- Agri-Food Biotechnology Laboratory, National Agri-Food Biotechnology Institute, Mohali, India
Wheat is a major cereal crop providing energy and nutrients to the billions of people around the world. Gluten is a structural protein in wheat, that is necessary for its dough making properties, but it is responsible for imparting certain intolerances among some individuals, which are part of this review. Most important among these intolerances is celiac disease, that is gluten triggered T-cell mediated autoimmune enteropathy and results in villous atrophy, inflammation and damage to intestinal lining in genetically liable individuals containing human leukocyte antigen DQ2/DQ8 molecules on antigen presenting cells. Celiac disease occurs due to presence of celiac disease eliciting epitopes in gluten, particularly highly immunogenic alpha-gliadins. Another gluten related disorder is non-celiac gluten-sensitivity in which innate immune-response occurs in patients along with gastrointestinal and non-gastrointestinal symptoms, that disappear upon removal of gluten from the diet. In wheat allergy, either IgE or non-IgE mediated immune response occurs in individuals after inhalation or ingestion of wheat. Following a life-long gluten-free diet by celiac disease and non-celiac gluten-sensitivity patients is very challenging as none of wheat cultivar or related species stands safe for consumption. Hence, different molecular biology, genetic engineering, breeding, microbial, enzymatic, and chemical strategies have been worked upon to reduce the celiac disease epitopes and the gluten content in wheat. Currently, only 8.4% of total population is affected by wheat-related issues, while rest of population remains safe and should not remove wheat from the diet, based on false media coverage.
Introduction
Cereal crops are recognized as a major food source for mankind. Wheat, rice and barley are some of the major cereal crops grown worldwide. Cereals are staple food for human nutrition and are considered as a major source of calories for human (1). The dietary components in cereals such as lipids, carbohydrates and proteins, play an instrumental role in processing and nutritional quality for food and feed. Among cereals, wheat is one of the major staple crops across the world and is unique due to its special bread-making properties. The estimated global wheat production for 2016 was 749.46 million metric tons (2). Furthermore, the global demand of wheat has increased incredibly after industrialization and urbanization due to its bread making properties and ability of being processed into different food products (3). The most commonly used wheat types are durum or tetraploid wheat (A and B genome) and hexaploid wheat (A, B, and D genome). Many species of wheat together make up the genus Triticum, amongst which the most widely used and grown is the hexaploid wheat (Triticum aestivum L. AABBDD) (4).
Wheat seed storage proteins are very important in determining the end products as they impart viscoelasticity and extensibility to dough which enables formation of a wide range of products such as bread, pasta, noodles, cakes, and pastries (3, 5). Seed storage proteins constitute about 8–15 percent of total flour weight and can be classified into albumins, globulins, gliadins, and glutenins on the basis of their solubility. Of these fractions, gliadins and glutenins constitute the gluten proteins and are stored together with starch in endosperm of the seed. Both gliadins and glutenins are involved in building the gluten polymer and determining bread-making properties of wheat (6). But, gluten present in wheat is the major factor responsible for causing certain disorders and allergies in some individuals. A wide variety of people are incapable to tolerate wheat consumption due to harmful immune response to gluten proteins present in wheat. Hence, despite of such large consumption of wheat worldwide, there are cases reported which show intolerance toward it (7). The most common wheat-related disorders associated with gluten ingestion are celiac disease (CD) and non-celiac gluten-sensitivity (NCGS), which result in impaired quality of life and significant morbidity in individuals (8). Wheat allergy is another condition arising from contact, inhalation or ingestion of wheat and is associated with gluten, other wheat proteins and carbohydrates present in wheat particularly fermentable, oligo, di, monosaccharides, and polyols (FODMAPs). Specific clinical manifestations can be observed in each of these disorders with some peculiar immunogenic pathways involved in their development (9). Adherence to gluten free foods is the only available remedy for patients with CD and NCGS. This manuscript provides detailed insight into the pathogenesis and mechanisms of gluten related disorders, particularly CD along with NCGS and wheat allergy; and different strategies to lower down wheat toxicity and gluten content in wheat.
Components of Wheat Involved in Intolerance
Different components of wheat which are responsible for eliciting immune response and gastrointestinal symptoms in certain individuals are:
Gluten
Gluten is the main storage protein found in wheat, rye and barley; and is important for dough formation (10). Gluten is classified as: (a) high molecular weight glutenin subunits (HMWGS); (b) low molecular weight glutenin subunits (LMWGS); (c) the S-poor prolamins (omega [ω]-gliadins); and (d) S-rich prolamins which include alpha (α), beta (β), and gamma (γ) gliadins (11–13). Gluten composition varies between both species as well as cultivars.
Glutens contain high contents of proline-rich polypeptide residues which make them resistant to proteolytic degradation by gastric, pancreatic, and intestinal juices containing digestive proteases (8, 14–17). When these proteins are consumed by genetically susceptible individuals, a cascade of immune reactions is triggered, which result in damage to the intestinal lining resulting in CD. Gluten is also responsible for causing other wheat related disorders such as NCGS, wheat allergy and contact urticaria (8, 9). The most widely prevalent of all is CD.
α-Amylase/Trypsin Inhibitors (ATIs) and Lectins
ATIs and lectins comprise of 2–4% of total proteins in modern hexaploid wheat. Wheat ATIs are disulphide linked, compact albumin proteins found in the endosperm of plant seeds and are resistant to degradation by the proteases (18). These proteins regulate starch metabolism during seed development and germination, and aid in providing defense to plants against parasites and insects (19, 20). ATIs have recently been implicated in wheat sensitivity. ATIs trigger innate immune response by activating toll-like receptor (TLR) 4 on myeloid cells and antigen presenting cells such as monocytes, macrophages, and dendritic cells in intestinal mucosa to produce inflammatory response by producing cytokines and chemokines, viz. interleukin (IL)-8, tumor necrosis factor (TNF)-α, or monocyte chemotactic protein-1 (21). Once antigen presenting cells are activated by ATIs, they migrate to peripheral lymph nodes and further enhance the ongoing immune response (22). ATIs mainly produce non-intestinal symptoms in NCGS (23–25) and also act as primary allergens in Baker's asthma (18). Experimental evidences show that ATIs serve as adjuvants in intestinal inflammatory diseases in mice (26).
Lectins are carbohydrate-binding proteins present in plants that provide defense to plants against pathogens (27, 28). Wheat germ agglutinin is a specific type of lectin which is described extensively in the literature for inducing adverse health effects. It binds to gut epithelium, damages intestinal cells, and results in reduced absorption of nutrients in the gut (29, 30).
FODMAPs
FODMAPs are short-chain carbohydrates comprising of fructans and galacto-oligosaccharides, and are present naturally in many foods in various forms viz. lactose in milk, free fructose in fruits like pears and apples, fructans in wheat and onions, galacto-oligosaccharides in legumes and sugar polyols viz. sorbitol and mannitol in stone fruit, some vegetables, and fermented foods (31). In wheat, FODMAPs viz. fructans, galacto-oligosaccharides, and mannitol are present. Human body, lacks enzymes for the breakdown of FODMAPs and hence these get absorbed slowly in the small intestine and pass undigested to reach large intestine where these are rapidly fermented by gut bacteria which produce gas and cause intestinal walls to stretch (32, 33). In most people, this process is asymptomatic, but in patients with irritable bowel syndrome and inflammatory bowel disease, this process is problematic and can produce various symptoms including flatulence, bloating, stomach pain, constipation, or diarrhea (34–36). Biesiekierski et al. (29) observed that removal of dietary FODMAPs caused improvement in gastrointestinal symptoms in 37 people with NCGS and irritable bowel syndrome.
Wheat Related Disorders
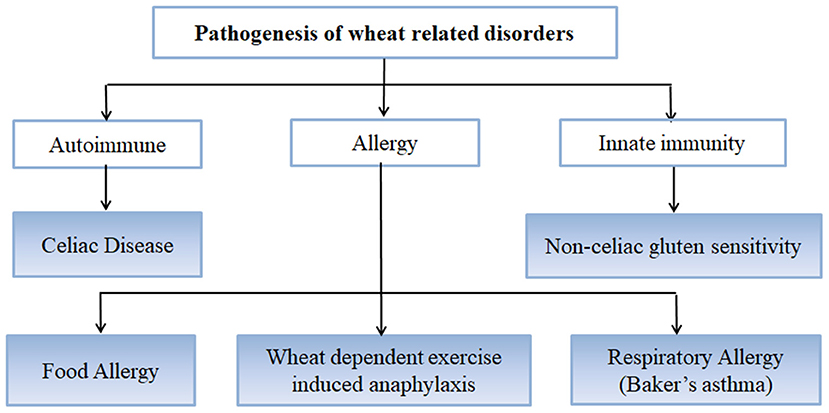
Wheat is the most important cereal crop worldwide and has been used as an important staple crop for centuries but in certain individuals and genetically predisposed people, it causes certain disorders and conditions arising mainly from the gluten fraction [Figure 1; (37)]. CD is caused by gluten proteins specifically gliadins but the problem lies not just with gluten protein but with other non-prolamins components of wheat as well which results in the onset of some other conditions as well (38). These include NCGS in which patients experience symptoms similar to CD but these get resolved when gluten is removed from the diet. However, patients do not test positive for CD (39). Other non-celiac wheat response is wheat allergy, which includes Baker's asthma, a respiratory allergic response caused by exposure to wheat flour dust (40–42); food allergy which is IgE-mediated immune response caused by wheat ingestion; and wheat-dependent exercise-induced anaphylaxis (WDEIA) which is exercise induced wheat allergic response (30). The most common wheat related disorders are:
CD
CD is a chronic immune-mediated enteropathy of the small intestine that develops in genetically susceptible individuals by exposure to gluten proteins found in certain cereals viz. wheat, rye and barley (43, 44). All these are close relatives of wheat, while distantly related species do not elicit CD. There are controversial studies related to involvement of oats in causing CD (45–48). Though oats do not contain gluten, these contain a small percentage (10–15% total protein content) of similar proline rich storage proteins called “avenins.” Few CD epitopes with different structures have been identified in oats which may cause intolerance in some individuals at low intensity (49, 50). But more serious problem is the production of pure oats free from gluten containing cereals in a conventional production chain as there are high chances of contamination with wheat, rye and barley during seed sowing, cultivation, harvesting, milling, and processing of oats (51). Figure 2 shows various gluten-rich cereals capable of eliciting CD immunogenicity in patients. Both glutenins and gliadins have certain amino acid sequences that act as epitopes for CD (52) and are termed as immunogenic peptides, antigenic peptides, T-cell epitopes, CD eliciting epitopes, or toxic peptides (53). These CD eliciting peptides resist degradation in gastrointestinal tract due to high content of amino acids viz. proline and glutamine.
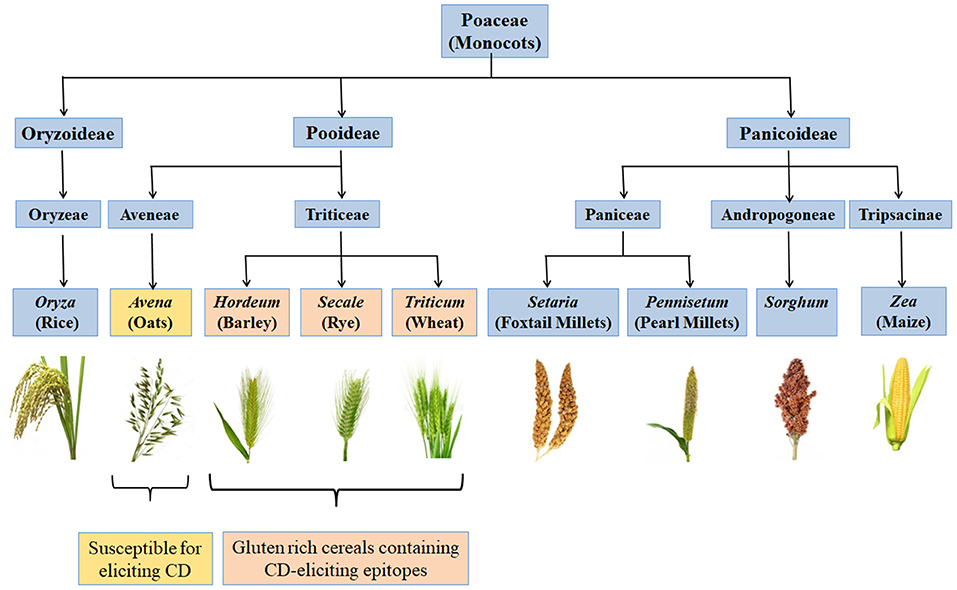
Figure 2. Classification of monocots and their CD eliciting potential. Gluten rich cereals viz. wheat, rye, and barley belonging to family Triticeae and containing CD-eliciting epitopes (pink boxes). Some cereals like oat are susceptible for eliciting CD (yellow box) whereas, many other cereals are safe for CD patients (blue boxes).
CD represents a chronic inflammatory condition affecting small intestine and jejunum, resulting in villous atrophy in intestine. The villi in small intestine get flattened and the surface area for nutrient absorption is highly reduced which leads to malnutrition, vitamin and mineral deficiencies and other gastrointestinal symptoms such as abdominal discomfort, bloating, loose bowel movements, and nausea (54, 55). Untreated patients develop risk of some chronic conditions and non-gastrointestinal symptoms such as anemia, osteoporosis, fatigue, infertility, eczema, and refractory CD which is associated with developing lymphoma (44, 56). Symptoms of CD are highly variable and may occur at any age. Diseases like type I diabetes, Hashimoto's thyroiditis, Grave's disease, Sjogren's syndrome, Down syndrome, Turner syndrome, primary biliary cirrhosis, and neurologic disorders like unexplained peripheral neuropathy, epilepsy, and ataxia are also sometimes associated with CD (57–63).
The global prevalence of CD was found to be 1.4% in 275,818 individuals studied using serological antibodies. According to biopsy based analysis, its prevalence was found to be 0.7% in ~1,40,000 individuals studied (53). In different countries, the range of disease varied as 0.5% in Africa and North America, 0.4% in South America, 0.6% in Asia, and 0.8% in Europe. The prevalence of disease varies with age, sex and presence of other autoimmune disorders (53). Currently, adhering to gluten free foods is the only option for individuals with CD (37, 64, 65).
CD Epitopes
CD eliciting epitopic sequences of glutens from hexaploid bread wheat (T. aestivum L.) are present in mainly six loci, with Gli-A1, Gli-B1, and Gli-D1 located on short arms of group 1 chromosomes (1AS, 1BS, and 1DS) and Gli-A2, Gli-B2, and Gli-D2 on the short arms of group 6 chromosomes (6AS, 6BS, and 6DS) [Figure 3; (11, 66)]. Gli-1 loci contain genes coding for γ, ω, or δ gliadins, whereas, Gli-2 loci contain genes coding for α-gliadins (66–68). HMWGS are encoded by three homoeologous loci (Glu-A1, Glu-B1, and Glu-D1) on the long arms of group 1 chromosomes, while LMWGS are encoded by Glu-A3, Glu-B3, and Glu-D3 loci on the short arms of group 1 chromosomes [Figure 3; (68, 69)].
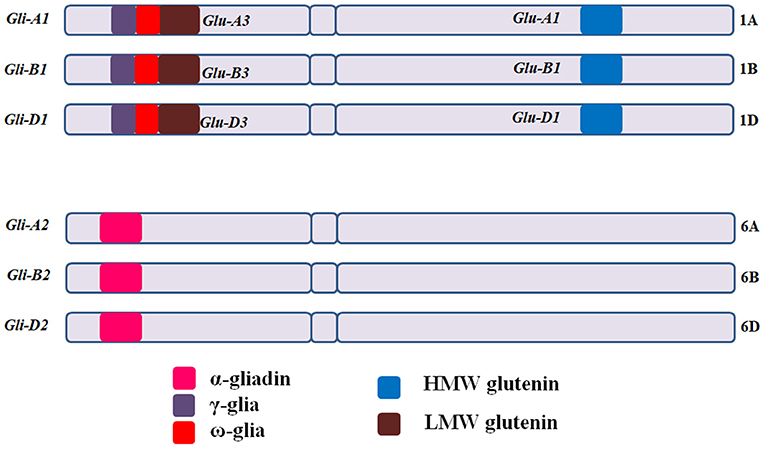
Figure 3. Gliadin and glutenin loci in Triticum aestivum (AABBDD), 2n = 6x = 42. The Gli-A1, Gli-B1, and Gli-D1 are located on the short arm of homoeologous group −1 chromosomes and comprise mainly of ω and γ-gliadins, while Gli-A2, Gli-B2, and Gli-D2 are located on short arm of group −6 chromosomes and comprise mainly of α-gliadins. HMWGS viz. Glu-A1, Glu-B1, and Glu-D1 are located on long arm of group −1 chromosomes, whereas, LMWGS viz. Glu-A3, Glu-B3, and Glu-D3 are located on the short arm of group −1 chromosomes. Gli, gliadin; Glu, glutenin.
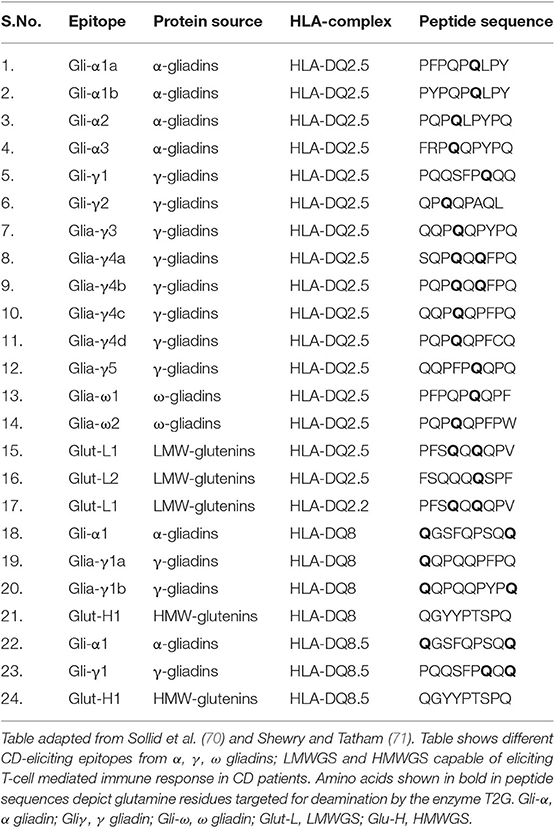
Till date many T-cell epitopes have been studied for inducing immunogenicity of CD. Both glutenins and gliadins are responsible for this disease. The major epitopes from wheat responsible for eliciting CD immunogenicity are listed in Table 1. All three α/β, ω and γ-gliadins are immunogenic for CD (52) but the 33-mer peptide sequence (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) of α-gliadins is found to be the most potent stimulator of T-cells and is the most immunogenic amongst all (52, 72, 73). The high importance of the 33-mer sequence in CD is evident by the production of two monoclonal antibodies (A1 and G12) against it (74). This 33-mer peptide fragment of gliadin α2 (57–67, 69–90) contains 6 partially overlapping copies of three highly immunogenic T cell epitopes, viz. PFPQPQLPY (DQ2.5-glia-α1a, 1 copy), PYPQPQLPY (DQ2.5-glia-α1b, 2 copies), and PQPQLPYPQ (DQ2.5-glia-α2, 3 copies) (70, 75, 76). The 33-mer sequence is rich in proline and glutamine residues and is a stimulator of T-cells after deamidation by the enzyme tissue transglutaminase (TG2) (77).
