- 1School of Life Sciences, Beijing University of Chinese Medicine, Beijing, China
- 2School of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing, China
- 3State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, Beijing, China
- 4Zhongcai Health (Beijing) Biological Technology Development Co., Ltd., Beijing, China
- 5Department of Biomedical Engineering, Shenzhen Research Institute, Beijing University of Chinese Medicine, Shenzhen, China
Screening functional food ingredients (FFI) from medicinal and edible plants (MEP) has still remained a great challenge due to the complexity of MEP and its obscure function mechanisms. Herein, an integrated strategy based on sequential metabolites identification approach, network pharmacology, molecular docking, and surface plasmon resonance (SPR) analysis was proposed for quickly identifying the active constituents in MEP. First, the sequential biotransformation process of MEP, including intestinal absorption and metabolism, and hepatic metabolism, was investigated by oral gavage, and intestinal perfusion with venous sampling method. Then the blood samples were analyzed by UPLC-Q Exactive Orbitrap HRMS. Second, the network pharmacology approach was used to explore the potential targets and possible mechanisms of the in vivo metabolites of MEP. Third, molecular docking and SPR approaches were used to verify the specific interactions between protein targets and representative ingredients. The proposed integrated strategy was successfully used to explore the heptoprotective components and the underlying molecular mechanism of Paeoniae Radix Alba (PRA). A total of 44 compounds were identified in blood samples, including 17 porotypes and 27 metabolites. The associated metabolic pathways were oxidation, methylation, sulfation, and glucuronidation. After further screening, 31 bioactive candidates and 377 related targets were obtained. In addition, the bioactive components contained in PRA may have therapeutic potentials for non-alcoholic fatty liver disease (NAFLD). The above results demonstrated the proposed strategy may provide a feasible tool for screening FFI and elaborating the complex function mechanisms of MEP.
Introduction
Medicinal and edible plants (MEP) has been extensively used for preventing and treating various diseases in China for centuries (1). After taken orally, MEP would be bio-transformed progressively and orderly from the gastrointestinal tract and liver, to the systemic blood stream (2, 3). However, current metabolic studies are just focus on characterizing the metabolites in different biological samples, like urine, blood, bile and other tissues/organs, and they could not comprehensively illustrate the dynamic biotransformation process of MEP (4). In addition, accumulating evidences have suggested that gut wall metabolism plays an important role in first pass metabolism of small molecules, while only a few in vitro studies using intestinal microsomes have been performed (5). Recently, many in situ approaches have been developed and demonstrated efficient to study drug intestinal absorption and metabolism, such as in situ closed-loop method (4), intestinal single-pass perfusion, intestinal recirculating perfusion, and intestinal perfusion with venous sampling (IPVS) (6). Among them, IPVS is recommended as it enables intestinal absorption/metabolism to occur at body temperature, and allows gut wall metabolism to be studied without interference by the confounding effects of liver metabolism (7, 8). Moreover, detecting and identifying the absorbed components and metabolites of MEP from complex plasma samples is often challenging, due to the extremely low concentrations of the interested compounds and the interference of the endogenous metabolites and proteins (9). Owing to the high sensitivity and selectivity, ultrahigh performance liquid chromatography coupled with hybrid Q-Exactive-Orbitrap high resolution mass spectrometry (UPLC-Q Exactive Orbitrap HRMS) has become a powerful tool for rapidly and accurately profiling the trace compounds in biological samples (10). Therefore, screening the prototypes and metabolites involved in the dynamic metabolic process using UPLC-Q Exactive Orbitrap HRMS and IPVS is the first step to systemically explicate material basis of MEP.
Unlike western medicine of “one target, one drug,” MEP is a complicated system with multi-component and multi-target characteristics, which achieves its therapeutic effects through targeting multiple physiological pathways (11). Due to the complicated chemical components in MEP, conventional approaches have great difficulties in delivering a systematic understanding of the synergistic effects of MEP for preventing and treating complex diseases (12). With bioinformatics' rapid progress, the newly emerging network pharmacology has greatly facilitated mechanistic studies into the synergistic actions of multi-component drugs at the proteome or systemic level (13). It emphasizes the concept of “network target, multi-component therapeutics,” which is consistent with the integrality and systematicness of traditional Chinese medicine (TCM) theory (14, 15). Up to date, this state-of-the-art method has been successfully used for elucidating the complex molecular mechanisms of TCM for the treatment of various diseases, such as Alzheimer's disease (2), cardiovascular disease (14, 16), cancer (17, 18), diabetes (19), asthma (20, 21), gastritis (22), acute ulcerative colitis (23), and acute mountain sickness (24).
Surface plasmon resonance (SPR) biosensor is a powerful tool for characterizing and quantifying the kinetics and binding affinities of biomolecular interactions (25, 26). When the analyte molecules in a liquid sample were in contact with the ligands, changes in the refractive index (RI) at the sensor surface were produced, which can be measured by the optical reader (27, 28). Hence, the major advantage of SPR is that the sample can be detected in real time and without the need of labeling (29, 30). Another advantage is that SPR can directly and specifically capture the bioactive candidates from complex matrices (31). Therefore, SPR has been increasingly penetrated into almost all fields of TCM research, such as bioactive compound screening and target fishing (32).
Paeoniae Radix Alba (PRA, Baishao in Chinese), the dried roots of Paeonia lactiflora Pall., is a famous herbal medicine or functional food in China and many other Asia countries (33). According to TCM theory, PRA has antispasmodic, tonic, astringent, and analgesic properties (34), which has been clinically used as an anticoagulant, antidepressant, antioxidant, and liver protectant agent for centuries (33). Recently, accumulating evidences indicate that PRA exerts significant effects in the prevention and treatment of liver diseases, such as acute liver injury (35), fatty liver diseases (36, 37), liver fibrosis (38, 39), cholestasis (40, 41), hepatitis (42), and liver cancer (43, 44). However, the bioactive components and the underlying molecular mechanisms are largely unknown.
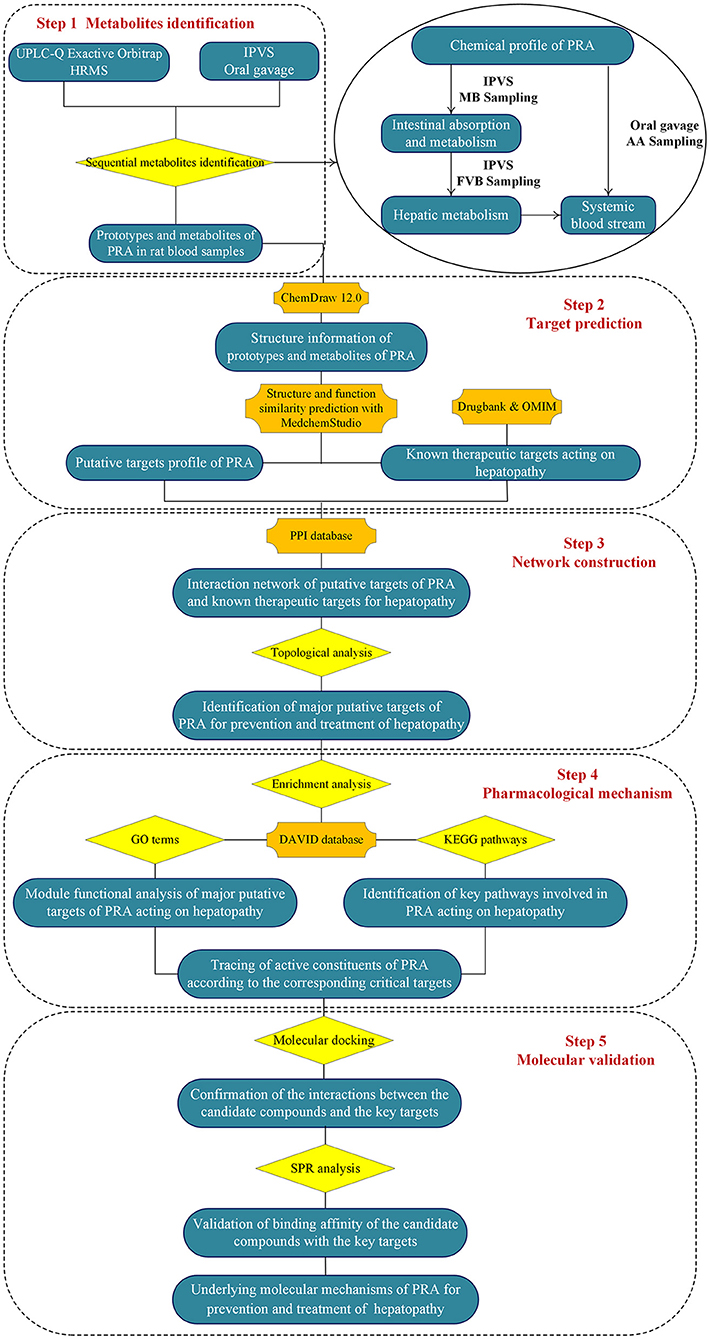
To address the above issues, an integrated strategy was developed for quickly identifying functional ingredients from MEP, using PRA as an example. First, the chemical profile of MEP extract was identified by UPLC-Q Exactive Orbitrap HRMS. Second, the intestinal absorption and metabolism of MEP was investigated by intestinal perfusion with mesenteric blood (MB) sampling. By comparing the components in mesenteric blood with the chemical components of MEP, we could identify the absorbed components and metabolites produced by gut wall metabolism. Third, the hepatic metabolism was investigated by intestinal perfusion with femoral venous blood (FVB) sampling. By comparing the components in mesenteric blood with the components in femoral venous blood, we could identify the components in the systemic blood stream and metabolites produced by hepatic metabolism. Fourth, to comprehensively screen the components in vivo, the major components and metabolites in rat plasma [blood was collected from the abdominal aorta (AA)] after oral administration of MEP extract were analyzed. Fifth, the network pharmacology strategy was used to screen the molecular targets and pathways involved in MEP for preventing and treating particular diseases. Finally, molecular docking and surface plasmon resonance methodologies were employed to confirm the binding abilities between the candidate compounds and their associated targets. We hope this integrated strategy would be helpful to identify the functional food ingredients (FFI) from MEP. The flowchart of the study design was illustrated in Figure 1.
