- 1Department of Nutrition, University of California, Davis, Davis, CA, United States
- 2USANA Health Sciences, Inc., Salt Lake City, UT, United States
Dietary fiber, a nutrient derived mainly from whole grains, vegetables, fruits, and legumes, is known to confer a number of health benefits, yet most Americans consume less than half of the daily recommended amount. Convenience and affordability are key factors determining the ability of individuals to incorporate fiber-rich foods into their diet, and many Americans struggle to access, afford, and prepare foods rich in fiber. The objective of this clinical study was to test the changes in microbial community composition, human metabolomics, and general health markers of a convenient, easy to use prebiotic supplement in generally healthy young participants consuming a diet low in fiber. Twenty healthy adults participated in this randomized, placebo-controlled, double-blind, crossover study which was registered at clinicaltrials.gov as NCT03785860. During the study participants consumed 12 g of a prebiotic fiber supplement and 12 g of placebo daily as a powder mixed with water as part of their habitual diet in randomized order for 4 weeks, with a 4-week washout between treatment arms. Fecal microbial DNA was extracted and sequenced by shallow shotgun sequencing on an Illumina NovaSeq. Plasma metabolites were detected using liquid chromatography–mass spectrometry with untargeted analysis. The phylum Actinobacteria, genus Bifidobacterium, and several Bifidobacterium species (B. bifidum, B. adolescentis, B. breve, B. catenulatum, and B. longum) significantly increased after prebiotic supplementation when compared to the placebo. The abundance of genes associated with the utilization of the prebiotic fiber ingredients (sacA, xfp, xpk) and the production of acetate (poxB, ackA) significantly changed with prebiotic supplementation. Additionally, the abundance of genes associated with the prebiotic utilization (xfp, xpk), acetate production (ackA), and choline to betaine oxidation (gbsB) were significantly correlated with changes in the abundance of the genus Bifidobacterium in the prebiotic group. Plasma concentrations of the bacterially produced metabolite indolepropionate significantly increased. The results of this study demonstrate that an easy to consume, low dose (12 g) of a prebiotic powder taken daily increases the abundance of beneficial bifidobacteria and the production of health-promoting bacteria-derived metabolites in healthy individuals with a habitual low-fiber diet.
Clinical Trial Registration: www.clinicaltrials.gov/, identifier: NCT03785860
Introduction
Many Americans consume diets that are deficient in fiber for reasons that range in nature from financial constraints to personal taste preferences. Dietary fibers are polymers of monosaccharides which are resistant to human digestive enzymes, and have been shown to have beneficial effects on metabolism including improvements in glucose and insulin levels, and reduction of blood cholesterol concentrations (1–3). Dietary fiber intake is also associated with increases in the overall diversity of the gut microbiome (4) and in the abundance of beneficial microbial taxa, i.e., Bifidobacterium (5). The presence of bifidobacteria in the gut microbiome is associated with multiple health benefits including short-chain fatty acid (SCFA) production (4, 6) and improved gut barrier functionality (7), which in turn are linked with reduced inflammation (8), reduced concentrations of circulating lipopolysaccharide (9, 10), and improved gut immune function (11, 12). Despite the fact that the daily consumption of dietary fiber is associated with beneficial health effects, the average daily intake of dietary fiber in the U.S. is only ~16 g; less than half of the amount recommended by the USDA dietary guidelines (13). There are many practical obstacles that individuals face when considering lifestyle changes that are necessary to increase the intake of dietary fiber. Many Americans do not have access to fresh produce, do not have the time or the knowledge to prepare meals containing fiber-rich foods, or simply cannot afford to do so (14–17). Thus, practical solutions, like fiber supplements, are needed to increase fiber intake in individuals who are consuming low-fiber diets, or who are not able to implement the necessary lifestyle changes that increase fiber intake through whole foods.
A prebiotic is defined as a substrate that is selectively utilized by host microorganisms conferring a health benefit (18). Prebiotics, including inulin, fructooligosaccharides, galactooligosaccharides, and resistant starch, are known to increase the abundance of beneficial gut microbiota, particularly bifidobacteria (19–22). The increased abundance of saccharolytic microbes is in turn associated with enhanced production of microbially derived secondary metabolites that improve overall gut and metabolic health (23). The beneficial effects of SCFA on gut barrier integrity, gut immune function, and overall metabolism have been extensively documented (23–30). Other microbially produced metabolites with potential beneficial or deleterious effects on human health have been identified utilizing targeted and untargeted metabolomic approaches. For example, the metabolite indolepropionate (IPA) has been associated with beneficial health effects (31–33), whereas the metabolite trimethylamine-N-oxide (TMAO) has been linked with an increased risk for cardiovascular disease (34, 35).
In this study, we tested the effects of a prebiotic dietary fiber supplement formulation on the gut microbiome and human metabolome in 20 participants using a randomized, double-blind, placebo-controlled, crossover study design. We hypothesized that 4 weeks of daily supplementation with a prebiotic fiber blend will: increase the abundance of bifidobacteria and the abundance of gut microbial genes associated with bacterial utilization of the prebiotic substrate, alter fecal SCFA concentrations and gut microbial genes related to the production of SCFA, modify plasma metabolites, specifically, increase IPA and decrease TMAO, and alter cardiometabolic profiles.
Materials and Methods
Participants
Twenty healthy men and women aged 18–45 y, BMI of 23.0–32.0 kg/m2, with a habitual diet low in fiber (<15 g/day) were enrolled at the Ragle Human Nutrition Center, University of California (UC), Davis. Recruitment began in April of 2019 and the study completion date was December of 2019. Exclusion criteria included for screening were: smoking, having anemia and difficulty with blood draws, use of probiotic or prebiotic formulations within 6 weeks of the study start date, use of antibiotics within 6 months prior to study commencement, use of medication such as statins, blood pressure medications, and other prescription medications, pregnancy, use of hormonal birth control in the last 6 months or plans to change or start use of hormonal birth control during the study period, allergies to any placebo or prebiotic ingredients, presence of illness (flu/cold in the last 2 weeks), presence of documented chronic diseases, presence of intestinal diseases (irritable bowel syndrome, celiac disease, or any inflammatory bowel disease including Crohn's disease and/or ulcerative colitis), presence of any immunosuppression symptoms at any point during the study or study enrollment, consumption of >1 alcoholic drink/day or frequent binge drinking (>3 alcoholic drinks in one episode) of >1 day/month, plans to change or recent significant changes in lifestyle (e.g., diet, exercise routine, or major travel), recent weight fluctuations (>10% in the last 6 months), regular use of over-the-counter pain medications (>1/week), use of prescription lipid medications or other supplements known to alter lipoprotein metabolism such as isoflavones, recent medical procedure such as surgery within the last 6 months, and any changes in the above during the course of the study.
The sample size was determined based on a previous study which included 25 healthy participants supplemented with an inulin-type fructan-diet, in which a significant increase in bifidobacteria was observed (36). The study was approved by the Institutional Review Board of UC Davis. Written consent was provided by all participants prior to entry into the protocol. One participant was withdrawn due to difficulty with blood draws, three participants were withdrawn due to non-compliance with inclusion criteria, and 20 participants (10 male and 10 female) completed the study (Figure 1). This clinical trial was registered at www.clinicaltrials.gov/ as NCT03785860.
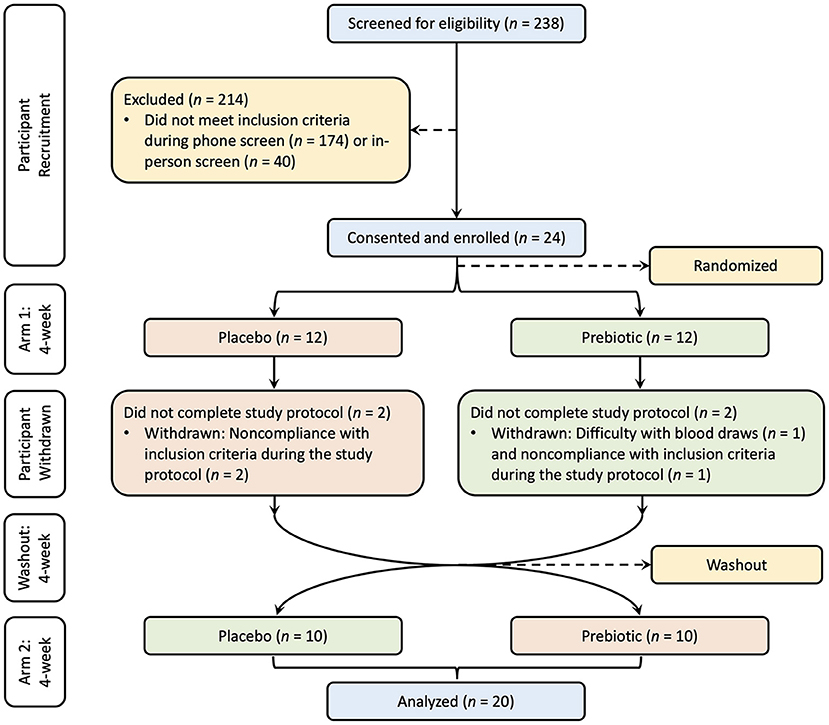
Figure 1. Study CONSORT diagram. Participants were recruited with screening, consent, and enrollment process. The treatment order was randomized into two groups, one group (red) supplemented with the placebo and the other group (green) supplemented with the prebiotic followed by a washout period and crossover to the other treatment for each group.
Study Design
The study was a randomized, double-blind, placebo-controlled, crossover design. All participants consumed a prebiotic fiber supplement and a matching placebo for a period of 4 weeks each, with a 4-week washout between intervention arms in random order. Twelve participants were randomly assigned to the prebiotic blend as their first intervention arm and 12 were assigned to the placebo blend as their first intervention arm. Four participants were withdrawn during the first intervention arm. After completion of the first intervention arm and the 4-week washout in which participants maintained their habitual diet without changing their regular lifestyle, the participants switched to the other intervention arm. Twenty subjects completed the study and were included in the analysis.
The prebiotic fiber blend used in this study was a dry mixture of fructooligosaccharides, resistant starch, sugarcane fiber, inulin, gum arabic, xanthan gum, and apple, raspberry, and blueberry fruit powders provided in single use packets of 12 g each. The placebo packets, also 12 g each, contained rice flour, xanthan gum and grape and plum fruit powders to match the prebiotic supplement in taste and appearance. Participants were provided the coded powder packet and instructed to mix with water for consumption. Participants were asked to record daily consumption of the powder packet and to bring back the empty packet for verification of compliance with study protocols.