Estimated copy number of α-gliadins ranges from 25 to 150 copies per haploid genome and consists of a highly diverse and complex gene family (78). α-gliadins from D-genome (specified by Gli-D2) are the most immunogenic while those from B-genome (specified by Gli-B2) are least immunogenic as these contain very few CD epitopes (44, 71, 79–82). After α-gliadins, γ-gliadins are found to be most immunogenic and their copy number ranges from 15 to 40 (83). HMWGS and LMWGS also contain CD epitopes but in less amount and present low immunogenicity (44, 71). Also, single or multiple amino acid substitutions in natural epitopic sequences result in the lack of immunogenicity. For example, proline to serine substitution in epitope core position p3 or p8, or proline to alanine substitution in epitope core position p5 in PFPQPQLPY sequence of DQ2.5 Gli-α1a resulted in lack of T-cell stimulation and reduced immunogenicity. Similarly, serine to phenylalanine substitution at p3, or glutamine to arginine substitution at p5 in QGSFQPSQQ sequence of DQ8.5 Gli-α1 completely removed the immunogenic potential of the epitope (84).
Mechanism of CD Toxicity
CD is an auto-immune disorder and its pathogenicity results from the interaction of gluten with genetic and environmental factors which initiate an immune response in the body. At genetic level, CD occurs in genetically predisposed individuals carrying specific major histocompatibility complex haplotype DQ2 or DQ8 in Human Leukocyte Antigen (HLA) in certain individuals (54, 85–87). In a study by Ciccocioppo et al. (88), it was found that gluten peptides involved in CD were able to specifically stimulate HLA-DQ2 or HLA-DQ8-restricted T-cell clones isolated from jejunal mucosa or peripheral blood of celiac patients. The HLA-DQ2 and HLA-DQ8 are cell surface receptors located on antigen presenting cells and contain positively charged pockets with a preference for binding negatively charged particles; and thus, have a strong binding affinity to these negatively charged gluten molecules (89). Over 95% of the affected CD patients express HLA-DQ2 molecules and the remaining express HLA-DQ8 (90, 91). Further in a process of CD, zonulin, a human protein and a modulator of intestinal tight junctions is involved in a reversible regulation of intestinal permeability (92). Its upregulation due to gluten exposure disrupts the integrity of tight junctions between epithelial cells of small intestine and increases the paracellular movement of protease resistant gluten fragments (93, 94). As a result, the undigested gluten peptide fragments pass through epithelial barrier of small intestine and enter lamina propria where these are deaminated by the enzyme tissue transglutaminase (TG2) which converts glutamine to glutamate thereby imparting negative charge to gluten fragments (95). Further, the activity of the enzyme TG2 is dependent on spacing between glutamine and proline. Glutamine in the sequence QXP is modified by the enzyme but in the sequence QXXP and QP it remains unmodified (X = any amino acid) (96). Deamination of gluten peptides by TG2 is the key pathogenic event that increases gliadin immunogenicity in CD and enhances the severity of disease (97). TG2 plays an important role in CD pathogenesis and anti-TG2 antibodies are used as the markers for CD diagnosis (95). The enzyme TG2 is located on the brush border epithelia of the small intestine or in extracellular space of sub-epithelial region (98). TG2 exists in inactivated form under normal oxidative conditions; but gets activated extracellularly under reducing conditions created by inflammatory response during CD (99). The negatively charged peptides produced through deamination by TG2 bind to the positively charged amino acids on HLA-DQ2 and DQ8 molecules and trigger adaptive as well as innate immune response in CD patients (88, 100). Adaptive immune response begins with the presentation of undigested gluten peptides by HLA molecules on antigen presenting cells to CD4+ T-cells in lamina propria. The recognition and binding of T-cell receptors to specific HLA-gliadin complexes lead to the production of high levels of pro-inflammatory cytokines dominated mainly by interferon (IFN)-γ (65, 101, 102). These cytokines either induce T-helper 1 cells to produce IL-15 and IL-21 which result in the activation of cytotoxic CD8+ intra epithelial lymphocytes (IELs) and promote CD8+ T-cell cytotoxic activity and contributes to intestinal lesions, inflammation and intestinal mucosal damage (95, 103) or T-helper 2 cells to cause B-lymphocytes differentiation for the secretion of anti-gliadin, anti-TG2, and anti-endomysium (EM) antibodies which are considered as the key characteristics of active CD (65, 104). An increased density of CD8+ IELs is also considered as an important characteristic of CD (90, 105). These immune responses disappear when gluten is excluded from the diet (65). Some gliadin peptides bind to TLR2 receptors which result in increased IL-1 production, through the mediation of MyD88, a key protein responsible for mediating the release of zonulin in response to gluten ingestion (106). Recently it has been claimed that the presence of HLA is not the only factor responsible for the onset of CD (107). The genome-wide association studies have identified 39 non-HLA loci affecting CD (108).
Several environmental factors also influence the occurrence of CD. Feeding patterns in the first year of life and the time at which gluten intake is initiated during infancy also determines the susceptibility to disease (64, 105). The initial intake of gluten before 4 months of age contributes to disease susceptibility whereas administration of gluten after 7 months denotes marginal risk to disease. Gluten intake along with breast feeding in infants reduces the risk of the disease (109). Again, this factor is just one aspect that can contribute to celiac susceptibility. Some recent studies have claimed that the potential viral infections caused by rotavirus might play an important role in the activation of this disease (110). The individuals having CD are found to have rotavirus infection but there is no confirmed proof for this aspect.
Diagnosis of CD
CD enteropathy occurs in 1.4% of population but remains undiagnosed in most of the cases despite having evidence of increasing rates of diagnosis (111). Various methods for the diagnosis of CD and other wheat related disorders are given in Figure 4. Study by Jansen et al. (112) showed that in a population of around 4,500 children aged around 6 years, 61% had sub-clinical CD when screened with TG2 IgA antibody. At this time, there are no confined tests that can precisely confirm the cause and occurrence of disease. Since there are so many factors controlling the occurrence of disease, there is a need for the standard test for its confirmation. Most physicians these days rely upon serological tests using specific antibodies or prefer the use of biopsies for checking the intestinal damage caused by the disease. Flow cytometric analysis of IELs is very helpful for the diagnosis of CD in few cases where serological tests and duodenal biopsies do not work (113).
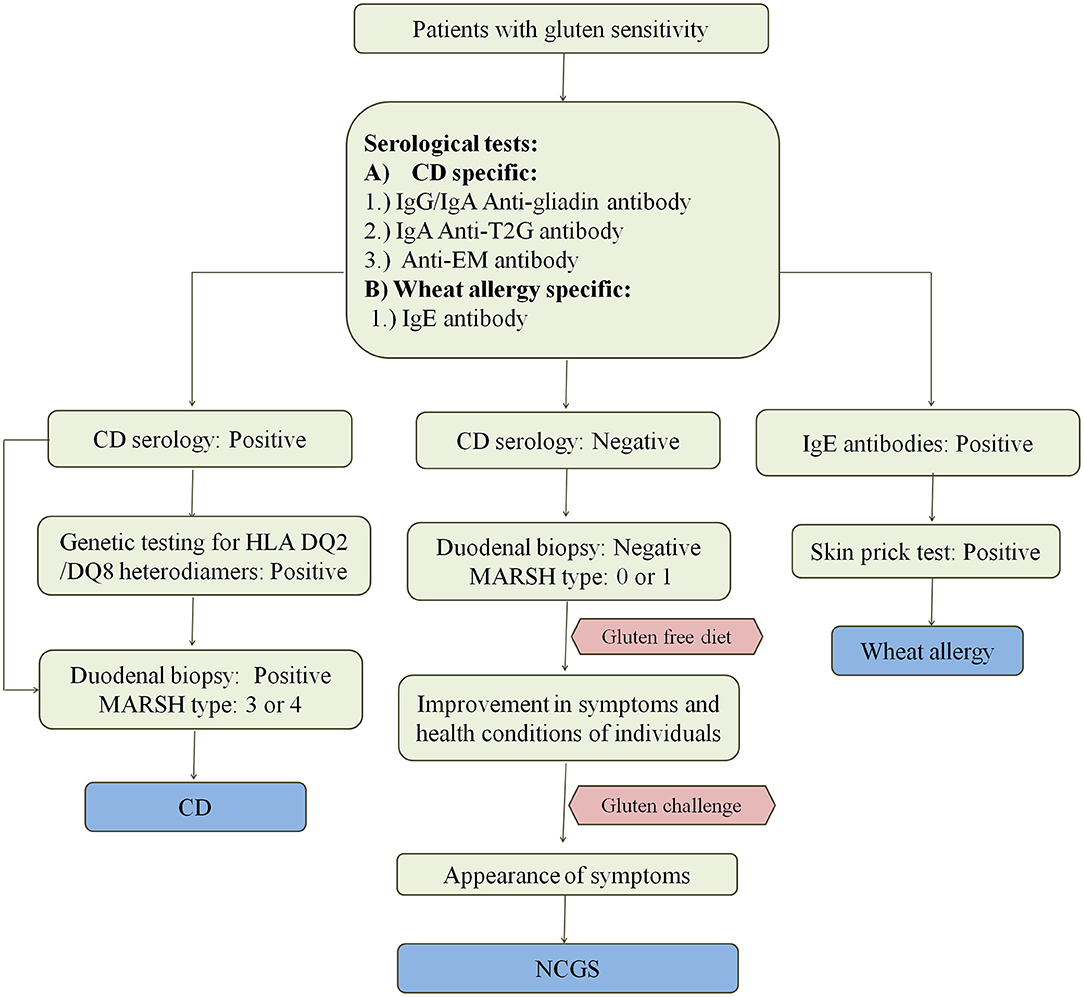
Figure 4. Diagnosis of wheat related disorders in patients. Figure shows different parameters for diagnosis of CD, NCGS, and wheat allergy. CD patients are confirmed on the basis of serological tests which include the detection of anti-gladin, anti-T2G, and anti-EM antibodies in the blood serum of patients. CD can be further confirmed by positive genetic testing for HLA DQ2/DQ8 heterodimers and MARSH type 3–4 on the basis of duodenal biopsy. Wheat allergy is detected on the basis of presence of IgE antibodies in the blood serum of patients and confirmed on the basis of positive skin prick test. NCGS is detected on the basis of negative serological tests for CD in patients showing gluten sensitivity, gastrointestinal symptoms and MARSH type 0–1. NCGS in patients is further confirmed by observing improvement in symptoms upon intake of gluten-free diet. If symptoms reappear upon gluten challenge, then NCGS is confirmed.
Serological testing is performed using anti-gliadin or anti-deaminated gliadin antibodies, anti-TG2 antibodies and anti-EM antibodies (114–119). Serological testing is performed on patients with some specific symptoms which include chronic diarrhea resulting in malabsorption of nutrients, iron deficiency anemia and deficiency of folates, vitamin E, D, K which result in apparent weight loss, osteoporosis, hypocalcemia, and unexplained elevation of transaminases (65, 118, 120–122).
In a study done using deaminated antibodies, it was found that anti-deaminated gliadin antibodies are better than conventional gliadin antibodies (123). The next in the list are TG2 specific antibodies which are found in serum as IgA and IgG isotypes, and are autoantigens for CD (124). Assaying for TG2-specific IgA is the most common clinical practice used in case of CD because of its highest specificity and sensitivity among all other methods (65, 125, 126). The sensitivity of these antibodies was tested on both humans as well as guinea pigs. Human TG2 antibodies were diagnosed in 64 out of 65 patients as compared to 58 out of 65 patients in case of guinea pigs. Hence, human TG2 antibodies are a better way to diagnose CD as compared to any other method (125). The next are anti-EM antibodies which were first studied on monkey esophagus. These antibodies react with EM of smooth muscles (127, 128). The specificity and sensitivity of IgA antibodies are 99 and 95%, respectively. A positive result of IgA EM antibodies is an indication of non-atrophic intestinal lesions (116). Prior to these serological methods, detection of only histological changes in small intestinal such as villous atrophy and presence of inflammatory markers were considered essential for the diagnosis of CD.
Genetic testing for CD follows the procedure for HLA-typing, but this test has low specificity. Determination of HLA-DQ2 and HLA-DQ8 types is useful along with histological findings (129). HLA-DQ2 and HLA-DQ8 molecules are associated with ~95 and 5% CD patients, respectively (59).
Serological testing by the use of anti-gliadin antibodies, TG2 or EM specific antibodies along with HLA-typing sometimes may not be successful and it cannot be considered as the confirmatory test. Individuals who are positive for serological testing are further confirmed with the help of biopsy. Sero-negative individuals also have to undergo biopsy if they have signs and symptoms highly suspicious for CD as none of the available serological tests have the sensitivity of 100% (130). Duodenal biopsy is the most preferred choice for the diagnosis of CD (131). The histological analysis of disease is performed using Mucosal Algorithmic Rules for Scoring Histology (MARSH) classification as discussed further. Endoscopic observation includes scalloping, fissuring, and the reduced villi folds in intestine. The pathologic lesion is a characteristic feature of disease. It is characterized by flat intestinal mucosa with infiltration of lymphocytes. Other symptoms include crypt hyperplasia and villous atrophy. Patient may suffer from anemia due to malabsorption (132). Sometimes due to poor results of both serological tests and biopsy, esophago gastro-duodenoscopy is done in CD subjects (116).
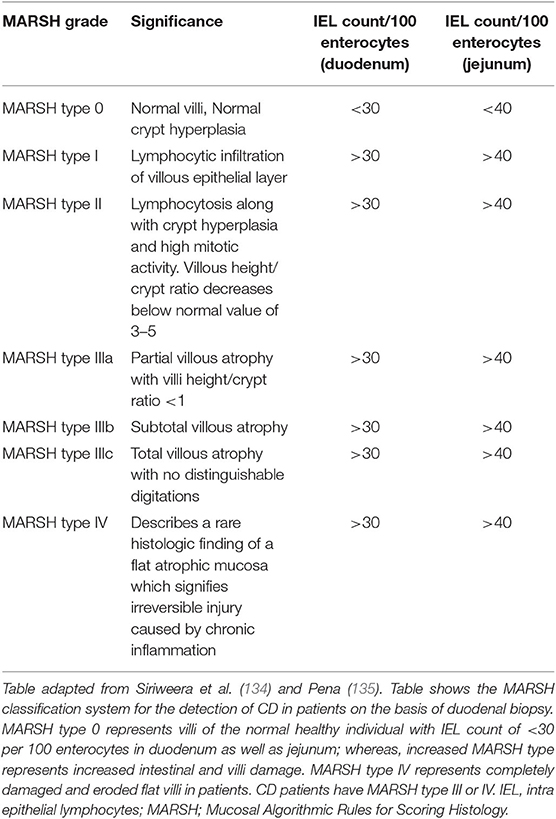
MARSH classification for histological diagnosis of CD
The recognition of histopathology of CD is done using MARSH classification system which describes the stages of damage in the small intestine and abnormalities in celiac mucosa during the progression of CD as seen under the microscope. MARSH classification system is useful because it avoids the misinterpretation of histo-pathological results of duodenum and hence, increases the sensitivity of the system (133). The modern MARSH classification system is given in Table 2.
In this classification system, a five-point scoring system is given by Catassi and Fasano (136), which is called 4 out of 5 rule system. Individuals who satisfy any four of these are considered to be susceptible for CD. But this criterion is not widely used by clinicians since the gain in sensitivity in this case is at the cost of its specificity. According to this, the five points are (i) symptoms of CD such as diarrhea, weight loss and iron deficiency anemia, (ii) positive CD serologies at high titer, (iii) presence of a DQ2 or DQ8 haplotype, (iv) characteristic histopathologic findings, and (v) serological or histological response to the gluten-free diet.
Strategies for Lowering Down CD Epitopes
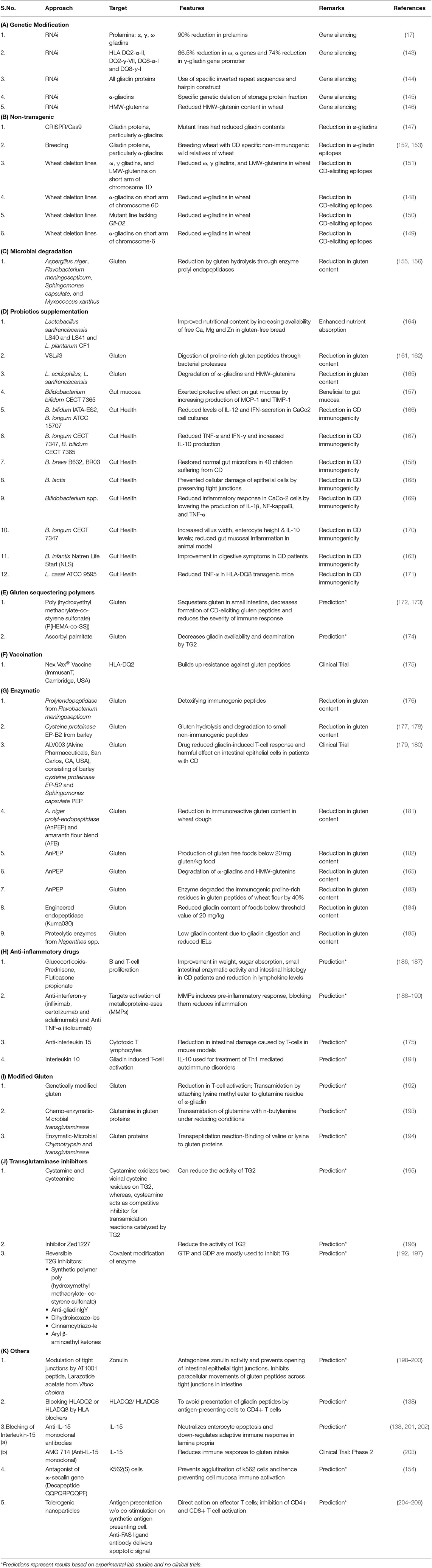
Following a life-long gluten-free diet is very difficult for CD patients. As a result, different strategies and therapies have been being discovered and explored to reduce the potential toxicity of gluten in CD (137). CD epitopes present in wheat can be lowered through various molecular biology, biotechnology, plant breeding, microbial, enzymatic, nano-technology, chemical, and pharmaceutical approaches (137–142).
Gene silencing by RNA-interference (RNAi) technology has been adopted for the successful down-regulation of α, γ, ω gliadins, and HMWGS in different studies (17, 143–146). Other non-transgenic approaches for lowering gluten epitopes in wheat include engineering gliadin epitopes in wheat with CRISPR/Cas9 (147); creating wheat deletion lines lacking α-gliadins on short arm of 6D chromosome (148–150); and wheat deletion lines lacking ω, γ gliadins, and LMWGS on short arm of chromosome 1D (151). Breeding hexaploid wheat varieties with less immunogenic wild relative of wheat Agropyron elongatum resulted in a significant reduction in immunogenic α-gliadin epitopes in subsequent generations (152, 153). In another study by Vita et al. (154), the ω-secalin gene encoding decapeptide QQPQRPQQPF from wheat-rye 1BL.1RS translocation line prevented K562 cells agglutination and mucosal cell immune activation in the presence of toxic gliadin epitopes. Another approach used in the reduction of CD-eliciting immunogenic gluten epitopes in wheat involves the use of microorganisms for the gluten hydrolysis, which occurs due to the presence of enzyme prolyl endopeptidases. Different microorganisms such as Aspergillus niger, Flavobacterium meningosepticum, Sphingomonas capsulate, and Myxococcus Xanthus have been used for gluten hydrolysis (155, 156).
