
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr., 20 May 2022
Sec. Food Chemistry
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.875765
This article is part of the Research TopicThe Nutritional Evidence and Research on TeaView all 10 articles
Coronavirus disease 2019 (COVID-19) is a contagious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that caused millions of deaths and lacks treatment. Although several studies have focused on the major component of green tea, epigallocatechin 3-gallate (EGCG), which is efficient in preventing COVID-19, systemic analyses of the anti-COVID-19 potential of green tea remain insufficient. Here, we co-analyzed the target genes of tea ingredients and COVID-19 signature genes and found that epigallocatechin 3-acetalbehyde was capable of reversing the major molecular processes of COVID-19 (MAPK and NF-κB activation). These findings were further supported by Western blotting (WB), immunofluorescence, and quantitative polymerase chain reaction (qPCR) in LPS-stimulated macrophages. Moreover, using molecular docking analysis, we identified three tea ingredients ((-)-catechin gallate, D-(+)-cellobiose, and EGCG) that may interact with the vital SARS-CoV-2 protein, 5R84, compared with the qualified 5R84 ligand WGS. Thus, our results indicated that tea ingredients have the potential to treat COVID-19 by suppressing the COVID-19 signature genes and interacting with the vital SARS-CoV-2 protein.
Coronavirus disease 2019 (COVID-19) is a disease with main manifestations involving the lungs and is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1). SARS-CoV-2 is rapidly spreading around the world, and the number of confirmed cases and infection-related deaths are increasing every day (2). The severity of COVID-19 is associated with increased inflammatory and chemokine factors; these factors also predict COVID-19 mortality (3). Although the pathogenesis of COVID-19 is not fully understood, the virus and host immune system play key roles in its development (4).
From Delta to Omicron, the new coronavirus is constantly mutating, the global epidemic is at a high level, and the number of infections continues to increase (5). While COVID-19 vaccines can greatly prevent the spread of the virus, they cannot treat patients infected with the virus (6). To treat patients with new coronavirus pneumonia, scientists have made considerable efforts in drug research and development; however, to date, there are still very few drugs that can treat COVID-19 (7). Although some neutralizing antibodies and small molecule inhibitors are being developed, there is uncertainty about their safety and efficacy (8). Therefore, we urgently need to explore new strategies to treat COVID-19.
Tea is popular all over the world as a food drink; in fact, tea has been used as an herbal medicine to prevent and treat various diseases (9). Tea and its characteristic polyphenols—catechins—have been shown to be active in preventing obesity, diabetes, cardiovascular disease, cancer, and other diseases (10–12). Tea ingredients have also been shown to have anti-viral activity as well as protective activity against diseases caused by oxidative stress and inflammation; many of these ingredients may help alleviate and treat COVID-19 (13, 14). Although several studies have focused on epigallocatechin gallate (EGCG), the major component of green tea, which has been shown to be effective in preventing COVID-19 (15), we focused on systematic research of the therapeutic potential of tea components for COVID-19, including inhibition of COVID-19 signature gene transcription and direct interactions with specific COVID-19 proteins. Systematic research about tea and COVID-19 currently remains insufficient. Systematic analyses of the anti-COVID-19 potential of green tea and other teas remains insufficient. In this study, we mainly used bioinformatics and computational network-based pharmacology to explore and determine the efficacy and possible therapeutic mechanisms of tea for the treatment of COVID-19 to reveal the potential uses of tea in the treatment of COVID-19. Using a network pharmacology strategy, we report the pharmacological targets and molecular pathways of tea ingredients. Therefore, in this bioinformatics report, we aimed to reveal the component-target-pathway network and pharmacological mechanisms of tea ingredients in the prevention and treatment of COVID-19.
Using effective tools such as Traditional Chinese Medicine Systems Pharmacology (TCMSP), Swiss Target Prediction, and SuperPred, the target genes of tea were screened from existing databases (16, 17). Other genes related to the occurrence of COVID-19 were obtained using the DisGeNET and GeneCards databases (18). In addition, these putative tea and COVID-19 genes were mapped using the UniProt tool prior to correction. After functional enrichment analysis using FunRich software, all anti-COVID-19 targets of tea were screened and identified.
After obtaining the targets of tea and COVID-19, the STRING database was used to further determine and construct a functional protein association network according to a specific algorithm (19, 20). In addition, based on the merged targets of tea and COVID-19, a protein-protein interaction (PPI) network was constructed using Cytoscape software (21, 22). Therefore, the key targets of tea in the treatment of COVID-19 were revealed, visualized, and determined with the topology parameters of the network analyzer tool (23, 24).
R language packages, such as ClusterProfiler, org.Hs.eg.Db, ReactomePA, and GOplot (3.6.1), have been used for enrichment analysis and visualization of the biological processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of intersection targets (25). In addition, gene annotation information from org.Hs.eg.Db (26, 27), a p-value cutoff = 0.05, and a q-value cutoff = 0.05 were used for enrichment before plotting the corresponding bubble chart, histogram, and Circos circle chart.
To screen and identify key targets for tea-based molecular docking assays, a chemical-protein binding approach was used (28, 29). After searching for a specific protein through the PDB database, the 5R84 protein was selected for docking with the tea compound. The three-dimensional structure of tea was drawn using ChemBio3D Draw in Chem Bio Office 2010 software before docking the molecular structure with AutoDock Vina software (30). The plausibility of the docking parameter settings was assessed by the root-mean-square deviation (RMSD) of the ligand molecules. An RMSD ≤ 4 Å is the threshold for ligand molecular conformation.
Murine macrophage RAW 264.7 cells were acquired from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were grown at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% penicillin streptomycin (Gibco, USA) in humidified incubators (Thermo, USA). Lipopolysaccharide (LPS, Escherichia coli 055: B5) and EGCG were purchased from Sigma Chemical Co. (St. Louis, USA). RAW 264.7 cells were treated with LPS, LPS+EGCG, or EGCG (for the concentrations of LPS and EGCG see the figure legend) for 24 h. For viability testing, the cells were starved for 24 h without serum before challenge and seeded at a density of 1 × 105 cells/mL in 96-well plates with four replications, and cell viability was analyzed with a CCK-8 cell counting kit (Vazyme, China).
The total RNA was isolated from cells using an RNA extraction kit (Vazyme, China). First-strand complementary DNA (cDNA) was synthesized using an iScript cDNA Synthesis Kit (Vazyme, China). Quantitative PCR was performed with SYBR green PCR Master Mix (Vazyme, China) using a ViiA 7 Real-Time PCR System (Applied Biosystems, CA). The primers are detailed in Table 1. The following cycle parameters were used: 55°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 30 s and 60°C for 30 s. The relative expressions of the target genes against that of the reference gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were calculated using the 2−ΔΔCT method. Cell samples were evaluated in triplicate, and every experiment was performed at least three times. The transcription levels of inducible nitric oxide synthase (iNOS), tumor necrosis factor alpha (TNF-α), interleukin (IL)-1β, IL-6, Arg-1, and GAPDH were determined.
Total cellular proteins were extracted using radioimmunoprecipitation assay buffer containing 1% sodium dodecyl sulfate (SDS); 40 mg of total lysate was separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a polyvinylidene fluoride membrane, blocked with 5% bovine serum albumin in tris-buffered saline for 90 min, and then incubated with the appropriate primary antibody overnight at 4°C. Membranes were incubated with secondary antibody for 90 min at room temperature after washing and then visualized using ECL Plus Western Blot Detection Reagent (Millipore, USA). The protein expression levels of extracellular signal-regulated kinase (ERK), p-ERK, c-Jun amino-terminal kinase (JNK) and p-JNK, and p38 and p-p38 were determined by Western blotting (WB). GAPDH was used as an internal control.
According to the manufacturer's instructions, the cell supernatant concentrations of IL-6, TNF-α, and IL-1β were determined using ELISA kits (ExCell Bio, China).
