
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 11 August 2021
Sec. Food Chemistry
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.705822
This article is part of the Research Topic Recent Breakthrough in Gluten Contamination View all 9 articles
Gluten related disorders, such as coeliac disease, wheat allergy and baker's asthma are triggered by proteins present in food products made from wheat and related cereal species. The only treatment of these medical illnesses is a strict gluten-free diet; however, gluten-free products that are currently available in the market can have lower nutritional quality and are more expensive than traditional gluten containing cereal products. These constraints have led to the development of gluten-free or gluten-reduced ingredients. In this vein, a non-GMO wheat flour that purports to contain “65% less allergenic gluten” was recently brought to market. The present study aims to understand the alteration of the proteome profile of this wheat flour material. Liquid chromatography-mass spectrometry was used to investigate the proteome profile of the novel wheat flour, which was contrasted to a wheat flour control. Using both trypsin and chymotrypsin digests and a combined database search, 564 unique proteins were identified with 99% confidence. These proteins and the specific peptides used to identify them were mapped to the wheat genome to reveal the associated chromosomal regions in the novel wheat flour and the mixed wheat control. Of note, several ω- and γ-gliadins, and low-molecular weight glutenins mapping to the short arm of chromosome 1, as well as α-gliadins from the chromosome 6 short arm were absent or expressed at lower levels in the novel wheat variety. In contrast, the high-molecular weight glutenins and α-amylase/trypsin inhibitors were notably more abundant in this variety. A targeted quantitation experiment was developed using multiple reaction monitoring assays to quantify 359 tryptic and chymotryptic peptides from gluten and related allergenic proteins revealing a 33% decrease of gluten protein content in the novel wheat flour sample in comparison to mixed wheat control. However, additional mapping of known allergenic epitopes showed the presence of 53% higher allergenic peptides. Overall, the current study highlights the importance of proteomic analyses especially when complemented by sequence analysis and epitope mapping for monitoring immunostimulatory proteins.
Wheat products account for some 20% of dietary calories and protein ingested globally (1). However, in susceptible people wheat proteins can elicit a range of health disorders including coeliac disease (CD), wheat allergy (WA), and non-coeliac wheat sensitivity (NCWS). The allergenic wheat proteins that cause these adverse immune reactions have been mapped to specific genes by The International Wheat Genome Sequencing Consortium (IWGSC) and collaborators (2, 3). CD is caused when dietary gluten reaches the small intestine of genetically predisposed individuals and stimulates an autoimmune response leading to localized damage and subsequent symptoms (4). WA, as well as baker's asthma (BA) and wheat-dependent exercise induced anaphylaxis (WDEIA), involve an IgE-mediated immune response to wheat proteins that are either ingested as food or occur via skin contact or inhalation. While these disorders can be triggered by gluten proteins, BA typically has a non-gluten protein trigger (5, 6). NCWS is diagnosed when symptoms develop in response to cereal grain consumption, but serological testing is negative for both an autoimmune response and the IgE-mediated allergic response, which contraindicates CD and WA, respectively (4, 7). While the term “non-celiac gluten sensitivity” has a history of use (8), NCWS better reflects the non-gluten wheat proteins (9) or non-protein wheat components like FODMAPs (10) that elicit similar symptoms (7). CD is estimated to afflict ~0.7–1.4% of the global population (11, 12), WA some 0.33–0.75% in adults (13–15), and NCWS being more variable but with prevalence estimated between 0.5 and 13% (16, 17). While gluten proteins are established antigens to those with CD and also contribute to various allergies, non-gluten wheat proteins are potential allergens and antigens capable of causing WA, BA, NCWS, as well as CD (3, 6, 18).
The only effective treatment for these wheat-related immune disorders is the exclusion of wheat and related crop species from the diet. This adds to demand for “gluten free” foodstuffs that resemble traditional wheat products, however wheat is replaced by substitute ingredients that contribute starch but without gluten or other cereal proteins. The absence of gluten proteins, however, can affect the consistency, texture, or taste of gluten-free products, and the substitute ingredients typically come at a higher cost and require recipe alterations (19).
Several approaches have aimed at reducing the gluten content while retaining the health benefits of whole grains or maintaining the unique functionality of cereal grains. One approach is to use ethyl methanesulfonate (EMS) mutagenesis to produce random mutations in genetic material by nucleotide substitution. EMS is often used as the technology base for “Targeting Induced Local Lesions in Genomes” (TILLING) which has proven effective at targeting key wheat enzymes to improve starch composition (20). It has been applied to wheat gluten genes (21), but is challenged by the sheer number of potential allergens and the fact that even low levels of expressed gliadins can elicit CD (22). In barley, ultra-low gluten levels (<5 mg/kg) were achieved in the variety Kebari® by using traditional breeding techniques to combine mutagenesis-derived barley varieties with decreased hordein content and composition (23, 24). Efforts to develop a low-allergen wheat variety have targeted genes that either exhibit a large immune response directly (25), or that conduct epigenetic regulation of downstream gluten protein genes (26), and have also made use of natural null-allele variants (27), CRISPR-Cas9 (28), and RNAi (29). A common phenotype is that downregulation of one or a subset of gluten protein encoding genes is accompanied by the compensatory upregulation of alternate storage proteins (30–34), with a change in technological properties (35). However, technology to characterize gluten in wheat products is the subject of ongoing research (36), as is the targeted removal of CD reactive epitopes from wheat (37).
Recently, a reduced gluten product was released that claims to be a non-GMO wheat variety which contains “65% less allergenic gluten than traditional flour.” It is clarified that the product is “developed for those with sensitive stomachs who don't have gluten or wheat allergies, but who want to reduce the amount of gluten in their diets” (38) and was developed using wheat prolamin box binding factor (PBF) mutants (21, US patents 9,150,839, 10,412,909, and 10,750,690). There are no reports of the proteome and overall characteristics of proteins present in the reduced gluten flour in comparison to commercial wheat varieties.
Wheat has more than 800 genes with potentially allergenic domains, and some 356 genes encoding reference food allergens are included in the “IWGSC v1.0 reference allergen map” (3). This includes gliadins (including α-, β-, γ-, and ω-subtypes) and glutenins (including low molecular weight (LMW) glutenins and high molecular weight (HMW) glutenin subunits), as well as avenin-like proteins (ALPs), α-amylase/trypsin inhibitors (ATIs), and lipid transfer proteins (LTPs) (3). The gluten proteins contain specific epitopes that are deamidated, recognized, and presented by MHC-II antigen presenting cells in the gastrointestinal tract, in doing so initiating the autoimmune response that characterizes CD (39). The canonical gluten proteins, the gliadins and glutenins, together make up some 80% of the protein content in the wheat endosperm, and the most potent contributors to CD toxicity are the chromosome 6D α-gliadins and chromosome 1D ω-gliadins (ω 1,2 sub-type), followed by the LMW glutenins and γ-gliadin (40). It is therefore important to precisely characterize protein groups and epitopes when quantifying “allergenic gluten” in new products. This present study aimed to understand the alterations to the proteome in this reduced gluten wheat product using LC-MS/MS in comparison to a mixed wheat control.
