- 1Department of Pharmacology, Graduate School of Medicine, The University of Tokyo, Bunkyo City, Bunkyo, Japan
- 2Graduate School of Brain Science, Doshisha University, Kyotanabe, Kyoto, Japan
To understand how neurons and neural circuits function during behaviors, it is essential to record neuronal activity in the brain in vivo. Among the various technologies developed for recording neuronal activity, molecular tools that induce gene expression in an activity-dependent manner have attracted particular attention for their ability to clarify the causal relationships between neuronal activity and behavior. In this review, we summarize recently developed activity-dependent gene expression tools and their potential contributions to the study of neural circuits.
1 Introduction
Uncovering the relationship between neural circuit activity and behavior is one of the most critical questions in neuroscience. Although only a small fraction of neurons are activated during a specific behavior or event, identifying and manipulating these active neurons provide valuable insights into how the neural circuits process information and execute actions.
One of the most powerful and prevalent technologies for investigating the relationship between neuronal activity and behavior is real-time imaging of cellular activity using genetically-encoded calcium indicators (GECIs) or genetically encoded voltage indicators (GEVIs) (Grewe and Helmchen, 2009; Grienberger et al., 2022). While this approach has significantly clarified the correlation between the two, its limitations include low time resolution, restricted fields of view, and the inability to conclusively prove causality between neuronal activity and behavior (Lecoq et al., 2019; Grienberger et al., 2022).
To overcome these limitations, tools have been developed to express exogenous genes in activated neurons. These tools enable the detection of individual neurons’ activity across the whole brain with single-cell resolution. Moreover, by manipulating neurons with opsins or Designer Receptors Exclusively Activated by Designer Drugs (DREADD) receptors expressed via these tools, it becomes possible to demonstrate the causal relationships between neural circuit activity and behavior through neuronal manipulation (Mayford, 2014; Josselyn and Tonegawa, 2020).
Several tools have been reported for the activity-dependent expression of exogenous molecules to label and manipulate activated neurons. These tools can mainly be classified into two types: those that utilize transcription systems associated with immediate early genes (IEGs), and those that convert the increase in calcium concentration, which accompanies neuronal activity, into reporter gene expression. This review describes recent techniques for labeling and manipulating activated neurons, primarily focusing on their application in the mammalian brain.
2 Advances in activity-dependent gene expressing systems
2.1 IEG-based systems
IEGs are a group of genes, including Arc and c-Fos, which undergo rapid transcription when neurons are activated. IEGs have been widely used as markers of activated neurons, visualized through techniques such as immunostaining. However, visualizing neurons with increased activity in association with a specific behavior for an extended period using IEG expression alone is challenging, due to the transient expression and the short lifetime of IEGs. To overcome this limitation of IEG staining, several methods have been developed to express inducible gene switches, including tetracycline transactivator (tTA) and Cre, under IEG promoters.
2.1.1 TetTag
In the TetTag system (Reijmers et al., 2007) (Figure 1A), tTA is expressed under the control of an IEG promoter. Upon neuronal activation, the IEG promoter drives the expression of tTA which then binds to the tetracycline response element (TRE) to induce transgene expression. As the presence of doxycycline (Dox) inhibits the binding of tTA to TRE, the concentration of Dox determines the time window for TetTag labeling. Reijmers et al. (2007) used c-Fos TetTag mice to label neurons in the basolateral amygdala which were activated during auditory fear conditioning. They demonstrated a strong correlation between the level of fear response during the second exposure to the context or the sound, and the number of IEG expressing neurons during the second exposure which were labeled with TetTag during the first conditioning. TetTag mice have also been used in combination with optogenetic or chemogenetic approaches. Liu et al. (2012) used TetTag mice to express channelrhodopsin-2 only in activated neurons in the dentate gyrus during contextual fear conditioning. By artificially activating these neurons with the light stimulation, mice exhibited freezing behavior, indicating the re-activation of a specific population of dentate gyrus neurons is sufficient for memory retrieval. In addition to these reports, TetTag mice have been widely used for the activity-labeling of neurons in the hippocampus (Ramirez et al., 2013; Tayler et al., 2013; Tanaka et al., 2014; Ohkawa et al., 2015; Okuyama et al., 2016; Pettit et al., 2022), cortex (Stegemann et al., 2023) and spinal cord (Groves et al., 2018).
