
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 07 April 2021
Sec. Food Chemistry
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.640100
This article is part of the Research Topic Milks Mean More: The Role of Milk in Nutrition, Digestion and Metabolism Across the Lifespan View all 26 articles
 Jason W. Arnold1,2†
Jason W. Arnold1,2† Hunter D. Whittington3†
Hunter D. Whittington3† Suzanne F. Dagher3
Suzanne F. Dagher3 Jeffery Roach2,4
Jeffery Roach2,4 M. Andrea Azcarate-Peril1,2
M. Andrea Azcarate-Peril1,2 Jose M. Bruno-Barcena3*
Jose M. Bruno-Barcena3*Complex dietary carbohydrate structures including β(1–4) galacto-oligosaccharides (GOS) are resistant to digestion in the upper gastrointestinal (GI) tract and arrive intact to the colon where they benefit the host by selectively stimulating microbial growth. Studies have reported the beneficial impact of GOS (alone or in combination with other prebiotics) by serving as metabolic substrates for modulating the assembly of the infant gut microbiome while reducing GI infections. N-Acetyl-D-lactosamine (LacNAc, Galβ1,4GlcNAc) is found in breast milk as a free disaccharide. This compound is also found as a component of human milk oligosaccharides (HMOs), which have repeating and variably branched lactose and/or LacNAc units, often attached to sialic acid and fucose monosaccharides. Human glycosyl-hydrolases do not degrade most HMOs, indicating that these structures have evolved as natural prebiotics to drive the proper assembly of the infant healthy gut microbiota. Here, we sought to develop a novel enzymatic method for generating LacNAc-enriched GOS, which we refer to as humanized GOS (hGOS). We showed that the membrane-bound β-hexosyl transferase (rBHT) from Hamamotoa (Sporobolomyces) singularis was able to generate GOS and hGOS from lactose and N-Acetyl-glucosamine (GlcNAc). The enzyme catalyzed the regio-selective, repeated addition of galactose from lactose to GlcNAc forming the β-galactosyl linkage at the 4-position of the GlcNAc and at the 1-position of D-galactose generating, in addition to GOS, LacNAc, and Galactosyl-LacNAc trisaccharides which were produced by two sequential transgalactosylations. Humanized GOS is chemically distinct from HMOs, and its effects in vivo have yet to be determined. Thus, we evaluated its safety and demonstrated the prebiotic's ability to modulate the gut microbiome in 6-week-old C57BL/6J mice. Longitudinal analysis of gut microbiome composition of stool samples collected from mice fed a diet containing hGOS for 5 weeks showed a transient reduction in alpha diversity. Differences in microbiome community composition mostly within the Firmicutes phylum were observed between hGOS and GOS, compared to control-fed animals. In sum, our study demonstrated the biological synthesis of hGOS, and signaled its safety and ability to modulate the gut microbiome in vivo, promoting the growth of beneficial microorganisms, including Bifidobacterium and Akkermansia.
Gut microbial communities play a critical role in the maintenance of host health (1, 2). Hence, beneficial modulation with probiotics (live microorganisms that when administered in adequate amounts provide a benefit to their hosts) (3) and prebiotics (selectively fermented dietary carbohydrate structures that promote the growth of beneficial microorganisms) (4, 5) is desirable and potentially effective translational therapeutics to treat gastrointestinal (GI) diseases linked to disrupted microbial communities (4, 6–9) [reviewed in (10)]. Synbiotics (combinations of prebiotics and probiotics) are also emerging as a focal point of GI biology research, as each component, individually and synergistically, could provide unique benefits reestablishing community resilience and host physiology (11, 12). In previous studies we evaluated highly pure β(1–4) galacto-oligosaccharides (GOS) formulations produced by the optimized version of the hexosyl-transferase gene from Hamamotoa (Sporobolomyces) singularis heterologously expressed in Komagataella (Pichia) pastoris (13, 14). This enzyme is one of the most promising catalysts in the field of glycobiology due to its high stability, highly desirable enzymatic properties, and the metabolism of its reaction products (GOS) by specific members of the gut microbial community, impacting its composition and function (15, 16). Beneficial members of the gut microbiota, including Lactobacillus and Bifidobacterium, hydrolyze GOS via β-galactosidases (17). Lactobacillus rhamnosus utilize PTS transporters to internalize GOS prior to hydrolysis (17), while other organisms like specific strains of Bifidobacterium (bifidum) secrete glycosyl hydrolases to break down complex carbohydrates, internalizing the products of hydrolysis (18). Short-chain fatty acids (SCFAs) generated as the result of GOS assimilation include acetate and lactate (17), which community members, including Roseburia and Faecalibacterium, can transform into butyrate (6, 19).
LacNAc is an essential component of human milk oligosaccharides (HMOs) and has been demonstrated to be a major bifidogenic factor in the 1950s (20–22). Several HMOs contain lactose (Galβ1-4Glc) at their reducing end, which can be elongated by the addition of β1-3- or β1-6-linked lacto-N-biose (Galβ1-3GlcNAc) or LacNAc (Galβ1-4GlcNAc). Lactose or the oligosaccharide can be then fucosylated by fucosyltransferases in α1-2, α1-3, or α1-4 linkage and/or sialylated by sialyltransferases in α2-3 or α2-6 linkage to yield a variety of terminal structures (23). The study by Yoshida et al. (24) characterized β-galactosidases of Bifidobacterium longum subsp. infantis to determine how this organism degrades type-1 (lacto-N-biose,) and type-2 (LacNAc,) isomers of HMOs. LacNAc has also been recognized as a building block of glycoproteins and glycolipids in the GI tract. These backbones serve to connect the core structure, which is directly linked to a protein or lipid aglycon with terminal sugars [reviewed in (25)]. LacNAc building blocks and terminal sugars also act as an important precursor of several blood group epitopes (Lewis A, Lewis B, sialyl Lewis A), which are involved in biological processes including fertilization (26), mediation of cell adhesion and pathogen adhesion to colonocytes (27–29).
Chemical and enzymatic synthesis processes have been the most frequently evaluated methods for LacNAc production (30). In recent years, glycoside hydrolases (EC 3.2.1.-) and β-galactosidases (EC 2.1.23) with both hydrolytic and transglycosylation activities, have gained special attention for regio- and stereo-selective synthesis of LacNAc oligosaccharides (https://www.cazypedia.org/index.php/Transglycosylases) (31–37). Enzymatic biosynthesis is considered the most efficient method for producing LacNAc due to specificity, synthesis in one-step reactions, low-cost substrates, sustainability, and overall low environmental impact, [reviewed in (38)]. Conversely, chemical methods to generate LacNAc require multiple reactive hydroxyl groups and laborious protocols for group protection and deprotection to control the stero- and regio-specificities (39, 40). Compared to enzymatic synthesis, generation of LacNAc by chemical synthesis has low yields, a cost-competitive disadvantage for industrial production, hindering the use of LacNAc as an additive in food products (30, 41, 42).
