- 1Department of Microbiology, School of Medicine, Iran University of Medical Sciences, Tehran, Iran
- 2Department of Bacteriology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
Background: Mycobacterium kansasii infection is one of the most common causes of non-tuberculosis mycobacterial (NTM) disease worldwide. However, accurate information on the global prevalence of this bacterium is lacking. Therefore, this study was conducted to investigate the prevalence of M. kansasii in clinical and environmental isolates.
Methods: Databases, including PubMed, Scopus, and the Web of Science, were utilized to gather articles on the prevalence of M. kansasii in clinical and environmental isolates. The collected data were analyzed using Comprehensive Meta-Analysis software.
Results: A total of 118 and 16 studies met the inclusion criteria and were used to analyze the prevalence of M. kansasii in clinical and environmental isolates, respectively. The prevalence of M. kansasii in NTM and environmental isolates were 9.4 and 5.8%, respectively. Subsequent analysis showed an increasing prevalence of M. kansasii over the years. Additionally, the results indicated a significant difference in the prevalence of this bacteria among different regions.
Conclusion: The relatively high prevalence of M. kansasii among NTM isolates suggests the need for further implementation of infection control strategies. It is also important to establish appropriate diagnostic criteria and management guidelines for screening this microorganism in environmental samples in order to prevent its spread, given its high prevalence in environmental isolates.
Introduction
The genus Mycobacterium comprises over 200 species, divided into the Mycobacterium tuberculosis (MTB) complex and non-tuberculosis mycobacteria (NTM) (Karami-Zarandi et al., 2019). NTM is a diverse group of opportunistic bacteria that are commonly found in water, soil, and dust. While tuberculosis (TB) is the most prevalent mycobacterial infection in developing countries, the incidence of NTM diseases is rising globally, surpassing tuberculosis in developed nations (Johansen et al., 2020; Pavlik et al., 2022).
Initially, NTMs were considered contaminants rather than pathogens due to their presence in environmental sources (Koh, 2017). However, the incidence of NTM diseases has increased, and the exact cause of this rise remains poorly understood. Factors such as an aging population, reduced immune function, and environmental exposure to mycobacteria have been suggested as possible explanations (Cowman et al., 2019).
Mycobacterium kansasii (M. kansasii) is a slow-growing NTM that causes pulmonary and extra-pulmonary infections, in immunocompromised and immunocompetent individuals (Khosravi et al., 2020). The disease caused by M. kansasii closely resembles pulmonary tuberculosis in terms of pathogenesis, clinical features, and treatment response, differing significantly from infections caused by other NTM, particularly the M. avium complex (Woods and Washington, 1987).
Traditionally, M. kansasii has been recognized as an NTM pathogen causing lung disease rather than a contaminant. The isolation of M. kansasii from sputum under appropriate conditions may be sufficient evidence to indicate disease and to initiate treatment (Matveychuk et al., 2012; Daley et al., 2020).
Global reports have identified M. kansasii as the sixth most commonly isolated NTM from clinical samples. Additionally, it has been reported as the leading cause of pulmonary NTM disease in sub-Saharan Africa and the third most prevalent NTM causing lung disease in Taiwan (Huang et al., 2017; Okoi et al., 2017).
There is a widely held belief that M. kansasii can be acquired from the environment and is present in various natural ecosystems, including water, soil, and dust. Numerous studies have documented the recovery of this organism from municipal water distribution systems, with isolates found in the same communities where M. kansasii disease patients have been identified (McSwiggan and Collins, 1974; Steadham, 1980). The epidemiology of M. kansasii primarily affects urban areas, particularly high-density, low-income communities in highly industrialized regions (Kwenda et al., 2015).
Considering the clinical importance of M. kansasii and the lack of a meta-analysis study examining the prevalence of M. kansasii in clinical and environmental samples, the aim of this study is to investigate its prevalence in both clinical and environmental samples. The information obtained from this study can contribute to the effective management of this bacterium.
Materials and methods
Search strategy
We conducted a search of journal articles in three databases (PubMed, Scopus, and Web of Science) until February 2023. All of these databases were searched using the following search strategy: “Mycobacterium kansasii” OR “M. kansasii.”
Eligibility criteria
All studies that provided the precise number of M. kansasii isolates - either as total isolates or as part of NTM isolates in clinical samples, as well as studies that reported the bacterial count in environmental samples, were included in this study.
All identified articles were collated using Endnote X20 Citation Manager Software, and duplicate articles were removed prior to review. The citations were then uploaded to Rayyan, a citation classification application (Ouzzani et al., 2016). Two independent reviewers screened the titles and abstracts, and removed irrelevant articles. Full texts of potentially relevant articles were independently collected and reviewed by two authors. If there was a disagreement about the inclusion of an article after screening, a third author determined its eligibility for full review.
Review articles, case report studies, short communications, conference papers, letters, book chapters, articles that did not mention the exact number of isolates, and articles written in languages other than English were excluded.
Data extraction
Two authors independently extracted all data from eligible articles. Any disagreements in data points were resolved through consensus and discussion. From each article, we collected information on the first author, publication year, sampling time, study country, continent, and sample size (total number of samples and number of M. kansasi in clinical and environmental samples). This study selection process was presented in a Preferred Reporting Item for Systematic Reviews and Meta-Analyses (PRISMA) flowchart (Figure 1).
Study quality assessment
The quality of included studies was evaluated using the Joanna Briggs Institute (JBI) Critical Appraisal Checklist (Munn et al., 2020). This checklist consists of nine questions that assess the quality of studies, focusing on appropriate sampling techniques, research objectives, and adequate data analysis. Each item is rated as “yes,” “no,” or “unclear.” A score of 1 point is given for each “yes” answer, and a score of 0 points is given for each “no” or “unclear” answer. Finally, the mean score of each paper was independently evaluated by two reviewers, and any disagreements were resolved through consensus between the two reviewers or by consulting a third author, if needed.
Data analysis
Data analysis on the prevalence of M. kansasi in clinics and the environment was conducted using Comprehensive Meta-analysis (CMA) software. The analysis included prevalence data for M. kansasi in NTM, clinical isolates, water isolates, and soil isolates.
Subgroup analyses were performed based on sampling period, country, continent, and method of detection for prevalence of M. kansasi in NTM clinical isolates. For water samples, subgroup analyses were conducted based on country, continent, and place of collection.
A random-effects model was utilized to estimate the pooled prevalence of M. kansasi in clinical and environmental samples with a 95% confidence interval. The heterogeneity among the studies in the meta-analysis was assessed using the I2 statistic. An I2 value of ≤25% indicates low homogeneity, 25% < I2 < 75% indicates moderate heterogeneity, and I2 > 75% indicates high heterogeneity.
Sensitivity analysis was performed to investigate the impact of individual studies on the prevalence of M. kansasi in NTM and environmental isolates. Funnel plots and Begg’s test were employed to assess the presence of publication bias. Results were considered to have publication bias if the p-value was <0.05.
Results
Search results
A total of 6,640 publications were identified. After removing duplicates using Endnote software, 2,192 articles were screened. Following the screening process, 1,640 studies were excluded, leaving 552 articles for full-text validation. After a thorough review, 118 published studies were used to analyze the prevalence of M. kansasi in clinical isolates (Wright et al., 1985; Debrunner et al., 1992; Rastogi and Goh, 1992; Shafer and Sierra, 1992; Parenti et al., 1995; Gamboa et al., 1997; Benjamin et al., 1998; Alcaide et al., 1999; Attorri et al., 2000; Ruiz et al., 2001; Mijs et al., 2002; Scarparo et al., 2002; Alcaide et al., 2003; Kontos et al., 2003; Rodriguez Dı́az et al., 2003; Tu et al., 2003; Martin-Casabona et al., 2004; Matos et al., 2004; Morita et al., 2005; Pierre-Audigier et al., 2005; Prammananan et al., 2005; Dailloux et al., 2006; Franco-Álvarez de Luna et al., 2006; Hillemann et al., 2006; Lai et al., 2006; Andréjak et al., 2007; Liao et al., 2007; Bodle et al., 2008; Pedro et al., 2008; Ryoo et al., 2008; al-Mahruqi et al., 2009; Shen et al., 2009; Sorlozano et al., 2009; Amorim et al., 2010; Bicmen et al., 2010; Moore et al., 2010; Shenai et al., 2010; al Houqani et al., 2011; Ani et al., 2011; Bicmen et al., 2011; Chae et al., 2011; del Giudice et al., 2011; Gitti et al., 2011; Hong et al., 2011; Hsiao et al., 2011; Jeong et al., 2011; Lan et al., 2011; Chen et al., 2012; Lucke et al., 2012; Matveychuk et al., 2012; Braun et al., 2013; Cortés-Torres et al., 2013; Hombach et al., 2013; Mirsaeidi et al., 2013; Saifi et al., 2013; Chou et al., 2014; Lin et al., 2014; Sookan and Coovadia, 2014; Wu et al., 2014; Bainomugisa et al., 2015; Chiang et al., 2015; Kim et al., 2015; Lee et al., 2015; Manika et al., 2015; Ng et al., 2015; Shao et al., 2015; Sheu et al., 2015; Blanc et al., 2016; Kodana et al., 2016; Nasr Esfahani et al., 2016; Ose et al., 2016; Riello et al., 2016; Agizew et al., 2017; Brown-Elliott and Wallace, 2017, 2021; Desikan et al., 2017; Pang et al., 2017; Park et al., 2017; Adzic-Vukicevic et al., 2018; Loizos et al., 2018; Luo et al., 2018; Naito et al., 2018; Nasiri et al., 2018; Tan et al., 2018; Davari et al., 2019; Fang et al., 2019; Gomathy et al., 2019; Marques et al., 2019; Modrá et al., 2019; Mortazavi et al., 2019; Pedrero et al., 2019; Xu et al., 2019; Feysia et al., 2020; Hara et al., 2020; Huang et al., 2020; López-Roa et al., 2020; Takenaka et al., 2020; Thomson et al., 2020; Abate et al., 2021; Ahn et al., 2021; Donohue, 2021; Huang et al., 2021; Kim M. J. et al., 2021; Kim Y. G. et al., 2021; Liu et al., 2021; Mahdavi et al., 2021; Ose et al., 2021; Thangavelu et al., 2021; Urabe et al., 2021; Chai et al., 2022; Das et al., 2022; Gaballah et al., 2022; Gao et al., 2022; He et al., 2022; Lee et al., 2022; Lin et al., 2022), while 16 published studies were used to analyze the prevalence of M. kansasi in environmental isolates (McSwiggan and Collins, 1974; Engel et al., 1980; Gruft et al., 1981; Wright et al., 1985; Kubalek and Mysak, 1996; Slosárek et al., 1996; Iivanainen et al., 1999; Santos et al., 2005; Ghaemi et al., 2006; Thomas et al., 2006; Sharma et al., 2007; Parashar et al., 2009; Adrados et al., 2011; Kwenda et al., 2015; Moghaddam et al., 2022).
Figure 1 illustrates the review and article selection process based on the Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) statement. Supplementary Table S1 provides the characteristics of the included studies and the quality control analysis score, while Supplementary Table S2 presents the details of the answers to the JBI checklist questions for quality control.
Meta-analysis
Funnel plots (Figure 2) showed publication bias for the prevalence result of M. kansasi in NTM clinical isolates and environmental isolates. Begg’s test was also used to indicate publication bias for the prevalence results (p = 0.11 for prevalence in NTM isolates and p = 0.079 for prevalence in environmental isolates).
