- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi Key Laboratory of Sugarcane Biology, College of Agriculture, Guangxi University, Nanning, China
- 2College of Life Science and Technology, Guangxi University, Nanning, China
- 3Key Laboratory of Integrated Management of Crop Diseases and Pests, Department of Plant Pathology, College of Plant Protection, Nanjing Agricultural University, Ministry of Education, Nanjing, China
Sugarcane (Saccharum ssp., Poaceae) provides enormous metabolites such as sugars, lipid, and other dietary metabolites to humans. Among them, lipids are important metabolites that perform various functions and have promising pharmacological value. However, in sugarcane, few studies are focusing on lipidomics and few lipid compounds were reported, and their pharmacological values are not explored yet. The transcriptomic and widely targeted lipidomics approach quantified 134 lipid compounds from the rind of six sugarcane genotypes. These lipid compounds include 57 fatty acids, 30 lysophosphatidylcholines, 23 glycerol esters, 21 lysophosphatidylethanolamines, 2 phosphatidylcholines, and 1 sphingolipid. Among them, 119 compounds were first time reported in sugarcane rind. Seventeen lipids compounds including 12 fatty acids, 2 glycerol lipids, LysoPC 16:0, LysoPE 16:0, and choline alfoscerate were abundantly found in the rind of sugarcane genotypes. From metabolic and transcriptomic results, we have developed a comprehensive lipid metabolic pathway and highlighted key genes that are differentially expressed in sugarcane. Several genes associated with α-linolenic acid and linoleic acid biosynthesis pathways were highly expressed in the rind of the ROC22 genotype. ROC22 has a high level of α-linolenic acid (an essential fatty acid) followed by ROC16. Moreover, we have explored pharmacological values of lipid compounds and found that the 2-linoleoylglycerol and gingerglycolipid C have strong binding interactions with 3CLpro of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) and these compounds can be utilized against SARS-CoV-2 as therapeutic agents. The transcriptome, metabolome, and bioinformatics analysis suggests that the sugarcane cultivars have a diversity of lipid compounds having promising therapeutic potential, and exploring the lipid metabolism will help to know more compounds that have promising cosmetic and pharmacological value.
Introduction
Sugarcane crop contributes to 80% of world sugar production and major feedstock for bioenergy production. Sugarcane belongs to the perennial grass family “Poaceae,” which is the fifth major cultivated crop in the world (http://faostat.fao.org/) (1). Brazil, India, and China are leading sugarcane-producing countries (2, 3). Sugarcane residues are also utilized as fodder, organic fertilizer, extraction of several essential color components, and several byproducts, and also burned for electricity generation (4). In addition, sugarcane contains health-promoting lipid compounds such as linoleic acid and α-linolenic acid also known as omega-6 fatty acid and omega-3 essential fatty acids, respectively (5). Some promising biologically active compounds such as policosanol that have unique nutritive and pharmacological values have been isolated from sugarcane (3, 6); recently, various phenolic compounds including phenolic acids (such as p-ferulic acid and coumaric acid) have been extracted from sugarcane rind which executes weight loss effect in rats (7, 8). Sugarcane rind is a storehouse of several health-promoting metabolites that have significant nutritive, cosmetic, and pharmacological values (7, 9). It is revealed that sugarcane rind possesses enormous kinds of metabolites such as lipids, carbohydrates, phenolics, alkaloids, aromatic amino acids, anthocyanins, phytosterols, flavonoids, triterpenoids, carotenoids, and other phytocompounds (10–12), which have diverse health-promoting properties such as anticancer, antiplatelet, antioxidant activity, antifungal, antibacterial, antiviral, anti-HIV activities quenching free radicals, and anti-inflammatory activities (13, 14).
Lipids are small amphiphilic or hydrophobic molecules that play important roles in signaling, structural component, and energy storage. Lipids include waxes, sterols, phospholipids, and fatty acids and they are highly soluble in an organic solvent but insoluble in water (14, 15). The various combination of isoprene and ketoacyl building blocks arise huge structural complexities that make lipids classification often challenging (15). The lipids are classified into the sub-categories such as free fatty acids, sphingolipids, glycerophospholipids, and glycerolipids (15, 16). The glycerophospholipids are further divided into phosphatidylcholine (PC) which is further converted to phosphatidylethanolamine (PE) (17). The lysophosphatidylcholines (LPCs) are derived from PC whereas the lysophosphatidylethanolamines (LPEs) are derived from PE. The LPCs and LPEs are found in the cell membrane of several plant species as minor phospholipids. The fatty acids compounds such as myristic acid, stearic acid, lauric acid, and palmitic acid are famous phytocompounds that significantly inhibit microbial activity (18, 19). Glycerophosphocholine (choline alfoscerate) is commonly utilized against various human neurodegenerative disorders such as Parkinson's disease and Alzheimer's disease (20). Besides this, the fat compounds are also combined with proteins known as lipoproteins. Moreover, lipids also act as light-absorbing pigments, enzyme cofactors, hydrophobic proteins, and electron carriers (14). Lipid compounds are widely utilized as a nutritive component, in food, and cosmetic industries and recently in nanotechnology (2, 21). The complex structures and identification of new lipid compounds will continue to be an imperious research field that opens new doors to explore plant lipidomics.
In sugarcane rind, only a few lipid compounds have been reported before. Previous studies mostly focused on the production of sugar and biofuel, whereas the diversity of lipid compounds and their pharmacological value is less explored. Sugarcane is the fifth largest cultivated crop in the world, thus its rind is widely available for extracting different valuable compounds that have potential pharmacological and cosmetic value. In this study, we used a transcriptomic and widely targeted liquid chromatography-mass with a tandem spectrometry-mass (LC-MS/MS) approach to explore the biosynthesis pathway and diversity of lipids in sugarcane genotypes. Additionally, we identify the major lipid compounds in the sugarcane and highlighted their distinctive pharmacological value which will be valuable for future lipidomics research.
Materials and methods
The rind samples were harvested from six cultivated sugarcane genotypes including ROC22 (ROC22), Yuetang93/159 (YT93/159), Taitang 172 (F172), Taitang134 (F134), ROC16 (ROC16), and Yuetang71/210 (YT71/210) which were planted in Guangxi University (experimental area), China. From each variety, rind samples were harvested (having three biological repeats) at 6 months stage (January 2021) and frozen immediately for transcriptome and LC-MS/MS analysis.
Transcriptomic analysis
TRIzol reagent procedure was adopted for total RNA extraction from sugarcane rind, referring to the company's guidelines (Invitrogen), followed by removal of genomic DNA by DNase I (Takara). From total RNA, library construction was achieved by the TruSeq RNA kit, and sequencing was performed according to the producer's procedures (Illumina, San Diego, CA). For sequencing, Illumina HiSeq 2500 was used for the production of a (paired-end) RNA-seq sequencing library. Annotation of unigenes was accomplished by using publicly available protein databases such as Pfam (protein family), Gene Ontology, and KEGG. GOseq R package was performed for enrichment analysis (gene ontology) of differentially expressed genes (DEGs), in which gene bias length was adjusted (22, 23). Saccharum spontaneum genome was used as a reference genome (publically available Saccharum Genome Database website; http://sugarcane.zhangjisenlab.cn/sgd/html/download.html). Gene ontology (GO) terms having <0.05 P-value were measured as significantly enriched by DEGs. For statistical enrichment of DEGs in KEGG (http://www.genome.jp/kegg/) pathways, KOBAS software was used. The transcriptomic data is submitted online and have PRJNA824938 accession number. The raw transcriptomic data of thirty-five genes accompanying lipid metabolism is represented in Supplementary File 1.
