
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 22 December 2022
Sec. Nutrition and Food Science Technology
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.1062006
 Yan He1,2
Yan He1,2 Yu Wang1,2
Yu Wang1,2 Mengzhuo Zhang1,2
Mengzhuo Zhang1,2 Guangsen Liu1,2
Guangsen Liu1,2 Cong Tian1,2
Cong Tian1,2 Xiangbin Xu1,2
Xiangbin Xu1,2 Yonggui Pan1
Yonggui Pan1 Xuequn Shi1,2
Xuequn Shi1,2 Zhengke Zhang1,2
Zhengke Zhang1,2 Lanhuan Meng1,2*
Lanhuan Meng1,2*Extensive data have demonstrated that carotenoid accumulation in tomato fruit is influenced by environmental cues and hormonal signals. However, there is insufficient information on the mechanism of its transcriptional regulation, as many molecular roles of carotenoid biosynthetic pathways remain unknown. In this work, we found that the silence of the BEL1-like family transcription factor (TF) BEL1-LIKE HOMEODOMAIN 11 (SlBEL11) enhanced carotenoid accumulation in virus induced gene silencing (VIGS) analysis. In its RNA interference (RNAi) transgenic lines, a significant increase in the transcription level for the lycopene beta cyclase 2 (SlLCY-b2) gene was detected, which encoded a key enzyme located at the downstream branch of the carotenoid biosynthetic pathway. In Electrophoretic mobility shift assay (EMSA), SlBEL11 protein was confirmed to bind to the promoter of SlLCY-b2 gene. In addition, the dual-luciferase reporter assay showed its intrinsic transcriptional repression activity. Collectively, our findings added a new member to the carotenoid transcriptional regulatory network and expanded the functions of the SlBEL11 transcription factor.
In tomato (Solanum lycopersicum), degradation of chlorophyll and accumulation of carotenoids result in the red color of ripe tomato fruits. Carotenoids are a group of important natural pigments, present not only in plants, but also in algae, fungi, and bacteria (1–3). In plants, carotenoids serve numerous crucial functions. Notably, carotenoids are essential for photosynthesis of photosynthetic organisms, and also play a vital role in protecting photosynthetic organisms under excessive light conditions (4). Carotenoids also are an important source of phytohormones including abscisic acid (ABA), which is involved in many aspects of plant developmental processes including inducing seed dormancy and responding to abiotic stresses (5). Studies have also provided evidence that carotenoid derivatives mediate plant development and act as signaling molecules in response to environmental factors including strigolactones, β-cyclocitral, and dihydroactinidiolide, a mobile bypass signal and karrikins with structural similarity to strigolactones (6–8). Carotenoids also exert biologically important functions in humans. Carotenoids are the main dietary source of vitamin A and also provide antioxidants to reduce the risk of cancer and alleviate chronic diseases (9, 10). In particular, β-carotene, lycopene, lutein and zeaxanthin have been widely identified as biological antioxidants and immune system modulators. The potential protective effect of high β-carotene intake in the development of cardiovascular diseases is also supported by earlier epidemiological studies (11). There have also been a large body of researches about carotenoids and their effects on cardiovascular diseases and infectious diseases such as HIV infections (12). As the main constituents of macular pigment, lutein and zeaxanthin are important for eye health. Their accumulation in the retina reduces the risk of macular degeneration, which is responsible for the main cause of blindness in the elderly (13).
Researches into carotenoids and their functions in human health have been paralleled by major advances in cloning core genes involved in carotenoid production. Taken together, it is these cognitive advancements that have ushered in a new era of elevated carotenoid levels in food. At present, highly conserved carotenoid biosynthetic pathway has been resolved in many plants (14). The major source of biosynthesis carotenoid molecules is methylerythritol-4-phosphate (MEP) pathway in plants (15). Geranylgeranyl diphosphate (GGPP) is ubiquitous and it is the isoprenoid precursor to generate carotenoids. GGPP is converted to phytoene, which is catalyzed by the phytoene synthase (PSY), a rate-limiting enzyme. This step has been subjected to genetic regulation (1). In tomato, both PSY1 and PSY2 are present in the fruit, but PSY1 functions in the formation of carotenoids during fruit ripening, while PSY2 primarily works in the formation of carotenoids in chloroplast-containing tissues (16). Then, the phytoene synthase catalyzed product, phytoene is catalyzed sequentially into ζ-carotene and prolycopene by the enzymes phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS), respectively (14). Isomerization of prolycopene to red-colored lycopene is carried out by carotene isomerase (CRTISO). Lycopene is further metabolized to produce β-carotene, zeaxanthin, violaxanthin, and neoxanthin, which is catalyzed by two classes of lycopene beta cyclases (LCY-bs, CYC-b). Specifically, LCYbs are thought to play a critical key role at the branching point of the carotenoid biosynthetic pathway, including the conversion of red lycopene to the downstream bright yellow or orange lutein (17). The existence of two types of LCY-bs genes have been confirmed in many plants. Notably, LCY-b1 is mainly expressed in leaf tissue, while LCY-b2 is primarily expressed in fruit tissue (18). Studies have demonstrated that altered expression levels of LCYbs cause abnormal lycopene accumulation in fruits or flowers (19). It has been reported that lycopene accumulation in red grapefruit (Citrus paradisi) results from downregulation of the citrus chromosomal-specific LCY-b2 (17). In citrus calli and tomato fruits, the overexpression of CsERF061 increases carotenoid accumulation and promotes fruits growth through directly binding to the promoter of LCY-b2 (20). Although the function of the promoter of LCY-b1 has been characterized in tomato, little information is available on the LCY-b2 promoter.
Numerous studies have proved that carotenoid biosynthesis is regulated by a variety of transcription factor families, including the R2R3-MYB, MADS-box, NAC, bHLH, SBP-box, AP2/ERF, HD-ZIP, NF-Y, and WRKY (21–24). The spontaneous ripening-related mutants in tomato, such as ripening-inhibitor (Rin), non-ripening (Nor), and colorless non-ripening (Cnr) promote our current understandings of the mechanisms underlying fruit ripening (25). Mutations at these loci can completely block normal ripening of tomato fruit. NOR-LIKE1 transcription factor is involved in lycopene accumulation by directly binding to SlSGR1 and SlGgpps2 genes and activating their expression (25). Overexpression of SlAPRR2 and SlGLK1 genes in tomato increased the number of plastids in fruit and carotenoid accumulation in fruit at ripening stage was higher than that of wild type (22). Silencing of SlNAC1 in tomato resulted in up-regulation of the expression of SIPSY1 and accumulation of total carotenoids. Besides, carotenoid accumulation was positively regulated by binding SlPSY1, SlZ-ISO, SlCRTISO promoters (23, 26, 27). However, the evaluation of RIN binding to promoters of target genes in ChIP experiments yielded inconsistent results (28) and the expression of SlPSY1 in FUL1-silenced and FUL2-silenced tomato fruits was not altered (29). Therefore, the roles of some transcription factors in regulating carotenoid biosynthesis still need to be further explored. A lot of regulatory factors of upstream products have been identified in the carotenoid metabolic pathway, while the research data of transcriptional regulation of downstream substances is insufficient.
In our experiment, the TF SlBEL11 gene had high expression level during ripening in tomato fruit. SlBEL11 silencing led to a pronounced immature phenotype, with fruit not reddening normally. The experimental results revealed that SlBEL11 was involved in the carotenoid accumulation of tomato fruit at the red stage by directly binding to the promoter of the LCY-b2 gene. Our results provided some evidence about the transcriptional regulation of tomato fruit carotenoid metabolism.
