
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr., 11 October 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.718389
 Sandeep Appunni1,2
Sandeep Appunni1,2 Muni Rubens3
Muni Rubens3 Venkataraghavan Ramamoorthy4
Venkataraghavan Ramamoorthy4 Raees Tonse5
Raees Tonse5 Anshul Saxena4,6
Anshul Saxena4,6 Peter McGranaghan3
Peter McGranaghan3 Adeel Kaiser5,6
Adeel Kaiser5,6 Rupesh Kotecha5,6*
Rupesh Kotecha5,6*Dietary factors have important role in modulating the gut microbiome, which in-turn regulates the molecular events in colonic mucosa. The composition and resulting metabolism of the gut microbiome are decisive factors in colorectal cancer (CRC) tumorigenesis. Altered gut microbiome is associated with impaired immune response, and the release of carcinogenic or genotoxic substances which are the major microbiome-induced mechanisms implicated in CRC pathogenesis. Diets low in dietary fibers and phytomolecules as well as high in red meat are important dietary changes which predispose to CRC. Dietary fibers which reach the colon in an undigested form are further metabolized by the gut microbiome into enterocyte friendly metabolites such as short chain fatty acid (SCFA) which provide anti-inflammatory and anti-proliferative effects. Healthy microbiome supported by dietary fibers and phytomolecules could decrease cell proliferation by regulating the epigenetic events which activate proto-oncogenes and oncogenic pathways. Emerging evidence show that predominance of microbes such as Fusobacterium nucleatum can predispose the colonic mucosa to malignant transformation. Dietary and lifestyle modifications have been demonstrated to restrict the growth of potentially harmful opportunistic organisms. Synbiotics can protect the intestinal mucosa by improving immune response and decreasing the production of toxic metabolites, oxidative stress and cell proliferation. In this narrative review, we aim to update the emerging evidence on how diet could modulate the gut microbial composition and revive colonic epithelium. This review highlights the importance of healthy plant-based diet and related supplements in CRC prevention by improving the gut microbiome.
The gut microbiome includes the collective genes and genome of all microorganisms residing in the gastrointestinal tract (GIT) (1). There are over 100 trillion microbes residing in the human GIT, and majority of them are located in the colon (2). Metagenomic studies demonstrate that there are ~1,952 uncultured bacterial species in the human gut, and many remain unclassified to date. This contributes to substantial diversity within the microbial ecosystem (3). The host-microbe relationship can be symbiotic or pathogenic. Several external factors, such as diet, medication, and lifestyle heavily influence the microbial ecosystem (4). Symbiotic relationships between host and microbes have a plethora of effects on physiological functions and overall health. The beneficial commensals have several functions such as providing essential micronutrients, regulating the immune response, modulating enterocyte function, influencing metabolism, and preventing colonization by pathogenic microorganisms (5). The gut ecosystem is highly dependent on the human diet, as well as its composition, as the microbes metabolize and thrive on the consumed foods. Dietary fibers, microbiota accessible carbohydrates (MAC), and certain plant-based proteins are metabolized to short chain fatty acids (SCFAs). SCFAs exhibit anti-inflammatory properties, maintain mucosal integrity, and retain microbial diversity (6, 7). Imbalances in ratios of vital nutrients to dangerous toxins are implicated for several diseases, including cancer. Altered microbial diversity, impaired immune response, and release of carcinogenic or genotoxic substances are the major microbiome-induced mechanisms implicated for cancer pathogenesis (8). In this study, we aim to present emerging evidence on the dietary factors associated with the development of colorectal cancer (CRC). In addition, we also explored how healthy dietary modifications can restore functional colonic epithelium and prevent CRC.
The gut microbiome can influence the development of CRC in several ways. Perturbations in the gut microbiota expose the GIT to inflammatory and genotoxic metabolites such as secondary bile salts, trimethylamine-N-oxide (TMAO), hydrogen sulfide (from sulfur containing amino acids), heme, nitrosamines, heterocyclic amines, and polyaromatic hydrocarbons, often resulting from consumption of red or processed meat and diet poor in fibers (9, 10). These dietary factors along with lifestyle factors such as smoking, alcohol and obesity increase the risk of oncogenic transformations in the colonic epithelial cells (1). Figure 1 shows a diet-outcome model that incorporates the host-microbe relationship and factors influencing their harmony. In the human body, most of the bacteria reside in the colon and an estimated 3.8 × 1013 bacteria could be found in a 70 kg “reference man” (11). In addition to natural gut defenses, human symbiotic microorganisms have additional roles in fighting pathogenic strains by stimulating the immune system. In turn, the immune system responds by producing a myriad of inflammatory mediators, chiefly consisting of anti-microbial peptides, inflammasomes, and cytokines [e.g., interleukin (IL)-22, IL-17, and IL-10] (12). Importantly, persistent activation of the immune system has its own adverse effects. Chronic inflammation can induce oxidative stress by producing reactive oxygen species (ROS), which have both cytotoxic and genotoxic effects, resulting in detrimental effects on intestinal mucosal cells (13). Inflammasomes produced by the innate immune system secondary to inflammation can lead to colitis and increase the risk for CRC (14). Moreover, inflammation-mediated persistent release of growth factors, suppression of apoptosis, and increased angiogenesis are additional factors which promote tumorigenesis (15). Carcinogenic metabolites or oncotoxins are produced due to alterations in microbial metabolism resulting from alerted dietary patterns such as consumption of processed and refined foods. These oncotoxins are implicated for promoting CRC (1). Yang et al. performed integrated metagenomic and metabolomic analysis and found that lower microbial diversity and increased production of cytotoxic polyamines such as cadaverine and putrescine are associated with increased risk for CRC (16). Diets higher in red and processed meats and lower in dietary fibers increases the predisposing factors for CRC (17). Indigestible dietary fibers reaching the lower GIT are metabolized by the gut microbiome into SCFAs such as acetate, propionate and butyrate, which has anti-inflammatory effect on the colonic mucosa (18). The mechanisms by which dysbiotic microbiome mediates CRC include increased microbial adherence to colon cells, downregulation of tumor suppressor genes, activation of oncogenes, induction of genotoxic effects on colonic enterocyte, and activation of angiogenesis (19). Thus, external factors can modulate the gut microbiome, resulting in either stimulatory or regulatory roles in priming the intestinal microenvironment toward or against tumorigenesis.
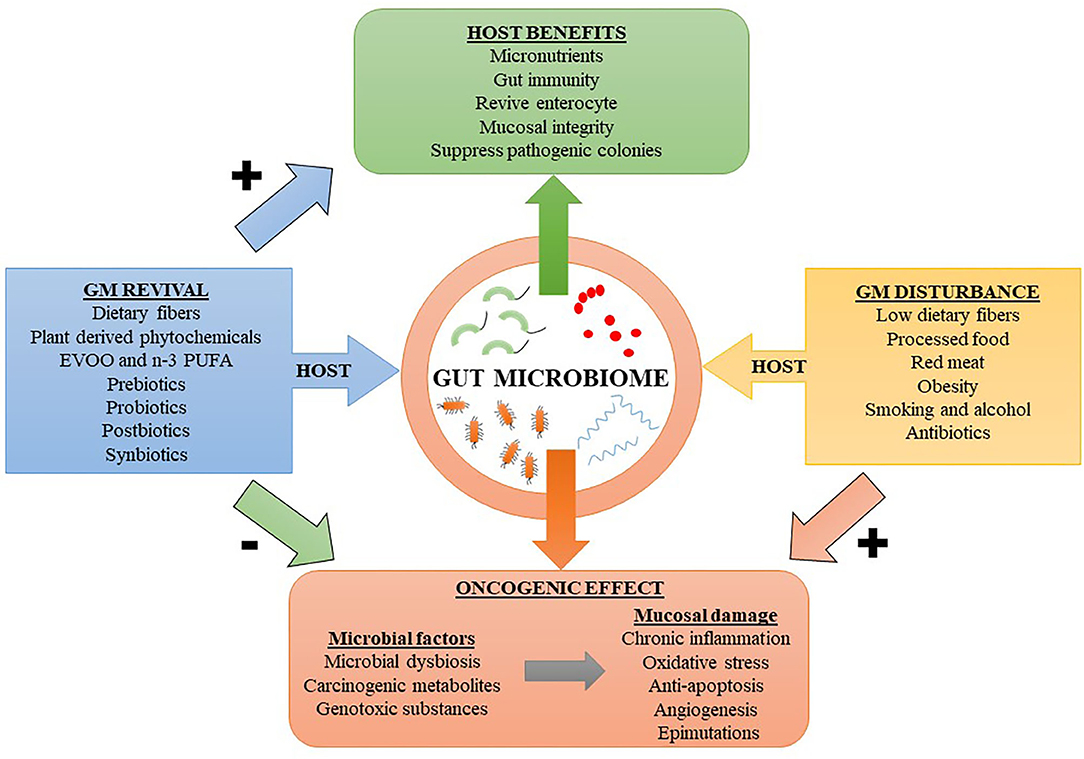
Figure 1. A host diet-outcome model showing the relation between host factors modulating the gut microbiome (GM). Dietary factors reviving the GM can result in beneficial effects to the host and prevent neoplastic changes in the colonic mucosa. While factors leading to GM disturbances can putatively increase the risk for colorectal cancer. EVOO, extra virgin olive oil; n-3 PUFA, omega-3 polyunsaturated fatty acid. Symbols: Positively associated (+); Negatively associated (–).
Poor microbial diversity is associated with increased risk for CRC (16, 20). Wu et al. reported abundance of Helicobacter spp. in right-sided and moderate to poorly differentiated CRCs, while Firmicutes phylum was higher in advanced CRCs with lymph node metastasis, compared to CRCs without lymph node metastasis (21). In colonic adenomatous polyposis (CAP), a precursor lesion to CRC, Bacteroides and Citrobacter taxa predominate, compared to Weissella and Lactobacillus, which are disproportionally low. The chief metabolites observed in the fecal samples of CAP patients were acetic acid and butyric acid, while healthy controls had higher levels of protective t10, c12-conjugated linoleic acid (not the same as dietary linoleic acid) (22). Conjugated linoleic acid such as c9, t11-CLA can be synthesized by natural gut colonizers such as probiotic Bifidobacteria species and strains of ruminal bacteria such as Megasphaera elsdenii produces t10, c12- conjugated linoleic acid (23, 24). Even though butyrate has pro-apoptotic and anti-proliferative role in CRC, it has paradoxically been shown to enhance polyp formation in Apcmin/+Msh2−/− (adenomatous polyposis colimin/+ and mutS homolog 2−/−) mice having defective mismatch repair (25, 26). Another bacterium that is highly implicated in CRC is Streptococcus gallolyticus subspecies gallolyticus (27). This bacterial strain carries a type VII expression system consisting of core genes expressing proteins which have potential pro-tumorigenic role in CRC through attachment to HT29 colon cancer cells and inducing subsequent proliferative changes. Deletion of the secretion system suppresses the protein expression related to bacterial attachment to the HT29 cells in vitro and decreased Streptococcus gallolyticus subspecies gallolyticus colonization in murine in vivo colon cancer models (27). This suggests that bacterial proteins produced by selective species can potentially exhibit pro-tumorigenic effects.
Certain pharmacological agents also modulate the colonic microbial diversity and alter the course of CRC. For example, Ternák et al. demonstrated that antibiotic therapy may have positive and negative correlation with development of several malignancies (28). In certain European regions, overconsumption of antibiotics such as penicillin and tetracyclines are associated with higher incidence of CRC, especially among females (28). Lee et al. reported that antibiotic therapy, either alone or in cocktail combinations, administered to murine colitis-associated cancer models decreased the bacterial load, suppressed inflammation, and impeded tumorigenesis in a drug-specific manner (29). This suggests that abnormal bacterial colonies can increase tumorigenesis and antibiotic therapies can plausibly modulate it. Anti-diabetic drug such as metformin produced significant changes in the gut microbiome of CRC patients with type 2 diabetes mellitus (T2DM). Comparative analysis among CRC patients with T2DM showed that metformin was associated with increase in Firmicutes and decrease in Bacteroidetes and Fusobacteria at the phyla level, while Bifidobacterium increased at genus level, whereas the abundance of pro-tumorigenic species F. nucleatum was decreased (30). Metformin treatment in T2DM has shown to increase the gut bacteria associated with enhanced production of SCFA such as butyrate and propionate which improve glucose metabolism and homeostasis (31). This suggest that metformin could improve gut microbiota that could have protective effects against CRC. Anti-diabetic drugs such as metformin induces changes in the gut microbiome of T2DM patients developing CRC. Comparative analysis showed that initiation of metformin in T2DM patients with CRC was associated with an increase in Firmicutes and decrease in Bacteroidetes and Fusobacteria at the phyla level while at genus level increase in Bifidobacterium with an associated decrease in F. nucleatum (30). Metformin treatment of T2DM has shown to enhance the microbial richness of the colon associated with enhanced production of SCFAs such as butyrate and propionate which potentially improves glucose metabolism and homeostasis (31). This suggest that initiation of metformin treatment in T2DM could be useful in recuperating altered gut microbiota, thereby imparting protective effects against CRC.
Five genera of microbes typically associated with CRC progression and reversed by metformin induction involves Bacteroides, Streptococcus, Achromobacter, Alistipes, and Fusobacterium (32). Passenger bacteria belonging to Fusobacterium genus, which reside in the oral cavity are associated with CRC progression (33, 34). Fusobacterium nucleatum, is mainly localized in the proximal colon and decreases in numbers from caecum to rectum, possibly due to anaerobic conditions. F. nucleatum is associated with more advanced and serrated forms of CRC (33–35). The abundance of F. nucleatum is affected by a number of environmental factors such as smoking, chronic periodontitis, and uncontrolled T2DM (36). In Apcmin/+ CRC model mice, metformin suppressed the tumor growth induced by F. nucleatum colonization (32). Yu et al. reported that F. nucleatum directly targeted the TLR4-MYD88 (toll-like receptor 4-myeloid differentiation primary response 88) axis of the innate immune system to activate autophagy. Autophagic activity mediated by enhanced ULK1 (unc-51 like autophagy activating kinase 1) and ATG7 (autophagy related 7) expression improved cell survival and decreased chemotherapy-induced cytotoxicity (37). Though abundance of F. nucleatum in stool samples was positively associated with CRC, no significant associations were observed for adenomas (38). Activation of various pro-oncogenic pathways and pro-inflammatory mediators are possibly linked to the pathophysiology of F. nucleatum mediated CRC (35). FadA is an amyloid like virulence factor released by F. nucleatum in diseased states (39, 40). In vitro co-culturing HCT-116 and HT29 colon cancer cells with wild type (WT) F. nucleatum increased cell proliferation and DNA damage induced by fadA mediated activation of checkpoint kinase 2 (CHK2). Knockout of the fadA gene in F. nucleatum subsequently attenuated these proliferative changes induced in co-cultured HCT-116 and HT29 colon cancer cells (41). Furthermore, cancer tissue from fadA−/− F. nucleatum-treated Apcmin/+ mice had decreased expression of pro-proliferative CHK2. Similarly, recombinant fadA enhanced the proliferation of SW480 colorectal cancer cells in vitro, which was dose and time dependent (42). Among human CRC patients Kashani et al. observed that all CRC tissue biopsies were colonized with fadA positive F. nucleatum, indicating a strong relationship (43). This shows that fadA secreted by F. nucleatum is a key virulence factor and has a pro-tumorigenic role in CRC. Moreover, low dietary fiber intake putatively enhances the risk for F. nucleatum mediated CRC (35). Mehta et al. reported that healthy diet consisting of whole grains and dietary fibers are associated with decreased risk of F. nucleatum positive CRC (44). This suggest that decreasing F. nucleatum colonization in the lower GIT through healthy dietary changes could be a preventive measure against CRC. Wang et al. identified that Eubacterium rectale initiated chronic inflammation by activating downstream NF-κB signaling, which increased chemokine and cytokine production (45). Upregulation of NF-κB signaling pathway in CRC has shown to promote cancer growth by inducing cell proliferation, angiogenesis, inflammation, metastasis, and drug resistance (15). Collectively, these mechanisms demonstrate that inflammation induced by altered gut microbiota could trigger potential oncogenic pathways.
