- 1Department of Microbiology, Quaid-i-Azam University, Islamabad, Pakistan
- 2National Agricultural Research Centre (NARC), National Institute for Genomics and Advanced Biotechnology (NIGAB), Islamabad, Pakistan
- 3Department of Biotechnology, University of Sialkot, Sialkot, Pakistan
- 4Department of Biology, College of Science, University of Hail, Hail, Saudi Arabia
- 5Molecular Diagnostics and Personalized Therapeutics Unit, University of Ha’il, Ha’il, Saudi Arabia
- 6Metabolic Research Group, Faculty of Science, School of Environmental and Life Sciences, The University of Newcastle, University Drive, Callaghan, NSW, Australia
- 7Department of Pharmacy, Quaid-i-Azam University, Islamabad, Pakistan
- 8Gomal Center of Biochemistry and Biotechnology, Gomal University, Dera Ismail Khan, Pakistan
- 9Genomics Research Hub (GENOMAC HUB), Ogbomoso, Nigeria
This review article addresses the strategic formulation of human probiotics and allows the reader to walk along the journey that metamorphoses commensal microbiota into target-based probiotics. It recapitulates what are probiotics, their history, and the main mechanisms through which probiotics exert beneficial effects on the host. It articulates how a given probiotic preparation could not be all-encompassing and how each probiotic strain has its unique repertoire of functional genes. It answers what criteria should be met to formulate probiotics intended for human use, and why certain probiotics meet ill-fate in pre-clinical and clinical trials? It communicates the reasons that taint the reputation of probiotics and cause discord between the industry, medical and scientific communities. It revisits the notion of host-adapted strains carrying niche-specific genetic modifications. Lastly, this paper emphasizes the strategic development of target-based probiotics using host-adapted microbial isolates with known molecular effectors that would serve as better candidates for bioprophylactic and biotherapeutic interventions in disease-susceptible individuals.
Introduction
The history of probiotics dates back over a 100 years, when Elie Metchnikoff, a Nobel laureate, theorized that “health and longevity could be achieved by manipulating intestinal microflora, i.e., replacing harmful microbes with beneficial microbes” (1–5). Since then, probiotics have gained substantial attention from researchers, clinicians, and the general public alike (6–8). The term probiotic originates from the Greek language and translates to “for life” (9).
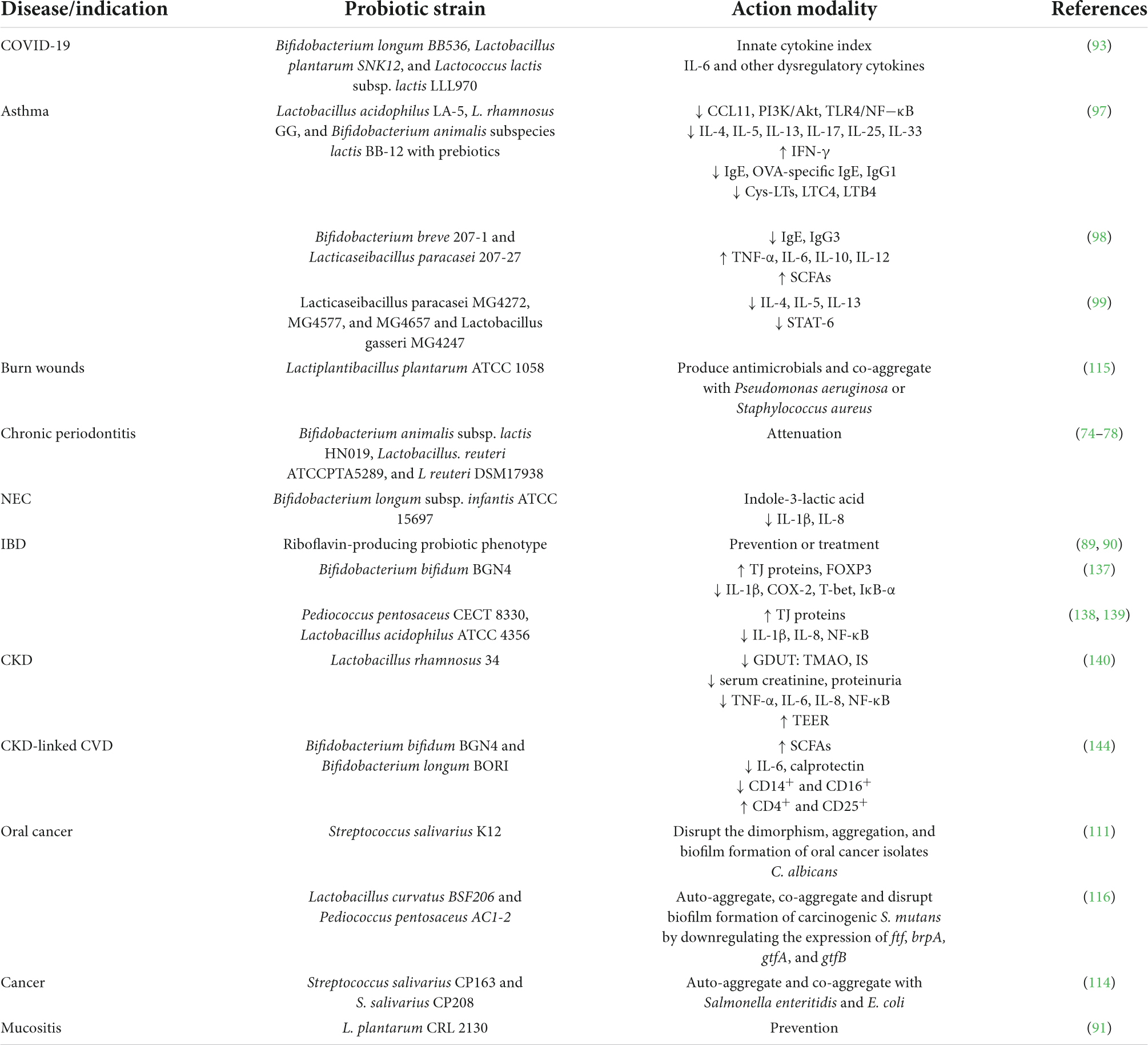
A brief history of probiotics is given in Figure 1 (10–12). The evolution of probiotics began around 1856–1864 when French microbiologist Louis Pasteur proved that food spoilage was caused by microorganisms (13–15). Following the discovery of food spoilage agents, around 1873, British Scientist Joseph Lister isolated Streptococcus lactis (now known as Lactococcus lactis) from rancid milk (16–19). Meanwhile French pediatrician Henry Tissier isolated Bacillus bifidum communis (now known as Bifidobacterium bifidum) in 1889 (20) and observed that Bifidobacteria were dominant among the microflora of breast-fed infants (12, 21, 22). Shortly after the discovery of Bifidobacteria, around 1900 Austrian pediatrician Ernst Moro isolated a gram-positive bacterium and characterized it as Bacillus acidophilus (now known as Lactobacillus acidophilus) (23–25); while Bulgarian scientist Stamen Grigorov isolated Lactobacillus bulgaricus (aka Lactobacillus delbrueckii subsp. bulgaricus) from Bulgarian yogurt in 1907 (10, 26, 27). The year 1923 marked the isolation of the first probiotic yeast, Saccharomyces boulardii from lychee and mangosteen by French scientist Henri Boulard (28, 29); while in 1930, a Japanese scientist Minoru Shirota isolated Lactobacillus casei strain shirota (now known as Lacticaseibacillus casei) from the human intestine and developed the first probiotic fermented product called Yakult (30–32).
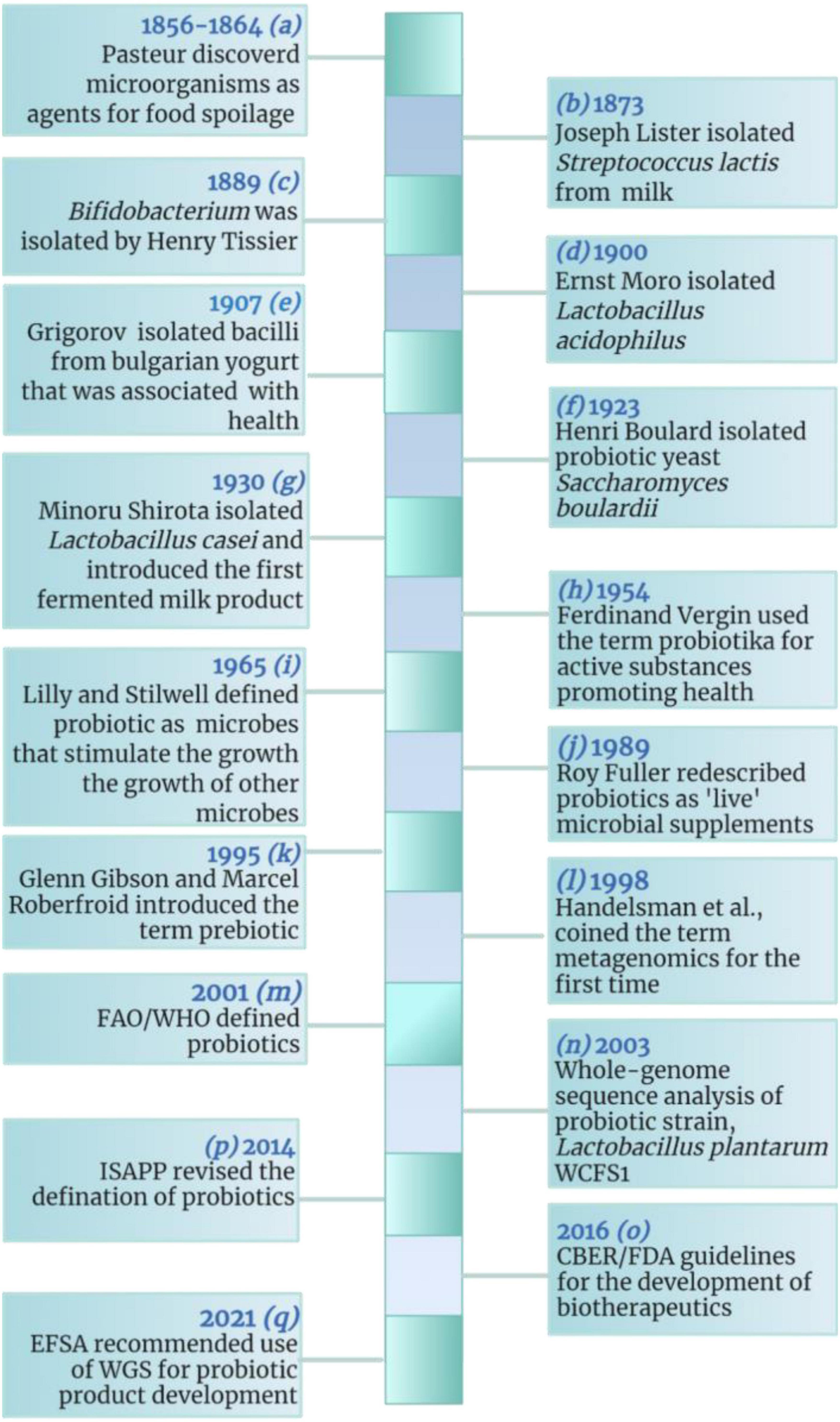
Figure 1. A brief history of probiotics. CBER, Center for Biologics Evaluation and Research; EFSA, European Food Safety Authority; FAO, Food and Agriculture Organization; FDA, Food and Drug Administration; ISAPP, International Scientific Association for Probiotics and Prebiotics; WHO, World Health Organization. The figure was drawn with BioRender.
In 1954, German scientist, Ferdinand Vergin used the term probiotika to describe “active substances that are essential for health” and he also emphasized the adverse effects of antibiotics on useful intestinal microbiota (33, 34). Later, in 1965, American scientists Lilly and Stillwell coined the term probiotic to describe “substances produced by one microorganism that stimulate the growth of another” (35). While in 1974, Parker described probiotics as “organisms and substances which contribute to intestinal microbial balance” (36–38). Then probiotics were redefined by Roy Fuller by 1980 as, “a live microbial feed supplement which beneficially affects the host by improving its intestinal microbial balance” (10). The concept of prebiotics was introduced by English scientists Gibson and Roberfroid in 1995 (39–41), while American researchers Handelsman et al. coined the term metagenomics in 1998 (42). Later, in 2003, whole-genome of Lactobacillus plantarum WCFS1 (now Lactiplantibacillus plantarum) was sequenced and analyzed (32, 43).
In 2014, the International Scientific Association for Probiotics and Prebiotics (ISAPP), defined probiotics as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (44); while in 2016, Center for Biologics Evaluation and Research/Food and Drug Administration (CBER/FDA) produced guidelines for the development of live biotherapeutic products. Recently, European Food Safety Authority (EFSA) recommended the use of whole-genome sequence (WGS) analysis for microorganisms intended to be used in the food chain (45).
Despite the ever-increasing gamut of probiotic-based products, microbiome-targeted therapies, and associated literature, the efficacy of probiotics in many disease indications remains a conundrum. In the present article, we aim to address this issue by highlighting the importance of the host-adapted strains while strategically formulating probiotics intended for human use.
How do probiotics exert their beneficial effects?
The action modality of probiotics is regulated by intricate and extensive molecular mechanisms that have not been well documented. However, various mechanisms have been hypothesized to explain the beneficial effects rendered by probiotics (46–49) including improvement of digestion, inhibition of pathogenic bacteria in the gastrointestinal tract, reduction of blood pressure and high blood glucose levels, improvement of intestinal health, reduction in serum cholesterol levels, toxin degradation, production of cofactors and vitamins, upregulation of the immune system, anti-inflammatory potential, and prevention of tumors and cancers (6–8, 11, 31, 46, 50–54). Probiotics-mediated molecular strategies involved in promoting health-associated phenotype and alleviating disease-associated phenotype are summarized in Tables 1, 2, respectively.
Competition for nutrients and prebiotics
One of the mechanisms imparted by probiotics to maintain health is to compete for nutrients that would otherwise be consumed by enteropathogenic microorganisms (55–57). Strain et al. reported that, unlike the distantly related species, the commensal symbionts and their phylogenetically related pathobionts often compete for similar microbial resources. These metabolites include vitamins, trace elements, carbon substrates, and secondary bile acids (58). Clostridium difficile is a potentially pathogenic bacteria that rely on monosaccharides for nutrition and growth. A study investigated the colonization resistance of C. difficile in a continuous flow culture model colonized with the mouse cecal flora. It was reported that an unidentified organism was highly efficient in taking up the monomeric glucose molecules, N-acetylneuraminic acid, and N-acetylglucosamine, thus limiting the growth of C. difficile (59). Similarly, Bifidobacterium adolescentis S2-1 outcompetes Porphyromonas gingivalis by readily utilizing vitamin K and other growth factors thus averting infection through nutrient limitation (60).
Augmenting the research mentioned above, Elli et al. reported the role of Lactobacilli in iron acquisition. Iron constitutes one of the micronutrients fundamental for the growth and functioning of most bacteria. Probiotic bacteria like L. delbrueckii and L. acidophilus bind ferric hydroxide at their cell surface, rendering thus, iron supply unavailable for pathobionts and pathogens (61).
