
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr., 28 June 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.689456
This article is part of the Research TopicRemodeling Composition and Function of Microbiome by Dietary Strategies - Functional Foods PerspectiveView all 8 articles
 Maria Carolina Rodríguez-Daza1,2†
Maria Carolina Rodríguez-Daza1,2† Elena C. Pulido-Mateos1,2†
Elena C. Pulido-Mateos1,2† Joseph Lupien-Meilleur1,2†
Joseph Lupien-Meilleur1,2† Denis Guyonnet3
Denis Guyonnet3 Yves Desjardins1,4
Yves Desjardins1,4 Denis Roy1,2*
Denis Roy1,2*The genome of gut microbes encodes a collection of enzymes whose metabolic functions contribute to the bioavailability and bioactivity of unabsorbed (poly)phenols. Datasets from high throughput sequencing, metabolome measurements, and other omics have expanded the understanding of the different modes of actions by which (poly)phenols modulate the microbiome conferring health benefits to the host. Progress have been made to identify direct prebiotic effects of (poly)phenols; albeit up to date, these compounds are not recognized as prebiotics sensu stricto. Interestingly, certain probiotics strains have an enzymatic repertoire, such as tannase, α-L-rhamnosidase, and phenolic acid reductase, involved in the transformation of different (poly)phenols into bioactive phenolic metabolites. In vivo studies have demonstrated that these (poly)phenol-transforming bacteria thrive when provided with phenolic substrates. However, other taxonomically distinct gut symbionts of which a phenolic-metabolizing activity has not been demonstrated are still significantly promoted by (poly)phenols. This is the case of Akkermansia muciniphila, a so-called antiobesity bacterium, which responds positively to (poly)phenols and may be partially responsible for the health benefits formerly attributed to these molecules. We surmise that (poly)phenols broad antimicrobial action free ecological niches occupied by competing bacteria, thereby allowing the bloom of beneficial gut bacteria. This review explores the capacity of (poly)phenols to promote beneficial gut bacteria through their direct and collaborative bacterial utilization and their inhibitory action on potential pathogenic species. We propose the term duplibiotic, to describe an unabsorbed substrate modulating the gut microbiota by both antimicrobial and prebiotic modes of action. (Poly)phenol duplibiotic effect could participate in blunting metabolic disturbance and gut dysbiosis, positioning these compounds as dietary strategies with therapeutic potential.
The gut microbiota plays a crucial role on host physiology. It is well-known that an intimate symbiotic relationship exists between the gut microbiome and the host, and that this association is complex and multi-dimensional, as it affects the gut-lung, gut-brain, gut-skin, gut-muscle, and gut-adipose tissue axes, among others (1–4). The tight connection between the gut ecosystem and host endogenous metabolic responses is receiving much attention lately, owing to its key involvement in the onset and progress of cardiometabolic diseases (4), intestinal inflammation (5), cancer (6), cognition (7), and neuropsychological disorders (1). The gut microbiota is now considered a relevant therapeutic target for many chronic societal diseases.
Diet plays a dominant role in modulating the intestinal microbiota (8). The fermentation of its components by commensal microbiota generates an array of cross-feeding networks which provide the host with nutrients and chemical signals affecting both immunity and metabolism (9–11). Many of these dietary substrates are prebiotics, which are selectively utilized by host colonic microbes conferring health benefits (12) by promoting the growth and the activity of beneficial bacterial strains. Until recently, the prebiotic concept was confined to selected non-digestible carbohydrates; however, phytochemicals, such as (poly)phenols, exert potentially prebiotic effects by selectively stimulating beneficial bacteria and reducing the incidence of diseases (12). (Poly)phenols are a diverse class of secondary plant metabolites found in most diets. Owing to their chemical structure, they are poorly absorbed and thus reach the colon, where they affect the resident microbiota (13, 14). Specifically, (poly)phenols can stimulate several of keystone bacterial species such as Akkermansia muciniphila, Bacteroides thetaiotaomicrom, Faecalibacterium prausnitzii, Bifidobacteria, and Lactobacilli (15–20).
The beneficial direct impact of (poly)phenols on the gut microbiota relies on two principal modes of action: a direct bacterial stimulatory effect and a direct antimicrobial effect. As a corollary to carbohydrate-associated enzymes (CAZymes) used by prebiotic stimulated bacteria, the genome of several beneficial bacteria encodes an array of (poly)phenol-associated enzymes (PAZymes) involved specifically in (poly)phenol metabolization (21, 22). In the presence of (poly)phenols, PAZymes-producing bacteria can utilize these compounds to improve their fitness and their persistence in intestinal niches. In addition, (poly)phenols can also selectively inhibit the development of potential pathogenic species often associated with metabolic disorders (5, 23–26).
The direct effects of (poly)phenols on the microbiota engender bacterial ecological shifts and favor syntrophic relationships that further modulate microbiota's composition and function. Indeed, collaterally to (poly)phenols antimicrobial effect, (poly)phenol resistant bacteria, such as Akkermansia muciniphila, are boosted after dietary (poly)phenol intake, highlighting their capacity to withstand the (poly)phenol's antimicrobial action and their ability to opportunistically occupy freed ecological niches. Furthermore, through successive catabolic reactions, PAZymes producing bacteria can provide phenolic metabolites used by other beneficial bacterial species in complex trophic cross-feeding chains. In this sense, emerging reports suggest that certain (poly)phenols can be utilized and transformed by beneficial gut microbes, such as Lactiplantibacillus plantarum (10, 27, 28), into bioactive phenolic metabolites that are later freely absorbed and transported to target organs. In this respect, it appears that some (poly)phenols may exert a genuine prebiotic effect by stimulating commensal bacteria and by inducing the production of (poly)phenol beneficial metabolites, further contributing to human health.
This review discusses how (poly)phenols interact directly with the gut microbiota to provide benefits to the host. To account for the two main (poly)phenols modes of action, we suggest the use of the term “duplibiotics” which broadens the scope of prebiotic activity to include and define a substrate able to modulate the gut microbiota's composition through a dual antimicrobial effect and beneficial bacteria stimulatory effect. To explore this concept, we consider the inhibitory activity of (poly)phenols on diet-induced inflammatory bacteria often associated with obesity and discuss their stimulatory action on several commensal bacterial species. We also provide insights on key bacterial enzymes involved in the potential prebiotic effects of (poly)phenols. These enzymes release (poly)phenol metabolites which participate in intricate microbial trophic networks in the intestinal environment. Finally, relying on A. muciniphila as a model organism, we describe how (poly)phenol-induced ecological changes can promote the growth of symbionts that are less metabolically adapted for the transformation of (poly)phenols.
The human gastrointestinal (GI) tract is inhabited by about 1014 metabolically active bacteria (29). The gut microbiota is mainly represented by a limited number of bacterial phyla: Firmicutes, Bacteroidota (30) (accounting for ~90% of the GI microbiota), Actinobacteria, Proteobacteria, and Verrucomicrobia (21, 22). Despite high inter-individual variability, Firmicutes is the largest phylum in humans and rodents, with more than 250 genera (22, 31). Moreover, the phylum Bacteroidota regroups around 20 genera, among which the genus Bacteroides is the most represented (31). The phylum Actinobacteria is less consistently detected as dominant and represents a lower percentage (5%) of the total bacteria; it includes the genus Bifidobacterium from which many probiotics have been identified (32). Finally, the phylum Verrucomicrobia includes the genus Akkermansia spp., which is present in low proportion (3–5%) and is now considered a next-generation probiotic (32, 33). On average, the gut microbiota encodes about 40 times more genes than its human host (34), providing an extended metabolic capacity largely oriented toward the catabolism of complex diet-acquired compounds.
The gut microbiota is particularly specialized in the degradation of plant constituents with thousands of genes dedicated to the digestion of complex carbohydrates (35). Apart from glycans, the gut microbiota also ferments and synthesizes proteins, transforms xenobiotics, as well as host bile acids, and provides the body with essential vitamins (36, 37). This intense activity leads to the production of beneficial bacterial metabolites such as short-chain fatty acids (SCFA, mainly acetate, propionate, and butyrate), indoles, neurotransmitters, and gasotransmitters (ex. H2S, NO) (38). The SCFA are tightly involved in energy homeostasis, insulin response, fat accumulation, and immune signaling, as reviewed elsewhere (39, 40). Among the SCFA, butyrate reinforces the intestinal mucosal barrier (41, 42), suppresses proinflammatory effectors in macrophages, and promotes the differentiation of dendritic cells (9). Moreover, gut microbial activity can also produce harmful metabolites such as several secondary bile acids, p-cresol, p-tyramine, trimethylamine-N-oxide [recently reviewed by McCarville et al. (38), Cortes-Martin et al. (43), and Saito et al. (44)].
The colonic epithelium is associated to an extensive lymphoid tissue representing the majority of the body's immunocytes (45) which constantly senses the commensal gut microbiota. The immune receptors on intestinal epithelial or on dendritic cells survey the microbial structures known as “pathogen recognition patterns,” and can signal the presence of pathogens to initiate an immune response (46). Under normal physiological conditions, the gut microbiota is maintained in a healthy homeostatic state (i.e., eubiosis) and preserves a balanced local gut immunological interaction with the host. An imbalance in the composition, diversity and metabolic capacity of the gut microbiota (dysbiosis) (47) can, however, negatively affect the genes (meta-transcriptomes), proteins (metaproteomes), functions (metagenomes), and metabolites (metabolomes) makeup, thereby influencing the whole intestinal environment (microbiome) and host health. Gnotobiotic murine models, fecal transplant experiments, or human twins' studies, have clearly demonstrated that an altered crosstalk between the gut microbiota and the host mucosal immune system is involved in the etiology of several chronic metabolic diseases (5, 25, 48, 49).
Numerous studies have highlighted the capacity of specific bacterial phylotypes in triggering beneficial immunological responses and reducing the severity of inflammatory diseases. This is the case of Bifidobacteria and Lactobacilli species, which attenuate intestinal inflammation and metabolic disturbances in ulcerative colitis (50, 51) and obesity (52, 53). For instance, certain probiotic strains, such as L. plantarum WCFS1, improve inflammation by lowering the levels of plasma proinflammatory cytokines (51). Other relevant microorganisms considered as next-generation probiotics like A. muciniphila, B. thetaiotaomicron, and F. prausnitzii are also intricately interacting with the host immune system and have been associated with beneficial effects on the host (54–56).
All in all, an increasing number of studies demonstrate the biological involvement of the gut microbiota in the onset, maintenance, or progress of different chronic diseases (6, 57–61). It is therefore of particular interest to identify the composition and the functional contribution of the gut microbiota to assess mechanisms by which they improve the host health. This knowledge is of paramount importance to develop microbiota-based therapies, including non-pharmaceutical interventions, such as dietary functional ingredients (prebiotics), live microbes (probiotics and fermented food), or microbial metabolites. The effect of diet on the gut microbiota and its influences on the host immunological responses and metabolic phenotype are summarized in Figure 1.
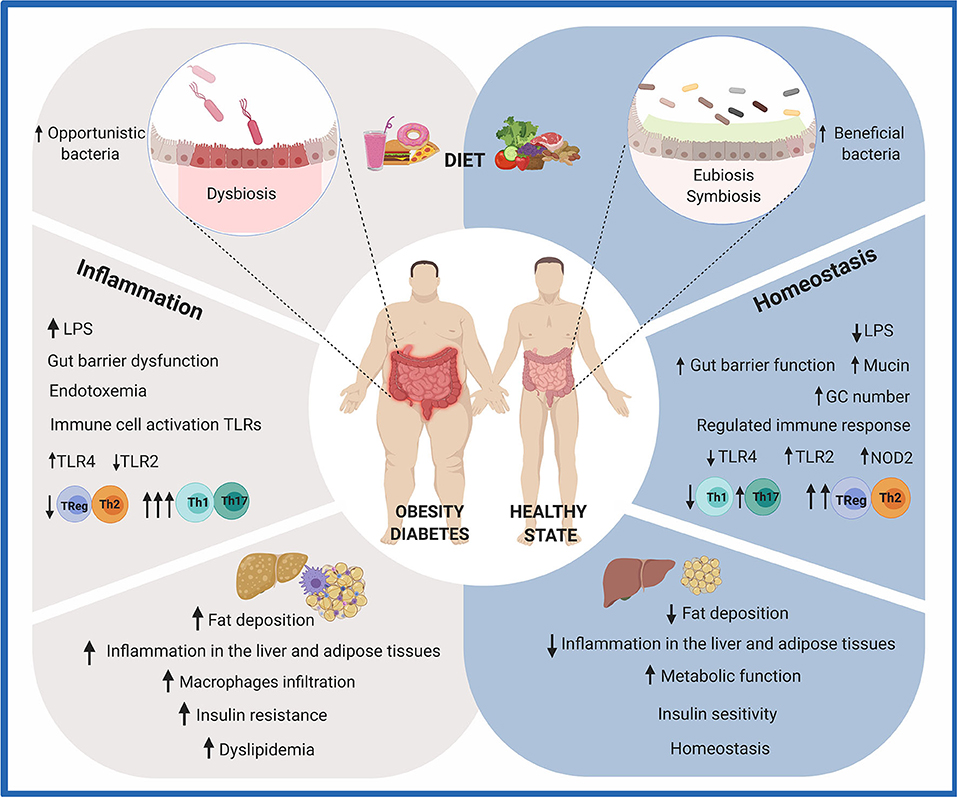
Figure 1. Effects of diet on gut health. The consumption of a diet rich in fat and sugar (HFHS) has been shown to negatively modulate the composition and metabolic activity of the human gut microbiota. Due to the crucial role it plays in human health, imbalances in gut microbiota composition and/or function (dysbiosis) are recognized as possible causes of intestinal, metabolic, and immune diseases. Particularly, HFHS leads to the translocation of bacterial lipopolysaccharides (LPS) and chronic inflammation. A group of metabolic abnormalities, including fat deposition, insulin resistance, hyperglycemia, and dyslipidemia, is exacerbated. On the other hand, a diet rich in fruits and vegetables contributes to gut homeostasis. Dietary functional ingredients have been shown to increase microbial diversity and functions, maintaining the gut microbiota composition in a eubiotic state. Intestinal and immune homeostasis is positively modulated in the host. This figure was created with BioRender.com.
(Poly)phenols are composed of an aromatic ring with at least one hydroxyl group; this structure can vary from monomers to complex polymers of high molecular weight. They are classified into two main groups: flavonoids and non-flavonoids (62). Their diversity and classification are illustrated in Figure 2. Only a small portion of (poly)phenols consumed are absorbed (5–10%), and a large proportion (90–95%), typically flavonoid aglycones and polymers, reach the colon (13, 14), where they interact with the microbiota and can exert their antimicrobial and prebiotic effects.
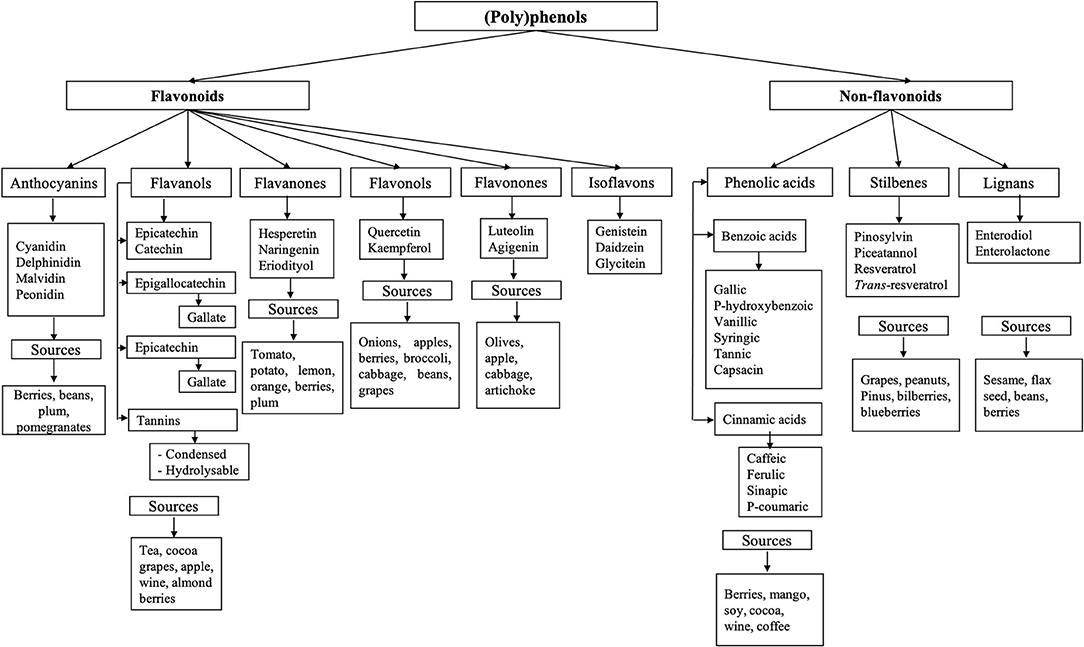
Figure 2. Classification of phenolic compounds. (Poly)phenols are classified into two main groups: flavonoids and non-flavonoids (62). Flavonoids are subclassified as anthocyanins, flavanol, flavanones, flavanols, flavonones, and isoflavones (62, 63). Non-flavonoid compounds include phenolic acids, stilbenes, and lignans.
Prebiotics are usually confined to specific non-digestible carbohydrates (i.e., inulin, fructooligosaccharides FOS, galactooligosaccharides GOS). However, this concept was revisited recently by the International Scientific Association for Probiotics and Prebiotics (ISAPP) (12), which pointed out that plant (poly)phenols can also be considered prebiotics. Indeed, based on the recent evidence of (poly)phenol trophic utilization by gut bacteria and their promoting action on species with health benefits to the host, several (poly)phenols may fit this category. Through these prebiotic-effects, (poly)phenol-rich foods can attenuate metabolic and inflammatory diseases (14), increase host intestinal mucus production (64, 65), induce the secretion of gut antimicrobial peptides (66, 67), modulate hepatic bile acids (68, 69), and gut immunoglobulins secretion (70).
