
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 11 August 2016
Sec. Molecular Recognition
Volume 3 - 2016 | https://doi.org/10.3389/fmolb.2016.00039
This article is part of the Research Topic Modulating prokaryotic lifestyle by DNA-binding proteins View all 25 articles
The proper initiation and occurrence of DNA synthesis depends on the formation and rearrangements of nucleoprotein complexes within the origin of DNA replication. In this review article, we present the current knowledge on the molecular mechanism of replication complex assembly at the origin of bacterial chromosome and plasmid replicon containing direct repeats (iterons) within the origin sequence. We describe recent findings on chromosomal and plasmid replication initiators, DnaA and Rep proteins, respectively, and their sequence-specific interactions with double- and single-stranded DNA. Also, we discuss the current understanding of the activities of DnaA and Rep proteins required for replisome assembly that is fundamental to the duplication and stability of genetic information in bacterial cells.
The replication of genetic material is one of the most fundamental processes that influence the proper functioning of each living cell. The synthesis of new DNA molecule, in case of both bacterial chromosomes and plasmids, starts at a well-defined place called origin and can be divided into the following steps: (1) origin recognition by replication initiation proteins and open complex formation (2) helicase loading, activation and primer synthesis (3) replisome assembly and DNA synthesis. Although these main steps during the DNA replication process are common, when considering replication of bacterial chromosomes and iteron plasmids replicated by theta mechanism, some differences can be observed (Table 1).
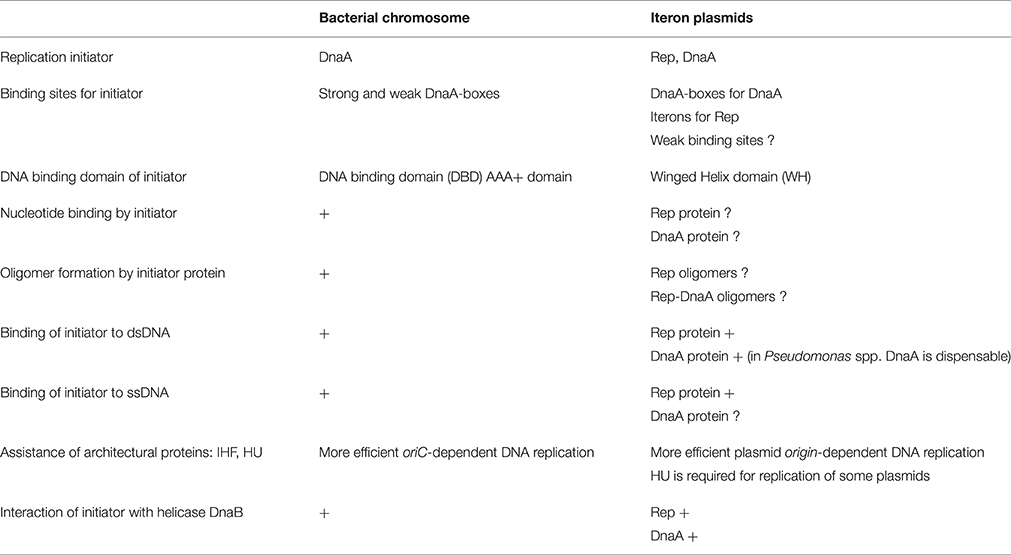
Table 1. Comparison of general features of iteron plasmid and chromosomal DNA replication initiation.
A DNA replication process of chromosome and plasmid DNA starts when Origin Binding Proteins (OBP) recognize and bind specific motifs located within origin region. Despite the differences in structure of bacterial and plasmid initiators, DnaA and Rep proteins, respectively, they have the same function. Binding of initiators results in a modulation of nearby DNA topology and opening of double-stranded helix structure in DNA unwinding element (DUE). A single-stranded DUE region becomes a place where helicase is loaded. In the next step the replisome is assembled and holoenzyme of DNA Polymerase III can play its role during DNA synthesis.
Despite many years of research on DNA replication, new aspects of this process are still being discovered. Recently, the novel activities of replication initiator proteins have been shown. However, especially in case of plasmid DNA replication, there are many questions concerning the replication initiation and replisome assembly that still need to be answered.
The very first step of replication initiation process is the recognition of specific motifs located within the origin region of DNA molecule (Figure 1) by replication initiation proteins (Figure 2, Stage I). The bacterial chromosome replication initiator DnaA protein consists of four domains, which play distinct roles (Sutton and Kaguni, 1997, Figure 3A). The best characterized DnaA is the Escherichia coli protein (EcDnaA), although structural data is limited only to domain I (resolved by NMR-analysis; Abe et al., 2007b) and IV (resolved in a nucleoprotein complex by crystallography; Fujikawa et al., 2003). Information concerning the structure of DnaA initiator is supplemented by structure of domains I and II of Mycoplasma genitalium DnaA (MgDnaA; Lowery et al., 2007), domains I and II of Helicobacter pylori DnaA (HpDnaA) in a complex with HobA protein (Natrajan et al., 2009), domains III and IV of Aquifex aeolicus DnaA (AaDnaA; Erzberger et al., 2002, 2006), domain III of Thermatoga maritima DnaA (TmDnaA; Ozaki et al., 2008), and domain IV of Mycobacterium tuberculosis (MtDnaA; Tsodikov and Biswas, 2011). Domain I of EcDnaA, located at the N-terminus of the protein, was shown to be involved in oligomerization of DnaA (Weigel et al., 1999; Simmons et al., 2003; Abe et al., 2007a), helicase loading (Sutton et al., 1998; Seitz et al., 2000), and interaction with DiaA (Keyamura et al., 2007), HU (Chodavarapu et al., 2008a), Dps (Chodavarapu et al., 2008b), and ribosomal protein L2 (Chodavarapu et al., 2011). The interaction with DiaA homologe, HobA protein, was shown for domains I and II of HpDnaA (Natrajan et al., 2007, 2009; Zawilak-Pawlik et al., 2007). In Bacillus subtilis, domain I of DnaA (BsDnaA) interacts with SirA, the sporulation-related protein (Rahn-Lee et al., 2011). However, the binding partner proteins can vary among DnaA orthologs, and replication initiator from one bacterium can interact with different partners compared to other orthologs, e.g., interaction of Thermoanerobacter tengcongensis DnaA with NusG protein, is not observed for BsDnaA (Liu et al., 2008). The second domain, forming a flexible linker, although it is not essential (Messer et al., 1999; Nozaki and Ogawa, 2008), was proposed to be involved in optimal helicase DnaB recruitment (Molt et al., 2009). The domain II, links domain I with domain III, which contains a common core structure of AAA+ proteins family members (Neuwald et al., 1999). Recent data showed that residues within this domain are engaged in interaction of DnaA (TmDnaA, EcDnaA, AaDnaA) with single-stranded DNA (ssDNA; Ozaki et al., 2008; Duderstadt et al., 2011). At the C-terminus of DnaA, domain IV (DNA Binding Domain, DBD) can be distinguished, which is responsible, via a helix-turn-helix motif (HTH), for interaction with double-stranded DNA (dsDNA) containing specific motifs named DnaA-boxes (Roth and Messer, 1995; Fujikawa et al., 2003). Interaction with these sequences is the very first step of the replication initiation process.
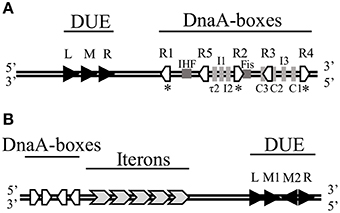
Figure 1. The minimal origins of DNA replication of (A) E. coli chromosome and (B) RK2 plasmid. (A) The genetic organization of E. coli oriC comprises 13-mers within the DNA Unwinding Element (DUE) and DnaA-boxes as well as binding sites of IHF and Fis proteins. Asterisks (*) below the oriC indicate strong DnaA-boxes. (B) The genetic organization of RK2 plasmid oriV consisting of DnaA-boxes, Iterons, and DUE. Black arrows mark 13-mers.
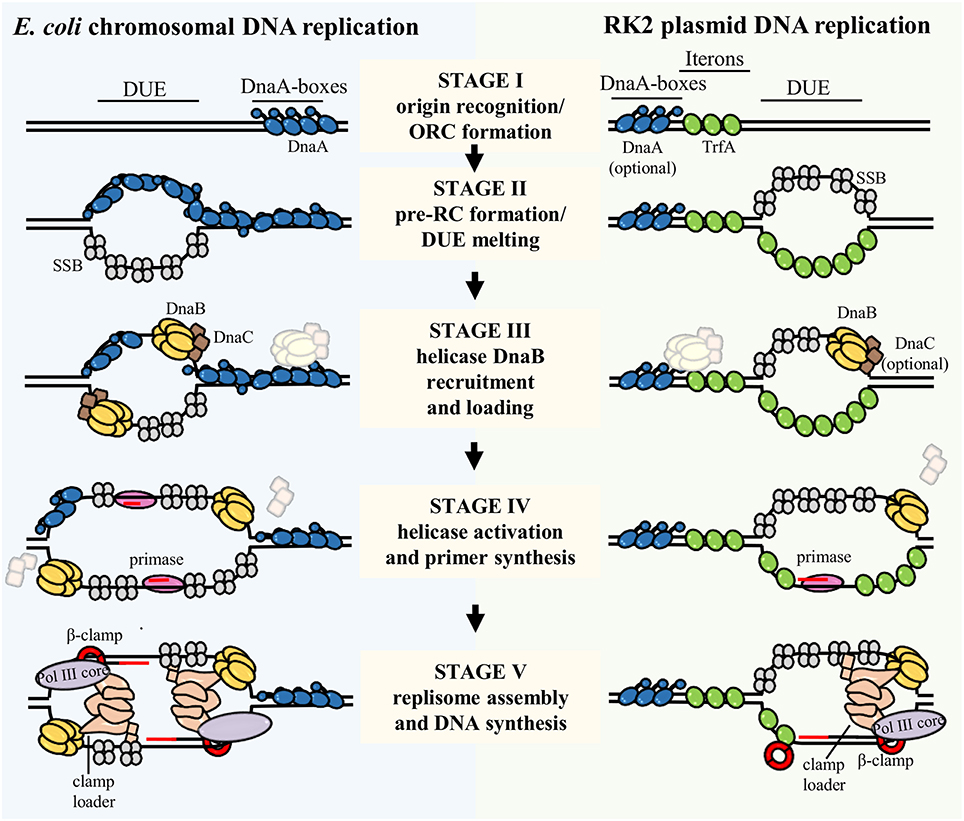
Figure 2. The process of bacterial chromosome and plasmid DNA replication initiation and replisome assembly. The scheme presents replication initiation and replisome assembly at chromosomal E. coli origin, oriC (left), and RK2 plasmid origin, oriV (right). The DNA replication initiation starts with binding a replication initiator(s) DnaA and TrfA to the DnaA boxes and Iterons, respectively (Stage I). Origin Recognition Complex (ORC) formation induces local destabilization and pre-Replication Complex (pre-RC) formation and melting of the DNA Unwinding Element (DUE) region (Stage II). Then, assisted by replication initiators and the DnaC helicase loader, the DnaB helicase is recruited and loaded onto the single-stranded DUE (Stage III). In case of plasmid DNA replication the requirement for DnaA and DnaC is optional as it depends on the host organism. Association of DnaG primase triggers the release of helicase loader, helicase activation and primers synthesis (Stage IV). Next, the holoenzyme of DNA Polymerase III, which comprises clamp loader, DNA Polymerase III core (Pol III core), and β-clamp is assembled and conducts DNA synthesis (Stage V). Lagging strand synthesis was omitted for simplicity. Proteins involved in described stages of DNA replication initiation and replisome assembly processes are depicted in the scheme. IHF and Fis were omitted in this scheme.
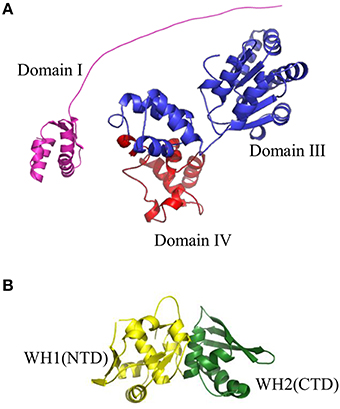
Figure 3. Structures of replication initiation proteins: (A) bacterial DnaA protein and (B) RepE protein from plasmid F. (A) Crystal structure of Domain I (shown in violet) of EcDnaA protein was obtained from the PDB database (2E0G). Domain III (shown in blue) and Domain IV (shown in red) were modeled using SWISS-MODEL server (http://swissmodel.expasy.org) basing on crystal structure of Domain IV obtained from PDB database (1J1V). The presented structure of EcDnaA does not include Domain II. (B) Crystal structure of RepE protein, comprising Winged Helix domain 1 (WH1), and Winged Helix domain 2 (WH2) (shown in yellow and green, respectively), were obtained from the PDB database (1REP).
