- 1Institute of Advanced Agricultural Science, Hubei University of Arts and Science, Xiangyang, Hubei, China
- 2College of Agriculture, Yangtze University, Jingzhou, China
- 3Guizhou Provincial Tobacco Company, Zunyi Branch, Zunyi, China
- 4Guizhou Provincial Academician Workstation of Microbiology and Health, Guizhou Academy of Tobacco Science, Guiyang, Guizhou, China
Piriformospora indica is an important endophytic fungus with broad potential for alleviating biotic and abiotic stress on host plants. This study monitored the growth dynamics of P. indica on five commonly used artificial media for microorganisms and analyzed its metabolic characteristics using Biolog Phenotype Microarray (PM) technology. The results showed that P. indica grew fastest on Potato Dextrose Agar (PDA), followed by Kidney Bean Agar (KBA), Alkyl Ester Agar (AEA), Oatmeal Agar (OA), and Luria-Bertani Agar (LB), and the most suitable medium for spore production was OA. Using Biolog PM1-10, 950 metabolic phenotypes of P. indica were obtained. P. indica could metabolize 87.89% of the tested carbon sources, 87.63% of the tested nitrogen sources, 96.61% of the tested phosphorus sources, and 100% of the tested sulfur sources. P. indica displayed 92 kinds of tested biosynthetic pathways, and it could grow under 92 kinds of tested osmotic pressures and 88 kinds of tested pH conditions. PM plates 1-2 revealed 43 efficient carbon sources, including M-Hydroxyphenyl acid, N-Acetyl-D-Glucosamine, Tyramine, Maltotrios, α-D-Glucosine, I-Erythritol, L-Valine, D-Melezitose, D-Tagatose, and Turanose. PM plates 3,6-8 indicated 170 efficient nitrogen sources, including Adenosine, Inosine Allantoin, D, L-Lactamide, Arg-Met, lle-Trp, Ala-Arg, Thr-Arg, Trp-Tyr, Val-Asn, Gly-Gly-D-Leu, Gly-Gly-Phe, and Leu-Leu-Leu. This study demonstrates that P. indica can metabolize a variety of substrates, such as carbon and nitrogen sources, and has a wide range of environmental adaptability. The growth dynamics on artificial culture media and metabolic phenotypes of P. indica can be used to investigate its biological characteristics, screen for more suitable growth and sporulation conditions, and elucidate the physiological mechanisms that enhance the stress resistance of host plants. This study provides a theoretical basis for its better application in agriculture.
Introduction
Piriformospora indica belongs to the genus Piriformospora, the family Sebacinaceae, the order Sebacinales, the class Hymenomycetes, and the phylum Basidiomycota (Waller et al., 2005; Hibbett et al., 2007). It is a root endophytic fungus that was isolated by Indian scientists from the rhizosphere of woody shrubs while collecting spores of the arbuscular mycorrhiza (AM) fungus Glomus mosseae in the interior desert of Rajasthan, India (Verma et al., 1998). It is named P. indica due to its pear-shaped chlamydospores (Varma et al., 2012). It shares similarities with some AM fungi, such as G. mosseae and Gigaspora margarita (Chen et al., 2007; Venice et al., 2020). They are both endophytic fungi that can coexist with the host plant (Singh et al., 2000; Kesharwani et al., 2022), promote the absorption of mineral elements and cycling substances in the soil, and enhance the plant's resistance to biotic and abiotic stress (Xu et al., 2018; Bandyopadhyay et al., 2022). Compared to the AM fungus, P. indica has notable advantages, such as its ability to obtain nutrition from non-living sources and its wide range of host plants (Varma et al., 2001; Deshmukh et al., 2006; Oelmüller et al., 2009). Therefore, it has gained widespread attention since its discovery (Kaldorf et al., 2005; Gill et al., 2016; Su et al., 2017). P. indica can interact with various plant species, including Gymnospermae and Angiospermae (Khalid et al., 2019), as well as lower plants such as Bryophyta and Pteridophyta (Varma et al., 1999; Manzar et al., 2021a). Currently, it is known to colonize over 200 plants, including monocotyledonous plants such as Oryza sativa (Ghorbani et al., 2021), Triticum aestivum (Li et al., 2022), Hordeum vulgare (Alikhani et al., 2013), and Zea mays (Xu et al., 2017), and dicotyledonous plants like Nicotiana tabacum (Hui et al., 2015), Passiflora edulis (Yan et al., 2021), Solanum tuberosum (Fakhro et al., 2010), and Cicer arietinum (Narayan et al., 2017). It can even colonize Arabidopsis thaliana, a cruciferous model plant that AM fungi cannot colonize (Abdelaziz et al., 2017; Manzar et al., 2021b). Many scholars have studied the colonization of P. indica in various plants, focusing on its effects on promoting host plant growth, improving disease resistance, and enhancing stress tolerance, including resistance to drought, cold, heat, salt, and flooding stresses (Ghabooli et al., 2013; Amani et al., 2021). However, research on the basic biological characteristics of P. indica, such as its growth dynamics on different culture media, has been limited and has only been focused on screening suitable media at a macro level. There have been no reports on the micro-metabolic phenotype profiling of P. indica up to now.
The metabolic phenotypes include the ability to utilize carbon, nitrogen, phosphorus, and sulfur sources, as well as growth tests under different osmotic pressure and pH conditions (Mazur et al., 2013; Narware et al., 2023). The Phenotype Microarray (PM) technology, developed by Biolog Company in the United States, can be used to determine various metabolic phenotypes of microorganisms. The PM is a novel technology that is parallel to genomics and proteomics and is widely used in the study of various microorganisms (Bochner, 2003). It involves fixing a dried cell culture medium and a colorimetric substance in each well of a 96-well microplate (PM plate). After adding microbial cell suspension and incubation, the cell metabolic phenotype can be observed through color changes. Finally, the dynamics of phenotypic reactions are obtained, and the strength of phenotypic reactions can be quantitatively measured (Li and Lu, 2007).
Based on these premises, this study aimed at screening the most suitable growth medium and sporulation medium for P. indica by analyzing its growth dynamics on five commonly used artificial culture media. The goal is to quickly provide the required biomass and spore quantities for production or scientific research. Additionally, the metabolic phenotype of P. indica was assessed using the Biolog PM technology, aiming to identify its characteristic nutrients and reveal its adaptability under different osmotic pressure and pH environments. This information will help to clarify the physiological reasons behind its enhancement of plant stress resistance and provide a theoretical basis for the development and utilization of P. indica as a biocontrol agent.
Materials and methods
Fungal strain, culture media, and reagents
The tested strain of Piriformospora indica was provided by the College of Life Sciences, Yangtze University (Dong et al., 2013). The five test media were formulated as follows: Potato Dextrose Agar (PDA): potato 200 g·L−1, glucose 20 g·L−1, and agar 20 g·L−1. Alkyl Ester Agar (AEA): yeast extract 5 g·L−1, glycerol 20 ml·L−1, MgSO4 0.25 g·L−1, NaNO3 6 g·L−1, KCl 0.5 g·L−1, KH2PO4 1.5 g·L−1, and agar 20 g·L−1. Oatmeal Agar (OA): raw oatmeal powder 30 g·L−1 and 20 g·L−1 agar. Kidney Bean Agar (KBA): kidney bean 150 g·L−1 and agar 20 g·L−1. Luria-Bertani (LB): tryptone 10 g·L−1, yeast extract 5 g·L−1, and NaCl 10 g·L−1. All culture media were sterilized at 116°C for 30 min.
PM1-10 metabolic plates (Grades: # 12111, # 12112, # 12121, # 12131, # 12141, # 12181, # 12182, # 12183, # 12161, and # 12162), sterile sampling tank (# 3002), FF-IF inoculum (# 72106), OmniLog PM system (# 91171), a turbidity meter (# 3531), and an 8-channel electric pipette (# 3501A) were produced by Biolog, Hayward, CA, United States.
Growth and sporulation monitoring of P. indica on five different culture media
The plates of P. indica preserved at 4°C were inoculated onto new PDA plates and incubated at 28°C for 1 week. Using sterile punchers (Ø = 6 mm), fungal blocks were made from the active edges of the fungal colonies and then injected into separate Petri dishes (Ø = 12 cm), containing different solid culture media. Fifteen plates were inoculated for each type of culture medium (PDA, AEA, OA, KBA, and LB), and all plates were placed in a dark incubator at 28°C. The diameter of the colony on each plate was measured daily using the vertical cross method; three repeats were set for each medium. The remaining plates were used for photography and microscopic examination to observe spore production and determine the quantity of spores produced. A small amount of mycelium was taken daily with a sterile toothpick, stained with 0.05% trypan blue for 5 min, washed twice with sterile water, and observed for sporulation and the amount of spore production under a 400× microscope (Nikon, Y-TV55, Japan).
Biolog PM metabolic phenotype detection of P. indica
The Biolog PM test involved 950 different conditions, including 190 carbon sources (PM1-2), 380 nitrogen sources (PM3, 6-8), 94 biosynthetic pathways (PM5), 59 phosphorus sources, 35 sulfur sources (PM4), 96 osmotic pressures (PM9), and 96 pH conditions (PM10). The phenotypic analysis of P. indica was conducted following the Biolog standard procedures (Bochner, 2003). P. indica was incubated on an OA plate under dark conditions at 28°C. After 5 days, spores were produced on the surface of the colony. Sterile cotton swabs moistened with sterile water were used to rotate and dip the spores on the colony's surface, and they were diluted to a 12-ml FF-IF inoculum. The concentration of the spore suspension was adjusted to 62% T (T is the Biolog standard concentration unit) (Bochner, 2003).
