- European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Cambridgeshire, United Kingdom
RNA-binding proteins (RBPs) play a key role in gene expression and post-transcriptional RNA regulation. As integral components of ribonucleoprotein complexes, RBPs are susceptible to genomic and RNA Editing derived amino acid substitutions, impacting functional interactions. This article explores the prevalent RNA Editing of RBPs, unravelling the complex interplay between RBPs and RNA Editing events. Emphasis is placed on their influence on single amino acid variants (SAAVs) and implications for disease development. The role of Proteogenomics in identifying SAAVs is briefly discussed, offering insights into the RBP landscape. RNA Editing within RBPs emerges as a promising target for precision medicine, reshaping our understanding of genetic and epigenetic variations in health and disease.
1 The ubiquitous role of RBPs in gene expression regulation
RNA-binding proteins (RBPs) are integral to gene expression regulation. Defined by their ability to bind RNA, many of them through modular RNA-binding domains (RBDs), they also contain conserved structural motifs enhancing RNA specificity and functional versatility. About 14%–32% of human protein-coding genes encode RBPs (Baltimore and Huang, 1970; Lunde et al., 2007; Cook et al., 2011; Mallam et al., 2019; Piovesan et al., 2019; Caudron-Herger et al., 2021). RBPs can be classified based on four interaction categories: RNA motif-dependent, RNA structure-dependent, RNA modification-dependent, and RNA guide-based interactions (Liu et al., 2020). These interactions form ribonucleoprotein (RNP) complexes, modulated by RNA through riboregulation, where RNA controls protein function by direct, specific binding.
Dysregulated RBPs are linked to diseases such as cardiovascular and peripheral vascular diseases, diabetes, cancer, neurodegenerative diseases, and autoimmune disorders (Gebauer et al., 2021; Kelaini et al., 2021; Hashimoto and Kishimoto, 2022; Liu and Cao, 2023; Naskar et al., 2023; Sanya et al., 2023). RBPs recognise and interact with numerous transcripts via RBDs, forming regulatory networks essential for controlling protein expression and maintaining cellular homeostasis. Acting as intermediaries, RBPs integrate genetic, epigenetic, transcriptional, post-transcriptional, translational, and environmental cues, leading to variations in protein expression among individuals (Glisovic et al., 2008) (Supplementary Figure S1A). Understanding the role of RBPs in these processes is crucial for elucidating mechanisms underlying phenotypic diversity and disease susceptibility.
Adenosine to Inosine (A-to-I) RNA Editing (RE) is predominantly mediated by double-stranded RNA-binding proteins (dsRBPs) with Adenosine Deaminase Acting on double-stranded (ds)RNA (ADAR) enzymatic activity, catalysing the hydrolytic deamination of adenine at the C6 position (Figure 1C). The most prevalent RE event involves the conversion of A-to-I, with less frequent changes such as cytosine-to-uracil (C-to-U), due to APOBEC1, acting exclusively on single stranded RNA (Bass et al., 1997; Blanc and Davidson, 2010). Specifically, enzymatic activity of ADAR1 and ADAR2, but not ADAR3, are central to this process (Savva et al., 2012), recognising “type A” stem-loop structures via dsRNA binding domains (Ryter and Schultz, 1998), with additional specificity derived from sequence-specific structural features within the dsRNA duplex. Recent studies show that editing-dependent functions of ADAR1 protect dsRNA from dsRNA-sensing molecules causing translation shut down (Chung et al., 2018) and inhibiting innate immunity and the interferon-mediated response. Deficiency in these ADAR1 functions due to 11 mutations affect either the catalytic domain or RNA binding (RB) loops (Wright and Vissel, 2012) and underlie the pathogenesis of autoinflammatory diseases, such as the type I interferonopathies Aicardi-Goutières syndrome.
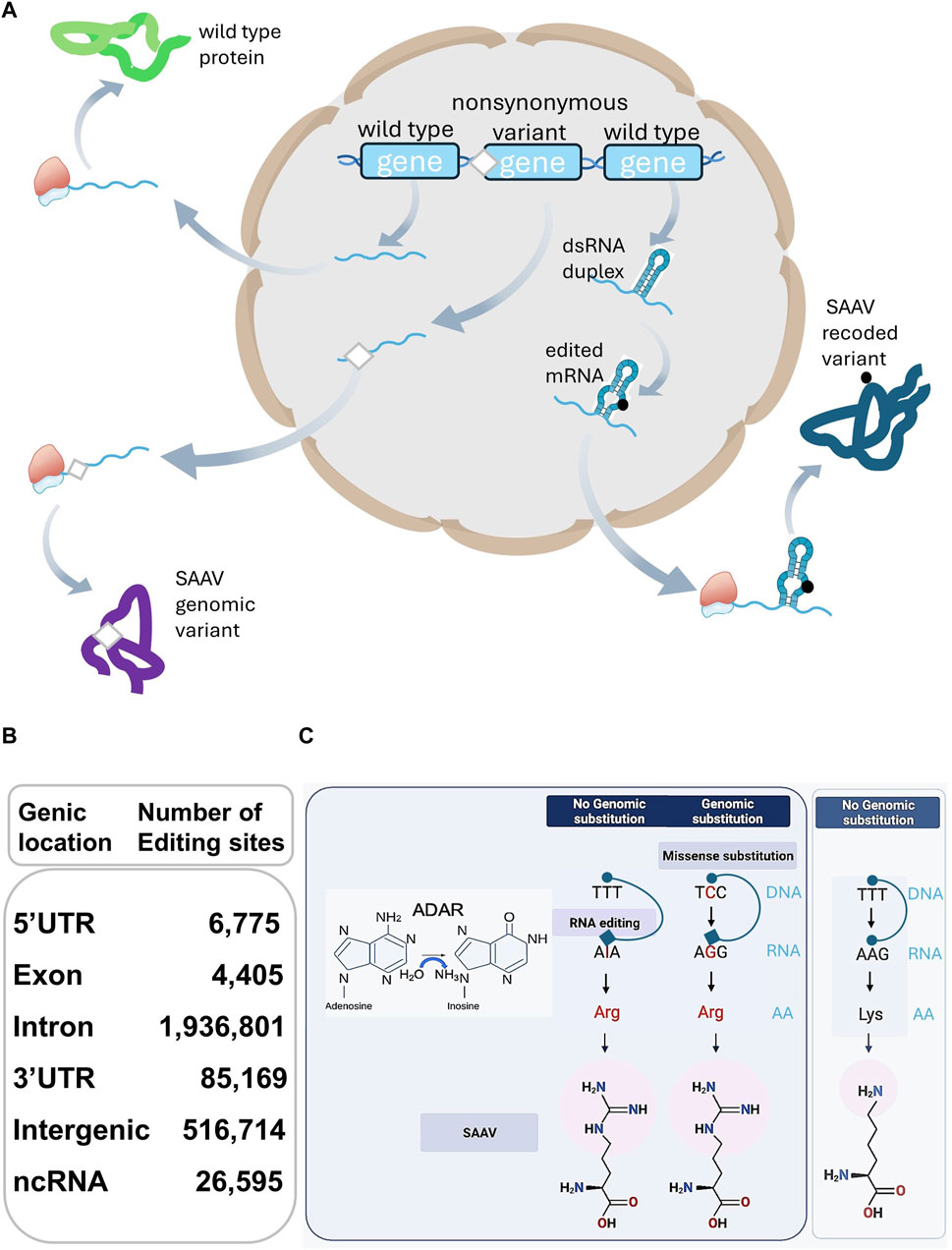
Figure 1. (A) SAAVs arise from genomic and RE substitutions, affecting protein sequences and structures differing from their corresponding wild-type counterparts. (B) The table describes the total number of human RE sites extracted from the RediPortal database. The total number of sites (2,572,054) present in non-protein coding genomic locations including 5′UTR (6,775), Intron (1,936,801), 3′UTR (85,169), Intergenic (516,714), and non-coding (nc)RNA (26,595) largely surpasses the number of nonsynonymous sites positioned in exons (4,405). (C) On the left side of the panel, Adenosine-to-Inosine substitution is catalysed by ADAR hydrolytic deamination activity at the C6 position of adenine. On the right side, a schematic representation of the differences between a genomic missense substitution and an RE-derived single AA substitution. Genomic missense single nucleotide substitution (C-to-G) or an RE event (A-to-I) with no genomic substitution, are both leading to a SAAV, represented as a Lysine-to-Arginine substitution. With no genomic substitution and in the absence of RNA editing, no SAAV but a Lysine is produced (far right side).
