- 1Department of Nutrition, Exercise and Sports, University of Copenhagen, Copenhagen, Denmark
- 2Novo Nordisk Foundation, Copenhagen, Denmark
- 3Department of Food Science, University of Copenhagen, Copenhagen, Denmark
- 4National Food Institute, Technical University of Denmark, Kongens Lyngby, Denmark
Background: Salivary amylase (AMY1) gene copy number (CN) and Prevotella abundance in the gut are involved in carbohydrate digestion in the upper and lower gastrointestinal tract, respectively; and have been suggested as prognostic biomarkers for weight loss among overweight individuals consuming diets rich in fiber and wholegrains.
Objective: We hypothesized that Prevotella abundance would be linked to greater loss of body fat after wholegrain consumption among individuals with low AMY1 CN, but not in those with high AMY1 CN.
Methods: We reanalyzed data from two independent randomized ad libitum wholegrain interventions (fiber intake ∼33 g/d for 6–8 weeks), to investigate the relationship between baseline Prevotella abundance and body fat loss among healthy, overweight participants stratified into two groups by median AMY1 CN. Individuals with no detected Prevotella spp. were excluded from the main analysis.
Results: In both studies, individuals with low AMY1 CN exhibited a positive correlation between baseline Prevotella abundance and fat loss after consuming the wholegrain diet (r > 0.5, P < 0.05), but no correlation among participants with high AMY1 CN (P ≥ 0.6). Following consumption of the refined wheat control diets, there were no associations between baseline Prevotella abundance and changes in body fat in any of the AMY1 groups.
Conclusion: These results suggest that Prevotella abundance together with AMY1 CN can help predict fat loss in response to ad libitum wholegrain diets, highlighting the potential of these biomarkers in personalized obesity management.
Introduction
Ad libitum diets rich in wholegrain (WG) have been found to reduce body weight compared with diets rich in refined grains (1–3). However, there is a large inter-individual variation in the weight loss success upon WG consumption. Given the role of the gut microbiota in digesting dietary polysaccharides (4), it is likely that individual differences in gut microbiota explain some of this variation in weight loss (5). Accordingly, we demonstrated that participants with high abundance of Prevotella lost more weight than participants with low abundance of Prevotella after 6 weeks of WG consumption (6), and that this loss could be associated with change in appetite regulation (7). Besides dietary fiber, WG is rich in starch, which is degraded enzymatically in humans by salivary amylase; the enzymatic ability to degrade starch differs considerably from individual to individual, as the copy number (CN) of the salivary amylase gene (AMY1) has been shown to vary from 1 to 27 (8). Consequently, AMY1 CN influences the amount of starch that remains undigested and made available to gut microbes (9), and can therefore influence the composition and function of gut microbial populations. For instance, participants with low AMY1 copy numbers have enhanced gut microbiome capacity to break down polysaccharides (10) and produce more methane (11). AMY1 CN has also been associated - both positively and negatively – with adiposity (12, 13), and we recently found that weight loss after consumption of a plant and fiber-rich diet for 26 weeks was positively associated with baseline Prevotella-to-Bacteroides ratio in overweight participants with low AMY CN, but not in participants with high AMY CN (14).
Here, we investigated the interaction between baseline Prevotella abundance and AMY1 CN in predicting weight loss and body fat loss in two independent but similar WG intervention trials. We hypothesized that Prevotella abundance would be linked to greater loss of body weight and fat after ad libitum intake of WG for 6–8 weeks in healthy, overweight participants with low AMY1 CN, but not in those with high AMY1 CN.
Materials and methods
Both WG intervention studies, study 1 [Vuholm et al. (15)] and study 2 [Roager et al. (2)], were conducted in accordance with the 1975 Declaration of Helsinki guidelines at the Department of Nutrition, Exercise, and Sports, University of Copenhagen, Denmark, in 2013 and 2014 respectively.
