- 1School of Marine Science and Engineering, Qingdao Agricultural University, Qingdao, China
- 2Key Laboratory of Sustainable Development of Polar Fishery, Ministry of Agriculture and Rural Affairs, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 3Key Laboratory of Bioorganic Synthesis of Zhejiang Province, College of Biotechnology and Bioengineering, Zhejiang University of Technology, Hangzhou, China
- 4Jiangsu Key Laboratory for Biomass-Based Energy and Enzyme Technology, Huaiyin Normal University, Huaian, China
- 5School of Environmental Science and Engineering, Suzhou University of Science and Technology, Suzhou, China
Oleaginous fungi (including fungus-like protists) are attractive in lipid production due to their short growth cycle, large biomass and high yield of lipids. Some typical oleaginous fungi including Galactomyces geotrichum, Thraustochytrids, Mortierella isabellina, and Mucor circinelloides, have been well studied for the ability to accumulate fatty acids with commercial application. Here, we review recent progress toward fermentation, extraction, of fungal fatty acids. To reduce cost of the fatty acids, fatty acid productions from raw materials were also summarized. Then, the synthesis mechanism of fatty acids was introduced. We also review recent studies of the metabolic engineering strategies have been developed as efficient tools in oleaginous fungi to overcome the biochemical limit and to improve production efficiency of the special fatty acids. It also can be predictable that metabolic engineering can further enhance biosynthesis of fatty acids and change the storage mode of fatty acids.
Introduction
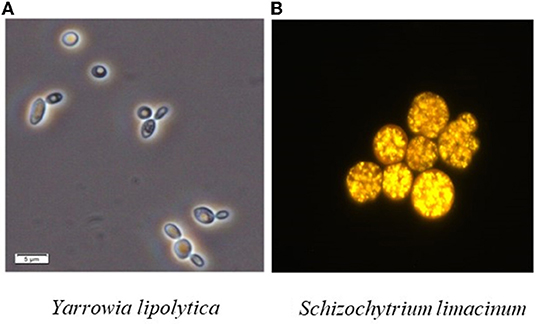
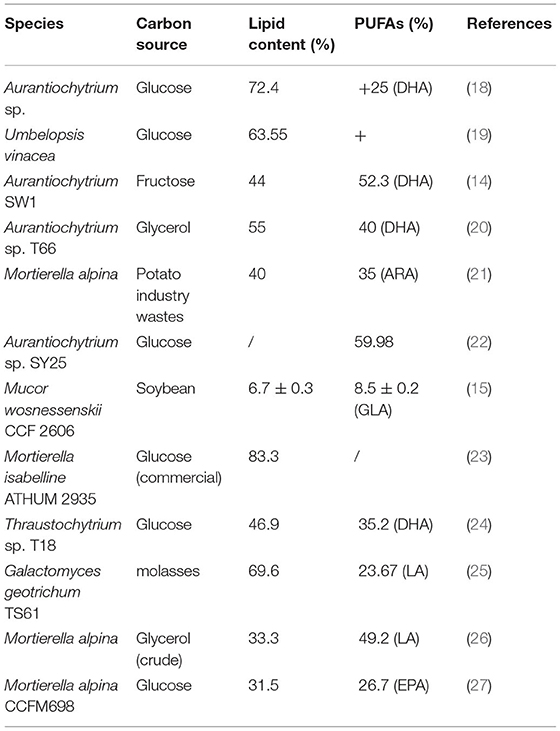
Similar to vegetable lipids, microbial lipids mainly include neutral fatty acids (FAs), free FAs and phospholipids (1, 2). Moreover, they share the same existence form with animal and plant lipids, i.e., existing in the cell structure such as membrane with constant content or forming lipid droplets or fat particles in the cytoplasm (3). The bright spheres in Figure 1 are lipid droplets in cells of different types of strains. Specifically, the outer layer of lipid droplets is a monolayer composed of phospholipids and specific proteins, and the inner core is mainly neutral lipids such as triacylglycerol (TAG) and sterol ester (SE) (4–7).
Microbial lipids are also widely used in the production of biodiesel. Excessive consumption and environmental damage caused by fossil fuels have hindered economic sustainable development (8, 9). Therefore, finding renewable and clean energy that can replace fossil energy is an important prerequisite for the development of green economy, energy conservation, emission reduction, and environmental protection (2). Methyl or ethyl fatty acid ester is obtained from methyl or ethyl esterification of FAs. The cellular lipids are mainly produced in the form of free FAs and acylglycerols (mostly as triglycerides) and are stored in the globular organelles called lipid bodies. Transesterification of microbial lipids is an essential step in microbial lipid production at both laboratory and commercial scale. Direct transesterification can considerably reduce costs, increase sample throughput and improve lipid yields (in particular fatty acid methyl esters, FAMEs). Fatty acid ethyl esters (FAEEs) are typically produced via the chemical transesterification of plant lipids and animal fats. Biosynthesis of FAEEs is limited by the supply of precursor lipids and acetyl-CoA (10–13).
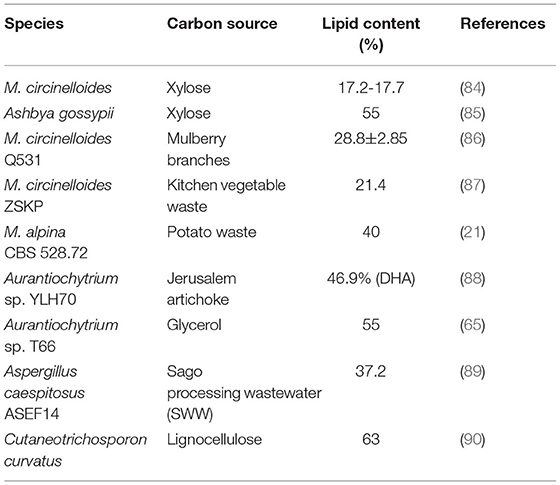
Oleaginous fungi, as lipid-producing microorganisms, are attractive in lipid production due to their short growth cycle, large amount of biomass and high yield of lipids (14, 15). Some filamentous fungi species have been reported able to accumulate polyunsaturated fatty acids (PUFAs), such as Mortierella isabellina, Mucor circinelloides, Pythium ultimum (2, 9). PUFAs play a vital role in human body; (PUFAs) belonging to the ω-3 and ω-6 classes are also substantial as precursors of eicosanoids or being structural components of various membrane phospholipids (9, 16, 17). Table 1 summarizes some high-yield fungi species and their lipid content.
As fungus-like protists, Thraustochytrids are progressively studied for his or her quicker growth rates and high lipid content (28). Thraustochytrids were first reported in 1936 and have attracted much attention since 1990 due to their high yield of FAs (29). The accumulated lipids account for more than 50% of dry cell weight (DCW), of which more than 25% is docosahexaenoic acid (DHA) with extremely high economic value. Aurantiochytrium sp. is a kind of thraustochytrids, which has a high yield of PUFAs, especially DHA (9). At present, the research on FA synthesis from Aurantiochytrium sp. is mainly focused on the optimization of culture conditions to increase the yield of unsaturated FAs, especially DHA (7).
Different from oleaginous yeasts, accumulation of fatty acids with important functions is the most attractive point of oleaginous fungi. However, unlike oleaginous yeasts, the recent developments of production, biosynthesis, and commercial applications of fatty acids from oleaginous fungi, have not been reviewed. In this article, we tried to summarize the studies of fatty acids from oleaginous fungi, and provided a point of reference.
Regulation of Fungal Fatty Acid Fermentation
Fungal lipid fermentation can be divided into two stages, i.e., the cell proliferation and the lipid accumulation (30). During the cell proliferation stage, cells proliferate and metabolize vigorously, with the nutrients in the medium consumed rapidly. During the fatty acid accumulation stage, the nitrogen source is exhausted but the carbon source is sufficient in the medium, which makes cells stop proliferating for the most part, with the lipid synthesis becoming the dominated metabolic activity (31–33).
According to the characteristics of fungal lipid fermentation production, controlling the nutrient composition of medium and regulating environmental conditions are a common strategy to promote lipid biosynthesis in lipid fermentation engineering. At present, three regulation strategies are widely adopted for lipid fermentation. The carbon-to-nitrogen (C/N) ratio, carbon and nitrogen sources, pH, incubation temperature and dissolved oxygen are the main factors influencing fatty acid production (18, 34). Nevertheless, other factors also play a crucial role in microbial activity, such as minerals (e.g., sulfur, zinc and phosphorus) and vitamins (e.g., thiamine and biotin) (35). Moreover, secondary metabolites (like citrate) are also an influencing factor for lipid production (34).
Promoting Lipid Accumulation by Nutrients Restriction
As for the de novo lipid accumulation, concentrations of nitrogen and carbon sources respectively determine the biomass content and the quantity of lipids in general (36). Accordingly, the C/N ratio is significant for the accumulated lipid content and the oleaginous microbial biomass (36–39). Previous studies demonstrate that the lipid accumulation is boosted at a C/N molar ratio of greater than 20. It is worth noting that the lipid production declines instead at the C/N ratio higher than 70 in some cases (40). Therefore, to achieve a high-level lipid accumulation, the initial C/N molar ratio should be optimized (41–43). For the lipid fermentation of the Thraustochytridae sp. PKU#Sw8, the increase in DHA production coincided with the up-regulation of gene expression under nitrogen-deficient culture conditions (44). Chen et al. (45) optimized the culture of Thraustochytrid sp. PKU#SW8 under optimal culture conditions (glycerol, 20 g/L; peptone, 2.5 g/L; 80% seawater; pH 4.0; 28°C), the cell mass, DHA concentration and yield of PKU#SW8 were increased to 7.5 ± 0.05 g/L, 2.14 ± 0.03 g/L and 282.9 ± 3.0 mg/g, respectively, on a 5-L scale fermentation.
Cellular lipid content and lipid yield were 62.2% and 0.205 g/g glucose, respectively, using a medium with a carbon to nitrogen (C/N) molar ratio of 6.1 and a C/P molar ratio of 9,552 (46), which means that the accumulation of lipid can also be regulated by limiting phosphorus in the medium. As a consequence, the regulation of phosphorus and sulfur limitation is of great significance for the production of lipids from nitrogen-rich crude materials (41, 45, 47, 48).
Promoting Lipid Accumulation by Small Molecules
Some small molecules can also regulate the accumulation of lipids (37, 48, 49). Li et al. (50) cultured Thraustochytriidae sp., a kind of marine oleaginous protists, by addition of different levels of sodium nitrate (1-50 mM) or urea (1-50 mM) in fermentation culture has a significant effect on fatty acid synthesis. They found that urine (50 mM) culture the cells accumulated 1.16 times the ω-3 PUFAs, of which DHA accounted for 49.49% and docosapentaenoic acid (DPA) was 5.28% compared with the original culture conditions. To sum up, it is easy to control lipid accumulation by small molecules, which is of great significance for the optimization of lipid production conditions (48, 51).
The accumulation of biomass and lipid-synthesizing fungi in any oily substance is highly affected by factors such as pH, temperature, light, and ventilation. Temperature change is also one of the factors affecting lipid accumulation (52, 53). They found that the low temperature has a significant impact on the formation of DHA, which can increase the DHA content from 43 to 65% of the total fatty acids. Low temperature may increase DHA content by facilitating a relatively large amount of substrates to enter the polyketide synthase (PKS) pathway (52).
Promoting Lipid Accumulation by Using Different Fermentation Modes
Generally speaking, there are three ways of microbial fermentation: batch culture, fed-batch culture and continuous culture (38, 54–57). The batch culture is the most widely used for lipid fermentation. Wang et al. (58) studied Schizochytrium sp. PKU#Mn4 and Thraustochytrid sp. PKU#Mn16, found that the largest DHA yields were 21% and 18.9%, and the yields were 27.6 mg/L-h and 31.9 mg/L-h, respectively in in 5-L bioreactor fermentation operated with optimal conditions and dual oxygen control strategy. The production of DHA increased by 3.4 times and 2.8 times (g/L) respectively. Rhizopus sp. using solid-state fermentation and submerged fermentation can produce valuable alternative feed ingredient due to their high protein and the well-balanced lipid content and amino acid profile (59).
In addition, electro-fermentation is a promising technology that can improve the performance of biological processes. When lipids are produced yeast R. toruloides under electro-fermentation conditions, the proportion of saturated FAs increases significantly from 37 to 50% (60).
