
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr., 17 November 2022
Sec. Nutrition and Food Science Technology
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.1039762
This article is part of the Research TopicAdvances on Innovative Protein-based Complexes with Promising Functionality, Nutrient Retention and Encapsulation CapacityView all 10 articles
Small-angle X-ray scattering (SAXS) is a method for examining the solution structure, oligomeric state, conformational changes, and flexibility of biomacromolecules at a scale ranging from a few Angstroms to hundreds of nanometers. Wide time scales ranging from real time (milliseconds) to minutes can be also covered by SAXS. With many advantages, SAXS has been extensively used, it is widely used in the structural characterization of biomacromolecules in food science and technology. However, the application of SAXS in charactering the structure of food biomacromolecules has not been reviewed so far. In the current review, the principle, theoretical calculations and modeling programs are summarized, technical advances in the experimental setups and corresponding applications of in situ capabilities: combination of chromatography, time-resolved, temperature, pressure, flow-through are elaborated. Recent applications of SAXS for monitoring structural properties of biomacromolecules in food including protein, carbohydrate and lipid are also highlighted, and limitations and prospects for developing SAXS based on facility upgraded and artificial intelligence to study the structural properties of biomacromolecules are finally discussed. Future research should focus on extending machine time, simplifying SAXS data treatment, optimizing modeling methods in order to achieve an integrated structural biology based on SAXS as a practical tool for investigating the structure-function relationship of biomacromolecules in food industry.
Bioactive macromolecules, including peptides, nuclear acids, proteins, carbohydrates, and lipids, are not only essential components of food but also are the dominant substances for food to realize its functions. Meantime, biomacromolecules are widely used as additives for beverages, yogurt, cereal products, nuts, snacks, etc. to improve the food nutrition (1). Besides, other functional properties, such as gelation, foamability, water retention and emulsification, typical functions induced by structural changes of biomacromolecules, play significant roles in food processing. Therefore, the structure-function relationship of biomacromolecules is one of the most important topics in food science and technology.
Several experimental techniques are available for the structural characterization of biomacromolecules. High-resolution structural techniques, including crystallography, electron microscopy (EM) and nuclear magnetic resonance (NMR) have yielded incredibly detailed structural information at the atomic level on highly populated static states (2–4). However, due to the requirement of good crystals for crystallography, requirement of solubilized and monodisperse sample, and the low molecular weight requirement of NMR, a significant fraction of food biomacromolecules cannot be analysed using these three high-resolution methods (5). Furthermore, because of the highly heterogeneous and polydisperse of most food biomacromolecules, it is also challenging for conventional techniques, Fourier Transform Infrared spectroscopy (FTIR), dynamic light scattering (DLS) to monitor the change in the structure of biomacromolecules, especially the dynamics of self-assemble and hydrolysis. Therefore, developing of alternative structural characterization techniques for biomacromolecules with rapid response, easy sample preparation, data collection under near-native conditions, and in situ capability, is needed in food science and technology.
Small-angle X-ray scattering (SAXS) is a powerful tool for structural characterization of samples under resolutions ranging from a few Angstroms to hundreds of nanometers. SAXS is sensitive to both ordered and not-ordered features in the sample and it has several advantages over direct characteristic techniques in that: a very small amount of sample for measurement, rapid data collection, no crystallization or fixation requirement, high-throughput screening model and multiple in situ capacities, etc. (6). These features make SAXS an interesting technique for academic and industrial applications of highly interdisciplinary field, including life science, biomedicine, and biomaterial engineering.
Since SAXS was first used to study the geometry of typical milk proteins β-lactoglobulin tetramer, there have been nearly 60 years of research on food biomacromolecules. Several studies have reviewed the advances and applications of SAXS in food field (7–13). Gilbert summarized the principle and the latest activities in the application of SAXS to food colloids (14). This work provides information for the SAXS expert who is interested in applying this method to food colloids and the food scientist that wishes to gain knowledge of the former.
However, a work concluding the recent development and applications of modeling programs and in situ capabilities of SAXS for the structural characterization of food biomacromolecules is not available yet. Therefore, in the present review, the SAXS principle, theoretical calculations and modeling programs are summarized, technical advances in the experimental setups of in situ capabilities: coupled with chromatography, time-resolved, temperature, pressure, flow-through, are elaborated. Recent applications of SAXS for studying the structural properties of food biomacromolecules including proteins, carbohydrates and lipids are highlighted. Moreover, the limitations and prospects of SAXS are also discussed. We hope this review will provide reference information for food scientists who investigate the relationship between the structure and function of biomacromolecules using SAXS.
A typical bio-SAXS measurement is performed using a sample concentration at least ∼0.5–10 mg ml–1 with a ∼15–30 μl of volume, and generally takes less than a few minutes on a synchrotron beamline or dozens of minutes to hours using an in-house instrument (15). The principle of SAXS is that a collision between a monochromatic incident X-ray beam and a surface particle results in scattering of the beam in all directions. The one-dimensional (1D) scattered intensity I(q) and the average of the various conformers present in the population of scattered particles are recorded using a two-dimensional (2D) detector. The magnitude of the scattering vector q = 4πsinθ/λ, where θ is the half of the angle between incident and scattered beams. At small angles (θ < 5°), the inhomogeneity in the electron clouds can be observed, which will provide information about the size and shape of biomacromolecules in the sample (16). The “background” scattering from the buffer is independently measured and subtracted from that of the solution (17).
The radius of gyration (Rg) of biomacromolecules can be estimated directly from small q values using Guinier approximation (18), , where I(q) is the scattering intensity and I(0) is the forward scattering intensity. The pair-distance distribution function p(r) (19), corresponding to the paired set of distances between all electrons within the scattered particle, can be generated via indirect Fourier transform by using the GNOM (20), PRIMUS (21), BioXTAS RAW (22), and BIFT programs (23). By comparing with molecular weight determined from I(0), the volume of the biomacromolecules can be calculated by the Porod approximation (24, 25). Moreover, the compactness or flexibility of biomacromolecules can be evaluated using the Kratky (26), dimensionless Kratky (27), and Porod-Debye plots (28). The principle, a typical SAXS measurement and derivative profiles can been seen from Figure 1.
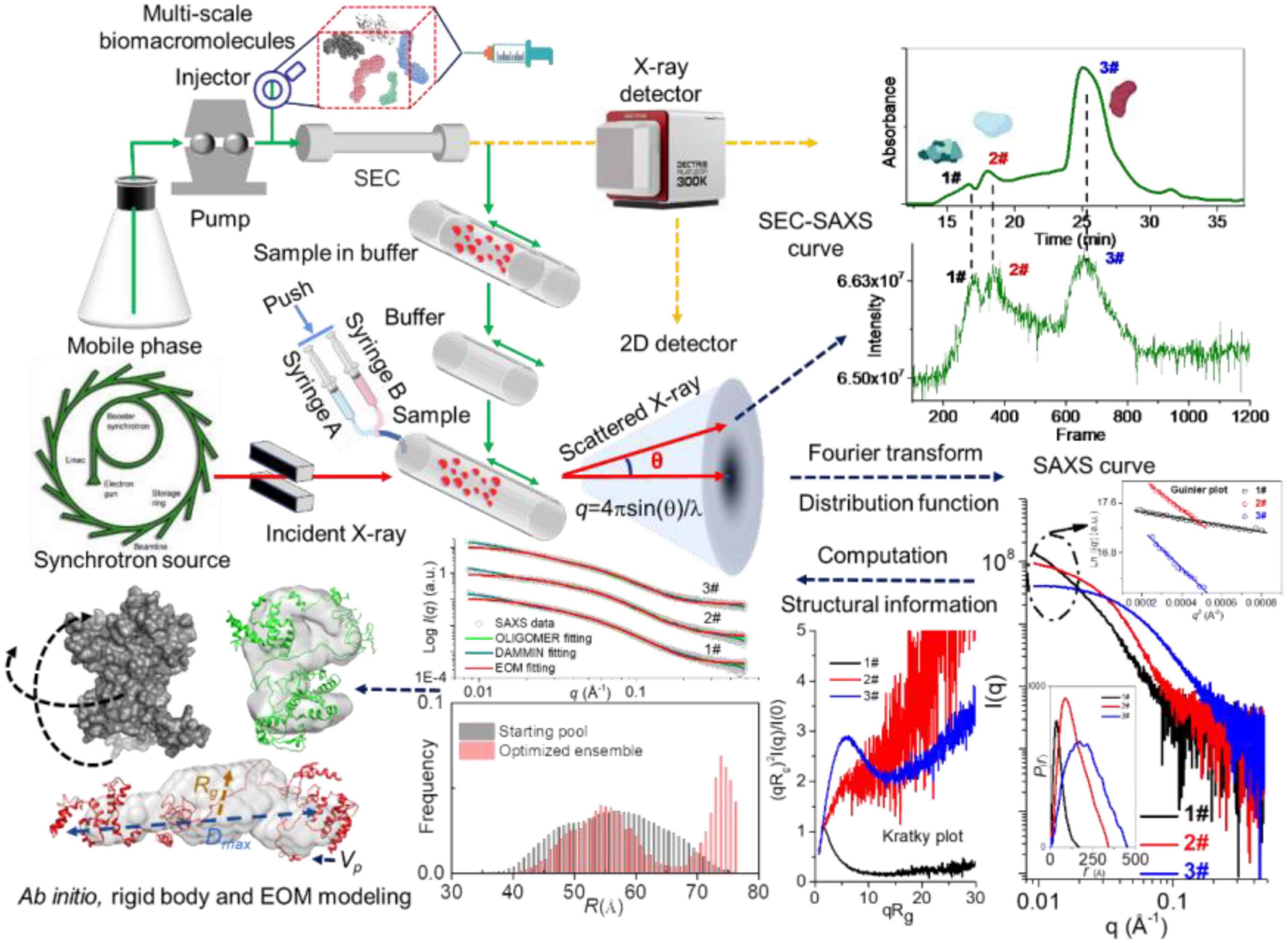
Figure 1. Principle, a typical small-angle X-ray scattering (SAXS) experiment and derivative profiles. Reproduced from Sun et al. (29) (Copyright 2021 Elsevier Publisher).
The net scattering intensity is critically dependent on the scale factor, especially at high q regions (as shown in Figure 2A), and any minor deviation from the accurate value may have a significant impact on the final results (30). As seen from Figures 2B–F, a sharp decrease in the net scattering intensity at high q regions is observed due to a slight increase in scale factor (1.000–1.001) for background buffer subtraction. SAXS allows the determination of the structural properties under a broad range from a few Angstroms to hundreds of nanometers, which covers the size of biomacromolecules and their complex (Figure 2G). The maximum distance within a scattered particle (Dmax) can be assessed by the distance r of the p(r) distribution equals zero (Figure 2H). When the p(r) curve with a maximum frequency at a distance less than half of the Dmax (olive curve in Figure 2H), the scattered particle adopts rather extended and elongated conformation in solution. Therefore, the change in conformation and structure of biomacromolecules can be monitored by p(r) profile. Moreover, an accurate determination of the scale factor for background subtraction has significant implications for obtaining the further reliable structural parameters of biomacromolecules (25).
