- 1College of Food Science and Technology, Shanghai Ocean University, Shanghai, China
- 2Laboratory for Quality and Safety Risk Assessment of Aquatic Products in Storage and Preservation of Ministry of Agriculture and Rural Affairs, Shanghai Ocean University, Shanghai, China
- 3Shanghai Professional Technology Service Platform on Cold Chain Equipment Performance and Energy Saving Evaluation, Shanghai Ocean University, Shanghai, China
- 4Shanghai Engineering Research Center of Aquatic Product Processing and Preservation, Shanghai Ocean University, Shanghai, China
- 5National Experimental Teaching Demonstration Center for Food Science and Engineering, Shanghai Ocean University, Shanghai, China
Shewanella putrefaciens is a special spoilage bacterium of seafood during cold storage, which is easy to form biofilm and bring serious hazard to the seafood quality. Life cycle of biofilm starts after bacterial adhesion, which is essential for the formation and development of biofilm. As a ubiquitous second messenger in bacteria, c-di-GMP regulates the conversion between bacterial planktonic state and biofilm state. In this study, the adhesion and biofilm formation of S. putrefaciens WS13 under 4°C were compared to those under 30°C. Atom force microscope and scanning electron microscope were used to study the bacterial adhesion. Biofilm was analyzed by Fourier transform infrared spectroscopy, Bradford assay and phenol-sulfuric acid method. High-performance liquid chromatographic-tandem mass spectrometric and quantitative real-time PCR were applied to study c-di-GMP level and genes encoding diguanylate cyclases in cells, respectively. Results showed that the swarming mobility of S. putrefaciens WS13 was weaker under 4°C, however, the adhesive force under 4°C was 4–5 times higher than that under 30°C. Biofilm biomass, extracellular polysaccharides and extracellular proteins were 2.5 times, 3 times, and 1.6 times more than those under 30°C, respectively, but biofilm composition formed under both temperatures were similar. c-di-GMP level in S. putrefaciens WS13 under 30°C was no more than half of that in the corresponding growth stage under 4°C. Quantitative real-time PCR analysis also showed that the expression of genes encoding diguanylate cyclases were significantly enhanced under 4°C than that under 30°C. S. putrefaciens WS13 adapted to the cold stress by enhancing the expression of genes encoding diguanylate cyclases to promote bacterial adhesion and biofilm formation. This study provides a theoretical foundation for the research on the cold adaptation mechanism of specific spoilage bacteria of seafood based on c-di-GMP, and also provides a new idea to control seafood quality from the perspective of microbial molecular biology.
Introduction
Shewanella putrefaciens is a specific Gram-negative bacterium in aquatic products such as Litopenaeus vannamei (1, 2), bigeye tuna (3), shellfish (4), and Paralichthys olivaceus (5). S. putrefaciens has strong biofilm-forming capability. The formation of biofilm enhances the antimicrobial resistance, antimicrobial tolerance, and stress tolerance of bacteria to strengthen their environmental tolerance, which is conducive to the survival of bacterial cells (6–8). Biofilm is a bacterial community that is formed when bacteria attach to biotic/abiotic surfaces, secret substances such as extracellular polysaccharides, extracellular proteins, extracellular DNA, and lipids, and encapsulate bacterial cells. Once formed, biofilm is difficult to remove (6, 9, 10). The formation of biofilm is divided into four stages: bacterial adhesion, bacterial aggregation, biofilm maturation, and detachment (11). Bacterial adhesion is the process by which bacteria in a planktonic state sense and adhere to biotic/abiotic surfaces. It has two stages, reversible adhesion and irreversible adhesion (12). At the stage of irreversible adhesion, bacteria cannot escape from biotic/abiotic surfaces, and irreversible adhesion starts the life cycle of biofilm formation. The adhesion of bacteria is the key to biofilm formation (12). Biofilm is the ubiquitous form of microbial cells in nature (13). The formation of biofilm is affected by factors such as temperature (14), metal ions (15), oxygen (16), and nutrients (17). A study on the mechanism of biofilm formation under environmental stress is helpful to control the hazard of biofilm.
To adapt to the complex and changeable environment, there are various signal transduction systems in bacterial cells to ensure the survival of bacteria, such as cyclic diguanylic acid (c-di-GMP) (18) and two-component systems (19). As the ubiquitous second messenger in bacteria, c-di-GMP modulates diverse biological phenotypes in bacteria such as virulence, flagellar motility, biofilm formation (17), and bacterial colonization (20). c-di-GMP was first discovered in Gluconacetobacter xylinus in 1987 (21), and its level in bacterial cells is regulated by diguanylate cyclases (DGCs) (22) and phosphodiesterases (PDEs) (22, 23). Most DGCs and PDEs have transmembrane helix structures which can sense various input signals such as oxygen, light, antibiotics (17), and temperature (24) to regulate the level of c-di-GMP. Previous studies (25, 26) have shown that the intracellular level of c-di-GMP can regulate the conversion between the planktonic state and biofilm state of bacteria. In addition, c-di-GMP can regulate the activity of flagella to control the movement ability of bacteria and then regulate bacterial adhesion, and also regulate the synthesis of extracellular polysaccharides and extracellular proteins to promote the formation of biofilm (25). However, few studies on the role of c-di-GMP in the formation of biofilm of specific spoilage organisms (SSOs) in seafood have been reported.
Based on the above, this study intended to reveal the correlation between c-di-GMP and the adhesion and biofilm formation of S. putrefaciens WS13 under cold stress. Our work showed the characteristic differences of adhesive force and biofilm of S. putrefaciens WS13 under 30 and 4°C and indicated the importance of c-di-GMP for bacterial adhesion and biofilm formation. This provides a theoretical basis for the study of c-di-GMP regulating the adhesion and biofilm formation of S. putrefaciens WS13 under cold stress.
Materials and methods
Strains and cultivation
S. putrefaciens WS13 strain, isolated from spoiled Litopenaeus vannamei, which was preserved by our group (NCBI No. CP028435.1) (27) was applied in this experiment. The strain was preserved in LB medium (Land Bridge Technology, Beijing, China) with 50% (v/v) glycerol (Sinopharm., AR, China) in a −80°C refrigerator and was activated to 8–9 log CFU/mL (OD600 value = 0.80) in fresh LB medium under 30°C for 150 rpm, and then, the culture was diluted at 0.1% ratio with LB medium for further use.
Growth curve and swarming mobility of S. putrefaciens WS13
The diluted culture was cultivated under oscillating conditions (150 rpm) under 4 and 30°C, respectively, and 200 μl of culture was taken out aseptically and added into 96-well polystyrene microtiter plates to measure bacterial growth curves with BioTek Synergy 2 (Winooski, VT, United States) at 600 nm.
The swarming mobility of bacteria was measured as described (28) previously with slightly modified. The culture was cultivated in semi-solid LB media containing 0.25% agar under 30 and 4°C for various times (12, 24, 48, 72, 96, 120, 144, and 168 h under 4 °C; 8, 12, 24, 36, 48, 60, 72, and 84 h under 30 °C), respectively. The sizes of bacterial colonies in the experiment were observed to analyze the swarming mobility of bacteria.
Atom force microscope
The adhesive force of S. putrefaciens WS13 was measured by AFM (Bioscope resolve, Bruker, Germany) as described (29).
Cell preparation
The diluted culture was cultivated in LB medium under 4 and 30°C to 8–9 log CFU/mL (OD600 value = 0.80), respectively. After centrifugation (9500 rpm, 8 min, 4°C), harvested cells were treated with 0.01 M sterile phosphate-buffered saline (PBS, pH 7.0) three times and suspended in Milli-Q grade water (OD600 value = 0.20–0.40). The suspension was applied to prepare cell probes (To minimize viability loss, cells could be treated with 2.5% (w/v) glutaraldehyde precooled under 4°C in advance and kept under 4°C).
