
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr., 15 July 2022
Sec. Nutritional Immunology
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.944390
This article is part of the Research TopicImmune-boosting Effects of Dietary Bioactive PolysaccharidesView all 10 articles
Early weaning increased the economic benefits of piglets. However, early weaning damages the intestinal barrier of piglets and causes immunological stress. The mechanism by which Hippophae rhamnoides polysaccharide (HRP) alleviates lipopolysaccharide (LPS)-induced intestinal porcine epithelial cells (IPEC-J2) inflammatory damage was investigated using proteomics in our previous studies. In this study we employed RNA-sequencing (RNA-seq) to determine the level and function of differentially expressed genes (DEGs) and further explore the mechanism of the HRP anti-inflammatory and immune process. The differential expression analysis indicated that 3622, 1216, and 2100 DEGs in the IPEC-J2 cells were identified in C vs. L, L vs. H6-L, and C vs. H6-L, respectively. The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis foundsix identified pathways related to the immune system. Additionally, we used the Science, Technology, Engineering, and Math (STEM) program to categorize the 3,134 DEGs that were differentially expressed in H2-L, H4-L and H6-L into eight possible expression profiles, in which 612 were clustered into two profiles. The accuracy and consistency of RNA-seq data were validated by the results of qRT-PCR of the nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (NFKB2), MAP kinase interacting serine/threonine kinase 2 (MKNK2), mitogen-activated protein kinase kinase 1 (MAP2K1), mitogen-activated protein kinase kinase kinase 8 (MAP3K8), Ras-related protein R-Ras (RRAS), TNF receptor-associated factor 1 (TRAF1), NF-kappa-B inhibitor alpha (NFKBIA), interleukin 8 (IL8), tumor necrosis factor, alpha-induced protein 3 (TNFAIP3), and transforming growth factor beta-1 (TGFB1). Transcriptome sequencing also indicated that HRP reduced the expression levels of related DEGs and inhibited the activation of the mitogen-activated protein kinase (MAPK)/nuclear factor kappa-B (NF-κB) signaling pathway. Our findings indicate that the application of HRP in piglet diets during the early weaning period can improve intestinal epithelial function and integrity, and relieve intestinal damage, and improve piglet health.
With the acceleration of the large-scale and intensified process of pig production, early weaning techniques for piglets have been gradually implemented (1). Weaned piglets are affected by stresses such as nutrition, immunity, and environment, and piglets have early weaning syndromes such as reduced feed utilization, poor growth, and diarrhea (2, 3). Piglet weaning is accompanied by the occurrence of intestinal inflammation, which causes a series of negative reactions in the incompletely developed intestinal tracts of piglets, such as intestinal mucosal injury, intestinal villi damage, and intestinal wall injury, Intestinal inflammation also affects the digestive and absorptive function of the intestinal tract in piglets and leads to sluggish growth, diarrhea, and even death, causing great economic losses to the swine industry (4). The intestinal mucosa is the host's first line of defense against pathogenic microorganisms. Intestinal epithelial cells (IECs) are an important part of the intestinal mucosal barrier (5, 6). Injury to IECs is an important pathological basis for intestinal dysfunction. IECs can produce severe immune stress in a variety of physiological, pathological, dietary, or environmental conditions, and the accumulation of excessive inflammatory cytokines can damage intestinal epithelial cells, resulting in intestinal dysfunction. Therefore, the key to reducing the diarrhea rate and improving the production performance of weaned piglets is to reduce weaning stress and protect the intestinal structure and function of piglets (7). Seeking immunostimulants to promote growth and reduce the intestinal inflammation caused by weaning has become a hot scientific issue.
In recent years, plant extracts as immunomodulators have received worldwide attention due to their nutritional and medicinal potential (8, 9). Hippophae rhamnoides L. (sea buckthorn) is a traditional medicinal plant (10). H.rhamnoides extracts have shown antioxidant, anti-inflammatory, and anti-viral effects by decreasing cytotoxicity and reactive oxygen species (ROS) generation (11, 12). A recent study showed that Hippophae rhamnoides polysaccharide (HRP) protected mice livers from CCl4-induced damage through its anti-inflammatory effect and the modulation of the balance between anti-inflammatory cytokines inimmune cells (13). The protective mechanism of chitosan oligosaccharide against LPS-induced inflammatory responses in IPEC-J2 and in mice with DSS dextran sulfate sodium-induced colitis is reported (14). In our previous study, the role of several key regulatory genes and proteins involved in HRP immunoregulation was examined (15, 16). However, the composition and mechanisms of the underlying global regulatory networks at the transcriptome levels are still poorly understood. To confirm the pre-protective effect of HRP on lipopolysaccharide (LPS)-induced intestinal porcine epithelial cells (IPEC-J2) in terms of anti-inflammatory or immunoregulatoryproperties, the pathways enriched by differentially expressed genes (DEGs) should be biologically validated. We employed RNA-sequencing (RNA-seq) to determine the abundance and function of genes (17, 18) and lay the foundation for further study of the anti-inflammatory immune response of HRP in order to provide a theoretical basis and technical support for the treatment and prevention of diseases such as diarrhea caused by intestinal damage in early-weaned piglets.
HRP, ≥98% (HPLC) purchased from Nanjing Zelang Biological Technology Co., Ltd., China. HRP was extracted by water decoction and alcohol precipitation as described previously (19, 20). The protein in the filtrate was removed primarily by the Sevage method, HRP was purified on a Sephadex G150 gel (Pharmacia) and the purity was also verified by HPLC. HRP consists of 70% carbohydrate and 14.2% uronic acid. The polysaccharide is composed of mannose, arabinose, glucose, galactose and rhamnose with a ratio of 2.02:1.02:4.24:1:9.22 as the indication of chromatographic analysis of HRP (19).
The IPEC-J2 cell line source and cells culture method was as described in our previous study (21). HRP was pretreated for 24 h. Subsequently, cells were exposed to LPS for 16 h. The cells were collected, and quickly frozen in liquid nitrogen, and stored at −80°C for future transcriptome analysis. This study included five treatments C represents the control group IPEC-J2 cells without treatment. L represents the treatment group of IPEC-J2 cells induced by 10 μg/mL LPS. H2-L, H4-L, and H6-L represent the pre-treatment of IPEC-J2 cells with 200, 400, and 600 μg/mL HRP, respectively, followed by treatment with 10 μg/mL LPS. The HRP and LPS concentrations used were based on the results of our previous articles (16, 17).
According to the manufacturer's instructions, TRIzol reagent was used to extract RNA from the IPEC-J2 cells in different groups. cDNA library construction and RNA-seq were as described by Shao et al. (17).