Production of antimicrobials
Another possible mechanism employed by probiotics for maintaining the health phenotype is the synthesis of antimicrobial compounds (62). Different species of Bifidobacteria and Lactobacilli produce various types of bacteriocins and other antimicrobial compounds that inhibit the proliferation of pathogens. Bacteriocins are defined as “compounds produced by bacteria that have a biologically active protein moiety and a bactericidal action” (63); these antibacterial peptides are small, cationic, and comprised of 30–60 amino acid molecules (64). Other biologically active antimicrobial compounds synthesized by Lactic acid bacteria (LAB) to inhibit the harmful microflora include short-chain fatty acids (SCFAs), diacetyl, and hydrogen peroxide (46, 65). SCFAs produced by gut microbes have long been associated with health benefits to the host. The commensal microbiota metabolizes indigestible carbohydrates from high-rich fiber diets into SCFAs. It has been reported elsewhere (66) that Bifidobacterium longum subsp. infantis 157F produces acetate that plays a significant part in the defense surveillance of the host intestinal epithelial cells (IECs). It prevents the translocation of E. coli O157:H7-induced shiga toxin from the gut lumen to blood thereby, protecting the host against the E. coli O157:H7 shiga toxin-induced death. Furthermore, an ATP-binding-cassette (ABC)-type promoter that facilitates the uptake of carbohydrates has been found that upregulates the acetate production by bifidobacteria strains (66).
Lactobacillus reuteri has been reported to protect keratinocytes by producing antimicrobials to restrict the growth of Staphylococcus aureus (67). Similarly, L. pentosus, L. rhamnosus I, L. paracasei subsp. paracasei, L. delbrueckii subsp. lactis I, and Streptococcus uberis II are reported to produce bacteriocin-like inhibitory substances (BLISs) against clinical isolates of Candida albicans and non-Candida albicans from women with vulvovaginitis (68). While whole-genome sequence analysis of a lung isolate Lactobacillus sakei JD10 reports a gene encoding bacteriocin carnocin that inhibit the proliferation of pathogenic bacteria (69). Another interesting study reported that Lactiplantibacillus plantarum PUK6 produces multiple bacteriocins including plantaricin NC8, A, and EF (70); while Diale et al. reported the production of multiple BLISs from Bacillus paranthracis MHSD3 with anti-proliferative potential against Staphylococcus aureus, S. epidermidis, S. saprophyticus, and Escherichia coli (71).
Bioconversion of substrates
Another interesting feat of probiotic microorganisms is their ability to modify substrates to the host’s benefit (46–49). Probiotics outcompete enteropathogens like Salmonella typhi, Helicobacter pylori, Entamoeba histolytica, Clostridium difficile, and E. coli by lowering the gut pH, this is brought by fermentation of substrates that result in the production of a large variety of organic acids and volatile fatty acids (72). The probiotic-mediated bioconversion (PMB) of metabolites is reported to have antimicrobial, anticancer, anti-inflammatory, and antioxidant activities. Furthermore, recent studies reported the positive effects of PMB of metabolites in periodontitis (73), the use of probiotic Bifidobacterium animalis subsp. lactis HN019, Lactobacillus reuteri ATCCPTA5289, and L. reuteri DSM17938 [now Limosilactobacillus reuteri (32)] alleviated chronic periodontitis (74–78).
Another interesting example of probiotic bioconversion is the biosynthesis of seleno amino acids from inorganic selenium. Saccharomyces boulardii and Saccharomyces cerevisiae are reported to metabolize selenate into selenomethionine and selenocysteine (79, 80). While other recent studies reported probiotic candidates including Streptococcus thermophilus ST8, Enterococcus faecium ST3, Lactiplantibacillus plantarum LP6 & LP9, Lactobacillus acidophilus LP16-2, Bifidobacterium animals ST20 as γ-aminobutyric acid (GABA) producing strains (81), and the use of probiotics for the biodegradation of mycotoxins (82).
Production of growth-promoting substrates
The probiotic microorganisms impart several health benefits to the host including the production of growth-promoting substrates (6–8, 11, 31, 46). Folate is required for DNA methylation, repair, and division, while its deficiency results in serious disorders (83). Lactiplantibacillus plantarum is reported to produce folate in the presence of para-aminobenzoic acid (pABA) (84) and another study reported the consumption of Bifidobacteria strains, B. adolescentis, and B. pseudocatenulatum resulted in a significant increase of folate concentration in fecal samples from 23 healthy individuals (85). Moreover, Liu et al. reported the potential of Latilactobacillus sakei LZ217 for folate production (83).
Riboflavin plays a crucial role in numerous cellular metabolic processes via its participation in oxidation-reduction reactions (86); its active forms flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) are used in pharmaceutical, feed, and food industries. LAB strains harboring the potential to produce riboflavin have been reported (87). A study reported screening of 60 Lactobacilli isolates for the presence of riboflavin biosynthesis genes through a polymerase chain reaction (PCR)-based technique; the Lactobacillus delbrueckii subsp. bulgaricus, L. mucosae, L. fermentum, and L. plantarum carried a complete set of riboflavin structural genes, while the other isolates carried partial genes or the absence of related genes. The Lactobacilli carrying a partial set of riboflavin structural genes were unable to grow in the riboflavin-deficient culture medium (86). In a separate study, a trinitrobenzene sulfonic (TNBS)-induced mouse model was used to study the effect of soy milk fermented with riboflavin-producing L. plantarum CRL 2130. It dramatically reduced TNBS-induced intestinal damage (88). Inflammatory bowel diseases (IBD) could be prevented or treated using the Riboflavin-producing phenotype (89, 90). Furthermore, the L. plantarum CRL 2130 could be beneficial in preventing mucositis during cancer treatment while having no effect on the primary treatment (91).
Immune stimulation
Immune stimulation is another aspect of probiotic modality; probiotics elicit immunomodulatory activity by downregulating inflammatory responses, activation of NK (Natural Killer) cells and DCs (Dendritic cells), modulating the expression of TLRs (Toll-like Receptors), secretion of specific Immunoglobulin A (IgA), regulating the proliferation of lymphocytes, and balancing the ratio of T-helper (Th1/Th2) cells (92). While the probiotic-induced immunomodulatory mechanisms are being discerned, their use as a biotherapeutic in preclinical and clinical trials is being evaluated. One such study evaluated the immunomodulatory potential of Bifidobacterium longum BB536, Lactobacillus plantarum SNK12, and Lactococcus lactis subsp. lactis LLL970 against COVID-19 using in vitro cytokine response assay followed by a single-arm, double-blind, prospective trial. Out of the three tested probiotics, L. plantarum SNK12 significantly increased the innate cytokine index and decreased the plasma levels of IL-6, a major constituent responsible for complex immune dysregulation in COVID-19 patients. The study concluded that L. plantarum SNK12 mimics the blood cytokine profile present in early viral infection and demonstrates a high immunomodulatory ability and could be taken as a bioprophylactic for COVID-19 (93).
The structural components of LAB are of great interest for bioprophylactics and biotherapeutic approaches as they exert immunostimulatory effects and can be used in lieu of antibiotics, as vaccine adjuvants, or as cognitive enhancements (94). The IgA inducing potential of Apilactobacillus kosoi 10HT was compared to twenty-nine other LAB strains using murine Peyer’s patch cells as a model elsewhere (95). It was observed that the species belonging to the genus Apilactobacillus (A. kosoi 10HT, A. kunkeei JCM16173T, and A. apinorum JCM30765T) displayed significantly higher IgA-inducing potential than the other LAB isolates. Subsequently, lipoteichoic acids (LTAs) were purified from the three immunostimulatory Apilactobacillus strains, and their IgA-inducing potential was compared with LTAs from Lacticaseibacillus rhamnosus GG and Lactiplantibacillus plantarum JCM1149T; while the significance of LTA lies in the fact that it is an important immunostimulatory constituent of Gram-positive bacterial cell wall as it interacts with host’s PRRs (Pattern Recognition Receptors) and elicits an immune response. The results disclosed that LTAs from Apilactobacillus spp. had significantly higher immunostimulatory potential than the other two probiotic strains (L. rhamnosus GG and L. plantarum JCM1149T). Later, structural analysis of the LTA glycolipid via matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) revealed that LTA from A. kosoi 10HT carried dihexosyl glycerol, whereas other LAB strains carried trihexosyl glycerol. This dissimilarity in LTA structure could be linked to different IgA-inducing capacities (95).
As probiotics and prebiotics have immunomodulatory effects recent research is investigating their potential to restore asthma associated-immune system dysregulation and alleviate allegro-inflammatory reactions (96). One such study investigated the activity of Lactobacillus acidophilus LA-5, L. rhamnosus GG, and Bifidobacterium animalis subspecies lactis BB-12 in regulating the toll-like receptor 4/nuclear factor-kappa light chain enhancer of activated B cells (TLR4/NF-κB) pathway and acute airway inflammation (97). The prebiotics used were FOS (fructo-oligosaccharides) and GOS (galacto-oligosaccharides) and the allergic asthma model of BALB/c mice was used. The results indicated that probiotics have the potential to downregulate the gene expression of the Chemokine (C-C motif) ligand (CCL11) and TLR4. Probiotics also controlled airway hyperresponsiveness, levels of immunoglobulins, eosinophil infiltration to perivascular and bronchoalveolar lavage fluid (BALF), Glutamic pyruvic transaminase, interleukin-17 (IL-17), mucus secretion, Eosinophil peroxidase activity (EPO), and goblet cell hyperplasia. While probiotics and prebiotics regulated gene expression of AKT (Protein kinase B), MUC5a (Mucin 5a), levels of cytokines (IL-4, 5, 13, 25, and 33), leukotrienes, NLRP3 (NOD-like receptor family pyrin domain containing 3), peribronchial inflammation, MyD88 (Myeloid differentiation primary response 88), NF-κB and increase in IL-38 gene expression. Whereas prebiotic controlled PI3K (Phosphoinositide 3-Kinase) gene expression. In short, the study concluded that probiotics-prebiotics-induced tolerance could attenuate allegro-inflammatory reactions (97).
Liang et al. investigated the anti-allergy effect of two probiotic strains L. paracasei 207-27 and B. breve 207-1 compared with L. rhamnosus GG using an allergic mouse model. It was seen that L. paracasei 207-27 possess a higher potential for downregulating serum immunoglobulin E (IgE) levels than L. rhamnosus GG while B. breve 207-1 displayed a weak anti-allergic effect. Furthermore, both probiotic strains reduced serum immunoglobulin G3 (IgG3) levels and induced tumor necrosis factor-alpha (TNF-α), IL-6, IL-10, and IL-12. In addition, L. paracasei 207-27 remarkedly upregulated the level of cecal SCFAs in allergic mouse models (98). Another recent study reported the potential of Lacticaseibacillus paracasei MG4272, MG4577, and MG4657 and Lactobacillus gasseri MG4247 to attenuate allergic inflammation by downregulating the expression of IL-4, IL-5, IL-13, and STAT6 phosphorylation (99).
Auto-aggregation, biofilm formation, and co-aggregation
The natural attachment and immobilization of microorganisms to abiotic or biotic surfaces in a submerged environment is termed as a biofilm. The presence of a high density of microbial cells in a biofilm ensures that microbes can withstand challenges such as osmotic stress, antibiotic stress, pH change, or starvation (100–105). LAB have been shown to form biofilms while displaying antagonistic activities against a variety of foodborne pathogens (106, 107); auto-aggregation of the bacterial cells and co-aggregation of bacteria with pathogens play a significant role in preventing pathogens from infecting, adhering to, and colonizing host cells, especially in the gastrointestinal tract (108). A study exploited the potential role of LAB biofilms and their naturally secreted compounds to control the biofilms of foodborne pathogens (109). It reported that isolates Enterococcus faecium C4, Enterococcus faecium F6, Pediococcus pentosaceus C3, Pediococcus pentosaceus C6, and Pediococcus pentosaceus C8 could auto-aggregate as well as co-aggregate with Bacillus cereus, Escherichia coli, and Salmonella typhimurium. Among all the tested strains, Enterococcus faecium C4 developed a significant amount of exopolysaccharide (EPS) and a biofilm with the highest cell density (109). It has been previously reported elsewhere (110) that EPS facilitates the colonization of probiotic bacteria on the intestinal epithelium.
In a study investigated the effect of probiotic Streptococcus salivarius K12 on pathogenic Candida albicans strains including oral cancer isolate ALC3, an acquired immunodeficiency syndrome (AIDS) isolate ALC2, and C. albicans ATCC MYA-4901. It was revealed that the auto-aggregation of C. albicans strains was categorized as high, whereas co-aggregation of C. albicans strains with S. salivarius K12 was categorized as low. S. salivarius K12 also disrupted the biofilm formation of the pathogenic yeast strains, evident by a significant decrease in the total cell count of C. albicans when co-cultured with S. salivarius K12 as compared with monocultured C. albicans. So, S. salivarius K12 efficiently disrupted the dimorphism, aggregation, and biofilm formation of C. albicans (111). In a separate study, probiotic yeast Saccharomyces boulardii and pathogenic Candida strains including C. glabrata ZIM 2369, C. krusei ATCC 6258, C. albicans ATCC 10261, and C. glabrata ZIM 2382 were evaluated for their potential to auto-aggregate and co-aggregate. It was revealed that all yeast strains harbored the potential to auto-aggregate and it increased significantly after 24 h of incubation, while co-aggregation between the probiotic yeast and the pathogenic yeasts was subjected to strain-specificity. However, it was indisputable that Saccharomyces boulardii significantly impeded the aggregation of all the tested pathogenic yeast strains except C. glabrata ZIM 2382 (112).
In another study, seven human breast milk-isolated LAB were characterized for their auto-aggregation, and co-aggregation potential with pathogenic Shigella flexneri and Enterococcus faecalis. Among the isolates, Levilactobacillus gastricus BDUMBT09 (MT774596), L. brevis BDUMBT11 (MW785062), L. brevis BDUMBT08 (MT673657), L. casei BDUMBT13 (MW785178), and L. casei BDUMBT12 (MW785063) demonstrated aggregation potential which increased with further incubation whereas L. paracasei BDUMBT10 (MT775430) and Brevibacillus brevis M2403 (MK371781) did not show such potential. Interestingly, all the isolates demonstrated better co-aggregation with Enterococcus faecalis as compared to Shigella flexneri (113). LAB isolates from human colostrum have been investigated for their bioprophylactic and biotherapeutic potential in another recent study. It reported Streptococcus salivarius CP163 and S. salivarius CP208 exhibit a higher anti-cancer potential among the 218 isolates. Both these probiotic bacteria displayed potential for auto-aggregation and co-aggregation with pathogens Salmonella enteritidis and E. coli (114).
The antibacterial potential of Lactiplantibacillus plantarum ATCC 1058 was evaluated by using in vitro simulated wound fluid (SWF) and in vivo wound models infected with two main burn wound-associated pathogens, i.e., Pseudomonas aeruginosa or Staphylococcus aureus. Results concluded both these pathogenic bacteria were susceptible to the L. plantarum-produced antimicrobial compounds and co-aggregation (115). Luan et al. evaluated the antimicrobial potential of Lactobacillus curvatus BSF206 and Pediococcus pentosaceus AC1-2 against the oral pathogen Streptococcus mutans (116). Both probiotic strains exhibited auto-aggregation and co-aggregation potential, these properties facilitate strains to adhere and colonize gingival epithelial cells and impede carcinogenic S. mutans from oral tissues. Moreover, the study reported that L. curvatus BSF206 and P. pentosaceus AC1-2 efficiently prevented S. mutans biofilm by downregulating the expression of key genes ftf, brpA, gtfA, and gtfB associated with biofilm formation (116).