Extraction of Fungal Fatty Acid
The conventional methods of wall breaking mainly include the following: grinding method, acid treatment, cell autolysis method, repeated freezing and thawing, ultrasonication and enzyme treatment (61–65). Among them, the autolysis method has simple steps and low cost, but has poor crushing result and low lipid yield; the enzymatic treatment method has mild conditions and no damage to intracellular substances, but is expensive and cannot be used for large-scale treatment. Ultrasonication is one in every of the additional normally used strategies. Using ultrasound to reinforce the synthesis of designer lipids, researchers have discovered an eco-friendly technique for enhancing the synthesis of designer lipids with numerous nutritional values (66).
The extraction of lipids is mostly done with low-boiling organic solvents. Commonly used solvents are ether, petroleum ether and chloroform. At present, the commonly used extraction methods of microbial lipids are as follows: acid heat method, Soxhlet extraction method, and supercritical CO2 extraction method (67–71). Among them, the Soxhlet extraction method is relatively accurate, but it is time-consuming and consumes too much organic solvent; the acid-heat method, although the yield is low, is fast and simple, and is suitable for the operation of multiple samples; the supercritical CO2 extraction method has high instrument requirements and requires Strictly control parameters (67, 71). The process of extracting lipid from fungi using acid-catalyzed predicament, microwave, and rapid ultrasonic-microwave treatment can create it have a high extraction rate, up to 70% (w/w) content (71). It is a novel green extraction method (63).
Cost Estimation of Fungal Fatty Acids
Take DHA as an example for cost estimation (72). If all the carbon sources needed to produce DHA were glucose, the amount of glucose required to produce 1 ton of biomass would be 2.78 tons. At the 2021 glucose price of us $903.9 per ton, it would cost US $2,512.84 to produce one ton of biomass. If there is only 50% lipid content in 1 ton of biomass, 40% of the lipid content is DHA (73). That's 0.2 tons of DHA. If one ton of DHA is produced, the calculated cost of glucose is $12,564.2. All this takes into account only glucose substrates, but if you add in other cost factors, including water and electricity, publicity, equipment, and so on, the cost of DHA increases further (72).
Recovery processes downstream of the fermenter typically contribute 60–80% to the cost of production of a fermentation product, therefore the fermentation step contributes only around 20–40% to the total production cost. The above analysis leads to the conclusion that the Schizochytrium limacinum grown on glucose cannot provide DHA cheaper than fish oil at present. If the biomass was used simply as an aquaculture feed additive, the downstream processing requirements would mostly disappear, although on the basis of equal DHA content, the biomass would still be more expensive than fish oil. Hence the need for cheaper nutrients for growing thraustochytrids (72).
In the future development, it is very important to improve production efficiency and reduce cost. First of all, in terms of carbon sources, the focus should be on replacing glucose while maintaining high biomass and lipid yields. In addition, lignocellulose hydrolysate may prove to be an inexpensive source of carbon for biomass production, which can efficiently metabolize xylose, and xylose metabolic engineering may help reduce fermentation costs (74). Metabolic engineering is also very important (75, 76). Therefore, strains should be improved, complete metabolic flux analysis should be carried out, and the protein engineering field should be evaluated with the goal of metabolic engineering, etc., in order to maximize the fatty acid production in the biomass.
Taking into consideration from another angle, developing new valuable products such as enzymes, and cell wall polysaccharides, during fungal fermentation besides fatty acids, would effectively reduce the cost. This “fungal-based biorefinery” strategy has not been applied in the studies, it may be the another choice to make the production of fatty acids more feasible (77).
Fatty Acid Production From Raw Materials
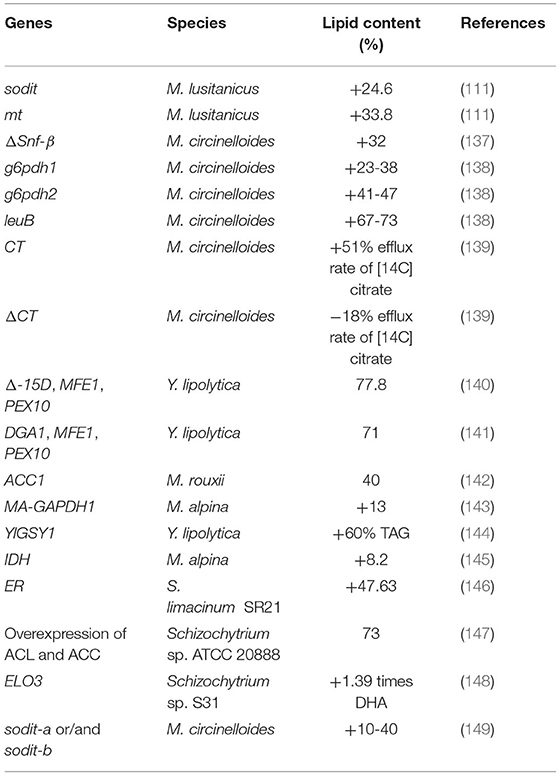
Glucose is the most basic carbon source of microorganisms. Many studies explore the glucose-based lipid accumulation of fungi, and the lipid content can reach higher than 70% (w/w) (41, 78–80). However, the large-scale production of microbial fatty acids with glucose as a raw material will face the problems of “competing with people for food” and “competing with food for land”, which necessitates the search for other suitable raw materials to reduce the costs (9, 81, 82). Recently, some cheap and available “raw materials” have been widely concerned, such as lignocellulose, non-grain sugar raw materials and commercial wastes (9, 79, 83). Table 2 summarizes some high-yielding fungal species that use non-glucose as substrates for lipid accumulation.
Lignocellulose
Lignocellulose is constituted by hemicellulose, cellulose and lignin (91). In recent years, lignocellulosic biomass has been recognized as a potential alternative feedstock to produce biofuels (9). The fatty acid production with lignocellulosic biomass includes the following two steps: (1) the degradation of biomass to corresponding monosaccharides by heat-acid treatment or enzyme hydrolysis, and (2) the biodegradable sugar fermentation by promising oleaginous microorganisms (92–95). Lignocellulose cannot be directly utilized, but must be hydrolyzed, which produces compounds that inhibit the growth of fungi, such as furan aldehydes, weak acids, and aromatic compounds, during the pretreatment process (96). The cumulative deleterious effects of some inhibitors (such as furfural, formic acid, acetic acid, and vanillin) on fatty acid accumulation in oleaginous fungi have been investigated (96, 97). Intasit et al. (98) used an integrated biotechnology, fungi and yeast to bioconvert lignocellulosic biomass into biodiesel, first pretreatment of the fungus, the fungus Aspergillus tubingensis TSIP 9 lipid yield 121.4 ± 2.7 mg/g-EFB (empty fruit bunch), the integrated biotechnology can greatly facilitate the conversion of lignocellulosic biomass to biodiesel feedstock is a cost-effective and sustainable biotransformation. Zhang et al. (84) deeply analyzed the effects of corn stover hydrolyzate on lipid accumulation by using xylose metabolism engineering strains of M. circinelloides strains. The results showed that the fatty acid contents of the engineered M. circinelloides strains were increased by 19.8% (in Mc-XI) and 22.3% (in Mc-XK), respectively, compared with the control strain.
Glucose and xylose coexist in lignocellulose hydrolysate. Lipid-producing yeasts consume glucose first and then xylose, and even some lipid-producing yeasts are unable to utilize xylose. Therefore, lignocellulose hydrolysate suffers from long fermentation cycle and low substrate utilization.
High-Carbonhydrate Plant Materials
Jerusalem artichoke is a kind of perennial plant resistant to barren, cold and drought. The planting of Jerusalem artichoke should not occupy cultivated land and other agricultural lands (99). The storage form of sugar in Jerusalem artichoke is inulin, which is a polyfructose linked by β-2,1 glycosidic bond with a glucose residue at the end (100). Yeast or molds can accumulate large amounts of lipids from inulin hydrolysates (101–103). In the medium containing inulin, fatty acids can be produced and the lipid content and biomass of cells can be changed. By converting the inulinase gene, the gene accumulates higher fatty acids (100). Aurantiochytrium sp. YLH70 can produce lipid in a medium with 695 mL/L hydrolysate of Jerusalem artichoke. The biomass higher biomass (32.71 g/L) and DHA content (46.9% of the total fatty acid) (88).
Commercial Organic Wastes
The combination of low-cost organic compounds contained in agro-commercial waste and the cultivation of lipid-producing microorganisms can effectively achieve the effect of accumulating lipids (104, 105). Lipid-producing microorganisms use some forms of carbon sources and nutrients for growth and fatty acid accumulation. Organic waste usually contains organic particles, which may be an ideal and inexpensive substrate for microbial fatty acid production, but the chemical composition of organic waste affects the lipid production of different species (104, 106). Lipid-producing yeasts can also transform commercial organic wastes into lipid (105, 107). Crude glycerol is a by-product of biodiesel industry, which is usually treated as commercial waste (64). The engineered strain of the filamentous fungus Ashbya gossypii can produce microbial lipids, whose efficiency is improved by three genomic manipulation methods. Using organic commercial waste as a raw material, the strain can accumulate lipid at about 40% of DCW (85). The use of commercial waste to produce lipid is also of great significance to environmental governance.
Synthesis Mechanism of Fatty Acids in Fungi
Essentially, the synthesis of microbial lipids is similar to that of animal and plant lipids. After the carboxylation of acetyl-CoA, saturated or unsaturated FAs are generated through chain extension and desaturation, and then triacylglycerols (TAGs) are formed.
General FA Biosynthesis
The synthesis of FAs in microbial cells requires acetyl-CoA acting as the precursor of FAs and a sufficient supply of NADPH to provide reducibility for the synthesis. It is generally believed that when nitrogen is depleted, the activity of AMP-deaminase increases. This can supplement for various metabolisms, decrease intracellular AMP level and the activity of isocitrate dehydrogenase (IDH) activated and cause the accumulation of isocitrate in mitochondria (108). Aconitase (AT) in mitochondria is able to catalyze the conversion of over-accumulated isocitrate to citric acid. The citric acid is then transported to the cytoplasm, where the ATP-citrate lyase (ACL) helps its cracking catalysis to acetyl-CoA and oxaloacetic acid (OAA). As a result, abundant acetyl-CoA is produced as the precursor of FAs (109). Acetyl-CoA directly participates in the FA synthesis, while oxaloacetate is first reduced to malate dehydrogenase (MD) and then undergoes oxidative decarboxylation in the presence of malic enzyme (ME) to release NADPH (110). Studies have shown that ME can regulate lipid accumulation in oleaginous microorganisms (111). Accordingly, if the activity of ME is inhibited, the lipid accumulation will decrease. This is because although many reactions in the cellular metabolic network can produce NADPH, the NADPH required by FA synthesis comes almost entirely from ME-catalyzed reactions (111). Catalyzed by acetyl-CoA carboxylase (ACC), acetyl-CoA and CO2 were transformed into monoyl-CoA. Multiple reactions can be continued in the presence of FAS. Acetyl-CoA combines with ACP to form acetyl-ACP, and malonyl-CoA and acetyl-CoA yield acyl-CoA via a condensation reaction. The three steps of reduction, dehydration and re-reduction are continued, and the FA chain extends by two carbon atoms. NADPH is taken as the reducing cofactor by FAS, and two NADPH molecules are required in each step of the acyl-CoA chain elongation. The chain is repeatedly extended to the desired length of the synthetic organism, and then some FAs are desaturated to form unsaturated FAs (112–114). The related reactions and enzymes are shown in Figure 2.
