- 1Wonju Severance Christian Hospital, Yonsei University Wonju College of Medicine, Wonju, South Korea
- 2Key Laboratory of Cerebral Microcirculation, Department of Neurology, Affiliated Hospital of Taishan Medical University, Universities of Shandong, Taian, China
- 3Department of Pathology, The Affiliated Hospital of Weifang Medical University, Weifang, China
- 4Department of Anesthesiology, The Affiliated Hospital of Weifang Medical University, Weifang, China
- 5Brain Research Centre, University of British Columbia, Vancouver, BC, Canada
Until now, large-scale genome-wide association studies have identified 94 genes associated with Alzheimer's disease, Parkinson's disease, and multiple sclerosis. Expression quantitative trait locus (eQTL) analysis showed that six genetic variants around six of these 94 genes could drive both disease susceptibility and altered expression of six nearby genes including CD33 (rs3865444), PILRB (rs1476679), NUP160 (rs10838725), LRRK2 (rs76904798), RGS1 (rs1323292), and METTL21B (rs701006). However, two of these six genetic variants rs1476679 and rs76904798 variants could regulate the expression of PILRB and LRRK2 only in the human monocyte-derived microglia-like (MDMi) cells, but not in human peripheral blood monocytes. Here, we aim to verify these findings using another two eQTL datasets in human peripheral blood immune cell CD14+ monocytes. The results that showed that rs1476679 and rs76904798 variants or their proxy variants could significantly regulate the expression of PILRB and LRRK2 in immune cell CD14+ monocytes and human peripheral blood. We believe that these findings provide important supplementary information about the regulatory mechanisms by which both variants influence PILRB and LRRK2 gene expression and neurodegenerative disease risk.
Introduction
It is known that Alzheimer's disease, Parkinson's disease and multiple sclerosis are three common neurodegenerative diseases (Liu et al., 2012, 2016, 2017b,c,d,e, 2018c; Beecham et al., 2013; Chang et al., 2017; Jun et al., 2017). In recent years, large-scale genome-wide association studies (GWAS) have identified some neurodegenerative disease risk variants (Liu et al., 2012, 2016, 2017b,c,d,e, 2018b; Beecham et al., 2013; Zhang et al., 2015, 2018; Li et al., 2016; Chang et al., 2017; Jiang et al., 2017; Jun et al., 2017). In 2013, the International Genomics of Alzheimer's Project (IGAP) conducted a meta-analysis of Alzheimer's disease GWAS datasets including 17,008 Alzheimer's disease cases and 37,154 controls in stage 1, and 8,572 Alzheimer's disease cases and 11,312 controls in stage 2. The meta-analysis of 74,046 individuals in stage 1 and stage 2 identified 11 new susceptibility loci for Alzheimer's disease (Lambert et al., 2013). In 2014, Nalls et al. conducted a large-scale meta-analysis of Parkinson's disease GWAS data including up to 13,708 PD cases and 95,282 control from 15 independent GWAS datasets of European descent, and identified six new Parkinson's disease risk loci (Nalls et al., 2014). In 2017, Chang et al. conducted a meta-analysis of Parkinson's disease GWAS data including 26,035 cases and 403,190 controls, and identified 17 novel Parkinson's disease risk loci (Chang et al., 2017).
Evidence shows that genetic variants could modify gene expression and cause disease risk (Liu et al., 2017d, 2018a,b,d). In a recent study, Ryan and colleagues developed a human monocyte-derived microglia-like (MDMi) cellular model to assess the effects of neurodegenerative disease variants (Ryan et al., 2017). Using this model system, Ryan and colleagues conducted an expression quantitative trait locus (eQTL) analysis to examine 94 genes associated with Alzheimer's disease, Parkinson's disease, and multiple sclerosis (Ryan et al., 2017). One eQTL analysis was performed using MDMi cells from 95 young, healthy subjects of European ancestry (Ryan et al., 2017). The other eQTL analysis was performed using human peripheral blood monocytes derived from 211 young, healthy subjects of European ancestry (Ryan et al., 2017). They identified that genetic variants could drive both disease susceptibility and altered expression of six nearby genes including CD33 (rs3865444), PILRB (rs1476679), NUP160 (rs10838725), LRRK2 (rs76904798), RGS1 (rs1323292), and METTL21B (rs701006) (Ryan et al., 2017). However, rs1476679 and rs76904798 variants could regulate the expression of PILRB and LRRK2 only in the MDMi cells, but not in human peripheral blood monocytes (Ryan et al., 2017). Here, we aim to further verify their findings using multiple eQTL datasets in human peripheral blood immune cell CD14+ monocytes.
