- 1Department of Physiology and Medical Physics, RCSI, University of Medicine and Health Sciences, Dublin, Ireland
- 2FutureNeuro SFI Research Centre, RCSI, University of Medicine and Health Sciences, Dublin, Ireland
- 3UCD School of Biomolecular and Biomedical Science, UCD Conway Institute, University College Dublin, Dublin, Ireland
RNA modifications have emerged as an additional layer of regulatory complexity governing the function of almost all species of RNA. N6-methyladenosine (m6A), the addition of methyl groups to adenine residues, is the most abundant and well understood RNA modification. The current review discusses the regulatory mechanisms governing m6A, how this influences neuronal development and function and how aberrant m6A signaling may contribute to neurological disease. M6A is known to regulate the stability of mRNA, the processing of microRNAs and function/processing of tRNAs among other roles. The development of antibodies against m6A has facilitated the application of next generation sequencing to profile methylated RNAs in both health and disease contexts, revealing the extent of this transcriptomic modification. The mechanisms by which m6A is deposited, processed, and potentially removed are increasingly understood. Writer enzymes include METTL3 and METTL14 while YTHDC1 and YTHDF1 are key reader proteins, which recognize and bind the m6A mark. Finally, FTO and ALKBH5 have been identified as potential erasers of m6A, although there in vivo activity and the dynamic nature of this modification requires further study. M6A is enriched in the brain and has emerged as a key regulator of neuronal activity and function in processes including neurodevelopment, learning and memory, synaptic plasticity, and the stress response. Changes to m6A have recently been linked with Schizophrenia and Alzheimer disease. Elucidating the functional consequences of m6A changes in these and other brain diseases may lead to novel insight into disease pathomechanisms, molecular biomarkers and novel therapeutic targets.
Introduction
The brain is the most complex and cellularly diverse organ in the body. Higher order brain functions, and those functions critical to sustain life are facilitated by the concerted activity of different cell types, each functionally driven by distinct, context dependent gene expression and gene expression regulation patterns (Hawrylycz et al., 2012; Mitra et al., 2021). Recently, the application of single cell RNA-sequencing technologies has begun to reveal at single cell resolution this dynamic and distinct gene expression patterns and how they respond to stimulus or activity (Agarwal et al., 2020; Pfisterer et al., 2020; Joglekar et al., 2021; Song et al., 2021).
Appropriate spatiotemporal gene expression in the brain involves several tightly monitored layers of regulation. Broad effector transcription factors direct transcription of genes via canonical binding sites throughout the genome, a process known to be critical for learning and memory and memory consolidation (Kaldun and Sprecher, 2019). Epigenetic mechanisms (DNA methylation, histone modifications etc.), often in response to transcription factor signals, modify DNA and associated histone structure to enhance or repress transcription (Kim et al., 2009; Conboy et al., 2021). Post-transcriptional mechanisms, including microRNAs, are also known to profoundly influence gene expression (or rather mRNA translation) in the brain adding an additional layer of regulatory complexity and fine-tuning of gene output (Bartel, 2018; Brennan and Henshall, 2020). The proteome is also tightly regulated and post-translational modifications such as phosphorylation, ubiquitylation and sumoylation dictate the efficacy or function of the resulting protein according to the needs of a given cell (Zhang et al., 2016; Czuba et al., 2018). These processes are not independent of each other and function cohesively, cumulatively forming a complex control over gene output.
RNA is now widely recognized to undergo complex post-transcriptional editing and modification (in addition to alternative splicing and polyadenylation) which confer additional information carrying capacity and profoundly influence its fate (Jain et al., 2019; Zaccara et al., 2019; He and He, 2021). Among the most well studied involves adenosine to inosine (A-to-I) conversion which is mediated by both ADAR I and II enzymes (Eisenberg and Levanon, 2018; Christofi and Zaravinos, 2019). A-I editing involves the hydrolytic deamination of adenosine to inosine. This recoding can result in non-synonymous amino acid substitutions, resulting in altered protein-coding sequences and potentially altered protein structure (Eisenberg and Levanon, 2018). Additionally, recoding of the RNA sequence within the 3′-UTR may alter mRNA-translational efficiency as it may affect microRNA-mediated targeting (Brummer et al., 2017; van der Kwast et al., 2020). Other modifications are more subtle than overt structural modification of bases, involving covalent modification of RNA such as the addition of methyl groups to specific nucleotides. Various forms of RNA were known to undergo extensive methylation as far back as the 1970s (Munns et al., 1977; Wei and Moss, 1977; Burke and Joh, 1982). However, recent advances in transcriptomic technologies and the development of antibodies, which specifically target these modifications have made it possible to reveal the extent and identity of these novel RNA modifications (Dominissini et al., 2012; Meyer et al., 2012; Dominissini et al., 2013). Critically, many of these modifications may be dynamic in nature undergoing context-dependent addition and removal.
The addition of methyl groups to the N6 position of adenosine [N6-methyladenosine (m6A)] is the most abundant internal modification in RNA and is prevalent in brain tissue. However, the function of m6A in brain is only beginning to emerge. In this review, we provide an overview of the mechanisms by which m6A is regulated and try to define its overall contribution to the gene expression landscape in brain cells. We also discuss recent reports, which detail the involvement of m6A in neurological diseases.
M6A Rna Methylation
It was identified in the 1970s that certain forms of RNA such as rRNA undergo extensive methylation (Munns et al., 1977; Wei and Moss, 1977). However, it was thought to be mainly structural and further interrogation was limited by the technology at this time (Furuichi et al., 1975). The development of an antibody which recognizes the m6A mark prompted transcriptome-wide investigations of N6-methyladenosine. This modification was found to be widespread and enriched near stop codons, the 5′ and 3′ UTR and internal long exons of mRNA, as well as rRNA, tRNA, snoRNA and lncRNA (Meyer et al., 2012; Berulava et al., 2015). A canonical m6A motif was identified and consists of RRACH with R =G/A and H =A/C/U (Balacco and Soller, 2019; Dominissini et al., 2012). The methyl group is catalyzed from a donor substrate S-adenosylmethionine (SAM) to an adenosine residue of an RNA moiety along a specific sequence as stated above (Bokar et al., 1997). The m6A modification plays a role in several diverse RNA mechanisms, most notably RNA stability and translational efficiency (Meyer et al., 2015; Chen X.Y. et al., 2019). Other studies have implicated m6A in the control of mRNA dynamics including alternative splicing (although there is considerable debate regarding this Ke et al., 2017) and subcellular localization. Moreover, the role of m6A may be dictated by the subcellular localization of the m6A-tagged RNA. In the nucleus, m6A deposited on nascent pre-mRNA may influence alternative splicing (Dominissini et al., 2012), and microRNA biogenesis (Alarcon et al., 2015), while in the cytoplasm, it is thought to regulate RNA stability (Batista et al., 2014; Wang X. et al., 2014), translational efficiency and RNA decay (Wang et al., 2015).
M6A Machinery
M6A is a dynamic modification, catalyzed by a distinct enzymatic complex (writers), identified and processed by several “reader” proteins and potentially removed by “eraser” proteins. In the following sections, we summarize what is known about the proteins associated with the deposition, identification, and removal of m6A (Figure 1).