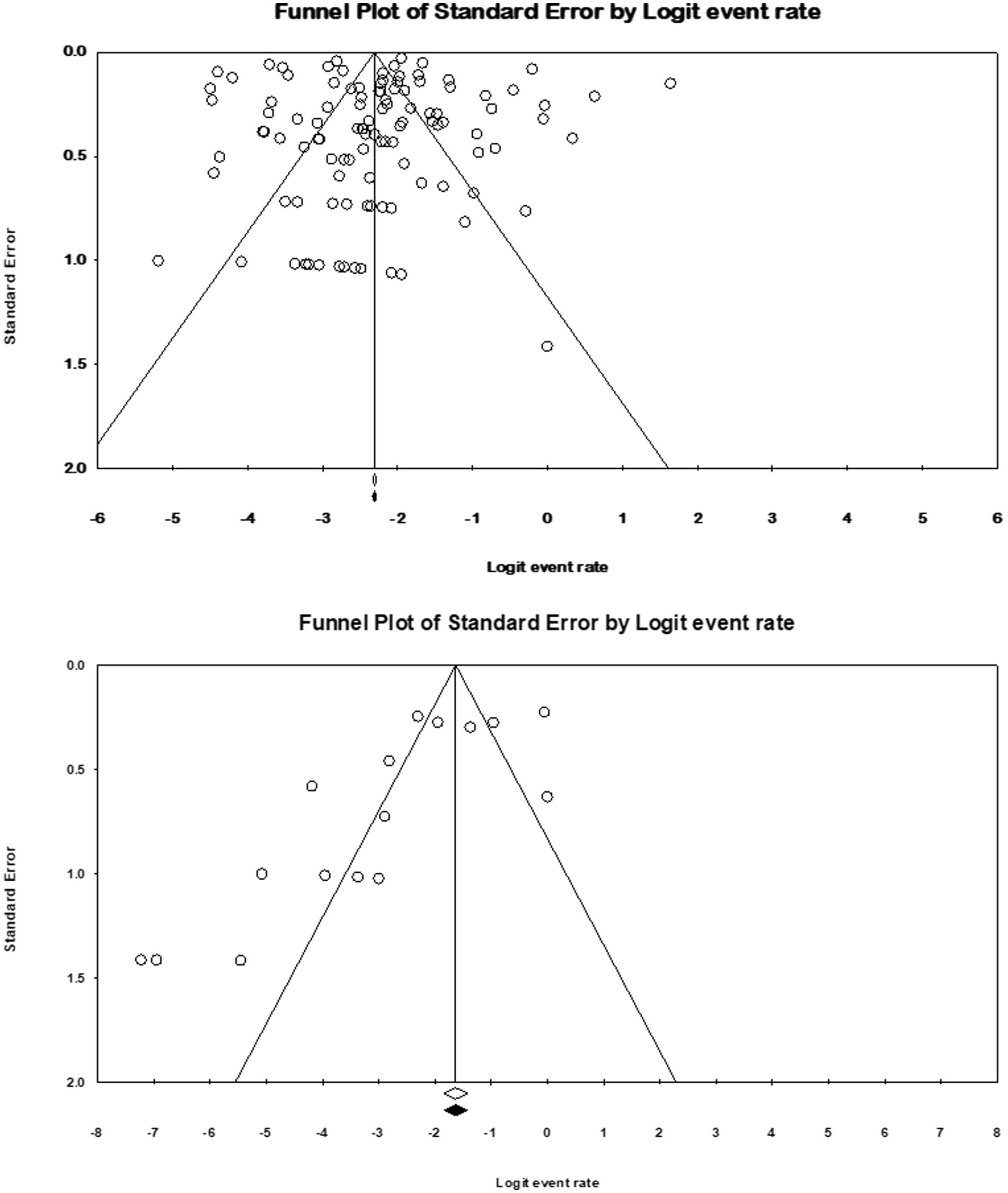
Figure 2. Funnel plots for identification of publication bias in clinical and environmental isolates.
Sensitivity analysis was conducted, and the results demonstrated that none of the studies influenced the prevalence of M. kansasi in NTM clinical isolates and environmental isolates.
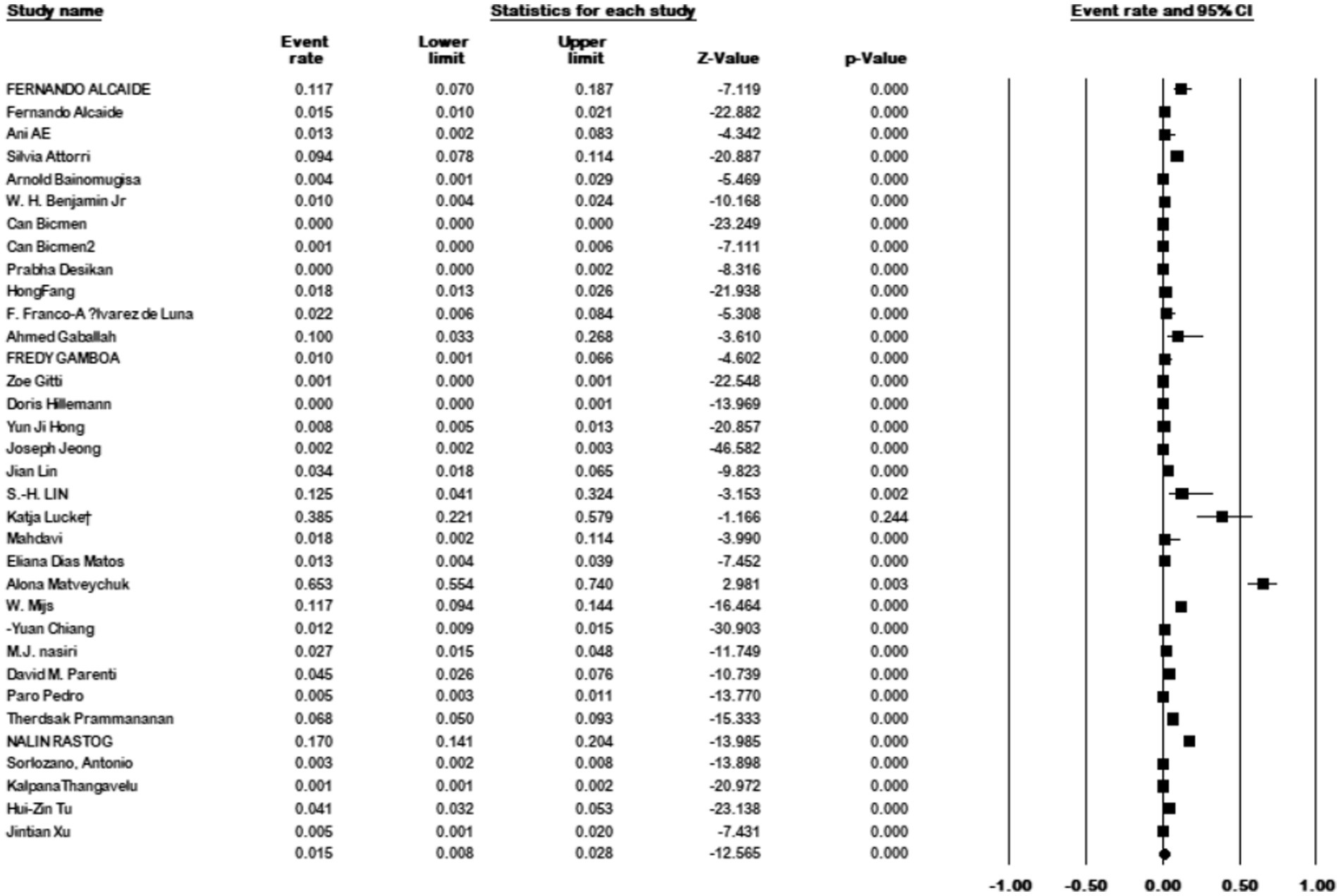
This study included 111 articles that reported the prevalence of M. kansasi in NTM isolates. The prevalence of M. kansasi in these isolates was 9.4% (95% CI: 0.07–0.11%; I2 = 97.75%; p < 0.001).
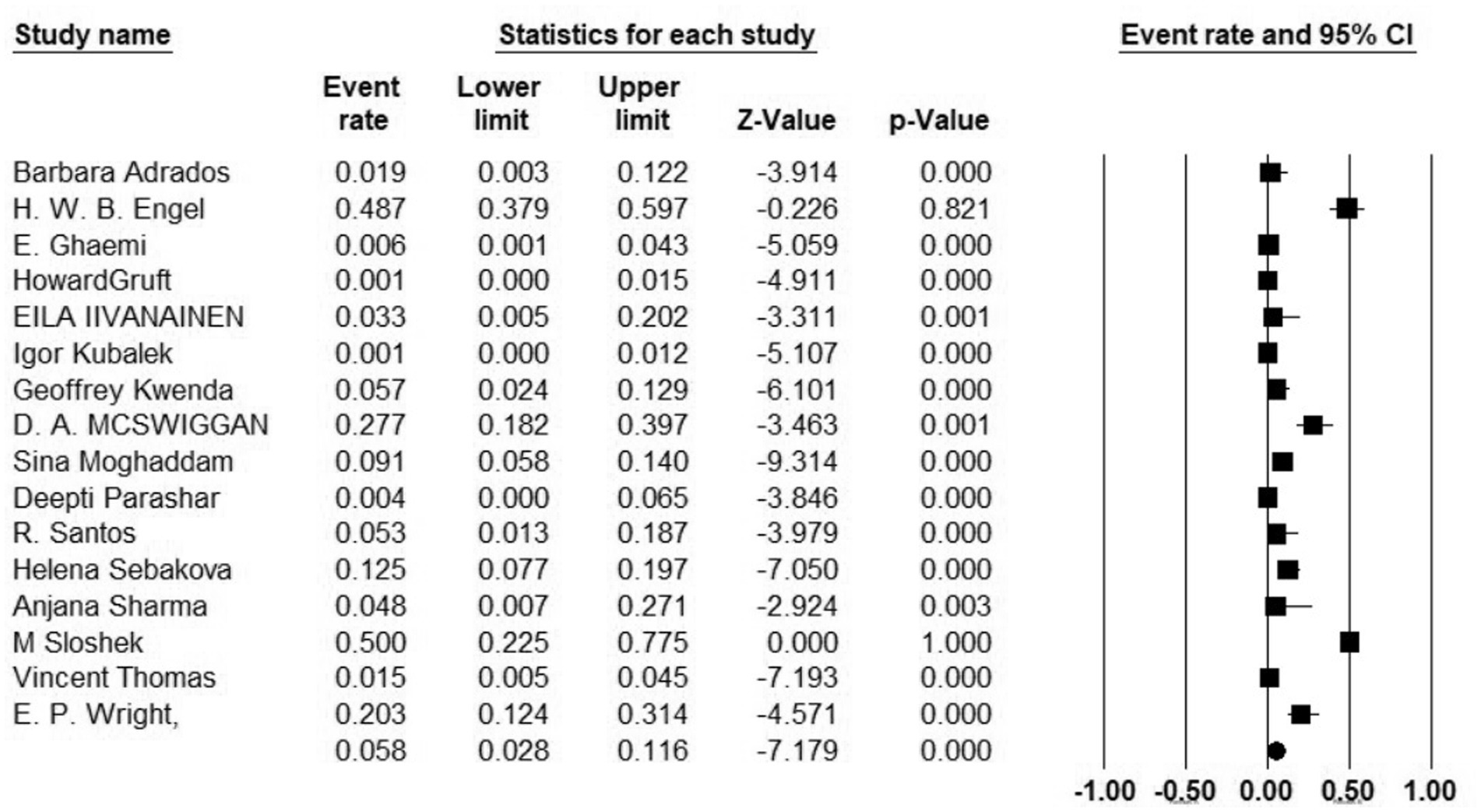
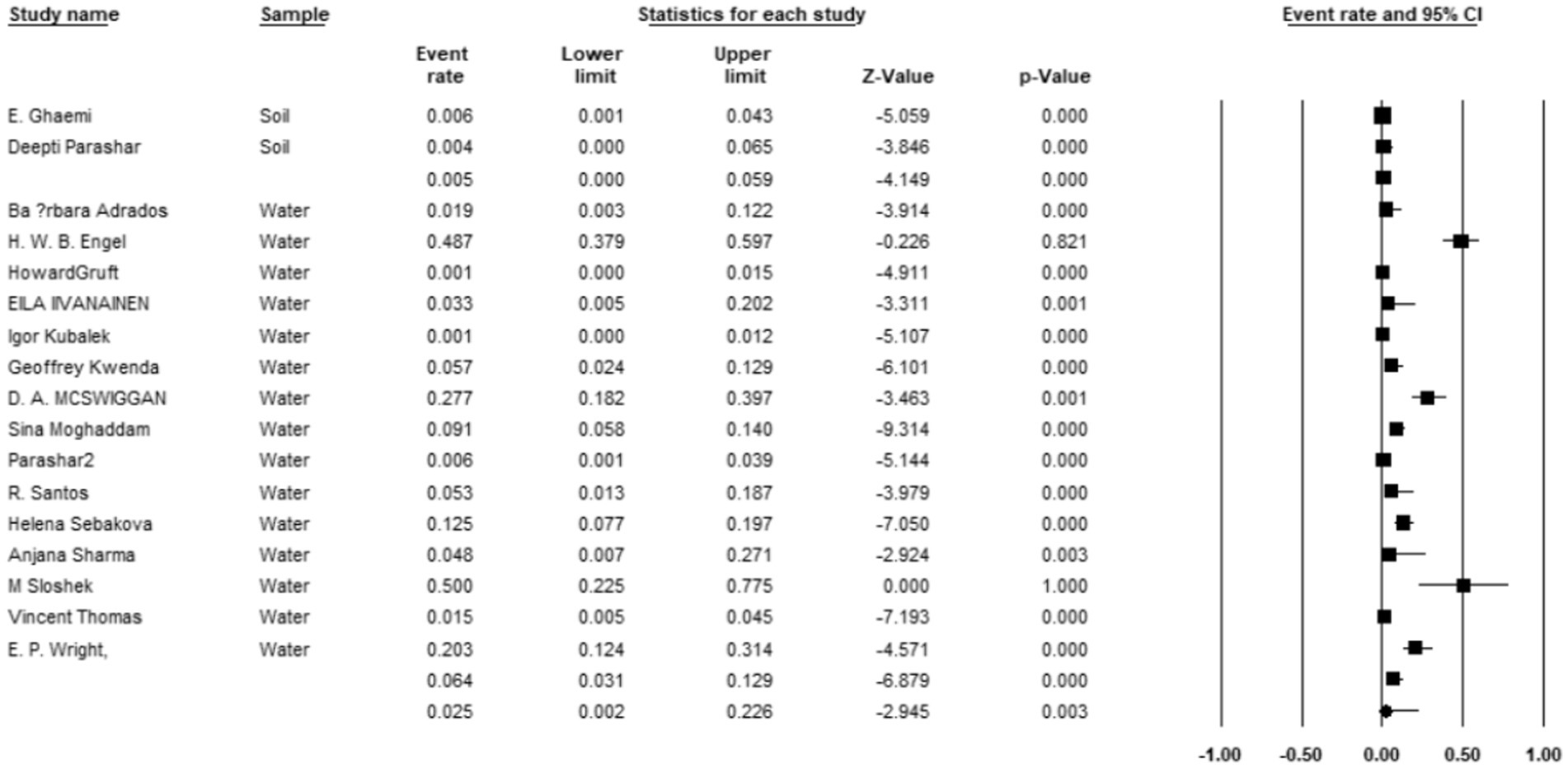
Additionally, 34 articles provided information on the total number of collected isolates, and the prevalence of M. kansasi was found to be 1.5% (95% CI: 0.08–0.028%; I2 = 98.44%; p < 0.001) (Figure 3). Sixteen articles investigated the prevalence of M. kansasi in environmental isolates, including water and soil. The prevalence of M. kansasi in these isolates was 5.8% (95% CI: 0.028–0.116%; I2 = 90.518%; p < 0.001) (Figure 4). Two articles examined the prevalence of M. kansasi in soil, resulting in a prevalence rate of 0.5% (95% CI: 0.000–0.059). Fifteen studies reported the prevalence of M. kansasi in water, which was found to be 6.4% (95% CI: 0.031–0.129; p < 0.001) (Figure 5).