Materials and Methods
Functional Analysis
It is described that enhancers are DNA regulatory sequences, and could regulate tissue-specific gene expression (Ong and Corces, 2011; Shlyueva et al., 2014). Hence, we first performed an enhancer analysis using HaploReg (version 4.1) to evaluate the overlap of these six variants with predicted enhancers in each reference epigenome data from Roadmap Epigenomics project (Ward and Kellis, 2016). HaploReg (version 4.1) defined the enhancers using four different methods including the 15-state core model, the 25-state model incorporating imputed epigenomes, the H3K4me1/H3K4me3 peaks and the H3K27ac/H3K9ac peaks (Ward and Kellis, 2016). In brief, both the core 15-state model and the 25-state model are based on the imputed marks, H3K4me1/H3K4me3 is based on peaks from H3K4me1 and H3K4me3, and H3K27ac/H3K9ac is based on peaks from H3K27ac and H3K9ac (Ward and Kellis, 2016). In addition to enhancer analysis, we also conducted a promoter analysis and DNAse analysis. More detailed information has been described in recent study (Liu et al., 2018b).
eQTL Analysis
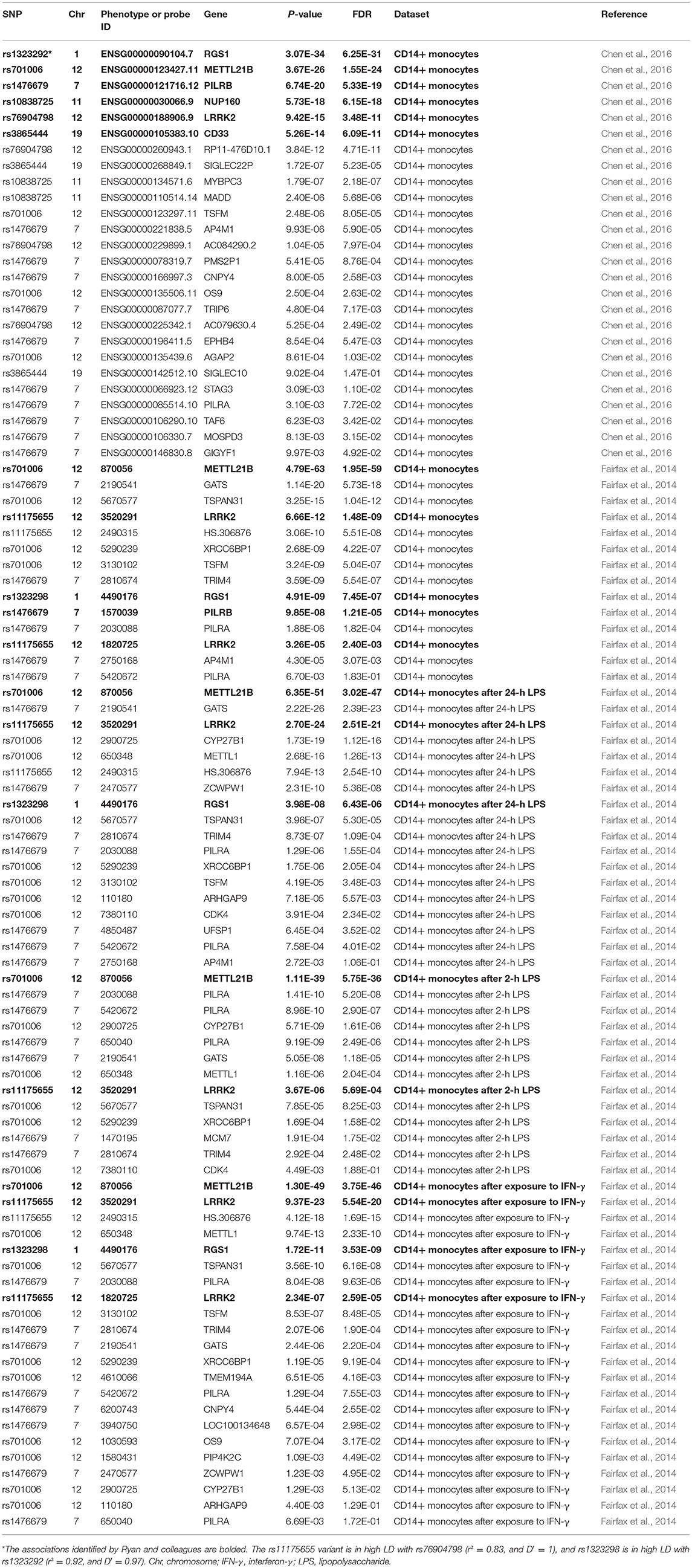
The first eQTL dataset in monocytes is from the International Human Epigenome Consortium (IHEC) (Chen et al., 2016). The IHEC consortium performed an eQTL analysis in three immune cell types (CD14+ monocytes, CD16+ neutrophils, and naive CD4+ T cells) from up to 197 individuals (Chen et al., 2016). Data from this project was produced in different institutes. Peripheral blood mononuclear cells were isolated from donors at University of Cambridge (Chen et al., 2016). The second eQTL dataset in monocytes (CD14+ monocytes) is from a recent study conducted by Fairfax et al. (2014). In the original study, primary monocytes from 432 healthy Europeans were exposed to interferon-γ (IFN-γ) or differing durations of lipopolysaccharide (LPS) (Fairfax et al., 2014). Then the impact of immune stimulation upon regulatory variant activity was systematically evaluated by an eQTL analysis (Fairfax et al., 2014). In brief, this eQTL analysis tested 609,704 genetic variants and expression data for 15,421 probes for 414 individuals in the naive state, 367 individuals after exposure to IFN-γ, 322 individuals after 24-h LPS, and 261 individuals after 2-h LPS (Fairfax et al., 2014).
