
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr., 17 November 2022
Sec. Food Chemistry
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.1012348
This article is part of the Research TopicResearch Advances of Modification and Nutrition Properties of Food Carbohydrates, volume IView all 13 articles
From Ziziphus Jujuba cv. Muzao fruit, a new polysaccharide (PZMP3-1) with high molecular weight was isolated. Constructional characterization revealed that PZMP3-1 comprized 2.56 rhamnose, 7.70 arabinoses, 3.73 galactose, and 6.73 galactose, and it has a 241 kDa average molecular weight. The principal structural components of PZMP3-1 were 1,2,4 and 1,4-linked GalpA, 1,4-linked Galp, 1,3 and 1,5-linked Araf, and 1-linked Rhap based on methylation and nuclear magnetic resonance spectroscopy (NMR) analyses. X-ray diffraction (XRD), Fourier transforms infrared spectroscopy (FT-IR), atomic force microscopy (AFM), and scanning electron microscopy (SEM) structural analysis of PZMP3-1 revealed a tangled and branching pattern. Overall, these structural results suggested that PZMP3-1 could have unique bioactivities and be widely used in nutritional supplements.
Recent studies have investigated the biological effects of polysaccharides (PZMP3-1) that include their abilities to suppress free radicals and exert anti-bacterial, anti-cancer, anti-tumor, anti-coagulant, anti-viral, and immunological effects, etc., (1, 2). The chemical composition and structural characteristics of polysaccharides could determine their biological effects (3). Numerous studies have demonstrated that sulfation produces excellent physiological effects that support health maintenance and disease prevention (4).
The fruit of Ziziphus Jujuba Mill., often known as jujube, is called Jujubae Fructus and is known by the Chinese names Dazao or Hongzao (5, 6). Jujube has been demonstrated as anti-oxidant, anti-provocative, anti-microbial, anti-cancer, cardiovascular, gastrointestinal protective, anti-HIV, neuroprotective, sedative-hypnotic, anxiolytic, and other bioactivities in vitro and in vivo technological research, demonstrating the fruit’s pharmacological potential (7, 8). Bioactive metabolites are responsible for these actions, containing polysaccharides, oligosaccharides, saponins, cyclopeptide alkaloids, minerals, triterpenoid acids, vitamins, and flavonoids, which are thought to be the distinctive and functional elements of the jujube fruit (9, 10).
The origin of Z. jujuba cv. Muzao fruit is mainly from Lüliang Shanxi Province and Yulin Shaanxi Province of China (11). Polysaccharides were found in jujube’s pharmacological components and have been linked to various health benefits, such as immunomodulation, anti-cancer, anti-oxidation, hypoglycemic, hepatoprotective, and gastrointestinal protection (12, 13). Numerous investigations on low molecular weight polysaccharides have emphasized their structural characteristics and pharmacological activities (14–16). Jujube polysaccharides with molecular weights varying between 104 and 106 Da have been detected in various experimental conditions, and these polysaccharides demonstrated anti-oxidant activities (2, 17). From Z. jujuba cv. Jinsixiaozao, the polysaccharides (four fractions) have Mw values ranging from 86 to 160 kDa, according to Li et al. (18, 19). However, the higher molecular weight fraction of Z. jujuba cv. Muzao polysaccharides is not present.
In the current study, from Z. jujuba, isolation and purification of a unique polysaccharide with high molecular weight, was given the designation PZMP3-1. The structural conformation and physicochemical properties of PZMP3-1 were detected by model analytical instruments [gas chromatography (GC), high-performance gel permeation chromatography (HPGPC), Fourier transforms infrared spectroscopy (FT-IR), methylation analysis, X-ray diffraction (XRD), nuclear magnetic resonance spectroscopy (NMR), atomic force microscopy (AFM), and scanning electron microscopy (SEM)]. The eventual objective of this research could be to offer a new scientific understanding of the composition of polysaccharides from jujube.
Ziziphus Jujuba cv. Muzao fruit was donated by Shaanxi Loess Plateau Experimental Orchard (China). GE Healthcare Life Sciences (Piscataway, NJ, USA) provided the Sephacryl S-300 gels and anion-exchange DEAE Sepharose Fast Flow. Sigma-Aldrich Co., Ltd (Sigma, St Louis, MO, USA) provided the standard monosaccharides. The analytical grade was used for all other compounds and reagents.
From Z. jujuba cv. Muzao, the unprocessed polysaccharides (ZMP) were isolated by water extraction, deproteinized, decolorized, precipitated by ethanol, and freeze-dried, as shown by the method of Ji et al. (20). The dissolved ZMP in deionized water was centrifuged and then filtered the supernatant through a membrane (0.45 μm), a 2.6 × 100 cm diethylaminoethyl (DEAE)-Sepharose Fast Flow column eluted with 0.3 M NaCl was loaded with ZMP. On a 2.6 × 100 cm Sephacryl S-300 column that was equilibrated with distilled water, a separated fraction was pooled, desalted, and further purified. PZMP3-1 and PZMP3-2 were pale yellow powders and derived from the fractions of the prominent peaks collected, concentrated, dialyzed, and lyophilized to get different parts (21). For further structural characterization, the PZMP3-1 fraction was employed.
The method (phenol-sulfuric acid) detected the total amount of carbohydrates in PZMP3-1, with glucose serving as the reference (22). Bradford’s technique assessed the protein content, with bovine serum albumin as the reference (23). The Folin-Ciocalteu test assessed the total phenolic content (24). At room temperature, the UV-vis spectra of the PZMP3-1 (1.0 mg/ml) in the wavelength (200–400 nm) were captured using a spectrophotometer (25).
