
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 01 May 2024
Sec. Terrestrial Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1384367
This article is part of the Research TopicInsights in Terrestrial Microbiology: 2023/2024View all 10 articles
 Xintong Dong1†
Xintong Dong1† Man Chen1†
Man Chen1† Qi Chen1
Qi Chen1 Kangfei Liu1
Kangfei Liu1 Jie Long1
Jie Long1 Yunzhou Li1
Yunzhou Li1 Yinuo Ren1
Yinuo Ren1 Tao Yang1
Tao Yang1 Jinxing Zhou2
Jinxing Zhou2 Saman Herath3
Saman Herath3 Xiawei Peng1,4,5*
Xiawei Peng1,4,5*Karst rocky desertification refers to the process of land degradation caused by various factors such as climate change and human activities including deforestation and agriculture on a fragile karst substrate. Nutrient limitation is common in karst areas. Moss crust grows widely in karst areas. The microorganisms associated with bryophytes are vital to maintaining ecological functions, including climate regulation and nutrient circulation. The synergistic effect of moss crusts and microorganisms may hold great potential for restoring degraded karst ecosystems. However, our understanding of the responses of microbial communities, especially abundant and rare taxa, to nutrient limitations and acquisition in the presence of moss crusts is limited. Different moss habitats exhibit varying patterns of nutrient availability, which also affect microbial diversity and composition. Therefore, in this study, we investigated three habitats of mosses: autochthonal bryophytes under forest, lithophytic bryophytes under forest and on cliff rock. We measured soil physicochemical properties and enzymatic activities. We conducted high-throughput sequencing and analysis of soil microorganisms. Our finding revealed that autochthonal moss crusts under forest had higher nutrient availability and a higher proportion of copiotrophic microbial communities compared to lithophytic moss crusts under forest or on cliff rock. However, enzyme activities were lower in autochthonal moss crusts under forest. Additionally, rare taxa exhibited distinct structures in all three habitats. Analysis of co-occurrence network showed that rare taxa had a relatively high proportion in the main modules. Furthermore, we found that both abundant and rare taxa were primarily assembled by stochastic processes. Soil properties significantly affected the community assembly of the rare taxa, indirectly affecting microbial diversity and complexity and finally nutrient acquisition. These findings highlight the importance of rare taxa under moss crusts for nutrient acquisition. Addressing this knowledge gap is essential for guiding ongoing ecological restoration projects in karst rocky desertification regions.
Land degradation associated with karst rocky desertification is caused by both natural processes and human activities on a fragile karst background (Meng et al., 2021). Rocky desertification has become a challenge for the sustainable ecological development of Southwest China (Yang et al., 2022a). Moss biocrusts are crucial for reducing and preventing soil erosion on rock surfaces and for supporting the long-term viability of the vegetation restoration process (Kidron and Drahorad, 2022). On the other hand, the microbial community in these moss biocrusts plays a significant ecological role (Cheng et al., 2022). Therefore, it is essential to comprehend the functions and mechanisms of these microorganisms in ecological functions.
Moss biocrusts in the karst areas contain a large number of highly diverse microorganisms. Microbial diversity is a fundamental aspect of supporting the services provided by the soil ecosystem (Fanin et al., 2018). These communities are influential in processes such as the turnover of soil organic matter (SOM), soil carbon (C) sequestration, and water acquisition (Xiao and Veste, 2017). Essentially, the presence of abundant and diverse soil microorganisms, particularly in biocrusts, can greatly benefit various soil processes, especially those related to C and nitrogen (N) acquisition (Bhattacharyya and Furtak, 2022). During nutrient deficiencies, microorganisms obtain nutrients by increasing secretion of extracellular enzymes that decompose SOM (Cui et al., 2018). For example, in N-limited regions, it has been shown that soil organic N mineralization is related to soil extracellular N-acquisition enzymes (Xiao et al., 2021). These interactions between soil microbes and soil enzymes are essential for understanding nutrient limitations in land restoration in degraded areas (Liu et al., 2023).
Assembly processes of microbial communities play crucial roles in determining the rate and efficiency of microbial growth (Anthony et al., 2020). Community assembly arises from the interaction of deterministic factors such as heterogeneous selection, homogenous selection, and stochastic processes such as dispersion limitations and homogeneous dispersal (Xue et al., 2018). Assembly processes drive ecosystem functions (Knelman and Nemergut, 2014); they are mainly reflected in aspects such as climate regulation, nutrient cycle, and plant growth (Gao et al., 2020; Hartmann and Six, 2022). Stochastic processes could bring new species from the regional pool which carry traits affecting ecosystem functioning but are not present in the initial community. In this way, stochastic processes could enhance the effect of biodiversity on functions through sampling effects (Knelman and Nemergut, 2014). Meanwhile, microbial communities commonly display an inclined distribution of species abundance with a large proportion of rare taxa coexisting with a small number of abundant taxa (Lynch and Neufeld, 2015; Jia et al., 2018). Under low-salt-stress environments, the abundant taxa play an important role in stabilizing ecological networks. However, the role of rare taxa becomes more and more important when salt stress increases (Li et al., 2023). Furthermore, although rare taxa have a low abundance, they are highly diverse and have functional redundancy, such as nitrogen fixation, sulfur oxidation, and accelerating organic matter breakdown (Peter et al., 2011; Sauret et al., 2014; Hua et al., 2015). The community composition of rare taxa is more stable under the influence of climate change and other disturbances, such as copper stress, freeze-thaw, and mechanical disturbances (Pedrós-Alió, 2011). The role of abundant and rare microorganisms under moss crusts in karst areas remains poorly understood. The problem of nutrient limitation is more prominent in karst areas (Zhou et al., 2020). Researchers have highlighted the important role of abundant and rare taxa on nutrient acquisition. In the karst area, microbes under moss crusts show inconsistent assembly processes. Based on this background, understanding the contribution of abundant and rare taxa and to nutrient acquisition is essential concerning the restoration of karst areas. This knowledge can offer novel insights into the microorganisms under bryophytes in karst areas.
In the present study, we focused on three different moss habitats: (i) lithophytic moss crust of forest, (ii) autochthonal moss crusts of forest, and (iii) lithophytic moss crust of cliff. We used high-throughput amplicon sequencing based on 16S rRNA genes to evaluate the community structure and network stability of bacteria under moss crust. Enzyme stoichiometric analysis (EEA) was performed. We also studied the abundant and rare taxa assembly process and analyzed the links with soil physicochemical properties and nutrient acquisition. We hypothesized that (i) different bryophyte habitats lead to different soil properties and soil enzyme activities; (ii) the microbial response to nutrient restriction differs in the three habitats; and (iii) the assembly processes of abundant and rare microorganisms are inconsistent in different habitats. Bryophytes and microorganisms are closely linked, and this study also aimed to provide recommendations for the restoration of karst areas in Southwest China using bryophytes combined with associated microbes.
Soil samples were collected from Guiyang and Anshun, Guizhou Province, China (94°37′9′′#x2013;103°31′9′′ E, 36°56′9′′–40°34′9′′ N). In total, 36 soil samples under bryophytes were collected from three different sites (site A, 11 samples from lithophytic moss crust under the forest; site B, 17 samples from autochthonal moss crusts under the forest; site C, 8 samples from lithophytic moss crust of cliff). The study area is situated in a warm and subtropical region characterized by a humid temperate landscape with a continental monsoon pattern. The annual average rainfall is 1,100 mm and the mean air temperature ranges from 15.3 to 19.8°C. There is very little soil under the lithophytic moss crust. We collected the soil using the five-point sampling method to be mixed together as a sample; samples from the same type of moss habitat were replications. There were 11 replications in site A, 17 replications in site B, and 8 replications in site C. The specific sampling method is as follows: for lithophytic moss crust, we used a sterile blade to shovel the moss crust tightly against the rock wall; then, we used a sterile brush to sweep the roots of the moss crust and collect the soil. For autochthonal moss crust, we randomly selected some 1 m × 1 m plots in the study area. The nearest distance between sampling points in the sample field was approximately 10 m, and the sampling depth of soil samples under the crust was 0–2 cm. The soil samples were sieved to a particle size of 2 mm eliminating any discernible roots or rock fragments. The soil samples were separated into three portions: the first portion was kept at a temperature of −80°C to extract microbial DNA from the soil; we put the second portion at a temperature of 4°C to measure the activity of soil enzymes within a week; and the third portion was dried in air for analysis physical and chemical properties of the soil.
Soil pH and electrical conductivity (EC) were measured using a pH-EC meter in the soil: water (1:5) extraction solution. Soil moisture content was determined gravimetrically. Soil samples were extracted with 2 M KCl and ammonium and nitrate nitrogen (NH4+-N and NO3–-N) were determined using an automatic flow injection analyzer (AutoAnalyzer-AA3, Sea Analytics, Norderstedt, Germany). Soil organic carbon (SOC) content was determined by the external heating method of K2Cr2O7 (Rayment and Lyons, 2011). Additionally, a Hanon Kjeltec 9840 analyzer (K9840, Hanon, CHN) was used to identify soil total nitrogen (TN) while a microplate reader (Infinite M200PRO, Tecan, CH) was used to measure the total phosphorus (TP) in the soil. We used a 0.5 M NaHCO3 solution and a microplate analyzer (Infinite M200PRO, Tecan, CH) to assess the available phosphorus (AP) for plants. Neutral ammonium acetate (1 M) was used to extract the soil available potassium (AK), and a flame spectrophotometer (FP6450, INESA, CHN) was used to measure the amount of AK.
