- 1Laboratory of Biotechnology, Department of Microbiology, Agricultural Research Center (ARC), Animal Health Research Institute (AHRI), Zagazig, Egypt
- 2Department of Pharmaceutics, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 3Botany and Microbiology Department, Faculty of Science, Benha University, Benha, Egypt
- 4Pharmaceutical Medicinal Chemistry and Drug Design Department, Faculty of Pharmacy (Boys), Al-Azhar University, Cairo, Egypt
- 5Epidemiological Surveillance Unit, Aweash El-Hagar Family Medicine Center, MOHP, Mansoura, Egypt
Background: Multidrug-resistant Klebsiella pneumoniae (MDR-KP) poses a significant global health threat due to its involvement in severe infections and high mortality rates. The emergence of MDR strains necessitates the exploration of alternative therapeutic strategies.
Methods: K. pneumoniae isolates were obtained from human and animal sources. Antibacterial susceptibility testing was performed, followed by the evaluation of essential oil activity through inhibition zone, MIC, and MBC determinations. Checkerboard assays were conducted to assess synergistic effects with amikacin. Gene expression analysis and transmission electron microscopy were employed to elucidate the mechanisms of action. Molecular docking studies were performed to identify potential binding targets of bioactive compounds.
Results: Klebsiella pneumoniae was isolated from 25 of the100 samples examined, representing a prevalence rate of 25%. All isolates were found to be multidrug-resistant. Tea tree and thyme essential oils exhibited potent antibacterial activity and synergistic effects with amikacin. Notably, these combinations significantly downregulated the expression of key capsule virulence genes (wcaG, rmpA, magA, uge, and wabG), suggesting a novel mechanism for enhancing amikacin efficacy. Transmission electron microscopy revealed disrupted cell integrity in MDR-KP cells treated with the combinations. Molecular docking analysis identified Terpinen-4-ol, Farnesol, 1,4-Dihydroxy-p-menth-2-ene, and 7-Oxabicyclo [4.1.0] heptane as potential bioactive compounds responsible for the observed effects.
Conclusion: By effectively combating MDR-KP, this research holds promise for reducing antibiotic resistance, improving treatment outcomes, and ultimately enhancing potential care.
Introduction
An opportunistic bacterium known as Klebsiella pneumoniae infects hospitalized or immunocompromised patients (Gorrie et al., 2017). Similarly, in animals, such as bovine, K. pneumoniae is a major causative agent of mastitis, leading to economic losses and public health concerns (Opoku-Temeng et al., 2019; Yang et al., 2019; El-Demerdash et al., 2023b). This rod-shaped, encapsulated, non-motile member of the Enterobacteriaceae family is gram-negative (Nasr et al., 2016).
In medical settings, multidrug-resistant Klebsiella pneumoniae (MDR-KP) is a significant contributor to the high rates of morbidity and death in patients with severe infections (Lee et al., 2016). Infections caused by MDR-KP strains are increasingly common worldwide (Abdelaziz et al., 2013). This issue may have originated in underdeveloped nations due to the overuse of antibiotics, incorrect prescriptions, unnecessary testing, and medication abuse (Nordmann et al., 2009; Shalaby et al., 2021; Megahed et al., 2023).
The global emergence of MDR-KP, often causing hospital-acquired infections, necessitates new therapeutic strategies (Davies and Davies, 2010; Essawi et al., 2020; Abd El-Emam et al., 2023; Hashem et al., 2024). The spread of strains producing extended-spectrum beta-lactamases (ESBLs) and carbapenems underscores the urgent need for alternative treatments (Hawkey et al., 2018; Ebrahem et al., 2023). Essential oils (EOs), volatile compounds extracted from plants, have emerged as promising antimicrobial agents due to their diverse mechanisms of action, including disruption of cell membranes, inhibition of protein synthesis, and interference with DNA replication (Birhan et al., 2011; Rybicki et al., 2012; Dhama et al., 2015).
Numerous investigations have demonstrated that specific phytochemicals, like those found in EOs, and antibiotics, when used together, produce synergistic effects. These combinations can inhibit folate biosynthesis, DNA/protein synthesis, and disrupt cell permeability and cell wall (Dhami, 2013). Additionally, EOs can further weaken bacteria by preventing them from synthesizing essential macromolecules like DNA, RNA, proteins, and polysaccharides (Konaté et al., 2012). This multi-pronged attack offered by the combined approach proves to be highly beneficial in combating multidrug-resistant (MDR) bacteria. By targeting multiple bacterial processes simultaneously, EOs and antibiotics can effectively overcome resistance mechanisms employed by MDR pathogens, leading to more efficient treatment (Abd El-Kalek and Mohamed, 2012).
Previous research has explored the antibacterial properties of EOs against Klebsiella pneumoniae (Mohamed et al., 2018; Diniz et al., 2023). However, no previous studies have examined the ability of essential oils to specifically target the capsule of K. pneumoniae, a crucial virulence factor associated with immune evasion.
The capsule that surrounds the surface of K. pneumoniae serves as the primary virulence factor associated with its viscous phenotype. It typically provides a defensive rather than offensive resistance to bactericidal activities (Zhu et al., 2021). K. pneumoniae utilizes capsules to prevent bacteria from binding, thus evading phagocytosis, complement, antimicrobial peptides, and specific antibodies. However, instances of active suppression and attack on immune cells through capsules are rarely reported (Paczosa and Mecsas, 2016). The chromosomal capsular polysaccharide synthesis (cps) region encodes the genes responsible for capsule production. The cps cluster comprises 27 genes, including rmpA, wcaG, magA, uge, and wabG (Ernst et al., 2020).
Therefore, this study addresses this gap by investigating the efficacy of EOs in downregulating capsule gene expression in MDR-KP. We employ a novel approach, utilizing specifically designed primers to quantify the downregulation of capsule genes in MDR-KP strains treated with essential oils. This study also incorporates microbiological assays and in silico analysis to provide a comprehensive understanding of the potential mechanisms involved. This approach offers a novel strategy to enhance antibiotic efficacy by targeting both bacterial survival and its ability to evade immune defenses.
Materials and methods
Essential oils
Ten different 100% pure essential oils were procured from the Medicinal and Aromatic Oils Unit at the National Research Center, Doki, Egypt: thyme, garlic, ginger, nigella, marjoram, moringa, tea tree, linseed, lemon, and sage.
Ethical approval and sampling
This study adhered to the Declaration of Helsinki principles with informed consent obtained from all human participants. A total of 100 samples were collected between January and April 2024, comprising 50 bovine mastitic milk and 50 human samples including blood, urine, and swabs. The animal study protocol was approved by the Faculty of Veterinary Medicine, Zagazig University (ZU-IACUC/2/F/285/2023), and the human study protocol was approved by the Faculty of Medicine, Zagazig University (ZU-IRB/409/2024). Both protocols adhered to ARRIVE guidelines (PLoS Biol 8(6), e1000412, 2010).
Isolation, identification and molecular confirmation of Klebsiella pneumoniae isolates
Klebsiella species isolates were obtained using the methods described by Garcia (2010). Bacterial cultures were grown on MacConkey agar and Eosin Methylene Blue then incubated at 37°C for 24 h. Molecular confirmation of K. pneumoniae was performed using a PCR assay targeting the 16S rRNA gene. DNA extraction was carried out using the QIAamp DNA Mini kit (Qiagen, Germany) followed by PCR amplification using specific primers (Table S2). A positive control (ATCC 13883) and a negative control (PCR mixture without DNA template) were included.
Antimicrobial susceptibility testing of the bacterial isolates
Disk diffusion assay
In vitro determination of susceptibility pattern of K. pneumoniae isolates to various antimicrobials was conducted adopting the disk diffusion method (Bauer et al., 1966) using Mueller Hinton agar and standard antimicrobial disks including: amikacin (30 μg), norfloxacin (5 μg), tetracycline (10 μg), cefotaxime (5 μg), ceftriaxone (5 μg), ceftazidime (30 μg), chloramphenicol (30 μg), aztreonam (30 μg), sulfamethoxazole-trimethoprim (25 μg), cephalexin (30 μg), erythromycin (5 μg), and meropenem (10 μg). Plates were incubated at 37°C for 24 h, and inhibition zone diameters were measured. The results were interpreted according to the Clinical & Laboratory Standards Institute (CLSI, 2020) guidelines.
Agar well diffusion assay of herbal oils
The agar well diffusion assay was employed to assess the antibacterial activity of 10 distinct essential oils against isolated Klebsiella isolates. Briefly, bacteria were grown in nutrient broth, adjusted to a concentration of 1.5 × 108 CFU/mL, and swabbed onto Muller-Hinton agar plates. After creating 7 mm-diameter wells in the agar, 100 μL of each essential oil, solubilized in 5% DMSO, was added. To account for the potential influence of DMSO on bacterial growth, control wells containing 5% DMSO alone were included. The plates were incubated at 37°C for 24 h. The diameter of inhibition zones surrounding the wells was measured. Inhibition zones less than 12 mm were considered indicative of no antibacterial activity based on previous studies of Durairaj et al. (2009) and El-Azzouny et al. (2018). Three replicates were performed for each sample, and the results were averaged.
Minimum inhibitory concentration and minimum bactericidal concentration determination
The experiment utilized 96 well plates (TPP, Switzerland) for the broth microdilution assay. The wells were injected with 1 × 105 CFU of bacteria in a final volume of 0.2 mL after the effective extracts and chosen drugs were diluted twofold in Luria broth (LB) broth® (Acumedia, Michigan, United States). Incubation took place for 24 h at 37°C. Following the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2020), MIC testing was conducted, with a concentration range of 0.062 to 1,024 μg/mL for each antimicrobial agent.
Subinhibitory concentration (SIC) refers to an antimicrobial agent’s concentration that is too low to completely inhibit microbial growth and replication, while the minimum inhibitory concentration (MIC) value is the lowest antimicrobial concentration that inhibits microbial growth. The minimum bactericidal concentration value (MBC) value was determined according to Khosravi and Malekan (2004) by subculturing colonies from wells exhibiting no visible growth onto fresh agar plates and incubating to assess bacterial viability.
Evaluation of synergistic effect
The checkerboard broth microdilution method was used to determine the synergy between antibiotics and plant extracts. Fractional inhibitory concentration (FIC) index was calculated to quantify the interaction. FIC value for each antimicrobial agent was calculated according to the formula described by van Vuuren and Viljoen (2011) and El-Demerdash et al. (2023d). The interactions were classified as being synergistic for ΣFIC values of ≤0.5, additive (≥0.5–1.0), indifferent (≥1.0 and ≤4.0) or antagonistic (ΣFIC > 4.0).
Gas chromatography–mass spectrometry analysis
At the Central Laboratories Network of the National Research Centre in Cairo, Egypt, a mass spectrometer detector (5977A) and gas chromatograph (7890B) were equipped with an Agilent Technologies GC–MS system. Essential oils were analyzed directly and diluted with hexane at a ratio of 1:19 (v/v). The GC was outfitted with a DB-WAX column measuring 30 m × 250 μm in internal diameter and 0.25 μm in film thickness. The temperature program for the analysis began at 40°C for 2 min, then increased to 250°C at a rate of 7°C/min. It was held at 250°C for 8 min while injecting 1 μL of hydrogen without splitting using a carrier gas flow rate of 3.0 mL/min. The injector and detector were maintained at 250°C. Mass spectra were generated by electron ionization (EI) at 70 eV with a spectral range of m/z 40–550 and solvent delay 3.5 min. By comparing the fragmentation of the spectrum with data from the Wiley and NIST Mass Spectral Library, many constituents could be identified.
In silico docking analysis
To investigate potential binding interactions, in silico docking studies were conducted using the extracted bioactive compounds and the crystal structures of FosAKP, K. pneumoniae K1 capsule, and OmpK36 (PDB IDs: 6C3U, 7W1E, 5o79). Crystal water molecules were eliminated, and hydrogen atoms were added to the protein structures prior to energy minimization using the MMFF94 force field. The 2D structures of the compounds were created using ChemBioDraw Ultra 14.0 and then energy minimized using the MMFF94 force field. Docking simulations were performed using LigandScout 2.0 (based on Autodock Vina), and the most favorable binding poses were chosen for analysis. The 3D and 2D binding modes were visualized using Biovia Discovery Studio Visualizer, and the results are summarized in Supplementary Table S1 (Al-Halbosy et al., 2023).