Subgroup analysis of prevalence of Mycobacterium kansasii in NTM isolates
Among the 111 studies that reported the prevalence of M. kansasi in NTM isolates, 64 were conducted in Asia, 25 in Europe, 10 in North America, 6 in Africa, 4 in South America, and 2 in Oceania. The prevalence of M. kansasi was highest in Europe with 12.1% (95% CI 0.08–0.17) and lowest in North America with 2.6% (95% CI 0.006–0.0103) (p < 0.001) (Figure 6).
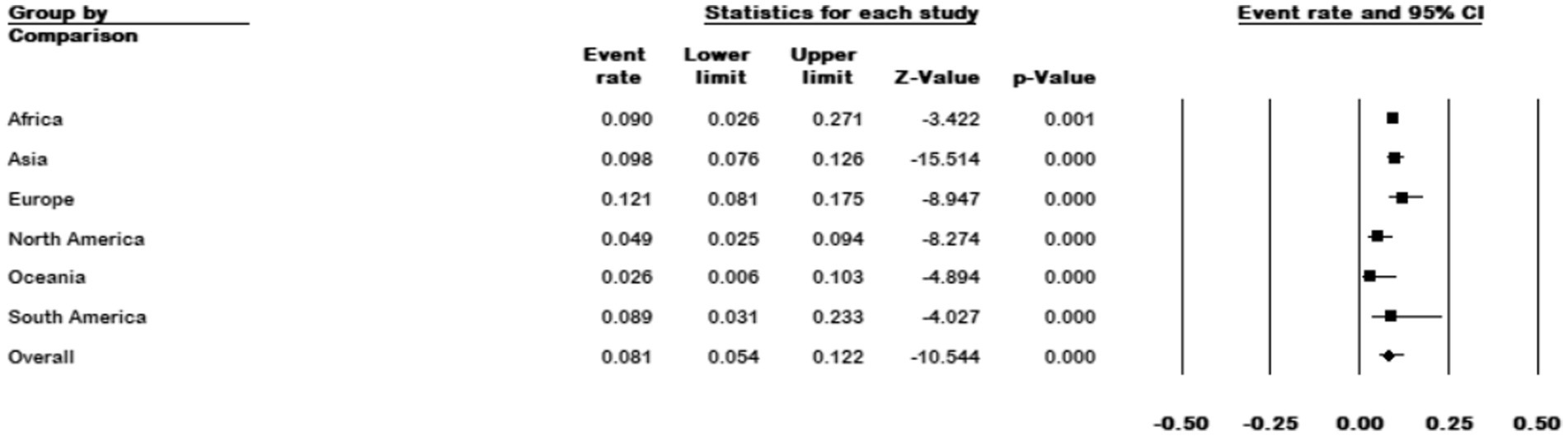
Figure 6. Forest plot showing the prevalence of M. kansasii in NTM isolates for different continents.
The results of the country subgroup meta-analysis showed that Israel had the highest prevalence of M. kansasi in NTM with 50% (95% CI 0.162–0.84), while Botswana had the lowest with 0.6% (95% CI 0.00–0.098) (p < 0.001) (Figure 7).
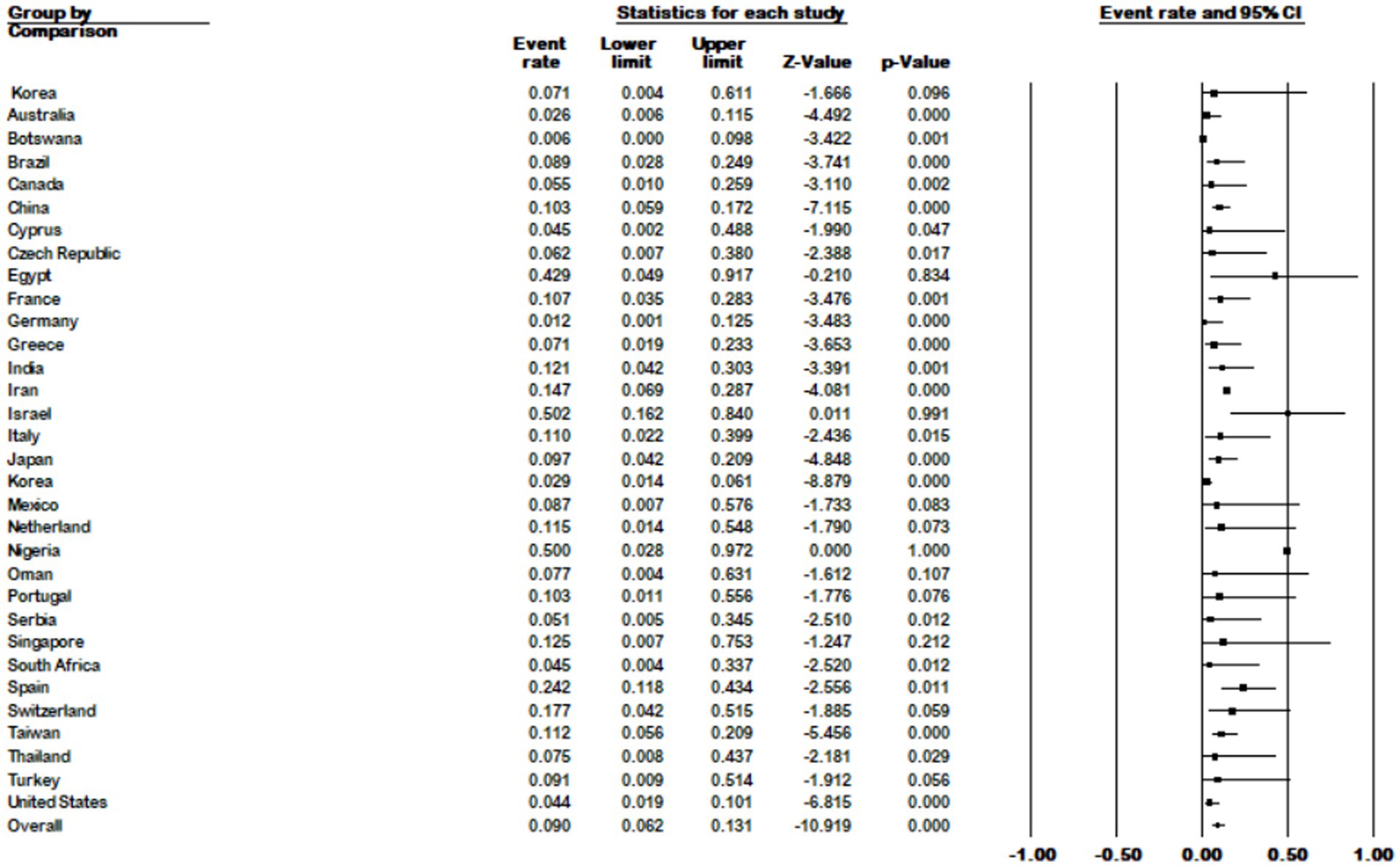
Figure 7. Forest plot showing the prevalence of M. kansasii in NTM isolates for different countries.
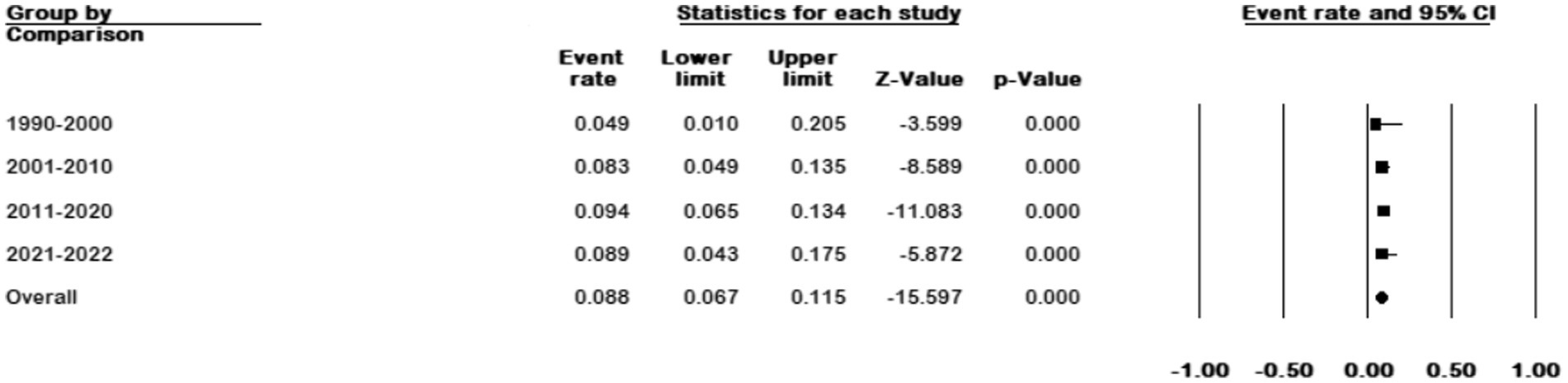
We divided the sample collection time into four periods and analyzed studies whose sample collection time matched our grouping. The results showed an increase in prevalence from 4.9% (95% CI 0.01–0.20) in 1990–2000 to 8.9% (95% CI 0.043–0.175) in 2021–2022 (p < 0.001) (Figure 8).
In general, we divided M. kansasi identification methods into two categories: phenotypic methods including culture characteristics, biochemical methods, MALDI-TOF, and HPLC; and genotypic methods such as sequencing, hybridization, and using probes. According to genotypic methods, the prevalence of M. kansasi in NTM isolates was 7.8% (95% CI 0.058–0.105), and according to phenotypic methods, it was 11% (95% CI 0.078–0.153) (p = 753) (Figure 9).