Quantification was made for the soil enzymes that are involved in carbon acquisition including α-1,4-glucosidase (AG), β-1,4-glucosidase (BG), xylosidase (XS), and β-D-cellobiohydrolase (CB); nitrogen acquisition enzymes including leucine aminopeptidase (LAP), and β-N-acetylglucosaminidase (NAG); phosphorus acquisition enzymes such as alkaline phosphatase (AP). The enzyme activities were evaluated using the microplate method described by Ai et al. (2012). All enzyme activities were standardized using Z-score to visually evaluate the differences between the three sites. We used Eq. 1 to calculate the Z-score:
where x, μ, and σ represent individual activity, average activity, and standard deviation of activity, respectively.
Two methods were used to investigate the microbial limitations. The first method was generating a scatter plot of the eco-enzymatic stoichiometry. The x-axis was determined by (LAP + NAG) / AP while the y-axis was determined by BG / (LAP + NAG). This strategy was based on the guidelines provided by Sinsabaugh et al. (2009). In this plot, four distinct categories of resource limitations were observed, as determined by deviations from the expected enzyme ratio of C:N (1:1) or N:P (1:1) as presented by Sinsabaugh et al. (2009). The soil enzyme activity ratio were calculated using Eqs 2–4.
The second method was calculating the lengths and angles of the vectors for enzymatic activity to quantify the microbial nutrient limitation. The vector length indicates the relative microbial C limitation; the larger the vector length greater the relative microbial C limitation degree (Ma et al., 2021). The vector angle represents the soil relative microbial N (or P) limitation degree. Vector angles <45° indicate microbial N limitation, while vector angles >45° indicate microbial P limitation. The vector length and angle were calculated using Eqs 5–8.
DNA Isolation Kit, which is produced by MP Biomedicals in Switzerland, was used to extract DNA from 0.5 g of fresh soil samples. The extraction process followed the manufacturer’s recommendations. The extracted DNA was assessed for its quality and quantity using a Nanodrop ND-2000 UV-vis spectrophotometer produced by Nanodrop Technologies in Wilmington, DE, USA. Primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to enhance the bacterial 16S rRNA gene V4 hypervariable region (Dong et al., 2022). PCR reactions were performed in triplicate (S1000 apparatus, Bio-Rad Laboratory, Hercules, CA, USA). The PCR conditions were 5 min at 94°C followed by 30 cycles of 94°C for 45 s, annealing at 54°C for 45 s, 72°C for 1 min followed by a final extension step of 10 min at 72°C (Katiraei et al., 2022). PCR products were purified using the Qiagen Gel Extraction Kit produced by Qiagen in Germany, following the instructions provided by the manufacturer. In the end, the library was sequenced using an Illumina Nova6000 platform, which produced paired end reads of 250 base pairs. The sequencing procedure was conducted by Guangdong Magigene Biotechnology Co., Ltd., situated in Guangzhou, China.
A series of standard processing steps, including demultiplexing, sample inference, read merging, quality filtering, and chimeric elimination, were applied to the raw FASTQ data. The DADA2 pipeline (version 1.20.0) was used to carry out these procedures. After that, an amplicon sequence variant (ASV) microbiological profile was generated (Deissová et al., 2023), ASVs with 100% sequence identity, which are more dependable and can be replicated, were employed to represent microbial taxonomic units. The Ribosomal Database Project (RDP) Classifier was utilized to determine the taxonomic details of every ASV,1 with an 80% level of confidence (Matheri et al., 2023).
Depending on their relative abundance and/or frequency, the microbial ASVs were divided into two groups: abundant and rare taxa. Rare taxa were defined as those with an average relative abundance of <0.1%, whereas an average relative abundance of >1% was classified as abundant taxa (Galand et al., 2009). Based on the copiotrophic-oligotrophic framework and additional documentation, those annotated identified phyla into copiotrophic and oligotrophic taxa (Fierer et al., 2007; Li H. et al., 2021; Li J. et al., 2021). To calculate the microbial copiotroph:oligotroph ratios, the relative abundance of known copiotrophic and oligotrophic members were summed, respectively (Ma et al., 2023).
Also, we evaluated the α-diversity of the microbial community using the vegan package in R version 4.1.2. We employed canonical principal coordinates analysis (PCoA) to investigate the bacterial community’s pattern (Cui et al., 2019). All networks were constructed based on Pearson correlations of log-transformed ASV abundances, followed by an RMT-based approach that determines the correlation cut-off threshold automatically (Yuan et al., 2021). The random forest tests in “RandomForest” package were used to forecast the significant factors (Chen et al., 2021). Additionally, the “rfPermute” package in R was used to analyze each predictor’s significance (Jiao et al., 2018). To assess the ecological processes taking place in microbial communities, we calculated β Nearest Taxon Index (βNTI) (Stegen et al., 2012). Through the integration of —βNTI— (2) and —RCbray— (0.95), we successfully elucidated underlying mechanisms governing community assembly processes. These mechanisms encompass heterogeneous selection, homogeneous selection, dispersal limitation, homogenous dispersal, and undominated processes (Zhou and Ning, 2017). Mantel tests comparing βNTI values with the Euclidean distance matrixes of physicochemical parameters were then performed in the “vegan” R package to explore the major factors influencing the assembly of abundant and rare taxa, and the relationship among soil enzyme activity ratio and network complexity standardized by Z-scores. The direct and indirect effects between soil physical and chemical properties, assembly processes of abundant and rare microbial communities, microbial diversity, microbial complexity, and nutrient acquisition were identified using structural equation models (SEMs) created with the “lavaan” package in R software.
The availability of SOC, NH4+-N, NO3–-N, and A-P in site B was higher, but the enzyme activities were lower compared with sites A and C (Table 1 and Figure 1A). The activity of BG, XS, NAG, and LAP showed significant differences in sites A and C (P < 0.05) (Figure 1A). The scatter plot of eco-enzymatic stoichiometry reveals that most microorganisms were limited by C&N and C&P (Figure 1B). Most microorganisms of the site A were limited by C&N, the microorganisms of sites B and C were susceptible to be limited by C&P (Figure 1B). Vector analysis showed that the site A had the lowest vector length among the three sites representing microbial C limitation. The vector angle at site A was below 45° whereas the vector angles at the other sites were above 45° (Figure 1C). Angles <45° are perceived as more constrained by N rather than P while the converse interpretation applies to angles >45°. This is consistent with the low content of SOC, NH4+-N, and NO3–-N in site A (Table 1). The soil microorganisms at site B were predominantly characterized by the r-strategy species, as indicated by the largest copiotroph/oligotroph ratio (Figure 1D). The high ratio reflected a more nutrient-rich environment.