Transmission electron microscopy assay
Bacterial samples were prepared for transmission electron microscopy (TEM) analysis following the methodology described by Amin et al. (2020). Briefly, Klebsiella pneumoniae cultures (both treated and control) were grown in nutrient broth for 24 h before centrifugation at 4,000 rpm for 10 min. The resulting pellet was washed with distilled water, fixed in 3% glutaraldehyde, and post-fixed in potassium permanganate for 5 min. Dehydration was achieved through a graded ethanol series (10–90%), culminating in absolute ethanol for 30 min. Samples were embedded in epoxy resin using an acetone gradient and ultrathin sections were cut and mounted on copper grids. Staining with uranyl acetate and lead citrate was followed by examination using a JEOL-JEM 1010 transmission electron microscope at 80 kV, located at the Regional Center for Mycology and Biotechnology (RCMB), Al-Azhar University, Egypt.
Capsule gene detection and expression analysis
This section details the methods used for conventional PCR detection of capsule genes and their quantitative expression analysis using real-time PCR.
Conventional PCR for capsule gene detection
Supplementary Table S2 lists the primer sequences for target genes, along with expected amplicon sizes and annealing temperatures. A 25 μL reaction mixture was prepared containing 12.5 μL of DreamTaq Green PCR Master Mix (2X) from Thermo Scientific, 1 μL of each primer (20 pmol), 5.5 μL of nuclease-free water, and 5 μL of DNA template. The PCR was carried out using a 2,720 thermal cycler (Applied Biosystems) following the manufacturer’s instructions.
The PCR products were separated by electrophoresis on a 1% agarose gel (Applichem, Germany) in 1x TBE buffer at room temperature with a 5 V/cm gradient. For analysis, 10 μL of PCR products were loaded into each well. Fragment sizes were determined using Gelpilot 100 bp Plus DNA ladder (Qiagen) and the Generuler 100 bp ladder (Thermo Scientific).
Quantitative analysis of capsule gene expression
RNA was extracted from bacterial cultures using the QIAamp RNeasy Mini Kit (Qiagen). RNA concentration for each sample was measured using a NanoDrop Eight Spectrophotometer (Thermo Scientific). 16S rRNA was used as the internal control (housekeeping gene). Primer sequences are listed in Table 1. New primers targeting the magA and wabG genes were designed using Primer3 and FastPCR software. These primers were optimized for specificity and sensitivity using “touchdown PCR” and were validated through experimental testing.
A 20 μL reaction mixture containing 10 μL of 2x HERA SYBR® Green RT-qPCR Master Mix (Willowfort, UK), 1 μL of RT Enzyme Mix (20X), 0.5 μL of each primer (20 pmol), 3 μL of nuclease-free water, and 5 μL of RNA template was prepared. The reaction was carried out using a StepOne™ real-time PCR system (Applied Biosystems) following the manufacturer’s protocol. The program included an initial denaturation step at 94°C for 15 min, followed by 40 cycles of denaturation (94°C for 15 s), annealing (60°C for 30 s), and extension (72°C for 30 s). A final extension step at 72°C for 10 min concluded the reaction.
The StepOne™ software calculated CT values and generated amplification curves. The ΔΔCt method (Yuan et al., 2006) was used to compare the CT of each sample with the positive control group using the formula (2−ΔΔCt) to assess the variation in gene expression among the RNA samples. All molecular work was performed in the Biotechnology Unit, Animal Health Research Institute, Zagazig Branch, Egypt.
Statistical analysis
Microsoft Excel (Microsoft Corporation, Redmond, WA, United States) was utilized to manipulate the data. Following the methodology of Razali and Wah (2011), a Shapiro–Wilk test was conducted to confirm normality. The One-way ANOVA (PROC ANOVA; Stokes et al., 2012) was employed to analyze the significant impacts of the treatments, with a significance level set at α = 0.05. The means ± SE of the results were then reported. In cases where a significant effect was observed. Tukey’s test was used to perform pairwise comparisons between means. A p-value threshold of less than 0.05 was established for determining statistical significance between means. The GraphPad Prism software 9.0 (GraphPad, United States) was used to create the figures.
Results
Prevalence rate
Bacteriological examination revealed that 25 out of 100 samples were positive for Klebsiella spp. resulting in an overall prevalence rate of 25%. All of the isolates were identified as Klebsiella pneumoniae through biochemical and genotypical assays. The prevalence of K. pneumoniae in human sources was significantly higher than in mastitic milk, being approximately twice as high (OR = 2.143, 95% CL = 1.635–2.748, Figure 1).
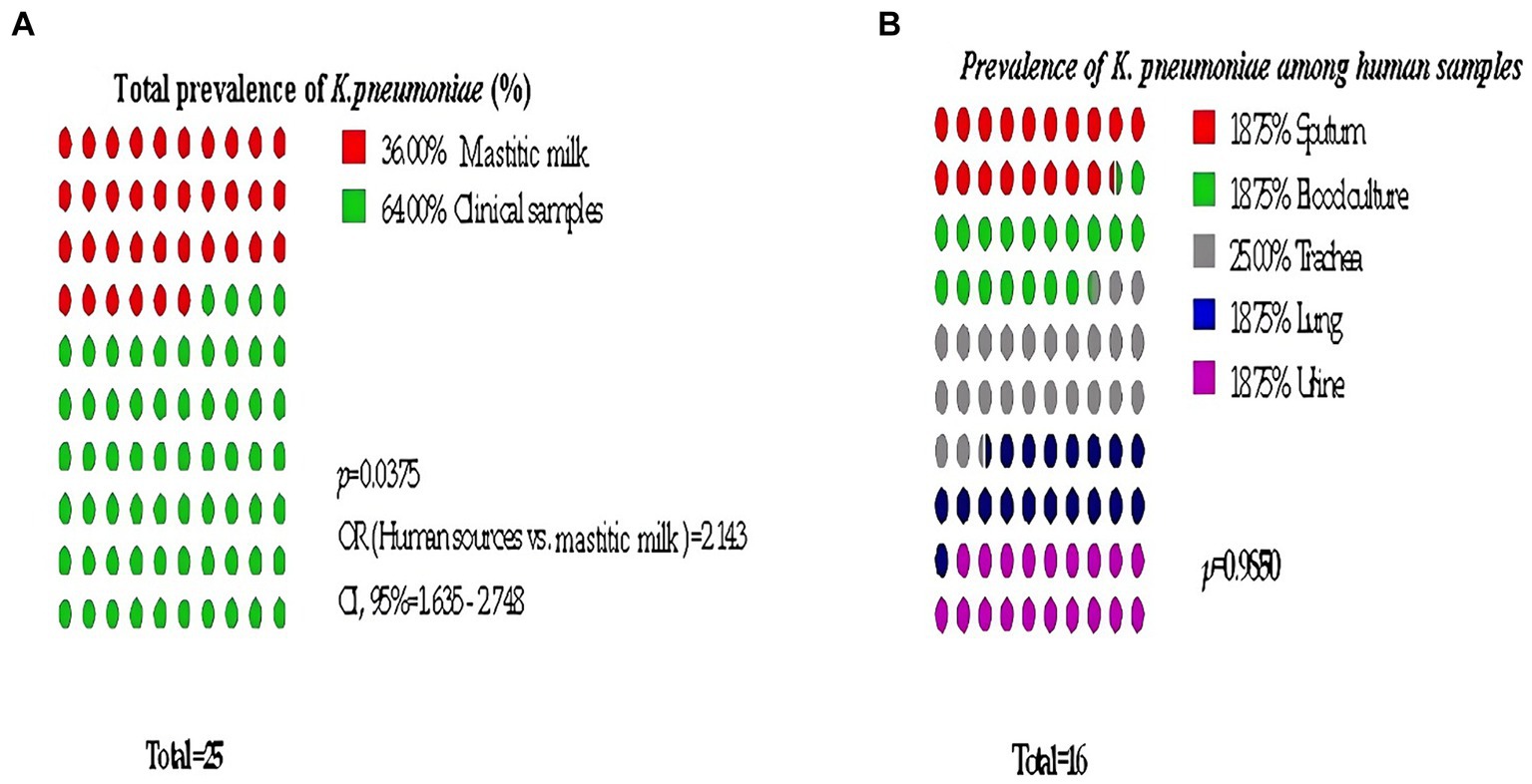
Figure 1. Significant prevalence of K. pneumoniae among examined samples. (A) Total prevalence, (B) Detailed Clinical samples prevalence.
Antibiogram pattern of Klebsiella pneumoniae isolates
The antibiotic resistance rate for each source and the whole sets of isolates are represented in (Supplementary Figures S1, S2). The antimicrobial resistance pattern showed absolute resistance to cefotaxime followed by ceftriaxone and cephalexin (92%). More than half of the isolates exhibited resistance the rest tested antibiotics except amikacin. Notably, all 25 isolates were multidrug-resistant (MDR) i.e., resistant to three or more groups of antibiotics.
Regarding K. pneumoniae isolates from human sources, absolute resistance to cefotaxime and erythromycin was detected while the erythromycin resistance percentage for those from animal source (mastitic milk) was 55.5%.
In contrast, K. pneumoniae isolates from animal sources showed a low resistance rate to meropenem and chloramphenicol (27 and 55%, respectively), however, their percentages were high in isolates recovered from human sources (80 and 62%, respectively).
In total, Klebsiella isolates from human sources displayed a significant pattern of resistance, with almost all isolates showing high frequencies of resistance to more than seven drugs.
Essential oil activity
The results of the study showed that tea tree oil (TTO) exhibited the strongest antimicrobial properties followed by thyme among the 10 oils tested. Both oils showed inhibition zones ranging from 14 to 32 mm against the tested Klebsiella isolates. On other hand, moringa and linseed oils did not show any antibacterial activity against the tested pathogens (Figure 2A).
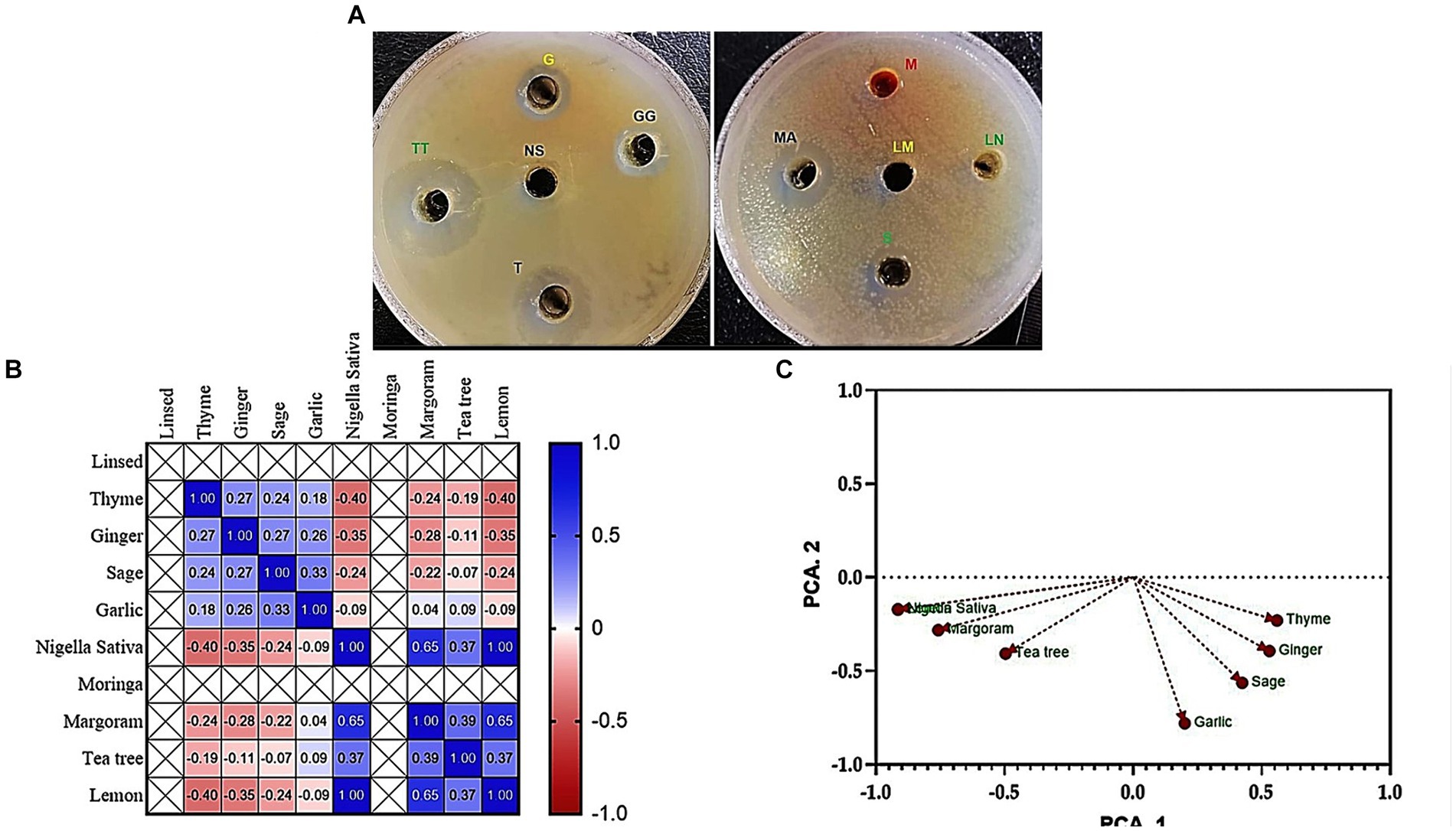
Figure 2. Antibacterial activity (A) Agar well diffusion assay shows the inhibition zones of K. pneumoniae (code No. KP11) caused by tested 10 oil extracts; TT, Tea tree; T, Thyme; MA, Margoram; LM, Lemon; NS, Nigella sativa; S, Sage; G, Garlic; GG, Ginger; M, Moringa; LN, Linseed. (B) Pearson’s correlation coefficient between inhibition zones (mm) of essential oils against MDR K. pneumoniae. (C) Principal component analysis (PCA) of inhibition zones (mm) of essential oils against MDR K. pneumoniae.
