- International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad, India
Peanut allergy is an important health concern among many individuals. As there is no effective treatment to peanut allergy, continuous monitoring of peanut-based products, and their sources is essential. Precise detection of peanut allergens is key for identification and development of improved peanut varieties with minimum or no allergens in addition to estimating the levels in peanut-based products available in food chain. The antibody based ELISA protocol along with sample preparation was standardized for Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8 to estimate their quantities in peanut seeds. Three different dilutions were optimized to precisely quantify target allergen proteins in peanut seeds such as Ara h 1 (1/1,000, 1/2,000, and 1/4,000), Ara h 2 and Ara h 3 (1/5,000, 1/10,000, and 1/20,000), Ara h 6 (1/40,000, 1/80,000, and 1/1,60,000), and Ara h 8 (1/10, 1/20, and 1/40). These dilutions were finalized for each allergen based on the accuracy of detection by achieving <20% coefficient of variation in three technical replicates. This protocol captured wide variation of allergen proteins in selected peanut genotypes for Ara h 1 (77–46,106 μg/g), Ara h 2 (265–5,426 μg/g), Ara h 3 (382–12,676 μg/g), Ara h 6 (949–43,375 μg/g), and Ara h 8 (0.385–6 μg/g). The assay is sensitive and reliable in precise detection of five major peanut allergens in seeds. Deployment of such protocol allows screening of large scale germplasm and breeding lines while developing peanut varieties with minimum allergenicity to ensure food safety.
Introduction
Food allergies have become the most important food safety concerns around the world. The prevalence of food allergy is more in North America, where 6% of young children and 4% of adults suffer from food allergies (1). Among all the food allergies, ~1.4% of Americans and 3% Australian are allergic to peanuts and peanut-based products (2, 3). Peanut allergy is very severe and sometimes causes death due to anaphylaxis (4, 5). It is reported that food allergies cause approximately 150 to 200 fatalities per year, based on data from a five year study of anaphylaxis in Minnesota from the Mayo Clinic. Fatal food anaphylaxis is most often caused by peanuts (50–62%) and tree nuts (15–30%) (6). Proteins are the major cause of food allergy and these allergenic proteins are usually highly resistant to heat and proteolysis (7). Peanut is the largest source for the IgE-mediated food allergies among eight major sources namely tree nuts, egg, cow's milk, fish, crustacean shellfish, soybean, and wheat. There is no effective treatment to peanut allergy and the allergic person is advised strictly to avoid consuming peanut or peanut-based products (8). Peanut being a common food ingredient in many food preparations, it is very challenging for the allergic person to know the composition of these preparations to avoid consumption (9). The threshold of allergen levels differ among the allergic population and even a minute dose of 100 μg of Ara h 1 can trigger an allergic reaction (10). The diagnosis of peanut allergy can be done using different methods such as double-blind, placebo-controlled food challenge (DBPCFC), the basophil activation test, the specific skin prick test (SPT), and the measurement of specific IgE (11–13).
Of the 32 different types of proteins present in peanut seeds, 16 of these proteins show allergenic property (14). Further, out of these 16 peanut allergen proteins, Ara h 1, 2, 3, and 6 are considered as major allergens because they are recognized by the IgE of a majority of allergic patients. Ara h 5, 7, 8, 9, 10, 11, and 12/13 are considered minor allergens and are not thought to be the causative agents in most of the life-threatening allergic reactions (anaphylaxis) (15). Nevertheless, if a person is already sensitive to Bet v 1 allergey caused due to birch pollen, then one of these minor peanut allergens, Ara h 8, shows cross-reactivity with IgE antibodies causing oral allergy syndrome (OAS) (16–18).
Allergic protein belongs to different protein families namely cupin (vicilin-type, 7S globulin, legumin-type, 11S globulin, glycinin), conglutin (2S albumin), profilin, nonspecific lipid-transfer protein 1, pathogenesis-related protein, PR-10, 14 kDa oleosin, and 16 kDa oleosin (19), particularly, Ara h 1, Ara h 2, Ara h 3, and Ara h 6 are the seed storage proteins. Many studies showed that Ara h 1 and Ara h 3 are abundant peanut proteins but, Ara h 2 and Ara h 6 strongly bind with IgE from peanut allergic patient and release mediators from basophils, which confirmed that both are the most important allergens in vitro (20–23) and in vivo (21, 24, 25) with regards to food allergy (23). Due to the fact that both Ara h 2 and Ara h 6 are recognized more prominently and more frequently than Ara h 1 and Ara h 3 by peanut allergic children with almost all of them showing IgE reactivity to these two allergens (26).
Immuno-assays are the most commonly preferred analytical techniques being used to detect the food allergens in many industries (27). Food industries commonly use ELISA to detect and quantify hidden allergens in food commodities as it is easy and less time-consuming technique (28–30). Further ELISA is highly sensitive, reliable, cost-effective, and accurate. In the past ELISA has been successfully used to detect allergens in several food products such as in meat and meat products, milk and milk products, fish and fish products, soybean, nuts and nut-based products, and fruit juices and their ingredients (31, 32). In ELISA, allergens can be detected by the colorimetric reaction after binding of the antigen with the specific enzyme-labeled antibody. Earlier studies reported the allergenic proteins quantification in peanut-based products through ELISA and HPLC technique (29, 33–35), however, no information on peanut seeds. The key issues in ELISA are the sample preparation and unacceptable variability in multiple measurements of the same sample. The best way of preparing a sample, right dilution of the sample used in the assay, and low coefficient of variation (CV) holds importance in achieving accuracy. As of now there is no standard protocol which can help to detect allergen proteins in peanut seeds on the basis of dilution factor and low coefficient of variation (CV). Further the genetic diversity available in peanut germplasm has not yet been fully exploited and the accumulation of favorable alleles may provide minimum allergen proteins in the improved peanut genotypes. At this juncture a highly sensitive, easy to use and reliable protocol for peanut seeds is required to screen various germplasm sets and breeding material. Therefore, the present study focused on developing a robust protocol to estimate major allergens in peanut seeds through ELISA based methods.
