
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr., 15 February 2022
Sec. Nutrition and Microbes
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.814269
Pancreatic-related disorders such as pancreatitis, pancreatic cancer, and type 1 diabetes mellitus (T1DM) impose a substantial challenge to human health and wellbeing. Even though our understanding of the initiation and progression of pancreatic diseases has broadened over time, no effective therapeutics is yet available for these disorders. Mounting evidence suggests that gut dysbiosis is closely related to human health and disease, and pancreatic diseases are no exception. Now much effort is under way to explore the correlation and eventually potential causation between the gut microbiome and the course of pancreatic diseases, as well as to develop novel preventive and/or therapeutic strategies of targeted microbiome modulation by probiotics, prebiotics, synbiotics, postbiotics, and fecal microbiota transplantation (FMT) for these multifactorial disorders. Attempts to dissect the intestinal microbial landscape and its metabolic profile might enable deep insight into a holistic picture of these complex conditions. This article aims to review the subtle yet intimate nexus loop between the gut microbiome and pancreatic diseases, with a particular focus on current evidence supporting the feasibility of preventing and controlling pancreatic diseases via microbiome-based therapeutics and therapies.
Pancreatic diseases, including pancreatitis, pancreatic cancer, and type 1 diabetes mellitus (T1DM), not only exert an outsized adverse effect on human health because of marked morbidity and mortality, but impose a heavy societal burden worldwide (1). Pancreatitis is one of the most common gastrointestinal (GI) disorders seen in U.S. hospitals, ranking third after GI hemorrhage and gallbladder disease (1). Globally, the annual incidence rates of acute pancreatitis (AP) and chronic pancreatitis (CP) are 34 cases and 10 cases per 100,000 person-years, respectively (2). Around 80–85% of patients with AP develop interstitial edematous pancreatitis, with a mortality rate of <2%. Other patients might evolve into necrotizing pancreatitis of varying severity, and, in severe cases, where patients develop persistent organ failure, the mortality rate rises to 15–20% (3). In the cases of CP, there is an 8–9-fold increased risk of developing pancreatic cancer (4, 5). Pancreatic cancer is a common and fatal malignancy of the pancreas and is considered to be the third leading cause of death in the U.S. According to the “Cancer Facts & Figure 2020” issued by the American Cancer Society, the estimated number of deaths for pancreatic cancer is 7.8%, which is secondary to that of lung cancer (22.4%) and colorectal cancer (8.8%) (6). Notably, the incidence rate for pancreatic cancer increased by 1% per year from 2007 to 2016 and is projected to be the second biggest cancer killer in the U.S. by 2030 (6, 7). Another pancreatic disease, T1DM, does not directly manifest as gastrointestinal symptoms, but polyuria, polydipsia, and polyphagia (8). As one of the most common chronic disorders among children and youths (9, 10), the global incidence of T1DM has an average annual increase of 3–4% over the past three decades (11).
Despite the substantial improvement of our understanding of the cause, pathogenesis, and treatment of pancreatic diseases over the past decade (12–15), no curative treatment is yet available (16, 17). The World Health Organization (WHO) underscored the fundamental role that prevention plays before an episode of a disease in the global action plan for the prevention and control of non-communicable diseases 2013–2020, including GI disorders and diabetes (18). An aetiology-oriented strategy to disease prevention appears to be one of the most effective approaches for preventing and controlling pancreatic diseases (Figure 1). For example, identifying and eliminating environmental insults, such as smoking cessation and abstinence from alcohol, help prevent from pancreatitis and pancreatic cancer. It has been thought that T1DM is unpreventable (19, 20), but a recent study suggested that teplizumab, an Fc receptor-non-binding anti-CD3 monoclonal antibody, could delay or even prevent progression of this disease (21, 22). Therefore, apart from applying a disease prevention approach in managing these refractory diseases, novel therapeutics and curative treatments are desperately needed.
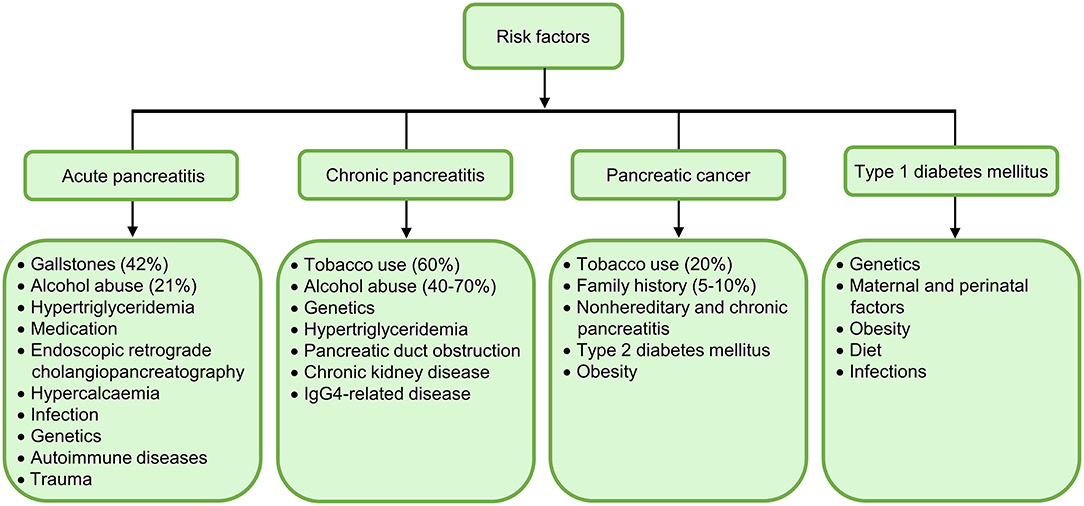
Figure 1. Summary of major aetiological risk factors for acute pancreatitis (AP), chronic pancreatitis (CP), pancreatic cancer, and type 1 diabetes mellitus (T1DM). Gallstones (42%) and alcohol abuse (21%) are the most frequent etiologies of AP; while tobacco use (60%) and alcohol abuse (40–70%) are the top aetiological factors of CP. Cigarette smoking (20%) and family history (5–10%) are dominant aetiological risk factors for pancreatic cancer. It is of note that alterations in oral microbiome predispose patients to pancreatic cancer. Genetics and environmental factors collectively contribute to the onset of T1DM, but other independent risk factors are unclear.
The human GI tract is home to trillions of microorganisms, including bacteria, archaea, fungi, viruses, and microeukaryotes, that are collectively known as the microbiome (23), forming a complex and unique ecosystem that influences human health and disease. Initially, our knowledge of the gut microbiome is largely reliant upon culture-dependent approaches. Breakthroughs in culture-independent approaches represented by high-throughput sequencing technologies, however, have further interrogated the intestinal microbial genomic blueprints and their functional potential in the past 16 years (24, 25). Likewise, the advent in mass spectrometry technology has facilitated the characterization and deciphering of gut microbial metabolite profiles (26). Accumulating evidence derived from metagenomics and metabolomics analyses suggests that gut microbiome composition and their metabolic activity are implicated in a multitude of conditions, including not only GI diseases (27, 28), but also extraintestinal disorders such as hepatic (29, 30), metabolic (31–40), respiratory (41–43), cardiovascular (44–47), neurologic (48–56), psychiatric (57–60), autoimmune (61–63), and oncologic components (64–69). There is no doubt that research into the gut microbiome and its role in human health and disease accompanied by tremendous technological advances in the past 16 years has taken center stage in biomedical science, and that most current microbiome research has revealed a close association between the gut microbiome and diseases, yet causative relationship has not been established in many cases. Given the bidirectionality of gut microbiome-disease interactions in human health, further research is needed in the future to dissect potential causality between them. In addition, gut dysbiosis is known to link to many disorders, and it thus seems feasible to manipulate the gut microbiome with products and treatments such as probiotics, prebiotics, synbiotics, postbiotics, and fecal microbiota transplantation (FMT) in clinical settings, irrespective of the stability and resilience of the intestinal microbiota (70–72).
This review summarizes existing evidence supporting the hypothesis that there is a close relationship between the gut microbiome and pancreatic diseases, and it further discusses the feasibility of preventing and controlling pancreatic diseases via microbiome-based therapeutics and therapies.
The fact that Helicobacter pylori-related disease (73–75) and recurrent Clostridium difficile-associated disease (76–78) are closely related to the gut microbiome has pushed the research into the functional interplay between the gut microbiome and human health/disease to a climax. An active role of the gut microbiome in influencing extraintestinal diseases has been increasingly recognized. More recently, research focus has thus shifted to investigate the relationship between the gut microbiome and extraintestinal manifestations like pancreatic diseases (79). Anatomically, the pancreas is located behind the stomach, where it connects the duodenum through the pancreatic duct and communicates with the liver through the common bile duct, forming a subtle gut-pancreas-liver axis for bidirectional communications. Experimental evidence also supports the existence of direct pancreas-gut microbiome interactions (Figure 2). For example, the production of cathelicidin-related antimicrobial peptide (CRAMP) by insulin-secreting beta-cells was controlled by gut microbiota-originated short-chain fatty acids (SCFA), highlighting a direct role of the gut microbiota in shaping the pancreatic immune microenvironment (80). On the other hand, Ahuja and colleagues demonstrated that ORAI calcium release-activated calcium modulator 1 (Orai1) produced by pancreatic acinar cells mediated secretion of antimicrobials that shaped the intestinal microbiota and intestinal immunity (81). These two seminal studies shed light on the existence of a gut-pancreatic axis, which warrants further preclinical and clinical investigations.
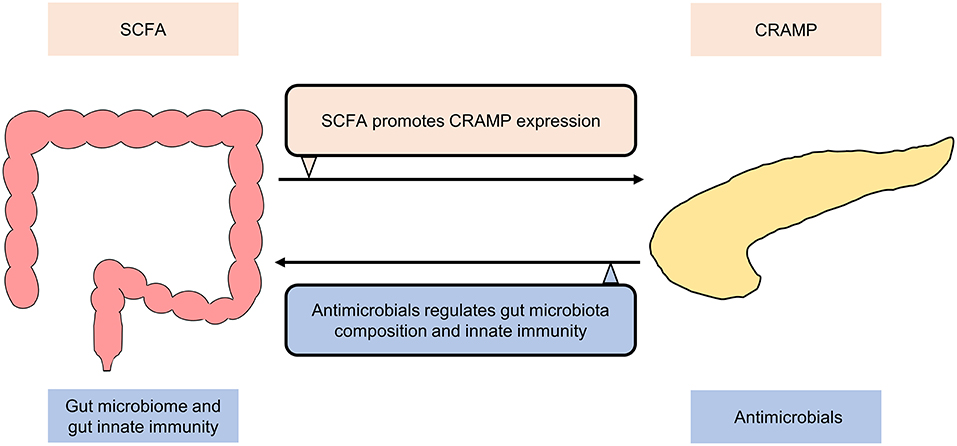
Figure 2. Bidirectional communication between gut and pancreas under normal conditions. Cathelicidin-related antimicrobial peptide (CRAMP) production by insulin-secreting beta-cells is controlled by short-chain fatty acids (SCFA) produced by the gut microbiota. ORAI calcium release activated calcium modulator 1 (Orai1) produced by pancreatic acinar cells mediates the secretion of antimicrobials, which shapes the gut microbiome and regulates gut innate immunity.
In addition, one caveat is that both normal and diseased individuals harbor a pancreatic microbiome (82), which might engage in disease initiation and progression (83–85). The landmark study by Pushalkar and colleagues discovered a link between pancreatic tumorigenesis and gut-originated intrapancreatic bacteria (84). It is notable that not only bacteria were the culprits, but also fungi. Gut-residing fungi that migrated from the intestine to pancreas might facilitate the progression of pancreatic ductal adenocarcinoma (PDAC) by driving the C3 cascade through activating mannose-binding lectin (85).
These studies challenged the conventional belief over the past decades that pancreas is a sterile organ and supported the presence of a pancreatic microbiome. Moreover, the results of various studies using traditional culture methods, real-time quantitative polymerase chain reaction (qPCR), 16S rRNA and shotgun metagenomics sequencing technologies consistently showed that the gut microbiota could migrate between the intestine and pancreas though the route of migration remains elusive. Three translocation pathways have been proposed: pancreatic duct reflux route, mesenteric venous drainage, and mesenteric lymphatic drainage (79). In-depth knowledge of the disease-associated microbial signatures, the exact mechanisms and trajectories of microbial migration, and the specificity of microbes capable of translocating through these routes may offer novel therapeutic targets and strategies.
Collectively, these works suggest that a gut-pancreatic axis may exist, which plays a role in determining the onset and severity of pancreatic diseases. Untangling this subtle yet intimate communication loop requires further preclinical and clinical studies.
A common inflammatory disorder of the pancreas is AP, in which pancreatic enzymes are activated locally due to a variety of etiologies, causing autodigestion, edema, hemorrhage, and even necrosis of pancreatic tissue, as well as dysfunction of remote organs and systems (86). Although the pathophysiology of AP has been well-described (87), none of these has pointed to the attribution to the gut microbiome. However, patients with AP, especially severe AP (SAP), are often accompanied by intestinal barrier dysfunction and subsequent bacterial translocation into the pancreas and/or peripheral blood (88, 89), and since the intestinal microbiome plays a vital role in maintaining intestinal epithelial barrier integrity and shaping intestinal mucosal immune system (90–92), the importance of the gut microbiome in the aetiology of AP should not be neglected.
It was not until recently that researchers began to apply high-throughput sequencing technologies in deciphering the role of gut dysbiosis in AP. For instance, gut dysbiosis and decreases in the expression of antimicrobial peptides in Paneth cells were observed in rats suffering from acute necrotizing pancreatitis induced by hypertriglyceridemia (93). 16S rRNA-based gut microbiota analysis revealed that patients had fewer commensal beneficial bacteria (such as Prevotella and Faecalibacterium) and more pathogenic bacteria (such as Escherichia-Shigella and Enterococcus) compared with healthy subjects (94). Moreover, a study found that patients with varying severity of AP had distinct gut microbiota signatures, characterized by more Bacteroides, Escherichis-Shigella, and Enterococcus in mild AP (MAP), moderately SAP (MSAP), and SAP, respectively (95). Similarly, another study reported that the intestinal microbiome signature in patients with SAP was distinct from those suffering from MAP and MSAP, characterized by a reduction in commensal bacteria such as Bacteroides, Alloprevotella, and Blautia (94).