High quality clean reads were obtained by removing reads containing adapters, more than 10% unknown nucleotides (N), and low-quality reads containing more than 50% low quality (Q-value ≤ 20) bases. The short reads alignment tool Bowtie2 (22) was used to map reads to the ribosome RNA (rRNA) database. The rRNA mapped reads were removed. The remaining reads were further used in the assembly and analysis of the transcriptome. Gene abundances were quantified by the RSEM software (23). Transcript abundance was normalized to fragments per kilobase of exon model per million mapped reads (FPKM). DEGs between groups were analyzed using the DEGs R package (http://www.rproject.org/). Genes with fold change (FC) >1.2 and a false discovery rate (FDR) <0.02 were considered significant. Significant enrichment of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways determined the most important biochemical metabolic pathways and signal transduction pathways that the protein participated in. The KEGG website (http://www.kegg.jp/kegg/) was used to query the immune-related signal pathways involved in significantly differently changed genes, and heat map analysis was performed on the significantly differentially expressed genes involved in immune-related signal pathways. For each treatment group, it is included three replicates to increase reliability of the data.
To validate the accuracy of the RNA-seq data, the DEG expressions for the nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (NFKB2), MAP kinase interacting serine/threonine kinase 2 (MKNK2), mitogen-activated protein kinase kinase 1 (MAP2K1), mitogen-activated protein kinase kinase kinase 8 (MAP3K8), Ras-related protein R-Ras (RRAS), TNF receptor-associated factor 1 (TRAF1), NF-kappa-B inhibitor alpha (NFKBIA), interleukin 8 (IL8), tumor necrosis factor, alpha-induced protein 3 (TNFAIP3), and transforming growth factor beta-1 (TGFB1) were determined by qRT-PCR analyses as described previously (16). We focused on the 600 μg/mL concentration of H. rhamnoides polysaccharide to pre-treat the IPEC-J2 cells in the following results (16, 21). The primers for the selected genes are listed in Supplementary Table S1.
For the qPCR analysis, all data analyses were performed by using SPSS software (SPSS version 20.0, Chicago, IL, USA). Tukey's multiple range test was used to compare the mean values (P < 0.05) to indicate significant differences and the results were expressed as mean ± SD.
The aggregate of 356,912,016 raw reads were accumulated by using the High-throughput RNA sequencing to do the paired-end sequencing of the fifteen constructed libraries. After quality control, assessment of contaminated rRNA and low-quality sequences, 356,207,308 qualified Illumina reads were obtained. Approximately 97.44% of the clean reads were mapped to the reference genome and then used for further gene expression analysis (Table 1). We focused on C, L, and H6-L samples in the following experiment.

Table 1. Summary statistics for sequence quality and alignment information of IPEC-J2 cells sample in every group.
DEGs were identified using digital gene expression tags. The expression analysis indicated that 3,622, 1,216, and 2,100 DEGs in the IPEC-J2 cells were identified in C vs. L, L vs. H6-L, and C vs. H6-L, respectively (FC >1.2 and FDR <0.02) The most DEGs were identified from C to L (Figures 1A–D and Supplementary Tables S2–S4). These results demonstrated that number of inflammatory cytokines were accentuated after LPS induction. However, number of inflammatory cytokines in HRP pre-treated groups were significantly reduced. The volcano plot shows that HRP plays an important role in the immune regulation of cellular inflammatory damage.
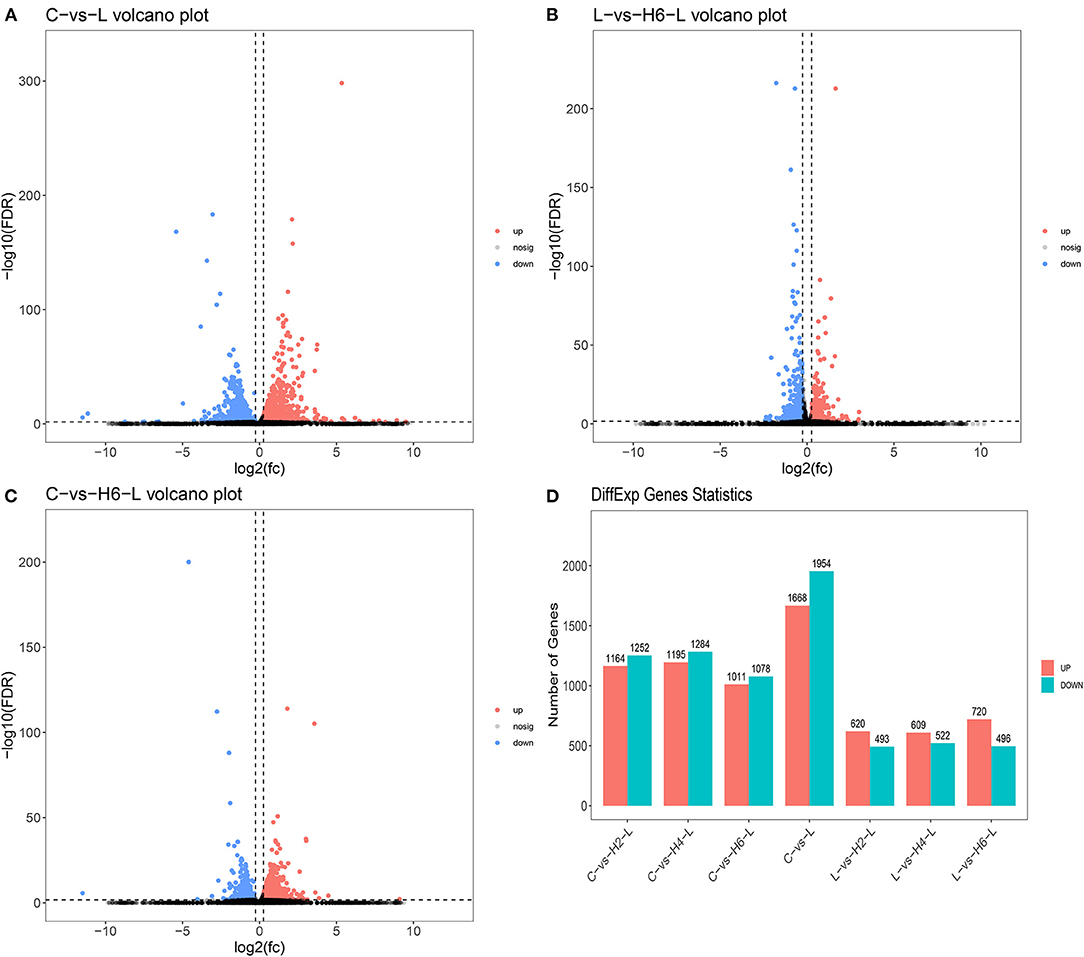
Figure 1. Analysis of the volcano plotof DEGs (A–C). Upregulated genes are shown in red, down-regulated genes are shown in blue, and genes with no significant difference in expression are indicated in black, significance was indicated by P < 0.05. DEGs statistics in the comparison group (D).
DEGs were categorized into three Gene Ontology (GO) groups: biological process, cellular component, and molecular function. In the biological process group, many DEGs were categorized as the cellular process, single-organism process, and metabolic process. In the cellular component group, many DEGs were categorized as the cell, cell part and organelle. In the molecular function group, many DEGs were categorized as the binding, catalytic activity, and transporter activity (Figures 2A–C and Supplementary Tables S5–S7).