GoodWheat™ (GW) white bread wheat flour was purchased directly from Arcadia Bioscience (Davis, CA, USA). Replicates of GW and of a mixed-wheat (MW) control flour sample were weighed out in quadruplicate. The MW control consisted of equal parts of flour from wheat cultivars: Alsen, Xiayan, Pastor, Westonia, Baxter, Chara, Yitpi, AC Barrie, and Volcania; selected to represent the diversity of wheat used in commercial production. Gluten proteins were specifically enriched from the wheat using an isopropanol/dithiothreitol (IPA/DTT) solvent as described previously (41). Flour (50mg) was weighed into a 1.5 mL micro-tube and 500 μL (10 μL/mg) of 55% IPA/2% DTT was added with vortex mixing until the flour was thoroughly combined with the solvent. The tubes were then sonicated for 5 min at room temperature and incubated in a thermo-mixer (400 rpm, 30 min, 50°C). The tubes were centrifuged for 15 min at 20,800 ×g. The solutions were centrifuged for 15 min at 20,800 ×g. Protein extracts (100 μL) were added to 10 kDa molecular weight cut off filters (Merck, Bayswater, Australia). The protein on the filter was washed twice with a buffer consisting of 8M urea in 0.1M Tris-HCl (pH 8.5) with centrifugation for 15 min at 20,800 ×g. Iodoacetamide (25mM; 100μL) prepared in 8 M urea and 100 mM Tris-HCl was added to the filters for cysteine alkylation with incubation in the dark for 20 min prior to centrifugation for 10 min at 20,800 ×g. The buffer was exchanged with 100 mM ammonium bicarbonate (pH 8.0) by two consecutive wash/centrifugation steps. The filters were transferred to fresh collection tubes and digestion enzyme, either trypsin or chymotrypsin (Promega, NSW, Australia), was prepared as 10 μg/mL in 100 mM ammonium bicarbonate, 50 mM calcium chloride and 200 μL was added to each filter with incubation for 16 h at 37°C. The filters were centrifuged for 15 min at 20,800 ×g. The filters were washed with 200 μL of 100 mM ammonium bicarbonate, and the combined filtrates were subsequently lyophilized.
The digested samples were reconstituted in 100 μL of 1% formic acid and the peptides (1 μL) were chromatographically separated on an Ekspert nanoLC415 (Eksigent, Dublin, CA, USA) coupled to a TripleTOF 6600MS (SCIEX, Redwood City, CA, USA). The peptides were desalted for 5 min on a ChromXP C18 (3 μm, 120 Å, 10 × 0.3 mm) trap column at a flow rate of 10 μL/min of 0.1% formic acid and separated on a ChromXP C18 (3 μm, 120 Å, 150 × 0.3 mm) column at a flow rate of 5 μL/min. The solvents used were (A) 5% DMSO, 0.1% formic acid, 94.9% water and (B) 5% DMSO, 0.1% formic acid, 90% acetonitrile, 4.9% water. A linear gradient from 3 to 25% solvent B over 68 min was employed followed by 25–35% B over 5 min, an increase to 80% B over 2 min, a 2 min hold at 90% B, return to 3% B over 1 min, and 8 min of re-equilibration. The eluent from the HPLC was directly coupled to the DuoSpray source of the TripleTOF 6600 MS. The ionspray voltage was set to 5,500 V; the curtain gas was set to 138 kPa (20 psi), and the ion source gas 1 and 2 (GS1 and GS2) were set to 103 and 138 kPa (15 and 20 psi). The heated interface was set to 150°C. The discovery data files of individual technical replicates of either trypsin or chymotrypsin digested GW and MW samples were searched using ProteinPilot v5.0.3 with Paragon Algorithm (SCIEX) against a FASTA file consisting of Triticeae tribe proteins from UniProt-KB [accessed 02/2021 supplemented with additional translated gene models from the IWGSC RefSeq v1 Assembly (2), as well as those listed on the common Repository of Adventitious Proteins (thegpm.org/crap)]. The FASTA file contained 817,698 protein sequences.
Reduced and alkylated tryptic and chymotryptic peptides were chromatographically separated on an Exion LC-40AD UHPLC system (SCIEX) and analyzed on a 6,500+ QTRAP mass spectrometer (SCIEX). Data acquisition was achieved using scheduled multiple reaction monitoring (sMRM) scanning experiments using a 60 s detection window for each MRM transition and a 0.3 s cycle time.
To build the MRM method, a FASTA file containing all identified proteins was imported into Skyline (42), all fully tryptic peptides sized between 6 and 30 amino acids were selected, and repeated peptides removed. All fully chymotryptic peptides between 6 and 30 amino acids were selected in independent experiments. Initially, five transitions were selected per peptide in an unscheduled MRM assay and assessed on both GW and MW samples. Those peptides where at least three transitions reproducibly co-eluted at the expected retention time (RT) without interference were then selected for inclusion in scheduled MRM assays. These were divided across several separate transition lists, such that all data was recorded with a 60 s detection window and maximum cycle time of 0.3 s. Precursor ions where three or more transitions had consistent RT, intensity over 1,000 cps, and a signal to noise ratio (S/N) >5 were kept and the three most intense transitions were selected for subsequent quantitative experiments. In this way, a total of 768 tryptic and 175 chymotryptic peptides that were unique to one protein were monitored, as well as 263 tryptic and 109 chymotryptic peptides that were present in more than one protein. Data was collected on four technical replicates of GW and MW. Peptide peak area data was exported from Skyline and analyzed (Graphpad Prism v8).
To quantify the relative abundance of individual gluten protein groups, peak areas of both unique and non-unique peptides were summed. Proteins were mapped to the wheat genome using the tBLASTn function of CLC Main Workbench v20.0.4 (Qiagen, Denmark), and multiple proteins mapping to the same gene were interpreted as different alleles of the same gene. Quantified MRM peptides were then allocated to protein groups according to the proteins in which they were found. Peptides occurring in proteins from multiple groups were labeled Multiple/Mixed. The significance and fold change of these protein groups were graphed using VolcaNoseR software (43).