Tomato plants (Solanum lycopersicum cv. Micro Tom) and BEL11 RNAi transgenic plants from the previous study (30) were grown in a greenhouse at 25°C with 16 h light and 8 h dark alternating. Flowers were tagged at anthesis. Clean and healthy fruit samples were immediately used for experiments or stored at –80°C after quick freezing in liquid nitrogen until they were used for measurement.
pTRV1 and pTRV2 vectors were subjected to this experiment described by Liu et al. (31). A 420-bp specific fragment of the SlBEL11 gene corresponding to bases 778–1197 of the SlBEL11 gene, a 540-bp fragment of the SlPDS gene located in bases 764–1303 of the PDS gene, a 340-bp fragment of the SlLCY-b1 gene located in bases 537–876 of the SlLCY-b1 gene, and a 339-bp fragment of the SlLCY-b2 gene located in bases 571–909 of the SlLCY-b2 gene were PCR amplified from tomato cDNA using Kod-plus polymerase (TOYOBO, Japan) and primers shown in Supplementary Table 1. The PCR amplified products then were ligated into the pTRV2 plasmids which were digested by SacI and XhoI. Then, pTRV1 vector plasmid and the recombinant plasmids identified by sequencing were transferred into Agrobacterium tumefaciens strain GV3101. The preparation of agrobacterium infection solution and the method of infecting tomato sprouts were described by Meng et al. (30). Tomato sprouts infiltrated with empty pTRV2 and pTRV2-PDS were set as negative and positive controls (31).
The extraction of total RNA was completed according to the manufacturers’ method described by the RNAprep Pure Plant Kit (TIANGEN Biotech, Hainan, China). In this experiment, containers and consumables were treated with RNase inactivation to ensure the integrity of the extracted RNA. The next steps were required to be completed on ice to confirm that the samples were not degraded. The RNA was subjected to cDNA synthesis with Fastking One Step RT-PCR Kit (TIANGEN Biotech, Hainan, China) following the instructions of the reagent manufacturer. qRT-PCR was conducted to examine the mRNA levels of the genes. In total, 1 μL of cDNA, together with 5 μL of SYBR green mix (TIANGEN Biotech, Hainan, China), 0.3 μL each forward and reverse primer, were added into the PCR mixtures (with the total volume of 10 μL). The PCR reactions were performed in A CFX Connect Detection System (Bio-Rad, CA, USA). Actin was selected as the reference gene. The method of 2–ΔΔCt was adopted for calculation of results. The primer sequences were provided in Supplementary Table 1.
Fresh samples were pulverized with liquid nitrogen. In total, 50 (±1) mg of sample powder was extracted for 24 h with 1 mL of extraction solution (methanol:water:formic acid = 75:23:2) and vortexed for 15 min. After centrifugation, add 0.5 ml extraction solution to the residue, repeat shaking and centrifugation, and combine the supernatant. Concentrate and dry in the concentrator at 4°C, reconstitute with 200 μL 80% methanol. Centrifuge again, store the supernatant in the bottle for LC-MS/MS analysis. The mobile phase consisted of 0.04% acetic acid in water (solvent A) and 0.04% acetic acid in acetonitrile (solvent B). The flow rate was 0.4 ml/min and the column temperature was 40°C. The parameters of the gradient separation were as follows: increase 5% B–95% B in 10 min, hold for 1 min and change back to a 5% B concentration in 6 s, hold for 2.9 min.
The coding sequence of the SlBEL11 HD-box was PCR amplified. Then, the coding sequence was cloned into pGEX-4T-1 which was fused with a glutathione S-transferase (GST) tag. The recombinant construct identified by sequencing was transformed into Escherichia coli strain Rosetta (DE3). Protein expression and purification were completed referring to the method reported by Xiao et al. (32).
The probes containing TTGACTTGACatagtGTCA motif sequence from the promoter of SlLCY-b2 were biotinylated using a DNA 3′ End Biotinylation Kit (Thermo Fisher Scientific, Waltham, USA). The same sequence without labeled DNA fragment was a competitor. The TTGACTTGACatagtGTCA DNA fragments were modified into TTTTTTTTTTatagtTTTT and used as a mutant probe in the assay. EMSA was performed according to the manufacturer’s protocol provided by A LightShift Chemiluminescent EMSA kit (Thermo Scientific).
In this assay, the SlLCY-b2 promoter was cloned into the pGreenII 0800-LUC dual reporter gene vector (33). The SlBEL11 gene was cloned into the pEAQ vector (34) as effector. The constructed SlLCY-b2-pro-LUC effector and pEAQ-HT-SlBEL11 reporter plasmids were co-transformation of tobacco with Agrobacterium tumefaciens strain GV3101. The method was subject to Yin et al. (35). The Agrobacterium cells that has been transformed into the corresponding expression vector were inserted into the LB liquid medium containing kanamycin and gentamicin for cultivation (28°C, 240 rpm, 12–24 h). The Agrobacterium cells were collected and suspended in osmotic solution (10 mM MES, 10 mM MgCl2, and 150 μM acetosyringone, pH 5.6), and adjusted to an appropriate concentration. The bacteria containing effector and reporter were mixed in a ratio of 9:1, and then the mixture was injected into tobacco leaves and cultured for 2–3 days. The ratio LUC/REN of two luciferase enzymes (LUC and REN) in leaves was determined by the dual luciferase reporting and analysis system (Promega). Each assay should contain at least 6 measurements.
The significance between the data was analyzed by t-test. The bar and line charts were plotted using GraphPad Prism 9 software.
The SlBEL11 belongs to the BEL1-like Homeodomain (BEL1-like) TFs family with high expression abundance in the red stage and involved in the regulation of chlorophyll synthesis and fruit pigmentation in tomato fruit (30). Compared with the control fruit, in addition to the observation that the SlBEL11 RNAi tomato fruits accumulate more chlorophyll at mature green stage, we also found that it still remained yellow at the red stage (Figure 1D). To further explore its possible role in tomato fruit, virus-induced gene silencing (VIGS) analysis was executed. In this experiment, a clearly demarcated red-yellow region was detected in fruits infected with the virus containing the TRV-SlPDS vector compared to tobacco rattle virus (TRV)-infected control and even red fruits. Among them, the tomato fruit injected with TRV-SlPDS was used as a positive control (Figure 1A). Similar to TRV-SlPDS tomato, tomato fruits injected with TRV-SlBEL11-containing vector virus also displayed mottled yellow and red areas, which were also separated by distinct borders. Gene expression analysis using quantitative real-time PCR (qRT-PCR) showed a reduction of approximately 80 and 90%, respectively, in SlPDS and SlBEL11 transcripts in the yellow areas of the TRV-SlPDS and TRV-SlBEL11 fruits, compared with the red TRV-Control fruits (Figures 1B, C). This mean that SlBEL11 may be involved in the regulation of carotenoid synthesis in tomato fruit.