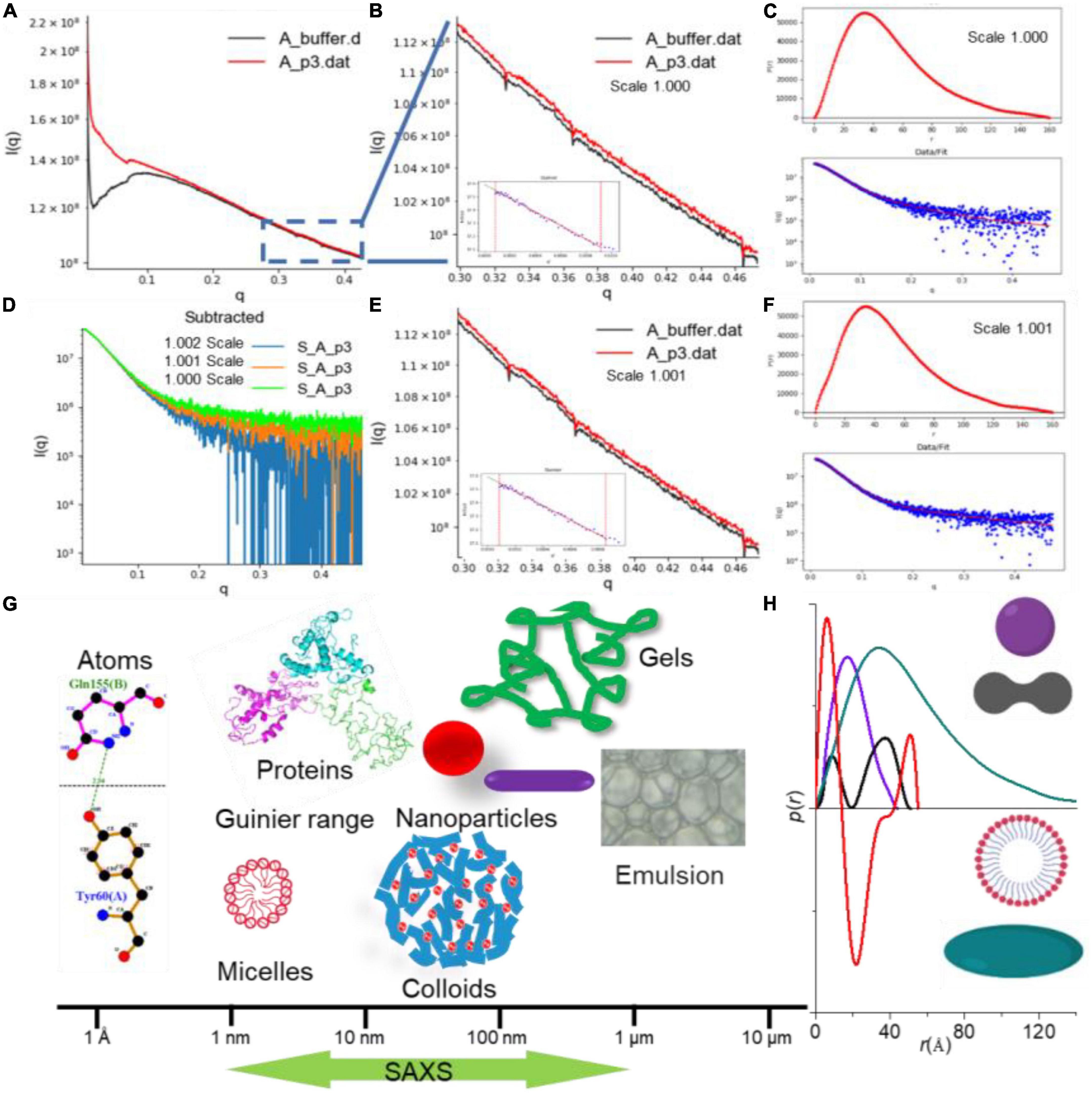
Figure 2. The small-angle X-ray scattering (SAXS) data analysis by RAW program (22). (A) Background subtraction of caseinate (29). (B–F) Scattering curves showing changes in net scattering intensity in the high q region with different scale factors (1.000 and 1.001), as well as the resulting p(r) in different scaling factor. (G) Size determination coverage of SAXS. (H) p(r) profile depends on the dimension and shape of the biomacromolecule.
Small-angle X-ray scattering profile can then be used for a further series of theoretical calculations to obtain structural information, ab initial model and ensemble structures. By translating scattering curves into bead representations consisting of a set of dummy atoms, an ab initio model of biomacromolecule with a resolution of 10–30 Å can be obtained via DAMMIN (31) and GASBOR programs (32). It is noted that DAMMIN is the most used in the modeling programs for reconstructing low-resolution shape of biomacromolecule. An iterative phase retrieval method, density from solution scattering (DENSS), has been expanded to analyze SAXS data, which may avoid many of the assumptions limiting the resolution and accuracy of modeling algorithms by explicitly calculating electron density (33). Deep learning methods, such as DecodeSAXS, have been reported used to classify and reconstruct the shapes with model parameters (34, 35). Critical Assessment of Protein Structure Prediction (CASP), a machine learning program, utilizes SAXS data to build improved models simulating the global shape of the target (36). Google’s AlphaFold 2 indisputably won the CASP14 competition. The results are so incredibly accurate that many have hailed this code as the solution to the long-standing protein structure prediction problem.
MONSA program (31) can be used to treat biomacromolecules containing multiple phases (e.g., intensities from protein/nucleic acid complexes) (6). SREFLEX (37), SASREF (38), and CORAL (39) programs can be used for high-resolution modeling of rigid bodies. SAXSDom has been utilized to model stable multi-domain proteins with flexible linker regions (40). Moreover, CRYSOL is used to compare the SAXS data with a PDB file with an X-ray or NMR structure of a protein or a protein-DNA(RNA) complex (41). SUPCOMB is a tool for superimposing one 3D structure onto another (42). In the case of flexible and mixed biomacromolecule systems (protein, DNA, and RNA), the structural ensembles of the biomacromolecule can be acquired by the ensemble optimization method (EOM) (43). For the oligomeric and transient state system, the volume fraction of components can be calculated using OLIGOMER, and the ab initio model of intermediate may be acquired by DAMMIX (21, 44). A hybrid resolution molecular dynamic (MD) method (hySAXS) has been used to create an ensemble of structures for intrinsically disordered proteins (IDPs), which is comparable to the experimental SAXS data (45). The extended experimental inferential structure determination method (X-EISD) (46) and Bayesian/Maximum entropy (BME) method (47) can be also used to calculate the maximum log-likelihood of an IDPs ensemble derived from SAXS. LIPMIX (48) and BILMIX programs (49) enable polydispersity of the model in multilamellar and asymmetric lipid vesicles and simultaneously generate the corresponding size distribution, respectively. The program ELLLIP can reconstruct the quasi-atomistic models of ellipsoidal liposomes (50) as well as bi-micellar systems involving proteins (51).
Several integrated docking methods by fitting the theoretical scattering curve of possible models to the experimental SAXS data have been developed to estimate the structure of complexes. Examples of such docking methods include SASREF (38), FoXSDock (52), HADDOCK (53), ClusPro FMFT-SAXS (54), pyDockSAXS (55), RosettaDockSAXS (56), PatchDock (57), and ATTRACT-SAXS (58). It has shown that iSPOT can filter docked structure and characterize a native-like model combined SAXS with foot-printing data by generating theoretical scattering of crystal structure (59). Molodenskiy et al. presented a PyMOL plugin, MPBuilder, which provided a set of adaptable routines for modeling membrane proteins (MPs), protein-detergent complex, bicelles, and lipid scaffold (saponin nanoparticles, nanodiscs) validated with SEC-SAXS data (60).
A comprehensive list of programs to reconstruct the structure and model of biomacromolecules based on SAXS data is shown in Table 1, and many of the programs are publicly available to academic users and moderately easy to operate. The details and applications of each program please see the corresponding reference. The popularity of SAXS has been propelled by novel data analysis and modeling algorithms. Developing user-friendly modeling programs will facilitate the utilization of SAXS for large-scale studies, which is also a major achievement in the community toward broader use of the method in combination with complementary techniques and enabling the cross-validation of structural data (61).
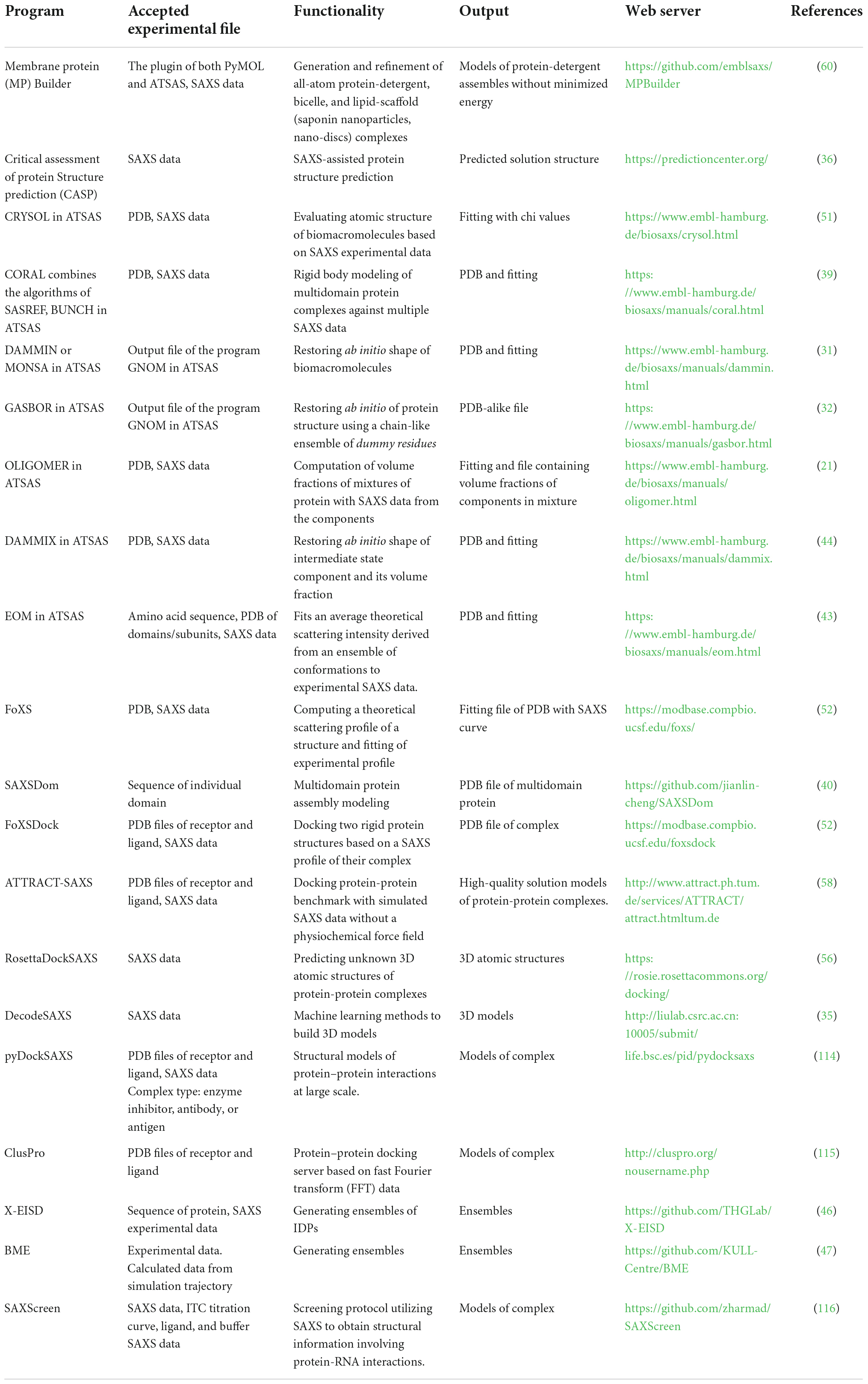
Table 1. List of some of the available software programs used in the analysis and reconstruction of models based on small-angle X-ray scattering (SAXS) data.
The online purification system coupled with SAXS, such as size-exclusion chromatography (SEC), gel filtration chromatography, and reversed-phase chromatography, is a standard approach for separating oligomeric species or components in a heterogeneous sample (62). The programs like CHROMIXS (63), DATASW (64), DELA (65), EFAMIX (66), and US-SOMO HPLC-SAXS module (67) have been developed to process chromatography-SAXS data. The scheme for SEC-SAXS setup is shown in Figures 1, 6, and applications of chromatography combined with SAXS for studying biomacromolecules are summarized in Table 2.
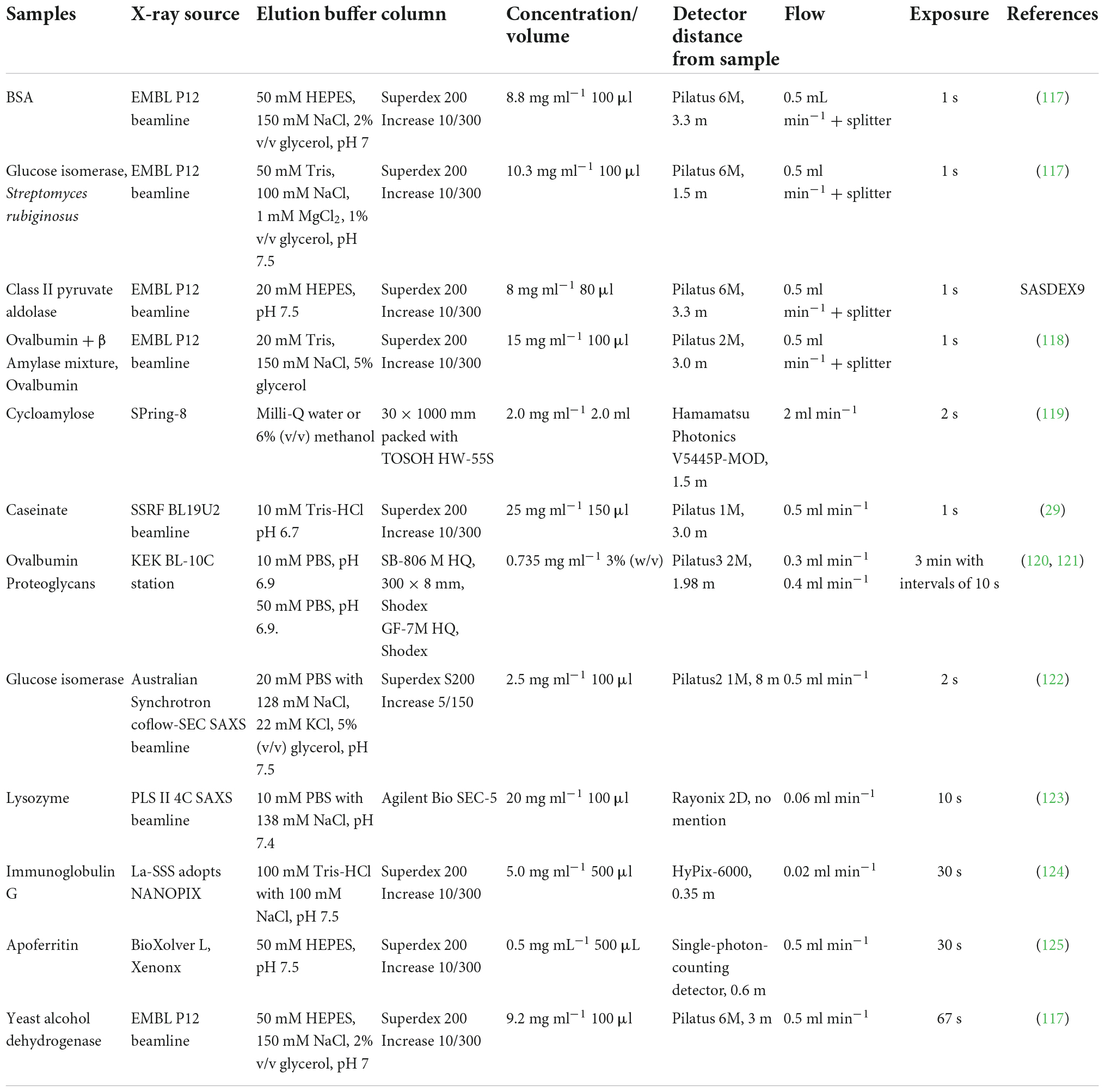
Table 2. Application of chromatography combined small-angle X-ray scattering (SAXS) for studying biomacromolecules.