Maintenance of barrier integrity
The gut-mucosal barrier consists of the mucus layer, IECs, lamina propria, i.e., loose connective tissue containing the immune cells at the sub-epithelial level, and intestinal microbiota. Probiotics positively modulate all these levels and thereby inhibit gut barrier permeability (117, 118); “leaky gut” is associated with the pathogenesis of several disorders (119, 120). Different underlying mechanisms have been proposed through which probiotics induce favorable modulations to maintain gut barrier integrity (121); one such mechanism includes probiotic bacteria competing for the cellular adhesion sites. The enteropathogens must adhere to these epithelial adhesion sites in the gastrointestinal tract (GIT) to colonize effectively (122) but the probiotic bacteria efficiently adhere to these sites thus acting as “colonization barriers” thereby effectively inhibiting the pathogens from adhering to the IECs (123, 124). A study reported the use of Lactobacillus rhamnosus strain GG and Lactobacillus plantarum 299 v to demonstrate this effect. Both of these strains were able to prevent adherence of Escherichia coli to human colon cells (125).
The probiotics use several other mechanisms for the maintenance of the gut mucosal barrier (124, 126) including improved trans-epithelial electric resistance (TEER), increase in the levels of butyrate, upregulation of tight-junction (TJ) proteins (ZO-1, occludin, and claudin-1), elevated mucus secretion (by upregulation of MUC1, MUC2, and MUC3 in epithelial cells of colons) and as well as modulation of gut microbiota (127). These physiological changes are brought by molecular effectors secreted by probiotic bacteria, for instance, L. plantarum produces 10-hydroxy-cis-12-octadecenoic acid (HYA) which has been shown to repress the downregulation of ZO-1 (zonula occludin), occludin, and claudin-1 and TJ permeability induced by TNF-α and interferon-gamma (IFN-γ) via regulation of TNF receptor 2 expression by the G-protein-coupled receptor (GPR)-40/mitogen-activated kinase (MEK)/extracellular-signal-regulated kinase (ERK) dependent pathway (127).
The potential role of Bifidobacterium dentium N8 in protecting the gut barrier was examined by using human colorectal adenocarcinoma cells (Caco-2) as a model of the intestinal epithelial barrier. Lipopolysaccharide (LPS) was used to increase the permeability of the cell line. B. dentium N8 suppressed the permeability of Caco-2 and elevated the TEER by upregulating the expression of TJ proteins (ZO-1, occludin, and claudin) and downregulating the expression of IL-1β, IL-6, and TNF-α. Thus, attenuating inflammation and improving the gut barrier integrity (128). Another study reported the role of surface-layer proteins (Slps) from different Lactobacillus strains in maintaining the robustness of the gut barrier. Four Slps significantly prevented the reduction of TEER and TNF-α-induced permeability in Caco-2 monolayers; interestingly, TNF-α-induced downregulation of ZO-1 and occludin was also ameliorated. Moreover, these four Slps also attenuated TNF-α-induced elevated levels of IL-8 and NF-κB activation (129).
Anti-inflammatory potential
Many studies have demonstrated that probiotics regulate inflammatory pathways, stimulate the expression of immune-related genes, and modulate the levels of immunological markers (130, 131). Recent studies have investigated the molecular mechanisms through which probiotics induce an anti-inflammatory response. One such study established a protective mechanism with breastmilk/probiotic use in necrotizing enterocolitis (NEC) (132); it is a serious gastrointestinal disease in premature infants caused by invasion of the pathogenic bacteria, the subsequent inflammation in the colon expedites to perforation and gut leakage, which leads to overwhelming infection and death. Whilst prevention is challenging, the best protection comes from the ingestion of maternal-expressed milk along with probiotics (133, 134). The study investigated the anti-inflammatory potential of secretions produced by Bifidobacterium longum subsp. infantis ATCC 15697 via ultra-high-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS). Indole-3-lactic acid (ILA), a B. infantis-secreted metabolite of breastmilk tryptophan was identified as an anti-inflammatory molecule and its potency was further investigated in a human immature small intestinal cell line, necrotizing colitis enterocytes, and mature enterocytes. ILA induced anti-inflammatory response via interacting with transcription factor aryl hydrocarbon receptor (AHR) to repress the IL-1β-induced transcription of IL-8 (132).
Inflammatory bowel diseases (IBD) arise from a wide spectrum of disorders that are marked by gut dysbiosis, chronic intestinal inflammation, mucosal ulceration, and ultimately loss of intestinal function (135, 136). Recent advances in research reveal a bidirectional relationship between gut dysbiosis and disease progression (136) therefore, current studies are focused on identifying novel therapies to modulate gut microbiota; one such study investigated the beneficial role of Bifidobacterium bifidum BGN4 as a probiotic (live) or parabiotic (heat or lysozyme treated) in alleviating dextran sulfate sodium (DSS)-induced colitis in mice (137). It was observed that the lysozyme-treated B. bifidum BGN4 group demonstrated better-preserved integrity of the intestinal barrier via upregulation of TJ protein expression probably by stimulating the NLRP-6/caspase-1/IL-18 signaling pathway. Moreover, the lysozyme treated group demonstrated immune tolerance by upregulating the expression of forkhead box P3 (FOXP3) and downregulating the pro-inflammatory molecules including IL-1β, cyclooxygenase-2 (COX-2), T-bet, and inhibitor of kappa light chain gene enhancer in B cells, alpha I-kappa-B-alpha (IκB-α), thereby exerting a preventive effect against DSS-induced colitis (137). Similarly, Dong et al. used Pediococcus pentosaceus CECT 8330 (138), while Li et al., used Lactobacillus acidophilus ATCC 4356 (139) and reported that the probiotic strains exhibited high anti-inflammatory potential and could be used to restore gut dysbiosis, preserve gut barrier integrity, and ameliorate intestinal inflammation.
Tungsanga et al. investigated the protective role of Lactobacillus rhamnosus 34 on enterocyte integrity and renal fibrosis. The study was carried out using 5/6-nephrectomy (5/6-Nx) mice, Caco-2 enterocytes, and HK2 proximal tubular cells. L. rhamnosus 34 demonstrated positive effects in 5/6-Nx mice evident by a significant decrease in GDUT (gut-derived uremic toxin) index including endotoxins, trimethylamine-N-oxide (TMAO), and indoxyl sulfate (IS) and reduction in renal injuries exhibited by lower serum creatinine and proteinuria levels in kidney fibrosis area and decrease in serum TNF-α when compared to 5/6-Nx controls. Furthermore, the L. rhamnosus 34-conditioned media downregulated the expression of IL-8, NF-κB and improved TEER value in Caco-2 enterocytes, and ameliorated the expression of IL-6, TNF-α, and collagen in HK2 proximal tubular cells, supporting L. rhamnosus 34 as a potential biotherapeutic for chronic kidney disease (CKD) (140). Recent advancements in multi-omics techniques revealed that gut dysbiosis is related to CKD and this phenomenon exacerbates in hemodialysis pediatric and adult patients. The gut dysbiosis-induced systemic inflammation contributes to cardiovascular diseases, like atherosclerosis. Additionally, the relation between gut dysbiosis and CKD is found to be bidirectional, a positive shift in the gut microbiome corresponds to a decrease in circulatory GDUT (141–143). Choi et al. investigated the potential of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI in reducing the inflammation and inflammation-associated cardiovascular risks in hemodialysis patients. The study concluded that there was a significant increase in fecal SCFAs and a significant decrease in IL-6 and serum calprotectin levels post-probiotic treatment. The anti-inflammatory potential of the Bifidobacteria strains was characterized by a significant decrease in the percentages of proinflammatory monocytes CD14+ and CD16+, and a significant increase in anti-inflammatory T regulatory cells CD4+ and CD25+. The study concluded that both Bifidobacteria strains possess the potential to mitigate systemic inflammation and cardiovascular diseases in hemodialysis patients (144). The probiotic action modality with associated molecular mediators is summarized below (Figure 2).
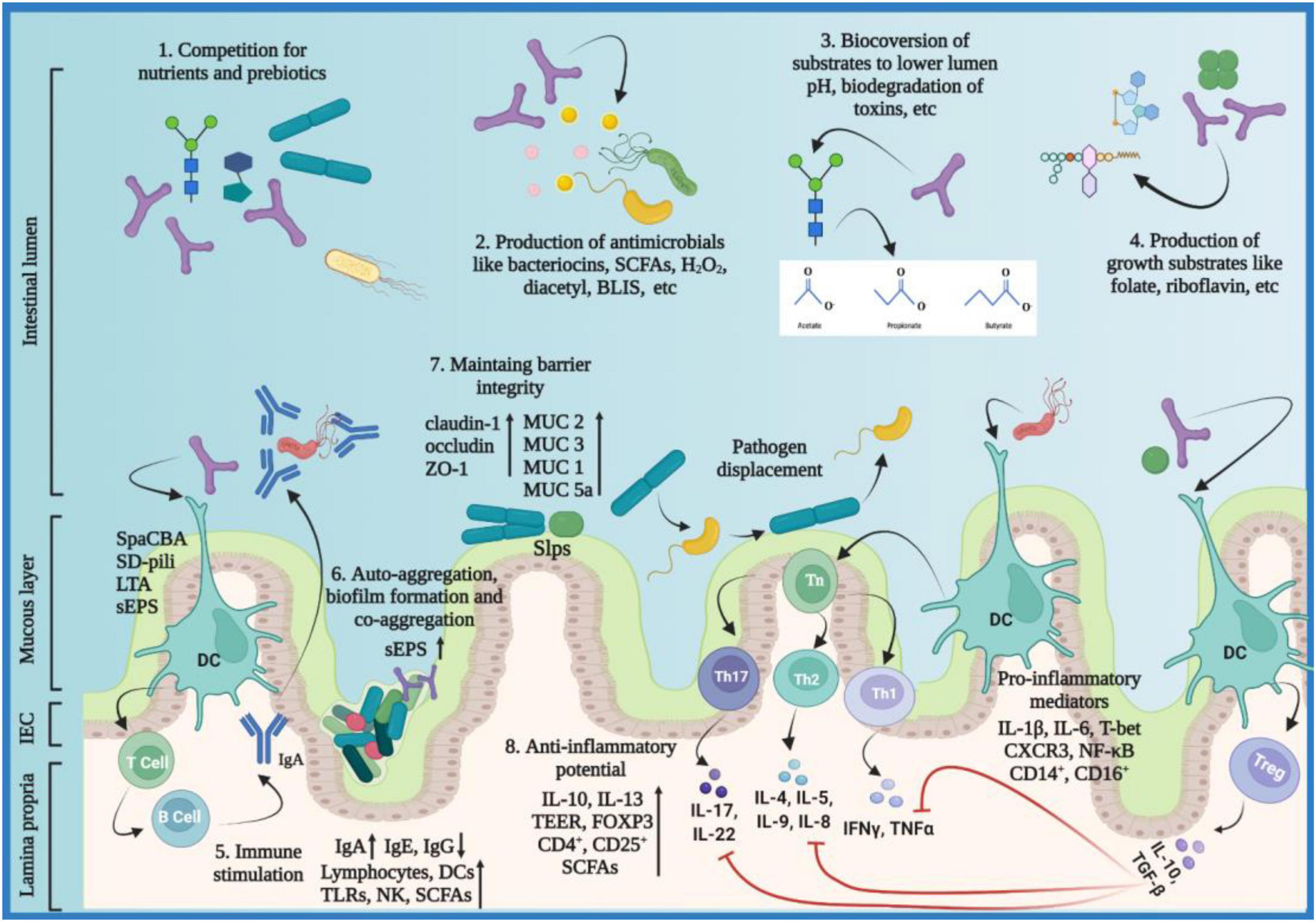
Figure 2. Probiotics action modality. Probiotics employ various mechanisms to exert their beneficial effects on the host including: (1) probiotics compete pathobionts and pathogens for microbial resources needed for growth and metabolism, e.g., acquisition of monosaccharides; (2) probiotics inhibit pathogens by producing antimicrobials, e.g., SCFAs, bacteriocins, antibiotics, microcins, etc.; (3) probiotics metabolize substrates into useful products, e.g., organic acids and volatile fatty acids; (4) probiotics produce growth substrates for beneficial microbiota and the host, e.g., folate and riboflavin; (5) probiotics induce immunomodulatory responses either by direct contact or surface molecules like SpaCBA, SD-pili, LTA, and sEPS; immune stimulation favors elevated expression of IgA and SCFAs in pathogenic infections and decreased expression of IgE and IgG in allegro-inflammatory responses via stimulating DCs, TLRs, NK cells, and lymphocytes; (6) sEPS facilitate probiotics to auto-aggregate and form protective biofilms and, co-aggregate with pathogens to prevent them from colonizing the host epithelial surfaces; (7) probiotics maintain barrier integrity by regulating TJ proteins (claudin-1, occludin, ZO-1) and by mucus formation via elevated expression of MUC 1,2,3 and 5a; Slps facilitate attachment of the probiotic strains to the gut epithelium; the colonization of pathogens is inhibited through pathogen displacement; (8) probiotics reduce inflammation by upregulating anti-inflammatory and downregulating pro-inflammatory mediators; CXCR3, C-X-C Motif Chemokine Receptor 3; BLIS, bacteriocin-like inhibitory substances; DC, dendritic cell; FOXP3, forkhead box p3; H2O2, hydrogen peroxide; IFN-γ, interferon-gamma; Ig, immunoglobulin; IL, interleukin; LTA, lipoteichoic acid; MUC, Mucin; NF-κB, nuclear factor-kappa light chain enhancer of activated B cells; NK, natural killer; SCFA, short chain fatty acids; SD-pili, sortase-dependent pili; sEPS, surface exopolysaccharides; Slps, surface-layer proteins; spaCBA, heterotrimeric pili complex made of protein subunits spa C, spa B and spa A; TEER, trans-epithelial electrical resistance; TGF-β, transforming growth factor-beta; Th, T-helper; TJ, tight junction; TLR, toll-like receptor, TNF-α, Tumor necrosis factor-alpha; Treg, T regulatory. The figure was drawn with BioRender.
What factors govern the efficacy of probiotics in given circumstances?
A wide spectrum of probiotics labeled with various health claims is available in the market. The production of probiotics has progressively surged as they are continuously prescribed by clinicians as biotherapeutics and consumed by the general public as over-the-counter bioprophylactics. Consequently, the global probiotic market was estimated to be valued at USD 61.1 billion in 2021 and is projected to reach USD 91.1 billion by 2026 (145). However, the ever-expanding probiotic product base is often mislabeled by the industry and misunderstood by the consumers. Probiotic manufacturing must avoid inconsistencies by encompassing correct labeling of the product, safety, and potency (146, 147); furthermore, the probiotic strains must endure the manufacturing processes and exposure to environmental factors to remain viable and retain the ability to colonize the gastrointestinal tract (148). Cell encapsulation improves the resistance of probiotic strains to adverse conditions (149, 150).