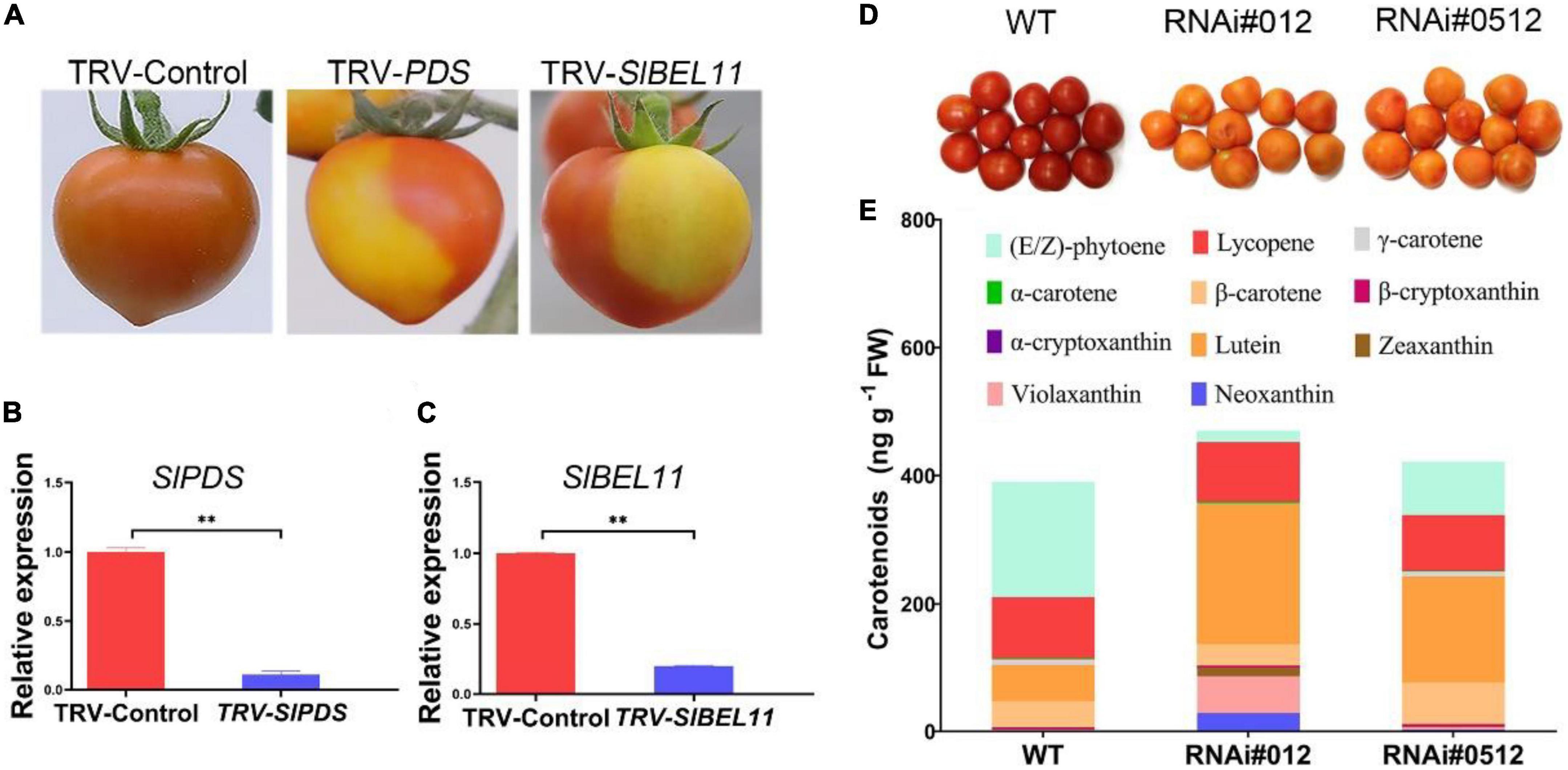
Figure 1. Silencing of the SlBEL11 gene affects carotenoid accumulation in tomato fruits. (A) Ripe fruit of plants infected with empty vector virus (TRV-Control), specific fragment of SlPDS gene (TRV-SlPDS), and specific fragment of SlBEL11 gene (TRV-SlBEL11). (B) Analysis of qRT-PCR for SlPDS gene in tomato fruits in TRV-Control (red sections) and TRV-SlPDS group (yellow sections). (C) qRT-PCR analysis for SlBEL11 gene in tomato fruits in TRV-Control (red sections) and TRV-SlBEL11 group (yellow sections). The internal reference was actin gene. The results were presented mean ± standard deviation (SD) of three independent replicates. **Indicated a P-value < 0.01 (Student’s t-test). (D) Tomato fruit phenotype of SlBEL11 RNAi and wild type (WT) at the red stage. The stage of tomato was defined by external and internal color of the tomato fruit. For the mature green stage, fruits will ripen to an acceptable level of horticultural quality. The entire surface of the fruits was either green or white, with no visible red. At the red stage, there was more than 90% of the surface showing red color. (E) Carotenoids content in SlBEL11 RNAi and WT tomato fruits determined at 5 days after breaker (Br + 5) stage.
In order to verify the hypothesis, we first carried out experiments to determine whether there was a difference in carotenoid content in the SlBEL11 RNAi and WT tomatoes. In this study, we found that the total carotenoid content in SlBEL11 RNAi tomatoes were much higher than that of WT at Br + 5 stage (Table 1). Compared to WT fruits, RNAi#012 and RNAi#0512 fruits showed a 90 and 54% reduction in phytoene, which was derived from upstream of the carotenoid synthesis pathway, respectively (Table 1). Whereas, the accumulation of substances downstream of carotenoid metabolism pathway was more than that of WT (Figure 1E). It is worth mentioning that large amounts of lutein accumulated in SlBEL11 silencing fruits and the content of lutein estimated in RNAi#012 (218.65 ng⋅g–1) and RNAi#0512 fruits (166.67 ng⋅g–1), was four times and three times that of WT fruits (56.67 ng⋅g–1), respectively. Though the SlBEL11 RNAi tomatoes showed comparable lycopene content to that of WT plants, the silencing of SlBEL11 gene resulted in a large increase of carotenoids including zeaxanthin, violaxanthin, and neoxanthin in SlBEL11 RNAi tomatoes. Remarkably, the most striking is that the accumulation of violaxanthin and neoxanthin in the RNAi#012 tomato fruits was detected to be approximately 42 and 35 times that of WT, respectively. Overall, the loss of SlBEL11 function tremendously affected carotenoid metabolism, which led to abnormalities in the color of the tomato fruit.
To explain how SlBEL11 regulated carotenoid synthesis at the genetic level, we examined the transcript levels of main enzyme genes from carotenoid biosynthesis pathway (PSY1, PSY2, PDS, ZDS, CRTISO, LCY-b1/b2, CYC-b, and LCY-e) by qRT-PCR (Figure 2). In the SlBEL11 RNAi tomato lines, the expression levels of PSY1, PSY2, PDS, and ZDS were much lower than those in the WT line (Figure 2B). These enzyme genes mainly came from the upstream of carotenoid biosynthesis pathway. On the contrary, the expression levels of LCY-b2 and LCY-e genes, which were derived from the downstream of the carotenoid metabolic pathway, were detected to be strikingly increased in the SlBEL11 RNAi tomato lines. Remarkably, the expression level of LCY-b2 transcript was elevated about nine and fivefold in RNAi#012 and RNAi#0512 tomato fruits respectively, compared to WT fruits. The obvious up-regulation of the LCY-b2 gene was consistent with the marked increase in the content of its catalytic products especially zeaxanthin, violaxanthin and neoxanthin. Similarly, the transcriptional level of LCY-e increased by threefold in both RNAi#012 and RNAi#0512 transgenic lines, which may be the reason for the massive accumulation of lutein downstream of the carotenoid metabolism pathway. Hence, silencing of SlBEL11 in tomato enhanced carotenoid content downstream, which may be caused by its transcriptional regulation of carotenoid pathway genes.