Despite the investigators have observed changes in intestinal microbiome in the setting of AP, few studies exist that have explored the effect of intestinal microbiome on AP. A few exceptions do exist, a seminal experimental study showed that antibiotic-treated mice and germ-free (GF) mice exhibited attenuated pancreatic injury after AP induction, and subsequent FMT worsened the severity of AP, demonstrating that the gut microbiota was a mediator in AP (94). This notion is underpinned by another recently published study where the authors reported that the gut microbiota and the NLRP3 inflammasome acted together to exacerbate the severity of AP (96). Specific intestinal species even appeared to be critical for the pathogenesis. For example, the gut commensal species, Escherichia coli, exacerbated acute necrotizing pancreatitis through targeting intestinal epithelial cells (97). These pioneering studies strongly support that the gut microbiota and dysbiosis are associated with the severity of AP.
Another common pancreatic disorder is CP. It is a progressive fibroinflammatory condition characterized by gradual replacement of pancreatic secretory parenchyma by fibrous tissues, resulting in endocrine and exocrine dysfunction (98). Similar to AP, the pathogenesis in CP might be associated with the gut microbiome (99–101). A seminal study systematically investigated the fecal microbiomes of patients with CP by 16S rRNA gene sequencing (102). The 16S rRNA microbiota in patients with CP had diminished gut microbial diversity and richness, and the dysbiosis was accompanied by alterations in the taxonomic microbiota profiles (102). A similar outcome was also observed for the first time in mice with CP where authors showed that CP mice had significantly reduced bacterial species richness and diversity (103). Specifically, the abundance of Bacteroides and Alloprevotella genera increased, while the abundance of Lachnospiraceae_NK4A136, Ruminiclostridium, and Roseburia decreased (103). A recent study also demonstrated a distinct difference in gut microbiota between CP mice and control mice (104). However, to our knowledge, no study to date has yet examined the role of intestinal microbiome on CP in humans or animals, which is worthy of future investigation.
Altogether, research on the gut microbiome of individuals with pancreatitis is still in its infancy, but current evidence does implicate that the host gut microbiome and pancreatitis are closely linked. Applications of GF mouse models and metagenomic sequencing approaches rather than relying on biomarker sequencing will likely provide a better insight into the intricate relationship between the gut microbiome and pancreatitis.
T1DM is an autoimmune disorder, characterized by T cell-mediated destruction of insulin-producing beta cells in the pancreas, resulting in a reliance on exogenous insulin throughout life (105). Genetic variation is a well-established risk factor for T1DM as more than 50 diverse genetic loci have been identified (106), many of which are located in the human leukocyte antigen (HLA) region (107). Apart from genetic predisposition to T1DM pathogenesis, environmental factors also play an integral role (108). In fact, genetic susceptibility and environmental events, conspire together to provide fertile ground for the initiation and progression of T1DM (109), and since both environmental factors (e.g., dietary habits) (110–113) and genetic risks (114–116) profoundly affect the human gut microbiome, it would be of interest to find out the role of the gut microbiome in T1DM.
In fact, the association between the gut microbiome and T1DM has been implicated in previous studies (117–122). 16S rRNA pyrosequencing analysis of fecal microbiota of children with and without beta-cell autoimmunity showed that children with autoimmunity had fewer lactate-producers, butyrate-producers, Bifidobacterium adolescentis, and Bifidobacterium pseudocatenulatum but more Bacteroides (123). Another study compared the gut microbiota of children with T1DM, maturity-onset diabetes of the young 2 (MODY2), and in healthy state in a case-control study by 16S rRNA pyrosequencing, and the results showed that children with T1DM had a significantly lower gut microbiota diversity; significantly more Bacteroides, Ruminococcus, Veillonella, Blautia, and Streptococcus genera; and significantly fewer Bifidobacterium, Roseburia, Faecalibacterium, and Lachnospira (124). Gut dysbiosis was observed in children aged 1–5 years with new-onset T1DM (125). Whole metagenome sequencing uncovered an increase in lipopolysaccharides-producing bacteria and a decrease in SCFA-producing bacteria in women with T1DM across pregnancy compared with healthy individuals as controls (126). Remarkably, the gut mycobiome signature is beginning to intrigue humans, as recently demonstrated in a pilot study assessing the gut mycobiome in adult patients with T1DM (127).
Indeed, the gut microbiome appears to be a principal driving force of the onset of T1DM, particularly in subjects genetically disposed to the disease. The hallmark study of Wen and colleagues first reported that GF but not specific pathogen-free non-obese diabetic (NOD) mice devoid of MyD88, a key intracellular component of multiple Toll-like receptor-mediated signaling pathways, did develop robust diabetes, and colonization of a defined microbial consortium could mitigate diabetes (128). A longitudinal study followed the serum conversion pattern of autoantibody of 33 HLA-matched infants from birth to 3 years of age (129). Four of the 11 seroconverted infants developed T1DM, accompanied by drastic reduction in intestinal microbiome diversity though no obvious change was observed in their major gut metabolites (129). On the other hand, The Environmental Determinants of Diabetes in the Young (TEDDY) study analyzed stool samples of 783 children collected monthly during their first 3 months of age until the clinical endpoint (130). The study found that early infant microbiome was dynamic and highly individualized both at the taxonomic and functional levels, and the colonic SCFA might influence early-onset T1DM (130). The gut microbiome not only drives T1DM development, but also has an impact on cognitive functions. For examples, depletion of acetate producing bacteria, caused by vancomycin exposure, resulted in cognitive impairment in T1DM mice (131). Moreover, gender bias in autoimmunity including T1DM was found to be influenced by the host gut microbiota, and the sex differences in the gut microbiome could drive hormone-dependent regulation of autoimmunity (132, 133). Thus, experimental and clinical evidence implicated that alterations in the gut microbiome and/or metabolome occurred prior to or even played an active role in driving the onset of T1DM in genetically susceptible infants, though the exact mechanisms are largely unclear. Yet, a previous study found that lipopolysaccharide from Bacteroides dorei might increase the risk of autoimmunity (134). Moreover, only very few studies applied metaproteomics in investigating the functional interactions between the host microbiota and T1DM disease risk (135).
Thus, it will be necessary to conduct more mechanistic studies to confirm the causal role of gut microbiome in the onset of T1DM not only by 16S rRNA gene sequencing, but also in combination with metagenomics, metabolomics, and metaproteomics approaches to decipher the functionality of the gut microbiome in T1DM.
Pancreatic cancer refers to tumors that start in the cells of the pancreas, including exocrine tumors. Generally, it is classified into PDAC (about 95% of pancreatic cancers) and endocrine tumors (about 5% of pancreatic cancers) (136). In PDAC, alterations in gut microbiota were observed both in humans and animals. For example, genetically engineered PDAC mice exhibited gut dysbiosis characterized with the predominance of Proteobacterial and Firmicutes as well as elevated serum polyamine metabolism (137). A previous study found that the 16S rRNA fecal microbiota of patients with pancreatic carcinoma (n = 85) exhibited significantly diminished alpha-diversity compared with matched healthy controls (n = 57), accompanied by increases in certain pathogens and lipopolysaccharides-producing bacteria, and decreases in probiotics and butyrate-producing bacteria (138). Patients with PDAC had an increased phylum Proteobacteria (such as Gammaproteobacteria) and a decreased phylum Firmicutes (such as butyrate-producing bacteria, including Eubacterium rectale, Faecalibacterium prausnitzii, and Roseburia intestinalis) when compared with healthy controls (139). Additionally, several recent review articles have underscored the importance of the gut microbiome in the development and progression of PDAC (140–142).
Evidence is also beginning to show that the gut microbiome is involved in pancreatic oncogenesis or tumor suppression (143). Thomas and colleagues demonstrated that the intestinal microbiome could accelerate pancreatic carcinogenesis through a long-distance mechanism in preclinical models (144). Ablation of the gut microbiome by oral antibiotics could diminish pancreatic malignancies burden through increasing infiltration of interferon gamma-producing T cells and reducing interleukin 17A and interleukin 10-producing T cells, further supporting the notion that the gut microbiome might promote the pathogenesis in pancreatic cancer (67).
Recent studies found that the pancreatic tissue harbors microorganisms under both normal and pathological conditions (82). A unique microbial signature has also been observed in patients with pancreatic cyst fluid, characterized by an enrichment of Bacteroides spp., Escherichia/Shigella spp., and Acidaminococcus spp. (145). The fact that the pancreas harbors microorganisms raises the question of whether the pancreatic microbiome, in addition to the gut microbiome, is tied to the disease process. Geller and colleagues showed by qPCR, 16S rRNA fluorescence in situ hybridization analysis, and immunohistochemistry that intratumor samples of pancreatic cancer patients contained more bacteria than matched tissues in control subjects (83). The intratumor microbiota of these human PDAC samples was dominated by Gammaproteobacteria, which conferred gemcitabine resistance in 14 of 15 of the human PDAC tumors (83). Collectively, these data showed that pancreatic tumor tissues contained bacteria that rendered resistance to gemcitabine and thereby affected the efficacy of cancer therapies (83). The study convincingly confirmed the role of intratumor bacteria in promoting gemcitabine resistance in pancreatic cancer.
More intriguingly, the observations of several recent studies suggested that the gut microbiome was in tight partnership with the intratumoral microbiome collectively contributing to carcinogenesis in the tumor microenvironment, implicating that there was gut-tumor microbial crosstalk and that such interactions could affect tumor outcome (84, 85). The tumor microbiome in PDAC patients also varied with tumor tempo; the 16S rRNA-tumor microbiome of patients of long-term survival (LTS) exhibited a higher alpha-diversity and more Pseudoxanthomonas, Saccharopolyspora, Streptomyces, and Bacillus clausii than patients of short-term survival (STS) (146). Mechanistically, the tumor microbiome shaped the antitumor immune responses through CD8+ T cell recruitment and activation (146). Furthermore, human gut microbiome was absent from normal tissues adjacent to tumor tissues, but it represented 25% of human tumor microbiome in matched tumor samples (146). The FMT of fecal samples of LTS but not STS or healthy donors significantly reduced tumor growth in antibiotics-fed mice with orthotopic syngeneic tumors, and the antitumor effect was attenuated by antibiotic treatment (146). These results suggested that the gut microbiota of pancreatic cancer patients had the capacity to migrate and colonize pancreatic tumor tissues and that the gut and/or tumor bacteria from LTS patients could confer a protective effect against tumor growth. It would thus be of interest to further elucidate the significance of gut-tumor microbial crosstalk in progression of pancreatic cancer. Moreover, since the oral microbiome is closely related to the gut microbiome, it is also worthy investigating if the oral microbiome is another driver for the progression of pancreatic cancer (147–149).
In summary, a subtle yet intimate nexus loop exists between the gut microbiome and pancreatic diseases, including pancreatitis, pancreatic cancer, and T1DM (Figure 3). Gut dysbiosis has been consistently reported in patients of pancreatic diseases; however, causal relationships between the gut microbiome and pancreatic disorders, as well as the mechanism of gut microbiome in contributing to the onset, progression, and pathogenesis of this spectrum of diseases, merit further elucidation. The expansion of our knowledge in the field will help identify novel treatment targets and deepen our insights into the development of personalized therapies.
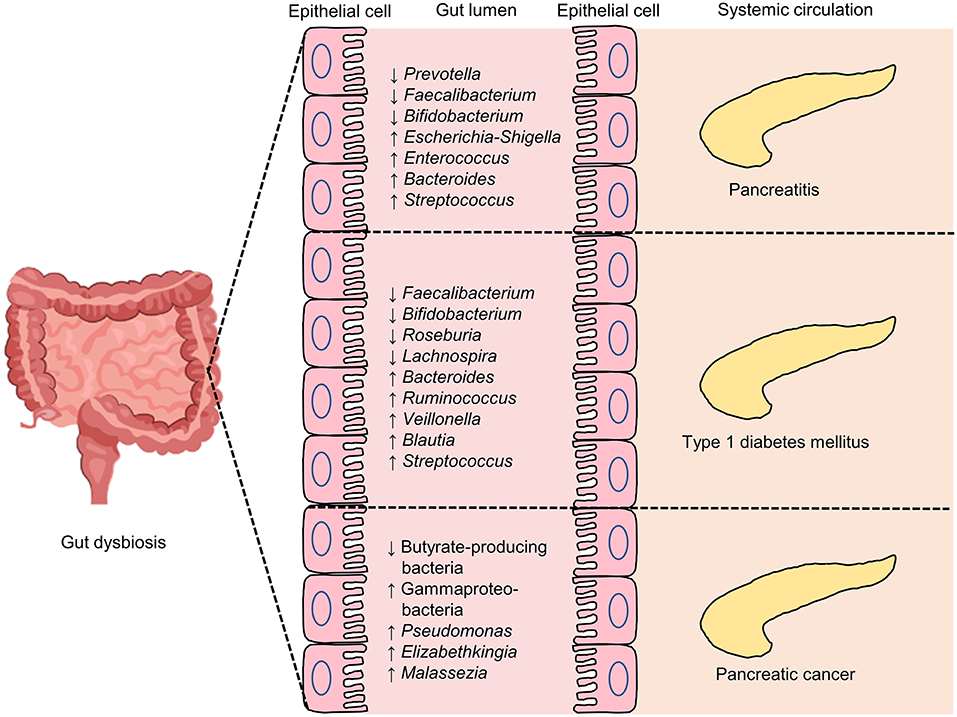
Figure 3. Some of the reported intestinal microbial genic features in pancreatic diseases, including pancreatitis, type 1 diabetes mellitus (T1DM) and pancreatic cancer. An overview of selected key gut microbiome features related to pancreatic diseases. ↓, lower abundance in pancreatic diseases when compared with control; ↑, higher levels in pancreatic diseases when compared with control.
