
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr. , 22 February 2022
Sec. Nutrition and Microbes
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.821900
This article is part of the Research Topic Next-Generation Prebiotics and Probiotics: Current Status and Future Development View all 7 articles
Allergy is a hypersensitivity reaction triggered by specific cell or antibody-mediated immune mechanisms. Allergies have increased in industrialized countries in recent decades. The rise in allergic respiratory diseases such as allergic rhinitis (AR) and allergic asthma (AA) is a potential threat to public health. Searches were conducted using PubMed, Google Scholar and Medline using the following key terms: allergic rhinitis OR asthma AND probiotics, allergic airway inflammation AND immune disorders, probiotics OR gut microbiota AND allergic disease, probiotics AND inflammatory. Studies from all years were included, specifically those published within the last 10 years. Some review articles and their reference lists were searched to identify related articles. The role of microbiota in respiratory allergic diseases has attracted more and more attention. Pieces of evidence suggested that the development of allergic diseases causes a possible imbalance in the composition of the gut microbiota. Compared to colonized mice, germ-free mice exhibit exaggerated allergic airway responses, suggesting that microbial host interactions play an important role in the development of allergic diseases. Probiotics modulate both the innate and adaptive inflammatory immune responses, often used as dietary supplements to provide health benefits in gastrointestinal disorders. Probiotics may serve as immunomodulators and activators of host defense pathways. Besides, oral probiotics can modulate the immune response in the respiratory system. Recently, studies in humans and animals have demonstrated the role of probiotic in RA and AA. To understand the characterization, microbiota, and the potential role of probiotics intervention of AA/AR, this review provides an overview of clinical features of AA and AR, probiotics for the prevention and treatment of AR, AA, changes in gut microbiota, and their mechanisms of action.
Respiratory tract allergic disease includes allergic asthma (AA) and allergic rhinitis (AR). Asthma is one of the most common chronic non-communicable diseases. Approximately 334 million people around the world suffer from asthma (1). Primary characterization of AA is airway inflammation, both reversible airway obstruction and intra-airway hyperresponsiveness. Nasal mucosa swelling, enhanced vascular permeability, mass watery secretions are features of AR. The identity in upper and lower airway makes AA associated with AR. AR is a common allergic airway disease that associated with allergic AA (2). The global prevalence of AR among children was 2~25%, adults were 10~40% (3).
Type I hypersensitivity reactions refer to the acute allergic reaction that takes place after an allergen challenge. The most important humoral player is allergen-specific immunoglobulin E (IgE), which is induced in a T helper-2 (Th2) milieu associated with diseases of the atopy spectrum, such as AR and AA. Allergen-specific IgE is bound via its Fc-tail to high-affinity Fc receptors (FcεRI) on mast cells in tissue and basophils in blood. Allergen binding to multiple IgE molecules on mast cells and basophils, so-called IgE crosslinking, will activate these cells and result in degranulation and immediate release of several pre-formed inflammatory mediators, including histamine, leukotrienes, and prostaglandins (4). In addition, mast cells and basophils will release inflammatory cytokines and chemokines (5). In both AA and AR, the inflammatory response involves innate immunity eosinophils, neutrophils, macrophages, mast cells, natural killer cells, γδ-T cells, innate lymphoid cells (ILCs) and dendritic cells (DCs) and adaptive immunity (T and B lymphocytes). Majority of asthma patients are atopic and have an allergic pattern of inflammation in their airways (6). T lymphocytes play a very important role in coordinating the inflammatory response in asthma through the release of specific patterns of cytokines, resulting in the recruitment and survival of eosinophils and in the maintenance of a mast cell population in the airways (7). Mast cells are key effector cells in asthma through their release of multiple bronchoconstrictor mediators, such as histamine, cysteinyl-leukotrienes and prostaglandin (PG)-D2 (8). The role of macrophages in asthma is currently uncertain as they may have pro-inflammatory or anti-inflammatory effects. Macrophages may be activated by allergens via low-affinity IgE receptors (9). DC takes up allergens, process them to peptides and migrate to local lymph nodes where they present the allergenic peptides to uncommitted T lymphocytes to programme the production of allergen-specific T cells. Immature DC in the respiratory tract promotes helper Th2 cell differentiation (10). The cytokine thymic stromal lymphopoietin released from epithelial cells in asthmatic patients programmes DC to release chemokines that attract Th2 cells into the airways (11). Eosinophilic inflammation is a characteristic feature of asthmatic airways (12). The mechanisms of neutrophilic inflammation in asthma are uncertain and could be related to the use of high doses of corticosteroids which prolong neutrophil survival in the airways or due to bacterial infection (13). The neutrophilic inflammation in severe asthma may be orchestrated by Th17 cells and increased expression of IL-17A and IL-17F is described in airways of patients with severe asthma (14). Allergic Rhinitis is a Th2-driven, IgE-mediated disease, is characterized by mucosal inflammation, driven by activated immune cells. Mast cells and Th2 cells might decrease epithelial barrier integrity in AR, maintaining a leaky epithelial barrier (15). Recent studies have demonstrated that increased IL-17 levels and Th17 cell number in nasal mucosa and peripheral blood are associated with clinical severity in patients with AR (16). An in-season study involving grass pollen-sensitized patients of allergic rhinoconjunctivitis showed a significant increase in circulating ILC2 numbers (17). ILC2s represent an abundant alternative source of Th2 cytokines and likely serve to amplify and maintain local Th2-driven allergic inflammation (18). The cardinal features of allergic rhinitis include increased allergen-specific IgE concentrations to clinically relevant allergens, IgE-dependent activation of mast cells, and local eosinophilia in target organs. Specific IgE can be synthesized and produced locally by B cells within the respiratory mucosa (19).
AA and AR are IgE mediated responses. Not only is ecological dysbiosis of the airway microbiota associated with AA and AR, but also there may be similar ecological dysbiosis of the gut microbiota (20, 21). The human microbiota starts from the mouth and crosses the esophagus, then the gastric, small intestine, and colon, and finally, reaches the rectum. The distribution surface area of microbiota in the human body is 150–200 m2, provides the opportunity for microbial colonization or transient occupation. Bacteria are found in many parts of the human body primarily on the external and internal surfaces, the vast majority of commensal bacteria reside in the colon, about 1014 bacteria, followed by the skin, about 1012 bacteria. Less than 1012 bacteria populate the rest of the body (22).
Asthma is related to gut microbiota (23). The research discovered that Lachnospira, Veillonella, Faecalibacterium, and Rothiawere reduced in stool samples from 3-month-old Canadian children with a high risk of asthma. Moreover, functional analysis of the bacterial community showed that the LPS pathway of biosynthesis was reduced across the microbiota of children at high risk of asthma (24). The study found the composition of Proteobacteria, Bacteroidetes, and Actinobacteria in allergic rhinitis were decreased (25). Thus, regulating gut microbiota may be an effective way to treat AA and AR.
Fecal microbiota transplantation (FMT) can be used to treat inflammatory bowel disease by reshaping the gut microbiota (26). Probiotics are microorganisms that promote human health by improving the intestinal micro-ecological balance, another choice to restore disorders of the microbiota may be probiotics. Interaction between a healthy microbiota and the immune system led to a well-balanced body (27). A dysfunctional microbiome and lack of diversity can lead to the evolution of disease (28). In this review, we discuss the characteristics of the microbiota in allergic airway inflammation (e.g., allergic rhinitis and asthma), the role of probiotics in experimental animal models and clinical studies.
In order to discuss the effect of probiotics on respiratory tract allergic disease and gut microbiota, we undertook a systematized literature search that included observational studies (cross-sectional, cohort, or case-control) and experimental studies. The following exclusion criteria were used to reduce possible relationships observed due to other comorbidities: Atopic dermatitis, food allergies, intestinal disorders, respiratory microbiota. Searches were conducted using PubMed, Google Scholar, and Medline using the following key terms: allergic rhinitis OR asthma AND probiotics, allergic airway inflammation AND immune disorders, probiotics OR gut microbiota AND allergic disease, probiotics AND inflammatory. Studies from all years were included, specifically those published within the last 10 years. Some review articles and their reference lists were also searched to identify related articles. The search was limited to in vivo studies.
AA is the most commonly diagnosed respiratory allergic disease in clinical trials. It is a disease of chronic inflammation of the airways that involves multiple cells and cellular components such as mast cells, eosinophils, and lymphocytes. Its pathophysiology is complex, and many aspects are not well understood. Asthma is a critical global health problem which affects people of all ages. Its prevalence is growing in many countries, especially among children. Despite decreases in hospitalizations and asthma deaths in some countries, asthma still places an unacceptable burden on the health care system and society, especially the devastating effects of childhood asthma on families (29).
The disease affects about 300 million people worldwide and could rise to 400 million by 2025, influencing 15 percent of the world's children and adolescents (30). Asthma is caused by a combination of poorly understood genetic and environmental factors. Studies have systematically mapped the effects of single nucleotide polymorphisms (SNPs) on the presence of childhood onset asthma by genome-wide association. Characterized more than 317,000 SNPs in DNA from 994 patients with childhood onset asthma and 1,243 non-asthmatics, using family and case-referent panels, confirmed that Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma (31). There is an association between IL33, IL1RL1/IL18R1, HLA-DQ, SMAD3, and IL2RB9 gene polymorphisms and the locus on chromosome 17q21 including ZPBP2, GSDMB, and ORMDL3 genes (32). In addition, these genes showed that abnormal epithelial barrier function, as well as innate and adaptive immune responses, can cause asthma.
