
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 13 October 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.726108
This article is part of the Research Topic Impact of Proteins, Peptides, Amino Acids and Food Additives on Gut Microbiota View all 18 articles
 Feng-Li Zhang1
Feng-Li Zhang1 Ya-Lin Yang2*
Ya-Lin Yang2* Zhen Zhang2
Zhen Zhang2 Yuan-Yuan Yao1
Yuan-Yuan Yao1 Rui Xia1
Rui Xia1 Chen-Chen Gao1
Chen-Chen Gao1 Dong-Dong Du1
Dong-Dong Du1 Juan Hu1
Juan Hu1 Chao Ran2
Chao Ran2 Zhen Liu3
Zhen Liu3 Zhi-Gang Zhou1*
Zhi-Gang Zhou1*Fatty liver and intestinal barrier damage were widespread in most farmed fish, which severely restrict the development of aquaculture. Therefore, there was an urgent need to develop green feed additives to maintain host liver and intestinal health. In this study, a probiotic pili-like protein, Amuc_1100 (AM protein), was anchored to the surface of Lactococcus lactis ZHY1, and the effects of the recombinant bacteria AM-ZHY1 on liver fat accumulation and intestinal health were evaluated. Zebrafish were fed a basal diet, high-fat diet, and high-fat diet with AM-ZHY1 (108 cfu/g) or control bacteria ZHY1 for 4 weeks. Treatment with AM-ZHY1 significantly reduced hepatic steatosis in zebrafish. Quantitative PCR (qPCR) detection showed that the expression of the lipogenesis [peroxisome-proliferator-activated receptors (PPARγ), sterol regulatory element-binding proteins-1c (SREBP-1c), fatty acid synthase (FAS), and acetyl-CoA carboxylase 1 (ACC1)] and lipid transport genes (CD36 and FABP6) in the liver were significantly downregulated (p < 0.05), indicating that AM-ZHY1 could reduce liver fat accumulation by inhibiting lipid synthesis and absorption. Moreover, supplementing AM-ZHY1 to a high-fat diet could significantly reduce serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels, indicating that liver injury caused by high-fat diets was improved. The expression of tumor necrosis factor (TNF)-a and interleukin (IL)-6 in the liver decreased significantly (p < 0.05), while IL-1β and IL-10 did not change significantly in the AM-ZHY1 group. Compared to the high-fat diet-fed group, the AM-ZHY1 group, but not the ZHY1 group, significantly increased the expression of intestinal tight junction (TJ) proteins (TJP1a, claudina, claudin7, claudin7b, claudin11a, claudin12, and claudin15a; p < 0.05). Compared to the high-fat diet group, the Proteobacteria and Fusobacteria were significantly reduced and increased in the AM-ZHY1 group, respectively. In conclusion, the recombinant bacteria AM-ZHY1 has the capacity to maintain intestinal health by protecting intestinal integrity and improving intestinal flora structure and improving fatty liver disease by inhibiting lipid synthesis and absorption. This study will lay a foundation for the application of AM protein in improving abnormal fat deposition and restoring the intestinal barrier in fish.
Fatty liver and intestinal barrier damage are widespread in most farmed fish, which severely restrict the development of aquaculture (1, 2). Therefore, there is an urgent need to develop green feed additives to maintain host liver and intestinal health. Akkermansia muciniphila is a human intestinal gram-negative anaerobic bacteria, with promising probiotic activities against many metabolic-related diseases such as obesity, diabetes, inflammatory bowel disease, etc. (3–7). Amuc_1100 (AM protein), a highly abundant pili-like membrane protein of A. muciniphila, partly recapitulates the beneficial effects of A. muciniphila (8, 9). Furthermore, AM protein can modulate host immune responses and specifically induce high cytokine production in peripheral blood mononuclear cells (PBMCs) likely via Toll-like receptor 2 (TLR2) signaling (10). Oral AM protein can activate the immune response of mice potentially via TLR2 (8). Other than that, AM protein can restore gut barriers and enhance the transepithelial resistance of Caco-2 cells (10). Specifically, oral AM protein enhanced the mouse intestinal barrier likely by acting on TLR2 and restoring the appropriate expression of tight junction proteins (8). AM protein can also alleviate metabolic endotoxemia, improve glucose and lipid metabolism, and reduce fat mass in high-fat diet-fed mice (8). These results indicate that A. muciniphila and AM protein have potential as a green feed additive in aquaculture.
As a strictly anaerobic bacteria, A. muciniphila is difficult to produce at an industrial scale, which limits its application in aquaculture. Compared with the bacteria, its probiotic element, AM protein, is easy to express heterologously and can be produced on a large scale, with great application potential. The applications of protein as a feed additive will face a series of problems such as a complex expression and purification process, poor stability, poor resistance, and high cost. A microbial [especially generally recognized as safe (GRAS) lactic acid bacteria] surface display can well circumvent the above problems, becoming attractive platforms for the surface display of heterologous proteins in various fields.
In this study, AM protein was anchored to the cell wall of fish-derived Lactococcus lactis ZHY1 using surface display technology. The lipid-lowering effect of recombinant bacteria on hepatic steatosis caused by a high-fat diet was tested, and the effects of the recombinant bacteria on intestinal barrier function and the inflammatory response of zebrafish were evaluated. This study will lay a foundation for the application of AM protein in improving abnormal fat deposition and restoring the intestinal barrier in fish.
In this experiment, L. lactis was isolated from the intestine of Acipenser sinensis. After the intestinal contents were suspended in 0.1 mM of phosphate-buffered saline (PBS), and then spread on a De Man, Rogosa and Sharpe (MRS) agar medium supplemented with calcium carbonate. Due to its high yield, we chose it as the carrier for the surface display, and it was identified as L. lactis by 16s rRNA sequencing, named ZHY1 (preservation number: CGMCC No. 22024). This strain was grown in a medium in M17 broth containing 0.5% glucose (HB0391, Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China) and was cultured statically at 30°C.
The 900-bp nucleotide coding sequence of AM was synthesized and cloned into the pET28a plasmid (Generay Biotech, Shanghai, China) and digested with XhoI (R0146S, NEB, Beijing, China) and XbaI (R0145V, NEB). The gene fragment of usp45 and AcmA was amplified by fusion PCR from the signal peptide genomic major lactococcal autolysin N-acetylmuramidase DNA of L. lactis MG1363 using a set of primers, usp45F (5′-CGAGCTCATATGAAAAAAAAGATTA-3′) and AcmAR (5′-GTCAGTATCTGCGAATAAAACTCGAG-3′). The amplification reaction was performed using the Q5 High-Fidelity DNA Polymerase following the recommendations of the manufacturer, and the amplicon was digested by SacI (R0156S, NEB) and XhoI. The two M0515, NEB digested products mentioned above were inserted into the shuttle vector pMG36e, and the ligation mixture was transformed into Escherichia coli MC1061 (TIANGEN, Beijing, China). The correct recombinant plasmid, pMG36e-usp45-AcmA-AM, was identified by DNA sequencing (Sangon Biotech, Shanghai, China) (Figure 1).