Pearson’s correlation
Pearson’s correlation coefficient (r) was performed to examine the relationship between the inhibition zones (mm) of essential oils and the prevalence of MDR K. pneumoniae. The results, presented in Figure 2B, showed a positive correlation between the inhibition zone values of tea tree oil and both thyme and margoram essential oils (r = 0.95 and 0.62, respectively; p < 0.05). Similarly, a highly positive correlation was observed between thyme and margoram (r = 0.61; p < 0.05). Conversely; a negative correlation coefficient was found between the other combinations of essential oils.
Principal component analysis
PCA was performed to identify associations between essential oil inhibition zones and the prevalence of MDR K. pneumoniae. The Varimax rotation method was used, and loadings greater than 0.50 or 0.60 were considered significant. Four principal components were extracted, explaining 87.98% of the total variance. The first component (17.77% variance) correlated with inhibition zones of tea tree, thyme, and marjoram oils. The second component (12.61% variance) was associated with sage and garlic oils, while the third component (9.51% variance) primarily represented ginger oil. The fourth component (8.50% variance) was associated with lemon oil (Table 2; Figure 2C).
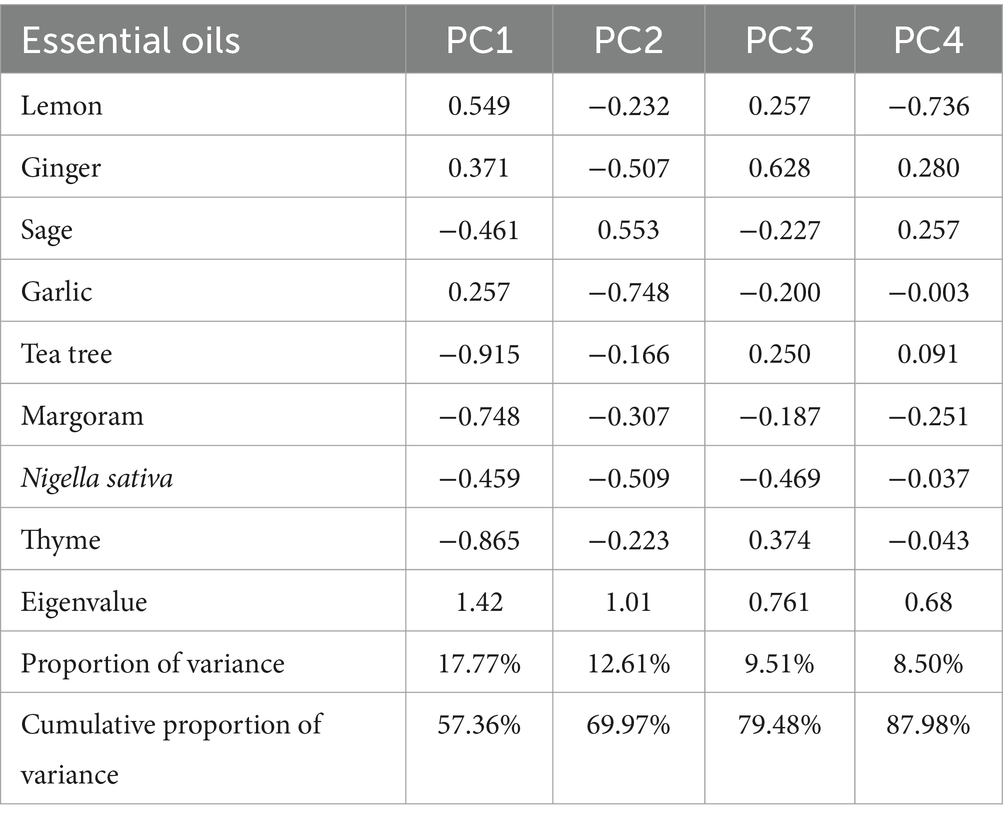
Table 2. Varimax rotated principal component analysis (PCA) of inhibition zones (mm) of essential oils against MDR K. pneumoniae (bold loadings are statistically significant).
Minimum inhibitory concentration and synergy with amikacin
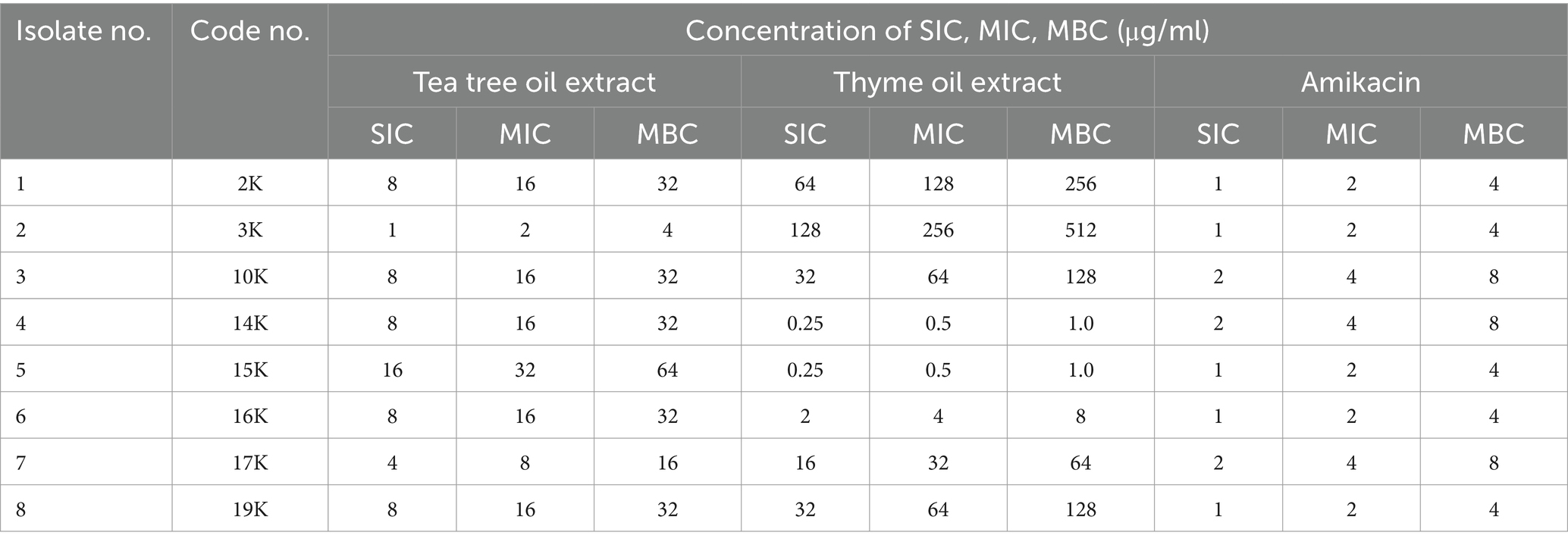
Tea tree oil displayed the most potent antibacterial activity against K. pneumoniae isolates, with MICs ranging from 2 to 32 μg/mL (Table 3; Supplementary Figure S3). Thyme oil also exhibited inhibitory effects (MICs: 0.5–256 μg/mL). Amikacin, a conventional antibiotic, showed good activity with MICs of 2–4 μg/mL.
Further investigations explored the synergistic potential of these essential oils with amikacin. Both tea tree and thyme oil combinations displayed synergistic effects against most isolates, with tea tree oil exhibiting a stronger synergy (FIC index: 0.1325–0.53) compared to thyme oil (FIC index: 0.15–0.53). Notably, the synergistic interaction was more pronounced for resistant isolates. Overall, these findings suggest that combining essential oils with amikacin could be a promising strategy to combat K. pneumoniae infections (Tables 4, 5).
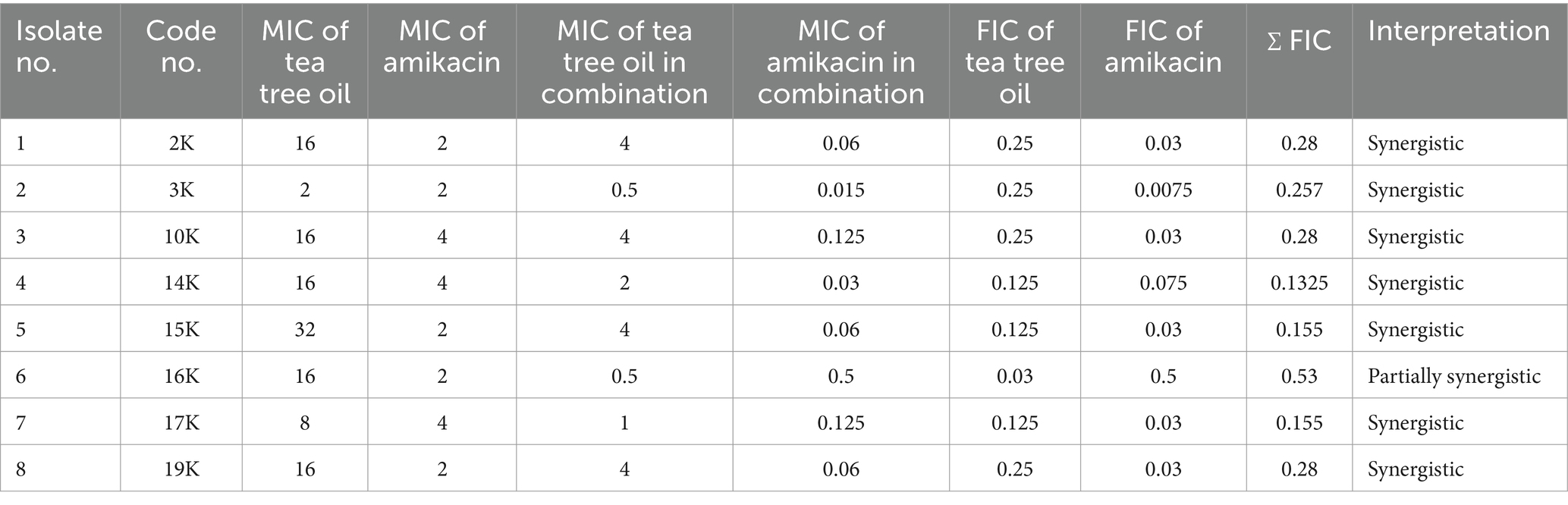
Table 4. MIC of tea tree oil and amikacin alone and in combination and FIC index against Klebsiella isolates by the checkerboard method.
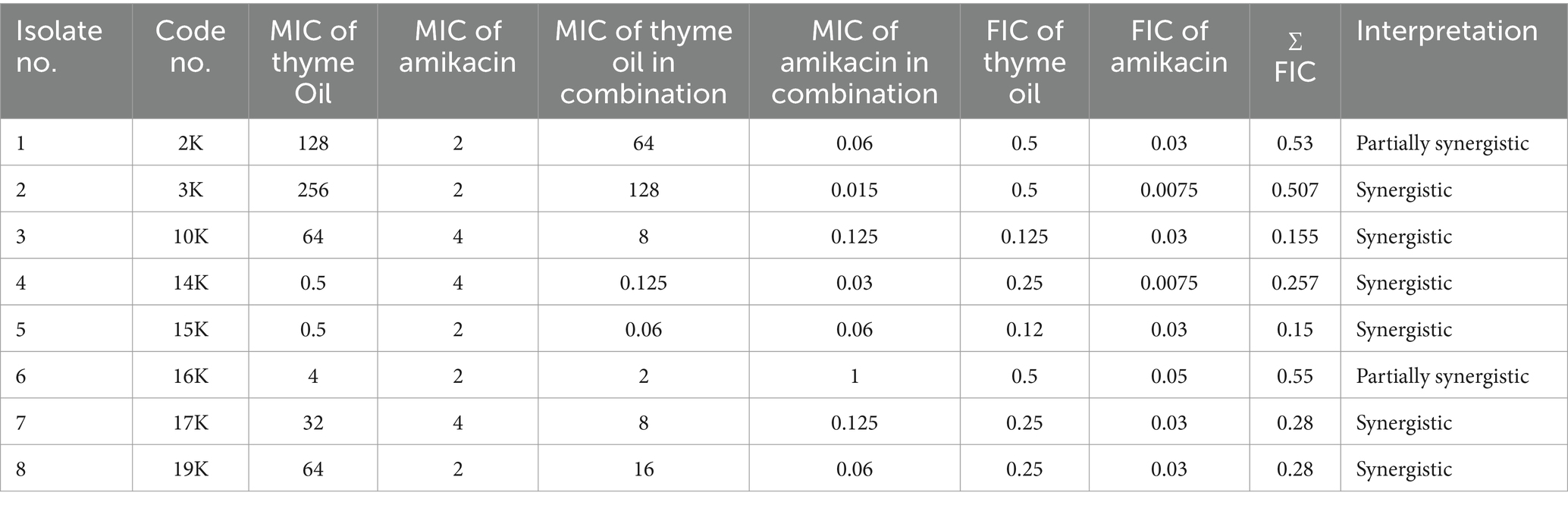
Table 5. MIC of thyme oil and amikacin alone and in combination and FIC index against Klebsiella isolates by the checkerboard method.