After an allergic response and consequent stimulation in the presence of dendritic cells (e.g., epithelial-derived thymic stromal lymphopoietin and other coactivators) adaptive T helper 2 cells produce interleukin (IL)-5, IL-4, and IL-13. In addition, some patients with predominantly neutrophilic disease from T helper 1 (Th1) cell (IL-2, interferon, tumor necrosis factor), T helper 17 (Th17) cells (IL-17), 21 or type 3 innate cytokine release from Th1 cell, Th17 cells (33), or three innate lymphocytes, as well as macrophage activation and the release of neutrophil chemokines, such as the C-X-C motif chemokine ligand eight (7). High morbidity and mortality as asthma account for one in 250 deaths worldwide. Currently, hormone therapy such as inhaled corticosteroids, oral corticosteroids or glucocorticolds are used to treat asthma, but it is not effective in treating the root cause, and the heavy medical costs can be financially stressful. Multiple large-scale cluster analyses of asthma patients have identified various asthma endotypes, the incidence of which depends on age, gender, BMI, or inflammation spectrum of asthma onset (34–36). In these groups, early-onset asthma patients (asthma that occurs between 0 and 12 years of age) usually exhibit atopic symptoms, predominantly male; however, after puberty, the population is predominantly female (37). Therefore, it is essential to improve knowledge in this field to discover new treatments. Probiotics are live microorganisms that have proven to be beneficial to human health. Many clinical trials have demonstrated that probiotics can modulate the gut microbiota and that the gut microbiota can regulate the systemic immune system response and play an important role in the development of asthma, thus promoting the health of the body (27).
The onset of probiotic protection dates from fetal life (38), probiotic bacteria settled in the gut wield an influence on respiratory allergies and can relieve the symptoms. Supplementation with probiotics in pre-and postnatal periods was likely to play an essential strategic role in the prevention of asthma (39). Therefore, probiotic supplementation may be a new way to prevent/treat allergic diseases.
The prevalence of chronic inflammatory diseases of the upper respiratory tract, such as AR and chronic rhinosinusitis (CRS), often occurring in conjunction with asthma and conjunctivitis which affects up to 50% of people in some countries (40), it is a global health problem with a major burden worldwide. Indeed, AR lead to unproductive time at work, sleep problems, and for children it also reduces participation in outdoor activities (41–43). Clinical symptoms of AR include nasal bleeding, sneezing, nasal congestion, itching, burning or redness of the eyes, and an itchy throat. AR is known as IgE immunoglobulins-mediated disease, and the main disease-modifying treatment is allergen immunotherapy. These immune responses involve inflammation of the mucosa controlled by type 2 cells (44). Available treatments include avoidance of allergens, H1-antihistamines or intranasal corticosteroids drug therapy, and allergen-specific immunotherapy (AIT) (45). However, the use of drugs for allergic rhinitis (such as histamine antagonists) can produce undesirable side effects, such as fatigue. Probiotics may be a new treatment for allergic rhinitis (46).
Probiotic interventions are potentially useful with low adverse effects such as Lactobacillus and Bifidobacterium. The consumption of probiotics may provide a more balanced gut microbiota in people with allergic rhinitis, which may limit the damage caused by inflammation. In addition, well-balanced gut microbiota may lead to less severe reactions to allergens (47). However, further research is needed to fully understand the potential mechanisms. The preventive effects of probiotics on allergic diseases have been reported, and although there was considerable variation between these studies, the results suggested that probiotics have important clinical and immunological effects in the treatment of allergic rhinitis (48).
The gut is the main site of interaction between the host immune system and both commensal, as well as pathogenic microbes during the first years of life. Immunity is shaped by commensal microbiota. From early life onwards, microbes colonize mucosal surfaces of the body and thereby trigger the establishment of immune homeostasis and defense mechanisms (49). Evidence reveals that the family of innate lymphoid cells (ILCs), which are mainly located in mucosal tissues, are essential in the maintenance of barrier functions as well as in the initiation of an appropriate immune response upon pathogenic infection. The formation of early lymphoid tissue inducer cells (LTi) seems to be independent of the presence of the commensal microbiota as germ-free (GF) mice harbor normal numbers of lymph nodes and Peyer's patches (50). Maternal dietary components, such as retinoic acid (RA), were shown to be crucial to maintain this LTi cell population during fetal development. Microbiota is directly regulating the availability of RA (51). CCR6+c-kithi LTi cells seed the intestinal lamina propria during fetal development, and low numbers of CCR6−c-kitlo ILC3s that expand strongly within the first 2–4 weeks of birth and that can acquire NK cell markers (52), which was shown to be dependent on signals originating from the maternal microbiota as shown in a model of reversible gestational colonization during pregnancy (53). Aryl hydrocarbon receptor (AhR) ligands produced by the maternal microbiota were transferred to the offspring postnatally through the milk and permanently increased the absolute number of NKp46+ILC3s in the offspring small intestinal lamina propria starting at postnatal day 14 (54). However, subject to controversy as other studies showed a negative influence of missing microbiota on the different ILC3 subsets (55). Natural ligands of the AhR include agonists derived from cruciferous vegetables in the diet as well as indoles produced by members of the microbiota. It is likely to speculate that AhR ligands derived from not only the maternal microbiota but also from the endogenous microbiota of the offspring contribute to the homeostasis of this ILC population. While the T-bet induced upregulation of NK cell receptors (NCRs) on ILC3s was dependent on the presence of the commensal microbiota as described above (56) the loss of RORyt expression in NCR+ILC3s and their switch to a more ILC1-like phenotype was prevented in colonized compared with GF mice (54). The role of microbiota in the differentiation of ILC1s and NK cells during early life has not been investigated in detail. A study suggested that colonization of the neonatal intestine is triggered through microbiota colonization after birth as this subset seems absent in the fetal intestine. In addition, they show that this ILC1 subset has the potential to differentiate into RORγt-ex- pressing ILC3 during adulthood in the presence of IL-23 and RA, the latter being partially regulated by the commensal microbiota (57). ILC1 and ILC2 subsets were most affected by the absence of commensal microbiota and acquired a phenotype that more closely resembled ILC3 subsets. In conclusion, while ILC3s seem to be mainly influenced by microbial signals during early life, ILC2s and ILC1s are more shaped through microbiota during adulthood.
Microbiota-induced IL-1β also induces the release of granulocyte monocyte colony-stimulating factor (GM-CSF) by ILC3, which in turn promotes mononuclear phagocytes to produce regulatory components, such as IL-10 and RA (58). The latter are important to promote regulatory T-cell (Treg) differentiation and expansion in the intestine to ensure intestinal homeostasis. In the absence of microbiota that functionally impaired DCs were unable to produce the NK priming cytokines IFN-I and IL-15. Behave as splenic NK cells in GF and antibiotic-treated mice are largely unresponsive when stimulated to produce IFN-c or exert specific lysis of target cells following Toll-like receptor (TLR) ligand exposure in vivo (59).
While ILC function is influenced by commensal microbiota, ILCs themselves can influence adaptive immune cells, which in return help keeping the host-microbial mutualism in check. The elimination of microbiota-specific T-cells was very important to prevent low-grade systemic and spontaneous intestinal inflammation (60). An independent study demonstrated that splenic but not intestinal MHC-II-expressing ILC3s can be activated through microbiota-derived IL-1b to express co-stimulator molecules and thus drive CD4+T-cell and B-cell responses in vivo (61). While microbiota diversity and phylum composition were unaltered in the absence of ILC3s, these mice exhibited higher levels of segmented filamentous bacteria (SFB) as well as Clostridiales species (62). A study demonstrated that Id2 expression in ILC3s was important for the generation of IL-22, which maintained a healthy microbiota that exhibited early colonization resistance to Citrobacter rodentium (62).
The gut microbiota is the normal microorganisms that colonize the human intestine are in a symbiotic relationship with the body and are diverse and abundant. There is evidence that more than 50 diseases are associated with dysbiosis of the gut microbiota, such as many infectious diseases, liver diseases, gastrointestinal malignancies, metabolic disorders, and allergic diseases (63). The colonization of gut microbiota early in life affects the immune status during childhood, and early gut microecological dysregulation due to various factors (mode of delivery, feeding practices, antibiotic use, environmental exposure, etc.) may disrupt the homeostatic regulatory mechanisms between Th1/Th2 cells, which in turn may negatively affect the development of immune tolerance and may eventually activate the allergic process, increase the risk of allergy, and thus induce and exacerbate allergic diseases (64).
The gut microbiota plays a crucial role in the maturation process of the postnatal immune system, especially in immune tolerance. It was found that germ-free mice failed to induce Th2-mediated immune tolerance, whereas oral immune tolerance developed after the re-establishment of gut microbiota. The role of gut microbiota on immune development is mediated by TLR2 through DCs. In addition, regulatory T cells play a key regulatory role in immune tolerance. The gut microbiota and its metabolites can induce regulatory T cells and participate in the formation of mucosal immune tolerance. The gut microbiota can also enhance the mucosal barrier effect by stimulating the secretion of sIgA. The gut microbiota protects the host from allergic reactions by acting on both intrinsic and adaptive immunity (65).