Although the nanocluster model for describing casein micelle structure is widely accepted, little direct evidence at the nanometer scale supported this model. Sun et al. (29) reported a method that can prove and quantify the conformation and the fine structure of the casein cluster based on SEC-SAXS. The SEC-SAXS results showed that casein cluster presented Rg values ranging from 39.45 to 40.77 Å with a Dmax of 180 Å. The dimensionless Kratky plot suggested a rather extended and elongated conformation of casein cluster in solution. The experimental Mw according to the Bayesian Interference analysis was 50.3–64.7 kD with a probability of 91.54%, indicating the presence of 2–3 casein monomers in the cluster. Further, the DAMMIX and OLIGOMER results indicated that the cluster consisted of four species, αs1-β-αs2-casein, αs1-casein, αs2-casein and αs1-αs2-casein with a volume fraction of 64.3, 22.8, 8.5, and 4.4 %, respectively. The results of EOM indicated the presence of two conformers in αs1-β-αs2-casein, the elongated one (∼60 Å of Rg) with 64.7% of volume fraction and the compact one (∼35 Å of Rg) with 35.3% of volume fraction. It is the first time to reveal the structural properties of casein cluster based on SEC-SAXS, which may help understand better for internal structure of casein micelles regarding their primary casein cluster. Therefore, SAXS has been proved to be a powerful tool to study the structure and dynamics of the flexible, disordered and mixed biomacromolecules.
Small-angle X-ray scattering measurements are performed over a set time period ranging from microseconds to hours to assess the time-resolved (TR-SAXS) and time-dependent (in situ or real time) changes in structure and function for protein, carbohydrate, fat or non-nutritive compounds, such as gelatinization, assembly, micellization or colloid formation as well as in digestion and hydrolysis (68). This method is particularly suited to differentiate triggers of structural changes, including optical excitation (69), electron transfer (70), temperature jump (T-jump) (71), pH-jump (72), photoreduction (73), and reactant concentration jump (74).
Kuang et al. investigated the lamellar structure change of waxy corn starch during gelatinization and reveal the gelatinization mechanism by TR-SAXS in the temperature range from 35 to 141.85 °C with a measurement of 60 s at each degree (75). Gilbert reviewed the latest activities in the application of time-dependent SAXS to food colloids (14). Hempt et al. reported a novel digestion model of milk using an integrated online flow-through TR-SAXS with an in vitro cell co-culture model (76). Krishnamoorthy et al. reported an approach based on the time-dependent SAXS from protein spherical nucleic acids to elucidate the enzymatic degradation of DNA, which should prove invaluable in probing other enzyme-catalyzed reactions on the nanoscale (77). The details of TR-SAXS equipped with the laser pulse recording as a function of the time delay between laser pulse and incident X-ray are shown in Figure 3.
The SAXS beam equipped with the microfluidic device (continuous flow and stopped-flow) may not only reduce the sample damage by radiation (78), but also monitor the dynamic structural alternations during interactions in real-time (79). Hsu et al. characterized the transient partially folded state of bovine α-lactalbumin (BLA) coupled with TR-SAXS following a T-jump (74). The structural responses of BLA after an 11.5°C T-jump from the initial temperatures of 60, 65, and 70°C were individually recorded from 20 μs to 70 ms delay. Three states were molten globule state and two terminal unfolded states, U1 and U2. The application of TR-SAXS for structural characterization of biomacromolecules in foods is listed in Table 3.
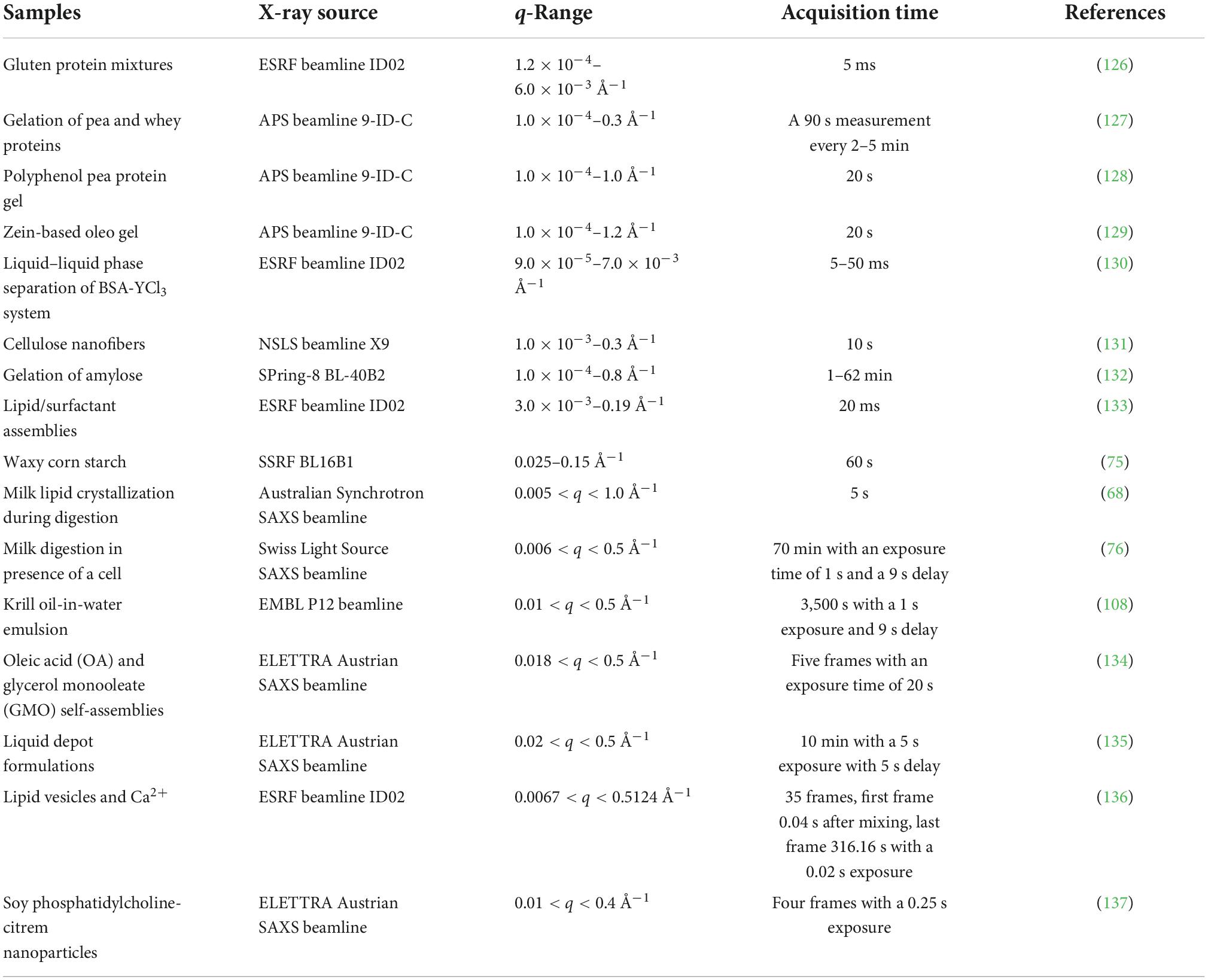
Table 3. Application of time-resolved small-angle X-ray scattering (TR-SAXS) at various synchrotron beamlines.
Temperature is one of the most important parameters controlling the formation, morphology, and structure of biomacromolecules, since much of biochemistry is thermally driven, functionally relevant conformational changes can also be triggered by changes in temperature. Generally, a trigger T-jump using a nanosecond laser pulse or an infrared (IR) light with a wavelength of 1450 nm (Figure 3, right upper) can be applied to most temperature-sensitive biomacromolecules to perturb the structural dynamics and reveal the changes in structural kinetics and association under various temperature conditions (80).
Berntsson et al. developed a CoSAXS beamline for millisecond T-jump experiments tracked by TR-SAXS with the Eiger2 and Mythen2 detectors and recorded the scattering of the solvent (80). A ∼15°C T-jump can be triggered by a 2 ms infrared laser light and maintained for several seconds with additional laser pulses. The structural changes in lysozyme induced by a T-jump were observed and the population of lysozyme structures differed at this temperature. Moreover, the data showed that IR radiation absorbed directly by the solvent did not show a significant effect compared with that induced by the thermal changes in the protein. Thus, the temperature induced change in structure of biomacromolecule and thermal dynamics of system can be monitored by SAXS effectively based on the scattering characteristic of sample in SAXS profile.
High-pressure (HP) food treatment including pasteurization, sterilization, and shelf-life extension, has widely been used to ensure food safety and preserve various thermally sensitive nutrients and bioactive compounds (81). HP-SAXS can also be used to track a wide range of structural changes of food biomacromolecules under pressure in real time (82). Moreover, the experimental setup of TR-SAXS studies of kinetic events induced by sub-millisecond timescale hydrostatic pressure jumps (P-jump, 1–5,000 bar) is also available in several synchrotron SAXS beamlines (83). Typically, the diamond anvil cell (DAC) covers the measurement of milk, solid powders, crystals and crystalline liquids (84). Hydrostatic pressure cell (HPC) is widely used to study phase diagrams of lipid, nano-assemblies, or pressure-dependent structure-function of biomacromolecules (85, 86).
Lehmkühler et al. reported the pressure-induced formation of super crystals from high-quality PEGylated colloidal nanoparticles using 5 ms P-jump SAXS (87). They demonstrated the crystallization pressure (pc) of the suspension by tracking SAXS patterns at pressures above 2 kbar in steps of 100 bar and verified pc between 2.9 and 3 kbar. They observed that the pressure (pf) jumped from 2.9 to 3.58 kbar averaging over 200 ms exposure time. The characteristic time (tw) decreased from 6.1 to 0.07 s with a reduction in Bragg reflection width from 0.138 to 0.0458 Å–1, suggesting the higher the pf, the faster the formation of nanoparticle structure. The results showed that a larger P-jump induced attractive interactions and thereby accelerated the formation of colloidal nanocrystal superlattices with enhanced crystal quality. Therefore, HP-SAXS can be utilized to track the structural change during interactions of biomacromolecules as well as to monitor the preparation of various biobased nanostructures. Exploiting easy operation setup will broad the applications of SAXS for complex biomacromolecule system.
As one of the most significant biomacromolecules in food, protein plays an essentially nutritional role in vivo. Meanwhile, protein-based ingredients fulfill several technical functions in food formulations and contribute to texture, color, flavor, and other properties such as solubility, stability emulsification, gelation and foaming (88). These researches involve studying protein structure-function relationships, optimizing the utilization of the components of the product, improving the quality, reducing costs, and developing novel protein application (89). SAXS is one of the most suitable techniques for protein structure and function relations study.