Certain pathogenic microbes residing in the proximal colon of CRC patients produces biofilm, which has pro-malignant potential (34). In familial adenomatous polyposis (FAP), which is a precursor lesion to CRC, colonization and invasion of the intestinal mucosa by carcinogenic toxin producing Escherichia coli and Bacteroides fragilis led to the formation of biofilms, which was associated with increase in the genes for bacterial toxins such as colibactin and enterotoxigenic Bacteroides fragilis (ETBF) toxin (46). Co-colonization of toxigenic Escherichia coli and Bacteroides fragilis into FAP model mice showed increased production of ETBF induced pro-inflammatory IL-17 and colibactin mediated DNA damage, which collectively accelerated the development of CRC (46). This shows that toxigenic bacterial species can enhance the risk of malignant transformation in benign colonic polyps. Therefore, evolving evidence show that altered gut microbiome could alter gut microbial crosstalk with the colonic mucosa and potentially enhanced the risk for CRC (Table 1).

Table 1. Plausible effect on the CRC tumorigenicity by relative abundance of diverse microbial taxa.
Dietary ingredients such as fibers, fat, and proteins fuel bacterial metabolisms which not only aid in the digestive process but also synthesize byproducts that have immense functional significance to the host. However, when this balance is impaired, toxic metabolites are generated by the gut microbes resulting in cytotoxic and genotoxic effects (Table 1). Moreover, diets rich in prebiotics and probiotics can increase the richness of microbiome by enhancing microbial diversity and nurturing the existing microbiota (65). Thus, the quality of diet delivered to the gut microbiota may be crucial for optimum health benefits. In the current era of processed food consumption, the gut biodiversity and chemical composition are profoundly affected, leading to chronic colonic inflammation, which increases the risk for CRC (9, 10, 66, 67). Processed meat consumption is associated with the risk of developing colorectal malignancies. Chemicals used for processing red meat react to form N-nitoso compounds (NOC), which are carcinogenic. This along with unhealthy dietary habits, obesity, heme iron, and associated alterations in gut microbiota enhance oncogenic changes in colonic mucosa (68). Viennois et al. reported that consumption of dietary emulsifiers such as carboxymethylcellulose (CMC) and polysorbate 80 (P80) alter the composition of the gut microbiome and increase the risk for intestinal inflammation and adenoma formation in Apcmin mice (69). Emulsifier consumption lowered the abundance of Clostridia (in both male and female mice) and increased the abundance of Proteobacteria (in male mice). Proteobacteria has been putatively linked to altered gut microbiome and risk for cancer (69, 70). In another study, Viennois et al. reported that CMC and polysorbate 80 induce low grade inflammation and alteration in the gut microbiota such as increase in Bacteroidales and decrease in Clostridiales (53). Long term consumption of emulsifiers increased fecal levels of bioactive products such as pro-inflammatory lipopolysaccharide and motility inducing flagellin. Consumption of food emulsifiers also increase the risk for developing metabolic syndrome and obesity, which are independent risk factors for CRC (71). This suggest that avoiding consumption of processed food could prevent colonization by harmful microorganisms, help in maintaining a healthy colonic mucosa, and decrease the risk for oncogenic changes.
Dietary factors such as higher levels of red meat, processed meat, refined sugar, alcohol, and high-fat diet, as well as lower levels of dietary fibers are implicated for mutagenic changes induced by the unhealthy microbiome and their metabolites (72, 73). Both red and processed meat are considered as potential risk factors for CRC because they alter the composition of gut microbiome (19). In a study done by Van Hecke et al. it was observed that nitrite-cured pork when cooked to very high temperatures increased the production of carcinogenic O6-carboxymethyl guanine DNA adduct when acted upon by fecal inoculums in an in vitro digestion model (74). In experimental rats, the heme iron from red meat decreased the number of operational taxonomical units (OTUs) in the colonic lumen, indicating altered gut microbiota. Firmicutes and Deferribacteres were specifically lowered, whereas Bacteroidetes and Proteobacteria counts were increased (52). Heme iron increases luminal lipid peroxidation, aldehydes, and ROS, leading to cytotoxic and genotoxic effects on colonic epithelium (52). Similarly, in a colitis model of mice fed with heme iron, Constante et al. reported depletion of Firmicutes phylum and overgrowth of the Proteobacteria. These mice had exacerbation of dextran sodium sulfate (DSS)-induced colitis and subsequently formed adenomas (75). However, clinical and epidemiological support for this outcome is limited. Based on a cohort study of over 49,000 women in Canada, there were no significant associations between iron, heme iron, or iron from meat and colorectal cancer (76). This evidence suggest that though red meat consumption could have detrimental effects on gut microbiota and colonic epithelium, epidemiological evidence is lacking, and further studies should explore this area.
Red meat is rich in sialic acid N-glycolylneuraminic acid (Neu5Gc), which following consumption gets incorporated into the cell surface glycans of endothelia and epithelia to behave as “xeno-autoantigens.” The human immune system produces anti-Neu5Gc antibodies against this antigen, triggering in chronic inflammation (77, 78). Neu5Gc is not expressed by human cells due to human-lineage specific genetic mutation in the enzyme cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH). However, mammalian red meat such as sheep, goat and dairy cow are rich in Neu5Gc (78, 79). Circulating antibodies against Neu5Gc could initiate inflammation, which could potentially lead to cancers. For example, Samraj et al. reported that among Cmah−/− deficient mice fed with Neu5Gc rich diet, there was higher levels of systemic inflammation and greater risk for hepatocellular carcinoma which was proportionate with levels of circulatory anti-Neu5Gc antibodies (80). Neu5Gc in diet increases the levels of Clostridium and Bacteroides, which efficiently express sialidases that release mucopolysaccharide from glycans (54). Although it is unclear whether this association is causational, microbes expressing Neu5Gc-specific sialidases can cleave Neu5Gc from red meat and prevent its incorporation into human tissue glycans (54). Such gut microbial species producing exo-sialidases may be protective for red meat consumers by potentially reducing Neu5Gc triggered inflammation. In addition, Firmicutes, Bacteroidetes, and Proteobacteria phyla can produces enzymes such as beta-glucuronidase and glycerol/diol dehydratase, which can metabolize heterocyclic amines from red meat into less toxic products, thus proving protective against CRC (55). These findings suggest that bacterial enzymes present in the microbiome could potentially metabolize pro-inflammatory substances and harmful toxins to lesser toxic metabolites, thereby decreasing the risk for inflammation and carcinogenesis.
Though evidence show that red meat may contribute to carcinogenesis via microbial alterations, the associations are weak. A review of 35 prospective studies showed that the association between red meat and colorectal cancer was minimal, and the highest relative risk was below 1.50 and not statistically significant (81). An alternate explanation is that specific combinations of foods may alter the beneficial vs. harmful effects associated with colonic microbiome. For example, when rats were concomitantly fed both red meat and high amylose-resistant starch, there was a shift in the gut metabolism from predominantly protein fermentation to a combination of both protein and carbohydrate fermentation. This was associated with increase in Ruminococcus bromii, Bifidobacteriales, Turicibacteraceae, and Lactobacillaceae in the gut microbiome (56). This change in taxonomical traits of the gut microbiome was associated with decreased expression of pro-oncogenic miR17-92, which imparted protective effects against CRC (56). Similarly, processed meats that were fortified with a prebiotic polysaccharide inulin, increased the abundance of Blautia genus, which increased the production of protective SCFAs such as propionate and butyrate (57). This was associated with decrease in colonic polyps in experimental rats. In addition, shifting to a fish-inclusive vegetarian diet (or pesco-vegetarian diet) might have potential benefit over a standard western diet due to favorable changes in gut microbiome taxonomical traits (82). Similarly, Orlich et al. reported that among all forms of mixed vegetarian and non-vegetarian diets, pesco-vegetarian diet holds the least risk for developing CRC (83). Collectively, it can be summarized that consumption of specific combination of foods could decrease the level of toxicity on the colonic epithelium and correspondingly decrease the risk for development of CRC.
Dietary constituents significantly modulate chronic inflammation by regulating the immune response. In a retrospective study, Liu et al. reported that CRC patients who consumed inflammatory foods had higher abundance of F. nucleatum in their cancer biopsies (48). The inflammatory potential of 18 food items were calculated based on the empirical dietary inflammatory pattern (EDIP) scoring system, which relies on the plasma levels of IL-6, C-reactive protein (CRP), and TNF receptor superfamily member 1B (TNFRSF1B) (84). Higher scores indicate inflammatory, while lower scores indicate anti-inflammatory effects. Foods with higher EDIP such as refined grains, red and processed meats, and carbonated beverages were associated with F. nucleatum positive CRC (48). Conversely, consumption of anti-inflammatory foods such as whole grains and fiber rich diets were associated with lower risk of developing F. nucleatum positive CRC (44). Fermented foods such as yogurts are protective to the colonic mucosa and maintain microbial diversity, which decreases the risk for CRC, especially in the proximal colon (85). Yogurts supplemented with lyophilized jabuticaba (Myrciaria jaboticaba) seed extract have strong prebiotic, antioxidant, and anti-cancer effects. These supplements, when fed to CRC rat models improved gut microbiome quality and increased the immune response and cytotoxic effects on colon cancer cells (86). This suggests that yogurt and other probiotics could be healthy supplements for the gut and its microbial ecosystem.
Antioxidant consumption is important for the survival of certain bacterial strains in the GIT. Decreased levels of antioxidants such as ascorbic acid, glutathione, and uric acids could be lethal for anaerobic gut bacterial species such as Clostridium sporogenes, Clostridium subterminale, and Romboutsia lituseburensis. Supplementation of these antioxidants in controlled aerobic condition in vitro resulted in production of protective SCFAs such as propanoic, butanoic, isobutanoic, and isopentanoic acids (58). This shows that supplementation of antioxidants decreases oxidative stress and enhances survival of anerobic microbes which produce SFCAs. SCFAs such as butyrate produced by the anaerobic species have protective effects in CRC (87). Among CRC survivors, consumption of legumes such as navy beans increased the production of beneficial metabolites such as such as piperidine, N-methylpipecolate, vanillate, and 2-aminoadipate. Gut microbes metabolized the indigestible substrates present in cooked navy beans and produced a total of 237 beneficial metabolites (88). In addition, navy bean consumers had 5.25 times higher levels of ophthalmic acid, which is associated with glutathione metabolism. Ophthalmic acid has an important role in detoxifying xenobiotics such as carcinogens and decreasing oxidative stress, thereby imparting protective effects against cancer (89–91). Individuals with diets deficient in dietary fibers and high in processed meat and sugary beverages show abundant colonization by sulfur-digesting bacteria such as Bilophila wadsworthia and Erysipelotrichaeceae. This is associated with an increase in risk for distal colon and rectal malignancies (59).
Glycyrrhiza uralensis polysaccharide (GCP) extracted from licorice impedes tumor growth and metastasis in mice inoculated with murine colon cancer (CT-26) cells. This is achieved by modifying the composition of gut microbiome such as increased levels of Enterorhabdus, Odoribacter, Ruminococcaceae_UCG_014, Ruminococcaceae_UCG_010, Enterococcus, and Ruminiclostridium_5 (92). Similarly, polysaccharides extracted from jujube was associated with decreased inflammation in mouse colon cancer models, most likely due to an associated decrease in Firmicutes and Bacteroidetes taxa in the gut microbiota (93). Similarly, combinations of Ganoderma lucidum polysaccharides and Gynostemma pentaphyllum saponins decreased colonic inflammatory and precancerous changes in Apcmin/+ mice. Together, they enhanced microbial richness by increasing SCFA producing microbes and decreasing sulfur digesting microbes (94). This suggests that certain plant and fungi-based products may be effective prebiotics and exert protective effects on the colonic epithelium.
Alcohol consumption is associated with alteration in gut microbiota that potentially accelerates CRC carcinogenesis. Alcohol is metabolized by the gut microbiota into toxic intermediates leading to colonic carcinogenesis via DNA-adducts, oxidative stress, epimutations, loss of epithelial barrier functions, and immunomodulations (95). These effects can be potentiated and aggravated by poor nutrition and chronic smoking status; covariates commonly associated with alcohol consumption. The microbiota in alcoholics have decreased levels of beneficial organisms such as Bacteroides and Ruminococcus and increased levels of harmful organisms such as Streptococcus (96). Integrated analysis using 16S rRNA (ribosomal RNA) gene analysis data and epidemiological characteristics by Kim et al. showed that alcohol consumption increased abundance of F. nucleatum in the gut which is associated with increased risk for CRC (97). Among alcoholics, deficiency of beneficial obligate anaerobe OTUs was demonstrated through decreased production of acetaldehyde in stool samples that were treated with specific quantities of ethanol under experimental conditions. This implies that restriction of alcohol consumption could potentially prevent genotoxic insults on colonic mucosa.
