- School of Food and Biological Engineering, Shaanxi University of Science & Technology, Xi'an, China
Large variations in the bioactivities and composition of milk fat globule membrane (MFGM) proteins were observed between Saanen dairy goat and Holstein bovine at various lactation periods. In the present study, 331, 250, 182, and 248 MFGM proteins were characterized in colostrum and mature milk for the two species by Q-Orbitrap HRMS-based proteomics techniques. KEGG pathway analyses displayed that differentially expressed proteins in colostrum involved in galactose metabolism and an adipogenesis pathway, and the differentially expressed proteins in mature milk associated with lipid metabolism and a PPAR signaling pathway. These results indicated that the types and functions of MFGM proteins in goat and bovine milk were different, and goat milk had a better function of fatty acid metabolism and glucose homeostasis, which can enhance our understanding of MFGM proteins in these two species across different lactation periods, and they provide significant information for the study of lipid metabolism and glycometabolism of goat milk.
Introduction
Milk fat is an important fraction of milk synthesized by the endoplasmic reticulum of mammary epithelial cells. It is a droplet composed of a neutral triglyceride core wrapped by a thin trilayered membrane. Milk fat globules containing cytoplasmic components are retained between the membrane layers as the fat droplets are released into milk. Therefore, milk fat is mainly composed of cholesterol, polar lipids, neutral lipids, and a protein group from the membrane and cytoplasmic crescents (1, 2). Protein, which accounts for 22–70% of the MFGM matter, not only provides protection to core milk fat but also has a series of biological functions, such as preventing infection of enteric pathogens, promoting immune and neurological functions, as well as the development of newborns (3, 4).
Due to the health benefits and nutritional values, MFGM proteins have attracted growing attention in dairy products. The major proteins in MFGM include lactadherin, mucin one, xanthine dehydrogenase/xanthine oxidase, fatty acid synthase, fatty acid-binding proteins lipophilin, and butyrophilin with physiological functions (5). For example, xanthine dehydrogenase/xanthine oxidase, one of the main MFGM enzyme proteins, has been reported to reveal antimicrobial properties and immuno-protective function. Xanthine dehydrogenase/xanthine oxidase in breast milk reacts with infant saliva to produce an effective combination of irritant and inhibitory metabolites that regulate the gut-microbiota (6). Lactadherin (milk fat globule-EGF factor 8) is a peripheral glycoprotein from human milk, which promotes the clearance and phagocytosis of apoptotic cells, and regulates the immune response (7).
With the accelerated development of proteomics technology in recent years, a large number of proteins have been identified and quantified in the milk of bovine (8), buffalo (9), donkey (10), and other mammals (11). Among them, because bovine milk is the major substitute for human milk, the comparative proteomics between bovine and human milk as well as bovine and human milk were extensively studied. Compared with bovine milk, goat milk-based dairy products may be less allergenic and more easily digested to the infants (12). In view of the unique economic significance, more and more studies have been absorbed in the nutritional and protective properties of proteins in goat milk. Chen et al. have studied the heat-dependent changes of goat milk protein, and found out that heat processing can improve protein digestibility, which was conducive to anti-atherosclerosis therapy (13). They also investigated the protein changes of goat milk during homogenization. The results showed that the homogenized goat milk proteome has changed significantly, which was mainly related to glycolysis/gluconeogenesis metabolism (14). These studies extend the understanding of protein composition in different processes. Major MFGM proteins of goat milk have been reported, which are significantly different from that of bovine. Sun et al. have characterized and compared the MFGM proteins of both Guanzhong goat and Holstein cow milk, using proteomic techniques (15). Furthermore, they analyzed and compared the MFGM proteomes of colostrum and mature milk of Xinong Saanen goat milk (16). However, the MFGM proteome is also affected by species.
Despite the poorly worldwide production of goat milk compared with the bovine, in the past years, there has been more and more interest in the in-depth characterization of its protein composition. This analysis was focused on MFGM proteins from two mammals (bovine and goat) and different lactation periods (colostrum and mature milk) to characterize the composition in conjunction with biological activity, localization, and molecular function of MFGM proteins differences related to lactation. The purpose was to reveal the differences of nutritional value and physiological states of these two species across different lactation periods to provide potential directions for infant formula and functional food development, as well as expand our current knowledge of MFGM proteome.
Materials and Methods
Sample Collection
The sample collection and preparation were shown in Supplementary Figure 1. The samples were collected, followed by the method reported by Sun et al. (16). The samples were collected at the Holstein bovine and Saanen dairy goat farm in Xi'an, Shaanxi province, China. Ten bovine colostrum (0–5 days postpartum), 10 mature-milk (1–6 months postpartum), 10 goat colostrum (0–5 days postpartum), and 10 mature-milk (1–6 months postpartum) samples were obtained from 20 healthy bovines and 20 healthy goats in the first lactation. All of the 40 animals were aged between 1 and 4 years old, and the animals of each species were under identical environmental conditions. Each sample of bovine and goat milk was collected twice a day and then mixed to dispel the effect of the sampling time of milk samples. These samples were transported to the laboratory on ice and stored at −80°C. Ten milk samples of each group were mixed to refrain from the influence of individual differences on MFGM protein in various lactation stages before the analysis. All handling practices involving animals carefully followed all the recommendations of the Directive 2010/63/EU of the European Parliament for the protection of animals for scientific purposes.
The extraction of MFGM proteins was conducted as described by Lu et al. (17) with minor modifications. Briefly, 50-ml milk samples were centrifuged at 12,000 × g for 40 min at 4°C. The supernatant (top layer) was transferred to another centrifuge tube and washed three times at 25°C for 10 min, with 0.1 mol L−1 PBS (pH 6.8) and centrifuged at 10,000 × g for 15 min at 4°C subsequently to remove residual whey proteins and caseins. Then we washed the cream twice, using ultrapure water to dislodge the residual salt ions. Finally, 0.4% SDS (1:1, v/v) was added to dilute the washed cream, sonicated for 1 min and centrifuged at 10,000 × g at 4°C for 40 min to separate the fat fraction. The MFGM proteins were collected in the aqueous phase (bottom layer), and their concentration was measured by BCA assay (Thermo Scientific Pierce BCA protein assay kit, USA).
Protein Digestion
The MFGM protein was reduced, alkylated, and digested, and followed the method reported by Lu et al. (18). For each milk sample, three independent biological replicates were made. First, 10-μL MFGM protein was dissolved in 100-μL, 50-mmol L−1 NH4HCO3, and then 10-μL, 100-mmol L−1 dithiothreitol was added and incubated at 56°C for 30 min. Subsequently, the MFGM protein was alkylated with 15 μL of 55-mmol L−1 iodoacetamide in dark for 30 min at room temperature and then adding sequencing grade-modified trypsin to digest the MFGM protein at a ratio of 1:100 enzyme/protein for 16–18 h at 37°C and terminated the reaction by adding 1% formic acid. Finally, the peptides mixture was desalting by Oasis HLB cartridges (Waters Cooperation, Milford, MA, USA), dried by a vacuum centrifuge and then resuspended in 40 μL of 0.1% (v/v) formic acid.
Liquid Chromatography Tandem Q-Orbitrap Mass Spectrometry
Peptide separation was performed by EASY-nLC 1000 system (Thermo Scientific, San Jose, CA), equipped with a C18-reverse phase column (75-μm inner diameter, 10-cm long, 3-μm resin; Thermo Scientific) at 200 nL/min and 35°C for a total run time of 100 min. Solution A (0.1% formic acid in water) and solution B (0.1% formic acid in 80% ACN) were used as eluents for the peptide separation according to the following elution gradients: 5–35% solution B for 50 min; 35–100% solution B for 25 min, followed by 15-min washing with 100% solution B, and return to 5% B in 0.1 min, re-equilibration during 9.9 min with 5% solution B.