According to Yang et al. SAXS was used to investigate the nanostructure of quinoa protein (Chenopodium quinoa) isolates (QPI), one of the emerging proteins native to South America with a well-balanced amino acid profile, and the effect of NaCl and CaCl2 on the heat-induced gelation of QPI (90). Thermal treatment increased the sample I(q) in low-q region and the scattering intensity remained almost the same in the high-q region, which suggested that heat-induced QPI aggregation and then gelation merely occurred on the micron scale, while the internal structure of QPI on the nanoscale changed little. A Guinier shoulder in the mid-q region (0.02 Å–1< q < 0.08 Å–1) of the Kratky plot suggested the existence of nanoscale protein particles or inhomogeneities in QPI gel containing 0–200 mM NaCl. By fitting with correlation length model (), the correlation length (ζ) or particle size of ∼32 Å was obtained for all the QPI gels containing 0–200 mM NaCl. Calcium binding or protein cross-linking induced minor protein inhomogeneities as indicated by substantial changes in the SAXS curve as well as a small peak at q∼0.2 Å–1 in SAXS patterns of QPI gel containing CaCl2.
Pohl et al. (91) reported a high-throughput SAXS screening approach to assess the conformational stability and initial dispersion state of Thermomyces lanuginosus (TLL) and Rhizomucor miehei (RML), important lipases used in the food industry. They found repulsion in nine different kinds of the buffer as indicated by the decreased intensity in the low q-region induced by interparticle diffraction, and a significantly reduced repulsion and reduced oligomerization in phosphate buffer. Salt (35, 70, 140 mM NaCl) had minimal impact on SAXS profiles of TLL in histidine buffer at pH 5.5 and pH 7.5. The major species in the solution in all conditions was found to be monomeric, which confirmed that the differences in SAXS data were related to protein-protein interaction, suggesting that SAXS is used more widely as a tool to gain in-depth knowledge especially for the later stages of protein formulation in the food industry (Figure 4).
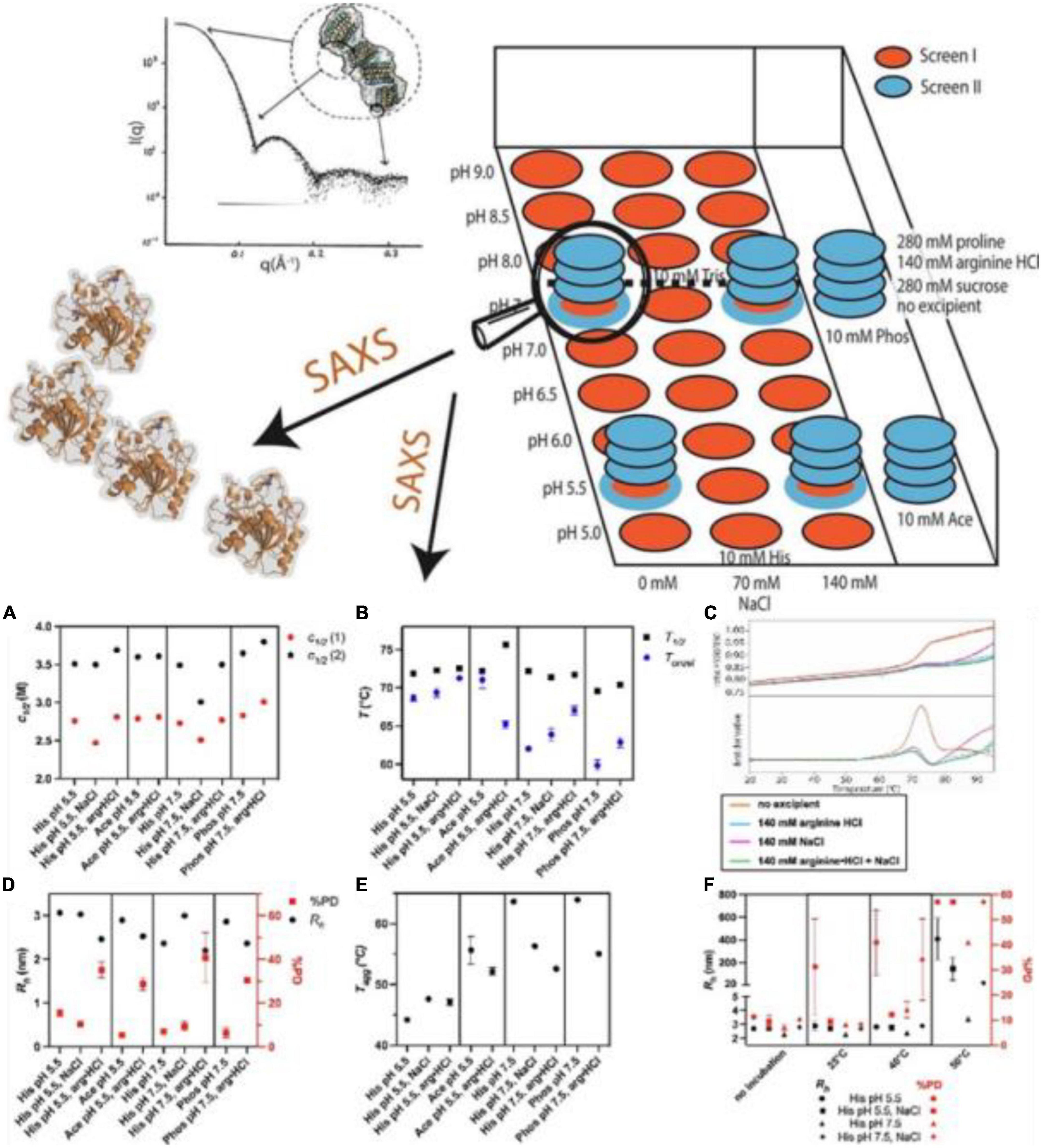
Figure 4. (Top) Combination of high throughput and structural screening to assess protein stability using small-angle X-ray scattering (SAXS). (Bottom) (A–F) High throughput stability screening of Thermomyces lanuginosus (TLL) (91) (Copyright 2022 Elsevier Publisher).
The structure of casein micelles contributes to the primary physicochemical and organoleptic properties of milk. Yang et al. (92) analyzed the changes in the internal structure of CNs under HHP (up to ∼1,000 MPa) using in situ HP-SAXS equipped with DAC at room temperature. They found a decrease in both scattering intensities at low q (∼0.003 Å–1) and high q (∼0.08 Å–1), suggesting the disruption of CNs and solubilization of the colloidal calcium phosphate (CCP) nanoclusters under HP treatment. The SAXS profiles under pressures ranging from 270 to 960 MPa showed two isosbestic points at q values of ∼0.013 and 0.03 Å–1, which confirmed the appearance of “sub-micelles” and dissociation of CCP. When the pressure returned to atmospheric pressure, the CNs structure reverted partially to the native one (Figure 5A). Similarly, Yang et al. (84) reported the hierarchical structure of milk at various lengths under a pressure of 200 or 400 MPa at 25, 40, or 60°C using HP-SAXS (Figure 5B). The changes in CNs nanostructures varied with pressure rather than time, and temperature played a central role during the HP process.
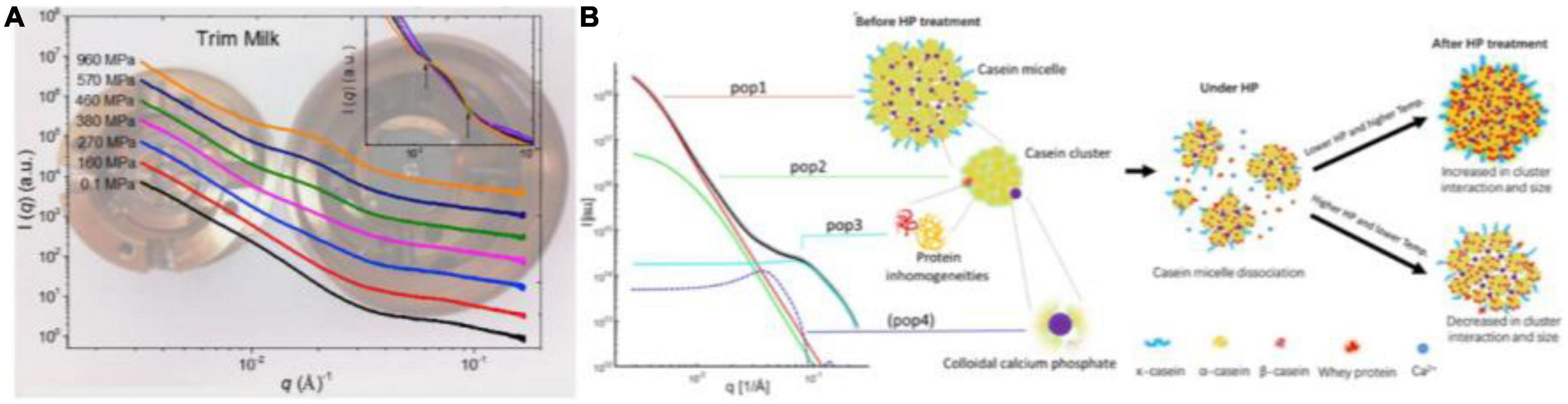
Figure 5. (A) In situ synchrotron small-angle X-ray scattering (SAXS) patterns of trim milk under different HHP using DAC (92) (Copyright 2018 Elsevier Publisher). (B) Graphic representation of the four contributions of CNs model at different length scales and structural dynamics during HHP from in situ SAXS, based on Yang et al. (84) (Copyright 2021 Elsevier Publisher).
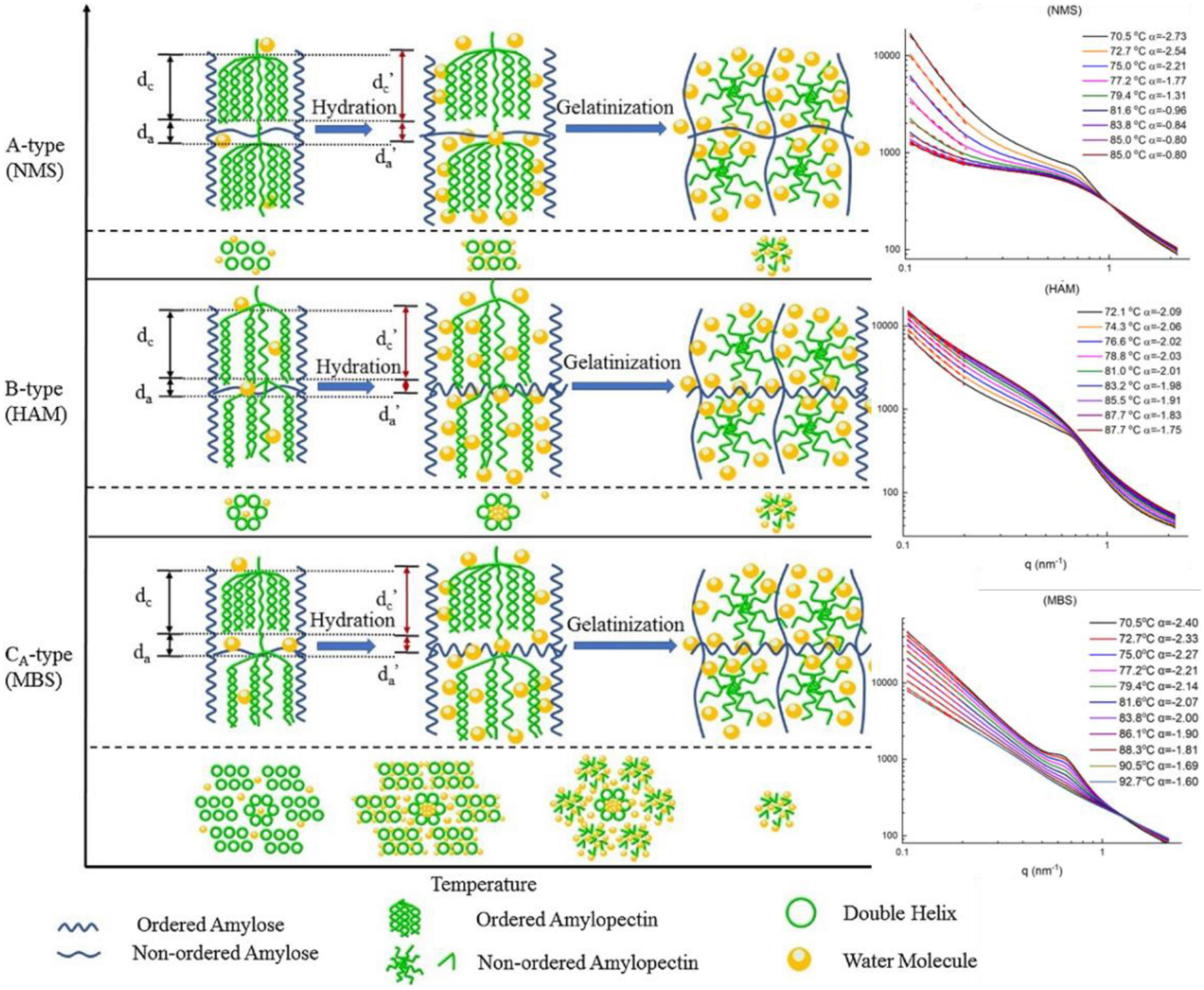
Figure 6. (Left) Schematic representation of lamellar and crystalline structural dynamics during heating. The two steps are shown: hydration, which is the uptake of water and swelling of starch granules; gelatinization, which is the disruption of starch crystal structure. (Right) Small-angle X-ray scattering (SAXS) profiles of HAM, NMS, MBS (95) (Copyright 2020 Elsevier Publisher).