Unconventional RBPs lacking canonical RBDs, often exhibit context-dependent RB, suggesting influence by cellular conditions or RNA structural features (Sternburg and Karginov, 2020; Van Nostrand et al., 2020; Gebauer et al., 2021; Ray et al., 2023; Sanya et al., 2023). Interacting through non-canonical domains including protein-protein interaction interfaces, enzymatic cores, intrinsically disordered regions (IDRs), these RBPs challenge conventional RNP complex understanding (Garcia-Moreno et al., 2018). RNA-driven interaction mechanisms leading to the formation of RNPs are frequent. This supports the idea that RNA-protein interactions can take place in the absence of classical RBDs in RBPs, emphasising RNA’s regulatory roles as scaffolds, in protein-networks driven cellular processes, or in RNP remodelling mediated by modifications (Lee and Lykke-Andersen, 2013; Garcia-Moreno et al., 2018). The expanding RBP repertoire within the human proteome suggests broader functional interplays and demands a precise and comprehensive catalogue, requiring experimental and computational efforts (Jin et al., 2023).
RBPs enhance network interactions by controlling transcription through regulatory RNAs. Like transcription factors (TFs), RBPs are associated with genome hotspots, particularly gene promoters, influencing transcriptional output (Xiao et al., 2019). Additionally, RBPs mediate post-transcriptional control of gene expression, including splicing, transport, modification, translation, and degradation, often involving novel RBPs like PRRC2B, with an essential role in translation required for cell cycle progression (Jiang et al., 2023). Leaky scanning, where ribosomes bypass upstream Open Reading Frames (uORFs) which slow down the translation of the main ORF, is also regulated by PRRC2 proteins. These RBPs bind translation initiation factors and preinitiation complexes, facilitating translation initiation on mRNAs with uORFs (Bohlen et al., 2023). Many mRNAs with uORFs promote translation reinitiation and bypass uORF-mediated suppression mediated by RBPs such as Drosophila Nocte. Nocte RBP is critical for Drosophila eye development, with disruptions leading to developmental defects and neurological disorders (Zhang et al., 2024).
RBPs undergo multifaceted modifications influenced by RE and genomic variants. Alterations affect expression levels, generating distinct isoforms and amino acid (AA) sequence modifications with significant implications, including post-translational modifications (PTMs), altered protein-protein interactions, and subcellular localisation changes (Gebauer et al., 2021) (Figure 2B). While most editing targets untranslated regions (Hundley and BASS, 2010; Ellingford et al., 2022), highlighting the potential changes occurring within coding regions is required, as genomic and RE missense substitutions can have pathogenic effects, supporting the exploration of underlying mechanisms.
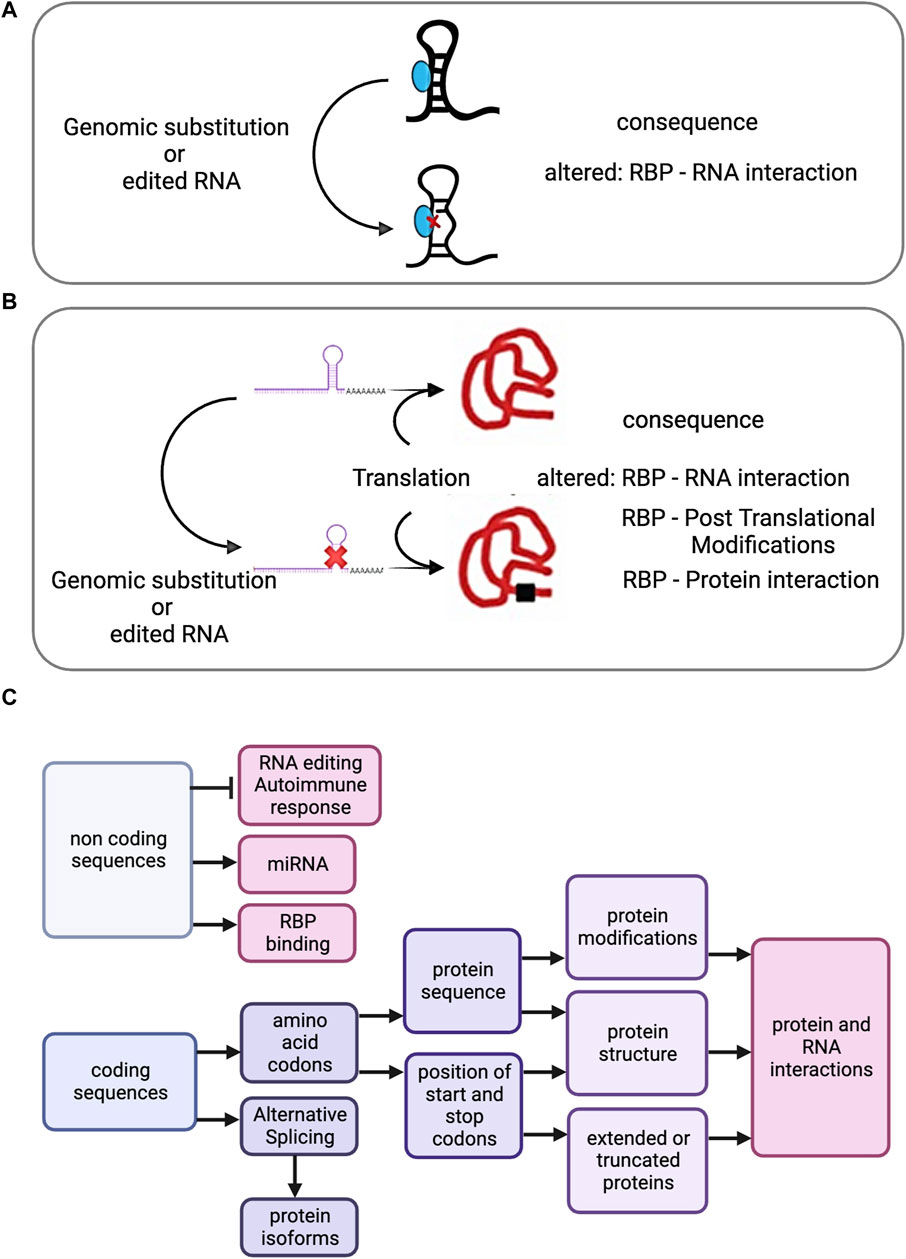
Figure 2. (A) Effect of genomic substitution or RNA recoding on Riboregulation. Either genomic substituted derived transcripts or recoded RNAs present in coding or non-coding RNAs may have implications for Riboregulation. A diagram showing a non-coding transcript with a dsRNA region, the recoded or genomic substituted derived transcript and its possible effect on the RNA - RBP interaction. (B) After translation, a recoded or genomic substituted derived RBP encoding transcript is represented, as well as the possible effects. (C) Cascade of potential effects propagated by SAAVs (derived from genomic or RNA editing substitutions) or by RNA editing recoding events in non-coding regions.