Dietary fibers provided by plant-based diets are not digested by the human intestinal enzymes and reach the colon unchanged. Colonic bacteria express enzymes that metabolize and ferment soluble dietary fibers into useful metabolites such as SCFAs which play a major role in decreasing colonic mucosal inflammation and lowering the risk for CRC (98, 99). Butyrate has an inhibitory effect over the histone deacetylases (HDAC) enzymes, resulting in enhanced expression of genes which arrest the cell cycle (100). Butyrate also serves as an energy source for normal enterocytes; however, rapidly dividing CRC cells are dependent on glycolysis rather than butyrate utilization for energy needs (101). Co-culturing certain bacterial strains in animal models enhanced production of butyrate and improved the SCFA-mediated protection against CRC. Faecalibacterium prausnitzii ATCC 27768 strain co-cultured with Bifidobacterium catenulatum KCTC 3221 and supplemented with fructooligosaccharides in anaerobic conditions significantly enhanced butyrate production (60). Exposing the co-culture supernatant to HT29 colon cancer cells and RAW 264.7 macrophages decreased the release of pro-inflammatory cytokine IL-8 in vitro (60). The supernatant from the co-cultured bacteria also enhanced acetate, propionate, and butyrate levels in the caecum of the DSS-induced colitis in mice model as well as decreased gene expression of IL-8, suggesting the anti-inflammatory effect of these bacterial species in vivo (60). Butyrate was also shown to increase the extracellular tight junction protein complexes in an Apcmin/+ mice model (102). This highlights the potential role of butyrate in decreasing the risk for CRC. Figure 2 illustrates the effects of dietary factors on the gut microbiome and their impact on CRC development.
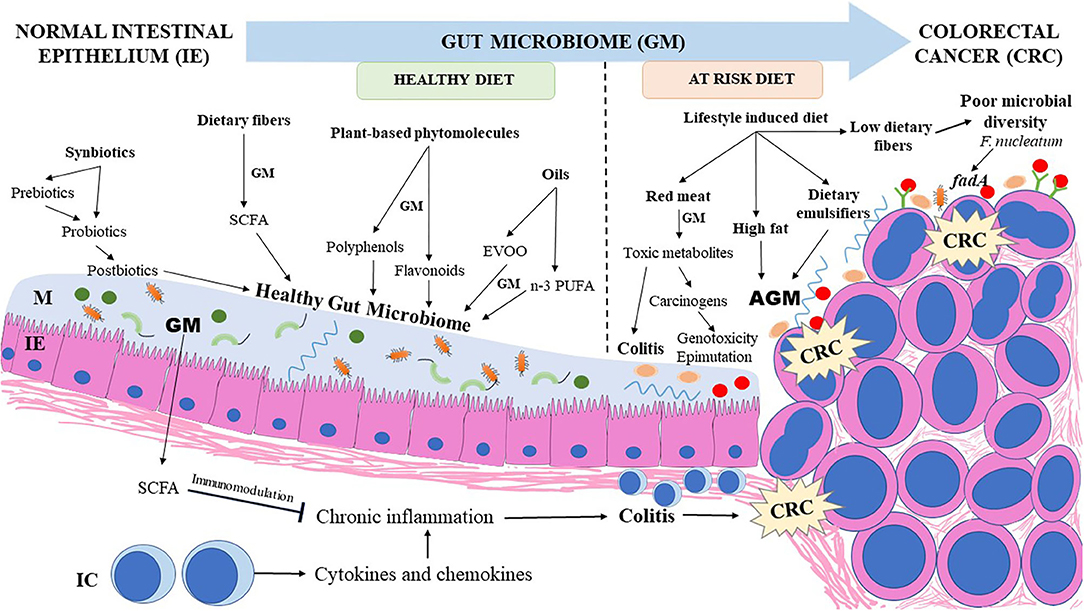
Figure 2. Influence of dietary factors on gut microbiome and its impact on CRC development. Dietary fibers, plant based phytomolecules, synbiotics, postbiotics, and essential oils such as n-3 PUFA (from marine sources) and EVOO chiefly support a healthy microbiome richness which provides useful metabolites protective to the colonic mucosa. Lifestyle induced changes in dietary pattern enhance the risk of DYB and aggravate colonic inflammation which progresses to CRC. CRC, colorectal cancer; AGM, altered gut microbiome; EVOO, extra virgin olive oil; GM, gut microbiome; IC, immune cells; IE, intestinal epithelium; M, mucus; n-3 PUFA, omega-3 polyunsaturated fatty acids; SCFA, short chain fatty acids.
Diet-derived phytochemicals, such as polyphenols and flavonoids, have protective effects on the colonic mucosa (103, 104). Most of the ingested polyphenols present in plant-based diets and their derivatives reach the colon unaltered and are metabolized by intestinal bacteria to more active substances which decrease oxidative stress, inflammation, and tumorigenesis (103). Polyphenols also act on the gut microbiota to enhance the proliferation of beneficial strains and inhibit pathogenic strains. Polyphenols increase the growth of probiotics and butyrate-producing microbiota such as Lactobacillus and Bifidobacterium, which inhibit inflammation, alleviates colitis, and decrease the risk for CRC (105). Polyphenols such as epigallocatechin-3-O-gallate and theaflavins present in tea extracts exert anti-inflammatory effects on F. nucleatum-induced inflammatory bowel disorders, thereby decreasing the risk for CRC (49). These anti-inflammatory effects are mediated by inactivation of NF-κB, as observed in U937-3xκB-LUC cell line (U937 cell line transfected with luciferase gene and coupled to three NF-κB binding sites). This results in decreased production of pro-inflammatory cytokines such as IL-1β, IL-6, tumor necrosis factor-α (TNF-α), and C-X-C motif chemokine ligand 8 (CXCL8). Polyphenols present in berries function as prebiotics and improve microbial richness in the form of Bifidobacterium, Lactobacillus and Akkermansia. Berry polyphenols also modulate the production of cytokines which alleviate inflammation and decrease the viability and proliferation of CRC cells (106). Polyphenols present in mango pulp such as gallotannins and gallic acid exhibit anti-inflammatory effects on the intestinal mucosa. In humans, consumption of mango pulp decreases pro-inflammatory cytokines such as IL-8, growth-regulated oncogene (GRO), and granulocyte macrophage colony-stimulating factor (GM-CSF). Mango polyphenols increase the abundance of Lactobacillus plantarum, Lactobacillus reuteri, and Lactobacillus lactis, as well as increase butyrate levels in feces (61). Date palms, another source of polyphenols and fibers, do not significantly alter gut microbiota or raise SCFAs in healthy volunteers, but decrease genotoxicity, fecal ammonia levels, and aid in bowel movements, thereby decreasing the risk for CRC (107). Similarly, administration of polyphenol-rich green tea extracts to human volunteers enhanced Firmicutes to Bacteroidetes ratio and SCFAs producing gut microbes (108). Thus, polyphenols are natural plant products which may have a potent role in reshaping the gut microbiome and thereby decrease the risk for CRC.
Curcumin, a natural product from Curcuma longa plant, is a polyphenol with a significant role in decreasing inflammation, oxidative stress, and alterations in gut microbiome (109). Similar to other polyphenols, curcumin is also subjected to bacterial metabolism, resulting in production of useful metabolites which have protective effects in CRC and also has key role in reviving beneficial microbial strains in the gut (110). IL-10-deficient CRC mice models on a curcumin-based diet demonstrated improved taxonomic profiles of gut microbiota, such as an abundance of Lactobacillales and lower levels of Coriobacterales. This was also associated with a reduction in tumor size and complete elimination of macroscopic lesions. In addition, there was restoration of β-catenin on plasma membranes, but the effect on mucosal inflammatory responses was limited (62). Farhana et al. reported that a combination of essential turmeric oil-curcumin and tocotrienol-rich fraction of vitamin E isomers effectively reduced the proliferation of colon cells (HCT-116 and HT-29 cells) in vitro and suppressed the growth of mice xenograft formed of HCT-116 cells in vivo (63). Bacterial 16S rRNA gene profiling revealed that turmeric oil-curcumin and tocotrienol treatment resulted in a significant increase in Lactobacillaceae and Bifidobacteriaceae, along with elevated Clostridium cluster XIVa, which produced an anti-inflammatory environment. In addition, the abundance of phylum Bacteroidetes and Firmicutes were relatively decreased. Collectively, this evidence supports the potential role of curcumin in combination with other natural substances in decreasing the risk for CRC.
Flavonoids are polyphenols abundant in fruits and vegetables and impart their natural colors (111). The gut microbiota and associated enzymes convert the flavonoids into bioactive metabolites, resulting in anti-inflammatory, antioxidant, and anti-tumor effects (111). Neohesperidin, a flavonoid which is abundant in citrus fruits, imparts tumoricidal activity in Apcmin/+ CRC mice models by inhibiting angiogenesis and promoting apoptosis (112). Fecal microbiota transplantation from neohesperidin treated mice decreased colonic tumorigenesis in vivo, mainly by modulating the gut microbiota composition. Neohesperidin treatment increased the abundance of phylum Firmicutes and Proteobacteria, while decreased Bacteroidetes (112). Black raspberry anthocyanins are another group of protective flavonoids that decreased tumorigenesis in colitis-associated CRC model mice by inducing epigenetic changes (50). Pan et al. reported that consumption of raspberry anthocyanin increased the abundance of anti-inflammatory bacterial genus such as Akkermansia and Desulfovibrio as well as butyrate producing Anaerostipes. However, alteration in the microbial composition was achieved only on consumption of whole raspberries (64). Thus, consumption of flavonoids abundant in plant-based diets improved microbial richness and decreased CRC growth.
Olive oil, an essential component of the Mediterranean diet, is high in monounsaturated fatty acids, squalene, phytosterols, and phenols (113). Phenolic derivatives of some of these nutrients are further metabolized by the gut microbiota into chemopreventive active substances. Consumption of extra virgin olive oil (EVOO) has beneficial effects on the mucosal cells, compared to sunflower and coconut oil. In an experimental mice model study, high-fat diet with sunflower and coconut oil produced perturbations in gut microbiota and inflammatory changes (73). EVOO also decreased harmful microbes from the genus Enterococcus, Staphylococcus, Neisseria, and Pseudomonas (73). This suggests that diets rich in EVOO could be protective against CRC, compared to other oils. N-3 polyunsaturated fatty acid (PUFA) in combination with fermentable dietary fibers have protective role in pathways related to programmed cell death and epigenetic dysregulation observed in CRC (114). In a randomized control trial, consumption of n-3 PUFA increased butyrate-producing bacteria such as Bifidobacterium, Roseburia, and Lactobacillus, suggesting that it had a role in reducing inflammation and CRC risk (115). However, it is noteworthy that PUFA and sphingolipids were altered in the fecal metabolomic profile of patients with adenoma-carcinoma and resulted in a preponderance of harmful species such as Firmicutes and Bacteroidetes (116). Moreover, Kraja et al. reported that n-3 PUFA from non-marine sources and reduced dietary fiber consumption increased the risk for CRC (117). It can be postulated that the possible benefits of n-3 PUFA on colonic mucosa are effective only when it is obtained from marine sources and consumed in combination with dietary fibers. Collectively, it suggests that careful selection of lipids in diet, especially EVOO and n-3 PUFA, is necessary for optimizing healthy colonic mucosa.
The combination of prebiotics and probiotics is known as synbiotics. Consumption of synbiotics is considered an active intervention to improve the quality of gut microbiome for preventing CRC. Synbiotics work by enriching the gut microbiome and the microbial strains which impart protective mucosal functions such as decreasing inflammation, preventing uncontrolled proliferation, preventing altered immune responses, lowering production of toxic metabolites, and lowering oxidative stress (118). In an experimental in vitro chip-based model (HuMiX gut-on-a-chip), synbiotics downregulated oncogenic signaling pathways (in Caco-2 cells). The synbiotics consists of probiotic Lactobacillus rhamnosus Gorbach-Goldin strain and a complex prebiotic formula simulating high fiber diet which was prepared from non-digestible carbohydrates and other prebiotics such as arabinogalactan, xylan and soy (119). Synbiotic administration not only decreased the production of oncometabolite lactate and suppressed drug resistance genes in colon cancer-derived cells, but also increased SCFAs such as acetate and formate (119). A new synbiotic combination of Lactobacillus gasseri 505 and Cudrania tricuspidata leaf extract in fermented milk decreased Staphylococcus and increased Lactobacillus, Bifidobacterium, and Akkermansia, thus increasing protective effects in DSS/azoxymethane (AOM) induced colitis-CRC model mice. This in vivo intervention decreased tumor proliferation and inflammation (marked by decreased levels of TNF-α, interferon (IFN)-γ, IL-1β, IL-6, inducible nitric oxide synthase, and cyclooxygenase-2) and led to upregulation of anti-inflammatory cytokines IL-4 and IL-10 (120). Praveen et al. developed raindrop candy consisting of polysaccharides extracted from Indian seaweed (S. wightii, E. compressa, and A. spicifera) and probiotic species L. plantarum NCIM 2083. These seaweed polysaccharides demonstrated anti-cancer effects on RAW 264.7 macrophage and HT-29 human colon cancer cell line in vitro (121). Thus, synbiotics could be novel therapeutic approaches to strengthen the gut microbiome for protective effects against CRC through alleviating inflammation and preventing tumorigenesis. However, due to the concerns of consuming live microbial species and uncertainties about the net effects of its colonization in the gut, postbiotics are gaining increasing attention as a paradigm shift (122, 123). Postbiotics are composed of inactivated microbial cells or their components with or without microbial metabolites, which shows beneficial effects on human health (124). Lactic acid bacteria (LAB) which are found in fermented foods are an essential source of postbiotic metabolites (PM) with physiological effects and health benefits including opposing effects on tumorigenesis (125). The cancer opposing effects of postbiotics are achieved through improving the gut microbiota, reprogramming the immune function, enhancing response to CRC treatment, and increasing antioxidant, anti-proliferative, and anti-inflammatory actions (126). Postbiotic metabolites (PMs) produced from Lactobacillus plantarum isolated from Malaysian fermented food, Tapai Ubihas, has remarkable anti-cancer effects such as enhanced cytotoxicity and anti-proliferative action (127, 128). PMs from six different strains of L. plantarum were evaluated for their anti-cancer effect on cells lines of five different cancers, which included breast, colorectal, cervical, liver, and leukemia. PMs obtained from two strains RG14 and I-UL4 effectively inhibited proliferation of HT29 colon cancer cells. Some PMs show anti-proliferative effects when added in specific microenvironments. For example, PM from the cell free supernatant of Streptococcus salivarius M18 cultured on prebiotic inulin displayed potent inhibitory effect on colon cancer cell proliferation through increasing extracellular acidity within the tumor (129). Postbiotics like SCFA and tryptophan have shown to downregulate both the adaptive and humoral immunity in inflammatory conditions such as inflammatory bowel diseases (130). This shows that postbiotics can not only prevent cancer proliferation, but also ameliorate mucosal inflammation. Thus, consumption of dietary fibers and diet-derived factors such as phytochemicals and essential fatty acids, as well as prebiotics, probiotics, and postbiotics could be considered a multifactorial protective approach against CRC. Table 2 presents the studies on dietary factors influencing the gut microbiome and its effect on the colonic mucosa and CRC progression.