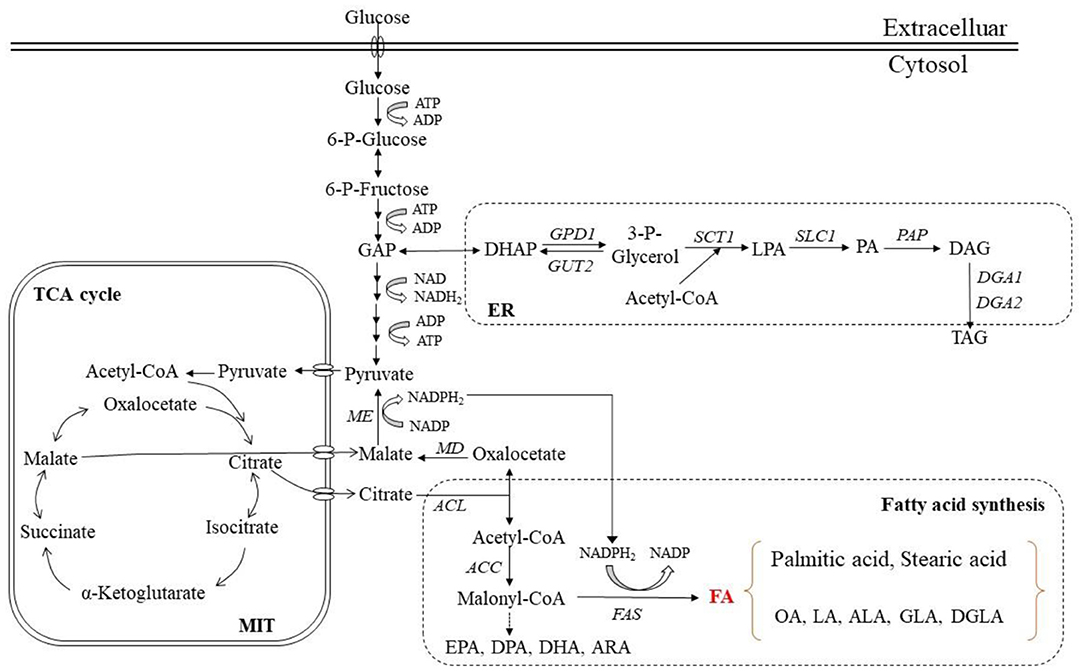
Figure 2. TAGs and fatty acid synthesis in microbial cells. MIT, Mitochondria; ER, Endoplasmic reticulum; ACL, ATP-citrate lyase; FAS, Fatty acid synthase; MD, Malic dehydrogenase; ME, Malic enzyme; ACC, Acetyl-CoA carboxylase; TCA, Tricarboxylic acid cycle; DAG, Diacylglycerol; PA, Phosphatidic acid; TAG, Triacylglycerol; FA, Fatty acid; OA, Oleic acid; LA, Linoleic acid; ALA, α-Linolenic acid; GLA, γ-Linolenic acid; DGLA, Dohomo-γ-linolenic acid; ARA, Arachidonic acid; EPA, Eicosapentaenoic acid; DPA, Docosapentaenoic acid; DHA, Docosahexaenoic acid.
PKS Pathway
The synthesis of some special FAs in microorganisms may be related to PKS. PKS is a sophisticated molecular machine responsible for synthesizing polyketides, which are natural products from the secondary metabolism with similarities to FA (115–117).
The DHA synthesis in Thraustochytrium, Schizochytrium limacinum, Aurantiochytrium sp. is considered to involve the PKS pathway. The successive condensation reactions of precursors catalyzed by PKS can form a variety of polyketides, and then numerous complex compounds are generated through modification reactions such as methylation, redox, glycosylation and hydroxylation (116, 118). In terms of structure and properties, PKS can be divided into three types: modular type (type I), repetition type (type II) and chalcone type (type III). The PKS found in fungi is mostly type I, which is large multifunctional proteins encoded by a single gene. It has multiple similar modules, and some domains are reused in the compound synthesis (117). The type I has a multidomain architecture whose active sites were distributed on large modules, while the type II is composed of monofunctional enzymes, with catalytic sites separated on different proteins (119). Type III polyketide synthases (PKSs) produce secondary metabolites with diverse biological activities, including antimicrobials (120, 121). In contrast to types I and II, type III PKSs are dimers of ketone synthases that undergo a series of reactions such as initiation of primer substrates, decarboxylation condensation of extended substrates, ring closure of growing polyketide chains and aromatization, and produce a variety of biologically active aromatic compounds (122). The PKS pathway of some species is shown in Figure 3 (119).
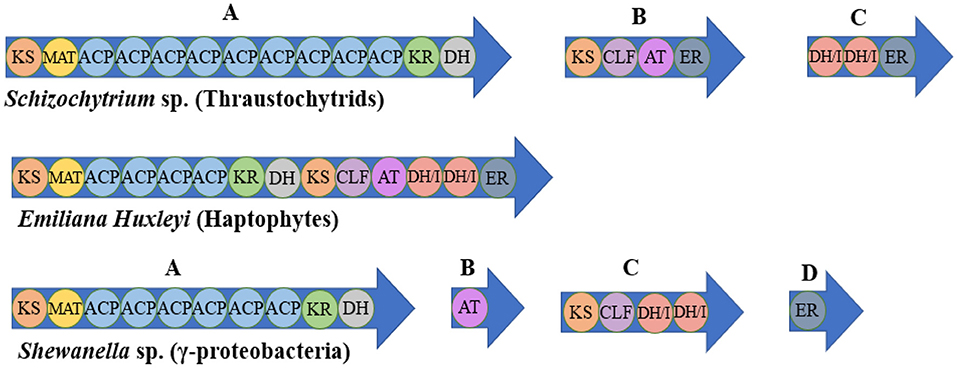
Figure 3. Examples of PUFA synthase organization in various representative organisms. KS, β-ketoacyl synthase; MAT, malonyl-CoA: ACP transacylase; ACP, acyl-carrier protein; KR, β-ketoreductase; DH, dehydratase; CLF, chain length factor; AT, acyl transferase; ER, enoyl-reductase; DH/I, dehydratase/isomerase.
TAG Synthesis
At present, TAGs are viewed as an important form of carbon source and energy storage unit in microorganisms. The TAG synthesis pathway is triggered at the point when carbon is abundant but nitrogen is depleted in the medium. The FA biosynthesis in cytosol involves several reactions which convert the precursor, acetyl-CoA, into long-chain FAs (123). The synthesized acyl-CoA has a typical chain length of 18 or 16 carbon atoms. These C18:0 and C16:0 molecules are then delivered to the endoplasmic reticulum (ER) to be further elongated and desaturated (124–126). The TAG synthesis requires a variety of enzymes, and phosphatidic acid (PA) and diglyceride are two key intermediates in anabolic metabolism.
Generally, the TAG synthesis involves the Kennedy pathway, with glycerol-3-phosphate (G3P) and acyl-CoA serving as the direct substrates in the process (126, 127). The first step of TAG assembly is the conversion of G3P into lysophosphatidic acid (LPA) with G3P acyltransferase (SCT1) as the catalyst (128). Subsequently, the LPA acylation occurs to generate PA in the presence of LPA acyltransferase (SLC1). Further, under the action of phosphatidic acid phosphatase (PAP), PA is dephosphorylated to produce diacylglycerol (DAG) (129). Finally, TAG is formed after the DAG acylation at the sn-3 position by an acyl-CoA-independent or acyl-CoA-dependent reaction (130, 131). Regarding the acyl-CoA-independent reaction, glycerophospholipid is the acyl group donor and phospholipid DAG acyl-transferase (LRO1) catalyzes the process. With respect to the acyl-CoA-dependent reaction, acyl-CoA acts as the final donor of acyl group and DAG acyltransferases, i.e., DGA1 or DGA2, are responsible for the catalysis. Furthermore, acting as the acyl-transferase of an acyl-CoA-dependent reaction, the steryl ester synthetase, which is encoded by ARE1, is proved able to promote the DAG acylation (123). The related enzymes and specific reactions are displayed in Figure 2.
TAG Degradation
The TAGs accumulated in cells store energy for them. Once carbon becomes insufficient, TAGs would be degraded with acetyl-CoA release so that the cellular metabolism can be maintained. Initially, free fatty acids (FFAs) can be produced from TAGs in the presence of intracellular lipases (TGL3 and TGL4) (132). These FFAs are activated by FAA1 to generate acyl-CoAs, which are then transported by specific transporters (Pxa1 and Pxa2) into the peroxisome (133). Alternatively, the transportation of the produced FFAs into the peroxisome takes place first, followed by the activation to acyl-CoAs therein by acyl/aryl-CoA ligase (AAL) (134). Afterward, acyl-CoAs are degraded in the peroxisome via the β-oxidation pathway to generate acetyl-CoA.
Metabolic Engineering of Oleaginous Fungi
Researchers have modified a variety of lipid-producing fungi to improve production efficiency of fatty acids. On the whole, these modifications can be divided into four categories: (1) enhancement of FA synthesis pathway, (2) enhancement of TAG synthesis pathway, (3) overexpression of key enzymes for providing cofactors, and (4) the blocking competitive pathway (32, 135, 136). Table 3 summarizes some studies on genetic modification of genes related to lipid synthesis.
Enhancement of FA Synthesis Pathway
Previous studies have shown that the expression of acetyl-CoA carboxylase (encoded by accA, accB, accC and accD) and thioesterase I (encoded by tesA) in Escherichia coli can speed up the FA synthesis by six times. This suggests that the catalytic reaction of acetyl-CoA carbohydrase is a rate-limiting step for FA synthesis (150–153). Overexpression of both heterologous Δ-15 desaturase (Δ-15D) sourced from flax and endogenous genes (SCD, ACC1, DGA1 and Δ-12D) along with the deletion of endogenous MFE1 and PEX10 can yield a superior lipid producer (140). It is able to produce lipid with content of 77.8% and titer of 50 g/L using glucose as the substrate in a 5 L stirred-tank bioreactor (140).
Han et al. (147) overexpressed in Schizochytrium sp. ATCC 20888 using the strong constitutive promoter ccg1, Schizochytrium ATP-citrate lyase (ACL) and acetyl-CoA carboxylase (ACC). The lipid content of overexpressed strains obtained by fermentation culture can reach a maximum of 73.0%, an increase of 38.3%. However, the ACC1 gene from mold M. rouxii is expressed in the Hansenula polymorpha, and the fat content is only 40% higher than the original (142), possibly because the fungus has a more powerful metabolic regulation system.
Enhancement of TAG Synthesis Pathway
Diacylglycerol acyltransferase (DGAT) catalyzes the conversion of DAG and acetyl-CoA to TAG, which is the last step in the TAG synthesis (32, 154, 155). When grown in batch conditions and minimal medium, the resulting strain consumes 12 g/L cellulose and accumulates 14% (DCW) lipids (156). Blazeck et al. (141) synergistically regulated multiple key genes related to the degradation and biosynthesis of lipids in Y. lipolytica with a combinatorial strategy, including MFE1, AMPD, PEX10, MAE, DGA1, ACL1, ACL2 and DGA2, with 57 distinct genotypes generated. The double deletion of MFE1 and PEX10 and the overexpression of DGA1 were most effective for modification. After the optimization of bioreaction conditions, the engineered strain had a lipid content of 71% (DCW) and a lipid titer of 25 g/L. Markedly, a 60-fold improvement was realized over the original strain.
Overexpression of Key Enzymes for Providing Cofactors
IDH and ME probably play a crucial role in the accumulation of lipids. When glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (PGD), IDH and ME are overexpressed in M. alpina, total FAs can be increased by 1.7 times; while ME2 is more effective in desaturation, and the content of arachidonic acid (AA) is increased by 1.5 times compared to the control (157). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is an enzyme highly conserved in the glycolytic pathway. The lipid-producing filamentous fungus M. alpina was used to characterize two copies of the gene encoding GAPDH, and the overexpression strain MA-GAPDH1 increases the lipid content by about 13% (143). First, the total lipid accumulation was increased by overexpressing a malic enzyme from Crypthecodinium cohnii to elevate NADPH supply. Then, the inhibition effect on acetyl-CoA carboxylase was relieved by overexpressing a codon-optimized ELO3 gene from M. alpina. After the above two-step engineering, contents of DHA was increased by 1.39-fold, reaching a level of 26.70% of dry cell weight, respectively (148).
The PKS cluster genes are supposed to synthesize PUFAs in S. limacinum. Ling et al. (146) improved lipid production domain expression by homologous recombination knocking out two enolate reductase (ER) genes located on the PKS cluster. The addition of triclosan as a modulator of the ER domain resulted in a 51.74% increase in PUFA production and a 47.63% increase in lipid production.
Blocking Competitive Pathways
The main competitive pathways blocking the lipid accumulation in microbial cells include β-oxidation of FAs, synthesis of phospholipids and conversion of phospho-enol-pyruvic acid (PEP) to oxalacetic acid. The β-oxidation occurs in peroxisomes, and peroxisome biogenesis is generally downregulated by the deletion of PEX3, PEX10 and PEX11 so that the degradation of TAGs can be prevented in commercial strains (158, 159). As an important intermediate metabolite, malate, its subcellular location and concentration have significant effects on fungal lipid metabolism. Yang et al. (149) deleted the two plasma membrane malate transporters “2-oxoglutarate:malate antiporter” (named SoDIT-a and SoDIT-b) of M. circinelloides WJ11 and analyzed their effects on growth ability, lipid accumulation and metabolism. Their results showed that the lipid content of the mutant was increased by ~10–40% compared to the control strain, indicating that defects in plasma membrane malate transport lead to an increase in malate for lipid synthesis.