The action of (poly)phenols on the gut microbiota often relies on dual antimicrobial and growth-stimulating (thus potentially prebiotics) effects. For example, red wine polyphenols increased the fecal abundance of Bifidobacteria, Lactobacilli, and F. prausnitzii, while inhibiting LPS producers, such as E. coli and Enterobacter cloacae in metabolic syndrome patients (16). In this sense, we propose the neologism duplibiotic to depict those inseparable substrate-induced modes of action in the gut, that is to promote beneficial bacterial species through concomitant antimicrobial and prebiotic effects (Figure 3). This term allows a more accurate and complete description of the dual effects of certain (poly)phenols.
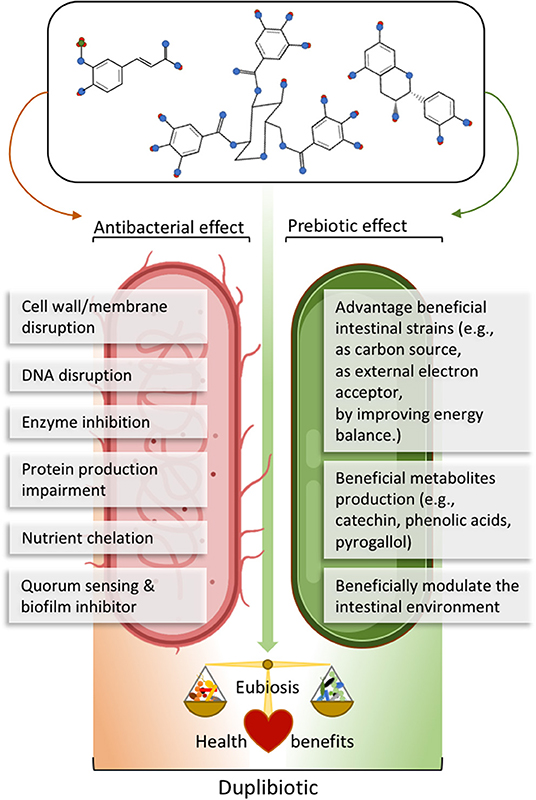
Figure 3. Description of (poly)phenols direct antibacterial and prebiotic effects. The antibacterial effect corresponds to the direct detrimental interaction between (poly)phenols or their metabolites and bacteria, while the prebiotic effect corresponds to the beneficial effect generated through direct bacterial metabolic utilization of (poly)phenols or their metabolites. The term duplibiotic design a non-digestible compound that, once reaching the colon, interacts with the gut microbiota through a dual antibacterial and prebiotic effect, favoring a eubiotic intestinal state and providing health benefits to the host.
Studies integrating multi-omic approaches have documented the pleiotropic antimicrobial mechanisms by which (poly)phenols modulate gut microbial communities (14, 71–75). For instance, (poly)phenols can interact with bacterial proteins to inhibit bacterial nucleic acid synthesis, alter cell membrane function and fluidity, modify cell wall integrity and synthesis, affect cell metabolism, and prevent biofilm formation (76). Likewise, (poly)phenols can inhibit quorum sensing (77, 78) and chelate essential metals like iron, copper, and zinc, which are essential to bacteria metabolism (79, 80). Through their antimicrobial actions, (poly)phenols can act as opportunistic pathogens inhibitors (73), protect the intestinal epithelium, and restore the microbiota homeostasis altered in several diseases.
(Poly)phenols antimicrobial action influences multiple microbial genetic responses, ranging from antibiotic resistance, metabolic responses, cell surface architecture, and stress response pathways (81). For instance, Firrman et al. (82) evaluated the growth patterns, cell morphology, and the genetic expression profiles of gut commensal bacteria exposed to different quercetin concentrations for 24 h and observed that it did not affect the growth of Ruminococcus gauvreauii, slightly repressed Bifidobacterium catenulatum, but inhibited the growth of Enterococcus caccae. The molecular analysis of the bacteria response to quercetin revealed putative mechanisms of tolerance/resistance. Interestingly, an increased expression of ABC transporters, a system associated with antibiotic resistance, was uncovered in R. gauvreauii and B. catenulatum. Particularly, these efflux pumps can expel harmful molecules out of the bacteria cell. In another study, a cranberry (poly)phenol extract down-regulated genes encoding for outer membrane proteins in Escherichia coli O157:H7 (83). Cranberry pomace and the flavonol-rich fraction had bactericidal effects against Salmonella strains (84); it reduced the expression of virulence genes and cell wall/membrane biogenesis after 20 h of incubation. Furthermore, (poly)phenols can also affect bacterial quorum sensing. In this case, cranberry proanthocyanins (PAC) reduced the production of QS-regulated virulence genes in Pseudomonas aeruginosa in a Drosophila melanogaster model (77, 84). Moreover, cranberry PAC polymers potentialized the action of conventional antibiotics by acting as quorum sensing inhibitors in bacteria (85, 86).
Metal ion-chelating properties of (poly)phenols can efficiently reduce pathogen colonization in the gut by decreasing the activity of metalloenzymes and by interfering with oxidative phosphorylation necessary for cytochrome's heme production (79). It has long been known that iron availability is crucial for bacterial growth, and iron sequestration can affect pathogens' growth and metabolism. For instance, iron-chelating gallotannins from mango inhibited Gram-positive food pathogens and Gram-negative E. coli, whereas they had no or little effects on lactic acid bacteria, as this last group was not heme dependent. Adding iron in the growing media decreased the heme-dependent inhibitory impact of polyphenols on bacteria (80).
In the context of obesity, the antimicrobial properties of (poly)phenols have been linked to the prevention of gut dysbiosis. Many gut opportunistic bacteria encountered in cases of obesity and IBD are inhibited by (poly)phenol-enriched diets (Table 1). In line with this, a black raspberry phenolic extract inhibited 10 bacterial genera from the phylum Firmicutes, and especially Ruminococcus, while the abundance of A. muciniphila was increased 157-fold in C57BL/6J mice fed an obesogenic diet (120). Also, a 4-week oolong tea (poly)phenols treatment repressed gut dysbiosis in HFD-induced obesity mouse model colonized with human gut microbiota; it increased the taxa of Prevotellaceae, and reduced Ruminococcaceae, Lachnospiraceae and, Veillonellaceae (74). The dietary administration of epigallocatechin-3-gallate (EGCG) to C57BL/6N mice fed an HFD, significantly lowered the abundance of the genera Mucispirillum, Ruminococcus, Lachnospiraceae, Desulfovibrionaceae and, Anaerotruncus, and significantly increased the abundance of the genera Adlercreutzia, Akkermansia, Allobaculum, and Parabacteroides (121). Those changes were associated with improvement in bile acid dysregulation (121).
(Poly)phenol-rich extracts exert a stimulatory effect on bacteria with crucial biological roles, such as probiotic species belonging to Lactobacilli and Bifidobacteria genera (32, 122, 123). This stimulation stems from either a (poly)phenol-induced shift in the microbial ecological niches, the reestablishment of the mucosal pro- and anti-inflammatory balance, the inhibition of potentially pathogenic bacteria, or from the direct utilization of (poly)phenols by gut bacteria. In this sense, it is important to distinguish the true prebiotic effect from indirect prebiotic-like effects induced by (poly)phenols. According to the most recent definition of prebiotics provided by the ISAPP, a compound will only be considered a prebiotic if its benefits to the host are brought about by its selective utilization by members of the gut microbiota (12). This criterion is of paramount importance to the concept of prebiotic as it excludes from its scope, the bacteriostatic, antimicrobial, and antibiotic compounds, as well as certain minerals and vitamins, even though they might beneficially impact the structure and activity of the microbiota, and thus contribute to host health. It is essential to evaluate the selective utilization of (poly)phenols by the gut microbiota since their antimicrobial activity can be improperly interpreted as a prebiotic effect. Indeed, the ISAPP experts highlighted that beneficial effects on the oral microbiota induced by cocoa and cranberry's polyphenols extracts (124, 125) are not be considered a prebiotic effect since it relies on bacteriostatic effects on susceptible pathogenic bacteria rather than a direct stimulatory effect on beneficial ones (12). Moreover, for similar reasons, bacterial (poly)phenol degradation without utilization—as in a detoxification process, for instance—cannot be considered a genuine prebiotic effect even if it leads to the liberation of metabolites able to produce beneficial effects or to participate in a trophic network. To illustrate this situation, quercetin degradation into dihydroxyphenylacetic acid by Eubacterium ramulus, a detoxification process protecting sugar transporters from inactivation (11), cannot be considered a prebiotic effect. In the same way, the bloom of beneficial bacteria resulting from a reduced microbial competition caused by (poly)phenol inhibitory action on co-exclusive opportunistic microbes would not be considered a prebiotic effect, but an indirect stimulation. Such a case is observed with the bloom of A. muciniphila, often prompted by (poly)phenols. However, as of now, its (poly)phenol-degrading ability has not been established
Several in vitro studies have demonstrated the direct positive impact of (poly)phenols on beneficial bacteria in pure culture (Table 1). Particularly, numerous strains from the genus Lacticaseibacillus, Lactiplantibacillus, and Lactobacillus grow faster when cultured with purified (poly)phenols. Furthermore, Bifidobacteria strains are also promoted by (poly)phenols in vitro. Both Lactobacilli (126) and Bifidobacteria have proven effects on health, and several species are recognized as probiotics with known modes of action (127–129). Certain modes of action by which (poly)phenols promote bacterial growth have been elucidated. (Poly)phenols provide carbon sources (after microbial deglycosylation), act as electronic acceptors (as is the case with hydroxycinnamic acids) or generate proton motive forces during their metabolization (as is the case with gallic acid) (Figure 3). (Poly)phenols also promote the growth of Bacteroides spp. in vivo (Table 1). The bloom of this genus has been related to its capacity to use (poly)phenols as a trophic substrate (130, 131). B. thetaiotaomicron, a bacterial keystone from this genus, is recognized as a next-generation probiotic (32) with carbohydrate-catabolic abilities. Among the bacterial species prompted by (poly)phenols, a bacterial consortium expressing PAZymes often stands out accompanied with co-occurring beneficial taxa after (poly)phenols intake (Table 1). However, there is still a paucity of results on the induction of PAZymes in several gut bacterial species promoted by dietary (poly)phenols.
The prebiotic action of (poly)phenols on the gut microbiota may directly stem from the activation of PAZymes like tannase, quercetinase (family of cupin-type dioxygenases: flavonol 2,4-dioxygenase, quercetin 2,3-dioxygenases), gallate decarboxylase, esterase, and phenolic acid decarboxylase enzymes (132–138), leading to both the generation of bioaccessible phenolic metabolites and microbial cross-feeding interactions in the gut. (Poly)phenol-rich powders and high molecular-weight polymeric fractions significantly increased the proportion of Eggerthellaceae and Coriobacteriaceae families (i.e., Adlercreutzia equolifaciens) in the gut microbiota of humans and animals (19, 72, 122, 139), two families displaying unique ability to break down (poly)phenols and transform them into trophic growth factors. Among the species belonging to these families, Gordonibacter spp., Eggerthella lenta, and A. equolifaciens have been identified as (poly)phenols-degrading gut bacteria (140). These species are known to metabolize ellagic acid and flavan-3-ols and have the capacity to cleave the C-ring of all (epi)catechin stereoisomers and other derivatives (141). Similarly, the growth-promoting activity of blueberry (poly)phenols on Bifidobacteria and Lactobacilli strains has been linked to their (poly)phenol-metabolizing abilities (132, 134–136, 142). Finally, Lactobacilli species, such as L. plantarum, Lacticaseibacillus casei, and Lactobacillus acidophilus thrive on (poly)phenolic-enriched media owing to their ability to metabolize tannins (27, 134, 143–145).
To date, the (poly)phenol metabolism of L. plantarum is one of the best-studied among probiotic bacteria. This ubiquitous microorganism colonizes several (poly)phenol-rich niches, such as plants phyllosphere and fermented vegetables, as well as dairy foods and the human gut. L. plantarum pan-genome includes several genes encoding enzymatic functions involved in the utilization of (poly)phenols (146, 147). Specifically, these include intracellular tannase (tanBLP), gallate decarboxylase (lpdB, lpdC), aryl glycosidase (10), rhamnosidases (rhaB1, rhaB2) (148), phenolic acid decarboxylase (hcrB, lp_3665), and vinylphenol reductase (lp_3125). Other genes are, however, highly strain-dependent: extracellular tannase (tanALP), a broad esterase (est_1092, with tannase and feruloyl esterase activity), and a non-identified enzyme(s) able to produce diphenylpropan-2-ol from catechins (87, 149). When exposed to plant environments, L. plantarum modifies its core metabolic pathways to save energy and utilizes alternative NAD+ generation routes (150). Interestingly, L. plantarum transcriptomic response to tannins (as well as tannic and gallic acid) involves the overexpression of genes related to its gastrointestinal survival (lp_2940 [cell surface protein] with gallic acid, but also argG [argininosuccinate synthase], copA [copper transporting ATPase], napA3 [Na(+)/H(+) antiporter] with tannic acid), indicating that this microorganism may be highly adapted to survive in the context of a tannin-rich diet (143). These metabolic characteristics make L. plantarum a prime candidate for the development of synbiotics aiming to potentialize the benefits of (poly)phenol-rich diets. The prebiotic potential of (poly)phenols thus appears dependent on the arsenal of PAZymes triggered by probiotics and commensal microbes.
Tannases or tannin acyl hydrolases represent a large esterase enzyme family presenting a broad range of substrates specificities that vary according to the enzyme molecular structure (133). Bacterial tannases can hydrolyze gallate and protocatechuate esters found in tannins and other phenolic compounds. However, it is unclear if gut bacterial tannases hydrolyze tannins with higher chemical complexity, such as ellagitannins (43).
Tannase activity has been mainly reported in lactic acid bacteria such as L. plantarum (151), Lactiplantibacillus pentosus, Lactiplantibacillus paraplantarum, L. acidophilus, Pediococcus acidilactici, and Pediococcus pentosaceus (152). The presence of this enzyme has also been noted in other gut bacteria like Roseburia intestinalis XB6B4 and Slackia heliotrinireducens (153). Gut microbial intracellular tannases may contribute to the transformation of compounds that potentially enter the microbial cell (e.g., methyl gallate, epigallocatechin gallate, and epicatechin gallate). Bacteria extracellular tannases may also transform larger molecules, such as gallotannins and, possibly, galloylated ellagitannins (154). In the colon, these gut-microbial extracellular tannases may play a key role in the prebiotic potential of tannins, releasing more bioaccessible molecules from non-absorbable polyphenols. Barnes et al. (155) found that lean individuals have higher microbial intestinal tannase activity than individuals with obesity, suggesting that gut microbial composition is likely to influence this enzyme activity. Gut tannases appear to stimulate the production of gallotannin-colonic metabolites, as higher concentrations of these metabolites (i.e., 4-O-methylgallic acid, 4-O-methylgallic acid-3-O-sulfate, and pyrogallol-O-sulfate metabolites) are later found in the systemic circulation of lean individuals eating a gallotannin-rich diet (400 g of mango pulp). Interestingly, subjects receiving an enriched gallotannin diet for six weeks tended to increase their tannase activity (155), which indicates that dietary strategies may reestablish this gut enzymatic function.
Gallate decarboxylases are microbial enzymes that transform gallic acid into pyrogallol. These PAZymes are highly present in lactic acid bacteria isolated from plant sources, including those belonging to the L. plantarum group and some Lactobacillus gasseri (103). Noteworthy, gallate decarboxylase favors energy uptake in L. plantarum, by generating a proton motive force during gallic acid metabolism (27). In the host, gallate decarboxylase-producing probiotic bacteria may enlarge the metabolic benefits induced by gallic acid and gallic acid-rich (poly)phenols. Fang et al. (51) observed that a gallate decarboxylase-producing L. plantarum strain (WCFS1), concomitantly administered with gallotannins (gallic acid glycosidic polymers), tended to potentialize the gallotannins antiobesity effects in a gnotobiotic mice model. The brown adipose tissue of L. plantarum-supplemented mice had higher mRNA levels of thermogenic genes when compared to mice receiving only gallotannins. Bacteria producing decarboxylase may thus boost pyrogallol production in the gut (51), entailing potential antiobesity and anticarcinogenic effects (156, 157). A high-throughput sequencing approach revealed that this enzymatic function is also present in different phylogenetic divisions of the gut microbiota. Gallate decarboxylase was predicted in strains of Erysipelotrichaceae bacterium, Enterobacter cloacae, Actinomyces glycerinitolerans, Anaerostipes hadrus, Enterococcus raffinosus, Pediococcus ethanolidurans, Klebsiella michiganensis, Blautia sp., Dorea longicatena, Clostridium butyricum (158).
Feruloyl esterases (also called hydroxycinnamoyl esterases) are a large family of PAZymes presenting different hydrolyzing capacities toward hydroxycinnamic acid esters (ferulic, p-coumaric, caffeic, sinapinic, or diferulic acids esters) (159), frequently found in plant cell walls, as in wheat bran (160), berries (161), and Jerusalem artichoke (162). Because they are usually covalently bound to fibers, they are not readily bioaccessible. Therefore, feruloyl esterases can improve the bioavailability of these functional compounds. This enzymatic activity has been reported so far in Bifidobacterium longum (99), Lactobacillus helveticus, Lactobacillus johnsonni, Limosilactobacillus reuteri, Lactobacillus acidophilus, Limosilactobacillus fermentum (100), and L. plantarum (163). However, substrate affinity may greatly differ between bacterial esterases. For example, L. fermentum's feruloyl esterase has a greater hydrolytic activity on methyl caffeate, than on trans p-coumarate or methyl ferulate (164). Feruloyl esterase-producing lactic acid bacteria have a broad spectrum of applications as they increase the bioaccessibility of ferulic and other hydroxycinnamic acids in fermented foods, such as fermented whole grain barley and oat groat (165). Additionally, supplementation with feruloyl esterase-producing probiotic strains, such as L. fermentum CRL1446, can greatly improve intestinal esterase activity and oxidative stress parameters of supplemented mice (166).