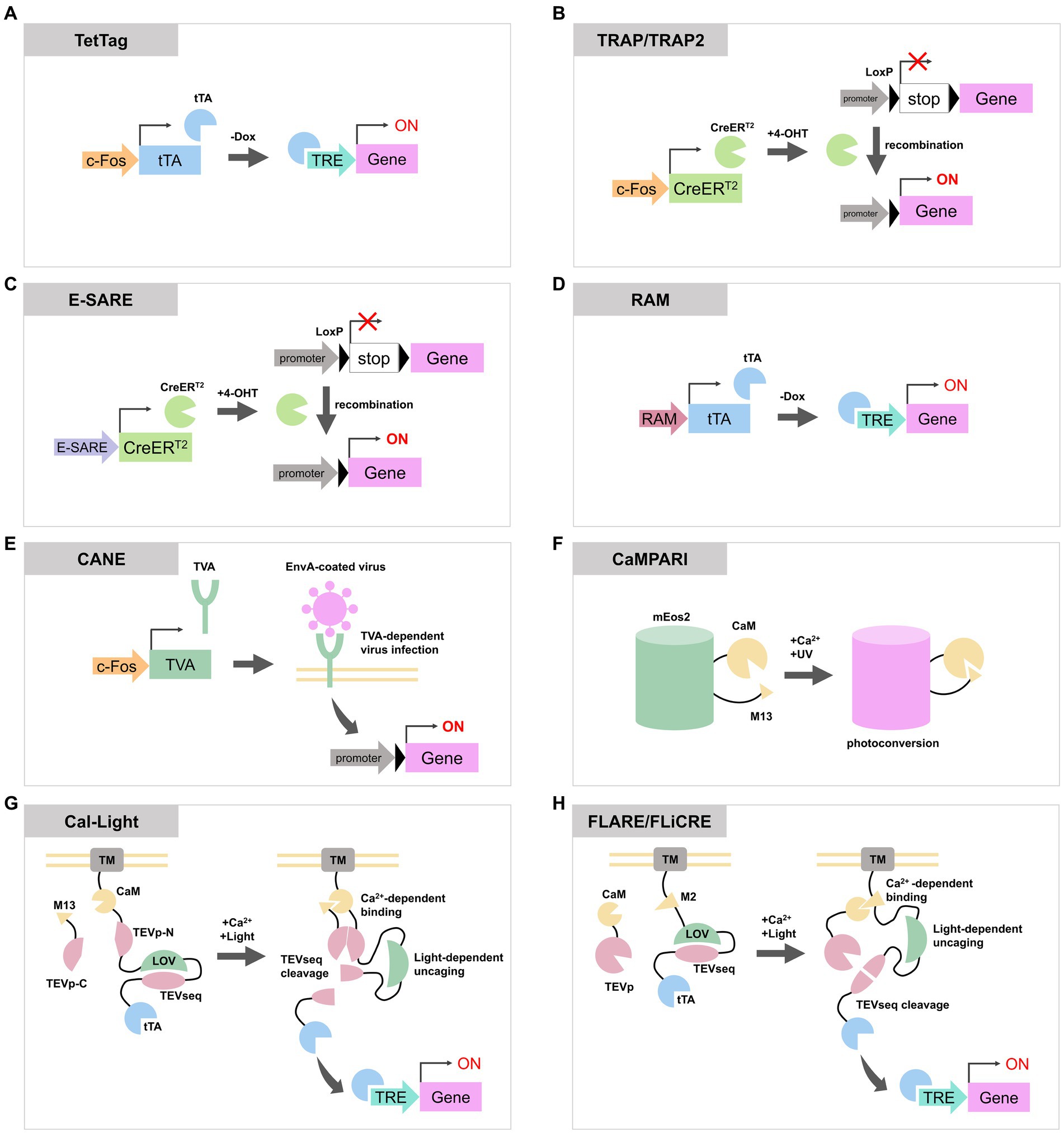
Figure 1. Schematic diagrams of activity-dependent labeling or gene expression tools reviewed in this article. (A) In TetTag system, c-Fos promoter expresses tTA, and tTA induces a transgene expression when the concentration of Dox is low. (B) TRAP and TRAP2 express CreERT2 via c-Fos promoter. CreERT2 recombine LoxP to enable the expression of a reporter gene. (C,D) Both E-SARE and RAM are artificial activity-dependent promoters. They can be combined with Tet-OFF system or CreERT2 to induce reporter expression. (E) In CANE system, activated neurons express destabilized TVA through c-Fos promoter. TVA is located on the plasma membrane and allows the entry of EnvA-coated virus containing the reporter gene. This process enables the expression of transgenes in activated neurons. (F) CaMPARI photoconverts from the green state to the red state in the presence of UV light and elevated Ca2+ concentration. (G,H) Cal-Light, FLARE, and FLiCRE are molecular tools with similar designs. When an increase in calcium ion concentration and exposure to blue light occur simultaneously, they cause cleavage of the TEV sequence to release tTA or Cre, inducing the expression of transgenes. tTA, tetracycline transactivator; Dox, doxycycline; TRE, tetracycline response element; 4-OHT, 4-hydroxytamoxifen; TVA, tumor Virus A receptor; EnvA, envelope protein A; CaM, calmodulin; TEVp, tobacco etch virus protease; TEVseq, TEV protease cleavage sequence; LOV, phototropin 1 light-oxygen-voltage 2 domain; TM, transmembrane domain.