In this study, we describe a novel biological synthesis solution to produce N-Acetyl-lactosamine (LacNAc)-enriched GOS (which we refer to as humanized GOS, hGOS) using optimized Hamamotoa (Sporobolomyces) singularis β-hexosyl transferase [rBHT (13, 14)]. The enzyme generates LacNAc-enriched GOS as the product of the reaction between lactose as a galactose donor and N-Acetylglucosamine as acceptor. We first evaluated the efficiency of a Komagataella (Pichia) pastoris cell line carrying membrane-bound β-hexosyl transferase on the generation of GOS and hGOS from lactose and N-Acetyl-glucosamine. Then, conventionally-raised 6-week-old C57BL/6J mice were fed a control diet or modified diets containing GOS or hGOS for 14 days to evaluate its safety and impact on fecal microbial diversity and composition.
Membrane-bound β-hexosyl transferase from Hamamotoa (Sporobolomyces) singularis in Komagataella (Pichia) pastoris was produced as previously described (13, 14). The standard transgalactosylation reaction utilizing Komagataella (Pichia) pastoris resting cells (harboring membrane-bound enzyme) was initiated by adding standardized amounts of enzyme (1 U g−1 lactose) in 5 mM sodium phosphate buffer (pH 5.0) to a similarly buffered solution containing lactose (200 g liter−1) and N-Acetylglucosamine (25 g liter−1) at 30°C. Reaction products and substrates were analyzed by high-performance liquid chromatography (HPLC) (Shimadzu Corporation, Kyoto, Japan) under isocratic conditions at 65°C and a 0.5-ml min−1 flow rate. The mobile phase was water, and separation was performed by two columns in tandem a Supelco gel Ca++ (Supelco, PA), and HPX-42A (Bio-Rad, CA) columns (300 mm by 7.8 mm) coupled to an SPD-20MA and ELSD-LT II detectors (Shimadzu Corporation, Kyoto, Japan). The column was calibrated using galactosyl-lactose (Carbosynth, Berkshire, United Kingdom), LacNAc, Lactose, N-Acetylglucosamine, Glucose, and Galactose (Sigma-Aldrich, St. Louis, MO). Enzymatic activity was determined using 4-nitrophenyl β-D- glucopyranoside or oNP-Glc as substrate as per the previously described methods (13, 14).
The human equivalent dose (HED) for LacNAc was calculated for the animal study using the methods highlighted by the United States Food and Drug Administration and is based on the approximate body weight of the subject (43). The equation used is as follows:
Where the Km factor is a number based on body surface area. For this study, we used an animal dose of 1,500 mg kg−1, based on ~30 mg LacNAc fed to a ~20 g mouse per day. Additionally, we used a Km factor of 3 for mice and 16 to represent a 5 kg human infant (43). Using the formula above, the HED for this study represent an equivalent of 281.25 mg LacNAc per kg body weight in infants. While for a 20 kg child (Km= 25), the HED would be 180 mg kg−1.
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina at Chapel Hill (Approved protocol number: 19-084).
A total of 50 6-week-old C57BL/6J mice were co-housed at random in groups of 5–6 animals and fed a defined diet (D17121301; Research Diets INC.) containing no prebiotics to normalize the gut microbiota for 2 weeks. After a 2-week standardization period, fresh stool samples were collected directly from the anus of each animal into a sterile tube. To avoid cage batch effects, animals were moved into paired housing such that no two animals from the same standardization group were co-housed. Each animal was considered one experimental unit. Upon reassigning housing (Figure 2A), animal pairs were split into three distinct groups, each of which began feeding on either the defined control diet (D17121301) (n = 17), GOS diet (D17121302) (n = 17) in which 71.8 g of cellulose per kilogram diet was replaced with 71.8 g of pure GOS, or hGOS (D18121401) (n = 16) where LacNAc represented a 1% (w/w). Composition of each diet is detailed in Supplementary Table 1. Each diet was offered ad libitum for 2 weeks prior to stool sample collection. Individual animal mass and dietary consumption were measured daily during the first 14 days of the dietary study to assess animal growth and food consumption (total food consumed in a cage/2 = individual animal food consumption) between diet groups. After day-14 sample collection, half of the animals in each treatment group were removed for a tangential study and all remaining animals (total=24, n = 8 per diet) continued to consume their respective diets ad libitum, with stool sample collections occurring at three additional time points each ~1-week apart prior to animal sacrifice. At the conclusion of the trial on day 38, each animal was euthanized via CO2 asphyxiation and cervical dislocation.
Total DNA was extracted from fecal pellets using the Qiagen ClearMag Extraction system on KingFisher Flex Magnetic Bead processing instrument as described (15). Briefly, stool samples were transferred to a screwcap tube containing 10 mg of sterile acid-washed glass beads (0.1–0.5 mm diameter) and 700 μl PM1 solution (Qiagen, Valencia, CA). Samples were homogenized for 5-min at 15 Hz in Qiagen Tissue Lyser II (Qiagen). Bead-beaten samples were treated with IRS-PCR inhibitor remover solution (Qiagen) (3:1; lysate:IRS ratio) overnight at 4°C and transferred to KingFisher Deep-well plate containing ClearMag magnetic beads and binding buffer (Qiagen). Sample plates were subsequently processed on KingFisher Flex instrument to isolate and wash DNA. DNA was quantified with Quant-iT™ PicoGreen® dsDNA reagent (Molecular Probes, Thermo Fisher Scientific, Waltham, MA) and stored at −20°C.
Total DNA was subject to amplification of the V4 variable region of the 16S rRNA gene using primers 515F and 806R (44) with Illumina adaptors. Amplicons were barcoded using Illumina dual-index barcodes [Index 1(i7) and Index 2(i5)], purified using Agencourt® AMPure® XP reagent (Beckman Coulter, Brea, CA) and quantified with Quant-iT™ PicoGreen™ dsDNA Reagent (Molecular Probes, Thermo Fisher Scientific). Libraries were pooled in equimolar amounts and sequenced on HiSeq2500 instrument (Illumina, San Diego, CA).
Analysis of 16S rRNA amplicon sequencing data was carried out using the QIIME2 pipeline as described (45). Briefly, sequences were grouped into Operational Taxonomic Units (OTUs) using UCLUST (46). OTU sequences were aligned, and phylogenetic trees were built (47). Before generating the phylogenetic tree, the overall OTU table was collapsed using the “taxa collapse” plugin in QIIME2. The set of representative sequences was then trimmed to include only one representative sequence for each collapsed OTU. The filtered set of representative sequences was then aligned using MAFFT, and a phylogenetic tree was generated from the alignment using RAxML (48, 49). The phylogenetic tree was finally annotated with presence/absence data using iToL and PhyloToAST (50–52). Alpha and beta diversity metrics were calculated in R 4.0.3 using the phyloseq and vegan packages (53–55). Only data from young animals (6 weeks old) were used in the calculation of diversity metrics. Both the Shannon entropy and inverse Simpson indexes were calculated to ensure an accurate estimation of the true alpha diversity of the samples. Beta diversity was calculated using principal coordinate analysis (PCoA) of the weighted UniFrac distances (56).