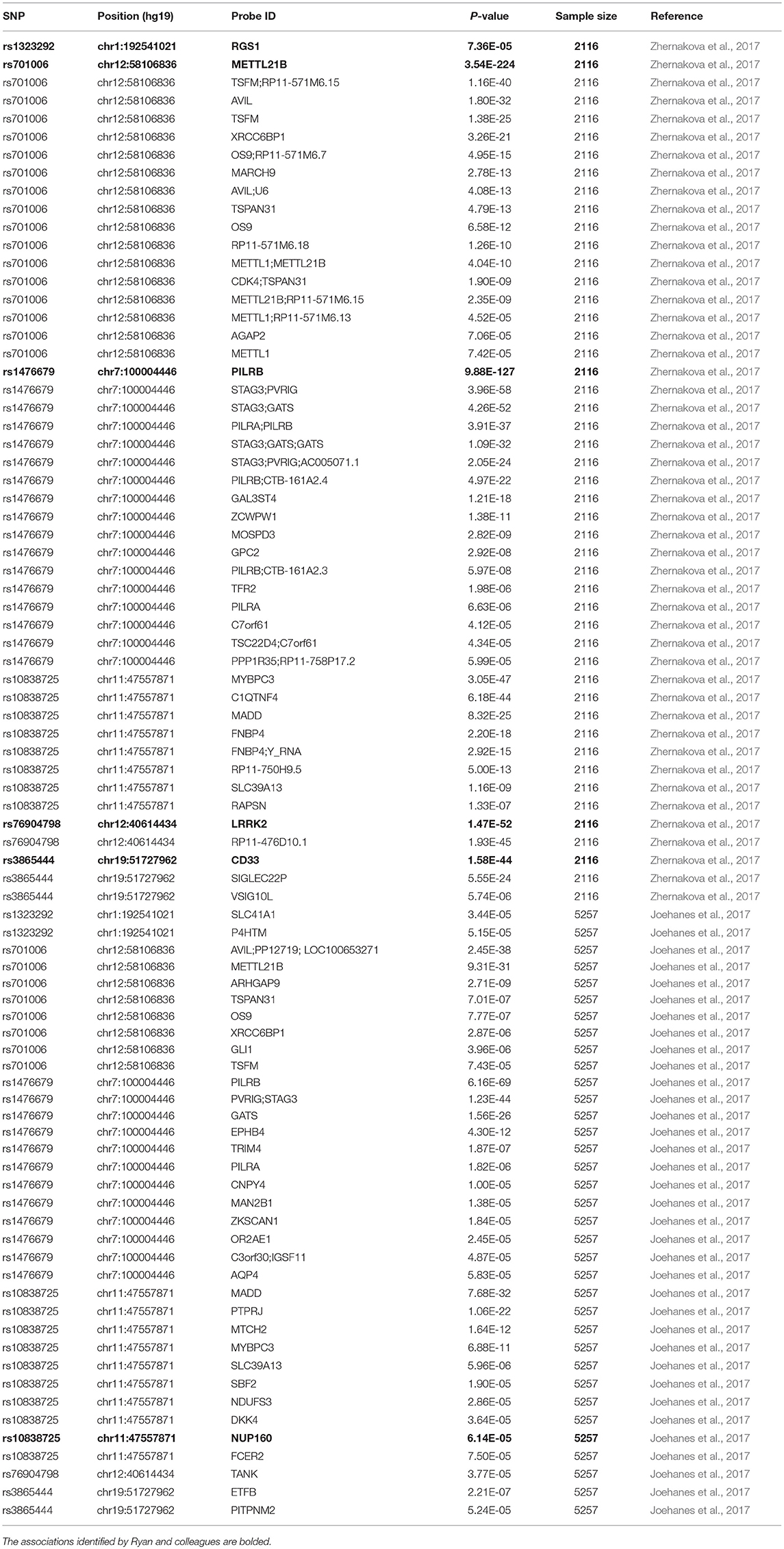
In both eQTL datasets, genetic variants mapping to within 1 Mb (on each side) of each tested gene, were tested their association with gene expression using a linear regression analysis (Fairfax et al., 2014; Chen et al., 2016). Here, we utilize the summary results from the corresponding studies to evaluate the potential association of six genetic variants with the expression of their nearby genes. If any variant is not available in above datasets, we used HaploReg (version 4) to identify it proxy variants based on the linkage disequilibrium (LD) information from the 1000 Genomes Project (EUR) (Ward and Kellis, 2012). We defined the genetic variants tagged by any genetic variant with r2> = 0.8 (Ward and Kellis, 2012). In addition to the monocytes, we also evaluated the association of six genetic variants with gene expression using two eQTL datasets in human peripheral blood including 2,116 and 5,257 samples, respectively (Joehanes et al., 2017; Zhernakova et al., 2017).
Results
Enhancer Analysis
The enhancer analysis using the 15-state core model, the 25-state model, the H3K4me1/H3K4me3 model and the H3K27ac/H3K9ac model showed that these 6 genetic variants are predicted to be mainly located in enhancers of blood cell types. These findings indicate that these 6 genetic variants are likely to regulate gene expression in the blood cell types, especially CD14+ monocytes. In brief, rs1476679 variant is predicted to be located in enhancers of monocytes-CD14+ RO01746 Primary Cells using the H3K4me1/H3K4me3 model and the H3K27ac/H3K9ac model (Table 1). The rs76904798 variant is predicted to be located in enhancers of monocytes-CD14+ RO01746 Primary Cells using the 15-state core model, the H3K4me1/H3K4me3 model and the H3K27ac/H3K9ac model. To validate the enhancer analysis, we then performed an eQTLs analysis using multiple datasets in CD14+ monocytes.