Study designs
Study 1: As described in Vuholm et al. (15), 75 healthy overweight adults were enrolled in a randomized, controlled, researcher-blinded, parallel 6-week trial. Subjects were assigned to either WG (rye and wheat) or refined wheat (RW) diets. At weeks 0 and 6, subjects collected a spot fecal sample, which was analyzed to determine gut microbiota; body composition measurements were performed by Dual-energy X-ray absorptiometry (DEXA). Study 2: As described in Roager et al. (2), 60 healthy overweight adults were enrolled in a randomized, controlled cross-over trial with two 8-week periods (WG diet or RW diet in random order, separated by a 6-week washout). Examinations were conducted at the beginning and the end of each period and included collection of a spot fecal sample, and body composition measurements performed by bioelectrical impedance analysis (QuadScan 4000, Bodystat Inc., Isle of Man, British Isles, United Kingdom).
Intervention diets
In both studies, diets were consumed ad libitum with no caloric restriction, and participants were instructed to replace all cereal products in their habitual diet with the provided WG products (while the control group received refined grain products). At baseline and during the last week of the interventions, participants completed a 4-day food record with a registration of the type and amount of all foods and beverages that they had consumed. Dietary compliance was evaluated by measuring plasma alkylresorcinols, as previously described (2, 15).
Fecal microbiota composition
Study 1: The microbiota composition of pretreatment fecal samples was analyzed by 16S rRNA gene (V3–V4 region) MiSeq-based (Illumina Inc.) sequencing. The Greengenes (version 13.8, Quantitative Insight Into Microbial Ecology) 16S rRNA gene collection was used as a reference database, and QIIME (versions 1.7.0 and 1.8.0) was used for analyzing the sequenced data with subsampling at 9,500 reads/sample, as described previously (15). Study 2: The pretreatment microbiota composition was analyzed by shotgun metagenomic sequencing with an average of seven gigabases readout per sample (Illumina 100 bp pair end), and the microbial sequences obtained from shotgun metagenomics were mapped to the integrated catalog of reference genes of the human gut microbiome (i.e., integrated gene catalog), as previously described (2). In both studies, genus-level baseline log-transformed Prevotella abundance was used as a surrogate of microbial enterotype (16). Subjects with no detected Prevotella spp. were excluded from the main analyses (but included in a secondary analysis), as previous studies indicate that these subjects respond differently than subjects with high and low Prevotella abundance, respectively, when consuming fiber-rich diets (6, 7).
Salivary amylase gene copy number
In both studies, the copy number variation of the AMY1 locus was analyzed from human buffy coat samples using droplet digital polymerase chain reaction (ddPCR), as previously described (14). Copy numbers of AMY1 were analyzed using QuantaSoft software version 1.7.4 (Bio-Rad Laboratories), assuming a diploid nature for the EIF2C1 reference gene. The median values of AMY1 CN was used as the cutoff to define low and high AMY1 CN groups in study 1 (median = 6.8) and study 2 (median = 6.4).
Statistics
Statistical analyses were conducted in R [R Core Team, (17)], version 3.4.2 (R Foundation). For each study, differences in baseline characteristics between the AMY1 groups were evaluated using an unpaired two sample t-test (normally distributed data), Wilcoxon rank-sum test (non-normally distributed data), and Pearson’s chi-square test (categorical data). Normality was assessed by visual inspection of residual plots, histograms and normal probability plots. Data not normally distributed including fecal microbiome data were log transformed before analysis. Correlations between baseline log-transformed Prevotella abundance and changes in body weight and fat (difference between endpoint and baseline) were analyzed by means of Pearson’s correlation coefficients in R and using the ppcor package when controlling for baseline body weight or baseline body fat, respectively. After combining the two studies, a linear multiple regression model was applied to assess the relationship between baseline log-transformed Prevotella abundance and change in body fat percentage while adjusting for baseline body fat percentage and study. Plots were created in R, version 3.4.2, and GraphPad Prism, version 8.3.1. The level of significance was set at P < 0.05.
