- 1Department of Chemistry, Sapienza University of Rome, Rome, Italy
- 2NMR-Based Metabolomics Laboratory (NMLab), Sapienza University of Rome, Rome, Italy
- 3Fondazione Policlinico Universitario 'Agostino Gemelli' IRCCS, Rome, Italy
- 4Department of Environmental Biology, Sapienza University of Rome, Rome, Italy
The global toll of type 1 diabetes (T1D) has steadily increased over the last decades. It is now widely acknowledged that T1D pathophysiology is more complex than expected. Indeed, a multifaceted interplay between genetic, metabolic, inflammatory and environmental factors exists that leads to heterogeneous clinical manifestations across individuals. Children with non-secretor phenotype and those affected by T1D share low abundance of bifidobacteria, low content of short-chain fatty acids, intestinal phosphatase alkaline and a high incidence of inflammatory bowel diseases. In this context, host-gut microbiota dyad may represent a relevant contributor to T1D development and progression due to its crucial role in shaping host immunity and susceptibility to autoimmune conditions. The FUT2 gene is responsible for the composition and functional properties of glycans in mucosal tissues and bodily secretions, including human milk. FUT2 polymorphisms may profoundly influence gut microbiota composition and host susceptibility to viral infections and chronic inflammatory disease. In this minireview, the possible interplay between mothers' phenotype, host FUT2 genetic background and gut microbiota composition will be discussed in perspective of the T1D onset. The study of FUT2-gut microbiota interaction may add a new piece on the puzzling T1D etiology and unveil novel targets of intervention to contrast T1D development and progression. Dietary interventions, including the intake of α-(1, 2)-fucosyl oligosaccharides in formula milk and the use of specific prebiotics and probiotics, could be hypothesized.
Introduction
Type 1 diabetes (T1D) is a multifactorial autoimmune disease characterized by the loss of pancreatic β-cells that leads to insulin deficiency and hyperglycemia (1). The incidence and prevalence of T1D have risen worldwide over the past 30 years, in particular in children younger than 5 years (1, 2). Several environmental and behavioral factors may have contributed to this trend (e.g., exposure to unhealthy diets, viral and bacterial infections, reduced gut microbiota diversity) (1).
Almost all new T1D diagnoses are characterized by measurable autoantibodies against one or more β-cells protein targets, including insulin, glutamic acid decarboxylase (GAD65), insulinoma-associated protein 2 (IA-2), zinc transporter 8 (ZnT8), and tetraspanin 7 (3, 4). Despite known genetic predisposition, the complex interaction between pancreatic insulin-producing cells and the immune system has not yet clearly established. Moreover, considerable heterogeneity in both symptom presentation and disease progression exist across individuals (1).
Over the last decades, the knowledge of potential T1D etiologic factors has rapidly progressed and novel candidate mechanisms have been identified. Among them, the host-gut microbiota mutual relationship may represent a crucial player in T1D development and progression (5, 6). Indeed, it is now widely recognized that early events associated with the establishment of gut microbiota (e.g., birth mode, breastfeeding, antibiotic use in the first 6 months of age) may shape both the innate and the adaptive immune system (7, 8). Moreover, host-gut microbiota interactions may influence susceptibility to pathogen infections as well as to the development of autoimmune and chronic inflammatory conditions (9).
The fucosyltransferase 2 (FUT2) gene encodes for a critical enzyme responsible for blood group secretor status and mucosal protective functions. FUT2 activities prime the composition of glycans on epithelial cell surfaces, on secretory glands and bodily fluids, including the human milk. As a result of the expression of FUT2 gene, four maternal phenotypes have been identified based on the content of fucosylated human milk oligosaccharides (HMOs) (10). The composition of HMOs content and intestinal mucus glycans may profoundly influence gut microbiota dynamics across all life stages and modulate host response to pathogens (10–12).
In addition, FUT2 polymorphisms have been associated with chronic inflammatory conditions and T1D (13). The FUT2 non-secretor phenotype was suggested as a factor linking genetic susceptibility to alterations in gut microbiota in T1D etiopathogenesis (Supplementary Material Table 1) (14). Yet, genome-wide association studies (GWASs) did not conclusively support a role of FUT2 gene variants in the crosstalk between the host genotype and gut microbiota in T1D development and progression.
In this mini review, the possible interaction between FUT2 gene variants and gut microbiota in T1D will be discussed. Interventions targeting the FUT2-gut microbiota dyad in T1D are also briefly summarized.
FUT2 Genetic Variants
The FUT2 gene is located on chromosome 19q13.33 and codes α-(1, 2)-fucosyltranferase enzyme, which catalyzes the transfer of fucose to galactose terminal residues on N-acetylglusamine or N-acetylglucosamine glycan chains. This leads to the expression of H antigen which is the precursor of ABH histo-blood group antigens (HBGAs) in body fluids, including the milk, and in the intestinal mucosa. The synthesis of HBGAs requires several glycosyltranferases acting on precursor oligosaccharides converted into H antigenic structures by fucosylation in α-(1, 2)-linkage. The secretion of H antigen also depends on other glycosyltransferases, such as FUT3 (Le) A/B enzymes, required to synthetize Lewis HBGAs, which are mainly expressed in colon. The FUT2 gene is characterized by several polymorphisms: individuals that are homozygous for the null FUT2 allele do not express ABH antigens in excreta and in the gastrointestinal tract, and are called non-secretors or se, whereas individuals carrying at least one functional allele are defined secretors or Se, as they express ABH on secretions (15).