2.1.2 TRAP and TRAP2
Targeted Recombination in Active Populations (TRAP) expresses tamoxifen-dependent Cre recombinase (CreERT2) under the c-Fos or Arc promoter (Guenthner et al., 2013; DeNardo et al., 2019) (Figure 1B). In this system, when a neuron is activated and tamoxifen is present, CreERT2 enter the nucleus and cause recombination of transgenes to enable permanent expression of reporter proteins. The tagging time frame of TRAP is determined by the lifetime of tamoxifen which is approximately one day. Its derivative, 4-hydroxytamoxifen (4-OHT), has a shorter lifetime and TRAP time window of 12 h but maintains the same efficiency as tamoxifen (Guenthner et al., 2013). The first generated TRAP mouse line, also known as TRAP1 or FosTRAP, had CreERT2 knocked into the endogenous c-Fos loci (Guenthner et al., 2013). Consequently, it could potentially impair the physiological function of Fos. In contrast, TRAP2 has CreERT2 fused to the C-terminus of endogenous c-Fos via a 2A peptide. This configuration enables the expression of CreERT2 under the regulation of the c-Fos promoter while maintaining the expression of c-Fos protein. Additionally, TRAP2 underwent codon optimization for Cre. These upgrades in TRAP2 led to improved labeling efficiency particularly in the striatum, amygdala and hypothalamus (DeNardo et al., 2019). TRAP or TRAP2 mice were also used to label auditory or motor cortex (Tasaka et al., 2018, 2020; Hwang et al., 2022), striatum (Girasole et al., 2018) and hypothalamus (Allen et al., 2017; Ishii et al., 2017). TRAP mice were employed for whole-brain analysis of activated neurons during specific events by crossing them with mice lines that express reporters via Cre recombination (Roy et al., 2022).
2.1.3 E-SARE and RAM
Both enhanced synaptic activity-responsive element (E-SARE) (Figure 1C) and Robust Activity Marking system (RAM) (Figure 1D) are synthetic promoters optimized to enhance efficiency and specificity for activity-dependent labeling. They comprise activity-responsive enhancer modules and minimal promoters of IEGs. E-SARE consists of five tandem repeats of the SARE enhancer region, which recruits activity-regulated transcription factors CREB, MEF2, and SRF (Kawashima et al., 2009), along with the minimal Arc promoter. In an assay stimulating cultured neurons, E-SARE has demonstrated higher reporter expression levels and a dynamic range compared to the c-Fos promoter (Kawashima et al., 2013). E-SARE was used to label activated neurons in cortex (Kawashima et al., 2013; Cummings et al., 2021) and hippocampus (Attardo et al., 2018).
RAM consists of four tandem repeats of enhancer modules, comprised of a consensus sequence of the Fos/Jun family called AP-1 and a binding motif of the IEG Npas4, and the human c-Fos minimal promoter. RAM promoter showed lower basal expression of reporters and a higher dynamic range compared to E-SARE (Sørensen et al., 2016). RAM can be employed in conjunction with the Tet-off expression system to regulate the timing of reporter expression. Sun et al. (2020) replaced enhancer modules of RAM with consensus DNA binding motifs of c-Fos or Npas4 to produce c-Fos-selective RAM (F-RAM) and Npas4-selective RAM (N-RAM). Using N-RAM and F-RAM, they were able to selectively tag ensembles of neurons in which the c-Fos-dependent pathway or Npas4-dependent pathway were enhanced during the contextual fear conditioning. They revealed that synaptic changes received by each ensemble are different, and each ensemble influences opposite roles of memory generalization and discrimination (Sun et al., 2020).
2.1.4 CANE
The capturing activated neuron ensembles (CANE) (Figure 1E) is used by a combination of a knockin mouse line, which contains 2A peptide-dsTVA (destabilized form of avian specific tumor virus A receptor) under the endogenous c-Fos loci, and envelope protein A (EnvA)-coated rabies or lentivirus (Sakurai et al., 2016). Activated c-Fos-positive neurons in the knockin mice express dsTVA. dsTVA allows the intracellular entry of EnvA-coated viruses, enabling the introduction of exogenous reporter genes exclusively into activated cells. The short half-life of dsTVA permits control over the timing for reporter expression by adjusting the interval between neuronal activation and the injection of EnvA-coated virus. CANE was used to label c-Fos positive neurons in hypothalamus (Sakurai et al., 2016; Jiang-Xie et al., 2019), periaqueductal grey (Tschida et al., 2019) and pons (Rodriguez et al., 2017). Since CANE can be combined with EnvA-coated rabies virus, it is possible to label presynaptic neurons of the activated neurons through trans-synaptic retrograde infection of the rabies virus (Michael et al., 2020).
2.2 Calcium and light dependent systems
IEG promoters and activity-dependent artificial promoters are used for labeling activated neurons, but they suffer from poor temporal resolutions, spanning several hours, and the fact that IEG expression does not always perfectly match cell firing (Fields et al., 1997). To overcome these limitations, systems have been developed in which cell labeling begins only when both calcium elevation and light exposure occur simultaneously. In these systems, experimenters can control the duration of labeling as desired.