Results
In study 1 [Vuholm et al. (15)] and 2 [Roager et al. (2)], 34 and 36 participants with detectable Prevotella abundance at baseline completed the WG interventions, respectively (Figure 1). Among these participants, AMY1 CN ranged from 2 to 12 (Supplementary Figures 1, 2). After stratifying participants by median AMY1 CN for each study, no differences in baseline characteristics, including body weight, body fat percentage, and Prevotella abundance were found between high and low AMY1 CN groups (Supplementary Tables 1, 2). During the ad libitum WG interventions, participants consumed on average 33 g (±11.4 g) of dietary fiber for 6 weeks (study 1) and 33 g (±10.0 g) of dietary fiber for 8 weeks (study 2).
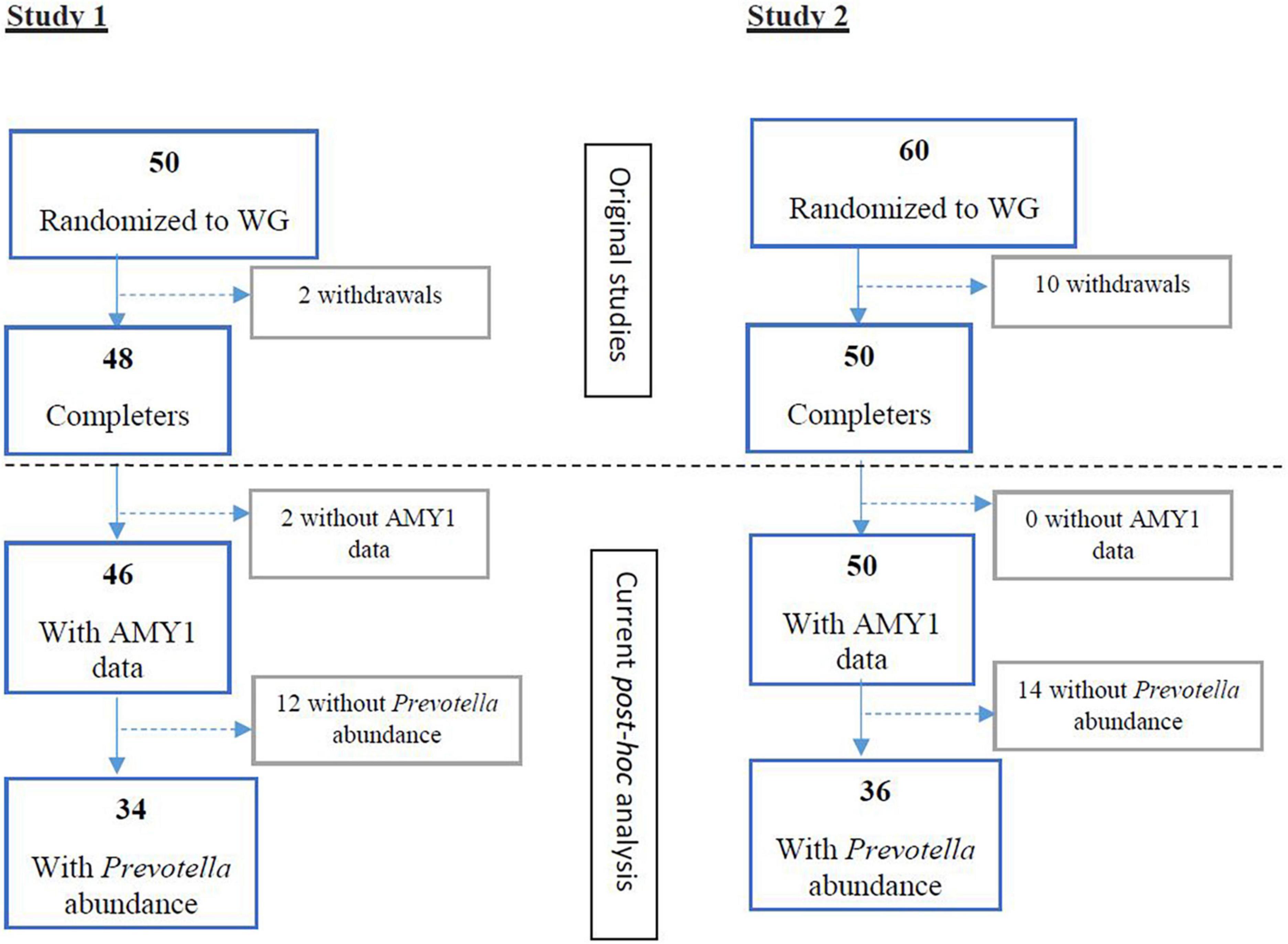
Figure 1. Flow of participants randomized to wholegrain (WG) diets and with baseline Prevotella abundance and AMY1 gene copy number data for study 1 (15) and study 2 (2). WG, wholegrain.
