- 1School of Public Health, Shenyang Medical College, Shenyang, China
- 2Central Hospital Affiliated to Shenyang Medical College, Shenyang, China
Cyanidin-3-O-glucoside (C3G) is the most widely distributed anthocyanin and it can reportedly reduce the risk of osteoporosis, but the molecular mechanism by which C3G promotes bone formation is poorly understood. In the current study, RNA sequencing (RNA-seq) was used to investigate the mechanism of action of C3G in osteogenesis. MC3T3-E1 mouse osteoblasts were divided into a C3G (100 μmol/L)-treated group and a vehicle-treated control group, and differentially expressed genes (DEGs) in groups were evaluated via RNA-seq analysis. The functions of the DEGs were evaluated by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, and the genes were validated by quantitative real-time PCR. The RNA-seq analysis identified 34 genes that were upregulated in C3G-treated cells compared to vehicle-treated cells, and 17 that were downregulated GO and KEGG pathway analyses indicated that these genes were highly enriched in functions related to lysosomes and glycolipid biosynthesis, among others. The differential expression of ATPase H+-transporting V0 subunit C (Atp6v0c), chemokine (C-X3-C motif) ligand 1 (Cx3cl1), and lymphocyte antigen 6 complex, locus A (Ly6a) genes was validated by quantitative real-time-PCR. Because these genes have been previously implicated in osteoporosis, they are potential target genes of C3G action in MC3T3-E1 cells. These results provide molecular level evidence for the therapeutic potential of C3G in the treatment of osteoporosis and other disorders of bone metabolism.
Introduction
Osteoporosis is a disorder of bone metabolism characterized by reduced bone mineral density and a high risk of bone fracture (1). It is a global public health problem (2) that will worsen with the increasing life expectancy of the human population. Osteoblasts and osteoclasts are responsible for bone remodeling, which maintains the integrity of the skeleton (3, 4). Primary cause of osteoporosis is dysfunctional osteoblasts and osteoclasts activity (5). Promoting the proliferation and differentiation of osteoblasts is an effective way to enhance bone mineral density and prevent osteoporosis (6).
Cyanidin-3-O-glucoside (C3G) is an anthocyanin that is widely distributed in nature (7, 8). C3G evidently has antioxidant and anti-inflammatory effects, therapeutic effects on disease, such as obesity, type 2 diabetes mellitus, and prostate cancer (9, 10), and promotes bone formation and reduces bone loss (11–16). In a study conducted in the United Kingdom, women with high dietary anthocyanin intake had higher bone mineral density (11, 12). In rodents, C3G supplementation can reportedly improve bone quality and reduce bone loss (13, 17, 18), and in other studies it has enhanced osteoblast differentiation and mineralization (19, 20). C3G may be a natural product supporting the prevention and treatment of osteoporosis (21).
To date most studies investigating the molecular etiology of osteoporosis have focused on components of signaling pathways related to bone development such as mitogen-activated protein kinase (MAPK) (22, 23), nuclear factor kappa B (NF-κB) (22), bone morphogenetic protein (BMP) (24), and Wnt (25) pathways. Recent studies have also revealed roles for the chemokine (C-X3-C motif) ligand 1 (CX3CL1)/chemokine (C-X3-C motif) receptor 1 (CX3CR1) signaling axis (26) and glycosylphosphatidylinositol-anchored proteins (27).
Anthocyanins from black rice, which is enriched in C3G (28) and blackberry (29) were shown to affect osteoblast proliferation and differentiation by modulating the expression of target genes including alkaline phosphatase (Alp), osteopontin (Opn), osterix (Osx), and bone gamma-carboxyglutamic acid-containing protein (Bglap) (20). C3G increased the mineralization capacity of osteoblasts via the extracellular signal regulated kinase 1/2 (ERK1/2) signaling pathway (19). Several molecular mechanisms of C3G have been investigated in bone cells, but its effects on gene regulation involved in bone formation remain largely unknown.
RNA sequencing (RNA-seq) technology combined with bioinformatics have enabled the large-scale identification of genes associated with normal biological processes and pathogenic processes (30). In current study, gene expression profiles of ME3T3-E1 osteoblast-like cells with and without C3G treatment were investigated by RNA-seq and a functional analyses of differentially expressed genes (DEGs) was conducted to identify those that potentially mediate the protective effects of C3G in osteoporosis.
Materials and methods
Chemicals and reagents
C3G with the purity over 98% was purchased from Meilunbio (Dalian, China). TRIzol reagent, primers of quantitative polymerase chain reaction (qRT-PCR), a reverse transcription kit and SYBR Green MasterMix were purchased from Takara Bio (Ostu, Japan). Pancreatin were purchased from Hyclone (Logan, UT, USA). Dimethylsulfoxide (DMSO), phosphate buffered saline (PBS), and other chemicals were purchased from Sigma-Aldrich (Sigma, USA).
Cell culture
Murine preosteoblast MC3T3-E1 cells obtained from the Cell Bank of the Chinese Academy of Science (Shanghai, China) were cultured in alpha-minimal essential medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (all from Hyclone, Logan, UT, USA) at 37°C and 5% CO2. In a previous study, 100 μmol/L C3G promoted cell proliferation of MC3T3-E1 cells (19), therefore, 7*105/ml MC3T3-E1 cells were seeded in 6-well plates in the present study. After the cells had adhere, the cells were synchronized for 24 h using serum-free medium. After completion of the synchronization treatment, the serum-free medium was replaced with complete medium with or without 100 μmol/L C3G, and the cells were culture for a further 24 h.
RNA-seq analysis
Total RNA was extracted from cells using TRIzol reagent and RNA integrity was evaluated via agarose gel electrophoresis and spectrophotometry using a NanoDrop ND-1000 instrument (Thermo Fisher Scientific, Waltham, MA, USA). An RNA library was constructed with the KAPA Stranded RNA-seq Library Prep Kit (Illumina, San Diego, CA, USA), and the quality of the library was assessed using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Quantification of the library was performed via quantitative real-time PCR (qRT-PCR). Sequencing was performed over 150 cycles using the Xten/NovaSeq system (Illumina). Raw RNA-seq data were submitted to NCBI Gene Expression Omnibus (accession number. GSE149731).