In bacterial chromosome origin, regions that are composed of a variable number of DnaA-boxes, can be identified (Ozaki and Katayama, 2009; Rajewska et al., 2012; Wolanski et al., 2014; Leonard and Grimwade, 2015). In the origin of E. coli chromosome (oriC), five 9-bp in length DnaA-boxes (R1–R5) were originally identified (Fuller et al., 1984; Matsui et al., 1985); in contrast, the origin of Caulobacter crescentus chromosome (Cori) possesses only two DnaA-boxes (named G-boxes; Shaheen et al., 2009). The studies with the use of in vivo and in vitro dimethylsulphate (DMS) footprinting as well as DNase I footprinting method showed that other, non–R DnaA binding sites are present in oriC, i.e., I (Grimwade et al., 2000; McGarry et al., 2004), C (Rozgaja et al., 2011), and τ sites (Kawakami et al., 2005). Such non-canonical sequences recognized by bacterial initiator were also found in oriC of C. crescentus (termed W-boxes; Taylor et al., 2011). The affinity of DnaA binding to R-boxes and non-R DnaA binding sites is different. Interestingly, binding of inititor to the DnaA-boxes in Cori of C. crescentus, both G-boxes and W-boxes, is lower compared to DnaA binding to the R-boxes in oriC of E. coli (Taylor et al., 2011), which might be characteristic for bacteria with a complex regulation of development. The DnaA binding sites, bound by initiator with affinity comparable only to interaction between DnaA and weak DnaA-boxes in E. coli oriC, were found in the origin of H. pylori (Zawilak-Pawlik et al., 2007; Charbon and Løbner-Olesen, 2011). In E. coli oriC three (named R1, R2, and R4) out of five DnaA-boxes are the widely separated, high affinity DnaA-boxes. They were found to be almost constantly bound by EcDnaA protein (Samitt et al., 1989; Nievera et al., 2006). The occupancy of only these three sites is insufficient for spontaneous origin opening and it was proposed that interaction of EcDnaA protein at high affinity binding sites may regulate conformation of the origin DNA (Kaur et al., 2014). Between the peripheral R1 and R4 sites, there are two arrays of low affinity binding sites, τ1 R5 τ2 I1 I2 and C3 C2 I3 C1, separated by one of high affinity—R2 (Rozgaja et al., 2011). EcDnaA molecules bound to the high affinity DnaA-boxes, termed bacterial Origin Recognition Complex (bORC), act as anchors and are required to assist in occupying weak sites by the EcDnaA protomers (Rozgaja et al., 2011; Kaur et al., 2014), and formation of replication-active pre-replication complex (pre-RC; Figure 2, Stage II). The binding affinity to particular sequences and replication activity of EcDnaA protein depend on nucleotide-bound state of protein. Although ADP-EcDnaA binds the high affinity DnaA-boxes and also R5 and C1 low affinity ones, the ATP-EcDnaA form is thought to be the replication-active one (Sekimizu et al., 1987; Leonard and Grimwade, 2011). ATP-EcDnaA form of initiator binds efficiently both high and low affinity binding sites (McGarry et al., 2004; Kawakami et al., 2005). Based on molecular docking, binding of ATP, instead of ADP, is presumed to cause changes in the EcDnaA protein conformation, thus leading to the formation of large oligomeric complex within the origin region (Saxena et al., 2015). The crystallographic data, when nonhydrolyzable ATP analog AMP-PCP was used, showed the formation of open-ended, right-handed helical filament of AaDnaA (Erzberger et al., 2006). Based on biochemical and genetic approaches it was found that there is an interaction between domain III (AAA + domain) of one DnaA (EcDnaA or AaDnaA) molecule and domain IV (DBD domain) of partner subunit (Duderstadt et al., 2010). It was proposed that during pORC and pre-RC complexes formation of the DBD domain is extended and the HTH motif is exposed, which results in the efficient binding of high and low affinity binding sites (Duderstadt et al., 2010). Occupation of the EcDnaA binding sites was shown to be sequential and polarized and DnaA protomers are released preferentially from the peripheral high affinity R1 and R4 boxes, through arrays of low affinity binding sites to the middle high affinity one—R2 (Rozgaja et al., 2011). The formation of DnaA oligomer within the oriC results in DNA destabilization in the DUE region (Speck and Messer, 2001; McGarry et al., 2004; Leonard and Grimwade, 2005, 2011; Duderstadt et al., 2010). Although two arrays of low affinity binding sites separated by high affinity sequences are occupied by EcDnaA protomers for efficient double-stranded DNA opening, binding of EcDnaA to a part of origin (containing only R1 high affinity box and τ1 R5 τ2 I1 I2 low affinity binding sites array) was shown to be active in DUE unwinding (Ozaki and Katayama, 2012). It was proposed that distinct DnaA multimers are formed on the left half (containing binding sites from R1 to I2) and the right half (containing binding sites from R2 to R4) of oriC (Ozaki and Katayama, 2012; Ozaki et al., 2012a).
The DUE melting is the consequence of DnaA binding to arrays of DnaA-boxes (Figures 1A, 2, Stage II). The location of particular binding sites suggests that DnaA, bound to sequences of the high affinity DnaA-boxes (R1, R2, R4), could cause the bending of DNA molecule via interaction through domain I of already bound three protomers (Rozgaja et al., 2011; Kaur et al., 2014; Leonard and Grimwade, 2015). The model of constrained loop formed by EcDnaA bound to the high affinity binding sites was proposed (Kaur et al., 2014). The bending of oriC containing DNA molecule is supported by accessory histone-like proteins HU and integration host factor (IHF). A binding site for IHF was found within the oriC region (Polaczek, 1990) and it was shown that IHF can enhance the unwinding of DNA by DnaA (Hwang and Kornberg, 1992; Ryan et al., 2002). It was demonstrated that HU has the same effect on DUE destabilization (Hwang and Kornberg, 1992), although its mechanism of action is different (Ryan et al., 2002). Data obtained with ELISA (Enzyme Linked Immunosorbent Assay) showed that HU interacts with domain I of EcDnaA, which was proposed as an interaction which stabilizes the DnaA oligomer (Chodavarapu et al., 2008a). The Fis protein, identified originally as factor for inversion stimulation in site-specific DNA recombination, was also shown to have an influence on DNA unwinding (Wold et al., 1996). Specific binding sites for Fis were identified in oriC (Gille et al., 1991). Although Fis, in contrast to IHF, negatively regulates DNA replication initiation, when the origin lacks some DnaA binding sites resulting in altered non-functional conformation of origin, both Fis and IHF can work together to correct these alterations (Kaur et al., 2014). This joint action is achieved by inducing bends in oriC and establishing functional origin conformation (Kaur et al., 2014).
The formation of DnaA oligomer with synergistic action of architectural proteins can introduce torsional strain into DUE, facilitating the melting of the double-stranded DNA helix. The binding of DnaA to DUE region was also thought to introduce DNA melting, and ATP-DnaA-boxes were distinguished within the oriC DUE sequence (Speck and Messer, 2001). Recent studies showed direct binding of EcDnaA and AaDnaA protein to formed single-stranded DNA within the DUE (Ozaki et al., 2008; Duderstadt et al., 2010, 2011; Cheng et al., 2015). Studies with DnaA mutants (Ozaki et al., 2008; Duderstadt et al., 2010), as well as crystallography (Duderstadt et al., 2011), showed that this interaction occurs through residues located within the AAA+ domain III of bacterial initiator. The AaDnaA protomers form a helical filament on ssDNA (Duderstadt et al., 2011), however, it differs from the filament formed on the dsDNA (Erzberger et al., 2006; Duderstadt et al., 2010). It was proposed that protomers in this oligomer are more compact when compared to the extended DnaA molecules in dsDNA-DnaA complex (Duderstadt et al., 2010, 2011). The binding of ssDNA concerns just one T-rich strand of DUE and depends on sequence of 13-nucleotide sequences, which can be distinguished within the DUE. In oriC three 13-mers are present (Bramhill and Kornberg, 1988a) and the binding of EcDnaA occurs at least two 13-mers. EcDnaA does not form a complex with ssDNA containing just one 13-mer (Ozaki et al., 2008). Formation of this nucleoprotein complex is achieved only by ATP-DnaA protein (Ozaki et al., 2008) and one AaDnaA protomer binds three nucleotides of ssDNA (Duderstadt et al., 2011; Cheng et al., 2015). Studies with the use of single-molecule fluorescence assays showed that the formation of this nucleoprotein complex is highly dynamic and that AaDnaA molecules assemble on ssDNA in the 3′ to 5′ direction (Cheng et al., 2015). The presence of dsDNA region containing DnaA-boxes, adjacent to ssDNA DUE, stabilizes the DnaA (EcDnaA and AaDnaA) filament on ssDNA (Ozaki and Katayama, 2012; Cheng et al., 2015). Recently published data revealed presence of a new origin element, termed DnaA-trio, composed of repeated trinucleotide motif that stabilizes DnaA filaments on the ssDNA (Richardson et al., 2016). What is important, binding single strand of DUE region is required for origin activity (Ozaki et al., 2008, 2012a,b; Duderstadt et al., 2011).
Similarly as during bacterial chromosome replication, the first step in open complex formation in many theta-replicating plasmids, especially in iteron-containing plasmids, is the binding of plasmid replication initiator, Rep protein, to specific sequences within origin region (Figure 2, Stage I). Rep proteins are structurally different from bacterial DnaA protein and consist of winged-helix (WH) domains (Figure 3B, Komori et al., 1999; Díaz-López et al., 2003; Sharma et al., 2004; Swan et al., 2006; Nakamura et al., 2007a,b; Pierechod et al., 2009). The crystal structures of nucleoprotein complexes of π protein from plasmid R6K (Swan et al., 2006), RepE protein from plasmid F (Komori et al., 1999; Nakamura et al., 2007b), and a DNA binding domain of Rep protein from ColE2–P9 plasmid (Itou et al., 2015) as well as N-terminal domain of RepA protein from plasmid pPS10 (Giraldo et al., 2003) were obtained. Furthermore, homological models for plasmid Rep proteins: RepA from P1 (Sharma et al., 2004), RepA from pSC101 (Sharma et al., 2004), and TrfA from RK2 (Pierechod et al., 2009) were shown. Plasmid Reps are composed of two WH domains, of which one is responsible for oligomerization and the role of a second one is the protein's interaction with DNA (Giraldo et al., 1998; Nakamura et al., 2004; Pierechod et al., 2009). Plasmid replication initiators are present as dimers in solution, however, an exception is known i.e., RepE protein from pAMβ1 plasmid is present as a monomer (Le Chatelier et al., 2001). Although the Rep dimers interact with DNA (Filutowicz et al., 1985; Ingmer et al., 1995; Komori et al., 1999), they are replication-active in the monomeric form (Kawasaki et al., 1990; Wickner et al., 1992; Sozhamannan and Chattoraj, 1993; Konieczny and Helinski, 1997). Conformational activation of plasmid replication initiators is carried out by chaperon proteins (Kawasaki et al., 1990; Wickner et al., 1992, 1994; Sozhamannan and Chattoraj, 1993; Konieczny and Helinski, 1997). In contrast to bacterial replication initiator DnaA, the domain responsible for binding of nucleotide was not identified in Reps' structures. There is also no evidence showing if Rep proteins can form helical filaments on DNA similar to that formed by the AaDnaA protein. For some Reps, e.g., TrfA protein from RK2 plasmid, two forms of protein, different in length, occur: the shorter 33 kDa (TrfA-33) and longer 44 kDa (TrfA-44). There are different requirements for each particular form depending on the host bacterium. In E. coli both forms of TrfA can initiate the plasmid replication, whereas in Pseudomonas aeruginosa only the longer form is active (Caspi et al., 2001; Jiang et al., 2003; Konieczny, 2003; Yano et al., 2013, 2016).
During plasmid replication initiation, monomers of Reps bind to specific repeated sequences, named iterons, present within origin region (Figures 1B, 2, Stage I). The number of iterons varies among plasmid origins, from two iterons in plasmids ColE2 and ColE3 (Yasueda et al., 1989), three iterons in pSC101 (Churchward et al., 1984), and some plasmids from IncQ incompatibility group (Loftie-Eaton and Rawlings, 2012), four iterons in origin of plasmid F and pPS10, up to five (origin of plasmids RK2 and P1) or even seven such sequences in oriƔ of plasmid R6K (Rajewska et al., 2012). Iterons are short sequences, in length ranging from 17-bp in RK2 plasmid (Stalker et al., 1981), 19-bp in plasmids F (Murotsu et al., 1981), and P1 (Abeles et al., 1984), to 22-bp in R6K (Filutowicz et al., 1987), and pPS10 (Nieto et al., 1992). But in some plasmids the iteron sequences which are present in one origin can differ in length and apart from short sequences, significantly longer iterons [up to even 76-bp in plasmid R478 from IncHI2 incompatibility group (Page et al., 2001)] can be present. The binding of Rep proteins to iterons is sequence-specific and mutations in these motifs disrupt binding of plasmid initiation protein. Changes in a sequence of iterons abolished binding of π protein within the oriƔ of plasmid R6K and thus replication activity in vivo (McEachern et al., 1985). Negative effects on replication was also observed for mutants in a sequence of P1 plasmid iterons (Brendler et al., 1997). The sequences separating particular iterons are also important for Rep nucleoprotein complexes formation and proper replication activity of origin. It was shown in case of the RK2 plasmid that in vitro the TrfA protein has a high preference for binding to DNA containing at least two out of five binding sites, when compared to the formation of nucleoprotein complex with DNA containing just one iteron (Perri et al., 1991). The requirement for the presence of more than just one iteron sequence for TrfA binding was also shown in vivo (Perri and Helinski, 1993). Rep proteins bind to iterons in a cooperative manner (Perri and Helinski, 1993; Xia et al., 1993; Bowers et al., 2007) and the cooperativity of binding depends on the spatial location of iterons, since separation of two iterons by a half helical turn abolished cooperativity (Bowers et al., 2007). These results suggest the possibility of formation of higher order nucleoprotein structure on plasmid iterons bound by Reps. It was shown that WH domains of Reps contact three nucleotides in DNA. In π protein from R6K plasmid, WH1 domain contacts wGwnCnT motif, and WH2 domian contacts GAG sequence (Swan et al., 2006). Similarly, the WH2 domain of RepE monomer also contacts three nucleotides of top (GTG sequence) and three nucleotide of bottom strand (GtCA sequence) of double-stranded molecule containing iteron sequence (Nakamura et al., 2007b). However, unlike for the bacterial DnaA protein, to date there are no evidence showing that strong and weak binding sites for Reps are present within plasmid origins. There were just predictions of potential binding sites, other than iterons, for π protein in R6K plasmid and suggestions on potential role of such sites (Rakowski and Filutowicz, 2013). Certainly like DnaA, Rep proteins can bind within single-stranded region of melted DUE, and this binding is sequence-specific, since binding concerns a particular strand. Nucleoprotein complexes formation with the ssDNA DUE was detected for TrfA (bound with A-rich strand) and RepE (bound with T-rich strand) proteins (Wegrzyn et al., 2014). Within the DUE of plasmid origins, repeated sequence, similar to 13-mers distinguishable in oriC, can be found. There are four 13-nucleotide sequences in plasmid RK2 DUE region (Doran et al., 1998) and all of them are required for TrfA-ssDNA DUE complex formation. Lack of even one 13-mer hinders plasmid replication (Wegrzyn et al., 2014). Also, even a point mutation within this region affects plasmid replication since the lack of DUE melting was observed for some of the changed sequences (Kowalczyk et al., 2005; Rajewska et al., 2008).