Chemical reagents for metabolic analysis and rind samples preparation and extraction
The MilliQ (Bradford, USA) water was used in all metabolic analysis processes. Hydrochloric acid and formic acid were obtained from Xinyang Chemical Reagent (Hunan, China) and Sigma-Aldrich (St. Louis, MO, USA), respectively. An internal standard from Sigma-Aldrich was purchased for quality control (http://www.sigmaaldrich.com/united-states.html) whereas the reagents methanol (MeOH, grade-HPLC) and acetonitrile were purchased from Merck (Darmstadt, Germany). The 1 mg/mL in 50% MeOH concentration of stock solutions were prepared for standards and stored at−20°C. The working solutions were made before analysis by dissolving the stock solutions with 50% MeOH.
For lipids quantitative analysis, multiple reactions monitoring (MRM) (http://en.metware.cn/) analysis was managed by Metware Co., Ltd., (Wuhan, China). The zirconia bead was used to crush (mixer mill, MM 400, Retsch) the rind freeze-dried samples at 30 Hz for 1.5 min. The 50 mg sugarcane rind was extracted with 1 mL methanol. After that, the extract was ultrasound and vortex for 5 min each followed by centrifuge at 4°C for 3 min at 12,000 × g and this step was repeated to extract the rind residue for a second time. Before LC-MS/MS analysis, the supernatants were together and filtrated (0.22 μm, Anpel).
Chromatography and instrument conditions and metabolic analysis
Ultra-performance liquid chromatography (UPLC) was used for sample extracts analysis combined with electrospray ionization tandem mass spectrometry (ESI-MS/MS) system (UPLC-ESI-MS/MS) (Triple Quadrupole Applied Biosystems AB4500, SCIEX brand, China; https://sciex.com.cn/) (24). The analytical conditions were as follows, methanol (0.1% formic acid); solvent system, water (0.1% formic acid): UPLC: column C18 (1.7 μm, 2.1 mm*100 mm); gradient program, 0 min 95:5V/V, 6 min 50:50 V/V, 12 min 5:95 V/V, wait 2 min, 14 min 95:5 V/V; wait 2 min; injection volume: 2 μL; temperature, 40°C; flow rate, 0.35 mL/min. On the other hand, the effluent was linked to electrospray ionization (ESI) triple quadrupole-linear ion trap (QTRAP)-MS.
ESI source operation parameters include temperature 550°C; turbo spray, ion source; ion spray 5,500 voltage (V) and negative 4,500 V mode; curtain (CUR) gas, ion source gas (GSI), and gas (GSII), and were set at 25, 50, and 60 psi, consistently. The MRM individual transitions were developed to further optimize the de-clustering potential and collision energy. According to the elution of metabolites, monitoring of MRM transition was specifically set for each period. The triple quadrupole (QQQ) and linear ion trap (LIT) were accomplished by QTRAP, equipped with an ESI Ion-Spray Turbo interface, operating in positive and negative ion mode, governed by Analyst 1.6.3 (AB Sciex, China) software.
For MRM metabolic analysis firstly, we eliminated the interference of the target (precursor ion) substance by screening, then the molecular weight of the parallel ion is omitted. Then, precursor ions were persuaded to ionize in the collision chamber to form several fragment ions (Supplementary Figure 1). Subsequently, to make the quantification more reliable, the fragment ions were filtered by using QQQ, to attain the specific or required (removing non-target ion interference) fragment ion. The individual metabolite peak area and mass spectrum peaks were integrated for all compounds and then matched in different samples (25). The retention time (RT) and secondary spectrum fragment of all samples were matched (one by one) with the RT and secondary spectra of the Metware (http://en.metware.cn/) database. For more precise widely targeted metabolomics results, a low-resolution mass spectrometer with higher sensitivity was used, and the lipids were characterized as RT, MS2 (secondary spectrum), Q1, and Q3).
Lipids pharmacological value assessment
The lipid compounds were explored for their pharmacological potential through docking against SARS-CoV-2. For molecular docking analyses, the same protocol was utilized as published in our previous study (26). A molecular operating environment (MOE) tool was used for this analysis (27). Briefly, the 3D structures of all lipid compounds were searched from PubChem (28) and the MPD3 database (29). Their energy was minimized and they were stored in the “.mdb” format database. From the protein data bank (PDB), the SARS-CoV-2 3D main protease (3CLpro) structure was copied (30) and processed for energy minimization and missing hydrogen atoms. Finally, the rigid docking algorithm of MOE was applied and the prepared lipids' database was screened against the ligand-binding site of SARS-CoV-2 3CLpro. PyMOL was used for the visualization of docked complexes (31).
Statistical analysis
The OmicShare tools (www.omicshare.com/tools) were used for heat map, hierarchical cluster, and principal component analysis (PCA) by using the hcluster algorithm as performed before (32). The peak area (from each variety with three replications) was used to perform the PCA and hierarchical cluster analysis (HCA) Data pre-processing was normalized and further details of normalization are represented (https://www.omicshare.com/forum/thread-407-1-1.html) a publically available online platform.
Results
Structure of major lipids compound
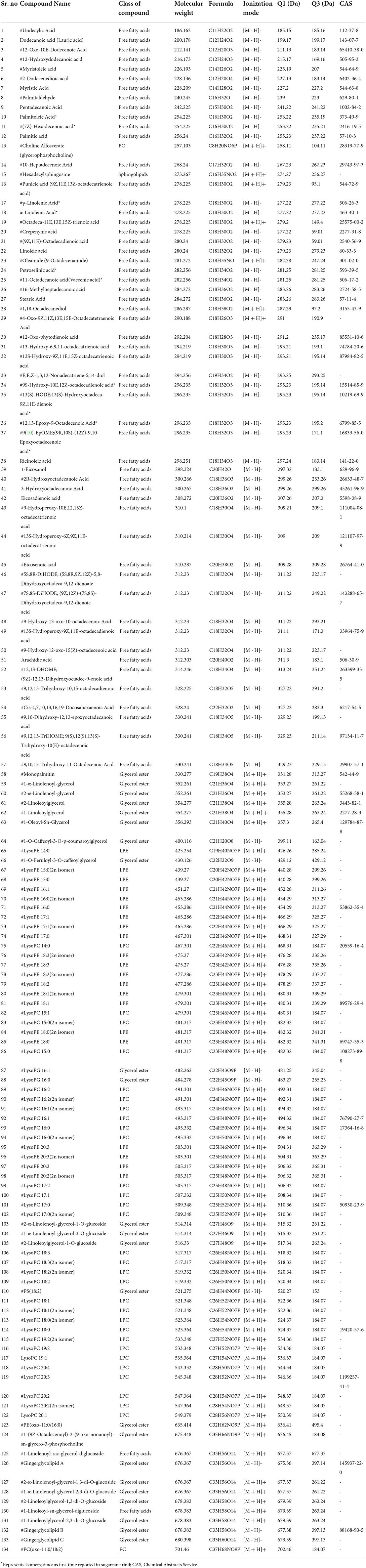
A total number of 134 lipids were identified from six sugarcane genotypes. These compounds belong to different sub-categories of lipids such as fatty acids, sphingolipids, phosphatidylcholine (PC), lysophosphatidylcholines (LPC), lysophosphatidylethanolamines (LPE), and glycerol esters (GE) (Table 1). The chemical formula, precursor ion Q1 (Da), daughter ion Q3 (Da), CAS-number, molecular weight, and ionization mode of lipids are present in Table 1. Among 134 lipid metabolism compounds, 119 compounds were first time identified in sugarcane genotypes (Table 1). Seventeen lipids compounds (including 12 fatty acids, 2 glycerol lipids, 1 LPC, 1 LPE, and 1 PC) were uniquely high in the rind of all sugarcane genotypes, among them, 11 compounds were reported first time in sugarcane rind (Figure 1; Table 1). The chemical structure of these 17 lipid compounds is represented (Figure 1). Among 17 major lipid compounds, myristic acid, linoleic acid, stearic acid, α-linolenic acid, arachidic acid, and petroselinic acid were reported before whereas the other compounds are reported first time in the rind of six sugarcane genotypes (Figure 1). Among 23 glycerol ester compounds (Table 1), gingerglycolipid A and gingerglycolipid B were uniquely high in the rind and these compounds are famous due to their promising pharmacological value (Figure 1).