The high-performance liquid chromatography (HPLC) measurement of PZMP3-1’s Mw was detected on an Agilent instrument (LC 1200, USA) with a 7.8 × 300 mm TSK-gel G3000PWxl column. Using a calibration curve, the Mw concerning dextran was calculated (26).
Utilizing a collection of monosaccharides as a standard, the Shimadzu GC (2014 C) with a high-performance capillary column DB-17 (30 ml × 0.25 mm ID; 0.25 μm film thickness, Agilent) measured the various monosaccharide by comparing retention durations and peak regions (27).
PZMP3-1 was combined with 100 mg of potassium bromide (KBr) powder before being crushed into granules for infrared spectral measurement between 4,000 and 400 cm–1. A spectrophotometer was used to measure the FT-IR spectra of polysaccharide (VERTEX 70, Bruker, Germany) (28).
To replace exchangeable protons, the freeze-dehydrated PZMP3-1 (50 mg) was dissolved in 2 ml of 99.9% D2O and freeze-dried three times. A Bruker-600 MHz NMR Spectrometer (Bruker, Rheinstetten, Germany) was used to record the one-dimensional NMR spectra for the 1H and 13C at 25°C. Using the standard Bruker NMR software, data were collected and examined (21).
As described in previous studies (29), PZMP3-1 was methylated, followed by hydrolysis, reduction, and acetylation, to analyze glycosyl bonds. The measurement of partially methylated alder aldehyde using gas chromatography/mass spectrometry (GC-MS) requires the use of GCMS-QP2010 Ultra apparatus and a DB-17MS capillary column (60.0 m × 0.25 mm × 0.25 μm).
PZMP3-1 used the XRD pattern to determine the crystal structure present. The angular range of the diffractometer was 5–50° (2θ), the step size was 0.01°, the scan speed was 15°/min, and tube pressure (40 kV) and tube flow (40 mA) were present. Using an SEM (S-4800, Japan), the morphological characteristics of PZMP3-1 were documented. We used the Cressington 208 HR Sputtering Coater with sputtering gold samples. We dissolved the polysaccharides with distilled water and dried the test samples dripping on the mica carrier surface at ambient pressure of 70°C. A 5500 atomic force microscope (Agilent) was used to create the AFM images (30).
Version 17.0 of SPSS was used for the statistical analysis. The data were reported as mean ± standard deviation (SD), with Duncan’s multiple-range test (p < 0.05) arriving after the analysis of variance (ANOVA) for each experiment, which was carried out in triplicate.
In the current study, the yield of about 3.82% of the fruit of the Z. jujuba cv. Muzao was used to produce crude ZMP. On a Sepharose Fast Flow column (DEAE), the ZMP was separated using 0.3 M NaCl solution (Figure 1). This fraction was then further purified using Sephacryl S-300 columns and yielded 3.04% of PZMP3-1, the same as the neutral polysaccharide PZMP1 from Z. jujuba cv. (2.95%) (31).
The number of carbohydrates in PZMP3-1, followed by the phenol-sulfuric acid method, was 95.35 ± 1.25%. The result was different with Z. jujuba cv. Hamidazao polysaccharides which Yang et al. reported that only 0.32% polyphenol and 2.11% protein were found (14). Figure 2A demonstrates that the lack of nucleic acids and proteins is shown by PZMP3-1’s UV spectrum (there is no absorption at 260–280 nm), which is in line with the research of chemical analysis as previously reported (32).
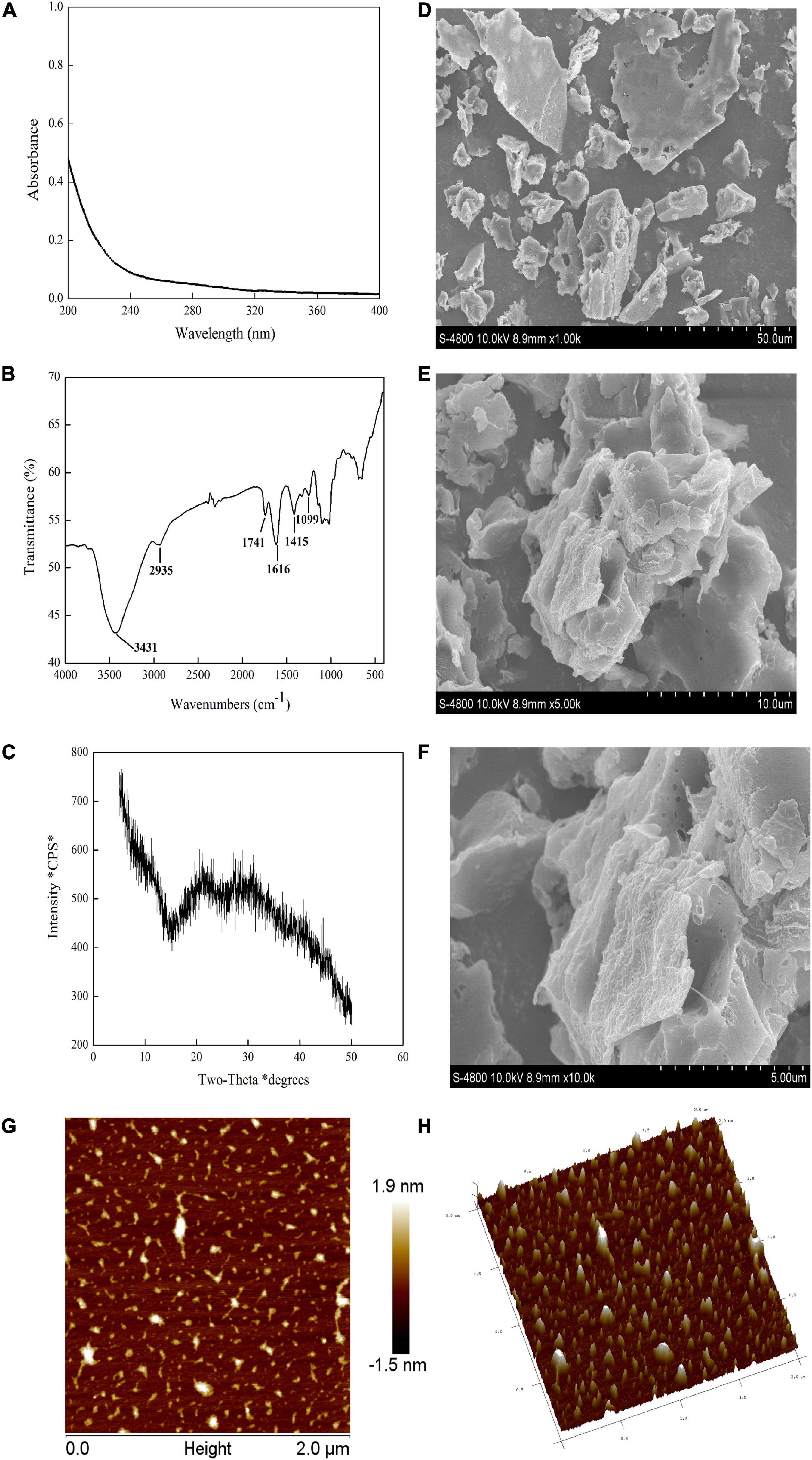