Additionally, the genes involved in the β-oxidation pathway are often the deletion targets to increase lipid accumulation (160). After the characterization and deletion of the YlGSY1 gene encoding glycogen synthase, Bhutada et al. (144) increased the TAG accumulation of the engineered strain by 60% as compared with the wild-type strains. This proves that glycogen synthesis is a competing pathway, and its elimination is beneficial for the production of neutral lipids.
Commercial Applications of Fungal FAs
Recently, many researchers make efforts to explore the applications of microbial lipids in various fields from the food and health industry to the production of plasticizers, lubricants, spices and pesticides. Additionally, they are also promising intermediates in fine chemicals and other industries. This part mainly introduces the applications of polyunsaturated fatty acids from fungi. Polyunsaturated fatty acids (PUFAs) have received increasing attention for their beneficial effects on human health. PUFAs refer to long-chain FAs containing two or more double bonds, mainly including linoleic acid (LA), conjugated linoleic acid (CLA), γ-linolenic acid (GLA), AA, eicosapentaenoic acid (EPA), DHA, which are mostly the precursors of bioactive substances. They have the capabilities of anti-aging, anti-oxidation and anti-inflammation and are able to inhibit the formation, proliferation and metastasis of tumor cells and treat heart disease, hypertension, etc. Table 4 summarizes the production of unsaturated fatty acids by some fungi and their application functions.
DHA
DHA is an important member of the ω-3 PUFA family and has a wide distribution. Among marine organisms such as fish, shrimps, crabs, and seaweeds, DHA is particularly abundant in lipids of deep-sea fish (161). It can be produced in the Thraustochytrium, Schizochytrium limacinum, Aurantiochytrium sp. and so on. Although DHA can be produced from α-linolenic acid, the reaction rate is low, which thus necessitates its intake from diet (7). It accumulates in retinal tissue and gray matter in general and plays a key role in early visual and neural development (162). Besides, it is conducive to the development of the retinal, neuronal and immune systems at embryonic and post-natal stages (163, 164) and is effective to prevent cardiovascular disease, maintain brain and learning functions and protect inflammation response systems in adulthood (165). As a nutrient, DHA can be used in maternal and infant products. DHA and other unsaturated FAs in microalgae can be fully digested and absorbed by some aquatic organisms to meet the growth and development of juvenile fish and improve their survival rate (166).
EPA
EPA belongs to the ω-3 series of PUFAs. Natural phospholipids containing EPA are mainly found in the eggs and muscle tissues of marine animals. EPA can be produced in Mucorales, M. alpina and so on. Studies have revealed that EPA is able to protect the heart against the deleterious effects of sepsis in female rats. The following two reasons account for this beneficial action: (1) The anti-inflammatory activity of EPA which reduces the oxidative stress and preserves the energy metabolism through an increase in UCP3; (2) the incorporation of EPA in membrane phospholipids that increases the vasodilator reserve of the coronary microvessels (167, 168). EPA and DHA have various physiological functions such as reducing cholesterol content, resisting arteriosclerosis, preventing Alzheimer's disease and vision loss and improving brain function (164, 169). EPA and DHA are usually added to health foods and baby foods. High levels of EPA and DHA can be used as drugs to treat cardiovascular and cerebrovascular diseases, e.g., hyperlipidemia and arteriosclerosis. The preparation of high-purity EPA and DHA is the current deep processing target of fish lipids (27, 170). EPA is also an important functional component of breast milk, which is essential for the development of the baby's brain and vision. Therefore, more and more researchers are committed to applying EPA to infant milk powder in hope of improving its nutritional value through simulating the nutrients in breast milk (171).
ARA
ARA is an important member of the ω-6 PUFA family and has a wide distribution. Like DHA, which plays an important role in the development of infants' brains and retinas, it is one of the important factors affecting the quality of infant milk powder (21, 172). Mortierella, a fungus of the order Mucor, is a good producer of ARA. Research shows that ARA and DHA together constitute 20% of the weight of the human brainstem and are mainly concentrated in the outer neuron membrane and iliac sheath (173, 174). Bieren et al. (175) discovered that ARA metabolites can promote the occurrence of acute inflammation and produced pro-inflammatory mediators, such as PGE2 and PGfc. Lipoxin A4 derived from ARA can promote the degradation of lipid mediators. ARA accounts for 15–17% of the total FAs in skeletal muscle, which benefits the growth and repair of skeletal muscle tissue (176). Studies have shown that ARA supplementation can stimulate prostaglandin release and induce skeletal muscle hypertrophy through COX-2 dependent pathways (177).
GLA
GLA, one of the essential FAs, is an important component of biofilm (15). The microbial sources of GLA are mainly fungi and microalgae. For example, the microbial sources of GLA mainly include Spirulina (S. maxima, S. arthrospirulina), Mucor (M. rucus and M. microflora), Rhizopus, and Crucifera (15, 178, 179). GLA plays a significant physiological role in cardiovascular, immune, reproductive and endocrine systems. It is important because of its nutritional value and medicinal applications (180). GLA can act on lipoprotein enzyme and lipase to affect the formation and expression of TAGs, total cholesterol and very-low-density and low-density lipoproteins, thus having the capacity to lower blood lipid (181).
High-Value Chemicals Production From Oleaginous Fungi
In addition to the above-mentioned polyunsaturated fatty acids, some of these strains can also produce high levels of squalene and carotenoids, two other compounds of commercial value with rapidly growing market potential (182). Squalene has antioxidant and anticancer activities with broad applications in food and cosmetics industries. Besides, squalene has been used as vaccine adjuvant in vaccine formulations (43, 183). Since, the demand for squalene has increased during the last decade, microbial production of squalene has been investigated as a promising alternative source for traditional extraction methods from shark liver or plant lipids (184). Microbial strains are capable of producing non-polluting, low-cost, high-quality and sustainable sources of squalene, which is the main direction of the lipid-based biofuel industry.
Aurantiochytrium strains have the potential to produce large amounts of squalene, and Aurantiochytrium is known for its potential to produce large amounts of polyunsaturated DHA on a large scale (185). Furthermore, Thraustochytrid reported the co-production of squalene and DHA from inexpensive feedstocks such as organic solvent pretreatment spruce hydrolysis (186).
Conclusion
Accumulation of fatty acids with important functions is the most attractive point of oleaginous fungi. However, the cost limits application of the functional fatty acids. It is very important to improve production efficiency and reduce cost of the fatty acids. Replacing glucose with raw materials, while maintaining high biomass and lipid yields, was considered a feasible strategy. More fundamentally, many metabolic engineering strategies have been developed as efficient tools in oleaginous fungi to overcome the biochemical limit and to improve production efficiency of fatty acids. Particularly, the special kind of functional fatty acid can be enhanced by modifying the biosynthetic pathway with much higher yield. It also can be predictable that metabolic engineering can change the storage mode of fatty acids, even simplify the extraction. Thus, oleaginous fungi can be developed as hosts for high-value fatty acids and fatty acid-derived chemicals.
Author Contributions
Z-PW and X-JY made important contributions to the study conception and design. X-YZ participated in the drafting and revision of the manuscript. BL and B-CH conducted data analysis and interpretation. F-BW, Y-QZ, S-GZ, and ML performed literature research and organization. X-YL, JJ, and H-YW revised the manuscript. All authors read and approved this manuscript and agreed to be responsible for all aspects of the research to ensure the data accuracy and integrity of this work.
Funding
This research was supported by the Natural Science Foundation of Shandong Province (ZR2020MC003), the First Class Fishery Discipline Programme in Shandong Province, a special talent programme One Thing One Decision (YishiYiyi) Programme in Shandong Province, China, the Zhejiang Provincial Natural Science Foundation of China (No. LY18C010004), and the Talent Research Foundation of Qingdao Agricultural University (663/1117023, 663/1120036, and 663/1120058).
Conflict of Interest
PW was employed by company Linyang Group.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Darvishi F, Fathi Z, Ariana M, Moradi H. Yarrowia lipolytica as a workhorse for biofuel production. Biochem Eng J. (2017) 127:87–96. doi: 10.1016/j.bej.2017.08.013
2. Jones AD, Boundy-Mills KL, Barla GF, Kumar S, Ubanwa B, Balan V. Microbial Lipid Alternatives to Plant Lipids. Methods Mol Biol. (2019) 1995:1–32. doi: 10.1007/978-1-4939-9484-7_1
3. Zeng SY, Liu HH, Shi TQ, Song P, Ren LJ, Huang H, et al. Recent Advances in Metabolic Engineering of Yarrowia lipolytica for Lipid Overproduction. Eur J Lipid Sci Technol. (2018) 120:1700352. doi: 10.1002/ejlt.201700352
4. Wang ZP, Xu HM, Wang GY, Chi Z, Chi ZM. Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Biochim Biophys Acta. (2013) 1831:675–82. doi: 10.1016/j.bbalip.2012.12.010
5. Fillet S, Adrio JL. Microbial production of fatty alcohols. World J Microbiol Biotechnol. (2016) 32:152. doi: 10.1007/s11274-016-2099-z
6. Kikukawa H, Sakuradani E, Ando A, Shimizu S, Ogawa J. Arachidonic acid production by the oleaginous fungus Mortierella alpina 1S-4: a review. J Adv Res. (2018) 11:15–22. doi: 10.1016/j.jare.2018.02.003
7. Patel A, Rova U, Christakopoulos P, Matsakas L. Mining of squalene as a value-added byproduct from DHA producing marine thraustochytrid cultivated on food waste hydrolysate. Science of The Total Environment. (2020) 139691. doi: 10.1016/j.scitotenv.2020.139691