Using 24-h diet records, participants recorded their diet for 3 days prior to each study visit at 2-week intervals for the duration of the study. The diet was patternized each week, meaning participants were asked to consume the same meals and foods for the 3 days prior to each test day without significantly changing their usual diet. This was done to stabilize the background diet during the days leading up to fecal sample collection, since it has been observed that dietary fluctuations over the preceding 3–4 days can significantly influence the gut microbiome (37, 38). Diet records were analyzed using the nutrition software, Food Processor SQL (version 11.7; ESHA).
Questionnaires and anthropometric measurement data were collected at each visit. The SF-12® Health questionnaire was collected to track the general health status of participants during the study intervention. Bowel movement type and frequency, which was rated by the participants throughout the study, was evaluated using the Bristol stool scale (39) and modified bowel movement questionnaire (40). Height was measured with a wall-mounted stadiometer (Ayrton Corp.) and body weight was measured with a calibrated electronic scale (Scale-Tronix; Welch Allyn). Blood pressure was measured with an automated sphygmomanometer (OxiMax; Welch Allyn) in a seated position. All measurements were performed in triplicate and the average was used for anthropometric measurement data.
Blood Sample Collection and Analysis of Cardiometabolic Profiles
Whole blood samples were collected after a 12-h overnight fast at the beginning and end of each intervention arm. Blood samples were collected in EDTA, SST, and PST tubes (Becton Dickinson) via venipuncture by a certified phlebotomist. The samples collected in EDTA tubes were immediately centrifuged (Sorvall-Legend RT) at 1,500 × g, 4°C for 10 min. Plasma samples were aliquoted immediately after centrifugation and stored at −80°C until further analyses. Blood samples collected in PST tubes were immediately centrifuged at 1,300 × g, 4°C for 10 min. The samples in SST tube were allowed to sit for 30 min for the blood to clot and then were centrifuged at 1,300 × g, 4°C for 10 min. Samples collected in PST and SST tubes were sent to UC Davis Medical Center Pathology Lab for analyses of glucose, lipid profiles, and insulin. Glucose was analyzed using the glucose oxidase method measuring absorbance at 652 nm after the peroxide catalyzed reaction (41). Insulin was analyzed with the Abbott Architect i1000 chemiluminescent microparticle immunoassay (42). A lipid panel [total cholesterol (TC), triacylglycerols (TG), HDL-cholesterol and calculated LDL-cholesterol] analysis was performed using a clinical analyzer (DXC 800; Beckman Coulter). TC, TG, and HDL-cholesterol were directly measured, and LDL-cholesterol was calculated using the Friedewald equation (43).
Stool Sample Collection
Fecal samples were self-collected by study participants. Prior to each stool sample collection day, participants were given stool collection kits consisting of a preservative-filled fecal collection tube (OMNIgeneGUT, OMR-200, DNA Genotek, Ottawa, ON, Canada), fecal collection sheet (Easy Sampler, EU version, GP Medical Devices, Holstebro, Denmark), and sanitizing kit. Trained study personnel educated participants on the stool collection protocol in person prior to the start of the study. Written instructions were also provided in the collection kits to ensure proper collection protocols were followed and to minimize sample contamination and deterioration. Participants were instructed to immediately transfer collected stool samples to a portable cooler packed with frozen icepacks and bring in the collected samples as soon as possible within 24 h of sample collection, including a transfer time of 4 h at most. Upon arrival to the laboratory, the samples were aliquoted into Eppendorf tubes and immediately stored at −80°C until analysis.
DNA Extraction, Sequencing, Preprocessing, and Assembly
Stool samples were processed at Diversigen (Houston, TX, USA) and shallow shotgun metagenomic sequencing was performed following DNA extraction. Briefly, DNA extraction from raw stool samples was performed with the DNeasy PowerSoil Pro Kit (Qiagen) automated for high throughput on the QiaCube HT (Qiagen) using Powerbead Pro (Qiagen) plates with 0.5 mm and 0.1 mm ceramic beads. Samples were quantified with Quant-iT Picogreen dsDNA Assay (Invitrogen). Libraries were prepared with a procedure adapted from the Nextera Library Prep kit (Illumina). Libraries were sequenced on an Illumina NovaSeq using single-end 1 × 100 cycles (Illumina). DNA sequences were filtered for low quality (Q-Score <30) and length (<50), and adapter sequences were trimmed using cutadapt (v1.15) (44). The sequences for each sample were assembled into contigs using SPAdes (v3.11.0) (45). The quality of assemblies for contigs >1,000 bases in length were assessed using QUAST (v4.5) (46).
Gene Annotation and Taxonomy Inference
Prokka (v1.12) (47) was used to annotate genes for each strain using the contigs > 1,000 bases as described above. Annotation files were parsed and combined to make gene content comparison tables. For taxonomy inference, trimmed and quality filtered sequences were aligned to every reference sequence in CoreBiome's Venti database at 97% identity using fully gapped alignment with BURST (v1.00) (48). Ties were broken by minimizing the overall number of unique gene, Operational Taxonomic Units (OTUs), hits. For taxonomy assignment, each input sequence was assigned the lowest common ancestor that was consistent across at least 80% of all reference sequences tied for best hits. The three most abundant taxa were reported for each strain.
Filtered taxonomy tables were generated. Briefly, samples with fewer than 10,000 sequences were discarded. OTUs accounting for less than one millionth of all strain-level markers and those with <0.01% of their unique genome regions covered (and <0.1% of the whole genome) at the species level were discarded. The number of counts for each OTU was normalized to the OTU's genome length for downstream analysis. The relative abundance of each sample was calculated by dividing OTU counts of each sample from the sum OTU counts of total sample using phyloseq (v1.36.0) (49).
Functional Annotation and Profiling
For functional annotation and profiling, Kyoto Encyclopedia of Genes and Genomes Orthology groups (KEGG KOs) were observed directly using alignment at 97% identity against a gene database derived from the strain database used above (Diversigen Venti database). The KEGG Orthology group table was filtered to the same subset of samples as the filtered taxonomic tables and used for downstream analysis.
Short-Chain Fatty Acid Analysis
Stool samples were processed at Microbiome Insights (University of British Columbia Vancouver, BC, Canada). Briefly, the SCFA extraction procedure is similar to that of Zhao et al. (50). Material was resuspended in MilliQ-grade H2O, and homogenized using MP Bio FastPrep, for 1 min at 4.0 m/s. Fecal suspensions were acidified with 5 M HCl to a final pH of 2.0. Acidified fecal suspensions were incubated and centrifuged at 10,000 rpm to separate the supernatant. Fecal supernatants were spiked with 2-ethylbutyric acid for a final concentration of 1 mM. Extracted SCFA supernatants were stored in 2-mL GC vials with glass inserts. Short-chain fatty acids were detected using gas chromatography (Thermo Trace 1310), coupled to a flame ionization detector (Thermo). The column used for SCFA detection was Thermo TG-WAXMS A GC Column (30 m, 0.32 mm, 0.25 μm) with a flow rate of 6.0 mL/min. The flame ionization detector was set to 240°C with controlled amounts of hydrogen: 35.0 mL/min, air: 350.0 mL/min, and makeup gas (Nitrogen): 40.0 mL/min. Short-chain fatty acid standards were acetic acid, propionic acid, isobutyric acid, butyric acid, isovaleric acid, valeric acid, and hexanoic acid from Sigma Aldrich.
Metabolomics
Untargeted metabolomic analysis was performed on the plasma samples at Metabolon (Morrisville, NC, USA) as previously described (51). All samples were maintained at −80°C until processing. Briefly, individual samples were subjected to methanol extraction then split into aliquots for analysis by ultra-high performance liquid chromatography-mass spectrometry (UHPLC-MS). The global biochemical profiling analysis comprised of four unique arms consisting of reverse phase chromatography positive ionization methods optimized for hydrophilic compounds (LC/MS Pos Polar) and hydrophobic compounds (LC/MS Pos Lipid), reverse phase chromatography with negative ionization conditions (LC/MS Neg), as well as a hydrophilic interaction liquid chromatography (HILIC) method coupled to positive and negative electrospray ionization modes (LC/MS Polar) (52). All the methods were alternated between full scan MS and data dependent MSn scans. The scan range varied slightly between methods but generally covered 70–1,000 m/z. Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries from Metabolon that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra and curated by visual inspection for quality control using software developed at Metabolon. Identification of known chemical entities was based on comparison to metabolomic library entries of purified standards (53).
Statistical Analysis
Differential expression analyses of OTU counts and gene counts were performed with the package DESeq2 (54) in R version 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria), which is based on a negative binomial model extended with Wald methods. To identify the OTU and gene differences between treatments over time, the count data was fitted into a generalized linear model with a design matrix: count ~ treatment * timepoint + subject. OTUs and genes that responded differently to the treatment relative to placebo at either timepoint were tested using the Wald test in the DESeq2 package (54). The primary outcome of the study was the change in the OTU counts of Bifidobacterium associated with the prebiotic supplement compared to the placebo. Therefore, we performed the differential expression analysis at the genus level. The secondary outcomes of the study were changes in other microbe abundances as well as gene counts, cardiometabolic profiles, SCFA concentrations, anthropometric measurements, and plasma metabolomic profiles. Specifically, we hypothesized that the gene counts of sacA, xfp, xpk, poxB, ackA, and buk genes would be altered on the prebiotic arm. Also, we hypothesized that the concentrations of IPA would increase, whereas TMAO would decrease on the prebiotic arm. The same data analysis pipeline was applied to gene count data. Linear mixed models with the previously mentioned design matrix were used to determine changes in cardiometabolic profiles, SCFA concentration, anthropometric measurements, and plasma metabolomic profiles with the R package limma (55). The Shapiro-Wilk normality test was performed prior to downstream analysis to check for variable normality. Log transformations were performed if variables did not fit a normal distribution. For any exploratory analyses, the Benjamini–Hochberg false discovery rate was calculated to adjust for multiple testing. The phyloseq R package was used to calculate the microbiome diversities (49). Kendall's correlation was performed to study novel relationships between changes in OTU counts of the genus Bifidobacterium and changes in gene counts of bacterial genes as well as metabolomic profiles. KEGG pathway based gene set enrichment analysis (GSEA) was performed and visualized using ClusterProfiler package in R (56) to test the effect of prebiotic treatment on metabolic pathways which include genes enriched by the treatment against the placebo. Partial least-squares discriminant analysis (PLS-DA) was performed to investigate the effect of the prebiotic treatment on overall metabolomic profiles using the pls and caret packages in R (57, 58).
