
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr., 21 January 2022
Sec. Nutritional Immunology
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.818902
This article is part of the Research TopicNutrients, Gut Microbiome, and Intestinal InflammationView all 35 articles
 Xue Guo1,2†
Xue Guo1,2† Chen Huang2†
Chen Huang2† Jing Xu1,2†
Jing Xu1,2† Haoming Xu2
Haoming Xu2 Le Liu3
Le Liu3 Hailan Zhao1,2
Hailan Zhao1,2 Jiaqi Wang1,2
Jiaqi Wang1,2 Wenqi Huang1,2
Wenqi Huang1,2 Wu Peng1,2
Wu Peng1,2 Ye Chen3
Ye Chen3 Yuqiang Nie1,2
Yuqiang Nie1,2 Yongjian Zhou1,2*
Yongjian Zhou1,2* Youlian Zhou1,2*
Youlian Zhou1,2*Inflammatory bowel disease (IBD), which includes ulcerative colitis (UC) and Crohn's disease (CD), is characterized by relapse and remission alternately. It remains a great challenge to diagnose and assess disease activity during IBD due to the lack of specific markers. While traditional biomarkers from plasma and stool, such as C-reactive protein (CRP), fecal calprotectin (FC), and S100A12, can be used to measure inflammation, they are not specific to IBD and difficult to determine an effective cut-off value. There is consensus that gut microbiota is crucial for intestinal dysbiosis is closely associated with IBD etiopathology and pathogenesis. Multiple studies have documented differences in the composition of gut microbiota between patients with IBD and healthy individuals, particularly regarding microbial diversity and relative abundance of specific bacteria. Patients with IBD have higher levels of Proteobacteria and lower amounts of Bacteroides, Eubacterium, and Faecalibacterium than healthy individuals. This review summarizes the pros and cons of using traditional and microbiota biomarkers to assess disease severity and treatment outcomes and addresses the possibility of using microbiota-focused interventions during IBD treatment. Understanding the role of microbial biomarkers in the assessment of disease activity and treatment outcomes has the potential to change clinical practice and lead to the development of more personalized therapies.
Inflammatory bowel disease (IBD) including ulcerative colitis (UC) and Crohn's disease (CD), is a chronic relapse-remitting disease characterized by intestinal inflammation. The conventional approach to treatment of IBD has mainly focused on symptom relief. However, treatment that only targets symptoms can be ineffective because symptoms do not consistently reflect the presence or severity of mucosal inflammation. Instead, a “treat-to-target” management approach aims to target mucosal healing in intestinal inflammation, especially histological healing, thereby improving patient prognosis (1). Thus, treatment would be improved through frequent, objective, and regular assessments of the inflammatory process (2). Biomarkers help to correctly categorize disease severity, distinguishing between patients who have exhibited improvement or resolution of their inflammation, and those who have had a recrudescence of inflammation after medically or surgically induced remission, even prior to recurrence of clinical symptoms. Previously, endoscopy was regarded as the gold standard for gastrointestinal diseases because this tool allows for visualization of the affected tissue and the opportunity for biopsy. However, endoscopy is limited in clinical application because of its invasiveness, high cost, reliance on anesthetics, and risk of intestinal perforation (3). In recent years, many serum and feces biomarkers have been proposed to help monitor disease and assess mucosal healing to effectively diagnose residual intestinal inflammation in patients with IBD.
Currently, the precise etiology of IBD is still not clear, but an inappropriate and persistent inflammatory response to commensal gut microbiomes in genetically susceptible individuals are currently thought to involve the pathogenesis of IBD. Moreover, epidemiological studies have showed that several environmental exposures such as diet, smoking, hygiene, antibiotics, mode of birth (vaginal vs. cesarean section), breast feeding, infections, and stress are known common risk factors for developing IBD. And all these aforementioned risk factors are well known to cause microbial perturbations, suggesting gut microbiota play a critical role in the pathogenesis of IBD (4–7). Indeed, studies show that the intestinal microbiota in patients with IBD is distinct from that found in healthy subjects (8–10). Microbial biomarkers, which are used to assess disease activity and treatment effectiveness, are promising method to be utilized in clinical practice and optimize personalized treatment strategies. There is also interest in the potential benefits of microbiome-modulating interventions, such as probiotics, prebiotics, antibiotics, fecal microbiota transplantation (FMT), and gene manipulation in IBD treatment. This review summarizes the characteristics of classical biomarkers commonly used in assessing IBD, describes research on gut microbiota (Figure 1), and discusses the advantages and disadvantages of bacteria as biomarkers for IBD.
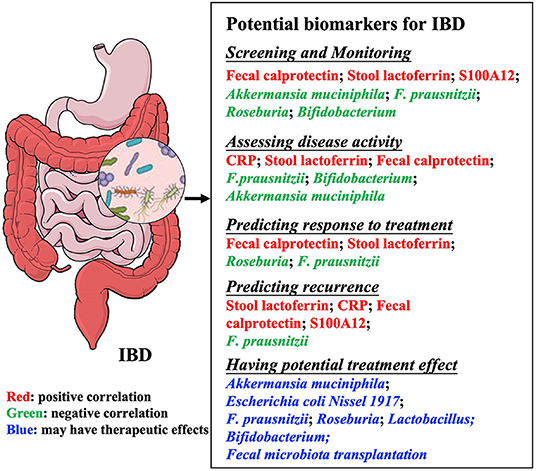
Figure 1. Summary of potential biomarkers in inflammatory bowel diseases. Microbial biomarkers are used for many purposes, including monitoring and evaluating disease activity, predicting recurrence or response to treatment, and treating diseases. IBD, Inflammatory bowel disease; CRP, C-reactive protein; F. prausnitzii, Faecalibacterium prausnitzii.
Laboratory tests that are fast, convenient, non-invasive, inexpensive, standardized, and repeatable, are widely used to assess disease progression. Some blood and stool biomarkers, including serum C-reactive protein (CRP), fecal calprotectin (FC), Calgranulin C (S100A12), stool lactoferrin (SL), and neutrophil-lymphocyte ratio (NLR) have been used as inflammatory indicators of IBD (Table 1). Some of these are used to distinguish IBD from irritable bowel syndrome (IBS) (24). For example, CRP is widely used in IBD screening and assessment of disease activity, clinical relapse and treatment responsiveness, serving as a predictor of surgical outcomes for a subgroup of patients with UC or CD (25, 26). In addition, baseline CRP levels, which reflect the persistence of severe disease and control of intestinal inflammation (27), can predict the response of patients with CD to anti-TNF and anti-adhesion molecule therapy (28). The high CRP group had a more severe clinical course (29) and a better response to infliximab and elevated CRP (>45 mg/L) in patients with IBD and can reliably predict the need for colon resection (30, 31). However, elevated serum CRP is not unique for intestinal inflammation, occurring in response to most systemic inflammatory disorders (14). In addition, there is considerable heterogeneity in CRP generation based on the genetics of individual patients (14). Plasma CRP is not a target for therapy, but rather a tool to monitor inflammation and guide radiology or endoscopy assessments.