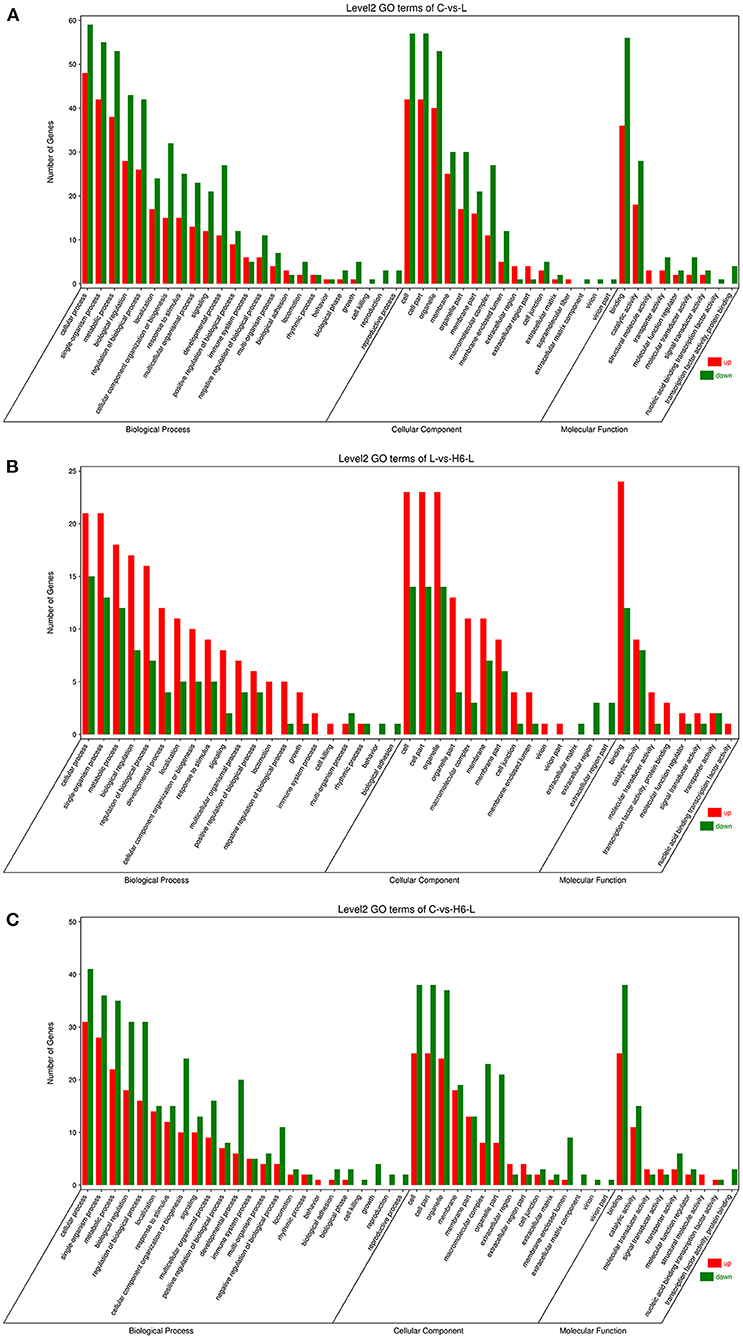
Figure 2. The GO analysis of the DEGs in IPEC-J2 cells. Classification of identified genes based on functional annotations using GO analysis are shown for comparisons between the L treatment and C (A), H6-L treatment and L alone (B), H6-L treatment and C (C).
As shown in Figures 3A–C and Supplementary Tables S8–S10, in C vs. L, 1,442 DEGs were mapped into 322 KEGG pathways. The key pathways were Apoptosis (ko04210) and the NF-kappa B signaling pathway (ko04064). In L vs. H6-L, 531 DEGs were mapped into 307 KEGG pathways. The key pathways were MAPK (ko04010), NF-kappa B (ko04064), TNF (ko04668), Apoptosis (ko04210), HIF-1 (ko04066), and the TGF-beta signaling pathway (ko04350). In C vs. H6-L, 892 DEGs were mapped into 318 KEGG pathways. The key pathway was the IL-17 signaling pathway (ko04657).
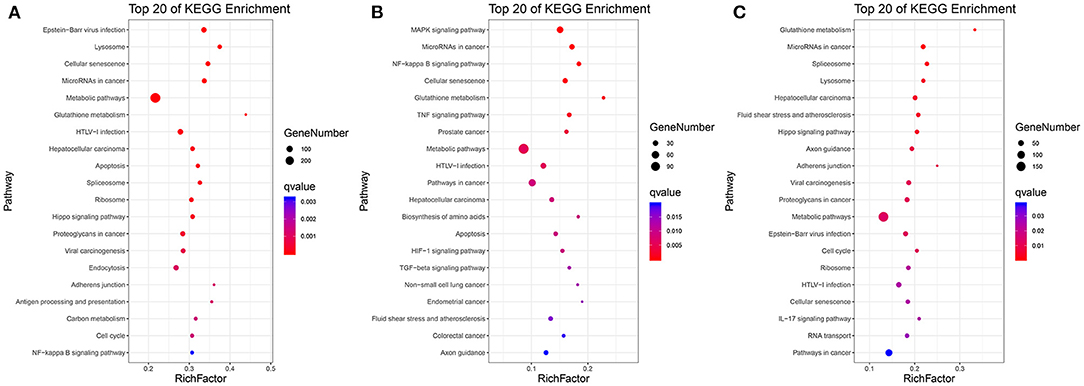
Figure 3. Top 20 pathway enrichment in KEGG pathway analysis. (A) C vs. L. (B) L vs. H6-L. (C) C vs. H6-L. The greater the rich factor, the higher the degree of enrichment. The Q value ranges from 0 to 1 and the closer it is to zero, the more significant.
In Figure 4 and Table 2, the sample gene expression of the C and L, L and H6-L, C and H6-L groups is shown using a pseudo color scale, with high levels of expression shown in red and low levels shown in blue. The results showed that most of the gene expression levels were down-regulated after HRP pre-treatment compared with the LPS group. The Log2-fold change and p-value statistics of DEGs related to the main immune pathways in the L-vs.-H6-L group are shown in Table 3. This data indicated that genes were significantly expressed in six immune pathways.