Functional analysis of identified genes
FastQC v0.11.8 was used to analyze the raw RNA-seq data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (31). Fragments per kilobase of gene/transcript model per million mapped fragments values for gene and transcript levels were calculated with the Ballgown package (https://www.bioconductor.org/packages/release/bioc/html/ballgown.html) of R v2.10.0 software. R was used to generate volcano plots and heatmaps to further analyze gene expression profiles.
Gene Ontology (GO) functional enrichment analysis of DEGs (www.geneontology.org/) was performed, and genes involved in biological process, cellular component, and molecular function GO categories were identified (32, 33). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (www.genome.jp/kegg/) was conducted to identify signaling pathways associated with the DEGs. All pathways were based on the KEGG database (33).
Quantitative real-time PCR
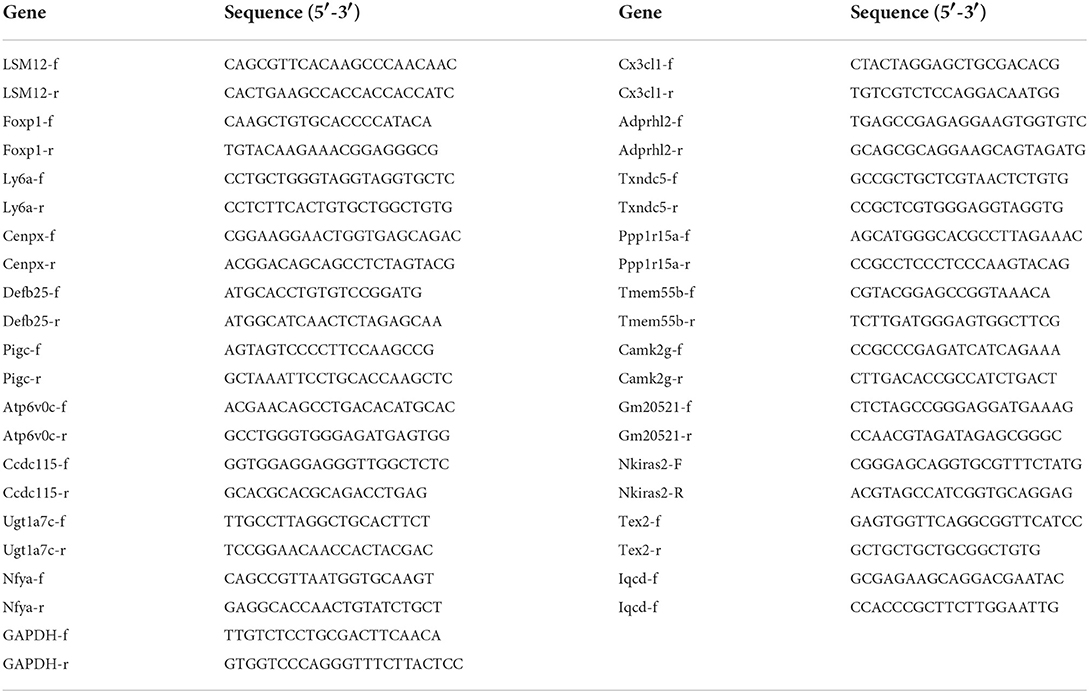
RNA was extracted from C3G-treated and untreated MC3T3-E1 cells (n = 3 replicates each) using TRIzol reagent. cDNA was synthesized using a reverse transcription kit, and SYBR Green MasterMix was used for qRT-PCR in a reaction volume of 20 μL. qRT-PCR was performed on a real-time PCR machine (ABI 7500, Applied Biosystem, Foster, California, USA). The primer sequences used are shown in Table 1. The glyceraldehyde 3-phosphate dehydrogenase (Gapdh) gene was used as the internal control to calculate target gene expression levels via the cycle threshold (2−ΔΔCt) method.
Statistical analysis
Data are presented as mean ± standard deviation. Statistical analyses were performed using SPSS v22.0 software (SPSS Inc, Chicago, IL, USA). Comparisons between two groups were performed with the student's t-tests, and p < 0.05 was deemed to indicate statistical significance.
Results
Identification of DEGs
Expression profiles of 11,238 genes in C3G-treated and untreated MC3T3-E1 cells were determined by RNA-seq, which yielded 51 DEGs (p < 0.05, fold change ≥1.2). A heatmap (Figure 1A) and a volcano plot (Figure 1B) were used to represent the abundance of these different transcripts, with expression levels ranging from high (red) to low (green), and there were 34 upregulated and 17 downregulated DEGs. Details of all DEGS are shown in the Supplementary Table 1.
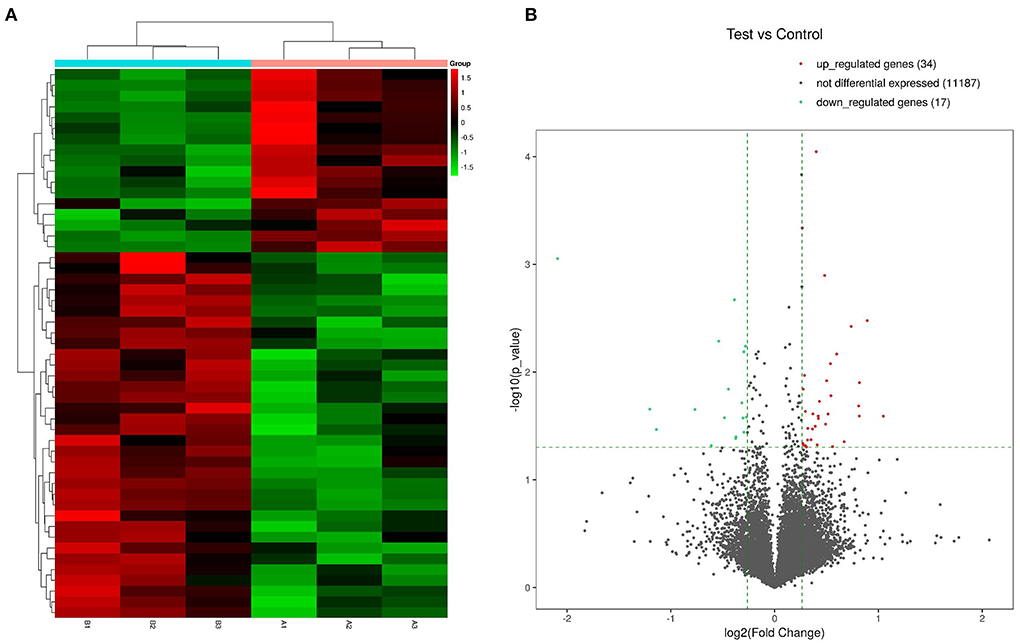
Figure 1. Heatmaps and volcano plot of DEGs in C3G-treated vs. untreated control MC3T3-E1 cells. (A) Heatmap of 34 upregulated and 17 downregulated DEGs. (B) Volcano plot of upregulated (red dots) and downregulated (green dots) DEGs and genes expressed at a normal level (gray dots).