This manuscript explores the connection between RBPs, post-transcriptional RE, and resulting Single Amino Acid Variants, SAAVs, influencing health and disease. Sections examine the RBP landscape, emphasising physiological recoding, RE, disease implications, and the evolving field of proteogenomics, offering a holistic perspective and insights from cutting-edge omics studies.
2 Genomic and RNA editing effects: exploring the human RNA encoding RBP editome
SAAVs in the proteome arise from the interplay of genomic and nonsynonymous RE missense events (Chen et al., 2020; Petrosino et al., 2021). While distinct, these mechanisms converge, influencing RBPs and impacting phenotypic outcomes (Figure 1A).
In contrast to genomic missense mutations, RE events are sporadic within transcripts encoding small ORFs and even rarer in the broader protein-encoding transcriptome (Picardi et al., 2017), constituting approximately less than 1% of nonsynonymous RE events (Figure 1B). This distinction lies in the preservation of the DNA sequence encoding the target transcripts during RE, in contrast to missense mutations (Bass, 2002), as depicted in Figure 1C.
The protein-coding transcriptome undergoing RE, documented in the REDIportal database (Mansi et al., 2021), includes diverse human RNA sequencing samples from Genotype-Tissue Expression (GTEx) and provides insights into RE prevalence. Beyond REDIportal, other databases (Kiran and Baranov, 2010; Ramaswami and Li, 2014; Mallam et al., 2019; Niu et al., 2019) have delineated physiological and aberrant RE in coding transcripts.
The potential translation of these edits into SAAV events, including missense mutations, at the protein level underscores their significance. The best-studied A-to-I editing substrates are the brain-specific transcripts coding for glutamate receptor (GluR) channels. AMPA-type (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate) glutamate receptors (AMPARs) are critical for fast excitatory transmission in the central nervous system. A key regulatory step involves RE of the GluA2 subunit transcript by the dsRBP ADAR2 (Lin and Chen, 2019). This editing event recodes glutamine to an arginine codon, altering the encoded AA within the AMPAR heterotetrameric channel pore region. This event is essential for proper AMPAR function, as unedited GluA2 subunits containing glutamine allow calcium influx, while edited subunits with arginine are calcium impermeable. Notably, nearly complete editing occurs at this specific site, highlighting its critical role. Disruption of this editing process, leading to unedited GluA2 subunits, can result in pathological calcium influx, potentially contributing to seizures and neuronal cell death (Lin and Chen, 2019).
High-throughput techniques, relying on UV-crosslinking, affinity purification, size-exclusion chromatography, and mass spectrometry (MS)-based proteomics, have identified RBPs, expanding the RBPome, a term encompassing the entire repertoire of RBPs (Sommer et al., 1991; Nechay and Kleiner, 2020; Gebauer et al., 2021). As methodologies evolve, elusive proteins with weak crosslinking or low expression become accessible. The surge in RBPs leads to diverse studies and databases (RBPbase (Gebauer et al., 2021), Mallam et al.’s database (Mallam et al., 2019), RBP2go (Caudron-Herger et al., 2021), EuRBPdb (Liao et al., 2020), RBPDB (Cook et al., 2011), RNAcompete (Ray et al., 2009; Van Nostrand et al., 2020)), highlighting the evolving RBP landscape.
A human superset of 2,650 RBPs, identified in at least two RNA interaction capture (RIC) studies, combined with curated lists from Gene Ontology (The Gene Ontology, 2019), RBPDB, and RNAcompete, serves as the comprehensive RBPbase resource - “humanRBPs-2021 RBPANNO000000078.1” - consisting of 3,470 RBPs, and here designated “humanRBPs-2021.” This comprehensive RBPbase resource (Resource 2) assists as a powerful tool to pinpoint RBPs implicated in genetic diseases (Gebauer et al., 2021) and it was selected as a basis for the analysis of RE events in RBPs, as it represents a combination identified in multiple high-throughput studies and a set of well-known and defined RBPs.
Leveraging editing events from REDIportal - a rich repository of human physiological RE events - reveals a substantial “human coding editome” defined by a pooled editing level exceeding 1% (2,414 editable proteins, Resource 1), where around one-fifth represents potentially recoded RBPs with RE events propagated to their AA sequences (hypergeometric test, p 6.6e-7, corrected for multiple testing), a previously unappreciated protein class target within the human protein-coding editome (Supplementary Figure S2A). This suggests a mechanism where RBP-encoding RNA undergo editing, propagating SAAVs diversifying the proteome, and impacting their function, localisation, and interactions (Eisenberg, 2021). Approximately 12% of the human protein-encoding genome, editable RBPs devoid of known RBDs, may engage with RNA through unconventional means, emphasising the importance of considering editable RNA-determined interactions in RNP formation (Beckmann et al., 2016). Approximately 3.4%, comparably lesser RBPs with RBD-Pfam domains are frequently editable, suggesting a higher functional diversity beyond RB. Statistical tests indicate a significant enrichment of gene ontology term “Ribosome” and protein classes such as “Ribosomal protein” and “translational protein” among editable RBPs (p < 0.05, corrected for multiple testing), hinting at a potential link to translational control (Supplementary Figure S2B).
3 Implications of RNA editing in disease and development
RE primarily occurs in non-coding regions, with intronic transcripts more frequently modified than intergenic counterparts (Figure 1B). A-to-I conversion by ADAR, marks dsRNA as “self,” preventing unsuitable immune responses (Lamers et al., 2019). ADAR safeguards against autoimmunity by suppressing pathways associated with cellular long dsRNA sensors, including antiviral RIG-I-like receptors, MDA5-MAVS pathway, Protein Kinase activated by RNA (PKR), and OAS-RNAses (Quinones-Valdez et al., 2019). ADAR-interacting RBPs regulate A-to-I editing (Reich and Bass, 2019). Without ADARs, long dsRNAs may be mistaken for viral dsRNA, eliciting abnormal immune responses (Shtrichman et al., 2012).
RE plays key roles in various biological processes, including embryonic development (Shtrichman et al., 2012), neuron generation (Greger et al., 2003), and somatic cell reprogramming (Guallar et al., 2020). The phenomenon of RNA recoding can be transient during early development, with varying tissue-specific levels of RE. Single-cell RNA sequencing analyses established dynamic changes in ADAR expression and stage-specific RE during early embryogenesis (Qiu et al., 2016). Spatiotemporal expression patterns of ADAR1 and ADAR2 examined during the development of mouse forebrain disclosed a broad distribution in most regions, with their precise colocalisation in neurons, and uncovered that editing for specific ADAR mRNA targets precede high levels of expression maintained into adulthood (Jacobs et al., 2009). Based on RNA sequencing, the transcriptional profiles of cloned and fertilised bovine embryos were compared. Cloned embryos were shown to lack RE sites, which may have resulted from a decreased ADAR expression. Consequently, the authors concluded that cloned embryo development may be affected by decreased ADAR expression and incomplete RE, and their analysis provided new data for further mechanistic studies of somatic nuclear reprogramming (Zhang et al., 2020).
Dysregulation of RE can trigger immune responses by forming dsRNA duplexes, potentially leading to autoimmune disorders like psoriasis, rheumatoid arthritis, systemic lupus erythematosus, and multiple sclerosis (Li et al., 2022). The human brain and cardiovascular tissues exhibit a high prevalence of RE events (Ma et al., 2021; Mann et al., 2023), with 30% of recoded transcripts encoding RBPs (“humanRBPs-2021”) such as COPA, PUM2, PDC7, SON, RHOA, RRNAD1, SRP9, PPIL3, FLNA, TSEN2, and NOP14, linking RE of RBPs to cardiovascular diseases (Chen et al., 2020).