Carbon metabolism phenotype analysis
A total of 0.05 ml of spore suspension was added to 23.95 ml of PM1 and PM2 inoculum, vortexed, and mixed evenly. An 8-channel electric pipette was used to add the mixed spore suspension to PM1 and PM2 metabolic plates, with 100 μl per well. The prepared phenotypic assay plate was incubated for 6 days at 28°C in an OmniLog constant-temperature incubator. The OmniLog software was set up to collect data. The carbon source metabolism phenotype of P. indica was analyzed based on its metabolic kinetics curve.
Nitrogen metabolism and pathway phenotype analysis
A total of 0.125 ml of spore suspension was added to 59.875 ml of PM3, PM5, PM6, PM7, and PM8 inoculum, vortexed, and mixed evenly. The spore suspension was added to the metabolic plates and incubated, and the data were collected as described above. The nitrogen source metabolism and pathway phenotype of P. indica were analyzed based on its metabolic kinetics curve.
Phosphorus and sulfur metabolism phenotype analysis
A total of 0.025 ml of spore suspension was added to 11.975 ml of PM4 inoculum, vortexed, and mixed evenly. The spore suspension was added to the metabolic plates and incubated, and the data were collected as described above. The phosphorus and sulfur metabolism phenotypes of P. indica were analyzed based on its metabolic kinetics curve.
Osmotic pressure and pH metabolism phenotype analysis
A total of 0.05 ml of spore suspension was added to 23.95 ml of PM9 and PM10 inoculum, vortexed, and mixed evenly. The spore suspension was added to the metabolic plates and incubated, and data were collected as described above. The osmotic pressure and pH metabolism phenotype of P. indica were analyzed based on its metabolic kinetics curve.
Data processing and metabolism phenotype analysis
Data analysis was performed using SPSS 17.0, and Excel 2007 was used to simulate the growth curve regression equation. The results were considered significant when means were assessed with Duncan's new multiple-range test at p < 0.05. Values are the means ± SD of three replications. The Biolog D5E_OKA_data.exe software was used to collect metabolic phenotype data for P. indica, and the Biolog OL_FM_1.2.exe software was used for data conversion. The size of the color area of the metabolic wells was used to analyze the metabolic phenotype characteristics of P. indica. Substrate utilization rate (%) = number of available substrate wells/(total number of wells with substrate – number of control) × 100%.
Results
Growth dynamics and sporulation results of P. indica on five different culture media
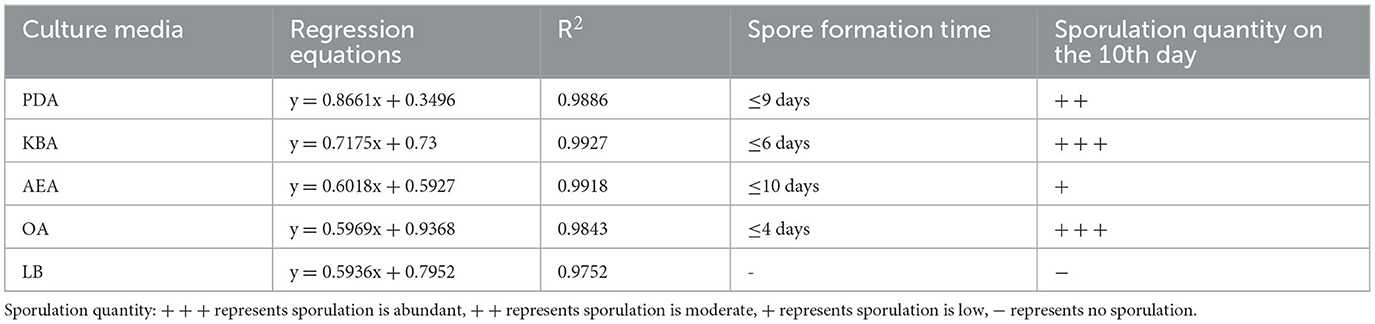
The results of the growth dynamics measurement of P. indica indicated a linear relationship between colony diameter (cm) and growth time (d) (Table 1). The growth rates in different cultural media were as follows: PDA > KBA > AEA > OA > LB (Figures 1, 2). Among them, it took 14 days, 17 days, 18 days, 19 days, and 20 days for P. indica to fully cover the culture dish (Ø = 12 cm) on PDA, KBA, AEA, OA, and LB, respectively. However, spores were first observed on the 4th day on the OA medium, followed by KBA on the 6th day, PDA on the 9th day, and AEA on the 10th day, and no spores were observed on the LB medium. Microscopic observations on the 10th day revealed that OA and KBA media had the highest spore yield (Figure 3). This indicated that among these five culture media, PDA was the most suitable for the growth of P. indica, while the OA medium was the most suitable for sporulation, followed by KBA. The OA medium required the shortest time for spore formation, and both the OA and KBA media had the highest spore yield.
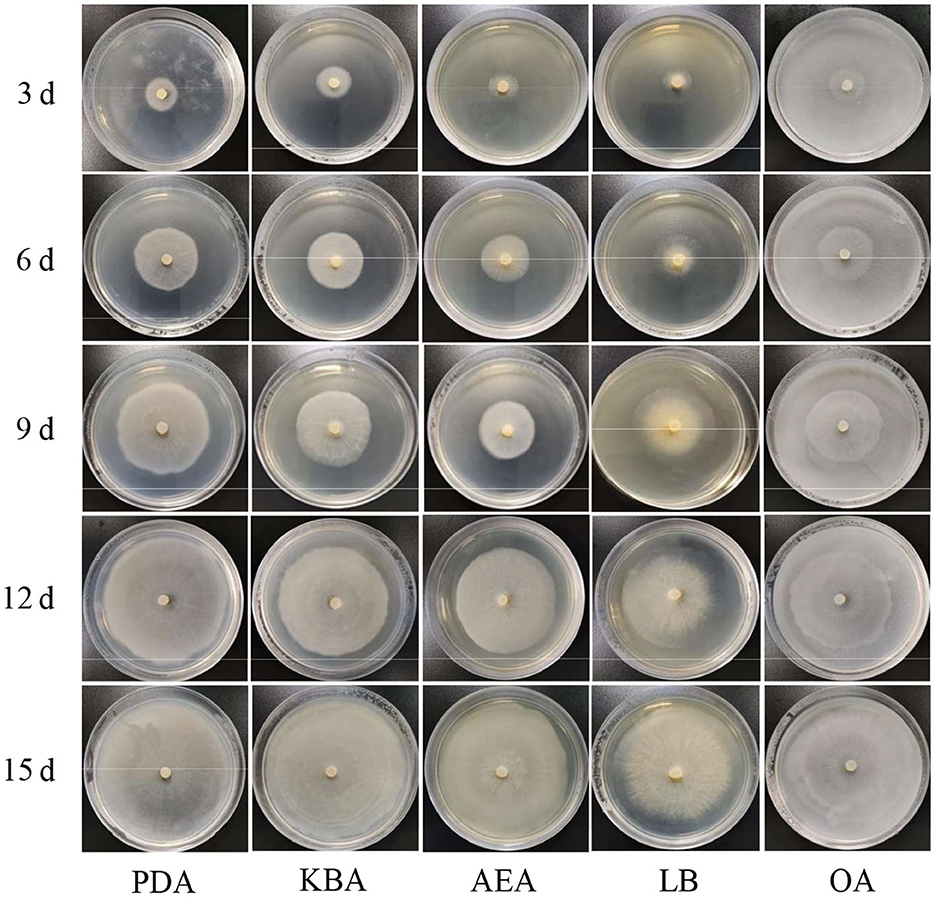
Figure 1. Morphology of Piriformospora indica cultured on five different media (PDA, KBA, AEA, LB, and OA) for 3, 6, 9, 12, and 15 days.
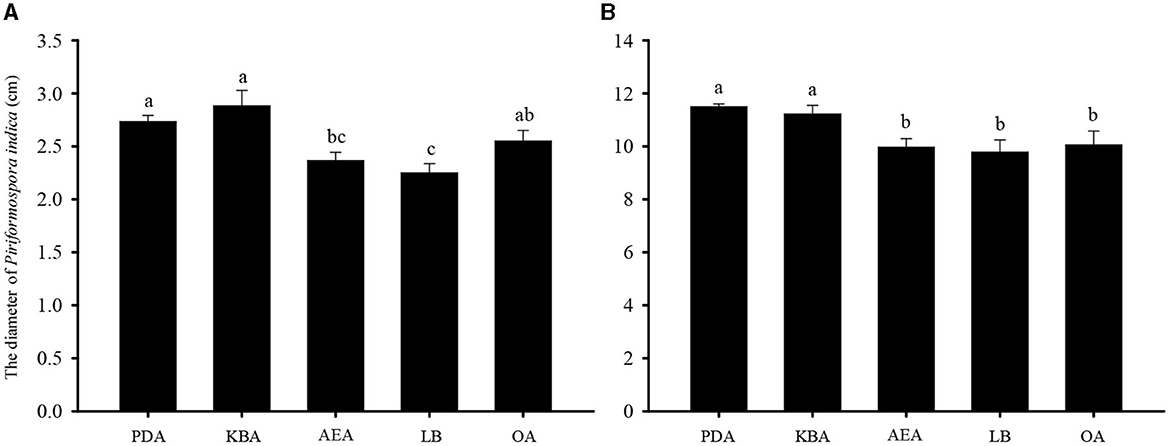
Figure 2. The diameter of Piriformospora indica cultured on five different media (PDA, KBA, AEA, LB, and OA) for 3 (A) and 15 (B) days.