8. Hu L, Xu J, Zhou S, He A, Tang X, Lin L, et al. Catalytic Advances in the Production and Application of Biomass-Derived 2,5-Dihydroxymethylfuran. ACS Catal. (2018) 8:2959–80. doi: 10.1021/acscatal.7b03530
9. Yang Y, Heidari F, Hu B. Fungi (Mold)-based lipid production. Methods Mol Biol. (2019) 1995:51–89. doi: 10.1007/978-1-4939-9484-7_3
10. Gao Q, Cao X, Huang YY, Yang JL, Chen J, Wei LJ, et al. Overproduction of fatty acid ethyl esters by the oleaginous yeast Yarrowia lipolytica through metabolic engineering and process optimization. ACS Synth Biol. (2018) 7:1371–80. doi: 10.1021/acssynbio.7b00453
11. Thangavelu K, Sundararaju P, Srinivasan N. Uthandi S. Bioconversion of sago processing wastewater into biodiesel: Optimization of lipid production by an oleaginous yeast, Candida tropicalis ASY2 and its transesterification process using response surface methodology. Microb Cell Fact. (2021) 20:167. doi: 10.1186/s12934-021-01655-7
12. Langseter AM, Dzurendova S, Shapaval V, Kohler A, Ekeberg D, Zimmermann B. Evaluation and optimisation of direct transesterification methods for the assessment of lipid accumulation in oleaginous filamentous fungi. Microb Cell Fact. (2021) 20:59. doi: 10.1186/s12934-021-01542-1
13. Zhang Y, Guo X, Yang H, Shi S. The studies in constructing yeast cell factories for the production of fatty acid alkyl esters. Front Bioeng Biotechnol. (2022) 9:799032. doi: 10.3389/fbioe.2021.799032
14. Nazir Y, Shuib S, Kalil MS, Song Y, Hamid AA. Optimization of culture conditions for enhanced growth, lipid and docosahexaenoic acid (DHA) production of aurantiochytrium SW1 by response surface methodology. Sci Rep. (2018) 8:8909. doi: 10.1038/s41598-018-27309-0
15. Klempová T, Slaný O, Šišmiš M, Marcinčák S, Certík M. Dual production of polyunsaturated fatty acids and beta-carotene with Mucor wosnessenskii by the process of solid-state fermentation using agro-commercial waste. J Biotechnol. (2020) 311:1–11. doi: 10.1016/j.jbiotec.2020.02.006
16. Patel A, Matsakas L, Hruzová K, Rova U, Christakopoulos P. Biosynthesis of Nutraceutical Fatty Acids by the Oleaginous Marine Microalgae Phaeodactylum tricornutum Utilizing Hydrolysates from Organosolv-Pretreated Birch and Spruce Biomass. Mar Drugs. (2019) 17:119. doi: 10.3390/md17020119
17. Poliner E, Farré EM, Benning C. Advanced genetic tools enable synthetic biology in the oleaginous microalgae Nannochloropsis sp. Plant Cell Rep. (2018) 37:1383–99. doi: 10.1007/s00299-018-2270-0
18. Yu XJ, Huang CY, Chen H, Wang DS, Chen JL Li HJ, Liu XY, et al. High-Throughput Biochemical Fingerprinting of Oleaginous Aurantiochytrium sp. Strains by Fourier Transform Infrared Spectroscopy (FT-IR) for Lipid and Carbohydrate Productions Molecules. (2019) 24:1593. doi: 10.3390/molecules24081593
19. Dzurendova S, Zimmermann B, Tafintseva V, Kohler A, Ekeberg D, Shapaval V. The influence of phosphorus source and the nature of nitrogen substrate on the biomass production and lipid accumulation in oleaginous Mucoromycota fungi. Appl Microbiol Biotechnol. (2020) 104:8065–76. doi: 10.1007/s00253-020-10821-7
20. Heggeset TMB, Ertesvåg H, Liu B, Ellingsen TE, Vadstein O, Aasen IM. Lipid and DHA-production in Aurantiochytrium sp. —Responses to nitrogen starvation and oxygen limitation revealed by analyses of production kinetics and global transcriptomes. Sci Rep. (2019) 9:19470. doi: 10.1038/s41598-019-55902-4
21. Goyzueta-Mamani LD, de Carvalho JC, Magalhães AI Jr, Soccol CR. Production of arachidonic acid by Mortierella alpina using wastes from potato chips industry. J Appl Microbiol. (2021) 130:1592–601. doi: 10.1111/jam.14864
22. Abdel-Wahab MA, El-Samawaty AEMA, Elgorban AM, Bahkali AH. Fatty acid production of thraustochytrids from Saudi Arabian mangroves. Saudi J Biol Sci. (2021) 28:855–64. doi: 10.1016/j.sjbs.2020.11.024
23. Papanikolaou S, Rontou M, Belka A, Athenaki M, Gardeli C, Mallouchos A, et al. Conversion of biodiesel-derived glycerol into biotechnological products of commercial significance by yeast and fungal strains. Eng Life Sci. (2017) 17:262–81. doi: 10.1002/elsc.201500191
24. Lowrey J, Armenta RE, Brooks MS. Recycling of lipid-extracted hydrolysate as nitrogen supplementation for production of biomass. J Ind Microbiol Biotechnol. (2016) 43:1105–15. doi: 10.1007/s10295-016-1779-x
25. Altu R, Esim N, Aykutoglu G, Baltaci MO, Adiguzel A, Taskin M. Production of linoleic acid-rich lipids in molasses-based medium by oleaginous fungus Galactomyces geotrichum TS61. J Food Process Preserv. (2020) 44:e14518. doi: 10.1111/jfpp.14518
26. Dedyukhina EG, Chistyakova TI, Mironov AA, Kamzolova SV, Morgunov IG, Vainshtein MB. Arachidonic acid synthesis from biodiesel-derived waste by Mortierella alpina. Eur J Lipid Sci Technol. (2014) 116:429–37. doi: 10.1002/ejlt.201300358
27. Huang M, Chen H, Tang X, Lu H, Zhao J, Zhang H, et al. Two-stage pH control combined with oxygen-enriched air strategies for the highly efficient production of EPA by Mortierella alpina CCFM698 with fed-batch fermentation. Bioprocess Biosyst Eng. (2020) 43:1725–33. doi: 10.1007/s00449-020-02367-9
28. Du F, Wang YZ, Xu YS, Shi TQ, Liu WZ, Sun XM, et al. Biotechnological production of lipid and terpenoid from thraustochytrids. Biotechnol Adv. (2021) 48:107725. doi: 10.1016/j.biotechadv.2021.107725
29. Fossier Marchan L, Lee Chang KJ, Nichols PD, Mitchell WJ, Polglase JL, Gutierrez T. Taxonomy, ecology and biotechnological applications of thraustochytrids: a review. Biotechnol Adv. (2018) 36:26–46. doi: 10.1016/j.biotechadv.2017.09.003
30. Tang M, Zhou W, Liu Y, Yan J, Gong Z, A. two-stage process facilitating microbial lipid production from N-acetylglucosamine by Cryptococcus curvatus cultured under non-sterile conditions. Bioresour Technol. (2018) 258:255–62. doi: 10.1016/j.biortech.2018.03.015
31. Dias KB, Oliveira NML, Brasil BSAF, Vieira-Almeida EC, Paula-Elias FC, Almeida AF. Simultaneous high nutritional single cell oil and lipase production by Candida viswanathii. Acta Sci Pol Technol Aliment. (2021) 20:93–102. doi: 10.17306/J.AFS.0856
32. Dourou M, Aggeli D, Papanikolaou S, Aggelis G. Critical steps in carbon metabolism affecting lipid accumulation and their regulation in oleaginous microorganisms. Appl Microbiol Biotechnol. (2018) 102:2509–23. doi: 10.1007/s00253-018-8813-z
33. Dourou M, Mizerakis P, Papanikolaou S, Aggelis G. Storage lipid and polysaccharide metabolism in Yarrowia lipolytica and Umbelopsis isabellina. Appl Microbiol Biotechnol. (2017) 101:7213–26. doi: 10.1007/s00253-017-8455-6
34. Carsanba E, Papanikolaou S, Erten H. Production of oils and fats by oleaginous microorganisms with an emphasis given to the potential of the nonconventional yeast Yarrowia lipolytica. Crit Rev Biotechnol. (2018) 38:1230–43. doi: 10.1080/07388551.2018.1472065
35. Dzurendova S, Zimmermann B, Tafintseva V, Kohler A, Horn SJ, Shapaval V. Metal and Phosphate Ions Show Remarkable Influence on the Biomass Production and Lipid Accumulation in Oleaginous Mucor circinelloides. J Fungi (Basel). (2020) 6:260. doi: 10.3390/jof6040260
36. Shafiq M, Zeb L, Cui G, Jawad M, Chi Z. High-Density pH-Auxostat Fed-Batch Culture of Schizochytrium limacinum SR21 with Acetic Acid as a Carbon Source. Appl Biochem Biotechnol. (2020) 192:1163–75. doi: 10.1007/s12010-020-03396-6
37. Zhang H, Cui Q, Song X. Research advances on arachidonic acid production by fermentation and genetic modification of Mortierella alpina. World J Microbiol Biotechnol. (2021) 37:4. doi: 10.1007/s11274-020-02984-2
38. Subramanian AM, Nanjan SE, Prakash H, Santharam L, Ramachandran A, Sathyaseelan V, et al. Biokinetics of fed-batch production of poly (3-hydroxybutyrate) using microbial co-culture. Appl Microbiol Biotechnol. (2020) 104:1077–95. doi: 10.1007/s00253-019-10274-7
39. Pomraning KR, Kim YM, Nicora CD, Chu RK, Bredeweg EL, Purvine SO, et al. Multi-omics analysis reveals regulators of the response to nitrogen limitation in Yarrowia lipolytica. BMC Genomics. (2016) 17:138. doi: 10.1186/s12864-016-2471-2
40. Papanikolaou S, Aggelis G. Lipids of oleaginous yeasts. Part II: Technology and potential applications. Eur J Lipid Sci Technol. (2011) 113:1052–73. doi: 10.1002/ejlt.201100015
41. Li YH, Liu B, Zhao ZB. Optimization of culture conditions for lipid production by Rhodosporidium toruloides. Chin J Biotech. (2006) 22: 650–6. doi: 10.1016/S1872-2075(06)60050-2
42. Bellou S, Triantaphyllidou IE, Mizerakis P, Aggelis G. High lipid accumulation in Yarrowia lipolytica cultivated under double limitation of nitrogen and magnesium. J Biotechnol. (2016) 234:116–26. doi: 10.1016/j.jbiotec.2016.08.001
43. Shakeri S, Khoshbasirat F, Maleki M. Rhodosporidium sp. DR37: a novel strain for production of squalene in optimized cultivation conditions. Biotechnol Biofuels. (2021) 14:95. doi: 10.1186/s13068-021-01947-5
44. Chen X, He Y, Ye H, Xie Y, Sen B, Jiao N, et al. Different carbon and nitrogen sources regulated docosahexaenoic acid (DHA) production of Thraustochytriidae sp. PKU#SW8 through a fully functional polyunsaturated fatty acid (PUFA) synthase gene (pfaB). Bioresour Technol. (2020) 318:124273. doi: 10.1016/j.biortech.2020.124273
45. Chen X, Sen B, Zhang S, Bai M, He Y, Wang G. Chemical and physical culture conditions significantly influence the cell mass and docosahexaenoic acid content of Aurantiochytrium limacinum strain PKU#SW8. Mar Drugs. (2021) 19:671. doi: 10.3390/md19120671
46. Wu S, Hu C, Jin G, Zhao X, Zhao ZK. Phosphate-limitation mediated lipid production by Rhodosporidium toruloides. Bioresour Technol. (2010) 101: 6124-6129. doi: 10.1016/j.biortech.2010.02.111
47. Wang Y, Zhang S, Zhu Z, Shen H, Lin X, Jin X, et al. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol Biofuels. (2018) 11:148. doi: 10.1186/s13068-018-1134-8
48. Kamal R, Shen H, Li Q, Wang Q, Yu X, Zhao ZK. Utilization of amino acid-rich wastes for microbial lipid production. Appl Biochem Biotechnol. (2020) 191:1594–604. doi: 10.1007/s12010-020-03296-9
49. Tchakouteu SS, Kopsahelis N, Chatzifragkou A, Kalantzi O, Stoforos NG, Koutinas AA, et al. Rhodosporidium toruloides cultivated in NaCl-enriched glucose-based media: adaptation dynamics and lipid production. Eng Life Sci. (2016) 17:237–48. doi: 10.1002/elsc.201500125
50. Li S, Hu Z, Yang X, Li Y. Effect of nitrogen sources on omega-3 polyunsaturated fatty acid biosynthesis and gene expression in Thraustochytriidae sp. Mar Drugs. (2020) 18:612. doi: 10.3390/md18120612
51. Wu SG, Zhao X, Hu CM, Zhang SF, Hua YY, Zhao ZB. Screening of fungi for microbial oil production using N-acetyl-D-glucosamine. China Biotechnol. (2008) 28: 58-62.