A high sequence similarity between gluten proteins meant many peptides were observed that were common to multiple gluten proteins, making it impossible quantify all proteins using unique peptides. To overcome this, peptides quantified via MRM were allocated to gluten protein groups that were quantified using unique peptides for GW and MW, revealing the relative abundance of protein groups in these samples. To do this, all peptides quantified were mapped to the wheat genome (2), and using a combination of sequence alignments, the presence of Pfam domains (PF13016, PF03157, PF00234), and manual checking of the matching proteins were allocated to one of the following protein groups: α-gliadins, ALPs, ATIs, γ-gliadin, HMW-GSs, LMW-GSs, and ω-gliadins. Where the proteins had two or more peptides from multiple protein groups they were defined as “mixed.” While the ATIs and ALPs are not canonical gluten proteins, several of the ALPs can function as nutrient reservoir proteins, and the ATIs exhibit some allergenicity making them relevant to this investigation.
To quantify protein groups, the monitored peptides were mapped to the identified gluten protein sequences and non-gluten protein families with immune-reactive properties using 100% sequence matching in the Motif search algorithm in CLC Genomic Workbench v21.0.3 (Qiagen, Denmark), and group specific peptides were identified. Quantitative data on all chymotryptic and tryptic peptides were combined, and the abundance of each peptide in each replicate was normalized to the average seen across all replicates from both GW and MW. Graphs were generated in Graphpad Prism v8.
GO enrichment analysis was performed to test for the downregulation of certain classes of proteins in GW. Those proteins present in both GW and MW were excluded so that only proteins unique to GW or MW were analyzed for enriched GO terms. GW- or MW-specific proteins were then mapped to the wheat genome using CLC Genomic Workbench v21.0.3 (Qiagen, Denmark), and lists of their corresponding wheat gene identifiers were pasted into g:Profiler (biit.cs.ut.ee/gprofiler/gost) for GO overrepresentation analysis.
The peptides identified at 1% FDR in discovery proteomics were searched for known CD related T cell epitopes [Ludvig M (44)], baker's asthma, and wheat allergy related epitopes collected from the Immune Epitope Database and Analysis Resource (www.iedb.org) using the Motif search algorithm in CLC Genomic Workbench v21.0.3 (Qiagen, Aarhus, Denmark). Additionally, peptides recognized by commercial ELISA kits using R5 and G12 monoclonal antibodies were also mapped to the protein and peptide sequences. Hits with 100% sequence identity were kept in the analysis. The peptides monitored in MRM assays were also mapped to the same protein list, and the overlap between CD epitopes and the monitored peptides were determined. Monitored peptides that contained an entire epitope in their sequence were selected and quantified in GW and MW to give a relative measure of potential immune reactivity.
Protein estimations were performed using a Coomassie dye binding protein assay using Bradford reagent (Sigma-Aldrich, St Louis, USA) following the manufacturer's instructions. Measurements were made at 595 nm using a Varioskan LUX microplate reader (Thermo Scientific, Scoresby, Australia). Bovine serum albumin (BSA) standard was used in the linear range 0.125–1.5 mg/mL.
Diluted wheat extracts were analyzed by sandwich ELISA using the Ridascreen Gliadin (R-Biopharm, Darmstadt, Germany). The analytical protocols provided by the kit manufacturer were strictly followed. Each of the samples was extracted using the extraction Cocktail (R7006/R7016, R-Biopharm) recommended by the manufacturer for optimal gluten extraction and measured on the using Varioskan LUX microplate reader (Thermo Scientific) in duplicate on a single ELISA plate alongside the supplied standards (representing a gluten concentration of 5–80 mg/kg). The results of absorbance readings were analyzed according to the kit manufacturer's instructions using cubic polynomial regression for the standard curve. The data analyses were performed using Microsoft Excel.
To identify the proteins in the GW and MW samples, combined database searches were performed on the discovery proteomics datasets. The numbers of distinct proteins identified at 1% global false discovery rate (FDR) excluding identifications against the common contaminants (cRAP database) are summarized in Table 1. This information is generated from the reports available at https://doi.org/10.25919/fr8e-k267, processed with the Protein Alignment Template v3.002 beta (SCIEX) and manual curation.
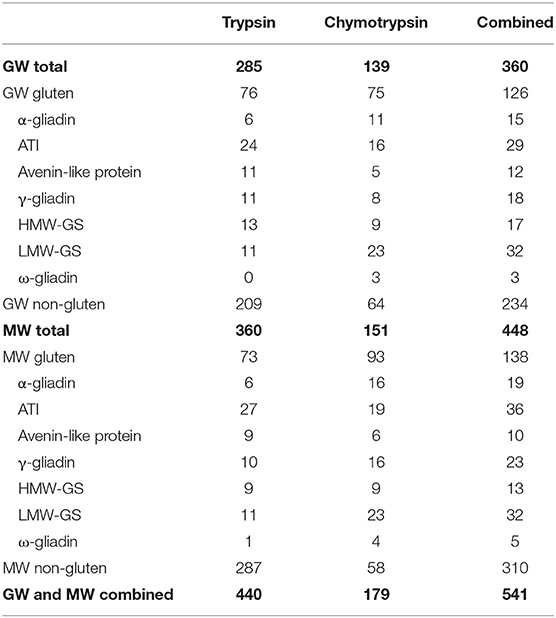
Table 1. Numbers of distinct proteins and of gluten and ATI proteins identified at 99% confidence in trypsin and chymotrypsin data.
Of the 541 proteins identified in both GW and MW (Table 1), more were identified in tryptic digests (440) than chymotryptic digests (179). Greater representation of α- and γ-gliadins and LMW-GS was achieved using chymotryptic digests, while more ATIs, ALP-derived, and non-gluten proteins were identified in tryptic digests (Table 1).
Considering trypsin and chymotrypsin data together enables a more complete comparison of the GW and MW proteomes. Together, there were 360 distinct proteins identified in GW and 448 in MW, with an overlap of 267. The higher number in MW reflects the genetic diversity of the multiple wheat varieties that are present. In GW, 126 of identified proteins were gluten-like proteins (35%) and in MW this number was 138 (31%). Notably, the GW and MW proteomes share 99 (60%) of the 165 detected gluten proteins. The numbers of proteins identified are compared in Figures 1A,B. To identify the chromosomal position of these proteins within the wheat genome protein, sequences were mapped to the IWGSC wheat genome assembly version 1 (2), and the number of peptides observed per 1 million base pairs (Mb) bins was determined. This revealed clusters of detected proteins in all known storage protein gene loci regions of the genome, corresponding to γ- and ω-gliadins, LMW-GS, and HMW-GS (3) on chromosome group 1 and α-gliadins on chromosome group 6. Figure 1C shows the location of the peptides detected superimposed on chromosomes 1A, B, and D, respectively. Though non-gluten proteins were also detected across all wheat chromosomes, there were no large-scale chromosome region changes observed in GW and MW, indicating the potential for gene expression of gluten proteins in GW.