Probe design and detection
The suspension prepared was set on the glass slide at room temperature and connected to polystyrene plastic spheres (PS spheres, diameter, 12 μm) by electrostatic attraction for 20 min. Bacteria-PS spheres prepared were tenderly cleaned with Milli-Q grade water to divide unattached cells. A biological microscope (DM1000 LED, Leica, Germany) at a magnification rate of 900X was used to ensure that cells were fixed on PS spheres. PS spheres with cells were dispersed on the carrier chip and bonded to the MLCT-F tipless cantilever of AFM with AB glue. The experiment which applied the system of Dimension Icon (Bruker, Santa Barbara, CA) was operated at room temperature. First, the polystyrene plastic (PS) substrate was probed with a bare probe to set a baseline for comparing the force measurements with the designed probe. The elastic constant of the probe cantilever was 0.1 N/m. Spring constants of the tips were in the range of 0.6 N/m and were measured for each probe. The loading force was set to 1 nN. The adhesive force was measured by the formula F = k*sensitivity*deflection error. K and sensitivity of MLCT-F tipless cantilever were 0.6 N/m and 20 nm/v, respectively. Nanoscope 1.8 software was used to quantify the adhesive force that was obtained from the force–distance curves.
Scanning electron microscope
According to bacterial growth curves, 1 mL of diluted culture was cultivated at 48-well polystyrene microtiter plates containing one piece of PS plastic sheet each under 30°C for 18 h and 4°C for 168 h, respectively. PS plastic sheets were taken out, washed with 0.01 M sterile phosphate-buffered saline (PBS, pH 7.0, ACMEC biochemical, China) three times, and treated with precooled 2.5 % glutaraldehyde (ACMEC biochemical, China) under 4°C for 4 h. After dehydrated with gradient ethanol (30, 50, 70, 90, and 100 %, v/v) for 10 min each and dried naturally, plastic sheets were treated with gold plating. Scanning electron microscope (SEM, S4500; Hitachi, Tokyo, Japan) at 3 kV was applied to obtain bacterial adhesive state (50 nm objective multiple) and morphology (5 nm objective multiple).
Analysis of biofilm
Biofilm biomass was measured quantitatively as described (6) previously with slightly modified. Briefly, 1 mL of diluted culture was added into 48-well polystyrene microtiter plates; then, the plates were covered by plastic to avoid evaporative loss. According to bacterial growth curves, after certain cultivation times (6, 12, 18, 24, 30, 36, and 42 h under 30°C; 24, 48, 72, 96, 120, 144, 168, 192, 216, 240, and 264 h under 4°C), biofilms in wells were carefully cleaned three times with 1 mL of 0.01 M sterile phosphate-buffered saline (PBS, pH 7.0) to remove unattached cells. After being dried for 25 min under 60°C, biofilms were stained with 1 mL of 0.2% (w/v) crystal violet (Sangon Biotech, Co., Ltd., Shanghai, China) for 15 min, then washed and dried as described above. Dye attached to the biofilm was released with 95% ethanol (v/v) for 10 min. Bio Tek Synergy 2 (Winooski, VT, United States) at 600 nm was applied to obtain biofilm content.
The components of biofilm were analyzed as described (30, 31) previously with slightly modified. The diluted culture was cultured at 30 and 4°C, respectively. According to bacterial growth curves, after certain cultivation times (10, 15, 20, 25, and 30 h under 30°C and 108, 132, 156, 180, and 204 h under 4°C), the culture was removed and biofilm was washed with 0.01 M sterile phosphate-buffered saline (PBS, pH 7.0) three times and was collected by scraping; then, the cells were transferred to 1 mL of 0.01 M KCl solution. After being treated with ultrasonic (20 kHz, five times, 10 s on/10 s off), the cells were filtered by 0.22 μm cellulose membrane (Sangon Biotech, Co., Ltd., Shanghai, China) and dried under vacuum (3–6 kPa) condition in the freeze dryer for 48 h to obtain anhydrous samples. The anhydrous samples were analyzed by Fourier transform infrared spectroscopy (FT-IR, Perkin-Elmer, Model 2000), Bradford assay, and phenol–sulfuric acid method to detect biofilm FT-IR characteristics, the content of extracellular proteins and extracellular polysaccharides, respectively. FT-IR studies were performed with up to 100 scans, a scan range of 4000 and 450 cm–1, and resolutions of4 cm–1.
High-performance liquid chromatography-tandem mass spectrometry
Cyclic diguanylic acid was analyzed as described (32) previously with slightly modified. The diluted culture was cultivated in LB medium under 30 and 4°C, respectively. According to bacterial growth curves, an equal volume of solution was extracted at the lag phase, early logarithmic phase (early log), middle logarithmic phase (mid-log), late logarithmic phase (late log), and stationary phase, respectively. Formaldehyde at a final concentration of 0.19% was added. After centrifugation (9500 rpm, 8 min, 4°C), harvested cells were treated with iced deionized water, centrifuged as described above, and then resuspended in 0.5 mL of iced deionized water to be heated at 96°C for 5 min. After adding 0.925 mL of ice anhydrous ethanol each, solutions were treated with vortex condition (30 s), centrifugation (13000 rpm, 4 min, 4°C), freeze-drying, and 50% (v/v) methanol to create resuspension solution. The samples were filtered by 0.22 μm cellulose membrane and analyzed by HPLC-MS-MS to quantify the intracellular c-di-GMP level.
Quantitative real-time PCR analysis
The diluted culture, under 4 and 30°C at 8–9 log CFU/mL (OD600 value = 0.80), respectively, was centrifuged (10000 rpm, 5 min, 4°C) to extract RNA. The RNA was obtained with the RNA extraction kit (ACMEC Biochemical Co., Ltd., Shanghai), and its concentration was evaluated with an ELISA reader (Labsystems, Multiskan EX). The prepared RNAs were reverse-transcribed into cDNA following M-MLV 4 First-Strand cDNA Synthesis Kit (Biomed, Beijing) which were then kept at – 20°C.
Quantitative real-time PCR was carried out with the QuantStudio™ 1 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA USA). The reaction system consisted of 10 μl of 2X SYBR Green qPCR Master Mix, 0.4 μl of 10 μM F Primer, 0.4 μl of 10 μM R Primer, 7.2 μl of dd H2O, and 2 μl of cDNA. The reaction system without cDNA served as a negative control. 16S rRNA was applied as the reference gene, and the gene-specific primers were designed and synthesized by Sangon (Sangon, Biotech Co., Ltd, Shanghai, China). The primers used in this experiment for quantitative real-time PCR are shown in Table 1.
Quantitative real-time PCR was operated with the following conditions: 50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 15 s; and 60°C for 1 min. Genes were selected (27), and the expression level of genes was analyzed by the 2−ΔΔCT method in triplicate. Statistical analysis was performed by Origin Pro 2021, and error bars represent the SD from the mean of measurements.
Data analysis
The experiments were carried out at least three times independently. Analysis of microbiological results was converted to OD and log CFU/mL. Origin Pro 2021 (Origin Lab Corp., Northampton, MA, United States) was applied to analyze the content of biofilm, biofilm components, and other data obtained. SigmaStat 3.5 was applied to analyze bacterial adhesive force. Differences at p-value <0.05 were regarded as statistically significant.
Results
Growth and swarming mobility of S. putrefaciens WS13
The growth and swarming mobility of S. putrefaciens WS13 are shown in Figure 1. The results showed that the lag phase under 30°C was < 2.5 h, and the lag phase under 4°C was significantly longer than 50 h. The maximum density value of cells under 4°C was higher than that under 30°C. At the stationary phase, the cell density of bacteria under 30°C was 3–5 log CFU/mL inferior to that under 4°C. The results of bacterial growth curves indicated that cold stress slowed down the bacterial growth rate and raised its density.