Results
Anthropometrics, Cardiometabolic Profiles, and Diet Records
Participant baseline characteristics and their cardiometabolic profiles pre and post the prebiotic and placebo arms are summarized in Table 1. No significant changes were found in anthropometric measurements. Cardiometabolic profiles showed no significant differences between prebiotic and placebo groups. Also, there were no significant changes in nutrient intake at any time point (Table 2). No significant changes were observed for bowel movement type and frequency during the study period (data not shown).
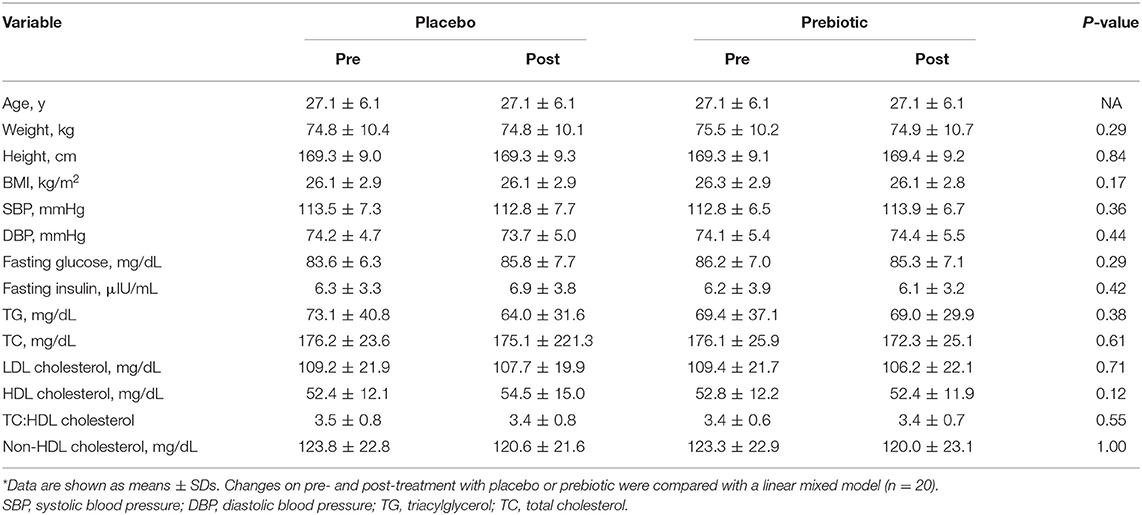
Table 1. Participant anthropometric and cardiometabolic characteristics pre- and post-treatment on placebo and prebiotic fiber*.
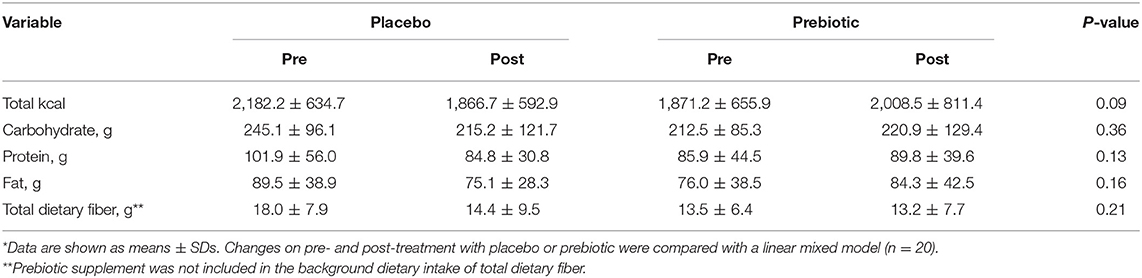
Table 2. The composition of the background diet of participants pre- and post-treatment with placebo and prebiotic fiber*.
Gut Microbial Composition
The overall gut microbial diversity was calculated using the Shannon diversity index to measure alpha diversity of microbiome species in samples and the Bray–Curtis dissimilarity index to evaluate beta diversity of species difference between the placebo and prebiotic intervention arms. There were no significant changes in microbial diversity after the prebiotic intervention compared to the placebo (Supplementary Figures 1A–C).
Further analyses on relative abundance of all gut microbiota in fecal samples were performed from phylum to species level. The relative abundance of the phylum Actinobacteria significantly increased after the prebiotic (P = 0.03) compared to the placebo arm (Figure 2A). Under the phylum Actinobacteria, the family Bifidobacteriaceae significantly increased after the prebiotic (P = 0.002) compared to the placebo arm (Figure 2B). Bifidobacterium counts were significantly increased after the prebiotic treatment (P = 0.005) compared to the placebo treatment (Figure 2C). In addition, several species belonging to the genus Bifidobacterium increased in the prebiotic arm compared to the placebo arm (Figures 2D–H), including Bifidobacterium bifidum (P = 0.01, unadjusted), Bifidobacterium adolescentis (P = 0.02, unadjusted), Bifidobacterium breve (P = 0.03, unadjusted), Bifidobacterium catenulatum (P = 0.03, unadjusted), and Bifidobacterium longum (P = 0.04, unadjusted).
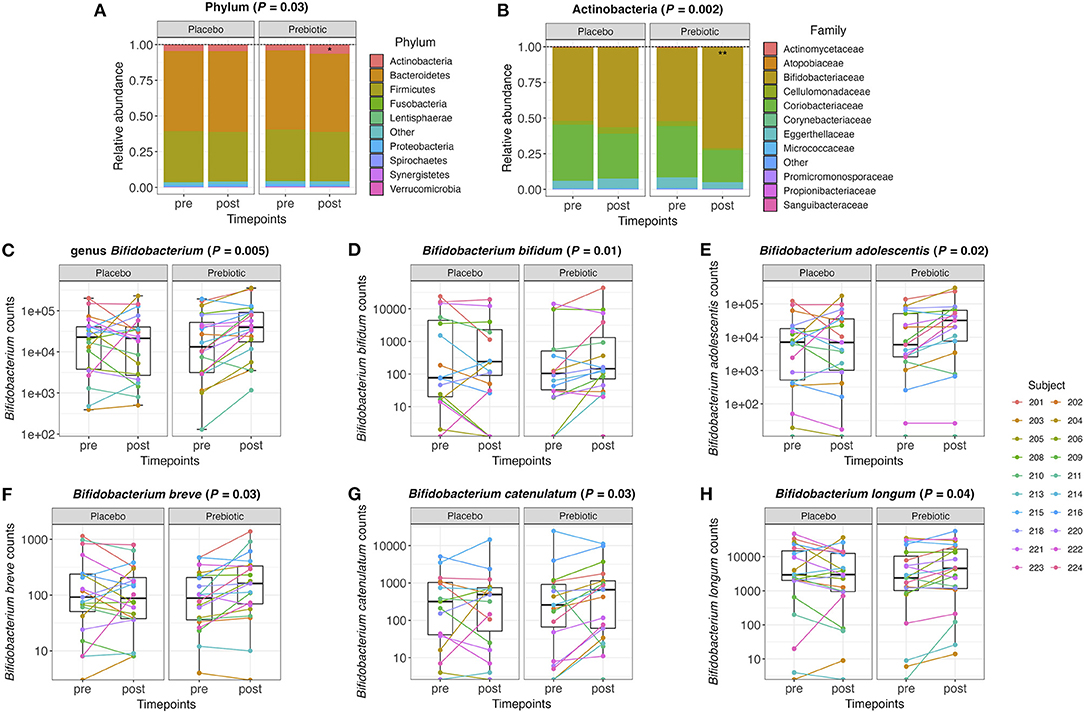
Figure 2. (A) Relative abundance of the gut microbiome at the phylum level and (B) the family level within the phylum Actinobacteria pre- and post-treatment with placebo or prebiotic. (C) Box plots of the genus Bifidobacterium counts pre- and post-treatment with placebo or prebiotic. (D–H) Box plots of Bifidobacterium species counts pre- and post-treatment with placebo or prebiotic (*P <0.05, **P < 0.01).
The overall composition of the gut microbiota that increased or decreased after the prebiotic supplement compared to the placebo is shown in the circular cladograms (Supplementary Figures 2A,B). The only phylum that significantly changed after the prebiotic supplement was Actinobacteria. At the genus level (Supplementary Figure 2A), 11 genera significantly (P ≤ 0.05, unadjusted) changed with the prebiotic treatment compared to the placebo. The genera Bifidobacterium, Anaerostipes, and Hungatella increased, while the eight other genera decreased. At the species level (Supplementary Figure 2B), 22 species significantly (P ≤ 0.05, unadjusted) changed with the prebiotic treatment compared to the placebo. However, none of these changes remained statistically significant after correction for multiple testing.
Gut Microbial Metagenome
The abundance of several genes increased or decreased after the prebiotic supplementation compared to the placebo (Figure 3A). Overall, 163 out of 2,718 genes significantly (P ≤ 0.05, unadjusted) increased or decreased after the prebiotic arm. Among the genes that changed, 49 decreased and 114 increased with the prebiotic treatment.