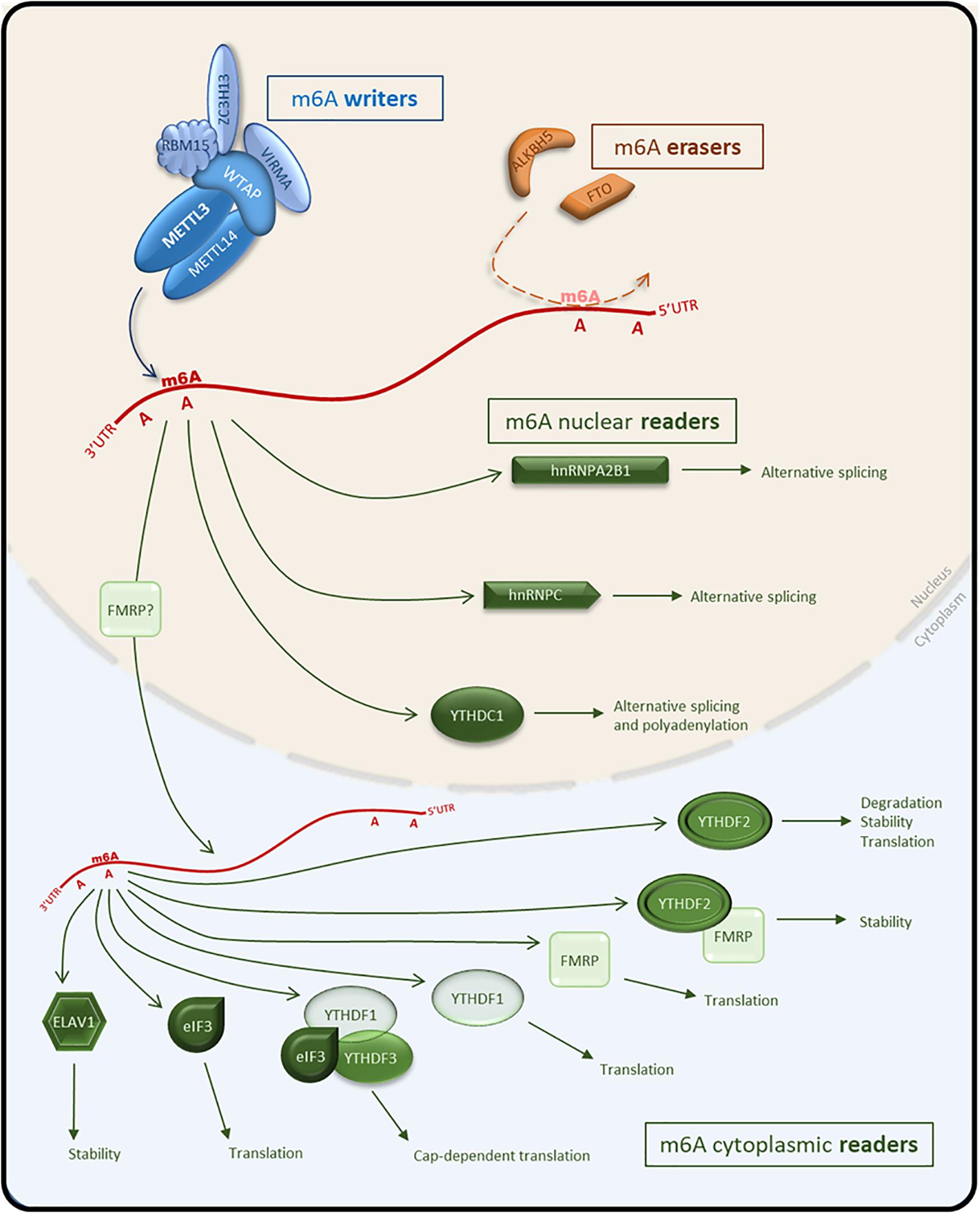
Figure 1. Schematic representation of the m6A pathway and effectors on mRNA. The MACOM complex composed of m6A writers (METTL3, METTL14, WTAP, VIRMA, ZC3H13, and RBM15) deposits m6A on target RNAs. M6A erasers (FTO and ALKBH5) remove the m6A mark. M6A nuclear readers (hnRNPC, hnRNPA2B1, and YTHDC1) facilitate alternative splicing or polyadenylation following recognition of m6A-tagged RNA. M6A-tagged RNA can be exported to the cytoplasm and bound by cytoplasmic readers (eIf3, ELAVL1, YTHDF1,2,3) to modulate stability, translational efficiency or the degradation of RNA. Blue, m6A writers; Orange, m6A erasers; Green, m6A readers.
M6A Writers
The methyltransferase complex which catalyzes m6A addition is composed of two distinct sub-complexes (Knuckles et al., 2018): the m6A-methyltransferase-like (METTL) complex (MAC) which is composed of METTL3 and METTL14 and the m6A-METTL associated complex (MACOM) which consists of RBM15, ZC3H13, WTAP, and VIRMA (Meyer and Jaffrey, 2014; Cao et al., 2016; Lence et al., 2019). Together MAC and MACOM function to catalyze the addition of methyl groups to adenosine.
MAC Complex
The MAC-associated proteins comprise the catalytic components of the methyltransferase complex, which co-transcriptionally deposit m6A on target mRNAs (Liu et al., 2014). There are two essential components of the MAC complex, METTL3 and METTL14 which form a conserved heterodimeric core. This dimerization is essential for their methylation function and provides a synergistic effect on the catalytic activity of the complex (Liu et al., 2014; Balacco and Soller, 2019). Crystallographic studies of the MAC complex have shed light on the mechanisms of m6A deposition on target mRNA molecules (Sledz and Jinek, 2016). Additionally, transcriptome-wide profiling of m6A has identified a specific sequence motif known as a RRACH with R = G/A and H = A/C/U sequence within which m6A is usually confined. This RRACH sequence is highly conserved and restricts m6A to a selection of conserved transcripts (Dominissini et al., 2012). In some species, such as C. elegans, the dimer is replaced by a prologue, METTL4. Furthermore, METTL16, which is not a prologue, can also methylate mRNA but its mechanism of action remains to be elucidated (Balacco and Soller, 2019).
METTL3 is the primary catalytic component of the MAC complex. Knockout of METTL3 has been reported to result in a significant reduction in global m6A levels (Batista et al., 2014; Wang Y. et al., 2014). METTL3 selectively targets mRNAs, depositing m6A on nascent RNA via its zinc-finger motifs (Batista et al., 2014; Sledz and Jinek, 2016). A mechanistic study in acute myeloid leukemia (AML) found that METTL3 is recruited by the transcription factor CHOP (CEBPζ) and methylates CHOP-regulated transcripts (Barbieri et al., 2017). METTL3 levels influence the expression of other m6A-associated writers. Indeed, both elevated and reduced levels of METTL3 lead to increased amounts of WTAP mRNA translation and promote the stabilization of the protein (Sorci et al., 2018). Furthermore, METTL3 can reportedly switch from writer to reader by translocating to the cytoplasm where it may regulate the translation of specific mRNAs by direct binding to RNA and recruitment of eIF3 (Lin et al., 2016).
METTL14 has a functional role in structural stabilization and RNA substrate recognition (Sledz and Jinek, 2016). METTL14 enhances METTL3 methyltransferase activity by binding the mRNA and orientating the SAM methyl group for the reaction (Schwartz et al., 2014; Balacco and Soller, 2019). In the brain, METTL14 also plays an important role in maintaining neuronal populations via targeting transcripts of transcription factors of cell cycle to promote their decay (Yoon et al., 2017; Balacco and Soller, 2019). While it does not possess catalytic activity, the depletion of METTL14 causes profound reduction in m6A levels in embryonic stem cells, is embryonic lethal in mice and affects cortical development suggesting a critical role in development and highlighting its necessity for normal MAC activity (Wang Y. et al., 2014; Yoon et al., 2017).
MACOM Complex
MACOM is an m6A-associated complex (Meyer and Jaffrey, 2014; Cao et al., 2016; Lence et al., 2019). It is thought that the proteins associated with the MACOM complex may regulate m6A deposition by integrating cellular signaling pathways and stimuli, dictating the repertoire of transcripts which undergo m6A-tagging (Livneh et al., 2020). The function of the MACOM is under intense investigation.
WTAP (Wilms’ tumor 1-associated protein) is a ubiquitously expressed protein and a regulatory subunit required for formation of a functional and stable MACOM complex (Ping et al., 2014). WTAP also modulates RNA processing, translation, and alternative splicing (Horiuchi et al., 2013). It was through the mass-spectrometric analysis of WTAP binding partners that several other methyltransferase enzymes were identified including METTL3-METTL14, RBM15 and VIRMA (Ping et al., 2014).
VIRMA (vir like m6A methyltransferase associated), also named KIAA1429, recruits the WTAP-METTL3-METTL14 complex via its binding to WTAP (Yue et al., 2018; Balacco and Soller, 2019). VIRMA may also interact with polyadenylation cleavage factors linking the machineries of m6A methylation and polyadenylation during mRNA processing (Yue et al., 2018).
RBM15 (RNA-binding motif protein 15) interacts with METTL3 via WTAP. It is particularly associated with the regulation of m6A levels on lncRNAs (Patil et al., 2016; Balacco and Soller, 2019). RBM15 acts as an adaptor protein recruiting the m6A methylosome to U-rich regions.
ZC3H13 (Zinc Finger CCCH-type containing 13), also named Flacc (Fl(2)d-associated complex component) (Wen et al., 2018; Balacco and Soller, 2019) may have a scaffolding function in the m6A methylosome. In Wen et al. (2018) found that ZC3H13 promotes pluripotency-associated gene expression and suppresses differentiation-associated genes in mESCs. Loss of ZC3H13 impairs WTAP-dependent 3′UTR m6A events. Moreover, Knuckles et al. have shown that ZC3H13 binds RBM15 and regulates the m6A pathway via the stabilization of the interaction between WTAP and RBM15 (Knuckles et al., 2018).
M6A Erasers
Two proteins have been shown to possess m6A-demethylase activity. FTO (Fat mass and obesity-associated protein) and ALKBH5 (AlkB homologue 5 protein). These proteins are [(α-ketoglutarate (α-KG)-dependent dioxygenases which are inhibited by D2-hydroxyglutarate (D2-HG)] (Fedeles et al., 2015; Chen X.Y. et al., 2019).