In humans, hydroxycinnamoyl esters hydrolysis first takes place in the small intestine, mainly by mucosal esterases (167). However, in many food sources, such as bran and aleurone, ferulic acid is mostly covalently bound to arabinoxylans and other cell wall polysaccharides able to resist digestion in the upper gastrointestinal tract (168). Once in the colon, the resident microbiota cooperatively breaks the cell wall polysaccharides and release ferulic acid from its chemical bonds. Using in vitro fermentation, Duncan et al. (169) pinpointed the role of butyrate-producing gut microbiota species, such as Bacteroidoita phylum, and their capacity to release ferulic acid from wheat bran fermentation. Bacteroidota species are primary degraders of arabinoxylan in the colon, deploying a differential enzymatic machinery according to the arabinoxylan type (soluble vs. complexed with ferulic acid side chains) (170). The genera of Bacteroides, Eubacterium, Roseburia, and Butyrivibrio (Lachnospiraceae) degrade xylans, while the genera of Ruminococcus and Faecalibacterium can degrade hemicelluloses or pectin. In line with this, Ruminococcus spp. and F. prausnitzii were prompted by wheat bran (arabinoxylan-rich phenolic acid-rich polysaccharides), resulting in the release of ferulic acid in the human gut microbiota (171). This phenolic compound was then rapidly reduced to produce phenylpropionic acids, known to benefit host health. Overall, many types of interactions between polysaccharide and feruloyl-esterase-producing bacteria may occur within the gut microbial ecology, potentially enriching the microbial diversity and metabolomes, including SCFA and phenolic metabolites. Ferulic acid is a highly bioavailable metabolite (172) that inhibits platelet activation (173), modulates insulin signaling (174), and reduces metabolic syndrome symptoms in diabetic rats (175). It mimics many of the beneficial effects of other (poly)phenols on cancer, cardiovascular, and cognitive diseases, among others (176).
Phenolic acid reductases are enzymes able to reduce hydroxycinnamic acids, and produce substituted phenylpropionic acids [phloretic, 3-(3-hydroxyphenyl) propionic acid (3-HPPA), dihydrocaffeic, and dihydroferulic acids]. These colonic microbial metabolites have been found in the urine and plasma of subjects following a supplementation of a green and roasted coffee blend as hydroxycinnamic acids source (177). Phenolic acid reductase activity has been reported in L. fermentum, Latilactobacillus curvatus. L. reuteri, Furfurilactobacillus rossiae, and L. plantarum (178). Interestingly, these PAZymes confers an energetic advantage for strict heterofermentative lactic acid bacteria, as they can use these phenolic acids as external electron acceptors in the pentose-phosphate pathway (28). The resulting (poly)phenol-derived metabolites have higher antiplatelet properties than the parental compounds (173). They also prevent monocyte endothelial adhesion (179) and may protect against pancreatic-β-cell dysfunction in vitro.
The α-L-rhamnosidases are enzymes able to release aglycones and glucose by cleaving the terminal α-L-rhamnoses present in glycosylated phenolic molecules (180). Since the human intestine possesses no rhamnosidase activity (168, 181), the vast majority of these rhamnose glycosides remain unabsorbed and reach the colon. The ability to transform specific rhamnose glycosides frequently found in citrus fruits, such as hesperidin and rutin, has been demonstrated in some probiotic bacteria. Hesperidin glycosides are converted, among others, by strains of Levilactobacillus brevis, Lacticaseibacillus paracasei, L. acidophilus (170), Bifidobacterium animalis, Bifidobacterium breve, B. catenulatum and Bifidobacterium pseudocatenulatum (171). However, the hydrolyzing capacity of these species is highly strain-dependent and varies according to the structure of the rhamnose glycoside. For instance, the rutin structure is less accessible to bacterial rhamnosidases, and only certain probiotics, such as L. fermentum, can degrade it to a small extent (109, 170). In addition, rhamnosidase activity may be influenced by certain carbon sources and other phenolics present in the media. Particularly, bacterial α-L-rhamnosidases were stimulated by narcissin and rhamnose (170) while inhibited by glucose (148). Other rhamnosidase-producing bacteria belonging to Lachnospiraceae, Enterobacteriaceae, Tannerellaceae, and Erysipelotricaceae, may be involved in the transformation of rhamnose glycosides, such as dietary rutin (170).
β-glucosidases are heterogeneous and widespread enzymes that enable bacteria to obtain carbon sources from glucosides catabolism (182). (Poly)phenol glucosides, occurring primarily as O-glucosides (OH-coupled), are deglycosylated by β-glucosidases, resulting in the release of sugars and aglycones: a free form in which (poly)phenols may passively diffuse across tissues (183, 184). C-Glycosides (C-C-coupled) are less frequent in nature and more difficult to degrade. Details of C-glycoside cleavage by intestinal bacteria have been recently reviewed by Wei et al. (185). β-glucosidases are produced in response to glucoside-rich environments by several intestinal bacteria such as B. thetaiotaomicron, Bifidobacterium spp., Blautia producta, Erysipelatoclostridium ramosum, E. coli, and L. plantarum (10, 186, 187). Zyzelewicz et al. (188) recently reported an increase in cecal β-glucosidase activity in high-fat-fed rats after a cocoa bean extract supplementation, indicating that diet composition may modulate this enzymatic activity. However, it is not clear if this (poly)phenol-induced activity stems from microbiota compositional changes or from alterations of the microbiome function, as only enzymatic activity was assessed.
Many lactic acid bacteria (Streptococcus thermophilus, L. acidophilus, Lactobacillus delbrueckii ssp. bulgaricus) are also able to produce β-glucosidases (182). Because of their safety, these β-glucosidase-producing-bacteria are used to increase the bioaccessibility of phenolic glucosides during food fermentation (189). One example is the hydrolysis of the glucoside daidzin, contained in soybeans, into its aglycone daidzein. This aglycone can be further converted into equol, a potent bioactive compound, by four subsequent enzymes (daidzein reductase, dihydrodaidzein reductase, tetrahydrodaidzein reductase, and dihydrodaidzein racemase). In addition to its anticancer, cardioprotective, and neuroprotective activities, equol has gained attention for its capacity to modulate estrogen levels, reducing menopause symptoms (190). Intestinal (poly)phenol-degrading bacteria such as Eggerthella spp. and certain strains of Adlercreutzia equolifaciens (141, 191, 192) produce equol from daidzein aglycone. This enzymatic machinery is also present in probiotic species such as B. breve, B. longum, Lactococcus garvieae, and Lactobacillus intestinalis (189). These observations have fostered the emergence of probiotic strategies and have resulted in better clinical outcomes in equol non-producers (193).
Microbial phenolic metabolites, like their native compounds, may exert antioxidant, antiproliferative, and anti-inflammatory activities (194, 195). The production of these beneficial metabolites is strongly dependent on the PAZyme production by the gut microbiota of each individual, a phenotype so-called metabotype. This important concept has been recently and thoughtfully reviewed elsewhere (43). Indeed, the notion of different metabotypes would explain, in part, the interindividual differences in the health outcomes observed after (poly)phenol intake. Therefore, the presence of (poly)phenol-degrading bacteria in the gut microbiota is crucial to potentiate the bioactivity of parent (poly)phenols. This is the case of bacteria producing equol from daidzin, enterolactones from lignans, 8-prenyl naringenin from xanthohumol, and urolithins from ellagitannins (196).
It is also believed that PAC bioactivity can be potentiated by gut bacteria to produce active isomers of valerolactones. Phenyl-γ-valerolactones and phenylvaleric acid derivatives, 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone, and 4-hydroxy-5-(3′,4′-dihydroxyphenyl)-valeric acids are flavan-3-ols derived metabolites from fecal fermentation of PACs. Using Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) and Variable Importance in Projection (VIP) analysis, Rocchetti et al. (197) reported fermentation markers linking the chemical structure of (poly)phenols to the colonic pathways in which they are involved. In that study, changes in the phenolic profiles after in vitro fermentation of (poly)phenol-rich nuts (high in PACs) were evaluated by both untargeted UHPLC-QTOF and targeted UHPLC-Orbitrap mass spectrometry. The OPLS-DA model revealed catabolic pathways involving PAC C-ring fission of the flavan-3-ol-backbone, leading to the formation of 5-(3′,4′-dihydroxyphenyl)-γ-valerolactones. Subsequent oxidation reactions are thought to produce lower-molecular-weight phenolic acids such as 3,4-dihydroxyphenylacetic acid and hydroxybenzoic acids. These reactions are driven by an enzymatic repertoire encoded by species belonging to Coriobacteriaceae (A. equolifaciens), Lactobacillaceae (L. plantarum), Ruminococcaceae (Flavonifractor plautii), and Eggerthellaceae (E. lenta, Paraeggerthella, Gordonibacter and Slackia equolifaciens) (140, 198, 199). The bioavailability and potential bioactivities of valerolactones are very topical and the focus of recent research (200, 201). Indeed, valerolactone isomers were recently shown to reduce monocyte adhesion in vascular endothelium, potentially preventing atherosclerosis (202). These metabolites appear to pass the blood-brain barrier and might thus be involved in neuroprotective effects of (poly)phenols-rich diets (203–205). Obviously, changes in gut microbiota functions may have repercussions on the production or degradation of certain metabolites, having marked effects on the well-being of the host. Studies have found that gut microbiota xenobiotic metabolism, particularly linked to (poly)phenols, is associated with reduced inflammation, promoting gut health in the context of obesity (206, 207).
The urolithin A, a metabolite derived from ellagitannins degradation, is considered a metabotype biomarker associated with reduced cardiometabolic risk (208, 209). It is worthy to note that urolithins also exert antimicrobial and anti-quorum sensing activities, for example, by inhibiting N-acyl homoserine lactones (AHL), a QS signal molecule involved in cell population density and biofilm formation (210). In fact, urolithins A and B can alter the expression levels of genes critically involved in the synthesis of lactones and swimming motility of the enteropathogenic Yersinia enterocolitica (210). Dietary derived (poly)phenols metabolites, like their parent compounds, may thus exert antimicrobial effects in the colon and suppress the colonization of opportunistic bacteria, contributing to reestablish gut dysbiosis. This inhibitory action on opportunistic bacteria, in addition to (poly)phenol-induced changes in the gut barrier and immune responses, is also a potential mechanism indirectly fostering gut bacteria with health benefits to the host.
(Poly)phenols can induce ecological shifts, that is, cause important changes in the structure, the functions of microbial ecosystems leading to alternative states reconfiguration (211, 212). Indeed, (poly)phenol-induced shifts manifest themselves into enterotype-like clustering of the gut microbiota. For instance, using CCA-ordinations based on the gut microbiota composition, mice fed an obesogenic diet supplemented either with berry (poly)phenols or fibers clustered in a distinct group. In this case, the obese mice were characterized by a Firmicutes/Ruminococcus enterotype enriched in Lachnospiraceae and Ruminococcaceae taxa (139), which are known to be opportunistic mucosa-associated bacteria. On the contrary, the gut microbiota of mice fed with a (poly)phenol-rich cranberry powder clustered in the contrasting enterotypes of Bacteroidetes/Muribaculaceae and Prevotella/Akkermansiaceae, which were enriched in Eggerthellaceae, lactobacilli, Akkermansia, and Lachnospiraceae_NKA136_group (139). Besides their duplibiotic action, these important changes in the gut microbiota can be linked to the prebiotic-like effects of (poly)phenols in the intestinal environment (20, 213, 214).
(Poly)phenols thus have the capacity (1) to free ecological niches by hindering the potential opportunistic pathogens, (2) to re-establish the normal function of the mucosal epithelial barrier and its immunological response, and (3) to reduce oxidative agents (reactive oxygen species and free radicals) (215). These three events favor an increased abundance of beneficial gut bacteria bearing health benefits to the host, which underly their indirect prebiotic-like effect. One typical example is the case of A. muciniphila.
Accumulating evidence confirm the impact of dietary (poly)phenols on A. muciniphila. Particularly, grape (poly)phenols, apple PACs, EGCG, puerarin, A-type PAC-rich cranberry extract have been shown to dramatically increase the growth of A. muciniphila and decreased the proportion of Firmicutes to Bacteroidetes in cecal and fecal samples of animals, in association with an improved host metabolic phenotype (14, 16, 17, 66, 216). Likewise, (poly)phenolic compounds such as chlorogenic acid, caffeic acid, quercetin, resveratrol, trans-resveratrol, and malvidin-3-galactoside promoted Akkermansia in dextran sulfate sodium (DSS) induced colitis C57BL6 mice model and in mice with liver cancer (217–220). As there is no precise evidence of the PAZymes-producing ability by A. muciniphila to utilize the aforementioned (poly)phenol compounds, this stimulating effect should be attributed to a prebiotic-like effect rather than a direct prebiotic one.
In analogy to the well-described promoting effect of antibiotics on Akkermansia, (poly)phenols induced blooms might be a collateral effect of (poly)phenol's antimicrobial action, in addition to their action on the synthesis of mucin by the gut epithelium (64, 65). Indeed, this bacterium can physiologically adapt and resist broad-spectrum antibiotics, conferring it a molecular advantage to colonize the colon (221–223). Akkermansia's resistance to phenolics, conjugated to a reduced competition from (poly)phenol-susceptible microbes, and the liberation of ecological niches occupied by opportunistic bacteria might explain its bloom in the presence of (poly)phenols. This outcome is illustrated, for example, by the suppression of the taxonomic abundance and activity of opportunistic bacteria colonizing the colonic mucosa such as Ruminococcus species (224), which in turn, reduce the microbial competitiveness and ability of mucosa-associated species to grow, as observed with A. muciniphila. The latter has a trophic preference to exploit the mucin glycoproteins and inhabit mucin-rich specialized niches, as the host epithelium (225), where it participates in a symbiotic crosstalk with the immune system (226, 227). Factors affecting mucin secretion and glycosylation, as well as the mucosa homeostasis thus have a major impact on the Akkermansia abundance. Indeed, a decrease of A. muciniphila in obesity and age-induced impaired mucus barrier is often observed (64, 228–231). In this sense, (poly)phenols have been shown to reinforce the mucosal epithelium by increasing the tight junction proteins (232), the mucus-secreting goblet cells, and improving the mucus thickness (19, 139), all factors favoring an optimal ecological niche for Akkermansia to thrive.
More studies are warranted to screen and identify the type of (poly)phenolic compounds contributing the most in prompting symbionts and modulating the functions of the gut microbiota. A recent report demonstrated a selective promoting effect of polymeric PACs rich fraction on A. muciniphila fecal abundance in mice fed an obesogenic diet, concomitantly with improved the colonic mucus thickness (19). Still, two questions remain unsolved; (1) does the Akkermansia bloom after (poly)phenols intake stems from its ability to degrade them, or (2) are the (poly)phenols boosting effects on Akkermansia, a consequence of the (poly)phenol-induced anti-inflammation and recovering of the intestinal mucosa-associated niche? To fill in part this gap in knowledge, we screened in-silico the sequenced Akkermansia's genome searching for genes involved in the degradation of (poly)phenols. Surprisingly, we found out that A. muciniphila (ATCC BAA-835) harbors two genes linked to flavonol quercetin degradation: a cupin domain-containing protein encoded by Amuc_0801 (NCBI Reference Sequence: WP_012419849.1) and γ- carboxymuconolactone decarboxylase encoded by Amuc_1806 (NCBI Reference Sequence: WP_081429203.1). The analysis of the conserved domains (CD) and protein architecture of those gene sequences led to the identification of potential matches and similarities against deposited sequences of quercetinases enzymes in the NCBI resources (Conserved domain search service). The quercetinases have been mostly described in plant-associated fungi; however, they have also been described in the gut bacterial species Bacillus subtilis and E. ramulus (104, 138, 233). Nevertheless, further in-depth characterization of the enzymatic activity coded by quercetinases-related genes in this species is required.
From a trophic perspective, the chemical structure and polymerization degree (DP) of (poly)phenols have a large influence on the yield of phenolic metabolites and subsequent stages of their catabolism by the gut microbiota. For instance, the human fecal fermentation of a grape seed fraction rich in flavan-3-ols monomers (70%) resulted in the generation of 3-(3,4-dihydroxyphenyl)-propionic acid, which was subsequently degraded by colonic bacteria (234). However, these phenolic metabolites yielded in higher proportion when flavan-3-ols polymers rich fraction was fermented, which was significantly linked with increased growth of Lactobacillus/Enterococcus taxa and the inhibition of H. histolytica.
(Poly)phenol polymers, which represent a large proportion of those ingested in the diet, are often intimately bound through hydrophobic and hydrogen bonds to plant cell wall matrix and therefore reach the colon in the form of (poly)phenolic fibers (235, 236). A clear case of fiber-associated (poly)phenols are polymeric flavan-3-ols, whose fermentation leads to SCFA biosynthesis while releasing the so-called “non-extractable (poly)phenols” (237, 238), thereby illustrating another example of trophic interactions in the gut environment. For instance, Mateos-Martin et al. (239) studied the fate and metabolism of grape dietary fiber rich in non-extractible PAC (14.8%), in female Sprague–Dawley rats for 24 h. The grape residues obtained after extraction with 70% acetone were rich sources of non-extractible PAC polymers. Once the colonic microbiota fermented the fibers, PAC were released and progressively depolymerized into epicatechin monomers and dimers and later metabolized into smaller units. Microbially derived PAC metabolites, such as valerolactones, phenylvaleric acids, phenylpropionic acids, phenylacetic acids, benzoic acid, and cinnamic acids, were detected only in the urine of the treated group. These fibers and (poly)phenol-derived metabolites could have an overall synergistic effect on the host. Dietary fibers and (poly)phenols have been shown to act synergistically in processes like antiradical activity and lipid oxidation in vivo (240), and to reduce hepatic cholesterol levels in apolipoprotein E-deficient mice with atherosclerosis (241). Another intervention study in diet-induced obese mice demonstrated the selectivity of PAC bound to cranberry fibers not only in inhibiting HFHS-induced opportunistic bacteria, but also boosting the (poly)phenol-degrading families of Coriobacteriaceae and Eggerthellaceae, as well as butyrate-producing bacteria belonging to Lachnospiraceae NK4A136 taxon; these (poly)phenolic-fiber induced microbial changes were reflected by reduced hepatic triglycerides in obese mice. Of particular interest, such a microbial modulatory effect and host health outcome were not reproduced by the blueberry PAC-free fibers (139).