Characterization of compounds present in the oily extracts (thyme and tea tree oils) by using GC–MS
Supplementary Tables S3, S4 and Supplementary Figures S4, S5 detail the characterization of compounds in the effective extracts. GC–MS analysis of tea tree oil identified seven bioactive compounds, with Terpinene-4-ol exhibiting the highest peak area. Other identified compounds included P-cymene, Alpha-Terpineol, Beta-pinene, Beta-myrcene, Limonene, and Farnesol. Similarly, thyme oil analysis revealed 10 chemical compounds, with Thymol being the principal bioactive component followed by p-cymene, Gamma terpinene, Linalool, and Eugenol.
Docking data
Molecular docking analysis was performed to investigate the potential binding interactions of identified bioactive compounds with target proteins in K. pneumoniae (OmpK36, K1 capsule, and FosAKP). The results revealed favorable binding energies for several compounds with each target protein. For example, Terpinen-4-ol exhibited strong binding affinity for all three targets (OmpK36: −6.25 kcal/mol; K1 capsule: −5.81 kcal/mol; FosAKP: −5.94 kcal/mol). Similarly, other identified compounds like 1,4-dihydroxy-p-menth-2-ene, 7-Oxabicyclo [4.1.0] heptane, and trans-Z-.α.-Bisabolene epoxide also demonstrated promising binding interactions (Figures 3–5).
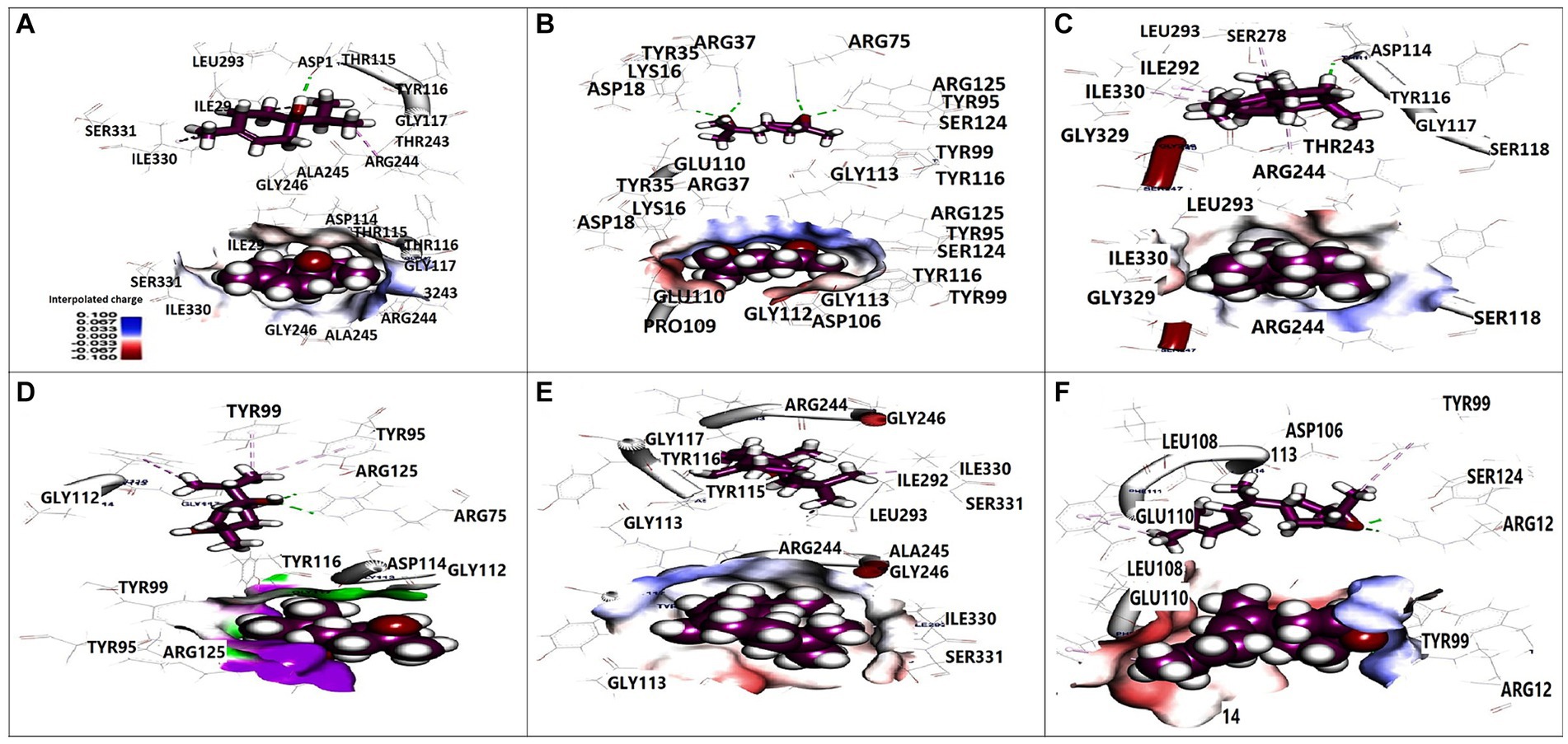
Figure 3. 3D orientation and surface mapping of active components against OmpK36 target site. (A) Terpinen-4-ol, (B) Farnesol, (C) (+) spathulenol, (D) 1,4-dihydroxy-p-menth-2-ene, (E) (−) spathulenol, (F) trans-Z-. alpha. -Bisabolene epoxide.
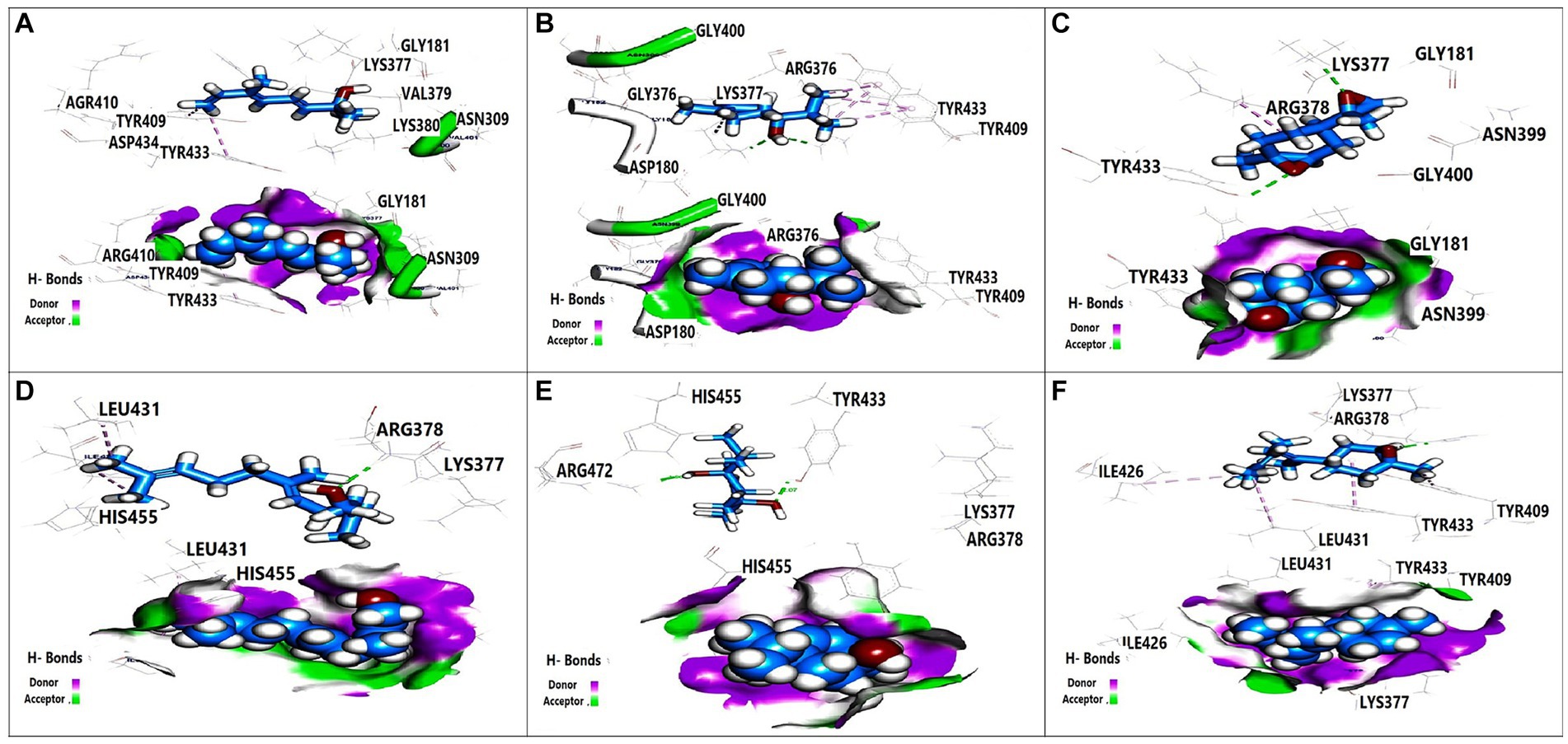
Figure 4. 3D orientation and surface mapping of active components against K. pneumoniae K1 target site. (A) 2,6-Dimethyl-3,5,7-octatriene-2, (B) Terpinen-4-ol, (C) 7-Oxabicyclo, (D) Farnesol, (E) 1,4-dihydroxy-p-menth-2-ene, (F) trans-Z alpha. -Bisabolene epoxide.
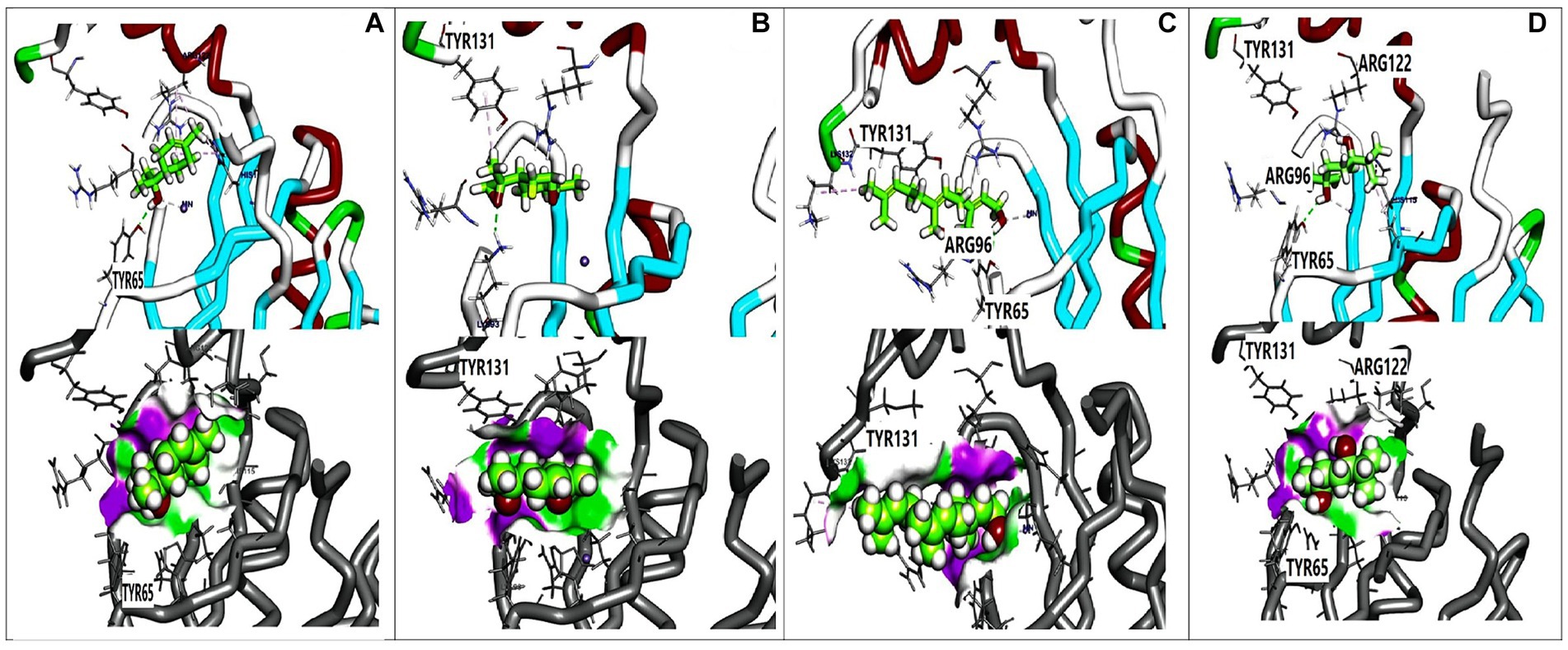
Figure 5. 3D orientation and surface mapping of active components against FosAKP target site. (A) Terpinen-4-ol (B) 7-oxabicyclo (C) Farnesol (D) 1,4-dihydroxy-p-menth-2-ene.