FTO is an AlkB-like 2-oxoglutarate-dependent nucleic acid demethylase (Gerken et al., 2007), initially identified for its role in diabetes and obesity (Lindgren and McCarthy, 2008; Loos and Bouchard, 2008). Since then it has been shown that, in the nucleus, FTO catalyzes the removal of m6A and m6Am (N6, 2’-O-dimethyladenosine) marks from mRNA although importantly shows a 100-fold greater affinity for m6Am than m6A (Mauer et al., 2017; Mauer and Jaffrey, 2018; Balacco and Soller, 2019). The m6Am mark is a terminal modification distinct from the internal m6A mark and follows the m7G (N7-methylguanosine) cap at the N6 position (Wei et al., 1975), where A or m6A are not found (Wei et al., 1976). Some studies have identified FTO as an important regulator of m6A-tagged transcripts, important for mRNA alternative splicing and gene expression, contributing to the regulation of adipogenesis (Zhao et al., 2014; Bartosovic et al., 2017; Chen X.Y. et al., 2019). However, some recent emerging data does not support the role of FTO as an m6A eraser and suggests that FTO under normal physiological conditions does not remove m6A and solely functions on m6Am (Mauer et al., 2017). Indeed, FTO is present almost exclusively in the nucleus, which suggests it is unlikely to dynamically regulate cytosolic mRNAs. Furthermore, studies by the Darnell group have shown that the levels of m6A are perhaps stable once deposited. Together, this calls into question the extent of m6A demethylation in vivo, at least under normal physiological conditions (Mauer et al., 2017; Mauer and Jaffrey, 2018). M6A removal by FTO or others may therefore be context and cell-type specific and/or developmentally regulated and this warrants further investigation.
ALKBH5 is an α-ketoglutarate dependent oxidase and nuclear demethylase only found in vertebrates which can bind single stranded nucleic acids (Zheng et al., 2013; Balacco and Soller, 2019). In Ensfelder et al. (2018) identified ALKBH5 as a ribosomal RNA m6A eraser. It has since been shown that ALKBH5 is also required for correct splicing and production of transcripts in spermatocytes via selective removal of m6A (Tang et al., 2018). Moreover, ALKBH5 is involved in glioblastoma to repress tumorigenesis (Zhang et al., 2017), in spermatogenesis and in male fertility (Zheng et al., 2013). Again, the relevance of ALKBH5 as an m6A eraser in somatic cells remains unclear. The study by Darnell did note that there was loss of m6A on a minority of transcripts suggesting a small amount of selective demethylation may occur under normal physiological conditions (Ke et al., 2017).
M6A Readers
M6A readers elicit different actions upon recognition and binding of the methylated RNA transcripts. Several reader proteins have now been identified.
HNRNPC & HNRNPA2B1 (Heterogeneous Nuclear Ribonucleoprotein C & A2B1) are both nuclear-confined m6A readers and are particularly involved in pre-mRNA processing and in maturation of pri-miRNA to pre-miRNA (Cao et al., 2016). HNRNPA2B1 promotes processing of METTL3-dependent microRNAs and plays a role in the regulation of the alternative splicing of exons (Alarcon et al., 2015).
YTH domain containing proteins (YTHDC1 and YTHDC2) and YTH domain-containing family proteins (YTHDF1, YTHDF2 and YTHDF3), a highly conserved family of proteins, have been shown to detect and bind m6A (Berlivet et al., 2019). The DC proteins are usually confined to the nucleus (Balacco and Soller, 2019) while the DF proteins are found primarily in the cytoplasm. The structure of m6A-bound by YTH-domain containing proteins was elucidated several years ago. The YTH domain is critical for this binding and is involved in the development of a tryptophan cage around the m6A mark (Theler et al., 2014; Xu et al., 2014). Recent iCLIP data from the Jaffrey lab, identified the binding sites for all YTH proteins. They found that DC1 binds m6A sites in mRNA and nuclear non-coding RNAs, while all three DF proteins bind most m6A sites (Patil et al., 2018). It has been found that DC1 contributes to extensive alternative polyadenylation and 3′UTR length alteration (Chen X.Y. et al., 2019) while DC2 is primarily localized in the testes where it has been found to have weak m6A affinity (Wojtas et al., 2017). In the mouse hippocampus, upon binding, YTHDF1 promotes the translation of m6A-tagged transcripts via interaction with a translation initiation factor (Wang et al., 2015) and the help of YTHDF3 (Shi et al., 2017) and of eIF1 (Chen X.Y. et al., 2019) to facilitate learning and memory (Shi et al., 2018). YTHDF2 recognition of m6A sites, contrarily, was reported to regulate the degradation of specific mRNAs via the recruitment of CCR4-NOT (Alarcon et al., 2015; Cao et al., 2016; Du et al., 2016; Chen X.Y. et al., 2019). This protein may also play a role in mRNA stability and translation (Wang X. et al., 2014; Chen X.Y. et al., 2019), targeting stop codon regions, 3′UTR and coding regions (Wang X. et al., 2014). Recent studies however, including iCLIP-sequencing datasets suggest that all three DF proteins do not selectively bind to m6A-tagged transcripts and all likely lead to degradation of bound transcripts. It is possible that context dependence is critical toward understanding the function of m6A-recognition by YTH-domain containing proteins.
METTL16, usually a writer enzyme, may also function as a reader when SAM levels are low, in order to stimulate the SAM synthetase MAT2A mRNA translation to increase the level of SAM. In contrast, when SAM levels are high, METTL16 methylates MAT2A mRNA, which promotes nuclear mRNA decay (Pendleton et al., 2017; Balacco and Soller, 2019).
elF3 (eukaryotic initiation Factor 3) is a component of the 43s translation pre-initiation complex. It has been found that this protein is an m6A reader, which promotes translation via two mechanisms. First, eIF3 can directly bind the 5′UTR-localised m6A to allow the recruitment of the 43s complex (Meyer et al., 2015; Cao et al., 2016). The second method is via its interaction with YTHDF1 which facilitates cap-dependent translation (Wang et al., 2015).
FMRP (Fragile X mental retardation protein) is an RNA-binding protein, loss of which is implicated in the neurodevelopmental condition Fragile X syndrome (FXS). It has been shown to be important for the stability of m6A-tagged mRNA (Zhang et al., 2018) and for their nuclear export (Hsu et al., 2019). FMRP has been found to interact with WTAP and recently suggested to bind m6A sites by competing with YTHDF2 to regulate specific mRNA turnover (Horiuchi et al., 2013; Balacco and Soller, 2019). Indeed, another study found that FMRP modulates the stability of its target through YTHDF2 (Zhang et al., 2018). FMRP was recently shown to directly bind to m6A-marked mRNA (Westmark et al., 2020) and negatively regulate their translation (Edupuganti et al., 2017). How m6A processing in FXS is affected has yet to be analyzed.
ELAVL1 (Drosophila homologue-like 1), also known as human antigen R (HuR), and IGF2BPs (Insulin-like growth factor 2 mRNA-binding proteins) has been identified as an m6A reader (Balacco and Soller, 2019). Several studies show that ELAVL1 increases the stability of transcripts via m6A-mRNA-binding. Indeed, in prostatic carcinoma cells, ELAVL1 plays a role in stabilizing integrin β1 mRNA (Li et al., 2020). It may also promote SOX2 mRNA stabilization to stimulate the maintenance of glioma stem-like cells (Visvanathan et al., 2018) as well as DRG1 in osteosarcoma human tissue (Ling et al., 2020).