It is worthy to note that (poly)phenols can also increase the production of SCFA. While the mechanisms explaining this production are not yet fully understood, this may be mediated by the physicochemical inhibition of the mouth amylase, leading to an increased concentration of complex carbohydrates reaching the gut (242), or by an increase in anaerobic bacteria (243, 244) through the reduction of oxidative molecules in the gut environment. The fermentation of glycosylated (poly)phenols reaching the gut could also represent a non-negligible source of carbohydrates for several members of the microbiota (140), potentially constituting a (poly)phenol prebiotic pathway leading to the production of SCFA. Not only (poly)phenols can favor the production of SCFA, but the latter can also affect the absorption of (poly)phenol metabolites. As a matter of fact, Van Rymenant et al. (245) have observed an increased translocation of secondary (poly)phenol metabolite from ferulic acid and hesperidin through the epithelium, in the presence of butyrate and propionate in an in-vitro model of Caco-2 cells. They proposed that those SCFA would increase the production of (poly)phenol transporters (MTC1, MTC4, and various ABC transporters), and that propionate could lead to the reduction of ferulic acids to dihydroferulic acids, increasing their transport across the membrane.
(Poly)phenol and carbohydrates substrates can potentiate further cooperative interactions between PAZymes-producing bacteria and those showing less or no PAZymes activity. This was observed by Rodriguez-Castano et al. (11), who evaluated in a simple co-culture model the interactions between the gut symbiont B. thetaiotaomicron and Eubacterium ramulus. The former gut symbiont has a wide capacity to degrade complex polysaccharides but is unable to degrade the flavonoid quercetin, while the latter is a quercetin-degrading species with limited capacity to breakdown polysaccharides. Indeed, E. ramulus has been shown to respond to the presence of quercetin in the human diet (246). In this model, the addition of starch increased E. ramulus quercetin degradation activity and the ensuing production of 3,4-dihydroxyphenylacetic acid, whereas high levels of butyrate were significantly induced when co-culturing with B. thetaiotaomicron. In contrast, when both species were cultured individually in starch and quercetin enriched media, no significant quercetin degradation nor butyrate production were observed. Hence, the release of glucose and maltose upon the starch fermentation by B. thetaiotaomicrom powered, in turn, the quercetin-degrading activity of E. ramulus. Such trophic interactions increase the potential generation of bio-active metabolites, such as SCFAs and phenolic metabolites. Particularly, the production of quercetin-derived metabolites including 3-(3,4-dihydroxyphenyl) propionic acid, 3,4-dihydroxyphenylacetic acid, 3,4-dihydroxybenzoic acid (i.e., protocatechuic acid), and phenylacetic acid (247, 248), can exert a potential role in the improvement of an impaired glucose metabolism (249, 250).
Deciphering the complex trophic relations taking place in the gut microbiota upon (poly)phenol-rich foods consumption is a huge challenge. Approaches involving simplified microbiotas or key microbial consortia co-cultures with different metabolic activities may help decrypt the ecological role of duplibiotics. Assessing the duplibiotic activity of a (poly)phenol requires determining both its antimicrobial activity and prebiotic effect. The simplest studies analyzing (poly)phenol effects on the gut microbiota are based on the direct challenge of individual bacterial strains by purified (poly)phenols. This approach allows identifying the metabolic activity and the associated PAZymes involved in the transformation of (poly)phenols, as well as the derived metabolites. Furthermore, (poly)phenol challenges provide information on anti/pro-microbial potential of a phenolic molecule and how bacteria resist its action (27, 132, 154, 251, 252). Studies on whole microbiota stabilized in intestinal in vitro systems—like the Simulator of Human Intestinal Microbiota (SHIME)—have been used to assess (poly)phenols' impact on the microbiota composition and function, to identify potential (poly)phenol-degrading bacteria and metabolites production (253). Given the complexity of the whole gut microbiota (254), co-culture experiments (11, 255) and the use of simplified synthetic gut microbial consortia combined with “omics” analyses (256, 257) may be suitable approaches to precisely assess symbiotic and trophic interactions induced by (poly)phenols. While in vitro models lack direct interactions with the host, they are complementary tools to elucidate mechanistic hypotheses involving the role of (poly)phenol-degrading bacterial consortia within the human gut and the downstream physiological outcomes. Indeed, transferring the metabolites-enriched supernatants (258) or the bacterial communities issued from these culture models (259) into in-vitro human-cell systems (253) or germ-free animals (260) can uncover the health benefits induced by (poly)phenol-rich food intake. Figure 4 summarizes the in vitro and animal approaches unveiling the duplibiotic nature of (poly)phenols. As stated by the definition of prebiotics, further well-designed and realistic human clinical studies are then essential to demonstrate the health benefits induced by the microbial utilization of (poly)phenols.
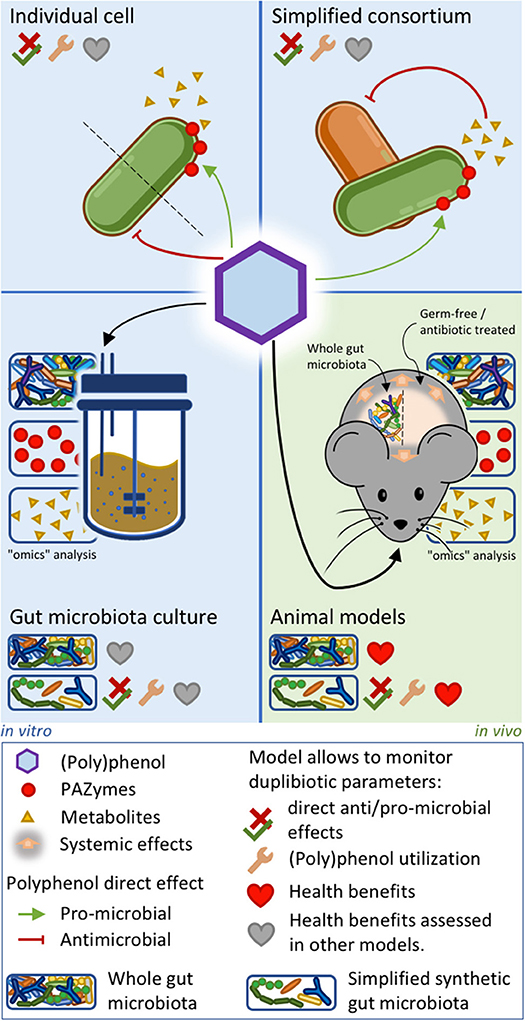
Figure 4. In vitro and animal approaches to evaluate duplibiotic nature of polyphenols. Individual bacterial strain or simplified bacterial consortium models allow determining (poly)phenol anti/pro-microbial effects, (poly)phenol transformation, and metabolite production. In addition, studies on simplified bacterial consortia can characterize symbiotic interactions. Gut microbiota culture and animal models combined with omics approaches provide insights on global microbial changes induced by (poly)phenols. Studies on conventionally grown, gnotobiotic, and antibiotic-treated animal models allow characterizing host health benefits. The health benefits stemming from the three in vitro approaches can be further demonstrated by transferring downstream metabolites into ex vivo systems. Synthetic microbiota inoculated in culture systems or in germ-free animals allows determining direct anti/pro-microbial effects, as well as demonstrating bacterial (poly)phenol utilization.
The literature strongly supports the beneficial effect of (poly)phenol-rich foods on the gut microbiota composition, contributing to the maintenance of host energy balance and mucosal immunological responses (5, 15–17, 134, 142, 258, 261–264). In this review, we have focused on illustrating the two opposing mechanisms of (poly)phenols directly affecting the microbiota composition: the antimicrobial effect and the prebiotic effect. Not all (poly)phenols can modulate the gut microbiota through these antagonistic modes of action; however, when they do, those effects cannot be assessed separately. For this purpose, we introduce the term duplibiotic that describes this equivocal activity of (poly)phenols on the microbial community and expands their potential beyond a prebiotic effect sensu stricto by considering their whole effects that could be greater than the sum of their parts.
While the antimicrobial effect of (poly)phenols has been well-described, the prebiotic concept has commonly been misused and undistinguished from the prebiotic-like effect. Indeed, a (poly)phenol prebiotic action is often attributed after observing a stimulation of gut beneficial bacteria without confirming a utilization of the phenolic compound, as denotes the ISAPP-updated prebiotic definition. In the consensus statement on the matter (12), the ISAPP stated that the expression “a substrate that is […] utilized” were chosen for the prebiotic concept to imply “growth through nourishment.” From this perspective, we illustrated three manners by which (poly)phenols would be used by the microbiota and thus exert a genuine prebiotic effect: (1) as carbon sources, (2) as electron acceptors, and (3) as generators of a proton motive force. The first (poly)phenol purpose fits the conventional microbial utilization of well-documented carbohydrate prebiotics, but the last two extend beyond the ISAPP revisited prebiotic concept by including energy production, which is also implied in bacterial nutrition. According to those considerations, very few (poly)phenols can yet be considered as prebiotics. Other prebiotic and duplibiotic compounds will certainly be identified using modern omics analytical technologies, with in vitro synthetic approaches and clinical studies targeting the gut microbiota.
To illustrate the prebiotic potential of (poly)phenols, we described a selection of PAZymes involved in the microbial utilization of these compounds. This PAZymes abbreviation was used in this paper to regroup and discuss (poly)phenol-associated enzymes. In analogy to CAZymes, PAZymes have noteworthy similarities, as they are diverse and target different compounds, having relevance in nutrition and health. From this perspective, an initiative to group PAZymes in sequence databases, as done for CAZymes (i.e., CAZypedia, CAZyDB) (265, 266), would be of great interest. This would facilitate the identification of novel PAZymes, as well PAZymes-harboring microorganisms by coupling omics outputs, including metagenomics. Insights on PAZymes-producing species have been obtained from members of Coribacteriaceae, Eggerthellaceae, and Lactobacillae, which have the capacity to release phenolic metabolites that can impact both commensal microbiota and host health. Here, we reviewed the role of several enzymes in breaking down different (poly)phenolic classes, including the tannases, quercetinases, feruloyl esterases, gallate decarboxylases, and phenolic acid reductases that support the direct prebiotic effects of (poly)phenols on lactobacilli species and other gut symbionts. Identifying potential probiotic bacteria harboring PAZymes is of high interest since it would promote the release of bioactive phenolic metabolites and possibly induce metabotype shifts in non-producer individuals.
Even though considerable progress has been made in identifying the bacterial enzymes and genes involved in (poly)phenol metabolism, much remains to be revealed. For example, the enzymes and the genes involved in the C-ring reductive cleavage of catechin isomers and the resulting release of diphenylpropan-2-ol have not yet been discovered. E. lenta, A. equolifaciens, and L. plantarum strains were shown to be involved in these transformations (88, 89). Likewise, F. plautii strains can utilize diphenylpropan-2-ol metabolites (89), leading to valerolactones production. The identification of the entire set of enzymes and genes involved in the catabolism of catechin is essential to pinpoint the bacterial consortia that favor valerolactones production (267, 268). Another representative example that remains to be investigated in greater depth is the identification of urolithin-producing PAZymes and their related genes. The ability to produce urolithins from ellagic acid has been described so far in Gordonibacter pamelaeae, Gordonibacter urolithinfaciens (93), Ellagibacter isourolithinifaciens (94), and B. pseudocatenulatum INIA P815 (95). Particularly, the latter strain is shown to produce urolithin A, but the related enzymatic activity is apparently absent in most B. pseudocatenulatum strains (95). Hence, precise identification of urolithins-producing enzymes and related genes would allow the discovery of potential symbionts that could be used as therapies in subjects with non-urolithin-producing metabotypes (or low-urolithin A producers) in order to favor the release of these antiobesity (269), anti-inflammatory (270), and neuroprotective metabolites (271).
It is suggested that PAZymes repertoire improve the fitness of certain beneficial species, contributing not only to detoxify the bacteria's environment but also enabling these microorganisms to access carbon sources (i.e., β-glucosides) or favoring them metabolically to produce more ATP (i.e., gallate decarboxylase or phenolic acid reductase). These gut bacteria (poly)phenol transformations release a variety of microbial metabolites that, contrary to large polyphenol polymers, easily cross cell membranes (through passive permeation or active transports), reach target tissues, and exert local anti-inflammatory activities. These (poly)phenol-derived metabolites are beneficial as they potentially modulate the immune system while hindering the growth of pro-inflammatory gut bacteria (Figure 5). Certainly, these metabolite-induced outcomes, as demonstrated for their parent compounds, impact the gut microbiota by generating ecological shifts. (Poly)phenol-induced changes in the microbiome (microbial functions, composition, and intestinal environment) (215, 217, 244) imply not only the duplibiotic nature of these molecules, but also other potential mechanisms of actions (i.e., as anti-inflammatory and antioxidant agents).
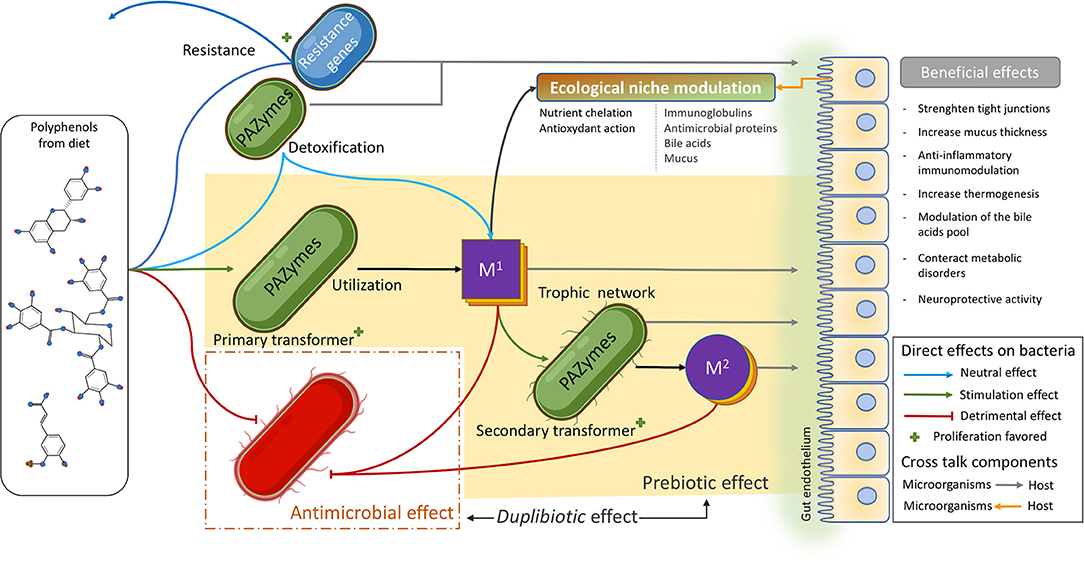
Figure 5. Polyphenol's mode of actions shaping the gut microbiota. (Poly)phenols reaching the gut can exert an important antimicrobial pressure (red lines) on several members of the microbiota. They can also be transformed by a vast and diverse array of microbial (poly)phenol-associated enzymes (PAZymes). Bacteria possessing the required PAZymes can improve their fitness by the metabolic utilization of a given polyphenol and thrive within the (poly)phenol-enriched gut environment (green arrow). This leads to the production of primary bioactive metabolites (M1) that can be involved in trophic interactions and further be metabolized by other microorganisms possessing the appropriated PAZymes, releasing secondary metabolites (M2), and so on. PAZymes can degrade (poly)phenols without conferring any direct advantage to the bacteria (light blue arrow) in a detoxifying-like effect. The residual product can become a primary bioactive metabolite (M1) and enter the trophic network. Other bacteria may not possess PAZymes required for the degradation of a specific (poly)phenolic compound but can instead resist its antimicrobial effect (dark blue arrow). This confers those strains an advantage to take over sensitive competitors. First and second transformers and resistant bacteria, as well as (poly)phenol metabolites (M1 and M2), can modulate the intestinal ecological niche and induce local and systemic beneficial effects by interacting with the host (gray arrows). In turn, those metabolic changes can also modulate the intestinal ecological niche (orange arrow) in a crosstalk fashion. (Poly)phenol metabolites (M1 and M2) can also induce an antimicrobial effect (red lines). The yellowish area highlights (poly)phenol prebiotic path process, while the red dashed square identifies the antimicrobial effect of (poly)phenol, both defining the duplibiotic effect.
Studies on microbiota-(poly)phenols interaction render insights into targeted mechanisms that can be modified with dietary approaches to counteract gut dysbiosis-associated diseases. Advances in in-vitro experimental colonic fermentation have helped to unveil the physiological roles, substrate preferences, and metabolites exchanges of key taxonomic members of the gut microbiota induced by (poly)phenols. In fact, analyses of bioactive phenolic metabolites generated from the action of bacterial PAZymes can predict and allow deciphering microbes-host crosstalk, underpinning the duplibiotic nature of dietary (poly)phenols in humans and animals.
In the future, the concept of duplibiotic could be extended to other compounds. Plants produce over 100,000 secondary metabolites intended—among other functions—to interact with their own microbiota (272). Thus, it would not be surprising to find in humans' omnivorous diet other phytochemicals which could exert antimicrobial effect or be metabolized by the human gut microbiota. These interactions between the plant molecules and the human gut microbiota can impact both, the microbiota composition, and its activity, as well as the human health itself. Already, several plant alkaloids—berberin (273, 274) and betalain (273, 274)—have been shown to exert antimicrobial and prebiotic-like actions and could be considered duplibiotics.
MR-D, EP-M, and JL-M have been responsible for the conception of this article and drafting the manuscript. JL-M and MR-D created the figures. EP-M and JL-M constructed the table. DR, YD, and DG mentored and critically reviewed this work. All authors read and approved the final manuscript.
This work was supported by a Collaborative Research and Development (CRD) program on the development of synergistic combinations of prebiotic polyphenols and probiotic bacteria, by the NSERC Industrial Research Chair (IRC) on the prebiotic effect of polyphenols in fruits and vegetables, and by the international research unit for chemical and biomolecular research on the microbiome and its impact on metabolic health and nutrition MICROMENU—Sentinel North. This manuscript includes content that appears in Rodríguez-Daza thesis 2020, as fulfillment to obtain a Ph.D. degree at Laval University, Quebec, QC, Canada.