Fecal calprotectin (FC) accounts for 60% of the cytoplasmic protein concentration of neutrophils and macrophages (32) and is considered the most reliable predictor of short-term recurrence and inflammatory activity in IBD (33–35). However, the cut-off value for fecal calprotectin has always been controversial and is affected by numerous factors including time of defecation (36), patient age (37), diet and lifestyle (38, 39), disease, medication usage (40), and storage conditions (36, 41). In addition, many studies have reported different cut-off values for fecal calprotectin, ranging from 100 to 250 μg/g (42, 43), such that only a moderate prediction of disease can be made for individual patients.
Calgranulin C, or S100A12, belongs to the S100 family of calcium-binding proteins. S100A12 is expressed as a cytoplasmic protein in neutrophils and has a pro-inflammatory function that includes potent chemotactic activity (44, 45). Both expression and secretion of S100A12 are increased in the serum and colon tissues from patients with IBD, and S100A12 levels in feces can be used as an indicator of mucosal healing and disease severity (15, 46). For example, S100A12 has 96% specificity for IBS vs. IBD when the threshold was set as 10 mg/kg. The odds ratio of stool S100A12 in IBD has a more accurate sensitivity, specificity, likelihood ratio, negative predictive value, and positive predictive value than FC and CRP even though studies set different thresholds (20, 47). S100A12 level can also increase in response to other diseases (48), however, the degree of daily change remains to be investigated. The lack of standardized detection methods may lead to different results (17, 49), which make it difficult to determine an accurate and effective cut-off value. In addition, there is no correlation between Crohn's Disease Activity Index (CDAI) and fecal S100A12 in patients with CD (47). Even though S100A12 has obvious advantages over FC, researchers have not yet reached a consensus, and the role of S100A12 in IBD development remains to be elucidated.
Stool lactoferrin (SL) is a multifunctional iron-binding glycoprotein. Inflammation in the intestinal lumen triggers the infiltration of pleomorphic granulocytes and neutrophils to the mucosal surface, which in turn causes a proportional increase in lactoferrin within the feces (50). There is a strong correlation between SL migration and the degree of mucosal inflammation determined by endoscopy (51). SL and FC have similar accuracy, sensitivity, and specificity (52). However, as compared with CRP and FC, SL testing has been primarily limited to research studies, likely as a result of the lower stability of lactoferrin at room temperature (53).
Of these biomarkers, fecal calprotectin (FC) correlates best with the number of active inflammation sites reported from biopsy than serum CRP and white blood cell count (15). Erythrocyte sedimentation rate (ESR), platelet count, and serum albumin level are also used to help assess IBD inflammation (54, 55), though they are not particularly accurate measures of the disease.
The gut microbiota harbor 100 times more genes than the host genome (56). The majority of gut microbiota belong to four phyla: Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria, with Firmicutes and Bacteroidetes predominating in healthy adults (57). Patients with UC and CD show a significant reduction in microbial diversity and alterations in some specific taxa (58) (Table 2). A prospective study of pediatric patients with CD (65) similarly concluded that gut dysbiosis is a marker for the presence and severity of inflammation. Multi-omics data supports the role of the microbiome in IBD by accurately describing the intestinal flora and its function, thus informing follow-up, diagnosis, prevention of recurrence or complications, and treatment (66). Understanding the role of microbial biomarkers in the assessment of disease activity and treatment outcomes is critical to the monitoring and treatment of IBD. A summary of recent findings relating to intestinal flora is described below.
Following the rapid advance of genomics, many studies found that patients with IBD can be distinguished from patients with IBS and healthy controls based on alterations in bacterial diversity and abundance of some particular bacterial communities (67–69). The main genera reduced in fecal samples of patients with CD are Bacteroides, Eubacterium, Faecalibacterium, and Ruminococcus (70, 71). Of these, extensive research has focused on Akkermansia muciniphila (71) and Faecalibacterium prausnitzii (72). Lopez-Siles et al. (73) assessed the presence of F. prausnitzii and E. coli in 28 healthy controls, 45 patients with CD, 28 patients with UC, and 10 patients with IBS. Findings confirmed that F prausnitzii was a specific indicator of IBD. The abundance of F. prausnitzii in patients with IBD was significantly lower than in IBS patients and healthy controls (P < 0.001). When F. prausnitzii is combined with E. coli, it can even distinguish colonic-CD from extensive colitis (73), suggesting that a combination of multiple bacteria may be more suitable biomarkers than individual bacteria for IBD. We previously found the gut microbiota is informative enough to distinguish HC from CD or UC samples with model accuracy of 89.5 and 93.2%, respectively (68). Similarly, the model built from RISK and PRISM biopsy samples (74) achieves high prediction accuracies as well, although Western fecal samples are less informative to classify IBD from HC (68).
The abundance of Actinobacteria and Proteobacteria was found to be relatively higher in patients with IBD than healthy subjects (68), while the abundance of Firmicutes was relatively lower. The degree of change to the flora was closely related to IBD severity. Recovery of Clostridiales is associated with disease remission and may be used to guide treatment. Fukuda and Fujita (72) obtained intestinal microflora from feces samples and used T-RFLP for OTU discriminant analysis, calculating standard discriminant function coefficients (Df) for each OTU. The sum of each OTU multiplied by the Df value is called the discriminant fraction (Ds). Patients in this experiment were divided into five groups, including patients with active UC (Group I), mild inflammation in the large intestine (Group IIa), no inflammation (Group IIb), consanguineous-healthy (Group III), and non-consanguineous-healthy (Group IV), The Ds values of each group decreased gradually from Group I to Group IV, suggesting that Ds may become a clinically relevant biomarker of disease activity in UC. He et al. (64) compared stool samples from patients with UC with different levels of inflammation and non-IBD controls and found lower diversity of microbiota in patients with UC, especially among those in the active stage. The abundance of Proteobacteria was significantly higher in patients with active UC and decreased significantly in patients in remission, while the reverse was true for Firmicutes. Klebsiella, Enterococcus, and Haemophilus were higher in patients with active UC, while Roseburia, Lachnospira, Blautia, and Faecalibacterium were higher in patients with UC who were in remission. A random-effects meta-analysis of 231 patients with CD and 392 patients with UC conducted by Prosberg et al. (75) showed that the abundance of F. prausnitzii was lower in patients with active CD and UC than those in remission, further suggesting that F. prausnitzii may be a reliable marker for assessing disease activity. Our previous study (68) also found that a relative increase in Actinobacteria and Proteobacteria (Enterobacteriaceae) and decrease in Firmicutes (Clostridiales) were strongly correlated with IBD severity by analyzing fecal microbiota in both UC and CD with mild, moderate or severe activity.
