
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr. , 12 July 2022
Sec. Nutrition and Food Science Technology
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.826131
This article is part of the Research Topic Wheat Biofortification to Alleviate Global Malnutrition View all 11 articles
 Om Prakash Gupta1†
Om Prakash Gupta1† Amit Kumar Singh2†
Amit Kumar Singh2† Archana Singh3
Archana Singh3 Gyanendra Pratap Singh1*
Gyanendra Pratap Singh1* Kailash C. Bansal4*
Kailash C. Bansal4* Swapan K. Datta5*
Swapan K. Datta5*Alleviating micronutrients associated problems in children below five years and women of childbearing age, remains a significant challenge, especially in resource-poor nations. One of the most important staple food crops, wheat attracts the highest global research priority for micronutrient (Fe, Zn, Se, and Ca) biofortification. Wild relatives and cultivated species of wheat possess significant natural genetic variability for these micronutrients, which has successfully been utilized for breeding micronutrient dense wheat varieties. This has enabled the release of 40 biofortified wheat cultivars for commercial cultivation in different countries, including India, Bangladesh, Pakistan, Bolivia, Mexico and Nepal. In this review, we have systematically analyzed the current understanding of availability and utilization of natural genetic variations for grain micronutrients among cultivated and wild relatives, QTLs/genes and different genomic regions regulating the accumulation of micronutrients, and the status of micronutrient biofortified wheat varieties released for commercial cultivation across the globe. In addition, we have also discussed the potential implications of emerging technologies such as genome editing to improve the micronutrient content and their bioavailability in wheat.
Micronutrient deficiency also known as “hidden hunger,” is one of the major global health problems afflicting more than 2 billion people globally (1). Micronutrient deficiency is common among population groups dependent on a single cereal (rice, wheat or maize) diet. These populations lack access to an adequate quantity of fruits, vegetables, dairy products, meats, etc. that are rich in essential minerals and vitamins. Globally, the human populations in Sub-Saharan Africa and South Asia bear the most significant burden of these micronutrient deficiencies (2). Among the micronutrient deficiencies, Iron (Fe), Zinc (Zn), Iodine (I), and Vitamin A deficiencies are the most widespread. Other micronutrient deficiencies include Calcium (Ca) and selenium (Se) that are relatively less widespread but could become more prevalent in the future if not addressed now. The daily intake of these micronutrients recommended dietary allowance (RDA) is vital to sustaining life as they are required for the proper physical and cognitive development, disease prevention, and overall human well-being. Children less than five years of age, pregnant women, and lactating women are amongst the most vulnerable group to such micronutrient deficiencies.
Fe is required for several processes in the human body, including oxygen transport, electron transport, and DNA synthesis (3). For example, it is a component of oxygen transport proteins, such as hemoglobin and myoglobin. Moreover, Fe is also required for many proteins and enzymes involved in the energy generation, synthesis of neurotransmitters, and proper functioning of the immune system (4). Deficiency of Fe is the leading cause of anemia; nearly 30% of the women of reproductive age (14-59 years of age) and 40% of the children under five years suffer from anemia, globally (5). Similarly, Zn is also a trace element required for proper growth and maintenance of the human body. It acts as an essential cofactor for over 300 enzymes that are involved in vital processes, including cell proliferation, healing of wounds, blood clotting, etc. (6, 7). Zn deficiency has also been associated with increased diarrheal diseases and acute respiratory infections in children under five years of age and is a critical factor contributing to disease burden in developing countries (8). In Africa, 58% of child deaths are estimated to be due to Zn deficiency (9). Se is another important essential trace element needed for a robust immune system, thyroid function and reproduction. It is an essential component of selenoproteins, which act as potent antioxidant protecting cellular components from free radicals (10). Globally, an estimated 1 billion population suffers from Se deficiency, and this number is expected to rise in coming decades, necessitating designing strategies to enhance its dietary intake (11). Ca has also been regarded as one of the essential micronutrients required for the proper growth and development of the human body. It is required for strong bones and teeth and is also involved in many fundamental processes, such as blood coagulation, muscular function, hormonal secretions, nerve impulse transmission, etc. (12). Its deficiency can cause many problems, such as rickets in children, and osteoporosis and osteopenia in adults.
For curbing hidden hunger in the developing world, increasing the content via biofortification vis-à-vis increasing the bioavailability of both Fe and Zn are the two major approaches. For the first approach i.e., biofortification, significant variation in the level of Fe (up to 88 mg kg–1) and Zn (14 to 190 mg kg–1) has been reported among wild wheat, especially wild emmer wheat (Triticum turgidum ssp. Dicoccoides) (13), which can be efficiently utilized by wheat breeders to transfer in the background of high yielding and disease resistant hexaploid wheat genotypes. A breeding target of > 59 μg g–1 Fe, and 38 μg g–1 Zn in wheat grains (14) against the baseline level of 30 μg g–1 Fe, and 25 μg g–1 Zn would be sufficient to meet the 30–40% of the average daily requirement of an adult. However, bioavailability of Fe and Zn in wheat is greatly limited due to the presence of phytic acid (PA, 0.4–2.0%), an anti-nutrient (15, 16). [PA]:[Fe and Zn] ratios are very vital in determining the potential bioavailability of the micronutrients and are inversely proportional i.e., higher the molar ratio, lesser the bioavailability and vice-versa. For [PA]:[Fe], the ratio should be < 1 (preferably < 0.4) to significantly improve Fe absorption (17), while for [PA]:[Zn] ratios of < 5, 5 to 15, and > 15 have been associated with high (50%), moderate (30%) and low (15%) Zn bioavailability, respectively (18). Therefore, it is desired that wheat genotypes be developed with suitable [PA]:[Fe and Zn] ratios for optimum bioavailability of Fe and Zn to humans and animals.
Generally, three major strategies i.e., dietary diversification, food fortification, and food supplementation were developed to address the problem of micronutrient deficiencies (19). Among these, dietary diversification focuses on modifying food consumption patterns at the individual household level, such as increasing the intake of more nutritious diets like fruits, vegetables, animal foods, etc. However, dietary diversification is not possible in many parts of the world due to poor socio-economic conditions and ethnic dietary choices. Other alternatives like food fortification and micronutrient supplementation for specific life stages and age groups can be considered stopgap measures for tackling micronutrient deficiencies. However, these strategies cannot provide a long-term sustainable solution for nutrient deficiencies in low and middle-income countries. These countries have a large population that lives in extreme poverty and does not have both physical as well as economic access to the adequate quantity of nutritious foods. Further, they cannot afford fortified food products or food supplements (20). Moreover, setting up the infrastructure to develop and distribute fortified foods or even food supplements would require a considerable investment that underdeveloped countries cannot afford. Agro-system diversification can assist local populations to expand their food baskets and solve the problem of micronutrient deficiencies, but it cannot be widely adopted in underdeveloped countries due to small landholdings. Therefore, in the past two decades, a greater emphasis has been laid on biofortification, which refers to increasing the bioavailable nutrient content of food crops either through conventional plant breeding or transgenic approaches. Biofortification is considered the most effective and sustainable approach for addressing the micronutrient deficiencies related problems in humans (21). A recent publication “Wheat and Barley Grain Biofortification” (Elsevier, United Kingdom), would serve as an important ready reckoner for different domains of wheat biofortification (22).
Wheat supplies approximately 20% of the human population’s total calories and protein intake worldwide. However, most of the commercial wheat cultivars grown across the world are deficient or have suboptimal levels of micronutrients. It is mainly due to the greater focus of national wheat breeding programs on increasing yield, which has resulted in the erosion of grain minerals and protein contents in improved varieties. For the development of nutrient-dense wheat varieties, the primary prerequisite is to identify donor lines with high concentrations of the targeted micronutrients. Therefore, the need of the hour is to explore natural genetic diversity among the landraces and wild wheat species for grain mineral content and utilize them in the breeding programs for developing biofortified varieties. In 2003, a program in this direction was initiated at CIMMYT, with the support from HarvestPlus. The breeding materials developed under this program have contributed to the development of few biofortified cultivars with higher grain Zn and Fe concentrations in India, Pakistan, Nepal, Mexico, and Bangladesh (23).
In wheat, conducting molecular studies, especially cloning of genes for any target trait, is considered a very challenging task due to the large and complex genome organization. Therefore, it took many years to clone a grain protein content (GPC) gene (Gpc-B1) from an accession of wild emmer wheat (Triticum turgidum, dicoccoides) (24). Even for the qualitative traits, such as disease resistance, which are generally controlled by a single major gene, cloning the resistance locus has never been easier due to the lack of information on the whole genome-level sequence as well as fully sequenced genes. However, in recent years, the availability of genomic resources such as the gold standard reference sequence of hexaploid wheat (25), reference genome sequences of its progenitor species (26, 27), transcriptome landscape of different tissues of wheat (28–31), single nucleotide polymorphism (SNP) genotyping arrays and genotyping by sequencing (GBS) methods (32) has led to a revolution in the field of wheat genomics. These tools have made it relatively easier to fine map and clone genes of essential traits in cultivated and wild wheat species (33). Further, in recent years, genome-wide association study (GWAS), which uses diverse germplasm lines or multiparent populations, has developed as a powerful tool for high-resolution trait mapping in crops and can be used to map nutritional quality traits using diverse association panels constituted from landraces, synthetics and wild species (34–36). Furthermore, recently, genome editing technologies, including prime editing and base editing have become promising targeted mutagenesis tools for crop improvement (37, 38). The first report of gene editing in wheat was the development of the targeted knockout for the Mlo gene that confers resistance against powdery mildew pathogen, Blumeria graminis f.sp. tritici (39). Since then, there are many reports of gene editing in wheat targeting genes associated with various agronomical and quality traits (40). Recently, nano-technology has also been explored for micronutrient biofortification in wheat. Khan et al. (41) have extensively reviewed the status of nano formulation-based wheat biofortification with a critical analysis of its merits and demerits.