The peptide eluted from the column was ionized by a Q-Exactive (Thermo Fisher Scientific, Waltham, USA) mass spectrometer in a positive mode. The spray voltage was operated at 3.8 kV. The m/z scan range of single MS scans of peptide precursors was m/z 300–1,700 at a resolution of 70,000 (at m/z 200). The AGC (automatic gain control) was 3 e6, and the maximum injection time was 200 ms. The top 20 most intense precursor ions with charge ≥2 determined by MS scan were used to obtain MS/MS data at a resolution of 17,500 by using a higher normalized collision energy of 27 eV. The AGC was 1 e5 and the maximum injection time was 50 ms. In order to avoid superfluous fragmentation, the dynamic exclusion time was set to 30 s.
Data Analysis
The raw LC-MS/MS files with three replicates were obtained for MFGM proteins of each milk group. Two proteins identified from three biological replicates of each milk group were used for subsequent analysis. The data analysis was carried out using the MaxQuant software (Max Planck Institute of Biochemistry, Martinsried, Germany, version 1.6.7.0), with Andromeda as a peptide search engine (Matrix Science, version 2.4), and searched against the database of Caprinae (67,040 entries, 02/08/2019) and Bos taurus (64,796 entries, 02/08/2019) organism group with reverse sequences generated by MaxQuant.
Search parameters were a first search peptide mass tolerance of 20 ppm and main search peptide mass tolerance of 4.5 ppm. Methionine oxidation and protein N-terminal acetylation were defined as variable modification and carbamidomethyl of cysteine defined as a fixed modification for both identification and quantification. A trypsin/P was adjusted as a proteolytic enzyme with a maximum of two missed cleavages. A maximum of 0.01 false discovery rates (FDRs) and at least two unique peptides for each protein were demanded for reliable identification and quantification. Label-free quantification (LFQ) was enabled in MaxQuant.
The MFGM proteins identified in at least two of the three biological replicates were used for subsequent statistical analysis. The LFQ intensity of identified proteins was analyzed using one-way ANOVA test. MFGM proteins with p < 0.05 and fold change >2 in the relative abundance ratios were considered to be differentially expressed MFGM proteins (DEMPs) between milk groups. Principal component analysis (PCA) and partial least squares regression-discriminant analysis (PLS-DA) were used to construct the recognition model and prediction models, respectively, among milk groups by using SIMCA 15 (19).
Bioinformatics Analyses
The molecular function, cellular component, and biological process of all identified MFGM proteins according to their gene ontology (GO) annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were achieved, using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/). Conversion of the genes of MFGM proteins was using Retrieve/ID mapping (https://www.uniprot.org/uploadlists/). Protein–protein interaction (PPI) network construction was performed, using STRING (https://string-db.org/), with the DEMPs displayed by proteomic data used as input. Statistical differences were proclaimed significantly if p ≤ 0.05.
Results and Discussion
Component Analysis of MFGM Proteins From Different Milk Groups
In this study, 331 MFGM proteins in goat colostrum (GC), 250 in goat mature milk (GM), 182 in bovine colostrum (BC), and 248 in bovine mature milk (BM) were identified and quantified using LC-Q-Orbitrap mass spectrometer (Supplementary Table 1). These proteins spanned more than six orders of magnitude. As shown in Figure 1, there were 136, 72, 37, and 78 uniquely expressed MFGM proteins identified in goat colostrum, goat mature milk, bovine colostrum, and bovine mature milk, respectively. The uniquely expressed proteins in goat colostrum included calreticulin, cofilin-1, and methanethiol oxidase in goat mature milk included FA complementation group I, phosphatidylethanol-amine binding protein 4, and perilipin-1. Approximately, 45 MFGM proteins, coupled with cream fractions, were identified in four milk groups, including the complement component 3, fatty acid synthase, sodium-dependent phosphate transport protein 2B, and pericentrin, indicating that goat milk was a substitute for bovine milk to some extent in the research and development of infant milk powder and functional food, and also providing an orientation for the further functional development of goat milk.
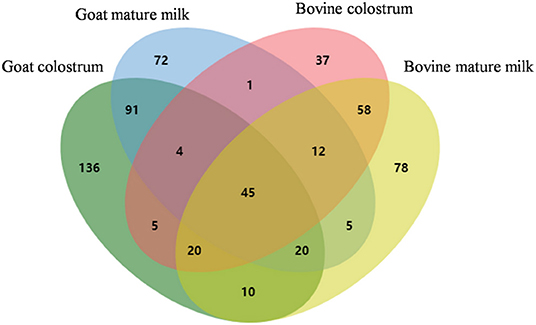
Figure 1. Venn diagram analysis of the identified MFGM protein components from bovine colostrum, bovine mature milk, goat colostrum, and goat mature milk.
In addition, proteins from caseins and whey, such as αs1-casein, αs2-casein, β-casein, κ-casein, lactoferrin, and β-lactoglobulin, were also identified in the MFGM fractions, which could be owing to the residual contamination of the proteins during MFGM extraction (20). In our study, polymeric immunoglobulin receptor (PIGR) was highly abundant in four milk groups. PIGR binds with polymerized immunoglobulin A and immunoglobulin M and transport them to perform immune functions across cell membranes (21). The other identified immunoglobulin proteins included IGL@ protein (IGL@), IGK protein (IGK), immunoglobulin heavy-constant mu (IGHM), and immunoglobulin J chain (JCHAIN). Previously observed major proteins, such as lactadherin, butyrophilin, and xanthine dehydrogenase/oxidase, were conserved in MFGM across four groups, which indicate the robustness of the methodology.
Chemometrics of MFGM Protein in Various Lactation Periods
Before chemometric analysis of MFGM protein, preprocessing of LC-MS/MS data was implemented. The correct normalization of each milk group can eliminate the systematically differences between features. However, the total peptide ion signals necessary to perform LC-MS/MS were distributed over several adjacent runs. Therefore, it was necessary to know the normalization coefficient (N) of each fraction to sum the peptide ion signal. Based on the least overall proteome variation, the quantities of proteins can be determined via a global optimization procedure after the intensities were normalized to a normalization factor as free variables. Hence, in sample A, the total intensity of a peptide ion P was defined as
where the index k covered all isotope patterns of peptide ion P in sample A.
A triangular matrix containing all paired protein ratios between any two samples was constructed. This matrix corresponds to the overdetermined system of equations for the protein abundance distributions in the sample. A subsequent least-squares analysis was performed to reconstruct the abundance profile based on the sum of squared differences in the matrix via the optimal satisfaction of individual protein ratios.
Then the whole profile was rescaled to the cumulative intensity of the samples, thereby retaining the total summed intensity of the protein over all samples, which was the “LFQ intensity” (22).