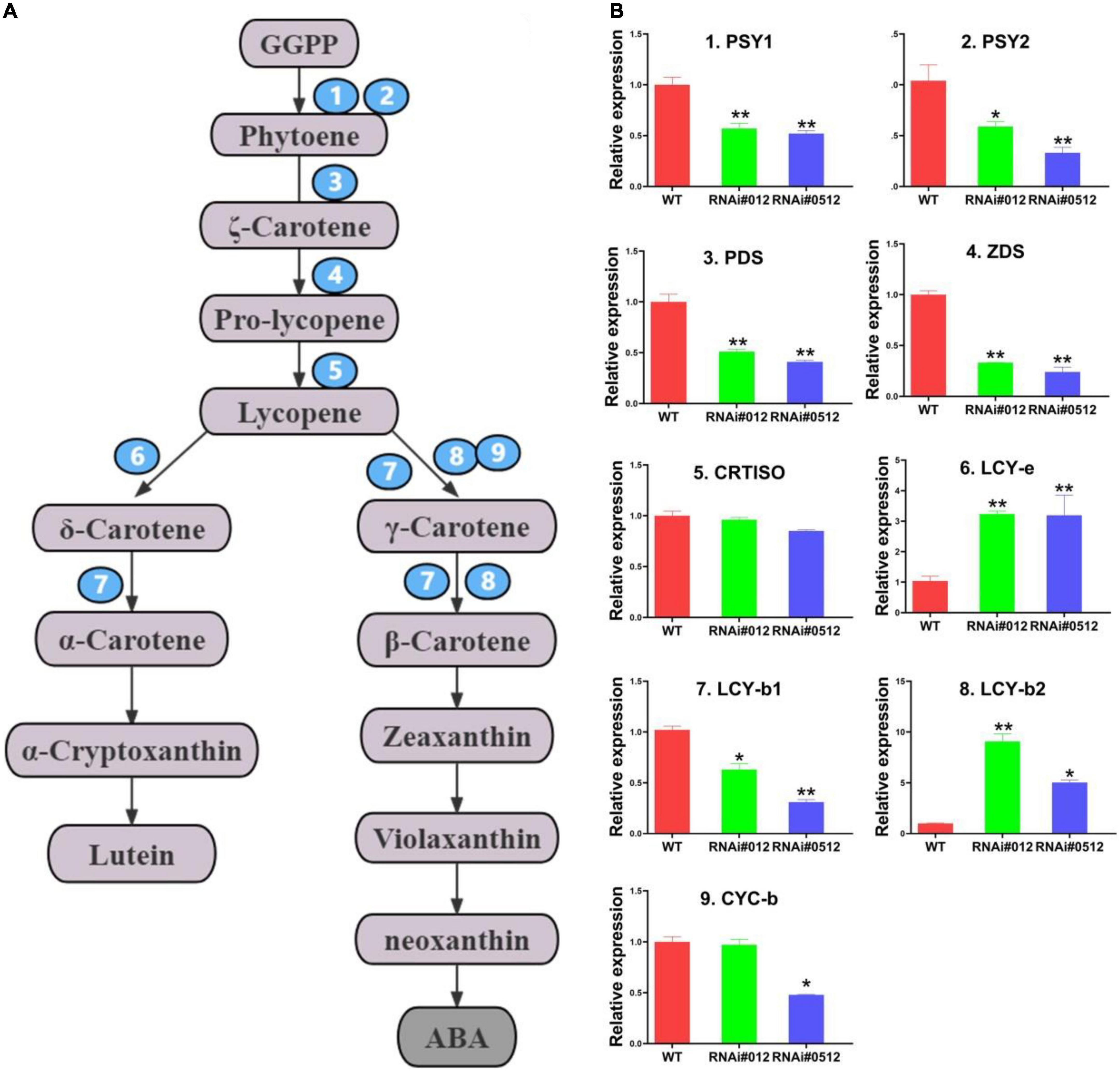
Figure 2. Relative transcript levels of genes related to carotenoids metabolic pathway from WT, RNAi#012, and RNAi#0512 tomato fruits in Br + 3 stage. (A) Schematic representation of the carotenoid metabolic pathway. Carotene isomerase; LCY-e. Abbreviations for other metabolites and enzymes have been described in the section “Introduction.” (B) Relative expression of core enzyme genes in carotenoids metabolic pathway in WT, RNAi#012, and RNAi#0512 tomatoes at Br + 3. Data contained three biological replicates, *representing P < 0.05 and **representing P < 0.01 (t-test).
To further explore the potential regulatory mechanisms of increased content of specific carotenoids and upregulation of specific genes from carotenoid biosynthetic pathway in SlBEL11 RNAi lines, we performed electrophoretic mobility-shift assay (EMSA). We found one possible motif TTGACTTGACatagtGTCA in the LCY-b2 promoter (Figure 3B). In EMSA, the capacity of the SlBEL11 protein of regulating the LCY-b2 promoter was analyzed. As shown in Figure 3, recombinant SlBEL11 protein physically bound to the LCY-b2 promoter fragment and caused changes in mobility. After adding more untagged competitor protein with the same sequence, the shifted band disappeared. In contrast, GST-SlBEL11 still showed bands with labeled probes when excess mutant probe was added (Figure 3B). Overall, in this experiment, SlBEL11 can directly bind to the TTGACTTGACatagtGTCA motif in the LCY-b2 promoter.
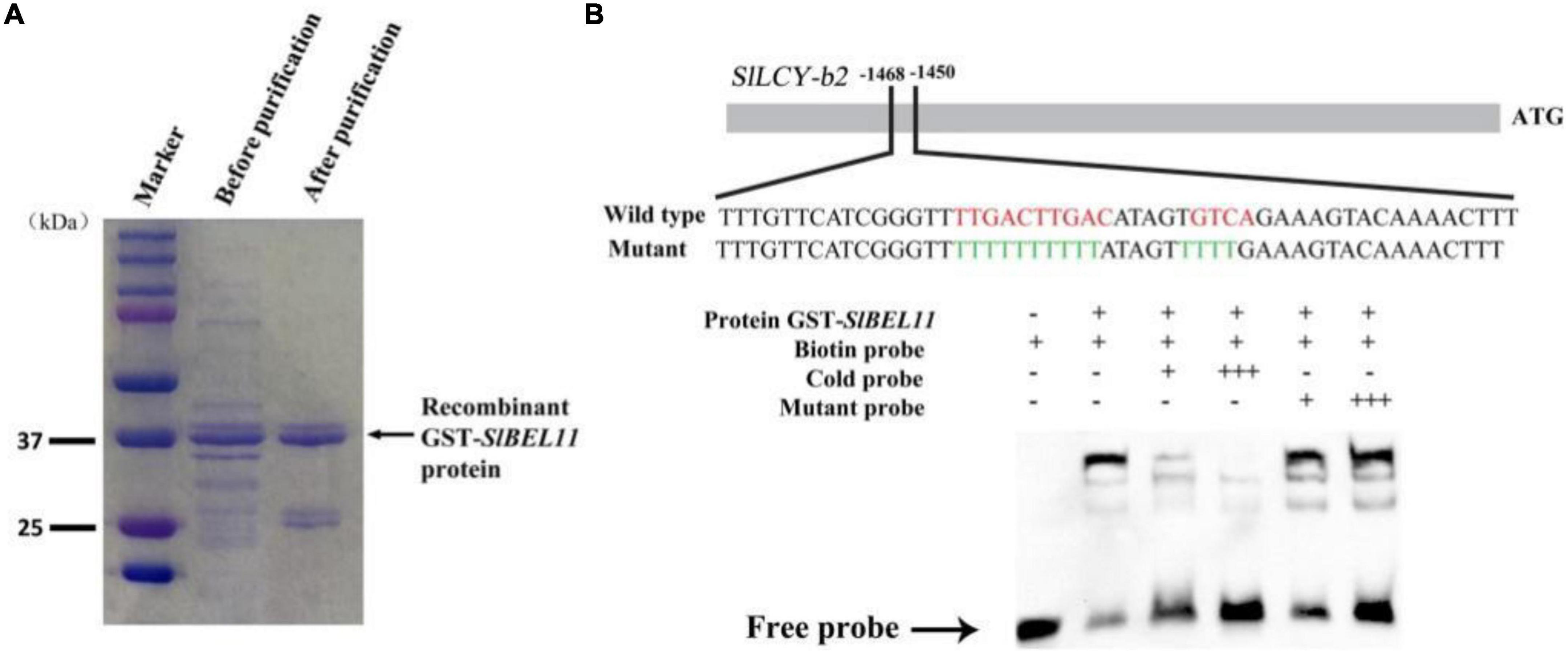
Figure 3. SlBEL11 bound to the promoter of SlLCY-b2 gene in vitro. (A) Affinity purification of recombinant GST-SlBEL11 protein by SDS-PAGE gel. (B) SlBEL11 bound to the promoter of SlLCY-b2 gene containing TTGACTTGACatagtGTCA element in EMSA. + and – indicate the presence and absence of the indicated probe or protein, respectively.
Through EMSA experiment, it has been determined that SlBEL11 can bind LCY-b2 promoter. But whether it has the activity of inhibiting or activating the LCY-b2 gene promoter needs to be further determined. To this end, the dual luciferase reporter assays were performed to examine whether it functioned to repress the LCY-b2 gene promoter. The dual luciferase reporter plasmids contained the SlLCY-b2 promoter were fused to LUC, which contained the TTGACTTGACatagtGTCA element. The CaMV35S promoter-driven REN served as an internal control, while the pEAQ-HT-SlBEL11 plasmid was used as an effector (Figure 4A). Compared with the control, expression of SlBEL11 significantly repressed the promoter activity of SlLCY-b2 (Figure 4B). Intriguingly, when the TTGACTTGACatagtGTCA element in promoter of SlLCY-b2 was mutated into TTTTTTTTTTatagtTTTT sequence (Figure 4C), there was no significant change in LUC/REN ratio (Figure 4D). From this we can conclude that the TTGACTTGACatagtGTCA element underlay the transcriptional repressive activity of SlBEL11 on the SlLCY-b2 gene promoter.