In the present review, we have discussed the current state of knowledge on the existing natural genetic variations for micronutrients content among cultivated and wild wheat germplasm, genes and genomic regions controlling the micronutrient traits, current status of biofortified wheat varieties released for commercial cultivation around the world and potential applications of genome editing tools in the improvement of nutritional quality traits in wheat.
Understanding the extent and magnitude of natural genetic variations for various essential nutrients in wheat and its wild species is critical for improving these traits through classical and modern breeding tools. In this context, extensive screening of germplasm collection of wheat and its wild species conserved in genebanks of national and international institutions can facilitate the discovery of novel germplasm donors for various essential nutrients. These donor germplasms can be further exploited in the breeding programs for the development of biofortified wheat varieties. Over the past two decades, several studies have explored cultivated and wild wheat germplasm for variations in grain micronutrient contents. The key findings of some of these studies are briefly presented below.
Several studies have reported variation in grain Fe and Zn concentrations of bread wheat cultivars, landraces and wild wheat (42–44). Generally, breeding lines and cultivars have low genetic variation for grain Fe and Zn concentrations compared to landraces, cultivated wheat progenitors and related wild wheat species (unadopted wheat) (45). Several studies show a negative correlation between mineral concentrations and yield, implying that increase in grain yield of wheat varieties was accompanied by a significant decrease in their grain mineral content (45, 46). Evaluation of eighty Iranian wheat cultivars bred over a period of 70 years revealed a significant decrease in grain Fe and Zn concentrations that ranged from 63.56 to 102.19 and 31.65 to 54.06 mg/kg, respectively (46). On the other hand, a wide range of variations for grain Fe and Zn concentrations have been reported in landraces and other unadopted germplasms (43, 44, 47, 48). Qury et al. (43) analyzed a diverse wheat genotype panel comprising of French landraces, elite breeding lines, modern varieties and a set of worldwide germplasm collection which showed variation in grain Zn and Fe concentrations ranging from 15 to 35 mg/kg, and 20 to 60 mg/kg, respectively. However, some unadopted lines of this panel had Fe and Zn concentrations as high as 88 and 43 mg/kg, respectively, which can be exploited to improve the wheat cultivar’s mineral concentrations. Another study on fifty landraces and ten varieties revealed higher Fe (24.93 to 66.51 mg/kg) and Zn (18.68 to 38.66 mg/kg) concentrations in landraces as compared to commercial cultivars (48). Recently, novel sources of variation for whole-grain Fe and Zn concentrations were identified in a panel of 245 diverse bread wheat lines derived from crosses between landraces of Watkin collections with a United Kingdom wheat cultivar Paragon (49). Further, the above studies have found that wide variation in grain Fe and Zn concentrations of wheat genotypes across different studies may not be solely due to genotypic differences since environment and soil nutrient status are known to significantly affect these traits, so promising Fe and Zn rich lines identified in these studies must be validated in multilocation trails.
Contrary to widely cultivated wheat, the primary and secondary gene pool of wheat such as Triticum monococcum, Triticum boeoticum, T. turgidum dicoccoides, Aegilops tauschii, T. spelta and Triticum polonicum are reported to contain wider variation for grain Fe and Zn concentrations (34, 45, 50–52). Among these species, T. turgidum ssp. dicoccoides is considered the most promising donor for the grain Fe and Zn content. Cakmak et al. (53) screened a large number of accessions of several wild wheat species and their relatives for grain Fe and Zn concentrations and observed unique variations among the T. dicoccoides accessions, which ranged from 14 to 190 mg/kg for Zn and from 15 to 109 mg/kg for Fe. Studies have also identified some accessions with high grain Fe and Zn concentrations in other species as well. Tiwari et al. (52) screened a large number of accessions of T. boeoticum and found one accession (pau5088) with higher levels of grain Fe and Zn concentration, 40.1 and 44.6 mg/kg, respectively.
Additionally, some non- progenitor, wild wheat species such as S, U and M genomes have been reported to contain 3-4 times more Fe and Zn than cultivated hexaploid and tetraploid species (51). Velu et al. (54) reported wide genetic variability for Fe and Zn concentration among the introgression lines derived from cultivated species’ crosses with wild wheat rich in grain Fe and Zn content. In addition to wild species of wheat, a few non-Triticum species such as rye and Leymus spp are also rich in mineral nutrient contents. The wheat-alien introgression lines derived from the crosses with Leymus racemosus, and also those carrying introgression of 2R, 3R chromosomes of rye contain a high level of Fe and Zn (55).
There are a few reports on the genetic variability of Se concentration in cultivated wheat and its wild relatives as compared to that of Fe and Zn (45, 56, 57). Lyons et al. (56) analyzed the Se concentration of ancestral and wild relatives of wheat, landraces, population, and cultivars grown in Mexico and Australia. The grain Se concentration of this set ranged from 5–720 μg/kg; however, much of this variation was attributed to variation in soil Se content across the locations. Nevertheless, they reported higher variation in diploid species Ae. tauschi and rye. Similarly, Zhao et al. (45) also reported limited genetic variability for grain Se concentration in commercial wheat cultivars. Apart from Fe, Zn, and Se, there are very limited genetic variability studies for Ca content in wheat. A study on Indian and Iranian wheat lines showed phenotypic variability for grain Ca content in the range of 104.3 to 663.5 mg/kg (58). Another study that analyzed a diverse panel of 353 wheat varieties, including winter and spring wheat varieties, reported wide variations for grain Ca content ranging from 288.2 to 647.5 mg/kg. Nirvana, a wheat variety from France, had a very high concentration of grain Ca (647.5 mg/kg DW) (59). The wide variability for grain Ca content in the above two studies suggest ample scope for developing biofortified Ca wheat varieties.
The phytic acid content in wheat grain can significantly affect the bioavailability of minerals such as Fe and Zn during digestion because of their strong ability to bind to metals. Therefore, wheat genotypes with low phytate and high mineral concentration could be immensely useful in breeding programs that aim to develop biofortified verities for essential mineral nutrients. Many studies have analyzed the variability of phytate content among wheat cultivars and germplasm lines (60–62). The analysis of phytic acid content of a set of 65 bread varieties of Pakistan showed variation in the range 0.706–1.113% (60). Another comprehensive study on the collection of global durum cultivars identified a 2-fold variation in phytic acid content ranging between 0.462 to 0.952% (62). Contrary, many other studies have identified higher values for phytic acids (more than 1%) in durum genotypes, which might be attributed to G × E effects (61, 63). The low phytate genotypes identified in various studies may be potentially used as parents for developing wheat varieties with enhanced bioavailable Fe and Zn levels.
Advancements in genomics, especially the availability of high-throughput genotyping assays such as whole genome re-sequencing, GBS, SNP arrays, etc., have made it easier to perform trait mapping in plant species like wheat with a large and complex genome. Both bi-parental and association mapping approaches have facilitated identifying several QTLs/genomic regions controlling grain minerals content. A brief description of the genomic regions/genes identified in the various studies is presented below.
In the past two decades, several studies have reported QTLs/candidate genes for grain Fe and Zn concentrations in wheat and its wild species using both QTL and association mapping methods (Table 1). Expectedly, most QTLs for Fe and Zn were identified from wild wheat and their relatives because there is a minimal variability for both these minerals in cultivated wheat germplasm. Tiwari et al. (52) were the first to report genetic mapping of grain Fe and Zn using an interspecific mapping population derived from the cross of T. boeoticum accession pau5088 (high grain Fe and Zn concentration) with T. monococcum accession pau14087. They identified two QTLs for grain Fe and one QTL for Zn. After that, many other studies have employed bi-parental mapping approach and identified QTLs for grain Fe and Zn concentrations on various chromosomes of wheat and its wild relatives (64–69). In the past few years, the association mapping approach has also facilitated identifying genomic regions/QTLs associated with grain Fe and Zn concentrations in diverse association panels constituted using diverse genotypes, including synthetic hexaploid wheat, advanced breeding lines, landraces and cultivars etc. However, most of the reported grain Fe and Zn QTLs have not been found stable across various locations suggesting profound effects of environment and genotype X environment on both these traits. Further, many identified regions have minor effects on grain Fe and Zn concentrations. Therefore, only the significant QTLs for Fe and Zinc identified in various studies should be focused on improving cultivated wheat’s mineral contents. Some of the significant QTLs for zinc concentration has been identified on chromosome 1B, 2B, 5A, 1B, 6B (65, 70). Moreover, some studies have identified common genomic regions for grain Fe and Zn concentrations, and even some are also associated with other valuable traits such as thousand-grain weight, protein etc. (65, 71). Tiwari et al. (65) had identified two major QTL for grain zinc concentration on 1B and 2B; of these, the QTL on 2B was colocalized with the QTL for grain Fe concentration. Similarly, a significant QTL for grain Zn on 2B co-located with the QTL for grain Fe concentration (66). These studies suggest that simultaneous improvement of both traits is possible using MAS. Compared to Fe and Zn, the QTL mapping studies for grain Se concentration in wheat are rare. A total of five QTLs for Se content were identified on chromosome 3D, 4A, 4D, 5B, and 7D, using two different RIL populations (72). Moreover, in a recent study, Wang et al. (73) identified nine Se concentration QTLs in a mapping population derived from the cross of winter wheat cultivars Tainong18 and Linmai6.