Anti-TNF treatment of IBD can trigger and maintain remission, improve quality of life, and alter the course of the disease (76, 77). However, nearly one-third of patients do not respond to anti-TNF treatment. Research indicates that changes in gut microbiota can reliably identify patients with CD responsiveness to TNF therapy, which can improve clinical management, reduce morbidity, and improve symptoms in these patients. Sanchis-Artero et al. (77) divided 27 CD patients initiating anti-TNF treatment into responders and non-responders to evaluate F. prausnitzii/Escherichia coli and F. prausnitzii/C. coccoides ratios before and after 6 months of treatment. Results suggest that the F. prausnitzii/Escherichia coli ratio can serve as a reliable early biomarker for identifying anti-TNF responsiveness in patients with CD. Kugathasan et al. (78) showed Veillonella was implicated in penetrating complications and Ruminococcus in stricturing CD in a prospective initial cohort study in children with CD. In our previous study, we also conclude that certain microbes, mainly Clostridiales, predicted the infliximab treatment effectiveness with 86.5% accuracy alone and 93.8% when combined with calprotectin levels and Crohn's disease activity index (CDAI) in a small CD cohort (68).
Approximately 50–75% of patients with CD need a bowel resection during the disease course (79), and 50% of patients have recurrent symptoms after surgery (80). To evaluate the risk of postoperative recurrence, ileal colonoscopy is currently recommended within 1 year after surgery to evaluate endoscopic recurrence using the Rutgeerts score. Currently, colonoscopy is still the gold standard for diagnosis and disease follow-up (81, 82), but a recent study suggests intestinal flora as a non-invasive marker. Pascal et al. (83) evaluated the predictive value of microbiota for postoperative recurrence using Kruskal-Wallis analysis of stool samples collected preoperatively from 54 patients and comparing them with the Rutgeerts score at 6 months after surgery. Streptococcus levels were higher in patients with postoperative recurrence, suggesting that the presence of these bacteria in preoperative stool samples may be a predictive marker of future recurrence. A reduction in F prausnitzii levels is related to a higher risk of recurrence in ileal CD. Soko et al. (84) analyzed the mucosal microflora of patients with CD at the time of surgical resection and 6 months post-surgery and found that a decrease in F. prausnitzii was related to endoscopic recurrence after 6 months. F. prausnitzii was positively correlated with the duration between flare-ups, with higher levels associated with longer remission times. Sokol et al. (85) compared the ileal microbiota of 201 patients in ileostomies at the time of resection and over the following year using 16S rRNA sequencing. Results showed that the abundance of ileal microbiota, including Gammaproteobacteria, Corynebacterium, and Ruminococcus gnavus, during resection, was significantly correlated with the risk of endoscopic recurrence. In addition, mounting evidence suggests that baseline gut microbiota may influence response to medication (86). Radhakrishnan et al. (87) conducted a systemic review by analyzing 19 articles that related to the association between anti-IBD drug treatment and gut microbiota. In this study, they concluded that compared to the IBD non-responders, those responders to enteral nutrition (EEN), infliximab, ustekinumab, and vedolizumab treatment have an increase in baseline microbial alpha diversity. Moreover, higher baseline Faecalibacterium levels can also predict responsiveness to ustekinumab and infliximab treatment.
During the last years, the therapeutic targets for IBD are focusing on “target” to treat that target the stage of mucosal healing in intestinal inflammation especially histological healing (88, 89). A significant proportion of patients with IBD are resistant to treatment using standard drugs. Even using optimal doses of 5-aminosalicylic acids (5-ASA) and/or azathioprine, annual recurrence rates can be as high as 25–40% (90). Introducing probiotics and prebiotics into the diet can help to restore the natural flora, prevent pathogenic bacteria infection by producing antimicrobial peptides and promote intestinal health by stimulating the growth and activity of beneficial bacteria through prebiotic fermentation. Bifidobacteria, lactobacillus, VSL#3, and butyric acid-producing bacteria have been used in clinical treatment. Probiotics effectively regulate the imbalance of intestinal flora, improve the microecological environment, enhance the intestinal mucosal barrier function, modulate the local and systemic immune responses, and provide new options for the treatment of diseases like IBD (91, 92).
Furthermore, researchers were also trying to address the potential of probiotic cocktails for IBD. For example, VSL3#, a mixture of eight probiotic cocktails, could upregulate the antagonists of NF-κB inflammatory pathway (93). Oral administration of 17 strains of bacteria to adult mice could attenuate colitis (94). Toumi et al. (95) reported that administration for a week of Ultrabiotique (Lactobacillus acidophilus, Bifidobacterium lactis, L. plantarum, and Bifidobacterium breve) can augment the production of intestinal mucus and goblet cells per crypt in mice, suggesting that probiotics may be a promising therapeutic intervention in situations that require immediate mucosal healing. Moreover, a four probiotic mixture containing L. plantarum, L. acidophilus, Enterococcus faecium, and L. rhamnosus can also exert positive effects on wound healing by increasing the wound healing rate of epithelial cells in vitro and enhancing the integrity of their tight junctions' formation (96). Dharmani et al. (97) reported that a mix of eight different probiotic bacteria promoted ulcer healing in rats through the induction of vascular endothelial growth factor (VEGF). These studies suggest probiotics have key roles in tissue repair or mucosal healing.
Currently, Fecal Microbiota Transplantation (FMT) is a novel UC treatment for intestinal microbiome disorders (98). Transplantation of functional bacteria from the stools of healthy people into the gastrointestinal tract of patients, and the reconstruction of new gut microbiota is an effective treatment for both intestinal and extra-intestinal disorders. FMT reduces intestinal permeability and disease severity by enhancing the production of short-chain fatty acids (SCFAs), particularly butyric acid, which helps maintain intestinal epithelial barrier integrity (99). FMT can also restore immune dysregulation by inhibiting Th1 differentiation, T cell activity, leukocyte adhesion, and the production of inflammatory factors (100, 101). Several multicenter, randomized, double-blind, placebo-controlled trials indicate that FMT induces remission in patients with active UC (100–103). In these studies, patients with active UC were divided into FMT and placebo groups. Remission rates were significantly higher in the FMT group than in the placebo group, with no difference in the incidence of adverse events. Treatment was most successful for newly diagnosed patients with UC, possibly because the microbiome was not as severely impacted by the disease. Although studies show that the intestinal flora of the host and donor are very similar approximately 2–4 weeks after FMT (104), it remains uncertain whether there are potential sequelae. Several questions still need to be addressed to optimize FMT treatment, including whether or not glucocorticoids should be discontinued during treatment, how time and intensity of intervention impact therapeutic effect, what inclusion criteria should be considered for the donor, whether single or multiple donors should be used for FMT, and whether aerobic or anaerobic treatment should be used for fecal treatment. Larger, long-term, multicenter studies with sufficient sample sizes and detailed microbiome analyses of both patients and donors will be needed to help inform UC treatment.