Figure 2. PZMP3-1’s physicochemical analysis. (A) The X-ray diffraction (XRD) pattern. (B) The spectrum of UV-vis. (C) The spectrum of Fourier transforms infrared spectroscopy (FT-IR). (D,E) Scanning electron microscopy (SEM) images [×1,000 in panel (D), ×5,000 in panel (E), and ×10,000 in panel (F)]. (G) Atomic force microscopy (AFM) images; (H) a three-dimensional image of AFM.
Biological activity depends on the Mw distribution of plant polysaccharides (33). The Mw of PZMP3-1 was determined to be 241 kDa based on the calibration curve of standards, which corresponded to a retention period of 17.10 min and might indicate a higher molecular weight polysaccharide. Li et al. observed that from Z. jujuba cv. Jinsixiaozao, the molecular weights of four polysaccharide fractions (ZSP1b, ZSP2, ZSP3c, and ZSP4b) varied between 86 and 160 kDa (19), and Cui et al. showed that Fructus Jujubae polysaccharides consisted mainly of low molecular weight (83.8 and 123.0 kDa) fractions (34). The PZMP3-1 obtained in this study significantly exceeded the polysaccharide Mw shown in previous studies. The Mw of Z. jujuba polysaccharide obtained by each research group was different. This may be due to the variety, extraction, purification process, and test method (13, 35).
The PMP-GC technique was used to examine the monosaccharides content of PZMP3-1. Rhamnose, arabinose, galactose, and galacturonic acid made up most of PZMP3-1, with the molar ratios being 2.56:7.70:3.73:6.73, with arabinose and galacturonic acid making up the most significant amount when compared to other monosaccharides, according to the monosaccharide standards. Previous reports (HJP1, the ratio of mannose to galacturonic acid to rhamnose to galactose to glucose to arabinose, was 1.3:6.7:27.6:13:3.7:47.6 and HJP3 was 0.6:16.7:16:21:6.5:39.2) reported by Wang et al. could be used to confirm the polysaccharide molar ratio and monosaccharide content found in Z. jujuba cv. Dongzao (36), which had rhamnose, arabinose, galactose, glucose, and xylose ratios of 1.0:3.6:1.0:0.5:0.2 (37) and had results that were distinct from those of PZMP3-1 in this work. The varietals, production conditions, and measurement techniques might play a role in the monosaccharide composition of jujube polysaccharides (13, 14).
Detecting distinctive organic groups in polysaccharides using FT-IR spectroscopy is a powerful technique (38, 39). The primary functional groups of plant polysaccharides may be better understood using the FT-IR spectrum. The FT-IR spectra showed that PZMP3-1 contained the typical absorption peaks of plant polysaccharides (Figure 2B; 40). Stretching vibration (O-H) caused the characteristic peak at 3,431 cm–1, and stretching vibration (C-H) caused the rise at 2,935 cm–1 (41). They were regarded as the defining bands for polymers comprized of plant polysaccharides because of their two significant absorption peaks. Stretching vibrations (carboxylic groups) were connected to the absorption peak at 1,741 cm–1 (29). PZMP3-1’s absorption peak at 1,616 cm–1 showed that symmetrical stretching vibrations (C = O) were present (42). The bands in the 1,415 cm–1 likely represented the bending and deformation of C-OH and C-H vibrations, respectively (43). The intense bands at 1,099 and neighboring 1,000 cm–1 revealed the pyranose form of galactosyl residues (44).