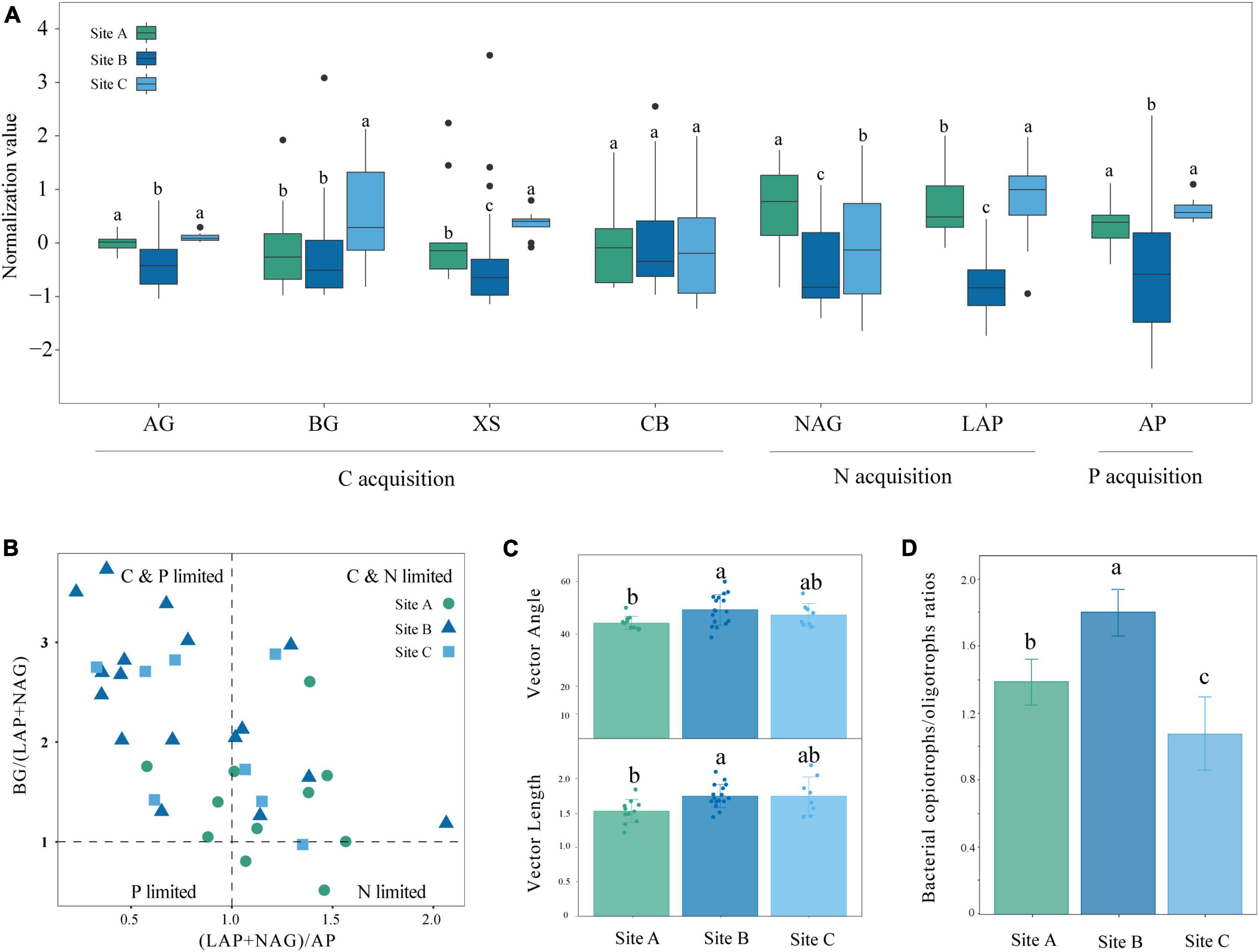
Figure 1. Enzyme activity and soil nutrient limitation in different moss biocrusts habitats: (A) soil enzyme activity of carbon, nitrogen, and phosphorus at three sites, (B) scatter plots of soil enzymatic stoichiometry for studied sites, (C) the vector length and vector angle of the studied sites, and (D) microbial copiotrophs and oligotrophic ratios at different sites. Different lowercase letters indicate significant differences among the three sites (P < 0.05). Different letters indicate statistically significant differences (one-way ANOVA, α = 0.05). AG, α-glucosidase; BG, β-glucosidase; XS, xylosidase; CB, β-D-cellobiohydrolase; NAG, N-acetyl-β-D-glucosidase; LAP, leucine aminopeptidase; AP, alkaline phosphatase.
β-Diversity reflects differences in species composition between the three sites. The microbial community structures can be observed according to the principal coordinates analysis (PCoA), and permutational analysis of variance by Adonis. The results showed significance for the whole (R2 = 0.48, P < 0.01), abundant (R2 = 0.48, P < 0.01), and rare (R2 = 0.66, P < 0.001) communities (Figures 2A–C). While the structure of the rare community was obviously differentiated in all three sites (Figure 2C). The Shannon index of site B was significantly higher than that of the other two sites (Table 2) (P < 0.05). The α-diversity of rare communities was significantly different in the three sites (P < 0.05); however, there was no significant difference between the site C and the other two sites in abundant communities (Table 2). We further analyzed the correlation between the Shannon index of abundant and rare taxa and soil C, N, and P and their stoichiometry (Supplementary Figures 1, 2) and observed a significant correlation between rare taxa diversity and soil C, N, and P nutrients (Supplementary Figures 1, 2). For example, the diversity of rare taxa was negatively correlated with SOC (R2 = −0.38, P = 0.021), TN (R2 = −0.52, P = 0.0012), and TP (R2 = −0.4, P = 0.016). However, the diversity of abundant taxa had no significant correlations with soil C, N, and P nutrients. Further, we analyzed the relationship between the dominant phyla of rare microorganisms and soil nutrients (Supplementary Figure 3), most of which showed negative correlation. There were significant negatively correlations between TN and Acidobacteriota, Actinobacteriota, Bacteroidota, and Desulfobacterota (P < 0.05), and significant positively correlations were observed between C:N ratio and Bacteroidota and Chloroflexi (P < 0.05). These bacterial groups play an important role in soil C, N, and P nutrient cycling.
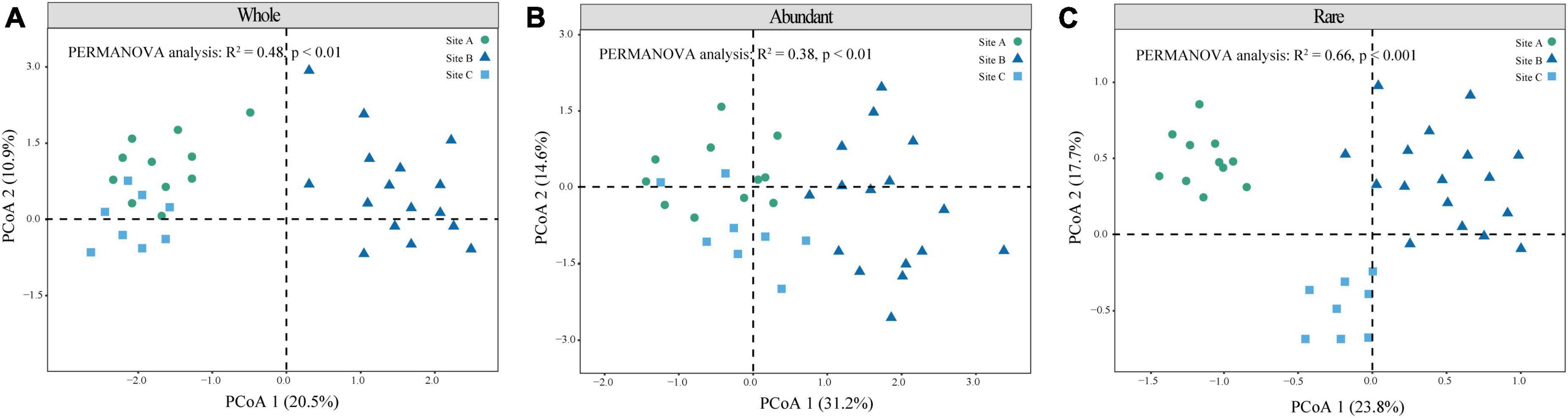
Figure 2. Microbial community structure of different moss crust habitats. Principal coordinate analysis (PCoA) of the whole (A), abundant (B), and rare (C) bacteria communities is given based on Bray–Curtis distances.
Most network nodes and edges were available in site B, resulting in highly clustered microbial network modules. Site C has the simplest network structure (Figure 3A). We used the network topological parameters of node and edge numbers, average degree, diameter, average clustering coefficient, and relative modularity to assess soil microbial network complexity, with higher topological properties representing greater network complexity. In the three sites, the network topology parameters of the site B were higher than those of the other two sites (Table 3). The key modules of the microbial co-occurrence network (major microbial clusters, modules 1 and 2) in the three habitats were dominated by rare taxa (Figure 3B). Our results showed that there was a significant positive correlation between Ln (BG)/Ln (NAG + LAP) and network complexity in sites A and B (P < 0.05) (Figure 3C). This indicates that the microbial network complexity increased as N limitation decreased in sites A and B. Further, there was a significant negative correlation between Ln (BG)/Ln (AP) and network complexity in site B (P < 0.05) (Figure 3D). This indicated that the complexity of microbial network was positively related to the level of P limitation in the site B. There was no significant relationship between network complexity and enzyme activity ratio in site C (Figures 3C, D).