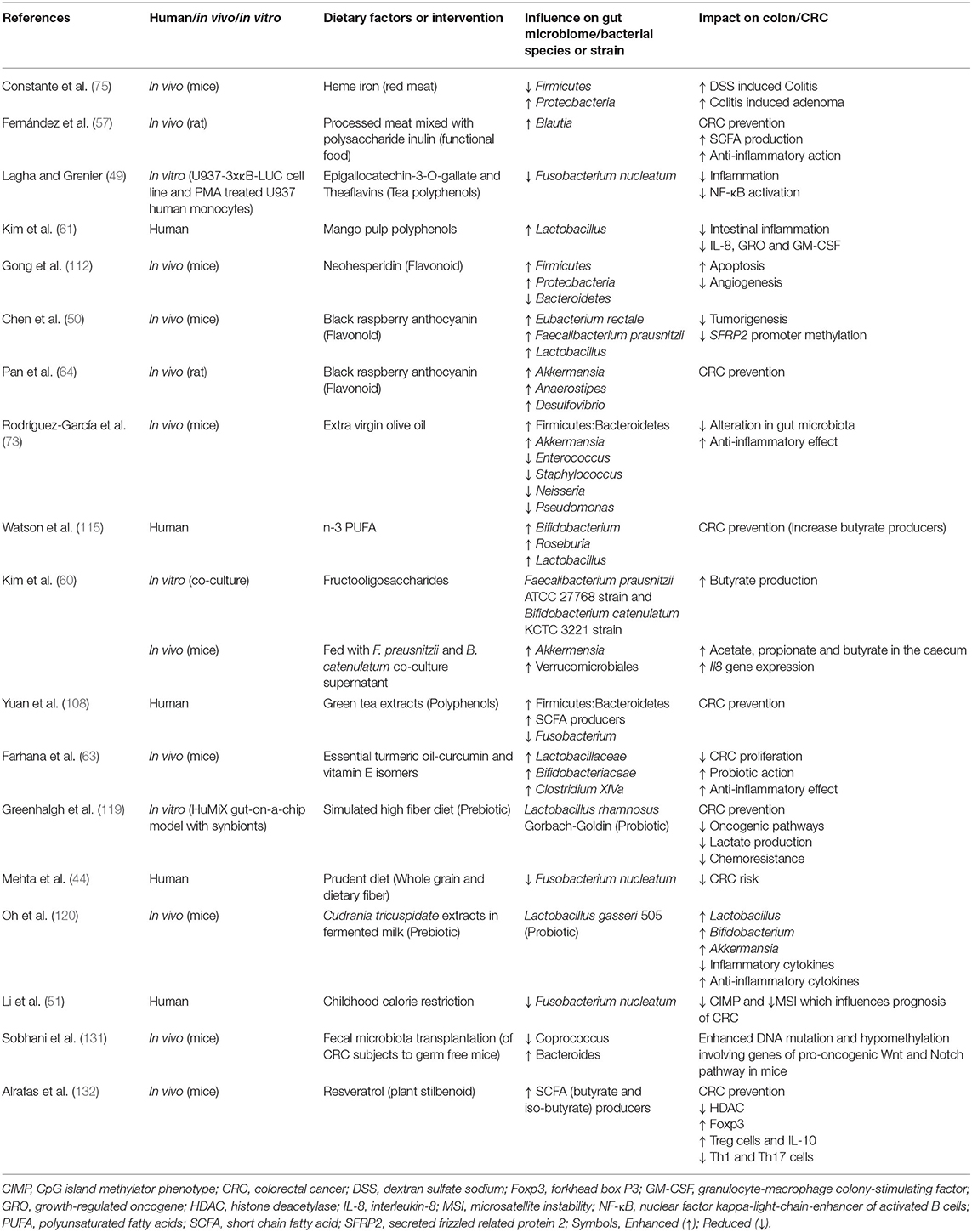
Table 2. Studies showing the effect of dietary factors influencing the gut microbiome and its impact on the colonic mucosa and CRC progression.
Cancer is triggered by a multitude of factors that destabilize the genetic regulatory mechanisms controlling the cell proliferation events. Apart from mutations occurring in the tumor suppressor genes, or protooncogenes leading to either loss or gain of resulting protein function, epigenetic changes also transform the transcriptomic profile and the genomic landscape resulting in oncogenic CRC traits (Figure 3). Epigenetic dysregulation, otherwise known as epimutations, commonly occur by promoter methylation/demethylation of CpG islands, histone acetylation/deacetylation, or by non-coding RNA such as miRNA which alter the expression of genes involved in cellular growth, differentiation, and metabolism (133). The gut microbiome is unique in the sense that it carries millions of genes which execute functions exotic to the human genome and their metabolic activities depend on the substrate presented to them by the host diet, thus establishing a symbiotic relationship. However, this symbiosis comes at a cost, as impaired nutrition can result in the synthesis of harmful metabolites which potentiates the susceptibility of the host genomic architecture to genotoxicity (133).

Figure 3. The host-gut microbiome influencing the CRC associated epigenetics. The resulting gut microbial metabolites can induce pro-oncogenic or onco-suppressive effects on CRC by modulating epigenomics. CRC, colorectal cancer; FFAR2, free fatty acid receptor 2; FMT, fecal microbiota transplantation; GM, gut microbiome; ncRNA, non-coding RNA; SCFA, short chain fatty acids; SFPR2, secreted frizzled related protein 2; TET3, ten eleven translocation 3. Symbols: Enhanced (↑); Reduced (↓); Activation (+).
The SCFAs, which are bacterial metabolites produced by digestion of dietary fibers by gut microbes, lower certain epigenetic alterations in enterocytes associated with CRC (98, 99). SCFAs, such as butyrate, protect the genetic and epigenetic architecture of enterocytes through multiple mechanisms (98, 134). The foremost includes its anti-inflammatory action, whereby it alleviates colonic mucosal inflammation and directly decreases the risk for CRC. Butyrate upregulates the activity of T-regulatory (T-reg) cells which exert an inhibitory effect on pro-inflammatory cytokine production and thereby blocking pro-oncogenic pathways (135). Butyrate has an inhibitory effect over the HDAC enzymes, which results in enhanced expression of genes arresting the cell cycle (100). Free fatty acid receptor 2 (FFAR2), activated by SCFAs such as butyrate, is known to suppress inflammation and prevent epigenetic dysregulation in CRC (136). Loss of FFAR2 in a DSS/AOM treated Apcmin/+ colitis-CRC model mice led to overexpression of HDAC, which is mediated through an upregulation of CREB (cAMP-response element binding protein) (136). This resulted in an epigenetic downregulation of immunomodulating genes such as Sfrp1, Dickkopf-related protein 3 (Dkk3), and suppression of cytokine signaling 1 (Socs1), which were collectively associated with enhanced infiltration of the colonic mucosa and tumor tissue by the neutrophils. The study demonstrated that the epigenetic dysregulation induced by loss of Ffar2 resulted in enhanced colonic inflammation, progressing into adenoma and adenocarcinoma formation (136). One carbon metabolism mediated by S-adenosyl methionine (SAM) transfers a methyl group to the CpG islands in the DNA promoter region which affects the gene expression and is of significance in CRC (137). Dietary consumption of methyl donors such as methionine, betaine, and choline as well as B vitamins (folate, B2, B6, and B12) have protective effects on the composition of gut microbiota (138). Deficiency of these nutrients decreases the abundance of beneficial organisms such as Akkermansia, Roseburia, and Faecalibacterium. Thus, the host genome is exposed to a number of dietary and microbial metabolites, which in turn would determine the beneficial vs. harmful effects on the pathogenesis of CRC.
The absence of caloric restriction during childhood may negatively impact microbial composition and may contribute to epigenetic dysregulation and development of CRC later in adulthood (51). Subjects who were energy restricted during their childhood had decreased abundance of pathogenic species such as F. nucleatum, Bacteroides fragilis, and Escherichia coli in later life, compared to non-restricted subjects (51). F. nucleatum is specifically associated with genetic and epigenetic defects such as microsatellite instability (MSI) and CpG island methylator phenotype (CIMP), respectively (51). Similarly, consumption of high caloric foods could lead to histone modifications such as methylation and acetylation of the active enhancers, thus augmenting the gene expression pertaining to CRC. Transplantation of colonic microbiota adapted to a high-fat diet into germ-free mice fed on high-caloric diet initiated the reoccurrence of these epigenetic changes (139). In another experiment, human fecal microbiota (from CRC subjects) transplanted to germ-free mice (treated with azoxymethane, CRC model) resulted in increased rate of DNA mutation and decreased DNA methylation involving the gene families of oncogenic Wnt and Notch pathway (131).
Plant-based derivatives along with microbiome can alter epigenomic changes associated with CRC. Anthocyanins present in freeze-dried black raspberries extracts have shown to induce demethylation of secreted frizzled related protein 2 (SFRP2) promoters, and revive useful probiotics such as Eubacterium rectale, Faecalibacterium prausnitzii, and Lactobacillus in DSS/AOM colitis-CRC mice model (50). Sfrp2 hypermethylation and subsequent downregulation are directly associated with development of hepatocellular carcinoma and CRC (140). Impaired gut microbiome activates ten-eleven translocation 3 (TET3) expression in colonocytes which induces demethylation of lamina-associated domains (LADs) leading to epigenetically programmed tumorigenesis and impaired chemotherapeutic response in CRC (141, 142). TET enzymes have active role in inducing demethylation of 5-methylcytosine (5mC) in the CpG islands. TET dioxygenases catalyze conversion of 5mC into oxidized forms, which are further converted to unmethylated cytosine by replication-dependent dilution or base excision repair (141). Resveratrol, a plant based stilbenoid, induces changes in the gut microbiome and is associated with an increased production of butyrate and isobutyrate producing taxa, causing release of anti-inflammatory cytokines (132). This is facilitated by resveratrol-induced inactivation of HDAC, which correlated with upregulation of transcription factor forkhead box P3 (Foxp3). This has several immunomodulatory functions, such as concomitant activation of T-regulatory (T-reg) cells, IL-10 synthesis, and reduction in pro-inflammatory Th1 and Th17m cells. This resulted in inhibition of inflammation in association with restoration of gut microbiome, thereby reducing the risk of colitis-associated CRC (132). Lactobacillus reuteri 6475, a commensal and probiotic producing a 2-carbon folate metabolite (5,10-ethenyl-tetrahydrofolyl polyglutamate), biochemically takes part in the transfer of 2 carbon atoms from acetate to homocysteine, resulting in formation of an exclusive amino acid ethionine, instead of conventional methionine. Incorporation of ethionine instead of methionine in proteins leads to decreased methylation and enhanced ethylation of lysine residues in histones (143). Tracking the source of the 2-carbon transporting form of folate by isotope labeling strikingly traces it to acetate in the culture medium of Lactobacillus reuteri 6475 (143). Dietary ethionine can result in immunomodulatory effects by suppressing cell mediated immunity and plausibly by NF-κB inhibition (143, 144). However, ethionine also carries carcinogenic potential, which can be reduced by supplementing sufficient methionine (145). This suggests that consumption of certain plant-based extracts and probiotics (such as Lactobacillus reuteri 6475) may help to prevent epigenetic alterations associated with CRC.
Finally, the non-coding RNA in the genome also regulates gene expression in CRC (133). Yuan et al. reported 76 differentially expressed microRNAs (miRNAs) in tumor samples, of which 55 were upregulated and 21 were downregulated. miR-182, miR-183, miR-503, and the miR-17~92 clusters were among the most consistently overexpressed miRNA in CRC (146). Genus Blautia was inversely correlated with miR-20a, miR-21, miR-96, miR-182, miR-183, and miR-7974, while positively correlated with miR-139, which is significantly expressed in normal tissues (146). However, enrichment analysis has shown that Akkermansia is the only genus associated with miRNA, which is linked to CRC pathway (146). This suggests that CRC associated alterations in gut microbiome often changes expression profiles of miRNA linked to cancer pathway. In the case of F. nucleatum, selective downregulation of miRNA, such as miR-18a and miR-4802, was shown to activate autophagy, inhibit apoptosis, and induce chemoresistance in HCT116 and HT29 CRC cells (37). miR-18a and miR-4802 post-transcriptionally regulated the expression of pro-autophagic proteins ULK1 and ATG7. However, F. nucleatum did not correlate significantly with miR-31 expression, previously shown to be upregulated in CRC with BRAF mutation (147, 148). Therefore, F. nucleatum-associated CRC plausibly has a key miRNA profile related to its pathogenesis.
The gut microbiome is also an enormous source of lipopolysaccharides (LPS), which flares inflammation and is associated with CRC progression. Exosomal miR-200c-3p notably impedes LPS-induced CRC invasion and migration by targeting zinc finger E-box-binding homeobox-1 (ZEB-1) as well as induces apoptosis in HCT116 cells in vitro (149). Bacterial small RNA (bsRNA), another microbial genomic component, are 50–400 nucleotides long non-coding RNA molecules which have a central role in regulating post-transcriptional gene expression in various bacterial cellular processes (150). Tarallo et al. using combined metagenomics and small RNA sequencing reported altered human and bsRNA profile in stools of CRC subjects showing alterations in the gut microbiota (151). Based on the bsRNA profile, 15 bacterial species were significantly altered in CRC. E. coli bsRNA levels were significantly overrepresented which also correlated with the high abundance of E. coli in stool of CRC subjects. Bacteroides ovatus bsRNA expression were lower in both CRC and adenomas (151). Stool samples from CRC patients also showed alterations in the gut microbiota, characterized by abundance of Alistipes putredinis species and Firmicutes phyla. Across human non-coding RNA (ncRNA), miR-378a-3p and piR-11481 were the most differentially expressed miRNA and small ncRNA, respectively. Mjelle et al. reported differential expression of small RNAs from Epstein-Barr virus and Fusobacterium nucleatum between colon cancer and adjacent normal mucosa (152). Differential expression of mRNA is also associated with mutational events associated with colon cancer such as microsatellite instability (MSI). microRNA such as miR-335, miR-26 and miR-625 are differentially expressed in association with MSI (152). Thus, it is suggestive that non-coding RNA expression affecting CRC pathogenesis correlates with the composition of the gut microbiome. Moreover, alteration in the expression of human and bacterial small RNA could be a useful tool in the diagnosis of CRC.