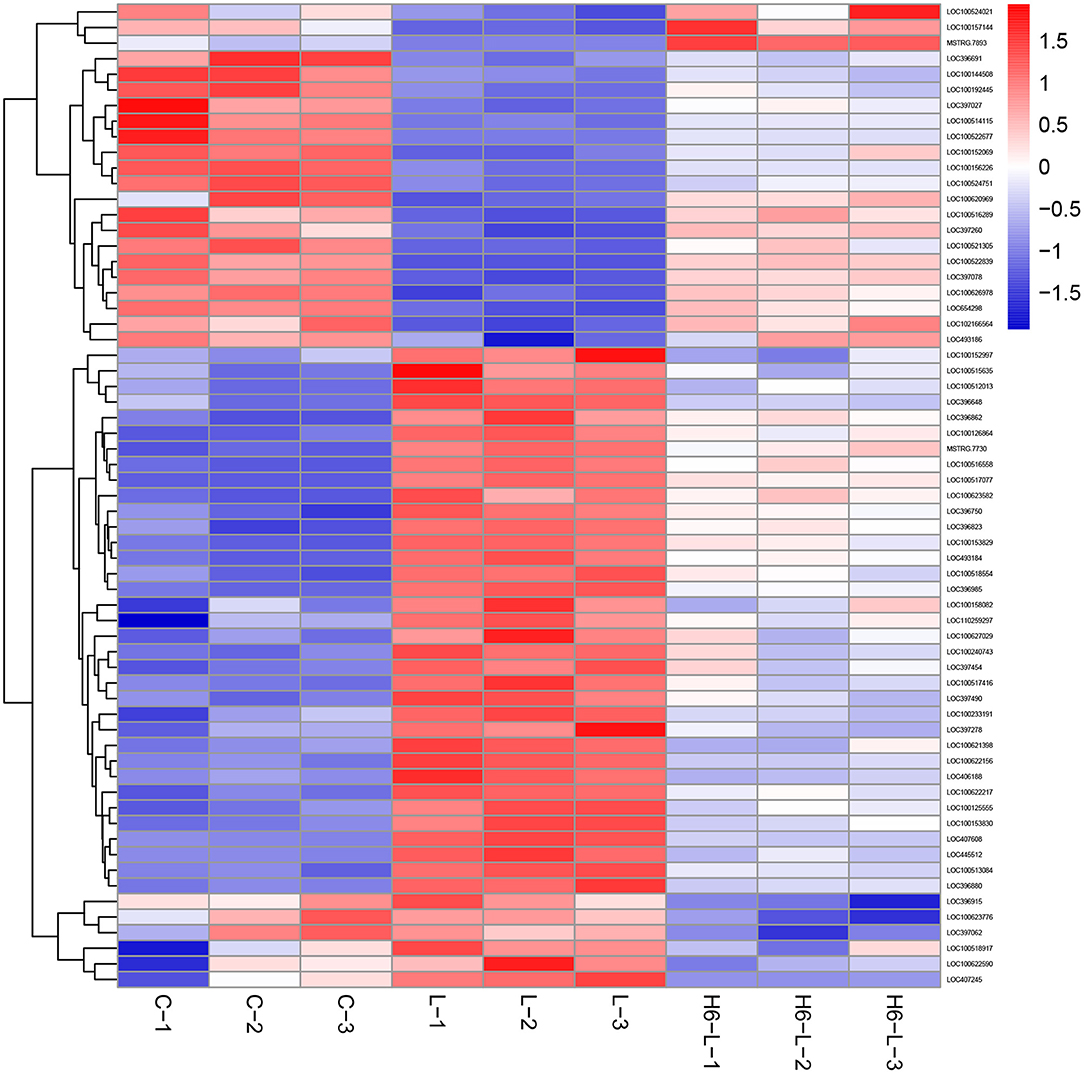
Figure 4. A heat map of the DEGs related to immune pathways in IPEC-J2 cells. The sample gene expression of the C (C1–C3) and L (L1–L3), L (L1–L3) and H6–L (H6-L-1-H6-L-3), C (C1–C3), and H6-L (H6-L-1-H6-L-3) groups is shown using a pseudo color scale, with high levels of expression shown in red and low levels shown in blue.
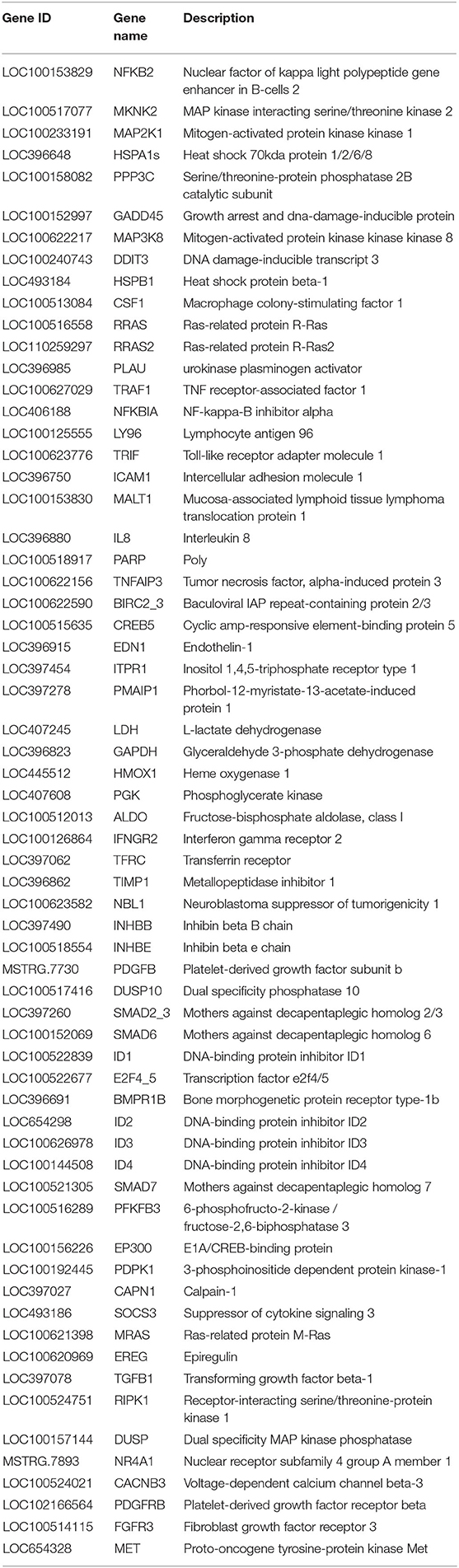
Table 2. Significant differentially expressed genes related to the main immune pathways in the L-vs-H6-L comparison group.
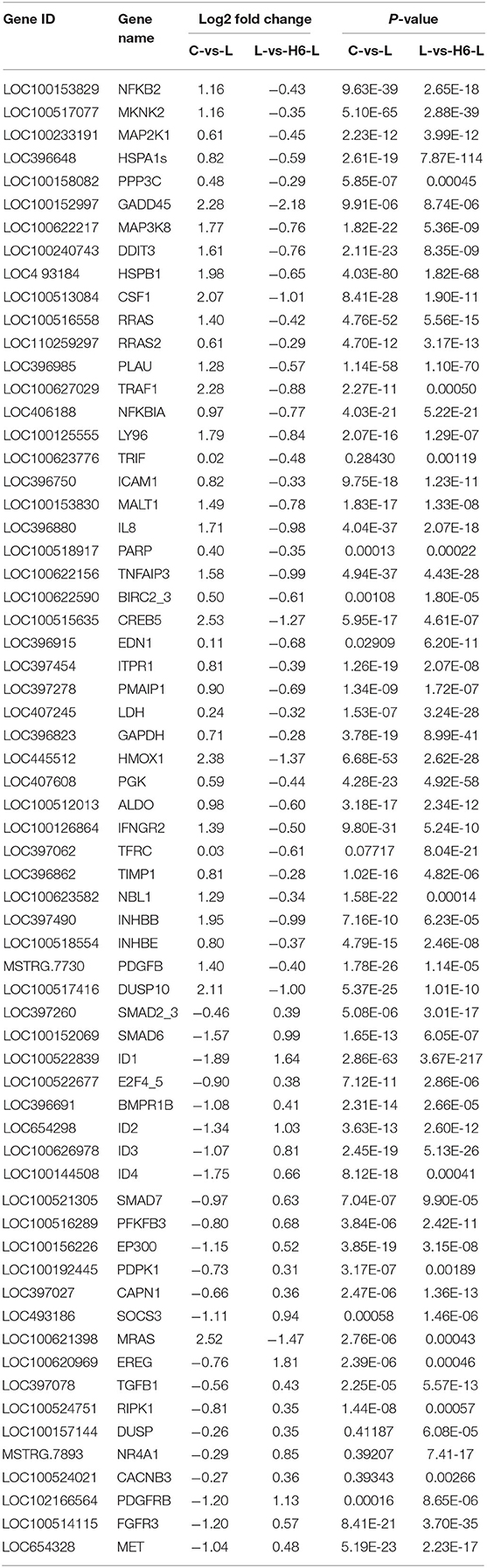
Table 3. Log2-fold change and p-value statistics of significant differentially expressed genes related to the main immune pathways in the L-vs-H6-L group.