Materials and Methods
Plant Material, Reagents and Chemicals
A total of 38 peanut genotypes (see Supplementary Table 1) were used while improving and standardizing the ELISA protocol. The seeds of these genotypes after harvest were stored at 10°C and were taken out only when needed for the experiment. ELISA components such as peanut allergen standards, monoclonal antibodies (MAbs; primary and secondary biotinylated antibody), and enzyme conjugates were purchased from INDOOR Biotechnologies Inc. (Charlottesville, VA, USA) for the estimation of five important peanut allergens namely Ara h 1, Ara h 2 Ara h 3, Ara h 6, and Ara h 8. These include monoclonal antibody 2C12 (clone 2C12 A11 A3) and biotinylated antibody 2F7 (clone 2F7 C12 D10) for Ara h 1, the monoclonal antibody 1E8 and biotinylated monoclonal antibody 4G9 for Ara h 3 while monoclonal antibody 3B8 (clone 3B8 B5) and biotinylated antibody 3E12 (clone 3E12 C4 B3) for Ara h 6. Similarly, the monoclonal antibody 1C4 (clone 1C4 G4 C8) and a polyclonal rabbit anti Ara h 2 antibody (AH2) for Ara h 2; capture antibody 4G6 and a polyclonal antiserum raised in rabbit for Ara h 8 were purchased from INDOOR Biotechnologies Inc. Conjugated Goat anti-Rabbit IgG was purchased from Jacksons Laboratories (Bar Harbor, USA Cat No. 111-036-046). ABTS™ (Cat No. 11204521001) and all other reagents and chemicals were purchased from Sigma-Aldrich Co. (Oakville, ON, Canada).
Allergen Standards and Enzyme Solution Preparation
The allergen standards were purchased from INDOOR Biotechnologies Inc. (Charlottesville, VA, USA). These standards were isolated from lightly roasted peanut flour (Runner cultivar) and purified by affinity chromatography. The purified standards were supplied in phosphate buffer, received on the ice and stored at −20°C until further use. Purified natural Ara h 1 (Lot 39285; Conc. 20,000 ng/ml), Ara h 2 (Lot 39158; Conc. 2,500 ng/ml), Ara h 3 (Lot 39051; Conc. 1,250 ng/ml) Ara h 6 (Lot 39198; Conc. 1,000 ng/ml), and Ara h 8 (Lot 39033; Conc. 2,500 ng/ml) were used as allergen standards for each assay. The standard concentration of each allergen ranged from 2000 - 4 ng/ml for Ara h 1, 250 - 0.5 ng/ml for Ara h 2, 125 - 0.24 ng/ml for Ara h 3, 100 - 0.2 ng/ml for Ara h 6 and 250 - 0.49 ng/ml for Ara h 8. ABTS Substrate is a water-soluble peroxidase substrate that yields a measurable green end product for use in ELISA methods. The ABTS™ (Sigma Aldrich Cat No. 11204521001) was dissolved in 1 mM ABTS solution. The ABTS solution contains 0.1M anhydrous citric acid and 0.2M Dibasic Sodium phosphate.7H202. The 274 mg ABTS™ were dissolved in 500 ml ABTS solution and store in an amber color bottle at 4°C until use.
Development of Protocol for Allergen Estimation
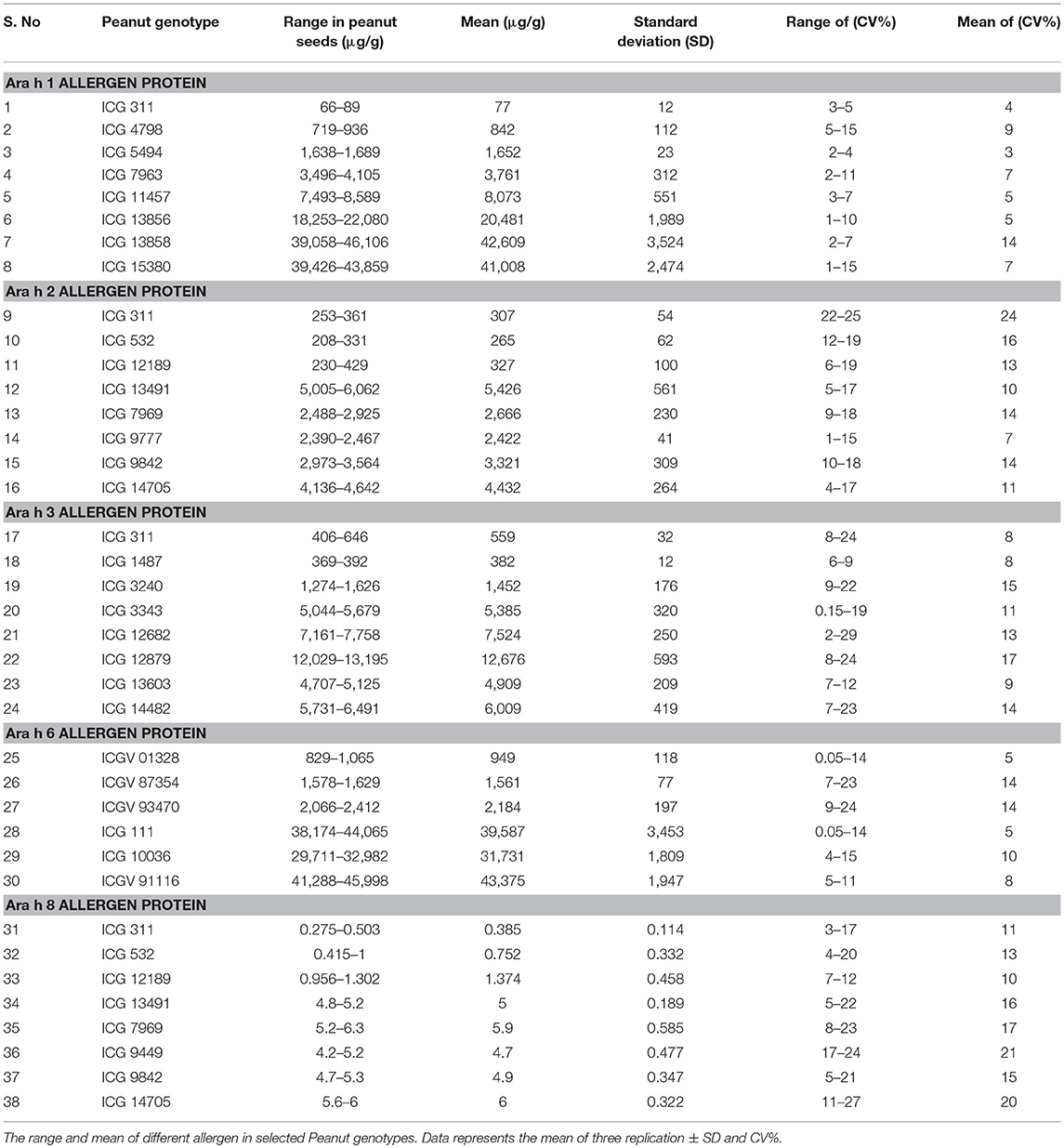
The ELISA is sandwich format having double-antibody based on the specific interaction between antigen and antibody. The peanut allergen proteins are sandwich between two antibodies such as capture antibody and conjugated antibody with streptavidin peroxidase (Figure 1). For color development ABTS™ was used. The color intensity depends on the concentration of allergen protein present in the specific sample and measured with an iMark microplate reader (Bio-Rad) at 405 nm. The Microplate Manager (MPM) software was used while analyzing the optical density values of standards and samples. The systemic representation showed the key steps involved to develop the ELISA protocol, to estimate the peanut allergens through peanut seeds (Figure 1).