Alterations in the gut microbiome pertaining to epigenetic landscape of CRC is highly dependent as well as regulated by our dietary pattern (98). Metabolic conditions like obesity are associated in epigenetic dysregulation and currently emphasis is driven toward identifying the epigenetic markers and the environmental factors deranging them as well as the effect of dietary or therapeutic intervention in regulating these epimutations (153). Paternal high-fat diet consumption leads to beta cell dysfunction in female rat offspring, especially associated with hypomethylation and increased expression of Il13ra2 (interleukin-13 receptor subunit alpha-2) gene (154). Together with scaffolding protein FAM120A (family with sequence similarity 120A), IL13RA2 activation enhances the survival, invasion, migration, and dissemination of colon cancer (155). Exposure to different dietary constituents, varying pH as well as humid and stable temperature leads to heterogeneity of the oral cavity microbiota, of which certain organisms like Fusobacterium are highly implicated in the pathogenesis of CRC (156, 157). F. nucleatum consistently increased the expression of miR21 in four colon cancer cell lines potentially via theTLR-4/MyD88/NF-κB signaling pathway. Interestingly, elevated levels of miR21 and F. nucleatum DNA in the colon tumor tissue was associated with advanced CRC and poor survival (47). miR21 has recognizable role in promoting proliferation of cancer stem cells as well as enhancing angiogenesis while its antagonism reversed the effects (158–160). Dietary fibers are essential source of SCFAs, which can regulate the epigenetic events associated with neoplastic events in human colon cells. Among the SCFAs, butyrate in comparison to acetate and propionate is the most potent metabolite in reducing the proliferation of HCT116 human colon cancer cells by strongly suppressing the cell cycle (G2 phase arrest) and inducing apoptosis (161). Altered gut microbiota in CRC is associated with decline in butyrate producing microbes with corresponding increases in harmful species (162). A prudent mechanism for suppression of CRC by butyrates is by regulating the expression of miRNA (163). In vitro experiment by Hu et al. has shown that butyrate's reduced the expression of pro-tumorigenic non-coding RNA such as precursor and mature miR-92a along with primary miR17-92a (164). Nielsen et al. reported that feeding experimental rat with high-amylose potato starch (HAPS), high-amylose maize starch (HAMS), and butyrylated high-amylose maize starch (HAMSB) shifted the gut microbiome toward metabolizing carbohydrates and consequently enhancing butyrate production. HAPS and HAMSB selectively decreased the expression of colonic oncogenic miR17-92 which is protective against CRC (56). This was associated with the downregulation of c-Myc and its overexpression increased the expression of primary miR17-92a. In colon adenoma LT97 cells, butyrate and trichostatin A as histone deacetylase inhibitors downregulated cancer specific miRNAs such as miR-135a, miR-135b, miR-24, miR-106b, and miR-let-7a (165). Butyrate and trichostatin A also upregulated p21 and cyclin D2 expression in non-transfected LT97 cells as compared to cells transfected with miR-106b and miR-135a, respectively. Also, in vitro transfection of miR106a mimics into HCT-116 CRC cells decreased the beneficial effect of p21 expression induced by butyrate (166). Thus, consumption of dietary fibers which modulates the gut microbiome to produce metabolites such as SCFAs are beneficial in regulating epigenetic events in CRC.
Overall, it appears that dietary patterns or habits regulating the gut microbiota heavily influences the epigenetic mechanism associated with the pathogenesis of CRC. Further clinical exploration on understanding the outcome of dietary intervention and its modulation of the gut microbiome and the host genome is warranted to validate the therapeutic potential of differential diet-based regimes in management of CRC.
Methodologically analyzing the gut microbiome and their metabolome is a very complex process and demands the use of advanced techniques to delineate the intricacy related to its composition and function. Culture based methods are the earliest measures to identify the bacterial species based on morphological and biochemical characteristics (167). But culturing underestimates the abundance and diversity of the microbial species in the gut and hence its utility is limited in microbiome research. The smaller 16S ribosome of bacteria contains the ribosomal RNA (rRNA) which is a phylogenetic marker representing the evolutionary divergence of bacteria and consist of several conserved constant regions and species-specific hypervariable regions (V1–V9) (167, 168). Molecular exploration of the hypervariable region is commonly employed to study the complex composition and diversity of the gut microbiome with relative accuracy (167, 168). Its variability at different taxonomic levels can be analyzed to estimate the bacterial genome and its relative abundances (169). These hypervariable regions are distributed between several highly conserved constant regions and variations in the nucleotide sequences within these hypervariable regions reflect evolutionary divergences of bacteria (167). Sequencing these hypervariable regions within the 16S rRNA gene is an essential technique for identification and classification of the bacteria with relative quantification of their abundance. Next generation sequencing is a powerful technique for efficient and cost-effective analysis of the 16S rRNA gene hypervariable regions which could not be effectively mapped using older methods (170). The sequenced data often contain poor quality reads which have to be removed using quality control filters and subsequently based on sequence similarities are clustered into operational taxonomic units or OTU. OTU represents the set of closely matched nucleotide sequences (>97% sequence similarity) which represent either a single or a group of bacterial species (88). The representative sequences from the OTU are mapped in a 16S rRNA gene sequence database for taxonomical classification and determination of diversity, relative abundance as well as annotation related to functional pathways (167). Even though 16S rRNA gene sequencing is a powerful technique in identifying the abundance of different bacterial taxa, it often has certain limitations. 16S rRNA gene sequencing is based on the assumption that >95% similarity is taxonomically associated with genus identification, while >98.7% sequence similarity is useful for distinguishing species (171, 172). But conventionally, the resolution is not enough for intraspecies strain identification, which are characterized by subtle changes in single nucleotide in the gene amplicon. Johnson et al. reported that sequencing the long reads or full-length amplicon of 16S rRNA gene can resolve minor nucleotide changes associated with intraspecies strain identification (173). Moreover, bacterial taxa analyzed by 16S rRNA gene sequencing is based on relative abundance than absolute count of the organism (174). Also, the 16S rRNA reference database is incomplete and does not cover all the reference sequences pertaining to specific microbial species from the diverse gut microbiota (167, 174). Amplification of erroneous sequences termed as chimera, which consist of a single amplicon originating from two different sources during the polymerase chain reaction (PCR) cycles could lead to wrong species identification and annotations (174). These drawbacks associated with inaccurate identification of bacterial species, incomplete 16S rRNA gene reference database and sequencing errors are major pitfalls associated with 16S rRNA gene sequencing and demand newer techniques to overcome these inadequacies and improve the resolution in identifying bacterial taxa to subspecies or strain levels. Shotgun metagenomic analysis using high throughput sequencing is another powerful technique, which collectively analyzes all the genes and genomes of the microbiota in the given specimen and is the next step for discretizing higher level of bacterial taxonomical characterization to achieve higher degree of sequence similarity and subspecies identification (167). This method circumvents the traditional 16S rRNA gene sequencing as well as provides better information on the functional capabilities of the microbiome (167). In this method the complete metagenome (cumulative genome of bacteria and host) obtained from the sample is fragmented into short segments and further sequenced using high throughput sequencing. Adequate filters are required for rarefaction of bacterial genomic component and remove the contaminants from host genome. The genome data is further analyzed using bioinformatic pipelines to delineate bacterial taxa. Compared to 16S rRNA gene sequencing, metagenomics reveals more information on low abundance genera and is more useful in determining microbial diversity when sufficient reads are available (175). Nanopore sequencing, using workflow, called Lathe (tool for generating bacterial genome from metagenomic data based on nanopore long read sequencing) could be used to assemble closed circular genome by incorporating long read assembly, short read error correction, and genome circularization (176, 177). It enhances binning assembly from assembled contigs as contiguity from short reads is often contaminated by repeat elements. This determines high-quality contiguous and circular bacterial genomes from diverse human gut microbiome. However, sequencing the closed circular genome from long reads requires extraction of high molecular weight DNA extraction which is technically challenging due to its low yield.
Newer modalities like metatranscriptomics, metaproteomics, and metabolomics together aid in understanding the functional role of the human gut microbiome (178, 179). Metatranscriptomics, which is based on RNA-seq further takes the bacterial taxonomical classification to the next level by determining the expressional significance of the proteins associated with altered gut microbiome (167). Longitudinally pairing metagenomics with metatranscriptomics will holistically elucidate the functional dynamics of the gut microbiome in specific diseases (180). Furthermore, metaproteomics and metabolomics unravels the functional significance of all expressed proteins and generated metabolites by the transforming gut microbiota and provides key information on its predictive or definitive causal relationship in different human diseases (181–183). Metaproteomics involves the study of the complete proteins produced by the gut microbiota in the provided specimen, which elucidates the functional aspects of microbial gene expression (167). Correspondingly, metabolomics deal with the complete metabolite profiling of the given specimen. The three useful and pivotal techniques for studying the metabolomic profile of the stool specimen commonly includes gas chromatography-mass spectrometry (GC–MS), liquid chromatography-mass spectrometry (LC-MS), and capillary electrophoresis-mass spectrometry (CE-MS) (181, 184). Metabolomic workflow involves sampling, sample processing, instrumental separation, metabolomic data analysis, and interpretation (181). Metabolomic pipelines can further characterize the metabolomic data in relation to gut microbiota composition. Recently, combined use of comprehensive metabolomic dataset and random forest-based machine learning algorithm have unraveled novel metabolic and biochemic pathways characterized to Bacteroides genus (185). This is a new milestone in the area of metabolomics as machine learning can incorporate technological edge in determining the metabolomic changes pertaining to specific microbe. Natural dietary components such as polyphenols are effective antioxidants and prebiotics which are processed by the gut microbiota into metabolites with immunomodulatory functions. A high throughput analysis known as foodomics utilizes various omics-based approach to identify suitable active polyphenols in the natural food source (186). This approach can also delineate the possible health benefits of consumption of dietary nutrients such as polyphenols. Microbiomics, nutri(epi)genomics, and metabolomics, collectively are high throughput “omics” technologies which give better understanding on the vital role of food bioactive substances in human health in a personalized manner, considering the interindividual differences in their metabolism, bioavailability, and bioefficiency (187). Overall, it can be summarized that omics-based approaches have pivotal role in understanding the human gut microbiota and the functional significance of its gene expression.
A solitary metagenomics analysis may be inadequate to establish association between the host, gut microbiome, and metabolite axis. Meta-analysis of metagenomics data is a powerful method for summarizing the impact of gut microbiota and its metabolites in CRC across different studies. Wirbel et al. in a meta-analysis based on eight fecal shotgun metagenomics studies (total 768 fecal metagenomes) identified potential microbial markers and altered metabolites associated with CRC (188). The analysis of diverse CRC metagenomes revealed a key set of 29 microbial markers, which were significantly associated with CRC. These also included genera such as Fusobacterium, Porphyromonas, Parvimonas, Peptostreptococcus, Gemella, Prevotella, and Solobacterium, which were previously associated with CRC (188). Functional analysis of the CRC metagenomes showed an upregulation of pathways related to protein, glycoprotein, and organic acid metabolism, while carbohydrate metabolizing genes were depleted (188). This suggested that the healthy gut microbiome which preferentially metabolized carbohydrate shifted toward an amino acid utilizing pattern in CRC. Interestingly, Thomas et al. in another meta-analysis involving large-scale CRC metagenomic datasets observed greater microbial richness in the gut microbiome of CRC subjects when compared to controls (189). This pattern was contributed by the overabundance of microbial species from the oral cavity in the gut microbiome of CRC subjects. CRC biomarker species such as F. nucleatum, Solobacterium moorei, Porphyromonas asaccharolytica, Parvimonas micra, Peptostreptococcus stomatis, and Parvimonas spp. were commonly observed in most of the individual studies. Analysis of the fecal metabolic profiles from the datasets show an abundance of the gene choline trimethylamine-lyase, which is required for the bacterial synthesis of trimethylamine, a potent carcinogen associated with CRC (189, 190). Thomas et al. also reported that predicting CRC based on an independent metagenomic dataset would incur errors and therefore performed alternative analysis based on leave one dataset out (LODO) predictive model which can be potentially useful in clinical scenario (189). Thus, meta-analysis-based approaches for comparing different metagenomic studies could be an effective method for identifying novel CRC associated microbial biomarker species and metabolites from large scale datasets.
Emerging evidence suggests a significant association between the gut microbiome and colorectal cancer. As a result, dietary constituents such as dietary fibers, phytomolecules, n-3 PUFAs, prebiotics, probiotics, and postbiotics may offer benefits in the prevention of CRC through favorable alterations in the gut microbiome. More specifically, dietary and lifestyle factors may enrich the growth of healthy microbes and suppress the non-beneficial strains. Beneficial strains of gut microbiome produce enterocyte-friendly metabolites, such as SCFAs, and may protect the mucosa against inflammation and induction of oncogenic pathways. At this time, prospective data examining this anti-cancer approach is lacking. Future studies should examine the microbiome impact of dietary risk factor modification in patients at high-risk for CRC.
RK and SA: conceptualization. MR: methodology. VR, PM, and RT: literature search. SA: writing—original draft preparation. MR, VR, AS, RT, PM, and AK: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Tözün N, Vardareli E. Gut microbiome and gastrointestinal cancer: les liaisons dangereuses. J Clin Gastroenterol. (2016) 50:S191–6. doi: 10.1097/MCG.0000000000000714
2. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. (2006) 124:837–48. doi: 10.1016/j.cell.2006.02.017
3. Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, et al. A new genomic blueprint of the human gut microbiota. Nature. (2019) 568:499–504. doi: 10.1038/s41586-019-0965-1
4. Malard F, Dore J, Gaugler B, Mohty M. Introduction to host microbiome symbiosis in health and disease. Mucosal Immunol. (2020) 14:1–8. doi: 10.1038/s41385-020-00365-4
5. Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host–bacterial symbiosis in health and disease. Adv Immunol. (2010) 107:243–74. doi: 10.1016/B978-0-12-381300-8.00008-3
6. Singh RK, Chang H-W, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. (2017) 15:1–17. doi: 10.1186/s12967-017-1175-y
7. Daniel CR, McQuade JL. Nutrition and cancer in the microbiome era. Trends Cancer. (2019) 5:521–4. doi: 10.1016/j.trecan.2019.07.003
8. Dahmus JD, Kotler DL, Kastenberg DM, Kistler CA. The gut microbiome and colorectal cancer: a review of bacterial pathogenesis. J Gastrointest Oncol. (2018) 9:769. doi: 10.21037/jgo.2018.04.07
9. Vipperla K, O'Keefe SJ. Diet, microbiota, and dysbiosis: a ‘recipe’for colorectal cancer. Food Funct. (2016) 7:1731–40. doi: 10.1039/C5FO01276G
10. Dalal N, Jalandra R, Bayal N, Yadav AK, Sharma M, Makharia GK, et al. Gut microbiota-derived metabolites in CRC progression and causation. J Cancer Res Clin Oncol. (2021) 1–15. doi: 10.1007/s00432-021-03729-w
11. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
12. Cheng H-Y, Ning M-X, Chen D-K, Ma W-T. Interactions between the gut microbiota and the host innate immune response against pathogens. Front Immunol. (2019) 10:607. doi: 10.3389/fimmu.2019.00607
13. Lucas C, Barnich N, Nguyen HTT. Microbiota, inflammation and colorectal cancer. Int J Mol Sci. (2017) 18:1310. doi: 10.3390/ijms18061310
14. Pandey A, Shen C, Man SM. Focus: organelles: inflammasomes in colitis and colorectal cancer: mechanism of action and therapies. Yale J Biol Med. (2019) 92:481.