2.2.1 CaMPARI
Calcium-modulated photoactivatable ratiometric integrator (CaMPARI) (Figure 1F) is a protein that undergoes photoconversion from the green state to the red state upon simultaneous occurrence of calcium ion concentration elevation and UV irradiation (Fosque et al., 2015). CaMPARI is derived from circularly permuted mEos2 (McKinney et al., 2009), which undergoes irreversible photoconversion upon violet light illumination. To confer calcium responsiveness to the color change, the N- and C-termini of mEos2 were, respectively, fused with the calcium-binding protein calmodulin (CaM) and its target peptide M13. Using CaMPARI, researchers have been able to label neurons dependent on both neuronal activity and UV light. CaMPARI has also been successfully employed in the larval zebrafish brain and the antennal lobe of Drosophila (Fosque et al., 2015).
It has been indicated that the photoconversion of CaMPARI occurs even in the absence of calcium ion, and that the efficiency of photoconversion is not high under stimulation conditions. Moeyaert et al. (2018) identified a mutant variant of CaMPARI, named CaMPARI2, through saturation mutagenesis at amino acid positions surrounding the fluorescent protein chromophore and protein interfaces between the fluorescent protein and the calcium-sensitive domains of CaMPARI. This new variant exhibited brighter fluorescence and reduced photoconversion under calcium-free conditions compared to CaMPARI. Although the photoconversion rate under high calcium conditions remained comparable to that of CaMPARI, CaMPARI2 showed slower color change in calcium-free conditions, leading to an improved rate contrast of color change (Moeyaert et al., 2018). Sha et al., (2020) also designed reversibly switchable CaMPARI (rsCaMPARI) which can be switched between bright and dark states. rsCaMPARI. rsCaMPARI shifts from a bright state to a dark state upon calcium binding and blue light exposure, and transitions back to the bright state from the dark state by violet light illumination.
CaMPARI labeling has predominantly been utilized for monitoring color changes in the soma of neurons. However, Perez-Alvarez et al. (2020) introduced SynTagMA, a method designed to localize CaMPARI2 specifically to synapses. By fusing CaMPARI2 to synaptophysin or FingR.PSD95, an intrabody against PSD95 (Gross et al., 2013), they successfully targeted CaMPARI2 to presynaptic and postsynaptic sites, respectively. This enabled them to map synaptic activity.
2.2.2 Cal-Light, FLARE, and FLiCRE
CaMPARI only provides “snapshots” of neurons with increased calcium ion concentration during violet light exposure, and it cannot manipulate the populations of neurons with increased activity using optogenetics or chemogenetics methods. Cal-Light (Lee et al., 2017) and Fast Light- and Activity-Regulated Expression (FLARE) (Wang et al., 2017) are both engineered transcription factors that induce the expression of proteins, including channelrhodopsin, in active neuron populations with high temporal precision (Figures 1G,H).
Cal-Light consists of two components. One component includes a transmembrane (TM) domain, CaM, the N-terminal part of tobacco etch virus (TEV) protease (TEVp-N), a TEV protease cleavage sequence (TEVseq) incorporated into an engineered Jα-helix of phototropin 1 light-oxygen-voltage 2 domains from Avena sativa (AsLOV2), and tTA. The other component is a fusion protein of M13 and C-terminal part of TEV protease (TEVp-C). When neurons become active, the concentration of calcium ions in the cytoplasm rises and the binding of calcium ions to CaM in the Cal-Light system increases. This calcium binding triggers a conformational change in CaM, enabling it to interact with M13, which brings TEVp-N and TEVp-C close together. When exposed to the blue light, the AsLOV2 domain undergoes a conformational change to uncover the TEVseq. The proximity of TEVp-N and TEVp-C reconstitutes the active TEV protease, which then cleaves the TEVseq within the AsLOV2 domain. This cleavage releases tTA, enabling the expression of a transgene through the Tet-OFF system in the nucleus (Lee et al., 2017). In the original version of Cal-Light, spontaneous gene expression is occasionally observed. This is attributed to the transient calcium influx at synapses and dendrites independent of action potentials, or the release of stored intracellular calcium. To eliminate the influence of these intrinsic, yet weakly related to neuronal activities, on background gene expression, an improved version called soma targeted Cal-Light (ST-Cal-Light) was developed (Hyun et al., 2022). In ST-Cal-Light, modifications were made to restrict the expression of Cal-Light to the soma, with the aim of minimizing the effect of intrinsic elevations in calcium concentration. The N-terminal 150 residues of the kainate receptor subunit 2, which serve as a soma localization sequence (Shemesh et al., 2017), were inserted between the cytosolic side of the TM domain and CaM (Hyun et al., 2022). In ST-Cal-Light, the expression level remains consistent when both light and calcium are present, while background expression of the reporter gene is suppressed. This resulted in a higher signal-to-noise ratio compared to the original version of Cal-Light in hippocampal primary culture neurons and cortical slices. By utilizing ST-Cal-Light, it was possible to express inhibitory opsins in hippocampal neurons activated during the formation of the contextual fear conditioning and to attenuate freezing responses upon re-exposure to the context (Hyun et al., 2022).