Diagnosis and management of IBD have evolved substantially in recent decades. Observational studies evaluating IBD disease outcomes have shown a reduction in surgery incidence, likely resulting from the development of novel therapeutics (105). While fecal calprotectin and other non-invasive markers of disease activity have allowed physicians to define disease activity more objectively, individual differences in response to specific drugs can affect treatment efficacy and/or toxicity. For example, 60% of patients with UC were shown to respond to mesalazine or 5-ASA treatment, suggesting that 40% of patients may not experience any benefit or may even have adverse drug effects (106). Primary non-responders have also been observed in 20–30% of patients with anti-TNFα treatment, vedolizumab, and/or ustekinumab treatment. This high rate of non-responsiveness may in part be explained by incomplete patient assessment preceding biological therapy. Therefore, it would be of great value to accurately predict responsiveness before therapy, but clinical predictors have proven insufficient and targeted markers are still lacking. For example, Hyams et al. (107) recruited pediatric patients aged 4–17 years with newly diagnosed UC from 29 centers in the USA and Canada. Findings supported the utility of assessing initial clinical activity and 4-week treatment responsiveness to mesalazine, showing 52-week glucocorticoid-free remission in children newly diagnosed with UC. Understanding individual clinical and biological disease features informs treatment decisions for UC. However, because up to one-third of patients without evidence of mucosal inflammation still present with gastrointestinal symptoms (108), there is clearly a need for more accurate markers to help guide clinical diagnosis.
Microbial biomarkers may also be useful to evaluate disease severity in individuals (64), helping to predict and monitor response to intervention. Endoscopic recurrence is linked to alterations of mucosa-associated microbiota in the ileal (109). Gut microbiota may aid clinicians in defining patient risk of postoperative relapse. Early prophylactic therapy can be initiated based on intestinal microbiome stratification (91). Current insights may also help in the design of microbiota modulation strategies to improve IBD outcomes (100). Studies described here present promising evidence that the intestinal bacteria may aid disease monitoring and enhance treatment efficacy for IBD like chemotherapy and immune checkpoint blockade improve cancer treatment (58, 66). Universally, higher baseline richness (without significant differences in baseline disease activity) and microbial diversity are linked to better outcomes, and less diverse gut microbiome is associated with more severe disease status.
In IBD, gut microbiota dysbiosis is associated with disease phenotypes and may be a causative or synergistic factor in prolonged or chronic inflammation. Thus, microbial treatment by restoring intestinal flora in patients with IBD has been attempted by an increasing number of scholars (110). For example, over the past decades, the frequency of clinical probiotic use has gradually increased, however, its efficacy has remained controversial. Several randomized clinical trials have shown that E. coli Nissle 1917 (111), VSL#3 (112), Bifidobacterium (113, 114) and other probiotics are effective in inducing remission or maintaining remission in IBD. Several studies have suggested that a decrease in the abundance of butyrate-producing bacteria is one of the key hallmarks of gut dysbiosis seen in IBD (60, 115, 116). Ideally, future research will focus on developing microbial-target treatments that restore gut microbial community homeostasis currently decreasing in IBD. There are promising outcomes for FMT in IBD therapy, with most patients achieving medication-free symptomatic control, and/or clinical remission (117).
The gut microbiome includes many potential biomarkers associated with disease activity and treatment outcomes in patients with IBD. Current insights may also help to design microbiota modulation strategies needed to improve IBD outcomes. Despite great advances in the microbiome field over the last decade, however, there are still a great number of issues that will need to be addressed before these findings are translated into effective therapeutic applications. While fecal flow research may be limited in its ability to detect microorganisms associated with the mucosal layer and thus directly involved in the initiation and continuation of disease, the application of this marker is far less invasive and allows for multiple and reproducible material collection, improving the monitoring of disease progression. First, most previous data were from cross-sectional rather than prospective longitudinal cohorts. Studies also differed in the information collection from IBD patients as many relevant important confounding information including diet, body mass index (BMI), antibiotic use, inflammation severity, and treatment interventions, etc. are determined to influence the gut microbiota (118). Second, the established studies mainly focus on the composition, especially the comparison of relative microbial abundances rather than the function of gut microbiota, which may be affected by total microbial counts and changes in the abundance of other species. Further, one major challenge with existing data is represented by the wide variability in the outcome definition. For example, the follow-up length, the number of outcomes assessed, and criteria met on the risk of bias assessment differed among the studies. Finally, an important limitation of existing studies is the lack of reproducibility of the microbiome in IBD. Most prediction studies did not include independent external validation cohorts. In addition, microbiome studies only focused on bacteria and did not explore other microorganisms such ad fungi and viruses, as well as how they interact with each other (10). Furthermore, studies have also varied in the depth of microbial sequencing, from PCR–based methods to 16s rRNA profiling to metagenomics sequencing and strain-level analysis. In short, this is still an open field and active research area for the investigation of gut microbiota in patients with IBD.
