- 1Key Laboratory of Aquatic Product Processing, Ministry of Agriculture and Rural Affairs, National R&D Center for Aquatic Product Processing, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2Co-Innovation Center of Jiangsu Marine Bio-industry Technology, Jiangsu Ocean University, Lianyungang, China
- 3College of Food Science and Engineering, Ocean University of China, Qingdao, China
- 4Collaborative Innovation Center of Seafood Deep Processing, Dalian Polytechnic University, Dalian, China
Angiotensin I converting enzyme (ACE) inhibitory peptides from fermented foods exhibit great potential to alleviate hypertension. In this study, the peptide extract from Chouguiyu exhibited a good inhibition effect on ACE, and the inhibition rate was significantly enhanced after fermentation for 8 days. The ACE inhibitory peptides were further identified, followed by their inhibition and formation mechanisms using microbiome technology and molecular docking. A total of 356 ACE inhibitory peptides were predicted using in silico, and most ACE inhibitory peptides increased after fermentation. These peptides could be hydrolyzed from 94 kinds of precursor proteins, mainly including muscle-type creatine kinase, nebulin, and troponin I. P1 (VEIINARA), P2 (FAVMVKG), P4 (EITWSDDKK), P7 (DFDDIQK), P8 (IGDDPKF), P9 (INDDPKIL), and P10 (GVDNPGHPFI) were selected as the core ACE inhibitory peptides according to their abundance and docking energy. The salt bridge and conventional hydrogen bond connecting unsaturated oxygen atoms in the peptides contributed most to the ACE inhibition. The cleavage proteases from the microbial genera in Chouguiyu for preparing these 7 core ACE inhibitory peptides were further analyzed by hydrolysis prediction and Pearson's correlation. The correlation network showed that P7, P8, and P9 were mainly produced by the proteases from LAB including Lactococcus, Enterococcus, Vagococcus, Peptostreptococcus, and Streptococcus, while P1, P2, P4, and P10 were mainly Produced by Aeromonas, Bacillus, Escherichia, and Psychrobacter. This study is helpful in isolating the proteases and microbial strains to directionally produce the responding ACE inhibitory peptides.
Introduction
Hypertension is a chronic health problem closely related to many diseases, such as stroke, heart attack, and heart failure (1). Angiotensin I converting enzyme (ACE) can elevate blood pressure by dilating the arterial blood vessels throughout the body (2). Many drugs have been designed to inhibit ACE, such as captopril and lisinopril, but synthetic drugs have many side effects (3). A large number of food-derived peptides have been identified as potential substitutes for synthetic drugs to treat hypertension (4, 5). These inhibitory peptides have distinct structural characteristics, such as possessing fewer amino acids, abundant hydrophobic amino acids, and aliphatic, aromatic, or positively charged amino acids in the N/C terminal (6, 7). These active sequences are collected on bioactive websites such as BIOPEP-UWM and AHTpin, which can predict the activity and characteristics of unknown peptides based on reliable models (6). Meanwhile, virtual screening is becoming an effective, accurate, and promising method to find novel active peptides and has been widely used in the screening of ACE inhibitory peptides (8). However, the ACE inhibitory peptides are usually prepared from protein hydrolysates using commercially available proteases. The restriction of their cleavage locations leads to the low diversity of ACE inhibitory peptides, thereby limiting the discovery of novel ACE inhibitory peptides with high bioactivity.
Fermented foods are the source of many bioactive peptides (9) and have significant advantages in the production of ACE inhibitory peptides. Plenty of fermented foods are focused on isolating ACE inhibitory peptides such as cheese (10), fermented goat milk (11), fish sauce (12), and bean paste (13). In fermented foods, the rich ACE inhibitory peptides benefit from the complex microorganisms that can produce various proteases (14). Chouguiyu is a typical fermented fish with rich peptides. Most research has focused on the formation mechanism of its distinctive flavor and taste (15, 16). However, few studies can discuss the role of microorganisms from Chouguiyu in the formation mechanism of ACE inhibitory peptides.
Microbiome technology facilitates the construction of a bridge between microorganisms and metabolites (17, 18), which can be applied to explore the formation mechanism of ACE inhibitory peptides. Shotgun metagenomics is the technology to obtain all the genetic information of microorganisms in the environment, using random fragments of microbial genes to splice small fragments into longer sequences for analysis (19). Peptidomics is an effective method to study the structure and abundance of bioactive peptides (20). With the aid of statistical methods, the metabolism formation of bioactive peptides by microorganisms can be illuminated through microbiome technology. However, there is a lack of study on the formation mechanism of bioactive peptides in fermented foods. Molecular docking, as a kind of computer-aided drug design technology, provides convenience for visualizing the activity of peptides and predicts the possible inhibition mechanisms between a small molecule ligand and a given protein receptor (21, 22). This technology has been widely used to screen ACE inhibitory peptides (23). However, the interaction at the atomic level between ACE inhibitory peptides and ACE has rarely been illustrated by molecular docking.
In this study, the microbial community and free peptides were analyzed by microbiome technology, including metagenomics and peptidomics, followed by molecular docking at the atomic level to explore the inhibition mechanism of ACE inhibitory peptides in Chouguiyu. The formation mechanism of ACE inhibitory peptides in Chouguiyu was further illustrated by the construction of a correlation network. This study is expected to illustrate the formation mechanism of the ACE inhibitory peptides in Chouguiyu from the perspective of microbial metabolism and to provide vital information on proteases and the related microbial strains for the production of ACE inhibitory peptides.
Materials and methods
Materials and chemicals
Chouguiyu was made from frozen mandarin fish (Siniperca chuatsi, av. 500 g) in Anhui, China according to the previous study (24). After fermentation at 15–20°C for 8 days, three samples of Chouguiyu were collected for each individual sample at 0 days (D0 group), 4 days (D4 group), and 8 days (D8 group) for further study. The reagents used for liquid chromatogram tandem mass were of mass pure grade, and all other reagents were of analytical grade.
Preparation of peptide extract from Chouguiyu
Peptide extract was obtained according to the previous study (25). In brief, after being peeled, the Chouguiyu (50.0 g) was smashed and homogenized for 0.5 h with 200 ml hydrochloric acid (0.01 mol/L). After centrifugation at 8,000 r/min and 4°C for 20 min, the supernatant was added with triploid ethanol for 20 min at 4°C and was then centrifuged to remove protein. The supernatant was lyophilized and stored at −80°C after desalination.
Analysis of ACE inhibitory activity of peptide extract in Chouguiyu
The assay of ACE inhibitory activity was performed using the ACE inhibitory assay kit (ACE kit-WST, A502, Dojindo, Kumamoto Ken, Japan) (13). In brief, the peptide extract of D0, D4, and D8 was dissolved in Milli-Q water (RephiLe Bioscience, Shanghai, China) with a concentration of 20 g/L. The sample (20 μl) was mixed with substrate buffer (20 μl) and enzyme working solution (20 μl). After incubation for 60 min at 37°C, the indicator working solution (200 μl) was added and incubated at room temperature for 10 min. Captopril was used as a positive control throughout the experiment. The absorbance of each sample was measured at 450 nm using an enzyme-labeled instrument (Sunrise-basic Tecan, Männedorf, Switzerland). The ACE inhibition rate (I) was calculated using the following equation:
where Ablank1 was the absorbance without sample, Ablank2 was the absorbance without enzyme working solution, and Asample was the absorbance with samples or positive control.
Identification of peptides in Chouguiyu by peptidomics
The peptidomics was conducted using the ultra-high performance liquid chromatography tandem mass as described previously (26) with some modifications. In brief, the peptide extract (4 mg) was dissolved in 0.1% trifluoroacetic acid and was then desalted with the Oasis HLB column (Waters, Milford, MA, USA). The eluents were dissolved by 50% acetonitrile with 0.5% trifluoroacetic acid and were desalted with the Oasis MCX column (Waters, Milford, MA, USA). After being freeze-dried, the samples were dissolved at 10 μg/μl using 2% acetonitrile and 0.1% formic acid. The samples (10 μl) were analyzed using the Easy-nLC 1200 UPLC-Q Exactive HF-X tandem mass spectrometer (Thermo, Waltham, MA, USA), coupled with a Thermo C-18 column (Thermo, Waltham, MA, USA). The instrument was operated in a single charge mode ([M+H]+) at the range of 350–1,500 (m/z). The peptide sequences were identified using Proteome DiscovererTM version 2.4 (Thermo, Waltham, MA, USA) according to the protein sequences of Siniperca chuatsi. False discovery rate <0.01, unique peptide ≥1, and Sequest HT >0 were used as the criteria to confirm the credible peptides. The peptides with ACE inhibitory activity from Chouguiyu were further predicted using AHTpin (http://crdd.osdd.net/raghava/ahtpin/).