MR-D is involved in a project funded by DianaFood. EP-M and JL-M research works are funded by a Collaborative Research and Development Grant (RDC), partly funded by Diana-Food and the Natural Sciences and Engineering Research Council of Canada (NSERC). DG is Scientific and Innovation Director at Diana Nova, part of Symrise Nutrition. YD holds an NSERC-DianaFood Industrial Chair on prebiotic effects of fruit and vegetable polyphenols. DR is a principal investigator within the RDC.
1. Martin CR, Osadchiy V, Kalani A, Mayer EA. The brain-gut-microbiome axis. Cell Mol Gastroenterol Hepatol. (2018) 6:133–48. doi: 10.1016/j.jcmgh.2018.04.003
2. Salem I, Ramser A, Isham N, Ghannoum MA. The gut microbiome as a major regulator of the gut-skin axis. Front Microbiol. (2018) 9:1459. doi: 10.3389/fmicb.2018.01459
3. Grosicki GJ, Fielding RA, Lustgarten MS. Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: biological basis for a gut-muscle axis. Calcif Tissue Int. (2018) 102:433–42. doi: 10.1007/s00223-017-0345-5
4. Xiao H, Kang S. The role of the gut microbiome in energy balance with a focus on the gut-adipose tissue axis. Front Genet. (2020) 11:297. doi: 10.3389/fgene.2020.00297
5. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027. doi: 10.1038/nature05414
6. Liu W, Zhang R, Shu R, Yu J, Li H, Long H, et al. Study of the relationship between microbiome and colorectal cancer susceptibility using 16SrRNA sequencing. Biomed Res Int. (2020) 2020:1–17. doi: 10.1155/2020/7828392
7. Philip P, Sagaspe P, Taillard J, Mandon C, Constans J, Pourtau L, et al. Acute intake of a grape and blueberry polyphenol-rich extract ameliorates cognitive performance in healthy young adults during a sustained cognitive effort. Antioxidants. (2019) 8:650. doi: 10.3390/antiox8120650
8. Alexander M, Turnbaugh PJ. Deconstructing mechanisms of diet-microbiome-immune interactions. Immunity. (2020) 53:264–76. doi: 10.1016/j.immuni.2020.07.015
9. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
10. Landete JM, Curiel JA, Rodríguez H, de las Rivas B, Muñoz R. Aryl glycosidases from Lactobacillus plantarum increase antioxidant activity of phenolic compounds. J Funct Food. (2014) 7:322–9. doi: 10.1016/j.jff.2014.01.028
11. Rodriguez-Castano GP, Dorris MR, Liu X, Bolling BW, Acosta-Gonzalez A, Rey FE. Bacteroides thetaiotaomicron starch utilization promotes quercetin degradation and butyrate production by Eubacterium ramulus. Front Microbiol (2019) 10:1145. doi: 10.3389/fmicb.2019.01145
12. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. (2017) 14:491–502. doi: 10.1038/nrgastro.2017.75
13. Clifford MN. Diet-derived phenols in plasma and tissues and their implications for health. Planta Med. (2004) 70:1103–14. doi: 10.1055/s-2004-835835
14. Masumoto S, Terao A, Yamamoto Y, Mukai T, Miura T, Shoji T. Non-absorbable apple procyanidins prevent obesity associated with gut microbial and metabolomic changes. Sci Rep-uk. (2016) 6:31208. doi: 10.1038/srep31208
15. Moreno-Indias I, Sanchez-Alcoholado L, Perez-Martinez P, Andres-Lacueva C, Cardona F, Tinahones F, et al. Red wine polyphenols modulate fecal microbiota and reduce markers of the metabolic syndrome in obese patients. Food Funct. (2016) 7:1775–87. doi: 10.1039/C5FO00886G
16. Roopchand DE, Carmody RN, Kuhn P, Moskal K, Rojas-Silva P, Turnbaugh PJ, et al. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet–induced metabolic syndrome. Diabetes. (2015) 64:2847–58. doi: 10.2337/db14-1916
17. Anhe FF, Roy D, Pilon G, Dudonne S, Matamoros S, Varin TV, et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. (2015) 64:872–83. doi: 10.1136/gutjnl-2014-307142
18. Liu YC, Li XY, Shen L. Modulation effect of tea consumption on gut microbiota. Appl Microbiol Biotechnol. (2020) 104:981–7. doi: 10.1007/s00253-019-10306-2
19. Rodriguez-Daza MC, Daoust L, Boutkrabt L, Pilon G, Varin T, Dudonne S, et al. Wild blueberry proanthocyanidins shape distinct gut microbiota profile and influence glucose homeostasis and intestinal phenotypes in high-fat high-sucrose fed mice. Sci Rep. (2020) 10:2217. doi: 10.1038/s41598-020-58863-1
20. Gonzalez-Sarrias A, Romo-Vaquero M, Garcia-Villalba R, Cortes-Martin A, Selma MV, Espin JC. The endotoxemia marker lipopolysaccharide-binding protein is reduced in overweight-obese subjects consuming pomegranate extract by modulating the gut microbiota: a randomized clinical trial. Mol Nutr Food Res. (2018) 62:e1800160. doi: 10.1002/mnfr.201800160
21. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473:174–80. doi: 10.1038/nature09944
22. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. (2008) 320:1647–51. doi: 10.1126/science.1155725
23. Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. (2015) 33:496–503. doi: 10.1016/j.tibtech.2015.06.011
24. Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. (2017) 23:859–68. doi: 10.1038/nm.4358
25. Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. (2007) 104:979–84. doi: 10.1073/pnas.0605374104
26. Beaumont M, Goodrich JK, Jackson MA, Yet I, Davenport ER, Vieira-Silva S, et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. (2016) 17:189. doi: 10.1186/s13059-016-1052-7
27. Reveron I, de las Rivas B, Matesanz R, Munoz R, Lopez de Felipe F. Molecular adaptation of Lactobacillus plantarum WCFS1 to gallic acid revealed by genome-scale transcriptomic signature and physiological analysis. Microb Cell Fact. (2015) 14:160. doi: 10.1186/s12934-015-0345-y
28. Filannino P, Gobbetti M, De Angelis M, Di Cagno R. Hydroxycinnamic acids used as external acceptors of electrons: an energetic advantage for strictly heterofermentative lactic acid bacteria. Appl Environ Microbiol. (2014) 80:7574–82. doi: 10.1128/AEM.02413-14
29. Zhu B, Wang X, Li L. Human gut microbiome: the second genome of human body. Protein Cell. (2010) 1:718–25. doi: 10.1007/s13238-010-0093-z
30. Whitman WB, Oren A, Chuvochina M, da Costa MS, Garrity GM, Rainey FA, et al. Proposal of the suffix -ota to denote phyla. Addendum to 'Proposal to include the rank of phylum in the International Code of Nomenclature of Prokaryotes'. Int J Syst Evol Microbiol. (2018) 68:967–9. doi: 10.1099/ijsem.0.002593
31. Graham C, Mullen A, Whelan K. Obesity and the gastrointestinal microbiota: a review of associations and mechanisms. Nutr Rev. (2015) 73:376–85. doi: 10.1093/nutrit/nuv004
32. Chang CJ, Lin TL, Tsai YL, Wu TR, Lai WF, Lu CC, et al. Next generation probiotics in disease amelioration. J Food Drug Anal. (2019) 27:615–22. doi: 10.1016/j.jfda.2018.12.011
33. Shin J, Noh JR, Chang DH, Kim YH, Kim MH, Lee ES, et al. Elucidation of Akkermansia muciniphila probiotic traits driven by mucin depletion. Front Microbiol. (2019) 10:1137. doi: 10.3389/fmicb.2019.01137
34. Rastelli M, Cani PD, Knauf C. The gut microbiome influences host endocrine functions. Endocr Rev. (2019) 40:1271–84. doi: 10.1210/er.2018-00280
35. Tap J, Furet JP, Bensaada M, Philippe C, Roth H, Rabot S, et al. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol. (2015) 17:4954–64. doi: 10.1111/1462-2920.13006
36. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464:59–65. doi: 10.1038/nature08821
37. Williams BA, Grant LJ, Gidley MJ, Mikkelsen D. Gut fermentation of dietary fibres: physico-chemistry of plant cell walls and implications for health. Int J Mol Sci. (2017) 18:2203. doi: 10.3390/ijms18102203
38. McCarville JL, Chen GY, Cuevas VD, Troha K, Ayres JS. Microbiota metabolites in health and disease. Annu Rev Immunol. (2020) 38:147–70. doi: 10.1146/annurev-immunol-071219-125715
39. Delzenne NM, Cani PD, Everard A, Neyrinck AM, Bindels LB. Gut microorganisms as promising targets for the management of type 2 diabetes. Diabetologia. (2015) 58:2206–17. doi: 10.1007/s00125-015-3712-7
40. Holscher HD. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes. (2017) 8:172–84. doi: 10.1080/19490976.2017.1290756
41. Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. (2011) 13:517–26. doi: 10.1016/j.cmet.2011.02.018
42. Yan H, Ajuwon KM. Butyrate modifies intestinal barrier function in IPEC-J2 cells through a selective upregulation of tight junction proteins and activation of the Akt signaling pathway. PLoS ONE. (2017) 12:e0179586. doi: 10.1371/journal.pone.0179586
43. Cortes-Martin A, Selma MV, Tomas-Barberan FA, Gonzalez-Sarrias A, Espin JC. Where to look into the puzzle of polyphenols and health? The postbiotics and gut microbiota associated with human metabotypes. Mol Nutr Food Res. (2020) 64:e1900952. doi: 10.1002/mnfr.201900952
44. Saito Y, Sato T, Nomoto K, Tsuji H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol Ecol. (2018) 94:1–11. doi: 10.1093/femsec/fiy125
45. Jung C, Hugot JP, Barreau F. Peyer's patches: the immune sensors of the intestine. Int J Inflam. (2010) 2010:823710. doi: 10.4061/2010/823710
46. Lavelle EC, Murphy C, O'Neill LA, Creagh EM. The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. (2010) 3:17–28. doi: 10.1038/mi.2009.124
47. Wilkins LJ, Monga M, Miller AW. Defining dysbiosis for a cluster of chronic diseases. Sci Rep. (2019) 9:12918. doi: 10.1038/s41598-019-49452-y
48. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. (2013) 341:1241214. doi: 10.1126/science.1241214
49. Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. (2004) 101:15718–23. doi: 10.1073/pnas.0407076101
50. Sheng K, He S, Sun M, Zhang G, Kong X, Wang J, et al. Synbiotic supplementation containing Bifidobacterium infantis and xylooligosaccharides alleviates dextran sulfate sodium-induced ulcerative colitis. Food Funct. (2020) 11:3964–74. doi: 10.1039/D0FO00518E
51. Fang C, Kim H, Yanagisawa L, Bennett W, Sirven MA, Alaniz RC, et al. Gallotannins and Lactobacillus plantarum WCFS1 mitigate high-fat diet-induced inflammation and induce biomarkers for thermogenesis in adipose tissue in gnotobiotic mice. Mol Nutr Food Res. (2019) 63:e1800937. doi: 10.1002/mnfr.201800937
52. Cano PG, Santacruz A, Trejo FM, Sanz Y. Bifidobacterium CECT 7765 improves metabolic and immunological alterations associated with obesity in high-fat diet-fed mice. Obesity. (2013) 21:2310–21. doi: 10.1002/oby.20330
53. Minami J, Kondo S, Yanagisawa N, Odamaki T, Xiao JZ, Abe F, et al. Oral administration of Bifidobacterium breve B-3 modifies metabolic functions in adults with obese tendencies in a randomised controlled trial. J Nutr Sci. (2015) 4:e17. doi: 10.1017/jns.2015.5
54. Naito Y, Uchiyama K, Takagi T. A next-generation beneficial microbe: Akkermansia muciniphila. J Clin Biochem Nutr (2018) 63:33–5. doi: 10.3164/jcbn.18-57
55. Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Microb Biotechnol. (2019) 12:1109–25. doi: 10.1111/1751-7915.13410
56. Leylabadlo HE, Ghotaslou R, Feizabadi MM, Farajnia S, Moaddab SY, Ganbarov K, et al. The critical role of Faecalibacterium prausnitzii in human health: an overview. Microb Pathog. (2020) 149:104344. doi: 10.1016/j.micpath.2020.104344
57. Ranjan R, Rani A, Finn PW, Perkins DL. Multiomic strategies reveal diversity and important functional aspects of human gut microbiome. Biomed Res Int. (2018) 2018:6074918. doi: 10.1155/2018/6074918
58. Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. (2009) 4:232–41. doi: 10.1038/ismej.2009.112
59. Zeng H, Ishaq SL, Liu Z, Bukowski MR. Colonic aberrant crypt formation accompanies an increase of opportunistic pathogenic bacteria in C57BL/6 mice fed a high-fat diet. J Nutr Biochem. (2018) 54:18–27. doi: 10.1016/j.jnutbio.2017.11.001
60. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. (2009) 457:480–4. doi: 10.1038/nature07540
61. de la Cuesta-Zuluaga J, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, Escobar JS. Gut microbiota is associated with obesity and cardiometabolic disease in a population in the midst of westernization. Sci Rep. (2018) 8:11356. doi: 10.1038/s41598-018-29687-x
62. Singla RK, Dubey AK, Garg A, Sharma RK, Fiorino M, Ameen SM, et al. Natural polyphenols: chemical classification, definition of classes, subcategories, and structures. J AOAC Int. (2019) 102:1397–400. doi: 10.5740/jaoacint.19-0133
63. Durazzo A, Lucarini M, Souto EB, Cicala C, Caiazzo E, Izzo AA, et al. Polyphenols: a concise overview on the chemistry, occurrence, and human health. Phytother Res. (2019) 33:2221–43. doi: 10.1002/ptr.6419
64. van der Lugt B, van Beek AA, Aalvink S, Meijer B, Sovran B, Vermeij WP, et al. Akkermansia muciniphila ameliorates the age-related decline in colonic mucus thickness and attenuates immune activation in accelerated aging Ercc1 (-/Delta7) mice. Immun Ageing. (2019) 16:6. doi: 10.1186/s12979-019-0145-z
65. Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. (2013) 11:61. doi: 10.1186/1741-7007-11-61
66. Collins B, Hoffman J, Martinez K, Grace M, Lila MA, Cockrell C, et al. A polyphenol-rich fraction obtained from table grapes decreases adiposity, insulin resistance and markers of inflammation and impacts gut microbiota in high-fat-fed mice. J Nutr Biochem. (2016) 31:150–65. doi: 10.1016/j.jnutbio.2015.12.021
67. Wan ML, Ling KH, Wang MF, El-Nezami H. Green tea polyphenol epigallocatechin-3-gallate improves epithelial barrier function by inducing the production of antimicrobial peptide pBD-1 and pBD-2 in monolayers of porcine intestinal epithelial IPEC-J2 cells. Mol Nutr Food Res. (2016) 60:1048–58. doi: 10.1002/mnfr.201500992
68. Chambers KF, Day PE, Aboufarrag HT, Kroon PA. Polyphenol effects on cholesterol metabolism via bile acid biosynthesis, CYP7A1: a review. Nutrients. (2019) 11:2588. doi: 10.3390/nu11112588
69. Anhe FF, Nachbar RT, Varin TV, Trottier J, Dudonne S, Le Barz M, et al. Treatment with camu camu (Myrciaria dubia) prevents obesity by altering the gut microbiota and increasing energy expenditure in diet-induced obese mice. Gut. (2019) 68:453–64. doi: 10.1136/gutjnl-2017-315565
70. Pierre JF, Heneghan AF, Feliciano RP, Shanmuganayagam D, Krueger CG, Reed JD, et al. Cranberry proanthocyanidins improve intestinal sIgA during elemental enteral nutrition. JPEN J Parenter Enteral Nutr. (2014) 38:107–14. doi: 10.1177/0148607112473654
71. Firrman J, Liu L, Argoty GA, Zhang L, Tomasula P, Wang M, et al. Analysis of temporal changes in growth and gene expression for commensal gut microbes in response to the polyphenol naringenin. Microbiol Insights. (2018) 11:1178636118775100. doi: 10.1177/1178636118775100
72. Lacombe A, Li RW, Klimis-Zacas D, Kristo AS, Tadepalli S, Krauss E, et al. Lowbush wild blueberries have the potential to modify gut microbiota and xenobiotic metabolism in the rat colon. PLoS ONE. (2013) 8:e67497. doi: 10.1371/journal.pone.0067497
73. Alvarez-Martinez FJ, Barrajon-Catalan E, Encinar JA, Rodriguez-Diaz JC, Micol V. Antimicrobial capacity of plant polyphenols against Gram-positive bacteria: a comprehensive review. Curr Med Chem. (2020) 27:2576–606. doi: 10.2174/0929867325666181008115650
74. Cheng M, Zhang X, Zhu J, Cheng L, Cao J, Wu Z, et al. A metagenomics approach to the intestinal microbiome structure and function in high fat diet-induced obesity mice fed with oolong tea polyphenols. Food Funct. (2018) 9:1079–87. doi: 10.1039/C7FO01570D
75. Daglia M. Polyphenols as antimicrobial agents. Curr Opin Biotechnol. (2012) 23:174–81. doi: 10.1016/j.copbio.2011.08.007
76. Makarewicz M, Drozdz I, Tarko T, Duda-Chodak A. The interactions between polyphenols and microorganisms, especially gut microbiota. Antioxidants. (2021) 10:188. doi: 10.3390/antiox10020188
77. Maisuria VB, Los Santos YL, Tufenkji N, Deziel E. Cranberry-derived proanthocyanidins impair virulence and inhibit quorum sensing of Pseudomonas aeruginosa. Sci Rep (2016) 6:30169. doi: 10.1038/srep30169