Many polymorphisms in FUT2 show population-specific patterns, yet frequencies of non-secretor phenotypes are similar across most populations (16). About 20% of the general population has a non-secretor status due to the presence of two inactive variants of the FUT2 gene. Specifically, the single nucleotide polymorphism (SNP) rs601338(A), which codes for a stop codon at position 143 (Trp–Ter), is predominant in Europeans, Iranians, and Africans. On the other hand, the SNP rs1047781(T), which results in the substitution of Ile for Phe at position 129, represents the East Asian counterpart (17). In a Swedish cohort of newborns, the frequencies of selected FUT2 and FUT3 SNPs were assessed and compared with data from the 1000 Genomes Project European Caucasian (EUR) population (18). The frequency of homozygotes for rs601338(A) was 25% and no statistically significant difference was found with the 1,000 Genomes Project cohort (18). Serpa et al. (19) examined FUT2 polymorphisms in a Caucasian population of non-secretor individuals from Northern Portugal and evaluated the functional properties of the mutant FUT2 enzymes. Two new polymorphisms in the FUT2 gene (FUT2-739G-A and FUT2-839T-C) were identified that lead to the expression of two inactive FUT2 variants (FUT2-247Gly-Ser and FUT2-280Phe-Ser). A recent study by Soejima and Koda (20) showed that the frequency of putative non-secretors was relatively low in Latin American populations except for Puerto Ricans. In the next sections, the role of FUT2 gene variants and associated phenotypes is discussed, with a particular focus on their putative effects on factors associated with T1D development (e.g., viral infections, autoimmune diseases, early life gut microbiota perturbations).
FUT2 in Pathogen Infections and Autoimmune Diseases
FUT2 gene variants have been associated with numerous conditions, including the susceptibility to bacterial, fungal or viral infections and (chronic) autoimmune diseases, although results are not univocal (21).
Rotavirus (RV) infection is strictly related to host genetics. Indeed, an epidemiological association between the secretor status and laboratory-confirmed RV gastroenteritis was observed (22). In various human RV strains, analyses of the carbohydrate binding properties of VP8* (the spike protein domain that mediates viral attachment) revealed a specific affinity for neutral oligosaccharides of the HBGAs (23). In vitro studies, using various glycan binding assays, have demonstrated that the most common human RVs recognize HBGAs through VP8* (24).
Regarding bacterial infections, links between FUT2 secretor status and Helicobacter pylori (H. pylori) ability to attach to human gastric mucosa were found. Mucosal glycans carrying blood group A, B or H antigens are known to be ligands for H. pylori and secretor individuals may be more susceptible to H. pylori infection (25). Recent findings by Rossez et al. (26) confirmed that gastric mucins from secretor people interact more efficiently with H. Pylori than those from non-secretors.
Noroviruses (NoVs) are the major causative agents of acute non-bacterial gastro-enteritis (27). Interestingly, some individuals do not develop infection, despite being exposed to high viral loads (28). A study performed on 77 volunteers showed that HBGAs were critical factors for NoV infection (27). In particular, the presence of inactive FUT2 alleles was responsible for this innate genetic resistance (27). Furthermore, the non-secretor phenotype has been associated to a slow progression of human immunodeficiency virus 1 (HIV-1) infection (29). Indeed, the lack of fucosylation may prevent the formation of glycolipid receptors for HIV-1 on the epithelial surface (29). Conversely, the non-secretor phenotype may confer higher susceptibility to Candida albicans (30), Streptococcus Pneumoniae (31), and mumps infection (32).
In addition, the non-secretor status was associated with self-reported kidney disease (32) and higher risk of celiac disease (33).
Autoimmune diseases are particularly burdensome due to their health, social and economic consequences (34). The FUT2 non-secretor phenotype has been associated with complex and multifactorial disorders, such as Behçet's disease, and inflammatory bowel diseases (IBDs), including Crohn's disease (CD) and ulcerative colitis (UC) (35, 36). CD is a chronic, relapsing inflammatory disease, mainly affecting the small intestine and colon. CD pathogenesis involves multiple interacting elements, such as genetic susceptibility factors, priming by the enteric microflora, and immune-mediated tissue injury (37). Results of a study by McGovern et al. (38) provided evidence that non-secretor status increased CD susceptibility in the European population. A study carried out on Caucasian individuals has shown that non-secretors are at high risk of developing CD, albeit no significant association between ABO variants and CD was detected (39).
Urine profiling analyses conducted by Rueedi et al. (40) on 835 individuals showed that urinary fucose was associated with a FUT2 gene variant linked to CD. Furthermore, elevated urinary concentrations of fucose, also observed in non-secretors, may reflect changes in gut flora activities shifting from a healthy status toward CD. Urinary fucose may thus represent an early biomarker of CD.
However, robust evidence linking FUT2 gene variants to autoimmune diseases is still lacking. A recent study on 635 people with CD did not find an association between rs601338(A) and disease phenotype, severity, or clinical outcomes, suggesting that lack of fucosylation may be involved in CD development rather than in disease progression (41). In contrast, in people with UC, functional FUT2 variants seem to contribute to the different clinical manifestations of the disease (42).
Although the current knowledge does not support a clear-cut association between FUT2 gene variants and the development and progression of autoimmune diseases, the interaction between FUT2 phenotype and the immune system is a promising target to be explored.
FUT2, Gut Mucus Glycans and Human Milk Oligosaccharides in Health and Disease
Genetics contribute to determining the chemical composition of the intestinal mucus layer, which, in turn, modulates gut microbiota composition and host innate immunity (43). In this context, a number of studies assessed whether FUT2 gene variants may influence the quality and quantity of microbiota in infants with conflicting results. The secretor status was associated with higher diversity, richness and abundance of gut Bifidobacterium and Bacteroides compared to non-secretor individuals during the first months of life (44–47). In both people with loss-of-function alleles of FUT2 gene and FUT2 null mice, specific microbiome, meta-proteome and meta-metabolome signatures were described (48). In particular, the gut microbiome in non-secretors was characterized by a depletion in amino acid metabolism pathways and an overall enrichment in genes encoding for carbohydrate and lipid metabolism, glycan biosynthesis, and catabolism (48). These features were paralleled by changes in metabolic profiles. Interestingly, the microbial compositional and functional signature associated with non-secretor phenotype was associated with sub-clinical intestinal inflammation (48).
However, the actual role of FUT2 gene variants in shaping the activity and composition of the gut microbiome is still disputed. Indeed, recent large observational studies as well as GWASs found no or weak association between FUT2 genotype and stool microbiome composition (49–52). In a large cohort of 1,190 healthy young adults, FUT2 genotype and secretor status were not associated with fecal microbial alpha diversity, composition or inferred microbial function (49). In 1,503 individuals from a twin cohort in the United Kingdom, a consistent link between the taxonomic composition of microbiota and the secretor status was not evident (50). Furthermore, in a large cohort from the Genetic Environmental Microbial (GEM) Project, GWAS of 3,727,707 SNPs found 58 SNPs associated with the relative abundance of specific microbial taxa (51). FUT2 gene variants were not present among the SNPs that showed an association with the gut microbiome (51). Finally, in fecal samples from two independent German cohorts, weak associations were found for FUT2 with Clostridium IV and unclassified Clostridiales (52), in partial agreement with Wacklin et al. (44). However, the contribution of FUT2 gene variants in terms of overall influence on microbial variation was negligible (52).