Microorganism determination by shot-gun metagenomics
The microorganisms in Chouguiyu were obtained after centrifugation of fermented liquid in each sample. Shot-gun metagenomics was adopted to analyze the microorganisms as in the previous study (27). In brief, the genomic DNA of microorganisms was fragmented to construct the DNA library using the NEXTFLEX® Rapid DNA-Seq (Bioo Scientific, Austin, TX, USA). The DNA library was then used for paired-end sequencing through the Illumina Hiseq Xten (Illumina Inc., San Diego, CA, USA). The abundance of microorganisms at the phylum and genus level was calculated according to the sum of all gene abundances. The genes were aligned to eggNOG database version 4.5.1 to obtain their functional annotation (28).
In silico analysis of ACE inhibitory peptides
The toxicity of peptides was obtained from ToxinPred (https://webs.iiitd.edu.in/raghava/toxinpred/multi_submit.php). The isoelectric point of peptides was obtained from Expasy (https://web.expasy.org/compute_pi/). The hydrophobicity was predicted from PepDraw (http://www.tulane.edu/~biochem/WW/PepDraw/). The instability of ACE inhibitory peptides was calculated from ProtParam (https://web.expasy.org/protparam/). PepBank was used to determine whether the peptide was reported on http://pepbank.mgh.harvard.edu/search/basic, containing the searches of Pubmed, PDF, ASPD, and UniProt. The water solubility was searched using http://www.innovagen.com/proteomics-tools. The predicted enzymes of core ACE inhibitory peptides were obtained from BIOPEP-UWM (https://biochemia.uwm.edu.pl/) (29), and their categories were searched from the enzyme database (https://www.brenda-enzymes.org/).
Molecular docking between ACE and ACE inhibitory peptides
The three-dimensional crystal structure of ACE (PDB ID: 1O8A) was downloaded from RCSB PDB (https://www.rcsb.org/) and prepared by Discovery Studio 2019 Client (San Diego, CA, USA) (30). The structures of ACE inhibitory peptides were generated using Chem Draw version 19.0 (Cambridge soft, Cambridge, MA, USA), and their energy was minimized under CHARMm using DS. The peptides were preloaded with hydrogen polar atoms, electric charges, and CFF force fields. The three-dimensional crystal structure of ACE was conducted with the clean process to delete water and former ligands, and its active sites of receptor cavities were further defined. The molecular docking between the ACE and ACE inhibitory peptides was conducted using the CDOCKER module. The hypotensive drug captopril was used as the positive control to dock with the ACE.
Statistical analysis
The principal component analysis (PCA) and abundance heatmaps of peptides were generated using Origin 9.5C (OriginLab Corp, Northampton, VA, USA). The peptide profile was drawn using Peptigram (http://bioware.ucd.ie/peptigram) (31). The correlation heatmap between the peptides and microorganisms was carried out using R (https://www.R-project.org/). The interaction network was constructed using Gephi (32).
Results and discussion
Identification of ACE inhibitory peptides from Chouguiyu
In this study, the ACE inhibition rates of the peptide extract from Chouguiyu at different fermentation times are shown in Supplementary Figure S1. Interestingly, the peptide extract in Chouguiyu exhibited good ACE inhibition activity as same as the positive drug captopril. The maximum ACE inhibition rate was observed in the D8 group, reaching 80.46%, significantly higher than that in the D0, D4, and positive control groups. To confirm which peptides contributed to the ACE inhibition activity, peptidomic analysis was used to identify the ACE inhibitory peptides. A previous study found that small-molecule peptides (generally ≤ 10 AAs) with various hydrophobic amino acids in the structure possessed good ACE inhibitory activity (33). A large number of ACE inhibitory peptides were found by virtual screening (8, 34). AHTpin was a common virtual method to predict and design efficient antihypertensive peptides (6, 35). In this study, a total of 356 potential ACE inhibitory peptides (AAs ≤ 10) were discovered by AHTpin. The principal component analysis (PCA) plot showed that the unfermented group (D0) was significantly different from the fermented groups (D4 and D8) (Figure 1A). Similar results were found in this study (Figure 1B), and more common ACE inhibitory peptides (120) were found in the groups D4 and D8. These bioactive peptides were named from P1 to P356 in order of their abundance (Figure 1B and Supplementary Table S1). Most of them increased with the increase of fermentation time, reaching their peaks in the D8 group. The most common ACE inhibitory peptides are rich in hydrophobic amino acids, and aromatic amino acids and aliphatic amino acids are conducive to enhancing ACE inhibitory activity (22, 36). In this study, plenty of hydrophobic amino acids were found in the ACE inhibitory peptides, of which 178 peptides possessed a proportion of hydrophobic amino acids in their sequences exceeding 50.00% (Supplementary Table S1). Valine, isoleucine, and proline were the main hydrophobic amino acids in the sequences of ACE inhibitory peptides (Supplementary Table S1).
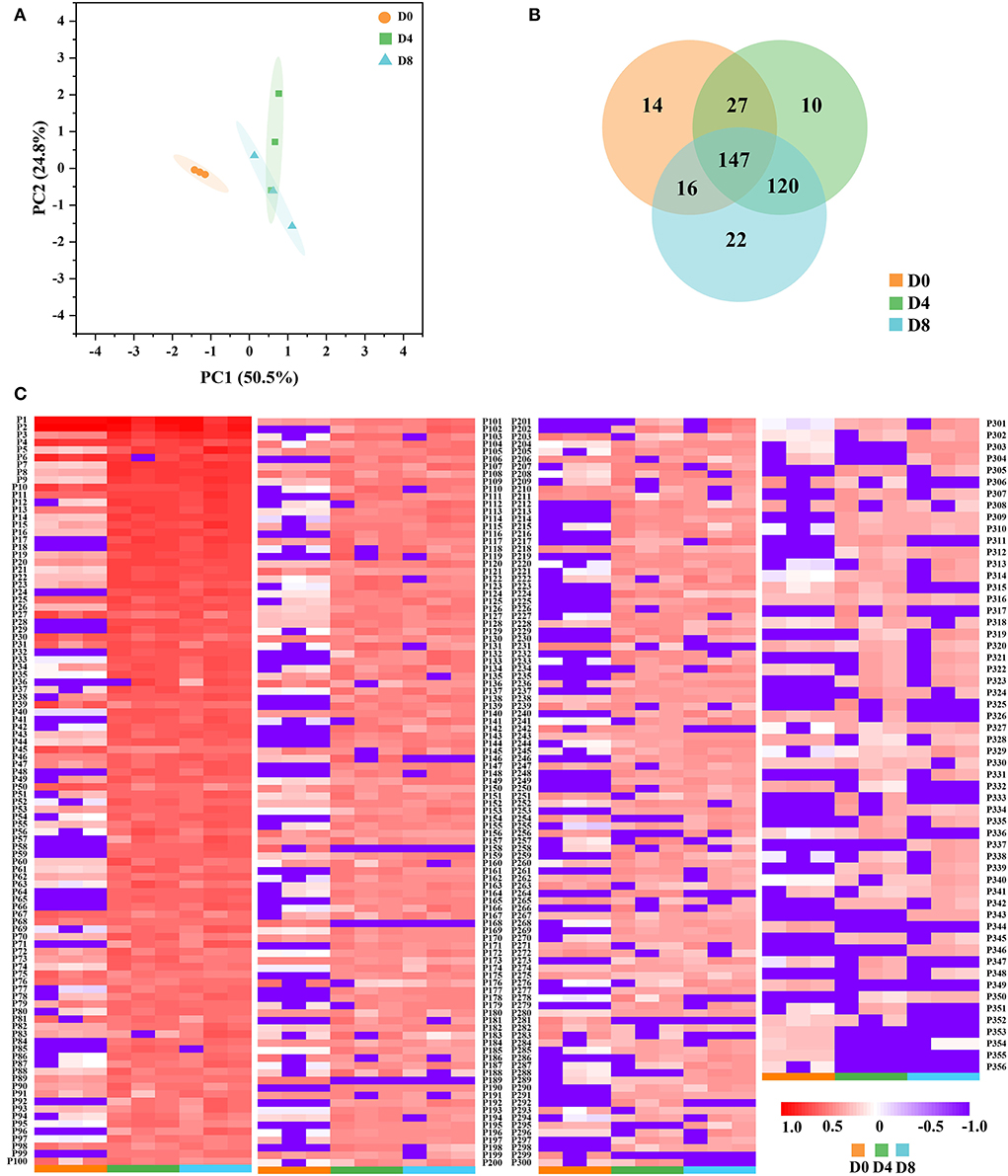
Figure 1. Identification of angiotensin I converting enzyme (ACE) inhibitory peptides from Chouguiyu at different fermentation times. (A) Principal component analysis (PCA), (B) Venn, and (C) heatmap analysis of ACE inhibitory peptides in the D0, D4, and D8 fermentation groups.