Less information about the detailed structure of jujube’s high molecular weight polysaccharides is available in the literature, especially the glycosidic bond types. Methylation analysis could determine the kind and number of glycosidic linkages in plant polysaccharide polymers (45). To determine PZMP3-1’s glycosidic bond types by GC-MS analysis, it was methylated, hydrolyzed, reduced, and converted into partially methylated alditol acetates (PMAAs) (46). The linkage patterns of PZMP3-1 were compiled, which are shown in Table 1, using a spectral database based on the PMAA standard date in the Complex Carbohydrate Research Center (CCRC) spectrum database, retention duration, and relevant literature. Six different methylated sugar derivatives were found, i.e., 1,2,4 and 1,4-linked GalpA, 1,4-linked Galp, 1,3 and 1,5-linked Araf, and 1-linked Rhap. The composition of monosaccharides and PZMP3-1 was discovered using methylation analysis (45). NMR spectra provided additional evidence supporting the structure of PZMP3-1.
To understand the structural characteristics of PZMP3-1, 1H and 13C NMR spectra (one-dimensional) were identified and resolved. The five residues of PZMP3-1 that GC-MS isolated were given chemical shifts (Table 2) based on 1D spectra (Figure 3) and information from the literature. Five anomeric signals were present in the 1H and 13C NMR spectra (Figures 3A,B) at 3.70–5.25 ppm and 60–110 ppm, respectively. The primary anomeric proton signals in PZMP3-1 were identified as A, B, C, D, and E in the 1H NMR spectra at 5.21, 5.21, 5.12, 5.12, and 4.94.
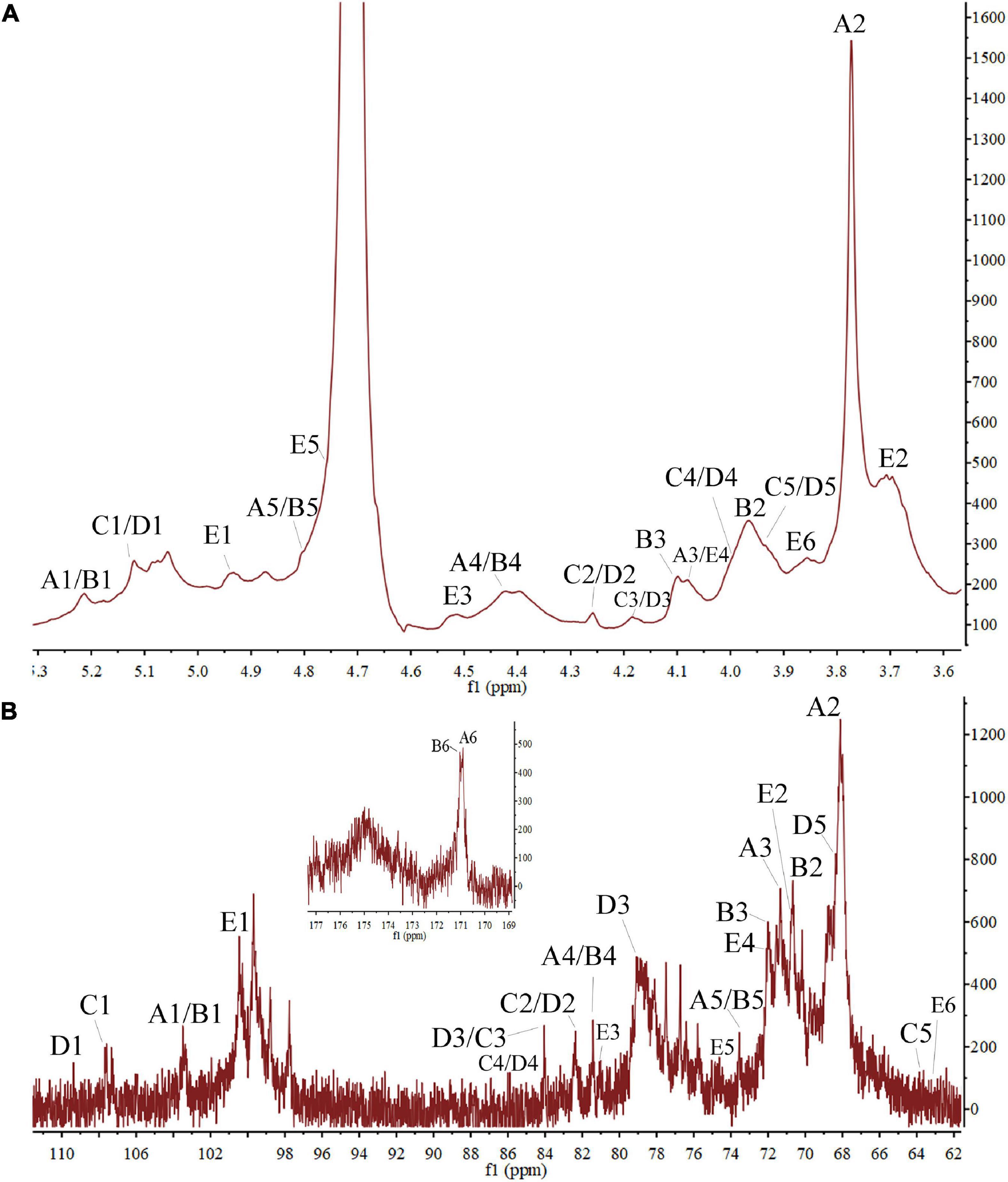
Figure 3. The PZMP3-1’s nuclear magnetic resonance spectroscopy (NMR) spectra in D2O. (A) 1H spectrum. (B) 13C spectrum.