Chemometric analysis is a technique involving statistical methods to comprehend chemical information generated by analytical instruments. To evaluate whether the data of proteomic analysis can be engaged to visually differentiate the four group milk samples, PCA analysis as an unsupervised data analysis was carried out by loading the LFQ intensities of all detected proteins as variables and the different MFGM matrices as observation points. In this case, the data from all the replicates were used. A clear separation of all MFGM matrices can be observed on the PCA score plot (Figure 2A) based on the first two principal components. Samples close to each other on the PCA score plot revealed similar properties, while the milk samples far from each other revealed dissimilar molecular weight in protein mass spectrographic analysis. The first two principal components accounted for 81.2% [R2X (1) + R2X (2)] of the total variation in the data. No outlier was observed by ellipse Hotelling's T2. The first principal component (45% of the total variation) clearly divided the samples into four well-separated clusters, corresponding to the four groups, showed that the distribution differences among the goat colostrum, goat mature milk, bovine colostrum, and bovine mature milk were mainly due to biological reasons. Meanwhile, R2X (cum) was cumulative R2X up to the specified component, where R2X was the fraction of X variation modeled in the component. Q2 (cum) was the cumulative Q2 up to the specified component, where Q2 was overall cross-validated R2X. R2 (cum) and Q2 (cum) were the critical parameters to evaluate the quality of the PCA model, which, respectively, reflect the degree of interpretation of the principal components to X variables and the predictive ability of the model. Figure 2A showed that R2X (cum) was 0.812 and Q2 (cum) was 0.966, illustrating that the PCA model was stable and predictable.
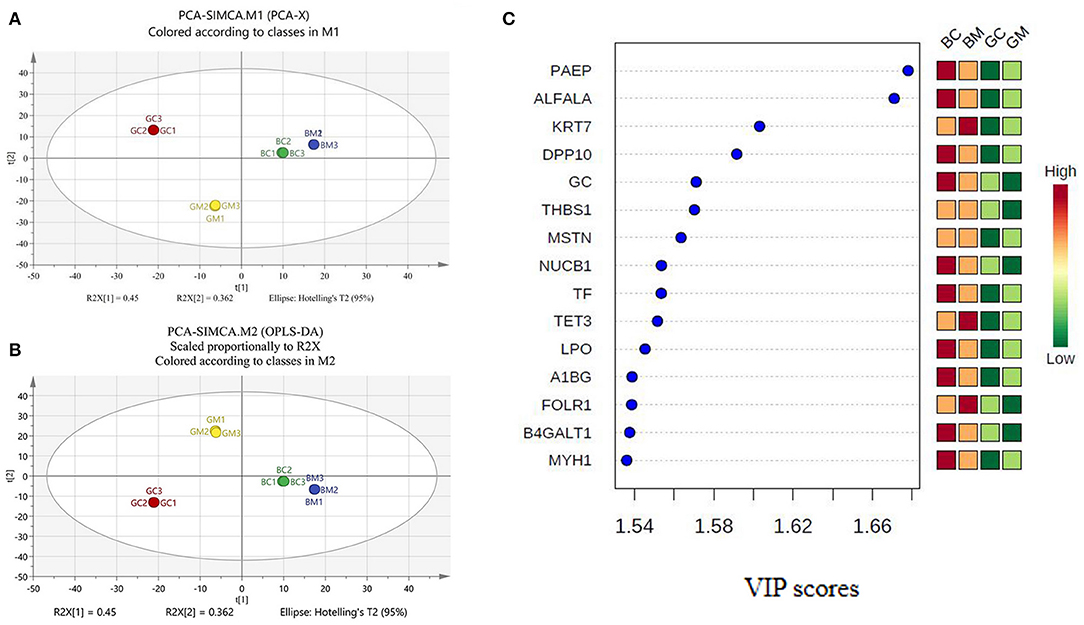
Figure 2. PCA and PLS-DA analysis of MFGM proteins in bovine and goat milk. (A) Principal components analysis (PCA) scores based on intensities of proteins. (B) Partial least squares discrimination analysis (PLS-DA) scores based on intensities of proteins. (C) VIP scores for differential protein.
PLS-DA is a chemical projection method that associates X and Y variable blocks via a linear multivariable model. The objective is to find the direction in X space that divides the classes according to the sample set with known class members (23, 24). In the built PLS-DA model, R2X (cum) was 0.812, R2Y (cum) was 1, and Q2 (cum) was 1, which denoted that the PLS-DA model has good fitting ability and prediction performance (Figure 2B). VIP scores were shown in Figure 2C, and variables with VIP scores higher than 1 were considered to have significantly contributed to the model. To evaluate the stability and reliability of PLS-DA model, cross-validation was adopted. The total correct classification rate of four MFGM proteins was 100%. The statistical significance of the predictive quality parameters in the built PLS-DA model was validated by 200 permutation tests (Figure 3). The Y-intercept of R2 and Q2 was 0.165 and −0.711, respectively, which ensured that the PLS-DA model was not overfitting.
DEMPs, Respectively, in Colostrum and Mature Milk of Goat and Bovine
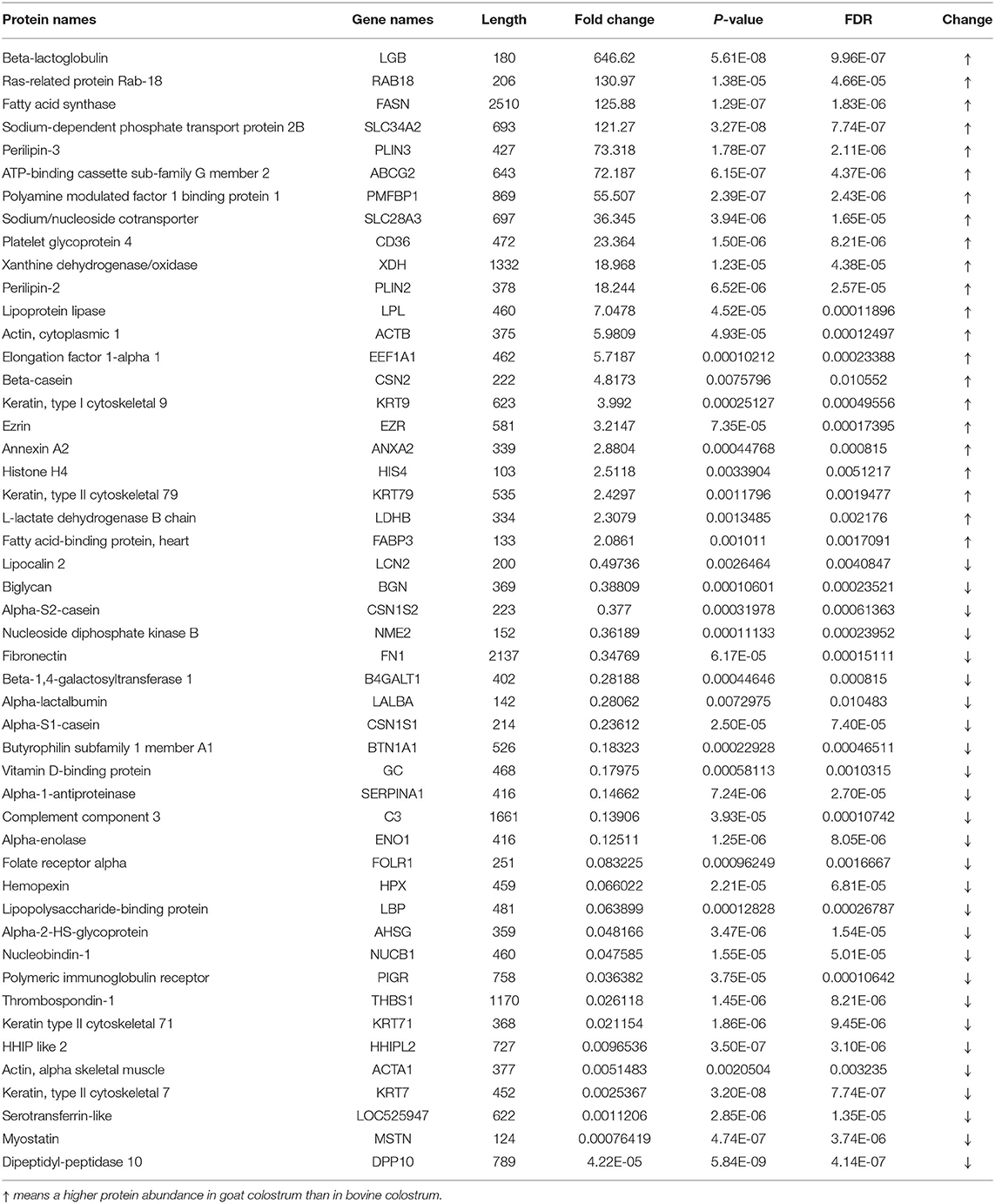
Three hundred and thirty-one MFGM proteins were identified in goat colostrum and 182 in bovine colostrum. T-test and fold change analysis were used to analyze the difference of the MFGM proteins in colostrum of goat and bovine, and the MFGM protein with p < 0.05 and at least two-fold was considered as the cutoff criteria of differential expression. As shown in Table 1, among 74 common proteins in colostrum of goat and bovine, 49 were differentially expressed. We found out that there were 22 upregulated and 27 downregulated MFGM proteins in goat colostrum. The levels of sodium/nucleoside cotransporter, lipoprotein lipase, sodium-dependent phosphate transport protein 2B, xanthine dehydrogenase/oxidase, and fatty acid synthase were higher in goat colostrum, while the levels of cathelicidin-1, lipopolysaccharide-binding protein, alpha-enolase, and vitamin D-binding protein were higher in bovine colostrum.