Precursor proteins and their cleavage locations of ACE inhibitory peptides from Chouguiyu
A total of 94 kinds of precursor proteins were identified after comparison with the proteins of Siniperca chuatsi (Supplementary Table S2). As shown in Figure 2, many ACE inhibitory peptides were derived from the same precursor protein at multiple locations, and most peptides shared the same cleavage locations with each other. The precursor proteins that could produce most ACE inhibitory peptides concentrated on muscle-type creatine kinase (27 peptides), nebulin (24 peptides), troponin I (16 peptides), myosin-binding protein H-like (14 peptides), fast skeletal muscle myosin heavy chain isoform 3 (14 peptides), nebulin isoform X8 (13 peptides), glyceraldehyde-3-phosphate dehydrogenase (11 peptides), troponin T (10 peptides), and nebulin (10 peptides), which were also important proteins of mandarin fish (Figure 2). The 10 peptides with high abundance including P1, P2, P3, P4, P5, P6, P7, P8, P9, and P10, derived from triosephosphate isomerase (location: 240–247), parvalbumin (location: 103–109), troponin I (location: 153–161), 14 kDa phosphohistidine phosphatase-like (location: 135–143), myosin heavy chain (location: 5–13), hemoglobin subunit alpha-B-like (location: 35–43), troponin T (location: 105–111), myosin-binding protein H-like (location: 424–430), nebulin isoform X8 (location: 364–371), and muscle-type creatine kinase (location: 60–69), respectively (Figure 2 and Supplementary Table S2).
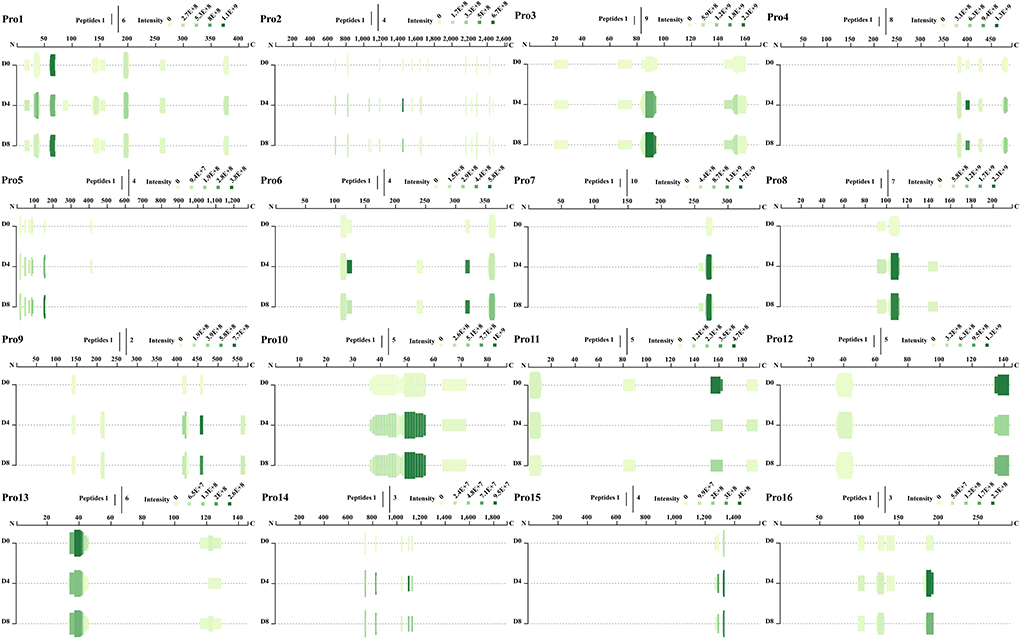
Figure 2. Angiotensin I converting enzyme inhibitory peptide profile of representative precursor proteins (Pro1–Pro16) during fermentation of Chouguiyu.
Characteristics of ACE inhibitory peptides from Chouguiyu by in silico analysis
The characteristics of thirty ACE inhibitory peptides with the highest abundance were further analyzed using in silico analysis, including isoelectric point, toxin, instability, hydrophobicity, water solubility, and aliphatic index (Supplementary Table S3). All the peptides were found to be novel peptides after searching from PepBank. All the ACE inhibitory peptides possessed non-toxicity, providing the possibility of safe use. A previous study showed that the hydrophobic peptides with small molecular weight from fish sauce had better ACE inhibition activity (12). In this study, the hydrophobicity of most ACE inhibitory peptides was more than 8.31 kcal/mol, indicating high hydrophobicity. At the same time, P1, P9, P25, P14, P20, P5, P24, P21, and P22 had high hydrophilicity index (>100), which indicated that they were highly amphiphilic and had the potential to be used in emulsion foods containing aqueous phase and lipid phase simultaneously (37). Most peptides had good water solubility and acidic isoelectric points. Similar studies found that many ACE inhibitory peptides exhibited acidic isoelectric points (37, 38). The stability of peptides plays a key role in maintaining their ACE inhibitory activities during digestion (39). The instability index ≥ 40 indicates that the peptides are considered to be unable to remain intact during digestion. In this study, a total of 18 peptides possessed low instability indexes (<40). According to the characteristics of ACE inhibitory peptides, P1, P2, P4, P7, P8, P9, and P10 were chosen as the core ACE inhibitory peptides for further analysis.
Intermolecular interactions between ACE and core ACE inhibitory peptides from Chouguiyu
Molecular docking technology has been widely used to study the ACE inhibition mechanism of peptides (4, 8). In this study, the energy of molecular docking between ACE and ACE inhibitory peptides is shown in Supplementary Table S4. Lower docking energy (ΔEdocking) indicates better affinity between the peptides and receptor (30, 40). In this study, the docking energy of seven ACE inhibitory peptides was less than −95 kcal/mol, far lower than that of positive control captopril (−15 kcal/mol) and the ACE inhibitory peptides in other reports (−3.86 kcal/mol) (13). The binding energy (ΔEbinding) of peptides was all lower than −145 kcal/mol, suggesting their stable binding with ACE. The electrostatic energy (ΔEele) of the peptides was much smaller than the van der Waals energy (ΔEvdw).