Materials and Methods
Materials and Reagents
Paeoniae Radix Alba was supplied by Lanzhou Foci Pharmaceutical Co., Ltd. (Qinghai, China) and authenticated by Professor Jingjuan Wang (Beijing University of Chinese Medicine, Beijing, China). Chromatographic-grade acetonitrile, menthol and formic acid was supplied by Fisher Scientific. Reference standards (purity ≥ 90%) of paeoniflorin, gallic acid, benzoic acid were purchased from National Institutes for Food and Drug Control (Beijing, China). Reference standards (purity ≥ 90%) of benzoylpaeoniflorin, oxypaeoniflorin, albiflorin, and 1,2,3,4,6-O-pentagalloylglucose were purchased from Shanghai Yuanye Biological Technology Co., Ltd. (Shanghai, China). Ultrapure water was purified by the Millipore Milli Q plus purification system. All other reagents used were of analytical grade and commercially available.
Preparation of PRA Solution
PRA was chopped into pieces, and then PRA pieces (about 300 g) were decocted with water twice (solid-liquid ratio: 1:10), 1 h for each time. The supernatant was then filtered and the two filtrates were mixed and condensed to obtain a PRA solution (1 g/mL) which was used for animal studies. For chemical analysis, the PRA solution (1 g/mL) was diluted to 10 mg/ml crude drug and then filtered with 0.22 μm membrane before UPLC-Q Exactive Orbitrap HRMS.
Preparation of Standard Solutions
Individual stock solutions of 7 reference standards were dissolved with methanol in a 10 mL volumetric flask and stored at 4°C. Then the standard solutions were filtered with 0.22 μm membrane before UPLC-Q Exactive Orbitrap HRMS.
Animals
Pathogen free male Sprague-Dawley rats (280–300 g) were purchased from Spfanimals Laboratory Animal Technology Co. Ltd (Beijing, China). Treatment for animals in this study was approved by the animal ethnic committee of Beijing University of Chinese Medicine. The rats had free access to water and standard diet, and were maintained in an environmentally controlled rearing room (temperature:23°C, the humidity: 60%) under a 12 h light/dark cycle. All animals were acclimated for at least a week, then fasted for ~12 h with water ad libitum before each experiment.
Surgical Procedures of IPVS
For animals undergoing IPVS experiments, the surgical procedures were conducted followed previously published reports (45). Briefly, prior to initiation of perfusion surgical operation, several rats were used for donor blood. Whole blood was drawn from the abdominal aorta using a 10-mL syringe (100 U of heparin added to 10 mL of donor blood) and incubated in a 37°C water bath until administered to the recipient rat. The recipient rat was anesthetized by intraperitoneal injection of chloral hydrate (400 mg/kg), fixed in a supine on the operating table, and kept warm by a heat lamp placed over the surgical area.
Upon verification of the loss of pain reflex, the left external jugular vein was exposed and cannulated with a 24-gauge i.v. catheter to transfuse blood from the donor blood reservoir. Then the abdominal cavity was opened along the abdominal line. The jejunum segment was located and the two ends were incised with surgical scissors for cannula. Two silicone tubes were inserted through the small slits and secured. The segment was then rinsed with warm isotonic saline until the effluent was clear. To collect venous outflow, a 24-gauge i.v. catheter filled with heparinized saline was intubated into the mesenteric vein (gut wall metabolism)/femoral vein (hepatic metabolism) and secured with instant glue. PRA solution (1 g/mL) was incubated in a 37°C water bath to maintain the temperature and pumped at a flow rate of 0.2 mL/min. The blood was pumped at the flow rate of 0.3 mL/min. At the end of the surgical procedure and throughout the experiment, the exposed intestinal segment was covered with a piece of sterilized gauze that had been moistened by frequent applications of warm isotonic saline.
Blood draining from the cannulated mesenteric vein was collected into heparinized centrifuge tubes within 2 h. Plasma samples were separated by centrifuging the blood samples at 4,000 rpm for 10 min and stored at −20°C.
Oral Drug Administration
Rats were randomly divided into eight groups (three animals each). Then, the four treatment group rats were administrated by oral gavage of 4 mL PRA solution (1 g/mL). The corresponding blank groups were given 4 mL saline instead. The rats were anesthetized by intraperitoneal injection of chloral hydrate (400 mg/kg). Then, blood samples were collected from the abdominal aorta at 0.5, 1, 1.5, and 2 h (three rats for each time), respectively. At the end of this study, all rats were sacrificed by conducting a bilateral thoracotomy.
Preparation of the Blood Samples
An aliquot of 1.5 mL of plasma sample was added into 1.5 mL of 4% phosphoric acid. Then the mixture was purified by solid phase extraction (SPE) on an Oasis PRiME HLB cartridge. The sample was loaded on the pretreated column, washed with 3 mL of water, and then eluted with 6 mL of acetonitrile: methanol (90:10 v/v). The eluate was collected and dried by nitrogen blowing instrument. The residue was dissolved in 1.5 mL of methanol and filtered with 0.22 μm membrane before LC/MS analysis.
LC/MS Analysis
For the LC/MS analysis, an Q Exactive Orbitrap high resolution mass spectrometer equipped with a heated electrospray ionization (HESI) source, was coupled to a Thermo Dionex Ultimate 3000 UPLC system (consisting of an autosampler, a diode array detector, a column oven and a dual pump connected to an online degasser). The data were recorded and processed using Xcalibur, Metworks and Mass Frontier 6.0 software packages (Thermo Fisher Scientific).
UPLC chromatographic separations were executed on a Waters CORTECS UPLC T3 (2.1 × 100 mm, 1.6 μm) column thermostated at 40°C. The mobile phases consisted of water with 0.1% formic acid (A) and acetonitrile (B), and the gradient program was conducted as follows: 0–1 min (5% B), 1–20 min (5–95% B), 20–21 min (95–95% B), 21–21.1 min (95–5% B), and 21.1–22 min (5–5% B). The sample flow rate was 0.3 mL/min and the injection volume was 2 μL.
The MS conditions were as follows: alternate switching (−)/(+) ESI full scan mode, the capillary temperature was 300°C, auxiliary temperature was 250°C, positive spray voltage was set at +3.5 kV, negative spray voltage was set at −3.0 kV, shealth gas (N2) flow was 35 Arb, aux gas flow rate was 10 Arb. Full MS scans were acquired in the range of m/z 100–1,500, the collision energy was set at 20, 30, 40 eV. The MS/MS experiments were set as data-dependent scans.
Predicting the PRA-Related Targets
As described in our previous studies (14, 22), a powerful drug similarity search tool named MedChem Studio (MedChem Studio, 3.0; Simulations Plus, Inc, Lancaster, CA, USA, 2012) was employed for the prediction of the PRA-related targets, with a similarity threshold of 0.60.
Collection of Hepatopathy (HP)-Related Targets
By using the query of “liver disease” and “hepatopathy,” and limiting the species with “Homo sapiens,” the HP-related targets were searched from DrugBank database (https://go.drugbank.com/, version 5.1.7, updated on July 2nd, 2020), and the Online Mendelian Inheritance in Man (OMIM) database (https://omim.org/, updated on July 1st, 2019). DrugBank is a public database which integrates the drug structures (including approved, investigational and withdrawn drugs), drug target proteins/genes, pathways and other information associated with human diseases (46). OMIM can provide detailed information on genetic disorders and their related human genes (47). To reduce the false positives, only the druggable proteins/genes links to the HP were retained.
Protein-Protein Interaction (PPI) Data
The PRA- and HP-related targets were input into STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (http://string-db.org/, version 11.0) to predict the possible PPI information. The STRING database is a versatile platform for the straightforward identification of direct or indirect functional interactions between proteins (48). Then, the PPI data whose confidence scores >0.4 would be reserved.
Network Construction and Analysis
To elaborate the connections among the components, target proteins/genes, and disease, a “component-target-disease” network was created by inputting the data of selected components of PRA, PRA-associated targets as well as the targets related to HP into Cytoscape software (version 3.6.0, Boston, MA, USA). Cytoscape is a valuable platform for the analysis and visualization of the complicated network involved in various biological processes (49). Then, node degree, a quantitative characteristic of the network, was calculated. Nodes with the degree values higher than twice the median degree of the whole nodes would be chosen as a hub (14). Next, the interaction network of hubs was established by using the direct links between hubs. Furthermore, the network analyzer function was used for the analysis of degree centrality (DC), betweenness centrality (BC), and closeness centrality (CC) to assess the topological importance of a node in the entire network, as described in our prior publications (22). To increase the reliability of the predicted results, the hubs with “degree” > median DC, “betweenness” > median BC and “closeness” > median CC were recognized as critical hubs.
Pathway Enrichment Performance
By using DAVID (Database for Annotation, Visualization and Integrated Discovery) bioinformatics resources (http://david.abcc.ncifcrf.gov/home.jsp/, version 6.7), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (KEGG, http://www.genome.jp/kegg/) and gene ontology (GO) enrichment evaluation were performed to clarify the pathways that were involved in PRA acting on liver disease. A P < 0.05 was considered high confidence.
Molecular Docking Simulation
To further confirm the interactions between the candidate compounds and the key targets, molecular docking analysis was performed by use of the CDOCKER module implemented in Discovery Studio 2016 (DS 2016). The crystallographic structures of the target proteins were retrieved from the protein data bank (PDB, http://www.rcsb.org/pdb/home/home.do), and prepared by adding hydrogens, deleting the ligands and water motifs. The three-dimensional (3D) structures of the potential active components were drawn with Chem3D Pro 12.0. CDOCKER interaction energies (CIEs) were adopt to evaluate the binding abilities between the vital target proteins and their corresponding constituents.
SPR Analysis
According to the molecular docking results, PRKAG1 and NFKB1 were chosen for SPR analysis. A Biacore 8K (GE Healthcare, Sweden) was used to perform the SPR experiments. A freshly prepared mixture of NHS and EDC (1:1, v/v) was first injected into the instrument to activate the carboxyl groups on the surface of the CM5 sensor (General Electric Company, USA). Then, proteins were diluted in sodium acetate solution (GE Healthcare) and immobilized on CM5 chips by using the Amine Coupling Kit (GE Healthcare), with immobilization levels of 2200 and 11800 RU (response units), respectively. The affinity measurement was carried out following the protocol provided by GE Healthcare. To exclude false-positive results, a reference channel without the conjugated protein was activated and blocked for each analysis, which served as a control to test for unspecific binding to the chip. Analytes were serially injected at a flow rate of 30 μL/min. The spontaneous association-dissociation process of the component on the protein was real-time monitored by the response value. The SPR curves were finally fitted by use of Biacore Insight Evaluation Software according to 1:1 Langmuir binding model, from which the binding constants and kinetic parameters were calculated.