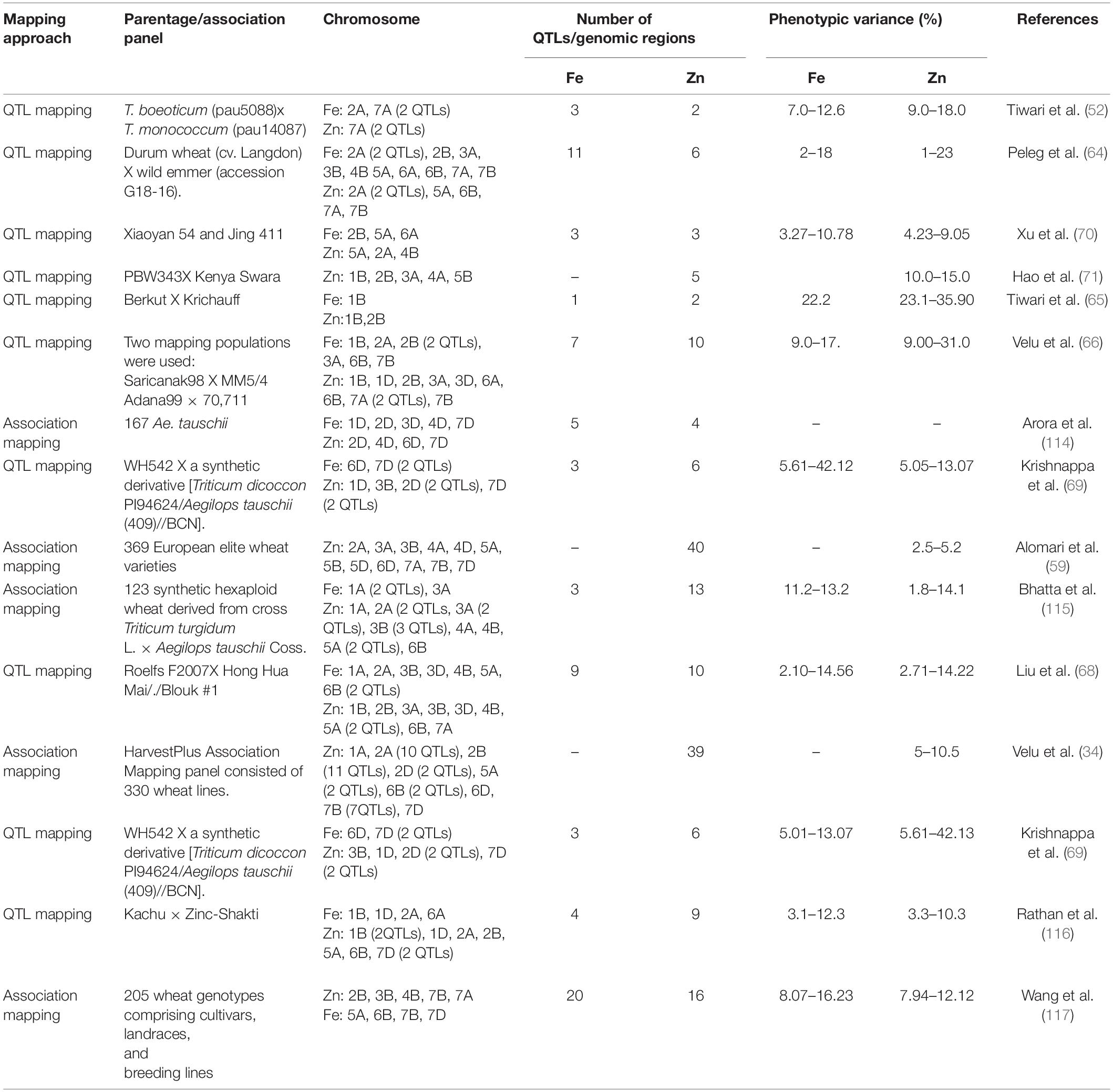
Table 1. Genomic regions/QTLs identified for grain Zn and Fe concentrations in cultivated and wild wheat using biparental and association analysis methods.
In contrast to grain Fe and Zn content, there is very limited information on genomic regions/QTLs for grain Ca accumulation. A total of 9 QTLs for grain Ca were reported in the RIL mapping population derived from durum and wild emmer wheat (64). In another study, association mapping using a diverse panel of European wheat accessions identified genomic regions for grain Ca accumulation on all the wheat chromosomes except 3D, 4B, and 4D (59). Recently, Alomari et al. (74) identified a major genomic region for grain Ca on the long arm of 5A, which overlapped with gene TraesCS5A02G542600 that encoded for a transmembrane protein.
Biofortification traits have complex regulations and are governed by many QTLs/genes, which are significantly affected by environment and genotype-environment interactions (75). The expression of some biofortification traits such as grain mineral content involves many processes such as mineral absorption, translocation, redistribution, and re-mobilization to sink, and each of these processes is controlled by many genes. This makes genetic dissection of such traits challenging by utilizing any single genetic or molecular analysis approach (69, 76). The conventional GWAS approach identifies a large number of genomic regions/QTLs that can not be directly utilized in a breeding program (34). Moreover, GWAS does not go beyond simple marker-trait correlation with no proof of causality; therefore, this approach alone may not provide insights on the functional basis of variation in biofortification traits. The above two limitations of GWAS can be overcome by incorporating functionome (multi-omics) data (77). In the past decade, significant technological improvements in the field of “omics” have made it feasible to generate large-scale omics data such as transcriptome, proteome, and metabolome, etc., from a large number of samples at a low cost (78). Integration of GWAS with various multi-omics data would enable a system-level understanding of biofortification traits and has great potential to precisely pinpoint the actual causal variant/candidate gene. There can be two approaches for integrating multi-omics data in GWAS analysis; 1) GWAS is independently performed using biofortification trait profiling data as well as associated omics data i.e., gene expression, proteome and metabolome data of association panel genotypes, and then marker-trait associations results are integrated to interpret pathways and identify causal variant/candidate genes associated with the traits; 2) GWAS is performed only using genome-wide DNA markers and biofortification traits, and then expression, proteome, and metabolite profiling data generated from a few contrasting genotypes of the association panel are mapped to genomic regions associated with the targeted trait to identify the candidate genes (Figure 1). The integration of functionome data in GWAS analysis of biofortification traits may not only help identify causal variants responsible for these traits but would also enable their comprehensive understanding at the cellular, biochemical and molecular levels.
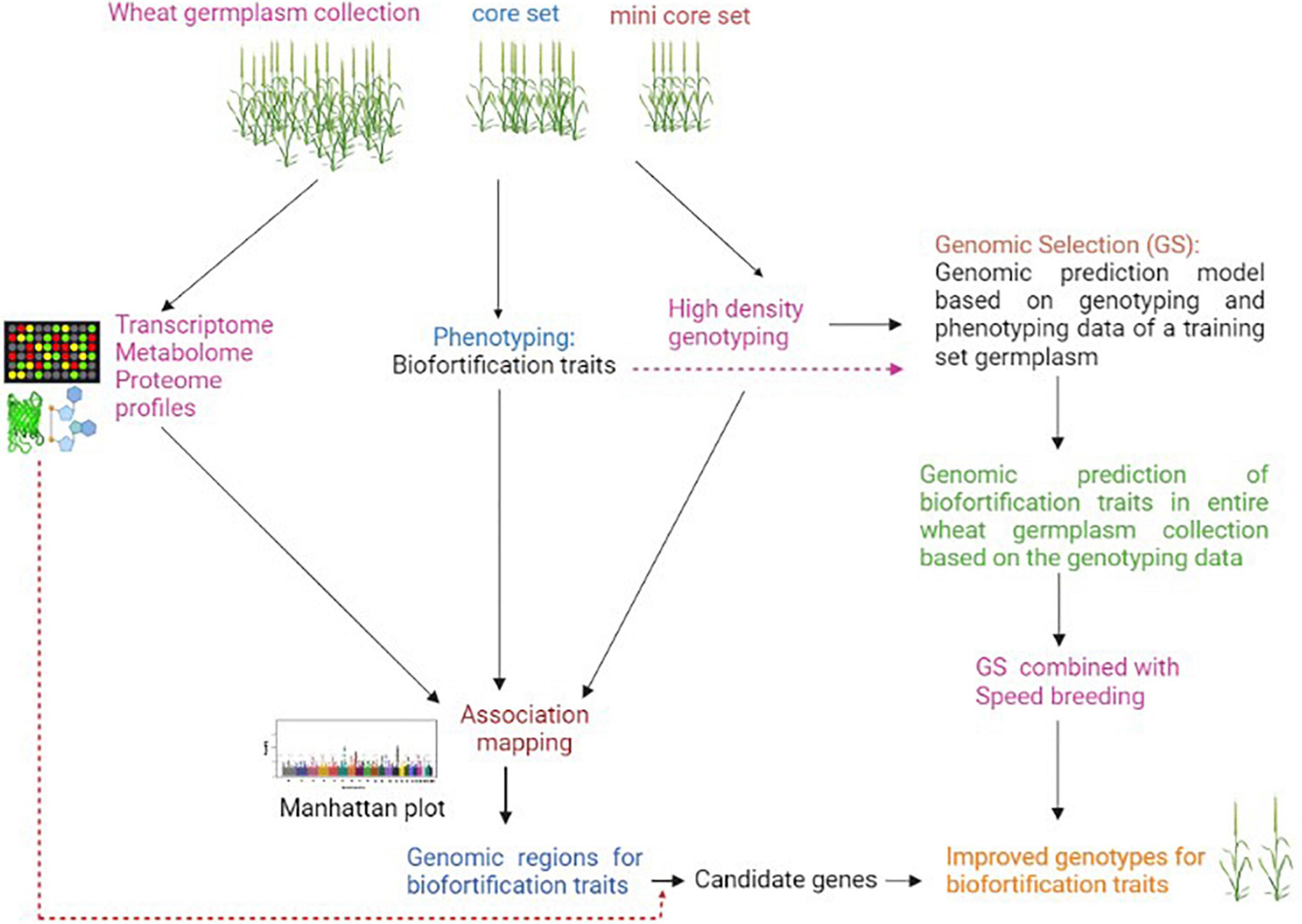
Figure 1. Scheme for integration of omics and genomic selection approaches for accelerating improvement of biofortification traits in wheat.