M6A in the Brain
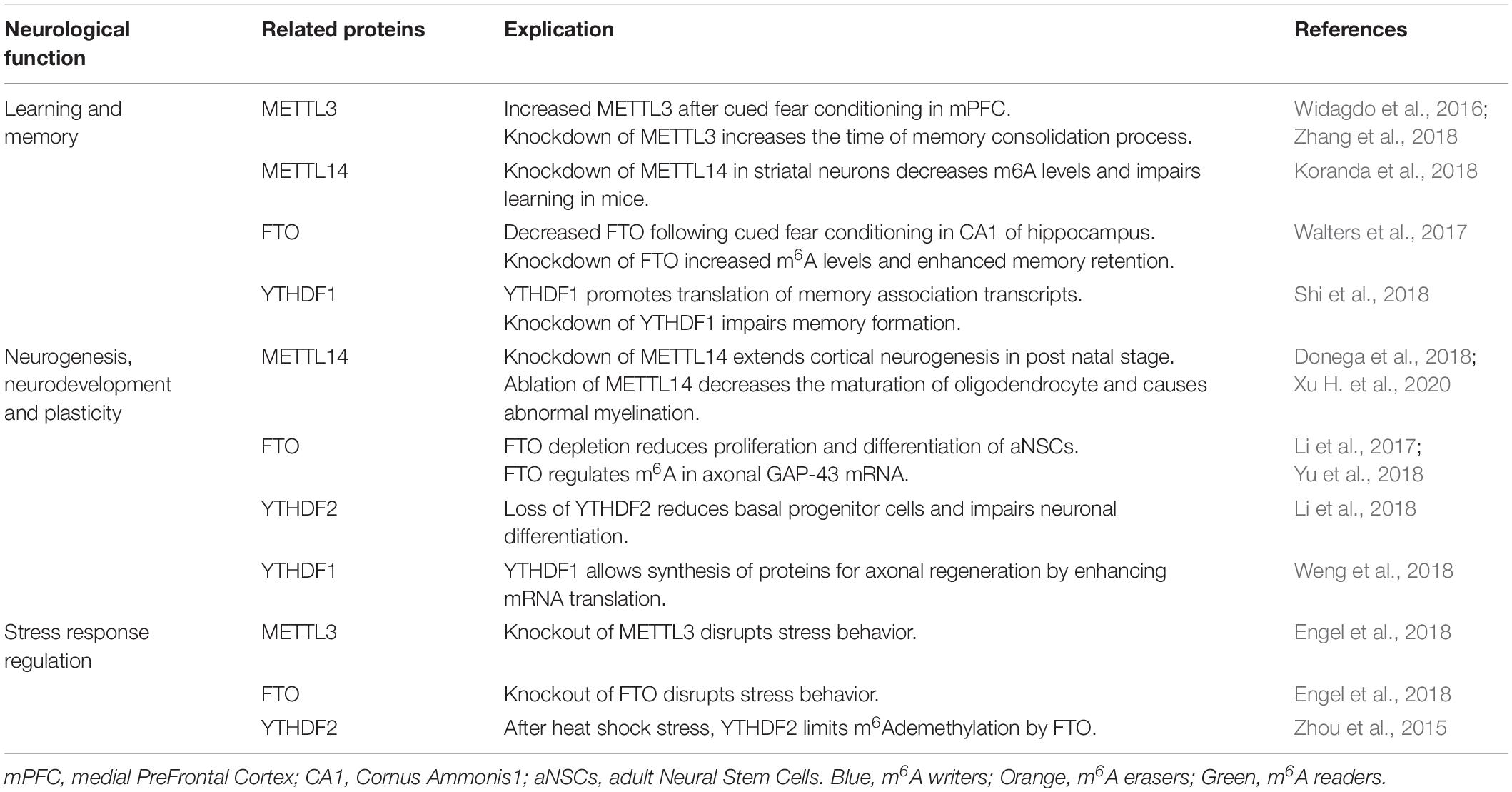
The m6A mark influences the behavior of RNA in several biological pathways and subsequently biological processes like cell differentiation and proliferation, development, sex determination and circadian rhythms (Chen et al., 2017). Here, we review the most important and studied cerebral processes impacted by m6A methylation such as learning and memory as well as neurogenesis, neurodevelopment, the stress response, myelination, and axon plasticity (Table 1).
Learning and Memory
In Widagdo et al. (2016) performed the first exploration of m6A in synaptic plasticity and memory processes. To assess whether m6A was regulated by experience, they used a cued-fear conditioning model in mice and found extensive experience-induced m6A changes using meRIP-Seq. Additionally, they found an increase in m6A marks on mRNA in the medial prefrontal cortex (mPFC) following the cued-fear conditioning paradigm as well as increased levels of METTL3 suggesting a critical role for m6A in behavioral adaptation. A subsequent study also using a fear-conditioning paradigm, found decreased levels of FTO in dorsal CA1 hippocampal neurons following fear-conditioning (Walters et al., 2017). Subsequent knock-down of FTO, in mPFC or in the CA1 increased m6A levels (Walters et al., 2017) and enhanced memory retention suggesting m6A plays a critical role in memory formation and consolidation (Widagdo et al., 2016; Walters et al., 2017). Zhang et al. found that conditional postnatal depletion of METTL3 in hippocampus of mice prolonged the process of memory consolidation but did not alter short-term plasticity. Furthermore, restitution of METTL3 improved learning while the overexpression of METTL3 with a mutated methyltransferase domain had no effect (Zhang et al., 2018). Together this suggests that METTL3 participates in the enhancement of long-term memory consolidation via its m6A methyltransferase function (Zhang et al., 2018). Moreover, another study showed that m6A methylation promotes learning and memory through YTHDF1, which boosts translation of memory-associated transcripts. Indeed, the depletion of YTHDF1 impairs long-term potentiation of hippocampal synapses leading to impairment of memory formation (Shi et al., 2018). The METTL3/YTHDF pathway is also required for memory formation in Drosophila. METTL3 knockdown in the mushroom body, impaired memory as assessed using an aversive conditioning paradigm to assess short-term memory. They further identified that YTHDF hemizygotes exhibited age-related memory impairments similar to METTL3 knockdown flies. Furthermore, METTL14 deletion in striatal neurons induces a decrease of m6A methylation and impairs learning in mice (Koranda et al., 2018).
Local supply of mRNA, microRNAs and translational machinery facilitate rapid synaptic alterations required for learning and memory. Recently, Merkurjev et al. (2018) demonstrated synaptic enrichment of several m6A-associated enzymes including METTL14, and YTHDF1-3 as well as m6A-tagged polyA RNA. The authors isolated and profiled synaptosomal m6A-tagged RNA using a low-input meRIP-Seq approach to reveal that the most significant biological processes represented by synaptic m6A-tagged RNAs were those critical for neuronal integrity and function, suggesting m6A-tagged synaptic RNAs critically modulate neuronal function. Furthermore, disruption of m6A-processing via knockdown of reader proteins YTHDF1 or YTHDF3 resulted in significant morphological disruptions including increased spine neck length and decreased spine head width, reduced PSD-95 clustering, and reduced surface expression of AMPA receptors (Merkurjev et al., 2018).
Other studies have also revealed a potential role for m6A-tagged RNAs in synaptic function. Indeed, several experimental paradigms have been shown to alter methylation of synapse associated RNAs including cocaine exposure, depression, and deep brain stimulation (Madugalle et al., 2020; Song et al., 2020; Xue et al., 2021).
Although further studies are required, these initial data suggest that m6A may represent a critical mechanism of mRNA translational priming and enable rapid sorting of mRNAs required for synaptic function.
Neurogenesis/Neuronal Development
Neurogenesis, the process by which new neurons are formed, is critical for correct neurodevelopment, function and repair. Furthermore, aberrant neurogenesis is associated with several neurological diseases including epilepsy (Danzer, 2008; Livneh et al., 2020). Selective deletion of METTL14 in embryonic mouse brains expands cortical neurogenesis into the postnatal stage, extends the cell cycle progression in neuronal progenitor cells and prolongs maintenance of radial glia cells (Yoon et al., 2017). This suggests m6A has an important role during brain development. During pre-natal cortical development m6A-tagging of developmental transcription factors Pax6, Sox2 and Neurogenin2 is enriched and promotes turnover allowing rapid and complex temporal regulation of gene expression critical for development (Donega et al., 2018). M6A levels and deposition are dynamically regulated during post-natal cerebellum development which correlates with spatiotemporal regulation of m6A-associated enzymes including METTL3, 14, and FTO (Ma et al., 2018). Importantly, transcripts essential for development in post-natal cerebellum were continuously and consistently methylated whereas time-specific processes were temporarily methylated inducing proper cerebellar development as disruption of either METTL3 or FTO caused developmental deficits. Furthermore, it has been shown that FTO deficiency reduces the proliferation and differentiation of adult neural stem cells (aNSCs) leading to a smaller brain in mice (Li et al., 2017). While m6A writers have emerged as important mediators of neuronal development, the role of m6A readers is less clear. Recently, Li and colleagues demonstrated that loss of YTHDF2 is embryonic lethal and found that neuronal development dysfunction resulting from YTHDF2 loss prevented viability. Analysis of embryonic brains from E12.5 to E14.5 mice identified that YTHDF2 loss resulted in reduced cortical layering and reduced cortical development. Further analysis revealed reduced basal progenitor cells and impaired neural differentiation (Li et al., 2018).
M6A methylation has been found to be essential for the maturation of oligodendrocytes and for central nervous system myelination. In mice, in the early postnatal stage, METTL14 ablation leads to a decrease of oligodendrocyte maturation and causes abnormal myelination. In vitro, METTL14 depletion induced a diminution of oligodendrocyte differentiation and a prolongation of the cell cycle progression. This indicates a role for m6A methylation in neuronal development, transmission and plasticity (Xu H. et al., 2020).
In addition to a role in neuronal development, m6A may also play a crucial role in neuronal repair and in axonal regeneration. In peripheral nerves, it was shown that m6A allows the translation of retrograde signaling molecule mRNAs which enables rapid axonal regeneration. Following axonal injury, m6A levels are increased, which promotes the synthesis of proteins involved in axon regeneration and recovery, through YTHDF1, to enhance mRNA translation (Weng et al., 2018). Moreover, FTO is enriched in axons leading to a regulation of m6A in axonal GAP-43 mRNA, which is required for axon elongation (Yu et al., 2018). Further research is now required to determine whether the m6A pathway may be exploited to improve nerve regeneration following physical injury.