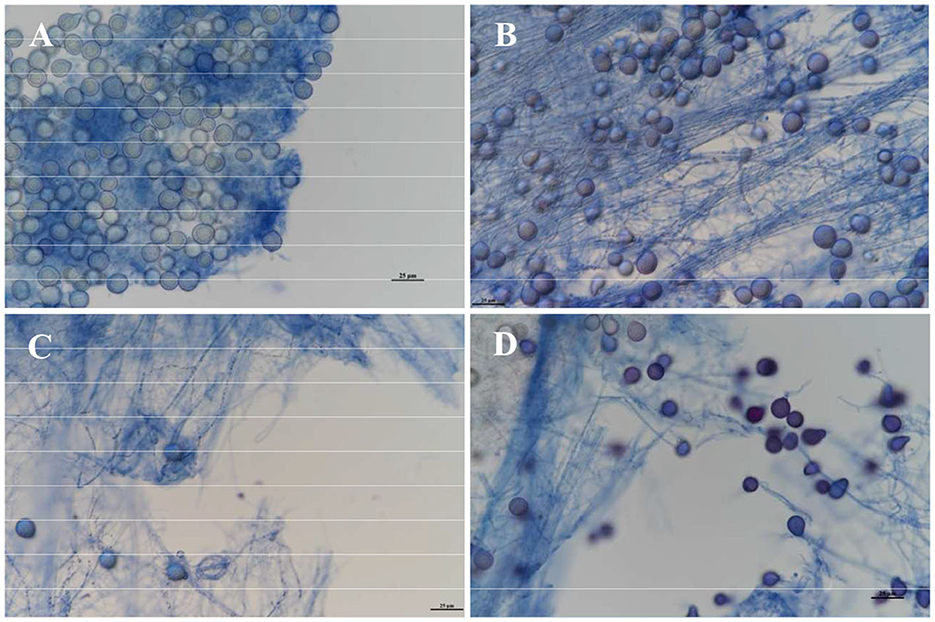
Figure 3. Observation of spore yield of Piriformospora indica cultured on different media on the 10th day under a microscope. (A) Stands for OA medium, (B) stands for KBA medium, (C) stands for AEA medium, and (D) stands for PDA medium.
Metabolic phenotypic features of Piriformospora indica
Fingerprinting patterns and metabolic data for P. indica were obtained, resulting in 950 pieces of information on metabolic phenotypes. Kinetic response curves reflecting microbial growth were generated for each well in PM1-10. Green areas represent the metabolic fingerprint of P. indica in each substrate; the larger the green proportion, the higher the efficiency of utilization (Figure 4). The results indicated that P. indica could utilize 87.89% of the tested carbon sources (76/95 in plate PM1 and 91/95 in plate PM2), 87.63% of the tested nitrogen sources (93/95 in plate PM3, 95/95 in plate PM6, 92/95 in plate PM7, and 53/95 in plate PM8), 96.61% of the tested phosphorus sources (57/59 in plate PM4), and 100% of the tested sulfur sources (35/35 in plate PM4), with 92 biosynthetic pathways tested. It was able to grow under 92 tested osmotic pressures and 88 tested pH values, demonstrating a wide range of substrate metabolism capability and strong environmental adaptability.

Figure 4. The metabolic fingerprint of microarray PM1-10 for Piriformospora indica. PM1-2 stands for carbon metabolic phenotype, PM3 and PM6-8 stands for nitrogen metabolic phenotype, PM4 Wells A01-E12 stands for phosphorous metabolic phenotype, PM4 Wells F01-H12 stands for sulfur metabolic phenotype, PM5 stands for biosynthetic pathway phenotype, PM9 stands for osmotic pressure phenotype, and PM10 stands for pH phenotype. Green areas represent the metabolic fingerprint of P. indica in each substrate; the larger the green proportion, the higher the efficiency of utilization.
Metabolic phenotypic analysis of Piriformospora indica on carbon sources
A total of 190 pieces of metabolic phenotypic information about carbon sources were obtained through metabolomics analysis. P. indica could metabolize 167 carbon sources, and 43 carbon sources were efficiently utilized (5/95 in plate PM1 and 38/95 in plate PM2; Table 2), including M-Hydroxyphenyl acetic acid, N-Acetyl-D-Glucosamine, Tyramine, Maltotrios, α-D-Glucose, N-Acetyl-D-Galactosamine, L-Valine, D-Melezitose, D-Tagatose, and Turanose. There were 75 moderately efficient carbon sources (39/95 in plate PM1 and 36/95 in plate PM2). The sporulation rate of P. indica on highly efficient carbon sources was 79.07% (4/5 in plate PM1 and 30/38 in plate PM2), and the overall sporulation rate among all metabolizable carbon sources was 70.66% (58/76 in plate PM1 and 60/91 in plate PM2). Only 23 carbon sources could not be metabolized by P. indica in all tested carbon substrates, including Methyl pyruvate, D-Glucose-1-Phosphate, L-Lyxose, D-Psicose, α-Methyl-D-Galactoside, D-Glucuronic acid, D-Lactic acid methyl ester, and 2-Hydroxybenzoic acid.
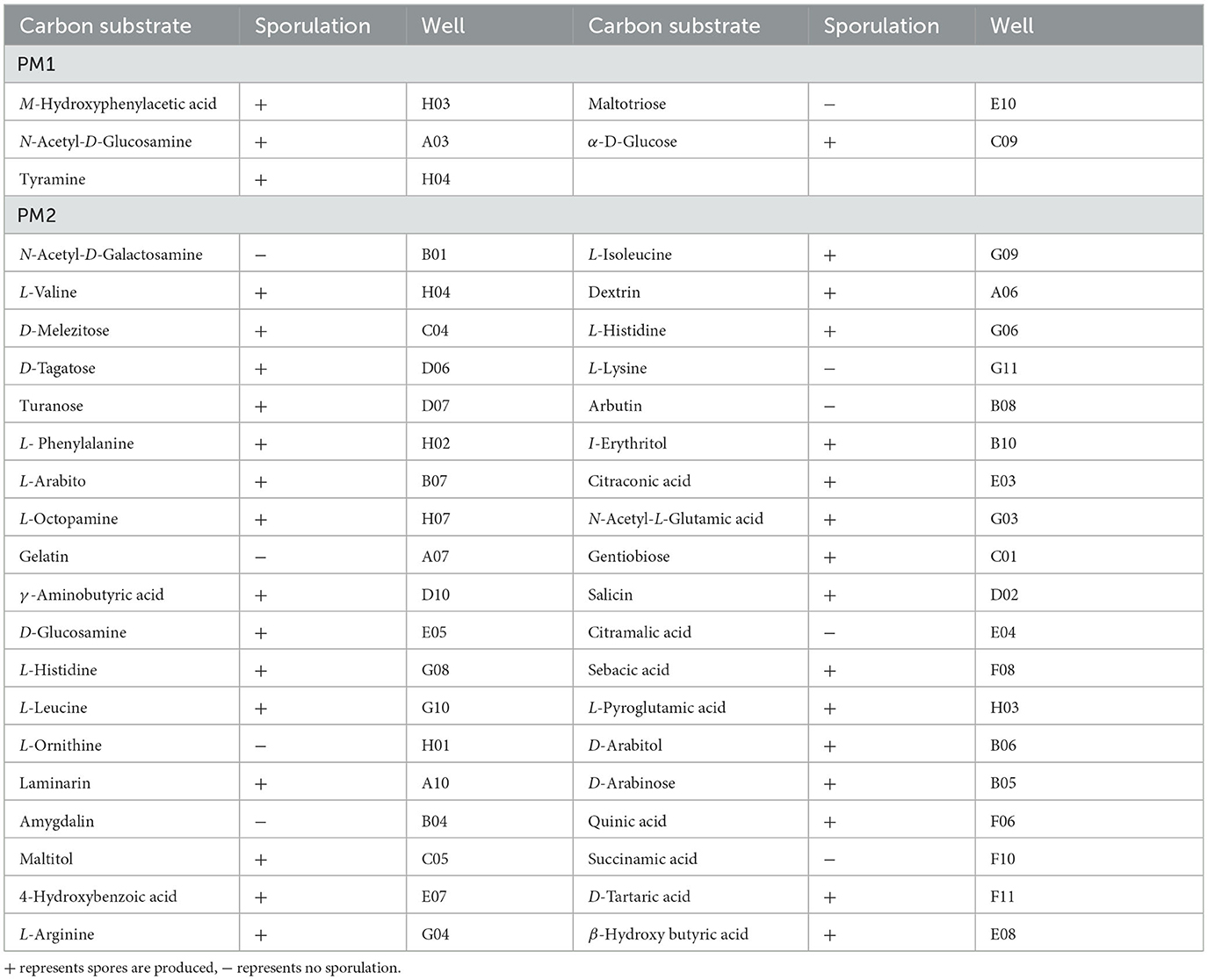
Table 2. Carbon substrates efficiently metabolized by Piriformospora indica and sporulation results in PM1-2.