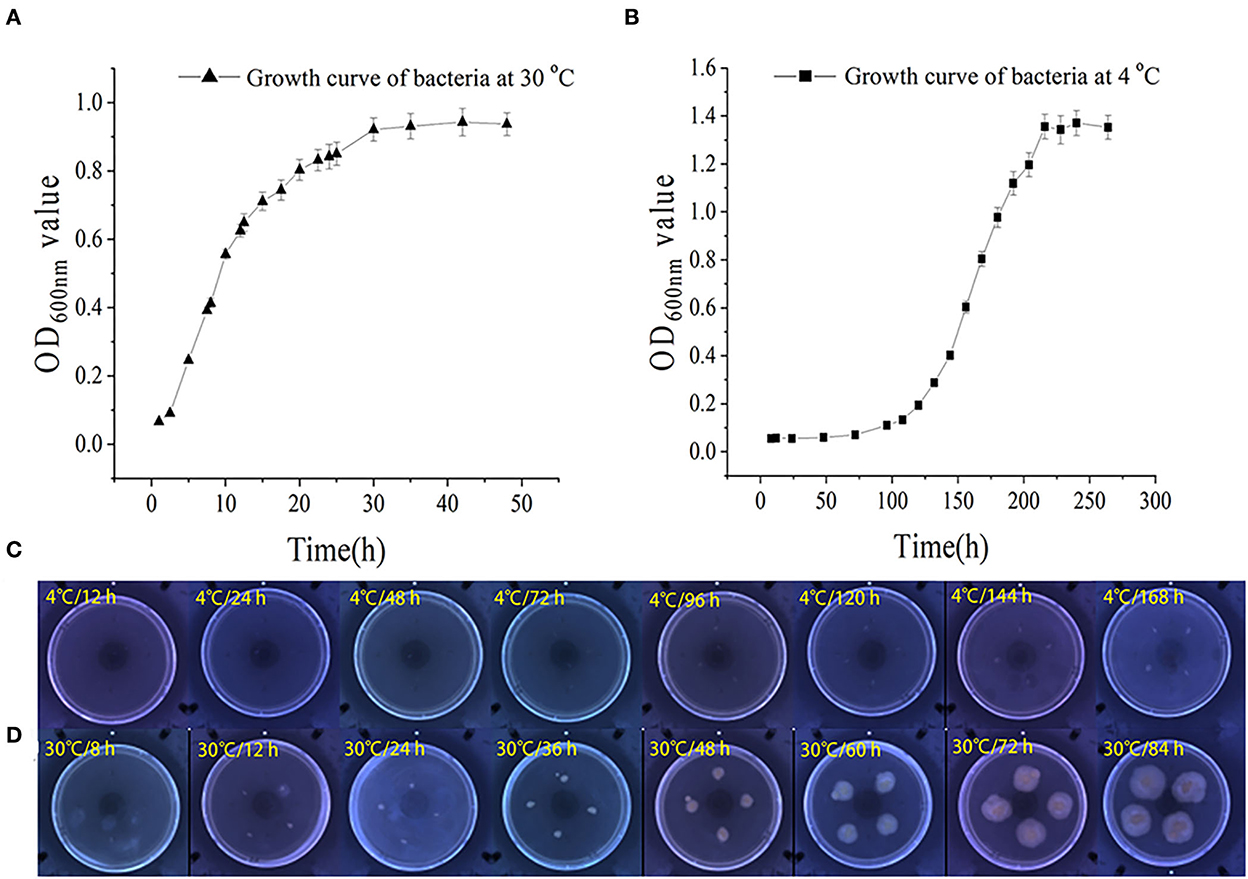
Figure 1. Growth and swarming mobility of S. putrefaciens WS13. (A) Growth curve of S. putrefaciens WS13 under 30°C. (B) Growth curve of S. putrefaciens WS13 under 4°C. (C) Swarming mobility of S. putrefaciens WS13 under 4°C. (D) Swarming mobility of S. putrefaciens WS13 under 30°C.
With the prolongation of incubation time, the sizes of colonies cultured under 4°C did not change significantly. However, the colonies cultured under 30°C appeared earlier, and the sizes of colonies increased with the extension of culture time. It showed that cold stress weakened bacterial swarming mobility (33) and made bacteria swarm in the smaller zone to form smaller colonies.
Atom force microscope
As shown in Figure 2, the horizontal baseline measured by the bare probe and the approach–retraction curve measured by sample probes were obtained. After calculation, the maximum difference between the two curves represented the adhesive force of the sample to the matrix. Under 4°C, the adhesive force of cells was four–five times higher than that under 30°C, demonstrating that the adhesive force of S. putrefaciens WS13 under 4°C was stronger than that under 30°C.
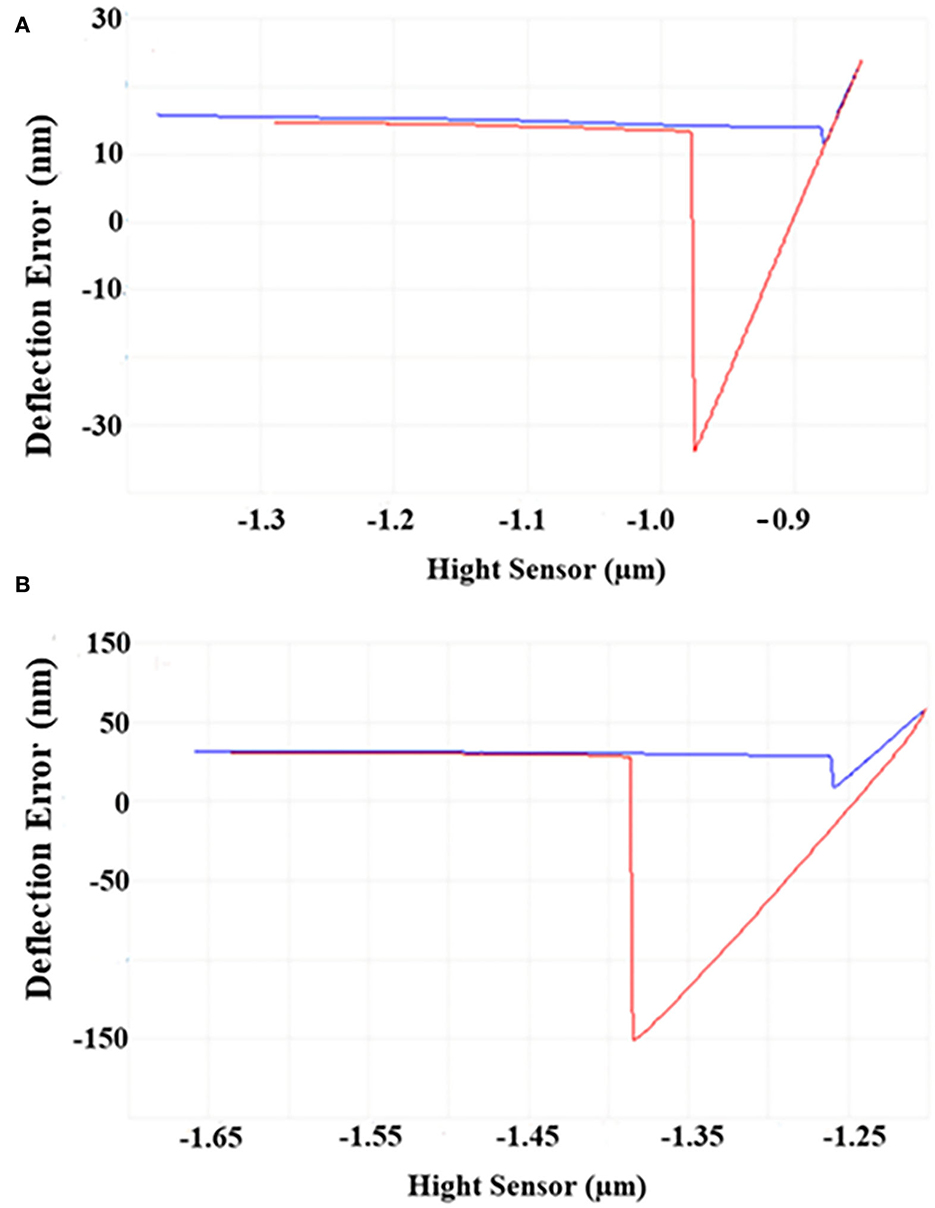
Figure 2. AFM images of S. putrefaciens WS13. (A) AFM image of S. putrefaciens WS13 under 30°C. (B) AFM image of S. putrefaciens WS13 under 4°C.
Scanning electron microscope
The morphological changes in the mature biofilm of S. putrefaciens WS13 under 30 and 4°C are shown in Figure 3. Cells in yellow rectangles in SEM images A and B were amplified as presented in C and D. SEM images indicated that the number of cells under 4°C was more than that under 30°C in matured biofilm. Bacterial morphology was affected by temperature, but the affection was tiny.