Disturbances in the gut microbiota also affect the development of immune tolerance in the respiratory mucosa. Gill et al. reported the relationship between pulmonary immunity, the mucosal immune system, and the gut microbiota group: changes in the gut microbiota lead to alterations in the downstream immune response, affecting the development of the immune system, which in turn affects the immune response in distal mucosal organs (e.g., the lung, which subsequently leads to the development of pulmonary inflammation); abnormalities in the immune system, In turn, affect the gut microbiota composition, which in turn affects the immune response of the distal mucosal system and triggers an immune imbalance in the lungs (66). The gut microbiota can enhance pulmonary immunity and clear pulmonary pathogens through the flora and its metabolites, reducing the development of pulmonary diseases; conversely, pulmonary diseases can affect the structural composition and diversity of the gut microbiota, causing the corresponding intestinal symptoms (67). Modern medicine refers to the interaction between the intestine and the lung as the “Gut-Lung axis”. The “Gut-Lung axis” helps us to better understand the relationship between gut microbiota and allergic diseases.
The human intestine starts from the mouth, passes through the esophagus, stomach, small intestine, colon, and finally reaches the rectum. Its huge surface area of 150–200 square meters provides an opportunity for microbes to colonize (4). They are mainly composed of the following five phyla: Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria (68, 69), and Bacteroides, Faecalibacterium, and Bifidobacterium are the most common genera in healthy adults (70). The bacterial composition of each part is as follows: the main colonizing bacteria of the oral cavity are: Streptococcaceae, Pasteurellaceae, Veillonellaceae, Prevotellaeace, and Neisseriaceae families and Gemella genus; the stomach mainly contains bacteria from the Lactobacillaceae family; the small intestine is dominated by Lactobacillaceae, Enterobacteriaceae, and Streptococcaceae; the large intestine contains Enterococcaceae, Clostridiaceae, Enterobacteriaceae, Bacteroidaceae, Bifidobacteriaceae, Fusobacteriaceae, Lactobacillaceae, Peptostreptococcaceae, Peptococcaceae, Prevotellaeace, Lachnospiraceae, Ruminococcaceae, Rikenelleacefrom families level, and the phylum Verrucomimicrobia (71, 72). Many functions of the host, including the production of vitamins, ion absorption, resistance to pathogens, histological development, immune function enhancement, and food fermentation all require the involvement of the gut microbiota (4).
Supplementation of feces from ovalbumin (OVA)-induced asthma mouse model to germ-free mice with representative species of the genera Lachnospira, Veillonella, Faecalibacterium, and Rothiawhich may induce airway inflammation (71). In another human birth cohort study, children with an increased relative abundance of Streptococcus and Bacteroides species and decrease Bifidobacterium species and Ruminococcusgnavus in fecal samples at 3 months of age have a higher risk of allergic reactions and wheezing at 5 years of age (6). Moreover, among American newborns classified into three groups based on gut microbiota composition, those with the lowest relative abundance of Bifidobacteria, Akkermansia, and Faecalibacterium and the highest relative abundance of Candida and Rhodotorula had the highest risk of developing asthma (71).
The effects of the gut microbiota on asthma are mediated, at least in part, by bacterial metabolites that may influence immune responses at distal sites in the body. The best-known metabolite is short-chain fatty acids (SCFA), which has protective properties in inflammation of the human respiratory tract. Children with high fecal butyrate and propionate levels at 1 year of age are significantly less likely to have atopic allergies and less likely to develop asthma at 3–6 years of age (24). In mice, SCFAs have been shown to increase the expression of the transcription factor FOXP3 by inhibiting histone deacetylation, thereby promoting the proliferation of Tregs and increasing IL-10 production (73). SCFAs have also been shown to be anti-inflammatory in models of airway inflammation induced by OVA and house dust mites (HDM) (74, 75). Recent studies have shown that human gut microbiota are capable of producing other pro and anti-inflammatory potential metabolites, like biogenic amines (including histamine) (76) and oxylipins such as 12, 13-diHOME (77). The numbers of histamine-secreting bacteria were significantly higher in asthma patients' fecal samples than non-asthmatic volunteers (78). Furthermore, the number of histamine-producing bacteria correlates with the severity of the disease. However, bacterial-derived histamine reduced the total number of cells in bronchoalveolar fluid (BAL) and the amount of IL-4, IL-5 and IL-13 in lung homogenates in an OVA-induced allergic airway inflammation model (79), illustrating the complexity of bacterial immune regulation. In contrast, intra-abdominal treatment of mice with 12, 13-diHOME reduced the number of Treg cells in the lung and increased lung inflammation in a cockroach antigen mouse model of airway inflammation (79). Metabolites secreted by gut microbiota may prove useful for other treatments.
In allergic rhinitis, the gut microbiota is emerging as a new target for early intervention in childhood atopic diseases. Recent studies suggested that a higher bacterial ratio between Klebsiella (an opportunistic pathogen) and Bifidobacterium (commensal bacteria of the gut microbiota) may predispose to allergic diseases (80), further studies suggest that the use of probiotics in infants may favorably alter this ratio and may prevent the future development of allergic diseases (81). Although the study did not find a significant correlation between first-year antibiotics and allergic rhinitis, it did find a significant correlation between first-year antibiotics and asthma, which is usually associated with allergic rhinitis (81).
High-throughput sequencing of bacteria in the fecal of 93 AR patients revealed that the diversity of the gut microbiota of AR patients was significantly reduced, with an increased relative abundance of Bacteroidetes at the phylum level and decreased relative abundance of Actinobacteria and Proteobacteria at the genus level, and significantly decreased relative abundance of Escherichia-Shigella. An analysis of LefSe showed that Escherichia-Shigella, Lachnoclostridium, Parabacteroides and Dialister were potential biomarkers of AR (82). Shannon index analysis showed a significant reduction in species richness and a reduction in chao1 diversity indices in AR patients. In the AR group, Bacteroidetes were significantly abundant. In contrast, the Firmicutes phylum was significantly less abundant. In addition, the abundance of Parabacteroides increased and the abundance of Oxalobacter and Clostridialesdecreased in AR adults (83). Another adult study showed that the AR group had significantly higher bacterial diversity than the non-AR group, Firmicutes, Fusobacteria, Actinobacteria, Cyanobacteria, and Chloroflexiwere the significantly differed phyla. Although bacterial diversity showed no remarkable differences between patients with moderate and severe AR, there was a significant correlation between nasal symptom scores or rhinopharyngitis levels and Butyricoccus and Eisenbergiella levels, revealing the potential for intervention through gut microbiota (84). Changes in the gut microbiota in allergic asthma and allergic rhinitis showed in Table 1.
Therefore, ameliorating AR through modulating gut microbiota may be a potential way, but the specific mechanism needs further study.
Probiotics are defined as “live microorganisms” that have a positive effect on host health when administered in sufficient quantities. It was discovered that the gut microbiota can be altered by oral beneficial bacteria and replace the harmful microorganisms, thus producing the concept of probiotics (85). Probiotics colonize and multiply in the intestine, enhance epithelial integrity, attach to the intestinal epithelium, increase adhesion to the intestinal mucosa, compete to exclude pathogenic microorganisms, resist the production of bactericidal substances, and maintain the gut microbiome's ecological balance (86).
Probiotics exhibited multiple mechanisms: probiotics can activate or inhibit type 1 T helper cells through altering the composition of the gut microbiota; probiotics are also stimulating interleukin 10, which suppresses the inflammatory response (87). In addition, probiotics may decrease an antigen-specific IgE levels in serum (88). Several probiotic strains, such as Lactobacillus rhamnosus GG, Streptococcus thermophilus, Lactobacillus plantarumMB452, and the gram-negative probiotic strain Escherichia coli Nissle 1917 was shown to increase the integrity of the epithelial barrier by enhancing tight junction-related genes or adhesion junction-related receptors through their microbiologically relevant molecular patterns with pattern recognition receptors in epithelial cell gene expression (89–91). In addition, it is essential that probiotic strains must be safe and effective for humans, maintain activity for the shelf life of the product, and not be pathogenic (92).
Research has shown that a particular probiotic strain has immunomodulatory functions, and can alleviate the symptoms of allergic airway inflammation (93). Clinical and laboratory evidence shows that there is a therapeutic effect of probiotics on allergic diseases by regulating gut microbiota and modulating host immunity, promoting the maintenance of normal immune tolerance (94).