Lactococcus lactis ZHY1 competent cells were prepared and transformed as described previously with slight modifications (11). The overnight culture of L. lactis was inoculated at a 5% ratio into a fresh GM17 medium with 2.5% glycine, and then cultivated until OD600 = 0.5. The bacteria were washed two times with a pre-cooled washing buffer (17.1% sucrose, 1% glycerol), and then resuspended in a pre-chilled washing buffer and mixed with the recombinant plasmid pMG36e-usp45-AcmA-AM. The electroporator was pulsed, and the cells were plated on a GM17 agar medium containing 10 ug/ml of erythromycin. The recombinant bacteria AM-ZHY1 were selected after 48 h of culture by PCR (MG36eF: 5′-ACTCTCTGGGGACTTTCG-3′; MG36eR: 5′-TCGCCTTTACCAACTGTC-3′).
The inoculation of AM-ZHY1 was performed into a GM17 medium containing 10 μg/ml erythromycin and cultured at 30°C for 48 h. The culture cells were then collected and resuspended in 1 ml of Tris-HCl (50 mM, pH 7), which contained 10 mM of magnesium chloride (MgCl2) and 4% SDS buffer, at 37°C for 2 h. The mixture was denatured by heating at 100°C before loading, and the supernatant was the lysed cell wall components. The samples were analyzed by 12% SDS-PAGE (BioRad, Hercules, CA, USA) and electrophoresis at a 120-V constant voltage for 2 h.
After electrophoresis, the protein bands were transferred to a polyvinylidene fluoride (PVDF) membrane (0.45 uM, IPVH00010, Millipore, Beijing, China). After the electrotransfer was completed, the membrane was blocked in 5% skim milk and incubated overnight at 4°C with an anti-His tag monoclonal antibody (1:1,000 dilution, AB102, TIANGEN). Then, Goat Anti-mouse IgG, HRP Conjugated (1:200 dilution, CW0102, cwbiotech, Beijing, China) was added at room temperature for 2 h after washing with 0.05% TBST (50 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20). Chemiluminescent HRP Substrate (WBKLS0100, Millipore) was used to visualize protein bands.
The experiment used zebrafish (1-month-old), AB strain. The zebrafish (n = 480, mean initial weight: 50 mg) were randomly allocated to 20 3-L tanks in the recirculation system. The zebrafish were fed for 4 weeks at 26°C, 12/12 light/dark cycle, dissolved oxygen ≥6 mg/L, total ammonia content ≤0.02 mg nitrogen/L, pH 7. All experimental and animal care procedures were approved by the Feed Research Institute of the Chinese Academy of Agricultural Sciences Animal Care Committee, under the auspices of the China Council for Animal Care (Assurance No. 2018-AF- FRI-CAAS-001).
The feed ingredients and formulas of the basal diet and high-fat diet are shown in Table 1. The recombinant bacteria, AM-ZHY1, and the control bacteria, ZHY1, were added to the high-fat diet at the amount of 108 cfu/g before being named the AM-ZHY1 group and ZHY1 group, respectively. Before preparation and after the feed was crushed (stored at −20°C), the viable cell numbers of L. lactis were determined by the plate counting method. The zebrafish were fed with the experimental diets two times a day (9:00, 17:00) at a ratio of 6% of body weight.
After 4 weeks of feeding, each fish in the tanks was weighed to assess body weight gain (WG), feed conversion ratio (FCR), and survival rate (SR) (12), which showed the following: WG: [100 × (Wt – W0)/W0], FCR: (Wf/Wd) × 100; SR = (N0/Nt) × 100; where W0 (g), Wt (g), Wf, Wd, N0, and Nt are the initial weight, final weight, total food intake, total weight gain, initial quantity, and final quantity of the zebrafish, respectively.
The determination of the triacylglycerol (TAG) content of the zebrafish livers mainly referred to the method of Zhang et al. (13). The liver tissues (5–10 mg) from five zebrafish were homogenized in a PBS buffer. About 5 ml of methanol/chloroform (1:2) solution was added to the homogenate, and it was centrifuged (3,000 rpm, 5 min) to avoid protein sticking to the wall of the centrifuge tube. The organic components were carefully dried at 70°C with nitrogen steam. Then, the dried lipids were emulsified in chloroform with 1% Triton X-100. Each of the dried emulsified lipids was dissolved in deionized water, and we then detected the amount of TAG contained in the unit weight of the sample using a triglyceride reagent (T2449, Sigma-Aldrich, Saint Louis, MO, USA) and a free glycerol reagent (F6428, Sigma-Aldrich).
The zebrafish liver was carefully removed and immediately fixed in 4% paraformaldehyde. After embedding in paraffin, the sections were cut and stained with hematoxylin-eosin (HE). The cellular lipid vacuoles of the liver were observed under a microscope (Leica DMIL-LED, Wetzlar, Germany).
Blood samples were collected from the zebrafish as previously described (14). The activities of zebrafish serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were detected using commercial diagnostic kits (Nanjing Jiancheng Bioengineering Institute, China) according to the instructions of the manufacturer. Serum ALT and AST activities were expressed as enzyme activity units per liter (U/L).
The total RNA of the zebrafish liver and intestine samples were isolated using a Trizol reagent (Invitrogen, Waltham, MA, USA) and reversed transcribed to cDNA using a FastKing-RT Supermix (KR118, TIANGEN). The expression of CD36, FABP6, sterol regulatory element-binding proteins-1c (SREBP-1c), peroxisome-proliferator-activated receptors (PPARγ), acetyl-CoA carboxylase 1 (ACC1), adipose triglyceride lipase (ATGL), and uncoupling protein 2 (UCP2) of the liver; tumor necrosis factor (TNF)-α, interleukin (IL)-6, IL-10, and IL-1β of the liver; and the intestinal tight junction (TJ) proteins (TJP1a, claudina, claudin7, claudin7b, claudin11a, claudin12, and claudin15a) were determined by quantitative real-time PCR (qRT-PCR) using the SYBR Green Supermix (FP204, TIANGEN) on a Light Cycler 480 system (Roche, Basel, Switzerland). The results were analyzed by the 2−ΔΔCt method (15). The primers of all detected genes are listed in Tables 2, 3, and the reference gene was Rps11.
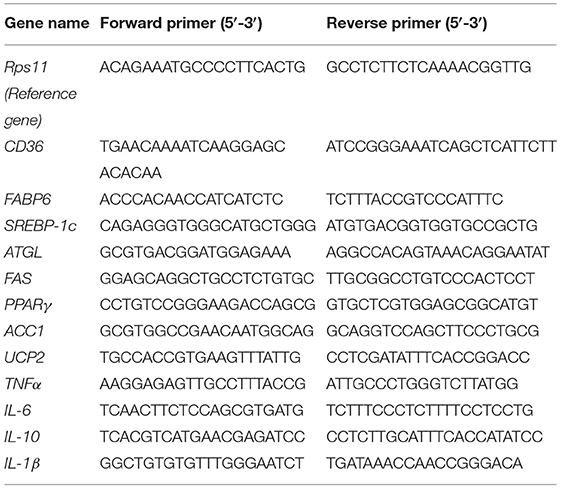
Table 2. The primer sequences for quantitative (qPCR) analysis of lipid metabolism and inflammation-related genes.