Transmission electron microscopy
Transmission electron micrographs revealed distinct morphological changes in K. pneumoniae cells following treatment with essential oils and amikacin. Untreated cells (Figure 6A) exhibited a normal rod-shaped morphology with a characteristically rough (rugose) and rigid cell surface. In contrast, cells treated with the combination of thyme oil and amikacin (Figure 6B) displayed significant alterations, including a crumpled and shrunken cell surface with irregular shapes. Notably, some cells exhibited outward openings and cleavages in the cell wall. Treatment with tea tree oil and amikacin (Figure 6C) resulted in even more severe damage, characterized by complete disruption of the cell envelope and leakage of cytoplasmic contents. These lysed cells appeared devoid of internal structures and possessed collapsed and fragmented cell walls.
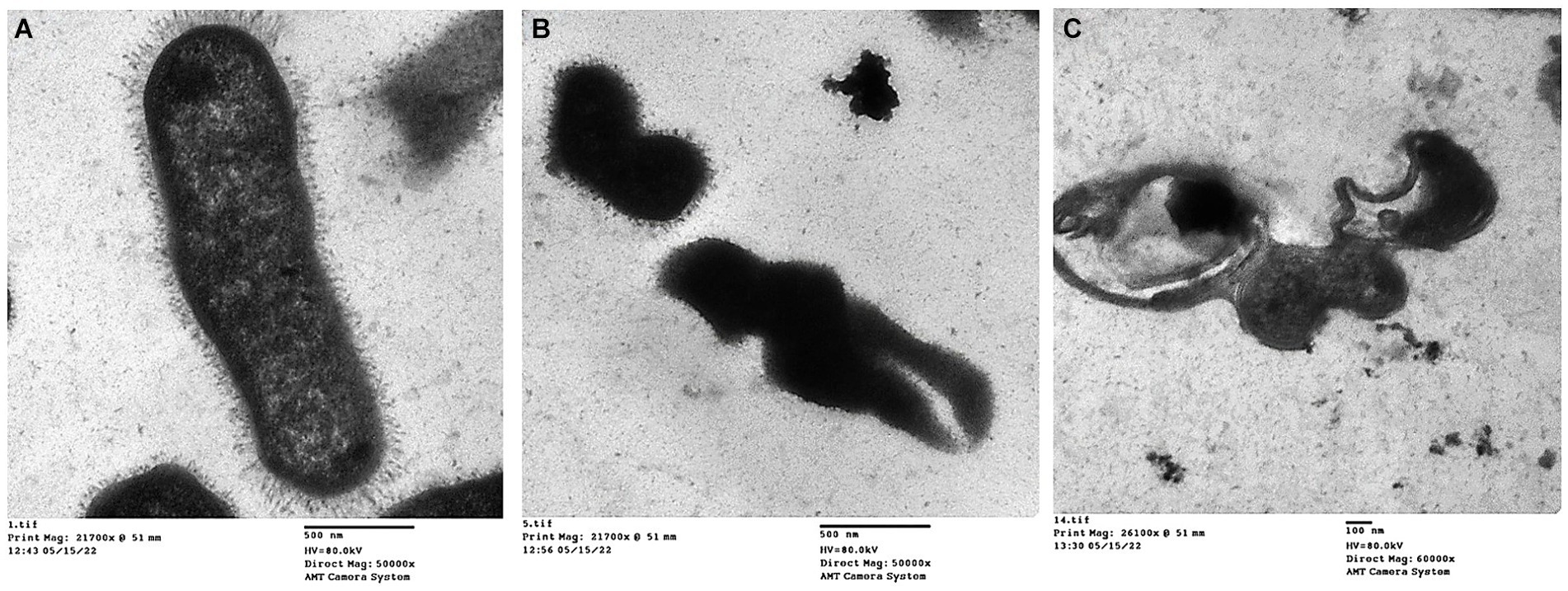
Figure 6. Transmission electron micrographs of K. pneumoniae cells; (A) Untreated K. pneumoniae cells; (B) K. pneumoniae cells treated with thyme oil and amikacin, and (C) K. pneumoniae cells treated with tea tree oil and amikacin.
Detection of capsule genes by conventional PCR
The detection of capsule genes in Klebsiella isolates was confirmed by the PCR. Five different primers which produced 169-bp, 516-bp, 1,282-bp, 534-bp, and 683-bp PCR products, enabled elucidating the presence of capsule genes in the genomes of Klebsiella isolates. This was obvious when the Klebsiella isolates compared to a positive control (Lane +C) as in Supplementary Figure S6.
Transcriptional analysis of capsule expression genes of Klebsiella pneumoniae
The relative expression of five virulence genes (wcaG, rmpA, magA, uge, and wabG) which are required for capsule formation, were measured in Klebsiella isolates treated with tea tree oil plus amikacin and thyme oil plus amikacin as illustrated in Figure 7. The 16 s rRNA housekeeping gene was used for qRT-PCR normalization.
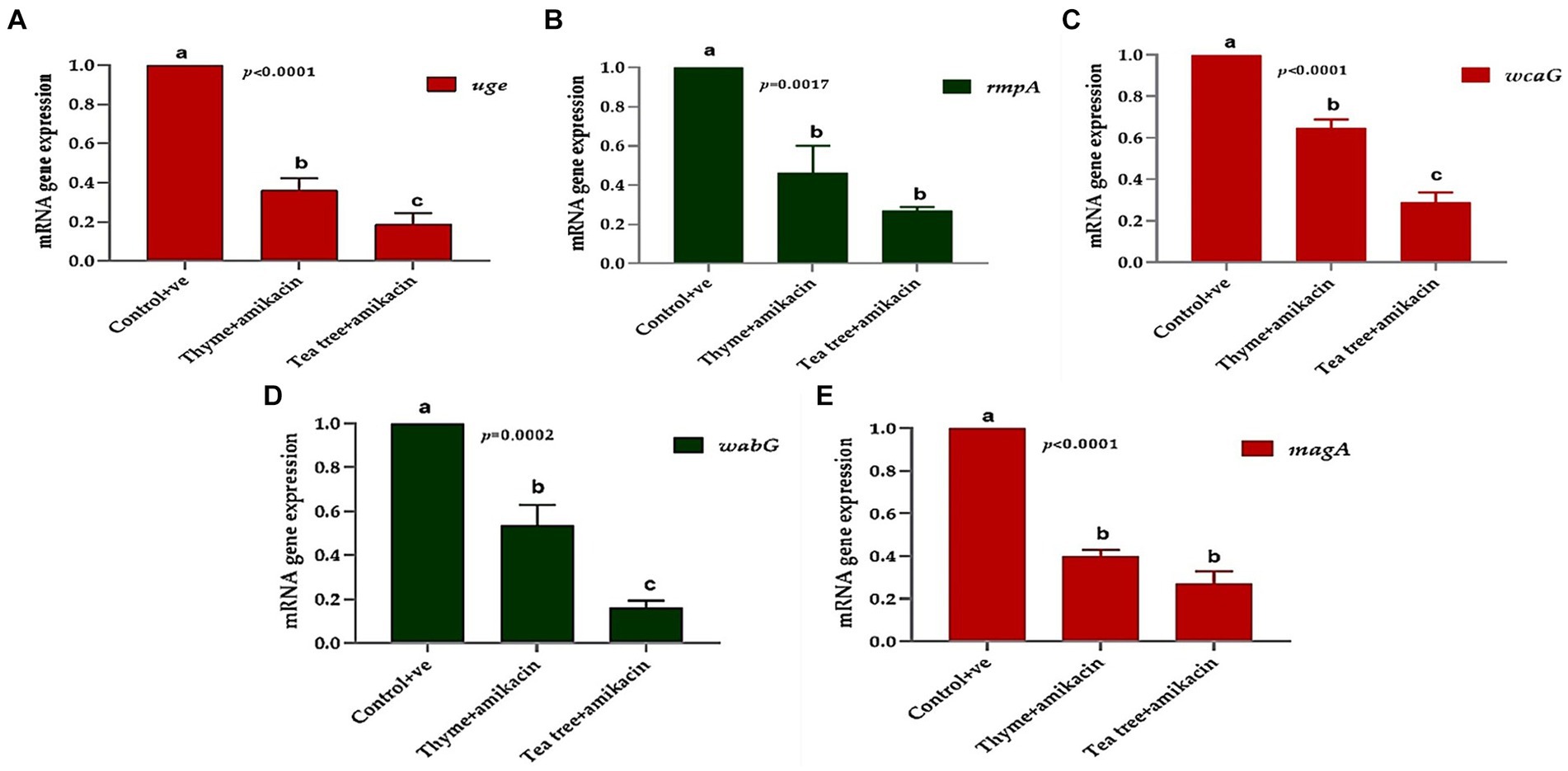
Figure 7. The relative mRNA expression levels of genes related to capsule formation through tested K. pneumoniae isolates before and after treatments; (A) uge, (B) rmpA, (C) wcaG, (D) wabG, and (E) magA.
Using RT-PCR, the amounts of the tested virulence gene products before and after treatment with thyme oil combined with amikacin can be compared. The fold change in the magA gene before treatment was 1 and after treatment, it was downregulated and reached 0.341 in the tested isolates.
The fold change in the wabG gene after treatment ranged from 0.353 to 0.683. The same downregulation occurred in the wcaG gene with a fold change of 0.562. Additionally, the rmpA and uge genes were downregulated with fold change ranges of 0.275–0.732 and 0.244–0.444, respectively.
Regarding the tea tree + amikacin-treated group, strong downregulation of the same virulence genes was detected. The magA gene exhibited fold change ranging from 0.15 to 0.34, the wabG gene had fold change ranges of 0.100–0.213, wcaG fold changes of 0.194–0.356, rmpA fold changes of 0.233–0.301 and uge gene had fold change ranging from 0.1060 to 0.299.
Overall, the treatments significantly affected the studied genes; the expression of uge, wcaG, and wabG was significantly downregulated (p < 0.05) in the tea tree + amikacin groups compared to thyme + amikacin treated groups (Figures 7A,C,D). Meanwhile, non-significant differences (p > 0.05) were observed between the aforementioned two groups for the expression of rmpA and magA genes (Figures 7B,E).
Discussion
Today, K. pneumoniae is considered the most common cause of hospital-acquired pneumonia in most countries worldwide (Anes et al., 2017). The emergence of multidrug resistant bacteria (MDR) isolates of K. pneumoniae is a global issue (Ripabelli et al., 2018). Numerous studies have isolated MDR K. pneumoniae from various animals and humans (Yang et al., 2019; El-Demerdash et al., 2023b). However, the correlation between capsule formation and antibiotic resistance in K. pneumoniae is not fully understood. Therefore, this study was conducted to investigate the prevalence of MDR Klebsiella isolates in animals and human in Egypt, as well as to evaluate possible solutions to overcome this antibiotic resistance and their ability to form capsules by using different herbal extracts.
In this study, K. pneumoniae was recovered from 25 out of 100 samples, yielding an overall prevalence of 25%. This finding is comparable to the prevalence range reported in previous studies on Klebsiella colonization in Western countries (5–35%) but notably higher than the reported rates in Europe and North America (5 and 3%, respectively) (Chang et al., 2021; Salari et al., 2023).
