- 1Agricultural Biotechnology Research Institute, Ayub Agricultural Research Institute, Faisalabad, Pakistan
- 2State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
- 3Department of Plant Breeding and Genetics, University of Agriculture, Faisalabad, Pakistan
- 4College of Life Sciences, Yan'an University, Yan'an, China
Puccinia striiformis (Pst) is a devastating biotrophic fungal pathogen that causes wheat stripe rust. It usually loves cool and moist places and can cause 100% crop yield losses in a single field when ideal conditions for disease incidence prevails. Billions of dollars are lost due to fungicide application to reduce stripe rust damage worldwide. Pst is a macrocyclic, heteroecious fungus that requires primary (wheat or grasses) as well as secondary host (Berberis or Mahonia spp.) for completion of life cycle. In this review, we have summarized the knowledge about pathogen life cycle, genes responsible for stripe rust resistance, and susceptibility in wheat. In the end, we discussed the importance of conventional and modern breeding tools for the development of Pst-resistant wheat varieties. According to our findings, genetic engineering and genome editing are less explored tools for the development of Pst-resistant wheat varieties; hence, we highlighted the putative use of advanced genome-modifying tools, i.e., base editing and prime editing, for the development of Pst-resistant wheat.
Introduction
Yellow rust caused by Puccinia striiformis f. sp. tritici (Pst) is an economically important disease of wheat. It is also called stripe rust because of the appearance of yellow streaks (pre-pustules), followed by small, bright yellow, elongated uredial pustules arranged in conspicuous rows on the leaves. Stripe rust is an epidemic fungal disease of spring as well as winter wheat. It has been frequently reported from over 60 countries in all continents except Antarctica where wheat is not grown (1). The latest entry to the stripe rust epidemic countries list is Zimbabwe (2). However, the epidemic occurs more frequently (2 or 3 years of every 5 years) in Pakistan, India, China, Nepal, Yemen, Ethiopia, Uzbekistan, Kenya, Australia, the United Kingdom, the United States, Chile, New Zealand, Ecuador, Peru, Colombia, and Mexico (1) (Figure 1).
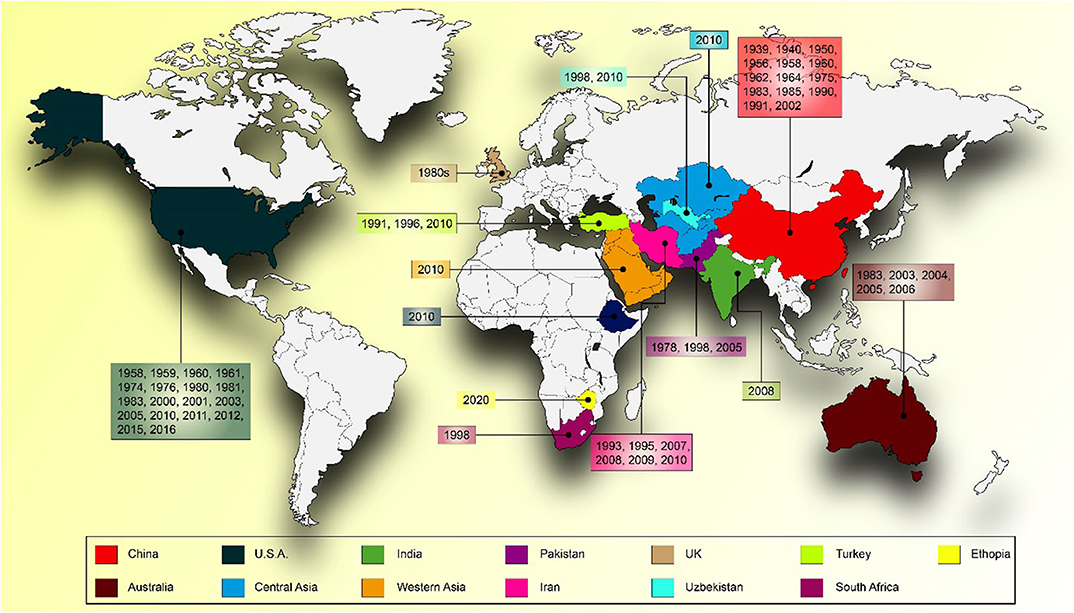
Figure 1. World map showing the epidemic years of stripe rust in different countries around the globe.
In the past couple of decades, Pst has devastated world wheat production, leaving 80% of wheat varieties susceptible to it. An annual loss of 5 million tons with an estimated cost of ~USD 1 billon is observed every year (3). So far, 51 major stripe rust epidemics have been observed globally from 1939 to 2016, varying in intensity from 2% yield losses to total crop failure (1). Up until now, more than 78 stripe rust resistance genes have been identified that are classified into phenotypically and mechanistically diverse classes, i.e., race-specific (seedling resistance genes) and race-non-specific [adult plant resistance (APR) genes]. Conventional breeding plays an essential role in crop improvement but usually entails growing and examining large populations of crops over multiple generations, a lengthy and labor-intensive process (4), whereas in modern era, gene hunting techniques and understanding about pathogen virulence pattern is increasing by introduction of modern genomics and breeding tools (5). Hence, we have summarized the latest developments in the field of genomics, genetic engineering, and genome editing in comparison to conventional breeding approaches and their role in the development of durable stripe rust-resistant wheat varieties.
Briefly, in this review, we have discussed the recent developments in genetics, genomics, and breeding of rust-resistant wheat varieties. Additionally, we have discussed the modern genomic tools, i.e., genetic engineering, RNAi, and genome editing, in comparison to conventional breeding and mutation breeding and their possible use in the development of Pst-resistant wheat varieties.
The Pst Pathogen
The Pst is an obligate fungal parasite that uses host plant photosynthetic machinery to fulfill its dietary requirements. Pst completes its life cycle in two different and unrelated host species, i.e., wheat and Berberis spp. (6). Three out of five different stages in the Pst life cycle, i.e., uredial, telial, and basidial phase, are completed on a primary host (wheat), whereas the pycnial and aecial phase needs an alternate host (1). Pst has a complex and specialized infection process including spore attachment to host plant, germination, formation of appressorium, and obtaining nutrients by haustorium formation and host invasion. Haustoria are surrounded by special “extrahaustorial membrane” and a gel-like matrix “extrahaustorial matrix” within the living host (7). Urediniospores usually grow when it rains freely with some moisture followed by a temperature of about 7–12°C minimum and 20–26°C maximum. With the passage of time, pathogens have adapted to high temperature, and severe outbreaks are occurring in subtropical areas as well. Pst as an airborne pathogen can travel thousands of miles to cause sudden disease epidemics (8).
Pst is evolving its race at rapid speed and is becoming a giant (9). However, race evolution and pathogenicity behavior are less explored. These unsolved issues provoked scientists and diverted their attention toward sequencing of the Pst genome to provide a deep insight into pathogenicity and host–pathogen interaction (10). To date, more than 15 Pst reference genomes are publicly available (11). Bioinformatics predictions have indicated that over 1,000 effector proteins exist in the Pst genome; however, their specific role during the infection process is yet to be explored. Advancements in the field of bioinformatics and genomics will help understand the pathogenicity process and race evolution of Pst (12). Draft of wheat genome is also available (13), which should also be explored for NBS-LRR and other gene families using bioinformatics tools to figure out putative Pst resistance genes for the development of durable Pst-resistant wheat cultivars (14).