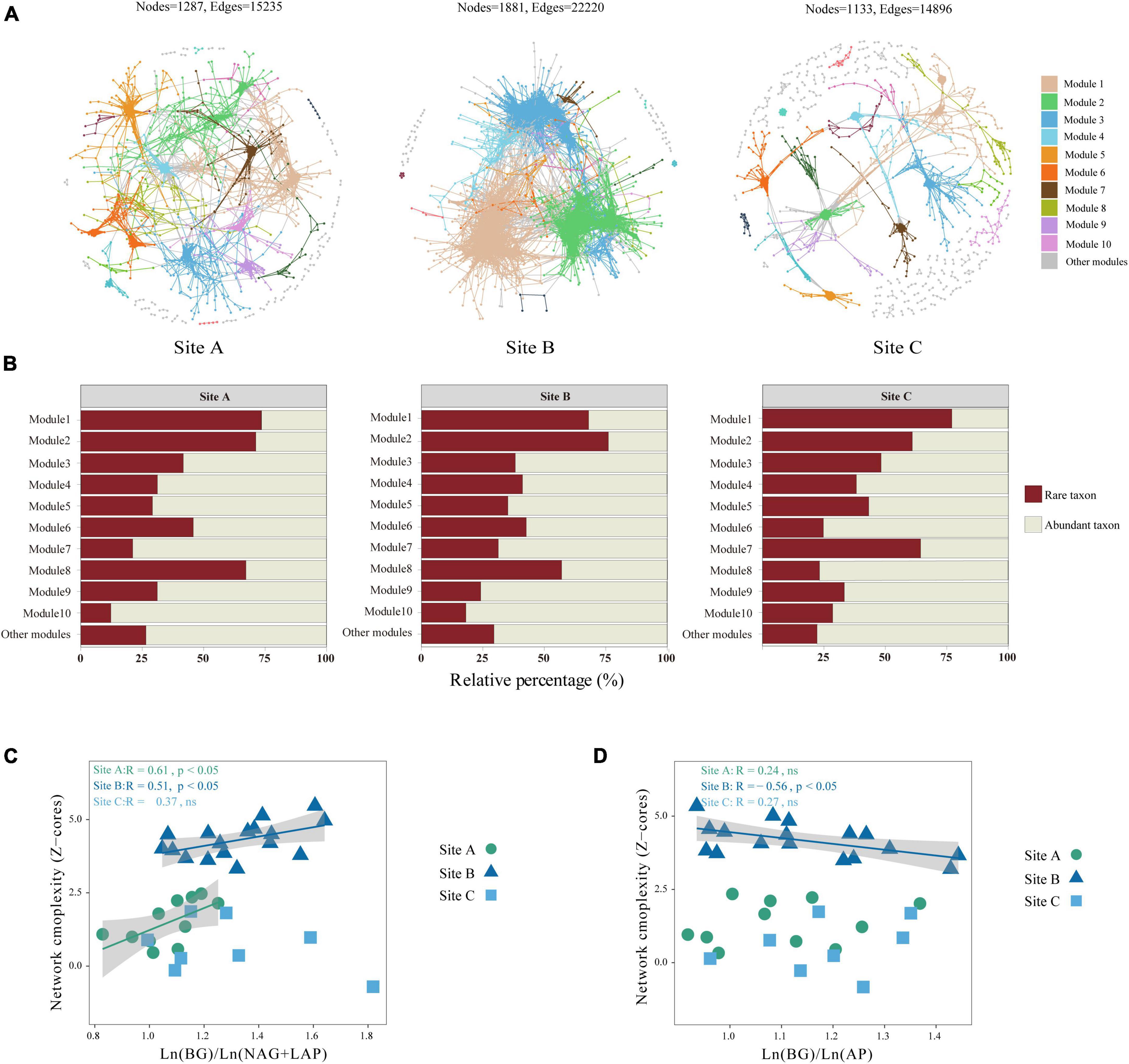
Figure 3. Co-occurrence pattern of bacteria in three sites: (A) the co-occurrence patterns among ASVs were revealed by network analysis. Large modules with ≥5 nodes are shown in different colors, and smaller modules are shown in gray. Details of network topological attributes are listed in Table 3: (B) ASVs abundance ratios of abundant and rare microorganisms in different modules; (C) relationships between network complexity standardized by Z-scores and Ln (BG)/Ln (NAG + LAP); and (D) relationships between network complexity standardized by Z-scores and Ln (BG)/Ln (AP). Different lowercase letters indicate significant differences among the three sites (P < 0.05). ns, not significant.
In the three different habitats, most of the beta-nearest taxon index (βNTI) values of abundant taxa and rare taxa were between −2 and 2 indicating that the assembly process of abundant and rare taxa was dominated by stochastic processes (Figure 4A). However, the dispersal limitation of stochastic processes dominated the assembly of abundant taxa (Figure 4B). We observed that the assembly process of rare taxa was significantly (P < 0.001) correlated with the changes in soil physical and chemical properties (Figure 4C). To identify the potential main contributors to the assembly processes of abundant and rare taxa, we applied random forest analysis which demonstrated that C/N, C/P, TN, SOC, and NH4+-N were the significant impact factors in determining the rare taxa assembly process. C/N and C/P were significant factors (P < 0.01) (Figure 4D). Overall, C/N, pH, SWC, and C/P were the significantly impacting factors in determining the abundant taxa assembly process, and among them, C/N was significant (P < 0.01) (Figure 4D).
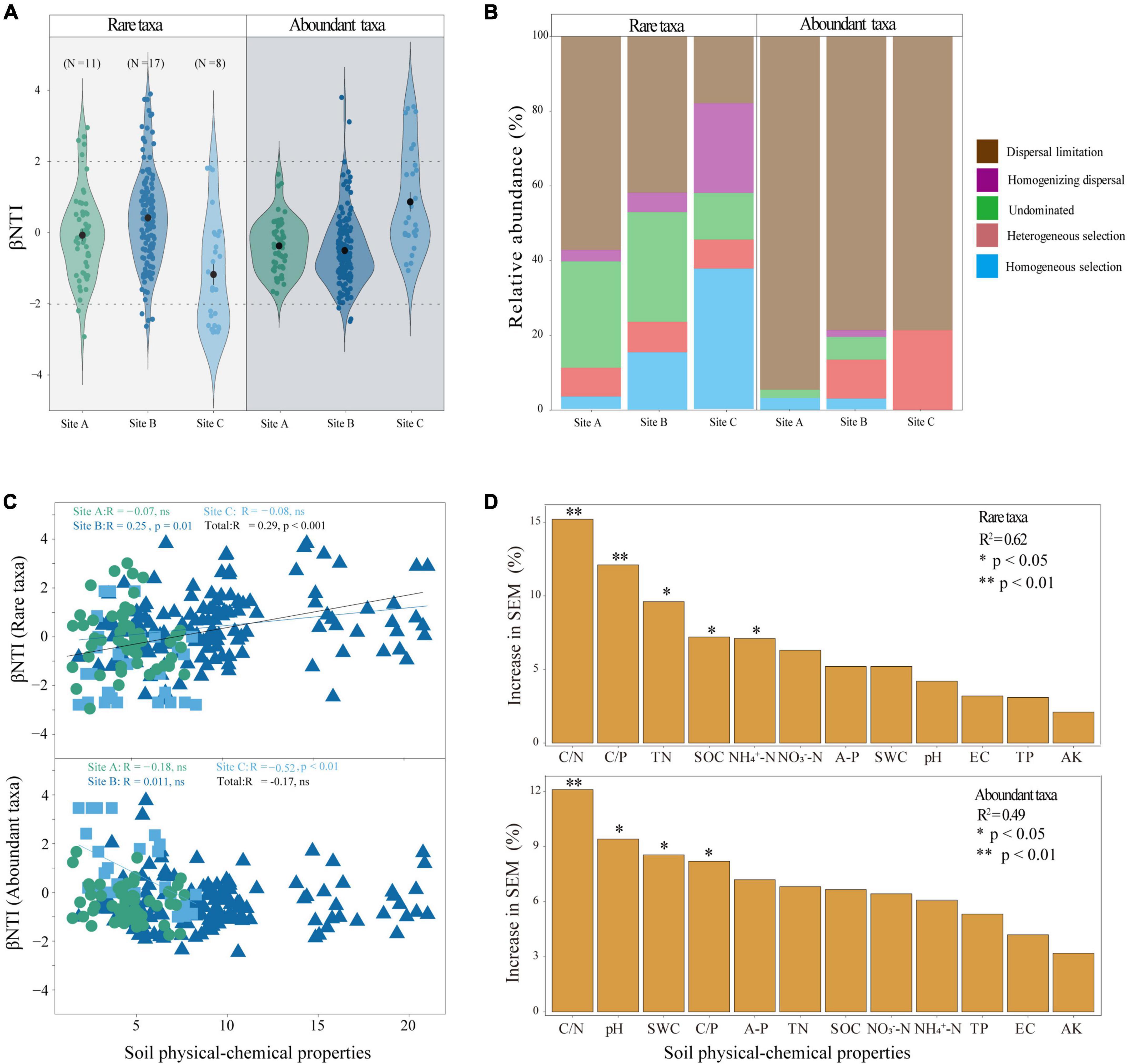
Figure 4. Assembly process and driving factors of abundant and rare taxa: (A) beta nearest taxon index (βNTI) value under different habitats. The horizontal dashed lines indicate the βNTI values of –2 and 2; (B) relative contribution of each ecological process to community assembly; (C) relationships between βNTI of the rare and abundant bacterial taxa and divergences of physical-chemical properties standardized; and (D) the random forest model identifies the major contributors to assembly processes of the abundant and rare taxa. ns, not significant. Asterisks denote significance levels. SWC, soil water content; EC, soil electric conductivity; TN, total nitrogen; AK, available potassium; A-P, available phosphorus; TP, total phosphorus; pH, potential of hydrogen; NO3–-N, nitrate nitrogen; NH4+-N, ammonium nitrogen; SOC, soil organic carbon; C/N, the ratio of soil organic carbon and total nitrogen; C/P, the ratio of soil organic carbon and total phosphorus.