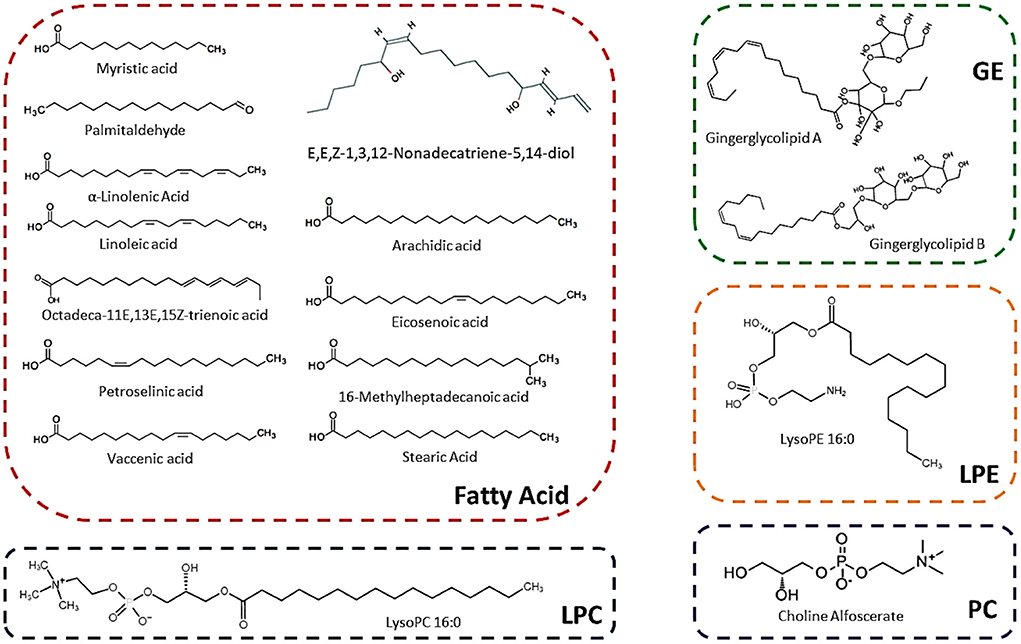
Figure 1. Structure of main lipids that are abundantly found in the rind of six sugarcane genotypes.
Principal component analysis of lipids compounds
The sugarcane variety-wise and individual lipid compounds-wise PCA (scatter plot) were performed (Figure 2). The lipids scatter plot was achieved through PCA (Figure 2A). In the scatter plot the lipids compounds showed two distinct clusters (Figure 2A). Most of the lipids compounds gathered in one cluster on the intersection point of the x-axis and y-axis while the second group (includes 6 lipid compounds) was clustered slightly below the first cluster (Figure 2A). The five lipid compounds including palmitaldehyde (L8), 16-Methylheptadecanoic acid (L26), stearic acid (L27), lysoPE 18:2 (2n isomer) (L78), and lysoPC 16:0 (L93) didn't fall in any cluster and showed their unique identity on the PCA scatter plot (Figure 2A). These five lipid compounds revealed maximum divergent appearances on PCA and displayed the highest absolute scores values in both PC1 98.5% (on the x-axis) and PC2 0.9% (on the y-axis) (Figure 2A). Moreover, these compounds were reported first time in this study except for stearic acid.
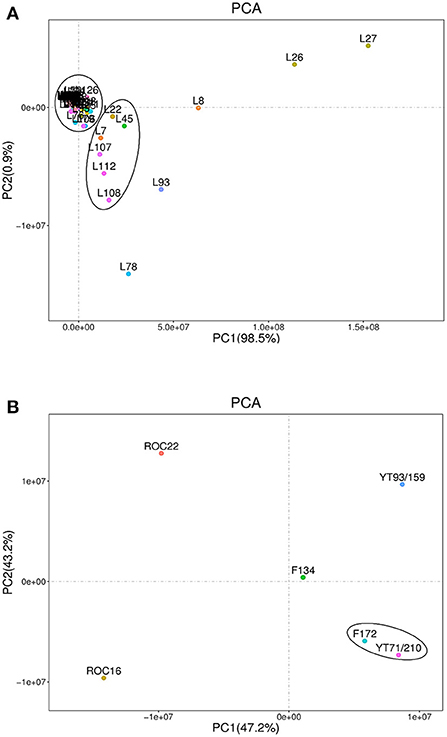
Figure 2. Principal component analyses of lipid compounds among six sugarcane genotypes. Each variety represents the data of three biological repeats. (A) Represents the metabolic-wise PCA (the L1-L134 numbers are corresponding to the sequence of lipid compounds in Table 1). (B) Represents the variety-wise PCA.
Sugarcane variety-wise scattered plot was attained using PCA (Figure 2B). The variety-wise PCA revealed that the F172 and YT71/210 varieties were very close to each other and made their distinct group from the other four varieties (Figure 2B). The other four sugarcane varieties such as F134, YT93/159, ROC16, and ROC22 were far away from each other and showed their distinct characteristics on scattered PCA plots and displayed higher absolute scores on both x-axis (PC1) and y-axis (PC2) (Figure 2B). Also, these four varieties were far away from F172 and YT71/210 clusters (Figure 2B). The PC1 accounted for 47.2% while the PC2 accounted for 43.2% of absolute scores values on variety-wise PCA scattered plots (Figure 2B). These outcomes revealed that most of the sugarcane genotypes bear unique characteristics regarding lipid metabolism and these results are interesting for the researchers and breeders who are engaged with lipid metabolism (sugarcane breeding programs) for enhancing the biodiesel and bioethanol production from sugarcane residues (such as rind) (33). Additionally, the lipid compounds have promising effects on the skin and have unique pharmacological effects (34, 35). Exploring the lipid metabolism of sugarcane will help to know more compounds that have promising cosmetic and pharmacological value.