The inhibition mechanisms of ACE inhibitory peptides were explored by intermolecular interactions at the atomic level (Figure 3, Supplementary Figure S2, and Supplementary Table S5). More kinds of interactions were found in seven ACE inhibitory peptides than in captopril with ACE, suggesting their stronger interactions, in accordance with the results of molecular docking energy. There were 14 kinds of interactions found in complexes of seven ACE inhibitory peptides and ACE, belonging to electrostatic interaction (attractive charge, pi-anion, and pi-cation), hydrogen bond (salt bridge, conventional hydrogen bond, carbon-hydrogen bond, and pi-donor hydrogen bond), hydrophobic interaction (pi-pi T-shaped, pi-sigma, alkyl, and pi-alkyl), and other interaction (pi-sulfur and sulfur-X). Hydrogen bond and hydrophobic interaction are considered the main interactions between ACE inhibitory peptides and ACE (4, 8). In this study, plenty of salt bridges, conventional hydrogen, carbon-hydrogen bond, and alkyl were found between the 7 ACE inhibitory peptides and ACE. The residues in ACE for the formation of the salt bridge were concentrated on Arg522, Arg124, and Lys118, while the residues Tyr62, Asn66, Asn70, Lys118, Asp121, Glu123, Ser219, Trp220, Tyr360, Arg402, Glu403, Gly404, Ser517, and Arg522 contributed to forming a conventional hydrogen bond. Similar residues in ACE were reported in the formation of conventional hydrogen bonds with ACE inhibitory peptides (30). Among these residues, Arg522 played an important role in the ACE activity because it acted as the chloride ion ligand for ACE activation for substrate hydrolysis (40). The conventional hydrogen bond forming between these residues and peptides might lead to the ACE inactivation (40). In this study, the interaction between the ACE inhibitory peptides and ACE was further analyzed at the atomic level. The unsaturated oxygen atoms in P1 (O17, O51, O94, and O127), P2 (O31, O64, O108, and O109), P4 (O13, O14, O16, O93, O94, O106, and O129), P7 (O11, O13, O115, O42, O43, O54, O55, O88, and O114), P8 (O36, O37, O39, O48, O49, O86, and O106), P9 (O20, O43, O44, O55, O56, O72, O93, and O132), and P10 (O24, O34, O141, and O142) as H-acceptors formed strong interactions with ACE, contributing to the formation of the salt bridge and conventional hydrogen bond. Residues Trp59, Glu123, Glu143, Met223, Glu403, Pro407, His410, Ser516, Ser517, and Pro519 in ACE played a key role in forming a carbon-hydrogen bond with ACE inhibitory peptides. Furthermore, plenty of alkyl interactions were found in P1 (C8-Met223, C39-Met223, and C39-Pro407), P2 (C38-Pro519 and C42-Pro519), P4 (C30-Val518), P7 (C64-Val518), P8 (P8-Val518 and C15-Met223), P9 (P9-Arg124), and P10 (C15-Met223, C15-Pro407, and C129-Leu139). There were also some pi-alkyl interactions in P1 (C65-Phe391 and C46-His410), P2 (C71-Trp59, C75-Trp59, and P2-Met223), P4 (C30-Phe512), P8 (C15-Phe570), P9 (C8-Trp59), and P10 (P10-Trp357 and C15-Phe570). In addition, there was an unfavorable bond found in P4 (H2-Arg124), suggesting the formation of repulsive charge interaction.
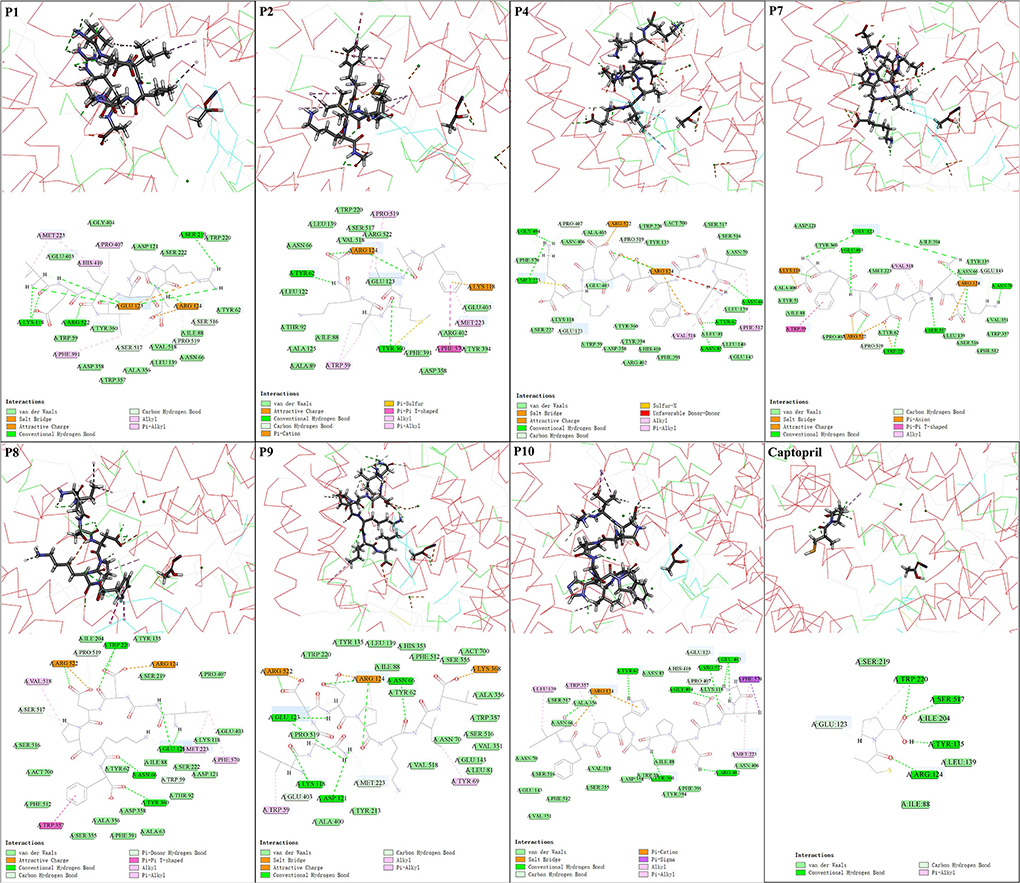
Figure 3. Molecular interactions of the core ACE inhibitory peptides (P1, P2, P4, P7, P8, P9, and P10) and captopril into the active site of ACE.
Correlation between core ACE inhibitory peptides and their cleavage proteases from microorganisms in Chouguiyu
Peptides from aquatic products exhibit different structures and inhibition functions to concurrently inhibit multiple enzymes involved in chronic diseases (40). As an aquatic product, Chouguiyu is produced under the metabolism of the complex microbial community. In this study, the ACE inhibitory peptides with high activity might be formed by the hydrolysis of proteases from the microbial community. The relative abundance of the microbial community in Chouguiyu during the fermentation process was first studied (Figure 4). Six bacterial phyla were identified according to the relative abundance of more than 0.1%, including Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Actinobacteria, and Uroviricota (Figure 4A). Among these, Firmicutes and Proteobacteria were the dominant phyla with a total proportion of over 84% in the whole fermentation process. Proteobacteria possessed the highest relative abundance at the beginning of fermentation (D0) reaching 70.09% and declined with the increasing fermentation time, while Firmicutes gradually occupied the main abundance and increased to 67.37% at the end of fermentation (D8). Similar dominant phyla were reported in other studies on the Chouguiyu (41). A total of 438 microbial genera with a relative abundance of more than 0.1% were found in Chouguiyu in the D0, D4, and D8 groups. The 20 genera with the highest total relative abundance of the D0, D4, and D8 groups are shown in Figure 4B. Few dominant bacteria were observed in the group D0, only including Escherichia (24.40%), Acinetobacter (11.09%), Psychrobacter (1.71%), and Bacillus (1.06%), while the relative abundance of other genera was all less than 1%. With the increasing fermentation time, the abundance of Escherichia and Bacillus significantly decreased, and more microbial genera with high abundance occurred. In the D4 group, there were 16 genera with an abundance of more than 1%, mainly including Vagococcus (18.79%), Acinetobacter (17.23%), Enterococcus (5.06%), Peptostreptococcus (5.06%), and Psychrobacter (4.63%). After fermentation for 8 days, the abundance of Vagococcus, Psychrobacter, and Enterococcus reached the maximum, respectively, reaching 33.77, 10.40, and 8.91%. Besides, the genera with an abundance of more than 1% in the D8 group also included Acinetobacter (8.63%), Peptostreptococcus (7.48%), Streptococcus (4.66%), Lactococcus (2.76%), and Carnobacterium (2.23%). Similar dominant genera such as Vagococcus, Psychrobacter, Lactococcus, Fusobacterium, Vibrio, and Psychrilyobacter were found in the late fermentation period in Chouguiyu (41).
