- Department of Chemistry, College of Science and Mathematics, Mindanao State University—Iligan Institute of Technology, Iligan, Philippines
Euphorbia hirta L. is a medicinal plant widely used in the Philippines and across tropical Asia against various diseases, including respiratory disorders. In this study, the phytochemical components of E. hirta were investigated in silico for their potential to inhibit the severe acute respiratory syndrome-coronavirus-2 main protease (SARS-CoV-2 Mpro), a coronavirus disease 2019 (COVID-19) drug target that plays a critical role in the infection process of SARS-CoV-2. Phytochemical mining in tandem with virtual screening (PM-VS) was the strategy implemented in this study, which allows efficient preliminary in silico assessment of the COVID-19 therapeutic potential of the reported phytochemicals from the plant. The main rationale for considering E. hirta in the investigation was its reported efficacy against respiratory disorders. It is very promising to investigate the phytochemicals of E. hirta for their potential efficacy against diseases, such as COVID-19, that also target the respiratory system. A total of 298 E. hirta phytochemicals were comprehensively collected from the scientific literature. One hundred seventy of these phytochemicals were computed through molecular docking and were shown to have comparable or better binding properties (promising inhibitors) toward SARS-CoV-2 Mpro than known in vitro inhibitors. In connection to our previous work considering different medicinal plants, antiviral compounds were also rediscovered from the phytochemical composition of E. hirta. This finding provides additional basis for the potential of the plant (or its phytochemicals) as a COVID-19 therapeutic directly targeting drug targets such as SARS-CoV-2 Mpro and/or addressing respiratory-system-related symptoms. The study also highlights the utility of PM-VS, which can be efficiently implemented in the preliminary steps of drug discovery and development.
1 Introduction
Euphorbia hirta L. (Euphorbiaceae) is a medicinal plant widely used in the Philippines and across tropical Asia, and it is commonly known by the following names: “asthma plant” (English), “tawa-tawa” (Filipino), and “mangagaw” (Cebuano). The extract of E. hirta is taken orally as an aqueous decoction for most of its folkloric uses. As its English common name suggests, the plant has been used for asthma and other respiratory difficulties (Ekpo and Pretorius, 2007; Ogunlesi et al., 2009; Rao et al., 2017). In addition, available studies conclusively suggest its potential against dengue (Guzman et al., 2016; Perera et al., 2018; Suganthi and Ravi, 2018); however, additional studies are required to validate the results (Perera et al., 2018). Nevertheless, the studies reveal E. hirta as a pool for compounds with interesting biological activities.
E. hirta is one of the medicinal plants currently being investigated in the Philippines for its potential against coronavirus (CoV) disease 2019 (COVID-19) (Luci-Atienza, 2021a, Luci-Atienza, 2021b; Tawa-Tawa Clinical Trial on COVID-19, 2021). The goal is to develop a formulation utilizing the plant as an adjuvant treatment for mild to moderate COVID-19. A recently published review article identified E. hirta as one of the Philippine medicinal plants with immunomodulatory effects and potential against severe acute respiratory syndrome-CoV-2 (SARS-CoV-2) (Dayrit et al., 2021), the virus responsible for COVID-19. In this connection, a parallel and complementary in silico study was conducted to investigate the potential of its phytochemicals against a specific COVID-19 drug target, SARS-CoV-2 main protease (Mpro). Mpro is seen as an important COVID-19 drug target because of the role it plays in the regulation of viral replication (Di Micco et al., 2021).
It was the reported activities of E. hirta or its phytochemicals against respiratory-related ailments that serve as the primary basis for considering it as a subject of the present investigation. This study was conducted in line with the ongoing effort to discover potential COVID-19 therapeutic chemicals from medicinal plants, starting first with those found in the Philippines (Philippine medicinal plants). A strategy called phytochemical mining in tandem with virtual screening (PM-VS) was implemented. PM-VS refers to the systematic and comprehensive collection of medicinal plant phytochemicals reported in the scientific literature (phytochemical mining) and subsequent in silico assessment of the potential efficacy of the phytochemicals against specific or multiple drug target(s) (virtual screening). PM-VS and its rationale have been elaborated elsewhere (Cayona and Creencia, 2021a; Cayona and Creencia, 2021b, Cayona and Creencia, 2022). Specifically focused in this study is E. hirta and automated targeted molecular docking as the medicinal plant and virtual screening tool, respectively. It is argued that PM-VS can be efficiently implemented in the preliminary steps of drug discovery and development.
2 Materials and Methods
2.1 Phytochemical Data Collection
The method implemented in this study is adapted from the method described in our previous papers (Cayona and Creencia, 2021a; Cayona and Creencia, 2021b) with slight modification. The Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) (Moher et al., 2009) protocol was implemented throughout the systematic data collection process. The sources of phytochemicals were peer-reviewed research and review articles from scientific journals deposited in the MEDLINE database by the US National Institutes of Health National Library of Medicine (https://pubmed.ncbi.nlm.nih.gov/). The Google Scholar (https://scholar.google.com/) search engine was utilized to find additional literature but other search engines were also consulted (i.e., Microsoft Academic and Semantic Scholar), similarly applying relevant search keys and filters when applicable.
The identified sources were then compared against each other to check for multiple entries and reference-checked to retrieve additional sources unintentionally omitted in the first part of literature gathering. Articles deposited in restricted repositories and which were not written in English were not included. Thereafter, the phytochemicals reported in every literature reference were trimmed down to unique chemical identities only (because one compound may have multiple reported names). For simplicity, the common names of the compounds were taken in cases where ambiguity does not manifest; otherwise, the International Union of Pure and Applied Chemistry (IUPAC) nomenclature was adopted. The study strictly adhered to the data collection protocol described in PRISMA (see Supplementary Materials).
2.2 Phytochemical Classification
The collected phytochemicals from E. hirta were classified according to the ClassyFire (Djoumbou Feunang et al., 2016) algorithm of chemical classification. This was done to gain insight that might be helpful in assessing the basic structure–activity relationship. The hierarchy of chemical taxonomic classification can be found in the Supplementary Materials.
2.3 Preparation of Ligands
Three-dimensional (3D) structure-data files (SDFs) of phytochemicals included in the final list were either conveniently collected from PubChem or manually generated whenever they are unavailable in the database. Hydrogen atoms were explicitly added to the structures. In some cases, two-dimensional (2D) SDFs were used but only for 2D compounds (linear or flat). In preparation for virtual screening and for future convenience, the SDFs of all the structures of the phytochemicals (the ligands) were combined into a single SDF using OpenBabel 2.4.1 (O’Boyle et al., 2011) to facilitate automated importing of the multiple structures into the virtual screening tool. The same preparation was done for the control compounds.
2.4 Receptor Preparation
The crystal structure at 2.16 Å of the SARS-COV-2 Mpro (PDB ID: 6LU7) in complex with the in vitro inhibitor N3 (Jin et al., 2020) was downloaded from the Protein Data Bank (http://www.rcsb.org/) in a PDB file format. The noninteracting atoms (e.g., water and buffer molecules) were removed, and hydrogen atoms were explicitly added to the enzyme and the native ligand.