Maternal FUT2 gene may also play a role in early phases of the colonization of the infant gut through its action on the composition of HMOs (53, 54). HMOs represent the third most abundant component of the breast milk. HMOs are minimally digested by the host, and are believed to influence biochemical processes in the infant's gut (10). The expression of the FUT2 gene, codingα-1,2-fucosyltransferase, and FUT3 gene, coding α-1,3/1,4-fucosyltransferase, affects directly HMOs content. Fucosylated oligosaccharydes such as 2′-fucosyllactose (2′ FL), 3′-fucosyllactose (3′ FL), and lacto-N-fucopentaose I/II/III (LNFP I/II/III) are higher in secretors compared to non-secretors (12, 55, 56) but, in general, the HMOs content changes during the lactation period (57). Chemical structures of HMOs are strictly related to their prebiotic effects and their putative role in inhibiting pathogen infections (58, 59). Free HMOs can mimic host receptors and bind directly to pathogens, thus allowing their elimination through feces. HMOs can also compete with pathogens for the binding to the host cell-surface glycan receptors (11, 12, 57).
Previous works showed that infants breastfed by secretor women exhibited a higher number of bifidobacteria, which commonly use HMOs as a carbon source, and in particular 2′ FL, 3′ FL and lactodifucotetraose (LDFT) (60). Some studies reported beneficial effects related to the intake of HMOs. For instance, 2′FL enhanced brain function, learning and memory in rats (61) and protected mice against necrotizing enterocolitis (NEC) (62). 2′ FL may also protect infants against Campylobacter-induced diarrhea (63, 64), while lacto-N-difucohexaose I (LNDFHI) has a positive effect in mitigating NV-related diarrhea (65). Milk from non-secretor mothers lacks 2′ FL and related fucosyl oligosaccharydes, and has been shown to exert a lower protection against enteropathogens that bind α-(1, 2)-fucose (66). A lower content in bifidobacteria taxa has been determined in gut microbiota of infant breastfed by non-secretor mothers. Intriguingly, a similar gut microbiota composition is also present in children with T1D (67), suggesting that a FUT2-dependent priming of gut microbiota may also be present in T1D (Figure 1). Several studies on newborn nutrition demonstrated that breastfeeding from secretor mothers is the most efficient way to transfer fucosylated HMOs to infants (33, 52, 53). Hence, investigations were conducted to evaluate if the addition of specific α-(1, 2)-fucosylated HMOs to formula milk may have the same beneficial effects on gut microbiota. Studies on specific HMOs supplementation in formula milk showed that 2′ FL resulted safe for infants up to 1 year of age (68) and led to a reduction in inflammatory cytokines (69). Yet, evidence correlating α-(1, 2)-fucosylated HMOs in formula milk and gut microbiota in the context of autoimmune diseases are presently lacking. Genetics contribute to determining the chemical composition of intestinal mucus layer, which, in turn, modulates gut microbiota composition and host innate immunity (70).
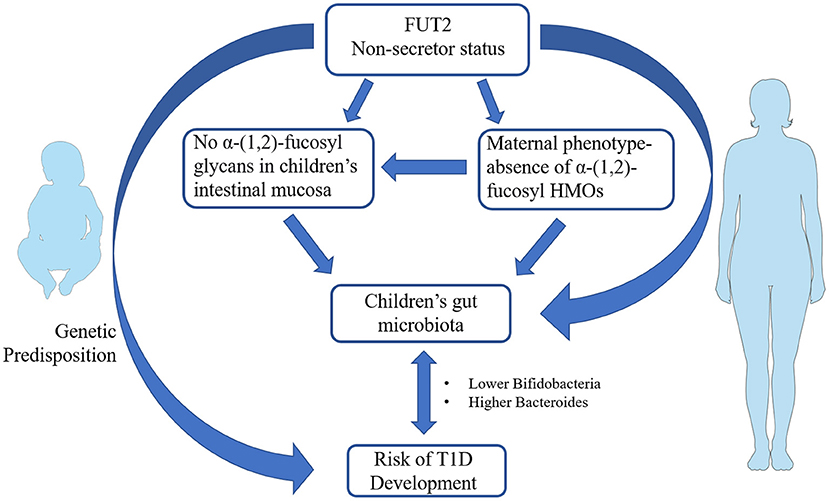
Figure 1. Schematic representation of the potential link between fucosyltransferase 2 (FUT2) non-secretor phenotype and infants' gut microbiota in the pathogenesis of type 1 diabetes (T1D). FUT2 genetic variants coding for non-secretor status has been associated with increased T1D susceptibility. Non-secretor status in mothers influence the composition of human milk oligosaccharides (HMOs) that in turn may prime the composition of newborns' gut microbiota. The occurrence of non-secretor status in infants modulates the glycan composition of intestinal mucosa and therein gut microbiota establishment. Non-secretor children and/or children breastfed by non-secretor mothers show a lower number of bifidobacteria and a higher content of Bacteroides in their gut. The same gut microbiota profiles are present in children with T1D. The figures were drawn using the freely available Servier Medical Art Resource (https://smart.servier.com/).
Through the modulation of infant mucosal glycans, HMOs may also affect intestinal cell maturation and barrier function (71). An increased intestinal permeability allows pathogens, toxins or food antigens to translocate from the intestinal lumen to extraintestinal compartments leading to intestinal disorders, allergies and diseases including T1D, IBD, irritable bowel syndrome, and metabolic syndrome (72, 73). HMOs produced by secretor mothers may stimulate the growth of specific bifidobacteria that in turn release peptides able to enhance the expression of tight-junction protein (74). Other milk-derived peptides have shown beneficial effects on intestinal barrier function, supporting the crucial role of lactation in promoting intestinal mucosal homeostasis (75).