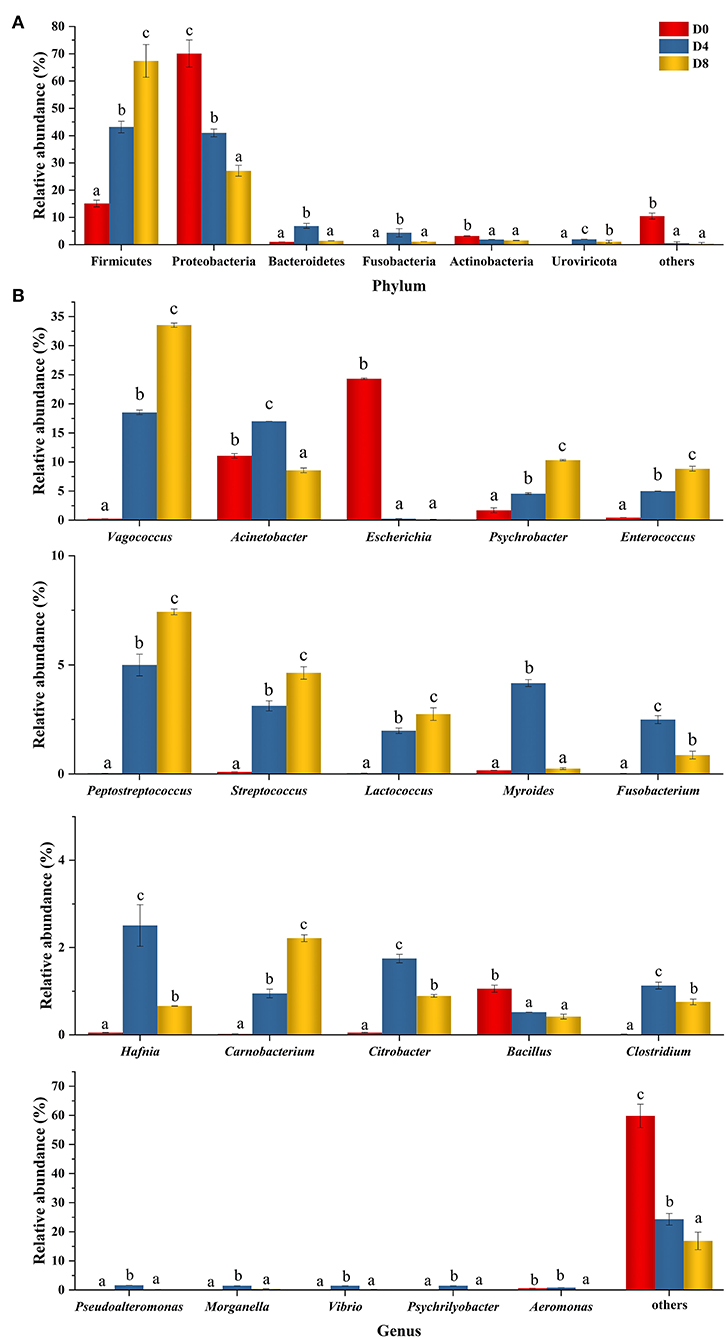
Figure 4. Changes in the relative abundance of the microbial community at the (A) phylum and (B) genus level in Chouguiyu during the fermentation process.
To explore the possible formation mechanism of core ACE inhibitory peptides, the correlation between 20 microbial genera and 7 core ACE inhibitory peptides was then analyzed by Pearson's correlation (Figure 5A). For microbial genera, three distinct clusters were observed, including microbial cluster 1 (Escherichia and Bacillus), microbial cluster 2 (Psychrobacter, Carnobacterium, Vagococcus, Enterococcus, Lactococcus, Peptostreptococcus, Streptococcus, Citrobacter, and Clostridium), and microbial cluster 3 (Aeromonas, Acinetobacter, Myroides, Pseudoalteromonas, Vibrio, Psychrilyobacter, Fusobacterium, Hafnia, and Morganella). Interestingly, most ACE inhibitory peptides in Chouguiyu showed negative relations with microbial cluster 1 but positive correlations with the genera in microbial cluster 2 and microbial cluster 3. The correlation network map was then built using the Pearson's correlation coefficient between the ACE inhibitory peptides and microbial genera (|r|>0.6 and p < 0.05) (Figure 5B). Meanwhile, the topological characteristics of the correlation network map were analyzed using R, including the betweenness centrality, closeness centrality, degree, neighborhood connectivity, topological coefficient, and average shortest path length (Supplementary Figure S3). Among these, the degree is the main metric to describe the centrality of nodes in the correlation network, and a higher degree indicates more importance in the network (42). Interestingly, P8, P7, P9, and P2 possessed the highest degree and were significantly positively correlated with the most microbial genera (16, 15, 14, and 13, respectively), while P1, P4, and P10 showed a significantly positive relation with less microbial genera, mainly including Escherichia and Bacillus.
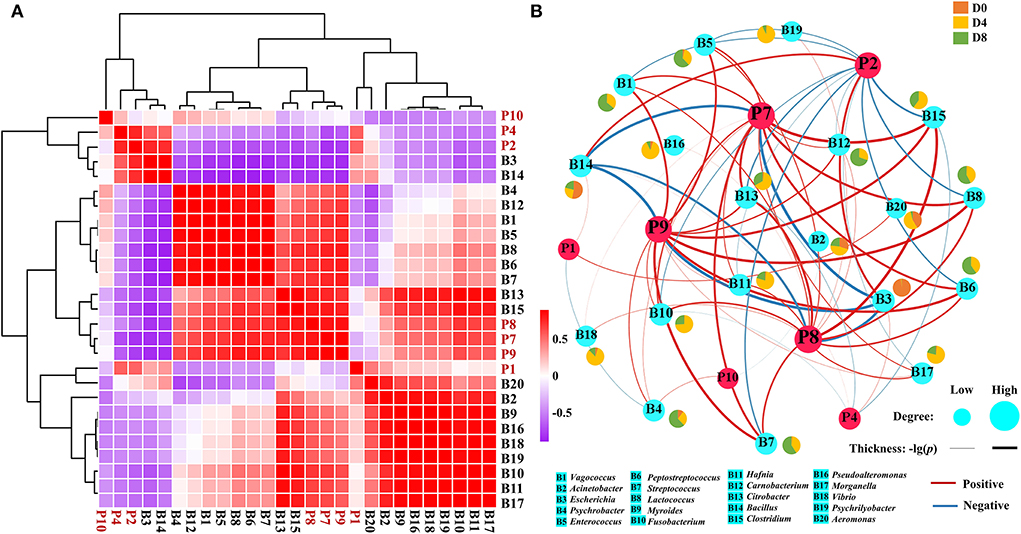
Figure 5. Correlation analysis between the ACE inhibitory peptides and microbial genera. (A) Clustering heatmap and (B) correlation network (|r| > 0.6 and p < 0.05) based on Pearson's correlation tests.
After prediction at BIOPEP-UWM, the cleavage proteases from the microbial genera in Chouguiyu for preparing 7 core ACE inhibitory peptides were further analyzed (Figure 6 and Supplementary Table S6). P1 could be produced by serine endopeptidases (COG1770, COG0466, COG1219, COG0265, and COG0681), cysteine endopeptidases (COG3672), aspartic endopeptidases (COG0616 and COG3577), and metalloendopeptidases (COG0826, COG0750, COG0501, COG4942, and COG1164) from Aeromonas (r = 0.69), as well as by serine endopeptidases (COG1506, COG0542, COG1404, COG1505, and ENOG410XP7P) and metalloendopeptidases (COG5504, COG3340, COG0465, COG3740, and COG0501) from Bacillus (r = 0.62). P2 could be prepared by serine endopeptidases (COG1770, ENOG410XP7P, COG0542, COG0265, and ENOG4111MMN), aspartic endopeptidases (COG0616), and metalloendopeptidases (COG0739, ENOG410XZ09, COG0465, and COG0339) from Escherichia (r = 0.84) and by serine endopeptidases (COG1506, COG0542, COG1404, COG1505, and ENOG410XP7P) and metalloendopeptidases (COG5504, COG3340, COG0465, COG3740, and COG0501) from Bacillus (r = 0.82). P4 could be obtained by serine endopeptidases (COG1770, NOG410XP7P, COG0542, COG0265, and ENOG4111MMN) and aspartic endopeptidases (COG0616) from Escherichia (r = 0.63), as well as by serine endopeptidases (COG1506, COG0542, COG1404, COG1505, and ENOG410XP7P) from Bacillus (r = 0.60). Many microbial genera could secrete various proteases for P7, P8, and P9 production. These proteases were concentrated on serine endopeptidases from Clostridium, Lactococcus, Peptostreptococcus, Citrobacter, Streptococcus, Vagococcus, Enterococcus, Fusobacterium, Morganella, Hafnia, Carnobacterium, and Psychrobacter and aspartic endopeptidases from Lactococcus, Citrobacter, Vagococcus, Enterococcus, Fusobacterium, Morganella, Hafnia, Carnobacterium, and Psychrobacter. In addition, fewer microbial genera could produce cysteine endopeptidases for P7, P8, and P9 production, mainly including Citrobacter (COG3672) and Psychrobacter (ENOG4111IPD). For P10, only Psychrobacter (r = 0.64) contributed most to the generation of P10 by serine endopeptidases (COG0466, COG0542, COG1219, COG1067, and COG0265), cysteine endopeptidases (ENOG4111IPD), and aspartic endopeptidases (COG0616).