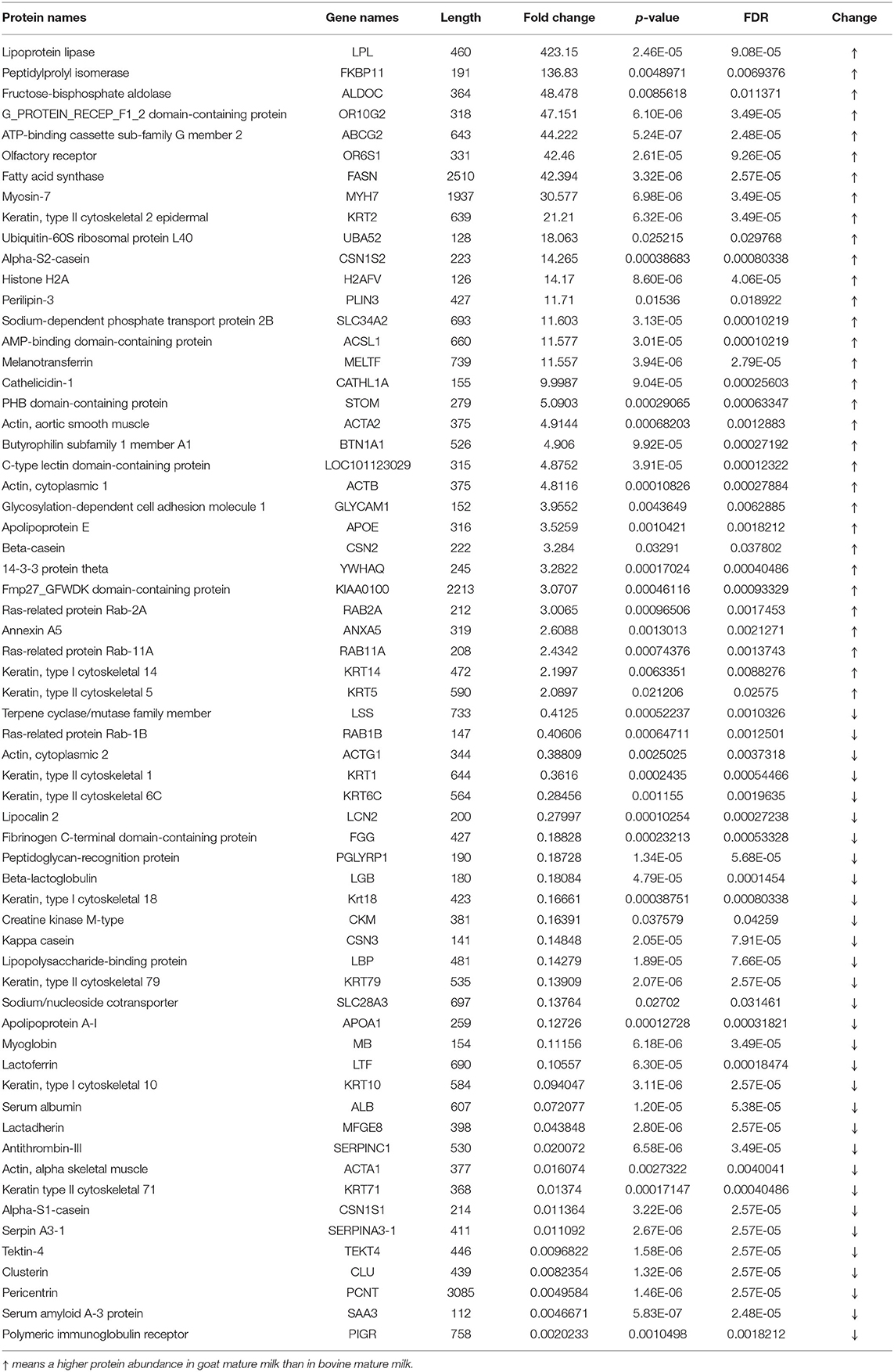
Two hundred and fifty MFGM proteins were identified in goat mature milk and 248 in bovine mature milk. As shown in Table 2, among 90 common proteins in mature milk of goat and bovine, 63 were differentially expressed on the basis of p < 0.05 and at least two-fold, with 32 upregulated and 31 downregulated MFGM proteins. The levels of lipoprotein lipase, fatty acid synthase, sodium-dependent phosphate transport protein 2B, and apolipoprotein E were higher in goat mature milk, while the levels of lipocalin 2, sodium/nucleoside cotransporter, apolipoprotein A-I, and polymeric immunoglobulin receptor were lower in bovine mature milk than in goat. The abundance of lipoprotein lipase and fatty acid synthase was higher in goat mature milk, which was the same as that in colostrum. Lipopolysaccharide-binding protein was higher in bovine mature milk in contrary. Lipocalin-2 can bind and eliminate enterochelin, a high-affinity siderophore, to reduce the accessibility of bacteria to iron and inhibit its growth, and participate in the modeling of immune response. Apolipoprotein A-I is a kind of lipoprotein expressed by glial cells, which is strongly induced in aging, injury or neurodegeneration, involved in the peripheral metabolic regulation and lipid processing of chylomicron, the occurrence and development of Parkinson's disease and Alzheimer's disease, and may play a neuroprotective role in the brain (16).
GO, KEGG Pathway, and PPI Analysis of the DEMPs
For the purpose of comparing the biological functions of MFGM proteins in colostrum, 49 DEMPs in colostrum of goat and bovine were analyzed by gene ontology (GO) functional annotation, which was divided into three categories of molecular function (MF), cellular components (CC), and biological process (BP). This was useful to further understand the colostrum MFGM proteins functions in goat and bovine. The most significant enrichment annotation information (p < 0.05) in each branch was shown in Figure 4A, in which the prevalent biological processes were a response to dehydroepiandrosterone, response to 11-deoxycorticosterone, response to estradiol, and response to progesterone. Others were involved in negative regulation of endopeptidase activity, acute-phase response, lactose biosynthetic process, and cell adhesion. The MFGM proteins were highly enriched in extracellular space, extracellular exosome, and blood microparticle, which illustrated that the majority of these proteins present due to leakage of the protein from the blood serum into the milk at the tight junctions in the cells in the mammary gland. Other enriched origin categories were Golgi lumen, Golgi apparatus, and extracellular region. Three prominent molecular functions of DEMPs in colostrum of goat and bovine were protein binding, transporter activity, and cytoskeletal protein binding, which was consistent with the result of Cunsolo et al. (25). Lactose synthase activity and structural molecule activity were also significantly represented.