Body fat loss is influenced by salivary amylase gene copy number and Prevotella abundance
Among participants with low AMY1 CN in study 1 (n = 17), baseline Prevotella abundance correlated with 6-wk changes in body weight (r = −0.59, P = 0.014), body fat mass (r = −0.72, P = 0.0012), and body fat percentage (r = −0.80, P = 0.0001) (Figure 2A). The association between Prevotella abundance and body fat percentage was also evident after controlling for baseline body fat percentage (P = 0.0002). On the contrary, no such relationship was observed among those with high AMY1 CN (n = 17, P ≥ 0.6) (Figure 2B).
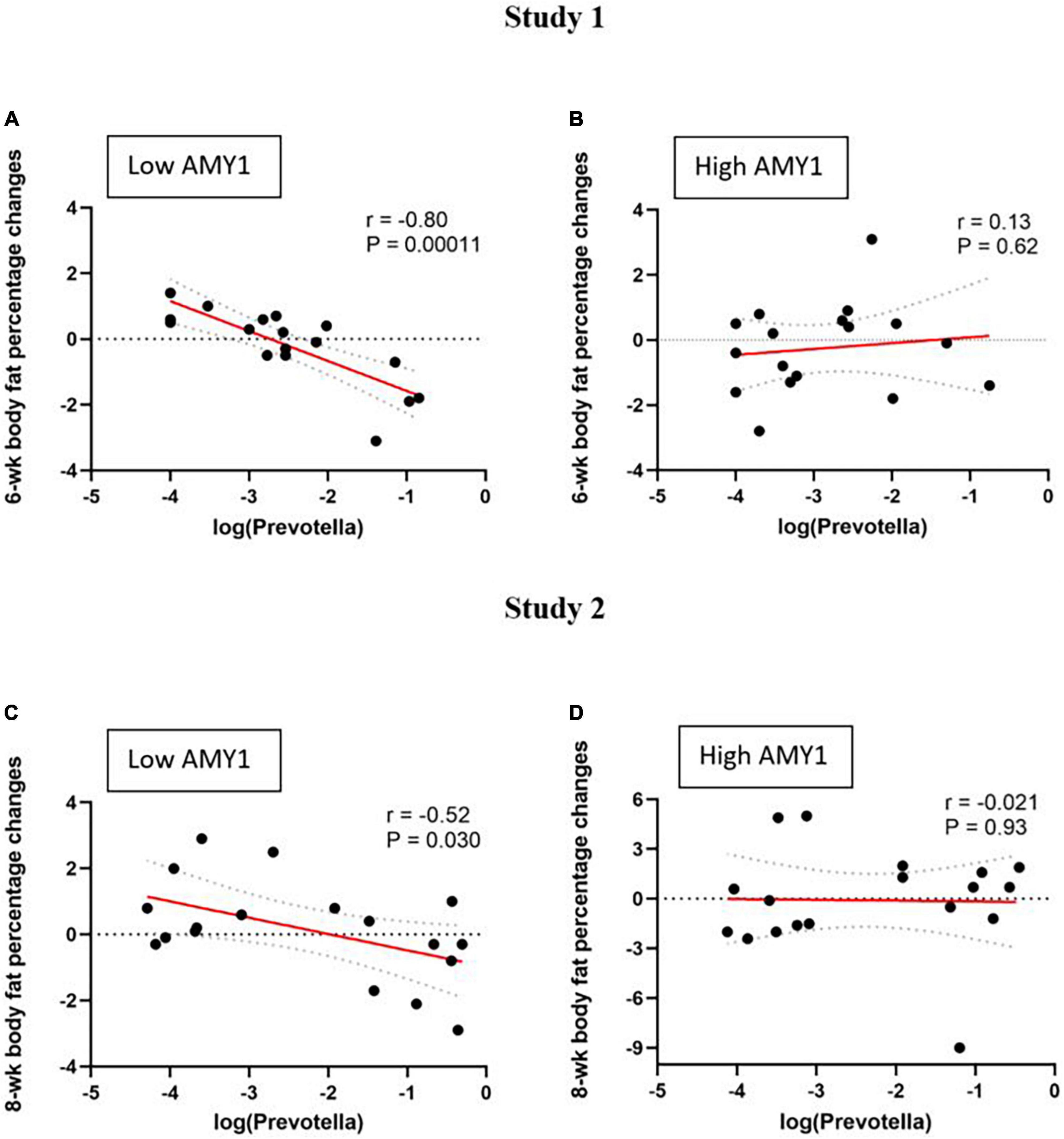
Figure 2. Correlations between baseline Prevotella abundance and change in body fat percentage (difference between endpoint and baseline) during two different wholegrain interventions in people with low and high AMY1 gene copy number. Study 1 (n = 34): (A) low AMY1 (n = 17) and (B) high AMY1 (n = 17) groups; and Study 2 (n = 36): (C) low AMY1 (n = 18) and (D) high AMY1 (n = 18) groups. Pearson’s correlation coefficients (r) and corresponding P-values are shown. Linear regression are depicted in solid red lines and respective 95% confidence intervals are drawn in dashed lines.