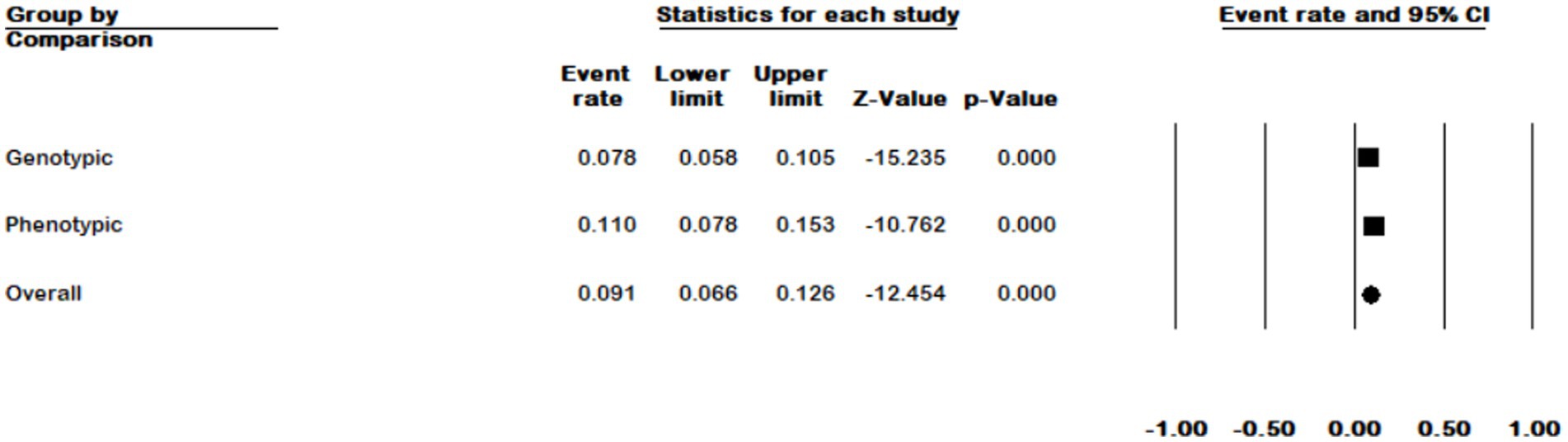
Figure 9. Forest plot showing the prevalence of M. kansasii in NTM isolates for two detection methods.
Subgroup analysis of prevalence of Mycobacterium kansasii in water
Among the 15 studies reporting the prevalence of M. kansasi in water, 3 were conducted in Asia, 10 in Europe, 1 in North America, and 1 in Africa. Europe exhibited the highest prevalence of M. kansasi at 9.7% (95% CI 0.042–0.21, p < 0.001) (Figure 10).
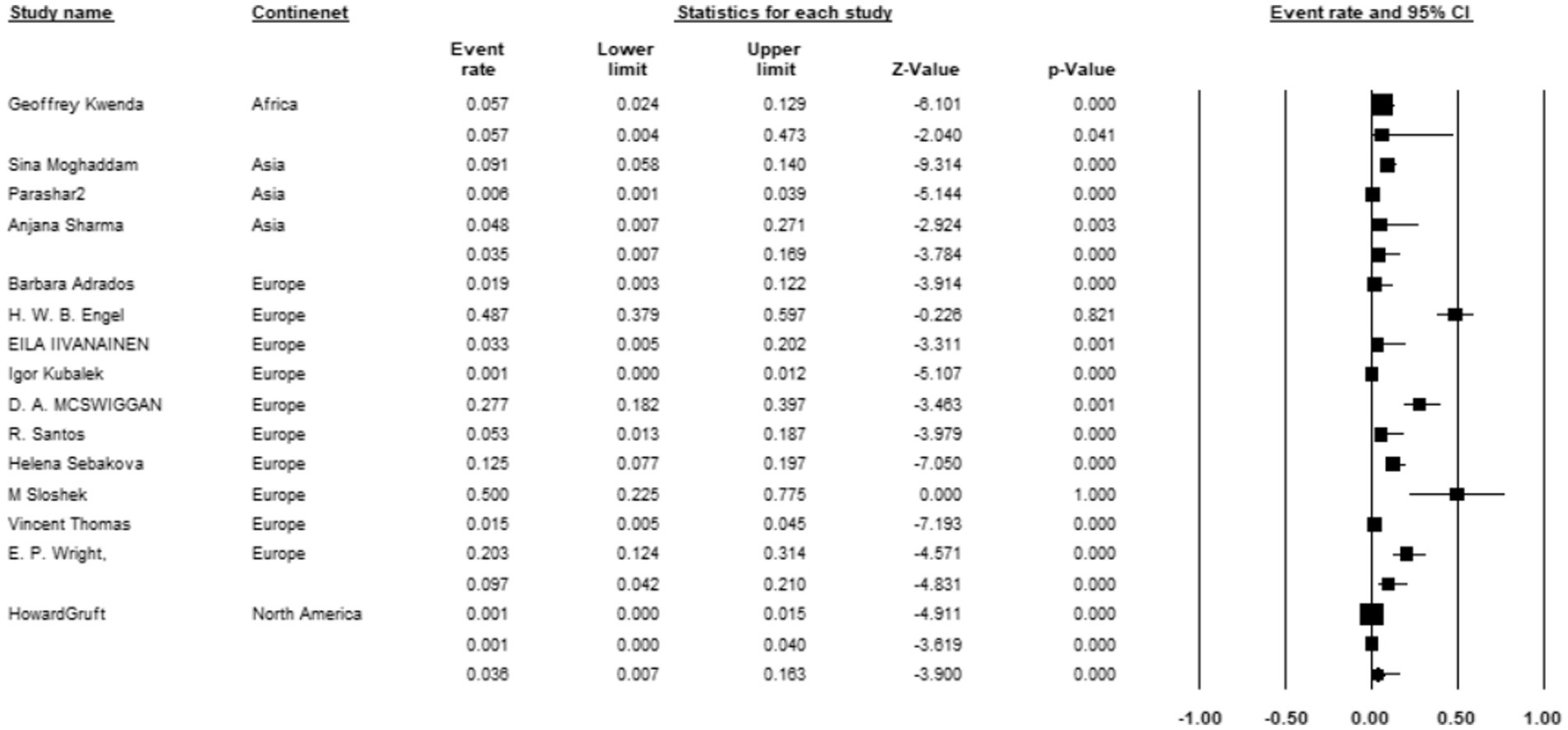
Figure 10. Forest plot showing the prevalence of M. kansasii in water sample in different continents.
The results of the country subgroup meta-analysis demonstrated that the Netherlands had the highest prevalence of M. kansasi in water, with a rate of 48.7% (95% CI 0.093–0.898, p < 0.001) (Figure 11). Furthermore, a subgroup analysis was conducted based on the location of water sample collection. The results indicated a particularly high prevalence of 17.9% (95% CI 0.048–0.484) in mine locations, compared to other sampling sites (p < 0.001) (Figure 12).

Figure 11. Forest plot showing the prevalence of M. kansasii in water sample in different countries.
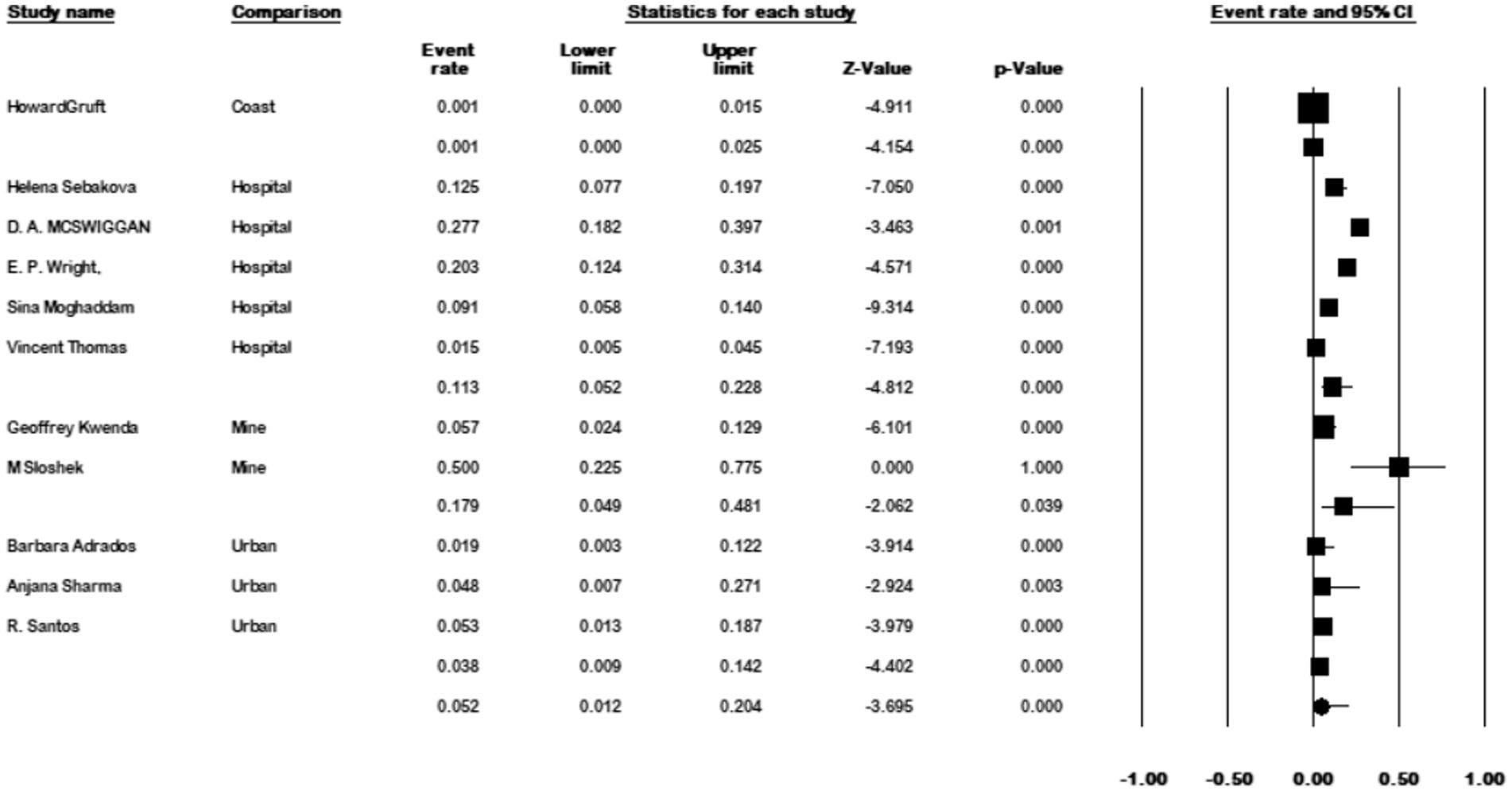
Figure 12. Forest plot showing the prevalence of M. kansasii in water sample in different locations.
Discussion
M. kansasii was among the first non-tuberculous mycobacteria (NTM) to be identified as a respiratory pathogen in humans (Akram and Rawla, 2023). It is the sixth most commonly encountered NTM globally, although there is limited information on its prevalence in various sources and countries (Bakuła et al., 2013). This systematic review and meta-analysis sought to investigate the prevalence of M. kansasii. Our findings revealed a global prevalence of 9.4% among NTM isolates.
Studies have documented distinct regional differences in the prevalence of M. kansasii pulmonary disease, ranging from 3 to 70% worldwide (Guo et al., 2022). Notably, certain regions exhibit a higher prevalence of this bacterium. Our data analysis highlighted that Europe had the highest prevalence at 12.1%, followed by Asia at 9.8%, while Oceania had the lowest prevalence at 2.6% among the continents examined. The reason for the higher prevalence of this bacterium in Europe may be due to the lack of advanced diagnostic facilities in developing countries along with a larger susceptible population due to the aging population in Europe.
Studies have revealed distinct regional differences in the prevalence of M. kansasii pulmonary disease, with a relatively high incidence observed in Brazil, Australia, Poland, and the United Kingdom (Zhang et al., 2023). The meta-analysis conducted by Okoi et al. identified M. kansasii as the most common cause of pulmonary NTM disease in a sub-Saharan African country (Okoi et al., 2017), with a prevalence rate of 69.2%. Additionally, Khosravi et al. reported frequencies ranging from 13 to 17% for this pathogen among all NTM isolates in Iran (Khosravi et al., 2020). Similarly, Morimoto et al. reported M. kansasii as the most prevalent form of NTM in Japan, with a prevalence rate of 43.6% (Morimoto et al., 2017).
Our analysis yielded the highest prevalence rates in Israel, Nigeria, and Egypt, with percentages of 50.2, 50, and 42.9%, respectively. On the other hand, Botswana and Germany exhibited the lowest prevalence rates at 0.06 and 1.2%, respectively.
