
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 19 August 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.713256
This article is part of the Research Topic Gut Microbial Response to Host Metabolic Phenotypes: Volume 1 View all 24 articles
Post-weaning diarrhea of piglets is associated with gut microbiota dysbiosis and intestinal pathogen infection. Recent studies have shown that Lactococcus lactis (L.lactis) could help suppress pathogen infection. This study aimed to investigate the effects of L.lactis on various factors related to growth and immunity in weaning piglets. The results showed that L.lactis improved the growth performance, regulated the amino acid profile (for example, increasing serum tryptophan and ileal mucosal cystine) and the intestinal GABAergic system (including inhibiting ileal gene expression of SLC6A13, GABAAρ1, π, θ, and γ1, and promoting ileal GABAAα5 expression). L.lactis also modulated intestinal immunity by promoting jejunal interleukin 17, 18, 22, ileal toll-like receptor 2, 5, 6, and myeloid differentiation primary response protein 88 gene expression while inhibiting jejunal interferon-γ and ileal interleukin 22 expressions. L.lactis highly affected the intestinal microbiota by improving the beta diversity of gut microbiota and the relative abundance of Halomonas and Shewanella. In conclusion, L.lactis improved the growth performance and regulated amino acid profiles, intestinal immunity and microbiota in weaning piglets.
Weaning is the most critical phase in pig production and is generally associated with intestinal infections and diarrhea (1). The biggest challenge for weaning piglets is diarrhea caused by weaning stress and pathogen infection such as enterotoxigenic Escherichia coli (ETEC). As an active player in host physiological activities, the gut microbiota plays a vital role in modulating pathogen infection and diarrhea in piglets (2). Weaning changes the gut microbiota in humans, piglets, and cows (3–5), which can result in immune system less development, insufficiency of physiological function (6), and increased risk of pathogen infection (7). Thus, appropriate strategies in microbiology could be used to relieve the stress of weaning and prevent infections.
In the past, antibiotics were wildly used as feed additives to promote growth and prevent pathogens in animal production and disease treatment (8–10). However, the overuse of antibiotics resulted in serious public health problems, such as antibiotic resistance gene transfer and an increase in antibiotic-resistance bacteria. Thus, animal producers in many countries have reduced or eliminated the use of antibiotics in feed (11).
There is a great opportunity to develop new strategies for preventing intestinal pathogen infection in weaning piglets. Probiotics can prevent infections caused by pathogens such as Clostridium difficile (12, 13). However, the ability of probiotics to prevent infection varies (14). Lactococcus lactis (L.lactis) was recently reported to prevent cholera (15, 16). Our previous research showed that L.lactis regulated the intestinal immune reaction via gamma-aminobutyric acid (GABA) production and prevented pathogen infections in piglets (17, 18). These findings suggest that L.lactis has great potential to prevent intestinal infections in piglets.
The current study aimed to evaluate the modulatory role of L.lactis in growth performance, amino acid profile, intestinal immunity, and gut microbiota in piglets.
Fifteen healthy piglets (Duroc × Landrace × Landrace, aged 21 days) were purchased from Hunan New Wellful Co., Ltd (Changsha, China). After an adaption period of 3 days, piglets were randomly assigned to the control group (n = 7) and the L.lactis group (n = 8). This study shared the data of the control group with our previous research (19). The piglets in the L.lactis group were orally dosed with L.lactis (2.0*109 CFU/ml, 20 ml) on days 1 and 8. All piglets were fed a corn-and soybean meal-based diet (Supplementary Table 1), and other feedings and environmental control conditions were the same as in our previous study (19). Body weight and feed intake were monitored weekly throughout the experiment, and average daily gain (ADG), average daily feed intake (ADFI), and feed conversion ratio (FCR) were calculated. At the end of week 3, piglets were sacrificed after anesthesia.
The blood, jejunum, jejunal mucosa, ileum, ileal mucosa, colon and luminal content were collected immediately, snap-frozen in liquid nitrogen, and stored at −80°C until further processing. All animal experiment procedures were approved by the Animal Welfare Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences (2016-4B).
L.lactis (ATCC 19435) was grown overnight in 5 ml of M17 medium (Thermo Fisher Scientific, Waltham, MA USA) broth at 37°C with gentle agitation (180 rpm/min). The next day, 3 ml of M17 medium was inoculated with 100 μl of the overnight culture for further amplification and culture.
The diarrhea index and diarrhea rate data of piglets were recorded daily according to the criterion of feces score (Supplementary Table 2). The E.coli loads in the jejunal mucosa, ileal mucosa, and colonic content were quantified by Maconkey Agar (Sigma-Aldrich, Burlington, United States) according to the previous work (17).
According to our previous report (19), the ileal mucosa and serum amino acid levels were measured using high-performance liquid chromatography. Authentic standards (Sigma-Aldrich, Burlington, United States) were used to quantify the amino acids in the samples.
Expression of the GABAergic system and immune-associated genes was analyzed by reverse transcriptase-polymerase chain reaction (RT-PCR), and primers (Supplementary Table 3) were selected according to our previous study (19). The samples were individually normalized to the housekeeping genes, β-actin (ACTB) and glyceraldehyde-3 phosphate dehydrogenase (GAPDH). The relative gene expression was calculated by formula 2−(ΔΔCT).