The Rep protein encoded by plasmids, can be accompanied by host DnaA initiator during open complex formation and DUE melting within a plasmid origin (Figure 2, Stage II). DnaA binding sites have been found in replication origin of many plasmids including plasmids P1 (Abeles et al., 1984, 1990; Abeles, 1986), F (Kline et al., 1986; Murakami et al., 1987; Kawasaki et al., 1996), RK2 (Doran et al., 1998; Caspi et al., 2000), pSC101 (Sutton and Kaguni, 1995). The number of DnaA-box sequences differs among plasmid origins, the position and orientation of these binding sites are as important as position and orientation of the iterons (Doran et al., 1998, 1999). The inversion of one out of four DnaA boxes in origin of RK2 plasmid abolished plasmid DNA replication, despite the fact that three remaining DnaA boxes were bound by the host initiator (Doran et al., 1999). Although the DnaA protein is not required for replication initiation for some plasmids, e.g., R1, binding of DnaA increased the plasmid replication efficiency (Bernander et al., 1991, 1992) and mutations within a binding site for DnaA decreased the R1 plasmid replication (Ortega-Jiménez et al., 1992). In bacteria, ATP-DnaA form is essential for chromosomal DNA replication (Sekimizu et al., 1987; Leonard and Grimwade, 2005, 2011). Interestingly, studies with ATP-binding mutant of DnaA, which was inactive in oriC replication, showed that bacterial initiator lacking an ability to bind a nucleotide was effective in open complex formation within plasmid R6K oriƔ (Lu et al., 1998). Also in the presence of ATPS, a non-hydrolyzable analog of ATP, the pattern of bands in KMnO4 footprinting assay with DnaA and TrfA proteins and plasmid RK2 DNA showed no significant differences, when compared to opening reaction containing ATP (Konieczny et al., 1997). Thus, the DnaA is suspected to play a different role in plasmid replication initiation, compared to its role in chromosome replication. A direct interaction between plasmid and host replication initiators was shown (Lu et al., 1998; Maestro et al., 2003) and the interaction was detected in the N-terminus of π (between 1 and 116 aa) protein of R6K plasmid (Lu et al., 1998) and RepA protein of pSC101 (Sharma et al., 2001) and domain I and IV of host initiator (Sharma et al., 2001). The mutations in RepA protein from pPS10 plasmid were introduced, which enhanced the interaction of RepA with DnaA protein and resulted in changes in host range of pPS10 plasmid (Maestro et al., 2003).
Similarly to bacterial chromosome replication initiation, the binding of DnaA protein to DnaA-boxes within plasmid origins can be enhanced by the presence of architectural proteins IHF, and HU (Shah et al., 1995; Fekete et al., 2006). The binding of IHF to its binding site in oriƔ region significantly enhanced binding of bacterial DnaA to R6K plasmid origin (Lu et al., 1998). In pSC101 plasmid binding IHF to its cognate binding site is required for plasmid replication initiation and mutations within this sequence disrupts plasmid replication (Stenzel et al., 1987). For plasmid P1 the binding of IHF to its site, located downstream of one out of two arrays of DnaA-boxes (the second array is located upstream of DUE) is required only when the nearby DUE array of DnaA-boxes is not active and the second DnaA-boxes array serves as a secondary origin compensating the function of the first one (Fekete et al., 2006). The P1-mini derivative was just slightly unstable in IHF E. coli mutant (Ogura et al., 1990). The mutations in gene for IHF protein did not affect plasmids F (Ogura et al., 1990) and RK2 (Shah et al., 1995) replication. In contrast, the lack of HU protein in vitro results in significant decrease in mini-F plasmid DNA synthesis (Zzaman et al., 2004) and in vivo KMnO4 reactivity of P1 plasmid origin (Park et al., 1998) as well as abolishment of plasmid F replication in vivo (Ogura et al., 1990). During plasmid RK2 replication initiation, HU could functionally replace DnaA protein, although it could not enhance DUE melting as efficiently as DnaA (Konieczny et al., 1997). It was proposed that one of the DnaA functions could be the stabilization of origin melting induced by Rep protein. The other DnaA role during replication initiation is its function in helicase loading. Interestingly, for some plasmids, e.g., RK2, DnaA assists Rep during plasmid replication initiation only in particular hosts, while in others DnaA is dispensable [DnaA P. aeruginosa is dispensable for RK2 plasmid replication initiation, but required in E. coli (Caspi et al., 2001; Konieczny, 2003)].
In bacteria the loading of DnaB helicase onto ssDNA of DUE is achieved by the action of replication initiation protein, DnaA, as well as the helicase loading factor, DnaC protein (Figure 2, Stage III). DnaB helicase is a two-tiered ring-shaped hexamer (Bailey et al., 2007b; Wang et al., 2008; Lo et al., 2009). Each monomer consists of N-terminal and C-terminal domain connected via linker helix (LH) region (Miron et al., 1992; Ingmer and Cohen, 1993; Komori et al., 1999). The N-terminal domain of helicase's monomers were shown to interact with ssDNA (observed in a crystal structure of Geobacillus kaustophilus helicase in a complex with ssDNA; Lo et al., 2009) which stabilizes the hexameric structure of DnaB (Biswas et al., 1994). The C-terminal domain, that contains RecA-like fold, is responsible for ATP binding and hydrolysis, interaction with DNA (Bailey et al., 2007a), and binding of DnaC loader factor (Lu et al., 1996). The helicase is positioned onto the ssDNA DUE in a single orientation with respect to the polarity of the sugar-phosphate backbone of DNA and the nucleic acid, bound primarily to one DnaB monomer (Jezewska et al., 1998a,b), passes through the cross-channel of helicase hexamer (Jezewska et al., 1998a). The hexamer of DnaB, when no ATP hydrolysis occurs, is bound to 20 (±3) nucleotides (Jezewska et al., 1996).
The binding of nucleotide as well as particular partner protein and DNA promotes helicase to adopt specific conformation. The X-ray crystal structure of A. aeolicus helicase revealed large conformational rearrangements, observed in N-terminal domain and the presence of at least two highly-distinct conformations: widened with broad central channel and a highly-constricted with a narrow pore (Strycharska et al., 2013). These conformations were also observed for E. coli DnaB, when analyzed in solution with the use of small-angle X-ray scattering (SAXS; Strycharska et al., 2013). Structural analysis with the use of negative-stain electron microscopy (EM) and SAXS of DnaB protein in complex with its loader, DnaC, showed that the hexamer of helicase interacts with helical arrangement of six DnaC monomers (Kobori and Kornberg, 1982; Arias-Palomo et al., 2013). However, it was argued that the active form of the DnaB-DnaC complex exists in 6:3 stoichiometry, which was studied by quantitative analysis of pre-priming complex (Makowska-Grzyska and Kaguni, 2010). Furthermore, the imbalance in level of DnaB and DnaC was shown to impair DNA replication (Allen and Kornberg, 1991; Brüning et al., 2016).
The concept of DnaC as a protein that loads DnaB helicase onto ssDNA of DUE, has been early established (Wickner and Hurwitz, 1975; Funnell et al., 1987; Bell and Kaguni, 2013). To further explain its exact function, the following models have been proposed: (1) DnaC breaks the helicase ring (Davey and O'Donnell, 2003; Arias-Palomo et al., 2013), (2) DnaC traps DnaB helicase as an open ring (Chodavarapu et al., 2016). Those hypotheses were tested by the SAXS method and deuterium exchange coupled to mass spectrometry, respectively. The ATPase activity of DnaC, a member of AAA+ proteins family, is not required for helicase hexamer opening and its loading by DnaC, hence the DnaB-binding domain of loader is sufficient for this process (Arias-Palomo et al., 2013). Yet the ATP hydrolysis by DnaC was proposed to occur during DnaB helicase activation, which results in DNA unwinding (Felczak et al., 2016).
Regarding the DnaC key contribution to helicase loading and activation in E. coli, it is particularly interesting to discuss replicons that are independent of helicase loader. The helicase loaders were identified only in few species and it is possible that in some bacteria the yet unidentified helicase loaders are present. The lack of DnaC orthologs can also arise from ability of self-loading by helicase (Costa et al., 2013) or it is possible that another protein of already assigned role, substitutes the DnaC function. Those hypotheses can be supported by complementation of dnaC temperature-sensitive mutant of E. coli by helicase from H. pylori (Soni et al., 2003). The dispensability for helicase loader was also shown during RK2 plasmid replication in Pseudomonas species (Jiang et al., 2003). In Pseudomonas sp. the helicase loading at plasmid RK2 origin is performed by the longer form of plasmid Rep protein, TrfA-44, which interacts with Pseudomonas helicase (Caspi et al., 2001; Jiang et al., 2003; Zhong et al., 2003). The shorter form of this plasmid initiator, TrfA-33, is not sufficient for helicase loading in P. aeruginosa. In Pseudomonas putida TrfA-33 can load helicase but only in the presence of DnaA (Caspi et al., 2001; Jiang et al., 2003). On the contrary, the DnaC helicase loader, together with DnaA, and Rep protein (either short or long form), is absolutely required for helicase loading at plasmid RK2 origin in E. coli (Caspi et al., 2001). It was shown that via interaction of DnaA with DnaBC, the helicase is first localized in DnaA-boxes and then via DnaA-DnaB and Rep-DnaB interactions translocated to ssDNA DUE (Pacek et al., 2001; Rajewska et al., 2008). Probably the Rep-DnaA interaction is also important in these processes. Apart from the proper protein-protein interaction, an efficiency of helicase translocation from DnaA-box position to DUE depends on the sequence of DUE region. It was shown via electron microscopy and in vitro experiments that even point mutations within the DUE of RK2 plasmid origin results in a decrease in helicase translocation and thus helicase DNA unwinding activity (Rajewska et al., 2008).
It was proposed that, upon DnaB-DnaC binding to ssDNA, DnaC dissociates, thus allowing DnaB to unwind double helix, and further to bind DnaG primase (Wahle et al., 1989, Figure 2, Stage IV). However, Makowska-Grzyska and Kaguni demonstrated, by performing molecular filtration of pre-priming complex at E. coli oriC, that the DnaG primase binds DnaB, synthesizes primer and in consequence, induces the release of DnaC from DnaB (Makowska-Grzyska and Kaguni, 2010). In E. coli, in further steps DnaG primase is associated with DnaB helicase and synthesizes primers on lagging strand (McHenry, 2011). Plasmid ColE2-P9 does not require DnaG primase in replication initiation (Takechi et al., 1995). Itoh group demonstrated that ColE2 origin and Rep protein as well as E. coli host DNA Polymerase I and SSB are sufficient for in vitro DNA synthesis (Itoh and Horii, 1989). Further studies revealed that the ColE2-Rep protein has joined functions, i.e., replication initiator and plasmid-specific primase (Takechi and Itoh, 1995).
Once activated, DnaB unwinds one nucleotide per one catalytic step in ATP-dependent manner (Lohman and Bjornson, 1996, Figure 2, Stage IV). It was shown that at 25°C the DnaB unwinds around 291 bp per second (Galletto et al., 2004) and it moves from 5′ to 3′ direction along the ssDNA (LeBowitz and McMacken, 1986). Because the replication of bacterial chromosome is bidirectional two helicases are loaded: one is loaded by DnaC on the top strand invaded by DnaA molecules and the other on the bottom strand. It was proposed that the helicase delivery to ssDNA DUE bottom A-rich strand occurs by direct interaction between DnaB and DnaA proteins (Mott et al., 2008; Soultanas, 2012). The Phe-46 of DnaA was shown to be important for this interaction (Keyamura et al., 2009). The order of helicase loading to a particular strand of DUE is not random but defined; first helicase is loaded onto the bottom/lower strand then the second onto the top/upper one (Weigel and Seitz, 2002). Such order of helicase loading probably supplies head-to-head orientation of unwound region of oriC and prevents back-to-back loading of the helicase. The basal level of DnaB activity in oriC is achieved when DnaA forms an oligomer in ssDNA DUE and dsDNA containing DnaA-boxes from R1 to I2 (called DAR-DF and DAR-LL). For the full activity of helicase the formation of DnaA filament on other DnaA-boxes (from R2 to R4; called DAR-RL and DAR-RE) is needed (Ozaki and Katayama, 2012).
The interaction between plasmid initiator Rep and helicase is an important factor for helicase activity on plasmid origin (Figure 2, Stage IV). It was shown for E. coli F plasmid that its initiator, RepE protein, cannot form a stable complex with Pseudomonas helicase and thus it does not replicate efficiently in Pseudomonas cells (Zhong et al., 2005). Interaction between plasmid Rep and host DnaB was also detected via ELISA and protein affinity chromatography for π protein of R6K (Ratnakar et al., 1996) and mutations within π were identified which decreased helicase binding and resulted in impaired plasmid DNA replication (Swan et al., 2006). A similar effect was observed for mutants of RepA protein form plasmid pSC101, invalid in the interaction with helicase (Datta et al., 1999). Although the Rep-DnaB interaction is required for helicase loading, the right balance in the strength of the interaction must be maintained. It was shown that too tight binding of Rep to DnaB is undesirable and the mutations within Rep, acquired by adaptation under antibiotic selection that decreased binding to helicase, result in the decrease in fitness cost and increase in plasmid copy number (Yano et al., 2016).