Given the close link between the gut microbiome and pancreatic disorders, it may be feasible to ameliorate pancreatic diseases by modulating the gut microbiome via application of products and treatments like probiotics (150), prebiotics (151), synbiotics (152), postbiotics (153), and FMT (154) (Figure 4). A better understanding of the causal relationship in gut microbiome-pancreatic disease interactions will improve the precision of novel target therapies.
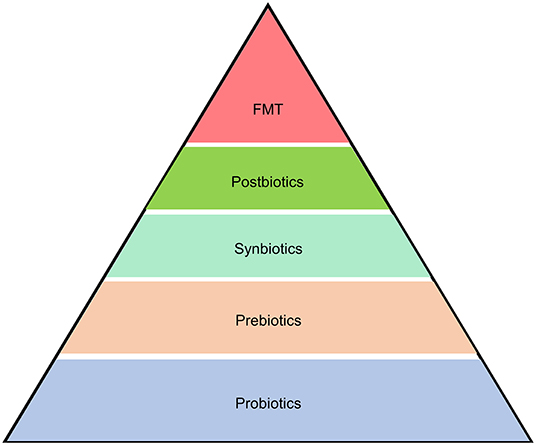
Figure 4. Microbiome-based therapeutics and therapies for pancreatic diseases. Strategies to alleviate pancreatic diseases by modulating the gut microbiome include application of products and treatments like probiotics, prebiotics, synbiotics, postbiotics, and fecal microbiota transplantation (FMT).
Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host,” according to the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus in 2013 (150). The role of probiotics in human health and disease has received increasing attention, especially in the prevention and treatment of acute gastroenteritis (155), Clostridium difficile-associated diarrhea (156–158), irritable bowel syndrome (159, 160), neonatal sepsis (161), and acute respiratory infection (162), which have shown beneficial clinical efficacy. Although less studied, clinical evidence also shows beneficial effects of administering probiotics in pancreatic-related disorders.
A number of preclinical studies have reported beneficial effects of probiotic application in mitigating AP (Table 1). For instance, ingesting Lactobacillus plantarum for 4 days before and after induction of AP could reduce microbial translocation in experimental pancreatitis (163). Consuming a microbial consortium comprising Streptococcus thermophilus, Lactobacillus acidophillus, Bifidobacterium lactis was capable of reducing the severity of experimental AP (164). A similar outcome was observed in rats that received Ecologic 641 probiotic formulations 5 days prior to the induction of AP (165). Modification of gut microbiota via administration of Ecologic 641 probiotic formulations reduced bacterial translocation, morbidity, and mortality in the course of experimental AP (166). In two independent studies, the authors observed that Saccharomyces boulardii could diminish bacterial infections and ameliorate pancreatitis (167, 168). A systematic review and meta-analysis strengthened the findings of the aforementioned studies by showing that probiotics did exhibit efficacy in animal models of AP (169). Although the probiotic product, Ecologic 641 probiotic formulations, has been widely promoted as beneficial for AP, a rat experimental model aiming to investigate the association between probiotic prophylaxis (by feeding Ecologic 641 probiotic formulations) and enteral nutrition with AP mortality did not find significant differences between groups in terms of histological severity of pancreatitis, degree of discomfort, weight loss, histological examination of small bowel and bacterial translocation, suggesting probiotic application was ineffectual nor harmful to AP-induced rats (170). Another study found that Clostridium butyricum and its major metabolite, butyrate, could reduce intestinal injury possibly by altering the functions of the intestinal mucosal barrier (171).
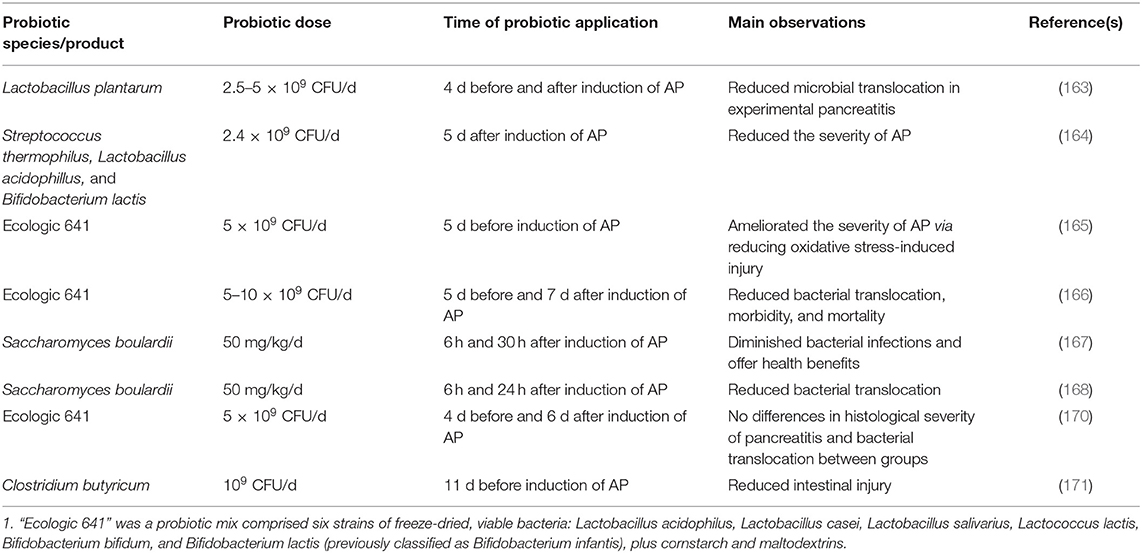
Table 1. Summary of preclinical studies investigating the effects of probiotic application in acute pancreatitis (AP).
It was not until 2002 that probiotics began to be used in clinical trials (Table 2). In Oláh et al. (172), 45 patients with AP were randomly assigned to receive either live or inactivated Lactobacillus plantarum (that is, postbiotics) with oat fiber for 7 days by nasojejunal tube. Significant differences were observed in the severity of infective pancreatic necrosis and abscesses between the two groups, but not the mean length of hospital stay (172). Similarly, Lactobacillus plantarum supplementation in patients with AP attenuated the severity of disease, improved intestinal permeability and clinical outcomes (173). Although beneficial effects of ingesting probiotics have been observed in some clinical studies in AP, other studies yielded neutral or even negative outcomes. For example, in a multicenter randomized, double-blind, placebo-controlled trial, 298 patients with SAP were randomly assigned to receive either Ecologic 641 probiotic formulations (n = 153) or placebo (n = 145), infectious complications occurred in 46 (30%) probiotic-receivers and 41 (28%) placebo-receivers, and the mortality rate of patients was higher in the probiotic group (24 patients; 16%) compared with (9 patients; 6%) the placebo group, suggesting that probiotic consumption did not reduce the risk of infectious complications but was associated with an increased risk of mortality (174). Probiotic supplementation did not affect gut integrity, infectious complications, mortality, and hospital stay in another double-blind randomized controlled trial published in 2011 (175). Furthermore, two meta-analyses reported that probiotics intake was neither beneficial nor harmful to patients with SAP (176, 177). A recent Cochrane review that included 84 randomized controlled trials found that, probiotics intake was not associated with the overall mortality in AP (178). Intriguingly, probiotic preparations consisting of Bifdobacterium longum, Lactobacillus acidophilus, and Enterococcus faecalis showed a curative effect in patients with SAP by lowering the levels of pro-inflammatory cytokines, restoring the gastrointestinal function sooner, decreasing complications such as infection, and shortening of hospital stay in patients with SAP (179). The consumption of a mixed probiotic preparation containing Bacillus subtilis and Enterococcus faecium reduced the percentage of pancreatic sepsis, multiple organ dysfunction syndrome, and mortality (180). Furthermore, the intake of the same probiotic mix reduced the length of hospital stay of patients with MAP, although no statistical difference was seen in recurrent abdominal pain between the probiotic and placebo group (181).
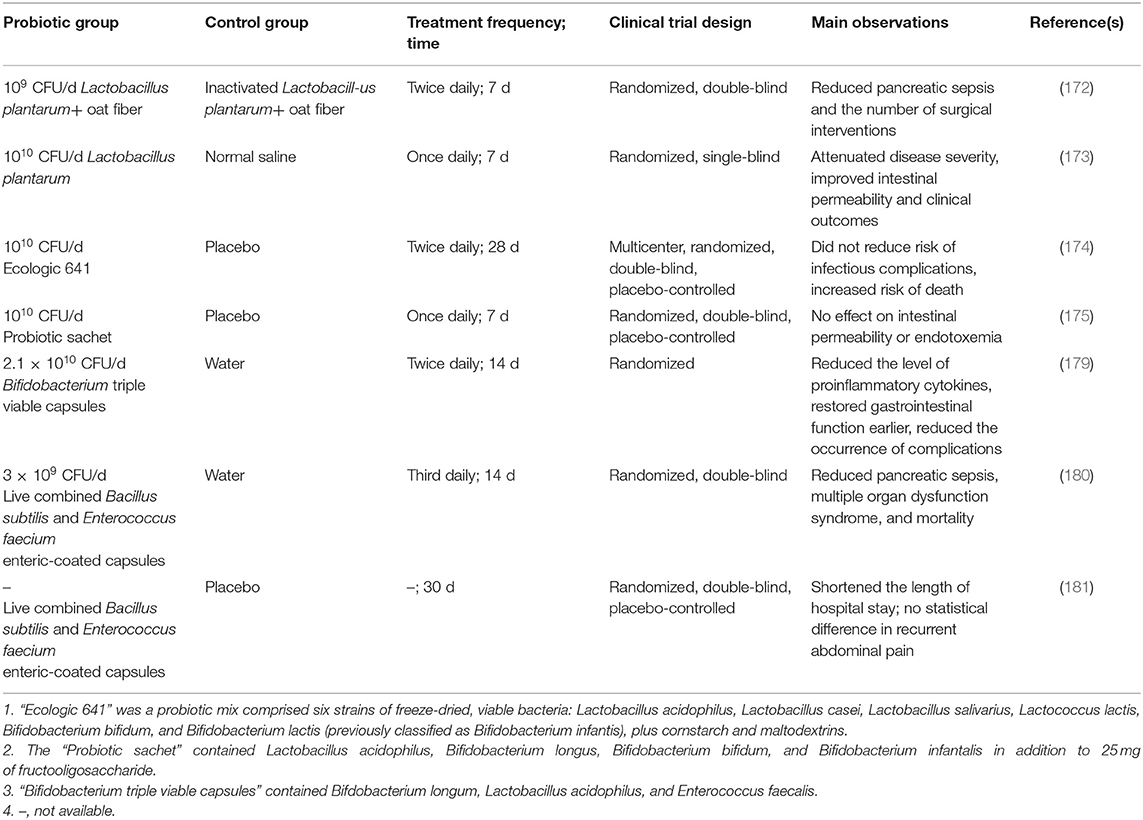
Table 2. Summary of clinical trials investigating the efficacy of probiotic application in pancreatitis.
Overall, there are discrepant results regarding the effects of probiotics on AP. Larger scale studies would be necessary to clarify whether probiotics intake could improve AP and if there are strain-specific effects in alleviation of AP-associated symptoms. It is noteworthy that there is scarce data on the effect of probiotics on CP in preclinical and clinical trials; thus, whether probiotic treatment has any clinical efficacy in CP remains to be confirmed.
The available literature reveals that the mechanisms of probiotic effects in AP are exerted via improving the intestinal barrier function, inducting inflammatory responses, and modulating the gut microbiome (Figure 5).
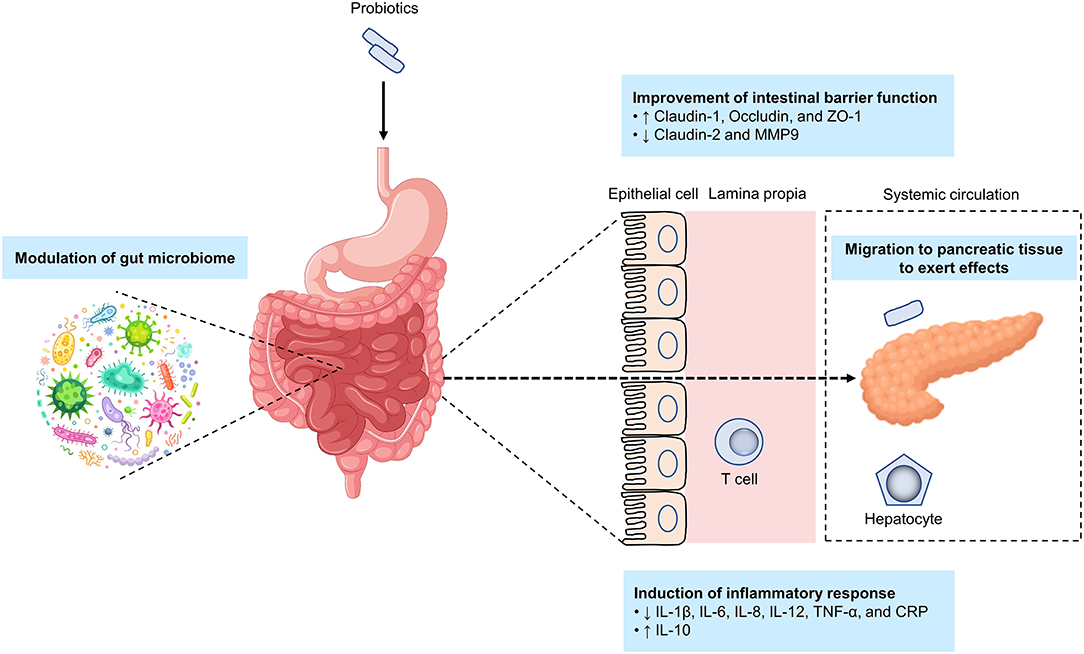
Figure 5. Potential mechanisms by which probiotics alleviate acute pancreatitis. The putative mechanisms by which probiotics alleviate acute pancreatitis include improving the intestinal barrier function, inducting inflammatory responses, modulating the gut microbiome, and targeting pancreatic tissue. ↑, indicates an increase; ↓, indicates a decrease.
