- 1Institute of Herbgenomics, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 2Traditional Chinese Medicine Health Industry Promotion Center of Dongxing District, Neijiang, China
- 3Neijiang Dongxing District Bureau of Health, Neijiang, China
- 4Committee of Education, Science, Culture and Health of Dongxing District, Neijiang, China
Asparagus cochinchinensis is a medicinal plant in China, which has gained attention owing its protective effect in human health. However, there are seldom studies to systematically reveal the rhizosphere bacterial community of A. cochinchinensis. In this study, we employed metagenomics and culturomics to analyze the bacterial community composition and diversity in continuous rhizosphere soil of A. cochinchinensis. Meanwhile, we assessed the effect of soil physicochemical properties on the bacterial community. Results showed that the most abundant TAXA is a taxon belonging to the family Streptomycetaceae, the genus Mycobacterium and the species Oligotropha carboxidovorans. The bacterial communities across various areas were similar. Significant differences of exchangeable magnesium and available phosphorus level were observed between three groups. Furthermore, bacterial community structure correlated closely with soil physicochemical properties. Additionally, a total of 103 strains were isolated and identified, representing 28 species. Based on this study, the rhizosphere bacterial community of A. cochinchinensis might influence its growth and development. The rhizosphere strains were isolated and their function request further investigation. This study firstly revealed the bacterial community in the A. cochinchinensis rhizosphere soil, providing valuable references for its quality improvement in practical cultivation process.
Introduction
Asparagus cochinchinensis is a renowned medicinal plant belonging to Liliaceae family. Its health benefits have elucidated wide attention in recent years, with application in treating coughs, promoting skin whiten, and anti-aging effects (Lee et al., 2017; Wang et al., 2022). Research by Zhang L. et al. (2021) highlighted the potential of exosome-like nanovesicles from A. cochinchinensis for antitumor therapy. Furthermore, A. cochinchinensis is utilized in the fermentation of wine (Kim M. et al., 2017; Kim N. G. et al., 2017). Therefore, the market demand of A. cochinchinensis shows an increasing trend in recent years. A. cochinchinensis was mainly distributed across east and south Asia including China, Vietnam, Japan, and Korea. Now, China has been one of the main producing countries and exporters worldwide. The first record of A. cochinchinensis in China could date back to more than 2000 years ago in the Han Dynasty in Shennong’s Classic of Materia Medica (Tianmendong in Chinese). Notably, it is listed in the Pharmacopoeia of the People’s Republic of China (2020 edition, Chinese Pharmacopeia Commission, 2020, pp. 56–57). As a result of the suitable environment, Neijiang city (in Sichuan province with latitude 29°1 “to 30°2” north and longitude 104°16 “to 105°26” east) has become one of the main production cities of A. cochinchinensis in China. It is essential to develop efficient strategy to ensure the quality improvement of A. cochinchinensis.
Increasing number studies underscore the pivotal role of soil microorganisms in the growth and development of medicinal plants (Yu et al., 2024). For instance, Feng et al. (2021) studied the effect of Bacillus on the phthalides accumulation in Angelica sinensis (Oliv.), and observed that the Bacillus strains, which were isolated from rhizosphere soils, directly stimulated plant growth and the biosynthesis of butylidenephthalide. Wei et al. (2020) reported that microbial inoculants combined utilization of microbial inoculant and with garbage enzyme effectively reduced cadmium (Cd) uptake in Salvia miltiorrhiza by 37.90%, compared with the control group. Moreover, they might also facilitate the microbial remediation of soil contaminated with Cd. Csorba et al. (2022) investigated the interaction between Alkanna tinctoria and soil microorganisms and found positive correlations between alkannin levels and the relative abundances of Labrys, Allorhizobium, Neorhizobium, Pararhizobium, Rhizobium, and Penicillium. The advancement of metagenomics and culturomics has facilitated comprehensive exploration of the rhizosphere microbiota and its interaction with medicinal plants. Lian et al. (2023) simultaneously used metagenomics and culturomics technology to reveal the structure of rhizosphere microorganisms in Sinai desert farming systems and indicated that ecosystem functions of rhizosphere microorganisms from different sampling areas were similar although the microbial community structure and diversity were different. However, systematic studies on the interaction between the rhizosphere bacterial community and A. cochinchinensis are notably scarce.
In this study, we applied metagenomics and culturomics technology to comprehensively characterize the rhizosphere bacterial community of A. cochinchinensis. Concurrently, we analyzed the physicochemical properties of the rhizosphere soil from A. cochinchinensis. Our findings firstly revealed the rhizosphere bacterial community composition and isolated the rhizosphere bacterial strains from the A. cochinchinensis rhizosphere soil samples. Meanwhile, the effect of physicochemical properties on the rhizosphere bacterial community was evaluated, which provided references for the application of synthetic community for A. cochinchinensis.
Materials and methods
Sample collection
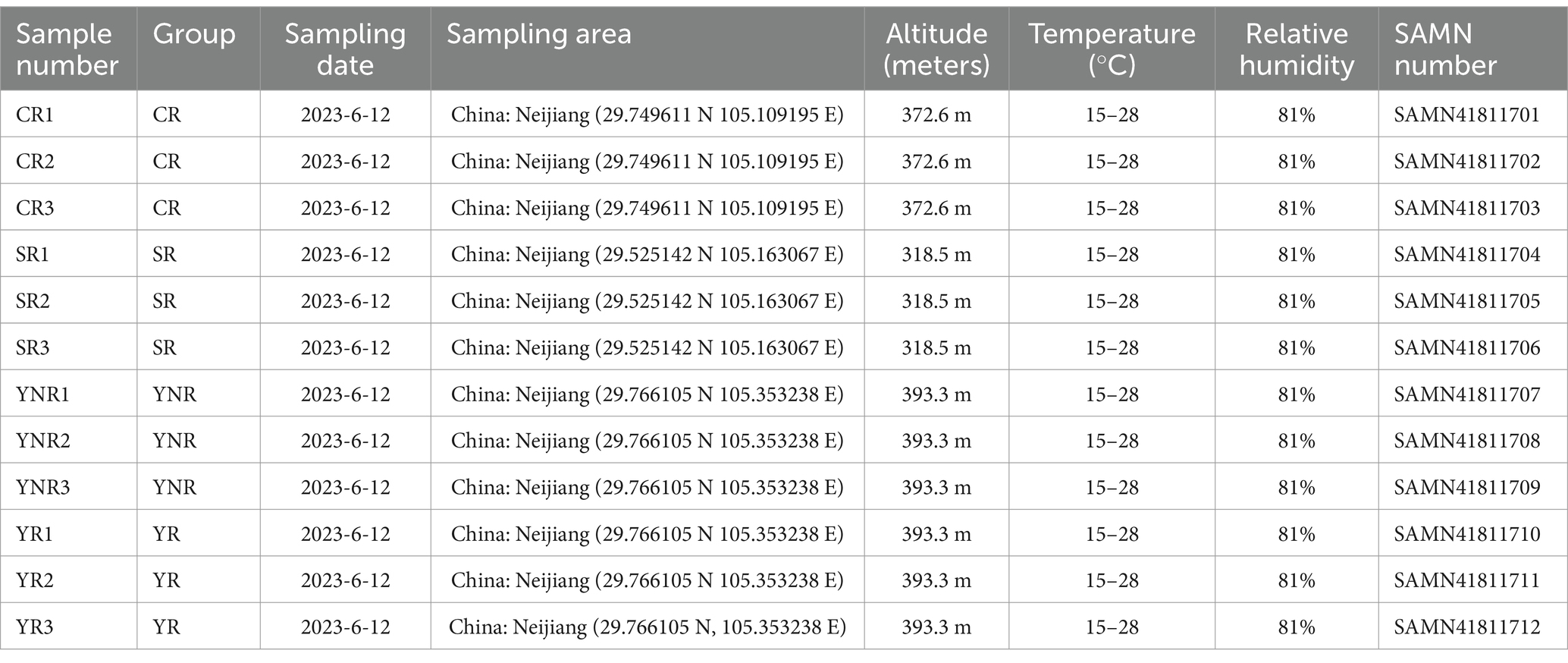
The soil samples were collected from the Neijiang City, Sichuan Province, China. Nine rhizosphere soil samples were collected from Yangjia Town (n = 3, YR), Shuangcai Town (n = 3, CR), and Guobei Town (n = 3, SR), and we also collected three non-rhizosphere soil samples from Yangjia Town (n = 3, YNR). The detailed collection information was listed in Table 1. The rhizosphere areas have a subtropical climate with an average annual temperature ranging from 15 to 28°C. The relative humidity annually averages around 80% and the annual rainfall is about 1,000 mm. The sampling procedure was as follows: approximately 2–3 cm of surface soil was initially removed to filter out plant detritus. Subsequently, rhizosphere soil located within 0.2 cm of the root was filtered through a 100-mesh screen and placed into sterilized bags. The samples were promptly submerged in liquid nitrogen immediately and stored at −80°C.