FLARE is a similarly functioning molecular tool consisting of two components. The transcription factor component includes the TM domain of Neurexin3β, the soma targeting sequence of Nav1.6, the CaM binding M2 domain derived from MK2, a TEVseq embedded in an engineered AsLOV2 domain called eLOV, and tTA. The protease component consists of CaM and a truncated TEV protease. Similar to Cal-Light, FLARE utilizes calcium and blue light to release tTA through TEVseq cleavage, which results in the expression of a reporter gene (Wang et al., 2017). As the expression efficiency of FLARE depends on the expression level or ratio of two FLARE components, Sanchez et al. (2020) developed single-chain FLARE (scFLARE). In scFLARE, they replaced CaM and the M2 domain in FLARE with their newly developed calcium-responsive TEV protease (CaTEV) to unite the two FLARE components into a single strand. FLARE and scFLARE were used to label activated neurons in the cortex, amygdala and hippocampus (Wang et al., 2017; Jung et al., 2023; Mocle et al., 2024).
Kim et al. (2020) reported Fast Light and Calcium-Regulated Expression (FLiCRE) which enables more efficient labeling compared to FLARE. In FLiCRE, the TEV protease was replaced with the ultra TEV protease called uTEVp (Sanchez and Ting, 2020), which possesses faster turnover of catalytic activity. Additionally, by combining it with the f-hLOV1, an improved version of LOV domain, obtained through rational engineering of eLOV, the reaction speed was increased, and a higher signal-to-noise ratio was achieved (Kim et al., 2020). It is reported that FLiCRE exhibits fewer leak expression under dark conditions and higher temporal resolution in response to light compared to the original version of Cal-Light (Kim et al., 2020).
3 Discussion
We have discussed recent technological advances in labeling and manipulating neurons based on their activity. These tools have made a significant contribution to elucidating the neuronal mechanisms of perception or action. However, there are still issues with these tools. First, particularly with IEG promoter-dependent systems, the temporal resolution of several hours is slower compared to the actual perception or behavior, which occurs within seconds to minutes (Wang et al., 2019). Due to this limitation, it is important to note that there is still a possibility that these neurons do not fire synchronously, although it can be considered that neurons labeled with activity-dependent systems are more likely to fire synchronously than randomly selected neurons. Second, these tools are not universally applicable to all brain regions and all cell types. While many studies have reported using these tools in the hippocampus, it has been noted that the labeling sensitivity of pyramidal neurons in the CA1 and CA3 regions is lower than that of the granule cells in the dentate gyrus when using TetTag or TRAP2 mice (Deng et al., 2013; Leake et al., 2021). Additionally, neurons belonging to functionally distinct circuits tend to express different IEGs (Mahringer et al., 2022).
Tools for activity-dependent labeling or manipulation of neurons so far have been mainly used for research focusing on investigating whether specific groups of neurons existing in particular regions are strongly associated with certain behaviors and whether artificial stimulations and inhibitions are necessary or sufficient for specific behaviors. However, research on how groups of activated neurons are interconnected to form circuits, as well as how the activation of circuits spanning multiple brain regions determines behaviors, has not yet been sufficiently conducted. Understanding how the activity of neural circuits influences behavior serves as an intermediary to explain the correlation between neuronal activity and behavior. In the future, combined with new genetic tools and measurement techniques, activity-dependent gene expression systems will clarify the causal relationship between the activity of neural circuits and behavior.
Author contributions
TO: Conceptualization, Writing – original draft, Writing – review & editing. KH: Writing – review & editing, Funding acquisition. TS: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work has been supported by JSPS KAKENHI (JP20H03427 and JP22K18358 to KH, JP20KK0171 and JP24K02125 to TS), JSPS Core-to-Core program A. Advanced Research Networks (JPJSCCA20220007 to TS) and Takeda Science Foundation (Bioscience and Specific Research Grants to TS).