Stress Response Regulation
Physical and emotional stressors are known to influence transcriptional and translational output and significantly impact cognitive function long-term (Short and Baram, 2019). Recently, studies have also shown that m6A-mediated gene regulation is significantly affected by stress. The dynamic nature of the stress response was recently demonstrated by Engel and colleagues, using a restraint stressor, a stressful stimulus for rodents. This elicited large-scale changes in the RNA methylome in a temporospatial manner (Engel et al., 2018). Restraint stress led to an increase in global methylation levels in amygdala and a decrease in prefrontal cortex. Mice deficient in METTL3 and FTO from excitatory hippocampal cells displayed lower resilience to stressful stimuli suggesting m6A-mediated mRNA regulation may improve adaptation to stress. These results highlight a role of m6A in stress response.
Heat Shock Stress Response
Zhou et al. found that m6A levels change in response to heat shock stress in both mouse embryonic fibroblasts (MEFs) and HeLa cells. They detected more m6A deposition in the 5′UTR of mRNAs, which promotes cap-independent translational initiation, allowing selective translation of heat-shock response proteins. YTHDF2 plays a key role in maintaining this 5′UTR methylation by preventing FTO-mediated demethylation (Zhou et al., 2015).
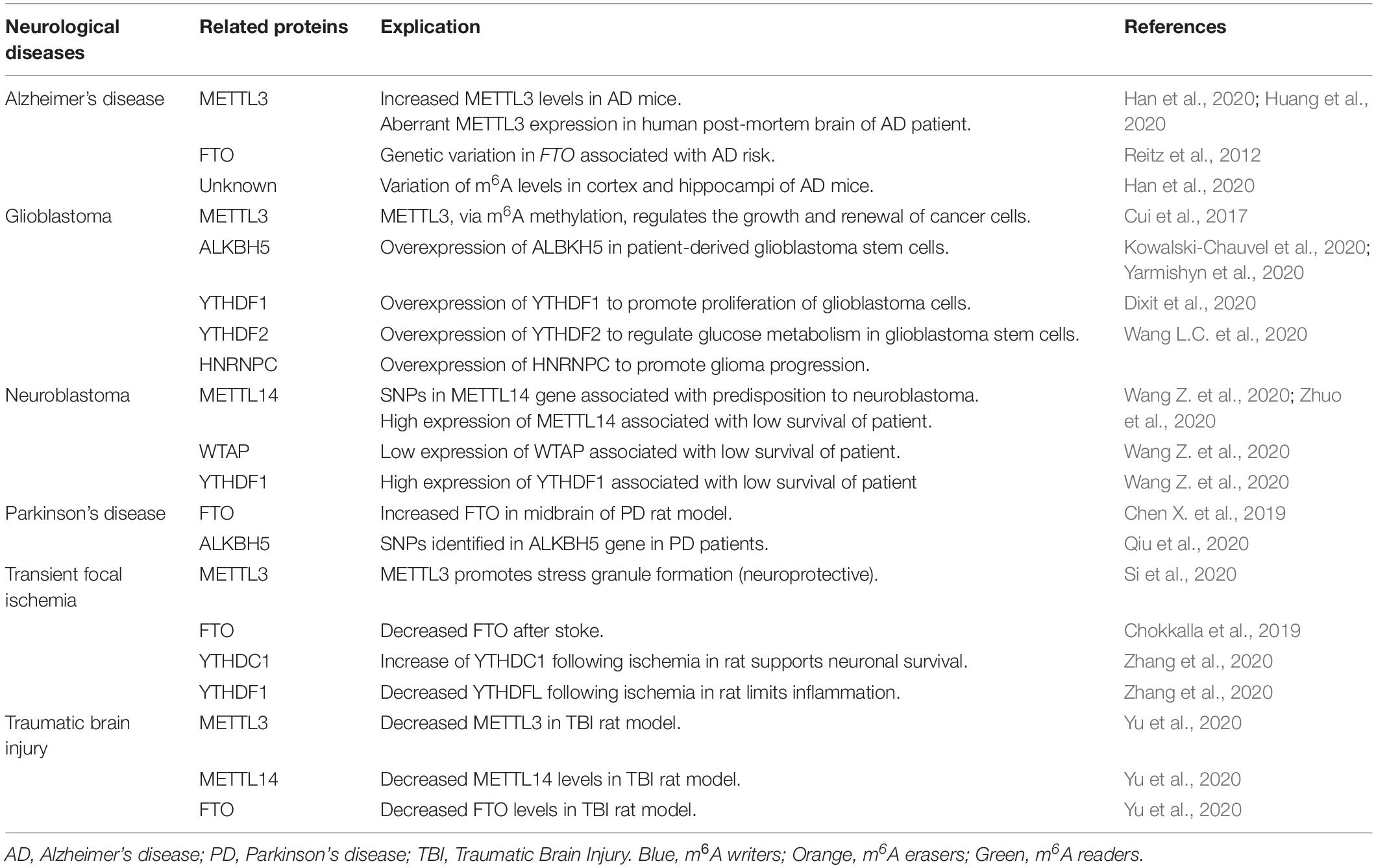
M6A in Brain Disorders
As described above, m6A RNA methylation is involved in many essential cerebral processes, so, unsurprisingly, this process is found to be altered in many brain disorders. Here we describe neurological disorders in which m6A, or m6A-associated proteins are affected such as Alzheimer’s disease, Parkinson’s disease, glioblastoma, schizophrenia, depression, transient focal ischemia, and traumatic brain injury (Table 2).
Neurodegenerative Diseases
Several studies in recent years have linked m6A involvement with neurodegenerative processes. Keller found that FTO is associated with dementia-like Alzheimer’s disease (AD) risk, suggesting that FTO may interact with the AD-risk factor gene APOE (Keller et al., 2011). Follow up studies have found that m6A is indeed dysregulated in AD. M6A levels were found to be disrupted in the cortex of APP/PS1 AD mice (Han et al., 2020), specifically they found gross changes in the RNA methylome with enhanced m6A levels on 659 gene transcripts and depleted m6A on 991 gene transcripts. Additionally, they found increased METTL3 expression in AD mice while FTO expression was decreased (Han et al., 2020). Aberrant METTL3 expression has been shown in human post-mortem brain of AD patients compared to non-AD autopsy tissue, further confirming mouse model data and strengthening the potential involvement of m6A-associated gene expression dysregulation in AD (Huang et al., 2020). Further investigations are needed however to determine whether aberrant m6A is a causal factor in AD development and progression or a consequence of altered physiological processes in the diseased brain.
M6A levels were reduced in the striatum of rats with 6-OHDA-induced Parkinson’s disease (PD) potentially due to an upregulation of ALKBH5. This reduction in m6A may promote the expression of N-methyl-D-aspartate receptor 1 (NMDA) and a subsequent elevation of oxidative stress and Ca2+ influx. This molecular cascade may then promote excitotoxic cell death of dopaminergic neurons (Chen X. et al., 2019). Since then, five Parkinson’s Disease-associated m6A-SNPs were identified in PD patients which perturbed this pathway, three of these SNPs were identified in the ALKBH5 gene (Qiu et al., 2020).
Together these data highlight the complex interplay amongst m6A associated proteins and suggest that further research investigating how m6A may actively contribute to perturbed gene expression regulation in neurodegenerative diseases is required.
CNS Tumors
M6A has been found to regulate glioblastoma stem cell tumorigenesis by controlling the expression of cancer-associated genes and processes. As in several other forms of cancer, m6A is thought to regulate the growth and self-renewal of cells via the methyltransferase catalytic activity of METTL3 (Cui et al., 2017). M6A readers may also play a critical role in glioblastoma tumorigenesis, indeed HNRNPC, YTHDF1 and YTHDF2 are all expressed at elevated levels in glioblastoma (Wang L.C. et al., 2020). YTHDF2 has been found to regulate glucose metabolism via the stabilization of the proto-oncogene MYC in glioblastoma stem cells (Dixit et al., 2020). Elevated YTHDF1 was associated with poor prognosis as it is thought to promote the proliferation and migration of glioblastoma cells (Yarmishyn et al., 2020). HNRNPC contributes to glioma progression although the mechanisms underlying this association are not yet known (Wang L.C. et al., 2020). The m6A-demethylase ALKBH5 has been shown to be overexpressed in patient-derived glioblastoma stem cells and may influence radio-resistance via regulation of DNA damage response genes including Chk1 (Kowalski-Chauvel et al., 2020).
M6A RNA modification has also been associated with other brain cancers such as neuroblastoma. Zhuo et al. identified several SNPs in the gene encoding METTL14, which may be associated with a predisposition to neuroblastoma development in a Chinese population (Zhuo et al., 2020). Moreover, m6A and associated proteins may also represent potential biomarkers of various brain cancers including neuroblastoma. High expression of METTL14 was correlated with low survival of neuroblastoma patients along with reduced expression of WTAP or a high expression of YTHDF1 (Wang Z. et al., 2020).