Hierarchical cluster analysis of lipids from rind samples
The heat map and hierarchical cluster analysis (HCA) of lipid compounds in the rind of six sugarcane genotypes are characterized (Figure 3). Figure 3A heat map represents the fatty acids and LPE compounds whereas the Figure 3B heat map signifies the LPC, PC, hexadecylsphingosine (sphingolipids), and glycerol ester compounds. The HCA analysis revealed that the YT71/210 and F172 stand near to each other and fall in the same group (Figure 3A). The F134 and YT93/159 are very close to each other and made the same cluster (Figure 3A). Additionally, the other two varieties ROC22 and ROC16 didn't make any cluster and represent their unique characteristics in hierarchical cluster analysis (Figure 3A); however, they were closer to each other (Figure 3A). According to the abundance of individual fatty acids and LPE compounds, all LPC and fatty acid compounds made two major groups and several sub-clusters in the rind of six sugarcane genotypes (Figure 3A). HCA analysis revealed that the ROC22 and ROC16 are closer to each other and fall in the same cluster whereas the F172 and YT71/210 again made the same cluster (Figure 3B) as in the first heat map (Figure 3A). Interestingly, the YT93/159 and F134 didn't make any clusters; it signifies that these two genotypes have unique characteristics (regarding LPC, PC, hexadecylsphingosine, and glycerol ester compounds) (Figure 3B).
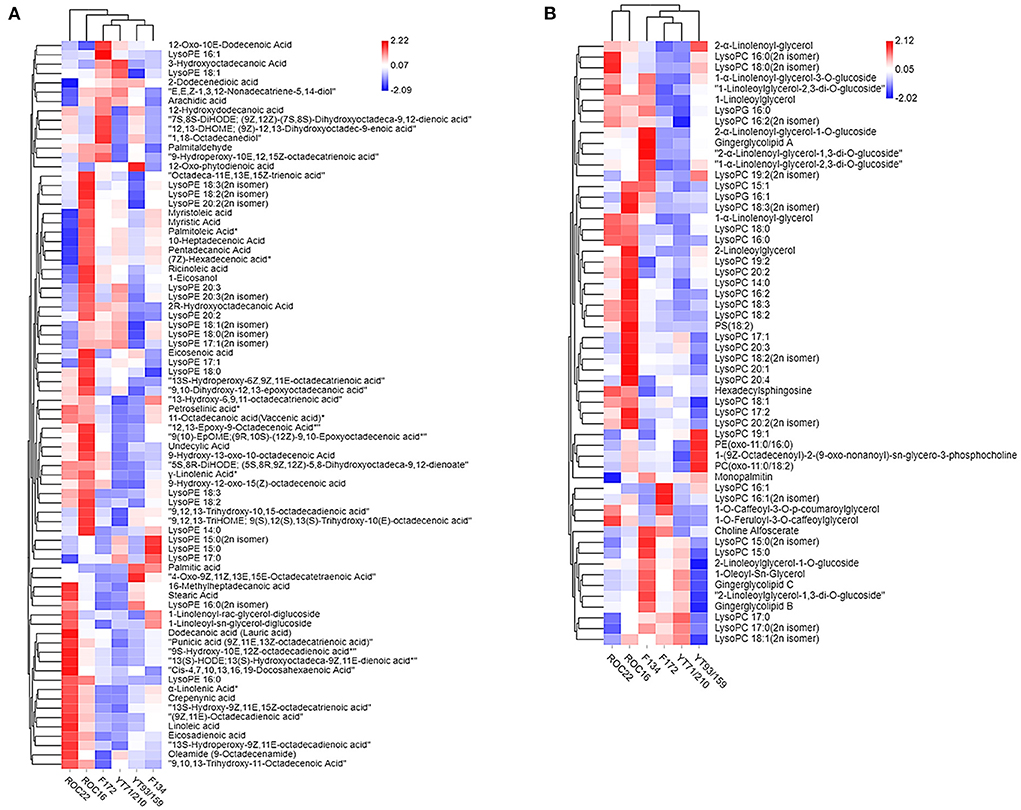
Figure 3. Hierarchical cluster analysis represents the distribution of lipid compounds in different sugarcane genotypes. The row represents lipid compounds and the column represents sugarcane genotypes. (A) Fatty acid and LPE. (B) LPC, PC, Glycerol ester, and hexadecylsphingosine. Red indicates high concentration and blue color means low concentration. Each variety represents the data of three biological repeats.
Some high-value fatty acids such as lauric acid, linoleic acid, eicosadienoic acid (an omega-6 fatty acid), punicic acid (an omega-5 fatty acid), crepenynic acid, α-linolenic acid, stearic acid, 16-methylheptadecanoic acid, petroselinic acid were uniquely high in the rind of ROC22 (Figure 3A). α-Linolenic acid (which can't be synthesized by humans, is an essential fatty acid) have strong anti-arrhythmic properties and it is associated with cancer and cardiovascular disease (34). Similarly, linoleic acid has also promising pharmacological value especially related to cardiovascular, cardio-metabolic effects, and brain function (35). Most of the LPE compounds were high in the rind of ROC16 and ROC22 whereas some LPE such as LysoPE 16:1 and LysoPE 15:0 were higher in F172 and F134, respectively (Figure 3A).
Among the major glycerol esters such as gingerglycolipid A, gingerglycolipid B, and gingerglycolipid C were uniquely high in the rind of the F134 genotype whereas the YT93/159 has the lowest concentration of these glycerol esters (Figure 3B). These glycerol esters have strong anti-cancer effects and several other health-promoting properties (36). The PC (oxo-11:0/18:2) and PE (oxo-11:0/16:0) compounds were uniquely high in the rind of YT93/159 whereas the maximum level of choline alfoscerate [effective choline compound for Alzheimer's disease treatment, also it is the precursor of acetylcholine (20)] was observed in the of F134 followed by F172 (Figure 3B). Among all sugarcane genotypes, the ROC16 rind showed a higher level of LPC compounds followed by ROC22 (Figure 3B). In short, the rind of different cultivated sugarcane genotypes is a promising source of lipid compounds that have auspicious nutritive and pharmacological value; however, the amount of these compounds varies broadly between sugarcane genotypes. Commonly, sugarcane rind is utilized for electricity generation, wasted, or used as organic fertilizer; however, we have reported several lipid compounds in sugarcane rind that have significant nutritive, cosmetic, and pharmacological value. These compounds can be extracted from the sugarcane rind to fulfill the increasing nutritive demands and can be used in the pharmacological and cosmetic industries.
The PCA and HCA results together revealed that the F172 and YT71/210 genotypes have quite similar levels of lipid compounds because in the PCA and HCA analysis they stand very close to each other and made almost the same cluster (Figures 2, 3). Besides this, the HCA and PCA results represented that the F134, YT93/159 group and ROC22, ROC16 group bears their unique characteristics regarding lipids compounds (Figures 2, 3); however, the F134, YT93/159 group and ROC22, ROC16 group are quite similar regarding fatty acid, LPE and LPC, PC, glycerol ester, and hexadecylsphingosine compounds in the first and second heat-map, respectively, (Figures 3A,B).