Despite having been conceived as homogenous mixtures of beneficial microorganisms, probiotic properties are both strain-specific and disease-specific (151), i.e., each probiotic strain has its unique repertoire of functional genes. It is imperative to understand that when it comes to the functional capacity of probiotics, a given preparation could not be all-encompassing in a given condition (7, 8, 44, 152–154). For a successful treatment, the selection of an appropriate probiotic must be based on probiotic-specific and host-specific factors (Figure 3). The probiotic-specific factors include: origin of the strain, strain-specific probiotic genetic markers, type of formulation, viability of the strain, and amount of dosage prescribed; while the host-specific factors include: type of disease or indication, composition of the gut microflora, diet, age, anthropometric measurements, and lifestyle of the host (7, 44, 151).
Figure 3. Venn diagram of factors affecting the efficacy of probiotics. The efficacy of a probiotic is a result of the interplay between numerous strain-specific and host-specific factors. The figure was drawn with BioRender.
“The efficacy of a probiotic is a measure of its ability to adapt to the host environment and exert its unique health-promoting effects; furthermore, the interplay between numerous strain-specific and host-specific factors conforms the efficacy of probiotics in a given circumstance.”
What criteria should be met for the formation of probiotics intended for human use?
The formulation of human probiotics requires evaluation of the health-promoting microorganisms by a global standard that is summarized as follows:
• As per World Health Organization (WHO)/Food and Agriculture Organization (FAO) recommendations, the origin of a probiotic strain must correlate to its site of action in the host; only isolates from human origin, viz. small intestine, large intestine, and breast milk have been approved to formulate probiotics for human use (155).
• The isolates must be carefully characterized and examined for their beneficial effects to be considered as a probiotic (44, 155).
• As per European Food Safety Authority (EFSA) recommendations, all microorganisms intended for human use must acquire genus, species, and strain levels of taxonomic identification (7, 8, 44, 45).
• Most commonly used probiotics are Lactic acid bacteria (LAB). They are identified as Generally Recognized as Safe (GRAS) by Food Drug Administration (FDA) and have Qualified Presumption of Safety (QPS) status by EFSA. Despite that, any strain intended as a probiotic for human use must undergo a rigorous safety assessment (44, 155–158).
• Moreover, as per recent EFSA guidelines, Whole-Genome Sequence (WGS)-based analysis of candidate strains is required for the formulation of probiotics (45).
• For the selection of a putative probiotic strain, its behavior under simulated GIT conditions has to be investigated. The probiotic candidates must tolerate acid and bile stress and osmotic variations to survive gastrointestinal transit (159).
• Finally, after satisfying the above-mentioned criteria, the health benefits proffered by probiotic candidates need validation through preclinical trials followed by double-blind and randomized human clinical trials (152).
• The recommended probiotic dose is between 108 (hundred million) and 1011 (hundred billion) viable colony forming units (CFU/mL/g) per day (152).
Why do certain probiotics perform inadequately in pre-clinical and clinical trials?
Irrespective of the WHO/FAO outlined recommendations, a plethora of scientific literature is based on identifying non-human lineage probiotic strains intended for human use. It not only taints the efficaciousness of probiotics but also causes discord between the industry, medical, and scientific communities. The in vitro probiotic properties displayed by non-human lineage strains often fail to extend in clinical trials due to the difference between their source of isolation and site of action (160); the ecological history of a probiotic strain, although often overlooked, plays a significant role in determining the fate of that strain i.e., to reach and then to survive at the required site of action in the host (161–164) (Figure 4). To understand the full extent of the dynamics between the source of isolated strain and the rate of success of the treatment, let us revise the concept of autochthonous vs. allochthonous strains. The autochthonous microbial strains are native to the host environment whereas the allochthonous microbial strains are non-native to the host environment. Some of the autochthonous and allochthonous strains that are frequently isolated from the human gastrointestinal tract and are used in probiotic formulations are listed below (165–171) (Table 3).
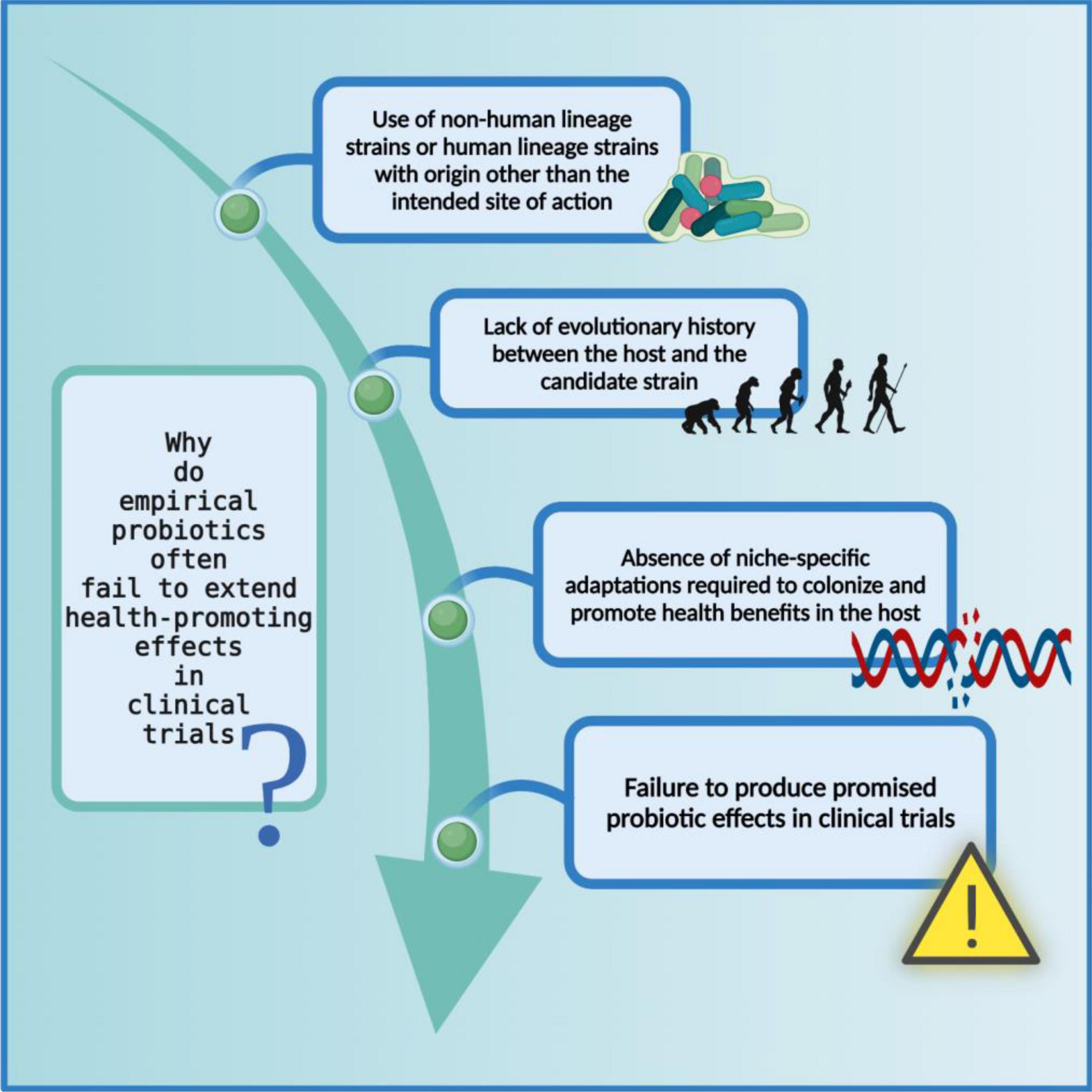
Figure 4. Limitations of non-human lineage strains as probiotics. The probiotic formulations based on non-human associated strain often fail to produce health-promoting effects in clinical trials possibly because they did not co-evolve with their respective host and consequently do not carry niche-specific genetic modifications. The figure was drawn with BioRender.
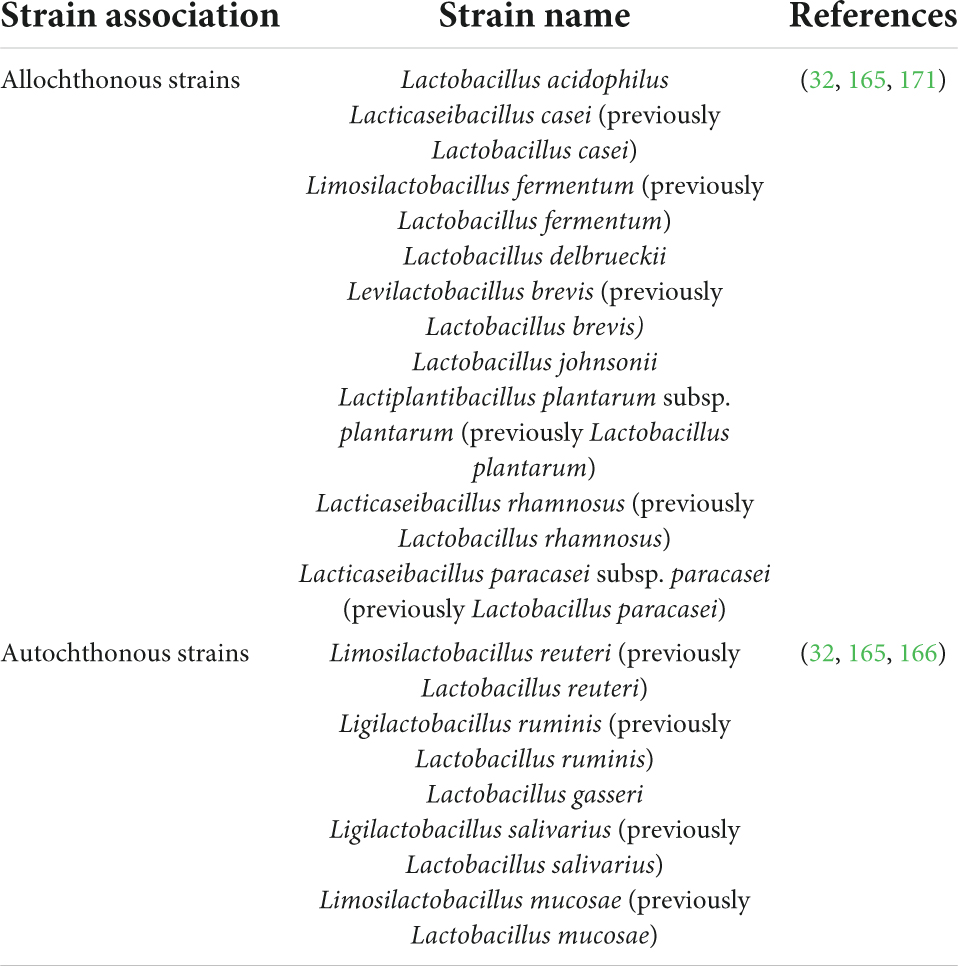
Table 3. Some frequently human gut-isolated allochthonous and autochthonous strains that are commonly used as probiotics with revised nomenclature.
Contributing to the significance of the ecological history of probiotics, a study reported the colonization potential of two strains Lactobacillus mucosae FSL-04 and Lactobacillus reuteri ATCC PTA 6475, both autochthonous to the human GIT compared with Lactobacillus acidophilus DDS-1, allochthonous to human GIT. The study concluded that autochthonous strains were established more efficiently in the human GIT than the allochthonous strain. However, when consumption was discontinued, all three strains became undetectable on the eighth day, highlighting the fact that probiotics persist in the gut only transiently (172). The potential of the probiotic strains to colonize the human GIT is dependent on the host microbiota. The colonization-resistant microbiome hinders the persistence of probiotic strains as opposed to the colonization-permissive microbiome (7, 173–175). Furthermore, studies have shown that even the autochthonous strains of one individual may not be able to colonize the GIT of another individual due to personalized differences (153).
“The rate of success of a probiotic intervention α establishment of a strain in the microecological niche of the host + production of microbial metabolites that positively influence the physiological pathway of the host and/or their gut microbiota.”
Why should host-adapted strains be favored for probiotic formulations?
Indifferent to the autochthonous vs. allochthonous origin of a probiotic isolate, the ability of an isolate to adapt to a specific ecological niche is accomplished by genome specialization. It is a process whereby, niche-specific phenotypic fitness is acquired through selective genome decay of unutilized genes and enrichment of the genes that facilitate adaptability in the respective habitat. Therefore, individual probiotic strains isolated from the same habitat carry the same niche-specific genetic signatures. This has been validated for Lactobacillus jensenii, and Lactobacillus gasseri both isolated from the human vagina as well as for Lactobacillus reuteri isolates from the GI tract of different vertebrates (43, 164). Similarly, Lactobacillus species including L. plantarum, L. rhamnosus, L. paracasei, and L. casei although not considered autochthonous in a classical sense, possess niche-specific adaptations to oral cavity and gut ecosystems that enable them to persist transiently (162). Studies have shown that host-adapted Lactobacilli strains show higher ecological fitness in their respective host (163, 176). Since host-adapted strains carry niche-specific genomic modifications, they display higher ecological fitness as opposed to the strains that do not share an evolutionary history with the host. The higher ecological fitness enables higher metabolic activity and higher resistance to enteropathogens (163). Also, host-adaptive strains establish immunotolerant profiles in host gut mucosa (177). This has been reported for L. reuteri strains as the human lineage autochthonous L. reuteri strain displayed anti-inflammatory effects while the poultry lineage L. reuteri strain displayed immune-stimulatory effects in human myeloid cells (161, 176).
Supporting the above-mentioned studies, Doron et al. reported that the attempt spanned over many years to colonize dairy-isolated Lactobacillus in the human gastrointestinal tract (GIT) is futile. The dairy-isolated Lactobacillus strains including L. acidophilus, L. casei, and L. bulgaricus do not harbor the necessary biological characteristics to successfully establish in the human host and thereby, are unlikely to demonstrate any beneficial effects (178). Similarly, Wong et al., reviewed differences in the physiological attributes (i.e., metabolic capabilities), and genetic attributes (i.e., comparative and functional genomics) of human residential and non-human residential Bifidobacteria strains (179). The study concluded that human residential strains possess better adaptive health attributes for their host (178) e.g., human infant-derived Bifidobacteria longum subsp. infantis has a gamut of glycoside hydrolases (GHs) and ABC transporters required for degrading human milk oligosaccharides (HMOs) and fol gene clusters for folate biosynthesis as opposed to non-human-associated Bifidobacteria (180) (Figure 5).