Similarly, among low AMY1 CN participants in study 2, we found a relationship between Prevotella abundance and 8-wk changes in body fat percentage (n = 18, r = −0.52, P = 0.030) (Figure 2C). This association was also evident after controlling for baseline body fat percentage (P < 0.04). Again, this association was not observed among the high AMY1 participants (n = 18, P ≥ 0.9) (Figure 2D). Also, in study 2, fat mass loss following WG consumption tended to correlate with baseline Prevotella abundance among the low AMY1 participants (r = 0.42, P = 0.08); however, weight loss did not correlate with Prevotella abundance in either AMY1 groups (P > 0.05).
When the studies were combined, we confirmed that baseline Prevotella abundance predicted change in body fat percentage (β = −0.59, P = 0.0005) among the participants with low AMY1 CN (n = 35) by multiple linear regression adjusted for baseline body fat percentage and study, while there was no relationship among the ones with high AMY1 CN (n = 35; β = −0.07, P = 0.84).
Notably, in agreement with our previous results (6), no correlations were observed between baseline Prevotella abundance and body weight and fat change in any of the studies when including the participants with no detectable Prevotella at baseline (Supplementary Figures 3, 4). Furthermore, none of the studies showed any correlations between baseline Prevotella abundance and changes in body weight and fat mass measures following consumption of the RW diets in any of the AMY1 groups (data not shown).