Metabolic phenotypic analysis of Piriformospora indica on nitrogen sources
A total of 380 pieces of nitrogen metabolic phenotypic information (PM3 and PM6-8) were obtained through metabolomics analysis. P. indica was able to metabolize 333 nitrogen sources, with 170 kinds of them being efficiently utilized (38/95 in plate PM3, 63/95 in plate PM6, 54/95 in plate PM7, and 15/95 in plate PM8; Table 3). This included substances such as Adenosine, Inosine Allantoin, D, L-Lactamide, Arg-Met, lle-Trp, Ala-Arg, Thr-Arg, Trp-Tyr, Val-Asn, Gly-Gly-D-Leu, Gly-Gly-Phe, and Leu-Leu-Leu. Interestingly, only Adenosine and D, L-Lactamide were found to produce spores among the highly efficient nitrogen sources. A total of 121 nitrogen sources were moderately utilized (36/95 in plate PM3, 30/94 in plate PM6, 33/94 in plate PM7, and 22/94 in plate PM8), and 39 nitrogen sources were weakly utilized (19/95 in plate PM3, 1/94 in plate PM6, 4/94 in plate PM7, and 15/94 in plate PM8). Only 47 nitrogen sources (12.37%) could not be metabolized by P. indica, including N-Acetyl-D-Mannosamine, Guanine, Lys-Pro, Met-Pro, Lys-Gly, Pro-Ile, D-Ala-Leu, Val-Pro, D-Leu-Gly, and D-Ala-Gly-Gly.
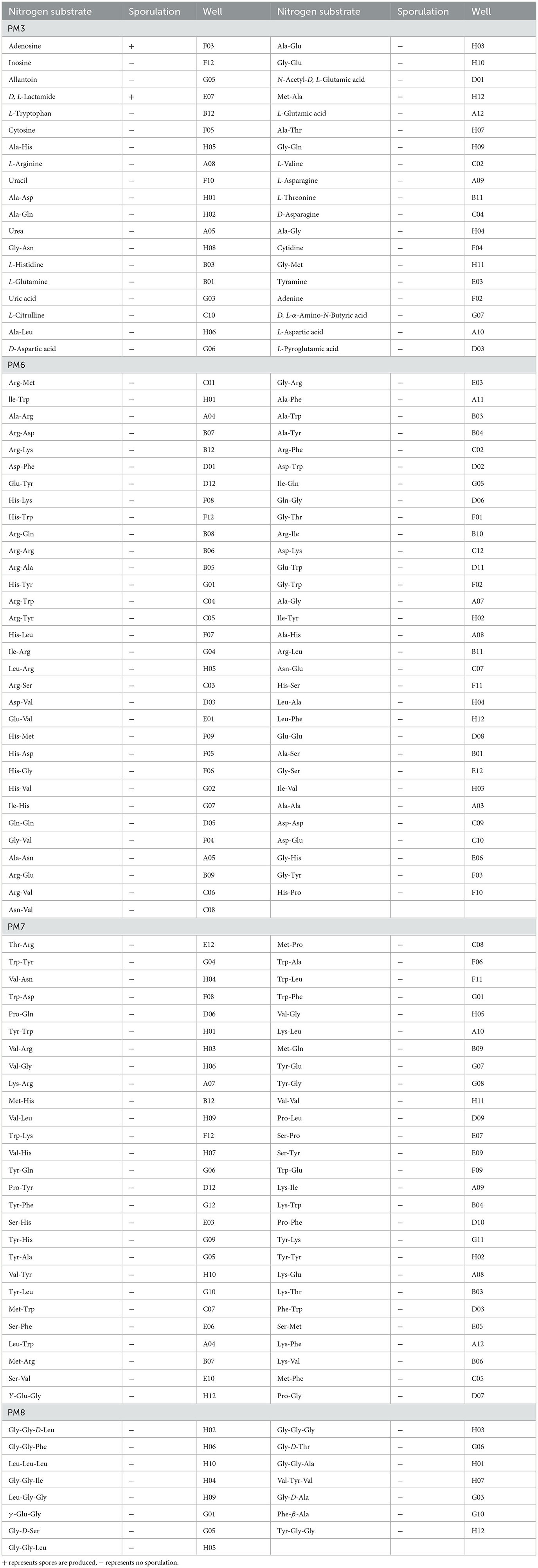
Table 3. Nitrogen substrates efficiently metabolized by Piriformospora indica and sporulation results in PM3, 6-8.
Metabolic phenotypic analysis of Piriformospora indica on phosphorus and sulfur sources
A total of 59 phosphorus sources and 35 sulfur sources metabolic phenotypic information were obtained through metabolomics analysis. P. indica could metabolize 57 phosphorus sources, with 42 efficiently utilized phosphorus sources (42/59 in plate PM4, Wells A02-E12; Table 4), including D-Mannose-1 phosphate, Adenosine-2′,3′-cyclic monophosphate, triple phosphate, phosphate, and trimetaphosphate. There were 12 moderately utilized phosphorus sources (12/59 in plate PM4) and 3 weakly utilized phosphorus sources (3/59 in plate PM4). Only two phosphorus sources (3.39%) could not be metabolized by P. indica, namely Thymidine 3′,5′-Cyclic monophosphate and Methylene diphosphonic acid. P. indica could metabolize 100% of the sulfur sources, with 34 efficiently utilized sulfur sources (34/35 in plate PM4, Wells F01-H12; Table 4), including Thiourea, N-Acetyl-L-Cysteine, 1-Thio-β-D-Glucose, Taurine, and p-Aminobenzene sulfonic acid. There was one moderately utilized sulfur source (1/35 in plate PM4, Wells F01-H12).
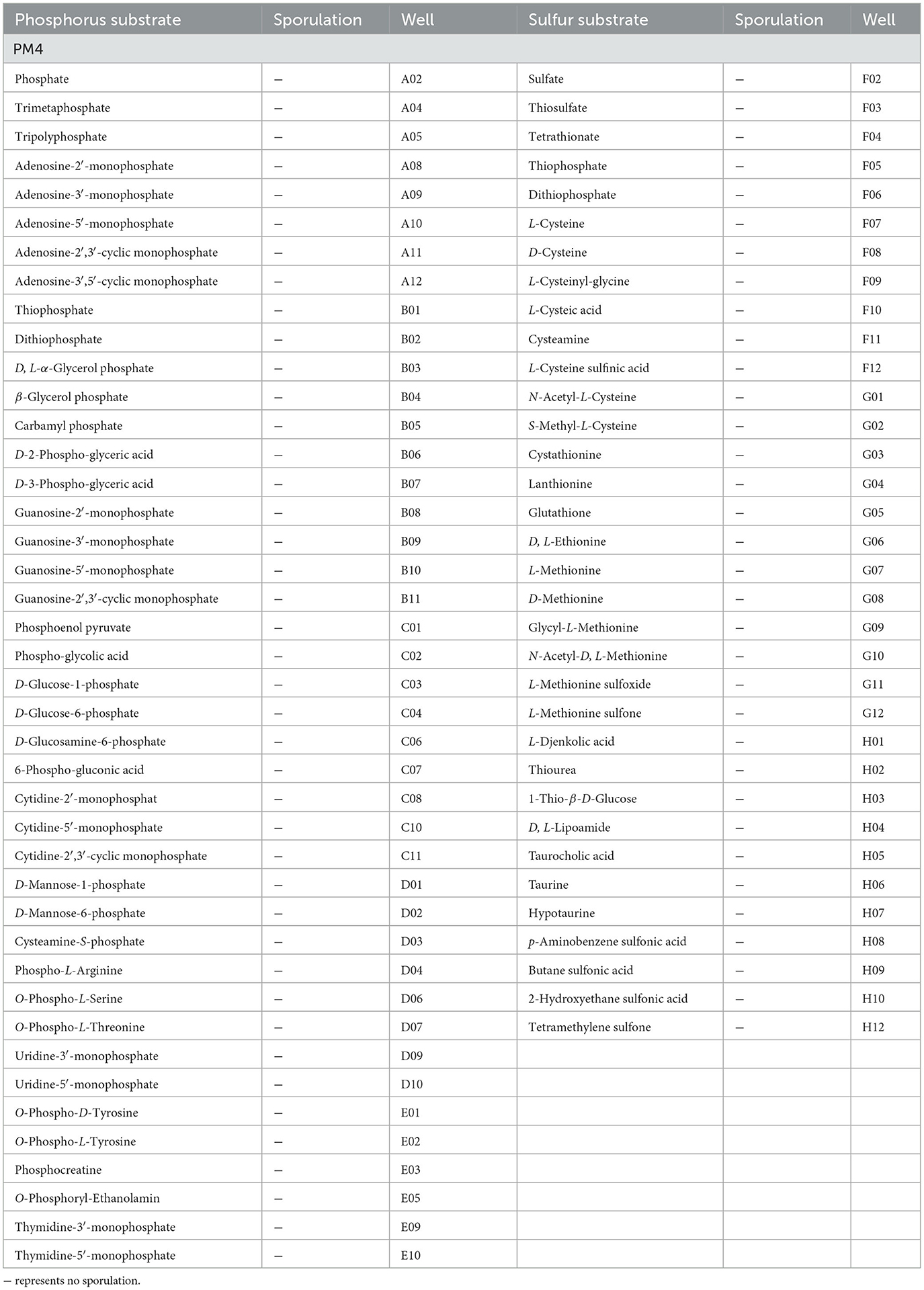
Table 4. Phosphorus and sulfur substrates efficiently metabolized by Piriformospora indica and sporulation results in PM4.