The most widely reported strategy in lowering CD pathogenicity involves the use of probiotics which grow optimally under the pH range present in gastrointestinal tract and exert protective effects on gut mucosa and microbiota (157–159). Some probiotics have the potential to digest gluten polypeptides or help in their alteration or breakdown into simpler non-immunogenic peptides. The commercialized probiotic preparation VSL#3 comprising of consortium of eight bacterial strains (Bifidobacterium breve, B. longum, B. infantis, Lactobacillus plantarum, L. acidophilus, L. casei, L. delbruecki subsp. bulgaricus, and Streptococcus thermophilus) (160) was found to hydrolyze gliadin epitopes in wheat flour including immunogenic 33-mer peptide sequence during prolonged fermentation (161, 162). Smecuol et al. (163) observed that probiotic administration of B. infantis Natren Life Start superstrain in CD patients resulted in the marked improvement in digestion and reduction in constipation after 3 weeks from the beginning of treatment. Other examples of the use of probiotic strains for protecting epithelial cells of small intestine against cellular damage and reducing the levels of inflammatory cytokines have been given in Table 3.
Use of therapeutic vaccine “NexVax2” developed by the biotechnology firm “ImmuSanT” is at the clinical trials stage. The vaccine consists of three gluten peptides and is supposed to induce tolerogenic response in CD patients by building up resistance against gluten peptides (175). “NexVax2” has recently been granted Fast Track designation by the U.S. Food and Drug Administration on January 2, 2019 (207). Other methods include use of gluten sequestering polymers such as poly (hydroxyethyl methacrylate-co-styrene sulfonate) (172, 173) and ascorbyl palmitate (174) which sequester gluten peptides and reduce their availability for subsequent immunogenic reactions. Chemo-enzymatic-microbial transglutaminase (193) and enzymatic-microbial chymotrypsin and transglutaminase (194) have also been used to lower gluten content in wheat flour. Various enzymes used for gluten hydrolysis involve prolylendopeptidase from F. meningosepticum (176), cysteine proteinase EP-B2 from barley (177, 178), ALV003 (Alvine Pharmaceuticals, San Carlos, CA, USA), consisting of barley cysteine proteinase EP-B2 and S. capsulate prolyl endoprotease (179, 180, 208), A. niger prolyl endoprotease (AnPEP) (165, 182, 183), and engineered endopeptidase (Kuma030) (184).
Other strategies involve modulation of tight junctions by targeting zonulins (198–200); blocking of IL-15 using anti-IL monoclonal antibodies (138, 201–203), blocking HLADQ2 or HLADQ8 by HLA blockers (138), use of genetically-modified gluten with modified glutamine residues (192), use of transglutaminase inhibitors (192, 195–197) and use of tolerogenic nanoparticles (204–206). Use of anti-inflammatory drugs (186, 187), anti-IFN-γ, anti-TNF-α (188–190), and anti-IL-15 agents (175) have also been suggested in reducing CD toxicity.
Many among these approaches are just hypotheses or predictions based on experimental data or some are at clinical trial levels. Also, none of these technologies ensure complete removal of CD-eliciting gluten epitopes from wheat and its safe consumption to avoid immunogenic response, but a good amount of research is being done in various spheres to find out promising strategies and alternatives to make gluten free foods in near future. Various strategies for lowering CD epitopes in wheat are given in Table 3.
Effect of Lowering CD Epitopes on Bread Quality
Gluten is a structural protein in wheat and is essential for dough making and preparing good quality baked products. Therefore, obtaining wheat varieties with reduced CD epitopes is a technological challenge due to the inability of gluten free flours to form dough with desired strength and visco-elastic properties (209). Furthermore, the baking and bread-making qualities of such flours may get adversely affected.
In a study by Van den Broeck et al. (148), altered dough mixing properties and dough rheology were observed in the hexaploid wheat cv. Chinese Spring deletion lines which were found to be less immunogenic as a result of missing short arm of chromosome 6D containing α-gliadin genes. The dough had reduced elasticity and higher stiffness. In contrast, the technological properties of wheat were retained in the deletion lines of wheat created by removing ω, γ-gliadins and LMWGS from the short arm chromosome 1D. Similarly, Piston et al. (210) found that no major effect on dough gluten strength was observed when γ-gliadins in bread wheat were down-regulated. In another study, Van den Broeck et al. (151) found increased dough elasticity and deteriorated dough quality in wheat having a reduced number of T-cell stimulatory epitopes (ω, γ-gliadins, and LMWGS) in short arm of chromosome 1D. Gil-Humanes et al. (211) reported that down-regulation of CD eliciting gliadin epitopes in different bread wheat varieties by RNAi provided flours with increased stability, different texture, less extensibility and less stickiness in comparison to dough from wild wheat. In a recent study by Zhang et al. (146), the silencing of HMWGS in wheat through RNAi and post-transcriptional gene silencing significantly reduced dough properties, wet gluten content, sedimentation value, and stability time of flour.
NCGS
In 1978, Ellis and Linaker (212) described a case with diarrhea and abdominal pain in the absence of CD (without any histological duodenal lesions or damage in the lining of small intestine) that improved with the elimination of gluten from the diet. Similarly, Cooper et al. (213) observed abdominal pain, diarrhea, and normal duodenal histology in patients. Their condition improved with intake of diet free from gluten but symptoms reoccurred following the gluten challenge. Since then, the terminology, NCGS is used where intestinal permeability and adaptive immune system have a less pronounced role than in CD (214).
NCGS is caused by gluten and carbohydrates present in wheat mainly the FODMAPs, ATIs and wheat-germ agglutinin (117, 215, 216). ATIs are capable of triggering TLR 4 present on myeloid cells, leading to the release of proinflammatory cytokines (21). The major symptoms in NCGS patients include abdominal bloating and pain in the upper or lower abdomen, diarrhea, nausea, aphthous stomatitis, and changing bowel habits. Other non-gastrointestinal symptoms include foggy mind, fatigue, tiredness, lack of well-being, headache, depression, anxiety, joint/muscle pain, numbness in legs or arms, and skin rash/dermatitis (214, 217, 218). These symptoms disappear when gluten is removed from the diet (219) and overlap with those of irritable bowel syndrome (220). Some of these symptoms are also present in CD but both these conditions differ in their genetics and immunological responses. NCGS can be separated from CD in terms of HLA index, specific immunological response; and the structure and function of small intestinal mucosa (221, 222). The differences between CD, NCGS and wheat allergy are given in Table 4.
Pathogenesis of NCGS
Although CD and NCGS share a common feature of gluten sensitivity; they differ with respect to immune response initiated during their onset. Unlike CD, NCGS is not related to autoimmune process involving adaptive immune response leading to intestinal epithelial cell damage (223). Many studies have thrown light on the underlying mechanisms involved in the pathogenesis NCGS but no confirmed mechanisms are still known. Sapone et al. (224) observed normal intestinal permeability and increased levels of IELα, IELβ, and TLR-2 expression in 26 NCGS patients in comparison to CD patients and controls. Different studies have shown the involvement of only intestinal innate immune system in NCGS as evident by the increase in TLR2, TNF-α, IL-8, and IL-12 expression (21, 225, 226). However, in some studies, an increase in anti-gliadin Ig antibodies and IFN-γ expression has also been observed (227–229). Uhde et al. (230) showed that individuals with NCGS displayed increased serum levels of intestinal fatty acid and lipopolysaccharide-binding proteins along with elevated levels of soluble cluster of differentiation 14 (CD14), all of which declined in response to the elimination of gluten from food. Carroccio et al. (227) reported basophil activation in 66% of patients with NCGS and found it to be associated with intraepithelial lymphocytosis of duodenum and infiltration of eosinophils in duodenum and colon. Some NCGS patients showed increased TLR2 levels in comparison to CD, and had dysbiosis similar to that observed in inflammatory bowel disease (231). Furthermore, in a study by Junker et al. (21), it was observed that wheat ATIs act as triggers of the innate immune response in intestinal monocytes, macrophages and dendritic cells by activating the TLR4-MD2-CD14 complex and eliciting the release of proinflammatory cytokines in cells from celiac and non-celiac patients.
Diagnostic Parameters for NCGS
At present, there are no known specific serological markers for NCGS. Unlike CD, diagnosis of NCGS is not dependent on the patient's response to different antibodies or biopsy analyses. Only anti-gliadin antibody can be used to test NCGS, while anti-TG2 and anti-EM antibodies are found to be negative in these patients (232, 233). Various studies have reported that 25–50% of NCGS patients have serum anti-gliadin antibodies, mainly IgG (225, 227, 234). Volta et al. (235) reported an increase in the levels of IgG antibodies specific to gliadins in NCGS patients. NCGS is diagnosed clinically on the basis of response to gluten free food, followed by gluten challenge (214, 236, 237). In a study by Catassi et al. (236), NCGS specific symptoms disappeared in patients after the removal of gluten from the diet and reappeared during oral gluten challenge performed after at least 3 weeks of gluten-free food in a double-blind placebo experiment. Gluten challenge is generally initiated once the patient's symptoms are reasonably under good control. The clinical response after gluten challenge might be variable but usually overlaps with symptoms of CD to a large degree (225, 238). Histological findings indicate duodenal biopsy to be between MARSH type 0-1 and there is also infiltration of CD3+ IELs (ranging from 25 to 40 IELs per 100 enterocytes) but not to an extent as seen in CD patients. Moreover, some levels of circulating basophils are also present (224, 227, 238–240). Verdu et al. (241) carried out a study on gluten sensitive transgenic mice with HLA-DQ8 genes and found that gliadin exposure resulted in the activation of innate immunity without any intestinal atrophy in these mice. However, Bucci et al. (242) carried out studies on NCGS patients and found that gliadin exposure did not result in the activation of mucosal inflammation or basophil production.
Wheat Allergy
Wheat allergy is characterized by IgE and non-IgE mediated immune response resulting in allergic reaction in certain individuals upon the uptake, contact, or inhalation of foods containing wheat (243, 244). IgE mediated allergic responses occur immediately after food consumption, are food-specific and reproducible (245) and result from the release of histamine, platelets activator factor, and leukotrienes from mast cells and basophils (246, 247). These allergic reactions can affect skin, respiratory or gastrointestinal tract and are characterized by TH2 lymphocytic inflammation which leads to the production of IL-4, IL-5, and IL-13; and causes B cells to produce IgE antibodies specific to certain foods (215, 248, 249). Genetic characteristics of individuals as well as environmental factors play an important role in the onset of such immune response (250, 251). Non-IgE mediated allergic responses are characterized by chronic infiltration of eosinophils and lymphocytes in the gastrointestinal tract. Different types of wheat allergies include:
Food Allergy
Food allergy is caused by the intake of wheat which triggers the IgE mediated immune response in certain individuals. Different components of wheat can cause food allergy, viz., ATIs, non-specific lipid transfer protein, gliadins, and LMWGS (40, 252, 253). Food allergy results in the development of different symptoms in patients, viz. urticaria, stomach cramps, asthma, allergic rhinitis, abdominal pain, vomiting and atopic dermatitis (42, 254). Remedy for food allergy includes the avoidance of wheat in the diet.
WDEIA
WDEIA is a type of food allergy which occurs after wheat ingestion followed by physical exercise but is not triggered by wheat ingestion alone. WDEIA was reported only few decades ago in 1985 (255) and its symptoms include anaphylactic reactions ranging from urticaria, angioedema, dyspnoea, hypotension, collapse, and shock. In wheat, ω5-gliadin, and HMWGS have been reported to be the major allergens that contribute to WDEIA (256–259). WDEIA is also induced by non-steroidal anti-inflammatory drugs, alcohol, and infections. The remedy suggested for this is to avoid having gluten rich diet before physical exercise (260). The exact mechanism of WDEIA is still not clear, but exercise enhances gastrointestinal osmolarity and permeability to allergens, increases blood flow and induces IgE-mediated mast cell degranulation (261, 262). Increased temperature also causes phosphorylation of tight junction proteins which results in increased absorption of allergens due to mucosal injury (263).
Baker's Asthma
Baker's asthma develops after inhalation of allergens, particularly cereal flour dust present in the work environment and affects 0.03–0.24% of bakery workers, confectioners, pastry factory workers, and cereal handlers. It is considered one of the most common types of occupational, cereal-induced allergic asthma and is mediated by IgE antibodies specific to cereal flour antigens: mainly proteins from wheat, rye, barley and rice (264–266). The primary cereal used in bread baking is wheat and it acts as a major allergen in 60–70% of symptomatic bakers (267). Bakers develop asthma as well as rhinitis with increased levels of specific IgE antibodies against flour dust from different sources (268). Incidences of bronchial hyper-responsiveness are also reported in bakery workers suffering from Baker's asthma (269, 270). Genetic factors also play a significant role in the onset of Baker's asthma with respiratory symptoms. In a study by Cho et al. (271) on Korean bakery workers, TLR4 gene polymorphism was found to be responsible for allergic sensitization to wheat flour. Similarly, Hur et al. (272) reported genetic polymorphisms of β2-adrenergic receptors to be responsible for the development of Baker's asthma in the workers exposed to wheat flour. ATIs along with non-specific lipid transfer protein are the major components of wheat resulting in the onset of Baker's asthma (40, 253). Other allergens include wheat peroxidase, thioredoxins, serine proteinase inhibitor, thaumatin-like proteins, gliadins, and LMWGS (41, 273–276).
Proper diagnostic measures for Baker's asthma are not present till date. The only available method is skin prick test using commercially available wheat extracts for serological analyses but this method is less sensitive and non-specific due to different concentration and composition of antigens in different wheat extracts, and implementation of non-standardized methods of preparation (117, 277). Hence, improved and standardized measures for the diagnosis of Baker's asthma are required.
Is Wheat Safe for Non-celiac Healthy Individuals?
Wheat is the most important cereal crop across the world and is rich source of variety of nutrients such as carbohydrates, proteins, fats, dietary fibers, lipids, B vitamins; minerals such as iron, calcium, zinc, magnesium, sodium, copper, and selenium; and many phytochemicals such as phenols and flavonoids (3). Though a number of diseases and conditions arise from wheat intake in certain individuals who cannot tolerate it but wheat consumption is safe for a wide percentage of individuals who can tolerate it. Globally 1.4% of individuals are reported for CD (53), 0.63–6.0% for NCGS (278), and 0.2–1.0% for wheat allergy (245, 249). Hence, at maximum, only 8.4% population is susceptible to wheat-related disorders and the remaining 91.6% population remains safe for the intake of wheat and should not remove it from diet based on false media coverage (Figure 5).
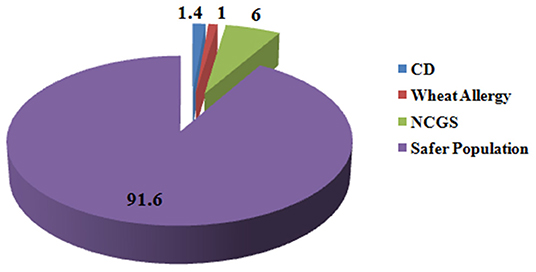
Figure 5. Percentage of people suffering from gluten-related disorders. Figure shows that in total 8.4% of people across the globe suffer from gluten-related disorders and 91.6% of population is safe from these disorders and is non-susceptible for gluten ingestion.
Is There Difference Between Diploid, Tetraploid, or Hexaploid Wheat Cultivars in Eliciting CD?
The diploid wheats (AA, 2n = 14; T. urartu, T. monococcum) and the tetraploid wheats (AABB, 2n = 28; T. durum, T. turgidum) were domesticated by man, about 10,000 years ago (279). Modern bread wheat is allohexaploid (AABBDD, 2n = 42; T. aestivum) species arising from hybridization between tetraploid T. turgidum having AB-genome and wild diploid species Aegilops tauschii having D genome (280). The introduction of D-genome in wheat has improved its bread-making properties (281–283). At genomic level, the genes encoding for immunogenic CD epitopes are located on the short arm of 6A and 6D chromosomes, while the short arm of 6B chromosome is mainly non-immunogenic. The highly immunogenic 33-mer CD epitopic sequence of α-gliadins is located on the 6D chromosome which can initiate a very strong immune response (70, 284). The literature suggests that the hexaploid wheat consists of more immunogenic CD epitopes and elicits a higher immunogenic response in CD patients than diploid and tetraploid wheat due to the presence of highly immunogenic D genome (79). Different studies have shown that tetraploid wheat are less immunogenic than hexaploid wheat (285–287). In one study, Schalk et al. (284) did not detect any 33-mer peptide sequence in two durum wheat cultivars and two emmer cultivars (genome AABB) by using liquid chromatography tandem mass spectrometry (LC-MS/MS) and attributed this to the absence of chromosome 6D. Similarly, Molberg et al. (288) showed absence of 33-mer peptide sequence encoded by α-gliadin genes in diploid einkorn (including T. monococcum, T. uraru, and A. speltoides). Kumar et al. (289) studied 34 tetraploid and hexaploid wheat varieties for their gliadin content and immunoreactivity with immunoglobulins (IgA) of CD and found tetraploid wheat varieties to be less immunoreactive than hexaploid wheat varieties. Ozuna and Barro (290) found in their study that durum wheat varieties tend to have lower gluten protein and CD eliciting epitopes in comparison to tetraploid and hexaploid wheat varieties.
Studies by De Vincenzi et al. (291); Pizzuti et al. (292), and Vincentini et al. (293) demonstrated that T. monococcum was non-immunogenic w.r.t. CD, while others studies showed that immunogenicity of diploids still exists, but is very less in comparison to hexaploid wheat varieties (79, 285, 286, 294, 295). On the contrary, in other studies, no difference in the immunogenic potential of diploid, tetraploid, and hexaploid wheat was observed (296–298). Ozuna et al. (286) reported A. tauschii to be highly immunogenic, while Escarnot et al. (295) showed lower immunogenicity of A. tauschii in comparison to bread wheat. Vaccino et al. (299) found the presence of 13 immunogenic CD eliciting peptides sequences in T. monococcum, thereby indicating that diploid wheat has the potential to trigger CD.
Is Breeding Responsible in Increasing CD Epitopes in Wheat?
At present, 95% of the wheat grown globally is hexaploid (utilized in bakery) and the remaining 5% is tetraploid (utilized for pasta making) (300). Studies have claimed that over the decades, the genetic improvement of wheat NexVax2 through breeding to improve yield, plant height, disease resistance, adaptation to climate changes, and bread-making characteristics by modifying wheat proteins particularly gluten content, may have led to a higher immunogenicity of wheat and therefore higher incidences of CD (279, 301).
Gluten constitutes 60–75% of total wheat proteins and is highly desirable in the food industry and is mainly responsible for imparting the desirable strength to the dough and contributes to its viscoelastic nature (302, 303). In addition, increase in wheat consumption, use of gluten in food processing, and consumption of processed foods has been seen over the years (304, 305). The preference of wheat varieties with higher gluten content has always been desirable but in spite of being an important wheat protein, gluten acts as a main source of immunogenic peptides triggering CD in certain individuals.