Future investigations are needed to clearly establish the role of FUT2 gene variants on gut microbiota composition. Moreover, further studies should determine if HMOs bifidogenic properties may be exploited by non-secretor mothers or mothers who are unable to breastfeed to promote a healthier gut environment in newborns.
FUT2, T1D and Microbial Composition and Metabolites
The multifactorial etiopathogenesis of T1D poses a major challenge for the implementation of personalized therapeutic strategies (76). As for T1D genetics, several susceptibility genes have been identified that vary across genders, racial groups, and geographical regions (1). Main genetic risk factors are located within the class II HLA region (77). In a meta-analysis of GWASs combining British and American cases and controls, FUT2 was not present among newly identified T1D loci (78). Additional T1D susceptibility genes were identified by researchers from the Type 1 Diabetes Genetics Consortium through the application of an extensive GWAS on larger sample of T1D cases (79, 80). Most candidate loci that passed the stringent genome-wide significance levels pertained to the inflammatory cytokine network (79, 80). Interestingly, the ABO blood group locus, that may influence the glycosylation patterns of cells at gastrointestinal mucosal lining, was associated with positivity to antibodies against gastric parietal cells (PCA) in T1D patients (80). However, in the same study, when FUT2 SNP rs601338(A) was tested for a potential association with PCA only unconvincing evidence was showed (80).
Although those GWASs and sequencing studies did not include FUT2 out of over 60 loci associated with T1D susceptibility, FUT2 SNPs have been retrieved among the numerous gene polymorphisms outside HLA regions associated with T1D risk (76). Smyth et al. (13) provided evidence of a recessive association between FUT2 SNP rs601338(A) and T1D in European populations. Further association between FUT2 gene variants and T1D was found in the study conducted by Ihara et al. (81), that examined the contribution of the FUT2 gene and ABO blood type in Japanese children. The SNP rs1047781(T) coding for non-secretor phenotype was found to confer susceptibility to T1D (81). Intriguingly, children with HLA-conferred disease susceptibility that are homozygous for the functional FUT2 allele G (coding for secretor phenotype) are predisposed to rapid disease progression (82).
Both genetic and environmental factors are involved in T1D pathogenesis (76). In particular, host genetics and gut microbiota may affect T1D development through their influence on metabolic and immune phenotypes (51, 83). Gut microbiota and bacterial-derived components are critical to educate immune system function after birth (84) and may be involved in the progression from β-cell autoimmunity to overt T1D (85). In non-obese diabetic (NOD) mice, specific gut microbial features and products influence the pancreatic immune environment and confer protection against T1D (86–88). In humans, T1D status has been associated with an increase in Bacteroides and Bacteroidetes-to-Firmicutes ratio (67, 89–91), and with a dearth of Bifidobacterium species (67, 90). In particular, a reduction in butyrate-producing species was found in children with β-cell autoimmunity (92). In The Environmental Determinants of Diabetes in the Young (TEDDY) study, the gut microbiome of children with islet autoimmunity and T1D showed fewer genes related to short-chain fatty acids (SCFA) biosynthesis than that of healthy controls, although taxonomic differences were modest and not significant (6). Bacterial-derived SCFA, such as acetate, propionate and butyrate, which originate from the microbial fermentation of several dietary substrates, including HMOs. SCFA are then absorbed at the colon epithelium level and influence both local and systemic metabolic and immune responses, as well as improving the intestinal barrier functionality by facilitating tight junction assembly (93, 94). Lactic acid bacteria belonging to Bifidobacterium genus are capable of breaking HMOs and are transferred from mother to child during delivery. In particular B. infantis are efficient at metabolizing HMOs into SCFA. Interestingly, a reduced abundance of lactate-producing bacteria has been observed in T1D patients (95).
In NOD mice, a 6-week supplementation with a mixture of HMOs in early life protected against T1D development and halted severe pancreatic insulitis later in life (89). HMOs effects were associated with beneficial changes in fecal microbiota composition and activity (e.g., high content of SCFA in stool and cecum) and reduction in pro-inflammatory diabetogenic cytokines (89). Moreover, HMOs combined with SCFA may modulate dendritic cells and prime functional regulatory T cells in vitro toward a tolerogenic phenotype (89).
Collectively, these findings suggest that perturbations in the gut microbial function may alter host-microbial metabolism and lead to the development of a “leaky” gut followed by autoimmunity (96). HMOs in human milk may confer protection to children at risk for T1D by priming immune and gut microbiota development in early life.
Timely interventions on gut microbiota may represent a promising strategy to reduce the risk of T1D (97). However, data on the effects of probiotics administration in children with T1D are scarce. In a multinational cohort study of children at increased genetic risk of T1D, reduced pancreatic islet autoimmunity (PIA) was observed in those who received probiotics before the first month of age (98). Early probiotic administration was associated with a 60% reduction in PIA risk in children at the highest genetic risk of T1D (98).
Further studies are needed to examine the association between Bacteroides and bifidobacterial species in non-secretors and T1D to identify specific prebiotic and probiotic mixtures with preventive and therapeutic function.
Several metabolites derived by host-microbiome interaction may modulate immune response to pathogens through the activation of numerous receptors in the intestinal epithelial cells (99).
In this context, significant changes in gut microbial metabolic pathways have been found in children with T1D. An increase in the multiple sugar transport system, which is involved in the utilization of D-galactose, D-xylose, L-arabinose, D-glucose, and D-mannose, and a decrease in the biosynthesis of a number of amino acids, in particular Tyr and Phe, have been observed (100). Furthermore, serum metabolic profiling studies in individuals who developed T1D showed reduced levels of succinic acid, phosphatidylcholine and ketoleucine and increased levels of glutamic acid months before autoantibody positivity. Autoimmunity may thus represent a later manifestation of early-life alterations, including metabolic perturbations (101).
In a recent study, people with T1D and IBD were found to exhibit shared intestinal inflammatory patterns. Individuals with T1D showed higher fecal calprotectin levels and lower fecal intestinal alkaline phosphatase (IAP) activities associated with lower levels of butyrate and propionate, as compared with non-diabetic controls (102). It is known that IAP plays an important role in the interactions among diet, gut microbiota composition, and inflammatory responses in the gastrointestinal tract (103). IAP regulates lipid transport through intestinal mucosa and interferes with bacterial translocation across the intestinal epithelium, thereby preventing the consequent inflammatory response. The expression and secretion of IAP also depend on diet and intestinal microbiota (103). Serum IAP activity was found to be interconnected with ABO blood groups and FUT2 secretor/non-secretor phenotypes (104). People with the FUT2 secretor phenotype also tend to have higher serum IAP activity levels after a meal compared with non-secretors (105).