However, direct comparisons between regions can be misleading due to variations in sample types, study populations, and methodological approaches. For instance, while the overall prevalence in Africa is reported as 45% (Salari et al., 2023), significant heterogeneity exists within the continent. To provide a more accurate context, future studies should focus on specific regional comparisons, considering factors such as healthcare settings, antibiotic usage, and socioeconomic conditions.
The abuse and misuse of antimicrobial medicines are leading to the emergence of resistance to these agents as a global crisis in the management of infections associated with K. pneumoniae (El-Demerdash et al., 2018; Ali et al., 2023; Ndlovu et al., 2023). This study highlights a concerning case of highly diversified antibiotic resistance. Data from the Chinese Antimicrobial Resistance Surveillance System (CARSS) indicates that K. pneumoniae is the second most common (20.2%) gram-negative infection among isolated bacteria.
The present study also investigated the low sensitivity of K. pneumoniae isolates to tetracycline. The low sensitivity of tetracycline is due to mutations in the chromosomes in the outer membrane of bacteria leading to a decrease in tetracycline penetration into the cell (Grossman, 2016).
Disk diffusion susceptibility testing revealed that all isolated K. pneumoniae strains (100%) were multidrug-resistant (MDR). This finding aligns with previous reports by (Del Prete et al., 2019; Ferreira et al., 2019) who also observed a high prevalence of MDR K. pneumoniae. Notably, the disk diffusion method likely identified high resistance to key β-lactam antibiotics such as cefotaxime and ceftriaxone, which are commonly used for treating Klebsiella infections. This resistance contributes significantly to the designation of MDR.
According to other sources, the MDR pattern may be attributed to the excessive use of antibiotics in Egyptian veterinary and human medicine, as well as the horizontal or vertical transmission of plasmids carrying antimicrobial resistance genes among different bacterial pathogens or between animals and humans (Witte, 1998; El Damaty et al., 2023; El-Demerdash et al., 2023a, 2023c; Saad et al., 2024). On the other hand, the best susceptibility (52%) was observed for amikacin which is an aminoglycoside antibiotic that prevents bacteria from synthesizing proteins by binding to the 30S ribosomal subunit mRNA and causing reading mistakes. Amikacin is also highly resistant to modification by bacterial enzymes leading many bacteria to be sensitive to this antibiotic (Ramirez and Tolmasky, 2017).
In the search for alternative antimicrobial agents, several studies have shown the potential of essential oils in fighting MDR bacteria. Terpenes and terpenoids are common volatile low molecular weight chemicals found in complex hydrophobic liquids known as essential oils (EOs). They are extracted from plants through solvent extraction, mechanical expression, or distillation. Previous studies have demonstrated the antibacterial properties of essential oils and their constituents against gastrointestinal and other infections (Millezi et al., 2016; El-Demerdash et al., 2023d). Building on this evidence, the present study evaluated the effectiveness of 10 essential oils against 25 Klebsiella isolates.
The antimicrobial activities of essential oils are attributed to their ability to penetrate microbial cells, causing structural and functional changes due to their hydrophobic nature. This disruption of the cytoplasmic membrane leads to cell lysis and the release of intracellular compounds (Lopez-Romero et al., 2015). Moreover, the diverse mechanisms of action and functional diversity of essential oils increase microbial sensitivity (Bakkali et al., 2008; Nazzaro et al., 2013). Among the tested plant extracts, tea tree and thyme oils exhibited high antimicrobial activities. Therefore, this study recommends the use of tea tree and thyme oils as antimicrobial agents and chemical preservatives due to their relatively lower toxicity and side effects (Santurio et al., 2014). Additionally, the multi-component nature of plant extracts makes it more difficult for bacteria to develop resistance compared to commonly used antibiotics, which have a single target site (El-Azzouny et al., 2018).
Without a doubt, the antibacterial action of essential oils is determined by their chemical structure which can vary depending on factors such as weather, soil type, and geographic location (Dardona, 2014; Dardona and Al-Hindi, 2019). Therefore, it is crucial to understand the chemical composition of essential oils in order to connect it to their antibacterial properties GC/MS was utilized to conduct chemical profiling of the oils extracted from tea trees and thyme.
This study uniquely investigates the efficacy of tea tree oil against MDR-KP strains, a significant contribution as it opens new avenues for alternative therapeutic strategies to address the growing challenge of antibiotic resistance. GC-mass analysis revealed the presence of various bioactive compounds in the effective tea tree oil, potentially contributing to the observed efficacy through a synergistic mechanism. The most abundant component, terpinen-4-ol, previously linked to biofilm inhibition in S. aureus (Cordeiro et al., 2020), along with limonene, another component with documented antibacterial activity (Slade et al., 2009; Matsuo et al., 2011), might contribute to the observed synergistic mechanism.
Similar to tea tree oil, thyme extract, analyzed using GC–MS, revealed a rich composition of bioactive compounds. Thymol, the primary component, is recognized for its broad spectrum of antimicrobial activity (Kowalczyk et al., 2020; Khan et al., 2021). Studies suggest that thymol disrupts bacterial membranes through interactions with membrane proteins and potentially by altering ATP levels (Tiwari et al., 2009). Additionally, thyme oil contains carvacrol, another well-documented antibacterial compound, known to inhibit flagella formation in bacteria (Burt et al., 2007). These findings highlight the potential mechanisms by which thyme extract might exert its antibacterial effects against MDR-KP strains.
However, a major limitation of essential oils is the high dosage required to combat resistant microorganisms, hindering their therapeutic application (Hyldgaard et al., 2012). This study investigated the potential of combining antibiotics with well-selected plant extracts, like tea tree and thyme oils, to address this challenge. Our findings demonstrate that this combination strategy offers several advantages: (i) overcoming resistance, (ii) reducing effective antibiotic dosages (leading to lower costs and minimized side effects), and (iii) broadening the spectrum of efficacy against MDR-KP strains (Hyldgaard et al., 2012; Purkait et al., 2020). Notably, the Fractional Inhibitory Concentration (FIC) index calculations revealed a strong synergistic effect between the antibiotics and herbal extracts, suggesting a combined action that is significantly more potent than their individual effects.
The observed synergy might be attributed to the ability of herbal extracts, particularly tea tree and thyme oil, to enhance the penetration of antibiotics, like amikacin, through the outer membrane of MDR-KP. In silico docking analysis further supported this hypothesis by revealing potential interactions between identified bioactive compounds within the extracts and key target proteins in K. pneumoniae. These targets included OmpK36, a porin crucial for outer membrane permeability, the K1 capsule polysaccharide essential for bacterial virulence, and FosAKP, an enzyme vital for lipopolysaccharide (LPS) biosynthesis.
Several compounds displayed promising binding affinities for OmpK36, suggesting they might disrupt its function. Terpinen-4-ol, with the strongest binding energy, interacted with hydrophobic residues and formed a hydrogen bond with Asp114 (Figure 3A). Similar interactions were observed with other compounds (Figures 3C,F). Disruption of OmpK36 could hinder nutrient uptake and antibiotic penetration by K. pneumoniae.
Docking analysis revealed favorable interactions between essential oil compounds and the K1 capsule biosynthesis machinery. Terpinen-4-ol once again demonstrated notable binding affinity for the K1 capsule target site (Figure 4B), suggesting potential interference with K1 capsule assembly. This could weaken the bacterial cell envelope and enhance antibiotic efficacy.
The essential oil compounds also showed binding potential for FosAKP.Terpinen-4-ol and 1,4-dihydroxy-p-menth-2-ene displayed favorable binding energies with interactions involving hydrophobic residues, hydrogen bonds, and metal ion interactions (Figures 5A,D). This suggests a potential mechanism for inhibiting LPS synthesis, another critical component of the bacterial outer membrane.
Overall, the docking analysis results provide compelling evidence for the potential mechanisms by which essential oil components might exert their antibacterial effects against K. pneumoniae. The observed interactions with key target proteins like OmpK36, K1 capsule components, and FosAKP suggest that these compounds might disrupt essential cellular processes, leading to membrane permeability alterations, reduced virulence, and ultimately, bacterial cell death. However, it is important to acknowledge that in silico results need to be validated through wet lab experiments to confirm the predicted binding modes and their functional consequences.
One of K. pneumoniae’s most significant virulence factors in terms of infection-causing capacity is capsular polysaccharide (CPS). Polymorphonuclear cells seen in CPS essentially form the pathogen’s outer coating and provide resistance to phagocytosis. By lowering the quantity of C3 on the bacteria and functioning as a barrier to prevent contact between macrophage receptors and their ligands on the bacterial surface, CPS reduces the connection between bacterial cells (Opoku-Temeng et al., 2019).
The transmission electron microscopy (TEM) results showed detailed cell damage of the Klebsiella isolate after treatment with a combination of essential oil and amikacin.
The TEM revealed that the size of capsules in treated cells, by both agents, was profoundly significantly reduced compared to that of control untreated cells. Overall, it has been documented that plant-derived compounds may be a promising resource for developing novel therapeutic approaches targeting bacterial capsule production (Upadhyay et al., 2014). Additionally, Dumlupinar et al. (2020) reported that the multicomponent structures of essential oils particularly tea tree oil reduce the potential for biosynthesis of bacterial capsular polysaccharides among virulent K. pneumoniae isolates.
In the current study, the use of essential oils (tea tree oil and thyme oil) in combination with amikacin downregulated the expression of genes responsible for capsule synthesis in Klebsiella pneumoniae.
The most important genes involved in capsule formation are magA, rmpA, uge, wcaG and wabG. The magA gene (muco viscosity associated gene A) was originally identified through transposon mutagenesis screening (Costa, 2019). MagA was identified based on its role in mucoviscosity, resistance to serum killing and phagocytosis, and virulence in mice (Fang et al., 2004). Mucoviscosity is indirectly related to magA because of its essential role in capsule production. While mucoviscosity might be mediated by capsule expression-promoting regulators such as rmpA (Hsu et al., 2011).
The production of capsule polysaccharides is regulated by the rmpA gene which has been identified in three forms: rmpA2 on a plasmid, rmpA located chromosomally, and rmpA on a plasmid (Shon et al., 2013). Hypercapsule production controlled by rmpA, inhibits opsonophagocytosis and phagocytosis of K. pneumoniae by the host’s immune cells. This also suppresses opsonization and lysis caused by complement. Ikeda et al. (2018) and Khan et al. (2021) have demonstrated that rmpA regulation is linked to K. pneumoniae’s ability to evade immune responses, emphasizing the critical role of the rmpA gene in the progression of infection.
Conversely, most isolates from urine include the uge gene (uridine diphosphate galacturonate-4epimerase). Studies have shown that K. pneumoniae strains lacking the uge gene are less pathogenic and unable to cause sepsis, pneumonia, or UTI. In experimental settings where urinary infections are induced, mutations in the uge gene have been found to decrease K. pneumoniae’s ability to colonize (Izquierdo et al., 2003; Ballén et al., 2021).
Similarly, Shu et al. (2009) have demonstrated that the wcaG virulence gene (GU325787) located in the transferable regions of the chromosome is responsible for K. pneumoniae capsule biosynthesis. It is needed for the conversion of mannose to fucose, which may enhance the ability of bacteria to evade phagocytosis by macrophages.
Additionally, almost all clinical isolates possess the significant gene wabG. Most research indicates that 88–100% of K. pneumoniae isolates harbor wabG, while one study only found wabG in 5.3% of isolates (Izquierdo et al., 2003). Furthermore, Jung et al. (2013) observed that in intraperitoneal, pneumonic, and UTI rodent infection models, K. pneumoniae strains lacking this gene are attenuated and unable to produce the Lipopolysaccharide (LPS) outer core or maintain capsular antigen.
qRT-PCR analysis revealed a dampening effect of the essential oil combinations on the fold change of the previously mentioned capsule gene expression. Notably, the tea tree oil combination exhibited a significantly stronger influence compared to thyme. These results suggest that tea tree oil might act as a more potent anti-virulence and antibiotic-resistance modifying agent when compared to thyme. Furthermore, the observed dampening effect on capsule gene expression by the essential oil combinations highlights a potentially novel mechanism by which they exert their antibacterial activity against K. pneumoniae.
These results indicate that essential oils were able to down-regulate the expression of the investigated genes which signifies their antagonistic properties against Klebsiella. Decreasing Klebsiella virulence through the synergistic effect of essential oils and antibiotics would reduce the pathogen’s ability to cause infection. This is an important concept in the era of antibiotic resistance.
In conclusion, this study highlights the promising potential of combining conventional antibiotics with essential oils derived from medicinal plants (such as tea tree and thyme) for developing novel antimicrobial therapies and mitigating drug resistance. The synergistic activity observed between the tested antibiotics and essential oils opens up new treatment strategies against various microbial infections and provides hope for combating the growing threat of antibiotic resistance.