The active site was taken as the region of the SARS-CoV-2 Mpro volume where the in vitro inhibitor N3 was attached. From the SARS-CoV-2 Mpro–N3 complex, the search space for the targeted molecular docking was then assigned with the help of BIOVIA Discovery Studio Visualizer v20.1.0.19295 (DSV, 2020). The interacting and the pocket amino acids (AAs) that lie within the 3.5 Å distance from the closest N3 atom were identified by visual inspection. The residues found within this region totaled 25 AAs. The interacting AAs were H41, M49, F140, N142, G143, H164, M165, E166, L167, P168, H172, Q189, T190, and T191; and the pocket AAs were T24, T25, T26, L27, Y54, L141, S144, C145, H163, D187, R188, and Q192. From this list of AAs, the H41–C145 catalytic dyad can be found (Wang YC. et al., 2020; Hakmi et al., 2020; Ullrich and Nitsche, 2020).
2.5 Virtual Screening Through Automated Molecular Docking
2.5.1 Molecular Docking Tools
The phytochemical ligands were virtually screened against SARS-CoV-2 Mpro (6LU7-neat) using PyRx0.8 (Dallakyan and Olson, 2015), a virtual screening tool that allows automated molecular docking of multiple ligands (or libraries) against target receptor (s). PyRx0.8 utilizes the enabling capabilities of AutoDock tools for receptor and ligand preparation just as in AutoDock 4 (Morris et al., 2009) and the earlier versions; AutoDock Vina for molecular docking (Trott and Olson, 2010); OpenBabel for file format interconversion (O’Boyle et al., 2011); and other open-source software. To save on computational cost, targeted molecular docking on the active site of Mpro was conducted.
2.5.2 Control Parameters
To enhance the accuracy, control parameters were set in molecular docking against Mpro. In addition to the phytochemical ligands, control ligand samples were also tested. Ten known inhibitors with established in vitro half-maximal effective concentration (EC50) against SARS-CoV-2 or half-maximal inhibitory concentration (IC50) against SARS-CoV-2 Mpro were used as positive controls. On the other hand, 10 small molecules that do not possess interesting pharmacological properties were also used as negative controls. The positive controls were N3 (6LU7 native ligand), efonidipine, bedaquiline, tideglusib, manidipine, N3, lercanidipine, boceprevir, shikonin, ebselen, and carmofur, whose inhibitory properties were reported elsewhere (Ghahremanpour et al., 2020; Jin et al., 2020; Ma et al., 2020). The negative controls chosen were anthracene, naphthalene, glycerol, decane, hexanol, benzene, cyclohexane, hexane, ethanol, and water. The positive controls (inhibitors) were expected to give satisfactory binding free energy (BFE) values towards the receptor SARS-CoV-2 Mpro because they are empirically established inhibitors. On this basis, their BFEs were taken as a reference in assigning promising phytochemicals against SARS-CoV-2 Mpro. In contrast, the negative controls should have unsatisfactory computed BFE towards the receptor. The control ligands provide a simple means to assess the reliability and performance of the virtual screening tool.
2.5.3 Automated Molecular Docking
The receptor (6LU7-neat) was loaded onto PyRx0.8 and set into the macromolecule (receptor) in the PDBQT format. The collective SDFs of the phytochemicals and positive and negative controls previously prepared using OpenBabel 2.4.1 were also loaded onto PyRx0.8 and subsequently extracted automatically to individual structures. The structures were then energy minimized by implementing suitable force fields. For most of the structures, MMFF94 was sufficient in energy minimization; however, UFF and/or Ghemical must be implemented for some ligands whose final structures were distorted under specified UFF minimization parameters. Thereafter, the ligands were converted into a docking-ready PDBQT file format.
Before docking, the search space for the targeted automated molecular docking was set. The interacting and the pocket AA residues of SARS-CoV-2 Mpro that were identified previously were selected, and the search space was adjusted manually in the PyRx0.8 interface so that all of the residues were included in the grid volume of the search space. The resulting grid dimensions are the following: center_x = −10.8864; center_y = 14.0407; center_z = 68.7458; size_x = 21.4856; size_y = 26.7715; size_z = 28.0882. The exhaustiveness of the most stable conformation search was set at 16. Finally, docking was commenced using the Vina (AutoDock Vina) tab in PyRx0.8.
2.5.4 Receptor–Ligand Interaction Analysis
Interactions of the ligands which have BFEs comparable to or better than those of the positive controls were analyzed. Those ligands whose most stable binding conformation (docking RMSD = 0) established interactions with the H41–C145 catalytic dyad (Jin et al., 2020; Khan et al., 2020; Menéndez et al., 2020; Mirza and Froeyen, 2020; Shitrit et al., 2020) of the SARS-CoV-2 Mpro and those with reported antiviral properties were given emphasis. Favorable computed BFE and catalytic dyad interaction(s) were considered as major criteria in identifying promising SARS-CoV-2 Mpro inhibitors.
2.5.5 Assessment of the Reliability of the Tools and Strategies
All the tools and strategies used in the study are well established throughout the scientific literature. The number of citations of the articles that report the tools and strategies partly establish their reputation in the field. For example, Google Scholar queries on PRISMA, ClassyFire, PyRx0.8, AutoDock Vina, and AutoDock 4 will reveal 7,156; 283; 873; 15,275; and 12,351 citations, respectively, as of July 17, 2021.
3 Results
3.1 Phytochemical Mining and Classification
Literature reports indicate that leaves, aerial parts, and whole plants are the sources of E. hirta phytochemicals. The relevant data collected from phytochemical mining (PM) E. hirta are presented in Table 1. Each phytochemical is provided with its molecular formula (MF), BFE value against SARS-CoV-2 Mpro, and chemical taxonomy grouping levels (ClassyFire Superclass, Class, and Subclass). The chemical structures of all E. hirta phytochemicals and the control samples (positive and negative) used in molecular docking can be found in the Supplementary Materials. In total, 298 phytochemical components of E. hirta were identified from verified sources. This is by far the most comprehensive data gathering for E. hirta phytochemicals. The phytochemicals gathered fall into 13 ClassyFire Superclass levels. Majority are lipids and lipid-like molecules (Lipids) (108, 36.2%); phenylpropanoids and polyketides (PPPKs) (82, 27.5%); organic oxygen compounds (OOCs) (29, 9.7%); lignan, neolignans, and related compounds (Lignans) (27, 9.1%); organic acids and derivatives (OADs) (16, 5.4%); benzenoids (15, 5.0%); and organoheterocyclic compounds (OHCCs) (13, 4.4%), comprising a total of 97.0%. The rest (*Miscellaneous) of the phytochemicals are hydrocarbons, organic 1,3-dipolar compounds, organic nitrogen, organohalogens, and organic salt.
3.2 Virtual Screening Through Automated Molecular Docking
The data obtained in Table 1 are graphically presented in Figure 1. The BFE values of the phytochemicals are described in 1A, and these are compared to the control compounds (positive and negative). It can be observed that the positive controls obtained more highly negative BFE values (thermodynamically stable receptor–ligand interaction) against SARS-CoV-2 Mpro than the negative controls (see entries in Table 1). The least negative in the group is that of carmofur with −6.0 kcal/mol computed BFE based on the AutoDock Vina docking algorithm. This value (−6.0 kcal/mol) was taken as the threshold for assigning promising inhibitors considering the fact that carmofur and the rest of the positive control compounds are actual in vitro inhibitors against SARS-CoV-2 Mpro. Phytochemicals having BFE values of ≤−6.0 kcal/mol qualify as promising inhibitors. In Figure 1A, these phytochemicals are represented by the points on and below the dashed horizontal line. Over this line are the non-promising inhibitors and the negative controls with less satisfactory BFE values. Overall, 170 (57.0%) of the phytochemicals found in E. hirta were identified as promising inhibitors against SARS-CoV-2 Mpro from a total of 298 phytochemicals.