15. Soleimani A, Rahmani F, Ferns GA, Ryzhikov M, Avan A, Hassanian SM. Role of the NF-κB signaling pathway in the pathogenesis of colorectal cancer. Gene. (2020) 726:144132. doi: 10.1016/j.gene.2019.144132
16. Yang Y, Misra BB, Liang L, Bi D, Weng W, Wu W, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. (2019) 9:4101. doi: 10.7150/thno.35186
17. Yang J, Yu J. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell. (2018) 9:474–87. doi: 10.1007/s13238-018-0543-6
18. Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. (2011) 3:858–76. doi: 10.3390/nu3100858
19. Abu-Ghazaleh N, Chua WJ, Gopalan V. Intestinal microbiota and its association with colon cancer and red/processed meat consumption. J Gastroenterol Hepatol. (2021) 36:75–88. doi: 10.1111/jgh.15042
20. Debesa-Tur G, Pérez-Brocal V, Ruiz-Ruiz S, Castillejo A, Latorre A, Soto JL, et al. Metagenomic analysis of formalin-fixed paraffin-embedded tumor and normal mucosa reveals differences in the microbiome of colorectal cancer patients. Sci Rep. (2021) 11:1–15. doi: 10.1038/s41598-020-79874-y
21. Wu Y, Shi L, Li Q, Wu J, Peng W, Li H, et al. Microbiota diversity in human colorectal cancer tissues is associated with clinicopathological features. Nutr Cancer. (2019) 71:214–22. doi: 10.1080/01635581.2019.1578394
22. Chen C, Niu M, Pan J, Du N, Liu S, Li H, et al. Bacteroides, butyric acid and t10, c12-CLA changes in colorectal adenomatous polyp patients. Gut Pathog. (2021) 13:1–9. doi: 10.1186/s13099-020-00395-0
23. Kim Y, Liu R, Rychlik J, Russell J. The enrichment of a ruminal bacterium (Megasphaera elsdenii YJ-4) that produces the trans-10, cis-12 isomer of conjugated linoleic acid. J Appl Microbiol. (2002) 92:976–82. doi: 10.1046/j.1365-2672.2002.01610.x
24. Raimondi S, Amaretti A, Leonardi A, Quartieri A, Gozzoli C, Rossi M. Conjugated linoleic acid production by bifidobacteria: screening, kinetic, and composition. Biomed Res Int. (2016). doi: 10.1155/2016/8654317
25. Clarke JM, Young GP, Topping DL, Bird AR, Cobiac L, Scherer BL, et al. Butyrate delivered by butyrylated starch increases distal colonic epithelial apoptosis in carcinogen-treated rats. Carcinogenesis. (2012) 33:197–202. doi: 10.1093/carcin/bgr254
26. Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. (2014) 158:288–99. doi: 10.1016/j.cell.2014.04.051
27. Taylor JC, Gao X, Xu J, Holder M, Petrosino J, Kumar R, et al. A type VII secretion system of Streptococcus gallolyticus subsp. gallolyticus contributes to gut colonization and the development of colon tumors. PLoS Pathog. (2021) 17:e1009182. doi: 10.1371/journal.ppat.1009182
28. Ternák G, Berényi K, Sümegi A, Szenczi Á, Fodor B, Németh B, et al. Antibiotic consumption patterns in european countries may be associated with the incidence of major carcinomas. Antibiotics. (2020) 9:643. doi: 10.3390/antibiotics9100643
29. Lee JG, Lee Y-R, Lee A-R, Park CH, Han DS, Eun CS. Role of the global gut microbial community in the development of colitis-associated cancer in a murine model. Biomed Pharmacother. (2021) 135:111206. doi: 10.1016/j.biopha.2020.111206
30. Huang Q-Y, Yao F, Zhou C-R, Huang X-Y, Wang Q, Long H, et al. Role of gut microbiome in regulating the effectiveness of metformin in reducing colorectal cancer in type 2 diabetes. World J Clin Cases. (2020) 8:6213. doi: 10.12998/wjcc.v8.i24.6213
31. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. (2015) 528:262–6. doi: 10.1038/nature15766
32. Huang X, Hong X, Wang J, Sun T, Yu T, Yu Y, et al. Metformin elicits antitumour effect by modulation of the gut microbiota and rescues Fusobacterium nucleatum-induced colorectal tumourigenesis. EBioMedicine. (2020) 61:103037. doi: 10.1016/j.ebiom.2020.103037
33. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver–passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. (2012) 10:575–82. doi: 10.1038/nrmicro2819
34. DeDecker L, Coppedge B, Avelar-Barragan J, Karnes W, Whiteson K. Microbiome distinctions between the CRC carcinogenic pathways. Gut Microbes. (2021) 13:1–12. doi: 10.1080/19490976.2020.1854641
35. Hussan H, Clinton SK, Roberts K, Bailey MT. Fusobacterium's link to colorectal neoplasia sequenced: a systematic review and future insights. World J Gastroenterol. (2017) 23:8626. doi: 10.3748/wjg.v23.i48.8626
36. Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. (2015) 23:141–7. doi: 10.1016/j.mib.2014.11.013
37. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. (2017) 170:548–63. e516. doi: 10.1016/j.cell.2017.07.008
38. Amitay EL, Werner S, Vital M, Pieper DH, Höfler D, Gierse I-J, et al. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis. (2017) 38:781–8. doi: 10.1093/carcin/bgx053
39. Li D-H, Li Z-P, Zhang Y, Zhou G-Z, Ren R-R, Zhao H-J, et al. Fecal Fusobacterium nucleatum harbored virulence gene fadA are associated with ulcerative colitis and clinical outcomes. Microb Pathog. (2021) 157:104964. doi: 10.1016/j.micpath.2021.104964
40. Meng Q, Gao Q, Mehrazarin S, Tangwanichgapong K, Wang Y, Huang Y, et al. Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep. (2021) 22:e52891. doi: 10.15252/embr.202152891
41. Guo P, Tian Z, Kong X, Yang L, Shan X, Dong B, et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J Exp Clin Cancer Res. (2020) 39:1–13. doi: 10.1186/s13046-020-01677-w
42. Dadashi M, Hajikhani B, Faghihloo E, Owlia P, Yaslianifard S, Goudarzi M, et al. Proliferative effect of FadA recombinant protein from Fusobacterium nucleatum on SW480 colorectal cancer cell line. Infect Disord Drug Targets. (2020) 21:623–8. doi: 10.2174/1871526520666200720113004
43. Kashani N, Abadi AB, Rahimi F, Forootan M. FadA-positive Fusobacterium nucleatum is prevalent in biopsy specimens of Iranian patients with colorectal cancer. New Microbes New Infect. (2020) 34:100651. doi: 10.1016/j.nmni.2020.100651
44. Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, et al. Association of dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. (2017) 3:921–7. doi: 10.1001/jamaoncol.2016.6374
45. Wang Y, Wan X, Wu X, Zhang C, Liu J, Hou S. Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis. Gut Pathog. (2021) 13:1–11. doi: 10.1186/s13099-020-00396-z
46. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. (2018) 359:592–7. doi: 10.1126/science.aah3648
47. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear Factor– κB, and up-regulating expression of microRNA-21. Gastroenterology. (2017) 152:851–66. e824. doi: 10.1053/j.gastro.2016.11.018
48. Liu L, Tabung FK, Zhang X, Nowak JA, Qian ZR, Hamada T, et al. Diets that promote colon inflammation associate with risk of colorectal carcinomas that contain Fusobacterium nucleatum. Clin Gastroenterol Hepatol. (2018) 16:1622–31. e1623. doi: 10.1016/j.cgh.2018.04.030
49. Lagha AB, Grenier D. Tea polyphenols inhibit the activation of NF-κB and the secretion of cytokines and matrix metalloproteinases by macrophages stimulated with Fusobacterium nucleatum. Sci Rep. (2016) 6:1–11. doi: 10.1038/srep34520
50. Chen L, Jiang B, Zhong C, Guo J, Zhang L, Mu T, et al. Chemoprevention of colorectal cancer by black raspberry anthocyanins involved the modulation of gut microbiota and SFRP2 demethylation. Carcinogenesis. (2018) 39:471–81. doi: 10.1093/carcin/bgy009
51. Li J, Wei Z, Cao S, Tan X, Liu S, Yao Z, et al. A pilot study on clinicopathological features and intestinal microflora changes in colorectal cancer patients born over a nine-year period encompassing three years before and after the Great Chinese famine. Cancer Epidemiol. (2019) 59:166–72. doi: 10.1016/j.canep.2019.02.001
52. Martin OC, Olier M, Ellero-Simatos S, Naud N, Dupuy J, Huc L, et al. Haem iron reshapes colonic luminal environment: impact on mucosal homeostasis and microbiome through aldehyde formation. Microbiome. (2019) 7:72. doi: 10.1186/s40168-019-0685-7
53. Viennois E, Merlin D, Gewirtz AT, Chassaing B. Dietary emulsifier–induced low-grade inflammation promotes colon carcinogenesis. Cancer Res. (2017) 77:27–40. doi: 10.1158/0008-5472.CAN-16-1359
54. Zaramela LS, Martino C, Alisson-Silva F, Rees SD, Diaz SL, Chuzel L, et al. Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates. Nat Microbiol. (2019) 4:2082–9. doi: 10.1038/s41564-019-0564-9
55. Zhang J, Lacroix C, Wortmann E, Ruscheweyh H-J, Sunagawa S, Sturla SJ, et al. Gut microbial beta-glucuronidase and glycerol/diol dehydratase activity contribute to dietary heterocyclic amine biotransformation. BMC Microbiol. (2019) 19:1–14. doi: 10.1186/s12866-019-1483-x
56. Nielsen TS, Bendiks Z, Thomsen B, Wright ME, Theil PK, Scherer BL, et al. High-amylose maize, potato, and butyrylated starch modulate large intestinal fermentation, microbial composition, and oncogenic mirna expression in rats fed a high-protein meat diet. Int J Mol Sci. (2019) 20:2137. doi: 10.3390/ijms20092137
57. Fernández J, Ledesma E, Monte J, Millán E, Costa P, de la Fuente VG, et al. traditional processed Meat products Re-designed towards inulin-rich functional foods Reduce polyps in two colorectal cancer Animal Models. Sci Rep. (2019) 9:1–17. doi: 10.1038/s41598-019-51437-w
58. Million M, Armstrong N, Khelaifia S, Guilhot E, Richez M, Lagier J-C, et al. The antioxidants glutathione, ascorbic acid and uric acid maintain butyrate production by human gut clostridia in the presence of oxygen in vitro. Sci Rep. (2020) 10:1–11. doi: 10.1038/s41598-020-64834-3
59. Nguyen LH, Ma W, Wang DD, Cao Y, Mallick H, Gerbaba TK, et al. Association between sulfur-metabolizing bacterial communities in stool and risk of distal colorectal cancer in men. Gastroenterology. (2020) 158:1313–25. doi: 10.1053/j.gastro.2019.12.029
60. Kim H, Jeong Y, Kang S, You HJ, Ji GE. Co-culture with Bifidobacterium catenulatum improves the growth, gut colonization, and butyrate production of Faecalibacterium prausnitzii: in vitro and in vivo studies. Microorganisms. (2020) 8:788. doi: 10.3390/microorganisms8050788
61. Kim H, Venancio VP, Fang C, Dupont AW, Talcott ST, Mertens-Talcott SU. Mango (Mangifera indica L.) polyphenols reduce IL-8. GRO, and GM-SCF plasma levels and increase Lactobacillus species in a pilot study in patients with inflammatory bowel disease. Nutr Res. (2020) 75:85–94. doi: 10.1016/j.nutres.2020.01.002
62. McFadden R-MT, Larmonier CB, Shehab KW, Midura-Kiela M, Ramalingam R, Harrison CA, et al. The role of curcumin in modulating colonic microbiota during colitis and colon cancer prevention. Inflamm Bowel Dis. (2015) 21:2483–94. doi: 10.1097/MIB.0000000000000522
63. Farhana L, Sarkar S, Nangia-Makker P, Yu Y, Khosla P, Levi E, et al. Natural agents inhibit colon cancer cell proliferation and alter microbial diversity in mice. PLoS ONE. (2020) 15:e0229823. doi: 10.1371/journal.pone.0229823
64. Pan P, Lam V, Salzman N, Huang Y-W, Yu J, Zhang J, et al. Black raspberries and their anthocyanin and fiber fractions alter the composition and diversity of gut microbiota in F-344 rats. Nutr Cancer. (2017) 69:943–51. doi: 10.1080/01635581.2017.1340491
65. Davis EC, Dinsmoor AM, Wang M, Donovan SM. Microbiome composition in pediatric populations from birth to adolescence: impact of diet and prebiotic and probiotic interventions. Dig Dis Sci. (2020) 65:706–22. doi: 10.1007/s10620-020-06092-x
66. De Almeida CV, de Camargo MR, Russo E, Amedei A. Role of diet and gut microbiota on colorectal cancer immunomodulation. World J Gastroenterol. (2019) 25:151. doi: 10.3748/wjg.v25.i2.151
67. Hills RD, Pontefract BA, Mishcon HR, Black CA, Sutton SC, Theberge CR. Gut microbiome: profound implications for diet and disease. Nutrients. (2019) 11:1613. doi: 10.3390/nu11071613
68. Oostindjer M, Alexander J, Amdam GV, Andersen G, Bryan NS, Chen D, et al. The role of red and processed meat in colorectal cancer development: a perspective. Meat Sci. (2014) 97:583–96. doi: 10.1016/j.meatsci.2014.02.011
69. Viennois E, Chassaing B. Consumption of select dietary emulsifiers exacerbates the development of spontaneous intestinal adenoma. Int J Mol Sci. (2021) 22:2602. doi: 10.3390/ijms22052602
70. Shin N-R, Whon TW, Bae J-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. (2015) 33:496–503. doi: 10.1016/j.tibtech.2015.06.011
71. Laster J, Bonnes SL, Rocha J. Increased use of emulsifiers in processed foods and the links to obesity. Curr Gastroenterol Rep. (2019) 21:1–6. doi: 10.1007/s11894-019-0723-4