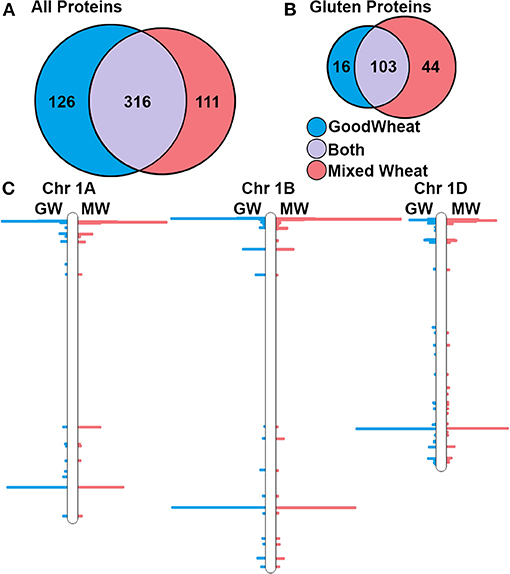
Figure 1. Summary of total and gluten-like proteins identifications at 1% FDR and their corresponding chromosomal locations. (A) Venn diagram showing the total number of proteins detected within the GW and MW datasets. (B) Detected gluten proteins in MW and GW. (C) Locations of genes for detected proteins on wheat chromosomes 1A, 1B, and 1D.
To investigate the quantitative changes across GW and MW wheat samples, LC-MRM-MS-based quantitative assays were developed for all peptides confidently identified in the discovery proteomics experiment (Figure 2). A total of 189 tryptic peptides and 170 chymotryptic were targeted. While 84 tryptic and 55 chymotryptic peptides were uniquely present, i.e., in only one protein isoform, many of the peptides monitored by MRM occur in multiple protein isoforms and therefore reflect the relative abundance of more than one protein. While LC-MRM-MS reveals peptide relative abundance, using this information to quantify proteins by combining the constituent peptides is confounded by both the presence of repeated peptides and differential ionization efficiency of various peptides. We therefore categorized peptides into groups that reflect the abundance of major allergen types and did not quantify specific proteins. This revealed the fold-change and significance of tryptic (Figure 2A) and chymotryptic peptides (Figure 2B) peptides between GW and MW. HMW-GS and ATI peptides tend to be higher in GW than MW, and many tryptic “non-gluten” peptides are higher in GW. Similarly, many LMW-GS, ALP, and α-gliadin peptides are lower in GW than MW.
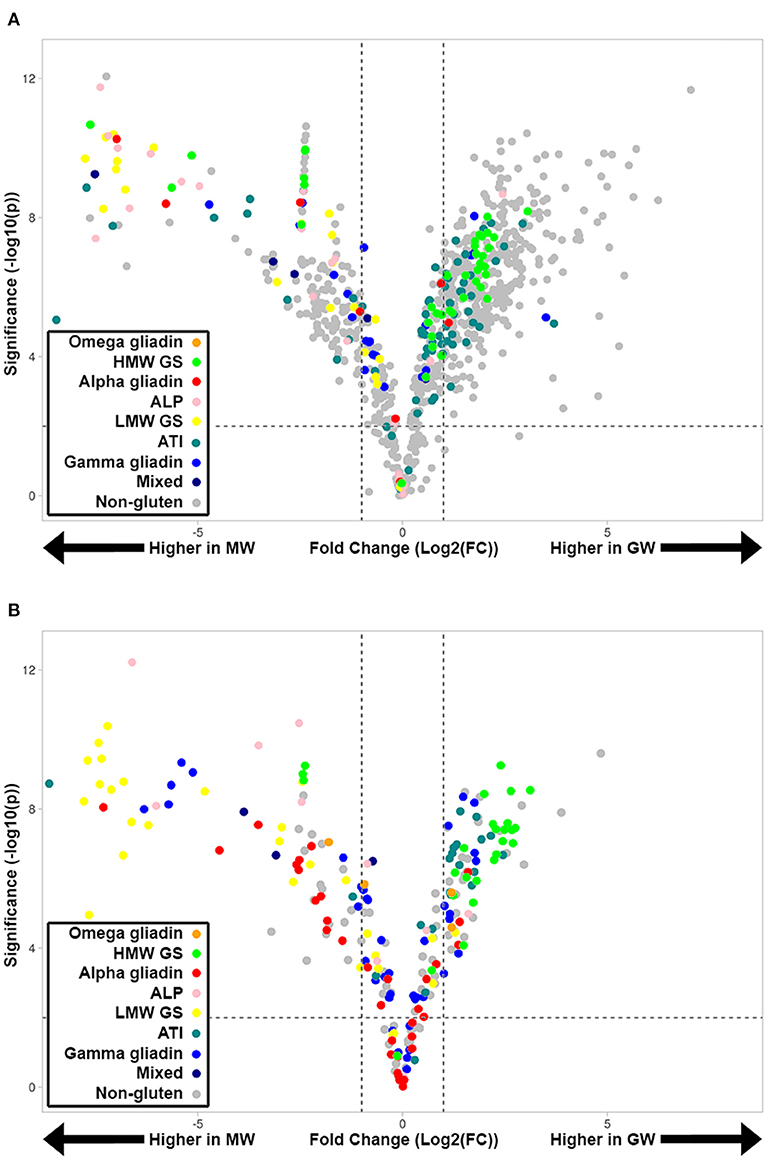
Figure 2. Volcano plots showing quantified tryptic (A) and chymotryptic (B) peptides in GW and MW samples colored according to gluten group. A fold-change of 2 is indicated by the dashed vertical lines [Log2(FC) = ±1]. Peptides above the horizontal dashed line have a significant change in abundance between GW and MW (p-value <0.01 [–log10(p) >2].
The normalized peak area for all peptides belonging to each protein group were then summed to compare the overall abundance of each protein group (Figure 3). Importantly, GW showed significantly lower abundance of LMW-glutenins, α-gliadins, and γ-gliadins, but showed an increase in HMW-glutenins relative to MW. GW also showed significant decreases in ALPs and increases in ATIs. Changes in net ω-gliadin abundance were not significant. The net change in canonical gluten content can be obtained by adding together the gliadins and glutenins (LMW-GS, HMW-GS, α-, γ-, ω-gliadins), showing that GW has 67% the relative gluten protein abundance as MW (Figure 3B).