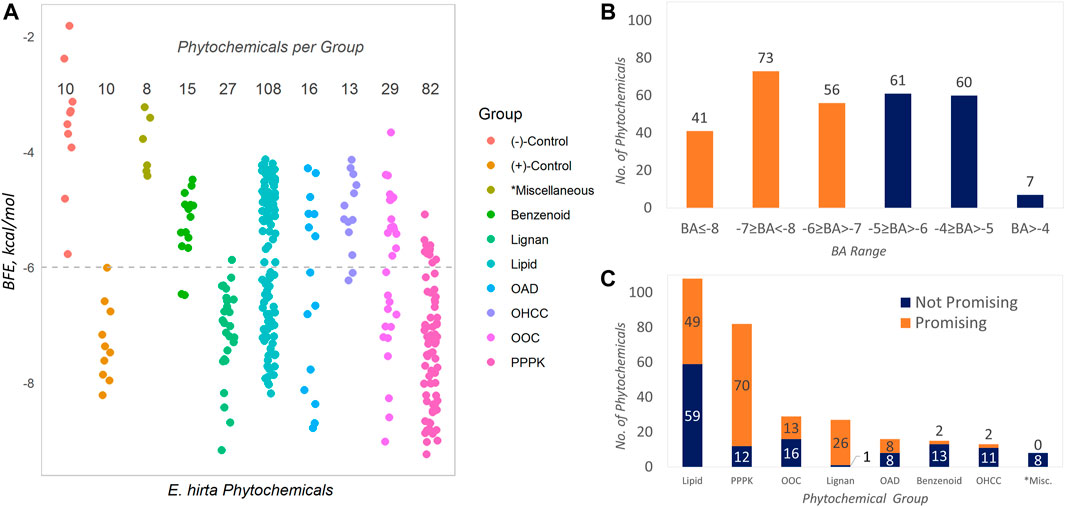
FIGURE 1. Binding properties of the phytochemicals from E. hirta against SARS-CoV-2 main protease. Benzenoids contain the benzene ring; hydrocarbons are composed of H and C atoms only; lignans contain dimeric phenylpropanoids; lipids contain isoprene moiety (terpene or terpenoids), fatty acyls, and derivatives; OADs contain the acyl group; OOCs contain oxygen atoms (e.g., alcohols and esters); OHCC, heterocyclic ring; PPPK, Ph-C3- and alternating—(C=O)-CH2-. *Miscellaneous groups are composed of the least abundant phytochemicals.
The distribution of the BFEs is shown in Figure 1B and that of the promising inhibitors is highlighted as orange bars. The BFE range with the most abundant phytochemicals is −7 ≥ BFE < −8 with 73 (24.5%) promising inhibitors. It can be seen in both Figure 1A and Figure 1C that the two most abundant groups are lipids (108, 36.2%) and PPPKs (82, 27.5%), collectively comprising 63.7% of the total. Interestingly, PPPKs have the most number of promising phytochemicals per group. There are 70 out of 82 (85.4%) PPPKs that are promising inhibitors. This value is 23.5% of the total number of E. hirta phytochemicals. This behavior by the PPPKs has been previously noted (Cayona and Creencia, 2021a, Cayona and Creencia, 2022). The relative numbers of promising and non-promising inhibitors with respect to chemical grouping are given in Figure 1C.
3.3 Antiviral Phytochemicals From E. hirta
Virtual screening revealed that E. hirta is an abundant source of promising inhibitors of SARS-CoV-2 Mpro. The list of promising inhibitors includes notable compounds with interesting biological and pharmacological properties. At least 12 of the promising inhibitors were established in vitro or in vivo antiviral compounds against various viruses. These are kaempferol (A), luteolin (B), quercetin (C), isoquercitrin (D), hyperoside (E), rutin (F), myricetin-3-O-rhamnoside (G), daphnoretin (H), digallic acid (I), epicatechin-3-gallate (J), trigallic acid (K), and corilagin (L). The chemical structures of the aforementioned compounds and their overlain conformations on the active site of SARS-CoV-2 Mpro represented by an H-bonding surface are shown in Figure 2. A–G all have a common molecular skeleton, of which A is the only one without an attached sugar moiety. The skeleton of H is an isomer of A–G skeleton, and I–L are gallic acid derivatives.
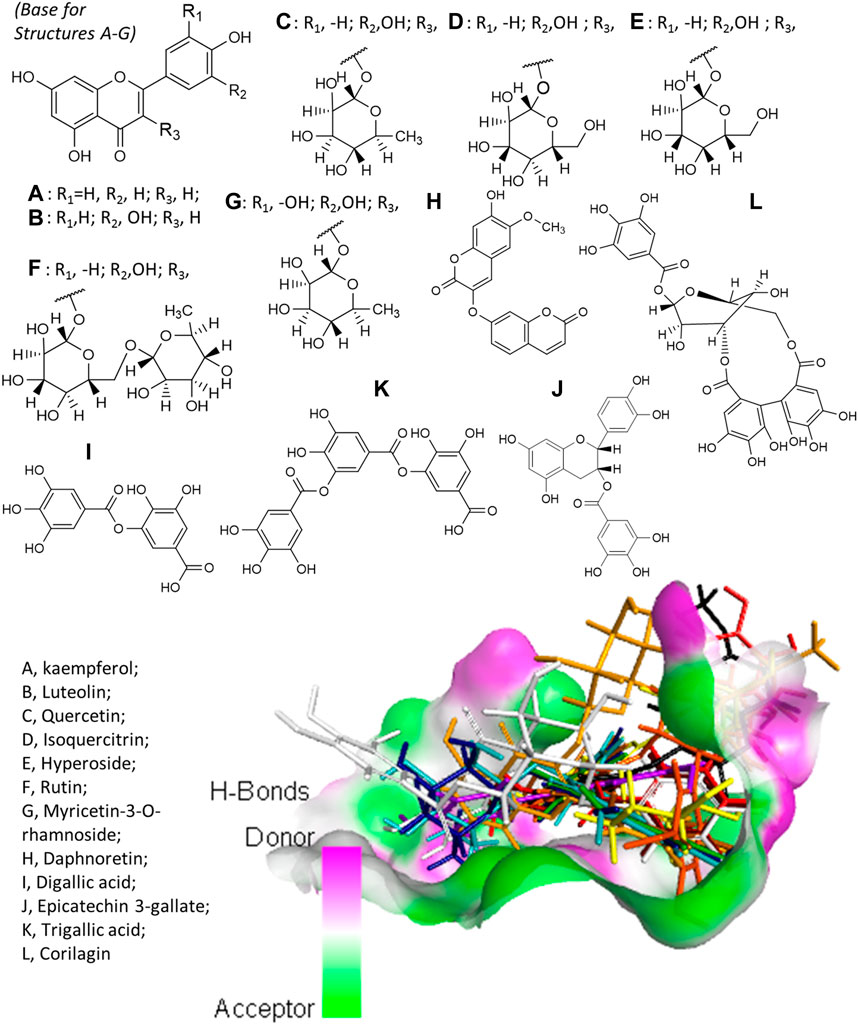
FIGURE 2. Structures of the antiviral compounds found in E. hirta and overlain stick representation of the ligands in complex with SARS-CoV-2 Mpro. (A) Brown; (B) purple; (C) mint green; (D) cyan; (E) blue; (F) gold; (G) yellow; (H) black; (I) maroon; (J) orange; (K) red; and (L) gray.