To determine the gene expression trajectories, we used the STEM program to categorize the 3,134 DEGs that were differentially expressed in H2-L, H4-L, and H6-L into eight possible expression profiles (P < 0.05) (Figures 5A,B, 6A–H, and Supplementary Table S11), in which 612 were clustered into two profiles (P ≤ 0.05), including two up-regulated patterns (Profile 6 and Profile 7). Profile 6 and 7 contained 452 and 160 DEGs, respectively. The consistent up-regulation of genes of Profile 6 in H2-L, H4-L, and H6-L indicated that DEGs may contribute to stimulatory functions during the polysaccharide anti-inflammatory function. The up-regulated genes of Profile 7 between only H2-L and H4-L revealed that these DEGs played a key role in the anti-inflammatory process. There was no significant difference in the up-regulated genes between H4-L and H6-L.

Figure 5. (A) Eight profiles of DEGs with unique expression alterations over H2-L, H4-L, and H6-L. The profile number and the number of genes are shown on top of each square. The number of genes assigned is used to order the profiles. The profiles with color (P < 0.05): significant enrichment trend. The profiles without color: non-significant enrichment trend. (B) Trend DEGs number and P-value histogram. X-axis indicates the eight profiles; Y-axis shows DEGs number of every profile. The color of the column represents P-value.
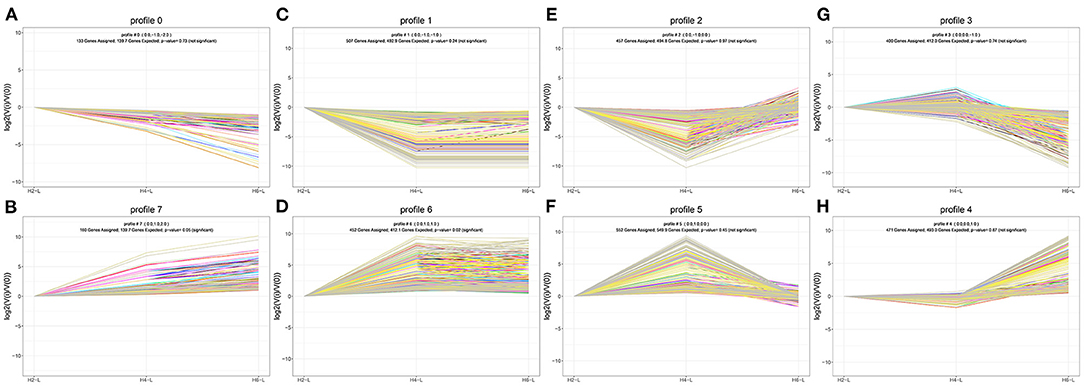
Figure 6. DEGs expression profiles (A–H) in the IPEC-J2 cells induced by LPS after HRP pre-treatment. (A,B), (C,D), (E,F), and (G,H) are two antagonistic profiles of DEGs curves, respectively. Each X-axis indicates the concentration of HRP pre-treated IPEC-J2 cells (H2-L, H4-L, and H6-L), Y-axis shows expression changes.
A total of 11.1% (940/8,499) of the DEGs could be annotated. As shown in Table 4, the metabolic pathways (ko01100), Cytokine-cytokine receptor interaction (ko04060), Neuroactive ligand-receptor interaction (ko04080), Pathways in cancer (ko05200), Olfactory transduction (ko04740), PI3K-Akt signaling pathway (ko04151), Calcium signaling pathway (ko04020), Human papillomavirus infection (ko05165), Jak-STAT signaling pathway (ko04630), and Systemic lupus erythematosus (ko05322) pathways were significantly enriched. The 13 genes among 125 DEGs (22.40%) in Profile 6, and five genes accounting for 11.63% of 43 DEGs in Profile 7 were annotated to the cytokine-cytokine receptor interaction. The three genes among 125 DEGs (2.40%) in Profile 6, and two genes accounting for 4.65% of 43 DEGs in Profile 7 were annotated to the PI3K-Akt signaling pathway. The seven genes among 125 DEGs (5.60%) in Profile 6, and two genes accounting for 4.65% of 43 DEGs in Profile 7 were annotated to the Jak-STAT signaling pathway.
Ten genes (NFKB2, MKNK, MAP2K1, MAP3K8, RRAS, TRAF1, NFKBIA, IL8, TNFAIP3, and TGFB1) were selected for qRT-PCR analysis.The results showed a strong correlation between the RNA sequencing data and the qRT-PCR data (Figure 7). This suggested that the expression results generated by RNA sequencing were reliable.
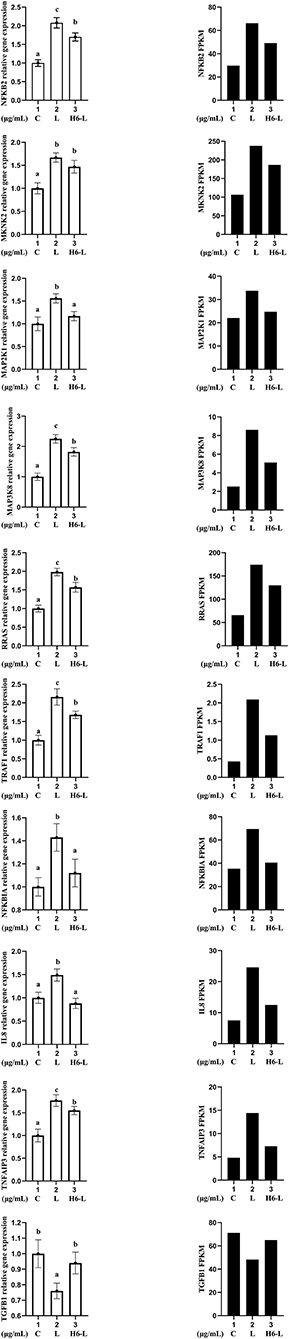
Figure 7. Candidate unigene expression levels revealed by qRT-PCR (left side) and RNA-seq (right side). Data from qRT-PCR are means of five replicates and bars represent SD.