In conclusion, host genetic factors, gut microbiota activities, and host-gut co-metabolism may all contribute to the development of β-cell autoimmunity and T1D. FUT2 may represent a plausible player in this complex puzzle. Yet, conflicting results exist on the relationship between FUT2-associated secretor phenotype and gut microbiota composition and function. Further studies are needed to clearly establish whether FUT2 variants shape the gut microbiome and affect T1D development and progression.
Conclusions
The interplay between FUT2 variants and associated glycan profiles, gut microbiota composition and function, and host inflammatory response may play a role in T1D pathogenesis. Recent studies have spurred interest on the subject for its possible clinical implications (14). FUT2-associated phenotypes may be associated with T1D pathogenesis at different levels and timeframes and this may offer new possibilities for both primary prevention started early in infancy and secondary prevention when islet autoimmunity has ensued. Maternal FUT2 status should be assessed and beneficial bifidogenic properties of supplemented HMOs formula may be exploited in case of non-secretor phenotypes. A comprehensive and conclusive analysis on the effects of FUT2 gene variants on children gut microbiota composition, inflammatory status and β-cell autoimmunity may provide relevant information for T1D disease risk stratification and early interventions (e.g., tailored diets, specific prebiotics, probiotics or symbiotic) to preserve gut integrity and impede autoimmune response. Gut microbiota of T1D children with FUT2 susceptibility SNPs could be targeted through the supplementation with α-(1, 2)-fucosyl-oligosaccharides enriched formula milk or specific prebiotics and probiotics to stimulate the intestinal production of SCFA or IAP.
However, conclusive evidence relating FUT2 status and T1D has yet to be achieved and more research is required in this area.
Author Contributions
OG, GC, RC, and AM designed the study, made the description plan, performed manuscript writing and figure charting, and actively participated to the discussions. RC and AM made arrangements on the manuscript according their discussions and reviewed critically this article. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2020.606171/full#supplementary-material
References
1. DiMeglio LA, Evans-Molina C, Oram RA. Type 1 diabetes. Lancet. (2018) 391:2449–62. doi: 10.1016/S0140-6736(18)31320-5
2. Chobot A, Polanska J, Brandt A, Deja G, Glowinska-Olszewska B, Pilecki O, et al. Updated 24-year trend of Type 1 diabetes incidence in children in Poland reveals a sinusoidal pattern and sustained increase. Diabet Med. (2017) 34:1252–8. doi: 10.1111/dme.13345
3. Achenbach P, Lampasona V, Landherr U, Koczwara K, Krause S, Grallert H, et al. Autoantibodies to zinc transporter 8 and SLC30A8 genotype stratify type 1 diabetes risk. Diabetologia. (2009) 52:1881–8. doi: 10.1007/s00125-009-1438-0
4. Purcell AW, Sechi S, DiLorenzo TP. The evolving landscape of autoantigen discovery and characterization in type 1 diabetes. Diabetes. (2019) 68:879–86. doi: 10.2337/dbi18-0066
5. Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. (2016) 387:2340–8. doi: 10.1016/S0140-6736(16)30507-4
6. Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. (2018) 562:589–94. doi: 10.1038/s41586-018-0620-2
7. Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, et al. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS ONE. (2016) 11:e0158498. doi: 10.1371/journal.pone.0158498
8. Stewart CJ, Ajami NJ, O'Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. (2018) 562:583–588. doi: 10.1038/s41586-018-0617-x
9. Grigg JB, Sonnenberg GF. Host-microbiota interactions shape local and systemic inflammatory diseases. J Immunol. (2017) 198:564. doi: 10.4049/jimmunol.1601621
10. Praticò G, Capuani G, Tomassini A, Baldassarre ME, Delfini M, Miccheli A. Exploring human breast milk composition by NMR-based metabolomics. Nat Prod Res. (2014) 28:95–101. doi: 10.1080/14786419.2013.843180
11. Monedero V, Buesa J, Rodríguez-Díaz J. The interactions between host glycobiology, bacterial microbiota, and viruses in the gut. Viruses. (2018) 10:96. doi: 10.3390/v10020096
12. Donovan SM, Comstock SS. Human milk oligosaccharides influence neonatal mucosal and systemic immunity. Ann Nutr Metab. (2016) 69:42–51. doi: 10.1159/000452818
13. Smyth DJ, Cooper JD, Howson JMM, Clarke P, Downes K, Mistry T, et al. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. (2011) 60:3081–4. doi: 10.2337/db11-0638
14. Yang P, Li H-L, Wang C-Y. FUT2 nonfunctional variant: a “missing link” between genes and environment in type 1 diabetes? Diabetes. (2011) 60:2685–7. doi: 10.2337/db11-1104
15. Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, Blancher A, et al. A natural history of FUT2 polymorphism in humans. Mol Biol Evol. (2009) 26:1993–2003. doi: 10.1093/molbev/msp108
16. Soejima M, Pang H, Koda Y. Genetic variation of FUT2 in a Ghanaian population: identification of four novel mutations and inference of balancing selection. Ann Hematol. (2007) 86:199–204. doi: 10.1007/s00277-006-0203-4
17. Ye BD, Kim BM, Jung S, Lee H-S, Hong M, Kim K, et al. Association of FUT2 and ABO with Crohn's disease in Koreans. J Gastroenterol Hepatol. (2020) 35:104–9. doi: 10.1111/jgh.14766
18. King JR, Varadé J, Hammarström L. Fucosyltransferase gene polymorphisms and Lewisb-negative status are frequent in Swedish Newborns, with implications for infectious disease susceptibility and personalized medicine. J Pediatr Infect Dis Soc. (2018) 8:507–18. doi: 10.1093/jpids/piy085