Promoter Analysis
All these four models including 15-state core model, the 25-state model, the H3K4me1/H3K4me3 model and the H3K27ac/H3K9ac model showed that the rs3865444 variant was predicted to be mainly located in promoter of blood cell types, especially in immune cell types. Three of these four models including the 15-state core model, the H3K4me1/H3K4me3 model and the H3K27ac/H3K9ac model indicated that the rs1323292 variant was predicted to be mainly located in promoter of blood cell types, especially in immune cell types. Table 2 provides the results from promoter analysis of six neurodegenerative disease variants using HaploReg (version 4.1).
DNAse Analysis
The DNAse analysis showed that rs1323292 variant was predicted to be mainly located in DNAse of blood cell types, especially in primary T cells from cord blood, primary T cells from peripheral blood, primary Natural Killer cells from peripheral blood, fetal Thymus, and small Intestine. The rs701006 variant was predicted to be mainly located in DNAse of Foreskin Melanocyte Primary Cells skin01 and Fetal Brain Female. The rs1476679 variant was predicted to be mainly located in DNAse of iPS DF 19.11 Cells, Breast variant Human Mammary Epithelial Cells (vHMEC), HSMM cell derived Skeletal Muscle Myotubes Cells. However, we did identify any rs10838725, rs76904798, rs3865444.
eQTLs Analysis in CD14+ Monocytes
In the first dataset including up to 197 individuals, the results showed significant association of all these six genetic variants (rs3865444, rs1476679, rs10838725, rs76904798, rs1323292, and rs701006) with the expression of nearby genes including CD33, PILRB, NUP160, LRRK2, RGS1, and METTL21B. In addition, there are also some other genes. Here, we list the significant results with P < 0.01 and the corresponding false discovery rate (FDR) in Table 3.