In recent years, with the availability of high throughput and cost-effective genotyping assays, genomic selection (GS) has emerged as a promising genomics-based tool for the improvement of complex traits in crops (79). In GS, the selection of elite genotypes is made using genomic estimated breeding values (GEBVs), which consider all marker effects across the genome. GS enhances breeding efficiency for quantitative traits by reducing breeding cycle duration and selection gain per unit time1. This approach has great potential for improving the quality traits with low genetic variance (80). Joukhadar et al. (81) have discussed in detail the potential application of GS in the biofortification of spring wheat. There are already some reports on genomic predictions of micronutrient traits in crops (82–84). Owens et al. (82) were among the first to estimate genomic prediction for a biofortification trait in a crop. They predicted pro-vitamin A content in maize using genome-wide as well as carotenoid pathway-based markers and identified a small number of candidate genes that can be targeted for conversion of elite genotype with low carotenoid content to one that has an orange color grain with higher levels of high pro-vitamin A. In wheat, Velu et al. (83) reported genome-wide predictions for grain Fe and Zn concentrations in a diverse panel of 330 genotypes with prediction accuracies ranging from 0.331 to 0.694 and 0.324 to 0.734, respectively. Another study in wheat also found moderate to high genomic prediction accuracies for various major and minor elements concentration in grains (85). The high genomic prediction accuracies for mineral nutrient traits suggest that GS holds great potential in accelerating breeding for biofortification traits. Further, speed breeding that enables taking up to six generations in one year under glasshouse can be very well combined with GS in different breeding schemes to accelerate genetic gain for biofortification traits (Figure 1).
In addition to basic research on micronutrients acquisition, the development of biofortified wheat varieties has recently upscaled with several successful examples. The conventional breeding approach has demonstrated great potential to biofortify hexaploid wheat genotypes by identifying suitable donor genetic resources such as synthetic, wild and primitive wheat genotypes for high Fe and Zn content with enhanced bioavailability. The most promising high Zn and Fe sources are diploid progenitors of hexaploid wheat (Aegilops tauschii), wild emmer (T. dicoccoides), einkorn (Triticum monococcum), T. spelta, T. polonicum, and T. aestivum landraces. Among wild wheat tested so far, the collections of wild emmer wheat, Triticum turgidum ssp. dicoccoides, showed a prominent genetic variation of Zn ranging from 14 to 190 mg kg–1 and Fe up to 88 mg kg–1 (13). Translocation from different Aegilops spp. and rye to Pavon 76 background at the International Maize and Wheat Improvement Center (CIMMYT, Mexico) has generated several synthetic hexaploids (SHW), T. spelta, and several pre-breeding lines having wider variation in Zn (38 to 72 mg kg–1) and Fe content (32 to 52 mg kg–1) (34).
With these concerted efforts, CIMMYT wheat breeders, in collaboration with other major institutions of India, Pakistan, Bangladesh, Nepal, and Bolivia, have facilitated the development and release of 40 biofortified wheat varieties for commercial cultivation (Table 2). Since 2014, A total of 24 biofortified wheat varieties (T. aestivum; 16 and T. durum; 8) for Fe, Zn and protein have been developed by ICAR-Indian Institute of Wheat and Barley Research, Karnal, Punjab Agricultural University, Ludhiana, ICAR- Indian Agricultural Research Institute, Delhi, Agharkar Research Institute, Pune, University of Agricultural Sciences, Dharwad, Banaras Hindu University, Varanasi and private seed companies and released to Indian farmers for common cultivation at different wheat growing zones of India. Similarly, CIMMYT, in collaboration with important wheat research institutions of Pakistan, Bangladesh, Bolivia, Nepal and Mexico, has developed and released 2, 1, 1, 2, and 1 biofortified wheat varieties, respectively (Table 2). Overall, utilization of wild relatives and SHW of wheat in conventional breeding programs have significantly impacted the development of micronutrient biofortified wheat varieties, which is expected to continue with increased bioavailability. Over the next two decades, developing and mainstreaming Zn and Fe in the wheat breeding program at CIMMYT and partner institutions across the globe would undoubtedly enable the release of high-yielding and Fe and Zn biofortified wheat varieties to a more significant percentage of farmers of South Asia to curb hidden hunger of children and pregnant and lactating mothers.
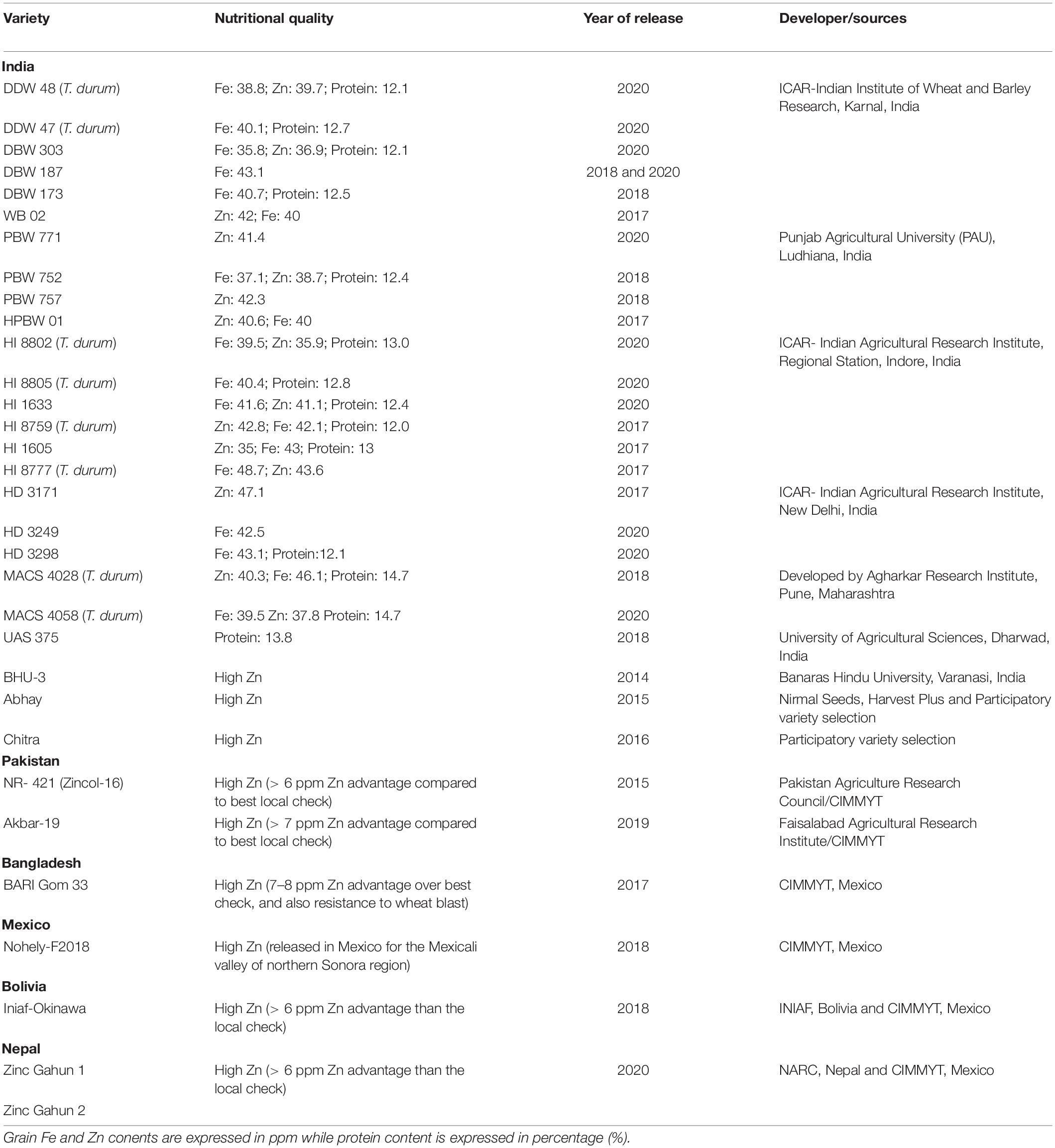
Table 2. List of biofortified wheat varieties developed through conventional breeding and released for commercial cultivation around the globe.
While conventional breeding is globally accepted, the absence of desired genetic diversity within the primary, secondary and tertiary gene pools for targeted traits within species (e.g., golden rice) or difficult to breed crops (e.g., banana) can efficiently be managed through genetic engineering technologies as a viable alternative. Also, the development of multi-nutrient cultivars by stacking multiple genes coupled with superior physiological and agronomic traits is often limited with conventional breeding, which can be circumvented by the genetic engineering approach (Figures 2A,B). However, wheat being hexaploid is comparatively challenging to transform and therefore needs the development of a robust transformation protocol to harness the full potential of the transgenic approach. As demonstrated in Figure 2B, the genetic engineering approach offers limitless cross-kingdom utilization and tacking of desired genes for multi-nutrients target traits improvement, including biotic and abiotic stresses, making it more attractive for farmers to adopt nutritionally improved nutrition wheat varieties. Moreover, it offers simultaneous biofortification of multi nutrients by metabolic engineering (86). However, the three significant bottlenecks of the transgenic approach are the lack of availability of suitable transformation protocol in polyploidy crop such wheat, fear of environmental escape of transgene and global genetically modified organisms (GMO) regulation. Knowledge gained in identifying and functional characterization of different genes actively associated with uptake and translocation of Fe and Zn can efficiently be used to increase Fe and Zn content in wheat by transgenic approach. Several proofs of concepts using the genetic engineering approaches have been tested with apparently stirring results in wheat for grain Fe and Zn. For example, The NAM-B1(Gpc-B1) transcription factor provides an entry point to increase Fe and Zn content. Knowing the critical control points, we can modify expression patterns, downstream targets or binding specificities to augment micronutrient content in grains. Wheat biofortification for Fe and Zn has been achieved using the transcription factor NAM-B1 (24), which was initially identified for increasing protein content in wild emmer (Triticum turgidum ssp dicoccoides). In recombinant substitution lines (RSL), the presence of NAM-B1 allele of T. dicoccoides increased Fe and Zn grain concentrations by 18 and 12%, respectively, in addition to 38% higher protein as compared with RSLs carrying the allele from cultivated wheat (Triticum durum) (87). Further, the increase in grain Fe and Zn content did not significantly correlate with yield reduction across the five environments (87). This gene is being widely used in breeding programs across several continents (88, 89).