Acknowledgments
We apologize to those whose work was not included here due to space limitations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allen, W. E., DeNardo, L. A., Chen, M. Z., Liu, C. D., Loh, K. M., Fenno, L. E., et al. (2017). Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155. doi: 10.1126/science.aan6747
Attardo, A., Lu, J., Kawashima, T., Okuno, H., Fitzgerald, J. E., Bito, H., et al. (2018). Long-term consolidation of ensemble neural plasticity patterns in hippocampal area CA1. Cell Rep. 25, 640–650.e2. doi: 10.1016/j.celrep.2018.09.064
Cummings, K. A., Bayshtok, S., Kenny, P. J., and Clem, R. L. (2021). Ensemble encoding of conditioned fear by prefrontal somatostatin interneurons 18, 452791. doi: 10.1101/2021.07.18.452791
DeNardo, L. A., Liu, C. D., Allen, W. E., Adams, E. L., Friedmann, D., Fu, L., et al. (2019). Temporal evolution of cortical ensembles promoting remote memory retrieval. Nat. Neurosci. 22, 460–469. doi: 10.1038/s41593-018-0318-7
Deng, W., Mayford, M., and Gage, F. H. (2013). Selection of distinct populations of dentate granule cells in response to inputs as a mechanism for pattern separation in mice. eLife 2:e00312. doi: 10.7554/eLife.00312
Fields, R. D., Eshete, F., Stevens, B., and Itoh, K. (1997). Action potential-dependent regulation of gene expression: temporal specificity in ca2+, cAMP-responsive element binding proteins, and mitogen-activated protein kinase signaling. J. Neurosci. Off. J. Soc. Neurosci. 17, 7252–7266. doi: 10.1523/JNEUROSCI.17-19-07252.1997
Fosque, B. F., Sun, Y., Dana, H., Yang, C.-T., Ohyama, T., Tadross, M. R., et al. (2015). Neural circuits labeling of active neural circuits in vivo with designed calcium integrators. Science 347, 755–760. doi: 10.1126/science.1260922
Girasole, A. E., Lum, M. Y., Nathaniel, D., Bair-Marshall, C. J., Guenthner, C. J., Luo, L., et al. (2018). A subpopulation of striatal neurons mediates levodopa-induced dyskinesia. Neuron 97, 787–795.e6. doi: 10.1016/j.neuron.2018.01.017
Grewe, B. F., and Helmchen, F. (2009). Optical probing of neuronal ensemble activity. Curr. Opin. Neurobiol. 19, 520–529. doi: 10.1016/j.conb.2009.09.003
Grienberger, C., Giovannucci, A., Zeiger, W., and Portera-Cailliau, C. (2022). Two-photon calcium imaging of neuronal activity. Nat. Rev. Methods Primers. 2:67. doi: 10.1038/s43586-022-00147-1
Gross, G. G., Junge, J. A., Mora, R. J., Kwon, H.-B., Olson, C. A., Takahashi, T. T., et al. (2013). Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 78, 971–985. doi: 10.1016/j.neuron.2013.04.017
Groves, A., Kihara, Y., Jonnalagadda, D., Rivera, R., Kennedy, G., Mayford, M., et al. (2018). A functionally defined in vivo astrocyte population identified by c-Fos activation in a mouse model of multiple sclerosis modulated by S1P dignaling: immediate-early astrocytes (ieAstrocytes). eNeuro 5:182018. doi: 10.1523/ENEURO.0239-18.2018
Guenthner, C. J., Miyamichi, K., Yang, H. H., Heller, H. C., and Luo, L. (2013). Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784. doi: 10.1016/j.neuron.2013.03.025
Hwang, F.-J., Roth, R. H., Wu, Y.-W., Sun, Y., Kwon, D. K., Liu, Y., et al. (2022). Motor learning selectively strengthens cortical and striatal synapses of motor engram neurons. Neuron 110, 2790–2801.e5. doi: 10.1016/j.neuron.2022.06.006
Hyun, J. H., Nagahama, K., Namkung, H., Mignocchi, N., Roh, S.-E., Hannan, P., et al. (2022). Tagging active neurons by soma-targeted Cal-light. Nat. Commun. 13:7692. doi: 10.1038/s41467-022-35406-y
Ishii, K. K., Osakada, T., Mori, H., Miyasaka, N., Yoshihara, Y., Miyamichi, K., et al. (2017). A labeled-line neural circuit for pheromone-mediated sexual behaviors in mice. Neuron 95, 123–137.e8. doi: 10.1016/j.neuron.2017.05.038
Jiang-Xie, L.-F., Yin, L., Zhao, S., Prevosto, V., Han, B.-X., Dzirasa, K., et al. (2019). A common neuroendocrine substrate for diverse general anesthetics and sleep. Neuron 102, 1053–1065.e4. doi: 10.1016/j.neuron.2019.03.033
Josselyn, S. A., and Tonegawa, S. (2020). Memory engrams: recalling the past and imagining the future. Science 367:eaaw4325. doi: 10.1126/science.aaw4325