Brain Injury
Ischemic stroke resulting from blood vessel occlusion deprives neurons and brain cells of oxygen and nutrients and causes pronounced neuronal damage in affected tissues. Recently, Chokkalla et al. found using immunoprecipitation of m6A followed by microarray analysis that following ischemic stroke in mice, there is an increase in m6A levels in the brain and levels are perturbed particularly on inflammation, apoptosis and transcription-associated transcripts suggesting involvement of m6A in the regulation of the molecular milieu initiated by stroke. Elevated levels of m6A following stroke may be explained by decreased expression of FTO levels (Chokkalla et al., 2019; Xu K. et al., 2020).
Using the middle cerebral artery occlusion (MCAO) model of ischemic stroke in rats, Si and colleagues found that METTL3-mediated methylation promoted maturation of miR-355 which then promoted stress granule formation which was neuroprotective (Si et al., 2020). A role for m6A readers as regulators of the brain response to stroke has also been proposed. YTHDC1 levels increased following ischemia in rats while YTHDF1, 2, and 3 levels were repressed (Zhang et al., 2020). Elevated YTHDC1 levels may support post-ischemic neuronal survival via the modulation of the Akt/PTEN pathway (Zhang et al., 2020). YTHDF1 reduction may be a protective or adaptive mechanism by limiting post-stroke inflammation (Zheng et al., 2020).
Following traumatic brain injury (TBI) in rats, METTL3, METTL14 and FTO expression are all downregulated in the cortex. m6A methylation levels are also significantly changed with upregulated m6A-tagging of some mRNA (370 transcripts) and a downregulation of others (552 transcripts) as detected using meRIP-Seq (Wang et al., 2019; Yu et al., 2020). The exact role of m6A following TBI has yet to be fully elucidated but initial reports suggest these investigations are warranted.
Enabling Technologies
As discussed, m6A was identified as a prevalent RNA modification as far back as the 1970s yet was largely ignored until recently due to a lack of resources available to profile and probe its function (Munns et al., 1977; Wei and Moss, 1977; Burke and Joh, 1982). The development of an m6A specific antibody coupled with the growing use of next generation sequencing in the last decade led to the development of methylated RNA immunoprecipitation sequencing (meRIP-Seq/m6A-Seq) approaches (Dominissini et al., 2013). This technology is an adaptation of a standard RIP-Seq approach whereby RNA is isolated from a sample and sheared to a specific length. RNA is ribosome depleted and/or polyA-selected and then subjected to immunoprecipitation to isolate m6A-tagged mRNAs. The resulting isolated m6A-tagged RNA is then used to prepare RNA-Seq libraries and subjected to next generation sequencing and whole transcriptome analysis. The development of this technique enabled widespread analysis of this modification in various tissues, organisms and disease settings and highlighted the extent of the epitranscriptomic modification as well as identify how it may contribute to disease. Certain limitations existed however including high RNA input requirements (300 (μg total RNA) and it also suffers from limited resolution although bioinformatic predictions do help to identify exact sites of m6A. These limitations have been overcome to some degree and meRIP-seq can now be performed with as little as 3 (μg starting material (Zeng et al., 2018). Furthermore, single base resolution of m6A can now be mapped using crosslinking and immunoprecipitation (miCLIP) approaches (Linder et al., 2015).
M6A can now be mapped in native full length RNA moieties without the need for reverse transcription, shearing or immunoprecipitation steps using nanopore long read sequencing technologies (Liu et al., 2019; McIntyre et al., 2019; Jenjaroenpun et al., 2021). Several robust bioinformatic pipelines to identify methylated bases from long sequencing reads have now also been established.
Future Directions
The complex functions of the mammalian brain are dependent upon precise control of gene expression and regulation in a temporospatial fashion. M6A represents an additional layer of gene regulation, which has now been shown to critically regulate neuronal development and function. Technically difficult to study, tools are now available to analyze m6A RNA methylation at a global level and at single base resolution and there is huge scientific interest in this modification at present (Dominissini et al., 2012; Linder and Jaffrey, 2019). The next hurdle lies in developing and adapting current applications to profile m6A profiles in discrete cell types and populations/brain regions to understand how it contributes to neurodevelopment at the single cell level as well as specific brain functions such as learning and memory. Further challenges lie in understanding the precise function of m6A-eraser proteins in a context-dependent manner as well as probing the regulatory mechanisms governing m6A-associated protein expression and function.
Understanding the role of m6A in physiological homeostasis and disease is critical as components of these pathways may represent therapeutic targets for the treatment of neurological disease including many underserved conditions like TBI and stroke. The development of specific pharmacological inhibitors and antisense oligonucleotide approaches will also enable more precise interrogation of transient manipulations of m6A-associated proteins and therapeutic viability.
Author Contributions
JM, DH, and GB researched and wrote the article. All authors contributed to the article and approved the submitted version.
Funding
This publication has emanated from research supported in part by a research grant from Science Foundation Ireland (SFI) under Grant nos. 18/SIRG/5646 (GPB) and 16/RC/3948 (DH) and co-funded under the European Regional Development Fund and by FutureNeuro industry partners.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agarwal, D., Sandor, C., Volpato, V., Caffrey, T. M., Monzon-Sandoval, J., Bowden, R., et al. (2020). A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat. Commun. 11:4183.
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. doi: 10.1038/nature14281
Balacco, D. L., and Soller, M. (2019). The m(6)A writer: rise of a machine for growing tasks. Biochemistry 58, 363–378. doi: 10.1021/acs.biochem.8b01166
Barbieri, I., Tzelepis, K., Pandolfini, L., Shi, J., Millan-Zambrano, G., Robson, S. C., et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131. doi: 10.1038/nature24678
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370. doi: 10.1093/nar/gkx778
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. doi: 10.1016/j.stem.2014.09.019
Berlivet, S., Scutenaire, J., Deragon, J. M., and Bousquet-Antonelli, C. (2019). Readers of the m(6)A epitranscriptomic code. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 329–342. doi: 10.1016/j.bbagrm.2018.12.008
Berulava, T., Rahmann, S., Rademacher, K., Klein-Hitpass, L., and Horsthemke, B. (2015). N6-adenosine methylation in MiRNAs. PLoS One 10:e0118438. doi: 10.1371/journal.pone.0118438
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247.
Brennan, G. P., and Henshall, D. C. (2020). MicroRNAs as regulators of brain function and targets for treatment of epilepsy. Nat. Rev. Neurol. 16, 506–519. doi: 10.1038/s41582-020-0369-8
Brummer, A., Yang, Y., Chan, T. W., and Xiao, X. (2017). Structure-mediated modulation of mRNA abundance by A-to-I editing. Nat. Commun. 8:1255.
Burke, W. J., and Joh, T. H. (1982). Effects of N6-methyladenosine on the synthesis of phenylethanolamine N-methyltransferase in cultured explants of rat adrenal medulla. J. Neurochem. 39, 92–96. doi: 10.1111/j.1471-4159.1982.tb04705.x
Cao, G., Li, H. B., Yin, Z., and Flavell, R. A. (2016). Recent advances in dynamic m6A RNA modification. Open Biol. 6:160003. doi: 10.1098/rsob.160003
Chen, X., Sun, Y. Z., Liu, H., Zhang, L., Li, J. Q., and Meng, J. (2017). RNA methylation and diseases: experimental results, databases, Web servers and computational models. Brief. Bioinform. 20, 896–917. doi: 10.1093/bib/bbx142
Chen, X., Yu, C., Guo, M., Zheng, X., Ali, S., Huang, H., et al. (2019). Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 10, 2355–2363. doi: 10.1021/acschemneuro.8b00657
Chen, X. Y., Zhang, J., and Zhu, J. S. (2019). The role of m(6)A RNA methylation in human cancer. Mol. Cancer 18:103.
Chokkalla, A. K., Mehta, S. L., Kim, T., Chelluboina, B., Kim, J., and Vemuganti, R. (2019). Transient focal ischemia significantly alters the m(6)A epitranscriptomic tagging of RNAs in the brain. Stroke 50, 2912–2921. doi: 10.1161/strokeaha.119.026433
Christofi, T., and Zaravinos, A. (2019). RNA editing in the forefront of epitranscriptomics and human health. J. Transl. Med. 17:319.
Conboy, K., Henshall, D. C., and Brennan, G. P. (2021). Epigenetic principles underlying epileptogenesis and epilepsy syndromes. Neurobiol. Dis. 148:105179. doi: 10.1016/j.nbd.2020.105179
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634. doi: 10.1016/j.celrep.2017.02.059
Czuba, L. C., Hillgren, K. M., and Swaan, P. W. (2018). Post-translational modifications of transporters. Pharmacol. Ther. 192, 88–99. doi: 10.1016/j.pharmthera.2018.06.013
Danzer, S. C. (2008). Postnatal and adult neurogenesis in the development of human disease. Neuroscientist 14, 446–458. doi: 10.1177/1073858408317008
Dixit, D., Prager, B. C., Gimple, R. C., Poh, H. X., Wang, Y., Wu, Q., et al. (2020). The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 11, 480–499. doi: 10.1158/2159-8290.cd-20-0331
Dominissini, D., Moshitch-Moshkovitz, S., Salmon-Divon, M., Amariglio, N., and Rechavi, G. (2013). Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 8, 176–189. doi: 10.1038/nprot.2012.148
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Donega, V., Marcy, G., Lo Giudice, Q., Zweifel, S., Angonin, D., Fiorelli, R., et al. (2018). Transcriptional dysregulation in postnatal glutamatergic progenitors contributes to closure of the cortical neurogenic period. Cell. Rep. 22, 2567–2574. doi: 10.1016/j.celrep.2018.02.030
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7:12626.