Consequently, SAXS can be used as an effective technique not only to track the dynamic properties of biomacromolecule interactions but also to monitor the internal structure of biomacromolecule assembles.
Carbohydrates contribute to the bulk of dietary energy and play a vital role due to their diverse biological properties and functionalities in the food industry, as a thickening, gelating, emulsifying, encapsulating, or bulking agent (93). Starch, as an important polysaccharide macronutrient, determines the processing and nutritional quality of starch-based foods (94). Increased attention is needed to identify significant opportunities for real-time monitoring of structural changes during starch processing, such as swelling, gelatinization, retrogradation, and digestibility of starch.
Liu group reported dynamic changes in lamellar structure and gelatinization of cereal starches with different amylose contents in real time using in situ SAXS (Figure 6; 75, 95, 96). In the low-q region, the curves fitted with a simple power law equation, I(q)∼q–α, where mass fractal dimension (0 < α < 3) was an indication of compactness, whereas the surface fractal dimension (3 < α < 4) was considered smooth. During gelatinization, all cereal starches showed a decreasing α value in the q-region between 0.01 and 0.02 Å–1, with the corresponding size of ∼30 to ∼60 nm with the temperature increasing from ∼70 to ∼90°C, implying a mass fractal structure of the starch gel. Interestingly, an isosbestic point in the middle q-region was observed for all samples, which confirmed a two-step gelatinization of starches, namely, two-correlation length (ξ) of particles in the paste/gel system. The structural paraments of lamellae, the average thickness of amorphous layers (da), crystalline and amorphous layer thickness (dc) and the long period distance (dac) parameters were calculated based on Lorentz-corrected SAXS profiles. For high-amylose maize starch (HAM), dc increased from 65°C with a decreased da value, demonstrating swelling of the lamellae following water uptake. For normal maize starch (NMS) and mung bean starch (MBS), dc increased from 60°C with a decreasing value of da. Both dac and dc rapidly decreased at 72.2, 70.2, and 69.4°C for the high amylopectin (HAP), normal rice starch (NS) and HAM samples, respectively (96).
Starch has a strong tendency to retrograde and undergoes syneresis on cooling, namely, retrogradation. The retrogradation starts with the self-assembly of amylose to form a double helix during the cooling and storage of starch gel, followed by the partial crystallization of branched polymers (amylopectin) after prolonged storage (97). Zeng et al. (94) reported the SAXS patterns of retrograde starch with α values of all samples ranging from 1.32 to 2.43, indicating the mass fractal structures of all retrograde starch samples. Compared with storage day 1, the fractal dimensions (Dm) on storage day 24 increased from 1.32 to 2.30, which was consistent with the formation of ordered crystalline structures in the long-range and an increase in the ordered structure of starch during storage.
Starch digestibility based on sustained dietary energy and low glycemic index (GI) of foods plays a vital role in public health (98). Yang et al. used SAXS to investigate thermally and enzymatically digested corn starches under various treatment times. The semi-crystalline lamellar structure of starch exhibited a scattering peak at a q-region of 0.06–0.07Å–1 with a size of 9-10 nm corresponding to the alternating crystalline and amorphous lamellar structure of amylopectin. The peak area of thermally treated and enzymatic digested starch was quantified by fitting SAXS data (0.02Å–1 < q < 0.2Å –1) with a power-law function combined with a Lorentzian peak with (99). The thermal treatment induced water uptake in the amorphous regions of the granule during heating, leading to an increase in the intensity of the low q-region. In contrast, enzymatically treated samples showed changes in the crystalline and amorphous regions within the semi-crystalline lamellar structure and the amorphous growth rings. However, both treatments had little impact on the mass fractal structures of starch as the power law exponent (P) and long period distance (d) were around ∼2 and ∼10 nm for all samples.
Overall, the structural parameters of biomacromolecules derived from SAXS data facilitated the determination of the structure-function relationship and the evolution of nanomaterials based on the role of carbohydrates in the food industry.
Fats and oils are important sources of energy and nutrition, and contribute to the desirable functionality, texture and palatability of foods (100). Chemically, fats consist mainly of triglycerides (TAGs) combined with free fatty acid moieties. Besides nutritional properties, lipids facilitate the delivery of lipophilic nutraceuticals. Lipid-based colloid formation or oleogelation is designed to simulate the structure and semi-solid rheological behavior, and is widely utilized in the food industry, including coating, bakery, dairy products, meat, plant-based and artificial meat products (101).
Clemente et al. (102) explored the water/oil/water interface of phospholipid 1,2-dimyristoyl-sn-glycerol-3-phosphocholine [DMPC; 1% (wt/wt)] dissolved in a mixture of volatile solvents such as cyclohexane/chloroform (volume ratio 2:1) using microfluidic devices and investigated the role of μ-SAXS (a sketch of the device provided in Figure 7 Top). The structural characteristics of oil/DMPC bilayers were indicated by the decay in the intensity of the SAXS pattern with an inflection at q = 0.074 Å–1 corresponding to the first minimum value in the form factor of flat objects extending over large distances (Figure 7 Bottom left), which was also reported in previous studies involving the liquid/liquid (L/L) interface of w/o emulsions generated by a microfluidic apparatus (103). The bilayer thickness was found to shrink under treatment at 50°C for 1 h, but not to increase the order of the lipid bilayers, as suggested by the minimum position (q = 0.074 Å–1) shifting toward higher q-region (q = 0.088 Å –1). The authors reported that the phase behavior and structural dynamics of phospholipids at the L/L interfaces can be detected well via micro-focusing SAXS (Figure 7 Bottom right), which may provide deeper insight into the role of double lipid emulsions in the food industry.
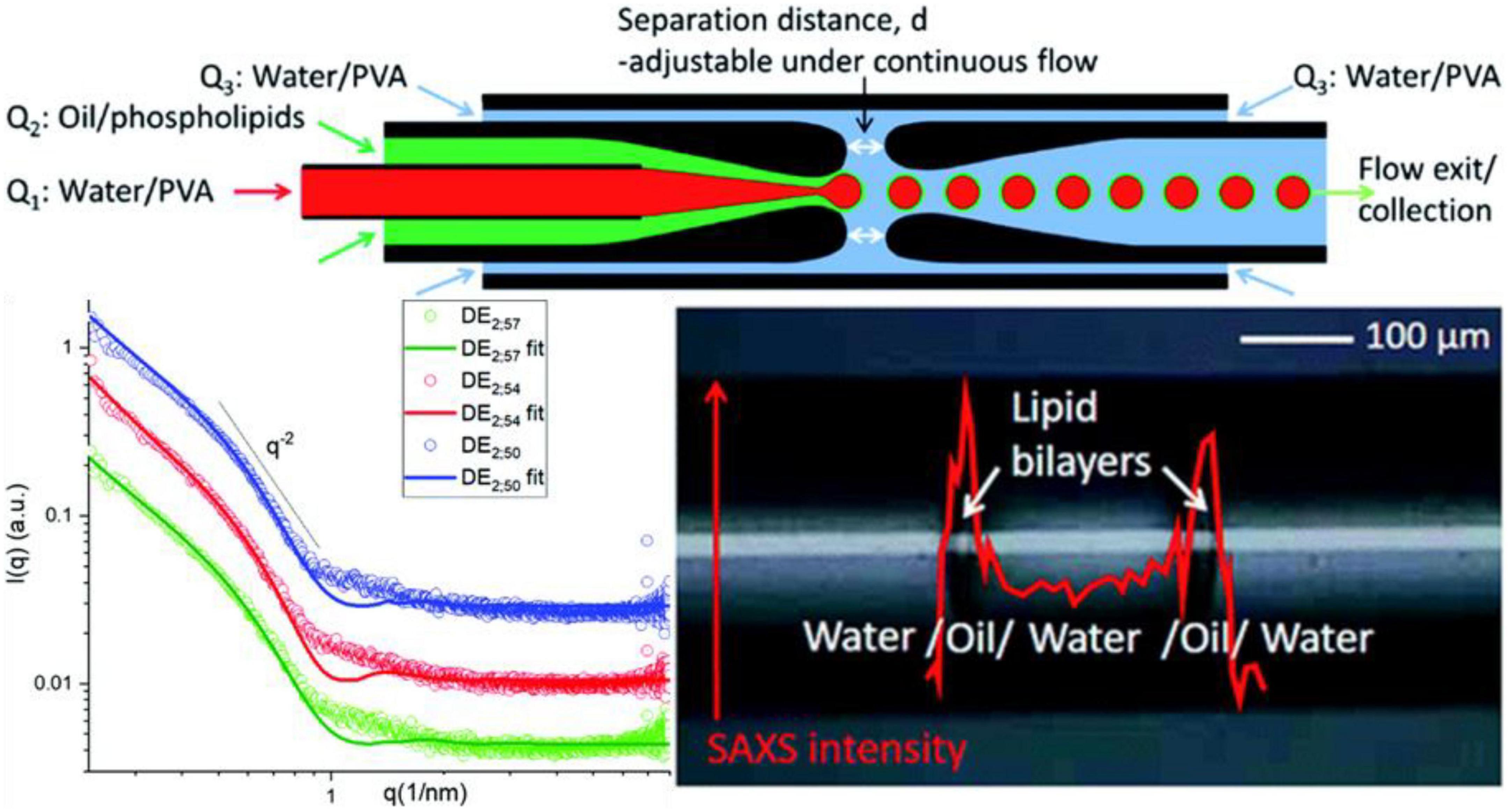
Figure 7. (Top) Setup of the microfluidic small-angle X-ray scattering (SAXS) device. (Bottom left) SAXS profiles of the w/o/w double emulsion interfaces obtained using the microfluidic device (RT; green), after storage at 50°C for 120 min (red) and for 240 min (blue), respectively. (Bottom right) Scheme of SAXS intensity at q = 0.9 nm– 1 recorded along an axial line covering a whole double emulsion unit. Reproduced from Clemente et al. (102).
Pham et al. (104) investigated the lipid self-assembly during in vitro digestion of bovine, human and goat milk using in situ TR-SAXS. The SAXS data revealed similar structural behavior during the early stages of the digestion of three types of milk, indicating lamellar (Lα), inverse hexagonal (H2), and continuous cubic (V2) phases (Figure 8 Top and Middle). All the milk tested self-assembled into non-lamellar liquid crystalline structures, with coexisting lamellar phases associated with calcium soap formation. By tracking the changes in Lα, H2, and V2 phases, the investigators concluded that different structures were formed during the digestion of all three infant formulas tested. During the digestion, soy and human milk that released long-chain fatty acids showed an inverse micellar cubic I2 phase at the oil–water interface, while bovine and goat milk yielded a greater proportion of medium-chain fatty acids tended to exhibit either the V2 or a H2 hexagonal phase. In addition, a TR-SAXS equipped with pH-stat or HHP system was used to monitor both the kinetics of lipolysis and structural behavior during the in vitro digestion of lipids in the presence of carbohydrates, such as chitosan (105), amylose (106), and other commercial supplements (as shown in Figure 8 Bottom).