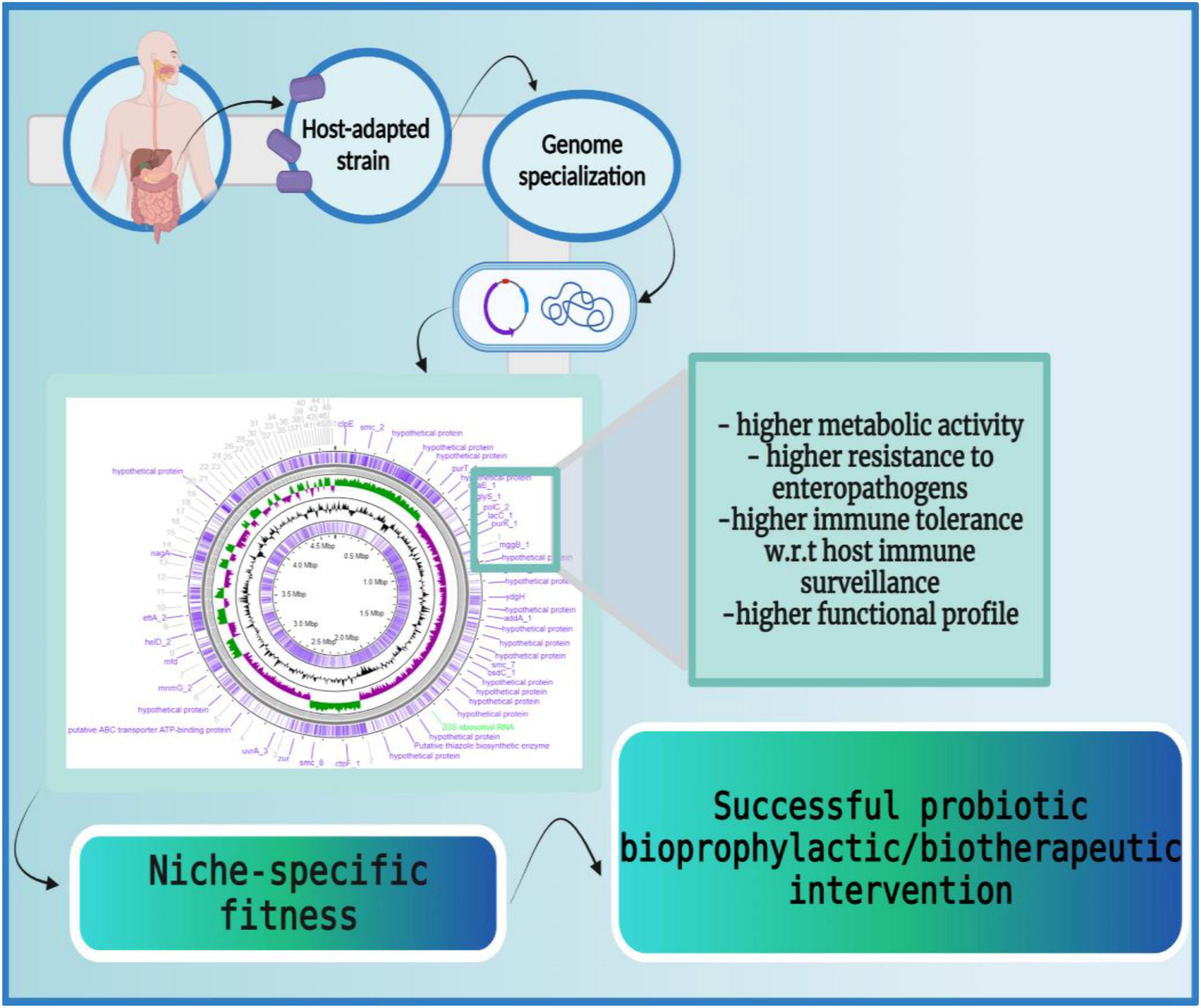
Figure 5. Host-adapted strains harbor niche-specific phenotypic fitness. The host-adapted microbial strains carry niche-specific genetic signatures which enable them to adapt to the host ecosystems efficiently. These genetic modifications enable higher metabolic activity (larger carbohydrate utilization cassettes, production of β-galactosidases, etc.), higher resistance to enteropathogens (production of lactic acids, synthesis of secondary metabolites, etc.), higher immune tolerance (induction of anti-inflammatory cytokines like IL-10, IL-13, etc.), and higher functional profiles (production of bile salt hydrolases to endure bile salts, accumulation of ATP synthesizing cassettes for conditional respiration, etc.). Thus, host-adapted microbial strains possess a large repertoire of niche-specific genes that facilitate their persistence in the host. The figure was drawn with BioRender.
“Thus, the ecological and evolutionary characteristics of a probiotic isolate are a measure of its functionality.”
Strategic formulation of target-based probiotics for human use
As has been discussed earlier, non-human lineage probiotics often fail to extend the in vitro-exhibited beneficial effects in clinical trials (160). We shall now discuss the use of host-adapted microbial isolates in the formulation of target-based probiotics intended for human use (163, 176) (Figure 6). As per standard guidelines, the source of isolation of a probiotic strain must correlate with its intended site of action. For instance, if the attenuation of gut dysbiosis is intended then the source of isolation should be the human GI tract. Similarly, if treatment for bacterial vaginosis or periodontitis is intended then corresponding probiotics must be formulated using vaginal or oral isolates, respectively (44).
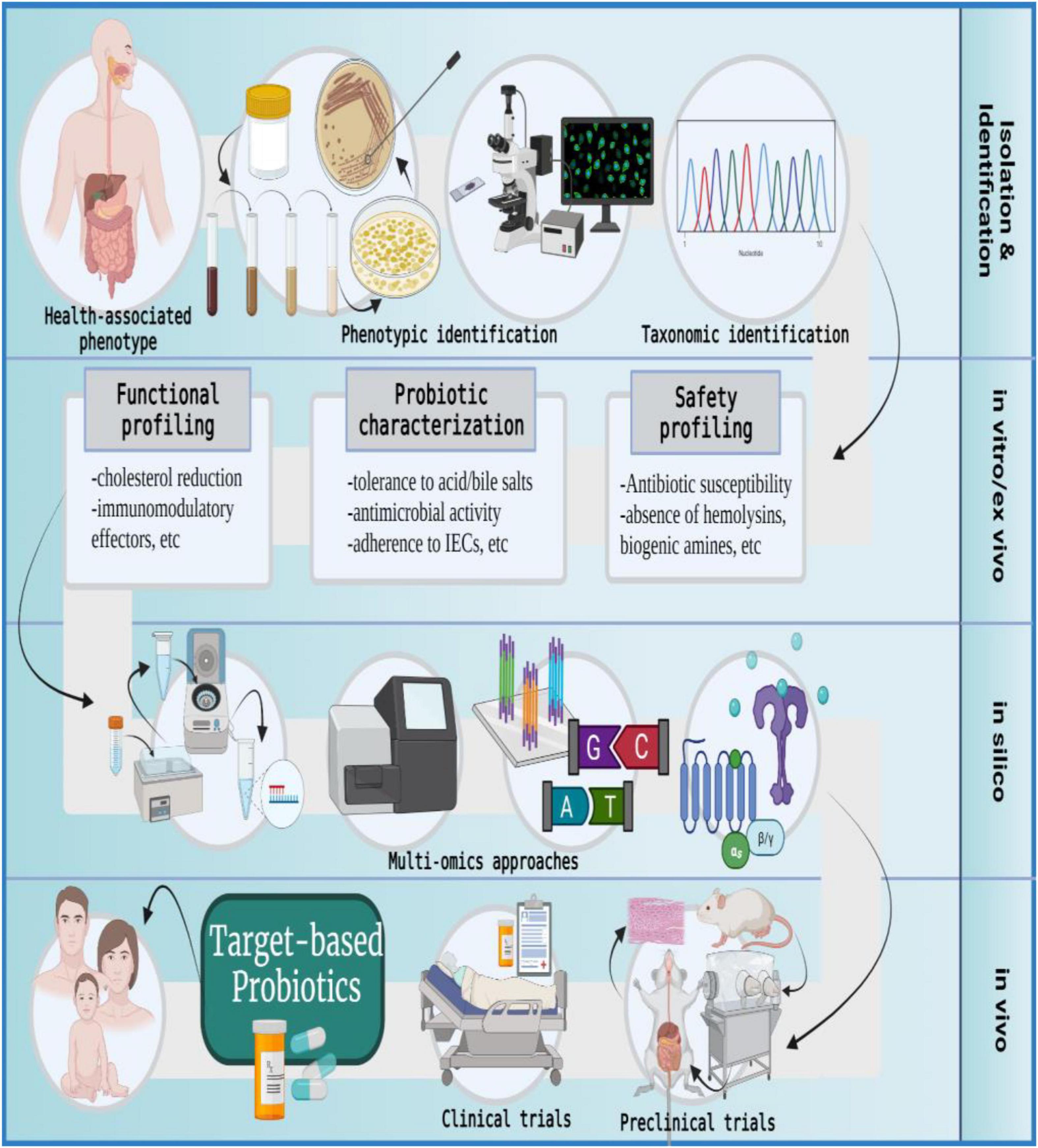
Figure 6. The formulation of target-based probiotics. The formulation of target-based probiotics is based on understanding the action modality of probiotic strains via a multi-omics approach and then tailoring a probiotic formulation that produces molecular effectors to ameliorate a specific disease or indication; IECs, intestinal epithelial cells. The figure was drawn with BioRender.
The metamorphoses of commensal microbiota into target-based probiotics involves the following procedure: The samples are collected from healthy individuals, and microbial strains are isolated using a specific growth medium. The isolates are subjected to phenotypic characterization and selected candidates are taxonomically identified on the genus-species-strain level (44, 45). The in vitro and ex vivo approaches evaluate the candidate probiotics in terms of safety and functionality. The in silico analysis uses multi-omics (genomics, transcriptomics, proteomics, and metabolomics) approaches to further identify molecular effectors (microbial metabolites) produced by the selected probiotic candidate that can potentially modify host or microbial pathways to promote health phenotype or ameliorate disease conditions. Post in silico analysis, the in vivo studies entail validation of proposed probiotic effects by using gnotobiotic animal models with metabolic, immune, microbial, or neuronal distinctions. Finally, the success of clinical trials culminates in the development of target-based probiotics for human use (10, 31, 152, 181–186).
Conclusion
Probiotics have gained substantial amount of attention over the past few decades and with the advancements in multi-omics techniques, we now understand better than ever, the role of probiotics in maintaining gut microbiota and health phenotype. A wide spectrum of probiotic products is available in the market and the probiotic industry is rejoicing in its exponential boom. However, the development of probiotics must be medical interest-driven rather than commercial interest-driven. Despite a large product base, there is heterogeneity between the claimed probiotic benefits in scientific literature and the efficacy of probiotics in clinical trials which can be resolved by understanding that the efficacy of a probiotic intervention depends on both the strain-specific and host-specific factors. The formulation of human probiotics requires a strict evaluation of candidate strains by a global standard. The harmony between the probiotic source of isolation and its intended site of action determines the success of a subsequent probiotic intervention. The host-adapted probiotic strains are likely to harbor selective genetic modifications that facilitate their niche-specific phenotypic fitness in the respective host. The evolutionary and ecological history of a probiotic strain, albeit often overlooked, heralds a significant outcome; ergo, the formulation of probiotics using host-adapted strains with known molecular effectors would serve as ideal candidates for target-based bioprophylactic and biotherapeutic interventions to maintain health and ameliorate diseased phenotypes.
Author contributions
MId and SG: conceptualization. MId: illustrations and writing—original draft preparation. MIm, RZ, NA, MA, MT, and AF: validation. MId, MIm, MA, OO, and SG: formal analysis. MIm, SG, RA, and AF: investigation. AA, TR, and OO: resources. MA, TR, RA, and AA: data curation. MId, MIm, OO, and MA: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Research Deanship in University of Ha’il—Saudi Arabia through project number MDR-22 012.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ABC, ATP-binding cassettes; AHR, aryl hydrocarbon receptor; AKT, protein kinase B; BALF, bronchoalveolar lavage fluid; BLIS, bacteriocin-like inhibitory substances; CBER, Center for Biologics Evaluation and Research; CFU, colony forming units; CKD, chronic kidney disease; COVID-19, coronavirus disease of 2019; COX-2, cyclooxygenase-2; CXCR3, C-X -C Motif Chemokine Receptor 3; DC, dendritic cell; DSS, dextran sulfate sodium; EFSA, European Food Safety Authority; EPO, eosinophil peroxidase activity; EPS, exopolysaccharides; ERK, extracellular-signal-regulated kinase; FAD, flavin adenine dinucleotide; FAO, Food and Agriculture Organization; FDA, Food and Drug Administration; FMN, flavin mononucleotide; FOXP3, forkhead box p3; GABA, gamma-aminobutyric acid; GDUT, gut-derived uremic toxin; GH, glycoside hydrolase; GIT, gastrointestinal tract; GPR, G-protein -coupled receptor; H2O2, hydrogen peroxide; HMO, human milk oligosaccharides; HYA, 10-hydroxy-cis-12-octadecenoic acid; IBD, inflammatory bowel disease; IEC, intestinal epithelial cell; IFN- γ, interferon-gamma; Ig, immunoglobulin; I κ B- α, inhibitor of kappa light chain gene enhancer in B cells, alpha I-kappa -B-alpha; IL, interleukin; ILA, indole lactic acid; IS, indoxyl sulfate; ISAPP, International Scientific Association for Probiotics and Prebiotics; LAB, lactic acid bacteria; LTA, lipoteichoic acid; MALDI-TOF MS, matrix-assisted laser desorption ionization–time-of-flight mass spectrometry; MEK, mitogen-activated kinase; MUC, Mucin; MyD88, Myeloid differentiation primary response 88; NEC, necrotizing enterocolitis; NF- κ B, nuclear factor-kappa light chain enhancer of activated B cells; NK, natural killer; NLRP3, NOD-like receptor family pyrin domain containing 3; PI3K, Phosphoinositide 3-Kinase; pABA, para-aminobenzoic acid; PCR, polymerase chain reaction; PMB, probiotics-mediated bioconversion; PRRs, Pattern recognition receptors; SCFA, short-chain fatty acids; SD-pili, sortase-dependent pili; sEPS, surface exopolysaccharides; Slps, surface-layer proteins; spaCBA, heterotrimeric pili complex made of protein subunits spa C spa B and spa A; TEER, trans-epithelial electrical resistance; TGF- β, transforming growth factor-beta; Th, T-helper; TJ, tight junction; TLR, toll-like receptor; TMAO, trimethylamine-N-oxide; TNBS, trinitrobenzene sulfonic; TNF- α, Tumor necrosis factor-alpha; Treg, T regulatory; UHPLC-MS/MS, ultra-high-performance liquid chromatography-tandem mass spectrometry; WGS, Whole-genome sequence; WHO, World Health Organization.
References
1. Metchnikoff E. The Prolongation of Life: Optimistic Studies. New York, NY: Springer Publishing Company (2004).
2. Underhill DM, Gordon S, Imhof BA, Núñez G, Bousso P. Élie Metchnikoff (1845–1916): celebrating 100 years of cellular immunology and beyond. Nat Rev Immunol. (2016) 16:651–6. doi: 10.1038/nri.2016.89
3. Santacroce L, Charitos IA, Bottalico L. A successful history: probiotics and their potential as antimicrobials. Exp Rev Anti Infect Ther. (2019) 17:635–45. doi: 10.1080/14787210.2019.1645597
4. Stambler I. Elie Metchnikoff—the founder of longevity science and a founder of modern medicine: in honor of the 170th anniversary. Adv Gerontol. (2015) 5:201–8. doi: 10.1134/S2079057015040219
5. Mackowiak P. Recycling Metchnikoff: probiotics, the intestinal microbiome and the quest for long life. Front Public Health. (2013) 1:52. doi: 10.3389/fpubh.2013.00052
6. Sanap DS, Garje MA, Godge GR. Probiotics, their health benefits and applications for development of human health: a review. J Drug Deliv Therap. (2019) 9:631–40.
7. Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. (2019) 25:716–29. doi: 10.1038/s41591-019-0439-x
8. Reid G, Gadir AA, Dhir R. Probiotics: reiterating what they are and what they are not. Front Microbiol. (2019) 10:424. doi: 10.3389/fmicb.2019.00424
9. Hamilton-Miller J, Gibson GR, Bruck W. Some insights into the derivation and early uses of the word ‘probiotic’. Br J Nutr. (2003) 90:845.
10. Fuller R. History and development of probiotics. In: R Fuller editor. Probiotics: The Scientific Basis. Dordrecht: Springer (1992). doi: 10.1007/978-94-011-2364-8
11. Reque PM, Brandelli A. Chapter 1 – An introduction to probiotics. In: A Brandelli editor. Probiotics. Cambridge, MA: Academic Press (2022). doi: 10.1016/B978-0-323-85170-1.00017-8
12. Ozen M, Dinleyici E. The history of probiotics: the untold story. Benef Microbes. (2015) 6:159–65. doi: 10.3920/BM2014.0103
13. Stillwell CR. The Wisdom of Cells: The Integrity of Elie Metchnikoff’s Ideas in Biology and Pathology. Notre Dame, IN: University of Notre Dame (1991).
14. Stackebrandt E, Teuber M. Molecular taxonomy and phylogenetic position of lactic acid bacteria. Biochimie. (1988) 70:317–24. doi: 10.1016/0300-9084(88)90204-0
15. Soomro A, Masud T, Anwaar K. Role of lactic acid bacteria (LAB) in food preservation and human health-a review. Pak J Nutr. (2002) 1:20–4. doi: 10.3923/pjn.2002.20.24
17. Sandine WE. New nomenclature of the non-rod-shaped lactic acid bacteria. Biochimie. (1988) 70:519–21. doi: 10.1016/0300-9084(88)90087-9
18. Shama G. The “Petri” dish: a case of simultaneous invention in bacteriology. Endeavour. (2019) 43:11–6. doi: 10.1016/j.endeavour.2019.04.001
19. DePaolo C. Sir Marc Armand Ruffer, MD: the early years, 1878–1896. J Med Biogr. (2019) 29:169–75. doi: 10.1177/0967772019883998
20. Gopal V, Dhanasekaran D. Chapter 22 – Probiotics as a growth promotant for livestock and poultry production. In: D Dhanasekaran, A Sankaranarayanan editors. Advances In Probiotics. Cambridge, MA: Academic Press (2021). p. 349–64. doi: 10.1016/B978-0-12-822909-5.00022-8
21. Mitsuoka T, Kaneuchi C. Ecology of the bifidobacteria. Am J Clin Nutr. (1977) 30:1799–810. doi: 10.1093/ajcn/30.11.1799
22. Kusumitha S, Aeron V, Gh Jeelani P, Chidambaram R. Prebiotics and probiotics in the formulation of infant foods. In: TJ Gutiérrez editor. Food Science, Technology and Nutrition for Babies and Children. Cham: Springer International Publishing (2020). p. 35–57. doi: 10.1007/978-3-030-35997-3_3
24. Weirich A, Hoffmann GF. Ernst Moro (1874–1951)—a great pediatric career started at the rise of university-based pediatric research but was curtailed in the shadows of Nazi laws. Eur J Pediatr. (2005) 164:599–606. doi: 10.1007/s00431-005-1703-2
25. Oh S. Lactobacillus acidophilus as a probiotics. J Dairy Sci Biotechnol. (2019) 37:155–66. doi: 10.22424/jmsb.2019.37.3.155
26. Anukam KC, Reid G. Probiotics: 100 years (1907–2007) after elie metchnikoff’s observation. In: A Méndez-Vilas editor. Communicating Current Research and Educational Topics and Trends in Applied Microbiology (Vol. 1), Badajoz: FORMATEX (2007). p. 466–74.
27. Stoilova E. Deconstructing the Authenticity: Who, When, and How Created the Bulgarian Yoghurt. Myth-Making and Myth-Breaking in History and the Humanities. (2011). p 125. Available online at: https://scholar.google.com/scholar?cluster=13654319592837750555&hl=en&as_sdt=0,5
28. Keikha M, Kamali H. The impact of Saccharomyces boulardii adjuvant supplementation on alternation of gut microbiota after H. pylori eradication; a metagenomics analysis. Gene Rep. (2022) 26:101499. doi: 10.1016/j.genrep.2022.101499
29. Obazelu P, Aruomaren I, Nwangwu C. Effects of probiotic Saccharomyces boulardii and co-trimoxazole on some haematological parameters in adult Wistar rats. Afr J Health Saf Environ. (2021) 2:55–62. doi: 10.52417/ajhse.v2i2.158
30. Yamashiro Y. Effects of enteral administration of Bifidobacterium breve on dysbiosis infants and children: a review. Br J Gastroenterol. (2020) 2:185–9. doi: 10.31488/bjg.1000119
31. Singh TP, Natraj BH. Next-generation probiotics: a promising approach towards designing personalized medicine. Crit Rev Microbiol. (2021) 47:479–98. doi: 10.1080/1040841X.2021.1902940
32. Zheng J, Wittouck S, Salvetti E, Franz C, Harris HMB, Mattarelli P, et al. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol. (2020) 70:2782–858. doi: 10.1099/ijsem.0.004107
33. Yadav MK, Kumari I, Singh B, Sharma KK, Tiwari SK. Probiotics, prebiotics and synbiotics: safe options for next-generation therapeutics. Appl Microbiol Biotechnol. (2022) 106:505–21. doi: 10.1007/s00253-021-11646-8
34. Gogineni VK, Morrow LE, Gregory PJ, Malesker MA. Probiotics: history and evolution. J Anc Dis Prev Rem. (2013) 1:1–7. doi: 10.4172/2329-8731.1000107
35. Lilly DM, Stillwell RH. Probiotics: growth-promoting factors produced by microorganisms. Science. (1965) 147:747–8. doi: 10.1126/science.147.3659.747
36. Fuller R. Probiotics in man and animals. J Appl Bacteriol. (1989) 66:365–78. doi: 10.1111/j.1365-2672.1989.tb05105.x
37. Arruda HS, Geraldi MV, Cedran MF, Bicas JL, Marostica Junior MR, Pastore GM. Chapter 4 – prebiotics and probiotics. In: CBB Cazarin, JL Bicas, GM Pastore, MR Marostica Junior editors. Bioactive Food Components Activity In Mechanistic Approach. Cambridge, MA: Academic Press (2022). p. 55–118. doi: 10.1016/B978-0-12-823569-0.00006-0
39. Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. (1995) 125:1401–12. doi: 10.1093/jn/125.6.1401
40. Panesar PS, Anal AK, Kaur R. Probiotics, prebiotics and synbiotics: opportunities, health benefits and industrial challenges. Probiotics Prebiotics Synbiotics. (2022) 1:1–3. doi: 10.1002/9781119702160
41. Chaudhari A, Dwivedi MK. Chapter 1 – The concept of probiotics, prebiotics, postbiotics, synbiotics, nutribiotics, and pharmabiotics. In: MK Dwivedi, N Amaresan, A Sankaranarayanan, EH Kemp editors. Probiotics in the Prevention and Management of Human Diseases. Cambridge, MA: Academic Press (2022). p. 1–11. doi: 10.1016/B978-0-12-823733-5.00013-1
42. Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. (1998) 5:R245–9. doi: 10.1016/S1074-5521(98)90108-9
43. Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U.S.A. (2003) 100:1990–5. doi: 10.1073/pnas.0337704100
44. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
45. EFSA. EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. (2021) 19:e06506. doi: 10.2903/j.efsa.2021.6506
46. Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A. Mechanisms of action of probiotics. Adv Nutr. (2019) 10(Suppl. 1):S49–66. doi: 10.1093/advances/nmy063
47. Ng S, Hart A, Kamm M, Stagg A, Knight SC. Mechanisms of action of probiotics: recent advances. Inflamm Bowel Dis. (2009) 15:300–10. doi: 10.1002/ibd.20602
48. Reque PM, Brandelli A. Chapter 1 – An introduction to probiotics. In: A Brandelli editor. Probiotics. Cambridge, MA: Academic Press (2022). p. 1–17. doi: 10.1016/B978-0-323-85170-1.00017-8
49. Ferreira RDS, Mendonca LABM, Ribeiro CFA, Calcas NC, Guimaraes RDCA, Nascimento VAD, et al. Relationship between intestinal microbiota, diet and biological systems: an integrated view. Crit Rev Food Sci Nutr. (2022) 62:1166–86. doi: 10.1080/10408398.2020.1836605
50. Nasr N. Psychological impact of probiotics and fermented foods on mental health of human in integrated healthy lifestyle. Int J Curr Microbiol Appl Sci. (2018) 7:2815–22. doi: 10.20546/ijcmas.2018.708.296
51. Hameed A, Condò C, Tauseef I, Idrees M, Ghazanfar S, Farid A, et al. Isolation and characterization of a cholesterol-lowering bacteria from Bubalus bubalis raw milk. Fermentation. (2022) 8:163. doi: 10.3390/fermentation8040163
52. Abid R, Waseem H, Ali J, Ghazanfar S, Muhammad Ali G, Elasbali AM, et al. Probiotic yeast saccharomyces: back to nature to improve human health. J Fungi. (2022) 8:444. doi: 10.3390/jof8050444
53. Abid S, Farid A, Abid R, Rehman MU, Alsanie WF, Alhomrani M, et al. Identification, biochemical characterization, and safety attributes of locally isolated Lactobacillus fermentum from Bubalus bubalis (buffalo) milk as a probiotic. Microorganisms. (2022) 10:954. doi: 10.3390/microorganisms10050954
54. Akmal U, Ghori I, Elasbali AM, Alharbi B, Farid A, Alamri AS, et al. Probiotic and antioxidant potential of the Lactobacillus spp. Isolated from artisanal fermented pickles. Fermentation. (2022) 8:328. doi: 10.3390/fermentation8070328
55. Bajaj BK, Claes IJ, Lebeer S. Functional mechanisms of probiotics. J Microbiol Biotechnol Food Sci. (2021) 2021:321–7.
56. Sharma H, Bajwa J. Potential role and mechanism of probiotics. Ann Roman Soc Cell Biol. (2021) 25:3616–24.
57. Vanderhoof JA, Young RJ. Use of probiotics in childhood gastrointestinal disorders. J Pediatr Gastroenterol Nutr. (1998) 27:323–32. doi: 10.1097/00005176-199809000-00011
58. Strain R, Stanton C, Ross RP. Effect of diet on pathogen performance in the microbiome. Microbiome Res Rep. (2022) 1:13. doi: 10.20517/mrr.2021.10
59. Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. (1988) 56:2610–4. doi: 10.1128/iai.56.10.2610-2614.1988
60. Hojo K, Nagaoka S, Murata S, Taketomo N, Ohshima T, Maeda N. Reduction of vitamin K concentration by salivary Bifidobacterium strains and their possible nutritional competition with Porphyromonas gingivalis. J Appl Microbiol. (2007) 103:1969–74. doi: 10.1111/j.1365-2672.2007.03436.x
61. Elli M, Zink R, Rytz A, Reniero R, Morelli L. Iron requirement of Lactobacillus spp. in completely chemically defined growth media. J Appl Microbiol. (2000) 88:695–703. doi: 10.1046/j.1365-2672.2000.01013.x
62. Rolfe RD. The role of probiotic cultures in the control of gastrointestinal health. J Nutr. (2000) 130:396S–402S. doi: 10.1093/jn/130.2.396S
63. Juven B, Meinersmann R, Stern N. A review: antagonistic effects of lactobacilli and pediococci to control intestinal colonization by human enteropathogens in live poultry. J Appl Bacteriol. (1991) 70:95–103. doi: 10.1111/j.1365-2672.1991.tb04433.x
64. Umu ÖC, Bäuerl C, Oostindjer M, Pope PB, Hernandez PE, Perez-Martinez G, et al. The potential of class II bacteriocins to modify gut microbiota to improve host health. PLoS One. (2016) 11:e0164036. doi: 10.1371/journal.pone.0164036
65. Lambert J, Hull R. Upper gastrointestinal tract disease and probiotics. Asia Pac J Clin Nutr. (1996) 5:31–5.
66. Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. (2011) 469:543–7. doi: 10.1038/nature09646
67. Prince T, McBain AJ, O’Neill CA. Lactobacillus reuteri protects epidermal keratinocytes from Staphylococcus aureus-induced cell death by competitive exclusion. Appl Environ Microbiol. (2012) 78:5119–26. doi: 10.1128/AEM.00595-12
68. Hefzy EM, Khalil MAF, Amin AAI, Ashour HM, Abdelaliem YF. Bacteriocin-like inhibitory substances from probiotics as therapeutic agents for Candida vulvovaginitis. Antibiotics. (2021) 10:306. doi: 10.3390/antibiotics10030306
69. Li Z, Li Y, Xiao C, Yan Z, Pan R, Gao Y, et al. Genomic and metabolic features of the Lactobacillus sakei JD10 revealed potential probiotic traits. Microbiol Res. (2022) 256:126954. doi: 10.1016/j.micres.2021.126954
70. Kawahara A, Murakami C, Hayashi R, Zendo T, Matsusaki H. Characterization of multiple bacteriocin-producing Lactiplantibacillus plantarum PUK6 isolated from misozuke-tofu. Food Sci Technol Res. (2022) 28:195–205. doi: 10.3136/fstr.FSTR-D-21-00294
71. Diale MO, Abrahams A, Serepa-Dlamini MH. Isolation and characterization of bacteriocin-like substances from Bacillus paranthracis Strain MHSD3, a potential probiotic. Res Sq. (2022). doi: 10.21203/rs.3.rs-1553916/v1 [Epub ahead of print].