Once DnaB helicase is loaded, DNA is unwound and primer is synthesized, the contribution of replication initiators becomes more enigmatic. Subsequent stages of DNA replication require building the replisome, i.e., the multiprotein replication machinery that synthesizes DNA (O'Donnell et al., 2013, Figure 2, Stage V). The replisome in bacteria is composed of DnaB helicase, DnaG primase, single-stranded DNA binding protein (SSB), and the holoenzyme of DNA Polymerase III (hPol III) (divided in three subcomplexes: Pol III core, clamp loader and β-clamp; O'Donnell, 2006). Reyes-Lamothe et al., suggested that both DnaA replication initiator and DnaC helicase loader are crucial for replisome assembly in E. coli (Reyes-Lamothe et al., 2008). This conclusion was drawn from studies that tracked the replisome components in living cells during the stages of DNA replication. However, it does not exclude the possibility that the role of replication initiator is limited to DUE destabilization and helicase loading, hence, indirect effects may be observed. Most studies regarding the mechanism of replisome assembly are performed using simplified experimental setup, e.g., primed ssDNA and replisome components, where replication initiators are omitted (Yuzhakov et al., 1996, 1999; Downey and McHenry, 2010; Cho et al., 2014).
Following the primer synthesis, clamp loader complex loads the ring-shaped β-clamp (discussed below in details), that encircles dsDNA, tethers DNA polymerase, and slides along dsDNA, thus significantly increasing speed (up to 100-fold), and processivity (up to 5000-fold) of DNA replication (Kelch et al., 2012). The E. coli clamp loader is composed of γ, τ, δ, δ', χ, and ψ subunits, albeit only γ, δ, δ' are crucial for β-clamp loading (reviewed in details in Kelch, 2016). The γ subunit is a truncated version of τ subunit, encoded by dnaX gene, and arises from translational frameshift (Flower and McHenry, 1990). Both γ and τ subunits have AAA+ domain, however, γ subunit lacks τ subunit domain responsible for DnaB helicase and Pol III core binding (Tsuchihashi and Kornberg, 1989; O'Donnell and Studwell, 1990; Flowers et al., 2003). Before clamp loader binds DNA, it adopts appropriate, ATP-driven conformational state that increases its affinity for the β-clamp (Podobnik et al., 2003). It is under debate whether the ring structure of β-clamp is actively opened or captured in open conformation. The T4 bacteriophage trimeric clamp is the least stable sliding clamp and it was found to dissociate from DNA by monomerization, thus no force in opening of the ring is required (Soumillion et al., 1998). The dimeric clamps (bacterial, e.g., E. coli) are regarded as stable, hence more active ring-opening mechanism is expected to be in demand. On the basis of a crystal structure of single subunit of E. coli clamp loader (namely δ subunit) in complex with β-clamp, it was proposed that δ subunit is a molecular wrench, that induces rearrangements of β-clamp at dimerization interface, albeit without ATP hydrolysis (Jeruzalmi et al., 2001). With the use of real-time fluorescence-based clamp binding and opening assays, it was shown that clamp loader binds closed β-clamp in solution, prior to β-clamp opening (Paschall et al., 2011). Yet, the deuterium exchange coupled to Mass Spectrometry experiments revealed that most sliding clamps are dynamic at their monomers' interfaces (Fang et al., 2011, 2014). Therefore, it is also probable that β-clamp is trapped in an open conformation by clamp loader.
The crystal structure of clamp loader complex was solved from T4 bacteriophage (Kelch et al., 2011), E. coli (Simonetta et al., 2009), and its eukaryotic homolog, Replication Factor C (RFC), from Saccharomyces cerevisiae (Bowman et al., 2004). Each of the clamp loader complex reveals pentameric structure. Since AAA + ATPases usually adopt circular hexamers, it was proposed that sixth subunit was lost during the evolution (Indiani and O'Donnell, 2006). Indeed, the gap between the first and fifth clamp loader subunits is favorable, because it provides the mechanism of specific accommodation of the primer-template junction structure (Kelch, 2016). It was suggested that clamp loader recognizes minor groove and thus it binds at the 3′ primer-template junction specifically. However, the crystal structure of the clamp loader:DNA complex revealed that clamp loader contacts template DNA exclusively (Bowman et al., 2005; Simonetta et al., 2009). Despite the fact that the DNA synthesis may be initiated only from 3′ OH primer end, the clamp loader can assemble in vitro at either 3′ or 5′ primer terminus forming a stable complex (Park and O'Donnell, 2009). While clamp loader binds only DNA template, β-clamp interacts with both RNA primer, and DNA template within the RNA-DNA hybrid and it was shown that the β-clamp distinguishes between the 5′ and 3′ primer end (Park and O'Donnell, 2009). Consistently, it was demonstrated that SSB hampers clamp loading on the 5′end of primer (Hayner et al., 2014). The ATPase activity of the clamp loader is lower when it is assembled at the 5′ terminus, comparing to the ATPase activity of clamp loader located at 3′ terminus (Park and O'Donnell, 2009). ATP hydrolysis triggers β-clamp closing on DNA and the release of clamp loader from β-clamp:DNA nucleoprotein complex (Pietroni and von Hippel, 2008). Thereby, the 3′ primed end loading preference also arises from the higher rate of clamp closure and clamp loader dissociation (Park and O'Donnell, 2009). The β-clamp must be closed in the ATP hydrolysis-dependent manner, to release clamp loader (Hayner et al., 2014). Clamp loader must free the β-clamp to allow the Pol III core to bind, since they accommodate the same binding site within the β-clamp, namely the hydrophobic cleft.
β-clamp crystal structures were obtained from various organisms i.e., E. coli (Oakley et al., 2003; Burnouf et al., 2004), P. aeruginosa (Wolff et al., 2014), Streptococcus pyogenes (Argiriadi et al., 2006), M. tuberculosis (Gui et al., 2011; Kukshal et al., 2012; Wolff et al., 2014), B. subtillis (Wolff et al., 2014), T. maritima (structure 1VPK), Eubacterium rectale (structure 3T0P), Streptococcus pneumoniae (Argiriadi et al., 2006). The crystal structures of β-clamp homologs—Proliferating Cell Nuclear Antigen (PCNA)—are also available from Eukaryotes and Archea (to name a few: Homo sapiens (Punchihewa et al., 2012), S. cerevisiae (Krishna et al., 1994), Sulfolobus solfaraticus (Williams et al., 2006). All of them adopt ring shaped homodimer (e.g., E. coli) or homotrimer (human PCNA, Pyrococcus furiosus PCNA), albeit the exception is PCNA of Archea, S. solfataricus, which exists as a heterotrimer (Dionne et al., 2003). β-clamp monomers bind in a head to tail manner (Kelman and O'Donnell, 1995). The β-clamp and PCNA structure is conserved among all kingdoms of life, in contrast to amino acid sequence (Jeruzalmi et al., 2001). However, the amino acid sequence of region termed hydrophobic cleft was found to be highly conserved (Jeruzalmi et al., 2001). The hydrophobic cleft is a site for interaction with β-clamp binding partners (Jeruzalmi et al., 2001). β-clamp forms a protein interaction hub and serves as a platform for multiple protein interactions crucial in various cellular processes, i.e., DNA elongation in every living organism (Hedglin et al., 2013), regulation of DNA replication in E. coli, B. subtilis, C. crescentus (Katayama et al., 2010), DNA repair in E. coli (Rangarajan et al., 1999), toxin-mediated replication fork collapse in C. crescentus (Aakre et al., 2013). All described β-clamp interaction partner proteins share similar motif, the Clamp Binding Motif (CBM; Dalrymple et al., 2001).
Interestingly, clamp binding motif was also identified in plasmid replication initiators, including RK2 plasmid initiator-TrfA (Kongsuwan et al., 2006; Dalrymple et al., 2007). It was shown that TrfA protein lacking the leucine 137 and phenyloalanine 138 within the clamp binding motif is unable to bind β-clamp (Kongsuwan et al., 2006). The TrfA ΔLF mutant facilitated the determination of biological relevance of this interaction. The complex of TrfA and β-clamp was found to be the key feature for replisome assembly and thereby for oriV-dependent DNA replication of both supercoiled dsDNA plasmid and ssDNA plasmid in vitro, albeit the clamp loader complex is still crucial (Wawrzycka et al., 2015). Hence, the question arises—how do the Rep and clamp loader cooperate to load the β-clamp at plasmid origin? Three hypothetical models to explain the mechanism of Rep-mediated β-clamp loading could be considered (Figure 4). In the “β-clamp hand-off model” TrfA binds to the bottom strand of ssDNA close to the 3′ end of synthesized primer, recruits β-clamp, and hands it off to the clamp loader complex (Figure 4A). Then, the δ subunit of clamp loader opens the β-clamp and clamp loader positions it onto primer-template junction, as it is thought to occur during replisome assembly at E. coli oriC. This model is consistent with the results of the in vitro DNA replication experiments performed with the use of ssDNA, containing sequence of RK2 plasmid oriV (Wawrzycka et al., 2015). It was demonstrated that TrfA interacts with specific strand of ssDNA of DUE, i.e., the bottom strand, which serves as the site for replisome assembly (Wegrzyn et al., 2014; Wawrzycka et al., 2015). It can be further speculated that TrfA may assist the clamp loader in recognition of the 3′ end of primer-template junction within the oriV. Another possible role of TrfA is illustrated in Figure 4B (second model, “β-clamp:clamp loader recruitment model”). Once TrfA is bound to bottom single strand of DUE, it recruits the β-clamp, which is in complex with clamp loader, to the RK2 plasmid origin. Thus, the local concentration of β-clamp:clamp loader complex increases, the clamp loader can assemble β-clamp onto the 3′ end of a primer within the plasmid origin. Because TrfA-β-clamp interaction was shown in the absence of DNA [using both ELISA and SPR (Surface Plasmon Resonance) technique (Kongsuwan et al., 2006; Wawrzycka et al., 2015)], the third model may also be justified (Figure 4C, “β-clamp directed to oriV model”). In the third model the TrfA that is not bound to DNA forms complex with β-clamp associated with the clamp loader, then directs it to the plasmid origin, oriV. Next, the clamp loader:β-clamp:TrfA complex binds to the bottom strand of DUE via TrfA. TrfA passes the β-clamp bound to clamp loader on the primer-template junction. Although ATP binding to clamp loader (namely γ and τ subunit) is required for β-clamp opening, it cannot be excluded that TrfA—whose ATPase activity has not been revealed—substitutes the clamp loader's function at this stage. TrfA may capture β-clamp in open conformation and load it onto primed DNA. Since ATP hydrolysis is required for β-clamp closing (Trakselis et al., 2001), here may participate the clamp loader.
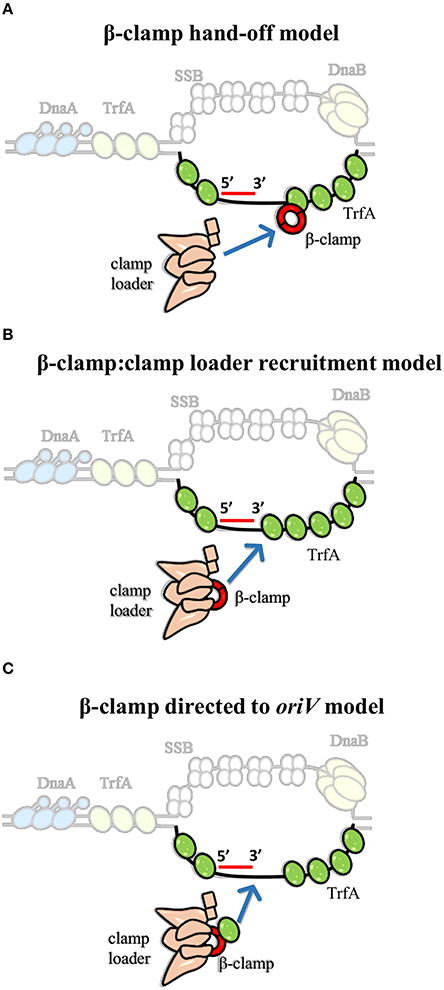
Figure 4. Models for the contribution of TrfA to β-clamp loading at RK2 plasmid origin (oriV). We propose three model mechanisms for β-clamp loading at oriV through cooperated action of TrfA and clamp loader. (A) The first model suggests that ssDNA-bound TrfA recruits and hands off the β-clamp to clamp loader. (B) The second model implies that ssDNA-bound TrfA recruits the β-clamp in complex with clamp loader, thereby increasing the local concentration of β-clamp:clamp loader complex at oriV. (C) The third model indicates that TrfA binds β-clamp that is in complex with the clamp loader and directs it to the ssDNA of oriV.
After the β-clamp closes around primer-template junction and clamp loader dissociates, the final replisome component arrives—the Pol III core, that is composed of three subunits: α (DNA polymerase), ε (3′–5′ proofreading exonuclease) and θ (ε subunit stabilizer; Kelman and O'Donnell, 1995; Taft-Benz and Schaaper, 2004). The number of Pol III cores within the replisome strictly depends on the clamp loader composition, since Pol III core is connected only through τ subunit to the clamp loader. Various clamp loader complexes were widely studied in the light of processivity of DNA replication and it was established that three ATPases (τ or γ subunit) must be included with δ and δ' subunits to form active pentameric structure (Kelch, 2016). Initially, it was thought that clamp loader contains two τ subunits (τ2γδδ'χψ), so that two Pol III cores could constitute the replisome and synthesize the leading strand and lagging strand at the same time (Maki et al., 1988). However, further reports have argued on the stoichiometry of hPol III subunits (McInerney et al., 2007; Reyes-Lamothe et al., 2010; Dohrmann et al., 2016). The millisecond single molecule fluorescence microscopy as well as in vitro biochemical experiments showed that active E. coli replisome contains three molecules of polymerase that are functional at replication fork (McInerney et al., 2007; Reyes-Lamothe et al., 2010). Both these studies assumed that trimeric polymerase is associated with three molecules of τ subunit. However, the very recent data indicated that in a bacterial cell there is predominately present Pol III2τ2γδδ'χψ complex (Dohrmann et al., 2016). Since plasmids do not encode all essential proteins required for a plasmid replication, it is implied that the stage of DNA synthesis is similar during chromosomal DNA replication.