Many studies have evaluated the role of breeding on the immunogenicity of wheat. Kaur et al. (306) studied variation in CD-eliciting epitopes of α-gliadins protein sequences in Indian wheat cultivars and found modern varieties (1971–2011) having a higher amount of intact T-cell stimulatory epitopes than old wheat varieties (1905–1970) which had comparatively higher variant epitopes. Van den Broeck et al. (307) analyzed the potential toxicity of several hexaploid wheat varieties including 36 modern and 50 landraces using Glia-α9 and Glia-α20 antibodies and found higher amounts of Glia-α9 epitopes in modern varieties. This suggests that modern wheat breeding practices may have led to a higher number of CD-triggering epitopes. De Santis et al. (308) explored the effect of breeding in the twentieth century in Italy and found an increase in gliadin and glutenin epitopes in modern durum wheat varieties. However, Kasarda (279) observed that breeding of wheat to improve its baking quality and dough strength did not influence antibody A1-G12 reactivity in old, mid and new wheat varieties from the 20th and twenty-first centuries in the United States of America. Also, the comparison of different landraces and wheat varieties did not show any significant difference in A1-G12 reactivity. But in some other studies, modern wheat varieties were found to have a reduced number of CD epitopes in comparison to old wheat varieties or landraces. Ribeiro et al. (298) showed that T. aestivum spp. vulgare landraces had higher reactivity to R5 antibody and presented higher amount of potential CD immunostimulatory epitopes than modern varieties; inferring that breeding practices were not responsible for the increase in CD immunostimulatory epitopes. Gelinas and McKinnon (287) found that the traditional wheat line available in the nineteenth century showed the highest reactivity to G12 antibody. Similarly, Schalk et al. (284) found that spelt cultivar Ober-kulmer had the highest amounts of the 33-mer peptide (523.4 μg/g flour), while other spelt cultivar Franckenkorn (353.9 μg/g flour) showed no significant difference from the common wheat cultivars used in the study. Similarly, no significant difference was observed in A1-G12 reactivity of tetraploid and hexaploid wheat varieties in a study by Escarnot et al. (295). Malalgoda et al. (309) quantified Glia-α9 and Glia-α20 by mass spectroscopy in 30 wheat varieties released between 1910 and 2013 in North Dakota but could not find any trend in the immunogenicity of these wheat varieties. Some other studies have also reported comparatively higher toxicity in ancient wheat varieties in comparison to modern wheat varieties. Prandi et al. (310) analyzed gluten peptides in old and modern Triticum varieties, using Liquid Chromatography Mass Spectroscopy (LC-MS) and found a significantly higher amount of CD-eliciting immunogenic peptides in old varieties than modern varieties. They concluded that old wheat varieties have the potential to trigger CD and are thus not safe for CD patients. Similarly, Gregorini et al. (311) and Colomba and Gregorini (312) found a higher percentage of immunogenic α-gliadin epitopes in ancient durum wheat varieties namely Graziella Ra and Kamut in comparison to modern wheat varieties and advised CD patients to avoid consuming these wheat varieties. All these studies suggest that there is no fixed trend in the results obtained due to immense variation in the immunogenic potential of different wheat varieties: old, modern or landraces. Thus wheat breeding did not cause any increase in CD toxicity (296, 310, 313, 314). This can be attributed to the fact that the genetic improvement of wheat through breeding has mainly focused on glutenins, which are mainly responsible for dough strength and are less immunogenic in comparison to gliadins. Ozuna and Barro (290) found in their study that breeding has infact contributed to the decrease in gliadin and total gluten content but not glutenin content in different wheat varieties belonging to Triticeae.
Gluten Free Diet (GFD)
At this point of time, therapies for CD patients are non-existent. The only solution to the problem is a stringent lifelong GFD. GFD means the absence of gluten or prolamine proteins from wheat, rye, barley and oats in natural as well as in processed foods (315–317). The limit of gluten in GFD has been set to 20 ppm by Codex Alimentarius Commission for International Food Standards of Food and Agriculture Organization of the United Nations [FAO]/World Health Organization (318). This cut-off limit is followed in many countries including Spain, Italy, UK, Canada, and USA but countries like Australia, New Zealand, and Chile have locally set the cut-off limit to 3 ppm, while Argentina has set the limit to 10 ppm (319).
In a double blind, placebo controlled study by Catassi et al. (320), it was observed that significant reduction in intestinal mucosal villous height/crypt depth ratio occurred upon intake of 50 mg gluten per day for 3 months but in a study by Lanzini et al. (321), intake of <10 mg gluten daily did not cause any considerable histological changes in the intestinal mucosa. Laurikka et al. (322) compared gastrointestinal symptoms in untreated, treated with GFD and healthy controls and observed higher diarrhea, indigestion, and abdominal pain in untreated CD patients than those on GFD and controls. Also, a very good response was observed in patients on GFD during a long-term follow-up. Akobeng and Thomas (323) observed that GFD with gluten concentration lower than 20 ppm required an intake of <50 mg gluten daily to ensure adequate safety.
GFD: Challenges
Correct quantification of gluten, its source, and proper labeling are necessary for the normal health of CD patients. Following a life-long GFD can be very challenging due to the presence of small amounts of hidden gluten in processed food products as contamination of foods is possible at different stages from farm to fork. Cross-contamination of GFD from gluten based foods can occur from grains from adjacent fields; harvesting, storing, and processing grains on shared equipment; making products on same equipment; food handling by unaware employees in various restaurants, and lack of awareness among people in general. Other challenges related to GFD are higher cost, less palatability, less availability, and less reliability in developing countries (324–326). Also, the likelihood of low income patients in developing countries to go on GFD is very low (327). For CD patients, eating out and traveling are major challenges because of poor availability of GFD and fear about gluten contamination. Producing low gluten-immunogenic wheat and food products will involve a great deal of efforts, as well as scientific and technological inputs. Educating people about GFD, promoting research and development for the production of cost-effective GFD and providing country-wide infrastructure are key steps to manage CD and gluten intolerance (328–330). This will help CD patients to follow a life-long GFD and will help to improve their quality of life.
Methods for Detection of Gluten in Food
The fast growth of CD diagnosis has sparked the investigation for the reliable methods for gluten estimation. Several methods have been proposed for gluten estimation in different food and food products. The most widely utilized method is enzyme-linked immunosorbent assay (ELISA); developed by utilizing monoclonal antibodies against toxic gluten peptides. Mainly, two monoclonal antibodies based (R5 and G12) ELISA kits are commercially available (331, 332). ELISA R5 Mendez Method has been approved by 2015 Codex Alimentarius Commission. In addition, polyclonal antibodies against gluten peptides have been prepared for potential use in ELISAs (333). For increasing the sensitivity of antibody based detection to picogram level, modifications like adsorption of food sources on small latex particles, fluorescent labeling, and flow cytometry have been investigated (334).
LC-MS/MS provides an alternative to ELISA based detection method. Protein extraction method has critical importance for efficacy of LC-MS/MS based methods (335, 336). Digestion of proteins with digestive enzyme or peptic-tryptic/chymotryptic treatment is required for simulating human gastrointestinal digestion of wheat products (337). For the adoption of this technology, the pre-requisite step is the identification of robust and sensitive CD triggering peptide markers as well as comprehensive and well-annotated sequence databases. GluPro V1.0 database of gluten proteins comprises of 630 discrete and unique full-length protein sequences (338) and is an alternative to the existing Viridiplantae database. LC-MS/MS has been documented to distinguish among different cereals and detect cereal contamination in different commercial flours (335, 339). Reverse phase-high performance liquid chromatography coupled with matrix-assisted laser desorption ionization time of flight mass spectrometry has been reported to assess prolamins from wheat (340). Peptide immobilized pH gradient-isoelectric focusing separation, coupled with LC-MS/MS, has been documented for detecting cereal contamination from beer (341). Two-dimensional electrophoresis and gel-permeation high performance liquid chromatography with fluorescence detection have also been reported for the detection of gliadins and glutenins (342, 343).
In order to increase the speed, sensitivity and decrease the cost of current methods, viz., ELISA and LC-MS/MS; several alternative electrochemical immunoassays like electronic tongue (e-tongue), nanorods, and immunochips have been proposed. The microfluidic e-tongue capable of detecting as low as 0.005 ppm of gliadin in foodstuffs has been investigated (344). The competitive and disposable amperometric immunosensor based on gliadin-functionalized carbon/nanogold screen-printed electrodes was developed for rapid gluten detection in processed food samples (345). The gliadin-immunochips, based on electrochemical impedance spectroscopy transduction method, capable of detecting 0.5 ppm of gliadin in beers and flours have been reported (346).
Conclusion
Wheat is one of the most important cereals and with the increase in incidences of wheat related diseases like CD, NCGS, it is imperative that these challenges are addressed now. Patients with CD must avoid foods containing gluten and should strictly follow GFD; patients with wheat allergy should avoid contact with wheat in any form; and patients with NCGS should also adhere to GFD. Much research advancements have happened in the diagnosis of CD and wheat allergy, but not in case of NCGS. Therefore, it is necessary to understand the underlying mechanism of pathogenicity in case of NCGS to develop more sensitive diagnostic markers. The fundamental understanding of the mechanisms of disease relevant pathways may come from the analysis of genome and gene expression.
More research and infrastructure are needed for the development of low-gluten wheat, and ultimately food products. This also requires changes in industrial food processing methods because low gluten content in wheat and its products would be an important trait for its commercialization in future. Plant breeding and biotechnological approaches could be used to make CD-safe wheat and food products with reduced immunogenic gluten fractions. Transferring genes containing less immunogenic CD epitopes from wild relatives of wheat to existing high yielding wheat cultivars can be done by using breeding and biotechnological approaches.
Nevertheless, only a small percentage of global population is affected by these wheat related disorders, hence, opting for GFD to improve well-being by the remaining population without any medical recommendation is an unhealthy option; wheat being a nutrient and dietary fiber rich cereal.
Author Contributions
MG drafted the manuscript layout and helped in overall supervision during manuscript preparation. NS, SB, and SK collected the literature and wrote the manuscript. MG, VC, SS, PK, and AK helped in manuscript and references editing. NS prepared the figures. NS and SB prepared the tables.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to National Agri-Food Biotechnology Institute for providing NABI—core grant to support this work. Contribution of Ms. Lovenpreet Kaur and Mr. Pargat Singh in manuscript editing is truly acknowledged.
References
1. Lafiandra D, Riccardi G, Shewry PR. Improving cereal grain carbohydrates for diet and health. J Cereal Sci. (2014) 59:312–26. doi: 10.1016/j.jcs.2014.01.001
2. Food and Agriculture Organization of the United Nations [FAO]. World Food and Agriculture-Statistical Pocketbook 2018 [FAO Statistics]. (2018). Available online at: http://www.fao.org/3/ca1796en/CA1796EN.pdf (accessed August 7, 2019).
3. Shewry PR, Hey SJ. The contribution of wheat to human diet and health. Food Energy Secur. (2015) 4:178–202. doi: 10.1002/fes3.64
4. Matsuoka Y. Evolution of polyploid Triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. (2011) 52:750–64. doi: 10.1093/pcp/pcr018
5. Sissons M. Role of durum wheat composition on the quality of pasta and bread. Food. (2008) 2:75–90.
6. Schofield JD. Wheat proteins: structure and functionality in milling and bread making. In: Bushuk W, Rasper VF, editors. Wheat: Production, Properties and Quality. Boston, MA: Springer (1994). p. 73–106. doi: 10.1007/978-1-4615-2672-8_7
7. Cabanillas B. Gluten-related disorders: Celiac disease, wheat allergy, and nonceliac gluten sensitivity. Crit Rev Food Sci Nutr. (2019) 12:1–16. doi: 10.1080/10408398.2019.1651689
8. Stamnaes J, Sollid LM. Celiac disease: autoimmunity in response to food antigen. Semin Immunol. (2015) 27:343–52. doi: 10.1016/j.smim.2015.11.001
9. Juhasz A, Belova T, Florides CG, Maulis C, Fischer I, Gell G, et al. Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat. Sci Adv. (2018) 4:eaar8602. doi: 10.1126/sciadv.aar8602
10. Shewry PR, Halford NG, Belton PS, Tatham AS. The structure and properties of gluten: an elastic protein from wheat grain. Philos Trans R Soc Lond B Biol Sci. (2002) 357:133–42. doi: 10.1098/rstb.2001.1024
11. Shewry PR, Halford NG, Lafiandra D. Genetics of wheat gluten proteins. Adv Genet. (2003) 49:111–84. doi: 10.1016/S0065-2660(03)01003-4
12. Biesiekierski JR. What is gluten? J Gastroenterol Hepatol. (2017) 32:78–81. doi: 10.1111/jgh.13703
13. Khan H. Genetic improvement for end-use quality in wheat. In: Qureshi A, Dar Z, Wani S, editors. Quality Breeding in Field Crops. Cham: Springer (2019). p. 239–53. doi: 10.1007/978-3-030-04609-5_12
14. Sollid LM. Molecular basis of celiac disease. Annu Rev Immunol. (2000) 18:53–81. doi: 10.1146/annurev.immunol.18.1.53
15. Arentz-Hansen H, McAdam SN, Molberg O, Fleckenstein B, Lundin KE, Jorgensen TJ, et al. Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology. (2002) 123:803–9. doi: 10.1053/gast.2002.35381
16. Dewar DH, Amato M, Ellis HJ, Pollock EL, Gonzalez-Cinca N, Wieser H, et al. The toxicity of high molecular weight glutenin subunits of wheat to patients with coeliac disease. Eur J Gastroenterol Hepatol. (2006) 18:483–91. doi: 10.1097/00042737-200605000-00005
17. Barro F, Iehisa JCM, Gimenez MJ, Garcia-Molina MD, Ozuna CV, Comino I, et al. Targeting of prolamins by RNAi in bread wheat: effectiveness of seven silencing-fragment combinations for obtaining lines devoid of coeliac disease epitopes from highly immunogenic gliadins. Plant Biotechnol J. (2016) 14:986–96. doi: 10.1111/pbi.12455
18. Tatham AS, Shewry PR. Allergens to wheat and related cereals. Clin Exp Allergy. (2008) 38:1712–26. doi: 10.1111/j.1365-2222.2008.03101.x
19. Franco OL, Rigden DJ, Melo FR, Grossi-De-Sa MF. Plant α-amylase inhibitors and their interaction with insect α-amylases: Structure, function and potential for crop protection. Eur J Biochem. (2002) 269:397–412. doi: 10.1046/j.0014-2956.2001.02656.x
20. Guo G, Lv D, Yan X, Subburaj S, Ge P, Li X, et al. Proteome characterization of developing grains in bread wheat cultivars (Triticum aestivum L.). BMC Plant Biol. (2012) 12:147. doi: 10.1186/1471-2229-12-147
21. Junker Y, Zeissig S, Kim SJ, Barisani D, Wieser H, Leffler DA, et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J Exp Med. (2012) 209:2395–408. doi: 10.1084/jem.20102660
22. Zevallos VF, Raker V, Tenzer S, Jimenez-Calvente C, Ashfaq-Khan M, Russel N, et al. Nutritional wheat amylase-trypsin inhibitors promote intestinal inflammation via activation of myeloid cells. Gastroenterology. (2017) 152:1100–13. doi: 10.1053/j.gastro.2016.12.006
23. Fasano A, Sapone A, Zevallos V, Schuppan D. Nonceliac gluten sensitivity. Gastroenterology. (2015) 148:1195–204. doi: 10.1053/j.gastro.2014.12.049
24. Schuppan D, Pickert G, Shfaq-Khan M, Zevallos V. Non-celiac wheat sensitivity: differential diagnosis, triggers and implications. Best Pract Res Clin Gastroenterol. (2015) 29:469–76. doi: 10.1016/j.bpg.2015.04.002
25. Leccioli V, Oliveri M, Romeo M, Berretta M, Rossi P. A New proposal for the pathogenic mechanism of non-coeliac/non-allergic gluten/wheat sensitivity: piecing together the puzzle of recent scientific evidence. Nutrients. (2017) 9:1203. doi: 10.3390/nu9111203
26. Bellinghausen I, Weigmann B, Zevallos V, Maxeiner J, Reibig S, Waisman A, et al. Wheat amylase-trypsin inhibitors exacerbate intestinal and airway allergic immune responses in humanized mice. J Allergy Clin Immunol. (2019) 143:201–12. doi: 10.1016/j.jaci.2018.02.041
27. Liu B, Bian HJ, Bao JK. Plant lectins: potential antineoplastic drugs from bench to clinic. Cancer Lett. (2010) 287:1–12. doi: 10.1016/j.canlet.2009.05.013
28. Van Buul VJ, Brouns FJPH. Health effects of wheat lectins: a review. J Cereal Sci. (2014) 59:112–7. doi: 10.1016/j.jcs.2014.01.010
29. Biesiekierski JR, Peters SL, Newnham ED, Rosella O, Muir JG, Gibson PR. No effects of gluten in patients with self-reported non-celiac gluten sensitivity after dietary reduction of fermentable, poorly absorbed, short-chain carbohydrates. Gastroenterology. (2013) 145:320–8. doi: 10.1053/j.gastro.2013.04.051
30. De Punder K, Pruimboom L. The dietary intake of wheat and other cereal grains and their role in inflammation. Nutrients. (2013) 5:771–87. doi: 10.3390/nu5030771
31. Varney J, Barrett J, Scarlata K, Catsos P, Gibson PR, Muir JG. FODMAPs: food composition, defining cutoff values and international application. J Gastroenterol Hepatol. (2017) 32:53–61. doi: 10.1111/jgh.13698
32. Mansueto P, Seidita A, D'Alcamo A, Carroccio A. Role of FODMAPs in patients with irritable bowel syndrome. Nutr Clin Pract. (2015) 30:665–82. doi: 10.1177/0884533615569886
33. Muir JG, Varney JE, Ajamian M, Gibson PR. Gluten-free and low-FODMAP sourdoughs for patients with coeliac disease and irritable bowel syndrome: a clinical perspective. Int J Food Microbiol. (2019) 290:237–46. doi: 10.1016/j.ijfoodmicro.2018.10.016
34. Tuck CJ, Muir JG, Barrett JS, Gibson PR. Fermentable oligo-, di-, monosaccharide and polyols: Role in irritable bowel syndrome. Expert Rev Gastroenterol Hepatol. (2014) 8:819–34. doi: 10.1586/17474124.2014.917956
35. De Giorgio R, Volta U, Gibson PR. Sensitivity to wheat, gluten and fodmaps in IBS: facts or fiction? Recent advances in clinical practice Gut. (2016) 65:169–78. doi: 10.1136/gutjnl-2015-309757
36. Altobelli E, Del Negro V, Angeletti PM, Latella G. Low-FODMAP diet improves irritable bowel syndrome symptoms: a meta-analysis. Nutrients. (2017) 9:940. doi: 10.3390/nu9090940
37. Gasbarrini GB, Mangiola F. Wheat-related disorders: a broad spectrum of ‘evolving' diseases. United Eur Gastroenterol J. (2014) 2:254–62. doi: 10.1177/2050640614535929
38. Clerx EM, Silvester J, Leffler D, De Groote M, Fishman LN. Sequence of acquisition of self-management skills to follow a gluten-free diet by adults with celiac disease. Dig Liver Dis. (2019) 51:1096–100. doi: 10.1016/j.dld.2019.02.008
39. Pinto-Sanchez M, Verdu EF. Non-celiac gluten or wheat sensitivity: it's complicated! Neurogastroenterol. Motil. (2018) 30:e13392. doi: 10.1111/nmo.13392
40. Pastorello EA, Farioli L, Conti A, Pravettoni V, Bonomi S, Iametti S, et al. Wheat IgE-mediated food allergy in European patients, α-amylase inhibitors, lipid transfer proteins and low-molecular-weight glutenins. Allergenic molecules recognized by double-blind, placebo-controlled food challenge. Int Arch Allergy Immunol. (2007) 144:10–22. doi: 10.1159/000102609
41. Bittner C, Grassau B, Frenzel K, Baur X. Identification of wheat gliadins as an allergen family related to baker's asthma. J Allergy Clin Immunol. (2008) 121:744–9. doi: 10.1016/j.jaci.2007.09.051
42. Salcedo G, Quirce S, Diaz-Perales A. Wheat allergens associated with Baker's asthma. J Invest Allergol Clin Immunol. (2011) 21:81–92; quiz 94.