Non-coding miRNA recoding affects miRNA binding and translation control, with dysregulated expression of recoded ncRNAs being a feature of cancer cells and the malignant microenvironment (Liu et al., 2014; Torsin et al., 2021). Large-scale analysis of A-to-I and C-to-U editing in human miRNAs across 13 tissues revealed hypo-editing of miRNA and downregulation of ADAR2 in glioblastoma samples (Paz-Yaacov et al., 2015). In osteosarcoma, up-regulated editing sites in the 3′UTR abolish miRNA binding, increasing the expression of EMP2 and several oncogenes, highlighting miRNA editing’s role in oncogenesis (Ge et al., 2023). A study of melanoma patients treated with immunotherapy found 34% of RE sites in the 3′UTR targeted miRNAs, and 66% in coding regions were non-synonymous mutations leading to AA substitutions (Lu et al., 2024).
4 Significance of proteogenomics in uncovering SAAVs and assessing alterations in the recoded proteome
Proteogenomics integrates DNA/RNA sequencing with MS-based proteomics to reveal frequently recoded human proteins, including ubiquitously expressed RBPs (Gabay et al., 2022; Levitsky et al., 2023). This approach enhances understanding of genomic and RE missense substitutions, and identifies unannotated non-canonical variants, driver genes, and novel therapeutic targets across cancers (Li et al., 2023; Liao et al., 2023; Wang et al., 2023). Advances in omics technologies, supported by AI-assisted bioinformatic tools, improve the identification of SAAVs and sequence alignment at RNA and protein levels (De Paoli-Iseppi et al., 2021; Dai and Shen, 2022). Deep learning models predict protein structures and assess missense substitutions’ pathogenicity, enhancing protein stability predictions based on SAAV data (Biswas and Chakrabarti, 2020; Alquraishi, 2021; Hagg and Kirschner, 2023; Jagota et al., 2023).
MS-based proteomics datasets show enrichment of A-to-I nonsynonymous RE events at PTM sites, such as acetylation, methylation, sumoylation, and ubiquitination, affecting protein functions like degradation and subcellular localisation (Li et al., 2022). Changes in PTMs within RBPs, identified through proteogenomics, may alter RNP complexes and interactomes (Figure 2B). Proteogenomics pinpoints SAAVs, while subsequent proteomics explores changes within the recoded proteome including PTMs and interconnected networks. These advancements deepen our understanding of genetic and epigenetic variations, enabling biomarker discovery and advancing precision medicine.
5 RNA editing, RBPs, and disease: a complex frontier
The complex relationship between RE and its impact on RBPs in disease development deserves comprehensive exploration.
RE is primarily catalysed by dsRBP ADAR1 (and ADAR2), and SAAVs stemming from genomic missense mutations are associated with rare genetic pigmentation disorders and autoimmune diseases, such as Dyschromatosis symmetrica hereditaria and Aicardi-Goutières syndrome (Wright and Vissel, 2012). In addition, recent data from REDIportal reports A-to-I editing of the human dsRBP ADAR transcript, yielding potential SAAVs within the human ADAR protein (Supplementary Figure S1B). Drosophila ADAR2 transcript likewise undergoes RE (Keegan et al., 2005), where serine is substituted by glycine near the deaminase active site. The level of this Adar S-to-G self-editing is low in embryos and increases to 40% in adult flies. The edited ADAR G isoform is less active than the genome-encoded ADAR S isoform, both in vivo and in vitro (Keegan et al., 2023).
Elevated A-to-I editing levels, mediated by ADAR1 and ADAR2 enzymes, significantly impact cancer (Zipeto et al., 2016; Jiang et al., 2019). ADAR1, a “double-edged sword”, promotes leukemic stem cell proliferation by influencing let-7 microRNA biogenesis and activating the JAK2 signalling pathway, highlighting its potential as a therapeutic target in leukaemia (Zhang and Slack, 2016). It also hyperedits cell-cycle regulatory and tumour suppressor mRNA, further promoting leukaemia development (Zhang and Slack, 2016). Conversely, reduced A-to-I editing, often involving ADAR2 downregulation, inhibits astrocytoma migration and proliferation (Cenci et al., 2008). This tumour-suppressive role also extends to oesophageal squamous cell carcinoma, where ADAR2 editing of IGFBP7 mRNA disrupts tumorigenic pathways.
5.1 Editing of RNA, RNA-RBP interactions and membrane-less organelles
Riboregulation signifies a paradigm shift in our understanding of RNA-RBP interactions, with RNA emerging as a regulator of RBP function (Hentze et al., 2018). This concept challenges the traditional view of RBPs as primary regulators of RNA, emphasising their reciprocal interplay. Examples include metabolic enzymes (Huppertz et al., 2022), dsRNA-binding biosensor proteins like PKR (Gal-Ben-Ari et al., 2018), and antiviral receptors such as PRR and MDA5 (Quinones-Valdez et al., 2019). RE, especially targeting non-coding region transcripts, or RBP-recoded variants can modulate RNA-RBP interactions, either amplifying or diminishing their regulatory effects (Figures 2A, B). A report surveyed the binding preferences of 150 RBPs to RE events, focusing on A-to-I editing in two human cell lines. It was deduced that changes in RE could alter RNA secondary structures, affecting RBP-binding preferences and influencing post-transcriptional processes like RNA splicing, structure formation, and decay (Hu et al., 2022).
RBPs possess IDRs (Huai et al., 2022), making them vulnerable to changes induced by genomic missense or RNA RE variants. These alterations can affect RBPs’ ability to undergo liquid-liquid phase separation (LLPS), crucial for forming dynamic RNP networks or membrane-less organelles (MLOs) like nucleoli, stress granules, P-bodies, and nuclear speckles. LLPS is essential for various cellular processes, including transcription, translation, and signal transduction (Tsang et al., 2020; Li et al., 2023; Dai and Yang, 2024). For example, nuclear speckles regulate gene expression by storing and modifying pre-mRNA splicing factors (Hofmann et al., 2021; King et al., 2024).
SON, an RBP, localises to nuclear speckles, which regulate gene expression by storing and modifying pre-mRNA splicing factors (Supplementary Figure S1B) (Kim et al., 2019; Ilik et al., 2020). SON acts as an mRNA splicing cofactor, facilitating the efficient splicing of numerous cell-cycle and DNA-repair transcripts. REDIportal has catalogued forty RE events in transcripts encoding SON RBP, with a notable increase of over 10% in bladder urothelial carcinoma compared to healthy tissue (Gabay et al., 2022). Of these forty substitutions, nineteen are within IDRs, some potentially having pathogenic implications.
Cross-referencing healthy editable RBPs from the RBPbase “humanRBPs-2021” with an MLO dataset (5,282 genes) reveals that approximately two-thirds of these RBPs may inhabit MLOs (p 1.2e-80) (documented in MLOsMetaDB - http://mlos.leloir.org.ar) (Supplementary Figure S2C). Disrupted LLPS is linked to various pathological conditions, including cancer, viral infections, and neurodegenerative disorders (Huai et al., 2022). Recoded RBPs within MLOs can remodel interactomes through PTMs and protein-protein interactions, influencing LLPS dynamics. For instance, multiple PTMs of FUS, such as serine and threonine phosphorylation and arginine methylation, significantly impact its aggregation and LLPS properties (Qamar et al., 2018).
5.2 Additional examples of prevalent recoded RBPs COPA, FLNA, and FLNB uncovered in recent proteogenomic analyses
The recoded COPA RBP (Supplementary Figure S1B) at position I164V within a WD40 repeat affects protein interactions and is highly expressed in diseases like bladder urothelial carcinoma, breast cancer, and liver adenocarcinoma (Slotkin and Nishikura, 2013; Lundin et al., 2020; Gabay et al., 2022). COPA is involved in transporting dilysine-tagged proteins from the endoplasmic reticulum (ER) to the Golgi apparatus. Its recoding may cause ER stress, contributing to unfolded protein responses seen in various diseases, including cancer, neurodegeneration, diabetes, and inflammatory disorders.