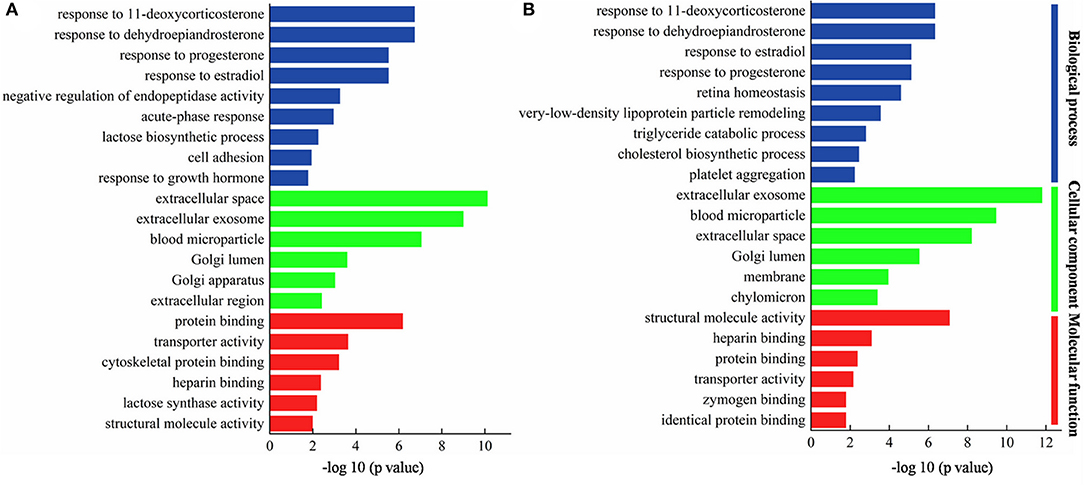
Figure 4. Gene ontology (GO) of DEMPs in colostrum of goat and bovine (A), in mature milk of goat and bovine (B).
The most significant enrichment annotation information (p < 0.05) in each branch of mature milk was shown in Figure 4B, in which the prevalent biological processes were response to dehydroepiandrosterone, response to 11-deoxycorticosterone, response to progesterone, and response to estradiol. It was consistent with the biological processes involved in colostrum, indicating that these biological processes occurred in the whole lactation period. In stressful situations, the adrenal cortex reacted to ACTH and began to secrete dehydroepiandrosterone that had been proved to exert anti-inflammatory and antioxidant effects, and played a protective and regenerative role (26). Others were involved in very-low-density lipoprotein particle remodeling, triglyceride catabolic process, and cholesterol biosynthetic process. The MFGM proteins were highly enriched in extracellular exosome, blood microparticle, and extracellular space, which was similar to the DEMPs in colostrum of goat and bovine. Other enriched origin categories were Golgi lumen, membrane, and chylomicron. The prominent molecular functions were structural molecule activity, heparin binding, and protein binding.
A KEGG pathway was employed to analyze the main pathways of MFGM protein differentially expressed in colostrum of goat and bovine. As shown in Figure 5A, the DEMPs mainly involved in the pathways of the phagosome, a PPAR signaling pathway, proteoglycans in cancer, ECM-receptor interaction, and galactose metabolism. As shown in Figure 5B, the DEMPs in mature milk mainly involved in the pathways of viral myocarditis, a PPAR signaling pathway, salmonella infection, and fatty acid biosynthesis.
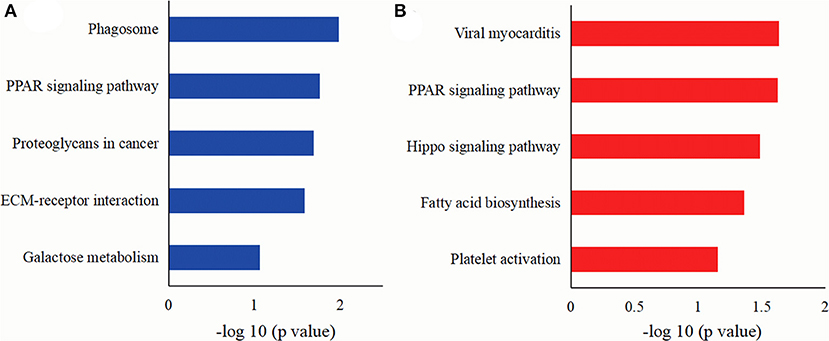
Figure 5. The KEGG pathways of DEMPs colostrum and mature milk of goat and bovine (A), in mature milk of goat and bovine (B).
PPI analysis of DEMPs in the colostrum and mature milk of goat and bovine was implemented to obtain a color-coded network, revealing the correlation between DEMPs (Supplementary Figure 2). The final network of colostrum consists of 49 nodes (proteins) and 99 edges (interactions). The avg. local clustering coefficient was 0.533, and the PPI enrichment p-value was lower than 1.0 e−16, indicating that the 49 DEMPs were biologically connected. Most highly interacting protein nodes in colostrum were divided into five communities, including a cellular process, cell adhesion, immune response, lactose synthase activity, and a metabolic process. Similarly, the final network of mature milk consists of 63 nodes (proteins) and 148 edges (interactions). The avg. local clustering coefficient and p-value were 0.575 and lower than 1.0 e−16, respectively, indicating that the 63 DEMPs were biologically connected. Furthermore, these highly interacting protein nodes were divided into three communities, including a cellular process, cell adhesion, and response to extracellular stimulus. Consequently, in light of the bioinformatics analysis of proteomics, the MFGM proteins in colostrum might be led to the intervention of related health issues, while the bioactivities of MFGM proteins between goat and bovine might be different.
Discussion
Protein Profile Analysis Based on Chemometrics
A label-free proteomic approach was employed to identify the MFGM proteins in goat and bovine milk. Compared with the previous reports, 102 MFGM proteins, mainly calcium-binding protein, cytoskeletal protein, and intercellular signal molecule were added in bovine milk, 114 in goat milk, mainly a metabolite interconversion enzyme, translational protein, cytoskeletal protein, and nucleic acid-binding protein [Supplementary Table 2; (15–17, 27)]. The nucleotide-binding proteins in this study were mainly DNA helicase, followed by several members of Ras super family (Rhos and Rabs). All these proteins are likely involved in the secretion of secreted milk components by vesicle (17). Furthermore, the results of PCA, PLS-DA, and VIP analysis found out that nucleobindin-1 (NUCB1), folate receptor alpha (FOLR1), vitamin D-binding protein (GC), thrombospondin-1 (THBS1), and beta-1,4-galactosyltransferase 1 (B4GALT1), with the lower abundance in goat mature milk than in other three milk groups, can be used as markers to distinguish the four milk groups. NUCB1 is Golgi-localized soluble protein, which contains multiple putative functional domains. NUCB1 localized in extracellular was considered to be a regulator of matrix maturation in bone (28, 29). FOLR1 can internalize folates into the cells, which is crucial to DNA repair and synthesis, and mediate the activation of pro-oncogene STAT3, which contributes to angiogenesis, tumor proliferation, and metastasis (30). THBS1 has been demonstrated to participate in mechano-signal transduction and is specific at the level of apoptosis induction (31). GC has the metabolic effects of influencing bone metabolism, chemotaxis, actin scavenging, innate immunity, modulation of inflammatory processes, and binding of fatty acids (32). As mentioned above, the data from our study not only provide a comprehensive understanding of the MFGM protein compositions among the four milk samples but also reveal the differences of MFGM proteins among different species of mammals. The results exhibited a scientific basis for the development of functional products, using goat milk.