Determination of the physicochemical properties of the Asparagus cochinchinensis rhizosphere soil
The analysis of pH, total nitrogen, total phosphorus, total potassium, organic carbon, available phosphorus, available potassium, exchangeable calcium, and exchangeable magnesium in the continuous rhizosphere soil samples was conducted as follows. Approximately 10 g of dried soil samples were placed into a 50 mL tube with 25 mL of distilled water and mixed for 1 min. After allowing it to settle for 30 min, pH was measured using a pH meter (PB-10, Sartorius, German). Approximately, 0.6 g of dried soil samples were analyzed using a Seal Auto Analyzer 3 Continuous Flow Analyzer (Germany) for total nitrogen, nitrate nitrogen, and ammonium nitrogen content. Total phosphorus and available phosphorus were quantified using an ultraviolet spectrophotometer (UV-2450, Shimadzu, Japan), while total potassium and available potassium levels were determined using a flame photometer (Shanghai, China). Exchangeable calcium and magnesium content were measured using an atomic absorption spectrophotometer (Hitachi Z-2000, Japan). The experimental procedures followed the methods outlined by Lu (2000).
DNA extraction, metagenomic sequencing and bioinformatic analysis
Approximately 1.0 g of frozen soil sample was transferred into a sterilized 15 mL centrifuge tube containing 1.5 g of glass grinding beads. Bacteria DNA products were extracted as the instruction of the EZNA® Soil DNA Kit (D5625, Omega Bio-Tek., Inc.) manufacturer. Purity and concentration of the extracted DNA were assessed using 2% agarose gel electrophoresis and Nanodrop (ND ONE, Genes Ltd.). A library was prepared using the NEB Next® UltraTM DNA Library Prep Kit for Illumina (NEB, United States). Qualified DNA samples were randomly fragmented into 350 bp fragments using a Covaris (Covaris S2 System, Massachusetts, United States) ultrasonic fragmentation instrument. A complete library was constructed as follows: terminus repair, polyA tailing, sequence linking, purification, and PCR amplification. Finally, the AMPure XP system was used to purify the PCR products, and an Agilent 2100 was used to determine the insert size of the library. The real-time PCR was used for quantitative analysis of the library concentration. The indexed coding samples were clustered on the cBot Cluster Generation System using the Illumina PE Cluster Kit (Illumina, United States) according to the manufacturer’s instructions. Purified DNA samples were stored at −20°C. Subsequently, DNA sequencing was performed on the Illumina NovaSeq 6000 platform. The raw data was deposited to the National Center for Biotechnology Information Sequence Read Archive database with accession numbers SAMN41811701- SAMN41811712.
The quality control and bioinformatics analysis were conducted as follows. The low-quality data was filtered using fastp and Samtools software. Clean sequences were assembled into contigs using MEGAHIT software (v. 1.2.9, −k-list 21, 29, 39, 59, 79, 99, 119, 141 –min-contig-len 500, v1.2.9, Li et al., 2015). The Prodigal package within Prokka (v. 1.14.6) software were employed to predict ORF genes (Seemann, 2014). Predicted proteins encoded by these genes were functionally annotated by blasting against the UniProt, KEGG, and GO database to infer their biological functions (Camacho et al., 2009; Finn et al., 2011). Clean reads were directly determined with Kraken2 (v. 2.1.2) gene sets to identify bacterial species (Wood et al., 2019). To compare the gene abundance differences among different groups, the number of genes were originally standardized. The TPM (Transcripts Per Million) algorithm is used to standardize gene abundance. The DEseq2 was used to analyze the differentially expressed genes between various groups. The criteria for between various groups DEGs were |log2 (FoldChange)| > 1.5 and FDR < 0.01. Relative abundances of these species were calculated using Bracken (Lu et al., 2017). Alpha diversity indices (Chao 1, Shannon, Simpson, and richness) were computed to evaluate bacterial community diversity and richness using USEARCH (v. 10.0.240, Edgar, 2010, 2013). Rarefaction curves were calculated using USEARCH (v. 10.0.240) and bacterial community composition diagrams were generated using R software v.3.3.1 (Jami et al., 2013). The beta diversity was analyzed using the prcomp function in R software (v.3.3.1, Jami et al., 2013) and visualized using the ggplot2 package. To explore the interaction between soil bacterial communities and physicochemical properties, we conducted mantel test (R software) to perform association analysis.
High-throughput isolation and identification of bacteria from the Asparagus cochinchinensis rhizosphere soil
Fresh soil samples from A. cochinchinensis were utilized to isolate and cultivate bacterial strains. Based on the previous reports by Zhang et al. (2018), we used multiple mediums (Tryptic soy broth medium, 02-34, Aobox Biotechnology, Beijing, China; Luria-Bertani medium; Nutrient Broth medium) for bacteria isolation and cultivation. The soil sample was transferred into a sterilized centrifuge tube with 9 mL of DEPC-treated water (cat. BL510B, LABGIC, Beijing, China) and subjected to oscillation cultivation at room temperature for 30 min with 30 rpm/min. Approximately 200 μL of culture supernatants were then diluted with 25 mL of 10 mM magnesium chloride solution and left at room temperature for 15 min to release bacteria. The diluted solution was cultivated into 96-well cell culture plates, with the concentration of diluted solution selected to ensure approximately 30% of the wells exhibited bacterial growth. Cultivation plates were subsequently incubated at room temperature for 14 days. After incubation, DNA products from bacterial strains were extracted using the TIANamp Bacteria DNA Kit (DP302, TIANGEN BIOTECH Co., Ltd., Beijing). Bacterial strains were identified by amplifying the 16S rRNA gene using designed primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGCTACCTTGTTACGACTT-3′, White et al., 1990). Identified high-quality strains were stored in 40% (vol/vol) glycerol at −80°C after three rounds of continuous streaking.
Results
Physicochemical properties of the Asparagus cochinchinensis rhizosphere soil
The results showed that the average organic carbon content in the YR group (7.18 g/kg) was higher than that in the SR (5.84 g/kg) and CR group (5.03 g/kg, Figure 1A). Significant difference in carbon content was observed between the YR and CR groups (p = 0.0093). The exchangeable calcium content was higher in the CR group (4.01 g/kg) than that in the SR (3.27 g/kg) and CR group (3.21 g/kg, Figure 1B). The exchangeable magnesium level in the YR group (0.21 g/kg) was significantly lower than that in the SR group (0.43 g/kg, p = 0.0049) and CR group (0.38 g/kg, p = 0.0014, Figure 1C). The CR group (20.00 mg/kg) had higher available phosphorus contents than the SR group (3.44 mg/kg, p = 0.0034) and CR group (181.00 mg/kg, Figure 1D). The available potassium content in the CR group was higher than that in the SR (122.67 mg/kg) and YR group (159.67 mg/kg, Figure 1E). The total phosphorus content was highest in the SR group (0.84 g/kg), followed by the CR group (0.75 g/kg) and YR group (0.67 g/kg, Figure 1F). The total potassium result showed that the YR group (25.49 g/kg) had the highest level, followed by the CR group (23.02 g/kg) and SR group (17.39 g/kg, Figure 1G). The pH results indicated that all CR and YR samples in this study were alkaline, and SR samples were acidic (Figure 1H). The total nitrogen content in the YR group (0.87 g/kg) was higher than that in the SR group (0.83 g/kg) and CR group (0.64 g/kg, Figure 1I).