The expression of phospho-p65 was detected by immunofluorescence assays using a fluorescence microscope. RAW 264.7 cells were cultured directly on glass coverslips in 6-well plates for 24 h. After stimulation with LPS in the presence or absence of EGCG, the cells were fixed with 4% paraformaldehyde in PBS. The membrane was permeabilized by treating the cells for 5 min with 0.1% Triton X-100 in PBS. After a brief washing in PBS, slides were blocked with 5% bovine serum albumin for 1 h and then incubated with rabbit polyclonal anti-human phopho-p65 antibody (dilution, 1:100) overnight at 4°C at room temperature. The next day, the specimens were rinsed with PBS three times. After washing, they were incubated with the secondary antibodies (Alexa Fluor® 594, Thermo Fisher Scientific, CA, USA) for 30 min and counterstained for nuclei with DAPI (Beijing Solarbio Science & Technology, Beijing, China) for 10 min. After a brief washing in PBS, slides were sealed using ProLong® Gold antifade reagent (Molecular Probes® by Life Technologies™, CA, USA). Fluorescence micrographs were acquired with a fluorescence microscope (Nikon ECLIPSE Ti-U, Nikon Co., Japan).
Normally distributed data were analyzed using Student's t-test (for two-group comparisons) or analysis of variance (for multiple-group comparisons). For non-normally distributed values (as determined by the Kolmogorov–Smirnov test), the Mann–Whitney's rank-sum test was used. All statistical tests were two-sided, and P < 0.05 was considered statistically significant. Data are presented as the mean ± standard error of the mean (SEM) and presented using GraphPad Prism 5 software (LaJolla, CA).
We first downloaded the ingredients and target genes of tea from the Traditional Chinese Medicine Integrated Database (TCMID) database (31). Eleven annotated ingredients and 931 target genes were reported, among which EGCG was the major ingredient and targeted 556 genes (Figures 1A,B). According to Gene Ontology (GO) enrichment analysis, the target genes of tea were involved in inflammation and chemokines (positive regulation of cytokine production, positive regulation of leukocyte migration, etc.), coagulation and cell death (neuronal death, extrinsic apoptotic signaling pathways, etc.) (Figure 1C), which were previously reported as the molecular characteristics of COVID-19, indicating that the tea has the potential to have anti-COVID-19 activity. The KEGG enrichment analysis also demonstrated that tea has a strong antiviral activity, with target genes that were functionally enriched in COVID-19 and influenza A, and represses inflammation (Figure 1D).
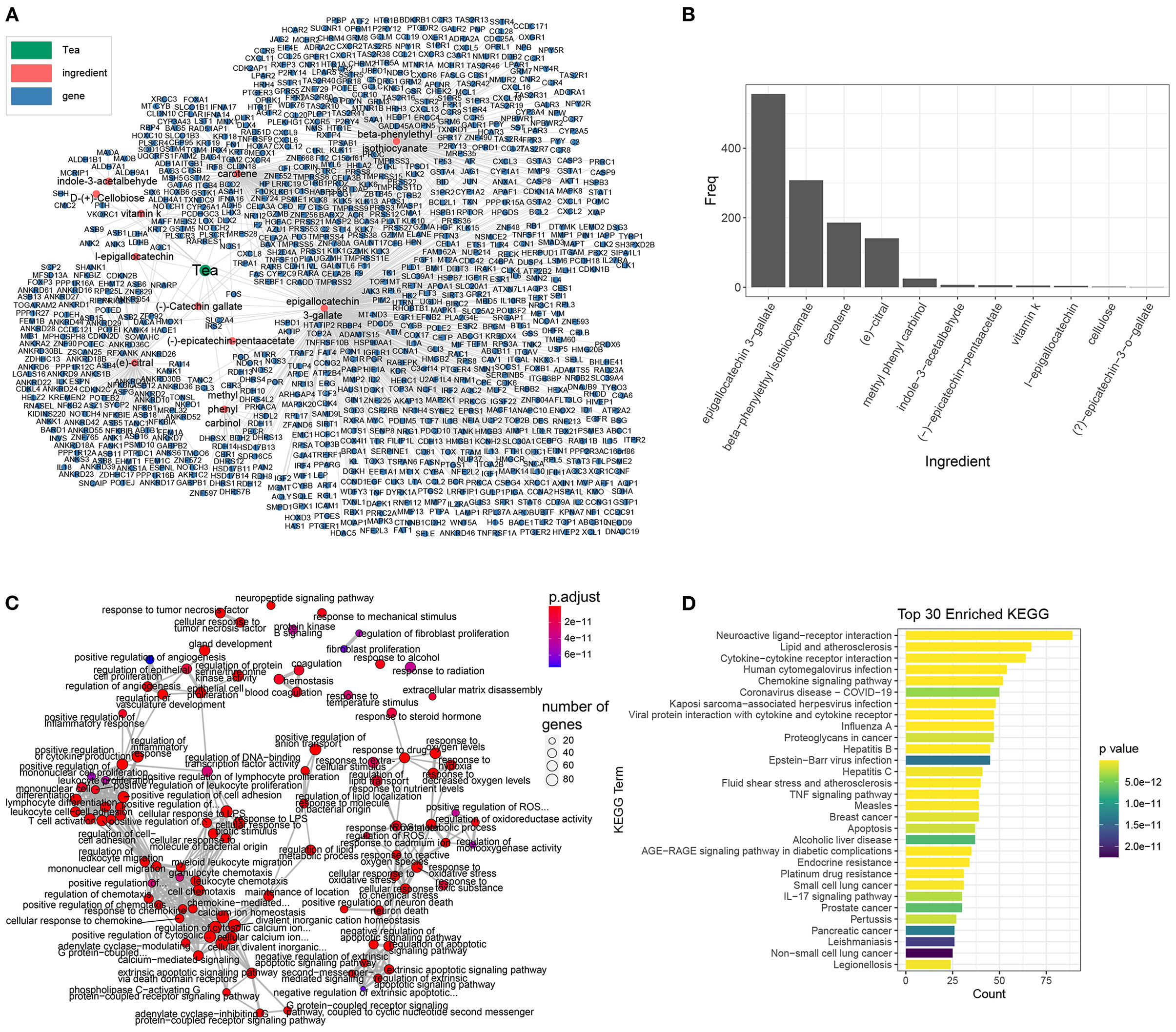
Figure 1. The ingredients and target genes of tea. (A) The network of tea ingredients and target genes, the dot color represents the components, while green, red, and blue represents the tea, tea ingredients, and target genes, respectively. (B) The target genes bar plot of each tea ingredient. (C) The GO enrichment map of tea target genes organized enriched terms into a network with edges connecting overlapping genes and easier to identify hub module. (D) The KEGG enrichment bar plot of the top 30 enriched terms, the bar color represents the P-value of each term.
In addition to EGCG, there are many additional components such as beta-phenylethyl isothiocyanate, carotene, and citral, that also have anti-inflammatory and anti-chemotactic effects for COVID-19. We intersected the targets of other tea ingredients with the signature genes of COVID-19 (Supplementary Figure 1) and found that the targets of EGCG covered the most signature genes of COVID-19. Furthermore, beta-phenylethyl isothiocyanate and carotene also covered some signature genes. The enrichment analysis of the corresponding intersected genes (Supplementary Figure 2) showed that in addition to EGCG, other tea ingredients can repress the corresponding pathological processes involved in COVID-19. For instance, the targets of beta-phenylethyl isothiocyanate are closely related to cell chemotaxis in COVID-19. Citral inhibits inflammation and NK-κB signaling in COVID-19. Finally, cartone is related to coagulation and cytokine secretion. In summary, these results imply that tea can suppress inflammation and prevent coronavirus disease.
We next employed the DisGeNET and KEGG databases to characterize the molecular signature of COVID-19 infection. There were 1,288 and 232 COVID-19 signature genes in the DisGeNET and KEGG databases, respectively, among which 87 genes were shared (Figure 2A). These genes were functionally enriched in response to viruses, innate and adaptive immune responses, inflammatory responses, and coagulation (Figure 2B). Notably, the signature genes of COVID-19 infection were also enriched in response to LPS, indicating a similar molecular pattern between COVID-19 infection and sepsis (Figure 2B). The similarity analysis of the enrichment results of the DisGeNET and KEGG COVID-19 genes revealed that cytokine and chemokine activity, endopeptidase activity, and phosphatidylinositol 3-kinase (PI3K) activity were the major processes of COVID-19 infection (Figure 2C). Furthermore, the enrichment of KEGG COVID-19 signature genes showed a MAPK signaling pathway specificity (Figure 3C). As for shared genes of the two databases, the PPI analysis implied that they were highly biologically relevant; among these genes, IL-6, TNF, and IL-1B were relevant to the highest degree (Figure 2D). In addition to cytokines, the Toll-like receptor (TLR2, TLR3, TLR7, and TLR8) and inflammatory signaling pathways (JAK-STAT, NF-κB, and MAPK signaling pathways) were also important components in the PPI network (Figure 2D). The functions of these genes included involvement in the antiviral process (COVID-19, influenza A, etc.) and responses to molecules of bacterial origin and inflammation (Figures 2E,F).