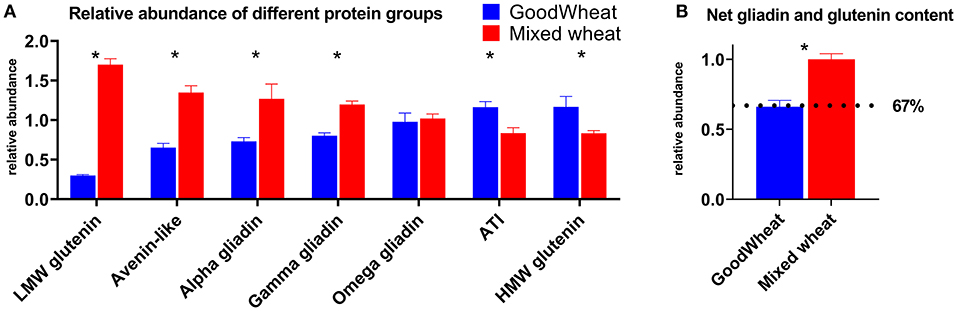
Figure 3. Relative abundance of different gluten or ATI protein groups (A). Quantitation was performed based on all detectable peptides from proteins classified to these groups. Error bars indicate SEM, and significant differences are indicated by asterisk. Adding together the LMW and HMW glutenins, and α-, γ-, ω-gliadins gives the net gliadin and glutenin content (B) which equated to GW having an estimated 67% of the gluten content of MW (dotted line).
To understand the enrichment of protein classes within the individual wheat samples, GO enrichment analysis was performed using g:Profiler on those proteins detected with a fold change ≥2 in MW and GW as shown in Figure 2. Proteins in MW showed predominant enrichment for nutrient reservoir activity (GO:0045735, Figure 4A). GW proteins showed enrichment of several classes of enzyme inhibitor and regulators, as well as enrichment of proteins localizing to the Extracellular Region (GO:0005576) cellular compartment indicating the compensation mechanism for the expression of non-gluten proteins. There was no enrichment of nutrient reservoir activity (Figure 4B).
To explore the potential immune reactive nature of proteins detected in GW compared to those in MW, peptides identified in the discovery data that contained full-length immune reactive epitopes were quantified (Figure 5). Known immunogenic regions within quantitated MRM peptides are quantified, including HLA-DQ T cell epitopes for CD patients (Figure 5A), baker's asthma epitopes (Figure 5B), and wheat allergy-related epitopes (Figure 5C). It should be noted that these represent a small subset of the known immune reactive epitopes. The discovery analysis results (Supplementary Table 1) indicate the presence of additional epitopes that were not quantified with MRM. There were six complete HLA-DQ T cell epitope sequences observed in a total of 25 peptides, nine BA epitopes in 12 peptides, two WA epitopes in 16 peptides, and one WDEIA epitope in one peptide. Overall, HLA-DQ reactive epitopes in GW were present at 67% the relative abundance of MW. BA-reactive epitopes were also more abundant in GW at 180% that of MW.WA-reactive epitopes were also more abundant in GW at 379% the level seen in MW. Only one wheat dependent exercise-induced anaphylaxis epitope was observed, which was notably lower in GW at 17.7% the level seen in MW.
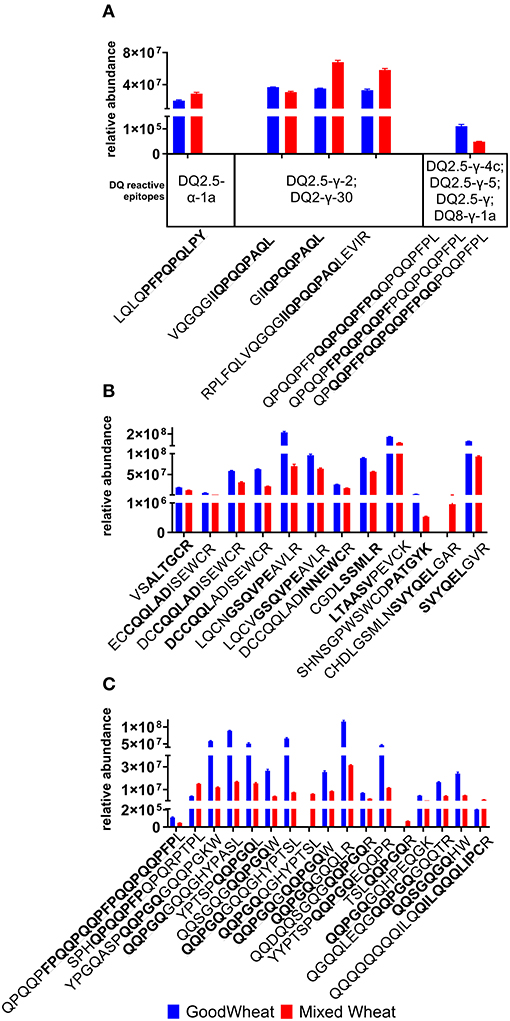
Figure 5. Relative abundance of immune reactive epitopes in peptides quantified in MRM assays, reported as mean and standard error. Epitope sequences are highlighted in bold within the peptide sequence: (A) Coeliac disease HLA-DQ reactive epitopes; (B) Bakers' asthma; (C) Wheat Allergy plus peptide QQQQQQQQILQQILQQQLIPCR which contains the epitope QILQQQLIPC antigenic for wheat-dependent exercise induced anaphylaxis. When calculating the total, duplicate peptides where multiple epitopes were detected were only counted once.
The protein content of MW and GW were evaluated and did not show a significant difference at 0.84 and 0.88 mg/mL, respectively. The gluten content was also evaluated by R5 ELISA and it was interesting to note that GW revealed a 39% higher gluten content than MW, an unexpected result given the overall decrease in gluten peptides detected by LC-MS.
The current study used complementary high sensitivity LC-MS techniques to identify gluten proteins and to monitor the relative abundance of gluten and allergenic wheat proteins in a recently developed wheat product (GoodWheat, GW) in comparison to a wheat sample mixed from equal amounts of nine commercial cultivars (Mixed wheat, MW). Peptides from gliadin and glutenin proteins were present in GW at 67% of the abundance of the MW control, indicating an average decrease of 33% (Figure 3B). This is complemented by our analysis of intact HLA-DQ reactive epitopes in the monitored peptides which were 67.3% as abundant in GW as MW (Figure 5A). While this may reduce but not remove the antigen content of GW, it is accompanied by an increase in peptides known to be related to Baker's asthma and wheat allergy, at 180 and 379%, respectively. The overlap between immunogenic DQ epitopes with peptides detected in discovery data, and quantified in MRM is presented in Supplementary Table 1. While the use of LC-MRM-MS in this work enabled the quantitation of gluten proteins and highlights its utility in grain protein research with specific reference to gluten, future studies should focus on using complementary extraction buffers to understand more about changes in the GW grain proteome.