To explore the microbial factors that affect nutrient acquisition, we applied a random forest model. The random forest analysis revealed that network complexity, network modules α-diversity, and the ratio of copiotrophs and oligotrophs significantly affected nutrient acquisition in the whole microbial community. βNTI, β-diversity, and α-diversity significantly affected the nutrient acquisition of the rare microbial community. In the abundant microbial community, α-diversity significantly affected nutrient acquisition (Figure 5A). We further employed SEMs to explore the interaction influence between microbial factors by combining the physical and chemical properties of soil. The findings demonstrated that soil physicochemical properties had a significant effect on the assembly process of rare taxa leading to an increase in the complexity of the microbial network and diversity of community composition, and further stimulated nutrient acquisition (Figure 5B).
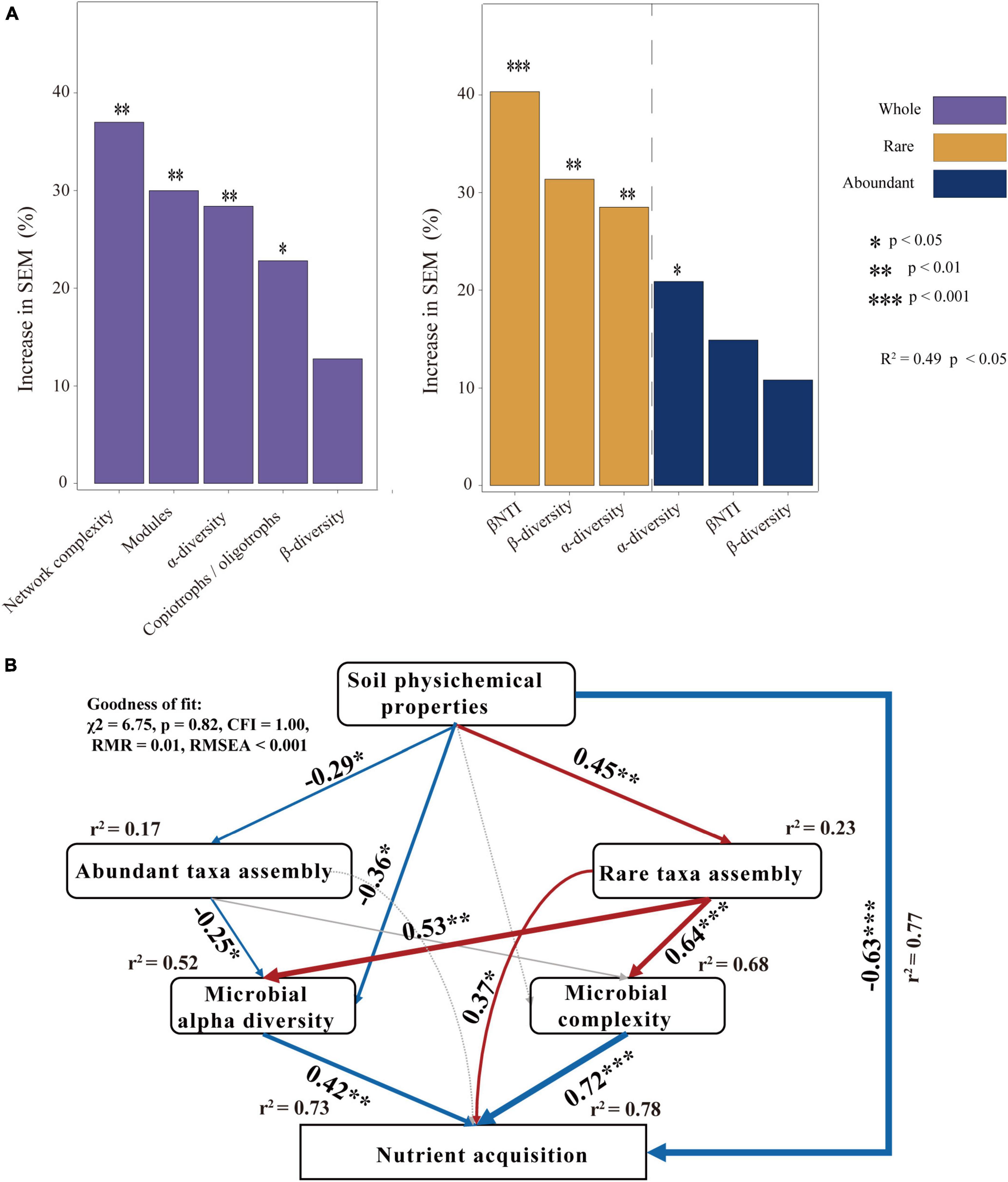
Figure 5. Analysis of factors affecting nutrient acquisition: (A) the random-forest model predicts the main factors for nutrient acquisition of bacterial communities and (B) structural equation model (SEM) revealing the direct and indirect effects of abundant taxon and rare taxon on nutrient acquisition. The numbers on the arrows are the path coefficients and are indicative of the standardized effect size of the relationship. Arrows in red and blue indicate positive and negative effects, respectively. R2 means the proportion of variance explained.
The availability of SOC, NH4+-N, NO3–-N, and A-P in sites A and C were lower but the enzyme activities were higher than those of site B (Tables 1, 4 and Figure 1A). This confirms that when the nutrient availability of the soil is low, microbes secrete more enzymes to meet the demand for nutrients (Sinsabaugh et al., 2002; Fierer et al., 2009; Wallenius et al., 2011; Xu et al., 2017). Significant differences were observed between site A and site C in the activity of BG, XS, NAG, and LAP (P < 0.05) (Table 4 and Figure 1A). Although sites A and C were both lithophytic moss crust habitats, site A was situated under forests, while site C was on the cliff. The difference made site A susceptible to shade from forests and site C susceptible to direct sunlight, resulting in varying soil moisture and nutrient conditions at the two sites. Soil nutrients have a significant influence on enzyme activities (Meier et al., 2020; Li et al., 2022). The scatter plot with eco-enzymatic stoichiometry reveals that microbial limitations in C&P, and C&N were prevalent across all locations (Figure 1B). It has been suggested that microbial C limitation is widespread (Schimel, 2003). N and P are mainly released from the decomposition of SOM (Pan et al., 2016; Alewell et al., 2020). Therefore, P or N limitation often coexists with C limitation in the three sites. Compared with site C, site A was not susceptible to C&P limitation (Figures 1B, C). This may be due to the higher diversity of rare taxa in site A (Table 2). Rare taxa exhibit diverse functions (Sauret et al., 2014), including microorganisms that can solubilize phosphorus and accelerate the breakdown of organic matter.
Site B had the higher nutrient availability and highest microbial diversity (Tables 1, 2), yet was still susceptible to significant nutrient limitations (Figures 1B, C). This can be explained by the nutritional strategies of microorganisms. Nutritional strategies involve a basic trade-off between the rate of growth and the efficiency of resource utilization (Fierer et al., 2007; Ho et al., 2017), enabling us to establish a direct correlation between microbial performance and environmental conditions. The soil microorganisms at site B were predominantly characterized as r-strategists, as indicated by the largest copiotroph/oligotroph ratio (Figure 1D). The r-strategy species (copiotrophic species) have a fast growth rate and a rapid response to available C and nutrient inputs, typically flourishing in environments enriched in nutrients (Yang et al., 2022c). In contrast, k-strategy species (oligotrophic species) are slow-growing and more common in oligotrophic environments (Yang et al., 2022c). The high ratio of copiotrophs results in significant nutrient demand, contributing to higher levels of nutrient limitations (Giovannoni et al., 2014). This further explains nutrient limitations of C, N, and P in site B (Figures 1B, C).