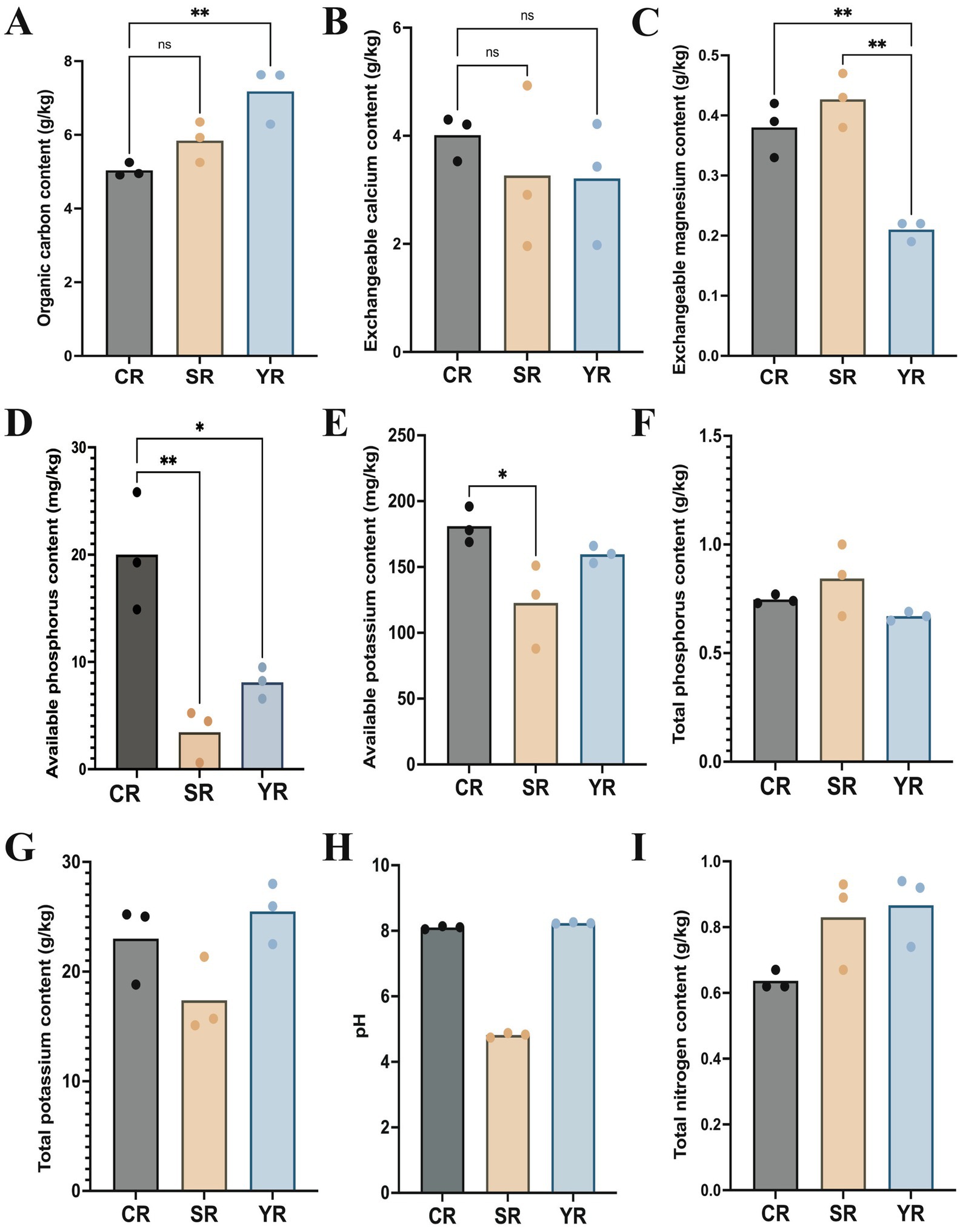
Figure 1. Physicochemical properties of the A. cochinchinensis rhizosphere soil. (A) Average organic carbon contents in the YR, SR, and CR group. (B) Average exchangeable calcium contents in the YR, SR, and CR group. (C) Average exchangeable magnesium contents in the YR, SR, and CR group. (D) Average available phosphorus contents in the YR, SR, and CR group. (E) Average available potassium contents in the YR, SR, and CR group. (F) Total phosphorus contents in the YR, SR, and CR group. (G) Total potassium contents in the YR, SR, and CR group. (H) pH in the YR, SR, and CR group. (I) Total nitrogen contents in the YR, SR, and CR group. * Represents p < 0.05, ** Represents p < 0.01.
Rhizosphere bacterial community in Asparagus cochinchinensis rhizosphere soil through metagenomics technology
Bacterial community composition in Asparagus cochinchinensis rhizosphere soil
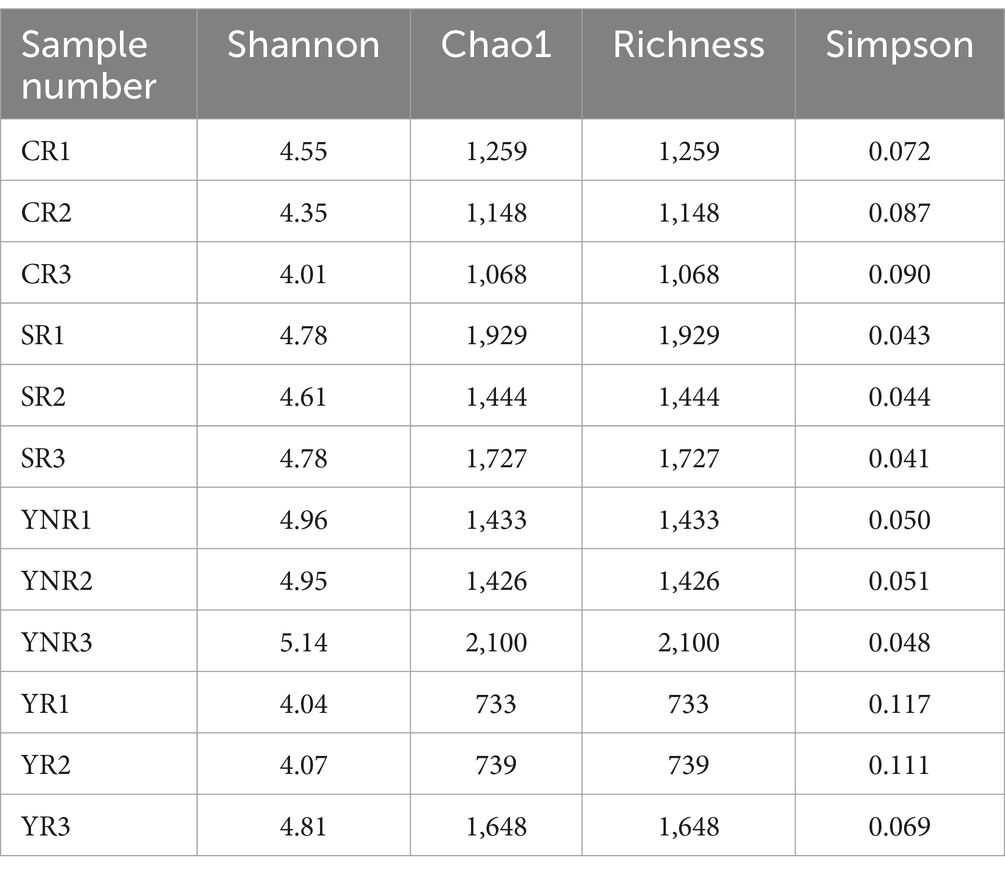
The bacterial community in 12 A. cochinchinensis rhizosphere soil samples was sequenced using high-throughput sequencing technology. A total of 586,290,474 clean reads were obtained from 12 rhizosphere soil samples. Each sample yielded 103,629 assembled contigs. The raw sequences were deposited in the National Center for Biotechnology Information Sequence Read Archive database with accession numbers. Gene assemble results indicated that the contig numbers in each sample ranged from 65,454 to 221,127, all with GC content exceeding 60%. Alpha diversity index (Shannon, Chao1, Richness, and Simpson index) statistical analysis showed that significant differences of Shannon and Simpson indices were observed between various groups based on Anova (Table 2). Taxonomic annotation results showed that a total of 75 phyla, 166 classes, 410 orders, 706 families, 1,565 genera, and 4,424 species were identified in this study. At the phylum level, Proteobacteria, Actinobacteria, and Pseudomonadota were the most abundant with the relative abundances of 0–71.5%, 0–40.8%, and 0–68.7% (Figure 2A). Actinobacteria (0–59.3%), Corynebacteriales (0–19.5%), Actinomycetes (0–43.6%), and Streptomycetaceae (0–12.2%) were dominant at the class, order, and family level, respectively (Figures 2B–D). Mycobacterium (0–34.6%) was the most abundant at the genus level (Figure 2E). At the species level, Oligotropha carboxidovorans (0–34.6%), Burkholderia cenocepacia (0–2.2%), Amycolatopsis mediterranei (0–8.8%), Achromobacter xylosoxidans (0–3.8%), Afipia carboxidovorans (0–11.6%), Sphingomonas wittichii (0–3.1%), Ensifer adhaerens (0–0.9%), Sinorhizobium fredii (0–2.8%), Mesorhizobium japonicum (0–2.0%), Variovorax paradoxus (0–0.2%), Variovorax paradoxus (0–0.2%), Pseudomonas aeruginosa (0–1.1%), Rhodanobacter denitrificans (0–0.3%), and Stenotrophomonas maltophilia (0–3.6%) were dominant among all species (Supplementary Table S1, Figure 2F). These findings provided a comprehensive overview of the bacterial composition in A. cochinchinensis rhizosphere soils.