GO analysis of DEGs
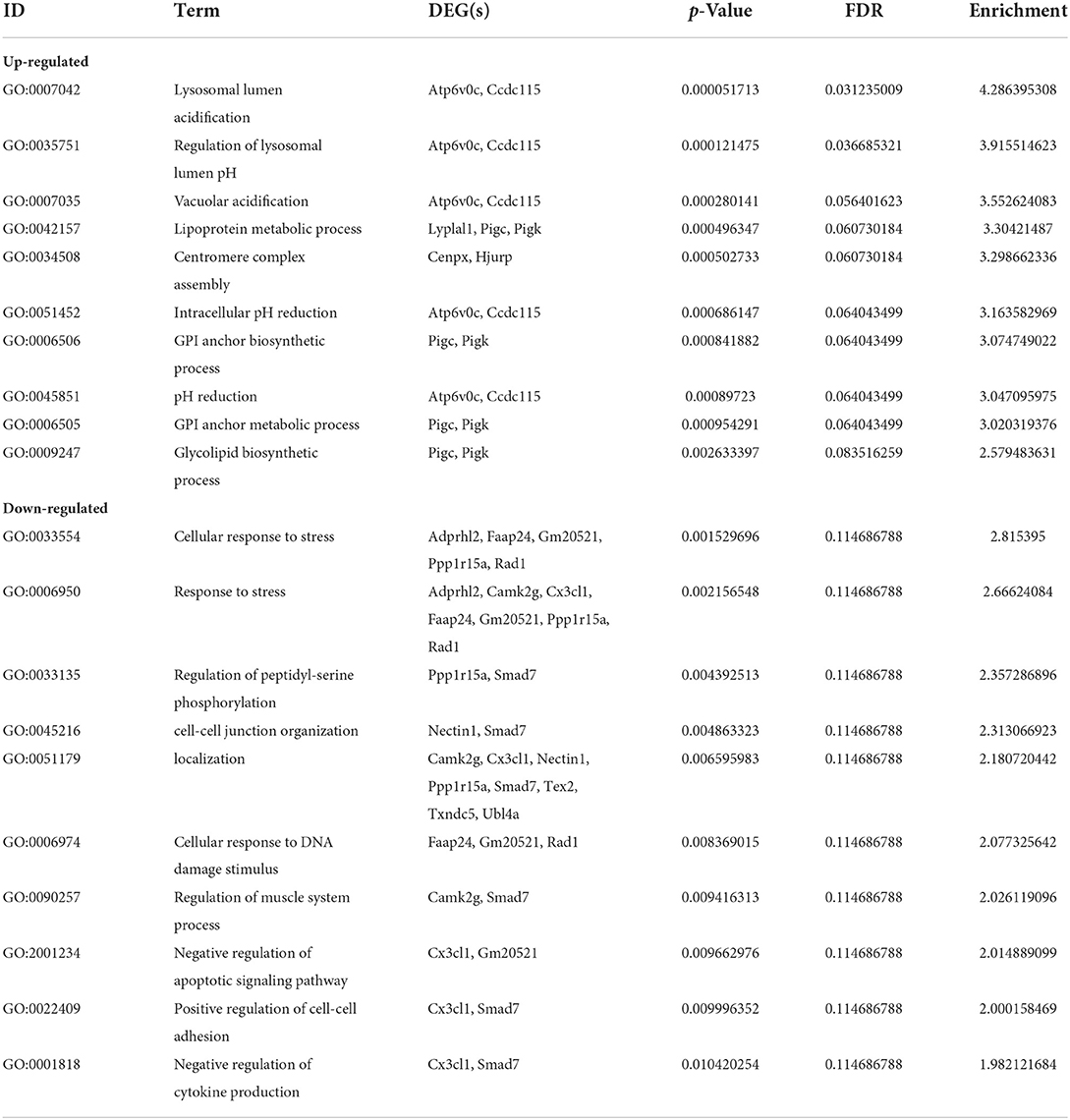
The top 10 biological process terms from the GO analysis of DEGs are presented in Table 2. All biological process terms are listed in Supplementary Table 2. The top three biological process terms for upregulated DEGs were lysosomal lumen acidification (GO:0007042; Atp6v0c, Ccdc115), regulation of lysosomal lumen pH (GO:0035751; Atp6v0c, Ccdc115), and lysosome organization (GO:0007040; Atp6v0c, Ccdc115). The top three biological process terms for downregulated DEGs were cellular responses to stress (GO:0033554; Adprhl2, Faap24, Gm20521, Ppp1r15a, Rad1), response to stress (GO:0006950; Adprhl2, Camk2g, Cx3cl1, Faap24, Gm20521, Ppp1r15a, Rad1), and vacuolar acidification (GO:0033135; Ppp1r15a, Smad7) for downregulated DEGs. Several significantly enriched entries were related to cellular responses to stress [GO:0033554; ADP-ribosylhydrolase-like 2 (Adprhl2), FA core complex-associated protein 24 (Faap24), Gm20521, protein phosphatase 1 regulatory subunit 15A (Ppp1r15a), Rad1], cell–cell junction organization [GO:0045216; Nectin1, Mothers against decapentaplegic homolog 7 (Smad7)], and cellular responses to DNA damage stimuli (GO:0006974; Faap24, Gm20521, Rad1). With respect to cellular components (Supplementary Table 3), the most significant terms were ATPase for upregulated DEGs (Figures 2A,B) and outer organelle membranes for downregulated DEGs (Figures 2C,D). The most significant terms pertaining to molecular function (Supplementary Table 4) were ubiquitin-like protein ligase binding for upregulated DEGs (Figures 2A,B) and cell adhesion molecule binding for downregulated DEGs (Figures 2C,D). Other significant GO terms included lipoprotein metabolic process [GO:0042157; lysophospholipase-like 1 (Lyplal1), phosphatidylinositol glycan anchor biosynthesis class C (Pigc), Pigk]; glycosylphosphatidylinositol (GPI) anchor biosynthetic process (GO:0006506; Pigc, Pigk); glycolipid biosynthetic process (GO:0009247; Pigc, Pigk); GPI-anchor metabolic process (GO:0006505; Pigc, Pigk); glycolipid biosynthetic process (GO:0009247; Pigc, Pigk); glycolipid metabolic process (GO:0006664; Pigc, Pigk); protein lipidation (GO:0006497; Pigc, Pigk); and lipoprotein biosynthetic process (GO:0042158; Pigc, Pigk).