We used 16S rDNA gene sequencing to analyze the V3–V4 region of ileal microbiota according to our previous study (19). The QIAGEN QIAamp DNA Stool Mini Kit (Qiagen, Hilden, NRW, Germany) was used to extract DNA from the ileal contents and Agarose gel electrophoresis was used to quantify the DNA. Sequencing libraries were then generated using the Ion Plus Fragment Library Kit (Thermo Fisher Scientific, Waltham, MA, USA), assessed on the Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), and sequenced on the Illumina MiSeq Sequencer. Under specific filtering conditions, the raw data were filtered to obtain high-quality clean reads according to the Cutadapt quality control process. Uparse software (Uparse v7.0.1001) was used for sequence analysis and operational taxonomic unit (OTU) clustering and the identity threshold was set to 97%. The species annotation was performed with the RDP Classifier (V2.2, Michigan State 14 University Board of Trustees, East Lansing MI) based on the GreenGene database. MUSCLE software (Version 3.8.31) was used for phylogenetic relationship analysis. Subsequently, we used R and QIIME software (V 1.7) on the normalized output data to analyze the alpha diversity, beta diversity, and environmental factor correlation (Spearman analysis). The FAPROTAX database was used for function prediction. Illumina MiSeq sequencing, processing of sequencing data, and bioinformatics analysis were performed by Beijing Novogene Bioinformatics Technology Co., Ltd. (Beijing, China).
According to the relative abundance at the phylum level, the top 28 taxa were used for correlation analysis with growth performance indicators, amino acid profiles, and intestinal immune factors.
The results were expressed as the mean ± standard error of the mean (SEM). All data were pre-processed with Excel 2019 (Microsoft, Redmond, Washington, USA). Word 2019 software (Microsoft, Redmond, Washington, USA) was used to prepare tables, and GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used to analyze statistics and generate figures. If the data followed a normal distribution, an unpaired t-test was used for the statistical analysis between the two groups; otherwise, the Wilcoxon signed-rank test was used for analysis. A P-value < 0.05 was considered statistically significant.
The body weight and average daily feed intake of piglets were similar between the control and L.lactis groups (Figures 1A,B). L.lactis increased average daily weight gain and reduced FCR in the 2nd week (P < 0.05), while did not affect them in the other 2 weeks (Figures 1C,D).
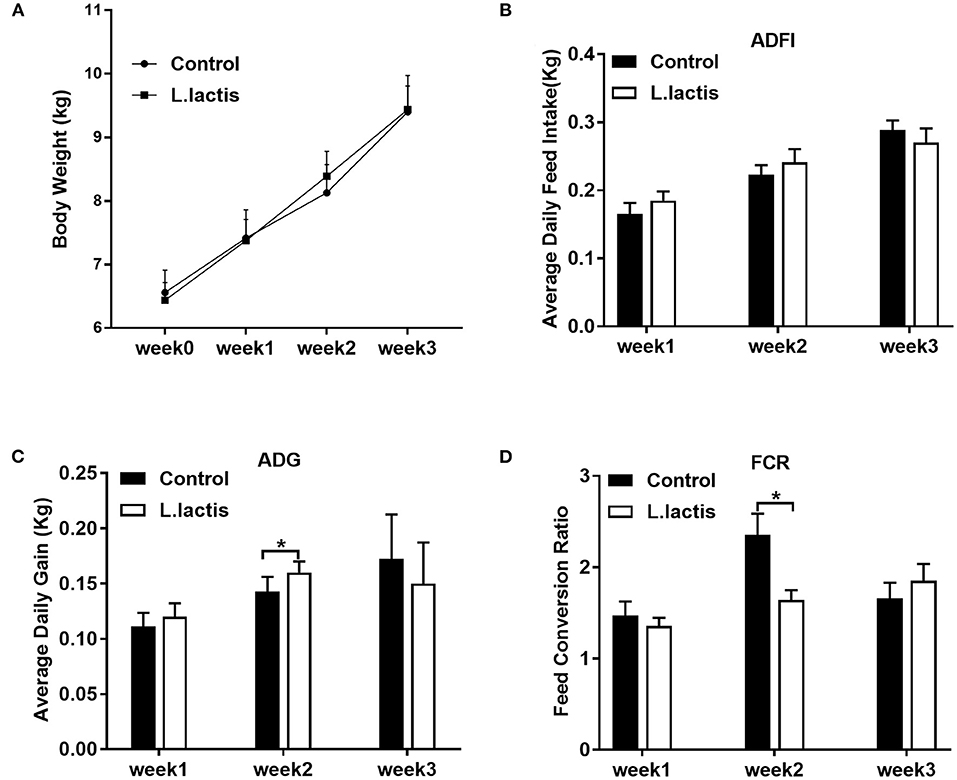
Figure 1. Effects of L.lactis on piglet growth performance. (A) Body weight; (B) average daily feed intake; (C) average daily gain; (D) feed conversion ratio (FCR). An unpaired t-test was used for analyzing the data (mean ± SEM). *P < 0.05.
Results of diarrhea index and diarrhea rate showed that L.lactis did not affect the diarrhea of piglets (Figures 2A,B). And L.lactis reduced E.coli load (P < 0.05) in jejunal mucosa but not ileal mucosa and colonic content (Figure 2C).