Genetics and Genomics of Wheat to Combat Pst
Many efforts are directed toward isolating rust resistance (R) genes in crop plants and understanding how to best deploy them for durable resistance. R genes are divided into two mechanistically and phenotypically diverse classes, i.e., seedling and APR genes. R genes encode immuno-receptors in plants to recognize specific pathogen effector proteins and start immune response to stop activity initiated by pathogens through hypersensitive response (15). Effectors are an array of proteins secreted by pathogens to target plant signaling and block immunity or may give rise to effector-triggered immunity (16).
In contrast to R genes, there are also susceptibility (S) genes in plants that promote and facilitate the proliferation of any disease or pathogen attack. These genes negatively regulate the plant resistance against disease and usually include sugar transporters, i.e., SWEET gene family that transport sugar out of the cell and made available to pathogens (17). Similarly, some members of the NAC gene family are also used as target by the host for their multiplication (18). Thus, durable and broad-spectrum disease resistance to Pst can be acquired by manipulating the S gene/s, i.e., TaSTP13, via the gene modification technique such as the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein (Cas) system. However, for a better understanding of wheat genetics and genomics for combating Pst, both R and S genes have been discussed below.
R Genes
Seedling resistance, also known as qualitative, vertical, or all stage resistance (ASR), counters one or a few Pst races. Seedling resistance genes (SRGs) express at seedling as well as adult plant stages and show phenotypes of major influence (5). SRGs are characterized by a strong to moderate immune response that fully halts pathogen sporulation and disease development. Majority of the SRGs encode nucleotide-binding site leucine-rich repeat (NBS-LRR) R proteins that identify effector proteins inside cytoplasm secreted by host and initiate defense response to curtail the growth of pathogens. However, variety becomes susceptible after a few years of release because of the evolution of new virulent dominant races (12). SRGs are easy to incorporate in any breeding programs and provide high-level resistance to Pst but are not durable. Many historic epidemics were observed in the past due to breakdown of resistance of SRGs, i.e., Yr2, Yr9, Yr17, and Yr27 (1, 19). A novel SRG, YrSP, was recently reported, which is a truncated variant of Yr5. Yr5, Yr7, and YrSP all belong to gene cluster on 2B encoding nucleotide-binding and leucine-rich repeat proteins (NLRs) with a non-canonical N-terminal zinc-finger BED domain (20).
APR, also known as quantitative resistance, combats more than one race of the pathogen. APR is race-nonspecific and is a durable type of resistance due to its polygenic nature (21), with each gene having partial influence. Characteristically, APR genes are slow rusting, have a long latent period, and have less and small-sized uredinia formed in 14 days post-inoculation as compared to susceptible wheat plants (22). APR genes slow down the fungal life cycle with reduced sporulation, fungal population size reduction, and loss of genetic diversity. APR is molecularly independent of NBS-LRR proteins as observed in the case of Yr18 and Yr46 that encode transporters. Similarly, Yr36 activates chloroplast-localized kinase, which activates ROS. Currently, the combination of classical R genes with non-classical R genes seems to be an effective strategy to combat Pst (12).
High-temperature adult-plant (HTAP) resistance functions under high temperatures at adult stages and is governed by quantitative trait loci (QTLs). Adequate levels of resistance are obtained by three or more genes because individual APR genes provide low levels of resistance (12). The most commonly used APR genes are Yr9, Yr17, and Yr18 (23). Genome-wide association studies (GWAS) identified a new APR resistance gene, Yr78 (QYr.ucw-6B located on chromosome 6B) (24). Similarly, suppressive subtractive hybridization (SSH) is also a useful approach for the isolation of APR genes (25). Cloning of numerous wheat adult plant resistant genes has given insight into the mechanism of minor gene resistance. A gene, Yr36, codes for a chloroplast-localized protein with wheat kinase and START (WKS) lipid binding domains, projected to less reactive oxygen detoxification by phosphorylation of ascorbate peroxidase as the resulting resistance increases (26). The details of SRG and APRG genes identified so far are summarized in Table 1.
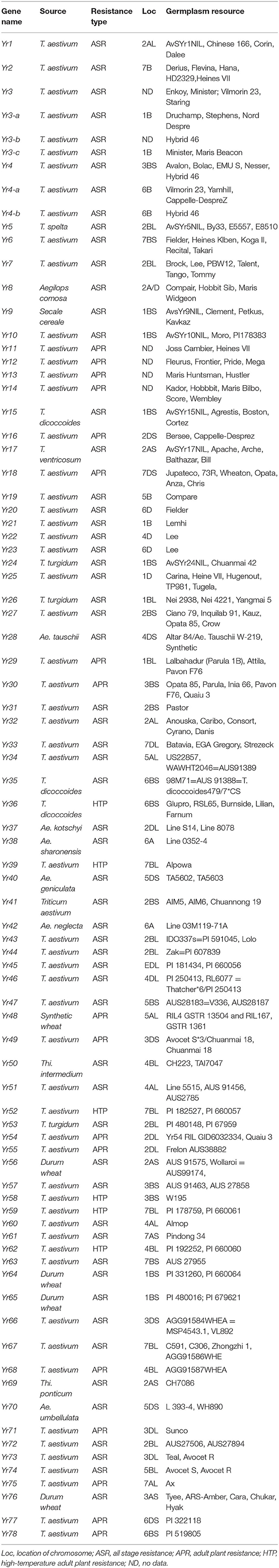
Table 1. List of seedling and adult plant yellow rust resistance and susceptibility genes with their source species, location on chromosomes, and germplasm resources.
Transcriptional factors (TFs) have gained significant importance during the past couple of decades due to their multiple stress responsive behavior. TFs contribute to Pst resistance by activation and repression of genes involved in various defense-associated metabolic pathways (27). Overexpression of TaWRKY62 provides high-temperature seedling-plant resistance to Pst. Interestingly, it was observed that switching on TaWRKY62 activated salicylic acid (SA)- and jasmonic acid (JA)-responsive genes TaPR1.1 and TaAOS, as well as ROS-associated genes TaCAT and TaPOD. On the other hand, ethylene (ET)-responsive gene TaPIE1 was downregulated (28). Similarly, TaLHY (a MYB TFs) reduces the negative impacts of the Pst infection by overexpressing in leaf blade and sheath (29).
TabZIP74 was activated in response to wounding due to Pst and starts resisting it. Silencing of TabZIP74 increases the susceptibility of wheat to Pst (30). TaNAC4 was overexpressed in response to Pst and by exogenously applied methyl Jasmonate (MeJA), ABA, and ethylene, indicating disease control through plant hormones (31). Above 50 TFs, families with thousands of genes have been reported in plants (http://planttfdb.cbi.pku.edu.cn) that play a direct and indirect role in stress tolerance and resistance. The role of majority of TFs is still unknown and needs further exploration.