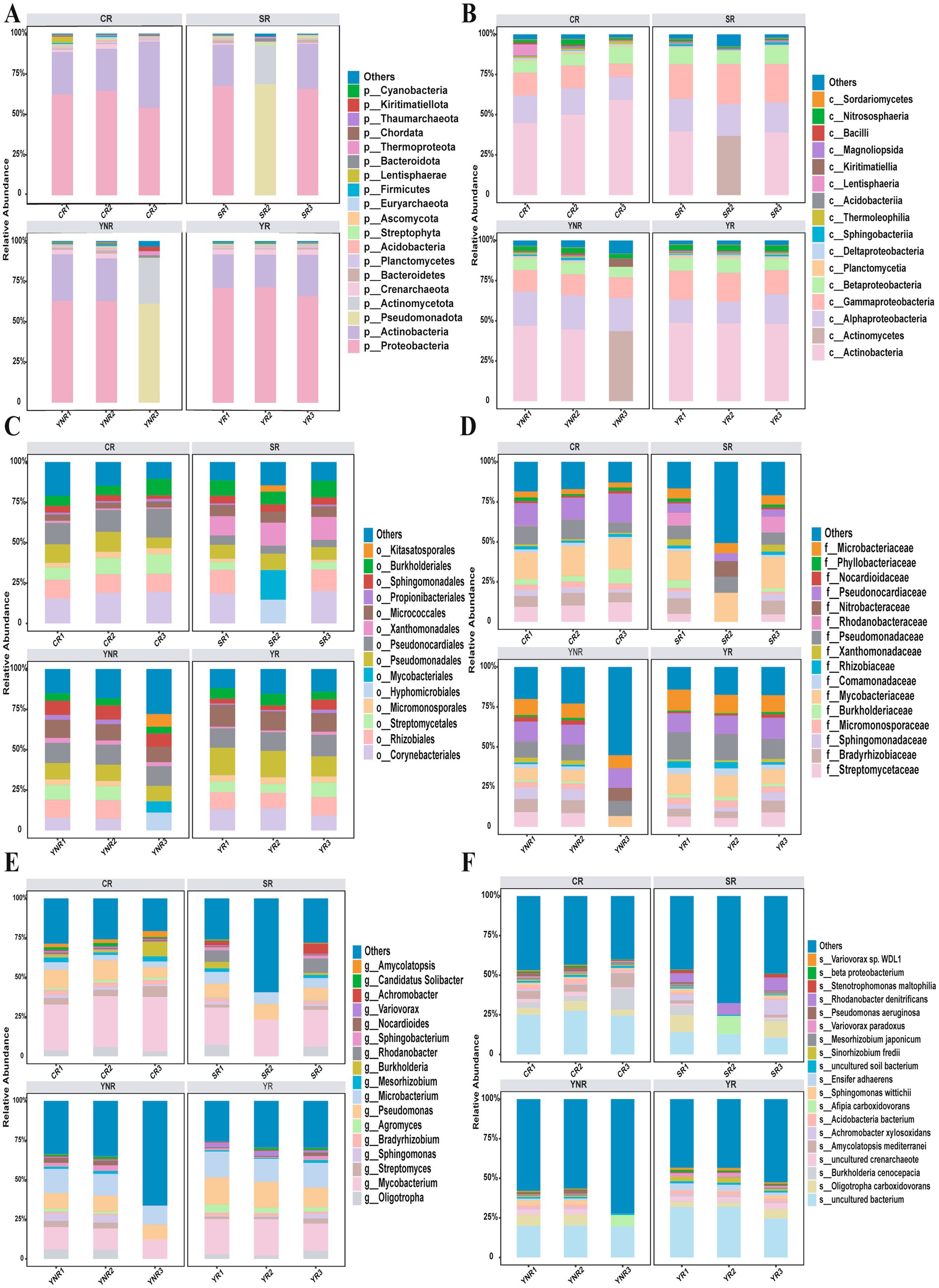
Figure 2. Bacterial community composition in the A. cochinchinensis rhizosphere soil at the (A) Phylum level. (B) Class level. (C) Order level. (D) Family level. (E) Genus level. (F) Species level.
Comparison of bacterial community in different groups based on Asparagus cochinchinensis rhizosphere soil
The rarefaction curve reflected that the sequencing data for each sample was sufficient to estimate the bacterial community (Figure 3A). Meanwhile, PCA and PCoA analysis further confirmed similarity of bacterial community composition across various areas (Figures 3B,D). Additionally, we compared differences of bacterial community between rhizosphere and non-rhizosphere soil groups. The average Chao 1 and Shannon indices in YNR group were higher than those in YR group (Table 2). The differences of bacterial composition were observed. At the genus level, the relative abundance of Escherichia in YR group was higher than those in YNR group. At the species level, YR group showed higher relative abundances of Mycolicibacterium fortuitum, Lactobacillus paracasei, Agrobacterium rhizogenes, Rhizobium tropici, Pseudomonas lini, Acinetobacter johnsonii, Pseudomonas mosselii, and Pseudomonas umsongensis (Supplementary Table S1). Gene expression difference analysis showed that there were 1,338 genes that expressed differently in YR and YNR group. One hundred and eighteen genes were up-regulated in YR group, and 1,220 genes were up-regulated in YNR group (Figure 3C). Differential gene enrichment analysis using KEGG Pathway showed that the genes of bacterial community function in YR group was mainly for plant-pathogen interaction, biotin metabolism, prodigiosin biosynthesis, meiosis-yeast, and pathogenic Escherichia coli infection. The function prediction result using Gene Ontology showed the bacterial community function in YR group was mainly for RNA–directed DNA polymerase activity, RNA–DNA hybrid ribonuclease activity, and cGMP binding (Figures 3E,F).
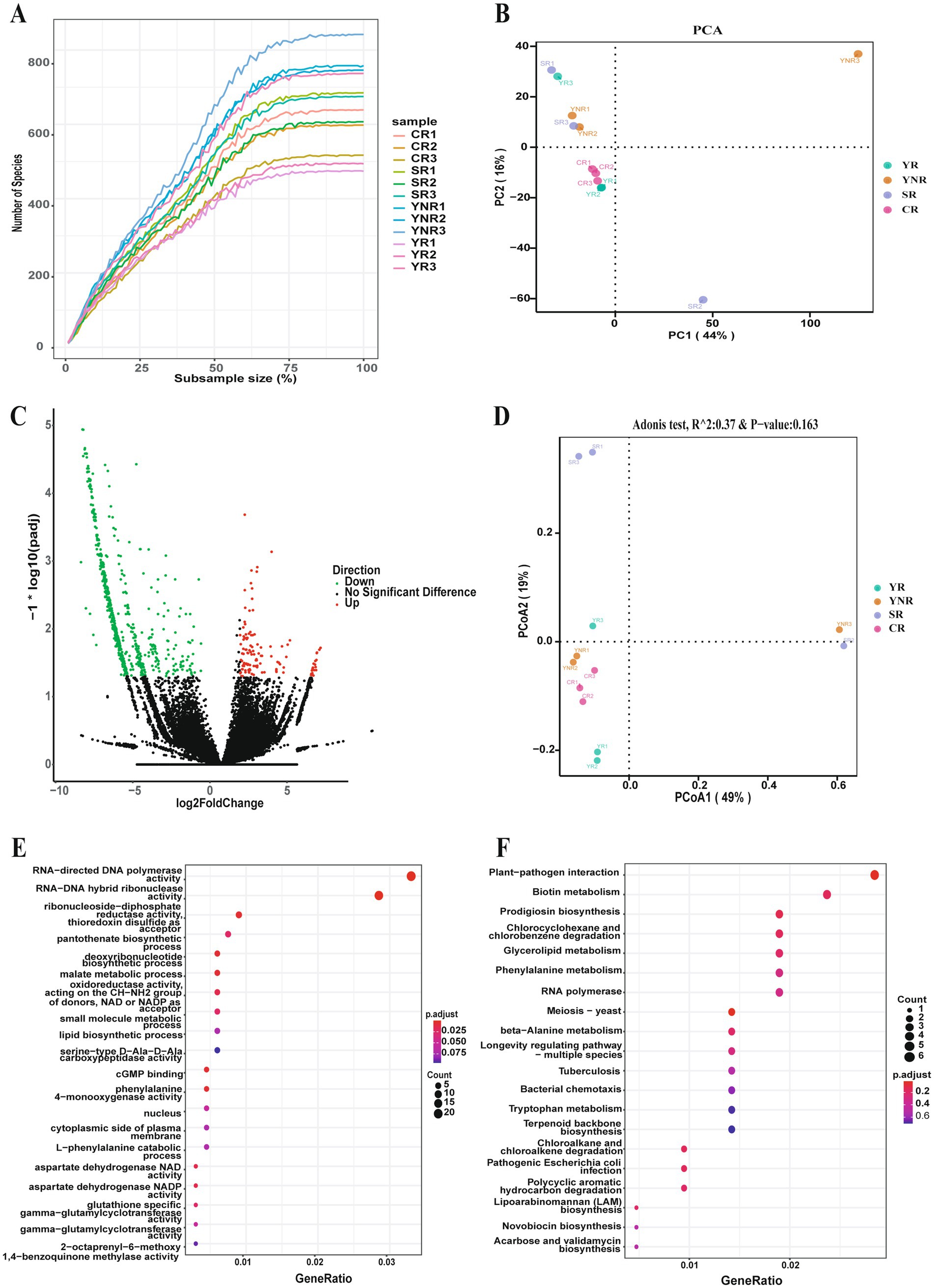
Figure 3. Comparison of the differences of bacterial community between various rhizosphere areas. (A) Rarefaction curve reveals the sequencing depth in each sample. (B) Principal Component Analysis of soil bacterial community among different groups. (C) Differentially expressed genes between various groups. (D) Principal co-ordinates analysis of soil bacterial community among different groups. (E) Gene function prediction of differentially expressed genes based on the Gene Ontology database. (F) Gene function prediction of differentially expressed genes based on the Kyoto Encyclopedia of Genes and Genomes database.