72. Bermudez-Brito M, Plaza-Díaz J, Muñoz-Quezada S, Gómez-Llorente C, Gil A. Probiotic mechanisms of action. Ann Nutr Metab. (2012) 61:160–74. doi: 10.1159/000342079
73. Lee Y, Yoon Y, Choi K-H. Probiotics-mediated bioconversion and periodontitis. Food Sci Anim Resour. (2021) 41:905–22. doi: 10.5851/kosfa.2021.e57
74. Schlagenhauf U, Rehder J, Gelbrich G, Jockel-Schneider Y. Consumption of Lactobacillus reuteri-containing lozenges improves periodontal health in navy sailors at sea: a randomized controlled trial. J Periodontol. (2020) 91:1328–38. doi: 10.1002/JPER.19-0393
75. Pelekos G, Acharya A, Eiji N, Hong G, Leung WK, McGrath C. Effects of adjunctive probiotic L. reuteri lozenges on S/RSD outcomes at molar sites with deep pockets. J Clin Periodontol. (2020) 47:1098–107. doi: 10.1111/jcpe.13329
76. Laleman I, Pauwels M, Quirynen M, Teughels W. A dual-strain Lactobacilli reuteri probiotic improves the treatment of residual pockets: a randomized controlled clinical trial. J Clin Periodontol. (2020) 47:43–53. doi: 10.1111/jcpe.13198
77. Grusovin MG, Bossini S, Calza S, Cappa V, Garzetti G, Scotti E, et al. Clinical efficacy of Lactobacillus reuteri-containing lozenges in the supportive therapy of generalized periodontitis stage III and IV, grade C: 1-year results of a double-blind randomized placebo-controlled pilot study. Clin Oral Invest. (2020) 24:2015–24. doi: 10.1007/s00784-019-03065-x
78. Invernici MM, Salvador SL, Silva PHF, Soares MSM, Casarin R, Palioto DB, et al. Effects of Bifidobacterium probiotic on the treatment of chronic periodontitis: a randomized clinical trial. J Clin Periodontol. (2018) 45:1198–210. doi: 10.1111/jcpe.12995
79. González-Salitre L, Alma Delia R-G, Rodríguez-Serrano G, Jaimez J, Bautista M, González-olivares L. Biotransformation mechanism of inorganic selenium into selenomethionine and selenocysteine by Saccharomyces boulardii in Silico Study. Res Sq. (2021). doi: 10.21203/rs.3.rs-359033/v1 [Epub ahead of print].
80. Martínez FG, Moreno-Martin G, Pescuma M, Madrid-Albarrán Y, Mozzi F. Biotransformation of selenium by lactic acid bacteria: formation of seleno-nanoparticles and seleno-amino acids. Front Bioeng Biotechnol. (2020) 8:506. doi: 10.3389/fbioe.2020.00506
81. Mousavi R, Mottawea W, Hassan H, Gomaa A, Audet MC, Hammami R. Screening, characterization and growth of γ−aminobutyric acid−producing probiotic candidates from food origin under simulated colonic conditions. J Appl Microbiol. (2022) 132:4452–65. doi: 10.1111/jam.15550
82. Akbar A, Khan MI, Khan GI. Probiotics in biodegradation of microbial toxins. In: PS Panesar, AK Anal editors. Probiotics, Prebiotics and Synbiotics. Hoboken, NJ: Wiley (2022). p. 161–71. doi: 10.1002/9781119702160.ch7
83. Liu M, Chen Q, Sun Y, Zeng L, Wu H, Gu Q, et al. Probiotic potential of a folate-producing strain Latilactobacillus sakei LZ217 and its modulation effects on human gut microbiota. Foods. (2022) 11:234. doi: 10.3390/foods11020234
84. Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. (2011) 3:118–34. doi: 10.3390/nu3010118
85. Strozzi GP, Mogna L. Quantification of folic acid in human feces after administration of Bifidobacterium probiotic strains. J Clin Gastroenterol. (2008) 42:S179–84. doi: 10.1097/MCG.0b013e31818087d8
86. Thakur K, Tomar SK, Brahma B, De S. Screening of riboflavin-producing lactobacilli by a polymerase-chain-reaction-based approach and microbiological assay. J Agric Food Chem. (2016) 64:1950–6. doi: 10.1021/acs.jafc.5b06165
87. Liu S, Hu W, Wang Z, Chen T. Production of riboflavin and related cofactors by biotechnological processes. Microbial Cell Fact. (2020) 19:31. doi: 10.1186/s12934-020-01302-7
88. Levit R, de Giori GS, de Moreno de LeBlanc A, LeBlanc JG. Evaluation of the effect of soymilk fermented by a riboflavin-producing Lactobacillus plantarum strain in a murine model of colitis. Benef Microbes. (2017) 8:65–72. doi: 10.3920/BM2016.0063
89. Levit R, de Giori GS, de Moreno de LeBlanc A, LeBlanc JG. Effect of riboflavin-producing bacteria against chemically induced colitis in mice. J Appl Microbiol. (2018) 124:232–40. doi: 10.1111/jam.13622
90. Levit R, de Giori GS, de Moreno de LeBlanc A, LeBlanc JG. Beneficial effect of a mixture of vitamin-producing and immune-modulating lactic acid bacteria as adjuvant for therapy in a recurrent mouse colitis model. Appl Microbiol Biotechnol. (2019) 103:8937–45. doi: 10.1007/s00253-019-10133-5
91. Levit R, de Giori GS, de Moreno de LeBlanc A, LeBlanc JG. Protective effect of the riboflavin-overproducing strain Lactobacillus plantarum CRL2130 on intestinal mucositis in mice. Nutrition. (2018) 54:165–72. doi: 10.1016/j.nut.2018.03.056
92. Tsai Y-T, Cheng P-C, Pan T-M. The immunomodulatory effects of lactic acid bacteria for improving immune functions and benefits. Appl Microbiol Biotechnol. (2012) 96:853–62. doi: 10.1007/s00253-012-4407-3
93. Kageyama Y, Nishizaki Y, Aida K, Yayama K, Ebisui T, Akiyama T, et al. Lactobacillus plantarum induces innate cytokine responses that potentially provide a protective benefit against COVID-19: a single-arm, double-blind, prospective trial combined with an in vitro cytokine response assay. Exp Ther Med. (2022) 23:20. doi: 10.3892/etm.2021.10942
94. dos Santos Freitas A, da Silva Fernandes LJ, Coelho-Rocha ND, de Jesus LCL, de Rezende Rodovalho V, da Silva TF, et al. Chapter 16 – Immunomodulatory and antiinflammatory mechanisms of probiotics. In: A Brandelli editor. Probiotics. Cambridge, MA: Academic Press (2022). p. 321–41. doi: 10.1016/B978-0-323-85170-1.00019-1
95. Matsuzaki C, Shiraishi T, Chiou T-Y, Nakashima Y, Higashimura Y, Yokota S-I, et al. Role of lipoteichoic acid from the genus Apilactobacillus in inducing a strong IgA response. Appl Environ Microbiol. (2022) 88:e0019022. doi: 10.1128/aem.00190-22
96. Gadhe ST, Patel VP. Role of probiotics in allergic diseases. In: RZ Sayyed, M Khan editors. Microbiome-Gut-Brain Axis: Implications on Health. Singapore: Springer (2022). p. 317–26. doi: 10.1007/978-981-16-1626-6_14
97. Wu Z, Mehrabi Nasab E, Arora P, Athari SS. Study effect of probiotics and prebiotics on treatment of OVA-LPS-induced of allergic asthma inflammation and pneumonia by regulating the TLR4/NF-kB signaling pathway. J Transl Med. (2022) 20:130. doi: 10.1186/s12967-022-03337-3
98. Liang H, Zhang Y, Miao Z, Cheng R, Jiang F, Ze X, et al. Anti-allergic effects of two potential probiotic strains isolated from infant feces in China. J Funct Foods. (2022) 92:105070. doi: 10.1016/j.jff.2022.105070
99. Lee JY, Kang J-H, Jung Y-R, Kang C-H. Lactobacillus gasseri MG4247 and Lacticaseibacillus paracasei MG4272 and MG4577 modulate allergic inflammatory response in RAW 264.7 and RBL-2H3 cells. Probiotics Antimicrob Proteins. (2022) 31:1–10. doi: 10.1007/s12602-022-09950-4
100. Todhanakasem T. Microbial biofilm in the industry. Afr J Microbiol Res. (2013) 7:1625–34. doi: 10.5897/AJMR12.2146
101. Alreshidi MM, Dunstan RH, Macdonald MM, Smith ND, Gottfries J, Roberts TK. Metabolomic and proteomic responses of Staphylococcus aureus to prolonged cold stress. J Proteomics. (2015) 121:44–55. doi: 10.1016/j.jprot.2015.03.010
102. Alreshidi MM. Selected metabolites profiling of Staphylococcus aureus following exposure to low temperature and elevated sodium chloride. Front Microbiol. (2020) 11:834. doi: 10.3389/fmicb.2020.00834
103. Alreshidi MM, Dunstan RH, Macdonald MM, Gottfries J, Roberts TK. The uptake and release of amino acids by Staphylococcus aureus at mid-exponential and stationary phases and their corresponding responses to changes in temperature, pH and osmolality. Front Microbiol. (2020) 10:3059. doi: 10.3389/fmicb.2019.03059
104. Alreshidi M, Dunstan H, Roberts T, Bardakci F, Badraoui R, Adnan M, et al. Changes in amino acid metabolism of Staphylococcus aureus following growth to the stationary phase under adjusted growth conditions. Microorganisms. (2022) 10:1503. doi: 10.3390/microorganisms10081503
105. Alreshidi MM, Dunstan RH, Macdonald MM, Singh VK, Roberts TK. Analysis of cytoplasmic and secreted proteins of Staphylococcus aureus revealed adaptive metabolic homeostasis in response to changes in the environmental conditions representative of the human wound site. Microorganisms. (2020) 8:1082. doi: 10.3390/microorganisms8071082
106. Kubota H, Senda S, Nomura N, Tokuda H, Uchiyama H. Biofilm formation by lactic acid bacteria and resistance to environmental stress. J Biosci Bioeng. (2008) 106:381–6. doi: 10.1263/jbb.106.381
107. Patel M, Siddiqui AJ, Hamadou WS, Surti M, Awadelkareem AM, Ashraf SA, et al. Inhibition of bacterial adhesion and antibiofilm activities of a glycolipid biosurfactant from Lactobacillus rhamnosus with its physicochemical and functional properties. Antibiotics. (2021) 10:1546. doi: 10.3390/antibiotics10121546
108. García-Cayuela T, Korany A, Bustos I, Gómez de Cadiñanos L, Requena T, Peláez C, et al. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res Int. (2014) 57:44–50. doi: 10.1016/j.foodres.2014.01.010
109. Tatsaporn T, Kornkanok K. Using potential lactic acid bacteria biofilms and their compounds to control biofilms of foodborne pathogens. Biotechnol Rep. (2020) 26:e00477. doi: 10.1016/j.btre.2020.e00477
110. German B, Schiffrin EJ, Reniero R, Mollet B, Pfeifer A, Neeser JR. The development of functional foods: lessons from the gut. Trends Biotechnol. (1999) 17:492–9. doi: 10.1016/S0167-7799(99)01380-3
111. Mokhtar M, Rismayuddin NAR, Mat Yassim AS, Ahmad H, Abdul Wahab R, Dashper S, et al. Streptococcus salivarius K12 inhibits Candida albicans aggregation, biofilm formation and dimorphism. Biofouling. (2021) 37:767–76. doi: 10.1080/08927014.2021.1967334
112. Tomièiæ R, Tomièiæ Z, Raspor P. Influence of culture conditions on co-aggregation of probiotic yeast Saccharomyces boulardii with Candida spp. and their auto-aggregation. Folia Microbiol. (2022) 67:507–15. doi: 10.1007/s12223-022-00956-7
113. Duraisamy S, Husain F, Balakrishnan S, Sathyan A, Subramani P, Chidambaram P, et al. Phenotypic assessment of probiotic and bacteriocinogenic efficacy of indigenous LAB strains from human breast milk. Curr Issues Mol Biol. (2022) 44:731–49. doi: 10.3390/cimb44020051
114. Srikham K, Thirabunyanon M. Bioprophylactic potential of novel human colostrum probiotics via apoptotic induction of colon cancer cells and cell immune activation. Biomed Pharmacother. (2022) 149:112871. doi: 10.1016/j.biopha.2022.112871
115. Moraffah F, Kiani M, Abdollahi M, Yoosefi S, Vatanara A, Samadi N. In vitro-in vivo correlation for the antibacterial effect of Lactiplantibacillus plantarum as a topical healer for infected burn wound. Probiotics Antimicrob Proteins. (2022) 14:675–89. doi: 10.1007/s12602-022-09934-4
116. Luan C, Jiang N, Zhou X, Zhang C, Zhao Y, Li Z, et al. Antibacterial and anti-biofilm activities of probiotic Lactobacillus curvatus BSF206 and Pediococcus pentosaceus AC1-2 against Streptococcus mutans. Microbial Pathog. (2022) 164:105446. doi: 10.1016/j.micpath.2022.105446
117. Hyland NP, Quigley EMM, Brint E. Microbiota-host interactions in irritable bowel syndrome: epithelial barrier, immune regulation and brain-gut interactions. World J Gastroenterol. (2014) 20:8859–66.