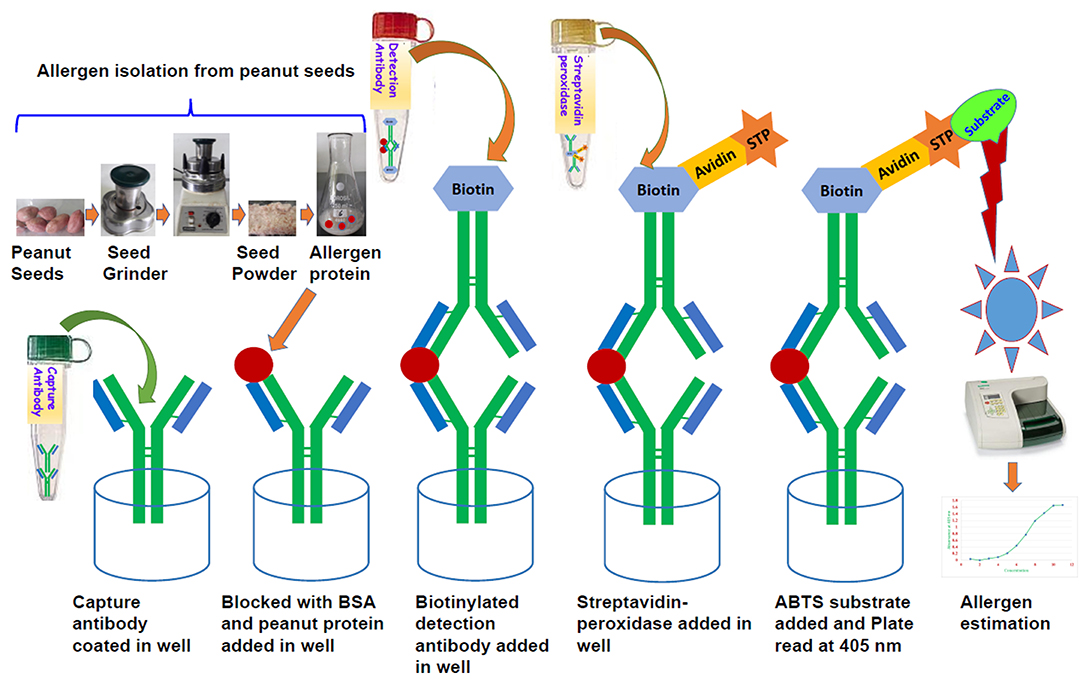
Figure 1. Systematic diagram showing the protocol for allergen estimation in peanut seed through sandwich ELISA.
Grinding of Seeds, Homogenization and Purification
Sample extracts were prepared by grinding two grams of peanut seeds in fine powder and then dissolved in 40 ml of PBS-T (0.05% Tween in phosphate buffered saline, pH 7.4) containing 1M NaCl in 50 ml falcon tubes (Sarstedt No:55.476). After 2 h of gentle stirring on a rocking platform at room temperature, the aqueous phase was collected by centrifugation at 2,500 rpm at 4°C for 20 min. The aqueous phase was subsequently centrifuged at 3,500 rpm for 10 min at room temperature to remove residual traces and insoluble particles. Protein extracts were stored at −20°C until use.
Dilution Factor for Different Peanut Allergens
Dilution of a sample extract is critical for an ELISA which in turn determines the values of detection range for antibody and target antigen concentrations. Subsequently, the concentration of that specific allergens sample was estimated by multiplying the concentration found from the graph by the dilution factor (36, 37). By using different dilutions, we estimated the detection range of target antigen and antibody concentration. We standardized the dilution factors to detect each allergen proteins presents in the peanut seeds. Each sample was diluted in three different dilutions to detect the allergic protein present in seeds. The major allergen proteins such as Ara h 1 (1/1,000, 1/2,000, and 1/4,000), Ara h 2 (1/5,000, 1/10,000, and 1/20,000), Ara h 3 (1/5,000, 1/10,000, and 1/20,000), and Ara h 6 (1/40,000, 1/80,000, and 1/160,000) were diluted on a high range as compared to minor allergen protein, Ara h 8, which was diluted in low dilution range (1/10, 1/20, and 1/40). The optimal concentration of HRP-conjugated streptavidin was determined in the same way.
Steps Involved in ELISA
Antibody coating and blocking
Polystyrene microtiter plates (NUNC Maxisorp, Roskilde, Denmark) were coated with 100 μL mAb at 10 μL /10 ml in 50 mM carbonate buffer (100 μL/well). After overnight incubation at 4°C, the coated wells were washed three times with washing buffer (phosphate buffer containing 0.05% Tween 20) and left to block with 1% BSA for 30 min at room temperature followed by three times washing with washing buffer.
Capture of allergen samples and standard
The standard and samples were diluted in washing buffer containing 1% bovine serum albumin fraction V (Sigma Aldrich Cat No. 10735086001). The standard of each allergen was diluted to make 10 serial doubling dilutions in dilution buffer. Subsequently, the allergen samples (100 μL/well) with three different dilutions were added in respective wells and incubated at room temperature for 1 h.
Adding of detection antibody in plate
After incubation plates were washed three times and specific biotinylated anti Ara h mAb were diluted to 1/1,000 in washing buffer containing BSA (1 mg/mL) was added to the wells (100 μL/well) and incubated for 1 h at room temperature.
Streptavidin-enzyme conjugate
After three washes, HRP-conjugated Streptavidin diluted to 1/1,000 in washing buffer containing BSA (1 mg/mL) was added to the wells and incubated for 1 h at room temperature.