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi: 10.1038/nsmb.3462
Eisenberg, E., and Levanon, E. Y. (2018). A-to-I RNA editing - immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490. doi: 10.1038/s41576-018-0006-1
Engel, M., Eggert, C., Kaplick, P. M., Eder, M., Roh, S., Tietze, L., et al. (2018). The Role of m(6)A/m-RNA methylation in stress response regulation. Neuron 99, 389.e9–403.e9.
Ensfelder, T. T., Kurz, M. Q., Iwan, K., Geiger, S., Matheisl, S., Muller, M., et al. (2018). ALKBH5-induced demethylation of mono- and dimethylated adenosine. Chem. Commun. 54, 8591–8593. doi: 10.1039/c8cc03980a
Fedeles, B. I., Singh, V., Delaney, J. C., Li, D., and Essigmann, J. M. (2015). The AlkB family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290, 20734–20742. doi: 10.1074/jbc.r115.656462
Furuichi, Y., Morgan, M., Shatkin, A. J., Jelinek, W., Salditt-Georgieff, M., and Darnell, J. E. (1975). Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl. Acad. Sci. U.S.A. 72, 1904–1908. doi: 10.1073/pnas.72.5.1904
Gerken, T., Girard, C. A., Tung, Y. C., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. doi: 10.1126/science.1151710
Han, M., Liu, Z., Xu, Y., Liu, X., Wang, D., Li, F., et al. (2020). Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front. Neurosci. 14:98. doi: 10.3389/fnins.2020.00098
Hawrylycz, M. J., Lein, E. S., Guillozet-Bongaarts, A. L., Shen, E. H., Ng, L., Miller, J. A., et al. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399.
He, P. C., and He, C. (2021). m(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 40:e105977.
Horiuchi, K., Kawamura, T., Iwanari, H., Ohashi, R., Naito, M., Kodama, T., et al. (2013). Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 288, 33292–33302. doi: 10.1074/jbc.m113.500397
Hsu, P. J., Shi, H., Zhu, A. C., Lu, Z., Miller, N., Edens, B. M., et al. (2019). The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J. Biol. Chem. 294, 19889–19895. doi: 10.1074/jbc.AC119.010078
Huang, H., Camats-Perna, J., Medeiros, R., Anggono, V., and Widagdo, J. (2020). Altered expression of the m6A methyltransferase METTL3 in Alzheimer’s disease. eNeuro 7:ENEURO.0125-20.2020.
Jain, M., Jantsch, M. F., and Licht, K. (2019). The Editor’s I on disease development. Trends Genet. 35, 903–913.
Jenjaroenpun, P., Wongsurawat, T., Wadley, T. D., Wassenaar, T. M., Liu, J., Dai, Q., et al. (2021). Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 49:e7. doi: 10.1093/nar/gkaa620
Joglekar, A., Prjibelski, A., Mahfouz, A., Collier, P., Lin, S., Schlusche, A. K., et al. (2021). A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 12:463.
Kaldun, J. C., and Sprecher, S. G. (2019). Initiated by CREB: resolving gene regulatory programs in learning and memory: switch in cofactors and transcription regulators between memory consolidation and maintenance network. Bioessays 41:e1900045.
Ke, S., Pandya-Jones, A., Saito, Y., Fak, J. J., Vagbo, C. B., Geula, S., et al. (2017). m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006. doi: 10.1101/gad.301036.117
Keller, L., Xu, W., Wang, H. X., Winblad, B., Fratiglioni, L., and Graff, C. (2011). The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: a prospective cohort study. J. Alzheimers Dis. 23, 461–469. doi: 10.3233/jad-2010-101068
Kim, J. K., Samaranayake, M., and Pradhan, S. (2009). Epigenetic mechanisms in mammals. Cell Mol. Life Sci. 66, 596–612.
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429. doi: 10.1101/gad.309146.117
Koranda, J. L., Dore, L., Shi, H., Patel, M. J., Vaasjo, L. O., Rao, M. N., et al. (2018). Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99, 283.e5–292.e5.
Kowalski-Chauvel, A., Lacore, M. G., Arnauduc, F., Delmas, C., Toulas, C., Cohen-Jonathan-Moyal, E., et al. (2020). The m6A RNA demethylase ALKBH5 promotes radioresistance and invasion capability of glioma stem cells. Cancers 13:40. doi: 10.3390/cancers13010040
Lence, T., Paolantoni, C., Worpenberg, L., and Roignant, J. Y. (2019). Mechanistic insights into m(6)A RNA enzymes. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 222–229. doi: 10.1016/j.bbagrm.2018.10.014
Li, E., Wei, B., Wang, X., and Kang, R. (2020). METTL3 enhances cell adhesion through stabilizing integrin beta1 mRNA via an m6A-HuR-dependent mechanism in prostatic carcinoma. Am. J. Cancer Res. 10, 1012–1025.
Li, L., Zang, L., Zhang, F., Chen, J., Shen, H., Shu, L., et al. (2017). Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411. doi: 10.1093/hmg/ddx128
Li, M., Zhao, X., Wang, W., Shi, H., Pan, Q., Lu, Z., et al. (2018). Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 19:69.
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345. doi: 10.1016/j.molcel.2016.03.021
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. doi: 10.1038/nmeth.3453
Linder, B., and Jaffrey, S. R. (2019). Discovering and mapping the modified nucleotides that comprise the epitranscriptome of mRNA. Cold Spring Harb. Perspect. Biol. 11:a032201. doi: 10.1101/cshperspect.a032201
Lindgren, C. M., and McCarthy, M. I. (2008). Mechanisms of disease: genetic insights into the etiology of type 2 diabetes and obesity. Nat. Clin. Pract. Endocrinol. Metab. 4, 156–163. doi: 10.1038/ncpendmet0723
Ling, Z., Chen, L., and Zhao, J. (2020). m6A-dependent up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth, migration, and colony formation in osteosarcoma. Biosci. Rep. 40:BSR20200282.
Liu, H., Begik, O., Lucas, M. C., Ramirez, J. M., Mason, C. E., Wiener, D., et al. (2019). Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat. Commun. 10:4079.
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Livneh, I., Moshitch-Moshkovitz, S., Amariglio, N., Rechavi, G., and Dominissini, D. (2020). The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 21, 36–51. doi: 10.1038/s41583-019-0244-z
Loos, R. J., and Bouchard, C. (2008). FTO: the first gene contributing to common forms of human obesity. Obes. Rev. 9, 246–250.
Ma, C., Chang, M., Lv, H., Zhang, Z. W., Zhang, W., He, X., et al. (2018). RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19:68.
Madugalle, S. U., Meyer, K., Wang, D. O., and Bredy, T. W. (2020). RNA N(6)-methyladenosine and the regulation of RNA localization and function in the brain. Trends Neurosci. 43, 1011–1023. doi: 10.1016/j.tins.2020.09.005
Mauer, J., and Jaffrey, S. R. (2018). FTO, m(6) Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 592, 2012–2022. doi: 10.1002/1873-3468.13092
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)Am in the 5′ cap controls mRNA stability. Nature 541, 371–375. doi: 10.1038/nature21022
McIntyre, A. B. R., Alexander, N., Grigorev, K., Bezdan, D., Sichtig, H., Chiu, C. Y., et al. (2019). Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat. Commun. 10:579.
Merkurjev, D., Hong, W. T., Iida, K., Oomoto, I., Goldie, B. J., Yamaguti, H., et al. (2018). Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014. doi: 10.1038/s41593-018-0173-6
Meyer, K. D., and Jaffrey, S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326. doi: 10.1038/nrm3785
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Mitra, S., Lee, W., Hayashi, K., Boyd, J., Milloy, M. J., Dong, H., et al. (2021). A gender comparative analysis of post-traumatic stress disorder among a community-based cohort of people who use drugs in Vancouver, Canada. Addict. Behav. 115:106793. doi: 10.1016/j.addbeh.2020.106793
Munns, T. W., Liszewski, M. K., and Sims, H. F. (1977). Characterization of antibodies specific for N6-methyladenosine and for 7-methylguanosine. Biochemistry 16, 2163–2168. doi: 10.1021/bi00629a019
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Patil, D. P., Pickering, B. F., and Jaffrey, S. R. (2018). Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 28, 113–127. doi: 10.1016/j.tcb.2017.10.001
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824.e14–835.e14.
Pfisterer, U., Petukhov, V., Demharter, S., Meichsner, J., Thompson, J. J., Batiuk, M. Y., et al. (2020). Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat. Commun. 11:5038.