43. Freeman HJ, Chopra A, Clandinin MT, Thomson AB. Recent advances in celiac disease. World J Gastroenterol. (2011) 17:2259–72. doi: 10.3748/wjg.v17.i18.2259
44. Scherf KA, Koehler P, Wieser H. Gluten and wheat sensitivities - an overview. J Cereal Sci. (2016) 67:2–11. doi: 10.1016/j.jcs.2015.07.008
45. Janatuinen EK, Kemppainen TA, Julkunen RJK, Kosma VM, Maki M, Heikkinen M, et al. No harm from five year ingestion of oats in coeliac disease. Gut. (2002) 50:332–5. doi: 10.1136/gut.50.3.332
46. Pulido OM, Gillespie Z, Zarkadas M, Dubois S, Vavasour E, Rashid M, et al. Introduction of oats in the diet of individuals with celiac disease: a systematic review. Adv Food Nutr Res. (2009) 57:235–85. doi: 10.1016/S1043-4526(09)57006-4
47. Fric P, Gabrovska D, Nevoral J. Celiac disease, gluten-free diet, and oats. Nutr Rev. (2011) 69:107–15. doi: 10.1111/j.1753-4887.2010.00368.x
48. Comino I, Moreno M, Sousa C. Role of oats in celiac disease. World J Gastroenterol. (2015) 21:11825–31. doi: 10.3748/wjg.v21.i41.11825
49. Hardy MY, Tye-Din JA, Stewart JA, Schmitz F, Dudek NL, Hanchapola I, et al. Ingestion of oats and barley in patients with celiac disease mobilizes cross-reactive T cells activated by avenin peptides and immuno-dominant hordein peptides. J Autoimmun. (2015) 56:56–65. doi: 10.1016/j.jaut.2014.10.003
50. Gilissen LJ, Van der Meer IM, Smulders MJ. Why oats are safe and healthy for celiac disease patients. Med Sci. (2016) 4:21. doi: 10.3390/medsci4040021
51. De Souza MCP, Deschenes ME, Laurencelle S, Godet P, Roy CC, Djilali-Saiah I. Pure oats as part of the canadian gluten-free diet in celiac disease: the need to revisit the issue. Can J Gastroenterol Hepatol. (2016) 2016:1576360. doi: 10.1155/2016/1576360
52. Camarca A, Anderson RP, Mamone G, Fierro O, Facchiano A, Costantini S, et al. Intestinal T cell responses to gluten peptides are largely heterogeneous: implications for a peptide-based therapy in celiac disease. J Immunol. (2009) 182:4158–66. doi: 10.4049/jimmunol.0803181
53. Singh P, Arora A, Strand TA, Leffler DA, Catassi C, Green PH, et al. Global prevalence of celiac disease: systematic review and meta-analysis. Clin Gastroenterol Hepatol. (2018) 16:823–36. doi: 10.1016/j.cgh.2017.06.037
54. Green PH, Cellier C. Celiac disease. N Engl J Med. (2007) 357:1731–43. doi: 10.1056/NEJMra071600
55. Nardecchia S, Auricchio R, Discepolo V, Troncone R. Extra-intestinal manifestations of coeliac disease in children: clinical features and mechanisms. Front Pediatr. (2019) 7:56. doi: 10.3389/fped.2019.00056
56. Hamer RJ. Coeliac disease: background and biochemical aspects. Biotechnol Adv. (2005) 23:401–8. doi: 10.1016/j.biotechadv.2005.05.005
57. Green PH, Rostami K, Marsh MN. Diagnosis of coeliac disease. Best Pract Res Clin Gastroenterol. (2005) 19:389–400. doi: 10.1016/j.bpg.2005.02.006
58. Stordal K, Bakken IJ, Suren P, Stene LC. Epidemiology of coeliac disease and comorbidity in Norwegian children. J Pediatr Gastroenterol Nutr. (2013) 57:467–71. doi: 10.1097/MPG.0b013e3182a455dd
59. Leffler DA, Green PHR, Fasano A. Extraintestinal manifestations of coeliac disease. Nat Rev Gastroenterol Hepatol. (2015) 12:561–71. doi: 10.1038/nrgastro.2015.131
60. Lundin KE, Wijmenga C. Coeliac disease and autoimmune disease-genetic overlap and screening. Nat Rev Gastroenterol Hepatol. (2015) 12:507–15. doi: 10.1038/nrgastro.2015.136
61. Canova C, Pitter G, Ludvigsson JF, Romor P, Zanier L, Zanotti R, et al. Celiac disease and risk of autoimmune disorders: a population based matched birth cohort study. J Pediatr. (2016) 174:146–52. doi: 10.1016/j.jpeds.2016.02.058
62. Assa A, Frenkel-Nir Y, Tzur D, Katz LH, Shamir R. Large population study shows that adolescents with celiac disease have an increased risk of multiple autoimmune and nonautoimmune comorbidities. Acta Paediatr. (2017) 106:967–72. doi: 10.1111/apa.13808
63. Hagopian W, Lee HS, Liu E, Rewers M, She JX, Ziegler AG, et al. Co-occurrence of type 1 diabetes and celiac disease autoimmunity. Pediatrics. (2017) 140:e20171305. doi: 10.1542/peds.2017-1305
64. Tack GJ, Verbeek WHM, Schreurs MWJ, Mulder CJJ. The spectrum of celiac disease: epidemiology, clinical aspects and treatment. Nat Rev Gastroenterol Hepatol. (2010) 7:204–13. doi: 10.1038/nrgastro.2010.23
65. Hardy MY, Tye-Din JA. Coeliac disease: a unique model for investigating broken tolerance in autoimmunity. Clin Transl Immunol. (2016) 5:e112. doi: 10.1038/cti.2016.58
66. Qi PF, Wei YM, Yue YW, Yan ZH, Zheng YL. Biochemical and molecular characterization of gliadins. J Mol Biol. (2006) 40:713–23. doi: 10.1134/S0026893306050050
67. Ruiz-Carnicer A, Comino I, Segura V, Ozuna CV, Moreno ML, Lopez-Casado MA, et al. Celiac immunogenic potential of α-gliadin epitope variants from Triticum and Aegilops species. Nutrients. (2019) 11:220. doi: 10.3390/nu11020220
68. Wang DW, Li D, Wang J, Zhao Y, Wang Z, Yue G, et al. Genome-wide analysis of complex wheat gliadins, the dominant carriers of celiac disease epitopes. Sci Rep. (2017) 7:44609. doi: 10.1038/srep44609
69. Kumar A, Kapoor P, Chunduri V, Sharma S, Garg M. Potential of Aegilops sp. for improvement of grain processing and nutritional quality in wheat (Triticum aestivum) Front Plant Sci. (2019) 10:308. doi: 10.3389/fpls.2019.00308
70. Sollid LM, Qiao SW, Anderson RP, Gianfrani C, Koning F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics. (2012) 64:455–60. doi: 10.1007/s00251-012-0599-z
71. Shewry PR, Tatham AS. Improving wheat to remove celiac epitopes but retain functionality. J Cereal Sci. (2016) 67:12–21. doi: 10.1016/j.jcs.2015.06.005
72. Shan L, Molberg O, Parrot I, Hausch F, Filiz F, Gray GM, et al. Structural basis for gluten intolerance in celiac sprue. Science. (2002) 297:2275–9. doi: 10.1126/science.1074129
73. Qiao SW, Bergseng E, Molberg O, Jung G, Fleckenstein B, Sollid LM. Refining the rules of gliadin T cell epitope binding to the disease-associated DQ2 molecule in celiac disease: importance of proline spacing and glutamine deamidation. J Immunol. (2005) 175:254–61. doi: 10.4049/jimmunol.175.1.254
74. Moron B, Bethune MT, Comino I, Manyani H, Ferragud M, Lopez MC, et al. Toward the assessment of food toxicity for celiac patients: characterization of monoclonal antibodies to a main immunogenic gluten peptide. PLoS ONE. (2008) 3:e2294. doi: 10.1371/journal.pone.0002294
75. Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AV. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med. (2000) 6:337–42. doi: 10.1038/73200
76. Arentz-Hansen H, Korner R, Molberg O, Quarsten H, Vader W, Kooy YMC, et al. The intestinal T cell response to α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med. (2000) 191:603–12. doi: 10.1084/jem.191.4.603
77. Qiao SW, Bergseng E, Molberg O, Xia J, Fleckenstein B, Khosla C, et al. Antigen presentation to celiac lesion-derived T cells of a 33-mer gliadin peptide naturally formed by gastrointestinal digestion. J Immunol. (2004) 173:1757–62. doi: 10.4049/jimmunol.173.3.1757
78. Anderson OD, Litts JC, Greene FC. The α-gliadin gene family. I. Characterization of ten new wheat α-gliadin genomic clones, evidence for limited sequence conservation of flanking DNA, and southern analysis of the gene family. Theor Appl Genet. (1997) 95:50–8. doi: 10.1007/s001220050531
79. Van Herpen TWJM, Goryunova SV, Van der Schoot J, Mitreva M, Salentijn E, Vorst O, et al. Alpha-gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes. BMC Genomics. (2006) 7:1. doi: 10.1186/1471-2164-7-1
80. Xie Z, Wang C, Wang K, Wang S, Li X, Zhang Z, et al. Molecular characterization of the celiac disease epitope domains in α-gliadin genes in Aegilops tauschii and hexaploid wheats (Triticum aestivum L.). Theor Appl Genet. (2010) 121:1239–51. doi: 10.1007/s00122-010-1384-8
81. Salentijn EM, Mitea DC, Goryunova SV, Van der Meer IM, Padioleau I, Gilissen LJWJ, et al. Celiac disease T-cell epitopes from gamma-gliadins: immunoreactivity depends on the genome of origin, transcript frequency, and flanking protein variation. BMC Genomics. (2012) 13:277. doi: 10.1186/1471-2164-13-277
82. Wang S, Shen X, Ge P, Li J, Subburaj S, Li X, et al. Molecular characterization and dynamic expression patterns of two types of γ-gliadin genes from Aegilops and Triticum species. Theor Appl Genet. (2012) 125:1371–84. doi: 10.1007/s00122-012-1917-4
83. Anderson OD, Hsia CC, Torres V. The wheat γ-gliadin genes: characterization of ten new sequences and further understanding of γ-gliadin gene family structure. Theor Appl Genet. (2001) 103:323–30. doi: 10.1007/s00122-001-0551-3
84. Mitea C, Salentijn EMJ, Van Veelen P, Goryunova SV, Van der Meer IM, Van den Broeck HC, et al. A universal approach to eliminate antigenic properties of alpha-gliadin peptides in celiac disease. PLoS ONE. (2010) 5:e15637. doi: 10.1371/journal.pone.0015637
85. Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, Van Rood JJ, et al. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci USA. (2003) 100:12390–5. doi: 10.1073/pnas.2135229100
86. Verdu EF, Galipeau HJ, Jabri B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat Rev Gastroenterol Hepatol. (2015) 12:497–506. doi: 10.1038/nrgastro.2015.90
87. Lindfors K, Ciacci C, Kurppa K, Lundin KEA, Makharia GK, Mearin ML, et al. Coeliac disease. Nat Rev Dis Primers. (2019) 5:3. doi: 10.1038/s41572-018-0054-z
88. Ciccocioppo R, Di Sabatino A, Corazza GR. The immune recognition of gluten in coeliac disease. Clin Exp Immunol. (2005) 140:408–16. doi: 10.1111/j.1365-2249.2005.02783.x
89. Kupfer SS, Jabri B. Pathophysiology of celiac disease. Gastrointest Endosc Clin N Am. (2012) 22:639–60. doi: 10.1016/j.giec.2012.07.003
90. Parzanese I, Qehajaj D, Patrinicola F, Aralica M, Chiriva-Internati M, Stifter S, et al. Celiac disease: From pathophysiology to treatment. World J Gastrointest Pathophysiol. (2017) 8:27–38. doi: 10.4291/wjgp.v8.i2.27
91. Malalgoda M, Manthey F, Simsek S. Reducing the celiac disease antigenicity of wheat. Cereal Chem. (2018) 95:49–58. doi: 10.1094/CCHEM-05-17-0100-FI
92. Fasano A. Intestinal permeability and its regulation by zonulin: diagnostic and therapeutic implications. Clin Gastroenterol Hepatol. (2012) 10:1096–100. doi: 10.1016/j.cgh.2012.08.012
93. Fasano A, Not T, Wang W, Uzzau S, Berti I, Tommasini A, et al. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet. (2000) 355:1518–9. doi: 10.1016/S0140-6736(00)02169-3
94. Menard S, Lebreton C, Schumann M, Matysiak-Budnik T, Dugave C, Bouhnik Y, et al. Paracellular versus transcellular intestinal permeability to gliadin peptides in active celiac disease. Am J Pathol. (2012) 180:608–15. doi: 10.1016/j.ajpath.2011.10.019
95. De Re V, Magris R, Cannizzaro R. New insights into the pathogenesis of celiac disease. Front Med. (2017) 4:137. doi: 10.3389/fmed.2017.00137
96. Vader W, Kooy Y, Van Veelen P, De Ru A, Harris D, Benckhuijsen W, et al. The gluten response in children with recent onset celiac disease: a highly diverse response towards multiple gliadin and glutenin derived peptides. Gastroenterology. (2002) 122:1729–37. doi: 10.1053/gast.2002.33606
97. Husby S, Murray JA. Diagnosing coeliac disease and the potential for serological markers. Nat Rev Gastroenterol Hepatol. (2014) 11:655–63. doi: 10.1038/nrgastro.2014.162
98. Sabatino AD, Vanoli A, Giuffrida P, Luinetti O, Solcia E, Corazza GR. The function of tissue transglutaminase in celiac disease. Autoimmun Rev. (2012) 11:746–53. doi: 10.1016/j.autrev.2012.01.007
99. Sollid LM, Jabri B. Celiac disease and transglutaminase 2: a model for posttranslational modification of antigens and HLA association in the pathogenesis of autoimmune disorders. Curr Opin Immunol. (2011) 23:732–8. doi: 10.1016/j.coi.2011.08.006
100. Costantini S, Rossi M, Colonna G, Facchiano AM. Modelling of HLA-DQ2 and its interaction with gluten peptides to explain molecular recognition in celiac disease. J Mol Graphics Modell. (2005) 23:419–31. doi: 10.1016/j.jmgm.2004.12.002
101. Qiao SW, Sollid LM, Blumberg RS. Antigen presentation in celiac disease. Curr Opin Immunol. (2009) 21:111–7. doi: 10.1016/j.coi.2009.03.004
102. Bodd M, Raki M, Tollefsen S, Fallang LE, Bergseng E, Lundin KE, et al. HLA-DQ2- restricted gluten-reactive T cells produce IL-21 but not IL-17 or IL-22. Mucosal Immunol. (2010) 3:594–601. doi: 10.1038/mi.2010.36
103. Gianfrani C, Auricchio S, Troncone R. Adaptive and innate immune responses in celiac disease. Immunol Lett. (2005) 99:141–5. doi: 10.1016/j.imlet.2005.02.017
104. Bjorck S, Lindehammer SR, Fex M, Agardh D. Serum cytokine pattern in young children with screening detected coeliac disease. Clin Exp Immunol. (2015) 179:230–5. doi: 10.1111/cei.12454
105. Troncone R, Jabri B. Coeliac disease and gluten sensitivity. J Intern Med. (2011) 269:582–90. doi: 10.1111/j.1365-2796.2011.02385.x
106. Palova-Jelinkova L, Danova K, Drasarova H, Dvorak M, Funda DP, Fundova P, et al. Pepsin digest of wheat gliadin fraction increases production of IL-1β via TLR4/MyD88/TRIF/MAPK/NF-κB signaling pathway and an NLRP3 inflammasome activation. PLoS ONE. (2013) 8:e62426. doi: 10.1371/journal.pone.0062426
107. Kuja-Halkola R, Lebwohl B, Halfvarson J, Wijmenga C, Magnusson PK, Ludvigsson JF. Heritability of non-HLA genetics in coeliac disease: a population-based study in 107000 twins. Gut. (2016) 65:1793–8. doi: 10.1136/gutjnl-2016-311713
108. Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. (2010) 42:295–302. doi: 10.1038/ng.543
109. Silano M, Agostoni C, Guandalini S. Effect of the timing of gluten introduction on the development of celiac disease. World J Gastroenterol. (2010) 16:193–1942. doi: 10.3748/wjg.v16.i16.1939
110. Troncone R, Auricchio S. Rotavirus and celiac disease: clues to the pathogenesis and perspectives on prevention. J Pediatr Gastroenterol Nutr. (2007) 44:527–8. doi: 10.1097/MPG.0b013e31804ca0ec
111. Rubio-Tapia A, Murray JA. Celiac disease. Curr Opin Gastroenterol. (2010) 26:116–22. doi: 10.1097/MOG.0b013e3283365263
112. Jansen M, Van Zelm M, Groeneweg M, Jaddoe V, Dik W, Schreurs M, et al. The identification of celiac disease in asymptomatic children: the Generation R study. J Gastroenterol. (2018) 53:377–86. doi: 10.1007/s00535-017-1354-x
113. Valle J, Morgado JMT, Ruiz-Martin J, Guardiola A, Lopes-Nogueras M, Garcia-Vela A, et al. Flow cytometry of duodenal intraepithelial lymphocytes improves diagnosis of celiac disease in difficult cases. United Eur Gastroenterol J. (2017) 5:819–26. doi: 10.1177/2050640616682181
114. Bhatnagar C, Tandon N. Diagnosis of celiac disease. Indian J Pediatr. (2006) 73:703–9. doi: 10.1007/BF02898449
115. Volta U, Granito A, Parisi C, Fabbri A, Fiorini E, Piscaglia M, et al. Deamidated gliadin peptide antibodies as a routine test for celiac disease: a prospective analysis. J Clin Gastroenterol. (2010) 44:186–90. doi: 10.1097/MCG.0b013e3181c378f6
116. Lebwohl B, Rubio-Tapia A, Assiri A, Newland C, Guandalini S. Diagnosis of celiac disease. Gastrointest Endosc Clin N Am. (2012) 22:661–77. doi: 10.1016/j.giec.2012.07.004
117. Elli L, Branchi F, Tomba C, Villalta D, Norsa L, Ferretti F, et al. Diagnosis of gluten related disorders: Celiac disease, wheat allergy and non-celiac gluten sensitivity. World J Gastroenterol. (2015) 21:7110. doi: 10.3748/wjg.v21.i23.7110
118. Tye-Din JA, Galipeau HJ, Agardh D. Celiac disease: a review of current concepts in pathogenesis, prevention, and novel therapies. Front Pediatr. (2018) 6:350. doi: 10.3389/fped.2018.00350
119. Al-Toma A, Volta U, Auricchio R, Castillejo G, Sanders DS, Cellier C, et al. European Society for the Study of Coeliac Disease (ESsCD) guideline for coeliac disease and other gluten-related disorders. United Eur Gastroenterol J. (2019) 7:583–613. doi: 10.1177/2050640619844125
120. Ransford RA, Hayes M, Palmer M, Hall MJ. A controlled, prospective screening study of celiac disease presenting as iron deficiency anemia. J Clin Gastroenterol. (2002) 35:228–33. doi: 10.1097/00004836-200209000-00006
121. Kalayci AG, Kanber Y, Birinci A, Yildiz L, Albayrak D. The prevalence of coeliac disease as detected by screening in children with iron deficiency anaemia. Acta Paediatr. (2005) 94:678–81. doi: 10.1111/j.1651-2227.2005.tb01964.x
122. Ludvigsson JF, Bai JC, Biagi F, Card TR, Ciacci C, Ciclitira PJ, et al. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut. (2014) 63:1210–28. doi: 10.1136/gutjnl-2013-306578
123. Rashtak S, Ettore MW, Homburger HA, Murray JA. Comparative usefulness of deamidated gliadin antibodies in the diagnosis of celiac disease. Clin Gastroenterol Hepatol. (2008) 6:426–32. doi: 10.1016/j.cgh.2007.12.030
124. Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO, et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. (1997) 3:797–801. doi: 10.1038/nm0797-797
125. Sblattero D, Berti I, Trevisiol C, Marzari R, Tommasini A, Bradbury A, et al. Human recombinant tissue transglutaminase ELISA: an innovative diagnostic assay for celiac disease. Am J Gastroenterol. (2000) 95:1253–7. doi: 10.1111/j.1572-0241.2000.02018.x
126. Lauret E, Rodrigo L. Celiac disease and autoimmune-associated conditions. BioMed Res Int. (2013) 2013. doi: 10.1155/2013/127589
127. Chorzelski TP, Sulej J, Tchorzewska H, Jablonska S, Beutner EH, Kumar V. IgA class endomysium antibodies in dermatitis herpetiformis and coeliac disease. Ann N Y Acad Sci. (1983) 420:325–34. doi: 10.1111/j.1749-6632.1983.tb22220.x
128. Chorzelski TP, Beutner EH, Sulej J, Tchorzewska H, Jablonska S, Kumar V, et al. IgA anti-endomysium antibody. A new immunological marker of dermatitis herpetiformis and coeliac disease. Br J Dermatol. (1984) 111:395–402. doi: 10.1111/j.1365-2133.1984.tb06601.x
129. Kaukinen K, Partanen J, Maki M, Collin P. HLA-DQ typing in the diagnosis of celiac disease. Am J Gastroenterol. (2002) 97:695–9. doi: 10.1111/j.1572-0241.2002.05471.x
130. Lebwohl B, Murray JA, Rubio-Tapia A, Green PH, Ludvigsson JF. Predictors of persistent villous atrophy in coeliac disease: a population-based study. Aliment Pharmacol Ther. (2014) 39:488–95. doi: 10.1111/apt.12621
131. Thijs WJ, Van Baarlen J, Kleibeuker JH, Kolkman JJ. Duodenal versus jejunal biopsies in suspected celiac disease. Endoscopy. (2004) 36:993–6. doi: 10.1055/s-2004-825954
132. Dewar DH, Ciclitira PJ. Clinical features and diagnosis of celiac disease. Gastroenterology. (2005) 128:S19–S24. doi: 10.1053/j.gastro.2005.02.010
133. Hahn M, Hagel AF, Hirschmann S, Bechthold C, Konturek P, Neurath M, et al. Modern diagnosis of celiac disease and relevant differential diagnoses in the case of cereal intolerance. Allergo J Int. (2014) 23:67–77. doi: 10.1007/s40629-014-0006-4
134. Siriweera E, Qi Z, Yong JLC. Validity of intraepithelial lymphocyte count in the diagnosis of celiac disease: a histopathological study. Int J Celiac Dis. (2015) 3:156–8. doi: 10.12691/ijcd-3-4-7
135. Pena AS. What is the best histopathological classification for celiac disease? Does it matter? Gastroenterol Hepatol Bed Bench. (2015) 8:239–43.