The viruses susceptible to compounds A–L are listed in Table 2 along with relevant details obtained from virtual screening (i.e., BFEs and interacting AAs). The susceptible viruses include herpes simplex virus (HSV), hepatitis, enterovirus, human immunodeficiency virus (HIV), and influenza. Interestingly, specific antivirals are effective against viruses that affect the respiratory tract, such as CoVs and respiratory syncytial viruses (RSVs). This property is clearly relevant when considering chemical therapy against respiratory tract-related diseases like COVID-19. Kaempferol is active against CoVs (Schwarz et al., 2014), and luteolin (Wang S. et al., 2020) and daphnoretin (Wang S. et al., 2020) are active against RSVs.
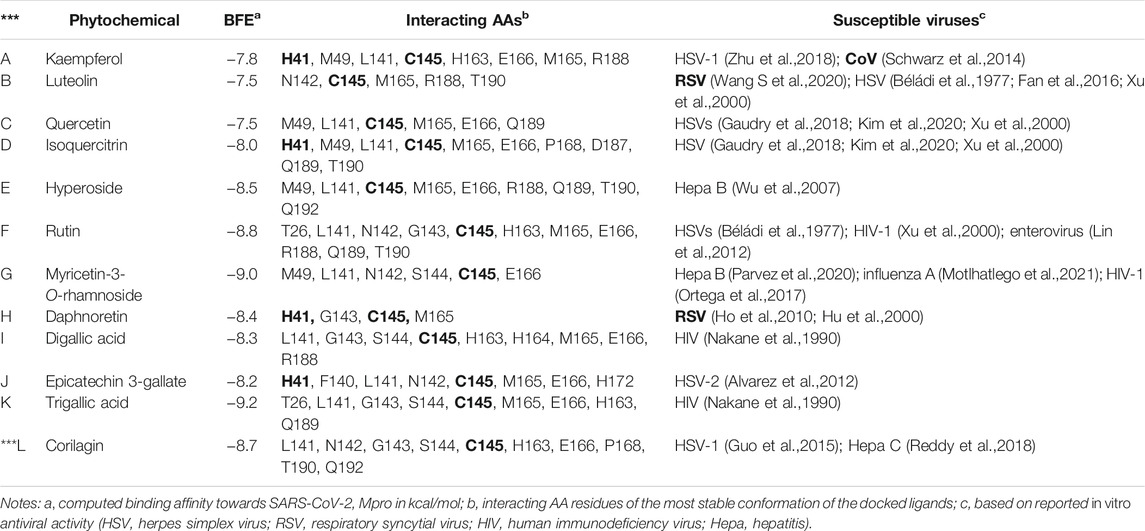
TABLE 2. In vitro antiviral phytochemicals rediscovered from the medicinal plants used in this study.
The interacting AAs are obtained from the most stable molecular docking conformation. These AAs are located at least 3.5 Å from the nearest atom of the docked ligands. It can be observed in Table 2 that H41 and/or C145 (in boldface) catalytic dyad residues in the active site of SARS-CoV-2 Mpro can interact with the promising inhibitors (identified using DSV 2020); however, molecular dynamics (MD) simulations are necessary to assess the stability of the receptor–ligand complex that can be formed. As stated previously, MD simulations are not covered in the scope of the present study and are reserved for future analyses. Nevertheless, the identification of these dyad residues in close proximity to the docked ligands provides a rationale for further studies. Figure 3 shows how the most stable docked conformations of kaempferol (one of the promising inhibitors) and N3 (known inhibitor) fit into the active site of SARS-CoV-2 Mpro. The AAs that are in close proximity to the ligands are also shown.
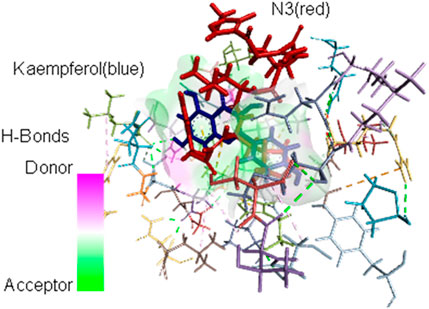
FIGURE 3. Most stable conformations of the docked kaempferol (blue) and the inhibitor N3 (red) into the active site of SARS-CoV-2 Mpro showing the AA residues near the ligands.
4 Discussion
This study provides the most comprehensive phytochemical gathering for E. hirta at present. It is argued in this the study that organized phytochemical composition will generate new information and enable meaningful analyses that may aid in understanding phytochemistry and plant metabolism. It was quite unexpected to discover an abundant cocktail of potential SARS-CoV-2 Mpro inhibitors from a single plant species. Clearly, lipids and PPPKs are among the most diverse groups of phytochemicals in E. hirta. These groups are also observed to be significantly more abundant in quantity than other phytochemical groups in E. hirta extracts (Sharma et al., 2014; Rao et al., 2017).
The molecular surface representation of the receptor reveals abundant hydrogen donor (purple) and acceptor (green) sites. This partly explains the observation that the ligands that can effectively establish H-bonding generally possess more negative BFEs than those that do not. Careful examination of the individual structures of the phytochemicals tested revealed that the capacity for H-bonding signifies direct correlation to favorable BFE. The ligands represented by the points below the dashed horizontal line in Figure 1A can H-bond more effectively and have more negative BFE towards SARS-CoV-2 Mpro than the ones above the line. Hydrocarbons (*Miscellaneous group), benzenoids, and OHCCs are obviously represented in the latter because they cannot (or can poorly) establish H-bonding with the receptor.
The molecular docking behavior of most PPPKs is interesting and deserves further investigation. Their docked conformations like the one presented in Figure 2 indicate the molecular skeleton that deeply buried and extended through the active site cavity of SARS-CoV-2 Mpro. The phenylpropanoids and their structurally related groups, the lignans, may feature pharmacophoric moieties.
Available preclinical information conclusively reveals that E. hirta possesses antiviral properties (Perera et al., 2018). In this study, some of these antiviral phytochemicals with established antiviral properties against various viruses, including those that affect the respiratory tract, were rediscovered through the PM-VS strategy. These properties are relevant in the effort to address a respiratory disease like COVID-19. More importantly, the strategy allowed the identification of many other promising inhibitors of SARS-CoV-2 Mpro despite its simplicity. Further studies are definitely necessary, but preliminary results gathered on the demonstration of the proof-of-principle for PM-VS provide a basis for exhaustive in silico investigations and future in vitro experiments. PM-VS can be efficiently implemented in the preliminary stages of drug discovery and development with minimal computational cost. Moving forward, other drug targets, not only COVID-19 drug targets, can also be investigated with PM-VS using different medicinal plants.