72. Bouvard V, Loomis D, Guyton KZ, Grosse Y, El Ghissassi F, Benbrahim-Tallaa L, et al. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. (2015) 16:1599–600. doi: 10.1016/S1470-2045(15)00444-1
73. Rodríguez-García C, Sánchez-Quesada C, Algarra I, Gaforio JJ. The high-fat diet based on extra-virgin olive oil causes dysbiosis linked to colorectal cancer prevention. Nutrients. (2020) 12:1705. doi: 10.3390/nu12061705
74. Van Hecke T, Vossen E, Hemeryck LY, Bussche JV, Vanhaecke L, De Smet S. Increased oxidative and nitrosative reactions during digestion could contribute to the association between well-done red meat consumption and colorectal cancer. Food Chem. (2015) 187:29–36. doi: 10.1016/j.foodchem.2015.04.029
75. Constante M, Fragoso G, Calvé A, Samba-Mondonga M, Santos MM. Dietary heme induces gut dysbiosis, aggravates colitis, and potentiates the development of adenomas in mice. Front Microbiol. (2017) 8:1809. doi: 10.3389/fmicb.2017.01809
76. Kabat G, Miller A, Jain M, Rohan T. A cohort study of dietary iron and heme iron intake and risk of colorectal cancer in women. Br J Cancer. (2007) 97:118–22. doi: 10.1038/sj.bjc.6603837
77. Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, Varki A, et al. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc Nat Acad Sci USA. (2003) 100:12045–50. doi: 10.1073/pnas.2131556100
78. Dhar C, Sasmal A, Varki A. From “serum sickness” to “xenosialitis”: Past, present, and future significance of the non-human sialic acid Neu5Gc. Front Immunol. (2019) 10:807. doi: 10.3389/fimmu.2019.00807
79. Bashir S, Fezeu LK, Ben-Arye SL, Yehuda S, Reuven EM, De Edelenyi FS, et al. Association between Neu5Gc carbohydrate and serum antibodies against it provides the molecular link to cancer: French NutriNet-Santé study. BMC Med. (2020) 18:1–19. doi: 10.1186/s12916-020-01721-8
80. Samraj AN, Pearce OM, Läubli H, Crittenden AN, Bergfeld AK, Banda K, et al. A red meat-derived glycan promotes inflammation and cancer progression. Proc Nat Acad Sci USA. (2015) 112:542–7. doi: 10.1073/pnas.1417508112
81. Alexander DD, Cushing CA. Red meat and colorectal cancer: a critical summary of prospective epidemiologic studies. Obesity Rev. (2011) 12:e472–93. doi: 10.1111/j.1467-789X.2010.00785.x
82. Sofi F, Dinu M, Pagliai G, Pierre F, Gueraud F, Bowman J, et al. Fecal microbiome as determinant of the effect of diet on colorectal cancer risk: comparison of meat-based versus pesco-vegetarian diets (the MeaTIc study). Trials. (2019) 20:1–9. doi: 10.1186/s13063-019-3801-x
83. Orlich MJ, Singh PN, Sabaté J, Fan J, Sveen L, Bennett H, et al. Vegetarian dietary patterns and the risk of colorectal cancers. JAMA Intern Med. (2015) 175:767–76. doi: 10.1001/jamainternmed.2015.59
84. Tabung FK, Smith-Warner SA, Chavarro JE, Wu K, Fuchs CS, Hu FB, et al. Development and validation of an empirical dietary inflammatory index. J Nutr. (2016) 146:1560–70. doi: 10.3945/jn.115.228718
85. Michels KB, Willett WC, Vaidya R, Zhang X, Giovannucci E. Yogurt consumption and colorectal cancer incidence and mortality in the Nurses' Health Study and the Health Professionals Follow-Up Study. Am J Clin Nutr. (2020) 112:1566–75. doi: 10.1093/ajcn/nqaa244
86. Fidelis M, Santos JS, Escher GB, Rocha RS, Cruz AG, Cruz TM, et al. Polyphenols of jabuticaba [Myrciaria jaboticaba (Vell.) O. Berg] seeds incorporated in a yogurt model exert antioxidant activity and modulate gut microbiota of 1, 2-dimethylhydrazine-induced colon cancer in rats. Food Chem. (2021) 334:127565. doi: 10.1016/j.foodchem.2020.127565
87. Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. (2002) 132:1012–7. doi: 10.1093/jn/132.5.1012
88. Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, et al. Defining operational taxonomic units using DNA barcode data. Philos Trans R Soc B Biol Sci. (2005) 360:1935–43. doi: 10.1098/rstb.2005.1725
89. Balendiran GK, Dabur R, Fraser D. The role of glutathione in cancer. Cell Biochem Function. (2004) 22:343–52. doi: 10.1002/cbf.1149
90. Hatem E, El Banna N, Huang M-E. Multifaceted roles of glutathione and glutathione-based systems in carcinogenesis and anticancer drug resistance. Antioxid Redox Signal. (2017) 27:1217–34. doi: 10.1089/ars.2017.7134
91. Chu Y, Lan RS, Huang R, Feng H, Kumar R, Dayal S, et al. Glutathione peroxidase-1 overexpression reduces oxidative stress, and improves pathology and proteome remodeling in the kidneys of old mice. Aging Cell. (2020) 19:e13154. doi: 10.1111/acel.13154
92. Zhang X, Zhao S, Song X, Jia J, Zhang Z, Zhou H, et al. Inhibition effect of glycyrrhiza polysaccharide (GCP) on tumor growth through regulation of the gut microbiota composition. J Pharmacol Sci. (2018) 137:324–32. doi: 10.1016/j.jphs.2018.03.006
93. Ji X, Hou C, Gao Y, Xue Y, Yan Y, Guo X. Metagenomic analysis of gut microbiota modulatory effects of jujube (Ziziphus jujuba Mill.) polysaccharides in a colorectal cancer mouse model. Food Funct. (2020) 11:163–73. doi: 10.1039/C9FO02171J
94. Khan I, Huang G, Li X-A, Liao W, Leong WK, Xia W, et al. Mushroom polysaccharides and jiaogulan saponins exert cancer preventive effects by shaping the gut microbiota and microenvironment in ApcMin/+ mice. Pharmacol Res. (2019) 148:104448. doi: 10.1016/j.phrs.2019.104448
95. Rossi M, Jahanzaib Anwar M, Usman A, Keshavarzian A, Bishehsari F. Colorectal cancer and alcohol consumption—populations to molecules. Cancers. (2018) 10:38. doi: 10.3390/cancers10020038
96. Tsuruya A, Kuwahara A, Saito Y, Yamaguchi H, Tsubo T, Suga S, et al. Ecophysiological consequences of alcoholism on human gut microbiota: implications for ethanol-related pathogenesis of colon cancer. Sci Rep. (2016) 6:1–12. doi: 10.1038/srep27923
97. Kim M, Lee S-T, Choi S, Lee H, Kwon SS, Byun JH, et al. Fusobacterium nucleatum in biopsied tissues from colorectal cancer patients and alcohol consumption in Korea. Sci Rep. (2020) 10:1–10. doi: 10.1038/s41598-020-76467-7
98. Bultman SJ. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol Nutr Food Res. (2017) 61:1500902. doi: 10.1002/mnfr.201500902
99. Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. (2018) 23:705–15. doi: 10.1016/j.chom.2018.05.012
100. Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. (2003) 133:2485S−93S. doi: 10.1093/jn/133.7.2485S
101. Wang G, Yu Y, Wang YZ, Wang JJ, Guan R, Sun Y, et al. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J Cell Physiol. (2019) 234:17023–49. doi: 10.1002/jcp.28436
102. Xia W, Khan I, Li X-A, Huang G, Yu Z, Leong WK, et al. Adaptogenic flower buds exert cancer preventive effects by enhancing the SCFA-producers, strengthening the epithelial tight junction complex and immune responses. Pharmacol Res. (2020) 159:104809. doi: 10.1016/j.phrs.2020.104809
103. Cueva C, Silva M, Pinillos I, Bartolomé B, Moreno-Arribas M. Interplay between dietary polyphenols and oral and gut microbiota in the development of colorectal cancer. Nutrients. (2020) 12:625. doi: 10.3390/nu12030625
104. Ganesan K, Jayachandran M, Xu B. Diet-derived phytochemicals targeting colon cancer stem cells and microbiota in colorectal cancer. Int J Mol Sci. (2020) 21:3976. doi: 10.3390/ijms21113976
105. Zhao Y, Jiang Q. Roles of the polyphenol–gut microbiota interaction in alleviating colitis and preventing colitis-associated colorectal cancer. Adv Nutr. (2020) 12:546–65. doi: 10.1093/advances/nmaa104
106. Lavefve L, Howard LR, Carbonero F. Berry polyphenols metabolism and impact on human gut microbiota and health. Food Funct. (2020) 11:45–65. doi: 10.1039/C9FO01634A
107. Eid N, Osmanova H, Natchez C, Walton G, Costabile A, Gibson G, et al. Impact of palm date consumption on microbiota growth and large intestinal health: a randomised, controlled, cross-over, human intervention study. Br J Nutr. (2015) 114:1226–36. doi: 10.1017/S0007114515002780
108. Yuan X, Long Y, Ji Z, Gao J, Fu T, Yan M, et al. Green tea liquid consumption alters the human intestinal and oral microbiome. Mol Nutr Food Res. (2018) 62:1800178. doi: 10.1002/mnfr.201800178
109. Pluta R, Januszewski S, Ułamek-Kozioł M. Mutual two-way interactions of curcumin and gut microbiota. Int J Mol Sci. (2020) 21:1055. doi: 10.3390/ijms21031055
110. Scazzocchio B, Minghetti L, D'Archivio M. Interaction between gut microbiota and curcumin: a new key of understanding for the health effects of curcumin. Nutrients. (2020) 12:2499. doi: 10.3390/nu12092499
111. Li Y, Zhang T, Chen GY. Flavonoids and colorectal cancer prevention. Antioxidants. (2018) 7:187. doi: 10.3390/antiox7120187
112. Gong Y, Dong R, Gao X, Li J, Jiang L, Zheng J, et al. Neohesperidin prevents colorectal tumorigenesis by altering the gut microbiota. Pharmacol Res. (2019) 148:104460. doi: 10.1016/j.phrs.2019.104460
113. Borzì AM, Biondi A, Basile F, Luca S, Vicari ESD, Vacante M. Olive oil effects on colorectal cancer. Nutrients. (2019) 11:32. doi: 10.3390/nu11010032
114. Chapkin RS, Navarro SL, Hullar MA, Lampe JW. Diet and gut microbes act coordinately to enhance programmed cell death and reduce colorectal cancer risk. Dig Dis Sci. (2020) 65:840–51. doi: 10.1007/s10620-020-06106-8
115. Watson H, Mitra S, Croden FC, Taylor M, Wood HM, Perry SL, et al. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut. (2018) 67:1974–83. doi: 10.1136/gutjnl-2017-314968
116. Kim M, Vogtmann E, Ahlquist DA, Devens ME, Kisiel JB, Taylor WR, et al. Fecal metabolomic signatures in colorectal adenoma patients are associated with gut microbiota and early events of colorectal cancer pathogenesis. MBio. (2020) 11. doi: 10.1128/mBio.03186-19
117. Kraja B, Muka T, Ruiter R, de Keyser CE, Hofman A, Franco OH, et al. Dietary fiber intake modifies the positive association between n−3 PUFA intake and colorectal cancer risk in a caucasian population. J Nutr. (2015) 145:1709–16. doi: 10.3945/jn.114.208462
118. Cruz BC, Sarandy MM, Messias AC, Gonçalves RV, Ferreira CL, Peluzio MC. Preclinical and clinical relevance of probiotics and synbiotics in colorectal carcinogenesis: a systematic review. Nutr Rev. (2020) 78:667–87. doi: 10.1093/nutrit/nuz087
119. Greenhalgh K, Ramiro-Garcia J, Heinken A, Ullmann P, Bintener T, Pacheco MP, et al. Integrated in vitro and in silico modeling delineates the molecular effects of a synbiotic regimen on colorectal-cancer-derived cells. Cell Rep. (2019) 27:1621–32. e1629. doi: 10.1016/j.celrep.2019.04.001
120. Oh NS, Lee JY, Kim YT, Kim SH, Lee JH. Cancer-protective effect of a synbiotic combination between Lactobacillus gasseri 505 and a Cudrania tricuspidata leaf extract on colitis-associated colorectal cancer. Gut Microbes. (2020) 12:1785803. doi: 10.1080/19490976.2020.1785803
121. Praveen MA, Parvathy KK, Patra S, Khan I, Natarajan P, Balasubramanian P. Cytotoxic and pharmacokinetic studies of Indian seaweed polysaccharides for formulating raindrop synbiotic candy. Int J Biol Macromol. (2020) 154:557–66. doi: 10.1016/j.ijbiomac.2020.03.086
122. Boyle RJ, Robins-Browne RM, Tang ML. Probiotic use in clinical practice: what are the risks? Am. J Clin Nutr. (2006) 83:1256–64. doi: 10.1093/ajcn/83.6.1256
123. Torres-Maravilla E, Boucard A-S, Mohseni AH, Taghinezhad-S S, Cortes-Perez NG, Bermúdez-Humarán LG. Role of gut microbiota and probiotics in colorectal cancer: onset and progression. Microorganisms. (2021) 9:1021. doi: 10.3390/microorganisms9051021
124. Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EM, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. (2021) 18:1–19. doi: 10.1038/s41575-021-00440-6
125. Rad AH, Maleki LA, Kafil HS, Zavoshti HF, Abbasi A. Postbiotics as promising tools for cancer adjuvant therapy. Adv Pharm Bull. (2021) 11:1. doi: 10.34172/apb.2021.007
126. Rad AH, Aghebati-Maleki L, Kafil HS, Abbasi A. Molecular mechanisms of postbiotics in colorectal cancer prevention and treatment. Crit Rev Food Sci Nutr. (2021) 61:1787–803. doi: 10.1080/10408398.2020.1765310
127. Chuah L-O, Foo HL, Loh TC, Alitheen NBM, Yeap SK, Mutalib NEA, et al. Postbiotic metabolites produced by Lactobacillus plantarum strains exert selective cytotoxicity effects on cancer cells. BMC Complement Altern Med. (2019) 19:1–12. doi: 10.1186/s12906-019-2528-2
128. Zakuan NM, Ling FH, Yazan LS. Anti-microbial, anti-cancer and immunomodulatory properties of proteinaceous postbiotic metabolite produced by Lactobacillus plantarum I-UL4. Mal J Med Health Sci. (2019) 15:81–4.