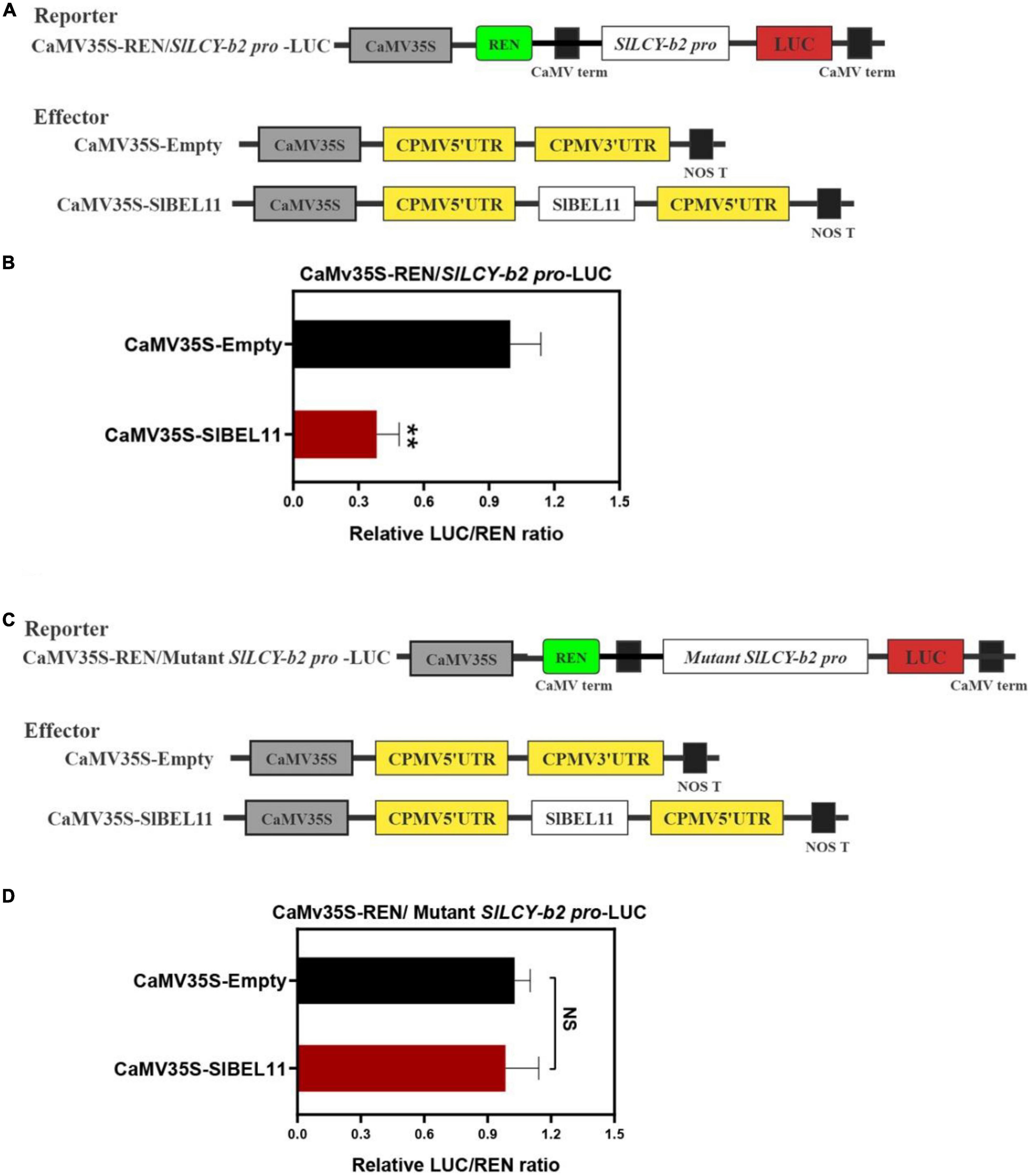
Figure 4. The dual luciferase reporter assay on SlBEL11 transcriptional repression of SlLCY-b2. (A,C), Schematic representation of reporters and effectors vector plasmids. (B) SlBEL11 suppressed the promoter activity of SlLCY-b2. (D) SlBEL11 can not suppress the activity of mutant SlLCY-b2 promoter. Values showed the means of six biological replicates, error bars represented SE. **Indicated that the difference between the values was extremely significant (P < 0.01) by Student’s t-test. NS, no significant difference.
Comparison of the amino acid sequence of SlLCY-b2 with known lycopene beta cyclase homologous protein sequences revealed a high degree of homology to other LCY-bs (Supplementary Figure 1). Particularly, SlLCY-b2 showed 87% identity with the amino acid sequence of SlLCY-b1, which predicted functional similarity between the two genes. Studies have reported that up-regulation of the SlLCY-b1 gene caused fruit orange pigmentation, which was accompanied by increased β-carotene content. Further VIGS experiment was conducted to investigate the possible function of SlLCY-b2. In this experiment, silencing of the positive control PDS gene in Micro-Tom tomato plants results in aberrant carotenoid biosynthesis, conferring a leaf photo-bleaching phenotype (Figure 5A), which ensured that TRV clones caused gene silencing. Tomato infected with TRV-SlLCY-b1 developed a yellow spotted phenotype in leaves about 6-week post-Agro-infiltration (Figure 5A). When SlLCY-b1 and SlLCY-b2 genes were silenced at the same time, tomato leaves developed a more pronounced yellow phenotype, suggesting that carotenoid synthesis was affected seriously (Figure 5A). However, no apparent phenotype was detected in tomato leaves infiltrated with TRV-SlLCY-b2. Then, RT-qPCR was performed to analyze the effect of VIGS on SlPDS, SlLCY-b1, SlLCY-b2. In tomato plants infiltrated with TRV-PDS, the transcript level of the PDS gene were detected to be reduced by 96% compared with the TRV infected controls, which was consistent with the photo-bleaching phenotype of PDS-silenced tomato plants (Figures 5A, B). Contrasted with TRV-Control plants, the gene expression of LCY-b1 in the leaves of TRV-LCYb1 tomato plants was down-regulated by about 99%, which was comparable to the expression level of LCY-b1 in TRV-LCYb1 + b2 plants (Figures 5C, E). However, in TRV-LCYb2 tomato plants, the expression level of the LCYb2 gene changed slightly, with a silencing level of 34%, which may result in invisible phenotype in its leaves (Figure 5D). In TRV-LCYb1 + b2 tomato plants, LCY-b1 and LCY-b2 genes were silenced to varying degrees, and the silencing efficiencies were 99% and 54%, respectively, relative to the control plants (Figures 5E, F). These results may suggest that down-regulation of the LCY-b2 gene, resulting in inhibition of carotenoid biosynthesis, was partly responsible for the yellowing phenotype of TRV-LCYb1 + b2 tomato plants.