After 4 weeks of feeding, the intestinal contents of the zebrafish from the four diet groups were obtained 4 h after the last feeding. The intestinal sample of each group contained six replicates, and six fish were pooled as a replicate. Afterward, DNA was extracted with a Fast DNA spin kit (MP, Biomedicals, Solon, OH, USA), according to the instructions of the manufacturer. The V3–V4 region of 16S rRNA was amplified with U341F (5′-CGGCAACGAGCGCAACCC-3′) and U806R (5′-CCATTGTAGCACGTGTGTAGCC-3′). The high-throughput sequencing of the gut microbes was performed at the MajorBio (Shanghai), using the Illumina HiSeq platform. Microbiota sequencing data of zebrafish intestinal flora are available from NCBI Sequence Read Archive with accession number PRJNA739824. The UPARSE-operational taxonomic unit (OTU) algorithm was used to control the quality of raw pair-end readings (16). The RDP classifier Bayesian algorithm was used to perform a taxonomic analysis at the 97% similar level (17). According to the results of the taxonomic analysis, a principal component analysis of different groups was performed using R 3.3.1 at (18).
The statistical data were analyzed by the GraphPad Prism 5 software (GraphPad Software Inc., San Diego, CA, USA). All statistics were from six repetitions. Differences among groups were assessed using Student's t-test. p < 0.05, p < 0.01 and p < 0.001 were considered significant differences.
As shown in Figure 2, the fusion protein AcmA-AM was successfully expressed in the surface of the recombinant bacteria AM-ZHY1. After 4 weeks of the feeding experiment, the WG, FCR, and SR of the four groups of zebrafish were calculated. As shown in Figure 3, both the WG of the AM-ZHY1 and ZHY1 groups showed 17.7 and 43.5% lower than high-fat diet group. The FCR of the AM-ZHY1 group was 25% higher than the high-fat diet group. The SRs was unaffected by the dietary treatments.
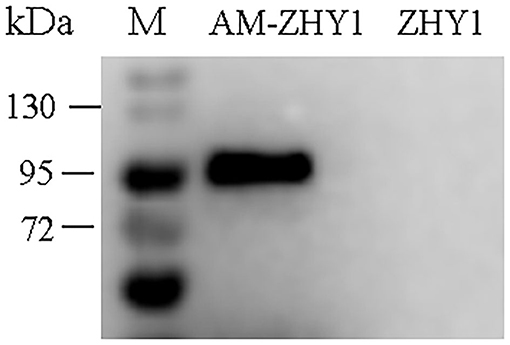
Figure 2. Western blotting detects the expression of Amuc_1100 (AM) on ZHY1. Line 1: protein marker (26630, Thermo Scientific), line 2: the whole cell wall protein of recombinant strain AM-ZHY1, line 3: the whole cell wall protein of control strain ZHY1.
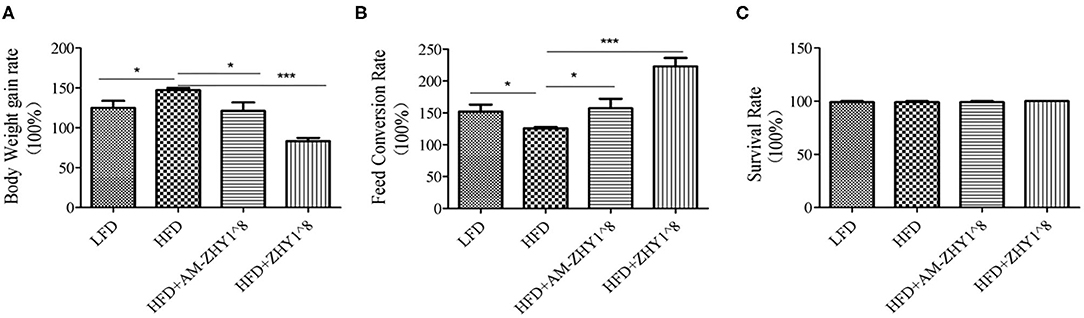
Figure 3. Effects of different diets on the (A) body weight gain, (B) feed conversion rate, and (C) survival rate of zebrafish. Data represent as the means ± SEM. The asterisks on the horizontal line represent significant differences between the two groups. *p < 0.05, ***p < 0.001.
The improvement of AM-ZHY1 on hepatic steatosis induced by the high-fat diet is shown in Figures 4A,B. The high-fat diet significantly increased liver fat accumulation compared with the basal diet (29.5% higher; p < 0.05). The supplementation of AM-ZHY1, but not ZHY1, significantly reduced TAG content in the liver compared with the high-fat diet group (by 31.6 and 7.6%, respectively; p < 0.05; p = 0.28). Moreover, the TAG content in zebrafish fed the high-fat diet with AM-ZHY1 was 26% lower than in the fish fed the high-fat diet with ZHY1 (p < 0.05). Accordingly, histopathologic analysis of the HE-stained liver sections showed hepatic steatosis in zebrafish fed high-fat diets, which was obviously reversed by the addition of AM-ZHY1. The effect of AM-ZHY1 on the lipid metabolism of the high-fat diet-fed zebrafish was investigated (Figure 5). The process of lipid metabolism mainly includes fat synthesis genes, lipolysis genes, and fat absorption-related genes at the genetic level. The expression of key enzymes involved in the de novo fatty acid synthesis pathway, including ACC1 and fatty acid synthase (FAS), was obviously lower in the AM-ZHY1 group than the high-fat diet group (by 71.9 and 71.9%, respectively; p < 0.05; p < 0.05). The mRNA expression of the transcription factors that regulate fatty acid and triglyceride synthesis, including PPARγ and SREBP-1c, was significantly downregulated in the AM-ZHY1 group (p < 0.05; p < 0.05). However, the expression of PPARγ and SREBP-1c was not significantly reduced in zebrafish fed with ZHY1 compared with those fed the high-fat diet (p = 0.12; p = 0.42). The AM-ZHY1 significantly inhibited the expression of the lipid absorption genes CD36 and FABP6 (p < 0.05; p < 0.05). However, the expression of these two genes in zebrafish fed with the high-fat diet supplemented with control bacteria ZHY1 did not decrease significantly (p = 0.58; p = 0.74). The addition of AM-ZHY1 and ZHY1 to the high-fat diet both inhibited the expression of the ATGL of zebrafish (p < 0.05; p = 0.07). Taken together, these results demonstrated that the supplement AM-ZHY1 with the high-fat diet led to the improvement of hepatic steatosis. Furthermore, this regulatory effect of AM-ZHY1 on liver lipid accumulation was through inhibiting lipid synthesis and absorption, rather than promoting lipolysis.