However, it is important to note that these findings are based on in vitro experiments. Further in vivo studies are necessary to confirm the observed synergy and evaluate the efficacy and safety of these combinations in a more complex biological system Additionally, understanding the molecular mechanisms underlying this synergistic interaction is crucial for the development of effective antibacterial drugs derived from medicinal plants.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the studies involving humans were approved by the Faculty of Medicine, Zagazig University. The studies were conducted in accordance with the local legislation and institutional requirements. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by The animal study was approved by the Faculty of Veterinary Medicine, Zagazig University. The study was conducted in accordance with the local legislation and institutional requirements. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AE-D: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. RA: Funding acquisition, Project administration, Writing – review & editing. FF: Methodology, Writing – original draft. MY: Conceptualization, Supervision, Writing – review & editing. AS: Data curation, Software, Validation, Visualization, Writing – original draft. GD: Conceptualization, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research project was supported by Researchers Supporting Project number (RSPD2024R1018), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1467460/full#supplementary-material
References
Abd El-Emam, M. M., Mostafa, M., Farag, A. A., Youssef, H. S., El-Demerdash, A. S., Bayoumi, H., et al. (2023). The potential effects of quercetin-loaded nanoliposomes on amoxicillin/clavulanate-induced hepatic damage: targeting the SIRT1/Nrf2/NF-$κ$B signaling pathway and microbiota modulation. Antioxidants 12:1487. doi: 10.3390/antiox12081487
Abd El-Kalek, H. H., and Mohamed, E. A. (2012). Synergistic effect of certain medicinal plants and amoxicillin against some clinical isolates of methicillin--resistant Staphylococcus aureus (MRSA). Int. J. Pharm. Appl. 3, 387–398.
Abdelaziz, M. O., Bonura, C., Aleo, A., Fasciana, T., and Mammina, C. (2013). NDM-1-and OXA-163-producing Klebsiella pneumoniae isolates in Cairo, Egypt, 2012. J. Glob. Antimicrob. Resist. 1, 213–215. doi: 10.1016/j.jgar.2013.06.003
Al-Halbosy, A. T. F., Hamada, A. A., Faihan, A. S., Saleh, A. M., Yousef, T. A., Abou-Krisha, M. M., et al. (2023). “Synthesis, spectroscopic, anti-microbial evaluation, ADMET, toxicity, and molecular docking studies of Nano-Thiourea derivative metal complexes” in ADMET, Toxicity, and Molecular Docking Studies of Nano-Thiourea Derivative Metal Complexes.
Ali, N. M., Mohamed, G. A. E., and El-Demerdash, A. S. (2023). Impact of Oral Administration of Chitosan--nanoparticles on oxidative stress index and gut microbiota of heat stressed broilers. J. Adv. Vet. Res. 13, 997–1003.
Amin, B. H., Abou-Dobara, M. I., Diab, M. A., Gomaa, E. A., El-Mogazy, M. A., El-Sonbati, A. Z., et al. (2020). Synthesis, characterization, and biological investigation of new mixed-ligand complexes. Appl. Organomet. Chem. 34:e5689. doi: 10.1002/aoc.5689
Anes, J., Hurley, D., Martins, M., and Fanning, S. (2017). Exploring the genome and phenotype of multi-drug resistant Klebsiella pneumoniae of clinical origin. Front. Microbiol. 8:1913. doi: 10.3389/fmicb.2017.01913
Bakkali, F., Averbeck, S., Averbeck, D., and Idaomar, M. (2008). Biological effects of essential oils – a review. Food Chem. Toxicol. 46, 446–475. doi: 10.1016/j.fct.2007.09.106
Ballén, V., Gabasa, Y., Ratia, C., Ortega, R., Tejero, M., and Soto, S. (2021). Antibiotic resistance and virulence profiles of Klebsiella pneumoniae strains isolated from different clinical sources. Front. Cell. Infect. Microbiol. 11:738223. doi: 10.3389/fcimb.2021.738223
Bauer, A. W., Kirby, W. M., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Birhan, W., Giday, M., and Teklehaymanot, T. (2011). The contribution of traditional healers’ clinics to public health care system in Addis Ababa, Ethiopia: a cross-sectional study. J. Ethnobiol. Ethnomed. 7, 1–7. doi: 10.1186/1746-4269-7-39
Burt, S. A., Fledderman, M. J., Haagsman, H. P., van Knapen, F., and Veldhuizen, E. J. A. (2007). Inhibition of Salmonella enterica serotype Enteritidis on agar and raw chicken by carvacrol vapour. Int. J. Food Microbiol. 119, 346–350. doi: 10.1016/j.ijfoodmicro.2007.05.002
Chang, D., Sharma, L., Dela Cruz, C. S., and Zhang, D. (2021). Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front. Microbiol. 12:750662. doi: 10.3389/fmicb.2021.750662
CLSI (2020). CLSI M100-ED29: 2021 performance standards for antimicrobial susceptibility testing, 30th Edition. CLSI 40, 50–51.
Cordeiro, L., Figueiredo, P., Souza, H., Sousa, A., Andrade-Júnior, F., Medeiros, D., et al. (2020). Terpinen-4-ol as an antibacterial and Antibiofilm agent against Staphylococcus aureus. Int. J. Mol. Sci. 21:4531. doi: 10.3390/ijms21124531
Costa, D. T. D. (2019). Virulence determinants and antibiotic resistance of extended-Spectrum Beta-lactamase (ESBL) producing Klebsiella pneumoniae isolated from hospital environment. Dhaka, Bangladesh: Brac University.
Dardona, Z. W. (2014). Evaluating the effect of selected midicinal plant extracts and their synergistic effect with metronidazole against entamoeba histolytica. EJBPS 6, 32–37.
Dardona, Z., and Al-Hindi, A. (2019). Evaluating the effect of selected medicinal plant extracts and their synergistic effect with metronidazole against Entamoeba histolytica. Eur. J. Biomed. Pharmaceutical Sci. 6, 32–37.
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Del Prete, R., Ronga, L., Addati, G., Magrone, R., Abbasciano, A., Decimo, M., et al. (2019). Trends in Klebsiella pneumoniae strains isolated from the bloodstream in a teaching hospital in southern Italy. Infez. Med. 27, 17–25
Dhama, K., Latheef, S. K., Mani, S., Samad, H. A., Karthik, K., Tiwari, R., et al. (2015). Multiple beneficial applications and modes of action of herbs in poultry health and production-a review. Int. J. Pharmacol. 11, 152–176. doi: 10.3923/ijp.2015.152.176
Dhami, N. (2013). Trends in Pharmacognosy: a modern science of natural medicines. J. Herb. Med. 3, 123–131. doi: 10.1016/j.hermed.2013.06.001
Diniz, A. F., Santos, B., Nóbrega, L., Santos, V. R. L., Mariz, W. S., Cruz, P. S. C., et al. (2023). Antibacterial activity of Thymus vulgaris (thyme) essential oil against strains of Pseudomonas aeruginosa, Klebsiella pneumoniae and Staphylococcus saprophyticus isolated from meat product. Braz. J. Biol. 83:e275306. doi: 10.1590/1519-6984.275306
Dumlupinar, B., Karatoprak, G., Celik, D. D., Gürer, Ü. S., Demirci, B., Gürbüz, B., et al. (2020). Synergic potential of Pelargonium endlicherianum Fenzl. Essential oil and antibiotic combinations against Klebsiella pneumoniae. S. Afr. J. Bot. 135, 117–126. doi: 10.1016/j.sajb.2020.08.022
Durairaj, S., Srinivasan, S., and Lakshmanaperumalsamy, P. (2009). In vitro antibacterial activity and stability of garlic extract at different pH and temperature. Electron. J. Biol. 5, 5–10.
Ebrahem, A. F., El-Demerdash, A. S., Orady, R. M., and Nabil, N. M. (2023). Modulatory effect of competitive exclusion on the transmission of ESBL E. coli in chickens. Probiotics Antimicrob proteins, 1–12.
El Damaty, H. M., El-Demerdash, A. S., Abd El-Aziz, N. K., Yousef, S. G., Hefny, A. A., Abo Remela, E. M., et al. (2023). Molecular characterization and antimicrobial susceptibilities of Corynebacterium pseudotuberculosis isolated from Caseous lymphadenitis of smallholder sheep and goats. Animals 13:2337. doi: 10.3390/ani13142337
El-Azzouny, M. M., El-Demerdash, A. S., Seadawy, H. G., and Abou-Khadra, S. H. (2018). Antimicrobial effect of garlic (Allium sativum) and thyme (Zataria multiflora Boiss) extracts on some food borne pathogens and their effect on virulence gene expression. Cell. Mol. Biol. 64, 79–86. doi: 10.14715/cmb/2018.64.10.13
El-Demerdash, A. S., Aggour, M. G., El-Azzouny, M. M., and Abou-Khadra, S. H. (2018). Molecular analysis of integron gene cassette arrays associated multi-drug resistant Enterobacteriaceae isolates from poultry. Cell. Mol. Biol. 64, 149–156. doi: 10.14715/cmb/2018.64.5.25
El-Demerdash, A. S., Al Atfeehy, N. M., Hamed, R. I., Bakry, N. R., Matter, A. A., and Eid, S. (2023a). Mobile Colistin resistance determinants among Enterobacteriaceae isolated from different poultry species. J. Adv. Vet. Res. 13, 1004–1010.
El-Demerdash, A. S., Bakry, N. R., Aggour, M. G., Elmasry, S. S., Mowafy, R. E., Erfan, A., et al. (2023b). Bovine mastitis in Egypt: bacterial etiology and evaluation of diagnostic biomarkers. Int. J. Vet. Sci. 12, 60–69. doi: 10.47278/journal.ijvs/2022.161
El-Demerdash, A. S., Mowafy, R. E., Fahmy, H. A., Matter, A. A., and Samir, M. (2023c). Pathognomonic features of Pasteurella multocida isolates among various avian species in Sharkia governorate, Egypt. World J. Microbiol. Biotechnol. 39:335. doi: 10.1007/s11274-023-03774-2
El-Demerdash, A. S., Orady, R. M., Matter, A. A., and Ebrahem, A. F. (2023d). An alternative approach using Nano-garlic emulsion and its synergy with antibiotics for controlling biofilm-producing multidrug-resistant Salmonella in chicken. Indian J. Microbiol. 63, 632–644. doi: 10.1007/s12088-023-01124-2
Ernst, C. M., Braxton, J. R., Rodriguez-Osorio, C. A., Zagieboylo, A. P., Li, L., Pironti, A., et al. (2020). Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat. Med. 26, 705–711. doi: 10.1038/s41591-020-0825-4
Essawi, W. M., El-Demerdash, A. S., El-Mesalamy, M. M., and Abonorag, M. A. (2020). Validation of camel’s fetal fluids as antimicrobial agents. Curr. Microbiol. 77, 1399–1404. doi: 10.1007/s00284-020-01945-0
Fang, C.-T., Chuang, Y.-P., Shun, C.-T., Chang, S.-C., and Wang, J.-T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Ferreira, R. L., Da Silva, B. C. M., Rezende, G. S., Nakamura-Silva, R., Pitondo-Silva, A., Campanini, E. B., et al. (2019). High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and $β$-lactamase encoding genes in a Brazilian intensive care unit. Front. Microbiol. 9:3198. doi: 10.3389/fmicb.2018.03198
Garcia, L. S. (2010). Clinical microbiology procedures handbook. Washington, D.C., USA: American Society for Microbiology Press.