52. Hu F, Clevenger AL, Zheng P, Huang Q, Wang Z. Low-temperature effects on docosahexaenoic acid biosynthesis in Schizochytrium sp. TIO01 and its proposed underlying mechanism. Biotechnol Biofuels. (2020) 13:172. doi: 10.1186/s13068-020-01811-y
53. Aussant J, Guihéneuf F, Stengel DB. Impact of temperature on fatty acid composition and nutritional value in eight species of microalgae. Appl Microbiol Biotechnol. (2018) 102:5279–97. doi: 10.1007/s00253-018-9001-x
54. Huang XF, Shen Y, Luo HJ, Liu JN, Liu J. Enhancement of extracellular lipid production by oleaginous yeast through preculture and sequencing batch culture strategy with acetic acid. Bioresour Technol. (2018) 247:395–401. doi: 10.1016/j.biortech.2017.09.096
55. Abeln F, Hicks RH, Auta H, Moreno-Beltrán M, Longanesi L, Henk DA, et al. Semi-continuous pilot-scale microbial oil production with Metschnikowia pulcherrima on starch hydrolysate. Biotechnol Biofuels. (2020) 13:127. doi: 10.1186/s13068-020-01756-2
56. Martínez-Avila O, Sánchez A, Font X, Barrena R. Fed-batch and sequential-batch approaches to enhance the bioproduction of 2-phenylethanol and 2-phenethyl acetate in solid-state fermentation residue-based systems. J Agric Food Chem. (2019) 67:3389–99. doi: 10.1021/acs.jafc.9b00524
57. Abeln F, Chuck CJ. Achieving a high-density oleaginous yeast culture: comparison of four processing strategies using Metschnikowia pulcherrima. Biotechnol Bioeng. (2019) 116:3200–14. doi: 10.1002/bit.27141
58. Wang Q, Ye H, Sen B, Xie Y, He Y, Park S, et al. Improved production of docosahexaenoic acid in batch fermentation by newly-isolated strains of Schizochytrium sp. and Thraustochytriidae sp through bioprocess optimization. Synth Syst Biotechnol. (2018) 3:121–9. doi: 10.1016/j.synbio.2018.04.001
59. Ibarruri J, Cebrián M, Hernández I. Valorisation of fruit and vegetable discards by fungal submerged and solid-state fermentation for alternative feed ingredients production. J Environ Manage. (2021) 281:111901. doi: 10.1016/j.jenvman.2020.111901
60. Arbter P, Sinha A, Troesch J, Utesch T, Zeng AP. Redox governed electro-fermentation improves lipid production by the oleaginous yeast Rhodosporidium toruloides. Bioresour Technol. (2019) 294:122122. doi: 10.1016/j.biortech.2019.122122
61. Kot AM, Gientka I, Bzducha-Wróbel A, Błazejak S, Kurcz A. Comparison of simple and rapid cell wall disruption methods for improving lipid extraction from yeast cells. J Microbiol Methods. (2020) 176:105999. doi: 10.1016/j.mimet.2020.105999
62. Garay LA, Boundy-Mills KL, German JB. Accumulation of high-value lipids in single-cell microorganisms: a mechanistic approach and future perspectives. J Agricult Food Chem. (2014) 62: 2709–27. doi: 10.1021/jf4042134
63. Martínez JM, Delso C, Aguilar DE, Álvarez I, Raso J. Organic-solvent-free extraction of carotenoids from yeast Rhodotorula glutinis by application of ultrasound under pressure. Ultrason Sonochem. (2019) 61:104833. doi: 10.1016/j.ultsonch.2019.104833
64. Kumar LR, Yellapu SK, Tyagi RD, Zhang X, A. review on variation in crude glycerol composition, bio-valorization of crude and purified glycerol as carbon source for lipid production. Bioresour Technol. (2019) 293:122155. doi: 10.1016/j.biortech.2019.122155
65. Jeevan Kumar SP, Banerjee R. Enhanced lipid extraction from oleaginous yeast biomass using ultrasound assisted extraction: a greener and scalable process. Ultrason Sonochem. (2019) 52:25–32. doi: 10.1016/j.ultsonch.2018.08.003
66. Jadhav HB, Gogate PR, Waghmare JT, Annapure US. Intensified synthesis of palm olein designer lipids using sonication. Ultrason Sonochem. (2021) 73:105478. doi: 10.1016/j.ultsonch.2021.105478
67. Ong CC, Chen YH. Investigation on cell disruption techniques and supercritical carbon dioxide extraction of Mortierella alpina Lipid. Foods. (2022) 11:582. doi: 10.3390/foods11040582
68. Breil C, Abert Vian M, Zemb T, Kunz W, Chemat F. “Bligh and Dyer” and folch methods for solid-liquid-liquid extraction of lipids from microorganisms. comprehension of solvatation mechanisms and towards substitution with alternative solvents. Int J Mol Sci. (2017) 18:708. doi: 10.3390/ijms18040708
69. Meullemiestre A, Breil C, Abert-Vian M, Chemat F. Microwave, ultrasound, thermal treatments, and bead milling as intensification techniques for extraction of lipids from oleaginous Yarrowia lipolytica yeast for a biojetfuel application. Biores Technol. (2016) 211: 190–199. doi: 10.1016/j.biortech.2016.03.040
70. Svenning JB, Dalheim L, Vasskog T, Matricon L, Vang B, Olsen RL. Lipid yield from the diatom Porosira glacialis is determined by solvent choice and number of extractions, independent of cell disruption. Sci Rep. (2020) 10:22229. doi: 10.1038/s41598-020-79269-z
71. Patel A, Arora N, Pruthi V, Pruthi PA, A. novel rapid ultrasonication-microwave treatment for total lipid extraction from wet oleaginous yeast biomass for sustainable biodiesel production. Ultrason Sonochem. (2019) 51:504–16. doi: 10.1016/j.ultsonch.2018.05.002
72. Chi G, Xu Y, Cao X, Li Z, Cao M, Chisti Y, et al. Production of polyunsaturated fatty acids by Schizochytrium (Aurantiochytrium) spp. Biotechnol Adv. (2022) 55:107897. doi: 10.1016/j.biotechadv.2021.107897
73. Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. (2001) 293:290–3. doi: 10.1126/science.1059593
74. Merkx-Jacques A, Rasmussen H, Muise DM, Benjamin JJR, Kottwitz H, Tanner K, et al. Engineering xylose metabolism in thraustochytrid T18. Biotechnol Biofuels. (2018) 11:248. doi: 10.1186/s13068-018-1246-1
75. Shene C, Paredes P, Flores L, Leyton A, Asenjo JA, Chisti Y. Dynamic flux balance analysis of biomass and lipid production by Antarctic thraustochytrid Oblongichytrium sp. RT2316-13. Biotechnol Bioeng. (2020) 117:3006-3017. doi: 10.1002/bit.27463
76. Simensen V, Voigt A, Almaas E. High-quality genome-scale metabolic model of Aurantiochytrium sp. T66 Biotechnol Bioeng. (2021) 118:2105–17. doi: 10.1002/bit.27726
77. Hu L, He A, Liu X, Xia J, Xu J, Zhou S, et al. Biocatalytic transformation of 5-hydroxymethylfurfural into high-value derivatives: recent advances and future aspects. ACS Sustain Chem Eng. (2018) 6:15915–35. doi: 10.1021/acssuschemeng.8b04356
78. Chatterjee S. Venkata Mohan S. Fungal biorefinery for sustainable resource recovery from waste. Bioresour Technol. (2022) 345:126443. doi: 10.1016/j.biortech.2021.126443
79. Spagnuolo M, Shabbir Hussain M, Gambill L, Blenner M. Alternative substrate metabolism in Yarrowia lipolytica. Front Microbiol. (2018) 9:1077. doi: 10.3389/fmicb.2018.01077
80. Zhu LY, Zong MH, Wu H. Efficient lipid production with Trichosporon fermentans and its use for biodiesel preparation. Bioresour Technol. (2008) 99: 7881-7885. doi: 10.1016/j.biortech.2008.02.033
81. Zhang Y, He Y, Yang W, Tan F, Li W. Wang Q. Sheng Wu Gong Cheng Xue Bao. (2022) 38:565–77. doi: 10.13345/j.cjb.210218
82. Ruan Z, Zanotti M, Zhong Y, Liao W, Ducey C, Liu Y. Co-hydrolysis of lignocellulosic biomass for microbial lipid accumulation. Biotechnol Bioeng. (2013) 110:1039–49. doi: 10.1002/bit.24773
83. Celińska E, Nicaud JM, Białas W. Hydrolytic secretome engineering in Yarrowia lipolytica for consolidated bioprocessing on polysaccharide resources: review on starch, cellulose, xylan, and inulin. Appl Microbiol Biotechnol. (2021) 105:975–89. doi: 10.1007/s00253-021-11097-1
84. Zhang Y, Song Y. Lipid Accumulation by Xylose Metabolism Engineered Mucor circinelloides Strains on Corn Straw Hydrolysate. Appl Biochem Biotechnol. (2021) 193:856–68. doi: 10.1007/s12010-020-03427-2
85. Díaz-Fernández D, Aguiar TQ, Martín VI, Romaní A, Silva R, Domingues L, et al. Microbial lipids from commercial wastes using xylose-utilizing Ashbya gossypii strains. Bioresour Technol. (2019) 293:122054. doi: 10.1016/j.biortech.2019.122054
86. Qiao W, Tao J, Luo Y, Tang T, Miao J, Yang Q. Microbial oil production from solid-state fermentation by a newly isolated oleaginous fungus, Mucor circinelloides Q531 from mulberry branches. R Soc Open Sci. (2018) 5:180551. doi: 10.1098/rsos.180551
87. Zininga JT, Puri AK, Govender A, Singh S, Permaul K. Concomitant production of chitosan and lipids from a newly isolated Mucor circinelloides ZSKP for biodiesel production. Bioresour Technol. (2019) 272: 545–51. doi: 10.1016/j.biortech.2018.10.035
88. Yu XJ, Liu JH, Sun J, Zheng JY, Zhang YJ, Wang ZX. Docosahexaenoic acid production from the acidic hydrolysate of Jerusalem artichoke by an efficient sugar-utilizing Aurantiochytrium sp. YLH70. Commercial Crops and Products. (2016) 83: 372-378. doi: 10.1016/j.indcrop.2016.01.013
89. Srinivasan N, Thangavelu K, Sekar A, Sanjeev B, Uthandi S. Aspergillus caespitosus ASEF14, an oleaginous fungus as a potential candidate for biodiesel production using sago processing wastewater (SWW). Microb Cell Fact. (2021) 20:179. doi: 10.1186/s12934-021-01667-3
90. Di Fidio N, Liuzzi F, Mastrolitti S, Albergo R, De Bari I. Single Cell Oil Production from Undetoxified Arundo donax L. hydrolysate by Cutaneotrichosporon curvatus. J Microbiol Biotechnol. (2019) 29:256–67. doi: 10.4014/jmb.1808.08015
91. Kim JY, Lee HW, Lee SM, Jae J, Park YK. Overview of the recent advances in lignocellulose liquefaction for producing biofuels, bio-based materials and chemicals. Bioresour Technol. (2019) 279:373–84. doi: 10.1016/j.biortech.2019.01.055
92. Huang C, Chen XF, Xiong L, Chen XD, Ma LL, Chen Y. Single cell oil production from low-cost substrates: the possibility and potential of its commercialization. Biotechnol Adv. (2013) 31:129–39. doi: 10.1016/j.biotechadv.2012.08.010
93. Kothri M, Mavrommati M, Elazzazy AM, Baeshen MN, Moussa TAA, Aggelis G. Microbial sources of polyunsaturated fatty acids (PUFAs) and the prospect of organic residues and wastes as growth media for PUFA-producing microorganisms. FEMS Microbiol Lett. (2020) 367:fnaa 028. doi: 10.1093/femsle/fnaa028
94. Xu J, Liu X, He J, Hu L, Dai B, Wu B. Enzymatic in situ saccharification of rice straw in aqueous-ionic liquid media using encapsulated Trichoderma aureoviride cellulase. J Chem Technol Biotechnol. (2014) 90:57–63. doi: 10.1002/jctb.4458
95. Zhao CH, Liu X, Zhan T, He J. Production of cellulase by Trichoderma reesei from pretreated straw and furfural residues. RSC Adv. (2018) 8:36233–8. doi: 10.1039/C8RA05936E
96. Konzock O, Zaghen S, Norbeck J. Tolerance of Yarrowia lipolytica to inhibitors commonly found in lignocellulosic hydrolysates. BMC Microbiol. (2021) 21:77. doi: 10.1186/s12866-021-02126-0
97. Chen X, Li Z, Zhang X, Hu F, Ryu DD, Bao J. Screening of oleaginous yeast strains tolerant to lignocellulose degradation compounds. Appl Biochem Biotechnol. (2009) 159:591–604. doi: 10.1007/s12010-008-8491-x
98. Intasit R, Cheirsilp B, Louhasakul Y, Boonsawang P. Consolidated bioprocesses for efficient bioconversion of palm biomass wastes into biodiesel feedstocks by oleaginous fungi and yeasts. Bioresour Technol. (2020) 315:123893. doi: 10.1016/j.biortech.2020.123893
99. Hughes SR, Qureshi N, López-Núñez JC, Jones MA, Jarodsky JM, Galindo-Leva LÁ, et al. Utilization of inulin-containing waste in commercial fermentations to produce biofuels and bio-based chemicals. World J Microbiol Biotechnol. (2017) 33:78. doi: 10.1007/s11274-017-2241-6
100. Zhao CH, Cui W, Liu XY, Chi ZM, Madzak C. Expression of inulinase gene in the oleaginous yeast Yarrowia lipolytica and single cell oil production from inulin-containing materials. Metab Eng. (2010) 12:510–7. doi: 10.1016/j.ymben.2010.09.001
101. Bao R, Wu X, Liu S, Xie T, Yu C, Lin X. Efficient Conversion of Fructose-Based Biomass into Lipids with Trichosporon fermentans Under Phosphate-Limited Conditions. Appl Biochem Biotechnol. (2018) 184:113–23. doi: 10.1007/s12010-017-2536-y
102. Wang G, Liu L, Liang W. Single Cell Oil Production from Hydrolysates of Inulin by a Newly Isolated Yeast Papiliotrema laurentii AM113 for Biodiesel Making. Appl Biochem Biotechnol. (2018) 184:168–81. doi: 10.1007/s12010-017-2538-9
103. Wang GY, Chi Z, Song B, Wang ZP, Chi ZM. High level lipid production by a novel inulinase-producing yeast Pichia guilliermondii Pcla22. Biores Technol. (2012) 124:77–82. doi: 10.1016/j.biortech.2012.08.024
104. Cho HU, Park JM. Biodiesel production by various oleaginous microorganisms from organic wastes. Bioresour Technol. (2018) 256:502–8. doi: 10.1016/j.biortech.2018.02.010
105. Diwan B, Parkhey P, Gupta P. From agro-commercial wastes to single cell oils: a step towards prospective biorefinery. Folia Microbiol (Praha). (2018) 63:547–68. doi: 10.1007/s12223-018-0602-7
106. Wang ZP, Wang PK, Ma Y, Lin JX, Wang CL, Zhao YX, et al. Laminaria japonica hydrolysate promotes fucoxanthin accumulation in Phaeodactylum tricornutum. Bioresour Technol. (2022) 344:126117. doi: 10.1016/j.biortech.2021.126117
107. Zhang L, Loh KC, Kuroki A, Dai Y, Tong YW. Microbial biodiesel production from commercial organic wastes by oleaginous microorganisms: Current status and prospects. J Hazard Mater. (2021) 402:123543. doi: 10.1016/j.jhazmat.2020.123543
108. Tang W, Zhang SF, Wang Q, Tan HD, Zhao ZK. The isocitrate dehydrogenase gene of oleaginous yeast Lipomyces starkeyi is linked to lipid accumulation. Can J Microbiol (2009) 55: 1062-1069. doi: 10.1139/W09-063
109. Yang J, Khan MAK, Zhang H, Zhang Y, Certik M, Garre V, et al. Mitochondrial citrate transport system in the fungus mucor circinelloides: identification, phylogenetic analysis, and expression profiling during growth and lipid accumulation. Curr Microbiol. (2020) 77:220–31. doi: 10.1007/s00284-019-01822-5
110. Zhao L, Cánovas-Márquez JT, Tang X, Chen H, Chen YQ, Chen W, et al. Role of malate transporter in lipid accumulation of oleaginous fungus Mucor circinelloides. Appl Microbiol Biotechnol. (2016) 100:1297–305. doi: 10.1007/s00253-015-7079-y
111. Wang X, Mohamed H, Bao Y, Wu C, Shi W, Song Y, et al. Heterologous expression of two malate transporters from an oleaginous fungus Mucor circinelloides improved the lipid accumulation in Mucor lusitanicus. Front Microbiol. (2021) 12:774825. doi: 10.3389/fmicb.2021.774825
112. Arhar S, Gogg-Fassolter G, Ogrizović M, Pačnik K, Schwaiger K, Žganjar M, et al. Engineering of Saccharomyces cerevisiae for the accumulation of high amounts of triacylglycerol. Microb Cell Fact. (2021) 20:147. doi: 10.1186/s12934-021-01640-0
113. Katja K Dove, Malika F Kadirova, Sara M Nowinski, Yeyun Ouyang, Jon G Van Vranken, Jared Rutter. Exploring the functional role of an ancient mitochondrial fatty acid synthesis pathway. The FASEB Journal (2019) 33: 660.5-660.5.