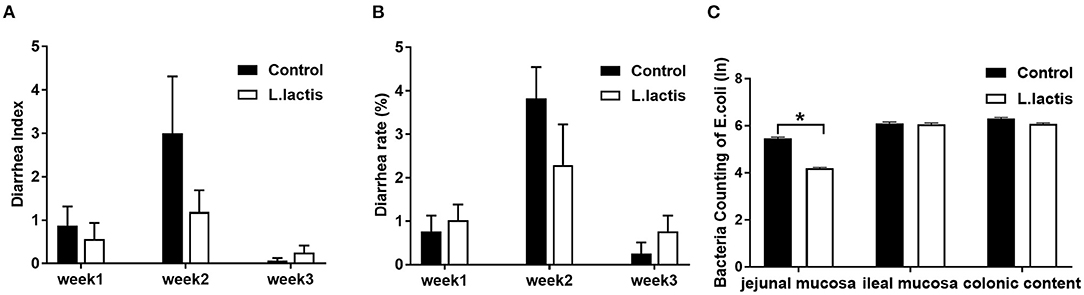
Figure 2. Effects of L.lactis on diarrhea and counting of E.coli. (A) Diarrhea index; (B) diarrhea rate; (C) E.coli count. Wilcoxon rank-sum test was used to analyze the data (mean ± SEM). *P < 0.05.
L.lactis significantly increased (P < 0.05) the concentrations of L-cystine and decreased (P < 0.05) the level of L-glutamic acid in ileal mucosa (Table 1). In peripheral circulation, the serum level of L-tryptophan (Trp) was improved (P < 0.05) due to L.lactis administration (Table 1).
To examine the effect of L.lactis on intestinal immunity, we used RT-PCR to measure the mRNA expression of jejunal and ileal immunity-related factors, including toll-like receptors (TLR)-2, 4, 5, and 6, myeloid differentiation primary response protein-88 (MyD88), tumor necrosis factor-alpha (TNF-α), interferon-gamma (IFN-γ), and interleukin (IL)-1, 2, 4, 6, 10, 17, 18, and 22. In the L.lactis group, jejunal IFN-γ (P < 0.01) and ileal IL-22 (P < 0.05) were reduced, and jejunal IL-17 (P < 0.05), 18 (P < 0.05), and 22 (P < 0.05), ileal TLR-2, 5, 6 (P < 0.01), and MyD88 (P < 0.05) were increased, while other factors was not changed, comparing with the controls (Figure 3).
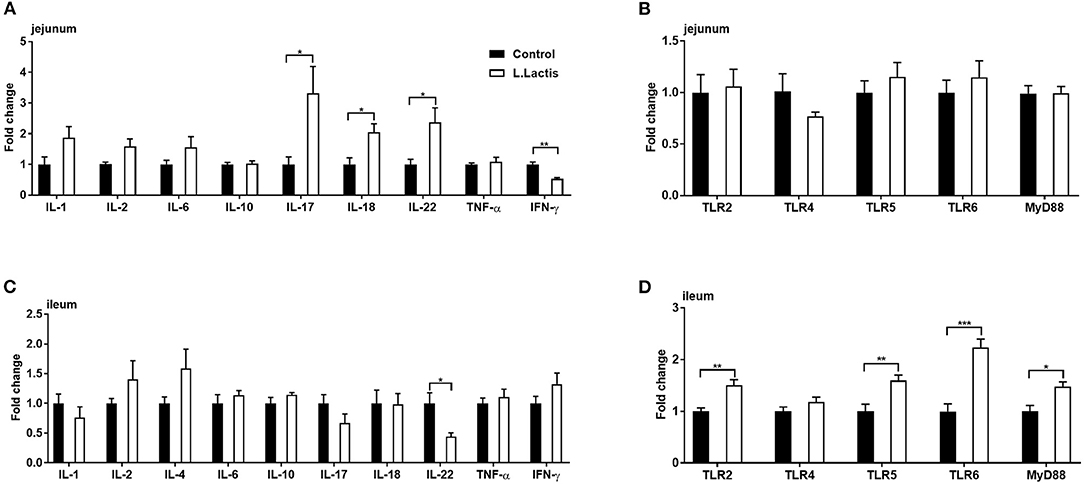
Figure 3. Jejunal and ileal mRNA expression of immune-related factors. Relative gene expression of (A,C) IL-1, 2, 4, 6, 8, 10, 17, 18, 22, TNF-α, and IFN-γ, and (B,D) TLR-2, 4, 5, 6, and MyD88 were analyzed by RT-PCR (jejunal IL-4 was undetected). An unpaired t-test was used for analyzing the data (mean ± SEM). *P < 0.05; **P < 0.01; ***P < 0.001.
The mRNA expression of the gut GABAergic system was analyzed using RT-PCR. The results showed that the expression of SLC6A13 was inhibited (P < 0.05) due to L.lactis treatment (Table 2). Analysis of the gene expressions of GABA receptors (GABAB1-2, GABAAα1-5, β2, γ1-2, δ, ε, π, θ, and ρ1) showed that L.lactis inhibited the expression of GABAAρ1, π, θ, and γ1 (P < 0.05), while it increased GABAAα5 expression (P < 0.05) (Table 2).