S Genes
On the other hand, S genes help pathogens in spreading disease, so by disrupting those genes, plant health can be improved (Figure 2). For example, knocking out TaSTP13 by barley stripe mosaic virus-induced gene silencing (VIGS) reduced wheat susceptibility to Pst. However, its overexpression in Arabidopsis enhanced susceptibility to powdery mildew through increased glucose production in the leaves. These results indicated that TaSTP13 is transcriptionally induced and contributes to wheat susceptibility to Pst by promoting cytoplasmic hexose accumulation for fungal sugar acquisition in wheat–Pst interactions (32). It was observed that Pst initiated ABA biosynthesis in wheat cells and upregulated TaSTP6 expression, which increases sugar supply and promotes fungal infection (33). TaWRKY49 inhibits the expression of Pst-responsive genes, and its expression increases when Pst attacks wheat plants (28). Similarly, expression of TaNAC30 increases in wheat plants attacked by a virulent race (CYR31) of the Pst. Knocking out of the TaNAC30 by VIGS reduced colonization of the virulent Pst isolate CYR31. Detailed histological analyses indicated that TaNAC30 restricts the accumulation of H2O2 and promotes disease development (18).
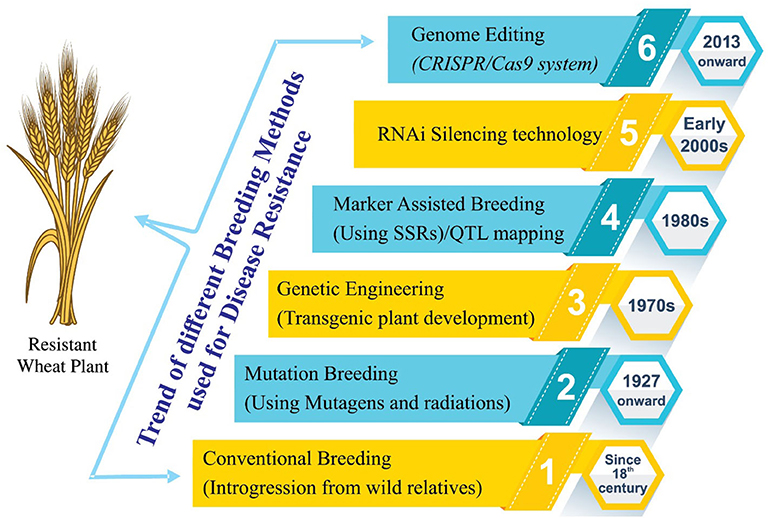
Figure 2. Historical view of different breeding techniques/procedures used for the development of stripe rust-resistant wheat cultivars. Before the advent of mutation breeding in the first quarter of the nineteenth century, conventional breeding approaches were used for resistance incorporation. Later on, advancement in the breeding approaches has brought us to the era of genome editing. Conventional breeding uses conservative breeding tools for the improvement of the trait of interest. Mutation breeding uses physical/chemical mutagens to introduce variation in a population followed by selection. Genetic engineering utilizes recombinant DNA technology for the alteration of the genetic makeup of plants and when the transfer of gene of interest is required from the distantly related organisms. Marker-assisted breeding uses DNA markers for selection of genes of interest and has the advantage of selecting the desirable plants using seedling or even seed sometimes. Genome editing is a way of making specific changes to the DNA of a cell or organism. An enzyme cuts the DNA at a specific sequence, and when this is repaired by the cell, a change or “edit” is made to the sequence.
Applications of Advanced Genomics Tools for Identification of Rust Resistance Genes
Advanced genomics tools such as GWAS and QTL-Seq are paving the way for fast-track genetic mapping of traits in various crops (34). GWAS is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait of interest (35), whereas QTL-Seq combines bulked segregant analysis (BSA) and high-throughput whole-genome re-sequencing to detect the major locus of a certain quantitative trait in a segregating population (36). GWAS was extensively used for searching of novel stripe rust resistance genes as described in the following examples. Two novel stripe rust resistance QTLs were identified using the GWAS approach on 5AS and 5AL wheat chromosomes (37). Similarly, many other GWAS-based studies have identified stripe rust resistance genes/QTLs (38–40). Keeping in view these highlighted examples for the use of GWAS and QTL-Seq in the identification of novel stripe rust resistance genes, both these approaches should be given primary importance in breeding programs focused on development of rust-resistant wheat varieties.
Breeding Strategies to Develop Pst-Resistant Wheat Varieties
A detailed breeding strategy has been proposed for effective control of stripe rust resistance in wheat. A pictorial history of the below-mentioned breeding methodologies is given in Figure 2.
Conventional Breeding
Conventional breeding approaches include, e.g., selection (pure line selection, mass selection), introduction, hybridization, wild hybridization, backcrossing, composite crossing, multi-line breeding, polyploidy, and heterosis breeding. Hybridization is followed by different breeding methods for the achievement of homozygosity in the filial generation such as pedigree method, bulk, single seed descent (SSD), ear to row, and many other methods that take 8–10 years for development and release of commercial cultivars for field testing as described briefly by Breseghello et al. (41). Phenotyping is done through visual observations under natural field conditions. Selection merely on phenotypic basis under natural infestation does not give satisfactory results and often leads toward selection of false-positive plants. Selection efficiency, however, can be improved by use of artificial inoculation. Still, this is a time-consuming and laborious task with underwhelming results. However, potential exists in wild hybridization. A detailed history of varieties developed through conventional breeding for Pst resistance is given in Table 1.
Marker-Assisted Breeding (MAB)
At the end of the twentieth century, MAB is increasingly used in different crop breeding programs with many advantages as compared to conventional breeding. Markers are selected on the basis of their linkage with gene of interest (GOI) (42, 43). MAB is frequently used for the development of disease-resistant varieties by probing the desirable markers (44). About 78 Pst resistance genes had been reported, many of which are race-specific in nature. A single resistant gene may become rapidly susceptible to new fungal races, causing breakdown of resistance; hence, gene pyramiding through MAB is a rational approach (5, 8). Stacking of multiple APR Pst resistance genes through MAB will result in durable stripe rust-resistant wheat genotypes (45). DNA markers are reported for many Pst resistance genes, i.e., Yr5, Yr9, Yr15, Yr17, Yr26, Yr33, and YrH52, as available on MAS wheat (https://maswheat.ucdavis.edu/) along with their complete protocol for MAB. SSR markers are widely used for Pst resistance gene pyramiding due to its highly polymorphic and co-dominant nature (46). Likewise, SNP markers using gene-chip technology provides a better-quality approach regarding Pst-resistant QTL mapping (47).
Mutation Breeding
Mutation increases biodiversity and also helps conventional plant breeding through the creation of novel variants that do not occur in nature (48). Continuous decline in genetic diversity has paved the way toward mutation breeding (49). Physical (irradiation) and chemical (alkylating agents, nitrous acid, base analogs, etc.) mutagens are used for the induction of mutation in plants. The molecular techniques of DNA fingerprinting, i.e., Random Amplified Polymorphic DNA (RAPD), Amplified Fragment Length Polymorphisms (AFLP), and Sequence-Tagged Microsatellite Sites (STMs), have helped in screening and analysis of desired mutants (50). Mutation breeding through physical or chemical means helps in the development of new varieties with exploitation of more genetic variability. In the present era, mutation breeding is important for modern plant breeding, recombination breeding, and transgenic breeding side by side as detailed in the following examples.