Discussion
Salivary amylase gene gene copy number and Prevotella abundance in the gut are both implicated in plant polysaccharide digestion in the upper and lower gastrointestinal tract, respectively (9, 18). We previously observed in study 1 that participants with greater Prevotella abundance lose more body weight in response to an ad libitum WG dietary intervention (6). Despite relatively small groups, we here found that combining Prevotella abundance with AMY1 CN leads to improved prediction of weight loss responses following WG consumption. More specifically, we found that the association between Prevotella abundance and fat loss was observed only among participants with low AMY1 CN. To validate this finding, we analyzed data from another independent WG intervention trial (2). Again, we showed that Prevotella abundance was positively correlated to fat loss among participants with low AMY1 CN. In both intervention studies, we found no associations between Prevotella and fat loss in the high AMY1 groups, or in either AMY1 group in response to the control RW-rich diets. Collectively, three independent studies in healthy, overweight Danish adults now link baseline Prevotella abundance to weight or fat loss success in individuals with low AMY1 participants consuming a fiber-rich diet (≥33 g/d) (14). However, it should be noted that Hjorth et al. (14) investigated a varied plant-based (New Nordic) diet with an even greater fiber intake (∼42 g/d), while the two present studies supplied WG rye and wheat products with 33 g/d of fibers and high abundance of arabinoxylan (AX) fibers. Previous studies suggest that P. copri, the main species of Prevotella (14), but also distinct Bacteroides species, are specialized in degrading AX fibers (19, 20). Yet, the functional capacity to degrade AX by P. copri depends on the abundance of different P. copri “clades” (21), which is a limitation in studies on the potential link between the abundance of the genus Prevotella and weight loss. Accordingly, this underlines the need to resolve microbial species and clades in future studies.
When considering that roughly 25–50% of caloric intake of typical diets comes from starch (22), and that the ability of the human host to degrade starch varies tremendously (8), it is likely that interactions between starch and amylase in the upper gastrointestinal tract have a substantial impact on glucose and body weight regulation, but also gut microbiota composition (8, 10–12, 23). With low amylase secretion due to low AMY1 CN, more starch from WG may escape digestion and propagate to the colon and thereby increase total availability of polysaccharide (9). This could potentially result in increased colonic fermentation depending on the individual’s gut microbiota composition and its capacity to metabolize starch. Besides dietary fiber, Prevotella has been linked to fermentation of starch in the gut (18), which lead to production of short-chain fatty acids (SCFAs) (24). SCFAs have been shown to promote production of appetite-regulating hormones in the gut, and may also enter the systemic circulation and affect adipose tissue and the brain (7).
In agreement with previous analyses (6, 25), we found individuals with no detectable Prevotella abundance at baseline independent of whether 16S rRNA sequencing (study 1) or shotgun metagenomics sequencing (study 2) were applied. Future studies will need to clarify whether these subjects do in fact have a distinct microbiota composition from low and high Prevotella subjects, or whether Prevotella in those subjects is simply below the detection limit of the sequencing methodologies (7). Other limitations of the current post-hoc analyses include the small sizes of the groups after stratification for AMY1 CN, and the relatively short duration of the studies, which is not optimal when investigating changes in body weight and body composition. Nonetheless, we analyzed changes in body fat, rather than only changes in body weight as done in previous similar studies (6, 20) in order to get insight into the tissue composition of the lost weight. In fact, we observed that body fat loss exhibited a stronger correlation with Prevotella abundance than body weight loss.
In conclusion, baseline Prevotella abundance was associated with greater fat loss in response to ad libitum wholegrain consumption among participants with low AMY1 CN, but not among participants with high AMY1 CN. This suggests that both baseline abundance of Prevotella and AMY1 CN may be used as biomarkers for predicting weight and fat loss responses to diets rich in fiber and wholegrains.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethical Committee of the Capital Region of Denmark. The patients/participants provided their written informed consent to participate in this study.
Author contributions
LC, MFH, and HR conceived the idea and designed the post-hoc analysis. LC performed lab work, analyzed data, and drafted the manuscript and had primary responsibility for the final content. All authors revised the manuscript critically for important intellectual content, and approved the final version.
Funding
The current study was supported by the Novo Nordisk Foundation (NNF19OC0056246; PRIMA—toward personalized dietary recommendations based on the interaction between diet, microbiome and abiotic conditions in the gut).