In contrast to the MRM analysis that revealed an overall lower gluten content in GW than MW (Figure 3), the R5 ELISA estimate of gluten content revealed a gluten content ~39% higher in GW than MW. The slightly elevated protein content (5%) in GW compared to MW would account for a minority of the observed difference. The elevated ELISA measurement likely reflects an overall increase in the ratio of R5 epitope per unit of protein. The choice of reference material, in particular the ratio of gliadin to glutenin, is known to affect measurements of gluten by ELISA even in simple food matrices (45), and kits that use different primary antibodies will yield different measurements of gluten (46) because of the specificities and sensitivities of the primary antibody (47). Future analyses should investigate the gluten content of GW using alternative ELISA kits or gluten protein quantitation employing fractionation (RP-HPLC or size-exclusion chromatography) protocols.
Important trends were seen in specific gluten protein types (Figures 2, 3), as the LMW-GSs, α-, and γ-gliadins are lower in GW, while the HMW-GSs were significantly more abundant. HMW glutenins contribute more to bread's elastic properties than other gluten proteins due to their relative size and ability to form large polymers (48). Their higher relative abundance in GW indicates that they in part compensate for the lower abundance of gliadins and LMW glutenins (49). Additionally, the level of immune response elucidated by HMW glutenins in CD is significantly lower compared to the α-, γ-, and ω-gliadins and LMW glutenins (40), making their increase less relevant to CD, however it has important implications for WA and BA. Along with ATIs, which were also significantly higher in GW, the higher HMW glutenin content in GW brings more allergenic epitopes related to WA and BA. This is reflected in Figure 5 as the allergenic epitopes recognized by different B and T cell types are increased by an overall 53.5%.
Interestingly, the ALPs were also present in significantly lower amounts in GW than MW. While named for their resemblance of oat avenins (50), these seed storage proteins share sequence similarity and secondary function with γ-gliadins and LMW glutenins (51). They contribute both to allergenicity (52) and bread dough quality (53), and contain one or two gliadin (PF13016) domains. ALPs also contain CD-related B cell epitopes (3), and their downregulation is important for CD toxicity.
The symmetry of the volcano plot (Figure 2) indicates the net decrease in gluten protein and ALP content is accompanied by compensatory expression of other proteins within the grain. GO enrichment analysis of the GW proteome revealed enzyme inhibitors and regulators that are enriched in GW which was also confirmed by the gene set enrichment analysis showing cysteine-rich proteins are overrepresented in the upregulated proteins in GW. Most of these proteins have a defense related function and were upregulated in lieu of proteins with a canonical “nutrient reservoir activity” GO MF annotation (Figure 4).
Our proteogenomic analysis indicates that there is no evidence of large-scale chromosome deletions or absence of storage protein gene clusters (Figures 1B,C) on chromosome group 1 and 6 in GW. While antibody-based assays or classical Osborne fractionation were not performed and thus represents a limitation of the present work, gluten proteins were present in both GW and MW and simply expressed at different levels (Figure 3). This would suggest the novel GW variety expresses less gluten proteins due to gene regulation at a transcriptional or post-transcriptional level. There are several known mechanisms implicated in seed development and gluten protein expression that may be at play. One is the LYS3 gene that encodes the transcription factor Prolamin Binding Factor (PBF). PBF is expressed early in seed development, and suppresses seed growth by reducing the expression of developmental and starch metabolism genes (54). Wheat lys3 mutants have been reported to contain lower levels of gliadins and LMW-GSs (21), which matches our results as shown in Figure 4. A barley variety with lys3a mutation causing it to not express C-hordein (a class of barley gluten) was used in a breeding program to derive an “ultra-low gluten” barley variety (30, 55), showing it is compatible with selective breeding. These low-gluten PBF mutant lines exhibit increased expression of lysine-rich genes that are otherwise related to developmental processes during germination (30). While it is possible that GW uses lys3 mechanisms to regulate gluten protein expression, using solely the proteomic information presented in this study we cannot conclusively determine the targeting of lys3 regulation.
In conclusion, the use of discovery and targeted proteomics-based experiments has enabled the detection and quantitation of gluten and additional allergenic proteins present in the GW and MW samples. This study revealed a 33% decrease in gluten-like proteins in GW and the compensatory expression of non-gluten proteins within MW samples that tend to have enzyme inhibitor or regulator activity GO terms. This study affirms that, as stated by the manufacturer, GW is not compatible with a gluten-free diet. Epitope mapping revealed a reduction in gluten protein-specific epitopes; however, there was an increase in epitopes related to baker's asthma and wheat allergy in GW wheat in comparison to MW. Additionally, the chromosomal level analysis of detected proteins showed no significant differences between GW and MW. Future studies focusing on integrating LC-MS/MS results with clinical measurements would be needed to investigate the nutritional benefits of GW. Overall, the current study exemplifies the use of proteogenomic approaches as a tool to explore the safety and/or health benefits of wheat varieties targeted toward consumers with wheat-related disorders.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.25919/fr8e-k267.
MN-W: sample preparation, LC-MS data collection, data analysis, and manuscript preparation. AJ and UB: data analysis and manuscript preparation. MC: project concept and design and manuscript preparation. All authors contributed to the article and approved the submitted version.
This study was partially supported by the ARC Centre of Excellence for Innovations in Peptide and Protein Science (CE200100012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer KS declared a past co-authorship with one of the authors, MC, to the handling editor.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.705822/full#supplementary-material
1. Shiferaw B, Smale M, Braun H-J, Duveiller E, Reynolds M, Muricho G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security. (2013) 5:291–317. doi: 10.1007/s12571-013-0263-y
2. IWGSC, Appels R, Eversole K, Stein N, Feuillet C, Keller B, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. (2018) 361:eaar7191. doi: 10.1126/science.aar7191
3. Juhász A, Belova T, Florides CG, Maulis C, Fischer I, Gell G, et al. Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat. Sci Adv. (2018) 4:eaar8602. doi: 10.1126/sciadv.aar8602
4. Ludvigsson JF, Leffler DA, Bai JC, Biagi F, Fasano A, Green PHR, et al. The Oslo definitions for coeliac disease and related terms. Gut. (2013) 62:43. doi: 10.1136/gutjnl-2011-301346
5. Cabanillas B. Gluten-related disorders: celiac disease, wheat allergy, and nonceliac gluten sensitivity. Crit Rev Food Sci Nutr. (2020) 60:2606–21. doi: 10.1080/10408398.2019.1651689
6. Salcedo G, Quirce S, Diaz-Perales A. Wheat allergens associated with Baker's asthma. J Investig Allergol Clin Immunol. (2011) 21:81–92.