The ileal microbiota was analyzed by 16S rDNA sequencing. According to the Venn diagram, 988 OTUs were clustered, in which 199 and 357 OTUs were unique in the control and L.lactis group, separately (Figure 4A). The Beta diversity analysis showed a remarkable difference between control and L.lactis groups (Figure 4B), while the Alpha diversity analysis showed no difference (Supplementary Table 4). At the phylum, family, genus, and species level, Firmicutes, Clostridiaceae_1, Clostridium_sensu_stricto_1, and Veillonella parvula were by far the dominative populations (Figures 4C–F). According to Linear discriminant analysis effect size (LEfSe) results, Oceanospirillales, Halomonas, and Halomonadaceae were enriched in the L.lactis group, while Burkholderiaceae and Clostridiales bacterium_canine_oral_taxon_219 were enriched in the controls (Figures 5A,B). L.lactis increased the relative abundance of Oceanospirillales, Halomonadaceae, Shewanellaceae, Halomonas, Shewanella, and Shewanella_algae (Figure 5C), and reduced the relative abundance of Burkholderiaceae (Figure 5D). Spearman correlation analysis indicated that ADG of the 2nd week was positively correlated with the relative abundance of Fusobacteria (Figure 5E). Ileal TLR-5 mRNA expression and the level of Trp in serum were positively correlated with the relative abundance of Thaumarchaeota. The serum level of Trp also was positively correlated with the relative abundance of Proteobacteria (Figure 5F).

Figure 4. Effects of L.lactis on ileal microbiota of the piglets. (A) The Venn diagram shows the common and unique OTUs of the control and L.lactis groups. (B) Rank abundance curves of beta diversity in the control and L.lactis groups. (C–F) Relative abundance of top 10 phyla (C), families (D), genera (E), and species (F) in the control and L.lactis groups. ***P < 0.001.
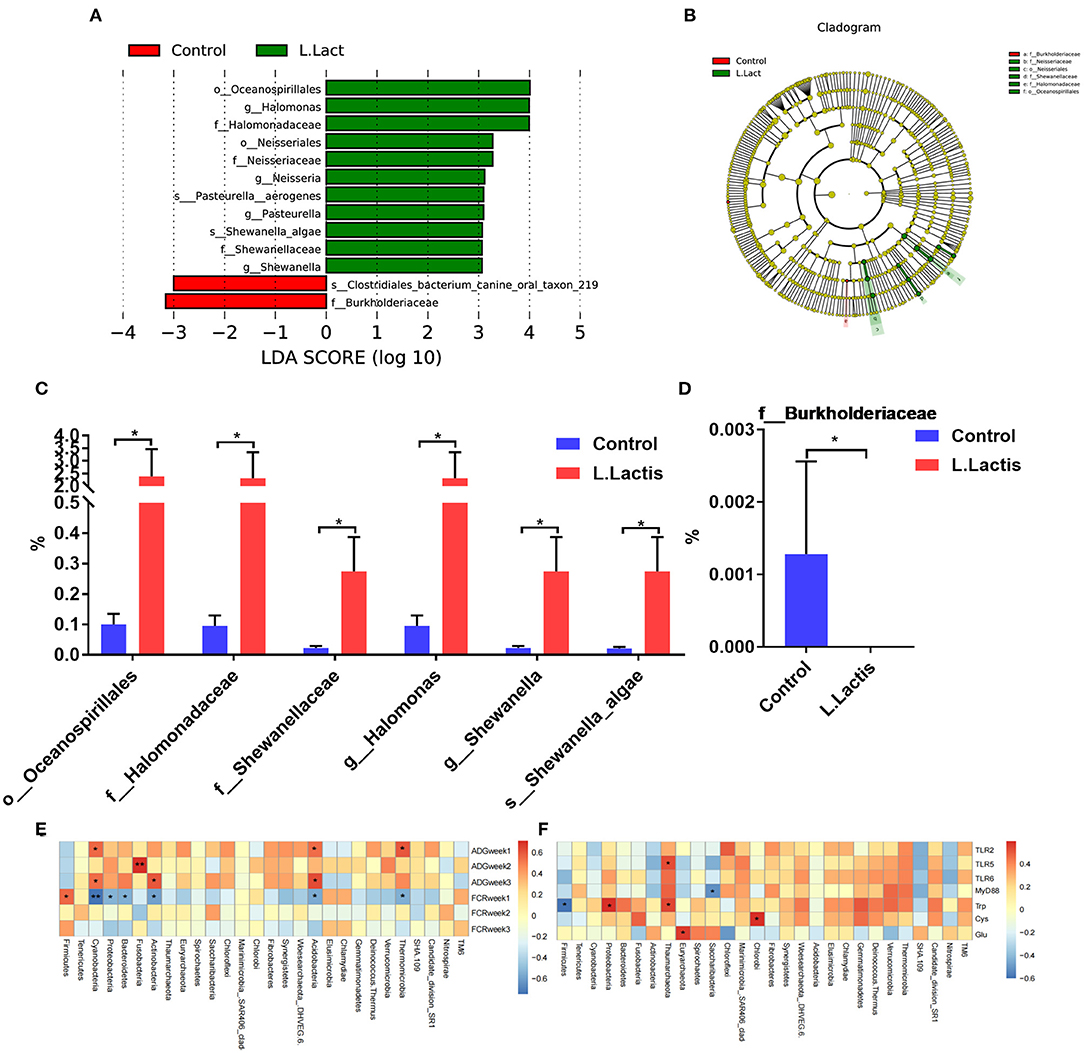
Figure 5. L.lactis regulated the ileal microbiota of the piglets. (A–D) LEfSe analysis demonstrated the significant members in the control and L.lactis groups. (E) Environmental factor correlation analysis indicated that ADG of week 2 was positively correlated with Fusobacteria. (F) Ileal TLR-5 mRNA expression and serum tryptophan (Trp) level were positively correlated with the relative abundance of Thaumarchaeota; serum Trp level was also positively correlated with the relative abundance of Proteobacteria. *P < 0.05; **P < 0.01.