Sugarcane lipids biosynthesis pathway and transcriptomic results
The sugarcane lipid biosynthesis pathway is characterized (Figure 4). In sugarcane, the lipid compounds are biosynthesized through glycerophospholipid and pyruvate metabolism. From glycerophospholipid metabolism, glycerophospholipids are formed which biosynthesized phosphatidylcholine (PC) (Figure 4). By the activity of the TAG lipase (TGL4) enzyme, the PC is concerted into α-linolenic acid and linoleic acid which are famous omega-3 and −6 fatty acids, respectively (Figure 4). The α-linolenic acid (essential fatty acids) further biosynthesized complex fatty acids conjugates and fatty acyls such as 13(s)-HpOTrE and 12-Oxo-phytodienoic acid, respectively (Figure 4). These two fatty acids are major fatty acids of sugarcane (Figure 1) and they are precursors of several fatty acyls, fatty acids conjugates, and octadecanoid (Figure 4). The pyruvate metabolism produces malonyl-CoA which further biosynthesized (after several steps) diverse groups of saturated fatty acids such as stearic acid, myristic acid, dodecanoic acid (lauric acid), palmitic acid, and some saturated fatty acids such as palmitoleic acid (Figure 4). These all saturated and unsaturated fatty acids are considered phytocompounds and their presence restrict microbial activity (18). From palmitic acid, palmitaldehyde is formed by the fatty acid degradation pathway (Figure 4).
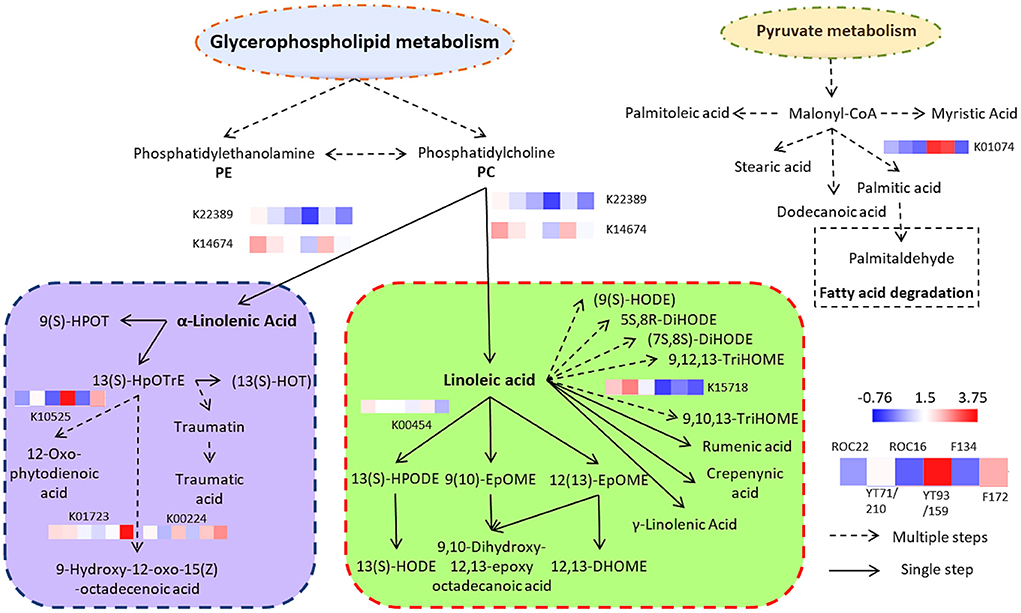
Figure 4. Complex sugarcane lipids biosynthesis pathway (thru Glycerophospholipid and Pyruvate metabolism) and expression of DEGs in the rind of six sugarcane genotypes. Each expression level is the mean of three biological repeats (the raw transcriptomic data is represented in Supplementary File 1).
The transcriptomic results revealed that eight genes were differentially expressed in the rind of six sugarcane genotypes associated with the α-linolenic acid and linoleic acid pathway (Figure 4). The α-linolenic acid and linoleic acid biosynthesis genes such as K22389 and K14674 were highly expressed in the rind of ROC22 followed by F134 whereas the YT93/159 revealed the lowest expression of these genes (Figure 4; Supplementary File 1). The metabolic results also showed that the rind of ROC22 has the highest level of α-linolenic acid and linoleic acid (Figure 3). The palmitic acid biosynthesis gene K01074 was expressed higher in the rind of YT93/159 and F134 followed by ROC22 whereas the other genotypes showed the lowest expression of this gene (Figure 4). The HCA analysis determined that the palmitic acid level was highest in YT93/159 followed by F134 (Figure 3). The expressions of K22389, K14674, and K01074 genes are highly correlated with the metabolic results (Figures 3, 4) which will beneficial for further study on the biosynthesis pathways of lipids in sugarcane.
Therapeutic potential of lipids against SARS-CoV-2
Molecular docking is a widely used computational approach to study the ligand binding dynamics with the protein (37). Lipid compounds therapeutic potential was assessed through molecular docking. SARS-CoV-2 3CLpro was the target in this study (PDB ID: 6LU7). The 3CLpro is vital for SARS-CoV-2 replication and inhibition of this enzyme can help to fight against COVID-19 (26). The lipids' were docked against the 3D structure of 3CLpro (Figure 5A). Previous studies reported that Cys-His catalytic dyad (Cys145 and His41) is the vital residue to target the 3CLpro (26, 38). Therefore, only those compounds were selected that were making strong binding interactions with the Cys145 and His41 residues. Results revealed that two compounds (Gingerglycolipid C and 2-Linoleoylglycerol) as potential therapeutic candidates for SARS-CoV-2 (Figure 5). Gingerglycolipid C bounds with 3CLpro with −12.80 kcal/mol binding energy score and made strong hydrogen bindings with Cys145, His41, Ser46, and Met49 (Figure 5B). While, 2-Linoleoylglycerol bound with 3CLpro having a −10.78 kcal/mol binding energy score and made strong hydrogen bindings with Cys145, His163, and Glu166, and strong polar interaction with His41 together with other binding site residues (Figure 5C).