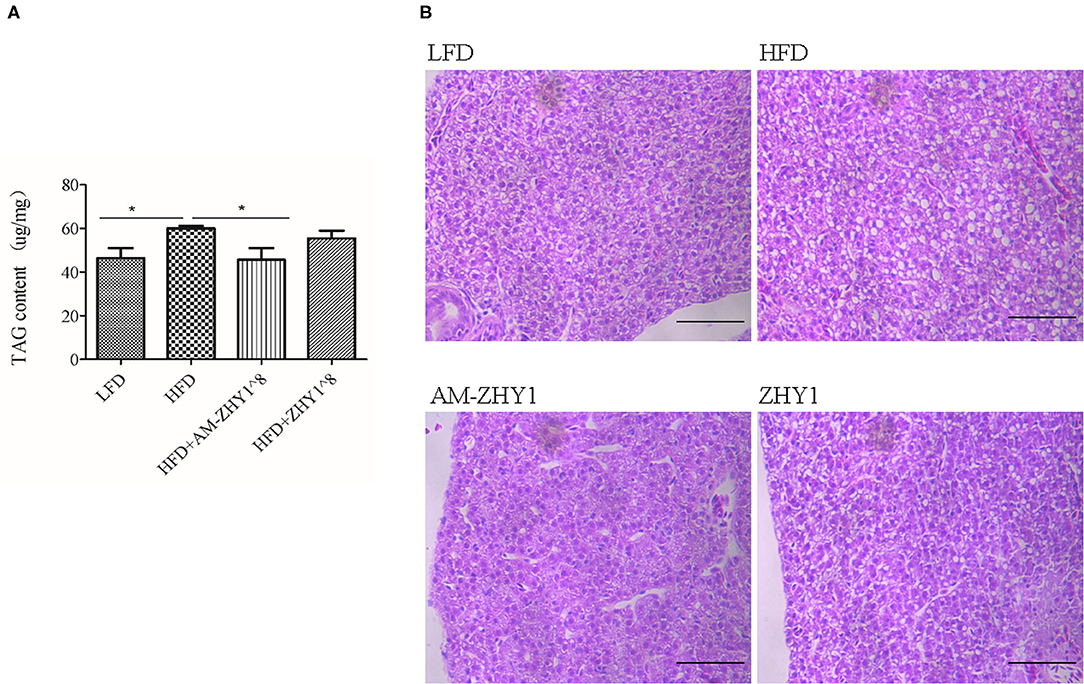
Figure 4. Triacylglycerol (TAG) content of the livers of zebrafish in different groups. (A) TAG content of the liver, the significance level was determined by a Student's t-test (*p < 0.05). (B) Histological changes of liver sections measured by HE staining (scale bar is 50 μm).
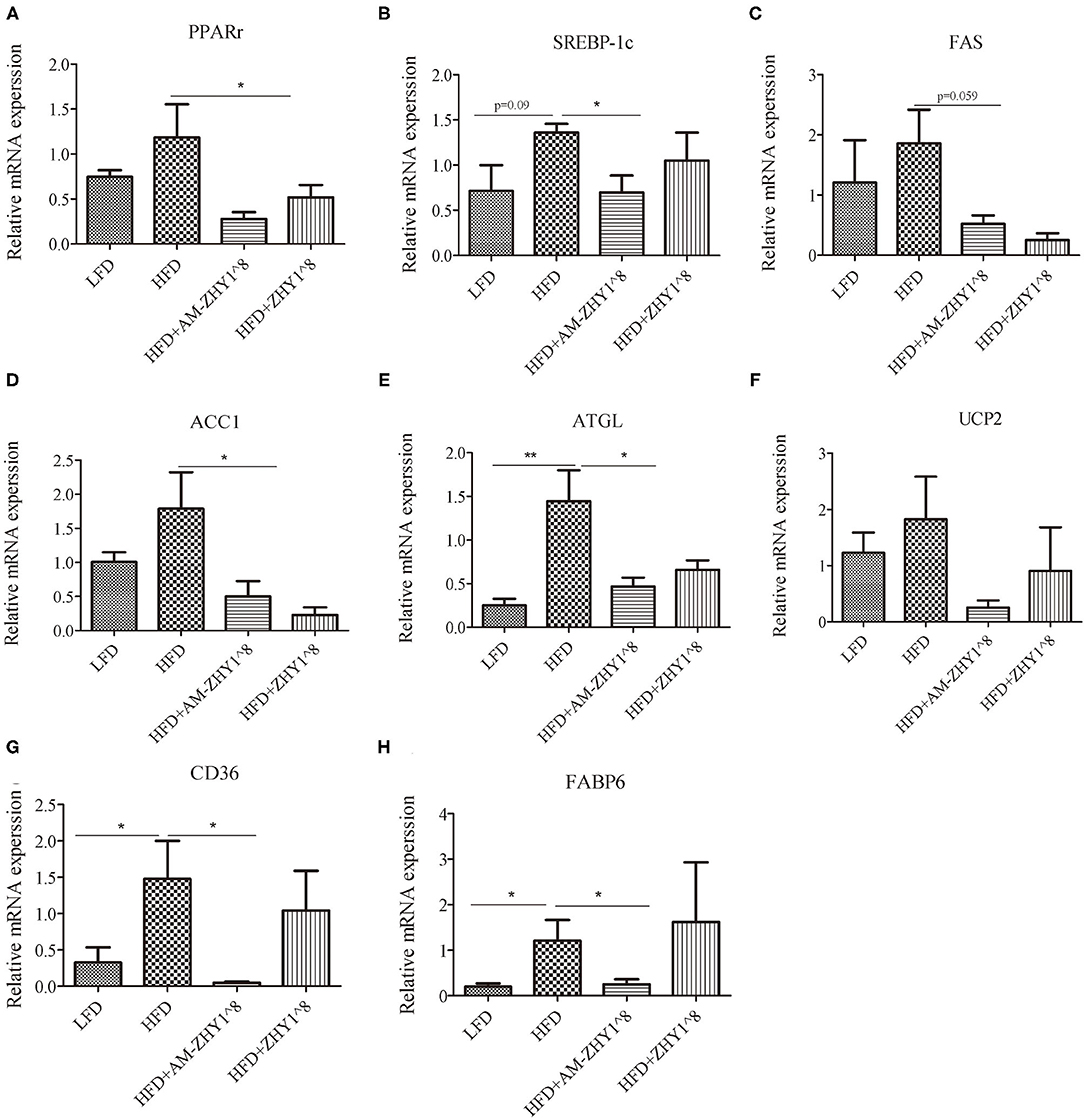
Figure 5. Relative mRNA expression of lipid metabolic-related genes PPARγ (A), SREBP-1c (B), FAS (C), ACC1 (D), ATGL (E), UCP2 (F), CD36 (G), FABP6 (H) of the livers of zebrafish in different groups. Data are presented as mean ± SEM (n = 6). The asterisks on the horizontal line represent significant differences between the two groups. *p < 0.05, **p < 0.01.
After the feeding experiment, the zebrafish serum ALT, AST, and liver inflammatory factor expression levels were tested to examine the effect of AM-ZHY1 on liver health. Compared with the high-fat diet group, the levels of ALT and AST were significantly reduced in the AM-ZHY1 group (p < 0.05, p < 0.05; Figure 6), and the expression level of TNF-a and IL-6 of the liver were significantly decreased in the AM-ZHY1 group (p < 0.05, p < 0.05; Figures 7A,B). The expression level of IL-1β and IL-10 had no significant changes in the AM-ZHY1 group (p = 0.24, p = 0.18; Figures 7C,D). These results indicated that an AM-ZHY1 treatment could alleviate liver injury and the inflammation caused by a high-fat diet.