7. Schuppan D, Pickert G, Ashfaq-Khan M, Zevallos V. Non-celiac wheat sensitivity: differential diagnosis, triggers and implications. Best Pract Res Clin Gastroenterol. (2015) 29:469–76. doi: 10.1016/j.bpg.2015.04.002
8. Ellis A, Linaker B. Non-coeliac gluten sensitivity? Lancet. (1978) 311:1358–9. doi: 10.1016/S0140-6736(78)92427-3
9. Gibson PR, Skodje GI, Lundin KE. Non-coeliac gluten sensitivity. J Gastroenterol Hepatol. (2017) 32:86–9. doi: 10.1111/jgh.13705
10. Skodje GI, Sarna VK, Minelle IH, Rolfsen KL, Muir JG, Gibson PR, et al. Fructan, rather than gluten, induces symptoms in patients with self-reported non-celiac gluten sensitivity. Gastroenterology. (2018) 154:529–39. doi: 10.1053/j.gastro.2017.10.040
11. Kang J, Kang A, Green A, Gwee K, Ho K. Systematic review: worldwide variation in the frequency of coeliac disease and changes over time. Aliment Pharmacol Ther. (2013) 38:226–45. doi: 10.1111/apt.12373
12. Singh P, Arora A, Strand TA, Leffler DA, Catassi C, Green PH, et al. Global prevalence of celiac disease: systematic review and meta-analysis. Clin Gastroenterol Hepatol. (2018) 16:823–36. doi: 10.1016/j.cgh.2017.06.037
13. Cabrera-Chávez F, Dezar GV, Islas-Zamorano AP, Espinoza-Alderete JG, Vergara-Jiménez MJ, Magaña-Ordorica D, et al. Prevalence of self-reported gluten sensitivity and adherence to a gluten-free diet in argentinian adult population. Nutrients. (2017) 9:81. doi: 10.3390/nu9010081
14. Ontiveros N, López-Gallardo JA, Vergara-Jiménez MJ, Cabrera-Chávez F. Self-reported prevalence of symptomatic adverse reactions to gluten and adherence to gluten-free diet in an adult Mexican population. Nutrients. (2015) 7:6000–15. doi: 10.3390/nu7075267
15. Ontiveros N, Rodríguez-Bellegarrigue CI, Galicia-Rodríguez G, Vergara-Jiménez MDJ, Zepeda-Gómez EM, Arámburo-Galvez JG, et al. Prevalence of self-reported gluten-related disorders and adherence to a gluten-free diet in Salvadoran adult population. Int J Environ Res Public Health. (2018) 15:786. doi: 10.3390/ijerph15040786
16. Catassi C, Bai JC, Bonaz B, Bouma G, Calabrò A, Carroccio A, et al. Non-celiac gluten sensitivity: the new frontier of gluten related disorders. Nutrients. (2013) 5:3839–53. doi: 10.3390/nu5103839
17. Fasano A, Sapone A, Zevallos V, Schuppan D. Nonceliac gluten sensitivity. Gastroenterology. (2015) 148:1195–204. doi: 10.1053/j.gastro.2014.12.049
18. Larre C, Lupi R, Gombaud G, Brossard C, Branlard G, Moneret-Vautrin D, et al. Assessment of allergenicity of diploid and hexaploid wheat genotypes: identification of allergens in the albumin/globulin fraction. J Proteomics. (2011) 74:1279–89. doi: 10.1016/j.jprot.2011.03.014
19. Fry L, Madden A, Fallaize R. An investigation into the nutritional composition and cost of gluten-free versus regular food products in the UK. J Human Nutr Diet. (2018) 31:108–20. doi: 10.1111/jhn.12502
20. Sestili F, Botticella E, Lafiandra D. TILLING for improved starch composition in wheat. In: Tuberosa R, Graner A, Frison E, editors. Genomics of Plant Genetic Resources: Volume 2. Crop Productivity, Food Security and Nutritional Quality. Dordrecht: Springer (2014). p. 467–87. doi: 10.1007/978-94-007-7575-6_20
21. Moehs CP, Austill WJ, Holm A, Large TA, Loeffler D, Mullenberg J, et al. Development of decreased-gluten wheat enabled by determination of the genetic basis of lys3a barley. Plant Physiol. (2019) 179:1692–703. doi: 10.1104/pp.18.00771
22. Frisoni M, Corazza GR, Lafiandra D, De Ambrogio E, Filipponi C, Bonvicini F, et al. Wheat deficient in gliadins: promising tool for treatment of coeliac disease. Gut. (1995) 36:375. doi: 10.1136/gut.36.3.375
23. Doll H, Køie B, Eggum BO. Induced high lysine mutants in barley. Radiat Bot. (1974) 14:73–80. doi: 10.1016/S0033-7560(74)90179-3
24. Tanner GJ, Blundell MJ, Colgrave ML, Howitt CA. Creation of the first ultra-low gluten barley (Hordeum vulgare L.) for coeliac and gluten-intolerant populations. Plant Biotechnol J. (2016) 14:1139–50. doi: 10.1111/pbi.12482
25. Sánchez-León S, Gil-Humanes J, Ozuna CV, Giménez MJ, Sousa C, Voytas DF, et al. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. (2018) 16:902–10. doi: 10.1111/pbi.12837
26. Wen S, Wen N, Pang J, Langen G, Brew-Appiah RAT, Mejias JH, et al. Structural genes of wheat and barley 5-methylcytosine DNA glycosylases and their potential applications for human health. Proc Natl Acad Sci USA. (2012) 109:20543–8. doi: 10.1073/pnas.1217927109
27. Carroccio A, Di Prima L, Noto D, Fayer F, Ambrosiano G, Villanacci V, et al. Searching for wheat plants with low toxicity in celiac disease: between direct toxicity and immunologic activation. Digest Liver Dis. (2011) 43:34–9. doi: 10.1016/j.dld.2010.05.005
28. Camerlengo F, Frittelli A, Sparks C, Doherty A, Martignago D, Larré C, et al. CRISPR-Cas9 multiplex editing of the α-amylase/trypsin inhibitor genes to reduce allergen proteins in durum wheat. Front Sustain Food Syst. (2020) 4:104. doi: 10.3389/fsufs.2020.00104
29. Kalunke RM, Tundo S, Sestili F, Camerlengo F, Lafiandra D, Lupi R, et al. Reduction of allergenic potential in bread wheat RNAi transgenic lines silenced for CM3, CM16 and 0.28 ATI genes. Int J Mol Sci. (2020) 21:5817. doi: 10.3390/ijms21165817
30. Bose U, Broadbent JA, Byrne K, Blundell MJ, Howitt CA, Colgrave ML. Proteome analysis of hordein-null barley lines reveals storage protein synthesis and compensation mechanisms. J Agric Food Chem. (2020) 68:5763–75. doi: 10.1021/acs.jafc.0c01410
31. García-Molina MD, Muccilli V, Saletti R, Foti S, Masci S, Barro F. Comparative proteomic analysis of two transgenic low-gliadin wheat lines and non-transgenic wheat control. J Proteomics. (2017) 165:102–12. doi: 10.1016/j.jprot.2017.06.010
32. Gil-Humanes J, Pistón F, Rosell CM, Barro F. Significant down-regulation of γ-gliadins has minor effect on gluten and starch properties of bread wheat. J Cereal Sci. (2012) 56:161–70. doi: 10.1016/j.jcs.2012.02.009
33. Gil-Humanes J, Pistón F, Tollefsen S, Sollid LM, Barro F. Effective shutdown in the expression of celiac disease-related wheat gliadin T-cell epitopes by RNA interference. Proc Natl Acad Sci USA. (2010) 107:17023–8. doi: 10.1073/pnas.1007773107
34. Schmidt D, Gaziola SA, Boaretto LF, Azevedo RA. Proteomic analysis of mature barley grains from C-hordein antisense lines. Phytochemistry. (2016) 125:14–26. doi: 10.1016/j.phytochem.2016.03.001
35. van den Broeck HC, van Herpen TW, Schuit C, Salentijn EM, Dekking L, Bosch D, et al. Removing celiac disease-related gluten proteins from bread wheat while retaining technological properties: a study with Chinese Spring deletion lines. BMC Plant Biol. (2009) 9:1–12. doi: 10.1186/1471-2229-9-41
36. Jouanin A, Borm T, Boyd LA, Cockram J, Leigh F, Santos BA, et al. Development of the GlutEnSeq capture system for sequencing gluten gene families in hexaploid bread wheat with deletions or mutations induced by γ-irradiation or CRISPR/Cas9. J Cereal Sci. (2019) 88:157–66. doi: 10.1016/j.jcs.2019.04.008
37. Jouanin A, Schaart JG, Boyd LA, Cockram J, Leigh FJ, Bates R, et al. Outlook for coeliac disease patients: towards bread wheat with hypoimmunogenic gluten by gene editing of α-and γ-gliadin gene families. BMC Plant Biol. (2019) 19:1–16. doi: 10.1186/s12870-019-1889-5
38. Arcadia Biosciencess. GoodWheat™. (2021). Available online at: https://arcadiabio.com/products/goodwheat/ (accessed July 26, 2021).