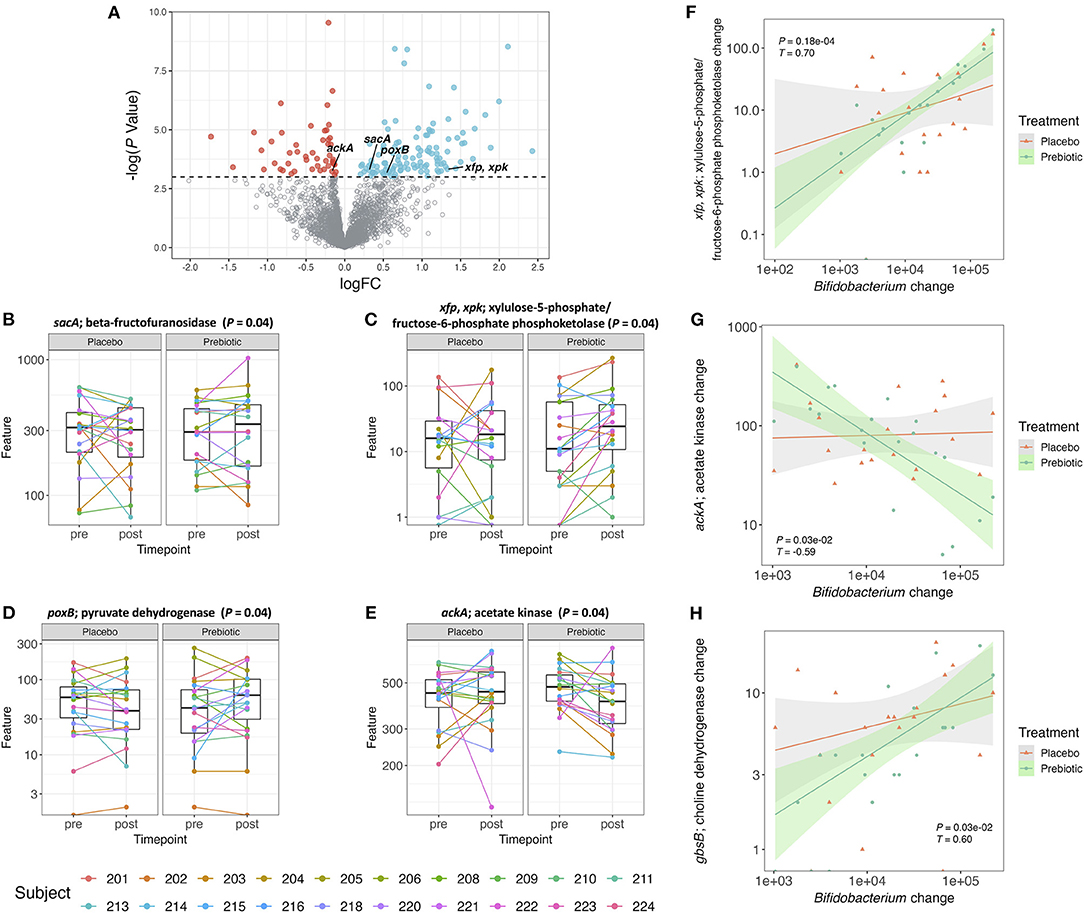
Figure 3. (A) Volcano plot of all detected genes. Genes with a logFC (post–pre treatment) > 0 and a –log(P-value) > 0.05 are colored blue and genes with a logFC <0 and a –log(P-value) > 0.05 are colored red. All the other genes are colored gray. (B–E) Box plot of gene counts pre- and post-treatment with placebo or prebiotic (P ≤ 0.05, unadjusted). (F–H) Correlation plot of changes (post–pre) in Bifidobacterium abundance against changes (post–pre) in gene counts for both placebo and prebiotic in 20 subjects (prebiotic: P ≤ 0.05, adjusted, Kendall T).
The pathways associated with the bacterial genes were analyzed by gene set enrichment analysis (GSEA) (Supplementary Figure 3A). There were 10 pathways that were significantly (P ≤ 0.05, unadjusted) enriched by the prebiotic supplementation, however, these differences did not remain statistically significant after correction for multiple testing. Starch and sucrose metabolism and the pentose phosphate pathway were two enriched pathways associated with the genes that we specifically hypothesized to increase in their counts after the prebiotic supplement compared to the placebo. The gene involved in starch and sucrose metabolism that showed significant increase after the prebiotic treatment was sacA coding for beta-fructofuranosidase (Figure 3B). This enzyme produces fructose and glucose from sucrose (59). The gene associated with fructose utilization was found to be significantly increased after the prebiotic supplement which was mostly fructose-based sugars (Figure 3C). The genes xfp, xpk, encoding for xylulose-5-phosphate/fructose-6-phosphate phosphoketolase, were shown to be associated with the pentose phosphate pathway converting D-xylulose-5-phosphate to D-glyceraldehyde-3-phosphate as well as D-fructose 6-phosphate to D-erythrose-4-phosphate (60). The phosphoketolase reaction on xylulose-5-phosphate and fructose-6-phosphate is one of the major reactions found in the bifidobacteria shunt (“bifid shunt”) (61, 62). Bifidobacteria utilize the hexose sugars to generate ATP producing SCFAs as byproducts (63).
As certain bifidobacterial species are known for their ability to produce SCFA such as acetate and butyrate, we specifically hypothesized genes associated with the production of these SCFAs would increase in gene counts after the prebiotic supplement compared to the placebo. Pyruvate to acetate conversion is mediated by the gene poxB encoding for pyruvate dehydrogenase (64). Acetyl phosphate to acetate conversion is mediated by the gene ackA encoding for acetate kinase/phosphotransacetylase (64). Butyryl phosphate to butyrate conversion is mediated by the gene buk encoding for butyrate kinase (65). In this study, the gene, poxB, related to SCFA production, specifically acetate formation (66), was increased after the prebiotic supplement (Figure 3D). The ackA gene counts were shown to decrease after the prebiotic supplement compared to the placebo (Figure 3E). The gene counts of other genes (pta, acs, and buk) in bacterial SCFA production pathways did not change after the prebiotic supplementation (data not shown).
A treatment-stratified correlation analysis between changes (post–pre) in OTU counts of Bifidobacterium and changes (post–pre) in gene counts (Supplementary Figure 3B) was performed to ascertain if any gene abundance changes coincided with the increase in bifidobacterial abundance. The changes in bifidobacterial OTU counts at the genus level were significantly correlated with the changes in gene counts from the prebiotic arm but not with the placebo arm. Among 30 genes, 29 genes were positively correlated, and 1 gene was negatively correlated with change in Bifidobacterium (Supplementary Figure 3B). In the correlation analysis of change in Bifidobacterium with gene count change, the gene sacA was not significant after multiple testing correction (Supplementary Figure 3B). However, the correlations between changes in Bifidobacterium and changes in the gene counts of xfp, xpk did remain statistically significant after multiple testing correction (Figure 3F). A negative correlation between changes in Bifidobacterium and changes in ackA gene counts also remained statistically significant after multiple testing correction (Figure 3G). Additionally, the gene counts for gbsB, a choline dehydrogenase, increased, although not significantly (P = 0.07) after the prebiotic treatment compared to the placebo. The changes in the abundance of the gbsB gene was positively correlated with changes in Bifidobacterium (P < 0.05) (Figure 3H).
Gut Microbe-Derived Metabolites
Plasma Metabolites
Forty-five of 889 detected metabolites significantly (P ≤ 0.05, unadjusted) increased or decreased with prebiotic supplementation (Figure 4A). The results of differential expression analysis of each metabolite (IPA and TMAO) are shown in Figures 4B,C. The prebiotic supplement formulation increased the total amount of IPA (P = 0.04) compared to the placebo. However, the supplement had no significant effect on the total amount of TMAO (P = 0.84) compared to the placebo.

Figure 4. (A) Volcano plot of all metabolites in human plasma samples. Metabolites with a logFC > 0 and a –log(P-value) > 0.05 are colored blue and metabolites with a logFC <0 and a –log(P-value) > 0.05 are colored red. All the other metabolites are colored gray. (B–J) Box plots of IPA, TMAO, choline, and acylcholines concentrations pre- and post-treatment with placebo or prebiotic (unadjusted P-value).
Significant decreases in choline and its acylated derivatives, the acylcholines were observed. Additionally, plasma concentrations of stearoylcholine, dihomo-linolenoyl-choline, linoleoylcholine, arachidonoylcholine, palmitoylcholine, oleoylcholine, and choline significantly decreased after the prebiotic intervention compared to the placebo (Figures 4D–J).
To determine whether any plasma metabolites were correlated with the abundance of Bifidobacterium a heatmap was generated to determine metabolites that are either positively or negatively associated with Bifidobacterium (Supplementary Figure 4A). Twenty-six metabolites were positively associated with Bifidobacterium and six metabolites were negatively associated with Bifidobacterium with adjusted P-values <0.05 after multiple testing correction. A total of six metabolites that significantly changed after the prebiotic treatment were also correlated with Bifidobacterium (Supplementary Figure 4A). Additionally, PLS-DA was performed to test if the prebiotic treatment had a discernible metabolomic signature. The prebiotic, while altering bifidobacterial abundance and increasing the concentration of IPA as well as decreasing the concentrations of several acylcholines, did not have an overall effect on the plasma metabolome, as shown by overlap of the time points in the scores plot (Supplementary Figure 4B).
Fecal Metabolites
SCFA concentrations in the stool samples were unchanged after the prebiotic treatment or placebo in healthy subjects (Supplementary Table 1).
Discussion
Low intake of dietary fiber in adults is associated with a number of deleterious health effects including an increased risk for metabolic disease and inflammation (67–72). The recommended intake for dietary fiber is 14 g/1,000 kcal per day (73). In this study, a prebiotic fiber supplement that has been shown to affect microbial community structure and metabolites in vitro (unpublished), was utilized to determine the effects in human participants. The objective was to determine if a low dose (12 g) prebiotic fiber supplement can have a measurable impact on gut microbial community composition and human plasma metabolites in participants consuming low-fiber diets (<15 g/day).
The prebiotic supplement did not change the overall composition of the microbiome, and a high degree of inter-individual variability in the microbiome was observed, as demonstrated extensively in previous studies (74–79). However, consuming the prebiotic supplement for 4 weeks led to significant changes in the specific bifidobacterial species, as hypothesized, likely due to the composition of fructan oligosaccharides, which are known to act as a selective substrate for bifidobacteria. The prebiotic increased the relative abundance of the genus Bifidobacterium as well as that of several bifidobacterial species and the phylum Actinobacteria. Bifidobacterium species that increased included B. bifidum, B. adolescentis, B. breve, B. catenulatum, and B. longum. These results align with the results of other studies, which demonstrate increases in bifidobacteria after the consumption of fructan-based oligosaccharides (19, 21, 22, 80). Healthy adults who consumed as little as 5 g of inulin for 21 days showed significant increases in Bifidobacterium species including B. adolescentis and B. bifidum (81). Treatment with 16 g/day of inulin-type fructans for 3 months in obese women resulted in a significant increase in the species B. adolescentis, B. pseudocatenulatum, and B. longum (21). Two other studies supplementing inulin-type fructan prebiotics (15–16 g/d) increased Bifidobacterium but did not significantly alter the overall microbial community due to large inter-individual variability (36, 82).