78. Vikram A, Jayaprakasha GK, Jesudhasan PR, Pillai SD, Patil BS. Suppression of bacterial cell-cell signalling, biofilm formation and type III secretion system by citrus flavonoids. J Appl Microbiol. (2010) 109:515–27. doi: 10.1111/j.1365-2672.2010.04677.x
79. Smith AH, Zoetendal E, Mackie RI. Bacterial mechanisms to overcome inhibitory effects of dietary tannins. Microb Ecol. (2005) 50:197–205. doi: 10.1007/s00248-004-0180-x
80. Engels C, Knodler M, Zhao YY, Carle R, Ganzle MG, Schieber A. Antimicrobial activity of gallotannins isolated from mango (Mangifera indica L.) kernels. J Agric Food Chem. (2009) 57:7712–8. doi: 10.1021/jf901621m
81. Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. (2013) 152:39–50. doi: 10.1016/j.cell.2012.10.052
82. Firrman J, Liu L, Zhang L, Arango Argoty G, Wang M, Tomasula P, et al. The effect of quercetin on genetic expression of the commensal gut microbes Bifidobacterium catenulatum, Enterococcus caccae and Ruminococcus gauvreauii. Anaerobe (2016) 42:130–41. doi: 10.1016/j.anaerobe.2016.10.004
83. Wu VC, Qiu X, de los Reyes BG, Lin CS, Pan Y. Application of cranberry concentrate (Vaccinium macrocarpon) to control Escherichia coli O157:H7 in ground beef and its antimicrobial mechanism related to the downregulated slp, hdeA and cfa. Food Microbiol. (2009) 26:32–8. doi: 10.1016/j.fm.2008.07.014
84. Das Q, Lepp D, Yin X, Ross K, McCallum JL, Warriner K, et al. Transcriptional profiling of Salmonella enterica serovar Enteritidis exposed to ethanolic extract of organic cranberry pomace. PLoS ONE. (2019) 14:e0219163. doi: 10.1371/journal.pone.0219163
85. Vadekeetil A, Alexandar V, Chhibber S, Harjai K. Adjuvant effect of cranberry proanthocyanidin active fraction on antivirulent property of ciprofloxacin against Pseudomonas aeruginosa. Microb Pathog (2016) 90:98–103. doi: 10.1016/j.micpath.2015.11.024
86. Ulrey RK, Barksdale SM, Zhou W, van Hoek ML. Cranberry proanthocyanidins have anti-biofilm properties against Pseudomonas aeruginosa. BMC Complement Altern Med (2014) 14:499. doi: 10.1186/1472-6882-14-499
87. Sanchez-Patan F, Tabasco R, Monagas M, Requena T, Pelaez C, Moreno-Arribas MV, et al. Capability of Lactobacillus plantarum IFPL935 to catabolize flavan-3-ol compounds and complex phenolic extracts. J Agric Food Chem. (2012) 60:7142–51. doi: 10.1021/jf3006867
88. Takagaki A, Nanjo F. Biotransformation of (–)-epicatechin, (+)-epicatechin, (–)-catechin, and (+)-catechin by intestinal bacteria involved in isoflavone metabolism. Biol Pharm Bull. (2015) 80:1–4. doi: 10.1080/09168451.2015.1079480
89. Takagaki A, Kato Y, Nanjo F. Isolation and characterization of rat intestinal bacteria involved in biotransformation of (-)-epigallocatechin. Arch Microbiol. (2014) 196:681–95. doi: 10.1007/s00203-014-1006-y
90. Pacheco-Ordaz R, Wall-Medrano A, Goni MG, Ramos-Clamont-Montfort G, Ayala-Zavala JF, Gonzalez-Aguilar GA. Effect of phenolic compounds on the growth of selected probiotic and pathogenic bacteria. Lett Appl Microbiol. (2018) 66:25–31. doi: 10.1111/lam.12814
91. Li S, Chen L, Yang T, Wu Q, Lv Z, Xie B, et al. Increasing antioxidant activity of procyanidin extracts from the pericarp of Litchi chinensis processing waste by two probiotic bacteria bioconversions. J Agric Food Chem. (2013) 61:2506–12. doi: 10.1021/jf305213e
92. Dey P, Olmstead BD, Sasaki GY, Vodovotz Y, Yu Z, Bruno RS. Epigallocatechin gallate but not catechin prevents nonalcoholic steatohepatitis in mice similar to green tea extract while differentially affecting the gut microbiota. J Nutr Biochem. (2020) 84:108455. doi: 10.1016/j.jnutbio.2020.108455
93. Selma MV, Beltran D, Garcia-Villalba R, Espin JC, Tomas-Barberan FA. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. (2014) 5:1779–84. doi: 10.1039/C4FO00092G
94. Beltran D, Romo-Vaquero M, Espin JC, Tomas-Barberan FA, Selma MV. Ellagibacter isourolithinifaciens gen. nov., sp. nov., a new member of the family Eggerthellaceae, isolated from human gut. Int J Syst Evol Microbiol. (2018) 68:1707–12. doi: 10.1099/ijsem.0.002735
95. Gaya P, Peirotén Á, Medina M, Álvarez I, Landete JM. Bifidobacterium pseudocatenulatum INIA P815: The first bacterium able to produce urolithins A and B from ellagic acid. J Funct Food. (2018) 45:95–9. doi: 10.1016/j.jff.2018.03.040
96. Bialonska D, Kasimsetty SG, Schrader KK, Ferreira D. The effect of pomegranate (Punica granatum L.) byproducts and ellagitannins on the growth of human gut bacteria. J Agric Food Chem. (2009) 57:8344–9. doi: 10.1021/jf901931b
97. Anitha P, Priyadarsini RV, Kavitha K, Thiyagarajan P, Nagini S. Ellagic acid coordinately attenuates Wnt/beta-catenin and NF-kappaB signaling pathways to induce intrinsic apoptosis in an animal model of oral oncogenesis. Eur J Nutr. (2013) 52:75–84. doi: 10.1007/s00394-011-0288-y
98. Atessahin A, Ceribasi AO, Yuce A, Bulmus O, Cikim G. Role of ellagic acid against cisplatin-induced nephrotoxicity and oxidative stress in rats. Basic Clin Pharmacol Toxicol. (2007) 100:121–6. doi: 10.1111/j.1742-7843.2007.00060.x
99. Kelly SM, O'Callaghan J, Kinsella M, van Sinderen D. Characterisation of a hydroxycinnamic acid esterase from the Bifidobacterium longum subsp. longum taxon. Front Microbiol. (2018) 9:2690. doi: 10.3389/fmicb.2018.02690
100. Fritsch C, Jansch A, Ehrmann MA, Toelstede S, Vogel RF. Characterization of cinnamoyl esterases from different Lactobacilli and Bifidobacteria. Curr Microbiol. (2017) 74:247–56. doi: 10.1007/s00284-016-1182-x
101. Esteban-Torres M, Reveron I, Santamaria L, Mancheno JM, de Las Rivas B, Munoz R. The Lp_3561 and Lp_3562 enzymes support a functional divergence process in the lipase/esterase toolkit from Lactobacillus plantarum. Front Microbiol (2016) 7:1118. doi: 10.3389/fmicb.2016.01118
102. Liu Z, Ma Z, Zhang H, Summah BS, Liu H, An D, et al. Ferulic acid increases intestinal Lactobacillus and improves cardiac function in TAC mice. Biomed Pharmacother. (2019) 120:109482. doi: 10.1016/j.biopha.2019.109482
103. Osawa R, Kuroiso K, Goto S, Shimizu A. Isolation of tannin-degrading lactobacilli from humans and fermented foods. Appl Environ Microbiol. (2000) 66:3093–7. doi: 10.1128/AEM.66.7.3093-3097.2000
104. Braune A, Gutschow M, Engst W, Blaut M. Degradation of quercetin and luteolin by Eubacterium ramulus. Appl Environ Microbiol (2001) 67:5558–67. doi: 10.1128/AEM.67.12.5558-5567.2001
105. Doan KV, Ko CM, Kinyua AW, Yang DJ, Choi YH, Oh IY, et al. Gallic acid regulates body weight and glucose homeostasis through AMPK activation. Endocrinology. (2015) 156:157–68. doi: 10.1210/en.2014-1354
106. Li Y, Xie Z, Gao T, Li L, Chen Y, Xiao D, et al. A holistic view of gallic acid-induced attenuation in colitis based on microbiome-metabolomics analysis. Food Funct. (2019) 10:4046–61. doi: 10.1039/C9FO00213H
107. Ulbrich K, Reichardt N, Braune A, Kroh LW, Blaut M, Rohn S. The microbial degradation of onion flavonol glucosides and their roasting products by the human gut bacteria Eubacterium ramulus and Flavonifractor plautii. Food Research International (2015) 67:349–55. doi: 10.1016/j.foodres.2014.11.051
108. Neyrinck AM, Van Hee VF, Bindels LB, De Backer F, Cani PD, Delzenne NM. Polyphenol-rich extract of pomegranate peel alleviates tissue inflammation and hypercholesterolaemia in high-fat diet-induced obese mice: potential implication of the gut microbiota. Br J Nutr. (2013) 109:802–9. doi: 10.1017/S0007114512002206
109. Mueller M, Zartl B, Schleritzko A, Stenzl M, Viernstein H, Unger FM. Rhamnosidase activity of selected probiotics and their ability to hydrolyse flavonoid rhamnoglucosides. Bioprocess Biosyst Eng. (2018) 41:221–8. doi: 10.1007/s00449-017-1860-5
110. Alapont CdG, Garcia-Domenech R, Gálvez J, Ros MJ, Wolski S, Garcia MD. Molecular topology: a useful tool for the search of new antibacterials. Bioorganic Med Chem Lett. (2000) 10:2033–6. doi: 10.1016/S0960-894X(00)00406-6
111. Yi Z, Yu Y, Liang Y, Zeng B. In vitro antioxidant and antimicrobial activities of the extract of Pericarpium Citri Reticulatae of a new Citrus cultivar and its main flavonoids. Lwt - Food Sci Technol. (2008) 41:597–603. doi: 10.1016/j.lwt.2007.04.008
112. Gwiazdowska D, Jus K, Jasnowska-Malecka J, Kluczynska K. The impact of polyphenols on Bifidobacterium growth. Acta Biochim Pol. (2015) 62:895–901. doi: 10.18388/abp.2015_1154
113. Duda-Chodak A. The inhibitory effect of polyphenols on human gut microbiota. J Physiol Pharmacol. (2012) 63:497–503.
114. Guirro M, Costa A, Gual-Grau A, Mayneris-Perxachs J, Torrell H, Herrero P, et al. Multi-omics approach to elucidate the gut microbiota activity: Metaproteomics and metagenomics connection. Electrophoresis. (2018) 39:1692–701. doi: 10.1002/elps.201700476
115. Guo K, Ren J, Gu G, Wang G, Gong W, Wu X, et al. Hesperidin protects against intestinal inflammation by restoring intestinal barrier function and up-regulating Treg cells. Mol Nutr Food Res. (2019) 63:e1800975. doi: 10.1002/mnfr.201800975
116. Estruel-Amades S, Massot-Cladera M, Perez-Cano FJ, Franch A, Castell M, Camps-Bossacoma M. Hesperidin effects on gut microbiota and gut-associated lymphoid tissue in healthy rats. Nutrients. (2019) 11:324. doi: 10.3390/nu11020324
117. Guirro M, Gual-Grau A, Gibert-Ramos A, Alcaide-Hidalgo JM, Canela N, Arola L, et al. Metabolomics elucidates dose-dependent molecular beneficial effects of hesperidin supplementation in rats fed an obesogenic diet. Antioxidants. (2020) 9:79. doi: 10.3390/antiox9010079
118. Zhang Z, Peng X, Li S, Zhang N, Wang Y, Wei H. Isolation and identification of quercetin degrading bacteria from human fecal microbes. PLoS ONE. (2014) 9:e90531. doi: 10.1371/journal.pone.0090531
119. Porras D, Nistal E, Martinez-Florez S, Pisonero-Vaquero S, Olcoz JL, Jover R, et al. Protective effect of quercetin on high-fat diet-induced non-alcoholic fatty liver disease in mice is mediated by modulating intestinal microbiota imbalance and related gut-liver axis activation. Free Radic Biol Med. (2017) 102:188–202. doi: 10.1016/j.freeradbiomed.2016.11.037
120. Tu P, Bian X, Chi L, Gao B, Ru H, Knobloch TJ, et al. Characterization of the functional changes in mouse gut microbiome associated with increased Akkermansia muciniphila population modulated by dietary black raspberries. ACS Omega. (2018) 3:10927–37. doi: 10.1021/acsomega.8b00064
121. Ushiroda C, Naito Y, Takagi T, Uchiyama K, Mizushima K, Higashimura Y, et al. Green tea polyphenol (epigallocatechin-3-gallate) improves gut dysbiosis and serum bile acids dysregulation in high-fat diet-fed mice. J Clin Biochem Nutr. (2019) 65:34–46. doi: 10.3164/jcbn.18-116
122. Boto-Ordóñez M, Urpi-Sarda M, Queipo-Ortuño M, Tulipani S, Tinahones FJ, Andres-Lacueva C. High levels of Bifidobacteria are associated with increased levels of anthocyanin microbial metabolites: a randomized clinical trial. Food Funct. (2014) 5:1932–8. doi: 10.1039/C4FO00029C
123. Gomez-Gallego C, Pohl S, Salminen S, De Vos WM, Kneifel W. Akkermansia muciniphila: a novel functional microbe with probiotic properties. Benef Microb. (2016) 7:571–84. doi: 10.3920/BM2016.0009
124. Percival RS, Devine DA, Duggal MS, Chartron S, Marsh PD. The effect of cocoa polyphenols on the growth, metabolism, and biofilm formation by Streptococcus mutans and Streptococcus sanguinis. Eur J Oral Sci (2006) 114:343–8. doi: 10.1111/j.1600-0722.2006.00386.x
125. Yamanaka A, Kimizuka R, Kato T, Okuda K. Inhibitory effects of cranberry juice on attachment of oral streptococci and biofilm formation. Oral Microbiol Immunol. (2004) 19:150–4. doi: 10.1111/j.0902-0055.2004.00130.x
126. Zheng J, Wittouck S, Salvetti E, Franz C, Harris HMB, Mattarelli P, et al. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol (2020) 70:2782–858. doi: 10.1099/ijsem.0.004107
127. Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. (1995) 125:1401–12. doi: 10.1093/jn/125.6.1401
128. Lebeer S, Vanderleyden J, De Keersmaecker SC. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. (2008) 72:728–64, Table of Contents. doi: 10.1128/MMBR.00017-08
129. O'Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. (2016) 7:925. doi: 10.3389/fmicb.2016.00925
130. Trosvik P, de Muinck EJ. Ecology of bacteria in the human gastrointestinal tract–identification of keystone and foundation taxa. Microbiome. (2015) 3:44. doi: 10.1186/s40168-015-0107-4
131. Banerjee S, Schlaeppi K, van der Heijden MGA. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. (2018) 16:567–76. doi: 10.1038/s41579-018-0024-1
132. Jimenez N, Curiel JA, Reveron I, de Las Rivas B, Munoz R. Uncovering the Lactobacillus plantarum WCFS1 gallate decarboxylase involved in tannin degradation. Appl Environ Microbiol. (2013) 79:4253–63. doi: 10.1128/AEM.00840-13
133. de Las Rivas B, Rodriguez H, Anguita J, Munoz R. Bacterial tannases: classification and biochemical properties. Appl Microbiol Biotechnol. (2019) 103:603–23. doi: 10.1007/s00253-018-9519-y
134. Tabasco R, Sánchez-Patán F, Monagas M, Bartolomé B, Moreno-Arribas VM, Peláez C, et al. Effect of grape polyphenols on lactic acid bacteria and bifidobacteria growth: resistance and metabolism. Food Microbiol. (2011) 28:1345–52. doi: 10.1016/j.fm.2011.06.005
135. Fritsch C, Heinrich V, Vogel RF, Toelstede S. Phenolic acid degradation potential and growth behavior of lactic acid bacteria in sunflower substrates. Food Microbiol. (2016) 57:178–86. doi: 10.1016/j.fm.2016.03.003
136. Filannino P, Di Cagno R, Gobbetti M. Metabolic and functional paths of lactic acid bacteria in plant foods: get out of the labyrinth. Curr Opin Biotechnol. (2018) 49:64–72. doi: 10.1016/j.copbio.2017.07.016
137. Kawabata K, Yoshioka Y, Terao J. Role of intestinal microbiota in the bioavailability and physiological functions of dietary polyphenols. Molecules. (2019) 24:370. doi: 10.3390/molecules24020370
138. Fetzner S. Ring-cleaving dioxygenases with a cupin fold. Appl Environ Microbiol. (2012) 78:2505–14. doi: 10.1128/AEM.07651-11
139. Rodriguez-Daza MC, Roquim M, Dudonne S, Pilon G, Levy E, Marette A, et al. Berry polyphenols and fibers modulate distinct microbial metabolic functions and gut microbiota enterotype-like clustering in obese mice. Front Microbiol. (2020) 11:2032. doi: 10.3389/fmicb.2020.02032
140. Braune A, Blaut M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes. (2016) 7:216–34. doi: 10.1080/19490976.2016.1158395
141. Takagaki A, Bulletin N-F. Bioconversion of (–)-epicatechin, (+)-epicatechin, (–)-catechin, and (+)-catechin by (–)-epigallocatechin-metabolizing bacteria. Biol Pharm Bull. (2015) 38:789–94. doi: 10.1248/bpb.b14-00813
142. Vendrame S, Guglielmetti S, Riso P, Arioli S, Klimis-Zacas D, Porrini M. Six-week consumption of a wild blueberry powder drink increases bifidobacteria in the human gut. J Agric Food Chem. (2011) 59:12815–20. doi: 10.1021/jf2028686
143. Reveron I, Rodriguez H, Campos G, Curiel JA, Ascaso C, Carrascosa AV, et al. Tannic acid-dependent modulation of selected Lactobacillus plantarum traits linked to gastrointestinal survival. PLoS ONE. (2013) 8:e66473. doi: 10.1371/journal.pone.0066473
144. Hervert-Hernandez D, Pintado C, Rotger R, Goni I. Stimulatory role of grape pomace polyphenols on Lactobacillus acidophilus growth. Int J Food Microbiol. (2009) 136:119–22. doi: 10.1016/j.ijfoodmicro.2009.09.016
145. Reveron I, Plaza-Vinuesa L, Franch M, de Las Rivas B, Munoz R, Lopez de Felipe F. Transcriptome-based analysis in Lactobacillus plantarum WCFS1 reveals new insights into resveratrol effects at system level. Mol Nutr Food Res. (2018) 62:e1700992. doi: 10.1002/mnfr.201700992