114. Nosheen S, Yang J, Naz T, Nazir Y, Ahmad MI, Fazili ABA Li S, et al. Annotation of AMP-activated protein kinase genes and its comparative transcriptional analysis between high and low lipid producing strains of Mucor circinelloides. Biotechnol Lett. (2021) 43:193–202. doi: 10.1007/s10529-020-02990-2
115. Schaub AJ, Moreno GO, Zhao S, Truong HV, Luo R, Tsai SC. Computational structural enzymology methodologies for the study and engineering of fatty acid synthases, polyketide synthases and nonribosomal peptide synthetases. Methods Enzymol. (2019) 622:375–409. doi: 10.1016/bs.mie.2019.03.001
116. Chen X, Xu M, Feng C, Hu C. Progress in fungal polyketide biosynthesis. Sheng Wu Gong Cheng Xue Bao. (2018) 34:151–64. doi: 10.13345/j.cjb.170219
117. Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep. (2007) 24:1041–72. doi: 10.1039/b603600g
118. Nicholson TP, Rudd BA, Dawson M, Lazarus CM, Simpson TJ, Cox RJ. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem Biol. (2001) 8:157–78. doi: 10.1016/S1074-5521(00)90064-4
119. Morabito C, Bournaud C, Maës C, Schuler M, Aiese Cigliano R, Dellero Y, et al. The lipid metabolism in thraustochytrids. Prog Lipid Res. (2019) 76:101007. doi: 10.1016/j.plipres.2019.101007
120. Navarro-Muñoz JC, Collemare J. Evolutionary histories of type III polyketide synthases in fungi. Front Microbiol. (2020) 10:3018. doi: 10.3389/fmicb.2019.03018
121. Larsen JS, Pearson LA, Neilan BA. Genome Mining and Evolutionary Analysis Reveal Diverse Type III Polyketide Synthase Pathways in Cyanobacteria. Genome Biol Evol. (2021) 13:evab056. doi: 10.1093/gbe/evab056
122. Manoharan G, Sairam T, Thangamani R, Ramakrishnan D. K Tiwari M, Lee JK, Marimuthu J. Identification and characterization of type III polyketide synthase genes from culturable endophytes of ethnomedicinal plants. Enzyme Microb Technol. (2019) 131:109396. doi: 10.1016/j.enzmictec.2019.109396
123. Wang J, Ledesma-Amaro R, Wei Y, Ji B, Ji XJ. Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica—a Review. Bioresour Technol. (2020) 313:123707. doi: 10.1016/j.biortech.2020.123707
124. Zoni V, Khaddaj R, Campomanes P, Thiam AR, Schneiter R, Vanni S. Pre-existing bilayer stresses modulate triglyceride accumulation in the ER versus lipid droplets. Elife. (2021) 10:e62886. doi: 10.7554/eLife.62886
125. Ledesma-Amaro R, Nicaud JM. Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog Lipid Res. (2016) 61:40–50. doi: 10.1016/j.plipres.2015.12.001
126. Rogers S, Henne WM. Analysis of Neutral Lipid Synthesis in Saccharomyces cerevisiae by Metabolic Labeling and Thin Layer Chromatography. J Vis Exp. (2021). doi: 10.3791/62201. [Epub ahead of print].
127. Papanikolaou S, Aggelis G. Modeling lipid accumulation and degradation in Yarrowia lipolytica cultivated on commercial fats. Curr Microbiol. (2003) 46 :0398-0402. doi: 10.1007/s00284-002-3907-2
128. Beopoulos A, Mrozova Z, Thevenieau F, Le Dall MT, Hapala I, Papanikolaou S, et al. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol. (2008) 74:7779–89. doi: 10.1128/AEM.01412-08
129. Silverman AM, Qiao K, Xu P, Stephanopoulos G. Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol Biotechnol. (2016) 100:3781–98. doi: 10.1007/s00253-016-7376-0
130. Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng. (2013) 15:1–9. doi: 10.1016/j.ymben.2012.08.007
131. Beopoulos A, Chardot T, Nicaud J-M. Yarrowia lipolytica: a model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie. (2009) 91:692–6. doi: 10.1016/j.biochi.2009.02.004
132. Dulermo T, Tréton B, Beopoulos A, Kabran Gnankon AP, Haddouche R, Nicaud JM. Characterization of the two intracellular lipases of Y. lipolytica encoded by TGL3 and TGL4 genes: new insights into the role of intracellular lipases and lipid body organisation. Biochim Biophys Acta Mol Cell Biol Lipids. (2013) 1831:1486–95. doi: 10.1016/j.bbalip.2013.07.001
133. Dulermo R, Gamboa-Meléndez H, Ledesma-Amaro R, Thévenieau F, Nicaud JM. Unraveling fatty acid transport and activation mechanisms in Yarrowia lipolytica. Biochim Biophys Acta Mol Cell Biol Lipids. (2015) 1851:1202–17. doi: 10.1016/j.bbalip.2015.04.004
134. Dulermo R, Gamboa-Meléndez H, Ledesma-Amaro R, Thevenieau F, Nicaud JM. Yarrowia lipolytica AAL genes are involved in peroxisomal fatty acid activation. Biochim Biophys Acta Mol Cell Biol Lipids. (2016) 1861:555–65. doi: 10.1016/j.bbalip.2016.04.002
135. Zhu X, Li S, Liu L, Li S, Luo Y, Lv C, et al. Genome Sequencing and Analysis of Thraustochytriidae sp. SZU445 provides novel insights into the polyunsaturated fatty acid biosynthesis pathway. Mar Drugs. (2020) 18:118. doi: 10.3390/md18020118
136. Abdel-Mawgoud AM, Markham KA, Palmer CM, Liu N, Stephanopoulos G, Alper HS. Metabolic engineering in the host Yarrowia lipolytica. Metab Eng. (2018) 50:192–208. doi: 10.1016/j.ymben.2018.07.016
137. Nosheen S, Naz T, Yang J, Hussain SA, Fazili ABA, Nazir Y, et al. Role of Snf-β in lipid accumulation in the high lipid-producing fungus Mucor circinelloides WJ11. Microb Cell Fact. (2021) 20:52. doi: 10.1186/s12934-021-01545-y
138. Tang X, Chen H, Gu Z, Zhang H, Chen YQ, Song Y, et al. Role of g6pdh and leuB on lipid accumulation in mucor circinelloides. J Agric Food Chem. (2020) 68:4245–51. doi: 10.1021/acs.jafc.9b08155
139. Yang W, Dong S, Yang J, Mohamed H, Shah AM, Nazir Y, et al. Molecular Mechanism of Citrate Efflux by the Mitochondrial Citrate Transporter CT in Filamentous Fungus Mucor circinelloides WJ11. Front Microbiol. (2021) 12:673881. doi: 10.3389/fmicb.2021.673881
140. Yan FX, Dong GR, Qiang S, Niu YJ, Hu CY, Meng YH. Overexpression of Δ12, Δ15-Desaturases for Enhanced Lipids Synthesis in Yarrowia lipolytica. Front. Microbiol. (2020) 11: 289. doi: 10.3389/fmicb.2020.00289
141. Blazeck J, Hill A, Liu L, Knight R, Miller J, Pan A, et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun. (2014) 5:3131. doi: 10.1038/ncomms4131
142. Ruenwai R, Cheevadhanarak S, Laoteng K. Overexpression of acetyl-CoA carboxylase gene of Mucor rouxii enhanced fatty acid content in Hansenula polymorpha. Mol Biotechnol. (2009) 42:327–32. doi: 10.1007/s12033-009-9155-y
143. Wang S, Chen H, Tang X, Zhang H, Hao G, Chen W, et al. The role of glyceraldehyde-3-phosphate dehydrogenases in NADPH supply in the oleaginous filamentous fungus Mortierella alpina. Front Microbiol. (2020) 11:818. doi: 10.3389/fmicb.2020.00818
144. Bhutada G, Kavscek M, Ledesma-Amaro R, Thomas S, Rechberger GN, Nicaud JM, et al. Sugar versus fat: elimination of glycogen storage improves lipid accumulation in Yarrowia lipolytica. FEMS Yeast Res. (2017) 17:fox020. doi: 10.1093/femsle/fox020
145. Tang X, Sun X, Wang X, Zhang H, Chen YQ, Zhao J, et al. Characterization of NAD+/NADP+-specific isocitrate dehydrogenases from Oleaginous Fungus Mortierella alpina involved in lipid accumulation. Front Nutr. (2021) 8:746342. doi: 10.3389/fnut.2021.746342
146. Ling X, Zhou H, Yang Q, Yu S, Li J, Li Z, et al. Functions of Enyolreductase (ER) Domains of PKS Cluster in Lipid Synthesis and Enhancement of PUFAs Accumulation in Schizochytrium limacinum SR21 Using Triclosan as a Regulator of ER. Microorganisms. (2020) 8:300. doi: 10.3390/microorganisms8020300
147. Han X, Zhao Z, Wen Y, Chen Z. Enhancement of docosahexaenoic acid production by overexpression of ATP-citrate lyase and acetyl-CoA carboxylase in Schizochytrium sp. Biotechnol Biofuels. (2020) 13:131. doi: 10.1186/s13068-020-01767-z
148. Wang F, Bi Y, Diao J, Lv M, Cui J, Chen L, et al. Metabolic engineering to enhance biosynthesis of both docosahexaenoic acid and odd-chain fatty acids in Schizochytrium sp. S31 Biotechnol Biofuels. (2019) 12:141. doi: 10.1186/s13068-019-1484-x
149. Yang J, Cánovas-Márquez JT Li P, Li S, Niu J, Wang X, Nazir Y, et al. Deletion of Plasma Membrane Malate Transporters Increased Lipid Accumulation in the Oleaginous Fungus Mucor circinelloides WJ11. J Agric Food Chem. (2021) 69:9632–41. doi: 10.1021/acs.jafc.1c03307
150. Satoh S, Ozaki M, Matsumoto S, Nabatame T, Kaku M, Shudo T, et al. Enhancement of fatty acid biosynthesis by exogenous acetyl-CoA carboxylase and pantothenate kinase in Escherichia coli. Biotechnol Lett. (2020) 42:2595–605. doi: 10.1007/s10529-020-02996-w
151. Wang J, Xu R, Wang R, Haque ME, Liu A. Overexpression of ACC gene from oleaginous yeast Lipomyces starkeyi enhanced the lipid accumulation in Saccharomyces cerevisiae with increased levels of glycerol 3-phosphate substrates. Biosci Biotechnol Biochem. (2016) 80:1214–22. doi: 10.1080/09168451.2015.1136883