129. Karaçam S, Tunçer S. Exploiting the acidic extracellular pH: evaluation of Streptococcus salivarius M18 postbiotics to target cancer cells. Probiotics Antimicrob Proteins. (2021) 1–17. doi: 10.1007/s12602-021-09806-3
130. Russo E, Giudici F, Fiorindi C, Ficari F, Scaringi S, Amedei A. Immunomodulating activity and therapeutic effects of short chain fatty acids and tryptophan post-biotics in inflammatory bowel disease. Front Immunol. (2019) 10:2754. doi: 10.3389/fimmu.2019.02754
131. Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Nat Acad Sci USA. (2019) 116:24285–95. doi: 10.1073/pnas.1912129116
132. Alrafas HR, Busbee PB, Chitrala KN, Nagarkatti M, Nagarkatti P. Alterations in the gut microbiome and suppression of histone deacetylases by resveratrol are associated with attenuation of colonic inflammation and protection against colorectal cancer. J Clin Med. (2020) 9:1796. doi: 10.3390/jcm9061796
133. Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. (2020) 17:111–30. doi: 10.1038/s41575-019-0230-y
134. Bishop KS, Xu H, Marlow G. Epigenetic regulation of gene expression induced by butyrate in colorectal cancer: involvement of MicroRNA. Genet Epigenet. (2017) 9:1179237X17729900. doi: 10.1177/1179237X17729900
135. Chen J, Vitetta L. Inflammation-modulating effect of butyrate in the prevention of colon cancer by dietary fiber. Clin Colorectal Cancer. (2018) 17:e541–4. doi: 10.1016/j.clcc.2018.05.001
136. Pan P, Oshima K, Huang YW, Agle KA, Drobyski WR, Chen X, et al. L oss of FFAR 2 promotes colon cancer by epigenetic dysregulation of inflammation suppressors. Int J Cancer. (2018) 143:886–96. doi: 10.1002/ijc.31366
137. Hanley MP, Rosenberg DW. One-carbon metabolism and colorectal cancer: potential mechanisms of chemoprevention. Curr Pharmacol Rep. (2015) 1:197–205. doi: 10.1007/s40495-015-0028-8
138. Gurwara S, Ajami NJ, Jang A, Hessel FC, Chen L, Plew S, et al. Dietary nutrients involved in one-carbon metabolism and colonic mucosa-associated gut microbiome in individuals with an endoscopically normal colon. Nutrients. (2019) 11:613. doi: 10.3390/nu11030613
139. Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, et al. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. (2018) 19:1–14. doi: 10.1186/s13059-018-1389-1
140. Yu J, Xie Y, Li M, Zhou F, Zhong Z, Liu Y, et al. Association between SFRP promoter hypermethylation and different types of cancer: a systematic review and meta-analysis. Oncol Lett. (2019) 18:3481–92. doi: 10.3892/ol.2019.10709
141. Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. (2017) 18:517. doi: 10.1038/nrg.2017.33
142. Zouggar A, Haebe JR, Benoit YD. Intestinal microbiota influences DNA methylome and susceptibility to colorectal cancer. Genes. (2020) 11:808. doi: 10.3390/genes11070808
143. Röth D, Chiang AJ, Hu W, Gugiu GB, Morra CN, Versalovic J, et al. Two-carbon folate cycle of commensal Lactobacillus reuteri 6475 gives rise to immunomodulatory ethionine, a source for histone ethylation. FASEB J. (2019) 33:3536–48. doi: 10.1096/fj.201801848R
144. Radix PM, Walters CS, Adkins JA. The influence of ethionine-supplemented soy protein diet on cell-mediated and humoral immunity. J Nutr. (1983) 113:159–64. doi: 10.1093/jn/113.1.159
145. Alix J. Molecular aspects of the in vivo and in vitro effects of ethionine, an analog of methionine. Microbiol Rev. (1982) 46:281. doi: 10.1128/mr.46.3.281-295.1982
146. Yuan C, Burns MB, Subramanian S, Blekhman R. Interaction between host MicroRNAs and the gut microbiota in colorectal cancer. MSystems. (2018) 3. doi: 10.1128/mSystems.00205-17
147. Nosho K, Igarashi H, Nojima M, Ito M, Maruyama R, Yoshii S, et al. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. (2014) 35:776–83. doi: 10.1093/carcin/bgt374
148. Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. (2015) 137:1258–68. doi: 10.1002/ijc.29488
149. Jiang Y, Ji X, Liu K, Shi Y, Wang C, Li Y, et al. Exosomal miR-200c-3p negatively regulates the migraion and invasion of lipopolysaccharide (LPS)-stimulated colorectal cancer (CRC). BMC Mol Cell Biol. (2020) 21:1–14. doi: 10.1186/s12860-020-00291-0
150. Nitzan M, Rehani R, Margalit H. Integration of bacterial small RNAs in regulatory networks. Annu Rev Biophys. (2017) 46:131–48. doi: 10.1146/annurev-biophys-070816-034058
151. Tarallo S, Ferrero G, Gallo G, Francavilla A, Clerico G, Luc AR, et al. Altered fecal small RNA profiles in colorectal cancer reflect gut microbiome composition in stool samples. Msystems. (2019) 4. doi: 10.1128/mSystems.00289-19
152. Mjelle R, Sjursen W, Thommesen L, Sætrom P, Hofsli E. Small RNA expression from viruses, bacteria and human miRNAs in colon cancer tissue and its association with microsatellite instability and tumor location. BMC Cancer. (2019) 19:1–12. doi: 10.1186/s12885-019-5330-0
153. Milagro F, Mansego M, De Miguel C, Martinez J. Dietary factors, epigenetic modifications and obesity outcomes: progresses and perspectives. Mol Aspects Med. (2013) 34:782–812. doi: 10.1016/j.mam.2012.06.010
154. Ng S-F, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. (2010) 467:963–6. doi: 10.1038/nature09491
155. Bartolomé RA, García-Palmero I, Torres S, López-Lucendo M, Balyasnikova IV, Casal JI. IL13 receptor α2 signaling requires a scaffold protein, FAM120A, to activate the FAK and PI3K pathways in colon cancer metastasis. Cancer Res. (2015) 75:2434–44. doi: 10.1158/0008-5472.CAN-14-3650
156. Klimesova K, Jiraskova Zakostelska Z, Tlaskalova-Hogenova H. Oral bacterial and fungal microbiome impacts colorectal carcinogenesis. Front Microbiol. (2018) 9:774. doi: 10.3389/fmicb.2018.00774
157. Ekine-Afolabi BA, Njan AA, Rotimi SO, RI A, Elbehi AM, Cash E, et al. The impact of diet on the involvement of non-coding RNAs, extracellular vesicles, and gut microbiome-virome in colorectal cancer initiation and progression. Front Oncol. (2020) 10:2545. doi: 10.3389/fonc.2020.583372
158. Song M-S, Rossi JJ. The anti-miR21 antagomir, a therapeutic tool for colorectal cancer, has a potential synergistic effect by perturbing an angiogenesis-associated miR30. Front Genet. (2014) 4:301. doi: 10.3389/fgene.2013.00301
159. Yu Y, Nangia-Makker P, Farhana L, Rajendra SG, Levi E, Majumdar AP. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. (2015) 14:1–11. doi: 10.1186/s12943-015-0372-7
160. Ma P, Pan Y, Li W, Sun C, Liu J, Xu T, et al. Extracellular vesicles-mediated noncoding RNAs transfer in cancer. J Hematol Oncol. (2017) 10:1–11. doi: 10.1186/s13045-017-0426-y
161. Zeng H, Hamlin SK, Safratowich BD, Cheng W-H, Johnson LK. Superior inhibitory efficacy of butyrate over propionate and acetate against human colon cancer cell proliferation via cell cycle arrest and apoptosis: linking dietary fiber to cancer prevention. Nutr Res. (2020) 83:63–72. doi: 10.1016/j.nutres.2020.08.009
162. Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, Medina JA, et al. The role of the gut microbiome in colorectal cancer development and therapy response. Cancers. (2020) 12:1406. doi: 10.3390/cancers12061406
163. Guz M, Jeleniewicz W, Malm A, Korona-Glowniak I. A Crosstalk between Diet, Microbiome and microRNA in Epigenetic Regulation of Colorectal Cancer. Nutrients. (2021) 13:2428. doi: 10.3390/nu13072428
164. Hu S, Liu L, Chang EB, Wang J-Y, Raufman J-P. Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol Cancer. (2015) 14:1–15. doi: 10.1186/s12943-015-0450-x
165. Schlörmann W, Naumann S, Renner C, Glei M. Influence of miRNA-106b and miRNA-135a on butyrate-regulated expression of p21 and Cyclin D2 in human colon adenoma cells. Genes Nutr. (2015) 10:1–11. doi: 10.1007/s12263-015-0500-4
166. Hu S, Dong TS, Dalal SR, Wu F, Bissonnette M, Kwon JH, et al. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS ONE. (2011) 6:e16221. doi: 10.1371/journal.pone.0016221
167. Sarangi AN, Goel A, Aggarwal R. Methods for studying gut microbiota: a primer for physicians. J Clin Exp Hepatol. (2019) 9:62–73. doi: 10.1016/j.jceh.2018.04.016
168. Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Nat Acad Sci USA. (1977) 74:5088–90. doi: 10.1073/pnas.74.11.5088
169. Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE. (2013) 8:e57923. doi: 10.1371/journal.pone.0057923
170. Zhang J, Zhu B, Xu C, Ding X, Li J, Zhang X, et al. Strategy of selecting 16S rRNA hypervariable regions for metagenome-phylogenetic marker genes based analysis. Ying yong sheng tai xue bao. (2015) 26:3545–53.
171. Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. (2005) 71:1501–6. doi: 10.1128/AEM.71.3.1501-1506.2005
172. Stackebrandt E. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. (2006) 33:152–5.
173. Johnson JS, Spakowicz DJ, Hong B-Y, Petersen LM, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. (2019) 10:1–11. doi: 10.1038/s41467-019-13036-1
174. Jo J-H, Kennedy EA, Kong HH. Research techniques made simple: bacterial 16S ribosomal RNA gene sequencing in cutaneous research. J Investigative Dermatol. (2016) 136:e23–7. doi: 10.1016/j.jid.2016.01.005
175. Durazzi F, Sala C, Castellani G, Manfreda G, Remondini D, De Cesare A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep. (2021) 11:1–10. doi: 10.1038/s41598-021-82726-y
176. Moss EL, Maghini DG, Bhatt AS. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat Biotechnol. (2020) 38:701–7. doi: 10.1038/s41587-020-0422-6
177. Maghini DG, Moss EL, Vance SE, Bhatt AS. Improved high-molecular-weight DNA extraction, nanopore sequencing and metagenomic assembly from the human gut microbiome. Nat Protoc. (2021) 16:458–71. doi: 10.1038/s41596-020-00424-x
178. Wang W-L, Xu S-Y, Ren Z-G, Tao L, Jiang J-W, Zheng S-S. Application of metagenomics in the human gut microbiome. World J Gastroenterol. (2015) 21:803. doi: 10.3748/wjg.v21.i3.803
179. Aguiar-Pulido V, Huang W, Suarez-Ulloa V, Cickovski T, Mathee K, Narasimhan G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis: supplementary issue: bioinformatics methods and applications for big metagenomics data. Evolutionary Bioinformatics. (2016) 12:EBO-S36436. doi: 10.4137/EBO.S36436
180. Lavelle A, Sokol H. Beyond metagenomics, metatranscriptomics illuminates microbiome functionality in IBD. Nat Rev Gastroenterol Hepatol. (2018) 15:193–4. doi: 10.1038/nrgastro.2018.15
181. Vernocchi P, Del Chierico F, Putignani L. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front Microbiol. (2016) 7:1144. doi: 10.3389/fmicb.2016.01144
182. Isaac NI, Philippe D, Nicholas A, Raoult D, Eric C. Metaproteomics of the human gut microbiota: challenges and contributions to other OMICS. Clin Mass Spectrometry. (2019) 14:18–30. doi: 10.1016/j.clinms.2019.06.001
183. Long S, Yang Y, Shen C, Wang Y, Deng A, Qin Q, et al. Metaproteomics characterizes human gut microbiome function in colorectal cancer. NPJ Biofilms Microbiomes. (2020) 6:1–10. doi: 10.1038/s41522-020-0123-4
184. Chen MX, Wang S-Y, Kuo C-H, Tsai I-L. Metabolome analysis for investigating host-gut microbiota interactions. J Formosan Med Assoc. (2019) 118:S10–22. doi: 10.1016/j.jfma.2018.09.007
185. Han S, Van Treuren W, Fischer CR, Merrill BD, DeFelice BC, Sanchez JM, et al. A metabolomics pipeline enables mechanistic interrogation of the gut microbiome. bioRxiv. (2021). doi: 10.1101/2021.05.25.445684
186. Zhang W, Qi S, Xue X, Al Naggar Y, Wu L, Wang K. Understanding the gastrointestinal protective effects of polyphenols using foodomics-based approaches. Front Immunol. (2021) 12:671150. doi: 10.3389/fimmu.2021.671150
187. Bayram B, González-Sarrías A, Istas G, Garcia-Aloy M, Morand C, Tuohy K, et al. Breakthroughs in the health effects of plant food bioactives: a perspective on microbiomics, nutri (epi) genomics, and metabolomics. J Agric Food Chem. (2018) 66:10686–92. doi: 10.1021/acs.jafc.8b03385
188. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. (2019) 25:679–89. doi: 10.1038/s41591-019-0406-6
189. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. (2019) 25:667–78. doi: 10.1038/s41591-019-0405-7
Keywords: dietary fibers, short chain fatty acid, gut microbiota, colorectal cancer prevention, epigenetics
Citation: Appunni S, Rubens M, Ramamoorthy V, Tonse R, Saxena A, McGranaghan P, Kaiser A and Kotecha R (2021) Emerging Evidence on the Effects of Dietary Factors on the Gut Microbiome in Colorectal Cancer. Front. Nutr. 8:718389. doi: 10.3389/fnut.2021.718389
Received: 31 May 2021; Accepted: 13 September 2021;
Published: 11 October 2021.
Edited by:
Alfonso Benítez-Páez, Principe Felipe Research Center (CIPF), SpainReviewed by:
Marc Heyndrickx, Institute for Agricultural, Fisheries and Food Research (ILVO), BelgiumCopyright © 2021 Appunni, Rubens, Ramamoorthy, Tonse, Saxena, McGranaghan, Kaiser and Kotecha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rupesh Kotecha, cnVwZXNoa0BiYXB0aXN0aGVhbHRoLm5ldA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.