Chemical mutation was induced in NN-Gandum-1, a wheat variety with 0.55% absolute resistance to yellow rust. This mutagenesis resulted in substitution of glutamic acid with alanine through SNP, which helped to alter protein structure. The maximum number of SNPs was found on chromosome 2B, and the minimum was found on chromosome 7D (51). A wheat mutant R39 showed APR to stripe rust, which was identified using specific length amplified fragment (SLAF) sequencing combined with bulk segregant analysis (BSA) and next-generation sequencing (NGS) (52). Gamma rays and electron beam (EBM) irradiation were used in two varieties, PBW343 and HD2967. Doses used were 250, 300, and 350 Gy and 150, 200, and 250 Gy of gamma rays and EBM, respectively. The EBM-derived population showed more mutants compared to the gamma ray population. Absence of sporulation indicates that resistance has been incorporated through mutation breeding (53).
Genetic Engineering
Genetic engineering is a novel plant breeding tool that involves production of transgenic plants by transformation of GOI in order to obtain desired results (54). Genetic engineering has wider applications for development of biotic and abiotic stress-tolerant crop plants and improving their nutritional quality (55). Genetic engineering has played a significant role in the development of fungal-resistant plants by producing pathogenesis-related protein, i.e., chitinase and β-1,3-glucanase. When fungal pathogens attack GM plants, chitinase is produced in large amounts to reduce the hyphal growth (56) as chitinase hydrolyzes fungal cell wall (57). Rice class I chitinase OsRC24 gene was transformed to genetic background of wheat for the development of Pst-resistant wheat plants. The OsRC24 possessing lines exhibited 27–36% more yield (58).
Similarly, transgenesis of barley chi26 gene in bread wheat by using biolistic bombardment was also effective in resisting Pst (57). Transformation of another gene, EuCHIT1, from Eucommia ulmoides in wheat had shown enhanced resistance to Pst (59). YrU1 was transformed to wheat from its progenitor Triticum urartu and transformed lines showed enhanced resistance against Pst (60). Sometimes, gene transcription becomes silent in transgenic plant families, which results in very limited or no expression (57). However, overall results of transgenic technology have shown good promise against rust infestation in wheat. In the future, genetic transformation could be used as a rapid source to obtain Pst wheat cultivars and minimize yield losses.
RNAi Silencing
RNA interference (RNAi) is a conserved mechanism in eukaryotic organisms with a very crucial role in defense mechanism against pathogenic infections and gene regulation (61). RNAi alters the gene function or silencing of crucial pathogenic genes of the pathogen. In order to silence any gene, RNAi uses double-stranded RNA (dsRNA), which is homologous to GOI (62). The natural pathway for RNAi silencing is cleaving double-stranded RNA (dsRNA) into 21- to 26-nucleotide-long small RNA (sRNA), which are short interfering RNAs (siRNAs) or microRNAs (miRNAs). These sRNAs are involved in various processes, i.e., maintaining RNA stability, processing, and response to various biotic stresses (63).
Pathogenesis-related genes in Pst were silenced using Barley stripe mosaic virus (BSMV) as a vector for expression of dsRNA homologous to the Pst target gene. The MAPK kinase PsFUZ7 gene, which is an important pathogenicity factor of Pst causing fungal infection and regulating hyphal morphology and development on host plant, was knocked down using RNAi. The RNAi constructs targeting the PsFUZ7 gene of Pst was successfully expressed in transgenic wheat lines and conferred strong and durable resistance (62). Similarly, the PsCPK1 gene (an important transcript of Pst expressed at early infection stage) was knocked down in transgenic wheat lines using RNAi (61). Other successful knockouts of Pst using transgenic wheat lines are PsHXT1 (64) and PstGSRE1 (65) genes, which are a Hexose transporter and an effector protein, respectively. Both these genes are required for pathogenicity. RNAi is an emerging genetic approach with great potential against Pst.
Genome Editing
Plant genome editing uses sequence-specific nucleases (SSNs) for stably inherited and predetermined gene modification by introducing favorable alleles into our crop of interest that results in a transgene-free desired genome (66). SSNs cause certain changes at the chromosomal level, which results in deletion, insertion, or substitution of specific nucleotide sequence at particular loci. Various types of SSNs, i.e., Zinc finger nucleases (ZFNs) and Transcription activator-like effector nucleases (TALENs), and the CRISPR-Cas system are being used for plant genome editing (67). Targeted genome editing has become the preferred genetic tool for resistance incorporation in plants against various pathogenic diseases (68). Susceptible genes of crop plant are edited and rewritten in such a way to transform them to resistant genes (69).
Nowadays, the CRISPR-Cas9 system, along with its variants, has more applications over other genome editing tools because it is easy to operate, time efficient, and cheap, and has a high success rate (70). For the sake of it, short guided RNA (sgRNA) are designed with which Cas9, an RNA-guided DNA endonuclease, makes a complex for exact targeting of a specific gene (71). In wheat, CRISPR-Cas9 has successfully shown resistance against powdery mildew by making TaEDR1 mutant wheat plants by simultaneous modification of TaEDR1 along with its three homologs (72). Similarly, CRISPR-Cas9 was also used for making a resistant, non-transgenic tomato variety against fungal mildew through deletion (67).
Another highlighted example of the CRISPR-Cas9 system is the successful editing of three targeted genes, i.e., TaABCC6, TaNFXL1, and TansLTP9, in the wheat protoplast system for activation of defense mechanism against Fusarium head blight (FHB) (73). The potential role of CRISPR-Cas9 in developing Pst-resistant plants is yet to be explored. Although different groups are working on the use of CRISPR-Cas9 for developing yellow rust resistance plants, no report has been published yet. However, a great variety of potentials exist for its use. This innovative technique could be used for stripe rust resistance in wheat by predetermined modification of defense genes that will be different from transgenic for increasing wheat production in a time-efficient manner. CRISPR-Cas09 figure in the article after this portion and re-labeled it as Figure 3.