The biggest challenge that weaning piglets faced is diarrhea caused by weaning stress and pathogen infection. During weaning, the gut microbiota of piglets is maladjusted due to diet and environmental changes (1), often leading to infection (2). Previous studies showed that the administration of probiotics could reduce weaning stress and pathogen infection by regulating the gut microbiota (20, 21). Our study showed that L.lactis improved growth performance and modulated intestinal immunity, ileal microbiota, and amino acid profiles of ileal mucosal and serum in weaning piglets.
Weaning stress impairs the feed intake and growth performance of the weaned piglets. Our previous research (18) showed that L.lactis promoted intestinal GABA production, and GABA was reported to enhance the growth performance (19) and inhibit the expression of cholecystokinin-related genes (22). We found that L.lactis partly increased the ADG and reduced the FCR in the 2nd week. The mechanism might be that GABA produced by L.lactis increased the secretion of hormones closely related to growth performance. The ADG and FCR of week 3 were similar between the control and L.lactis groups. The possible reason is that the L.lactis transplantation has a time-limited effect on piglets. Our results contradicted a previous finding that the administration of L.lactis reduced body weight (23). The difference may be explained by different animal models or different dosages of L.lactis. However, the mechanism under the improvement of growth performance driven by L.lactis needs to be further studied.
The weaning stress of piglets usually causes diarrhea, slowing down the growth of piglets. Weaning piglets are susceptible to diarrhea caused by pathogenic E.coli (e.g., ETEC) infection. Growing studies indicate that probiotics prevent pathogenic bacteria colonization and proliferation (12, 13). Manuela et al. (24) showed that probiotics inhibited pathogenic bacteria by producing antimicrobial metabolites and competing for energy substances. Lactic acid bacteria have been found to produce various antimicrobial substances (e.g., lactic acid, bacteriocins, and hydrogen peroxide) to inhibit pathogenic bacteria colonization (25–27). Similarly, we found that L.lactis reduced the jejunal mucosal E.coli load, which helps prevent diarrhea caused by harmful E.coli colonization.
The GABAergic system plays vital role in intestinal health and disease, partly relying on hormone secretion and intestinal immunity (28, 29). Therefore, we analyzed the effect of L.lactis on the intestinal GABAergic system. An increasing number of studies have illustrated the critical roles of GABA transporters (GAT) on health and diseases. Xia et al. (30) showed that GAT2 (SLC6A13) sustained IL-1β production in macrophages, and Ren et al. (18) identified the role of GAT2 in the defense against pathogen infection. This study found that L.lactis reduced GAT2 expression and regulated GABA receptors. Thus, L.lactis transplantation caused significant regulation of the intestinal GABAergic system. L.lactis can regulate the function of immune cells by regulating the intestinal GABAergic system, thus maintaining intestinal homeostasis. However, these results are limited in clarifying the relationship between L.lactis, intestinal GABAergic system and intestinal immune responses.
Amino acids play an essential role as reactive substances in peptide and protein biosynthesis. Moreover, recent studies have shown that amino acids (e.g., tryptophan, cysteine) contribute to the metabolic reprogramming of immune cells such as T cells and macrophages (31). For example, tryptophan is required for T cell proliferation and activation, and tryptophan metabolism is enhanced in activated immune cells. Our study showed that L.lactis transplantation increased the tryptophan level of serum, which might subsequently activate immune cells to resist pathogenic infection. Cysteine, by facilitating glutathione synthesis, plays a vital role in maintaining redox balance to support the function of immune cells (32). In this study, the improved level of ileal mucosal cysteine facilitated by L.lactis might further generate glutathione to counter the production of reactive oxygen species that can cause cell death at high concentrations. Many amino acids such as tryptophan, cysteine and glutamic acid are regulators of growth performance, intestinal immunity, and gut microbiota, indicating that L.lactis promotes growth performance and regulates intestinal immunity might partly by affecting amino acids.
The intestinal tract is the primary organ for food digestion and nutrient absorption and is also the largest immune organ. The intestinal immune system is essential to resisting pathogen infection (33). Our study showed that L.lactis promoted the ileal expression of TLR-2, 5, and 6, as well as MyD88, in weaning piglets. These TLRs recognize different pathogenic components and activate immune cells to kill pathogens (34). Therefore, L.lactis could activate immune cells by activating TLRs signaling pathways, thus resisting intestinal infection. According to our previous research, ETEC-infection increased the abundance of L.lactis, promoting the T helper cell 17 (Th17) immune response via GABA production (18). Indeed, L.lactis-promoted jejunal IL-17 gene expression was also observed in this study. Our previous study showed that GABA supplementation could increase the expression of intestinal SLC6A13 during ETEC infection (35). Thus, the glutamate in the intestine might be used for GABA production in this study. It was reported that the glutamine-glutamate-GABA metabolic pathway supports the Th17 immune reaction to IL-17 production (36). According to these results, the intestinal GABA derived from host glutamate metabolism and L.lactis might support the Th17 immune reaction. Our study also found that L.lactis increased the jejunal gene expression of IL-18 and IL-22 and reduced the ileal gene expression of IL-22. IL-18 can induce the intestinal epithelium to produce antimicrobial proteins (37), while, intestinal IL-22 signaling was positively correlated with the differentiation and antimicrobial effect of Paneth cells (38), and IL-22 has been reported to promote intestinal stem cell-mediated epithelial regeneration (39). Thus, L.lactis transplantation regulates intestinal immune response, which would help maintain intestinal immune homeostasis. For example, Lactobacillus could mitigate colitis by producing aryl hydrocarbon receptor agonists (AHR) (40).