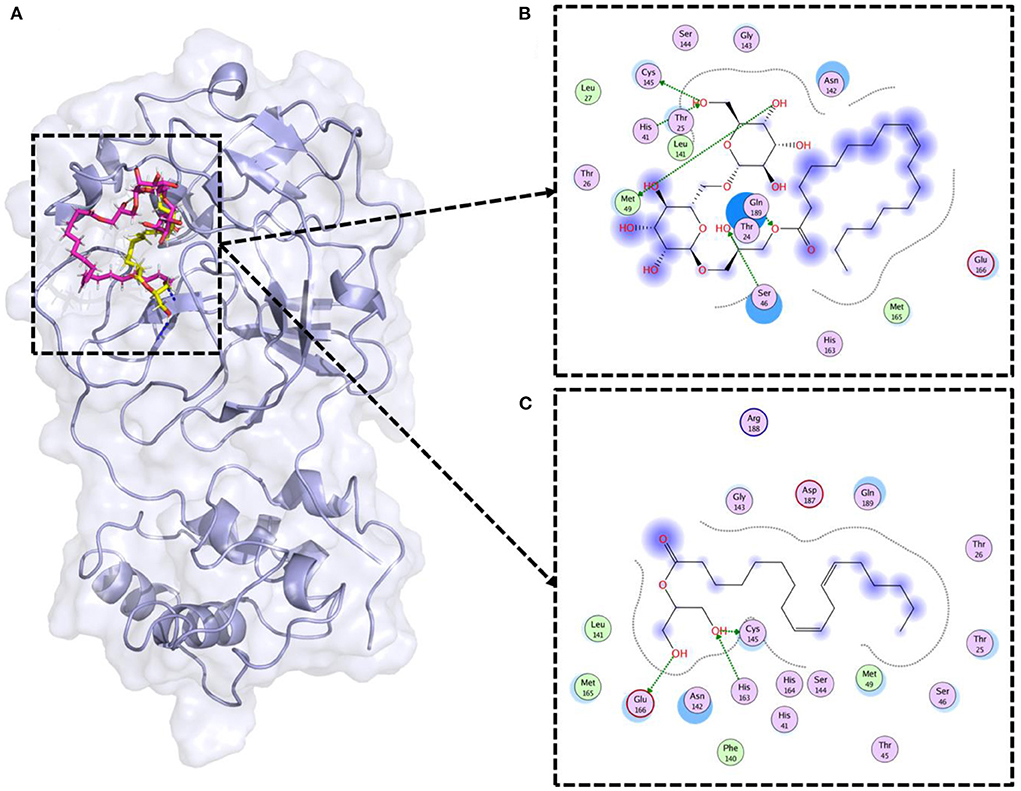
Figure 5. Molecular docking of lipid compounds against SARS-CoV-2 main protease. (A) 3D structural representation of top lipids bound 3CLpro (PDB ID: 6LU7). The pink color ligand is Gingerglycolipid C, while the yellow color ligand is 2-Linoleoylglycerol. (B) Binding mode of Gingerglycolipid C inside the binding pocket of 3CLpro. (C) Binding mode of 2-Linoleoylglycerol inside the binding pocket of 3CLpro.
Discussion
The rind of sugarcane is a potential source for primary and secondary metabolites that have wide-ranging functions and are significant for cosmetic and pharmacological value (6, 39, 40). Lipids are an essential constituent of all plant cells (14, 15), providing a hydrophobic barrier that allows the separation among the subcellular structures of the cell. Lipids also activate several enzymes such as stearyl CoA desaturase, β-hydroxybutyric dehydrogenase, and glucose-6-phosphatase. Recently, it is revealed that lipids play a central role in cell membranes remodeling during drought and cold stress in Arabidopsis thaliana (41). Compared with bagasse, the sugarcane rind is mostly used to make hardboards, burned to produce energy, or as animal feed (7). In this study, we have reported new lipids compounds first time in sugarcane rinds and highlighted novel pharmaceutical values of sugarcane lipids that are worth paying attention for future lipidomics studies (Table 1).
The fatty acids and their derivatives are known for antifungal, antibacterial, and antimalarial activity (18, 42). Myristic acid has antifungal activity against several pathogenic funguses such as Alternaria solani (causing early blight disease in tomato and potato plants), Penicillium italicum (cause citrus blue mold), Aspergillus niger, Fusarium oxysporum (Causing Pokkah Boeng disease in sugarcane), Emericella nidulans (cause disease in humans), Penicillium glabrum and Candida albicans (43). In plants, linoleic acid (polyunsaturated fatty acid) has strong antifungal activity against various pathogenic fungi (43). The rind of ROC22 contains the highest level of linoleic acid and α-linolenic acid (essential fatty acids) whereas the F172 and YT71/210 possess the lowest level of these compounds (Figure 3).
Undecylic Acid can inhibit the activity of Trichophyton rubrum (a dermatophytic fungus that causes athlete's foot and ringworm) and Saccharomyces cerevisiae (44). Palmitic acid also showed resilient activity against Alernaria spp., Cucumerinum lagenarium, Fusarium oxysporum, and Emericella nidulans (43). Additionally, palmitic acid, linoleic acid, stearic acid, and linoleic acid help to acclimatize the salt stress in cucumber (45). These all fatty acids are abundantly present in the rind and are considered as major fatty acids of sugarcane (Figure 1). Some lipid compounds have promising effects on different human diseases. The lipid glycerol ester compounds such as gingerglycolipid A, gingerglycolipid B, and gingerglycolipid C have strong anti-ulcer and anti-tumor activities (13). The 2-Linoleoylglycerol can inhibit the activity of anandamide and 2-arachidonolyglycerol in humans (46). The rind of F134 showed a high concentration of gingerglycolipid A, B, and C followed by YT71/210 (Figure 3) whereas the ROC16 rind possesses a high level of 2-Linoleoylglycerol compound (Figure 3).
Gingerglycolipid C belongs to the class of organic compounds known as glycosylmonoacylglycerols. These glycosylglycerols form an ester linkage between the fatty acyl chain and the glycerol moiety. Previously, it has been reported that gingerglycolipid C compound has anti-tumor and anti-ulcer activities (13). Previously, it is reported that 2-linoleoylglycerol is a partial agonist at the human cannabinoid type 1 receptor and can modulate the activity of the established endocannabinoid (46). Phytochemicals such as 2-linoleoylglycerol from Scolymus maculatus L. have broad-spectrum antioxidant and antimicrobial activities to cure different diseases (47). 2-Linoleoylglycerol exhibits strong activity against Salmonella typhimurium (Gram-negative bacterium), Staphylococcus aureus (staphylococcal bacteria), and Candida albicans (cause fungal infection in humans) (47). Cultivated sugarcane showed dissimilar levels of therapeutic and medicinal lipid compounds such as ROC22 have a high level of essential fatty acid ‘α-linolenic acid' whereas the F134 and ROC16 showed a high level of lipid-glycerol ester compounds (Figure 3). The F134 and ROC16 rinds are better for therapeutic compounds, however, ROC22 rinds have a high level of essential fatty acid required for maintaining normal human health. In short, the lipid compounds have promising health-promoting activities that can be utilized as therapeutic agents to protect humans from several chronic.
Earlier studies have explored the potential therapeutic roles of lipids in human health (48). To access the pharmacological value of sugarcane lipid compounds we have examined their activity against SARS-CoV-2. COVID-19 (coronavirus disease 2019) pandemic caused by positive-stranded RNA virus SARS-CoV-2 (49), infected >583 million with >6.4 million people died worldwide according to the World Health Organization COVID-19 dashboard (https://covid19.who.int). Few vaccines are approved for COVID-19 but due to rapid mutations and novel variants, they are not fully effective against all types of SARS-CoV-2 infections. Herein, our results revealed two lipid compounds from sugarcane rind, gingerglycolipid C and 2-linoleoylglycerol, as potential SARS-CoV-2 therapeutic agents. Both compounds showed strong binding interactions with 3CLpro of SARS-CoV-2 catalytic dyad residues Cys145 and His41. Gingerglycolipid C and 2-linoleoylglycerol compounds attained stable conformations inside the binding pocket of 3CLpro with docking scores of−12.80 kcal/mol and−10.78 kcal/mol. Both compounds also have good drug-like properties. Thus, our findings warrant further in vivo and in vitro studies to explore their therapeutic potential in detail to fight against COVID-19.