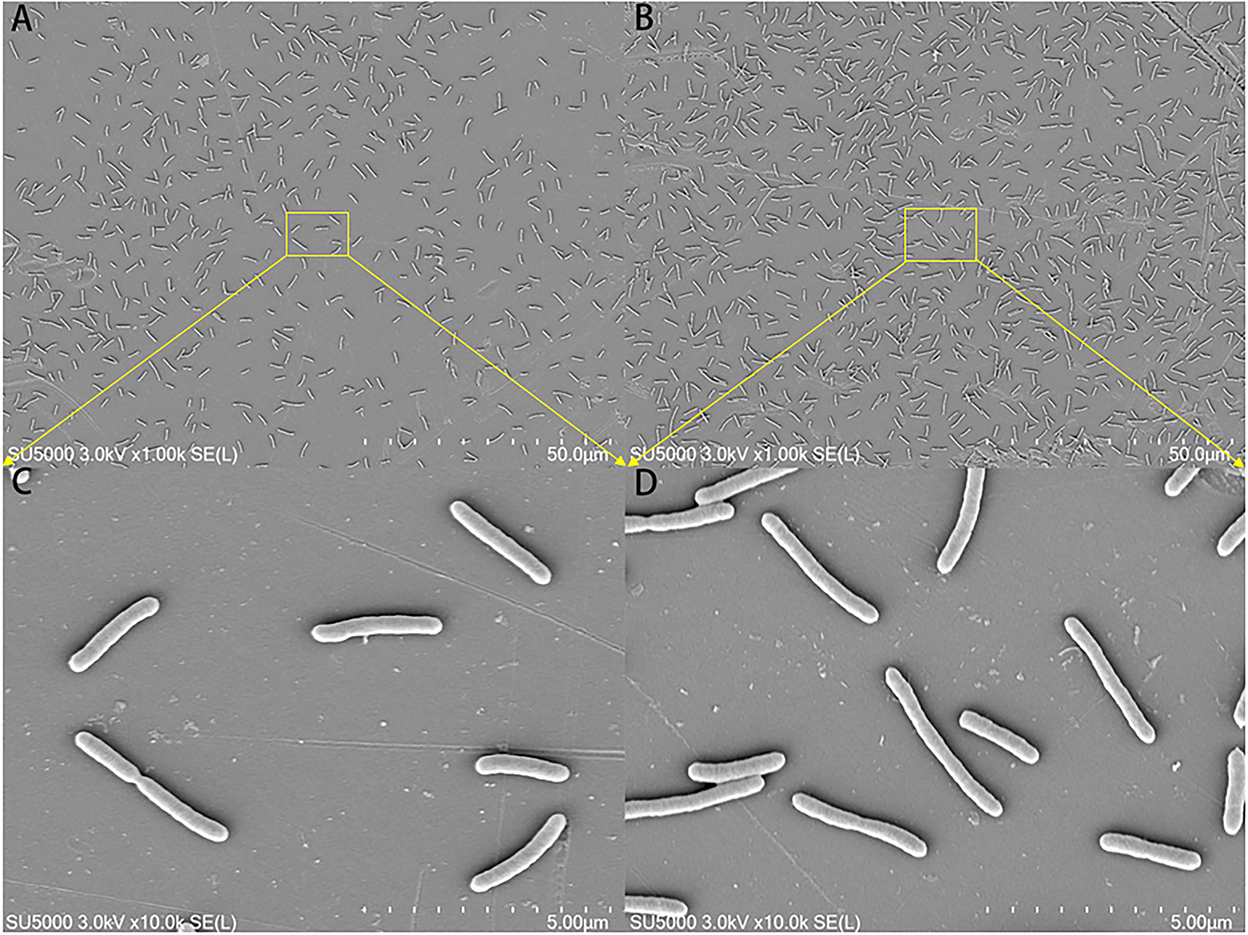
Figure 3. SEM images of the mature biofilm of S. putrefaciens WS13 under 30 and 4°C, respectively. The scale bar in (A,B) represents 50.0 μm. The scale bar in (C,D) represents 5.00 μm.
Analysis of biofilm
The results of biofilm biomass and components are shown in Figure 4. The biofilm matured under 30 and 4°C at 18 and 168 h, respectively (Figures 4A,B). S. putrefaciens WS13 produced more biofilm under cold stress. The results of Fourier transform infrared spectroscopy (FT-IR) characteristics of the mature biofilm formed under 30°C at 18 h and 4°C at 168 h are shown in Figure 4C, respectively. The characteristic wave numbers are shown in Table 2. Bands of extracellular polysaccharides were at 920 cm−1, 1100–1260 cm−1, and 3300–3400 cm−1, respectively. The peak at 1450 cm−1, 1690–1700 cm−1, and 3300–3400 cm−1 indicated the existence of extracellular proteins in biofilm, respectively. Other biomacromolecules such as lipids were verified in the biofilm, which is consistent with the previous studies (30, 34). FT-IR spectra of biofilm formed under 30 and 4°C were roughly the same. Extracellular polysaccharides and extracellular proteins reached the maximum value at 20 h and 180 h under 30 and 4°C, respectively. The maximum value of extracellular polysaccharides and extracellular proteins under 4°C was three and 1.5 times higher than that under 30°C, respectively.
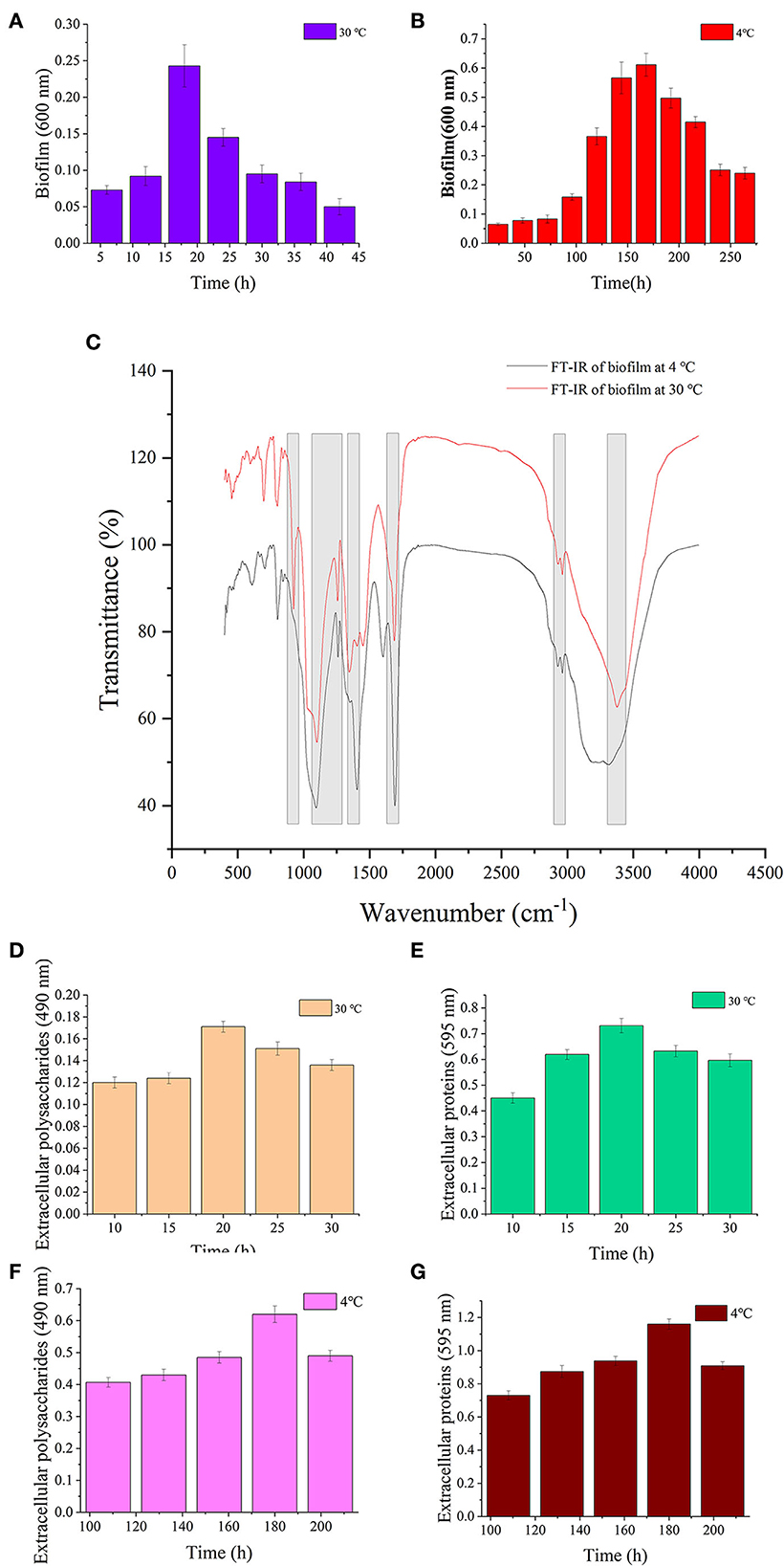
Figure 4. Biofilm analysis of S. putrefaciens WS13. (A) Biofilm biomass formed under 30°C. (B) Biofilm biomass formed under 4°C. (C) FT-IR spectra of S. putrefaciens WS13 under 30°C (red line) and 4°C (black line). (D) The extracellular polysaccharides content of EPS under 30°C. (E) Extracellular proteins of EPS content under 30°C. (F) The extracellular polysaccharides content of EPS under 4°C. (G) Extracellular proteins of EPS content under 4°C.