Figure 2. Schematic representation of conventional (A) genetic engineering (B) and genome editing (C) approaches in wheat for targeted biofortifcation of micronutrients. Genetic engineering and genome editing approaches in combination with conventional breeding offers simultaneous incorporation of multi-nutrient (minerals and vitamins) traits along with improved physiological and agronomic features.
Transgenic approaches involving endosperm-specific expression of wheat ferritin, TaFer1-A (90) or soybean ferritin (91) led to 1.5- to 1.9-fold and 1.1- to 1.6-fold increase in grain Fe, respectively alongside increased phytase activity (92). However, the stability of wheat TaFer1-A in subsequent generations remains a question. Two independent workers have depicted that the overexpression of NICOTIANAMINE SYNTHASE 2 (OsNAS2) gene in wheat produced Fe up to 93.1 mg g–1 (93) and 80 mg g–1 (94) under greenhouse and field conditions, respectively. Connorton et al. (95) demonstrated the doubling of total Fe content in wheat flour by using VACUOLAR IRON TRANSPORTER 2 (TaVIT2) gene, which effectively enhances vacuolar Fe and manganese (Mn) transport in the endosperm. In addition to micronutrients, progress has been made to discourse the challenges of most deficient nutrients like vitamin A and quality proteins in wheat. The provitamin A content of wheat has been enhanced by expressing bacterial Phytoene synthase (CrtB) and Carotene desaturase gene (CrtI) (96, 97). To increase Fe bioavailability, phytase activity was increased by expressing the Phytochrome (phyA) gene (98), while phytic acid content was decreased by silencing the wheat ABCC13 transporter gene (99). Protein content, especially essential amino acids lysine, methionine, cysteine, and tyrosine contents in wheat grains, were also attempted to enhance using Amaranthus albumin gene ama1 (100). Wheat has also been targeted to improve the antioxidant activity by expressing maize regulatory genes C1, B-peru involved in anthocyanin production (101). The development of biofortified crop varieties either by conventional breeding or transgenic methods is considered a sustainable solution to the problem of micronutrient deficiency. The advantage of this strategy over others like dietary diversification, food fortification, and food supplementation is that once the initial research and development is completed, the benefits of the nutritionally enhanced crops will be sustainable with little further investment. With the advent of powerful reverse genome editing tools such as transcription activator-like effector nucleases (TALENs) and Clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) coupled with fully sequenced genomes of wheat can be tested as a proof-of-concept for multiple micronutrients biofortification by targeting genes associated with micronutrients uptake and redistribution in different tissues. This will increase the biochemical and physiological pathway’s efficiency system biology (pathway reconstruction) and decrease the anti-nutritional factor to increase the bioavailability.
Genome editing technologies have emerged as advanced biotechnological, new plant breeding techniques, which offer efficient, target-specific and accurate approaches to engineer genome of a plant. The recent development of CRISPR/Cas9 based genome editing technologies in wheat has shown a ray of hope for improving the nutritional quality of wheat grains (Figure 2C). Edited gene constructs generated by these techniques have been delivered to the host genome by PEG-mediated protoplast fusion, particle gun bombardment, or ribonucleoprotein complex. Since wheat carries a complex hexaploid genome, CRISPR/Cas9—mediated wheat geminiviral based DNA replicons, delivering RNPs by biolistic method and multiplex editing has proved to be a more appropriate method of genome editing (102, 103). The TALEN and CRISPR/Cas9 systems have already demonstrated their utility for generating abiotic and biotic stress-resistant engineered wheat plants (37). For instance, the mildew resistance locus O (TaMLO) gene was edited by CRISPR/Cas9 and TALEN through PEG-mediated protoplast fusion method (39, 104), to achieve resistance to the fungal pathogen. Further, CRISPR/Cas9 genome editing system was applied to engineer dehydration responsive element-binding protein 2 (TaDREB2) and wheat ethylene-responsive factor 3 (TaERF3) to increase abiotic stress tolerance (105). A more sophisticated technique of multiplexed genome editing with CRISPR/Cas9 has also been demonstrated for wheat using TaGW, TaLpx and TaMLO genes (103). The gene editing approach has been deployed to improve wheat’s grain traits by utilizing the CRISPR/Cas9 RNP delivery of TaGW2 and TaGASR7 (negative regulators of grain traits and kernel weight) for increasing kernel weight (102). However, some recent studies have highlighted the use of genome editing for breeding varieties with improved grain quality and increased nutritional value in wheat. CRISPR/Cas9 system was applied to obtain a wheat variety with hypoimmunogenic gluten content by editing α-gliadin genes (40, 106). Similarly, high-amylose modern wheat varieties, needed for better human health, were developed through targeted mutagenesis of the gene TaSBEIIa by CRISPR/Cas 9 system (107). Further, the CRISPR/Cas9 editing tool has been demonstrated to be effective in simultaneous editing of multiple genes such as large α- and γ-gliadin gene families in the polyploid bread wheat (106). Other grain quality characteristics such as hardness, starch composition and dough color have been altered in wheat by targeting the pinb, waxy, ppo and psy genes (108).
Application of gene-editing techniques is required to be utilized furthermore for biofortification of wheat for enhancing Fe, Zn, Se, Ca and other micronutrient contents. Through QTL mapping and association mapping, information on genomic regions/QTLs responsible for grain Zn and Fe concentrations in different wheat varieties is now available, which can enhance Fe and Zn content in the high yielding varieties. Transgenic technology has been used to generate genetically modified plants with enhanced Fe content as well as better absorption by deploying different genes such as NAM-B1 transcription factor gene (24), TaFer1-A (90, 91), NAS2 (93, 94), TaVIT2 (95), and Phytochrome (phyA) (98). However, it is recommended that genome editing by CRISPR/Cas9 strategy be applied to provide marker- and foreign DNA- free genetically engineered plants with high Fe content and increased absorption. Attempts are needed to identify and validate genomic regions/QTLs contributing to phytic acid level in wheat grain which can then be engineered to modulate its level by the gene-editing system. For instance, ABCC13 can be knocked out to reduce the quantity of phytic acid to enhance the concentration of bioavailable minerals. Similarly, future research should focus on enhancing essential amino acid concentration and vitamin levels in wheat. Moreover, CRISPR/cas9—based genome editing systems can also be utilized to trim the unwanted sequences like marker gene, T-DNA region, etc. from the transgenic plants. It is highly recommended that the genome-edited wheat genotypes with improved grain quality characteristics be deployed regularly in wheat breeding programs to enrich the agronomically superior varieties with high nutritional value. It is also envisioned that more recently developed techniques like precise genome editing through base editors, and prime editors are utilized to improve grain quality efficiently and effectively (38, 109).
Wheat is the most widely cultivated, prominent food crop. There have been several attempts to improve wheat quality and crop yield after the “green revolution”. However, the main focus of wheat improvement programs has been on high yield, resulting in high yielding wheat varieties over time but with suboptimal levels of minerals and micronutrients. In order to achieve food and nutritional security, and to provide an adequate supply of calories and nutrients, it is imperative to improve both qualitative and quantitative traits of wheat. Biofortification of wheat can be achieved by exploring natural genetic diversity in wheat and its wild species for higher minerals and phytonutrients through utilizing advanced genomic tools such as QTL mapping and genome-wide association study (GWAS) for mapping nutritional quality traits in wheat, molecular breeding approaches, genomic selection, and genome engineering by transgenic technology and genome editing strategies. Genetic diversity studies have been performed, and few wild relatives of wheat, T. dicoccoides, Aegilops tauschii, T. dicoccoides, T. boeoticun, T. spelta, T. polonicum, and T. aestivum landraces have been identified that carry relatively higher concentrations of Fe, Zn and Mn. Similarly, QTL mapping and GWAS studies on wheat led to identifying loci responsible for grain Zn and Fe concentrations. Many attempts have been made using transgenic technology to generate wheat with better mineral and micronutrient content and transgenic wheat lines with higher content of Fe, Mn and Vit A have been reported. However, more recently, genome engineering tools like gene editing via CRISPR/Cas system, prime editors and base editors have gained much more popularity among scientists over conventional breeding and transgenic technology because of their efficacy, precision, simplicity and robustness.
The significant advantage of genome editing is that it eliminates the foreign DNA/transgene from the final engineered plants. Genome editing by TALEN and CRISPR/Cas 9 system has been employed in wheat to improve stress tolerance and grain yield; however, this system has not been explored as much for biofortification of wheat. Therefore, considering that biofortification of wheat is essential for improving grain quality, genome editing needs to be deployed to improve the content of Fe, Zn, Se, Ca, essential amino acids and decrease the concentration of antinutrients such as phytic acid. Other methods such as multiplex gene editing, transiently expressing CRISPR/Cas9, base editing, prime editing and CRISPR/Cas9 ribonucleoproteins are also promising and should be considered for future research. Exploring natural genetic diversity and broadening the narrow genetic base of hexaploid cultivated wheat varieties is essential and warrants greater attention through whole-genome sequencing of large number of accessions (e.g., the composite core set), and functional genomics for gene discovery associated with agronomic and nutritional traits (110, 111). Such efforts could help generate useful genetic information and genomic resources for accelerating wheat improvement through genome editing (112). Gene editing in germline cells and the CRISPR system carrying RNA interference elements need to be explored in wheat. Similarly, epigenetic genome modifications deserve attention. Also, simulation model-based prediction of superior wheat quality traits under different environmental conditions (113) might accelerate the global wheat nutritional quality program. Hence, utilization of these techniques to improve the nutritional quality of wheat grains and combine them with high yielding traits is emphasized.