Acknowledgments
We thank Stine Vuholm, Mette Kristensen, and colleagues at University of Copenhagen for conducting study 1, and members of the 3G Center, Technical University of Denmark and colleagues at University of Copenhagen for conducting study 2.
Conflict of interest
MFH is co-inventor on a pending provisional patent application for the use of biomarkers to predict responses to weight-loss diets.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.947349/full#supplementary-material
Abbreviations
AMY1, salivary amylase gene; CN, copy number; SCFAs, short-chain fatty acids; RW, refined wheat; WG, wholegrain.
References
1. Suhr J, Vuholm S, Iversen KN, Landberg R, Kristensen M. Wholegrain rye, but not wholegrain wheat, lowers body weight and fat mass compared with refined wheat: A 6-week randomized study. Eur J Clin Nutr. (2017) 71:959–67.
2. Munch Roager H, Vogt JK, Kristensen M, Hansen LBS, Ibrügger S, Maerkedahl RB, et al. Whole grain-rich diet reduces body weight and systemic low-grade inflammation without inducing major changes of the gut microbiome: A randomised cross-over trial. Gut. (2019) 68:83–93. doi: 10.1136/gutjnl-2017-314786
3. Iversen KN, Carlsson F, Andersson A, Michaëlsson K, Langton M, Risérus U, et al. A hypocaloric diet rich in high fiber rye foods causes greater reduction in body weight and body fat than a diet rich in refined wheat: A parallel randomized controlled trial in adults with overweight and obesity (the RyeWeight study). Clin Nutr. (2021) 45:155–69. doi: 10.1016/j.clnesp.2021.07.007
4. Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. (2016) 535:56–64. doi: 10.1038/nature18846
5. Roager HM, Christensen LH. Personal diet-microbiota interactions and weight loss. Proc Nutr Soc. (2022) 1–28. doi: 10.1017/S0029665122000805
6. Christensen L, Vuholm S, Roager HM, Nielsen DS, Krych L, Kristensen M, et al. Prevotella abundance predicts weight loss success in healthy, overweight adults consuming a whole-grain diet ad libitum: A post hoc analysis of a 6-Wk randomized controlled trial. J Nutr. (2019) 149:2174–81. doi: 10.1093/jn/nxz198
7. Christensen L, Roager HM, Astrup A, Hjorth MF. Microbial enterotypes in personalized nutrition and obesity management. Am J Clin Nutr. (2018) 108:1–7.
8. Fernández CI, Wiley AS. Rethinking the starch digestion hypothesis for AMY1 copy number variation in humans. Am J Phys Anthropol. (2017) 163:645–57. doi: 10.1002/ajpa.23237
9. Elder PJ, Ramsden DB, Burnett D, Weickert MO, Barber TM. Human amylase gene copy number variation as a determinant of metabolic state. Expert Rev Endocrinol Metab. (2018) 13:193–205.
10. Poole AC, Goodrich JK, Youngblut ND, Luque GG, Ruaud A, Sutter JL, et al. Human salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe. (2019) 25:553–564.e7. doi: 10.1016/j.chom.2019.03.001
11. Atkinson FS, Hancock D, Petocz P, Brand-Miller JC. The physiologic and phenotypic significance of variation in human amylase gene copy number. Am J Clin Nutr. (2018) 108:737–48.