The gut microbiota affects many physiological functions of the host and is linked to the pathogenesis of various diseases such as inflammatory bowel disease (41), cancer (42), and obesity (43). Numerous studies have reported that health and disease markers highly correlate with the gut microbiome (44), and the occurrence of various diseases is associated with the decrease of intestinal microbial diversity (45). Recent studies demonstrated that many probiotics regulated gut microbiota and inhibited intestinal diseases (46). As a promising non-colonizing probiotic, it is reasonable that L.lacatis was undetectable after short-term and low dosage administration. Although it did not change the relative abundance of intestinal L.lactis after treatment for 2 weeks, L.lactis was found sifted and regulated the gut microbiota, such as enriching some beneficial bacteria and suppressing potential pathogenic bacteria. L.lactis treatment reduced the relative abundance of Burkholderia, which is highly related to inflammatory bowel disease and intestinal infection (47, 48). And L.lactis transplantation increased the relative abundance of Shewanella which benefits pancreatic beta cell expansion and insulin production (49). Li et al. (50) showed that transplantation of fecal bacteria from healthy pigs improved the growth status of the recipient pigs, although the overall composition of intestinal bacteria could not be changed, some potential probiotics were significantly enriched. Derrien et al. (51) showed that probiotics do not significantly alter the composition of fecal microbiota in healthy adults but can help maintain the dynamic balance of gut microbiota and reduce the adverse effects of intestinal microbial disorders. Therefore, the function of L.lactis may be more dependent on maintaining the dynamic balance of gut microbiota and microbial metabolic activities. For example, tryptophan metabolites of gut microbiota can improve intestinal barrier function and alleviate dextran sulfate sodium (DSS)-induced colitis in mice (52). Spearman correlation analysis of our data indicated that the effect of L.lactis on the gut microbiota was closely related to amino acid profiles, growth performance, and intestinal immunity. Thus, L.lactis may influence the intestinal microbiota to help regulate these factors in weaning piglets.
L.lactis improved the growth performance, regulated amino acid profiles and intestinal immunity in weaning piglets, which might be associated with changing the intestinal microbiota. These results would help evaluate the feasibility of L.lactis in pig production to reduce the negative health effects of weaning.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: 16S rDNA gene profiling data were available in the NCBI database under BioProject PRJNA745933.
The animal study was reviewed and approved by the Animal Welfare Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences.
SC, BT, and LG designed the experiment and reviewed and revised the manuscript. DY, SC, and YX conducted the experiment. DY and SC analyzed the data. DY and LG prepared tables and figures. DY prepared the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Key Research and Development Program of China (2017YFD0500503) and the Innovation Province Project (Grant: 2019RS3021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.713256/full#supplementary-material
L.lactis, Lactococcus lactis; MyD88, myeloid differentiation primary response protein 88; ADFI, average daily feed intake; ADG, average daily weight gain; E.coli, Escherichia coli; ETEC, Enterotoxigenic E.coli; GABA, gamma-aminobutyric acid; GAD, glutamic acid decarboxylase; GAT, GABA transporter; SLC, solute carriers; IFN-γ, interferences-gamma; IL, interleukin; AHR, aryl hydrocarbon receptor; OTU, operational taxonomic unit; RT-PCR, reverse transcription-polymerase chain reaction; SEM, standard error of the mean; TNF-α, tumor necrosis factor-alpha; DSS, dextran sulfate sodium.
1. Gresse R, Chaucheyras-Durand F, Fleury MA, Van de Wiele T, Forano E, Blanquet-Diot S. Gut microbiota dysbiosis in postweaning piglets: understanding the Keys to Health. Trends Microbiol. (2017) 25:851–73. doi: 10.1016/j.tim.2017.05.004
2. Bin P, Tang Z, Liu S, Chen S, Xia Y, Liu J, et al. Intestinal microbiota mediates enterotoxigenic Escherichia coli-induced diarrhea in piglets. BMC Vet Res. (2018) 14:385. doi: 10.1186/s12917-018-1704-9
3. Khine WWT, Rahayu ES, See TY, Kuah S, Salminen S, Nakayama J, et al. Indonesian children fecal microbiome from birth until weaning was different from microbiomes of their mothers. Gut Microbes. (2020) 12:e1761240. doi: 10.1080/19490976.2020.1761240
4. Hu J, Nie Y, Chen J, Zhang Y, Wang Z, Fan Q, et al. Gradual changes of gut microbiota in weaned miniature piglets. Front Microbiol. (2016) 7:1727. doi: 10.3389/fmicb.2016.01727