Addition of the substrate
The colorimetric substrate was added to the wells and which formed a colored solution when catalyzed by the enzyme. The wells were washed three times and 100 μL of 1 mM ABTS was added to each well. After 5 min, the color development was observed.
Detection through ELISA reader
The optical density (OD) was measured at 405 nm using BioRad Microplate Reader and the data were processed using Microplate Manager V 6.1 (Bio-Rad Laboratories).
Spike and Recovery Studies
To test the accuracy of peanut allergens estimation, known amount of each allergen was spiked in the peanut extracts. The spike standard concentration for each allergen was ranging from 100 to 1,600 ng/ml for Ara h 1, 25 to 200 ng/ml for Ara h 2, and 6.25 to 100 ng/ml for Ara h 3, 10 to 160 for Ara h 3, and from 10 to 100 ng/ml for Ara h 8. These known amounts of individual standard (Ara h 1, Ara 2, Ara 3 Ara h 6, and Ara h 8) allergens were spiked in peanut extract and later calculating their final content in extract. The recovery was calculated as (B–C)/(A × 100), Where A = Known amount of peanut standard, B = Concentration of spiked standard, C = Concentration of peanut extract.
Statistical Analysis and Sample Analysis Design
All experiments were conducted with three replications based on dilution factor (DF). Mean ELISA plate reading values (Optical Density; OD) for each standard and sample were used to plot a standard curve by placing each allergen standard concentration values on Y axis and respective OD values on X axis on an Excel spreadsheet on the computer. Using regression equation values, we estimated specific allergen concentration in each sample. Data are expressed as means ± standard deviation (SD). Allergen standards and samples were placed in the 96- well format plate as given in Figure 2. The statistical analysis of data was performed using SigmaPlot 11.0.
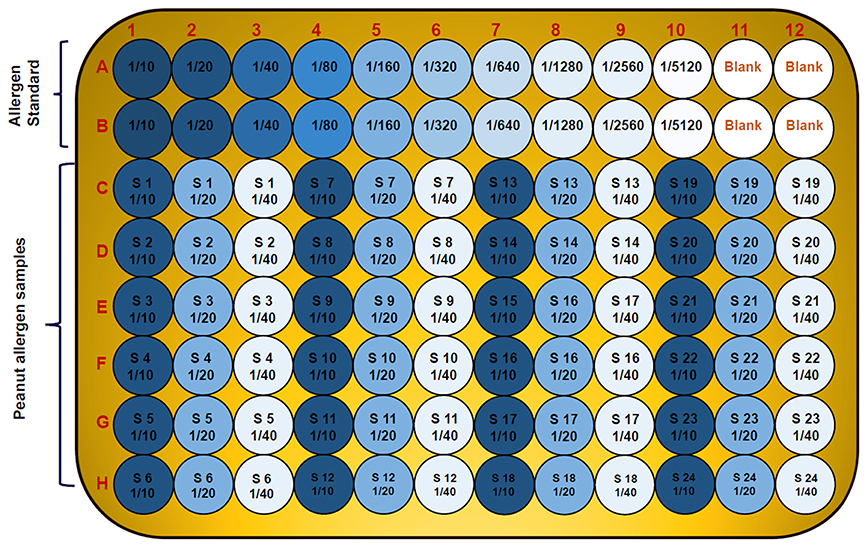
Figure 2. Diagram showing 96-well plate design and the serial dilution of allergen standard and peanut samples in three replicates and three dilutions. The wells A1 to A10 and B1 to B10 contain serial dilution of specific allergen standard; A11 to A12 and B11 to B12 are blank; while the C, D, E, F, G, and H wells are with unknown peanut samples in three different dilutions for specific allergen.
Results
Optimization of Dilution Factors (DFs) for Different Allergens From Peanut Seeds
To find the concentration of the different types of unknown allergens in the peanut seed samples, dilutions were performed on the different scale for specific allergens so that the relative concentrations fall according to their standard graph. Finally, after the absorbance of the specific allergens was determined, the concentrations of these specific allergens were determined by finding the location on the graph that corresponds to the absorbance of the standard. The concentrations of the allergens sample were estimated by multiplying samples concentration with dilution factor. For Ara h 1, the standard ranges from 0.004 to 2 μg/mL (Figure 3A), while it ranged from 0.0005 to 0.25 μg/mL and 0.00024 to 0.125 μg/mL for Ara h 2 and Ara h 3 (Figures 3C,E), respectively. The standard graph range of Ara h 6 and Ara h 8 was showing range between 0.0002 to 0.10 μg/mL and 0.0005 to 0.250 μg/mL (Figures 4A,C).
Figure 3. Standard curve and optimized dilutions for Ara h 1, Ara h 2, and Ara h 3 allergens in peanut seeds. (A) standard curve for Ara h 1 allergen and (B) standardization of dilution factor to estimate Ara h 1. (C) standard curve for Ara h 2 allergen and (D) standardization of dilution factor to estimate Ara h 2. (E) standard curve for Ara h 3 allergen, and (F) standardization of dilution factor to estimate Ara h 3.
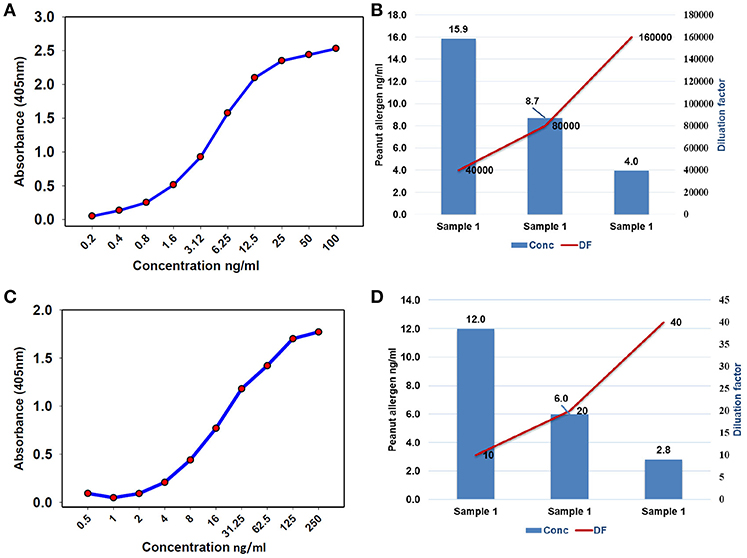
Figure 4. Standard curve and optimized dilutions for Ara h 6 and Ara h 8 allergens in peanut seeds. (A) standard curve for Ara h 6 allergen by sandwich ELISA, and (B) standardization of dilution factor to estimate Ara h 6. (C) standard curve for Ara h 8 allergen by sandwich ELISA, and (D) standardization of dilution factor to estimate Ara h 8.