Gene function annotation using various databases
The function of the bacterial community in A. cochinchinensis plantation soil was predicted using the KEGG and GO database. According to enrichment annotation, the bacterial community primarily contributes to global and overview maps, translation processes, and the metabolism of cofactors and vitamins. Gene enrichment annotation revealed that at the biological level, the bacterial community is involved in translation, phosphorelay signal transduction systems, and transmembrane transport. At the cellular level, integral components of membranes, cytoplasm, and plasma membrane played crucial roles. At the molecular level, ATP binding, metal ion binding, and DNA binding were the main function. Based on the COG database, the bacterial community function was predicted to be engaged in amino acid transport and metabolism, as well as energy production and conversion (Figures 4A,B).
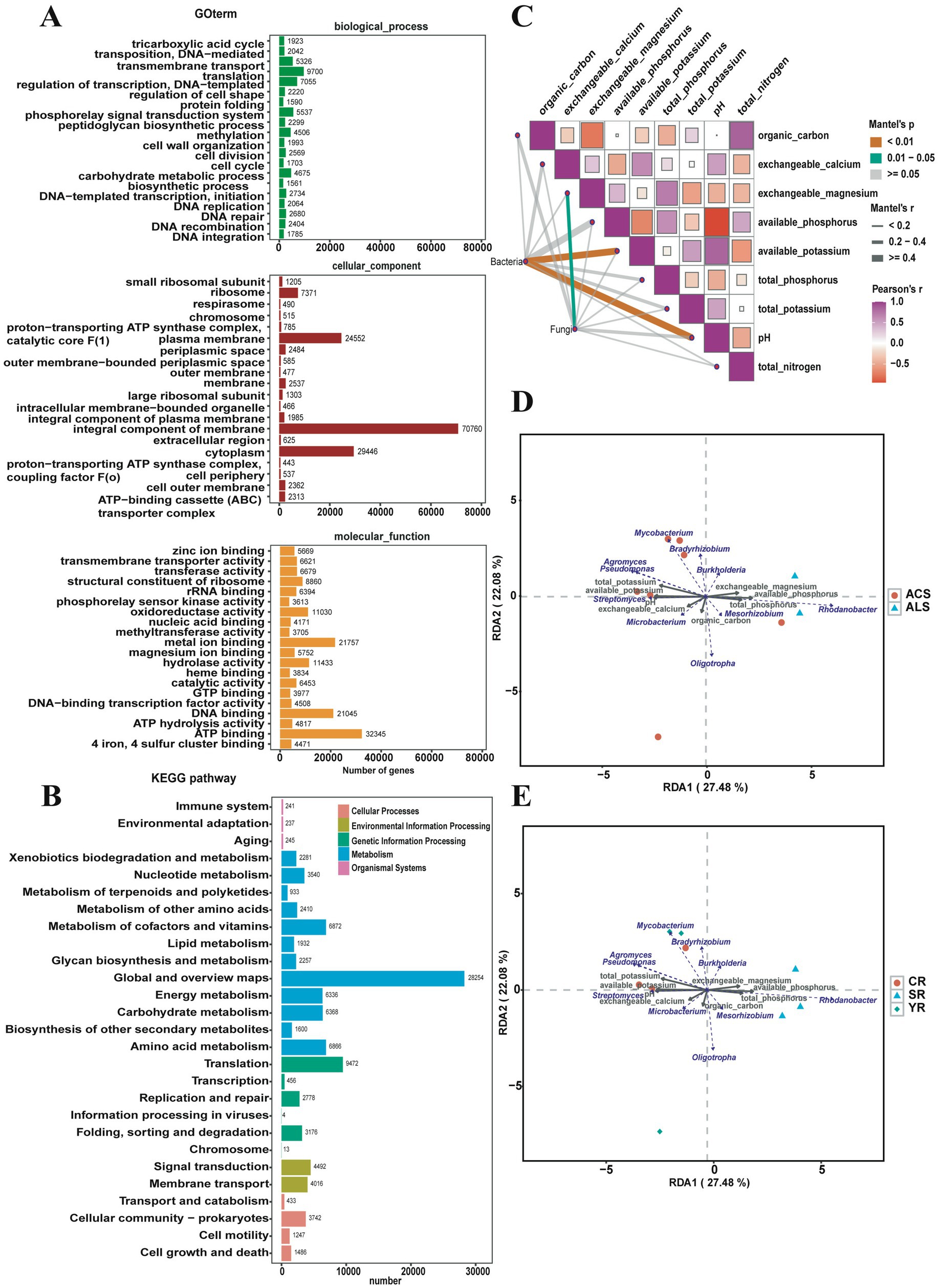
Figure 4. Function prediction of bacterial community and association analysis of physicochemical properties and soil bacterial community. (A) Function prediction of soil bacterial community based on the Gene Ontology database. (B) Function prediction of soil bacterial community based on the Kyoto Encyclopedia of Genes and Genomes database. (C) Association analysis between various physicochemical properties. (D) Mantel test for the analysis of relevance of pH and soil bacterial community. (E) Redundancy Analysis (RDA) of relationships between the relative abundance of top 10 bacterial genera and soil physicochemical factors in the YR, SR, and CR group.
Association analysis between physicochemical properties factors and soil bacterial community composition
To explore the interaction between soil bacterial communities and physicochemical properties, we conducted mantel test (R software) to perform association analysis. The results revealed significant positive correlations (p < 0.01) between bacterial communities and pH, as well as available potassium activities. Additionally, Pearson’s correlation analysis indicated positive relationships among various physicochemical properties. Specifically, total nitrogen correlated positively with organic carbon and available phosphorus indices. pH levels showed positive associations with exchangeable calcium, available potassium, and total potassium indices. Furthermore, total phosphorus levels were positively correlated with exchangeable magnesium levels (Figure 4C). We employed Redundancy Analysis (RDA) to explore relationships between the relative abundance of top 10 bacterial genera and soil physicochemical factors. The results showed that the relative abundance of Pseudomonas exhibited a strong positive correlation with cumulative total potassium and available potassium. The relative abundance of Streptomyces was notably associated with cumulative pH levels, while the relative abundance of Rhodanobacter correlated with cumulative available phosphorus. Additionally, the relative abundance of Burkholderia showed a significant relationship with cumulative exchangeable magnesium levels (Figures 4D,E).
Isolation and identification of bacterial strains from the Asparagus cochinchinensis rhizosphere soil
In this study, we used TSB, LB, and NB medium to isolate and identify the bacterial strains from the A. cochinchinensis rhizosphere soil. The strains were further identified through amplifying 16S rDNA region using the 27F/1492R primer. A total of 103 strains were isolated and identified, encompassing 30 species including Agromyces binzhouensis, Arthrobacter pascens, Bordetella petrii, Aeromicrobium kwangyangensis, Rhodanobacter lindaniclasticus, Pseudarthrobacter oxydans, Marmoricola scoriae, Lysobacter brunescens, Arthrobacter pokkalii, Rhodanobacteraceae bacterium, Caulobacter zeae, Flavobacterium johnsoniae, Roseateles chitosanitabidusm, Caulobacter radicis, Pseudomonas lini, Flavobacterium ajazii, Pseudomonas resinovorans, Pseudoxanthomonas mexicana, Pseudomonas prosekii, Paenibacillus purispatii, Pseudomonas oleovorans, Flavobacterium anhuiense, Caulobacter flavus, Bacillus coreaensis, Shinella kummerowiae, Flavobacterium glycines, Flavobacterium tructae, Arthrobacter humicola, Pseudarthrobacter siccitolerans, and Pseudarthrobacter bacteria (Figure 5; Supplementary Figure S1). All these strains were stored in institute of Herbgenomics, Chengdu University of Traditional Chinese Medicine for further experimentation.