Results and Discussion
Sequential Metabolism of PRA
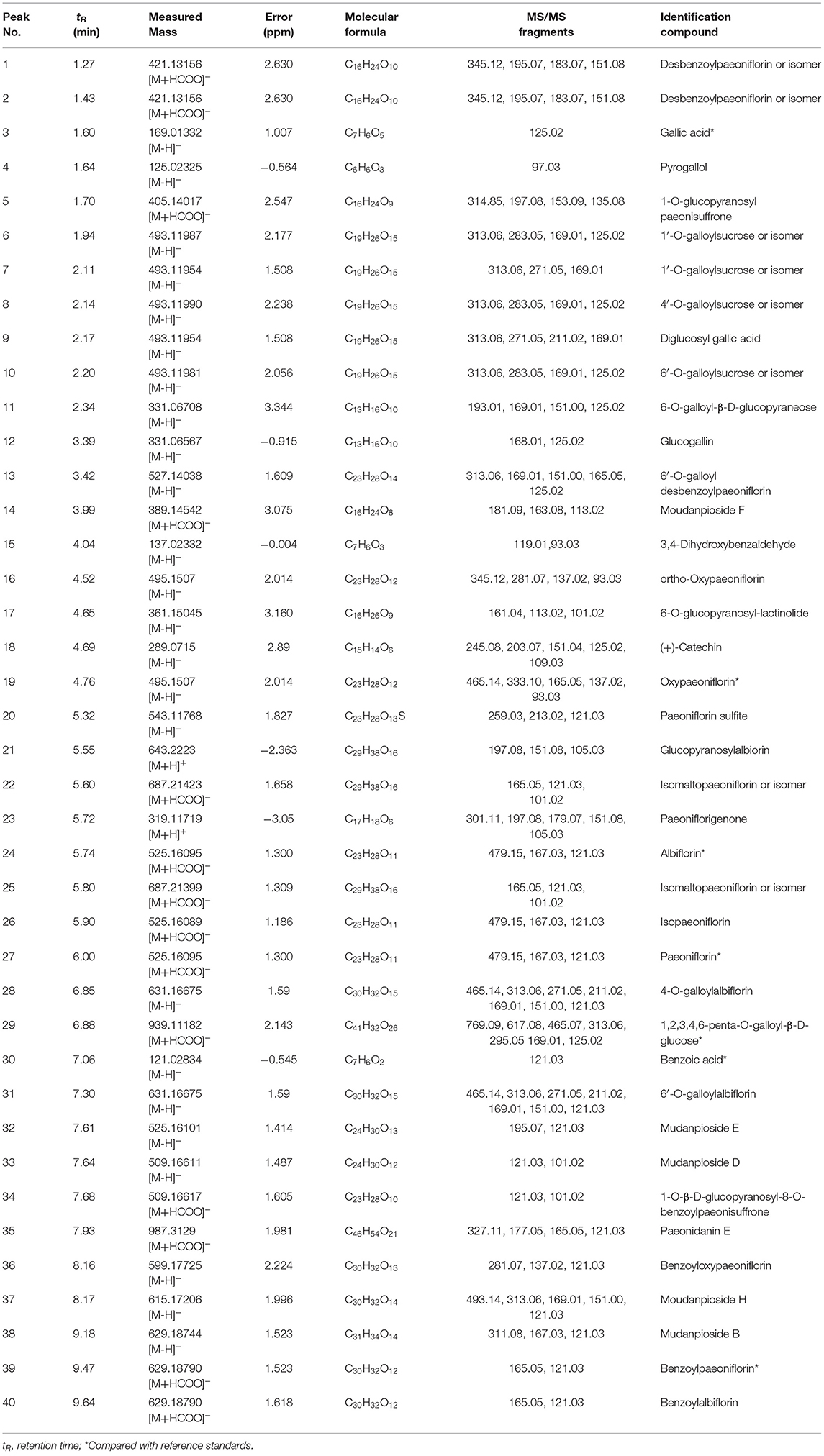
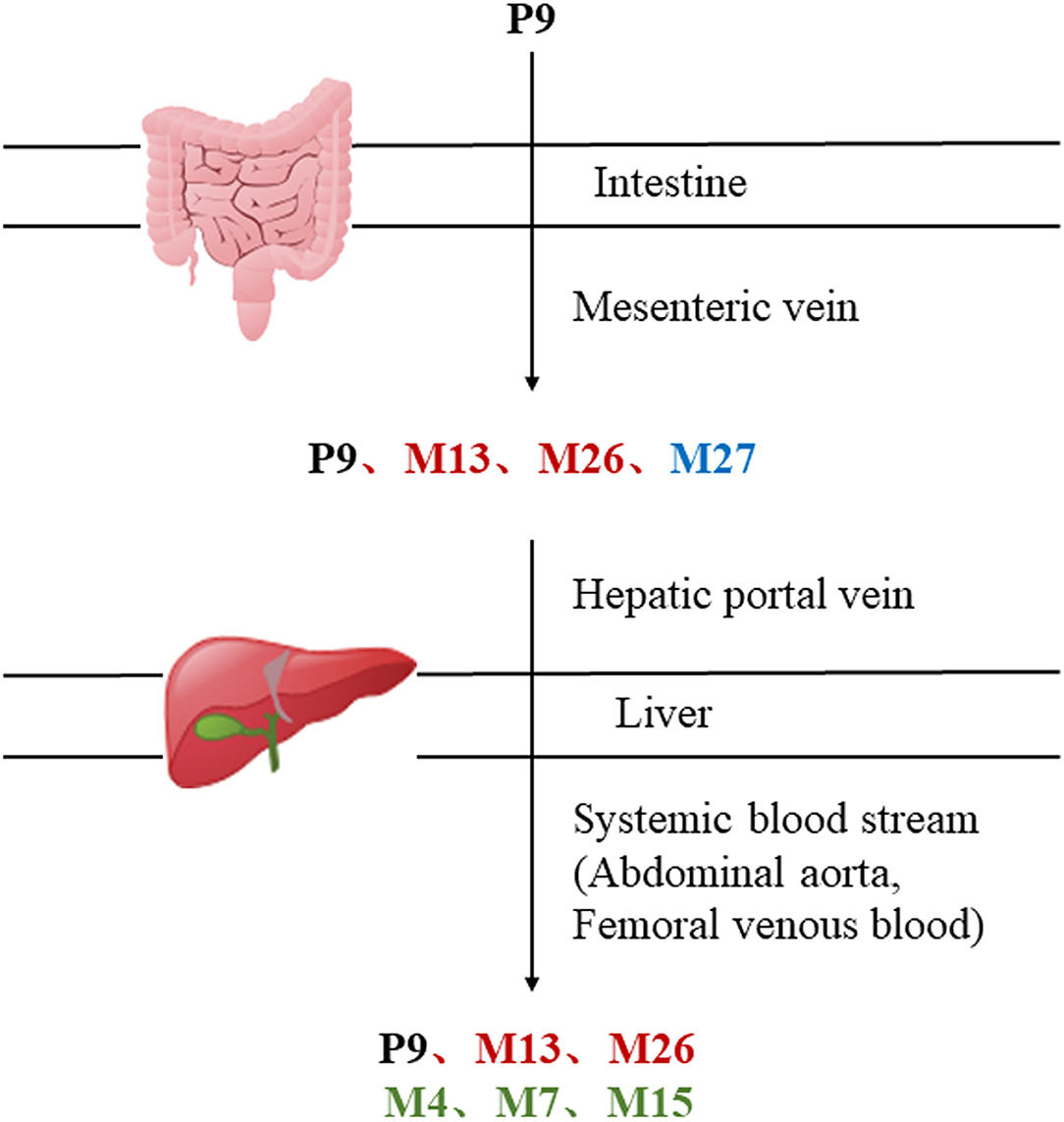
MEP taken by oral administration pass sequentially from the gastrointestinal lumen, through the gut wall and liver, and then reach the systemic circulation (50), which are subject to extensive “first-pass” elimination in many cases (51). In this work, the detection and identification of components in blank plasma sample, drug-treated plasma sample and PRA sample were conducted by UPLC-Q Exactive Orbitrap HRMS. As depicted in Supplementary Figures 1, 2 and Table 1, a total of 40 components were first identified from the water extract of PRA, mainly including monoterpenes and their glycosides, phenolic compounds, and tannins. We selected oxypaeoniflorin (peak 19), gallic acid (peak 3) and 1,2,3,4,6-penta-O-galloyl-β-D-glucose (peak 29) as the representative components to illustrate the characteristic fragmentation rules of monoterpenes, phenolic compounds and tannins, respectively (Supplementary Figures 3–5). Further, 44 compounds (Table 2), including 17 prototypes and 27 metabolites were identified from drug-treated plasma sample through comparing their molecular formulas, fragment ions, and retention times with those of the parent compounds. Among them, 32 of them were from MB group, 21 were from FVB group, and 29 were from AA group. The main metabolic pathways of PRA were found, including oxidation, methylation, sulfation, glucuronidation (Supplementary Figures 6–12). It should be noted that 15 metabolites were produced after passing the intestine, which may be mainly due to the metabolizing enzymes present in gut wall, suggesting that the intestine may play an important role in the first-pass metabolism of MEP. In addition, most metabolites of PRA were generated after hepatic metabolism, indicating that liver was the major metabolic site of PRA. The sequential metabolism of PRA was clearly characterized and paeoniflorin (P9) was selected as an example, as shown in Figure 2. Taken together, the above study could give a comprehensive map of the dynamic metabolic process of PRA, which would effectively narrow the range of potentially bioactive components of PRA.