Data were evaluated for homogeneity of variance using Levene's test. Statistical significance of alpha diversity was evaluated using a repeated-measures ANOVA followed by Tukey's Honest Significant Difference test to separate means. The 95% confidence ellipses for beta diversity plots were calculated in R 4.0.3 using ggplot2 (57). Beta diversity statistical analyses were performed using the PERMANOVA and PERMDISP functions of the vegan package in R 4.0.3. All statistical analysis results for the alpha and beta diversity analyses can be found in Supplementary Table 2. The α for all statistical tests was fixed at 0.05.
All sequencing data has been submitted to the NCBI repository and can be accessed via the following accession number: PRJNA681811.
The reactions catalyzed by the rBHT enzyme were regio-selective, forming the β-galactosyl linkage at the 4-position of the GlcNAc and the 1-position of D-galactose, synthesizing various glycoconjugates directly from soluble GlcNAc. The obtained products, in addition to GOS, included Gal-β(1, 4)GlcNAc (LacNAc, Figure 1A, panel B) disaccharides and Galβ-(1, 4)Galβ-(1, 4)GlcNAc (Galactosyl-LacNAc, Figure 1A, panel C) trisaccharides, which were produced by two sequential transgalactosylations. Figure 1B shows the kinetics of the reaction performed during 8 days of incubation using rBHT polypeptides (e.g., whole cells displaying membrane-bound enzyme). The enrichment of GOS with LacNAc at a ratio lactose/N-Acetylglucosamine of 8:1 performed for these experiments (200 g/L lactose and 25 g/L GlcNAc) generated 25 g/L of LacNAc and 100 g/L hGOS after 48 h of incubation. At this time point, the reaction was terminated, and the products of the reaction (hGOS) were freeze-dried.
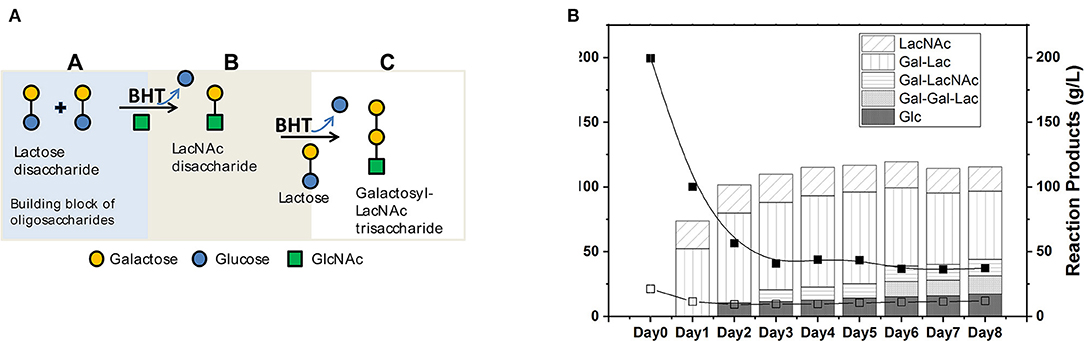
Figure 1. (A) Illustrative representation of catalysis of hGOS from carbohydrate constituents. (B) Time course evolution in g/L of dietary carbohydrate structures generation and residual substrates in g/L over 8 days; Lactose (Lac), N-Acetylglucosamine (GlcNAc), and Glucose (Glc). The products of the reaction were N-Acetyl-lactosamine (LacNAc), Galactosyl-β(1–4)lactose (Gal-Lac), Galactosyl-β(1–4)N-Acetyl-lactosamine (Gal-LacNAc), and Galactosyl-β(1–4)Galactosyl-β(1–4)lactose (Gal-Gal-Lac). The reactions were performed using whole cells membrane-bound protein (1 U rBHT.g−1 lactose). The initial conditions of the reactions contained 200 g/L lactose; 25 g/L N-Acetylglucosamine (GlcNAc), in 5 mM sodium phosphate buffer (pH 5.0) and incubated at 30°C. Samples were removed periodically and separated by HPLC and quantified with sequential ELSD and PDA detectors.
We conducted an animal experiment with conventional 6-week-old C57BL/6J mice fed a control diet, or modified diets containing GOS or hGOS to assess their impact on the gut microbiome (Figure 2A). Results showed no impact of GOS or hGOS diets on weight or daily food consumption. Each mouse consumed ~3 g of food per day, with no significant differences between the diets (Supplementary Figure 1). Therefore, based on the formulation of each prebiotic diet, we calculated that the dose of prebiotic consumed per day by each mouse was 0 mg/day (on control diet), and ~190 mg/day of total prebiotic (GOS, hGOS). This estimate translates to ~171 mg/day of GOS (GOS diet) and ~30 mg/day of LacNAc (hGOS diet) based on prebiotic formulation data.
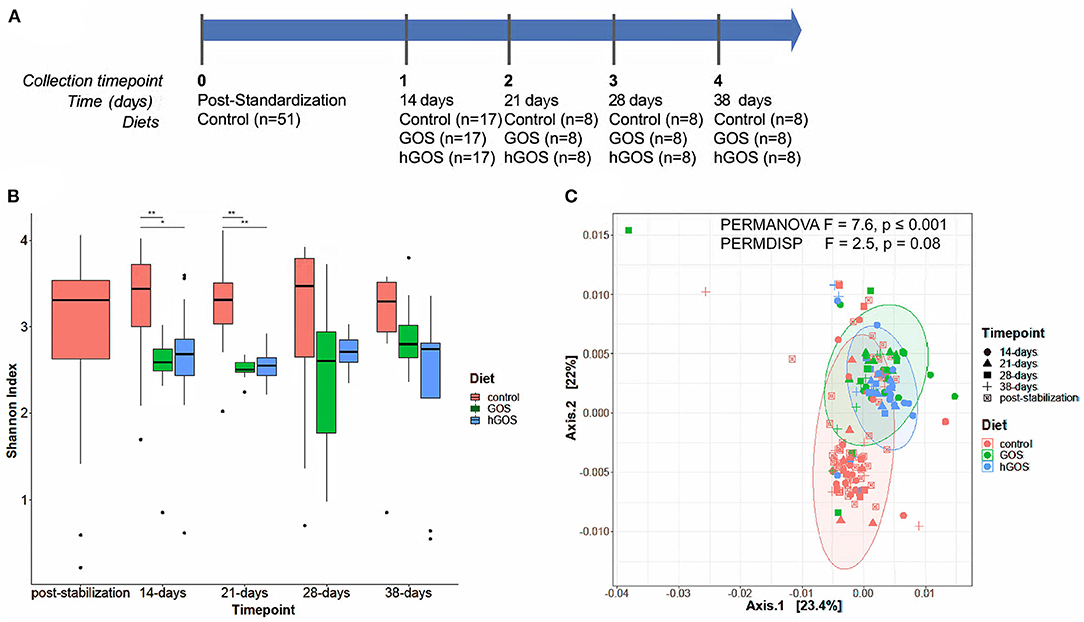
Figure 2. (A) Experimental timeline of the animal study, delineating the number of animals in each experimental group, and the duration between sample collection and analysis. (B) Box plots show changes in Shannon diversity values between groups fed control (red), GOS (green), or hGOS (blue) diets over time. Significant differences in Shannon diversity between diets are indicated by bars and asterisks, with *indicating p < 0.05 and **indicating p < 0.01. Only statistically significant differences are shown. (C) PCoA, PERMANOVA, and PERMDISP analyses of samples between time points show significant differences in clustering as a function of the diet.