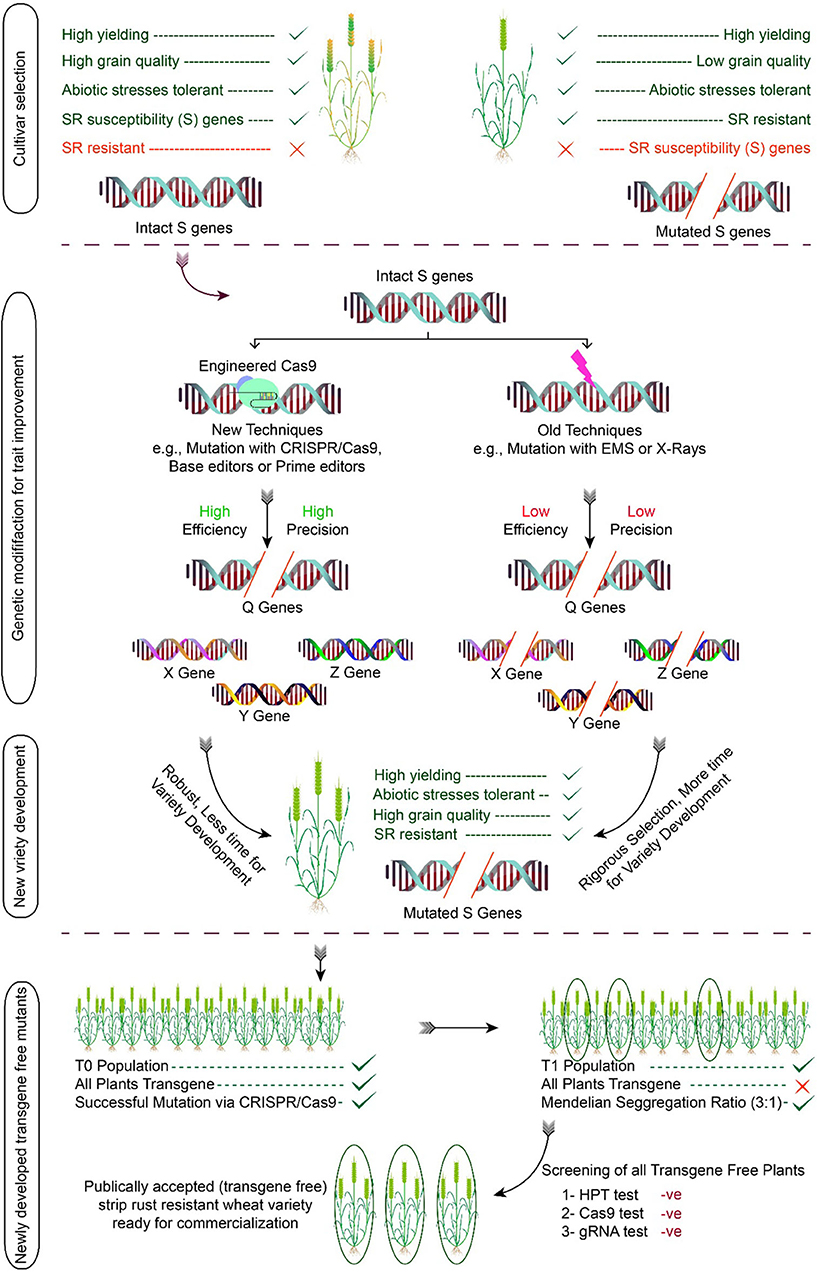
Figure 3. Development of stripe rust-resistant wheat variety using the CRISPR/Cas system. The process begins by selecting the cultivars having the S gene to be modified followed by trait improvement through genome editing. The modified plants are used in the breeding program for the development of variety, and in later generation, transgene-free plants are selected. The illustration also discusses the pros and cons of conventional breeding vs. genome editing. The efficiency of conventional breeding techniques is low as compared to genome editing. Similarly, the off-targeting effects of conventional mutations tools are relatively high in comparison to genome editing.
Speed Breeding
Speed breeding is a novel plant breeding technique that shortens the harvest time of crops by using optimal light intensity, light quality, day length, and controlled temperature for accelerated photosynthesis, flowering, early seed harvest, and shortened generation time. The technology can be used to obtain up to six generations per year of spring and durum wheat, barley, peas, and chickpea and up to four generations of canola under normal greenhouse conditions (74). The technology accelerates plant development in fully enclosed, controlled-environment growth chambers. The light supplementation through LEDs in a glasshouse environment allows rapid generation cycling through SSD and the potential for adaptation to larger-scale crop improvement programs. Unlike double haploid technology, speed breeding does not require specialized labs and can be used for diverse germplasm. The innovations in the LEDs and extended photoperiods coupled with early seed harvest technology have further reduced the generation period (75). Speed breeding was widely exploited in wheat for aboveground (plant height, tolerance to crown rot, and resistance to leaf rust) and belowground (seminal root angle and seminal root number) trait improvement (76, 77). Similarly, it was applied in barley for leaf rust, net blotch, and spot blotch resistance incorporation through modified backcross strategy using multi-trait phenotypic screens (78). These examples highlighted that potential exists for the development of yellow rust-resistant wheat varieties in a shorter time span.
Conclusion and Future Prospects
The frequent outbreak of Pst epidemics and worldwide distribution highlighted the need for an effective control strategy. The pattern of evolution of Pst, population structures, migration routes, and reproduction modes are well-known. The advent of genome editing and RNAi silencing technologies has provided a great platform for plant breeders and pathologists to develop resistance varieties. Although breeding resistant cultivars should remain the primary focus, some alternative measures, i.e., integrating fungicides, shifting planting date, changing crop nutrition pattern, eradicating volunteer plants, cultivar mixing, and intercropping, may be adopted as a temporary solution. Side-by-side development and distribution of resistant wheat cultivars to provide cost-effective and environment-friendly solution should be continued. Efforts of breeders and pathologists have borne fruit, and to date, ~78 Yr genes have been identified, some of which are seedlings and others are APR genes (8). Marker systems have been optimized for efficient identification of stripe rust resistance genes, which provide the opportunity for pyramiding of multiple Pst resistance genes in a cultivar for durable resistance.
Focus should be diverted toward identification of more Pst resistance genes from wild relatives. More than 2,500 members of the NBS-LRR gene family have been reported in wheat, 570 in Triticum urata, 842 in Aegilops tauschii, 316 in Brachypodium distachyon, and 420 in Hordeum vulgare (79), and majority of these have potential for disease resistance (80). Use of bioinformatics tools will help to understand the functions of the NBS-LRR gene family for hunting putative Pst resistance genes. Sequencing of the Pst genome provides the opportunity for functional annotation of genes involved in biology and pathogenicity of Pst. This has opened new horizons and provided opportunities to understand virulence variation in pathogens and the mechanism of resistance in hosts. In silico-based studies should be devised to identify genes responsible for the pathogenicity of Pst and mutations responsible for the development of new races. Simultaneous surveying of wheat genome for durable resistance genes from NBS-LRR or other gene families, i.e., TaLTPs (80), needs to be taken into consideration.
The future of plant breeding efforts for durable resistance against Pst should not be limited to conventional breeding approaches; instead, novel knowledge-generating tools, i.e., bioinformatics (81), phenomics (82), advanced genomics technologies (83), and novel genome modification techniques, i.e., CRISPR and RNAi (68), should be combined to evolve durable resistance wheat varieties. Similarly, to shorten the breeding time span, double haploid breeding, shuttle breeding, and speed breeding may be incorporated (74, 75). Currently, there are four classes of CRISPR-Cas-derived genome editing agents, including nucleases, base editors, transposases, and prime editors (84). The latest breakthrough in genome editing, i.e., base editing (the irreversible conversion of a base at the target site without involving donor templates, double-stranded breaks, and dependency on NHEJ and HDR), prime editing [the introduction of indels and all 12 base-to-base conversions without inducing a DNA double-strand break using prime editing guide RNA (pegRNA) that drives the Cas9 endonuclease], and genome editing using rice zygote (which overcomes the problem in delivery of macromolecule to the host cells and tissues and difficulty in transformation and regeneration), has opened new horizons for biotechnologists and plant pathologists (84). In this regard, the identification of new S genes is highly needed. In the future, researchers should focus on the identification of new genes that facilitate the proliferation of strip rust pathogen. Later, those genes can be used to improve plant health against strip rust by using genome-editing tools. Briefly, these cutting-edge advancements have paved the way for the fast-track development of stripe rust-resistant wheat varieties.
Author Contributions
SJ and RS conceived the idea. RS, RF, RZ, and MA drafted the manuscript. SA prepared illustrations. XW provided the literature and technical assistance. SJ, RS, SA, MZI, and XW reviewed and improved the draft. All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
The publication of the present work was supported by the National Key Research and Development Program of China (Grant No. 2017YFC0504704) and the National Natural Science Foundation of China (51669034, 41761068, and 51809224).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are highly thankful to colleagues at Maize and Millets Research Institute (Yusafwala, Sahiwal) and Ayub Agricultural Research Institute (Faisalabad) for guidance and moral support and to Mr. Baber Ali, lab assistant, for technical support. Apologies to the authors whose work has not been cited due to space limitations.