OPG, AKS, GPS, KCB, and SKD conceived the idea and designed the outline. OPG, AKS, AS, and KCB drafted the manuscript. OPG, AKS, and AS prepared the illustrations. KCB, GPS, and SKD reviewed and improved the draft. All authors collected, compiled, analyzed, and interpreted the literature and contributed significantly to the article and approved the final version.
OPG and GPS are grateful to the Indian Council of Agricultural Research, Department of Agricultural Research and Education, Government of India for providing financial help under grant no. 1006422, institutional project (CRSCIIWBRSIL 201500900190). AKS, GPS, and OPG acknowledge the financial support received from DBT under the grant BT/Ag/Network/Wheat/2019-20.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the reviewers for critically reviewing the manuscript for its improvement.
1. Gillespie S, Hodge J, Yosef S, Pandya-Lorch R. Nourishing Millions: Stories of Change in Nutrition. Washington, DC: International Food Policy Research Institute (2016).
2. WHO (2017). Prevalence of Anaemia in Non-Pregnant Women Estimates by WHO Region. Available online at: https://apps.who.int/gho/data/view.main.ANAEMIAWOMENNPWREG (accessed October 20, 2021).
3. Abbaspour N, Hurrell R, Kelishadi R. Review on iron and its importance for human health. J Res Med Sci. (2014) 19:164–74.
5. World Health Organization [WHO].Global Anemia Estimates. (2021). Available online at: https://www.who.int/data/gho/data/themes/topics/anaemia_in_women_and_children (accessed October 3, 2021).
7. Lin PH, Sermersheim M, Li H, Lee P, Steinberg SM, Ma J. Zinc in wound healing modulation. Nutrients. (2017) 10:16. doi: 10.3390/nu10010016
8. Bailey RL, West KP Jr, Black RE. The epidemiology of global micronutrient deficiencies. Ann Nutr Metab. (2015) 66:22–33. doi: 10.1159/000371618
9. Walker CF, Ezzati M, Black RE. Global and regional child mortality and burden of disease attributable to zinc deficiency. Eur J Clin Nutr. (2009) 63:591–7. doi: 10.1038/ejcn.2008.9
10. Mehri A. Trace elements in human nutrition (II)–an update. Int J Prev Med. (2020) 11:2. doi: 10.4103/ijpvm.IJPVM_48_19
11. Jones GD, Droz B, Greve P, Gottschalk P, Poffet D, McGrath SP, et al. Selenium deficiency risk predicted to increase under future climate change. Proc Natl Acad Sci U S A. (2017) 114:2848–53. doi: 10.1073/pnas.1611576114
12. Pravina P, Sayaji D, Avinash M. Calcium and its role in human body. Int J Res Pharm Biomed Sci. (2013) 4:659–68.
13. Cakmak I, Kalayci M, Kaya Y, Torun AA, Aydin N, Wang Y, et al. Biofortification and localization of zinc in wheat grain. J Agric Food Chem. (2010) 58:9092–102. doi: 10.1021/jf101197h
14. Ibrahim S, Saleem B, Naeem MK, Arain SM, Khan MR. Next-generation technologies for iron and zinc biofortification and bioavailability in cereal grains. Crop Pasture Sci. (2021) 73:77–92.
15. Das A., Raychaudhuri U., Chakraborty R. (2011). Cereal based functional food of Indian subcontinent: a review. J. Food Sci. Tech. 49, 665–672. doi: 10.1007/s13197-011-0474-1
16. Vashishth A, Ram S, Beniwal V. Cereal phytases and their importance in improvement of micronutrients bioavailability. 3 Biotech. (2017) 7:42. doi: 10.1007/s13205-017-0698-5
17. Hurrell R, Egli I. Iron bioavailability and dietary reference values. Am J Clin Nutr. (2010) 91:1461S–7S.
18. Gibson RS. Zinc: the missing link in combating micronutrient malnutrition in developing countries. Proc Nutr Soc. (2006) 65:51–60. doi: 10.1079/pns2005474
19. Bhutta ZA, Salam RA, Das JK. Meeting the challenges of micronutrient malnutrition in the developing world. Br Med Bull. (2013) 106:7–17. doi: 10.1093/bmb/ldt015
20. Yadava DK, Hossain F, Mohapatra T. Nutritional security through crop biofortification in India: status & future prospects. Indian J Med Res. (2018) 148:621–31. doi: 10.4103/ijmr.IJMR_1893_18
21. Welch RM, Graham RD. Breeding for micronutrients in staple food crops from a human nutrition perspective. J Exp Bot. (2004) 55:353–64. doi: 10.1093/jxb/erh064
22. Gupta OP, Pandey V, Narwal S, Sharma P, Ram S, Singh GP editors. Wheat and Barley Grain Biofortification. 1st ed. Sawston: Woodhead Publishing (2020). p. 364. doi: 10.1016/C2018-0-04173-0
23. Singh R, Velu G. Zinc-Biofortified Wheat: Harnessing Genetic Diversity for Improved Nutritional Quality. Bonn: CIMMYT, HarvestPlus, and the Global Crop Diversity Trust (2017).
24. Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. (2006) 314:1298–301. doi: 10.1126/science.1133649
25. International Wheat Genome Sequencing Consortium [IWGSC]. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. (2018) 361:eaar7191. doi: 10.1126/science.aar7191
26. Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. (2017) 357:93–7. doi: 10.1126/science.aan0032
27. Ling H-Q, Ma B, Shi X, Liu H, Dong L, Sun H, et al. Genome sequence of the progenitor of wheat a subgenome Triticum urartu. Nature. (2018) 557:424–8. doi: 10.1038/s41586-018-0108-0
28. Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, et al. The transcriptional landscape of polyploid wheat. Science. (2018) 361:aar6089. doi: 10.1126/science.aar6089
29. Gupta OP, Pandey V, Saini R, Narwal S, Malik VK, Khandale T, et al. Identifying transcripts associated with efficient transport and accumulation of Fe and Zn in hexaploid wheat (T. aestivum L.). J Biotechnol. (2020) 316:46–55. doi: 10.1016/j.jbiotec.2020.03.015
30. Gupta OP, Pandey V, Saini R, Narwal S, Malik VK, Khandale T, et al. Transcriptomic dataset reveals the molecular basis of genotypic variation in hexaploid wheat (T. aestivum L.) in response to Fe/Zn deficiency. Data Brief. (2020) 31:105995. doi: 10.1016/j.dib.2020.105995
31. Gupta OP, Pandey V, Saini R, Khandale T, Singh A, Malik VK, et al. Comparative physiological, biochemical and transcriptomic analysis of hexaploid wheat (T. aestivum L.) roots and shoots identifies potential pathways and their molecular regulatory network during Fe and Zn starvation. Genomics. (2021) 113:3357–72. doi: 10.1016/j.ygeno.2021.07.029
32. Jia JZ, Zhao G. Wheat 660 SNP Array Developed by CAAS. (2016). Available online at: https://wheat.pw.usda.gov/ggpages/topics/Wheat660_SNP_array_developed_by_CAAS.pdf (accessed October 15, 2021).
33. Gadaleta A, Colasuonno P, Giove SL, Blanco A, Giancaspro A. Map-based cloning of QFhb.mgb-2A identifies a WAK2 gene responsible for Fusarium head blight resistance in wheat. Sci Rep. (2019) 9:6929. doi: 10.1038/s41598-019-43334-z
34. Velu G, Singh RP, Crespo-Herrera L, Juliana P, Dreisigacker S, Valluru R, et al. Genetic dissection of grain zinc concentration in spring wheat for mainstreaming biofortification in CIMMYT wheat breeding. Sci Rep. (2018) 8:13526. doi: 10.1038/s41598-018-31951-z
35. Nigro D, Gadaleta A, Mangini G, Colasuonno P, Marcotuli I, Giancaspro A, et al. Candidate genes and genome-wide association study of grain protein content and protein deviation in durum wheat. Planta. (2019) 249:1157–75. doi: 10.1007/s00425-018-03075-1
36. Muhu-Din Ahmed HG, Sajjad M, Zeng Y, Iqbal M, Habibullah Khan S, Ullah A, et al. Genome-wide association mapping through 90k SNP array for quality and yield attributes in bread wheat against water-deficit conditions. Agriculture. (2020) 10:392. doi: 10.3390/agriculture10090392
37. Sharma SK, Gupta OP, Pathaw N, Sharma D, Maibam A, Sharma P, et al. CRISPR-Cas-led revolution in diagnosis and management of emerging plant viruses: new avenues toward food and nutritional security. Front Nutr. (2021) 8:751512. doi: 10.3389/fnut.2021.751512
38. Molla KA, Sretenovic S, Bansal KC, Qi Y. Precise plant genome editing using base editors and prime editors. Nat Plants. (2021) 7:1166–87. doi: 10.1038/s41477-021-00991-1
39. Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. (2014) 32:947–51. doi: 10.1038/nbt.2969
40. Sánchez-León S, Gil-Humanes J, Ozuna CV, Giménez MJ, Sousa C, Voytas DF, et al. Low-gluten, non-transgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. (2018) 16:902–10. doi: 10.1111/pbi.12837
41. Khan MK, Pandey A, Hamurcu M, Gezgin S, Athar T, Rajput VD, et al. Insight into the prospects for nanotechnology in wheat biofortification. Biology. (2021) 10:1123. doi: 10.3390/biology10111123
42. Balint AF, Kovacs G, Erdei L, Sutka J. Comparison of the Cu, Zn, Fe, Ca and Mg contents of the grains of wild, ancient and cultivated wheat species. Cereal Res Commun. (2001) 29:375–82.
43. Qury FX, Leenhardt F, Remesy C, Chanliaud E, Duperrier B, Balfourier F, et al. Genetic variability and stability of grain magnesium, zinc and iron concentrations in bread wheat. Eur J Agron. (2006) 25:177–85.