A number of MFGM proteins were obviously differentially expressed in the goat and bovine milk, whose functions may associate with the flavor and protection of the lamb or calf from infections. Milk fatty acids are extracted from the arterial blood or synthesized de novo in the mammary gland and involve many kinds of mammary enzymes, including lipoprotein lipase and fatty acid synthetase (33). The abundance of lipoprotein lipase was higher in colostrum of goat milk than that of bovine milk. Lipoprotein lipase was more closely bound with fat globules and has a better correlation with spontaneous lipolysis in goat milk. It can release fatty acids from lipoproteins and chyle particles, which may relate to the differences in the flavor of goat and bovine milk. Zhu et al. (34) have clarified that inhibition at the gene-expression level of fatty acid synthetase restrained the accumulation of TAG and the formation of lipid droplet by reducing esterification and lipogenesis and promoting lipolysis in goat milk. In addition, fatty acid synthetase also helped generate toll-like receptor 4 (TLR4), which is vital for lipid metabolism regulation. Lipopolysaccharide-binding protein, a 58–60 kDa protein, catalyzed the transfer of bacterial lipopolysaccharide to CD14, which exists in soluble form and facilitates lipopolysaccharide presentation to TLR4 as a cell surface receptor. This activates the intracellular signaling pathways and promotes the upregulation of adhesion molecules and proinflammatory cytokines, which are participated in the innate immune response (35). The low abundance of lipopolysaccharide-binding protein in goat colostrum may be related to its low sensitization. Cathelicidin-1 found in epithelial and neutrophils cells is an antimicrobial peptide and has profound impacts on wound healing, inflammation, and the regulation of adaptive immunity (36). The existence of antimicrobial proteins in the colostrum of goat and bovine milk explained that the protection from milk was necessary for newborn mammals and varies across species.
DEMPs Analysis Based on Bioinformatics
Bioinformatics analysis showed that the response to dehydroepiandrosterone, 11-deoxycorticosterone, estradiol, and progesterone were the most abundant biological processes in colostrum and mature milk. Previous researches showed that the interaction between low-density lipoprotein and heparin results in irreversible structural changes in apolipoprotein B and affects low-density lipoprotein oxidation, phospholipolysis, and fusion (37). Dehydroepiandrosterone, 11-deoxycorticosterone, estradiol, and progesterone are endogenous steroid hormones, which are often conjugated with proteins, secreted by endocrine glands and endocrine cells, dispersed in other organs, and then transported to various organs through blood, provided with the function of coordinating and controlling tissue, organ metabolism, and physiological function (38). Progesterone is a good marker to determine the milk production function status, which may affect the milk production of goats and make it lower than that of bovines. Cholesterol-rich plaques accumulated in the arteries, preventing enough blood flow to the heart, causing cardiovascular disease, bring about many medical suggestions, and require a reduction of the cholesterol intake (39). However, throughout the lactation periods, the cholesterol levels in milk decreased sharply, and the cholesterol concentration in mature milk of goat (11.64 ± 1.09 mg/dL) was lower than that in bovine (20.58 ± 4.21 mg/dL), which is consistent with the results of the GO analysis of the DEMPs in mature milk of goat and bovine. The result of the most abundant biological processes in colostrum and mature milk indicated that goat milk is more comfortable for the human body. The extent of a triglyceride catabolic process, which occurs via acid lipolysis in the lysosome and neutral lipolysis in the cytoplasm, can regulate lipid droplet size (40). LPL and APOE as the critical enzymes that participate in the triglyceride catabolic process were higher in mature milk of goat than that in bovine goat milk, which confirmed that the regulating lipid metabolism in MFGM proteins of the goat was superior to that of bovine MFGM proteins.
The phagosome pathway is the important process of catabolism and transport, which has been reported in the Guanzhong goat and Holstein cow mature milk (16). Yang et al. (41) revealed that the phagosome pathway was a complicated process of organic uptake and elimination of apoptotic cells and pathogens, which contributes to inflammation, host defense, and homeostasis in tissues. Thus, the MFGM proteins may be crucial to the immune system of newborn mammals. The ECM-receptor interaction pathway has been found to be associated with depot-specific adipogenesis in cow and overrepresented in specific cattle breeds related to the adaptive immune response after virus inoculation in Holstein cattle (42). The structural functional diversity of proteoglycans makes them as key mediators of the interaction between tumor cells and host microenvironment, and directly participates in the tissue and dynamic remodeling of the milieu. As constituents of the ECM or extracellular milieu, proteoglycans may invariably participate in the control of a variety of oncogenic events in a multivalent manner (43). KEGG pathway analysis also displayed that the differential MFGM proteins in colostrum goat and bovine significantly regulated glycometabolism through the pathway of galactose metabolism and revealed that the MFGM proteins of goat and bovine milk possessed different effects on glucose metabolism. Peroxisome proliferator-activated receptors (PPARs) are the members of the nuclear hormone receptor superfamily, which has three member isotypes: PPARα, PPARβ/δ, and PPARγ, and is ligand-activated transcription factors. PPARs govern the expression of the crux molecules in fatty acid metabolic pathway, including the absorption, oxidation, and storage of fatty acids. Meanwhile, PPARγ maintains glucose homeostasis by activating glucose transporter 2 and glucokinase in the pancreas and liver (44). LPL, ACSL1, FABP3, and CD36 play key roles in the PPAR signaling pathway, and the abundance of these proteins in goat milk is higher than that in bovine milk, which indicated that goat milk has better function of fatty acid metabolism and glucose homeostasis. Due to the different functions of MFGM protein, the MFGM protein expressed in different lactation periods of bovine and goat provided significant information for functional food and infant formula. According to the nutritional needs of different lactating infants, functional proteins can be added to the corresponding formula, which is conducive to the health of infants.
In a previous study, the efficiency of preventing a series of enteric inflammatory and infectious diseases by milks has been documented. Viral myocarditis and salmonella infection are the subcategory of cardiovascular disease and infectious disease, respectively. The MFGM protein in mature milk significantly inhibited the internalization and binding of salmonella during the growth of mammals, and the inhibitory effect of goat milk was stronger than that of bovine milk, which was in line with our data (45). Sun et al. (15) reported that most of goat MFGM proteins were related to metabolism processes, including lipid metabolism and carbohydrate metabolism. According to the KEGG analysis results, the difference of fatty acid synthesis pathway between bovine and goat could mainly be a consequence of fatty acid synthetase activity differences between bovine and goat. Fatty acid synthases are located on the surface of endoplasmic reticulum and on the cytoplasmic lipid droplets in mammary epithelial cells. The endoplasmic reticulum possesses a series of membrane enzymes, such as palmitoyl-CoA and glycerol 3-phosphate, which can synthesize triglycerides from activated fatty acids. Palmitoyl-CoA, a major product of fatty acid synthases, and fatty acid synthases were directly connected in the fatty acid synthesis pathway (46). In addition, glucose and fructose promote the accumulation of triglycerides in milk to convert sugar to fat, the allosteric inhibition of fatty acid oxidation via increased availability of triose phosphate precursors and acetyl-CoA and metabolites, such as malonyl-CoA, for fatty acid formation via glycerol-3-phosphate biosynthesis and de novo lipogenesis (47). Regulation of fatty acid biosynthetic could inhibit hepatic steatosis and lipid accumulation.