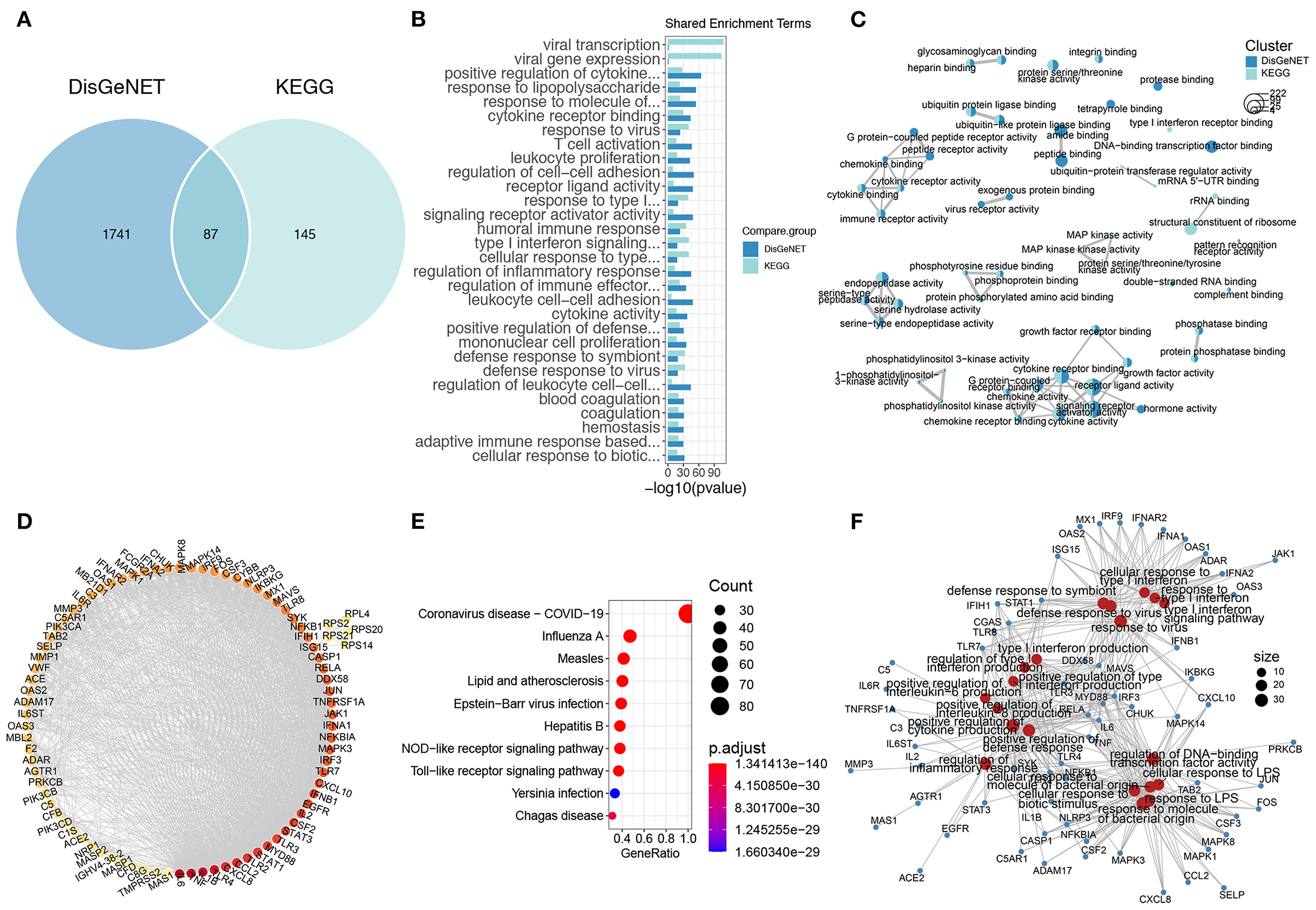
Figure 2. The gene signature of COVID-19. (A) The Venn plot of COVID-19 gene signature of DisGeNET and KEGG. (B) The shared COVID-19 gene signature GO terms of DisGeNET and KEGG. (C) The comparison GO enrichment network of DisGeNET and KEGG, the number of circles in the bottom left corner represents the gene number of each enriched term, the proportion of clusters in the pie chart is determined by the number of genes. (D) The PPI of shared genes of DisGeNET and KEGG COVID-19 gene signature, the dot color represents the connectivity of each gene, while from yellow to red represents from low to high. (E) The top 10 KEGG enriched terms of shared COVID-19 signature genes. (F) The genes and GO enriched terms network of shared COVID-19 signature genes, the red dots represent the GO enriched terms while blue dots are the related genes.
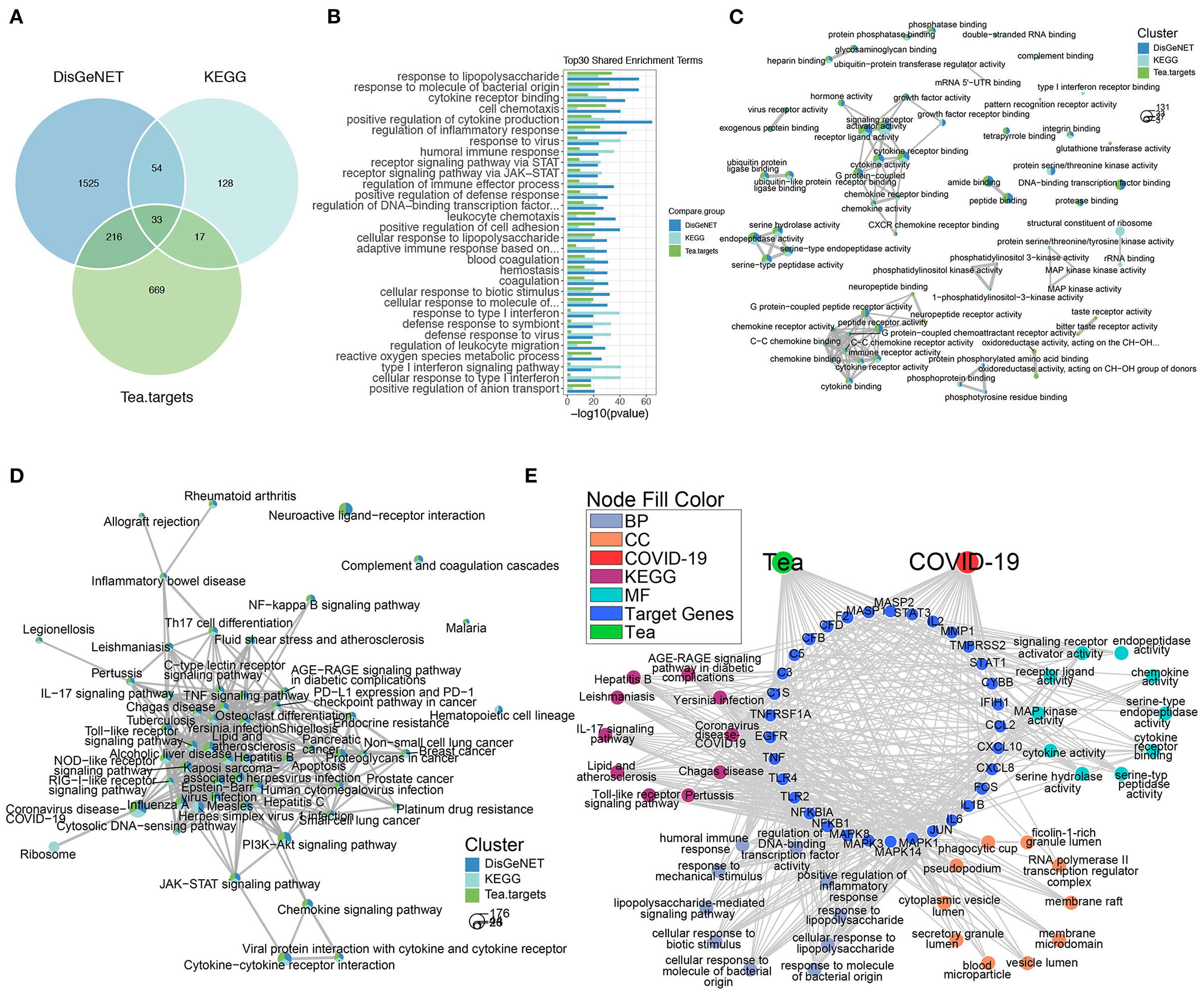
Figure 3. The anti-COVID-19 potential of tea. (A) The Venn plot of COVID-19 signature genes and tea target genes. (B) The shared GO enriched terms of COVID-19 signature genes and tea target genes. The comparison GO (C) and KEGG (D) enrichment network of COVID-19 signature genes and tea target genes, the bottom left circles stand for the gene number of each enriched term, the proportion of clusters in the pie chart indicates the number of genes. (E) The network of shared COVID-19 signatures genes and tea target genes along with the GO and KEGG enrichment terms.