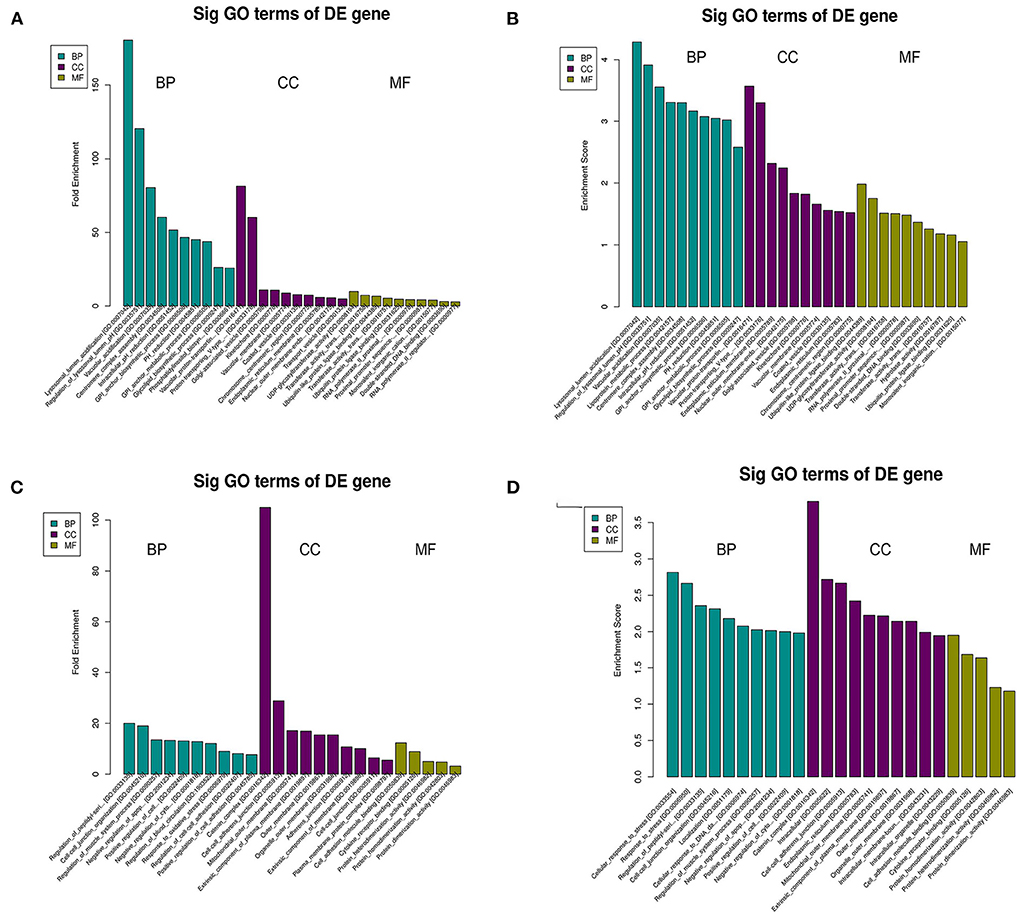
Figure 2. GO analysis of DEGs in C3G-treated MC3T3-E1 cells and untreated control MC3T3-E1 cells. Enriched GO terms corresponding to DEGs in the biological process (BP), cellular component (CC), and molecular function (MF) categories are shown. (A–D) GO terms based on upregulated (A,B) and downregulated (C,D) DEGs.
KEGG pathway analysis of DEGs
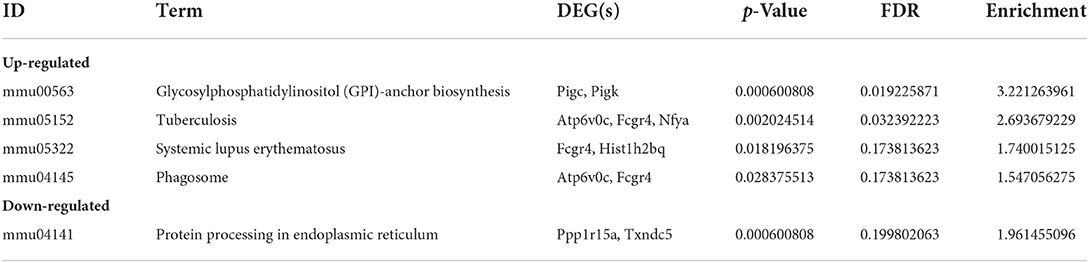
KEGG pathway analysis revealed five significantly enriched signaling pathways (Table 3). Four were upregulated including GPI-anchored protein (GPI-AP) biosynthesis signaling pathway (mmu00563; Pigc, Pigk), tuberculosis [mmu05152; Atp6v0c, Fc receptor, IgG, low affinity IV (Fcgr4), nuclear transcription factor Y subunit alpha (Nfya)], Systemic lupus erythematosus [mmu05322; Fcgr4, histone H2B (Hist1h2bq)], and Phagosome (mmu04145; Atp6v0c, Fcgr4) (Figure 3), and one was downregulated protein processing in endoplasmic reticulum [mmu04141; Ppp1r15a, thioredoxin domain-containing 5 (Txndc5)] (Figure 3).