5 Conclusion
A method described as phytochemical mining allowed the systematic collection and organization of phytochemical components from E. hirta. A total of 298 E. hirta phytochemicals collected from the literature represent the most comprehensive phytochemical data collection for the plant. Virtual screening through molecular docking of the phytochemicals revealed an abundant cocktail of 170 promising inhibitors against SARS-CoV-2 Mpro. Twelve of the promising inhibitors are also prominent natural products with reported antiviral property against diverse viruses including respiratory CoV and RSVs. Finally, PM-VS was successfully implemented in this study, and the preliminary results obtained so far suggest further investigations.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author Contributions
The authors (RC and EC) contributed equally to the conceptualization, data gathering, analysis, and manuscript preparation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the Drug Design and Development Related Research Lab (DDDRRL) of the Premier Research Institute of Science and Mathematics (PRISM), MSU-IIT, for supporting this study. RC thanks DOST-SEI for the scholarship grant under ASTHRDP-NSC.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.801401/full#supplementary-material
References
Ali, M. Z., Mehmood, M. H., Saleem, M., and Gilani, A.-H. (2020). The Use of Euphorbia Hirta L. (Euphorbiaceae) in Diarrhea and Constipation Involves Calcium Antagonism and Cholinergic Mechanisms. BMC Complement. Med. Ther. 20 (1), 14. doi:10.1186/s12906-019-2793-0
Álvarez, Á. L., Dalton, K. P., Nicieza, I., Diñeiro, Y., Picinelli, A., Melón, S., et al. (2012). Bioactivity-Guided Fractionation of Phyllanthus Orbicularis and Identification of the Principal Anti HSV-2 Compounds. Phytother. Res. 26 (10), 1513–1520. doi:10.1002/ptr.4608
Amos Samkumar, R., Premnath, D., and David Paul Raj, R. S. (2019). Strategy for Early Callus Induction and Identification of Anti-snake Venom Triterpenoids from Plant Extracts and Suspension Culture of Euphorbia Hirta L. 3 Biotech. 9 (7), 266. doi:10.1007/s13205-019-1790-9
Bach, L. T., Dung, L. T., Quetin-Leclereq, J., Kestermont, P., Scippo, M.-L., Phuong, T. N., et al. (2017). The Flavonoid Isolation and Antioxidant Activity of Euphorbia Hirta L. Extracts. Vietnam J. Chem. 55 (5E34), 568–573. Retrieved from: https://aquabioactive.ctu.edu.vn/images/upload/publications/bai-bao-chi-bach.pdf
Bach, L. T., Hue, B. T. B., Tram, N. T. T., Thu, D. N. A., and Dung, L. T. (2020). Chemical Constituents from N-Hexane and Ethyl Acetate Extracts of Euphorbia Hirta L. Grown in Vietnam. IOP Conf. Ser. Mater. Sci. Eng. 736, 022083. doi:10.1088/1757-899X/736/2/022083
Béládi, I., Pusztai, R., Mucsi, I., Bakay, M., and Gábor, M. (1977). Activity of Some Flavonoids against Viruses. Ann. NY Acad. Sci. 284 (1 Third Confere), 358–364. doi:10.1111/j.1749-6632.1977.tb21971.x
Cayona, R., and Creencia, E. (2021a). Discovery of a "Cocktail" of Potential SARS-COV-2 Main Protease Inhibitors through Virtual Screening of Known Chemical Components of Vitex Negundo L. ("Lagundi"). Med. Chem. 18 (3), 364381. doi:10.2174/1573406417666210618132003
Cayona, R., and Creencia, E. (2021b). Phytochemical Mining Of SARS-COV-2 Main Protease Inhibitors From the Philippine Medicinal Plants: Vitex Negundo, Blumea Balsamifera, Euphorbia Hirta, Catharanthus Roseus, Mentha Cordifolia, Ehretia Microphylla, Combretum Indicum, Lagerstroemia Speciosa, Peperomia Pellucida, and Senna alata [Dissertation]. Iligan City: Mindanao State University-Iligan Institute of Technology.
Cayona, R., and Creencia, E. (2022). Phytochemical Mining of Potential SARS-CoV-2 Main Protease Inhibitors from Blumea Balsamifera (L.) DC. Philippine J. Sci. 151 (1), 235–261. Retrieved from: https://philjournalsci.dost.gov.ph/111-vol-151-no-1-february-2022/1555-phytochemical-mining-of-potential-sars-cov-2-main-protease-inhibitors-from-blumea-balsamifera-l-dc
Chi, S.-M., Wang, Y., Zhao, Y., Pu, J.-X., Du, X., Liu, J.-P., et al. (2012). A New Cyclopentanone Derivative from Euphorbia Hirta. Chem. Nat. Compd. 48 (4), 577–579. doi:10.1007/s10600-012-0315-0
Dallakyan, S., and Olson, A. J. (2015). “Small-Molecule Library Screening by Docking with PyRx,” in Chemical Biology: Methods and Protocols. Editors J. E. Hempel, C. H. Williams, and C. C. Hong (Springer), 243–250. doi:10.1007/978-1-4939-2269-7_19
Dayrit, F. M., Jr, A. M. G., and Gloriani, N. G. (2021). Philippine Medicinal Plants with Potential Immunomodulatory and Anti-SARS-CoV-2 Activities. Philippine J. Sci. 150 (5), 17. Retrieved from: https://philjournalsci.dost.gov.ph/108-vol-150-no-5-october-2021/1467-philippine-medicinal-plants-with-potential-immunomodulatory-and-anti-sars-cov-2-activities
Di Micco, S., Musella, S., Scala, M. C., Sala, M., Campiglia, P., Bifulco, G., et al. (2021). In Silico Analysis Revealed Potential Anti-SARS-CoV-2 Main Protease Activity by the Zonulin Inhibitor Larazotide Acetate. Front. Chem. 8, 628609. doi:10.3389/fchem.2020.628609