To further verify the anti-COVID-19 activity of tea ingredients, we co-analyzed the target genes of tea with COVID-19 signature genes. There were 249 and 50 shared targets genes of tea with DisGeNET and KEGG COVID-19 gene signatures, respectively, and 33 shared genes in all conditions (Figure 3A). The shared GO enrichment items were focused on the response to bacteria and viruses, inflammation (cytokines and chemokines), immune responses, and coagulation; these cover the majority of signature genes of COVID-19 that were enriched (Figures 2B, 3B), suggesting that the ingredients of tea might act as anti-COVID-19 components. The comparison of the GO enrichment results also showed that tea could target the critical pathological processes involved in COVID-19 infection including cytokine and chemokine activity, endopeptidase activity, and the MAPK signaling pathway (Figure 3C). The comparison of the KEGG enrichment results also showed a similar pattern that covered the major inflammatory signaling pathways including the JAK-STAT, NF-κB, and MAPK signaling pathways (Figure 3D). Furthermore, the shared 33 genes in all three conditions were functionally involved in inflammation and immune responses, which are similar to the major pathological processes of COVID-19, which involve the Toll-like receptor signaling pathway, IL-17 signaling pathway, and cytokine and chemokine activity (Figure 3E). Notably, the shared 33 genes were similar to the high degree genes in PPI, such as IL-6, TNF, and IL-1β, revealing that they were centrally involved in the pathological status of COVID-19 infection. Thus, the results demonstrated that the target genes of tea covered the critical processes involved in COVID-19 infection and might serve as anti-COVID-19 components.
Previous studies have reported that small molecules are able to block the COVID-19 virus through interaction with vital virus proteins, such as 5R84 (32). We next examined the interaction of tea ingredients with the COVID-19 protein 5R84 by molecular docking (33). Six of 11 tea ingredients were capable of interacting with 5R84 and had lower free binding energies than the qualified 5R84 ligand WGS; these were (–)-catechin gallate, carotene, l-epigallocatechin, (–)-epicatechin-pentaacetate, D-(+)-cellobiose, and epigallocatechin 3-gallate (Figure 4A). Among these 6 ingredients, (–)-catechin gallate had the lowest binding free energy (−8.8 kcal/mol) and formed 5 hydrogen bonds with the ARG40, TYR54, GLU55, ASN-180, and ARG-188 residues of 5R84 (Figure 4B). The other 5 ingredients shared comparable binding free energies (~-7.26 kcal/mol) and formed 0 to 5 hydrogen bonds with residues (Figure 4B). Notably, although carotene had a low binding free energy, it could not form hydrogen bonds with 5R84, implying that the interaction of carotene with 5R84 was not stable. In summary, we identified three tea ingredients ((–)-catechin gallate, D-(+)-cellobiose, and epigallocatechin 3-gallate) that were sufficient to block COVID-19 by interacting with 5R84 protein.
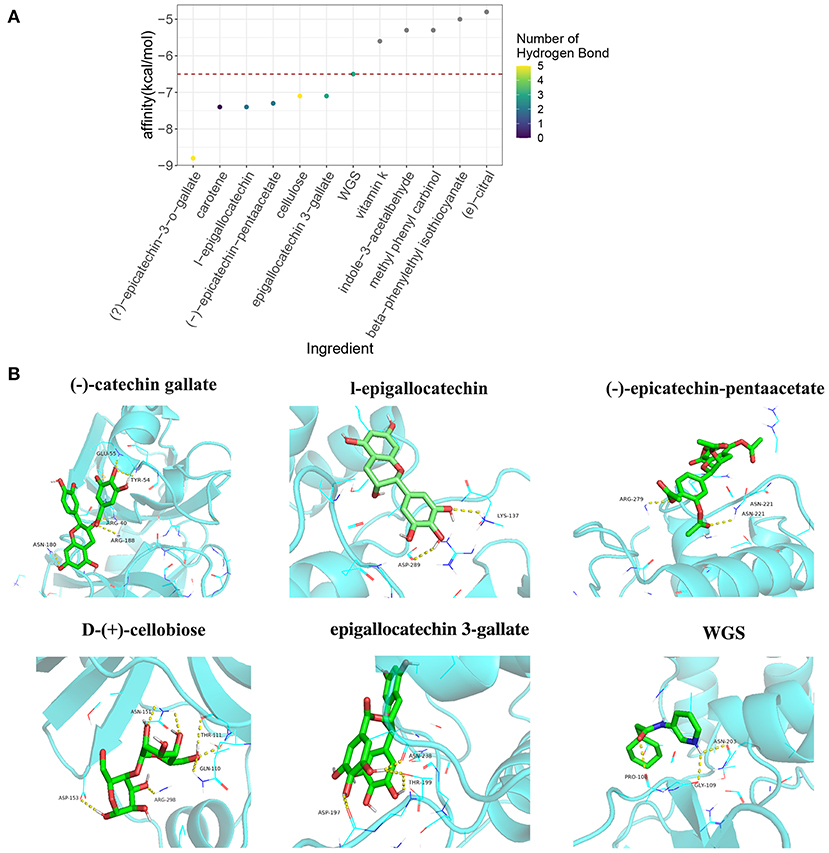
Figure 4. The molecular docking analysis of tea ingredients with COVID-19 5R84 protein. (A) The binding-free energy and hydrogen bond numbers of tea ingredients with 5R84 protein with qualified 5R84 ligand WGS as a reference, the red dot horizontal line indicates the binding-free energy of WGS with 5R84. (B) The representative interaction of tea ingredients and WGS with 5R84, the yellow dot lines indicate the hydrogen bonds of the specific ligand with 5R84.
Based on the abovementioned biometric analysis results, it is reasonable to hypothesize that EGCG is involved in inflammation in COVID-19. To ascertain whether EGCG can protect the body from inflammatory injury, we conducted a CCK8 assay. The results revealed that cell viability began to decline when the concentration of EGCG exceeded 50 nM (Figure 5A). Subsequently, we analyzed the effect of EGCG on macrophage polarizations. The LPS (100 ng/mL)-induced mRNA expression of M1 marker genes including iNOS, TNF-α, Il-1β, and IL-6 was significantly reduced by EGCG (Figures 5B–E). On the other hand, EGCG showed an increased effect on the level of induction of the M2 marker gene Arg1 stimulated by LPS in RAW264.7 cells (Figure 5F). Then, we collected RAW264.7 cell supernatants after LPS stimulation in a culture system with or without EGCG to measure the secretion of inflammatory factors by ELISA. The results showed that EGCG significantly reduced the production of IL-6, TNF-α, and IL-1β compared with the LPS stimulation group (Figures 5G–I). Moreover, we also detected the inflammatory factor IL-17A secreted by macrophages and the expression of TLR4 and PI3K, which were previously screened (Figure 3E); EGCG significantly suppressed the production of IL-17A and the mRNA levels of TLR4 and PI3K compared with the LPS stimulation group (Supplementary Figure 3). These results indicate that EGCG reduces the secretion of inflammatory factors in vitro.