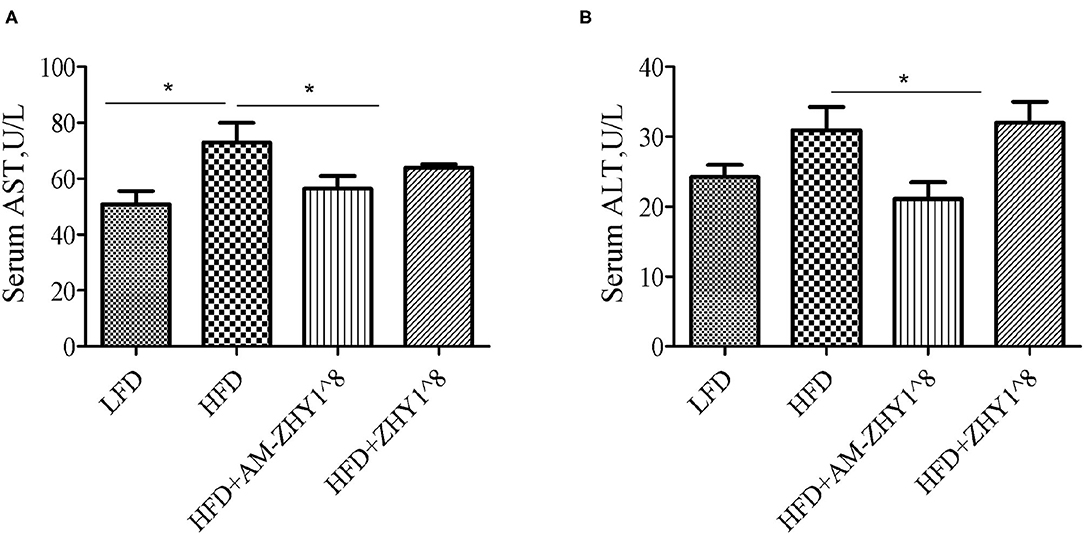
Figure 6. Serum alanine aspartate aminotransferase [AST, (A)] and alanine aminotransferase [ALT, (B)] levels of zebrafish fed with a basal diet and high-fat diet, with AM-ZHY1(108cfu/g) and ZHY1(108cfu/g) supplemented with the high-fat diet. Data are presented as mean ± SEM (n = 6). The asterisks on the horizontal line represent significant differences between the two groups. *p < 0.05.
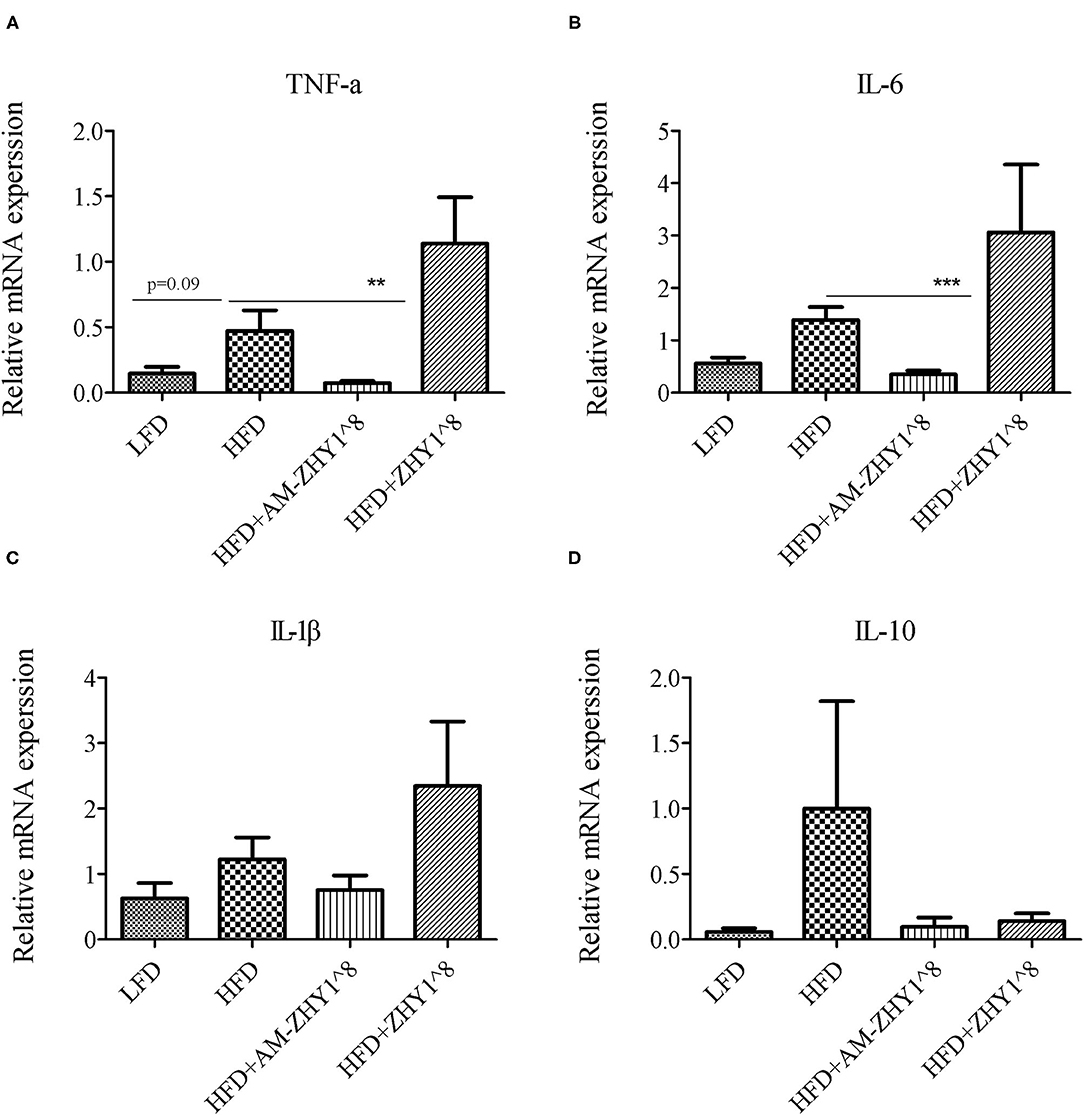
Figure 7. Relative mRNA expression of inflammation related genes: TNFα (A), IL-6 (B), IL-1β (C), IL-10 (D) of liver of zebrafish of different groups. Data are presented as mean ± SEM (n = 6). The asterisks on the horizontal line represent significant differences between the two groups. **p < 0.01, ***p < 0.001.
A total of 1,519,923 high-quality 16S rRNA sequences were generated from 30 samples. After subsampling each sample to an equal sequencing depth of 33,004 reads per sample, sequences were clustered into 1,706 OTUs. Treating with AM-ZHY1 could significantly change the structure of zebrafish intestinal microbiota at both the genus and phylum level (Figure 8). Compared to the high-fat diet group, Proteobacteria were significantly reduced, and Fusobacteria were significantly increased in the AM-ZHY1 group (p < 0.05; p < 0.05; Table 4). At the genus level, the relative content of Aeromonas was significantly reduced in the AM-ZHY1 group compared with the high-fat diet group (p < 0.05). The relative abundance of Cetobacterium was significantly increased in the AM-ZHY1 group (p < 0.05), which is higher than any other group. The relative content of Plesiomonas in the AM-ZHY1 group was consistent with that in the basal diet group. The relative contents of Acinetobacter and Bacillus did not change significantly among these groups (Table 5).