Microbial diversity depends on the availability of resources (Zhu L. et al., 2023). Varying ecosystems exhibit distinct soil temperatures, moisture levels, pH levels, and nutrient content which can influence the diversity and composition of soil microorganisms (Bahram et al., 2018). Site B had higher available nutrient content, such as NH4+-N, NO3–-N, and A-P than the other two sites (Table 1). However, sites A and C were characterized by lithophytic moss environments that exhibited low nutrient levels and supported oligotrophic microorganisms. The copiotroph/oligotroph ratio also showed the nutrient availability in the three sites (Figure 1D). This further explains the effect of different bryophyte habitats on microbial diversity. In addition, the abundant and rare microbial diversity of the three sites also showed distinct differences, rare microbes show a higher diversity (Table 2). Qin et al. (2022) indicated that rare biosphere possesses higher taxonomic diversity. This depended on multiple ecological principles behind the assembly of microbial communities (Lynch and Neufeld, 2015). The tremendous diversity of the rare biosphere is subjected to more complicated ecological processes such as speciation, drift, and extinction (Mo et al., 2018). Abundant communities have higher niche breadth and hence show strong resistance and adaptability to different environments, and spread in different ecosystems (Yang et al., 2022b). This explains the lack of clear structure of abundant communities in the three sites (Figure 2B). This study analyzed the relationship between abundant and rare microbial α-diversity and soil C, N, and P nutrients. There is usually a positive correlation between microbial diversity and soil nutrients (Chen et al., 2020); however, we found a negative relationship between diversity and soil nutrients in rare taxa (Supplementary Figure 1), inconsistent with previous findings. This can be explained by the competition for scarce nutrients occurs between bacteria and dominant plants, in nutrient-limited karst ecosystems (Wang et al., 2020). Furthermore, lower microbial metabolic activity may lead to the negative relationship between microbial diversity and soil nutrients (Hu et al., 2021). The karst rocky desertification ecosystem may have more dormant or inactive microorganisms, which explains that the metabolic activities of microorganisms are not strong, resulting in a slow turnover of resources. In addition, the soil C:N ratio in this study was much lower than the normal value, hindering the metabolic activities of soil microorganisms (Liu et al., 2018; Hu et al., 2021). Therefore, the dominant phyla of rare taxa were positively correlated with C/N, but negatively correlated with TN (Supplementary Figure 3). According to a previous study, there was a difference in structure and co-occurrence networks of the soil bacteria between different habitats (Zhong et al., 2022). Site B exhibited the highest complexity and community stability of the microbial network (Figure 3A). The complexity and stability of microbial networks are related to the network structure. As listed in Table 3, the network topology parameter of site B was the highest among the three sites. A previous study has reported a positive relationship between network complexity and microbial diversity (Ma et al., 2020). The microbial diversity at site B was also the highest (Table 2). We further found that rare taxa were the main component of microbial network modules 1 and 2 in the three sites (Figure 3B). The results indicated that rare taxa may play crucial roles in maintaining the structure of the co-occurrence network. Rare taxa form complex interactions and relationships with other taxa to maintain the network structure (Zhu M. et al., 2023; He et al., 2024). Reduced N limitation or enhanced P limitation increased microbial network complexity in site B (Figures 3C, D). Microorganisms in site B were more susceptible to P limitation than N limitation (Figure 1B). In this condition, soil microbial communities of site B can adapt their strategies in response to resource availability. Microbial strategies enhance their complexity to maintain a steady state (Yang et al., 2022c).
Community assembly describes how different ecological processes shape the composition and structure of microbial communities (Ning et al., 2024). Studying the effects of environmental factors on the assembly of microbial communities is critical to understanding microbial biodiversity and ecological function (Kang et al., 2023). The present study found that the community assembly of both abundant and rare microbes was mostly influenced by stochastic processes. Further, it was noted that the assembly process of abundant taxa was dominated by dispersal limitations of stochastic processes (Figure 4B). More individuals of abundant taxa are likely to be involved in dispersal events but vulnerable to physical barriers, distance and other factors (Liu et al., 2015). Diverse assembly processes contribute to the rare taxa (Figure 4B). Rare taxa occupy narrower niches, are closer in phylogenetic clustering, and are more likely to be filtered and dispersed by diverse assembly processes (He J. et al., 2022). Diverse assembly processes can drive high diversity and versatility of rare taxa (Zhou and Ning, 2017). From this perspective, the rare taxa may be the main driving force for maintaining ecosystem functional stability and diversity of the soil microbial communities (Gobet et al., 2012; Hugoni et al., 2013; Alonso-Sáez et al., 2015). Determining the elements that lead to the occurrence of different processes in the formation of microbial communities is essential in the study of community ecology. Prior studies have demonstrated a correlation between assembly processes and soil attributes such as pH and NH4+-N (Jiang et al., 2019; Wan et al., 2021). We found that soil C/N and C/P together significantly affected the community assembly of both abundant and rare taxa (P < 0.05) (Figure 4D). Especially, our findings indicated that TN, SOC and NH4+-N significantly influenced the assembly processes of the rare taxon. The assembly processes of the abundant taxa were significantly influenced by soil pH and SWC (P < 0.05) (Figure 4D). Soil C, N, and P are essential nutrients in soil ecosystems, and the stoichiometric ratio of soil C, N, and P affected the assembly process of microorganisms by affecting microbial investment in nutrient acquisition (Li J. et al., 2021; Xu et al., 2022; Duan et al., 2023). Therefore, both abundant and rare microbes were affected by soil C/N and C/P. It was worth distinguishing that abundant taxa were more susceptible to the non-nutrient properties of soil, such as pH and SWC. Soil pH and SWC can affect the cellular homeostasis of microorganisms and thus affect the exchange with external substances. The versatility of rare microbes allows for their ability to withstand substantial variances and be less affected by changes in environmental pH and SWC (He Z. et al., 2022). In total, the SEM demonstrated that the physical and chemical properties of soil mainly significantly affected the community assembly of the rare taxa, indirectly affecting microbial diversity and complexity and finally nutrient acquisition (Figure 5B). This suggested that rare microbial taxa were major drivers of nutrient.
This study aimed to determine the response of microorganisms under moss crust to nutrient acquisition in karst area. There were nutrient limitations in the three habitats. Our study demonstrated that nutrient acquisition was mainly driven by microbial assembly, diversity, and complexity. The microbial diversity and complexity were higher in the autochthonal moss crust of forest compared to the lithophytic moss crust. The microbial responses were influenced by the specific habitats of bryophytes, especially differences in soil nutrients. To enhance the efficiency of ecological restoration initiatives in karst rocky desertification areas, it is imperative to understand the influence of soil microbial communities on ecosystem processes. Further, with a full understanding of local nutrient limitations, the practice of cultivating bryophytes alongside rare microorganisms serves as a significant approach to managing and mitigating the process of rocky desertification. By addressing these technological limitations, it is possible to enhance the effectiveness and precision of the restoration activities aimed at mitigating karst rocky desertification and land degradation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
XD: Conceptualization, Investigation, Methodology, Writing – original draft. MC: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft. QC: Formal analysis, Software, Writing – review & editing. KL: Data curation, Methodology, Writing – review & editing. JL: Formal analysis, Software, Writing – review & editing. YL: Investigation, Methodology, Writing – review & editing. YR: Data curation, Investigation, Writing – review & editing. TY: Software, Writing – review & editing. JZ: Funding acquisition, Writing – review & editing. SH: Supervision, Writing – review & editing. XP: Funding acquisition, Supervision, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (31971729 and 31870707) and the Innovation and Entrepreneurship Training Program for College Students of Beijing Forestry University (S202310022059).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1384367/full#supplementary-material