XG, CH, and JX are involved in the designing and drafting of the manuscript. HX, LL, HZ, JW, WH, and WP are responsible for searching and screening papers, and manuscript revision. YN and YC interpretation of the data and revision of the article. YouZ and YonZ concept and design of the study, interpretation of the data, and revision of the article. All authors contributed to the paper and approved the submitted version.
This work was supported by the grants from National Natural Science Foundation of China (No. NSFC 81700487), Natural Science Foundation of Guangdong Province (No. 2020A1515011000), Guangzhou Planned Project of Science and Technology (Nos. 202002030293 and 202002020012), Innovative Clinical Technique of Guangzhou (No. 2019GX05), and Projection of building a state-level synthesize hospital demonstrating traditional Chinese medicine and west medicine (No. 3012000000261).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Maaser C, Sturm A, Vavricka SR, Kucharzik T, Fiorino G, Annese V, et al. ECCO-ESGAR guideline for diagnostic assessment in IBD Part 1: initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis. (2019) 13:144–64. doi: 10.1093/ecco-jcc/jjy113
2. Bouguen G, Levesque BG, Feagan BG, Kavanaugh A, Peyrin-Biroulet L, Colombel JF, et al. Treat to target: a proposed new paradigm for the management of Crohn's disease. Clin Gastroenterol Hepatol. (2015) 13:1042–50. doi: 10.1016/j.cgh.2013.09.006
3. Ikhtaire S, Shajib MS, Reinisch W, Khan WI. Fecal calprotectin: its scope and utility in the management of inflammatory bowel disease. J Gastroenterol. (2016) 51:434–46. doi: 10.1007/s00535-016-1182-4
4. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. (2015) 12:205–17. doi: 10.1038/nrgastro.2015.34
5. Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. (2014) 16:2891–904. doi: 10.1111/1462-2920.12238
6. Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. (2014) 159:514–29. doi: 10.1016/j.cell.2014.09.048
7. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
8. Vaga S, Lee S, Ji B, Andreasson A, Talley NJ, Agreus L, et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci Rep. (2020) 10:14977. doi: 10.1038/s41598-020-71939-2
9. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. (2012) 9:599–608. doi: 10.1038/nrgastro.2012.152
10. Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. (2017) 14:573–84. doi: 10.1038/nrgastro.2017.88
11. Mendoza JL, Abreu MT. Biological markers in inflammatory bowel disease: practical consideration for clinicians. Gastroenterol Clin Biol. (2009) 33:S158–73. doi: 10.1016/S0399-8320(09)73151-3
12. Kathiresan S, Larson MG, Vasan RS, Guo CY, Gona P, Keaney JF, et al. Contribution of clinical correlates and 13 C-reactive protein gene polymorphisms to interindividual variability in serum C-reactive protein level. Circulation. (2006) 113:1415–23. doi: 10.1161/CIRCULATIONAHA.105.591271
13. Langley BO, Guedry SE, Goldenberg JZ, Hanes DA, Beardsley JA, Ryan JJ. Inflammatory bowel disease and neutrophil-lymphocyte ratio: a systematic scoping review. J Clin Med. (2021) 10:4219. doi: 10.3390/jcm10184219
14. Jones J, Loftus EV Jr, Panaccione R, Chen LS, Peterson S, et al. Relationships between disease activity and serum and fecal biomarkers in patients with Crohn's disease. Clin Gastroenterol Hepatol. (2008) 6:1218–24. doi: 10.1016/j.cgh.2008.06.010
15. Kyle BD, Agbor TA, Sharif S, Chauhan U, Marshall J, Halder SLS, et al. Fecal calprotectin, CRP and leucocytes in IBD patients: comparison of biomarkers with biopsy results. J Can Assoc Gastroenterol. (2021) 4:84–90. doi: 10.1093/jcag/gwaa009
16. Henderson P, Anderson NH, Wilson DC. The diagnostic accuracy of fecal calprotectin during the investigation of suspected pediatric inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. (2014) 109:637–45. doi: 10.1038/ajg.2013.131
17. Sidler MA, Leach ST, Day AS. Fecal S100A12 and fecal calprotectin as noninvasive markers for inflammatory bowel disease in children. Inflamm Bowel Dis. (2008) 14:359–66. doi: 10.1002/ibd.20336
18. Mosli MH, Zou G, Garg SK, Feagan SG, MacDonald JK, Chande N, et al. C-reactive protein, fecal calprotectin, and stool lactoferrin for detection of endoscopic activity in symptomatic inflammatory bowel disease patients: a systematic review and meta-analysis. Am J Gastroenterol. (2015) 110:802–19. doi: 10.1038/ajg.2015.120
19. Menees SB, Powell C, Kurlander J, Goel A, Chey WD. A meta-analysis of the utility of C-reactive protein, erythrocyte sedimentation rate, fecal calprotectin, and fecal lactoferrin to exclude inflammatory bowel disease in adults with IBS. Am J Gastroenterol. (2015) 110:444–54. doi: 10.1038/ajg.2015.6
20. Kaiser T, Langhorst J, Wittkowski H, Becker K, Friedrich AW, Rueffer A, et al. Faecal S100A12 as a non-invasive marker distinguishing inflammatory bowel disease from irritable bowel syndrome. Gut. (2007) 56:1706–13. doi: 10.1136/gut.2006.113431
21. Ordás I, Feagan BG, Sandborn WJ. Therapeutic drug monitoring of tumor necrosis factor antagonists in inflammatory bowel disease. Clin Gastroenterol Hepatol. (2012) 10:1079–87. doi: 10.1016/j.cgh.2012.06.032
22. Parsi MA, Shen B, Achkar JP, Remzi FF, Goldblum JR, Boone J, et al. Fecal lactoferrin for diagnosis of symptomatic patients with ileal pouch-anal anastomosis. Gastroenterology. (2004) 126:1280–6. doi: 10.1053/j.gastro.2004.02.012
23. Dulai PS, Peyrin-Biroulet L, Danese S, Sands BE, Dignass A, Turner D, et al. Approaches to integrating biomarkers into clinical trials and care pathways as targets for the treatment of inflammatory bowel diseases. Gastroenterology. (2019) 157:1032–43. doi: 10.1053/j.gastro.2019.06.018
24. Liu F, Lee SA, Riordan SM, Zhang L, Zhu L. Global studies of using fecal biomarkers in predicting relapse in inflammatory bowel disease. Front Med. (2020) 7:580803. doi: 10.3389/fmed.2020.580803
25. Henderson P, Kennedy NA, Van Limbergen JE, Cameron FL, Satsangi J, Russell RK, et al. Serum C-reactive protein and CRP genotype in pediatric inflammatory bowel disease: influence on phenotype, natural history, and response to therapy. Inflamm Bowel Dis. (2015) 21:596–605. doi: 10.1097/MIB.0000000000000296
26. Sandor Kiss L, Papp M, Dorottya Lovasz B, Vegh Z, Anna Golovics P, Janka E, et al. High-sensitivity C-reactive protein for identification of disease phenotype, active disease, and clinical relapses in Crohn's disease: a marker for patient classification? Inflamm Bowel Dis. (2012) 18:1647–54. doi: 10.1002/ibd.21933