In the current study, in addition to assessing the increase in relative abundance of bifidobacteria, we also sought to determine the gene functional changes at the level of the metagenome that occur in response to supplementation, to elucidate the specific mechanisms by which an increase in bifidobacterial abundance in the gut is associated with functional changes that may confer benefits to the host. We hypothesized that counts of genes that code for proteins involved in the transport and metabolism of the oligosaccharides contained within the prebiotic supplement would increase on the prebiotic arm. Specifically, we hypothesized that sacA, coding for the enzyme beta-fructofuranosidase, and xfp, xpk, coding for xylulose-5-phosphate/fructose-6-phosphate phosphoketolase, would both increase in response to the supplement. The beta-fructofuranosidase enzyme converts sucrose into glucose and fructose (59), which is involved in the utilization of glucose and fructose for growth or energy sources by bifidobacteria (83–86). We found that changes in OTU counts of Bifidobacterium and changes in abundance of the sacA gene were not significantly correlated. This is likely because many gut bacteria other than bifidobacteria also express beta-fructofuranosidase to utilize fructose as a substrate (87, 88), and thus the increase in the gene counts of this enzyme could not be attributed solely to the increase in bifidobacteria (84, 89). The enzymes xylulose-5-phosphate/fructose-6-phosphate phosphoketolase each convert D-xylulose-5-phosphate to D-glyceraldehyde-3-phosphate and D-fructose 6-phosphate to D-erythrose-4-phosphate (60). The dual enzymatic reaction of these phosphoketolases are related to bifidobacterial utilization of hexose sugars in the “bifid shunt”, producing acetyl phosphate as well as SCFAs as byproducts while generating ATP (62, 63). Bifidobacterial species are specifically known for their ability to produce acetate from fructooligosaccharides (90, 91). Under anaerobic conditions, such as the human gut, acetyl phosphate is converted to acetate, which is facilitated by acetate kinase encoded by the ackA gene (92). Another enzyme, pyruvate dehydrogenase, which produces acetate from pyruvate is encoded by the poxB gene (93). In the current study the gene counts of ackA decreased and those of poxB increased after the prebiotic treatment. This could result in an overall increase in the acetate pool, providing substrate for acetyl-CoA (94). The acetyl-CoA may in turn enter the TCA cycle for complete oxidation of sugar molecules in cellular respiration (95). Acetyl-CoA may also be involved in the production of intracellular butyrate (65, 96). However, gene counts of the buk gene associated with butyrate production and other genes related to the metabolic pathways of SCFA production did not change.
Furthermore, acetate, butyrate, and other SCFA concentrations in the stool samples were unchanged. It is not clear why the increase in acetate production genes was not associated with changes in acetate measured directly in fecal samples. The increases in these gene counts may not have been sufficient to be directly reflected in the plasma and fecal concentrations of acetate (97, 98). Similar results were found in clinical studies supplementing prebiotic (inulin-type fructans) in healthy individuals (36, 82). The utilization of acetate and butyrate by the host as energy sources, particularly in the colon (99, 100), may be one of the contributing factors for the lack of measurable effects on acetate and butyrate concentrations. Alternatively, the prebiotic supplement at a dosage of 12 g/day may not be sufficient to produce detectable changes in fecal SCFA concentrations. Lastly, discrepancies in the water content of each fecal sample may have decreased the signal to noise ratio. A recent paper demonstrated that lyophilization of fecal samples reduces detection errors from water content and improves SCFA stability (101). Thus, implementing fecal lyophilization may be able to capture more accurate SCFA concentrations in future studies.
In this study we sought to further elucidate the relationships between gene count changes and the increase in bifidobacterial abundance by performing a treatment-stratified correlation analysis between the change in the OTU counts of the genus Bifidobacterium and changes in counts of genes. Two genes, xfp, xpk and ackA, were found to have a positive and a negative association with Bifidobacterium, respectively. These genes were also differentially expressed after the prebiotic supplement compared to the placebo. The genes xfp, xpk are related to a unique carbohydrate metabolism pathway utilizing the phosphoketolase enzyme in bifidobacteria, known as the bifid shunt (102). The positive correlation between changes in bifidobacterial OTU counts and changes in counts of xfp, xpk genes may be due to the unique utilization of the prebiotic supplement during the intervention. The ackA-pta pathway and poxB pathway are known to have an important role during the exponential growth phase and stationary phase, respectively, in E. coli for producing acetate (64). In E. coli, a small RNA, SdhX, regulates encoding enzymes of the TCA cycle and represses the expression of ackA while pta is not significantly affected (103). Several papers show significance of the acetate fermentation genes ackA-pta in bifidobacteria (103–105).
An interesting gene shown to be positively correlated with bifidobacteria was gbsB encoding for choline dehydrogenase. Not much is known about the role of choline dehydrogenase in bifidobacteria, and this may be the first report of a positive correlation between gbsB gene and Bifidobacterium. A paper showed decreased levels of choline in rats treated with certain strains of Bifidobacterium and Lactobacillus (106). The most well-studied links between choline and gut microbiota are associated with trimethylamine (TMA) production (107–109). The cut gene cluster, including choline-TMA lyase, encoded by the cutC gene is expressed by diverse taxa of Firmicutes and Proteobacteria converting choline into TMA (110). Prebiotic supplementation may modulate the gut microbiota that utilize choline and thus affect plasma TMA or TMAO concentrations, as has been shown in obese children (111) and in mice (112). However, the concentration of TMAO did not change after the prebiotic compared to the placebo, confirming a similar lack of effect in other studies (35, 38). On the other hand, choline metabolites were found to uniformly decrease after the prebiotic treatment including choline itself and several acylcholines including stearoylcholine, dihomo-linolenoyl-choline, linoleoylcholine, arachidonoylcholine, palmitoylcholine, and oleoylcholine. In colon cancer patients the consumption of rice bran results in a decrease in palmitoylcholine, linoleoylcholine, and oleoylcholine (113). In contrast, infants supplemented with rice bran showed increases in palmitoylcholine, oleoylcholine, linoleoylcholine, and stearoylcholine (114). Long-chain acylcholines have been found to have associations with a number of disease states (115), including elevated concentrations in endometrial cancer patients (116) and patients at high risk of pulmonary embolism (117), and lower concentrations in patients with myalgic encephalomyelitis/chronic fatigue syndrome (118) and chronic thromboembolic pulmonary hypertension (119). Acylcholines are known to have cholinergic signaling properties, with important implications for signaling through the nicotinic acetylcholine receptor (α7 nAChR), and effects on cytokine synthesis in macrophages and T cells (115, 120).
In addition, we hypothesized that the metabolite IPA would increase after the prebiotic intervention, and indeed, an increase in IPA was observed. Previous studies have shown that the concentration of IPA in the blood was positively correlated with dietary fiber intake (121). A clinical study in healthy individuals who consumed a high-fiber Mediterranean diet also showed an increase in IPA (38). IPA is produced by gut microbes from tryptophan and has been determined to play a crucial role in sustaining mucosal barrier function (23, 122, 123). Evidence on the beneficial health effects of IPA is growing (124–126), highlighting the therapeutic potential of probiotics and prebiotics that increase IPA production.
In this tightly controlled dietary intervention study, great care was taken to maximize the stability of the background diet as much as possible in order to maximize the ability to detect an effect of the prebiotic supplement. Participants were instructed to maintain their habitual diet throughout the course of the 12-week study protocol. We confirmed through 3-day diet record analysis that there were no significant changes in the background diet. This intervention in young, healthy participants who consume a low-fiber diet, but who were nonetheless metabolically healthy, did not affect cardiometabolic profiles. Fasting glucose, insulin, and lipid panels were not significantly altered after the prebiotic supplementation compared to the placebo. Several studies show clinically meaningful decreases in fasting or postprandial blood glucose and insulin concentrations in response to fiber supplementation (1, 127, 128). However, many of these studies were conducted in participants with elevated baseline values of these cardiometabolic parameters, such as individuals with metabolic syndrome (127) or type 2 diabetes (129), or in healthy participants (130, 131) but at much higher fiber doses (e.g., 38 vs. 12 g/d in this study) (1). Other studies show results that are in line with this study demonstrating no significant differences in cardiometabolic profiles between groups supplemented with fiber diet vs. control diet (132, 133).
The prebiotic supplement did not have gender-specific differences on the overall gut microbial composition and diversity as well as other secondary outcomes. The prebiotic supplement was well-tolerated by the participants without any abdominal discomfort reported during the study period. The strength of this study is in the study design, which was randomized, placebo-controlled, double-blind, crossover. A weakness of this study is the relatively small number of subjects and relatively short intervention period. However, a crossover design of the study increases the power to detect changes in response to the treatment with a small number of subjects and a shorter period. Additionally, several studies showed even a short-term intervention could result in a distinct change in the gut microbiome (36, 134–137). A previous study showed a significant increase in bifidobacteria after daily consumption of a mean intake of inulin-type fructans (15 g) in 26 healthy individuals in just 2 weeks (36). In this study, a 4-week intervention period was additionally chosen because it would allow for sampling at the same phase of the menstrual cycle for the female participants, eliminating the potential confounding effects of hormonal fluctuations on the gut microbiome.
Taking into account the fact that the prebiotic was hypothesized to be tested as a daily supplement to healthy individuals, the results that we found may be beneficial to the general population and suggest a simple way to increase the relative abundance of bifidobacteria, which is well-known for its beneficial health effects. On the other hand, the population of bifidobacteria not only decreases as we age but becomes absent in the gut of patients with certain diseases. Thus, maintaining the relative abundance of bifidobacteria throughout the life span may be crucial even in healthy individuals. However, other beneficial health effects of consuming prebiotics (e.g., reductions in triglycerides, LDL-cholesterol) were not shown in this study, which is likely due to the fact that the participants were already healthy at the start of the study.
In conclusion, supplementation with 12 g/day of a diverse, prebiotic dietary fiber blend resulted in measurable increases in beneficial Bifidobacterium species, changes in counts of genes associated with the utilization of the prebiotic as well as acetate production, and changes in plasma IPA, choline, and acylcholines in generally healthy individuals who consume a low-fiber diet. These results demonstrate a tangible benefit of a relatively low dose of a prebiotic fiber supplement in individuals who do not consume recommended amounts of dietary fiber, highlighting that even small, easy to incorporate changes in dietary intake can have beneficial effects on gut microbiome-mediated metabolism.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: ENA; PRJEB52881.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board of University of California, Davis. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JK, CW, MB, RB, RM, and AZ conceived and designed the study. JK, JZ, and JA conducted the clinical trial. JK and XT processed and analyzed the data. JK wrote the manuscript and created figures and tables. JK, XT, CW, MB, RB, RM, JZ, JA, and AZ contributed to editing and reviewing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the USANA Health Sciences Inc.
