseases. Front Mol Biosci. (2021) 8:718222.
20. Si J, Lee C, Ko G. Oral microbiota: microbial biomarkers of metabolic syndrome independent of host genetic factors. Front Cell Infect Microbiol. (2017) 7:516. doi: 10.3389/fcimb.2017.00516
21. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. (2012) 486:207–14. doi: 10.1038/nature11234
22. Li Y, Cui J, Liu Y, Chen K, Huang L, Liu Y. Oral, Tongue-coating microbiota, and metabolic disorders: a novel area of interactive research. Front Cardiovasc Med. (2021) 8:730203. doi: 10.3389/fcvm.2021.730203
23. de Jesus V, Khan M, Mittermuller B, Duan K, Hu P, Schroth R, et al. Characterization of supragingival plaque and oral swab microbiomes in children with severe early childhood caries. Front Microbiol. (2021) 12:683685. doi: 10.3389/fmicb.2021.683685
24. McLean J. Advancements toward a systems level understanding of the human oral microbiome. Front Cell Infect Microbiol. (2014) 4:98. doi: 10.3389/fcimb.2014.00098
25. Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. (2019) 18:1533033819867354. doi: 10.1177/1533033819867354
26. Chen Y, Chen X, Yu H, Zhou H, Xu S. Oral microbiota as promising diagnostic biomarkers for gastrointestinal cancer: a systematic review. Onco Targets Ther. (2019) 12:11131–44. doi: 10.2147/OTT.S230262
27. Peng X, Cheng L, You Y, Tang C, Ren B, Li Y, et al. Oral microbiota in human systematic diseases. Int J Oral Sci. (2022) 14:1–11. doi: 10.1038/s41368-022-00163-7
28. Peven J, Jakicic J, Rogers R, Lesnovskaya A, Erickson K, Kang C, et al. The effects of a 12-month weight loss intervention on cognitive outcomes in adults with overweight and obesity. Nutrients. (2020) 12:1–15. doi: 10.3390/nu12102988
29. Simpson K, Thomas J. Oral microbiome: contributions to local and systemic infections. Curr Oral Heal Rep. (2016) 3:45–55. doi: 10.1007/s40496-016-0079-x
30. Parahitiyawa N, Jin L, Leung W, Yam W, Samaranayake L. Microbiology of odontogenic bacteremia: beyond endocarditis. Clin Microbiol Rev. (2009) 22:46–64, Table of Contents. doi: 10.1128/CMR.00028-08
31. Iida Y, Honda K, Suzuki T, Matsukawa S, Kawai T, Shimahara T, et al. Brain abscess in which Porphyromonas gingivalis was detected in cerebrospinal fluid. Br J Oral Maxillofac Surg. (2004) 42:180. doi: 10.1016/S0266-4356(03)00190-6
32. Ewald C, Kuhn S, Kalff R. Pyogenic infections of the central nervous system secondary to dental affections–a report of six cases. Neurosurg Rev. (2006) 29:163–7. doi: 10.1007/s10143-005-0009-1
33. Wang F, Wang W, Yin P, Liu Y, Liu J, Wang L, et al. Mortality and years of life lost in diabetes mellitus and its subcategories in china and its provinces, 2005-2020. J Diabetes Res. (2022) 2022:1609267. doi: 10.1155/2022/1609267
34. Xiao E, Mattos M, Vieira G, Chen S, Corrêa J, Wu Y, et al. Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe. (2017) 22:120–8.e4. doi: 10.1016/j.chom.2017.06.014
35. Zhou W, Xu H, Zhan L, Lu X, Zhang L. Dynamic development of fecal microbiome during the progression of diabetes mellitus in zucker diabetic fatty rats. Front Microbiol. (2019) 10:232. doi: 10.3389/fmicb.2019.00232
36. Naseer M, Bibi F, Alqahtani M, Chaudhary A, Azhar E, Kamal M, et al. Role of gut microbiota in obesity, type 2 diabetes and Alzheimer’s disease. CNS Neurol Disord Drug Targets. (2014) 13:305–11. doi: 10.2174/18715273113126660147
37. Almeida-Santos A, Martins-Mendes D, Gayà-Vidal M, Pérez-Pardal L, Beja-Pereira A. Characterization of the oral microbiome of medicated type-2 diabetes patients. Front Microbiol. (2021) 12:610370. doi: 10.3389/fmicb.2021.610370
38. Lee J, Lee Y, Choi S, Bae E, Lee D. Correlation analysis of the oral mucosal microbiome and diabetes mellitus using microbial DNA in elderly male subjects. Oral Biol Res. (2022) 46:10–20. doi: 10.21851/obr.46.01.202203.10
39. Ali T, Rumnaz A, Urmi U, Nahar S, Rana M, Sultana F, et al. Type-2 diabetes mellitus individuals carry different periodontal bacteria. Pesqui Bras Odontopediatria Clin Integr. (2021) 21:1–10. doi: 10.1590/pboci.2021.049
40. Sun X, Li M, Xia L, Fang Z, Yu S, Gao J, et al. Alteration of salivary microbiome in periodontitis with or without type-2 diabetes mellitus and metformin treatment. Sci Rep. (2020) 10:15363. doi: 10.1038/s41598-020-72035-1