Based on the standard graph, the dilution factor was optimized on the basis of OD value and dilution of the peanut samples. Each allergen protein was estimated at different dilution factors (DF) in same samples. Here we used a number of dilutions in the peanut samples to detect the specific allergic protein in seeds. The Ara h 1 was detected on three serial doubling dilutions, 1:1,000, 1:2,000, and 1:4,000 (Figure 3B) while Ara h 2 and Ara h 3 detected on same dilution 1: 5,000, 1:10,000, and 1: 20,000 (Figures 3D,F). In peanut seeds, the Ara h 6 was detected in the high range (1:40,000, 1:80,000, and 1:160,000) dilution factor (Figure 4B), while Ara h 8 detected in a low range of dilution i.e., 1:10, 1:20, and 1:40 (Figure 4D). The result showed that in 1,000 dilution, the amount of Ara h 1 were detected 617 ng/ml while in 4000 dilution it decreased upto 152 ng/ml (Figures 3B). Likewise other peanut allergens Ara h 2, Ara h 3, Ara h 6, and Ara h 8 also showing similar pattern in their optimized result. The optimized dilution graphs (Figures 3D,F; 4B,D) showing that when we increasing the dilution, peanut allergen concentration is decreasing accordingly.
Allergens Estimation in Peanut Seeds
The specificity of the sandwich ELISA was investigated by testing sample extracts of 38 peanut genotypes using different dilution factors for different allergens. Here, we used a single sample with three different dilutions and estimated four major (Ara h 1, Ara h 2, Ara h 3, and Ara h 6) and one minor (Ara h 8) allergen proteins in peanut seeds using specific antibodies. Supplementary Table 1 showed the selected genotypes on the basis of minimum and the maximum amount of allergens present in specific genotypes. The minimum and maximum amount of Ara h 1 was detected in ICG 311 (12.5 μg/g) and ICG 15380 (42733.87 μg/g) genotypes. The low and high level of Ara h 2 was detected in ICG 532 (22.2 μg/g) and ICG 14482 (20600.3 μg/g) while ICG 311 (157.8 μg/g) and ICG 12879 (14556.4 μg/g) showing low and high amount of Ara h 3, respectively. Genotype ICG 3992 (5767.6 μg/g) contained low amount of allergen while ICGV 91116 (42317.6 μg/g) contained high amount of Ara h 6 allergen among the genotypes. Ara h 8 showed low range of allergen ranging from lowest in ICG 311(0.084 μg/g) to highest in ICG 14705 (6.8 μg/g).
Repeatability and Reproducibility
To validate the protocol, we repeated our experiments in three technical replicates with three dilutions optimized for each specific allergen. The assay had a linearity of r2 ranges from 0.987 to 0.999 for all the specific allergen concentration present in peanut seeds. The coefficient of variation (CV) of each peanut sample in triplicates was consistently <20% (Table 1). Assay repeatability and reproducibility was <15% CV as measured by using the three peanut samples (Table 1). We found that dilution factor and CV are the key features of this protocol. This method is fairly simple to estimate specific allergen protein present in a single peanut seed. The validation experiments showed linearity of the method and each specific allergen showing different range in different peanut seeds. By using this protocol, we identified wide range of specific allergens, Ara h 1 (77–42,609 μg/g) (Figure 5A, Table 1), Ara h 2 (265–5,426 μg/g) (Figure 5B, Table 1), Ara h 3 (382–12,676 μg/g) (Figure 5C, Table 1), Ara h 6 (949–43,375 μg/g) (Figure 5D, Table 1), and Ara h 8 (0.385–6 μg/g) (Figure 5E, Table 1) in peanut genotypes.
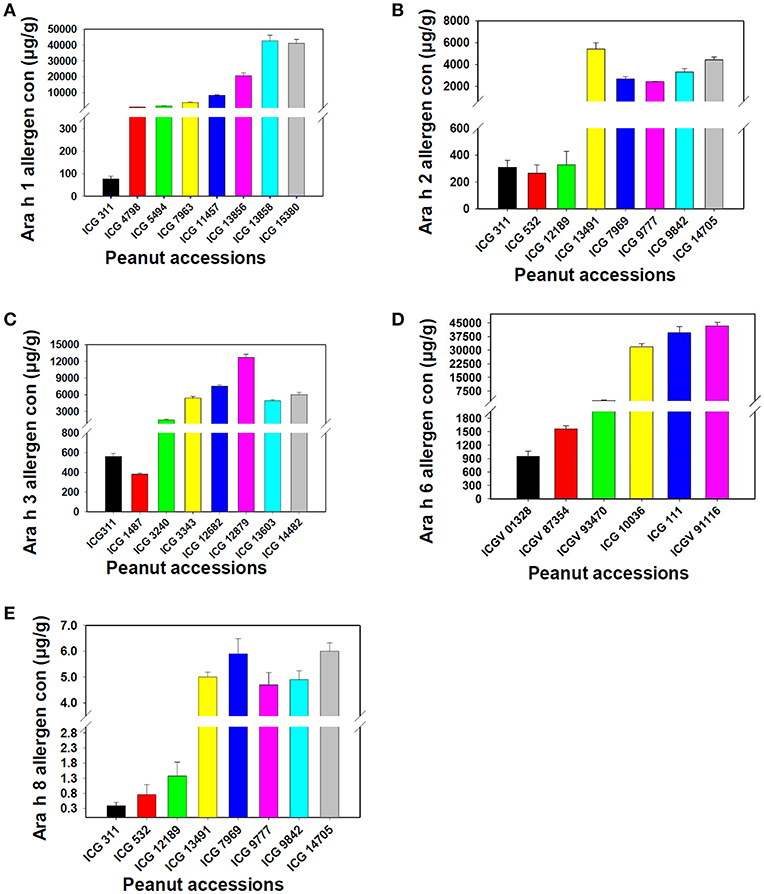
Figure 5. Assessment of the new protocol for its reproducibility for Ara h 1, Ara h 2, Ara h 3, Ara h 6 and Ara h 8 allergen proteins. (A) Ara h 1 allergen proteins, (B) Ara h 2 allergen proteins, (C): Ara h 3 allergen proteins, (D): Ara h 6 allergen proteins and (E) Ara h 8 allergen protein. The figure shows variation in the presence of allergen protein in low to high range in different peanut genotypes.