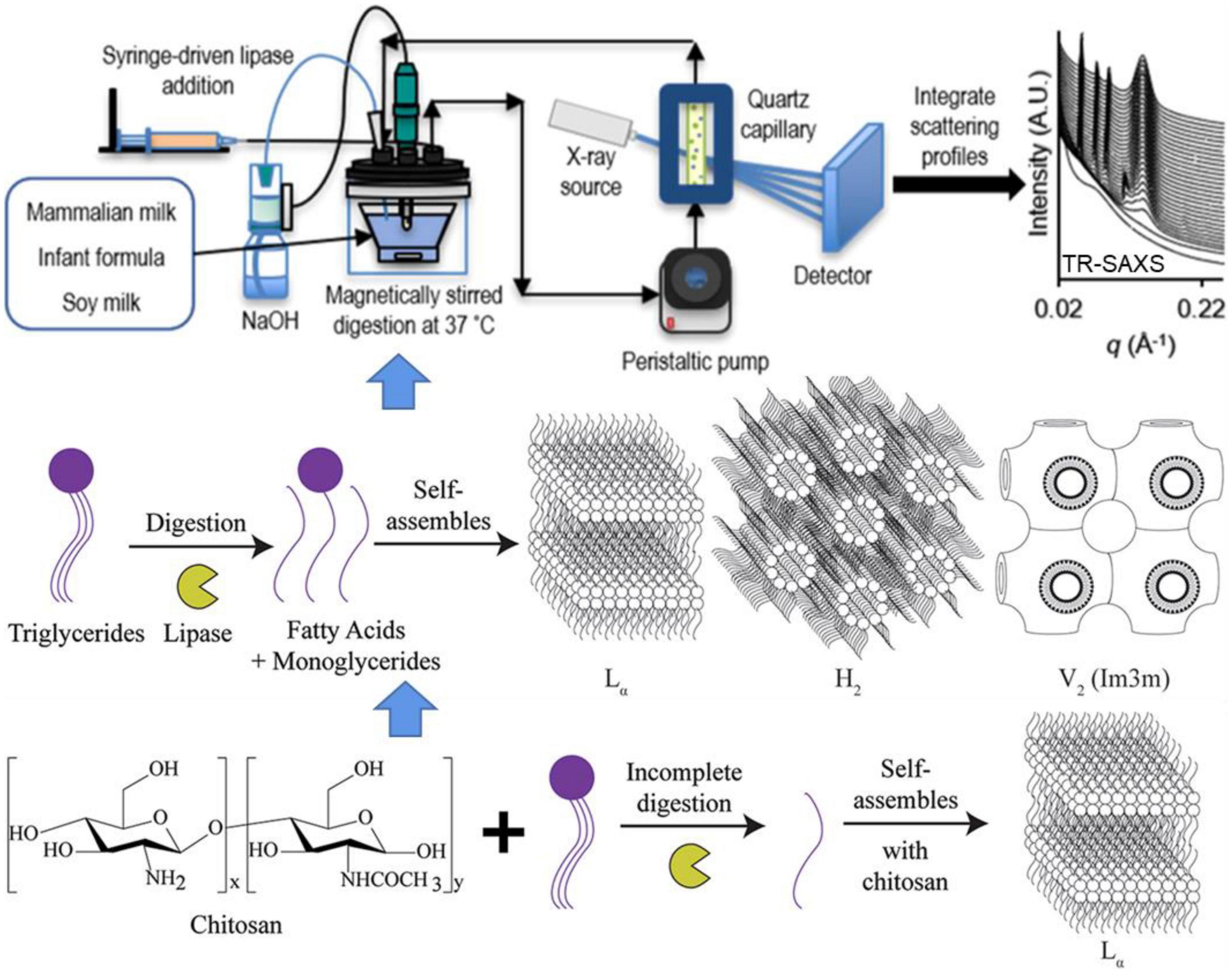
Figure 8. Schematic diagram showing the detection of self-assembled structures of lipids during the digestion of milk using a lipolysis device coupled with in situ TR-SAXS. Reprinted from Pham et al. (104).
Undoubtedly, the TR-SAXS (106–108) facilitates real-time monitoring of the crystallization of milk lipids under different treatments, but also assessment of the lipid self-assembly during in vitro digestion or under varying buffer conditions, which play a critical role in industrial applications involving the processing and storage of lipid-based foods.
Traditionally, natural sausage casings are made from collagen-rich intestinal submucosa derived from bovine, porcine and ovine sources. Collagen arrangement in the intestinal submucosa provides strength to animal tissues, although the relationship between structure and strength is not well-characterized. Gunn et al. (109) demonstrated collagen fibril orientation, orientation index (OI) and d-spacing of bovine, porcine and ovine sausage casings using SAXS. The d-spacing was calculated in the range of 64.8–65.2 nm, and the collagen fibrils were arranged in planar layers with OI values of 0.86–0.91 based on the X-ray adsorption edge energies of all samples.
Pectin has been approved as Generally Recognized as Safe (GRAS) and as a food emulsifier, stabilizer, thickener, and gelling agent (110). Mendez et al. (111) elucidated the different emulsification mechanisms of three pectin sources (watermelon, citrus and apple) using SAXS. The two shoulder-like peaks in the low-q (q < ∼0.03 Å–1) and high-q regions were as attributed to intermolecular interactions and chain clusters, and the scattering of rod-like pectin chains, respectively. The Rg3 value of all samples with cross-sections of rod-like pectin chains ranged from 1.4 to 2.3 nm. The P2 corresponding to the smallest structural level ranged from 1.8 to 2.8, suggesting the existence of flexible folded chains rather than ideal rigid rods. The Rg2 values, referring to the size of the molecular clusters originating as a result of chain bending, determined for pectin solutions (20–38 nm) fall within the range previously determined by SAXS (6.3–42 nm) for pectin with different degrees of esterification.
Food-grade surfactants, like lauric arginate (LAE), have been used widely as a preservative against a wide range of food pathogens and spoilage organisms such as processed meats, dairy products and fruit juices (112). Nallamilli et al. (113) reported a coacervation complex of LAE with λ-carrageenan consisting of loosely packed and disordered LAE molecules with an internal bilayer-like structure indicated by a scattering intensity peak at q ≈ 0.161 Å–1, based on which, a periodicity value of d = 2π/q∼3.9 nm was calculated. This was consistent with the quantitative analysis involving fitting a Teubner-Strey structure factor yielding d-spacing of the lamellae from 3.75 to 4.01 nm. Formation of bilayer coacervates was observed at the LAE/carrageenan weight ratio of 2 and the maximum coacervation was detected at a ratio of 5.
In the current review, recent advances in the structural characterization of food biomacromolecules using SAXS are summarized, including the principle, theoretical calculation methods, in situ capabilities, and applications. The unique feature of SAXS not only provides direct and rapid structural information of biomolecules in their native state but also facilitates the elucidation of conformational dynamics in real-time. SAXS combined with online chromatography represents a fascinating tool for separating and detecting mixtures and flexible systems synchronously, such as disordered fragments in proteins, long-chain ribonucleic acid and IDPs, which significantly widens the range of SAXS applications. Microfluidics installed in SAXS facilitate the study of binding kinetics by reducing the required sample volume to the sub-microliter level. A combination of T-jump or P-jump pump coupled to TR-SAXS allows direct tracking of the structural dynamics triggered by changes in temperature or pressure over a set time period ranging from microseconds to hours. Based on the volume of studies reported, SAXS is becoming a promising tool for monitoring the structure, conformation, interaction, kinetics, and reaction of biomacromolecules to provide molecular insights into the structure-function relationship of biomolecules in different food processing applications.
In the next decade, the application of the fourth-generation of high-brilliance synchrotron facilities will provide insight into the biomacromolecules at the atomic and molecular levels, and promote cutting-edge research via high-resolution imaging, ultrafast process exploration and advanced structural analysis based on SAXS. Furthermore, with the development of artificial intelligence (AI), research that was previously inconceivable or wildly impractical, especially involving protein structure prediction, is now feasible. We have reason to anticipate potential future applications of AI coupled with high-brilliance SAXS in understanding not merely the individual biomacromolecules and complexes in the food industry, but entire cells or even tissues in life science.
YS: conceptualization, methodology, software, writing—review and editing, revise, and supervision. XL and RC: software and writing—review and editing. FL: writing and editing. SW: writing—review and editing, revise, and supervision.
We acknowledge financial support from the Basic Research Program of Yunnan Province (202201AT070031) and High-level Talent of Yunnan Normal University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Okolie CL, Akanbi TO, Mason B, Udenigwe CC, Aryee ANA. Influence of conventional and recent extraction technologies on physicochemical properties of bioactive macromolecules from natural sources: a review. Food Res Int. (2019) 116:827–39. doi: 10.1016/j.foodres.2018.09.018
2. Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. (2009) 109:4108–39. doi: 10.1021/cr900033p
3. Trampari S, Neumann C, Hjorth-Jensen SJ, Shahsavar A, Quistgaard EM, Nissen P. Insights into the mechanism of high lipid-detergent crystallization of membrane proteins. J Appl Crystallogr. (2021) 54:1775–83. doi: 10.1073/pnas.0606149103
4. Wehbie M, Onyia KK, Mahler F, Le Roy A, Deletraz A, Bouchemal I, et al. Maltose-based fluorinated surfactants for membrane-protein extraction and stabilization. Langmuir. (2021) 37:2111–22. doi: 10.1021/acs.langmuir.0c03214
5. Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys. (2003) 66:1735–82.
6. Gräwert TW, Svergun DI. Structural modeling using solution small-angle X-ray scattering (SAXS). J Mol Biol. (2020) 432:3078–92. doi: 10.1016/j.jmb.2020.01.030
7. de Kruif CG. The structure of casein micelles: a review of small-angle scattering data. J Appl Crystallogr. (2014) 47:1479–89.
8. de Kruif CG, Huppertz T, Urban VS, Petukhov AV. Casein micelles and their internal structure. Adv Colloid Interface Sci. (2012) 171:36–52.
9. Huang J, Wang Z, Fan L, Ma S. A review of wheat starch analyses: methods, techniques, structure and function. Int J Biol Macromol. (2022) 203:130–42.
10. Lopez-Rubio A, Hernandez-Munoz P, Catala R, Gavara R, Lagaron JM. Improving packaged food quality and safety. Part 1: synchrotron X-ray analysis. Food Addit Contam. (2005) 22:988–93. doi: 10.1080/02652030500246370
11. Rostamabadi H, Falsafi SR, Assadpour E, Jafari SM. Evaluating the structural properties of bioactive-loaded nanocarriers with modern analytical tools. Compr Rev Food Sci Food Saf. (2020) 19:3266–322. doi: 10.1111/1541-4337.12653
12. Smith G, Brok E, Jensen GV, Midtgaard SR, Skar-Gislinge N, Arleth L. Casein studied by X-ray and neutron scattering. Abstracts of Papers of the American Chemical Society. Vol. 256. Washington, DC: American Chemical Society (2018).
13. Smith GN, Brok E, Christiansen MV, Ahrne L. Casein micelles in milk as sticky spheres. Soft Matter. (2020) 16:9955–63.
14. Gilbert EP. Small-angle X-Ray and neutron scattering in food colloids. Curr Opin Colloid Interface Sci. (2019) 42:55–72.
15. Schneidman-Duhovny D, Hammel M, Sali A. Macromolecular docking restrained by a small angle X-ray scattering profile. J Struct Biol. (2011) 173:461–71. doi: 10.1016/j.jsb.2010.09.023
16. Singh AK. Chapter 4 - Experimental methodologies for the characterization of nanoparticles. In: AK Singh editor. Engineered Nanoparticles. Boston, MA: Academic Press (2016). p. 125–70.
17. Molodenskiy DS, Svergun DI, Kikhney AG. Artificial neural networks for solution scattering data analysis. Structure. (2022) 30:900–8.e2.
18. Guinier A. La diffraction des rayons X aux très petits angles : application à l’étude de phénomènes ultramicroscopiques. Ann Phys. (1939) 11:161–237.
19. Glatter O. A new method for the evaluation of small-angle scattering data. J. Appl Crystallogr. (1977) 10:415–21.
20. Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. (1992) 25:495–503.
21. Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. (2003) 36:1277–82.
22. Hopkins JB, Gillilan RE, Skou S. BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J Appl Crystallogr. (2017) 50:1545–53. doi: 10.1107/S1600576717011438
23. Hansen S. Bayesian estimation of hyperparameters for indirect Fourier transformation in small-angle scattering. J Appl Crystallogr. (2000) 33:1415–21.
24. Porod G. Die Röntgenkleinwinkelstreuung von dichtgepackten kolloiden Systemen. Kolloid Zeitschrift. (1952) 125:51–7.
25. Trewhella J. Small angle scattering and structural biology: data quality and model validation. In: H Nakamura, G Kleywegt, SK Burley, JL Markley editors. Integrative Structural Biology with Hybrid Methods. Berlin: Springer (2018). p. 77–100.
27. Bizien T, Durand D, Roblina P, Thureau A, Vachette P, Pérez J. A brief survey of state-of-the-art BioSAXS. Protein Pept Lett. (2016) 23:217–31. doi: 10.2174/0929866523666160106153655
28. Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the porod-debye law. Biopolymers. (2011) 95:559–71. doi: 10.1002/bip.21638
29. Sun Y, Tai Z, Yan T, Dai Y, Hemar Y, Li N. Unveiling the structure of the primary caseinate particle using small-angle X-ray scattering and simulation methodologies. Food Res Int. (2021) 149:110653. doi: 10.1016/j.foodres.2021.110653
30. Wang G, Xu L-F, Shen J-L, Yao G-B, Ge Z-L, Li W-Q, et al. Iterative and accurate determination of small angle X-ray scattering background. Nucl Sci Tech. (2016) 27:105.
31. Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. (1999) 76:2879–86. doi: 10.1016/S0006-3495(99)77443-6
32. Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. (2001) 80:2946–53. doi: 10.1016/S0006-3495(01)76260-1
33. Grant TD. Ab initio electron density determination directly from solution scattering data. Nat Methods. (2018) 15:191–3.
34. Franke D, Jeffries CM, Svergun DI. Machine learning methods for X-ray scattering data analysis from biomacromolecular solutions. Biophys J. (2018) 114:2485–92. doi: 10.1016/j.bpj.2018.04.018
35. He H, Liu C, Liu H. Model reconstruction from small-angle X-ray scattering data using deep learning methods. iScience. (2020) 23:100906. doi: 10.1016/j.isci.2020.100906
36. Hura GL, Hodge CD, Rosenberg D, Guzenko D, Duarte JM, Monastyrskyy B, et al. Small angle X-ray scattering-assisted protein structure prediction in CASP13 and emergence of solution structure differences. Proteins. (2019) 87:1298–314. doi: 10.1002/prot.25827
37. Panjkovich A, Svergun DI. Deciphering conformational transitions of proteins by small angle X-ray scattering and normal mode analysis. Phys Chem Chem Phys. (2016) 18:5707–19. doi: 10.1039/c5cp04540a
38. Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. (2005) 89:1237–50. doi: 10.1529/biophysj.105.064154
39. Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. (2012) 45:342–50.