5. Meale SJ, Li SC, Azevedo P, Derakhshani H, DeVries TJ, Plaizier JC, et al. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves. Sci Rep. (2017) 7:198. doi: 10.1038/s41598-017-00223-7
6. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, et al. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity. (2019) 50:1276–88. doi: 10.1016/j.immuni.2019.02.014
7. Jayaraman B, Nyachoti CM. Husbandry practices and gut health outcomes in weaned piglets: a review. Anim Nutr. (2017) 3:205–11. doi: 10.1016/j.aninu.2017.06.002
8. Chatterjee A, Modarai M, Naylor NR, Boyd SE, Atun R, Barlow J, et al. Quantifying drivers of antibiotic resistance in humans: a systematic review. Lancet Infect Dis. (2018) 18:e368–78. doi: 10.1016/S1473-3099(18)30296-2
9. Van den Bergh B, Michiels JE, Wenseleers T, Windels EM, Boer PV, Kestemont D, et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol. (2016) 1:16020. doi: 10.1038/nmicrobiol.2016.20
10. Van Boeckel TP, Pires J, Silvester R, Zhao C, Song J, Criscuolo NG, et al. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science. (2019) 365:eaaw1944. doi: 10.1126/science.aaw1944
11. Laxminarayan R, Sridhar D, Blaser M, Wang M, Woolhouse M. Achieving global targets for antimicrobial resistance. Science. (2016) 353:874–5. doi: 10.1126/science.aaf9286
12. Goldenberg JZ, Mertz D, Johnston BC. Probiotics to prevent clostridium difficile infection in patients receiving antibiotics. Jama. (2018) 320:499–500. doi: 10.1001/jama.2018.9064
13. Mullish BH, Marchesi JR, McDonald JAK, Pass DA, Masetti G, Michael DR, et al. Probiotics reduce self-reported symptoms of upper respiratory tract infection in overweight and obese adults: should we be considering probiotics during viral pandemics? Gut Microbes. (2021) 13:1–9. doi: 10.1080/19490976.2021.1900997
14. Kandasamy S, Vlasova AN, Fischer DD, Chattha KS, Shao L, Kumar A, et al. Unraveling the differences between gram-positive and gram-negative probiotics in modulating protective immunity to enteric infections. Front Immunol. (2017) 8:334. doi: 10.3389/fimmu.2017.00334
15. Satchell KJF. Engineered bacteria for cholera prophylaxis. Cell Host Microbe. (2018) 24:192–4. doi: 10.1016/j.chom.2018.07.013
16. Mao N, Cubillos-Ruiz A, Cameron DE, Collins JJ. Probiotic strains detect and suppress cholera in mice. Sci Transl Med. (2018) 10:eaao2586. doi: 10.1126/scitranslmed.aao2586
17. Liu G, Ren W, Fang J, Hu CA, Guan G, Al-Dhabi NA, et al. L-Glutamine and L-arginine protect against enterotoxigenic Escherichia coli infection via intestinal innate immunity in mice. Amino Acids. (2017) 49:1945–54. doi: 10.1007/s00726-017-2410-9
18. Ren W, Yin J, Xiao H, Chen S, Liu G, Tan B, et al. Intestinal microbiota-derived GABA mediates interleukin-17 expression during enterotoxigenic Escherichia coli infection. Front Immunol. (2016) 7:685. doi: 10.3389/fimmu.2016.00685
19. Chen S, Tan B, Xia Y, Liao S, Wang M, Yin J, et al. Effects of dietary gamma-aminobutyric acid supplementation on the intestinal functions in weaning piglets. Food Funct. (2019) 10:366–78. doi: 10.1039/C8FO02161A
20. Zhang W, Zhu YH, Yang GY, Liu X, Xia B, Hu X, et al. Lactobacillus rhamnosus GG affects microbiota and suppresses autophagy in the intestines of pigs challenged with Salmonella infantis. Front Microbiol. (2017) 8:2705. doi: 10.3389/fmicb.2017.02705
21. Barba-Vidal E, Castillejos L, Roll VFB, Cifuentes-Orjuela G, Moreno Muñoz JA, Martín-Orúe SM. The probiotic combination of Bifidobacterium longum subsp. infantis CECT 7210 and Bifidobacterium animalis subsp. lactis BPL6 reduces pathogen loads and improves gut health of weaned piglets orally challenged with Salmonella typhimurium. Front Microbiol. (2017) 8:1570. doi: 10.3389/fmicb.2017.01570
22. Wang DM, Chacher B, Liu HY, Wang JK, Lin J, Liu JX. Effects of γ-aminobutyric acid on feed intake, growth performance and expression of related genes in growing lambs. Animal. (2015) 9:445–8. doi: 10.1017/S1751731114002651
23. Naudin CR, Maner-Smith K, Owens JA, Wynn GM, Robinson BS, Matthews JD, et al. Lactococcus lactis subspecies cremoris elicits protection against metabolic changes induced by a western-style diet. Gastroenterology. (2020) 159:639–51. doi: 10.1053/j.gastro.2020.03.010
24. Raffatellu M. Learning from bacterial competition in the host to develop antimicrobials. Nat Med. (2018) 24:1097–103. doi: 10.1038/s41591-018-0145-0
25. Zamfir M, Callewaert R, Cornea PC, De Vuyst L. Production kinetics of acidophilin 801, a bacteriocin produced by Lactobacillus acidophilus IBB 801. FEMS Microbiol Lett. (2000) 190:305–8. doi: 10.1111/j.1574-6968.2000.tb09303.x
26. Mahrous H, Mohamed A, El-Mongy MA, El-Batal AI, Hamza HAJF. Study bacteriocin production and optimization using new isolates of Lactobacillus spp. isolated from some dairy products under different culture conditions. Food Nutr Sci. (2013) 4:342–56. doi: 10.4236/fns.2013.43045