Intestinal barrier dysfunction is closely linked to the course of AP (89). Bacterial translocation is triggered by an imbalance in intestinal barrier function, especially bacterial migration from the small intestine into the pancreatic tissue and/or in peripheral blood is a major cause of infection in the pancreas or peripancreatic tissue (88, 182). Probiotics may act by improving intestinal barrier function that in turn prevents bacterial translocation. Oral gavage of Ecologic 641 probiotic formulations once daily for 2 days to mice prior to induction of AP by intraperitoneal injections with cerulein could prevent intestinal barrier dysfunction in the late phase of AP; however, such effect was not seen if probiotics were administered after induction of AP, indicating that the efficacy of probiotics was related with the timing of probiotic application (183). In a randomized, placebo-controlled, multicenter trial, probiotic application showed a capacity for preventing bacterial translocation (184). Another study found that the probiotic mechanism of improvement of intestinal barrier function was via increasing tight-junction proteins (including claudin-1, occludin, and ZO-1) while reducing claudin-2 and MMP9 (171). It is worth noting that the metabolites of probiotics, also known as postbiotics similarly have the effect of improving intestinal barrier function and thus preventing bacterial translocation (185–187).
As discussed earlier, bacteria and fungi are able to migrate from the gut into the pancreatic tissue, where they are responsible for the course of disease (84, 85). Translocation of a gut pathobiont, Enterococcus gallinarum, to the liver and other systemic tissues triggered autoimmune responses in genetically predisposed hosts provides a prime example (188). Moreover, the dissemination of Escherichia coli from primary colorectal cancer (CRC) via the gut vascular barrier allowed the bacteria to migrate to the liver to form a premetastatic niche, paving a way for CRC to metastasize to the liver in advance (189). Despite the aforementioned two studies are beyond the scope of this review, they have provided compelling evidence of dissemination of gut-originated bacteria in relation to onset and development of diseases. In our laboratory, we found that Lactobacillus rhamnosus Probio-M9 could transmit from the gut to mammary tissue to alleviate symptoms of Staphylococcus aureus-induced mastitis in rats. Here, we hypothesize that the mechanism of action of probiotics resembles that seen in several examples mentioned above, albeit we do not know whether probiotics will promote or alleviate the development of disease (Figure 5).
Another probiotic mechanism is via reducing inflammatory responses in pancreatitis. Ingesting probiotics could lower proinflammatory cytokines, such as interleukin (IL)-8, tumor necrosis factor (TNF)-α, and C-reactive protein (179). Meanwhile, the species Clostridium butyricum has been shown to suppress IL-6, IL-12, IL-1β, and TNF-α production (171), while another study found that probiotics intake could reduce TNF-α and IL-6 expression and enhanced IL-10 expression in SAP (180).
Finally, probiotics might serve as a gut microbiota modifier, regulating the gut function and homeostasis (190), particularly in the presence of pancreas-gut microbiota cross-talk and interactions (143). Notably, few studies have investigated the effects and mechanisms of action of probiotics on the gut microbiome in AP. Large-scale animal models and high-quality clinical trials are therefore needed to determine the interactions between probiotics, host gut microbiome, and AP development and pathogenesis.
The NOD mouse model is one of the most important animal models in T1DM research (191). A myriad of evidence regarding the role of probiotics in T1DM stems from NOD mouse models and human studies (Table 3). Female NOD mice receiving the probiotic mix, VSL#3, orally could prevent spontaneous autoimmune diabetes via an IL-10-dependent mechanism and gut microbiota modulation (192, 193). A previous study in NOD mice showed that Lactococcus lactis could serve as a potential immunotherapeutics for autoimmune T1DM (194). As one of the examples, oral administration of Lactococcus lactis secreting the beta-cell antigen glutamic acid decarboxylase (GAD)-65 along with anti-inflammatory cytokine IL-10 to newly diabetic NOD mice led to a significant diabetes reversal, which was fortified up to 67% when in combination with low-dose anti-CD3 antibody (195). Importantly, this effect that persisted after treatment discontinuation (195). Clostridium butyricum has been reported to protect against T1DM by modulating intestinal immune homeostasis and inducing pancreatic regulatory T cells (196). The development of autoimmune diabetes in NOD mice could be alleviated by taking a probiotic combination comprising five bacterial strains that acted by reducing gut permeability, inducing gut-homing Treg cells, and reducing Th1 polarization (197). Akkermansia muciniphila, a candidate of next-generation probiotics, appeared to slow the progression of T1DM in NOD mice (198, 199).
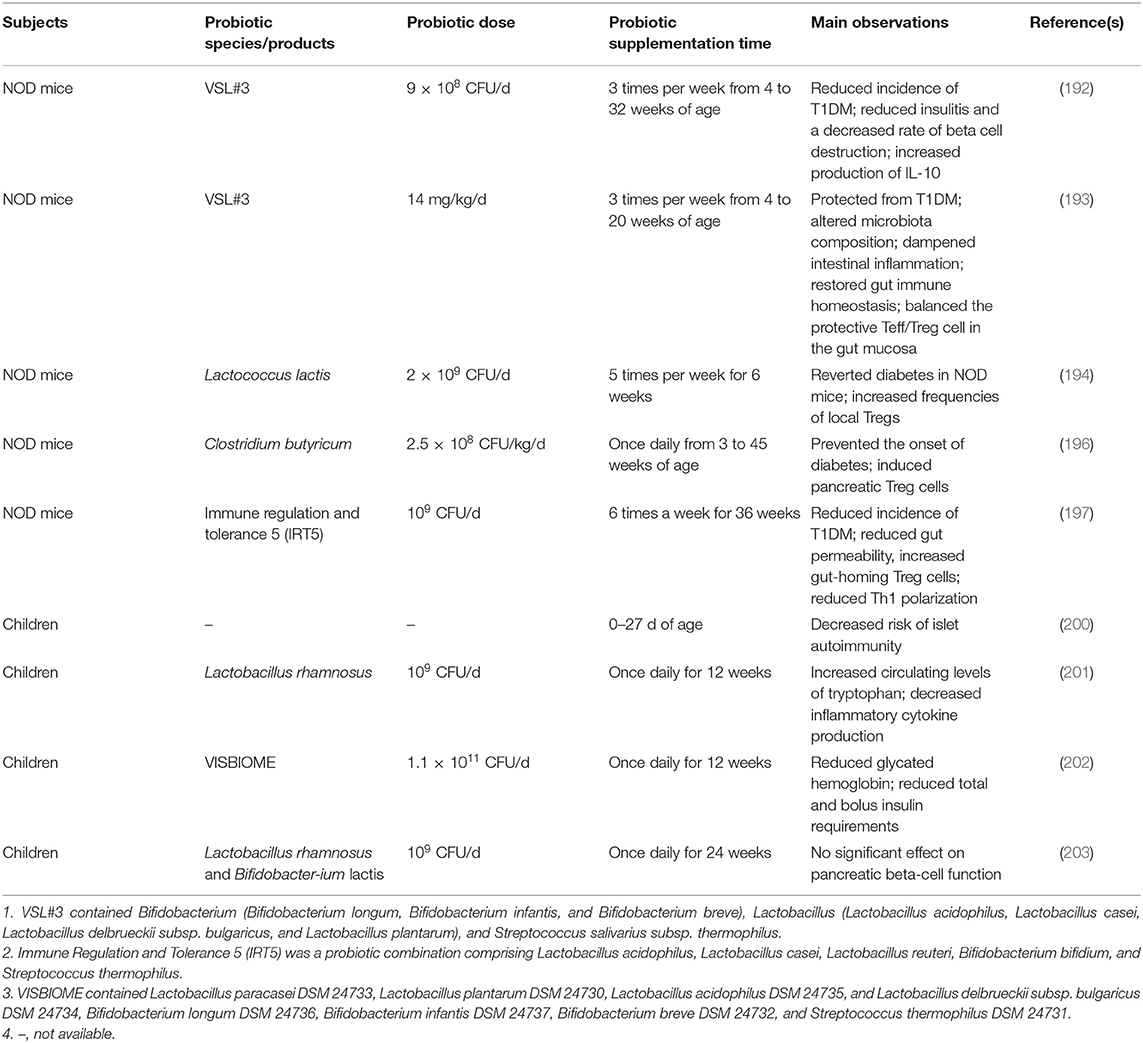
Table 3. Studies investigating the efficacy of probiotics in preventing and alleviating murine and human type 1 diabetes mellitus (T1DM).
A prospective cohort study of 7,473 children showed that early probiotic supplementation could reduce the risk of islet autoimmunity in children at the highest genetic risk of T1DM (200). Another study of 61 young T1DM patients given either Lactobacillus rhamnosus GG or placebo daily for 12 weeks showed that intake of probiotics significantly increased circulating levels of tryptophan and decreased inflammatory cytokine production (201). The results of a recent randomized, double-blind, and placebo-controlled pilot study assessing treatment with a multispecies probiotic preparation, in 90 children with new-onset T1DM showed that better glycemic control and a decrease in insulin requirements (202). Notably, a double-blind, randomized controlled trial found that the intake of Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 had no effect on beta-cell function in children newly diagnosed with T1DM (203).
Similar to AP, contradicting results have been obtained regarding the clinical efficacy of probiotics in management of T1DM. The discrepancies should be taken into account in future studies and in design of novel treatment strategies.
Studies of the beneficial effects of probiotic consumption on pancreatic cancer seem to be just beginning, and most published works were conducted in animal models. Probiotic Lactobacillus could enhance gemcitabine-mediated antitumor efficacy in mice with pancreatic cancer (204). This finding is strengthened by another recently published study where authors further observed that probiotics could reduce gemcitabine-induced side effects by restoring a favorable microbiota (205). In addition, probiotic-treated super-charged NK cells could prevent the growth of pancreatic tumors through lysis and differentiation in humanized-BLT mice (206). The MAPK-p38 and transforming growth factor-β (TGF-β) signaling pathways have been hypothesized to be the mechanisms by which probiotics exert their anti-tumor effects (207, 208). Whether probiotics have a role in slowing or inhibiting the progression of pancreatic cancer needs to be further investigated and warrants future preclinical and clinical studies.
The ISAPP consensus statement (2016) defined the term prebiotics as “a substrate that is selectively utilized by host microorganisms conferring a health benefit,” including conjugated linoleic acids, polyunsaturated fatty acids, oligosaccharides such as fructooligosaccharides, inulin, galactooligosaccharides, mannanoligosaccharides, xylooligosaccharides, human milk oligosaccharides, phenolics, and phytochemicals (151). Administration of chitosan oligosaccharides to mice for 4 weeks prior to induction of SAP significantly reduced the severity of pancreatic injury by reducing oxidative stress and modulating the gut microbiota (209). Correlations between prebiotic fiber supplementation and hospital stay, duration nutrition therapy, acute phase response and overall complications were identified in patients with SAP in a randomized prospective double-blind controlled clinical trial (210). Inulin-type fructans were thought to play a role in preventing the development of AP and T1DM (211, 212). Supplementation of low-methoxyl pectin in NOD mice alleviated T1DM by increasing colonic SCFA production (213, 214). Similar results have been reported in a separate study investigating the effect of human milk oligosaccharides on T1DM in NOD mice (215). A randomized, placebo-controlled trial showed that the administration of prebiotics (oligofructose-enriched inulin) could improve glycemic control in children with T1DM (216). A series of investigations have also shown that a diet rich in resistant starch was significantly associated with decreased risk of experimental pancreatic malignancies (217, 218).
The definition of symbiotic is “a mixture comprising live microorganisms and substrate(s) selectively utilized by host microorganisms that confers a health benefit on the host,” according to the ISAPP consensus statement issued in 2019 (152). Enteral feeding with Synbiotic 2000 not only reduced organ dysfunctions in patients with SAP, but also improved intestinal barrier function (219, 220). Administration of synbiotics composed of Lactobacillus casei, Lactobacillus rhamnosus, Lactobacillus acidophilus, Bifidobacterium bifidum, and fructooligosaccharides in patients with CP increased the serum concentrations of hemoglobin, hematocrit, erythrocytes, total count of lymphocytes, magnesium, and albumin, and meanwhile reduced the total cholesterol values, without altering the nutritional status of the patients (221). In another single-blind prospective randomized control trial, synbiotics containing Streptococcus faecalis, Clostridium butyricum, Bacillus mesentericus, Lactobacillus sporogenes, and fructooligosaccharides significantly reduced septic complications, hospital stay, and antibiotic requirement in patients undergoing pancreatic surgery for CP (222). A randomized, double-blind, placebo-controlled trial showed that synbiotic supplementation in patients with T1DM might be effective in improving fasting blood glucose, hemoglobin A1c, insulin, hypersensitive C-reactive protein, and total antioxidant capacity (223).
The nomenclature of postbiotics was not clear until 2021 when the ISAPP defined a postbiotic as a “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host” (153). Evidence is emerging that postbiotics have multiple health benefits and are considered to be the next horizon in microbial therapeutics and functional foods (224). In a cerulein-induced AP mouse, oral administration of probiotic Lactobacillus brevis SBL88-derived polyphosphate for 24 days prior to induction of AP mitigated the severity of AP, which was not only associated with modulation of the intestinal microbiome but also enhancement of the gut barrier integrity (225). A postbiotic, heat-killed Lactococcus chungangensis CAU 1447, could facilitate wound healing in type I diabetic mice; however, the authors did not investigate its effect on T1DM (226). Ferrichrome, derived from Lactobacillus casei ATCC334, has been shown to inhibit the growth of pancreatic cancer cells (227).
Fecal microbiota transplantation is a procedure in which stool from a healthy donor is placed into another patient's intestine (228). FMT to both antibiotic-treated mice and GF mice resulted in an aggravated AP (94). Heparanase-transgenic mice had more severe AP than wild-type mice; however, transfer of feces from the former to the latter worsened the disease (229). Parallels have also been observed in Western-type diet in combination with acute necrotizing pancreatitis (ANP) mice (230). However, a case report showed that FMT could be an effective therapeutic strategy in MSAP patients (231). The exact implication of FMT in the onset of pancreatitis remains to be proven in animal and human studies. In specific pathogen-free NOD mice, females have 1.3–4.4 times higher incidence of T1DM. Gavage transfer of gut microbiota from adult males to immature females altered the recipient's microbiota, elevated the testosterone, caused metabolomic changes, reduced islet inflammation and autoantibody production, and protected against T1DM (133). Transplanting fecal samples from diabetes-protected MyD88-deficient NOD mice to wild type female NOD/LtJ mice led to a delayed onset of diabetes and a reduced insulitis (119). Early-life antibiotic exposure not only perturbed the intestinal microbiota, but also accelerated the development of T1DM in the NOD mouse model; however, maternal cecal microbiota transfer to antibiotic-induced NOD mice restored the enhanced disease risk to baseline levels (232, 233). A randomized controlled trial published in 2021 found that FMT halted the onset of T1DM in human (234). Some other human FMT trials are ongoing for T1DM (NCT04124211; NCT04749030). A paucity of studies have investigated the application of FMT in pancreatic cancer, except those mentioned earlier.