Metabolic phenotypic analysis of the biosynthetic pathways of Piriformospora indica
The experimental results of P. indica in PM5 showed that it had 92 kinds of tested biosynthetic pathways (92/94 tested, plate PM5, Wells A03-H12). Typical substrates for biosynthetic pathways included Tween 40, D, L-Mevalonic acid, Tween 20, Orotic acid, Butyric acid, α-Ketobutyric acid, D, L-Carnitine, D, L-α-Lipoic acid (oxidized form), and Caprylic acid.
Metabolic phenotypic analysis of Piriformospora indica at different osmotic pressures
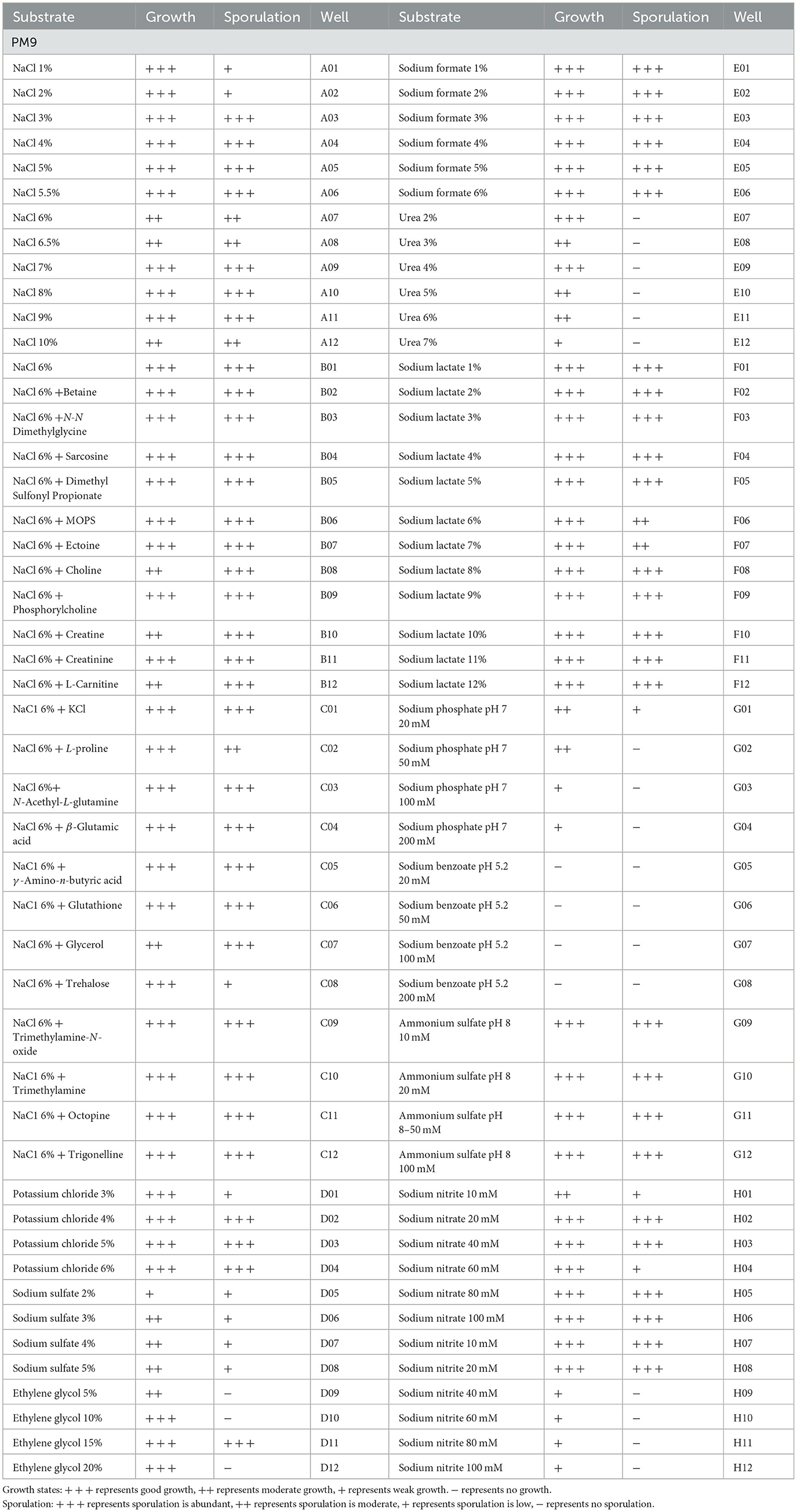
A total of 96 pieces of information on osmotic pressure metabolic phenotypes were obtained. P. indica was able to grow in 92 osmotic pressure environments (92/96 tested, plate PM9, Wells A01-H12; Table 5), including 1%−10% NaCl, 3%−6% potassium chloride, 2%−5% sodium sulfate, 5%−20% ethylene glycol, 1%−6% sodium formate, 2%−7% urea, 1%−12% sodium lactate, 20–200 mM sodium phosphate (pH 7), 10–100 mM ammonium sulfate (pH 8), 10–100 mM sodium nitrate, and 10–100 mM sodium nitrite. Under the synergistic effect of 6% NaCl, P. indica could still grow under various other osmotic substances (plate PM9, Wells B01-C12). It could not grow under the condition of 20–200 mM sodium benzoate (pH 5.2). Spore production circumstances accounted for 82.61% of all osmotic pressure conditions under which growth was normal.
Metabolic phenotypic analysis of Piriformospora indica under different pH values
A total of 96 pieces of information on pH metabolic phenotypes were obtained, and P. indica could grow normally in 88 pH environments (88/96 tested, plate PM10, Wells A01-H12; Table 6). The pH range for P. indica growth was 3.5–10, with the optimal pH for growth being approximately 5.0. In a pH 4.5 environment, P. indica was able to grow normally in all amino acids (PM10, B01-D12). In contrast, in a pH 9.5 environment, it could not grow normally in eight tested amino acids. Among the pH conditions where growth was normal, the conditions that detected spore production accounted for 53.41%. The amino acid decarboxylase and deaminase activities of microorganisms were tested under pH 4.5 and 9.5 conditions using the B1-D12 and E1-G12 wells of the PM10 plate, respectively. The results showed that P. indica has very strong amino acid decarboxylase activity (100% substrate utilization) and relatively strong deaminase activity (77.78% substrate utilization).
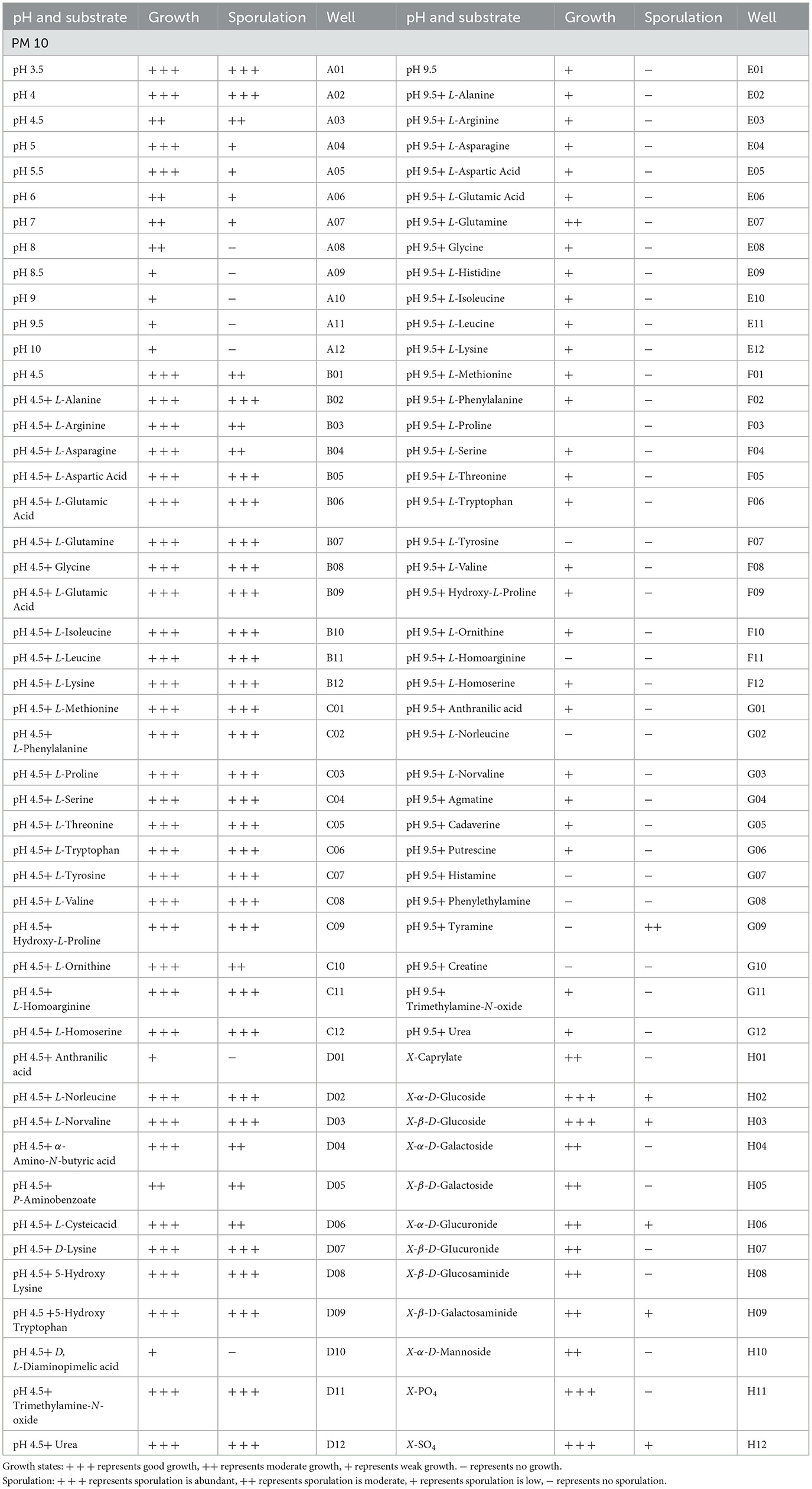
Table 6. Metabolic phenotypic characteristics of Piriformospora indica under different pH environments in PM10.