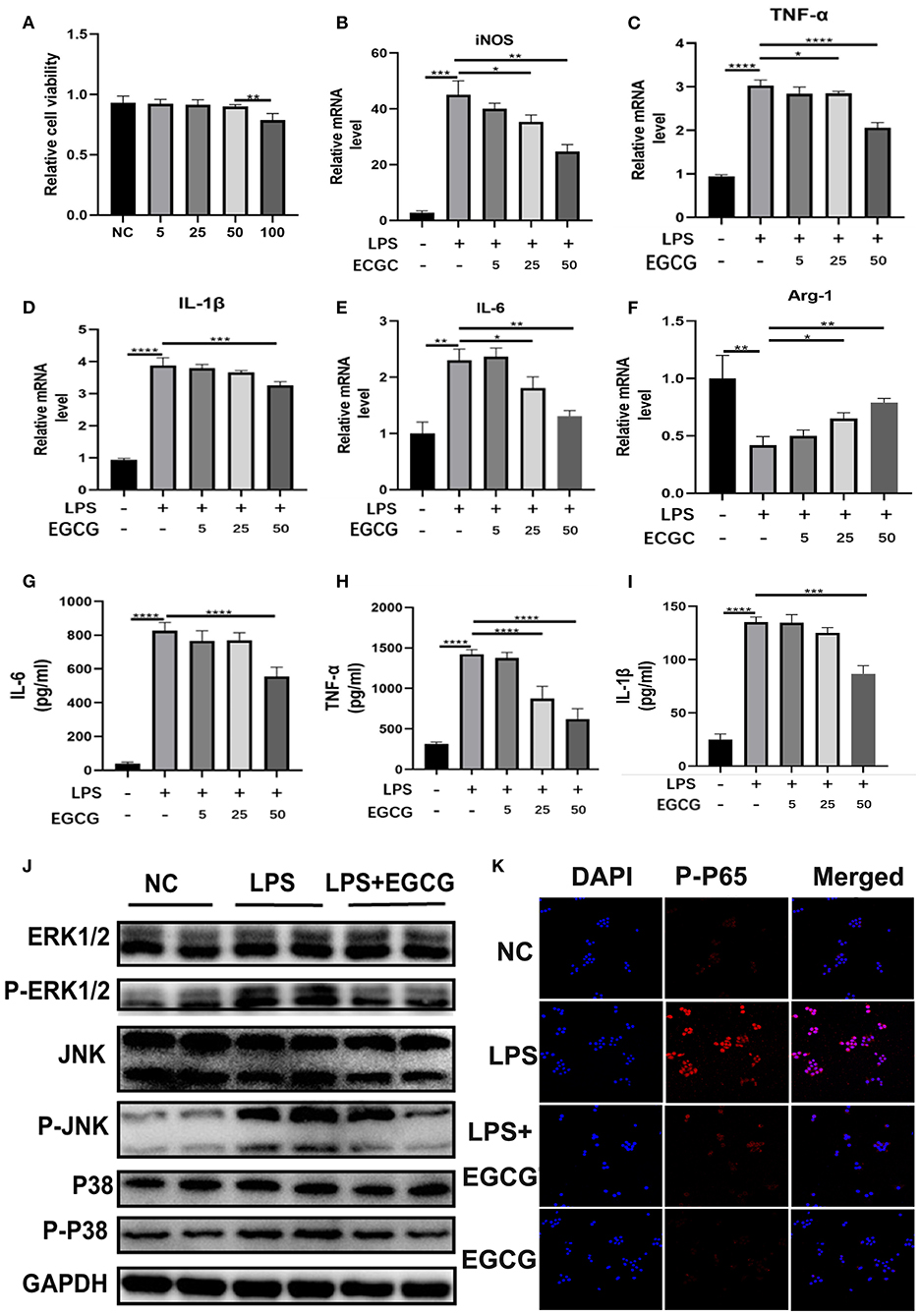
Figure 5. EGCG suppressed secretion of inflammatory factors, macrophage polarization, and MAPK/NF-κB signaling in vitro. (A) RAW 264.7 cells were incubated with EGCG (50 mM) for 24 h. Cell viability was determined by CCK8 assay (n = 5). (B–F) The mRNA levels of iNOS, TNF-α, Il-1β, IL-6, and Arg1 in the RAW 264.7 cells with LPS (100 ng/ml) and EGCG (50 nM) for 24 h were detected by q-PCR (n = 3). (G–I) The concentrations of IL-6, TNF-α, and IL-1β in RAW 264.7 cell supernatant after LPS and EGCG treatment for 24 h were determined by ELISA kits (n = 4). (J) The protein levels of ERK1/2, P-ERK1/2, JNK, P-JNK, P38, and p-p38 in the RAW 264.7 cells treated with LPS (100 ng/ml) and EGCG (50 nM) for 24 h were detected by Western blotting. (K) The expressions of p-p65 (red) and DAPI (blue) in RAW 264.7 cells were detected by using an immunofluorescence staining assay (scale bar: 50 μm). *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To further explore the mechanism by which EGCG alleviates inflammatory damage to cells, we investigated the inflammation pathway in vitro. We measured the activation of the MAPK pathway. As shown in Figure 5J, phosphorylation of p-ERK, p-JNK, and p-p38 in macrophages was significantly increased after LPS challenge; this effect was suppressed by EGCG in vitro as determined by WB (Figure 5J). This demonstrates that EGCG could effectively inhibit the MAPK pathway. Furthermore, we investigated the suppressive effect of EGCG treatment on the NF-κB signaling cascade in RAW264.7 macrophages. Our investigations indicated that the phosphorylation of p65 was significantly increased after LPS challenge, and this was suppressed by EGCG (Figure 5K). This finding confirms that EGCG suppressed inflammation by inhibiting MAPK/NF-κB signaling.
Tea is one of the three most consumed beverages in the world and is known as the beverage of the twenty-first century, not only because of the long history of tea culture but also because of its nutritional value and health care functions (34, 35). Studies have shown that tea contains numerous active ingredients, mainly tea polyphenols, tea pigments, tea polysaccharides, γ-aminobutyric acid, tea saponins, alkaloids, vitamins, pyrroloquinoline quinone, pantothenic acid, minerals, and other ingredients (36, 37). Tea polyphenols are the most abundant soluble components in tea, and they are also the most important substances in tea that exert biological effects (35) that can reduce the incidence of cardiovascular disease, decrease blood lipids, decrease body fat formation, and change the intestinal flora ecology (35, 38). Studies have shown that after drinking a cup of tea for half an hour, the antioxidant capacity (ability to fight oxygen free radicals) in the blood increases by 41% to 48% and can last for one and a half hours at a high level (39).
In our work, we first screened the main ingredient of tea, EGCG, in databases, suggesting that EGCG may play an important role in the treatment of COVID-19. EGCG is the main component of green tea polyphenols and is a catechin monomer isolated from tea (40). Studies have shown that EGCG has several functions including significant anti-oxidation, involvement in scavenging free radicals, reduction of inflammation and allergic reactions, anti-mutagenic effects, inhibition of tumor growth, and strong inhibitory effects on dysentery, typhoid fever, Staphylococcus aureus, and other bacteria (41–43). EGCG also has the functions of anti-aging, lowering blood lipids, improving low-density lipoprotein, inhibiting the growth of liver fat and cholesterol, preventing atherosclerosis, and enhancing immunity (44–46). In addition, EGCG can inhibit the proliferation of glomerular cell membranes and improve renal function (47). Several studies have reported the potential of EGCG to prevent COVID-19. For instance, EGCG inhibits the angiotensin-converting enzyme 2 (ACE2) receptor (the cellular receptor for SARS-CoV-2) and TMPRSS2, which mediate viral entry into cells, by activating Nrf2 (48, 49). By inhibiting the main protease of SARS-CoV-2, EGCG may inhibit viral reproduction (48). EGCG protects against SARS-CoV-2-induced mitochondrial reactive oxygen species (ROS) (promoting SARS-CoV-2 replication) and ROS bursts caused by neutrophil extracellular traps through its broad antioxidant activity (48, 50). EGCG can potentially inhibit the SARS-CoV-2 life cycle by inhibiting ER-resident GRP78 activity and expression (51, 52). EGCG has also been shown to protect against (1) cytokine storm-related acute lung injury/acute respiratory distress syndrome (48, 53), (2) thrombosis through inhibition of tissue factor and activation of platelets (54), (3) inactivation of redox-sensitive HMGB1-induced sepsis (55), and (4) pulmonary fibrosis by increasing Nrf2 and inhibiting NF-κB (13). However, these activities remain to be further confirmed in animals and humans.