27. Abo-Amer AE. Optimization of bacteriocin production by Lactobacillus acidophilus AA11, a strain isolated from Egyptian cheese. Ann Microbiol. (2011) 61:445–52. doi: 10.1007/s13213-010-0157-6
28. Auteri M, Zizzo MG, Serio R. GABA and GABA receptors in the gastrointestinal tract: from motility to inflammation. Pharmacol Res. (2015) 93:11–21. doi: 10.1016/j.phrs.2014.12.001
29. Vlainić JV, Šuran J, Vlainić T, Vukorep AL. Probiotics as an adjuvant therapy in major depressive disorder. Curr Neuropharmacol. (2016) 14:952–8. doi: 10.2174/1570159X14666160526120928
30. Xia Y, He F, Wu X, Tan B, Chen S, Liao Y, et al. GABA transporter sustains IL-1β production in macrophages. Sci Adv. (2021) 7:eabe9274. doi: 10.1126/sciadv.abe9274
31. Kelly B, Pearce EL. Amino Assets: how amino acids support immunity. Cell Metab. (2020) 32:154–75. doi: 10.1016/j.cmet.2020.06.010
32. Vene R, Delfino L, Castellani P, Balza E, Bertolotti M, Sitia R, et al. Redox remodeling allows and controls B-cell activation and differentiation. Antioxid Redox Sign. (2010) 13:1145–55. doi: 10.1089/ars.2009.3078
33. Ren W, Chen S, Zhang L, Liu G, Hussain T, Hao X, et al. Interferon tau affects mouse intestinal microbiota and expression of IL-17. Mediat Inflamm. (2016) 2016:2839232. doi: 10.1155/2016/2839232
34. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. doi: 10.1016/j.cell.2006.02.015
35. Chen S, Wu X, Xia Y, Wang M, Liao S, Li F, et al. Effects of dietary gamma-aminobutyric acid supplementation on amino acid profile, intestinal immunity, and microbiota in ETEC-challenged piglets. Food Funct. (2020) 11:9067–74. doi: 10.1039/D0FO01729A
36. Yang G, Xia Y, Ren W. Glutamine metabolism in Th17/Treg cell fate: applications in Th17 cell-associated diseases. Sci China Life Sci. (2021) 64:221–33. doi: 10.1007/s11427-020-1703-2
37. Jarret A, Jackson R, Duizer C, Healy ME, Zhao J, Rone JM, et al. Enteric nervous system-derived IL-18 orchestrates mucosal barrier immunity. Cell. (2020) 180:50–63. doi: 10.1016/j.cell.2019.12.016
38. Gaudino SJ, Beaupre M, Lin X, Joshi P, Rathi S, McLaughlin PA, et al. IL-22 receptor signaling in Paneth cells is critical for their maturation, microbiota colonization, Th17-related immune responses, and anti-Salmonella immunity. Mucosal Immunol. (2021) 14:389–401. doi: 10.1038/s41385-020-00348-5
39. Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. (2015) 528:560–4. doi: 10.1038/nature16460
40. Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. (2016) 22:598–605. doi: 10.1038/nm.4102
41. Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. (2018) 9:3294. doi: 10.1038/s41467-018-05470-4
42. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. (2014) 12:661–72. doi: 10.1038/nrmicro3344
43. Anhe FF, Roy D, Pilon G, Dudonne S, Matamoros S, Varin TV, et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. (2015) 64:872–83. doi: 10.1136/gutjnl-2014-307142
44. Manor O, Dai CL, Kornilov SA, Smith B, Price ND, Lovejoy JC, et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun. (2020) 11:5206. doi: 10.1038/s41467-020-18871-1
45. Kriss M, Hazleton KZ, Nusbacher NM, Martin CG, Lozupone CA. Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery. Curr Opin Microbiol. (2018) 44:34–40. doi: 10.1016/j.mib.2018.07.003
46. Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastro Hepat. (2019) 16:605–16. doi: 10.1038/s41575-019-0173-3
47. Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe. (2017) 21:603–10. doi: 10.1016/j.chom.2017.04.010
48. Schieber AM, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, et al. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science. (2015) 350:558–63. doi: 10.1126/science.aac6468
49. Hill JH, Franzosa EA, Huttenhower C, Guillemin K. A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. eLife. (2016) 5:e20145. doi: 10.7554/eLife.20145
50. Li Y, Wang X, Wang XQ, Wang J, Zhao J. Life-long dynamics of the swine gut microbiome and their implications in probiotics development and food safety. Gut microbes. (2020) 11:1824–32. doi: 10.1080/19490976.2020.1773748
51. Derrien M, van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. (2015) 23:354–66. doi: 10.1016/j.tim.2015.03.002
Keywords: Lactococcus lactis, amino acid, weaning piglet, intestinal immunity, gut microbiota
Citation: Yu D, Xia Y, Ge L, Tan B and Chen S (2021) Effects of Lactococcus lactis on the Intestinal Functions in Weaning Piglets. Front. Nutr. 8:713256. doi: 10.3389/fnut.2021.713256
Received: 22 May 2021; Accepted: 19 July 2021;
Published: 19 August 2021.
Edited by:
Yong Su, Nanjing Agricultural University, ChinaReviewed by:
Mingliang Jin, Northwestern Polytechnical University, ChinaCopyright © 2021 Yu, Xia, Ge, Tan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangpeng Ge, Z2VsaWFuZ3BlbmcxOTgyQDE2My5jb20=; Bie Tan, YmlldGFuQGlzYS5hYy5jbg==; Shuai Chen, Y2hlbnNodWFpQG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.