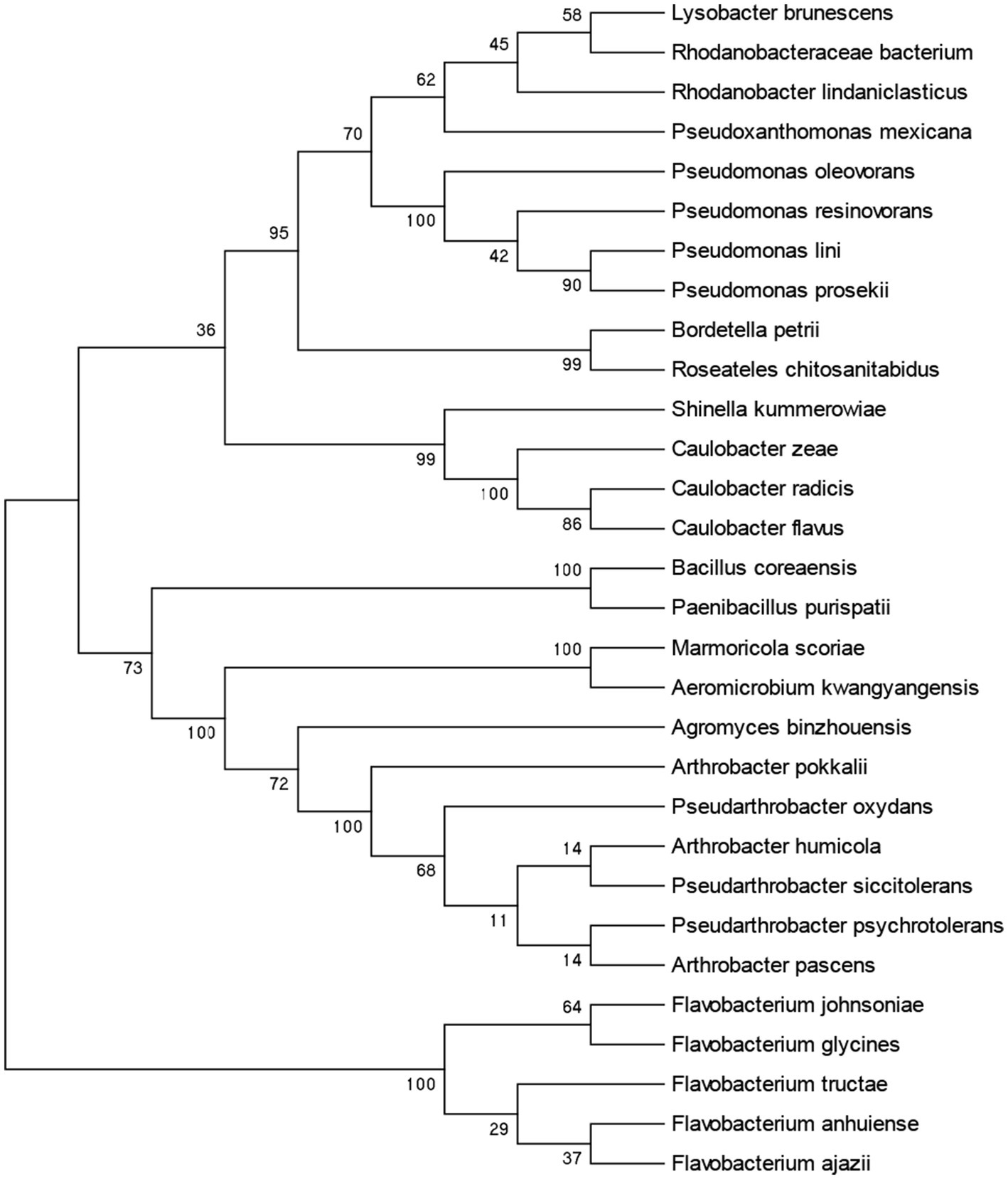
Figure 5. Phylogenetic tree based on 16S rRNA of bacterial strains isolated from A. cochinchinensis rhizosphere soil.
Discussion
Recent studies have extensively investigated the interaction between medicinal plants and soil bacterial communities. It has been reported that soil bacterial community might influence the metabolism of natural products in medicinal plants. However, the detailed mechanisms of these interactions require further exploration. Su et al. (2023) investigated the contribution of soil condition and microbiome in the accumulation of monoterpenes in the fruit of Citrus reticulata “Chachi” and they indicated that the rhizosphere microbiome enhanced terpene synthesis and increased the monoterpene level by affecting the host immune system. Zhang W. et al. (2024) conducted amplicon sequencing and metabolic profiling to analyze the relationship between soil microbiome and bisbenzylisoquinoline alkaloids in Phellodendron amurense. Results showed that there were seven bisbenzylisoquinoline alkaloids positively correlated with Nectriaceae and Sphingobacteriaceae. Similarly, Liu et al. (2022) reported direct regulation of glycyrrhizin and glycyrrhizic acid accumulation in licorice plants by the rhizosphere microbiome. Secondly, the soil microbiome might assist host medicinal plant in growth and resistance to biotic and abiotic stress. Astragalus mongolicus Bunge is a well-known medicinal plant and its cultivation requires amounts of nutrients. Shi et al. (2023) isolated and identified 86 strains form the root and rhizosphere soil samples of A. mongolicus and demonstrated that Bacillus sp. J1, Arthrobacter sp. J2, and Bacillus sp. G4 promoted the accumulation of nitrogen, phosphorus, and potassium content in A. mongolicus. Li et al. (2024) evaluated the effect of Paenibacillus polymyxa 7250 in combination with the symbiotic bacteria on ginseng. Results showed that this treatment improved ginseng yield and disease resistance through increasing Rhodanobacter, Pseudolabrys, and Gemmatimonas. Under low nitrogen conditions, Jiang et al. (2024) reported that the beneficial plant growth-promoting rhizobacteria improved the nitrogen utilization efficiency and regulated the synthesis of target furanocoumarins of Angelica dahurica var. formosana. In this study, we revealed the bacterial community of A. cochinchinensis rhizosphere soil through Illumina NovaSeq 6000 platform. Based on our results, Mycobacterium, Pseudomonas, Microbacterium, Agromyces, and Bradyrhizobium were the main genera in the soil samples. KEGG function prediction results showed that these genera were related to the immune system and environmental adaption. Several of these genera have been reported as beneficial microorganisms for plants (Cao et al., 2023; De Maria et al., 2011; Gao et al., 2024; Tsavkelova et al., 2024). Thus, to deepen our understanding of the interaction between these beneficial microorganisms, pathogens, and A. cochinchinensis, we isolated these strains from the A. cochinchinensis rhizosphere soil samples for further investigation. Based on the previous studies, 11 of these strains have been reported as functional rhizosphere bacterial strains. Benmrid et al. (2024) investigated the beneficial effect of multiple rhizobacteria on the wheat growth under drought conditions, and results showed that the incubation with A. pascens increased dry weight of shoot and root of wheat plants. Additionally, A. pascens had the ability to assisting host plants to resist soil salinity stress and enhance physicochemical properties (Guan et al., 2023; Ullah and Bano, 2015). A. kwangyangensis, A. humicola, C. flavus, P. oxydans, F. anhuiense, and P. lini were reported as biocontrol strains to promote the growth of multiple plants (Elshafie and Camele, 2022; Giannelli et al., 2024; Kim M. et al., 2017; Kim N. G. et al., 2017; Jeong et al., 2019; Muñoz Torres et al., 2021; Tian et al., 2024). Meanwhile, A. pokkalii, P. lini, C. flavus and B. coreaensis had the potential to resisting biotic and abiotic stresses (Anand et al., 2023; Barbaccia et al., 2022; Krishnan et al., 2016; Zhou et al., 2021).
In this study, we used a high-throughput isolation and identification method to enrich the bacterial strains from the A. cochinchinensis rhizosphere soil. This method was initially constructed based on the reports by Zhang J. et al. (2021) we further optimized and adjusted this method based on the characteristics of A. cochinchinensis rhizosphere soil. Previous applications of this method have successfully been adapted for diverse multiple conditions such as deep vadose zone and plant roots (Zhang L. et al., 2024; Zhang W. et al., 2024; Jin et al., 2022). However, the disadvantages of this method should be optimized in the further studies. On one hand, single medium was difficult to comprehensively isolate bacterial strains in the soil sample, therefore, we employed three media to isolate and identify strains in this work. We considered to use more media in further studies. On the other, we mainly focused on the aerobic bacteria, while the isolation and identification of anaerobic bacteria requires to be considered in the further study. In conclusion, continuous optimization of this method continuously to explore the mechanisms is crucial for advancing our understanding of soil microorganisms and their impact on the growth and development of medicinal plants.