Conflict of Interest
JK and AZ have received research support from USANA Health Sciences, Inc. CW, MB, RB, and RM are employees of USANA Health Sciences, Inc. These interests have been reviewed and managed by the University of California, Davis in accordance with its Conflict-of-Interest policies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that this study received funding from USANA Health Sciences, Inc. The funder had the following involvement in the study: study conceptualization and manuscript review.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors highly appreciate undergraduate interns Neroli Huiyao Xie, Frances M Gross, and Emily Sabedra from Department of Nutrition, University of California, Davis for their contribution to preparing and running the clinical study as well as inputting data for dietary records.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.908534/full#supplementary-material
References
1. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. (2015) 22:971–82. doi: 10.1016/j.cmet.2015.10.001
2. Surampudi P, Enkhmaa B, Anuurad E, Berglund L. Lipid lowering with soluble dietary fiber. Curr Atheroscler Rep. (2016) 18:75. doi: 10.1007/s11883-016-0624-z
3. Jenkins DJA, Kendall CWC, Vuksan V, Vidgen E, Parker T, Faulkner D, et al. Soluble fiber intake at a dose approved by the US Food and Drug Administration for a claim of health benefits: serum lipid risk factors for cardiovascular disease assessed in a randomized controlled crossover trial. Am J Clin Nutr. (2002) 75:834–9. doi: 10.1093/ajcn/75.5.834
4. Sonnenburg JL, Bäckhed F. Diet–microbiota interactions as moderators of human metabolism. Nature. (2016) 535:56–64. doi: 10.1038/nature18846
5. So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JT, et al. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am J Clin Nutr. (2018) 107:965–83. doi: 10.1093/ajcn/nqy041
6. Markowiak-Kopeć P, Slizewska K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients. (2020) 12:1107. doi: 10.3390/nu12041107
7. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. (2016) 167:1339–53.e21. doi: 10.1016/j.cell.2016.10.043
8. Ruiz L, Delgado S, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and their molecular communication with the immune system. Front Microbiol. (2017) 8:2345. doi: 10.3389/fmicb.2017.02345
9. Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. (2007) 50:2374–83. doi: 10.1007/s00125-007-0791-0
10. Khokhlova EV, Smeianov VV, Efimov BA, Kafarskaia LI, Pavlova SI, Shkoporov AN. Anti-inflammatory properties of intestinal Bifidobacterium strains isolated from healthy infants. Microbiol Immunol. (2012) 56:27–39. doi: 10.1111/j.1348-0421.2011.00398.x
11. Lee WJ, Hase K. Gut microbiota–generated metabolites in animal health and disease. Nat Chem Biol. (2014) 10:416–24. doi: 10.1038/nchembio.1535
12. Tojo R, Suárez A, Clemente MG., de los Reyes-Gavilán CG, Margolles A, Gueimonde M, et al. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis.s World J Gastroenterol. (2014) 20:15163–76. doi: 10.3748/wjg.v20.i41.15163
13. Rosalie Bliss,. Online Nutrition Resources at Your Fingertips. (2017). Available from: https://www.usda.gov/media/blog/2015/03/31/online-nutrition-resources-your-fingertips (accessed December 01, 2021).
14. Clemens R, Kranz S, Mobley AR, Nicklas TA, Raimondi MP, Rodriguez JC, et al. Filling America's fiber intake gap: summary of a roundtable to probe realistic solutions with a focus on grain-based foods. J Nutr. (2012) 142:1390S−401S. doi: 10.3945/jn.112.160176
15. Deehan EC, Walter J. The fiber gap and the disappearing gut microbiome: implications for human nutrition. Trends Endocrinol Metab. (2016) 27:239–42. doi: 10.1016/j.tem.2016.03.001
16. Storey M, Anderson P. Income and race/ethnicity influence dietary fiber intake and vegetable consumption. Nutr Res. (2014) 34:844–50. doi: 10.1016/j.nutres.2014.08.016
17. Hsiao B sek, Sibeko L, Troy LM. A systematic review of mobile produce markets: facilitators and barriers to use, and associations with reported fruit and vegetable intake. J Acad Nutr Diet. (2019) 119:76–97.e1. doi: 10.1016/j.jand.2018.02.022
18. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. (2017) 14:491–502. doi: 10.1038/nrgastro.2017.75
19. Liu F, Li P, Chen M, Luo Y, Prabhakar M, Zheng H, et al. Fructooligosaccharide (FOS) and Galactooligosaccharide (GOS) increase bifidobacterium but reduce butyrate producing bacteria with adverse glycemic metabolism in healthy young population. Sci Rep. (2017) 7:11789. doi: 10.1038/s41598-017-10722-2
20. Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. (2012) 6:1535–43. doi: 10.1038/ismej.2012.4
21. Salazar N, Dewulf EM, Neyrinck AM, Bindels LB, Cani PD, Mahillon J, et al. Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin Nutr. (2015) 34:501–7. doi: 10.1016/j.clnu.2014.06.001
22. Cardelle-Cobas A, Corzo N, Olano A, Peláez C, Requena T, Ávila M. Galactooligosaccharides derived from lactose and lactulose: influence of structure on Lactobacillus, Streptococcus and Bifidobacterium growth. Int J Food Microbiol. (2011) 149:81–7. doi: 10.1016/j.ijfoodmicro.2011.05.026
23. Kang JW, Zivkovic AM. The potential utility of prebiotics to modulate Alzheimer's Disease: a review of the evidence. Microorganisms. (2021) 9:2310. doi: 10.3390/microorganisms9112310
24. Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, et al. GPR109A is a G-protein–coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. (2009) 69:2826–32. doi: 10.1158/0008-5472.CAN-08-4466
25. Docampo MD, Stein-Thoeringer CK, Lazrak A, Burgos da Silva MD, Cross J, van den Brink MRM. Expression of the butyrate/niacin receptor, GPR109a on T cells plays an important role in a mouse model of graft versus host disease. Blood. (2018) 132(Suppl. 1):61. doi: 10.1182/blood-2018-99-118783
26. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. (2014) 40:128–39. doi: 10.1016/j.immuni.2013.12.007
27. Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients. (2015) 7:2839–49. doi: 10.3390/nu7042839
28. Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol. (2013) 13:869–74. doi: 10.1016/j.coph.2013.08.006
29. Campos-Perez W, Martinez-Lopez E. Effects of short chain fatty acids on metabolic and inflammatory processes in human health. Biochim Biophys Acta BBA Mol Cell Biol Lipids. (2021) 1866:158900. doi: 10.1016/j.bbalip.2021.158900
30. Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. Chapter three - the role of short-chain fatty acids in health and disease. In: Alt FW, editor. Advances in Immunology. Academic Press (2014). p. 91–119.
31. Sakurai T, Odamaki T, Xiao J. Production of indole-3-lactic acid by Bifidobacterium strains isolated fromhuman infants. Microorganisms. (2019) 7:340. doi: 10.3390/microorganisms7090340
32. Qi Q, Li J, Yu B, Moon JY, Chai JC, Merino J, et al. Host and gut microbial tryptophan metabolism and type 2 diabetes: an integrative analysis of host genetics, diet, gut microbiome and circulating metabolites in cohort studies. Gut. (2021) 71:1095–105. doi: 10.1136/gutjnl-2021-324053
33. Zambrana LE, McKeen S, Ibrahim H, Zarei I, Borresen EC, Doumbia L, et al. Rice bran supplementation modulates growth, microbiota and metabolome in weaning infants: a clinical trial in Nicaragua and Mali. Sci Rep. (2019) 9:13919. doi: 10.1038/s41598-019-50344-4
34. Simó C, García-Cañas V. Dietary bioactive ingredients to modulate the gut microbiota-derived metabolite TMAO. New opportunities for functional food development. Food Funct. (2020) 11:6745–76. doi: 10.1039/D0FO01237H
35. Baugh ME, Steele CN, Angiletta CJ, Mitchell CM, Neilson AP, Davy BM, et al. Inulin supplementation does not reduce plasma trimethylamine N-oxide concentrations in individuals at risk for type 2 diabetes. Nutrients. (2018) 10:793. doi: 10.3390/nu10060793
36. Hiel S, Bindels LB, Pachikian BD, Kalala G, Broers V, Zamariola G, et al. Effects of a diet based on inulin-rich vegetables on gut health and nutritional behavior in healthy humans. Am J Clin Nutr. (2019) 109:1683–95. doi: 10.1093/ajcn/nqz001
37. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
38. Zhu C, Sawrey-Kubicek L, Beals E, Rhodes CH, Houts HE, Sacchi R, et al. Human gut microbiome composition and tryptophan metabolites were changed differently by fast food and Mediterranean diet in 4 days: a pilot study. Nutr Res. (2020) 77:62–72. doi: 10.1016/j.nutres.2020.03.005
39. Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. (1997) 32:920–4. doi: 10.3109/00365529709011203
40. Everhart JE, Go VL, Johannes RS, Fitzsimmons SC, Roth HP, White LR, et al. longitudinal survey of self-reported bowel habits in the United States. Dig Dis Sci. (1989) 34:1153–62. doi: 10.1007/BF01537261
41. Tao Z, Raffel RA, Souid AK, Goodisman J. Kinetic studies on enzyme-catalyzed reactions: oxidation of glucose, decomposition of hydrogen peroxide and their combination. Biophys J. (2009) 96:2977–88. doi: 10.1016/j.bpj.2008.11.071
42. Yao P, Liu Z, Tung S, Dong Z, Liu L. Fully automated quantification of insulin concentration using a microfluidic-based chemiluminescence immunoassay. J Lab Autom. (2016) 21:387–93. doi: 10.1177/2211068215578822
43. Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. (1972) 18:499–502. doi: 10.1093/clinchem/18.6.499
44. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. (2011) 17:10–2. doi: 10.14806/ej.17.1.200
45. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinforma. (2020) 70:e102. doi: 10.1002/cpbi.102
46. Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST quality assessment tool for genome assemblies. Bioinforma Oxf Engl. (2013) 29:1072–5. doi: 10.1093/bioinformatics/btt086
47. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinforma Oxf Engl. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
48. Al-Ghalith G, Knights D. BURST enables mathematically optimal short-read alignment for big data. Bioinformatics. (2020). doi: 10.1101/2020.09.08.287128 Available online at: https://github.com/knights-lab/burst
49. McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
50. Zhao G, Nyman M, Jönsson JÅ. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed Chromatogr. (2006) 20:674–82. doi: 10.1002/bmc.580
51. Hatano T, Saiki S, Okuzumi A, Mohney RP, Hattori N. Identification of novel biomarkers for Parkinson's disease by metabolomic technologies. J Neurol Neurosurg Psychiatry. (2016) 87:295–301. doi: 10.1136/jnnp-2014-309676
52. Evans AM, Bridgewater BR, Liu Q, Mitchell MW, Robinson RJ, Dai H. High resolution mass spectrometry improves data quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. Metabolomics. (2014) 04(02):2153–0769. doi: 10.4172/2153-0769.1000132
53. DeHaven CD, Evans AM Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminformatics. (2010) 2:9. doi: 10.1186/1758-2946-2-9
54. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
55. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
56. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. (2021) 2:100141. doi: 10.1016/j.xinn.2021.100141
57. Mevik BH, Wehrens R. The pls package: Principal component and partial least squares regression in R. J Stat Soft. (2007) 18:1–23. doi: 10.18637/jss.v018.i02