In the second dataset including 414 samples in the naive state, two genetic variants rs3865444 and rs10838725, as well as their proxy variants (r2 ≥ 0.8), are not available. The other four genetic variants or their proxy variants were significantly associated with the expression of nearby genes including PILRB, LRRK2, RGS1, and METTL21B. In brief, rs11175655 is in high LD with rs76904798 (r2 = 0.83, and D′ = 1), and rs1323298 is in high LD with rs1323292 (r2 = 0.92, and D′ = 0.97). Meanwhile, there also some other genes, as provided in Table 2. In addition to the naive state, some findings were also observed in other three states including 367 individuals after exposure to IFN-γ, 322 individuals after 24-h LPS, and 261 individuals after 2-h LPS, as provided in Table 3.
QTLs Analysis in Human Peripheral Blood
In the two eQTL datasets in human peripheral blood, we again found significant association of all these six genetic variants (rs3865444, rs1476679, rs10838725, rs76904798, rs1323292, and rs701006) with the expression of nearby genes including CD33, PILRB, NUP160, LRRK2, RGS1, and METTL21B, as well as other genes (Table 4).
Discussion
In recent years, large-scale GWAS datasets have identified 94 genes associated with Alzheimer's disease, Parkinson's disease, or multiple sclerosis (Ryan et al., 2017). However, it is still unclear how these variants functionally affect the underlying neurodegenerative disease pathogenesis. Growing evidence shows that genetic variants may affect disease risk by regulating gene expression (Bao et al., 2015; Liu et al., 2015, 2016, 2017a; Hu et al., 2017). Ryan and colleagues applied a human MDMi cellular model, and conducted an eQTL analysis to evaluate the effects of these neurodegenerative disease variants (Ryan et al., 2017).
In summary, Ryan et al. identified that six neurodegenerative disease variants were associated with disease susceptibility, and could alter the expression of six nearby genes. Both rs1476679 and rs76904798 variants could only regulate the expression of PILRB and LRRK2 in the MDMi cells, but not in human peripheral blood monocytes (Ryan et al., 2017). Ryan et al. concluded that the differentiation of monocytes into microglia-like cells could cause the acquisition of a cellular state, which could reveal the functional consequences of certain genetic variants (Ryan et al., 2017). Ryan et al. provided an in vitro translational tool to generate microglia-like cells quickly and easily from adult blood, and could be useful for exploring microglia function and dysfunction (Ryan et al., 2017). However, Ryan et al. just selected single eQTL dataset in human peripheral blood monocytes without any replication.
Here, we first performed an enhancer analysis of these six neurodegenerative disease variants. The results showed that these six genetic variants were predicted to be mainly located in enhancers of blood cell types, especially in CD14+ monocytes. The promoter analysis showed that rs3865444 and rs1323292 variants were predicted to be mainly located in promoter of blood cell types, especially in immune cell types. Hence, we further evaluated these six genetic variants using multiple eQTL datasets in human peripheral blood immune cell CD14+ monocytes. The results that showed that rs1476679 and rs76904798 variants or their proxy variants could significantly regulate the expression of PILRB and LRRK2 in immune cell CD14+ monocytes and human peripheral blood.
In summary, we believe that these findings provide important supplementary information about the regulatory mechanisms by which both variants influence PILRB and LRRK2 gene expression and neurodegenerative disease risk.
Author Contributions
BS and HY conceived and initiated the project. JS and YH analyzed the data, and wrote the first draft of the manuscript. YZ, LW, and LL contributed to the interpretation of the results and critical revision of the manuscript for important intellectual content and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the International Human Epigenome Consortium (IHEC), Fairfax and colleagues for the eQTL datasets. This work was supported in part by funds from the National Natural Science Foundation of China (Grant No. 81870938) and Fund of Taishan scholar project (to BS).