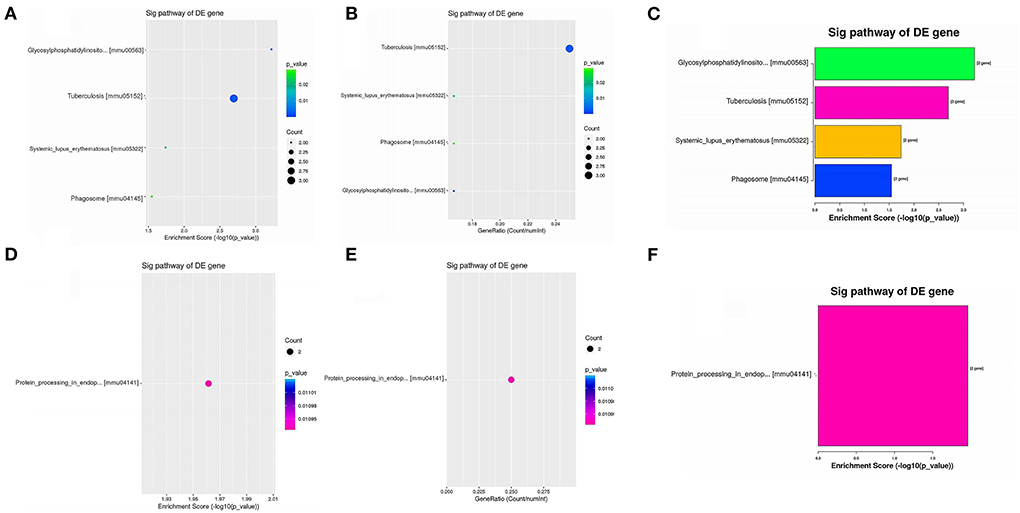
Figure 3. Enriched KEGG pathways of upregulated DEGs and downregulated DEGs. (A,B) Dot plots showing the enrichment score (–log10[p-value]) of significantly enriched pathways (A) and the gene ratio of the top 10 most significantly enriched pathways (B). (C) Bar plot showing the top 10 enrichment scores (–log10[p-value]) of significantly enriched pathways. (D,E) Dot plots showing the enrichment score (–log10[p-value]) of the significantly enriched pathways (D) and the gene ratios of the top 10 most significantly enriched pathways (E). (F) Bar plot showing the top 10 enrichment scores (–log10[p-value]) of significantly enriched pathways.
qRT-PCR analysis of DEGs
Among the top 10 upregulated genes identified by RNA-seq, seven [lymphocyte antigen 6 complex, locus A (Ly6a); centromere protein X (Cenpx), defensin beta 125 (Defb25), Pigc, Atp6v0c, UDP glucuronosyltransferase family 1 member A7 (Ugt1a7c), and Nfya] exhibited significant differences in expression in comparative qRT-PCR analysis of C3G-treated and untreated MC3T3-E1 cells, whereas LSM12 homolog (Lsm12), forkhead box P1 (Foxp1), and Ccdc115 mRNA levels did not differ significantly between the groups (Figure 4A). Among the top 10 downregulated DEGs, four [Cx3cl1, ADP-ribosylserine hydrolase 12 (Adprh12), Txndc5, and calcium/calmodulin-dependent protein kinase II gamma (Camk2g)] were expressed at significantly lower levels in C3G-treated MC3T3-E1 cells as determined by qRT-PCR, whereas phosphatidylinositol-4,5-bisphosphate 4-phosphatase (Tmem55b), Gm20521, Nkieas2, and testis expressed 2 (Tex2) were upregulated. There were no significant differences in Ppp1r15a or IQ motif-containing D [Iqcd] expression between treated and untreated cells (Figure 4B).
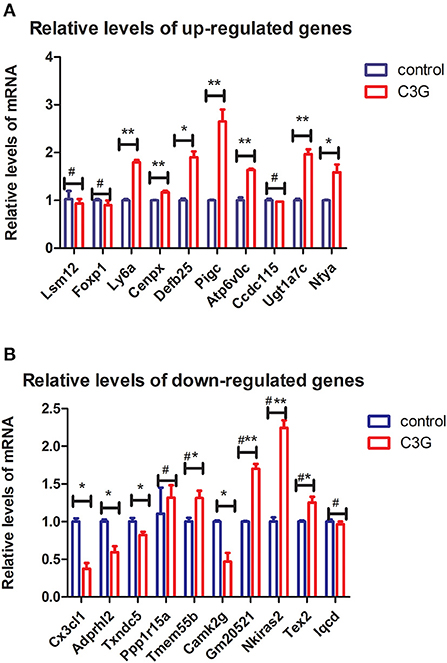
Figure 4. qRT-PCR validation of top DEGs. (A,B) Expression levels of top 10 upregulated (A) and downregulated (B) DEGs evaluated by qRT-PCR (n = 3 replicates). *p < 0.05, **p < 0.01.
Discussion
Osteoblasts are specialized fibroblasts that secrete and mineralize bone matrix and play a critical role in osteoporosis. MC3T3-E1 cells can be induced to differentiate into osteoblasts and are used as in vitro models to investigate the molecular mechanisms of osteogenesis (34). In current study, RNA-seq of MC3T3-E1 cells treated with the anthocyanin C3G revealed many DEGs as well as GO terms related to lysosomes—e.g., lysosomal lumen acidification, regulation of lysosomal lumen pH, and lysosome organization. This is consistent with previous report that lysosomes play an important role in biogenesis, mineralization, and trafficking of nanovesicles in osteoblasts (35).