The increasing evidence of a relationship between alterations in the microbiota and asthma supports the idea that the microbiota could be harnessed to treat allergic airway inflammation. Probiotics and gut microbiota interact and confer an array of positive effects on the epithelial layer to maintain gastrointestinal and systemic health, by interacting with the gut-associated lymphoid tissues (GALT) which mediate oral tolerance and mucosal immunity (95). Both probiotics and commensal bacteria enforce the functions of the mucosal barrier of the GIT epithelia, induce mucus secretion, and stimulate secretion of IgA which neutralizes pathogens inside the lumen (96). Correspondingly, cross-talk between epithelial cells and residing epithelial immune cells are mediated and enforced by probiotics, and contribute to their effector functions (97). Probiotics are also able to induce the expression of adhesion molecules, stimulate the innate immune system, antigen-presenting cells (APC) and natural killer (NK) cells in both mice and humans (98). Adaptive immunity is stimulated by probiotics, such that IgG and IgA antibodies are produced in response to probiotic consumption (99). In addition, probiotics can stimulate CD8+ T cells, Treg cells and cytokines (i.e., interferon (IFN)-gamma and IL-10 (100). APCs exposed to probiotics present harmless peptides to T cells and subsequently induce Treg cells, to produce anti-inflammatory cytokines including, transforming growth factor beta (TGF-β), IL-10 and retinoic acid. In addition, Treg cells suppress effector Th1, Th17 and cytotoxic T (Tc) cells and IgA secretion (101). Thus, probiotic bacteria can control the “on/off” switch of immune responses in a strain-dependant manner, modulating the host immune system at the mucosal level. Lacking of sufficient probiotic bacteria and their subsequent stimulatory impact on the immune system leads to inadequate or inappropriate immune modulation. Insufficient probiotic bacteria alone, or inadequacy, together with stimulation of the immune system by invasive pathogens, mediate a range of immunopathogenic disorders such as AA and AR.
Probiotics confer immunological protection to the host through the regulation, stimulation, and modulation of immune responses, and have been widely promoted due to their positive effect on the attenuation of abnormal immune responses such as asthma. The phenotype of allergic airway disease is influenced by host genetics and interactions between the gut microbiota, may be regulated by probiotics. The phenotype of allergic airway disease is influenced by host genetics and interactions between the gut microbiota, the gut microbiota may be regulated by probiotics.
The study gavaged Lactobacillus paracasei L9 for OVA-induced acute asthma mice, and found that Lactobacillus paracasei L9 reduced airway hyperresponsiveness and the proportion of eosinophils in bronchoalveolar lavage fluid by correcting the Th2/Th1 immune imbalance in the lungs of mice, thereby alleviating asthma (102). In another study, oral administration of Lactobacilli to asthmatic rats, resulted in decreased levels of inflammatory infiltrates (IL-4, IL-5), eosinophils, and serum allergen-specific IgE, IgA, TGF-β, mucin-2, and tight junction protein were increased, and intestinal morphological changes were normalized (103).
In addition, administration of Bifidobacterium breve M-16V (B. breve M-16V) to pregnant mice exposed to residual oil fly ash also resulted in a reduction in eosinophils in bronchoalveolar lavage fluid and reduced allergic lung inflammation in neonatal mice, besides decreased expression of Th2 cytokines (IL-5 and IL-13). Also, oral administration of B. breve M-16V reduced Firmicutesin. Besides, several bacterial strains of the mouse fecal microbiota were strongly associated with Th2 cytokine and histological scores (104). Moreover, studies using Bifidobacterium longum 7952 (Bl7952) in OVA-sensitized mice showed allergy-reducing properties and inhibition of airway inflammation, as well as reduced eosinophil levels in bronchoalveolar lavage fluid (BALF). Systemic and local levels of Th2-related cytokines and OVA-specific IgE antibodies were decreased by Bl7952 (105). The oral administration of Bifidobacterium longum 51A to both male A/J and C57BL/6 mice with airway inflammation revealed that only the A/J mice showed reduced inflammation, which was partly due to acetate production. Acetate-producer probiotics modulated the abundance of specific bacterial (e.g., Akkermansia and Allistipes) genera in the mouse strain. The gut microbiota diversity was increased in C57BL/6 mice given probiotics compared to A/J mice at baseline. To understand the relationship between microbial composition and the development of allergic disease, implantation of A/J embryos into female C57BL/6 mice showed an increased diversity of the gut microbiota and a decreased eosinophilic inflammation in A/J mice (106). Furthermore, oral administration of Feacalibacterium prausnitzii to house dust mite-induced asthma mice reduced inflammatory cell infiltration and cytokine secretion, alleviated pathological changes, and increased the proportion of Treg. It improved microbial ecological dysregulation and enhanced SCFA production and raised Faecalibaculum, Dubosiella, Streptococcus, and Lachnoclostridium abundance. It also normalized pathways involved in carbohydrate and lipid metabolism, which may relate to SCFA production. Besides correlation analysis showed that immune indicators are closely related to the production of SCFA (107). Oral administration of Lactobacillus rhamnosus diminished the characteristics of allergic asthma in a mouse model and induced immunomodulatory effects by a CD4 (+) CD25 (+) Foxp3 (+) Treg cell-mediated mechanism (108). Oral administration of Lactobacillus rhamnosus GG prevents OVA-induced allergic airway inflammation by amplifying mesenteric CD103+ DCs and accumulating mucosal Tregs cells. The diversity of the gut microbiota was increased, the Shannon and Simpson index and the abundance of Bacteroidetes, Alloprevotella, and Oscillibacter increased, while the abundance of Firmicutes decreased (109). Thus, single probiotics play a beneficial role in the prevention of allergic diseases.
Probiotic-induced protection is associated with gut microbiota and provides new evidence for the use of probiotics in allergic airway inflammation. The association between allergic asthma and microbiota varies by diet and has been extensively studied, implying the potential of oral alternative medicine supplements for the control of asthma. According to Song et al. three probiotic mixtures (Lactobacillus plantarum GREEN CROSS Wellbeing 1001, Lactobacillus rhamnosus GCWB 1,085, and Lactobacillus rhamnosus GCWB 1,156) were orally administered to mice in a diesel exhaust-induced BALB/c asthma model to reduce inflammatory cell levels. In addition, IgE and matrix metalloproteinase (MMP)-9 activities was significantly reduced in serum and BAL (110). In another study, six mixed probiotics (Lactobacillus gasseri LK001; Lactobacillus salivarius LK002; Lactobacillus johnsonii LK003; Lactobacillus paracasei LK004; Lactobacillus reuteri LK005, and Bifidobacterium animals LK011) alleviate OVA-induced asthma in mice by reducing irritant DC surface moleculesand increasing CD103+ DC-induced mucosal irritant Treg in the mesentery (111). Probiotics enhanced gut microbial diversity and Bacteroidetes/Firmicutes (B/F) ratios, and increased levels of Lactobacillus in mice (112).
Therefore, targeting the gut microbiota would be an effective approach to treating allergic airway diseases. Mixed probiotics are of particular interest for their beneficial effects on the host, but their potential mechanisms need to be further explored. There are fewer studies of mixed probiotics in mice and more research is needed to confirm their beneficial effects in asthma treatment.
A meta-analysis of randomized controlled trials (RCTs) showed that supplemented with Lactobacillus rhamnosus GG reduced the incidence of asthma, and probiotic supplementation before and after delivery may play an important strategic role in asthma prevention (113). However, a randomized, controlled, double-blind study of 159 newborns found that supplementation with Lactobacillus rhamnosus GG for the first 6 months of life seems not to prevent the development of asthma at age two (114). In addition, using of probiotics for asthma early in life demonstrated a significant reduction intotal IgE and atopic allergy in a meta-analysis (115). Moreover, patients with asthma have elevated blood levels of TNF-α, interferon-δ, and IL-12. Chen et al. found there was a positive effect on clinical symptoms and cytokine levels in asthmatic children 6 to 12 years old on daily doses of Lactobacillus for 2 months (39). In four meta-analyses of RCTs, probiotics had no benefit for asthma treatment (116). The use of probiotics in the clinical treatment of asthma is still controversial, and a large number of clinical studies are still needed to confirm their effectiveness. Effect of probiotics on allergic asthma showed in Figure 1.
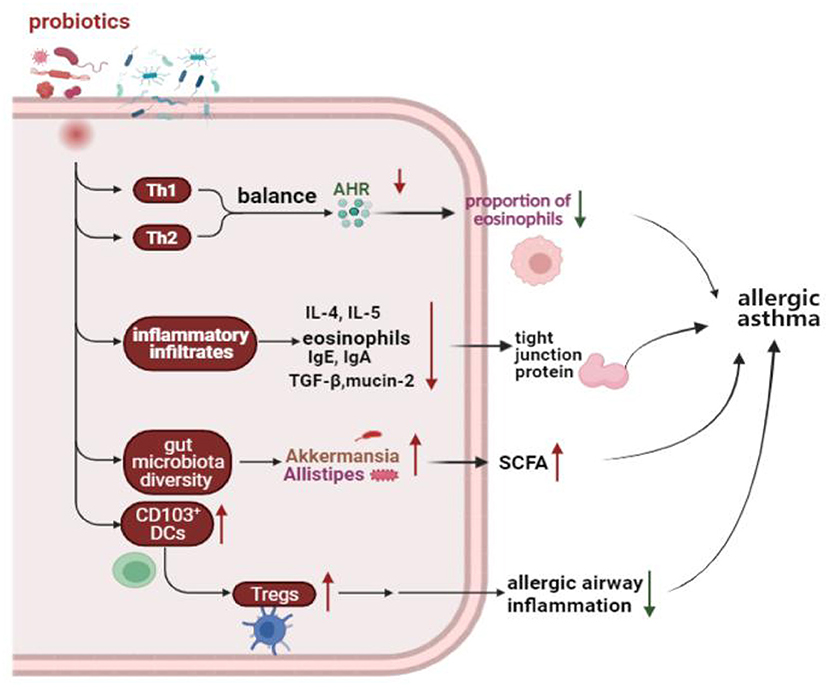
Figure 1. Effect of probiotics on allergic asthma. Probiotics protect the homeostasis of the immune system by regulating the Th1 and Th2 balance, reducing the inflammatory response, increasing the number of Tregs, and modulating the gut microbiota. Th1, T helper 1; Th2, T helper 2; AHR, Airway hyperresponsiveness; IgE, Immunoglobulin E; IgA, Immunoglobulin A; IL-4, Interleukin 4; IL-5, Interleukin 5; TGF-β, Transforming growth factor-beta; DCs, Dendritic cells; Tregs, Regulatory T cells; SCFA, Short-chain-fatty acids.