Studies have shown that macrophages play an important role in COVID-19 (56). Cytokine storm syndrome (CSS) refers to the continuous activation and expansion of lymphocytes and macrophages caused by the infection of microorganisms, and a variety of cytokines such as TNF-α, IL-1, IL-6, IL-12, interferon (IFN)-α, IFN-β, IFN-γ, monocyte chemoattractant protein (MCP)-1, and IL-8 are rapidly produced in large quantities (57). CSS is an excessive immune phenomenon of the body to external stimuli and is an important cause of acute respiratory distress syndrome and multiple organ failure (58). Studies have shown that cytokine storms play a key role in the transition to severe and critical illness in most coronavirus-infected patients (59). In addition, one study found that there is a highly pro-inflammatory macrophage microenvironment in the lungs of severely ill patients with the new strain, which the researchers said may help to elucidate the underlying mechanism behind the immune response triggered by the new coronavirus (60). Therefore, we focused on the role of EGCG in regulating changes in macrophage function to improve COVID-19. The inflammatory response in COVID-19 is much more complex than that in LPS-induced RAW264.7 cells, and it is extremely important to distinguish the inflammatory subtypes of different diseases. However, the inflammatory response in COVID-19 still shares some common signatures with the inflammatory response in LPS-induced RAW264.7 cells, among which the most typical are TLR4, NF-κB, and other signaling pathways and their corresponding cytokines (including IL-6, TNF, IL-1β, etc.) (Figure 2D). Through the LPS-stimulated macrophage model, we attempted to demonstrate the possibility of EGCG molecules indirectly inhibiting COVID-19 inflammation.
EGCG has been reported to alleviate acute lung injury, regulate the polarization of macrophages to M2 (61), and inhibit secretion of inflammatory factors, and its protective mechanism may be related to the inhibition of the MAPK and NF-κB signaling pathway (62–64). In addition, EGCG derivatives have anti-inflammatory activity in LPS-stimulated mouse macrophages (65). Furthermore, EGCG-modified collagen membranes have been shown to downregulate the expression of inflammatory factors and promote M2 (CD163 and CD206) macrophages (66). EGCG also stimulates LC3-II production and autophagosome formation and inhibits LPS-induced upregulation and extracellular release of HMGB1 (67). Our results are consistent with those described above; however, the origins of the abovementioned research and our study are different. There is some heterogeneity in the inflammatory responses of different diseases and different states of certain diseases. Starting from the gene signature of COVID-19, we co-analyzed the target genes of each component of tea in an attempt to identify the potential of specific components of tea for the treatment of COVID-19. The results showed that the intersection of COVID-19 signature genes and tea target genes was highly focused on the response to LPS stimulation (Figure 3B). This phenomenon itself is an important discovery. Among the different components of tea, EGCG is obviously an important molecule regulating this process in COVID-19; furthermore, it has the most target genes and is the major active ingredient in tea. We then indirectly verified our findings in LPS-stimulated macrophages in vitro to examine the suppression effects of EGCG on the LPS-like responses in COVID-19. Finally, our study is slightly different from the abovementioned literature (61–67) in terms of molecular signaling pathways. Based on the results of the bioinformatics analysis, we focused on the most credible MAPK (ERK1/2-JNK-P38) signaling pathway. In addition, EGCG reduced the secretion of inflammatory factors and regulated macrophage polarization (from M1 to M2) in vitro. These cell experiments verified the results of our bioinformatics analysis; namely, the active ingredient of tea, ECGC, can directly act on macrophages in the cytokine storm environment of COVID-19, and inhibit the secretion of inflammatory factors and the activation of the MAPK and NF-κB signaling pathways, improving the prognosis of COVID-19.
Moreover, Douangamath et al. (68) performed a large-scale electrophilic and non-covalent fragment screening of the major proteases of SARS-CoV-2 by combined mass spectrometry and X-ray and found that 5R84 is one of two cysteine viral proteases essential for viral replication. We therefore examined the interaction of tea components with the COVID-19 protein 5R84. Through molecular docking analysis, we identified three tea ingredients ((–)-epicatechin-3-o-gallate, D-(+)-cellobiose, and EGCG) that likely interact with the vital SARS-CoV-2 protein, 5R84, compared with the qualified 5R84 ligand WGS. According to the description in PubChem (https://pubchem.ncbi.nlm.nih.gov/compound/24802025#section=Household-Products), D-(+)-cellobiose is indeed insoluble in water and cannot be absorbed by the human body; thus, it is nearly impossible to inhibit SARS-CoV-2 through absorption from the gastrointestinal tract and into circulation. However, considering the droplet transmission and fecal-oral transmission of SARS-CoV-2, namely, that SARS-CoV-2 exists on the surfaces of the respiratory tract and digestive tract, D-(+)-cellobiose may directly interact with SARS-CoV-2 on the corresponding surfaces. However, the roles of (–)-epicatechin-3-o-gallate and D-(+)-cellobiose in COVID-19 should be studied further in cell and animal experiments.
In summary, our research systematically analyzed the active ingredients of tea, namely, (–)-epicatechin-3-o-gallate, D-(+)-cellobiose and EGCG, which have the potential to treat COVID-19 by suppressing the target genes and signaling pathways of COVID-19 and interacting with the vital SARS-CoV-2 protein. In addition, we validated the above results in macrophages. Our study analyzed the anti-COVID-19 effects of the active ingredients of tea and provided new ideas for the prevention and treatment of COVID-19.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
LW and QT: conception and design, collection and/or assembly of data, data analysis and interpretation, visualization, and manuscript writing and final approval of the manuscript. ZW and JS: collection and/or assembly of data. WY and LZ: collection and/or assembly of data. XY, LW, and YS: financial support, administrative support, provision of study material, supervision, data analysis and interpretation, visualization, manuscript writing, and final approval of the manuscript. All authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Key Research and Development Program of China (No: 2020YFC2005300), the Natural Science Youth Foundation of the Jiangsu Province (Grant BK20210074), the Introduction Program of high-level innovative and entrepreneurial talents in Jiangsu province, Wuxi first Double hundred Young and middle-aged Top-notch Medical and health talents Program (HB2020108), and Wuxi Health Commission scientific research project youth project (Q202059).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.875765/full#supplementary-material
1. Girija ASS, Shankar EM, Larsson M. Could SARS-CoV-2-induced hyperinflammation magnify the severity of coronavirus disease (CoViD-19) leading to acute respiratory distress syndrome? Front Immunol. (2020) 11:1206. doi: 10.3389/fimmu.2020.01206
2. Zhang B, Liu S, Tan T, Huang W, Dong Y. L., Chen, et al. Treatment with convalescent plasma for critically ill patients with severe acute respiratory syndrome Coronavirus 2 infection chest. (2020) 158:e9–9158:tment 1016/j.chest.2020.03.039
3. Shang Y, Liu T, Wei Y, Li J, Shao L. M. Liu, et al. Scoring systems for predicting mortality for severe patients with COVID-19 E. Clin Med. (2020) 24:100426. doi: 10.1016/j.eclinm.2020.100426
4. Lotfi M. and Rezaei N, SARS-CoV-2:a comprehensive review from pathogenicity of the virus to clinical consequences. J Med Virol. (2020) 92:1864–864:0Viroli1002/jmv.26123
5. Khandia R, Singhal S, Alqahtani T, Kamal MA, El-Shall NA. F. Nainu, et al. Emergence of SARS-CoV-2 Omicron (B11529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ Res. (2022) 209:112816. doi: 10.1016/j.envres.2022.112816
6. Chen JM. Should the world collaborate imminently to develop neglected live-attenuated vaccines for COVID-19? J Med Virol. (2022) 94:82irolould th1002/jmv.27335
7. Yin J, Li C, Ye C, Ruan Z, Liang Y. Y. Li, et al. Advances in the development of therapeutic strategies against COVID-19 and perspectives in the drug design for emerging SARS-CoV-2 variants. Comput Struct Biotechnol J. (2022) 20:824ol JJhnol JJ1016/j.csbj.2022.01.026
8. Ghosh A, Kar PK, Gautham A, Gupta R, Singh R, Chakravarthi R, et al. An insight into SARS-CoV2 structure, Pathogenesis, target hunting for drug development and vaccine initiatives. RSC Med Chem. (2022). doi: 10.1039/D2MD00009A
9. Bag S, Mondal A, Majumder A, Banik A. Tea and its phytochemicals:Hidden health benefits & modulation of signaling cascade by phytochemicals. Food Chem. (2022) 371:131098. doi: 10.1016/j.foodchem.2021.131098
10. Mitra S, Tareq AM, Das R, Emran TB, Nainu F, Chakraborthy AJ, et al., Polyphenols: A first evidence in the synergism and bioactivities. Food Rev Int. (2022) 1–23. doi: 10.1080/87559129.2022.2026376
11. Truong V-L, Jeong W-S. Antioxidant and anti-inflammatory roles of tea polyphenols in inflammatory bowel diseases. Food Sci Hum Wellness. (2022) 11:502–11. doi: 10.1016/j.fshw.2021.12.008
12. Hidalgo I, Ortiz-Flores M, Villarreal F, Fonseca-Coronado S, Ceballos G, Meaney E, et al. Is it possible to treat nonalcoholic liver disease using a flavanol-based nutraceutical approach? Basic and clinical data. J Basic Clin Physiol Pharmacol. (2022) doi: 10.1515/jbcpp-2021-0285
13. Hong M, Cheng L, Liu Y, Wu Z, Zhang P, Zhang X, et al. Natural plant source-tea polyphenols, a potential drug for improving immunity and combating virus. Nutrients. (2022) 14:550. doi: 10.3390/nu14030550
14. Khalil A. and D. Tazeddinova the upshot of polyphenolic compounds on immunity amid COVID-19 pandemic and other emerging communicable diseases:an appraisal. Nat Prod Bioprospect. (2020) 10:411ectoprospect1007/s13659-020-00271-z
15. Lanka CNS. Antiviral Properties of Tea: Black Tea May Become the Unique Brew of Choice With No Side Effects to Fight Against Corona Virus? Talawakelle: Tea Research Institute of Sri Lanka (2022).