Djoumbou Feunang, Y., Eisner, R., Knox, C., Chepelev, L., Hastings, J., Owen, G., et al. (2016). ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminform. 8 (1), 61. doi:10.1186/s13321-016-0174-y
Ekpo, O. E., and Pretorius, E. (2007). Asthma, Euphorbia Hirta and its Anti-Inflammatory Properties. South Afr. J. Sci. 103 (5), 201–203. Retrieved from: https://www.researchgate.net/publication/286838495_Asthma_Euphorbia_hirta_and_its_anti-inflammatory_properties
Fan, W., Qian, S., Qian, P., and Li, X. (2016). Antiviral Activity of Luteolin against Japanese Encephalitis Virus. Virus. Res. 220, 112–116. doi:10.1016/j.virusres.2016.04.021
Gaudry, A., Bos, S., Viranaicken, W., Roche, M., Krejbich-Trotot, P., Gadea, G., et al. (2018). The Flavonoid Isoquercitrin Precludes Initiation of Zika Virus Infection in Human Cells. Int. J. Mol. Sci. 19 (4), 1093. doi:10.3390/ijms19041093
Ghahremanpour, M. M., Tirado-Rives, J., Deshmukh, M., Ippolito, J. A., Zhang, C.-H., Cabeza de Vaca, I., et al. (2020). Identification of 14 Known Drugs as Inhibitors of the Main Protease of SARS-CoV-2. ACS Med. Chem. Lett. 11 (12), 2526–2533. doi:10.1021/acsmedchemlett.0c00521
Gopi, K., Anbarasu, K., Renu, K., Jayanthi, S., Vishwanath, B. S., and Jayaraman, G. (2016). Quercetin-3-O-rhamnoside from Euphorbia Hirta Protects Against Snake Venom Induced Toxicity. Biochim. Biophys. Acta (Bba) - Gen. Subjects. 1860 (7), 1528–1540. doi:10.1016/j.bbagen.2016.03.031
Guo, Y.-J., Luo, T., Wu, F., Liu, H., Li, H.-R., Mei, Y.-W., et al. (2015). Corilagin Protects Against HSV1 Encephalitis through Inhibiting the TLR2 Signaling Pathways In Vivo and In Vitro. Mol. Neurobiol. 52 (3), 1547–1560. doi:10.1007/s12035-014-8947-7
Guzman, G., Dacanay, A., Andaya, B., and Alejandro, G. (2016). Ethnopharmacological Studies on the Uses of Euphorbia Hirta in the Treatment of Dengue in Selected Indigenous Communities in Pangasinan (Philippines). J. Intercult Ethnopharmacol. 5 (3), 239–243. doi:10.5455/jice.20160330124637
Hakmi, M., Bouricha, E. M., Kandoussi, I., Harti, J. E., and Ibrahimi, A. (2020). Repurposing of Known Anti-Virals as Potential Inhibitors for SARS-CoV-2 Main Protease Using Molecular Docking Analysis. Bioinformation. 16 (4), 301. doi:10.6026/97320630016301
Ho, W.-S., Xue, J.-Y., Sun, S. S. M., Ooi, V. E. C., and Li, Y.-L. (2010). Antiviral Activity of Daphnoretin Isolated from Wikstroemia Indica. Phytother. Res. 24 (5), 657–661. doi:10.1002/ptr.2935
Hu, K., Kobayashi, H., Dong, A., Iwasaki, S., and Yao, X. (2000). Antifungal, Antimitotic and Anti-HIV-1 Agents from the Roots of Wikstroemia Indica. Planta Med. 66 (6), 564–567. doi:10.1055/s-2000-8601
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020). Structure of Mpro from SARS-CoV-2 and Discovery of its Inhibitors. Nature. 582 (7811), 289–293. doi:10.1038/s41586-020-2223-y
Karki, S., Shrestha, K., Gautam, R., and Narayan, R. (2019). Phytochemical Screening, FT-IR and GC-MS Analysis of Euphorbia Hirta. J. Pharmacognosy Phytochemistry. 9 (1), 1883–1889. Retrieved from: https://www.researchgate.net/publication/349124683_Phytochemical_screening_FT-IR_and_GC-MS_analysis_of_Euphorbia_hirta
Khan, A., Ali, S. S., Khan, M. T., Saleem, S., Ali, A., Suleman, M., et al. (2020). Combined Drug Repurposing and Virtual Screening Strategies with Molecular Dynamics Simulation Identified Potent Inhibitors for SARS-CoV-2 Main Protease (3CLpro). J. Biomol. Struct. Dyn. 39 (0), 4659–4670. doi:10.1080/07391102.2020.1779128
Kim, C. H., Kim, J.-E., and Song, Y.-J. (2020). Antiviral Activities of Quercetin and Isoquercitrin Against Human Herpesviruses. Molecules. 25 (10), 2379. doi:10.3390/molecules25102379
Li, E.-T., Liu, K.-H., Zang, M.-H., Zhang, X.-L., Jiang, H.-Q., Zhou, H.-L., et al. (2015). Chemical Constituents from Euphorbia Hirta. Biochem. Syst. Ecol. 62, 204–207. doi:10.1016/j.bse.2015.09.007
Lin, Y.-J., Chang, Y.-C., Hsiao, N.-W., Hsieh, J.-L., Wang, C.-Y., Kung, S.-H., et al. (2012). Fisetin and Rutin as 3C Protease Inhibitors of Enterovirus A71. J. Virol. Methods. 182 (1), 93–98. doi:10.1016/j.jviromet.2012.03.020
Linfang, H., Shilin, C., and Meihua, Y. (2012). Euphorbia Hirta (Feiyangcao): A Review on its Ethnopharmacology, Phytochemistry and Pharmacology. J. Med. Plants Res. 6 (39), 5176–5185. doi:10.5897/JMPR12.206
Liu, Y., Murakami, N., Ji, H., Abreu, P., and Zhang, S. (2007). Antimalarial Flavonol Glycosides fromEuphorbia Hirta. Pharm. Biol. 45 (4), 278–281. doi:10.1080/13880200701214748
Luci-Atienza, C. (2021a). 68 COVID-19 Patients Already Recruited to Participate in PH’s Tawa-Tawa Clinical Trial. Manila Bulletin. Available at: https://mb.com.ph/2021/07/02/68-covid-19-patients-already-recruited-to-participate-in-phs-tawa-tawa-clinical-trial/.
Luci-Atienza, C. (2021b). Recruitment of Volunteers for PH Clinical Trial for Tawa-Tawa as COVID-19 Treatment Still Ongoing—DOST. Manila Bulletin. Available at: https://mb.com.ph/2021/08/27/recruitment-of-volunteers-for-ph-clinical-trial-for-tawa-tawa-as-covid-19-treatment-still-ongoing-dost/.