146. Plaza-Vinuesa L, Hernandez-Hernandez O, Moreno FJ, de Las Rivas B, Munoz R. Unravelling the diversity of glycoside hydrolase family 13 alpha-amylases from Lactobacillus plantarum WCFS1. Microb Cell Fact. (2019) 18:183. doi: 10.1186/s12934-019-1237-3
147. Siezen R, Boekhorst J, Muscariello L, Molenaar D, Renckens B, Kleerebezem M. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics. (2006) 7:126. doi: 10.1186/1471-2164-7-126
148. Avila M, Jaquet M, Moine D, Requena T, Pelaez C, Arigoni F, et al. Physiological and biochemical characterization of the two alpha-L-rhamnosidases of Lactobacillus plantarum NCC245. Microbiology. (2009) 155:2739–49. doi: 10.1099/mic.0.027789-0
149. Barroso E, Sanchez-Patan F, Martin-Alvarez PJ, Bartolome B, Moreno-Arribas MV, Pelaez C, et al. Lactobacillus plantarum IFPL935 favors the initial metabolism of red wine polyphenols when added to a colonic microbiota. J Agric Food Chem. (2013) 61:10163–72. doi: 10.1021/jf402816r
150. Filannino P, Di Cagno R, Crecchio C, De Virgilio C, De Angelis M, Gobbetti M. Transcriptional reprogramming and phenotypic switching associated with the adaptation of Lactobacillus plantarum C2 to plant niches. Sci Rep. (2016) 6:27392. doi: 10.1038/srep27392
151. Vaquero I, Marcobal A, Munoz R. Tannase activity by lactic acid bacteria isolated from grape must and wine. Int J Food Microbiol. (2004) 96:199–204. doi: 10.1016/j.ijfoodmicro.2004.04.004
152. Nishitani Y, Sasaki E, Fujisawa T, Osawa R. Genotypic analyses of lactobacilli with a range of tannase activities isolated from human feces and fermented foods. Syst Appl Microbiol. (2004) 27:109–17. doi: 10.1078/0723-2020-00262
153. Lopez de Felipe F, de Las Rivas B, Munoz R. Bioactive compounds produced by gut microbial tannase: implications for colorectal cancer development. Front Microbiol. (2014) 5:684. doi: 10.3389/fmicb.2014.00684
154. Jimenez N, Esteban-Torres M, Mancheno JM, de Las Rivas B, Munoz R. Tannin degradation by a novel tannase enzyme present in some Lactobacillus plantarum strains. Appl Environ Microbiol. (2014) 80:2991–7. doi: 10.1128/AEM.00324-14
155. Barnes RC, Kim H, Fang C, Bennett W, Nemec M, Sirven MA, et al. Body massindex as a determinant of systemic exposure to gallotannin metabolites during 6-week consumption of mango (Mangifera indica L.) and modulation of intestinal microbiota in lean and obese individuals. Mol Nutr Food Res. (2019) 63:e1800512. doi: 10.1002/mnfr.201800512
156. Nemec MJ, Kim H, Marciante AB, Barnes RC, Talcott ST, Mertens-Talcott SU. Pyrogallol, an absorbable microbial gallotannins-metabolite and mango polyphenols (Mangifera Indica L.) suppress breast cancer ductal carcinoma in situ proliferation in vitro Food Funct. (2016) 7:3825–33. doi: 10.1039/C6FO00636A
157. Fang C, Kim H, Noratto G, Sun Y, Talcott ST, Mertens-Talcott SU. Gallotannin derivatives from mango (Mangifera indica L.) suppress adipogenesis and increase thermogenesis in 3T3-L1 adipocytes in part through the AMPK pathway. J Funct Food. (2018) 46:101–9. doi: 10.1016/j.jff.2018.04.043
158. Esteban-Torres M, Santamaria L, Cabrera-Rubio R, Plaza-Vinuesa L, Crispie F, de Las Rivas B, et al. A diverse range of human gut bacteria have the potential to metabolize the dietary component gallic acid. Appl Environ Microbiol. (2018) 84:e01558–18. doi: 10.1128/AEM.01558-18
159. Hunt CJ, Antonopoulou I, Tanksale A, Rova U, Christakopoulos P, Haritos VS. Insights into substrate binding of ferulic acid esterases by arabinose and methyl hydroxycinnamate esters and molecular docking. Sci Rep. (2017) 7:17315. doi: 10.1038/s41598-017-17260-x
160. Wu H, Li H, Xue Y, Luo G, Gan L, Liu J, et al. High efficiency co-production of ferulic acid and xylooligosaccharides from wheat bran by recombinant xylanase and feruloyl esterase. Biochem Eng J. (2017) 120:41–8. doi: 10.1016/j.bej.2017.01.001
161. Patras MA, Jaiswal R, McDougall GJ, Kuhnert N. Profiling and quantification of regioisomeric caffeoyl glucoses in berry fruits. J Agric Food Chem. (2018) 66:1096–104. doi: 10.1021/acs.jafc.7b02446
162. Aguirre Santos EA, Schieber A, Weber F. Site-specific hydrolysis of chlorogenic acids by selected Lactobacillus species. Food Res Int. (2018) 109:426–32. doi: 10.1016/j.foodres.2018.04.052
163. Esteban-Torres M, Landete JM, Reveron I, Santamaria L, de las Rivas B, Munoz R. A Lactobacillus plantarum esterase active on a broad range of phenolic esters. Appl Environ Microbiol. (2015) 81:3235–42. doi: 10.1128/AEM.00323-15
164. Xu Z, He H, Zhang S, Guo T, Kong J. Characterization of feruloyl esterases produced by the four Lactobacillus Species: L. amylovorus, L. acidophilus, L. farciminis and L. fermentum, isolated from ensiled corn stover. Front Microbiol. (2017) 8:941. doi: 10.3389/fmicb.2017.00941
165. Hole AS, Rud I, Grimmer S, Sigl S, Narvhus J, Sahlstrom S. Improved bioavailability of dietary phenolic acids in whole grain barley and oat groat following fermentation with probiotic Lactobacillus acidophilus, Lactobacillus johnsonii, and Lactobacillus reuteri. J Agric Food Chem (2012) 60:6369–75. doi: 10.1021/jf300410h
166. Russo M, Fabersani E, Abeijon-Mukdsi MC, Ross R, Fontana C, Benitez-Paez A, et al. ctobacillus fermentum CRL1446 ameliorates oxidative and metabolic parameters by increasing intestinal feruloyl esterase activity and modulating microbiota in caloric-restricted mice. Nutrients. (2016) 8:415. doi: 10.3390/nu8070415
167. Andreasen MF, Kroon PA, Williamson G, Garcia-Conesa MT. Esterase activity able to hydrolyze dietary antioxidant hydroxycinnamates is distributed along the intestine of mammals. J Agric Food Chem. (2001) 49:5679–84. doi: 10.1021/jf010668c
168. Day AJ, DuPont MS, Ridley S, Rhodes M, Rhodes MJC, Morgan MRA, et al. Deglycosylation of flavonoid and isoflavonoid glycosides by human small intestine and liver β-glucosidase activity. Febs Lett. (1998) 436:71–5. doi: 10.1016/S0014-5793(98)01101-6
169. Duncan SH, Russell WR, Quartieri A, Rossi M, Parkhill J, Walker AW, et al. Wheat bran promotes enrichment within the human colonic microbiota of butyrate-producing bacteria that release ferulic acid. Environ Microbiol. (2016) 18:2214–25. doi: 10.1111/1462-2920.13158
170. Riva A, Kolimar D, Spittler A, Wisgrill L, Herbold CW, Abranko L, et al. Conversion of rutin, a prevalent dietary llavonol, by the human gut microbiota. Front Microbiol. (2020) 11:585428. doi: 10.3389/fmicb.2020.585428
171. Amaretti A, Raimondi S, Leonardi A, Quartieri A, Rossi M. Hydrolysis of the rutinose-conjugates flavonoids rutin and hesperidin by the gut microbiota and bifidobacteria. Nutrients. (2015) 7:2788–800. doi: 10.3390/nu7042788
172. Konishi Y, Shimizu M. Transepithelial transport of ferulic acid by monocarboxylic acid transporter in Caco-2 cell monolayers. Biosci Biotechnol Biochem. (2003) 67:856–62. doi: 10.1271/bbb.67.856
173. Baeza G, Bachmair EM, Wood S, Mateos R, Bravo L, de Roos B. The colonic metabolites dihydrocaffeic acid and dihydroferulic acid are more effective inhibitors of in vitro platelet activation than their phenolic precursors. Food Funct. (2017) 8:1333–42. doi: 10.1039/C6FO01404F
174. Narasimhan A, Chinnaiyan M, Karundevi B. Ferulic acid exerts its antidiabetic effect by modulating insulin-signalling molecules in the liver of high-fat diet and fructose-induced type-2 diabetic adult male rat. Appl Physiol Nutr Metab. (2015) 40:769–81. doi: 10.1139/apnm-2015-0002
175. Senaphan K, Kukongviriyapan U, Sangartit W, Pakdeechote P, Pannangpetch P, Prachaney P, et al. Ferulic acid alleviates changes in a rat model of metabolic syndrome induced by high-carbohydrate, high-fat diet. Nutrients. (2015) 7:6446–64. doi: 10.3390/nu7085283
176. Mancuso C, Santangelo R. Ferulic acid: pharmacological and toxicological aspects. Food Chem Toxicol. (2014) 65:185–95. doi: 10.1016/j.fct.2013.12.024
177. Gomez-Juaristi M, Martinez-Lopez S, Sarria B, Bravo L, Mateos R. Bioavailability of hydroxycinnamates in an instant green/roasted coffee blend in humans. identification of novel colonic metabolites. Food Funct. (2018) 9:331–43. doi: 10.1039/C7FO01553D
178. Santamaria L, Reveron I, Lopez de Felipe F, de Las Rivas B, Munoz R. Unravelling the reduction pathway as an alternative metabolic route to hydroxycinnamate decarboxylation in Lactobacillus plantarum. Appl Environ Microbiol (2018) 84:e01123–18. doi: 10.1128/AEM.01123-18
179. Krga I, Monfoulet LE, Konic-Ristic A, Mercier S, Glibetic M, Morand C, et al. Anthocyanins and their gut metabolites reduce the adhesion of monocyte to TNFalpha-activated endothelial cells at physiologically relevant concentrations. Arch Biochem Biophys. (2016) 599:51–9. doi: 10.1016/j.abb.2016.02.006
180. Mantziari A, Tölkkö S, Ouwehand AC, Löyttyniemi E, Isolauri E, Salminen S, et al. The effect of donor human milk fortification on the adhesion of probiotics in vitro. Nutrients. (2020) 12:182. doi: 10.3390/nu12010182
181. Nielsen IL, Chee WS, Poulsen L, Offord-Cavin E, Rasmussen SE, Frederiksen H, et al. Bioavailability is improved by enzymatic modification of the citrus flavonoid hesperidin in humans: a randomized, double-blind, crossover trial. J Nutr. (2006) 136:404–8. doi: 10.1093/jn/136.2.404
182. Michlmayr H, Kneifel W. beta-Glucosidase activities of lactic acid bacteria: mechanisms, impact on fermented food and human health. FEMS Microbiol Lett. (2014) 352:1–10. doi: 10.1111/1574-6968.12348
183. Day AJ, Cañada FJ, Diaz JC, Kroon PA, McLauchlan R, Faulds CB, et al. Dietary flavonoid and isoflavone glycosides are hydrolysed by the lactase site of lactase phlorizin hydrolase. Febs Lett. (2000) 468:166–70. doi: 10.1016/S0014-5793(00)01211-4
184. Lancon A, Delmas D, Osman H, Thenot JP, Jannin B, Latruffe N. Human hepatic cell uptake of resveratrol: involvement of both passive diffusion and carrier-mediated process. Biochem Biophys Res Commun. (2004) 316:1132–7. doi: 10.1016/j.bbrc.2004.02.164
185. Wei B, Wang YK, Qiu WH, Wang SJ, Wu YH, Xu XW, et al. Discovery and mechanism of intestinal bacteria in enzymatic cleavage of C-C glycosidic bonds. Appl Microbiol Biotechnol. (2020) 104:1883–90. doi: 10.1007/s00253-019-10333-z
186. Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol. (2008) 66:487–95. doi: 10.1111/j.1574-6941.2008.00520.x
187. Hur HG, Lay JO Jr, Beger RD, Freeman JP, Rafii F. Isolation of human intestinal bacteria metabolizing the natural isoflavone glycosides daidzin and genistin. Arch Microbiol. (2000) 174:422–8. doi: 10.1007/s002030000222
188. Zyzelewicz D, Bojczuk M, Budryn G, Jurgonski A, Zdunczyk Z, Juskiewicz J, et al. Influence of diet enriched with cocoa bean extracts on physiological Indices of laboratory rats. Molecules. (2019) 24:825. doi: 10.3390/molecules24050825
189. Heng Y, Kim MJ, Yang HJ, Kang S, Park S. Lactobacillus intestinalis efficiently produces equol from daidzein and chungkookjang, short-term fermented soybeans. Arch Microbiol. (2019) 201:1009–17. doi: 10.1007/s00203-019-01665-5
190. Chen LR, Ko NY, Chen KH. Isoflavone supplements for menopausal women: a systematic review. Nutrients. (2019) 11:2649. doi: 10.3390/nu11112649
191. Toh H, Oshima K, Suzuki T, Hattori M, Morita H. Complete genome sequence of the equol-producing bacterium Adlercreutzia equolifaciens DSM 19450T. Genome Announc. (2013) 1:e00742–13. doi: 10.1128/genomeA.00742-13
192. Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol. (2008) 58:1221–7. doi: 10.1099/ijs.0.65404-0
193. Ishiwata N, Melby MK, Mizuno S, Watanabe S. New equol supplement for relieving menopausal symptoms: randomized, placebo-controlled trial of Japanese women. Menopause. (2009) 16:141–8. doi: 10.1097/gme.0b013e31818379fa
194. Alakomi HL, Puupponen-Pimia R, Aura AM, Helander IM, Nohynek L, Oksman-Caldentey KM, et al. Weakening of salmonella with selected microbial metabolites of berry-derived phenolic compounds and organic acids. J Agric Food Chem. (2007) 55:3905–12. doi: 10.1021/jf070190y
195. Saha P, Yeoh BS, Singh R, Chandrasekar B, Vemula PK, Haribabu B, et al. Gut microbiota conversion of dietary ellagic acid into bioactive phytoceutical urolithin A inhibits heme peroxidases. PLoS ONE. (2016) 11:e0156811. doi: 10.1371/journal.pone.0156811
196. Espin JC, Gonzalez-Sarrias A, Tomas-Barberan FA. The gut microbiota: a key factor in the therapeutic effects of (poly)phenols. Biochem Pharmacol. (2017) 139:82–93. doi: 10.1016/j.bcp.2017.04.033
197. Rocchetti G, Bhumireddy SR, Giuberti G, Mandal R, Lucini L, Wishart DS. Edible nuts deliver polyphenols and their transformation products to the large intestine: an in vitro fermentation model combining targeted/untargeted metabolomics. Food Res Int. (2019) 116:786–94. doi: 10.1016/j.foodres.2018.09.012
198. Cho GS, Ritzmann F, Eckstein M, Huch M, Briviba K, Behsnilian D, et al. Quantification of Slackia and Eggerthella spp. in human feces and adhesion of representatives strains to caco-2 cells. Front Microbiol. (2016) 7:658. doi: 10.3389/fmicb.2016.00658
199. Gupta RS, Chen WJ, Adeolu M, Chai Y. Molecular signatures for the class Coriobacteriia and its different clades; proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov Int J Syst Evol Microbiol. (2013) 63:3379–97. doi: 10.1099/ijs.0.048371-0
200. Mele L, Carobbio S, Brindani N, Curti C, Rodriguez-Cuenca S, Bidault G, et al. Phenyl-gamma-valerolactones, flavan-3-ol colonic metabolites, protect brown adipocytes fromoxidative stress without affecting their differentiation or function. Mol Nutr Food Res. (2017) 61:1700074. doi: 10.1002/mnfr.201700074
201. Mena P, Bresciani L, Brindani N, Ludwig IA, Pereira-Caro G, Angelino D, et al. Phenyl-gamma-valerolactones and phenylvaleric acids, the main colonic metabolites of flavan-3-ols: synthesis, analysis, bioavailability, and bioactivity. Nat Prod Rep. (2019) 36:714–52. doi: 10.1039/C8NP00062J
202. Lee CC, Kim JH, Kim JS, Oh YS, Han SM, Park JHY, et al. 5-(3',4'-Dihydroxyphenyl-gamma-valerolactone), a major microbial metabolite of proanthocyanidin, attenuates THP-1 monocyte-endothelial adhesion. Int J Mol Sci. (2017) 18:1363. doi: 10.3390/ijms18071363
203. Marquez Campos E, Stehle P, Simon MC. Microbial metabolites of flavan-3-Ols and their biological activity. Nutrients. (2019) 11:2260. doi: 10.3390/nu11102260
204. Mena P, González de Llano D, Brindani N, Esteban-Fernández A, Curti C, Moreno-Arribas MV, et al. 5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone and its sulphate conjugates, representative circulating metabolites of flavan-3-ols, exhibit anti-adhesive activity against uropathogenic Escherichia coli in bladder epithelial cells. J Funct Food. (2017) 29:275–80. doi: 10.1016/j.jff.2016.12.035
205. Angelino D, Carregosa D, Domenech-Coca C, Savi M, Figueira I, Brindani N, et al. 5-(Hydroxyphenyl)-gamma-valerolactone-sulfate, a key microbial metabolite of flavan-3-ols, is able to reach the brain: evidence from different in silico, in vitro and in vivo experimental models. Nutrients. (2019) 11:2678. doi: 10.3390/nu11112678
206. Fernandez-Millan E, Ramos S, Alvarez C, Bravo L, Goya L, Martin MA. Microbial phenolic metabolites improve glucose-stimulated insulin secretion and protect pancreatic beta cells against tert-butyl hydroperoxide-induced toxicity via ERKs and PKC pathways. Food Chem Toxicol. (2014) 66:245–53. doi: 10.1016/j.fct.2014.01.044