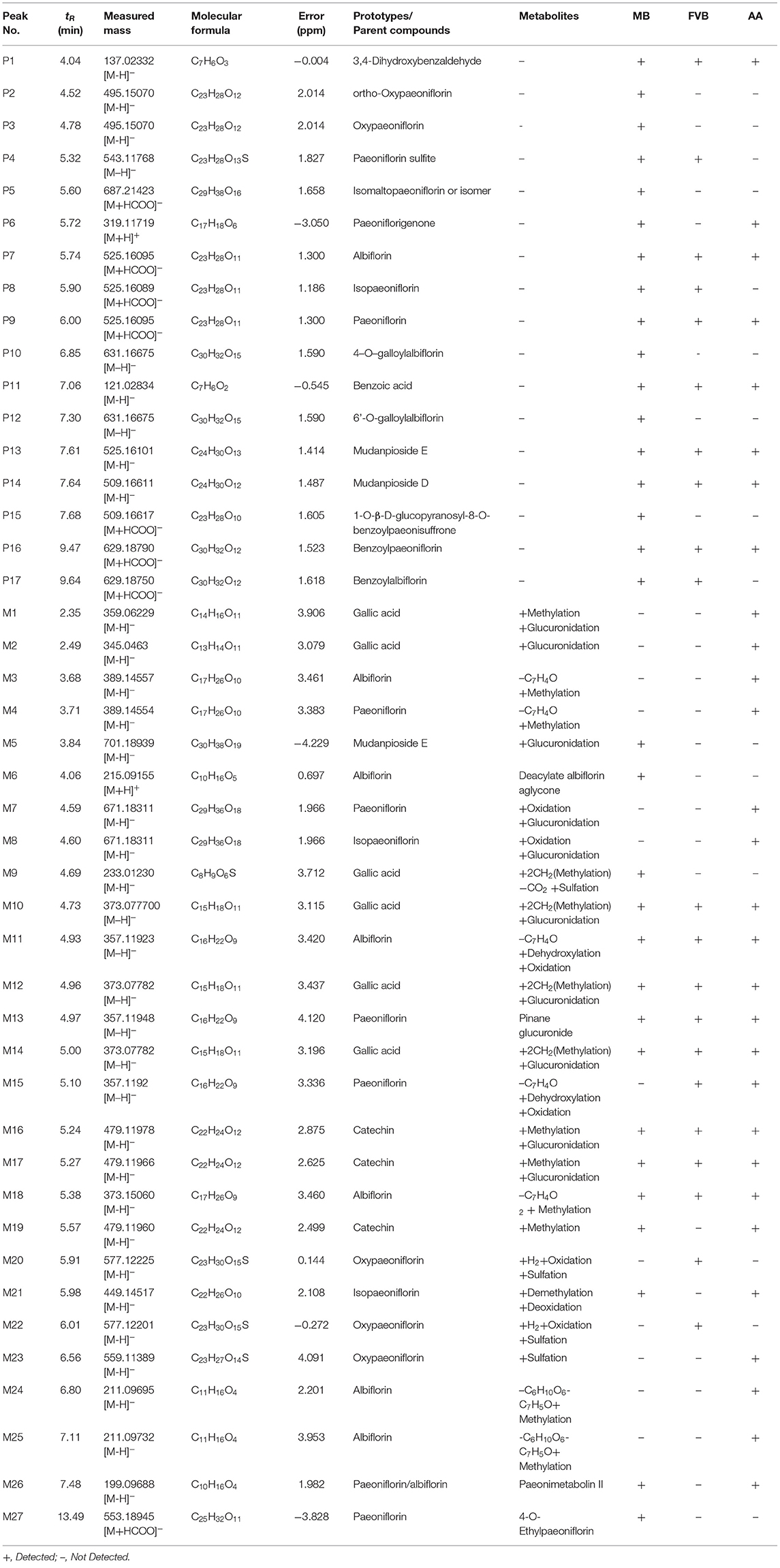
Table 2. Identification of prototypes and metabolites of PRA in different plasma samples by UPLC-Q Exactive Orbitrap HRMS.
Putative Targets for PRA
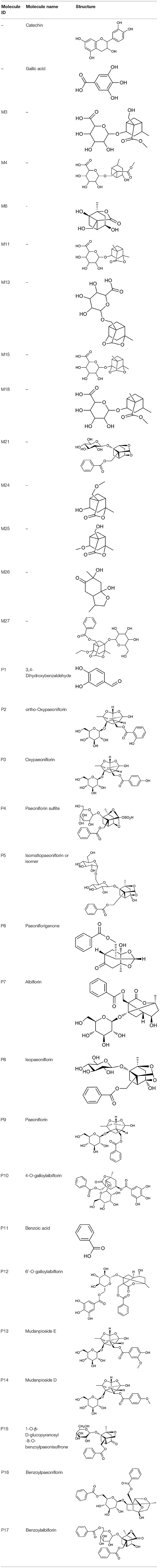
All the identified prototypes and metabolites of PRA in rat blood samples were used for network pharmacology analysis. For the phase II metabolites, their corresponding prototypes or phase I metabolites were selected (52). The chemical structures of 31 components used for target screening were summarized in Table 3. By use of MedChem Studio, 377 putative targets for PRA were obtained as listed in Supplementary Table 1.
HP-Related Targets
After eliminating redundant entries, 75 HP-related targets were acquired (Supplementary Table 2), in which 57 were from Drugbank and 18 were from OMIM.
Network and Pathway Analysis
In order to clearly distinguish the relationship between PRA and HP, a “compound-target-disease” interaction network was constructed by connecting the major components in the blood, the PRA-associated targets as well as the HP-associated targets according to the PPI data from the STRING database. In the PPI network, the node with a higher score may be more important in the central correlation. Therefore, 408 nodes and 4,073 edges, ranked by the scores (>0.4), were included in the network (Supplementary Table 3).
To identify the nodes which were highly interconnected within the network, the DC, BC, and CC values of all nodes in the drug target-disease network were calculated. According to the screen condition, 133 hubs were considered as the critical targets accordingly (DC > 16, BC > 0.0013, and CC > 0.3882). Among them, 111 hubs were PRA-associated targets, 10 hubs were HP-associated targets, and 12 hubs were both associated with PRA and HP. The details were listed in Supplementary Table 4.
To understand the biological functions, pathways or cell localization of the major hubs, GO enrichment analysis was performed, including biological process (BP), cell component (CC), and molecular function (MF). The results indicated that hundreds of GO entries were enriched, and the top 10 significant entries in the BP, MF, and CC categories were listed in Figures 3A–C. The enriched BP ontologies were dominated by response to hypoxia, platelet activation, signal transduction, inflammatory response, transcription from RNA polymerase II promoter, apoptotic process, platelet degranulation, oxidation-reduction process, protein phosphorylation, fibrinolysis (Figure 3A). And the enriched MF ontologies were dominated by enzyme binding, heme binding, protein binding, oxygen binding, steroid hormone receptor activity, drug binding, protein heterodimerization activity, NADP binding, protein heterodimerization activity, chromatin binding (Figure 3B). Furthermore, platelet alpha granule lumen, extracellular exosome, plasma membrane, extracellular space, cytosol, protein kinase complex, protein complex, cell surface, receptor complex, and extracellular region were ranked as top 10 CC ontologies (Figure 3C).
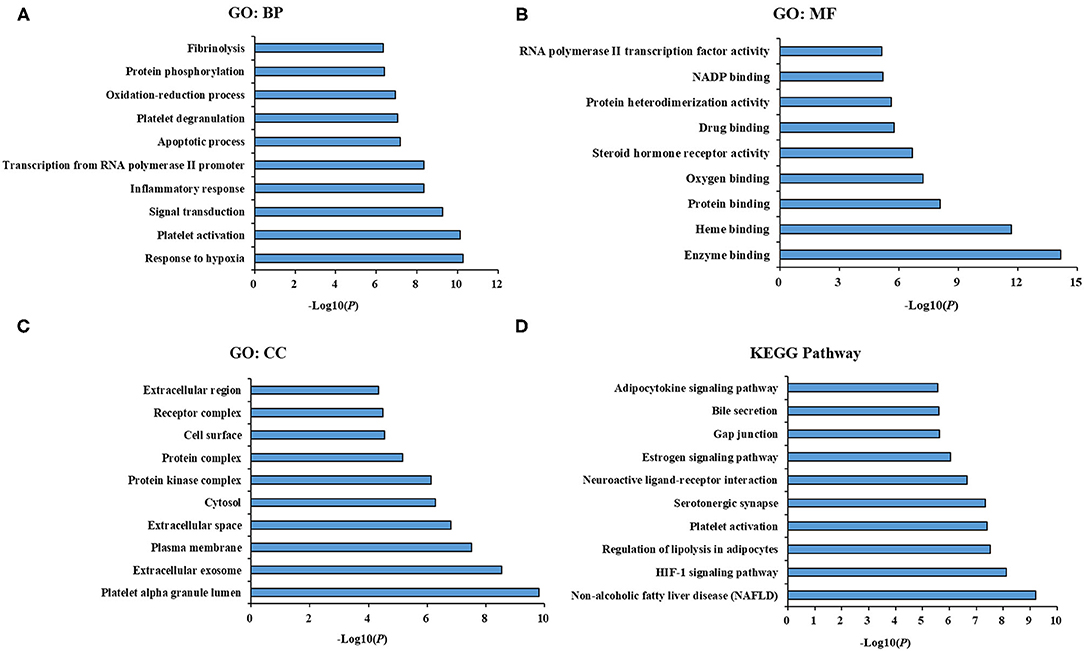
Figure 3. GO term performance and pathway enrichment analysis of major hubs. (A) GO term performance by a biological process (BP); (B) GO term performance by a molecular function (MF); (C) GO term performance by a cellular component (CC); (D) pathway enrichment analysis by KEGG. The ordinate stands for GO terms or main pathways, the primary abscissa stands for minus log10(P).
To further excavate the significance of the major hubs, KEGG pathway enrichment analysis were conducted and resulted in 108 pathways with significant enrichment (P < 0.05). The top 10 pathways were listed in Figure 3D, which could be categorized into three major functional groups: metabolism (such as non-alcoholic fatty liver disease, regulation of lipolysis in adipocytes, estrogen signaling pathway, adipocytokine signaling pathway), blood circulation system (such as HIF-1 signaling pathway, platelet activation and serotonergic synapse), and signal transduction (such as neuroactive ligand-receptor interaction, gap junction and bile secretion).
Then we built a network which connects the interaction among the components of PRA in the blood, main hubs, and significant pathways to achieve a comprehensive understanding of the action mechanism (Figure 4). Interestingly, the pathway of non-alcoholic fatty liver disease (NAFLD) was highly enriched in main KEGG pathways, suggesting that PRA might provide bright prospects for the prevention and treatment of NAFLD.