In the current piglet production system, early weaning is an important means to improve the efficiency of pig production (24, 25). However, early weaning is very stressful to piglets, and can easily cause piglet immune stress and affect the healthy growth of piglets. How to alleviate the immune stress of piglets has become an area of concerning the piglet industry (26, 27). Hippophae rhamnoides extracts are widely used to enhancing immunity in both healthy and diseased animals (28). Polysaccharide is the main active ingredient of H.rhamnoides. HRP has been shown to have immunomodulatory effects (10). No research has been conducted on the molecular mechanism of HRP in piglets by transcriptome sequencing. Variation in gene expression may provide a key to uncovering the mechanisms of diseases. Transcriptome sequencing, also called RNA-seq, provides a new technique to quantify whole-genome expression profiling in any organism. It promises digital transcriptome profiling with high resolution and is rapidly replacing microarray technology (29, 30). Studies have used RNA-seq technology to explore the protective mechanism of IPEC-J2 cells stimulated by LPS after astragalus polysaccharide (APS) pretreatment. APS relieves cell damage by inhibiting the activation of the MAPK and NF-κB inflammatory pathways, thereby reducing intestinal inflammation (31). Cluster analysis revealed 3134 DEGs that were differentially expressed in H2-L, H4-L, and H6-L into eight possible expression profiles, in which 168 were clustered into two profiles. The up-regulated genes of Profile 6, only between H2-L and H4-L, revealed that these DEGs played a key role in the anti-inflammatory process. The consistent up-regulation of genes of the Profile 7 in H2-L, H4-L, and H6-L indicated that DEGs may contribute to stimulatory functions during the polysaccharide anti-inflammatory process. KEGG enrichment analysis found that the six identified pathways were related to the immune system. Among the six identified pathways related to the immune system, the MAPK signaling pathway and NF-κB signaling pathway play important immunomodulatory roles in our study. Finally, we selected 10 DEGs (NFKB2, MKNK2, MAP2K1 (MEK1), MAP3K8, RRAS, TRAF1, NFKBIA, IL8, TNFAIP3, and TGFB1) related to the main immune pathways to validate the RNA-Seq data using qRT-PCR.
The downstream signal transduction pathways mediated by LPS mainly include the NF-κB signal transduction pathway and the MAPK signal transduction pathway (32). Extracellular regulated protein kinases (ERK), c-Jun N-terminal kinase (JNK), and p38 mitogen-activated protein kinase (p38 MAPK) belong to the three subtypes of the MAPK signaling pathway (33). The Ras/Mitogen-activated protein kinase kinase (MEK)/ERK pathway is one of the most important signal transduction pathways among the MAPK pathways. The Ras/MEK/ERK pathway involves the regulation of a variety of physiological functions of cells and plays a key role in the pathogenesis and pathophysiology of various diseases (34). The activation of ERK is a key step in transferring signals from surface receptors to the nucleus. The activation of ERK induced by LPS leads to the secretion of large amounts of tumor necrosis factor-alpha (TNF-α), as well as interleukin-6 (IL-6) and IL-8, and increases the expression of inducible nitric oxide synthase and nitric oxide. Son of Sevenless (SOS) binds to RRAS-Guanosine diphosphate (GDP), prompting guanosine triphosphate (GTP) to replace GDP on RRAS and activate RRAS protein, then activate MEK and ERK sequentially. In recent years, some initial reports on targeting SOS to inhibit the activation of RRAS, thereby inhibiting the activation of the MAPK signaling pathway, have also achieved satisfactory results (35). The activation of ERK promotes the secretion of MKNK. MKNK is an important downstream protein kinase of ERK that has an important immunomodulatory function. MKNK dysfunction can inhibit the inflammatory signal of upstream ERK and affect downstream eIf4E and CREB and other effector proteins, thereby preventing cell inflammation. MAP3K8 is essential for the activation of the intracellular MAPK/ERK pathway induced by LPS in cells (36). Therefore, MAP3K8 is a critical factor for the production of pro-inflammatory cytokines during immune responses (37). Therefore, in our study, the results showed that MKNK2, MAP2K1, MAP3K8, and RRAS gene expression levels were down-regulated after HRP pre-treatment compared with the LPS group. The reduction of MKNK2, MAP2K1, MAP3K8, and RRAS gene levels plays an important immuno-regulatory role in HRP alleviating LPS-induced cell damage.
LPS, a trigger of inflammation, can activate the NF-κB signaling pathway (38). NFKBIA is a specific inhibitor of NF-κB that binds to NF-κB at a resting state to cause NF-κB to enter an inactive state. Phosphorylated NFKBIA is separated from NF-κB, and NF-κB is activated. Activated NF-κB migrates to the nucleus, where NF-κB nuclear transcription factor can up-regulate the levels of inflammation-related genes TNF, IL-6, and IL-8 and down-regulate the level of TGFB1 (39). Studies have proven that LPS promotes the degradation of NFKBIA, activates the DNA binding ability of NF-κB, and regulates the gene expression level of cytokines (40). Moreover, TRAFs are key regulatory proteins in NF-κB signaling pathways. TRAF1 enhances the activation of TNF-R2 induced by NF-κB (41), therefore promoting the release of a large number of inflammatory cytokines. A previous study showed that TRAF1 is over-expressed in a variety of lymphoma and leukemia cell lines and is a crucial mediator of diverse oncogenic signaling in the development of lymphoid malignancies. TNFAIP3 is a cytokine-induced protein that inhibits apoptosis and activates NF-κB (42). The main function of TNFAIP3 is to inhibit the activity of NF-κB and inhibit TNF-mediated apoptosis, thereby having an important impact on immune regulation and inflammatory processes (43, 44). The release of NFKB2, TRAF1, NFKBIA, IL8, and TNFAIP3, as important regulatory genes of NF-κB, can activate the upstream pathway NF-κB. Jayashankar et al. found that the intervention of supercritical carbon dioxide extract from seabuckthorn leaves can inhibit the expression levels of TNF-α and IL-6 after LPS-induced inflammatory damage, inhibit the activation of the MAPK/NF-κB signaling pathway, reduce inflammation, and play a immunomodulatory role (45). Our study indicated that NFKB2, TRAF1, NFKBIA, IL8, and TNFAIP3were increased and TGFB1 was reduced after LPS induction. However, after HRP pre-treatment, the gene expression level showed the opposite trend and they played an important immune-regulatory role in HRP alleviating LPS-induced cell damage, which provided more targets and prevention directions for theoretical and basic research on intestinal health. Studies have shown that APS may block radiation-induced bystander effects (RIBE) in bone mesenchymal stem cells (BMSCs) induced by the irradiated A549 through regulating the MAPK/NF-κB pathway (46).
This study was the first using a RNA-Seq technique to establish a dynamic transcriptomic profile of three stages (C, L, and H6-L) related to pre-treatment with HRP followed by challenge with LPS in IPEC-J2 cells. Subsequently, bioinformatics analysis (GO, KEGG, and series cluster) helped us to identify key regulatory genes (IL8 and NFKB2, among others.) related IPEC-J2 cellular immune regulation. Transcriptome analysis also showed that HRP protected IPEC-J2 cells from LPS-induced inflammation and decreased the expression of inflammatory cytokines by mainly inhibiting the MAPK/NF-κB signaling pathway. This study does not only provide the useful transcriptomic reference for HRP to effectively protect LPS-induced inflammatory damage in IPEC-J2 cells, but also provides a benchmark for the discovery of biomarkers related to HRP immune regulation.