High-performance liquid chromatography-tandem mass spectrometry
Figure 5 shows that the intracellular levels of c-di-GMP in S. putrefaciens WS13 varied at different growth stages under 30 and 4°C. The level of c-di-GMP in bacterial cells increased first and then decreased both under 30 and 4°C. Mostly, the c-di-GMP level in cells was higher under 4°C than that under 30°C. Under the same culture stage, the intracellular c-di-GMP level under 30°C was < 50 % of that under 4°C.
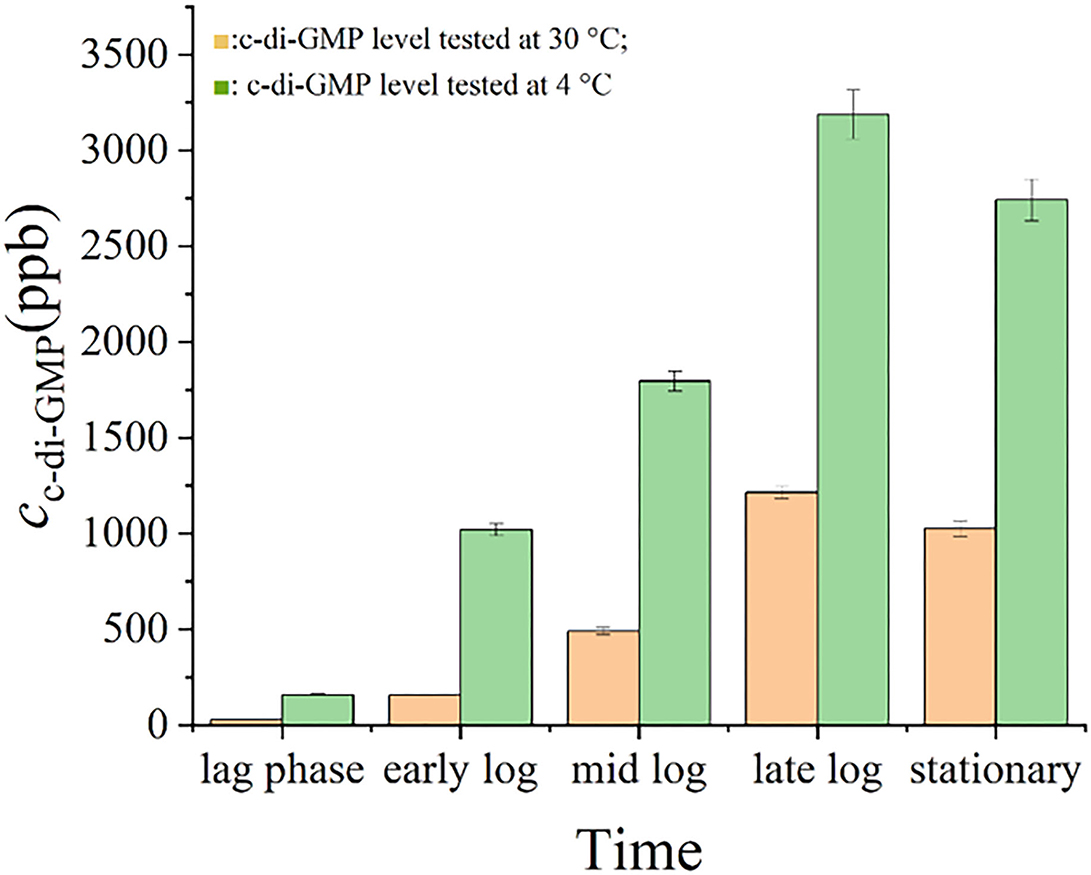
Figure 5. c-di-GMP levels in S. putrefaciens WS13. c-di-GMP levels in S. putrefaciens WS13 under 30 ( ) and 4°C (
) and 4°C ( ).
).
Quantitative real-time PCR analysis
The impact of temperature on the expression level of genes encoding diguanylate cyclases was evaluated by quantitative real-time PCR analysis, and the result is shown in Figure 6. Under 4°C, the expression levels of all tested genes were higher than those under 30°C, and the change in gene expression under cold stress was different. The expression levels of genes AVV82427.1, AVV82583.1, AVV82979.1, and AVV83156.1 under 4°C were upregulated by < 2 times (1.72, 1.08, 1.07, and 1.64, respectively) than that under 30°C, and the expression levels of genes AVV84424.1, AVV85746.1, AVV86051.1, AVV84237.1, AVV84755.1, and AVV85279.1 were upregulated much more (2.22, 2.54, 2.52, 6.27, 6.31, and 4.11,respectively).
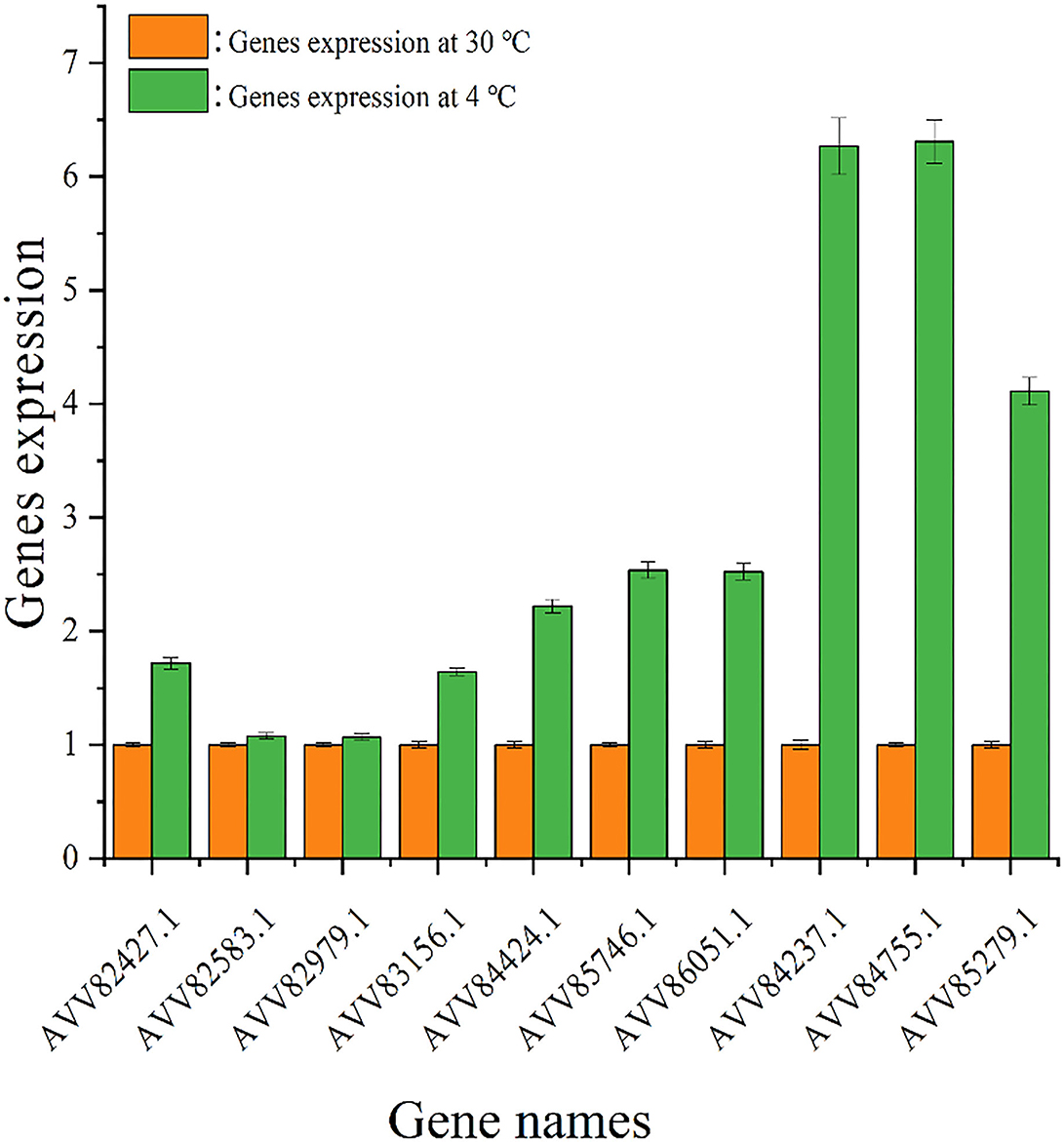
Figure 6. Expression of genes encoding diguanylate cyclases. The expression of genes encoding diguanylate cyclases under 30 ( ) and 4°C (
) and 4°C ( ).