Overall, NTM diseases are increasingly prevalent worldwide, potentially due to a rising population susceptible to weakened immune systems, organ transplantation, aging, changes in the environment favoring NTM development, and reduced anti-mycobacterial immunity following failed tuberculosis treatment (Chin et al., 2020). To date, there is no comprehensive global study investigating the temporal prevalence of M. kansasii. However, existing studies that have examined its prevalence over time have reported a significant increase in the prevalence of this bacterium. We divided the time of sample collection into four periods (1990–2000, 2001–2010, 2011–2020, and 2021–2022). The results of data analysis showed prevalence rates of 49, 83, 94, and 89%, respectively. Overall, our results demonstrated a significant increase in the prevalence of M. kansasii over time.
In this study, we analyzed the prevalence of M. kansasii in water samples. The prevalence of M. kansasii in water samples was found to be 5.8%, while the prevalence in all clinical isolates was 1.5%. This indicates a high prevalence of this microorganism in water samples, suggesting that water serves as a reservoir for this bacterium. Furthermore, the ability of M. kansasii to form biofilms may result in the release of this microorganism into water, posing a risk to consumers through drinking or inhalation of aerosols from showers, swimming pools, spas, and other water systems (Muñoz-Egea et al., 2023). Therefore, monitoring water samples is crucial for infection control.
Mining has long been associated with diseases caused by NTM, implying that exposure to mining dust may contribute to NTM transmission (Marras and Daley, 2002). As a result, miners may face a higher risk of exposure to potentially pathogenic environmental mycobacteria compared to workers in other occupations. In our study, water samples isolated from the mine had the highest prevalence rate at 17.9%, underscoring the significance of this location as a potential risk factor for M. kansasii transmission.
Although we made efforts to conduct a comprehensive search, it is possible that not all of the relevant existing literature was included. One potential limitation of this meta-analysis is that gray literature was not included in the search strategy. As a result, relevant studies or data that may have been available through gray literature channels, such as conference proceedings or unpublished dissertations, might have been inadvertently overlooked. This limitation could potentially introduce selection bias and limit the comprehensiveness of the findings. The relatively high heterogeneity between studies was another limitation of the present study. To address this, we conducted subgroup analysis to explore the sources of heterogeneity and minimize its impact on the results.
Conclusion
Mycobacterium kansasii is a prevalent causative agent of nontuberculous mycobacterial lung disease globally. Our results have highlighted a substantial prevalence of M. kansasii in clinical isolates, emphasizing the urgent need for heightened attention from health authorities, physicians, and microbiologists.
Furthermore, our investigation into the prevalence over time has revealed a significant increase in the occurrence of this bacterium, underscoring the importance of enhanced identification and control measures within infection control strategies to mitigate its further spread. Additionally, our study has demonstrated a higher prevalence of M. kansasii in water samples, further accentuating the significance of screening these samples for this microorganism as a preventive measure against the associated disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
NN: Conceptualization, Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing. NB: Data curation, Writing – review & editing. FG: Data curation, Writing – review & editing. SR: Investigation, Methodology, Writing – review & editing. FJ: Supervision, Writing – review & editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by Iran University of Medical Sciences (Tehran, Iran).
Acknowledgments
The authors thank Iran University of Medical Sciences (Tehran, Iran) for supporting this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1321273/full#supplementary-material
References
Abate, G., Stapleton, J. T., Rouphael, N., Creech, B., Stout, J. E., el Sahly, H. M., et al. (2021). Variability in the Management of Adults with Pulmonary Nontuberculous Mycobacterial Disease. Clin. Infect. Dis. 72, 1127–1137. doi: 10.1093/cid/ciaa252
Adrados, B., Julián, E., Codony, F., Torrents, E., Luquin, M., and Morató, J. (2011). Prevalence and concentration of non-tuberculous mycobacteria in cooling towers by means of quantitative PCR: a prospective study. Curr. Microbiol. 62, 313–319. doi: 10.1007/s00284-010-9706-2
Adzic-Vukicevic, T., Barac, A., Blanka-Protic, A., Laban-Lazovic, M., Lukovic, B., Skodric-Trifunovic, V., et al. (2018). Clinical features of infection caused by non-tuberculous mycobacteria: 7 years' experience. Infection 46, 357–363. doi: 10.1007/s15010-018-1128-2
Agizew, T., Basotli, J., Alexander, H., Boyd, R., Letsibogo, G., Auld, A., et al. (2017). Higher-than-expected prevalence of non-tuberculous mycobacteria in HIV setting in Botswana: implications for diagnostic algorithms using Xpert MTB/RIF assay. PLoS One 12:e0189981. doi: 10.1371/journal.pone.0189981
Ahn, K., Kim, Y. K., Hwang, G. Y., Cho, H., and Uh, Y. (2021). Continued upward trend in non-tuberculous mycobacteria isolation over 13 years in a tertiary Care Hospital in Korea. Yonsei Med. J. 62, 903–910. doi: 10.3349/ymj.2021.62.10.903
Akram, S. M., and Rawla, P., Mycobacterium kansasii Infection”, in: StatPearls. Treasure Island (FL): StatPearls Publishing. (2023).
al Houqani, M., Jamieson, F., Chedore, P., Mehta, M., May, K., and Marras, T. K. (2011). Isolation prevalence of pulmonary nontuberculous mycobacteria in Ontario in 2007. Can. Respir. J. 18, 19–24. doi: 10.1155/2011/865831
Alcaide, F., Benítez, M. A., and Martín, R. (1999). Epidemiology of Mycobacterium kansasii. Ann. Intern. Med. 131, 310–311. doi: 10.7326/0003-4819-131-4-199908170-00016
Alcaide, F., Galí, N., Domínguez, J., Berlanga, P., Blanco, S., Orús, P., et al. (2003). Usefulness of a new mycobacteriophage-based technique for rapid diagnosis of pulmonary tuberculosis. J. Clin. Microbiol. 41, 2867–2871. doi: 10.1128/JCM.41.7.2867-2871.2003
al-Mahruqi, S. H., van Ingen, J., al-Busaidy, S., Boeree, M. J., al-Zadjali, S., Patel, A., et al. (2009). Clinical relevance of nontuberculous mycobacteria, Oman. Emerg. Infect. Dis. 15, 292–294. doi: 10.3201/eid1502.080977
Amorim, A., Macedo, R., Lopes, A., Rodrigues, I., and Pereira, E. (2010). Non-tuberculous mycobacteria in HIV-negative patients with pulmonary disease in Lisbon, Portugal. Scand. J. Infect. Dis. 42, 626–628. doi: 10.3109/00365541003754485
Andréjak, C., Lescure, F. X., Douadi, Y., Laurans, G., Smail, A., Duhaut, P., et al. (2007). Non-tuberculous mycobacteria pulmonary infection: management and follow-up of 31 infected patients. J. Infect. 55, 34–40. doi: 10.1016/j.jinf.2007.01.008
Ani, A. E., Diarra, B., Dahle, U. R., Lekuk, C., Yetunde, F., Somboro, A. M., et al. (2011). Identification of mycobacteria and other acid fast organisms associated with pulmonary disease. Asian Pac. J. Trop. Dis. 1, 259–262. doi: 10.1016/S2222-1808(11)60061-3
Attorri, S., Dunbar, S., and Clarridge, J. E. (2000). Assessment of morphology for rapid presumptive identification of Mycobacterium tuberculosis and Mycobacterium kansasii. J. Clin. Microbiol. 38, 1426–1429. doi: 10.1128/JCM.38.4.1426-1429.2000
Bainomugisa, A., Wampande, E., Muchwa, C., Akol, J., Mubiri, P., Ssenyungule, H., et al. (2015). Use of real time polymerase chain reaction for detection of M. tuberculosis, M. Avium and M. kansasii from clinical specimens. BMC Infect. Dis. 15:181. doi: 10.1186/s12879-015-0921-0
Bakuła, Z., Safianowska, A., Nowacka-Mazurek, M., Bielecki, J., and Jagielski, T. (2013). Subtyping of Mycobacterium kansasii by PCR-restriction enzyme analysis of the hsp65 gene. Biomed. Res. Int. 2013, 1–4. doi: 10.1155/2013/178725
Benjamin, W. H., Waites, K. B., Beverly, A., Gibbs, L., Waller, M., Nix, S., et al. (1998). Comparison of the MB/BacT system with a revised antibiotic supplement kit to the BACTEC 460 system for detection of mycobacteria in clinical specimens. J. Clin. Microbiol. 36, 3234–3238. doi: 10.1128/JCM.36.11.3234-3238.1998
Bicmen, C., Coskun, M., Gunduz, A. T., Senol, G., Cirak, A. K., and Tibet, G. (2010). Nontuberculous mycobacteria isolated from pulmonary specimens between 2004 and 2009: causative agent or not? New Microbiol. 33, 399–403.