58. Kuhn M. The Caret Package. Available from: https://topepo.github.io/caret/ (accessed March 3, 2022).
59. Nadeem H, Rashid MH, Siddique MH, Azeem F, Muzammil S, Javed MR, et al. Microbial invertases: a review on kinetics, thermodynamics, physiochemical properties. Process Biochem. (2015) 50:1202–10. doi: 10.1016/j.procbio.2015.04.015
60. Roopashri AN, Varadaraj MC. Molecular characterization of native isolates of lactic acid bacteria, bifidobacteria and yeasts for beneficial attributes. Appl Microbiol Biotechnol. (2009) 83:1115–26. doi: 10.1007/s00253-009-1991-y
61. O'Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. (2016) 7:925. doi: 10.3389/fmicb.2016.00925
62. Meile L, Rohr LM, Geissmann TA, Herensperger M, Teuber M. Characterization of the d-Xylulose 5-Phosphate/d-Fructose 6-Phosphate Phosphoketolase Gene (xfp) from Bifidobacterium lactis. J Bacteriol. (2001) 183:2929–36. doi: 10.1128/JB.183.9.2929-2936.2001
63. Devika NT, Raman K. Deciphering the metabolic capabilities of Bifidobacteria using genome-scale metabolic models. Sci Rep. (2019) 9:18222. doi: 10.1038/s41598-019-54696-9
64. Dittrich CR, Bennett GN, San KY. Characterization of the acetate-producing pathways in Escherichia coli. Biotechnol Prog. (2005) 21:1062–7. doi: 10.1021/bp050073s
65. Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. 5:e00889–14. doi: 10.1128/mBio.00889-14
66. Pinhal S, Ropers D, Geiselmann J, de Jong H. Acetate metabolism and the inhibition of bacterial growth by acetate. J Bacteriol. 201:e00147–19. doi: 10.1128/JB.00147-19
67. Liu S, Buring JE, Sesso HD, Rimm EB, Willett WC, Manson JE, et al. prospective study of dietary fiber intake and risk of cardiovascular disease among women. J Am Coll Cardiol. (2002) 39:49–56. doi: 10.1016/S0735-1097(01)01695-3
68. Anderson JW, Deakins DA, Floore TL, Smith BM, Whitis SE. Dietary fiber and coronary heart disease. Crit Rev Food Sci Nutr. (1990) 29:95–147. doi: 10.1080/10408399009527518
69. Lattimer JM, Haub MD. Effects of dietary fiber and its components on metabolic health. Nutrients. (2010) 2:1266–89. doi: 10.3390/nu2121266
70. Grooms KN, Ommerborn MJ, Pham DQ, Djoussé L, Clark CR. Dietary fiber intake and cardiometabolic risks among US adults, NHANES 1999-2010. Am J Med. (2013) 126:1059–67.e4. doi: 10.1016/j.amjmed.2013.07.023
71. Zhang C, Liu S, Solomon CG, Hu FB. Dietary fiber intake, dietary glycemic load, and the risk for gestational diabetes mellitus. Diabetes Care. (2006) 29:2223–30. doi: 10.2337/dc06-0266
72. Wei B, Liu Y, Lin X, Fang Y, Cui J, Wan J. Dietary fiber intake and risk of metabolic syndrome: A meta-analysis of observational studies. Clin Nutr. (2018) 37(6, Part A):1935–42. doi: 10.1016/j.clnu.2017.10.019
73. U.S. Department of Agriculture and U.S. Department of Health and Human Services. Dietary Guidelines for Americans, 2020–2025. 9th ed. Washington, DC: U.S Department of Agriculture. Available online at: https://www.dietaryguidelines.gov/sites/default/files/2020-12/Dietary_Guidelines_for_Americans_2020-2025.pdf
74. Healey GR, Murphy R, Brough L, Butts CA, Coad J. Interindividual variability in gut microbiota and host response to dietary interventions. Nutr Rev. (2017) 75:1059–80. doi: 10.1093/nutrit/nux062
75. Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. (2014) 8:2218–30. doi: 10.1038/ismej.2014.63
76. Zhang C, Derrien M, Levenez F, Brazeilles R, Ballal SA, Kim J, et al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. (2016) 10:2235–45. doi: 10.1038/ismej.2016.13
77. Magne F, Abély M, Boyer F, Morville P, Pochart P, Suau A. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. (2006) 57:128–38. doi: 10.1111/j.1574-6941.2006.00097.x
78. Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE. (2010) 5:e15046. doi: 10.1371/journal.pone.0015046
79. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, et al. Personalized nutrition by prediction of glycemic responses. Cell. (2015) 163:1079–94. doi: 10.1016/j.cell.2015.11.001
80. Franco-Robles E, López MG. Implication of fructans in health: immunomodulatory and antioxidant mechanisms. Sci World J. (2015) 2015:289267. doi: 10.1155/2015/289267
81. Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. (2008) 101:541–50. doi: 10.1017/S0007114508019880
82. Healey G, Murphy R, Butts C, Brough L, Whelan K, Coad J. Habitual dietary fibre intake influences gut microbiota response to an inulin-type fructan prebiotic: a randomised, double-blind, placebo-controlled, cross-over, human intervention study. Br J Nutr. (2018) 119:176–89. doi: 10.1017/S0007114517003440
83. Gu J, Mao B, Cui S, Tang X, Liu Z, Zhao J, et al. Bifidobacteria exhibited stronger ability to utilize fructooligosaccharides, compared with other bacteria in the mouse intestine. J Sci Food Agri. (2022) 102:2413–23. doi: 10.1002/jsfa.11580
84. Lincoln L, More SS. Bacterial invertases: occurrence, production, biochemical characterization, and significance of transfructosylation. J Basic Microbiol. (2017) 57:803–13. doi: 10.1002/jobm.201700269
85. Ryan SM, Fitzgerald GF, van Sinderen D. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl Environ Microbiol. (2005) 71:3475–82. doi: 10.1128/AEM.71.7.3475-3482.2005
86. Ehrmann MA, Korakli M, Vogel RF. Identification of the gene for β-Fructofuranosidase of Bifidobacterium lactis DSM10140T and characterization of the enzyme expressed in Escherichia coli. Curr Microbiol. (2003) 46:0391–7. doi: 10.1007/s00284-002-3908-1
87. Goh YJ, Zhang C, Benson AK, Schlegel V, Lee JH, Hutkins RW. Identification of a putative operon involved in fructooligosaccharide utilization by Lactobacillus paracasei. Appl Environ Microbiol. (2006) 72:7518–30. doi: 10.1128/AEM.00877-06
88. Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, et al. Substrate-driven gene expression in Roseburia inulinivorans: Importance of inducible enzymes in the utilization of inulin and starch. Proc Natl Acad Sci USA. (2011) 108(Suppl. 1):4672–9. doi: 10.1073/pnas.1000091107
89. Reid SJ, Abratt VR. Sucrose utilisation in bacteria: genetic organisation and regulation. Appl Microbiol Biotechnol. (2005) 67:312–21. doi: 10.1007/s00253-004-1885-y
90. Palframan RJ, Gibson GR, Rastall RA, Vriers D. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr Issues Intest Microbiol. (2003) 4:71–5.
91. Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, et al. Fermentation of Fructooligosaccharides and Inulin by Bifidobacteria: a comparative study of pure and fecal cultures. Appl Environ Microbiol. (2005) 71:6150–8. doi: 10.1128/AEM.71.10.6150-6158.2005
92. Margolles A, Sánchez B. Selection of a Bifidobacterium animalis subsp. lactis Strain with a Decreased Ability To Produce Acetic Acid. Appl Environ Microbiol. (2012) 78:3338–42. doi: 10.1128/AEM.00129-12
93. Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. (2005) 69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005
94. Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. (2006) 72:3593–9. doi: 10.1128/AEM.72.5.3593-3599.2006
95. Bernal V, Castaño-Cerezo S, Cánovas M. Acetate metabolism regulation in Escherichia coli: carbon overflow, pathogenicity, and beyond. Appl Microbiol Biotechnol. (2016) 100:8985–9001. doi: 10.1007/s00253-016-7832-x
96. Morrison DJ, Mackay WG, Edwards CA, Preston T, Dodson B, Weaver LT. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br J Nutr. (2006) 96:570–7.
97. Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, et al. Dissecting the in vivo metabolic potential of two human gut acetogens *. J Biol Chem. (2010) 285:22082–90. doi: 10.1074/jbc.M110.117713
98. Nogal A, Louca P, Zhang X, Wells PM, Steves CJ, Spector TD, et al. Circulating levels of the short-chain fatty acid acetate mediate the effect of the gut microbiome on visceral fat. Front Microbiol. (2021) 12:711359. doi: 10.3389/fmicb.2021.711359
99. Meyer D, Stasse-Wolthuis M. The bifidogenic effect of inulin and oligofructose and its consequences for gut health. Eur J Clin Nutr. (2009) 63:1277–89. doi: 10.1038/ejcn.2009.64
100. Belenguer A, Duncan SH, Holtrop G, Flint HJ, Lobley GE. Quantitative analysis of microbial metabolism in the human large intestine. Curr Nutr Food Sci. (2008) 4:109–26. doi: 10.2174/157340108784245957
101. Hsu YL, Chen CC, Lin YT, Wu WK, Chang LC, Lai CH, et al. Evaluation and optimization of sample handling methods for quantification of short-chain fatty acids in human fecal samples by GC–MS. J Proteome Res. (2019) 18:1948–57. doi: 10.1021/acs.jproteome.8b00536
102. Gupta RS, Nanda A, Khadka B. Novel molecular, structural and evolutionary characteristics of the phosphoketolases from bifidobacteria and Coriobacteriales. PLoS ONE. (2017) 12:e0172176. doi: 10.1371/journal.pone.0172176
103. Mets FD, Melderen LV, Gottesman S. Regulation of acetate metabolism and coordination with the TCA cycle via a processed small RNA. Proc Natl Acad Sci U. S. A. (2019) 116:1043–52. doi: 10.1073/pnas.1815288116
104. Arzamasov AA, van Sinderen D, Rodionov DA. Comparative genomics reveals the regulatory complexity of bifidobacterial arabinose and arabino-oligosaccharide utilization. Front Microbiol. (2018) 9:776. doi: 10.3389/fmicb.2018.00776
105. Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in bifidobacteria. Genes Nutr. (2011) 6:285–306. doi: 10.1007/s12263-010-0206-6
106. Wang M, Chen Y, Wang Y, Li Y, Zhang X, Zheng H, et al. Beneficial changes of gut microbiota and metabolism in weaned rats with Lactobacillus acidophilus NCFM and Bifidobacterium lactis Bi-07 supplementation. J Funct Foods. (2018) 48:252–65. doi: 10.1016/j.jff.2018.07.008
107. Wortmann SB, Mayr JA. Choline-related-inherited metabolic diseases—A mini review. J Inherit Metab Dis. (2019) 42:237–42. doi: 10.1002/jimd.12011
108. Zeisel SH, Warrier M. Trimethylamine N-oxide, the microbiome, and heart and kidney disease. Annu Rev Nutr. (2017) 37:157–81. doi: 10.1146/annurev-nutr-071816-064732
109. Day-Walsh P, Shehata E, Saha S, Savva GM, Nemeckova B, Speranza J, et al. The use of an in-vitro batch fermentation (human colon) model for investigating mechanisms of TMA production from choline, l-carnitine and related precursors by the human gut microbiota. Eur J Nutr. (2021) 60:3987–99. doi: 10.1007/s00394-021-02572-6
110. Rath S, Heidrich B, Pieper DH, Vital M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome. (2017) 5:54. doi: 10.1186/s40168-017-0271-9
111. Zhang C, Yin A, Li H, Wang R, Wu G, Shen J, et al. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine. (2015) 2:968–84. doi: 10.1016/j.ebiom.2015.07.007
112. Li Q, Wu T, Liu R, Zhang M, Wang R. Soluble dietary fiber reduces trimethylamine metabolism via gut microbiota and co-regulates host AMPK pathways. Mol Nutr Food Res. (2017) 61:1700473. doi: 10.1002/mnfr.201700473
113. Zarei I, C Oppel R, C Borresen E, J Brown R, P Ryan E. Modulation of plasma and urine metabolome in colorectal cancer survivors consuming rice bran. Integr Food Nutr Metab. (2019) 6:1–14. doi: 10.15761/IFNM.1000252
114. Zambrana LE, Weber AM, Borresen EC, Zarei I, Perez J, Perez C, et al. Daily rice bran consumption for 6 months influences serum glucagon-like peptide 2 and metabolite profiles without differences in trace elements and heavy metals in weaning nicaraguan infants at 12 months of age. Curr Dev Nutr. (2021) 5:nzab101. doi: 10.1093/cdn/nzab101
115. Kinchen JM, Mohney RP, Pappan KL. Long-chain acylcholines link butyrylcholinesterase to regulation of non-neuronal cholinergic signaling. J Proteome Res. (2022) 21:599–611. doi: 10.1021/acs.jproteome.1c00538
116. Audet-Delage Y, Villeneuve L, Grégoire J, Plante M, Guillemette C. Identification of metabolomic biomarkers for endometrial cancer and its recurrence after surgery in postmenopausal women. Front Endocrinol. (2018) 9:87. doi: 10.3389/fendo.2018.00087
117. Zeleznik OA, Poole EM, Lindstrom S, Kraft P, Van Hylckama Vlieg A, Lasky-Su JA, et al. Metabolomic analysis of 92 pulmonary embolism patients from a nested case–control study identifies metabolites associated with adverse clinical outcomes. J Thromb Haemost. (2018) 16:500–7. doi: 10.1111/jth.13937
118. Germain A, Barupal DK, Levine SM, Hanson MR. Comprehensive circulatory metabolomics in ME/CFS reveals disrupted metabolism of acyl lipids and steroids. Metabolites. (2020) 10:34. doi: 10.3390/metabo10010034
119. Heresi GA, Mey JT, Bartholomew JR, Haddadin IS, Tonelli AR, Dweik RA, et al. Plasma metabolomic profile in chronic thromboembolic pulmonary hypertension. Pulm Circ. (2020) 10:2045894019890553. doi: 10.1177/2045894019890553
120. Akimov MG, Kudryavtsev DS, Kryukova EV, Fomina-Ageeva EV, Zakharov SS, Gretskaya NM, et al. Arachidonoylcholine and other unsaturated long-chain acylcholines are endogenous modulators of the acetylcholine signaling system. Biomolecules. (2020) 10:283. doi: 10.3390/biom10020283
121. Tuomainen M, Lindström J, Lehtonen M, Auriola S, Pihlajamäki J, Peltonen M, et al. Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals. Nutr Diabetes. (2018) 8:1–5. doi: 10.1038/s41387-018-0046-9
122. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. (2013) 39:372–85. doi: 10.1016/j.immuni.2013.08.003
123. Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, et al. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS ONE. (2013) 8:e80604. doi: 10.1371/journal.pone.0080604
124. Hwang IK, Yoo KY Li H, Park OK, Lee CH, Choi JH, et al. Indole-3-propionic acid attenuates neuronal damage and oxidative stress in the ischemic hippocampus. J Neurosci Res. (2009) 87:2126–37. doi: 10.1002/jnr.22030
125. Zhao ZH, Xin FZ, Xue Y, Hu Z, Han Y, Ma F, et al. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp Mol Med. (2019) 51:1–14. doi: 10.1038/s12276-019-0304-5
126. Negatu DA, Gengenbacher M, Dartois V, Dick T. Indole propionic acid, an unusual antibiotic produced by the gut microbiota, with anti-inflammatory and antioxidant properties. Front Microbiol. (2020) 11:2654. doi: 10.3389/fmicb.2020.575586
127. Cicero AFG, Derosa G, Bove M, Imola F, Borghi C, Gaddi AV. Psyllium improves dyslipidaemia, hyperglycaemia and hypertension, while guar gum reduces body weight more rapidly in patients affected by metabolic syndrome following an AHA Step 2 diet. Mediterr J Nutr Metab. (2010) 3:47–54. doi: 10.1007/s12349-009-0056-1
128. Gibb RD, McRorie JW Jr, Russell DA, Hasselblad V, D'Alessio DA. Psyllium fiber improves glycemic control proportional to loss of glycemic control: a meta-analysis of data in euglycemic subjects, patients at risk of type 2 diabetes mellitus, and patients being treated for type 2 diabetes mellitus. Am J Clin Nutr. (2015) 102:1604–14. doi: 10.3945/ajcn.115.106989
129. Chandalia M, Garg A, Lutjohann D, von Bergmann K, Grundy SM, Brinkley LJ. Beneficial effects of high dietary fiber intake in patients with type 2 diabetes mellitus. N Engl J Med. (2000) 342:1392–8. doi: 10.1056/NEJM200005113421903
130. Aller R, de Luis DA, Izaola O, La Calle F., del Olmo L, Fernandez L, et al. Effect of soluble fiber intake in lipid and glucose leves in healthy subjects: a randomized clinical trial. Diabetes Res Clin Pract. (2004) 65:7–11. doi: 10.1016/j.diabres.2003.11.005
131. Kristensen M, Jensen MG, Aarestrup J, Petersen KE, Søndergaard L, Mikkelsen MS, et al. Flaxseed dietary fibers lower cholesterol and increase fecal fat excretion, but magnitude of effect depend on food type. Nutr Metab. (2012) 9:8. doi: 10.1186/1743-7075-9-8
132. Crimarco A, Springfield S, Petlura C, Streaty T, Cunanan K, Lee J, et al. A randomized crossover trial on the effect of plant-based compared with animal-based meat on trimethylamine-N-oxide and cardiovascular disease risk factors in generally healthy adults: Study With Appetizing Plantfood—Meat Eating Alternative Trial (SWAP-MEAT). Am J Clin Nutr. (2020) 112:1188–99. doi: 10.1093/ajcn/nqaa203
133. Brown L, Rosner B, Willett WW, Sacks FM. Cholesterol-lowering effects of dietary fiber: a meta-analysis. Am J Clin Nutr. (1999) 69:30–42. doi: 10.1093/ajcn/69.1.30
134. She J, Wong CC, Yu J. Targeted prebiotics alter the obese gut microbiome in humans. Signal Transduct Target Ther. (2021) 6:1–2. doi: 10.1038/s41392-021-00758-2
135. Delannoy-Bruno O, Desai C, Raman AS, Chen RY, Hibberd MC, Cheng J, et al. Evaluating microbiome-directed fibre snacks in gnotobiotic mice and humans. Nature. (2021) 595:91–5. doi: 10.1038/s41586-021-03671-4
136. Costabile A, Deaville ER, Morales AM, Gibson GR. Prebiotic potential of a maize-based soluble fibre and impact of dose on the human gut microbiota. PLoS ONE. (2016) 11:e0144457. doi: 10.1371/journal.pone.0144457
137. Chung WSF, Walker AW, Bosscher D, Garcia-Campayo V, Wagner J, Parkhill J, et al. Relative abundance of the Prevotella genus within the human gut microbiota of elderly volunteers determines the inter-individual responses to dietary supplementation with wheat bran arabinoxylan-oligosaccharides. BMC Microbiol. (2020) 20:283. doi: 10.1186/s12866-020-01968-4
Keywords: prebiotic, gut microbiome, bifidobacteria, indolepropionate, cholines
Citation: Kang JW, Tang X, Walton CJ, Brown MJ, Brewer RA, Maddela RL, Zheng JJ, Agus JK and Zivkovic AM (2022) Multi-Omic Analyses Reveal Bifidogenic Effect and Metabolomic Shifts in Healthy Human Cohort Supplemented With a Prebiotic Dietary Fiber Blend. Front. Nutr. 9:908534. doi: 10.3389/fnut.2022.908534
Received: 30 March 2022; Accepted: 20 May 2022;
Published: 17 June 2022.
Edited by:
Balamurugan Ramadass, All India Institute of Medical Sciences Bhubaneswar, IndiaReviewed by:
Pugazhendhi Srinivasan, University of Kansas Medical Center, United StatesNanda Kumar Navalpur Shanmugam, Massachusetts General Hospital and Harvard Medical School, United States
Copyright © 2022 Kang, Tang, Walton, Brown, Brewer, Maddela, Zheng, Agus and Zivkovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela M. Zivkovic, YW16aXZrb3ZpY0B1Y2RhdmlzLmVkdQ==
 Jea Woo Kang
Jea Woo Kang Xinyu Tang1
Xinyu Tang1 Charles J. Walton
Charles J. Walton Rachel A. Brewer
Rachel A. Brewer Rolando L. Maddela
Rolando L. Maddela Jack Jingyuan Zheng
Jack Jingyuan Zheng Angela M. Zivkovic
Angela M. Zivkovic