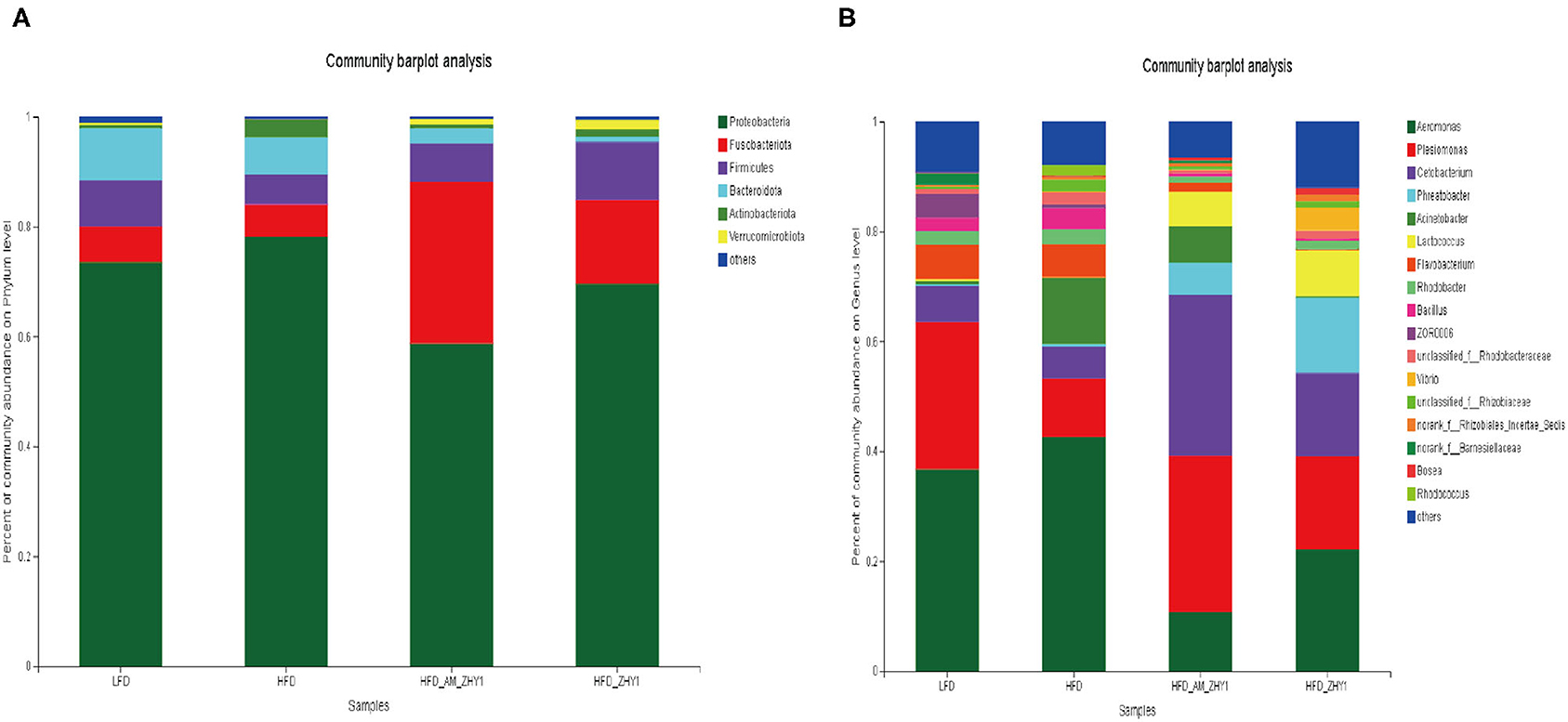
Figure 8. The effect of different diets on the relative abundance of intestinal flora at the phylum in zebrafish (A) and the genus (B) level. LFD, basal diet; HFD, high-fat diet; HFD_AM_ZHY1, supplement 108cfu/g AM-ZHY1 based on a high-fat diet; HFD_ZHY1, supplement 108cfu/g ZHY1 based on a high-fat diet.
The effect of AM-ZHY1 supplementation on intestinal health was elucidated by detecting the gene expression of TJP1a, claudina, claudin7, claudin7b, claudin11a, claudin12, and claudin15a. The AM-ZHY1 group, but not the ZHY1 group, significantly increased the expression of the above-mentioned intestinal TJ proteins vs. the high-fat diet group (p < 0.05; Figure 9). The above results indicated that AM-ZHY1 could improve zebrafish intestinal barrier structure.
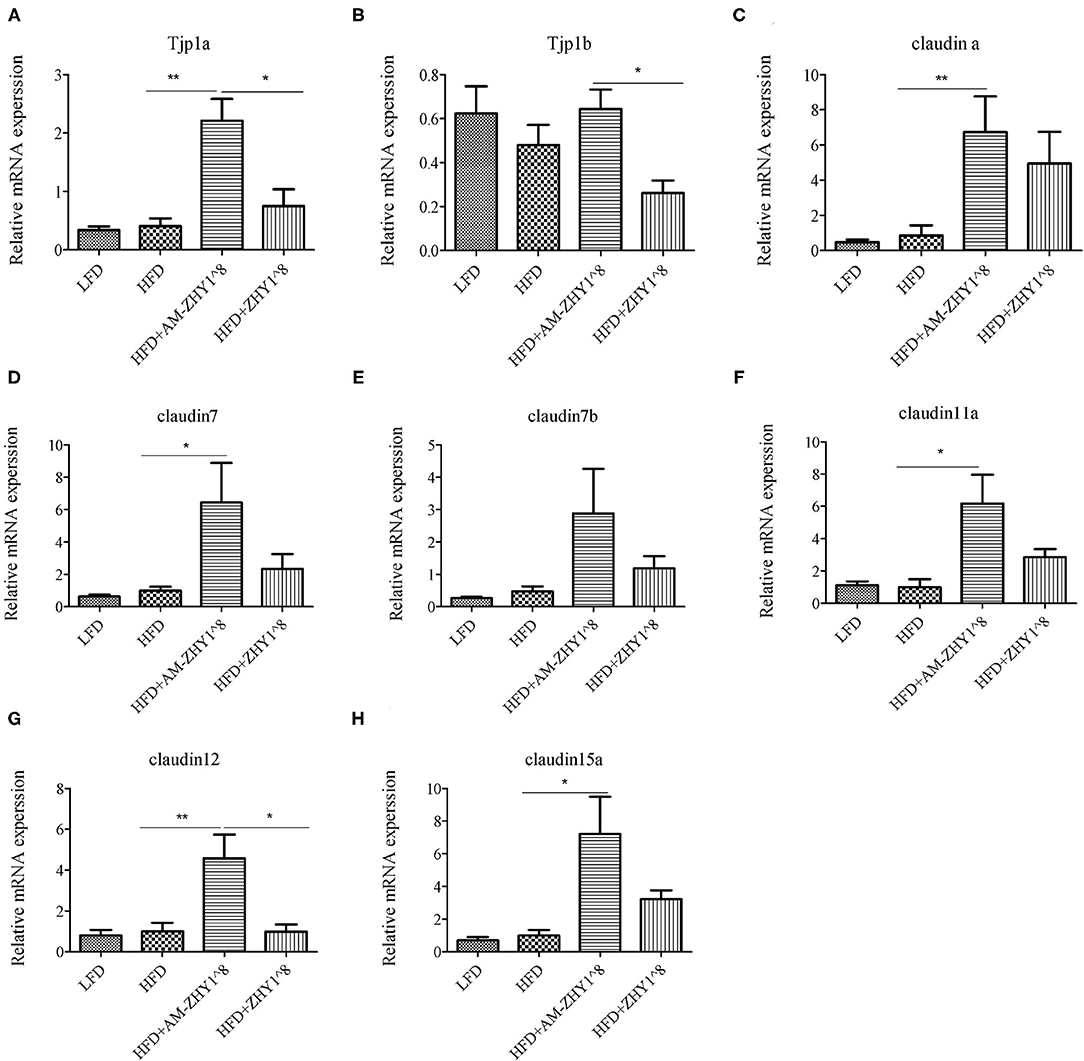
Figure 9. Relative mRNA expression of the tight junction protein genes TJP1a (A), TJP1b (B), Claudina (C), Claudin7 (D), Claudin7b (E), Claudin11a (F), Claudin12 (G), Claudin15a (H) of the intestines of zebrafish in different groups. Data are presented as mean ± SEM (n = 6). The asterisks on the horizontal line represent significant differences between the two groups. (*p < 0.05,**p < 0.01).