16. Manivannan J, Silambarasan T, Kadarkarairaj R, Raja B. Systems pharmacology and molecular docking strategies prioritize natural molecules as cardioprotective agents. RSC Adv. (2015) 5:77042–70425v. tec1039/C5RA10761J
17. Wu C, Huang ZH, Meng ZQ, Fan XT, Lu S, Tan YY, et al. A network pharmacology approach to reveal the pharmacological targets and biological mechanism of compound kushen injection for treating pancreatic cancer based on WGCNA and in vitro experiment validation. Chin Med. (2021) 16:121. doi: 10.1186/s13020-021-00534-y
18. Lin H, Wang X, Liu M, Huang M, Shen Z. J. Feng, et al. Exploring the treatment of COVID-19 with Yinqiao powder based on network pharmacology. Phytother Res. (2021) 35:2651–651:1her Re1002/ptr.7012
19. Yan F, Zhao Q, Gao H, Wang X, Xu K, Wang Y, et al. Exploring the mechanism of (–)-Epicatechin on premature ovarian insufficiency based on network pharmacology and experimental evaluation. Biosci Re. (2021) 41:2. doi: 10.1042/BSR20203955
20. Veselkov K, Gonzalez G, Aljifri S, Galea D, Mirnezami R, Youssef J, et al. Hyper foods:machine intelligent mapping of cancer-beating molecules in foods. Sci Re. (2019) 9:9237. doi: 10.1038/s41598-019-45349-y
21. Yang H, Yue GGL, Leung PC, Wong CK, Lau CBS. A review on the molecular mechanisms, the therapeutic treatment including the potential of herbs and natural products, and target prediction of obesity-associated colorectal cancer. Pharmacol Res. (2022) 175:106031. doi: 10.1016/j.phrs.2021.106031
22. Wang B, Ding Y, Zhao P, Li W, Li M. Zhu J, et al. Systems pharmacology-based drug discovery and active mechanism of natural products for coronavirus pneumonia (COVID-19):An example using flavonoids. Comput Biol Med. (2022) 143:105241. doi: 10.1016/j.compbiomed.2022.105241
23. Mhatre S, Naik S, Patravale V. A. molecular docking study of EGCG and theaflavin digallate with the druggable targets of SARS-CoV-2. Comput Biol Med. (2021) 129:104137. doi: 10.1016/j.compbiomed.2020.104137
24. Maiti S. Banerjee Epigallocatechin gallate and theaflavin gallate interaction in SARS-CoV-2 spike-protein central channel with reference to the hydroxychloroquine interaction:Bioinformatics and molecular docking study. Drug Dev Res. (2021) 82:86Dev Res dock1002/ddr.21730
25. Qin X, Huang C, Wu K, Li Y, Liang X, Su M, et al. Anti-coronavirus disease (2019) (COVID-19) targets and mechanisms of puerarin. J Cell Mol Med. (2021) 25:677–85. doi: 10.1111/jcmm.16117
26. Mah N, Taškova K. hgu133plus2CellScore 1.15.0: Standard Expression Set for CellScore [hgu133plus2]. (2021). doi: 10.18129/B9.bioc.hgu133plus2CellScore
27. Hu Q, Zhang Z-G. An online system for functional relationship analysis of genome-wide gene products. In: (2010) 4th International Conference on Bioinformatics and Biomedical Engineering. (2010).
28. Zhu MZ, Wen BB, Wu H, Li J, Lin HYQ. Li, et al. The quality control of tea by near-infrared reflectance (nir) spectroscopy and chemometrics. J Spectrosc. (2019) 2019: 8129648. doi: 10.1155/2019/8129648
29. Bharadwaj S, Lee KE, Dwivedi VD. Yadavac U, Kang SG, Computational aided mechanistic understanding of Camellia sinensis bioactive compounds against co-chaperone p23 as potential anticancer agent. J Cell Biochem. (2019) 120:19064–9064:ml Bio1002/jcb.29229
30. Wu FX. Chen C, Peng FL. Potential association between asthma helicobacter pylori infection gastric cancer. Front. Oncol. (2021) 11:498. doi: 10.3389/fonc.2021.630235
31. Fang J, Liu C, Wang Q, Lin P, Cheng F. In silico polypharmacology of natural products. Brief Bioinform. (2018) 19:1153–153:8orminfo1093/bib/bbx045
32. Nand M, Maiti P, Joshi T, Chandra S, Pande V, Kuniyal JC, et al. Virtual screening of anti-HIV1 compounds against SARS-CoV-2: machine learning modeling, chemoinformatics and molecular dynamics simulation based analysis. Sci Re. (2020) 10:20397. doi: 10.1038/s41598-020-77524-x
33. El-Hawary SS, Mohammed R, Bahr HS, Attia EZ, El-Katatny MH. Abelyan N, et al. Soybean-associated endophytic fungi as potential source for anti-COVID-19 metabolites supported by docking analysis. J Appl Microbiol. (2021) 131:1193–1931:iollbio1111/jam.15031
34. Thielecke F. Boschmann The potential role of green tea catechins in the prevention of the metabolic syndrome - a review. Phytochemistry. (2009) 70:11emistryyion 1016/j.phytochem.2008.11.011
35. Xing L, Zhang H, Qi R, Tsao R, Mine Y. Recent advances in the understanding of the health benefits and molecular mechanisms associated with green tea polyphenols. J Agric Food Chem. (2019) 67:1029–029:9Chem C1021/acs.jafc.8b06146
36. Huang Y, Xing K, Qiu L, Wu Q, Wei H. Therapeutic implications of functional tea ingredients for ameliorating inflammatory bowel disease:a focused review. Crit Rev Food Sci Nutr. (2021) 26:1-15. doi: 10.1080/10408398.2021.1884532
37. Feng MY, Zheng X, Wan J, Pan WJ, Xie XY, Hu BZ, et al. Research progress on the potential delaying skin aging effect and mechanism of tea for oral and external use. Food Funct. (2021) 12:2814–814:1unct p1039/D0FO02921A
38. Chen T, Yang CS. Biological fates of tea polyphenols and their interactions with microbiota in the gastrointestinal tract: implications on health effects. Crit Rev Food Sci Nutr. (2020) 60:2691–709. doi: 10.1080/10408398.2019.1654430
39. McKay DL, Blumberg JB. The role of tea in human health: an update. J Am Coll Nutr. (2002) 21:1–13. doi: 10.1080/07315724.2002.10719187
40. Rahim AA, Nofrizal S, Saad B. Rapid tea catechins and caffeine determination by HPLC using microwave-assisted extraction and silica monolithic column. Food Chem. (2014) 147:262eine determ1016/j.foodchem.2013.09.131
41. Fan FY, Sang LX, Jiang M. Catechins and their therapeutic benefits to inflammatory bowel disease. Molecules. (2017) 22:484. doi: 10.3390/molecules22030484
42. Schramm L. Going green:the role of the green tea component egcg in chemoprevention. J Carcinog Mutagen. (2013) 4:1000142. doi: 10.4172/2157-2518.1000142
43. Rafieian-Kopaei M. Movahedi Breast cancer chemopreventive and chemotherapeutic effects of Camellia Sinensis (green tea): an updated review. Electron Physician. (2017) 9:3838–83817tron P19082/3838
44. Gad SB, Zaghloul DM. Beneficial effects of green tea extract on liver and kidney functions, ultrastructure, lipid profile and hematological parameters in aged male rats. Glob Veter. (2013) 11:191–205. doi: 10.5829/idosi.gv.2013.11.2.7472