19. Serpa J, Mendes N, Reis CA, Santos Silva LF, Almeida R, Le Pendu J, et al. Two new FUT2. (fucosyltransferase 2 gene) missense polymorphisms, 739G → A and 839T → C, are partly responsible for non-secretor status in a Caucasian population from Northern Portugal. Biochem J. (2004) 383:469–74. doi: 10.1042/BJ20040803
20. Soejima M, Koda Y. FUT2 polymorphism in Latin American populations. Clin Chim Acta. (2020) 505:1–5. doi: 10.1016/j.cca.2020.02.011
21. Goto Y, Uematsu S, Kiyono H. Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol. (2016) 17:1244–51. doi: 10.1038/ni.3587
22. Payne DC, Currier RL, Staat MA, Sahni LC, Selvarangan R, Halasa NB, et al. Epidemiologic association between FUT2 secretor status and severe rotavirus gastroenteritis in children in the United States. JAMA Pediatr. (2015) 169:1040. doi: 10.1001/jamapediatrics.2015.2002
23. Imbert-Marcille B-M, Barbé L, Dupé M, Le Moullac-Vaidye B, Besse B, Peltier C, et al. A FUT2 gene common polymorphism determines resistance to rotavirus A of the P[8] genotype. J Infect Dis. (2014) 209:1227–30. doi: 10.1093/infdis/jit655
24. Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, et al. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. (2012) 485:256–9. doi: 10.1038/nature10996
25. Boren T, Falk P, Roth K, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. (1993) 262:1892. doi: 10.1126/science.8018146
26. Rossez Y, Maes E, Lefebvre Darroman T, Gosset P, Ecobichon C, Joncquel Chevalier Curt M, et al. Almost all human gastric mucin O-glycans harbor blood group A, B or H antigens and are potential binding sites for Helicobacter pylori. Glycobiology. (2012) 22:1193–206. doi: 10.1093/glycob/cws072
27. Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. (2003) 9:548–53. doi: 10.1038/nm860
28. Matsui SM, Greenberg HB. Immunity to calicivirus infection. J Infect Dis. (2000) 181:S331–5. doi: 10.1086/315587
29. Kindberg E, Hejdeman B, Bratt G, Wahren B, Lindblom B, Hinkula J, et al. A nonsense mutation. (428G → A) in the fucosyltransferase FUT2 gene affects the progression of HIV-1 infection. AIDS. (2006) 20:685–9. doi: 10.1097/01.aids.0000216368.23325.bc
30. Thom SM, Blackwell CC, MacCallum CJ, Weir DM, Brettle RP, Kinane DF, et al. Non-secretion of blood group antigens and susceptibility to infection by Candida species. FEMS Microbiol Lett. (1989) 47:401–6. doi: 10.1111/j.1574-6968.1989.tb02428.x
31. Blackwell CC, Jónsdóttir K, Hanson M, Todd WTA, Chaudhuri AKR, Mathew B, et al. Non-secretion of ABO antigens predisposing to infection by Neisseria meningitidis and Streptococcus pneumoniae. Lancet. (1986) 328:284–5. doi: 10.1016/S0140-6736(86)92103-3
32. Azad M, Wade K, Timpson N. FUT2 secretor genotype and susceptibility to infections and chronic conditions in the ALSPAC cohort. Wellcome Open Res. (2018) 3:65. doi: 10.12688/wellcomeopenres.14636.2
33. Parmar AS, Alakulppi N, Paavola-Sakki P, Kurppa K, Halme L, Färkkilä M, et al. Association study of FUT2 (rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue Antigens. (2012) 80:488–93. doi: 10.1111/tan.12016
34. Rioux JD, Abbas AK. Paths to understanding the genetic basis of autoimmune disease. Nature. (2005) 435:584–9. doi: 10.1038/nature03723
35. Franke A, McGovern DPB, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. (2010) 42:1118–25. doi: 10.1038/ng.717
36. Xavier JM, Shahram F, Sousa I, Davatchi F, Matos M, Abdollahi BS, et al. FUT2: filling the gap between genes and environment in Behçet's disease? Ann Rheum Dis. (2015) 74:618. doi: 10.1136/annrheumdis-2013-204475
38. McGovern DPB, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. (2010) 19:3468–76. doi: 10.1093/hmg/ddq248
39. Forni D, Cleynen I, Ferrante M, Cassinotti A, Cagliani R, Ardizzone S, et al. ABO histo-blood group might modulate predisposition to Crohn's disease and affect disease behavior. J Crohns Colitis. (2014) 8:489–94. doi: 10.1016/j.crohns.2013.10.014
40. Rueedi R, Ledda M, Nicholls AW, Salek RM, Marques-Vidal P, Morya E, et al. Genome-wide association study of metabolic traits reveals novel gene-metabolite-disease links. PLoS Genet. (2014) 10:1–10. doi: 10.1371/journal.pgen.1004132
41. Wang W, Melinamani S, Alexander C, Bermea R, Sakuraba A, Dalal SR, et al. 696 FUT2 genotype is not associated with disease phenotype or outcomes in patients with Crohn's Disease. Am J Gastroenterol. (2019) 114:S407–9. doi: 10.14309/01.ajg.0000592320.06348.62
42. Aheman A. Association of fucosyltransferase 2 gene variants with ulcerative colitis in Han and Uyghur patients in China. World J Gastroenterol. (2012) 18:4758. doi: 10.3748/wjg.v18.i34.4758
43. van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. (2008) 9:593–601. doi: 10.1038/ni.f.203
44. Wacklin P, Mäkivuokko H, Alakulppi N, Nikkilä J, Tenkanen H, Räbinä J, et al. Secretor Genotype (FUT2 gene) Is Strongly Associated with the Composition of Bifidobacteria in the Human Intestine. PLoS ONE. (2011) 6:e20113. doi: 10.1371/journal.pone.0020113
45. Cabrera-Rubio R, Kunz C, Rudloff S, García-Mantrana I, Crehuá-Gaudiza E, Martínez-Costa C, et al. Association of maternal secretor status and human milk oligosaccharides with milk microbiota: an observational pilot study. J Pediatr Gastroenterol Nutr. (2019) 68:256–63. doi: 10.1097/MPG.0000000000002216
46. Sela DA. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol. (2011) 149:58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025