Ai, C., Liang, G., Sun, J., Wang, X., and Zhou, W. (2012). Responses of extracellular enzyme activities and microbial community in both the rhizosphere and bulk soil to long-term fertilization practices in a fluvo-aquic soil. Geoderma 173, 330–338. doi: 10.1016/j.geoderma.2011.07.020
Alewell, C., Ringeval, B., Ballabio, C., Robinson, D. A., Panagos, P., and Borrelli, P. (2020). Global phosphorus shortage will be aggravated by soil erosion. Nat. Commun. 11:4546. doi: 10.1038/s41467-020-18326-7
Alonso-Sáez, L., Díaz-Pérez, L., and Morán, X. G. (2015). The hidden seasonality of the rare biosphere in coastal marine bacterioplankton. Environ. Microbiol. 17, 3766–3780. doi: 10.1111/1462-2920.12801
Anthony, M. A., Crowther, T. W., Maynard, D. S., Van Den Hoogen, J., and Averill, C. (2020). Distinct assembly processes and microbial communities constrain soil organic carbon formation. One Earth 2, 349–360. doi: 10.1016/j.oneear.2020.03.006
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Bhattacharyya, S. S., and Furtak, K. (2022). Soil–plant–microbe interactions determine soil biological fertility by altering rhizospheric nutrient cycling and biocrust formation. Sustainability 15:625. doi: 10.3390/su15010625
Chen, Q.-L., Ding, J., Zhu, D., Hu, H.-W., Delgado-Baquerizo, M., Ma, Y.-B., et al. (2020). Rare microbial taxa as the major drivers of ecosystem multifunctionality in long-term fertilized soils. Soil Biol. Biochem. 141:107686. doi: 10.1016/j.soilbio.2019.107686
Chen, Q.-L., Hu, H.-W., Yan, Z.-Z., Li, C.-Y., Nguyen, B. T., Sun, A.-Q., et al. (2021). Deterministic selection dominates microbial community assembly in termite mounds. Soil Biol. Biochem. 152:108073. doi: 10.1016/j.soilbio.2020.108073
Cheng, C., Li, Y., Long, M., Gao, M., Zhang, Y., Lin, J., et al. (2022). Moss biocrusts buffer the negative effects of karst rocky desertification on soil properties and soil microbial richness. Plant Soil 475, 163–168. doi: 10.1007/s11104-020-04602-4
Cui, Y., Bing, H., Fang, L., Wu, Y., Yu, J., Shen, G., et al. (2019). Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the eastern Tibetan Plateau. Geoderma 338, 118–127. doi: 10.1016/j.geoderma.2018.11.047
Cui, Y., Fang, L., Guo, X., Wang, X., Zhang, Y., Li, P., et al. (2018). Ecoenzymatic stoichiometry and microbial nutrient limitation in rhizosphere soil in the arid area of the northern Loess Plateau, China. Soil Biol. Biochem. 116, 11–21. doi: 10.1016/j.soilbio.2017.09.025
Deissová, T., Zapletalová, M., Kunovský, L., Kroupa, R., Grolich, T., Kala, Z., et al. (2023). 16S rRNA gene primer choice impacts off-target amplification in human gastrointestinal tract biopsies and microbiome profiling. Sci. Rep. 13:12577. doi: 10.1038/s41598-023-39575-8
Dong, W., Yang, Q., George, T. S., Yin, H., Wang, S., Bi, J., et al. (2022). Investigating bacterial coupled assimilation of fertilizer-nitrogen and crop residue-carbon in upland soils by DNA-qSIP. Sci. Total Environ. 845:157279. doi: 10.1016/j.scitotenv.2022.157279
Duan, P., Fu, R., Nottingham, A. T., Domeignoz-Horta, L. A., Yang, X., Du, H., et al. (2023). Tree species diversity increases soil microbial carbon use efficiency in a subtropical forest. Glob. Change Biol. 29, 7131–7144. doi: 10.1111/gcb.16971
Fanin, N., Gundale, M. J., Farrell, M., Ciobanu, M., Baldock, J. A., Nilsson, M. C., et al. (2018). Consistent effects of biodiversity loss on multifunctionality across contrasting ecosystems. Nat. Ecol. Evol. 2, 269–278. doi: 10.1038/s41559-017-0415-0
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A., and Cleveland, C. C. (2009). Global patterns in belowground communities. Ecol. Lett. 12, 1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x
Galand, P. E., Casamayor, E. O., Kirchman, D. L., and Lovejoy, C. (2009). Ecology of the rare microbial biosphere of the Arctic Ocean. Proc. Am. Natl. Sci. U. S.A. 106, 22427–22432. doi: 10.1073/pnas.0908284106
Gao, G.-F., Peng, D., Tripathi, B., Zhang, Y., and Chu, H. (2020). Distinct community assembly processes of abundant and rare soil bacteria in coastal wetlands along an inundation gradient. mSystems 5:e01150-20. doi: 10.1128/mSystems.01150-20
Giovannoni, S. J., Cameron Thrash, J., and Temperton, B. (2014). Implications of streamlining theory for microbial ecology. ISME J. 8, 1553–1565. doi: 10.1038/ismej.2014.60
Gobet, A., Böer, S. I., Huse, S. M., Van Beusekom, J. E., Quince, C., Sogin, M. L., et al. (2012). Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 6, 542–553. doi: 10.1038/ismej.2011.132
Hartmann, M., and Six, J. (2022). Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 4, 4–18. doi: 10.1038/s43017-022-00366-w
He, J., Zhang, N., Muhammad, A., Shen, X., Sun, C., Li, Q., et al. (2022). From surviving to thriving, the assembly processes of microbial communities in stone biodeterioration: A case study of the West Lake UNESCO World heritage area in China. Sci. Total Environ. 805:150395. doi: 10.1016/j.scitotenv.2021.150395
He, X., Xiao, X., Wei, W., Li, L., Zhao, Y., Zhang, N., et al. (2024). Soil rare microorganisms mediated the plant cadmium uptake: The central role of protists. Sci. Total Environ. 908:168505. doi: 10.1016/j.scitotenv.2023.168505
He, Z., Liu, D., Shi, Y., Wu, X., Dai, Y., Shang, Y., et al. (2022). Broader environmental adaptation of rare rather than abundant bacteria in reforestation succession soil. Sci. Total Environ. 828:154364. doi: 10.1016/j.scitotenv.2022.154364
Ho, A., Di Lonardo, D. P., and Bodelier, P. L. (2017). Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93:fix006. doi: 10.1093/femsec/fix006
Hu, W., Ran, J., Dong, L., Du, Q., Ji, M., Yao, S., et al. (2021). Aridity-driven shift in biodiversity–soil multifunctionality relationships. Nat. Commun. 12:5350. doi: 10.1038/s41467-021-25641-0
Hua, Z.-S., Han, Y.-J., Chen, L.-X., Liu, J., Hu, M., Li, S.-J., et al. (2015). Ecological roles of dominant and rare prokaryotes in acid mine drainage revealed by metagenomics and metatranscriptomics. ISME J. 9, 1280–1294. doi: 10.1038/ismej.2014.212
Hugoni, M., Taib, N., Debroas, D., Domaizon, I., Jouan Dufournel, I., Bronner, G., et al. (2013). Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc. Am. Natl. Sci. U.S.A. 110, 6004–6009. doi: 10.1073/pnas.1216863110
Jia, X., Dini-Andreote, F., and Salles, J. F. (2018). Community assembly processes of the microbial rare biosphere. Trends Microbiol. 26, 738–747. doi: 10.1016/j.tim.2018.02.011
Jiang, Y., Song, H., Lei, Y., Korpelainen, H., and Li, C. (2019). Distinct co-occurrence patterns and driving forces of rare and abundant bacterial subcommunities following a glacial retreat in the eastern Tibetan Plateau. Biol. Fertil. 55, 351–364. doi: 10.1007/s00374-019-01355-w
Jiao, S., Chen, W., Wang, J., Du, N., Li, Q., and Wei, G. (2018). Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome 6, 1–13. doi: 10.1186/s40168-018-0526-0
Kang, Y., Wu, H., Zhang, Y., Wu, Q., Guan, Q., Lu, K., et al. (2023). Differential distribution patterns and assembly processes of soil microbial communities under contrasting vegetation types at distinctive altitudes in the Changbai Mountain. Front. Microbiol. 14:1152818. doi: 10.3389/fmicb.2023.1152818
Katiraei, S., Anvar, Y., Hoving, L., Berbée, J. F., Van Harmelen, V., and Willems Van Dijk, K. (2022). Evaluation of full-length versus V4-region 16S rRNA sequencing for phylogenetic analysis of mouse intestinal microbiota after a dietary intervention. Curr. Microbiol. 79:276. doi: 10.1007/s00284-022-02956-9
Kidron, G. J., and Drahorad, S. L. (2022). Biocrust-induced extracellular polymeric substances are responsible for dune stabilization in the Negev. Land. Degrad. Dev. 33, 425–438. doi: 10.1002/ldr.4146
Knelman, J. E., and Nemergut, D. R. (2014). Changes in community assembly may shift the relationship between biodiversity and ecosystem function. Front. Microbiol. 5:424. doi: 10.3389/fmicb.2014.00424
Li, C., Jin, L., Zhang, C., Li, S., Zhou, T., Hua, Z., et al. (2023). Destabilized microbial networks with distinct performances of abundant and rare biospheres in maintaining networks under increasing salinity stress. iMeta 2:e79. doi: 10.1002/imt2.79
Li, H., Yang, S., Semenov, M. V., Yao, F., Ye, J., Bu, R., et al. (2021). Temperature sensitivity of SOM decomposition is linked with a K-selected microbial community. Glob. Change Biol. 27, 2763–2779. doi: 10.1111/gcb.15593
Li, J., Pei, J., Dijkstra, F. A., Nie, M., and Pendall, E. (2021). Microbial carbon use efficiency, biomass residence time and temperature sensitivity across ecosystems and soil depths. Soil Biol. Biochem. 154:108117. doi: 10.1016/j.soilbio.2020.108117
Li, M., You, Y., Tan, X., Wen, Y., Yu, S., Xiao, N., et al. (2022). Mixture of N2-fixing tree species promotes organic phosphorus accumulation and transformation in topsoil aggregates in a degraded karst region of subtropical China. Geoderma 413:115752. doi: 10.1016/j.geoderma.2022.115752