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Qiu, X., He, H., Huang, Y., Wang, J., and Xiao, Y. (2020). Genome-wide identification of m(6)A-associated single-nucleotide polymorphisms in Parkinson’s disease. Neurosci. Lett. 737:135315. doi: 10.1016/j.neulet.2020.135315
Reitz, C., Tosto, G., Mayeux, R., Luchsinger, J. A., Group, N.-L. N. F. S., and Alzheimer’s Disease Neuroimaging, I. (2012). Genetic variants in the Fat and Obesity Associated (FTO) gene and risk of Alzheimer’s disease. PLoS One 7:e50354. doi: 10.1371/journal.pone.0050354.
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell. Rep. 8, 284–296. doi: 10.1016/j.celrep.2014.05.048
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Shi, H., Zhang, X., Weng, Y. L., Lu, Z., Liu, Y., Lu, Z., et al. (2018). m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253. doi: 10.1038/s41586-018-0666-1
Short, A. K., and Baram, T. Z. (2019). Early-life adversity and neurological disease: age-old questions and novel answers. Nat. Rev. Neurol. 15, 657–669. doi: 10.1038/s41582-019-0246-5
Si, W., Li, Y., Ye, S., Li, Z., Liu, Y., Kuang, W., et al. (2020). Methyltransferase 3 mediated miRNA m6A methylation promotes stress granule formation in the early stage of acute ischemic stroke. Front. Mol. Neurosci. 13:103. doi: 10.3389/fnmol.2020.00103
Sledz, P., and Jinek, M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 5:e18434.
Song, L., Pan, S., Zhang, Z., Jia, L., Chen, W. H., and Zhao, X. M. (2021). STAB: a spatio-temporal cell atlas of the human brain. Nucleic Acids Res. 49, D1029–D1037.
Song, N., Du, J., Gao, Y., and Yang, S. (2020). Epitranscriptome of the ventral tegmental area in a deep brain-stimulated chronic unpredictable mild stress mouse model. Transl. Neurosci. 11, 402–418. doi: 10.1515/tnsci-2020-0146
Sorci, M., Ianniello, Z., Cruciani, S., Larivera, S., Ginistrelli, L. C., Capuano, E., et al. (2018). METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 9:796.
Tang, C., Klukovich, R., Peng, H., Wang, Z., Yu, T., Zhang, Y., et al. (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U.S.A. 115, E325–E333.
Theler, D., Dominguez, C. Blatter, M., Boudet, J., and Allain, F. H. (2014). Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 42, 13911–13919. doi: 1093/nar/gku1116
van der Kwast, R., Parma, L., van der Bent, M. L., van Ingen, E., Baganha, F., Peters, H. A. B., et al. (2020). Adenosine-to-inosine editing of vasoactive MicroRNAs alters their targetome and function in ischemia. Mol. Ther. Nucleic Acids 21, 932–953. doi: 10.1016/j.omtn.2020.07.020
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533. doi: 10.1038/onc.2017.351
Walters, B. J., Mercaldo, V., Gillon, C. J., Yip, M., Neve, R. L., Boyce, F. M., et al. (2017). The role of The RNA demethylase FTO (fat mass and obesity-associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology 42, 1502–1510. doi: 10.1038/npp.2017.31
Wang, L. C., Chen, S. H., Shen, X. L., Li, D. C., Liu, H. Y., Ji, Y. L., et al. (2020). M6A RNA methylation regulator HNRNPC contributes to tumorigenesis and predicts prognosis in glioblastoma multiforme. Front. Oncol. 10:536875. doi: 10.3389/fonc.2020.536875
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198. doi: 10.1038/ncb2902
Wang, Y., Mao, J., Wang, X., Lin, Y., Hou, G., Zhu, J., et al. (2019). Genome-wide screening of altered m6A-tagged transcript profiles in the hippocampus after traumatic brain injury in mice. Epigenomics 11, 805–819. doi: 10.2217/epi-2019-0002
Wang, Z., Cheng, H., Xu, H., Yu, X., and Sui, D. (2020). A five-gene signature derived from m6A regulators to improve prognosis prediction of neuroblastoma. Cancer Biomark. 28, 275–284. doi: 10.3233/cbm-191196
Wei, C., Gershowitz, A., and Moss, B. (1975). N6, O2’-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253. doi: 10.1038/257251a0
Wei, C. M., Gershowitz, A., and Moss, B. (1976). 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15, 397–401. doi: 10.1021/bi00647a024
Wei, C. M., and Moss, B. (1977). Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16, 1672–1676. doi: 10.1021/bi00627a023
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 69, 1028.e6–1038.e6.
Weng, Y. L., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018). Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313.e6–325.e6.
Westmark, C. J., Maloney, B., Alisch, R. S., Sokol, D. K., and Lahiri, D. K. (2020). FMRP regulates the nuclear export of Adam9 and Psen1 mRNAs: secondary Analysis of an N6-methyladenosine dataset. Sci. Rep. 10:10781. doi: 10.1038/s41598-020-66394-y
Widagdo, J., Zhao, Q. Y., Kempen, M. J., Tan, M. C., Ratnu, V. S., Wei, W., et al. (2016). Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777. doi: 10.1523/JNEUROSCI.4053-15.2016
Wojtas, M. N., Pandey, R. R., Mendel, M., Homolka, D., Sachidanandam, R., and Pillai, R. S. (2017). Regulation of m(6)A transcripts by the 3′–>5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell. 68, 374.e12–387.e12.
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929. doi: 10.1038/nchembio.1654
Xu, H., Dzhashiashvili, Y., Shah, A., Kunjamma, R. B., Weng, Y. L., Elbaz, B., et al. (2020). m(6)A mRNA methylation is essential for oligodendrocyte maturation and CNS myelination. Neuron 105, 293.e5–309.e5.
Xu, K., Mo, Y., Li, D., Yu, Q., Wang, L., Lin, F., et al. (2020). N(6)-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther. Adv. Chronic Dis. 11:2040622320916024.
Xue, A., Huang, Y., Li, M., Wei, Q., and Bu, Q. (2021). Comprehensive analysis of differential m6A RNA methylomes in the hippocampus of cocaine-conditioned mice. Mol. Neurobiol. [Epub ahead of print].
Yarmishyn, A. A., Yang, Y. P., Lu, K. H., Chen, Y. C., Chien, Y., Chou, S. J., et al. (2020). Musashi-1 promotes cancer stem cell properties of glioblastoma cells via upregulation of YTHDF1. Cancer Cell. Int. 20:597.
Yoon, K. J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171, 877.e17–889.e17.
Yu, J., Chen, M., Huang, H., Zhu, J., Song, H., Zhu, J., et al. (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423. doi: 10.1093/nar/gkx1182
Yu, J., Zhang, Y., Ma, H., Zeng, R., Liu, R., Wang, P., et al. (2020). Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol. Brain 13:11.
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10.
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624.
Zeng, Y., Wang, S., Gao, S., Soares, F., Ahmed, M., Guo, H., et al. (2018). Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biol. 16:e2006092. doi: 10.1371/journal.pbio.2006092
Zhang, P., Torres, K., Liu, X., Liu, C. G., and Pollock, R. E. (2016). An overview of chromatin-regulating proteins in cells. Curr. Protein Pept. Sci. 17, 401–410.
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591.e6–606.e6.
Zhang, Z., Wang, M., Xie, D., Huang, Z., Zhang, L., Yang, Y., et al. (2018). METTL3-mediated N(6)-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 28, 1050–1061.
Zhang, Z., Wang, Q., Zhao, X., Shao, L., Liu, G., Zheng, X., et al. (2020). YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 11:977.
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419.
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49, 18–29.
Zheng, L., Tang, X., Lu, M., Sun, S., Xie, S., Cai, J., et al. (2020). microRNA-421-3p prevents inflammatory response in cerebral ischemia/reperfusion injury through targeting m6A Reader YTHDF1 to inhibit p65 mRNA translation. Int. Immunopharmacol. 88:106937.
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., and Qian, S. B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594.
Keywords: m6A (N6-methyladenosine), brain function and brain diseases, brain development, epitranscriptomics, METTL3
Citation: Mathoux J, Henshall DC and Brennan GP (2021) Regulatory Mechanisms of the RNA Modification m6A and Significance in Brain Function in Health and Disease. Front. Cell. Neurosci. 15:671932. doi: 10.3389/fncel.2021.671932
Received: 24 February 2021; Accepted: 19 April 2021;
Published: 19 May 2021.
Edited by:
Daniela Tropea, Trinity College Dublin, IrelandReviewed by:
Francesca Ruberti, Institute of Biochemistry and Cell Biology, National Research Council, Consiglio Nazionale delle Ricerche, ItalyChristina Gross, Cincinnati Children’s Hospital Medical Center, United States
Copyright © 2021 Mathoux, Henshall and Brennan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gary P. Brennan, gary.brennan@ucd.ie
 Justine Mathoux
Justine Mathoux