References
1. Chen X. Pathogens which threaten food security: Puccinia striiformis, the wheat stripe rust pathogen. Food Sec. (2020) 12:239–51. doi: 10.1007/s12571-020-01016-z
2. Boshoff W, Visser B, Lewis C, Adams T, Saunders D, Terefe T, et al. First report of Puccinia striiformis f. sp tritici, causing stripe rust of wheat. Zimbabwe Plant Dis. (2020) 104:290. doi: 10.1094/PDIS-07-19-1395-PDN
3. Beddow JM, Pardey PG, Chai Y, Hurley TM, Kriticos DJ, Braun H-J, et al. Research investment implications of shifts in the global geography of wheat stripe rust. Nat Plants. (2015) 1:1–5. doi: 10.1038/nplants.2015.132
4. Ellis JG, Lagudah ES, Spielmeyer W, Dodds PN. The past, present and future of breeding rust resistant wheat. Front Plant Sci. (2014) 5:641. doi: 10.3389/fpls.2014.00641
5. Chen W, Wellings C, Chen X, Kang Z, Liu T. Wheat stripe (yellow) rust caused by P uccinia striiformis f. sp tritici. Mol Plant Pathol. (2014) 15:433–46. doi: 10.1111/mpp.12116
6. Zhao J, Wang M, Chen X, Kang Z. Role of alternate hosts in epidemiology and pathogen variation of cereal rusts. Ann Rev Phytopathol. (2016) 54:207–28. doi: 10.1146/annurev-phyto-080615-095851
7. Garnica DP, Nemri A, Upadhyaya NM, Rathjen JP, Dodds PN. The ins and outs of rust haustoria. PLoS Pathog. (2014) 10:e1004329. doi: 10.1371/journal.ppat.1004329
8. Wang M, Chen X. Stripe rust resistance. In: Chen X and Kang Z, editors. Stripe Rust. Dordrecht: Springer (2017). p. 353–558.
9. Liu T, Wan A, Liu D, Chen X. Changes of races and virulence genes in Puccinia striiformis f. sp. tritici, the wheat stripe rust pathogen, in the United States from 1968 to 2009. Plant Dis. (2017) 101:1522–32. doi: 10.1094/PDIS-12-16-1786-RE
10. Li Y, Xia C, Wang M, Yin C, Chen X. Whole-genome sequencing of Puccinia striiformis f. sp tritici mutant isolates identifies avirulence gene candidates. BMC Genom. (2020) 21:1–22. doi: 10.1186/s12864-020-6677-y
11. Li Y, Xia C, Wang M, Yin C, Chen X. Genome sequence resource of a Puccinia striiformis isolate infecting wheatgrass. Phytopathology. (2019) 109:1509–12. doi: 10.1094/PHYTO-02-19-0054-A
12. Schwessinger B. Fundamental wheat stripe rust research in the 21st century. New Phytologist. (2017) 213:1625–31. doi: 10.1111/nph.14159
13. Eversole K, Rogers J, Caugant I. The international wheat genome sequencing consortium In: Plant and Animal Genome XXVI Conference (January 13-17, 2018): PAG. San Diego (2020).
14. Wang Z, Ren J, Du Z, Che M, Zhang Y, Quan W, et al. Identification of a major QTL on chromosome arm 2AL for reducing yellow rust severity from a Chinese wheat landrace with evidence for durable resistance. Theore Appl Genet. (2019) 132:457–71. doi: 10.1007/s00122-018-3232-1
15. Dobon A, Bunting DC, Cabrera-Quio LE, Uauy C, Saunders DG. The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genom. (2016) 17:380. doi: 10.1186/s12864-016-2684-4
16. Cantu D, Segovia V, Maclean D, Bayles R, Chen X, Kamoun S, et al. Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp triticireveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genom. (2013) 14:270. doi: 10.1186/1471-2164-14-270
17. Dong OX, Ronald PC. Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol. (2019). 180:26–38. doi: 10.1104/pp.18.01224
18. Wang B, Wei J, Song N, Wang N, Zhao J, Kang Z. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J Integr Plant Biol. (2018) 60:432–43. doi: 10.1111/jipb.12627
19. Bux H, Ashraf M, Chen X. Expression of high-temperature adult-plant (HTAP) resistance against stripe rust (Puccinia striiformis f. sp tritici) in Pakistan wheat landraces. Can J Plant Pathol. (2012) 34:68–74. doi: 10.1080/07060661.2012.662998
20. Marchal C, Zhang J, Zhang P, Fenwick P, Steuernagel B, Adamski NM, et al. BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants. (2018) 4:662–8. doi: 10.1038/s41477-018-0236-4
21. Saleem K, Arshad HMI, Shokat S, Atta BM. Appraisal of wheat germplasm for adult plant resistance against stripe rust. J Plant Protect Res. (2015) 55:405–14. doi: 10.1515/jppr-2015-0055
22. Chen X, Line R. Gene action in wheat cultivars for durable, high-temperature, adult-plant resistance and interaction with race-specific, seedling resistance to Puccinia striiformis. Phytopathology. (1995) 85:567–57. doi: 10.1094/Phyto-85-567
23. Zeng Q-D, Han D-J, Wang Q-L, Yuan F-P, Wu J-H, Zhang L, et al. Stripe rust resistance and genes in Chinese wheat cultivars and breeding lines. Euphytica. (2014) 196:271–84. doi: 10.1007/s10681-013-1030-z
24. Dong Z, Hegarty JM, Zhang J, Zhang W, Chao S, Chen X, et al. Validation and characterization of a QTL for adult plant resistance to stripe rust on wheat chromosome arm 6BS (Yr78). Theor Appl Genet. (2017) 130:2127–37. doi: 10.1007/s00122-017-2946-9
25. Huang X-L, Ma J-B, Chen X, Wang X-J, Ding K, Han D-J, et al. Genes involved in adult plant resistance to stripe rust in wheat cultivar Xingzi 9104. Physiol Mol Plant Pathol. (2013) 81:26–32. doi: 10.1016/j.pmpp.2012.10.004
26. Periyannan S, Milne RJ, Figueroa M, Lagudah ES, Dodds PN. An overview of genetic rust resistance: from broad to specific mechanisms. PLoS Pathog. (2017) 13:e1006380. doi: 10.1371/journal.ppat.1006380
27. Baillo EH, Kimotho RN, Zhang Z, Xu P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes. (2019) 10:771. doi: 10.3390/genes10100771
28. Wan A, Wang X, Kang Z, Chen X. Variability of the stripe rust pathogen. In: Chen X and Kang, editors. Stripe Rust. Dordrecht: Springer (2017). p. 35−154.