After 2 weeks of feeding on diets containing prebiotics, animals exhibited a significant (repeated measures ANOVA p < 0.05) reduction in alpha diversity (Figure 2B). Over the length of the study, diversity of prebiotic-fed animals returned to values comparable to the control diet with no statistically significant differences between groups at day 28. PCoA plots revealed distinct clustering of hGOS-fed animals, which displayed a much tighter dispersion pattern compared to control animals (PERMDISP p = 0.009), suggesting a higher similarity between communities within hGOS-fed than control-fed animals (Figure 2C). Spatial medians were significantly different between groups (PERMANOVA F = 7.6463, p ≤ 0.001).
Taxonomy plots of relative microbial abundance revealed the genus-level variability between animals fed control diets, and those consuming either GOS or hGOS diets over time (Figure 3A). The most dramatic changes in the assembled microbial communities were observed between timepoint 0 and 14 days after introducing prebiotic diets. Changes between the communities within prebiotic-fed animals after 14 days were minimal. Analysis of Composition of Microbiome (ANCOM) used to further explore microbial abundance changes within the communities of prebiotic-fed animals across all time points revealed an increased relative abundance of beneficial microorganisms including Akkermansia, Bifidobacterium, and Bacteroides, along with Allobaculum in both GOS and hGOS diets. The dietary interventions reduced the relative abundance of Butyricicoccus, Clostridium, Turicibacter, and Lachnospiraceae across all time points (Figures 3A,B, Supplementary Figure 2).
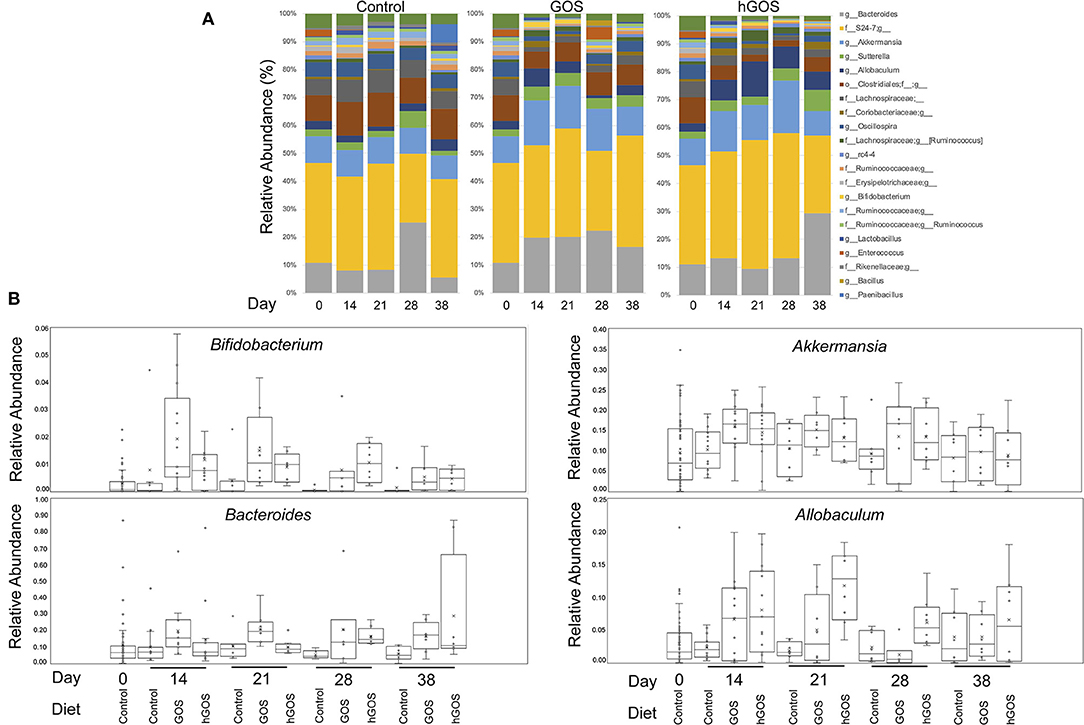
Figure 3. (A) Genus-level taxonomy plots reveal highly abundant taxa in each group at each time point in the study. Changes associated with diet were observed immediately (day 14) and were persistent throughout the trial. (B) Bifidobacterium, Akkermansia, Bacteroides, and Allobaculum were significantly increased in prebiotic-fed animals at 14 days compared to controls.
Figure 4 shows a phylogenetic tree generated using PhyloToAST that includes 117 unique OTUs detected in at least one of the three diets examined. Of the 117 unique OTUs detected, 76 OTUs were detected in all conditions (Control, GOS, hGOS), and 40 were detected in control and GOS but not in hGOS fed mice, while none was detected in control and hGOS but not GOS fed mice. The majority of the OTUs detected in the control and GOS mice, but not in hGOS mice, belonged to the Firmicutes phylum. Taxa not detected in the hGOS group compared to control and GOS group within the Phylum Firmicutes and Class Bacilli included Lactobacillus reuteri, L. zeae, species of Enterococcus, Brevibacillus, Paenibacillus, Anaerobacillus, Virgibacillus, Facklamia, Unclassified Lactobacillales, Bacillales, Enterococcaceae, and Planococcaceae. Within the Phylum Firmicutes and Class Clostridia, the following were not detected in the hGOS group: Veillonella dispar, Ruminococcus flavefaciens, Faecalibacterium prausnitzi, species of Butyrivibrio, Pseudobutyrivibrio, Lachnospira, Oxobacter, Roseburia, Dialister, Veillonella, Phascolarctobacterium, Anaerotruncus, Blautia and Unclassified Clostridiales, Clostridiaceae, and Veillonellaceae. Only 3 OTUs corresponding to the Phylum Bacteroidetes (Bacteroides eggerthii, B. caccae, and B. fragilis) and 7 Proteobacteria (Burkholderia bryophila, Acinetobacter, Bilophila, Hydrogenophaga, Pseudomonas, Halomonas, and Unclassified Enterobacteriaceae) were not detected in the hGOS group. Supplementary Table 1 presents the mean relative abundance values for each observed OTU in each of the experimental conditions.