Ren et al. investigated the benefits of oral administration of Bifidobacterium short (B. breve) in allergic rhinitis and observed that B. breve exert its anti-allergic function by suppressing type II helper T cell immune responses and enhancing CD4+CD25+Treg activity. Allergic rhinitis symptoms such as sneezing were reduced when B. breve was administered at doses of 107 CFU or higher (117). Cui et al. found that oral administration of Lactobacillus Plantarum CJLP133 or CJLP243 in a mice model of AR improved allergic rhinitis by reducing airway hyperreactivity and histological scores, as well as the number of infiltrating cells in the nasal cavity and lungs. Samples of bronchoalveolar lavage fluid and draining lymph nodes from mice showed reduced immune cell counts and increased secretion of Th1 and Th2 type cytokines, decreased IL-4, IL-5, IL-13, serum IgE, and specific serum IgG1 levels, and increased secretion of IFN-γ and specific serum IgG2a (118). Our research in oral administration of Clostridium butyricum (C. butyricum) CGMCC0313-1 which reduced allergic airway inflammation induced by ovalbumin in mice, and found C. butyricumsignificantly reduced lung resistance, pulmonary airway inflammation, mast cell degranulation, airway remodeling, and OVA-specific IgE/G1 expression in asthmatic mice. In addition, it reversed the Th1/Th2 imbalance and increased the anti-inflammatory cytokine IL-10 (119).
Kim et al. revealed that treatment with a probiotic mixture (PM) of Bifidobacterium longum IM55 and Lactobacillus Plantarum IM55 isolated from human feces and kimchi could alleviate AR by restoring Th2/Treg imbalance (inhibiting Th cell differentiation to Th2 cells and inducing differentiation to Treg cells) and gut microbiota disorders (significantly suppressing Proteobacteria and increasing the composition of Bacteroidetes and Actinobacteria) (120).
A randomized, double-blind, placebo-controlled study in 40 children worldwide discovered significant improvements in allergic rhinitis symptoms and quality of life in children treated with a probiotic mixture (Bifidobacterium longum BB536, Bifidobacterium infantis M-63, Bifidobacterium shortum M-16V), all parameters significantly better than the placebo group (121). Similarly, in a double-blind, placebo-controlled, parallel, randomized clinical trial study, Dennis-Wall et al. suggested that the ingestion of Lactobacillus garciaeKS-13, Bifidobacterium bifidum G9-1, and Bifidobacterium longum MM-2 improved the overall scores of patients with rhinoconjunctivitis on the quality of life questionnaire, the percentage of Tregs in fasting whole-blood samples increased. However, the mechanisms involved are not clear (87).
A prospective trial study found that adjuvant conventional treatment with Enterococcus faecalis reduced the number and duration of rhinitis attacks in children and adolescents (122). Ye et al. systematically reviewed and meta-analyzed the therapeutic effects of probiotics on AR, there were 16 clinical trials (randomized controlled trials), 1,374 patients involved, and found that probiotics were highly effective in reducing symptoms in patients with AR compared to the placebo group (123). Ivory et al. showed that volunteers treated with Lactobacillus casei Shirota (LcS) reduced antigen-induced production of cytokines show that probiotic supplementation modulates immune responses in AR and may have the potential to alleviate the severity of symptoms (124). However, the same group Ivory et al. in 2013 showed that changes of the immune status did not have any effects on the clinical symptoms (125). Lack of changes in the clinical symptoms could be explained by an inability of a single nasal allergen challenge to reproduce the chronicity of a natural allergen exposure (126), it does not represent a real-life situation where individuals are exposed to lower concentrations of allergen over a prolonged period of time. Both allergen dose and multiplicity of challenge can influence the outcome. Effect of probiotics on allergic rhinitis showed in Figure 2. Probiotics used in allergic asthma and allergic rhinitis showed in Table 2.
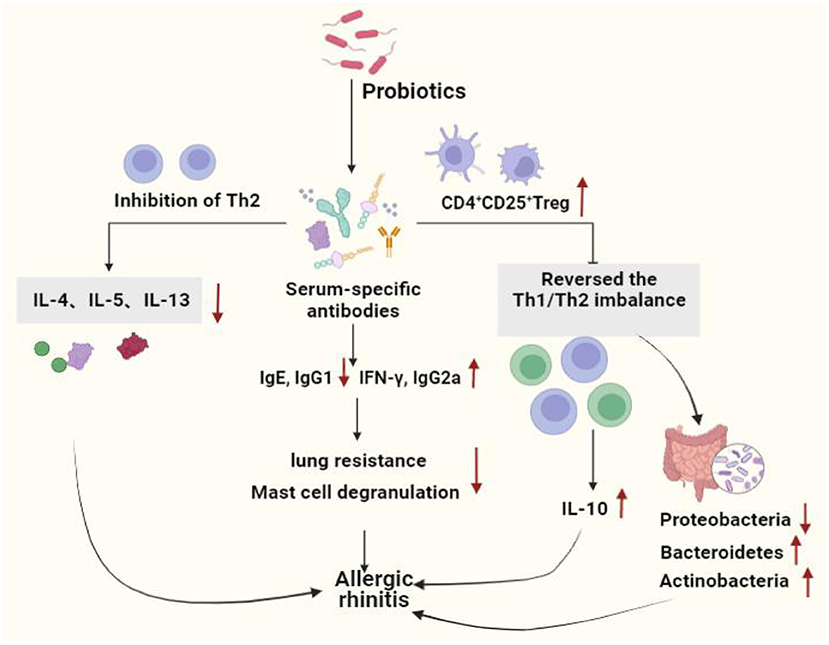
Figure 2. Effect of probiotics on allergic rhinitis. Probiotics protect the allergic rhinitis by decreasing inflammation in serum and lung tissue, increasing the number of immune cells, immune system homeostasis by regulating the Th1 and Th2 balance and increasing the number of Tregs. Moreover, probiotics regulated intestinal stability by increasing the level of beneficial bacteria, hence improving AR. Th1, T helper 1; Th2, T helper 2; IL-4, Interleukin 4; IL-5, Interleukin 5;IL-10, Interleukin 10;IL-13, Interleukin13; IgG1, Immunoglobulin G1; IgE, Immunoglobulin E; IgG2a, Immunoglobulin G2a; IFN-γ, Interferon-γ.
In conclusion, this study summarized the role of probiotics as an intervention or treatment for common allergic diseases. Probiotics reduced allergen-induced hyperreactivity and inflammation as well as reduced cytokine release, among other beneficial effects. In several studies, no significant differences were found between the probiotic-treated and placebo-treated. Although probiotics did not eliminate allergies, their administration may reduce the incidence and duration of allergy symptoms. However, the effects of probiotics depend on their species or strain, their derived metabolites, and the patient's gut microbiota. This review provided a broader understanding of new alternatives for the treatment of allergies, in both pediatric patients and adults, showing that probiotics can reduce symptoms in some cases as well as the severity of such disorders. However, there are limitations to some studies, such as small sample sizes, randomization processes, and the use of small numbers of bacterial populations.
Although numerous studies have been conducted, future basic and human studies that must be done to solidify the role of probiotics in treating allergies. Most current studies have found preliminary effects of probiotics in the treatment of allergic diseases in the clinic. However, some studies have found that probiotic treatment did not significantly improve clinical symptoms, possibly because clinical trials include many variables such as concomitant disease, age, gender and many other factors. Inclusion of patients and standardization of probiotic administration should be standardized in future studies to provide sound evidence and detailed information for the beneficial effects of probiotics. In conclusion, the administration of probiotics in respiratory allergic diseases seems to be promising. In addition, probiotics as a vehicle to reshape the gut microbiota may be a new therapy in the future.
JLH conducted the literature search and wrote the first draft of the manuscript. XS and JZ reviewed and XZW, ZHJ, and PPZ commented on several drafts. All authors have read and agree to the published version of the manuscript.
This work was supported by the National Natural Science Foundation of China (82170026) and the Discipline Promotion Project of Xijing Hospital (XJZT18MJ23).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are thankful to the National Natural Science Foundation of China (82170026) and the Discipline Promotion Project of Xijing Hospital (XJZT18MJ23) supported this work, thankful to Department of Pediatrics, Xijing Hospital, the Fourth Military Medical University for providing the various resources for completion of the current article, and thankful to JZ providing language help, thanks to XS writing assistance and proof reading the article.