27. Langhorst J, Elsenbruch S, Koelzer J, Rueffer A, Michalsen A, Dobos GJ. Noninvasive markers in the assessment of intestinal inflammation in inflammatory bowel diseases: performance of fecal lactoferrin, calprotectin, and PMN-elastase, CRP. and clinical indices. Am J Gastroenterol. (2008) 103:162–9. doi: 10.1111/j.1572-0241.2007.01556.x
28. Wright EK, Kamm MA, De Cruz P, Hamilton AL, Selvaraj F, Princen F, et al. Anti-TNF therapeutic drug monitoring in postoperative Crohn's Disease. J Crohns Colitis. (2018) 12:653–61. doi: 10.1093/ecco-jcc/jjy003
29. Yang RX, Song WJ, Wu ZQ, Goyal H, Xu HG. Association of serum neuron-specific enolase and C-reactive protein with disease location and endoscopic inflammation degree in patients with Crohn's disease. Front Med. (2021) 8:663920. doi: 10.3389/fmed.2021.663920
30. Koelewijn CL, Schwartz MP, Samsom M, Oldenburg B. C-reactive protein levels during a relapse of Crohn's disease are associated with the clinical course of the disease. World J Gastroenterol. (2008) 14:85–9. doi: 10.3748/wjg.14.85
31. Iaculli E, Agostini M, Biancone L, Fiorani C, Di Vizia A, Montagnese F, et al. C-reactive protein levels in the perioperative period as a predictive marker of endoscopic recurrence after ileo-colonic resection for Crohn's disease. Cell Death Discov. (2016) 2:1–8. doi: 10.1038/cddiscovery.2016.32
32. Amitai MM, Zarchin M, Lahat A, Yablecovitch D, Neuman S, Levhar N, et al. Structural bowel damage in quiescent Crohn's disease. Dig Liver Dis. (2017) 49:490–4. doi: 10.1016/j.dld.2017.02.001
33. Kostas A, Siakavellas SI, Kosmidis C, Takou A, Nikou J, Maropoulos G, et al. Fecal calprotectin measurement is a marker of short-term clinical outcome and presence of mucosal healing in patients with inflammatory bowel disease. World J Gastroenterol. (2017) 23:7387–96. doi: 10.3748/wjg.v23.i41.7387
34. Jukic A, Bakiri L, Wagner EF, Tilg H, Adolph TE. Calprotectin: from biomarker to biological function. Gut. (2021) 70:1978–88. doi: 10.1136/gutjnl-2021-324855
35. Wright EK, Kamm MA, De Cruz P, Hamilton AL, Ritchie KJ, Krejany EO, et al. Measurement of fecal calprotectin improves monitoring and detection of recurrence of Crohn's disease after surgery. Gastroenterology. (2015) 148:938–47. doi: 10.1053/j.gastro.2015.01.026
36. Lasson A, Stotzer PO, Ohman L, Isaksson S, Sapnara M, Strid H. The intra-individual variability of faecal calprotectin: a prospective study in patients with active ulcerative colitis. J Crohns Colitis. (2015) 9:26–32. doi: 10.1016/j.crohns.2014.06.002
37. Joshi S, Lewis SJ, Creanor S, Ayling RM. Age-related faecal calprotectin, lactoferrin and tumour M2-PK concentrations in healthy volunteers. Ann Clin Biochem. (2010) 47:259–63. doi: 10.1258/acb.2009.009061
38. Poullis A, Foster R, Shetty A, Fagerhol MK, Mendall MA. Bowel inflammation as measured by fecal calprotectin: a link between lifestyle factors and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. (2004) 13:279–84. doi: 10.1158/1055-9965.EPI-03-0160
39. Konikoff MR, Denson LA. Role of fecal calprotectin as a biomarker of intestinal inflammation in inflammatory bowel disease. Inflamm Bowel Dis. (2006) 12:524–34. doi: 10.1097/00054725-200606000-00013
40. Weis S, Schwiertz A, Unger MM, Becker A, Fassbender K, Ratering S, et al. Effect of Parkinson's disease and related medications on the composition of the fecal bacterial microbiota. NPJ Parkinsons Dis. (2019) 5:28. doi: 10.1038/s41531-019-0100-x
41. Oyaert M, Van den Bremt S, Boel A, Bossuyt X, Van Hoovels L. Do not forget about pre-analytics in faecal calprotectin measurement! Clin Chim Acta. (2017) 473:124–6. doi: 10.1016/j.cca.2017.08.025
42. Suchismita A, Jha A. IDDF2019-ABS-0129 Optimal cut-off value of fecal calprotectin for the evaluation of inflammatory bowel disease: an unsolved issue? Gut. (2019) 68:A85–6. doi: 10.1136/gutjnl-2019-IDDFAbstracts.160
43. Lee SH, Mainman H, Borthwick H, Dhar A. PTH-094 faecal calprotectin testing in primary and secondary care – are the current manufacturer's cut-off levels clinically useful? Gut. (2013) 62:A249.2-A. doi: 10.1136/gutjnl-2013-304907.581
44. Vrabie R, Kane S. Noninvasive markers of disease activity in inflammatory bowel disease. Gastroenterol Hepatol. (2014) 10:576–84.
45. Foell D, Kucharzik T, Kraft M, Vogl T, Sorg C, Domschke W, et al. Neutrophil derived human S100A12 (EN-RAGE) is strongly expressed during chronic active inflammatory bowel disease. Gut. (2003) 52:847–53. doi: 10.1136/gut.52.6.847
46. Däbritz J, Langhorst J, Lugering A, Heidemann J, Mohr M, Wittkowski H, et al. Improving relapse prediction in inflammatory bowel disease by neutrophil-derived S100A12. Inflamm Bowel Dis. (2013) 19:1130–8. doi: 10.1097/MIB.0b013e318280b1cd
47. de Jong NS, Leach ST, Day AS. Fecal S100A12: a novel noninvasive marker in children with Crohn's disease. Inflamm Bowel Dis. (2006) 12:566–72. doi: 10.1097/01.ibd.0000227626.72271.91
48. Leach ST, Yang Z, Messina I, Song C, Geczy CL, Cunningham AM, et al. Serum and mucosal S100 proteins, calprotectin (S100A8/S100A9) and S100A12, are elevated at diagnosis in children with inflammatory bowel disease. Scand J Gastroenterol. (2007) 42:1321–31. doi: 10.1080/00365520701416709
49. Whitehead SJ, Ford C, Gama RM, Ali A, McKaig B, Waldron JL, et al. Effect of faecal calprotectin assay variability on the management of inflammatory bowel disease and potential role of faecal S100A12. J Clin Pathol. (2017) 70:1049–56. doi: 10.1136/jclinpath-2017-204340
50. Kopylov U, Rosenfeld G, Bressler B, Seidman E. Clinical utility of fecal biomarkers for the diagnosis and management of inflammatory bowel disease. Inflamm Bowel Dis. (2014) 20:742–56. doi: 10.1097/01.MIB.0000442681.85545.31
51. Klimczak K, Lykowska-Szuber L, Eder P, Krela-Kazmierczak I, Stawczyk-Eder K, Szymczak A, et al. The diagnostic usefulness of fecal lactoferrin in the assessment of Crohn's disease activity. Eur J Intern Med. (2015) 26:623–7. doi: 10.1016/j.ejim.2015.06.015
52. D'Incà R, Dal Pont E, Di Leo V, Ferronato A, Fries W, Vettorato MG, et al. Calprotectin and lactoferrin in the assessment of intestinal inflammation and organic disease. Int J Colorectal Dis. (2007) 22:429–37. doi: 10.1007/s00384-006-0159-9
53. Silberer H, Kuppers B, Mickisch O, Baniewicz W, Drescher M, Traber L, et al. Fecal leukocyte proteins in inflammatory bowel disease and irritable bowel syndrome. Clin Lab. (2005) 51:117–26.