The data presented in the study are deposited in the following repository: https://www.ncbi.nlm.nih.gov/Traces/study/, accession number PRJNA854604.
ML, HS, and LZ contributed to conception, design of study, drafting the manuscript, and critical revision. YZ conducted acquisition of data. LC conducted analysis of data. All authors read and approved the final manuscript.
This project was supported by grant from the National Key Research and Development Program of China (2017YFD0500506) and the Personnel Foundation of Heilongjiang Bayi Agricultural University (NO. XYB202015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.944390/full#supplementary-material
1. He Q, Tang H, Ren P, Kong X, Wang Y. Dietary supplementation with l-arginine partially counteracts serum metabonome induced by weaning stress in piglets. J Prot Res. (2011) 10:5214–21. doi: 10.1021/pr200688u
2. Shanahan F. Probiotics in inflamatory bowel disease. Gut. (2001) 48:609. doi: 10.1136/gut.48.5.609
3. Hoque MA, Skerratt LF, Rahman MA, Rabiul Alam Beg ABM, Debnath NC. Factors limiting traditional household duck production in Bangladesh. Trop Ani Health Prod. (2010) 42:1579–87. doi: 10.1007/s11250-010-9609-z
4. Lakatos L, Rednik A. Astroenterologic sub-acute care unit at the hospital department of internal medicine letter. Orvosi Hetilap. (1997) 138:1668.
5. Berschneider M. Development of a normal cultured small intestinal epithelial cell line which transport na and cl. Gastroenterology. (1989) 96:A41.
6. Hermes RG, Manzanilla EG, Susana M Martín-Orúe José F Pérez . Influence of dietary ingredients on in vitro inflammatory response of intestinal porcine epithelial cells challenged by an enterotoxigenic escherichia coli (k88). Compar Immunol Microbiol Infect Dis. (2011) 34:479–88. doi: 10.1016/j.cimid.2011.08.006
7. Pan L, Qin G, Zhao Y, Wang J, Liu F, Che D. Effects of soybean agglutinin on mechanical barrier function and tight junction protein expression in intestinal epithelial cells from piglets. Int J Molecul. (2013) 14:21689–704. doi: 10.3390/ijms141121689
8. Mlot C. Antidotes for antibiotic use on the farm. BioScience. (2000) 50, 955–960. doi: 10.1641/0006-3568(2000)050(0955:AFAUOT)2.0.CO;2
9. VanCott JL, Kobayashi T, Yamamoto M, Pillai S, McGhee JR, Kiyono H. Induction of pneumococcal polysaccharide-specific mucosal immune responses by oral immunization. Vaccine. (1996) 14:392–8. doi: 10.1016/0264-410X(95)00198-A
10. Zhang A, Sun H, Wang X. Recent advances in natural products from plants for treatment of liver diseases. Eur J Med Chem. (2013) 63:570–7. doi: 10.1016/j.ejmech.2012.12.062
11. Wang X, Liu J, Zhang X, Zhao S, Zou K, Xie J. Seabuckthorn berry polysaccharide extracts protect against acetaminophen induced hepatotoxicity in mice via activating the Nrf-2/HO-1-SOD-2 signaling pathway. Phytomedicine. (2018) 38:90–7. doi: 10.1016/j.phymed.2017.11.007
12. Suryakumar G, Gupta A. Medicinal and therapeutic potential of sea buckthorn (hippophae rhamnoides l.). J Ethnopharmacol. (2011) 138:268–78. doi: 10.1016/j.jep.2011.09.024
13. Zhang W Zhang X, Zou K, Xie J, Zhao S, Liu J. Seabuckthorn berry polysaccharide protects against carbon tetrachloride-induced hepatotoxicity in mice via anti-oxidative and anti-inflammatory activities. Food Func. (2017) 8:3130–38. doi: 10.1039/C7FO00399D
14. Shi L, Fang B, Yong Y, Li X, Gong D, Li J. Chitosan oligosaccharide-mediated attenuation of LPS-induced inflammation in IPEC-J2 cells is related to the TLR4/NF-κB signaling pathway. Carbohyd Poly. (2019) 219:269–79. doi: 10.1016/j.carbpol.2019.05.036
15. Zhao L, Li M, Su S, Geng T, Sun H. Hippophae rhamnoides Linn polysaccharide enhances antioxidant enzyme activity, cytokine level and related mRNA expression in intestinal porcine epithelial cells. Canadian J Ani Sci. (2020) 100:193–204. doi: 10.1139/cjas-2019-0134
16. Zhao L, Li M, Sun K, Su S, Geng T, Sun H. Hippophae rhamnoides polysaccharides protect ipec-j2 cells from lps-induced inflammation, apoptosis and barrier dysfunction in vitro via inhibiting tlr4/nf-κb signaling pathway. Int J Biol Macromol. (2020) 155:1202–15. doi: 10.1016/j.ijbiomac.2019.11.088
17. Shao D, Hu Y, Wang Q, Tong H, Shi S. Transcriptome sequencing reveals genetic mechanisms of reproduction performance stimulated by dietary daidzein in laying breeder Hens Theriogenol. (2019) 142:120–30. doi: 10.1016/j.theriogenology.2019.09.040
18. Gao Y, Li S, Bao X, Luo C, Yang H, Wang J. Transcriptional and proteomic analysis revealed a synergistic effect of aflatoxin m1 and ochratoxin a mycotoxins on the intestinal epithelial integrity of differentiated human Caco-2 cells. J Prot Res. (2018) 17:3128–42. doi: 10.1021/acs.jproteome.8b00241
19. Liu H, Zhang W, Dong S. Protective effects of sea buckthorn polysaccharide extracts against LPS/d-GalN-induced acute liver failure in mice via suppressing TLR4-NF-κB signaling. J Ethnopharmacol. (2015) 176(Complete):69–78. doi: 10.1016/j.jep.2015.10.029
20. Ni W, Gao T, Wang H, Du Y, Li J, Li C. Anti-fatigue activity of polysaccharides from the fruits of four Tibetan plateau indigenous medicinal plants. J Ethnopharmacol. (2013) 150:529–535. doi: 10.1016/j.jep.2013.08.055
21. Zhao L, Geng T, Sun K, Su S, Zhao Y, Bao N. Proteomic analysis reveals the molecular mechanism of Hippophae rhamnoides polysaccharide intervention in LPS-induced inflammation of IPEC-J2 cells in piglets. Int J Biol Macromol. (2020) 164:3294–304. doi: 10.1016/j.ijbiomac.2020.08.235
22. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. (2012) 9:357–9. doi: 10.1038/nmeth.1923
23. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. (2011) 12:1. doi: 10.1186/1471-2105-12-323
24. Bomba L, Minuti A, Moisá SJ, Trevisi E, Eufemi E, Lizier M. Gut response induced by weaning in piglet features marked changes in immune and inflammatory response. Func Int Genom. (2014) 14:657–71. doi: 10.1007/s10142-014-0396-x