In the 1H NMR spectrum, residues C-2, 3, 4, and 5’s protons received chemical shifts between 3.7 and 4.28. Five anomeric signals were resonated at 103.39, 103.39, 107.53, 109.35, and 100.52 after labeling the corresponding signal (allogeneic carbon) tagged in the 13C NMR spectra. Based on findings from the literature, Table 2 displays the results; the signals of all the tagged residues in the 1H and 13C NMR spectra are assigned. The heteropoly proton of residue A’s chemical shift caused the signal at δH 5.21, and in the heterogeneous carbon, the corresponding signal was seen at δC 103.39. The signals were created by the residue A’s C-2, 3, 4, 5, and 6 at δC 68.08/δH 3.77, δC 71.26/δH 4.08, δC 73.49/δH 4.80, and δC 170.91, respectively. According to the NMR data, this residue A’s chemical shifts were the same as those of →4)-GalpA-(1→ (17). Similar to how residue B was identified as →2,4)-GalpA-(1→, the signals at δC 2 70.62/δH 2 3.97, δC 3 71.98/δH 3 4.10, and δC 6 171.01 matched the residue B (anomeric carbons and protons) (47).
Araf was initially given credit for the cross-peak in the anomeric area at 107–110 ppm (residues C and D). The chemical shifts of H-1, 2, 3, 4, and 5 were determined from the 1H spectra at 5.12, 4.28, 4.18, 3.98, and 3.92 ppm, respectively (Figure 3A), in agreement with previous reports. From the proton chemical shifts in the C spectra, the carbon chemical shifts of the residue C from C-1, 2, 3, 4, and 5 were found (Table 2). The carbons and anomeric protons of residue D were identified by the signals at δC-1 109.35/δH-1 5.12, δC-3 79.06/δH-3 4.17, and δC-5 68.34/δH-5 3.93. These outcomes corroborated FT-IR and methylation analysis results, demonstrating that residues C/D were (1→3)-linked Araf/(1→5)-linked Araf (29, 31). According to the NMR data, the similar alternate signals for carbon and hydrogen were 100.52 (4.94), 70.56 (3.71), 80.96 (4.50), 72.01 (4.07), 74.39 (4.77), and 63.67 (3.85), this residue’s chemical changes were the same as those of 1,4-linked Galp (48, 49).
It is generally known that plant polysaccharides could be crystallized using XRD technology (50). Figure 2C depicts the X-ray diffraction pattern of PZMP3-1. PZMP3-1 had a prominent peak that appeared at around 20 and 30°, and its X-ray diffraction curves were “bun-shaped” (30, 51). The semicrystalline structure we obtained earlier could explain that PZMP3-1 might have some semicrystalline structures (an abundant polysaccharide in galacturonic acid from Z. jujuba cv. Muzao) (21, 52).
The findings of the SEM analysis of PZMP3-1’s surface morphology are shown in Figures 2D–F. PZMP3-1 was formed in an aggregation condition with an unsteady surface. The formation of intermolecular and intramolecular hydrogen bonds between polysaccharides requires the polysaccharide to have a higher molecular weight (30, 53). For this reason, PZMP3-1 revealed a tangled structure that is folded with each other.
Observing the three-dimensional structure of biologically active macromolecules requires using AFM and nanoscale microstructures, especially the form of plant polysaccharides (54, 55). PZMP3-1’s 3-dimensional and planar AFM pictures are displayed in Figures 2G,H, respectively. PZMP3-1 had a linear or branched structure, and the chain of PZMP3-1 featured a helical shape. PZMP3-1 aggregation might be explained by intermolecular and intramolecular hydrogen bonding on its surface, which acted as a catalyst to produce the potent intramolecular and intermolecular connections and the interactions with the water molecule (56, 57). Considering these morphological traits, we could conclude that the entangled and branched structure existed in PZMP3-1 molecules, which could significantly affect the bioactivity, structure, and distribution.
In the current work, Z. jujuba cv. Muzao fruit was used to create a new polysaccharide PZMP3-1 with high molecular weight, and its characteristics were elucidated through physicochemical and experimental investigations using modern analytical instruments. Rhamnose, galactose, arabinose, and galacturonic acid were the main components of PZMP3-1, which was 241 kDa in weight and had a molar ratio of 2.56:3.73:7.70:6.73. PZMP3-1’s primary linkage types included →2,4)-GalpA-(1→, →4)-GalpA-(1→, →4)-Galp-(1→, →5)-Araf-(1→, →3)-Araf-(1→ and Rhap-(1→ based on the findings of the methylation and NMR analyses. According to studies on chain conformation, PZMP3-1 was entangled with itself. The biological functions of PZMP3-1 and the links between its structure and activity are the subject of in-depth research.
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
XJ was involved in the study’s idea, design, and funding. ZW and XH set the database into order. YZ and YL wrote the initial draft of the document. XG participated in writing – review and editing. GL provided funding for this manuscript. All authors approved and reviewed the article’s submission.
This work was supported by the National Natural Science Foundation of China (32201969 and 82204668), Hebei Natural Science Foundation (H2022423376), Young and Middle-Aged Top Talent Project in Chongqing Three Georges Medical College, Chongqing Talent Project (cstc2022ycjh-bgzxm0226), Natural Science Foundation of Henan Province (212300410297), the Basic Research Plan of Higher Education School Key Scientific Research Project of Henan Province (21A550014), the Doctoral Research Foundation of Zhengzhou University of Light Industry (2020BSJJ015), and the Science and Technology Research Project of Higher Education in Hebei Province (QN2020233) are all acknowledged.