).
Discussion
S. putrefaciens WS13 is a specific spoilage organism (SSO) of seafood under cold storage and is related to aquatic product disease named shewanellosis (35). S. putrefaciens has a strong biofilm-forming capability. Our group has found that the formation of biofilm of S. putrefaciens WS13 is conducive to its survival under cold stress (6, 36). Further study found that genes such as motA, ppsA, and fabs family expressed in S. putrefaciens WS13 under 4°C help attenuate bacterial motility, enhance bacterial adhesion, and increase biofilm fluidity (36, 37), which was beneficial for bacteria to form biofilm under cold stress. Biofilm formation began after bacteria adhered to the surface of the object, and the adhesion of bacteria was regulated by c-di-GMP (11, 25). However, the characteristics of adhesion and biofilm of S. putrefaciens WS13 under cold stress, as well as intracellular c-di-GMP level, which regulated these two, needed further study. Therefore, the refrigeration temperature (4°C) and an optimum growth temperature (30°C) were selected in this work to investigate the difference between those features of S. putrefaciens WS13.
The swarming mobility, intracellular c-di-GMP level, biofilm, cell morphology, and adhesion, as well as gene expression of S. putrefaciens WS13 under 30 and 4°C, were investigated in this work. S. putrefaciens WS13 grew slower under 4°C, but its final density was higher than that under 30°C. The swarming mobility of S. putrefaciens WS13 under 4°C was weaker than that under 30°C. Studies on Listeria monocytogenes and Ralstonia solanacearum strains showed that bacteria under cold stress achieved a greater proliferation rate and a lower growth rate, and bacterial mobility on growing colonies was attenuated (38, 39), which were consistent with our work. Researchers have reported that the c-di-GMP level could affect bacterial swarming mobility through flagella (40) or fimbriae (41), and bacterial swarming mobility affected bacterial self-aggregation (42), which in turn affected bacterial biofilm formation (43). The high c-di-GMP level in bacterial cells would bind the proteins such as YcgR to affect the flagellar motor, weakening bacterial swarming mobility, which improved bacterial self-aggregation. Those results would promote biofilm formation too (43, 44). Our work found that the intracellular c-di-GMP level in S. putrefaciens WS13 increased and weakened the ability of swarming mobility of S. putrefaciens WS13 under cold stress compared to that under 30°C, which was consistent with the previous study.
Bacterial adhesion is affected by many factors such as cellular appendages and extracellular substances (45–47). Our work found that S. putrefaciens WS13 cells had stronger adhesive force under 4°C than that under 30°C. The biomass of EPS of S. putrefaciens WS13 under 4°C was higher, and extracellular proteins and extracellular polysaccharides were higher than those that synthesized under 30°C. According to the research, EPS affected the adhesion of bacteria (48–50). This might indicate that S. putrefaciens WS13 would strengthen bacterial adhesion by increasing the content of EPS under cold stress. However, the details for those remained for further analysis.
The content of EPS and its components (such as extracellular proteins and extracellular polysaccharides) are regulated by many factors, such as c-di-GMP (51–53). The increase in the intracellular c-di-GMP level under cold stress may help to promote the synthesis and secretion of extracellular polysaccharides and extracellular proteins (51–53) and enhance bacterial adhesion, which is conducive for bacteria to survive under cold stress. It was found that the extracellular polysaccharides Pel and/or Psl contributed to the adhesion of Pseudomonas Aeruginosa, and the synthesis and secretion of Pel and/or Psl were regulated by c-di-GMP (25). In addition, it was found that c-di-GMP in Pseudomonas putida modulated the transcription factor such as FleQ, affecting the synthesis and secretion of adhesin LapA, which regulated the adhesion of P. putida (50). The increase in the intracellular c-di-GMP level under cold stress was identified to promote the synthesis and secretion of extracellular polysaccharides and proteins of EPS, which would strengthen bacterial adhesive force (51–53). This study also confirmed that c-di-GMP regulates extracellular polymers to affect the adhesion of bacteria.
Biofilm formed by S. putrefaciens WS13 under cold stress was also investigated. The results showed that the FT-IR characteristics, morphology, and structure of bacterial biofilm formed under 30 and 4°C were similar, which meant the composition of biofilm formed under both temperatures was almost the same. The intracellular c-di-GMP level affects biofilm formation (54). The high c-di-GMP level would increase bacterial extracellular matrix level and accelerate biofilm formation (55). Compared to that under 30°C, we found that the intracellular c-di-GMP level increased under cold stress, which was consistent with the conclusion that c-di-GMP regulated bacterial living lifestyle between biofilm state and planktonic state (56, 57). However, the effects of c-di-GMP under low temperature affecting bacterial biofilm composition and biofilm formation remained to be elucidated.
The level of c-di-GMP in bacterial cells affects the activity of proteases in the cytoplasm, which could hydrolyze the substances such as extracellular proteins that were located on the outer membrane of bacteria, changing the composition and structure of biofilm (58, 59). In P. fluorescens Pf0-1, the high level of c-di-GMP would bind to LapD and promote LapD to interact with the periplasmic protease LapG, which inhibited LapD to cleave the N-terminus of adhesin LapA and enhance the ability of bacterial adhesion (58, 59). Mostly, intracellular c-di-GMP at the high level would promote the secretion of EPS by controlling the regulatory factors to promote gene transcription. CuxR, the c-di-GMP-responsive protein in plant symbiotic α-proteobacteria, would stimulate the transcription of the gene cluster that synthesized EPS at high c-di-GMP levels, assisting bacterial adhesion (54). Also, researchers found that in S. putrefaciens, the high level of c-di-GMP could bind to the transcription regulator FlrA, which would strengthen the synthesis of adhesin BpfA, and reinforce bacterial adhesion (52). It explained the difference in FT-IR characteristics of bacterial biofilm formed under both temperatures. Biofilm biomass formed by S. putrefaciens WS13 under 4°C was higher than that under 30°C, which was consistent with our previous work (6). It demonstrated that cold stress promoted S. putrefaciens WS13 to form more biofilms. However, further work on how cold stress affected biofilm formation was needed. Our work found that under cold stress, the intracellular c-di-GMP level of S. putrefaciens WS13 was higher under 4°C than that under 30°C at the same growth stage, which affected the swarming mobility, biofilm, cell morphology, and adhesion of bacteria. The intracellular c-di-GMP level was regulated by DGCs and PDEs (22, 23), while environmental signals such as light, oxygen, (17) and temperature (24) would affect the expression of DGCs and PDEs and then regulate the level of c-di-GMP in cells. Quantitative real-time PCR analysis showed the expression of genes that encoded diguanylate cyclases was upregulated under cold stress, and the increased expression of genes contributed to the increase in c-di-GMP in bacterial cells. The high c-di-GMP level reduced bacterial swarming mobility, promoted bacterial adhesion and biofilm formation, and also promoted the bacterial ability to produce extracellular polysaccharides and extracellular proteins under 4°C, which directly improved bacterial adhesion, which made biofilm more than that under 30°C. This study evaluated the effects of c-di-GMP on the bacterial adhesion and biofilm formation of SSO in seafood under cold stress. However, the specific signal pathways of c-di-GMP regulating bacterial adhesion and biofilm formation need to be further studied. This study provides a theoretical basis for studying the effect of c-di-GMP on the adhesion and biofilm formation of S. putrefaciens WS13 under cold stress.