44. Goel S, Singh B, Grewal S, Jaat RS, Singh NK. Variability in Fe and Zn content among Indian wheat landraces for improved nutritional quality. Indian J Genet Plant Breed. (2018) 78:426–32.
45. Zhao FJ, Su YH, Dunham SJ, Rakszegi M, Bedo Z, McGrath SP, et al. Variation in mineral micronutrient concentrations in grain of wheat lines of diverse origin. J Cereal Sci. (2009) 49:290–5.
46. Amiri R, Bahraminejad S, Sasani S, Jalali-Honarmand S, Fakhri R. Bread wheat genetic variation for grain’s protein, iron and zinc concentrations as uptake by their genetic ability. Eur J Agron. (2015) 67:20–6.
47. Guzman C, Peña RJ, Singh R, Autrique E, Dreisigacker S, Crossa J, et al. Wheat quality improvement at CIMMYT and the use of genomic selection on it. Appl Transl Genomics. (2016) 11:3–8. doi: 10.1016/j.atg.2016.10.004
48. Heidari P, Etminan A, Azizinezhad R, Khosroshahli M. Genomic variation studies in durum wheat (Triticum turgidum ssp. durum) using CBDP, SCoT and ISSR markers. Indian J Genet. (2017) 77:379–86.
49. Khokhar JS, King J, King IP, Young SD, Foulkes MJ, De Silva J, et al. Novel sources of variation in grain Zinc (Zn) concentration in bread wheat germplasm derived from Watkins landraces. PLoS One. (2020) 15:e0229107. doi: 10.1371/journal.pone.0229107
50. Chhuneja P, Dhaliwal HS, Bains NS, Singh K. Aegilops kotschyi and Aegilops tauschii as sources for higher levels of grain Iron and Zinc. Plant Breed. (2006) 125:529–31.
51. Rawat N, Tiwari VK, Singh N, Randhawa GS, Singh K, Chhuneja P, et al. Evaluation and utilization of Aegilops and wild Triticum species for enhancing iron and zinc content in wheat. Genet Resour Crop Evol. (2008) 56:53–64.
52. Tiwari VK, Rawat N, Chhuneja P, Neelam K, Aggarwal R, Randhawa GS, et al. Mapping of quantitative trait loci for grain iron and zinc concentration in diploid A genome wheat. J Hered. (2009) 100:771–6. doi: 10.1093/jhered/esp030
53. Cakmak I, Torun A, Millet E, Feldman M, Fahima T, Korol AB, et al. Triticum dicoccoides: an important genetic resource for increasing zinc and iron concentration in modern cultivated wheat. Soil Sci Plant Nutr. (2004) 50:1047–54.
54. Velu G, Crespo Herrera L, Guzman C, Huerta J, Payne T, Singh RP. Assessing genetic diversity to breed competitive biofortified wheat with enhanced grain Zn and Fe concentrations. Front Plant Sci. (2019) 9:1971. doi: 10.3389/fpls.2018.01971
55. Johansson E, Henriksson T, Prieto-Linde ML, Andersson S, Ashraf R, Rahmatov M. Diverse wheat-alien introgression lines as a basis for durable resistance and quality characteristics in bread wheat. Front Plant Sci. (2020) 11:1067. doi: 10.3389/fpls.2020.01067
56. Lyons G, Ortiz-Monasterio I, Stangoulis J, Graham R. Selenium concentration in wheat grain: Is there sufficient genotypic variation to use in breeding? Plant Soil. (2005) 269:369–80.
57. Lee S, Woodard HJ, Doolittle JJ. Selenium uptake response among selected wheat (Triticum aestivum) varieties and relationship with soil selenium fractions. Soil Sci Plant Nutr. (2011) 57:823–32.
58. Pandey A, Khan MK, Hakki EE, Thomas G, Hamurcu M, Gezgin S, et al. Assessment of genetic variability for grain nutrients from diverse regions: potential for wheat improvement. Springerplus. (2016) 5:1912. doi: 10.1186/s40064-016-3586-2
59. Alomari DZ, Eggert K, von Wirén N, Pillen K, Röder MS. Genome-wide association study of calcium accumulation in grains of European wheat cultivars. Front Plant Sci. (2017) 8:1797. doi: 10.3389/fpls.2017.01797
60. Hussain S, Maqsood M, Miller L. Bioavailable zinc in grains of bread wheat varieties of Pakistan. Cereal Res Commum. (2012) 40:62–73.
61. Branković G, Dragičević V, Dodig D, Knežević D, Kandić V, Šurlan-Momirović G, et al. Phytic acid, inorganic phosphorus, antioxidants in bread and durum wheat and their associations with agronomic traits. Agric Food Sci. (2015) 24:183–94.
62. Magallanes-López AM, Hernandez-Espinosa N, Velu G, Posadas-Romano G, Ordñez-Villegas V, Crossa J, et al. Variability in iron, zinc and phytic acid content in a worldwide collection of commercial durum wheat cultivars and the effect of reduced irrigation on these traits. Food Chem. (2017) 237:499–505. doi: 10.1016/j.foodchem.2017.05.110
63. Tabekha MM, Donnelly BJ. Phytic acid in durum wheat and its milled products. Cereal Chem. (1982) 59:105–7.
64. Peleg Z, Cakmak I, Ozturk L, Yazici A, Jun Y, Budak H, et al. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat x wild emmer wheat RIL population. Theor Appl Genet. (2009) 19:353–69. doi: 10.1007/s00122-009-1044-z
65. Tiwari C, Wallwork H, Arun B, Mishra VK, Velu G, Stangoulis J. Molecular mapping of quantitative trait loci for zinc, iron and protein content in the grains of hexaploid wheat. Euphytica. (2016) 207:563–70. doi: 10.1371/journal.pone.0179851
66. Velu G, Tutus Y, Gomez-Becerra HF, Hao Y, Demir L, Kara R, et al. QTL mapping for grain zinc and iron concentrations and zinc efficiency in a tetraploid and hexaploid wheat mapping populations. Plant Soil. (2017) 411:81–99. doi: 10.1007/s11104-016-3025-8
67. Krishnappa G, Singh AM, Chaudhary S, Ahlawat AK, Singh SK, Shukla RB, et al. Molecular mapping of the grain iron and zinc concentration, protein content and thousand kernel weight in wheat (Triticum aestivum L.). PLoS one. (2017) 12:e0174972. doi: 10.1371/journal.pone.0174972
68. Liu J, Wu B, Singh RP, Velu G. QTL mapping for micronutrients concentration and yield component traits in a hexaploid wheat mapping population. J Cereal Sci. (2019) 88:57–64. doi: 10.1016/j.jcs.2019.05.008
69. Krishnappa G, Rathan ND, Sehgal D, Ahlawat AK, Singh SK, Singh SK, et al. Identification of novel genomic regions for biofortification traits using an SNP marker-enriched linkage map in wheat (Triticum aestivum L.). Front Nutr. (2021) 8:669444. doi: 10.3389/fnut.2021.669444
70. Xu Y, An D, Liu D, Zhang A, Xu H, Li B. Molecular mapping of QTLs for grain zinc, iron and protein concentration of wheat across two environments. Food Crop Res. (2012) 138:57–62. doi: 10.1016/j.fcr.2012.09.017
71. Hao Y, Velu G, Peña RJ, Singh S, Singh RP. Genetic loci associated with high grain zinc concentration and pleiotropic effect on kernel weight in wheat (Triticum aestivum L.). Mol Breed. (2014) 34:1893–902. doi: 10.1007/s11032-014-0147-7
72. Pu CX, Han YF, Zhu S, Song FY, Zhao Y, Wang CY, et al. The rice receptor-like kinases DWARF AND RUNTISH SPIKELET1 and 2 repress cell death and affect sugar utilization during reproductive development. Plant Cell. (2017) 29:70–89. doi: 10.1105/tpc.16.00218
73. Wang C, Ji J, Zhu F. Characterizing Se transfer in the soil-crop systems under field condition. Plant Soil. (2017) 415:535–48. doi: 10.1002/0471142913.faf0102s00
74. Alomari DZ, Alqudah AM, Pillen K, von Wirén N, Röder MS. Toward identification of a putative candidate gene for nutrient mineral accumulation in wheat grains for human nutrition purposes. J Exp Bot. (2021) 72:6305–18. doi: 10.1093/jxb/erab297
75. Upadhyaya H, Bajaj D, Das S, Kumar V, Gowda CL, Sharma S, et al. Genetic dissection of seed-iron and zinc concentrations in chickpea. Sci Rep. (2016) 6:24050. doi: 10.1038/srep24050
76. Grusak MA, Cakmak I. Methods to improve the crop-delivery of minerals to humans and livestock. In: MR Broadley, PJ White editors. Plant Nutritional Genomics. Oxford: Blackwell Publishing Ltd (2005). p. 265–6.