40. Hou J, Adhikari B, Tanner JJ, Cheng J. SAXSDom: modeling multidomain protein structures using small-angle X-ray scattering data. Proteins. (2020) 88:775–87. doi: 10.1002/prot.25865
41. Svergun D, Barberato C, Koch MHJ. CRYSOL– a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. (1995) 28:768–73.
42. Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. (2001) 34:33–41.
43. Tria G, Mertens HDT, Kachala M, Svergun DI. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. (2015) 2:207–17. doi: 10.1107/S205225251500202X
44. Konarev PV, Svergun DI. Direct shape determination of intermediates in evolving macromolecular solutions from small-angle scattering data. IUCrJ. (2018) 5:402–9. doi: 10.1107/S2052252518005900
45. Hermann MR, Hub JS. SAXS-restrained ensemble simulations of intrinsically disordered proteins with commitment to the principle of maximum entropy. J Chem Theory Comput. (2019) 15:5103–15. doi: 10.1021/acs.jctc.9b00338
46. Lincoff J, Haghighatlari M, Krzeminski M, Teixeira JMC, Gomes G-NW, Gradinaru CC, et al. Extended experimental inferential structure determination method in determining the structural ensembles of disordered protein states. Commun Chem. (2020) 3:74. doi: 10.1038/s42004-020-0323-0
47. Bottaro S, Bengtsen T, Lindorff-Larsen K. Integrating molecular simulation and experimental data: a bayesian/maximum entropy reweighting approach. In: Gáspári Z editor. Structural Bioinformatics: Methods and Protocols. New York, NY: Springer US (2020). p. 219–40. doi: 10.1007/978-1-0716-0270-6_15
48. Konarev PV, Gruzinov AY, Mertens HDT, Svergun DI. Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data. J Appl Crystallogr. (2021) 54:169–79. doi: 10.1107/S1600576720015368
49. Konarev PV, Petoukhov MV, Dadinova LA, Fedorova NV, Volynsky PE, Svergun DI, et al. BILMIX: a new approach to restore the size polydispersity and electron density profiles of lipid bilayers from liposomes using small-angle X-ray scattering data. J Appl Crystallogr. (2020) 53:236–43.
50. Petukhov MV, Konarev PV, Dadinova LA, Fedorova NV, Volynsky PE, Svergun DI, et al. Quasi-atomistic approach to modeling of liposomes. Crystallogr Rep. (2020) 65:258–63.
51. Manalastas-Cantos K, Konarev PV, Hajizadeh NR, Kikhney AG, Petoukhov MV, Molodenskiy DS, et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J Appl Crystallogr. (2021) 54:343–55. doi: 10.1107/S1600576720013412
52. Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. (2016) 44:W424–9. doi: 10.1093/nar/gkw389
53. Karaca E, Bonvin AMJJ. On the usefulness of ion-mobility mass spectrometry and SAXS data in scoring docking decoys. Acta Crystallogr D Biol Crystallogr. (2013) 69:683–94. doi: 10.1107/S0907444913007063
54. Ignatov M, Kazennov A, Kozakov D. ClusPro FMFT-SAXS: ultra-fast filtering using small-angle X-ray scattering data in protein docking. J Mol Biol. (2018) 430:2249–55. doi: 10.1016/j.jmb.2018.03.010
55. Jiménez-García B, Bernadó P, Fernández-Recio J. Structural characterization of protein–protein interactions with pyDockSAXS. In: Gáspári Z editor. Structural Bioinformatics: Methods and Protocols. New York, NY: Springer US (2020). p. 131–44.
56. Sønderby P, Rinnan Å, Madsen JJ, Harris P, Bukrinski JT, Peters GHJ. Small-angle X-ray scattering data in combination with Rosettadock improves the docking energy landscape. J Chem Inf Model. (2017) 57:2463–75. doi: 10.1021/acs.jcim.6b00789
57. Schneidman-Duhovny D, Hammel M. Modeling structure and dynamics of protein complexes with SAXS profiles. Methods Mol Biol. (2018) 1764:449–73.
58. Schindler, Christina EM, de Vries SJ, Sasse A, Zacharias M. SAXS data alone can generate high-quality models of protein-protein complexes. Structure. (2016) 24:1387–97. doi: 10.1016/j.str.2016.06.007
59. Huang W, Ravikumar KM, Parisien M, Yang S. Theoretical modeling of multiprotein complexes by iSPOT: integration of small-angle X-ray scattering, hydroxyl radical footprinting, and computational docking. J Struct Biol. (2016) 196:340–9. doi: 10.1016/j.jsb.2016.08.001
60. Molodenskiy DS, Svergun DI, Mertens HDT. MPBuilder: a PyMOL plugin for building and refinement of solubilized membrane proteins against small angle X-ray scattering data. J Mol Biol. (2021) 433:166888. doi: 10.1016/j.jmb.2021.166888
61. Mahieu E, Gabel F. Biological small-angle neutron scattering: recent results and development. Acta Crystallogr D Struct Biol. (2018) 74:715–26.
62. Brennich M, Hutin S, Weinhaupl K, Schanda P, Pernot P. Beyond size exclusion: online liquid chromatography for BioSAXS. Acta Crystallogr A Found Adv. (2017) 73:A83–83.
63. Panjkovich A, Svergun DI. CHROMIXS: automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics. (2018) 34:1944–6. doi: 10.1093/bioinformatics/btx846
64. Shkumatov AV, Strelkov SV. DATASW, a tool for HPLC–SAXS data analysis. Acta Crystallogr D Biol Crystallogr. (2015) 71:1347–50. doi: 10.1107/S1399004715007154
65. Malaby AW, Chakravarthy S, Irving TC, Kathuria SV, Bilsel O, Lambright DG. Methods for analysis of size-exclusion chromatography–small-angle X-ray scattering and reconstruction of protein scattering. J Appl Crystallogr. (2015) 48:1102–13. doi: 10.1107/S1600576715010420
66. Konarev PV, Graewert MA, Jeffries CM, Fukuda M, Cheremnykh TA, Volkov VV, et al. EFAMIX, a tool to decompose inline chromatography SAXS data from partially overlapping components. Protein Sci. (2022) 31:269–82. doi: 10.1002/pro.4237
67. Brookes E, Vachette P, Rocco M, Perez J. US-SOMO HPLC-SAXS module: dealing with capillary fouling and extraction of pure component patterns from poorly resolved SEC-SAXS data. J Appl Crystallogr. (2016) 49:1827–41. doi: 10.1107/S1600576716011201
68. Clulow AJ, Salim M, Hawley A, Boyd BJ. A closer look at the behaviour of milk lipids during digestion. Chem Phys Lipids. (2018) 211:107–16. doi: 10.1016/j.chemphyslip.2017.10.009
69. Ravishankar H, Nors Pedersen M, Sitsel A, Li C, Duelli A, Levantino M, et al. Tracking Ca2+ATPase intermediates in real-time by X-ray solution scattering. Sci Adv. (2020) 6:eaaz0981. doi: 10.1126/sciadv.aaz0981
70. Heyes DJ, Hardman SJO, Pedersen MN, Woodhouse J, De La Mora E, Wulff M, et al. Light-induced structural changes in a full-length cyanobacterial phytochrome probed by time-resolved X-ray scattering. Commun Biol. (2019) 2:1. doi: 10.1038/s42003-018-0242-0
71. Thompson MC, Barad BA, Wolff AM, Sun Cho H, Schotte F, Schwarz DMC, et al. Temperature-jump solution X-ray scattering reveals distinct motions in a dynamic enzyme. Nat Chem. (2019) 11:1058–66. doi: 10.1038/s41557-019-0329-3
72. Rimmerman D, Leshchev D, Hsu DJ, Hong J, Abraham B, Henning R, et al. Revealing fast structural dynamics in ph-responsive peptides with time-resolved X-ray scattering. J Phys Chem B. (2019) 123:2016–21. doi: 10.1021/acs.jpcb.9b00072
73. Kim TW, Lee SJ, Jo J, Kim JG, Ki H, Kim CW, et al. Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering. Proc Natl Acad Sci USA. (2020) 117:14996–5005. doi: 10.1073/pnas.1913442117
74. Hsu DJ, Leshchev D, Kosheleva I, Kohlstedt KL, Chen LX. Unfolding bovine α-lactalbumin with T-jump: characterizing disordered intermediates via time-resolved x-ray solution scattering and molecular dynamics simulations. J Chem Phys. (2021) 154:105101. doi: 10.1063/5.0039194
75. Kuang Q, Xu J, Liang Y, Xie F, Tian F, Zhou S, et al. Lamellar structure change of waxy corn starch during gelatinization by time-resolved synchrotron SAXS. Food Hydrocoll. (2017) 62:43–8.
76. Hempt C, Gontsarik M, Buerki-Thurnherr T, Hirsch C, Salentinig S. Nanostructure generation during milk digestion in presence of a cell culture model simulating the small intestine. J Colloid Interface Sci. (2020) 574:430–40. doi: 10.1016/j.jcis.2020.04.059
77. Krishnamoorthy K, Kewalramani S, Ehlen A, Moreau LM, Mirkin CA, de la Cruz MO, et al. Enzymatic degradation of DNA probed by in situ X-ray scattering. ACS Nano. (2019) 13:11382–91.
78. Narayanan T, Konovalov O. Synchrotron scattering methods for nanomaterials and soft matter research. Materials. (2020) 13:752.
79. Ilhan-Ayisigi E, Yaldiz B, Bor G, Yaghmur A, Yesil-Celiktas O. Advances in microfluidic synthesis and coupling with synchrotron SAXS for continuous production and real-time structural characterization of nano-self-assemblies. Colloids Surf B Biointerfaces. (2021) 201:111633. doi: 10.1016/j.colsurfb.2021.111633
80. Berntsson O, Terry AE, Plivelic TS. A setup for millisecond time-resolved X-ray solution scattering experiments at the CoSAXS beamline at the MAX IV Laboratory. J Synchrotron Radiat. (2022) 29:555–62. doi: 10.1107/S1600577522000996
81. Balasubramaniam VM. Process development of high pressure-based technologies for food: research advances and future perspectives. Curr Opin Food Sci. (2021) 42:270–277.
82. Ferreira Zielinski AA, Sanchez-Camargo ADP, Benvenutti L, Ferro DM, Dias JL, Salvador Ferreira SR. High-pressure fluid technologies: recent approaches to the production of natural pigments for food and pharmaceutical applications. Trends Food Sci Technol. (2021) 118:850–69.
83. Harish B, Gillilan RE, Zou J, Wang J, Raleigh DP, Royer CA. Protein unfolded states populated at high and ambient pressure are similarly compact. Biophys J. (2021) 120:2592–8. doi: 10.1016/j.bpj.2021.04.031
84. Yang S, Tyler AII, Ahrné L, Kirkensgaard JJK. Skimmed milk structural dynamics during high hydrostatic pressure processing from in situ SAXS. Food Res Int. (2021) 147:110527. doi: 10.1016/j.foodres.2021.110527
85. Rai DK, Gillilan RE, Huang Q, Miller R, Ting E, Lazarev A, et al. High-pressure small-angle X-ray scattering cell for biological solutions and soft materials. J Appl Crystallogr. (2021) 54:111–22. doi: 10.1107/S1600576720014752
86. Vella J, Hemar Y, Gu Q, Wu ZR, Li N, Söhnel T. In-situ SAXS investigation of high-pressure triglyceride polymorphism in milk cream and anhydrous milk fat. LWT. (2021) 135:110174.
87. Lehmkühler F, Schroer MA, Markmann V, Frenzel L, Möller J, Lange H, et al. Kinetics of pressure-induced nanocrystal superlattice formation. Phys Chem Chem Phys. (2019) 21:21349–54. doi: 10.1039/c9cp04658e
88. Loveday SM. Food proteins: technological, nutritional, and sustainability attributes of traditional and emerging proteins. Annu Rev Food Sci Technol. (2019) 10:311–39. doi: 10.1146/annurev-food-032818-121128
89. Alrosan M, Tan TC, Easa AM, Gammoh S, Alu’datt MH. Molecular forces governing protein-protein interaction: structure-function relationship of complexes protein in the food industry. Crit Rev Food Sci Nutr. (2022) 62:4036–52. doi: 10.1080/10408398.2021.1871589
90. Yang Z, de Campo L, Gilbert EP, Knott R, Cheng L, Storer B, et al. Effect of NaCl and CaCl2 concentration on the rheological and structural characteristics of thermally-induced quinoa protein gels. Food Hydrocoll. (2022) 124:107350.