118. Liu Z, Qin H, Yang Z, Xia Y, Liu W, Yang J, et al. Randomised clinical trial: the effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery – a double-blind study. Aliment Pharmacol Therap. (2011) 33:50–63. doi: 10.1111/j.1365-2036.2010.04492.x
119. Kocot AM, Jarocka-Cyrta E, Drabiñska N. Overview of the importance of biotics in gut barrier integrity. Int J Mol Sci. (2022) 23:2896. doi: 10.3390/ijms23052896
120. Martel J, Chang S-H, Ko Y-F, Hwang T-L, Young JD, Ojcius DM. Gut barrier disruption and chronic disease. Trends Endocrinol Metab. (2022) 33:247–65. doi: 10.1016/j.tem.2022.01.002
121. Nourizadeh R, Sepehri B, Abbasi A, Sayyed RZ, Khalili L. Impact of probiotics in modulation of gut microbiome. In: RZ Sayyed, M Khan editors. Microbiome-Gut-Brain Axis: Implications on Health. Singapore: Springer (2022). p. 401–9. doi: 10.1007/978-981-16-1626-6_20
123. Fuller R, Gibson GR. Modification of the intestinal microflora using probiotics and prebiotics. Scand J Gastroenterol Suppl. (1997) 222:28–31. doi: 10.1080/00365521.1997.11720714
124. Rose EC, Odle J, Blikslager AT, Ziegler AL. Probiotics, prebiotics and epithelial tight junctions: a promising approach to modulate intestinal barrier function. Int J Mol Sci. (2021) 22:6729. doi: 10.3390/ijms22136729
125. Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol. (1999) 276:G941–50. doi: 10.1152/ajpgi.1999.276.4.G941
126. Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. (2010) 298:G807–19. doi: 10.1152/ajpgi.00243.2009
127. Miyamoto J, Mizukure T, Park SB, Kishino S, Kimura I, Hirano K, et al. A gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40-MEK-ERK pathway. J Biol Chem. (2015) 290:2902–18. doi: 10.1074/jbc.M114.610733
128. Zhao L, Xie Q, Etareri Evivie S, Liu D, Dong J, Ping L, et al. Bifidobacterium dentium N8 with potential probiotic characteristics prevents LPS-induced intestinal barrier injury by alleviating the inflammatory response and regulating the tight junction in Caco-2 cell monolayers. Food Funct. (2021) 12:7171–84. doi: 10.1039/D1FO01164B
129. Zhang J, Li Q, Wu L, Xu S, Lu R. Protective effect of surface-layer proteins from four Lactobacillus strains on tumor necrosis factor-α-induced intestinal barrier dysfunction. J Sci Food Agric. (2022) 102:4446–53. doi: 10.1002/jsfa.11798
130. Sadiq MB, Azhar F-U-A, Ahmad I. Probiotic and prebiotic interactions and their role in maintaining host immunity. In: RZ Sayyed, M Khan editors. Microbiome-Gut-Brain Axis: Implications on Health. Singapore: Springer (2022). p. 425–43. doi: 10.1007/978-981-16-1626-6_22
131. Thipe VC, Mentor S, Lima CSA, Freitas LF, Fonseca ACM, Nogueira KM, et al. Chapter 3 – The role of probiotics in maintaining immune homeostasis. In: MK Dwivedi, N Amaresan, A Sankaranarayanan, EH Kemp editors. Probiotics in the Prevention and Management of Human Diseases. Cambridge, MA: Academic Press (2022). p. 41–58. doi: 10.1016/B978-0-12-823733-5.00024-6
132. Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, et al. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. (2020) 88:209–17. doi: 10.1038/s41390-019-0740-x
133. Arnold M, Moore SW, Nadler EP. Necrotizing enterocolitis. In: EA Ameh, SW Bickler, K Lakhoo, BC Nwomeh, D Poenaru editors. Pediatric Surgery: A Comprehensive Textbook For Africa. Cham: Springer International Publishing (2020). p. 727–45. doi: 10.1007/978-3-030-41724-6_70
134. Mustapha M, Wilson KA, Barr S. Optimising nutrition of preterm and term infants in the neonatal intensive care unit. Paediatr Child Health. (2021) 31:38–45. doi: 10.1016/j.paed.2020.10.008
135. Luo H, Cao G, Luo C, Tan D, Vong CT, Xu Y, et al. Emerging pharmacotherapy for inflammatory bowel diseases. Pharmacol Res. (2022) 178:106146. doi: 10.1016/j.phrs.2022.106146
136. Shan Y, Lee M, Chang EB. The gut microbiome and inflammatory bowel diseases. Annu Rev Med. (2022) 73:455–68. doi: 10.1146/annurev-med-042320-021020
137. Lee SY, Lee BH, Park JH, Park MS, Ji GE, Sung MK. Bifidobacterium bifidum BGN4 paraprobiotic supplementation alleviates experimental colitis by maintaining gut barrier and suppressing nuclear factor kappa B activation signaling molecules. J Med Food. (2022) 25:146–57. doi: 10.1089/jmf.2021.K.0150
138. Dong F, Xiao F, Li X, Li Y, Wang X, Yu G, et al. Pediococcus pentosaceus CECT 8330 protects DSS-induced colitis and regulates the intestinal microbiota and immune responses in mice. J Transl Med. (2022) 20:33. doi: 10.1186/s12967-022-03235-8
139. Li P, Chen G, Zhang J, Pei C, Chen Y, Gong J, et al. Live Lactobacillus acidophilus alleviates ulcerative colitis via the SCFAs/mitophagy/NLRP3 inflammasome axis. Food Funct. (2022) 13:2985–97. doi: 10.1039/D1FO03360C
140. Tungsanga S, Katavetin P, Panpetch W, Udompornpitak K, Saisorn W, Praditpornsilpa K, et al. Lactobacillus rhamnosus L34 attenuates chronic kidney disease progression in 5/6 nephrectomy mouse model through the excretion of anti-inflammatory molecules. Nephrol Dial Transplant. (2022). gfac032. doi: 10.1093/ndt/gfac032
141. Gupta S, Kanwar S. Intestinal dysbiosis induced chronic inflammation that risks up oxalate nephropathy. J Cell Sci Ther. (2022) 13:342.
142. Noce A, Marchetti M, Marrone G, Di Renzo L, Di Lauro M, Di Daniele F, et al. Link between gut microbiota dysbiosis and chronic kidney disease. Eur Rev Med Pharmacol Sci. (2022) 26:2057–74.
143. Yoo JY, Sniffen S, McGill Percy KC, Pallaval VB, Chidipi B. Gut dysbiosis and immune system in Atherosclerotic Cardiovascular Disease (ACVD). Microorganisms. (2022) 10:108. doi: 10.3390/microorganisms10010108
144. Choi E, Yang J, Ji G-E, Park MS, Seong Y, Oh SW, et al. The effect of probiotic supplementation on systemic inflammation in dialysis patients. Kidney Res Clin Pract. (2022) 41:89–101. doi: 10.23876/j.krcp.21.014
145. Mehra. A. Probiotics Market. by Application (Functional Food & Beverages (Dairy Products, Non-dairy Beverages, Infant Formula, Cereals), Dietary Supplements, Feed), Ingredient (Bacteria, Yeast), Form (Dry, Liquid), End User, & Region – Global Forecast to 2026. (2022). Available online at: https://www.marketsandmarkets.com/PressReleases/probiotics.asp (accessed June 2, 2022).
146. Sanders ME, Merenstein D, Merrifield CA, Hutkins R. Probiotics for human use. Nutr Bull. (2018) 43:212–25. doi: 10.1111/nbu.12334
147. Sharifi-Rad J, Rodrigues CF, Stojanoviæ-Radiæ Z, Dimitrijeviæ M, Aleksiæ A, Neffe-Skociñska K, et al. Probiotics: versatile bioactive components in promoting human health. Medicina (Kaunas). (2020) 56:433. doi: 10.3390/medicina56090433
148. Huys G, Botteldoorn N, Delvigne F, De Vuyst L, Heyndrickx M, Pot B, et al. Microbial characterization of probiotics–advisory report of the working group “8651 probiotics” of the Belgian Superior Health Council (SHC). Mol Nutr Food Res. (2013) 57:1479–504. doi: 10.1002/mnfr.201300065
149. Rodrigues FJ, Cedran MF, Bicas JL, Sato HH. Encapsulated probiotic cells: relevant techniques, natural sources as encapsulating materials and food applications – A narrative review. Food Res Int. (2020) 137:109682. doi: 10.1016/j.foodres.2020.109682
150. Chen J, Wang Q, Liu C-M, Gong J. Issues deserve attention in encapsulating probiotics: critical review of existing literature. Crit Rev Food Sci Nutr. (2017) 57:1228–38. doi: 10.1080/10408398.2014.977991
151. Sniffen JC, McFarland LV, Evans CT, Goldstein EJC. Choosing an appropriate probiotic product for your patient: an evidence-based practical guide. PLoS One. (2018) 13:e0209205. doi: 10.1371/journal.pone.0209205
152. Binda S, Hill C, Johansen E, Obis D, Pot B, Sanders ME, et al. Criteria to qualify microorganisms as “probiotic” in foods and dietary supplements. Front Microbiol. (2020) 11:1662. doi: 10.3389/fmicb.2020.01662
153. Siezen RJ, van Hylckama Vlieg JE. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb Cell Fact. (2011) 10(Suppl. 1):S3. doi: 10.1186/1475-2859-10-S1-S3
154. Garcia-Gonzalez N, Battista N, Prete R, Corsetti A. Health-promoting role of Lactiplantibacillus plantarum isolated from fermented foods. Microorganisms. (2021) 9:349. doi: 10.3390/microorganisms9020349
155. Food and Agriculture Organization of the United Nations.Probiotics in Food: HEALTH and Nutritional Properties and Guidelines for Evaluation: Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria, Cordoba, Argentina, 1-4 October 2001 [and] Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food, London, Ontario, Canada, 30 April -1 May 2002. Rome: Food and Agriculture Organization of the United Nations (2006).
156. Seddik HA, Bendali F, Gancel F, Fliss I, Spano G, Drider D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob Proteins. (2017) 9:111–22. doi: 10.1007/s12602-017-9264-z
157. Bourdichon F, Laulund S, Tenning P. Inventory of microbial species with a rationale: a comparison of the IDF/EFFCA inventory of microbial food cultures with the EFSA Biohazard Panel qualified presumption of safety. FEMS Microbiol Lett. (2019) 366:fnz048. doi: 10.1093/femsle/fnz048
158. Koutsoumanis K, Allende A, Alvarez−Ordóñez A, Bolton D, Bover−Cid S, Chemaly M, et al. Scientific opinion on the update of the list of QPS−recommended biological agents intentionally added to food or feed as notified to EFSA (2017–2019). EFSA J. (2020) 18:e05966. doi: 10.2903/j.efsa.2020.5966
159. Papadimitriou K, Alegría Á, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, et al. Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev. (2016) 80:837–90. doi: 10.1128/MMBR.00076-15
160. Suryavanshi MV, Paul D, Doijad SP, Bhute SS, Hingamire TB, Gune RP, et al. Draft genome sequence of Lactobacillus plantarum strains E2C2 and E2C5 isolated from human stool culture. Stand Genomic Sci. (2017) 12:15. doi: 10.1186/s40793-017-0222-x
161. Spinler JK, Sontakke A, Hollister EB, Venable SF, Oh PL, Balderas MA, et al. From prediction to function using evolutionary genomics: human-specific ecotypes of Lactobacillus reuteri have diverse probiotic functions. Genome Biol Evol. (2014) 6:1772–89. doi: 10.1093/gbe/evu137
162. Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, et al. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. (2017) 41(Supp_1):S27–48. doi: 10.1093/femsre/fux030
163. Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, et al. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. (2010) 4:377–87. doi: 10.1038/ismej.2009.123
164. Mendes-Soares H, Suzuki H, Hickey RJ, Forney LJ. Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment. J Bacteriol. (2014) 196:1458–70. doi: 10.1128/JB.01439-13
165. Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol. (2008) 74:4985–96. doi: 10.1128/AEM.00753-08
166. Tannock GW, Munro K, Harmsen HJM, Welling GW, Smart J, Gopal PK. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol. (2000) 66:2578–88. doi: 10.1128/AEM.66.6.2578-2588.2000
167. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. (1977) 31:107–33. doi: 10.1146/annurev.mi.31.100177.000543
168. Azcarate-Peril MA, Altermann E, Goh YJ, Tallon R, Sanozky-Dawes RB, Pfeiler EA, et al. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl Environ Microbiol. (2008) 74:4610–25. doi: 10.1128/AEM.00054-08
169. Forde BM, Neville BA, O’ Donnell MM, Riboulet-Bisson E, Claesson MJ, Coghlan A, et al. Genome sequences and comparative genomics of two Lactobacillus ruminis strains from the bovine and human intestinal tracts. Microbial Cell Fact. (2011) 10:S13. doi: 10.1186/1475-2859-10-S1-S13
170. Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. (2011) 7:e1001314. doi: 10.1371/journal.pgen.1001314
171. Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr Issues Intest Microbiol. (2001) 2:43–53.
172. Frese S, Hutkins R, Walter J. Comparison of the colonization ability of autochthonous and allochthonous strains of lactobacilli in the human gastrointestinal tract. Adv Microbiol. (2012) 2:399–409. doi: 10.4236/aim.2012.23051
173. van den Nieuwboer M, van Hemert S, Claassen E, de Vos WM. Lactobacillus plantarum WCFS1 and its host interaction: a dozen years after the genome. Microb Biotechnol. (2016) 9:452–65. doi: 10.1111/1751-7915.12368
174. Zhang C, Derrien M, Levenez F, Brazeilles R, Ballal SA, Kim J, et al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. (2016) 10:2235–45. doi: 10.1038/ismej.2016.13
175. Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. (2018) 174:1388–405.e21. doi: 10.1016/j.cell.2018.08.041
176. Duar RM, Frese SA, Lin XB, Fernando SC, Burkey TE, Tasseva G, et al. Experimental evaluation of host adaptation of Lactobacillus reuteri to different vertebrate species. Appl Environ Microbiol. (2017) 83:e00132–17. doi: 10.1128/AEM.00132-17
177. Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci U.S.A. (2011) 108(Suppl. 1):4645–52. doi: 10.1073/pnas.1000099107
178. Doron S, Snydman DR, Gorbach SL. Lactobacillus GG: bacteriology and clinical applications. Gastroenterol Clin North Am. (2005) 34:483–98, ix. doi: 10.1016/j.gtc.2005.05.011
179. Wong CB, Sugahara H, Odamaki T, Xiao JZ. Different physiological properties of human-residential and non-human-residential bifidobacteria in human health. Benef Microbes. (2018) 9:111–22. doi: 10.3920/BM2017.0031
180. Katayama T. Host-derived glycans serve as selected nutrients for the gut microbe: human milk oligosaccharides and bifidobacteria. Biosci Biotechnol Biochem. (2016) 80:621–32. doi: 10.1080/09168451.2015.1132153
181. Kumar R, Sood U, Gupta V, Singh M, Scaria J, Lal R. Recent advancements in the development of modern probiotics for restoring human gut microbiome dysbiosis. Indian J Microbiol. (2020) 60:12–25. doi: 10.1007/s12088-019-00808-y
182. Koll P, Mändar R, Smidt I, Hütt P, Truusalu K, Mikelsaar R-H, et al. Screening and evaluation of human intestinal lactobacilli for the development of novel gastrointestinal probiotics. Curr Microbiol. (2010) 61:560–6. doi: 10.1007/s00284-010-9653-y
183. Nyholm L, Koziol A, Marcos S, Botnen AB, Aizpurua O, Gopalakrishnan S, et al. Holo-omics: integrated host-microbiota multi-omics for basic and applied biological research. iScience. (2020) 23:101414. doi: 10.1016/j.isci.2020.101414
184. Veiga P, Suez J, Derrien M, Elinav E. Moving from probiotics to precision probiotics. Nat Microbiol. (2020) 5:878–80. doi: 10.1038/s41564-020-0721-1
185. Yan F, Polk DB. Probiotics and probiotic-derived functional factors—mechanistic insights into applications for intestinal homeostasis. Front Immunol. (2020) 11:1428. doi: 10.3389/fimmu.2020.01428
Keywords: lactic acid bacteria, host-adapted strains, target-based probiotics, human probiotics formulation, multi-omics, bioprophylactics, biotherapeutics
Citation: Idrees M, Imran M, Atiq N, Zahra R, Abid R, Alreshidi M, Roberts T, Abdelgadir A, Tipu MK, Farid A, Olawale OA and Ghazanfar S (2022) Probiotics, their action modality and the use of multi-omics in metamorphosis of commensal microbiota into target-based probiotics. Front. Nutr. 9:959941. doi: 10.3389/fnut.2022.959941
Received: 03 June 2022; Accepted: 24 August 2022;
Published: 16 September 2022.
Edited by:
Mitesh Patel, Parul University, IndiaReviewed by:
Tarun Kumar Upadhyay, Parul University, IndiaKartik Patel, Veer Narmad South Gujarat University, India
Copyright © 2022 Idrees, Imran, Atiq, Zahra, Abid, Alreshidi, Roberts, Abdelgadir, Tipu, Farid, Olawale and Ghazanfar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maryam Idrees, bWFyeWFtaWRyZWVzQGJzLnFhdS5lZHUucGs=; orcid.org/0000-0001-6948-841X; Mousa Alreshidi, bW8uYWxyZXNoaWRpQHVvaC5lZHUuc2E=; Shakira Ghazanfar, c2hha2lyYV9ha21hbEB5YWhvby5jb20=
 Maryam Idrees
Maryam Idrees Muhammad Imran
Muhammad Imran Naima Atiq1
Naima Atiq1 Rabaab Zahra
Rabaab Zahra Rameesha Abid
Rameesha Abid Tim Roberts
Tim Roberts Muhammad Khalid Tipu
Muhammad Khalid Tipu Arshad Farid
Arshad Farid Shakira Ghazanfar
Shakira Ghazanfar