Gorrie, C. L., Mirčeta, M., Wick, R. R., Edwards, D. J., Thomson, N. R., Strugnell, R. A., et al. (2017). Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215. doi: 10.1093/cid/cix270
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6:a025387. doi: 10.1101/cshperspect.a025387
Hartman, L. J., Selby, E. B., Whitehouse, C. A., Coyne, S. R., Jaissle, J. G., Twenhafel, N. A., et al. (2009). Rapid real-time PCR assays for detection of Klebsiella pneumoniae with the rmpA or magA genes associated with the hypermucoviscosity phenotype: screening of nonhuman primates. J. Mol. Diagn. 11, 464–471. doi: 10.2353/jmoldx.2009.080136
Hashem, N. M., Essawi, W. M., El-Demerdash, A. S., and El-Raghi, A. A. (2024). Biomolecule-producing probiotic bacterium Lactococcus lactis in free or Nanoencapsulated form for endometritis treatment and fertility improvement in buffaloes. J. Funct. Biomater. 15:138. doi: 10.3390/jfb15060138
Hawkey, P. M., Warren, R. E., Livermore, D. M., McNulty, C. A. M., Enoch, D. A., Otter, J. A., et al. (2018). Treatment of infections caused by multidrug-resistant gram-negative bacteria: report of the British Society for Antimicrobial Chemotherapy/healthcare infection society/british infection association joint working party. J. Antimicrob. Chemother. 73, iii2–iii78. doi: 10.1093/jac/dky027
Hsu, C.-R., Lin, T.-L., Chen, Y.-C., Chou, H.-C., and Wang, J.-T. (2011). The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology (N.Y.) 157, 3446–3457. doi: 10.1099/mic.0.050336-0
Hyldgaard, M., Mygind, T., and Meyer, R. L. (2012). Essential oils in food preservation: mode of action, synergies, and interactions with food matrix components. Front. Microbiol. 3:12. doi: 10.3389/fmicb.2012.00012
Ikeda, M., Mizoguchi, M., Oshida, Y., Tatsuno, K., Saito, R., Okazaki, M., et al. (2018). Clinical and microbiological characteristics and occurrence of Klebsiella pneumoniae infection in Japan. Int. J. Gen. Med. 11, 293–299. doi: 10.2147/IJGM.S166940
Izquierdo, L., Coderch, N., Piqué, N., Bedini, E., Corsaro, M. M., Merino, S., et al. (2003). The Klebsiella pneumoniae WabG gene: role in biosynthesis of the core lipopolysaccharide and virulence. J. Bacteriol. 185, 7213–7221. doi: 10.1128/JB.185.24.7213-7221.2003
Jung, S. W., Chae, H. J., Park, Y. J., Yu, J. K., Kim, S. Y., Lee, H. K., et al. (2013). Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol. Infect. 141, 334–340. doi: 10.1017/S0950268812000933
Khan, M. R., Huang, C., Zhao, H., Huang, H., Ren, L., Faiq, M., et al. (2021). Antioxidant activity of thymol essential oil and inhibition of polyphenol oxidase enzyme: a case study on the enzymatic browning of harvested longan fruit. Chem. Biol. Technol. Agric. 8, 1–10. doi: 10.1186/s40538-021-00259-y
Khosravi, A., and Malekan, M. A. (2004). Determination of alcoholic and aqueous extract of lavender Astvkas on Staphylococcus aureus and other gram-negative bacteria. J. Qazvin Univ. Med. Sci. 29, 3–9.
Konaté, K., Mavoungou, J. F., Lepengué, A. N., Aworet-Samseny, R. R. R., Hilou, A., Souza, A., et al. (2012). Antibacterial activity against $β$-lactamase producing methicillin and ampicillin-resistants Staphylococcus aureus: fractional inhibitory concentration index (FICI) determination. Ann. Clin. Microbiol. Antimicrob. 11, 18–12. doi: 10.1186/1476-0711-11-18
Kowalczyk, A., Przychodna, M., Sopata, S., Bodalska, A., and Fecka, I. (2020). Thymol and thyme essential oil—new insights into selected therapeutic applications. Molecules 25:4125. doi: 10.3390/molecules25184125
Lee, C.-R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., and Lee, S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. doi: 10.3389/fmicb.2016.00895
Lopez-Romero, J. C., González-Ríos, H., Borges, A., and Simões, M. (2015, 2015). Antibacterial effects and mode of action of selected essential oils components against Escherichia coli and Staphylococcus aureus. Evid. Based Complement. Alternat. Med. 2015:795435. doi: 10.1155/2015/795435
Matsuo, A. L., Figueiredo, C. R., Arruda, D. C., Pereira, F. V., Scutti, J. A. B., Massaoka, M. H., et al. (2011). $α$-Pinene isolated from Schinus terebinthifolius Raddi (Anacardiaceae) induces apoptosis and confers antimetastatic protection in a melanoma model. Biochem. Biophys. Res. Commun. 411, 449–454. doi: 10.1016/j.bbrc.2011.06.176
Megahed, M. M. M., El-Nagar, A. M. A., El-Demerdash, A. S., Ayoub, M. A., and Tolba, H. M. N. (2023). Evaluation and development of diagnostic tools for rapid detection of Riemerella anatipestifer and Pasteurella multocida in ducks. J. Adv. Vet. Anim. Res. 10, 211–221. doi: 10.5455/javar.2023.j671
Millezi, A. F., Piccoli, R. H., Oliveira, J. M., and Pereira, M. O. (2016). Anti-biofim and antibacterial effect of essential oils and their major compounds. J. Essent. Oil Bear. Plants 19, 624–631. doi: 10.1080/0972060X.2014.960262
Mohamed, S. H., Salem, D., Azmy, M., and Fam, N. S. (2018). Antibacterial and antibiofilm activity of cinnamaldehyde against carbapenem-resistant Acinetobacter baumannii in Egypt: in vitro study. J. Appl. Pharm. Sci. 8, 151–156. doi: 10.7324/JAPS.2018.81121
Nasr, N. N., Babiker, H. E., and Saeed, H.A. (2016). Phenotypic and molecular characterization of community-acquired bacterial pneumonia in Khartoum state: Sudan University of Science and Technology. Int. J. Adv. Sci. Tech. 6, 268–283.
Nazzaro, F., Fratianni, F., De Martino, L., Coppola, R., and De Feo, V. (2013). Effect of essential oils on pathogenic bacteria. Pharmaceuticals 6, 1451–1474. doi: 10.3390/ph6121451
Ndlovu, T., Kgosietsile, L., Motshwarakgole, P., and Ndlovu, S. I. (2023). Evaluation of potential factors influencing the dissemination of multidrug-resistant Klebsiella pneumoniae and alternative treatment strategies. Trop. Med. Infect. Dis. 8:381. doi: 10.3390/tropicalmed8080381
Nimnoi, P., and Pongsilp, N. (2022). Identification, characterization, and virulence gene expression of marine enterobacteria in the upper gulf of Thailand. Microorganisms 10:511. doi: 10.3390/microorganisms10030511
Nordmann, P., Cuzon, G., and Naas, T. (2009). The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9, 228–236. doi: 10.1016/S1473-3099(09)70054-4
Opoku-Temeng, C., Kobayashi, S. D., and DeLeo, F. R. (2019). Klebsiella pneumoniae capsule polysaccharide as a target for therapeutics and vaccines. Comput. Struct. Biotechnol. J. 17, 1360–1366. doi: 10.1016/j.csbj.2019.09.011
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Purkait, S., Bhattacharya, A., Bag, A., and Chattopadhyay, R. R. (2020). Synergistic antibacterial, antifungal and antioxidant efficacy of cinnamon and clove essential oils in combination. Arch. Microbiol. 202, 1439–1448. doi: 10.1007/s00203-020-01858-3
Ramirez, M. S., and Tolmasky, M. E. (2017). Amikacin: uses, resistance, and prospects for inhibition. Molecules 22:2267. doi: 10.3390/molecules22122267
Razali, N. M., and Wah, Y. B. (2011). Power comparisons of shapiro-wilk, kolmogorov-smirnov, lilliefors and Anderson-darling tests. J. Stat. Model. Anal. 2, 21–33.
Ripabelli, G., Tamburro, M., Guerrizio, G., Fanelli, I., Flocco, R., Scutellà, M., et al. (2018). Tracking multidrug-resistant Klebsiella pneumoniae from an Italian hospital: molecular epidemiology and surveillance by PFGE, RAPD and PCR-based resistance genes prevalence. Curr. Microbiol. 75, 977–987. doi: 10.1007/s00284-018-1475-3
Rybicki, E. P., Chikwamba, R., Koch, M., Rhodes, J. I., and Groenewald, J.-H. (2012). Plant-made therapeutics: an emerging platform in South Africa. Biotechnol. Adv. 30, 449–459. doi: 10.1016/j.biotechadv.2011.07.014
Saad, M. F., Elsayed, M. M., Khder, M., Abdelaziz, A. S., and El-Demerdash, A. S. (2024). Biocontrol of multidrug resistant pathogens isolated from fish farms using silver nanoparticles combined with hydrogen peroxide insight to its modulatory effect. Sci. Rep. 14:7971. doi: 10.1038/s41598-024-58349-4
Salari, N., Kanjoori, A. H., Hosseinian-Far, A., Hasheminezhad, R., Mansouri, K., and Mohammadi, M. (2023). Global prevalence of drug-resistant tuberculosis: a systematic review and meta-analysis. Infect. Dis. Poverty 12, 1–12. doi: 10.1186/s40249-023-01107-x
Santurio, D. F., de Jesus, F. P. K., Zanette, R. A., Schlemmer, K. B., Fraton, A., and Fries, L. L. M. (2014). Antimicrobial activity of the essential oil of thyme and of thymol against Escherichia coli strains. Acta Sci. Vet. 42, 1–4.
Shalaby, A. G., Bakry, N. R., and El-Demerdash, A. S. (2021). Virulence attitude estimation of Pasteurella multocida isolates in embryonated chicken eggs. Arch. Microbiol. 203, 6153–6162. doi: 10.1007/s00203-021-02579-x
Shon, A. S., Bajwa, R. P. S., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Shu, H.-Y., Fung, C.-P., Liu, Y.-M., Wu, K.-M., Chen, Y.-T., Li, L.-H., et al. (2009). Genetic diversity of capsular polysaccharide biosynthesis in Klebsiella pneumoniae clinical isolates. Microbiology (N.Y.) 155, 4170–4183. doi: 10.1099/mic.0.029017-0
Slade, D., Galal, A. M., Gul, W., Radwan, M. M., Ahmed, S. A., Khan, S. I., et al. (2009). Antiprotozoal, anticancer and antimicrobial activities of dihydroartemisinin acetal dimers and monomers. Bioorg. Med. Chem. 17, 7949–7957. doi: 10.1016/j.bmc.2009.10.019
Stokes, M. E., Davis, C. S., and Koch, G. G. (2012). Categorical data analysis using SAS. Cary, North Carolina, USA: SAS Institute.
Tiwari, B. K., Valdramidis, V. P., O’Donnell, C. P., Muthukumarappan, K., Bourke, P., and Cullen, P. J. (2009). Application of natural antimicrobials for food preservation. J. Agric. Food Chem. 57, 5987–6000. doi: 10.1021/jf900668n
Turton, J. F., Perry, C., Elgohari, S., and Hampton, C. V. (2010). PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 59, 541–547. doi: 10.1099/jmm.0.015198-0
Upadhyay, A., Upadhyaya, I., Kollanoor-Johny, A., and Venkitanarayanan, K. (2014). Combating pathogenic microorganisms using plant-derived antimicrobials: a Minireview of the mechanistic basis. Biomed. Res. Int. 2014, 1–18. doi: 10.1155/2014/761741
van Vuuren, S., and Viljoen, A. (2011). Plant-based antimicrobial studies--methods and approaches to study the interaction between natural products. Planta Med. 77, 1168–1182. doi: 10.1055/s-0030-1250736
Witte, W. (1998). Medical consequences of antibiotic use in agriculture. Science 279, 996–997. doi: 10.1126/science.279.5353.996
Yang, F., Deng, B., Liao, W., Wang, P., Chen, P., and Wei, J. (2019). High rate of multiresistant Klebsiella pneumoniae from human and animal origin. Infect. Drug Resist. 12, 2729–2737. doi: 10.2147/IDR.S219155
Yuan, J. S., Reed, A., Chen, F., and Stewart, C. N. (2006). Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85
Keywords: antibacterial activity, bioactive compounds, capsule gene expression, Klebsiella pneumoniae , tea tree oil, thyme oil
Citation: El-Demerdash AS, Alfaraj R, Farid FA, Yassin MH, Saleh AM and Dawwam GE (2024) Essential oils as capsule disruptors: enhancing antibiotic efficacy against multidrug-resistant Klebsiella pneumoniae. Front. Microbiol. 15:1467460. doi: 10.3389/fmicb.2024.1467460
Edited by:
Petar Knezevic, University of Novi Sad, SerbiaReviewed by:
Gospava Lazic, Scientific Veterinary Institute Novi Sad, SerbiaAhmed S. Khairalla, University of Regina, Canada
Copyright © 2024 El-Demerdash, Alfaraj, Farid, Yassin, Saleh and Dawwam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Azza SalahEldin El-Demerdash, ZHIuYXp6YXNhbGFoQHlhaG9vLmNvbQ==,;ZHIuYXp6YUBhaHJpLmdvdi5lZw==; Rihaf Alfaraj, cmFsZmFyYWpAa3N1LmVkdS5zYQ==
†ORCID: Azza SalahEldin El-Demerdash, orcid.org/0000-0002-0825-9776
 Azza SalahEldin El-Demerdash
Azza SalahEldin El-Demerdash Rihaf Alfaraj
Rihaf Alfaraj Faten A. Farid3
Faten A. Farid3 Abdulrahman M. Saleh
Abdulrahman M. Saleh Ghada E. Dawwam
Ghada E. Dawwam