12. Falchi M, El-Sayed Moustafa JS, Takousis P, Pesce F, Bonnefond A, Andersson-Assarsson JC, et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat Genet. (2014) 46:492–7. doi: 10.1038/ng.2939
13. Bonnefond A, Yengo L, Dechaume A, Canouil M, Castelain M, Roger E, et al. Relationship between salivary/pancreatic amylase and body mass index: A systems biology approach. BMC Med. (2017) 15:37. doi: 10.1186/s12916-017-0784-x
14. Hjorth MF, Christensen L, Larsen TM, Roager HM, Krych L, Kot W, et al. Pretreatment Prevotella-to-Bacteroides ratio and salivary amylase gene copy number as prognostic markers for dietary weight loss. Am J Clin Nutr. (2020) 111:1079–86. doi: 10.1093/ajcn/nqaa007
15. Vuholm S, Nielsen DS, Iversen KN, Suhr J, Westermann P, Krych L, et al. Whole-grain rye and wheat affect some markers of gut health without altering the fecal microbiota in healthy overweight adults: A 6-week randomized trial. J Nutr. (2017) 147:2067–75. doi: 10.3945/jn.117.250647
16. Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. Microbial enterotypes, inferred by the Prevotella-to-Bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new Nordic diet. Appl Environ Microbiol. (2014) 80:1142–9. doi: 10.1128/AEM.03549-13
17. R Core Team. R: A Language and Environment for Statistical Computing. (2022). Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
18. Costea PI, Hildebrand F, Manimozhiyan A, Bäckhed F, Blaser MJ, Bushman FD, et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. (2017) 3:8–16.
19. Fehlner-Peach H, Magnabosco C, Raghavan V, Scher JU, Tett A, Cox LM, et al. Distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe. (2019) 26:680–90. doi: 10.1016/j.chom.2019.10.013
20. Christensen L, Sørensen CV, Wøhlk FU, Kjølbæk L, Astrup A, Sanz Y, et al. Microbial enterotypes beyond genus level: Bacteroides species as a predictive biomarker for weight change upon controlled intervention with arabinoxylan oligosaccharides in overweight subjects. Gut Microbes. (2020) 12:1–16. doi: 10.1080/19490976.2020.1847627
21. Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. (2019) 26:666–79. doi: 10.1016/j.chom.2019.08.018
22. Cust AE, Skilton MR, van Bakel MME, Halkjær J, Olsen A, Agnoli C, et al. Total dietary carbohydrate, sugar, starch and fibre intakes in the European prospective investigation into cancer and nutrition. Eur J Clin Nutr. (2009) 63:37–60.
23. Farrell M, Ramne S, Gouinguenet P, Brunkwall L, Ericson U, Raben A, et al. Effect of AMY1 copy number variation and various doses of starch intake on glucose homeostasis: Data from a cross-sectional observational study and a crossover meal study. Genes Nutr. (2021) 16:21. doi: 10.1186/s12263-021-00701-8
24. Hald S, Schioldan AG, Moore ME, Dige A, Lærke HN, Agnholt J, et al. Effects of arabinoxylan and resistant starch on intestinal microbiota and short-chain fatty acids in subjects with metabolic syndrome: A randomised crossover study. PLoS One. (2016) 11:e0159223. doi: 10.1371/journal.pone.0159223
Keywords: weight loss, obesity, enterotypes, microbiota, Prevotella, AMY1, wholegrains, dietary fiber
Citation: Christensen L, Hjorth MF, Krych L, Licht TR, Lauritzen L, Magkos F and Roager HM (2022) Prevotella abundance and salivary amylase gene copy number predict fat loss in response to wholegrain diets. Front. Nutr. 9:947349. doi: 10.3389/fnut.2022.947349
Received: 18 May 2022; Accepted: 02 August 2022;
Published: 22 August 2022.
Edited by:
Rikard Landberg, Chalmers University of Technology, SwedenReviewed by:
Johan Dicksved, Swedish University of Agricultural Sciences, SwedenEugeni Belda, Institut de Recherche pour le Development (IRD), France
Copyright © 2022 Christensen, Hjorth, Krych, Licht, Lauritzen, Magkos and Roager. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lars Christensen, bGFjaEBuZXhzLmt1LmRr
 Lars Christensen
Lars Christensen Mads F. Hjorth2
Mads F. Hjorth2 Lukasz Krych
Lukasz Krych Tine Rask Licht
Tine Rask Licht Lotte Lauritzen
Lotte Lauritzen Faidon Magkos
Faidon Magkos Henrik M. Roager
Henrik M. Roager