Conclusion
This study analyzed the variation of c-di-GMP level and the expression of genes encoding diguanylate cyclases in S. putrefaciens WS13 under 30 and 4°C, as well as the effects of c-di-GMP on bacterial adhesion and biofilm formation. The expression of genes encoding diguanylate cyclases and the levels of c-di-GMP increased in S. putrefaciens WS13 under cold stress, which reduced the swarming ability of bacteria, enhanced the ability of cells to adhere, and promoted bacteria to form more biofilm compared to that under 30°C. In addition, the amount of EPS such as extracellular polysaccharides and extracellular proteins also increased, which also enhanced bacterial adhesion and biofilm biomass. This study provided valuable clues to better understand the effect of c-di-GMP on the bacterial adhesion and biofilm formation of S. putrefaciens WS13 under cold stress. Research on the mechanism of adhesion and biofilm formation of S. putrefaciens WS13 regulated by c-di-GMP under cold stress also laid a theoretical foundation for the development of technology to control the hazard caused by biofilm.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
RX: data curation, writing—original draft preparation, writing—review and editing, and visualization. JYan: conceptualization, methodology, validation, investigation, resources, data curation, writing—review and editing, and visualization. JM and JYe: visualization. JX: conceptualization, methodology, validation, investigation, resources, data curation, writing—review and editing, visualization, supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31972142) and the Science and Technology Commission of Shanghai Municipality (20DZ2292200 and 19DZ1207503).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gao X, Li PY, Mei J, Xie J. TMT-based quantitative proteomics analysis of the fish-borne spoiler Shewanella putrefaciens subjected to cold stress using LC-MS/MS. J Chem-NY. (2021) 2021:8876986. doi: 10.1155/2021/8876986
2. Ye JX, Yang SP, Qian YF, Xie J. Effect of cell-free supernatant from Aeromonas sobria on the spoilage of Shewanella putrefaciens in Pacific white shrimp (Litopenaeus vannamei) with the influence of temperature fluctuation. Appl Sci-Basel. (2019) 9:587. doi: 10.3390/app9030587
3. Yi Z, Xie J. Assessment of spoilage potential and amino acids deamination & decarboxylation activities of Shewanella putrefaciens in bigeye tuna (Thunnus obesus). LWT-Food Sci Technol. (2021) 156:113016. doi: 10.1016/j.lwt.2021.113016
4. Kang CH, Shin Y, Jeon H, Choi JH, Jeong S, So JS. Antibiotic resistance of Shewanella putrefaciens isolated from shellfish collected from the west sea in Korea. Mar Pollut Bull. (2013) 76:85–8. doi: 10.1016/j.marpolbul.2013.09.025
5. Li M, Lin H, Khan MN, Wang JX, Kong LH. Effects of bacteriophage on the quality and shelf life of Paralichthys olivaceus during chilled storage. J Sci Food Agric. (2014). (6475) 94:1657–62. doi: 10.1002/jsfa.6475
6. Yan J, Xie J. Comparative proteome analysis of Shewanella putrefaciens WS13 mature biofilm under cold stress. Front Microbiol. (2020) 11:1225. doi: 10.3389/fmicb.2020.01225
7. Sahli C, Moya SE, Lomas JS, Gravier-Pelletier C, Briandet R, Hemadi M, et al. Recent advances in nanotechnology for eradicating bacterial biofilm. Theranostics. (2022) 12:2383–405. doi: 10.7150/thno.67296
8. Singh BP, Ghosh S, Chauhan A. Development, dynamics and control of antimicrobial-resistant bacterial biofilms: a review. Environ Chem Lett. (2021) 19:1983–93. doi: 10.1007/s10311-020-01169-5
9. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. (2010) 8:623–33. doi: 10.1038/nrmicro2415
10. Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S, et al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. (2016) 14:563–75. doi: 10.1038/nrmicro.2016.94
11. Feng Q, Luo L, Chen X, Zhang K, Fang F, Xue Z, et al. Facilitating biofilm formation of Pseudomonas aeruginosa via exogenous N-Acy-L-homoserine lactones stimulation: Regulation on the bacterial motility, adhesive ability and metabolic activity. Bioresource Technol. (2021) 341:125727. doi: 10.1016/j.biortech.2021.125727
12. Monds RD, O'Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. (2009) 17:73–87. doi: 10.1016/j.tim.11, 001.
13. Soares A, Gomes LC, Mergulhao FJ. Comparing the recombinant protein production potential of planktonic and biofilm cells. Microorganisms. (2018) 6:48. doi: 10.3390/microorganisms6020048
14. Bisht K, Moore JL, Caprioli RM, Skaar EP, Wakeman CA. Impact of temperature-dependent phage expression on Pseudomonas aeruginosa biofilm formation. NPJ Biofilms Microbi. (2021) 7:22. doi: 10.1038/s41522-021-00194-8
15. Sun SN, Pandelia ME. HD-[HD-GYP] Phosphodiesterases: Activities and evolutionary diversification within the HD-GYP family. Biochemistry. (2020) 59:2340–50. doi: 10.1021/acs.biochem.0c00257
16. Wu C, Cheng YY, Yin H, Song XN Li WW, Zhou XX, et al. Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci Rep. (2013) 3:1945. doi: 10.1038/srep01945
17. Liu XB, Cao B, Yang L, Gu JD. Biofilm control by interfering with c-di-GMP metabolism and signaling. Biotechnol Adv. (2022) 56:107915. doi: 10.1016/j.biotechadv.2022.107915
18. Fu Y, Yu ZQ, Zhu L, Li Z, Yin W, Shang XD, et al. The multiple regulatory relationship between RNA-chaperone Hfq and the second messenger c-di-GMP. Front Microbiol. (2021) 12:689619. doi: 10.3389/fmicb.2021.689619
19. Van Hoek ML, Hoang KV, Gunn JS. Two-component systems in francisella species. Front Cell Neurosci. (2019) 9:198. doi: 10.3389/fcimb.2019.00198
20. Isenberg RY, Christensen DG, Visick KL, Mandel MJ. High levels of cyclic diguanylate interfere with beneficial bacterial colonization. MBio. (2022) 13:22. doi: 10.1128/mbio.01671-22
21. Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. (1987) 325:279–81. doi: 10.1038/325279a0
22. Sondermann H, Shikuma NJ, Yildiz FH. You've come a long way: c-di-GMP signaling. Curr Opin Microbiol. (2012) 15:140–6. doi: 10.1016/j.mib.12, 008.
23. Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. (2017) 15:271–84. doi: 10.1038/nrmicro.2016.190
24. Townsley L, Yildiz FH. Temperature affects c-di-GMP signaling and biofilm formation in Vibrio cholerae. Environ Microbiol. (2015) 17:4290–305. doi: 10.1111/1462-2920.12799
25. Ha DG, O'Toole GA. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr. (2015) 3:14. doi: 10.1128/microbiolspec.MB-0003-2014
26. Liu C, Sun D, Liu JW, Chen Y, Zhou XG, Ru YR, et al. cAMP and c-di-GMP synergistically support biofilm maintenance through the direct interaction of their effectors. Nat Commun. (2022) 13:1493. doi: 10.1038/s41467-022-29240-5
27. Chen L, Yang S, Qian Y, Xie J. Sequencing and analysis of the S. putrefaciens WS13 genome J Biob Mater Bioener. (2019) 13:182–7. doi: 10.1166/jbmb.2019.1850
28. Yang A, Tang WS, Si TY, Tang JX. Influence of physical effects on the swarming motility of Pseudomonas aeruginosa. Biophys J. (2017) 112:1462–71. doi: 10.1016/j.bpj.02, 019.