39. Sollid LM. The roles of MHC class II genes and post-translational modification in celiac disease. Immunogenetics. (2017) 69:605–16. doi: 10.1007/s00251-017-0985-7
40. Tye-Din JA, Stewart JA, Dromey JA, Beissbarth T, van Heel DA, Tatham A, et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci Transl Med. (2010) 2:41ra51. doi: 10.1126/scitranslmed.3001012
41. Bose U, Broadbent JA, Byrne K, Hasan S, Howitt CA, Colgrave ML. Optimisation of protein extraction for in-depth profiling of the cereal grain proteome. J Proteomics. (2019) 197:23–33. doi: 10.1016/j.jprot.2019.02.009
42. MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. (2010) 26:966–8. doi: 10.1093/bioinformatics/btq054
43. Goedhart J, Luijsterburg MS. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci Rep. (2020) 10:20560. doi: 10.1038/s41598-020-76603-3
44. Sollid LM, Tye-Din JA, Qiao S-W, Anderson RP, Gianfrani C, Koning F. Update 2020: nomenclature and listing of celiac disease–relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics. (2020) 72:85–8. doi: 10.1007/s00251-019-01141-w
45. Schalk K, Koehler P, Scherf KA. Targeted liquid chromatography tandem mass spectrometry to quantitate wheat gluten using well-defined reference proteins. PLoS ONE. (2018) 13:e0192804. doi: 10.1371/journal.pone.0192804
46. Thompson T, Méndez E. Commercial assays to assess gluten content of gluten-free foods: why they are not created equal. J Am Diet Assoc. (2008) 108:1682–7. doi: 10.1016/j.jada.2008.07.012
47. Scherf KA. Gluten analysis of wheat starches with seven commercial ELISA test kits—Up to six different values. Food Anal Methods. (2017) 10:234–46. doi: 10.1007/s12161-016-0573-8
48. Delcour JA, Joye IJ, Pareyt B, Wilderjans E, Brijs K, Lagrain B. Wheat gluten functionality as a quality determinant in cereal-based food products. Annu Rev Food Sci Technol. (2012) 3:469–92. doi: 10.1146/annurev-food-022811-101303
49. Gianibelli M, Larroque O, MacRitchie F, Wrigley C. Biochemical, genetic, and molecular characterization of wheat endosperm proteins. Cereal Chem. (2001) 78:635–46. doi: 10.1094/CCHEM.2001.78.6.635
50. Kan Y, Wan Y, Beaudoin F, Leader DJ, Edwards K, Poole R, et al. Transcriptome analysis reveals differentially expressed storage protein transcripts in seeds of Aegilops and wheat. J Cereal Sci. (2006) 44:75–85. doi: 10.1016/j.jcs.2006.04.004
51. Colgrave ML, Goswami H, Byrne K, Blundell M, Howitt CA, Tanner GJ. Proteomic profiling of 16 cereal grains and the application of targeted proteomics to detect wheat contamination. J Proteome Res. (2015) 14:2659–68. doi: 10.1021/acs.jproteome.5b00187
52. Zhang Y, Hu X, Juhasz A, Islam S, Yu Z, Zhao Y, et al. Characterising avenin-like proteins (ALPs) from albumin/globulin fraction of wheat grains by RP-HPLC, SDS-PAGE, and MS/MS peptides sequencing. BMC Plant Biol. (2020) 20:45. doi: 10.1186/s12870-020-2259-z
53. Ma F, Li M, Li T, Liu W, Liu Y, Li Y, et al. Overexpression of avenin-like b proteins in bread wheat (Triticum aestivum L.) improves dough mixing properties by their incorporation into glutenin polymers. PLoS ONE. (2013) 8:e66758. doi: 10.1371/journal.pone.0066758
54. Orman-Ligeza B, Borrill P, Chia T, Chirico M, DoleŽel J, Drea S, et al. LYS3 encodes a prolamin-box-binding transcription factor that controls embryo growth in barley and wheat. J Cereal Sci. (2020) 93:102965. doi: 10.1016/j.jcs.2020.102965
Keywords: gluten, wheat, celiac disease, allergy, food safety, proteomics, mass spectrometry
Citation: Nye-Wood MG, Juhász A, Bose U and Colgrave ML (2021) Proteome Analysis and Epitope Mapping in a Commercial Reduced-Gluten Wheat Product. Front. Nutr. 8:705822. doi: 10.3389/fnut.2021.705822
Received: 06 May 2021; Accepted: 16 July 2021;
Published: 11 August 2021.
Edited by:
Anil K. Verma, Marche Polytechnic University, ItalyReviewed by:
Francisco Barro, Spanish National Research Council, SpainCopyright © 2021 Nye-Wood, Juhász, Bose and Colgrave. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle L. Colgrave, m.colgrave@ecu.edu.au
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.