References
Bao, X., Liu, G., Jiang, Y., Jiang, Q., Liao, M., Feng, R., et al. (2015). Cell adhesion molecule pathway genes are regulated by cis-regulatory SNPs and show significantly altered expression in Alzheimer's disease brains. Neurobiol. Aging 36, 2904. e1–7. doi: 10.1016/j.neurobiolaging.2015.06.006
Beecham, A. H., Patsopoulos, N. A., Xifara, D. K., Davis, M. F., Kemppinen, A., Cotsapas, C., et al. (2013). Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 45, 1353–1360. doi: 10.1038/ng.2770
Chang, D., Nalls, M. A., Hallgrimsdottir, I. B., Hunkapiller, J., Van Der Brug, M., Cai, F., et al. (2017). A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat. Genet. 49, 1511–1516. doi: 10.1038/ng.3955
Chen, L., Ge, B., Casale, F. P., Vasquez, L., Kwan, T., Garrido-Martin, D., et al. (2016). Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398–1414e24. doi: 10.1016/j.cell.2016.10.026
Fairfax, B. P., Humburg, P., Makino, S., Naranbhai, V., Wong, D., Lau, E., et al. (2014). Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343:1246949. doi: 10.1126/science.1246949
Hu, Y., Jin, S., Cheng, L., Liu, G., and Jiang, Q. (2017). Autoimmune disease variants regulate GSDMB gene expression in human immune cells and whole blood. Proc. Natl. Acad. Sci. U.S.A. 114, E7860–E7862. doi: 10.1073/pnas.1712127114
Jiang, Q., Jin, S., Jiang, Y., Liao, M., Feng, R., Zhang, L., et al. (2017). Alzheimer's disease variants with the genome-wide significance are significantly enriched in immune pathways and active in immune cells. Mol. Neurobiol. 54, 594–600. doi: 10.1007/s12035-015-9670-8
Joehanes, R., Zhang, X., Huan, T., Yao, C., Ying, S. X., Nguyen, Q. T., et al. (2017). Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biol. 18:16. doi: 10.1186/s13059-016-1142-6
Jun, G. R., Chung, J., Mez, J., Barber, R., Beecham, G. W., Bennett, D. A., et al. (2017). Transethnic genome-wide scan identifies novel Alzheimer's disease loci. Alzheimers Dement. 13, 727–738. doi: 10.1016/j.jalz.2016.12.012
Lambert, J. C., Ibrahim-Verbaas, C. A., Harold, D., Naj, A. C., Sims, R., Bellenguez, C., et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458. doi: 10.1038/ng.2802
Li, Y., Song, D., Jiang, Y., Wang, J., Feng, R., Zhang, L., et al. (2016). CR1 rs3818361 polymorphism contributes to Alzheimer's disease susceptibility in Chinese population. Mol. Neurobiol. 53, 4054–4059. doi: 10.1007/s12035-015-9343-7
Liu, G., Bao, X., and Wang, R. (2015). Expression quantitative trait loci regulate HNF4A and PTBP1 expression in human brains. Proc. Natl. Acad. Sci. U.S.A. 112:E3975. doi: 10.1073/pnas.1509048112
Liu, G., Hu, Y., Jin, S., and Jiang, Q. (2017a). Genetic variant rs763361 regulates multiple sclerosis CD226 gene expression. Proc. Natl. Acad. Sci. U.S.A. 114, E906–E907. doi: 10.1073/pnas.1618520114
Liu, G., Jiang, Y., Wang, P., Feng, R., Jiang, N., Chen, X., et al. (2012). Cell adhesion molecules contribute to Alzheimer's disease: multiple pathway analyses of two genome-wide association studies. J. Neurochem. 120, 190–198. doi: 10.1111/j.1471-4159.2011.07547.x
Liu, G., Liu, Y., Jiang, Q., Jiang, Y., Feng, R., Zhang, L., et al. (2016). Convergent genetic and expression datasets highlight TREM2 in Parkinson's disease susceptibility. Mol. Neurobiol. 53, 4931–4938. doi: 10.1007/s12035-015-9416-7
Liu, G., Sun, J. Y., Xu, M., Yang, X. Y., and Sun, B. L. (2017b). SORL1 variants show different association with early-onset and late-onset Alzheimer's disease risk. J. Alzheimers. Dis. 58, 1121–1128. doi: 10.3233/JAD-170005
Liu, G., Wang, T., Tian, R., Hu, Y., Han, Z., Wang, P., et al. (2018a). Alzheimer's disease risk variant rs2373115 regulates GAB2 and NARS2 expression in human brain tissues. J. Mol. Neurosci. 66, 37–43. doi: 10.1007/s12031-018-1144-9