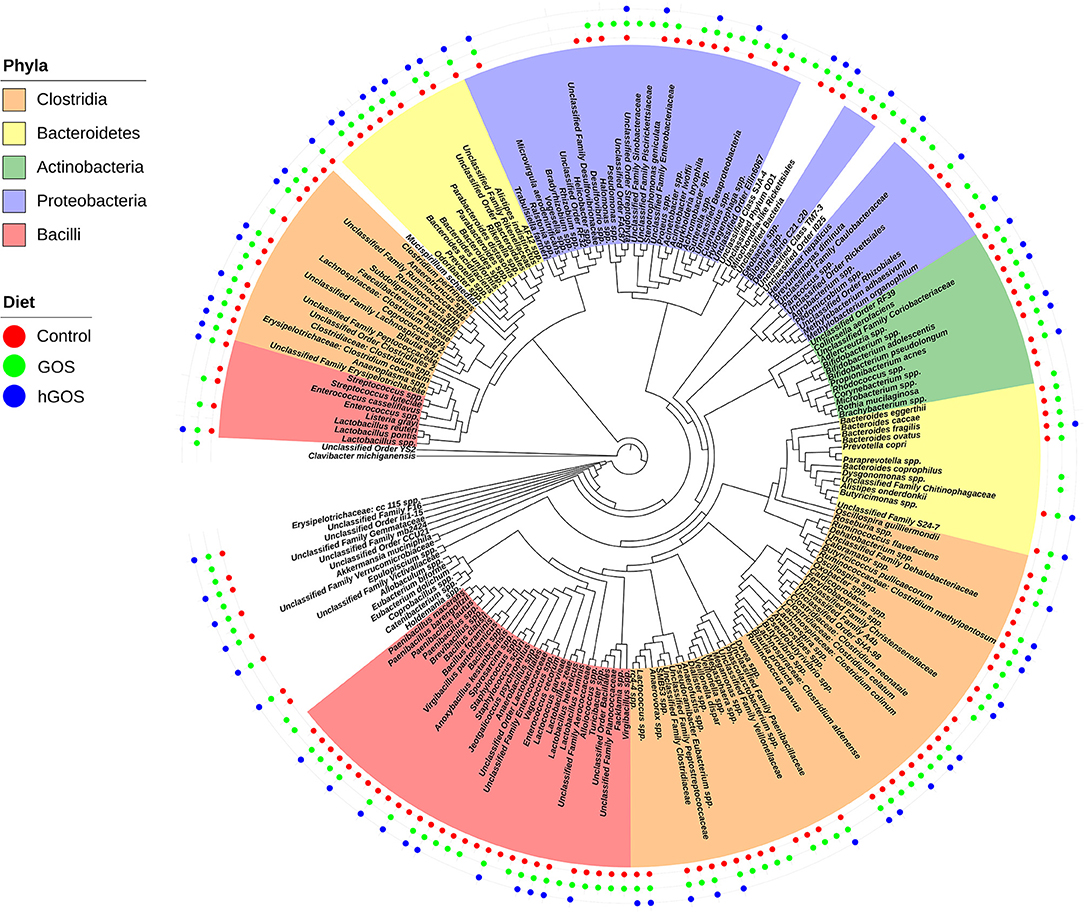
Figure 4. Phylogenetic tree shows the taxa that are present or absent in animals fed each experimental or control diet. Taxa that are highlighted by phyla and presence is designated by adding a colored dot for each diet in which the taxa were observed.
Prebiotics, including GOS, are selectively fermented by gut microorganisms and promote the growth of beneficial microorganisms when consumed in adequate amounts (16, 17). In this study, we report the biological synthesis of hGOS enriched in LacNAc and determined its lack of adverse effects by determining the impact of feeding on the gut microbiome of healthy 6-week-old C57BL/6J mice in comparison with defined control and GOS-containing diets.
Traditionally, higher values of gut microbiota diversity has been associated with good health (58–60). In our study, feeding of both GOS and hGOS-enriched diets initially reduced diversity, even when constituents of the gut microbial community considered beneficial (Bifidobacterium, Akkermansia, and species of Bacteroides) increased. Diversity increased at 28 and 38 days, suggesting that sustained hGOS feeding would lead to a diversity comparable to the control group. In addition, we did not observe differences in dietary consumption or weight in prebiotic-fed animals compared to the control group. We have recently reported an initial decreased diversity in 6- and 60-week old GOS-fed C57BL/6J mice after 2 weeks (15), which is in accordance with studies of GOS-supplemented infant formula (61) but contrast with other studies on human adults (6, 62) and young or adult BALB/c mice that showed no changes on diversity due to GOS feeding (63, 64). Considering the biochemical structure of GOS and hGOS and their similarity to HMOs, it makes sense that these prebiotics exert a restrictive selection of microorganisms to only microbes capable of establishing a mutualistic relationship with the host as observed in breastfed infants (65). The restrictive colonization effect leads in babies to the successive establishment of different bacterial groups, from aerotolerant bacteria to progressively stricter anaerobes (66, 67), and could provide in adults and older adults a strategy to beneficially modulate the gut microbiome by the subsequent introduction of microbial network units (68).
Members of the gut microbial community including strains of Bifidobacterium and Lactobacillus encode galactosidases genes that hydrolyze complex carbohydrates including GOS, as demonstrated in our previous and current studies (15–17, 19, 69) generating products which other members of the gut microbiota can further utilize through cross-feeding (19, 70). Due to structural similarities between the dietary carbohydrate structures contained in GOS and hGOS, it can be expected that their hydrolysis will result in similar molecules, including lactate and acetate, which could subsequently be utilized to generate other SCFAs of biological relevance, including butyrate. We anticipated that the additional LacNAc residues in hGOS would provide an additional substrate for bacterial enzymatic systems, allowing for different microorganisms to utilize these compounds compared to GOS. However, our study was not able to detect bacterial groups that used hGOS but not GOS. Further experiments will be required to characterize the gut bacterial metabolism of hGOS. Among other changes in the gut microbiota, feeding GOS and hGOS increased the abundance of Akkermansia muciniphila, a microorganism that predominantly utilizes mucin as its energy source. GOS enrichment of Akkermansia is likely a consequence of increased mucin production (15). However, hGOS (containing LacNAc) may be utilized directly by Akkermansia due to a similar LacNAc structure found in hGOS and mucin (71). These findings are consistent with our previous animal studies (15, 16); however, animal models have significant limitations due to fundamental differences between human and mouse-originated microorganisms (72). Further studies are currently underway to better assess the impacts of hGOS on human bacterial isolates, with the ultimate goal of developing a prebiotic optimized for human consumption.