Author XH was employed by company Heilongjiang Feihe Dairy Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kadam SU, Tiwari BK, O’Donnell CP. Extraction, structure and biofunctional activities of laminarin from brown algae. Int J Food Sci Technol. (2015) 50:24–31. doi: 10.1111/ijfs.12692
2. Li J, Shang W, Si X, Bu D, Strappe P, Zhou Z, et al. Carboxymethylation of corn bran polysaccharide and its bioactive property. Int J Food Sci Technol. (2017) 52:1176–84. doi: 10.1111/ijfs.13382
3. Zong A, Cao H, Wang F. Anticancer polysaccharides from natural resources: a review of recent research. Carbohydrate Polymers. (2012) 90:1395–410. doi: 10.1016/j.carbpol.2012.07.026
4. Wang J, Hu S, Nie S, Yu Q, Xie M. Reviews on mechanisms of in vitro antioxidant activity of polysaccharides. Oxidative Med Cell Longevity. (2016) 2016:1–13. doi: 10.1155/2016/5692852
5. Liu S, Lv Y, Tang Z, Zhang Y, Xu H, Zhang D, et al. Ziziphus jujuba mill., a plant used as medicinal food: a review of its phytochemistry, pharmacology, quality control and future research. Phytochemistry Rev. (2021) 2s0:507–41. doi: 10.1007/s11101-020-09709-1
6. Ji X, Hou C, Gao Y, Xue Y, Yan Y, Guo X. Metagenomic analysis of gut microbiota modulatory effects of jujube (Ziziphus jujuba mill.) polysaccharides in a colorectal cancer mouse model. Food Function. (2020) 11:163–73. doi: 10.1039/C9FO02171J
7. Lu Y, Bao T, Mo J, Ni J, Chen W. Research advances in bioactive components and health benefits of jujube (Ziziphus jujuba mill.) fruit. J Zhejiang Univ Sci B. (2021) 22:431–49. doi: 10.1631/jzus.B2000594
8. Chen J, Tsim KW. Review of edible jujube, the Ziziphus jujuba fruit: a heath food supplement for anemia prevalence. Front Pharmacol. (2020) 11:593655. doi: 10.3389/fphar.2020.593655
9. Wang M, Gao Q, Shen J, Wang X, Ji X. The jujube (Ziziphus jujuba mill.) fruit: a review of current knowledge of fruit composition and health benefits. J Agric Food Chem. (2013) 61:3351–63. doi: 10.1021/jf4007032
10. Rashwan AK, Karim N, Shishir MRI, Bao T, Lu Y, Chen W. Jujube fruit: a potential nutritious fruit for the development of functional food products. J Funct Foods. (2020) 75:104205. doi: 10.1016/j.jff.2020.104205
11. Zhang L, Liu X, Wang Y, Liu G, Zhang Z, Zhao Z, et al. In vitro antioxidative and immunological activities of polysaccharides from Zizyphus jujuba cv. Muzao. Int J Biol Macromol. (2017) 95:1119–25. doi: 10.1016/j.ijbiomac.2016.10.102
12. Xie J, Tang W, Jin M, Li J, Xie M. Recent advances in bioactive polysaccharides from Lycium barbarum L., Zizyphus jujuba mill, Plantago spp., and morus spp.: structures and functionalities. Food Hydrocolloids. (2016) 60:148–60. doi: 10.1016/j.foodhyd.2016.03.030
13. Ji X, Peng Q, Yuan Y, Shen J, Xie X, Wang M. Isolation, structures and bioactivities of the polysaccharides from jujube fruit (Ziziphus jujuba mill.): a review. Food Chem. (2017) 227:349–57. doi: 10.1016/j.foodchem.2017.01.074
14. Yang Y, Qiu Z, Li L, Vidyarthi SK, Zheng Z, Zhang R. Structural characterization and antioxidant activities of one neutral polysaccharide and three acid polysaccharides from Ziziphus jujuba cv. hamidazao: a comparison. Carbohydrate Polymers. (2021) 261:117879. doi: 10.1016/j.carbpol.2021.117879
15. Zhu Y, He Z, Bao X, Wang M, Yin S, Song L, et al. Purification, in-depth structure analysis and antioxidant stress activity of a novel pectin-type polysaccharide from Ziziphus Jujuba cv. Muzao residue. J Funct Foods. (2021) 80:104439. doi: 10.1016/j.jff.2021.104439
16. Ji X, Hou C, Yan Y, Shi M, Liu Y. Comparison of structural characterization and antioxidant activity of polysaccharides from jujube (Ziziphus jujuba Mill.) fruit. Int J Biol Macromol. (2020) 149:1008–18. doi: 10.1016/j.ijbiomac.2020.02.018
17. Ji X, Hou C, Shi M, Yan Y, Liu Y. An insight into the research concerning Panax ginseng CA meyer polysaccharides: a review. Food Rev Int. (2022) 38:1149–65. doi: 10.1080/87559129.2020.1771363
18. Li J, Fan L, Ding S. Isolation, purification and structure of a new water-soluble polysaccharide from Zizyphus jujuba cv. jinsixiaozao. Carbohydrate Polymers. (2011) 83:477–82. doi: 10.1016/j.carbpol.2010.08.014
19. Li J, Liu Y, Fan L, Ai L, Shan L. Antioxidant activities of polysaccharides from the fruiting bodies of Zizyphus jujuba cv. jinsixiaozao. Carbohydrate Polymers. (2011) 84:390–4. doi: 10.1016/j.carbpol.2010.11.051
20. Ji X, Cheng Y, Tian J, Zhang S, Jing Y, Shi M. Structural characterization of polysaccharide from jujube (Ziziphus jujuba mill.) fruit. Chem Biol Technol Agric. (2021) 8:1–7. doi: 10.1186/s40538-021-00255-2