In summary, research on the application of probiotics in pancreatic diseases has spawned a great deal of data; however, the data from prebiotics, synbiotics, postbiotics, and FMT is rather limited. Although heterogeneity exists in some studies, the application of probiotics, prebiotics, synbiotics, postbiotics, and FMT for improving pancreatic diseases remains promising.
Recent metagenomics and metabolomics studies have highlighted the complex interplay between the gut microbiome and pancreatic-related disorders. However, these approaches have several limitations. Firstly, they are association studies rather than causal research, which is needed to show whether alterations in the gut microbiome and its metabolites are a cause or a consequence of the disease. Secondly, although it is possible to describe the full spectrum of microorganisms through metagenomics studies, targeting strains like traditional culture approaches is difficult. Thirdly, metabolomics has almost become a golden standard used to depict metabolic profiles of the gut microbiome, but it is not easy to identify sources of specific metabolites on the species or strain level. To address these constraints, GF animal model and culturomics have become essential tools for exploring potential causality between host-microbial interactions (235, 236). The GF animal model is an attractive model, which is devoid of microorganisms per se. Excluding interference introduced by the host indigenous microbiome makes GF animal an indispensable key model for causality research. Culturomics has enabled laboratory cultivation and characterization of the vast majority of intestinal microbes, which is integral to research into microbiome (237). Additionally, application of other complementary technologies, such as metatranscriptomics and metaproteomics, will expand our understanding of the gut microbiome and its function.
Modulation of the gut microbiome by probiotics, prebiotics, synbiotics, postbiotics, and FMT has reinforced the potential for improving pancreatic diseases. However, animal and human data with respect to probiotics are equivocal in some cases, which could be due to several reasons. Firstly, discrepancies in probiotic strains, dosage, duration and frequency of administration in studies investigating effects of probiotic application are likely responsible for the inconsistent findings. Secondly, probiotics are supposed to act on the gut microbiome; however, the large variation in the indigenous gut microbial landscape between individuals is another major confounder. Consistency of the baseline microbiota should be taken into account given that environmental factors [such as diet (238–240), age (241), geography (241), and birth mode (242, 243)] that determine the gut microbiome differ between individuals. Finally, heterogeneity in the host per se exists. Collectively, these limitations should be taken into account in future study design (Box 1).
Box 1. Strategies and considerations in design of microbiome and probiotics studies in the future.
• Determining potential causality between host-microbial interactions by combining GF animal model, culturomics, and multi-omic technologies, e.g., metagenomics, metabolomics, metatranscriptomics, and metaproteomics.
• Assessing the consistency of the baseline microbiota prior to the start of preclinical and clinical trials.
• Constructing standardized protocols for trial conduction, e.g., probiotic strains, doses, duration and frequency of administration, to ensure comparability between studies and results generated by different laboratories.
• Constructing a standardized probiotic effect evaluation protocol to improve comparability of clinical outcomes between studies.
There is growing appreciation of the fact that the gut microbiome imprints pancreatic diseases including pancreatitis, pancreatic cancer, and T1DM based on a multitude of preclinical and clinical studies. Evidence of the intestinal microbiome in driving pancreatic diseases further supports the significance of the extensive gut-pancreas crosstalk, leading to intense interest in microbiome characterization and engineering via application of probiotics, prebiotics, synbiotics, postbiotics, or FMT. Researches over the last decades have advanced our understanding of the relationship between the gut microbiome and pancreatic diseases. Such advent would also bring about novel strategies in improving pancreatic diseases via modulating the gut microbiome. Despite this breakthrough, potential causality between the gut microbiome and pancreatic diseases remains obscure. Efforts to untie the causality and microbiota-mediated mechanisms of pancreatic diseases have critical clinical implications, enabling us to better constitute a new and targeted approach to modulate the gut microbiome. Moreover, a body of preclinical research and high-quality clinical trials are needed to further elucidate mechanisms by which the gut microbiome influences pancreatic diseases, as well as mechanisms of probiotics, prebiotics, synbiotics, postbiotics, and FMT in improving disease outcomes. Ultimately, future development in the field will aim to harness microbiome engineering in clinical practice on a personalized level.
GG and HS: conceptualization. TZ: writing—original draft preparation. L-YK and ZS: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
This project was supported by the Science and Technology Major Projects of Inner Mongolia Autonomous Region (2021ZD0014) and China Agriculture Research System of MOF and MARA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Peery AF, Crockett SD, Murphy CC, Lund JL, Dellon ES, Williams JL, et al. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2018. Gastroenterology. (2019) 156:254–72 e211. doi: 10.1053/j.gastro.2018.08.063
2. Xiao AY, Tan ML, Wu LM, Asrani VM, Windsor JA, Yadav D, et al. Global incidence and mortality of pancreatic diseases: a systematic review, meta-analysis, and meta-regression of population-based cohort studies. Lancet Gastroenterol Hepatol. (2016) 1:45–55. doi: 10.1016/S2468-1253(16)30004-8
3. Forsmark CE, Vege SS, Wilcox CM. Acute pancreatitis. N Engl J Med. (2016) 375:1972–81. doi: 10.1056/NEJMra1505202
4. Rijkers AP, Bakker OJ, Ahmed Ali U, Hagenaars J, van Santvoort HC, Besselink MG, et al. Risk of pancreatic cancer after a primary episode of acute pancreatitis. Pancreas. (2017) 46:1018–22. doi: 10.1097/MPA.0000000000000879
5. Kirkegård J, Mortensen FV, Cronin-Fenton D. Chronic pancreatitis and pancreatic cancer risk: a systematic review and meta-analysis. Am J Gastroenterol. (2017) 112:1366–72. doi: 10.1038/ajg.2017.218
6. American Cancer Society. Cancer Facts & Figure 2020. American Cancer Society (2020). Available online at: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2020.html
7. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. (2014) 74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155
8. Adolph TE, Mayr L, Grabherr F, Schwärzler J, Tilg H. Pancreas-microbiota cross talk in health and disease. Annu Rev Nutr. (2019) 39:249–66. doi: 10.1146/annurev-nutr-082018-124306
9. Patterson CC, Dahlquist GG, Gyürüs E, Green A, Soltész G. Incidence trends for childhood type 1 diabetes in Europe during 1989-2003 and predicted new cases 2005-20: a multicentre prospective registration study. Lancet. (2009) 373:2027–33. doi: 10.1016/S0140-6736(09)60568-7
10. Mayer-Davis EJ, Lawrence JM, Dabelea D, Divers J, Isom S, Dolan L, et al. Incidence trends of type 1 and type 2 diabetes among youths, 2002-2012. N Engl J Med. (2017) 376:1419–29. doi: 10.1056/NEJMoa1610187
11. Norris JM, Johnson RK, Stene LC. Type 1 diabetes-early life origins and changing epidemiology. Lancet Diabetes Endocrinol. (2020) 8:226–38. doi: 10.1016/S2213-8587(19)30412-7
12. Mederos MA, Reber HA, Girgis MD. Acute pancreatitis: a review. JAMA. (2021) 325:382–90. doi: 10.1001/jama.2020.20317
13. Singh VK, Yadav D, Garg PK. Diagnosis and management of chronic pancreatitis: a review. JAMA. (2019) 322:2422–34. doi: 10.1001/jama.2019.19411
14. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. (2016) 388:73–85. doi: 10.1016/S0140-6736(16)00141-0
15. Bluestone JA, Herold K, Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. (2010) 464:1293–300. doi: 10.1038/nature08933
16. Katsarou A, Gudbjörnsdottir S, Rawshani A, Dabelea D, Bonifacio E, Anderson BJ, et al. Type 1 diabetes mellitus. Nat Rev Dis Primers. (2017) 3:17016. doi: 10.1038/nrdp.2017.16
17. Vege SS, DiMagno MJ, Forsmark CE, Martel M, Barkun AN. Initial medical treatment of acute pancreatitis: American Gastroenterological Association Institute Technical review. Gastroenterology. (2018) 154:1103–139. doi: 10.1053/j.gastro.2018.01.031
18. World Health Organization. Global Action Plan for the Prevention and Control of Noncommunicable Diseases 2013-2020. World Health Organization (2013). Available online at: https://apps.who.int/iris/handle/10665/94384
19. World Health Organization. Diabetes. World Health Organization (2021). Available online at: https://www.who.int/news-room/fact-sheets/detail/diabetes
20. The Lancet. Type 1 diabetes-progress and prospects. Lancet. (2014) 383:2. doi: 10.1016/S0140-6736(13)62715-4
21. Dayan CM, Korah M, Tatovic D, Bundy BN, Herold KC. Changing the landscape for type 1 diabetes: the first step to prevention. Lancet. (2019) 394:1286–96. doi: 10.1016/S0140-6736(19)32127-0
22. Herold KC, Bundy BN, Long SA, Bluestone JA, DiMeglio LA, Dufort MJ, et al. An anti-CD3 antibody, teplizumab, in relatives at risk for type 1 diabetes. N Engl J Med. (2019) 381:603–13. doi: 10.1056/NEJMoa1902226
23. Libertucci J, Young VB. The role of the microbiota in infectious diseases. Nat Microbiol. (2018) 4:35–45. doi: 10.1038/s41564-018-0278-4
24. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464:59–65. doi: 10.1038/nature08821
25. Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. (2014) 32:834–41. doi: 10.1038/nbt.2942
26. Donia MS, Fischbach MA. Human microbiota. Small molecules from the human microbiota. Science. (2015) 349:1254766. doi: 10.1126/science.1254766
27. Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, et al. Precision editing of the gut microbiota ameliorates colitis. Nature. (2018) 553:208–11. doi: 10.1038/nature25172
28. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. (2019) 25:968–76. doi: 10.1038/s41591-019-0458-7
29. Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol. (2019) 16:235–46. doi: 10.1038/s41575-018-0099-1
30. Trebicka J, Bork P, Krag A, Arumugam M. Utilizing the gut microbiome in decompensated cirrhosis and acute-on-chronic liver failure. Nat Rev Gastroenterol Hepatol. (2021) 18:167–80. doi: 10.1038/s41575-020-00376-3
31. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. (2021) 19:55–71. doi: 10.1038/s41579-020-0433-9
32. Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol. (2020) 20:40–54. doi: 10.1038/s41577-019-0198-4
33. Aron-Wisnewsky J, Vigliotti C, Witjes J, Le P, Holleboom AG, Verheij J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. (2020) 17:279–97. doi: 10.1038/s41575-020-0269-9
34. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027–31. doi: 10.1038/nature05414
35. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. (2012) 490:55–60. doi: 10.1038/nature11450
36. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. (2013) 498:99–103. doi: 10.1038/nature12198
37. Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. (2017) 25:1054–62.e1055. doi: 10.1016/j.cmet.2017.04.001
38. Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. (2013) 62:1787–94. doi: 10.1136/gutjnl-2012-303816
39. Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. (2014) 510:417–21. doi: 10.1038/nature13421
40. Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. (2012) 487:477–81. doi: 10.1038/nature11228
41. Depner M, Taft DH, Kirjavainen PV, Kalanetra KM, Karvonen AM, Peschel S, et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat Med. (2020) 26:1766–75. doi: 10.1038/s41591-020-1095-x
42. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. (2021) 70:698–706. doi: 10.1136/gutjnl-2020-323020
43. Lai HC, Lin TL, Chen TW, Kuo YL, Chang CJ, Wu TR, et al. Gut microbiota modulates COPD pathogenesis: role of anti-inflammatory Parabacteroides goldsteinii lipopolysaccharide. Gut. (2021) 71:309–21. doi: 10.1136/gutjnl-2020-322599
44. Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM, et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell. (2020) 180:862–77.e822. doi: 10.1016/j.cell.2020.02.016
45. Chen PB, Black AS, Sobel AL, Zhao Y, Mukherjee P, Molparia B, et al. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol. (2020) 38:1288–97. doi: 10.1038/s41587-020-0549-5
46. Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, et al. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation. (2018) 138:2486–98. doi: 10.1161/CIRCULATIONAHA.118.033714
47. Bartolomaeus H, Balogh A, Yakoub M, Homann S, Markó L, Höges S, et al. Short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation. (2019) 139:1407–21. doi: 10.1161/CIRCULATIONAHA.118.036652
48. Cryan JF, O'Riordan KJ, Sandhu K, Peterson V, Dinan TG. The gut microbiome in neurological disorders. Lancet Neurol. (2020) 19:179–94. doi: 10.1016/S1474-4422(19)30356-4
49. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. (2016) 167:1469–80.e1412. doi: 10.1016/j.cell.2016.11.018
50. Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. (2019) 572:474–80. doi: 10.1038/s41586-019-1443-5
51. Wang X, Sun G, Feng T, Zhang J, Huang X, Wang T, et al. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer's disease progression. Cell Res. (2019) 29:787–803. doi: 10.1038/s41422-019-0216-x
52. Kim MS, Kim Y, Choi H, Kim W, Park S, Lee D, et al. Transfer of a healthy microbiota reduces amyloid and tau pathology in an Alzheimer's disease animal model. Gut. (2020) 69:283–94. doi: 10.1136/gutjnl-2018-317431
53. Li B, He Y, Ma J, Huang P, Du J, Cao L, et al. Mild cognitive impairment has similar alterations as Alzheimer's disease in gut microbiota. Alzheimers Dement. (2019) 15:1357–66. doi: 10.1016/j.jalz.2019.07.002
54. Sharon G, Cruz NJ, Kang DW, Gandal MJ, Wang B, Kim YM, et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell. (2019) 177:1600–18.e1617. doi: 10.1016/j.cell.2019.05.004
55. Cignarella F, Cantoni C, Ghezzi L, Salter A, Dorsett Y, Chen L, et al. Intermittent fasting confers protection in CNS autoimmunity by altering the gut microbiota. Cell Metab. (2018) 27:1222–35.e1226. doi: 10.1016/j.cmet.2018.05.006
56. Stanley D, Mason LJ, Mackin KE, Srikhanta YN, Lyras D, Prakash MD, et al. Translocation and dissemination of commensal bacteria in post-stroke infection. Nat Med. (2016) 22:1277–84. doi: 10.1038/nm.4194