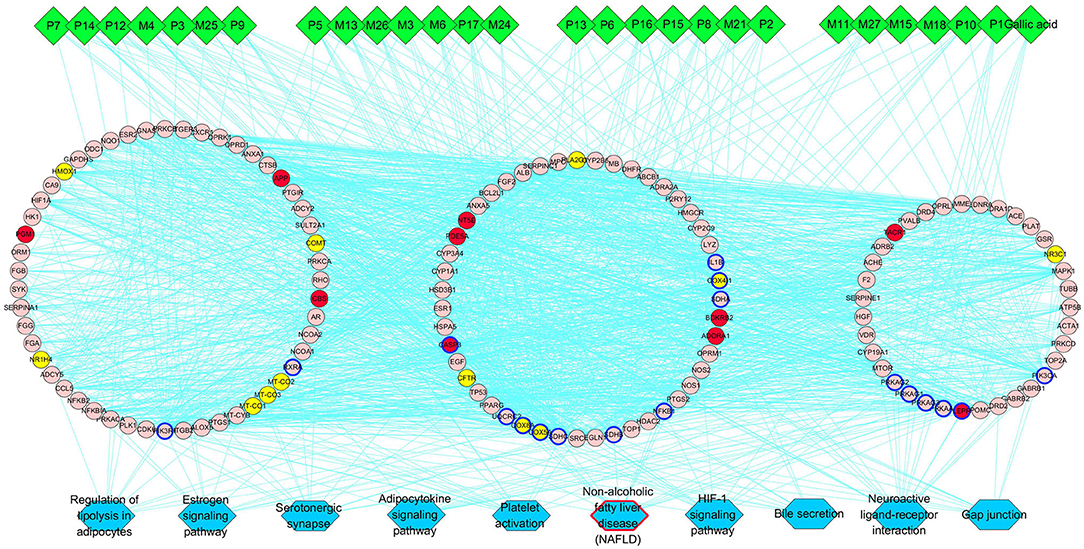
Figure 4. PRA key components-major hubs–main pathways network. Green diamonds represent each component (prototype/metabolite) in PRA; round pink nodes represent putative targets of PRA components; round red nodes represent known therapeutic targets for HP; round yellow nodes represent both putative targets of PRA components and known therapeutic targets for HP, and targets circled in blue represent those important hubs involved in the progression of PRA acting on HP; blue rectangles represent top 10 pathways from enrichment analysis of major targets, and blue rectangle circled in red represent the most significant pathway; edges represent interactions among PRA key components, putative targets, known therapeutic targets for the treatment of HP, and main pathways.
Potential Mechanisms of PRA in Treating NAFLD
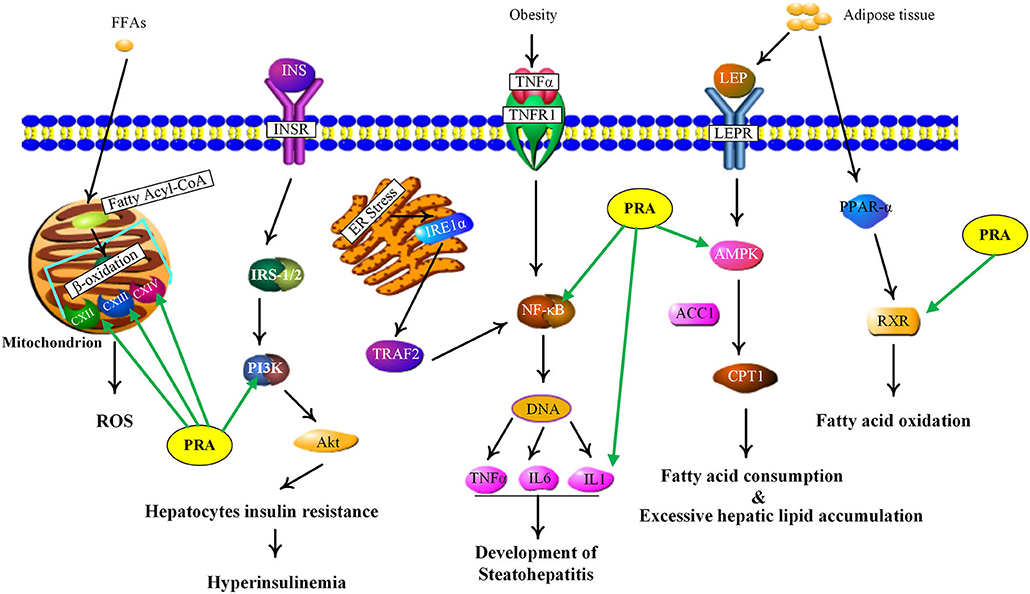
NAFLD, defined as excessive hepatic fat accumulation in the absence of alcohol consumption (53), has become the leading cause of chronic liver disease worldwide (54). Pathologically, NAFLD includes several subgroups, such as simple steatosis, non-alcoholic steatohepatitis (NASH) and non-alcoholic steatofibrosis (NASF), which may progress to irreversible cirrhosis and hepatocellular carcinoma (55, 56). As shown in Supplementary Table 5, the PRA putative targets involved in NAFLD include adenosine 5'-monophosphate (AMP)-activated protein kinases (AMPKs, including PRKAG1, PRKAG2, PRKAG3, and PRKAA1), retinoid X receptor alpha (RXRA), nuclear factor kappa B subunit 1 (NFKB1), interleukin 1 beta (IL1B), mitochondrial electron transport chain complexes (UQCRC2, COX8A, COX4I1, COX5A, SDHA, SDHB, and SDHC), phosphoinositide-3-kinase (PI3K, including PIK3CA and PIK3R1). Figure 5 showed the main NAFLD pathogenesis concerning the PRA putative targets and the details were discussed below.
Current evidence indicates that adipose tissue inflammation could drive the progression of NAFLD in obesity (57). Leptin and adiponectin, two important adipokines secreted from adipose tissue, could cause the phosphorylation and activation of AMPK via binding with their corresponding receptors, called leptin receptor (LEPR) and adiponectin receptor (ADIPOR) (58). AMPK is a highly conserved master regulator of energy metabolism in the liver and its activation would lead to fatty acid oxidation, the increase of glucose uptake and the suppression of lipogenesis through multiple metabolic pathways (59). However, inactivated AMPK was observed in NAFLD, which triggers the inhibition of carnitine palmitoyl transferase 1 (CPT1) via dephosphorylating acetyl-CoA carboxylase 1 (ACC1), leading to the reduction of fatty acid consumption and excessive hepatic lipid accumulation (56). Therefore, AMPK activation could be a potential candidate for treating NAFLD. According to our predicted results, the PRA constituents targeting AMPK are two phenolics, including gallic acid and 3,4-dihydroxybenzaldehyde (P1), which are well-known natural activators of AMPK. For example, gallic acid has been reported to exert beneficial effects on body weight and glucose homeostasis via AMPK activation (60). 3,4-dihydroxybenzaldehyde has also been demonstrated to inhibit the activity of glucose-6-phosphatase through stimulating AMPK phosphorylation (61). Taken together, the anti-NAFLD function of PRA may be partially attributed to the activation of AMPKs (PRKAG1, PRKAG2, PRKAG3, and PRKAA1).
RXR, an obligate heterodimeric partner of many nuclear receptors (NR), occupies a central place in NR signaling and plays a critical role in maintaining energy homeostasis (62). In human liver, RXR could promote fatty acid oxidation by forming a heterodimer with peroxisome proliferator-activated receptor alpha (PPARα) which could be activated by adiponectin to magnify this signaling (63). According to our predicted results, nine compounds (P2, P5, P8, P9, P12, P13, P16, P17, and M27) were considered as RXR agonists and they may be partially responsible for the anti-NAFLD effect of PRA.
NF-κB, an integrator of inflammatory pathway networks, is also essential in the occurrence and development of NAFLD (64). In response to endoplasmic reticulum (ER) stress, serine/threonine-protein kinase/endoribonuclease (IRE1) is activated, which sequentially triggers the activation of tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) and NF-κB, leading to the expression and release of inflammatory cytokines (such as IL-1, IL-6, and TNF-α) via binding to DNA. Additionally, in response to obesity, transduction of TNF-α signal by TNF receptor superfamily member 1A (TNFR1) also involves activation of NF-κB (65, 66). Hence, aberrant activation of NF-κB is one of the major risk factors in the pathological process of steatohepatitis. It has been evident that P1 and gallic acid could alleviate inflammation through the down-regulation of NF-κB (67, 68). Therefore, NF-κB can be speculated as the potential therapeutic target of PRA in NAFLD.
Mitochondria, whose most important function is to generate adenosine triphosphate (ATP), plays a fundamental role in lipid metabolism and oxidative stress (69). Upon entering the mitochondria, free fatty acids (FFAs) are converted into fatty acyl-CoA and then undergo the process of β-oxidation, which is catalyzed by a series of mitochondrial enzymes and generates acetyl-coA, NADH (nicotinamide adenine dinucleotide) and FADH2 (flavine adenine dinucleotide, reduced) (70). NADH and FADH2 could transfer their electrons to the electron transport chain (ETC) to induce an oxidation-reduction reaction at each step and ATP will be produced by oxidative phosphorylation, a process which needs the action of five ETC complexes (71, 72). However, impaired electron transport within the ETC induces electron leakage from the ETC complex, thus resulting in reactive oxygen species (ROS) production. The increased oxidative stress would induce the release of inflammatory cytokines and affect the activity of major enzymes associated with lipid metabolism, which may contribute to the pathological process of NAFLD (73). Many components have been shown to play antioxidative roles by reducing ROS levels, such as paeoniflorin (74) and albiflorin (75). Therefore, PRA may modify the NAFLD status by decreasing ROS formation and ameliorating mitochondrial dysfunction.
PI3K, a member of lipid kinases family, is associated with an extraordinarily diverse group of cellular functions (76). In human liver, transduction of insulin signal by insulin receptor (InsR) involves activation of insulin receptor substrate-1/2 (IRS-1/2), which in turn activates PI3K, leading to the activation of protein kinase B (Akt). Finally, the activated Akt mediates the diverse pharmacological functions (like glucose uptake and lipid metabolism) of insulin via transmitting the biological signal to the downstream targets (77, 78). An increasing body of evidence suggests that the deregulation of PI3K/Akt signaling pathway in hepatocytes may lead to insulin resistance and further induce NAFLD development (77). The two abundant components in PRA, albiflorin, and paeoniflorin, have been reported to exert beneficial effects on NAFLD and obesity via regulating PI3K/Akt pathway (79, 80). Therefore, the restoration of PI3K activity may be involved in the anti-NAFLD function of PRA.
Taken together, PRA might exert curative effects on NAFLD through acting on multiple targets involved in multiple signaling pathways. The results also pointed out the huge potential for the clinical use of MEP with heptoprotective activity for preventing and treating NAFLD.