136. Catassi C, Fasano A. Celiac disease diagnosis: simple rules are better than complicated algorithms. Am J Med. (2010) 123:691–3. doi: 10.1016/j.amjmed.2010.02.019
137. Yoosuf S, Makharia GK. Evolving therapy for celiac disease. Front Pediatr. (2019) 7:193. doi: 10.3389/fped.2019.00193
138. Kaukinen K, Lindfors K, Maki M. Advances in the treatment of coeliac disease: an immunopathogenic perspective. Nat Rev Gastroenterol Hepatol. (2014) 11:36–44. doi: 10.1038/nrgastro.2013.141
139. Jouanin A, Gilissen LJWJ, Boyd LA, Cockram J, Leigh FJ, Wallington EJ, et al. Food processing and breeding strategies for coeliac-safe and healthy wheat products. Food Res Int. (2018) 110:11–21. doi: 10.1016/j.foodres.2017.04.025
140. Meijer C, Shamir R, Szajewska H, Mearin L. Celiac disease prevention. Front Pediatr. (2018) 6:368. doi: 10.3389/fped.2018.00368
141. Ribeiro M, Nunes FM, Rodriguez-Quijano M, Carrillo JM, Branlard G, Igrejas G. Next-generation therapies for celiac disease: the gluten-targeted approaches. Trends Food Sci Technol. (2018) 75:56–71. doi: 10.1016/j.tifs.2018.02.021
142. Scherf KA, Wieser H, Koehler P. Novel approaches for enzymatic gluten degradation to create high-quality gluten-free products. Food Res Int. (2018) 110:62–72. doi: 10.1016/j.foodres.2016.11.021
143. Gil-Humanes J, Piston F, Tollefsen S, Sollid LM, Barro F. Effective shutdown in the expression of celiac disease-related wheat gliadin T-cell epitopes by RNA interference. Proc Natl Acad Sci USA. (2010) 107:17023–8. doi: 10.1073/pnas.1007773107
144. Piston F, Gil-Humanes J, Barro F. Integration of promoters, inverted repeat sequences and proteomic data into a model for high silencing efficiency of coeliac disease related gliadins in bread wheat. BMC Plant Biol. (2013) 13:136. doi: 10.1186/1471-2229-13-136
145. Zorb C, Becker D, Hasler M, Muhling KH, Godde V, Niehaus K, et al. Silencing of the sulfur rich α-gliadin storage protein family in wheat grains (Triticum aestivum L.) causes no unintended side-effects on other metabolites. Front Plant Sci. (2013) 4:369. doi: 10.3389/fpls.2013.00369
146. Zhang X, Zhang B, Wu H, Lu C, Lu G, Liu D, et al. Effect of high-molecular-weight glutenin subunit deletion on soft wheat quality properties and sugar-snap cookie quality estimated through near-isogenic lines. J Integr Agric. (2018) 17:1066–73. doi: 10.1016/S2095-3119(17)61729-5
147. Sanchez-Leon S, Gil-Humanes J, Ozuna CV, Gimenez MJ, Sousa C, Voytas DF, et al. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. (2018) 16:902–10. doi: 10.1111/pbi.12837
148. Van den Broeck HC, Van Herpen TW, Schuit C, Salentijn EM, Dekking L, Bosch D, et al. Removing celiac disease-related gluten proteins from bread wheat while retaining technological properties: a study with Chinese Spring deletion lines. BMC Plant Biol. (2009) 9:41. doi: 10.1186/1471-2229-9-41
149. Camerlengo F, Sestili F, Silvestri M, Colaprico G, Margiotta B, Ruggeri R, et al. Production and molecular characterization of bread wheat lines with reduced amount of α-type gliadins. BMC Plant Biol. (2017) 17:248. doi: 10.1186/s12870-017-1211-3
150. Wang Z, Li Y, Yang Y, Liu X, Qin H, Dong Z, et al. New insight into the function of wheat glutenin proteins as investigated with two series of genetic mutants. Sci Rep. (2017) 7:3428. doi: 10.1038/s41598-017-03393-6
151. Van den Broeck HC, Gilissen LJWJ, Smulders MJM, Van der Meer IM, Hamer RJ. Dough quality of bread wheat lacking α-gliadins with celiac disease epitopes and addition of celiac-safe avenins to improve dough quality. J Cereal Sci. (2011) 53:206–16. doi: 10.1016/j.jcs.2010.12.004
152. Liu S, Zhao S, Chen F, Xia G. Generation of novel high quality HMW-GS genes in two introgression lines of Triticum aestivum/Agropyron elongatum. BMC Evol Biol. (2007) 7:76. doi: 10.1186/1471-2148-7-76
153. Chen F, Xu C, Chen M, Wang Y, Xia G. A new α-gliadin gene family for wheat breeding: Somatic introgression line II-12 derived from Triticum aestivum and Agropyron elongatum. Mol Breed. (2008) 22:675–85. doi: 10.1007/s11032-008-9208-0
154. Vita PD, Ficco DB, Luciani A, Vincentini O, Pettoello-Mantovani M, Silano M, et al. A ω-secalin contained decamer shows a celiac disease prevention activity. J Cereal Sci. (2012) 55:234–42. doi: 10.1016/j.jcs.2011.12.006
155. Shan L, Marti T, Sollid LM, Gray GM, Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J. (2004) 383:311–8. doi: 10.1042/BJ20040907
156. Mitea C, Havenaar R, Drijfhout JW, Edens L, Dekking L, Koning FC. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: implications for coeliac disease. Gut. (2008) 57:25–32. doi: 10.1136/gut.2006.111609
157. Cinova J, De Palma G, Stepankova R, Kofronova O, Kverka M, Sanz Y, et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS ONE. (2011) 6:e16169. doi: 10.1371/journal.pone.0016169
158. Quagliariello A, Aloisio I, Cionci NB, Luiselli D, D'Auria G, Martinez-Priego L, et al. Effect of Bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: a pilot study. Nutrients. (2016) 8:E660. doi: 10.3390/nu8100660
159. Cristofori F, Indrio F, Miniello VL, De Angelis M, Francavilla R. Probiotics in celiac disease. Nutrients. (2018) 10:e1824. doi: 10.3390/nu10121824
160. Chapman TM, Plosker G, Figgitg DP. VSL#3 probiotic mixture: a review of its use in chronic inflammatory bowel diseases. Drugs. (2006) 66:1371–87. doi: 10.2165/00003495-200666100-00006
161. De Angelis M, Rizzelo CG, Fasano A, Clemente MG, De Simone C, Silano M, et al. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for celiac sprue. Biochim Biophys Acta. (2006) 1762:80–93. doi: 10.1016/j.bbadis.2005.09.008
162. Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. (2010) 51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e
163. Smecuol E, Hwang HJ, Sugai E, Corso L, Chernavsky AC, Bellavite FP, et al. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacterium infantis Natren Life Start strain super strain in active celiac disease. J Clin Gastroenterol. (2013) 47:139–47. doi: 10.1097/MCG.0b013e31827759ac
164. Cagno RD, Rizzello CG, Angelis MD, Cassone A, Giuliani G, Benedusi A, et al. Use of selected sourdough strains of Lactobacillus for removing gluten and enhancing the nutritional properties of gluten-free bread. J Food Prot. (2008) 71:1491–5. doi: 10.4315/0362-028X-71.7.1491
165. Brzozowski B. Immunoreactivity of wheat proteins modified by hydrolysis and polymerisation. Eur Food Res Technol. (2016) 242:1025–40. doi: 10.1007/s00217-015-2608-6
166. De Palma G, Cinova J, Stepankova R, Tuckova L, Sanz Y. Pivotal advance: bifidobacteria and Gram-negative bacteria differentially influence immune responses in the proinflammatory milieu of celiac disease. J Leukocyte Biol. (2010) 87:765–78. doi: 10.1189/jlb.0709471
167. Medina M, De Palma G, Ribes-Koninckx C, Calabuig M, Sanz Y. Bifidobacterium strains suppress in vitro the pro-inflammatory milieu triggered by the large intestinal microbiota of coeliac patients. J Inflamm. (2008) 5:19. doi: 10.1186/1476-9255-5-19
168. Lindfors K, Blomqvist T, Juuti-Uusitalo K, Stenman S, Venalainen J, Maki M, et al. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin Exp Immunol. (2008) 152:552–8. doi: 10.1111/j.1365-2249.2008.03635.x
169. Laparra JM, Sanz Y. Bifidobacteria inhibit the inflammatory response induced by gliadins in intestinal epithelial cells via modifications of toxic peptide generation during digestion. J Cell Biochem. (2010) 109:801–7. doi: 10.1002/jcb.22459
170. Laparra JM, Olivares M, Gallina O, Sanz Y. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS ONE. (2012) 7:e30744. doi: 10.1371/journal.pone.0030744
171. D'Arienzo R, Stefanile R, Maurano F, Mazzarella G, Ricca E, Troncone R, et al. Immunomodulatory effects of Lactobacillus casei administration in a mouse model of gliadin-sensitive enteropathy. Scand J Immunol. (2011) 74:335–41. doi: 10.1111/j.1365-3083.2011.02582.x
172. Liang L, Pinier M, Leroux JC, Subirade M. Interaction of α-gliadin with poly (HEMA-co-SS): structural characterization and biological implication. Biopolymers. (2009) 91:169–78. doi: 10.1002/bip.21109
173. Pinier M, Verdu EF, Nasser-Eddine M, David CS, Vezina A, Rivard N, et al. Polymeric binders suppress gliadin-induced toxicity in the intestinal epithelium. Gastroenterology. (2009) 136:288–98. doi: 10.1053/j.gastro.2008.09.016
174. Engstrom N, Saenz-Mendez P, Scheers J, Scheers N. Towards celiac-safe foods: decreasing the affinity of transglutaminase 2 for gliadin by addition of ascorbyl palmitate and ZnCl2 as detoxifiers. Sci Rep. (2017) 7:77. doi: 10.1038/s41598-017-00174-z
175. Makharia GK. Current and emerging therapy for celiac disease. Front Med. (2014) 1:6. doi: 10.3389/fmed.2014.00006
176. Osorio C, Wen N, Gemini R, Zemetra R, Von Wettstein D, Rustgi S. Targeted modification of wheat grain protein to reduce the content of celiac causing epitopes. Funct Integr Genomics. (2012) 12:417–38. doi: 10.1007/s10142-012-0287-y
177. Bethune MT, Strop P, Tang Y, Sollid LM, Khosla C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem Biol. (2006) 13:637–47. doi: 10.1016/j.chembiol.2006.04.008
178. Gass J, Vora H, Bethune MT, Gray GM, Khosla C. Effect of barley endoprotease EP-B2 on gluten digestion in the intact rat. J Pharmacol Exp Ther. (2006) 318:1178–86. doi: 10.1124/jpet.106.104315
179. Gass J, Bethune MT, Siegel M, Spencer A, Khosla C. Combination enzyme therapy for gastric digestion of dietary gluten in patients with celiac sprue. Gastroenterology. (2007) 133:472–80. doi: 10.1053/j.gastro.2007.05.028
180. Tye-Din JA, Anderson RP, Ffrench RA, Brown GJ, Hodsman P, Siegel M, et al. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin Immunol. (2010) 134:289–95. doi: 10.1016/j.clim.2009.11.001
181. Heredia-Sandoval NG, De La Barca AMC, Carvajal-Millan E, Islas-Rubio AR. Amaranth addition to enzymatically modified wheat flour improves dough functionality, bread immunoreactivity and quality. Food Funct. (2018) 9:534–40. doi: 10.1039/C7FO01332A
182. Walter T, Wieser H, Koehler P. Degradation of gluten in wheat bran and bread drink by means of a proline-specific peptidase. J Nutr Food Sci. (2014) 4:1000293. doi: 10.4172/2155-9600.1000293
183. Rees D, Holtrop G, Chope G, Moar KM, Cruickshank M, Hoggard N. A randomised, double-blind, cross-over trial to evaluate bread, in which gluten has been pre-digested by prolyl endoprotease treatment, in subjects self-reporting benefits of adopting a gluten-free or low-gluten diet. Br J Nutr. (2018) 119:496–506. doi: 10.1017/S0007114517003749
184. Wolf C, Siegel JB, Tinberg C, Camarca A, Gianfrani C, Paski S, et al. Engineering of Kuma030: A gliadin peptidase that rapidly degrades immunogenic gliadin peptides in gastric conditions. J Am Chem Soc. (2015) 137:13106–13. doi: 10.1021/jacs.5b08325
185. Rey M, Yang ML, Lee L, Zhang Y, Sheff JG, Sensen CW, et al. Addressing proteolytic efficiency in enzymatic degradation therapy for celiac disease. Sci Rep. (2016) 6:30980. doi: 10.1038/srep30980
186. Wall AJ, Douglas AP, Booth CC, Pearse AGE. Response of the jejunal mucosa in adult coeliac disease to oral prednisolone. Gut. (1970) 11:7–14. doi: 10.1136/gut.11.1.7
187. Mitchison HC, Al Mardini H, Gillespie S, Laker M, Zaitoun A, Record CO. A pilot study of fluticasone propionate in untreated coeliac disease. Gut. (1991) 32:260–5. doi: 10.1136/gut.32.3.260
188. Przemioslo RT, Lundin KEA, Sollid LM, Nelufer J, Ciclitira PJ. Histological changes in bowel mucosa induced by gliadin sensitive T lymphocytes can be blocked by interferon γ antibody. Gut. (1995) 36:874–9. doi: 10.1136/gut.36.6.874
189. Gillett HR, Arnott ID, McIntyre M, Campbell S, Dahele A, Priest M, et al. Successful infliximab treatment for steroid-refractory celiac disease: a case report. Gastroenterology. (2002) 122:800–5. doi: 10.1053/gast.2002.31874
190. Sollid LM, Khosla C. Novel therapies for coeliac disease. J Intern Med. (2011) 269:604–13. doi: 10.1111/j.1365-2796.2011.02376.x
191. Salvati VM, Mazzarella G, Gianfrani C, Levings MK, Stefanile R, De Giulio B, et al. Recombinant human interleukin 10 suppresses gliadin dependent T cell activation in ex vivo cultured coeliac intestinal mucosa. Gut. (2005) 54:46–53. doi: 10.1136/gut.2003.023150
192. Bakshi A, Stephen S, Borum ML, Doman DB. Emerging therapeutic options for celiac disease: potential alternatives to a gluten-free diet. Gastroenterol Hepatol. (2012) 8:582–588.
193. Ribeiro M, Nunes FM, Guedes S, Domingues P, Silva AM, Carrillo JM, et al. Efficient chemo-enzymatic gluten detoxification: reducing toxic epitopes for celiac patients improving functional properties. Sci Rep. (2015) 5:18041. doi: 10.1038/srep18041
194. Heredia-Sandoval NG, Islas-Rubio AR, Cabrera-Chavez F, De La Barca AMC. Transamidation of gluten proteins during the bread-making process of wheat flour to produce breads with less immunoreactive gluten. Food Funct. (2014) 5:1813–8. doi: 10.1039/C4FO00118D
195. Jeitner TM, Pinto JT, Cooper A. Cystamine and cysteamine as inhibitors of transglutaminase activity in vivo. Biosci Rep. (2018) 38:BSR20180691. doi: 10.1042/BSR20180691
196. Ventura MAE, Sajko K, Hils M, Pasternack R, Greinwald R, Tewes B, et al. The oral transglutaminase 2 (TG2) inhibitor Zed1227 blocks TG2 activity in a mouse model of intestinal inflammation. Gastroenterology. (2018) 154:S-490. doi: 10.1016/S0016-5085(18)31861-4
197. Gujral N, Freeman HJ, Thomson ABR. Celiac disease: prevalence, diagnosis, pathogenesis and treatment. World J Gastroenterol. (2012) 18:6036–59. doi: 10.3748/wjg.v18.i42.6036
198. Di Pierro M, Lu R, Uzzau S, Wang W, Margaretten K, Pazzani C, et al. Zonula occludens toxin structure-function analysis. Identification of the fragment biologically active on tight junctions and of the zonulin receptor binding domain. J Biol Chem. (2001) 276:19160–5. doi: 10.1074/jbc.M009674200
199. Drago S, El Asmar R, Di Pierro M, Clemente MG, Tripathi A, Sapone A, et al. Gliadin, zonulin and gut permeability: effects on celiac and non-celiac intestinal mucosa and intestinal cell lines. Scand J Gastroenterol. (2006) 41:408–19. doi: 10.1080/00365520500235334
200. Paterson B, Lammers K, Arrieta M, Fasano A, Meddings J. The safety, tolerance, pharmacokinetic and pharmacodynamic effects of single doses of AT-1001 in coeliac disease subjects: a proof of concept study. Aliment Pharmacol Ther. (2007) 26:757–66. doi: 10.1111/j.1365-2036.2007.03413.x
201. Hue S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J, et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. (2004) 21:367–77. doi: 10.1016/j.immuni.2004.06.018
202. Sarra M, Cupi ML, Monteleone I, Franze E, Ronchetti G, Di Sabatino A, et al. IL-15 positively regulates IL-21 production in celiac disease mucosa. Mucosal Immunol. (2013) 6:244–55. doi: 10.1038/mi.2012.65
203. Lahdeaho ML, Scheinin M, Pesu M, Kivela L, Lovro Z, Keisala J, et al. 618 - AMG 714 (Anti-IL-15 MAB) ameliorates the effects of gluten consumption in celiac disease: a phase 2A, randomized, double-blind, placebo-controlled study evaluating AMG 714 in adult patients with celiac disease exposed to a high-dose gluten challenge. Gastroenterology. (2018) 154:S-130. doi: 10.1016/S0016-5085(18)30861-8
204. Maldonado RA, La Mothe RA, Ferrari JD, Zhang AH, Rossi RJ, Kolte PN, et al. Polymeric synthetic nanoparticles for the induction of antigen-specific immunological tolerance. Proc Natl Acad Sci USA. (2015) 112:E156–65. doi: 10.1073/pnas.1408686111
205. Clemente-Casares X, Blanco J, Ambalavanan P, Yamanouchi J, Singha S, Fandos C, et al. Expanding antigen-specific regulatory networks to treat autoimmunity. Nature. (2016) 530:434–40. doi: 10.1038/nature16962
206. Kishimoto TK, Maldonado RA. Nanoparticles for the induction of antigen-specific immunological tolerance. Front Immunol. (2018) 9:230. doi: 10.3389/fimmu.2018.00230
207. Immusan T. ImmusanT secures Fast-Track Designation for Lead Therapeutic Vaccine Candidate nexvax2 for Patients with Celiac Disease (CeD). (2019). Avaialble online at: http://www.immusant.com/ImmusanT%20Nexvax2%20Fast%20Track%20-%20FINAL.pdf (accessed August 19, 2019).