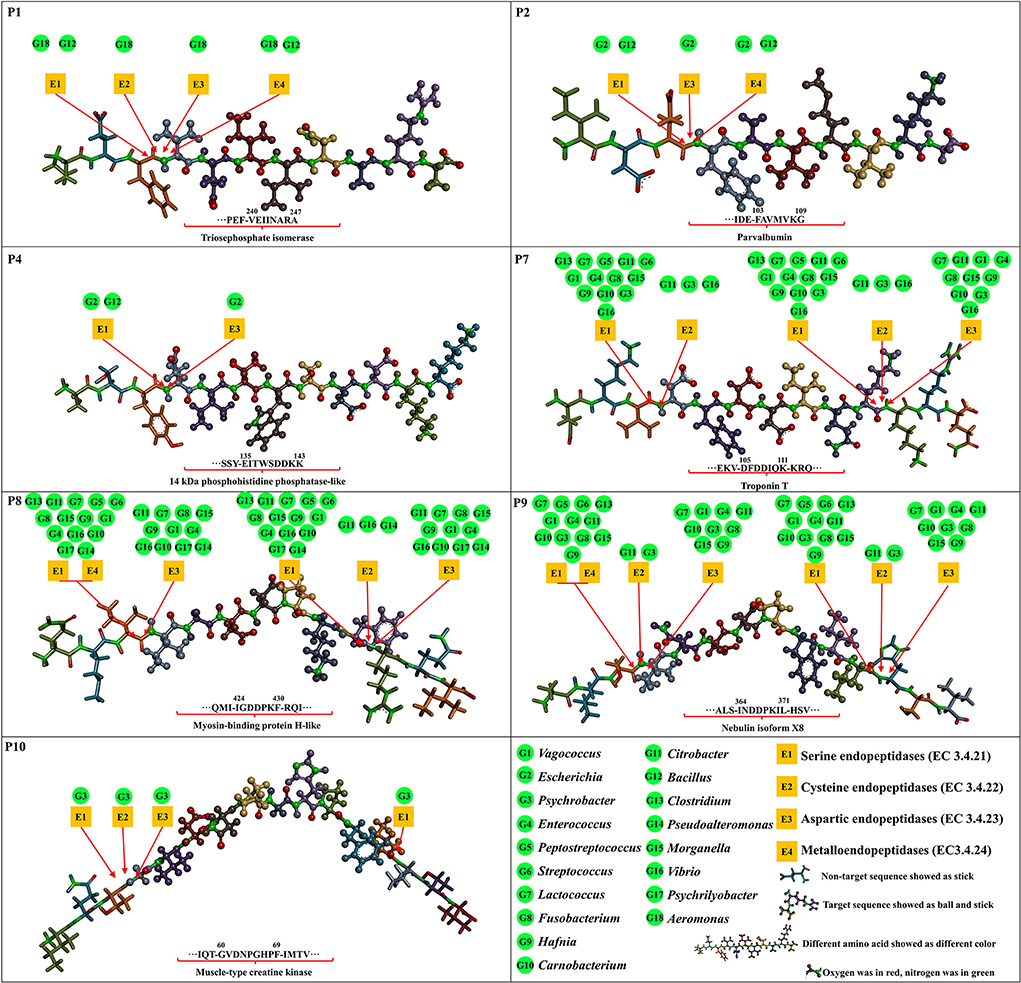
Figure 6. Formation of the core ACE inhibitory peptides (P1, P2, P4, P7, P8, P9, and P10) through cleavage proteases from the microbial genera based on hydrolysis prediction and correlation network.
Formation and inhibition mechanism of core ACE inhibitory peptides from Chouguiyu
In this study, the peptide extract from Chouguiyu exhibited good ACE inhibition, and the inhibition rate was significantly enhanced after fermentation for 8 days (Supplementary Figure S1). To identify the ACE inhibitory peptides in Chouguiyu, peptidomics and AHTpin were combined, and 356 ACE inhibitory peptides were obtained (Figure 1). P1, P2, P4, P7, P8, P9, and P10 were selected as the core ACE inhibitory peptides according to their abundance, characteristics, and docking energy. Their precursor proteins were triosephosphate isomerase, parvalbumin, 14 kDa phosphohistidine phosphatase-like, troponin T, myosin-binding protein H-like, nebulin isoform X8, and muscle-type creatine kinase, respectively (Figure 2 and Supplementary Table S2). Based on the hydrolysis prediction, these 7 core peptides could be hydrolyzed from various proteases derived from plenty of microorganisms (Supplementary Table S6). Lactic acid bacteria (LAB) were considered an important group of microorganisms in Chouguiyu, contributing greatly to its aroma formation (43). After the further correlation analysis between these proteases and ACE inhibitory peptides, the P7, P8, and P9 were mainly produced by the proteases from LAB including Lactococcus, Enterococcus, Vagococcus, Peptostreptococcus, and Streptococcus. These genera were also reported for the formation of umami peptides in Chouguiyu (26). Differently, the P1, P2, P4, and P10 were mainly produced by Aeromonas, Bacillus, Escherichia, and Psychrobacter. Bacillus is famous for its ability to produce various high-activity proteases (44). The high abundance of these genera at the beginning of fermentation time was helpful in producing these four ACE inhibitory peptides. The high abundance of Vagococcus, Lactococcus, Fusobacterium, Streptococcus, Escherichia, Bacillus, and Pseudoalteromonas contributed to the production of the core ACE inhibitory peptides. The high abundance of these genera was also reported in Chouguiyu in other studies (41, 45, 46). The isolation of specific microorganisms is expected to directionally produce the responding ACE inhibitory peptides.
Intermolecular interaction analysis by molecular docking is considered an effective technology to reveal the inhibition mechanism of ACE inhibitory peptides on ACE (8). In this study, molecular docking between ACE inhibitory peptides and ACE showed that the amino acid residues at the end of peptides played an important role in their ACE inhibitory activity. Previous studies showed that the amino acid residues at N-terminal in ACE inhibitory peptides could form strong interactions with ACE (22, 47). Similar results were also observed in this study that V- in P1 formed 3 conventional hydrogen bonds, 1 carbon-hydrogen bond, and 1 alkyl; E- in P4 formed 1 salt bridge and 1 conventional hydrogen bond; D- in P7 formed 1 salt bridge, 2 conventional hydrogen bond, and 1 carbon-hydrogen bond; I- in P8 formed 1 conventional hydrogen bond and 1 alkyl; I- in P9 formed 1 conventional hydrogen bond, 2 carbon-hydrogen bond, and 1 pi-alkyl; G- in P10 formed 1 conventional hydrogen bond and 1 carbon-hydrogen bond (Figure 3 and Supplementary Table S5). Meanwhile, the positively charged (13, 48) and hydrophobic (49) amino acid residues at C-terminal were beneficial to their ACE inhibitory activity. Similar structures were observed in this study, including the positively charged residues (-A in P1, -K in P4, and -K in P7) and hydrophobic residues (-F in P8, -L in P9, and -I in P10). These residues could perform a good ACE inhibitory activity, probably due to the formation of plenty of salt bridges, conventional hydrogen bonds, carbon-hydrogen bonds, and alkyl with ACE (Figure 3 and Supplementary Table S5). The illustration of the formation and inhibition mechanism of ACE inhibitory peptides in Chouguiyu provides important technical support for the isolation of the proteases and microbial strains to directionally produce the responding ACE inhibitory peptides and new ideas to screen novel ACE inhibitory peptides from other fermented foods.
Conclusion
Peptide extract of Chouguiyu exhibited a good inhibition effect on ACE, especially in the D8 group. A total of 356 ACE inhibitory peptides were predicted using in silico, and most ACE inhibitory peptides increased with the increase of fermentation time. These peptides could be obtained from 94 kinds of precursor proteins which were concentrated on muscle-type creatine kinase, nebulin, and troponin I. Seven core ACE inhibitory peptides with high abundance, stable structure, high hydrophobicity, and low docking energy were selected, including P1 (VEIINARA), P2 (FAVMVKG), P4 (EITWSDDKK), P7 (DFDDIQK), P8 (IGDDPKF), P9 (INDDPKIL), and P10 (GVDNPGHPFI). Molecular docking showed that the inhibition of ACE mainly resulted from unsaturated oxygen atoms in the seven peptides forming a salt bridge and conventional hydrogen bond with ACE as H-acceptors, followed by active carbon atoms forming alkyl and pi-alkyl interactions. The N-terminal and C-terminal amino acid residues of these peptides played an important role in their ACE inhibitory activity. P7, P8, and P9 were mainly produced by the proteases from LAB including Lactococcus, Enterococcus, Vagococcus, Peptostreptococcus, and Streptococcus, while P1, P2, P4, and P10 were mainly produced by Aeromonas, Bacillus, Escherichia, and Psychrobacter after analysis by hydrolysis prediction and correlation network. This study provides a theoretical basis for understanding the formation and inhibition mechanism of ACE inhibitory peptides in Chouguiyu and important information for the related microbial strains and proteases to produce the responding ACE inhibitory peptides.