Discussion
P. indica possesses most of the functions of AM fungi for host plants, but the key difference is its ability to be cultured in vitro. In this study, we monitored the growth dynamics of P. indica on five common microbial culture media, including four fungal culture media and one bacterial culture medium. According to a regression equation that was derived to describe the relationship between colony diameter and growth days on different culture media, P. indica showed the fastest hyphal growth rate in the PDA culture medium. Previous studies have mainly used PDA medium and Kaefer medium (Kumar et al., 2011; Kashyap et al., 2023) for P. indica cultivation. However, the Kaefer medium, despite being considered optimal for growth, sporulation, and preservation of P. indica, is complex to prepare and expensive due to its requirement of more than 20 components. This poses challenges for its widespread application in laboratory or production settings (Attri and Varma, 2018; Kumar et al., 2023). Our study findings indicate that although PDA culture medium could induce spore production of P. indica, the sporulation time was long and the spore yield was low, making it difficult to meet the needs of strain preservation, metabolic phenotype testing, or other basic research. In contrast, this study demonstrates that the OA culture medium is the most suitable for sporulation, with the shortest sporulation time and the highest spore yield. The optimum medium for sporulation is coincident with some fungi, for instance, Epicoccum latusicollum (Li et al., 2023a) and Valdensinia heterodoxa (Zhao and Shamoun, 2006), but it is contrary to other fungi, including Alternaria alternata (Masangkay et al., 2000). OA culture medium, made from simple and inexpensive raw materials, contains a suitable carbon-to-nitrogen ratio and abundant vitamins and minerals. At the same time, the nutrients, especially nitrogen sources, in OA are not as abundant as those in PDA, YD, AEA, etc., making it favorable for the spore production of P. indica. The most suitable medium for P. indica can be used to provide the required biomass and spore quantity quickly for production or scientific research.
As a consequence, we employed an OA culture medium for subsequent Biolog PM metabolic phenotype experiments on P. indica. Compared with traditional microbial metabolism measurement methods, this high-throughput metabolic phenomics technology offers the advantages of high throughput, fast speed, and high accuracy (Khatri et al., 2013). It can dynamically reflect the biochemical processes of microbial cell metabolism in real-time (Kashyap et al., 2021). Biolog PM metabolic phenotype experiments have been used in the metabolic phenotype research of various microorganisms, including Helicobacter pylori (Lee et al., 2017), A. alternate (Wang H. C. et al., 2015), and Phytophthora parasitica (Wang M. S. et al., 2015). Our study obtained metabolic phenotype information from P. indica on various metabolic substrates and different environmental conditions, revealing a wide metabolic range of available carbon sources (87.89%), which is much higher than that of some plant pathogens, such as Botrytis cinerea (Wang et al., 2018), Rhizopus oryzae (Li et al., 2023b), and E. latusicollum (Li et al., 2023a). The available carbon sources accounted for 17%, 54.21%, and 61.58% individually. This characteristic allows P. indica to be co-cultivated with plant seedlings, colonizing their roots during the seedling stage. At this stage, due to the limited types of carbon sources in plants, some pathogenic bacteria are difficult to grow normally. However, P. indica can utilize a wider range of carbon sources, which enhances its growth and spore production. After successful colonization, P. indica has great potential to enhance the plant's disease resistance. In the carbon sources of PM1-2, it is interesting that some of the carbon sources that cannot be metabolized by P. indica are isomers of metabolizable substrates, for example, α-Methyl-D-Galactoside cannot be utilized, while β-Methyl-D-Galactoside can be moderately utilized. However, isomers of some substrates, such as α-Methyl-D-Glucoside and β-Methyl-D-Glucoside, can be moderately utilized.
Furthermore, P. indica exhibits a relatively extensive range of metabolizable nitrogen sources (87.63%), including amino acids, dipeptides, or tripeptides, which is higher than that of some plant pathogens, such as B. cinerea (63% of available amino acid nitrogen substrates and 80% of available peptide nitrogen substrates) (Wang et al., 2018). Its metabolic ability on phosphorus sources (96.61%) and sulfur sources (100%) is significantly higher than that of certain plant pathogens. For instance, Pseudomonas syringae showed active metabolism with 10.17% phosphorus sources and metabolized none of the tested sulfur sources (Guo et al., 2017; Manzar et al., 2022a). It is worth noting that the sporulation rate of P. indica was 70.66% among all available carbon sources, while among all available nitrogen sources, only specific substances, such as Adenosine, D, L-Lactamide, Xanthosine, Ethanolamine, Ethylamine, and N-Butylamine induced spore production. This may be attributed to excessive nitrogen sources in the nitrogen source metabolism experiments, resulting in promoting mycelium growth but hindering sporulation. Therefore, it is necessary to select suitable types of nitrogen sources and control the appropriate carbon-to-nitrogen ratio to facilitate the production of P. indica spores in cultivation.
P. indica demonstrated a wide range of tested biosynthetic pathways, reflecting its ability to utilize various substrates to produce complex products or to metabolize complex substrates into simpler products. These biosynthetic pathways may play a crucial role in its life activities. For example, they are involved in the production of energy, such as the tricarboxylic acid cycle, and the synthesis of substances such as amino acids, nucleotides, and fatty acids. The diversity of biosynthetic pathways allows microbes to utilize substrates and energy more efficiently, thereby increasing resilience and competitiveness. The wide range of biosynthetic pathways in P. indica coincides with its powerful capability of substrate utilization. P. indica can grow in various tested osmotic environments, but it cannot grow under the conditions of 20–200 mM sodium benzoate (pH 5.2). The osmotic pressure adaptability of P. indica is stronger than that of some pathogens, such as R. oryzae could not survive in 3–6% sodium formate, 4–7% urea, and 20–200 mM sodium benzoate (pH 5.2). The extensive osmotic pressure adaptability of P. indica allows it to grow normally under various conditions of dryness and watering (Wang H. C. et al., 2015). P. indica had a wide range of pH tolerance, with the activity of decarboxylase being higher than that of deaminase. Decarboxylase of the microbe generates alkaline amines by the catabolism of amino acids, which help counteract the low pH condition (Maurer et al., 2005). Deaminase generates ketonic acid through the catabolism of amino acids, which helps counteract high pH. The deaminase activity is significantly higher than that of A. alternata (Wang H. C. et al., 2015). Additionally, P. indica could produce spores in most of the growable osmotic pressure and pH conditions. Obviously, P. indica has a wide range of environmental adaptability, which is consistent with its ability to colonize a variety of plant hosts (Qiang et al., 2012; Manzar et al., 2022b; Mahawer et al., 2023).
The growth dynamics and the Biolog PM metabolic phenotype information obtained in this study for P. indica can be used to optimize the culture conditions, spore formation, and germination conditions of artificial culture media for P. indica. These findings aid in understanding its colonization ability on various plants and enhance the host's resilience, enabling better cultivation and utilization of its colonization on various plants and maximizing its function in enhancing resistance to plant diseases and stress after colonization. Simultaneously, it provides a reference for the metabolic phenotype of other species within the genus Piriformospora.
Conclusion
This study indicates that among the five commonly used microbial media tested for P. indica, PDA was found to be the most suitable medium for mycelium growth, while OA was the most suitable medium for spore production. Biolog PM1-10 was used to assess the metabolic phenotype of P. indica, revealing its ability to metabolize 87.89% of the tested carbon sources, 87.63% of the tested nitrogen sources, 96.61% of the tested phosphorus sources, and 100% of the tested sulfur sources, with 92 kinds of tested biosynthetic pathways. P. indica could grow under 92 kinds of test osmotic pressures and 88 kinds of test pH conditions, indicating a wide range of substrate metabolism in terms of carbon, nitrogen, phosphorus, and sulfur sources, as well as a wide range of environmental adaptability. This study provides a theoretical basis for screening more suitable growth and sporulation conditions for P. indica, elucidating its adaptive mechanisms in symbiosis with plants, understanding the physiological reasons behind enhancing the resistance of host plants, and establishing preferable application strategies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
J-rH: Conceptualization, Funding acquisition, Software, Writing – original draft, Writing – review & editing. J-mL: Conceptualization, Writing – original draft. H-yW: Conceptualization, Writing – original draft, Software. M-lS: Data curation, Writing – original draft. C-yH: Conceptualization, Writing – original draft. H-cW: Writing – original draft, Methodology, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Key R&D Project of the Zunyi Company of Guizhou Tobacco Company (2022XM09), the National Natural Science Foundation (Grant Nos. 32302343 and 32172400), the China National Tobacco Corporation [110202101048 (LS-08)], the Hundred Level Innovative Talent Foundation of Guizhou Province [GCC(2022)028-1], the Guizhou Science Technology Foundation (ZK[2021]Key036), and the Guizhou Province Applied Technology Research and Development Funding Postsubsidy Project. The authors declare that this study received funding from the Zunyi Company of Guizhou Tobacco Company and the China National Tobacco Corporation. The funders were not involved in the study design, collection, analysis, and interpretation of data, the writing of this article, or the decision to submit it for publication.