Conclusions
To conclude, we have reported 119 lipid compounds for the first time in the rind of six cultivated sugarcane genotypes. By using transcriptomic and metabolomic data, we have found that several genes associated with α-linolenic acid and linoleic acid biosynthesis were highly expressed in the rind of ROC22 whereas YT93/159 revealed the lowest expression. Among six sugarcane cultivars, the ROC22 and ROC16 showed a high level of lipid compounds. Moreover, we also found that the two lipid-glycerol ester compounds (2-linoleoylglycerol and gingerglycolipid C) have strong binding interactions with 3CLpro of SARS-CoV-2 and these compounds can be potentially utilized as therapeutic agents against SARS-CoV-2. The rind of ROC16 revealed a high level of 2-linoleoylglycerol whereas the F134 rind showed a high concentration of gingerglycolipid C. In short, sugarcane is a storehouse of promising lipid compounds that have auspicious cosmetic and pharmacological value. Further studies should be conducted to screen more lipid-glycerol ester compounds and investigate their action as a therapeutic against COVID-19 in humans. Moreover, simplified extraction protocols should be developed to harvest more valuable compounds from sugarcane rinds.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI with accession PRJNA824938.
Author contributions
MR: conceptualization, project administration, visualization, and roles/writing–original draft. MR, QA, and MT: data curation. MR, DW, MT, and MD: formal analysis. LW: funding acquisition. MR, MD, LM, and MT: investigation. MR, MD, QA, and MT: methodology. MR, SH, LM, MD, and LW: resources. MR and MT: software. LH and LW: supervision. MR, MT, and MD: validation. MT, LH, and LW: writing–review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Guangxi (2020GXNSFDA238027), a Postdoctoral Project from GXU, Guangxi Natural Science Foundation Youth Fund (2022GXNSFBA035665), and the National Natural Science Foundation of China (31771775 and 31171524).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.988249/full#supplementary-material
Supplementary File 1. Raw metabolomic and transcriptomic data.
Supplementary Figure 1. Representing the methodology used for the detection of different lipid compounds in the rind of six sugarcane varieties.
References
1. Garcia Tavares R, Lakshmanan P, Peiter E, O'Connell A, Caldana C, Vicentini R, et al. ScGAI is a key regulator of culm development in sugarcane. J Exp Bot. (2018) 69:3823–37. doi: 10.1093/jxb/ery180
2. Sindhu R, Gnansounou E, Binod P, Pandey A. Bioconversion of sugarcane crop residue for value added products–an overview. Renew Energy. (2016) 98:203–15. doi: 10.1016/j.renene.2016.02.057
3. de Oliveira Bordonal R, Carvalho JLN, Lal R, de Figueiredo EB, de Oliveira BG, La Scala N. Sustainability of sugarcane production in Brazil. a review. Agron Sustain Dev. (2018) 38:13. doi: 10.1007/s13593-018-0490-x
4. Wei YA, Li YR. Status and trends of sugar industry in China. Sugar Tech. (2006) 8:10–5. doi: 10.1007/BF02943735
5. Attard TM, McElroy CR, Rezende CA, Polikarpov I, Clark JH, Hunt AJ. Sugarcane waste as a valuable source of lipophilic molecules. Ind Crops Prod. (2015) 76:95–103. doi: 10.1016/j.indcrop.2015.05.077
6. Singh A, Lal UR, Mukhtar HM, Singh PS, Shah G, Dhawan RK. Phytochemical profile of sugarcane and its potential health aspects. Pharmacogn Rev. (2015) 9:45. doi: 10.4103/0973-7847.156340
7. Geng P, Fang Y, Xie R, Hu W, Xi X, Chu Q, et al. Separation of phenolic acids from sugarcane rind by online solid-phase extraction with high-speed counter-current chromatography. J Sep Sci. (2017) 40:991–8. doi: 10.1002/jssc.201600887
8. Zhang X, Jin Y, Wu Y, Zhang C, Jin D, Zheng Q, et al. Anti-hyperglycemic and anti-hyperlipidemia effects of the alkaloid-rich extract from barks of Litsea glutinosa in ob/ob mice. Sci Rep. (2018) 8:1–10. doi: 10.1038/s41598-018-30823-w
9. Rao MJ, Duan M, Wei X, Zuo H, Ma L, Tahir ul Qamar M, et al. LC-MS / MS -based metabolomics approach revealed novel phytocompounds from sugarcane rind having promising pharmacological value. J Sci Food Agric. (2022) 10:8–23. doi: 10.1002/jsfa.12030
10. Duarte-Almeida JM, Salatino A, Genovese MI, Lajolo FM. Phenolic composition and antioxidant activity of culms and sugarcane (Saccharum officinarum L.) products. Food Chem. (2011) 125:660–4. doi: 10.1016/j.foodchem.2010.09.059
11. Rao MJ, Duan M, Yang M, Fan H, Shen S, Hu L, et al. Novel Insights into anthocyanin metabolism and molecular characterization of associated genes in sugarcane rinds using the metabolome and transcriptome. Int J Mol Sci. (2022) 23:338. doi: 10.3390/ijms23010338
12. Rao MJ, Duan M, Yang M, Li M, Wang L. Sugarcane rind secondary metabolites and their antioxidant activities in eleven cultivated sugarcane varieties. Sugar Tech. (2022) 7:1–14. doi: 10.1007/s12355-021-01097-w
13. Yoshikawa M, Yamaguchi S, Kunimi K, Matsuda H, Okuno Y, Yamahara J, et al. Stomachic principles in ginger. III. An anti-ulcer principle, 6-gingesulfonic acid, and three monoacyldigalactosylglycerols, gingerglycolipids A, B, and C, from Zingiberis Rhizoma originating in Taiwan. Chem Pharm Bull. (1994) 42:1226–30. doi: 10.1248/cpb.42.1226
14. Nakamura Y. Plant phospholipid diversity: emerging functions in metabolism and protein–lipid interactions. Trends Plant Sci. (2017) 22:1027–40. doi: 10.1016/j.tplants.2017.09.002
15. Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill Jr AH, Murphy RC, et al. A comprehensive classification system for lipids. Eur J lipid Sci Technol. (2005) 107:337–64. doi: 10.1002/ejlt.200405001
16. Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CRH, Shimizu T, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. (2009) 50:S9–14. doi: 10.1194/jlr.R800095-JLR200
17. Ecker J, Liebisch G. Application of stable isotopes to investigate the metabolism of fatty acids, glycerophospholipid and sphingolipid species. Prog Lipid Res. (2014) 54:14–31. doi: 10.1016/j.plipres.2014.01.002
18. Kachroo A, Kachroo P. Fatty acid–derived signals in plant defense. Annu Rev Phytopathol. (2009) 47:153–76. doi: 10.1146/annurev-phyto-080508-081820
19. Ruffell SE, Müller KM, McConkey BJ. Comparative assessment of microalgal fatty acids as topical antibiotics. J Appl Phycol. (2016) 28:1695–704. doi: 10.1007/s10811-015-0692-4
20. Catanesi M, d'Angelo M, Antonosante A, Castelli V, Alfonsetti M, Benedetti E, et al. Neuroprotective potential of choline alfoscerate against β-amyloid injury: Involvement of neurotrophic signals. Cell Biol Int. (2020) 44:1734–44. doi: 10.1002/cbin.11369
21. Mashaghi S, Jadidi T, Koenderink G, Mashaghi A. Lipid nanotechnology. Int J Mol Sci. (2013) 14:4242–82. doi: 10.3390/ijms14024242
22. Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. (2010) 11:R14. doi: 10.1186/gb-2010-11-2-r14