152. Qiao K, Imam Abidi SH, Liu H, Zhang H, Chakraborty S, Watson N, Kumaran Ajikumar P, Stephanopoulos G. Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. (2015) 29: 56-65. doi: 10.1016/j.ymben.2015.02.005
153. Hardie DG, Pan DA. Regulation of fatty acid synthesis and oxidation by the AMP-activated protein kinase. Biochem Soc Trans. (2002) 30:1064–70. doi: 10.1042/bst0301064
154. Xu Y, Caldo KMP, Pal-Nath D, Ozga J, Lemieux MJ, Weselake RJ, et al. Properties and Biotechnological Applications of Acyl-CoA: diacylglycerol acyltransferase and phospholipid: diacylglycerol acyltransferase from terrestrial plants and microalgae. Lipids. (2018) 53:663–88. doi: 10.1002/lipd.12081
155. Friedlander J, Tsakraklides V, Kamineni A, Greenhagen EH, Consiglio AL, MacEwen K, et al. Engineering of a high lipid producing Yarrowia lipolytica strain. Biotechnol Biofuels. (2016) 9:77. doi: 10.1186/s13068-016-0492-3
156. Guo ZP, Robin J, Duquesne S, O'Donohue MJ, Marty A, Bordes F. Developing cellulolytic Yarrowia lipolytica as a platform for the production of valuable products in consolidated bioprocessing of cellulose. Biotechnol Biofuels. (2018) 11:141. doi: 10.1186/s13068-018-1144-6
157. Hao G, Chen H, Gu Z, Zhang H, Chen W, Chen YQ. Metabolic engineering of mortierella alpina for enhanced arachidonic acid production through the NADPH-supplying strategy. Appl Environ Microbiol. (2016) 82:3280–8. doi: 10.1128/AEM.00572-16
158. Dulermo R, Dulermo T, Gamboa-Meléndez H, Thevenieau F, Nicaud JM. Role of Pex11p in lipid homeostasis in Yarrowia lipolytica. Eukaryot Cell. (2015) 14 :511-525. doi: 10.1128/EC.00051-15
159. Xie D, Jackson EN, Zhu Q. Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: from fundamental research to commercial production. Appl Microbiol Biotechnol. (2015) 99:1599–610. doi: 10.1007/s00253-014-6318-y
160. Lazar Z, Liu N, Stephanopoulos G. Holistic approaches in lipid production by Yarrowia lipolytica. Trends Biotechnol. (2018) 36:1157–70. doi: 10.1016/j.tibtech.2018.06.007
161. Yang RL, Lu N, Zhang H, Zhou SN, Zhang Q, Xue CH, Tang QJ. Improvement of lipid metabolism by DHA liposomes. Modern Food Science and Technology. (2021) 37: 11-20.
162. Lazzarin N, Vaquero E, Exacoustos C, Bertonotti E, Romanini ME, Arduini D. Low-dose aspirin and omega-3 fatty acids improve uterine artery blood flow velocity in women with recurrent miscarriage due to impaired uterine perfusion. Fertil. Steril. (2009) 92, 296-300. doi: 10.1016/j.fertnstert.2008.05.045
163. Swanson D, Block R, Mousa S. Omega-3 fatty acids EPA and DHA: Health benefits throughout life. Adv Nutr An Int Rev J. (2012) 3:1–7. doi: 10.3945/an.111.000893
164. Calder PC. Very long-chain n-3 fatty acids and human health: fact, fiction and the future. Proc Nutr Soc. (2018) 77:52–72. doi: 10.1017/S0029665117003950
165. Abdelhamid AS, Brown TJ, Brainard JS, Biswas P, Thorpe GC, Moore HJ, Deane KH, AlAbdulghafoor FK, Summerbell CD, Worthington HV, Song F, Hooper L. Omega-3 fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst Rev. (2018) 7:CD003177. doi: 10.1002/14651858.CD003177.pub4
166. Zhang XM, Zhao L, Hu XW. Progress on Nutritional Value of Schizochytrium limacinum and its application in animal production. J Henan Agricult Sci. (2021) 50:1–10. doi: 10.15933/j.cnki.1004-3268.2021.03.001
167. Peng Z, Zhang C, Yan L, Zhang Y, Yang Z, Wang J, et al. is More effective than DHA to improve depression-like behavior, glia cell dysfunction and hippcampal apoptosis signaling in a chronic stress-induced rat model of depression. Int J Mol Sci. (2020) 21:1769. doi: 10.3390/ijms21051769
168. Leger T, Jouve C, Hininger-Favier I, Rigaudiere JP, Capel F, Sapin V, et al. is Cardioprotective in male rats subjected to sepsis, but ALA is not beneficial. Antioxidants (Basel). (2020) 9:371. doi: 10.3390/antiox9050371
169. Riediger ND, Othman RA, Suh M, Moghadasian MH. A systemic review of the roles of n-3 fatty acids in health and disease. J Am Dietetic Assoc. (2009) 109: 668–79. doi: 10.1016/j.jada.2008.12.022
170. Byelashov OA, Sinclair AJ, Kaur G. Dietarysources, current intakesand nutritional role of omega-3 docosapentaenoic acid. Lipid Technology. (2015) 27: 79-82. doi: 10.1002/lite.201500013
171. Qin SY. Analysis of patent application of EPA in infant milk powder. Guangdong Chem. (2020) 17:261–3.
172. Tounian P, Bellaïche M, Legrand P, ARA. or no ARA in infant formulae, that is the question. Arch Pediatr. (2021) 28:69–74. doi: 10.1016/j.arcped.2020.10.001
173. Cao G, Guan Z, Liu FG, Liao X, Cai Y. Arachidonic acid production by Mortierella alpina using raw crop materials. Acta Sci Pol Technol Aliment. (2015) 14:133–43. doi: 10.17306/J.AFS.15
174. Salem N Jr, Wegher B, Mena P, Uauy R. Arachidonic and docosahexaenoic acids are biosynthesized from their 18-carbon precursors in human infants. Proc Natl Acad Sci USA. (1996) 93:49–54. doi: 10.1073/pnas.93.1.49
175. Bieren J. Esser-von. Immune-regulation and functions of eicosanoid lipid mediators. Biol Chem. (2017) 398:1177–91. doi: 10.1515/hsz-2017-0146
176. Tallima H, Hadley K, Ridi RE. Praziquantel and arachidonic acid combination. Innovative approach to the treatment of Schistosomiasis. Intech Opens. (2015) 145–72. doi: 10.5772/61185
177. Markworth JF, Mitchell CJ, D'Souza RF, Aasen KMM, Durainayagam BR, Mitchell SM, et al. Arachidonic acid supplementation modulates blood and skeletal muscle lipid profile with no effect on basal inflammation in resistance exercise trained men. Prostaglandins Leukot Essent Fatty Acids. (2018) l:74–86. doi: 10.1016/j.plefa.2017.12.003
178. Certik M, Adamechova Z, Laoteng K. Microbial production of gamma-linolenic acid: Submerged versus solid-state fermentations. Food Sci Biotechno. (2012) 21:921–6. doi: 10.1007/s10068-012-0121-2
179. Mohamed H, El-Shanawany AR, Shah AM, Nazir Y, Naz T, Ullah S, et al. Comparative analysis of different isolated oleaginous mucoromycota fungi for their γ-linolenic acid and carotenoid production. Biomed Res Int. (2020) 2020:3621543. doi: 10.1155/2020/3621543
180. Zhang Y, Luan X, Zhang H, Garre V, Song Y, Ratledge C. Improved γ-linolenic acid production in Mucor circinelloides by homologous overexpressing of delta-12 and delta-6 desaturases. Microb Cell Fact. (2017) 16:113. doi: 10.1186/s12934-017-0723-8
181. Del Gobbo LC, Imamura F, Aslibekyan S, Marklund M, Virtanen JK, Wennberg M, et al. ω-3 polyunsaturated fatty acid biomarkers and coronary heart disease: pooling project of 19 cohort studies. JAMA internal medicine (2016) 176: 1155-1166. doi: 10.1001/jamainternmed.2016.2925
182. Aasen IM, Ertesvåg H, Heggeset TM, Liu B, Brautaset T, Vadstein O, et al. Thraustochytrids as production organisms for docosahexaenoic acid (DHA), squalene, and carotenoids. Appl Microbiol Biotechnol. (2016) 100:4309–21. doi: 10.1007/s00253-016-7498-4
183. Tegenge MA, Von Tungeln LS, Mitkus RJ, Anderson SA, Vanlandingham MM, Forshee RA, et al. Pharmacokinetics and biodistribution of squalene-containing emulsion adjuvant following intramuscular injection of H5N1 influenza vaccine in mice. Regul Toxicol Pharmacol. (2016) 81:113–9. doi: 10.1016/j.yrtph.2016.08.003
184. Gohil N, Bhattacharjee G, Khambhati K, Braddick D, Singh V. Engineering strategies in microorganisms for the enhanced production of squalene: advances, challenges and opportunities. Front Bioeng Biotechnol. (2019) 7:50. doi: 10.3389/fbioe.2019.00050
185. Shakeri S, Amoozyan N, Fekrat F, Maleki M. Antigastric cancer bioactive Aurantiochytrium oil rich in docosahexaenoic acid: from media optimization to cancer cells cytotoxicity assessment. J Food Sci. (2017) 82:2706–18. doi: 10.1111/1750-3841.13925
Keywords: oleaginous fungi, triacylglycerols, regulation strategy, fatty acids, Commercial application
Citation: Zhang X-Y, Li B, Huang B-C, Wang F-B, Zhang Y-Q, Zhao S-G, Li M, Wang H-Y, Yu X-J, Liu X-Y, Jiang J and Wang Z-P (2022) Production, Biosynthesis, and Commercial Applications of Fatty Acids From Oleaginous Fungi. Front. Nutr. 9:873657. doi: 10.3389/fnut.2022.873657
Received: 11 February 2022; Accepted: 31 March 2022;
Published: 19 May 2022.
Edited by:
Yuanda Song, Shandong University of Technology, ChinaReviewed by:
Heitor Bento, São Paulo State University, BrazilAlok Patel, Luleå University of Technology, Sweden
Copyright © 2022 Zhang, Li, Huang, Wang, Zhang, Zhao, Li, Wang, Yu, Liu, Jiang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Jun Yu, eGp5dUB6anV0LmVkdS5jbg==; Zhi-Peng Wang, d2FuZ3pwbWJpb0AxNjMuY29t; Jing Jiang, amlhbmdqaW5nQHVzdHMuZWR1LmNu
 Xin-Yue Zhang1
Xin-Yue Zhang1 Hai-Ying Wang
Hai-Ying Wang Xin-Jun Yu
Xin-Jun Yu Xiao-Yan Liu
Xiao-Yan Liu Jing Jiang
Jing Jiang Zhi-Peng Wang
Zhi-Peng Wang