The DNA synthesis is facilitated by SSB, especially on the lagging strand (where the DNA synthesis is performed discontinuously) and is present in organisms from all domains of life (Shereda et al., 2008). Primary function of SSB is to protect ssDNA against degradation and melting secondary structures (Mackay and Linn, 1976; Meyer et al., 1979). SSB is linked to the clamp loader via χ subunit (Glover and McHenry, 1998), which was shown to be important for DnaG primase displacement (Yuzhakov et al., 1999). Yet, SSB was termed the organizer of genome maintenance complexes and was shown to interact with at least 14 proteins, thus implying its diverse functions (reviewed in details in Shereda et al., 2008). The SSB interactions with proteins involves the C-terminal region of SSB, that is highly conserved among eubacterial SSB proteins. Some plasmids also encode SSB-like proteins, i.e., plasmid F, ColIb-P9, and RK2 (Chase et al., 1983; Howland et al., 1989; Thomas and Sherlock, 1990). While SSB of plasmid F and ColIb-P9 have similar structural domains, the RK2 SSB, termed P116, is smaller and contains only the N-terminal domain, which is responsible for DNA binding. P116 lacks the C-terminal protein binding-tail (Curth et al., 1996; Naue et al., 2013; Su et al., 2014), which may suggest that the role of P116 limits to ssDNA protection against nucleases.
The ground-breaking model of DNA replication initiation, introduced by Bramhill and Kornberg is still valid today (Bramhill and Kornberg, 1988b). They proposed that first the DnaA binds to DnaA-boxes to form an initial complex, then DnaA melts the AT-rich region (DUE) to form an open complex. Finally, DnaA directs the DnaB:DnaC complex into the open complex, thus forming a pre-priming complex, which marks the future forks of DNA replication (Bramhill and Kornberg, 1988a,b). In this concept the chromosomal replication initiator, DnaA triggers the DNA replication initiation and is further required at each stage of the replication initiation process. Iteron plasmids also encode replication initiators that drive their replication initiation machinery. Despite the fact that plasmid and chromosomal replicons use overlapping set of proteins, there seems to be some subtle differences that may largely affect the whole process. Recent reports describe novel functions of replication initiators, both plasmid and chromosomal, that outreach the replication initiation process. The contribution of plasmid Rep protein to replisome assembly by providing direct Rep-β-clamp interaction, shed a new light on how far-reaching activities replication initiators have i.e., determination of direction of DNA replication (Wawrzycka et al., 2015). DnaA is also involved in a regulation of DNA replication initiation by a process termed RIDA (Regulatory Inactivation of DnaA; Katayama et al., 2010). One may ask if there is any other unanticipated activity of replication initiators to be discovered? What other processes are influenced by replication initiators? Described model mechanisms and unsolved questions of the structure-function relation of replication initiators in DNA replication and beyond this process await to be experimentally challenged.
KW and MG wrote the manuscript and prepared figures, UU prepared the model of E. coli DnaA and figures, IK discussed and corrected the text of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Magdalena Rajewska for critical reading of the manuscript. This work was supported by the Polish National Science Centre (Grant2012/04/A/NZ1/ 00048) and Polish Ministry of Science and Higher Education (DS/530-M040-D094-16). UU was supported by the European Commission from the FP7 Project Centre of Molecular Biotechnology for Healthy Life (MOBI4Health).
Aakre, C. D., Phung, T. N., Huang, D., and Laub, M. T. (2013). A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell 52, 617–628. doi: 10.1016/j.molcel.2013.10.014
Abe, Y., Jo, T., Matsuda, Y., Matsunaga, C., Katayama, T., and Ueda, T. (2007a). Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282, 17816–17827. doi: 10.1074/jbc.M701841200
Abe, Y., Watanabe, N., Yoshida, Y., Ebata, F., Katayama, T., and Ueda, T. (2007b). Assignment of 1H, 13C and 15N resonances of N-terminal domain of DnaA protein. Biomol. NMR Assign. 1, 57–59. doi: 10.1007/s12104-007-9015-2
Abeles, A. L. (1986). P1 plasmid replication. Purification and DNA-binding activity of the replication protein RepA. J. Biol. Chem. 261, 3548–3555.
Abeles, A. L., Reaves, L. D., and Austin, S. J. (1990). A single DnaA box is sufficient for initiation from the P1 plasmid origin. J. Bacteriol. 172, 4386–4391.
Abeles, A. L., Snyder, K. M., and Chattoraj, D. K. (1984). P1 plasmid replication: replicon structure. J. Mol. Biol. 173, 307–324. doi: 10.1016/0022-2836(84)90123-2
Allen, G. C. Jr., and Kornberg, A. (1991). Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J. Biol. Chem. 266, 22096–22101.
Argiriadi, M. A., Goedken, E. R., Bruck, I., O'Donnell, M., and Kuriyan, J. (2006). Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium. BMC Struct. Biol. 6:2. doi: 10.1186/1472-6807-6-2
Arias-Palomo, E., O'Shea, V. L., Hood, I. V., and Berger, J. M. (2013). The bacterial DnaC helicase loader is a DnaB ring breaker. Cell 153, 438–448. doi: 10.1016/j.cell.2013.03.006
Bailey, S., Eliason, W. K., and Steitz, T. A. (2007a). The crystal structure of the Thermus aquaticus DnaB helicase monomer. Nucleic Acids Res. 35, 4728–4736. doi: 10.1093/nar/gkm507
Bailey, S., Eliason, W. K., and Steitz, T. A. (2007b). Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318, 459–463. doi: 10.1126/science.1147353
Bell, S. P., and Kaguni, J. M. (2013). Helicase loading at chromosomal origins of replication. Cold Spring Harb Perspect. Biol. 5:a010124. doi: 10.1101/cshperspect.a010124
Bernander, R., Dasgupta, S., and Nordström, K. (1991). The E. coli cell cycle and the plasmid R1 replication cycle in the absence of the DnaA protein. Cell 64, 1145–1153. doi: 10.1016/0092-8674(91)90269-5
Bernander, R., Krabbe, M., and Nordström, K. (1992). Mapping of the in vivo start site for leading strand DNA synthesis in plasmid R1. EMBO J. 11, 4481–4487.
Biswas, S. B., Chen, P. H., and Biswas, E. E. (1994). Structure and function of Escherichia coli DnaB protein: role of the N-terminal domain in helicase activity. Biochemistry 33, 11307–11314. doi: 10.1021/bi00203a028
Bowers, L. M., Krüger, R., and Filutowicz, M. (2007). Mechanism of origin activation by monomers of R6K-encoded pi protein. J. Mol. Biol. 368, 928–938. doi: 10.1016/j.jmb.2007.02.074
Bowman, G. D., Goedken, E. R., Kazmirski, S. L., O'Donnell, M., and Kuriyan, J. (2005). DNA polymerase clamp loaders and DNA recognition. FEBS Lett. 579, 863–867. doi: 10.1016/j.febslet.2004.11.038
Bowman, G. D., O'Donnell, M., and Kuriyan, J. (2004). Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730. doi: 10.1038/nature02585
Bramhill, D., and Kornberg, A. (1988a). Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52, 743–755. doi: 10.1016/0092-8674(88)90412-6
Bramhill, D., and Kornberg, A. (1988b). A model for initiation at origins of DNA replication. Cell 54, 915–918. doi: 10.1016/0092-8674(88)90102-X
Brendler, T. G., Abeles, A. L., Reaves, L. D., and Austin, S. J. (1997). The iteron bases and spacers of the P1 replication origin contain information that specifies the formation of a complex structure involved in initiation. Mol. Microbiol. 23, 559–567. doi: 10.1046/j.1365-2958.1997.d01-1869.x
Brüning, J. G., Myka, K. K., and McGlynn, P. (2016). Overexpression of the replicative helicase in Escherichia coli inhibits replication initiation and replication fork reloading. J. Mol. Biol. 428, 1068–1079. doi: 10.1016/j.jmb.2016.01.018
Burnouf, D. Y., Olieric, V., Wagner, J., Fujii, S., Reinbolt, J., Fuchs, R. P., et al. (2004). Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J. Mol. Biol. 335, 1187–1197. doi: 10.1016/j.jmb.2003.11.049
Caspi, R., Helinski, D. R., Pacek, M., and Konieczny, I. (2000). Interactions of DnaA proteins from distantly related bacteria with the replication origin of the broad host range plasmid RK2. J. Biol. Chem. 275, 18454–18461. doi: 10.1074/jbc.M000552200
Caspi, R., Pacek, M., Consiglieri, G., Helinski, D. R., Toukdarian, A., and Konieczny, I. (2001). A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO J. 20, 3262–3271. doi: 10.1093/emboj/20.12.3262
Charbon, G., and Løbner-Olesen, A. (2011). A role for the weak DnaA binding sites in bacterial replication origins. Mol. Microbiol. 82, 272–274. doi: 10.1111/j.1365-2958.2011.07840.x
Chase, J. W., Merrill, B. M., and Williams, K. R. (1983). F sex factor encodes a single-stranded DNA binding protein (SSB) with extensive sequence homology to Escherichia coli SSB. Proc. Natl. Acad. Sci. U.S.A. 80, 5480–5484. doi: 10.1073/pnas.80.18.5480
Cheng, H. M., Gröger, P., Hartmann, A., and Schlierf, M. (2015). Bacterial initiators form dynamic filaments on single-stranded DNA monomer by monomer. Nucleic Acids Res. 43, 396–405. doi: 10.1093/nar/gku1284
Cho, W. K., Jergic, S., Kim, D., Dixon, N. E., and Lee, J. B. (2014). Loading dynamics of a sliding DNA clamp. Angew. Chem. Int. Ed Engl. 53, 6768–6771. doi: 10.1002/anie.201403063
Chodavarapu, S., Felczak, M. M., and Kaguni, J. M. (2011). Two forms of ribosomal protein L2 of Escherichia coli that inhibit DnaA in DNA replication. Nucleic Acids Res. 39, 4180–4191. doi: 10.1093/nar/gkq1203
Chodavarapu, S., Felczak, M. M., Yaniv, J. R., and Kaguni, J. M. (2008a). Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 67, 781–792. doi: 10.1111/j.1365-2958.2007.06094.x
Chodavarapu, S., Gomez, R., Vicente, M., and Kaguni, J. M. (2008b). Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol. Microbiol. 67, 1331–1346. doi: 10.1111/j.1365-2958.2008.06127.x
Chodavarapu, S., Jones, A. D., Feig, M., and Kaguni, J. M. (2016). DnaC traps DnaB as an open ring and remodels the domain that binds primase. Nucleic Acids Res. 44, 210–220. doi: 10.1093/nar/gkv961
Churchward, G., Linder, P., and Caro, L. (1984). Replication functions encoded by the plasmid pSC101. Adv. Exp. Med. Biol. 179, 209–214. doi: 10.1007/978-1-4684-8730-5_21
Costa, A., Hood, I. V., and Berger, J. M. (2013). Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 82, 25–54. doi: 10.1146/annurev-biochem-052610-094414
Curth, U., Genschel, J., Urbanke, C., and Greipel, J. (1996). In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 24, 2706–2711. doi: 10.1093/nar/24.14.2706
Dalrymple, B. P., Kongsuwan, K., and Wijffels, G. (2007). Identification of putative DnaN-binding motifs in plasmid replication initiation proteins. Plasmid 57, 82–88. doi: 10.1016/j.plasmid.2006.07.005
Dalrymple, B. P., Kongsuwan, K., Wijffels, G., Dixon, N. E., and Jennings, P. A. (2001). A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. U.S.A. 98, 11627–11632. doi: 10.1073/pnas.191384398
Datta, H. J., Khatri, G. S., and Bastia, D. (1999). Mechanism of recruitment of DnaB helicase to the replication origin of the plasmid pSC101. Proc. Natl. Acad. Sci. U.S.A. 96, 73–78. doi: 10.1073/pnas.96.1.73
Davey, M. J., and O'Donnell, M. (2003). Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13, R594–R596. doi: 10.1016/S0960-9822(03)00523-2
Díaz-López, T., Lages-Gonzalo, M., Serrano-López, A., Alfonso, C., Rivas, G., Diaz-Orejas, R., et al. (2003). Structural changes in RepA, a plasmid replication initiator, upon binding to origin DNA. J. Biol. Chem. 278, 18606–18616. doi: 10.1074/jbc.M212024200
Dionne, I., Nookala, R. K., Jackson, S. P., Doherty, A. J., and Bell, S. D. (2003). A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Cell 11, 275–282. doi: 10.1016/S1097-2765(02)00824-9
Dohrmann, P. R., Correa, R., Frisch, R. L., Rosenberg, S. M., and McHenry, C. S. (2016). The DNA polymerase III holoenzyme contains gamma and is not a trimeric polymerase. Nucleic Acids Res. 44, 1285–1297. doi: 10.1093/nar/gkv1510
Doran, K. S., Helinski, D. R., and Konieczny, I. (1999). A critical DnaA box directs the cooperative binding of the Escherichia coli DnaA protein to the plasmid RK2 replication origin. J. Biol. Chem. 274, 17918–17923. doi: 10.1074/jbc.274.25.17918
Doran, K. S., Konieczny, I., and Helinski, D. R. (1998). Replication origin of the broad host range plasmid RK2. Positioning of various motifs is critical for initiation of replication. J. Biol. Chem. 273, 8447–8453. doi: 10.1074/jbc.273.14.8447
Downey, C. D., and McHenry, C. S. (2010). Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader. Mol. Cell 37, 481–491. doi: 10.1016/j.molcel.2010.01.013
Duderstadt, K. E., Chuang, K., and Berger, J. M. (2011). DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213. doi: 10.1038/nature10455
Duderstadt, K. E., Mott, M. L., Crisona, N. J., Chuang, K., Yang, H., and Berger, J. M. (2010). Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 285, 28229–28239. doi: 10.1074/jbc.M110.147975
Erzberger, J. P., Mott, M. L., and Berger, J. M. (2006). Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13, 676–683. doi: 10.1038/nsmb1115
Erzberger, J. P., Pirruccello, M. M., and Berger, J. M. (2002). The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21, 4763–4773. doi: 10.1093/emboj/cdf496
Fang, J., Engen, J. R., and Beuning, P. J. (2011). Escherichia coli processivity clamp beta from DNA polymerase III is dynamic in solution. Biochemistry 50, 5958–5968. doi: 10.1021/bi200580b
Fang, J., Nevin, P., Kairys, V., Venclovas, C., Engen, J. R., and Beuning, P. J. (2014). Conformational analysis of processivity clamps in solution demonstrates that tertiary structure does not correlate with protein dynamics. Structure 22, 572–581. doi: 10.1016/j.str.2014.02.001
Fekete, R. A., Venkova-Canova, T., Park, K., and Chattoraj, D. K. (2006). IHF-dependent activation of P1 plasmid origin by dnaA. Mol. Microbiol. 62, 1739–1751. doi: 10.1111/j.1365-2958.2006.05479.x
Felczak, M. M., Sage, J. M., Hupert-Kocurek, K., Aykul, S., and Kaguni, J. M. (2016). Substitutions of conserved residues in the C-terminal region of DnaC cause thermolability in helicase loading. J. Biol. Chem. 291, 4803–4812. doi: 10.1074/jbc.M115.708586
Filutowicz, M., Davis, G., Greener, A., and Helinski, D. R. (1985). Autorepressor properties of the pi-initiation protein encoded by plasmid R6K. Nucleic Acids Res. 13, 103–114. doi: 10.1093/nar/13.1.103
Filutowicz, M., McEachern, M. J., Mukhopadhyay, P., Greener, A., Yang, S. L., and Helinski, D. R. (1987). DNA and protein interactions in the regulation of plasmid replication. J. Cell Sci. Suppl. 7, 15–31. doi: 10.1242/jcs.1987.Supplement_7.2
Flower, A. M., and McHenry, C. S. (1990). The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 87, 3713–3717. doi: 10.1073/pnas.87.10.3713
Flowers, S., Biswas, E. E., and Biswas, S. B. (2003). Conformational dynamics of DnaB helicase upon DNA and nucleotide binding: analysis by intrinsic tryptophan fluorescence quenching. Biochemistry 42, 1910–1921. doi: 10.1021/bi025992v
Fujikawa, N., Kurumizaka, H., Nureki, O., Terada, T., Shirouzu, M., Katayama, T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086. doi: 10.1093/nar/gkg309
Fuller, R. S., Funnell, B. E., and Kornberg, A. (1984). The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 38, 889–900. doi: 10.1016/0092-8674(84)90284-8
Funnell, B. E., Baker, T. A., and Kornberg, A. (1987). In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 262, 10327–10334.