57. Zheng P, Zeng B, Liu M, Chen J, Pan J, Han Y, et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci Adv. (2019) 5:eaau8317. doi: 10.1126/sciadv.aau8317
58. Zhu F, Guo R, Wang W, Ju Y, Wang Q, Ma Q, et al. Transplantation of microbiota from drug-free patients with schizophrenia causes schizophrenia-like abnormal behaviors and dysregulated kynurenine metabolism in mice. Mol Psychiatry. (2020) 25:2905–18. doi: 10.1038/s41380-019-0475-4
59. Zheng P, Yang J, Li Y, Wu J, Liang W, Yin B, et al. Gut microbial signatures can discriminate unipolar from bipolar depression. Adv Sci. (2020) 7:1902862. doi: 10.1002/advs.201902862
60. Simpson CA, Diaz-Arteche C, Eliby D, Schwartz OS, Simmons JG, Cowan CSM. The gut microbiota in anxiety and depression - a systematic review. Clin Psychol Rev. (2021) 83:101943. doi: 10.1016/j.cpr.2020.101943
61. Choi SC, Brown J, Gong M, Ge Y, Zadeh M, Li W, et al. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci Transl Med. (2020) 12:eaax2220. doi: 10.1126/scitranslmed.aax2220
62. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab. (2020) 31:837–51.e810. doi: 10.1016/j.cmet.2020.03.003
63. Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. (2018) 67:108–19. doi: 10.1136/gutjnl-2016-312135
64. Kadosh E, Snir-Alkalay I, Venkatachalam A, May S, Lasry A, Elyada E, et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature. (2020) 586:133–8. doi: 10.1038/s41586-020-2541-0
65. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. (2018) 360:eaan5931. doi: 10.1126/science.aan5931
66. Loo TM, Kamachi F, Watanabe Y, Yoshimoto S, Kanda H, Arai Y, et al. Gut microbiota promotes obesity-associated liver cancer through PGE(2)-mediated suppression of antitumor immunity. Cancer Discov. (2017) 7:522–38. doi: 10.1158/2159-8290.CD-16-0932
67. Sethi V, Kurtom S, Tarique M, Lavania S, Malchiodi Z, Hellmund L, et al. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology. (2018) 155:33–7.e36. doi: 10.1053/j.gastro.2018.04.001
68. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. (2018) 359:91–7. doi: 10.1126/science.aan3706
69. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359:97–103. doi: 10.1126/science.aan4236
70. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. (2012) 489:220–30. doi: 10.1038/nature11550
71. Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. (2015) 350:663–6. doi: 10.1126/science.aad2602
72. Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. (2017) 15:630–8. doi: 10.1038/nrmicro.2017.58
73. Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. (1984) 1:1311–5. doi: 10.1016/S0140-6736(84)91816-6
74. Fukase K, Kato M, Kikuchi S, Inoue K, Uemura N, Okamoto S, et al. Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open-label, randomised controlled trial. Lancet. (2008) 372:392–7. doi: 10.1016/S0140-6736(08)61159-9
75. Choi IJ, Kim CG, Lee JY, Kim YI, Kook MC, Park B, et al. Family history of gastric cancer and Helicobacter pylori treatment. N Engl J Med. (2020) 382:427–36. doi: 10.1056/NEJMoa1909666
76. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. (2013) 368:407–15. doi: 10.1056/NEJMoa1205037
77. Drekonja D, Reich J, Gezahegn S, Greer N, Shaukat A, MacDonald R, et al. Fecal microbiota transplantation for Clostridium difficile infection: a systematic review. Ann Intern Med. (2015) 162:630–8. doi: 10.7326/M14-2693
78. Bagdasarian N, Rao K, Malani PN. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA. (2015) 313:398–408. doi: 10.1001/jama.2014.17103
79. Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol. (2020) 17:53–64. doi: 10.1038/s41575-019-0242-7
80. Sun J, Furio L, Mecheri R, van der Does AM, Lundeberg E, Saveanu L, et al. Pancreatic beta-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity. (2015) 43:304–17. doi: 10.1016/j.immuni.2015.07.013
81. Ahuja M, Schwartz DM, Tandon M, Son A, Zeng M, Swaim W, et al. Orai1-mediated antimicrobial secretion from pancreatic acini shapes the gut microbiome and regulates gut innate immunity. Cell Metab. (2017) 25:635–46. doi: 10.1016/j.cmet.2017.02.007
82. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
83. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. (2017) 357:1156–60. doi: 10.1126/science.aah5043
84. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. (2018) 8:403–16. doi: 10.1158/2159-8290.CD-17-1134
85. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. (2019) 574:264–7. doi: 10.1038/s41586-019-1608-2
86. Petrov MS, Yadav D. Global epidemiology and holistic prevention of pancreatitis. Nat Rev Gastroenterol Hepatol. (2019) 16:175–84. doi: 10.1038/s41575-018-0087-5
87. Lee PJ, Papachristou GI. New insights into acute pancreatitis. Nat Rev Gastroenterol Hepatol. (2019) 16:479–96. doi: 10.1038/s41575-019-0158-2
88. Wu LM, Sankaran SJ, Plank LD, Windsor JA, Petrov MS. Meta-analysis of gut barrier dysfunction in patients with acute pancreatitis. Br J Surg. (2014) 101:1644–56. doi: 10.1002/bjs.9665
89. Liu J, Huang L, Luo M, Xia X. Bacterial translocation in acute pancreatitis. Crit Rev Microbiol. (2019) 45:539–47. doi: 10.1080/1040841X.2019.1621795
90. Sharkey KA, Beck PL, McKay DM. Neuroimmunophysiology of the gut: advances and emerging concepts focusing on the epithelium. Nat Rev Gastroenterol Hepatol. (2018) 15:765–84. doi: 10.1038/s41575-018-0051-4
91. Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. (2020) 38:23–48. doi: 10.1146/annurev-immunol-070119-115104
92. Li XY, He C, Zhu Y, Lu NH. Role of gut microbiota on intestinal barrier function in acute pancreatitis. World J Gastroenterol. (2020) 26:2187–93. doi: 10.3748/wjg.v26.i18.2187
93. Huang C, Chen J, Wang J, Zhou H, Lu Y, Lou L, et al. Dysbiosis of intestinal microbiota and decreased antimicrobial peptide level in Paneth cells during hypertriglyceridemia-related acute necrotizing pancreatitis in rats. Front Microbiol. (2017) 8:776. doi: 10.3389/fmicb.2017.00776
94. Zhu Y, He C, Li X, Cai Y, Hu J, Liao Y, et al. Gut microbiota dysbiosis worsens the severity of acute pancreatitis in patients and mice. J Gastroenterol. (2019) 54:347–58. doi: 10.1007/s00535-018-1529-0
95. Yu S, Xiong Y, Xu J, Liang X, Fu Y, Liu D, et al. Identification of dysfunctional gut microbiota through rectal swab in patients with different severity of acute pancreatitis. Dig Dis Sci. (2020) 65:3223–37. doi: 10.1007/s10620-020-06061-4
96. Li X, He C, Li N, Ding L, Chen H, Wan J, et al. The interplay between the gut microbiota and NLRP3 activation affects the severity of acute pancreatitis in mice. Gut Microbes. (2020) 11:1774–89. doi: 10.1080/19490976.2020.1770042
97. Zheng J, Lou L, Fan J, Huang C, Mei Q, Wu J, et al. Commensal Escherichia coli aggravates acute necrotizing pancreatitis through targeting of intestinal epithelial cells. Appl Environ Microbiol. (2019) 85:e00059–19. doi: 10.1128/AEM.00059-19
98. Braganza JM, Lee SH, McCloy RF, McMahon MJ. Chronic pancreatitis. Lancet. (2011) 377:1184–97. doi: 10.1016/S0140-6736(10)61852-1
99. Wang W, Xiao Y, Wang X, Zhou Y, Wang T, Xv C, et al. Disordered gut microbiota in children who have chronic pancreatitis and different functional gene mutations. Clin Transl Gastroenterol. (2020) 11:e00150. doi: 10.14309/ctg.0000000000000150
100. Frost F, Weiss FU, Sendler M, Kacprowski T, Rühlemann M, Bang C, et al. The gut microbiome in patients with chronic pancreatitis is characterized by significant dysbiosis and overgrowth by opportunistic pathogens. Clin Transl Gastroenterol. (2020) 11:e00232. doi: 10.14309/ctg.0000000000000232
101. Wu C, Li M, Chen W. Characteristics of gut microbiota in cerulein-induced chronic pancreatitis. Diabetes Metab Syndr Obes. (2021) 14:285–94. doi: 10.2147/DMSO.S291822
102. Zhou CH, Meng YT, Xu JJ, Fang X, Zhao JL, Zhou W, et al. Altered diversity and composition of gut microbiota in Chinese patients with chronic pancreatitis. Pancreatology. (2020) 20:16–24. doi: 10.1016/j.pan.2019.11.013
103. Han MM, Zhu XY, Peng YF, Lin H, Liu DC, Li L. The alterations of gut microbiota in mice with chronic pancreatitis. Ann Transl Med. (2019) 7:464. doi: 10.21037/atm.2019.08.18
104. Tao J, Cheema H, Kesh K, Dudeja V, Dawra R, Roy S. Chronic pancreatitis in a caerulein-induced mouse model is associated with an altered gut microbiome. Pancreatology. (2022) 22:30–42. doi: 10.1016/j.pan.2021.12.003
105. Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. (2016) 12:154–67. doi: 10.1038/nrendo.2015.218
106. Robertson CC, Rich SS. Genetics of type 1 diabetes. Curr Opin Genet Dev. (2018) 50:7–16. doi: 10.1016/j.gde.2018.01.006
107. Pociot F, Lernmark Å. Genetic risk factors for type 1 diabetes. Lancet. (2016) 387:2331–9. doi: 10.1016/S0140-6736(16)30582-7
108. Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. (2016) 387:2340–8. doi: 10.1016/S0140-6736(16)30507-4
109. Gillespie KM. Type 1 diabetes: pathogenesis and prevention. CMAJ. (2006) 175:165–70. doi: 10.1503/cmaj.060244
110. Ang QY, Alexander M, Newman JC, Tian Y, Cai J, Upadhyay V, et al. Ketogenic diets alter the gut microbiome resulting in decreased intestinal Th17 cells. Cell. (2020) 181:1263–75.e1216. doi: 10.1016/j.cell.2020.04.027
111. Reynolds AN, Akerman AP, Mann J. Dietary fibre and whole grains in diabetes management: systematic review and meta-analyses. PLoS Med. (2020) 17:e1003053. doi: 10.1371/journal.pmed.1003053
112. Kolodziejczyk AA, Zheng D, Elinav E. Diet-microbiota interactions and personalized nutrition. Nat Rev Microbiol. (2019) 17:742–53. doi: 10.1038/s41579-019-0256-8
113. Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. (2018) 362:776–80. doi: 10.1126/science.aau5812
114. Kubinak JL, Stephens WZ, Soto R, Petersen C, Chiaro T, Gogokhia L, et al. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun. (2015) 6:8642. doi: 10.1038/ncomms9642
115. Silverman M, Kua L, Tanca A, Pala M, Palomba A, Tanes C, et al. Protective major histocompatibility complex allele prevents type 1 diabetes by shaping the intestinal microbiota early in ontogeny. Proc Natl Acad Sci USA. (2017) 114:9671–6. doi: 10.1073/pnas.1712280114
116. Russell JT, Roesch LFW, Ördberg M, Ilonen J, Atkinson MA, Schatz DA, Triplett EW, et al. Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat Commun. (2019) 10:3621. doi: 10.1038/s41467-019-11460-x
117. Alkanani AK, Hara N, Gottlieb PA, Ir D, Robertson CE, Wagner BD, et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. (2015) 64:3510–20. doi: 10.2337/db14-1847
118. Miani M, Le Naour J, Waeckel-Enée E, Verma SC, Straube M, Emond P, et al. Gut microbiota-stimulated innate lymphoid cells support β-defensin 14 expression in pancreatic endocrine cells, preventing autoimmune diabetes. Cell Metab. (2018) 28:557–72.e556. doi: 10.1016/j.cmet.2018.06.012
119. Peng J, Narasimhan S, Marchesi JR, Benson A, Wong FS, Wen L. Long term effect of gut microbiota transfer on diabetes development. J Autoimmun. (2014) 53:85–94. doi: 10.1016/j.jaut.2014.03.005
120. Costa FR, Françozo MC, de Oliveira GG, Ignacio A, Castoldi A, Zamboni DS, et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J Exp Med. (2016) 213:1223–39. doi: 10.1084/jem.20150744
121. Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, McRitchie S, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. (2016) 1:16140. doi: 10.1038/nmicrobiol.2016.140
122. Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. (2017) 18:552–62. doi: 10.1038/ni.3713
123. de Goffau MC, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. (2013) 62:1238–44. doi: 10.2337/db12-0526
124. Leiva-Gea I, Sanchez-Alcoholado L, Martin-Tejedor B, Castellano-Castillo D, Moreno-Indias I, Urda-Cardona A, et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case-control study. Diabetes Care. (2018) 41:2385–95. doi: 10.2337/dc18-0253
125. de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. (2014) 57:1569–77. doi: 10.1007/s00125-014-3274-0
126. Roth-Schulze AJ, Penno MAS, Ngui KM, Oakey H, Bandala-Sanchez E, Smith AD, et al. Type 1 diabetes in pregnancy is associated with distinct changes in the composition and function of the gut microbiome. Microbiome. (2021) 9:167. doi: 10.21203/rs.3.rs-376289/v1
127. Salamon D, Sroka-Oleksiak A, Gurgul A, Arent Z, Szopa M, Bulanda M, et al. Analysis of the gut mycobiome in adult patients with type 1 and type 2 diabetes using next-generation sequencing (NGS) with increased sensitivity-pilot study. Nutrients. (2021) 13:1066. doi: 10.3390/nu13041066
128. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. (2008) 455:1109–13. doi: 10.1038/nature07336
129. Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen AM, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. (2015) 17:260–73. doi: 10.1016/j.chom.2015.01.001
130. Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. (2018) 562:589–94. doi: 10.1038/s41586-018-0620-2
131. Zheng H, Xu P, Jiang Q, Xu Q, Zheng Y, Yan J, et al. Depletion of acetate-producing bacteria from the gut microbiota facilitates cognitive impairment through the gut-brain neural mechanism in diabetic mice. Microbiome. (2021) 9:145. doi: 10.1186/s40168-021-01088-9
132. Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. (2013) 39:400–12. doi: 10.1016/j.immuni.2013.08.013
133. Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. (2013) 339:1084–8. doi: 10.1126/science.1233521
134. Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. (2016) 165:842–53. doi: 10.1016/j.cell.2016.04.007
135. Gavin PG, Mullaney JA, Loo D, Cao KL, Gottlieb PA, Hill MM, et al. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care. (2018) 41:2178–86. doi: 10.2337/dc18-0777
136. Columbia Surgery. Pancreatic Cancer. New York, NY: Columbia University Irving Medical Center; Department of Surgery (2020). Available online at: https://columbiasurgery.org/conditions-and-treatments/pancreatic-cancer
137. Mendez R, Kesh K, Arora N, Di Martino L, McAllister F, Merchant N, et al. Microbial dysbiosis and polyamine metabolism as predictive markers for early detection of pancreatic cancer. Carcinogenesis. (2020) 41:561–70. doi: 10.1093/carcin/bgz116
138. Ren Z, Jiang J, Xie H, Li A, Lu H, Xu S, et al. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget. (2017) 8:95176–91. doi: 10.18632/oncotarget.18820
139. Zhou W, Zhang D, Li Z, Jiang H, Li J, Ren R, et al. The fecal microbiota of patients with pancreatic ductal adenocarcinoma and autoimmune pancreatitis characterized by metagenomic sequencing. J Transl Med. (2021) 19:215. doi: 10.1186/s12967-021-02882-7
140. Kazmierczak-Siedlecka K, Stachowska E, Folwarski M, Przewłócka K, Makarewicz W, Bryl E. The potential of gut microbiome as a non-invasive predictive biomarker for early detection of pancreatic cancer and hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. (2021) 25:7275–84.