Recoding of the FLNA RBP (Supplementary Figure S1B), specifically glutamine to arginine exchange, regulates angiogenesis in tumours. Hyper-editing reduces angiogenesis, while hypo-editing enhances it, potentially by altering VEGFR2 turnover. This modification also influences tumour metastasis through extracellular matrix interactions, highlighting FLNA editing’s role in angiogenesis, tumour growth, and metastasis (Jain et al., 2022).
FLNB RBP (Supplementary Figure S1B) recoding, specifically the M2293V edit, appears to suppress the growth- and invasion-inhibiting functions of the protein by affecting its nuclear localisation, thereby diminishing its EMT-suppressive role. This recoding likely disrupts FLNB RBP function by interfering with binding partners, impacting both its function and localisation (Levitsky et al., 2023).
5.3 Frequent RBP-derived neoantigens identified via immunopeptidomics
In a noteworthy study involving cancer patients, the integration of whole exome sequencing (WES), RNA sequencing (RNA-seq), and MS-based Immunopeptidomics revealed predominant RNA-derived neoantigens originating from coding regions, with approximately one-third of protein-coding genes exhibiting missense substitutions correlating with the RBPbase database “humanRBPs-2021.” Immunopeptidomics-driven identification of tumour neoantigens holds promise for personalised anti-cancer vaccines, making far-reaching changes in treatment strategies based on individual genomic and proteomic landscapes (Tretter et al., 2023; Zhang and Bassani-Sternberg, 2023).
5.4 Role of RE in disease and therapeutic potential
Genetic variants linked to RE, i.e., edQTL analyses, underscore the role of RNA secondary structure in dictating RE levels at specific sites, impacting complex traits and diseases by modulating the stability and levels of crucial RNA molecules (Park et al., 2021). mRNA editing’s involvement in various diseases, such as glioblastomas, inflammatory and autoimmune disorders, and its positive correlation with somatic point mutation burden in cancer, including the development of a novel metric, RE load, underscores its significance (Paz-Yaacov et al., 2015; Paul et al., 2017; Baker et al., 2022).
The impact of cellular stress-induced hyper-editing extends beyond cancer to cardiovascular diseases, highlighting mRNA recoding’s role in diverse pathologies (Mann et al., 2023). These collective findings expand our understanding of the interplay of genetic and epigenetic variations in health and disease, potentially catalysing a paradigm shift in medicine. RE emerges as a therapeutic target across a spectrum of diseases, showcasing its versatility in modulating life-threatening pathologies (Khosravi and Jantsch, 2021; Booth et al., 2023).
One potential RE therapeutic approach to rectifying G-to-A nonsense mutations involves the utilisation of endogenous ADAR. For instance, in scenarios involving a TAG stop codon disease variant, a guide RNA can be engineered to complement the mutated region, recruiting ADAR to edit the mRNA UAG stop codon back to UGG. This corrective action effectively eradicates the nonsense mutation, allowing mRNA translation to proceed unimpeded. These transformative developments highlight the profound influence of RE in shaping the landscape of genetic and epigenetic variation, offering new avenues for precision medicine tailored to individual patient’s unique genetic profiles (Pfeiffer and Stafforst, 2023).
6 Concluding remarks
The relationship between RE, RBPs, and disease unveils a significant area for research and therapeutic innovation. Understanding this interplay will enhance our comprehension of genetic and epigenetic variations underlying health and disease.
Proteogenomics, by elucidating RE events and identifying SAAVs in RBPs, serves as a potent tool for discovering disease biomarkers and developing tailored diagnostics and treatments. While the precise mechanisms of RE variants influencing RBPs in disease remain unclear, further investigation will clarify the complex interactions between edited and unedited dsRNAs and RBPs.
Deciphering the impact of RBP RE variants on key RNAs in disease mechanisms promises to provide biomarkers and therapeutic insights. Actively researching these interactions is crucial for understanding disease mechanisms and developing novel therapies.
RE emerges as a target for interventions across various life-threatening conditions, extending beyond cancer to a broader range of diseases. This highlights RE’s potential to advance precision medicine and enable personalised treatments, significantly transforming healthcare.
Author contributions
IF-M: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 945405 (ARISE programme).
Acknowledgments
My gratitude to Ernesto Picardi and Eli Eisenberg (ELIXIR, Integrating epitranscriptomic data into the ELIXIR ecosystem), and Matthias Hentze (EMBL), for relevant comments and teamwork. Special thanks to Alexander Schmidt and colleagues (Biozentrum of the University of Basel) for providing an excellent scientific and collegial environment.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
During the preparation of this work the author used Wordtune to proofread and improve language. After using this tool, the author reviewed and edited the content as needed and takes full responsibility for the content of the publication.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2024.1454241/full#supplementary-material
References
Alquraishi, M. (2021). Machine learning in protein structure prediction. Curr. Opin. Chem. Biol. 65, 1–8. doi:10.1016/j.cbpa.2021.04.005
Baker, A. R., Miliotis, C., Ramirez-Moya, J., Marc, T., Vlachos, I. S., Santisteban, P., et al. (2022). Transcriptome profiling of ADAR1 targets in triple-negative breast cancer cells reveals mechanisms for regulating growth and invasion. Mol. Cancer Res. 20, 960–971. doi:10.1158/1541-7786.MCR-21-0604
Baltimore, D., and Huang, A. S. (1970). Interaction of HeLa cell proteins with RNA. J. Mol. Biol. 47, 263–273. doi:10.1016/0022-2836(70)90301-3
Bass, B. L. (2002). RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846. doi:10.1146/annurev.biochem.71.110601.135501
Bass, B. L., Nishikura, K., Keller, W., Seeburg, P. H., Emeson, R. B., O'Connell, M. A., et al. (1997). A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3, 947–949.