The accuracy of the developed method was determined by performing the spike and recovery test. The mean recoveries (n = 3) for all the five allergens, individually tested, were in the range of 81–115% (Table 2). This indicates that the developed method is accurate for all five peanut allergens through peanut extract.
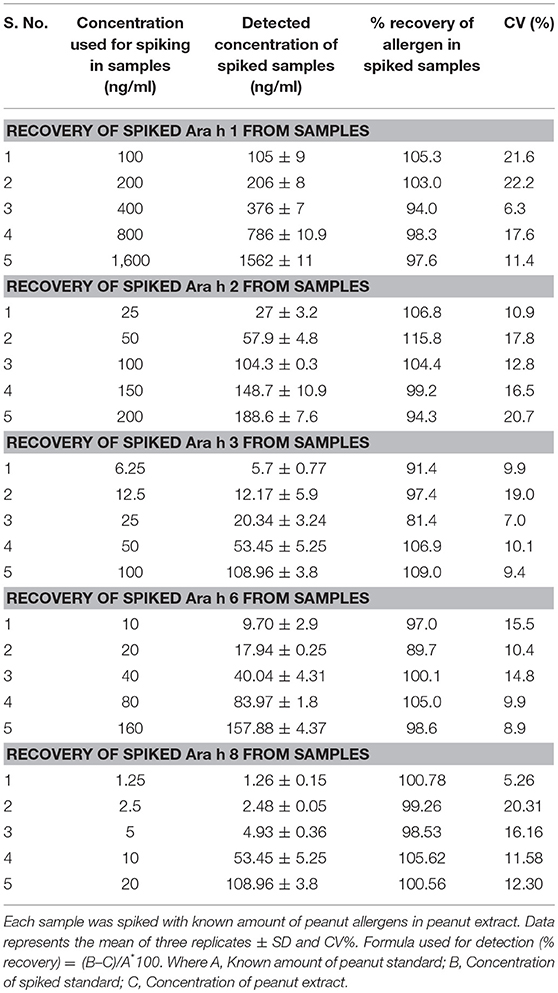
Table 2. Recovery of five major allergens (Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8) from artificial spiked peanut allergens through ELISA in peanut seeds.
Discussion
Peanut allergy is one of the major food allergies and leads to death in case of severe allergenic reaction. In order to tackle this problem, efforts toward the development of peanut varieties with minimum or nil allergens needs to be initiated in addition to efforts for developing cure for the allergy caused due to peanut consumption. Peanut genetic improvement program for this particular trait require a reliable, robust and cost-effective assay to screen the large scale germplasm and breeding material for major allergens. In this context, this article reports development of a cost-effective and reliable ELISA-based protocol to estimate five major allergens (Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8) in peanut seeds.
In serological tests, two-fold differences in measurements of replicates of the same sample are considered as acceptable (38). Current studies describe the standardization of dilution factor for peanut seed sample to analyse the specific allergen protein concentration through ELISA method. It has been observed that the concentration and dilution factors are inverse means and the concentration of the allergens decreased with the increasing dilution of the samples. All major allergen proteins (Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8) concentration was gradually decreased while increasing the dilution. For instance, the concentration of Ara h 1 was 617 ng/mL in 1,000 dilution, and in 2,000 dilution the concentration of Ara h 1 was detected as 296 ng/mL while in 4,000 dilution the concentration of Ara h 1 was detected as 152 ng/mL (Figure 3B). On the basis of optimized dilution factors, major allergen proteins were estimated in peanut seeds with increased precision. The results from our study show the importance of standard curve and dilution as an important parameter for quantifying the ELISA results (39). Dilution factor is an important step in ELISA before analyzing the results. Dilution assay apprises the actual concentration would be taken to experiment. To attained highly sensitive results for protein concentration, optimum values for dilution assay would provide a valid conclusion (39, 40). Earlier few attempts were made to quantify the levels of allergens in peanut seed using immuno assays and chromatography methods. But majority of them were expressed only in terms of percentages out of total protein present in the raw peanut seed. In one of the earlier studies, it was concluded that quantification of a single protein is not possible from a sample mixture of allergenic and non-allergenic proteins (41). Later in another study, using human sera from patients with documented history of peanut allergy, allergens from different peanut introductions were quantified and expressed in terms of ELISA optical density readings (4). In a recent study, Ara h 3 was quantified and found that it is the most abundant protein followed by Ara h 1 and Ara h 2 (34). However, in our study, Ara h 1 found in larger quantities followed by Ara h 6, Ara h 2, Ara h 3, and Ara h 8. Extraction method and proper dilutions of extracts plays crucial role in the precise quantification of these allergen proteins. Here in our study, we have thoroughly studied series of dilutions for each allergen and their impact on the quantification of allergens. These results clearly indicate three different dilutions for each major peanut allergen are required for precise detection in peanut seeds. Further the suggested 96-well plate design can allow the analysis of 24 samples in three replicates.
The newly developed ELISA protocol was validated on assay linearity, accuracy, repeatability and reproducibility. On the basis of primary results, few peanut genotypes were selected with low, medium and higher range of allergen proteins present in seeds. Our results indicated a different level of specific allergen profiles among peanut genotypes. Marked differences in specific peanut allergen profile were observed in peanut flour and peanut-based products such as peanut butter and other confectionary preparations for clinical use (42). In the serological assay, the variability produces continuous value were summarized by the coefficient of variation (CV), often reported in percentage (42, 43). The low CV observed in the current study while preparing dilutions provided more confidence in results produced for multiple genotypes using this protocol. Therefore, this protocol can provide accurate estimation for targeted five allergens and hence, can be very useful in accelerating the peanut research leading to detection and development of hypo-allergic peanuts.
The sample value obtained from an ELISA is dependent upon the interaction between the protein of interest and the ELISA's antibodies, and comparison of this interaction relative to a recombinant protein standard curve. We have validated this newly developed ELISA protocol for estimation of the major peanut allergens in peanut seeds by conducting the recovery experiments. The validation of ELISA protocol depends upon the recovery experiment which allow for a well-ordered presentation of the results (42).