Molecular Docking
Molecular docking studies were used to verify the interactions between the PRA components and the targets related to NAFLD (PRKAG3, UQCRC2, RXRA, PRKAG1, COX8A, PRKAG2, COX4I1, NFKB1, COX5A, SDHA, SDHB, SDHC, IL1B, PIK3CA, PRKAA1, and PIK3R1). The results indicated that the PRA components had been docked successfully with PRKAG1, PRKAA1, NFKB1, PIK3CA, PIK3R1, SDHC, and COX4I1, as listed in Supplementary Table 6. Therefore, PRA may treat NAFLD mainly through these seven targets.
SPR Assays for Affinity
SPR assay was used to verify the direct binding activities of the target proteins and their corresponding compounds according to the results of molecular docking, including gallic acid-PRKAG1, 3,4-Dihydroxybenzaldehyde-PRKAG1, gallic acid-NFKB1, 3,4-Dihydroxybenzaldehyde-NFKB1. As depicted in Figure 6, we found two compound–target pairs that exhibited relatively better affinities, including gallic acid-NFKB1 (KD = 28.6 μM) and gallic acid-PRKAG1 (KD = 14 μM). Interestingly, gallic acid had relatively slow association and dissociation rates at NFKB1 and PRKAG1 (81), suggesting that gallic acid may be a promising ligand for in vivo targeting NFKB1 and PRKAG1. The above results demonstrated the validity of network pharmacology and molecular docking approaches, and the in-depth biological functions of these ligand–target pairs were worth exploring.
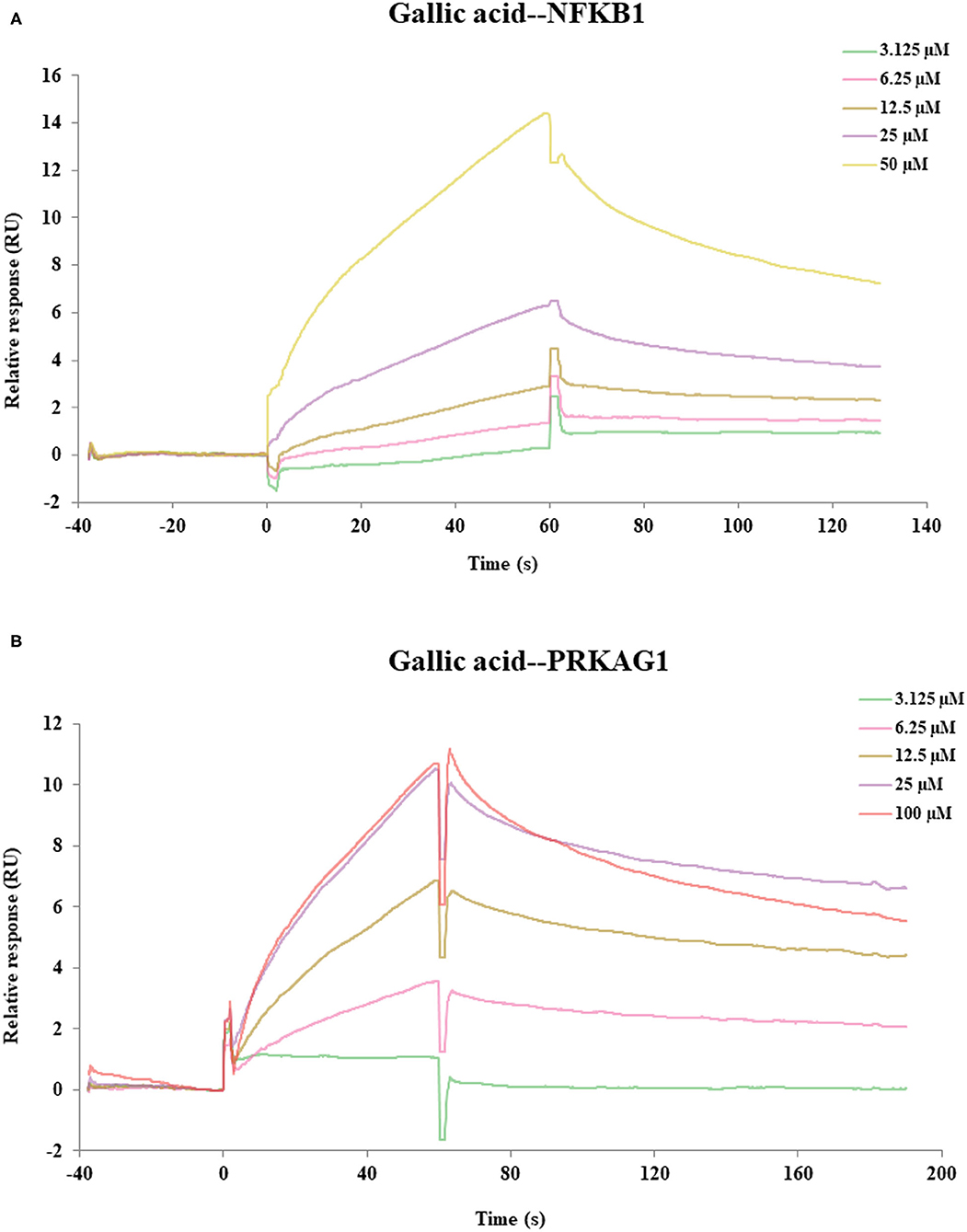
Figure 6. The SPR assay of the interaction of gallic acid with NFKB1, gallic acid with PRKAG1. (A) The SPR curves of gallic acid binding to NFKB1. (B) The SPR curves of gallic acid binding to PRKAG1.
Conclusions
Compared with conventional metabolite identification method for partial metabolism in the whole metabolic route, we developed a sequential metabolites identification approach by integrating IPVS and LC/MS to characterize the dynamic biotransformation process of MEP. Using this strategy, the metabolic profile of PRA was described rapidly and comprehensively, including 17 prototypes and 27 metabolites, which were bio-transformed via oxidation, methylation, sulfation, and glucuronidation. Next, the network and pathway analysis were performed based on the identified metabolites and the results indicated that the pathway of non-alcoholic fatty liver disease (NAFLD) was highly enriched and 16 key targets were found, suggesting that PRA may have beneficial effects in the prevention and treatment of NAFLD. Moreover, the molecular docking and SPR experiments showed that several constituents exhibited good affinity to specific targets (gallic acid-NFKB1, KD = 28.6 μM; gallic acid-PRKAG1, KD = 14 μM). Collectively, this work provides a systems perspective to study the chemical and functional basis of PRA for preventing and treating liver disease, which demonstrated that the proposed strategy may be a powerful tool in screening potential active compounds and their corresponding targets from other MEPs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethnic Committee of Beijing University of Chinese Medicine.
Author Contributions
YL, GY, ZL, and YS conceived and designed the experiments. ZL, XH, and WY performed the experiments. GW, JW, XJ, MS, and XL contributed to data analysis and manuscript preparation. ZL and GY wrote the paper. All authors have read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (No. 2019YFC1710105), the National Traditional Chinese Medicine Standardization Project (ZYBZH-Y-HUB-20), and National Natural Science Foundation of China (No. 81973295).
Conflict of Interest
GW was employed by company Zhongcai Health (Beijing) Biological Technology Development Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Lei Zhang from Peking University for manuscript review in this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.677659/full#supplementary-material
References
1. Guo R, Luo X, Liu J, Liu L, Wang X, Lu H. Omics strategies decipher therapeutic discoveries of traditional Chinese medicine against different diseases at multiple layers molecular-level. Pharmacol Res. (2020) 152:104627. doi: 10.1016/j.phrs.2020.104627
2. Feng G, Sun Y, Liu S, Song F, Pi Z, Liu Z. Stepwise targeted matching strategy from in vitro to in vivo based on ultra–high performance liquid chromatography tandem mass spectrometry technology to quickly identify and screen pharmacodynamic constituents. Talanta. (2019) 194:619–26. doi: 10.1016/j.talanta.2018.10.074
3. Annunziata G, Maisto M, Schisano C, Ciampaglia R, Daliu P, Narciso V, et al. Colon bioaccessibility and antioxidant activity of white, green and black tea polyphenols extract after in vitro simulated gastrointestinal digestion. Nutrients. (2018) 10:1711. doi: 10.3390/nu10111711
4. Luo Z, Ma X, Liu Y, Lu L, Yang R, Yu G, et al. An approach to characterizing the complicated sequential metabolism of salidroside in rats. Molecules. (2016) 21:706. doi: 10.3390/molecules21060706
5. Zhang L, Zhao H, Liu Y, Dong H, Lv B, Fang M, et al. Metabolic routes along digestive system of licorice: multicomponent sequential metabolism method in rat. Biomed Chromatogr. (2016) 30:902–12. doi: 10.1002/bmc.3626
6. Schanker LS, Tocco DJ, Brodie BB, Hogben CA. Absorption of drugs from the rat small intestine. J Pharmacol Exp Ther. (1958) 123:81–88. PubMed PMID:13539795.