The combined utilization of metagenomics and culturomics technologies revealed the rhizosphere bacterial community of A. cochinchinensis. In recent years, more and more reports have applied both technologies simultaneously to study the microbiome in various aspects including gut microbiome, environmental microbiome, and food microbiome (Lee et al., 2022; Li et al., 2023; Vacca et al., 2024). Compared with using metagenomics or culturomics technology alone, the combined application overcomes certain disadvantages. Firstly, the metagenomics sequencing data may provide references for the selection of mediums of culturomics. Almost all microorganisms (culture-dependent and culture-independent) might be identified through the metagenomics sequencing technology, researchers can select the relevant mediums to isolate and identify the specific strain for further studies. In this study, the selection of mediums was also based on the metagenomic results, which Mycobacterium, Pseudomonas, Microbacterium, Agromyces, and Bradyrhizobium were the main genera in this study. Moreover, the interaction network analysis and community function prediction result based on metagenome is an important indicator for the construction of synthetic microbial community in culturomics. Li et al. (2021) analyzed the rhizosphere and root bacterial community in Astragalus mongholicus and constructed a disease-resistant bacterial community based on the metagenome result through the culturomics platform. Results demonstrated that the synthetic bacterial community suppressed the root rot disease of A. mongholicus infected by Fusarium oxysporum and promoted the growth of A. mongholicus. Secondly, due to bias in metagenomic sequencing data, culturomics has the ability of correcting this bias. Culturomics can not only be used to isolate and identify microorganisms, but it also has the potential to discovering new taxa and reduce unassigned operational taxonomy units based on metagenomic analysis (Lagier et al., 2018). Additionally, the strains isolated through the culturomics platform can be studied through multiple detection methods (e.g., genomics, transcriptomics, proteomics, and metabolomics technology). Therefore, the combined utilization of metagenomics and culturomics technologies in microbiome studies has been widely recognized. The application of these two technologies provides both genome data and strain resources simultaneously. In future, the combined utilization of both technologies will provide more comprehensive and systematical insights for microbiome studies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JY: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. SY: Formal analysis, Investigation, Writing – original draft. XZ: Funding acquisition, Methodology, Resources, Writing – original draft. XL: Funding acquisition, Methodology, Resources, Writing – original draft. XT: Funding acquisition, Methodology, Resources, Writing – original draft. LW: Funding acquisition, Methodology, Resources, Writing – original draft. JC: Funding acquisition, Methodology, Resources, Writing – original draft. HL: Funding acquisition, Methodology, Resources, Writing – original draft. CL: Formal analysis, Writing – original draft. CS: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by introduces the talented person scientific research start funds subsidization project of Chengdu University of Traditional Chinese Medicine (030040015).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1467864/full#supplementary-material
References
Anand, V., Kaur, J., Srivastava, S., Bist, V., Dharmesh, V., Kriti, K., et al. (2023). Potential of methyltransferase containing Pseudomonas oleovorans for abatement of arsenic toxicity in rice. Sci. Total Environ. 856:158944. doi: 10.1016/j.scitotenv.2022.158944
Barbaccia, P., Gaglio, R., Dazzi, C., Miceli, C., Bella, P., Lo, P., et al. (2022). Plant growth-promoting activities of Bacteria isolated from an anthropogenic soil located in Agrigento Province. Microorganisms 10:2167. doi: 10.3390/microorganisms10112167
Benmrid, B., Ghoulam, C., Ammar, I., Nkir, D., Saidi, R., Staropoli, A., et al. (2024). Drought-tolerant rhizobacteria with predicted functional traits enhanced wheat growth and P uptake under moderate drought and low P-availability. Microbiol. Res. 285:127795. doi: 10.1016/j.micres.2024.127795
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Cao, Y., Du, P., Zhang, J., Ji, J., Xu, J., and Liang, B. (2023). Dopamine alleviates cadmium stress in apple trees by recruiting beneficial microorganisms to enhance the physiological resilience revealed by high-throughput sequencing and soil metabolomics. Hortic. Res. 10:uhad112. doi: 10.1093/hr/uhad112
Chinese Pharmacopeia Commission (2020). Pharmacopoeia of People’s Republic of China. (2020). Part 1. Beijing: Chemical Industry Press, 56–57.
Csorba, C., Rodić, N., Zhao, Y., Antonielli, L., Brader, G., Vlachou, A., et al. (2022). Metabolite production in Alkanna tinctoria links plant development with the recruitment of individual members of microbiome thriving at the root-soil interface. mSystems 7:e0045122. doi: 10.1128/msystems.00451-22
De Maria, S., Rivelli, A. R., Kuffner, M., Sessitsch, A., Wenzel, W. W., Gorfer, M., et al. (2011). Interactions between accumulation of trace elements and macronutrients in Salix caprea after inoculation with rhizosphere microorganisms. Chemosphere 84, 1256–1261. doi: 10.1016/j.chemosphere.2011.05.002
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Elshafie, H. S., and Camele, I. (2022). Rhizospheric Actinomycetes revealed antifungal and plant-growth-promoting activities under controlled environment. Plants (Basel) 11:1872. doi: 10.3390/plants11141872
Feng, W. M., Liu, P., Yan, H., Zhang, S., Shang, E. X., Yu, G., et al. (2021). Impact of Bacillus on phthalides accumulation in Angelica sinensis (Oliv.) by stoichiometry and microbial diversity analysis. Front. Microbiol. 11:611143. doi: 10.3389/fmicb.2020.611143
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39 (Web Server issue), W29–W37. doi: 10.1093/nar/gkr367
Gao, H., Guo, Z., He, X., Yang, J., Jiang, L., Yang, A., et al. (2024). Stress mitigation mechanism of rice leaf microbiota amid atmospheric deposition of heavy metals. Chemosphere 362:142680. doi: 10.1016/j.chemosphere.2024.142680
Giannelli, G., del Vecchio, L., Cirlini, M., Gozzi, M., Gazza, L., Galaverna, G., et al. (2024). Exploring the rhizosphere of perennial wheat: potential for plant growth promotion and biocontrol applications. Sci. Rep. 14:22792. doi: 10.1038/s41598-024-73818-6
Guan, T. K., Wang, Q. Y., Li, J. S., Yan, H. W., Chen, Q. J., Sun, J., et al. (2023). Biochar immobilized plant growth-promoting rhizobacteria enhanced the physicochemical properties, agronomic characters and microbial communities during lettuce seedling. Front. Microbiol. 14:1218205. doi: 10.3389/fmicb.2023.1218205
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Jeong, J. J., Sajidah, S., Oh, J. Y., Sang, M. K., Kim, K. S., and Kim, K. D. (2019). Complete genome sequence data of Flavobacterium anhuiense strain GSE09, a volatile-producing biocontrol bacterium isolated from cucumber (Cucumis sativus) root. Data Brief 25:104270. doi: 10.1016/j.dib.2019.104270
Jiang, Y., Zhang, Y., Liu, Y., Zhang, J., Jiang, M., Nong, C., et al. (2024). Plant growth-promoting rhizobacteria are key to promoting the growth and furanocoumarin synthesis of Angelica dahurica var. formosana under low-nitrogen conditions. J. Agric. Food Chem. 72, 6964–6978. doi: 10.1021/acs.jafc.3c09655
Jin, X., Bai, Y., Khashi, U., Rahman, M., Kang, X., Pan, K., et al. (2022). Biochar stimulates tomato roots to recruit a bacterial assemblage contributing to disease resistance against Fusarium wilt. iMeta 1:e37. doi: 10.1002/imt2.37
Kim, M., Kim, W. B., Koo, K. Y., Kim, B. R., Kim, D., Lee, S., et al. (2017). Optimal fermentation conditions of hyaluronidase inhibition activity on Asparagus cochinchinensis Merrill by Weissella cibaria. J. Microbiol. Biotechnol. 27, 701–708. doi: 10.4014/jmb.1611.11051