Conclusions
This study reports successful development and validation of an accurate ELISA method for allergen estimation in peanut seeds. The method developed here is a simple, fast and cost-effective method and can be applied to a large number of samples for specific allergen estimation. Our results clearly indicates three different dilutions for each major peanut allergen are required for precise detection in peanut seeds. Further the suggested 96-well plate design can allow the analysis of 24 samples in three replicates. This protocol can be very useful in accelerating the research in identifying peanut genotypes with minimum allergen proteins. Furthermore, the high level of phenotypic variation in a selected set of peanut germplasm showed very positive indication and hope toward efforts of developing allergen-free peanut genotypes in coming years using genomics-assisted breeding including genome editing.
Author Contributions
MP conceived the idea. AP performed the experiment. MP, HS, RV, and AP designed the experiments, analyzed the data, and wrote the manuscript.
Funding
This work has been undertaken as part of the CGIAR Research Program on Grain Legumes and Dryland Cereals (GLDC). ICRISAT is a member of CGIAR Consortium.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are thankful to Dr. Hari D. Upadhyaya for providing seeds of genotypes included in the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2019.00068/full#supplementary-material
References
1. Waserman S, Watson W. Food allergy. Allergy Asthma Clin Immunol. (2011) 7:S7. doi: 10.1186/1710-1492-7-S1-S7
2. Sicherer SH, Sampson HA. Peanut allergy: emerging concepts and approaches for an apparent epidemic. J Allergy Clin Immunol. (2007) 120:491–503. doi: 10.1016/j.jaci.2007.07.015
3. Sicherer SH, Sampson HA. Food allergy: epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. (2014) 133:291–307. doi: 10.1016/j.jaci.2013.11.020
4. Dodo H, Marsic D, Callender M, Cebert E, Viquez O. Screening 34 peanut introductions for allergen content using ELISA. Food Agri Immunol. (2002) 14:147–54. doi: 10.1080/09540100220145179
5. Sicherer SH, Sampson HA. Food allergy. J Allergy Clin Immunol. (2010) S116–25. doi: 10.1016/j.jaci.2009.08.028
6. Lanser BJ, Wright BL, Orgel KA, Vickery BP, Fleischer DM. Current options for the treatment of food allergy. Pediatric Clin. (2015) 62:1531–49. doi: 10.1016/j.pcl.2015.07.015
7. Cabanillas B, Maleki SJ, Rodríguez J, Burbano C, Muzquiz M, Jiménez MA, Pedrosa MM, Cuadrado C, Crespo JF (2012). Heat and pressure treatments effects on peanut allergenicity. Food chemistry. 132, 360–6. doi: 10.1016/j.foodchem.2011.10.093
8. Wen HW, Borejsza-Wysocki W, DeCory TR, Durst RA. Peanut allergy, peanut allergens, and methods for the detection of peanut contamination in food products. Compr Rev Food Sci Food Safety. (2007) 6:47–58. doi: 10.1111/j.1541-4337.2007.00017.x
9. Maleki SJ, Chung SY, Champagne ET, Raufman JP. The effects of roasting on the allergenic properties of peanut proteins. J Allergy Clin Immunol. (2000) 106:763–8. doi: 10.1067/mai.2000.109620
10. Lehmann K, Schweimer K, Reese G, Randow S, Suhr M, Becker WM, et al. Structure and stability of 2S albumin-type peanut allergens: implications for the severity of peanut allergic reactions. Biochem J. (2006) 395:463–72. doi: 10.1042/BJ20051728
11. Hamilton RG, MacGlashan DW, Saini SS. IgE antibody-specific activity in human allergic disease. Immunol Res. (2010) 47:273–84. doi: 10.1007/s12026-009-8160-3
12. Lieberman JA, Sicherer SH. Diagnosis of food allergy: epicutaneous skin tests, in vitro tests, and oral food challenge. Curr Allergy Asthma Rep. (2011) 11:58–64. doi: 10.1007/s11882-010-0149-4
13. Nicolaou N, Murray C, Belgrave D, Poorafshar M, Simpson A, Custovic A. Quantification of specific IgE to whole peanut extract and peanut components in prediction of peanut allergy. J Allergy Clin Immunol. (2011) 127:684–5. doi: 10.1016/j.jaci.2010.12.012
14. Palladino C, Breiteneder H. Peanut allergens. Mol Immunol. (2018) 100:58–70. doi: 10.1016/j.molimm.2018.04.005
15. Hurlburt BK, Offermann LR, McBride JK, Majorek KA, Maleki SJ, Chruszcz M. Structure and function of the peanut panallergen Ara h 8. J Biol Chem. (2013) 288:36890–901. doi: 10.1074/jbc.M113.517797
16. Mittag D, Akkerdaas J, Ballmer-Weber BK, Vogel L, Wensing M, Becker WM, et al. Ara h 8, a Bet v 1–homologous allergen from peanut, is a major allergen in patients with combined birch pollen and peanut allergy. J Allergy Clin Immunol. (2004) 114:1410–7. doi: 10.1016/j.jaci.2004.09.014
17. Riecken S, Lindner B, Petersen A, Jappe U, Becker WM. Purification and characterization of natural Ara h 8, the Bet v 1 homologous allergen from peanut, provides a novel isoform. Biol Chem. (2008) 389:415–23. doi: 10.1515/BC.2008.038
18. Kondo Y, Urisu A. Oral allergy syndrome. Allergol Int. (2009) 58:485–91. doi: 10.2332/allergolint.09-RAI-0136
19. Pele M. Peanut allergens. Roman Biotechnol Lett. (2010) 15:5204–12. Available online at: https://e-repository.org/rbl/vol.15/iss.2/19.pdf
20. de Jong EC, van Zijverden M, Spanhaak S, Koppelman SJ, Pellegrom H, Penninks AH. Identification and partial characterization of multiple major allergens in peanut proteins. Clin Exp Allergy. (1998) 28:743–51. doi: 10.1046/j.1365-2222.1998.00301.x
21. Koppelman SJ, De Jong GAH, Laaper-Ertmann MD, Peeters KABM, Knulst AC, Hefle SL, et al. Purification and immunoglobulin E-binding properties of peanut allergen Ara h 6: evidence for cross-reactivity with Ara h 2. Clin Exp Allergy. (2005) 35:490–7. doi: 10.1111/j.1365-2222.2005.02204.x