23. Anders S, Huber W. Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg Ger Eur Mol Biol Lab. (2012) 10:F1000Res.
24. Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant. (2013) 6:1769–80. doi: 10.1093/mp/sst080
25. Fraga CG, Clowers BH, Moore RJ, Zink EM. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography– mass spectrometry, XCMS, and chemometrics. Anal Chem. (2010) 82:4165–73. doi: 10.1021/ac1003568
26. ul Qamar MT, Alqahtani SM, Alamri MA, Chen L-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. (2020) 10:313–9. doi: 10.1016/j.jpha.2020.03.009
27. Vilar S, Cozza G, Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. (2008) 8:1555–72. doi: 10.2174/156802608786786624
28. Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. (2016) 44:1202–13. doi: 10.1093/nar/gkv951
29. Mumtaz A, Ashfaq UA, ul Qamar MT, Anwar F, Gulzar F, Ali MA, et al. MPD3: a useful medicinal plants database for drug designing. Nat Prod Res. (2017) 31:1228–36. doi: 10.1080/14786419.2016.1233409
30. Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, et al. The protein data bank. Acta Crystallogr Sect D Biol Crystallogr. (2002) 58:899–907. doi: 10.1107/S0907444902003451
31. DeLano WL. Pymol: an open-source molecular graphics tool. CCP4 Newsl protein Crystallogr. (2002) 40:82–92.
32. Liao J, Huang H, Meusnier I, Adreit H, Ducasse A, Bonnot F, et al. Pathogen effectors and plant immunity determine specialization of the blast fungus to rice subspecies. Elife. (2016) 5:1–18. doi: 10.7554/eLife.19377
33. Bezerra TL, Ragauskas AJ. A review of sugarcane bagasse for second-generation bioethanol and biopower production. Biofuel Bioprod Bioref. (2016) 10:634–47. doi: 10.1002/bbb.1662
34. Rajaram S. Health benefits of plant-derived a -linolenic acid. Am J Clin Nutr. (2014) 3:1–3. doi: 10.3945/ajcn.113.071514
35. Marangoni F, Agostoni C, Borghi C, Catapano AL, Cena H, Ghiselli A, et al. Dietary linoleic acid and human health: focus on cardiovascular and cardiometabolic effects. Atherosclerosis. (2020) 292:90–8. doi: 10.1016/j.atherosclerosis.2019.11.018
36. Ramakrishnan R. Anticancer properties of Zingiber officinale–Ginger: a review. Int J Med Pharm Sci. (2013) 3:11–20.
37. Alamri MA, ul Qamar MT, Mirza MU, Alqahtani SM, Froeyen M, Chen L-L. Discovery of human coronaviruses pan-papain-like protease inhibitors using computational approaches. J Pharm Anal. (2020) 10:546–59. doi: 10.1016/j.jpha.2020.08.012
38. Alamri MA, Tahir Ul Qamar M, Mirza MU, Bhadane R, Alqahtani SM, Muneer I, et al. Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CLpro. J Biomol Struct Dyn. (2020) 1–13. doi: 10.20944/preprints202002.0308.v1
39. Rao MJ, Ahmed U, Ahmed MH, Duan M, Wang J, Wang Y, et al. Comparison and quantification of metabolites and their antioxidant activities in young and mature leaves of sugarcane. ACS Food Sci Technol 1. (2021) 3:362–73. doi: 10.1021/acsfoodscitech.0c00133
40. Rao MJ, Duan M, Wang J, Han S, Ma L, Mo X, et al. Transcriptomic and widely targeted metabolomic approach identified diverse group of bioactive compounds, antiradical activities, and their associated genes in six sugarcane varieties. Antioxidants. (2022) 11:1319. doi: 10.3390/antiox11071319
41. Tarazona P, Feussner K, Feussner I. An enhanced plant lipidomics method based on multiplexed liquid chromatography-mass spectrometry reveals additional insights into cold- and drought-induced membrane remodeling. Plant J. (2015) 84:621–33. doi: 10.1111/tpj.13013
42. Pohl CH, Kock JLF, Thibane VS. Antifungal free fatty acids: a review. Sci against Microb Pathog Commun Curr Res Technol Adv. (2011) 3:61–71.
43. Liu S, Ruan W, Li J, Xu H, Wang J, Gao Y, et al. Biological control of phytopathogenic fungi by fatty acids. Mycopathologia. (2008) 166:93–102. doi: 10.1007/s11046-008-9124-1
44. Peres NTA, Cursino-Santos JR, Rossi A, Martinez-Rossi NM. In vitro susceptibility to antimycotic drug undecanoic acid, a medium-chain fatty acid, is nutrient-dependent in the dermatophyte Trichophyton rubrum. World J Microbiol Biotechnol. (2011) 27:1719–23. doi: 10.1007/s11274-010-0613-2
45. Radhakrishnan R, Lee I-J. Regulation of salicylic acid, jasmonic acid and fatty acids in cucumber (Cucumis sativus L) by spermidine promotes plant growth against salt stress. Acta Physiol Plant. (2013) 35:3315–22. doi: 10.1007/s11738-013-1364-0
46. Lu L, Williams G, Doherty P. 2-Linoleoylglycerol is a partial agonist of the human cannabinoid type 1 receptor that can suppress 2-arachidonolyglycerol and anandamide activity. Cannabis Cannabinoid Res. (2019) 4:231–9. doi: 10.1089/can.2019.0030
47. Abu-Lafi S, Rayan M, Masalha M, Abu-Farich B, Al-Jaas H, Abu-Lafi M, et al. Phytochemical composition and biological activities of wild Scolymus maculatus L. Medicines. (2019) 6:53. doi: 10.3390/medicines6020053
48. Li Y, Wang C, Shang H, Hu R, Fu H, Xiao X. A high-throughput and untargeted lipidomics approach reveals new mechanistic insight and the effects of salvianolic acid B on the metabolic profiles in coronary heart disease rats using ultra-performance liquid chromatography with mass spectrometry. RSC Adv. (2020) 10:17101–13. doi: 10.1039/D0RA00049C
Keywords: sugarcane rind, metabolomics approach, lipidomics, transcriptomic, pharmacological value, COVID-19
Citation: Rao MJ, Tahir ul Qamar M, Wang D, Ali Q, Ma L, Han S, Duan M, Hu L and Wang L (2022) A high-throughput lipidomics and transcriptomic approach reveals novel compounds from sugarcane linked with promising therapeutic potential against COVID-19. Front. Nutr. 9:988249. doi: 10.3389/fnut.2022.988249
Received: 07 July 2022; Accepted: 11 August 2022;
Published: 02 September 2022.
Edited by:
Iftikhar Ali Khan, Shenzhen University, ChinaReviewed by:
Muhammad Irfan, University of Sargodha, PakistanValeria Villanova, University of Verona, Italy
Copyright © 2022 Rao, Tahir ul Qamar, Wang, Ali, Ma, Han, Duan, Hu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingqiang Wang, bHF3YW5nQGd4dS5lZHUuY24=; Lihua Hu, bGlodWFodUBneHUuZWR1LmNu
 Muhammad Junaid Rao
Muhammad Junaid Rao Muhammad Tahir ul Qamar
Muhammad Tahir ul Qamar Dongxin Wang
Dongxin Wang Qurban Ali
Qurban Ali Li Ma1
Li Ma1 Mingzheng Duan
Mingzheng Duan Lingqiang Wang
Lingqiang Wang