Bicmen, C., Gunduz, A. T., Coskun, M., Senol, G., Cirak, A. K., and Ozsoz, A. (2011). Molecular detection and identification of mycobacterium tuberculosis complex and four clinically important nontuberculous mycobacterial species in smear-negative clinical samples by the genotype mycobacteria direct test. J. Clin. Microbiol. 49, 2874–2878. doi: 10.1128/JCM.00612-11
Blanc, P., Dutronc, H., Peuchant, O., Dauchy, F. A., Cazanave, C., Neau, D., et al. (2016). Nontuberculous mycobacterial infections in a French hospital: a 12-year retrospective study. PLoS One 11:e0168290. doi: 10.1371/journal.pone.0168290
Bodle, E. E., Cunningham, J. A., Della-Latta, P., Schluger, N. W., and Saiman, L. (2008). Epidemiology of nontuberculous mycobacteria in patients without HIV infection, New York City. Emerg. Infect. Dis. 14, 390–396. doi: 10.3201/eid1403.061143
Braun, E., Sprecher, H., Davidson, S., and Kassis, I. (2013). Epidemiology and clinical significance of non-tuberculous mycobacteria isolated from pulmonary specimens. Int. J. Tuberc. Lung Dis. 17, 96–99. doi: 10.5588/ijtld.12.0237
Brown-Elliott, B. A., and Wallace, R. J. (2017). In vitro susceptibility testing of Tedizolid against nontuberculous mycobacteria. J. Clin. Microbiol. 55, 1747–1754. doi: 10.1128/JCM.00274-17
Brown-Elliott, B. A., and Wallace, R. J. (2021). Comparison of in vitro susceptibility of Delafloxacin with ciprofloxacin, moxifloxacin, and other comparator antimicrobials against isolates of nontuberculous mycobacteria. Antimicrob. Agents Chemother. 65:e0007921. doi: 10.1128/AAC.00079-21
Chae, D. R., Kim, Y. I., Kee, S. J., Kim, Y. H., Chi, S. Y., Ban, H. J., et al. (2011). The impact of the 2007 ATS/IDSA diagnostic criteria for nontuberculous mycobacterial disease on the diagnosis of nontuberculous mycobacterial lung disease. Respiration 82, 124–129. doi: 10.1159/000320254
Chai, J., Han, X., Mei, Q., Liu, T., Walline, J. H., Xu, J., et al. (2022). Clinical characteristics and mortality of non-tuberculous mycobacterial infection in immunocompromised vs Immunocompetent Hosts. Front. Med. 9:884446. doi: 10.3389/fmed.2022.884446
Chen, C. Y., Chen, H. Y., Chou, C. H., Huang, C. T., Lai, C. C., and Hsueh, P. R. (2012). Pulmonary infection caused by nontuberculous mycobacteria in a medical center in Taiwan, 20052008. Diagn. Microbiol. Infect. Dis. 72, 47–51. doi: 10.1016/j.diagmicrobio.2011.09.009
Chiang, C. Y., Yu, M. C., Yang, S. L., Yen, M. Y., and Bai, K. J. (2015). Surveillance of tuberculosis in Taipei: the influence of nontuberculous mycobacteria. PLoS One 10:e0142324. doi: 10.1371/journal.pone.0142324
Chin, K. L., Sarmiento, M. E., Alvarez-Cabrera, N., Norazmi, M. N., and Acosta, A. (2020). Pulmonary non-tuberculous mycobacterial infections: current state and future management. Eur. J. Clin. Microbiol. Infect. Dis. 39, 799–826. doi: 10.1007/s10096-019-03771-0
Chou, M. P., Clements, A. C. A., and Thomson, R. M. (2014). A spatial epidemiological analysis of nontuberculous mycobacterial infections in Queensland, Australia. BMC Infect. Dis. 14:14. doi: 10.1186/1471-2334-14-279
Cortés-Torres, N., González-y-Merchand, J. A., González-Bonilla, C., and García-Elorriaga, G. (2013). Molecular analysis of mycobacteria isolated in Mexican patients with different Immunodeficiencies in a tertiary care hospital. Arch. Med. Res. 44, 562–569. doi: 10.1016/j.arcmed.2013.09.002
Cowman, S., van Ingen, J., Griffith, D. E., and Loebinger, M. R. (2019). Non-tuberculous mycobacterial pulmonary disease. Eur. Respir. J. 54:1900250. doi: 10.1183/13993003.00250-2019
Dailloux, M., Abalain, M. L., Laurain, C., Lebrun, L., Loos-Ayav, C., Lozniewski, A., et al. (2006). Respiratory infections associated with nontuberculous mycobacteria in non-HIV patients. Eur. Respir. J. 28, 1211–1215. doi: 10.1183/09031936.00063806
Daley, C. L., Iaccarino, J. M., Lange, C., Cambau, E., Wallace, R. J. Jr., Andrejak, C., et al. (2020). Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin. Infect. Dis. 71, 905–913. doi: 10.1093/cid/ciaa1125
Das, S., Mishra, B., Mohapatra, P. R., Preetam, C., and Rath, S. (2022). Clinical presentations of nontuberculous mycobacteria as suspected and drug-resistant tuberculosis: experience from a tertiary care center in eastern India. Int. J. Mycobacteriol. 11, 167–174. doi: 10.4103/ijmy.ijmy_68_22
Davari, M., Irandoost, M., Sakhaee, F., Vaziri, F., Sepahi, A. A., Rahimi Jamnani, F., et al. (2019). Genetic diversity and prevalence of nontuberculous mycobacteria isolated from clinical samples in Tehran, Iran. Microb. Drug Resist. 25, 264–270. doi: 10.1089/mdr.2018.0150
Debrunner, M., Salfinger, M., Brandli, O., and von Graevenitz, A. (1992). Epidemiology and clinical significance of nontuberculous mycobacteria in patients negative for human immunodeficiency virus in Switzerland. Clin. Infect. Dis. 15, 330–345. doi: 10.1093/clinids/15.2.330
del Giudice, G., Iadevaia, C., Santoro, G., Moscariello, E., Smeraglia, R., and Marzo, C. (2011). Nontuberculous mycobacterial lung disease in patients without HIV infection: a retrospective analysis over 3 years. Clin. Respir. J. 5, 203–210. doi: 10.1111/j.1752-699X.2010.00220.x
Desikan, P., Tiwari, K., Panwalkar, N., Khaliq, S., Chourey, M., Varathe, R., et al. (2017). Public health relevance of non-tuberculous mycobacteria among AFB positive sputa. Germs 7, 10–18. doi: 10.18683/germs.2017.1103
Donohue, M. J. (2021). Epidemiological risk factors and the geographical distribution of eight Mycobacterium species. BMC Infect. Dis. 21:258. doi: 10.1186/s12879-021-05925-y
Engel, H., Berwald, L., and Havelaar, A. (1980). The occurrence of Mycobacterium kansasii in tapwater. Tubercle 61, 21–26. doi: 10.1016/0041-3879(80)90055-0
Fang, H., Shangguan, Y., Wang, H., Ji, Z., Shao, J., Zhao, R., et al. (2019). Multicenter evaluation of the biochip assay for rapid detection of mycobacterial isolates in smear-positive specimens. Int. J. Infect. Dis. 81, 46–51. doi: 10.1016/j.ijid.2019.01.036
Feysia, S. G., Hasan-Nejad, M., Amini, S., Hamzelou, G., Kazemian, H., Kardan-Yamchi, J., et al. (2020). Incidence, clinical manifestation, treatment outcome, and drug susceptibility pattern of nontuberculous mycobacteria in HIV patients in Tehran, Iran. Ethiop. J. Health Sci. 30, 75–84. doi: 10.4314/ejhs.v30i1.10
Franco-Álvarez de Luna, F., Ruiz, P., Gutiérrez, J., and Casal, M. (2006). Evaluation of the GenoType mycobacteria direct assay for detection of Mycobacterium tuberculosis complex and four atypical mycobacterial species in clinical samples. J. Clin. Microbiol. 44, 3025–3027. doi: 10.1128/JCM.00068-06
Gaballah, A., Ghazal, A., Almiry, R., Emad, R., Sadek, N., Abdel Rahman, M., et al. (2022). Simultaneous detection of Mycobacterium tuberculosis and atypical mycobacteria by DNA-microarray in Egypt. Med. Princ. Pract. 31, 246–253. doi: 10.1159/000524209
Gamboa, F., Manterola, J. M., Lonca, J., Matas, L., Viñado, B., Giménez, M., et al. (1997). Detection and identification of mycobacteria by amplification of RNA and DNA in pretreated blood and bone marrow aspirates by a simple lysis method. J. Clin. Microbiol. 35, 2124–2128. doi: 10.1128/jcm.35.8.2124-2128.1997
Gao, C. H., Zhang, Y. A., and Wang, M. S. (2022). Performance of interferon-γ release assays in patients with Mycobacterium kansasii infection. Infect. Drug Resist. 15, 7727–7732. doi: 10.2147/IDR.S385570
Ghaemi, E., Ghazisaidi, K., Koohsari, H., and Mansoorian, A. (2006). Environmental mycobacteria in areas of high and low tuberculosis prevalence in the Islamic Republic of Iran. East. Mediterr. Health J. 12, 280–285.
Gitti, Z., Mantadakis, E., Maraki, S., and Samonis, G. (2011). Clinical significance and antibiotic susceptibilities of nontuberculous mycobacteria from patients in Crete, Greece. Future Microbiol. 6, 1099–1109. doi: 10.2217/fmb.11.91
Gomathy, N. S., Padmapriyadarsini, C., Silambuchelvi, K., Nabila, A., Tamizhselvan, M., Banurekha, V. V., et al. (2019). Profile of patients with pulmonary non-tuberculous mycobacterial disease mimicking pulmonary tuberculosis. Indian J. Tuberc. 66, 461–467. doi: 10.1016/j.ijtb.2019.04.013
Gruft, H., Falkinham, J. O. III, and Parker, B. C. (1981). Recent experience in the epidemiology of disease caused by atypical mycobacteria. Rev. Infect. Dis. 3, 990–996. doi: 10.1093/clinids/3.5.990
Guo, Y., Cao, Y., Liu, H., Yang, J., Wang, W., Wang, B., et al. (2022). Clinical and microbiological characteristics of Mycobacterium kansasii pulmonary infections in China. Microbiol. Spect. 10, e01475–e01421. doi: 10.1128/spectrum.01475-21
Hara, R., Kitada, S., Iwai, A., Kuge, T., Oshitani, Y., Kagawa, H., et al. (2020). Diagnostic validity of gastric aspirate culture in nontuberculous mycobacterial pulmonary disease. Ann. Am. Thorac. Soc. 17, 1536–1541. doi: 10.1513/AnnalsATS.201911-852OC
He, G., Wu, L., Zheng, Q., and Jiang, X. (2022). Antimicrobial susceptibility and minimum inhibitory concentration distribution of common clinically relevant non-tuberculous mycobacterial isolates from the respiratory tract. Ann. Med. 54, 2500–2510. doi: 10.1080/07853890.2022.2121984
Hillemann, D., Richter, E., and Rüsch-Gerdes, S. (2006). Use of the BACTEC mycobacteria growth indicator tube 960 automated system for recovery of mycobacteria from 9,558 extrapulmonary specimens, including urine samples. J. Clin. Microbiol. 44, 4014–4017. doi: 10.1128/JCM.00829-06
Hombach, M., Somoskövi, A., Hömke, R., Ritter, C., and Böttger, E. C. (2013). Drug susceptibility distributions in slowly growing non-tuberculous mycobacteria using MGIT 960 TB eXiST. Int. J. Med. Microbiol. 303, 270–276. doi: 10.1016/j.ijmm.2013.04.003
Hong, Y. J., Chung, Y. H., Kim, T. S., Song, S. H., Park, K. U., Yim, J. J., et al. (2011). Usefulness of three-channel multiplex real-time PCR and melting curve analysis for simultaneous detection and identification of the Mycobacterium tuberculosis complex and nontuberculous mycobacteria. J. Clin. Microbiol. 49, 3963–3966. doi: 10.1128/JCM.05662-11
Hsiao, C. H., Tsai, T. F., and Hsueh, P. R. (2011). Characteristics of skin and soft tissue infection caused by non-tuberculous mycobacteria in Taiwan. Int. J. Tuberc. Lung Dis. 15, 811–817. doi: 10.5588/ijtld.10.0481
Huang, H.-L., Cheng, M. H., Lu, P. L., Shu, C. C., Wang, J. Y., Wang, J. T., et al. (2017). Epidemiology and predictors of NTM pulmonary infection in Taiwan-a retrospective, five-year multicenter study. Sci. Rep. 7:16300. doi: 10.1038/s41598-017-16559-z
Huang, J. J., Li, Y. X., Zhao, Y., Yang, W. H., Xiao, M., Kudinha, T., et al. (2020). Prevalence of nontuberculous mycobacteria in a tertiary hospital in Beijing, China, January 2013 to December 2018. BMC Microbiol. 20:158. doi: 10.1186/s12866-020-01840-5
Huang, M., Lin, Y., Chen, X., and Wu, D. (2021). The value of gene chip detection of bronchoalveolar lavage fluid in the diagnosis of nontuberculous mycobacterial lung disease. Ann. Palliat. Med. 10, 6438–6445. doi: 10.21037/apm-21-1205
Iivanainen, E., Northrup, J., Arbeit, R. D., Ristola, M., Katila, M. L., and von reyn, C. F. (1999). Isolation of mycobacteria from indoor swimming pools in Finland. APMIS 107, 193–200. doi: 10.1111/j.1699-0463.1999.tb01544.x
Jeong, J., Kim, S. R., Lee, S. H., Lim, J. H., Choi, J. I., Park, J. S., et al. (2011). The use of high performance liquid chromatography to speciate and characterize the epidemiology of mycobacteria. Lab. Med. 42, 612–617. doi: 10.1309/LMDDEHPSYE6ZDM3C
Johansen, M. D., Herrmann, J.-L., and Kremer, L. (2020). Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 18, 392–407. doi: 10.1038/s41579-020-0331-1
Karami-Zarandi, M., Bahador, A., Gizaw Feysia, S., Kardan-Yamchi, J., Hasan-Nejad, M., Mosavari, N., et al. (2019). Identification of non-tuberculosis mycobacteria by line probe assay and determination of drug resistance patterns of isolates in Iranian patients. Arch. Razi Inst. 74, 375–384. doi: 10.22092/ari.2019.127144.1372
Khosravi, A. D., Asban, B., Hashemzadeh, M., and Nashibi, R. (2020). Molecular identification, and characterization of Mycobacterium kansasii strains isolated from four tuberculosis regional reference laboratories in Iran during 2016–2018. Infect. Drug Resist. 13, 2171–2180. doi: 10.2147/IDR.S245295
Kim, M. J., Kim, K. M., Shin, J. I., Ha, J. H., Lee, D. H., Choi, J. G., et al. (2021). Identification of nontuberculous mycobacteria in patients with pulmonary diseases in Gyeongnam, Korea, using multiplex PCR and multigene sequence-based analysis. Can. J. Infect. Dis. Med. Microbiol. 2021:8844306. doi: 10.1155/2021/8844306
Kim, Y. G., Lee, H. Y., Kwak, N., Park, J. H., Kim, T. S., Kim, M. J., et al. (2021). Determination of clinical characteristics of Mycobacterium kansasii-derived species by reanalysis of isolates formerly reported as M. kansasii. Ann. Lab. Med. 41, 463–468. doi: 10.3343/alm.2021.41.5.463
Kim, J., Seong, M. W., Kim, E. C., Han, S. K., and Yim, J. J. (2015). Frequency and clinical implications of the isolation of rare nontuberculous mycobacteria. BMC Infect. Dis. 15:9. doi: 10.1186/s12879-014-0741-7
Kodana, M., Tarumoto, N., Kawamura, T., Saito, T., Ohno, H., Maesaki, S., et al. (2016). Utility of the MALDI-TOF MS method to identify nontuberculous mycobacteria. J. Infect. Chemother. 22, 32–35. doi: 10.1016/j.jiac.2015.09.006
Koh, W.-J. (2017). Nontuberculous mycobacteria—overview. Microbiol. Spect. 5, 10–128. doi: 10.1128/microbiolspec.TNMI7-0024-2016
Kontos, F., Petinaki, E., Nicolaou, S., Gitti, Z., Anagnostou, S., Maniati, M., et al. (2003). Multicenter evaluation of the fully automated Bactec MGIT 960 system and three molecular methods for the isolation and the identification of mycobacteria from clinical specimens. Diagn. Microbiol. Infect. Dis. 46, 299–301. doi: 10.1016/S0732-8893(03)00078-6
Kubalek, I., and Mysak, J. (1996). The prevalence of environmental mycobacteria in drinking water supply systems in a demarcated region in Czech Republic, in the period 1984–1989. Eur. J. Epidemiol. 12, 471–474. doi: 10.1007/BF00143998
Kwenda, G., Churchyard, G. J., Thorrold, C., Heron, I., Stevenson, K., Duse, A. G., et al. (2015). Molecular characterisation of clinical and environmental isolates of Mycobacterium kansasii isolates from south African gold mines. J. Water Health 13, 190–202. doi: 10.2166/wh.2014.161
Lai, C., Lee, L., Ding, L., Yu, C., Hsueh, P., and Yang, P. (2006). Emergence of disseminated infections due to nontuberculous mycobacteria in non-HIV-infected patients, including immunocompetent and immunocompromised patients in a university hospital in Taiwan. J. Infect. 53, 77–84. doi: 10.1016/j.jinf.2005.10.009
Lan, R., Yang, C., Lan, L., Ou, J., Qiao, K., Liu, F., et al. (2011). Mycobacterium tuberculosis and non-tuberculous mycobacteria isolates from HIV-infected patients in Guangxi, China. Int. J. Tuberc. Lung Dis. 15, 1669–1675. doi: 10.5588/ijtld.11.0036
Lee, E. H., Chin, B. S., Kim, Y. K., Yoo, J. S., Choi, Y. H., Kim, S., et al. (2022). Clinical characteristics of nontuberculous mycobacterial disease in people living with HIV/AIDS in South Korea: a multi-center, retrospective study. PLoS One 17:e0276484. doi: 10.1371/journal.pone.0276484
Lee, M. R., Yang, C. Y., Shu, C. C., Lin, C. K., Wen, Y. F., Lee, S. W., et al. (2015). Factors associated with subsequent nontuberculous mycobacterial lung disease in patients with a single sputum isolate on initial examination. Clin. Microbiol. Infect. 21, 250.e1–250.e7. doi: 10.1016/j.cmi.2014.08.025
Liao, C. H., Lai, C. C., Ding, L. W., Hou, S. M., Chiu, H. C., Chang, S. C., et al. (2007). Skin and soft tissue infection caused by non-tuberculous mycobacteria. Int. J. Tuberc. Lung Dis. 11, 96–102.