1. Testa D, DI Bari M, Nunziata M, Cristofaro G, Massaro G, Marcuccio G, et al. Allergic rhinitis and asthma assessment of risk factors in pediatric patients: a systematic review. Int J Pediatr Otorhinolaryngol. (2020) 129:109759. doi: 10.1016/j.ijporl.2019.109759
2. Brozek JL, Bousquet J, Agache I, Agarwal A, Bachert C, Bosnic-Anticevich S, et al. Allergic rhinitis and its impact on asthma (ARIA) guidelines-2016 revision. J Allergy ClinImmunol. (2017) 140:950–8. doi: 10.1016/j.jaci.2010.06.047
3. Zhang Y, Zhang L. Increasing prevalence of allergic rhinitis in china. Allergy Asthma Immunol Res. (2019) 11:156–69. doi: 10.4168/aair.2019.11.2.156
4. Kay AB. Allergy and allergic diseases. First of two parts. N Engl J Med. (2001) 344:30–7. doi: 10.1056/NEJM200101043440106
5. Varricchi G, Raap U, Rivellese F, Marone G, Gibbs BF. Human mast cells and basophils-how are they similar how are they different? Immunol Rev. (2018) 282:8–34. doi: 10.1111/imr.12627
6. Barnes PJ. Pathophysiology of allergic inflammation. Immunol Rev. (2011) 242:31–50. doi: 10.1111/j.1600-065X.2011.01020.x
7. Lambrecht BN, Hammad H. The immunology of asthma. Nat. Immunol. (2015)16: 45–56. doi: 10.1038/ni.3049
8. Arthur G, Bradding P. New developments in mast cell biology: clinical implications. Chest. (2016) 150:680–93. doi: 10.1016/j.chest.2016.06.009
9. Brightling CE, Bradding P, Symon FA, Holgate ST, Wardlaw AJ, Pavord ID. Mast-cell infiltration of airway smooth muscle in asthma. N Engl J Med. (2002) 346:1699–705. doi: 10.1056/NEJMoa012705
10. Upham JW, Xi Y. Dendritic cells in human lung disease: recent advances. Chest. (2016) 151:668–73. doi: 10.1016/j.chest.2016.09.030
11. Hammad H, Lambrecht BN. Barrier epithelial cells and the control of type 2 immunity. Immunity. (2015) 43:29–40. doi: 10.1016/j.immuni.2015.07.007
12. Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. (2013)13:9–22. doi: 10.1038/nri3341
13. Barnes PJ. Therapeutic approaches to asthma-chronic obstructive pulmonary disease overlap syndromes. J Allergy Clin Immunol. (2015) 136:531–45. doi: 10.1016/j.jaci.2015.05.052
14. Al-Ramli W, Prefontaine D, Chouiali F, Martin JG, Olivenstein R, Lemiere C, et al. T(H)17-associated cytokines (IL-17A and IL-17F) in severe asthma. J Allergy Clin Immunol. (2009) 123:1185–7. doi: 10.1016/j.jaci.2009.02.024
15. Steelant B, Seys SF, Gerven LV, Woensel MV, Farre R, Wawrzyniak P, et al. Histamine and T helper cytokine-driven epithelial barrier dysfunction in allergic rhinitis. J Allergy Clin Immunol. (2018)141:951–63.e8. doi: 10.1016/j.jaci.2017.08.039
16. Carr VM, Obinson AM, Kern RC. Tissue-specific effects of allergic rhinitis in mouse nasal epithelia. Chem Senses. (2012) 37:655–68. doi: 10.1093/chemse/bjs048
17. Lombardi V, Beuraud C, Neukirch C, Moussu H, Morizur L, Horiot S, et al. Circulating innate lymphoid cells are differentially regulated in allergic and nonallergic subjects. J Allergy Clin Immunol. (2016) 138:305–8. doi: 10.1016/j.jaci.2015.12.1325
18. Bal SM, Bernink JH, Nagasawa M, Groot J, Shikhagaie MM, Golebski K, et al. IL-1beta, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat Immunol. (2016) 17:636–45. doi: 10.1038/ni.3444
19. Shamji MH, Durham SR. Mechanisms of allergen immunotherapy for inhaled allergens and predictive biomarkers. J Allergy Clin Immunol. (2017) 140:1485–98. doi: 10.1016/j.jaci.2017.10.010
20. Msz A, Bz B, Zlg A, Zheng RP, Marcellin DFHM, Saro A, et al. Altered diversity and composition of gut microbiota in patients with allergic rhinitis. Microb Pathog. (2021) 161:105272. doi: 10.1016/j.micpath.2021.105272
21. Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity. (2020) 52:241–55. doi: 10.1016/j.immuni.2020.01.007
22. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. (2016) 164:337–40. doi: 10.1016/j.cell.2016.01.013
23. Sozańska B. Microbiome in the primary prevention of allergic diseases and bronchial asthma. Allergol Immunopathol. (2019) 47:79–84. doi: 10.1016/j.aller.2018.03.005
24. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. SciTransl Med. (2015) 7:152r−307r. doi: 10.1126/scitranslmed.aab2271
25. Kim WG, Kang GD, Kim HI, Han MJ, Kim DH. Bifidobacteriumlongum IM55 and Lactobacillus plantarum IM76 alleviate allergic rhinitis in mice by restoring Th2/Treg imbalance and gut microbiota disturbance. Benef Microbes. (2019) 10:55–67. doi: 10.3920/BM2017.0146
26. Ooijevaar RE, Terveer EM, Verspaget HW, Kuijper EJ, Keller JJ. Clinical application and potential of fecal microbiotatransplantation. Annu Rev Med. (2019) 70:335–51. doi: 10.1146/annurev-med-111717-122956
27. Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. (2017) 20:145–55. doi: 10.1038/nn.4476
28. Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. (2017) 356:j831. doi: 10.1136/bmj.j831
29. Bateman ED, Hurd SS, Barnes PJ, Bousquet J, Drazen JM, FitzGerald JM, et al. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J. (2008) 31:143–78. doi: 10.1183/09031936.00138707
30. Christiansen SC, Zuraw BL. Treatment of hypertension in patients with asthma. N Engl J Med. (2019) 381:1046–57. doi: 10.1056/NEJMra1800345
31. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. (2007) 448:470–3. doi: 10.1038/nature06014
32. Portelli MA, Hodge E, Sayers I. Genetic risk factors for the development of allergic disease identified by genome-wide association. Clin Exp Allergy. (2015) 45:21–31. doi: 10.1111/cea.12327
33. Doe C, Bafadhel M, Siddiqui S, Desai D, Mistry V, Rugman P, et al. Expression of the T helper 17-associated cytokines IL-17A and IL-17F in asthma and COPD. Chest. (2010) 138:1140–7. doi: 10.1378/chest.09-3058
34. Haldar P, Pavord LD, Shaw DE, Berry MA, Thomas M, Brightling CE, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. (2008) 178:218–24. doi: 10.1164/rccm.200711-1754OC
35. Lefaudeux D, De Meulder B, Loza MJ, Peffer N, Rowe A, Baribaud F, et al. U-BIOPRED clinical adult asthma clusters linked to a subset of sputum omics. J Allergy Clin Immunol. (2017) 139:1797–807. doi: 10.1016/j.jaci.2016.08.048
36. Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the severe asthma research program. Am J Respir Crit Care Med. (2010) 181:315–23. doi: 10.1164/rccm.200906-0896OC
37. Turner S, Upham JW. Editorial: asthma in children and adults-what are the differences and what can they tell us about asthma? Front Pediatr. (2020) 8:141. doi: 10.3389/fped.2020.00141
38. Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med. (2020) 26:599–607. doi: 10.1038/s41591-020-0761-3
39. Du XZ, Wang L, Wu SY, Yuan L, Tang S, Xiang Y, et al. Efficacy of probiotic supplementary therapy for asthma, allergic rhinitis, and wheeze: a meta-analysis of randomized controlled trials. Allergy Asthma Proc. (2019) 40:250–60. doi: 10.2500/aap.2019.40.4227
40. Bousquet PJ, Leynaert B, Neukirch F, Sunyer J, Janson CM, Anto J, et al. Geographical distribution of atopic rhinitis in the European Community Respiratory Health Survey I. Allergy. (2008) 63:1301–9. doi: 10.1111/j.1398-9995.2008.01824.x
41. Vandenplas O, Vinnikov D, Blanc PD, Agache I, Bachert C, Bewick M, et al. Impact of rhinitis on work productivity: a systematic review. J Allergy Clin Immunol Pract. (2018) 6:1274–86. doi: 10.1016/j.jaip.2017.09.002
42. Devillier P, Bousquet J, Salvator H, Naline E, Grassin-Delyle S, de Beaumont O. In allergic rhinitis, work, classroom and activity impairments are weakly related to other outcome measures. Clin Exp Allergy. (2016) 46:1456–64. doi: 10.1111/cea.12801
43. Bousquet J, Khaltaev N, Cruz AA, Denburg J, Fokkens WJ, Togias A, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2)LEN and AllerGen). Allergy. (2008) 63(Suppl 86):8–160. doi: 10.1111/j.1398-9995.2007.01620.x
44. Bousquet J, Anto JM, Bachert C, Baiardini I, Bosnic-Anticevich S, Walter CG, et al. Allergic rhinitis. Nat Rev Dis Primers. (2020) 6:95. doi: 10.1038/s41572-020-00227-0
45. Greiner AN, Hellings PW, Rotiroti G, Scadding GK. Allergic rhinitis. Lancet. (2011) 378:2112–22. doi: 10.1016/S0140-6736(11)60130-X
46. Schuler Iv CF, Montejo JM. Allergic rhinitis in children and adolescents. Pediatr Clin North Am. (2019) 66:981–993. doi: 10.1016/j.pcl.2019.06.004