Conclusions
Our study described a more specific strategy to provide insights into proteome differences between GC, GM, BC, and BM. Bioinformatics analysis displayed that these DEMPs in colostrum significantly regulated glycometabolism through the pathway of galactose metabolism, and the DEMPs in mature milk regulated lipid metabolism through the pathway of fatty acid biosynthesis. These trials and results could reveal the differences of nutritional value and physiological states between bovine and goat at various lactation periods, and provide direction for the application of goat milk in infant formula food and functional food. Further study on the exact role of these significant difference proteins and the specific mechanism of activating the regulation pathway of galactose and lipid is necessary and could potentially help to determine the new biomarkers or establish the optimized formula of goat milk-based functional food.
Data Availability Statement
The original contributions generated for the study are publicly available. This data can be found here: https://zenodo.org/record/4638812#.YHbYPOhKiUm.
Author Contributions
WJ: conceptualization, methodology, software, writing- original draft preparation, and supervision. RZ: data curation and writing- original draft preparation. ZZ: reviewing and editing. LS: software. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the National Natural Science Foundation of China (No. 31801643), Scientific Research Program Funded by Shaanxi Province Education Department, China (No. 20JC004), Key Research and Development Program of Shaanxi (No. 2019NY-117), and Science and Technology Project of Weiyang District, Xi'an City (No. 201936).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.680683/full#supplementary-material
References
1. Calvo MV, Martin-Hernandez MC, Garcia-Serrano A, Castro-Gomez MP, Alonso-Miravalles L, Garcia-Martin R, et al. Comprehensive characterization of neutral and polar lipids of buttermilk from different sources and its milk fat globule membrane isolates. J Food Compos Anal. (2020) 86:103386. doi: 10.1016/j.jfca.2019.103386
2. Priyanka M, Sabine D, Aman P, Koen D, Christophe B. The interfacial properties of various milk fat globule membrane components using Langmuir isotherms. Food Biosci. (2017) 20:96–103. doi: 10.1016/j.fbio.2017.08.008
3. Spitsberg VL. Invited review: bovine milk fat globule membrane as a potential nutraceutical. J Dairy Sci. (2005) 88:2289–94. doi: 10.3168/jds.S0022-0302(05)72906-4
4. Li TE, Gao J, Du M, Mao XY. Milk fat globule membrane supplementation modulates the gut microbiota and attenuates metabolic endotoxemia in high-fat diet-fed mice. J Fun Foods. (2018) 47:56–65. doi: 10.1016/j.jff.2018.05.038
5. Cebo C, Caillat H, Bouvier F, Martin P. Major proteins of the goat milk fat globule membrane. J Dairy Sci. (2010) 93:868–76. doi: 10.3168/jds.2009-2638
6. Ozturk G, Shah IM, Mills DA, German JB, Bell JMLND. The antimicrobial activity of bovine milk xanthine oxidase. Int Dairy J. (2020) 102:104581. doi: 10.1016/j.idairyj.2019.104581
7. Chen ZL, Chopp M, Zacharek A, Li W, Venkat P, Wang FJ, et al. Brain-Derived Microparticles (BDMPs) contribute to neuroinflammation and lactadherin reduces BDMP induced neuroinflammation and improves outcome after stroke. Front Immunol. (2019) 10:2747. doi: 10.3389/fimmu.2019.02747
8. Chopra A, Ali SA, Bathla S, Rawat P, Vohra V, Kumar S, et al. High-resolution mass spectrometer-based ultra-deep profile of milk whey proteome in Indian Zebu (Sahiwal) cattle. Front Nutr. (2020) 7:150. doi: 10.3389/fnut.2020.00150
9. Nguyen HTH, Ong L, Hoque A, Kentish SE, Williamson N, Ang CS, et al. A proteomic characterization shows differences in the milk fat globule membrane of buffalo and bovine milk. Food Biosci. (2017) 19:7–16. doi: 10.1016/j.fbio.2017.05.004
10. Li WX, Li MH, Cao XY, Han HJ, Kong FH, Yue XQ. Comparative analysis of whey proteins in donkey colostrum and mature milk using quantitative proteomics. Food Res Int. (2020) 127:108741. doi: 10.1016/j.foodres.2019.108741
11. Cebo C, Rebours E, Henry C, Makhzami S, Cosette P, Martin P. Identification of major milk fat globule membrane proteins from pony mare milk highlights the molecular diversity of lactadherin across species. J Dairy Sci. (2012) 95:1085–98. doi: 10.3168/jds.2011-4455
12. Jia W, Dong X, Shi L, Chu X. Discrimination of milk from different animal species by a foodomics approach based on high-resolution mass spectrometry. J Agric Food Chem. (2020) 68:6638–45. doi: 10.1021/acs.jafc.0c02222
13. Chen D, Li XY, Zhao X, Qin YS, Wang JM, Wang CF. Comparative proteomics of goat milk during heated processing. Food Chem. (2019) 275:504–14. doi: 10.1016/j.foodchem.2018.09.129
14. Chen D, Li XY, Zhao X, Qin YS, Zhang XX, Li J, et al. Proteomics and microstructure profiling of goat milk protein after homogenization. J Dairy Sci. (2019) 102:3839–50. doi: 10.3168/jds.2018-15363
15. Sun YX, Wang CN, Sun XM, Guo MR. Comparative proteomics of whey and milk fat globule membrane proteins of Guanzhong goat and Holstein cow mature milk. J Food Sci. (2019) 84:244–53. doi: 10.1111/1750-3841.14428
16. Sun YX, Wang CN, Sun XM, Jiang SL, Guo MR. Characterization of the milk fat globule membrane proteome in colostrum and mature milk of Xinong Saanen goats. J Food Sci. (2020) 103:3017–24. doi: 10.3168/jds.2019-17739
17. Lu J, Liu L, Pang XY, Zhang SW, Jia ZH, Ma CL, et al. Comparative proteomics of milk fat globule membrane in goat colostrums and mature milk. Food Chem. (2016) 209:10–16. doi: 10.1016/j.foodchem.2016.04.020
18. Lu J, Wang XY, Zhang WQ, Liu L, Pang XY, Zhang SW, et al. Comparative proteomics of milk fat globule membrane in different species reveals variations in lactation and nutrition. Food Chem. (2016) 196:665–72. doi: 10.1016/j.foodchem.2015.10.005
19. Jia W, Zhang R, Shi L, Zhang F, Chang J, Chu XG. Effects of spices on the formation of biogenic amines during the fermentation of dry fermented mutton sausage. Food Chem. (2020) 321:126723. doi: 10.1016/j.foodchem.2020.126723
20. Ji XX, Li XS, Ma Y, Li D. Differences in proteomic profiles of milk fat globule membrane in yak and cow milk. Food Chem. (2017) 221:1822–7. doi: 10.1016/j.foodchem.2016.10.097
21. Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. (2005) 206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x
22. Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide Label-free quantification by delayed normalization and maximal peptide ratio extraction, Termed MaxLFQ. Mol Cell Proteomics. (2014) 13:2513–26. doi: 10.1074/mcp.M113.031591