47. Kumbhare SV, Kumar H, Chowdhury SP, Dhotre DP, Endo A, Mättö J, et al. A cross-sectional comparative study of gut bacterial community of Indian and Finnish children. Sci Rep. (2017) 7:10555. doi: 10.1038/s41598-017-11215-y
48. Tong M, McHardy I, Ruegger P, Goudarzi M, Kashyap PC, Haritunians T, et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J. (2014) 8:2193–206. doi: 10.1038/ismej.2014.64
49. Turpin W, Bedrani L, Espin-Garcia O, Xu W, Silverberg MS, Smith MI, et al. FUT2 genotype and secretory status are not associated with fecal microbial composition and inferred function in healthy subjects. Gut Microbes. (2018) 9:357–68. doi: 10.1080/19490976.2018.1445956
50. Davenport ER, Goodrich JK, Bell JT, Spector TD, Ley RE, Clark AG. ABO antigen and secretor statuses are not associated with gut microbiota composition in 1,500 twins. BMC Genomics. (2016) 17:941. doi: 10.1186/s12864-016-3290-1
51. GEM Project Research Consortium, Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. (2016) 48:1413–7. doi: 10.1038/ng.3693
52. Wang J, Thingholm LB, Skiecevičiene J, Rausch P, Kummen M, Hov JR, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. (2016) 48:1396–406. doi: 10.1038/ng.3695
53. Newburg DS, Morelli L. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr Res. (2015) 77:115–20. doi: 10.1038/pr.2014.178
54. Newburg DS, Ko JS, Leone S, Nanthakumar NN. Human milk oligosaccharides and synthetic galactosyloligosaccharides contain 3′-, 4-, and 6′-Galactosyllactose and attenuate inflammation in human T84, NCM-460, and H4 cells and intestinal tissue ex vivo. J Nutr. (2016) 146:358–67. doi: 10.3945/jn.115.220749
55. Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr. (2014) 34:143–69. doi: 10.1146/annurev-nutr-071813-105721
56. Kunz C, Meyer C, Collado MC, Geiger L, García-Mantrana I, Bertua-Ríos B, et al. Influence of gestational age, secretor, and Lewis blood group status on the oligosaccharide content of human milk. J Pediatr Gastroenterol Nutr. (2017) 64:789–98. doi: 10.1097/MPG.0000000000001402
57. De Leoz MLA, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. (2015) 14:491–502. doi: 10.1021/pr500759e
58. Hong P, Ninonuevo MR, Lee B, Lebrilla C, Bode L. Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN). Br J Nutr. (2008) 101:482–86. doi: 10.1017/S0007114508025804
59. Huang P, Xia M, Tan M, Zhong W, Wei C, Wang L, et al. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J Virol. (2012) 86:4833. doi: 10.1128/JVI.05507-11
60. Yu Z-T, Chen C, Kling DE, Liu B, McCoy JM, Merighi M, et al. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology. (2013) 23:169–77. doi: 10.1093/glycob/cws138
61. Vazquez E, Barranco A, Ramirez M, Gruart A, Delgado-Garcia JM, Jimenez ML, et al. Dietary 2'-fucosyllactose enhances operant conditioning and long-term potentiation via gut-brain communication through the vagus nerve in rodents. PLoS ONE. (2016) 11:e0166070. doi: 10.1371/journal.pone.0166070
62. Good M, Sodhi CP, Yamaguchi Y, Jia H, Lu P, Fulton WB, et al. The human milk oligosaccharide 2′-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. Br J Nutr. (2016) 116:1175–87. doi: 10.1017/S0007114516002944
63. Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. (2003) 278:14112–20. doi: 10.1074/jbc.M207744200
64. Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, et al. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr. (2004) 145:297–303. doi: 10.1016/j.jpeds.2004.04.054
65. Newburg DS. Glycobiology of human milk. Biochem Mosc. (2013) 78:771–85. doi: 10.1134/S0006297913070092
66. Newburg DS, Ruiz-Palacios GM, Altaye M, Chaturvedi P, Guerrero ML, Meinzen-Derr JK, et al. Human milk alphal,2-linked fucosylated oligosaccharides decrease risk of diarrhea due to stable toxin of E. coli in breastfed infants. Adv Exp Med Biol. (2004) 554:457–61. doi: 10.1007/978-1-4757-4242-8_64
67. de Goffau MC, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. (2013) 62:1238. doi: 10.2337/db12-0526
68. EFSA Panel on Dietetic Products Nutrition and Allergies (NDA). Safety of 2′-O-fucosyllactose as a novel food ingredient pursuant to Regulation (EC) No 258/97. EFSA J. (2015) 13:4184. doi: 10.2903/j.efsa.2015.4184
69. Goehring KC, Marriage BJ, Oliver JS, Wilder JA, Barrett EG, Buck RH. Similar to those who are breastfed, infants fed a formula containing 2′-fucosyllactose have lower inflammatory cytokines in a randomized controlled trial. J Nutr. (2016) 146:2559–66. doi: 10.3945/jn.116.236919
70. Gampa A, Engen PA, Shobar R, Mutlu EA. Relationships between gastrointestinal microbiota and blood group antigens. Physiol Genomics. (2017) 49:473–83. doi: 10.1152/physiolgenomics.00043.2017
71. Sánchez de Medina F, Romero-Calvo I, Mascaraque C, Martínez-Augustin O. Intestinal inflammation and mucosal barrier function. Inflamm Bowel Dis. (2014) 20:2394–404. doi: 10.1097/MIB.0000000000000204
72. Bibbò S, Dore MP, Pes GM, Delitala G, Delitala AP. Is there a role for gut microbiota in type 1 diabetes pathogenesis? Ann Med. (2017) 49:11–22. doi: 10.1080/07853890.2016.1222449
73. Natividad JMM, Verdu EF. Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res. (2013) 69:42–51. doi: 10.1016/j.phrs.2012.10.007
74. Deja S, Barg E, Młynarz P, Basiak A, Willak-Janc E. 1H NMR-based metabolomics studies of urine reveal differences between type 1 diabetic patients with high and low HbAc1 values. J Pharm Biomed Anal. (2013) 83:43–8. doi: 10.1016/j.jpba.2013.04.017
75. Tanabe S. Short peptide modules for enhancing intestinal barrier function. Curr Pharm Des. (2012) 18:776–81. doi: 10.2174/138161212799277653