45. Deng X, Hou Y, Zhou H, Li Y, Xue Z, Xue X, et al. Hypolipidemic, anti-inflammatory, and anti-atherosclerotic effects of tea before and after microbial fermentation. Food Sci Nutr. (2021) 9:1160–16021trci N1002/fsn3.2096
46. Wichansawakun S, Buttar HS. Antioxidant diets and functional foods promote healthy aging and longevity through diverse mechanisms of action. In: The Role of Functional Food Security in Global Health. Elsevier (2019). p. 541–63. doi: 10.1016/B978-0-12-813148-0.00032-3
47. Kanlaya R. Thongboonkerd protective effects of epigallocatechin-3-gallate from green tea in various kidney diseases. Adv Nutr. (2019) 10:112ive effects 1093/advances/nmy077
48. Zhang Z, Zhang X, Bi K, He Y, Yan W, Yang CS, et al. Potential protective mechanisms of green tea polyphenol EGCG against COVID-19. Trends Food Sci Technol. (2021) 114:11nol Sci Tech1016/j.tifs.2021.05.023
49. McCord JM, Hybertson BM, Cota-Gomez A, Geraci KP, Gao B. Nrf2 Activator PB125((R)) as a potential therapeutic agent against COVID-19. Antioxidants (Basel). (2020) 9:518. doi: 10.3390/antiox9060518
50. Hong S, Seo SH, Woo SJ, Kwon Y. M. Song, Ha NC, Epigallocatechin gallate inhibits the uridylate-specific endoribonuclease Nsp15 and efficiently neutralizes the SARS-CoV-2 strain. J Agric Food Chem. (2021) 69:5948–948:1Chem C1021/acs.jafc.1c02050
51. Siri M, Dastghaib S, Zamani M, Rahmani-Kukia N, Geraylow KR. S. Fakher, et al. Autophagy unfolded protein response, and neuropilin-1 cross-talk in SARS-CoV-2 infection: what can be learned from other Corona viruses. Int J Mol Sci. (2021) 22:5992. doi: 10.3390/ijms22115992
52. Ahmed A., et al. (2019). The combination of quercetin and bromelain with zinc, egcg, retinoic acid, vitamin c and vitamin d for the potential symptom reducer, prevention, and treatment for coronavirus disease 2019 (COVID-19). doi: 10.6084/m9.figshare.15153186
53. Yang CC, Wu CJ, Chien CY. Chien C-T. Green tea polyphenol catechins inhibit coronavirus replication and potentiate the adaptive immunity and autophagy-dependent protective mechanism to improve acute lung injury in mice. Antioxidants (Basel). (2021) 10:928. doi: 10.3390/antiox10060928
54. Schr92 K. J. Stampfuss, Weber A-A Green tea catechins containing a galloyl group in the 3n the 3group in the 3 in the 3te the adaptive immunity and a. Thromb Haemost. (2017) 93:1200–200:7ttaem1055/s-0037-1616631
55. Tang D, Kang R, Zeh HJ, Lotze MT. High-mobility group box 1, oxidative stress, and disease. Antioxid Redox Signal. (2011) 14:1315–315:1 Signa1089/ars.2010.3356
56. Brake SJ, Eapen MS, McAlinden KD, Markos J, Haug GJ. Larby, et al. SARS-CoV-2 (COVID-19) adhesion site protein upregulation in small airways type 2 pneumocytes, and alveolar macrophages of smokers and copd - possible implications for interstitial fibrosis. Int J Chron Obstruct Pulmon Dis. (2022) 17:101isPulmon Dis2147/COPD.S329783
57. Aitong W, Leisheng Z, Hao Y. Visualized analyses of investigations upon mesenchymal stem/stromal cell-based cytotherapy and underlying mechanisms for COVID-19 associated ARDS. Curr Stem Cell Res Ther. (2022) 17:2Cell Res The2174/1574888X16666210712212421
58. Zanza C, Romenskaya T, Manetti AC, Franceschi F, La Russa RG. Bertozzi, et al. Cytokine storm in COVID-19:immunopathogenesis and therapy. Medicina (Kaunas). (2022) 58:144. doi: 10.3390/medicina58020144
59. Sen A, Nigam A, Vachher M. Role of polypeptide inflammatory biomarkers in the diagnosis and monitoring of COVID-19. Int J Pept Res Ther. (2022) 28:59. doi: 10.1007/s10989-022-10366-5
60. Vaka R, Khan S, Ye B, Risha Y, Parent SD Courtman. Direct comparison of different therapeutic cell types susceptibility to inflammatory cytokines associated with COVID-19 acute lung injury. Stem Cell Res Ther. (2022) 13:20. doi: 10.1186/s13287-021-02699-7
61. Azambuja JH, Mancuso RI, Via FID, Torello CO, Saad STO. Protective effect of green tea and epigallocatechin-3-gallate in a LPS-induced systemic inflammation model. J Nutr Biochem. (2022) 101:108920. doi: 10.1016/j.jnutbio.2021.108920
62. He Y, Yang Z, Pi J, Cai T, Xia YX Cao. EGCG attenuates the neurotoxicity of methylglyoxal via regulating MAPK and the downstream signaling pathways and inhibiting advanced glycation end products formation. Food Chem. (2022) 384:132358. doi: 10.1016/j.foodchem.2022.132358
63. Wang M, Zhong H, Zhang X, Huang X, Wang J, Li Z, et al. EGCG promotes PRKCA expression to alleviate LPS-induced acute lung injury and inflammatory response. Sci Re. (2021) 11:11014. doi: 10.1038/s41598-021-90398-x
64. Dai W, Lou N, Xie D, Hu Z, Song H, Lu M, et al. N-Ethyl-2-pyrrolidinone-substituted flavan-3-ols with anti-inflammatory activity in lipopolysaccharide-stimulated macrophages are storage-related marker compounds for green tea. J Agric Food Chem. (2020) 68:12164–21640Chem C. doi: 10.1021/acs.jafc.0c03952
65. Wang Q, Huang J, Zheng Y, Guan X, Lai C, Gao H, et al. Selenium-enriched oolong tea (Camellia sinensis) extract exerts anti-inflammatory potential via targeting NF-κ and MAPK pathways in macrophages. Food Sci Hum Wellness. (2022) 11:635 Hum hways. doi: 10.1016/j.fshw.2021.12.020
66. Pore SK, Hahm -ER. Phytochemicals in breast cancer-induced osteoclastogenesis and bone resorption:mechanism and future perspective. Curr Pharmacol Rep. (2022):1–19. doi: 10.1007/s40495-021-00279-0
67. Brodzikowska A, Ciechanowska M, Kopka M, Stachura A, Wlodarski PK. Role of lipopolysaccharide derived from various bacterial species, in pulpitis-a systematic review. Biomolecules. (2022) 12:138. doi: 10.3390/biom12010138
Keywords: COVID-19, molecular docking, network pharmacology, tea ingredients, macrophage, key targets
Citation: Wang L, Tao Q, Wang Z, Shi J, Yan W, Zhang L, Sun Y and Yao X (2022) Tea Ingredients Have Anti-coronavirus Disease 2019 (COVID-19) Targets Based on Bioinformatics Analyses and Pharmacological Effects on LPS-Stimulated Macrophages. Front. Nutr. 9:875765. doi: 10.3389/fnut.2022.875765
Received: 14 February 2022; Accepted: 14 March 2022;
Published: 20 May 2022.
Edited by:
Minhao Xie, Nanjing University of Finance and Economics, ChinaReviewed by:
Jianan Zhang, University of North Carolina at Chapel Hill, United StatesCopyright © 2022 Wang, Tao, Wang, Shi, Yan, Zhang, Sun and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoxiang Sun, c3RhZmYyMDM3QHl4cGguY29t; Xiaoming Yao, eWFveG03M0BzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.