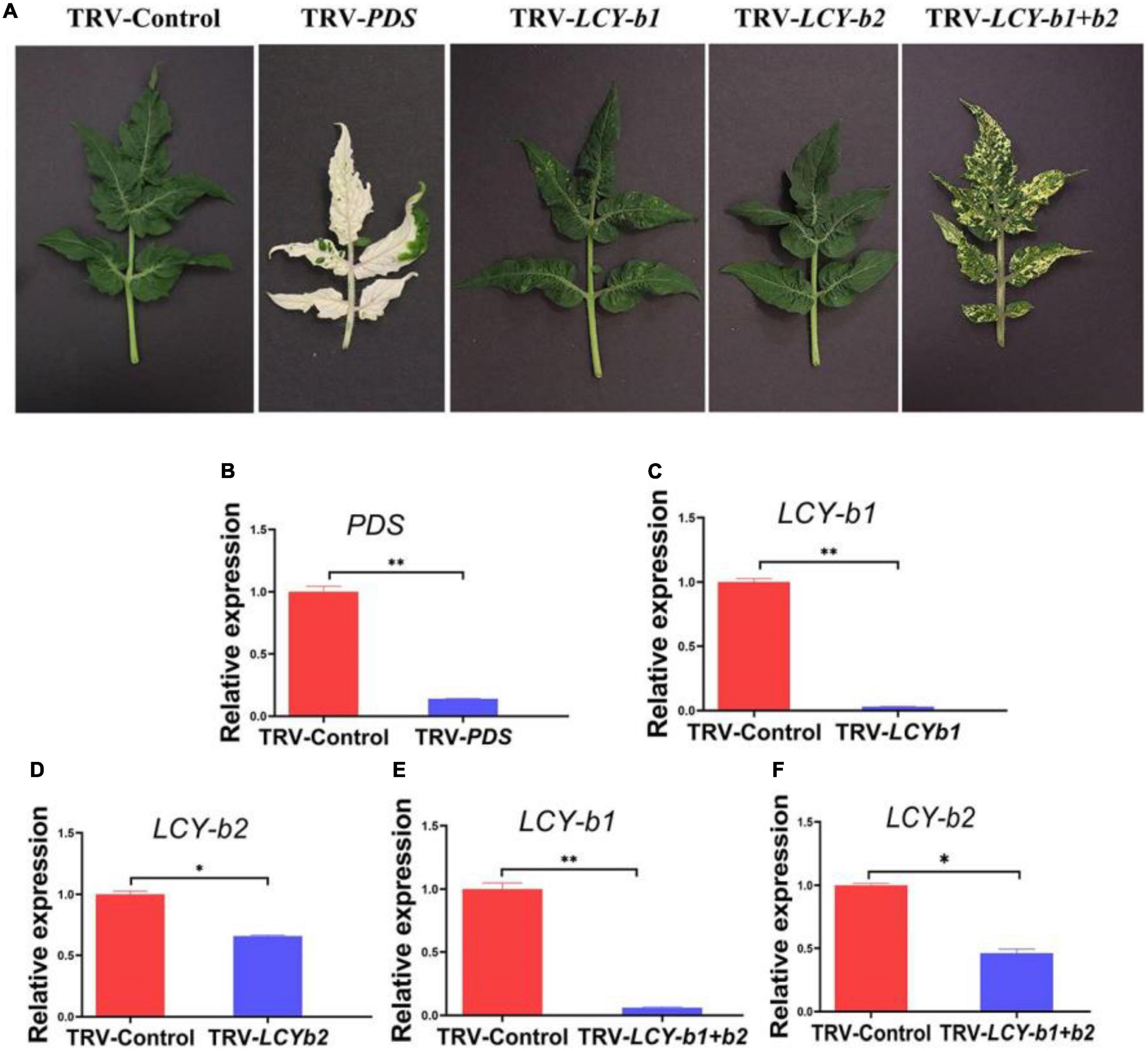
Figure 5. VIGS analysis of LCY-bs. (A) The excised leaves of 6-week-old tomato plants infected separately with vectors containing no insert (TRV-Control), specific sequences (TRV-PDS; TRV-SlLCY-b1; TRV-SlLCY-b2: TRV-SlLCY-b1 + b2). (B) Analysis of relative gene expression for PDS gene of tomato leaves in TRV-Control and TRV-PDS groups. (C) Analysis of relative gene expression for LCY-b1 gene in tomato leaves in TRV-Control and TRV-LCY-b1 groups. (D) Analysis of relative gene expression for LCY-b2 gene in tomato leaves in TRV-Control and TRV-LCY-b2 groups. (E) Analysis of relative gene expression for LCY-b1 gene in tomato leaves in TRV-Control and TRV- LCY-b1 + b2 groups. (F) Analysis of relative gene expression for LCY-b2 gene in tomato leaves in TRV-Control and TRV-LCY-b1 + b2 groups. The internal reference was actin gene. The values were expressed as the mean ± SD of three independent repeats. *Indicated a P-value < 0.05; **Indicated a P-value < 0.01 (t-test).
The BEL1-like family (BELL) of transcription factors is ubiquitous in plant and typically plays important roles in cell wall formation, flower and leaf development (36). Previous studies reported that the silence of SlBEL11 resulted in the dark green phenotype of immature tomato fruit, which explained its transcriptional regulation role in chloroplast development and chlorophyll synthesis (30). However, there is no evidence that SlBEL11 regulates carotenoid accumulation of tomato fruits. A total of 13 BEL1-like transcription factors expressed in various organs and developmental stages in tomato, among which SlBEL11 has a higher expression level during ripening stage, which indicates that it has a certain function at the ripening process. In our experiment, it was observed that SlBEL11 gene distinctly affected fruit coloration when the ripening candidate transcription factor SlBEL11 was silenced by VIGS approach. Therefore, it was speculated that SlBEL11 might be involved in the regulation of genes related to pigment formation, or the expression of ripening genes, or the hormone signal transduction process. Later transgenic tomatoes confirmed SlBEL11 as a transcriptional regulator of tomato fruit carotenoid accumulation.
In our work, we found that SlBEL11 RNAi tomato fruits were yellow at the red stage, while WT tomato fruits were all red, suggesting that silencing of SlBEL11 gene affected fruit pigment accumulation (Figure 1D). Further experiments revealed that the total amount of major carotenoids was elevated in SlBEL11 RNAi tomato fruits (Table 1). There were many kinds of carotenoids, among which red lycopene and yellow β-carotene are dominant in tomato fruit (37, 38). Our results demonstrate that silencing of SlBEL11 gene resulted in a decrease in upstream carotenoids and an increase in downstream carotenoids (Figure 1E). It has been reported that the accumulation of fruit pigments was determined by the expression of genes related to carotenoid synthesis (39). Compared to WT, phytoene was detected considerably dropped in BEL11 RNAi tomato fruits (Table 1), which may be due to the remarkable decrease in the expression of PSY1 and PSY2 in the carotenoid synthesis pathway. The increased lutein, zeaxanthin, violaxanthin and neoxanthin in SlBEL11 RNAi fruits may provide explanation for the yellow phenotype of SlBEL11 RNAi fruits. Lutein, the product of lycopene catalyzed by LCY-e enzyme, increased threefold in SlBEL11 RNAi fruits relative to WT (Figure 2). The increase of zeaxanthin, violaxanthin and neoxanthin may be caused by the elevation of LCY-b2, which were nine and fivefold higher in RNAi#012 and RNAi#0512 than that in WT, respectively. Previous studies have demonstrated the down-regulation of SlLCY-b1 gene was the mechanism responsible for the massive accumulation of lycopene during fruit ripening, which turned the tomato’s red fruit to orange (40, 41). To date, however, the roles of SlLCYb2 in carotenoid biosynthesis are still unclear. SlLCY-b2 showed 87% identity with the amino acid sequence of SlLCY-b1 (Supplementary Figure 1), which predicted similar functions between SlLCY-b1 and SlLCY-b2. In VIGS experiment, the co-silencing of SlLCY-b1 and SlLCY-b2 genes severely affected the production of carotenoids in tomato leaves, resulting in yellow phenotype in leaves, which demonstrated that SlLCY-b2 was a key node controlling the carotenoid metabolic pathway. Moreover, this speculation was also confirmed by further EMSA experiment that SlBEL11 directly binds to the promoter of SlLCY-b2 containing TTGACTTGACatagtGTCA element (Figure 3). Furthermore, in this work, we showed that SlBEL11 possessed transcriptional repression activity of SlLCY-b2.
In addition to the regulation of transcription factors at the molecular level, the accumulation of carotenoids during fruit ripening is also affected by different hormones such as ethylene, auxin, ABA and brassinolide (42, 43). Therefore, it is also possible that SlBEL11 may regulate production of carotenoids including lutein by combined effect of ABA and ethylene. In this study, we also observed the phenomenon of premature senescence of tomato fruits, manifesting in accelerated fruit shedding. This may be due to the increased accumulation of downstream carotenoids providing the raw materials for ABA synthesis, but further experiments are needed to verify the specific mechanism (23, 44).