Jung, J. H., Wang, Y., Rashid, A. J., Zhang, T., Frankland, P. W., and Josselyn, S. A. (2023). Examining memory linking and generalization using scFLARE2, a temporally precise neuronal activity tagging system. Cell Rep. 42:113592. doi: 10.1016/j.celrep.2023.113592
Kawashima, T., Kitamura, K., Suzuki, K., Nonaka, M., Kamijo, S., Takemoto-Kimura,, et al. (2013). Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat. Methods 10, 889–895. doi: 10.1038/nmeth.2559
Kawashima, T., Okuno, H., Nonaka, M., Adachi-Morishima, A., Kyo, N., Okamura, M., et al. (2009). Synaptic activity-responsive element in the arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc. Natl. Acad. Sci. USA 106, 316–321. doi: 10.1073/pnas.0806518106
Kim, C. K., Sanchez, M. I., Hoerbelt, P., Fenno, L. E., Malenka, R. C., Deisseroth, K., et al. (2020). A molecular calcium integrator reveals a striatal cell type driving aversion. Cell 183, 2003–2019.e16. doi: 10.1016/j.cell.2020.11.015
Leake, J., Zinn, R., Corbit, L. H., Fanselow, M. S., and Vissel, B. (2021). Engram size varies with learning and reflects memory content and precision. J. Neurosci. Off. J. Soc. Neurosci. 41, 4120–4130. doi: 10.1523/JNEUROSCI.2786-20.2021
Lecoq, J., Orlova, N., and Grewe, B. F. (2019). Wide. Fast. Deep: recent advances in multiphoton microscopy of in vivo neuronal activity. J. Neurosci. 39, 9042–9052. doi: 10.1523/JNEUROSCI.1527-18.2019
Lee, D., Hyun, J. H., Jung, K., Hannan, P., and Kwon, H.-B. (2017). A calcium- and light-gated switch to induce gene expression in activated neurons. Nat. Biotechnol. 35, 858–863. doi: 10.1038/nbt.3902
Liu, X., Ramirez, S., Pang, P. T., Puryear, C. B., Govindarajan, A., Deisseroth, K., et al. (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385. doi: 10.1038/nature11028
Mahringer, D., Zmarz, P., Okuno, H., Bito, H., and Keller, G. B. (2022). Functional correlates of immediate early gene expression in mouse visual cortex. Peer Community J. 2:e45. doi: 10.24072/pcjournal.156
Mayford, M. (2014). The search for a hippocampal engram. Phil. Trans. R. Soc. B 369:20130161. doi: 10.1098/rstb.2013.0161
McKinney, S. A., Murphy, C. S., Hazelwood, K. L., Davidson, M. W., and Looger, L. L. (2009). A bright and photostable photoconvertible fluorescent protein. Nat. Methods 6, 131–133. doi: 10.1038/nmeth.1296
Michael, V., Goffinet, J., Pearson, J., Wang, F., Tschida, K., and Mooney, R. (2020). Circuit and synaptic organization of forebrain-to-midbrain pathways that promote and suppress vocalization. eLife 9:e63493. doi: 10.7554/eLife.63493
Mocle, A. J., Ramsaran, A. I., Jacob, A. D., Rashid, A. J., Luchetti, A., Tran, L. M., et al. (2024). Excitability mediates allocation of pre-configured ensembles to a hippocampal engram supporting contextual conditioned threat in mice. Neuron 112, 1487–1497.e6. doi: 10.1016/j.neuron.2024.02.007
Moeyaert, B., Holt, G., Madangopal, R., Perez-Alvarez, A., Fearey, B. C., Trojanowski, N. F., et al. (2018). Improved methods for marking active neuron populations. Nat. Commun. 9:4440. doi: 10.1038/s41467-018-06935-2
Ohkawa, N., Saitoh, Y., Suzuki, A., Tsujimura, S., Murayama, E., Kosugi, S., et al. (2015). Artificial association of pre-stored information to generate a qualitatively new memory. Cell Rep. 11, 261–269. doi: 10.1016/j.celrep.2015.03.017
Okuyama, T., Kitamura, T., Roy, D. S., Itohara, S., and Tonegawa, S. (2016). Ventral CA1 neurons store social memory. Science 353, 1536–1541. doi: 10.1126/science.aaf7003
Perez-Alvarez, A., Fearey, B. C., O’Toole, R. J., Yang, W., Arganda-Carreras, I., Lamothe-Molina, P. J., et al. (2020). Freeze-frame imaging of synaptic activity using SynTagMA. Nat. Commun. 11:2464. doi: 10.1038/s41467-020-16315-4
Pettit, N. L., Yap, E.-L., Greenberg, M. E., and Harvey, C. D. (2022). Fos ensembles encode and shape stable spatial maps in the hippocampus. Nature 609, 327–334. doi: 10.1038/s41586-022-05113-1
Ramirez, S., Liu, X., Lin, P.-A., Suh, J., Pignatelli, M., Redondo, R. L., et al. (2013). Creating a false memory in the hippocampus. Science 341, 387–391. doi: 10.1126/science.1239073
Reijmers, L. G., Perkins, B. L., Matsuo, N., and Mayford, M. (2007). Localization of a stable neural correlate of associative memory. Science 317, 1230–1233. doi: 10.1126/science.1143839