91. Pohl C, Mahapatra S, Kulakova A, Streicher W, Peters GHJ, Nørgaard A, et al. Combination of high throughput and structural screening to assess protein stability – A screening perspective. Eur J Pharm Biopharm. (2022) 171:1–10. doi: 10.1016/j.ejpb.2021.08.018
92. Yang Z, Gu Q, Banjar W, Li N, Hemar Y. In situ study of skim milk structure changes under high hydrostatic pressure using synchrotron SAXS. Food Hydrocoll. (2018) 77:772–6.
93. Comerford KB, Papanikolaou Y, Jones JM, Rodriguez J, Slavin J, Angadi S, et al. Toward an evidence-based definition and classification of carbohydrate food quality: an expert panel report. Nutrients. (2021) 13:2667. doi: 10.3390/nu13082667
94. Zeng X, Zheng B, Li T, Chen L. How to synchronously slow down starch digestion and retrogradation: a structural analysis study. Int J Biol Macromol. (2022) 212:43–53. doi: 10.1016/j.ijbiomac.2022.05.099
95. Xu J, Blennow A, Li X, Chen L, Liu X. Gelatinization dynamics of starch in dependence of its lamellar structure, crystalline polymorphs and amylose content. Carbohydr Polym. (2020) 229:115481. doi: 10.1016/j.carbpol.2019.115481
96. Xu J, Li Z, Zhong Y, Zhou Q, Lv Q, Chen L, et al. The effects of molecular fine structure on rice starch granule gelatinization dynamics as investigated by in situ small-angle X-ray scattering. Food Hydrocoll. (2021) 121:107014.
97. Díaz-Calderón P, Simone E, Tyler AII, Enrione J, Foster T. A structural study of the self-association of different starches in presence of bacterial cellulose fibrils. Carbohydr Polym. (2022) 288:119361. doi: 10.1016/j.carbpol.2022.119361
98. Li C, Gong B, Hu Y, Liu X, Guan X, Zhang B. Combined crystalline, lamellar and granular structural insights into in vitro digestion rate of native starches. Food Hydrocoll. (2020) 105:105823.
99. Yang Z, Swedlund P, Hemar Y, Mo G, Wei Y, Li Z, et al. Effect of high hydrostatic pressure on the supramolecular structure of corn starch with different amylose contents. Int J Biol Macromol. (2016) 85:604–14.
100. Bascuas S, Morell P, Hernando I, Quiles A. Recent trends in oil structuring using hydrocolloids. Food Hydrocoll. (2021) 118:106612.
101. Wang G, Chen H, Wang L, Zou Y, Wan Z, Yang X. Formation of protein oleogels via capillary attraction of engineered protein particles. Food Hydrocoll. (2022) 133:107912.
102. Clemente I, Torbensen K, Di Cola E, Rossi F, Ristori S, Abou-Hassan A. Exploring the water/oil/water interface of phospholipid stabilized double emulsions by micro-focusing synchrotron SAXS. RSC Adv. (2019) 9:33429–35. doi: 10.1039/c9ra05894j
103. Di Cola E, Torbensen K, Clemente I, Rossi F, Ristori S, Abou-Hassan A. Lipid-stabilized water–oil interfaces studied by microfocusing small-angle X-ray scattering. Langmuir. (2017) 33:9100–5. doi: 10.1021/acs.langmuir.7b02076
104. Pham AC, Peng K-Y, Salim M, Ramirez G, Hawley A, Clulow AJ, et al. Correlating digestion-driven self-assembly in milk and infant formulas with changes in lipid composition. ACS Appl Bio Mater. (2020) 3:3087–98. doi: 10.1021/acsabm.0c00131
105. May KL, Tangso KJ, Hawley A, Boyd BJ, Clulow AJ. Interaction of chitosan-based dietary supplements with fats during lipid digestion. Food Hydrocoll. (2020) 108:105965.
106. Jia X, Sun S, Chen B, Zheng B, Guo Z. Understanding the crystal structure of lotus seed amylose–long-chain fatty acid complexes prepared by high hydrostatic pressure. Food Res Int. (2018) 111:334–41. doi: 10.1016/j.foodres.2018.05.053
107. Dyett B, Zychowski L, Bao L, Meikle TG, Peng S, Yu H, et al. Crystallization of Femtoliter Surface Droplet Arrays Revealed by Synchrotron Small-Angle X-ray Scattering. Langmuir. (2018) 34:9470–6. doi: 10.1021/acs.langmuir.8b01252
108. Yaghmur A, Lotfi S, Ariabod SA, Bor G, Gontsarik M, Salentinig S. Internal lamellar and inverse hexagonal liquid crystalline phases during the digestion of krill and astaxanthin oil-in-water emulsions. Front Bioeng Biotechnol. (2019) 7:384. doi: 10.3389/fbioe.2019.00384
109. Gunn S, Sizeland KH, Wells HC, Haverkamp RG. Collagen arrangement and strength in sausage casings produced from natural intestines. Food Hydrocoll. (2022) 129:107612.
110. Li Z, Xiong Y, Wang Y, Zhang Y, Luo Y. Low density lipoprotein-pectin complexes stabilized high internal phase pickering emulsions: the effects of pH conditions and mass ratios. Food Hydrocoll. (2023) 134:108004.
111. Mendez DA, Fabra MJ, Martínez-Abad A, Martínez-Sanz M, Gorria M, López-Rubio A. Understanding the different emulsification mechanisms of pectin: comparison between watermelon rind and two commercial pectin sources. Food Hydrocoll. (2021) 120:106957.
112. Hu X, Huang E, Barringer SA, Yousef AE. Factors affecting Alicyclobacillus acidoterrestris growth and guaiacol production and controlling apple juice spoilage by lauric arginate and ϵ-polylysine. LWT. (2020) 119:108883.
113. Nallamilli T, Ketomaeki M, Prozeller D, Mars J, Morsbach S, Mezger M, et al. Complex coacervation of food grade antimicrobial lauric arginate with lambda carrageenan. Curr Res Food Sci. (2021) 4:53–62. doi: 10.1016/j.crfs.2021.01.003
114. Jiménez-García B, Pons C, Svergun DI, Bernadó P, Fernández-Recio J. pyDockSAXS: protein–protein complex structure by SAXS and computational docking. Nucleic Acids Res. (2015) 43:W356–61.
115. Xia B, Vajda S, Kozakov D. Accounting for pairwise distance restraints in FFT-based protein–protein docking. Bioinformatics. (2016) 32:3342–4. doi: 10.1093/bioinformatics/btw306
116. Chen P-C, Masiewicz P, Rybin V, Svergun D, Hennig J. A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein–RNA Interactions. ACS Comb Sci. (2018) 20:197–202. doi: 10.1021/acscombsci.8b00007
117. Graewert M, Vela SD, Grwert TW, Molodenskiy DS, Jeffries CMJC. Adding size exclusion chromatography (SEC) and light scattering (LS) devices to obtain high-quality small angle X-ray scattering (SAXS) data. Crystals. (2020) 10:975.
118. Graewert MA, Franke D, Jeffries CM, Blanchet CE, Ruskule D, Kuhle K, et al. Automated pipeline for purification, biophysical and X-ray analysis of biomacromolecular solutions. Sci Rep. (2015) 5:10734. doi: 10.1038/srep10734
119. Takemasa M, Yuguchi Y, Kitamura S. Size and shape of cycloamylose estimated using column chromatography coupled with small-angle X-ray scattering. Food Hydrocoll. (2020) 108:105948.
120. Watanabe Y. Size-exclusion chromatography combined with solution X-ray scattering measurement of the heat-induced aggregates of water-soluble proteins at low ionic strength in a neutral solution. J Chromatogr A. (2019) 1603:190–8. doi: 10.1016/j.chroma.2019.06.042
121. Watanabe Y, Inoko Y. Characterization of a large glycoprotein proteoglycan by size-exclusion chromatography combined with light and X-ray scattering methods. J Chromatogr A. (2013) 1303:100–4. doi: 10.1016/j.chroma.2013.06.048
122. Ryan TM, Trewhella J, Murphy JM, Keown JR, Casey L, Pearce FG, et al. An optimized SEC-SAXS system enabling high X-ray dose for rapid SAXS assessment with correlated UV measurements for biomolecular structure analysis. J Appl Crystallogr. (2018) 51:97–111.
123. Kim JH, Min B, Yun YD, Choi HJ, Jin KS. Size-exclusion chromatography coupled with small-angle X-ray scattering on the 4C small-angle X-ray scattering beamline at pohang light source II. Bull Korean Chem Soc. (2020) 41:1052–5.
124. Inoue R, Nakagawa T, Morishima K, Sato N, Okuda A, Urade R, et al. Newly developed Laboratory-based Size exclusion chromatography Small-angle x-ray scattering System (La-SSS). Sci Rep. (2019) 9:12610. doi: 10.1038/s41598-019-48911-w
125. Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B. Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument. J Appl Crystallogr. (2018) 51:1623–32. doi: 10.1107/S1600576718014462
126. Banc A, Pincemaille J, Costanzo S, Chauveau E, Appavou M-S, Morel M-H, et al. Phase separation dynamics of gluten protein mixtures. Soft Matter. (2019) 15:6160–70.
127. Chen D, Kuzmenko I, Ilavsky J, Pinho L, Campanella O. Structural evolution during gelation of pea and whey proteins envisaged by time-resolved ultra-small-angle x-ray scattering (USAXS). Food Hydrocoll. (2022) 126:107449.
128. Chen D, Zhu X, Ilavsky J, Whitmer T, Hatzakis E, Jones OG, et al. Polyphenols Weaken Pea Protein Gel by Formation of Large Aggregates with Diminished Noncovalent Interactions. Biomacromolecules. (2021) 22:1001–14. doi: 10.1021/acs.biomac.0c01753
129. Tsung K-L, Ilavsky J, Padua GW. Formation and characterization of zein-based oleogels. J Agric Food Chem. (2020) 68:13276–81. doi: 10.1021/acs.jafc.0c00184
130. Da Vela S, Braun MK, Dörr A, Greco A, Möller J, Fu Z, et al. Kinetics of liquid–liquid phase separation in protein solutions exhibiting LCST phase behavior studied by time-resolved USAXS and VSANS. Soft Matter. (2016) 12:9334–41. doi: 10.1039/c6sm01837h
131. Mao Y, Su Y, Hsiao BS. Probing structure and orientation in polymers using synchrotron small- and wide-angle X-ray scattering techniques. Eur Polym J. (2016) 81:433–46.
132. Yamamoto K, Suzuki S, Kitamura S, Yuguchi Y. Gelation and structural formation of amylose by in situ neutralization as observed by small-angle X-ray scattering. Gels. (2018) 4:57.
133. Royes J, Bjørnestad VA, Brun G, Narayanan T, Lund R, Tribet C. Transition kinetics of mixed lipid:photosurfactant assemblies studied by time-resolved small angle X-ray scattering. J Colloid Interface Sci. (2022) 610:830–41. doi: 10.1016/j.jcis.2021.11.133
134. Gontsarik M, Yaghmur A, Salentinig S. Dispersed liquid crystals as pH-adjustable antimicrobial peptide nanocarriers. J Colloid Interface Sci. (2021) 583:672–82. doi: 10.1016/j.jcis.2020.09.081
135. Yaghmur A, Rappolt M, Jonassen ALU, Schmitt M, Larsen SW. In situ monitoring of the formation of lipidic non-lamellar liquid crystalline depot formulations in synovial fluid. J Colloid Interface Sci. (2021) 582:773–81.
136. Komorowski K, Schaeper J, Sztucki M, Sharpnack L, Brehm G, Köster S, et al. Vesicle adhesion in the electrostatic strong-coupling regime studied by time-resolved small-angle X-ray scattering. Soft Matter. (2020) 16:4142–54.
137. Khaliqi K, Ghazal A, Azmi IDM, Amenitsch H, Mortensen K, Salentinig S, et al. Direct monitoring of lipid transfer on exposure of citrem nanoparticles to an ethanol solution containing soybean phospholipids by combining synchrotron SAXS with microfluidics. Analyst. (2017) 142:3118–26. doi: 10.1039/c7an00860k
Keywords: SAXS, structural characterization, food biomacromolecules, modeling, in situ capabilities, time-resolved, chromatography, integrated structural biology
Citation: Sun Y, Li X, Chen R, Liu F and Wei S (2022) Recent advances in structural characterization of biomacromolecules in foods via small-angle X-ray scattering. Front. Nutr. 9:1039762. doi: 10.3389/fnut.2022.1039762
Received: 08 September 2022; Accepted: 03 November 2022;
Published: 17 November 2022.
Edited by:
Xiao Feng, Nanjing University of Finance and Economics, ChinaReviewed by:
Mohammad Alrosan, Universiti Sains Malaysia (USM), MalaysiaCopyright © 2022 Sun, Li, Chen, Liu and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song Wei, NTk3NTg2NjQxQHFxLmNvbQ==; Yang Sun, c3VueWFuZ0BoYnVhcy5lZHUuY24=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.