Data availability statement
The data presented in the study are deposited in ProteomeXchange, accession number PXD033251.
Ethics statement
Ethical review and approval was not required for the animal study because Frozen mandarin fish were used in this study.
Author contributions
DY: methodology and writing – original draft. LL: conceptualization. CL: writing – review and editing and conceptualization. SC and JD: visualization. SY: data curation. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the National Key R&D Program of China (2019YFD0901903), the China Agriculture Research System of MOF and MARA (CARS-46), the Guangdong Basic and Applied Basic Research Foundation (2021A1515010872), the Young S&T Talent Training Program of Guangdong Provincial Association for S&T, China (SKXRC202210), and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (2020TD69).
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this study.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.920945/full#supplementary-material
References
1. Paisansak S, Sangtanoo P, Srimongkol P, Saisavoey T, Reamtong O, Choowongkomon K, et al. Angiotensin-I converting enzyme inhibitory peptide derived from the shiitake mushroom (Lentinula edodes). J Food Sci Technol. (2021) 58:85–97. doi: 10.1007/s13197-020-04517-z
2. Rubak YT, Nuraida L, Iswantini D, Prangdimurti E. Angiotensin-I-Converting enzyme inhibitory peptides in goat milk fermented by lactic acid bacteria isolated from fermented food and breast milk. Food Sci Anim Resour. (2022) 42:46–60. doi: 10.5851/kosfa.2021.e55
3. Ishak NH, Shaik MI, Yellapu NK, Howell NK, Sarbon NM. Purification, characterization and molecular docking study of angiotensin-I converting enzyme (ACE) inhibitory peptide from shortfin scad (Decapterus macrosoma) protein hydrolysate. J Food Sc Technol. (2021) 58:4567–77. doi: 10.1007/s13197-020-04944-y
4. Yang G, Qin S, Li W. Purification and characterization of a novel angiotensin I-converting enzyme-inhibitory peptide derived from Alaska pollack skins. J Food Sci. (2021) 86:2457–67. doi: 10.1111/1750-3841.15754
5. Kircheva N, Dobrev S, Yakimova B, Stoineva I, Angelova S. Molecular insights into the interaction of angiotensin I-converting enzyme (ACE) inhibitors and HEXXH motif. Biophys Chem. (2021) 276:106626. doi: 10.1016/j.bpc.2021.106626
6. Zhang P, Roytrakul S, Sutheerawattananonda M. Production and purification of glucosamine and angiotensin-I converting enzyme (ACE) inhibitory peptides from mushroom hydrolysates. J Funct Foods. (2017) 36:72–83. doi: 10.1016/j.jff.2017.06.049
7. Lo WMY, Li-Chan ECY. Angiotensin i converting enzyme inhibitory peptides from in vitro pepsin–pancreatin digestion of soy protein. J Agr Food Chem. (2005) 53:3369–76. doi: 10.1021/jf048174d
8. Chen J, Yu X, Chen Q, Wu Q, He Q. Screening and mechanisms of novel angiotensin-I-converting enzyme inhibitory peptides from rabbit meat proteins: A combined in silico and in vitro study. Food Chem. (2022) 370:131070. doi: 10.1016/j.foodchem.2021.131070
9. Chaudhary A, Bhalla S, Patiyal S, Raghava GPS, Sahni G. FermFooDb: A database of bioactive peptides derived from fermented foods. Heliyon. (2021) 7:e6668. doi: 10.1016/j.heliyon.2021.e06668
10. Yousefi L, Habibi Najafi MB, Edalatian Dovom MR, Mortazavian AM. Production of angiotensin-converting enzyme inhibitory peptides in Iranian ultrafiltered white cheese prepared withLactobacillus brevis KX572382. Int J Food Sci Technol. (2021) 56:2530–8. doi: 10.1111/ijfs.14891
11. Parmar H, Hati S, Panchal G, Sakure AA. Purification and production of novel angiotensin I-Converting enzyme (ACE) inhibitory bioactive peptides derived from fermented goat milk. Int J Pept Res Ther. (2020) 26:997–1011. doi: 10.1007/s10989-019-09902-7
12. Khositanon P, Panya N, Roytrakul S, Krobthong S, Chanroj S, Choksawangkarn W. Effects of fermentation periods on antioxidant and angiotensin I-converting enzyme inhibitory activities of peptides from fish sauce by-products. LWT-Food Sci Technol. (2021) 135:110122. doi: 10.1016/j.lwt.2020.110122
13. Li M, Fan W, Xu Y. Identification of angiotensin converting enzyme (ACE) inhibitory and antioxidant peptides derived from Pixian broad bean paste. LWT-Food Sci Technol. (2021) 151:112221. doi: 10.1016/j.lwt.2021.112221
14. Majumdar RK, Roy D, Bejjanki S, Bhaskar N. An overview of some ethnic fermented fish products of the Eastern Himalayan region of India. Journal of Ethnic Foods. (2016) 3:276–83. doi: 10.1016/j.jef.2016.12.003
15. Wang Y, Shen Y, Wu Y, Li C, Li L, Zhao Y, et al. Comparison of the microbial community and flavor compounds in fermented mandarin fish (Siniperca chuatsi): Three typical types of Chinese fermented mandarin fish products. Food Res Int. (2021) 144:110365. doi: 10.1016/j.foodres.2021.110365
16. Li C, Wu J, Li Y, Dai Z. Identification of the aroma compounds in stinky mandarin fish (Siniperca chuatsi) and comparison of volatiles during fermentation and storage. Int J Food Sci Technol. (2013) 48:2429–37. doi: 10.1111/ijfs.12254
17. Zhao Y, Wang Y, Li C, Li L, Yang X, Wu Y, et al. Novel insight into physicochemical and flavor formation in naturally fermented tilapia sausage based on microbial metabolic network. Food Res Int. (2021) 141:110122. doi: 10.1016/j.foodres.2021.110122
18. Parmanand BA, Kellingray L, Le Gall G, Basit AW, Fairweather-Tait S, Narbad A, et al. decrease in iron availability to human gut microbiome reduces the growth of potentially pathogenic gut bacteria; An in vitro colonic fermentation study. J Nutr Biochem. (2019) 67:20–7. doi: 10.1016/j.jnutbio.2019.01.010
19. Wang Y, Li C, Li L, Yang X, Chen S, Wu Y, et al. Application of UHPLC-Q/TOF-MS-based metabolomics in the evaluation of metabolites and taste quality of Chinese fish sauce (Yu-lu) during fermentation. Food Chem. (2019) 296:132–41. doi: 10.1016/j.foodchem.2019.05.043
20. Sánchez-López F, Robles-Olvera VJ, Hidalgo-Morales M, Tsopmo A. Angiotensin-I converting enzyme inhibitory activity of Amaranthus hypochondriacus seed protein hydrolysates produced with lactic bacteria and their peptidomic profiles. Food Chem. (2021) 363:130320. doi: 10.1016/j.foodchem.2021.130320
21. Li X, Xie X, Wang J, Xu Y, Yi S, Zhu W, et al. Identification, taste characteristics and molecular docking study of novel umami peptides derived from the aqueous extract of the clam meretrix meretrix Linnaeus. Food Chem. (2020) 312:126053. doi: 10.1016/j.foodchem.2019.126053
22. Fan H, Liao W, Wu J. Molecular interactions, bioavailability, and cellular mechanisms of angiotensin-converting enzyme inhibitory peptides. J Food Biochem. (2019) 43:e12572. doi: 10.1111/jfbc.12572
23. Shanmugam VP, Kapila S, Kemgang TS, Reddi S, Kapila R, Muthukumar S, et al. Isolation and characterization of angiotensin converting enzyme inhibitory peptide from buffalo casein. Int J Pept Res Ther. (2021) 27:1481–91. doi: 10.1007/s10989-021-10185-0