77. Weckwerth W, Ghatak A, Bellaire A, Chaturvedi P, Varshney RK. PANOMICS meets germplasm. Plant Biotechnol J. (2020) 7:1507–25. doi: 10.1111/pbi.13372
78. Gupta OP, Deshmukh R, Kumar A, Singh SK, Sharma P, Ram S, et al. From gene to biomolecular networks: a review of evidences for understanding complex biological function in plants. Curr Opin Biotechnol. (2022) 74:66–74. doi: 10.1016/j.copbio.2021.10.023
79. Massman JM, Jung HJG, Bernardo R. Genome-wide selection versus marker-assisted recurrent selection to improve gain yield and stover-quality traits for cellulosic ethanol in maize. Crop Sci. (2013) 53:58–66. doi: 10.2135/cropsci2012.02.0112
80. Gaikwad KB, Rani S, Kumar M, Gupta V, Babu PH, Bainsla NK, et al. Enhancing the nutritional quality of major food crops through conventional and genomics-assisted breeding. Front Nutr. (2020) 7:533453. doi: 10.3389/fnut.2020.533453
81. Joukhadar R, Thistlethwaite R, Trethowan RM, Hayden MJ, Stangoulis J, Cu S, et al. Genomic selection can accelerate the biofortification of spring wheat. Theor Appl Genet. (2021) 134:3339–50. doi: 10.1007/s00122-021-03900-4
82. Owens BF, Gore MA, Magallanes-Lundback M, Tiede T, Diepenbrock CH, Kandianis CB, et al. A foundation for provitamin a biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels. Genetics. (2014) 198:1699–716. doi: 10.1534/genetics.114.169979
83. Velu G, Crossa J, Singh RP, Hao Y, Dreisigacker S, Perez-Rodriguez P, et al. Genomic prediction for grain zinc and iron concentrations in spring wheat. Theor Appl Genet. (2016) 129:1595–605. doi: 10.1007/s00122-016-2726-y
84. Prasanna BM, Palacios-Rojas N, Hossain F, Muthusamy V, Menkir A, Dhliwayo T, et al. Molecular breeding for nutritionally enriched maize: status and prospects. Front Genet. (2020) 10:1392. doi: 10.3389/fgene.2019.01392
85. Manickavelu A, Hattori T, Yamaoka S, Yoshimura K, Kondou Y, Onogi A, et al. Genetic nature of elemental contents in wheat grains and its genomic prediction: toward the effective use of wheat landraces from Afghanistan. PLoS One. (2017) 12:e0169416. doi: 10.1371/journal.pone.0169416
86. Ludwig Y, Slamet-Loedin IH. Genetic biofortification to enrich rice and wheat grain iron: from genes to product. Front Plant Sci. (2019) 10:833. doi: 10.3389/fpls.2019.00833
87. Distelfeld A, Cakmak I, Peleg Z, Ozturk L, Yazici AM, Budak H, et al. Multiple QTL-efects of wheat Gpc-B1 locus on grain protein and micronutrient concentrations. Physiol Plant. (2007) 129:635–43.
88. Randhawa HS, Asif M, Pozniak C, Clarke JM, Graf RJ, Fox SL, et al. Application of molecular markers to wheat breeding in Canada. Plant Breed. (2013) 132:458–71.
89. Tabbita F, Lewis S, Vouilloz JP, Ortega MA, Kade M, Abbate PE, et al. Effects of the Gpc-B1 locus on high grain protein content introgressed into Argentinean wheat germplasm. Plant Breed. (2013) 132:48–52.
90. Borg S, Brinch-Pedersen H, Tauris B, Madsen LH, Darbani B, Noeparvar S, et al. Wheat ferritins: improving the iron content of the wheat grain. J Cereal Sci. (2012) 56:204–13.
91. Xiaoyan S, Yan Z, Shubin W. Improvement Fe content of wheat (Triticum aestivum) grain by soybean ferritin expression cassette without vector backbone sequence. J Agric Biotechnol. (2012) 20:766–73.
92. Holm PB, Kristiansen KN, Pedersen HB. Transgenic approaches in commonly consumed cereals to improve iron and zinc content and bioavailability. J Nutr. (2002) 132:514S–6S. doi: 10.1093/jn/132.3.514S
93. Singh D, Geat N, Rajawat MVS, Mahajan MM, Prasanna R, Singh S, et al. Deciphering the mechanisms of endophyte-mediated biofortification of Fe and Zn in wheat. J Plant Growth Regul. (2017) 37:174–82.
94. Beasley JT, Bonneau JP, Sanchez-Palacios JT, Moreno-Moyano LT, Callahan DL, Tako E, et al. Metabolic engineering of bread wheat improves grain iron concentration and bioavailability. Plant Biotechnol J. (2019) 17:1514–26. doi: 10.1111/pbi.13074
95. Connorton JM, Jones ER, Rodríguez-Ramiro I, Fairweather-Tait S, Uauy C, Balk J. Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. (2017) 174:2434–44. doi: 10.1104/pp.17.00672
96. Cong L, Wang C, Chen L, Liu H, Yang G, He G. Expression of phytoene synthase1 and carotene desaturase crtI genes result in an increase in the total carotenoids content in transgenic elite wheat (Triticum aestivum L.). J Agric Food Chem. (2009) 57:8652–60. doi: 10.1021/jf9012218
97. Wang C, Zeng J, Li Y, Hu W, Chen L, Miao Y, et al. Enrichment of provitamin A content in wheat (Triticum aestivum L.) by introduction of the bacterial carotenoid biosynthetic genes CrtB and CrtI. J Exp Bot. (2014) 65:2545–56. doi: 10.1093/jxb/eru138
98. Brinch-Pederson H, Olesen A, Rasmussen SK, Holm PB. Generation of transgenic wheat (Triticum aestivum L.) for constitutive accumulation of an Aspergillus phytase. Mol Breed. (2000) 6:195–206.
99. Bhati KK, Alok A, Kumar A, Kaur J, Tiwari S, Pandey AK. Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J Exp Bot. (2016) 67:4379–89. doi: 10.1093/jxb/erw224
100. Tamas C, Kisgyorgy BN, Rakszegi M, Wilkinson MD, Yang MS, Lang L, et al. Transgenic approach to improve wheat (Triticum aestivum L.) nutritional quality. Plant Cell Rep. (2009) 28:1085–94. doi: 10.1007/s00299-009-0716-0
101. Doshi KM, Eudes F, Laroche A, Gaudet D. Transient embryo specific expression of anthocyanin in wheat. In Vitro Cell Dev Biol. (2006) 42:432–8.
102. Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. (2016) 7:12617. doi: 10.1038/ncomms12617
103. Wang W, Pan Q, He F, Akhunova A, Chao S, Trick H, et al. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. (2018) 1:65–74. doi: 10.1089/crispr.2017.0010
104. Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. (2014) 9:2395–410. doi: 10.1038/nprot.2014.157
105. Kim D, Alptekin B, Budak H. CRISPR/Cas9 genome editing in wheat. Funct Integr Genomics. (2018) 18:31–41.
106. Jouanin A, Schaart JG, Boyd LA, Cockram J, Leigh FJ, Bates R, et al. Outlook for coeliac disease patients: towards bread wheat with hypoimmunogenic gluten by gene editing of α-and γ-gliadin gene families. BMC Plant Biol. (2019) 19:333. doi: 10.1186/s12870-019-1889-5
107. Li J, Jiao G, Sun Y, Chen J, Zhong Y, Yan L, et al. Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotechnol J. (2021) 19:937–51. doi: 10.1111/pbi.13519
108. Zhang S, Zhang R, Gao J, Song G, Li J, Li W, et al. CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnol J. (2021) 19:1684–6. doi: 10.1111/pbi.13647
109. Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. (2017) 35:438–40. doi: 10.1038/nbt.3811
110. Dutta M, Phogat BS, Kumar S, Kumar N, Kumari J, Pandey AC, et al. Development of core set of wheat (Triticum spp.) germplasm conserved in the National Genebank in India. In: Y Ogihara, S Takumi, H Handa editors. Advances in Wheat Genetics: From Genome to Field. Berlin: Springer (2015). p. 33–45. doi: 10.1007/978-4-431-55675-6_4
111. Phogat BS, Kumar S, Kumari J, Kumar N, Pandy AC, Singh TP, et al. Characterization of wheat germplasm conserved in the Indian National Genebank and establishment of a composite core collection. Crop Sci. (2020) 61:604–20. doi: 10.1002/csc2.20285
112. Li S, Zhang C, Li J, Yan L, Wang N, Xia L. Present and future prospects for wheat improvement through genome editing and advanced technologies. Plant Commun. (2021) 2:100211. doi: 10.1016/j.xplc.2021.100211
113. Mohan D, Sendhil R, Gupta OP, Pandey V, Gopalareddy K, Singh GP. Wheat quality index: new holistic approach to identify quality superior genotypes. Cereal Res Commun. (2022). doi: 10.1007/s42976-022-00254-5
114. Arora S, Cheema J, Poland J, Uauy C, Chhuneja P. Genome-wide association mapping of grain micronutrients concentration in Aegilops tauschii. Front Plant Sci. (2019) 10:54. doi: 10.3389/fpls.2019.00054
115. Bhatta M, Baenziger PS, Waters BM, Poudel R, Belamkar V, Poland J, et al. Genome-wide association study reveals novel genomic regions associated with 10 grain minerals in synthetic hexaploid wheat. Int J Mol Sci. (2018) 19:3237. doi: 10.3390/ijms19103237
116. Rathan ND, Sehgal D, Thiyagarajan K, Singh R, Singh A-M, Govindan V. Identification of genetic loci and candidate genes related to grain zinc and iron concentration using a zinc-enriched wheat ‘zinc-shakti’. Front Genet. (2021) 12:652653. doi: 10.3389/fgene.2021.652653
Keywords: micronutrients, hidden hunger, phytate, QTLs, genome editing
Citation: Gupta OP, Singh AK, Singh A, Singh GP, Bansal KC and Datta SK (2022) Wheat Biofortification: Utilizing Natural Genetic Diversity, Genome-Wide Association Mapping, Genomic Selection, and Genome Editing Technologies. Front. Nutr. 9:826131. doi: 10.3389/fnut.2022.826131
Received: 30 November 2021; Accepted: 06 June 2022;
Published: 12 July 2022.
Edited by:
Miroslav Nikolic, University of Belgrade, SerbiaReviewed by:
Roi Ben-David, Agricultural Research Organization (ARO), IsraelCopyright © 2022 Gupta, Singh, Singh, Singh, Bansal and Datta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gyanendra Pratap Singh, Z3Auc2luZ2hAaWNhci5nb3YuaW4=; Kailash C. Bansal, a2NiYW5zYWwyN0BnbWFpbC5jb20=; Swapan K. Datta, c3dwbmRhdHRhQHlhaG9vLmNvbQ==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.