In this experiment, the AM protein was displayed on the surface of L. lactis ZHY1, and its probiotic effects, including relieving high-fat diet-induced liver steatosis and improving the barrier function of the intestinal tract, were confirmed in zebrafish. For the first time, AM protein was used to treat fish fatty liver in aquaculture, and this research, combined with surface display technology, advanced its practical application.
Studies have shown that AM protein could decrease the body weight and fat content of high-fat diet-fed mice (8). In this experiment, AM-ZHY1 reduced the high-fat diet-induced weight gain to the basal diet level, similar to the same function in mammals. However, the ZHY1 group could also significantly reduce weight gain. This may be due to the fact that lactic acid bacteria themselves could play an anti-obesity effect by regulating the lipid metabolism of the body and changing the composition of intestinal microbes (9, 19–22). Next, we tested the content of liver TAG and found that the content of liver TAG in the ZHY1 group did not decrease compared with the high-fat diet group, but the AM-ZHY1 group significantly reduced the accumulation of liver fat. This further confirmed that AM protein-induced weight loss by reducing liver fat accumulation, while ZHY1-induced weight loss may be due to the reduction of fat content at mesenteric, perirenal white adipose tissues, or other body mass, similar to other Lactobacillus species (22–24).
Furthermore, AM-ZHY1 significantly reduced the expression of lipogenic genes (PPARγ, SREBP-1c, ACC1, and FAS) and fatty acid transport-related proteins (CD36, FABP6) in high-fat diet-fed zebrafish. However, compared with high-fat diet-fed zebrafish treated with ZHY1, AM-ZHY1 had no significant effect on the expression of ATGL and UCP2 in high-fat diet-fed zebrafish. These results indicated that AM-ZHY1 mainly reduces high-fat diet-induced fat accumulation by reducing fat synthesis and inhibiting fat absorption.
The intestinal barrier function was important as a defense against foreign infections. The permeability and stability of the epithelial barrier maybe dependent on the TJs and adherens junctions between epithelial cells (25). The TJs of the intestinal epithelium were a specialized structure, mainly composed of transmembrane proteins, including occludin, trillulin and claudin families, and zonula occludens (ZO-1) (26, 27). These TJ proteins maintained dynamic balance through continuous remodeling and renewal under the strict regulation of extracellular and intracellular factors. The destruction of this dynamic balance would lead to intestinal epithelial barrier dysfunction which would increase intestinal epicellular paracellular permeability, and cause the translocation of luminal toxic substances and bacteria to the interior organization (28). It was reported that, as the TJ expression levels in the intestines of mice with high-fat diet-induced obesity decreased, the permeability of the intestine increased and even caused endotoxemia (29–31). We explored whether AM-ZHY1 had a protective effect on the destruction of intestinal barrier function caused by the high-fat diet. This study had shown that AM-ZHY1 could maintain intestinal health by increasing the expression of a series of TJ proteins (TJP1a, claudina, claudin7, claudin7b, claudin11a, claudin12, and claudin15a).
The homeostasis of the intestinal microbial community was vital to the gut histologic structure, metabolism, and gut immunity of fish (32, 33). A high-fat diet could cause a bad tilt of intestinal flora and reduce the abundance and diversity of microorganisms (34, 35). In our previous studies, the increase in the abundance of Fusobacteria and the decrease in the abundance of Proteobacteria indicated that the intestinal health of the zebrafish had improved (36). In this study, the addition of the recombinant bacteria AM-ZH1 could significantly increase the abundance of Fusobacteria (Cetobacterium) in the guts of high-fat diet zebrafish.
In conclusion, AM protein was successfully anchored to the cell surface of L. lactis ZHY1. This recombinant L. lactis AM-ZHY1 could reduce hepatic fat accumulation, relieve liver injury, and inhibit liver inflammation in high-fat diet-fed zebrafish. Furthermore, AM-ZHY1 could maintain the barrier function of the intestinal tract by increasing the expression of intestinal TJ proteins. Moreover, the relative abundance of beneficial commensal bacterium (Cetobacterium) was increased in the AM-ZHY1 group.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA739824.
The animal study was reviewed and approved by Feed Research Institute of the Chinese Academy of Agricultural Sciences Animal Care Committee. Written informed consent was obtained from the owners for the participation of their animals in this study.
F-LZ, Z-GZ, Y-LY, and ZZ participated in the research design. F-LZ, Z-GZ, and ZZ conducted the experiments and performed the data analysis. F-LZ, Y-YY, RX, C-CG, D-DD, JH, and CR, ZL wrote the manuscript or contributed to the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 32172958, 3180131599, and 31925038), the earmarked fund for the Modern Agro-industry Technology Research System (SCGWZJ20211104-4), and the Innovation Capability Support Program of Shaanxi (2018TD-021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Zhao H, Luo Y, Zhang Y, Chen X, Wu Z. Effects of Bacillus subtilis on hepatic lipid metabolism and oxidative stress response in grass carp (Ctenopharyngodon idellus) fed a high-fat diet. Mar Life Sci Technol. (2020) 2:50–9. doi: 10.1007/s42995-019-00005-2
2. Liu L, Zhou R, Li ZZ, Xiao RX, Li PF, Sun XG, et al. Luteolin alleviates non-alcoholic fatty liver disease in rats via restoration of intestinal mucosal barrier damage and microbiota imbalance involving in gut-liver axis. Arch Biochem Biophys. (2021) 711:109019. doi: 10.1016/j.abb.2021.109019
3. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. (2013) 110: 9066–71. doi: 10.1073/pnas.1219451110
4. Shen J, Tong X, Sud N, Khound R, Song YY, Maldonado-Gomez MX, et al. Low-density lipoprotein receptor signaling mediates the triglyceride-lowering action of Akkermansia muciniphila in genetic-induced hyperlipidemia. Arteriosclerosis. (2016) 36:1448–56. doi: 10.1161/ATVBAHA.116.307597
5. Rao Y, Kuang ZQ, Li C, Guo SY, Xu YY, Zhao DD, et al. Gut Akkermansia muciniphila ameliorates metabolic dysfunction-associated fatty liver disease by regulating the metabolism of L-aspartate via gut-liver axis. Gut Microbes. (2021) 13:1–19. doi: 10.1080/19490976.2021.1927633
6. Yoon HS, Cho CH, Yun MS, Sung JJ, You HJ, Jun-hyeong K, et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat Microbiol. (2021) 6:563–73. doi: 10.1038/s41564-021-00880-5
7. Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+T. Gut. (2020) 69:1988–97. doi: 10.1136/gutjnl-2019-320105
8. Plovier H, Everard A, Druart C, Depommier C, Van HM, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. (2016) 23:107–13. doi: 10.1038/nm.4236
9. Wang H, Ni X, Xiaodan Q, Zeng D, Min L, Lei L, et al. Live probiotic Lactobacillus johnsonii BS15 promotes growth performance and lowers fat deposition by improving lipid metabolism, intestinal development, and gut microflora in broilers. Front Microbiol. (2017) 8:e1073. doi: 10.3389/fmicb.2017.01073
10. Ottman N, Reunanen J, Meijerink M, Pietilä TE, Kainulainen V, Klievink J, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE. (2017) 12:e0173004. doi: 10.1371/journal.pone.0173004
11. Mathiesen G, Verland L, Kuczkowska K, Eijsink V. Anchoring of heterologous proteins in multiple Lactobacillus species using anchors derived from Lactobacillus plantarum. Sci Rep. (2020) 10:9640. doi: 10.1038/s41598-020-66531-7
12. Guo X, Ran C, Zhang Z, He S, Jin M, Zhou Z. The Growth-promoting effect of dietary nucleotides in fish is associated with an intestinal microbiota-mediated reduction in energy expenditure. Journal of Nutrition. (2017) 147:781–8. doi: 10.3945/jn.116.245506
13. Zhang Z, Zhou Z, Li Y, Zhou LK, Ding QW, Xu L. Isolated exopolysaccharides from Lactobacillus rhamnosus GG alleviated adipogenesis mediated by TLR2 in mice. Sci Rep. (2016) 2016:e36083. doi: 10.1038/srep36083
14. Pedroso GL, Hammes TO, Escobar T, Fracasso LB, Forgiarini LF, Silveira T. Blood collection for biochemical analysis in adult zebrafish. J Vis Exp. (2012) 63:e3865. doi: 10.3791/3865
15. Zhang YL, Duan XD, Feng L, Jiang WD, Wu P, Liu Y, et al. Soybean glycinin impaired immune function and caused inflammation associated with PKC-ζ/NF-κb and mTORC1 signaling in the intestine of juvenile grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. (2020) 106:393–403. doi: 10.1016/j.fsi.2020.08.008
16. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996. doi: 10.1038/nmeth.2604
17. Maidak BL, Olsen GJ, Niels L, Ross OB, McCaughey MJ, Woese CR. The RDP (Ribosomal Database Project). Nucleic Acids Res. (1997) 25:109–10. doi: 10.1093/nar/25.1.109
19. Yadav H, Lee JH, Lloyd J, Walter P, Rane SG. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J Biol Chem. (2013) 288:25088–97. doi: 10.1074/jbc.M113.452516
20. Kondo S, Xiao JZ, Satoh T, Odamaki T, Takahashi S, Sugahara H, et al. Antiobesity effects of Bifidobacterium breve strain B-3 supplementation in a mouse model with high-fat diet-induced obesity. Biosci Biotechnol Biochem. (2010) 74:1656–61. doi: 10.1271/bbb.100267
21. Kang JH, Yun SI, Park MH, Park JH, Jeong SY, Park H. Anti-obesity effect of Lactobacillus gasseri BNR17 in high-Sucrose diet-induced obese Mice. PLoS ONE. (2013) 8:e54617. doi: 10.1371/journal.pone.0054617
22. Xie N, Yi C, Yin YN, Xin Z, Lu FG. Effects of two Lactobacillus strains on lipid metabolism and intestinal microflora in rats fed a high-cholesterol diet. BMC Complement Altern Med. (2011) 11:1–11. doi: 10.1186/1472-6882-11-53
23. Grompone G, Vidal DR, Guerola PM, Martinez SG, Soriano PO, Pla SL, et al. Lactobacillus Rhamnosus Strain for Reducing Body fat Accumulation. US9855304 (2018).
24. Jang HR, Park HJ, Kang D, Chung H, Lee HY. A protective mechanism of probiotic Lactobacillus against hepatic steatosis via reducing host intestinal fatty acid absorption. Exp Mol Med. (2019) 51:1–14. doi: 10.1038/s12276-019-0293-4
25. Yang H, Rao JN, Wang JY. Posttranscriptional regulation of intestinal epithelial tight junction barrier by RNA-binding proteins and microRNAs. Tissue Barriers. (2014) 2:e28320. doi: 10.4161/tisb.28320
26. Furuse M, Izumi Y, Oda Y, Higashi T, Iwamoto N. Molecular organization of tricellular tight junctions. Tissue Barriers. (2014) 2:615–21. doi: 10.4161/tisb.28960
27. Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, et al. JunD represses transcription and translation of the tight junction protein zona occludens−1 modulating intestinal epithelial barrier function. Mol Biol Cell. (2008) 19:3701–12. doi: 10.1091/mbc.e08-02-0175
28. Cao S, Xiao L, Rao JN, Zou T, Liu L, Zhang D, et al. Inhibition of Smurf2 translation by miR-322/503 modulates TGF-β/Smad2 signaling and intestinal epithelial homeostasis. Mol Biol Cell. (2014) 25:1234–43. doi: 10.1091/mbc.e13-09-0560
29. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced diet-induced obesity and diabetes in mice. Diabetes. (2008) 57:1470–81. doi: 10.2337/db07-1403
30. Serre C, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE, et al. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol. (2010) 299:G440–8. doi: 10.1152/ajpgi.00098.2010
31. Oliveira RB, Canuto LP, Collares-Buzato CB. Intestinal luminal content from high-fat-fed prediabetic mice changes epithelial barrier function in vitro. Life Sci. (2018) 216:10–21. doi: 10.1016/j.lfs.2018.11.012
32. Guerreiro I, Enes P, Rodiles A, Merrifield D, Oliva T. Effects of rearing temperature and dietary short-chain fructooligosaccharides supplementation on allochthonous gut microbiota, digestive enzymes activities and intestine health of turbot (Scophthalmus maximus L) juveniles. Aquacult Nutr. (2016) 22:631–42. doi: 10.1111/anu.12277
33. Stagaman K, Burns AR, Guillemin K, Bohannan BJ. The role of adaptive immunity as an ecological filter on the gut microbiota in zebrafish. ISME J. (2017) 11:1630–9. doi: 10.1038/ismej.2017.28
34. Wong S, Stephens WZ, Burns AR, Stagaman K, David LA, Bohannan B, et al. Ontogenetic differences in dietary fat influence microbiota assembly in the zebrafish gut. MBio. (2015) 6:e00687. doi: 10.1128/mBio.00687-15
35. Arias-Jayo N, Abecia L, Alonso-Sáez Ramirez-Garcia A, Rodriguez A, Pardo MA. High-Fat diet consumption induces microbiota dysbiosis and intestinal inflammation in zebrafish. Microb Ecol. (2018) 76:1089–101. doi: 10.1007/s00248-018-1198-9
Keywords: Amuc_1100, Lactococcus lactis, fatty liver, intestinal health, microbiota, inflammation, zebrafish
Citation: Zhang F-L, Yang Y-L, Zhang Z, Yao Y-Y, Xia R, Gao C-C, Du D-D, Hu J, Ran C, Liu Z and Zhou Z-G (2021) Surface-Displayed Amuc_1100 From Akkermansia muciniphila on Lactococcus lactis ZHY1 Improves Hepatic Steatosis and Intestinal Health in High-Fat-Fed Zebrafish. Front. Nutr. 8:726108. doi: 10.3389/fnut.2021.726108
Received: 16 June 2021; Accepted: 07 September 2021;
Published: 13 October 2021.
Edited by:
Xi Ma, China Agricultural University, ChinaReviewed by:
Jianmin Wu, China Agricultural University, ChinaCopyright © 2021 Zhang, Yang, Zhang, Yao, Xia, Gao, Du, Hu, Ran, Liu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya-Lin Yang, eWFuZ3lhbGluQGNhYXMuY24=; Zhi-Gang Zhou, emhvdXpoaWdhbmcwM0BjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.