21. Ji X, Yan Y, Hou C, Shi M, Liu Y. Structural characterization of a galacturonic acid-rich polysaccharide from Ziziphus jujuba cv. Muzao. Int J Biol Macromol. (2020) 147:844–52. doi: 10.1016/j.ijbiomac.2019.09.244
22. Dubois M, Gilles KA, Hamilton JK, Rebers PAT, Smith F. Colorimetric method for determination of sugars and related substances. Analy Chem. (1956) 28:350–6. doi: 10.1021/ac60111a017
23. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analy Biochem. (1976) 72:248–54. doi: 10.1016/0003-2697(76)90527-3
24. Guo Y, Deng G, Xu X, Wu S, Li S, Xia E, et al. Antioxidant capacities, phenolic compounds and polysaccharide contents of 49 edible macro-fungi. Food Funct. (2012) 3:1195–205. doi: 10.1039/C2FO30110E
25. He B, Zheng Q, Guo L, Huang J, Yun F, Huang S, et al. Structural characterization and immune-enhancing activity of a novel high-molecular-weight polysaccharide from Cordyceps militaris. Int J Biol Macromol. (2020) 145:11–20. doi: 10.1016/j.ijbiomac.2019.12.115
26. Liu X, Liu H, Yan Y, Fan L, Yang J, Wang X, et al. Structural characterization and antioxidant activity of polysaccharides extracted from jujube using subcritical water. LWT Food Sci Technol. (2020) 117:108645. doi: 10.1016/j.lwt.2019.108645
27. Wang X, Zhang L, Wu J, Xu W, Wang X, Lü X. Improvement of simultaneous determination of neutral monosaccharides and uronic acids by gas chromatography. Food Chem. (2017) 220:198–207. doi: 10.1016/j.foodchem.2016.10.008
28. Wang L, Zhao Z, Zhao H, Liu M, Lin C, Li L, et al. Pectin polysaccharide from flos magnoliae (xin yi, magnolia biondii pamp. flower buds): hot-compressed water extraction, purification and partial structural characterization. Food Hydrocolloids. (2022) 122:107061. doi: 10.1016/j.foodhyd.2021.107061
29. Ji X, Zhang F, Zhang R, Liu F, Peng Q, Wang M. An acidic polysaccharide from Ziziphus jujuba cv. Muzao: purification and structural characterization. Food Chem. (2019) 274:494–9. doi: 10.1016/j.foodchem.2018.09.037
30. Tan M, Zhao Q, Zhao B. Physicochemical properties, structural characterization and biological activities of polysaccharides from quinoa (Chenopodium quinoa willd.) seeds. Int J Biol Macromol. (2021) 193:1635–44. doi: 10.1016/j.ijbiomac.2021.10.226
31. Ji X, Liu F, Peng Q, Wang M. Purification, structural characterization, and hypolipidemic effects of a neutral polysaccharide from Ziziphus jujuba cv. Muzao. Food Chem. (2018) 245:1124–30. doi: 10.1016/j.foodchem.2017.11.058
32. Jia X, Zhang C, Qiu J, Wang L, Bao J, Wang K, et al. Purification, structural characterization and anticancer activity of the novel polysaccharides from Rhynchosia minima root. Carbohydrate Polymers. (2015) 132:67–71. doi: 10.1016/j.carbpol.2015.05.059
33. Ji X, Peng B, Ding H, Cui B, Nie H, Yan Y. Purification, structure and biological activity of pumpkin polysaccharides: a review. Food Rev Int. (2021) 2021:1–13. doi: 10.1080/87559129.2021.1904973
34. Cui G, Zhang W, Wang Q, Zhang A, Mu H, Bai H, et al. Extraction optimization, characterization and immunity activity of polysaccharides from fructus jujubae. Carbohydrate Polymers. (2014) 111:245–55. doi: 10.1016/j.carbpol.2014.04.041
35. Hou C, Yin M, Lan P, Wang H, Nie H, Ji X. Recent progress in the research of angelica sinensis (oliv.) diels polysaccharides: extraction, purification, structure and bioactivities. Chem Biol Technol Agric. (2021) 8:1–14. doi: 10.1186/s40538-021-00214-x
36. Wang Y, Liu X, Zhang J, Liu G, Liu Y, Wang K, et al. Structural characterization and in vitro antitumor activity of polysaccharides from Zizyphus jujuba cv. Muzao. RSC Adv. (2015) 5:7860–7. doi: 10.1039/C4RA13350A
37. Zhao Z, Liu M, Tu P. Characterization of water soluble polysaccharides from organs of Chinese jujube (Ziziphus jujuba mill. cv. Dongzao). Eur Food Res Technol. (2008) 226:985–9. doi: 10.1007/s00217-007-0620-1
38. He Y, Liu C, Chen Y, Ji A, Shen Z, Xi T, et al. Isolation and structural characterization of a novel polysaccharide prepared from Arca subcrenata lischke. J Biosci Bioeng. (2007) 104:111–6. doi: 10.1263/jbb.104.111
39. Yang C, He N, Ling X, Ye M, Zhang C, Shao W, et al. The isolation and characterization of polysaccharides from longan pulp. Separation Purificat Technol. (2008) 63:226–30. doi: 10.1016/j.seppur.2008.05.004
40. Gao X, Qi J, Ho C, Li B, Xie Y, Chen S, et al. Purification, physicochemical properties, and antioxidant activities of two low-molecular-weight polysaccharides from Ganoderma leucocontextum fruiting bodies. Antioxidants. (2021) 10:1145. doi: 10.3390/antiox10071145