Finding the proper dose of a new therapeutic compound is vital not only to ensure safety and efficacy in clinical trials but is also necessary to ensure the economic feasibility of the new product. For GOS, a low dose (below 2 g per day) may not elicit the desired modulatory effect (6, 73), while an excessively high dose (over 15 grams per day) may induce undesired GI effects (6, 74). Studies have shown the importance of translating the dose of a compound validated in animal models to the HED [reviewed in (75)]. Here, we demonstrated the lack of adverse effects of a HMO mimetic composition, LacNAc-enriched GOS, and its ability to modulate the gut microbiome at a HED of 180 mg kg−1 day−1. The values tested during our experiments are in accordance with the recommended values by the EFSA panel (76) for 2′-O-fucosyllactose (2′-FL) and lacto-N-neotetraose (LNnT). The tested HED was six times higher than the calculated average amount of LacNAc consumed in a day by a 5 kg infant (1,400 vs. 232 mg day−1), potentially highlighting the lack of adverse effects of LacNAc, even at a higher-than-physiological doses (77, 78).
Breast milk is undoubtedly the optimal source of nutrition for the human infant (79) and, until recently, the HMOs present in mother's milk could not be replicated in enough quantities to add to infant formulas. Five years ago, the study by Marriage et al. (80) showed that weight, length, head circumference growth and uptake of 2′FL, measured in the blood and urine, were similar to those of breastfed babies and today, some infant formulas have already incorporated this HMO. As a major building block of HMOs, the addition of LacNAc to the existing list of prebiotic compounds is of paramount importance for the further development of safe, nutritionally, and immunologically complete formulas. Hence, our study represents the first step in evaluating the safety and efficacy of enzymatically produced hGOS in an animal model of weaned human infants.
All sequencing data has been submitted to NCBI repository and can be accessed via the following accession number: PRJNA681811.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill (Approved protocol number: 19-084).
JA performed and analyzed animal experiments. HW and JR curated and analyzed the amplicon sequencing data presented in the manuscript. SD produced the humanized GOS for the animal experiments. MA-P and JB-B designed the experiments and edited the manuscript. All authors contributed to the writing of the manuscript.
This project was supported by the Chancellors Innovation Fund (CIF) 2018 and the Department of Plant and Microbial Biology at North Carolina State University. The UNC Microbiome Core was supported in part by P30 DK034987 Center for Gastrointestinal Biology and Disease (CGIBD) and P30 DK056350 UNC Nutrition Obesity Research Center (NORC). HW was supported by the NC State Graduate Student Support Plan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.640100/full#supplementary-material
Supplementary Figure 1. Bodyweight and daily dietary consumption datasets are included to show that animal growth and intake were not inhibited by the introduction of either experimental diet (GOS and hGOS).
Supplementary Figure 2. Bacterial taxa shown to have a significantly reduced relative abundance in the presence of hGOS.
Supplementary Table 1. Composition of diets used in the animal study.
1. Arnold JW, Roach J, Azcarate-Peril MA. emerging technologies for gut microbiome research. Trends Microbiol. (2016) 24:887–901. doi: 10.1016/j.tim.2016.06.008
2. Madupu R, Szpakowski S, Nelson K. Microbiome in human health and disease. Sci Prog. (2013) 96:153–70. doi: 10.3184/003685013X13683759820813
3. Reid G, Jass J, Sebulsky MT, McCormick JK. Potential uses of probiotics in clinical practice. Clin Microbiol Rev. (2003) 16:658–72. doi: 10.1128/CMR.16.4.658-672.2003
4. de Vrese M, Schrezenmeir J. Probiotics, prebiotics, and synbiotics. Adv Biochem Eng Biotechnol. (2008) 111:1–66. doi: 10.1007/10_2008_097
5. Roberfroid M, Gibson GR, Hoyles L, McCartney AL, Rastall R, Rowland I, et al. Prebiotic effects: metabolic and health benefits. Br Nutr J. (2010) 104(Suppl 2):S1–63. doi: 10.1017/S0007114510003363
6. Azcarate-Peril MA, Ritter AJ, Savaiano D, Monteagudo-Mera A, Anderson C, Magness ST, et al. Impact of short-chain galactooligosaccharides on the gut microbiome of lactose-intolerant individuals. Proc Natl Acad Sci USA. (2017) 114:E367–75. doi: 10.1073/pnas.1606722113
7. Duncan SH, Flint HJ. Probiotics and prebiotics and health in ageing populations. Maturitas. (2013) 75:44–50. doi: 10.1016/j.maturitas.2013.02.004
8. Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, et al. New scientific paradigms for probiotics and prebiotics. J Clin Gastroenterol. (2003) 37:105–18. doi: 10.1097/00004836-200308000-00004
9. Vitetta L, Briskey D, Alford H, Hall S, Coulson S. Probiotics, prebiotics and the gastrointestinal tract in health and disease. Inflammopharmacology. (2014) 22:135–54. doi: 10.1007/s10787-014-0201-4
10. Belorkar SA, Gupta AK. Oligosaccharides: a boon from nature's desk. AMB Express. (2016) 6:82. doi: 10.1186/s13568-016-0253-5
11. Markowiak P, Slizewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. (2017) 9:1021. doi: 10.3390/nu9091021
12. West NP, Pyne DB, Cripps A, Christophersen CT, Conlon MA, Fricker PA. Gut balance, a synbiotic supplement, increases fecal Lactobacillus paracasei but has little effect on immunity in healthy physically active individuals. Gut Microbes. (2012) 3:221–7. doi: 10.4161/gmic.19579
13. Dagher SF, Azcarate-Peril MA, Bruno-Barcena JM. Heterologous expression of a bioactive beta-hexosyltransferase, an enzyme producer of prebiotics, from Sporobolomyces singularis. Appl. Environ Microbiol. (2013) 79:1241–9. doi: 10.1128/AEM.03491-12
14. Dagher SF, Bruno-Bárcena JM. A novel N-terminal region of the membrane β-hexosyltransferase: its role in secretion of soluble protein by Pichia pastoris. Microbiology. (2016) 162:23–34. doi: 10.1099/mic.0.000211