207. Fang C, Kim H, Barnes RC, Talcott ST, Mertens-Talcott SU. Obesity-associated diseases biomarkers are differently modulated in lean and obese individuals and inversely correlated to plasma polyphenolic metabolites after 6 weeks of mango (Mangifera indica L.) consumption. Mol Nutr Food Res. (2018) 62:e1800129. doi: 10.1002/mnfr.201800129
208. Li Z, Henning SM, Lee R-P, Lu Q-Y, Summanen PH, Thames G, et al. Pomegranate extract induces ellagitannin metabolite formation and changes stool microbiota in healthy volunteers. Food Funct. (2015) 6:2487–95. doi: 10.1039/C5FO00669D
209. Tomás-Barberán FA, González-Sarrías A, García-Villalba R, Núñez-Sánchez MA, Selma MV, García-Conesa MT, et al. Urolithins, the rescue of “old” metabolites to understand a “new” concept: metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol Nutr Food Res. (2017) 61:1500901. doi: 10.1002/mnfr.201500901
210. Mendez-Vilas A, Giménez JA, Truchado P, Larrosa M, Espín JC, Tomás-Barberán F, et al. Urolithins, metabolites produced by human colonic microflora, act as quorum sensing inhibitors of Yersinia enterocolitica affecting its gene expression. In: Méndez-Vilas A, , editor. Science and Technology Against Microbial Pathogens. World Scientific Publishing Co. Pte. Ltd. (2011). p. 202–6. doi: 10.1142/9789814354868_0039
211. Andersen T, Carstensen J, Hernandez-Garcia E, Duarte CM. Ecological thresholds and regime shifts: approaches to identification. Trends Ecol Evol. (2009) 24:49–57. doi: 10.1016/j.tree.2008.07.014
212. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. (2012) 336:1255–62. doi: 10.1126/science.1224203
213. Kim Y, Choi Y, Ham H, Jeong H-S, Lee J. Protective effects of oligomeric and polymeric procyanidin fractions from defatted grape seeds on tert-butyl hydroperoxide-induced oxidative damage in HepG2 cells. Food Chem. (2013) 137:136–41. doi: 10.1016/j.foodchem.2012.10.006
214. Ogura K, Ogura M, Shoji T, Sato Y, Tahara Y, Yamano G, et al. Oral administration of apple procyanidins ameliorates insulin resistance via suppression of pro-inflammatory cytokines expression in liver of diabetic ob/ob mice. J Agric Food Chem. (2016) 64:8857–65. doi: 10.1021/acs.jafc.6b03424
215. Magrone T, Magrone M, Russo MA, Jirillo E. Recent advances on the anti-inflammatory and antioxidant properties of red grape polyphenols: in vitro and in vivo studies. Antioxidants. (2019) 9:35. doi: 10.3390/antiox9010035
216. Sheng L, Jena PK, Liu HX, Hu Y, Nagar N, Bronner DN, et al. Obesity treatment by epigallocatechin-3-gallate-regulated bile acid signaling and its enriched Akkermansia muciniphila. FASEB J. (2018) 32:6371–84. doi: 10.1096/fj.201800370R
217. Zhang Z, Wu X, Cao S, Cromie M, Shen Y, Feng Y, et al. Chlorogenic acid ameliorates experimental colitis by promoting growth of Akkermansia in mice. Nutrients. (2017) 9:677. doi: 10.3390/nu9070677
218. Zhang Z, Wu X, Cao S, Wang L, Wang D, Yang H, et al. Caffeic acid ameliorates colitis in association with increased Akkermansia population in the gut microbiota of mice. Oncotarget. (2016) 7:31790–9. doi: 10.18632/oncotarget.9306
219. Etxeberria U, Arias N, Boque N, Macarulla MT, Portillo MP, Martinez JA, et al. Reshaping faecal gut microbiota composition by the intake of trans-resveratrol and quercetin in high-fat sucrose diet-fed rats. J Nutr Biochem. (2015) 26:651–60. doi: 10.1016/j.jnutbio.2015.01.002
220. Cheng Z, Wang Y, Li B. Illumina MiSeq reveals the influence of blueberry malvidin-3-galactoside on fecal microbial community structure and metabolizes of liver cancer mice. Peerj Prepr. (2018) 6:e27429v2. doi: 10.7287/peerj.preprints.27429
221. Caputo A, Dubourg G, Croce O, Gupta S, Robert C, Papazian L, et al. Whole-genome assembly of Akkermansia muciniphila sequenced directly from human stool. Biol Direct. (2015) 10:5. doi: 10.1186/s13062-015-0041-1
222. Guo X, Li S, Zhang J, Wu F, Li X, Wu D, et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC Genomics. (2017) 18:800. doi: 10.1186/s12864-017-4195-3
223. Dubourg G, Lagier JC, Armougom F, Robert C, Audoly G, Papazian L, et al. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int J Antimicrob Agents. (2013) 41:149–55. doi: 10.1016/j.ijantimicag.2012.10.012
224. Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. (2010) 105:2420–8. doi: 10.1038/ajg.2010.281
225. van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One. (2011) 6:e16876. doi: 10.1371/journal.pone.0016876
226. Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. (2019) 364:1179–84. doi: 10.1126/science.aaw7479
227. Greig CJ, Alper A, Goodman AL, Cowles RA. Mucosal homeostasis is altered in the ileum of gnotobiotic mice. J Surg Res. (2018) 231:331–7. doi: 10.1016/j.jss.2018.05.055
228. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc National Acad Sci USA. (2013) 110:9066–71. doi: 10.1073/pnas.1219451110
229. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. (2013) 500:541–6. doi: 10.1038/nature12506
230. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. (2020) 51:102590. doi: 10.1016/j.ebiom.2019.11.051
231. Ellekilde M, Krych L, Hansen CH, Hufeldt MR, Dahl K, Hansen LH, et al. Characterization of the gut microbiota in leptin deficient obese mice - correlation to inflammatory and diabetic parameters. Res Vet Sci. (2014) 96:241–50. doi: 10.1016/j.rvsc.2014.01.007
232. Chen T, Yang CS. Biological fates of tea polyphenols and their interactions with microbiota in the gastrointestinal tract: implications on health effects. Crit Rev Food Sci Nutr. (2020) 60:2691–709. doi: 10.1080/10408398.2019.1654430
233. Schaab MR, Barney BM, Francisco WA. Kinetic and spectroscopic studies on the quercetin 2,3-dioxygenase from Bacillus subtilis. Biochemistry (2006) 45:1009–16. doi: 10.1021/bi051571c
234. Cueva C, Sanchez-Patan F, Monagas M, Walton GE, Gibson GR, Martin-Alvarez PJ, et al. In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: changes in microbial groups and phenolic metabolites. FEMS Microbiol Ecol. (2013) 83:792–805. doi: 10.1111/1574-6941.12037
235. Jakobek L, Matić P. Non-covalent dietary fiber - Polyphenol interactions and their influence on polyphenol bioaccessibility. Trends Food Sci Technol. (2019) 83:235–47. doi: 10.1016/j.tifs.2018.11.024
236. Liu X, Le Bourvellec C, Renard C. Interactions between cell wall polysaccharides and polyphenols: effect of molecular internal structure. Compr Rev Food Sci Food Saf. (2020) 19:3574–617. doi: 10.1111/1541-4337.12632
237. Curko N, Tomasevic M, Cvjetko Bubalo M, Gracin L, Radojcic Redovnikovic I, Kovacevic Ganic K. Extraction of proanthocyanidins and anthocyanins from grape skin by using ionic liquids. Food Technol Biotechnol. (2017) 55:429–37. doi: 10.17113/ftb.55.03.17.5200
238. Ding Y, Morozova K, Scampicchio M, Ferrentino G. Non-eextractable polyphenols from food by-products: current knowledge onrecovery, characterisation, and potential applications. Process. (2020) 8:925. doi: 10.3390/pr8080925
239. Mateos-Martin ML, Perez-Jimenez J, Fuguet E, Torres JL. Non-extractable proanthocyanidins from grapes are a source of bioavailable (epi)catechin and derived metabolites in rats. Br J Nutr. (2012) 108:290–7. doi: 10.1017/S0007114511005678
240. Celik EE, Gokmen V, Skibsted LH. Synergism between soluble and dietary fiber bound antioxidants. J Agric Food Chem. (2015) 63:2338–43. doi: 10.1021/acs.jafc.5b00009
241. Auclair S, Silberberg M, Gueux E, Morand C, Mazur A, Milenkovic D, et al. Apple polyphenols and fibers attenuate atherosclerosis in apolipoprotein E-deficient mice. J Agric Food Chem. (2008) 56:5558–63. doi: 10.1021/jf800419s
242. Nyambe-Silavwe H, Villa-Rodriguez JA, Ifie I, Holmes M, Aydin E, Jensen JM, et al. Inhibition of human α-amylase by dietary polyphenols. J Funct Food. (2015) 19:723–32. doi: 10.1016/j.jff.2015.10.003
243. Liu J, Hao W, He Z, Kwek E, Zhao Y, Zhu H, et al. Beneficial effects of tea water extracts on the body weight and gut microbiota in C57BL/6J mice fed with a high-fat diet. Food Funct. (2019) 10:2847–60. doi: 10.1039/C8FO02051E
244. Li J, Wu T, Li N, Wang X, Chen G, Lyu X. Bilberry anthocyanin extract promotes intestinal barrier function and inhibits digestive enzyme activity by regulating the gut microbiota in aging rats. Food Funct. (2019) 10:333–43. doi: 10.1039/C8FO01962B
245. Van Rymenant E, Abranko L, Tumova S, Grootaert C, Van Camp J, Williamson G, et al. Chronic exposure to short-chain fatty acids modulates transport and metabolism of microbiome-derived phenolics in human intestinal cells. J Nutr Biochem. (2017) 39:156–68. doi: 10.1016/j.jnutbio.2016.09.009
246. Simmering R, Pforte H, Jacobasch G, Blaut M. The growth of the flavonoid-degrading intestinal bacterium, Eubacterium ramulus, is stimulated by dietary flavonoids in vivo. FEMS Microbiol Ecol. (2002) 40:243–8. doi: 10.1111/j.1574-6941.2002.tb00957.x
247. Peng X, Zhang Z, Zhang N, Liu L, Li S, Wei H. In vitro catabolism of quercetin by human fecal bacteria and the antioxidant capacity of its catabolites. Food Nutr Res. (2017) 58:23406. doi: 10.3402/fnr.v58.23406
248. Serra A, Macià A, Romero M-P, Reguant J, Ortega N, Motilva M-J. Metabolic pathways of the colonic metabolism of flavonoids (flavonols, flavones and flavanones) and phenolic acids. Food Chem. (2012) 130:383–93. doi: 10.1016/j.foodchem.2011.07.055
249. Kakkar S, Bais S. A review on protocatechuic acid and its pharmacological potential. ISRN Pharmacol. (2014) 2014:952943. doi: 10.1155/2014/952943
250. Carrasco-Pozo C, Gotteland M, Castillo RL, Chen C. 3,4-Dihydroxyphenylacetic acid, a microbiota-derived metabolite of quercetin, protects against pancreatic beta-cells dysfunction induced by high cholesterol. Exp Cell Res. (2015) 334:270–82. doi: 10.1016/j.yexcr.2015.03.021
251. Reveron I, Jimenez N, Curiel JA, Penas E, Lopez de Felipe F, de Las Rivas B, et al. Differential gene expression by Lactobacillus plantarum WCFS1 in response to phenolic compounds reveals new genes involved in tannin degradation. Appl Environ Microbiol. (2017) 83:e03387–16. doi: 10.1128/AEM.03387-16
252. Curiel JA, Rodriguez H, de Las Rivas B, Anglade P, Baraige F, Zagorec M, et al. Response of a Lactobacillus plantarum human isolate to tannic acid challenge assessed by proteomic analyses. Mol Nutr Food Res. (2011) 55:1454–65. doi: 10.1002/mnfr.201000621
253. Garcia-Villalba R, Vissenaekens H, Pitart J, Romo-Vaquero M, Espin JC, Grootaert C, et al. Gastrointestinal simulation model TWIN-SHIME shows differences between human urolithin-metabotypes in gut microbiota composition, pomegranate polyphenol metabolism, and transport along the intestinal tract. J Agric Food Chem. (2017) 65:5480–93. doi: 10.1021/acs.jafc.7b02049
254. Vrancken G, Gregory AC, Huys GRB, Faust K, Raes J. Synthetic ecology of the human gut microbiota. Nat Rev Microbiol. (2019) 17:754–63. doi: 10.1038/s41579-019-0264-8
255. Riviere A, Gagnon M, Weckx S, Roy D, De Vuyst L. Mutual cross-feeding interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl Environ Microbiol. (2015) 81:7767–81. doi: 10.1128/AEM.02089-15
256. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. (2016) 167:1339–53 e21. doi: 10.1016/j.cell.2016.10.043
257. Schape SS, Krause JL, Engelmann B, Fritz-Wallace K, Schattenberg F, Liu Z, et al. The Simplified Human Intestinal Microbiota (SIHUMIx) shows high structural and functional resistance against changing transit times in in vitro bioreactors. Microorganisms. (2019) 7:641. doi: 10.3390/microorganisms7120641
258. Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio. (2014) 5:e01438–14. doi: 10.1128/mBio.01438-14
259. Aoki-Yoshida A, Saito S, Fukiya S, Aoki R, Takayama Y, Suzuki C, et al. Lactobacillus rhamnosus GG increases Toll-like receptor 3 gene expression in murine small intestine ex vivo and in vivo. Benef Microbes. (2016) 7:421–9. doi: 10.3920/BM2015.0169
260. Chassaing B, Van de Wiele T, De Bodt J, Marzorati M, Gewirtz AT. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut. (2017) 66:1414–27. doi: 10.1136/gutjnl-2016-313099
261. Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ Res. (2015) 117:817–24. doi: 10.1161/CIRCRESAHA.115.306807
262. Nie Y, Stürzenbaum SR. Proanthocyanidins of natural origin: molecular mechanisms and implications for lipid disorder and aging-associated diseases. Adv Nutr. (2019) 10:464–78. doi: 10.1093/advances/nmy118
263. Henning SM, Summanen PH, Lee R-P, Yang J, Finegold SM, Heber D, et al. Pomegranate ellagitannins stimulate the growth of Akkermansia muciniphila in vivo. Anaerobe. (2017) 43:56–60. doi: 10.1016/j.anaerobe.2016.12.003
264. Lacombe A, Wu VCH, White J, Tadepalli S, Andre EE. The antimicrobial properties of the lowbush blueberry (Vaccinium angustifolium) fractional components against foodborne pathogens and the conservation of probiotic Lactobacillus rhamnosus. Food Microbiol. (2012) 30:124–31. doi: 10.1016/j.fm.2011.10.006
265. Consortium CA. Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology. (2018) 28:3–8. doi: 10.1093/glycob/cwx089
266. Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. (2014) 42(Database issue):D490–5. doi: 10.1093/nar/gkt1178
267. Takagaki A, Nanjo F. Effects of metabolites produced from (-)-epigallocatechin gallate by rat intestinal bacteria on angiotensin I-converting enzyme activity and blood pressure in spontaneously hypertensive rats. J Agric Food Chem. (2015) 63:8262–6. doi: 10.1021/acs.jafc.5b03676
268. Unno K, Pervin M, Nakagawa A, Iguchi K, Hara A, Takagaki A, et al. Blood-brain barrier permeability of green tea catechin metabolites and their neuritogenic activity in human neuroblastoma SH-SY5Y cells. Mol Nutr Food Res. (2017) 61:1700294. doi: 10.1002/mnfr.201700294
269. Xia B, Shi XC, Xie BC, Zhu MQ, Chen Y, Chu XY, et al. Urolithin A exerts antiobesity effects through enhancing adipose tissue thermogenesis in mice. PLoS Biol. (2020) 18:e3000688. doi: 10.1371/journal.pbio.3000688
270. Ishimoto H, Shibata M, Myojin Y, Ito H, Sugimoto Y, Tai A, et al. In vivo anti-inflammatory and antioxidant properties of ellagitannin metabolite urolithin A. Bioorg Med Chem Lett. (2011) 21:5901–4. doi: 10.1016/j.bmcl.2011.07.086
271. Casedas G, Les F, Choya-Foces C, Hugo M, Lopez V. The metabolite urolithin-A ameliorates oxidative stress in neuro-2a cells, becoming a potential neuroprotective agent. Antioxidants. (2020) 9:177. doi: 10.3390/antiox9020177
272. Dixon RA. Natural products and plant disease resistance. Nature. (2001) 411:843–7. doi: 10.1038/35081178
273. Song H, Chu Q, Yan F, Yang Y, Han W, Zheng X. Red pitaya betacyanins protects from diet-induced obesity, liver steatosis and insulin resistance in association with modulation of gut microbiota in mice. J Gastroenterol Hepatol. (2016) 31:1462–9. doi: 10.1111/jgh.13278
Keywords: polyphenols, prebiotics, antimicrobial action, polyphenol-associated enzymes, Akkermansia muciniphila, duplibiotic, Lactiplantibacillus plantarum, trophic interactions
Citation: Rodríguez-Daza MC, Pulido-Mateos EC, Lupien-Meilleur J, Guyonnet D, Desjardins Y and Roy D (2021) Polyphenol-Mediated Gut Microbiota Modulation: Toward Prebiotics and Further. Front. Nutr. 8:689456. doi: 10.3389/fnut.2021.689456
Received: 31 March 2021; Accepted: 27 May 2021;
Published: 28 June 2021.
Edited by:
Silvia Turroni, University of Bologna, ItalyReviewed by:
Franck Carbonero, Washington State University Health Sciences Spokane, United StatesCopyright © 2021 Rodríguez-Daza, Pulido-Mateos, Lupien-Meilleur, Guyonnet, Desjardins and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Denis Roy, ZGVuaXMucm95QGZzYWEudWxhdmFsLmNh
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.