47. Wang H, Lee IS, Braun C, Enck P. Effect of probiotics on central nervous system functions in animals and humans: a systematic review. J Neurogastroenterol Motil. (2016) 22:589–605. doi: 10.5056/jnm16018
48. Eslami M, Bahar A, Keikha M, Karbalaei M, Kobyliak NM, Yousefi B. Probiotics function and modulation of the immune system in allergic diseases. Allergol Immunopathol. (2020) 48:771–88. doi: 10.1016/j.aller.2020.04.005
49. Ganal-Vonarburg SC, Duerr CU. The interaction of intestinal microbiota and innate lymphoid cells in health and disease throughout life. Immunology. (2020) 159:39–51. doi: 10.1111/imm.13138
50. Van de Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat Rev Immunol. (2010) 10:664–74. doi: 10.1038/nri2832
51. van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. (2014) 508:123–7. doi: 10.1038/nature13158
52. Klose CSN, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. (2013) 494:261–5. doi: 10.1038/nature11813
53. Aguero MGD, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. (2016) 351:1296–302. doi: 10.1126/science.aad2571
54. Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. (2010) 33:736–51. doi: 10.1016/j.immuni.2010.10.017
55. Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. (2011) 12:320–6. doi: 10.1038/ni.2002
56. Klose CSN, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. (2014) 157:340–56. doi: 10.1016/j.cell.2014.03.030
57. Bernink JH, Krabbendam L, Germar K, Jong ED, Gronke K, Kofoed-Nielsen M, et al. Interleukin-12 and−23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity. (2015) 43:146–60. doi: 10.1016/j.immuni.2015.06.019
58. Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. (2014) 343:1249–88. doi: 10.1126/science.1249288
59. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. (2012) 37:171–86. doi: 10.1016/j.immuni.2012.05.020
60. Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, et al. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. (2015) 348:1031–5. doi: 10.1126/science.aaa4812
61. von Burg N, Chappaz S, Baerenwaldt A, Horvath E, Dasgupta SB, Ashok D, et al. Activated group 3 innate lymphoid cellsvpromote T-cell-mediated immune responses. Proc Natl Acad Sci USA. (2014) 111:12835–40. doi: 10.1073/pnas.1406908111
62. Guo X, Liang Y, Zhang Y, Lasorella A, Kee BL, Fu XY. l Innate lymphoid cells control early colonization resistance against intestinal pathogens through ID2-dependent regulation of the microbiota. Immunity. (2015) 42:731–43. doi: 10.1016/j.immuni.2015.03.012
63. Pickard JM, Zeng MY, Caruso R, Nunez J. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. (2017) 279:70–89. doi: 10.1111/imr.12567
64. Stiemsma LT, Turvey SE. Asthma and the microbiome: defining the critical window in early life. Allergy Asthma Clin Immunol. (2017) 13:3. doi: 10.1186/s13223-016-0173-6
65. Tanaka M, Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int. (2017) 66:515–22. doi: 10.1016/j.alit.2017.07.010
66. Gill N, Wlodarska M, Finlay BB. The future of mucosal immunology: studying an integrated system-wide organ. Nature immunology. (2010) 11:558–60. doi: 10.1038/ni0710-558
67. Zhou A, Lei Y, Tang L, Hu S, Yang M, Wu LY, et al. Gut microbiota: the emerging link to lung homeostasis and disease. J Bacteriol. (2021) 203:e00454–20. doi: 10.1128/JB.00454-20
68. Desai M, Oppenheimer J. Elucidating asthma phenotypes and endotypes: progress towards personalized medicine. Ann Allergy Asthma Immunol. (2016) 116:394–401. doi: 10.1016/j.anai.2015.12.024
69. Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, et al. A new genomic blueprint of the human gut microbiota. Nature. (2019) 568:499–504. doi: 10.1038/s41586-019-0965-1
70. Marsland BJ, Trompette A. GollwitzerES. The Gut-Lung axis in respiratory disease. Ann Am Thorac Soc. (2015) 12(Suppl 2):S150–6.
71. Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial ecology along the gastrointestinal tract. Microbes Environ. (2017) 32:300–13. doi: 10.1264/jsme2.ME17017
72. Pereira FC, Berry D. Microbial nutrient niches in the gut. Environ Microbiol. (2017) 19:1366–78. doi: 10.1111/1462-2920.13659
73. Arrieta MC, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. (2018) 142:424–34. doi: 10.1016/j.jaci.2017.08.041
74. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitizedatopy and T cell differentiation. Nat Med. (2016) 22:1187–91. doi: 10.1038/nm.4176
75. Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. (2019) 74:799–809. doi: 10.1111/all.13660
76. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, DeRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
77. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. (2014) 20:159–66. doi: 10.1038/nm.3444
78. Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. (2018) 11:785–95. doi: 10.1038/mi.2017.75
79. Pugin B, Barcik W, Westermann P, Heider A, Wawrzyniak M, Hellings P, et al. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb Ecol Health Dis. (2017) 28:1353881. doi: 10.1080/16512235.2017.1353881
80. Levan SR, Stamnes KA, Lin DL, Panzer AR, Fukui E, McCauley K, et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol. (2019) 4:1851–61. doi: 10.1038/s41564-019-0498-2
81. Barcik W, Pugin B, Brescó MS, Westermann P, Rinaldi A, Groeger D, et al. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy. (2019) 74:899–909. doi: 10.1111/all.13709
82. Low J, Soh SE, Lee YK, Kwek K, Holbrook JD, Van der Beek EM, et al. Ratio of Klebsiella/Bifidobacterium in early life correlates with later development of paediatric allergy. Benef Microbes. (2017) 8:681–95. doi: 10.3920/BM2017.0020
83. Candy D, Van Ampting M, Oude NM, Wopereis H, Butt AM, Peroni DG, et al. A synbiotic-containing amino-acid-based formula improves gut microbiota in non-IgE-mediated allergic infants. Pediatr Res. (2018) 83:677–86. doi: 10.1038/pr.2017.270
84. Kulig M, Bergmann R, Klettke U, Wahn V, Tacke U, Wahn U. Natural course of sensitization to food and inhalant allergens during the first 6 years of life. J Allergy Clin Immunol. (1999) 103:1173–9. doi: 10.1016/S0091-6749(99)70195-8
85. Krzych-Fałta E, Furmańczyk K, Tomaszewska A, Olejniczak D, Samoliński B, Samolińska-Zawisza U. Probiotics: myths or facts about their role in allergy prevention. Adv Clin Exp Med. (2018) 27:119–24. doi: 10.17219/acem/65476
86. Martens K, Pugin B, De Boeck I, Spacova I, Steelant B, Seys SF, et al. Probiotics for the airways: Potential to improve epithelial and immune homeostasis. Allergy. (2018) 73:1954–63. doi: 10.1111/all.13495
87. Dennis-Wall JC, Culpepper T, Nieves CJ, Rowe CC, Burns AM, Rusch CT, et al. Probiotics (Lactobacillus gasseri KS-13, Bifidobacteriumbifidum G9-1, and Bifidobacteriumlongum MM-2) improve rhinoconjunctivitis-specific quality of life in individuals with seasonal allergies: a double-blind, placebo-controlled, randomized trial. Am J Clin Nutr. (2017) 105:758–67. doi: 10.3945/ajcn.116.140012
88. Hoyte F, Nelson HS. Recent advances in allergic rhinitis. F1000Res. (2018) 7:F1000. doi: 10.12688/f1000research.15367.1
89. Blackwood BP, Yuan CY, Wood DR, Nicolas JD, Grothaus JS, Hunter CJ. Probiotic lactobacillus species strengthen intestinal barrier function and tight junction integrity in experimental necrotizing enterocolitis. J Probiotics Health. (2017) 5:159. doi: 10.4172/2329-8901.1000159
90. Secher T, Maillet I, Mackowiak C, Le Bérichel J, Philippeau A, Panek C, et al. The probiotic strain Escherichia coli Nissle 1917 prevents papain-induced respiratory barrier injury and severe allergic inflammation in mice. Sci Rep. (2018) 8:11245. doi: 10.1038/s41598-018-29689-9
91. Chen Y, Zhang M, Ren F. A role of exopolysaccharide produced by streptococcus thermophilus in the intestinal inflammation and mucosal barrier in caco-2 monolayer and dextran sulphate Sodium-Induced experimental murine colitis. Molecules. (2019) 24:513. doi: 10.3390/molecules24030513
92. Vanderhoof JA, Young R. Probiotics in the United States. Clin Infect Dis. (2008) 46(Suppl 2):S67–72. doi: 10.1086/523339