Liu, G., Wang, H., Yan, G., Wang, M., Jiang, S., Wang, X., et al. (2023). Soil enzyme activities and microbial nutrient limitation during the secondary succession of boreal forests. Catena 230:107268. doi: 10.1016/j.catena.2023.107268
Liu, L., Yang, J., Yu, Z., and Wilkinson, D. M. (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 9, 2068–2077. doi: 10.1038/ismej.2015.29
Liu, X., Zhang, W., Wu, M., Ye, Y., Wang, K., and Li, D. (2018). Changes in soil nitrogen stocks following vegetation restoration in a typical karst catchment. Land. Degrad. Dev. 30, 60–72. doi: 10.1002/ldr.3204
Lynch, M. D., and Neufeld, J. D. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
Ma, L., Zhang, J., Li, Z., Xin, X., Guo, Z., Wang, D., et al. (2020). Long-term phosphorus deficiency decreased bacterial-fungal network complexity and efficiency across three soil types in China as revealed by network analysis. Appl. Soil Ecol. 148:103506. doi: 10.1016/j.apsoil.2020.103506
Ma, S., Zhu, W., Wang, W., Li, X., and Sheng, Z. (2023). Microbial assemblies with distinct trophic strategies drive changes in soil microbial carbon use efficiency along vegetation primary succession in a glacier retreat area of the southeastern Tibetan Plateau. Sci. Total Environ. 867:161587. doi: 10.1016/j.scitotenv.2023.161587
Ma, Z., Zhang, X., Zheng, B., Yue, S., Zhang, X., Zhai, B., et al. (2021). Effects of plastic and straw mulching on soil microbial P limitations in maize fields: Dependency on soil organic carbon demonstrated by ecoenzymatic stoichiometry. Geoderma 388:114928. doi: 10.1016/j.geoderma.2021.114928
Matheri, F., Kambura, A. K., Mwangi, M., Ongeso, N., Karanja, E., Adamtey, N., et al. (2023). Composition, structure, and functional shifts of prokaryotic communities in response to co-composting of various nitrogenous green feedstocks. BMC Microbiol. 23:50. doi: 10.1186/s12866-023-02798-w
Meier, I. C., Tuckmantel, T., Heitkotter, J., Muller, K., Preusser, S., Wrobel, T. J., et al. (2020). Root exudation of mature beech forests across a nutrient availability gradient: The role of root morphology and fungal activity. New Phytol. 226, 583–594. doi: 10.1111/nph.16389
Meng, W., Dai, Q., Ren, Q., Tu, N., and Leng, T. (2021). Ecological stoichiometric characteristics of soil-moss C, N, and P in restoration stages of karst rocky desertification. PLoS One 16:e0252838. doi: 10.1371/journal.pone.0252838
Mo, Y., Zhang, W., Yang, J., Lin, Y., Yu, Z., and Lin, S. (2018). Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J. 12, 2198–2210. doi: 10.1038/s41396-018-0153-6
Ning, D., Wang, Y., Fan, Y., Wang, J., Van Nostrand, J. D., Wu, L., et al. (2024). Environmental stress mediates groundwater microbial community assembly. Nat. Microbiol. 9, 490–501. doi: 10.1038/s41564-023-01573-x
Pan, F., Liang, Y., Zhang, W., Zhao, J., and Wang, K. (2016). Enhanced nitrogen availability in karst ecosystems by oxalic acid release in the rhizosphere. Front. Plant Sci. 7:687. doi: 10.3389/fpls.2016.00687
Pedrós-Alió, C. (2011). The rare bacterial biosphere. Annu. Rev. Mar. Sci. 4, 449–466. doi: 10.1146/annurev-marine-120710-100948
Peter, H., Beier, S., Bertilsson, S., Lindström, E. S., Langenheder, S., and Tranvik, L. J. (2011). Function-specific response to depletion of microbial diversity. ISME J. 5, 351–361. doi: 10.1038/ismej.2010.119
Qin, Z., Zhao, Z., Xia, L., Wang, S., Yu, G., and Miao, A. (2022). Responses of abundant and rare prokaryotic taxa in a controlled organic contaminated site subjected to vertical pollution-induced disturbances. Sci. Total Environ. 853:158625. doi: 10.1016/j.scitotenv.2022.158625
Rayment, G. E., and Lyons, D. J. (2011). Soil chemical methods: Australasia. Clayton, VIC: CSIRO publishing.
Sauret, C., Séverin, T., Vétion, G., Guigue, C., Goutx, M., Pujo-Pay, M., et al. (2014). Rare biosphere’ bacteria as key phenanthrene degraders in coastal seawaters. Environ. Pollut. 194, 246–253.
Schimel, J. (2003). The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: A theoretical model. Soil Biol. Biochem. 35, 549–563. doi: 10.1016/s0038-0717(03)00015-4
Sinsabaugh, R., Carreiro, M., and Repert, D. (2002). Allocation of extracellular enzymatic activity in relation to litter composition, N deposition, and mass loss. Biogeochemistry 60, 1–24. doi: 10.1023/A:1016541114786
Sinsabaugh, R. L., Hill, B. H., and Follstad Shah, J. J. (2009). Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 462, 795–798. doi: 10.1038/nature08632
Stegen, J. C., Lin, X., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Wallenius, K., Rita, H., Mikkonen, A., Lappi, K., Lindström, K., Hartikainen, H., et al. (2011). Effects of land use on the level, variation and spatial structure of soil enzyme activities and bacterial communities. Soil Biol. Biochem. 43, 1464–1473. doi: 10.1016/j.soilbio.2011.03.018
Wan, W., Gadd, G. M., Yang, Y., Yuan, W., Gu, J., Ye, L., et al. (2021). Environmental adaptation is stronger for abundant rather than rare microorganisms in wetland soils from the Qinghai-Tibet Plateau. Mol. Ecol. 30, 2390–2403. doi: 10.1111/mec.15882
Wang, X., Li, F. Y., Wang, Y., Liu, X., Cheng, J., Zhang, J., et al. (2020). High ecosystem multifunctionality under moderate grazing is associated with high plant but low bacterial diversity in a semi-arid steppe grassland. Plant Soil 448, 265–276. doi: 10.1007/s11104-020-04430-6
Xiao, B., and Veste, M. (2017). Moss-dominated biocrusts increase soil microbial abundance and community diversity and improve soil fertility in semi-arid climates on the Loess Plateau of China. Appl. Soil Ecol. 117-118, 165–177. doi: 10.1016/j.apsoil.2017.05.005
Xiao, L., Liu, G., Li, P., and Xue, S. (2021). Ecological stoichiometry of plant-soil-enzyme interactions drives secondary plant succession in the abandoned grasslands of Loess Plateau, China. Catena 202:105302. doi: 10.1016/j.catena.2021.105302
Xu, Z., Wang, Y., Sun, D., Li, H., Dong, Y., Wang, Z., et al. (2022). Soil nutrients and nutrient ratios influence the ratios of soil microbial biomass and metabolic nutrient limitations in mountain peatlands. Catena 218:106528. doi: 10.1016/j.catena.2022.106528
Xu, Z., Yu, G., Zhang, X., He, N., Wang, Q., Wang, S., et al. (2017). Soil enzyme activity and stoichiometry in forest ecosystems along the North-South Transect in eastern China (NSTEC). Soil Biol. Biochem. 104, 152–163. doi: 10.1016/j.soilbio.2016.10.020
Xue, Y., Chen, H., Yang, J. R., Liu, M., Huang, B., and Yang, J. (2018). Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 12, 2263–2277. doi: 10.1038/s41396-018-0159-0
Yang, T., Chen, Q., Yang, M., Wang, G., Zheng, C., Zhou, J., et al. (2022a). Soil microbial community under bryophytes in different substrates and its potential to degraded karst ecosystem restoration. Int. Biodeter. Biodegr. 175:105493. doi: 10.1016/j.ibiod.2022.105493
Yang, Y., Dou, Y., Wang, B., Xue, Z., Wang, Y., An, S., et al. (2022c). Deciphering factors driving soil microbial life-history strategies in restored grasslands. iMeta 2:e66. doi: 10.1002/imt2.66
Yang, Y., Cheng, K., Li, K., Jin, Y., and He, X. (2022b). Deciphering the diversity patterns and community assembly of rare and abundant bacterial communities in a wetland system. Sci. Total Environ. 838:156334. doi: 10.1016/j.scitotenv.2022.156334
Yuan, M. M., Guo, X., Wu, L., Zhang, Y., Xiao, N., Ning, D., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348. doi: 10.1038/s41558-021-00989-9
Zhong, Y., Sorensen, P. O., Zhu, G., Jia, X., Liu, J., Shangguan, Z., et al. (2022). Differential microbial assembly processes and co-occurrence networks in the soil-root continuum along an environmental gradient. iMeta 1:e18. doi: 10.1002/imt2.18
Zhou, J., and Ning, D. (2017). Stochastic community assembly: Does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81, e00002–e00017. doi: 10.1128/mmbr.00002-17
Zhou, L., Wang, X., Wang, Z., Zhang, X., Chen, C., and Liu, H. (2020). The challenge of soil loss control and vegetation restoration in the karst area of southwestern China. Int. Soil Water Conserv. 8, 26–34. doi: 10.1016/j.iswcr.2019.12.001
Zhu, L., Chen, Y., Sun, R., Zhang, J., Hale, L., Dumack, K., et al. (2023). Resource-dependent biodiversity and potential multi-trophic interactions determine belowground functional trait stability. Microbiome 11:95. doi: 10.1186/s40168-023-01539-5
Keywords: land degradation, bryophytes, extracellular enzyme stoichiometry, assembly processes, co-occurrence networks
Citation: Dong X, Chen M, Chen Q, Liu K, Long J, Li Y, Ren Y, Yang T, Zhou J, Herath S and Peng X (2024) Rare microbial taxa as the major drivers of nutrient acquisition under moss biocrusts in karst area. Front. Microbiol. 15:1384367. doi: 10.3389/fmicb.2024.1384367
Received: 09 February 2024; Accepted: 08 April 2024;
Published: 01 May 2024.
Edited by:
Jeanette M. Norton, Utah State University, United StatesReviewed by:
Weidong Kong, Chinese Academy of Sciences (CAS), ChinaCopyright © 2024 Dong, Chen, Chen, Liu, Long, Li, Ren, Yang, Zhou, Herath and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiawei Peng, eGlhd2VpcGVuZ0AxNjMuY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.