208. Siegel M, Garber ME, Spencer AG, Botwick W, Kumar P, Williams RN, et al. Safety, tolerability, and activity of ALV003: results from two phase 1 single, escalating-dose clinical trials. Dig Dis Sci. (2012) 57:440–50. doi: 10.1007/s10620-011-1906-5
209. Capriles VD, Areas JAG. Novel approaches in gluten-free breadmaking: interface between food science, nutrition, and health. Compr Rev Food Sci Food Saf. (2014) 13:871–90. doi: 10.1111/1541-4337.12091
210. Piston F, Gil-Humanes J, Rodriguez-Quijano M, Barro F. Down-regulating γ-gliadins in bread wheat leads to non-specific increases in other gluten proteins and has no major effect on dough gluten strength. PLoS ONE. (2011) 6:e24754. doi: 10.1371/journal.pone.0024754
211. Gil-Humanes J, Piston F, Barro F, Rosell CM. The shutdown of celiac disease-related gliadin epitopes in bread wheat by RNAi provides flours with increased stability and better tolerance to over-mixing. PLoS ONE. (2014) 9:e91931. doi: 10.1371/journal.pone.0091931
212. Ellis A, Linaker BD. Non-coeliac gluten sensitivity? Lancet. (1978) 1:1358–9. doi: 10.1016/S0140-6736(78)92427-3
213. Cooper BT, Holmes GK, Ferguson R, Thompson RA, Allan RN, Coole MT. Gluten-sensitive diarrhea without evidence of celiac disease. Gastroenterology. (1980) 79:801–6. doi: 10.1016/0016-5085(80)90432-1
214. Sapone A, Bai JC, Ciacci C, Dolinsek J, Green PH, Hadjivassiliou M, et al. Spectrum of gluten-related disorders: consensus on new nomenclature and classification. BMC Med. (2012) 10:13. doi: 10.1186/1741-7015-10-13
215. Aziz I, Hadjivassiliou M, Sanders DS. The spectrum of noncoeliac gluten sensitivity. Nat Rev Gastroenterol Hepatol. (2015) 12:516–26. doi: 10.1038/nrgastro.2015.107
216. Skodje GI, Sarna VK, Minelle IH, Rolfsen KL, Muir JG, Gibson PR, et al. Fructan, rather than gluten, induces symptoms in patients with self-reported non-celiac gluten sensitivity. Gastroenterology. (2018) 154:529–39. doi: 10.1053/j.gastro.2017.10.040
217. Biesiekierski JR, Iven J. Non-coeliac gluten sensitivity: piecing the puzzle together. United Eur Gastroenterol J. (2015) 3:160–5. doi: 10.1177/2050640615578388
218. Casella G, Villanacci V, Di Bella C, Bassotti G, Bold J, Rostami K. Non celiac gluten sensitivity and diagnostic challenges. Gastroenterol Hepatol Bed Bench. (2018) 11:197–202.
219. Elli L, Tomba C, Branchi F, Roncoroni L, Lombardo V, Bardella M, et al. Evidence for the presence of non-celiac gluten sensitivity in patients with functional gastrointestinal symptoms: results from a multicenter randomized double-blind placebo-controlled gluten challenge. Nutrients. (2016) 8:84. doi: 10.3390/nu8020084
220. Esvaran S, Goel A, Cley WD. What role does wheat play in the symptoms of Irritable Bowel Syndrome? J Gastroenterol Hepatol. (2013) 9:85–91.
221. Czaja-Bulsa G. Non coeliac gluten sensitivity – a new disease with gluten intolerance. Clin Nutr. (2015) 34:189–94. doi: 10.1016/j.clnu.2014.08.012
222. Krigel A, Lebwohl B. Nonceliac gluten sensitivity. Adv Nutr. (2016) 7:1105–10. doi: 10.3945/an.116.012849
223. Elli L, Roncoroni L, Bardella MT. Non-celiac gluten sensitivity: time for sifting the grain. World J Gastroenterol. (2015) 21:8221–6. doi: 10.3748/wjg.v21.i27.8221
224. Sapone A, Lammers KM, Mazzarella G, Mikhailenko I, Carteni M, Casolaro V, et al. Differential mucosal IL-17 expression in two gliadin-induced disorders: gluten sensitivity and the autoimmune enteropathy celiac disease. Int Arch Allergy Immunol. (2010) 152:75–80. doi: 10.1159/000260087
225. Sapone A, Lammers KM, Casolaro V, Cammarota M, Giuliano MT, De Rosa M, et al. Divergence of gut permeability and mucosal immune gene expression in two gluten-associated conditions: celiac disease and gluten sensitivity. BMC Med. (2011) 9:23. doi: 10.1186/1741-7015-9-23
226. Vazquez-Roque MI, Camilleri M, Smyrk T, Murray JA, Marietta E, O'Neill J, et al. A controlled trial of gluten-free diet in patients with irritable bowel syndrome-diarrhea: effects on bowel frequency and intestinal function. Gastroenterology. (2013) 144:903–11. doi: 10.1053/j.gastro.2013.01.049
227. Carroccio A, Mansueto P, Iacono G, Soresi M, D'Alcamo A, Cavataio F. Non Celiac Wheat Sensitivity diagnosed by double-blind placebo controlled challenge: Exploring a new clinical entity. Am J Gastroenterol. (2012) 107:1898–906. doi: 10.1038/ajg.2012.236
228. Brottveit M, Beitnes AC, Tollefsen S, Bratlie JE, Jahnsen FL, Johansen FE, et al. Mucosal cytokine response after short-term gluten challenge in celiac disease and non-celiac gluten sensitivity. Am J Gastroenterol. (2013) 108:842–50. doi: 10.1038/ajg.2013.91
229. Volta U, Bardella MT, Calabro A, Troncone R, Corazza GR. An Italian prospective multicenter survey on patients suspected of having non-celiac gluten sensitivity. BMC Med. (2014) 12:85. doi: 10.1186/1741-7015-12-85
230. Uhde M, Ajamian M, Caio G, De Giorgio R, Indart A, Green PH, et al. Intestinal cell damage and systemic immune activation in individuals reporting sensitivity to wheat in the absence of coeliac disease. Gut. (2016) 65:1930–7. doi: 10.1136/gutjnl-2016-311964
231. Losurdo G, Giorgio F, Piscitelli D, Montenegro L, Covelli C, Fiore MG, et al. May the assessment of baseline mucosal molecular pattern predict the development of gluten related disorders among microscopic enteritis? World J Gastroenterol. (2016) 22:8017–25. doi: 10.3748/wjg.v22.i35.8017
232. Volta U, Caio G, De Giorgio R, Henriksen C, Skodje G, Lundin KE. Non-celiac gluten sensitivity: a work-in-progress entity in the spectrum of wheat-related disorders. Best Pract Res Clin Gastroenterol. (2015) 29:477–91. doi: 10.1016/j.bpg.2015.04.006
233. Igbinedion SO, Ansari J, Vasikaran A, Gavins FN, Jordan P, Boktor M, et al. Non-celiac gluten sensitivity: all wheat attack is not celiac. World J Gastroenterol. (2017) 23:7201–10. doi: 10.3748/wjg.v23.i40.7201
234. Caio G, Volta U, Tovoli F, De Giorgio R. Effect of gluten free diet on immune response to gliadin in patients with non-celiac gluten sensitivity. BMC Gastroenterol. (2014) 14:26. doi: 10.1186/1471-230X-14-26
235. Volta U, Tovoli F, Cicola R, Parisi C, Fabbri A, Piscaglia M, et al. Serological tests in gluten sensitivity (non-celiac gluten intolerance). J Clin Gastroenterol. (2012) 46:680–5. doi: 10.1097/MCG.0b013e3182372541
236. Catassi C, Bai JC, Bonaz B, Bouma G, Calabro A, Carroccio A, et al. Non-celiac gluten sensitivity: the new frontier of gluten related disorders. Nutrients. (2013) 5:3839–53. doi: 10.3390/nu5103839
237. Volta U, Caio G, Karunaratne TB, Alaedini A, De Giorgio R. Non-coeliac gluten/wheat sensitivity: advances in knowledge and relevant questions. Expert Rev Gastroenterol Hepatol. (2017) 11:9–18. doi: 10.1080/17474124.2017.1260003
238. Hollon J, Puppa EL, Greenwald B, Goldberg E, Guerrerio A, Fasano A. Effect of gliadin on permeability of intestinal biopsy explants from celiac disease patients and patients with non-celiac gluten sensitivity. Nutrients. (2015) 7:1565–76. doi: 10.3390/nu7031565
239. Biesiekierski JR, Newnham ED, Irving PM, Barrett JS, Haines M, Doecke JD, et al. Gluten causes gastrointestinal symptoms in subjects without celiac disease: a double-blind randomized placebo-controlled trial. Am J Gastroenterol. (2011) 106:508–14. doi: 10.1038/ajg.2010.487
241. Verdu EF, Huang X, Natividad J, Lu J, Blennerhassett PA, David CS, et al. Gliadin-dependent neuromuscular and epithelial secretory responses in gluten-sensitive HLA-DQ8 transgenic mice. Am J Physiol Gastrointest Liver Physiol. (2008) 294:G217–25. doi: 10.1152/ajpgi.00225.2007
242. Bucci C, Zingone F, Russo C, Morra I, Tortora R, Pogna N, et al. Gliadin does not induce mucosal inflammation or basophil activation in patients with non celiac gluten sensitivity. Clin Gastroenterol Hepatol. (2013) 11:1294–9. doi: 10.1016/j.cgh.2013.04.022
243. Czaja-Bulsa G, Bulsa M. What do we know now about IgE-mediated wheat allergy in children? Nutrients. (2017) 9:35. doi: 10.3390/nu9010035
244. Jin Y, Acharya HG, Acharya D, Jorgensen R, Gao H, Secord J, et al. Advances in molecular mechanisms of wheat allergenicity in animal models: a comprehensive review. Molecules. (2019) 24:E1142. doi: 10.3390/molecules24061142
245. Cianferoni A. Wheat allergy: diagnosis and management. J Asthma Allergy. (2016) 9:13–25. doi: 10.2147/JAA.S81550
246. Sampson HA. Food allergy. Part 1: immunopathogenesis and clinical disorders. J Allergy Clin Immunol. (1999) 103:717–28. doi: 10.1016/S0091-6749(99)70411-2
247. Lee LA, Burks AW. Food allergies: prevalence molecular characterization, and treatment/prevention strategies. Annu Rev Nutr. (2006) 26:539–65. doi: 10.1146/annurev.nutr.26.061505.111211
248. Cianferoni A, Spergel JM. Food allergy: review, classification and diagnosis. Allergol Int. (2009) 58:457–66. doi: 10.2332/allergolint.09-RAI-0138
249. Ortiz C, Valenzuela R, Lucero YA. Celiac disease, non celiac gluten sensitivity and wheat allergy: comparison of 3 different diseases triggered by the same food. Rev Chil Pediatr. (2017) 88:417–23. doi: 10.4067/S0370-41062017000300017
250. Romagnani P, Annunziato F, Piccinni MP, Maggi E, Romagnani S. Th1/ Th2 cells, their associated molecules and role in pathophysiology. Eur Cytokine Netw. (2000) 11:510–1.
251. Lack G. Epidemiologic risks for food allergy. J Allergy Clin Immunol. (2008) 121:1331–6. doi: 10.1016/j.jaci.2008.04.032
252. Mittag D, Niggemann B, Sander I, Reese I, Fiedler EM, Worm M, et al. Immunoglobulin E-reactivity of wheat-allergic subjects (baker's asthma, food allergy, wheat-dependent, exercise-induced anaphylaxis) to wheat protein fractions with different solubility and digestibility. Mol Nutr Food Res. (2004) 48:380–9. doi: 10.1002/mnfr.200400016
253. Pahr S, Selb R, Weber M, Focke-Tejkl M, Hofer G, Dordic A, et al. Biochemical, biophysical and IgE-epitope characterization of the wheat food allergen, Tri a 37. PLoS ONE. (2014) 9:e111483. doi: 10.1371/journal.pone.0111483
254. Cianferoni A, Muraro A. Food-induced anaphylaxis. Immunol Allergy Clin North Am. (2012) 32:165–95. doi: 10.1016/j.iac.2011.10.002
255. Kushimoto H, Aoki T. Masked type I wheat allergy: relation to exercise-induced anaphylaxis. Arch Dermatol. (1985) 121:355–60. doi: 10.1001/archderm.1985.01660030077023
256. Palosuo K, Alenius H, Varjonen E, Koivuluhta M, Mikkola J, Keskinen H, et al. A novel wheat gliadin as a cause of exercise-induced anaphylaxis. J Allergy Clin Immunol. (1999) 103:912–7. doi: 10.1016/S0091-6749(99)70438-0
257. Morita E, Matsuo H, Mihara S, Morimoto K, Savage AW, Tatham AS. Fast ω-gliadin is a major allergen in wheat-dependent exercise induced anaphylaxis. J Dermatol Sci. (2003) 33:99–104. doi: 10.1016/S0923-1811(03)00156-7
258. Lombardo C, Bolla M, Chignola R, Senna G, Rossin G, Caruso B, et al. Study on the immunoreactivity of Triticum monococcum (Einkorn) wheat in patients with wheat-dependent exercise-induced anaphylaxis for the production of hypoallergenic foods. J Agric Food Chem. (2015) 63:8299–306. doi: 10.1021/acs.jafc.5b02648
259. Asaumi T, Manabe T, Yanagida N, Sato S, Ebisawa M. Wheat-dependent exercise-induced anaphylaxis. Curr Treat Options Allergy. (2017) 4:291–302. doi: 10.1007/s40521-017-0135-3
260. Scherf KA, Brockow K, Biedermann T, Koehler P, Wieser H. Wheat-dependent exercise-induced anaphylaxis. Clin Exp Allergy. (2016) 46:10–20. doi: 10.1111/cea.12640
261. Khamnei S, Alipour MR, Ahmadiasl N. The combined effects of exercise and post dehydration water drinking on plasma argenine vasopressin, plasma osmolality and body temperature in healthy males. Int J Endocrinol Metab. (2005) 2:80–6.