Beckmann, B. M., Castello, A., and Medenbach, J. (2016). The expanding universe of ribonucleoproteins: of novel RNA-binding proteins and unconventional interactions. Pflugers Arch. 468, 1029–1040. doi:10.1007/s00424-016-1819-4
Biswas, N., and Chakrabarti, S. (2020). Artificial intelligence (AI)-Based systems biology approaches in multi-omics data analysis of cancer. Front. Oncol. 10, 588221. doi:10.3389/fonc.2020.588221
Blanc, V., and Davidson, N. O. (2010). APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 594–602. doi:10.1002/wsbm.82
Bohlen, J., Roiuk, M., Neff, M., and Teleman, A. A. (2023). PRRC2 proteins impact translation initiation by promoting leaky scanning. Nucleic Acids Res. 51, 3391–3409. doi:10.1093/nar/gkad135
Booth, B. J., Nourreddine, S., Katrekar, D., Savva, Y., Bose, D., Long, T. J., et al. (2023). RNA editing: expanding the potential of RNA therapeutics. Mol. Ther. 31, 1533–1549. doi:10.1016/j.ymthe.2023.01.005
Caudron-Herger, M., Jansen, R. E., Wassmer, E., and Diederichs, S. (2021). RBP2GO: a comprehensive pan-species database on RNA-binding proteins, their interactions and functions. Nucleic Acids Res. 49, D425–D436. doi:10.1093/nar/gkaa1040
Cenci, C., Barzotti, R., Galeano, F., Corbelli, S., Rota, R., Massimi, L., et al. (2008). Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 283, 7251–7260. doi:10.1074/jbc.M708316200
Chen, J., Wang, L., Wang, F., Liu, J., and Bai, Z. (2020). Genomic identification of RNA editing through integrating omics datasets and the clinical relevance in hepatocellular carcinoma. Front. Oncol. 10, 37. doi:10.3389/fonc.2020.00037
Chung, H., Calis, J. J. A., Wu, X., Sun, T., Yu, Y., Sarbanes, S. L., et al. (2018). Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824. doi:10.1016/j.cell.2017.12.038
Cook, K. B., Kazan, H., Zuberi, K., Morris, Q., and Hughes, T. R. (2011). RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 39, D301–D308. doi:10.1093/nar/gkq1069
Dai, X., and Shen, L. (2022). Advances and trends in omics Technology development. Front. Med. (Lausanne) 9, 911861. doi:10.3389/fmed.2022.911861
Dai, Z., and Yang, X. (2024). The regulation of liquid-liquid phase separated condensates containing nucleic acids. FEBS J. 291, 2320–2331. doi:10.1111/febs.16959
De Paoli-Iseppi, R., Gleeson, J., and Clark, M. B. (2021). Isoform age - splice isoform profiling using long-read technologies. Front. Mol. Biosci. 8, 711733. doi:10.3389/fmolb.2021.711733
Eisenberg, E. (2021). Proteome diversification by RNA editing. Methods Mol. Biol. 2181, 229–251. doi:10.1007/978-1-0716-0787-9_14
Ellingford, J. M., Ahn, J. W., Bagnall, R. D., Baralle, D., Barton, S., Campbell, C., et al. (2022). Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 14, 73. doi:10.1186/s13073-022-01073-3
Gabay, O., Shoshan, Y., Kopel, E., Ben-Zvi, U., Mann, T. D., Bressler, N., et al. (2022). Landscape of adenosine-to-inosine RNA recoding across human tissues. Nat. Commun. 13, 1184. doi:10.1038/s41467-022-28841-4
Gal-Ben-Ari, S., Barrera, I., Ehrlich, M., and Rosenblum, K. (2018). PKR: a Kinase to remember. Front. Mol. Neurosci. 11, 480. doi:10.3389/fnmol.2018.00480
Garcia-Moreno, M., Jarvelin, A. I., and Castello, A. (2018). Unconventional RNA-binding proteins step into the virus-host battlefront. Wiley Interdiscip. Rev. RNA 9, e1498. doi:10.1002/wrna.1498
Gebauer, F., Schwarzl, T., Valcarcel, J., and Hentze, M. W. (2021). RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 22, 185–198. doi:10.1038/s41576-020-00302-y
Ge, F., Cao, X., and Jiang, Y. (2023). A-to-I RNA editing shows dramatic up-regulation in osteosarcoma and broadly regulates tumor-related genes by altering microRNA target regions. J. Appl. Genet. 64, 493–505. doi:10.1007/s13353-023-00777-5
Glisovic, T., Bachorik, J. L., Yong, J., and Dreyfuss, G. (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986. doi:10.1016/j.febslet.2008.03.004
Greger, I. H., Khatri, L., Kong, X., and Ziff, E. B. (2003). AMPA receptor tetramerization is mediated by Q/R editing. Neuron 40, 763–774. doi:10.1016/s0896-6273(03)00668-8
Guallar, D., Fuentes-Iglesias, A., Souto, Y., Ameneiro, C., Freire-Agulleiro, O., Pardavila, J. A., et al. (2020). ADAR1-Dependent RNA editing promotes MET and iPSC reprogramming by alleviating ER stress. Cell Stem Cell 27, 300–314. doi:10.1016/j.stem.2020.04.016
Hagg, A., and Kirschner, K. N. (2023). Open-Source machine learning in computational chemistry. J. Chem. Inf. Model 63, 4505–4532. doi:10.1021/acs.jcim.3c00643
Hashimoto, S., and Kishimoto, T. (2022). Roles of RNA-binding proteins in immune diseases and cancer. Semin. Cancer Biol. 86, 310–324. doi:10.1016/j.semcancer.2022.03.017
Hentze, M. W., Castello, A., Schwarzl, T., and Preiss, T. (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341. doi:10.1038/nrm.2017.130
Hofmann, S., Kedersha, N., Anderson, P., and Ivanov, P. (2021). Molecular mechanisms of stress granule assembly and disassembly. Biochim. Biophys. Acta Mol. Cell Res. 1868, 118876. doi:10.1016/j.bbamcr.2020.118876
Huai, Y., Mao, W., Wang, X., Lin, X., Li, Y., Chen, Z., et al. (2022). How do RNA binding proteins trigger liquid-liquid phase separation in human health and diseases? Biosci. Trends 16, 389–404. doi:10.5582/bst.2022.01449
Hundley, H. A., and Bass, B. L. (2010). ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 35, 377–383. doi:10.1016/j.tibs.2010.02.008
Huppertz, I., Perez-Perri, J. I., Mantas, P., Sekaran, T., Schwarzl, T., Russo, F., et al. (2022). Riboregulation of Enolase 1 activity controls glycolysis and embryonic stem cell differentiation. Mol. Cell 82, 2666–2680 e11. doi:10.1016/j.molcel.2022.05.019
Hu, X., Zou, Q., Yao, L., and Yang, X. (2022). Survey of the binding preferences of RNA-binding proteins to RNA editing events. Genome Biol. 23, 169. doi:10.1186/s13059-022-02741-8
Ilik, I. A., Malszycki, M., Lubke, A. K., Schade, C., Meierhofer, D., and Aktas, T. (2020). SON and SRRM2 are essential for nuclear speckle formation. Elife 9, e60579. doi:10.7554/eLife.60579
Jacobs, M. M., Fogg, R. L., Emeson, R. B., and Stanwood, G. D. (2009). ADAR1 and ADAR2 expression and editing activity during forebrain development. Dev. Neurosci. 31, 223–237. doi:10.1159/000210185
Jagota, M., Ye, C., Albors, C., Rastogi, R., Koehl, A., Ioannidis, N., et al. (2023). Cross-protein transfer learning substantially improves disease variant prediction. Genome Biol. 24, 182. doi:10.1186/s13059-023-03024-6
Jain, M., Manjaly, G., Maly, K., De Vries, M. R., Janisiw, M., Konig, L., et al. (2022). Filamin A pre-mRNA editing modulates vascularization and tumor growth. Mol. Ther. Nucleic Acids 30, 522–534. doi:10.1016/j.omtn.2022.11.004
Jiang, F., Hedaya, O. M., Khor, E., Wu, J., Auguste, M., and Yao, P. (2023). RNA binding protein PRRC2B mediates translation of specific mRNAs and regulates cell cycle progression. Nucleic Acids Res. 51, 5831–5846. doi:10.1093/nar/gkad322