15. Arnold JW, Roach J, Fabela S, Moorfield E, Ding S, Blue E, et al. The pleiotropic effects of prebiotic galacto-oligosaccharides on the aging gut. Microbiome. (2021) 9:31. doi: 10.1186/s40168-020-00980-0
16. Monteagudo-Mera A, Arthur JC, Jobin C, Keku TO, Bruno Barcena JM, Azcarate-Peril MA. High purity galacto-oligosaccharides enhance specific Bifidobacterium species and their metabolic activity in the mouse gut microbiome. Benef Microb. (2016) 3:1–18. doi: 10.3920/BM2015.0114
17. Arnold JW, Simpson JB, Roach J, Bruno-Barcena JM, Azcarate-Peril MA. Prebiotics for lactose intolerance: variability in galacto-oligosaccharide utilization by intestinal Lactobacillus rhamnosus. Nutrients. (2018) 10:1517. doi: 10.3390/nu10101517
18. Møller PL, Jørgensen F, Hansen OC, Madsen SM, Stougaard P. Intra- and extracellular beta-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, comparative characterization. Appl Environ Microbiol. (2001) 67:2276–83. doi: 10.1128/AEM.67.5.2276-2283.2001
19. Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. (2016) 7:979. doi: 10.3389/fmicb.2016.00979
20. György P. N-Containing saccharides in human milk. In: Wolstenholme GEW, O'Connor M, editors. Ciba Foundation Symposium - Chemistry and Biology of Mucopolysaccharides. J. and A. Churchill Ltd (1958). p. 140–56. doi: 10.1002/9780470719060.ch9
21. György P, Norris RF, Rose CS. Bifidus factor. I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch Biochem Biophys. (1954) 48:193–201. doi: 10.1016/0003-9861(54)90323-9
22. György P, Rose CS. Microbiological studies on growth factor for L. bifidus var. pennsylvanicus. Exp Biol Med. (1955) 90:219–23. doi: 10.3181/00379727-90-21988
23. Bode L. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J Nutrit. (2006) 136:2127–30. doi: 10.1093/jn/136.8.2127
24. Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, et al. Bifidobacterium longum subsp. infantis uses two different beta-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. (2012) 22:361–8. doi: 10.1093/glycob/cwr116
25. van den Eijnden DH. On the origin of oligosaccharide species—glycosyltransferases in action. In: Ernst B, Hart GW, Sinaý P, . Carbohydrates in Chemistry Biology. Weinheim: Wiley-VCH (2000). p. 589–624. doi: 10.1002/9783527618255.ch23
26. Kerr CL, Hanna WF, Shaper JH, Wright WW. Lewis X-containing glycans are specific and potent competitive inhibitors of the binding of ZP3 to complementary sites on capacitated, acrosome-intact mouse sperm1. Biol Reprod. (2004) 71:770–7. doi: 10.1095/biolreprod.103.023812
27. Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. (2012) 22:1147–62. doi: 10.1093/glycob/cws074
28. Du J, Yarema KJ. Carbohydrate engineered cells for regenerative medicine. Adv Drug Deliv Rev. (2010) 62:671–82. doi: 10.1016/j.addr.2010.01.003
29. Lane JA, Mehra RK, Carrington SD, Hickey RM. The food glycome: a source of protection against pathogen colonization in the gastrointestinal tract. Int J Food Microbiol. (2010) 142:1–13. doi: 10.1016/j.ijfoodmicro.2010.05.027
30. Krasnova L, Wong C.-H. Oligosaccharide synthesis and translational innovation. J Am Chem Soc. (2019) 141:3735–54. doi: 10.1021/jacs.8b11005
31. Arreola SL, Intanon M, Wongputtisin P, Kosma P, Haltrich D, Nguyen TH, et al. Transferase activity of Lactobacillal and Bifidobacterial β-Galactosidases with various sugars as galactosyl acceptors. J Agric Food Chem. (2016) 64:2604–11. doi: 10.1021/acs.jafc.5b06009
32. Black BA, Lee VSY, Zhao YY, Hu Y, Curtis JM, Gänzle MG. Structural identification of novel oligosaccharides produced by Lactobacillus bulgaricus and Lactobacillus plantarum. J Agric Food Chem. (2012) 60:4886–94. doi: 10.1021/jf300917m
33. Bridiau N, Maugard T. A comparative study of the regioselectivity of the β-galactosidases from Kluyveromyces lactis and Bacillus circulans in the enzymatic synthesis of N-Acetyl-lactosamine in aqueous media. Biotechnol Prog. (2011) 27:386–94. doi: 10.1002/btpr.542
34. Fischöder T, Laaf D, Dey C, Elling L. Enzymatic synthesis of N-Acetyllactosamine (LacNAc) Type 1 oligomers and characterization as multivalent galectin ligands. Molecules. (2017) 22:1320. doi: 10.3390/molecules22081320
35. Li W, Sun Y, Ye H. Synthesis of oligosaccharides with lactose and N-acetylglucosamine as substrates by using β-D-galactosidase from Bacillus circulans. Eur Food Res Technol. (2010) 231:55–63. doi: 10.1007/s00217-010-1254-2
36. Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. (2014) 42:D490–D5. doi: 10.1093/nar/gkt1178
37. The CC. Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology. (2018) 28:3–8. doi: 10.1093/glycob/cwx089
38. Karimi Alavijeh M, Meyer AS, Gras SL, Kentish SE. Simulation and economic assessment of large-scale enzymatic N-acetyllactosamine manufacture. Biochem Eng J. (2020) 154:107459. doi: 10.1016/j.bej.2019.107459
39. Mong TKK, Huang C-Y, Wong C-H. A new reactivity-based one-pot synthesis of N-Acetyllactosamine oligomers. J Org Chem. (2003) 68:2135–42. doi: 10.1021/jo0206420
40. Romanò C, Oscarson S. Synthesis of lactosamine-based building blocks on a practical scale and investigations of their assembly for the preparation of 19F-labelled LacNAc oligomers. Organ Biomol Chem. (2019) 17:2265–78. doi: 10.1039/C8OB03066A
41. Benkoulouche M, Fauré R, Remaud-Siméon M, Moulis C, André I. Harnessing glycoenzyme engineering for synthesis of bioactive oligosaccharides. J R Soc Interface Focus. (2019) 9:20180069. doi: 10.1098/rsfs.2018.0069
42. Martins GN, Ureta MM, Tymczyszyn EE, Castilho PC, Gomez-Zavaglia A. Technological aspects of the production of fructo and galacto-oligosaccharides. enzymatic synthesis and hydrolysis. Front Nutrit. (2019) 6:78. doi: 10.3389/fnut.2019.00078
43. U.S. Department of Health and Human Services Food and Drug Administration. (2005). Guidance for Industry: Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers. Rockville, MD: Center for Drug Evaluation and Research (CDER), 7.
44. Khan F. Review of Soil Taxonomy Information for Submitted Environmental Fate Studies of Ametroctradin. Washington, DC: U.S. Environmental Protection Agency (2012).