41. Wang M, Chen G, Chen D, Ye H, Sun Y, Zeng X, et al. Purified fraction of polysaccharides from Fuzhuan brick tea modulates the composition and metabolism of gut microbiota in anaerobic fermentation in vitro. Int J Biol Macromol. (2019) 140:858–70. doi: 10.1016/j.ijbiomac.2019.08.187
42. Liu Z, Jiao Y, Lu H, Shu X, Chen Q. Chemical characterization, antioxidant properties and anticancer activity of exopolysaccharides from Floccularia luteovirens. Carbohydrate Polymers. (2020) 229:115432. doi: 10.1016/j.carbpol.2019.115432
43. Zhang J, Chen M, Wen C, Zhou J, Gu J, Duan Y, et al. Structural characterization and immunostimulatory activity of a novel polysaccharide isolated with subcritical water from Sagittaria sagittifolia L. Int J Biol Macromol. (2019) 133:11–20. doi: 10.1016/j.ijbiomac.2019.04.077
44. Kpodo FM, Agbenorhevi JK, Alba K, Bingham RJ, Oduro IN, Morris GA, et al. Pectin isolation and characterization from six okra genotypes. Food Hydrocolloids. (2017) 72:323–30. doi: 10.1016/j.foodhyd.2017.06.014
45. Pei J, Wang Z, Ma H, Yan J. Structural features and antitumor activity of a novel polysaccharide from alkaline extract of Phellinus linteus mycelia. Carbohydrate Polymers. (2015) 115:472–7. doi: 10.1016/j.carbpol.2014.09.017
46. Yuan Q, Zhao L, Li Z, Harqin C, Peng Y, Liu J. Physicochemical analysis, structural elucidation and bioactivities of a high-molecular-weight polysaccharide from Phellinus igniarius mycelia. Int J Biol Macromol. (2018) 120:1855–64. doi: 10.1016/j.ijbiomac.2018.09.192
47. Xiong Q, Luo G, Zheng F, Wu K, Yang H, Chen L, et al. Structural characterization and evaluation the elicitors activity of polysaccharides from Chrysanthemum indicum. Carbohydrate Polymers. (2021) 263:117994. doi: 10.1016/j.carbpol.2021.117994
48. Cheng X, Shi S, Su J, Xu Y, Ordaz-Ortiz JJ, Li N, et al. Structural characterization of a heteropolysaccharide from fruit of Chaenomelese speciosa (sweet) nakai and its antitumor activity. Carbohydrate Polymers. (2020) 236:116065. doi: 10.1016/j.carbpol.2020.116065
49. Zhang S, An L, Li Z, Wang H, Shi L, Zhang J, et al. An active heteropolysaccharide from the rinds of Garcinia mangostana linn.: structural characterization and immunomodulation activity evaluation. Carbohydrate Polymers. (2020) 235:115929. doi: 10.1016/j.carbpol.2020.115929
50. Milani A, Jouki M, Rabbani M. Production and characterization of freeze-dried banana slices pretreated with ascorbic acid and quince seed mucilage: physical and functional properties. Food Sci Nutr. (2020) 8:3768–76. doi: 10.1002/fsn3.1666
51. Contreras-Jiménez B, Torres-Vargas OL, Rodríguez-García ME. Physicochemical characterization of quinoa (Chenopodium quinoa) flour and isolated starch. Food Chem. (2019) 298:124982. doi: 10.1016/j.foodchem.2019.124982
52. Qian J, Chen W, Zhang W, Zhang H. Adulteration identification of some fungal polysaccharides with SEM, XRD, IR and optical rotation: a primary approach. Carbohydrate Polymers. (2009) 78:620–5. doi: 10.1016/j.carbpol.2009.05.025
53. Chen Z, Zhao Y, Zhang M, Yang X, Yue P, Tang D, et al. Structural characterization and antioxidant activity of a new polysaccharide from Bletilla striata fibrous roots. Carbohydrate Polymers. (2020) 227:115362. doi: 10.1016/j.carbpol.2019.115362
54. Zhang L, Zhao S, Lai S, Chen F, Yang H. Combined effects of ultrasound and calcium on the chelate-soluble pectin and quality of strawberries during storage. Carbohydrate Polymers. (2018) 200:427–35. doi: 10.1016/j.carbpol.2018.08.013
55. Zhang Y, Xu X, Xu J, Zhang L. Dynamic viscoelastic behavior of triple helical lentinan in water: effects of concentration and molecular weight. Polymer. (2007) 48:6681–90. doi: 10.1016/j.polymer.2007.09.005
56. Wu Z, Li H, Wang Y, Yang D, Tan H, Zhan Y, et al. Optimization extraction, structural features and antitumor activity of polysaccharides from Ziziphus jujuba cv. Ruoqiangzao seeds. Int J Biol Macromol. (2019) 135:1151–61. doi: 10.1016/j.ijbiomac.2019.06.020
Keywords: jujube, high-molecular-weight polysaccharide, structural characterization, NMR, GC-MS
Citation: Ji X, Wang Z, Hao X, Zhu Y, Lin Y, Li G and Guo X (2022) Structural characterization of a new high molecular weight polysaccharide from jujube fruit. Front. Nutr. 9:1012348. doi: 10.3389/fnut.2022.1012348
Received: 05 August 2022; Accepted: 25 October 2022;
Published: 17 November 2022.
Edited by:
Jianhua Xie, Nanchang University, ChinaReviewed by:
Hua-Min Liu, Henan University of Technology, ChinaCopyright © 2022 Ji, Wang, Hao, Zhu, Lin, Li and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoli Li, bGlndW9saTkyOUAxNjMuY29t; Xudan Guo, Z3VveHVkYW4xMjNAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.