7. Luo Z, Liu Y, Zhao B, Tang M, Dong H, Zhang L, et al. Ex vivo and in situ approaches used to study intestinal absorption. J Pharmacol Toxicol Methods. (2013) 68:208–16. doi: 10.1016/j.vascn.2013.06.001
8. Gong J, Jiang Z, Yang T, Zhu Y. In vitro and in vivo pharmacokinetics and metabolism of MK-8353 by liquid chromatography combined with diode array detector and Q-Exactive-Orbitrap tandem mass spectrometry. J Pharm Biomed Anal. (2019) 168:64–74. doi: 10.1016/j.jpba.2019.02.012
9. Han F, Li Y, Ma L, Liu T, Wu Y, Xu R, et al. A rapid and sensitive UHPLC-FT-ICR MS/MS method for identification of chemical constituents in Rhodiola crenulata extract, rat plasma and rat brain after oral administration. Talanta. (2016) 160:183–93. doi: 10.1016/j.talanta.2016.07.014
10. Liu R, Liu Q, Li B, Liu L, Cheng D, Cai X, et al. Pharmacokinetics, bioavailability, excretion, and metabolic analysis of Schisanlactone E, a bioactive ingredient from Kadsura heteroclita (Roxb) Craib, in rats by UHPLC–MS/MS and UHPLC-Q-Orbitrap HRMS. J Pharm Biomed Anal. (2020) 177:112875. doi: 10.1016/j.jpba.2019.112875
11. Liu Y, Xing H, Jiang X, Chen Y, Huang M, Yu S. Network pharmacology-based preventive effect of XZF on cutaneous toxicities induced by EGFR inhibitor. Biomed Pharmacother. (2020) 123:109755. doi: 10.1016/j.biopha.2019.109755
12. Wang YL, Cui T, Li YZ, Liao ML, Zhang HB, Hou WB, et al. Prediction of quality markers of traditional Chinese medicines based on network pharmacology. Chinese Herbal Medicines. (2019) 11:349–56. doi: 10.1016/j.chmed.2019.08.003
13. Zeng Q, Li L, Siu W, Jin Y, Cao M, Li W, et al. A combined molecular biology and network pharmacology approach to investigate the multi-target mechanisms of Chaihu Shugan San on Alzheimer's disease. Biomed Pharmacother. (2019) 120:109370. doi: 10.1016/j.biopha.2019.109370
14. Yu G, Luo Z, Zhou Y, Zhang L, Wu Y, Ding L, et al. Uncovering the pharmacological mechanism of Carthamus tinctorius L. on cardiovascular disease by a systems pharmacology approach. Biomed Pharmacother. (2019) 117:109094. doi: 10.1016/j.biopha.2019.109094
15. Luo Z, Yu G, Chen X, Liu Y, Zhou Y, Wang G, et al. Integrated phytochemical analysis based on UHPLC-LTQ-Orbitrap and network pharmacology approaches to explore the potential mechanism of Lycium ruthenicum Murr. for ameliorating Alzheimer's disease. Food Funct. (2020) 11:1362–72. doi: 10.1039/C9FO02840D
16. Wang J, Zhang Y, Liu YM, Yang XC, Chen YY, Wu GJ, et al. Uncovering the protective mechanism of Huoxue Anxin Recipe against coronary heart disease by network analysis and experimental validation. Biomed Pharmacother. (2020) 121:109655. doi: 10.1016/j.biopha.2019.109655
17. Yang B, Wang N, Wang S, Li X, Zheng Y, Li M, et al. Network-pharmacology-based identification of caveolin-1 as a key target of Oldenlandia diffusa to suppress breast cancer metastasis. Biomed Pharmacother. (2019) 112:108607. doi: 10.1016/j.biopha.2019.108607
18. Ren W, Luo Z, Pan F, Liu J, Sun Q, Luo G, et al. Integrated network pharmacology and molecular docking approaches to reveal the synergistic mechanism of multiple components in Venenum Bufonis for ameliorating heart failure. PeerJ. (2020) 8:e10107. doi: 10.7717/peerj.10107
19. Qin H, Chen H, Zou Y, Zhang X, Wei C, Chen W, et al. Systematic investigation of the mechanism of Cichorium glandulosum on type 2 diabetes mellitus accompanied with non-alcoholic fatty liver rats. Food Funct. (2019) 10:2450–60. doi: 10.1039/C8FO02284D
20. Yu G, Zhang Y, Ren W, Dong L, Li J, Geng Y, et al. Network pharmacology-based identification of key pharmacological pathways of Yin-Huang-Qing-Fei capsule acting on chronic bronchitis. Int J Chron Obstruct Pulmon Dis. (2016) 12:85–94. doi: 10.2147/COPD.S121079
21. Lv X, Xu Z, Xu G, Li H, Wang C, Chen J, et al. Investigation of the active components and mechanisms of Schisandra chinensis in the treatment of asthma based on a network pharmacology approach and experimental validation. Food Funct. (2020) 11:3032–42. doi: 10.1039/D0FO00087F
22. Yu G, Wang W, Wang X, Xu M, Zhang L, Ding L, et al. Network pharmacology-based strategy to investigate pharmacological mechanisms of Zuojinwan for treatment of gastritis. BMC Complement Altern Med. (2018) 18:292. doi: 10.1186/s12906-018-2356-9
23. Zhang W, Chen Y, Jiang H, Yang J, Wang Q, Du Y, et al. Integrated strategy for accurately screening biomarkers based on metabolomics coupled with network pharmacology. Talanta. (2020) 211:120710. doi: 10.1016/j.talanta.2020.120710
24. Ou C, Geng T, Wang J, Gao X, Chen X, Luo X, et al. Systematically investigating the pharmacological mechanism of Dazhu Hongjingtian in the prevention and treatment of acute mountain sickness by integrating UPLC/Q-TOF-MS/MS analysis and network pharmacology. J Pharm Biomed Anal. (2020) 179:113028. doi: 10.1016/j.jpba.2019.113028
25. Yang Y, Wang Q, Guo D. A novel strategy for analyzing RNA-protein interactions by surface plasmon resonance biosensor. Mol Biotechnol. (2008) 40:87–93. doi: 10.1007/s12033-008-9066-3
26. Fisher RJ, Fivash M. Surface plasmon resonance based methods for measuring the kinetics and binding affinities of biomolecular interactions. Curr Opin Biotechnol. (1994) 5:389–95. doi: 10.1016/0958-1669(94)90047-7
27. He Q, Chen Y, Shen D, Cui X, Zhang C, Yang H, et al. Development of a surface plasmon resonance immunosensor and ELISA for 3-nitrotyrosine in human urine. Talanta. (2019) 195:655–61. doi: 10.1016/j.talanta.2018.11.110
28. Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem Rev. (2008) 108:462–93. doi: 10.1021/cr068107d
29. Koyun S, Akgönüllü S, Yavuz H, Erdem A, Denizli A. Surface plasmon resonance aptasensor for detection of human activated protein C. Talanta. (2019) 194:528–33. doi: 10.1016/j.talanta.2018.10.007
30. Sanders M, McPartlin D, Moran K, Guo Y, Eeckhout M, O'Kennedy R, et al. Comparison of enzyme-linked immunosorbent assay, surface plasmon resonance and biolayer interferometry for screening of deoxynivalenol in wheat and wheat dust. Toxins. (2016) 8:103. doi: 10.3390/toxins8040103
31. Zhang X, Li G, Wu D, Yu Y, Hu N, Wang H, et al. Emerging strategies for the activity assay and inhibitor screening of alpha-glucosidase. Food Funct. (2020) 11:66–82. doi: 10.1039/C9FO01590F
32. Chen L, Wang D, Lv D, Wang X, Liu Y, Chen X, et al. Identification of eupatilin and ginkgolide B as p38 ligands from medicinal herbs by surface plasmon resonance biosensor-based active ingredients recognition system. J Pharm Biomed Anal. (2019) 171:35–42. doi: 10.1016/j.jpba.2019.03.029
33. Hu XL, Li YB, Qi S, Zhang Q, Ren TS, Meng WH, et al. 4-O-galloylalbiflorin discovered from Paeonia lactiflora Pall. is a potential β-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitor. J Funct Foods. (2016) 27:517–25. doi: 10.1016/j.jff.2016.10.012
34. Shu X, Duan W, Liu F, Shi X, Geng Y, Wang X, et al. Preparative separation of polyphenols from the flowers of Paeonia lactiflora Pall. by high-speed counter-current chromatography. J Chromatogr B. (2014) 947-948:62–7. doi: 10.1016/j.jchromb.2013.12.004
35. Qin Y, Tian YP. Protective effects of total glucosides of paeony and the underlying mechanisms in carbon tetrachloride-induced experimental liver injury. Arch Med Sci. (2011) 7:604–12. doi: 10.5114/aoms.2011.24129
36. Zhang L, Yang B, Yu B. Paeoniflorin protects against nonalcoholic fatty liver disease induced by a high-fat diet in mice. Biol Pharm Bull. (2015) 38:1005–11. doi: 10.1248/bpb.b14-00892
37. Ma Z, Chu L, Liu H, Li J, Zhang Y, Liu W, et al. Paeoniflorin alleviates non-alcoholic steatohepatitis in rats: involvement with the ROCK/NF-κB pathway. Int Immunopharmacol. (2016) 38:377–84. doi: 10.1016/j.intimp.2016.06.023
38. Wang H, Wei W, Wang NP, Wu CY, Yan SX, Yue L, et al. Effects of total glucosides of peony on immunological hepatic fibrosis in rats. World J Gastroenterol. (2005) 11:2124–9. doi: 10.3748/wjg.v11.i14.2124
39. Zhao Y, Ma X, Wang J, Zhu Y, Li R, Wang J, et al. Paeoniflorin alleviates liver fibrosis by inhibiting HIF-1α through mTOR-dependent pathway. Fitoterapia. (2014) 99:318–27. doi: 10.1016/j.fitote.2014.10.009
40. Ma X, Zhao YL, Zhu Y, Chen Z, Wang JB, Li RY, et al. Paeonia lactiflora Pall. protects against ANIT-induced cholestasis by activating Nrf2 via PI3K/Akt signaling pathway. Drug Des Devel Ther. (2015) 9:5061–74. doi: 10.2147/DDDT.S90030
41. Chen Z, Ma X, Zhu Y, Zhao Y, Wang J, Li R, et al. Paeoniflorin ameliorates ANIT-induced cholestasis by activating Nrf2 through an PI3K/Akt-dependent pathway in rats. Phytother Res. (2015) 29:1768–75. doi: 10.1002/ptr.5431
42. Shen M, Men R, Fan X, Wang T, Huang C, Wang H, et al. Total glucosides of paeony decreases apoptosis of hepatocytes and inhibits maturation of dendritic cells in autoimmune hepatitis. Biomed Pharmacother. (2020) 124:109911. doi: 10.1016/j.biopha.2020.109911
43. Song SS, Yuan PF, Li PP, Wu HX, Ni WJ, Lu JT, et al. Protective effects of total glucosides of paeony on N-nitrosodiethylamine-induced hepatocellular carcinoma in rats via down-regulation of regulatory B cells. Immunol Invest. (2015) 44:521–35. doi: 10.3109/08820139.2015.1043668