141. Sammallahti H, Kokkola A, Rezasoltani S, Ghanbari R, Asadzadeh Aghdaei H, Knuutila S, et al. Microbiota alterations and their association with oncogenomic changes in pancreatic cancer patients. Int J Mol Sci. (2021) 22:12978. doi: 10.3390/ijms222312978
142. Abdul Rahman R, Lamarca A, Hubner RA, Valle JW, McNamara MG. The microbiome as a potential target for therapeutic manipulation in pancreatic cancer. Cancers. (2021) 13:3779. doi: 10.3390/cancers13153779
143. Akshintala VS, Talukdar R, Singh VK, Goggins M. The gut microbiome in pancreatic disease. Clin Gastroenterol Hepatol. (2019) 17:290–5. doi: 10.1016/j.cgh.2018.08.045
144. Thomas RM, Gharaibeh RZ, Gauthier J, Beveridge M, Pope JL, Guijarro MV, et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis. (2018) 39:1068–78. doi: 10.1093/carcin/bgy073
145. Li S, Fuhler GM, Bn N, Jose T, Bruno MJ, Peppelenbosch MP, et al. Pancreatic cyst fluid harbors a unique microbiome. Microbiome. (2017) 5:147. doi: 10.1186/s40168-017-0363-6
146. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806.e712. doi: 10.1016/j.cell.2019.07.008
147. Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. (2012) 61:582–8. doi: 10.1136/gutjnl-2011-300784
148. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. (2018) 67:120–7. doi: 10.1136/gutjnl-2016-312580
149. Gaiser RA, Halimi A, Alkharaan H, Lu L, Davanian H, Healy K, et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut. (2019) 68:2186–94. doi: 10.1136/gutjnl-2018-317458
150. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
151. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. (2017) 14:491–502. doi: 10.1038/nrgastro.2017.75
152. Swanson KS, Gibson GR, Hutkins R, Reimer RA, Reid G, Verbeke K, et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol. (2020) 17:687–701. doi: 10.1038/s41575-020-0344-2
153. Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. (2021) 18:649–67. doi: 10.1038/s41575-021-00440-6
154. Cammarota G, Ianiro G, Tilg H, Rajilić-Stojanović M, Kump P, Satokari R, et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. (2017) 66:569–80. doi: 10.1136/gutjnl-2016-313017
155. Allen SJ, Martinez EG, Gregorio GV, Dans LF. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst Rev. (2010) 2010:CD003048. doi: 10.1002/14651858.CD003048.pub3
156. Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R, et al. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA. (2012) 307:1959–69. doi: 10.1001/jama.2012.3507
157. Johnston BC, Ma SS, Goldenberg JZ, Thorlund K, Vandvik PO, Loeb M, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. Ann Intern Med. (2012) 157:878–88. doi: 10.7326/0003-4819-157-12-201212180-00563
158. Shen NT, Maw A, Tmanova LL, Pino A, Ancy K, Crawford CV, et al. Timely use of probiotics in hospitalized adults prevents Clostridium difficile infection: a systematic review with meta-regression analysis. Gastroenterology. (2017) 152:1889–900.e1889. doi: 10.1053/j.gastro.2017.02.003
159. Moayyedi P, Ford AC, Talley NJ, Cremonini F, Foxx-Orenstein AE, Brandt LJ, et al. The efficacy of probiotics in the treatment of irritable bowel syndrome: a systematic review. Gut. (2010) 59:325–32. doi: 10.1136/gut.2008.167270
160. Andresen V, Gschossmann J, Layer P. Heat-inactivated Bifidobacterium bifidum MIMBb75 (SYN-HI-001) in the treatment of irritable bowel syndrome: a multicentre, randomised, double-blind, placebo-controlled clinical trial. Lancet Gastroenterol Hepatol. (2020) 5:658–66. doi: 10.1016/S2468-1253(20)30056-X
161. Singer JR, Blosser EG, Zindl CL, Silberger DJ, Conlan S, Laufer VA, et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat Med. (2019) 25:1772–82. doi: 10.1038/s41591-019-0640-y
162. Hao Q, Dong BR, Wu T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst Rev. (2015) CD006895. doi: 10.1002/14651858.CD006895.pub3
163. Mangiante G, Colucci G, Canepari P, Bassi C, Nicoli N, Casaril A, et al. Lactobacillus plantarum reduces infection of pancreatic necrosis in experimental acute pancreatitis. Dig Surg. (2001) 18:47–50. doi: 10.1159/000050096
164. Muftuoglu MA, Isikgor S, Tosun S, Saglam A. Effects of probiotics on the severity of experimental acute pancreatitis. Eur J Clin Nutr. (2006) 60:464–8. doi: 10.1038/sj.ejcn.1602338
165. Lutgendorff F, Trulsson LM, van Minnen LP, Rijkers GT, Timmerman HM, Franzen LE, et al. Probiotics enhance pancreatic glutathione biosynthesis and reduce oxidative stress in experimental acute pancreatitis. Am J Physiol Gastrointest Liver Physiol. (2008) 295:G1111–21. doi: 10.1152/ajpgi.00603.2007
166. van Minnen LP, Timmerman HM, Lutgendorff F, Verheem A, Harmsen W, Konstantinov SR, et al. Modification of intestinal flora with multispecies probiotics reduces bacterial translocation and improves clinical course in a rat model of acute pancreatitis. Surgery. (2007) 141:470–80. doi: 10.1016/j.surg.2006.10.007
167. Sahin T, Aydin S, Yüksel O, Bostanci H, Akyürek N, Memiş L, et al. Effects of the probiotic agent Saccharomyces boulardii on the DNA damage in acute necrotizing pancreatitis induced rats. Hum Exp Toxicol. (2007) 26:653–61. doi: 10.1177/0960327107077596
168. Karen M, Yuksel O, Akyürek N, Ofluoglu E, Caglar K, Sahin TT, et al. Probiotic agent Saccharomyces boulardii reduces the incidence of lung injury in acute necrotizing pancreatitis induced rats. J Surg Res. (2010) 160:139–44. doi: 10.1016/j.jss.2009.02.008
169. Hooijmans CR, de Vries RB, Rovers MM, Gooszen HG, Ritskes-Hoitinga M. The effects of probiotic supplementation on experimental acute pancreatitis: a systematic review and meta-analysis. PLoS ONE. (2012) 7:e48811. doi: 10.1371/journal.pone.0048811
170. van Baal MC, van Rens MJ, Geven CB, van de Pol FM, van den Brink IW, Hannink G, et al. Association between probiotics and enteral nutrition in an experimental acute pancreatitis model in rats. Pancreatology. (2014) 14:470–7. doi: 10.1016/j.pan.2014.10.002
171. Zhao HB, Jia L, Yan QQ, Deng Q, Wei B. Effect of Clostridium butyricum and butyrate on intestinal barrier functions: study of a rat model of severe acute pancreatitis with intra-abdominal hypertension. Front Physiol. (2020) 11:561061. doi: 10.3389/fphys.2020.561061
172. Oláh A, Belágyi T, Issekutz A, Gamal ME, Bengmark S. Randomized clinical trial of specific lactobacillus and fibre supplement to early enteral nutrition in patients with acute pancreatitis. Br J Surg. (2002) 89:1103–7. doi: 10.1046/j.1365-2168.2002.02189.x
173. Qin HL, Zheng JJ, Tong DN, Chen WX, Fan XB, Hang XM, et al. Effect of Lactobacillus plantarum enteral feeding on the gut permeability and septic complications in the patients with acute pancreatitis. Eur J Clin Nutr. (2008) 62:923–30. doi: 10.1038/sj.ejcn.1602792
174. Besselink MG, van Santvoort HC, Buskens E, Boermeester MA, van Goor H, Timmerman HM, et al. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet. (2008) 371:651–9. doi: 10.1016/S0140-6736(08)60207-X
175. Sharma B, Srivastava S, Singh N, Sachdev V, Kapur S, Saraya A. Role of probiotics on gut permeability and endotoxemia in patients with acute pancreatitis: a double-blind randomized controlled trial. J Clin Gastroenterol. (2011) 45:442–8. doi: 10.1097/MCG.0b013e318201f9e2
176. Sun S, Yang K, He X, Tian J, Ma B, Jiang L. Probiotics in patients with severe acute pancreatitis: a meta-analysis. Langenbecks Arch Surg. (2009) 394:171–7. doi: 10.1007/s00423-008-0379-2
177. Gou S, Yang Z, Liu T, Wu H, Wang C. Use of probiotics in the treatment of severe acute pancreatitis: a systematic review and meta-analysis of randomized controlled trials. Crit Care. (2014) 18:R57. doi: 10.1186/cc13809
178. Moggia E, Koti R, Belgaumkar AP, Fazio F, Pereira SP, Davidson BR, et al. Pharmacological interventions for acute pancreatitis. Cochrane Database Syst Rev. (2017) 4:CD011384. doi: 10.1002/14651858.CD011384.pub2
179. Cui LH, Wang XH, Peng LH, Yu L, Yang YS. The effects of early enteral nutrition with addition of probiotics on the prognosis of patients suffering from severe acute pancreatitis. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. (2013) 25:224–8. doi: 10.3760/cma.j.issn.2095-4352.2013.04.011
180. Wang G, Wen J, Xu L, Zhou S, Gong M, Wen P, et al. Effect of enteral nutrition and ecoimmunonutrition on bacterial translocation and cytokine production in patients with severe acute pancreatitis. J Surg Res. (2013) 183:592–7. doi: 10.1016/j.jss.2012.12.010
181. Wan YD, Zhu RX, Bian ZZ, Sun TW. Effect of probiotics on length of hospitalization in mild acute pancreatitis: a randomized, double-blind, placebo-controlled trial. World J Gastroenterol. (2021) 27:224–32. doi: 10.3748/wjg.v27.i2.224
182. Fritz S, Hackert T, Hartwig W, Rossmanith F, Strobel O, Schneider L, et al. Bacterial translocation and infected pancreatic necrosis in acute necrotizing pancreatitis derives from small bowel rather than from colon. Am J Surg. (2010) 200:111–7. doi: 10.1016/j.amjsurg.2009.08.019
183. Rychter JW, van Minnen LP, Verheem A, Timmerman HM, Rijkers GT, Schipper ME, et al. Pretreatment but not treatment with probiotics abolishes mouse intestinal barrier dysfunction in acute pancreatitis. Surgery. (2009) 145:157–67. doi: 10.1016/j.surg.2008.09.011
184. Besselink MG, van Santvoort HC, Renooij W, de Smet MB, Boermeester MA, Fischer K, et al. Intestinal barrier dysfunction in a randomized trial of a specific probiotic composition in acute pancreatitis. Ann Surg. (2009) 250:712–9. doi: 10.1097/SLA.0b013e3181bce5bd
185. Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. (2020) 72:558–77. doi: 10.1016/j.jhep.2019.10.003
186. Gao J, Li Y, Wan Y, Hu T, Liu L, Yang S, et al. A novel postbiotic from Lactobacillus rhamnosus GG with a beneficial effect on intestinal barrier function. Front Microbiol. (2019) 10:477. doi: 10.3389/fmicb.2019.00477
187. Morniroli D, Vizzari G, Consales A, Mosca F, Giannì ML. Postbiotic supplementation for children and newborn's health. Nutrients. (2021) 13:781. doi: 10.3390/nu13030781
188. Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. (2018) 359:1156–61. doi: 10.1126/science.aar7201
189. Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Lo Cascio A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell. (2021) 39:708–24.e711. doi: 10.1016/j.ccell.2021.03.004
190. Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. (2019) 16:605–16. doi: 10.1038/s41575-019-0173-3
191. Pearson JA, Wong FS, Wen L. The importance of the Non Obese Diabetic (NOD) mouse model in autoimmune diabetes. J Autoimmun. (2016) 66:76–88. doi: 10.1016/j.jaut.2015.08.019
192. Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S, et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia. (2005) 48:1565–75. doi: 10.1007/s00125-005-1831-2
193. Dolpady J, Sorini C, Di Pietro C, Cosorich I, Ferrarese R, Saita D, et al. Oral probiotic VSL#3 prevents autoimmune diabetes by modulating microbiota and promoting indoleamine 2,3-dioxygenase-enriched tolerogenic intestinal environment. J Diabetes Res. (2016) 2016:7569431. doi: 10.1155/2016/7569431
194. Takiishi T, Korf H, Van Belle TL, Robert S, Grieco FA, Caluwaerts S, et al. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. J Clin Invest. (2012) 122:1717–25. doi: 10.1172/JCI60530
195. Robert S, Gysemans C, Takiishi T, Korf H, Spagnuolo I, Sebastiani G, et al. Oral delivery of glutamic acid decarboxylase (GAD)-65 and IL10 by Lactococcus lactis reverses diabetes in recent-onset NOD mice. Diabetes. (2014) 63:2876–87. doi: 10.2337/db13-1236