22. Palmer GW, Dibbern DA Jr, Burks AW, Bannon GA, Bock SA, Porterfield HS, et al. Comparative potency of Ara h 1 and Ara h 2 in immunochemical and functional assays of allergenicity. Clin Immunol. (2005) 115:302–12. doi: 10.1016/j.clim.2005.02.011
23. Porterfield HS, Murray KS, Schlichting DG, Chen X, Hansen KC, Duncan MW, et al. Effector activity of peanut allergens: a critical role for Ara h 2, Ara h 6, and their variants. Clin Exp Allergy. (2009) 39:1099–108. doi: 10.1111/j.1365-2222.2009.03273.x
24. Koppelman SJ, Knol EF, Vlooswijk RAA, Wensing M, Knulst AC, Hefle SL, et al. Peanut allergen Ara h 3: isolation from peanuts and biochemical characterization. Allergy. (2003) 58:1144–51. doi: 10.1034/j.1398-9995.2003.00259.x
25. Peeters KA, Koppelman SJ, Van Hoffen E, Van Der Tas CW, den Hartog Jager CF, Penninks AH, et al. Does skin prick test reactivity to purified allergens correlate with clinical severity of peanut allergy?. Clin Exp Allergy. (2007) 37:108–15. doi: 10.1111/j.1365-2222.2006.02628.x
26. Flinterman AE, Van Hoffen E, den Hartog Jager CF, Koppelman S, Pasmans SG, Hoekstra MO, et al. Children with peanut allergy recognize predominantly Ara h2 and Ara h6, which remains stable over time. Clin Exp Allergy. (2007) 37:1221–8. doi: 10.1111/j.1365-2222.2007.02764.x
27. Wu X, Li Y, Liu B, Feng Y, He W, Liu Z, et al. Two-site antibody immunoanalytical detection of food allergens by surface plasmon resonance. Food Anal Methods. (2016) 9:582–8. doi: 10.1007/s12161-015-0232-5
28. Hefle SL, Lambrecht DM. Validated sandwich enzyme-linked immunosorbent assay for casein and its application to retail and milk-allergic complaint foods. J Food Prot. (2004) 67:1933–8. doi: 10.4315/0362-028X-67.9.1933
29. Jayasena S, Smits M, Fiechter D, De Jong A, Nordlee J, Baumert J, et al. Comparison of six commercial ELISA kits for their specificity and sensitivity in detecting different major peanut allergens. J Agric Food Chem. (2015) 63:1849–55. doi: 10.1021/jf504741t
30. Stephan O, Vieths S. Development of a real-time PCR and a sandwich ELISA for detection of potentially allergenic trace amounts of peanut (Arachis hypogaea) in processed foods. J Agric Food Chem. (2004) 52:3754–60. doi: 10.1021/jf035178u
31. Asensio L, González I, García T, Martín R. Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA). Food Control. (2008) 19:1–8. doi: 10.1016/j.foodcont.2007.02.010
32. You J, Li D, Qiao S, Wang Z, He P, Ou D, et al. Development of a monoclonal antibody-based competitive ELISA for detection of β-conglycinin, an allergen from soybean. Food Chem. (2008) 106:352–60. doi: 10.1016/j.foodchem.2007.05.040
33. Koppelman SJ, Vlooswijk RAA, Knippels LMJ, Hessing M, Knol EF, Van Reijsen FC, et al. Quantification of major peanut allergens Ara h 1 and Ara h 2 in the peanut varieties Runner, Spanish, Virginia, and Valencia, bred in different parts of the world. Allergy. (2001) 56:132–7. doi: 10.1034/j.1398-9995.2001.056002132.x
34. Koppelman SJ, Jayasena S, Luykx D, Schepens E, Apostolovic D, De Jong GA, et al. Allergenicity attributes of different peanut market types. Food Chem Toxicol. (2016) 91:82–90. doi: 10.1016/j.fct.2016.02.016
35. Chen H, Zou Z, Tao A. A quantitative method for detecting Ara h 2 by generation and utilization of monoclonal antibodies. J Immunol Res. (2018) 2018:4894705. doi: 10.1155/2018/4894705
36. Engvall E. Quantitative enzyme immunoassay (ELISA) in microbiology. Med Biol. (1977) 55:193–200.
37. Voller A, Bartlett A, Bidwell DE. Enzyme immunoassays with special reference to ELISA techniques. J Clin Pathol. (1978) 31:507–20. doi: 10.1136/jcp.31.6.507
39. Thakur K, Sharma S, Prabhakar S, Gupta P, Anand A. Revisiting the dilution factor as vital parameter for sensitivity of ELISA assay in CSF and Plasma. Ann Neurosci. (2015) 22:37. doi: 10.5214/ans.0972.7531.220108
40. Andreasson U, Perret-Liaudet A, van Waalwijk van Doorn LJ, Blennow K, Chiasserini D, Engelborghs S, et al. A practical guide to immunoassay method validation. Front Neurol. (2015) 6:179. doi: 10.3389/fneur.2015.00179
41. Kleber-Janke T, Crameri R, Appenzeller U, Schlaak M, Becker WM. Selective cloning of peanut allergens, including profilin and 2S albumins, by phage display technology. Int Arch Allergy Immunol. (1999) 265:74. doi: 10.1159/000024203
42. Filep S, Block DS, Smith BR, King EM, Commins S, Kulis M, et al. Specific allergen profiles of peanut foods and diagnostic or therapeutic allergenic products. J Allergy Clin Immunol. (2018) 141:626–31. doi: 10.1016/j.jaci.2017.05.049
Keywords: protocol, peanut allergens, Ara h 1, Ara h 2, Ara h 3, Ara h 6, Ara h 8, ELISA
Citation: Pandey AK, Varshney RK, Sudini HK and Pandey MK (2019) An Improved Enzyme-Linked Immunosorbent Assay (ELISA) Based Protocol Using Seeds for Detection of Five Major Peanut Allergens Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8. Front. Nutr. 6:68. doi: 10.3389/fnut.2019.00068
Received: 28 February 2019; Accepted: 25 April 2019;
Published: 05 June 2019.
Edited by:
José M. Alvarez-Suarez, University of the Americas, EcuadorReviewed by:
Carmen Cuadrado, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), SpainIsabel Ballesteros, University of the Americas, Ecuador
Copyright © 2019 Pandey, Varshney, Sudini and Pandey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hari K. Sudini, aC5zdWRpbmlAY2dpYXIub3Jn; Manish K. Pandey, bS5wYW5kZXlAY2dpYXIub3Jn
 Arun K. Pandey
Arun K. Pandey Rajeev K. Varshney
Rajeev K. Varshney Hari K. Sudini
Hari K. Sudini Manish K. Pandey
Manish K. Pandey