54. El Amrousy D, El Ashry H, Hodeib H, Hassan S. Vitamin D in children with inflammatory bowel disease: a randomized controlled clinical trial. J Clin Gastroenterol. (2021) 55:815–20. doi: 10.1097/MCG.0000000000001443
55. Matsubayashi M, Kobayashi T, Okabayashi S, Nakano M, Sagami S, Ozaki R, et al. Determining the usefulness of capsule scoring of ulcerative colitis in predicting relapse of inactive ulcerative colitis. J Gastroenterol Hepatol. (2021) 36:943–50. doi: 10.1111/jgh.15220
56. Bäckhed F. Programming of host metabolism by the gut microbiota. Ann Nutr Metab. (2011) 58:44–52. doi: 10.1159/000328042
57. Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. (2015) 21:8787–803. doi: 10.3748/wjg.v21.i29.8787
58. Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. (2004) 53:685–93. doi: 10.1136/gut.2003.025403
59. Kabeerdoss J, Sankaran V, Pugazhendhi S, Ramakrishna BS. Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol. (2013) 13:20. doi: 10.1186/1471-230X-13-20
60. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. (2014) 63:1275–83. doi: 10.1136/gutjnl-2013-304833
61. Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. (2011) 60:631–7. doi: 10.1136/gut.2010.223263
62. Fite A, Macfarlane S, Furrie E, Bahrami B, Cummings JH, Steinke DT, et al. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J Clin Microbiol. (2013) 51:849–56. doi: 10.1128/JCM.02574-12
63. Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. (2010) 105:2420–8. doi: 10.1038/ajg.2010.281
64. He XX, Li YH, Yan PG, Meng XC, Chen CY, Li KM, et al. Relationship between clinical features and intestinal microbiota in Chinese patients with ulcerative colitis. World J Gastroenterol. (2021) 27:4722–37. doi: 10.3748/wjg.v27.i28.4722
65. Di Biase AR, Marasco G, Ravaioli F, Dajti E, Colecchia L, Righi B, et al. Gut microbiota signatures and clinical manifestations in celiac disease children at onset: a pilot study. J Gastroenterol Hepatol. (2021) 36:446–54. doi: 10.1111/jgh.15183
66. Lacroix V, Cassard A, Mas E, Barreau F. Multi-omics analysis of gut microbiota in inflammatory bowel diseases: what benefits for diagnostic, prognostic and therapeutic tools? Int J Mol Sci. (2021) 22:11255. doi: 10.3390/ijms222011255
67. de Meij TGJ, de Groot EFJ, Peeters CFW, de Boer NKH, Kneepkens CMF, Eck A, et al. Variability of core microbiota in newly diagnosed treatment-naive paediatric inflammatory bowel disease patients. PLoS ONE. (2018) 13:e0197649. doi: 10.1371/journal.pone.0197649
68. Zhou Y, Xu ZZ, He Y, Yang Y, Liu L, Lin Q, et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems. (2018) 3:e00188–17. doi: 10.1128/mSystems.00188-17
69. Wills ES, Jonkers DM, Savelkoul PH, Masclee AA, Pierik MJ, Penders J. Fecal microbial composition of ulcerative colitis and Crohn's disease patients in remission and subsequent exacerbation. PLoS ONE. (2014) 9:e90981. doi: 10.1371/journal.pone.0090981
70. Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn's disease. Digestion. (2016) 93:59–65. doi: 10.1159/000441768
71. Yu Y, Yang W, Li Y, Cong Y. Enteroendocrine cells: sensing gut microbiota and regulating inflammatory bowel diseases. Inflamm Bowel Dis. (2020) 26:11–20. doi: 10.1093/ibd/izz217
72. Fukuda K, Fujita Y. Determination of the discriminant score of intestinal microbiota as a biomarker of disease activity in patients with ulcerative colitis. BMC Gastroenterol. (2014) 14:49. doi: 10.1186/1471-230X-14-49
73. Lopez-Siles M, Martinez-Medina M, Busquets D, Sabat-Mir M, Duncan SH, Flint HJ, et al. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish Irritable Bowel Syndrome and inflammatory bowel disease phenotypes. Int J Med Microbiol. (2014) 304:464–75. doi: 10.1016/j.ijmm.2014.02.009
74. Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. (2014) 15:382–92. doi: 10.1016/j.chom.2014.02.005
75. Prosberg M, Bendtsen F, Vind I, Petersen AM, Gluud LL. The association between the gut microbiota and the inflammatory bowel disease activity: a systematic review and meta-analysis. Scand J Gastroenterol. (2016) 51:1407–15. doi: 10.1080/00365521.2016.1216587
76. Monif GR. Understanding therapeutic concepts in Crohn's disease. Clin Med Insights Gastroenterol. (2018) 11:1179552218815169. doi: 10.1177/1179552218815169
77. Sanchis-Artero L, Martinez-Blanch JF, Manresa-Vera S, Cortes-Castell E, Valls-Gandia M, Iborra M, et al. Evaluation of changes in intestinal microbiota in Crohn's disease patients after anti-TNF alpha treatment. Sci Rep. (2021) 11:10016. doi: 10.1038/s41598-021-88823-2
78. Kugathasan S, Denson LA, Walters TD, Kim MO, Marigorta UM, Schirmer M, et al. Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet. (2017) 389:1710–8. doi: 10.1016/S0140-6736(17)30317-3
79. Cosnes J, Nion-Larmurier I, Beaugerie L, Afchain P, Tiret E, Gendre JP. Impact of the increasing use of immunosuppressants in Crohn's disease on the need for intestinal surgery. Gut. (2005) 54:237–41. doi: 10.1136/gut.2004.045294
80. de Buck van Overstraeten A, Eshuis EJ, Vermeire S, Van Assche G, Ferrante M, D'Haens GR, et al. Short- and medium-term outcomes following primary ileocaecal resection for Crohn's disease in two specialist centres. Br J Surg. (2017) 104:1713–22. doi: 10.1002/bjs.10595
81. De Cruz P, Kamm MA, Hamilton AL, Ritchie KJ, Krejany EO, Gorelik A, et al. Crohn's disease management after intestinal resection: a randomised trial. Lancet. (2015) 385:1406–17. doi: 10.1016/S0140-6736(14)61908-5
82. Regueiro M, Feagan BG, Zou B, Johanns J, Blank MA, Chevrier M, et al. Infliximab reduces endoscopic, but not clinical, recurrence of Crohn's disease after ileocolonic resection. Gastroenterology. (2016) 150:1568–78. doi: 10.1053/j.gastro.2016.02.072
83. Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, et al. A microbial signature for Crohn's disease. Gut. (2017) 66:813–22. doi: 10.1136/gutjnl-2016-313235
84. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. (2008) 105:16731–6. doi: 10.1073/pnas.0804812105
85. Sokol H, Brot L, Stefanescu C, Auzolle C, Barnich N, Buisson A, et al. Prominence of ileal mucosa-associated microbiota to predict postoperative endoscopic recurrence in Crohn's disease. Gut. (2020) 69:462–72. doi: 10.1136/gutjnl-2019-318719
86. Oancea I, Movva R, Das I, Aguirre de Carcer D, Schreiber V, Yang Y, et al. Colonic microbiota can promote rapid local improvement of murine colitis by thioguanine independently of T lymphocytes and host metabolism. Gut. (2017) 66:59–69. doi: 10.1136/gutjnl-2015-310874
87. Radhakrishnan ST, Alexander JL, Mullish BH, Gallagher KI, Powell N, Hicks LC, et al. Systematic review: the association between the gut microbiota and medical therapies in inflammatory bowel disease. Aliment Pharmacol Ther. (2022) 55:26–48. doi: 10.1111/apt.16656