262. Torres-Atencio I, Ainsua-Enrich E, De Mora F, Picado C, Martin M. Prostaglandin E2 prevents hyperosmolar-induced human mast cell activation through prostanoid receptors EP2 and EP4. PLoS ONE. (2014) 9:e110870. doi: 10.1371/journal.pone.0110870
263. Munoz-Cano R, Pascal M, Araujo G, Goikoetxea MJ, Valero AL, Picado C, et al. Mechanisms, cofactors, and augmenting factors involved in anaphylaxis. Front Immunol. (2017) 8:1193. doi: 10.3389/fimmu.2017.01193
264. Brant A. Baker's asthma. Curr Opin Allergy Clin Immunol. (2007) 7:152–5. doi: 10.1097/ACI.0b013e328042ba77
265. Kim JH, Kim JE, Choi GS, Hwang EK, An S, Ye YM, et al. A case of occupational rhinitis caused by rice powder in the grain industry. Allergy Asthma Immunol Res. (2010) 2:141–3. doi: 10.4168/aair.2010.2.2.141
266. Quirce S, Diaz-Perales A. Diagnosis and management of grain-induced asthma. Allergy Asthma Immunol Res. (2013) 5:348–56. doi: 10.4168/aair.2013.5.6.348
267. Raulf M, Quirce S, Vandenplas O. Addressing molecular diagnosis of occupational allergies. Curr Allergy Asthma Rep. (2018) 18:6. doi: 10.1007/s11882-018-0759-9
268. Baur X, Degens PO, Sander I. Baker's asthma: still among the most frequent occupational respiratory disorders. J Allergy Clin Immunol. (1998) 102:984–97. doi: 10.1016/S0091-6749(98)70337-9
269. Marraccini P, Brass DM, Hollingsworth JW, Maruoka S, Garantziotis S, Schwartz DA. Bakery flour dust exposure causes non-allergic inflammation and enhances allergic airway inflammation in mice. Clin Exp Allergy. (2008) 38:1526–35. doi: 10.1111/j.1365-2222.2008.03038.x
270. Baatjies R, Lopata AL, Sander I, Raulf-Heimsoth M, Bateman ED, Meijster T, et al. Determinants of asthma phenotypes in supermarket bakery workers. Eur Respir J. (2009) 34:825–33. doi: 10.1183/09031936.00164408
271. Cho HJ, Kim SH, Kim JH, Choi H, Son JK, Hur GY, et al. Effect of Toll-like receptor 4 gene polymorphisms on work-related respiratory symptoms and sensitization to wheat flour in bakery workers. Ann Allergy Asthma Immunol. (2011) 107:57–64. doi: 10.1016/j.anai.2011.04.003
272. Hur GY, Koh DH, Kim HA, Park HJ, Ye YM, Kim KS, et al. Prevalence of work-related symptoms and serum-specific antibodies to wheat flour in exposed workers in the bakery industry. Respir Med. (2008) 102:548–55. doi: 10.1016/j.rmed.2007.11.015
273. Kobrehel K, Wong JH, Balogh A, Kiss F, Yee BC, Buchanan BB. Specific reduction of wheat storage proteins by thioredoxin H. Plant Physiol. (1992) 99:919–24. doi: 10.1104/pp.99.3.919
274. Weichel M, Glaser AG, Ballmer-Weber BK, Schmid-Grendelmeier P, Crameri R. Wheat and maize thioredoxins: a novel cross-reactive cereal allergen family related to baker's asthma. J Allergy Clin Immunol. (2006) 117:676–81. doi: 10.1016/j.jaci.2005.11.040
275. Constantin C, Quirce S, Grote M, Touraev A, Swoboda I, Stoecklinger A, et al. Molecular and immunological characterization of a wheat serine proteinase inhibitor as a novel allergen in baker's asthma. J Immunol. (2008) 180:7451–60. doi: 10.4049/jimmunol.180.11.7451
276. Lehto M, Airaksinen L, Puustinen A, Tillander S, Hannula S, Nyman T, et al. Thaumatin-like protein and baker's respiratory allergy. Ann Allergy Asthma Immunol. (2010) 104:139–46. doi: 10.1016/j.anai.2009.11.062
277. Sander I, Merget R, Degens PO, Goldscheid N, Bruning T, Raulf-Heimsoth M. Comparison of wheat and rye flour skin prick test solutions for diagnosis of baker's asthma. Allergy. (2004) 59:95–8. doi: 10.1046/j.1398-9995.2003.00349.x
278. Aufiero R, Fasano A, Mazzarella G. Non-celiac gluten sensitivity: how its gut immune activation and potential dietary management differ from celiac disease. Mol Nutr Food Res. (2018) 62:1700854. doi: 10.1002/mnfr.201700854
279. Kasarda DD. Can an increase in celiac disease be attributed to an increase in the gluten content of wheat as a consequence of wheat breeding? J Agric Food Chem. (2013) 61:1155–9. doi: 10.1021/jf305122s
280. Gupta PK, Mir RR, Mohan A, Kumar J. Wheat genomics: present status and future prospects. Int J Plant Genomics. (2008) 2008. doi: 10.1155/2008/896451
281. Kerber ER, Tipples KH. Effects of the D genome on milling and baking properties of wheat. Can J Plant Sci. (1969) 49:255–63. doi: 10.4141/cjps69-046
282. Payne PI. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu Rev Plant Physiol. (1987) 38:141–53. doi: 10.1146/annurev.pp.38.060187.001041
283. Branlard G, Dardevet M, Saccomano R, Lagoutte F, Gourdon J. Genetic diversity of wheat storage proteins and bread wheat quality. Euphytica. (2001) 119:59–67. doi: 10.1023/A:1017586220359
284. Schalk K, Lang C, Wieser H, Koehler P, Scherf KA. Quantitation of the immunodominant 33-mer peptide from α-gliadin in wheat flours by liquid chromatography tandem mass spectrometry. Sci Rep. (2017) 7:45092. doi: 10.1038/srep45092
285. Suligoj T, Gregorini A, Colomba M, Ellis HJ, Ciclitira PJ. Evaluation of the safety of ancient strains of wheat in coeliac disease reveals heterogeneous small intestinal T cell responses suggestive of coeliac toxicity. Clin Nutr. (2013) 32:1043–9. doi: 10.1016/j.clnu.2013.02.003
286. Ozuna CV, Iehisa JC, Gimenez MJ, Alvarez JB, Sousa C, Barro F. Diversification of the celiac disease α-gliadin complex in wheat: a 33-mer peptide with six overlapping epitopes, evolved following polyploidization. Plant J. (2015) 82:794–805. doi: 10.1111/tpj.12851
287. Gelinas P, McKinnon C. Gluten weight in ancient and modern wheat and the reactivity of epitopes towards R5 and G12 monoclonal antibodies. Int J Food Sci Technol. (2016) 51:1801–10. doi: 10.1111/ijfs.13151
288. Molberg O, Uhlen AK, Jensen T, Flaete NS, Fleckenstein B, Arentz-Hansen H, et al. Mapping of gluten T-cell epitopes in the bread wheat ancestors: implications for celiac disease. Gastroenterology. (2005) 128:393–401. doi: 10.1053/j.gastro.2004.11.003
289. Kumar MBV, Prasada Rao UJS, Prabhasankar P. Immunogenicity characterization of hexaploid and tetraploid wheat varieties related to celiac disease and wheat allergy. Food Agric Immunol. (2017) 28:888–903. doi: 10.1080/09540105.2017.1319342
290. Ozuna CV, Barro F. Characterization of gluten proteins and celiac disease-related immunogenic epitopes in the Triticeae: cereal domestication and breeding contributed to decrease the content of gliadins and gluten. Mol Breed. (2018) 38:22. doi: 10.1007/s11032-018-0779-0
291. De Vincenzi M, Luchetti R, Giovannini C, Pogna NE, Saponaro C, Galterio G, et al. In vitro toxicity testing of alcohol-soluble proteins from diploid wheat Triticum monococcum in celiac disease. J Biochem Toxicol. (1996) 11:313–8. doi: 10.1002/(SICI)1522-7146(1996)11:6<313::AID-JBT7>3.0.CO;2-N
292. Pizzuti D, Buda A, D'Odorico A, D'Inca R, Chiarelli S, Curioni A, et al. Lack of intestinal mucosal toxicity of Triticum monococcum in celiac disease patients. Scand J Gastroenterol. (2006) 41:1305–11. doi: 10.1080/00365520600699983
293. Vincentini O, Maialetti F, Gazza L, Silano M, Dessi M, De Vincenzi M, et al. Environmental factors of celiac disease: cytotoxicity of hulled wheat species Triticum monococcum, T. turgidum ssp dicoccum and T. aestivum ssp spelta. J Gastroenterol Hepatol. (2007) 22:1816–22. doi: 10.1111/j.1440-1746.2006.04680.x
294. Gianfrani C, Maglio M, Aufiero VR, Camarca A, Vocca I, Iaquinto G, et al. Immunogenicity of monococcum wheat in celiac patients. Am J Clin Nutr. (2012) 96:1339–45. doi: 10.3945/ajcn.112.040485
295. Escarnot E, Gofflot S, Sinnaeve G, Dubois B, Bertin P, Mingeot D. Reactivity of gluten proteins from spelt and bread wheat accessions towards A1 and G12 antibodies in the framework of celiac disease. Food Chem. (2018) 268:522–32. doi: 10.1016/j.foodchem.2018.06.094
296. Spaenij-Dekking L, Kooy-Winkelaar Y, Van Veelen P, Drijfhout JW, Jonker H, Van Soest L, et al. Natural variation in toxicity of wheat: potential for selection of nontoxic varieties for celiac disease patients. Gastroenterology. (2005) 129:797–806. doi: 10.1053/j.gastro.2005.06.017
297. Salentijn EM, Goryunova SV, Bas N, Van der Meer IM, Van den Broeck HC, Bastien T, et al. Tetraploid and hexaploid wheat varieties reveal large differences in expression of alpha-gliadins from homoeologous Gli-2 loci. BMC Genomics. (2009) 10:48. doi: 10.1186/1471-2164-10-48
298. Ribeiro M, Rodriguez-Quijano M, Nunes FM, Carrillo JM, Branlard G, Igrejas G. New insights into wheat toxicity: breeding did not seem to contribute to a prevalence of potential celiac disease's immunostimulatory epitopes. Food Chem. (2016) 213:8–18. doi: 10.1016/j.foodchem.2016.06.043
299. Vaccino P, Becker HA, Brandolini A, Salamini F, Kilian B. (2009). A catalogue of Triticum monococcum genes encoding toxic and immunogenic peptides for celiac disease patients. Mol. Genet. Genomics. 281, 289–300. doi: 10.1007/s00438-008-0412-8
301. Summers RW, Brown JKM. Constraints on breeding for disease resistance in commercially competitive wheat cultivars. Plant Pathol. (2013) 62:115–21. doi: 10.1111/ppa.12165
302. Gallagher E, Gormley TR, Arendt EK. Recent advances in the formulation of gluten-free cereal-based products. Trends Food Sci Technol. (2004) 15:143–52. doi: 10.1016/j.tifs.2003.09.012
303. Battais F, Richard C, Jacquenet S, Denery-Papini S, Moneret-Vautrin DA. Wheat grain allergies: an update on wheat allergens. Eur Ann Allergy Clin Immunol. (2008) 40:67–76.
304. Day L, Augustin MA, Batey IL, Wrigley CW. Wheat-gluten uses and industry needs. Trends Food Sci Technol. (2006) 17:82–90. doi: 10.1016/j.tifs.2005.10.003
305. Atchison J, Head L, Gates A. Wheat as food, wheat as industrial substance; comparative geographies of transformation and mobility. Geoforum. (2010) 41:236–46. doi: 10.1016/j.geoforum.2009.09.006
306. Kaur A, Bains NS, Sood A, Yadav B, Sharma P, Kaur S, et al. Molecular characterization of α-gliadin gene sequences in Indian wheat cultivars vis-a‘-vis celiac disease eliciting epitopes. J Plant Biochem Biotechnol. (2016) 26:106–12. doi: 10.1007/s13562-016-0367-5
307. Van den Broeck HC, De Jong HC, Salentijn ELM, Dekking L, Bosch D, Hamer RJ, et al. Presence of celiac disease epitopes in modern and old hexaploid wheat varieties: wheat breeding may have contributed to increased prevalence of celiac disease. Theor Appl Genet. (2010) 121:1527–39. doi: 10.1007/s00122-010-1408-4
308. De Santis MA, Giuliani MM, Giuzio L, De Vita P, Lovegrove A, Shewry PR, et al. Differences in gluten protein composition between old and modern durum wheat genotypes in relation to 20th century breeding in Italy. Eur J Agron. (2017) 87:19–29. doi: 10.1016/j.eja.2017.04.003
309. Malalgoda M, Meinhardt SW, Simsek S. Detection and quantitation of immunogenic epitopes related to celiac disease in historical and modern hard red spring wheat cultivars. Food Chem. (2018) 264:101–7. doi: 10.1016/j.foodchem.2018.04.131
310. Prandi B, Tedeschi T, Folloni S, Galaverna G, Sforza S. Peptides from gluten digestion: a comparison between old and modern wheat varieties. Food Res Int. (2017) 91:92–102. doi: 10.1016/j.foodres.2016.11.034
311. Gregorini A, Colomba M, Ellis HJ, Ciclitira PJ. Immunogenicity characterization of two ancient wheat α-gliadin peptides related to celiac disease. Nutrients. (2009) 1:276–90. doi: 10.3390/nu1020276
312. Colomba MS, Gregorini A. Are ancient durum wheats less toxic to celiac patients? A study of α-gliadin from Graziella Ra and Kamut. Sci World J. (2012) 2012. doi: 10.1100/2012/837416
313. Shewry PR. Do ancient types of wheat have health benefits compared with modern bread wheat? J Cereal Sci. (2018) 79:469–76. doi: 10.1016/j.jcs.2017.11.010
314. Ribeiro M, Nunes FM. We might have got it wrong: modern wheat is not more toxic for celiac patients. Food Chem. (2019) 278:820–2. doi: 10.1016/j.foodchem.2018.12.003
315. Kupper C. Dietary guidelines and implementation for celiac disease. Gastroenterology. (2005) 128:S121–7. doi: 10.1053/j.gastro.2005.02.024
316. Hischenhuber C, Crevel R, Jarry B, Maki M, Moneret-Vautrin DA, Romano A, et al. Safe amounts of gluten for patients with wheat allergy or coeliac disease. Aliment Pharmacol Ther. (2006) 23:559–75. doi: 10.1111/j.1365-2036.2006.02768.x
317. Cohen IA, Day AS, Shaoul R. Gluten in celiac disease-more or less? Rambam maimonides. Med J. (2019) 10:e0007. doi: 10.5041/RMMJ.10360
318. CODEX Alimentarius Commission for International Food Standards of Food and Agriculture Organization of the United Nations [FAO]/World Health Organization. Standard for Foods for Special Dietary Use for Persons Intolerant to Gluten. (2008). Available online at: www.fao.org/input/download/standards/291/CXS_118e_2015.pdf (accessed August 17, 2019).
319. Bascunan KA, Vespa MC, Araya M. Celiac disease: understanding the gluten-free diet. Eur J Nutr. (2017) 56:449–59. doi: 10.1007/s00394-016-1238-5
320. Catassi C, Fabiani E, Iacono G, D'Agate C, Francavilla R, Biagi F, et al. A prospective, double-blind, placebo-controlled trial to establish a safe gluten threshold for patients with celiac disease. Am J Clin Nutr. (2007) 85:160–6. doi: 10.1093/ajcn/85.1.160
321. Lanzini A, Lanzarotto F, Villanacci V, Mora A, Bertolazzi S, Turini D, et al. Complete recovery of intestinal mucosa occurs very rarely in adult coeliac patients despite adherence to gluten-free diet. Aliment Pharmacol Ther. (2009) 29:1299–308. doi: 10.1111/j.1365-2036.2009.03992.x
322. Laurikka P, Salmi T, Collin P, Huhtala H, Maki M, Kaukinen K, et al. Gastrointestinal symptoms in celiac disease patients on a long-term gluten-free diet. Nutrients. (2016) 8:429. doi: 10.3390/nu8070429
323. Akobeng AK, Thomas AG. Systematic review: tolerable amount of gluten for people with coeliac disease. Aliment Pharmacol Ther. (2008) 27:1044–52. doi: 10.1111/j.1365-2036.2008.03669.x
324. Leonard MM, Cureton P, Fasano A. Indications and use of the gluten contamination elimination diet for patients with non-responsive celiac disease. Nutrients. (2017) 9:E1129. doi: 10.3390/nu9101129
325. Wolf RL, Lebwohl B, Lee AR, Zybert P, Reilly NR, Cadenhead J, et al. Hypervigilance to a gluten-free diet and decreased quality of life in teenagers and adults with celiac disease. Dig Dis Sci. (2018) 63:1438–48. doi: 10.1007/s10620-018-4936-4
326. Lee AR, Wolf RL, Lebwohl B, Ciaccio EJ, Green PHR. Persistent economic burden of the gluten free diet. Nutrients. (2019) 11:E399. doi: 10.3390/nu11020399
327. Rajpoot P, Makharia GK. Problems and challenges to adaptation of gluten free diet by Indian patients with celiac disease. Nutrients. (2013) 5:4869–79. doi: 10.3390/nu5124869
328. Makharia GK, Mulder CJ, Goh KL, Ahuja V, Bai JC, Catassi C, et al. Issues associated with the emergence of coeliac disease in the Asia-Pacific region: a working party report of the World Gastroenterology Organization and the Asian Pacific Association of Gastroenterology. J Gastroenterol Hepatol. (2014) 29:666–77. doi: 10.1111/jgh.12514
329. See JA, Kaukinen K, Makharia GK, Gibson PR, Murray JA. Practical insights into gluten-free diets. Nat Rev Gastroenterol Hepatol. (2015) 12:580–91. doi: 10.1038/nrgastro.2015.156
330. Khoury DE, Balfour-Ducharme S, Joye IJ. A Review on the gluten-free diet: technological and nutritional challenges. Nutrients. (2018) 10:1410. doi: 10.3390/nu10101410
331. Haas-Lauterbach S, Immer U, Richter M, Koehler P. Gluten fragment detection with a competitive ELISA. J AOAC Int. (2012) 95:377–81. doi: 10.5740/jaoacint.SGE_Haas-Lauterbach
332. Halbmayr-Jech E, Rogers A, Don C, Prinster M. Gluten in rice flour and baked rice products by G12 sandwich ELISA: first action 2014.03. J AOAC Int. (2015) 98:103–11. doi: 10.5740/jaoacint.14-197
333. Sharma GM, Rallabhandi P, Williams KM, Pahlavan A. Characterization of antibodies for grain-specific gluten detection. J Food Sci. (2016) 81:T810–6. doi: 10.1111/1750-3841.13241
334. Capparelli R, Ventimiglia I, Longobardo L, Iannelli D. Quantification of gliadin levels to the picogram level by flow cytometry. Cytometry. (2005) 63A:108–13. doi: 10.1002/cyto.a.20109
335. Manfredi A, Mattarozzi M, Giannetto M, Careri M. Multiplex liquid chromatography-tandem mass spectrometry for the detection of wheat, oat, barley and rye prolamins towards the assessment of gluten-free product safety. Anal Chim Acta. (2015) 895:62–70. doi: 10.1016/j.aca.2015.09.008
336. Fallahbaghery A, Zou W, Byrne K, Howitt CA, Colgrave ML. Comparison of gluten extraction protocols assessed by LC-MS/MS analysis. J Agric Food Chem. (2017) 65:2857–66. doi: 10.1021/acs.jafc.7b00063
337. Prandi B, Faccini A, Tedeschi T, Cammerata A, Sgrulletta D, D'Egidio MG, et al. Qualitative and quantitative determination of peptides related to celiac disease in mixtures derived from different methods of simulated gastrointestinal digestion of wheat products. Anal Bioanal Chem. (2014) 406:4765–75. doi: 10.1007/s00216-014-7858-9
338. Bromilow S, Gethings LA, Buckley M, Bromley M, Shewry PR, Langridge JI, et al. A curated gluten protein sequence database to support development of proteomics methods for determination of gluten in gluten-free foods. J Proteomics. (2017) 163:67–75. doi: 10.1016/j.jprot.2017.03.026
339. Colgrave ML, Byrne K, Blundell M, Howitt CA. Identification of barley-specific peptide markers that persist in processed foods and are capable of detecting barley contamination by LC-MS/MS. J Proteomics. (2016) 147:169–76. doi: 10.1016/j.jprot.2016.03.045
340. Mejias JH, Lu X, Osorio C, Ullman JL, Von Wettstein D, Rustgi S. Analysis of wheat prolamins, the causative agents of celiac sprue, using reversed phase high performance liquid chromatography (RP-HPLC) and matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF-MS). Nutrients. (2014) 6:1578–97. doi: 10.3390/nu6041578
341. Grochalova M, Konecna H, Stejskal K, Potesil D, Fridrichova D, Srbova E, et al. Deep coverage of the beer proteome. J Proteomics. (2017) 162:119–24. doi: 10.1016/j.jprot.2017.05.001
342. Scherf KA, Wieser H, Koehler P. Improved quantitation of gluten in wheat starch for celiac disease patients by gel-permeation high-performance liquid chromatography with fluorescence detection (GP-HPLC-FLD). J Agric Food Chem. (2016) 64:7622–31. doi: 10.1021/acs.jafc.6b02512
343. Garcia-Molina MD, Muccilli V, Saletti R, Foti S, Masci S, Barro F. Comparative proteomic analysis of two transgenic low-gliadin wheat lines and non-transgenic wheat control. J Proteomics. (2017) 165:102–12. doi: 10.1016/j.jprot.2017.06.010
344. Daikuzono CM, Shimizu FM, Manzoli A, Riul A, Piazzetta MHO, Gobbi AL, et al. Information visualization and feature selection methods applied to detect gliadin in gluten-containing foodstuff with a microfluidic electronic tongue. ACS Appl Mater Interfaces. (2017) 9:19646–52. doi: 10.1021/acsami.7b04252
345. Manfredi A, Giannetto M, Mattarozzi M, Costantini M, Mucchino C, Careri M. Competitive immunosensor based on gliadin immobilization on disposable carbon-nanogold screen-printed electrodes for rapid determination of celiotoxic prolamins. Anal Bioanal Chem. (2016) 408:7289–98. doi: 10.1007/s00216-016-9494-z
Keywords: wheat, gluten, celiac disease, non-celiac gluten sensitivity, wheat allergy, gluten-free diet
Citation: Sharma N, Bhatia S, Chunduri V, Kaur S, Sharma S, Kapoor P, Kumari A and Garg M (2020) Pathogenesis of Celiac Disease and Other Gluten Related Disorders in Wheat and Strategies for Mitigating Them. Front. Nutr. 7:6. doi: 10.3389/fnut.2020.00006
Received: 04 November 2019; Accepted: 20 January 2020;
Published: 07 February 2020.
Edited by:
Wilson Savino, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Nicolò Merendino, Università degli Studi della Tuscia, ItalyClaudio Nicoletti, University of Florence, Italy
Copyright © 2020 Sharma, Bhatia, Chunduri, Kaur, Sharma, Kapoor, Kumari and Garg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monika Garg, bWthamdhcmdAZ21haWwuY29t; bW9uaWthZ2FyZ0BuYWJpLnJlcy5pbg==; bWdhcmcxMDBAeWFob28uY29t
 Natasha Sharma
Natasha Sharma Simran Bhatia
Simran Bhatia Venkatesh Chunduri
Venkatesh Chunduri Satveer Kaur
Satveer Kaur Saloni Sharma
Saloni Sharma Payal Kapoor
Payal Kapoor Anita Kumari
Anita Kumari Monika Garg
Monika Garg