GPI metabolism and lipidation were implicated in the effects of C3G on osteoblasts, as revealed by functional enrichment analyses of DEGs. Phosphorylation of ERK1/2 in osteoblasts promotes the expression of GPI-anchored proteins that maintain cell membrane integrity, which affects osteoblast proliferation (27). Other biological processes including cellular responses to stress (36), cell–to-cell junction organization (19) and cellular responses to DNA damage (37) have been linked to the effects of C3G on osteoblasts. The most significantly enriched KEGG pathways were involved in GPI-anchored protein biosynthesis (upregulated), lupus erythematosus (upregulated), and phagosome (upregulated), and protein processing in endoplasmic reticulum (downregulated). GPI is a ubiquitous glycolipid in eukaryotes (38) that anchors proteins to cell surfaces (39). To date, more than 150 GPI-APs have been identified in mammals (40) including some that are related to bone formation such as ALP, acetylcholinesterase (AChE) (41), and Ly6a (42). The related enzymes PIGC and PIGK play critical roles in GPI synthesis (43). One of the functions of GPI-anchor biosynthesis signaling is to synthesize GPI-Aps. Whether C3G can influence the proliferation and differentiation of osteoblasts by promoting the production of GPI-APs warrants further investigation.
ATP6V0C is a member of the V-ATPase family of enzymes that plays an important role in osteogenesis. Loss of function of V-ATPases results in an osteopetrorickets phenotype due to reduced bone formation (44), and Atp6v1h+/− mice exhibit impaired osteoblast growth as well as abnormal ALP levels (45). V-ATPase deficiency also inhibits osteogenic differentiation and stimulates adipogenic differentiation (46). The CX3CL1/CX3CR1 axis—which has been linked to various diseases including rheumatoid arthritis, spinal cord injury, and osteoarthritis (47)—was identified as a possible target for osteoporosis immunotherapy (48). Cx3cr1 is expressed in osteoclast precursors, implying that CX3CL1/CX3CR1 signaling can regulate osteoclast differentiation and thus affect the development of osteoporosis (49). Ly6a is involved in skull development, fat formation, osteogenesis, and chondrogenesis (50). Ly6a-deficient mice exhibit reduced bone formation and osteoclast counts and increased mineralization of trabecular bone, and develop features of consistent with age-related osteoporosis in humans including low bone mass, brittleness, and changes in the mechanical properties of bone (51). Inspired by the results described above, we will perform further studies of investigating the roles of the genes identified in present study,for example via the generation of stable osteoblastic cell lines lacking or overexpressing specific genes. That will be necessary to clarify the molecular mechanisms underlying osteoblast mineralization.
However, the present study had several limitations. Firstly, we did not examine the expression profile of long non-coding RNAs or microRNAs that may be involved in the effects of C3G on osteogenesis. Secondly, while qRT-PCR validation of RNA-seq results is essential, the methods used differ in terms of sensitivity and specificity, which may explain the differences in DEGs that were observed in our two datasets. In order to identify more sensitive targets, we did not use a higher concentration of C3G but instead selected the minimum concentration that was shown to promote MC3T3-E1 cell proliferation in previous experiments. This may be the reason why Runx2 and other known osteoporosis-related genes were not identified as DEGs in our RNA-seq analysis (34). In future work, immunoblotting and other experimental approaches should be used to validate biological targets of C3G in MC3T3-E1 cells.
Conclusion
In summary, we identified 51 genes and 5 signaling pathways (GPI-anchor biosynthesis, tuberculosis, systemic lupus erythematous, phagosome, and protein processing in the endoplasmic reticulum) via RNA-seq that may mediate the effects of C3G in osteogenesis. Based on their known involvement in osteoporosis, Atp6v0c, Cx3cl1, and Ly6a are the most promising targets of C3G action in osteoblasts. These findings provide evidence for the therapeutic potential of C3G in the treatment of osteoporosis and other disorders of bone metabolism.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
BZ designed the study. LC, BH, YC, and XW performed the experiments. LC and BH analyzed the data. LC and BZ wrote the manuscript. All authors have read and agreed to the publication of this version of the manuscript.
Funding
This work was supported by the Liaoning Science and Technology Program (Grant No. 20170540879), Shenyang Medical College Masters Science and Technology Innovation Fund Project (Grant No. Y20190508), Shenyang Science and Technology Project (Grant No. 19-112-4-024), and Shenyang Medical College Science and Technology Fund (Grant No. 20182045).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.995643/full#supplementary-material
References
1. Jin X, Zhou B, Zhang D. Replication study confirms the association of the common rs1800629 variant of the TNF α gene with postmenopausal osteoporosis susceptibility in the han Chinese population. Genet Test Mol Biomarkers. (2018) 22:246–51. doi: 10.1089/gtmb.2017.0204
2. Salari N, Darvishi N, Bartina Y, Larti M, Kiaei A, Hemmati M, et al. Global prevalence of osteoporosis among the world older adults: a comprehensive systematic review and meta-analysis. J Orthop Surg Res. (2021) 16:669. doi: 10.1186/s13018-021-02821-8
3. Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. (2011) 377:1276–87. doi: 10.1016/S0140-6736(10)62349-5
4. Luigi G, Merlotti A, Roberta C, Iacopo C. Emerging therapeutic targets for osteoporosis. Expert Opin Ther Targets. (2020) 2018:115–30. doi: 10.1080/14728222.2020.1726889
5. Oton-Gonzalez L, Mazziotta C, Iaquinta MR, Mazzoni E, Nocini R, Trevisiol L, et al. (2022). Genetics and epigenetics of bone remodeling and metabolic bone diseases. Int J Mol Sci. 23:1500. doi: 10.3390/ijms23031500
6. Drake MT, Clarke BL, Lewiecki EM, Clin T. The pathophysiology and treatment of osteoporosis. Clin Ther. (2015) 37:1837–50. doi: 10.1016/j.clinthera.2015.06.006
7. Blesso CN. Dietary anthocyanins and human health. Nutrients. (2019) 11:2107. doi: 10.3390/nu11092107
8. Millar CL, Duclos Q, Blesso CN. Effects of dietary flavonoids on reverse cholesterol transport, HDL metabolism, and HDL function. Adv Nutr. (2017) 8:226–39. doi: 10.3945/an.116.014050
9. Rózańska D, Regulska-Ilow B. The significance of anthocyanins in the prevention and treatment of type 2 diabetes. Adv Clin Exp Med. (2018) 27:135–42. doi: 10.17219/acem/64983
10. Mottaghipisheh J, Doustimotlagh AH, Irajie C, Tanideh N, Barzegar A, Iraji A. The promising therapeutic and preventive properties of anthocyanidins/anthocyanins on prostate cancer. Cells. (2022) 11:1070. doi: 10.3390/cells11071070
11. Welch A, MacGregor A, Jennings A, Fairweather-Tait S, Spector T, Cassidy A. Habitual flavonoid intakes are positively associated with bone mineral density in women. J Bone Miner Res. (2012) 27:1872–8. doi: 10.1002/jbmr.1649
12. Hardcastle AC, Aucott L, Reid DM, Macdonald HM. Associations between dietary flavonoid intakes and bone health in a scottish population. J Bone Miner Res. (2011) 26:941–7. doi: 10.1002/jbmr.285
13. Zheng X, Mun S, Lee SG, Vance TM, Hubert P, Koo SI, et al. Anthocyanin-rich blackcurrant extract attenuates ovariectomy-induced bone loss in mice. J Med Food. (2016) 19:390–7. doi: 10.1089/jmf.2015.0148
14. Dou C, Li J, Kang F, Cao Z, Yang X, Jiang H, et al. Dual effect of cyanidin on RANKL-Induced differentiation and fusion of osteoclasts. J Cell Physiol. (2016) 231:558–67. doi: 10.1002/jcp.24916
15. Nagaoka M, Maeda T, Chatani M, Handa K, Yamakawa T, Kiyohara S, et al. A delphinidin-enriched maqui berry extract improves bone metabolism and protects against bone loss in osteopenic mouse models. Antioxidants. (2019) 8:386. doi: 10.3390/antiox8090386
16. Qi S, He J, Han H, Zheng H, Jiang H, Hu CY, et al. Anthocyanin-rich extract from black rice (Oryza sativa L. Japonica) ameliorates diabetic osteoporosis in rats. Food Funct. (2019) 10:5350–60. doi: 10.1039/C9FO00681H
17. Jang W, Seo C, Jang HH, Song N, Kim J, Ahn J, et al. Black rice (Oryza sativa L.) extracts induce osteoblast differentiation and protect against bone loss in ovariectomized rats. Food Funct. (2015) 6:264–74. doi: 10.1039/C4FO00836G
18. Melough MM, Sun X, Chun OK. The role of AOPP in age-related bone loss and the potential benefits of berry anthocyanins. Nutrients. (2017) 9:789. doi: 10.3390/nu9070789
19. Hu BS, Chen L, Chen Y, Zhang Z, Wang XH, Zhou B. Cyanidin-3-glucoside regulates osteoblast differentiation via the ERK1/2 signaling pathway. ACS Omega. (2021) 6:4759–66. doi: 10.1021/acsomega.0c05603
20. Park KH, Gu DR, So HS, Kim KJ, Lee SH. Dual role of cyanidin-3-glucoside on the differentiation of bone cells. J Dent Res. (2015) 94:1676–83. doi: 10.1177/0022034515604620
21. Mao W, Huang G, Chen H, Xu L, Qin S, Li A. Research progress of the role of anthocyanins on bone regeneration. Front Pharmacol. (2021) 12:773660. doi: 10.3389/fphar.2021.773660
22. Yin Z, Zhu W, Wu Q, Zhang Q, Guo S, Liu T, et al. Glycyrrhizic acid suppresses osteoclast differentiation and postmenopausal osteoporosis by modulating the NF-κB, ERK, and JNK signaling pathways. Eur J Pharmacol. (2019) 859:172550. doi: 10.1016/j.ejphar.2019.172550
23. Zhang X, Zhang Y, Zhao G, Wang Y, Wang J, Cheng L, et al. Activation of JNK signaling in osteoblasts is inversely correlated with collagen synthesis in age-related osteoporosis. Biochem Bioph Res Co. (2018) 504:771–6. doi: 10.1016/j.bbrc.2018.08.094
24. Gooding S, Olechnowicz SWZ, Morris EV, Armitage AE, Arezes J, Frost J, et al. Transcriptomic profiling of the myeloma bone-lining niche reveals BMP signalling inhibition to improve bone disease. Nat Commun. (2019) 10. doi: 10.1038/s41467-019-12296-1
25. Xu ML, Bi CWC, Liu EYL, Dong TTX, Tsim KWK. Wnt3a induces the expression of acetylcholinesterase during osteoblast differentiation via the Runx2 transcription factor. J Biol Chem. (2017) 292:12667–78. doi: 10.1074/jbc.M117.777581
26. Wojdasiewicz P, Turczyn P, Dobies-Krzesniak B, Frasunska J, Tarnacka B. Role of CX3CL1/CX3CR1 signaling axis activity in osteoporosis. Mediat Inflamm. (2019) 2019:1–9. doi: 10.1155/2019/7570452
27. Xing Y, Gu Y, Xu LC, Siedlecki CA, Donahue HJ, You J. Effects of membrane cholesterol depletion and GPI-anchored protein reduction on osteoblastic mechanotransduction. J Cell Physiol. (2011) 226:2350–9. doi: 10.1002/jcp.22579
28. Sakaki J, Melough M, Lee S, Kalinowski J, Koo S, Lee S, et al. Blackcurrant supplementation improves trabecular bone mass in young but not aged mice. Nutrients. (2018) 10:1671. doi: 10.3390/nu10111671
29. Kaume L, Gilbert W, Smith BJ, Devareddy L. Cyanidin 3-O-β-d-glucoside improves bone indices. J Med Food. (2015) 18:690–7. doi: 10.1089/jmf.2014.0029
30. Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. (2019) 20:631–56. doi: 10.1038/s41576-019-0150-2