88. Le Berre C, Peyrin-Biroulet L, Group S-Is. Selecting end points for disease-modification trials in inflammatory bowel disease: the SPIRIT consensus from the IOIBD. Gastroenterology. (2021) 160:1452–60. doi: 10.1053/j.gastro.2020.10.065
89. Turner D, Ricciuto A, Lewis A, D'Amico F, Dhaliwal J, Griffiths AM, et al. STRIDE-II: an update on the selecting therapeutic targets in inflammatory bowel disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): determining therapeutic goals for treat-to-target strategies in IBD. Gastroenterology. (2021) 160:1570–83. doi: 10.1053/j.gastro.2020.12.031
90. Madarame A, Fukuzawa M, Yamauchi Y, Kono S, Sugimoto A, Yamaguchi H, et al. Predictive factors of relapse after dose reduction of oral 5-aminosalicylic acid in patients with ulcerative colitis in the remission phase. PLoS ONE. (2021) 16:e0255620. doi: 10.1371/journal.pone.0255620
91. Balakrishnan M, Floch MH. Prebiotics, probiotics and digestive health. Curr Opin Clin Nutr Metab Care. (2012) 15:580–5. doi: 10.1097/MCO.0b013e328359684f
92. Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, et al. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. (2000) 289:1560–3. doi: 10.1126/science.289.5484.1560
93. Reiff C, Delday M, Rucklidge G, Reid M, Duncan G, Wohlgemuth S, et al. Balancing inflammatory, lipid, and xenobiotic signaling pathways by VSL#3, a biotherapeutic agent, in the treatment of inflammatory bowel disease. Inflamm Bowel Dis. (2009) 15:1721–36. doi: 10.1002/ibd.20999
94. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. (2013) 500:232–6. doi: 10.1038/nature12331
95. Toumi R, Abdelouhab K, Rafa H, Soufli I, Raissi-Kerboua D, Djeraba Z, et al. Beneficial role of the probiotic mixture Ultrabiotique on maintaining the integrity of intestinal mucosal barrier in DSS-induced experimental colitis. Immunopharmacol Immunotoxicol. (2013) 35:403–9. doi: 10.3109/08923973.2013.790413
96. Ghyselinck J, Verstrepen L, Moens F, Van den Abbeele P, Said J, Smith B, et al. A 4-strain probiotic supplement influences gut microbiota composition and gut wall function in patients with ulcerative colitis. Int J Pharm. (2020) 587:119648. doi: 10.1016/j.ijpharm.2020.119648
97. Dharmani P, De Simone C, Chadee K. The probiotic mixture VSL#3 accelerates gastric ulcer healing by stimulating vascular endothelial growth factor. PLoS ONE. (2013) 8:e58671. doi: 10.1371/journal.pone.0058671
98. Shen ZH, Zhu CX, Quan YS, Yang ZY, Wu S, Luo WW, et al. Relationship between intestinal microbiota and ulcerative colitis: mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J Gastroenterol. (2018) 24:5–14. doi: 10.3748/wjg.v24.i1.5
99. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. (2015) 8:80–93. doi: 10.1038/mi.2014.44
100. Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. (2017) 389:1218–28. doi: 10.1016/S0140-6736(17)30182-4
101. Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. (2019) 321:156–64. doi: 10.1001/jama.2018.20046
102. Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. (2015) 149:102–9. doi: 10.1053/j.gastro.2015.04.001
103. Sood A, Mahajan R, Singh A, Midha V, Mehta V, Narang V, et al. Role of faecal microbiota transplantation for maintenance of remission in patients with ulcerative colitis: a pilot study. J Crohns Colitis. (2019) 13:1311–7. doi: 10.1093/ecco-jcc/jjz060
104. Aroniadis OC, Brandt LJ. Fecal microbiota transplantation: past, present and future. Curr Opin Gastroenterol. (2013) 29:79–84. doi: 10.1097/MOG.0b013e32835a4b3e
105. Singh S, Al-Darmaki A, Frolkis AD, Seow CH, Leung Y, Novak KL, et al. Postoperative mortality among patients with inflammatory bowel diseases: a systematic review and meta-analysis of population-based studies. Gastroenterology. (2015) 149:928–37. doi: 10.1053/j.gastro.2015.06.001
106. Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R, et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet. (1991) 338:771–4. doi: 10.1016/0140-6736(91)90663-A
107. Hyams JS, Davis Thomas S, Gotman N, Haberman Y, Karns R, Schirmer M, et al. Clinical and biological predictors of response to standardised paediatric colitis therapy (PROTECT): a multicentre inception cohort study. Lancet. (2019) 393:1708–20. doi: 10.1016/S0140-6736(18)32592-3
108. Gracie DJ, Williams CJ, Sood R, Mumtaz S, Bholah MH, Hamlin PJ, et al. Negative effects on psychological health and quality of life of genuine irritable bowel syndrome-type symptoms in patients with inflammatory bowel disease. Clin Gastroenterol Hepatol. (2017) 15:376–84. doi: 10.1016/j.cgh.2016.05.012
109. Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe. (2017) 21:603–10. doi: 10.1016/j.chom.2017.04.010
110. Loddo I, Romano C. Inflammatory bowel disease: genetics, epigenetics, and pathogenesis. Front Immunol. (2015) 6:551. doi: 10.3389/fimmu.2015.00551
111. Kruis W, Frič P, Pokrotnieks J, Lukáš M, Fixa B, Kaščák M, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. (2004) 53:1617–23. doi: 10.1136/gut.2003.037747
112. Sood A, Midha V, Makharia GK, Ahuja V, Singal D, Goswami P, et al. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol. (2009) 7:1202–9. doi: 10.1016/j.cgh.2009.07.016
113. Matsuoka K, Uemura Y, Kanai T, Kunisaki R, Suzuki Y, Yokoyama K, et al. Efficacy of Bifidobacterium breve fermented milk in maintaining remission of ulcerative colitis. Dig Dis Sci. (2018) 63:1910–9. doi: 10.1007/s10620-018-4946-2
114. Kato K, Mizuno S, Umesaki Y, Ishii Y, Sugitani M, Imaoka A, et al. Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment Pharmacol Ther. (2004) 20:1133–41. doi: 10.1111/j.1365-2036.2004.02268.x
115. Dasgupta S, Kasper DL. Relevance of commensal microbiota in the treatment and prevention of inflammatory bowel disease. Inflamm Bowel Dis. (2013) 19:2478–89. doi: 10.1097/MIB.0b013e318297d884
116. Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. (2014) 52:398–406. doi: 10.1128/JCM.01500-13
117. Anderson JL, Edney RJ, Whelan K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment Pharmacol Ther. (2012) 36:503–16. doi: 10.1111/j.1365-2036.2012.05220.x
Keywords: inflammatory bowel disease, ulcerative colitis, Crohn's disease, biomarkers, gut microbiome, fecal microbiota transplantation
Citation: Guo X, Huang C, Xu J, Xu H, Liu L, Zhao H, Wang J, Huang W, Peng W, Chen Y, Nie Y, Zhou Y and Zhou Y (2022) Gut Microbiota Is a Potential Biomarker in Inflammatory Bowel Disease. Front. Nutr. 8:818902. doi: 10.3389/fnut.2021.818902
Received: 20 November 2021; Accepted: 29 December 2021;
Published: 21 January 2022.
Edited by:
Yangchun Cao, Northwest A&F University, ChinaReviewed by:
Shanlin Ke, Harvard Medical School, United StatesCopyright © 2022 Guo, Huang, Xu, Xu, Liu, Zhao, Wang, Huang, Peng, Chen, Nie, Zhou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youlian Zhou, ZXl5b3VsaWFuemhvdUBzY3V0LmVkdS5jbg==; Yongjian Zhou, ZXl6aG91eW9uZ2ppYW5Ac2N1dC5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.