Galletto, R., Jezewska, M. J., and Bujalowski, W. (2004). Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: the effect of the 3′ arm and the stability of the dsDNA on the unwinding activity of the Escherichia coli DnaB helicase. J. Mol. Biol. 343, 101–114. doi: 10.1016/j.jmb.2004.07.056
Gille, H., Egan, J. B., Roth, A., and Messer, W. (1991). The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 19, 4167–4172. doi: 10.1093/nar/19.15.4167
Giraldo, R., Andreu, J. M., and Díaz-Orejas, R. (1998). Protein domains and conformational changes in the activation of RepA, a DNA replication initiator. EMBO J. 17, 4511–4526. doi: 10.1093/emboj/17.15.4511
Giraldo, R., Fernandez-Tornero, C., Evans, P. R., Diaz-Orejas, R., and Romero, A. (2003). A conformational switch between transcriptional repression and replication initiation in the RepA dimerization domain. Nat. Struct. Biol. 10, 565–571. doi: 10.1038/nsb937
Glover, B. P., and McHenry, C. S. (1998). The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 273, 23476–23484. doi: 10.1074/jbc.273.36.23476
Grimwade, J. E., Ryan, V. T., and Leonard, A. C. (2000). IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 35, 835–844. doi: 10.1046/j.1365-2958.2000.01755.x
Gui, W. J., Lin, S. Q., Chen, Y. Y., Zhang, X. E., Bi, L. J., and Jiang, T. (2011). Crystal structure of DNA polymerase III beta sliding clamp from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 405, 272–277. doi: 10.1016/j.bbrc.2011.01.027
Hayner, J. N., Douma, L. G., and Bloom, L. B. (2014). The interplay of primer-template DNA phosphorylation status and single-stranded DNA binding proteins in directing clamp loaders to the appropriate polarity of DNA. Nucleic Acids Res. 42, 10655–10667. doi: 10.1093/nar/gku774
Hedglin, M., Kumar, R., and Benkovic, S. J. (2013). Replication clamps and clamp loaders. Cold Spring Harb. Perspect. Biol. 5:a010165. doi: 10.1101/cshperspect.a010165
Howland, C. J., Rees, C. E., Barth, P. T., and Wilkins, B. M. (1989). The ssb gene of plasmid ColIb-P9. J. Bacteriol. 171, 2466–2473.
Hwang, D. S., and Kornberg, A. (1992). Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 267, 23083–23086.
Indiani, C., and O'Donnell, M. (2006). The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 7, 751–761. doi: 10.1038/nrm2022
Ingmer, H., and Cohen, S. N. (1993). Excess intracellular concentration of the pSC101 RepA protein interferes with both plasmid DNA replication and partitioning. J. Bacteriol. 175, 7834–7841.
Ingmer, H., Fong, E. L., and Cohen, S. N. (1995). Monomer-dimer equilibrium of the pSC101 RepA protein. J. Mol. Biol. 250, 309–314. doi: 10.1006/jmbi.1995.0378
Itoh, T., and Horii, T. (1989). Replication of ColE2 and ColE3 plasmids: in vitro replication dependent on plasmid-coded proteins. Mol. Gen. Genet. 219, 249–255. doi: 10.1007/BF00261184
Itou, H., Yagura, M., Shirakihara, Y., and Itoh, T. (2015). Structural basis for replication origin unwinding by an initiator primase of plasmid ColE2-P9: duplex DNA unwinding by a single protein. J. Biol. Chem. 290, 3601–3611. doi: 10.1074/jbc.M114.595645
Jeruzalmi, D., O'Donnell, M., and Kuriyan, J. (2001). Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell 106, 429–441. doi: 10.1016/S0092-8674(01)00463-9
Jezewska, M. J., Kim, U. S., and Bujalowski, W. (1996). Binding of Escherichia coli primary replicative helicase DnaB protein to single-stranded DNA. Long-range allosteric conformational changes within the protein hexamer. Biochemistry 35, 2129–2145. doi: 10.1021/bi952345d
Jezewska, M. J., Rajendran, S., Bujalowska, D., and Bujalowski, W. (1998a). Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB Helicase? Fluorescence energy transfer studies. J. Biol. Chem. 273, 10515–10529. doi: 10.1074/jbc.273.17.10515
Jezewska, M. J., Rajendran, S., and Bujalowski, W. (1998b). Functional and structural heterogeneity of the DNA binding site of the Escherichia coli primary replicative helicase DnaB protein. J. Biol. Chem. 273, 9058–9069. doi: 10.1074/jbc.273.15.9058
Jiang, Y., Pacek, M., Helinski, D. R., Konieczny, I., and Toukdarian, A. (2003). A multifunctional plasmid-encoded replication initiation protein both recruits and positions an active helicase at the replication origin. Proc. Natl. Acad. Sci. U.S.A. 100, 8692–8697. doi: 10.1073/pnas.1532393100
Katayama, T., Ozaki, S., Keyamura, K., and Fujimitsu, K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170. doi: 10.1038/nrmicro2314
Kaur, G., Vora, M. P., Czerwonka, C. A., Rozgaja, T. A., Grimwade, J. E., and Leonard, A. C. (2014). Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 91, 1148–1163. doi: 10.1111/mmi.12525
Kawakami, H., Keyamura, K., and Katayama, T. (2005). Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430. doi: 10.1074/jbc.M502764200
Kawasaki, Y., Matsunaga, F., Kano, Y., Yura, T., and Wada, C. (1996). The localized melting of mini-F origin by the combined action of the mini-F initiator protein (RepE) and HU and DnaA of Escherichia coli. Mol. Gen. Genet. 253, 42–49. doi: 10.1007/s004380050294
Kawasaki, Y., Wada, C., and Yura, T. (1990). Roles of Escherichia coli heat shock proteins DnaK, DnaJ and GrpE in mini-F plasmid replication. Mol. Gen. Genet. 220, 277–282. doi: 10.1007/BF00260494
Kelch, B. A. (2016). Review: the lord of the rings: structure and mechanism of the sliding clamp loader. Biopolymers 105, 532–546. doi: 10.1002/bip.22827
Kelch, B. A., Makino, D. L., O'Donnell, M., and Kuriyan, J. (2011). How a DNA polymerase clamp loader opens a sliding clamp. Science 334, 1675–1680. doi: 10.1126/science.1211884
Kelch, B. A., Makino, D. L., O'Donnell, M., and Kuriyan, J. (2012). Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 10:34. doi: 10.1186/1741-7007-10-34
Kelman, Z., and O'Donnell, M. (1995). DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64, 171–200. doi: 10.1146/annurev.bi.64.070195.001131
Keyamura, K., Abe, Y., Higashi, M., Ueda, T., and Katayama, T. (2009). DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 284, 25038–25050. doi: 10.1074/jbc.M109.002717
Keyamura, K., Fujikawa, N., Ishida, T., Ozaki, S., Su'etsugu, M., Fujimitsu, K., et al. (2007). The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 21, 2083–2099. doi: 10.1101/gad.1561207
Kline, B. C., Kogoma, T., Tam, J. E., and Shields, M. S. (1986). Requirement of the Escherichia coli dnaA gene product for plasmid F maintenance. J. Bacteriol. 168, 440–443.
Kobori, J. A., and Kornberg, A. (1982). The Escherichia coli dnaC gene product. II. Purification, physical properties, and role in replication. J. Biol. Chem. 257, 13763–13769.
Komori, H., Matsunaga, F., Higuchi, Y., Ishiai, M., Wada, C., and Miki, K. (1999). Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 A resolution. EMBO J. 18, 4597–4607. doi: 10.1093/emboj/18.17.4597
Kongsuwan, K., Josh, P., Picault, M. J., Wijffels, G., and Dalrymple, B. (2006). The plasmid RK2 replication initiator protein (TrfA) binds to the sliding clamp beta subunit of DNA polymerase III: implication for the toxicity of a peptide derived from the amino-terminal portion of 33-kilodalton TrfA. J. Bacteriol. 188, 5501–5509. doi: 10.1128/JB.00231-06
Konieczny, I. (2003). Strategies for helicase recruitment and loading in bacteria. EMBO Rep. 4, 37–41. doi: 10.1038/sj.embor.embor703
Konieczny, I., Doran, K. S., Helinski, D. R., and Blasina, A. (1997). Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J. Biol. Chem. 272, 20173–20178. doi: 10.1074/jbc.272.32.20173
Konieczny, I., and Helinski, D. R. (1997). The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc. Natl. Acad. Sci. U.S.A. 94, 14378–14382. doi: 10.1073/pnas.94.26.14378
Kowalczyk, L., Rajewska, M., and Konieczny, I. (2005). Positioning and the specific sequence of each 13-mer motif are critical for activity of the plasmid RK2 replication origin. Mol. Microbiol. 57, 1439–1449. doi: 10.1111/j.1365-2958.2005.04770.x
Krishna, T. S., Kong, X. P., Gary, S., Burgers, P. M., and Kuriyan, J. (1994). Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243. doi: 10.1016/0092-8674(94)90014-0
Kukshal, V., Khanam, T., Chopra, D., Singh, N., Sanyal, S., and Ramachandran, R. (2012). M. tuberculosis sliding beta-clamp does not interact directly with the NAD+-dependent DNA ligase. PLoS ONE 7:e35702. doi: 10.1371/journal.pone.0035702
LeBowitz, J. H., and McMacken, R. (1986). The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 261, 4738–4748.
Le Chatelier, E., Janniere, L., Ehrlich, S. D., and Canceill, D. (2001). The RepE initiator is a double-stranded and single-stranded DNA-binding protein that forms an atypical open complex at the onset of replication of plasmid pAMbeta 1 from Gram-positive bacteria. J. Biol. Chem. 276, 10234–10246. doi: 10.1074/jbc.M010118200
Leonard, A. C., and Grimwade, J. E. (2005). Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol. Microbiol. 55, 978–985. doi: 10.1111/j.1365-2958.2004.04467.x
Leonard, A. C., and Grimwade, J. E. (2011). Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 65, 19–35. doi: 10.1146/annurev-micro-090110-102934
Leonard, A. C., and Grimwade, J. E. (2015). The orisome: structure and function. Front. Microbiol. 6:545. doi: 10.3389/fmicb.2015.00545
Liu, J., Pei, H., Mei, S., Li, J., Zhou, L., and Xiang, H. (2008). Replication initiator DnaA interacts with an anti-terminator NusG in T. tengcongensis. Biochem. Biophys. Res. Commun. 371, 573–577. doi: 10.1016/j.bbrc.2008.04.131
Lo, Y. H., Tsai, K. L., Sun, Y. J., Chen, W. T., Huang, C. Y., and Hsiao, C. D. (2009). The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Res. 37, 804–814. doi: 10.1093/nar/gkn999
Loftie-Eaton, W., and Rawlings, D. E. (2012). Diversity, biology and evolution of IncQ-family plasmids. Plasmid 67, 15–34. doi: 10.1016/j.plasmid.2011.10.001
Lohman, T. M., and Bjornson, K. P. (1996). Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169–214. doi: 10.1146/annurev.bi.65.070196.001125
Lowery, T. J., Pelton, J. G., Chandonia, J. M., Kim, R., Yokota, H., and Wemmer, D. E. (2007). NMR structure of the N-terminal domain of the replication initiator protein DnaA. J. Struct. Funct. Genomics 8, 11–17. doi: 10.1007/s10969-007-9022-7
Lu, Y. B., Datta, H. J., and Bastia, D. (1998). Mechanistic studies of initiator-initiator interaction and replication initiation. EMBO J. 17, 5192–5200. doi: 10.1093/emboj/17.17.5192
Lu, Y. B., Ratnakar, P. V., Mohanty, B. K., and Bastia, D. (1996). Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc. Natl. Acad. Sci. U.S.A. 93, 12902–12907. doi: 10.1073/pnas.93.23.12902
Mackay, V., and Linn, S. (1976). Selective inhibition of the dnase activity of the recBC enzyme by the DNA binding protein from Escherichia coli. J. Biol. Chem. 251, 3716–3719.