24. Yang D, Li C, Li L, Chen S, Hu X, Xiang H. Taste mechanism of umami peptides from Chinese traditional fermented fish (Chouguiyu) based on molecular docking using umami receptor T1R1/T1R3. Food Chem. (2022) 389:133019. doi: 10.1016/j.foodchem.2022.133019
25. Escudero E, Aristoy M, Nishimura H, Arihara K, Toldrá F. Antihypertensive effect and antioxidant activity of peptide fractions extracted from Spanish dry-cured ham. Meat Sci. (2012) 91:306–11. doi: 10.1016/j.meatsci.2012.02.008
26. Yang D, Li C, Li L, Wang Y, Wu Y, Chen S, et al. Novel insight into the formation mechanism of umami peptides based on microbial metabolism in Chouguiyu, a traditional Chinese fermented fish. Food Res Int. (2022) 157:111211. doi: 10.1016/j.foodres.2022.111211
27. Wang Y, Li C, Zhao Y, Li L, Yang X, Wu Y, et al. Novel insight into the formation mechanism of volatile flavor in Chinese fish sauce (Yu-lu) based on molecular sensory and metagenomics analyses. Food Chem. (2020) 323:126839. doi: 10.1016/j.foodchem.2020.126839
28. Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. EggNOG 45: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. (2016) 44:D286–93. doi: 10.1093/nar/gkv1248
29. Minkiewicz P, Iwaniak A, Darewicz M. BIOPEP-UWM database of bioactive peptides: current opportunities. Int J Mol Sci. (2019) 20:5978. doi: 10.3390/ijms20235978
30. Tu M, Wang C, Chen C, Zhang R, Liu H, Lu W, et al. Identification of a novel ACE-inhibitory peptide from casein and evaluation of the inhibitory mechanisms. Food Chem. (2018) 256:98–104. doi: 10.1016/j.foodchem.2018.02.107
31. Manguy J, Jehl P, Dillon ET, Davey NE, Shields DC, Holton TA. Peptigram: A Web-Based application for peptidomics data visualization. J Proteome Res. (2017) 16:712–9. doi: 10.1021/acs.jproteome.6b00751
32. Li C, Li W, Li L, Chen S, Wu Y, Qi B. Microbial community changes induced by a newly isolated salt-tolerant Tetragenococcus muriaticus improve the volatile flavor formation in low-salt fish sauce. Food Res Int. (2022) 156:111153. doi: 10.1016/j.foodres.2022.111153
33. Tu M, Qiao X, Wang C, Liu H, Cheng S, Xu Z, et al. In vitro and in silico analysis of dual-function peptides derived from casein hydrolysate. Food Sci Hum Wellness. (2021) 10:32–7. doi: 10.1016/j.fshw.2020.08.014
34. Zhao W, Xue S, Yu Z, Ding L, Li J, Liu J. Novel ACE inhibitors derived from soybean proteins using in silico and in vitro studies. J Food Biochem. (2019) 43:e12975. doi: 10.1111/jfbc.12975
35. Kumar R, Chaudhary K, Singh Chauhan J, Nagpal G, Kumar R, Sharma M, et al. An in silico platform for predicting, screening and designing of antihypertensive peptides. Sci Rep-Uk. (2015) 5:1–10. doi: 10.1038/srep12512
36. Zhang Y, Pan D, Yang Z, Gao X, Dang Y. Angiotensin I-Converting enzyme (ACE) inhibitory and dipeptidyl Peptidase-4 (DPP-α) inhibitory activity of umami peptides from Ruditapes philippinarum. LWT-Food Sci Technol. (2021) 144:111265. doi: 10.1016/j.lwt.2021.111265
37. Zheng Y, Shi P, Li Y, Zhuang Y, Linzhang Y, Liu L, et al. A novel ACE-inhibitory hexapeptide from camellia glutelin-2 hydrolysates: Identification, characterization and stability profiles under different food processing conditions. LWT-Food Sci Technol. (2021) 147:111682. doi: 10.1016/j.lwt.2021.111682
38. Undhad Trupti J, Das S, Solanki D, Kinariwala D, Hati S. Bioactivities and ACE-inhibitory peptides releasing potential of lactic acid bacteria in fermented soy milk. Food Product Process Nutr. (2021) 3:1–14. doi: 10.1186/s43014-021-00056-y
39. Wu N, Xu W, Liu K, Xia Y. Shuangquan Angiotensin-converting enzyme inhibitory peptides from Lactobacillus delbrueckii QS306 fermented milk. J Dairy Sci. (2019) 102:5913–21. doi: 10.3168/jds.2018-15901
40. Zhang Y, He S, Rui X, Simpson BK. Interactions of C Frondosa-derived inhibitory peptides against angiotensin I-converting enzyme (ACE), α-amylase and lipase. Food Chem. (2022) 367:130695. doi: 10.1016/j.foodchem.2021.130695
41. Yang Z, Wu R, Wei X, Zhang Z, Wang W, Liu A, et al. Moderate fermentation contributes to the formation of typical aroma and good organoleptic properties: A study based on different brands of Chouguiyu. LWT- Food Sci Technol. (2021) 152:112325. doi: 10.1016/j.lwt.2021.112325
42. Huang CH, Chen TH, Ng KL. Graph theory and stability analysis of protein complex interaction networks. Iet Syst Biol. (2016) 10:64–75. doi: 10.1049/iet-syb.2015.0007
43. Dai Z, Li Y, Wu J, Zhao Q. Diversity of lactic acid bacteria during fermentation of a traditional chinese fish product, Chouguiyu (Stinky mandarin fish). J Food Sci. (2013) 78:M1778–83. doi: 10.1111/1750-3841.12289
44. Zeng X, Xia W, Jiang Q, Yang F. Chemical and microbial properties of Chinese traditional low-salt fermented whole fish product Suan yu. Food Control. (2013) 30:590–5. doi: 10.1016/j.foodcont.2012.07.037
45. Yang Z, Liu S, Lv J, Sun Z, Xu W, Ji C, et al. Microbial succession and the changes of flavor and aroma in Chouguiyu, a traditional Chinese fermented fish. Food Biosci. (2020) 37:100725. doi: 10.1016/j.fbio.2020.100725
46. Ji C, Zhang J, Lin X, Han J, Dong X, Yang S, et al. Metaproteomic analysis of microbiota in the fermented fish, Siniperca chuatsi. LWT-Food Sci Technol. (2017) 80:479–84. doi: 10.1016/j.lwt.2017.03.022
47. Chen R, Miao Y, Hao X, Gao B, Ma M, Zhang JZH, et al. Investigation on the characteristics and mechanisms of ACE inhibitory peptides by a thorough analysis of all 8000 tripeptides via binding free energy calculation. Food Sci Nutr. (2021) 9:2943–53. doi: 10.1002/fsn3.2253
48. Lu Y, Wang Y, Huang D, Bian Z, Lu P, Fan D, et al. Inhibitory mechanism of angiotensin-converting enzyme inhibitory peptides from black tea. J Zhejiang Univ-Sc B. (2021) 22:575–89. doi: 10.1631/jzus.B2000520
Keywords: Chouguiyu, ACE inhibitory peptide, metagenomics, peptidomics, molecular docking, correlation network
Citation: Yang D, Li L, Li C, Chen S, Deng J and Yang S (2022) Formation and inhibition mechanism of novel angiotensin I converting enzyme inhibitory peptides from Chouguiyu. Front. Nutr. 9:920945. doi: 10.3389/fnut.2022.920945
Received: 15 April 2022; Accepted: 27 June 2022;
Published: 22 July 2022.
Edited by:
Elena Mudura, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Neena Puri, Guru Nanak Khalsa College Yamuna Nagar Haryana, IndiaSubrota Hati, Kamdhenu University, India
Yali Dang, Ningbo University, China
Copyright © 2022 Yang, Li, Li, Chen, Deng and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunsheng Li, bGljaHVuc2hlbmdAc2NzZnJpLmFjLmNu; Laihao Li, bGFpaGFvbGlAMTYzLmNvbQ==
 Daqiao Yang
Daqiao Yang Laihao Li
Laihao Li Chunsheng Li
Chunsheng Li Shengjun Chen
Shengjun Chen Jianchao Deng
Jianchao Deng Shaoling Yang
Shaoling Yang