Taken together, our experimental results clarified that the transcription factor SlBEL11 could directly bind to the promoter of the target gene LCY-b2 and suppress its expression. Total or partial silencing of SlBEL11 will promote the expression of LCY-b2 gene. In addition, SlBEL11 gene silencing resulted in changes in the expression of core enzyme genes in the carotenoid metabolic pathway, including PSY1, PSY2, PDS, ZDS, LCY-b1/b2, CYC-b, and LCY-e. Among them, the relative gene expression of LCY-b2 gene was up-regulated most obviously. The upregulation of LCY-b2 expression contributed to increased carotenoid content in tomato fruit including zeaxanthin, violaxanthin and neoxanthin (Table 1). These evidences suggested one way in which the SlBEL11 transcription factor regulated the accumulation of carotenoids in tomato fruit by directly binding to the SlLCY-b2 gene to suppress its expression (Figure 6).
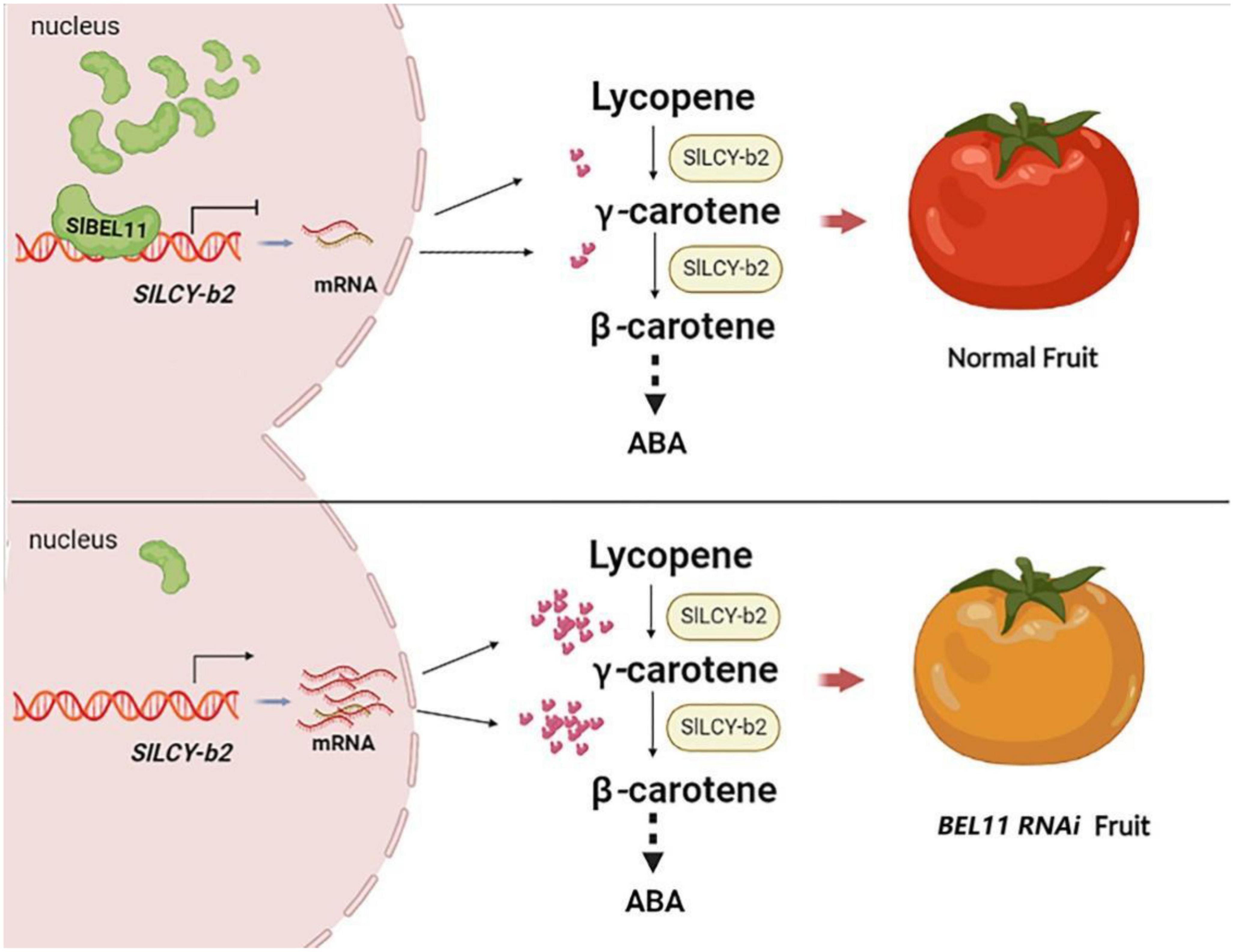
Figure 6. Working principle of the role of SlBEL11 for carotenoid biosynthesis in tomato fruit. SlBEL11 was a transcription factor that acted directly on the promoter of SlLCY-b2 and suppressed its transcription. When SlBEL11 was silenced, the gene encoding the SlLCY-b2 enzyme that catalyzed lycopene was upregulated, resulting in increased downstream carotenoid accumulation. Ultimately, SlBEL11 RNAi tomatoes yielded yellow fruits instead of red ones because of abnormal carotenoid accumulation.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author. Sequence data from this article can be found in GenBank data library under the following numbers: SlBEL11, Solanum lycopersicum (gene ID: 543692), Slactin (gene ID: 101260631), SlPDS (gene ID: 544073), SlPSY1 (gene ID: 543988), SlPSY2 (gene ID: 543964), SlZDS (Gene ID: 543629), SlCRTISO (Gene ID: 101267857), SlLCY-e (Gene ID: 544129), SlCYC-b (Gene ID: 543649), SlLCY-b1 (gene ID:544104), SlLCY-b2 (gene ID:101267662), AtLCY-b, Arabidopsis thaliana (U50739), OsLCY-b, Oryza sativa (XM_015771748).
YH: data acquisition and analysis, writing and modification of manuscript, and conduct of experiments. YW: assistance of software installation and use. MZ: instrument appointment. GL and CT: data collection and sorting. XX, YP, XS, and ZZ: methodology. LM: supervision, review of manuscript, acquisition of funds, and guidance for submission. All authors agreed with the above contribution description.
This research was supported by the National Natural Science Foundation of China (31901739 and 32260792), Hainan Provincial Key Research and Development Program of China (ZDYF2020044), Hainan Provincial Natural Science Foundation of China (322QN252), and the Starting Research Fund from the Hainan University, China [KYQD(2R)1956].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.1062006/full#supplementary-material
1. Fraser P, Bramley P. The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res. (2004) 43:228–65. doi: 10.1016/j.plipres.2003.10.002
2. Sun T, Yuan H, Cao H, Yazdani M, Tadmor Y, Li L. Carotenoid metabolism in plants: the role of plastids. Mol Plant. (2018) 11:58–74. doi: 10.1016/j.molp.2017.09.010
3. Yuan H, Zhang J, Nageswaran D, Li L. Carotenoid metabolism and regulation in horticultural crops. Hortic Res. (2015) 2:15036. doi: 10.1038/hortres.2015.36
4. Hashimoto H, Uragami C, Cogdell R. Carotenoids and photosynthesis. SubcellularBiochemistry. (2016) 79:111–3. doi: 10.1007/978-3-319-39126-7_4
5. Liotenberg S, North H, Marion-Poll A. Molecular biology and regulation of abscisic acid biosynthesis in plants. Plant Physiol Biochem. (1999) 37:341–50. doi: 10.1016/s0981-9428(99)80040-0
6. Adhikari E, Lee D, Giavalisco P, Sieburth L. Long-distance signaling in bypass1 mutants: bioassay development reveals the bps signal to be a metabolite. Mol Plant. (2013) 6:164–73. doi: 10.1093/mp/sss129
7. Lv F, Zhou J, Zeng L, Xing D. β-cyclocitral upregulates salicylic acid signalling to enhance excess light acclimation in Arabidopsis. J Exp Bot. (2015) 66:4719–32. doi: 10.1093/jxb/erv231
8. Cui X. Strigolactones, karrikins, and more: newly discovered molecules light up plant signaling. Mol Plant. (2014) 7:579–81. doi: 10.1093/mp/ssu026
9. Joanna F, Květoslava B. Potential role of carotenoids as antioxidants in human health and disease. Nutrients. (2014) 6:466–88. doi: 10.3390/nu6020466
10. Tanaka T, Shnimizu M, Moriwaki H. Cancer chemoprevention by carotenoids. Molecules. (2012) 17:3202. doi: 110.3390/molecules17033202
11. Poppel G. Epidemiological evidence for beta-carotene in prevention of cancer and cardiovascular disease. Eur J Clin Nutr. (1996) 50(Suppl. 3):S57–61.