45. Allali I, Arnold JW, Roach J, Cadenas MB, Butz N, Hassan HM, et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. (2017) 17:194. doi: 10.1186/s12866-017-1101-8
46. Edgar RC. Search and clustering orders of magnitude faster than BLAS. Bioinformatics. (2010) 26:2460–1. doi: 10.1093/bioinformatics/btq461
47. Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. (2010) 5:e9490. doi: 10.1371/journal.pone.0009490
48. Katoh K, Standley DM. MAFFT multiple sequence alignment software Version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/molbev/mst010
49. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
50. Dabdoub SM, Fellows ML, Paropkari AD, Mason MR, Huja SS, Tsigarida AA, et al. PhyloToAST: bioinformatics tools for species-level analysis and visualization of complex microbial datasets. Sci Rep. (2016) 6:29123. doi: 10.1038/srep29123
51. Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. (2011) 39:W475–8. doi: 10.1093/nar/gkr201
52. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. (2016) 44:W242–5. doi: 10.1093/nar/gkw290
53. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
54. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package. (2.5-5 ed.) San Francisco, CA (2019).
55. Team RC. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing (2018).
56. Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. (2011) 5:169–72. doi: 10.1038/ismej.2010.133
57. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag (2009). doi: 10.1007/978-0-387-98141-3
58. Jones RM. The influence of the gut microbiota on host physiology: in pursuit of mechanisms. Yale J Biol Med. (2016) 89:285–97.
59. Larsen OFA, Claassen E. The mechanistic link between health and gut microbiota diversity. Sci Rep. (2018) 8:2183. doi: 10.1038/s41598-018-20141-6
60. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. (2019) 7:14. doi: 10.3390/microorganisms7010014
61. Matsuki T, Tajima S, Hara T, Yahagi K, Ogawa E, Kodama H. Infant formula with galacto-oligosaccharides (OM55N) stimulates the growth of indigenous bifidobacteria in healthy term infants. Benef Microb. (2016) 7:453–61. doi: 10.3920/BM2015.0168
62. So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JT, et al. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am J Clin Nutr. (2018) 107:965–83. doi: 10.1093/ajcn/nqy041
63. Cheng W, Lu J, Li B, Lin W, Zhang Z, Wei X, et al. Effect of functional oligosaccharides and ordinary dietary fiber on intestinal microbiota diversity. Front Microbiol. (2017) 8:1750. doi: 10.3389/fmicb.2017.01750
64. Szklany K, Wopereis H, de Waard C, van Wageningen T, An R, van Limpt K, et al. Supplementation of dietary non-digestible oligosaccharides from birth onwards improve social and reduce anxiety-like behaviour in male BALB/c mice. Nutr Neurosci. (2019) 23:896–910. doi: 10.1080/1028415X.2019.1576362
65. Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun. (2018) 9:4169. doi: 10.1038/s41467-018-06473-x
66. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. (2005) 307:1915–20. doi: 10.1126/science.1104816
67. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. (2008) 320:1647–51. doi: 10.1126/science.1155725
68. Van den Abbeele P, Verstraete W, El Aidy S, Geirnaert A, Van de Wiele T. Prebiotics, faecal transplants and microbial network units to stimulate biodiversity of the human gut microbiome. Microb Biotechnol. (2013) 6:335–40. doi: 10.1111/1751-7915.12049
69. Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de los Reyes-Gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. (2016) 7:185. doi: 10.3389/fmicb.2016.00185
70. Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett. (2015) 362:fnv176. doi: 10.1093/femsle/fnv176
71. Leclaire C, Lecointe K, Gunning PA, Tribolo S, Kavanaugh DW, Wittmann A, et al. Molecular basis for intestinal mucin recognition by galectin-3 and C-type lectins. FASEB J. (2018) 32:3301–20. doi: 10.1096/fj.201700619R
72. Fouladi F, Glenny EM, Bulik-Sullivan EC, Tsilimigras MCB, Sioda M, Thomas SA, et al. Sequence variant analysis reveals poor correlations in microbial taxonomic abundance between humans and mice after gnotobiotic transfer. ISME J. (2020) 14:1809–20. doi: 10.1038/s41396-020-0645-z
73. Davis LM, Martinez I, Walter J, Hutkins R. A dose dependent impact of prebiotic galactooligosaccharides on the intestinal microbiota of healthy adults. Int J Food Microbiol. (2010) 144:285–92. doi: 10.1016/j.ijfoodmicro.2010.10.007
74. Liu F, Li P, Chen M, Luo Y, Prabhakar M, Zheng H, et al. Fructooligosaccharide (FOS) and Galactooligosaccharide (GOS) increase bifidobacterium but reduce butyrate producing bacteria with adverse glycemic metabolism in healthy young population. Sci Rep. (2017) 7:11789. doi: 10.1038/s41598-017-10722-2
75. Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. (2008) 22:659–61. doi: 10.1096/fj.07-9574LSF
76. EFSA Panel on Dietetic Products Nutrition and Allergies (NDA). Safety of 2′-O-fucosyllactose as a novel food ingredient pursuant to Regulation (EC) No 258/97. EFSA J. (2015) 13:4184. doi: 10.2903/j.efsa.2015.4184
77. Balogh R, Jankovics P, Béni S. Qualitative and quantitative analysis of N-acetyllactosamine and lacto-N-biose, the two major building blocks of human milk oligosaccharides in human milk samples by high-performance liquid chromatography–tandem mass spectrometry using a porous graphitic carbon column. J Chromatograp A. (2015) 1422:140–6. doi: 10.1016/j.chroma.2015.10.006
78. Dewey KG, Lönnerdal B. Milk and nutrient intake of breast-fed infants from 1 to 6 months: relation to growth and fatness. J Pediatric Gastroenterol Nutrit. (1983) 2:497–506. doi: 10.1097/00005176-198302030-00018
79. Thompson AL, Monteagudo-Mera A, Cadenas MB, Lampl ML, Azcarate-Peril MA. Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front Cell Infect Microbiol. (2015) 5:3. doi: 10.3389/fcimb.2015.00003
Keywords: galactooligosaccharide (GOS), N-Acetyl-D-lactosamine (LacNAc), safety, human milk oligosaccharides (HMOs), LacNAc synthesis, mouse models
Citation: Arnold JW, Whittington HD, Dagher SF, Roach J, Azcarate-Peril MA and Bruno-Barcena JM (2021) Safety and Modulatory Effects of Humanized Galacto-Oligosaccharides on the Gut Microbiome. Front. Nutr. 8:640100. doi: 10.3389/fnut.2021.640100
Received: 10 December 2020; Accepted: 11 March 2021;
Published: 07 April 2021.
Edited by:
Nicole Clemence Roy, University of Otago, New ZealandReviewed by:
Gerald Tannock, University of Otago, New ZealandCopyright © 2021 Arnold, Whittington, Dagher, Roach, Azcarate-Peril and Bruno-Barcena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jose M. Bruno-Barcena, amJiYXJjZW5AbmNzdS5lZHU=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.