25. Khafipour E, Munyaka PM, Nyachoti CM, Krause DO, Rodriguez-Lecompte JC. Effect of crowding stress and escherichia coli k88+ challenge in nursery pigs supplemented with anti-escherichia coli k88+ probiotics. J Animal Sci. (2014) 92:2017–29. doi: 10.2527/jas.2013-7043
26. Pluske JR, Hampson DJ, Williams IH. Factors influencing the structure and function of the small intestine in the weaned pig: a review. Livestock Prod Sci. (1997) 51:215–36. doi: 10.1016/S0301-6226(97)00057-2
27. Xiao T. Innate immune recognition of nucleic acids. Immunol Res. (2009) 43:98–108. doi: 10.1007/s12026-008-8053-x
28. Tanwar H, Shweta Singh D, Singh SB, Ganju L. Anti-inflammatory activity of the functional groups present in Hippophae rhamnoides (Seabuckthorn) leaf extract. Inflammopharmacology. (2017) 26:291–301. doi: 10.1007/s10787-017-0345-0
29. Herwig R. Predictive network modelling with toxicogenomics data. Toxicol Lett. (2014) 229:S4–S21. doi: 10.1016/j.toxlet.2014.06.043
30. Davoli R, Zambonelli P, Hedeegard J, Hornshoj H, Nanni Costa L, Stella A. Transcriptome analysis of skeletal muscle tissue to identify genes involved in pre-slaughter stress response in pigs. Italian J Anil Sci. (2009) 8:69–71. doi: 10.4081/ijas.2009.s2.69
31. Dong N, Li X, Xue C, Zhang L, Wang C, Xu X. Astragalus polysaccharides alleviates lps-induced inflammation via the nf-κb/mapk signaling pathway. J Cell Physiol. (2020) 235:1–16. doi: 10.1002/jcp.29452
32. Wu CX, Sun H, Liu Q, Guo H, Gong JP. Lps induces hmgb1 relocation and release by activating the nf-κb-cbp signal transduction pathway in the murine macrophage-like cell line raw264.7. J Surg Res. (2012) 175:88–100. doi: 10.1016/j.jss.2011.02.026
33. Costa AP, Lopes MW, Rieger DK, Barbosa SGR, Gonçalves FM, Xikota JC. Differential activation of mitogen-activated protein kinases, ERK 1/2, 38MA.PK and JNK p54/p46 during postnatal development of rat hippocampus. Neurochem Res. (2015) 41:1160–9. doi: 10.1007/s11064-015-1810-z
34. Okano JI, Snyder LC, Rustgi AK. Paclitaxel induces prolonged activation of the ras/mek/erk pathway independently of activating the programmed cell death machinery. Gastroenterology. (2011) 120:A661–2. doi: 10.1016/S0016-5085(01)83290-X
35. Zhao H, Li YY, Fucini RV, Ross SE, Pessin JE, Koretzky GA. T cell receptor-induced phosphorylation of sos requires activity of cd45, lck, and protein kinase c, but not erk. J Biol Chem. (1997) 272:21625–34. doi: 10.1074/jbc.272.34.21625
36. Santag S, Siegel F, Wengner AM, Lange C, Petersen K. Abstract 341: preclinical mode of action and anti-tumor efficacy of the selective mknk1 inhibitor bay 11,43,269 in nsclc models. Cancer Res. (2016) 76(14 Supplement):341. doi: 10.1158/1538-7445.AM2016-341
37. Wei R, Yang Q, Han B, Li Y, Yao K, Yan X. Microrna-375 inhibits colorectal cancer cells proliferation by downregulating jak2/stat3 and map3k8/erk signaling pathways. Oncotarget. (2017) 8:16633–41. doi: 10.18632/oncotarget.15114
38. Shishodia S, Koul D, Aggarwal BB. Cyclooxygenase (cox)-2 inhibitor celecoxib abrogates tnf-induced nf-κb activation through inhibition of activation of iκbα kinase and akt in human non-small cell lung carcinoma: correlation with suppression of cox-2 synthesis. J Immunol. (2004) 173:2011–22. doi: 10.4049/jimmunol.173.3.2011
39. Koh Y. Inhibition of SRC Tyrosine kinases suppresses activation of nuclear factor-κB, and Serine and Tyrosine Phosphorylation of IκB-α in Lipopolysaccharide-stimulated Raw 264.7 macrophages. J Toxicol Environ Health, Part A. (2005) 68:1643–62. doi: 10.1080/15287390500192114
40. Li MY, Sun L, Niu XT, Chen XM, Tian JX, Kong YD, et al. Astaxanthin protects lipopolysaccharide-induced inflammatory response in Channa argus through inhibiting NF-κB and MAPKs signaling pathways. Fish Shellfish Immunol. (2019) 86:280–6. doi: 10.1016/j.fsi.2018.11.011
41. Tang X, Zhang L, Wei W. Roles of TRAFs in NF-κB signaling pathways mediated by BAFF. Immunol Lett. (2018) 196:113–8. doi: 10.1016/j.imlet.2018.01.010
42. Rhee L, Murphy SF, Kolodziej LE, Grimm WA, Weber CR, Lodolce JP. Expression of TNFAIP3 in intestinal epithelial cells protects from DSS- but not TNBS-induced colitis. Am J Physiol-Gastroint Liver Physiol. (2012) 303:G220–7. doi: 10.1152/ajpgi.00077.2012
43. Gui J, Yue Y, Chen R, Xu W, Xiong S. A20 (TNFAIP3) alleviates CVB3-induced myocarditis via inhibiting NF-κB signaling. PLoS ONE. (2012) 7:e46515. doi: 10.1371/journal.pone.0046515
44. Varfolomeev E, Goncharov T, Maecker H, Zobel K, Komuves LG, Deshayes K, et al. Cellular inhibitors of apoptosis are global regulators of NF- B and MAPK activation by members of the TNF family of receptors. Sci Signal. (2012) 5:ra22. doi: 10.1126/scisignal.2001878
45. Jayashankar B, Mishra KP, Kumar MSY, Udayasankar K, Misra K, Ganju L, et al. A supercritical CO2 extract from seabuckthorn leaves inhibits pro-inflammatory mediators via inhibition of mitogen activated protein kinase p38 and transcription factor nuclear factor-κB. Int Immunopharmacol. (2012) 13:461–7. doi: 10.1016/j.intimp.2012.05.011
Keywords: IPEC-J2 cells, HRP, anti-inflammatory, MAPK/NF-κB signaling pathway, transcriptome
Citation: Li M, Chen L, Zhao Y, Sun H and Zhao L (2022) Research on the Mechanism of HRP Relieving IPEC-J2 Cells Immunological Stress Based on Transcriptome Sequencing Analysis. Front. Nutr. 9:944390. doi: 10.3389/fnut.2022.944390
Received: 15 May 2022; Accepted: 13 June 2022;
Published: 15 July 2022.
Edited by:
Baojun Xu, United International College, ChinaReviewed by:
Utoomporn Surayot, Chiang Mai University, ThailandCopyright © 2022 Li, Chen, Zhao, Sun and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Zhao, emxobGpieWF1QDEyNi5jb20=; Hui Sun, c2hqbGF1QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.