44. Wu JJ, Sun WY, Hu SS, Zhang S, Wei W. A standardized extract from Paeonia lactiflora and Astragalus membranaceus induces apoptosis and inhibits the proliferation, migration and invasion of human hepatoma cell lines. Int J Oncol. (2013) 43:1643–51. doi: 10.3892/ijo.2013.2085
45. Li H, Dong L, Liu Y, Wang G, Wang G, Qiao Y. Biopharmaceutics classification of puerarin and comparison of perfusion approaches in rats. Int J Pharm. (2014) 466:133–8. doi: 10.1016/j.ijpharm.2014.03.014
46. Liu B, He H, Luo H, Zhang T, Jiang J. Artificial intelligence and big data facilitated targeted drug discovery. Stroke Vasc Neurol. (2019) 4:206–13. doi: 10.1136/svn-2019-000290
47. Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. (2005) 33:D514–7. doi: 10.1093/nar/gki033
48. Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. (2009) 37:D412–6. doi: 10.1093/nar/gkn760
49. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
50. Cho HJ, Kim JE, Kim DD, Yoon IS. In vitro–in vivo extrapolation (IVIVE) for predicting human intestinal absorption and first-pass elimination of drugs: principles and applications. Drug Dev Ind Pharm. (2014) 40:989–98. doi: 10.3109/03639045.2013.831439
51. Mudra DR, Borchardt RT. Absorption barriers in the rat intestinal mucosa: 1. application of an in situ perfusion model to simultaneously assess drug permeation and metabolism. J Pharm Sci. (2010) 99:982–98. doi: 10.1002/jps.21912
52. Yu S, Liu H, Li K, Qin Z, Qin X, Zhu P, et al. Rapid characterization of the absorbed constituents in rat serum after oral administration and action mechanism of Naozhenning granule using LC–MS and network pharmacology. J Pharm Biomed Anal. (2019) 166:281–90. doi: 10.1016/j.jpba.2019.01.020
53. Poynard T, Ratziu V, Charlotte F, Messous D, Munteanu M, Imbert-Bismut F, et al. Diagnostic value of biochemical markers (NashTest) for the prediction of non alcoholo steato hepatitis in patients with non-alcoholic fatty liver disease. BMC Gastroenterol. (2006) 6:34. doi: 10.1186/1471-230X-6-34
54. Delik A, Dinçer S, Akkiz H. The role of genetic and epigenetic factors in non alcoholic fatty liver disease (NAFLD) pathogenesis. Meta Gene. (2020) 24:100647. doi: 10.1016/j.mgene.2020.100647
55. Lee J, Kim Y, Friso S, Choi SW. Epigenetics in non-alcoholic fatty liver disease. Mol Aspects Med. (2017) 54:78–88. doi: 10.1016/j.mam.2016.11.008
56. Song K, Zhang Y, Ga Q, Bai Z, Ge RL. High-altitude chronic hypoxia ameliorates obesity-induced non-alcoholic fatty liver disease in mice by regulating mitochondrial and AMPK signaling. Life Sci. (2020) 252:117633. doi: 10.1016/j.lfs.2020.117633
57. Cordeiro A, Costa R, Andrade N, Silva C, Canabrava N, Pena MJ, et al. Does adipose tissue inflammation drive the development of non-alcoholic fatty liver disease in obesity? Clin Res Hepatol Gastroenterol. (2020) 44:394–402. doi: 10.1016/j.clinre.2019.10.001
58. Lago F, Gómez R, Gómez-Reino JJ, Dieguez C, Gualillo O. Adipokines as novel modulators of lipid metabolism. Trends Biochem Sci. (2009) 34:500–10. doi: 10.1016/j.tibs.2009.06.008
59. Garcia D, Hellberg K, Chaix A, Wallace M, Herzig S, Badur MG, et al. Genetic liver-specific AMPK activation protects against diet-induced obesity and NAFLD. Cell Rep. (2019) 26:192–208. doi: 10.1016/j.celrep.2018.12.036
60. Vazirian M, Nabavi SM, Jafari S, Manayi A. Natural activators of adenosine 5'-monophosphate (AMP)-activated protein kinase (AMPK) and their pharmacological activities. Food Chem Toxicol. (2018) 122:69–79. doi: 10.1016/j.fct.2018.09.079
61. Kasangana PB, Eid HM, Nachar A, Stevanovic T, Haddad PS. Further isolation and identification of anti-diabetic principles from root bark of Myrianthus arboreus P. Beauv.: The ethyl acetate fraction contains bioactive phenolic compounds that improve liver cell glucose homeostasis. J Ethnopharmacol. (2019) 245:112167. doi: 10.1016/j.jep.2019.112167
62. Ziouzenkova O, Plutzky J. Retinoid metabolism and nuclear receptor responses: new insights into coordinated regulation of the PPAR–RXR complex. FEBS Lett. (2008) 582:32–8. doi: 10.1016/j.febslet.2007.11.081
63. Bais S, Patel NJ. Role of adiponectin and its target receptors to control deposition of fat in obesity related disorders. Obesity Medicine. (2019) 16:100148. doi: 10.1016/j.obmed.2019.100148
64. Chen Z, Yu R, Xiong Y, Du F, Zhu S. A vicious circle between insulin resistance and inflammation in nonalcoholic fatty liver disease. Lipids Health Dis. (2017) 16:203. doi: 10.1186/s12944-017-0572-9
65. Lee Y, Lee JY. Chapter 8 - protective actions of polyphenols in the development of nonalcoholic fatty liver disease. In: Watson RR, Preedy VR, editors. Dietary Interventions in Liver Disease. London: Academic Press (2019). p. 91–9. doi: 10.1016/B978-0-12-814466-4.00008-2
66. Chávez-Tapia NC, Uribe M, Ponciano-Rodríguez G, Medina-Santillán R, Méndez-Sánchez N. New insights into the pathophysiology of nonalcoholic fatty liver disease. Ann Hepatol. (2009) 8:S9–7. doi: 10.1016/S1665-2681(19)31821-6
67. Zhou Z, Liu Y, Miao AD, Wang SQ. Protocatechuic aldehyde suppresses TNF-alpha-induced ICAM-1 and VCAM-1 expression in human umbilical vein endothelial cells. Eur J Pharmacol. (2005) 513:1–8. doi: 10.1016/j.ejphar.2005.01.059
68. Tanaka M, Kishimoto Y, Sasaki M, Sato A, Kamiya T, Kondo K, et al. Terminalia bellirica (Gaertn.) Roxb. extract and gallic acid attenuate LPS-induced inflammation and oxidative stress via MAPK/NF-κB and Akt/AMPK/Nrf2 pathways. Oxid Med Cell Longev. (2018) 2018:9364364. doi: 10.1155/2018/9364364
69. Grattagliano I, de Bari O, Bernardo TC, Oliveira PJ, Wang DQH, Portincasa P. Role of mitochondria in nonalcoholic fatty liver disease-from origin to propagation. Clin Biochem. (2012) 45:610–8. doi: 10.1016/j.clinbiochem.2012.03.024
70. Stanley WC, Sabbah HN. Metabolic therapy for ischemic heart disease: the rationale for inhibition of fatty acid oxidation. Heart Fail Rev. (2005) 10:275–9. doi: 10.1007/s10741-005-7542-4
71. Gu F, Chauhan V, Kaur K, Brown WT, LaFauci G, Wegiel J, et al. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl Psychiatry. (2013) 3:e299. doi: 10.1038/tp.2013.68
72. Marquez J, Flores J, Kim AH, Nyamaa B, Nguyen ATT, Park N, et al. Rescue of TCA cycle dysfunction for cancer therapy. J Clin Med. (2019) 8:2161. doi: 10.3390/jcm8122161
73. Stevanović J, Beleza J, Coxito P, Ascensão A, Magalhães J. Physical exercise and liver “fitness”: role of mitochondrial function and epigenetics-related mechanisms in non-alcoholic fatty liver disease. Mol Metab. (2020) 32:1–14. doi: 10.1016/j.molmet.2019.11.015
74. Song S, Xiao X, Guo D, Mo L, Bu C, Ye W, et al. Protective effects of Paeoniflorin against AOPP-induced oxidative injury in HUVECs by blocking the ROS-HIF-1α/VEGF pathway. Phytomedicine. (2017) 34:115–26. doi: 10.1016/j.phymed.2017.08.010
75. Xu YJ, Mei Y, Shi XQ, Zhang YF, Wang XY, Guan L, et al. Albiflorin ameliorates memory deficits in APP/PS1 transgenic mice via ameliorating mitochondrial dysfunction. Brain Res. (2019) 1719:113–23. doi: 10.1016/j.brainres.2019.05.037
76. Ren L, Zhou X, Huang X, Wang C, Li Y. The IRS/PI3K/Akt signaling pathway mediates olanzapine-induced hepatic insulin resistance in male rats. Life Sci. (2019) 217:229–36. doi: 10.1016/j.lfs.2018.12.015
77. Dai B, Wu Q, Zeng C, Zhang J, Cao L, Xiao Z, et al. The effect of Liuwei Dihuang decoction on PI3K/Akt signaling pathway in liver of type 2 diabetes mellitus (T2DM) rats with insulin resistance. J Ethnopharmacol. (2016) 192:382–9. doi: 10.1016/j.jep.2016.07.024
78. Fan Y, He Z, Wang W, Li J, Hu A, Li L, et al. Tangganjian decoction ameliorates type 2 diabetes mellitus and nonalcoholic fatty liver disease in rats by activating the IRS/PI3K/AKT signaling pathway. Biomed. Pharmacother. (2018) 106:733–7. doi: 10.1016/j.biopha.2018.06.089
79. Jeong MY, Park J, Youn DH, Jung Y, Kang J, Lim S, et al. Albiflorin ameliorates obesity by inducing thermogenic genes via AMPK and PI3K/AKT in vivo and in vitro. Metabolism. (2017) 73:85–99. doi: 10.1016/j.metabol.2017.05.009
80. Ma Z, Chu L, Liu H, Wang W, Li J, Yao W, et al. Beneficial effects of paeoniflorin on non-alcoholic fatty liver disease induced by high-fat diet in rats. Sci Rep. (2017) 7:44819. doi: 10.1038/srep44819
Keywords: Paeoniae Radix Alba, non-alcoholic fatty liver disease, sequential metabolism, UPLC-Q Exactive Orbitrap HRMS, surface plasmon resonance, network pharmacology
Citation: Luo Z, Liu Y, Han X, Yang W, Wang G, Wang J, Jiang X, Sen M, Li X, Yu G and Shi Y (2021) Mechanism of Paeoniae Radix Alba in the Treatment of Non-alcoholic Fatty Liver Disease Based on Sequential Metabolites Identification Approach, Network Pharmacology, and Binding Affinity Measurement. Front. Nutr. 8:677659. doi: 10.3389/fnut.2021.677659
Received: 08 March 2021; Accepted: 23 August 2021;
Published: 16 September 2021.
Edited by:
Daniel Cozzolino, University of Queensland, AustraliaReviewed by:
Giuseppe Annunziata, University of Naples Federico II, ItalySheng Guo, Nanjing University of Chinese Medicine, China
Copyright © 2021 Luo, Liu, Han, Yang, Wang, Wang, Jiang, Sen, Li, Yu and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Liu, liuyang@bucm.edu.cn; Guohua Yu, ghyu@bucm.edu.cn; Yuanyuan Shi, yshi@bucm.edu.cn
 Zhiqiang Luo
Zhiqiang Luo