196. Jia L, Shan K, Pan LL, Feng N, Lv Z, Sun Y, et al. Clostridium butyricum CGMCC0313.1 protects against autoimmune diabetes by modulating intestinal immune homeostasis and inducing pancreatic regulatory T cells. Front Immunol. (2017) 8:1345. doi: 10.3389/fimmu.2017.01345
197. Kim TK, Lee JC, Im SH, Lee MS. Amelioration of autoimmune diabetes of NOD mice by immunomodulating probiotics. Front Immunol. (2020) 11:1832. doi: 10.3389/fimmu.2020.01832
198. Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. (2012) 55:2285–94. doi: 10.1007/s00125-012-2564-7
199. Hanninen A, Toivonen R, Poysti S, Belzer C, Plovier H, Ouwerkerk JP, et al. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. (2018) 67:1445–53. doi: 10.1136/gutjnl-2017-314508
200. Uusitalo U, Liu X, Yang J, Aronsson CA, Hummel S, Butterworth M, et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr. (2016) 170:20–8. doi: 10.1001/jamapediatrics.2015.2757
201. Mondanelli G, Orecchini E, Volpi C, Panfili E, Belladonna ML, Pallotta MT, et al. Effect of probiotic administration on serum tryptophan metabolites in pediatric type 1 diabetes patients. Int J Tryptophan Res. (2020) 13:1178646920956646. doi: 10.1177/1178646920956646
202. Kumar S, Kumar R, Rohilla L, Jacob N, Yadav J, Sachdeva N. A high potency multi-strain probiotic improves glycemic control in children with new-onset type 1 diabetes mellitus: a randomized, double-blind, and placebo-controlled pilot study. Pediatr Diabetes. (2021) 22:1014–22. doi: 10.1111/pedi.13244
203. Groele L, Szajewska H, Szalecki M, Swiderska J, Wysocka-Mincewicz M, Ochocińska A, Stelmaszczyk-Emmel A, et al. Lack of effect of Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 on beta-cell function in children with newly diagnosed type 1 diabetes: a randomised controlled trial. BMJ Open Diabetes Res Care. (2021) 9:e001523. doi: 10.1136/bmjdrc-2020-001523
204. Chen SM, Chieng WW, Huang SW, Hsu LJ, Jan MS. The synergistic tumor growth-inhibitory effect of probiotic Lactobacillus on transgenic mouse model of pancreatic cancer treated with gemcitabine. Sci Rep. (2020) 10:20319. doi: 10.1038/s41598-020-77322-5
205. Panebianco C, Pisati F, Ulaszewska M, Andolfo A, Villani A, Federici F, et al. Tuning gut microbiota through a probiotic blend in gemcitabine-treated pancreatic cancer xenografted mice. Clin Transl Med. (2021) 11:e580. doi: 10.1002/ctm2.580
206. Kaur K, Kozlowska AK, Topchyan P, Ko M-W, Ohanian N, Chiang J, et al. Probiotic-treated super-charged NK cells efficiently clear poorly differentiated pancreatic tumors in Hu-BLT mice. Cancers. (2019) 12:63. doi: 10.3390/cancers12010063
207. Konishi H, Isozaki S, Kashima S, Moriichi K, Ichikawa S, Yamamoto K, et al. Probiotic Aspergillus oryzae produces anti-tumor mediator and exerts anti-tumor effects in pancreatic cancer through the p38 MAPK signaling pathway. Sci Rep. (2021) 11:11070. doi: 10.1038/s41598-021-90707-4
208. Chen SM, Hsu LJ, Lee HL, Lin CP, Huang SW, Lai CJL, et al. Lactobacillus attenuate the progression of pancreatic cancer promoted by porphyromonas gingivalis in K-ras(G12D) transgenic mice. Cancers. (2020) 12:3522. doi: 10.3390/cancers12123522
209. Mei QX, Hu JH, Huang ZH, Fan JJ, Huang CL, Lu YY, et al. Pretreatment with chitosan oligosaccharides attenuate experimental severe acute pancreatitis via inhibiting oxidative stress and modulating intestinal homeostasis. Acta Pharmacol Sin. (2021) 42:942–53. doi: 10.1038/s41401-020-00581-5
210. Karakan T, Ergun M, Dogan I, Cindoruk M, Unal S. Comparison of early enteral nutrition in severe acute pancreatitis with prebiotic fiber supplementation versus standard enteral solution: a prospective randomized double-blind study. World J Gastroenterol. (2007) 13:2733–37. doi: 10.3748/wjg.v13.i19.2733
211. He Y, Wu C, Li J, Li H, Sun Z, Zhang H, et al. Inulin-type fructans modulates pancreatic-gut innate immune responses and gut barrier integrity during experimental acute pancreatitis in a chain length-dependent manner. Front Immunol. (2017) 8:1209. doi: 10.3389/fimmu.2017.01209
212. Chen K, Chen H, Faas MM, de Haan BJ, Li J, Xiao P, et al. Specific inulin-type fructan fibers protect against autoimmune diabetes by modulating gut immunity, barrier function, and microbiota homeostasis. Mol Nutr Food Res. (2017) 61:1601006. doi: 10.1002/mnfr.201601006
213. Wu C, Pan LL, Luo Y, Niu W, Fang X, Liang W, et al. Low methoxyl pectin protects against autoimmune diabetes and associated caecal dysfunction. Mol Nutr Food Res. (2019) 63:e1900307. doi: 10.1002/mnfr.201900307
214. Wu C, Pan LL, Niu W, Fang X, Liang W, Li J, et al. Modulation of gut microbiota by low methoxyl pectin attenuates type 1 diabetes in non-obese diabetic mice. Front Immunol. (2019) 10:1733. doi: 10.3389/fimmu.2019.01733
215. Xiao L, Van't Land B, Engen PA, Naqib A, Green SJ, Nato A, et al. Human milk oligosaccharides protect against the development of autoimmune diabetes in NOD-mice. Sci Rep. (2018) 8:3829. doi: 10.1038/s41598-018-22052-y
216. Ho J, Nicolucci AC, Virtanen H, Schick A, Meddings J, Reimer RA, et al. Effect of prebiotic on microbiota, intestinal permeability, and glycemic control in children with type 1 diabetes. J Clin Endocrinol Metab. (2019) 104:4427–40. doi: 10.1210/jc.2019-00481
217. Panebianco C, Villani A, Pazienza V. High levels of prebiotic resistant starch in diet modulate gene expression and metabolomic profile in pancreatic cancer xenograft mice. Nutrients. (2019) 11:709. doi: 10.3390/nu11040709
218. Trivieri N, Panebianco C, Villani A, Pracella R, Latiano TP, Perri F, et al. High levels of prebiotic resistant starch in diet modulate a specific pattern of miRNAs expression profile associated to a better overall survival in pancreatic cancer. Biomolecules. (2020) 11:26. doi: 10.3390/biom11010026
219. Oláh A, Belágyi T, Pótó L, Romics L Jr, Bengmark S. Synbiotic control of inflammation and infection in severe acute pancreatitis: a prospective, randomized, double blind study. Hepatogastroenterology. (2007) 54:590–4.
220. Plaudis H, Pupelis G, Zeiza K, Boka V. Early low volume oral synbiotic/prebiotic supplemented enteral stimulation of the gut in patients with severe acute pancreatitis: a prospective feasibility study. Acta Chir Belg. (2012) 112:131–8. doi: 10.1080/00015458.2012.11680811
221. Dos Santos PQ, Guedes JC, de Jesus RP, Santos RRD, Fiaconne RL. Effects of using symbiotics in the clinical nutritional evolution of patients with chronic pancreatitis: study prospective, randomized, controlled, double blind. Clin Nutr ESPEN. (2017) 18:9–15. doi: 10.1016/j.clnesp.2017.01.005
222. Rammohan A, Sathyanesan J, Rajendran K, Pitchaimuthu A, Perumal SK, Balaraman K, et al. Synbiotics in surgery for chronic pancreatitis: are they truly effective? A single-blind prospective randomized control trial. Ann Surg. (2015) 262:31–7. doi: 10.1097/SLA.0000000000001077
223. Zare Javid A, Aminzadeh M, Haghighi-Zadeh MH, Jamalvandi M. The effects of synbiotic supplementation on glycemic status, lipid profile, and biomarkers of oxidative stress in type 1 diabetic patients. A placebo-controlled, double-blind, randomized clinical trial. Diabetes Metab Syndr Obes. (2020) 13:607–17. doi: 10.2147/DMSO.S238867
224. Nataraj BH, Ali SA, Behare PV, Yadav H. Postbiotics-parabiotics: the new horizons in microbial biotherapy and functional foods. Microbial Cell Factor. (2020) 19:168. doi: 10.1186/s12934-020-01426-w
225. Takauji S, Konishi H, Fujiya M, Ueno N, Tanaka H, Sato H, et al. Polyphosphate, derived from Lactobacillus brevis, modulates the intestinal microbiome and attenuates acute pancreatitis. Dig Dis Sci. (2021) 66:3872–84. doi: 10.1007/s10620-020-06747-9
226. Nam Y, Kim J, Baek J, Kim W. Improvement of cutaneous wound healing via topical application of heat-killed Lactococcus chungangensis CAU 1447 on diabetic mice. Nutrients. (2021) 13:2666. doi: 10.3390/nu13082666
227. Kita A, Fujiya M, Konishi H, Tanaka H, Kashima S, Iwama T, et al. Probiotic-derived ferrichrome inhibits the growth of refractory pancreatic cancer cells. Int J Oncol. (2020) 57:721–32. doi: 10.3892/ijo.2020.5096
228. Gupta A, Khanna S. Fecal microbiota transplantation. JAMA. (2017) 318:102. doi: 10.1001/jama.2017.6466
229. Lei Y, Tang L, Liu S, Hu S, Wu L, Liu Y, et al. Parabacteroides produces acetate to alleviate heparanase-exacerbated acute pancreatitis through reducing neutrophil infiltration. Microbiome. (2021) 9:115. doi: 10.1186/s40168-021-01065-2
230. van den Berg FF, van Dalen D, Hyoju SK, van Santvoort HC, Besselink MG, Wiersinga WJ, et al. Western-type diet influences mortality from necrotising pancreatitis and demonstrates a central role for butyrate. Gut. (2021) 70:915–27. doi: 10.1136/gutjnl-2019-320430
231. Hu Y, Xiao HY, He C, Lv NH, Zhu L. Fecal microbiota transplantation as an effective initial therapy for pancreatitis complicated with severe Clostridium difficile infection: a case report. World J Clin Cases. (2019) 7:2597–604. doi: 10.12998/wjcc.v7.i17.2597
232. Zhang XS, Li J, Krautkramer KA, Badri M, Battaglia T, Borbet TC, et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. eLife. (2018) 7:e37816. doi: 10.7554/eLife.37816
233. Zhang XS, Yin YS, Wang J, Battaglia T, Krautkramer K, Li WV, et al. Maternal cecal microbiota transfer rescues early-life antibiotic-induced enhancement of type 1 diabetes in mice. Cell Host Microbe. (2021) 29:1249–65.e9. doi: 10.1016/j.chom.2021.06.014
234. de Groot P, Nikolic T, Pellegrini S, Sordi V, Imangaliyev S, Rampanelli E, et al. Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut. (2021) 70:92–105. doi: 10.1136/gutjnl-2020-322630
235. Bhattarai Y, Kashyap PC. Germ-free mice model for studying host-microbial interactions. Methods Mol Biol. (2016) 1438:123–35. doi: 10.1007/978-1-4939-3661-8_8
236. Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, et al. Culturing the human microbiota and culturomics. Nat Rev Microbiol. (2018) 16:540–50. doi: 10.1038/s41579-018-0041-0
237. Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature. (2016) 533:543–6. doi: 10.1038/nature17645
238. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. (2012) 488:178–84. doi: 10.1038/nature11319
239. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
240. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. (2019) 16:35–56. doi: 10.1038/s41575-018-0061-2
241. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
242. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. (2010) 107:11971–75. doi: 10.1073/pnas.1002601107
Keywords: gut microbiome, pancreatic diseases, probiotics, prebiotics, synbiotics, postbiotics, fecal microbiota transplantation (FMT)
Citation: Zhang T, Gao G, Sakandar HA, Kwok L-Y and Sun Z (2022) Gut Dysbiosis in Pancreatic Diseases: A Causative Factor and a Novel Therapeutic Target. Front. Nutr. 9:814269. doi: 10.3389/fnut.2022.814269
Received: 25 November 2021; Accepted: 21 January 2022;
Published: 15 February 2022.
Edited by:
Silvia Turroni, University of Bologna, ItalyReviewed by:
Derya Onal Darilmaz, Aksaray University, TurkeyCopyright © 2022 Zhang, Gao, Sakandar, Kwok and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihong Sun, c3VuemhpaG9uZzc4QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.