Ma, C., Hu, Y., Townsend, J. A., Lagarias, P. I., Marty, M. T., Kolocouris, A., et al. (2020). Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors. ACS Pharmacol. Transl. Sci. 3 (6), 1265–1277. doi:10.1021/acsptsci.0c00130
Mahomoodally, M. F., Dall’Acqua, S., Sinan, K. I., Sut, S., Ferrarese, I., Etienne, O. K., et al. (2020). Phenolic Compounds Analysis of Three Euphorbia Species by LC-DAD-MSn and Their Biological Properties. J. Pharm. Biomed. Anal. 189, 113477. doi:10.1016/j.jpba.2020.113477
Mallavadhani, U. V., and Narasimhan, K. (2009). Two Novel Butanol Rhamnosides from an Indian Traditional Herb,Euphorbia Hirta. Nat. Product. Res. 23 (7), 644–651. doi:10.1080/14786410802214009
Martínez-Vázquez, M., Apan, T. O. R., Lazcano, M. E., and Bye, R. (1999). Anti-inflammatory Active Compounds from the N-Hexane Extract of Euphorbia Hirta. J. Mexican Chem. Soc. 43 (3,4), 103–105. Retrieved from: https://www.redalyc.org/pdf/475/47543410.pdf
Mekam, P. N., Martini, S., Nguefack, J., Tagliazucchi, D., and Stefani, E. (2019). Phenolic Compounds Profile of Water and Ethanol Extracts of Euphorbia Hirta L. Leaves Showing Antioxidant and Antifungal Properties. South Afr. J. Bot. 127, 319–332. doi:10.1016/j.sajb.2019.11.001
Menéndez, C. A., Byléhn, F., Perez-Lemus, G. R., Alvarado, W., and de Pablo, J. J. (2020). Molecular Characterization of Ebselen Binding Activity to SARS-CoV-2 Main Protease. Sci. Adv. 6 (37), eabd0345. doi:10.1126/sciadv.abd0345
Mirza, M. U., and Froeyen, M. (2020). Structural Elucidation of SARS-CoV-2 Vital Proteins: Computational Methods Reveal Potential Drug Candidates Against Main Protease, Nsp12 Polymerase and Nsp13 Helicase. J. Pharm. Anal. 10 (4), 320–328. doi:10.1016/j.jpha.2020.04.008
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., and Group, T. P. (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Plos Med. 6 (7), e1000097. doi:10.1371/journal.pmed.1000097
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 30 (16), 2785–2791. doi:10.1002/jcc.21256
Motlhatlego, K. E., Mehrbod, P., Fotouhi, F., Abdalla, M. A., Eloff, J. N., and McGaw, L. J. (2021). Anti-influenza A Virus Activity of Two Newtonia Species and the Isolated Compound Myricetin-3-O-Rhamnoside. BMC Complement. Med. Ther. 21 (1), 92. doi:10.1186/s12906-021-03250-0
Nakane, H., Fukushima, M., and Ono, K. (1990). Differential Inhibition of Reverse Transcriptase and Various DNA Polymerases by Digallic Acid and its Derivatives. J. Nat. Prod. 53 (5), 1234–1240. doi:10.1021/np50071a015
Nomoto, Y., Sugimoto, S., Matsunami, K., and Otsuka, H. (2013). Hirtionosides A-C, Gallates of Megastigmane Glucosides, 3-hydroxyoctanoic Acid Glucosides and a Phenylpropanoid Glucoside from the Whole Plants of Euphorbia Hirta. J. Nat. Med. 67 (2), 350–358. doi:10.1007/s11418-012-0692-5
Nugroho, A., Heryani, H., and Istikowati, W. T. (2020). Quantitative Determination of Quercitrin and Myricitrin in Three Different Parts of Euphorbia Hirta as Bioflavonoid Source for Functional Food. IOP Conf. Ser. Earth Environ. Sci. 443, 012042–012046. doi:10.1088/1755-1315/443/1/012042
O'Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., and Hutchison, G. R. (2011). Open Babel: An Open Chemical Toolbox. J. Cheminform 3 (1), 33. doi:10.1186/1758-2946-3-33
Ogunlesi, M., Okiei, W., Ofor, E., and Osibote, A. E. (2009). Analysis of the Essential Oil from the Dried Leaves of Euphorbia Hirta Linn (Euphorbiaceae), a Potential Medication for Asthma. Afr. J. Biotechnol. 8 (24), 7042–7050. doi:10.4314/AJB.V8I24.68792
Olaoluwa, O., Moronkola, D., Taiwo, O., and Iganboh, P. (2018). Volatile Oil Composition, Antioxidant and Antimicrobial Properties of Boerhavia Erecta L. And Euphorbia Hirta L. Trends Phytochem. Res. 2 (3), 171–178. Retrieved from: http://tpr.iau-shahrood.ac.ir/article_543327.html
Ortega, J. T., Suárez, A. I., Serrano, M. L., Baptista, J., Pujol, F. H., and Rangel, H. R. (2017). The Role of the Glycosyl Moiety of Myricetin Derivatives in Anti-HIV-1 Activity In Vitro. AIDS Res. Ther. 14 (1), 57. doi:10.1186/s12981-017-0183-6
Parvez, M. K., Al-Dosari, M. S., Arbab, A. H., Al-Rehaily, A. J., and Abdelwahid, M. A. S. (2020). Bioassay-Guided Isolation of Anti-Hepatitis B Virus Flavonoid Myricetin-3-O-Rhamnoside Along with Quercetin from Guiera Senegalensis Leaves. Saudi Pharm. JournalThe Official Publ. Saudi Pharm. Soc. 28 (5), 550–559. doi:10.1016/j.jsps.2020.03.006
Perera, S. D., Jayawardena, U. A., and Jayasinghe, C. D. (2018). Potential Use ofEuphorbia Hirtafor Dengue: A Systematic Review of Scientific Evidence. J. Trop. Med. 2018, 7. doi:10.1155/2018/2048530
Perumal, S., and Mahmud, R. (2013). Chemical Analysis, Inhibition of Biofilm Formation and Biofilm Eradication Potential of Euphorbia Hirta L. Against Clinical Isolates and Standard Strains. BMC Complement. Altern. Med. 13, 346. doi:10.1186/1472-6882-13-346
Perumal, S., Mahmud, R., and Mohamed, N. (2018). Combination of Epicatechin 3-Gallate from Euphorbia Hirta and Cefepime Promotes Potential Synergistic Eradication Action against Resistant Clinical Isolate of Pseudomonas aeruginosa. Evidence-Based Complement. Altern. Med. 2018, 1–7. doi:10.1155/2018/5713703
Perumal, S., Mahmud, R., and Ramanathan, S. (2015). Anti-Infective Potential of Caffeic Acid and Epicatechin 3-gallate Isolated from Methanol Extract ofEuphorbia hirta(L.) againstPseudomonas Aeruginosa. Nat. Product. Res. 29 (18), 1766–1769. doi:10.1080/14786419.2014.999242
Prachi, S., and Pradeep, T. (2014). 13α-methyl-27-norolean-14-en-3β-ol, a Triterpeneisolated from the Stem of Euphorbia Hirta (Linn) Possess an Anti-asthmatic Properties. Res. J. Chem. Sci. 4 (3), 21–26. Retrieved from: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.1082.2070andrep=rep1andtype=pdf
Ragasa, C. Y., and Cornelio, K. B. (2013). Triterpenes from Euphorbia Hirta and Their Cytotoxicity. Chin. J. Nat. Medicines. 11 (5), 528–533. doi:10.1016/S1875-5364(13)60096-5
Rao, C., Gupta, S., Azmi, L., and Mohapatra, P. (2017). Flavonoids from Whole Plant of Euphorbia Hirta and Their Evaluation against Experimentally Induced Gastroesophageal Reflux Disease in Rats. Phcog Mag. 13 (Suppl. 1), 127–S134. doi:10.4103/0973-1296.203987