Conflict of interest
C-yH was employed by Guizhou Provincial Tobacco Company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1301743/full#supplementary-material
References
Abdelaziz, M. E., Kim, D., Ali, S., Fedoroff, N. V., and Al-Babili, S. (2017). The endophytic fungus Piriformospora indica enhances Arabidopsis thaliana growth and modulates Na+/K+ homeostasis under salt stress conditions. Plant Sci. 263, 107–115. doi: 10.1016/j.plantsci.2017.07.006
Alikhani, M., Khatabi, B., Sepehri, M., Nekouei, M. K., Mardi, M., and Salekdeh, G. H. (2013). A proteomics approach to study the molecular basis of enhanced salt tolerance in barley (Hordeum vulgare L.) conferred by the root mutualistic fungus Piriformospora indica. Mol. Biosyst. 9, 1498–1510. doi: 10.1039/c3mb70069k
Amani, S., Mohebodini, M., Khademvatan, S., Jafari, M., and Kumar, V. (2021). Piriformospora indica based elicitation for overproduction of phenolic compounds by hairy root cultures of Ficus carica. J. Biotechnol. 327, 43–53. doi: 10.1016/j.jbiotec.2020.12.015
Attri, M. K., and Varma, A. (2018). Comparative study of growth of Piriformospora indica by using different sources of jaggery. J. Pure Appl. Microbio. 12, 933–942. doi: 10.22207/JPAM.12.2.56
Bandyopadhyay, P., Yadav, B. G., Kumar, S. G., Kumar, R., Kogel, K. H., and Kumar, S. (2022). Piriformospora indica and Azotobacter chroococcum consortium facilitates higher acquisition of N, P with improved carbon allocation and enhanced plant growth in Oryza sativa. J. Fungi 8, 453. doi: 10.3390/jof8050453
Bochner, B. R. (2003). New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4, 309–314. doi: 10.1038/nrg1046
Chen, B. D., Zhu, Y. G., Duan, J., Xiao, X. Y., and Smith, S. E. (2007). Effects of the arbuscular mycorrhizal fungus Glomus mosseae on growth and metal uptake by four plant species in copper mine tailings. Environ. Pollut. 147, 374–380. doi: 10.1016/j.envpol.2006.04.027
Deshmukh, S., Hückelhoven, R., Schäfer, P., Imani, J., Sharma, M., and Weiss, M. (2006). The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. PNAS. 103, 18450–18457. doi: 10.1073/pnas.0605697103
Dong, S. Q., Tian, Z. H., Chen, P. J., Senthil Kumar, R., Shen, C. H., Cai, D. G., et al. (2013). The maturation zone is an important target of Piriformospora indica in Chinese cabbage roots. J. Exp. Bot. 64, 4529–4540. doi: 10.1093/jxb/ert265
Fakhro, A., Andrade-Linares, D. R., von Bargen, S., Bandte, M., Büttner, C., and Grosch, R. (2010). Impact of Piriformospora indica on tomato growth and on interaction with fungal and viral pathogens. Mycorrhiza 20, 191–200. doi: 10.1007/s00572-009-0279-5
Ghabooli, M., Khatabi, B., Ahmadi, F. S., Sepehri, M., Mirzaei, M., Amirkhani, A., et al. (2013). Proteomics study reveals the molecular mechanisms underlying water stress tolerance induced by Piriformospora indica in barley. J. Proteom. 94, 289–301. doi: 10.1016/j.jprot.2013.09.017
Ghorbani, A., Tafteh, M., Roudbari, N., Pishkar, L., Zhang, W., and Wu, C. (2021). Piriformospora indica augments arsenic tolerance in rice (Oryza sativa) by immobilizing arsenic in roots and improving iron translocation to shoots. Ecotoxicol. Environ. Saf. 209, 111793. doi: 10.1016/j.ecoenv.2020.111793
Gill, S. S., Gill, R., Trivedi, D. K., Anjum, N. A., Sharma, K. K., and Ansari, M. W. (2016). Piriformospora indica: potential and significance in plant stress tolerance. Front. Microbiol. 7, 332. doi: 10.3389/fmicb.2016.00332
Guo, Y. S., Su, X. K., Cai, L. T., and Wang, H. C. (2017). Phenotypic characterization of Pseudomonas syringae pv. tabaci, the causal agent of tobacco wildfire. J. Plant Pathol. 99, 499–504. Available online at: https://www.jstor.org/stable/44686798
Hibbett, D. S., Binder, M., Bischoff, J. F., Blackwell, M., Cannon, P. F., Eriksson, O. E., et al. (2007). A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111, 509–547. doi: 10.1016/j.mycres.2007.03.004
Hui, F., Liu, J., Gao, Q., and Lou, B. (2015). Piriformospora indica confers cadmium tolerance in Nicotiana tabacum. J. Environ. Sci. 37, 184–191. doi: 10.1016/j.jes.2015.06.005
Kaldorf, M., Koch, B., Rexer, K. H., Kost, G., and Varma, A. (2005). Patterns of interaction between Populus Esch5 and Piriformospora indica: a transition from mutualism to antagonism. Plant Biol. 7, 210–218. doi: 10.1055/s-2005-837470
Kashyap, A. S., Manzar, N., Meshram, S., and Sharma, P. K. (2023). Screening microbial inoculants and their interventions for cross kingdom management of wilt disease of solanaceous crop- A step towards sustainable agriculture. Front. Microbiol. 14, 1174532. doi: 10.3389/fmicb.2023.1174532
Kashyap, A. S., Manzar, N., Rajawat, M. V. S., Kesharwani, A. K., Singh, R. P., Dubey, S. C., et al. (2021). Screening and biocontrol potential of rhizobacteria native to gangetic plains and hilly regions to induce systemic resistance and promote plant growth in chilli against bacterial wilt disease. Plants 10, 2125. doi: 10.3390/plants10102125
Kesharwani, A. K., Singh, D., Kulshreshtha, A., Kashyap, A. S., Avasthi, A. S., and Geat, N. (2022). Black rot disease incited by Indian race 1 of Xanthomonas campestris pv. campestris in Brassica juncea ‘Pusa Bold' in India. Plant Dis. 107, 212. doi: 10.1094/PDIS-04-22-0738-PDN
Khalid, M., Rahman, S. U., and Huang, D. (2019). Molecular mechanism underlying Piriformospora indica-mediated plant improvement/protection for sustainable agriculture. Acta Bioch. Bioph. Sin. 51, 229–242. doi: 10.1093/abbs/gmz004
Khatri, B., Fielder, M., Jones, G., Newell, W., Abu-Oun, M., and Wheeler, P. R. (2013). High throughput phenotypic analysis of Mycobacterium tuberculosis and Mycobacterium bovis strains' metabolism using biolog phenotype microarrays. PLoS ONE 8, e52673. doi: 10.1371/journal.pone.0052673
Kumar, S., Korra, T., Thakur, R., Arutselvan, R., Kashyap, A. S., Nehela, Y., et al. (2023). Role of plant secondary metabolites in defence and transcriptional regulation in response to biotic stress. Plant Stress 8, 100154. doi: 10.1016/j.stress.2023.100154
Kumar, V., Sahai, V., and Bisaria, V. S. (2011). High-density spore production of Piriformospora indica, a plant growth-promoting endophyte, by optimization of nutritional and cultural parameters. Bioresour. Technol. 102, 3169–3175. doi: 10.1016/j.biortech.2010.10.116
Lee, W. C., Goh, K. L., Loke, M. F., and Vadivelu, J. (2017). Elucidation of the metabolic network of Helicobacter pylori J99 and Malaysian clinical strains by phenotype microarray. Helicobacter 22, e12321. doi: 10.1111/hel.12321
Li, L., Guo, N., Feng, Y., Duan, M., and Li, C. (2022). Effect of Piriformospora indica-induced systemic resistance and basal immunity against Rhizoctonia cerealis and Fusarium graminearum in wheat. Front. Plant Sci. 13, 836940. doi: 10.3389/fpls.2022.836940
Li, W., and Lu, C. D. (2007). Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J. Bacteriol. 189, 5413–5420. doi: 10.1128/JB.00432-07
Li, Z., Hu, J. R., Li, W. H., Wang, H. C., Guo, Z. N., Cheng, X., et al. (2023a). Characteristics of Epicoccum latusicollum as revealed by genomic and metabolic phenomic analysis, the causal agent of tobacco Epicoccus leaf spot. Front. Plant Sci. 14, 1199956. doi: 10.3389/fpls.2023.1199956
Li, Z., Shi, C. H., Huang, Y., Wang, H. C., Li, W. H., and Cai, L. T. (2023b). Phenotypic analysis and genome sequence of Rhizopus oryzae strain Y5, the causal agent of tobacco pole rot. Front. Microbiol. 13, 1031023. doi: 10.3389/fmicb.2022.1031023