29. Zhang Z, Chen J, Su Y, Liu H, Chen Y, Luo P, et al. TaLHY, a 1R-MYB transcription factor, plays an important role in disease resistance against stripe rust fungus and ear heading in wheat. PLoS ONE. (2015) 10:e0127723. doi: 10.1371/journal.pone.0127723
30. Wang F, Lin R, Li Y, Wang P, Feng J, Chen W, et al. TabZIP74 acts as a positive regulator in wheat stripe rust resistance and involves root development by mRNA splicing. Front Plant Sci. (2019) 10:1551. doi: 10.3389/fpls.2019.01551
31. Xia N, Zhang G, Liu X-Y, Deng L, Cai G-L, Zhang Y, et al. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol Biol Rep. (2010) 37:3703–12. doi: 10.1007/s11033-010-0023-4
32. Huai B, Yang Q, Wei X, Pan Q, Kang Z, Liu J. TaSTP13 contributes to wheat susceptibility to stripe rust possibly by increasing cytoplasmic hexose concentration. BMC Plant Biol. (2020) 20:49. doi: 10.1186/s12870-020-2248-2
33. Huai B, Yang Q, Qian Y, Qian W, Kang Z, Liu J. ABA-induced sugar transporter tastp6 promotes wheat susceptibility to stripe rust. Plant Physiol. (2019) 181:1328–43. doi: 10.1104/pp.19.00632
34. Arora S, Steuernagel B, Gaurav K, Chandramohan S, Long Y, Matny O, et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol. (2019) 37:139–43. doi: 10.1038/s41587-018-0007-9
35. Srivastava RK, Singh RB, Srikanth B, Satyavathi CT, Yadav R, Gupta R. Genome-wide association studies (GWAS) and genomic selection (GS) in pearl millet: advances and prospects. Front Genet. (2019) 10:1389. doi: 10.3389/fgene.2019.01389
36. Zhang X, Zhang K, Wu J, Guo N, Liang J, Wang X, et al. QTL-Seq and sequence assembly rapidly mapped the gene BrMYBL2. 1 for the purple trait in brassica rapa. Sci Rep. (2020) 10:1–9. doi: 10.1038/s41598-020-58916-5
37. Ye X, Li J, Cheng Y, Yao F, Long L, Yu C, et al. Genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp tritici) in Sichuan wheat. BMC Plant Biol. (2019) 19:1–15. doi: 10.1186/s12870-019-1764-4
38. Singh S, Ledesma-Ramírez L, Solís-Moya E, Iturriaga G, Sehgal D, Reyes-Valdes MH, et al. GWAS to identify genetic loci for resistance to yellow rust in wheat pre-breeding lines derived from diverse exotic crosses. Front Plant Sci. (2019) 10:1390. doi: 10.3389/fpls.2019.01390
39. Cheng B, Gao X, Cao N, Ding Y, Gao Y, Chen T, et al. Genome-wide association analysis of stripe rust resistance loci in wheat accessions from southwestern China. J Appl Genet. (2020) 61:37–50. doi: 10.1007/s13353-019-00533-8
40. Kumar D, Kumar A, Chhokar V, Gangwar OP, Bhardwaj SC, Sivasamy M, et al. Genome-wide association studies in diverse spring wheat panel for stripe, stem and leaf rust resistance. Front Plant Sci. (2020) 11:748. doi: 10.3389/fpls.2020.00748
41. Breseghello F, Coelho ASG. Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). J Agric Food Chem. (2013) 61:8277–86. doi: 10.1021/jf305531j
42. Osei MK, Prempeh R, Adjebeng-Danquah J, Opoku JA, Danquah A, Danquah E, et al. Marker-assisted selection (MAS): a fast-track tool in tomato breeding. In: Nyaku ST and Danquah A, editors. Recent Advances in Tomato Breeding and Production. London: IntechOpen (2018).
43. Agrawal N, Tripathi R, Jain M. Molecular marker tools for breeding program in crops. In: Andersen SB, editor. Plant Ecophysiology Adaptation Under Climate Change: Mechanisms Perspectives II. London: Springer (2020). p. 567–82.
44. Toth J, Pandurangan S, Burt A, Mitchell Fetch J, Kumar S. Marker-assisted breeding of hexaploid spring wheat in the Canadian prairies. Can J Plant Sci. (2018) 99:111–27. doi: 10.1139/cjps-2018-0183
45. Waqar A, Khattak SH, Begum S, Rehman T, Shehzad A, Ajmal W, et al. Stripe rust: a review of the disease, yr genes and its molecular markers. Sarhad J Agric. (2018) 34:188–201. doi: 10.17582/journal.sja/2018/34.1.188.201
46. Zhang P, Li X, Gebrewahid T-W, Liu H, Xia X, He Z, et al. QTL mapping of adult-plant resistance to leaf and stripe rust in wheat cross SW 8588/Thatcher using the wheat 55K SNP array. Plant Dis. (2019) 103:3041–9. doi: 10.1094/PDIS-02-19-0380-RE
47. Lu Y, Lan C, Liang S, Zhou X, Liu D, Zhou G, et al. QTL mapping for adult-plant resistance to stripe rust in italian common wheat cultivars libellula and strampelli. Theor Appl Genet. (2009) 119:1349–59. doi: 10.1007/s00122-009-1139-6
48. Amin R, Laskar RA, Khan S. Assessment of genetic response and character association for yield and yield components in Lentil (Lens culinaris L.) population developed through chemical mutagenesis. Cogent Food Agric. (2015) 1:1000715. doi: 10.1080/23311932.2014.1000715
49. Raina A, Laskar RA, Khursheed S, Amin R, Tantray YR, Parveen K, et al. Role of mutation breeding in crop improvement-past, present and future. Asian Res J Agric. (2016) 2:1−13. doi: 10.9734/ARJA/2016/29334
50. Oladosu Y, Rafii MY, Abdullah N, Hussin G, Ramli A, Rahim HA, et al. Principle and application of plant mutagenesis in crop improvement: a review. Biotechnol Biotechnol Equip. (2016) 30:1–16. doi: 10.1080/13102818.2015.1087333
51. Hussain M, Iqbal MA, Till BJ, Rahman M-U. Identification of induced mutations in hexaploid wheat genome using exome capture assay. PLoS ONE. (2018) 13:e0201918. doi: 10.1371/journal.pone.0201918
52. Yin J-L, Fang Z-W, Sun C, Zhang P, Zhang X, Lu C, et al. Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific locus amplified fragment (SLAF) sequencing. Sci Rep. (2018) 8:1–9. doi: 10.1038/s41598-018-21489-5
53. Shu Q-Y, Forster BP, Nakagawa H, Nakagawa H. Plant Mutation Breeding Biotechnology. Wallingford: CABI (2012).
54. Ouyang B, Gu X, Holford P. Plant genetic engineering and biotechnology: a sustainable solution for future food security and industry. Plant Growth Regul. (2017) 83:171–3. doi: 10.1007/s10725-017-0300-5
55. Nalluri N, Karri VR. Recent advances in genetic manipulation of crops: A promising approach to address the global food and industrial applications. Plant Sci Today. (2020) 7:70–92. doi: 10.14719/pst.2020.7.1.659
56. Dan Z. Application of modern biotechnology in the control of plant diseases and insect pests. Revista de la Facultad de Agronomia de la Universidad del Zulia, 37.