Maestro, B., Sanz, J. M., Diaz-Orejas, R., and Fernandez-Tresguerres, E. (2003). Modulation of pPS10 host range by plasmid-encoded RepA initiator protein. J. Bacteriol. 185, 1367–1375. doi: 10.1128/JB.185.4.1367-1375.2003
Maki, H., Maki, S., and Kornberg, A. (1988). DNA Polymerase III holoenzyme of Escherichia coli. IV. The holoenzyme is an asymmetric dimer with twin active sites. J. Biol. Chem. 263, 6570–6578.
Makowska-Grzyska, M., and Kaguni, J. M. (2010). Primase directs the release of DnaC from DnaB. Mol. Cell 37, 90–101. doi: 10.1016/j.molcel.2009.12.031
Matsui, M., Oka, A., Takanami, M., Yasuda, S., and Hirota, Y. (1985). Sites of dnaA protein-binding in the replication origin of the Escherichia coli K-12 chromosome. J. Mol. Biol. 184, 529–533. doi: 10.1016/0022-2836(85)90299-2
McEachern, M. J., Filutowicz, M., and Helinski, D. R. (1985). Mutations in direct repeat sequences and in a conserved sequence adjacent to the repeats result in a defective replication origin in plasmid R6K. Proc. Natl. Acad. Sci. U.S.A. 82, 1480–1484. doi: 10.1073/pnas.82.5.1480
McGarry, K. C., Ryan, V. T., Grimwade, J. E., and Leonard, A. C. (2004). Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U.S.A. 101, 2811–2816. doi: 10.1073/pnas.0400340101
McHenry, C. S. (2011). DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 80, 403–436. doi: 10.1146/annurev-biochem-061208-091655
McInerney, P., Johnson, A., Katz, F., and O'Donnell, M. (2007). Characterization of a triple DNA polymerase replisome. Mol. Cell 27, 527–538. doi: 10.1016/j.molcel.2007.06.019
Messer, W., Blaesing, F., Majka, J., Nardmann, J., Schaper, S., Schmidt, A., et al. (1999). Functional domains of DnaA proteins. Biochimie 81, 819–825. doi: 10.1016/S0300-9084(99)00215-1
Meyer, R. R., Glassberg, J., and Kornberg, A. (1979). An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc. Natl. Acad. Sci. U.S.A. 76, 1702–1705. doi: 10.1073/pnas.76.4.1702
Miron, A., Mukherjee, S., and Bastia, D. (1992). Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance. EMBO J. 11, 1205–1216.
Molt, K. L., Sutera, V. A. Jr., Moore, K. K., and Lovett, S. T. (2009). A role for nonessential domain II of initiator protein, DnaA, in replication control. Genetics 183, 39–49. doi: 10.1534/genetics.109.104760
Mott, M. L., Erzberger, J. P., Coons, M. M., and Berger, J. M. (2008). Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 135, 623–634. doi: 10.1016/j.cell.2008.09.058
Murakami, Y., Ohmori, H., Yura, T., and Nagata, T. (1987). Requirement of the Escherichia coli dnaA gene function for ori-2-dependent mini-F plasmid replication. J. Bacteriol. 169, 1724–1730.
Murotsu, T., Matsubara, K., Sugisaki, H., and Takanami, M. (1981). Nine unique repeating sequences in a region essential for replication and incompatibility of the mini-F plasmid. Gene 15, 257–271. doi: 10.1016/0378-1119(81)90135-9
Nakamura, A., Komori, H., Kobayashi, G., Kita, A., Wada, C., and Miki, K. (2004). The N-terminal domain of the replication initiator protein RepE is a dimerization domain forming a stable dimer. Biochem. Biophys. Res. Commun. 315, 10–15. doi: 10.1016/j.bbrc.2004.01.018
Nakamura, A., Wada, C., and Miki, K. (2007a). Expression and purification of F-plasmid RepE and preliminary X-ray crystallographic study of its complex with operator DNA. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 346–349. doi: 10.1107/S1744309107012894
Nakamura, A., Wada, C., and Miki, K. (2007b). Structural basis for regulation of bifunctional roles in replication initiator protein. Proc. Natl. Acad. Sci. U.S.A. 104, 18484–18489. doi: 10.1073/pnas.0705623104
Natrajan, G., Hall, D. R., Thompson, A. C., Gutsche, I., and Terradot, L. (2007). Structural similarity between the DnaA-binding proteins HobA (HP1230) from Helicobacter pylori and DiaA from Escherichia coli. Mol. Microbiol. 65, 995–1005. doi: 10.1111/j.1365-2958.2007.05843.x
Natrajan, G., Noirot-Gros, M. F., Zawilak-Pawlik, A., Kapp, U., and Terradot, L. (2009). The structure of a DnaA/HobA complex from Helicobacter pylori provides insight into regulation of DNA replication in bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 21115–21120. doi: 10.1073/pnas.0908966106
Naue, N., Beerbaum, M., Bogutzki, A., Schmieder, P., and Curth, U. (2013). The helicase-binding domain of Escherichia coli DnaG primase interacts with the highly conserved C-terminal region of single-stranded DNA-binding protein. Nucleic Acids Res. 41, 4507–4517. doi: 10.1093/nar/gkt107
Neuwald, A. F., Aravind, L., Spouge, J. L., and Koonin, E. V. (1999). AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43.
Nieto, C., Giraldo, R., Fernández-Tresguerres, E., and Díaz, R. (1992). Genetic and functional analysis of the basic replicon of pPS10, a plasmid specific for Pseudomonas isolated from Pseudomonas syringae patovar savastanoi. J. Mol. Biol. 223, 415–426. doi: 10.1016/0022-2836(92)90661-3
Nievera, C., Torgue, J. J., Grimwade, J. E., and Leonard, A. C. (2006). SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell 24, 581–592. doi: 10.1016/j.molcel.2006.09.016
Nozaki, S., and Ogawa, T. (2008). Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154, 3379–3384. doi: 10.1099/mic.0.2008/019745-0
Oakley, A. J., Prosselkov, P., Wijffels, G., Beck, J. L., Wilce, M. C., and Dixon, N. E. (2003). Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III. Acta Crystallogr. D Biol. Crystallogr. 59, 1192–1199. doi: 10.1107/S0907444903009958
O'Donnell, M. (2006). Replisome architecture and dynamics in Escherichia coli. J. Biol. Chem. 281, 10653–10656. doi: 10.1074/jbc.R500028200
O'Donnell, M., Langston, L., and Stillman, B. (2013). Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 5:a010108. doi: 10.1101/cshperspect.a010108
O'Donnell, M., and Studwell, P. S. (1990). Total reconstitution of DNA polymerase III holoenzyme reveals dual accessory protein clamps. J. Biol. Chem. 265, 1179–1187.
Ogura, T., Niki, H., Kano, Y., Imamoto, F., and Hiraga, S. (1990). Maintenance of plasmids in HU and IHF mutants of Escherichia coli. Mol. Gen. Genet. 220, 197–203. doi: 10.1007/BF00260482
Ortega-Jiménez, S., Giraldo-Suárez, R., Fernández-Tresguerres, M. E., Berzal-Herranz, A., and Diaz-Orejas, R. (1992). DnaA dependent replication of plasmid R1 occurs in the presence of point mutations that disrupt the dnaA box of oriR. Nucleic Acids Res. 20, 2547–2551. doi: 10.1093/nar/20.10.2547
Ozaki, S., and Katayama, T. (2009). DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid 62, 71–82. doi: 10.1016/j.plasmid.2009.06.003
Ozaki, S., and Katayama, T. (2012). Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665. doi: 10.1093/nar/gkr832
Ozaki, S., Kawakami, H., Nakamura, K., Fujikawa, N., Kagawa, W., Park, S. Y., et al. (2008). A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362. doi: 10.1074/jbc.M708684200
Ozaki, S., Noguchi, Y., Hayashi, Y., Miyazaki, E., and Katayama, T. (2012a). Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 287, 37458–37471. doi: 10.1074/jbc.M112.372052
Ozaki, S., Noguchi, Y., Nishimura, M., and Katayama, T. (2012b). Stable nucleotide binding to DnaA requires a specific glutamic acid residue within the AAA+ box II motif. J. Struct. Biol. 179, 242–250. doi: 10.1016/j.jsb.2012.05.001
Pacek, M., Konopa, G., and Konieczny, I. (2001). DnaA box sequences as the site for helicase delivery during plasmid RK2 replication initiation in Escherichia coli. J. Biol. Chem. 276, 23639–23644. doi: 10.1074/jbc.M100255200
Page, D. T., Whelan, K. F., and Colleran, E. (2001). Characterization of two autoreplicative regions of the IncHI2 plasmid R478: RepHI2A and RepHI1A((R478)). Microbiology 147, 1591–1598. doi: 10.1099/00221287-147-6-1591
Park, K., Mukhopadhyay, S., and Chattoraj, D. K. (1998). Requirements for and regulation of origin opening of plasmid P1. J. Biol. Chem. 273, 24906–24911. doi: 10.1074/jbc.273.38.24906
Park, M. S., and O'Donnell, M. (2009). The clamp loader assembles the beta clamp onto either a 3′ or 5′ primer terminus: the underlying basis favoring 3′ loading. J. Biol. Chem. 284, 31473–31483. doi: 10.1074/jbc.M109.050310
Paschall, C. O., Thompson, J. A., Marzahn, M. R., Chiraniya, A., Hayner, J. N., O'Donnell, M., et al. (2011). The Escherichia coli clamp loader can actively pry open the beta-sliding clamp. J. Biol. Chem. 286, 42704–42714. doi: 10.1074/jbc.M111.268169
Perri, S., and Helinski, D. R. (1993). DNA sequence requirements for interaction of the RK2 replication initiation protein with plasmid origin repeats. J. Biol. Chem. 268, 3662–3669.
Perri, S., Helinski, D. R., and Toukdarian, A. (1991). Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host range plasmid RK2. J. Biol. Chem. 266, 12536–12543.
Pierechod, M., Nowak, A., Saari, A., Purta, E., Bujnicki, J. M., and Konieczny, I. (2009). Conformation of a plasmid replication initiator protein affects its proteolysis by ClpXP system. Protein Sci. 18, 637–649. doi: 10.1002/pro.68
Pietroni, P., and von Hippel, P. H. (2008). Multiple ATP binding is required to stabilize the “activated” (clamp open) clamp loader of the T4 DNA replication complex. J. Biol. Chem. 283, 28338–28353. doi: 10.1074/jbc.M804371200
Podobnik, M., Weitze, T. F., O'Donnell, M., and Kuriyan, J. (2003). Nucleotide-induced conformational changes in an isolated Escherichia coli DNA polymerase III clamp loader subunit. Structure 11, 253–263. doi: 10.1016/S0969-2126(03)00027-3
Polaczek, P. (1990). Bending of the origin of replication of E. coli by binding of IHF at a specific site. New Biol. 2, 265–271.
Punchihewa, C., Inoue, A., Hishiki, A., Fujikawa, Y., Connelly, M., Evison, B., et al. (2012). Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. J. Biol. Chem. 287, 14289–14300. doi: 10.1074/jbc.M112.353201
Rahn-Lee, L., Merrikh, H., Grossman, A. D., and Losick, R. (2011). The sporulation protein SirA inhibits the binding of DnaA to the origin of replication by contacting a patch of clustered amino acids. J. Bacteriol. 193, 1302–1307. doi: 10.1128/JB.01390-10
Rajewska, M., Kowalczyk, L., Konopa, G., and Konieczny, I. (2008). Specific mutations within the AT-rich region of a plasmid replication origin affect either origin opening or helicase loading. Proc. Natl. Acad. Sci. U.S.A. 105, 11134–11139. doi: 10.1073/pnas.0805662105
Rajewska, M., Wegrzyn, K., and Konieczny, I. (2012). AT-rich region and repeated sequences - the essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 36, 408–434. doi: 10.1111/j.1574-6976.2011.00300.x
Rakowski, S. A., and Filutowicz, M. (2013). Plasmid R6K replication control. Plasmid 69, 231–242. doi: 10.1016/j.plasmid.2013.02.003
Rangarajan, S., Woodgate, R., and Goodman, M. F. (1999). A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 9224–9229. doi: 10.1073/pnas.96.16.9224
Ratnakar, P. V., Mohanty, B. K., Lobert, M., and Bastia, D. (1996). The replication initiator protein pi of the plasmid R6K specifically interacts with the host-encoded helicase DnaB. Proc. Natl. Acad. Sci. U.S.A. 93, 5522–5526. doi: 10.1073/pnas.93.11.5522
Reyes-Lamothe, R., Possoz, C., Danilova, O., and Sherratt, D. J. (2008). Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133, 90–102. doi: 10.1016/j.cell.2008.01.044
Reyes-Lamothe, R., Sherratt, D. J., and Leake, M. C. (2010). Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328, 498–501. doi: 10.1126/science.1185757
Richardson, T. T., Harran, O., and Murray, H. (2016). The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature 534, 412–416. doi: 10.1038/nature17962
Roth, A., and Messer, W. (1995). The DNA binding domain of the initiator protein DnaA. EMBO J. 14, 2106–2111.
Rozgaja, T. A., Grimwade, J. E., Iqbal, M., Czerwonka, C., Vora, M., and Leonard, A. C. (2011). Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 82, 475–488. doi: 10.1111/j.1365-2958.2011.07827.x
Ryan, V. T., Grimwade, J. E., Nievera, C. J., and Leonard, A. C. (2002). IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 46, 113–124. doi: 10.1046/j.1365-2958.2002.03129.x
Samitt, C. E., Hansen, F. G., Miller, J. F., and Schaechter, M. (1989). In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 8, 989–993.