76. Ilonen J, Lempainen J, Veijola R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat Rev Endocrinol. (2019) 15:635–50. doi: 10.1038/s41574-019-0254-y
77. Robertson CC, Rich SS. Genetics of type 1 diabetes. Curr Opin Genet Dev. (2018) 50:7–16. doi: 10.1016/j.gde.2018.01.006
78. Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. (2008) 40:1399–401. doi: 10.1038/ng.249
79. Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. (2009) 41:703–7. doi: 10.1038/ng.381
80. Plagnol V, Howson JMM, Smyth DJ, Walker N, Hafler JP, Wallace C, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. (2011) 7:e1002216. doi: 10.1371/journal.pgen.1002216
81. Ihara K, Fukano C, Ayabe T, Fukami M, Ogata T, Kawamura T, et al. FUT2 non-secretor status is associated with Type 1 diabetes susceptibility in Japanese children. Diabet Med. (2017) 34:586–9. doi: 10.1111/dme.13288
82. Pöllänen PM, Lempainen J, Laine A-P, Toppari J, Veijola R, Vähäsalo P, et al. Characterisation of rapid progressors to type 1 diabetes among children with HLA-conferred disease susceptibility. Diabetologia. (2017) 60:1284–93. doi: 10.1007/s00125-017-4258-7
83. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. (2014) 159:789–99. doi: 10.1016/j.cell.2014.09.053
84. Zhao Q, Elson CO. Adaptive immune education by gut microbiota antigens. Immunology. (2018) 154:28–37. doi: 10.1111/imm.12896
85. Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. (2016) 12:154–67. doi: 10.1038/nrendo.2015.218
86. Xiao L, van't Land B, Engen PA, Naqib A, Green SJ, Nato A, et al. Human milk oligosaccharides protect against the development of autoimmune diabetes in NOD-mice. Sci Rep. (2018) 8:3829. doi: 10.1038/s41598-018-22052-y
87. Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. (2017) 18:552–62. doi: 10.1038/ni1117-1271c
88. Sun J, Furio L, Mecheri R, van der Does AM, Lundeberg E, Saveanu L, et al. Pancreatic β-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity. (2015) 43:304–17. doi: 10.1016/j.immuni.2015.07.013
89. Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. (2011) 5:82–91. doi: 10.1038/ismej.2010.92
90. Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. (2013) 11:46. doi: 10.1186/1741-7015-11-46
91. de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. (2014) 57:1569–77. doi: 10.1007/s00125-014-3274-0
92. Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE. (2011) 6:e25792. doi: 10.1371/journal.pone.0025792
93. Peng L, Li Z-R, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. (2009) 139:1619–25. doi: 10.3945/jn.109.104638
94. Han H, Li Y, Fang J, Liu G, Yin J, Li T, et al. Gut microbiota and type 1 diabetes. Int J Mol Sci. (2018) 19:995. doi: 10.3390/ijms19040995
95. Zheng P, Li Z, Zhou Z. Gut microbiome in type 1 diabetes: a comprehensive review. Diabetes Metab Res Rev. (2018) 34:e3043. doi: 10.1002/dmrr.3043
96. Davis-Richardson AG, Triplett EW. A model for the role of gut bacteria in the development of autoimmunity for type 1 diabetes. Diabetologia. (2015) 58:1386–93. doi: 10.1007/s00125-015-3614-8
97. Insel R, Knip M. Prospects for primary prevention of type 1 diabetes by restoring a disappearing microbe. Pediatr Diabetes. (2018) 19:1400–6. doi: 10.1111/pedi.12756
98. Uusitalo U, Liu X, Yang J, Aronsson CA, Hummel S, Butterworth M, et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr. (2016) 170:20–8. doi: 10.1001/jamapediatrics.2015.2757
99. Li Z, Quan G, Jiang X, Yang Y, Ding X, Zhang D, et al. Effects of metabolites derived from gut microbiota and hosts on pathogens. Front Cell Infect Microbiol. (2018) 8:314. doi: 10.3389/fcimb.2018.00314
100. Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A-M, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. (2015) 17:260–73. doi: 10.1016/j.chom.2015.01.001
101. Dedrick S, Sundaresh B, Huang Q, Brady C, Yoo T, Cronin C, et al. The role of gut microbiota and environmental factors in type 1 diabetes pathogenesis. Front Endocrinol. (2020) 11:78. doi: 10.3389/fendo.2020.00078
102. Lassenius MI, Fogarty CL, Blaut M, Haimila K, Riittinen L, Paju A, et al. Intestinal alkaline phosphatase at the crossroad of intestinal health and disease – a putative role in type 1 diabetes. J Intern Med. (2017) 281:586–600. doi: 10.1111/joim.12607
103. Estaki M. Interplay between intestinal alkaline phosphatase, diet, gut microbes and immunity. World J Gastroenterol. (2014) 20:15650. doi: 10.3748/wjg.v20.i42.15650
104. Matsushita M, Irino T, Stigbrand T, Nakajima T, Komoda T. Changes in intestinal alkaline phosphatase isoforms in healthy subjects bearing the blood group secretor and non-secretor. Clin Chim Acta. (1998) 277:13–24. doi: 10.1016/S0009-8981(98)00102-8
Keywords: FUT2 gene, T1D, gut microbiota, secretor, non-secretor, HMOs, bifidobacteria, short-chain fatty acids (SCFA)
Citation: Giampaoli O, Conta G, Calvani R and Miccheli A (2020) Can the FUT2 Non-secretor Phenotype Associated With Gut Microbiota Increase the Children Susceptibility for Type 1 Diabetes? A Mini Review. Front. Nutr. 7:606171. doi: 10.3389/fnut.2020.606171
Received: 14 September 2020; Accepted: 07 December 2020;
Published: 23 December 2020.
Edited by:
Annalisa Terranegra, Sidra Medicine, QatarReviewed by:
Jukka Partanen, Finnish Red Cross Blood Service, FinlandLaura Bordoni, University of Camerino, Italy
Copyright © 2020 Giampaoli, Conta, Calvani and Miccheli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alfredo Miccheli, YWxmcmVkby5taWNjaGVsaUB1bmlyb21hMS5pdA==
†These authors have contributed equally to this work
 Ottavia Giampaoli
Ottavia Giampaoli Giorgia Conta
Giorgia Conta Riccardo Calvani
Riccardo Calvani Alfredo Miccheli
Alfredo Miccheli