Rodriguez, E., Sakurai, K., Xu, J., Chen, Y., Toda, K., Zhao, S., et al. (2017). A craniofacial-specific monosynaptic circuit enables heightened affective pain. Nat. Neurosci. 20, 1734–1743. doi: 10.1038/s41593-017-0012-1
Roy, D. S., Park, Y.-G., Kim, M. E., Zhang, Y., Ogawa, S. K., DiNapoli, N., et al. (2022). Brain-wide mapping reveals that engrams for a single memory are distributed across multiple brain regions. Nat. Commun. 13:1799. doi: 10.1038/s41467-022-29384-4
Sakurai, K., Zhao, S., Takatoh, J., Rodriguez, E., Lu, J., Leavitt, A. D., et al. (2016). Capturing and manipulating activated neuronal ensembles with CANE delineates a hypothalamic social-fear circuit. Neuron 92, 739–753. doi: 10.1016/j.neuron.2016.10.015
Sanchez, M. I., Nguyen, Q.-A., Wang, W., Soltesz, I., and Ting, A. Y. (2020). Transcriptional readout of neuronal activity via an engineered Ca2+—activated protease. Proc. Natl. Acad. Sci. USA 117, 33186–33196. doi: 10.1073/pnas.2006521117
Sanchez, M. I., and Ting, A. Y. (2020). Directed evolution improves the catalytic efficiency of TEV protease. Nat. Methods 17, 167–174. doi: 10.1038/s41592-019-0665-7
Sha, F., Abdelfattah, A. S., Patel, R., and Schreiter, E. R. (2020). Erasable labeling of neuronal activity using a reversible calcium marker. eLife 9:e57249. doi: 10.7554/eLife.57249
Shemesh, O. A., Tanese, D., Zampini, V., Linghu, C., Piatkevich, K., Ronzitti, E., et al. (2017). Temporally precise single-cell-resolution optogenetics. Nat. Neurosci. 20, 1796–1806. doi: 10.1038/s41593-017-0018-8
Sørensen, A. T., Cooper, Y. A., Baratta, M. V., Weng, F.-J., Zhang, Y., Ramamoorthi, K., et al. (2016). A robust activity marking system for exploring active neuronal ensembles. eLife 5:e13918. doi: 10.7554/eLife.13918
Stegemann, A., Liu, S., Retana Romero, O. A., Oswald, M. J., Han, Y., Beretta, C. A., et al. (2023). Prefrontal engrams of long-term fear memory perpetuate pain perception. Nat. Neurosci. 26, 820–829. doi: 10.1038/s41593-023-01291-x
Sun, X., Bernstein, M. J., Meng, M., Rao, S., Sørensen, A. T., Yao, L., et al. (2020). Functionally distinct neuronal ensembles within the memory engram. Cell 181, 410–423.e17. doi: 10.1016/j.cell.2020.02.055
Tanaka, K. Z., Pevzner, A., Hamidi, A. B., Nakazawa, Y., Graham, J., and Wiltgen, B. J. (2014). Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron 84, 347–354. doi: 10.1016/j.neuron.2014.09.037
Tasaka, G.-I., Feigin, L., Maor, I., Groysman, M., DeNardo, L. A., Schiavo, J. K., et al. (2020). The temporal association cortex plays a key role in auditory-driven maternal plasticity. Neuron 107, 566–579.e7. doi: 10.1016/j.neuron.2020.05.004
Tasaka, G.-I., Guenthner, C. J., Shalev, A., Gilday, O., Luo, L., and Mizrahi, A. (2018). Genetic tagging of active neurons in auditory cortex reveals maternal plasticity of coding ultrasonic vocalizations. Nat. Commun. 9:871. doi: 10.1038/s41467-018-03183-2
Tayler, K. K., Tanaka, K. Z., Reijmers, L. G., and Wiltgen, B. J. (2013). Reactivation of neural ensembles during the retrieval of recent and remote memory. Curr. Biol. 23, 99–106. doi: 10.1016/j.cub.2012.11.019
Tschida, K., Michael, V., Takatoh, J., Han, B.-X., Zhao, S., Sakurai, K., et al. (2019). A specialized neural circuit gates social vocalizations in the mouse. Neuron 103, 459–472.e4. doi: 10.1016/j.neuron.2019.05.025
Wang, W., Kim, C. K., and Ting, A. Y. (2019). Molecular tools for imaging and recording neuronal activity. Nat. Chem. Biol. 15, 101–110. doi: 10.1038/s41589-018-0207-0
Keywords: neuronal activity, immediately early gene, activity-dependent tools, molecular tools, neural circuits
Citation: Onishi T, Hirose K and Sakaba T (2024) Molecular tools to capture active neural circuits. Front. Neural Circuits. 18:1449459. doi: 10.3389/fncir.2024.1449459
Edited by:
Haruyuki Kamiya, Hokkaido University, JapanReviewed by:
Masayuki Sakamoto, Kyoto University, JapanCopyright © 2024 Onishi, Hirose and Sakaba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kenzo Hirose, a2Vuem9oQG0udS10b2t5by5hYy5qcA==; Takeshi Sakaba, dHNha2FiYUBtYWlsLmRvc2hpc2hhLmFjLmpw
 Taichi Onishi1
Taichi Onishi1 Takeshi Sakaba
Takeshi Sakaba