93. Ozdemir O. Various effects of different probiotic strains in allergic disorders: an update from laboratory and clinical data. Clin Exp Immunol. (2010) 160:295–304. doi: 10.1111/j.1365-2249.2010.04109.x
94. McKenzie C, Tan J, Macia L, Mackay CR. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol Rev. (2017) 278:277–95. doi: 10.1111/imr.12556
95. Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. (2020) 38:23–48. doi: 10.1146/annurev-immunol-070119-115104
96. D'Amelio P, Sassi F. Gut microbiota, immune system, and bone. Calcif Tissue Int. (2018) 2:415–25. doi: 10.1007/s00223-017-0331-y
97. Hardy H, Harris J, Lyon E, Beal J, Foey AD. Probiotics, prebiotics and immunomodulation of gut mucosal defences: homeostasis and immunopathology. Nutrients. (2013) 5:1869–912. doi: 10.3390/nu5061869
98. Mortha A, Diefenbach A. Natural killer cell receptor-expressing innate lymphocytes: more than just NK cells. Cell Mol Life Sci. (2011) 68:3541–55. doi: 10.1007/s00018-011-0803-6
99. Delgado S, Sanchez B, Margolles A, Ruas-Madiedo P, Ruiz L. Molecules produced by probiotics and intestinal microorganisms with immunomodulatory activity. Nutrients. (2020) 12:391. doi: 10.3390/nu12020391
100. Fink LN, Zeuthen LH, Christensen HR, Morandi B, Frokiær H, Ferlazzo G. Distinct gut-derived lactic acid bacteria elicit divergent dendritic cell-mediated NK cell responses. Int Immunol. (2007) 19:1319–27. doi: 10.1093/intimm/dxm103
101. Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, et al. Extrathymically generated regulatory T cells control mucosal T H 2 inflammation. Nature. (2012) 482:395–9. doi: 10.1038/nature10772
102. Liu X, Tao J, Li J, Cao X, Li Y, Gao X, et al. Dysbiosis of fecal microbiota in allergic rhinitis patients. Am J Rhinol Allergy. (2020) 34:650–60. doi: 10.1177/1945892420920477
103. Watts AM, West NP, Zhang P, Smith PK, Cripps AW, Cox AJ. The gut microbiome of adults with allergic rhinitis is characterised by reduced diversity and an altered abundance of key microbial taxa compared to controls. Int Arch Allergy Immunol. (2021) 182:94–105. doi: 10.1159/000510536
104. Zhu L, Xu F, Wan W, Yu B, Tang L, Yang Y, et al. Gut microbial characteristics of adult patients with allergy rhinitis. Microb Cell Fact. (2020) 19:171. doi: 10.1186/s12934-020-01430-0
105. Wang X, Hui Y, Zhao L, Hao Y, Guo H, Ren F. Oral administration of Lactobacillus paracasei L9 attenuates PM25-induced enhancement of airway hyperresponsiveness and allergic airway response in murine model of asthma. Plos ONE. (2017) 12:e171721. doi: 10.1371/journal.pone.0171721
106. Cervantes-García D, Jiménez M, Rivas-Santiago CE, Gallegos-Alcalá P, Hernández-Mercado A, Santoyo-Payán LS, et al. Lactococcuslactis NZ9000 prevents asthmatic airway inflammation and remodelling in rats through the improvement of intestinal barrier function and systemic TGF-β production. Int Arch Allergy Immunol. (2021) 182:277–91. doi: 10.1159/000511146
107. Terada-Ikeda C, Kitabatake M, Hiraku A, Kato K, Yasui S, Imakita N, et al. Maternal supplementation with Bifidobacteriumbreve M-16V prevents their offspring from allergic airway inflammation accelerated by the prenatal exposure to an air pollutant aerosol. PLoS ONE. (2020) 15:e238923. doi: 10.1371/journal.pone.0238923
108. Pyclik MJ, Srutkova D, Razim A, Hermanova P, Svabova T, Pacyga K, et al. Viability Status-Dependent Effect of Bifidobacteriumlongum ssp. Longum CCM 7952 on Prevention of Allergic Inflammation in Mouse Model. Front Immunol(2021) 12: 707728. doi: 10.3389/fimmu.2021.707728
109. Casaro MB, Thomas AM, Mendes E, Fukumori C, Ribeiro WR, Oliveira FA, et al. A probiotic has differential effects on allergic airway inflammation in A/J and C57BL/6 mice and is correlated with the gut microbiome. Microbiome. (2021) 9:134. doi: 10.1186/s40168-021-01116-8
110. Hu W, Lu W, Li L, Zhang H, Lee YK, Chen W, et al. Both living and dead Faecalibacteriumprausnitzii alleviate house dust mite-induced allergic asthma through the modulation of gut microbiota and short-chain fatty acid production. J Sci Food Agric. (2021) 101:5563–73. doi: 10.1002/jsfa.11207
111. Li L, Fang Z, Liu X, Hu W, Lu W, Lee YK, et al. Lactobacillus reuteri attenuated allergic inflammation induced by HDM in the mouse and modulated gut microbes. PLoS ONE. (2020) 15:e0231865. doi: 10.1371/journal.pone.0231865
112. Jang SO, Kim HJ, Kim YJ, Kang MJ, Kwon JW, Seo JH, et al. Asthma prevention by Lactobacillus rhamnosus in a mouse model is associated with CD4(+)CD25(+)Foxp3(+) t cells. Allergy Asthma Immunol Res. (2012) 4:150–6. doi: 10.4168/aair.2012.4.3.150
113. Zhang J, Ma JY Li QH, Su H, Sun X. Lactobacillus rhamnosus GG induced protective effect on allergic airway inflammation is associated with gut microbiota. Cell Immunol. (2018) 332:77–84. doi: 10.1016/j.cellimm.2018.08.002
114. Jin SW, Lee GH, Jang MJ, Hong GE, Kim JY, Park GD, et al. Lactic acid bacteria ameliorate diesel exhaust particulate Matter-Exacerbated allergic inflammation in a murine model of asthma. Life. (2020) 10:260. doi: 10.3390/life10110260
115. Zhang J, Ma J, Li Q, Su H, Sun X. Exploration of the effect of mixed probiotics on microbiota of allergic asthma mice. Cell Immunol. (2021) 367:104399. doi: 10.1016/j.cellimm.2021.104399
116. Cabana MD, McKean M, Caughey AB, Fong L, Lynch S, Wong A, et al. Early probiotic supplementation for eczema and asthma prevention: a randomized controlled trial. Pediatrics. (2017) 140:e20163000. doi: 10.1542/peds.2016-3000
117. Ta V, Laubach S. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. (2014) 134(Suppl 3):S141. doi: 10.1542/peds.2014-1817O
118. Chen YS, Jan RL, Lin YL, Chen HH, Wang JY. Randomized placebo-controlled trial of lactobacillus on asthmatic children with allergic rhinitis. Pediatr Pulmonol. (2010) 45:1111–20. doi: 10.1002/ppul.21296
119. Wopereis H, Sim K, Shaw A, Warner JO, Knol J, Kroll JS. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J Allergy Clin Immunol. (2018) 141:1334–42. doi: 10.1016/j.jaci.2017.05.054
120. Ren J, Zhao Y, Huang S, Lv D, Yang F, Lou L, et al. Immunomodulatory effect of Bifidobacteriumbreve on experimental allergic rhinitis in BALB/c mice. Exp Ther Med. (2018) 16:3996–4004. doi: 10.3892/etm.2018.6704
121. Choi SP, Oh HN, Choi CY, Ahn H, Yun HS, Chung YM, et al. Oral administration of Lactobacillus plantarum CJLP133 and CJLP243 alleviates birch pollen-induced allergic rhinitis in mice. J Appl Microbiol. (2018) 124:821–8. doi: 10.1111/jam.13635
122. Kitz R, Martens U, Zieseniß E, Enck P, Rose M. Probiotic E.faecalis— adjuvant therapy in children with recurrent rhinosinusitis. Cent Eur J Med. (2012) 7:362–5. doi: 10.2478/s11536-011-0160-8
123. Ye SF, Liu Z, Wang YF, He P, Wen LJ, Teng B. [the therapeutic effect of probiotics on allergic rhinitis:a Meta analysis]. Lin Chung Er Bi Yan HouTou Jing WaiKeZaZhi. (2017) 31:467–74. doi: 10.13201/j.issn.1001-1781.2017.06.014
124. Ivory K, Chambers SJ, Pin C, Prieto E, Arques JL, Nicoletti C. Oral delivery of Lactobacillus casei shirota modifies allergen-induced immune responses in allergic rhinitis. Clin Exp Allergy. (2008) 38:1282–9. doi: 10.1111/j.1365-2222.2008.03025.x
125. Ivory K, Wilson AM, Sankaran P, Westwood M, Carville JM, Brockwell C, et al. Oral delivery of a probiotic induced changes at the nasal mucosa of seasonal allergic rhinitis subjects after local allergen challenge: a randomised clinical trial. PLoS ONE. (2013) 8:e78650. doi: 10.1371/journal.pone.0078650
Keywords: probiotics, allergic airway inflammation, asthma, allergic rhinitis, gut microbiota
Citation: Huang J, Zhang J, Wang X, Jin Z, Zhang P, Su H and Sun X (2022) Effect of Probiotics on Respiratory Tract Allergic Disease and Gut Microbiota. Front. Nutr. 9:821900. doi: 10.3389/fnut.2022.821900
Received: 15 December 2021; Accepted: 31 January 2022;
Published: 22 February 2022.
Edited by:
Satya Prakash, McGill University, CanadaReviewed by:
Claudio Nicoletti, University of Florence, ItalyCopyright © 2022 Huang, Zhang, Wang, Jin, Zhang, Su and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Su, SHVpc3UyMDE0QDE2My5jb20=; Xin Sun, c3VueGluNkBmbW11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.