Rautela, I., Joshi, P., Thapliyal, P., Pant, M., Dheer, P., Bisht, S., et al. (2020). Comparative GC-MS Analysis of Euphorbia Hirta and Euphorbia Milli for Therapeutic Potential Utilities. Plant Arch. 20 (2), 3515–3512. Retrieved from: http://plantarchives.org/20-2/3515-3522%20(6354).pdf
Reddy, B. U., Mullick, R., Kumar, A., Sharma, G., Bag, P., Roy, C. L., et al. (2018). A Natural Small Molecule Inhibitor Corilagin Blocks HCV Replication and Modulates Oxidative Stress to Reduce Liver Damage. Antiviral Res. 150, 47–59. doi:10.1016/j.antiviral.2017.12.004
Schwarz, S., Sauter, D., Wang, K., Zhang, R., Sun, B., Karioti, A., et al. (2014). Kaempferol Derivatives as Antiviral Drugs Against the 3a Channel Protein of Coronavirus. Planta Med. 80 (2–3), 177–182. doi:10.1055/s-0033-1360277
Selin-Rani, S., Senthil-Nathan, S., Thanigaivel, A., Vasantha-Srinivasan, P., Edwin, E.-S., Ponsankar, A., et al. (2016). Toxicity and Physiological Effect of Quercetin on Generalist Herbivore, Spodoptera Litura Fab. And a Non-Target Earthworm Eisenia fetida Savigny. Chemosphere. 165, 257–267. doi:10.1016/j.chemosphere.2016.08.136
Shah, A. P., Parmar, G. R., Sailor, G. U., and Seth, A. K. (2019). Antimalarial Phytochemicals Identification from Euphorbia Hirta against Plasmepsin Protease: An In Silico Approach. Folia Med. 61 (4), 584–593. doi:10.3897/folmed.61.e47965
Sharma, N., Samarakoon, K., Gyawali, R., Park, Y.-H., Lee, S.-J., Oh, S., et al. (2014). Evaluation of the Antioxidant, Anti-Inflammatory, and Anticancer Activities of Euphorbia Hirta Ethanolic Extract. Molecules. 19 (9), 14567–14581. doi:10.3390/molecules190914567
Sheliya, M. A., Rayhana, B., Ali, A., Pillai, K. K., Aeri, V., Sharma, M., et al. (2015). Inhibition of α-glucosidase by New Prenylated Flavonoids from euphorbia Hirta L. Herb. J. Ethnopharmacology. 176, 1–8. doi:10.1016/j.jep.2015.10.018
Shitrit, A., Zaidman, D., Kalid, O., Bloch, I., Doron, D., Yarnizky, T., et al. (2020). Conserved Interactions Required for Inhibition of the Main Protease of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Sci. Rep. 10 (1), 20808. doi:10.1038/s41598-020-77794-5
Suganthi, A., and Ravi, T. K. (2018). Estimation of Anti-Dengue Phytochemical Markers Gallic Acid, Rutin and Quercetin in Methanolic Extract of Euphorbia Hirta (L.) and Tawa-Tawa Capsule Formulation by Validated RP-HPLC Method. Chem. Methodologies. 3 (1), 43–54. doi:10.22034/chemm.2018.129381.1051
Tawa-tawa Clinical Trial on COVID-19 (2021). Philippine Health Research Registry. Available at: https://registry.healthresearch.ph/index.php/registry?view=research&layout=details&cid=3475.
Tayone, W. C., Ishida, K., Goto, S., Tayone, J. C., Arakawa, M., Morita, E., et al. (2020). Anti-Japanese Encephalitis Virus (JEV) Activity of Triterpenes and Flavonoids from Euphorbia Hirta. Philippine J. Sci. 149 (3), 603–613. Retrieved from: https://philjournalsci.dost.gov.ph/publication/regular-issues/past-issues/98-vol-149-no-3-september-2020/1211-anti-japanese-encephalitis-virus-jev-activity-of-triterpenes-and-flavonoids-from-euphorbia-hirta
Trott, O., and Olson, A. J. (2009). AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 31 (2), 455–461. doi:10.1002/jcc.21334
Ullrich, S., and Nitsche, C. (2020). The SARS-CoV-2 Main Protease as Drug Target. Bioorg. Med. Chem. Lett. 30 (17), 127377. doi:10.1016/j.bmcl.2020.127377
Wang, S., Ling, Y., Yao, Y., Zheng, G., and Chen, W. (2020). Luteolin Inhibits Respiratory Syncytial Virus Replication by Regulating the MiR-155/SOCS1/STAT1 Signaling Pathway. Virol. J. 17 (1), 187. doi:10.1186/s12985-020-01451-6
Wang, Y. C., Yang, W. H., Yang, C. S., Hou, M. H., Tsai, C. L., Chou, Y. Z., et al. (2020). Structural Basis of SARS-CoV-2 Main Protease Inhibition by a Broad-Spectrum Anti-coronaviral Drug. Am. J. Cancer Res. 10 (8), 2535–2545. Retrieved from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7471349/pdf/ajcr0010-2535.pdf
Wu, L.-l., Yang, X.-b., Huang, Z.-m., Liu, H.-z., and Wu, G.-x. (2007). In Vivo and In Vitro Antiviral Activity of Hyperoside Extracted from Abelmoschus Manihot (L) Medik. Acta Pharmacologica Sinica. 28 (3), 404–409. doi:10.1111/j.1745-7254.2007.00510.x
Wu, Y., Qu, W., Geng, D., Liang, J.-Y., and Luo, Y.-L. (2012). Phenols and Flavonoids from the Aerial Part of Euphorbia Hirta. Chin. J. Nat. Medicines. 10 (1), 40–42. doi:10.1016/S1875-5364(12)60009-0
Xu, H.-X., Wan, M., Dong, H., But, P. P.-H., and Foo, L. Y. (2000). Inhibitory Activity of Flavonoids and Tannins against HIV-1 Protease. Biol. Pharm. Bull. 23 (9), 1072–1076. doi:10.1248/bpb.23.1072
Yan, S., Ye, D., Wang, Y., Zhao, Y., Pu, J., Du, X., et al. (2011). Ent-Kaurane Diterpenoids from Euphorbia hirta. Rec. Nat. Prod. 5 (4), 247–251.
Yang, Z.-N., Su, B.-J., Wang, Y.-Q., Liao, H.-B., Chen, Z.-F., and Liang, D. (2020). Isolation, Absolute Configuration, and Biological Activities of Chebulic Acid and Brevifolincarboxylic Acid Derivatives from Euphorbia Hirta. J. Nat. Prod. 83 (4), 985–995. doi:10.1021/acs.jnatprod.9b00877
Zhang, L., Wang, X.-L., Wang, B., Zhang, L.-T., Gao, H.-M., Shen, T., et al. (2020). Lignans from Euphorbia Hirta L. Nat. Product. Res. 36, 26–36. doi:10.1080/14786419.2020.1761358
Keywords: Euphorbia hirta, COVID-19, molecular docking, phytochemical mining, medicinal plant, Philippine medicinal plant, SARS-CoV-2 Mpro, virtual screening
Citation: Cayona R and Creencia E (2022) Phytochemicals of Euphorbia hirta L. and Their Inhibitory Potential Against SARS-CoV-2 Main Protease. Front. Mol. Biosci. 8:801401. doi: 10.3389/fmolb.2021.801401
Received: 25 October 2021; Accepted: 30 December 2021;
Published: 04 February 2022.
Edited by:
Arvind Ramanathan, Argonne National Laboratory (DOE), United StatesReviewed by:
Debsindhu Bhowmik, Oak Ridge National Laboratory (DOE), United StatesLorane Hage-Melim, Universidade Federal do Amapá, Brazil
Copyright © 2022 Cayona and Creencia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruel Cayona, cnVlbC5jYXlvbmFAZy5tc3VpaXQuZWR1LnBo; Evelyn Creencia, ZXZlbHluLmNyZWVuY2lhQGcubXN1aWl0LmVkdS5waA==
 Ruel Cayona
Ruel Cayona Evelyn Creencia
Evelyn Creencia