29. Dunker K, Canny SGD, Nordgard CT, Dague E, Formosa-Dague C, Bakke I, et al. Elucidating bacterial adhesion to mucosal surface by an original AFM approach. BMC Microbiol. (2021) 21:244. doi: 10.1186/s12866-021-02303-1
30. Gieroba B, Krysa M, Wojtowicz K, Wiater A, Pleszczyriska M, Tomczyk M, et al. The FT-IR and Raman spectroscopies as tools for biofilm characterization created by Cariogenic streptococci. Int J Mol Sci. (2020) 21:3811 doi: 10.3390/ijms21113811
31. Li YF, Tan L, Guo X, Zhang P, Malakar PK, Ahmed F, et al. Acidic electrolyzed water more effectively breaks down mature Vibrio parahaemolyticus biofilm than DNase I. Food Control. (2020) 117:107312. doi: 10.1016/j.foodcont.2020.107312
32. Pérez-Mendoza D, Aragón IM, Prada-Ramírez HA, Romero-Jiménez L, Ramos C, Gallegos MT, et al. Responses to elevated c-di-GMP levels in mutualistic and pathogenic plant-interacting bacteria. PLoS ONE. (2014) 9:e91645. doi: 10.1371/journal.pone.0091645
33. Paksanont S, Sintiprungrat K, Yimthin T, Pumirat P, Peacock SJ, Chantratita N, et al. Effect of temperature on Burkholderia pseudomallei growth, proteomic changes, motility and resistance to stress environments. Sci Rep. (2018) 8:9167. doi: 10.1038/s41598-018-27356-7
34. Bosch A, Serra D, Prieto C, Schmitt J, Naumann D, Yantorno O. Characterization of bordetella pertussis growing as biofilm by chemical analysis and FT-IR spectroscopy. Appl Microbiol Biotechnol. (2006) 71:736–47. doi: 10.1007/s00253-005-0202-8
35. Pazdzior E. Shewanella putrefaciens-a new opportunistic pathogen of freshwater fish. J Vet Res. (2016) 60:429–34. doi: 10.1515/jvetres-2016-0064
36. Yan J, Yang Z, Xie J. Comparative transcriptome analysis of Shewanella putrefaciens WS13 biofilms under cold stress. Front Cell Infect Microbiol. (2022) 12:851521. doi: 10.3389/fcimb.2022.851521
37. Chen L, Yu H, Yang SP, Qian YF, Xie J. Study on the mechanism of cold tolerance of the strain Shewanella putrefaciens WS13 through fatty acid metabolis. Nanosci Nanotech Let. (2020) 11:1718–23. doi: 10.1166/nnl.2019.3055
38. Bocsanczy AM, Achenbach UCM, Mangravita-Novo A, Yuen JMF, Norman DJ. Comparative effect of low temperature on virulence and twitching motility of Ralstonia solanacearum strains resent in Florida. Phytopathology. (2012) 102:185–94. doi: 10.1094/PHYTO-05-11-0145
39. Cordero N, Maza F, Navea-Perez H, Aravena A, Marquez-Fontt B, Navarrete P, et al. Different transcriptional responses from slow and fast growth rate strains of Listeria monocytogenes adapted to low temperature. Front Microbiol. (2016) 7:229. doi: 10.3389/fmicb.2016.00229
40. Xin LY, Zeng YK, Sheng S, Chea RA, Liu Q, Li HY, et al. Regulation of flagellar motor switching by c-di-GMP phosphodiesterases in Pseudomonas aeruginosa. J Biol Chem. (2019) 294:13789–99. doi: 10.1074/jbc.RA119.009009
41. Raterman EL, Shapiro DD, Stevens DJ, Schwartz KJ, Welch RA. Genetic analysis of the role of yfiR in the ability of Escherichia coli CFT073 to control cellular cyclic dimeric GMP levels and to persist in the urinary tract. Infect Immun. (2013) 81:3089–98. doi: 10.1128/IAI.01396-12
42. Wang RJ, Wang FB, He R, Zhang RJ, Yuan JH. The second messenger c-di-GMP adjusts motility and promotes surface aggregation of bacteria. Biophys J (2018) 115. (2018) 2242–9. doi: 10.1016/j.bpj.10020
43. Wang XY, Xie J. Comparison of physicochemical changes and water migration of Acinetobacter johnsonii, Shewanella putrefaciens, and Cocultures from spoiled Bigeye tuna (Thunnus obesus) during cold storage. Front Microbiol. (2021) 12:727333. doi: 10.3389/fmicb.2021.727333
44. Nieto V, Partridge JD, Severin GB, Lai RZ, Waters CM, Parkinson JS, et al. Under elevated c-di-GMP in Escherichia coli, YcgR alters flagellar motor bias and speed sequentially, with additional negative control of the flagellar regulon via the adaptor protein RssB. J Bacteriol. (2020) 202:00578–19. doi: 10.1128/JB.00578-19
45. Asker D, Awad TS, Baker P, Howell PL, Hatton BD. Non-eluting, surface-bound enzymes disrupt surface attachment of bacteria by continuous biofilm polysaccharide degradation. Biomaterials. (2018) 167:168–76. doi: 10.1016/j.biomaterials.2018.03.016
46. Laganenka L, Lopez ME, Colin R, Sourjik V. Flagellum-mediated mechanosensing and RflP control motility state of pathogenic Escherichia coli. MBio. (2020) 11:e02269–19. doi: 10.1128/mBio.02269-19
47. Li Q, Becker T, Zhang RY, Xiao TF. Investigation on adhesion of Sulfobacillus thermosulfidooxidans via atomic force microscopy equipped with mineral probes. Colloids Surf B. (2019) 173:639–46. doi: 10.1016/j.colsurfb.10, 046.
48. Ji CC, Zhou H, Deng SK, Chen KY, Dong XY, Xu XH, et al. Insight into the adhesion propensities of extracellular polymeric substances (EPS) on the abiotic surface using XDLVO theory. J Environ Chem Eng. (2021) 9:106563. doi: 10.1016/j.jece.2021.106563
49. Ruas-Madiedo P, Gueimonde M. de los Reyes-Gavilan CG, Salminen S. Short communication: Effect of exopolysaccharide isolated from “viili” on the adhesion of probiotics and pathogens to intestinal mucus. J Dairy Sci. (2006) 89:2355–8. doi: 10.3168/jds.S0022-0302(06)72307-4
50. Xiao Y, Chen H, Nie L, He M, Peng Q, Zhu W, et al. Identification of c-di-GMP/FleQ-regulated new target genes, including cyaA, encoding adenylate cyclase, in Pseudomonas putida, mSystems. (2021) 6:e00295–21. doi: 10.1128/mSystems.00295-21
51. Ahmad I, Cimdins A, Beske T, Romling U. Detailed analysis of c-di-GMP mediated regulation of csgD expression in Salmonella typhimurium. BMC Microbiol. (2017) 17:27. doi: 10.1186/s12866-017-0934-5
52. Schaper S, Steinchen W, Krol E, Altegoer F, Skotnick D, Sogaard-Andersen L, et al. AraC-like transcriptional activator CuxR binds c-di-GMP by a PilZ-like mechanism to regulate extracellular polysaccharide production. P Natl Acad Sci USA. (2017) 114:E4822–31. doi: 10.1073/pnas.1702435114
53. Yang SS, Wu YC, Qu CC, Fein JB, He YZ, Huang QY, et al. et a. Quantitative analysis of the surficial and adhesion properties of the Gram-negative bacterial species Comamonas testosteroni modulated by c-di-GMP. Colloid Surface B. (2021) 198:111497. doi: 10.1016/j.colsurfb.2020.111497
54. Cheng YY, Wu C, Wu JY, Jia HL, Wang MY, Wang HY, et al. FlrA represses transcription of the biofilm-associated bpfA operon in Shewanella putrefaciens. Appl Environ Microb. (2017) 83:e02410–16. doi: 10.1128/AEM.02410-16
55. Diaz M, Castro M, Copaja S, Guiliani N. Biofilm formation by the Acidophile Bacterium Acidithiobacillus thiooxidans involves c-di-GMP pathway and pelexopolysaccharide. Genes. (2018) 9:113. genes9020113 doi: 10.3390/genes9020113
56. Almblad H, Randall TE, Liu F, Leblanc K, Groves RA, Kittichotirat W, et al. Bacterial cyclic diguanylate signaling networks sense temperature. Nat Commun. (2021) 12:1986. doi: 10.1038/s41467-021-22176-2
57. Purcell EB, Tamayo R. Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiol Rev. (2016) 40:753–73. doi: 10.1093/femsre/fuw013
58. Newell PD, Boyd CD, Sondermann H, O'Toole GAA. c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. (2011) 9:e1000587. doi: 10.1371/journal.pbio.1000587
Keywords: Shewanella putrefaciens, cold stress, c-di-GMP, adhesion, biofilm
Citation: Xiong R, Yan J, Mei J, Ye J and Xie J (2022) The enhanced expression of genes encoding diguanylate cyclases under cold stress contributes to the adhesion and biofilm formation of Shewanella putrefaciens WS13. Front. Nutr. 9:1076932. doi: 10.3389/fnut.2022.1076932
Received: 22 October 2022; Accepted: 15 November 2022;
Published: 01 December 2022.
Edited by:
Yueqi Wang, Chinese Academy of Fishery Sciences (CAFS), ChinaReviewed by:
Yangtai Liu, University of Shanghai for Science and Technology, ChinaJibing Li, Chinese Academy of Sciences (CAS), China
Copyright © 2022 Xiong, Yan, Mei, Ye and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Xie, anhpZUBzaG91LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Ruheng Xiong1,2,3,4,5†
Ruheng Xiong1,2,3,4,5† Jun Yan
Jun Yan Jing Xie
Jing Xie