Kim, N. G., Seo, E. Y., Han, S. H., Gong, J. S., Park, C. N., Park, H. S., et al. (2017). Pseudomonas oleovorans strain KBPF-004 culture supernatants reduced seed transmission of cucumber green mottle mosaic virus and pepper mild mottle virus, and remodeled aggregation of 126 kDa and subcellular localization of movement protein of pepper mild mottle virus. Plant Pathol. J. 33, 393–401. doi: 10.5423/PPJ.OA.03.2017.0047
Krishnan, R., Menon, R. R., Tanaka, N., Busse, H. J., Krishnamurthi, S., and Rameshkumar, N. (2016). Arthrobacter pokkalii sp nov, a novel plant associated Actinobacterium with plant beneficial properties, isolated from saline tolerant Pokkali Rice, Kerala, India. PLoS One 11:e0150322. doi: 10.1371/journal.pone.0150322
Lagier, J. C., Dubourg, G., Million, M., Cadoret, F., Bilen, M., Fenollar, F., et al. (2018). Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550. doi: 10.1038/s41579-018-0041-0
Lee, W. J., Ryu, S., Kang, A. N., Song, M., Shin, M., Oh, S., et al. (2022). Molecular characterization of gut microbiome in weaning pigs supplemented with multi-strain probiotics using metagenomic, culturomic, and metabolomic approaches. Anim. Microbiome 4:60. doi: 10.1186/s42523-022-00212-w
Lee, H. A., Song, B. R., Kim, H. R., Kim, J. E., Yun, W. B., Park, J. J., et al. (2017). Butanol extracts of Asparagus cochinchinensis fermented with Weissella cibaria inhibit iNOS-mediated COX-2 induction pathway and inflammatory cytokines in LPS-stimulated RAW264.7 macrophage cells. Exp. Ther. Med. 14, 4986–4994. doi: 10.3892/etm.2017.5200
Li, Z., Bai, X., Jiao, S., Li, Y., Li, P., Yang, Y., et al. (2021). A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant-induced systemic resistance. Microbiome 9:217. doi: 10.1186/s40168-021-01169-9
Li, S., Lian, W. H., Han, J. R., Ali, M., Lin, Z. L., Liu, Y. H., et al. (2023). Capturing the microbial dark matter in desert soils using culturomics-based metagenomics and high-resolution analysis. NPJ Biofilms Microbiomes 9:67. doi: 10.1038/s41522-023-00439-8
Li, X., Liu, Q., Gao, Y., Zang, P., and Zheng, T. (2024). Effects of a co-bacterial agent on the growth, disease control, and quality of ginseng based on rhizosphere microbial diversity. BMC Plant Biol. 24:647. doi: 10.1186/s12870-024-05347-3
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Lian, W. H., Mohamad, O. A. A., Dong, L., Zhang, L. Y., Wang, D., Liu, L., et al. (2023). Culturomics- and metagenomics-based insights into the microbial community and function of rhizosphere soils in Sinai desert farming systems. Environ. Microbiome 18:4. doi: 10.1186/s40793-023-00463-3
Liu, Y., Li, D., Gao, H., Li, Y., Chen, W., Jiao, S., et al. (2022). Regulation of soil micro-foodwebs to root secondary metabolites in cultivated and wild licorice plants. Sci. Total Environ. 828:154302. doi: 10.1016/j.scitotenv.2022.154302
Lu, R. K. (2000). Soil agricultural chemistry analytical methods. Beijing: Chinese Agricultural Science and Technology Press.
Lu, J., Breitwieser, F., Thielen, P., and Salzberg, S. (2017). Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3:e104. doi: 10.7717/peerj-cs.104
Muñoz Torres, P., Cárdenas, S., Arismendi Macuer, M., Huanacuni, N., Huanca-Mamani, W., Cifuentes, D., et al. (2021). The Endophytic Pseudomonas sp. S57 for plant-growth promotion and the biocontrol of Phytopathogenic Fungi and nematodes. Plants (Basel) 10:1531. doi: 10.3390/plants10081531
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shi, Z., Guo, X., Lei, Z., Wang, Y., Yang, Z., Niu, J., et al. (2023). Screening of high-efficiency nitrogen-fixing bacteria from the traditional Chinese medicine plant Astragalus mongolicus and its effect on plant growth promotion and bacterial communities in the rhizosphere. BMC Microbiol. 23:292. doi: 10.1186/s12866-023-03026-1
Su, J., Wang, Y., Bai, M., Peng, T., Li, H., Xu, H. J., et al. (2023). Soil conditions and the plant microbiome boost the accumulation of monoterpenes in the fruit of Citrus reticulata 'Chachi'. Microbiome 11:61. doi: 10.1186/s40168-023-01504-2
Tian, T., Gheysen, G., Kyndt, T., Mo, C., Xiao, X., Lv, Y., et al. (2024). Pepper root exudate alleviates cucumber root-knot nematode infection by recruiting a rhizobacterium. Plant Commun. 30:101139. doi: 10.1016/j.xplc.2024
Tsavkelova, E. A., Volynchikova, E. A., Potekhina, N. V., Lavrov, K. V., and Avtukh, A. N. (2024). Auxin production and plant growth promotion by Microbacterium albopurpureum sp. nov. from the rhizoplane of leafless Chiloschista parishii Seidenf. Orchid. Front. Plant Sci. 15:1360828. doi: 10.3389/fpls.2024.1360828
Ullah, S., and Bano, A. (2015). Isolation of plant-growth-promoting rhizobacteria from rhizospheric soil of halophytes and their impact on maize (Zea mays L.) under induced soil salinity. Can. J. Microbiol. 61, 307–313. doi: 10.1139/cjm-2014-0668
Vacca, M., Celano, G., Serale, N., Costantino, G., Calabrese, F. M., Calasso, M., et al. (2024). Dynamic microbial and metabolic changes during Apulian Caciocavallo cheesemaking and ripening produced according to a standardized protocol. J. Dairy Sci. 107, 6541–6557. doi: 10.3168/jds.2023-24049
Wang, M., Wang, S., Hu, W., Wang, Z., Yang, B., and Kuang, H. (2022). Asparagus cochinchinensis: a review of its botany, traditional uses, phytochemistry, pharmacology, and applications. Front. Pharmacol. 13:1068858. doi: 10.3389/fphar.2022.1068858
Wei, X., Cao, P., Wang, G., and Han, J. (2020). Microbial inoculant and garbage enzyme reduced cadmium (cd) uptake in Salvia miltiorrhiza (Bge.) under cd stress. Ecotoxicol. Environ. Saf. 192:110311. doi: 10.1016/j.ecoenv.2020.110311
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR protocols a guide to methods and applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Wood, D. E., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0
Yu, J., Zheng, Y., Song, C., and Chen, S. (2024). New insights into the roles of fungi and bacteria in the development of medicinal plant. J. Adv. Res. 65, 137–152. doi: 10.1016/j.jare.2023.12.007
Zhang, W., Gao, R., Tian, L., and Xu, Z. (2024). Integrated microbiome and metabolomics analysis reveal the relationship between plant-specialized metabolites and microbial community in Phellodendron amurense. Front. Plant Sci. 15:1363063. doi: 10.3389/fpls.2024.1363063
Zhang, L., He, F., Gao, L., Cong, M., Sun, J., Xu, J., et al. (2021). Engineering exosome-like nanovesicles derived from Asparagus cochinchinensis can inhibit the proliferation of hepatocellular carcinoma cells with better safety profile. Int. J. Nanomedicine 16, 1575–1586. doi: 10.2147/IJN.S293067
Zhang, J., Liu, Y. X., Guo, X., Qin, Y., Garrido-Oter, R., Schulze-Lefert, P., et al. (2021). High-throughput cultivation and identification of bacteria from the plant root microbiota. Nat. Protoc. 16, 988–1012. doi: 10.1038/s41596-020-00444-7
Zhang, J., Zhang, N., Liu, Y. X., Zhang, X., Hu, B., Qin, Y., et al. (2018). Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci. China Life Sci. 61, 613–621. doi: 10.1007/s11427-018-9284-4
Zhang, L., Zhao, H., Qin, S., Hu, C., Shen, Y., Qu, B., et al. (2024). Genome-resolved metagenomics and denitrifying strain isolation reveal new insights into microbial denitrification in the deep vadose zone. Environ. Sci. Technol. 58, 2323–2334. doi: 10.1021/acs.est.3c06466
Keywords: medicinal plant, Asparagus cochinchinensis, soil bacterial community, metagenomics, culturomics
Citation: Yu J, Yang S, Zhang X, Liu X, Tang X, Wang L, Chen J, Luo H, Liu C and Song C (2024) Integrating metagenomics and culturomics to uncover the soil bacterial community in Asparagus cochinchinensis cultivation. Front. Microbiol. 15:1467864. doi: 10.3389/fmicb.2024.1467864
Edited by:
Durgesh K. Jaiswal, Graphic Era University, IndiaReviewed by:
Yong-Xin Liu, Agricultural Genomics Institute at Shenzhen (CAAS), ChinaSudipta Sankar Bora, Assam Agricultural University, India
Copyright © 2024 Yu, Yang, Zhang, Liu, Tang, Wang, Chen, Luo, Liu and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chi Song, c29uZ2NoaUBjZHV0Y20uZWR1LmNu
†These authors have contributed equally to this work
 Jingsheng Yu
Jingsheng Yu Shuai Yang
Shuai Yang Xiaoyong Zhang2,3
Xiaoyong Zhang2,3 Chi Song
Chi Song