31. Andrews S. (2018). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed October 04, 2018).
32. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. (2000) 25:25–9. doi: 10.1038/75556
33. Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, et al. A systems biology approach for pathway level analysis. Genome Res. (2007) 17:1537–45. doi: 10.1101/gr.6202607
34. Genetos DC, Geist DJ, Liu D, Donahue HJ, Duncan RL. Fluid Shear-Induced ATP secretion mediates prostaglandin release in MC3T3-E1 osteoblasts. J Bone Miner Res. (2005) 20:41–9. doi: 10.1359/JBMR.041009
35. Iwayama T, Okada T, Ueda T, Tomita K, Matsumoto S, Takedachi M, et al. Osteoblastic lysosome plays a central role in mineralization. Sci Adv. (2019) 5:x672. doi: 10.1126/sciadv.aax0672
36. Domazetovic V, Marcucci G, Falsetti I, Bilia AR, Vincenzini MT, Brandi ML, et al. Blueberry juice antioxidants protect osteogenic activity against oxidative stress and improve long-term activation of the mineralization process in human osteoblast-like SaOS-2 cells: involvement of SIRT1. Antioxidants. (2020) 9:125. doi: 10.3390/antiox9020125
37. Raut N, Wicks SM, Lawal TO, Mahady GB. Epigenetic regulation of bone remodeling by natural compounds. Pharmacol Res. (2019) 147:104350. doi: 10.1016/j.phrs.2019.104350
38. Müller GA. Membrane insertion and intercellular transfer of glycosylphosphatidylinositol-anchored proteins: potential therapeutic applications. Arch Physiol Biochem. (2018) 1–18. doi: 10.1080/13813455.2018.1498904
39. Kinoshita T, Fujita M, Maeda Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem. (2008) 144:287–94. doi: 10.1093/jb/mvn090
40. Homans SW, Ferguson MA, Dwek RA, Rademacher TW, Anand R, Williams AF. Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature. (1988) 333:269–72. doi: 10.1038/333269a0
41. Ren X, Hu H, Jiang W, Ma X, Ma Y, Li G, et al. Three GPI-anchored alkaline phosphatases are involved in the intoxication of Cry1Ca toxin to spodoptera exigua larvae. J Invertebr Pathol. (2018) 151:32–40. doi: 10.1016/j.jip.2017.10.009
42. Karam N, Lavoie J, St-Jacques B, Bouhanik S, Franco A, Ladoul N, et al. Bone-specific overexpression of PITX1 induces senile osteoporosis in mice through deficient self-renewal of mesenchymal progenitors and wnt pathway inhibition. Sci Rep. (2019) 9:3544. doi: 10.1038/s41598-019-40274-6
43. Nguyen T, Murakami Y, Sheridan E, Ehresmann S, Rousseau J, St-Denis A, et al. Mutations in GPAA1, encoding a GPI transamidase complex protein, cause developmental delay, epilepsy, cerebellar atrophy, and osteopenia. Am J Hum Genet. (2017) 101:856–65. doi: 10.1016/j.ajhg.2017.09.020
44. Scimeca JC, Franchi A, Trojani C, Parrinello H, Grosgeorge J, Robert C, et al. The gene encoding the mouse homologue of the human osteoclast-specific 116-kDa V-ATPase subunit bears a deletion in osteosclerotic (oc/oc) mutants. Bone. (2000) 26:207–13. doi: 10.1016/S8756-3282(99)00278-1
45. Duan X, Liu J, Zheng X, Wang Z, Zhang Y, Hao Y, et al. Deficiency of ATP6V1H causes bone loss by inhibiting bone resorption and bone formation through the TGF-beta1 pathway. Theranostics. (2016) 6:2183–95. doi: 10.7150/thno.17140
46. Li L, Yang S, Zhang Y, Ji D, Jin Z, Duan X. ATP6V1H regulates the growth and differentiation of bone marrow stromal cells. Biochem Biophys Res Commun. (2018) 502:84–90. doi: 10.1016/j.bbrc.2018.05.124
47. Hamann I, Unterwalder N, Cardona AE, Meisel C, Zipp F, Ransohoff RM, et al. Analyses of phenotypic and functional characteristics of CX3CR1-expressing natural killer cells. Immunology. (2011) 133:62–73. doi: 10.1111/j.1365-2567.2011.03409.x
48. Chen Y, Huang C, Liu H, Yao W, Wu W, Lu Y, et al. Serum CX3CL1/fractalkine concentrations are positively associated with disease severity in postmenopausal osteoporotic patients. Brit J Biomed Sci. (2016) 73:121–8. doi: 10.1080/09674845.2016.1209897
49. Koizumi K, Saitoh Y, Minami T, Takeno N, Tsuneyama K, Miyahara T, et al. Role of CX3CL1/Fractalkine in osteoclast differentiation and bone resorption. J Immunol. (2009) 183:7825–31. doi: 10.4049/jimmunol.0803627
50. Steenhuis P, Pettway GJ, Ignelzi MA. Cell surface expression of stem cell antigen-1 (Sca-1) distinguishes osteo-, chondro-, and adipoprogenitors in fetal mouse calvaria. Calcified Tissue Int. (2008) 82:44–56. doi: 10.1007/s00223-007-9083-4
Keywords: cyanidin-3-O-glucoside, osteogenesis, RNA-sequencing, MC3T3-E1 cells, Atp6v0c, Cx3cl1, Ly6a
Citation: Chen L, Hu B, Wang X, Chen Y and Zhou B (2022) Functional role of cyanidin-3-O-glucoside in osteogenesis: A pilot study based on RNA-seq analysis. Front. Nutr. 9:995643. doi: 10.3389/fnut.2022.995643
Received: 16 July 2022; Accepted: 25 August 2022;
Published: 30 September 2022.
Edited by:
Er Sheng Gong, Gannan Medical University, ChinaReviewed by:
Yuehua Wang, Shenyang Agricultural University, ChinaXiaohui Hua, Anhui Medical University, China
Copyright © 2022 Chen, Hu, Wang, Chen and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Zhou, emhvdWJvNjNAaG90bWFpbC5jb20=
 Lin Chen1
Lin Chen1 Bo Zhou
Bo Zhou