Jiang, Q., Isquith, J., Zipeto, M. A., Diep, R. H., Pham, J., Delos Santos, N., et al. (2019). Hyper-editing of cell-cycle regulatory and tumor suppressor RNA promotes malignant progenitor propagation. Cancer Cell 35, 81–94. doi:10.1016/j.ccell.2018.11.017
Jin, W., Brannan, K. W., Kapeli, K., Park, S. S., Tan, H. Q., Gosztyla, M. L., et al. (2023). HydRA: deep-learning models for predicting RNA-binding capacity from protein interaction association context and protein sequence. Mol. Cell 83, 2595–2611 e11. doi:10.1016/j.molcel.2023.06.019
Keegan, L. P., Brindle, J., Gallo, A., Leroy, A., Reenan, R. A., and O'Connell, M. A. (2005). Tuning of RNA editing by ADAR is required in Drosophila. EMBO J. 24, 2183–2193. doi:10.1038/sj.emboj.7600691
Keegan, L. P., Hajji, K., and O'Connell, M. A. (2023). Adenosine deaminase acting on RNA (ADAR) enzymes: a journey from weird to wondrous. Acc. Chem. Res. 56, 3165–3174. doi:10.1021/acs.accounts.3c00433
Kelaini, S., Chan, C., Cornelius, V. A., and Margariti, A. (2021). RNA-binding proteins hold key roles in function, dysfunction, and disease. Biol. (Basel) 10, 366. doi:10.3390/biology10050366
Khosravi, H. M., and Jantsch, M. F. (2021). Site-directed RNA editing: recent advances and open challenges. RNA Biol. 18, 41–50. doi:10.1080/15476286.2021.1983288
Kim, J., Han, K. Y., Khanna, N., Ha, T., and Belmont, A. S. (2019). Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 132, jcs226563. doi:10.1242/jcs.226563
King, M. R., Ruff, K. M., Lin, A. Z., Pant, A., Farag, M., Lalmansingh, J. M., et al. (2024). Macromolecular condensation organizes nucleolar sub-phases to set up a pH gradient. Cell 187, 1889–1906 e24. doi:10.1016/j.cell.2024.02.029
Kiran, A., and Baranov, P. V. (2010). DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics 26, 1772–1776. doi:10.1093/bioinformatics/btq285
Lamers, M. M., Van Den Hoogen, B. G., and Haagmans, B. L. (2019). ADAR1: “Editor-in-Chief” of cytoplasmic innate immunity. Front. Immunol. 10, 1763. doi:10.3389/fimmu.2019.01763
Lee, S. R., and Lykke-Andersen, J. (2013). Emerging roles for ribonucleoprotein modification and remodeling in controlling RNA fate. Trends Cell Biol. 23, 504–510. doi:10.1016/j.tcb.2013.05.001
Levitsky, L. I., Ivanov, M. V., Goncharov, A. O., Kliuchnikova, A. A., Bubis, J. A., Lobas, A. A., et al. (2023). Massive proteogenomic reanalysis of publicly available proteomic datasets of human tissues in search for protein recoding via adenosine-to-inosine RNA editing. J. Proteome Res. 22, 1695–1711. doi:10.1021/acs.jproteome.2c00740
Liao, J. Y., Yang, B., Zhang, Y. C., Wang, X. J., Ye, Y., Peng, J. W., et al. (2020). EuRBPDB: a comprehensive resource for annotation, functional and oncological investigation of eukaryotic RNA binding proteins (RBPs). Nucleic Acids Res. 48, D307–D313. doi:10.1093/nar/gkz823
Liao, Y., Savage, S. R., Dou, Y., Shi, Z., Yi, X., Jiang, W., et al. (2023). A proteogenomics data-driven knowledge base of human cancer. Cell Syst. 14, 777–787 e5. doi:10.1016/j.cels.2023.07.007
Li, C., Li, Z., Wu, Z., and Lu, H. (2023b). Phase separation in gene transcription control. Acta Biochim. Biophys. Sin. (Shanghai) 55, 1052–1063. doi:10.3724/abbs.2023099
Li, H., Wang, J., and Tu, J. (2022b). A-to-I nonsynonymous RNA editing was significantly enriched in the ubiquitination site and correlated with clinical features and immune response. Sci. Rep. 12, 15079. doi:10.1038/s41598-022-18926-x
Lin, C. H., and Chen, S. C. (2019). The cancer editome atlas: a resource for exploratory analysis of the adenosine-to-inosine RNA editome in cancer. Cancer Res. 79, 3001–3006. doi:10.1158/0008-5472.CAN-18-3501
Li, Q., Gloudemans, M. J., Geisinger, J. M., Fan, B., Aguet, F., Sun, T., et al. (2022a). RNA editing underlies genetic risk of common inflammatory diseases. Nature 608, 569–577. doi:10.1038/s41586-022-05052-x
Liu, H., Ma, C. P., Chen, Y. T., Schuyler, S. C., Chang, K. P., and Tan, B. C. (2014). Functional Impact of RNA editing and ADARs on regulation of gene expression: perspectives from deep sequencing studies. Cell Biosci. 4, 44. doi:10.1186/2045-3701-4-44
Liu, J., and Cao, X. (2023). RBP-RNA interactions in the control of autoimmunity and autoinflammation. Cell Res. 33, 97–115. doi:10.1038/s41422-022-00752-5
Liu, S., Li, B., Liang, Q., Liu, A., Qu, L., and Yang, J. (2020). Classification and function of RNA-protein interactions. Wiley Interdiscip. Rev. RNA 11, e1601. doi:10.1002/wrna.1601
Li, Y., Dou, Y., Da Veiga Leprevost, F., Geffen, Y., Calinawan, A. P., Aguet, F., et al. (2023a). Proteogenomic data and resources for pan-cancer analysis. Cancer Cell 41, 1397–1406. doi:10.1016/j.ccell.2023.06.009
Lunde, B. M., Moore, C., and Varani, G. (2007). RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490. doi:10.1038/nrm2178
Lundin, E., Wu, C., Widmark, A., Behm, M., Hjerling-Leffler, J., Daniel, C., et al. (2020). Spatiotemporal mapping of RNA editing in the developing mouse brain using in situ sequencing reveals regional and cell-type-specific regulation. BMC Biol. 18, 6. doi:10.1186/s12915-019-0736-3
Lu, Q., Zhou, W., Fan, L., Ding, T., Wang, W., and Zhang, X. (2024). Tumor neoantigens derived from RNA editing events show significant clinical relevance in melanoma patients treated with immunotherapy. Anticancer Drugs 35, 305–314. doi:10.1097/CAD.0000000000001565
Mallam, A. L., Sae-Lee, W., Schaub, J. M., Tu, F., Battenhouse, A., Jang, Y. J., et al. (2019). Systematic discovery of endogenous human ribonucleoprotein complexes. Cell Rep. 29, 1351–1368. doi:10.1016/j.celrep.2019.09.060
Mann, T. D., Kopel, E., Eisenberg, E., and Levanon, E. Y. (2023). Increased A-to-I RNA editing in atherosclerosis and cardiomyopathies. PLoS Comput. Biol. 19, e1010923. doi:10.1371/journal.pcbi.1010923
Mansi, L., Tangaro, M. A., Lo Giudice, C., Flati, T., Kopel, E., Schaffer, A. A., et al. (2021). REDIportal: millions of novel A-to-I RNA editing events from thousands of RNAseq experiments. Nucleic Acids Res. 49, D1012–D1019. doi:10.1093/nar/gkaa916
Ma, Y., Dammer, E. B., Felsky, D., Duong, D. M., Klein, H. U., White, C. C., et al. (2021). Atlas of RNA editing events affecting protein expression in aged and Alzheimer's disease human brain tissue. Nat. Commun. 12, 7035. doi:10.1038/s41467-021-27204-9
Naskar, A., Nayak, A., Salaikumaran, M. R., Vishal, S. S., and Gopal, P. P. (2023). Phase separation and pathologic transitions of RNP condensates in neurons: implications for amyotrophic lateral sclerosis, frontotemporal dementia and other neurodegenerative disorders. Front. Mol. Neurosci. 16, 1242925. doi:10.3389/fnmol.2023.1242925
Nechay, M., and Kleiner, R. E. (2020). High-throughput approaches to profile RNA-protein interactions. Curr. Opin. Chem. Biol. 54, 37–44. doi:10.1016/j.cbpa.2019.11.002
Niu, G., Zou, D., Li, M., Zhang, Y., Sang, J., Xia, L., et al. (2019). Editome Disease Knowledgebase (EDK): a curated knowledgebase of editome-disease associations in human. Nucleic Acids Res. 47, D78–D83. doi:10.1093/nar/gky958