Mahawer, A. K., Dubey, A. K., Awasthi, O. P., Singh, D., Dahuja, A., Sevanthi, A. M., et al. (2023). Elucidation of physio-biochemical changes in citrus spp. incited by Xanthomonas citri pv. citri. Horticulturae 9, 324. doi: 10.3390/horticulturae9030324
Manzar, N., Kashyap, A. S., Goutam, R. S., Rajawat, M. V. S., Sharma, P. K., Sharma, S. K., et al. (2022b). Trichoderma: advent of versatile biocontrol agent, its secrets and insights into mechanism of biocontrol potential. Sustainability 14, 12786. doi: 10.3390/su141912786
Manzar, N., Kashyap, A. S., Maurya, A., Rajawat, M. V. S., Sharma, P. K., Srivastava, A. K., et al. (2022a). Multi-gene phylogenetic approach for identification and diversity analysis of Bipolaris maydis and Curvularia lunata isolates causing foliar blight of Zea mays. J. Fungi 8, 802. doi: 10.3390/jof8080802
Manzar, N., Kashyap, A. S., Sharma, P. K., and Saxena, A. K. (2021b). First report of leaf spot of maize caused by Curvularia geniculata in India. Plant Dis. 105, 4155. doi: 10.1094/PDIS-03-21-0637-PDN
Manzar, N., Singh, Y., Kashyap, A. S., Sahu, P. K., Rajawat, S. M. V., Bhowmik, A., et al. (2021a). Biocontrol potential of native Trichoderma spp. against anthracnose of great millet (Sorghum bicolour L.) from Tarai and hill regions of India. Biol. Control 152, 104474. doi: 10.1016/j.biocontrol.2020.104474
Masangkay, R. F., Paulitz, T. C., Hallett, S. G., and Watson, A. K. (2000). Characterization of sporulation of Alternaria alternata f. sp. sphenocleae. Biocontrol. Sci. Techn. 10, 385–397. doi: 10.1080/09583150050114981
Maurer, L. M., Yohannes, E., Bondurant, S. S., Radmacher, M., and Slonczewski, J. L. (2005). pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187, 304–319. doi: 10.1128/JB.187.1.304-319.2005
Mazur, A., Stasiak, G., Wielbo, J., Koper, P., Kubik-Komar, A., and Skorupska, A. (2013). Phenotype profiling of Rhizobium leguminosarum bv. trifolii clover nodule isolates reveal their both versatile and specialized metabolic capabilities. Arch. Microbiol. 195, 255–267. doi: 10.1007/s00203-013-0874-x
Narayan, O. P., Verma, N., Singh, A. K., Oelmüller, R., Kumar, M., and Prasad, D. (2017). Antioxidant enzymes in chickpea colonized by Piriformospora indica participate in defense against the pathogen Botrytis cinerea. Sci. Rep. 7, 13553. doi: 10.1038/s41598-017-12944-w
Narware, J., Singh, S. P., Manzar, N., and Kashyap, A. S. (2023). Biogenic synthesis, characterization, and evaluation of synthesized nanoparticles against the pathogenic fungus Alternaria solani. Front. Microbiol. 14, 1159251. doi: 10.3389/fmicb.2023.1159251
Oelmüller, R., Sherameti, I., Tripathi, S., and Varma, A. (2009). Piriformospora indica, a cultivable root endophyte with multiple biotechnological applications. Symbiosis 49, 1–17. doi: 10.1007/s13199-009-0009-y
Qiang, X., Weiss, M., Kogel, K. H., and Schäfer, P. (2012). Piriformospora indica-a mutualistic basidiomycete with an exceptionally large plant host range. Mol. Plant Pathol. 13, 508–518. doi: 10.1111/j.1364-3703.2011.00764.x
Singh, A., Sharma, J., Rexer, K. H., and Varma, A. (2000). Plant productivity determinants beyond minerals, water and light: Piriformospora indica–A revolutionary plant growth promoting fungus. Curr. Sci. 79, 1548–1554. Available online at: https://www.jstor.org/stable/24104847
Su, Z. Z., Wang, T., Shrivastava, N., Chen, Y. Y., Liu, X., and Sun, C. (2017). Piriformospora indica promotes growth, seed yield and quality of Brassica napus L. Microbiol. Res. 199, 29–39. doi: 10.1016/j.micres.2017.02.006
Varma, A., Bakshi, M., Lou, B. G., Hartmann, A., and Oelmueller, R. (2012). Piriformospora indica: a novel plant growth-promoting mycorrhizal fungus. Agric. Res. 1, 117–131. doi: 10.1007/s40003-012-0019-5
Varma, A., Singh, A., Sudha, Sahay, N. S., Sharma, J., Roy, A., et al. (2001). Piriformospora indica: an axenically culturable mycorrhiza-like endosymbiotic fungus. Fung. Assoc. 9, 125–150. doi: 10.1007/978-3-662-07334-6_8
Varma, A., Verma, S., Sudha, S. N., Bütehorn, B., and Franken, P. (1999). Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl. Environ. Microb. 65, 2741–2744. doi: 10.1128/AEM.65.6.2741-2744.1999
Venice, F., Ghignone, S., Salvioli di Fossalunga, A., Amselem, J., Novero, M., Xie, X. N., et al. (2020). At the nexus of three kingdoms: the genome of the mycorrhizal fungus Gigaspora margarita provides insights into plant, endobacterial and fungal interactions. Environ. Microbial. 22, 122–141. doi: 10.1111/1462-2920.14827
Verma, S., Varma, A., Rexer, K. H., Hassel, A., Kost, G., Sarbhoy, A., et al. (1998). Piriformospora indica, gen. et sp. nov., a new root-colonizing fungus. Mycologia 90, 896–903. doi: 10.1080/00275514.1998.12026983
Waller, F., Achatz, B., Baltruschat, H., Fodor, J., Becker, K., Fischer, M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. PNAS 102, 13386–13391. doi: 10.1073/pnas.0504423102
Wang, H. C., Huang, Y. F., Xia, H. Q., Wang, J., Wang, M. S., Zhang, C. Q., et al. (2015). Phenotypic analysis of Alternaria alternata, the causal agent of tobacco brown spot. Plant Pathol. J. 14, 79–85. doi: 10.3923/ppj.2015.79.85
Wang, H. C., Li, L. C., Cai, B., Cai, L. T., Chen, X. J., Yu, Z. H., et al. (2018). Metabolic phenotype characterization of Botrytis cinerea, the causal agent of gray mold. Front. Microbiol. 9, 470. doi: 10.3389/fmicb.2018.00470
Wang, M. S., Wang, H. C., Huang, Y. F., Wang, J., Zhang, C. Q., and Lu, H. X. (2015). Phenotypic analysis of Phytophthora parasitica by using high throughput phenotypic microarray. Acta Microbiol. Sin. 55, 1356–1363. doi: 10.13343/j.cnki.wsxb.20150036
Xu, L., Wang, A., Wang, J., Wei, Q., and Zhang, W. (2017). Piriformospora indica confers drought tolerance on Zea mays L. through enhancement of antioxidant activity and expression of drought-related genes. Crop J. 5, 251–258. doi: 10.1016/j.cj.2016.10.002
Xu, L., Wu, C., Oelmüller, R., and Zhang, W. (2018). Role of phytohormones in Piriformospora indica-induced growth promotion and stress tolerance in plants: more questions than answers. Front. Microbiol. 9, 1646. doi: 10.3389/fmicb.2018.01646
Yan, C., Rizwan, H. M., Liang, D., Reichelt, M., Mithöfer, A., Scholz, S. S., et al. (2021). The effect of the root-colonizing Piriformospora indica on passion fruit (Passiflora edulis) development: Initial defense shifts to fitness benefits and higher fruit quality. Food Chem. 359, 129671. doi: 10.1016/j.foodchem.2021.129671
Keywords: Piriformospora indica, growth and sporulation, Biolog Phenotype Microarray, metabolic profiling, culture media
Citation: Hu J-r, Li J-m, Wang H-y, Sun M-l, Huang C-y and Wang H-c (2024) Analysis of growth dynamics in five different media and metabolic phenotypic characteristics of Piriformospora indica. Front. Microbiol. 14:1301743. doi: 10.3389/fmicb.2023.1301743
Received: 25 September 2023; Accepted: 13 December 2023;
Published: 08 January 2024.
Edited by:
Mario Serrano, National Autonomous University of Mexico, MexicoReviewed by:
Khan Mohd. Sarim, Rudjer Boskovic Institute, CroatiaAbhijeet Shankar Kashyap, National Bureau of Agriculturally Important Microorganisms (ICAR), India
Copyright © 2024 Hu, Li, Wang, Sun, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai-yan Wang, eGRueTIwMjBAc2luYS5jb20=; Chun-yang Huang, aHVhbmcuaGN5XzA3MDFAMTI2LmNvbQ==; Han-cheng Wang, eGlhb2JhaXlhbmcxMjZAaG90bWFpbC5jb20=
 Jing-rong Hu
Jing-rong Hu Jin-meng Li2
Jin-meng Li2 Hai-yan Wang
Hai-yan Wang Mei-li Sun
Mei-li Sun Han-cheng Wang
Han-cheng Wang