Liu, G., Xu, Y., Jiang, Y., Zhang, L., Feng, R., and Jiang, Q. (2017c). PICALM rs3851179 variant confers susceptibility to Alzheimer's disease in Chinese population. Mol. Neurobiol. 54, 3131–3136. doi: 10.1007/s12035-016-9886-2
Liu, G., Zhang, F., Hu, Y., Jiang, Y., Gong, Z., Liu, S., et al. (2017d). Genetic variants and multiple sclerosis risk gene SLC9A9 expression in distinct human brain regions. Mol. Neurobiol. 54, 6820–6826. doi: 10.1007/s12035-016-0208-5
Liu, G., Zhang, F., Jiang, Y., Hu, Y., Gong, Z., Liu, S., et al. (2017e). Integrating genome-wide association studies and gene expression data highlights dysregulated multiple sclerosis risk pathways. Mult. Scler. 23, 205–212. doi: 10.1177/1352458516649038
Liu, G., Zhang, Y., Wang, L., Xu, J., Chen, X., Bao, Y., et al. (2018b). Alzheimer's disease rs11767557 variant regulates EPHA1 gene expression specifically in human whole blood. J. Alzheimers Dis. 61, 1077–1088. doi: 10.3233/JAD-170468
Liu, G., Zhao, Y., Jin, S., Hu, Y., Wang, T., Tian, R., et al. (2018c). Circulating vitamin E levels and Alzheimer's disease: a Mendelian randomization study. Neurobiol Aging 72, 189.e1–189.e9. doi: 10.1016/j.neurobiolaging.2018.08.008
Liu, G., Zhao, Y., Sun, J. Y., and Sun, B. L. (2018d). Parkinson's disease risk variant rs1109303 regulates the expression of INPP5K and CRK in human brain. Neurosci. Bull. doi: 10.1007/s12264-018-0289-4. [Epub ahead of print].
Nalls, M. A., Pankratz, N., Lill, C. M., Do, C. B., Hernandez, D. G., Saad, M., et al. (2014). Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 46, 989–993. doi: 10.1038/ng.3043
Ong, C. T., and Corces, V. G. (2011). Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293. doi: 10.1038/nrg2957
Ryan, K. J., White, C. C., Patel, K., Xu, J., Olah, M., Replogle, J. M., et al. (2017). A human microglia-like cellular model for assessing the effects of neurodegenerative disease gene variants. Sci. Transl. Med. 9:eaai7635. doi: 10.1126/scitranslmed.aai7635
Shlyueva, D., Stampfel, G., and Stark, A. (2014). Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286. doi: 10.1038/nrg3682
Ward, L. D., and Kellis, M. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934. doi: 10.1093/nar/gkr917
Ward, L. D., and Kellis, M. (2016). HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881. doi: 10.1093/nar/gkv1340
Zhang, S., Zhang, D., Jiang, Y., Wu, L., Shang, H., Liu, J., et al. (2015). CLU rs2279590 polymorphism contributes to Alzheimer's disease susceptibility in Caucasian and Asian populations. J. Neural. Transm. 122, 433–439. doi: 10.1007/s00702-014-1260-9
Zhang, Y., Wang, L., Jia, H., Liao, M., Chen, X., Xu, J., et al. (2018). Genetic variants regulate NR1H3 expression and contribute to multiple sclerosis risk. J. Neurol. Sci. 390, 162–165. doi: 10.1016/j.jns.2018.04.037
Keywords: genome-wide association study, neurodegenerative disease, eQTLs, CD14+ monocytes, Alzheimer's disease
Citation: Sun J, Hou Y, Zhang Y, Wang L, Liu L, Sun B and Yuan H (2018) Genetic Variants Associated With Neurodegenerative Diseases Regulate Gene Expression in Immune Cell CD14+ Monocytes. Front. Genet. 9:666. doi: 10.3389/fgene.2018.00666
Received: 06 October 2018; Accepted: 04 December 2018;
Published: 18 December 2018.
Edited by:
Yan Huang, Harvard Medical School, United StatesCopyright © 2018 Sun, Hou, Zhang, Wang, Liu, Sun and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-liang Sun, YmxzdW44OEAxNjMuY29t
Hui Yuan, eXVhbmh1aTAzMTRAMTYzLmNvbQ==
†These authors share first authorship
 Jing-yi Sun1†
Jing-yi Sun1† Bao-liang Sun
Bao-liang Sun