Saxena, R., Vasudevan, S., Patil, D., Ashoura, N., Grimwade, J. E., and Crooke, E. (2015). Nucleotide-induced conformational changes in Escherichia coli DnaA protein are required for bacterial ORC to Pre-RC conversion at the chromosomal origin. Int. J. Mol. Sci. 16, 27897–27911. doi: 10.3390/ijms161126064
Seitz, H., Weigel, C., and Messer, W. (2000). The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 37, 1270–1279. doi: 10.1046/j.1365-2958.2000.02096.x
Sekimizu, K., Bramhill, D., and Kornberg, A. (1987). ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50, 259–265. doi: 10.1016/0092-8674(87)90221-2
Shah, D. S., Cross, M. A., Porter, D., and Thomas, C. M. (1995). Dissection of the core and auxiliary sequences in the vegetative replication origin of promiscuous plasmid RK2. J. Mol. Biol. 254, 608–622. doi: 10.1006/jmbi.1995.0642
Shaheen, S. M., Ouimet, M. C., and Marczynski, G. T. (2009). Comparative analysis of Caulobacter chromosome replication origins. Microbiology 155, 1215–1225. doi: 10.1099/mic.0.025528-0
Sharma, R., Kachroo, A., and Bastia, D. (2001). Mechanistic aspects of DnaA-RepA interaction as revealed by yeast forward and reverse two-hybrid analysis. EMBO J. 20, 4577–4587. doi: 10.1093/emboj/20.16.4577
Sharma, S., Sathyanarayana, B. K., Bird, J. G., Hoskins, J. R., Lee, B., and Wickner, S. (2004). Plasmid P1 RepA is homologous to the F plasmid RepE class of initiators. J. Biol. Chem. 279, 6027–6034. doi: 10.1074/jbc.M310917200
Shereda, R. D., Kozlov, A. G., Lohman, T. M., Cox, M. M., and Keck, J. L. (2008). SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318. doi: 10.1080/10409230802341296
Simmons, L. A., Felczak, M., and Kaguni, J. M. (2003). DnaA Protein of Escherichia coli: oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol. Microbiol. 49, 849–858. doi: 10.1046/j.1365-2958.2003.03603.x
Simonetta, K. R., Kazmirski, S. L., Goedken, E. R., Cantor, A. J., Kelch, B. A., McNally, R., et al. (2009). The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell 137, 659–671. doi: 10.1016/j.cell.2009.03.044
Soni, R. K., Mehra, P., Choudhury, N. R., Mukhopadhyay, G., and Dhar, S. K. (2003). Functional characterization of Helicobacter pylori DnaB helicase. Nucleic Acids Res. 31, 6828–6840. doi: 10.1093/nar/gkg895
Soultanas, P. (2012). Loading mechanisms of ring helicases at replication origins. Mol. Microbiol. 84, 6–16. doi: 10.1111/j.1365-2958.2012.08012.x
Soumillion, P., Sexton, D. J., and Benkovic, S. J. (1998). Clamp subunit dissociation dictates bacteriophage T4 DNA polymerase holoenzyme disassembly. Biochemistry 37, 1819–1827. doi: 10.1021/bi972526a
Sozhamannan, S., and Chattoraj, D. K. (1993). Heat shock proteins DnaJ, DnaK, and GrpE stimulate P1 plasmid replication by promoting initiator binding to the origin. J. Bacteriol. 175, 3546–3555.
Speck, C., and Messer, W. (2001). Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20, 1469–1476. doi: 10.1093/emboj/20.6.1469
Stalker, D. M., Thomas, C. M., and Helinski, D. R. (1981). Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol. Gen. Genet. 181, 8–12. doi: 10.1007/BF00338997
Stenzel, T. T., Patel, P., and Bastia, D. (1987). The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell 49, 709–717. doi: 10.1016/0092-8674(87)90547-2
Strycharska, M. S., Arias-Palomo, E., Lyubimov, A. Y., Erzberger, J. P., O'Shea, V. L., Bustamante, C. J., et al. (2013). Nucleotide and partner-protein control of bacterial replicative helicase structure and function. Mol. Cell 52, 844–854. doi: 10.1016/j.molcel.2013.11.016
Su, X. C., Wang, Y., Yagi, H., Shishmarev, D., Mason, C. E., Smith, P. J., et al. (2014). Bound or free: interaction of the C-terminal domain of Escherichia coli single-stranded DNA-binding protein (SSB) with the tetrameric core of SSB. Biochemistry 53, 1925–1934. doi: 10.1021/bi5001867
Sutton, M. D., Carr, K. M., Vicente, M., and Kaguni, J. M. (1998). Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 273, 34255–34262. doi: 10.1074/jbc.273.51.34255
Sutton, M. D., and Kaguni, J. M. (1995). Novel alleles of the Escherichia coli dnaA gene are defective in replication of pSC101 but not of oriC. J. Bacteriol. 177, 6657–6665.
Sutton, M. D., and Kaguni, J. M. (1997). Novel alleles of the Escherichia coli dnaA gene. J. Mol. Biol. 271, 693–703. doi: 10.1006/jmbi.1997.1209
Swan, M. K., Bastia, D., and Davies, C. (2006). Crystal structure of pi initiator protein-iteron complex of plasmid R6K: implications for initiation of plasmid DNA replication. Proc. Natl. Acad. Sci. U.S.A. 103, 18481–18486. doi: 10.1073/pnas.0609046103
Taft-Benz, S. A., and Schaaper, R. M. (2004). The theta subunit of Escherichia coli DNA polymerase III: a role in stabilizing the epsilon proofreading subunit. J. Bacteriol. 186, 2774–2780. doi: 10.1128/JB.186.9.2774-2780.2004
Takechi, S., and Itoh, T. (1995). Initiation of unidirectional ColE2 DNA replication by a unique priming mechanism. Nucleic Acids Res. 23, 4196–4201. doi: 10.1093/nar/23.20.4196
Takechi, S., Matsui, H., and Itoh, T. (1995). Primer RNA synthesis by plasmid-specified Rep protein for initiation of ColE2 DNA replication. EMBO J. 14, 5141–5147.
Taylor, J. A., Ouimet, M. C., Wargachuk, R., and Marczynski, G. T. (2011). The Caulobacter crescentus chromosome replication origin evolved two classes of weak DnaA binding sites. Mol. Microbiol. 82, 312–326. doi: 10.1111/j.1365-2958.2011.07785.x
Thomas, C. M., and Sherlock, S. (1990). Broad host range plasmid RK2 encodes a polypeptide related to single-stranded DNA binding protein (SSB) of Escherichia coli. Nucleic Acids Res. 18, 2812. doi: 10.1093/nar/18.9.2812
Trakselis, M. A., Alley, S. C., Abel-Santos, E., and Benkovic, S. J. (2001). Creating a dynamic picture of the sliding clamp during T4 DNA polymerase holoenzyme assembly by using fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 98, 8368–8375. doi: 10.1073/pnas.111006698
Tsodikov, O. V., and Biswas, T. (2011). Structural and thermodynamic signatures of DNA recognition by Mycobacterium tuberculosis DnaA. J. Mol. Biol. 410, 461–476. doi: 10.1016/j.jmb.2011.05.007
Tsuchihashi, Z., and Kornberg, A. (1989). ATP interactions of the tau and gamma subunits of DNA polymerase III holoenzyme of Escherichia coli. J. Biol. Chem. 264, 17790–17795.
Wahle, E., Lasken, R. S., and Kornberg, A. (1989). The dnaB-dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J. Biol. Chem. 264, 2469–2475.
Wang, G., Klein, M. G., Tokonzaba, E., Zhang, Y., Holden, L. G., and Chen, X. S. (2008). The structure of a DnaB-family replicative helicase and its interactions with primase. Nat. Struct. Mol. Biol. 15, 94–100. doi: 10.1038/nsmb1356
Wawrzycka, A., Gross, M., Wasaznik, A., and Konieczny, I. (2015). Plasmid replication initiator interactions with origin 13-mers and polymerase subunits contribute to strand-specific replisome assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E4188–E4196. doi: 10.1073/pnas.1504926112
Wegrzyn, K., Fuentes-Perez, M. E., Bury, K., Rajewska, M., Moreno-Herrero, F., and Konieczny, I. (2014). Sequence-specific interactions of Rep proteins with ssDNA in the AT-rich region of the plasmid replication origin. Nucleic Acids Res. 42, 7807–7818. doi: 10.1093/nar/gku453
Weigel, C., Schmidt, A., Seitz, H., Tungler, D., Welzeck, M., and Messer, W. (1999). The N-terminus promotes oligomerization of the Escherichia coli initiator protein DnaA. Mol. Microbiol. 34, 53–66. doi: 10.1046/j.1365-2958.1999.01568.x
Weigel, C., and Seitz, H. (2002). Strand-specific loading of DnaB helicase by DnaA to a substrate mimicking unwound oriC. Mol. Microbiol. 46, 1149–1156. doi: 10.1046/j.1365-2958.2002.03232.x
Wickner, S., Gottesman, S., Skowyra, D., Hoskins, J., McKenney, K., and Maurizi, M. R. (1994). A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. U.S.A. 91, 12218–12222. doi: 10.1073/pnas.91.25.12218
Wickner, S., and Hurwitz, J. (1975). Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc. Natl. Acad. Sci. U.S.A. 72, 921–925. doi: 10.1073/pnas.72.3.921
Wickner, S., Skowyra, D., Hoskins, J., and McKenney, K. (1992). DnaJ, DnaK, and GrpE heat shock proteins are required in oriP1 DNA replication solely at the RepA monomerization step. Proc. Natl. Acad. Sci. U.S.A. 89, 10345–10349. doi: 10.1073/pnas.89.21.10345
Williams, G. J., Johnson, K., Rudolf, J., McMahon, S. A., Carter, L., Oke, M., et al. (2006). Structure of the heterotrimeric PCNA from Sulfolobus solfataricus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 944–948. doi: 10.1107/S1744309106034075
Wolanski, M., Donczew, R., Zawilak-Pawlik, A., and Zakrzewska-Czerwinska, J. (2014). oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 5:735. doi: 10.3389/fmicb.2014.00735
Wold, S., Crooke, E., and Skarstad, K. (1996). The Escherichia coli Fis protein prevents initiation of DNA replication from oriC in vitro. Nucleic Acids Res. 24, 3527–3532. doi: 10.1093/nar/24.18.3527
Wolff, P., Amal, I., Oliéric, V., Chaloin, O., Gygli, G., Ennifar, E., et al. (2014). Differential modes of peptide binding onto replicative sliding clamps from various bacterial origins. J. Med. Chem. 57, 7565–7576. doi: 10.1021/jm500467a
Xia, G., Manen, D., Yu, Y., and Caro, L. (1993). In vivo and in vitro studies of a copy number mutation of the RepA replication protein of plasmid pSC101. J. Bacteriol. 175, 4165–4175.
Yano, H., Rogers, L. M., Knox, M. G., Heuer, H., Smalla, K., Brown, C. J., et al. (2013). Host range diversification within the IncP-1 plasmid group. Microbiology 159, 2303–2315. doi: 10.1099/mic.0.068387-0
Yano, H., Wegrzyn, K., Loftie-Eaton, W., Johnson, J., Deckert, G. E., Rogers, L. M., et al. (2016). Evolved plasmid-host interactions reduce plasmid interference cost. Mol. Microbiol. doi: 10.1111/mmi.13407. [Epub ahead of print].
Yasueda, H., Horii, T., and Itoh, T. (1989). Structural and functional organization of ColE2 and ColE3 replicons. Mol. Gen. Genet. 215, 209–216. doi: 10.1007/BF00339719
Yuzhakov, A., Kelman, Z., and O'Donnell, M. (1999). Trading places on DNA–a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96, 153–163. doi: 10.1016/S0092-8674(00)80968-X
Yuzhakov, A., Turner, J., and O'Donnell, M. (1996). Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell 86, 877–886. doi: 10.1016/S0092-8674(00)80163-4
Zawilak-Pawlik, A., Kois, A., Stingl, K., Boneca, I. G., Skrobuk, P., Piotr, J., et al. (2007). HobA–a novel protein involved in initiation of chromosomal replication in Helicobacter pylori. Mol. Microbiol. 65, 979–994. doi: 10.1111/j.1365-2958.2007.05853.x
Zhong, Z., Helinski, D., and Toukdarian, A. (2003). A specific region in the N terminus of a replication initiation protein of plasmid RK2 is required for recruitment of Pseudomonas aeruginosa DnaB helicase to the plasmid origin. J. Biol. Chem. 278, 45305–45310. doi: 10.1074/jbc.M306058200
Zhong, Z., Helinski, D., and Toukdarian, A. (2005). Plasmid host-range: restrictions to F replication in Pseudomonas. Plasmid 54, 48–56. doi: 10.1016/j.plasmid.2004.11.001
Keywords: replication initiation, DnaA, Rep, iteron plasmids, replisome assembly
Citation: Wegrzyn KE, Gross M, Uciechowska U and Konieczny I (2016) Replisome Assembly at Bacterial Chromosomes and Iteron Plasmids. Front. Mol. Biosci. 3:39. doi: 10.3389/fmolb.2016.00039
Received: 21 June 2016; Accepted: 25 July 2016;
Published: 11 August 2016.
Edited by:
Tatiana Venkova, The University of Texas Medical Branch at Galveston, USAReviewed by:
Ramon Diaz Orejas, Spanish National Research Council, SpainCopyright © 2016 Wegrzyn, Gross, Uciechowska and Konieczny. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katarzyna E. Wegrzyn, a2F0YXJ6eW5hLndlZ3J6eW5AYmlvdGVjaC51Zy5lZHUucGw=
Igor Konieczny, aWdvci5rb25pZWN6bnlAYmlvdGVjaC51Zy5lZHUucGw=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.