23. Jia W, Shi QY, Zhang R, Shi L, Chu XG. Unraveling proteome changes of irradiated goat meat and its relationship to off-flavor analyzed by high-throughput proteomics analysis. Food Chem. (2021) 337:127806. doi: 10.1016/j.foodchem.2020.127806
24. Lefebvre G, Shevlyakova M, Charpagne A, Marquis J, Vogel M, Kirsten T, et al. Time of lactation and maternal fucosyltransferase genetic polymorphisms determine the variability in human milk oligosaccharides. Front Nutr. (2020) 7:574459. doi: 10.3389/fnut.2020.574459
25. Cunsolo V, Fasoli E, Di Francesco A, Saletti R, Muccilli V, Gallina S, et al. Polyphemus, Odysseus and the ovine milk proteome. J Proteomics. (2017) 152:58–74. doi: 10.1016/j.jprot.2016.10.007
26. Fustini M, Galeati G, Gabai G, Mammi LE, Bucci D, Baratta M, et al. Overstocking dairy cows during the dry period affects dehydroepiandrosterone and cortisol secretion. J Dairy Sci. (2017) 100:620–8. doi: 10.3168/jds.2016-11293
27. Zhao LL, Du M, Gao J, Zhan BY, Mao XY. Label-free quantitative proteomic analysis of milk fat globule membrane proteins of yak and cow and identification of proteins associated with glucose and lipid metabolism. Food Chem. (2019) 275:59–68. doi: 10.1016/j.foodchem.2018.09.044
28. Pacheco-Fernandez N, Pakdel M, Blank B, Sanchez-Gonzalez I, Weber K, Tran ML, et al. Nucleobindin-1 regulates ECM degradation by promoting intra-Golgi trafficking of MMPs. J Cell Biol. (2020) 219:e201907058. doi: 10.1083/jcb.201907058
29. Tsukumo Y, Tsukahara S, Saito S, Tsuruo T, Tomida A. A novel endoplasmic reticulum export signal proline at the +2-position from the signal peptide cleavage site. J Biol Chem. (2009) 284:27500–10. doi: 10.1074/jbc.M109.021592
30. Holm J, Hansen SI. Characterization of soluble folate receptors (folate binding proteins) in humans. Biological roles and clinical potentials in infection and malignancy. BBA Proteins Proteomics. (2020) 1868:140466. doi: 10.1016/j.bbapap.2020.140466
31. Luo CC, Zhao SG, Dai WT, Zheng N, Wan JQ. Proteomic analysis of lysosomal membrane proteins in bovine mammary epithelial cells illuminates potential novel lysosome functions in lactation. J Agric Food Chem. (2018) 66:13041–9. doi: 10.1021/acs.jafc.8b04508
32. Delanghe JR, Speeckaert R, Speeckaert MM. Behind the scenes of vitamin D binding protein: more than vitamin D binding. Best Pract Res Clin Endocrinol Metab. (2005) 29:773–86. doi: 10.1016/j.beem.2015.06.006
33. Bernard L, Rouel J, Leroux C, Ferlay A, Faulconnier Y, Legrand P, et al. Mammary lipid metabolism and milk fatty acid secretion in alpine goats fed vegetable lipids. J Dairy Sci. (2005) 88:1478–89. doi: 10.3168/jds.S0022-0302(05)72816-2
34. Zhu JJ, Luo J, Sun YT, Shi HB, Li J, Wu M, et al. Effect of inhibition of fatty acid synthase on triglyceride accumulation and effect on lipid metabolism genes in goat mammary epithelial cells. J Dairy Sci. (2015) 98:3485–91. doi: 10.3168/jds.2014-8202
35. Zeng R, Bequette BJ, Vinyard BT, Bannerman DD. Determination of milk and blood concentrations of lipopolysaccharide-binding protein in cows with naturally acquired subclinical and clinical mastitis. J Dairy Sci. (2009) 92:980–9. doi: 10.3168/jds.2008-1636
36. Chen JL, Zhai ZY, Long HR, Yang GM, Deng BC, Deng JP. Inducible expression of defensins and cathelicidins by nutrients and associated regulatory mechanisms. Peptides. (2020) 123:170177. doi: 10.1016/j.peptides.2019.170177
37. Jayaraman S, Chavez OR, Pérez A, Miñambres I, Sánchez-Quesada JL, Gursky O. Binding to heparin triggers deleterious structural and biochemical changes in human low-density lipoprotein, which are amplified in hyperglycemia. BBA Mol Cell Biol Lipids. (2020) 1865:158712. doi: 10.1016/j.bbalip.2020.158712
38. Xu BZ, Zhang F, Xu XL, Li XM, He MY, Nie XM, et al. Effect of physiological and production activities on the concentration of naturally occurring steroid hormones in raw milk. Int J Dairy Technol. (2020) 73:471–8. doi: 10.1111/1471-0307.12672
39. Larsen T. Enzymatic-fluorometric quantification of cholesterol in bovine milk. Food Chem. (2012) 135:1261–7. doi: 10.1016/j.foodchem.2012.05.061
40. El-Tarabany MS, El-Tarabany AA, Roushdy EM. Impact of lactation stage on milk composition and blood biochemical and hematological parameters of dairy Baladi goats. Saudi J Biol Sci. (2018) 25:1632–8. doi: 10.1016/j.sjbs.2016.08.003
41. Yang M, Deng W, Cao XY, Wang LJ, Yu N, Zheng Y, et al. Quantitative phosphoproteomics of milk fat globule membrane in human colostrum and mature milk: new insights into changes in protein phosphorylation during lactation. J Agric Food Chem. (2020) 68:4546–56. doi: 10.1021/acs.jafc.9b06850
42. Criscitiello MF, Kraev I, Lange S. Post-translational protein deimination signatures in serum and serum-extracellular vesicles of Bos taurus reveal immune, anti-pathogenic, anti-viral, metabolic and cancer-related pathways for deimination. Int J Mol Sci. (2020) 21:2861. doi: 10.3390/ijms21082861
43. Garusi E, Rossi S, Perris R. Antithetic roles of proteoglycans in cancer. Cell Mol Life Sci. (2012) 69:553–79. doi: 10.1007/s00018-011-0816-1
44. Variya BC, Bakrania AK, Patel SS. Antidiabetic potential of gallic acid from Emblica officinalis: improved glucose transporters and insulin sensitivity through PPAR-γ and Akt signaling. Phytomedicine. (2020) 73:152906. doi: 10.1016/j.phymed.2019.152906
45. Guri A, Griffiths M, Khursigara CM, Corredig M. The effect of milk fat globules on adherence and internalization of Salmonella enteritidis to HT-29 cells. J Dairy Sci. (2012) 95:6937–6945. doi: 10.3168/jds.2012-5734
46. Moriya H, Uchida K, Okajima T, Matsuda T, Nadano D. Secretion of three enzymes for fatty acid synthesis into mouse milk in association with fat globules, and rapid decrease of the secreted enzymes by treatment with rapamycin. Archiv Biochem Biophys. (2011) 508:87–92. doi: 10.1016/j.abb.2011.01.015
Keywords: milk fat globule membrane protein, goat milk, bovine milk, Q-Orbitrap, colostrum, mature milk
Citation: Jia W, Zhang R, Zhu Z and Shi L (2021) A High-Throughput Comparative Proteomics of Milk Fat Globule Membrane Reveals Breed and Lactation Stages Specific Variation in Protein Abundance and Functional Differences Between Milk of Saanen Dairy Goat and Holstein Bovine. Front. Nutr. 8:680683. doi: 10.3389/fnut.2021.680683
Received: 15 March 2021; Accepted: 06 May 2021;
Published: 28 May 2021.
Edited by:
Junli Xu, University College Dublin, IrelandReviewed by:
Cunfang Wang, Qilu University of Technology, ChinaXiaohui Lin, University College Dublin, Ireland
Adriano Rangel, Federal University of Rio Grande Do Norte, Brazil
Copyright © 2021 Jia, Zhang, Zhu and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Jia, amlhd2VpQHN1c3QuZWR1LmNu; Lin Shi, NDUwOUBzdXN0LmVkdS5jbg==
 Wei Jia
Wei Jia Rong Zhang
Rong Zhang