Lin, S. H., Lai, C. C., Huang, S. H., Hung, C. C., and Hsueh, P. R. (2014). Mycobacterial bone marrow infections at a medical Centre in Taiwan, 2001-2009. Epidemiol. Infect. 142, 1524–1532. doi: 10.1017/S0950268813002707
Lin, J., Zhao, Y., Wei, S., Dai, Z., and Lin, S. (2022). Evaluation of the MeltPro Myco assay for the identification of non-tuberculous mycobacteria. Infect. Drug Resist. 15, 3287–3293. doi: 10.2147/IDR.S369160
Liu, C. F., Song, Y. M., He, W. C., Liu, D. X., He, P., Bao, J. J., et al. (2021). Nontuberculous mycobacteria in China: incidence and antimicrobial resistance spectrum from a nationwide survey. Infect. Dis. Poverty 10:59. doi: 10.1186/s40249-021-00844-1
Loizos, A., Soteriades, E. S., Pieridou, D., and Koliou, M. G. (2018). Lymphadenitis by non-tuberculous mycobacteria in children. Pediatr. Int. 60, 1062–1067. doi: 10.1111/ped.13708
López-Roa, P., Aznar, E., Cacho, J., Cogollos-Agruña, R., Domingo, D., García-Arata, M. I., et al. (2020). Epidemiology of non-tuberculous mycobacteria isolated from clinical specimens in Madrid, Spain, from 2013 to 2017. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1089–1094. doi: 10.1007/s10096-020-03826-7
Lucke, K., Hombach, M., Friedel, U., Ritter, C., and Böttger, E. C. (2012). Automated quantitative drug susceptibility testing of non-tuberculous mycobacteria using MGIT 960/EpiCenter TB eXiST. J. Antimicrob. Chemother. 67, 154–158. doi: 10.1093/jac/dkr399
Luo, J., Yu, X., Jiang, G., Fu, Y., Huo, F., Ma, Y., et al. (2018). In vitro activity of Clofazimine against nontuberculous mycobacteria isolated in Beijing, China. Antimicrob. Agents Chemother. 62:e00072-18. doi: 10.1128/AAC.00072-18
Mahdavi, N., Gholizadeh, P., Maleki, M. R., Esfahani, A., Chavoshi, S. H., Nikanfar, A., et al. (2021). Isolation and identification of nontuberculous mycobacteria from specimens of lower respiratory tract of transplanted patients based on the evaluation of 16SrRNA gene. Ann. Ig. 33, 189–197. doi: 10.7416/ai.2021.2424
Manika, K., Tsikrika, S., Tsaroucha, E., Karabela, S., Karachaliou, I., Bosmi, I., et al. (2015). Distribution of nontuberculous mycobacteria in treated patients with pulmonary disease in Greece - relation to microbiological data. Future Microbiol. 10, 1301–1306. doi: 10.2217/FMB.15.50
Marques, L. R. M., Ferrazoli, L., and Chimara, É. (2019). Pulmonary nontuberculous mycobacterial infections: presumptive diagnosis based on the international microbiological criteria adopted in the state of São Paulo, Brazil, 2011-2014. J. Bras. Pneumol. 45:e20180278. doi: 10.1590/1806-3713/e20180278
Marras, T. K., and Daley, C. L. (2002). Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin. Chest Med. 23, 553–567. doi: 10.1016/S0272-5231(02)00019-9
Martin-Casabona, N., Bahrmand, A. R., Bennedsen, J., Østergaard Thomsen, V., Curcio, M., Fauville-Dufaux, M., et al. (2004). Non-tuberculous mycobacteria: patterns of isolation. A multi-country retrospective survey. Int. J. Tuberc. Lung Dis. 8, 1186–1193.
Matos, E. D., Santana, M. A., Santana, M. C., Mamede, P., Bezerra, B. L., Panão, E. D., et al. (2004). Nontuberculosis mycobacteria at a multiresistant tuberculosis reference center in Bahia: clinical epidemiological aspects. Braz. J. Infect. Dis. 8, 296–304. doi: 10.1590/S1413-86702004000400005
Matveychuk, A., Fuks, L., Priess, R., Hahim, I., and Shitrit, D. (2012). Clinical and radiological features of Mycobacterium kansasii and other NTM infections. Respir. Med. 106, 1472–1477. doi: 10.1016/j.rmed.2012.06.023
McSwiggan, D., and Collins, C. (1974). The isolation of M. Kansasii and M. xenopi from water systems. Tubercle 55, 291–297. doi: 10.1016/0041-3879(74)90038-5
Mijs, W., Vreese, K. D., Devos, A., Pottel, H., Valgaeren, A., Evans, C., et al. (2002). Evaluation of a commercial line probe assay for identification of mycobacterium species from liquid and solid culture. Eur. J. Clin. Microbiol. Infect. Dis. 21, 794–802. doi: 10.1007/s10096-002-0825-y
Mirsaeidi, M., Hadid, W., Ericsoussi, B., Rodgers, D., and Sadikot, R. T. (2013). Non-tuberculous mycobacterial disease is common in patients with non-cystic fibrosis bronchiectasis. Int. J. Infect. Dis. 17, e1000–e1004. doi: 10.1016/j.ijid.2013.03.018
Modrá, H., Ulmann, V., Caha, J., Hübelová, D., Konečný, O., Svobodová, J., et al. (2019). Socio-economic and environmental factors related to spatial differences in human non-tuberculous mycobacterial diseases in the Czech Republic. Int. J. Environ. Res. Public Health 16:3969. doi: 10.3390/ijerph16203969
Moghaddam, S., Nojoomi, F., Dabbagh Moghaddam, A., Mohammadimehr, M., Sakhaee, F., Masoumi, M., et al. (2022). Isolation of nontuberculous mycobacteria species from different water sources: a study of six hospitals in Tehran, Iran. BMC microbiology 22:261. doi: 10.1186/s12866-022-02674-z
Moore, J. E., Kruijshaar, M. E., Ormerod, L. P., Drobniewski, F., and Abubakar, I. (2010). Increasing reports of non-tuberculous mycobacteria in England, Wales and Northern Ireland, 1995-2006. BMC Public Health 10:612. doi: 10.1186/1471-2458-10-612
Morimoto, K., Hasegawa, N., Izumi, K., Namkoong, H., Uchimura, K., Yoshiyama, T., et al. (2017). A laboratory-based analysis of nontuberculous mycobacterial lung disease in Japan from 2012 to 2013. Ann. Am. Thorac. Soc. 14, 49–56. doi: 10.1513/AnnalsATS.201607-573OC
Morita, H., Nakamura, A., Kato, T., Kutsuna, T., Niwa, T., Katou, K., et al. (2005). Isolation of nontuberculous mycobacteria from patients with pneumoconiosis. J. Infect. Chemother. 11, 89–92. doi: 10.1007/s10156-004-0368-5
Mortazavi, Z., Bahrmand, A., Sakhaee, F., Hosseini Doust, R., Vaziri, F., Siadat, S. D., et al. (2019). Evaluating the clinical significance of nontuberculous mycobacteria isolated from respiratory samples in Iran: an often overlooked disease. Infect. Drug Resist. 12, 1917–1927. doi: 10.2147/IDR.S214181
Munn, Z., Barker, T. H., Moola, S., Tufanaru, C., Stern, C., McArthur, A., et al. (2020). Methodological quality of case series studies: an introduction to the JBI critical appraisal tool. JBI Evid. Synth. 18, 2127–2133. doi: 10.11124/JBISRIR-D-19-00099
Naito, M., Kurahara, Y., Yoshida, S., Ikegami, N., Kobayashi, T., Minomo, S., et al. (2018). Prognosis of chronic pulmonary aspergillosis in patients with pulmonary non-tuberculous mycobacterial disease. Respir. Investig. 56, 326–331. doi: 10.1016/j.resinv.2018.04.002
Nasiri, M. J., Dabiri, H., Fooladi, A. A. I., Amini, S., Hamzehloo, G., and Feizabadi, M. M. (2018). High rates of nontuberculous mycobacteria isolation from patients with presumptive tuberculosis in Iran. New Microbes New Infect. 21, 12–17. doi: 10.1016/j.nmni.2017.08.008
Nasr Esfahani, B., Moghim, S., Ghasemian Safaei, H., Moghoofei, M., Sedighi, M., and Hadifar, S. (2016). Phylogenetic analysis of prevalent tuberculosis and non-tuberculosis mycobacteria in Isfahan, Iran, based on a 360 bp sequence of the rpoB gene. Jundishapur J. Microbiol. 9:e30763. doi: 10.5812/jjm.30763
Ng, S. S. Y., Tay, Y. K., Koh, M. J. A., Thoon, K. C., and Sng, L. H. (2015). Pediatric cutaneous nontuberculous Mycobacterium infections in Singapore. Pediatr. Dermatol. 32, 488–494. doi: 10.1111/pde.12575
Okoi, C., Anderson, S. T. B., Antonio, M., Mulwa, S. N., Gehre, F., and Adetifa, I. M. O. (2017). Non-tuberculous mycobacteria isolated from pulmonary samples in sub-Saharan Africa-a systematic review and meta analyses. Sci. Rep. 7:12002. doi: 10.1038/s41598-017-12175-z
Ose, N., Maeda, H., Takeuchi, Y., Susaki, Y., Kobori, Y., Taniguchi, S., et al. (2016). Solitary pulmonary nodules due to non-tuberculous mycobacteriosis among 28 resected cases. Int. J. Tuberc. Lung Dis. 20, 1125–1129. doi: 10.5588/ijtld.15.0819
Ose, N., Takeuchi, Y., Kitahara, N., Matumura, A., Kodama, K., Shiono, H., et al. (2021). Analysis of pulmonary nodules caused by nontuberculous mycobacteriosis in 101 resected cases: multi-center retrospective study. J. Thorac. Dis. 13, 977–985. doi: 10.21037/jtd-20-3108
Ouzzani, M., Hammady, H., Fedorowicz, Z., and Elmagarmid, A. (2016). Rayyan—a web and mobile app for systematic reviews. Syst. Rev. 5, 1–10. doi: 10.1186/s13643-016-0384-4
Pang, Y., Zheng, H., Tan, Y., Song, Y., and Zhao, Y. (2017). In vitro activity of Bedaquiline against nontuberculous mycobacteria in China. Antimicrob. Agents Chemother. 61:e02627-16. doi: 10.1128/AAC.02627-16
Parashar, D., das, R., Chauhan, D. S., Sharma, V. D., Lavania, M., Yadav, V. S., et al. (2009). Identification of environmental mycobacteria isolated from Agra, North India by conventional & molecular approaches. Indian J. Med. Res. 129, 424–431.