12. Milani A, Basirnejad M, Shahbazi S, Bolhassani A. Carotenoids: biochemistry, pharmacology and treatment. Br J Pharmacol. (2017) 174:1290–324. doi: 10.1111/bph.13625
13. Bronwyn E, Sharon N, Gerald L, Victoria F. Lutein and zeaxanthin—food sources, bioavailability and dietary variety in age-related macular degeneration protection. Nutrients. (2017) 9:120. doi: 10.3390/nu9020120
14. Yuan Y, Ren S, Liu X, Su L, Wu Y, Zhang W, et al. SlWRKY35 positively regulates carotenoid biosynthesis by activating the MEP pathway in tomato fruit. New Phytol. (2022) 234:164–78. doi: 10.1111/nph.17977
15. Liu L, Shao Z, Zhang M, Wang Q. Regulation of carotenoid metabolism in tomato. Mol Plant. (2015) 8:28–39. doi: 10.1016/j.molp.2014.11.006
16. Fraser P, Kiano J, Truesdale M, Schuch W, Bramley P. Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid synthesis in ripening fruit. Plant Mol Biol. (1999) 40:687–98. doi: 10.1023/a:1006256302570
17. Lu S, Yin Z, Zheng X, Zhu K, Xu Q, Deng X. Isolation and functional characterization of a lycopene β-cyclase gene promoter from citrus. Front Plant Sci. (2016) 7:1367. doi: 10.3389/fpls.2016.01367
18. Berta A, Lorenzo Z, Rodrigo M. Molecular and functional characterization of a novel chromoplast-specific lycopene β-cyclase from Citrus and its relation to lycopene accumulation. J Exp Bot. (2009) 60:1783–97. doi: 10.1093/jxb/erp048
19. Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Nat Acad Sci USA. (2000) 97:11102–7. doi: 10.1073/pnas.190177497
20. Zhu K, Sun Q, Chen H, Mei X, Lu S, Ye J, et al. Ethylene activation of carotenoid biosynthesis by a novel transcription factor CsERF061. J Exp Bot. (2021) 72:3137–54. doi: 10.1093/jxb/erab047
21. Powell AL, Nguyen C, Hill T, Cheng K, Figueroa-Balderas R, Aktas H, et al. Uniform ripening encodes a golden 2-like transcription factor regulating tomato fruit chloroplast development. Science. (2012) 336:1711–5. doi: 10.1126/science.1222218
22. Pan Y, Bradley G, Pyke K, Ball G, Lu C, Fray R, et al. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. (2013) 161:1476–85. doi: 10.1104/pp.112.212654
23. Stanley L, Yuan Y. Transcriptional regulation of carotenoid biosynthesis in plants: so many regulators, so little consensus. Front Plant Sci. (2019) 10:1017. doi: 10.3389/fpls.2019.01017
24. Zhu F, Luo T, Liu C, Wang Y, Yang H, Yang W, et al. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. (2017) 216:178–92. doi: 10.1111/nph.14684
25. Gao Y, Wei W, Zhao X, Tan X, Fan Z, Zhang Y, et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Horticult Res. (2018) 216:178–92.
26. Fujisawa M, Shima A, Nakagawa A, Kitagawa A, Kimbara B. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell. (2014) 26:89–101. doi: 10.1105/tpc.113.119453
27. Shima Y, Kitagawa M, Fujisawa M, Nakano T, Kato H, Kimbara J, et al. Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol Biol. (2013) 82:427–38. doi: 10.1007/s11103-013-0071-y
28. Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. (2013) 25:371–86. doi: 10.1105/tpc.112.108118
29. Bemer M, Karlova R, Ballester A, Tikunov Y, Bovy A, Wolters-Arts M, et al. The tomato fruitfull homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell. (2012) 24:4437–51. doi: 10.1105/tpc.112.103283
30. Meng L, Fan Z, Zhang Q, Wang C, Gao Y, Deng Y, et al. BEL1-like homeodomain 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. Plant J. (2018) 94:1126–40. doi: 10.1111/tpj.13924
31. Liu Y, Schiff M, Dinesh-Kumar S. Virus-induced gene silencing in tomato. Plant J. (2002) 31:777–86. doi: 10.1046/j.1365-313x.2002.01394.x
32. Xiao Y, Kuang J, Qi X, Ye Y, Wu Z, Chen J, et al. A comprehensive investigation of starch degradation process and identification of a transcriptional activator MabHLH6 during banana fruit ripening. Plant Biotechnol J. (2018) 16:151–64. doi: 10.1111/pbi.12756
33. Hellens R, Allan A, Friel E, Bolitho K, Grafton K, Templeton M, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. (2005) 1:13. doi: 10.1186/1746-4811-1-13
34. Sainsbury F, Thuenemann E, Lomonossoff G. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J. (2009) 7:682–93. doi: 10.1111/j.1467-7652.2009.00434.x
35. Yin X, Allan A, Chen K, Ferguson I. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. (2010) 153:1280–92. doi: 10.1104/pp.110.157081
36. Liu Y, You S, Taylor-Teeples M, Li W, Schuetz M, Brady S, et al. BEL1-like homeodomain6 and knotted Arabidopsis thaliana 7 interact and regulate secondary cell wall formation via repression of revoluta. Plant Cell. (2014) 26:4843–61. doi: 10.1105/tpc.114.128322
37. Bruno A, Wetzel C. The early light-inducible protein (ELIP) gene is expressed during the chloroplast-to-chromoplast transition in ripening tomato fruit. J Exp Bot. (2004) 55:2541–8. doi: 10.1093/jxb/erh273
38. Li L, Yuan H. Chromoplast biogenesis and carotenoid accumulation. Arch Biochem Biophys. (2013) 539:102–9. doi: 10.1016/j.abb.2013.07.002
39. Fraser P, Enfissi E, Halket J, Truesdale M, Yu D, Gerrish C, et al. Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell. (2007) 19:3194–211. doi: 10.1105/tpc.106.049817
40. Pecker I, Gabbay R, Cunningham F, Hirschberg J. Cloning and characterization of the cDNA for lycopene β-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol Biol. (1996) 30:807–19. doi: 10.1007/bf00019013
41. Ronen G, Cohen M, Zamir D, Hirschberg J. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant delta. Plant J. (1999) 17:341–51. doi: 10.1046/j.1365-313x.1999.00381.x
42. Liu L, Jia C, Zhang M, Chen D, Chen S, Guo R, et al. Ectopic expression of a BZR1-1D transcription factor in brassinosteroid signalling enhances carotenoid accumulation and fruit quality attributes in tomato. Plant Biotechnol J. (2014) 12:105–15. doi: 10.1111/pbi.12121
43. Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, et al. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol. (2015) 15:114. doi: 10.1186/s12870-015-0495-4
Keywords: BEL1-LIKE HOMEODOMAIN 11, carotenoids, transcription factor, tomato fruit, virus induced gene silencing
Citation: He Y, Wang Y, Zhang M, Liu G, Tian C, Xu X, Pan Y, Shi X, Zhang Z and Meng L (2022) SlBEL11 affects tomato carotenoid accumulation by regulating SlLCY-b2. Front. Nutr. 9:1062006. doi: 10.3389/fnut.2022.1062006
Received: 05 October 2022; Accepted: 23 November 2022;
Published: 22 December 2022.
Edited by:
Lei Chen, Guangdong Ocean University, ChinaReviewed by:
Zhongqi Fan, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 He, Wang, Zhang, Liu, Tian, Xu, Pan, Shi, Zhang and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanhuan Meng, ✉ aHVhbmh1YW5haW5pMTAxMkAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.