41. Omori M, Kato-Kogoe N, Sakaguchi S, Kamiya K, Fukui N, Gu Y, et al. Characterization of salivary microbiota in elderly patients with type 2 diabetes mellitus: a matched case-control study. Clin Oral Investig. (2022) 26:493–504. doi: 10.1007/s00784-021-04027-y
42. Chen B, Wang Z, Wang J, Su X, Yang J, Zhang Q, et al. The oral microbiome profile and biomarker in Chinese type 2 diabetes mellitus patients. Endocrine. (2020) 68:564–72. doi: 10.1007/s12020-020-02269-6
43. Fouhy F, Clooney A, Stanton C, Claesson M, Cotter P. 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. (2016) 16:123. doi: 10.1186/s12866-016-0738-z
44. Negrini T, Carlos I, Duque C, Caiaffa K, Arthur R. Interplay among the oral microbiome, oral cavity conditions, the host immune response, diabetes mellitus, and its associated-risk factors-an overview. Front Oral Health. (2021) 2:697428. doi: 10.3389/froh.2021.697428
45. Krajmalnik-Brown R, Ilhan Z, Kang D, DiBaise J. Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract. (2012) 27:201–14. doi: 10.1177/0884533611436116
46. Donin A, Nightingale C, Owen C, Rudnicka A, Jebb S, Ambrosini G, et al. Dietary energy intake is associated with type 2 diabetes risk markers in children. Diabetes Care. (2014) 37:116–23. doi: 10.2337/dc13-1263
47. Zhou Y, Zhi F. Lower level of Bacteroides in the gut microbiota is associated with inflammatory bowel disease: a meta-analysis. Biomed Res Int. (2016) 2016:5828959. doi: 10.1155/2016/5828959
48. Tsalamandris S, Antonopoulos A, Oikonomou E, Papamikroulis G, Vogiatzi G, Papaioannou S, et al. The role of inflammation in diabetes: current concepts and future perspectives. Eur Cardiol. (2019) 14:50–9. doi: 10.15420/ecr.2018.33.1
49. Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P, et al. The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients. (2020) 12:1474. doi: 10.3390/nu12051474
Keywords: oral microbiome, biodetector, diabetes, T2DM, salivary bacterial
Citation: Hardinsyah H, Nurkolis F, Kurniawan R, Gunawan WB, Augusta PS, Setyawardani A, Agustianto RF, Al Mahira MFN, Praditya GN, Lailossa DG, Yudisthira D, Farradisya S and Barazani H (2023) Can salivary microbiome become a biodetector for type-2 diabetes? Opinion for future implications and strategies. Front. Nutr. 10:1113591. doi: 10.3389/fnut.2023.1113591
Received: 01 December 2022; Accepted: 05 January 2023;
Published: 19 January 2023.
Edited by:
Hani El-Nezami, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Tien Sy Dong, UCLA Health System, United StatesCopyright © 2023 Hardinsyah, Nurkolis, Kurniawan, Gunawan, Augusta, Setyawardani, Agustianto, Al Mahira, Praditya, Lailossa, Yudisthira, Farradisya and Barazani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hardinsyah Hardinsyah,  aGFyZGluc3lhaDIwMTBAZ21haWwuY29t
aGFyZGluc3lhaDIwMTBAZ21haWwuY29t
†These authors have contributed equally to this work and share senior authorship
‡ORCID: Hardinsyah Hardinsyah, orcid.org/0000-0002-0748-4373; Fahrul Nurkolis, orcid.org/0000-0003-2151-0854; Rudy Kurniawan, orcid.org/0000-0002-4811-1696; William Ben Gunawan, orcid.org/0000-0003-0633-4477; Piko Satria Augusta, orcid.org/0000-0002-0620-486X; Astuti Setyawardani, orcid.org/0000-0003-4783-9834; Rafiv Fasya Agustianto, orcid.org/0000-0003-4262-7656; Msy Firyal Nadya Al Mahira, orcid.org/0000-0002-7272-8135; Ghevira Naila Praditya, orcid.org/0000-0002-5464-2645; Deogifta Graciani Lailossa, orcid.org/0000-0003-2335-2466; Dewangga Yudisthira, orcid.org/0000-0002-3317-1602; Salsabila Farradisya, orcid.org/0000-0002-6153-1521; Hero Barazani, orcid.org/0000-0003-2171-2800
 Hardinsyah Hardinsyah1*†‡
Hardinsyah Hardinsyah1*†‡ Fahrul Nurkolis
Fahrul Nurkolis Rudy Kurniawan
Rudy Kurniawan William Ben Gunawan
William Ben Gunawan Astuti Setyawardani
Astuti Setyawardani Rafiv Fasya Agustianto
Rafiv Fasya Agustianto Msy Firyal Nadya Al Mahira
Msy Firyal Nadya Al Mahira Ghevira Naila Praditya
Ghevira Naila Praditya Deogifta Graciani Lailossa
Deogifta Graciani Lailossa Dewangga Yudisthira
Dewangga Yudisthira Salsabila Farradisya
Salsabila Farradisya Hero Barazani
Hero Barazani