Park, E., Jiang, Y., Hao, L., Hui, J., and Xing, Y. (2021). Genetic variation and microRNA targeting of A-to-I RNA editing fine tune human tissue transcriptomes. Genome Biol. 22, 77. doi:10.1186/s13059-021-02287-1
Paul, D., Sinha, A. N., Ray, A., Lal, M., Nayak, S., Sharma, A., et al. (2017). A-to-I editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme. Sci. Rep. 7, 2466. doi:10.1038/s41598-017-02397-6
Paz-Yaacov, N., Bazak, L., Buchumenski, I., Porath, H. T., Danan-Gotthold, M., Knisbacher, B. A., et al. (2015). Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 13, 267–276. doi:10.1016/j.celrep.2015.08.080
Petrosino, M., Novak, L., Pasquo, A., Chiaraluce, R., Turina, P., Capriotti, E., et al. (2021). Analysis and interpretation of the impact of missense variants in cancer. Int. J. Mol. Sci. 22, 5416. doi:10.3390/ijms22115416
Pfeiffer, L. S., and Stafforst, T. (2023). Precision RNA base editing with engineered and endogenous effectors. Nat. Biotechnol. 41, 1526–1542. doi:10.1038/s41587-023-01927-0
Picardi, E., D'Erchia, A. M., Lo Giudice, C., and Pesole, G. (2017). REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 45, D750–D757. doi:10.1093/nar/gkw767
Piovesan, A., Antonaros, F., Vitale, L., Strippoli, P., Pelleri, M. C., and Caracausi, M. (2019). Human protein-coding genes and gene feature statistics in 2019. BMC Res. Notes 12, 315. doi:10.1186/s13104-019-4343-8
Qamar, S., Wang, G., Randle, S. J., Ruggeri, F. S., Varela, J. A., Lin, J. Q., et al. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell 173, 720–734. doi:10.1016/j.cell.2018.03.056
Qiu, S., Li, W., Xiong, H., Liu, D., Bai, Y., Wu, K., et al. (2016). Single-cell RNA sequencing reveals dynamic changes in A-to-I RNA editome during early human embryogenesis. BMC Genomics 17, 766. doi:10.1186/s12864-016-3115-2
Quinones-Valdez, G., Tran, S. S., Jun, H. I., Bahn, J. H., Yang, E. W., Zhan, L., et al. (2019). Regulation of RNA editing by RNA-binding proteins in human cells. Commun. Biol. 2, 19. doi:10.1038/s42003-018-0271-8
Ramaswami, G., and Li, J. B. (2014). RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42, D109–D113. doi:10.1093/nar/gkt996
Ray, D., Kazan, H., Chan, E. T., Pena Castillo, L., Chaudhry, S., Shaheynoor, T., et al. (2009). Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 27, 667–670. doi:10.1038/nbt.1550
Ray, D., Laverty, K. U., Jolma, A., Nie, K., Samson, R., Pour, S. E., et al. (2023). RNA-binding proteins that lack canonical RNA-binding domains are rarely sequence-specific. Sci. Rep. 13, 5238. doi:10.1038/s41598-023-32245-9
Reich, D. P., and Bass, B. L. (2019). Mapping the dsRNA world. Cold Spring Harb. Perspect. Biol. 11, a035352. doi:10.1101/cshperspect.a035352
Ryter, J. M., and Schultz, S. C. (1998). Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17, 7505–7513. doi:10.1093/emboj/17.24.7505
Sanya, D. R. A., Cava, C., and Onesime, D. (2023). Roles of RNA-binding proteins in neurological disorders, COVID-19, and cancer. Hum. Cell 36, 493–514. doi:10.1007/s13577-022-00843-w
Savva, Y. A., Rieder, L. E., and Reenan, R. A. (2012). The ADAR protein family. Genome Biol. 13, 252. doi:10.1186/gb-2012-13-12-252
Shtrichman, R., Germanguz, I., Mandel, R., Ziskind, A., Nahor, I., Safran, M., et al. (2012). Altered A-to-I RNA editing in human embryogenesis. PLoS One 7, e41576. doi:10.1371/journal.pone.0041576
Slotkin, W., and Nishikura, K. (2013). Adenosine-to-inosine RNA editing and human disease. Genome Med. 5, 105. doi:10.1186/gm508
Sommer, B., Kohler, M., Sprengel, R., and Seeburg, P. H. (1991). RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19. doi:10.1016/0092-8674(91)90568-j
Sternburg, E. L., and Karginov, F. V. (2020). Global approaches in studying RNA-binding protein interaction networks. Trends Biochem. Sci. 45, 593–603. doi:10.1016/j.tibs.2020.03.005
The Gene Ontology, C. (2019). The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338. doi:10.1093/nar/gky1055
Torsin, L. I., Petrescu, G. E. D., Sabo, A. A., Chen, B., Brehar, F. M., Dragomir, M. P., et al. (2021). Editing and chemical modifications on non-coding RNAs in cancer: a new tale with clinical significance. Int. J. Mol. Sci. 22, 581. doi:10.3390/ijms22020581
Tretter, C., De Andrade Kratzig, N., Pecoraro, M., Lange, S., Seifert, P., Von Frankenberg, C., et al. (2023). Proteogenomic analysis reveals RNA as a source for tumor-agnostic neoantigen identification. Nat. Commun. 14, 4632. doi:10.1038/s41467-023-39570-7
Tsang, B., Pritisanac, I., Scherer, S. W., Moses, A. M., and Forman-Kay, J. D. (2020). Phase separation as a missing mechanism for interpretation of disease mutations. Cell 183, 1742–1756. doi:10.1016/j.cell.2020.11.050
Van Nostrand, E. L., Freese, P., Pratt, G. A., Wang, X., Wei, X., Xiao, R., et al. (2020). A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719. doi:10.1038/s41586-020-2077-3
Wang, X. Y., Xu, Y. M., and Lau, A. T. Y. (2023). Proteogenomics in cancer: then and now. J. Proteome Res. 22, 3103–3122. doi:10.1021/acs.jproteome.3c00196
Wright, A., and Vissel, B. (2012). The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain. Front. Mol. Neurosci. 5, 34. doi:10.3389/fnmol.2012.00034
Xiao, R., Chen, J. Y., Liang, Z., Luo, D., Chen, G., Lu, Z. J., et al. (2019). Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121. doi:10.1016/j.cell.2019.06.001
Zhang, W. C., and Slack, F. J. (2016). ADARs edit MicroRNAs to promote leukemic stem cell activity. Cell Stem Cell 19, 141–142. doi:10.1016/j.stem.2016.07.012
Zhang, B., and Bassani-Sternberg, M. (2023). Current perspectives on mass spectrometry-based immunopeptidomics: the computational angle to tumor antigen discovery. J. Immunother. Cancer 11, e007073. doi:10.1136/jitc-2023-007073
Zhang, L., Yu, M., Xu, H., Wei, X., Liu, Y., Huang, C., et al. (2020). RNA sequencing revealed the abnormal transcriptional profile in cloned bovine embryos. Int. J. Biol. Macromol. 150, 492–500. doi:10.1016/j.ijbiomac.2020.02.026
Zhang, T., Xue, Y., Su, S., Altouma, V., Ho, K., Martindale, J. L., et al. (2024). RNA-binding protein Nocte regulates Drosophila development by promoting translation reinitiation on mRNAs with long upstream open reading frames. Nucleic Acids Res. 52, 885–905. doi:10.1093/nar/gkad1122
Keywords: RNA editing, RNA-binding proteins (RBPs), single amino acid variants (SAAV), missense variants, proteogenomics, recoding, proteomics
Citation: Fierro-Monti I (2024) RBPs: an RNA editor’s choice. Front. Mol. Biosci. 11:1454241. doi: 10.3389/fmolb.2024.1454241
Received: 25 June 2024; Accepted: 25 July 2024;
Published: 06 August 2024.
Edited by:
Alessio Colantoni, Sapienza University of Rome, ItalyReviewed by:
Davide Mariani, Italian Institute of Technology (IIT), ItalyFeng Jiang, Stanford University, United States
Kaalak Reddy, University at Albany, United States
Copyright © 2024 Fierro-Monti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivo Fierro-Monti, aXZmaW1vQGViaS5hYy51aw==
†Present Address: Ivo Fierro-Monti, Biozentrum of the University of Basel, Basel, Switzerland
 Ivo Fierro-Monti
Ivo Fierro-Monti