Parenti, D. M., Symington, J. S., Keiser, J., and Simon, G. L. (1995). Mycobacterium kansasii bacteremia in patients infected with human immunodeficiency virus. Clin. Infect. Dis. 21, 1001–1003. doi: 10.1093/clinids/21.4.1001
Park, S., Jo, K. W., Lee, S. D., Kim, W. S., and Shim, T. S. (2017). Clinical characteristics and treatment outcomes of pleural effusions in patients with nontuberculous mycobacterial disease. Respir. Med. 133, 36–41. doi: 10.1016/j.rmed.2017.11.005
Pavlik, I., Ulmann, V., Hubelova, D., and Weston, R. T. (2022). Nontuberculous mycobacteria as sapronoses: a review. Microorganisms 10:1345. doi: 10.3390/microorganisms10071345
Pedrero, S., Tabernero, E., Arana-Arri, E., Urra, E., Larrea, M., and Zalacain, R. (2019). Changing epidemiology of nontuberculous mycobacterial lung disease over the last two decades in a region of the Basque country. ERJ Open Res. 5, 00110–02018. doi: 10.1183/23120541.00110-2018
Pedro, H. D. S. P., Pereira, M. I. F., Goloni, M. R. A., Ueki, S. Y. M., and Chimara, E. (2008). Nontuberculous mycobacteria isolated in São José do Rio Preto, Brazil between 1996 and 2005. J. Bras. Pneumol. 34, 950–955. doi: 10.1590/S1806-37132008001100010
Pierre-Audigier, C., Ferroni, A., Sermet-Gaudelus, I., le Bourgeois, M., Offredo, C., Vu-Thien, H., et al. (2005). Age-related prevalence and distribution of nontuberculous mycobacterial species among patients with cystic fibrosis. J. Clin. Microbiol. 43, 3467–3470. doi: 10.1128/JCM.43.7.3467-3470.2005
Prammananan, T., Cheunoy, W., Na-Ubol, P., Tingtoy, N., Srimuang, S., and Chaiprasert, A. (2005). Evaluation of polymerase chain reaction and restriction enzyme analysis for routine identification of mycobacteria: accuracy, rapidity, and cost analysis. Southeast Asian J. Trop. Med. Public Health 36, 1252–1260.
Rastogi, N., and Goh, K. S. (1992). Effect of pH on radiometric MICs of clarithromycin against 18 species of mycobacteria. Antimicrob. Agents Chemother. 36, 2841–2842. doi: 10.1128/AAC.36.12.2841
Riello, F. N., Brígido, R. T. S., Araújo, S., Moreira, T. A., Goulart, L. R., and Goulart, I. M. B. (2016). Diagnosis of mycobacterial infections based on acid-fast bacilli test and bacterial growth time and implications on treatment and disease outcome. BMC Infect. Dis. 16:142. doi: 10.1186/s12879-016-1474-6
Rodriguez Dı́az, J. C., López, M., Ruiz, M., and Royo, G. (2003). In vitro activity of new fluoroquinolones and linezolid against non-tuberculous mycobacteria. Int. J. Antimicrob. Agents 21, 585–588. doi: 10.1016/S0924-8579(03)00048-7
Ruiz, M., Rodriguez, J. C., Escribano, I., Garcia‐Martinez, J., Rodriguez‐Valera, F., and Royo, G. (2001). Application of molecular biology techniques to the diagnosis of nontuberculous mycobacterial infections. APMIS 109, 857–864. doi: 10.1034/j.1600-0463.2001.091208.x
Ryoo, S. W., Shin, S., Shim, M. S., Park, Y. S., Lew, W. J., Park, S. N., et al. (2008). Spread of nontuberculous mycobacteria from 1993 to 2006 in Koreans. J. Clin. Lab. Anal. 22, 415–420. doi: 10.1002/jcla.20278
Saifi, M., Jabbarzadeh, E., Bahrmand, A. R., Karimi, A., Pourazar, S., Fateh, A., et al. (2013). HSP65-PRA identification of non-tuberculosis mycobacteria from 4892 samples suspicious for mycobacterial infections. Clin. Microbiol. Infect. 19, 723–728. doi: 10.1111/j.1469-0691.2012.04005.x
Santos, R., Oliveira, F., Fernandes, J., Gonçalves, S., Macieira, F., and Cadete, M. (2005). Detection and identification of mycobacteria in the Lisbon water distribution system. Water Sci. Technol. 52, 177–180. doi: 10.2166/wst.2005.0258
Scarparo, C., Piccoli, P., Rigon, A., Ruggiero, G., Ricordi, P., and Piersimoni, C. (2002). Evaluation of the BACTEC MGIT 960 in comparison with BACTEC 460 TB for detection and recovery of mycobacteria from clinical specimens. Diagn. Microbiol. Infect. Dis. 44, 157–161. doi: 10.1016/S0732-8893(02)00437-6
Shafer, R. W., and Sierra, M. F. (1992). Mycobacterium xenopi, Mycobacterium fortuitum, Mycobacterium kansasii, and other nontuberculous mycobacteria in an area of endemicity for AIDS. Clin. Infect. Dis. 15, 161–162. doi: 10.1093/clinids/15.1.161
Shao, Y., Chen, C., Song, H., Li, G., Liu, Q., Li, Y., et al. (2015). The epidemiology and geographic distribution of nontuberculous mycobacteria clinical isolates from sputum samples in the eastern region of China. PLoS Negl. Trop. Dis. 9:e0003623. doi: 10.1371/journal.pntd.0003623
Sharma, A., Chandraker, S. K., and Bharti, M. (2007). Nontubercular mycobacteria in drinking water of some educational institutes in Jabalpur (MP), India. Indian J. Microbiol. 47, 233–240. doi: 10.1007/s12088-007-0044-4
Shen, G. H., Hung, C. H., Hu, S. T., Wu, B. D., Lin, C. F., Chen, C. H., et al. (2009). Combining polymerase chain reaction restriction enzyme analysis with phenotypic characters for mycobacteria identification in Taiwan. Int. J. Tuberc. Lung Dis. 13, 472–479.
Shenai, S., Rodrigues, C., and Mehta, A. (2010). Time to identify and define non-tuberculous mycobacteria in a tuberculosis-endemic region. Int. J. Tuberc. Lung Dis. 14, 1001–1008.
Sheu, L. C., Tran, T. M., Jarlsberg, L. G., Marras, T. K., Daley, C. L., and Nahid, P. (2015). Non-tuberculous mycobacterial infections at San Francisco general hospital. Clin. Respir. J. 9, 436–442. doi: 10.1111/crj.12159
Slosárek, M., Alugupalli, S., Kaustová, J., and Larsson, L. (1996). Rapid detection of Mycobacterium kansasii in water by gas chromatography/mass spectrometry. J. Microbiol. Methods 27, 229–232. doi: 10.1016/S0167-7012(96)00954-2
Sookan, L., and Coovadia, Y. M. (2014). A laboratory-based study to identify and speciate non-tuberculous mycobacteria isolated from specimens submitted to a central tuberculosis laboratory from throughout KwaZulu-Natal Province, South Africa. S. Afr. Med. J. 104, 766–768. doi: 10.7196/SAMJ.8017
Sorlozano, A., Soria, I., Roman, J., Huertas, P., Soto, M. J., Piedrola, G., et al. (2009). Comparative evaluation of three culture methods for the isolation of mycobacteria from clinical samples. J. Microbiol. Biotechnol. 19, 1259–1264. doi: 10.4014/jmb.0901.0059
Steadham, J. E. (1980). High-catalase strains of Mycobacterium kansasii isolated from water in Texas. J. Clin. Microbiol. 11, 496–498. doi: 10.1128/jcm.11.5.496-498.1980
Takenaka, S., Ogura, T., Oshima, H., Izumi, K., Hirata, A., Ito, H., et al. (2020). Development and exacerbation of pulmonary nontuberculous mycobacterial infection in patients with systemic autoimmune rheumatic diseases. Mod. Rheumatol. 30, 558–563. doi: 10.1080/14397595.2019.1619220
Tan, Y., Su, B., Shu, W., Cai, X., Kuang, S., Kuang, H., et al. (2018). Epidemiology of pulmonary disease due to nontuberculous mycobacteria in southern China, 2013-2016. BMC Pulm. Med. 18:168. doi: 10.1186/s12890-018-0728-z
Thangavelu, K., Krishnakumariamma, K., Pallam, G., Dharm Prakash, D., Chandrashekar, L., Kalaiarasan, E., et al. (2021). Prevalence and speciation of non-tuberculous mycobacteria among pulmonary and extrapulmonary tuberculosis suspects in South India. J. Infect. Public Health 14, 320–323. doi: 10.1016/j.jiph.2020.12.027
Thomas, V., Herrera-Rimann, K., Blanc, D. S., and Greub, G. (2006). Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 72, 2428–2438. doi: 10.1128/AEM.72.4.2428-2438.2006
Thomson, R. M., Furuya-Kanamori, L., Coffey, C., Bell, S. C., Knibbs, L. D., and Lau, C. L. (2020). Influence of climate variables on the rising incidence of nontuberculous mycobacterial (NTM) infections in Queensland, Australia 2001-2016. Sci. Total Environ. 740:139796. doi: 10.1016/j.scitotenv.2020.139796
Tu, H. Z., Chang, S. H., Huaug, T. S., Huaug, W. K., Liu, Y. C., and Lee, S. S. (2003). Microscopic morphology in smears prepared from MGIT broth medium for rapid presumptive identification of Mycobacterium tuberculosis complex, Mycobacterium avium complex and Mycobacterium kansasii. Ann. Clin. Lab. Sci. 33, 179–183.
Urabe, N., Sakamoto, S., Ito, A., Sekiguchi, R., Shimanuki, Y., Kanokogi, T., et al. (2021). Bronchial brushing and diagnosis of pulmonary nontuberculous mycobacteria infection. Respiration 100, 877–885. doi: 10.1159/000515605
Woods, G. L., and Washington, J. A. (1987). Mycobacteria other than Mycobacterium tuberculosis: review of microbiologic and clinical aspects. Rev. Infect. Dis. 9, 275–294. doi: 10.1093/clinids/9.2.275
Wright, E. P., Collins, C. H., and Yates, M. D. (1985). Mycobacterium xenopi and Mycobacterium kansasii in a hospital water supply. J. Hosp. Infect. 6, 175–178. doi: 10.1016/S0195-6701(85)80095-5
Wu, J., Zhang, Y., Li, J., Lin, S., Wang, L., Jiang, Y., et al. (2014). Increase in nontuberculous mycobacteria isolated in Shanghai, China: results from a population-based study. PLoS One 9:e109736. doi: 10.1371/journal.pone.0109736
Xu, J., Li, P., Zheng, S., Shu, W., and Pang, Y. (2019). Prevalence and risk factors of pulmonary nontuberculous mycobacterial infections in the Zhejiang Province of China. Epidemiol. Infect. 147:e269. doi: 10.1017/S0950268819001626
Keywords: Mycobacterium kansasii , meta-analysis, CMA, prevalence, NTM
Citation: Narimisa N, Bostanghadiri N, Goodarzi F, Razavi S and Jazi FM (2024) Prevalence of Mycobacterium kansasii in clinical and environmental isolates, a systematic review and meta-analysis. Front. Microbiol. 15:1321273. doi: 10.3389/fmicb.2024.1321273
Edited by:
Matt Johansen, University of Technology Sydney, AustraliaReviewed by:
Beatriz E. Ferro, Universidad Icesi, ColombiaDereje Abate Negatu, Center for Discovery and Innovation, Hackensack Meridian Health, United States
Copyright © 2024 Narimisa, Bostanghadiri, Goodarzi, Razavi and Jazi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Faramarz Masjedian Jazi, Masjedian.f@iums.ac.ir
 Negar Narimisa
Negar Narimisa