57. Eissa HF, Hassanien SE, Ramadan AM, El-Shamy MM, Saleh OM, Shokry AM, et al. Developing transgenic wheat to encounter rusts and powdery mildew by overexpressing barley chi26 gene for fungal resistance. Plant Methods. (2017) 13:41. doi: 10.1186/s13007-017-0191-5
58. Huang X, Wang J, Du Z, Zhang C, Li L, Xu Z. Enhanced resistance to stripe rust disease in transgenic wheat expressing the rice chitinase gene RC24. Transgenic Res. (2013) 22:939–47. doi: 10.1007/s11248-013-9704-9
59. Ding Y, Dong X, Zhao D. Transformed EuCHIT1 gene in wheat (Triticum aestivum) enhances resistance to stripe rust (Puccinia striiformis f. sp tritici). J Agric Biotechnol. (2017) 25:1013–22.
60. Wang H, Zou S, Li Y, Lin F, Tang D. An ankyrin-repeat and WRKY-domain-containing immune receptor confers stripe rust resistance in wheat. Nat Commun. (2020) 11:1–11. doi: 10.1038/s41467-020-15139-6
61. Qi T, Zhu X, Tan C, Liu P, Guo J, Kang Z, et al. Host-induced gene silencing of an important pathogenicity factor P s CPK 1 in Puccinia striiformis f. sp tritici enhances resistance of wheat to stripe rust. Plant Biotechnol J. (2018) 16:797–807. doi: 10.1111/pbi.12829
62. Zhu X, Qi T, Yang Q, He F, Tan C, Ma W, et al. Host-induced gene silencing of the MAPKK gene PsFUZ7 confers stable resistance to wheat stripe rust. Plant Physiol. (2017) 175:1853–63. doi: 10.1104/pp.17.01223
63. Wang B, Sun Y, Song N, Zhao M, Liu R, Feng H, et al. Puccinia striiformis f. sp. tritici mi croRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytologist. (2017) 215:338–50. doi: 10.1111/nph.14577
64. Chang Q, Lin X, Yao M, Liu P, Guo J, Huang L, et al. Hexose transporter PsHXT1-mediated sugar uptake is required for pathogenicity of wheat stripe rust. Plant Biotechnol J. (2020) 1–13. doi: 10.1111/pbi.13398
65. Qi T, Guo J, Liu P, He F, Wan C, Islam MA, et al. Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol Plant. (2019) 12:1624–38. doi: 10.1016/j.molp.2019.09.010
66. Khlestkina E, Shumny V. Prospects for application of breakthrough technologies in breeding: the CRISPR/Cas9 system for plant genome editing. Russian J Genet. (2016) 52:676–87. doi: 10.1134/S102279541607005X
67. Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. (2017) 7:1–6. doi: 10.1038/s41598-017-00578-x
68. Ahmad S, Wei X, Sheng Z, Hu P, Tang S. CRISPR/Cas9 for development of disease resistance in plants: recent progress, limitations and future prospects. Brief Func Genom. (2020) doi: 10.1093/bfgp/elz041
69. Borrelli VM, Brambilla V, Rogowsky P, Marocco A, Lanubile A. The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front Plant Sci. (2018) 9:1245. doi: 10.3389/fpls.2018.01245
70. Satheesh V, Zhang H, Wang X, Lei M. Precise editing of plant genomes–prospects and challenges. In: Seminars in Cell & Developmental Biology. Academic Press (2019) 96:115–23.
71. Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V. Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol. (2015) 32:76–84. doi: 10.1016/j.copbio.2014.11.007
72. Zhang Y, Bai Y, Wu G, Zou S, Chen Y, Gao C, et al. Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. (2017) 91:714–24. doi: 10.1111/tpj.13599
73. Cui X. Targeted Gene Editing Using CRISPR/Cas9 in a Wheat Protoplast System. Ottawa: Université d'Ottawa/University of Ottawa (2017).
74. Watson A, Ghosh S, Williams MJ, Cuddy WS, Simmonds J, Rey M-D, et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat Plants. (2018) 4:23–9. doi: 10.1038/s41477-017-0083-8
75. Hickey LT, Hafeez AN, Robinson H, Jackson SA, Leal-Bertioli SC, Tester M, et al. Breeding crops to feed 10 billion. Nat Biotechnol. (2019) 37:744–54. doi: 10.1038/s41587-019-0152-9
76. Bassi FM, Bentley AR, Charmet G, Ortiz R, Crossa J. Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci. (2016) 242:23–36. doi: 10.1016/j.plantsci.2015.08.021
77. Alahmad S, Dinglasan E, Leung KM, Riaz A, Derbal N, Voss-Fels KP, et al. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods. (2018) 14:36. doi: 10.1186/s13007-018-0302-y
78. Hickey LT, Germán SE, Pereyra SA, Diaz JE, Ziems LA, Fowler RA, et al. Speed breeding for multiple disease resistance in barley. Euphytica. (2017) 213:64. doi: 10.1007/s10681-016-1803-2
79. Gu L, Si W, Zhao L, Yang S, Zhang X. Dynamic evolution of NBS–LRR genes in bread wheat and its progenitors. Mol Genet Genom. (2015) 290:727–38. doi: 10.1007/s00438-014-0948-8
80. Liu J, Qiao L, Zhang X, Li X, Zhan H, Guo H, et al. Genome-wide identification and resistance expression analysis of the NBS gene family in Triticum urartu. Genes Genom. (2017) 39:611–21. doi: 10.1007/s13258-017-0526-7
81. Kushwaha UKS, Deo I, Jaiswal JP, Prasad B. Role of bioinformatics in crop improvement. Glob J Sci Front Res D Agric Vet. (2017) 17:13−24. Available online at: https://globaljournals.org/GJSFR_Volume17/3-Role-of-Bioinformatics-in-Crop.pdf
82. Simko I, Jimenez-Berni JA, Sirault XR. Phenomic approaches and tools for phytopathologists. Phytopathology. (2017) 107:6–17. doi: 10.1094/PHYTO-02-16-0082-RVW
83. Hussain M. Bridging Mutational and Genomic Approaches for the Development of Novel Wheat Germplasm. Islamabad: Pakistan Institute of Engineering & Applied Sciences (2019).
Keywords: fungal pathogen, Puccina striiformis, yellow rust, wheat, resistance genes, new breeding strategies
Citation: Jamil S, Shahzad R, Ahmad S, Fatima R, Zahid R, Anwar M, Iqbal MZ and Wang X (2020) Role of Genetics, Genomics, and Breeding Approaches to Combat Stripe Rust of Wheat. Front. Nutr. 7:580715. doi: 10.3389/fnut.2020.580715
Received: 06 July 2020; Accepted: 19 August 2020;
Published: 06 October 2020.
Edited by:
Rakesh Bhardwaj, National Bureau of Plant Genetic Resources (ICAR), IndiaReviewed by:
Palmiro Poltronieri, National Research Council (CNR), ItalyAmit Singh, National Bureau of Plant Genetic Resources (ICAR), India
Copyright © 2020 Jamil, Shahzad, Ahmad, Fatima, Zahid, Anwar, Iqbal and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shakra Jamil, shakrajamil29@gmail.com; Rahil Shahzad, rahilshahzad91@gmail.com; Xiukang Wang, wangxiukang@yau.edu.cn
 Shakra Jamil
Shakra Jamil Rahil Shahzad
Rahil Shahzad Shakeel Ahmad
Shakeel Ahmad Rida Fatima
Rida Fatima Rameesha Zahid3
Rameesha Zahid3 Muhammad Zaffar Iqbal
Muhammad Zaffar Iqbal Xiukang Wang
Xiukang Wang